Abstract
Our aim was to determine the frequency of 12 common respiratory viruses in patients admitted to intensive care units with respiratory symptoms, evaluate the clinical characteristics and to compare the results to routine microbiological diagnostics. Throat swabs from 122 intensive care‐patients >18 years with acute respiratory symptoms were collected upon admission and analysed with multiplex real‐time polymerase chain reaction, for 12 community respiratory viruses. Blood and respiratory tract specimens were analysed for bacteria and fungi upon clinicians' request. Clinical and paraclinical data were collected. Viruses were detected in 19 (16%) of the 122 study patients. Five virus‐positive patients (26%) had possible clinically relevant bacteria or fungi co‐detected. Patients with exacerbation in COPD were associated with a viral infection (p = 0.02). Other comorbidities, clinical and paraclinical parameters, and death were independent of a viral infection or co‐detection of bacteria/fungi. In conclusion, respiratory viruses were frequently detected in the patients. The investigated clinical and paraclinical parameters were not different in viral infections compared to other agents, thus respiratory viruses likely have similar impact on the clinical course as other agents. In 25% of the virus‐positive patients, polymicrobial aetiology was identified. Comprehensive and sensitive diagnostic methods should be emphasized to enhance respiratory diagnostics.
Keywords: Respiratory viruses, intensive care, acute respiratory disease, influenza, PCR
Abbreviations
- COPD
Chronic Obstructive Pulmonary Disease
- PCR
Polymerase Chain Reaction
- ICU
Intensive Care Units
- RSV
Respiratory Syncytial Virus
- hMPV
Human Metapneumovirus
- CoV
Coronavirus
- SAPS II
Simplified Acute Physiology Score II
- CRP
C‐reactive protein
- CoNS
Coagulase‐Negative Staphylococci
- ARDS
Acute Respiratory Distress Syndrome
- MODS
Multiple Organ Dysfunction Syndrome
- DIC
Disseminated Intravascular Coagulation
Community respiratory viruses are frequent causes of acute respiratory tract infections and community‐acquired pneumonia 1, 2, 3, 4, 5. Respiratory viruses may also trigger acute exacerbations in pre‐existing chronic conditions such as asthma, chronic obstructive pulmonary disease (COPD) and congestive heart failure 6, 7. Some of these patients need hospitalization and even intensive care treatment. Still, respiratory viruses are generally not considered of clinical relevance when monitoring critically ill patients 8, 9.
Today, a large number of pathogens, mainly viruses, can be identified with rapid, highly sensitive and specific molecular methods, such as polymerase chain reaction (PCR). Clinically, it is difficult to determine the microbiological aetiology of respiratory tract infections. The microbiological findings are often influenced by antibiotic treatment before admission to intensive care units (ICU), which makes the microbiological test results difficult to interpret, and may disguise significant pathogens. Furthermore, the differentiation between pathogens causing invasive infection and colonization is often impossible. This has implications for the patients, as infections with different pathogens have different prognosis and response to treatment. Viral diagnostics are often used as second‐line diagnostics and saved for cases where no significant bacterial pathogen has been revealed during the initial analyses, or for cases with deterioration in clinical symptoms despite treatment of revealed bacteria. This approach can lead to a delay in potentially beneficially antiviral treatment. The aetiology of severe acute respiratory disease remains undetermined in more than 50% of the patients 10, 11, 12. Virological molecular methods have improved markedly during the recent years by the introduction of molecular techniques 2, 13, 14. However, there are still few studies using these methods for detection of a large number of viruses in ICU‐patients 10, 11, 12.
The purpose of this study was to determine the frequency of 12 respiratory viruses in adult ICU‐patients without selection for predisposing conditions, describe the clinical characteristics and epidemiology, and to compare the results of current practice respiratory diagnostics in these patients. This study is one of few which include both comprehensive virological PCR‐analyses and conventional microbiological analyses, while mimicking the clinical everyday reality at an ICU.
Materials and Methods
Study population
All adult patients >18 years of age admitted successively with clinical suspicion of acute respiratory infection at the intensive care units of two University Hospitals in Copenhagen, Denmark, were tested for the presence of 12 respiratory viruses on throat swabs. One ICU, with 10 beds (ICU‐A), collected swabs from 17 December 2008 to 30 November 2009, and the other, with six beds (ICU‐B), collected swabs from 22 January 2009 to 30 August 2009. Only throat swabs taken upon admission were included, and no additional virological analyses were requested from sample material sent for bacterial diagnostics. Throat swabs were chosen for the study due to the non‐invasive nature of sample collection, combined with minimal discomfort for the patients and an acceptable sensitivity. The inclusion of patients was based upon the assessment of the attending physicians. Inclusion criteria were wide, as respiratory viruses do not always appear as a uniform clinical picture. A patient was included if any sign of respiratory distress or failure was apparent, including exacerbation in COPD or asthma, and pneumonia. Patients with fever, if accompanied by uncompensated congestive heart failure or multiple organ dysfunction syndrome, metabolic acidosis, sepsis or cardiac arrest, were also included.
Microbiological analyses
All throat swabs were analysed consecutively for the presence of respiratory viruses upon arrival at Department of Virology, Statens Serum Institut, Copenhagen, Denmark. Analyses included influenza A and B, respiratory syncytial virus (RSV), human metapneumovirus (hMPV), parainfluenza virus 1, 2 and 3, coronaviruses OC43 (CoV OC43), 229E (CoV 229E) and NL‐63 (CoV NL‐63), rhinovirus and adenovirus, using a modified version of the multiplex real‐time PCR‐assay published previously by Brittain‐Long et al. 15, which also included analyses of enterovirus, Chlamydophila pneumoniae and Mycoplasma pneumonia. For technical purposes, these were omitted from our assay. The amplifications were performed on a Stratagene MX3005 thermocycler with MXpro software, (Stratagene, La Jolla, California, USA), following the manufacturer's instructions. The subtyping of influenza A was based on an in‐house accredited PCR‐assay, detecting the haemagglutinin‐ and neuraminidase‐regions of H1N1, (H1N1)pdm09 and H3N2.
