Summary
Cellular iron homeostasis is dominated by FBXL5-mediated degradation of iron regulatory protein 2 (IRP2), which is dependent on both iron and oxygen. However, how the physical interaction between FBXL5 and IRP2 is regulated remains elusive. Here we show that the C-terminal substrate-binding domain of FBXL5 harbors a [2Fe2S] cluster in the oxidized state. A cryo-EM structure of the IRP2-FBXL5-SKP1 complex reveals that the cluster organizes the FBXL5 C-terminal loop responsible for recruiting IRP2. Interestingly, IRP2 binding to FBXL5 hinges on the oxidized state of the [2Fe2S] cluster maintained by ambient oxygen, which could explain hypoxia-induced IRP2 stabilization. Steric incompatibility also allows FBXL5 to physically dislodge IRP2 from iron-responsive element-RNA to facilitate its turnover. Taken together, our studies have identified an iron-sulfur cluster within FBXL5, which promotes IRP2 polyubiquitination and degradation in response to both iron and oxygen concentrations.
Graphical Abstract
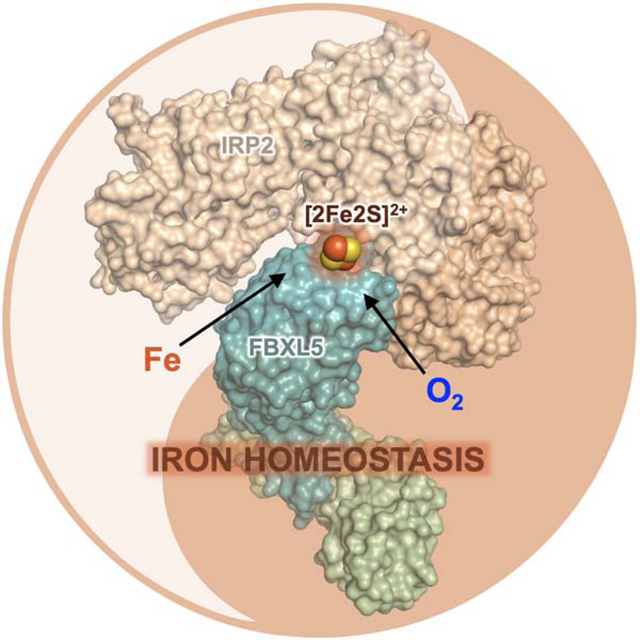
eTOC Blurb
Wang et al. discovered an unexpected [2Fe2S] cluster in FBXL5, which enables the F-box protein to promote iron- and oxygen-sensitive degradation of IRP2 in iron homeostasis regulation. The authors further determined the cryo-EM structure of the FBXL5-IRP2 complex and revealed the structural basis of [2Fe2S] cluster-mediated IRP2 recognition by FBXL5.
Introduction
Iron, an essential element of most life forms, is widely utilized by a variety of critical biological processes. These include, but are not limited to, respiration, DNA synthesis, oxygen transport, and reactive oxygen species (ROS) sensing (Abbaspour et al., 2014; Lieu et al., 2001). Iron is naturally exploited by the cell through its incorporation into numerous proteins, either directly or via cofactors such as heme and Fe-S clusters (Beard, 2001). Iron deficiency, which impairs the functions of iron-containing proteins, affects billions of people worldwide and leads to cognitive defects in children and anemia in adults. Iron overload, on the other hand, generates reactive radicals that oxidatively damage cellular components and is associated with hemochromatosis and neurodegenerative disorders, including Parkinson’s and Alzheimer’s diseases (Crielaard et al., 2017). Cellular iron levels, therefore, must be strictly maintained (Hentze et al., 2004; Hentze et al., 2010).
In mammals, cellular iron homeostasis is predominantly regulated at the post-transcriptional level. In response to low iron levels, Iron Regulatory Proteins 1 and 2 (IRP1 and 2) control the expression of a cohort of iron metabolism genes by binding to the iron-responsive elements (IREs) found in their mRNA transcripts (Anderson et al., 2012; Rouault, 2006; Wallander et al., 2006). Interestingly, despite the high sequence homology shared between the two IRPs, their IRE-binding activities are distinctively regulated. IRP1 is switched from an IRE-binding protein to a cytosolic aconitase upon the insertion of a [4Fe4S] cluster, whereas IRP2 is primarily regulated by protein stability. With limiting iron or oxygen, IRP2 is stable and binds IREs. Under iron- and oxygen-enriched conditions, IRP2 undergoes ubiquitin-dependent proteasomal degradation (Guo et al., 1995; Haile et al., 1992; Hanson et al., 1999; Hanson et al., 2003; Iwai et al., 1995).
A decade ago, F-box and leucine-rich repeats protein 5 (FBXL5) was identified as the substrate receptor subunit of an SKP1-CUL1-F box (SCF) ubiquitin ligase complex that specifically recognizes IRP2 and promotes its iron- and oxygen-dependent degradation (Salahudeen et al., 2009; Vashisht et al., 2009). Distinct from other F-box proteins, FBXL5 contains an N-terminal hemerythrin-like (Hr) domain that can directly bind iron (Chollangi et al., 2012; Salahudeen et al., 2009; Shu et al., 2012; Thompson et al., 2012; Vashisht et al., 2009). Topological changes of the FBXL5 Hr domain triggered by iron depletion have been shown to destabilize the F-box protein, thereby stabilizing IRP2. Structural studies of the FBXL5 Hr domain have revealed a di-iron center as the basis for iron sensing. However, the oxygen sensing mechanism of FBXL5 remains unresolved (Shu et al., 2012; Thompson et al., 2012). At the physiological level, loss of FBXL5 in mice leads to embryonic death, which can be rescued by co-deletion of IRP2 (Moroishi et al., 2011). The FBXL5-IRP2 axis, therefore, plays a central role in iron homeostasis in vivo.
Despite recent advances, FBXL5 research remains in its infancy with some fundamental questions poorly addressed. In particular, the mechanism by which the F-box protein specifically recognizes IRP2 has not been explored. Whether binding of IRP2 to FBXL5 is regulated by cellular signals, a phenomenon that has been frequently observed for other F-box protein-substrate interactions, remains an open question. In fact, emerging evidence hints at additional iron-sensing mechanisms that mediate FBXL5-IRP2 interaction independent of the FBXL5 Hr domain (Salahudeen et al., 2009; Thompson and Bruick, 2012; Vashisht et al., 2009). By means of integrated biochemical, biophysical, structural, and cellular analyses, we herein report the identification of a unique Fe-S cluster in FBXL5, which serves as a critical cofactor of the ubiquitin ligase dictating the recruitment of IRP2 for its iron- and oxygen-dependent degradation.
Results
FBXL5 harbors a [2Fe2S] cluster
To probe IRP2 recognition by FBXL5, we set out to reconstitute their interactions with purified recombinant proteins. FBXL5 recruits IRP2 via a 492 amino acids region C-terminal to its Hr domain, which consists of an F-box motif and a leucine-rich repeat (LRR) domain (Salahudeen et al., 2009; Vashisht et al., 2009) (Figure S1A). Early studies have shown that IRP2 differs from IRP1 by a protease-sensitive 73 amino acids insertion, which is dispensable for FBXL5 binding (Dycke et al., 2007; Salahudeen et al., 2009; Vashisht et al., 2009). We accordingly constructed and purified the internally truncated IRP2 protein (IRP2Δ73) and FBXL5 lacking the Hr domain (FBXL5C492) together with the SCF adaptor SKP1. Despite being soluble, FBXL5C492-SKP1 was aggregation-prone and only weakly interacted with IRP2Δ73 (hereafter referred to as IRP2) (Figures 1A and 1B). Based on sequence analysis, we further optimized the F-box protein by deleting an LRR domain loop region that is less conserved and predicted to be structurally disordered (Figure S1B). In contrast to the original construct, this FBXL5 core fragment (FBXL5C492Δ) is not only mono-dispersed, but also highly competent in binding IRP2 (Figures 1A and 1B).
Figure 1. FBXL5 possesses a [2Fe2S] cluster.
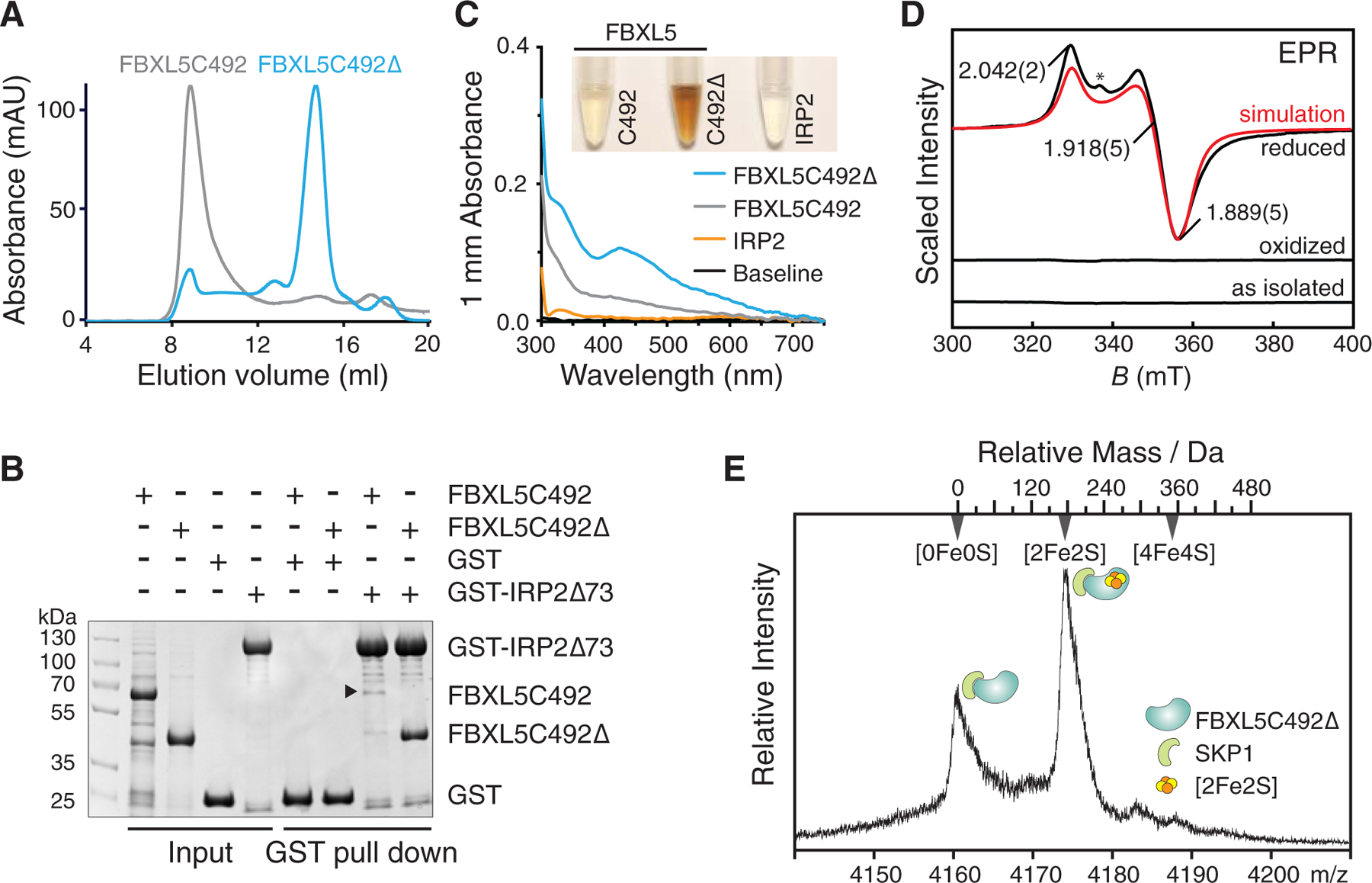
(A) Size-exclusion chromatography analysis of two FBXL5 constructs bound to SKP1. Elution profiles of the aggregation-prone FBXL5C492 and mono-dispersed FBXL5C492Δ samples are shown in gray and blue, respectively.
