Sperm cells with damaged DNA cause mitotic errors in the fertilized embryo, leading to a diverse spectrum of genomic abnormalities.
Abstract
Genomic instability is common in human embryos, but the underlying causes are largely unknown. Here, we examined the consequences of sperm DNA damage on the embryonic genome by single-cell whole-genome sequencing of individual blastomeres from bovine embryos produced with sperm damaged by γ-radiation. Sperm DNA damage primarily leads to fragmentation of the paternal chromosomes followed by random distribution of the chromosomal fragments over the two sister cells in the first cell division. An unexpected secondary effect of sperm DNA damage is the induction of direct unequal cleavages, which include the poorly understood heterogoneic cell divisions. As a result, chaotic mosaicism is common in embryos derived from fertilizations with damaged sperm. The mosaic aneuploidies, uniparental disomies, and de novo structural variation induced by sperm DNA damage may compromise fertility and lead to rare congenital disorders when embryos escape developmental arrest.
INTRODUCTION
In early embryonic development, there is reduced activity of cell cycle checkpoints and apoptotic pathways until the embryonic genome becomes activated (1–3). As a consequence, mitotic errors such as chromosome missegregations and spindle abnormalities are frequently tolerated in the first cleavage divisions with aneuploidies and subchromosomal aberrations, involving one or multiple chromosomes as a consequence. The result of this genomic instability is genetic mosaicism, i.e., the phenomenon that cleavage-stage embryos are composed of multiple genetic lineages. Mosaicism affects approximately three-quarters of the human day 3 cleavage-stage embryos and is considered to be the leading cause for the high miscarriage rates and failed implantations that underlie the low success rate of in vitro fertilization (IVF) (4–8). On rare occasions, chromosomally abnormal cells may develop to molar pregnancies or contribute to parthenogenetic, androgenetic chimeric, and mixoploid lineages in live-born humans (9–12). Thus, genetically distinct cell lineages within an embryo can participate in development and contribute to disease. Despite the immediate relevance for human health and fertility, the causes for the high mitotic error rates in human preimplantation embryos are largely unknown (5, 13). Mosaicism is prevalent in human spontaneous abortions of natural pregnancies (14), indicating that the causes for the high mitotic error rate in embryos are unrelated to the IVF procedures such as the ovarian stimulation regime, fluctuations in oxygen tension or temperature, and composition of the culture medium (15–17). While advanced maternal age increases, the risk for meiotic errors leading to whole-embryo aneuploidies, mitotic errors, and embryo mosaicism is not correlated with female age (18). A genome-wide association study has identified a common haplotype (~30% global minor allele frequency) spanning the polo-like kinase 4 (PLK4) gene that is associated with mitotic errors in development. PLK4 is involved in centriole duplication, and the minor allele is correlated with tripolar chromosome segregations (19). However, PLK4 polymorphisms alone cannot explain the high prevalence of mosaicism in human embryos. Thus, the causes for the high mitotic error rates in human preimplantation embryos are still largely unknown (5, 13, 20).
The role of the sperm cell in embryonic mosaicism has thus far been frequently underrecognized (21), possibly because paternal effects on the embryonic genome are presumed to be mostly restricted to the zygote stage. A plethora of factors can cause sperm DNA damage, including protamine imbalances, abortive apoptosis, advanced male age, oxidative stress, storage temperatures, and infections (22), while sperm DNA damage itself does not necessarily influence seminal parameters, sperm morphology, and motility or impair fertilization of the oocyte (23). The aim of this study is to investigate the consequences of sperm DNA damage on embryonic genome integrity. To address this question, we used bovine IVF and embryo culture, which is a highly valuable model for those countries where the creation of human embryos for research purposes is forbidden. It is also a recognized model system to study genomic instability in early development, because the degree of mosaicism is comparable to that observed in human IVF, while mitotic errors are rarely observed in cultured mouse embryos (24–26). Single-cell whole-genome sequencing (WGS) of individual blastomeres of two-cell– and eight-cell–stage bovine embryos revealed that sperm DNA damage results in reciprocal gains and losses of chromosomes and chromosomal segments in individual blastomeres at the two-cell stage. In addition to these immediate consequences, sperm DNA damage causes genomic instability, leading to chaotic mosaicism with a broad variety of genomic aberrations in eight-cell–stage embryos.
RESULTS
Sperm DNA damage causes mirrored mosaicism in two-cell–stage embryos
Early bovine and human embryo development is a near deterministic process regulated by maternally deposited factors until the embryonic genome becomes activated at the four- to eight-cell stage (1, 27). To examine the consequences of sperm DNA damage on the developmental competence of embryos, bovine IVF was performed with sperm subjected to γ-radiation. The advantage of γ-radiation as exogenous source of DNA damage is that its effects on DNA damage are well known and the dosage can be strictly controlled (28). Furthermore, in contrast with other DNA-damaging reagents such as doxorubicin and camptothecin, γ-radiation also induces DNA damage on noncycling cells such as sperm cells in a dose-dependent manner (23). In agreement with previous findings (23), exposing sperm cells to increasing levels of γ-radiation greatly reduced blastocyst formation rates (Fig. 1A) while having a limited effect on cleavage rates (control group, 80.4% ± 3.4; 10 Gy, 72% ± 1.8). The main effect of sperm radiation was developmental arrest at around the eight-cell stage, which coincides with the activation of the embryonic genome (23, 27). The absence of strong selective forces until the eight-cell stage of development allows the formation of genomic aberrations that are nonviable at later stages, and therefore, these early embryonic stages provide a window of opportunity to naively study genomic instability in the absence of selection.
Fig. 1. Characteristics of blastocysts and single-cell libraries of analyzed bovine embryos.
