Abstract
This study reports the prevalence of potential faecal pathogens in the microbiome detected in a cohort of cats and dogs with diarrhoea in Perth, Western Australia. Records from a commercial diagnostic laboratory using faecal PCR testing between July 2014 and August 2015 were reviewed.Of 289 feline faecal samples reviewed, Salmonella spp. (1.7%), Campylobacter spp. (47.6%), Clostridium perfringens (81.3%), Giardia spp. (11.1%), Toxoplasma gondii (1.2%), Tritrichomonas foetus (4.8%), panleukopenia virus (6.5%) and coronavirus (39.5%) were detected. In dogs, Salmonella spp. (5.4%), Campylobacter spp. (36.3%), C. perfringens (85.4%), Giardia spp. (6.2%), parvovirus (9.4%), coronavirus (4.7%) and distemper virus (1.5%) were detected.
Keywords: cats, diarrhoea, dogs, faecal microbiome
Abbreviations
- CCV
canine coronavirus
- CPV
canine parvovirus
Faecal PCR panels are frequently assessed in the investigation of diarrhoea in cats and dogs. The interpretation of these results can be difficult when assessing causality and involves both clinical and epidemiological assessment. It is often uncertain if infectious diarrhoea is a result of a single pathogen or due to the concurrent presence of viral, protozoal or bacterial pathogens; therefore, identification of potential infectious agents may assist in strategic case management. This retrospective review investigated the prevalence of enteric bacterial, protozoal and viral microorganisms and whether co‐infection was present in a cohort of diarrhoeic dogs and cats in Perth, Western Australia.
Faecal samples from dogs and cats were submitted to a laboratory for investigation of diarrhoea. Samples were assessed from July 2014 to August 2015 and were from domestic pets and shelter inhabitants. Faecal samples were analysed by PCR utilising multiplex tandem reverse‐transciptase (RT‐PCR) methods† to allow the screening of a panel of multiple infectious agents. For dogs, the panel included: Campylobacter spp., Clostridium perfringens alpha‐toxin gene, Salmonella spp., canine parvovirus (CPV), Giardia lamblia, Cryptosporidium (parvum and hominis), canine coronavirus (CCV) and canine distemper virus. For cats, the panel included: Campylobacter spp., C. perfringens alpha‐toxin gene, Salmonella spp., feline panleukopenia virus, Toxoplasma gondii, Tritrichomonas foetus, G. lamblia, Cryptosporidium (parvum and hominis) and feline coronavirus. Results were retrospectively reviewed.
Data from 289 feline faecal samples were reviewed, which included 59 entire females, 66 spayed females, 66 entire males and 65 neutered males. The genders from 33 felines were unclassified. There were samples from 27 various breeds (Table 1), and there were 25 individuals whose breed was not classified. Ages ranged from 25 days to 20 years, with a median of 1 year.
Table 1.