The bacterial and fungal analyses were performed at the Department of Clinical Microbiology, Herlev Hospital, 2730 Herlev, Denmark, serving both hospitals. The analyses consisted of culture, identification and antibiotic susceptibility testing. PCR for Legionella pneumophila, Chlamophila pneumoniae and Mycoplasma pneumoniae was performed upon request from the clinicians. The material included blood, tracheal aspirations, bronchoalveolar lavage, sputum and pleural fluid. Samples collected within 5 days before or after the viral sample date were included. Evaluation of bacterial and fungal content of the sample was done according to the normal procedure for the department of Clinical Microbiology. Gram‐staining of each respiratory secretion was evaluated by technicians and microbiologists. Secretions containing epithelium from the deeper parts of the airways by microscopic evaluation were considered sufficient materials. Bacteria in association with such epithelium were considered as being possible pathogens. Culture of these bacteria was done to distinguish between usually non‐pathogenic and possible pathogenic organisms. Blood cultures were performed according to well recognized methods 16, 17.
Clinical parameters
Clinical information regarding the hospitalization, epidemiology and comorbidity was recorded from the medical records. The information included the following: age, gender, underlying comorbidity, use of immunosuppressant drugs, respiratory symptoms, diagnoses on admission, diagnoses on discharge, length of hospital stay, ICU stay and intubation, Simplified Acute Physiology Score II (SAPS II)‐scores, administration of antibiotics, non‐invasive ventilation, chest x‐ray, laboratory analyses and results of the physical examination, which included temperature, saturation, stethoscopic findings and clinical signs of respiratory infection or distress.
The SAPS II‐score is a severity of disease classification system applied during the first 24 h of admission to ICU. The resulting point score interval is 0–163, and the predicted mortality between 0 and 100%. The patients at ICU‐A were routinely scored according to the SAPS II‐guidelines at admission. This was not an established routine at ICU‐B during the study period. In this case, patients were scored retrospectively according to the international SAPS II‐guidelines by one anaesthesiologist, blinded as regards other results of the study.
Immunosuppression was defined either by a malignant disease, by administration of chemotherapy or radiotherapy within one year before admission, or by the use of corticosteroids at doses exceeding the equivalent of 10 mg/day of prednisolone for at least 2 months, or 1 mg/kg/day for at least a week during the last 3 months before admission 11, 18.
Statistical analyses
Intergroup characteristics were compared using the Wilcoxon rank sum test for numerical variables, and chi‐square test or Fisher's exact test for dichotomous variables. For continuous variables, median and interquartile ranges (IQR) were estimated. Multiple groups with numerical variables were compared using the Kruskal‐Wallis test. Multiple groups with dichotomous variables were compared using chi‐square test or Fisher's exact test, where appropriate. The p‐values <0.05 were considered significant. EpiData, version 3.1 (EpiData Association Denmark), was used for data entry, and all statistical analyses were performed using STATA/IC, version 11.1 (Statacorp LP, USA).
Ethics
The National Board of Health, Denmark, and the Danish Data Protection Agency approved the data collection (ref.no 7–604–04–02/97/HKR and ref.no 2007–54–0364). The regional Committee on Biomedical Research Ethics in Copenhagen was informed about the study (ref.no H–B–2008–FSP/33). According to Danish Law, and given the observational nature of the study without deviation from current medical practice, an approval was not necessary, and informed consent was waived.
Results
Sample inclusion
A total of 73 throat swabs were collected at ICU‐A, and 97 throat swabs were collected at ICU‐B (Fig. 1). Of these, five patients were incorrectly registered at the laboratory, six medical records were unavailable, and 37 patients were retrospectively found not to meet the inclusion criteria when assessing the admission. These patients were excluded. The final study group consisted of 122 patients, all suspected of respiratory infection upon admission to ICU‐A (n = 56) and ICU‐B (n = 66). Geographical location, patient population and medical care of the two units were comparable.
Figure 1.
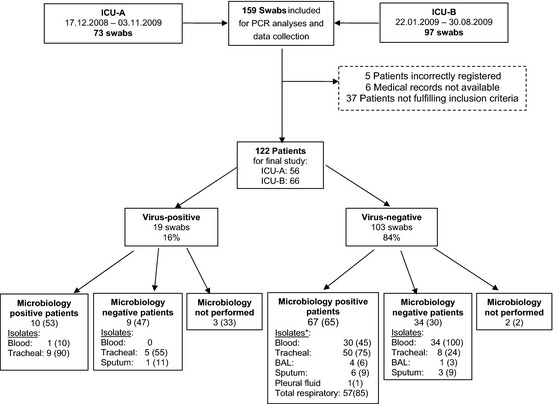
Inclusion of samples and patients *Some patients have positive isolates from more than one type of specimen ICU, Intensive care unit; PCR, Polymerase chain reaction; BAL, bronchoalveolar lavage. The flow chart shows inclusion, exclusion, and results of virological and microbiological analyses in number of patients. Data are presented as frequencies and percentages. Only the throat swab collected at admission was kept for study purposes. Retrospective reviewing of the sample indications led to the exclusion of 37 patients.
Patient characteristics
The median age of the final study group was 69 years (IQR, 63–76 years), and 64 (52%) were men. A total of 19 patients (16%) had a virus detected (Fig. 1). Clinical and paraclinical characteristics are shown in Table 1. The frequency of patients admitted to hospital with exacerbation in chronic obstructive pulmonary disease (COPD) was higher in the virus‐positive group: five patients (26%) vs eight patients (8%) (X 2‐test, p = 0.02). Pneumonia was the most frequent reason for admission in both groups. Among the virus‐positive, pneumonia occurred in nine of 10 influenza‐patients, and was also observed for patients with hMPV, RSV and adenovirus. The plasma concentrations of C‐reactive protein (CRP) were independent of a viral infection, with large interquartile ranges also detected in virus‐positive patients.