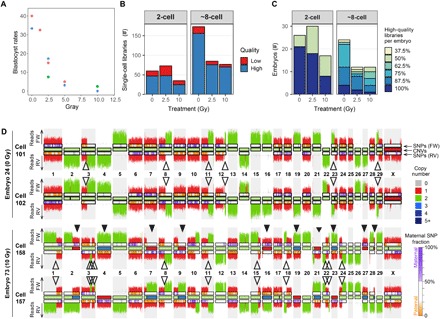
(A) Percentage of blastocysts that develop from fertilization with sperm treated with different doses of γ-radiation. Forty to 268 maturated oocytes per treatment group were fertilized with sperm radiated with 0, 1.25, 2.5, 5, 10, and 25 Gy. The number of blastocysts was counted at day 8 after fertilization. The results were obtained from three independent fertilization experiments indicated by dot color. (B) Number of successfully sequenced single-cell libraries per developmental stage. High-quality libraries have more than 100,000 nonduplicate reads with a mapping quality of more than 10 and 10 or less filtered chromosomal or segmental abnormalities. Strand-seq libraries additionally required the typical strand inheritance patterns. Sequencing results for low-quality libraries with more than 10,000 reads are also included in the karyograms, because they can be informative for identifying sister cells, but they are excluded for further quantitative analyses. (C) Percentage of sequenced high-quality single-cell libraries per embryo. (D) Strand-seq–derived copy number profiles for the Watson strand and the Crick strand plotted above and below chromosome ideograms, respectively. Top: Two sister cells from a diploid control embryo. Bottom: Two sister cells from a mosaic embryo produced with 10 Gy–treated sperm. Closed arrowheads highlight mitotic errors. Mitotic errors are reciprocal, where a gain in one cell is lost in its sister cell. Open arrowheads designate SCE events. SCEs have occurred at positions where the template strands switch from Watson-Crick to Watson-Watson or Crick-Crick. SCEs are fully mirrored between sister cells. FW, forward/Crick strand; RV, reverse/Watson strand. The inside of the chromosome ideograms is color-coded to indicate copy number (see legend for the copy number states). The ideograms for the Watson and the Crick strands are color-coded to indicate the contribution of maternal SNVs (see legend for details).
To study the consequences of sperm DNA damage on the embryonic genome, we first performed Strand-seq, a single-cell WGS technique in which the DNA strands that were used as templates during DNA replication before cell division are selectively sequenced. For Strand-seq, cells are grown in the presence of bromodeoxyuridine (BrdU) for one cell cycle. The newly synthesized DNA strands, which have BrdU incorporated during replication, are degraded by treatment with Hoechst 33258 and ultraviolet light, while the remaining template DNA strands are used for the preparation of sequencing libraries (29). For each chromosome, Strand-seq libraries of diploid cells display either Watson-Watson, Watson-Crick, or Crick-Crick inheritance patterns of the template strands that were inherited by the daughter cell after cell division (29). Strand-seq was originally developed to detect sister chromatid exchanges (SCEs), but Strand-seq is also suitable for the genome-wide detection of copy number changes based on read depth and the detection of copy neutral structural variants based on the strand inheritance patterns between daughter cells (29–31). Strand-seq was performed on both blastomeres of two-cell–stage embryos [~28 hours post-fertilization (hpf)] produced with sperm that was untreated or subjected to a low (2.5 Gy) or high (10 Gy) dose of radiation. All IVF experiments were performed with cryopreserved sperm that was derived from the same bull to minimize the variation between IVF experiments. The sequenced libraries of 47 individual blastomeres derived from 26 two-cell–stage control embryos and 73 individual blastomeres of 47 two-cell–stage embryos produced with damaged sperm passed quality control and were further analyzed (Fig. 1, B and C, and table S1). Sister cells displayed the expected complementary strand inheritance patterns, with SCE events visible as template switches that are mirrored between both sister cells (Fig. 1D). Read depth–based copy number analysis revealed that ~10% of embryos produced with untreated sperm contained one or more copy number change due to a meiotic error, identified as variants shared between both sister cells (Figs. 2 and 3A), which is consistent with previous reports (24, 32). Approximately 29% of the two-cell embryos produced with untreated sperm showed defects due to mitotic errors, i.e., variants that are not shared between both sister cells (Fig. 3A). Strikingly, most of the embryos produced with damaged sperm showed multiple whole chromosome and segmental gains and losses, with the number of aberrations increasing in a dose-dependent manner (Figs. 1D and 3B). As a result, there were significantly more mosaic embryos in the 2.5-Gy and 10-Gy groups compared with the controls (respective P values of 0.0007 and 0.004, two-tailed Fisher’s exact test). Most of the detected abnormalities were reciprocal between sister cells, with chromosomes or chromosomal segments gained in one cell being lost in its sister cell. These reciprocal aberrations result in an average disomic copy number state in the embryo as a whole, a phenomenon we refer to as mirrored mosaicism (Figs. 1D and 3C). As a consequence, most variants would have been missed if the cells had not been sequenced individually but in “bulk” instead. Bulk WGS of the sperm DNA enabled identification of paternal single-nucleotide variants (SNVs) and haplotyping of the embryonic single-cell sequencing data (Fig. 3D). This analysis revealed that mitotically derived copy number alterations in the treatment groups were, as expected, strongly biased toward the γ-radiation–exposed paternally derived chromosomes (Fig. 3, D to G). In contrast, meiotic errors leading to aneuploidies shared by all blastomeres from the same embryo were biased toward maternal chromosomes (Fig. 3H), which is in line with previous observations in human embryos (33). Together, these results indicate that postmeiotic sperm DNA damage mainly affects the paternally derived chromosomes in the embryo, which become fragmented followed by the distribution of the chromosomal fragments over both daughter cells during the first embryonic cell division.
Fig. 2. Meiotic errors in early embryos.
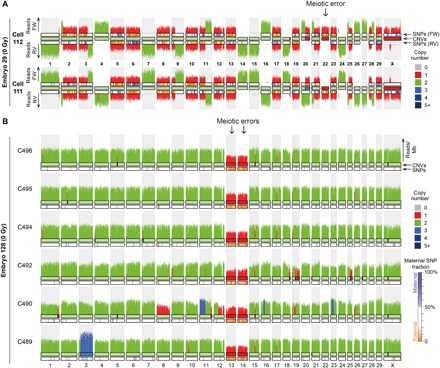
(A) Example of a two-cell embryo (E29) analyzed with Strand-seq containing a loss of chromosome 22 due to a meiotic error in the maternal germline. The remaining chromosome 22 is enriched for paternal SNVs. The embryo also contains a reciprocal mitotic copy number change on chromosome 20. (B) Karyogram of six sequenced cells from one embryo showing meiotic losses of chromosomes 13 and 14. Only the paternally inherited copies of chromosome 13 and 14 are present, indicating that the meiotic errors occurred on the maternal alleles.
Fig. 3. Sperm DNA damage leads to fragmentation of paternally derived chromosomes in early embryos.
