Abstract
Objective A real‐time polymerase chain reaction (PCR)/high‐resolution melt (HRM) curve analysis protocol was developed in our laboratory to differentiate infectious bronchitis (IB) virus reference strains. In the current study, this method was used to detect and classify IB viruses in field submissions.
Procedure Over an 11‐month period samples from 40 cases of suspected IB virus were received and 17 submissions were positive for IB virus by polymerase chain reaction. HRM curve analysis classified each strain as subgroup 1, 2 or 3 strain (12 submissions) or a strain that was unable to be classified (5 submissions). The 3′ untranslated region (UTR) and partial S1 gene nucleotide sequences for the 17 IB virus strains were determined and their identity with those of the relative reference strains compared to confirm the classifications generated using the HRM curve analysis.
Results Of the 12 IB field viruses classified as subgroup 1, 2, or 3 using HRM curve analysis, the 3′UTR and S1 gene nucleotide sequences had identities ≥99% with the respective subgroup reference strain. Analysis of the 3′ UTR and S1 gene nucleotide sequences for the five IB virus strains that could not be classified indicated that four belonged to one of the subgroups, and one was a potential recombinant strain (between strains from subgroups 2 and 3). A novel recombinant strain was also detected.
Conclusion HRM curve analysis can rapidly assign the majority of IB viruses present in field submissions to known subgroups. Importantly, HRM curve analysis also identified variant genotypes that require further investigation.
Keywords: coronavirus, high‐resolution melt, infectious bronchitis virus, recombinant viruses, strain identification
Abbreviations
- bp
base pairs
- GCP
genotype confidence percentage
- HRM
high‐resolution melt
- IB
infectious bronchitis
- NSW
New South Wales
- PCR
polymerase chain reaction
- QLD
Queensland
- SPF
specific‐pathogen free
- UTR
untranslated region
- VIC
Victoria
- WA
Western Australia
A type 3 coronavirus is the causative agent of infectious bronchitis (IB), which is a disease of major significance throughout the international poultry industry. Infection in chickens causes tracheitis and/or interstitial nephritis, 1 , 2 the extent of which is dependent on a number of factors, including the virulence of the strain. In layers, infection with IB virus has also been implicated in declines in egg quality and production rate, 3 , 4 caused by cytopathological changes in the oviduct, 5 which may or may not be accompanied by characteristic respiratory signs. 6
Recently, we demonstrated the application of an emerging technology, high‐resolution melt (HRM) curve analysis following real‐time polymerase chain reaction (PCR), to differentiate IB virus strains in Australia against known reference and vaccine strains. 7 This method could successfully and objectively classify IB virus strains as subgroup 1, 2 or 3, based on differences in their 3′ untranslated region (3′UTR).
However, the ability of HRM analyses to differentiate IB virus strains in field submissions and/or detect novel and/or recombinant strains has not been studied to date. Recombination of coronaviruses has been demonstrated in the group 2 coronavirus mouse hepatitis virus. 8 Recombination of coronaviruses that infect different species of animals has also been detected. 9 The ability of the IB virus to generate new strains through recombination has been suggested and examined in a number of investigations, 10 , 11 , 12 , 13 and a number of recombination sites have been detected throughout the length of the IB virus genome.
The purpose of this study was to assess the capacity of HRM curve analysis to differentiate IB virus strains in field submissions, in comparison with S1 gene sequencing, as a routine method for diagnosis.
Materials and methods
IB virus strains
The viral strains used in this study included vaccine strain I (subgroup 1 reference strain) (Fort Dodge Australia), N1/88 (subgroup 2 reference strain) and N1/03 (subgroup 3 reference strain). The S1 gene GenBank 14 accession numbers are FJ235187, DQ490207 and FJ235194, respectively. The 3′UTR GenBank accession numbers are FJ235181, DQ490207 and FJ235186, respectively.