Table 1.
Different parameters of ICU‐patients with (n = 19) and without (n = 103) respiratory viruses
| General characteristics | Virus‐positive | n = | Virus‐negative | n = | p‐values |
|---|---|---|---|---|---|
| Age (years) | 69 (64–78) | 19 | 70 (63–75) | 103 | 0.76 |
| Gender male | 8 (42) | 19 | 56 (54) | 103 | 0.33 |
| Smoker, present and former | 11 (91) | 12 | 61 (72) | 85 | 0.18 |
| Comorbidity | |||||
| Respiratory disease, incl. COPD | 9 (47) | 19 | 40 (39) | 102 | 0.51 |
| Cardiac disease | 6 (32) | 19 | 43 (42) | 102 | 0.39 |
| Immunosuppression | 8 (42) | 19 | 30 (29) | 102 | 0.27 |
| Diabetes mellitus | 2 (11) | 19 | 21 (21) | 102 | 0.52 |
| Reasons for admission to hospital | |||||
| COPD‐exacerbation | 5 (26) | 19 | 8 (8) | 103 | 0.02 |
| Pneumonia | 7 (37) | 19 | 33 (33) | 103 | 0.68 |
| Respiratory failure | 2 (11) | 19 | 21 (20) | 103 | 0.52 |
| Cardiac disease | 2 (11) | 19 | 17 (17) | 103 | 0.74 |
| Infection | 3 (16) | 19 | 19 (18) | 103 | 1.0 |
| Surgery | 1 (5) | 19 | 11 (11) | 103 | 0.69 |
| Unconsciousness | 1 (5). | 19 | 16 (16) | 103 | 0.47 |
| Sepsis | 3 (16) | 19 | 9 (9) | 103 | 0.40 |
| Clinical and paraclinical parameters | |||||
| CRP (mg/L) | 65 (34–112) | 19 | 119 (39–213) | 103 | 0.16 |
| Leukocytes (109 cells/L) | 11.2 (7.3–15.9) | 19 | 11.4 (7.9–17.4) | 103 | 0.62 |
| Neutrophils (109 cells/L) | 8.5 (5.8–13.1) | 19 | 8.1 (5.9–14.9) | 93 | 1.0 |
| Lymphocytes (109 cells/L) | 0.6 (0.5–1.2) | 19 | 0.8 (0.6–1.6) | 93 | 0.11 |
| Temperature, °C | 38.0 (36.8–39.3) | 12 | 37.7 (37.2–38.3) | 57 | 0.51 |
| Fever >37.5 °C | 8 (57) | 14 | 39 (51) | 76 | 0.69 |
COPD, Chronic obstructive respiratory disease; Cardiac disease includes Ischaemic heart disease, congestive heart failure, arterial hypertension and angina pectoris, ICU, Intensive care unit; CRP, C‐reactive protein; Clinical and paraclinical characteristics upon admission of ICU‐patients admitted with respiratory disease. Numerical variables are presented as medians and interquartile range. Categorical data are presented as frequencies and percentages. Wilcoxon rank sum was used for numerical data, and Chi‐square test or Fisher's exact test, for categorical data.
Clinical intervention and diagnoses upon discharge are presented in Table 2. All 19 virus‐positive patients received antibiotics, compared to 98 of the virus‐negative patients (97%). Twelve (63%) of the virus‐positive patients received combination antibiotic therapy, which was also the case for 88 (87%) of virus‐negative patients (X 2‐test, p = 0.010). Oseltamivir was administered to three virus‐positive patients (16%) during the period of the 2009 pandemic. Additional clinical parameters, comorbidity, intervention and discharge diagnoses did not differ significantly between the patient groups. In the virus‐positive group, 16 of 19 patients (84%) were transferred to ICU 4 days or less after hospital admittance. In the virus‐negative group, this was the case for 77 of 103 patients (76%).
Table 2.
Intervention and diagnoses of ICU‐patients with (n = 19) and without (n = 103) respiratory viruses
| Characteristic | Virus‐positive | n = | Virus‐negative | n = | p‐value |
|---|---|---|---|---|---|
| Clinical intervention | |||||
| Hospital stay (days) | 9.5 (5–23) | 18 | 15.5 (6–30.5) | 92 | 0.25 |
| ICU stay (days) | 5 (2–14) | 19 | 6 (2–13) | 103 | 0.96 |
| Days before transfer to ICU | 1 (0–4) | 19 | 1 (0–4) | 101 | 0.78 |
| SAPS II‐score | 54 (35–73) | 17 | 47 (35–57) | 101 | 0.36 |
| Mechanical ventilation (days) | 4 (2–13) | 18 | 6 (3–13) | 71 | 0.42 |
| Mechanical ventilation | 18 (95) | 19 | 81 (79) | 103 | 0.12 |
| Non‐invasive ventilation | 1 (5) | 19 | 10 (10) | 101 | 1.0 |
| Antibiotic therapy | 19 (100) | 19 | 98 (97) | 101 | 0.68 |
| Combination antibiotic therapy | 12 (63) | 19 | 88 (87) | 101 | 0.01 |
| Discharge diagnoses | |||||
| COPD‐exacerbation | 3 (16) | 19 | 8 (8) | 103 | 0.38 |
| Pneumonia | 12 (63) | 19 | 56 (54) | 103 | 0.48 |
| Sepsis incl. septic shock | 7 (39) | 18 | 52 (52) | 100 | 0.40 |
| Respiratory failure | 15 (79) | 19 | 68 (66) | 103 | 0.27 |
| MODS, DIC or ARDS | 5 (26) | 19 | 15 (15) | 103 | 0.20 |
| 30‐day mortality | 11 (58) | 19 | 48 (47) | 103 | 0.37 |
ICU, Intensive care unit; SAPS II, Simplified Acute Physiology Score II; COPD, Chronic obstructive respiratory disease; MODS, Multiple organ dysfunction syndrome; DIC, Disseminated intravascular coagulation; ARDS, Acute respiratory distress syndrome; Clinical intervention and diagnoses upon discharge in virus‐positive and virus‐negative ICU‐patients admitted with acute respiratory symptoms. Numerical variables are presented as medians and interquartile range. Categorical data are presented as frequencies and percentages. Wilcoxon rank sum was used for numerical data, and Chi‐square test or Fisher's exact test, for categorical data.
