Despite their opposing actions on food intake, POMC and NPY/AgRP neurons in the arcuate nucleus of the hypothalamus (ARH) are derived from the same progenitors that give rise to ARH neurons. However, the mechanism whereby common neuronal precursors subsequently adopt either the anorexigenic (POMC) or the orexigenic (NPY/AgRP) identity remains elusive. We hypothesize that POMC and NPY/AgRP cell fates are specified and maintained by distinct intrinsic factors.
Keywords: development, feeding, hypothalamus, melanocortin, Prdm12, transcription
Abstract
Despite their opposing actions on food intake, POMC and NPY/AgRP neurons in the arcuate nucleus of the hypothalamus (ARH) are derived from the same progenitors that give rise to ARH neurons. However, the mechanism whereby common neuronal precursors subsequently adopt either the anorexigenic (POMC) or the orexigenic (NPY/AgRP) identity remains elusive. We hypothesize that POMC and NPY/AgRP cell fates are specified and maintained by distinct intrinsic factors. In search of them, we profiled the transcriptomes of developing POMC and NPY/AgRP neurons in mice. Moreover, cell-type-specific transcriptomic analyses revealed transcription regulators that are selectively enriched in either population, but whose developmental functions are unknown in these neurons. Among them, we found the expression of the PR domain-containing factor 12 (Prdm12) was enriched in POMC neurons but absent in NPY/AgRP neurons. To study the role of Prdm12 in vivo, we developed and characterized a floxed Prdm12 allele. Selective ablation of Prdm12 in embryonic POMC neurons led to significantly reduced Pomc expression as well as early-onset obesity in mice of either sex that recapitulates symptoms of human POMC deficiency. Interestingly, however, specific deletion of Prdm12 in adult POMC neurons showed that it is no longer required for Pomc expression or energy balance. Collectively, these findings establish a critical role for Prdm12 in the anorexigenic neuron identity and suggest that it acts developmentally to program body weight homeostasis. Finally, the combination of cell-type-specific genomic and genetic analyses provides a means to dissect cellular and functional diversity in the hypothalamus whose neurodevelopment remains poorly studied.
SIGNIFICANCE STATEMENT POMC and NPY/AgRP neurons are derived from the same hypothalamic progenitors but have opposing effects on food intake. We profiled the transcriptomes of genetically labeled POMC and NPY/AgRP neurons in the developing mouse hypothalamus to decipher the transcriptional codes behind the versus orexigenic neuron identity. Our analyses revealed 29 transcription regulators that are selectively enriched in one of the two populations. We generated new mouse genetic models to selective ablate one of POMC-neuron enriched transcription factors Prdm12 in developing and adult POMC neurons. Our studies establish a previously unrecognized role for Prdm12 in the anorexigenic neuron identity and suggest that it acts developmentally to program body weight homeostasis.
Introduction
Serving as the interface between the nervous and endocrine systems, the hypothalamus is a critical regulator of many physiological processes essential for life. Recent advances in optogenetic, chemogenetic, and neuroimaging techniques have enabled the isolation of individual hypothalamic circuits that control satiety (Atasoy et al., 2012), thirst (Oka et al., 2015), aggression (H. Lee et al., 2014), and many more. These studies further reveal that each anatomically defined hypothalamic nucleus consists of functionally heterogeneous neuronal subtypes that dictate distinct survival behaviors.
On the other hand, how cellular and functional diversity is established in the developing hypothalamus remains poorly understood. The hypothalamus is enormously complex and consists of hundreds of neuronal and non-neuronal cell types (Chen et al., 2017; Romanov et al., 2017). However, much is unknown regarding the early patterning events that produce hypothalamic progenitors (Bedont et al., 2015). Moreover, postmitotic neuronal identities in the hypothalamus were traditionally defined by the expression of peptidergic markers. The axonal and dendritic distribution of neuropeptides makes it difficult to track cell bodies of peptidergic neurons, therefore impeding the search for cell-specific markers. Furthermore, hypothalamic neurodevelopment spans an extended period of time, during which gene expression and synaptic connectivity are highly dynamic (Bouret et al., 2004; Padilla et al., 2010). What's more, cell fate remains plastic as adult hypothalamic neurons can still switch their transmitter identities in response to environmental stimuli (Dulcis et al., 2013). For these reasons, while fate specification and maintenance have been thoroughly investigated in other parts of the nervous system, it remains an understudied subject within the hypothalamus.
Notably, the fate choice between the anorexigenic pro-opiomelanocortin (POMC) and the orexigenic NPY/AgRP neurons in the ARH presents a unique challenge. Alpha-melanocyte-stimulating hormone, a proteolytic product of POMC, activates the melanocortin 4 receptor (Mc4r) to suppress food intake, whereas the orexigenic agouti-related peptide (AgRP) serves as an inverse agonist (Cone, 1999). Despite their opposing actions on food intake, both neurons are derived from the same Rax+/Nkx2.1+ progenitors that give rise to ARH neurons (Kimura et al., 1996; Lu et al., 2013). Earlier studies uncovered a transcriptional cascade that sequentially specifies postmitotic precursors (Kimura et al., 1996; McNay et al., 2006; Lu et al., 2013; Nasif et al., 2015). However, the mechanism whereby common neuronal precursors subsequently adopt either the anorexigenic (POMC) or the orexigenic (NPY/AgRP) identity remains elusive. We speculate that the orexigenic and anorexigenic cell fates are specified and maintained by distinct intrinsic factors. However, transcripts that are differentially expressed by developing POMC and NPY/AgRP neurons remain largely unknown. Since cell bodies of these neurons are closely intermingled with each other, existing ISH databases (e.g., Genepaint and Allen Brain Library) offer few clues for cell-type-specific gene expression. Furthermore, while single-cell RNA sequencing (scRNA-seq) has recently been used to probe cellular heterogeneity and cell-type-specific gene expression in the developing hypothalamus (Huisman et al., 2019; D. W. Kim et al., 2019), given that POMC and NPY/AgRP neurons constitute only a small fraction of all hypothalamic cells, low sequencing depth and coverage rates remain a limitation to fully reveal POMC- and NPY/AgRP neuron-enriched transcripts (Ding et al., 2019). To uncover potential regulators of the orexigenic versus anorexigenic neuron identity, we purified genetically labeled POMC and NPY/AgRP neurons from the developing ARH and profiled their transcriptomes using whole-transcriptome RNA sequencing.
Materials and Methods
Mice
All experimental procedures were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center. Mice were housed in a barrier facility with a 12 h light/dark cycle (lights 7:00 A.M. to 7:00 P.M.). Unless otherwise noted, mice were fed standard chow (Envigo 2016 Teklad) and had ad libitum access to water. Littermates of both males and females were used in all studies.
The Prdm12fl mice were generated using CRISPR-Cas9-based genome editing. Two guide RNA sequences (CATGACCCGCTGCGCCCTTG; CAGCAGCTTTTCGTTACCGA) were designed in silico using CRISPOR (www.crispor.tefor.net) flanking the Exon 5. Single-stranded DNA repair templates containing 80 bp homology arms and a loxP sequence were synthesized as IDT ultramers. Guides, tracRNA, ssDNA repair template, and Cas9 protein (IDT) were mixed by the University of Texas Southwestern Transgenic Core for pronuclear injection. We used the following primers to genotype the Prdm12fl (248 bp) and the Prdm12+ (200 bp) alleles: 5′-GGTGCAAGGTCCAAATGTCT-3′ and 5′-CACACTGCAGCTGGCTAAAC-3′. Prdm12Δ (401 bp) was detected using 5′-TTCGGATGAGATGGCTAAGG-3′ and 5′-CCAATGTGCCACAGTACAGG-3′. POMCeGFP (#009593), NPYhGFP (#006417), R26RAi14 (#007908), and POMC-Cre (#010714) mice are available at The Jackson Laboratory.
Flow cytometry and total RNA isolation
Under a fluorescent dissecting microscope, we extracted the arcuate nucleus from PomceGFP and NpyhrGFP embryos at E15.5. Cells were disassociated as described previously with minor modifications (Wyler et al., 2016). Specifically, dissected tissue was collected in solution consisting of Hybernate E (Invitrogen) supplemented with 2% B27 (Thermo Fisher Scientific) and 0.25% Glutamax (Invitrogen). Tissue was washed with PBS and incubated on a shaker at 37°C for 30 min in 20 U/ml papain, 1 mm L-cystine, 0.5 mm EDTA, and 0.05 mg/ml DNaseI in Earle's Balanced Salt Solution using the Papain Disassociation System (Worthington). Tissue was then washed 3 times in PBS and resuspended in Hybernate E/B27/Glutamax solution described above. Cells were disassociated with a fire-polished glass pipette. Digested cells were sorted using a Becton Dickinson FACS Aria digital cell sorter with an argon laser (200 mW at 488 nm). Each biological replicate was defined as an independent sort using 5-12 pooled GFP+ embryos collected from multiple dams. Total RNA from ∼5000-10,000 cells/replicate was isolated using chloroform extraction with 10 µg of Glycoblue (Ambion) used as a carryover. RNA was cleaned and DNase I treated using the RNA Clean & Concentrator-5 kit (Zymo). RNA quantity and quality were assessed using an Agilent 2100 bioanalyzer. All samples had an RNA integrity number ≥ 9.3. Libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit version 2 (Clonetech) using 1 ng of total RNA and sequenced using an Illumina HiSeq 2500.