Specimens from 40 cases of suspected IB virus infection, which were submitted to the University of Melbourne between 1 June 2008 and 30 April 2009, were examined by PCR to detect IB virus and positive specimens were further examined using HRM curve analysis. Table 1 shows the location of each farm, the age of the flock, the type of tissue submitted and the age at which the chickens were vaccinated against IB virus (in all cases with subgroup 1 vaccines) for each IB‐virus‐positive field submission. All IB virus strains identified in field submissions and not classified within a subgroup were designated ‘unclassified’.
Table 1.
Field submissions from broiler chickens determined to be positive for infectious bronchitis (IB) virus
| ID | Submission type | Farm location | Age of chicken (days) | Age vaccinated against IB virus (days) b |
|---|---|---|---|---|
| 013 | Tissue (trachea) | VIC | 28 | 1 and 7 |
| 015 | Tissue (trachea) | NSW | 54 | 1 and 21 |
| 018 | Tissue (trachea) | NSW | 25 | 1 and 7 |
| 020 | Tissue (kidney) | QLD | 21 | 1 |
| 025 | Tissue (kidney) | VIC | 8 | 1 |
| 026 | Tissue (kidney) | VIC | 12 | 1 |
| 027 | Tissue (trachea) a | VIC | 7 | 1 |
| 028 | Tissue (kidney) a | VIC | 18 | 1 |
| 031 | Tissue (kidney) a | VIC | 8 | 1 |
| 032 | Swabs (trachea) | NSW | 7 | 1 |
| 038a | Allantoic fluid (trachea, passage 2) | WA | 7 | 1 |
| 038b | Allantoic fluid (unknown, passage 2) | WA | 21 | 1 |
| 040 | Tissue (trachea) | NSW | 40 | 1 |
| 041 | Tissue (kidney) | QLD | 14 | 1 |
| 043 | Tissue (trachea) | NSW | 31 | 1 and 19 |
| 044 | Tissue (kidney) | VIC | 11 | 1 (VicS) |
| 050 | Tissue (trachea) | NSW | 47 | 1 |
Tissue specimen chosen for analysis because both trachea and kidney specimens tested positive for IB viruses with similar HRM curves and GCPs.
Subgroup 1 vaccines I, S and B were used.
GCP, genotype confidence percentage; HRM, high‐resolution melt; NSW, New South Wales; QLD, Queensland; VIC, Victoria; WA, Western Australia.
The specimens submitted were from Queensland (QLD), New South Wales (NSW), Victoria (VIC) and Western Australia (WA) and most were trachea and/or kidney tissue, but swabs and allantoic fluid (from the second passage through specific‐pathogen free (SPF) eggs) were also submitted.
Extraction of RNA
Total extracted RNA was available in our laboratory for all reference and vaccine strains. 7 For field submissions, 400 µL of Qiagen RLT lysis buffer (Qiagen, VIC, Australia) with 5% β‐mercaptoethanol was added to a small section (approximately 5 × 5 × 5 mm) of kidney, or a tracheal scraping or swab. For cultured viruses, 100 µL of allantoic fluid was mixed with the lysis buffer. Total RNA was extracted and complementary DNA prepared from each of the field submissions as previously described. 7 Diethyl pyrocarbonate‐treated water was used as a negative extraction control.
Real‐time PCR and HRM curve analysis of the 3′UTR of IB viruses
All primers used in this study are shown in Table 2. Real‐time PCR and HRM curve analysis was performed as previously described, 7 using a Rotorgene 6000 (Corbett Life Science, NSW, Australia), and the conventional melt and HRM curves were used to classify each field submission as subgroup 1, 2 or 3. The genotype confidence percentage (GCP) and subsequent classification recorded at the time of initial processing (or most recent repeat analysis) were retrieved.
Table 2.
Oligonucleotide primers used in study of infectious bronchitis viruses in field specimens
| Primer | Target/location a | Sequence (5′–3′) | Citation no. |
|---|---|---|---|
| Poly‐F1 | S1 gene/20070‐20090 | GATTGTGCATGGTGGACAATG | 15 |
| PP2‐R | S1 gene/22169‐22145 | GTTTGTATGTACTCATCTGTAACAG | 7 |
| All1‐F | 3′UTR/26930‐26948 | CAGCGCCAAAACAACAGCG | 7 |
| Del1‐R | 3′UTR/27362‐27344 | CATTTCCCTGGCGATAGAC | 24 |
Location of the primer on the reference IB virus strain Beaudette (GenBank accession number NC001451).