Microbiology
Viruses
Of the 122 patients included in the study group, 19 (16%) were positive for a virus, of which the most frequently detected were influenza A (n = 9) and RSV (n = 3, Fig. 2). Regarding influenza A, the subtypes were H3N2 in four patients and (H1N1)pdm09 in three. The remaining two patients were positive for influenza A during season 2008‐2009, but the subtype could not be determined.
Figure 2.
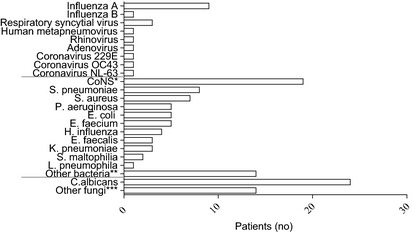
Distribution of viruses and the most frequently isolated microorganism in the study population The influenza A subtypes included 4 H3N2, 3 (H1N1)pdm09, and 2 untyped strains. *Coagulase‐negative staphylococci **M. catharrhalis, S. marcescens, E. cloacae, K. oxytoca, P. vulgaris, P. acnes, Pseudomonas‐ species, Micrococcus‐species, Bacillus‐species ***C. dubliniensis, C. koseri, C. glabrata, C. tropicalis, C. krusei, unspecified Candida‐species A total of 122 patients were tested for viruses, including parainfluenza viruses 1–3. Nineteen were virus‐positive. Co‐infections with clinically significant microbiological pathogens were found in four virus‐positive patients. Of the virus‐negative patients, 67 were positive for either bacteria or fungi, and two patients were not tested. Thirty‐four were negative for bacteria/fungi. Agents of considered relevance for the respiratory symptoms were isolated in 23 of the 67 patients; seven from blood and 21 from tracheal aspiration.
Bacteria and fungi
A negative viral swab was found in 103 patients (Fig. 1). Of these, 67 patients (65%) had bacteria or fungi in their respiratory specimens (n = 37), blood (n = 8) or both (n = 22) within 5 days of the viral sample collection date. Overall, the isolate most frequently detected in respiratory specimens was Candida albicans (n = 19, Fig. 2). The most frequently detected isolates in blood were coagulase‐negative staphylococci (CoNS) (n = 13). Of the 67 patients with any positive culture, 23 (34%) had culture results of considered clinical significance for the respiratory disease. Of the 59 patients with a positive respiratory culture, bacteria of considered clinical relevance were isolated in respiratory specimens from 21 patients (36%). The isolates included Streptococcus pneumoniae (n = 6), Staphylococcus aureus (n = 4), Pseudomonas aeruginosa (n = 5), Haemophilus influenza (n = 4), Klebsiella pneumoniae (n = 2) and Legionella pneumophila (n = 1).
Of 30 virus‐negative patients with a positive blood culture, bacteria of likely clinical relevance for the respiratory disease were isolated from seven patients (23%). The isolates included S. pneumoniae (n = 5), S. aureus (n = 1) and P. aeruginosa (n = 1).
For the remaining 36 of the 103 virus‐negative patients (35%) without positive microbiology, tracheal aspirations were performed in eight patients, sputum in three, bronchoalveolar lavage in one and blood culture in 34 patients. Two patients did not have respiratory or blood cultures collected.
Co‐detection of microbiological agents
Of the above‐mentioned 19 virus‐positive patients, a total of 10 patients (53%) were found to have a coinfection or colonization with bacteria or fungi. Five culture results (50%) were considered to be of clinical relevance to the respiratory symptoms (Table 3). The five coinfections were detected in tracheal aspirations, and represented 4% of the total study population. In addition, one patient, had hMPV and both CoNS and Candida dubliniensis of unknown clinical significance isolated in blood cultures. Nearly half of the 10 patients with a co‐detection of a microbe, including those with a clinically significant agent, were admitted directly to ICU, and only one patient was admitted more than 3 days after hospital admission. The most prevalent virus in the 10 patients with coinfections or colonizations was influenza A (two H3N2, three H1N1pdm09). The other viruses were influenza B, CoV OC43, CoV 229E, hMPV and rhinovirus, each detected once. The most frequent microbiological isolate in tracheal aspirations considered to be of possible clinical significance was S. aureus (n = 3), while P. aeruginosa and K. pneumoniae were detected once.
Table 3.
Microbiological agents in virus‐positive patients with co‐detection of bacteria/fungi (n = 10)
| Virus | Microbiological isolate | Material | Admission days prior to ICU |
|---|---|---|---|
| Influenza A (H1N1)pdm09 | S. aureus | Tracheal aspiration | 0 |
| Influenza A (H1N1)pdm09 | C. albicans a | Tracheal aspiration | 0 |
| Influenza A (H1N1)pdm09 | C. albicans a | Tracheal aspiration | 3 |
| Influenza A | K. pneumoniae | Tracheal aspiration | 1 |
| Influenza A | P. aeruginosa and CoNS b | Tracheal aspiration | 0 |
| Influenza B | S. aureus | Tracheal aspiration | 3 |
| Rhinovirus | C. albicans a | Tracheal aspiration | 1 |
| Coronavirus OC43 | S. aureus | Tracheal aspiration | 10 |
| Coronavirus 229E | C. albicans a | Tracheal aspiration | 0 |
| Human metapneumovirus | CoNS b and C. dubliniensis a | Blood culture | 2 |
Clinical significance unknown.