RNA-Seq data analysis
Data were analyzed in R using Bioconductor package. Reads were aligned to mouse reference genome GRCm38/mm10 assembly using HISAT2. We obtained between 71.9 and 87.6 million mapped reads/sample (average of 77.8 million). Duplicates were removed using differential expression analysis performed using Bioconductor version 3.8 packages rSamtools, edgeR version 3.8 (Li et al., 2009; Robinson et al., 2010), using a cutoff of 25 counts per million and a fold-change cutoff of 2 and an FDR of 5%, q value (Benjamin-Hochberg correction). A list of transcription factors (TFs) was generated by combining databases from Gene ontology (GO) term (0140110), Riken Institute (genome.gsc.riken.jp/TFdb), and the TFCheckpoint database (www.tfcheckpoint.org) (Chawla et al., 2013). This yielded a correlated list of 2476 TFs in the mouse genome. GO was performed with WebGestalt 2019 (Zhang et al., 2005) using overrepresentation enrichment analysis with a minimum of three genes per GO category and an FDR of 5% (hypergeometric test, with a BH correction). Data files are deposited in Gene Expression Omnibus (GEO) accession number:GSE146816.
Immunohistochemistry, ISH, and qPCR analyses
Details for performing these experiments were described previously (Liu et al., 2010).
The primary antibodies used include anti-GFP (#ab13970, Abcam), anti-Nkx2.1 (#sc-13 040, Santa Cruz Biotechnology), anti-Tcf7l2 (#2569, Cell Signaling Technology), anti-ACTH (#AFP-156102789, National Hormone & Peptide Program), and anti-β-endorphin (#H-022-33, Phoenix Pharmaceutical). Secondary antibodies (AlexaFluor-488 or -594) were from Thermo Fisher Scientific.
The following primer sets were used to generated PCR templates for synthesizing digoxigenin (DIG)-labeled RNA probes. Pomc_T3S: 5′-GAATTAACCCTCACTAAAGGGGCTTGCAAACTCGACCTCTC-3′; Pomc_T7AS: 5′-GTAATACGACTCACTATAGGGCTCTTGATGATGGCGTTCTTG-3′; Agrp_T3S: 5′-GAATTAACCCTCACTAAAGGGCCCAAGAATGGACTGAGCA-3′; Agrp_T7AS: 5′-GTAATACGACTCACTATAGGGCGCAAAAGGCATTGAAGAAGC-3′; Npy_T3S: 5′-GAATTAACCCTCACTAAAGGGCCGGTGGATCTCTTCTCTCA-3′; and Npy_T7AS: 5′-GTAATACGACTCACTATAGGGCGATGAGGGTGGAAACTTGGA-3′.
RNAscope was performed using ACD RNAscope Multiplex Fluorescent Detection Kit version 2 following the manufacturer's (Advanced Cell Diagnostics) protocol with the following probes Mm-Pomc, Mm-Npy, Mm-Prdm12-C2, and Mm-Nr5a1-C2.
qPCR was performed using a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) and TaqMan Universal PCR MasterMix (4364338). TaqMan probes for Agrp (Mm00475829_g1), Npy (Mm01410146_m1), Pomc (Mm00435874_m1), Cartpt (Mm04210469_m1), Kiss1 (Mm03058560_m1), Sst (Mm00436671_m1), Th (Mm00447557_m1), Lepr (Mm00440181_m1), Gad1 (Mm04207432_g1), and Gad2 (Mm00484623_m1) were from Thermo Fisher Scientific.
Cell counting
We manually counted fluorescently labeled cells from every fifth coronal section (25 µm) throughout the entire rostral-caudal axis of the arcuate nucleus. For calculating the percentage of overlapping POMC-eGFP/TdTomato cells, the numbers of duel-labeled cells were divided by the total number of POMC-GFP cells for each mouse (n = 4). Male mice were between 4 and 8 weeks of age.
Sox binding sites analyses
Sequences of mouse nPE1 (chr12:3941972-3942627, 656 bp) and nPE2 (chr12:3944600-3944917, 318 bp) defined by de Souza et al. (2005), were obtained using the ECR Genome Browser (https://ecrbrowser.dcode.org). Sequence alignment was conducted using the ConTra V3 web server (http://bioit2.irc.ugent.be/contra/v3/). Matrices used to predict Sox binding motifs include taipale-TCAATWNCATTGA-Sox1-DBD for Sox1, MA0143.3 for Sox2, M1592_1.02 for Sox3, and taipale-ACAATANCATTG-Sox14-DBD for Sox4 with the stringency setting (core = 1.00, similarity matrix = 0.95).
Metabolic analysis
Details for performing body weight, length, body composition, glucose tolerance test (GTT), insulin tolerance test (ITT), blood chemistry, and metabolic chamber analyses in 10-week-old male and female mice were described previously (Liu et al., 2014).
Experimental design and statistical analyses
Data are presented as mean ± SEM. Statistical differences were analyzed using paired/unpaired two-tailed Student's t test or two-way ANOVA with Sidak's post hoc testing where applicable (Prism 7.0). Statistical analyses for RNA-seq experiments can be found in RNA-seq data analysis. The statistical parameters (i.e., the exact n numbers, p values, and the number of biological repeats) can be found in Results or figure legends.
Data availability
The data that support the findings of this study are available from the corresponding author on request. RNA sequencing data reported in this paper have been deposited into NCBI's Gene Expression Omnibus under accession number GEO:
Results
Transcriptomic profiling of developing POMC and NPY/AgRP neurons
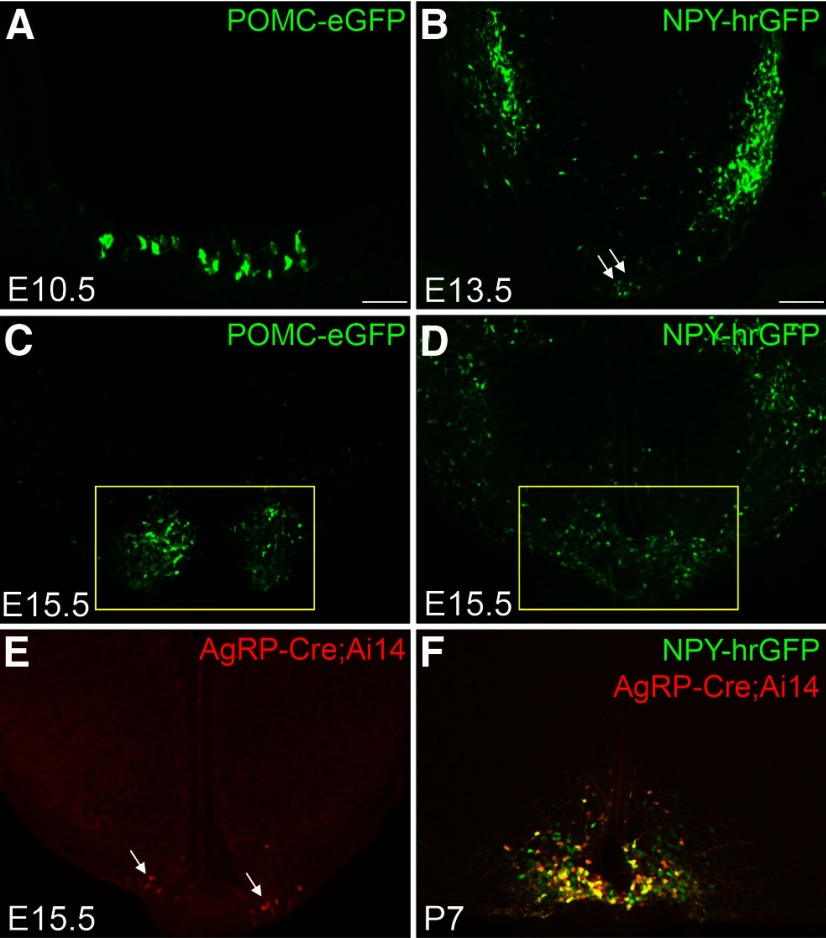
To track POMC and NPY/AgRP neurons in the developing hypothalamus, we took advantage of two well-characterized transgenic mice (POMCeGFP and NPYhrGFP) in which POMC and NPY/AgRP neurons are genetically labeled by the expression of GFPs (Cowley et al., 2001; Partridge et al., 2009). Despite a shared birthdate (Padilla et al., 2010), POMC and NPY/AgRP neurons launch their peptidergic marker expression at distinct developmental stages. Pomc mRNA first appears in the hypothalamic ventricular zone at embryonic (E) day E10.5 (McNay et al., 2006). It is transiently expressed in other neurons before being restricted to prospective POMC neurons after E14.5 (Padilla et al., 2012). In comparison, Npy mRNA is not present in the presumptive ARH until E13.5 (Padilla et al., 2010). Consistent with endogenous Pomc and Npy, we observed the onset of POMC-eGFP and NPY-hrGFP expression at E10.5 and E13.5, respectively (Fig. 1A,B). By E15.5, comparable numbers of POMCeGFP and NPYhrGFP neurons are present in the developing ARH (Fig. 1C,D). Since NPY/AgRP neurons coexpress Npy and Agrp in the adult brain (Hahn et al., 1998), we investigated the onset of Agrp expression in the developing ARH. Using AgRP-iresCre activity as a surrogate (Tong et al., 2008), we detected the Cre-activated tomato reporter (Rosa26Ai14) expression in the ARH as early as E15.5 (Fig. 1E). The number of tomato-positive neurons gradually increases over time, and by postnatal day 7 (P7), reporter expression is found only in a subset of NPY-hrGFP neurons (Fig. 1F), suggesting that the acquisition of the AgRP transmitter identity spans an extended postnatal period (Nilsson et al., 2005).
