Structure of the Drosophila melanogaster Hedgehog release protein Dispatched reveals an open conformation of its ectodomain.
Abstract
The Hedgehog (Hh) signaling pathway controls embryonic development and adult tissue homeostasis in multicellular organisms. In Drosophila melanogaster, the pathway is primed by secretion of a dually lipid-modified morphogen, Hh, a process dependent on a membrane-integral protein Dispatched. Although Dispatched is a critical component of the pathway, the structural basis of its activity has, so far, not been described. Here, we describe a cryo–electron microscopy structure of the D. melanogaster Dispatched at 3.2-Å resolution. The ectodomains of Dispatched adopt an open conformation suggestive of a receptor-chaperone role. A three-dimensional reconstruction of Dispatched bound to Hh confirms the ability of Dispatched to bind Hh but using a unique mode distinct from those previously observed in structures of Hh complexes. The structure may represent the state of the complex that precedes shedding of Hh from the surface of the morphogen-releasing cell.
INTRODUCTION
The Hedgehog (Hh) signaling pathway controls embryonic development and adult tissue homeostasis in multicellular organisms. The Hh pathway is a key homeostatic regulator in embryogenic development, tissue regeneration, and stem cell maintenance (1, 2). Disruption of this pathway leads to deregulation of tissue homeostasis, developmental diseases, and cancer (3, 4). The key players in the Hh pathway are highly conserved across species (5), which has led the Drosophila melanogaster Hh system to being one of the main sources of insights into the fundamental mechanisms of Hh signaling regulation. The D. melanogaster Hh is synthesized as a preprotein that is autoproteolytically processed by its C-terminal domain and dually lipidated with an N-terminal palmitoyl and a C-terminal cholesterol moieties (the processed form of Hh, HhN-P, is here referred to as HhN) (6–8). The lipid modifications presumably anchor the Hh ligand to the biological membranes and extracellular matrix, ensuring that its short- and long-range actions are tightly controlled (9, 10). Binding of HhN to Patched on the surface of the Hh-responding cells alleviates the inhibitory action of Patched on Smoothened, a G protein–coupled receptor-like membrane protein (11). This leads to phosphorylation of the Smoothened C terminus and recruitment of Costal 2 and Fused, with subsequent activation of the Cubitus interruptus transcription factor that induces the transcription of the Hh target genes (11).
The Hh pathway is initiated by the release of the HhN from the producing cell (Fig. 1A), and this process is facilitated by vertebrate and invertebrate Dispatched homologs via an undefined mechanism. Drosophila Dispatched is a 139-kDa protein predicted to contain 12 transmembrane (TM) helices and two extracellular domains (ECDs, also called ectodomains) (12). Similar to Patched, Dispatched belongs to the Resistance-Nodulation-Division (RND) family of membrane transporters. The TM2 to TM6 region of Dispatched is annotated as the sterol-sensing domain (SSD) (13), similar to Patched and other eukaryotic RND proteins. The functional significance of the Dispatched SSD for interaction with HhN is unclear (14). The vertebrate Dispatched homolog has been proposed to synergize with Scube2 in facilitating the release of the cholesterol-modified sonic Hh (Shh) (15, 16). Early experiments showed that a possible transporter-like activity of Dispatched may be necessary for its role in Hh release (17). Furthermore, a recent report indicated that Shh release by mouse Dispatched requires processing of the latter by a convertase Furin (18). A similar proteolytic activation was proposed for the D. melanogaster Dispatched (18). Recent structural work on mammalian RND transporters, such as PTCH1 (the human homologue of Patched) and Niemann-Pick disease protein 1 (NPC1), revealed the architecture of these proteins and provided snapshots of these proteins in various states (19–23). Despite these recent breakthroughs, the precise role of Dispatched in Hh release has remained unclear.
Fig. 1. Structure of the D. melanogaster Dispatched.
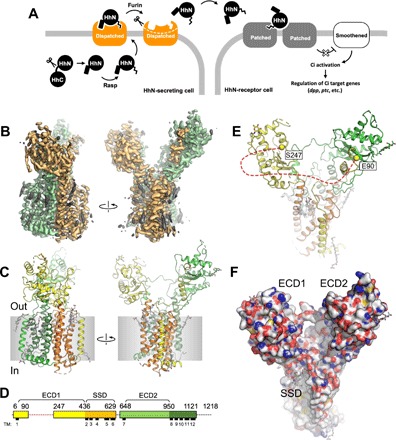
(A) Schematic representation of Hh ligand release, which initiates the Hh pathway. (B) A cryo-EM map of Dispatched reconstructed at 3.2-Å resolution; orange and green colors indicate the N- and C-terminal halves of the protein; elements of the density map assigned to bound sterols or unassigned (i.e., corresponding to the unstructured components of the detergent micelle) are colored gray. (C) Atomic model of Dispatched, built using the 3.2-Å map shown in (B). (D) Sequence coverage of the atomic model and annotation of the sequence. The colors of the linear representation of Dispatched sequence in (D) correspond to the colors of the secondary structure elements in (C). Dashed line indicates a portion of the protein not resolved in the cryo-EM map. (E) The view of the protein model from the extracellular side indicates the Cα atoms of the residues E90 and S247; the red dashed line indicates an unresolved loop between these two residues. (F) The space-filling representation of the Dispatched model reveals a large cavity formed by the two ECDs, with the SSD in close proximity.