Listed breeds producing faecal samples analysed
| Feline faecal amples | Canine faecal samples | ||
|---|---|---|---|
| Feline breed | Number | Canine breed | Number |
| Domestic short hair | 137 | Crossbreed | 80 |
| Ragdoll | 27 | German shepherd | 36 |
| Burmese | 10 | Cavalier King Charles | 23 |
| Maine Coone | 9 | Poodle | 17 |
| Tonkingese | 9 | Golden retriever | 17 |
| British Shorthair | 7 | Labrador | 13 |
| Domestic long hair | 6 | Staffordshire terrier | 16 |
| Birman | 6 | Maltese | 12 |
| Russian Blue | 6 | Border collie | 12 |
| Bengal | 5 | Greyhound | 10 |
| Siamese | 4 | Cocker spaniel | 10 |
| Persian | 4 | Miniature schnauzer | 8 |
| Chinchilla | 2 | French bulldog | 7 |
| Tiffany | 2 | Siberian husky | 7 |
| Burmilla | 1 | Jack Russell terrier | 7 |
| Snowshoe | 1 | Rottweiler | 7 |
| Cornish rex | 1 | Boxer | 6 |
| Somali | 1 | Shih Tzu | 5 |
| Oriental | 1 | Bernese mountain dog | 4 |
| Devon rex | 1 | Great Dane | 4 |
| Exotic | 1 | Doberman | 3 |
| Sphinx | 1 | Weimaraner | 3 |
| Scottish fold | 1 | Chihuahua | 3 |
| Siberian | 1 | Kelpie | 3 |
| Dog de Bordeaux | 3 | ||
Organisms detected in feline faecal samples are shown in Figure 1. Coronavirus was found to have 39.5% prevalence in the cats tested. Cryptosporidium spp. was not detected in any sample. T. gondii was detected in three samples (1.2%), and all had co‐presence of C. perfringens. Of the samples assessed, 57.4% had more than one organism, as demonstrated in Figure 2. All 14 samples in which T. foetus was detected had co‐infection with other pathogens. C. perfringens was present in 235 (81.3%) samples and as a single faecal pathogen in 77 cats, or 32.8% of all cats with this pathogen detected, and as a dual infection in 158 (68.2%). No breed predisposition was detected for any infectious agent.
Figure 1.
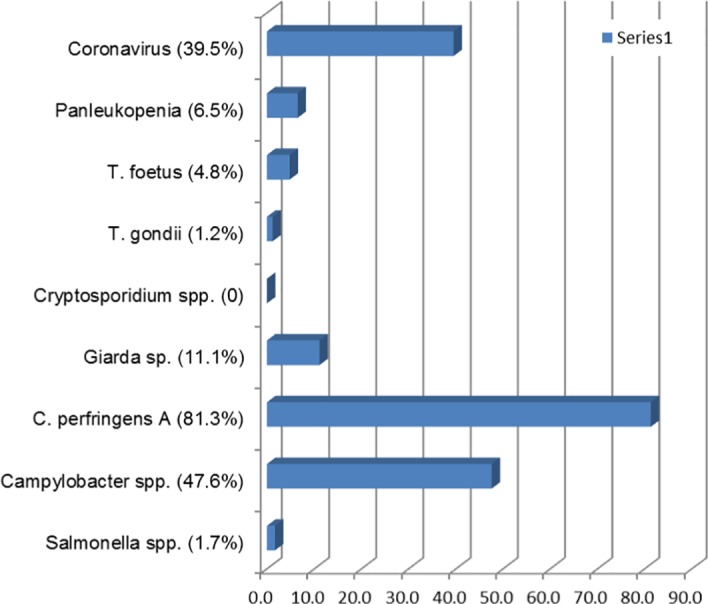
Enteropathogens detected in faecal samples from cats. Expressed as a percentage of faecal microorganisms detected.
Figure 2.
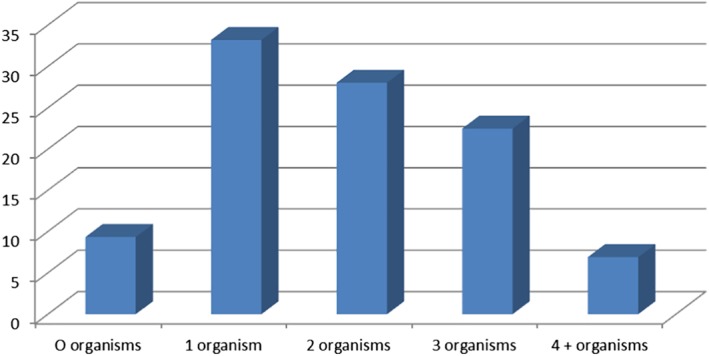
Number of organisms detected in feline diarrhoeic faecal samples. Expressed as a percentage of faecal microorganisms detected.