Figure 1.
The onset of POMC-eGFP, NPY-hrGFP, and AgRPCre;Ai14 in the developing ARH. A, Immunohistochemistry for POMC-eGFP in E10.5 hypothalamus. B–D, Native fluorescence of POMC-eGFP or NPY-hrGFP. Yellow box represents the ARH. E, Native fluorescence of tdTomato. White arrows point to tdTomato-expressing neurons. F, Double-labeling between tdTomato and NPY-hrGFP. Scale bars: A, 50 µm; B–F, 100 µm.
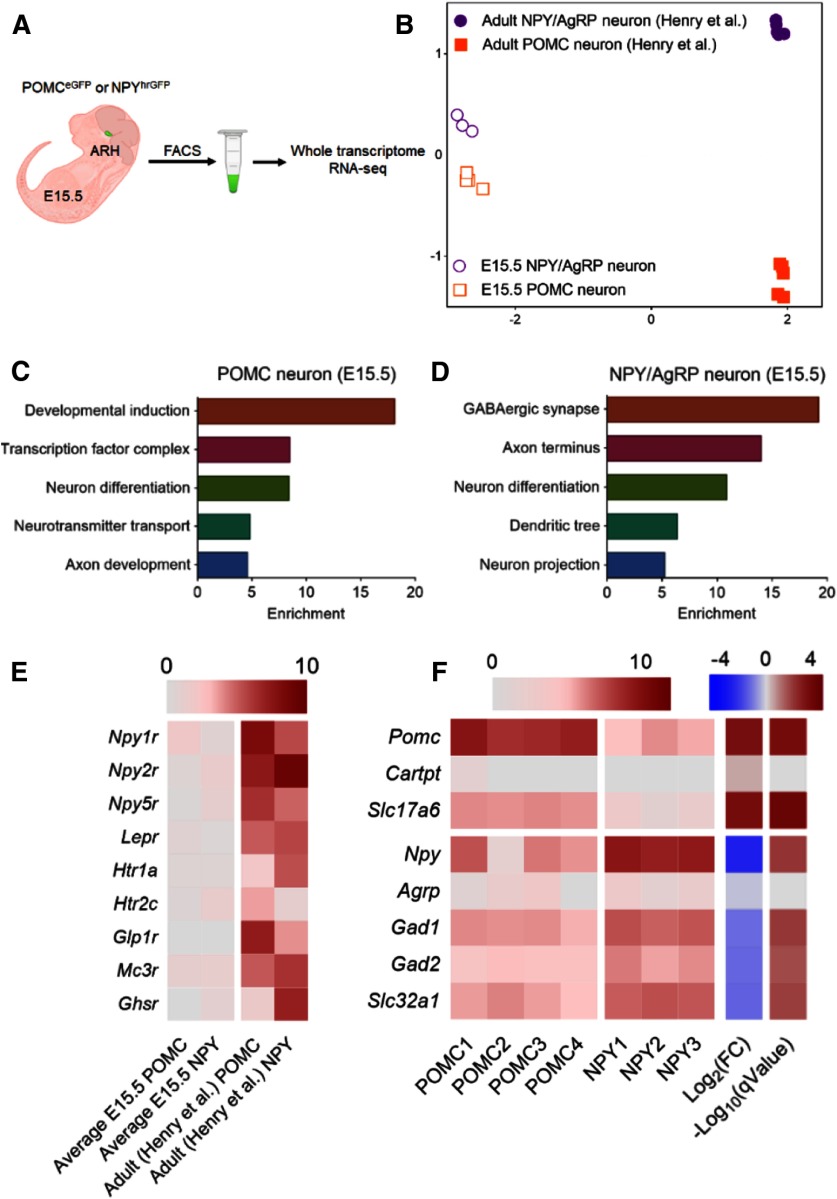
We profiled the transcriptomes of E15.5 POMC and NPY/AgRP neurons while they were acquiring their adult neurotransmitter identities. Under a fluorescent dissecting microscope, we dissected the ARH from POMCeGFP or NPYhrGFP embryos and purified GFP-positive neurons through FACS (Fig. 2A). We collected 3 or 4 biological replicates per genotype. Each biological replicate was defined as an independent sort of pooled GFP+ embryos from multiple dams. We extracted total RNAs from ∼5000 to 10,000 neurons per replicate and subjected them to whole-transcriptome RNA sequencing (Illumina HiSeq 2500). We obtained an average of 77.8 million mapped reads/sample. Reads were aligned to the mouse reference genome GRCm38/mm10 assembly using HISAT2 (D. Kim et al., 2015). From there, we detected 10,317 genes present in at least one of the two populations with a minimum expression of 10 transcripts per million mapped reads (TPM; Fig. 2-1).
Figure 2.
Transcriptomic profiling of embryonic POMC and NPY/AgRP neurons. A, Schematic of isolation and profiling transcriptomes of E15.5 POMC and NPY/AgRP neurons (Fig. 2-1). B, Multidimensional scaling projection of distance (1-correlation coefficient) of E15.5 POMC and NPY/AgRP neurons compared with data extracted from previously published adult (fed only) POMC and NPY/AgRP neurons (Henry et al., 2015). C, D, Gene ontology for enriched genes in POMC or NPY/AgRP neurons, respectively (Fig. 2-2). E, F, Heatmap of receptors and cell fate marker expression in POMC and NPY/AgRP neurons. Rows are replicates for each gene. Log2(FC), Log fold change of gene expression in POMC neurons over that in NPY/AgRP neurons (Fig. 2-3). Scale: log2(TPM + 1) q value: adjusted p value; FDR ≤ 5% (Benjamin-Hochberg correction).
List of genes and expression values in E15.5 POMC and NPY/AgRP neurons. LogFC, log2 fold change; LogCPM, log2 counts per million; F, F-statistic (negative binomial test). Download Figure 2-1, XLSX file (2.2MB, xlsx)
Gene ontology of pathways enriched in E15.5 POMC and NPY/AgRP neurons. Genes enriched (overrepresentation enrichment analysis) in Pomc or NPY neurons with a FDR of 5% and a minimum of 3 genes per GO category (hypergeometric test, with a BH correction). Genes are segregated by Biological Process, Cellular Component, and Molecular Function. Download Figure 2-2, XLSX file (17.9KB, xlsx)
List of genes enriched in E15.5 POMC and NPY/AgRP neurons. List of genes significantly enriched in E15.5 Pomc neurons over NPY neurons with a False Discovery Rate of 5% and ≥2.0 fold change. LogFC, log2 fold change; LogCPM, log2 counts per million; F, F-statistic (negative binomial test). Download Figure 2-3, XLSX file (23KB, xlsx)
Multidimensional scaling analyses revealed that developing POMC and NPY/AgRP neurons possess distinct gene expression profiles compared with their adult counterparts (Fig. 2B). Pathway analyses showed an enrichment of genes involved in neuronal growth, differentiation, and axon development in both groups at E15.5 (Fig. 2C,D; Fig. 2-2). Notably, adult POMC and NPY/AgRP express receptors for multiple metabolic hormones, such as leptinz, ghrelin, and serotonin (Henry et al., 2015). However, their receptors were barely detectable at E15.5 (Fig. 2E), suggesting that these neurons do not respond to these metabolic cues at this stage. Moreover, the transcriptomes of adult POMC and AgRP neurons differ substantially from each other in the adult brain, with 694 differentially expressed genes between the two groups (fold change > 2, q < 0.05) (Henry et al., 2015). In comparison, our analysis found 120 differentially expressed genes at E15.5 (Fig. 2-3), which likely reflects an early stage of divergence of gene expression between the two groups. As expected, transcripts for Pomc and Npy were highly enriched in POMC and NPY/AgRP neurons, respectively (Fig. 2F). Other than the peptidergic markers, we found an enrichment of Slc17a6 (vGLUT2), a marker for glutamatergic neurons in POMC neurons, whereas those for GABAergic neurons, such as Slc32a1 (vGAT), Gad1, and Gad2, were overrepresented in NPY/AgRP neurons (Fig. 2F). Other identified differentially expressed genes include TFs, ion channels, and axon guidance molecules, whose developmental roles in either neuron remain to be determined.
