Microbial life is surprisingly abundant and diverse in global desert ecosystems. In these environments, microorganisms endure a multitude of physicochemical stresses, including low water potential, carbon and nitrogen starvation, and extreme temperatures. In this review, we summarize our current understanding of the energetic mechanisms and trophic dynamics that underpin microbial function in desert ecosystems. Accumulating evidence suggests that dormancy is a common strategy that facilitates microbial survival in response to water and carbon limitation.
KEYWORDS: desert, dormancy, energetics, energy reserve, photosynthesis, trace gas
ABSTRACT
Microbial life is surprisingly abundant and diverse in global desert ecosystems. In these environments, microorganisms endure a multitude of physicochemical stresses, including low water potential, carbon and nitrogen starvation, and extreme temperatures. In this review, we summarize our current understanding of the energetic mechanisms and trophic dynamics that underpin microbial function in desert ecosystems. Accumulating evidence suggests that dormancy is a common strategy that facilitates microbial survival in response to water and carbon limitation. Whereas photoautotrophs are restricted to specific niches in extreme deserts, metabolically versatile heterotrophs persist even in the hyper-arid topsoils of the Atacama Desert and Antarctica. At least three distinct strategies appear to allow such microorganisms to conserve energy in these oligotrophic environments: degradation of organic energy reserves, rhodopsin- and bacteriochlorophyll-dependent light harvesting, and oxidation of the atmospheric trace gases hydrogen and carbon monoxide. In turn, these principles are relevant for understanding the composition, functionality, and resilience of desert ecosystems, as well as predicting responses to the growing problem of desertification.
INTRODUCTION
Drylands cover ∼40% of the terrestrial land surface area, with arid and hyper-arid regions constituting 11.5% and 6.4%, respectively (1). Projections based on current global warming trends suggest that drylands will constitute more than half of land surfaces by the end of the century (2). Organisms living in these ecosystems face prolonged and severe water deprivation, which curtail cellular and metabolic activities. Without extracellular water, nutrients and substrates cannot be mobilized in a dissolved form for cellular uptake (3) and microbes themselves are unable to move to find resources, leading to starvation (4). Cellular metabolism is further restricted by environmental stressors, such as low organic carbon and nitrogen availability, high UV irradiation, dryland salinity, and temperature extremes (5–7). In particular, the distribution of key primary producers, such as oxygenic phototrophs, is limited by these cumulative pressures (Fig. 1). Processes such as cyanobacterial photosynthesis in soil biocrusts (8) or water uptake by plant roots (9) generally cease below matrix water potentials of −3 MPa; however, the matrix water potential in desiccated soils is typically between −40 and −95 MPa (10). As a result, in hyper-arid desert soils, photosynthetic organisms are generally restricted to specific lithic refugia, such as the pore spaces of coarse-grained rocks (endoliths) and the ventral surfaces of translucent minerals, such as quartz (hypoliths). Here, they are protected from UV radiation and buffered against extreme temperature and desiccation, while benefiting from sufficient incident light for photosynthesis (11, 12).
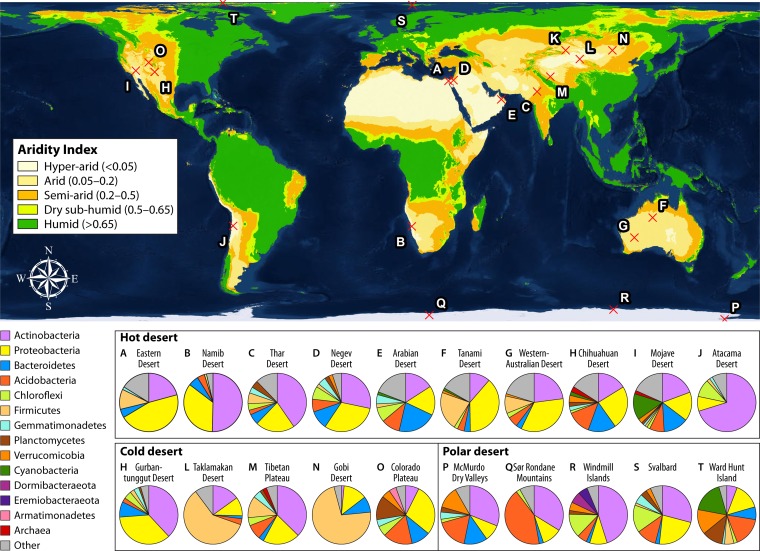
FIG 1.
Microbial community structure of global desert soils. The map is generated by ArcGIS 10.6 and shaded by global aridity index, a ratio of mean annual precipitation to potential evapotranspiration (160) modeled by Antonio Trabucco and Robert Zomer (161). The relative abundances of major microbial groups in 20 desert (nonbiocrust) soils from Africa (162, 163), Antarctica (77, 132, 164), Asia (165–170), Australia (171), Europe (172), North America (47, 77, 173), and South America (13) are displayed in pie charts and in Table S1 in the supplemental material. Phyla with a <1% relative abundance were grouped into the category “Other.” Actinobacteria is the most abundant phylum detected in bare soils (25.5%), followed by Proteobacteria (21%), Acidobacteria (6.5%), Bacteroidetes (6%), Chloroflexi (2.5%), and Firmicutes (2%) (median values of the 20 samples are shown in Table S1). Cyanobacteria, though abundant in soil biocrusts and lithic niches, are present in less than 1% in most bare soil samples.
Information on published samples of the 20 global desert sites used to construct microbial community profiles. Download Table S1, XLSX file, 0.02 MB (20.1KB, xlsx) .
Copyright © 2020 Leung et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Despite photoautotrophs typically being low in abundance, diverse and viable microbial communities are present in the topsoils of most deserts, including the hyper-arid soils of the Atacama Desert (13) and Antarctic Dry Valleys (14). As summarized in Fig. 1, deserts globally are usually dominated by heterotrophic bacteria from phyla such as Actinobacteria, Proteobacteria, and Chloroflexi. In these environments, given that there can be years without precipitation, heterotrophs face extreme starvation for their preferred organic energy and carbon sources. Currently, researchers lack a holistic understanding of the energetic basis of their growth and persistence in desert environments. However, through a combination of culture-based and culture-independent studies, several metabolic mechanisms that may allow these microbes to survive desiccation and associated stresses have been uncovered.
DORMANCY AS A GENERAL STRATEGY TO REDUCE ENERGY EXPENDITURE
Desert soil microorganisms routinely experience extended periods of desiccation, during which they are subjected to extreme energy limitation and other environmental stresses. In response to these harsh conditions, some microbes reversibly enter a metabolically less active state termed dormancy (15). Different groups of microorganisms may adopt different dormancy strategies. Some species form morphologically distinct resting structures that are commonly characterized by thickened cell walls or accumulations of extracellular polymeric substances (15). For instance, members of Actinobacteria and Firmicutes, Gram-positive bacterial phyla widely found in drylands (7, 13, 14, 16), are well known for their ability to form highly stress-resistant spores (17, 18). Consistently, these sporulating taxa are among the most common groups identified in desert soils (19–22) based on both conventional cultivation studies and modern molecular phylogenetic analyses. Common Cyanobacteria isolated from desert biocrusts, such as Anabaena, Nostoc, and Cylindrospermum (23, 24), can differentiate into specific spore-like structures termed akinetes, which tend to be much larger in size than their vegetative structures (25). Gram-negative Proteobacteria isolated from arid soils, such as Azotobacter (26) and Ramlibacter (27), are likewise able to transit into multilayered cysts. Under favorable conditions, these resting stages can germinate to produce vegetative cells.
A substantial proportion of microorganisms, such as the common arid soil actinobacterium Arthrobacter (28), do not undergo extensive morphological differentiation during the transition to the dormant state. However, they still share core strategies with sporulators, including reduction or cessation of growth; reduction of cell size; repression of energetically expensive activities, such as motility and the synthesis of macromolecules; alteration of the composition of membrane lipids and cell wall components; and upregulation of macromolecular repair machinery (15, 29, 30). The highly radiation- and desiccation-resistant genus Deinococcus provides an extreme example of how microorganisms can minimize energy expenditure during desiccation persistence. This bacterium tolerates substantial DNA damage, including numerous double-strand breaks (31), without apparently initiating repair in its desiccated state (32). The chromosome is reassembled from the fragments only upon rehydration (33). Accumulation of antioxidants, such as carotenoids, small peptides, and manganese complexes, offers a protective environment for proteins involved in recovery, thereby reducing the energy costs of repair (34, 35). It has been suggested that members of the Rubrobacteria (36), a highly abundant desert actinobacterial class (37, 38), adopt similar survival strategies.
Regardless of the form that it takes, dormancy increases cellular resistance to external stresses while reducing energy expenditure. However, dormancy does not completely eliminate the requirement for energy, given that a basal energy supply is required to maintain cellular integrity. Although the exact maintenance energy for microbial communities in soils with low water content has not been reported, modeling studies suggest that microorganisms in moist nutrient-deficient soils may metabolize between 10 and 100 μg of carbon per gram of biomass carbon per hour for maintenance (39), and one experiment demonstrated that desiccated Arthrobacter in laboratory conditions consumed 0.0005% of cellular carbon per hour (40). This indicates that dormancy cannot be sustained indefinitely without external energy input. Exacerbating energy demands, hot desert microorganisms are subjected to high levels of oxidative stress due to the high gas permeability of sandy desert soils, desiccation-induced reactive oxygen species formation, Maillard reactions, and extreme temperature-accelerated damage (30, 41–43). The accumulation of excessive damage to nucleic acids, proteins, and cell membranes, if not repaired, will eventually lead to mortality. Therefore, even dormant cells may require basal levels of energy to repair damaged cellular components either during and/or following quiescence. Energy is also required to maintain a minimum membrane potential for ATP synthesis and metabolite transport (44).