We now report cryo–electron microscopy (cryo-EM) structures of the D. melanogaster Dispatched in the apo state (unbound to ligand) and in the presence of the Hh ligand. The ectodomain (ECD) of Dispatched splays open in a new conformation. Furthermore, the structure of the complex highlights a new interface of binding for the morphogen. The structure likely represents the state of the complex that precedes shedding of Hh from the surface of the morphogen-releasing cell. The ability of the Dispatched ECD to form a bowl-shaped cavity may indicate that homologous proteins of the RND family of membrane transporters, such as Patched (PTCH1) or NPC1, may adopt similar conformations.
RESULTS
Cryo-EM structure of Dispatched
To gain insight into the structure and function of Dispatched, we expressed the Drosophila Dispatched homolog in human embryonic kidney (HEK) 293F cells (fig. S1A), purified the protein by affinity chromatography, and collected a single-particle cryo-EM dataset (fig. S2A). The excellent quality of the sample allowed us to reconstruct the cryo-EM density map of Dispatched at a resolution of 3.2 Å (Fig. 1B and fig. S2, B to D). Using the density map of Dispatched, we built a model of the protein, including the TM domain bundle and the two ECDs, covering the majority (~69%) of the Dispatched amino acid sequence (Fig. 1, C and D).
The structure revealed the expected TM domain arrangement characteristic of the RND transporters (Figs. 1, C and D, and 2). The density map of Dispatched showed a number of density elements that we interpreted as molecules of bound cholesteryl hemisuccinate (CHS), which was present during protein purification (Fig. 1B and fig. S3). The SSD of Dispatched seems to have the ability to accommodate several sterol molecules within its pocket region, similar to the SSD of PTCH1 bound to ShhNC24II. We have modeled a total of nine sterol molecules bound to the inner and outer leaflet regions at the protein-lipid bilayer interface of Dispatched (fig. S2). Our reconstruction also revealed a notable feature of the extracellular portion of the protein: the ECDs of the Dispatched splayed apart creating a large bowl-shaped cavity (Fig. 1, E and F). The loop between the residues E90 and S247, which belongs to ECD1, is not resolved in our reconstruction (Fig. 1E). This loop is predicted to be mostly unstructured, which may explain the lack of interpretable cryo-EM density corresponding to this region of the protein. The absence of a connecting protein density between the two ECDs creates an opening into the Dispatched ECD cavity in close proximity to the protein’s membrane-embedded SSD region (Fig. 1F).
Fig. 2. The conformation of Dispatched is unique among the RND proteins with known structures.
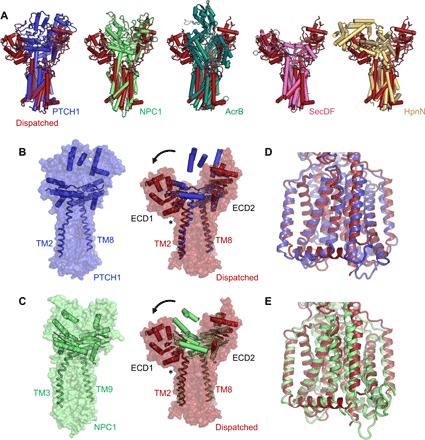
(A) Structural alignment of Dispatched with PTCH1 [root mean square deviation (RMSD) of 4.5 Å over 520 residues; PDB ID: 6d4h], NPC1 (RMSD of 4.2 Å over 544 residues; PDB ID: 5u74), AcrB (RMSD of 4.7 Å over 216 residues; PDB ID: 2hrt), SecDF (RMSD of 5.1 Å over 176 residues; PDB ID: 5aqp), and HpnN (RMSD of 4.6 Å over 256 residues; PDB ID: 5khn) reveals a remarkably diverse arrangement of the ECDs in these proteins. RMSD values were calculated using cealign in PyMOL. (B and C) Comparison to the closest homologs of Dispatched, NPC1 (B) and PTCH1 (C), shows that the ECD1 of Dispatched swings out to create the ECD cavity. The hinge region for this rearrangement appears to be at the boundary between the C terminus of ECD1 and the N-terminal portion of TM2 of Dispatched (indicated by an asterisk). The major secondary structure elements, α helices and β sheets, present in the ECDs of Dispatched, PTCH1 and NPC1, are shown in cartoon representation. The connecting TM domains TM2 and TM8 (TM3 and TM9 in NPC1) are shown as ribbons. (D and E) Despite the large differences in the relative position of ECD1, the TM domains of Dispatched, PTCH1 and NPC1, are very well aligned.