Of the 80 shelter cats assessed with diarrhoea, 6 had no microorganisms detected. Most commonly identified were Campylobacter spp. (42/80), C. perfringens (65/80) and coronavirus (43/80). No Salmonella spp. was found in the faecal samples of any shelter cat. Giardia spp. and panleukopenia virus were occasionally detected (7/80 and 11/80, respectively).
There were 405 canine faecal samples assessed. Canine faecal samples were obtained from 96 entire males, 104 neutered males, 75 entire females and 105 neutered females, and gender was unclassified in 25 individuals. Breeds from which faecal samples were analysed are listed in Table 1, with an additional 35 other breeds, with fewer than three samples from each breed identified. Ages of dogs ranged from 10 days to 15.5 years, with a median 2 years and 3 months. Similar to cats, no breed predisposition was noted for any potential infectious pathogen.
Of the 405 samples, 7 were from shelter animals and 2 from racing greyhounds. Eight other greyhound samples were from either dogs retired from racing or puppies. The remainder of the submitted samples were from pets. No samples tested positive for Cryptosporidium spp. (Figure 3). Typically, there were one to two organisms detected in samples, and 35 (8.6%) dogs with diarrhoea had no potential pathogens detected (Figure 4). C. perfringens was present in 85.4% of canine faecal samples and as a single infection in 180 dogs, or 52% of those identified with the organism and as a dual infection in 166 (48%).
Figure 3.
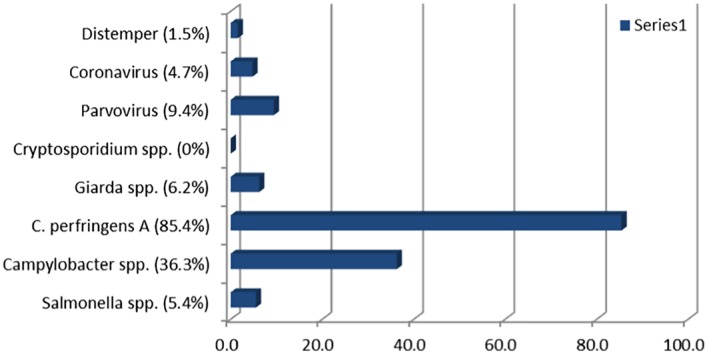
Enteropathogens detected in faecal samples from dogs. Expressed as a percentage of faecal microorganisms detected.
Figure 4.
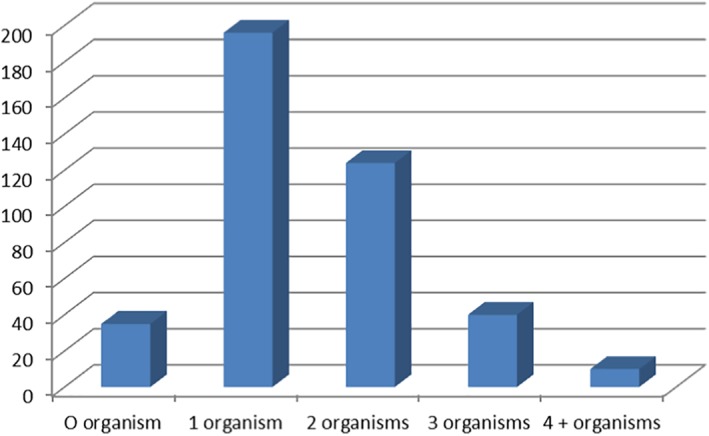
Number of organisms detected in canine diarrhoeic faecal samples. Expressed as a percentage of faecal microorganisms detected.