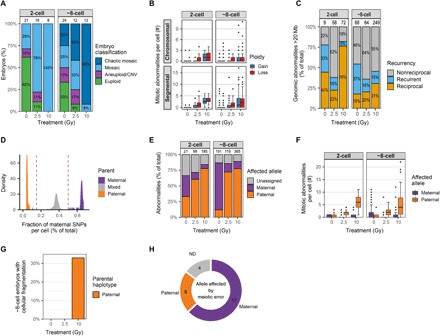
(A) Classification of all the embryos for the different treatment groups. Most of the embryos derived from fertilization with damaged sperm are mosaic, containing more than a single genotype. Embryos containing cells with a mix of more than three different genotypes are considered chaotic mosaic. The number of analyzed embryos per group is indicated above the bars. (B) Radiation dosage-dependent increase in the number of whole-chromosome and segmental gains and losses per cell in two- and eight-cell–stage embryos. (C) Many genomic abnormalities (>20 Mb in size) are mirrored between cells, showing reciprocal gains and losses between at least two cells. The relatively high number of nonreciprocal abnormalities, i.e., restricted to one cell per embryo, is largely due to variation in copy number variant (CNV) calls between cells and, in some cases, due to missing or excluded cells. Numbers above the bars indicate the number of unique CNVs per group. (D) Bulk WGS enabled the detection of homozygous SNV positions in the genome of the bull whose sperm was used for all IVF experiments. SNVs in the blastomeres that are different from the SNVs in the father are considered to be maternally inherited SNVs. SNVs overlapping between the father and the blastomeres can be paternally or maternally inherited (if the mother has the same SNV). Figure represents aggregated data from all embryos. (E) Most of the copy number changes (>10 Mb) in embryos derived from fertilization with damaged sperm are located on alleles inherited from the father. Numbers above the bars indicate the number of analyzed copy number abnormalities per group. (F) Sperm DNA damage leads to genomic abnormalities (>10 Mb) on the paternally inherited chromosomes. (G) Quantification of the eight-cell–stage embryos containing fragmented cells. Around a third of the eight-cell–stage embryos produced with 10 Gy–treated sperm contain fragmented cells with only paternally inherited chromosomes. (H) Number of meiotic errors (>10 Mb) on maternally and paternally inherited chromosomes. The copy number changes on the maternal alleles are largely caused by meiotic errors and are shared among all blastomeres from the same embryo. ND, not determined.
Sperm DNA damage leads to chaotic mosaicism
To examine the fate of the sperm-induced gains and losses, we performed single-cell WGS (30) of individual blastomeres at the about eight-cell stage of development (~48 hpf). Strand-seq is less efficient on eight-cell–stage embryos, because it depends on BrdU incorporation for exactly one cell cycle, but the cell cycles of all the blastomeres within a four-cell–stage embryo may not be precisely synchronized. Therefore, we initially performed regular single-cell WGS, which is equally suited for read depth–based copy number analysis. In total, the genomes of 302 individual blastomeres of 48 embryos were successfully sequenced (Fig. 1, B and C, and table S1). Copy number alterations were frequently observed, and many were either shared or, as observed in the two-cell–stage embryos, reciprocal between blastomeres from the same embryo (Figs. 3C and 4A). Embryos derived from fertilization with irradiated sperm contained fewer euploid cells and more cells with complex rearrangements, i.e., affecting at least three chromosomes. For each condition, the average number of chromosomal aberrations per cell was similar between the two-cell stage and the eight-cell stage (Fig. 3F), which indicates no further fragmentation of chromosomes from the two-cell stage onward. However, many eight-cell embryos contained cells representing more than three different genotypes, particularly in the embryos derived from damaged sperm (Fig. 4, A and B). In the control group, most of the embryos were composed of blastomeres that shared the same karyotype, while most of the embryos from the experimental conditions contained four or more different karyotypes (Fig. 4B). These results indicate that sperm DNA damage induces progressive genomic instability through mitotic errors.
Fig. 4. Chaotic mosaicism in eight-cell–stage embryos produced with damaged sperm.
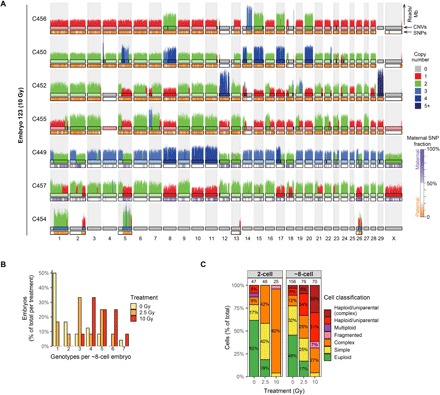
(A) Standard single-cell WGS derived copy number profiles of seven cells from embryo E123 showing complex genomic abnormalities. Cells C450, C452, C455, and C456 are uniparental with only paternal chromosomes, whereas C449 and C457 are biparental. C454 is a fragmented cell containing chromosomal fragments that are complementary to copy number losses in C449 and C452. The insides of the top chromosome ideograms are color-coded to indicate copy number (see legend for the copy number states). The ideograms for the bottom chromosome ideograms are color-coded to indicate the contribution of maternal SNVs (see legend for details). (B) Embryos produced with damaged sperm frequently show more than three different genetic lineages around the eight-cell stage of development, indicative of genomic instability through mitotic errors. (C) Classification of all the cells for the different treatment groups. The proportion of cells with multiple genomic abnormalities increases with sperm radiation dose. Cells without chromosomal or segmental abnormalities are classified as euploid. Cells with one or two chromosomal or segmental abnormalities are classified as simple. Cells with three or more chromosomal or segmental abnormalities are classified as complex. Fragmented cells only contain a few chromosomal fragments. Numbers above the bars indicate the number of analyzed cells per group.
Chaotic mosaicism, where blastomeres of the same embryo have distinct karyotypes, was common in embryos produced with irradiated sperm, with significantly more chaotically mosaic embryos in the 10-Gy groups compared with the controls (P = 0.0002, two-tailed Fisher’s exact test; Fig. 3A). The variety of genomic abnormalities ranged from aneuploidies, segmental changes, abnormal ploidy states, to cells containing minimal chromosomal content restricted to a few chromosomal fragments (Fig. 4, A and C). To further investigate the processes that contribute to chaotic mosaicism, we performed Strand-seq on individual blastomeres of 12 about eight-cell–stage embryos produced with damaged sperm. Although Strand-seq is less efficient on eight-cell–stage embryos, successful Strand-seq libraries on several eight-cell–stage embryos could be produced facilitating lineage reconstruction of chaotically mosaic embryos. From the strand inheritance patterns of two of these embryos, we could deduce that seven cells were formed by direct unequal cleavage of both blastomeres of a two-cell–stage embryo that cleaved into three and four cells, respectively (Fig. 5). These observations indicate that sperm DNA damage can cause aberrant cleavage divisions at the two-cell–stage embryo, resulting in chaotic mosaicism at later stages. In the Strand-seq libraries, we also observed sister cells that inherited complementary acentric fragments, suggesting that these fragments have been translocated to centromere-containing chromosomes, enabling their segregation upon replication (e.g., the fragments of acrocentric chromosome 11 in cells 828 and 833 of embryo 170 in Fig. 5A and the fragments of chromosome 16 in cells 806 and 807 of embryo 167 in Fig. 5B).
Fig. 5. Direct unequal cell divisions at the two-cell stage cause chaotic mosaicism.