UTR, untranslated region.
The 3′UTR amplicons from each field submission were separated on a 1% agarose gel, the band was excised and the DNA extracted using the Qiaquick Gel Extraction Spin Kit (Qiagen) and sequenced (Applied Genetic Diagnostics, University of Melbourne, Parkville, VIC, Australia) using 5 µmol/L of each PCR primer (All1‐F and Del1‐R). The resulting sequences were aligned with subgroup reference strains using ClustalW2 (http://www.ebi.ac.uk/clustalw) and the highest identity with the reference sequence was recorded.
Amplification and sequencing of the S1 gene from each field submission
All field submissions were subjected to PCR using primers Poly‐F1 15 and PP2‐R, as previously described, 16 to amplify a product of approximately 2100 base pairs (bp). The amplified PCR products were separated on a 1.2% agarose gel, the band was excised and sequenced as described above, using 5 µmol/L each of primer Poly‐F1 and PP2‐R. The sequences obtained were aligned using ClustalW2 and the nucleotide identity to reference strains recorded.
Results
Identification of IB viruses in field submissions using HRM curve analysis
Of the 40 field submissions received for IB virus detection, 17 were found to contain virus by PCR–HRM curve analysis. All these samples were from broiler flocks. In cases where both tracheal and kidney tissues had been submitted and determined to be positive for IB virus, the results of the HRM curve analysis from both organs were identical (data not shown) and only one tissue sample was used for subsequent analyses.
Two IB viruses in submissions from QLD (020 and 041), six in submissions from VIC (013, 025, 026, 027, 028 and 031), and one each from NSW (032) and WA (038a) were classified as subgroup 1 (vaccine or vaccine‐related strains). Two IB viruses (018 and 040), both from NSW, were classified as subgroup 3. These classifications were based on both their conventional melt curve shape and the derivation of a GCP > 80.13 with the relevant reference strain 7 (Table 3) from the normalised HRM curves. Three IB viruses detected in submissions from NSW (015, 043 and 050) and one each from VIC (044) and WA (038b) were deemed ‘unclassified’ as they had GCP values <80.13 with each of the subgroup reference strains. The conventional and normalised melt curves for all 17 IB virus field strains are shown in Figure 1.
Table 3.
Classification of field submissions into subgroups of infectious bronchitis virus
| ID | HRM classification (subgroup) | GCP with subgroup reference | S1 gene length used to generate identities e | Nucleotide sequence identity with subgroup reference (%) | |
|---|---|---|---|---|---|
| S1 gene | 3′UTR | ||||
| 013 | 1 a | 95.66 | 1717 f | 99 | 100 |
| 015 | Unclassified b | NA d | 1723 f | Subgroup 2 h (93) | Subgroup 3 (98) |
| 018 | 3 c | 84.95 | 1705 f | 99 | 99 |
| 020 | 1 | 97.41 | − g | − | 99 |
| 025 | 1 | 93.32 | 1138 | 99 | 99 |
| 026 | 1 | 94.14 | 1717 f | 99 | 99 |
| 027 | 1 | 92.43 | 1684 | 99 | 99 |
| 028 | 1 | 87.94 | 1717 f | 99 | 99 |
| 031 | 1 | 95.86 | 1425 | 100 | 99 |
| 032 | 1 | 89.99 | 1666 | 99 | 99 |
| 038a | 1 | 86.19 | 1294 | 99 | 99 |
| 038b | Unclassified | NA | 1225 | Subgroup 1 (99) | Subgroup 1 (99) |
| 040 | 3 | 94.30 | 925 | 99 | 99 |
| 041 | 1 | 82.80 | − | − | 99 |
| 043 | Unclassified | NA | 800 | Subgroup 3 (99) | Subgroup 3 (99) |
| 044 | Unclassified | NA | 1717 f | Subgroup 1 (99) | Subgroup 1 (99) |
| 050 | Unclassified | NA | − | − | Subgroup 3 (99) |
Vaccine I used as the genotype/reference.
b HRM profile did not match any of the class reference strains.
c Reference strain N1/03 used as the genotype/reference.
d Highest GCP below cut‐off.