TFs differentially enriched in developing POMC and NPY/AgRP neurons
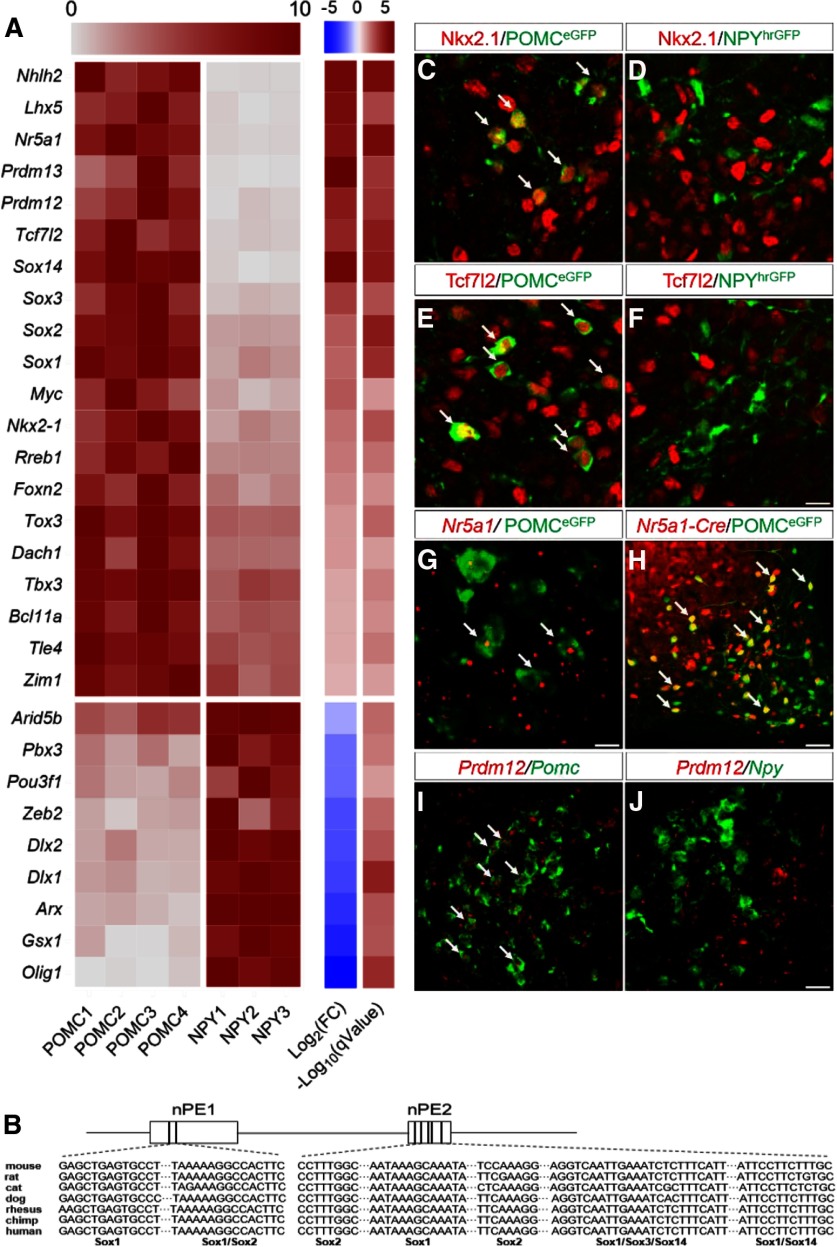
We focused our analyses on TFs as they play a critical role in neuronal differentiation, fate specification, and maintenance. We detected transcripts of 1106 TFs in either POMC or NPY/AgRP neurons at E15.5 (Fig. 3-1). While the vast majority of these factors were expressed at equivalent levels in both neurons, 29 of them showed differential enrichment in one of the two groups (Fig. 3A). As expected, the basic helix-loop-helix factor Nhlh2 is enriched in POMC neurons and controls the levels of prohormone convertase I (Pcsk1), the enzyme that converts POMC to alpha-melanocyte-stimulating hormone (Jing et al., 2004). Moreover, TFs, such as Nkx2.1, Tbx3, Dlx1, and Dlx2, are initially expressed in a broad domain of progenitors during early differentiation (Kimura et al., 1996; Eisenstat et al., 1999; Pontecorvi et al., 2008). We found that their expression persists in postmitotic neurons and are selectively enriched in either POMC (Nkx2.1 and Tbx3) or NPY/AgRP (Dlx1 and Dlx2) neurons (Fig. 3A,C,D). These findings are consistent with recent reports that these factors continue to play a role in POMC or NPY/AgRP neurons after neurogenesis (B. Lee et al., 2018; Orquera et al., 2019; Quarta et al., 2019).
Figure 3.
TFs selectively enriched in either POMC or NPY/AgRP neurons at E15.5. A, Heatmap of 29 differently expressed TFs at E15.5 (Fig. 3-1). Each row is normalized to the maximum expression value for each replicate sample. Log2(FC), Log fold change of gene expression in POMC neurons over that in NPY/AgRP neurons. q value: adjusted p value; FDR ≤ 5% (Benjamin-Hochberg correction). B, Conserved Sox binding motifs in Pomc neuronal enhancers nPE1 and nPE2. C–F, Double-staining of Nkx2.1 (red), Tcf7l2 (red), and GFP (green) in the ARH at E15.5. G, Double FISH (RNAscope) of Nr5a1 mRNA (red) and Pomc mRNA (green) at E15.5. H, Native fluorescence of tomato (red) and immunofluorescence of GFP (green) in the adult ARH. I, J, Double FISH (RNAscope) of Prdm12 mRNA (red), Pomc mRNA (green), or Npy mRNA (green) in the ARH at E15.5. Arrowheads indicate neurons that express both epitopes or mRNAs. Scale bars: G, 5 µm; C–F, 10 µm; I, J, 20 µm; H, 50 µm.
List of all transcription factors expressed in either POMC or NPY/AgRP neurons. All 1106 transcription factors expressed in Either E15.5 POMC or E15.5 NPY/AgRP neurons with a minimal average expression of 10 counts per million in either Pomc or Npy neurons. Download Figure 3-1, XLSX file (154.5KB, xlsx)
The majority (20 of 29) of the differentially expressed TFs are found in the anorexigenic POMC neurons. For example, several members of the Sox proteins (Sox1, Sox2, Sox3, and Sox14) are enriched in POMC neurons (Fig. 3A). de Souza et al. (2005) previously characterized upstream enhancer elements (nPE1 and nPE2) that were both necessary and sufficient to drive reporter gene expression in hypothalamic POMC neurons. Analyses of these sequences revealed several conserved Sox binding sites across multiple mammalian species (Fig. 3B). These findings therefore raise the possibility that Sox proteins may directly interact with upstream enhancer sequences to regulate Pomc transcription. Moreover, developing POMC neurons, but not NPY/AgRP neurons, express mRNAs and proteins of the TF 7-like 2 (Tcf7l2; Fig. 3A,E,F). Polymorphisms of Tcf7l2 are among the most significant genetic markers associated with Type 2 diabetes across multiple ethnic groups (Grant et al., 2006). Previous studies largely focused on its role in peripheral organs, such as the pancreatic islets and liver (Jin, 2016). However, given the importance of POMC neurons in coordinating glucose metabolism (Caron et al., 2018), it is possible that these neurons may serve as a critical node where Tcf7l2 programs glucose homeostasis at critical developmental periods. Furthermore, the nuclear receptor Nr5a1 (also known as steroidogenic factor 1, sf1) has been known as a selective marker for VMH neurons during development (K. W. Kim et al., 2009). The data on its expression in the developing ARH, however, have been inconsistent. While an earlier study reported Nr5a1 proteins in developing POMC neurons (McNay et al., 2006), it was not corroborated by a subsequent analysis (Shimogori et al., 2010). Using cell-type-specific RNA-seq, we detected Nr5a1 mRNAs in POMC neurons at E15.5 (Fig. 3A,G). In support of this finding, fate-mapping of Nr5a1-Cre-expressing neurons (Dhillon et al., 2006) revealed that a subset of adult POMC neurons (37.6 ± 5.5%, n = 4 mice) express the Cre-activated tomato reporter (Fig. 3H). Since Nr5a1 is absent from adult POMC neurons (Henry et al., 2015), we conclude that Nr5a1 is transiently expressed in POMC neurons during development. However, a developmental role for Nr5a1 within POMC neurons remains to be determined. Furthermore, whether its expression demarcates a functionally unique subset of POMC neurons warrants further investigations. Finally, our analyses uncovered an enrichment of two PR-domain containing factors Prdm12 and Prdm13 in developing POMC neurons (Fig. 3A,I,J). Both factors have recently been implicated in fate specification in the developing sensory ganglia and spinal cord (Mona et al., 2017; Bartesaghi et al., 2019; Desiderio et al., 2019). However, their roles in the hypothalamus remain largely unknown.
Generation of a conditional Prdm12 allele
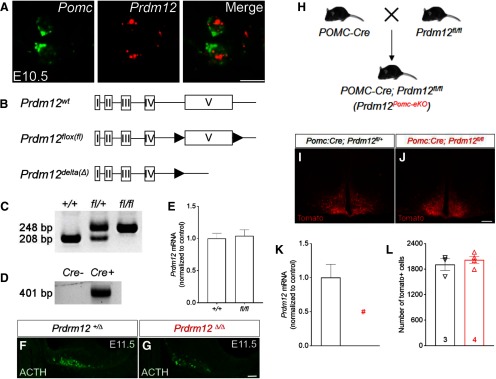
To test whether POMC-enriched TFs are necessary for the anorexigenic neuron identity, we turned to mouse genetics to interrogate their functions in vivo. We first targeted Prdm12 since it has not been implicated in the development of hypothalamic neurons or in energy homeostasis. Before the first appearance of Pomc, we detected Prdm12 mRNA in the ventral hypothalamic neuroepithelium at E9.5. Moreover, it is present within POMC neurons when they first emerge at E10.5 (Fig. 4A). To investigate the physiological role of Prdm12 in vivo, we developed a conditional Prdm12 allele (Prdm12flox) by inserting two loxP sequences flanking the endogenous exon that encodes the DNA-binding domain (Fig. 4B). The floxed exon 5 (the last exon) could be removed in the presence of Cre recombinase, resulting in a deleted Prdm12 allele (Prdm12Δ; Fig. 4B,C,D). Similar to Prdm12 null mice, pups homozygous for the deleted Prdm12 allele (Prdm12Δ/Δ) die quickly after birth (Desiderio et al., 2019), suggesting that Prdm12Δ likely encodes a functionally null allele. Meanwhile, qPCR analyses detected comparable levels of Prdm12 mRNAs within the hypothalamus between Prdm12+/+ and Prdm12fl/fl littermates, suggesting that the introduced loxP sites did not perturb endogenous Prdm12 transcription (n = 5–7, p = 0.7617, unpaired t test; Fig. 4E).