A previous study identified one or more organisms in 68.3% of diarrhoeic dogs, of which 54.9% were single and 45.1% had co‐infections.1 Co‐infections have previously been reported to occur in 44% and 57.4% of diarrhoeic cats by faecal PCR.2, 3 One or more pathogens were identified in >90% of cats and dogs in this study; however, this difference may reflect colonisation rather than a causal pathogen and may better be termed co‐carriage of the organisms. C. perfringens was identified in 83.5 and 85.4% of cat and dog faeces, respectively, as both singular and dual organism presence. The high prevalence of C. perfringens in healthy animals is a major limitation of faecal PCR assays, particularly those assays such as the assay used here, that target the alpha‐toxin gene. The alpha‐toxin gene is of questionable virulence and is present in all C. perfringens strains. The pathogen that was detected as a co‐infection in 67.2% and 48% of cats and dogs, respectively, may represent a secondary or commensal carriage as the PCR is not specific for clostridial enteropathy. Combining C. perfringens enterotoxin detection by ELISA with PCR detection of enterotoxogenic strains is currently recommended to facilitate the diagnosis of C. perfringens‐associated diarrhoea.4
A study of feline shelters in California, USA identified the prevalence of Giardia spp. and Cryptosporidium spp. by faecal floatation, immunoassay and direct immunofluorescence as 9.8% and 4.7%, respectively, in diarrhoeic and non‐diarrhoeic cats.5 Faecal PCR in diarrhoeic cats has previously been reported to demonstrate Giardia spp. in 20.6%, and in a study of diarrhoeic dogs with faecal PCR, a prevalence of 13.5% was found.1, 2 Giardia spp. was detected in 6.2% and 11.1% of diarrhoeic dogs and cats, respectively, in this study. Cryptosporidium spp. was not detected in any sample. This may reflect an absence of infection, the effect of antigenic diversity on the performance of the assay or a lack of sensitivity to detect Cryptosporidium felis or Cryptosporidium canis that infects dogs and cats.6 The collection method of the faeces is unknown, and if a combined sample or increased sample numbers are assessed, the predictive value of the test increases, which has unknown impact on the results of this study.5, 7
Giardia spp. is considered a potential zoonotic pathogen and is therefore of public health interest. It was detected in 6.2% of cats, of which 13 of 18 with known ages were <1 year old, and 11.1% of dogs (5 weeks 5 days to 4 years 2 months, median 6 months). This is similar to a meta‐analysis of 15.2% and 12% in dogs and cats, respectively, and is less than a UK study that detected 20.6% prevalence through a faecal PCR.2, 8 While direct immunofluorescent coproscopy has been adopted as the gold standard for identifying faecal Giardia cysts, immunoassays have been developed to detect excreted Giardia antigen with generally poor positive predictive value, possibly in part due to variable faecal shedding.9 In people, comparison of microscopy, ELISA and real‐time PCR for the detection of Giardia intestinalis in faecal specimens has been assessed, with an RT‐PCR (79.8%) much more sensitive and beneficial for rapid and accurate diagnosis compared to ELISA (46.8%).10 Considering the zoonotic potential of this pathogen, further assessment of the faecal PCR compared to other techniques would seem prudent to determine if more rapid identification, and thus environmental control measures, could be performed.