Maximum length of sequence obtained from S1 gene sequencing that was used to generate the nucleotide sequence identities (full amplicon approximately 1720 bp).
f Complete S1 gene sequence obtained.
g Amplification/sequencing not successful.
h Reference strain N1/88 used as the genotype/reference.
bp, base pairs; GCP, genotype confidence percentage; HRM, high‐resolution melt; NA, not available; UTR, untranslated region.
Figure 1.
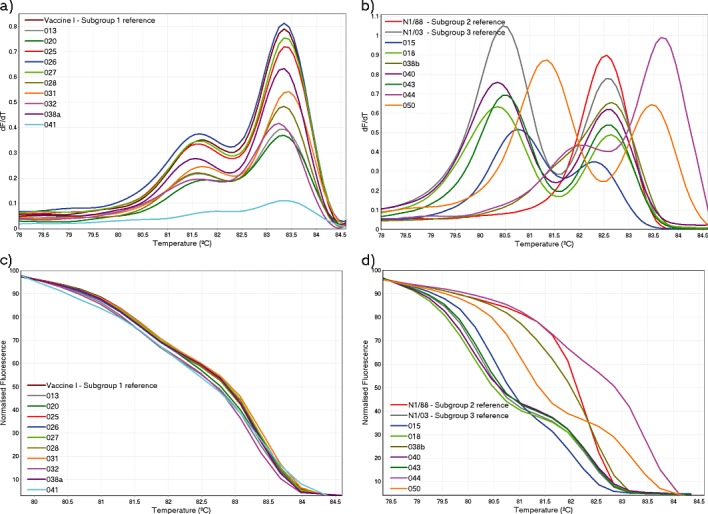
Melt curves generated using HRM curve analysis. The conventional and normalised melt curves generated for all field submissions classified as subgroup 1, including the subgroup 1 reference strain, vaccine I, are shown in (a) and (c) respectively. The conventional and normalized melt curves for all remaining field submissions, including subgroup 2 and subgroup 3 reference strains (N1/88 and N1/03 respectively), are shown in (b) and (d) respectively.
The unclassified field strains 015 and 038b produced conventional melt curves that were distinct from the subgroup reference strains. Both 043 and 050 produced melt curve profiles that were similar to the subgroup 3 reference strain. However, a slight variation in the shape of the curves resulted in GCP values below the cut‐off with the subgroup 3 reference strain (data not shown).
Submission 044 was a kidney tissue submitted from an 11‐day‐old SPF chicken, vaccinated at 1 day old with a subtype 1 vaccine (VicS). The conventional melt curve for strain 044 was similar in shape to that produced by subgroup 1 strains, but a melt curve shift to a higher temperature resulted in a GCP value below the cut‐off with the subgroup 1 reference strain.
3′UTR sequences and HRM curve analysis classification
The 3′UTR of the genome was sequenced for all 17 field submissions that were positive for IB virus. These sequences were aligned with the subgroup reference strains and the highest identity with the reference strains for each of the 17 IB virus field strains was determined (Table 3). The 3′UTR nucleotide sequences for all 10 IB field viruses classified as subgroup 1 by HRM had identities of 99% and 100% with the Australian vaccine strains I and S, respectively. The 3′UTR of the unclassified field IB virus 038b contained a 40‐bp deletion in its 3′UTR (data not shown), but the remaining sequence had 99% identity with vaccine strains I and S. There was a G to A base substitution in the 3′UTR of IB field virus 044 compared with subgroup 1 strains (data not shown).
IB field viruses 018 and 040 (both classified as subgroup 1) had 3′UTR nucleotide sequence identity scores of 99%, whereas the unclassified strain 015 had an identity score of 98% with the subgroup 3 reference strain N1/03 (Table 3). The unclassified IB field viruses 043 and 050 both had 3′UTR nucleotide sequence identity scores of 99% with subgroup 3 reference strain N1/03 (Table 3), with both containing the same C to T base substitution at position 353 of the amplicon (data not shown), which explained the variations in the melt curves.