Figure 4.
Generation and validation of a floxed Prdm12 allele. A, FISH for Pomc and Prdm12 mRNAs at E10.5. B, Schematics of the WT (wt), floxed (fl), and δ(Δ) Prdm12 alleles. C, PCR genotyping of Prdm12+/+, Prdm12fl/+, and Prdm12fl/fl mice. D, Detection of the Prdm12Δ allele by PCR. E, qPCR analysis of Prdm12 mRNA in the hypothalamus. F, G, Immunostaining for ACTH. H. Mating scheme for generating Prdm12POMCeKO mice. I, J, Native fluorescence of tomato. K, qPCR analysis of Prdm12 in FACS-sorted tomato-positive neurons. n = 3 replicates from pooled embryos. #Not detectable. L, Cell counts of tomato-positive neurons; n = 3 or 4. Scale bars: A, 5 µm; F, G, 50 µm; I, J, 200 µm.
Prdm12 is required for hypothalamic Pomc expression in the developing ARH
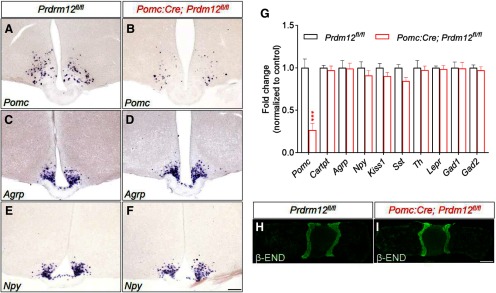
To determine whether Prdm12 is necessary for the onset of ARH Pomc expression, we examined proteins of ACTH, a proteolytic product of POMC, in E11.5 Prdm12Δ/Δ embryos that lack functional Prdm12 in all cells. Immunohistochemistry revealed a significant loss of ACTH in the ARH of Prdm12Δ/Δ mice compared with the controls (Prdm12+/Δ; Fig. 4F,G), suggesting that Prdm12 is needed for the specification of POMC neuron identity. To study the role of Prdm12 specifically in postmitotic POMC neurons, we bred POMC-Cre mice (McHugh et al., 2007) with Prdm12fl/fl mice to generate POMC-Cre; Prdm12fl/fl mice in which Prdm12 was selectively deleted in embryonic POMC neurons (designated hereafter as Prdm12POMCeKO mice; Fig. 4H). A Cre-dependent tdTomato reporter (Rosa26Ai14) was used to mark POMC-Cre expressing neurons in Prdm12POMCeKO mice as well as those in mice heterozygous for the floxed Prdm12 allele (POMC-Cre; Prdm12fl/+; Fig. 4I,J). We verified the loss of Prdm12 mRNAs in FACS-sorted tdTomato+ neurons in Prdm12POMCeKO mice (n = 3, p < 0.0001, unpaired t test; Fig. 4K). Meanwhile, cell counting analyses found similar numbers of tdTomato+ cells present in the adult brains of both genotypes, suggesting that Prdm12 is dispensable for the survival of POMC neurons (n = 3 or 4, p = 0.5093, unpaired t test; Fig. 4L). Remarkably, ISH uncovered a significant loss of Pomc mRNAs in the ARH of Prdm12POMCeKO mice (Fig. 5A,B). In contrast, the expression of Npy and Agrp remained unaffected (Fig. 5C–F). Consistent with these findings, qPCR analyses showed a selective loss of Pomc transcripts among several genes known to be expressed in the ARH (n = 11 or 12, p = 0.0002, unpaired t test; Fig. 5G). Importantly, immunohistochemistry for β-endorphin, a proteolytic product of POMC, suggested that Pomc expression was intact in the pituitary gland (Fig. 5H,I). Collectively, these findings showed that Prdm12 is necessary for hypothalamic Pomc expression in the developing ARH.
Figure 5.
Prdm12 is necessary for arcuate Pomc expression. A–F, ISH for Pomc, Agrp, and Npy in 6-week-old control and Prdm12POMCeKO mice. G, qPCR analysis of gene expression in the hypothalamus (n = 11 or 12, 6-8 weeks old). ***p = 0.0002 (unpaired t test). H, I, Immunostaining for pituitary β-endorphin (END). Scale bars: A–F, 200 µm; H, I, 1 mm.
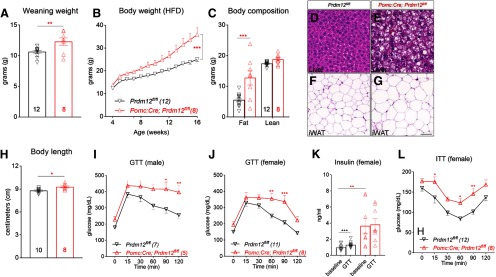
Severe obesity and glucose intolerance in mice lacking Prdm12 in POMC-Cre neurons
POMC deficiency leads to severe early-onset obesity in humans (Farooqi et al., 2003). We investigated whether selective loss of Prdm12 in POMC-Cre neurons was sufficient to cause similar metabolic deficits (Yeo et al., 1998). We found that female Prdm12POMCeKO mice had increased body weight at the time of weaning (n = 8-12, p = 0.0083, unpaired t test; Fig. 6A). These mice gained significantly more weight when fed a high-fat diet (HFD; n = 8-12, F(12,192) = 9.494, p < 0.0001, two-way ANOVA; Fig. 6B; HFD, 60 kcal% fat, D12492i, Research Diets). Nuclear MRI revealed that the elevated body weight was largely due to an increase in fat mass, along with a modest gain in lean mass (n = 8-12, F(1,18) = 13.07, p = 0.002, two-way ANOVA; Fig. 6C). In support of this finding, H&E staining revealed significantly increased lipid accumulation in both the white adipose tissue and liver of Prdm12POMCeKO mice (Fig. 6D–G). Moreover, similar to human POMC/MC4R deficiency, Prdm12POMCeKO mice had an increased body length (nose to anus) compared with their littermate controls (n = 8–10, p = 0.01, unpaired t test; Fig. 6H). Concomitant with obesity were perturbations in glucose homeostasis. For example, both male and female Prdm12POMCeKO mice showed a deficit in glucose clearance during a GTT (n = 5–7, two-way ANOVA, F(5,50) = 3.106, p = 0.0161, Fig. 6I; n = 8–11, F(5,85) = 4.308, p = 0.0015, Fig. 6J). In control mice, plasma insulin levels rose in response to the glucose challenge during the GTT. In contrast, insulin levels were significantly higher in Prdm12POMCeKO mice at the baseline but did not increase during the GTT (n = 8–11, F(1,17) = 13.22, p = 0.002, two-way ANOVA; Fig. 6K). The impairment in insulin sensitivity was further corroborated by an ITT (n = 8–12, F(1,18) = 9.067, p = 0.0075, two-way ANOVA; Fig. 6L).
Figure 6.
Early-onset obesity, increased linear growth, and glucose intolerance in Prdm12POMCeKO mice. A–C, Body weight and body composition (16-week-old) of female Prdm12POMCeKO mice. D–G, H&E staining in the liver and in inguinal white adipose tissue (iWAT) of HFD-fed female mice. Scale bars: D, E, 50 µm; F, G, 100 µm. Body length of female Prdm12POMCeKO mice. I, J, GTT of HFD-fed mice. K, Serum insulin levels during GTT. L, ITT of HFD-fed mice. *p < 0.05; **p < 0.01; ***p < 0.001; unpaired t test (A, H); two-way ANOVA with Sidak's tests for (B, C, I–L).