Coagulase‐negative staphylococci, clinical significance unknown. Of the 19 virus‐positive patients, nine were found to have microbial coinfections or colonizations in respiratory specimens. Five of these patients had potentially fatal bacteria present. An additional patient had coagulase‐negative staphylococci and Candida‐species in a blood culture, without having any tracheal aspiration‐analyses performed. Influenza A was the virus most frequently detected.
Clinical implications of infections with different microbiological agents
Table 4 summarizes clinical parameters in four groups of patients detected with and without viruses and with and without bacteria/fungi identified in blood or respiratory specimens within five days from the viral sample date. Chronic obstructive pulmonary disease (COPD) was a frequent predisposing comorbidity in the group of patients with a virus‐only infection, compared to the other patients. The COPD‐patients were also frequently observed with a virus‐only infection when admitted with exacerbation. The SAPS‐II score was independent of microbiological agent detected. Biochemical and radiological results could not reliably differ between the groups. However, it appeared that the virus‐only patients had a lower median level of C‐reactive protein in comparison with the Bacteria/Fungi‐only patients (47 mg/L compared to 122 mg/L, Wilcoxon rank sum, p = 0.035) when separated from the other groups. The same two groups also differed regarding median duration of mechanical ventilation, which appeared to be shorter among patients with a virus‐only infection (3 days compared to 9 days in Bacteria/Fungi‐only patients, Wilcoxon rank sum, p = 0.022). In all groups, positive for any microbiology, there was a high frequency of development of pneumonia, also for the patients with a virus‐only infection.
Table 4.
Clinical implications for infections with different microbiological agents (n = 122)
| Characteristic | Virus + Bacteria or fungi−(n = 9) | n | Virus + Bacteria or fungi + (n = 10) | n | Virus−Bacteria or fungi + (n = 67) | n | Virus−Bacteria or fungi−(n = 36) | n | p |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 72 (66–78) | 9 | 69 (50–76) | 10 | 71 (63–77) | 67 | 65 (61–74) | 36 | 0.32 |
| Gender male | 3 (33) | 9 | 5 (50) | 10 | 38 (57) | 67 | 18 (50) | 36 | 0.59 |
| Comorbidity | |||||||||
| COPD | 7 (88) | 8 | 1 (10) | 10 | 22 (34) | 64 | 10 (29) | 35 | 0.01 |
| Cardiac disease | 3 (33) | 9 | 3 (30) | 10 | 29 (44) | 66 | 14 (39) | 36 | 0.83 |
| Diagnoses at admission | |||||||||
| COPD exacerbation | 4 (44) | 9 | 1 (10) | 10 | 5 (7) | 67 | 3 (8) | 36 | 0.03 |
| Pulmonary infection | 2 (22) | 9 | 5 (50) | 10 | 24 (36) | 67 | 6 (17) | 36 | 0.09 |
| Parameters at admission | |||||||||
| SAPS II‐score | 54 (33–65) | 7 | 51 (35–76) | 10 | 47.5 (36–56) | 66 | 46 (34–65) | 35 | 0.79 |
| Body temperature, °C | 38.9 (38.2–39.4) | 6 | 37.1 (36.4–37.8) | 6 | 37.8 (37.4–38.3) | 36 | 37.4 (36.7–38.5) | 21 | 0.10 |
| CRP (mg/L) | 47a (29–57) | 9 | 74 (65–190) | 10 | 122a (47–232) | 67 | 85 (24–172) | 36 | 0.19 |
| Leukocytes (109 cell/L) | 13.7 (8.9–15.9) | 9 | 7.5 (6.6–13.2) | 10 | 10.8 (7.9–15.2) | 67 | 13.2 (8.4–17.8) | 36 | 0.23 |
| Infiltrates, chest X‐ray | 3 (38) | 8 | 4 (44) | 9 | 34 (62) | 55 | 20 (67) | 30 | 0.37 |
| Intervention and clinical course | |||||||||
| ICU‐stay (days) | 4 (2–6) | 9 | 10 (3–20) | 10 | 8 (3–17) | 67 | 4 (2–9) | 36 | 0.06 |
| Days until ICU‐transfer | 1 (0–4) | 9 | 1 (0–3) | 10 | 1 (0–5) | 66 | 1 (0–3) | 35 | 0.95 |
| Intubation (days) | 3b (2–4) | 8 | 8.5 (3–17) | 10 | 9b (3–14) | 46 | 4 (2–8) | 25 | 0.04 |
| Pneumonia | 5 (56) | 9 | 7 (70) | 10 | 43 (64) | 67 | 13 (36) | 36 | 0.037 |
| Respiratory failure | 8 (89) | 9 | 7 (70) | 10 | 47 (70) | 67 | 21 (31) | 36 | 0.33 |
| 30‐day mortality | 5 (56) | 9 | 6 (60) | 10 | 34 (51) | 67 | 14 (39) | 36 | 0.54 |
COPD, chronic obstructive respiratory disease; ICU, Intensive care unit; SAPS II, Simplified Acute Physiology Score II; CRP, C‐reactive protein.
Cardiac disease includes congestive heart failure, former myocardial infarction, angina pectoris or arterial hypertension.
The difference is significant when comparing these two groups only, p = 0.035.
The difference is significant when comparing these two groups only, p = 0.022.
Numerical data are presented as medians and interquartile range, categorical data as frequencies and percentages.
Fatal outcome
Of the 122 patients included in the study, 59 (48%) died within 30 days of the viral sample date. The non‐survivors had a median age of 72 years (IQR 64–79 years), which was higher than that of survivors, who had a median age of 66 years (IQR 58–73 years, Wilcoxon rank sum, p = 0.002). Immunosuppression was seen in 18 (31%) of the non‐survivors, and in nine (14%) of the survivors (X 2‐test, p = 0.027). The median SAPS II‐score in non‐survivors was 52 (IQR 41–61), which was higher than that in the survivors (score 43, IQR 34‐54, Wilcoxon rank sum, p = 0.001.)