Comparisons of Dispatched to RND transporters show a novel open conformation
Comparison of the Dispatched atomic model to those of other RND transporters solved previously by either x-ray crystallography or cryo-EM, including human PTCH1 [Protein Data Bank (PDB) ID: 6d4h], human NPC1 (PDB ID: 5u74), Escherichia coli AcrB (PDB ID: 2hrt), Thermus thermophilus SecDF (PDB ID: 3aqp), and Burkholderia multivorans HpnN (PDB ID: 5khn), revealed a distinct arrangement of the ECD region of Dispatched (Fig. 2A). The closest structural homologs of Dispatched, PTCH1 and NPC1, adopt a closed conformation of the ECD (Fig. 2, B and C). The open conformation of Dispatched is established by a splaying motion of the ECD1 relative to ECD2 and the TM domain bundle, at a hinge point residing in the region of the protein at the TM2-ECD1 boundary (Fig. 2, C and D). Despite this marked difference in the position of the ECD1, the TM domains of Dispatched and PTCH1 or NPC1 can be accurately aligned (Fig. 2, D and E).
Structure of Dispatched-Hh complex reveals protein-based interactions between Dispatched and the morphogen ligand
Given the role of Dispatched in Hh release, we reasoned that the ECD cavity may have a role in binding of the ligand. To test this experimentally, we performed cryo-EM analysis and three-dimensional (3D) reconstruction of a complex formed by Dispatched and a recombinant HhNC85II fragment (Fig. 3, A and C, and figs. S1B, S5, and S6). The modified ligand, lacking a sterol modification and featuring two Ile residues mimicking the palmitoylated C85 residue, was used in analogous fashion to the human ShhNC24II (24). In comparison to the transfected upstream activating sequence (UAS)–HhN construct or the lipidated recombinant human ShhN fragment, the HhNC85II did not elicit Hh pathway activation under cell culture conditions (Fig. 3A) when inoculated in culture medium. It did, however, show binding to Dispatched in vitro as measured by microscale thermophoresis (MST), with an apparent affinity of 6.2 ± 2.5 μM (Fig. 3A). Our cryo-EM analysis confirmed the binding of the ligand at the interface between ECD1 and ECD2 of Dispatched (Fig. 3, B and C). The resolution of the 3D reconstruction was limited to 4.8 Å, but it was sufficient to place and orient a model of HhN into the map (Fig. 3C and figs. S5 and S6, A and B). The N terminus of HhN is close to the base of the Dispatched ECD cavity, and the C terminus of HhN is adjacent to the SSD (Fig. 3, D and E), consistent with a possible role of the SSD in cholesterol interaction. On the basis of the presence of sterol-like density elements in several sites accessible to the SSD in our apo-Dispatched reconstruction (Fig. 1B and figs. S3 and S4), the C terminus of the cholesterol-modified HhN may interact with the protein via more than one site. The functional role of the different sterol interaction sites at the protein-lipid interface of Dispatched will require careful future investigation.
Fig. 3. Structure of Dispatched bound to a modified Hh ligand.
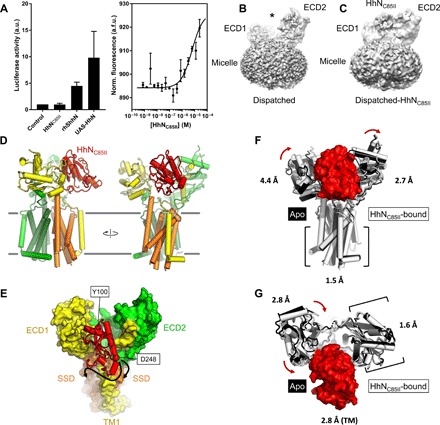
(A) Biochemical activity of HhNC85II ligand, assessed by luciferase luminescence (left) and MST (right). The data correspond to values of luciferase activity normalized to the values measured for the “Control” sample (medium without the ligand); n = 4 [for recombinant human sonic hedgehog (rhShhN), n = 2]. The affinity of HhNC85II for Dispatched–yellow fluorescent protein (YFP), determined by MST, was 6.2 ± 2.5 μM (n = 3). a.u., arbitrary units; Norm., normalized; a.f.u., arbitrary fluorescence units. (B and C) Comparison of the density maps of Dispatched (B) and Dispatched-HhNC85II complex (C); the asterisk (B) indicates the cavity between the ECD1 and ECD2 regions, occupied by HhNC85II density in the map shown in (C). (D) Overview of the Dispatched-HhNC85II complex model; color coding for Dispatched is the same as in Fig. 1 (C and D); the Hh ligand is colored red. (E) View of the complex from the luminal/extracellular side of the membrane shows the orientation of the HhNC85II ligand. The Cα atoms of N-terminal residue Y100 and the C-terminal residue D248 in the model are shown as red spheres. Arrows indicate possible proximity of the cholesterol-modified C-terminal region of HhN to the SSD region of Dispatched. (F and G) Structural alignment of the ligand-free and ligand-bound Dispatched; the complex aligned to apo-Dispatched using the TM domain (F) or the ECD2 domain (G). The numbers correspond to RMSD between the atoms of the individual domains. The red arrows indicate the whole domain structural rearrangements that accompany HhN ligand binding.
Only relatively few residues, S48, Q73, H77-H78, R250, E252-Y253, E296, A404-R408, G434 of Dispatched ECD1, and N755 of the ECD2, contribute to the interaction with HhN (fig. S6). This may explain the relatively low affinity of this interaction and the poor density corresponding to the bound HhN. It is likely that the processed, dually lipidated natural form of HhN engages Dispatched at additional sites within and near the ligand-binding cleft. For example, the palmitoylated N terminus of the protein may be in contact with the surface of ECD2, whereas the C-terminal cholesteryl moiety may interact with membrane-embedded SSD of Dispatched.