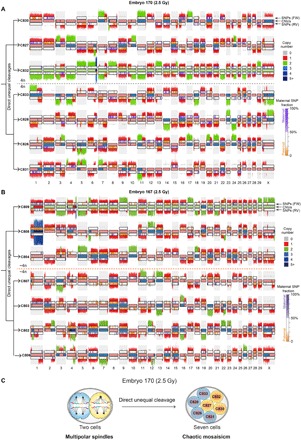
(A) Strand-seq karyogram of seven-cell embryo (E170) showing chaotic mosaicism after direct unequal cleavage divisions at the two-cell stage. The strand inheritance patterns detected by Strand-seq enable the identification of sister cells, the deduction of the preceding division, and the distribution of the chromosomal fragments. This analysis reveals that both blastomeres at the two-cell stage performed a multipolar division; a tripolar division generated the sister cells C830, C827, and C832; and a tetrapolar division generated the sister cells C833, C828, C826, and C831. This led to the random distribution of the tetraploid set of chromosomes over the sister cells. As a consequence, the DNA fragments distributed over the sister cells sum up to a 4n copy number state. (B) Karyogram of a seven-cell embryo (E167) analyzed by Strand-seq showing the results of direct unequal cell divisions at the two-cell stage. The strand inheritance patterns indicate that cells C809, C808, and C804 are sister cells, as are cells C807, C803, C802, and C806. The DNA fragments distributed over the sister cells sum up to a 4n copy number state. (C) Schematic reconstruction of the direct unequal cleavage divisions of a two-cell–stage embryo. The blastomeres of a two-cell–stage embryo [embryo 170 produced with 2.5 Gy–treated sperm, whose Strand-seq ideogram is shown in (A)] cleaved directly into three and four daughter cells. Sister cells are depicted with the same color.
Sperm DNA damage induces heterogoneic and direct unequal cleavage divisions
Strikingly, a large number of cells from eight-cell–stage embryos produced with damaged sperm lacked X chromosomes or contained nullisomies, indicating that these cells are (near) haploid or uniparental (Figs. 4A and 6). To accurately quantify the number of haploid and uniparental cells, we screened for cells that lack heterozygous SNVs. Only a few cells are haploid and/or uniparental in two-cell–stage embryos and in control eight-cell–stage embryos (Figs. 3D and 6B). In contrast, the proportion of haploid/uniparental cells in eight-cell–stage embryos produced with damaged sperm increased with radiation dose, amounting to nearly two-thirds of the cells in the embryos produced with 10 Gy–treated sperm (Figs. 3D and 6B). Of these embryos with haploid cells, most of the embryos were either fully haploid/uniparental or half of the blastomeres (Fig. 6C).
Fig. 6. Sperm DNA damage induces heterogoneic and direct unequal cleavage divisions.
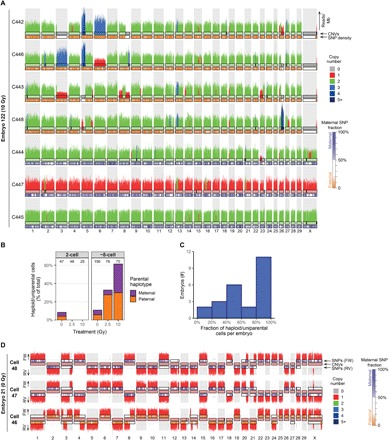
(A) Example karyogram of a seven-cell embryo (E122) produced with damaged sperm containing a segregation of uniparental maternal and paternal cells, suggesting a heterogoneic cell division of the zygote. Copy number changes are mostly present in the cells containing a paternal genome (C442, C443, C446, and C448), and cells with maternally inherited chromosomes are relatively unaffected by copy number changes, in contrast to the cells with paternally inherited chromosomes. (B) Embryos generated with damaged sperm contain more haploid and uniparental cells having a genomic content from either the father or the mother, indicating that sperm DNA damage causes heterogoneic cell divisions. Numbers above the bars indicate the number of analyzed cells per group. (C) In many embryos, only half of the cells are haploid/uniparental, suggesting that, in some cases, haploid/uniparental cells may arise after the two-cell stage. (D) Example of three haploid cells originating from a heterogoneic cell division at the zygote stage, which lead to segregation of the paternal and maternal genomes. Sister cells C47 and C48 contain a haploid maternal genome. The sister cell of C46 may have been lost during collection of the single blastomeres.
A study on bovine IVF embryos described complete segregation of the haploid maternal and paternal genomes through so-called heterogoneic cell divisions, which were hypothesized to be the result of direct unequal cleavage of the zygote (24). To examine whether this process can lead to heterogoneic cell divisions, we sequenced all blastomeres from nine embryos containing three cells that were formed after a direct unequal division of the zygote. In three of nine three-cell embryos (two control embryos and one from irradiated sperm), all sequenced blastomeres were haploid (Fig. 6D). These observations indicate that unequal cleavages of zygotes can lead to heterogoneic cell divisions yielding uniparental lineages.
DISCUSSION
Our study demonstrates that sperm DNA damage leads to the fragmentation and random distribution of paternal chromosome segments over both sister cells of two-cell–stage embryos. In addition, embryos that have been derived from fertilization with damaged sperm are prone to direct unequal cleavage divisions, leading to the formation of haploid and uniparental cells. This leads to chaotic mosaicism at the eight-cell stage, with blastomeres displaying a variety of genomic abnormalities ranging from aneuploidies, segmental changes, abnormal ploidy states, to cells containing minimal chromosomal content restricted to a few chromosomal fragments, thereby covering the broad spectrum of chromosomal aberrations that have been previously described in human, primate, and bovine embryos (6, 24, 32). Since sperm DNA damage has adverse effects on fertility (34), human IVF embryos may also be biased toward being produced with damaged sperm. Notably, in a recent study in rhesus macaque embryos, chaotic aneuploidy was correlated with one particular sperm donor (32), which may indicate that sperm DNA damage is the underlying cause. Chaotic mosaicism is also common in human embryos produced with sperm from men with nonobstructive azoospermia, a condition that is also associated with high levels of sperm DNA damage (35). Complex abnormal mosaic embryos have reduced implantation and clinical pregnancy rates and reduced chances to develop to term (8). Chaotic mosaicism thus appears to be the responsible intermediate step for the well-established correlation between sperm DNA damage and reduced fertility (34, 36).
The sperm that was used in the current study is from a single breeding bull of proven fertility and good health characteristics. This is reflected by the relatively high number of euploid cells in two-cell–stage control embryos (Fig. 3A), allowing to detect a signal of the treatment above the background noise. In theory, it could be argued that the observed effects may not be representative for the effects of γ-radiation on sperm of other bulls. Although we cannot formally exclude this possibility, we consider this unlikely because sperm cells lack transcription, translation, and DNA repair activity, which makes it unlikely that sperm cells from another bull would respond differently to this type of physical DNA damage. The observed effects of broken chromosomes at the two-cell stage are also in line with the well-known consequences of ionizing radiation on the DNA, leading to double-strand breaks, as has been demonstrated in a plethora of cell types including sperm cells. The genomic instability observed in later developmental stages is probably caused by the response of the zygote to the damaged paternal chromatin and not the result of bull-specific effects of radiation of the sperm cells.