Of the 19 virus‐positive patients, 11 (58%) died, out of which four patients were detected with influenza A and two patients were detected with RSV. The remaining five had positive tests for adenovirus, rhinovirus, hMPV, CoV 229E and CoV OC43. In total, four of the nine influenza A‐positive patients (44%) in the study population, of which two had the 2009 pandemic strain, and two of the three RSV‐positive patients (67%) died. Of the 11 virus‐positive non‐survivors, five (56%) had a co‐detection of bacteria/fungi in a respiratory specimen, of which two patients were detected with influenza A and C. albicans, one with rhinovirus and C. albicans, one with CoV OC43 and S. aureus and one with CoV 229E, E. coli and C. albicans. There were no significant differences between the virus‐positive survivors and non‐survivors in demographics or clinical characteristics to predict fatal outcome.
Of the above‐mentioned 67 virus‐negative patients with positive respiratory or blood cultures for bacteria or fungi, 34 (51%) died. The most frequent isolates were C. albicans (n = 10), enterococci (n = 5), S. aureus (n = 3), S. pneumoniae (n = 3), P. aeruginosa (n = 2), K. pneumoniae (n = 2), and H. influenzae (n = 2). Fatal outcome was independent of infection with different microbiological agents.
Discussion
This study is to our knowledge one of a few studies 2, 8, 12, 19, 20, which compares comprehensive virological PCR methods and microbiological findings in adult ICU‐patients without selecting for predisposing conditions. The study mimics the clinical everyday reality at an ICU in an attempt to ease the comparison between the results of the study and clinical practice. The virological analyses were based on multiplex real‐time PCR‐methods, detecting a large number of respiratory viruses, which now gradually are replacing the previous, less sensitive methods for rapid diagnostics of viruses.
ICU‐patients are a heterogeneous group, often with several concurrent diseases and complex medical histories. Although there are major guidelines on the management of community‐acquired pneumonia, consensus regarding the microbiological diagnostic approach is still lacking 1. There are for instance different approaches and opinions on which sample material is most suitable for identification of respiratory agents, and also on how proper sample collection is verified. Previously administered antibiotics may influence the results markedly, eventually disguising pathogens and promoting selection of other microorganisms present in the patient. The presence of bacteria or fungi in samples from the airways in ICU‐settings will only rarely be considered diagnostic for a given infection, as most of the isolations could represent both an infection and a colonizing organism. Even in blood cultures, the microorganism detected may not necessarily represent an infection. The results in our study illustrate both issues, as only 36% of the respiratory isolates and 23% of isolates detected in blood cultures from virus‐negative patients were considered to be clinically relevant for the respiratory disease.
The previously estimated frequencies of respiratory viruses in ICU‐patients vary from 9 to 30% 2, 12, 13, 14. The detection rate of 16% viruses in our study population confirms together with other studies that respiratory viruses are common in ICU‐patients, and confirms the validity of the PCR method used. Influenza A was the virus most frequently detected, followed by RSV, which supports previous results 12, 21, 22. The detection of respiratory viruses such as influenza, parainfluenza viruses and coronaviruses in the presence of acute respiratory symptoms have previously been shown to cause severe pneumonia, requiring intensive care treatment and mechanical ventilation 5, 8, 22, 23. The other respiratory viruses detected in our study, such as rhinoviruses, RSV and hMPV, have also been shown by several studies to cause significant symptoms, morbidity and potential need of intensive care treatment 6, 24. Most of the respiratory viruses detected in the multiplex PCR presented show seasonal variations of frequency 25, 26, 27. Although some weeks were missing at the beginning of the influenza season in our study, the main seasons for the different viruses were covered. Any seasonal variations of the viruses during the study period should be accommodated. The timing of the sample collection could have interfered in the number of positive samples if performed late in the clinical course of the infection, but 75% of the samples were taken within 4 days of admission with acute respiratory disease.
Regarding the clinical impact of the respiratory viruses detected, we found no parameters which reliably could distinguish virus‐positive and virus‐negative patients. There were no differences in the results of the clinical or paraclinical analyses. This underlines the importance of comprehensive and sensitive diagnostic measures when accurate diagnoses are sought, as clinical judgment alone cannot distinguish between the different aetiologies.
Clinical intervention and outcome between virus‐positive and virus‐negative patients were also similar in both groups, even when considering co‐detection of other microbiological agents. The only exception was a slightly shorter ventilation time for the patients with a virus‐only infection. The results may suggest that respiratory viruses have a similar influence on the clinical course as bacteria and fungi. The diagnoses upon discharge showed a large amount of patients developing pneumonia among both virus‐positive and virus‐negative patients. Although most virus‐positive patients were treated with antibiotics, only a third of the influenza‐positive patients with pneumonia received oseltamivir, which for all these cases occurred during the 2009 pandemic when the focus on influenza was high.
Coincident detection of a virus and one or more respiratory microbiological agents constituted more than 25% of the viral infections in our study, and 4% of the total study population, which fits well with the results of similar studies 3, 8, 12. Several studies throughout the last century have demonstrated the association between respiratory viruses and bacterial infections 3, 28, 29, 30. The most frequent combination of virus and bacteria in our study was influenza A and S. aureus, which is a well‐described relationship. Most patients were admitted either directly at the ICU or less than 3 days after hospital admission, which makes nosocomial infection or selection of opportunistic agents less likely. Influenza‐positive patients frequently had a coincident finding in respiratory cultures, of which two thirds of the microorganisms were considered clinically significant and likely to represent a severe super‐infection. A recent study reported similar results; approximately half of influenza‐patients presenting with a coinfection 12. These findings demonstrate that neither bacteriological nor virological analyses should stand alone. In this study, patients with a virus plus a microbiological agent tended to be admitted and ventilated for a similar period as the virus‐negative, bacteria/fungi‐positive patients. This could indicate a more severe morbidity caused by the combination of agents. We found no association between the number of deaths and different combinations of microbiological agents, but a large frequency of pneumonia was noted even among virus‐only infected patients. The findings are in line with some recent studies 12, 31, 32, and partly supported by a study on nearly 210.000 ICU‐patients which found a strong association between being infected with viruses and bacteria during the same hospitalization and developing septic shock or multiple organ dysfunction syndrome 8. As opposed to our results, this study also demonstrated a strong association with mortality in these patients 8.