We analyzed the interfaces that the HhN ligand presents to different binding partners, in the context of our Dispatched-HhN complex (fig. S7). Structural alignment of the available structures featuring the insect or mammalian Hh ligand bound to its receptors showed that Dispatched interacts with HhN in a unique way (fig. S7A). One site in the sequence of the Hh ligands, corresponding to residues Y100-Q106 in the Drosophila HhN, participated in interactions with all of the compared receptor proteins (fig. S7, B and C). This suggests that this site in the morphogen may have a critically important role in the Hh pathway, facilitating the interactions between HhN and a wide range of Hh receptor proteins.
DISCUSSION
The structures of Dispatched alone and in a complex with HhN protein fragment point to exciting new possibilities for the role of Dispatched at the early stage of the Hh signaling and provide the structural basis for understanding the activity of Dispatched (Fig. 4). The conserved arrangement of the core residues previously shown to be important for Dispatched activity (17) and the presence of multiple sterols at binding sites surrounding the membrane domain of Dispatched point to a notable similarity between the membrane-spanning regions of the eukaryotic RND proteins (Fig. 4 and figs. S3 and S4). The open conformation of the Dispatched ECD capable of accommodating HhN fragment in the cleft between the ECD1 and ECD2 suggests that Dispatched may act directly as a receptor for HhN protein, independently of lipid modification. The loop between the TM1 and the ECD1 region of dispatched, missing in our 3D reconstruction (Fig. 4B), may play a molecular gating role, regulating the access of the Hh ligand to or its release from Dispatched. Although this region is not visible in our reconstruction, it is possible that under some conditions, the loop may occupy a defined position within the ECD cavity. This site may also play a role in the interactions between Dispatched and yet unknown components of the Hh release machinery. Although cholesterol-based interactions have been suggested to be critical for ligand release by Dispatched, it is now clear that HhN can engage Dispatched via protein-protein interactions. This interaction may be relevant during the endosomal Dispatched-dependent recycling of HhN (25, 26). Regardless of the precise location where Dispatched acts to facilitate Hh release, the conformation observed in our reconstruction likely represents a prerelease state of the complex.
Fig. 4. Structural basis of Dispatched activity.
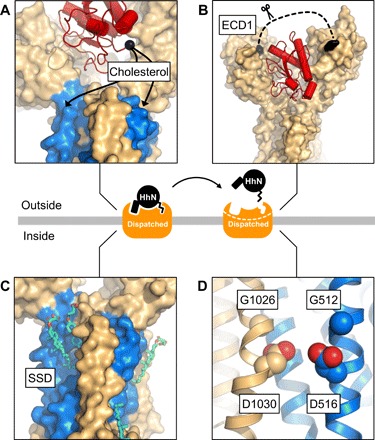
The cryo-EM structures of Dispatched alone and in a complex with HhNC85II provide the framework for understanding the structural basis of Dispatched-assisted HhN release. (A to D) The key features of Dispatched involved in the activity of Dispatched include that (A) the orientation of the C-terminal region of Hh coincides with the sterol modification in close proximity to the SSD (blue); (B) the unstructured region of the ECD1 (residues E90-S247; dashed line), which may represent a molecular gate for access or dissociation of the Hh ligand; proteolytic processing of this region has been suggested as a prerequisite for Dispatched activation (18), (C) multiple sterols identified the model of apo-Dispatched and (D) the GxxxD motifs in TM4 and TM10, presumed to be involved in transport-like activity of Dispatched (17). The release of the Hh ligand is likely dependent on the interplay between these structural elements of Dispatched.
The selection of the HhNC85II version of the ligand for this study was made by analogy with the similar modification of ShhN (ShhNC24II) that retains the ability to bind PTCH1 (24). However, it is worth noting the evolutionary divergence between the fly and the vertebrate Hh signaling pathways. Key differences exist between the invertebrate and vertebrate ligands, such as the conservation of zinc-binding site in mammalian ShhN (important for interactions between ShhN with the PTCH1 co-receptors, such as Cdo/Boc, GAS1, and Hhip), missing in the fly HhN (27). There may also be some divergence in the Hh secretion mechanism. For example, the protein Scube2 is required for ShhN secretion in vertebrates but is absent in Drosophila (15). Although these evolutionary differences do not affect our interpretation of the interaction between Dispatched and HhN, they have to be considered to understand the implications of our structures for the invertebrate and vertebrate Hh signaling.
A number of questions pertaining to the structure of Dispatched remain open. For example, our preparations of the protein show that it is monomeric in detergent micelle, yet it has previously been suggested to form oligomers (18, 28). In light of the variety of stable oligomeric states observed recently by cryo-EM for PTCH1, including monomers (19, 21, 24), dimers (20, 22), and tetramers (29), it is possible that Dispatched is capable of oligomerizing in a ligand-dependent or ligand-independent manner. On the other hand, although the structural features and the biological role of Dispatched appear to be unique among the RND transporters, it is also possible that homologous proteins (e.g., PTCH1 and NPC1) may, under certain conditions, adopt open conformations similar to that of Dispatched.