Coronavirus was found to have 39.5% prevalence in the cats tested, which is less than that observed in previous studies. A recent study in the United Kingdom reported a prevalence of 56.9% by faecal PCR in diarrhoeic cats.2 Coronavirus was detected in 34% of pet cats (25 days to 16 years old, median 1 year old) and 43% of shelter cats. Faecal antigen can be shed for 2–10 months post‐infection and is more prevalent in kittens and older cats as seen with the increased incidence in cats <1 year here. In addition, as cats with increased serological titres are more likely to shed faecal antigen than cats with reduced titres, the faecal PCR test may prove useful to identify infected cats and reduce transmission in the cattery/shelter environment.11
CCV was detected in 4.7% of dogs and was mainly identified in younger dogs (range 6 weeks to 6 years, median 22 weeks). In 5 (26%) of 19 of these dogs, there was co‐infection with CPV. Given that co‐infection with CCV can potentially enhance the severity of CPV and hence the prognosis or prolong hospitalisation, the faecal PCR may provide important prognostic information.12
Salmonella spp. was identified in 1.7% of feline and 5.4% of canine faeces, significantly lower than a previous study of cats in the UK, where Salmonella enterica had 20.6% prevalence through faecal PCR.2 While the pathogen was isolated, this may reflect colonisation or carriage rather than infection. Its presence in healthy dogs and cats can be similar to those with diarrhoea. Furthermore, in animals that hunt and eat raw meat, there is more likely to be increased colonisation and shedding of Salmonella spp. The prevalence of Salmonella spp.in diarrhoeic dogs and cats ranges from 0% to 3.5% and from 0% to 8.6%, respectively, whereas the prevalence range for Salmonella spp. in stray or shelter dogs and cats is 0%–51.4%.4 It was not identified in any dog or cat from a shelter in this study. It was only identified in young cats (range 14 weeks to 2 years and 10 months, median 1 year old), which may reflect a naïve host immune response or a type II error given that only 289 samples were assessed. There was no age cluster in dogs, and it was detected in dogs aged 14 weeks to 15 years. While Salmonella spp. detection is of uncertain pathogenicity, salmonellosis is a major zoonosis, and all positive PCR samples should be cultured using selective enrichment media to isolate and identify the infecting organism.
PCR has previously been identified to have a sensitivity of 94% for the detection of T. foetus and is a commonly used test.13 T. foetus was identified in 4.8% (14/289) of cat stools, of which only one was identified as being from a shelter environment. This compared to a previous study in Australia where no cats in shelters or catteries were identified to have this infection.14 Trichomonad infections have been notably identified to occur as co‐infections, most notably Giardia spp. and Coccidia spp.15 The presence of Coccidia spp. was not assessed; however, five cats (35.7%) were identified to have co‐infection with Giardia spp. All cats had co‐infection, with variable presence of Giardia spp., coronavirus, C. perfringens and Campylobacter spp. Given that more than half of the cats that go into clinical remission will have PCR evidence of trichomonad infection and therefore be asymptomatic carriers,15 comment cannot be made on the causality and detection of this pathogen. Diagnosis should be made with respect to clinical presentation.
Campylobacter spp. was detected in 47.6% and 36.3% of cats and dogs, respectively. Some Campylobacter species are non‐pathogenic and can form part of the normal intestinal microbiota. Campylobacter spp. has variably been reported, with 58%–97% detected by PCR in dog faeces and the same study identifying 58% of healthy dogs and 97% of diarrhoeic dogs shedding detectable levels of Campylobacter spp. with Campylobacter coli, Campylobacter concisus, Campylobacter fetus, Campylobacter gracilis, Campylobacter helveticus, Campylobacter jejuni, Campylobacter lari, Campylobacter mucosalis, Campylobacter showae, Campylobacter sputorum and Campylobacter upsaliensis levels significantly higher in the diarrhoeic population.16 Detection of Campylobacter spp. in a faecal PCR suggests that further molecular techniques combined with the interpretation of clinical signs should be used to characterise the infection.
Dogs and cats that are housed under crowded conditions such as kennels or shelters are more likely to be culture positive for Campylobacter spp. than household animals.4 Over 50% of cats (42/80) had Campylobacter spp. detected, and of the seven dogs from shelters, none had Campylobacter spp. detected, although this is likely a type II error. The larger number of cats may reflect, for example, the detection of C. helveticus and C. upsaliensis, which may form part of the normal microbiota.
Of this cohort, Campylobacter spp. and C. perfringens were commonly detected in dogs and cats, in addition to coronavirus in cats. Similar to previous studies, multiple potential infectious agents were detected in cats and dogs, which supports the concept of pathogen interdependence. Further studies on the specific presence of types of co‐infection or co‐carriage of organisms and the severity of diarrhoea and response to specific treatment and management may assist in advancing treatment strategies in cases of infectious diarrhoea. The diagnostic utility of the multiplex PCR faecal panel should be optimised by undertaking other laboratory methods such as culture, microscopy and ELISA‐based assays.