S1 gene nucleotide sequences for classified field viruses
The complete S1 gene sequence (approximately 1720 bp excluding the flanking sequences for the polymerase and S2 genes) was determined for submissions 013, 015, 018, 026, 028 and 044. Only partial S1 gene sequences could be obtained for IB field viruses 025, 027, 031, 032, 038a, 038b and 040 because of the small amount of template amplified (relative to the viruses for which the complete S1 gene was successfully sequenced). The length of the partial S1 gene sequences ranged from 800 bp (043) to 1684 bp (027). The S1 gene nucleotide sequence identities of the field submissions with the most closely related reference strain are shown in Table 3. As the partial S1 gene sequences did not all span the same area, the initial alignment was used to determine the reference strain to which each partial sequence was most related. Each partial S1 gene sequence was then aligned separately with its respective reference strain and the nucleotide sequence identity recorded. Amplification of the S1 genes from field submissions 020, 041, and 050 was either unsuccessful or did not produce sufficient template for sequencing.
The IB virus field strain 015 had the highest S1 gene nucleotide sequence identity score (93%) with the subgroup 2 reference strain N1/88, and nucleotide sequence identities ≤ 67% with all other strains. This contrasted with its 3′UTR nucleotide sequence identity score of 98% with the subgroup 3 reference strain N1/03, suggesting that this IB field virus is likely to be a recombinant virus.
IB field viruses 013, 025, 026, 027, 028, 031, 032 and 038a had S1 gene nucleotide sequence identities ≥ 99% with the subgroup 1 reference strain vaccine I, correlating with their 3′UTR nucleotide sequence identity scores with this strain and the HRM curve analyses. IB field viruses 038b and 044 also had S1 gene nucleotide sequence identities ≥ 99% with the subgroup 1 reference strain vaccine I correlating with their 3′UTR nucleotide sequence identity scores with this strain.
IB field viruses 018 and 040 had nucleotide sequence identities of 99% with the subgroup 3 reference strain N1/03, correlating with their 3′UTR nucleotide sequence identity scores with this strain and the HRM curve analyses. The S1 gene nucleotide sequence identity of IB field virus 043 was 99% with the subgroup 3 reference strain N1/03, correlating with its 3′UTR nucleotide sequence identity with this strain.
Discussion
HRM curve analyses of field submissions containing IB viruses identified strains of the virus that required further investigation, and enabled the detection of an Australian recombinant IB virus strain. The classification of field submissions as either subgroup 1 or 3 using HRM curve analysis correlated with both S1 gene and 3′UTR nucleotide sequence comparisons. No IB field viruses were classified by HRM curve analysis as being related to the subgroup 2 reference strain N1/88.
IB field virus 015 had contrasting nucleotide sequence identity matches for its 3′UTR and S1 gene sequences, and also produced a distinct melt curve profile. Its 3′UTR sequence suggested a close relationship with the subgroup 3 reference strain N1/03, whereas its S1 gene was most similar to the subgroup 2 reference strain N1/88. Strains belonging to subgroup 2 are distinct from strains belonging to subgroup 3. 20 This suggests that IB field virus 015 was a recombination of two viruses from different subgroups. Recombination involving the S1 gene has been extensively documented 11 , 13 , 17 , 18 , 19 , 20 , 21 , 22 and has even been suggested as a basis for the virulence of turkey coronavirus. 23
The nucleotide sequences of the 3′UTR and the S1 gene of the unclassified IB field virus 038b were 99% identical to that of the subgroup 1 reference strain, but this virus generated a distinct conventional melt curve. The 3′UTR nucleotide sequencing revealed that this was because of a 40‐bp deletion compared with the sequences for subgroup 1 viruses. Further investigation revealed that this deletion was immediately upstream of the 58‐bp deletion that exists in the 3′UTR of all subgroup 1 strains (vaccine and vaccine‐related IB virus strains). 24 Thus, the HRM curve analysis was able to differentiate this IB virus when S1 gene sequencing alone would not have detected a difference. This 40‐bp deletion has been detected previously 7 in our laboratory when VicS is amplified using primers All1‐F and Del1‐R. Interestingly, the flock from which the submission originated had been vaccinated with VicS vaccine at 1 day old.