MATERIALS AND METHODS
Expression and purification of Drosophila Dispatched
The synthetic DNA fragment encoding the D. melanogaster Dispatched (Genewiz; UniProt ID: Q9VNJ5) was cloned into a pACMV vector (30) in frame with a C-terminal 3C-YFP-twinStrep tag fusion. The plasmid was transfected into HEK293F cells, and a stable clone of the cells constitutively expressing Dispatched was selected by treatment of the cells with G418 according to the established protocol (30). Protein expression was performed in 2-liter Erlenmeyer flasks, with 800-ml culture per flask, at 37°C in protein expression medium in the presence of 5% fetal bovine serum, 1× penicillin/streptomycin mixture, 1× GlutaMAX, and 1× nonessential amino acids. At cell density of ~2 × 106 cells/ml, the cultures were collected by centrifugation at 500g, resuspended in phosphate-buffered saline (40 ml/1 liter of culture), frozen, and stored at −80°C until purification.
For protein purification, the frozen pellets corresponding to 4 liters of culture of Dispatched-expressing HEK293F cells were thawed, resuspended in 150 ml of buffer A [50 mM tris (pH 8), 150 mM NaCl, and 10% glycerol], in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) and one tablet per 50 ml of Roche cOmplete protease inhibitor cocktail (EDTA-free). The resuspended cells were homogenized using a Dounce homogenizer. Following an incubation on a rotating wheel in the presence of 1% dodecyl-β-d-maltoside (DDM) and 0.2% CHS at 4°C for 1 hour, the cell lysates were clarified by ultracentrifugation (rotor Ti45, 35,000 rpm at 4°C for 30 min). The supernatants were added to 3 ml of preequilibrated CNBr-sepharose resin coupled with purified anti–green fluorescent protein nanobody (31) and incubated for 1 hour on a rotating wheel at 4°C. The sepharose resin was collected by gravity flow into a glass Econo-Column (Bio-Rad), washed with 30 column volumes (CVs) of buffer B [50 mM tris (pH 8) and 150 mM NaCl] containing DDM/CHS mixture (0.025% DDM/0.005% CHS) and with 10 CVs of buffer B containing 0.1% digitonin. The protein was eluted from the resin using overnight cleavage by 3C protease at 4°C, and the eluted protein-containing solution was subjected to a negative Ni-NTA purification step (using 1 ml of Ni-NTA resin preequilibrated with buffer B) to remove the 3C protease. The final purified protein sample was isolated using size exclusion chromatography (SEC) using Superose 6 Increase 10/300 GL column equilibrated in buffer B in the presence of 0.1% digitonin (fig. S1). The selected fractions of the SEC peak corresponding to the purified Dispatched were concentrated to 4 mg/ml using an Amicon Ultra-4 concentrator (100-kDa cutoff; Millipore).
For purification of Dispatched–yellow fluorescent protein (YFP), the frozen pellets of Dispatched-expressing cells (2 liters of culture) were thawed and resuspended in 60 ml of buffer A in the presence of 1 mM PMSF and one tablet of Roche cOmplete protease inhibitor cocktail (EDTA-free). The resuspended cells were homogenized using a Dounce homogenizer. Following an incubation on a rotating wheel in the presence of 1% DDM and 0.2% CHS at 4°C for 1 hour, the cell lysates were clarified by centrifugation (rotor A8.24; 19,000 rpm at 4°C for 30 min). The supernatant was incubated with 5 ml of equilibrated Strep-Tactin Superflow resin (IBA Lifesciences). The resin was washed and equilibrated with buffer A containing 0.02% DDM and 0.004% CHS. Soluble lysate was mixed with the resin and incubated for 30 min and then loaded on a gravity column. Resin was washed with 3 × 10 CVs of buffer A containing 0.02% DDM and 0.004% CHS and 2 × 5 CVs of buffer A containing 0.1% digitonin. Elution was performed in the same buffer supplemented with 5 mM desthiobiotin. The protein was further purified using SEC on a Superose 6 Increase 10/300 GL column equilibrated in buffer B [50 mM tris (pH 8) and 150 mM NaCl) in the presence of 0.1% digitonin. Selected fractions were concentrated to 1 mg/ml using Amicon Ultra-4 concentrator (100-kDa cutoff; Millipore) and flash-frozen in 10% glycerol.
Expression and purification of Drosophila hedgehog N-terminal fragment HhN
The synthetic DNA fragment encoding the modified D. melanogaster hedgehog N-terminal fragment, referred to as HhN (residues 85 to 257, modified at the N terminus by replacing the Cys85 residue with two Ile residues; UniProt ID: Q02936), fused with the N-terminal 6×His-SUMO tag, was cloned into pET28a plasmid. For expression of the HhN protein, the plasmid was transformed into BL21(DE3) RIPL E. coli cells. The transformed cultures were grown in a shaking incubator at 37°C until OD600 (optical density at 600 nm) of 0.8, at which point the temperature was switched to 30°C, and the expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 hours. The induced cells were harvested by centrifugation, frozen, and stored at −80°C until the day of experiment.