Conflicts of interest and sources of funding
The authors declare no conflicts of interest or sources of funding for the work presented here.
Acknowledgments
The authors thank the reception and laboratory staff of Vetpath Laboratory Services for their assistance with data collection and sample processing.
Footnotes
Vetpath Laboratory Services Canine and Feline Faecal Multiplex PCR Panel.
References
- 1. da Rocha Gizzi AB, Tostes Oliveira S, Leutenegger LM et al. Presence of infectious agents and co‐infections in diarrheic dogs determined with a real‐time polymerase chain reaction‐based panel. BMC Vet Res 2014;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paris JK, Wills S, Balzer HJ et al. Enteropathogen co‐infection in UKcats with diarrhoea. BMC Vet Res 2014;10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sabshin SJ, Levy JK, Tupler T et al. Enteropathogens identified in cats entering a Florida animal shelter with normal feces or diarrhea. J Am Vet Med Assoc 2012;241:331–337. [DOI] [PubMed] [Google Scholar]
- 4. Marks SL, Rankin SC, Byrne BA et al. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med 2011;25:1195–1208. [DOI] [PubMed] [Google Scholar]
- 5. Mekaru SR, Marks SL, Felley AJ et al. Comparison of direct immunofluorescence, immunoassays, and fecal flotation for detection of Cryptosporidium spp. and Giardia spp. in naturally exposed cats in 4 Northern California animal shelters. J Vet Intern Med 2007;21:959–965. [DOI] [PubMed] [Google Scholar]
- 6. Fayer R, Trout JM, Xiao L et al. Cryptosporidium canis n. sp. from domestic dogs. J Parasitol 2001;87:1415–1422. [DOI] [PubMed] [Google Scholar]
- 7. Marks SL, Hanson TE, Melli AC Comparison of direct immunofluorescence, modified acid‐fast staining, and enzyme immunoassay techniques for detection of Cryptosporidium spp in naturally exposed kittens. J Am Vet Med Assoc 2004;225:1549–1553. [DOI] [PubMed] [Google Scholar]
- 8. Bouzid M, Halai K, Jeffreys D et al. The prevalence of Giardia infection in dogs and cats, a systematic review and meta‐analysis of prevalence studies from stool samples. Vet Parasitol 2015;207:181–202. [DOI] [PubMed] [Google Scholar]
- 9. Rishniw M, Liotta J, Bellosa M et al. Comparison of 4 giardia diagnostic tests in diagnosis of naturally acquired canine chronic subclinical giardiasis. J Vet Intern Med 2010;24:293–297. [DOI] [PubMed] [Google Scholar]
- 10. Beyhan YE, Taş Cengiz Z Comparison of microscopy, ELISA, and real‐time PCR for detection of Giardia intestinalis in human stool specimens. Turk J Med Sci 2017;47:1295–1299. [DOI] [PubMed] [Google Scholar]
- 11. Pederson NC, Allen CE, Lyons LA Pathogenesis of feline enteric coronavirus infection. J Feline Med Surg 2008;10:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCaw DL, Hoskins JD. Canine viral enteritis In: Greene CE, editor. Infectious diseases of the dog and cat. 3rd edn. Saunders Elsevier, 2006;63–73. [Google Scholar]
- 13. Gookin JL, Stebbins ME, Hunt E et al. Prevalence of and risk factors for feline Tritrichomonas foetus and giardia infection. J Clin Microbiol 2004;42:2707–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bissett S, Stone BS, Malik R et al. Observed occurrence of Tritrichomonas foetus and other enteric parasites in Australian cattery and shelter cats. J Feline Med Surg 2009;11:803–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao C, Köster LS Tritrichomonas foetus infection, a cause of chronic diarrhea in the domestic cat. Vet Res 2015;46:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaban B, Ngeleka M, Hill JE Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol 2010;10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]