(B) GST pull-down assay using recombinant GST-IRP2Δ73 and purified FBXL5C492 and FBXL5C492Δ with GST as a negative control. The black arrow indicates a specific band of FBXL5C492 in small amount.
(C) Colors and UV/vis absorption spectra (300–750 nm range) of the purified FBXL5C492, FBXL5C492Δ and IRP2 protein samples. All protein samples recorded for UV spectra and color display are at the concentration of 8.0 mg/mL.
(D) 9.425 GHz EPR spectra of FBXL5C492Δ in oxidized, reduced and as-isolated states. Simulation (red) parameters: g values 2.042, 1.918, and 1.889, g-strain widths 0.0001, 0.0006, and 0.0006; 1 mT Lorentzian FWHM line broadening. The asterisk indicates an organic radical.
(E) Native mass spectrometry analysis of the FBXL5C492Δ-SKP1 complex. The region assigned to the 13+ ions exhibits two features corresponding to the apo and the [2Fe2S] cluster-containing complexes. The top axis shows the mass relative to the apo complex.
See also Figures S1 and S2.
Unexpectedly, a concentrated sample of highly purified FBXL5C492Δ was brown in color (Figure 1C), suggesting the presence of an iron-containing prosthetic group. Its UV/vis absorption spectrum revealed peaks at 330 nm and 425 nm, as well as a broad shoulder at longer wavelengths (Figure 1C). These characteristics match the signature spectrum of a typical Fe-S cluster (Freibert et al., 2018). By contrast, aggregation prone FBXL5C492 was faintly colored with less pronounced UV/vis absorption features. Notably, purified IRP2 protein at the same concentration was clear and showed no featured Fe-S cluster spectrum, which is in line with the earlier observation that IRP2 does not contain an iron sulfur cluster by nature (Phillips et al., 1996) (Figure 1C).
To identify the Fe-S cluster in FBXL5, we analyzed the FBXL5C492Δ sample using electron paramagnetic resonance (EPR). Both untreated and potassium ferricyanide oxidized samples were EPR silent and retained their brownish color, whereas sodium dithionite-reduced sample rapidly became pale-yellow and showed a prominent signal due to a spin-1/2 species with principal g values of 2.042(2), 1.918(5), and 1.889(5) and an average g value of 1.950(4) (Figure 1D). These g-values are consistent with a four-cysteine coordinated ferredoxin-type [2Fe2S]+ cluster (Beinert et al., 1997; Liu et al., 2014; Shubin and Dikanov, 2006). Hence, isolated FBXL5 possesses a [2Fe2S] cluster in its oxidized [2Fe2S]2+ state. Moreover, this FBXL5-bound [2Fe2S] cluster is redox-active as evidenced by its reduction by dithionite. To further validate the identity of the Fe-S cluster, we subjected the SKP1-complexed FBXL5C492Δ sample to native protein mass spectrometry analysis. The spectra of FBXL5C492Δ-SKP1 exhibited two distinct populations, representing the apo and the co-factor-bound forms. The mass difference between the two species is ~176 Da, which matches the molecular weight of a [2Fe2S] cluster (Figures 1E and S2). Collectively, we conclude that FBXL5 is a Fe-S protein containing a redox-active [2Fe2S] cluster.
Overall structure of IRP2-FBXL5-SKP1 complex
To elucidate the structural basis of FBXL5-IRP2 interaction and the role of the [2Fe2S] cluster, we assembled and determined the structure of an IRP2-FBXL5C492Δ-SKP1 complex bound to CUL1 N-terminal domain (NTD) at a resolution of 3.0 Å by single-particle cryo-electron microscopy (cryo-EM) (Figures 2 and S3; Table 1). With homology models of IRP2 and FBXL5C492Δ, the intermediate map obtained with unmasked 3D refinement readily unveiled the overall architecture of the quaternary complex (Figure 2A). Focused 3D refinement further improved the quality of the map, which enabled us to manually rebuild the model of FBXL5C492Δ (hereafter referred to as FBXL5) and three of the four IRP2 domains (Figures 2B, 2C, and S4).
Figure 2. Cryo-EM structure of the IRP2-FBXL5-SKP1 complex.
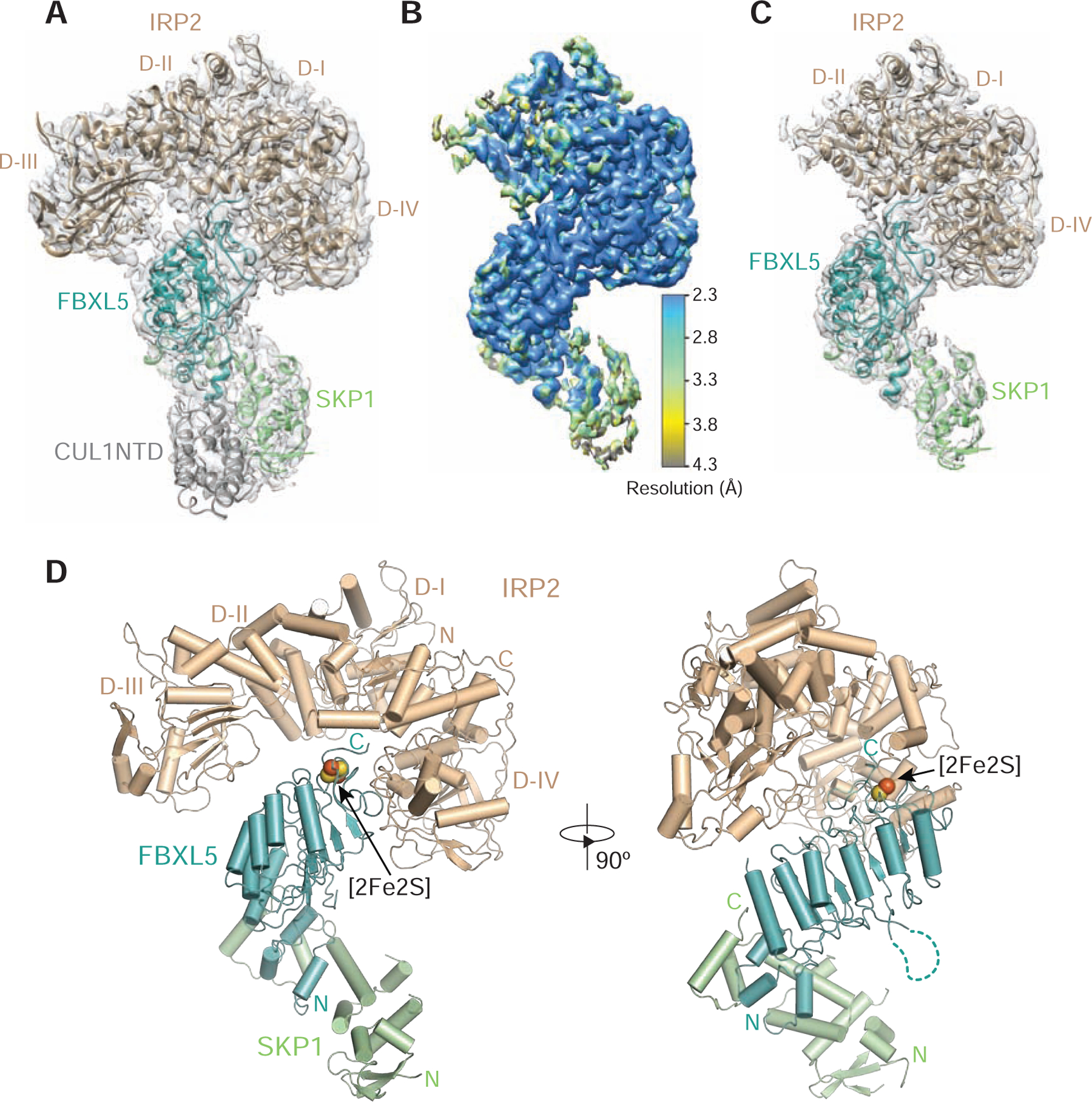
(A) A 3.9 Å electron microscopy map fit with structural models of IRP2 (wheat), FBXL5 (teal), SKP1 (green) and CUL1NTD (gray).
(B) 3D reconstruction of the IRP2-FBXL5-SKP1-CUL1NTD complex at an overall resolution of 3.0 Å is colored according to local resolution estimated by ResMap.
(C) The structure of the IRP2-FBXL5-SKP1 complex modeled with the 3.0 Å density map shown in (B).
(D) Two orthogonal views of the IRP2-FBXL5-SKP1 complex shown in ribbon diagram with the same color scheme as shown in (A). The [2Fe2S] cluster is shown in spheres. The N and C termini of different proteins are labeled N and C in corresponding colors. The dashed line in teal indicates the deleted internal loop of FBXL5.
See also Figures S1, S3, S4, and S5.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| IRP2-FBXL5-SKP1 | |
|---|---|
| Data collection and processing | |
| Microscope | Titan Krios |
| Detector | K2 Summit |
| Magnification | 130,000 |
| Voltage (kV) | 300 |
| Electron exposure (e-/Å2) | 73.8 |
| Defocus range (μm) | 1.5 to 3 |
| Pixel size (Å) | 1.056 (physical) |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 2,832,595 |
| Final particle images (no.) | 955,060 |
| Resolution at 0.143 FSC threshold (Å) | 3.0 |
| Map resolution range (Å) | 2.3–4.0 |
| Refinement | |
| Resolution at 0.5 FSC threshold (Å) | 3.1 |
| Map sharpening B factor (Å2) | −78 |
| Model composition | |
| Non-hydrogen atoms | 8082 |
| Protein residues | 1033 |
| [2Fe2S] | 1 |
| B-factors (Å2) | |
| Protein | 41.74 |
| [2Fe2S] | 23.88 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.644 |
| Validation | |
| MolProbity score | 2.05 |
| Clashscore | 9.28 |
| Rotamer outliers (%) | 0.11 |
| CaBLAM outliers (%) | 4.73 |
| EMRinger score | 3.72 |
| Ramachandran plot | |
| Favored (%) | 90.23 |
| Allowed (%) | 9.77 |
| Outliers (%) | 0 |
The IRP2-FBXL5-SKP1 complex adopts an overall structure reminiscent of a dragon, whose bulky head is formed by the multi-domain IRP2 protein with a winding neck represented by FBXL5-SKP1 (Figure 2D). IRP2 notably is in an L-shaped open conformation with its domain III and domain IV widely separated on two sides of the protein complex (Figures 2D and S5A–S5C). IRP2 is recruited to the F-box protein predominantly through its domain IV. Like other F-box proteins, FBXL5 utilizes its three-helix F-box motif to engage the SCF adaptor protein SKP1 and folds its C-terminal LRR domain into a slender curved solenoid. Unlike most known LRR-containing F-box proteins, which recognize their substrates via either the concave or top surface of their LRR domains (Hao et al., 2005; Xing et al., 2013), FBXL5 captures IRP2 through the C-terminal end of its LRR domain. In close vicinity of the FBXL5-IRP2 interface is the striking [2Fe2S] cluster, which is cradled by a highly conserved and intertwined FBXL5 C-terminal loop (Figures 2D and S1).