Infections inevitably have an important influence on the clinical course in patients admitted to ICU. However, current microbiological diagnostic practices leave many patients undiagnosed, and the interpretation of the bacterial results are often difficult. Adding a comprehensive multiplex real‐time PCR increased the sensitivity of the respiratory diagnostics in our study, and results on microbiological agents were provided for 70% of the patients, though not all agents were likely to be of further clinical interest.
The results for the virus‐positive patients with co‐detected microorganisms were equally difficult to interpret, with only 50% of the isolates being of clinical interest. A potential pathogenic effect of presumably colonizing organisms in the remaining culture‐positive patients cannot be ruled out, though. This supports the fact that interpreting current practice microbiological analyses is difficult in ICU‐patients. This was also recently pointed out in a study investigating biomarkers in bronchoalveolar lavages for diagnosing respiratory viral infections 33. The clinical picture of respiratory tract infections in ICU is further complicated by the fact that severe infections with focus outside of the respiratory system may also result in pulmonary congestion and infiltrates due to ARDS. These changes may resemble pneumonia, but the pathogen may not be found in either the respiratory secretions or in the blood. Improving the sensitivity of microbiological analyses with molecular methods, or adding other detection methods or biomarker analyses may reduce the number of severely ill patients with undetermined aetiology of their respiratory disease.
The virus‐negative patients received combination antibiotic therapy significantly more frequently, which might be connected to a generally worse clinical condition. It is noteworthy that all of the virus‐positive patients received antibiotics, and 63% even combination therapy. This may further cloud interpretation of bacterial analyses, and contribute to the lack of diagnoses in some patients. A more comprehensive diagnostic programme than today's practice, including a wider use of molecular methods in bacteriology, could possibly result in more accurate diagnoses and a reduction in the use of empirical broad‐spectrum antibiotic treatment. In greater perspectives, this could contribute to relieving the increasing issues with antibiotic resistance. There is still a need for great awareness, refined clinical guidelines and implementation of comprehensive microbiological and virological analyses to address these issues.
We are aware of the limitations of this study, of which the main are the low number of virus‐positive patients and the lack of a control group of patients without respiratory symptoms. The SAPS‐II‐scores were performed retrospectively at one of the hospitals, although performed blinded regarding the other results; this may have influenced the scores. More than 75% of the patients were admitted either directly at the ICU or were transferred less than 4 days after hospital admission. Still, antibiotic treatment, nosocomial infections and selection of opportunistic agents may have happened prior to entering the study. Unfortunately, information regarding this was not available for a suitable amount of the patients.
Conclusions
In this study, death was independent of the presence of viruses, and a fairly large proportion of the virus‐positive patients developed pneumonia. The investigated clinical and paraclinical parameters were not different in viral infections compared to other agents. This may suggest that respiratory viruses have a similar impact on the clinical course as other pathogens. Interpretation of microbiological results in ICU‐settings is difficult, as several diseases and conditions may be combined in each patient and the results might be influenced by previously administered antibiotics. In more than 25% of the virus‐positive patients in our study, respiratory bacteria or fungi considered to be clinically significant were identified, which demonstrates that neither virological nor bacteriological analyses should stand alone. The patients should be tested with optimized, sensitive and comprehensive methods, and interpretations should preferably be done in collaboration with a clinical microbiologist or specialists in infectious diseases to enhance the diagnostic process.
Conflict of Interests
The authors declare that they have no competing interest of any nature.
We thank Dr. med Asger Bendtsen (Department of Anaesthesiology, Glostrup Hospital, Glostrup, Denmark), MD Kirsten Gani (Department of Anaesthesiology, Herlev Hospital, Herlev, Denmark) and Prof., Dr. med Henrik Birgens (Department of Haematology, Herlev Hospital, Herlev, Denmark) for invaluable contribution to the study concept and execution, Chief Nursing Officer Marlene Klarskov Fleischer (Department of Anaesthesiology, Glostrup Hospital, Glostrup, Denmark) for execution of the study, and Jens Nielsen (Department of Epidemiology, Statens Serum Institut, Copenhagen, Denmark) for advice on statistics. Furthermore, we are greatly indebted to nursing staff and attending physicians of the two ICUs for their kind cooperation in this study.
Østby A‐C , Gubbels S , Baake G , Nielsen LP , Riedel C , Arpi M . Respiratory virology and microbiology in intensive care units: a prospective cohort study. APMIS 2013; 121: 1097–1108.