ENERGY RESERVE HYPOTHESIS
In desert ecosystems, a transient water supply can be provided in various ways: occasional precipitation events, condensation of dew or fog, and ice or snow melts in polar deserts. Desert microorganisms may depend on such brief “metabolic windows” to generate biomass and accumulate reserve compounds in preparation for long periods of water scarcity and the consequent requirement of maintenance energy. This constitutes the energy reserve hypothesis, adapted from the “pulse-reserve” paradigm proposed by Noy-Meir in 1973 (45) for plant adaptation in desert ecosystems.
In plant-free desert soils, oxygenic photosynthetic organisms are key primary producers. Such organisms are the keystone taxa of biocrust communities (46, 47) and photosynthetic lithic communities (11, 12, 47). Biocrusts cover up to 70% of semiarid and arid zones on all seven continents (46) and comprise a global area of over 1.3 billion hectares (48). In these environments, water is often provided in the form of early-morning dew, quickly followed by desiccation as the day breaks with rising temperatures and declining relative humidity (49, 50). Microbial communities within the biocrusts must therefore rapidly respond to wetting by resuming respiration and photosynthesis for biomass synthesis and then just as rapidly shut off these systems. For example, simulation of dew hydration in Leptolyngbya ohadii, a dominant cyanobacterium in desert sand biocrusts, causes a rapid resumption of photosynthesis (51). Upon the onset of desiccation, Microcoleus vaginatus, a keystone cyanobacterial species in biocrusts, channels energy into the synthesis of energy and carbon storage compounds, such as polyhydroxyalkanoates, polyphosphates, and cyanophycins (10). Many other cyanobacteria, such as Scytonema and Aphanizomenon, are also known to accumulate energy reserves in response to water stress or during the transition to dormancy (25, 52, 53).
For heterotrophs, metabolic substrates become available at the instance when soil is wetted. Increases in soil water potential cause the mobilization of extracellular soil organic carbon (54). In addition, due to osmotic changes, hydration is thought to stimulate the release of organic carbon from microorganisms through either inducing cellular lysis (55, 56) or stimulating the secretion of intracellular osmoprotectants, such as trehalose and glycine betaine (57, 58); note, however, that in situ evidence for osmolyte release remains lacking (59–61). The sudden availability of metabolizable substrates supports the idea that heterotrophs consume them rapidly, with respiration rates peaking within minutes of soil hydration and then gradually declining (62, 63). This phenomenon is termed the “Birch effect” (64). Upon depletion of these carbon sources, heterotrophs rely on cross-feeding by exometabolite exchange with phototrophs and other heterotrophs (Fig. 2). Cyanobacteria in biocrusts release a large range of photosynthates and exudates, such as hexose sugars, while heterotrophs excrete a smaller subset of metabolites (65, 66). Additionally, biocrust microorganisms, most prominently M. vaginatus, produce large amounts of complex extracellular polymeric substances, such as polysaccharides (67–69), which can potentially be digested by associated specialized heterotrophs like Bacteroidetes (70), as observed, for example, in marine consortia (70, 71). The sharing and differential partitioning of this exometabolite pool allows the rapid buildup and accumulation of biomass. In response to xeric stress, heterotrophs upregulate the synthesis of reserve molecules, such as glycogen (72), wax esters (73), and lipids (74).
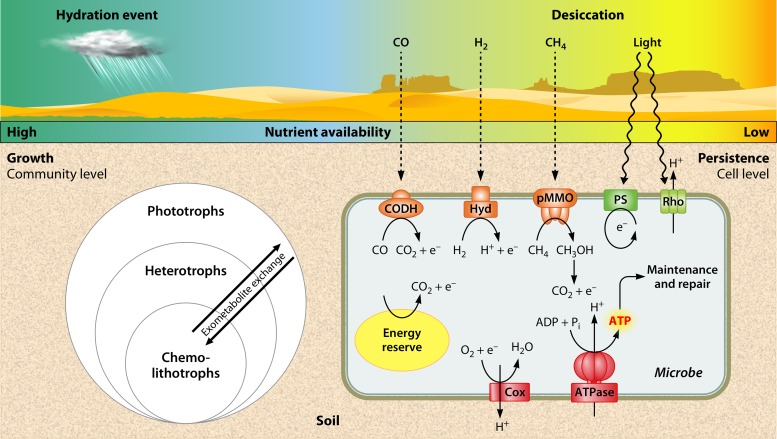
FIG 2.
Conceptual diagram representing the model lifestyle of a microbial community in a desert in response to hydration-desiccation cycles. It is proposed that organic carbon reserves (energy reserve hypothesis), light (light-dependent continual-energy-harvesting hypothesis), and trace gases (air-dependent continual-energy-harvesting hypothesis) are the major energy sources that allow dormant microorganisms to persist during prolonged desiccation. Abbreviations: CODH, carbon monoxide dehydrogenase; Hyd, group 1h [NiFe] hydrogenase; pMMO, particulate methane monooxygenase; PS, photosystem of aerobic anoxygenic phototroph; Rho, microbial rhodopsin; and Cox, terminal oxidase.
Chemolithoautotrophs also benefit from trophic interactions with phototrophs and heterotrophs. Diazotrophic Cyanobacteria fix nitrogen to ammonia, a portion of which is excreted or leaked from cells (6, 75, 76). This supply of fixed nitrogen supports nitrifying microorganisms, such as Thaumarchaeota, which are typically the dominant archaea in desert soils (77–79). In addition, the anaerobic conditions resulting from rapid respiration by heterotrophs after a wetting event can create microenvironments for chemolithoautotrophic anaerobes to flourish (80, 81). This is exemplified by the detection of methanogens in arid soils containing biocrusts, albeit in low abundance (82). Methanogenesis can also be activated by wetting of arid soils, whereby acetoclastic/hydrogenotrophic Methanosarcina and hydrogenotrophic Methanocella organisms consume fermentative end products produced by heterotrophs as substrates for methane production (78, 83, 84). Chemolithoautotrophs are also known to accumulate energy reserves when under stressed conditions (85–89).
While the energy reserve hypothesis provides a feasible mechanism for maintaining microbial cell energy requirements in desert soils, it is not without limitations. Some deserts may receive insufficient water input over long periods to support this mechanism. Hyper-arid regions in the central Atacama Desert can receive less than 5 mm of rainfall per year (90) and may experience decades without precipitation (6, 13). Likewise, annual precipitation of less than 10 mm in certain areas of the Antarctic McMurdo Dry Valleys is common (91), and a significant fraction of this moisture sublimates (5). The combined effects of other environmental factors may reduce the capacity to generate sufficient energy during one short “water pulse.” For instance, salt accumulated in hyper-arid soils reduces water bioavailability (92–94), the low mean temperatures in Antarctic soils reduce cellular metabolism (95), and the highly limited organic carbon and bioavailable nitrogen in hyper-arid soils may restrict heterotrophic processes (5). Overall, the ability of xerotolerant microorganisms inhabiting desert soils to accumulate and utilize long-term energy storage compounds requires more extensive study, especially in situ.
CONTINUAL-ENERGY-HARVESTING HYPOTHESIS
It is increasingly realized that heterotrophic microorganisms in desert environments possess hidden metabolic flexibility. As elaborated below, they may meet energy demands during starvation by continually harvesting atmospheric trace gases (lithoheterotrophy) or sunlight (photoheterotrophy) as alternative energy sources. These mechanisms are likely to be particularly important in the bare soils of deserts, which are typically dominated by heterotrophic bacterial taxa (Fig. 1), with relatively low numbers of photoautotrophs, such as Cyanobacteria (13, 47, 96).
Light-dependent energy harvesting (photoheterotrophy).
Oxygenic photosynthesis is limited by the availability of its electron donor: water. Water limitation, together with damage of photosystems due to desiccation-induced reactive oxygen species (42) and salt stress (97), is thought to primarily limit the abundance of Cyanobacteria in hyper-arid soils. However, it is possible for heterotrophs to derive energy from light by using photons to generate a membrane potential independently of the photosynthetic dark reactions. Two variants of such a light-harvesting mechanism that are dependent on bacteriochlorophyll or rhodopsin (Rho) have been identified (98) (Fig. 2). These processes have sometimes been referred to as “aerobic anoxygenic phototrophy.”
Bacteriochlorophyll (BChl)-dependent light harvesting has been observed in four bacterial phyla: Proteobacteria (99), Chloroflexi (100), Acidobacteria (101), and Gemmatimonadetes (102). Unlike Cyanobacteria, these bacteria contain only one photosystem, with bacteriochlorophyll a being the major photosynthetic pigment (103). The electron transport chain in BChl-dependent light harvesting can operate in a cyclic fashion without exogenous electron donors (103), and therefore, energy can be generated continually with solar input. While BChl-dependent light harvesting is a widespread mechanism for harvesting supplemental energy, particularly in bacteria inhabiting oligotrophic aquatic environments (104, 105), its ecological role in arid soils has not been explored. A culture-dependent study confirmed the presence of soil crust bacterial strains capable of BChl-dependent light harvesting (106). BChl-dependent light harvesting may also be important in hyper-arid Antarctic desert soils. Amplicon sequence screening for genetic determinants of BChl-dependent light harvesting in oligotrophic soils from the Sør Rondane Mountains identified diverse bacteria with this capacity, which were affiliated primarily with the class Alphaproteobacteria (107, 108). A subsequent isolation campaign recovered nearly 1,000 isolates, many affiliated with known Alphaproteobacteria, harboring BChl-dependent light-harvesting capacity (22). Likewise, a Hymenobacter strain (phylum Bacteriodetes) was found to have BChl-dependent light-harvesting potential, a trait not previously observed in members from this phylum (22).
Rhodopsin-based light harvesting (Rho-light harvesting) is a minimalistic light energy-harvesting mechanism consisting of a single ion-pumping protein (type I opsin) with a retinal chromophore cofactor (109). This process generates an ion-motive force for ATP synthesis (but not reducing power), potentially providing a survival advantage for microorganisms during nutrient deprivation (110–112). The minimal genetic determinants of this process, namely, a single opsin gene and another gene for retinal synthesis from carotenoid (109), facilitate horizontal gene transfer (113, 114); this has likely enabled the dissemination and diversification of microbial rhodopsins across archaea and bacteria. A global metagenomic survey focused on marine environments estimated that rhodopsin genes are carried by half of prokaryotic taxa and are 3-fold-more abundant than genes for photochemical reaction centers (115). Despite the possible importance of this physiology as a survival strategy in dry oligotrophic habitats, the ecological relevance of Rho-light harvesting in arid soils has received little attention. Analyses of soil crust metagenomes by Finkel et al. indicated that up to half of microbial genomes encode rhodopsins (115). Several more recent studies have focused on Antarctic deserts: while PCR amplification failed to detect rhodopsin genes in soils from the Sør Rondane Mountains (107, 108), a metagenomic study of hypolithic communities from the Miers Valley (McMurdo Dry Valley region) indicated that 20% of bacterial taxa harbored rhodopsin genes (116).
Atmospheric trace gas oxidation (lithoheterotrophy).
While light-dependent energy harvesting strategies are clearly important physiological processes in desert soil habitats (5, 6, 11, 46), such processes are always constrained by light penetration. Atmospheric trace gases may provide a viable alternative energy source for desert soil microorganisms residing within and below the photic zone. Trace gases, such as hydrogen (H2), carbon monoxide (CO), and methane (CH4), are ubiquitous, diffusive, and high-energy electron donors. The porosity of dry desert soils, due to their coarse texture and low water retention, may also facilitate trace gas permeation (117). The possibility that these substrates support respiration in desert soil microbial communities should therefore be considered (Fig. 2).
Dihydrogen, as the most fundamental molecule, can serve as an energy source for microorganisms from a wide range of taxa and ecosystems (118, 119). Soil microorganisms scavenge H2, which is present at atmospheric mixing ratios of 530 ppbv (120), as an electron donor for aerobic respiration (121). While this process was inferred some four decades ago, the organisms and enzymes responsible for this process have only recently been characterized (122, 123). Genetic and biochemical studies have shown that this process is catalyzed by the group 1h [NiFe]-hydrogenases linked to the respiratory chain; synthesis of this enzyme is induced during nutrient starvation and is critical for long-term survival (124–128). It is now established that atmospheric H2 oxidation is a broadly distributed trait among major soil microbial phyla, having been experimentally verified in Actinobacteria (126, 128–130), Acidobacteria (125, 131), and Chloroflexi (124). The genetic determinants of this activity were found to be carried by at least five additional cultured microbial phyla (123) and two candidate phyla (132).
Carbon monoxide, present at ∼90 ppbv in the atmosphere (133), is also aerobically respired by soil microbial communities. Physiological studies have shown that the enzyme responsible for this process, carbon monoxide dehydrogenase (134), is also induced during carbon limitation and enhances survival during starvation (124, 135–138). At least four microbial phyla can scavenge atmospheric CO (135), namely, Actinobacteria (135, 139), Proteobacteria (140, 141), Chloroflexi (124, 142), and Euryarchaeota (143, 144). Moreover, a recent genomic survey identified putative CO dehydrogenase genes in 16 microbial phyla, encompassing most of the dominant taxa detected in soils (135).
Increasing evidence suggests that oxidation of atmospheric H2 and CO is a feasible continuous-energy-harvesting strategy for microorganisms living in desert ecosystems. Indeed, the genetic determinants for these reactions are consistently detected in desert surveys. Analysis of metagenomic and metatranscriptomic sequence data from the Colorado Desert and Tarim Basin revealed that the genes encoding these enzymes are both abundant and expressed by the soil microbial communities (135). Metagenome-assembled genomes of bacteria with trace gas oxidation potentials, including Pseudonocardia from the high-elevation Atacama Desert (145) and “Candidatus Dormibacteraeota,” “Candidatus Eremiobacteraeota,” Actinobacteria, Chloroflexi, and Verrucomicrobia from Robinson Ridge, Antarctica (132), have been recovered from hyper-arid mineral soils. Experimentally, the rapid consumption of H2 and CO to subatmospheric concentrations was demonstrated in microcosm experiments using Antarctic soils (132). In addition, calculations of theoretical energy yield from trace gas oxidation suggest that this process is sufficient to support the maintenance energy requirement of soil microbial communities (132, 146). Trace gas oxidation may explain why Actinobacteria is the dominant bacterial phylum in desert soils. The relative abundance of this phylum increases with aridity (147, 148), and it typically accounts for 30 to 80% of the microbial community in hyper-arid sites (7, 13, 132, 145, 147, 149) (Fig. 1). Concomitantly, genome-mining data suggest that group 1h [NiFe] hydrogenase genes (123) and carbon monoxide dehydrogenase genes (135) are universally distributed within this phylum. Importantly, CO and H2 oxidation can remain active at low water potentials, with the slow uptake of atmospheric CO detectable at water potentials between −41 MPa and −117 MPa (143, 150), comparable to values in hyper-arid desert soils.
Methane, present at 1.9 ppmv, is the most abundant reduced gas in the troposphere (151). Unlike atmospheric H2 and CO oxidation, it is likely that atmospheric CH4 oxidation has a limited role in supporting microbial persistence in desert environments. This compound is oxidized to methanol by particulate and soluble methane monooxygenases, which is further oxidized for energy production or carbon assimilation (152). To date, taxa with the ability to oxidize CH4 at atmospheric concentrations are exclusively found in specific lineages of the Alphaproteobacteria and Gammaproteobacteria (153). In desert soil ecosystems, atmospheric CH4 oxidation has been reported (154–157). However, the observation of detectable methane oxidation and methane monooxygenase genes in different samples is highly sporadic, especially in more arid soils (145, 155, 156, 158). Moreover, the activity of CH4 oxidation and the abundance of methanotrophs appear to decline dramatically at low water content (117, 150, 159). However, it is known that some methylotrophs are in high relative abundance in some desert soils; an example is Methylobacterium radiotolerans, which dominates the microbial communities at depths below 5 meters in the Playa of the Atacama Desert soils (94).
CONCLUSIONS
Recent advances in “omics” techniques, in combination with pure culture studies and sensitive biogeochemical measurements, have enabled a rapid expansion of knowledge of the diversity and function of organisms living in water-scarce environments. It is now acknowledged that surprisingly diverse microbial communities survive in even the most arid and oligotrophic soils, such as the Antarctic cold deserts and the Atacama Desert. In the absence of macrophytic phototrophs, these microorganisms are the predominant contributors to primary productivity and biogeochemical activities. However, our understanding of how these organisms survive during long periods of water deficiency and how biodiversity in arid soil environments is maintained and shaped remains incomplete. Here, we have presented two strategies for microbial survival in arid ecosystems that can sustain dormancy: the energy reserve hypothesis and the continual-energy-harvesting hypothesis. The strategies are certainly not mutually exclusive, but their degree of relative importance is likely to vary according to the severity of different environmental parameters, such as light availability, oligotrophy, and water availability (Fig. 2). A deeper understanding of these mechanisms is likely to contribute substantially to our capacity to predict how ecosystems, as well as the services that they provide, are affected by the projected global desertification.
ACKNOWLEDGMENTS
This work was supported by an ARC DECRA fellowship (DE170100310; awarded to C.G.), a Swiss National Science Foundation Early Postdoc Mobility fellowship (P2EZP3_178421; awarded to E.C.), an Australian Government Research training stipend (awarded to P.M.L.), Monash University Ph.D. scholarships (awarded to P.M.L. and S.K.B.), and a European Research Council (ERC) starting grant funded by the ERC under the European Union’s Horizon 2020 research and innovation program (636928; awarded to D.W.).
Biographies

Pok Man Leung graduated from the International Research Enrichment Program at the Hong Kong University of Science and Technology, where he received intensive research training on environmental microbiology. He then worked as a research intern at the Scripps Institution of Oceanography, investigating extremophiles from the subseafloor environment. Fascinated by the largely untapped potentials and capabilities of microorganisms, he started his Ph.D. candidature in this area under the supervision of Associate Professor Chris Greening at Monash University in 2018. His current research focuses on the energetic mechanisms that support microbial growth and persistence in different ecosystems using both culture-dependent and culture-independent approaches.

Sean K. Bay completed his Honours in Evolutionary Biology degree at the University of Exeter before spending time working as an environmental field officer down under. He thereafter completed a Masters in Environmental Monitoring, Modelling, and Management at King’s College London, learning computer- and field-based techniques to query environments undergoing change. He is currently a final-year Ph.D. student at Monash University investigating the structure and basis of soil microbial biodiversity under the primary supervision of Associate Professor Chris Greening. Over the past 3 years Sean has used molecular, biogeochemical, and in situ approaches to understand how microbial communities are structured and how they remain energized in aerated surface soils.

Dimitri V. Meier completed Bachelors and Masters degrees at Georg-August University of Göttingen, Göttingen, Germany. He subsequently completed a Ph.D. on microbial communities of hydrothermal vents at the Max Planck Institute for Marine Microbiology, Bremen, Germany, in 2016. In 2017, he moved to the University of Vienna, Vienna, Austria, to work on microbial ecology and survival in desert soil crusts and hypersaline microbial mats. He is fascinated by microbial survival in seemingly extreme conditions and aims to gain a holistic understanding of these ecosystems. In his current work, he uses largely culture-independent methods, such as metagenomics, metatranscriptomics, and microscopy, to obtain a comprehensive picture of microbial lifestyles and strategies that enable survival at very low water availability while maintaining ecosystem functions.

Eleonora Chiri attained her Ph.D. in Science at the Swiss Federal Institute of Technology ETH Zurich in 2016, investigating the ecological role of methane-oxidizing bacteria in natural and anthropogenic soil chronosequences. Since then, she has extended her field-based studies of microbial methane and hydrogen oxidation to Australian termite mound and savanna soil ecosystems during her postdoctoral experience at Charles Darwin University, the University of Melbourne, and, most recently, Monash University. As a Swiss National Science Foundation Fellow, her current research focuses primarily on understanding the ecological role of microbial atmospheric trace gas oxidation by investigating the establishment and development of these biogeochemical processes in oligotrophic and extreme soil ecosystems. Uncovering the identity, activity, and ecosystem function of soil microorganisms that “live from air” represent the core goals of her scientific career.

Don A. Cowan was educated (B.Sc., M.Sc., Ph.D.) at the University of Waikato (Hamilton, New Zealand) and completed a 4-year period of postdoctoral research under the supervision of Professor Roy Daniel before moving to a Lectureship at University College London (UK) in 1985. After 16 years in London, he was appointed to the Chair of Microbiology and Head of the Department of Biotechnology at the University of the Western Cape (Cape Town, Republic of South Africa), where he established the Institute for Microbial Ecology and Metagenomics. In 2012, he moved to the University of Pretoria, where he is a professor in the Department of Biochemistry, Genetics, and Microbiology, and is currently the director of both the Genomics Research Institute and the Centre for Microbial Ecology and Genomics. Much of Don’s research focuses on the diversity and function of microbial communities in extreme environments, particularly hot (Namib) and cold (Antarctic) desert soils.

Osnat Gillor attained her Ph.D. in 2002 from the Department of Environmental Sciences at the Hebrew University of Jerusalem. She investigated the causes of harmful cyanobacterial blooms in freshwater and seawater systems. During her postdoc, she explored toxin-mediated bacterial interactions in the lab and murine gastrointestinal tract at Yale University and the University of Massachusetts, Amherst, MA. In 2006, she joined the faculty of the Zuckerberg Institute for Water Research at Ben Gurion University of the Negev, where she studies microbial interactions in water and soil systems. She has a broad set of research interests, ranging from the role of antimicrobials in biofilm formation to the diversity, composition, and function of microbial communities in arid soils, dust, and rocks. What unites these disparate topics is the study of processes and patterns that control microbial interactions from the most complex habitat, the soil, to a simplified laboratory model system.

Dagmar Woebken studied biology at the Leibniz University, Hannover, Germany, and conducted her Ph.D. research at the Max Planck Institute for Marine Microbiology in Bremen, Germany (2007). After her postdoctoral work at Stanford University in collaboration with the NASA Ames Research Center and the Lawrence Livermore National Laboratory in California, she relocated to Austria in 2012 for a group leader position at the University of Vienna. She currently holds an assistant professorship and is a member of the Young Academy of the Austrian Academy of Sciences. Dagmar’s group explores the function of microorganisms in soils, which are among the most diverse microbial communities on Earth. In her ERC-funded project, she has focused on the survival strategies of soil microorganisms. Dagmar is particularly fascinated by the physiologies that allow the survival of microorganisms under unfavorable conditions, as it is a key factor in maintaining the high diversity in soil.

Chris Greening studied molecular and cellular biochemistry at the Bachelor and Master levels (University of Oxford, 2010). For his doctoral degree in molecular microbiology (University of Otago, 2014), he investigated the physiological role of mycobacterial hydrogenases under the mentorship of Professor Gregory Cook. He subsequently gained postdoctoral experience at the University of Otago, CSIRO, and the Australian National University before setting up his group at Monash University in 2016. Chris’ team is dedicated to understanding the metabolic mechanisms that allow environmental and pathogenic bacteria to adapt to resource limitation. This has led to a range of key findings, for example, that atmospheric trace gases serve as alternative energy sources for bacteria in nutrient-starved environments, including desert ecosystems. He uses his diverse experiences to connect findings at the molecular, cellular, and ecosystem scales.
This minireview went through the journal's normal peer review process. DayTwo sponsored the minireview and its associated video but had no editorial input on the content.
REFERENCES
- 1.Cherlet M, Hutchinson C, Reynolds J, Hill J, Sommer S, von Maltitz G (ed). 2018. World atlas of desertification, 3rd ed Publication Office of the European Union, Luxembourg. [Google Scholar]
- 2.Huang JP, Yu HP, Guan XD, Wang GY, Guo RX. 2016. Accelerated dryland expansion under climate change. Nat Clim Chang 6:166–171. doi: 10.1038/nclimate2837. [DOI] [Google Scholar]
- 3.Wang G, Or D. 2013. Hydration dynamics promote bacterial coexistence on rough surfaces. ISME J 7:395–404. doi: 10.1038/ismej.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schimel JP. 2018. Life in dry soils: effects of drought on soil microbial communities and processes. Annu Rev Ecol Evol Syst 49:409–432. doi: 10.1146/annurev-ecolsys-110617-062614. [DOI] [Google Scholar]
- 5.Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol 8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 6.Pointing SB, Belnap J. 2012. Microbial colonization and controls in dryland systems. Nat Rev Microbiol 10:551–562. doi: 10.1038/nrmicro2831. [DOI] [PubMed] [Google Scholar]
- 7.Makhalanyane TP, Valverde A, Gunnigle E, Frossard A, Ramond JB, Cowan DA. 2015. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev 39:203–221. doi: 10.1093/femsre/fuu011. [DOI] [PubMed] [Google Scholar]
- 8.Brock TD. 1975. Effect of water potential on a Microcoleus (Cyanophyceae) from a desert crust. J Phycol 11:316–320. doi: 10.1111/j.1529-8817.1975.tb02786.x. [DOI] [Google Scholar]
- 9.Tardieu F, Draye X, Javaux M. 2017. Root water uptake and ideotypes of the root system: whole-plant controls matter. Vadose Zone J 16. doi: 10.2136/vzj2017.05.0107. [DOI] [Google Scholar]
- 10.Rajeev L, da Rocha UN, Klitgord N, Luning EG, Fortney J, Axen SD, Shih PM, Bouskill NJ, Bowen BP, Kerfeld CA, Garcia-Pichel F, Brodie EL, Northen TR, Mukhopadhyay A. 2013. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J 7:2178–2191. doi: 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker JJ, Pace NR. 2007. Endolithic microbial ecosystems. Annu Rev Microbiol 61:331–347. doi: 10.1146/annurev.micro.61.080706.093302. [DOI] [PubMed] [Google Scholar]
- 12.Wierzchos J, de los Ríos A, Ascaso C. 2012. Microorganisms in desert rocks: the edge of life on Earth. Int Microbiol 15:173–183. doi: 10.2436/20.1501.01.170. [DOI] [PubMed] [Google Scholar]
- 13.Schulze-Makuch D, Wagner D, Kounaves SP, Mangelsdorf K, Devine KG, de Vera J-P, Schmitt-Kopplin P, Grossart H-P, Parro V, Kaupenjohann M, Galy A, Schneider B, Airo A, Frösler J, Davila AF, Arens FL, Cáceres L, Cornejo FS, Carrizo D, Dartnell L, DiRuggiero J, Flury M, Ganzert L, Gessner MO, Grathwohl P, Guan L, Heinz J, Hess M, Keppler F, Maus D, McKay CP, Meckenstock RU, Montgomery W, Oberlin EA, Probst AJ, Sáenz JS, Sattler T, Schirmack J, Sephton MA, Schloter M, Uhl J, Valenzuela B, Vestergaard G, Wörmer L, Zamorano P. 2018. Transitory microbial habitat in the hyperarid Atacama Desert. Proc Natl Acad Sci U S A 115:2670–2675. doi: 10.1073/pnas.1714341115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambrechts S, Willems A, Tahon G. 2019. Uncovering the uncultivated majority in Antarctic soils: toward a synergistic approach. Front Microbiol 10:242. doi: 10.3389/fmicb.2019.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 16.Angel R, Conrad R. 2013. Elucidating the microbial resuscitation cascade in biological soil crusts following a simulated rain event. Env Microbiol 15:2799–2815. doi: 10.1111/1462-2920.12140. [DOI] [PubMed] [Google Scholar]
- 17.Galperin MY. 2013. Genome diversity of spore-forming firmicutes. Microbiol Spectr 1. doi: 10.1128/microbiolspectrum.TBS-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensign JC. 1978. Formation, properties, and germination of actinomycete spores. Annu Rev Microbiol 32:185–219. doi: 10.1146/annurev.mi.32.100178.001153. [DOI] [PubMed] [Google Scholar]
- 19.Nunes da Rocha U, Cadillo-Quiroz H, Karaoz U, Rajeev L, Klitgord N, Dunn S, Truong V, Buenrostro M, Bowen BP, Garcia-Pichel F, Mukhopadhyay A, Northen TR, Brodie EL. 2015. Isolation of a significant fraction of non-phototroph diversity from a desert biological soil crust. Front Microbiol 6:277. doi: 10.3389/fmicb.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maza F, Maldonado J, Vásquez-Dean J, Mandakovic D, Gaete A, Cambiazo V, González M. 2019. Soil bacterial communities from the Chilean Andean highlands: taxonomic composition and culturability. Front Bioeng Biotechnol 7:10. doi: 10.3389/fbioe.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fatima A, Aftab U, Shaaban KA, Thorson JS, Sajid I. 2019. Spore forming actinobacterial diversity of Cholistan Desert Pakistan: polyphasic taxonomy, antimicrobial potential and chemical profiling. BMC Microbiol 19:49. doi: 10.1186/s12866-019-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tahon G, Willems A. 2017. Isolation and characterization of aerobic anoxygenic phototrophs from exposed soils from the Sor Rondane Mountains, East Antarctica. Syst Appl Microbiol 40:357–369. doi: 10.1016/j.syapm.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Temraleeva AD. 2018. Cyanobacterial diversity in the soils of Russian dry steppes and semideserts. Microbiology 87:249–260. doi: 10.1134/S0026261718020169. [DOI] [Google Scholar]
- 24.Rehakova K, Johansen JR, Casamatta DA, Xueson L, Vincent J. 2007. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia 46:481–502. doi: 10.2216/06-92.1. [DOI] [Google Scholar]
- 25.Kaplan-Levy RN, Hadas O, Summers ML, Rücker J, Sukenik A. 2010. Akinetes: dormant cells of Cyanobacteria, p 5–27. In Lubzens E, Cerda J, Clark M (ed), Dormancy and resistance in harsh environments. Springer, Berlin, Germany. [Google Scholar]
- 26.Fuller WH, Hanks K. 1982. Distribution of Azotobacter in arid soils. Plant Soil 64:355–361. [Google Scholar]
- 27.De Luca G, Barakat M, Ortet P, Fochesato S, Jourlin-Castelli C, Ansaldi M, Py B, Fichant G, Coutinho PM, Voulhoux R, Bastien O, Maréchal E, Henrissat B, Quentin Y, Noirot P, Filloux A, Méjean V, DuBow MS, Barras F, Barbe V, Weissenbach J, Mihalcescu I, Verméglio A, Achouak W, Heulin T. 2011. The cyst-dividing bacterium Ramlibacter tataouinensis TTB310 genome reveals a well-stocked toolbox for adaptation to a desert environment. PLoS One 6:e23784. doi: 10.1371/journal.pone.0023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cacciari I, Lippi D. 1987. Arthrobacters: successful arid soil bacteria: a review. Arid Soil Res Rehab 1:1–30. doi: 10.1080/15324988709381125. [DOI] [Google Scholar]
- 29.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43(Spec No):93–100. [PubMed] [Google Scholar]
- 30.Lebre PH, De Maayer P, Cowan DA. 2017. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol 15:285–296. doi: 10.1038/nrmicro.2017.16. [DOI] [PubMed] [Google Scholar]
- 31.Slade D, Radman M. 2011. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattimore V, Battista JR. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol 178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569–573. doi: 10.1038/nature05160. [DOI] [PubMed] [Google Scholar]
- 34.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, Lee DY, Wehr NB, Viteri GA, Berlett BS, Levine RL. 2010. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, Wackett LP, Fredrickson JK, Ghosal D. 2004. Accumulation of Mn(II) in, Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 36.Egas C, Barroso C, Froufe HJC, Pacheco J, Albuquerque L, da Costa MS. 2014. Complete genome sequence of the radiation-resistant bacterium Rubrobacter radiotolerans RSPS-4. Stand Genomic Sci 9:1062–1075. doi: 10.4056/sigs.5661021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes AJ, Bowyer J, Holley MP, O'Donoghue M, Montgomery M, Gillings MR. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol Ecol 33:111–120. doi: 10.1111/j.1574-6941.2000.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 38.Crits-Christoph A, Robinson CK, Barnum T, Fricke WF, Davila AF, Jedynak B, McKay CP, DiRuggiero J. 2013. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 1:28. doi: 10.1186/2049-2618-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price PB, Sowers T. 2004. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci U S A 101:4631–4636. doi: 10.1073/pnas.0400522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boylen CW. 1973. Survival of Arthrobacter crystallopoietes during prolonged periods of extreme desiccation. J Bacteriol 113:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol Rev 58:755–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billi D, Potts M. 2002. Life and death of dried prokaryotes. Res Microbiol 153:7–12. doi: 10.1016/s0923-2508(01)01279-7. [DOI] [PubMed] [Google Scholar]
- 43.Georgiou CD, Sun HJ, McKay CP, Grintzalis K, Papapostolou I, Zisimopoulos D, Panagiotidis K, Zhang G, Koutsopoulou E, Christidis GE, Margiolaki I. 2015. Evidence for photochemical production of reactive oxygen species in desert soils. Nat Commun 6:7100. doi: 10.1038/ncomms8100. [DOI] [PubMed] [Google Scholar]
- 44.Hoehler TM, Jorgensen BB. 2013. Microbial life under extreme energy limitation. Nat Rev Microbiol 11:83–94. doi: 10.1038/nrmicro2939. [DOI] [PubMed] [Google Scholar]
- 45.Noy-Meir I. 1973. Desert ecosystems: environment and producers. Annu Rev Ecol Syst 4:25–51. doi: 10.1146/annurev.es.04.110173.000325. [DOI] [Google Scholar]
- 46.Belnap J, Lange OL. 2001. Biological soil crusts: structure, function, and management. Springer, Berlin, Germany. [Google Scholar]
- 47.Lee KC, Archer SDJ, Boyle RH, Lacap-Bugler DC, Belnap J, Pointing SB. 2016. Niche filtering of bacteria in soil and rock habitats of the Colorado Plateau Desert, Utah, USA. Front Microbiol 7:1489. doi: 10.3389/fmicb.2016.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbert W, Weber B, Burrows S, Steinkamp J, Budel B, Andreae MO, Poschl U. 2012. Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nature Geosci 5:459–462. doi: 10.1038/ngeo1486. [DOI] [Google Scholar]
- 49.Rao BQ, Liu YD, Wang WB, Hu CX, Dunhai L, Lan SB. 2009. Influence of dew on biomass and photosystem II activity of cyanobacterial crusts in the Hopq Desert, northwest China. Soil Biol Biochem 41:2387–2393. doi: 10.1016/j.soilbio.2009.06.005. [DOI] [Google Scholar]
- 50.Li BN, Wang LX, Kaseke KF, Vogt R, Li L, Seely MK. 2018. The impact of fog on soil moisture dynamics in the Namib Desert. Adv Water Resour 113:23–29. doi: 10.1016/j.advwatres.2018.01.004. [DOI] [Google Scholar]
- 51.Raanan H, Oren N, Treves H, Berkowicz SM, Hagemann M, Pade N, Keren N, Kaplan A. 2016. Simulated soil crust conditions in a chamber system provide new insights on cyanobacterial acclimation to desiccation. Environ Microbiol 18:414–426. doi: 10.1111/1462-2920.12998. [DOI] [PubMed] [Google Scholar]
- 52.Allen MM. 1984. Cyanobacterial cell inclusions. Annu Rev Microbiol 38:1–25. doi: 10.1146/annurev.mi.38.100184.000245. [DOI] [PubMed] [Google Scholar]
- 53.Page-Sharp M, Behm CA, Smith GD. 1998. Cyanophycin and glycogen synthesis in a cyanobacterial Scytonema species in response to salt stress. FEMS Microbiol Lett 160:11–15. doi: 10.1111/j.1574-6968.1998.tb12883.x. [DOI] [Google Scholar]
- 54.Appel T. 1998. Non-biomass soil organic N—the substrate for N mineralization flushes following soil drying-rewetting and for organic N rendered CaCl2-extractable upon soil drying. Soil Biol Biochem 30:1445–1456. doi: 10.1016/S0038-0717(97)00230-7. [DOI] [Google Scholar]
- 55.Vangestel M, Merckx R, Vlassak K. 1993. Microbial biomass responses to soil drying and rewetting—the fate of fast-growing and slow-growing microorganisms in soils from different climates. Soil Biol Biochem 25:109–123. doi: 10.1016/0038-0717(93)90249-B. [DOI] [Google Scholar]
- 56.Blazewicz SJ, Schwartz E, Firestone MK. 2014. Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil. Ecology 95:1162–1172. doi: 10.1890/13-1031.1. [DOI] [PubMed] [Google Scholar]
- 57.Halverson LJ, Jones TM, Firestone MK. 2000. Release of intracellular solutes by four soil bacteria exposed to dilution stress. Soil Sci Soc Am J 64:1630–1637. doi: 10.2136/sssaj2000.6451630x. [DOI] [Google Scholar]
- 58.Warren CR. 2014. Response of osmolytes in soil to drying and rewetting. Soil Biol Biochem 70:22–32. doi: 10.1016/j.soilbio.2013.12.008. [DOI] [Google Scholar]
- 59.Williams MA, Xia K. 2009. Characterization of the water soluble soil organic pool following the rewetting of dry soil in a drought-prone tallgrass prairie. Soil Biol Biochem 41:21–28. doi: 10.1016/j.soilbio.2008.08.013. [DOI] [Google Scholar]
- 60.Kakumanu ML, Cantrell CL, Williams MA. 2013. Microbial community response to varying magnitudes of desiccation in soil: a test of the osmolyte accumulation hypothesis. Soil Biol Biochem 57:644–653. doi: 10.1016/j.soilbio.2012.08.014. [DOI] [Google Scholar]
- 61.Boot CM, Schaeffer SM, Schimel JP. 2013. Static osmolyte concentrations in microbial biomass during seasonal drought in a California grassland. Soil Biol Biochem 57:356–361. doi: 10.1016/j.soilbio.2012.09.005. [DOI] [Google Scholar]
- 62.Kieft TL, Soroker E, Firestone MK. 1987. Microbial biomass response to a rapid increase in water potential when dry soil is wetted. Soil Biol Biochem 19:119–126. doi: 10.1016/0038-0717(87)90070-8. [DOI] [Google Scholar]
- 63.Sponseller RA. 2007. Precipitation pulses and soil CO2 flux in a Sonoran Desert ecosystem. Global Change Biol 13:426–436. doi: 10.1111/j.1365-2486.2006.01307.x. [DOI] [Google Scholar]
- 64.Birch HF, Friend MT. 1956. Humus decomposition in East African soils. Nature 178:500–501. doi: 10.1038/178500a0. [DOI] [Google Scholar]
- 65.Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, Da Rocha UN, Chakraborty R, Bowen BP, Karaoz U, Cadillo-Quiroz H, Garcia-Pichel F, Northen TR. 2015. Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun 6:8289. doi: 10.1038/ncomms9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swenson TL, Karaoz U, Swenson JM, Bowen BP, Northen TR. 2018. Linking soil biology and chemistry in biological soil crust using isolate exometabolomics. Nat Commun 9:19. doi: 10.1038/s41467-017-02356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazor G, Kidron GJ, Vonshak A, Abeliovich A. 1996. The role of cyanobacterial exopolysaccharides in structuring desert microbial crusts. FEMS Microbiol Ecol 21:121–130. doi: 10.1111/j.1574-6941.1996.tb00339.x. [DOI] [Google Scholar]
- 68.Hokputsa S, Hu CX, Paulsen BS, Harding SE. 2003. A physico-chemical comparative study on extracellular carbohydrate polymers from five desert algae. Carbohydr Polym 54:27–32. doi: 10.1016/S0144-8617(03)00136-X. [DOI] [Google Scholar]
- 69.Rossi F, De Philippis R. 2015. Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats. Life (Basel) 5:1218–1238. doi: 10.3390/life5021218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lapebie P, Lombard V, Drula E, Terrapon N, Henrissat B. 2019. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat Commun 10:2043. doi: 10.1038/s41467-019-10068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang SX, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 72.Cytryn EJ, Sangurdekar DP, Streeter JG, Franck WL, Chang WS, Stacey G, Emerich DW, Joshi T, Xu D, Sadowsky MJ. 2007. Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress. J Bacteriol 189:6751–6762. doi: 10.1128/JB.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Finkelstein DB, Brassell SC, Pratt LM. 2010. Microbial biosynthesis of wax esters during desiccation: adaptation for colonization of the earliest terrestrial environments? Geology 38:247–250. doi: 10.1130/G30398.1. [DOI] [Google Scholar]
- 74.Hauschild P, Röttig A, Madkour MH, Al-Ansari AM, Almakishah NH, Steinbüchel A. 2017. Lipid accumulation in prokaryotic microorganisms from arid habitats. Appl Microbiol Biotechnol 101:2203–2216. doi: 10.1007/s00253-017-8149-0. [DOI] [PubMed] [Google Scholar]
- 75.Mayland HF, Mcintosh TH. 1966. Availability of biologically fixed atmospheric nitrogen-15 to higher plants. Nature 209:421–422. doi: 10.1038/209421a0. [DOI] [Google Scholar]
- 76.Belnap J, Harper KT. 1995. Influence of cryptobiotic soil crusts on elemental content of tissue of 2 desert seed plants. Arid Soil Res Rehab 9:107–115. doi: 10.1080/15324989509385879. [DOI] [Google Scholar]
- 77.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aschenbach K, Conrad R, Rehakova K, Dolezal J, Janatkova K, Angel R. 2013. Methanogens at the top of the world: occurrence and potential activity of methanogens in newly deglaciated soils in high-altitude cold deserts in the Western Himalayas. Front Microbiol 4:359. doi: 10.3389/fmicb.2013.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Magalhaes CM, Machado A, Frank-Fahle B, Lee CK, Cary SC. 2014. The ecological dichotomy of ammonia-oxidizing archaea and bacteria in the hyper-arid soils of the Antarctic Dry Valleys. Front Microbiol 5:515. doi: 10.3389/fmicb.2014.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.GarciaPichel F, Belnap J. 1996. Microenvironments and microscale productivity of cyanobacterial desert crusts. J Phycol 32:774–782. doi: 10.1111/j.0022-3646.1996.00774.x. [DOI] [Google Scholar]
- 81.Šťovíček A, Kim M, Or D, Gillor O. 2017. Microbial community response to hydration-desiccation cycles in desert soil. Sci Rep 7:45735. doi: 10.1038/srep45735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peters V, Conrad R. 1995. Methanogenic and other strictly anaerobic-bacteria in desert soil and other oxic soils. Appl Environ Microbiol 61:1673–1676. doi: 10.1128/AEM.61.4.1673-1676.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Angel R, Matthies D, Conrad R. 2011. Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS One 6:e20453. doi: 10.1371/journal.pone.0020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Angel R, Claus P, Conrad R. 2012. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J 6:847–862. doi: 10.1038/ismej.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murray PA, Zinder SH. 1987. Polysaccharide reserve material in the acetotrophic methanogen, Methanosarcina-Thermophila strain Tm-1—accumulation and mobilization. Arch Microbiol 147:109–116. doi: 10.1007/BF00415270. [DOI] [Google Scholar]
- 86.Anderson KL, Apolinario EE, Sowers KR. 2012. Desiccation as a long-term survival mechanism for the archaeon Methanosarcina barkeri. Appl Environ Microbiol 78:1473–1479. doi: 10.1128/AEM.06964-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orell A, Navarro CA, Rivero M, Aguilar JS, Jerez CA. 2012. Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 16:573–583. doi: 10.1007/s00792-012-0457-9. [DOI] [PubMed] [Google Scholar]
- 88.Jasso-Chavez R, Santiago-Martinez MG, Lira-Silva E, Pineda E, Zepeda-Rodriguez A, Belmont-Diaz J, Encalada R, Saavedra E, Moreno-Sanchez R. 2015. Air-adapted Methanosarcina acetivorans shows high methane production and develops resistance against oxygen stress. PLoS One 10:0117331. doi: 10.1371/journal.pone.0117331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Liu QH, Wu X, Huang Y, Wise MJ, Liu ZZ, Wang W, Hu JF, Wang CY. 2019. Bioinformatics analysis of metabolism pathways of archaeal energy reserves. Sci Rep 9:1034. doi: 10.1038/s41598-018-37768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McKay CP, Friedmann EI, Gomez-Silva B, Caceres-Villanueva L, Andersen DT, Landheim R. 2003. Temperature and moisture conditions for life in the extreme arid region of the Atacama Desert: four years of observations including the El Nino of 1997–1998. Astrobiology 3:393–406. doi: 10.1089/153110703769016460. [DOI] [PubMed] [Google Scholar]
- 91.Fountain AG, Nylen TH, Monaghan A, Basagic HJ, Bromwich D. 2010. Snow in the McMurdo Dry Valleys, Antarctica. Int J Climatol 30:633–642. [Google Scholar]
- 92.Claridge GCG, Campbell BI. 1977. The salts in Antarctic soils, their distribution and relationship to soil processes. Soil Sci 123:377–384. doi: 10.1097/00010694-197706000-00006. [DOI] [Google Scholar]
- 93.Ewing SA, Sutter B, Owen J, Nishiizumi K, Sharp W, Cliff SS, Perry K, Dietrich W, McKay CP, Amundson R. 2006. A threshold in soil formation at Earth’s arid-hyperarid transition. Geochim Cosmochim Acta 70:5293–5322. doi: 10.1016/j.gca.2006.08.020. [DOI] [Google Scholar]
- 94.Warren-Rhodes KA, Lee KC, Archer SDJ, Cabrol N, Ng-Boyle L, Wettergreen D, Zacny K, Pointing SB, NASA Life in the Atacama Project Team. 2019. Subsurface microbial habitats in an extreme desert Mars-analog environment. Front Microbiol 10:69. doi: 10.3389/fmicb.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrett JE, Virginia RA, Parsons AN, Wall DH. 2006. Soil carbon turnover in the McMurdo Dry Valleys, Antarctica. Soil. Biol Biochem 38:3065–3082. doi: 10.1016/j.soilbio.2006.03.025. [DOI] [Google Scholar]