The frozen E. coli pellets (corresponding to 4 liters of cultures) were thawed, resuspended in 120 ml of buffer C [50 mM tris (pH 7.5) and 200 mM NaCl] containing 25 mM imidazole, disrupted by sonication, and clarified by centrifugation at 18,000g for 30 min at 4°C. The supernatant was added to 2 ml of Ni-NTA resin and incubated at 4°C with rotation for 1 hour. The Ni-NTA resin was collected into a gravity column and washed with 30 CVs of buffer C containing 25 mM imidazole and 10 CVs of buffer C containing 50 mM imidazole. The protein was eluted using buffer C containing 200 mM imidazole. The eluted fractions were pooled, diluted eightfold to reduce the concentration of imidazole, and incubated with 250 μg of purified SUMO protease Ulp1 (ubiquitin-like specific protease-1) overnight at 4°C. The mixture was passed through 2 ml of immobilized Ni-NTA resin preequilibrated with buffer C containing 25 mM imidazole to remove the uncleaved HhN, the cleaved off SUMO tag, and the Ulp1. The flow-through was concentrated to 1 ml and applied to a Superdex 200 Increase 10/300 GL column (fig. S1). The fractions corresponding to HhN were pooled, supplemented with 10% glycerol, flash-frozen in liquid nitrogen, and stored at −80°C until the day of experiment.
Microscale thermophoresis
For MST analysis, the dilution series of purified HhNC85II were prepared in buffer A containing 0.1% digitonin (16 concentration points, ranging from 50 μM to 1.52 nM). Dispatched-YFP was mixed with each solution at a 1:1 ratio (final Dispatched-YFP concentration, 20 nM), and the mixtures were incubated for about 20 min at room temperature. Monolith NT 1.115 Premium Capillaries (NanoTemper) were filled with each reaction mixture, loaded on a cartridge into the Monolith NT 1.115 (NanoTemper), and MST analysis was performed using the following parameters: MST power, medium; excitation power, 100%; excitation type, “blue”; temperature, 22°C. The PALMIST software (32) was used for data analysis and model fitting, using weighted fitting for the three sets of data, and with cold values measured before laser firing and hot values after thermophoresis. Analysis results were exported and plotted with Prism.
Cryo-EM sample preparation and data collection
The concentrated sample of Dispatched (4 mg/ml) was kept on ice and was immediately used for cryo-EM grid preparation. The Dispatched-HhN sample was prepared by mixing the concentrated Dispatched (4 mg/ml) with a twofold molar excess of the concentrated HhN (desalted into the buffer B containing 0.1% digitonin) and incubating the mixture for ~30 min on ice before cryo-EM grid freezing. For cryo-EM sample preparation, UltrAuFoil 1.2/1.3 Au 300-mesh grids were glow discharged using a PELCO easiGlow (Ted Pella) plasma cleaning system for 30 s at 25 mA in air. The protein samples (3.5 μl) were deposited on the surface of the grid immediately before blotting for 3 s on a Vitrobot Mark IV (FEI) set to operation at 4°C with 100% humidity and with a blotting force of 2 and plunging the grid into liquid ethane. The grids were stored in liquid nitrogen until the day of data collection.
For Dispatched, a cryo-EM dataset was collected at the European Molecular Biology Laboratory (EMBL) Heidelberg using a Titan Krios microscope equipped with a Gatan K2 Summit detector and an energy filter. A total of 9216 micrographs were automatically collected in counting mode using SerialEM at a magnification of ×165,000 (resulting in the micrograph pixel size of 0.81 Å per pixel), with a total exposure of 8 s over 40 frames and a total dose of 50.42 e/Å2.
The cryo-EM dataset for Dispatched-HhN was collected at Netherlands Centre for Electron Nanoscopy (NeCEN), Leiden using a Cs-corrected Titan Krios microscope equipped with a Falcon 3 camera. A total of 5609 movies were collected in counting mode using EPU software, at a nominal magnification of ×96,000 (resulting in the micrograph pixel size of 0.88 Å per pixel), with a total exposure of 60 s over 49 frames and a total dose of 51.5 e/Å2.
Cryo-EM data analysis
Drift correction and alignment of the cryo-EM movies were performed using MotionCor2 (33), and the defocus values of the aligned micrographs were estimated using Gctf (34). Only micrographs with an estimated resolution below 4 Å were kept for further processing using relion-2.1 and relion-3.0 (35, 36). After manual picking and 2D classification using a small subset of data (35,554 particles), about 2.2 M particles were autopicked from all selected micrographs using the best 2D classes as references. Several rounds of 2D classification of twofold downsampled particles reduced the dataset to 531,859 particles. For the first round of 3D classification, we used an initial model generated in relion-2.1 using data collected with the TVIPS (Tietz Video and Image Processing Systems) camera–equipped JEOL JEM-2200 microscope at Paul Scherrer Institut (PSI). Briefly, a small dataset of 12 micrographs was acquired (2.24 Å per pixel), and 20,208 particles were autopicked in relion-2.0. Following several rounds of 2D classification, an initial model was generated using 4000 particle images in relion-3.0. This model was used for 3D classification with four classes, resulting in one class (7680 particles) that was subsequently used as an initial model for processing of the cryo-EM data collected using Titan Krios. The best resulting 3D class was used for downstream image processing steps. Following additional rounds of 3D classification, the particles belonging to the best 3D class could be refined to 3.9 Å [fourier shell correlation (FSC) cutoff, 0.143]. Particles were then reextracted and rescaled to the original pixel size and refined with a mask to 3.2 Å. Bayesian particle polishing and contrast transfer function (CTF) refinement in relion-3.0 resulted in the postprocessed map of Dispatched at 3.15-Å resolution (using b factor of −50 for map sharpening; the b factor value was determined empirically to obtain a well-interpretable density map). The complete data processing workflow is shown in fig. S2.