References
- 1. Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious diseases society of America/American thoracic society consensus guidelines on the management of community‐acquired pneumonia in adults. Clin Infect Dis 2007;44:S27–S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Legoff J, Guerot E, Ndjoyi‐Mbiguino A, Matta M, Si‐Mohamed A, Gutmann L, et al. High prevalence of respiratory viral infections in patients hospitalized in an intensive care unit for acute respiratory infections as detected by nucleic acid‐based assays. J Clin Microbiol 2005;43:455–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Roux AD, Marco MA, Garcia E, Mensa J, Ewig S, Lode H, et al. Viral community‐acquired pneumonia in nonimmunocompromised adults. Chest 2004;125:1343–51. [DOI] [PubMed] [Google Scholar]
- 4. Jennings LC, Anderson TP, Beynon KA, Chua A, Laing RTR, Werno AM, et al. Incidence and characteristics of viral community‐acquired pneumonia in adults. Thorax 2008;63:42–s8. [DOI] [PubMed] [Google Scholar]
- 5. Luyt C‐E, Combes A, Nieszkowska A, Trouillet J‐L, Chastre J. Viral infections in the ICU. Curr Opin Crit Care 2008;14:605–8. [DOI] [PubMed] [Google Scholar]
- 6. Wedzicha JA. Role of viruses in exacerbations of chronic obstructive pulmonary disease. Proc Am Thorac Soc 2004;1:115–20. [DOI] [PubMed] [Google Scholar]
- 7. Jacoby DB. Virus‐induced asthma attacks. Jama‐Journal of the American Medical Association 2002;287:755–61. [DOI] [PubMed] [Google Scholar]
- 8. Miggins M, Hasan A, Hohmann S, Southwick F, Casella G, Schain D, et al. The Potential Influence of Common Viral Infections Diagnosed during Hospitalization among Critically Ill Patients in the United States. PLoS ONE. 2011;6: e18890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazzulli T. Value of RVP in clinical settings: intensive care. J Clin Virol 2007;40:S55–S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woodhead M, Welch CA, Harrison DA, Bellingan G, Ayres JG. Community‐acquired pneumonia on the intensive care unit: secondary analysis of 17 869 cases in the ICNARC Case Mix Programme Database. Crit Care. 2006;10(Suppl 2): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson PA, Ferguson J. Severe community‐acquired pneumonia: an Australian perspective. Inter Med J 2005;35:699–705. [DOI] [PubMed] [Google Scholar]
- 12. Cillóniz C, Ewig S, Ferrer M, Polverino E, Gabarrús A, de la Bellacasa JP, et al. Community‐acquired polymicrobial pneumonia in the intensive care unit: aetiology and prognosis. Crit Care 2011;15:R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sandrock C, Stollenwerk N. Acute febrile respiratory illness in the ICU–reducing disease transmission. Chest 2008;133:1221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daubin C, Parienti J‐J, Vincent S, Vabret A, du Cheyron D, Ramakers M, et al. Epidemiology and clinical outcome of virus‐positive respiratory samples in ventilated patients: a prospective cohort study. Crit Care. 2006;10: R142. doi: 10.1186/cc5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brittain‐Long R, Nord S, Olofsson S, Westin J, Anderson LM, Lindh M. Multiplex real‐time PCR for detection of respiratory tract infections. J Clin Virol 2008;41:53–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinstein MP, Reller LB, Murphy JR, Lichtenstein KA. The clinical‐significance of positive blood cultures – a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults .1. laboratory and epidemiologic observations. Rev Infect Dis 1983;5:35–53. [DOI] [PubMed] [Google Scholar]
- 17. Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett′s Principles and Practice of Infectious Diseases. 7th edition USA: Churchill Livingstone; 2010. [Google Scholar]
- 18. Cardoso TC, Lopes LM, Carneiro AH. A case‐control study on risk factors for early‐onset respiratory tract infection in patients admitted in ICU. BMC pulmon med 2007;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bauer TT, Ewig S, Rodloff AC, Mueller EE. Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin Infect Dis 2006;43:748–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daubin C, Vincent S, Vabret A, du Cheyron D, Parienti JJ, Ramakers M, et al. Nosocomial viral ventilator‐associated pneumonia in the intensive care unit: a prospective cohort study. Intensive Care Med 2005;31:1116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cameron RJ, de Wit D, Welsh TN, Ferguson J, Grissell TV, Rye PJ. Virus infection in exacerbations of chronic obstructive pulmonary disease requiring ventilation. Intensive Care Med 2006;32:1022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garbino J, Gerbase MW, Wunderli W, Kolarova L, Nicod LP, Rochat T, et al. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest 2004;125:1033–9. [DOI] [PubMed] [Google Scholar]
- 23. Oliveira EC, Lee B, Colice GL. Influenza in the intensive care unit. J Int Care Med 2003;18:80–91. [DOI] [PubMed] [Google Scholar]
- 24. Carman WF, Mahoney JB. The pathogens. J Clin Virol 2007;40:S5–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brittain‐Long R, Andersson LM, Olofsson S, Lindh M, Westin J. Seasonal variations of 15 respiratory agents illustrated by the application of a multiplex polymerase chain reaction assay. Scand J Infect Dis 2012;44:9–17. [DOI] [PubMed] [Google Scholar]
- 26. Monto AS. Occurrence of respiratory virus: time, place and person. Pediatr Infect Dis J 2004;23:S58–64. [DOI] [PubMed] [Google Scholar]
- 27. Garbino J, Soccal PM, Aubert JD, Rochat T, Meylan P, Thomas Y, et al. Respiratory viruses in bronchoalveolar lavage: a hospital‐based cohort study in adults. Thorax 2009;64:399–404. [DOI] [PubMed] [Google Scholar]
- 28. Metersky ML, Masterton RG, Lode H, File TM Jr, Babinchak T. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases 2012;16:e321–31. [DOI] [PubMed] [Google Scholar]
- 29. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008;198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thorburn K, Harigopal S, Reddy V, Taylor N, van Saene HK. High incidence of pulmonary bacterial co‐infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006;61:611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin‐Loeches I, Sanchez‐Corral A, Diaz E, Maria Granada R, Zaragoza R, Villavicencio C, et al. Community‐Acquired respiratory coinfection in critically iii patients with pandemic 2009 influenza A(H1N1) virus. Chest 2011;139:555–62. [DOI] [PubMed] [Google Scholar]
- 32. De Serres G, Lampron N, La Forge J, Rouleau I, Bourbeau J, Weiss K, et al. Importance of viral and bacterial infections in chronic obstructive pulmonary disease exacerbations. J Clin Virol 2009;46:129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sumino KC, Walter MJ, Mikols CL, Thompson SA, Gaudreault‐Keener M, Arens MQ, et al. Detection of respiratory viruses and the associated chemokine responses in serious acute respiratory illness. Thorax 2010;65:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