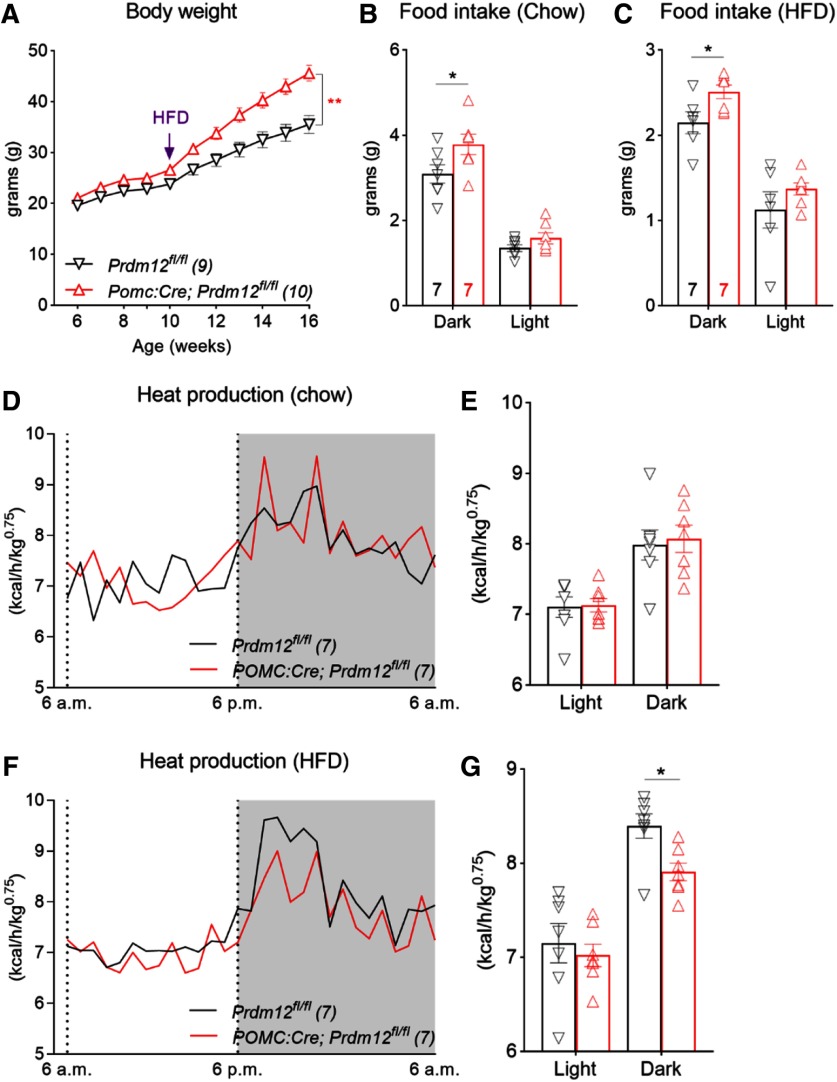
Altered food intake and energy expenditure in mice lacking Prdm12 in POMC-Cre neurons
Similar to females, male Prdm12POMCeKO mice exhibited severe obesity and glucose intolerance when fed an HFD from weaning (data not shown). We selected weight-matched control and Prdm12POMCeKO male mice for another study, in which they were initially maintained on a chow diet before being switched to an HFD. We found that body weight diverged slowly on the chow diet. However, obesity in Prdm12POMCeKO mice was exacerbated when they were fed the HFD (n = 9 or 10, F(10,170) = 19.47, p < 0.0001, two-way ANOVA; Fig. 7A). To further unravel the physiological changes behind the excessive weight gain, we conducted indirect calorimetry analyses using the fully automated TSE metabolic chambers. Male mice were initially maintained on the chow diet during acclimation and the first 3 d of recording before being switched to the HFD for the next 7 d. Under both dietary conditions, Prdm12POMCeKO mice consumed more food during the dark phase of the day (n = 7, F(1,12) = 4.787, p = 0.04, Fig. 7B; n = 7, F(1,12) = 5.697, p = 0.03, Fig. 7C; two-way ANOVA). On the other hand, heat production was similar between chow-fed Prdm12POMCeKO and control mice (n = 7, F(1,12) = 0.07371, p = 0.7906, two-way ANOVA; Fig. 7D,E) but was significantly lower in Prdm12POMCeKO mice during HFD feeding (n = 7, F(1,12) = 5.545, p = 0.03, two-way ANOVA, Fig. 7F,G). Collectively, these results suggest that the HFD-induced exacerbation of weight gain results from a combination of increased energy intake and reduced expenditure.
Figure 7.
Pomc Prdm12 regulates food intake and adaptation to HFD. A, Body weight of male Prdm12POMCeKO mice. B, C, Food intake during light (12 h) and dark (12 h) phases of a day. D, E, Heat production of chow-fed mice. F, G, Heat production of HFD-fed mice. *p < 0.05; **p < 0.01; two-way ANOVA with Sidak's tests (A–G).
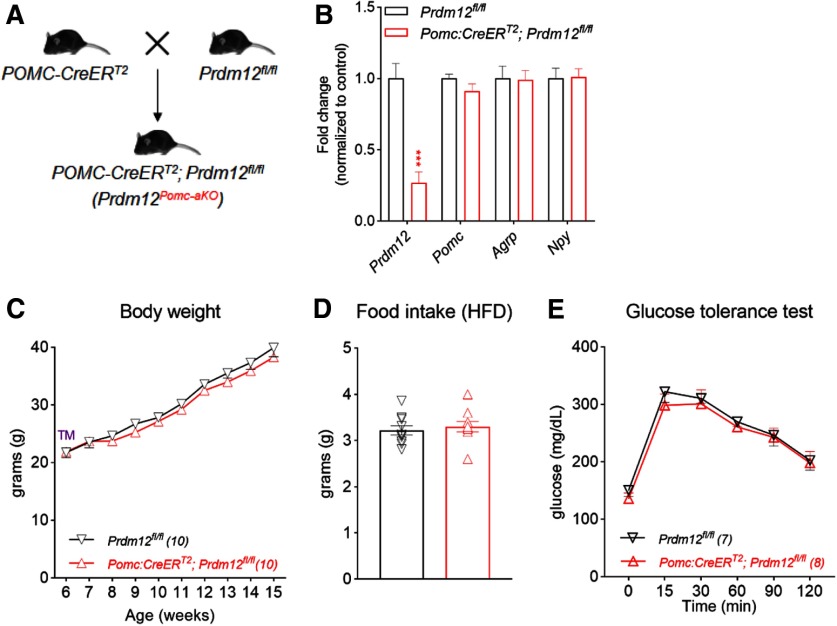
Prdm12 is dispensable for Pomc expression in the adult brain
Transcriptional profiling of adult POMC neurons showed that Prdm12 expression persists in POMC neurons (Henry et al., 2015). The persistent expression of Prdm12 in adult POMC neurons therefore raises the possibility that Prdm12 is required for the maintenance of Pomc expression and energy balance in the adult brain. To test this hypothesis, we selectively ablated Prdm12 in adult POMC neurons using the tamoxifen (TM)-inducible POMC-CreERT2 mice that we previously generated and characterized (Fig. 8A) (Berglund et al., 2013). Inducible Cre activity was detected in >95% POMC neurons following adult TM treatments but was absent from other neurons or in the absence of TM (Caron et al., 2018). We treated both Prdm12fl/fl and POMC-CreERT2; Prdm12fl/fl mice (designated hereafter as Prdm12POMCaKO mice) with TM at 6 weeks of age (three single daily oral gavages, 150 mg/kg). We then harvested ARH from both mice 2 weeks after the last TM dose. qPCR analyses showed that, although Prdm12 mRNAs were significantly decreased in the ARH of Prdm12POMCaKO mice, Pomc levels remained comparable between the two genotypes (n = 11, p = 0.0003, unpairedt test; Fig. 8B). Moreover, HFD-fed Prdm12fl/fl and Prdm12POMCaKO mice gained similar weights after TM treatments (n = 10, F(9,162) = 0.4906, p = 0.8794, two-way ANOVA; Fig. 8C). Consistent with this finding, adult deletion of Prdm12 in POMC neurons did not alter food intake (n = 10, p = 0.5904, unpaired t test; Fig. 8D) or normal glucoregulatory responses in a GTT (n = 7–10, F(5,75) = 0.2265, p = 0.9499, two-way ANOVA; Fig. 8E). Collectively, these findings showed that Prdm12 acts during development to regulate Pomc expression and energy balance.
Figure 8.
Prdm12 is no longer required in adulthood. A, Mating scheme for B–E. B, qPCR analysis of hypothalamic genes from adult male mice. ***p < 0.001 (unpaired t test). C, Body weight. D, Food intake. E, GTT for HFD-fed Prdm12POMCaKO mice.
Discussion
While much has been learned about how POMC and NPY/AgRP regulate food intake in the adult brain, relatively little is known about how these neurons emerge in the developing hypothalamus.
The combination of cell-type-specific transcriptomics and mouse genetics provided a means to dissect cellular and functional diversity in the developing hypothalamus and allowed us to decipher the transcriptional codes behind the orexigenic versus anorexigenic identity. In addition to the control of cell fate, our profiling revealed genes involved in other developmental milestones, such as axonal growth, pathfinding, and synapse formation. As a result, our dataset will serve as a resource to study how functional melanocortin feeding circuits are assembled during development.
The establishment and maintenance of the anorexigenic POMC neuron identity are critical for energy homeostasis. Indeed, mutations in Pomc lead to severe early-onset obesity in humans (Krude et al., 1998). Before the first appearance of Pomc expression, a cascade of TFs, such as Ascl1 (McNay et al., 2006), Ngn3 (Pelling et al., 2011), Rax (Lu et al., 2013), and Nkx2.1 (Kimura et al., 1996), sequentially specifies hypothalamic progenitors that give rise to the ARH. Moreover, de Souza et al. (2005) previously characterized the upstream cis-regulatory sequences that drive hypothalamic Pomc expression. By identifying TFs that bind these sequences, these investigators recently reported that two homeodomain TFs Isl1 and Nkx2.1 transactivate Pomc transcription in the developing hypothalamus. Loss of either factor in hypothalamic progenitors prevents the onset of Pomc expression in the developing ARH (Nasif et al., 2015; Orquera et al., 2019). As a result, these findings establish a critical role for both genes in specifying the POMC neuron identity. Moreover, the expression of Isl1 and Nkx2.1 persists in postmitotic POMC neurons. It is conceivable that both factors may continue to play a role in the maintenance of POMC neuron identity. However, while early deletion of Nkx2.1 led to a complete loss of POMC neurons, its selective deletion in POMC-Cre neurons did not change the number of Pomc-immunoreactive cells and only caused a mild reduction in Pomc levels (Orquera et al., 2019; and unpublished observation by our group). On the other hand, Isl1 is expressed by other ARH neurons and is necessary for the expression of multiple ARH markers, including Npy, Agrp, GHRH, and Sst. Importantly, it is required for the postnatal survival of ARH neurons (B. Lee et al., 2016). Although selective deletion of Isl1 in POMC-Cre neurons led to a significant loss of Pomc mRNAs in the adult brain, it is less clear whether a potential loss of POMC neurons could have contributed to the observed deficit. Collectively, these findings raise the possibility that the transcriptional maintenance of Pomc in postmitotic POMC neurons requires additional transcription regulators that have yet been identified.