We observed a marked increase in the number of haploid/uniparental cells in the embryos produced with damaged sperm. In four embryos of the 2.5-Gy group (E150, E158, E159, and E160), we observed haploid cells of paternal origin, but not of maternal origin. Possibly, these are the result of dispermic fertilizations, where one of the paternal genomes has been allocated to a separate lineage. We did not find indications for an elevated level of dispermic fertilizations in the 10-Gy group, which contains a 50/50 contribution of maternal and paternal haploid cells, while dispermic embryos are expected to be biased toward paternal haploid cells. In contrast, this increase in haploid/uniparental cells appears to result from direct unequal cleavages through so-called heterogoneic cell divisions. The number of direct unequal cleavage divisions at the zygote stage seems to be insufficient to account for all the observed haploid and uniparental cells at the eight-cell–stage embryos derived from fertilizations with damaged sperm. Because parental genomes still occupy distinct territories at the two-cell stage (37, 38), uniparental and haploid cells may also be formed by unequal cleavages at this stage of development. The observation that frequently only half of the cells of eight-cell–stage embryos were haploid/uniparental may indicate heterogoneic cell divisions of two-cell–stage blastomeres. Although we did not find further evidence to support heterogoneic divisions of two-cell–stage blastomeres, our Strand-seq data of eight-cell–stage embryos provides strong evidence for direct unequal cleavage divisions at the two-cell stage, which is in line with observations in live-cell imaging experiments (32).
In mouse zygotes, two spindles are formed, one for each haploid parental genome, and the dual spindles are aligned before the first cleavage division (37). Sperm DNA damage may interfere with this process, thereby inducing heterogoneic cell divisions of the zygote (24). As an alternative explanation, γ-radiation of sperm may damage the centrioles, which are exclusively paternally inherited in bovine and human zygotes (39). While we cannot formally exclude this possibility, we think that it is unlikely that this could explain the high frequency of haploid cells observed at the eight-cell stage. First, our observations demonstrate that γ-radiation almost exclusively affects the paternal chromosomes in the embryo, while centriolar defects would also lead to segregation defects of maternal chromosomes. Second, γ-radiation itself does not impair other aspects of sperm function such as sperm morphology and motility or the capacity to fertilize oocytes (23). In our opinion, there is no good argument for specific γ-radiation–induced damage to the relatively small centrioles, while other essential organelles remain unaffected. Chromatin constitutes the largest volume of the sperm cell, and γ-radiation is well known for its DNA-damaging properties. The sperm DNA damage itself may then, after fertilization, induce centrosome amplification via the DNA damage response, as is the case in cancer cells that have been exposed to ionizing radiation or cytostatic drugs (40). Another possibility is that sperm DNA damage may induce chromosome misalignments, which may disturb the integrity of the spindle poles without centrosome amplification (41). It is also possible that the haploid phenotypes and chromosomal rearrangements are mediated through yet unknown processes.
Mature sperm cells lack mechanisms of DNA repair and depend on maternal factors for repair that are only available after fertilization (42). By the absence of homologous templates, zygotic repair of paternal double-strand breaks depends on nonhomologous repair mechanisms that are considered error-prone, generating structural variation when originally distal fragments are joined. Pathogenic de novo structural variation that causes severe intellectual disability and other congenital anomalies is mostly of paternal origin (43), as is the case for the copy number alterations that were observed in the current study. While our single-cell sequencing data are not suited for the reliable detection of balanced structural variation, we observed sister cells that inherited acentric fragments in the Strand-seq libraries, suggesting that these fragments have been translocated to other chromosomes. These observations are in line with previous studies that described structural changes to the paternal genome upon DNA damage (44).
Several techniques are available to measure the degree of DNA damage in a population of sperm cells such as the comet assay, the sperm chromatin dispersion assay, terminal deoxyuridine nick end labeling (TUNEL) assay, and sperm chromatin structure assay (45). However, the sensitivity of most of these assays is limited. Even the comet assay, which is considered the most sensitive method to detect sperm DNA damage, has an estimated lower bound of 100 double-strand breaks per cell (45). On the basis of the number of foci of the phosphorylated form of histone H2AX, mouse zygotes produced with 3-Gy γ-irradiated sperm have, on average, about five double-strand breaks (46). Our results from the 2.5-Gy group demonstrate that this mild induction of sperm DNA damage can induce genomic instability, although the number of double-strand breaks is far below the detection limit of the comet assay. In addition, note that it is inherently impossible to know the degree of DNA damage of the individual sperm cell that was used to fertilize an oocyte. Further experiments will be required to establish if our bovine results are replicated in human to understand whether fertilizations with damaged sperm contribute to the widespread genomic instability in human embryos (6).
Here, we have shown that sperm DNA damage induces fragmentation of chromosomes and segregation errors through direct unequal cleavages. A consequence of these two processes is chaotic mosaicism of embryos. In support with our findings, chaotic mosaicism is also common in human embryos produced with sperm from men with nonobstructive azoospermia, a condition that is also associated with high levels of sperm DNA damage (7). Complex abnormal mosaic embryos have reduced implantation and clinical pregnancy rates and reduced chances to develop to term (8). Chaotic mosaicism thus appears to be responsible for the well-established correlation between sperm DNA damage and reduced fertility (34, 36). The chromosomal aberrations that are induced by damaged sperm include de novo structural variation, uniparental disomies, mosaic aneuploidies, and mixoploidy, all of which have been observed in human embryos. In addition to their negative impact on fertility, these aberrations may contribute to congenital disease when embryos escape developmental arrest (47–49). To improve embryo selection, time-lapse microscopy (32) can be used to identify embryos that undergo direct unequal cleavages, induced either by sperm DNA damage or by other factors.
MATERIALS AND METHODS
The aim of this study was to determine the consequences of sperm DNA damage on the embryonic genome. Bovine embryos were produced with sperm damaged by different doses of γ-radiation. At the two-cell and eight-cell stages of development, the individual blastomeres of the resulting embryos were collected and subjected to single-cell WGS. Two different single-cell sequencing technologies were used: standard WGS and Strand-seq. Both techniques were used to detect read depth–based differences in copy number state. Strand-seq was also used for the accurate reconstruction of multipolar cell divisions.