- 96.Bay S, Ferrari B, Greening C. 2018. Life without water: how do bacteria generate biomass in desert ecosystems? Microbiol Aust 39:28–32. [Google Scholar]
- 97.Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N. 2000. Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056. doi: 10.1104/pp.123.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bryant DA, Frigaard NU. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 99.Yurkov V, Csotonyi JT. 2009. New light on aerobic anoxygenic phototrophs, p 31–55. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), The purple phototrophic bacteria. Springer, Dordrecht, Netherlands. [Google Scholar]
- 100.Gest H, Favinger JL. 1983. Heliobacterium-Chlorum, an anoxygenic brownish-green photosynthetic bacterium containing a new form of bacteriochlorophyll. Arch Microbiol 136:11–16. doi: 10.1007/BF00415602. [DOI] [Google Scholar]
- 101.Bryant DA, Costas AMG, Maresca JA, Chew AGM, Klatt CG, Bateson MM, Tallon LJ, Hostetler J, Nelson WC, Heidelberg JF, Ward DM. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526. doi: 10.1126/science.1143236. [DOI] [PubMed] [Google Scholar]
- 102.Zeng YH, Feng FY, Medova H, Dean J, Koblizek M. 2014. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci U S A 111:7795–7800. doi: 10.1073/pnas.1400295111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yurkov V, Hughes E. 2017. Aerobic anoxygenic phototrophs: four decades of mystery, p 193–214. In Hallenbeck PC. (ed), Modern topics in the phototrophic prokaryotes: environmental and applied aspects. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 104.Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 105.Lami R, Cottrell MT, Ras J, Ulloa O, Obernosterer I, Claustre H, Kirchman DL, Lebaron P. 2007. High abundances of aerobic anoxygenic photosynthetic bacteria in the South Pacific Ocean. Appl Environ Microbiol 73:4198–4205. doi: 10.1128/AEM.02652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Csotonyi JT, Swiderski J, Stackebrandt E, Yurkov V. 2010. A new environment for aerobic anoxygenic phototrophic bacteria: biological soil crusts. Environ Microbiol Rep 2:651–656. doi: 10.1111/j.1758-2229.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- 107.Tahon G, Tytgat B, Stragier P, Willems A. 2016. Analysis of cbbL, nifH, and pufLM in soils from the Sor Rondane Mountains, Antarctica, reveals a large diversity of autotrophic and phototrophic bacteria. Microb Ecol 71:131–149. doi: 10.1007/s00248-015-0704-6. [DOI] [PubMed] [Google Scholar]
- 108.Tahon G, Tytgat B, Willems A. 2016. Diversity of phototrophic genes suggests multiple bacteria may be able to exploit sunlight in exposed soils from the Sor Rondane Mountains, East Antarctica. Front Microbiol 7:2026. doi: 10.3389/fmicb.2016.02026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pinhassi J, DeLong EF, Béjà O, González JM, Pedrós-Alió C. 2016. Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology, and ecology. Microbiol Mol Biol Rev 80:929–954. doi: 10.1128/MMBR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gomez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, Gonzalez JM, Pinhassi J. 2010. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. 2011. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Z, O’Shaughnessy TJ, Soto CM, Rahbar AM, Robertson KL, Lebedev N, Vora GJ. 2012. Function and regulation of Vibrio campbellii proteorhodopsin: acquired phototrophy in a classical organoheterotroph. PLoS One 7:e38749. doi: 10.1371/journal.pone.0038749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frigaard NU, Martinez A, Mincer TJ, DeLong EF. 2006. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 114.McCarren J, DeLong EF. 2007. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ Microbiol 9:846–858. doi: 10.1111/j.1462-2920.2006.01203.x. [DOI] [PubMed] [Google Scholar]
- 115.Finkel OM, Beja O, Belkin S. 2013. Global abundance of microbial rhodopsins. ISME J 7:448–451. doi: 10.1038/ismej.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guerrero LD, Vikram S, Makhalanyane TP, Cowan DA. 2017. Evidence of microbial rhodopsins in Antarctic Dry Valley edaphic systems. Environ Microbiol 19:3755–3767. doi: 10.1111/1462-2920.13877. [DOI] [PubMed] [Google Scholar]
- 117.von Fischer JC, Butters G, Duchateau PC, Thelwell RJ, Siller R. 2009. In situ measures of methanotroph activity in upland soils: a reaction-diffusion model and field observation of water stress. J Geophys Res 114:G01015. [Google Scholar]
- 118.Schlegel HG. 1974. Production, modification, and consumption of atmospheric trace gases by microorganisms. Tellus 26:11–20. doi: 10.3402/tellusa.v26i1-2.9732. [DOI] [Google Scholar]
- 119.Piche-Choquette S, Constant P. 2019. Molecular hydrogen, a neglected key driver of soil biogeochemical processes. Appl Environ Microbiol 85:e02418-18. doi: 10.1128/AEM.02418-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Novelli PC, Lang PM, Masarie KA, Hurst DF, Myers R, Elkins JW. 1999. Molecular hydrogen in the troposphere: global distribution and budget. J Geophys Res 104:30427–30444. doi: 10.1029/1999JD900788. [DOI] [Google Scholar]
- 121.Schmidt U. 1974. Molecular hydrogen in the atmosphere. Tellus 26:78–90. doi: 10.1111/j.2153-3490.1974.tb01954.x. [DOI] [Google Scholar]
- 122.Constant P, Chowdhury SP, Pratscher J, Conrad R. 2010. Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ Microbiol 12:821–829. doi: 10.1111/j.1462-2920.2009.02130.x. [DOI] [PubMed] [Google Scholar]
- 123.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Islam ZF, Cordero PRF, Feng J, Chen YJ, Bay SK, Jirapanjawat T, Gleadow RM, Carere CR, Stott MB, Chiri E, Greening C. 2019. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J 13:1801–1813. doi: 10.1038/s41396-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Greening C, Carere CR, Rushton-Green R, Harold LK, Hards K, Taylor MC, Morales SE, Stott MB, Cook GM. 2015. Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc Natl Acad Sci U S A 112:10497–10502. doi: 10.1073/pnas.1508385112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Greening C, Berney M, Hards K, Cook GM, Conrad R. 2014. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci U S A 111:4257–4261. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liot Q, Constant P. 2016. Breathing air to save energy—new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. Microbiologyopen 5:47–59. doi: 10.1002/mbo3.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meredith LK, Rao D, Bosak T, Klepac-Ceraj V, Tada KR, Hansel CM, Ono S, Prinn RG. 2014. Consumption of atmospheric hydrogen during the life cycle of soil-dwelling actinobacteria. Environ Microbiol Rep 6:226–238. doi: 10.1111/1758-2229.12116. [DOI] [PubMed] [Google Scholar]
- 129.Constant P, Poissant L, Villemur R. 2008. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H-2. ISME J 2:1066–1076. doi: 10.1038/ismej.2008.59. [DOI] [PubMed] [Google Scholar]
- 130.Constant P, Chowdhury SP, Hesse L, Pratscher J, Conrad R. 2011. Genome data mining and soil survey for the novel group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H2-oxidizing bacteria. Appl Environ Microbiol 77:6027–6035. doi: 10.1128/AEM.00673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Myers MR, King GM. 2016. Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat. Int J Syst Evol Microbiol 66:5328–5335. doi: 10.1099/ijsem.0.001516. [DOI] [PubMed] [Google Scholar]
- 132.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, Montgomery K, Lines T, Beardall J, van Dorst J, Snape I, Stott MB, Hugenholtz P, Ferrari BC. 2017. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552:400–403. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 133.Novelli PC, Steele LP, Tans PP. 1992. Mixing ratios of carbon-monoxide in the troposphere. J Geophys Res 97:20731–20750. doi: 10.1029/92JD02010. [DOI] [Google Scholar]