For Dispatched-HhN dataset, the initial steps were similar to those described above. After several rounds of 2D classification in relion-3.0, a dataset of 162,907 particles was selected. One round of 3D classification with two classes was performed, and the best 3D class (98,623 particles) could be refined to 7.04-Å resolution. This class was subjected to a 3D classification with a single class (40 iterations, T20, E7). The particles in this class were imported into cisTEM (37), and local searches were performed using an initial resolution cutoff of 15 Å, followed by the same procedure with a 10-Å cutoff. A mask excluding the micelle but covering Dispatched and the bound HhN density was used (a soft edge of 10 Å was applied during processing in cisTEM). The density outside the mask was low-pass filtered to 30 Å, and the weight outside the mask was set to 0.3. The refinement converged after several iterations to an overall resolution of 4.3 Å (FSC cutoff, 0.143); b factor sharpening resulted in a final map at 4.76-Å resolution.
Model building and validation
Model building was performed in Coot (38) using the postprocessed map of Dispatched. Building of the Dispatched ECD1 and ECD2 was assisted by homology models generated using Swiss-Model server using PTCH1 (PDB ID: 6d4h) and NPC1 (PDB ID: 5u74) as templates. The density elements corresponding to bound detergents/lipids were interpreted as molecules of CHS. The atomic model of Dispatched was refined using phenix.real_space_refine (39) implemented in Phenix (1.16-3549) (39). For model validation, the atom coordinates in the refined model were randomly displaced by a maximum of 0.5 Å using the PDB tools in Phenix. The derived model was subjected to real-space refinement using one of the refined half maps (half-map1). Map versus model FSC comparison was made for the model against the corresponding half-map1 used in the refinement job and for the same model versus the half-map2 (not used during refinement) (40). The geometry of the Dispatched model was validated using MolProbity (41). For modeling of the Dispatched-HhN complex, a D. melanogaster Hh model (PDB ID: 2ibg) and our model of Dispatched were manually docked into the density map in University of California, San Francisco (UCSF) Chimera and jiggle fit as a rigid body in coot, followed by real-space refinement in Phenix. Figures featuring the models and density maps were prepared using PyMOL (42) and UCSF Chimera (43).
Dual-luciferase activity assay
Clone 8 cells were grown on 10-cm plates in Shields and Sang M3 insect medium (Sigma-Aldrich, no. S8398) supplemented with insulin (0.5 mg/ml; Sigma-Aldrich, no. I6634), 2% fetal bovine serum (JRH, no. 12103-78P), 2.5% fly extract (Drosophila Genomics Resource Center, Indiana), and penicillin/streptomycin (Gibco, no. 15070-063). A day before transfection, cells were seeded into a 24-well plate at a density of 0.4 × 106 cells per well. On the day of transfection, cells were transfected using the Effectene kit (QIAGEN) following the standard protocol. A DNA Master Mix was prepared for each assay, with a 5:1 ratio of ptc Firefly luciferase:tub Renilla luciferase constructs. Transfected luciferase or Hh plasmids were generated as described previously (44); the UAS-HhN construct is constitutively expressed in the transfected cells through regulation of the UAS, by Gal4. For experiments involving purified ligands, purified HhNC85II or recombinant human sonic hedgehog, rhShhN (R&D Systems, no. 8908-SH/CF), was sterile-filtered and added to the medium during transfection to a final concentration of 200 nM. Cells were lysed using the Passive Lysis Buffer from the Dual-Luciferase Kit (Promega) after 24 hours in the case of stimulation with purified ligand or 48 hours in the case of ligand transfection. Cell lysates were used immediately upon preparation or stored at −80°C. Luciferase activity was measured according to the Dual-Luciferase Kit protocol using PHERAstar FSX microplate reader (BMG LABTECH).