Visual observation of the melt curves and the S1 gene and 3′UTR nucleotide sequences for 043 and 044 suggested that they were subgroup 3 and subgroup 1 IB viruses, respectively, but the base changes discovered in their 3′UTRs most likely resulted in the melt curve changes and, ultimately, GCPs below the cut‐off. This further demonstrates the sensitivity of HRM curve analysis for detecting differences between IB viruses and its capacity for non‐subjective classification using GCP values. It is interesting to note that field submission 044 was from an 11‐day‐old SPF chicken that had been vaccinated at 1 day old with VicS vaccine, but subsequently developed interstitial nephritis. It is not known if the IB virus present in field submission 044 was derived from the VicS vaccine.
Based on the age of detection, the age of vaccination (Table 1) and their S1 gene and 3′UTR nucleotide sequence identities, IB field viruses 025, 027, 031, 032 and 038b, which were classified by HRM curve analysis as subgroup 1 strains (vaccine or vaccine‐related), have presumably been re‐isolations of the vaccine virus.
The failure to amplify S1 gene sequences from 020, 041, and 050 was most probably related to the large size of the target amplicon (approximately 1720 bp) and the limited quantity of viral RNA available. This, together with the detection of novel strains 015 and 038b by HRM curve analysis, further emphasises that nucleotide sequencing of the S1 gene alone is not sufficient or reliable for IB virus classification. 25 , 26 , 27 , 28 Sequencing alone of the S1 gene of field strains 015 and 038b would have resulted in incorrect characterisation of these two IB field viruses.
Further analysis of field strain 015 is necessary to establish the origin of this strain and to determine its virulence for chickens. It would also be of interest to assess whether the current Australian vaccines protect against this recombinant strain.
Acknowledgments
The primary author was supported by scholarships from The University of Melbourne and the Australian Poultry Cooperative Research Centre. The authors thank Denise O'Rourke for technical assistance.
References
- 1. Cook JKA, Chesher J, Baxendale W et al Protection of chickens against renal damage caused by a nephropathogenic infectious bronchitis virus. Avian Pathol 2001;30:423–426. [DOI] [PubMed] [Google Scholar]
- 2. Ignjatovic J, Reece R, Ashton F. Susceptibility of three genetic lines of chicks to infection with a nephropathogenic T strain of avian infectious bronchitis virus. J Comp Pathol 2003;128:92–98. [DOI] [PubMed] [Google Scholar]
- 3. Muneer MA, Newman JA, Halvorson DA, Sivanandan V, Coon CN. Effects of avian infectious bronchitis virus (arkansas strain) on vaccinated laying chickens. Avian Dis 1987;31:820–828. [PubMed] [Google Scholar]
- 4. Sevoian M, Levine PP. Effects of infectious bronchitis on the reproductive tracts, egg production, and egg quality of laying chickens. Avian Dis 1957;1:136–164. [Google Scholar]
- 5. Chousalkar KK, Roberts JR. Ultrastructural study of infectious bronchitis virus infection in infundibulum and magnum of commercial laying hens. Vet Microbiol 2007;122:223–236. [DOI] [PubMed] [Google Scholar]
- 6. Cavanagh D, Naqi SA. Infectious bronchitis In: Saif YM, Barnes HJ, Glisson JR. et al, editors. Diseases of poultry. 11th edn. Iowa State Press, 2003;101–119. [Google Scholar]
- 7. Hewson K, Noormohammadi A, Devlin J, Mardani K, Ignjatovic J. Rapid detection and non‐subjective characterisation of infectious bronchitis virus isolates using high‐resolution melt curve analysis and a mathematical model. Arch Virol 2009;154:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Masters PS, Margniorosch K, Murphy FA, Shatkin AJ. Reverse genetics of the largest RNA viruses. Adv Virus Res 1999;53:245–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herrewegh AAPM, Smeenk I, Horzinek MC, Rottier PJM, De Groot RJ. Feline coronavirus type II strains 79‐1683 and 79‐1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol 1998;72:4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kusters JG, Jager EJ, Niesters HGM, Van Der Zeijst BAM. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine 1990;8:605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L, Junker D, Collisson EW. Evidence of natural recombination within the S1 genes of infectious bronchitis virus. Virology 1993;192:710–716. [DOI] [PubMed] [Google Scholar]