- 134.King GM, Weber CF. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- 135.Cordero PRF, Bayly K, Leung PM, Huang C, Islam ZF, Schittenhelm RB, King GM, Greening C. 2019. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J 13:2868–2881. doi: 10.1038/s41396-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Patrauchan MA, Miyazawa D, LeBlanc JC, Aiga C, Florizone C, Dosanjh M, Davies J, Eltis LD, Mohn WW. 2012. Proteomic analysis of survival of Rhodococcus jostii RHA1 during carbon starvation. Appl Environ Microbiol 78:6714–6725. doi: 10.1128/AEM.01293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Christie-Oleza JA, Fernandez B, Nogales B, Bosch R, Armengaud J. 2012. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J 6:124–135. doi: 10.1038/ismej.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.King GM. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl Environ Microbiol 69:4067–4075. doi: 10.1128/aem.69.7.4067-4075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gadkari D, Schricker K, Acker G, Kroppenstedt RM, Meyer O. 1990. Streptomyces-Thermoautotrophicus sp. nov., a thermophilic CO-oxidizing and H2-oxidizing obligate chemolithoautotroph. Appl Environ Microbiol 56:3727–3734. doi: 10.1128/AEM.56.12.3727-3734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.King GA. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl Environ Microbiol 69:7257–7265. doi: 10.1128/aem.69.12.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weber CF, King GM. 2017. Volcanic soils as sources of novel CO-oxidizing Paraburkholderia and Burkholderia: Paraburkholderia hiiakae sp. nov., Paraburkholderia metrosideri sp. nov., Paraburkholderia paradisi sp. nov., Paraburkholderia peleae sp. nov., and Burkholderia alpina sp. nov., a member of the Burkholderia cepacia complex. Front Microbiol 8:207. doi: 10.3389/fmicb.2017.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.King CE, King GM. 2014. Description of Thermogemmatispora carboxidivorans sp nov., a carbon-monoxide-oxidizing member of the class Ktedonobacteria isolated from a geothermally heated biofilm, and analysis of carbon monoxide oxidation by members of the class Ktedonobacteria. Int J Syst Evol Microbiol 64:1244–1251. doi: 10.1099/ijs.0.059675-0. [DOI] [PubMed] [Google Scholar]
- 143.King GM. 2015. Carbon monoxide as a metabolic energy source for extremely halophilic microbes: implications for microbial activity in Mars regolith. Proc Natl Acad Sci U S A 112:4465–4470. doi: 10.1073/pnas.1424989112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McDuff S, King GM, Neupane S, Myers MR. 2016. Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinders, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3(T), sp. nov. FEMS Microbiol Ecol 92:fiw028. doi: 10.1093/femsec/fiw028. [DOI] [PubMed] [Google Scholar]
- 145.Lynch RC, Darcy JL, Kane NC, Nemergut DR, Schmidt SK. 2014. Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria. Front Microbiol 5:698. doi: 10.3389/fmicb.2014.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Conrad R. 1999. Soil microorganisms oxidizing atmospheric trace gases (CH4, CO, H2, NO). Indian J Microbiol 39:193–203. [Google Scholar]
- 147.Neilson JW, Califf K, Cardona C, Copeland A, van Treuren W, Josephson KL, Knight R, Gilbert JA, Quade J, Caporaso JG, Maier RM. 2017. Significant impacts of increasing aridity on the arid soil microbiome. mSystems 2:e00195-16. doi: 10.1128/mSystems.00195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rego A, Raio F, Martins TP, Ribeiro H, Sousa AGG, Seneca J, Baptista MS, Lee CK, Cary SC, Ramos V, Carvalho MF, Leao PN, Magalhaes C. 2019. Actinobacteria and cyanobacteria diversity in terrestrial Antarctic microenvironments evaluated by culture-dependent and independent methods. Front Microbiol 10:1018. doi: 10.3389/fmicb.2019.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC. 2012. The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities. ISME J 6:1046–1057. doi: 10.1038/ismej.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.King GM. 2017. Water potential as a master variable for atmosphere-soil trace gas exchange in arid and semiarid ecosystems, p 31–45. In Steven B. (ed), The biology of arid soils. De Gruyter, Berlin, Germany. [Google Scholar]
- 151.Stocker TF, Qin D, Plattner GK, Tignor MMB, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM. 2013. IPCC, 2013: Climate change 2013: the physical science basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 152.Ross MO, Rosenzweig AC. 2017. A tale of two methane monooxygenases. J Biol Inorg Chem 22:307–319. doi: 10.1007/s00775-016-1419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dunfield PF. 2007. The soil methane sink, p 152–170. In Reay D, Hewitt CN, Smith K, Grace J (ed), Greenhouse gas sinks. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 154.Striegl RG, Mcconnaughey TA, Thorstenson DC, Weeks EP, Woodward JC. 1992. Consumption of atmospheric methane by desert soils. Nature 357:145–147. doi: 10.1038/357145a0. [DOI] [Google Scholar]
- 155.Angel R, Conrad R. 2009. In situ measurement of methane fluxes and analysis of transcribed particulate methane monooxygenase in desert soils. Environ Microbiol 11:2598–2610. doi: 10.1111/j.1462-2920.2009.01984.x. [DOI] [PubMed] [Google Scholar]
- 156.Hall SJ, Silver WL, Amundson R. 2012. Greenhouse gas fluxes from Atacama Desert soils: a test of biogeochemical potential at the Earth’s arid extreme. Biogeochemistry 111:303–315. doi: 10.1007/s10533-011-9650-7. [DOI] [Google Scholar]
- 157.Hou LY, Wang ZP, Wang JM, Wang B, Zhou SB, Li LH. 2012. Growing season in situ uptake of atmospheric methane by desert soils in a semiarid region of northern China. Geoderma 189:415–422. doi: 10.1016/j.geoderma.2012.05.012. [DOI] [Google Scholar]
- 158.Sullivan BW, Selmants PC, Hart SC. 2013. Does dissolved organic carbon regulate biological methane oxidation in semiarid soils? Glob Chang Biol 19:2149–2157. doi: 10.1111/gcb.12201. [DOI] [PubMed] [Google Scholar]
- 159.Chiri E, Nauer PA, Henneberger R, Zeyer J, Schroth MH. 2015. Soil-methane sink increases with soil age in forefields of Alpine glaciers. Soil Biol Biochem 84:83–95. doi: 10.1016/j.soilbio.2015.02.003. [DOI] [Google Scholar]
- 160.Middleton N, Thomas DSG (ed). 1992. World atlas of desertification. Edward Arnold, London, United Kingdom. [Google Scholar]
- 161.Trabucco A, Zomer RJ. 2018. Global aridity index and potential evapotranspiration (ET0) Climate Database v2 figshare https://figshare.com/articles/Global_Aridity_Index_and_Potential_Evapotranspiration_ET0_Climate_Database_v2/7504448/3. [DOI] [PMC free article] [PubMed]
- 162.Koberl M, Muller H, Ramadan EM, Berg G. 2011. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS One 6:e24452. doi: 10.1371/journal.pone.0024452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ronca S, Ramond JB, Jones BE, Seely M, Cowan DA. 2015. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front Microbiol 6:845. doi: 10.3389/fmicb.2015.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tahon G, Tytgat B, Willems A. 2018. Diversity of key genes for carbon and nitrogen fixation in soils from the Sor Rondane Mountains, East Antarctica. Polar Biol 41:2181–2198. doi: 10.1007/s00300-018-2353-y. [DOI] [Google Scholar]
- 165.Liu RY, Li K, Zhang HX, Zhu JG, Joshi D. 2014. Spatial distribution of microbial communities associated with dune landform in the Gurbantunggut Desert, China. J Microbiol 52:898–907. doi: 10.1007/s12275-014-4075-3. [DOI] [PubMed] [Google Scholar]
- 166.An S, Couteau C, Luo F, Neveu J, DuBow MS. 2013. Bacterial diversity of surface sand samples from the Gobi and Taklamaken Deserts. Microb Ecol 66:850–860. doi: 10.1007/s00248-013-0276-2. [DOI] [PubMed] [Google Scholar]
- 167.Chu HY, Sun HB, Tripathi BM, Adams JM, Huang R, Zhang YJ, Shi Y. 2016. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ Microbiol 18:1523–1533. doi: 10.1111/1462-2920.13236. [DOI] [PubMed] [Google Scholar]
- 168.Rao S, Chan Y, Bugler-Lacap DC, Bhatnagar A, Bhatnagar M, Pointing SB. 2016. Microbial diversity in soil, sand dune and rock substrates of the Thar Monsoon Desert, India. Indian J Microbiol 56:35–45. doi: 10.1007/s12088-015-0549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baubin C, Farrell AM, Šťovíček A, Ghazaryan L, Giladi I, Gillor O. 2019. Seasonal and spatial variability in total and active bacterial communities from desert soil. Pedobiologia (Jena) 74:7–14. doi: 10.1016/j.pedobi.2019.02.001. [DOI] [Google Scholar]
- 170.Abed RMM, Tamm A, Hassenruck C, Al-Rawahi AN, Rodriguez-Caballero E, Fiedler S, Maier S, Weber B. 2019. Habitat-dependent composition of bacterial and fungal communities in biological soil crusts from Oman. Sci Rep 9:6468. doi: 10.1038/s41598-019-42911-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reith F, Brugger J, Zammit CM, Gregg AL, Goldfarb KC, Andersen GL, DeSantis TZ, Piceno YM, Brodie EL, Lu ZM, He ZL, Zhou JZ, Wakelin SA. 2012. Influence of geogenic factors on microbial communities in metallogenic Australian soils. ISME J 6:2107–2118. doi: 10.1038/ismej.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ferrari BC, Bissett A, Snape I, van Dorst J, Palmer AS, Ji MK, Siciliano SD, Stark JS, Winsley T, Brown MV. 2016. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environ Microbiol 18:1834–1849. doi: 10.1111/1462-2920.13034. [DOI] [PubMed] [Google Scholar]
- 173.Steven B, Lionard M, Kuske CR, Vincent WF. 2013. High bacterial diversity of biological soil crusts in water tracks over permafrost in the High Arctic Polar Desert. PLoS One 8:e71489. doi: 10.1371/journal.pone.0071489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Trabucco A, Zomer RJ. 2018. Global aridity index and potential evapotranspiration (ET0) Climate Database v2 figshare https://figshare.com/articles/Global_Aridity_Index_and_Potential_Evapotranspiration_ET0_Climate_Database_v2/7504448/3. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Information on published samples of the 20 global desert sites used to construct microbial community profiles. Download Table S1, XLSX file, 0.02 MB (20.1KB, xlsx) .
Copyright © 2020 Leung et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.