Bovine IVF and blastomere collection
Fertilization and embryo culture were performed, according to previously described procedures (50). In short, bovine cumulus oocyte complexes were aspirated from 2- to 8-mm antral follicles of slaughterhouse ovaries from 2- to 6-year-old adult cows. Subsequently, germinal vesicle stage oocytes with an intact multilayered cumulus were selected and matured in M199 supplemented with 26.2 mM NaHCO3, recombinant human follicle-stimulating hormone (0.05 IU/ml; Organon, Oss, The Netherlands), 10% fetal bovine serum (Gibco-BRL), and 1% (v/v) penicillin-streptomycin (Gibco-BRL) at 39°C in a humidified atmosphere of 5% CO2 in air. IVF was performed at 23 hours after maturation with 0.5 × 106 sperm cells/ml sperm. To obtain sperm with damaged DNA, sperm straws were subjected to ionizing radiation from Gammacell 1000 (Atomic Energy of Canada Limited, Mississauga, Southern Ontario, Canada) before IVF. Ionizing radiation allows induction of DNA damage on noncycling sperm cells while maintaining accurate control over the dosage. Untreated sperm from the same bull was used for the control group. All experiments were performed with sperm from the same donor bull to control for the potential natural variation in DNA damage between individuals. At 18 to 22 hours after sperm addition, the cumulus cells and adhering sperm cells were removed, and the denuded zygotes were further cultured in synthetic oviductal fluid (SOF) in a humidified incubator at 39°C with 5% CO2 and 7% O2. To obtain blastocysts, cleaved embryos were transferred to fresh SOF at day 5 and cultured until day 8. For Strand-seq experiments at the two-cell stage, BrdU was added to the fertilization medium and the embryo culture medium from the start of the embryo culture. Blastomeres were collected from 28 hpf onward. For Strand-seq experiments at the eight-cell stage, four-cell–stage embryos (at 29 to 33 hpf) were transferred to medium containing BrdU and cultured until the eight-cell stage (at 48 hpf) when individual blastomeres were collected. For single-cell WGS of eight-cell–stage embryos, embryos were cultured in medium without BrdU and blastomeres were collected from 48 hpf onward. To collect individual blastomeres, embryos were placed in a droplet of 0.1% protease in phosphate-buffered saline (PBS) with 0.05% polyvinyl alcohol. After the zona pellucida was dissolved, the embryos were transferred to a droplet of trypsin-EDTA to dissociate the blastomeres. Blastomeres were transferred to single wells from a 96-well plate containing 5-μl cryoprotectant consisting of 50% PBS, 42.5% ProFreeze (Lonza), and 7.5% dimethyl sulfoxide. Full plates were stored at −80°C until further processing. Initially, plates contained no-cell, empty well negative controls and multiple cell positive controls. The negative controls resulted in empty libraries with no reads mapping to the bovine genome. All plates also contained some cell-containing wells that yielded empty libraries. These also served as negative controls, because they confirmed that the ingredients for the library preps were clean.
Single-cell genome sequencing and primary data processing
Strand-seq and single-cell WGS libraries were generated as previously described by Falconer et al. (29) and van den Bos et al. (30), respectively. Libraries were pooled (192 libraries per rapid run flow cell lane) and sequenced on the Illumina HiSeq 2500 sequencing platform. Raw sequencing reads were mapped to the Bos taurus UMD3.1 (bt8) reference genome using Bowtie2 (51), and BamUtil was used to filter duplicated reads. The median read count was 692,678 reads per cell after primary data processing (table S1).
Single-cell copy number variant calling and filtering
The BAM files for all single-cell libraries were merged to generate a composite BAM file using Samtools merge. Bedtools intersect was used to calculate the coverage per 100-kb genomic bins. A blacklist for copy number variant (CNV) calling (included with the scripts) was generated by selecting the 3% bins with the highest and 2% of the bins with the lowest read counts on the autosomes and the bins with the top 5% and bottom 3% read counts on the X chromosome. The R package AneuFinder (v1.8.0) was used to count the reads (with a minimal mapping quality of 10) in fixed-width bins of 1 Mb and to call CNVs using the “edivisive” method (36). The genomic sequence provided by the R package BSgenome.Btaurus.UCSC.bosTau8 (v1.4.2) was used for GC correction applied by AneuFinder. CNV calls with a limited change in read count compared to the median read count per bin per cell were excluded (decrease of <25% for presumed losses and increase of <25% for gains). Subsequently, CNV calls for each cell were merged on the basis of a variable overlap threshold dependent on CNV size into one CNV call set per embryo (e.g., larger CNVs require a higher percentage of overlap to merge than smaller CNVs). CNV calls occurring in more than 15% of the high-quality control libraries (with more than 200,000 reads), which likely correspond to common population variants or reference genome artifacts, were removed from the call sets. CNVs were considered to be reciprocal if there is at least one gain and one loss at a genomic location in the embryo. Sequenced libraries with more than 100,000 reads, 10 or less filtered chromosomal or segmental abnormalities, less than 80 genomic segments detected by AneuFinder, and, if applicable, alternating Watson/Crick strand inheritance patterns (whose mother cell incorporated BrdU during replication and underwent mitosis) were used as high-quality libraries for further analyses (table S1).
Bulk WGS of bovine sperm DNA
Sperm DNA was extracted with the guanidine thiocyanate method (52). A Covaris sonicator was used to shear the isolated DNA to fragments of 400 to 500 base pairs. Libraries for WGS were prepared using the TruSeq DNA Nano Library Prep Kit (Illumina) according to the manufacturer’s protocol. Paired-end 2 × 150–base pair read WGS (2 × 150–base pair reads) was performed on an Illumina HiSeq X sequencer to a mean genome coverage depth of 34×. Reads were aligned to the B. taurus UMD3.1 reference genome using BWA-0.7.5a with settings BWA-MEM -t 12 -c 100 -M -R (53). Reads were realigned with GATK IndelRealigner (54), and duplicate reads were flagged with Sambamba markdup (55).
SNV genotyping of sperm and blastomere DNA
All nonreference SNVs were called from the composite BAM file (containing all the reads from the sequenced single-cell libraries) using bcftools mpileup and bcftools call (56). All heterozygous SNVs with more than two reference and two alternative allele counts and with a maximum coverage depth of 50 were selected to generate a list of 2,626,948 embryonic SNVs. Subsequently, the paternal sperm WGS data were genotyped for the embryonic SNV positions using bcftools. To enable classification of SNVs in single embryonic cells as maternal (nonpaternal), only the SNV positions that are homozygous in the father (with a coverage depth between 10 and 75 in the sperm WGS data) were selected. All the single cells were genotyped for these 912,144 homozygous sperm SNV positions using bcftools. An SNV was classified as maternally inherited if the genotype is different from the homozygous genotype in the father.