Supplementary Material
Acknowledgments
We thank the Electron Microscopy Facility at PSI, Villigen (E. Mueller-Gubler and T. Ishikawa) for support. We thank the EMBL Heidelberg cryo-EM service platform (F. Weis) for the support and expertise in cryo-EM data collection. We thank the staff at the NeCEN (C. Diebolder) for the support and expertise in cryo-EM data collection. We thank D. Ozerov, S. Bliven, and M. Caubet Serrabou (PSI) for the computational expertise and IT support. Funding: This study has been supported by the Swiss National Science Foundation grant to V.M.K. (SNF Professorship, 150665 and 176992). Author contributions: F.C. designed and performed the experiments, analyzed the data, and wrote the manuscript. C.Q. performed the experiments. J.F. performed the experiments and analyzed the data. G.H. designed the experiments and analyzed the data. K.B. designed the experiment and analyzed the data. V.M.K. designed and performed the experiments, analyzed the data, and wrote the manuscript. Competing interests: The authors declare that they have no competing financial or nonfinancial interests. Data and materials availability: Coordinates and cryo-EM maps have been deposited in the PDB (accession codes 6TBU and 6TD6) and Electron Microscopy Data Bank (accession codes EMD-10452 and EMD-10464). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaay7928/DC1
REFERENCES AND NOTES
- 1.Briscoe J., Thérond P. P., The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Ingham P. W., Nakano Y., Seger C., Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Pak E., Segal R. A., Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev. Cell 38, 333–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasca di Magliano M., Hebrok M., Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3, 903–911 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Hausmann G., von Mering C., Basler K., The Hedgehog signaling pathway: Where did it come from? PLOS Biol. 7, e1000146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., Taylor F. R., Wang E. A., Galdes A., Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037–14045 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Porter J. A., Ekker S. C., Park W.-J., von Kessler D. P., Young K. E., Chen C.-H., Ma Y., Woods A. S., Cotter R. J., Koonin E. V., Beachy P. A., Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Guerrero I., Chiang C., A conserved mechanism of Hedgehog gradient formation by lipid modifications. Trends Cell Biol. 17, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Callejo A., Torroja C., Quijada L., Guerrero I., Hedgehog lipid modifications are required for Hedgehog stabilization in the extracellular matrix. Development 133, 471–483 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Peters C., Wolf A., Wagner M., Kuhlmann J., Waldmann H., The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 8531–8536 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham P. W., McMahon A. P., Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Burke R., Nellen D., Bellotto M., Hafen E., Senti K.-A., Dickson B. J., Basler K., Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803–815 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Kuwabara P. E., Labouesse M., The sterol-sensing domain: Multiple families, a unique role? Cell 18, 193–201 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Blassberg R., Jacob J., Lipid metabolism fattens up hedgehog signaling. BMC Biol. 15, 95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creanga A., Glenn T. D., Mann R. K., Saunders A. M., Talbot W. S., Beachy P. A., Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev. 26, 1312–1325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tukachinsky H., Kuzmickas R. P., Jao C. Y., Liu J., Salic A., Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2, 308–320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y., Erkner A., Gong R., Yao S., Taipale J., Basler K., Beachy P. A., Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63–75 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Stewart D. P., Marada S., Bodeen W. J., Truong A., Sakurada S. M., Pandit T., Pruett-Miller S. M., Ogden S. K., Cleavage activates dispatched for Sonic Hedgehog ligand release. eLife 7, e31678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong X., Qian H., Cao P., Zhao X., Zhou Q., Lei J., Yan N., Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361, eaas8935 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Qi X., Schmiege P., Coutavas E., Li X., Two patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, eaas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi X., Schmiege P., Coutavas E., Wang J., Li X., Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Bulkley D. P., Xin Y., Roberts K. J., Asarnow D. E., Sharma A., Myers B. R., Cho W., Cheng Y., Beachy P. A., Structural basis for cholesterol transport-like activity of the Hedgehog receptor patched. Cell 175, 1352–1364.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong X., Qian H., Zhou X., Wu J., Wan T., Cao P., Huang W., Zhao X., Wang X., Wang P., Shi Y., Gao G. F., Zhou Q., Yan N., Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 165, 1467–1478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi C., Di Minin G., Vercellino I., Wutz A., Korkhov V. M., Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci. Adv. 5, eaaw6490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Angelo G., Matusek T., Pizette S., Thérond P. P., Endocytosis of Hedgehog through dispatched regulates long-range signaling. Dev. Cell 32, 290–303 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Callejo A., Bilioni A., Mollica E., Gorfinkiel N., Andrés G., Ibáñez C., Torroja C., Doglio L., Sierra J., Guerrero I., Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc. Natl. Acad. Sci. U.S.A. 108, 12591–12598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beachy P. A., Hymowitz S. G., Lazarus R. A., Leahy D. J., Siebold C., Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 24, 2001–2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etheridge L. A., Crawford T. Q., Zhang S., Roelink H., Evidence for a role of vertebrate Disp1 in long-range Shh signaling. Development 137, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian H., Cao P., Hu M., Gao S., Yan N., Gong X., Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. bioRxiv, 461491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhary S., Pak J. E., Gruswitz F., Sharma V., Stroud R. M., Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat. Protoc. 7, 453–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubala M. H., Kovtun O., Alexandrov K., Collins B. M., Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19, 2389–2401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuermann T. H., Padrick S. B., Gardner K. H., Brautigam C. A., On the acquisition and analysis of microscale thermophoresis data. Anal. Biochem. 496, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. D66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amunts A., Brown A., Bai X.-c., Llácer J. L., Hussain T., Emsley P., Long F., Murshudov G., Scheres S. H. W., Ramakrishnan V., Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- 43.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Chen C. H., von Kessler D. P., Park W., Wang B., Ma Y., Beachy P. A., Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98, 305–316 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaay7928/DC1