- 12. Casais R, Davies M, Cavanagh D, Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol 2005;79:8065–8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bochkov YA, Tosi G, Massi P, Drygin V. Phylogenetic analysis of partial S1 and N gene sequences of infectious bronchitis virus isolates from Italy revealed genetic diversity and recombination. Virus Genes 2007;35:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benson DA, Karsch‐Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res 2008;36:D25–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mardani K, Noormohammadi AH, Ignatovic J, Browning GF. Typing infectious bronchitis virus strains using reverse transcription‐polymerase chain reaction and restriction fragment length polymorphism analysis to compare the 3′ 7.5 kb of their genomes. Avian Pathol 2006;35:63–69. [DOI] [PubMed] [Google Scholar]
- 16. Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol 2003;32:567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ammayappan A, Upadhyay C, Gelb J, Vakharia V. Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination. Virol J 2008;5:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H‐W, Huang Y‐P, Wang C‐H. Identification of Taiwan and China‐like recombinant avian infectious bronchitis viruses in Taiwan. Virus Res 2009;140:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dolz R, Pujols J, Ordóñez G, Porta R, Majó N. Antigenic and molecular characterization of isolates of the Italy 02 infectious bronchitis virus genotype. Avian Pathol 2006;35:77–85. [DOI] [PubMed] [Google Scholar]
- 20. Ignjatovic J, Gould G, Sapats S. Isolation of a variant infectious bronchitis virus in Australia that further illustrates diversity among emerging strains. Arch Virol 2006;151:1567–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu S, Han Z, Chen J et al S1 gene sequence heterogeneity of a pathogenic infectious bronchitis virus strain and its embryo‐passaged, attenuated derivatives. Avian Pathol 2007;36:231–234. [DOI] [PubMed] [Google Scholar]
- 22. Sapats SI, Ashton F, Wright PJ, Ignjatovic J. Sequence analysis of the S1 glycoprotein of infectious bronchitis viruses: identification of a novel genotypic group in Australia. J Gen Virol 1996;77:413–418. [DOI] [PubMed] [Google Scholar]
- 23. Jackwood MW, Paterson AH, Kissinger JC et al Comparative genomics on avian coronaviruses: origin and divergence associated with host and pathogenic shifts In: Heffels‐Redmann U, Sommer D, Kaleta EF, editors. VI International Symposium on Avian Corona and Pneumoviruses and Complicating Pathogens. Rauischholzhausen, Germany, 2009. [Google Scholar]
- 24. Mardani K, Browning GF, Ignjatovic J, Noormohammadi AH. Rapid differentiation of current infectious bronchitis virus vaccine strains and field isolates in Australia. Aust Vet J 2006;84:59–62. [DOI] [PubMed] [Google Scholar]
- 25. Ladman BS, Loupos AB, Gelb Jr J. Infectious bronchitis virus S1 gene sequence comparison is a better predictor of challenge of immunity in chickens than serotyping by virus neutralization. Avian Pathol 2006;35:127–133. [DOI] [PubMed] [Google Scholar]
- 26. Liu HJ, Lee LH, Shih WL, Lin MY, Liao MH. Detection of infectious bronchitis virus by multiplex polymerase chain reaction and sequence analysis. J Virol Methods 2003;109:31–37. [DOI] [PubMed] [Google Scholar]
- 27. McFarlane R, Verma R. Sequence analysis of the gene coding for the S1 glycoprotein of infectious bronchitis virus (IBV) strains from New Zealand. Virus Genes 2008;37:351–357. [DOI] [PubMed] [Google Scholar]
- 28. Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol 2000;145:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]