Integrity of Fe-S cluster is essential for FBXL5 to recognize IRP2
The FBXL5 LRR domain is composed of six complete LRRs (LRR1–LRR6), which are capped by an additional β-strand preceding the C-terminal loop region (Figure 3A). The FBXL5 LRRs pack in tandem to form an arched assembly with a canonical β-strand-loop-α-helix structure in each repeating unit. Distinct from other β-strands in LRRs, the capping β-strand is led by two extra residues in sequence that form a horn at the basal ridge of the LRR fold (Figures S5D–S5G). This feature is echoed at the apical ridge by the sequence succeeding the capping β-strand, which tilts moderately and opens up a nest-like area. The subsequent FBXL5 loop packs against the α-helix of LRR6 and winds back to the nest area, creating a hook-like structure (Figure 3A). As discussed below, this structural element plays a vital role in docking IRP2 and, therefore, is named “interface loop”. Following the “interface loop”, the F-box protein terminates with a circled loop covering the nest area. We named it “lid loop” (Figure 3A).
Figure 3. Integrity of the [2Fe2S] cluster is essential for the FBXL5-IRP2 interaction.
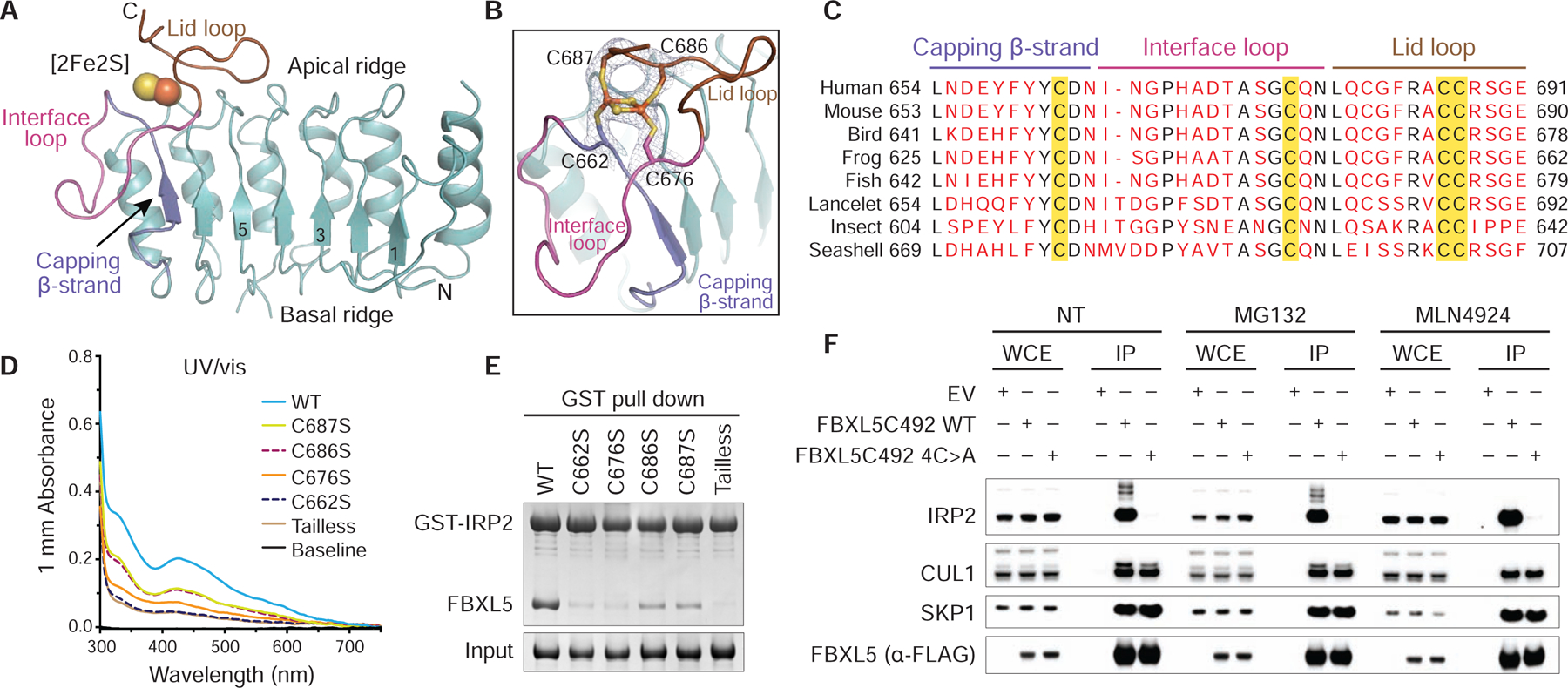
(A) Ribbon diagram of the LRR domain of FBXL5 (teal) containing the [2Fe2S] cluster (spheres). The capping β-strand (slate), interface loop (magenta) and lid loop (brown) are labeled and colored. Select LRRs are numbered and labeled.
(B) A close-up view of the [2Fe2S] cluster (spheres) ligated by four cysteines (sticks) from the capping β-strand (slate), interface loop (magenta) and lid loop (brown). A density map at 3.0 Å resolution covering the [2Fe2S] cluster and its cysteine ligands is shown in gray mesh at contour level of 2.0 σ. For clarity, the residues in the utmost C-terminus of FBXL5 are not shown.
(C) Sequence alignment of the C-terminal loop region in FBXL5 orthologs from human (Homo sapiens), mouse (Mus musculus), bird (Taeniopygia guttata), frog (Xenopus tropicalis), fish (Danio rerio), lancelet (Branchiostoma belcheri), insect (Zootermopsis nevadensis), and seashell (Oyster). Strictly conserved residues are colored black with key structural elements at the C-terminal loop region labeled. Cysteines as [2Fe2S] cluster ligands are highlighted in yellow.
(D) UV/vis absorption spectra (300–750 nm) of FBXL5 WT and mutants measured at a protein concentration of 15.0 mg/mL.
(E) Analysis of IRP2-FBXL5 interaction by GST pull-down assay using recombinant GST-IRP2 and FBXL5 WT and mutants.
(F) HEK293T cells were transiently transfected with pcDNA5/FRT/TO as an empty vector (EV) or the indicated FLAG-tagged FBXL5 plasmids. Twenty-four hours after transfection, cells were harvested and lysed. Where indicated, cells were treated with 10 μM MG132 or 2.5 μM MLN4924 for 4 hours before collection. Whole-cell extracts (WCE) were subjected to immunoprecipitation (IP) using anti-FLAG agarose beads and immunoblotting against the antibodies of FLAG, IRP2, CUL1, and SKP1.
See also Figure S5.
A close examination of the nest area sequestered by the intertwining C-terminal fold reveals an extra rhombus-shaped density with a well-defined plane (Figures 3A and 3B). It is coordinated by four strictly conserved cysteine residues in the C-terminal loop region, including Cys662 following the capping β-strand, Cys676 in the “interface loop”, and Cys686 and Cys687 in the “lid loop” (Figures 3B and 3C). In addition, a network of contacts surrounding the extra density helps maintain a generally hydrophobic environment of the site (Figure S5H). These features strongly suggest that the density belongs to the [2Fe2S] cluster identified in our biophysical studies. To validate this [2Fe2S] cluster coordination site, we purified four mutants of the F-box protein with each cysteine residue substituted by serine and a C-terminally truncated mutant lacking the last six C-terminal residues including the two consecutive cysteines (Cys686 and Cys687). As expected, the UV/vis absorption spectra of the C676S, C686S and C687S mutants displayed substantially diminished [2Fe2S] cluster peaks, while the C662S and tailless mutants lost all the spectral features of a Fe-S protein (Figure 3D). Together, our structural and mutational results unambiguously identify the location of the [2Fe2S] cluster-binding site and highlight the important role of the C-terminal loop region with the four ligand cysteine residues in stabilizing the cofactor.
While incorporation of the [2Fe2S] cluster is cooperatively mediated by the FBXL5 capping β-strand and C-terminal loop region, it is conceivable that the cluster cofactor would reciprocally organize and stabilize the local structure of the C-terminal end of the FBXL5 LRR domain. Because of its physical proximity to the FBXL5-IRP2 interface, compromised integrity of the [2Fe2S] cluster is expected to directly impact the interaction between the ubiquitin ligase and its substrate. In consistence with this notion, individual mutation of all four cysteine residues to serine either pronouncedly weakened or largely abrogated the ability of FBXL5 to interact with IRP2 in vitro (Figure 3E). Similarly, the tailless F-box protein missing half of the “lid loop” lost its IRP2-binding activity. To further validate the functional importance of these [2Fe2S] cluster-coordinating cysteines, we tested the binding of endogenous IRP2 to the ectopically expressed FBXL5C492 WT and a cysteine mutant, which had all four cysteine residues replaced by alanine and was expected to completely abolish [2Fe2S] cluster incorporation. Through co-immunoprecipitation, we show that unlike the WT protein, the FBXL5C492 4C>A mutant was indeed unable to bind IRP2 (Figure 3F). Its defect in engaging IRP2 was further manifested by its inability to destabilize exogenous IRP2 in comparison to the WT protein (Figure S5I). Incorporation of the [2Fe2S] cluster, therefore, is crucial for FBXL5 to recruit IRP2 competently.
Interface between FBXL5 and IRP2
Consistent with previous mutational analysis, IRP2 predominantly uses its domain IV to dock to the distal end of the FBXL5 LRR fold (Wang et al., 2008). The complementary FBXL5-IRP2 interface is surprisingly compact and is primarily mediated by loop structures from both sides (Figures 4A and 4B). The region in IRP2 domain IV responsible for binding FBXL5 is characterized by two spatially adjacent loops, Loop1 and Loop2, that are nucleated by four central hydrophobic residues, Ile761, Pro778, Phe781 and Tyr784 (Figures 4B and S6). Protruding out from Loop1 is a strictly conserved IRP2 residue, Arg779, which inserts its side chain deeply into the center of the FBXL5 “interface loop” (Figures 4B, 4D, and S6). In addition to a cation-π interaction with Tyr661 on the capping β-strand of FBXL5, IRP2 Arg779 is locked to the “interface loop” by donating hydrogen bonds to three backbone carbonyl groups of the F-box protein (Figure S5J). In Loop2, an auxiliary arginine residue, Arg763, stabilizes the peripheral interface by interacting with the negatively charged FBXL5 basal ridge horn and packing against Pro778 of Loop1 from IPR2 (Figures 4B and S5J). Together, Arg779 and Arg763 coordinate to rivet IRP2 onto the lower portion of FBXL5 C-terminal loop region (Figure 4A). Meanwhile, the “lid loop” of FBXL5 is embraced by an α-helix (“arm helix”) in domain I of IRP2, which forms a concave surface at the junction of IRP2 domain I and domain IV (Figures 4A, 4C, and S6). This engagement features a close packing between Arg224 of IRP2 and Arg688 of FBXL5, both of which are neutralized by the surrounding residues via a cluster of polar interactions (Figures 4C, S1, and S6).
Figure 4. Interface between FBXL5 and IRP2.