To identify these factors, we profiled the transcriptomes of genetically labeled POMC and NPY/AgRP neurons using whole-genome RNA sequencing. At E15.5, Pomc and Npy expression had just been established while both neurons were still acquiring other markers (e.g., Cart or Agrp) that define their adult neurotransmitter identity. Our analysis discovered >1000 TFs in either population. However, only 29 of them are selectively enriched in one of the two groups. The differential expression patterns suggest that these factors likely play important roles in programming the orexigenic versus anorexigenic identity and function. Indeed, the POMC-enriched TFs provide new candidate genes for the transcriptional maintenance of Pomc. For example, the enrichment of Sox proteins, as well as the presence of phylogenetically conserved Sox binding sites, suggests that Sox proteins may directly interact with Pomc enhancer sequences to regulate Pomc transcription. Moreover, both Prdm12 and Prdm13 are necessary for cell fate specification in sensory ganglia and spinal cord, respectively (Mona et al., 2017; Bartesaghi et al., 2019), and therefore could have similar functions in the developing hypothalamus. Indeed, we found that selective deletion of Prdm12 in POMC-Cre neurons led to a ∼75% loss of Pomc mRNAs in the hypothalamus. Furthermore, indirect calorimetry analyses revealed that the excessive weight gain in HFD-fed Prdm12POMCeKO mice was the result of a combination of increased energy intake and decreased expenditure, which further supports a loss of POMC neuron function in these mice. Of note, obesity in Prdm12POMCeKO mice is comparable with other mutant mouse models that yield a similar loss of hypothalamic Pomc expression. Together, these findings support a recently proposed threshold of Pomc transcription (∼30%–40% of normal values) on physiology, below which hyperphagia and obesity become evident (Lam et al., 2015). On the other hand, we believe the role of Prdm12 in POMC neurons may be different from that of Isl1, Nkx2.1, and Tbx3 (Nasif et al., 2015; Orquera et al., 2019; Quarta et al., 2019). Unlike Isl1, Prdm12 is not required for cell survival. Moreover, ablation of Prdm12 in POMC-Cre neurons led to a more severe loss of Pomc and obesity than similar deletion of Nkx2.1. Finally, different from Tbx3, loss of Prdm12 in POMC-Cre neurons did not alter the expression of Npy or Agrp. Collectively, these findings support a cell-autonomous role for Prdm12 in Pomc expression in developing POMC neurons.
Among several genes examined in the ARH, we found a selective loss of Pomc mRNA in Prdm12POMCeKO mice. However, Pomc is unlikely the only transcriptional target of Prdm12. Rather, we expect Prdm12 regulates many genes other than the anorexigenic neuron marker. Future work comparing the transcriptomes of Prdm12-deficient and WT POMC neurons will help reveal a Prdm12-dependent transcriptional network. Moreover, the observation that Prdm12 is necessary for Pomc expression in the hypothalamus, but not in the pituitary gland, raises the possibility that it directly interacts with previously identified enhancer sequences that drive neuronal Pomc expression (de Souza et al., 2005). However, the consensus DNA binding sequences for Prdm12 remain unknown. Moreover, the lack of an antibody for chromatin immunoprecipitation prevented us from testing this hypothesis in the current study. Of note, in addition to the zinc-finger DNA binding domain, all PRDM proteins possess a PR domain that resembles the catalytic domain of histone methyltransferases (Di Zazzo et al., 2013). Indeed, some PRDM factors can regulate gene expression through their intrinsic methyltransferase activity, or by recruiting other chromatin remodeling complexes (Gyory et al., 2004; Hayashi et al., 2005). Whether or not this is the case for Prdm12 remains to be determined. However, it is formally possible that Prdm12 may promote Pomc expression through epigenetic mechanisms.
TFs that maintain normal Pomc levels in the adult brain remain unknown. The use of the TM-inducible POMC-CreERT2 mice allowed us to interrogate the function of Prdm12 in adult POMC neurons after it has already fulfilled its role during development. To our surprise, Prdm12 is no longer needed for the maintenance of normal Pomc levels or body weight in the adult brain. It is possible that other TFs may compensate for Prdm12 loss of function in adult POMC neurons. Alternatively, Prdm12 may adopt new functions and no longer participate in Pomc transcription in the adult. A shift in transcriptional targets between developmental and adult stages has been reported for TFs in other neurotransmitter systems (Wyler et al., 2016). Regardless, the observation that Prdm12 is dispensable for Pomc expression in the adult raises the possibility that different TFs regulate Pomc transcription during distinct developmental and adult periods. Notably, previous work demonstrated that the adipocyte-derived hormone leptin exerts its neurotrophic actions on ARH neurons only during a critical developmental period (Bouret et al., 2004; Kamitakahara et al., 2018). Similarly, Prdm12 may act within a defined developmental window to program energy intake and expenditure. Therefore, future studies are warranted to temporally delineate such a critical period during which Prdm12 dysfunction poses a major risk for obesity.
Footnotes
This work was supported by National Institutes of Health Grants R01 DK114036 to C.L., F32DK116427 to S.C.W., and F31 NS111796 to M.A.L. H.C.L was supported by the Rita Allen Foundation Award in Pain. M.A.L. was supported by the William F. and Grace H. Kirkpatrick Award. C.L. was also supported by an American Heart Association Scientist Development Grant 16SDG27260001, a University of Texas Southwestern Pilot & Feasibility Award, and a Grossman Endowment Award for Diabetes Research. We thank members of the Next Generation Sequencing Core, the Transgenic Core, the Histo Pathology Core, the Flow Cytometry Core, and the Metabolic Phenotyping Core at University of Texas Southwestern.
The authors declare no competing financial interests.
References
- Atasoy D, Betley JN, Su HH, Sternson SM (2012) Deconstruction of a neural circuit for hunger. Nature 488:172–177. 10.1038/nature11270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi L, Wang Y, Fontanet P, Wanderoy S, Berger F, Wu H, Akkuratova N, Bouçanova F, Médard JJ, Petitpré C, Landy MA, Zhang MD, Harrer P, Stendel C, Stucka R, Dusl M, Kastriti ME, Croci L, Lai HC, Consalez GG, et al. (2019) PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Rep 26:3484–3492.e4. 10.1016/j.celrep.2019.02.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, Newman EA, Blackshaw S (2015) Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip Rev Dev Biol 4:445–468. 10.1002/wdev.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund ED, Liu C, Sohn JW, Liu T, Kim MH, Lee CE, Vianna CR, Williams KW, Xu Y, Elmquist JK (2013) Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis. J Clin Invest 123:5061–5070. 10.1172/JCI70338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouret SG, Draper SJ, Simerly RB (2004) Trophic action of leptin on hypothalamic neurons that regulate feeding. Science 304:108–110. 10.1126/science.1095004 [DOI] [PubMed] [Google Scholar]
- Caron A, Dungan Lemko HM, Castorena CM, Fujikawa T, Lee S, Lord CC, Ahmed N, Lee CE, Holland WL, Liu C, Elmquist JK (2018) POMC neurons expressing leptin receptors coordinate metabolic responses to fasting via suppression of leptin levels. Elife 7:e33710 10.7554/eLife.33710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla K, Tripathi S, Thommesen L, Lægreid A, Kuiper M (2013) TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 29:2519–2520 [DOI] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, Zhang Y (2017) Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Rep 18:3227–3241. 10.1016/j.celrep.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone RD. (1999) The central melanocortin system and energy homeostasis. Trends Endocrinol Metab 10:211–216. 10.1016/s1043-2760(99)00153-8 [DOI] [PubMed] [Google Scholar]
- Cowley MA, Smart JL, Rubinstein M, Cerdan MG, Diano S, Horvath TL, Cone RD, Low MJ (2001) Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411:480–484. 10.1038/35078085 [DOI] [PubMed] [Google Scholar]
- de Souza FS, Santangelo AM, Bumaschny V, Avale ME, Smart JL, Low MJ, Rubinstein M (2005) Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol Cell Biol 25:3076–3086. 10.1128/MCB.25.8.3076-3086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S, Vermeiren S, Van Campenhout C, Kricha S, Malki E, Richts S, Fletcher EV, Vanwelden T, Schmidt BZ, Henningfeld KA, Pieler T, Woods CG, Nagy V, Verfaillie C, Bellefroid EJ (2019) Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Rep 26:3522–3536. e5. 10.1016/j.celrep.2019.02.097 [DOI] [PubMed] [Google Scholar]
- Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA, Tang V, Kenny CD, Christiansen LM, White RD, Edelstein EA, Coppari R, Balthasar N, Cowley MA, Chua S, Elmquist JK, Lowell BB (2006) Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 49:191–203. 10.1016/j.neuron.2005.12.021 [DOI] [PubMed] [Google Scholar]
- Di Zazzo E, De Rosa C, Abbondanza C, Moncharmont B (2013) PRDM proteins: molecular mechanisms in signal transduction and transcriptional regulation. Biology (Basel) 2:107–141. 10.3390/biology2010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T, Nguyen LT, Kwon JYH, Barak B, Ge W, Kedaigle AJ, Carroll S, Li S, Hacohen N, Rozenblatt-Rosen O, Shalek AK, Villani A-C, Regev A, Levin JZ, (2019) Systematic comparative analysis of single cell RNA-sequencing methods. BioRxiv. doi: 10.1101/632216. [DOI] [Google Scholar]
- Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC (2013) Neurotransmitter switching in the adult brain regulates behavior. Science 340:449–453. 10.1126/science.1234152 [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL (1999) DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol 414:217–237. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O'Rahilly S (2003) Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med 348:1085–1095. 10.1056/NEJMoa022050 [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, et al. (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323. 10.1038/ng1732 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejer G, Seto E, Wright KL (2004) PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5:299–308. 10.1038/ni1046 [DOI] [PubMed] [Google Scholar]