Determination of the ploidy status of single blastomeres
Haploid and uniparental cells were identified on the basis of several parameters. First, all cells were genotyped for the 2,626,948 variable embryonic SNV positions in the composite BAM file (see above). Cells were considered to be uniparental if less than 15% or more than 50% of the called SNVs in the cell were different from the homozygous SNVs in the father (Fig. 3D). In addition, haploid cells were detected by a loss of heterozygous SNV positions. Haploid/uniparental cells with more than 3000 covered SNVs were required to have less than one heterozygous REF/ALT SNV (excluding SNVs overlapping copy number gains) per 1000 called homozygous SNVs. Strand-seq libraries of haploid cells were recognized by the absence of bins with reads on both the Watson and Crick strands (haploid cells should only contain reads on one strand per bin after Strand-seq). Cells classified as haploid/uniparental were considered to be haploid (with a copy number state of one) if the majority (>80%) of called copy number losses are nullisomies.
Classifications of individual blastomeres and embryos
Cells were classified on the basis of their ploidy status and the number of segmental and whole-chromosome copy number changes. Cells containing three or more segmental or whole-chromosome abnormalities were classified as complex. Cells with more than 10,000 reads and more than 25% of their reads on a single chromosome were considered to be fragmented. To determine the presence of different genotypes within each embryo, copy number changes (>20 Mb) were compared between cells. Cells sharing more than 75% of their CNVs are considered to be of the same genotype. Embryos containing more than one or more than three different genotypes are classified as mosaic and chaotic mosaic, respectively.
Supplementary Material
Acknowledgments
We thank W. Kloosterman for helpful discussions and R. Janssen for bioinformatics support. We would also like to thank the Hartwig Medical Foundation for WGS. Funding: This work was supported by the funding provided by the Netherlands Science Foundation (NWO) Vici grant (865.12.004) to E.C. and provided by De Snoo-van ‘t Hoogerhuijs Stichting to E.W.K. Author contributions: H.T.A.v.T. and E.W.K. performed wet-lab experiments. D.C.J.S., V.G., and P.M.L. performed single-cell sequencing. S.M., H.T.A.v.T., D.C.J.S., and E.W.K. performed data analysis. S.M., H.T.A.v.T., D.C.J.S., B.A.J.R., P.M.L., E.C., and E.W.K. were involved in the conceptual design of the study. P.M.L., E.C., and E.W.K. acquired financial support for the study. S.M., E.C., and E.W.K. wrote the manuscript with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All sequencing data have been deposited in the European Nucleotide Archive (www.ebi.ac.uk/ena) under accession number PRJEB32696. Custom code used in this study is available on GitHub (https://github.com/UMCUGenetics/Bovine_Embryo/).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaaz7602/DC1
REFERENCES AND NOTES
- 1.Kiessling A. A., Bletsa R., Desmarais B., Mara C., Kallianidis K., Loutradis D., Evidence that human blastomere cleavage is under unique cell cycle control. J. Assist. Reprod. Genet. 26, 187–195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braude P., Bolton V., Moore S., Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Toyoshima M., Analysis of p53 dependent damage response in sperm-irradiated mouse embryos. J. Radiat. Res. 50, 11–17 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Taylor T. H., Gitlin S. A., Patrick J. L., Crain J. L., Wilson J. M., Griffin D. K., The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum. Reprod. Update 20, 571–581 (2014). [DOI] [PubMed] [Google Scholar]
- 5.van Echten-Arends J., Mastenbroek S., Sikkema-Raddatz B., Korevaar J. C., Heineman M. J., van der Veen F., Repping S., Chromosomal mosaicism in human preimplantation embryos: A systematic review. Hum. Reprod. Update 17, 620–627 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Vanneste E., Voet T., le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F., Fryns J.-P., Verbeke G., D’Hooghe T., Moreau Y., Vermeesch J. R., Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15, 577–583 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Macklon N. S., Geraedts J. P. M., Fauser B. C. J. M., Conception to ongoing pregnancy: The ‘black box’ of early pregnancy loss. Hum. Reprod. Update 8, 333–343 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Spinella F., Fiorentino F., Biricik A., Bono S., Ruberti A., Cotroneo E., Baldi M., Cursio E., Minasi M. G., Greco E., Extent of chromosomal mosaicism influences the clinical outcome of in vitro fertilization treatments. Fertil. Steril. 109, 77–83 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Kaiser-Rogers K. A., McFadden D., Livasy C. A., Dansereau J., Jiang R., Knops J. F., Lefebvre L., Rao K. W., Robinson W. P., Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J. Med. Genet. 43, 187–192 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makrydimas G., Sebire N. J., Thornton S. E., Zagorianakou N., Lolis D., Fisher R. A., Complete hydatidiform mole and normal live birth: A novel case of confined placental mosaicism: Case report. Hum. Reprod. 17, 2459–2463 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Robinson W. P., Lauzon J. L., Innes A. M., Lim K., Arsovska S., McFadden D. E., Origin and outcome of pregnancies affected by androgenetic/biparental chimerism. Hum. Reprod. 22, 1114–1122 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Strain L., Warner J. P., Johnston T., Bonthron D. T., A human parthenogenetic chimaera. Nat. Genet. 11, 164–169 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Schneider I., Ellenberg J., Mysteries in embryonic development: How can errors arise so frequently at the beginning of mammalian life? PLOS Biol. 17, e3000173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vorsanova S. G., Kolotii A. D., Iourov I. Y., Monakhov V. V., Kirillova E. A., Soloviev I. V., Yurov Y. B., Evidence for high frequency of chromosomal mosaicism in spontaneous abortions revealed by interphase FISH analysis. J. Histochem. Cytochem. 53, 375–380 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Baart E. B., Martini E., Eijkemans M. J., Van Opstal D., Beckers N. G. M., Verhoeff A., Macklon N. S., Fauser B. C. J. M., Milder ovarian stimulation for in-vitro fertilization reduces aneuploidy in the human preimplantation embryo: A randomized controlled trial. Hum. Reprod. 22, 980–988 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Bean C. J., Hassold T. J., Judis L., Hunt P. A., Fertilization in vitro increases non-disjunction during early cleavage divisions in a mouse model system. Hum. Reprod. 17, 2362–2367 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Verpoest W., Fauser B. C., Papanikolaou E., Staessen C., van Landuyt L., Donoso P., Tournaye H., Liebaers I., Devroey P., Chromosomal aneuploidy in embryos conceived with unstimulated cycle IVF. Hum. Reprod. 23, 2369–2371 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Antonarakis S. E., Avramopoulos D., Blouin J.-L., Talbot C. C. Jr., Schinzel A. A., Mitotic errors in somatic cells cause trisomy 21 in about 4.5% of cases and are not associated with advanced maternal age. Nat. Genet. 3, 146–150 (1993). [DOI] [PubMed] [Google Scholar]
- 19.McCoy R. C., Demko Z., Ryan A., Banjevic M., Hill M., Sigurjonsson S., Rabinowitz M., Fraser H. B., Petrov D. A., Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 348, 235–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vázquez-Diez C., FitzHarris G., Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction 155, R63–R76 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Colaco S., Sakkas D., Paternal factors contributing to embryo quality. J. Assist. Reprod. Genet. 35, 1953–1968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González-Marín C., Gosálvez J., Roy R., Types, causes, detection and repair of DNA fragmentation in animal and human sperm cells. Int. J. Mol. Sci. 13, 14026–14052 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fatehi A. N., Bevers M. M., Schoevers E., Roelen B. A. J., Colenbrander B., Gadella B. M., DNA damage in bovine sperm does not block fertilization and early embryonic development but induces apoptosis after the first cleavages. J. Androl. 27, 176–188 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Destouni A., Zamani Esteki M., Catteeuw M., Tšuiko O., Dimitriadou E., Smits K., Kurg A., Salumets A., van Soom A., Voet T., Vermeesch J. R., Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 26, 567–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tšuiko O., Catteeuw M., Zamani Esteki M., Destouni A., Bogado Pascottini O., Besenfelder U., Havlicek V., Smits K., Kurg A., Salumets A., D’Hooghe T., Voet T., Van Soom A., Robert Vermeesch J., Genome stability of bovine in vivo-conceived cleavage-stage embryos is higher compared to in vitro-produced embryos. Hum. Reprod. 32, 2348–2357 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Bolton H., Graham S. J. L., van der Aa N., Kumar P., Theunis K., Fernandez Gallardo E., Voet T., Zernicka-Goetz M., Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 7, 11165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graf A., Krebs S., Heininen-Brown M., Zakhartchenko V., Blum H., Wolf E., Genome activation in bovine embryos: Review of the literature and new insights from RNA sequencing experiments. Anim. Reprod. Sci. 149, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Lomax M. E., Folkes L. K., O’Neill P., Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin. Oncol. 25, 578–585 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Falconer E., Hills M., Naumann U., Poon S. S. S., Chavez E. A., Sanders A. D., Zhao Y., Hirst M., Lansdorp P. M., DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 9, 1107–1112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Bos H., Bakker B., Taudt A., Guryev V., Colomé-Tatché M., Lansdorp P. M., Foijer F., Spierings D. C. J., Quantification of aneuploidy in mammalian systems. Methods Mol. Biol. 1896, 159–190 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Bakker B., Taudt A., Belderbos M. E., Porubsky D., Spierings D. C. J., de Jong T. V., Halsema N., Kazemier H. G., Hoekstra-Wakker K., Bradley A., de Bont E. S. J. M., van den Berg A., Guryev V., Lansdorp P. M., Colomé-Tatché M., Foijer F., Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 17, 115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daughtry B. L., Rosenkrantz J. L., Lazar N. H., Fei S. S., Redmayne N., Torkenczy K. A., Adey A., Yan M., Gao L., Park B., Nevonen K. A., Carbone L., Chavez S. L., Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 29, 367–382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassold T., Hunt P., To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Evenson D. P., Darzynkiewicz Z., Melamed M. R., Relation of mammalian sperm chromatin heterogeneity to fertility. Science 210, 1131–1133 (1980). [DOI] [PubMed] [Google Scholar]
- 35.Magli M. C., Gianaroli L., Ferraretti A. P., Gordts S., Fredericks V., Crippa A., Paternal contribution to aneuploidy in preimplantation embryos. Reprod. Biomed. Online 18, 536–542 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Zhao J., Zhang Q., Wang Y., Li Y., Whether sperm deoxyribonucleic acid fragmentation has an effect on pregnancy and miscarriage after in vitro fertilization/intracytoplasmic sperm injection: A systematic review and meta-analysis. Fertil. Steril. 102, 998–1005.e8 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Reichmann J., Nijmeijer B., Hossain M. J., Eguren M., Schneider I., Politi A. Z., Roberti M. J., Hufnagel L., Hiiragi T., Ellenberg J., Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 361, 189–193 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Mayer W., Smith A., Fundele R., Haaf T., Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 148, 629–634 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fishman E. L., Jo K., Nguyen Q. P. H., Kong D., Royfman R., Cekic A. R., Khanal S., Miller A. L., Simerly C., Schatten G., Loncarek J., Mennella V., Avidor-Reiss T., A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 9, 2210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Löffler H., Fechter A., Liu F. Y., Poppelreuther S., Krämer A., DNA damage-induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene 32, 2963–2972 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Maiato H., Logarinho E., Mitotic spindle multipolarity without centrosome amplification. Nat. Cell Biol. 16, 386–394 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Marchetti F., Essers J., Kanaar R., Wyrobek A. J., Disruption of maternal DNA repair increases sperm-derived chromosomal aberrations. Proc. Natl. Acad. Sci. U.S.A. 104, 17725–17729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hehir-Kwa J. Y., Rodríguez-Santiago B., Vissers L. E., de Leeuw N., Pfundt R., Buitelaar J. K., Pérez-Jurado L. A., Veltman J. A., De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J. Med. Genet. 48, 776–778 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Searle A. G., Ford C. E., Evans E. P., Beechey C. V., Burtenshaw M. D., Clegg H. M., Papworth D. G., The induction of translocations in mouse spermatozoa I. Kinetics of dose response with acute x-irradiation. Mutat. Res. 22, 157–174 (1974). [DOI] [PubMed] [Google Scholar]
- 45.Zini A., Sigman M., Are tests of sperm DNA damage clinically useful? Pros and cons. J. Androl. 30, 219–229 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Derijck A. A., van der Heijden G. W., Giele M., Philippens M. E., van Bavel C. C., de Boer P. V., γH2AX signalling during sperm chromatin remodelling in the mouse zygote. DNA Repair 5, 959–971 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Conlin L. K., Thiel B. D., Bonnemann C. G., Medne L., Ernst L. M., Zackai E. H., Deardorff M. A., Krantz I. D., Hakonarson H., Spinner N. B., Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 19, 1263–1275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kloosterman W. P., Cuppen E., Chromothripsis in congenital disorders and cancer: Similarities and differences. Curr. Opin. Cell Biol. 25, 341–348 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Pellestor F., Gatinois V., Puechberty J., Geneviève D., Lefort G., Chromothripsis: Potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil. Steril. 102, 1785–1796 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Aardema H., van Tol H. T. A., Wubbolts R. W., Brouwers J. F. H. M., Gadella B. M., Roelen B. A. J., Stearoyl-CoA desaturase activity in bovine cumulus cells protects the oocyte against saturated fatty acid stress. Biol. Reprod. 96, 982–992 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffin J., Methods of sperm DNA extraction for genetic and epigenetic studies. Methods Mol. Biol. 927, 379–384 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/16/eaaz7602/DC1