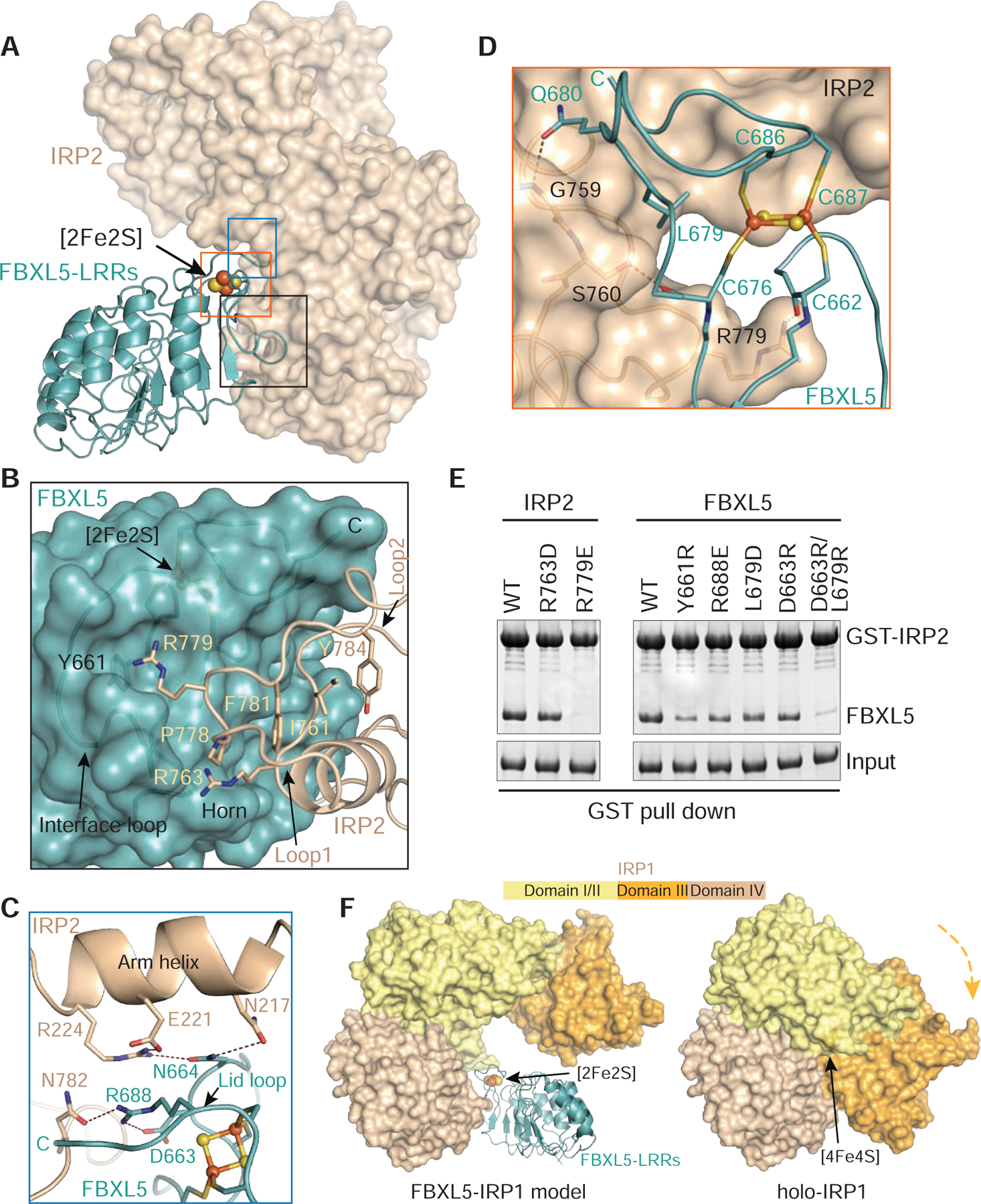
(A) An overall view of the interface formed by FBXL5-LRRs (teal ribbons) containing the [2Fe2S] cluster (spheres) and IRP2 (wheat surface). For clarity, domain III of IRP2 is not shown.
(B–D) Close-up views of FBXL5-IRP2 interface around Loop1/2 of IRP2, Interface loop of FBXL5, Arm helix of IRP2, and the [2Fe2S] cluster. FBXL5 is shown in teal ribbon or surface, while IRP2 is shown in wheat ribbon or surface. The [2Fe2S] cluster is shown in spheres. Select interface residues are shown in sticks. Dashed lines in brown represent hydrogen bonds and polar interactions. The C terminus of FBXL5 is labeled C in teal or black.
(E) GST pull-down assay assessing the ability of FBXL5 WT and mutants to bind IRP2.
(F) Comparison of IRP1 structures in the FBXL5-IRP1 model and the [4Fe4S] cluster bound holo-aconitase (PDB: 2B3X). FBXL5-LRRs is shown in ribbon, while IRP1 is shown in surface with domain I/II in yellow, domain III in orange, and domain IV in wheat. The [2Fe2S] and [4Fe4S] clusters are shown in spheres. The orange arrow indicates the movement of IRP1 domain III for incorporating the [4Fe4S] cluster.
See also Figures S1, S5, and S6.
Although the [2Fe2S] cluster of FBXL5 is not situated at the immediate FBXL5-IRP2 interface, it contributes to IRP2 recognition by securing several critical interactions. First, the backbone carbonyls of two cluster ligand cysteines, Cys662 and Cys676, are tied to IRP2 by hydrogen bonding with Arg779 and Ser760, respectively (Figure 4D). Second, Leu679 of FBXL5, which guards the nonpolar environment of the cluster, directly packs against IRP2 Loop1 (Figure 4D). This local intermolecular contact is further strengthened by its nearby FBXL5 residue, Gln680, whose side chain forms a hydrogen bond with IRP2 Gly759. Above all, the central role of the [2Fe2S] cluster in shaping the complementary surface of FBXL5 manifests its indispensability in IRP2 recruitment.
To map the key structural elements supporting FBXL5-IRP2 interaction, we tested a series of FBXL5 and IRP2 mutants based on our structural analysis. Substitution of IRP2 Arg763 modestly affected complex formation (Figure 4E), indicating an accessary role of the residue at the interface. By contrast, FBXL5 binding was completely abolished by substitution of IRP2 Arg779 (R779E), which represents a “hot spot” for FBXL5 association (Figure 4E). Correspondingly, alteration of FBXL5 Tyr661 (Y661R) that directly contacts IRP2 Arg779 markedly compromised IRP2 recognition (Figure 4E). Interestingly, none of the remaining single amino acid substitution mutants showed detrimental effect on the FBXL5-IRP2 interaction, suggesting that the FBXL5-IRP2 interface is multivalent and distributive. Indeed, a combination of mutations introduced at more than one region of the interface severely impaired IRP2 association, as exemplified by the FBXL5 L679R/D663R double mutants (Figure 4E).
Previously studies have shown that, distinct from the wild-type protein, an IRP1 mutant defective in binding [4Fe4S] cluster can be recognized by FBXL5 and undergo proteasomal degradation (Salahudeen et al., 2009; Vashisht et al., 2009). In light of our structure, this can be rationalized by all the key residues at the FBXL5-IRP2 interface, which are also conserved in IRP1, and the open conformation of apo-IRP1 (Walden et al., 2006) (Figures S5A–S5C and S6). A structural comparison between the holo-IRP1 protein (Dupuy et al., 2006) and an FBXL5-IRP1 model generated from our structure indicates that FBXL5 would clash with IRP1 domain III, which has been shown to rotate towards domain IV upon [4Fe4S] cluster insertion (Figure 4F). By incorporating the cluster cofactor, IRP1 effectively shields its potential interface for FBXL5 binding. Our structure of the FBXL5-IRP2 complex, therefore, provides a structural explanation for the mechanism by which cellular holo-IRP1 is spared from FBXL5-mediated degradation.
Ability of FBXL5 to dislodge IRP2 from IRE
The IRP proteins have been previously shown to bind IREs with high affinities (Guo et al., 1994; Samaniego et al., 1994). How IRP2 is effectively decoupled from IREs and degraded has been poorly understood. The overall resemblance of IRPs in the FBXL5-IRP2 and IRE-IRP1 structures allowed us to derive a model of the IRP2-IRE complex. A side-by-side comparison of IRP2 in the FXBL5- and IRE-complexed forms readily reveals that IRP2 domain IV is equally exploited by the two binding partners (Figure 5A). The last three LRRs and the C-terminal loop region of FBXL5 occupy the same space next to IRP2 domain IV as the helical region of IRE does. In particular, the [2Fe2S] cluster organized C-terminal loop of FBXL5 shares the same IRP2 binding pocket with the bulge C of the IRE, precluding IRP2 from simultaneous engagement with the ubiquitin ligase and the RNA element (Figure S7A). Moreover, FBXL5 and IRE have developed their distinctive means to further immobilize the multi-domain IRP2 protein. The “lid loop” of the F-box protein hooks the “arm helix” of IRP2 domain I, whereas the terminal loop of IRE specifically clinches IRP2 domain III (Figure 5A). These interactions fix IPR2 in a similar L-shaped open conformation in both structures. Altogether, these structural comparisons raise the possibility that FBXL5 might be able to directly compete with IRE for IRP2 binding.
Figure 5. Functional analysis of FBXL5-IRP2 interaction.
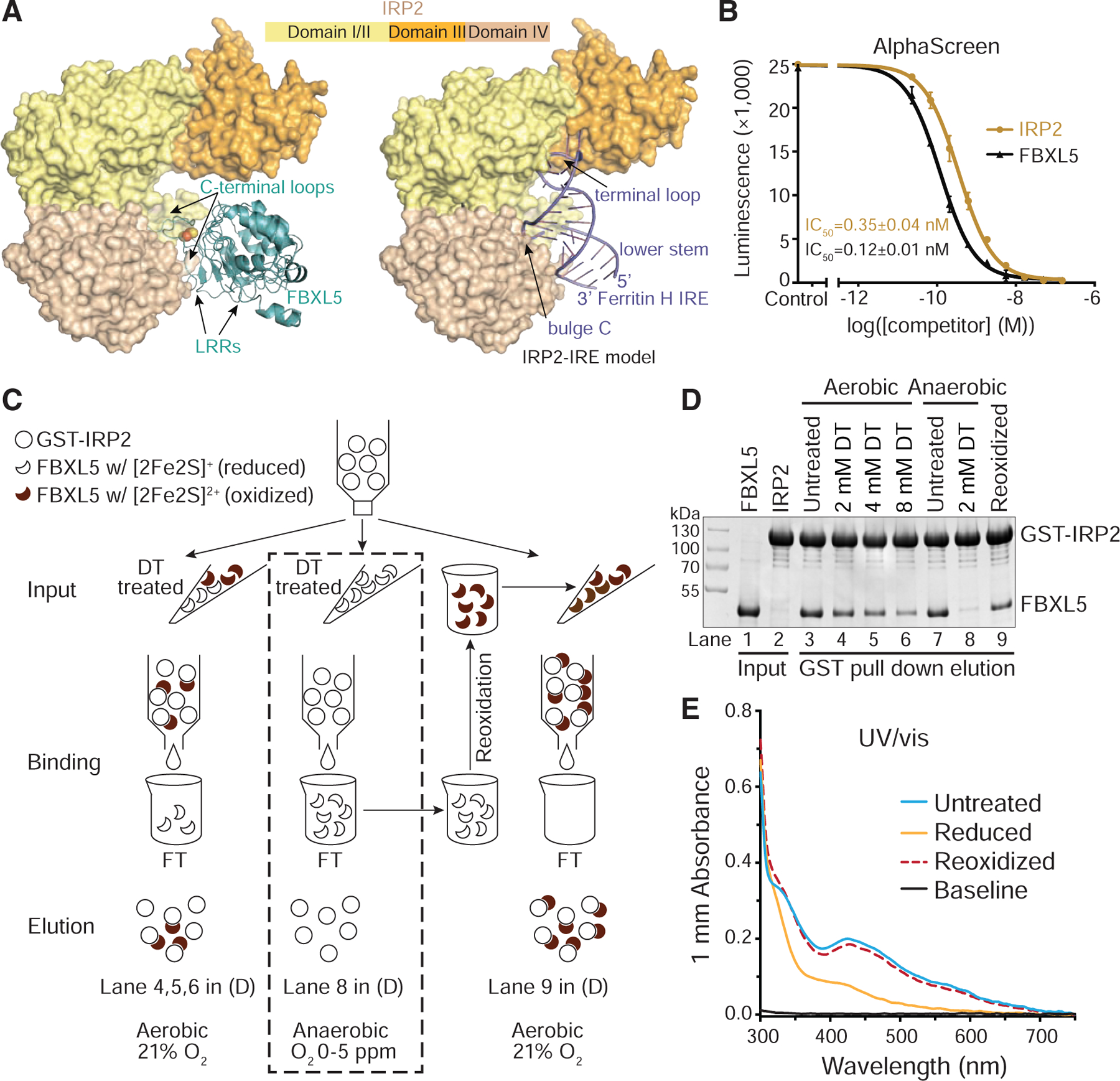
(A) Structural comparison of IRP2-FBXL5 complex and the modeled IRP2-IRE complex. FBXL5 (teal) and Ferritin H IRE (slate) are shown in ribbon, while IRP2 is shown in surface with domain I/II in yellow, domain III in orange, and domain IV in wheat. The [2Fe2S] cluster is shown in spheres. The IRP2-IRE model is generated on the basis of the known structure of the IRP1-Ferritin H IRE complex (PDB: 3SNP). Critical IRP2-contacting elements of FBXL5 and IRE are labeled correspondingly.
(B) AlphaScreen assay assessing the ability of FBXL5 to compete with Ferritin H IRE for binding IRP2. Data were measured in triplicate and plotted as mean ± SD.
(C) A schematic diagram showing the GST pull-down assays for assessing the IRP2-binding abilities of untreated and sodium dithionite-treated FBXL5 with [2Fe2S] cluster in different redox states under aerobic or anaerobic conditions.
(D) Experimental results of (C) as analyzed by SDS-PAGE.
(E) UV/vis spectra (300–750 nm) of the FBXL5 samples tested in (C) and (D) at the same concentration.
See also Figure S7.
To test this idea, we developed an Amplified Luminescence Proximity Homogeneous Assay (AlphaScreen) to quantitatively assess the ability of FBXL5 to displace IRE from IRP2 in vitro. Biotinylated IRE and GST-fused IRP2 were immobilized on the donor and acceptor beads, respectively, which yielded a robust luminescence signal due to stable complex formation. By titrating the concentration of label-free IRP2 and FBXL5, we were able to obtain dose-response curves and the half maximal inhibitory concentration (IC50) for the competitors. Remarkably, IRP2 abolished the luminescent signal with an IC50 of 0.35 nM, while FBXL5 achieved the same effect with an IC50 of 0.12 nM (Figure 5B). Therefore, FBXL5 is equally, if not more, potent than IRP2 in disrupting an existing IRE-IRP2 complex. Overall, these results suggest that the FBXL5-IRP2 and IRE-IRP2 complexes are comparable in affinity and the F-box protein is capable of physically dislodging IRP2 from IRE.
Oxygen-dependent FBXL5-IRP2 interaction
The critical role of the FBXL5 [2Fe2S] cluster in binding IRP2 and its redox active property prompted us to next investigate whether changes in the redox state of the cluster affect the association between the two proteins. To reduce the [2Fe2S] cluster, we pretreated FBXL5 samples with sodium dithionite at increasing concentrations and monitored their binding to IRP2 (Figure 5C). In a dose-dependent manner, dithionite was able to reduce the cluster to its [2Fe2S]+ state and compromise the ability of FBXL5 to interact with IRP2 (Figures 1D and 5D, lane 3–6). To prevent FBXL5-bound [2Fe2S] cluster from being rapidly re-oxidized by oxygen in the ambient air, we repeated the assay under the anaerobic condition. Surprisingly, untreated FBXL5 retained the ability to bind IRP2, whereas FBXL5 treated with 2 mM dithionite essentially lost its IRP2-binding activity (Figures 5C and 5D, lane 7, 8). Notably, the same concentration of dithionite only weakened but not abolished the interaction under aerobic condition (Figure 5D, lane 3, 4). To rule out the possibility that the [2Fe2S] cluster was permanently damaged during anaerobic reduction, we monitored the dithionite-treated and subsequently re-oxidized sample by UV/vis spectrum. In contrast to the less featured spectrum of FBXL5 in its reduced state, the re-oxidized sample regained the characteristic peaks, indicating that both oxidation and reduction of the [2Fe2S] cluster are reversible (Figure 5E). Remarkably, after we transferred the anaerobically reduced FBXL5 sample, which was deficient for IRP2 binding and had passed through the column with immobilized IRP2, back to aerobic condition, the interaction between FBXL5 and IRP2 was largely restored (Figures 5C and 5D, lane 9). Altogether, these experiments establish FBXL5-bound [2Fe2S] cluster as a reversible redox switch, which confers oxygen-dependence for IRP2 binding.
Discussion
In this study, we uncover a [2Fe2S] cluster as an FBXL5 cofactor, which plays a critical role in controlling the recruitment and polyubiquitination of IRP2 in iron homeostasis. In conjunction with the previous reports of iron binding to the N-terminal Hr domain of the F-box protein, our findings unveil a plausible two-tiered mechanism by which FBXL5 senses the levels of iron to dictate IRP2 stability (Figures 6 and S7B). Although the relative abundance of labile iron and [2Fe2S] cluster remains to be determined, it is generally thought that the cellular concentration of labile iron is kept at a minimal level due to its toxicity. It is conceivable that the two sensor domains of FBXL5, namely the N-terminal Hr domain and the C-terminal LRR domain, perceive iron at two different levels. While the Hr domain governs the stability of the ubiquitin ligase upon iron depletion, the LRR domain with its iron-sulfur cluster might determine IRP2 turnover rate in response to increasing iron levels as reflected by the availability of the [2Fe2S] cluster. The importance of the FBXL5-bound [2Fe2S] cluster in IRP2 regulation is manifested by the pronounced accumulation of IRP2 observed when Fe-S cluster biogenesis is impaired (Tong and Rouault, 2006; Ye et al., 2010).
Figure 6. A model for the FBXL5-IRP2 axis regulation by iron and oxygen.
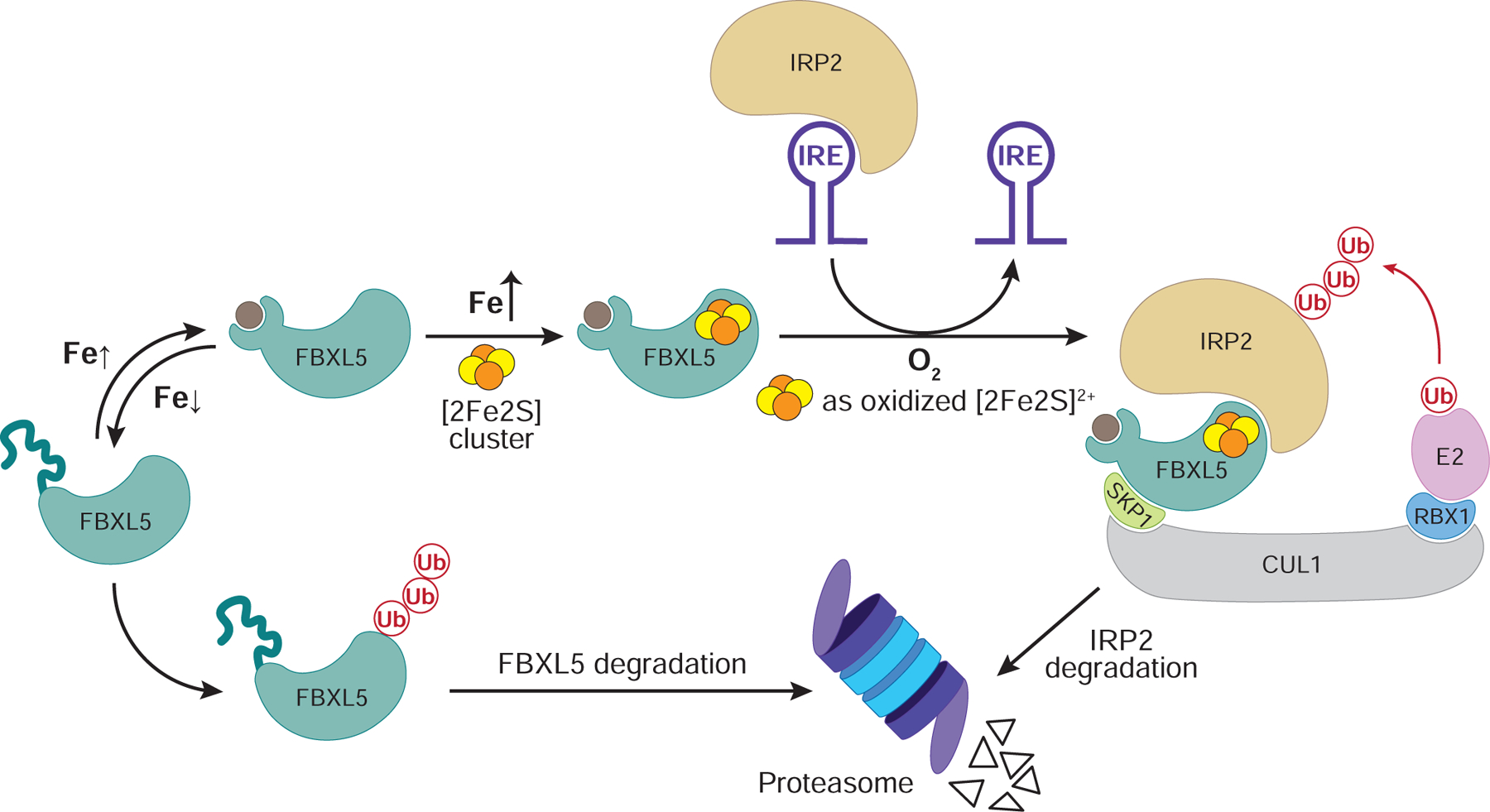
In iron-depleted cells, the N-terminal Hr domain of FBXL5 cannot bind iron and undergoes conformational changes that destabilizes the entire protein. When iron is present at low levels, FBXL5 is stabilized. Increasing levels of iron facilitate the production of [2Fe2S] cluster, which is incorporated into the C-terminal LRR domain of FBXL5. Only when oxygen level is high enough to maintain the cluster in its oxidized [2Fe2S]2+ state could the SCFFBXL5 E3 ligase recruits IRP2 as a substrate for polyubiquination and degradation. The physical interaction between IRP2 and FBXL5 might help release IRP2 from IREs to alter the translation of iron metabolism genes.
See also Figure S7.
Besides coupling IRP2-FBXL5 interaction with iron availability, our biochemical studies suggest that the [2Fe2S] cluster cofactor of FBXL5 is capable of linking IRP2 binding to oxygen, which has an intricate relationship with iron. By monitoring FBXL5-IRP2 interaction with highly purified recombinant proteins, we demonstrate that the [2Fe2S] cluster is reversibly redox active and only supports IRP2 binding to FBXL5 when it is oxidized by oxygen to the [2Fe2S]2+ state. This special property might allow the iron-sulfur cluster to fine-tune FBXL5-IRP2 complex formation, thereby tailoring IRP2 degradation based on the oxygen level in different tissues and under various physiological conditions. Iron-sulfur clusters have long been documented to play critical roles in a variety of biological regulatory mechanisms by sensing the levels of different physiological factors (Johnson et al., 2005; Lill, 2009; Rouault, 2015). Recently, a growing body of evidence suggest that, in response to the changes of the cellular redox states, the Fe-S cluster bound to DNA metabolism enzymes or transcription regulators can act as a switch for their DNA binding activities (Barton et al., 2019; Munnoch et al., 2016). Our findings on the [2Fe2S] cluster in FBXL5 reinforce the role of an iron-sulfur cluster as a redox sensor and further expand its functions to regulating protein-protein interaction and protein degradation.
Our cryo-EM structural analyses suggest that the molecular basis for the redox-dependent FBXL5-IRP2 interaction might reside in the unique coordination of the [2Fe2S] cluster in FBXL5 by two adjacent cysteines. Upon reduction, the geometry of these cysteines and their nearby residues might be conformationally re-arranged, a phenomenon that has been described for the N2 Fe-S cluster from respiratory complex I (Berrisford and Sazanov, 2009). Such structural changes could lead to re-organization of the FBXL5 C-terminal loop region, which blocks IRP2 binding. Further studies of FBXL5 with its [2Fe2S] cluster in the reduced state will shed light on the underlying structural mechanism.
Recent studies have revealed an interaction between FBXL5 and the cytosolic iron-sulfur cluster assembly (CIA)-targeting complex, which is sensitive to oxygen and promotes IRP2 ubiquitination (Mayank et al., 2019; Tan et al., 2013). While it is tempting to speculate that the [2Fe2S] cluster is loaded onto FBXL5 by the CIA-targeting complex, their functional relationship calls for further investigation. In contrast to the four [2Fe2S]-coordinating cysteines, the FBXL5 sequence responsible for binding the CIA-targeting-complex is only conserved in mammalian FBXL5 orthologs. Moreover, abrogation of FBXL5-CIA interaction only partially compromised IRP2 binding to the F-box protein, which is inconsistent with an essential role in delivering the [2Fe2S] cluster to the E3 ligase. It remains possible that the FBXL5-CIA-targeting complex interaction might have evolved as a separate and redundant mechanism for regulating FBXL5-mediated IRP2 degradation.
Substrate recognition by ubiquitin E3 ligases is susceptible to regulation by a variety of cellular signals (Ravid and Hochstrasser, 2008; Zheng and Shabek, 2017). FBXL5 represents the first example of a human ubiquitin ligase that sports an ancient protein co-factor, which usually mediates oxidation-reduction catalytic reactions, but is repurposed by evolution to control oxygen-responsive protein-protein interactions, and, indirectly, RNA-protein interactions. Our discovery not only underscores the functional versatility of ubiquitin ligases as signaling hubs, but also the multifaceted role of iron-sulfur clusters in biology.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ning Zheng (nzheng@uw.edu). Unique and stable reagents generated in this study are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For DNA extraction, E. coli DH5α was used. For bacmid production, E. coli DH10Bac was used. For baculovirus production and amplification, Sf9 insect cells were used. For protein expression, both E. coli BL21(DE3) and HighFive insect cells were used. For immunoprecipitation in mammalian cells, HEK293T and HEK293 cells were used.
METHOD DETAILS
Protein expression and purification
The human IRP2 protein was expressed as a glutathione S-transferase (GST) N-terminal fusion protein in HighFive monolayer insect cells and isolated by glutathione affinity and subsequent anion exchange chromatography after off-column cleavage by tobacco etch virus (TEV) protease. The human FBXL5 and SKP1 proteins were co-expressed and produced in BL21 (DE3) E. coli cells in media supplemented with cysteine and Ferric ammonium citrate (FAC) at the concentrations of 121 mg/L and 25 mg/L, respectively. The plasmid encoding operon suf was co-transformed when needed. The FBXL5-SKP1 complex was purified by nickel affinity and subsequent anion exchange chromatography after off-column cleavage by TEV protease. Human CUL1NTD was expressed as a glutathione S-transferase (GST) N-terminal fusion protein in BL21 (DE3) E. coli cells and purified similarly to IRP2. To assemble the complex of IRP2-FBXL5-SKP1-CUL1NTD for cryo-EM study, the individually isolated proteins were mixed in stoichiometric amounts and subsequently applied to the Superdex-200 gel filtration column (GE Healthcare) in a buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 5 mM DTT (dithiothreitol). The mono-dispersive peak in the elution profile contained the tetrameric complex with a mass of ~200 kDa. All the purification procedures were performed at 4°C. The mutants of FBXL5 and IRP2 were expressed and isolated in the same way as the WT proteins. The affinity tag may be left on the proteins for the purposes of different assays.
Cryo-EM sample preparation and data collection
To prepare grids for cryo-EM data collection, an UltraAuFoil R1.2/1.3 grid (Quantifoil Micro Tools GmbH) was glow discharged for 2 minutes at 20 mA with a glow discharge cleaning system (PELCO easiGlow). 3.0 μL of the purified IRP2-FBXL5-SKP1-CUL1NTD complex at 0.5 mg/mL was applied to a freshly glow-discharged grid. After incubating in the chamber at 4 °C and 100% relative humidity, grids were blotted for 4 s with a blotting force of zero, then immediately plunge-frozen in liquid ethane using a Vitrobot Mark IV system (Thermo Fisher Scientific). Data collection was carried out on a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV. Automation scheme was implemented using the Leginon software (Suloway et al., 2005) at a nominal magnification of 130 K, resulting a physical pixel size of 1.056 Å. Zero-loss-energy images were acquired on a Gatan K2 Summit direct detector operated in super-resolution counting mode (pixel size in super-resolution mode is 0.528 Å), with the slit width of post-column Gatan Quantum GIF energy filter set to be 20 eV. The dose rate was adjusted to 8.2 electrons per Å2 per second, and a total dose of 73.8 electrons per Å2 for each image were fractionated into 60 frames. Data were collected in four sessions with a defocus range of 1.5–3 μm. In total, 5,768 movies were collected with CompuStage in the microscope non-tilted, and a set of 1172 movies were acquired when the CompuStage was tilted 40 degrees.
Image processing and 3D reconstruction
Alignment of movie frames was performed using MotionCor2 (Zheng et al., 2017) through the RELION3.0 pipeline (Kimanius et al., 2016) with images binned 2 in both dimensions by Fourier cropping, resulting in a pixel size of 1.056 Å of the summed images. Dose-weighted summed images were imported into cisTEM (Grant et al., 2018) for manual inspection to remove bad images. The final set of good images contained 5,309 non-tilted and 475 40-degree-tilted images. Around 114,000 particles were selected automatically on a subset of both non-tilted and tilted images after estimation of contrast transfer function using CTFFIND4 (Rohou and Grigorieff, 2015) within cisTEM (Grant et al., 2018) and were subjected to 2D classification to generate 2D averages, which were used later as templates for automatic particle picking. Later steps of data processing were all implemented within the RELION3.0 pipeline (Kimanius et al., 2016). The parameters of contrast transfer function were estimated using cleaned non-dose-weighted motion-corrected sums by GCTF (Zhang, 2016). Template-based automatic particle picking resulted in a set of 2,832,595 particles. GCTF (Zhang, 2016) was used again to perform per-particle estimation of the parameters of contrast transfer function on the automatically picked particles. Particles were then extracted from dose-weighted motion-corrected sums and 3X binned to a box size of 108 pixels. After 3 rounds of reference-free 2D classification, a total of 1,716,405 particles were selected from the original pool. A 3D density map was obtained by employing the 3D initial model job-type within RELION3.0 from a subset of 2D-cleaned-up particles, which was low-pass-filtered to 60 Å and was used as the initial model for 3D classification. A pool of 955,060 particles belonging to the best class from 3D classification were selected, extracted without binning, and subjected to 3D auto-refine without applying a mask, which gave rise to a reconstruction of 3.9 Å. The post-processing procedure with a soft mask generated in RELION3.0 reported an estimated resolution of 3.2 Å. To improve the resolution, CTF refinement (Zivanov et al., 2018) was executed to refine the defocus values for each particle and to calculate the beam-tilt parameters for each separate data-collection session. The resulting particles were polished through the Bayesian polishing approach in RELION3.0 (Zivanov et al., 2019). These polished shiny particles went through another round of focused 3D refinement by imposing a soft mask surrounding the region of interest, which comprises most of mass of the complex, including SKP1, FBXL5, and domains I, II and IV of IRP2. The final reconstruction was measured to be at a resolution of 3.0 Å, and the map was post-processed in RELION3.0, with correction for the modulation transfer function and sharpened by applying a global B-factor of −78 Å2 that was estimated in the post-process protocol. Reported resolutions are based on the gold-standard FSC (Fourier shell correlation) using the 0.143 criterion (Rosenthal and Henderson, 2003). ResMap (Kucukelbir et al., 2014) was used to determine local resolution.
Model building and refinement
Initial model building was performed on the basis of the resulting map at 3.0 Å after local refinement using a mask to exclude the less-defined flexible regions. Considering that the crystal structures of IRP1 were determined in two different conformations (as an aconitase with PDB ID: 2B3X and as an IRE-binding protein with PDB ID: 3SNP, 3SN2), we used the structures of individual domains in IRP1 as the templates to build the model of IRP2. Models of four domains of IRP1 were fitted into the map as rigid bodies in Chimera (Pettersen et al., 2004) with domain I, II, IV well docked and domain III unassigned due to the lack of density. All the amino acids were changed to the sequence of IRP2 and the model was manually rebuilt in COOT (Emsley et al., 2010). The model of FBXL5 was built de novo in COOT based on the map showing clear side chain densities of the residues in majority, while the crystal structure of SKP1 in GGTase3-FBXL2-SKP1 complex (PDB ID: 6O60) was used as the template for the model building of SKP1. Iterative rounds of real-space refinement in PHENIX (Adams et al., 2010) and manual adjustment in COOT were carried out for model improvement. The final model was evaluated using MolProbity and the cryo-EM data collection, refinement and validation statistics are summarized in Table 1.
Affinity pull-down assay
The GST pull-down assay was performed using ~500 μg of purified GST or GST-tagged IRP2 WT or mutant proteins as the bait and ~400 μg of His-tagged FBXL5 WT and mutant proteins. Reaction mixtures were incubated with 100 μL GST beads (GE Healthcare) at 4°C for 1 hour in the binding buffer with 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 5 mM DTT. After extensive wash with binding buffer, the protein complexes on the beads were eluted by 5 mM glutathione. The eluted samples were resolved by SDS-PAGE and analyzed by Coomassie staining. For the pull-down assays with reduced FBXL5, the purified His-tagged FBXL5 WT proteins were treated with 2 mM, 4 mM and 8 mM DT (sodium dithionite) in ambient air or 2 mM DT in an anaerobic glove box (McCoy), and immediately applied to the binding reaction following the same protocol described above. The IRP2-binding-deficient FBXL5 FT (flow through) sample was re-oxidized overnight in ambient air and reapplied to the binding reaction next day (Figure 5C). Inputs represent 3–5% of the total amount of proteins used for each reaction.
UV/vis absorption spectrometry
UV/vis absorption spectra were recorded with 1 μL of protein samples under aerobic conditions at RT on a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific) in a range from 220 to 750 nm. The protein samples were at the concentrations of 8.0 mg/mL or 15.0 mg/mL in the buffer containing 20 mM Tris pH 8.0, 150 mM NaCl and 5 mM DTT. The reduced sample of FBXL5 was prepared anaerobically prior to the aerobic UV/vis spectrum and subsequently re-oxidized overnight in the ambient air. The UV/vis spectrum of the re-oxidized sample was recorded next day.
EPR
The EPR samples were prepared from 300 μM FBXL5C492Δ-SKP1 protein complex in 20 mM Tris pH 8.0, 150 mM NaCl, 5 mM DTT and 20% (v/v) Glycerol. The reduced EPR sample was prepared in an anaerobic box (McCoy) by the addition of 1.5 mM freshly prepared DT (sodium dithionite). The oxidized sample was prepared aerobically by the addition of 1 mM K3Fe(CN)6. About 100 μL of solutions were transferred to 4 mm O.D. quartz tubes (Wilmad) and flash-frozen in liquid nitrogen. Atmosphere was removed by freeze-pump-thawing and the samples were flame sealed. X-band EPR spectra were collected on a Bruker EMX spectrometer equipped with an SHQE resonator. The sample temperature was set to 20 K utilizing an Oxford ESR900 liquid-helium flow cryostat. The modulation amplitude was 15 G, microwave power was 32 mW, and the sweep rate was 4 mT s−1. The magnetic-field axis was calibrated with a teslameter, and the frequency was measured using a frequency counter. The spectrum was simulated using EasySpin (Stoll and Schweiger, 2006).
Native mass spectrometry
FBXL5C492Δ-SKP1 protein complex was buffer exchanged into aqueous 200 mM ammonium acetate at pH 7.0 using centrifugal concentrators (10 kDa MWCO, Spin-X UF, Corning, Inc.) and a centrifuge operated at 4°C. The protein sample used in positive ion mode was also heated using a heating block at ~52°C for one minute prior to analysis. The protein sample was loaded into glass capillaries with inner diameters of 0.78 mm that were pulled to approximately 1 to 3 μm on one end using a micropipette puller (P-97, Sutter Instruments, Novato, CA) for nano electrospray ionization. 10 μM of sample was loaded onto the glass capillary. Electrical contact with the solution was attained by inserting a platinum wire electrode into the wide end of the capillary (Davidson et al., 2017). To minimize impurity carryover between experiments, the electrode was washed with aqueous 25% HCl (v:v) and rinsed with ultrapure (18.2 MΩ) water between samples. Data were acquired using a Waters Synapt G2 HDMS hybrid mass spectrometer (Giles et al., 2011) (Waters Co., Wilmslow, UK) in which the traveling-wave ion mobility cell was replaced with a radio-frequency (RF) confining drift cell (Allen et al., 2016) that contained approximately 1.7 Torr He. The following MS parameters were used for positive ion mode spectrum: capillary voltage, less than 1.0 kV; sampling cone, 70 V; extraction cone, 5 V; source temperature, ~30°C; trap collision energy, 50 V. The following MS parameters were used for negative ion mode spectrum: source temperature, ~30°C; extraction cone, 2 V; trap collision energy, 30 V; sampling cone: 50 V; trap gas flow: 1 ml/min. Mass spectra were analyzed using MassLynx v4.1 (Waters, Co., Milford, MA).
Mammalian expression plasmids
Genestrings of FBXL5C492 WT and 4C>A mutant carrying HA and FLAG tag, BamHI and XhoI sites were custom synthesized and cloned initially into Zero Blunt™ TOPO™ PCR Cloning Kit (Catalog number: 451245). The insert was then restricted from TOPO vector using BamHI and XhoI restriction enzymes and subcloned into in pcDNA5/FRT/TO (Addgene) vector. FLAG tagged IRP2 cloned in pcDNA5/FRT/TO (Addgene) as described in previous study (Vashisht et al., 2009) was used for the indicated experiments.
Mammalian cell culture
Cell lines were purchased from ATCC and routinely monitored for Mycoplasma contamination using the Universal Mycoplasma Detection Kit (ATCC 30–1012K). HEK293T/HEK293 (ATCC CRL-3216/ATCC CRL-1573) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Corning Life Sciences) and 1% penicillin/streptomycin/L-glutamine (Corning Life Sciences). Cell lines were maintained at 37°C and 5% CO2 in a humidified atmosphere.
Immunoprecipitation and immunoblotting
HEK293T/HEK293 cells were transiently transfected with the indicated plasmids using Polyethylenimine (Polysciences) or Lipofectamine 2000 (Invitrogen). Where indicated, 24 hours after transfection, cells were treated with proteasome inhibitor MG132 (10 μM), or neddylation inhibitor MLN4924 (2.5 μM) for 4 hours before collection. The treatment of MG132 was extended to overnight in antibiotics free media when needed. Cells were harvested and lysed in lysis buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1mM EGTA, 5mM MgCl2, and 0.2% NP-40), supplemented with protease inhibitors (Complete ULTRA, Roche), phosphatase inhibitors (PhosSTOP, Roche), and 1 mM 1,4-Dithiothreitol (DTT) (Sigma-Aldrich). The insoluble fraction was removed by centrifugation (20,000×g) for 15 min at 4°C. Immunoprecipitations of FLAG-tagged proteins were carried out using FLAG-M2 agarose beads (Sigma-Aldrich) for 2 hours at 4°C. The beads were then washed four times in lysis buffer. Immunoprecipitates were eluted in NuPAGE® LDS sample buffer (Thermo Fisher Scientific) supplemented with β-mercaptoethanol (Sigma-Aldrich) and incubated at 95°C for 5 minutes. Whole cell lysates and immunoprecipitates were separated by SDS-PAGE and transferred to either 0.45 μm Immobilon-P PVDF membranes (Millipore Sigma) or nitrocellulose membrane for Western blotting. Transfer efficiency was checked by Ponceau S (Sigma-Aldrich) staining. Membranes were then blocked in 5% milk/TBST for 1 hour at room temperature and incubated with the indicated primary antibodies at 4°C overnight. The detection of proteins was accomplished using the appropriate secondary antibodies conjugated to horseradish peroxidase or for fluorescence (GE Healthcare/Invitrogen) in 5% milk/TBST. Western blots were developed using SuperSignal enhanced chemiluminescence (Thermo Fisher Scientific) using the ImageQuant LAS 4000 (GE Healthcare) image analyzer or LICOR Odyssey imager (LI-COR, Lincoln, NE, USA).
AlphaScreen luminescence proximity assay
AlphaScreen assays for determining and measuring protein-protein and protein-RNA interactions were performed using EnSpire reader (PerkinElmer). Biotinylated Ferritin H IRE (pre-treated as described below) was immobilized to streptavidin coated AlphaScreen donor beads. GST-tagged IRP2 was attached to anti-GST AlphaScreen acceptor beads. The donor and acceptor beads were brought into proximity by the interaction between IRE and IRP2. When excited by a laser beam of 680 nm, the donor beads emit singlet oxygen that activates thioxene derivatives in the acceptor beads, which then release photons of 520–620 nm as the binding signal. Competition assays were performed by titrating the concentrations of the tag-free IRP2 and FBXL5 as competitors in the pre-mixed IRE-IRP2 complex and measuring the half maximal inhibitory concentration (IC50) for competitors based on the dose response curves showing signal recession.
The experiments were conducted in triplicates with 0.15 nM biotinylated Ferritin H IRE RNA and 0.15 nM GST-IRP2 in the presence of 5 μg/ml donor and acceptor beads in a buffer of 25mM Hepes pH 7.7, 150 mM NaCl, 5 mM DTT and 0.1% BSA. The concentrations of IRP2 and FBXL5 as competitors ranged from 0.02 nM to 150 nM. IC50 values were determined using non-linear curve fitting of the dose response curves generated with Prism 8 (GraphPad).
The biotinylated Ferritin H IRE RNA was synthesized from Integrated DNA Technologies (IDT). The lyophilized RNA was dissolved in RNase-free water and diluted to 200 μM in concentration. This stock solution was heated at 95°C for 5 min and then cooled in an ice bath for 10 min before being aliquoted. All the aliquots were stored at −80°C to avoid degradation and directly applied to assays after thawing.
QUANTIFICATION AND STATISTICAL ANALYSIS
Protein quantification was done using the Bradford Protein Assay protocol and Bio-rad Protein Assay Dye on a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific) at room temperature.
DATA AND CODE AVAILABILITY
Cryo-EM density map of the IRP2-FBXL5-SKP1 complex has been deposited in the Electron Microscopy Data Bank (EMDB) under the accession code: EMD-21149. Atomic coordinates have been deposited to the Protein Data Bank (PDB) with the accession number PDB: 6VCD.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-FLAG (1:2,000) | Sigma-Aldrich | Cat. #F7425; RRID:AB_439687 |
| Rabbit monoclonal anti-IRP2 (1:1,000/1:250) | Cell Signaling Technology | Cat. #37135S; RRID:AB_2799110 |
| Rabbit polyclonal anti-CUL1 (1:1,000) | Thermo Fisher Scientific | Cat. #71–8700; RRID:AB_2534002 |
| Anti-SKP1 (1:5,000) | Michele Pagano’s lab | N/A |
| Anti-rabbit, HRP-conjugated (1:5,000) | GE Healthcare | Cat. #NA934; RRID:AB_2722659 |
| Mouse monoclonal anti-β-Actin (1:1,000) | Cell Signaling Technology | Cat. #3700S |
| Mouse monoclonal anti-Flag (1:1,000) | Cell Signaling Technology | Cat. #8146S |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor Plus 800 (1:10,000) | Invitrogen | Cat. #A32735 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor 680 | Invitrogen | Cat. #A28183 |
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | NEB | Cat. #C2527I |
| E. coli DH5α | NEB | Cat. #C2987I |
| E. coli DH10Bac | Life Technologies | Cat. #10361012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ammonium ferric citrate (FAC) | Sigma-Aldrich | Cat. #F5879 |
| L-cysteine free base | MP Biomedicals | Cat. #101444 |
| Sodium Dithionite | Acros Organics | Cat. #169590250 |
| Potassium hexacyanoferrate (K3Fe(CN)6) | Sigma-Aldrich | Cat. #31253 |
| Lipofectamine 2000 | Invitrogen | Cat. #11668019 |
| Polyethylenimine | Polysciences | Cat. # 24765–1 |
| MG132 | Peptides International | Cat. #IZL-3175v |
| MLN4924 | Active Biochem | Cat. # A-1139 |
| TEV protease | In house | N/A |
| Critical Commercial Assays | ||
| SuperSignal West Pico Chemiluminescent Substrate | Thermo Fisher Scientific | Cat. #PI34580 |
| SuperSignal West Femto Chemiluminescent Substrate | Thermo Fisher Scientific | Cat. #PI34095 |
| Deposited Data | ||
| IRP2-FBXL5-SKP1 complex model coordinates | This paper | PDB: 6VCD |
| IRP2-FBXL5-SKP1 complex cryo-EM map | This paper | EMDB: EMD-21149 |
| Experimental Models: Cell Lines | ||
| Spodoptera frugiperda: Sf9 | Life Technologies | Cat. #B825–01 |
| Trichoplusia ni: HighFive | Life Technologies | Cat. #B85502 |
| Human: HEK293 | ATCC | Cat. #CRL-1573 |
| Human: HEK293T | ATCC | Cat. #CRL-3216 |
| Oligonucleotides | ||
| Biotinylated Ferritin H IRE RNA | IDT | N/A |
| Recombinant DNA | ||
| pET-FBXL5-SKP1 | This paper | N/A |
| pET-FBXL5 Mutants-SKP1 | This paper | N/A |
| pFB-GST-IRP2 | This paper | N/A |
| pFB-GST-IRP2 Mutants | This paper | N/A |
| pGEX-CUL1NTD | This paper | N/A |
| pcDNA5/FRT/TO Empty Vector | Addgene | N/A |
| pcDNA5/FRT/TO-HA-FLAG-FBXL5C492 | This paper | N/A |
| pcDNA5/FRT/TO-HA-FLAG-FBXL5C492 4C>A | This paper | N/A |
| pcDNA5/FRT/TO-FLAG-IRP2 | Vashisht et al., 2009 | https://science.sciencemag.org/content/326/5953/718/tab-figures-data |
| Software and Algorithms | ||
| Phenix | Adams et al., 2012 | https://www.phenix-online.org/ |
| Coot | Emsley and Cowtan, 2004 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | Pymol | http://pymol.org/2/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Relion-3.0 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| cisTEM | Grant et al., 2018 | https://cistem.org |
| GCTF | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/ |
| MATLLAB 2019a | MathWorks | N/A |
| EasySpin | Stoll and Schweiger, 2016 | http://easyspin.org |
| Image Studio | LI-COR | https://www.licor.com/bio/image-studio-lite/ |
| Prism 8 | GraphPad | N/A |
| Other | ||
| EnSpire reader | PerkinElmer | N/A |
| Anaerobic glove box | McCoy | N/A |
| ANTI-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat. #A2220 |
| Glutathione Sepharose 4B | GE healthcare | Cat. #17-0756-05 |
| Ni Sepharose excel | GE healthcare | Cat. #17-3712-02 |
| HiTrap Q-HP, 1mL | GE healthcare | Cat. #17-1153-01 |
| Superdex 200 Increase 10/300 GL | GE healthcare | Cat. #28990944 |
| AlphaScreen Streptavidin-coated donor beads | PerkinElmer | Cat. #6760002S |
| AlphaLISA anti-GST AlphaScreen acceptor beads | PerkinElmer | Cat. #AL110C |
| 4 mm O.D. quartz tubes | Wilmad-LabGlass | Cat. #707-SQ-250M |
| EMX X-band spectrometer | Bruker | N/A |
| ESR900 helium cryostat | OxfordInstruments | N/A |
Highlights.
FBXL5 harbors an [2Fe2S] cluster, which is indispensable for IRP2 recruitment
Structural basis of [2Fe2S] cluster-mediated FBXL5-IRP2 interaction
The [2Fe2S] cluster confers FBXL5 oxygen-dependent IRP2 recognition
FBXL5 dislodges IRP2 from IRE to facilitate its turnover
Acknowledgments
We thank the cryo-EM facility at the University of Washington and J. D. Quispe for technical support in electron microscopy data acquisition, J. Kollman and M. C. Johnson at University of Washington, W. Huang at Case Western Reserve University for help in data processing, P. Cresswell in Yale for the gift of suf operon plasmid, J. A. Leigh and L. Gu at the University of Washington for the access to the anaerobic glove boxes in their labs, T. R. Hinds for help on data analysis of AlphaScreen assay, and the members of Zheng lab for help. This work was supported by funds from the National Science Foundation (CHE-1807382 to M.F.B.), National Institutes of Health (T32GM008268 to E.R.C., R01DK107712 to E.A.L., and R01GM57587 to M.P.), and the AAI Careers in Immunology Fellowship awarded to M.R.. N.Z. and M.P. are Howard Hughes Medical Institute Investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
N.Z. is a member of the scientific advisory board of Kymera Therapeutics and a co-founder of Coho Therapeutics. M.P. is a consultant for BeyondSpring Pharmaceutics and a member of the scientific advisory board of Kymera Therapeutics and Cullgen Therapeutics. The authors declare no other competing interests.
References
- Abbaspour N, Hurrell R, and Kelishadi R (2014). Review on iron and its importance for human health. J Res Med Sci 19, 164–174. [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SJ, Giles K, Gilbert T, and Bush MF (2016). Ion mobility mass spectrometry of peptide, protein, and protein complex ions using a radio-frequency confining drift cell. Analyst 141, 884–891. [DOI] [PubMed] [Google Scholar]
- Anderson CP, Shen M, Eisenstein RS, and Leibold EA (2012). Mammalian iron metabolism and its control by iron regulatory proteins. Biochim Biophys Acta 1823, 1468–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton JK, Silva RMB, and O’Brien E (2019). Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annu Rev Biochem 88, 163–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard JL (2001). Iron biology in immune function, muscle metabolism and neuronal functioning. J Nutr 131, 568S–579S; discussion 580S. [DOI] [PubMed] [Google Scholar]
- Beinert H, Holm RH, and Munck E (1997). Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Berrisford JM, and Sazanov LA (2009). Structural basis for the mechanism of respiratory complex I. J Biol Chem 284, 29773–29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollangi S, Thompson JW, Ruiz JC, Gardner KH, and Bruick RK (2012). Hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5) communicates cellular iron and oxygen availability by distinct mechanisms. J Biol Chem 287, 23710–23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard BJ, Lammers T, and Rivella S (2017). Targeting iron metabolism in drug discovery and delivery. Nat Rev Drug Discov 16, 400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson KL, Oberreit DR, Hogan CJ, and Bush MF (2017). Nonspecific aggregation in native electrokinetic nanoelectrospray ionization. Int. J. Mass Spectrom 420, 35–42. [Google Scholar]
- Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, and Fontecilla-Camps JC (2006). Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 14, 129–139. [DOI] [PubMed] [Google Scholar]
- Dycke C, Bougault C, Gaillard J, Andrieu JP, Pantopoulos K, and Moulis JM (2007). Human iron regulatory protein 2 is easily cleaved in its specific domain: consequences for the haem binding properties of the protein. Biochem J 408, 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibert SA, Weiler BD, Bill E, Pierik AJ, Muhlenhoff U, and Lill R (2018). Biochemical Reconstitution and Spectroscopic Analysis of Iron-Sulfur Proteins. Methods Enzymol 599, 197–226. [DOI] [PubMed] [Google Scholar]
- Giles K, Williams JP, and Campuzano I (2011). Enhancements in travelling wave ion mobility resolution. Rapid Commun Mass Spectrom 25, 1559–1566. [DOI] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Phillips JD, Yu Y, and Leibold EA (1995). Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J Biol Chem 270, 21645–21651. [DOI] [PubMed] [Google Scholar]
- Guo B, Yu Y, and Leibold EA (1994). Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem 269, 24252–24260. [PubMed] [Google Scholar]
- Haile DJ, Rouault TA, Harford JB, Kennedy MC, Blondin GA, Beinert H, and Klausner RD (1992). Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc Natl Acad Sci U S A 89, 11735–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson ES, Foot LM, and Leibold EA (1999). Hypoxia post-translationally activates iron-regulatory protein 2. J Biol Chem 274, 5047–5052. [DOI] [PubMed] [Google Scholar]
- Hanson ES, Rawlins ML, and Leibold EA (2003). Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem 278, 40337–40342. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, and Pavletich NP (2005). Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell 20, 9–19. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, and Andrews NC (2004). Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, and Camaschella C (2010). Two to tango: regulation of Mammalian iron metabolism. Cell 142, 24–38. [DOI] [PubMed] [Google Scholar]
- Iwai K, Klausner RD, and Rouault TA (1995). Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J 14, 5350–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, and Johnson MK (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74, 247–281. [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu PT, Heiskala M, Peterson PA, and Yang Y (2001). The roles of iron in health and disease. Mol Aspects Med 22, 1–87. [DOI] [PubMed] [Google Scholar]
- Lill R (2009). Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838. [DOI] [PubMed] [Google Scholar]
- Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, Bhagi A, and Lu Y (2014). Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem Rev 114, 4366–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayank AK, Pandey V, Vashisht AA, Barshop WD, Rayatpisheh S, Sharma T, Haque T, Powers DN, and Wohlschlegel JA (2019). An Oxygen-Dependent Interaction between FBXL5 and the CIA-Targeting Complex Regulates Iron Homeostasis. Mol Cell 75, 382–393 e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Nishiyama M, Takeda Y, Iwai K, and Nakayama KI (2011). The FBXL5-IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab 14, 339–351. [DOI] [PubMed] [Google Scholar]
- Munnoch JT, Martinez MT, Svistunenko DA, Crack JC, Le Brun NE, and Hutchings MI (2016). Characterization of a putative NsrR homologue in Streptomyces venezuelae reveals a new member of the Rrf2 superfamily. Sci Rep 6, 31597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Phillips JD, Guo B, Yu Y, Brown FM, and Leibold EA (1996). Expression and biochemical characterization of iron regulatory proteins 1 and 2 in Saccharomyces cerevisiae. Biochemistry 35, 15704–15714. [DOI] [PubMed] [Google Scholar]
- Ravid T, and Hochstrasser M (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Rouault TA (2006). The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2, 406–414. [DOI] [PubMed] [Google Scholar]
- Rouault TA (2015). Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol 16, 45–55. [DOI] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, and Bruick RK (2009). An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaniego F, Chin J, Iwai K, Rouault TA, and Klausner RD (1994). Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. Structure, function, and post-translational regulation. J Biol Chem 269, 30904–30910. [PubMed] [Google Scholar]
- Shu C, Sung MW, Stewart MD, Igumenova TI, Tan X, and Li P (2012). The structural basis of iron sensing by the human F-box protein FBXL5. Chembiochem 13, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin AA, and Dikanov SA (2006). Variations of g-tensor principal values in reduced [2Fe–2S] cluster of iron-sulfur proteins. Appl. Magn. Reson 30, 399–416. [Google Scholar]
- Stoll S, and Schweiger A (2006). EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson 178, 42–55. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Tan MK, Lim HJ, Bennett EJ, Shi Y, and Harper JW (2013). Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell 52, 9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JW, and Bruick RK (2012). Protein degradation and iron homeostasis. Biochim Biophys Acta 1823, 1484–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JW, Salahudeen AA, Chollangi S, Ruiz JC, Brautigam CA, Makris TM, Lipscomb JD, Tomchick DR, and Bruick RK (2012). Structural and molecular characterization of iron-sensing hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5). J Biol Chem 287, 7357–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong WH, and Rouault TA (2006). Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab 3, 199–210. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009). Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, and Volz K (2006). Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314, 1903–1908. [DOI] [PubMed] [Google Scholar]
- Wallander ML, Leibold EA, and Eisenstein RS (2006). Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta 1763, 668–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen G, Lee J, and Pantopoulos K (2008). Iron-dependent degradation of IRP2 requires its C-terminal region and IRP structural integrity. BMC Mol Biol 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, and Zheng N (2013). SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, and Rouault TA (2010). Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest 120, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, and Shabek N (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 86, 129–157. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, and Scheres SHW (2019). A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density map of the IRP2-FBXL5-SKP1 complex has been deposited in the Electron Microscopy Data Bank (EMDB) under the accession code: EMD-21149. Atomic coordinates have been deposited to the Protein Data Bank (PDB) with the accession number PDB: 6VCD.