- Hahn TM, Breininger JF, Baskin DG, Schwartz MW (1998) Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci 1:271–272. 10.1038/1082 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y (2005) A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438:374–378. 10.1038/nature04112 [DOI] [PubMed] [Google Scholar]
- Henry FE, Sugino K, Tozer A, Branco T, Sternson SM (2015) Cell-type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife 4:09800 10.7554/eLife.09800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman C, Cho H, Brock O, Lim SJ, Youn SM, Park Y, Kim S, Lee SK, Delogu A, Lee JW (2019) Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nat Commun 10:3696. 10.1038/s41467-019-11667-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T. (2016) Current understanding on role of the Wnt signaling pathway effector TCF7L2 in glucose homeostasis. Endocr Rev 37:254–277. 10.1210/er.2015-1146 [DOI] [PubMed] [Google Scholar]
- Jing E, Nillni EA, Sanchez VC, Stuart RC, Good DJ (2004) Deletion of the Nhlh2 transcription factor decreases the levels of the anorexigenic peptides alpha melanocyte-stimulating hormone and thyrotropin-releasing hormone and implicates prohormone convertases I and II in obesity. Endocrinology 145:1503–1513. 10.1210/en.2003-0834 [DOI] [PubMed] [Google Scholar]
- Kamitakahara A, Bouyer K, Wang CH, Simerly R (2018) A critical period for the trophic actions of leptin on AgRP neurons in the arcuate nucleus of the hypothalamus. J Comp Neurol 526:133–145. 10.1002/cne.24327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Washington PW, Wang ZQ, Lin S, Sun C, Jiang L, Blackshaw S (2019) Single cell RNA-Seq analysis identifies molecular mechanisms controlling hypothalamic patterning and differentiation. BioRxiv 657148. doi: 10.1101/657148. [DOI] [Google Scholar]
- Kim KW, Zhao L, Parker KL (2009) Central nervous system-specific knockout of steroidogenic factor 1. Mol Cell Endocrinol 300:132–136. 10.1016/j.mce.2008.09.026 [DOI] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ (1996) The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 10:60–69. 10.1101/gad.10.1.60 [DOI] [PubMed] [Google Scholar]
- Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A (1998) Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet 19:155–157. 10.1038/509 [DOI] [PubMed] [Google Scholar]
- Lam DD, de Souza FS, Nasif S, Yamashita M, López-Leal R, Otero-Corchon V, Meece K, Sampath H, Mercer AJ, Wardlaw SL, Rubinstein M, Low MJ (2015) Partially redundant enhancers cooperatively maintain Mammalian pomc expression above a critical functional threshold. PLoS Genet 11:e1004935. 10.1371/journal.pgen.1004935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Lee S, Lee SK, Lee JW (2016) The LIM-homeobox transcription factor Isl1 plays crucial roles in the development of multiple arcuate nucleus neurons. Development 143:3763–3773. 10.1242/dev.133967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Kim J, An T, Kim S, Patel EM, Raber J, Lee SK, Lee S, Lee JW (2018) Dlx1/2 and Otp coordinate the production of hypothalamic GHRH- and AgRP-neurons. Nat Commun 9:2026. 10.1038/s41467-018-04377-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim DW, Remedios R, Anthony TE, Chang A, Madisen L, Zeng H, Anderson DJ (2014) Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature 509:627–632. 10.1038/nature13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup.. (2009) The Sequence alignment/map (SAM) format and SAMtools, Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Bookout AL, Lee S, Sun K, Jia L, Lee C, Udit S, Deng Y, Scherer PE, Mangelsdorf DJ, Gautron L, Elmquist JK (2014) PPARγ in vagal neurons regulates high-fat diet induced thermogenesis. Cell Metab. 19:722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, Deneris ES (2010) Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 13:1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Kar D, Gruenig N, Zhang ZW, Cousins N, Rodgers HM, Swindell EC, Jamrich M, Schuurmans C, Mathers PH, Kurrasch DM (2013) Rax is a selector gene for mediobasal hypothalamic cell types. J Neurosci 33:259–272. 10.1523/JNEUROSCI.0913-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S (2007) Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science 317:94–99. 10.1126/science.1140263 [DOI] [PubMed] [Google Scholar]
- McNay DE, Pelling M, Claxton S, Guillemot F, Ang SL (2006) Mash1 is required for generic and subtype differentiation of hypothalamic neuroendocrine cells. Mol Endocrinol 20:1623–1632. 10.1210/me.2005-0518 [DOI] [PubMed] [Google Scholar]
- Mona B, Uruena A, Kollipara RK, Ma Z, Borromeo MD, Chang JC, Johnson JE (2017) Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 6:e25787 10.7554/eLife.25787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasif S, de Souza FS, Gonzalez LE, Yamashita M, Orquera DP, Low MJ, Rubinstein M (2015) Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proc Natl Acad Sci USA 112:E1861–E1870. 10.1073/pnas.1500672112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, Johansen JE, Schalling M, Hokfelt T, Fetissov SO (2005) Maturation of the hypothalamic arcuate agouti-related protein system during postnatal development in the mouse. Brain Res Dev Brain Res 155:147–154. 10.1016/j.devbrainres.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Oka Y, Ye M, Zuker CS (2015) Thirst driving and suppressing signals encoded by distinct neural populations in the brain. Nature 520:349–352. 10.1038/nature14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orquera DP, Tavella MB, de Souza FS, Nasif S, Low MJ, Rubinstein M (2019) The homeodomain transcription factor NKX2.1 is essential for the early specification of melanocortin neuron identity and activates Pomc expression in the developing hypothalamus. J Neurosci 39:4023–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla SL, Carmody JS, Zeltser LM (2010) Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med 16:403–405. 10.1038/nm.2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla SL, Reef D, Zeltser LM (2012) Defining POMC neurons using transgenic reagents: impact of transient Pomc expression in diverse immature neuronal populations. Endocrinology 153:1219–1231. 10.1210/en.2011-1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JG, Janssen MJ, Chou DY, Abe K, Zukowska Z, Vicini S (2009) Excitatory and inhibitory synapses in neuropeptide Y-expressing striatal interneurons. J Neurophysiol 102:3038–3045. 10.1152/jn.00272.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling M, Anthwal N, McNay D, Gradwohl G, Leiter AB, Guillemot F, Ang SL (2011) Differential requirements for neurogenin 3 in the development of POMC and NPY neurons in the hypothalamus. Dev Biol 349:406–416. 10.1016/j.ydbio.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Pontecorvi M, Goding CR, Richardson WD, Kessaris N (2008) Expression of Tbx2 and Tbx3 in the developing hypothalamic-pituitary axis. Gene Expr Patterns 8:411–417. 10.1016/j.gep.2008.04.006 [DOI] [PubMed] [Google Scholar]
- Quarta C, Fisette A, Xu Y, Colldén G, Legutko B, Tseng YT, Reim A, Wierer M, De Rosa MC, Klaus V, Rausch R, Thaker VV, Graf E, Strom TM, Poher AL, Gruber T, Le Thuc O, Cebrian-Serrano A, Kabra D, Bellocchio L, et al. (2019) Functional identity of hypothalamic melanocortin neurons depends on Tbx3. Nat Metab 1:222–235. 10.1038/s42255-018-0028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, Alpár A, Mulder J, Clotman F, Keimpema E, Hsueh B, Crow AK, Martens H, Schwindling C, Calvigioni D, Bains JS, Máté Z, Szabó G, Yanagawa Y, Zhang MD, et al. (2017) Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 20:176–188. 10.1038/nn.4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S (2010) A genomic atlas of mouse hypothalamic development. Nat Neurosci 13:767–775. 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB (2008) Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci 11:998–1000. 10.1038/nn.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler SC, Spencer WC, Green NH, Rood BD, Crawford L, Craige C, Gresch P, McMahon DG, Beck SG, Deneris E (2016) Pet-1 switches transcriptional targets postnatally to regulate maturation of serotonin neuron excitability. J Neurosci 36:1758–1774. 10.1523/JNEUROSCI.3798-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, O'Rahilly S (1998) A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20:111–112. 10.1038/2404 [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes and expression values in E15.5 POMC and NPY/AgRP neurons. LogFC, log2 fold change; LogCPM, log2 counts per million; F, F-statistic (negative binomial test). Download Figure 2-1, XLSX file (2.2MB, xlsx)
Gene ontology of pathways enriched in E15.5 POMC and NPY/AgRP neurons. Genes enriched (overrepresentation enrichment analysis) in Pomc or NPY neurons with a FDR of 5% and a minimum of 3 genes per GO category (hypergeometric test, with a BH correction). Genes are segregated by Biological Process, Cellular Component, and Molecular Function. Download Figure 2-2, XLSX file (17.9KB, xlsx)
List of genes enriched in E15.5 POMC and NPY/AgRP neurons. List of genes significantly enriched in E15.5 Pomc neurons over NPY neurons with a False Discovery Rate of 5% and ≥2.0 fold change. LogFC, log2 fold change; LogCPM, log2 counts per million; F, F-statistic (negative binomial test). Download Figure 2-3, XLSX file (23KB, xlsx)
List of all transcription factors expressed in either POMC or NPY/AgRP neurons. All 1106 transcription factors expressed in Either E15.5 POMC or E15.5 NPY/AgRP neurons with a minimal average expression of 10 counts per million in either Pomc or Npy neurons. Download Figure 3-1, XLSX file (154.5KB, xlsx)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request. RNA sequencing data reported in this paper have been deposited into NCBI's Gene Expression Omnibus under accession number GEO: