Abstract
The inaccessibility of living bone marrow hampers the study of its pathophysiology under myelotoxic stress induced by drugs, radiation or genetic mutations. Here, we show that a vascularized human bone-marrow-on-a-chip supports the differentiation and maturation of multiple blood-cell lineages over 4 weeks while improving CD34+ cell maintenance, and that it recapitulates aspects of marrow injury, including myeloerythroid toxicity after clinically relevant exposures to chemotherapeutic drugs and ionizing radiation as well as marrow recovery after drug-induced myelosuppression. The chip comprises a fluidic channel filled with a fibrin gel in which CD34+ cells and bone-marrow-derived stromal cells are co-cultured, a parallel channel lined by human vascular endothelium and perfused with culture medium, and a porous membrane separating the two channels. We also show that bone-marrow chips containing cells from patients with the rare genetic disorder Shwachman–Diamond syndrome reproduced key haematopoietic defects and led to the discovery of a neutrophil-maturation abnormality. As an in vitro model of haematopoietic dysfunction, the bone-marrow-on-a-chip may serve as a human-specific alternative to animal testing for the study of bone-marrow pathophysiology.
The human BM is the site where all adult blood cells originate and thus BM injury and dysfunction causes significant patient morbidity and mortality. BM injury commonly occurs due to drug- and radiation-related toxicities as a result of its high cell proliferation rates and abnormal hematopoietic function plays a significant role in various genetic disorders, including congenital marrow failure syndromes. While these abnormalities can be diagnosed and managed by monitoring peripheral blood counts, it is the proliferation and differentiation of hematopoietic cells in the marrow that is directly targeted in these disease states. Aside from invasive biopsies, there are no methods to study these responses in situ over time in human patients. In vitro models of human hematopoiesis offer the opportunity to better understand marrow pathophysiology through controlled experimentation. Various culture methods for human hematopoietic cells have been described, including culturing CD34+ hematopoietic progenitors in suspension (including methylcellulose-based assays)1,2 or on two-dimensional (2D) stromal cell monolayers (e.g., Dexter culture and assays to assess long-term culture-initiating cells and cobblestone area-forming cells)3,4. Newer hematopoietic culture methods utilizing three-dimensional (3D) gels and scaffolds as well as a variety of dynamic culture setups (e.g., perfused devices5–9) have also been developed (Supplementary Table 1).
The use of in vitro culture systems and animal models have yielded fundamental insight into the biology of hematopoiesis1,2,10. They also have been useful for the expansion of CD34+ progenitors and differentiation of specific hematopoietic lineages for potential uses in cell therapy6,11–15. However, their use in modeling human marrow injury and other non-neoplastic disorders for translational purposes, such as drug development, has been more limited. A system capable of predicting drug-induced hematotoxicity in patients when exposed to drugs with clinically relevant pharmacokinetics (PK), for example, would be highly valuable for the later stages of drug development, particularly when designing human clinical trials, as well as for regulatory drug safety assessments. Existing hematopoietic toxicity assays are largely based on static methylcellulose colony cultures3,16, in which cells are bathed in drug for extended times, and they are unsuited for this purpose. For these reasons, current in vitro BM models have a limited ability to recapitulate marrow injury and recovery at human-relevant exposures to hematotoxic stressors, such as drugs as well as radiation exposure. Improved methods of doing so would expand their applications to human health and the development of therapeutics, in addition to helping to expedite their regulatory approval.
Results
Human BM Chip supports hematopoiesis in vitro
The hematopoietic compartment of the marrow contains stem and progenitor cells that proliferate and differentiate into mature white and red blood cells through interactions with a variety of surrounding stromal cells (including BMSCs)10,17–19 and it interfaces directly with an endothelium-lined vasculature. The vascular compartment also contributes to BM function by producing angiocrine factors, supplying nutrients, and removing waste10,20. To model this organ functional unit, we adapted a previously described 2-channel, microfluidic Organ-on-a-Chip (Organ Chip) culture device21,22 (Fig. 1a). The top ‘hematopoietic’ channel was filled with a 3D co-culture of human CD34+ cells and BMSCs in a fibrin gel while the bottom ‘vascular’ channel was lined by human umbilical vein endothelial cells (HUVECs) (Fig. 1b). The BM Chip was fed exclusively via perfusion through the vascular channel using medium containing progenitor-supporting cytokines (stem cell factor, Flt-3 ligand, and thrombopoietin) as well as G-CSF and erythropoietin to drive myeloerythroid differentiation.
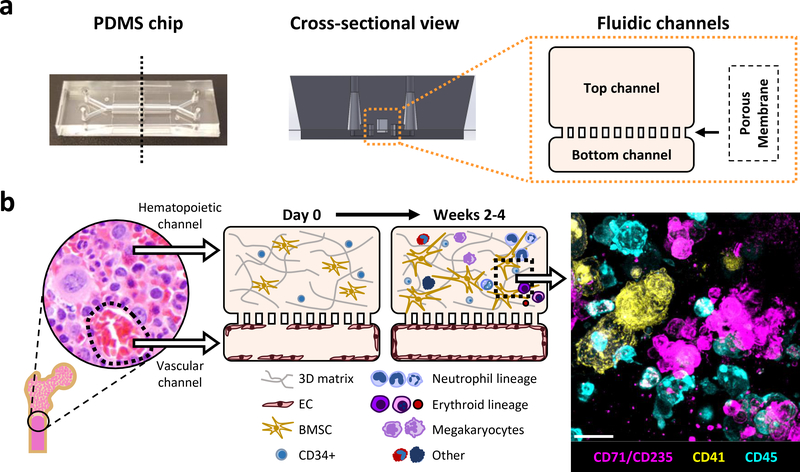
Fig. 1 |. Primary human BM Chip supports in vitro hematopoiesis over 4 weeks in culture and improves CD34+ progenitor survival and colony forming capacity.
a, Photograph of an optically clear PDMS Organ Chip (left) used to create the human BM Chip along with a schematic of the vertical cross-section of the chip (middle) and a magnified diagram of the fluidic channels. b, Schematic of human bone with a micrograph showing normal human BM histology (left) and a schematic cross-sectional view of the human BM Chip on day 0 after seeding showing singly dispersed CD34+ progenitors and BMSCs in a gel in the top channel and an incomplete vascular lining (seeded on either day 0 or day 8) in the bottom channel (left middle). Within 2 weeks of culture initiation, endothelial cells grow to cover all four sides of the lower channel and create a vascular lumen while CD34+ cells undergo expansion and multilineage differentiation (right middle), as illustrated by the immunofluorescence image of a vertical cross section through the gel in the upper channel of the BM Chip taken at day 14 (magenta: erythroid lineage; yellow: megakaryocyte lineage; blue: neutrophil and other hematopoietic lineages; Scale bar, 20 μm).
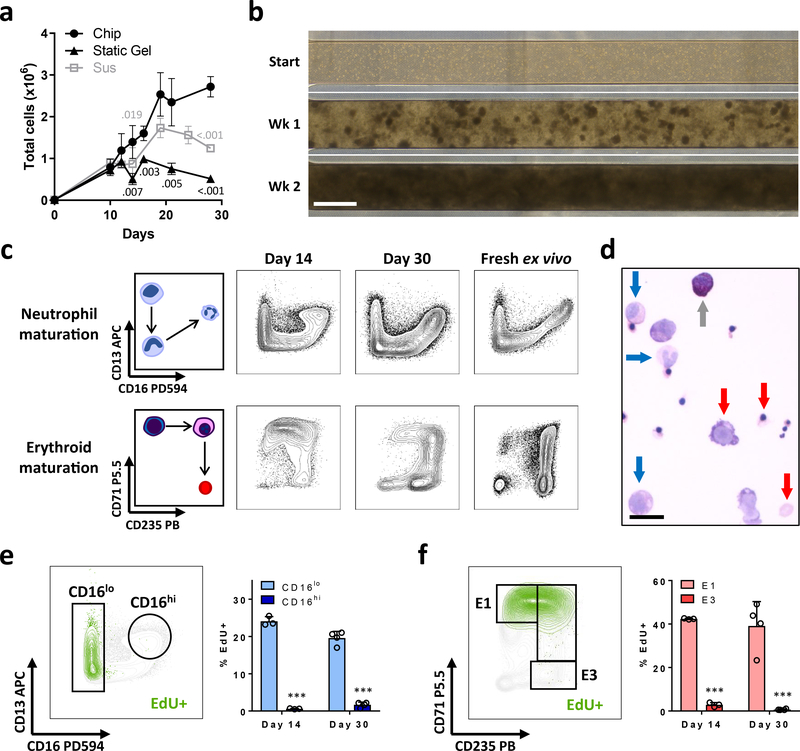
Starting with 1 × 104 CD34+ progenitors, the total number of hematopoietic cells within the BM Chip increased approximately 300-fold over 28 days of culture (Fig. 2a,b), forming a dense cellular microenvironment composed of multiple hematopoietic lineages within 2 weeks, analogous to that observed in living BM (Fig. 1b, 2b and Supplementary Fig. 1a). In comparison to CD34+ cells cultured alone in suspension or with BMSCs in static fibrin gels, BM Chips generated more total cells over time (Fig. 2a). BM Chip cultures also retained higher numbers of CD34+ cells and more colony forming progenitors than suspension and static gel co-cultures, although these numbers eventually decreased after ~ 3 weeks (Supplementary Fig. 1b). Quantification of myeloid versus erythroid colony forming units (CFUs) showed that BM Chips tended to have higher numbers of both colony types as compared to CD34+ cell suspension cultures or static gel co-cultures of CD34+ cells and BMSCs, with and without HUVECs (Supplementary Fig. 1c). Immunophenotypically immature CD34+CD38low cells were also retained at higher levels in the BM Chip (Supplementary Fig. 1d), although the biological significance of CD38 expression after extended in vitro culture remains unclear23–25. We did not detect multipotential colony forming units − granulocyte, erythroid, macrophage, megakaryocyte in any of the conditions at day 14. Addition of HUVECs to static gel co-cultures of CD34+ cells and BMSCs did not increase total, CD34+, neutrophil, or erythroid cell numbers so endothelial cell-derived soluble factors cannot account for the enhanced hematopoiesis observed in the BM Chip (Supplementary Fig. 1e). Taken together, these results show that the BM Chip with dynamic flow increases in vitro hematopoietic output and improves myeloerythroid progenitor numbers and function compared to static suspension and 3D gel culture models.
Fig. 2 |. Continuous myeloerythroid proliferation and differentiation in the BM Chip.
a, Numbers of total cells measured over time in the BM Chip, standard 96-well plate suspension cultures (CD34+ cells alone), or static 3D gel co-cultures (CD34+ cells with BMSCs) as measured by flow cytometry (symbol and error bars represent the mean ± s.d. of n=3–14 chips or 3–6 wells per timepoint; data pooled from 5 independent experiments; numbers indicate P values for static gel [black] and suspension [gray] cultures compared to BM Chips using two-tailed Student’s t-test). b, Bright field micrographs of the top channel of the BM Chip as viewed from above, highlighting the development of cell clusters at week 1 that coalesce to form a dense cellular microenvironment by week 2. Representative image of at least 4 replicates. Scale bar, 1mm. c, Schematic diagrams (left) and representative flow cytometry plots of neutrophil and erythroid maturation (right) showing similar phenotypic profiles between cells extracted from the BM Chip and from fresh human bone marrow. d, Wright-Giemsa stain of cells from a BM Chip at day 21 showing multiple cell types at varying stages of maturation (blue arrows, neutrophil lineage; red arrows, erythroid lineage; gray arrow, non-neutrophil granulocyte; Scale bar, 20 μm). e, Cell proliferation within the neutrophil lineage was assessed by a 2-hour EdU pulse immediately prior to cell harvesting. Flow plot illustrates representative gating strategy for immature CD16lo and mature CD16hi neutrophil subpopulations while EdU+ neutrophil lineage cells are highlighted in green. The percentages of immature and mature neutrophil cells that were EdU+ were quantified by flow cytometry at days 14 and 30. f, Cell proliferation within the erythroid lineage was similarly assessed in the same BM Chips. Flow plot illustrates representative gating strategy for immature CD71+CD235- (E1) and mature CD71-CD235+ (E3) erythroid subpopulations while EdU+ erythroid lineage cells are highlighted in green. The percentages of immature and mature erythroid cells that were EdU+ were quantified by flow cytometry at days 14 and 30. (e and f, n=3–4 chips per timepoint; error bars, s.d.; data pooled from 2 independent experiments; ***P < 0.001 for % EdU+ cells in mature populations compared to immature populations using two-tailed Student’s t-test).
Extended myeloerythroid proliferation and differentiation in the BM Chip
Flow cytometric analysis and Wright-Giemsa staining showed that cells within the BM Chip were predominantly of the neutrophil and erythroid lineages, and they displayed the full spectrum of maturation states (Fig. 2c,d and Supplementary Fig. 1a). Fewer numbers of other blood cell types were also produced, including megakaryocytes and macrophages (Supplementary Fig. 1a), which also occurs in human BM in vivo. Lymphoid-specific growth factors were not included in the culture medium and, as expected, no lymphoid differentiation was observed.
To better assess the dynamics of on-chip hematopoiesis, cells were labeled with a 2 hour pulse of EdU just before they were harvested and analyzed at days 14 and 30. EdU+ cells within the neutrophil lineage appropriately resided largely within the immature CD16lo population as opposed to the mature CD16hi population (Fig. 2e). Similarly, EdU+ cells of the erythroid lineage primarily exhibited an immature CD71+CD235- (E1) phenotype as opposed to a mature CD71-CD235+ phenotype (E3) (Fig. 2f). Importantly, the immature CD16lo neutrophil and E1 erythroid cells maintained their proliferative nature over time (Fig. 2e,f), and higher numbers of these neutrophil and erythroid precursors were found in the BM Chip than in suspension or static gel cultures (Supplementary Fig. 1f). We observed that total neutrophil lineage cell numbers increased steadily in the BM Chip, while total erythroid lineage cell numbers were largely stable in all 3 culture methods, suggesting that there was rapid erythroid turnover in our cultures (Supplementary Fig. 1g). And while the proliferative fraction of total cells decreased as the BM Chip evolved to contain an increasing percentage of mature post-mitotic cells, the CD34+ population retained significantly higher EdU labeling, indicating continued CD34+ progenitor cell proliferation (Supplementary Fig. 1h). These data, in concert with detection of higher numbers of CD34+ cells and CFUs, show that the BM Chip improves in vitro modeling of myeloerythroid proliferation and development over an extended time period compared to conventional static cultures.
In the course of these studies, we observed that a small fraction of cells spontaneously migrated or “intravasated” into the vascular channel over the first 2 weeks of culture, and these cells were markedly biased towards the myeloid lineage relative to the erythroid lineage when compared to cells remaining in the hematopoietic channel (Supplementary Fig. 2a). These intravasating cells also contained a 5-fold higher proportion of mature CD16hi neutrophils (Supplementary Fig. 2a), which is reminiscent of the selective migration from marrow into peripheral blood by mature, but not immature, granulocytes that occurs in vivo. Plate-based cultures of CD34+ cells cannot mimic these effects and they also have known limitations with respect to diffusion-limited exchange of oxygen and nutrients, as well as removal of waste and soluble inhibitory factors26. Indeed, we found that oxygen levels dropped precipitously to near anoxic levels in static suspension or fibrin gel co-cultures with BMSCs (Supplementary Fig. 2b). In contrast, when we fabricated BM Chips using microfluidic devices that contained embedded oxygen sensors27, we found that oxygen levels remained constant at ~75% of atmospheric level (Supplementary Fig. 2b). These findings confirm that the BM Chip significantly improves oxygen delivery (and likely nutrient and waste exchange as well) compared to static plate-based cultures. Having established a model system that supports stable and robust myeloerythroid proliferation and differentiation over time, we next sought to assess its ability to model human bone marrow pathophysiology in response to various stressors in vitro.
BM Chip predicts clinically observed hematotoxicities at patient-relevant drug exposures
BM toxicity is a significant dose-limiting side effect for many drugs. Hence, there is a great need for a human in vitro preclinical model that can predict drug-induced myelosuppression at patient-relevant exposures. 5-fluorouracil (5-FU) is a widely used chemotherapeutic with well-known myelosuppressive effects and it acts predominantly via inhibition of thymidylate synthase, resulting in DNA damage and death in proliferating cells28. Because it is included within multiple chemotherapeutic regimens, there is significant variation in the use of 5-FU with regards to both dosage and schedule, though it is commonly administered via continuous infusion to overcome its short plasma half-life (<15 minutes)29. And while plasma levels vary widely in actual practice, multiple 5-FU-based regimens (e.g., FOLFOX and FOLFIRI) deliver the drug over 2-day infusions to reach target Area Under the Curve (AUC) exposures of 20 – 30 mg.h/L, which corresponds to plasma concentrations ~4 μM (3.2 – 4.8 μM)29,30. These levels of 5-FU over 2 days cause myelosuppression in patients and effective in vitro models should therefore exhibit 5-FU toxicity with a comparable treatment regimen31,32.
We therefore measured the effects of the clinically relevant 2 day exposure to 5-FU on hematopoietic cells in static suspension, 3D gel, and BM Chip cultures, all of which were matured for 10–12 days before drug treatment and thus contained cells of different lineages and maturation stages. BM Chips infused with 5-FU through the vascular channel for 2 days displayed the predicted hematotoxicity at clinically relevant low micromolar concentrations while analogously treated suspension and static gel co-cultures did not (Fig. 3a, Supplementary Fig. 3a). Furthermore, the response at 4 μM in the BM Chip was consistently observed across 6 different donors of CD34+ progenitor cells, demonstrating the robustness of the system for hematopoietic toxicity testing purposes (Fig. 3b).
Fig. 3 |. BM Chip predicts clinically observed hematotoxicities at patient-relevant drug exposures.
a, BM Chips, suspension cultures, and static gel co-cultures were allowed to mature for 10–12 days and then treated for 48 hours with various doses of 5-FU. Cells were harvested immediately afterwards and analyzed by flow cytometry to quantitate total and neutrophil lineage cells (symbol and error bars represent the mean ± s.d. of n=3–9 BM Chips or wells per concentration, data pooled from 3 independent experiments; numbers indicate P values for static gel [black] and suspension [gray] cultures compared to BM Chips using two-tailed Student’s t-test; ***P < 0.001). The clinical target range of patient plasma 5-FU concentrations for a 2-day infusion is highlighted in orange for reference (exposure levels are known to cause myelosuppression)29,30. b, BM Chips were created using CD34+ cells from 6 different donors and treated at day 12 with 4μM 5-FU for 2 days. Total cell numbers were quantified at day 14 by flow cytometry (each color represents an individual donor; n=3–4 replicates per donor; error bars, s.d.; ***P < 0.001 using two-tailed Student’s t-test). c, BM Chips were matured for 10 days, treated with 2-hour or 48-hour infusions of AZD2811, and then cultured again in drug-free medium. AZD2811 concentrations were measured by mass spectrometry in BM Chip outlet medium (circles) and used to fit a PK model of BM Chip drug exposure (black line). For reference, in vivo plasma levels of AZD2811 (gray) were simulated for an average patient at a range of clinical doses35,37 for 2-hour (left) and 48-hour (right) infusions based on the known PK characteristics of AZD2811. Drug levels were below the detection limit (ND = not detectable) at 48 and 96 hours after the start of infusion for the 2-hour and 48-hour regimens, respectively. Dotted line represents the detection limit of mass spectrometry during these experiments (symbol and error bars represent the mean ± s.d. of n=4–8 BM Chips per concentration). d, Graphs showing the effects of infusing the BM Chips on day 10 of culture with varying doses of AZD2811 for 2 hours (left) versus 48 hours (right). Total neutrophil (blue) and erythroid (red) cell numbers were quantified on day 12 by flow cytometry (n=6 chips per condition; error bars, s.d.; data pooled from 2 independent experiments; numbers indicate P values of drug-treated versus control chips using two-tailed Student’s t-test; ***P < 0.001).
While previous studies using methylcellulose colony assays reported in vitro toxicity in the expected low micromolar range16,33, these studies did not use patient-relevant drug exposure durations. In fact, cells in these studies were treated with 5-FU for up to 2 weeks, which represents significant overexposure compared to what patients experience, and we confirmed that a constant 2 week exposure to 5-FU in our static suspension cultures reproduced these published results (Supplementary Fig. 3b). We observed that static cultures treated with the 2-day 5-FU treatment regimen only showed toxicity beginning at ~40 μM, which is an order of magnitude higher than target in vivo patient plasma levels (Fig. 3a). Moreover, when we exposed suspension cultures to the 2-day 5-FU regimen beginning at day 3, we observed a decrease in neutrophils, but an unexpected increase in erythroid cells, with no change in total cell numbers (Supplementary Fig. 3c). Taken together, these data clearly show that static hematopoietic cultures do not reproduce clinical toxicities when exposed to patient-relevant dosing, whereas the human BM Chip does.
To probe the potential value of the BM Chip for preclinical drug safety testing of new compounds, we challenged the model with AZD2811, a selective small molecule inhibitor of Aurora B kinase that is currently in Phase II clinical development as an encapsulated polymer nanoparticle. Aurora B kinase is necessary for proper alignment and segregation of chromosomes during mitosis as well as for completion of cytokinesis34. Aurora B kinase inhibition is being investigated for potential use against both solid and hematologic cancers35–37 and, while it induces polyploidy and apoptotic cell death in leukemia cell lines38, the effect on normal human marrow cells has not been well characterized. When AZD2811 was previously investigated in Phase I clinical trials, administered by intravenous infusion as its rapidly converting phosphate prodrug barasertib (also known as AZD1152) rather than as a nanoparticle, it was found to produce lineage-selective, target-related hematotoxicity that was schedule-dependent. A 2-hour infusion regimen at higher concentrations resulted in neutropenia and anemia while a 48-hour infusion at lower concentrations resulted in more severe neutropenia but no anemia despite similar total exposures to barasertib35,37. Thus, this regimen-dependent side effect profile was unexpected and the ability to use the BM Chip to predict these types of toxicity and recovery behaviors would help to explore their mode of action, which will be critical for advancement of this drug through clinical trials and the regulatory assessment process.
To mimic the infusion schedules used in the phase I studies35,37, we infused the vascular channels of 10 day old BM Chips with clinically relevant amounts of AZD2811 over 2 or 48 hours (total AUC of 0.5, 1, or 2 μM.h in both cases). These doses were calculated to deliver a range of drug exposures that reach those observed in the plasma of human patients who exhibited cytopenias35,37 (see Methods for details). Importantly, use of microfluidics and dynamic fluid flow through the vascular channel of the BM Chip allowed us to reproduce patient-relevant dosing in terms of the AUC and overall shape of the drug exposure profiles (Fig. 3c, Supplementary Fig. 3d). This was confirmed by comparing direct measurements of AZD2811 concentrations in BM Chip outlet medium, pharmacokinetic (PK) models of BM Chip exposures based on those measurements, and the free AZD2811 concentration profiles in the plasma calculated for an average patient at a range of clinical doses (based on its known PK properties)35,37.
Flow cytometric analysis revealed that the 2-hour and 48-hour infusions produced dose-dependent toxicities with these clinical infusion regimens (Supplementary Fig. 3e). Furthermore, dose-dependent toxicity in the erythroid lineage was much greater following the 2-hour infusion regimen than the relatively mild effects seen with the 48-hour regimen (Fig. 3d), which closely aligns with clinical data that only reported anemia (CTCAE grade ˂ 3) in patients receiving the 2-hour infusion of barasertib35,37. Conversely, dose-dependent toxicity of the neutrophil lineage in the BM Chip was greater with the 48-hour infusion (Fig. 3d, Supplementary Fig. 4b) than the 2-hour infusion (68% vs. 48% decrease at the highest tested dose; p < 0.001). This is also consistent with published clinical data showing a higher incidence of severe neutropenia (CTCAE grade ≥ 3) with 48-hour versus 2-hour infusions at a given AUC (Supplementary Fig. 4a)35,37. In contrast, suspension cultures of the same CD34+ cells failed to reproduce the dose regimen-dependent erythroid toxicity (anemia) profile that was successfully mimicked by the human BM Chip (Supplementary Fig. 4c). These data show that for both 5-FU and AZD2811, the BM Chip more accurately predicts hematologic side effects experienced by patients at clinically relevant drug exposures than other BM culture models.
Modeling maturation-dependent cytotoxicity and marrow recovery with the BM Chip
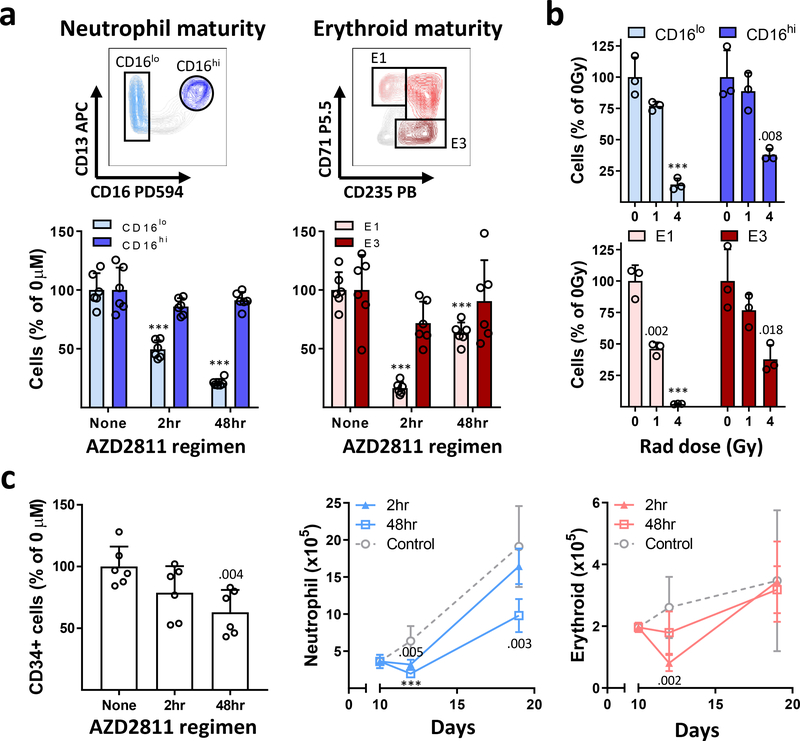
In the marrow, Aurora B kinase inhibition by AZD2811 should selectively target dividing neutrophil and erythroid precursors while sparing their surrounding post-mitotic mature blood cell neighbors. This is likely how it induces peripheral cytopenias, but this is difficult to prove without carrying out a marrow biopsy soon after AZD2811 infusion. In contrast, we could easily examine this hypothesis in the human BM Chip, and we found that, indeed, immature CD16lo neutrophil precursors decreased significantly while mature CD16hi neutrophils were essentially unchanged with both dosing regimens (Fig. 4a, Supplementary Fig. 4b). Similarly, the anemia-inducing 2-hour infusion regimen markedly reduced early E1 erythroid progenitors while the more mature E3 erythroid cells were minimally affected (Fig. 4a). Interestingly, the effect of AZD2811 on CD34+ cell numbers was less striking, suggesting its toxicity is somehow mitigated in the CD34+ progenitor population (Fig. 4c).
Fig. 4 |. Confirmation of maturation-dependent cytotoxicity and modeling marrow recovery with the BM Chip.
a, BM Chips were treated on day 10 with the highest area under the curve (AUC) of AZD2811 (2 μM.h) over 2 or 48 hours and analyzed on day 12 by flow cytometry. Representative flow cytometry gating (top) and graphs (bottom) showing the selective decrease in immature CD16lo neutrophil lineage and E1 erythroid cells compared to their more mature CD16hi neutrophil and E3 erythroid cell counterparts (n=6 chips per condition; error bars, s.d.; data pooled from 2 independent experiments; ***P < 0.001 for drug-treated versus control chips using two-tailed Student’s t-test). b, BM Chips were matured for 10 days before exposure to human-relevant doses of ionizing radiation (0,1, or 4 Gy), and then analyzed on day 14 by flow cytometry. Graphs depict human-appropriate dose responsiveness (1 Gy = mild; 4 Gy = severe) and the preferential loss of immature CD16lo neutrophil lineage and E1 erythroid cells (n=3 chips; error bars, s.d.; data representative of 2 independent experiments; numbers indicate P values for irradiated versus control chips using two-tailed Student’s t-test; ***P < 0.001). c, BM Chips were infused on day 10 with the highest AUC of AZD2811 (2μM.h) over 2 or 48 hours and subsequently allowed to recover in drug-free medium until day 19. CD34+ cell numbers (left) were minimally affected when quantified at day 12. Neutrophil (middle) and erythroid (right) numbers were quantified by flow cytometry at days 10, 12, and 19 to assess the ability of BM Chips to recover after injury (n=6 chips per condition at each timepoint; error bars, s.d.; data pooled from 2 independent experiments; numbers indicate P values for drug-treated versus control chips using two-tailed Student’s t-test; ***P < 0.001).
This maturation-dependent toxicity also would be predicted to occur in response to other myelotoxic stressors that preferentially act upon proliferating cells, such as ionizing gamma (γ)-radiation. To test this, we exposed BM Chips to ionizing radiation doses ranging from 1 to 4 Gray (Gy). Total body irradiation exposure to 1 Gy in humans results in mild recoverable cytopenias, whereas 4 Gy causes 50% lethality over 60 days in the absence of supportive care39. BM Chips exposed to these levels of γ-radiation showed moderately decreased cell numbers at 1 Gy and severe toxicity at 4 Gy, closely matching human radiation sensitivities (Supplementary Fig. 5a). They also displayed the predicted preferential toxicity in immature neutrophil and erythroid precursors (Fig. 4b).
Unlike AZD2811, however, ionizing radiation decreased the numbers of mature CD16hi neutrophils and E3 erythroid cells and had a striking effect on CD34+ cells (Fig. 4b, Supplementary Fig. 5a). This may be due to reactive oxygen species generation and other radiation-associated mechanisms of cell death that a targeted kinase inhibitor like AZD2811 would not trigger. This type of detailed analysis can be examined using the BM chip, whereas it would be difficult to study in conventional methylcellulose colony-based toxicity assays. The response of the BM Chip to ionizing radiation was similar across 5 individual donors of CD34+ progenitor cells, again highlighting the robustness of the system for assessing marrow injury (Supplementary Fig. 5b). The ability of the human BM Chip to quantitatively match in vivo human radiosensitivity while enabling detailed studies of the effects on hematopoietic cells is important for preclinical testing of radiation medical countermeasures, as small animal models are limited in their ability to recapitulate human acute radiation syndrome and often require radiation doses that deviate significantly from those relevant for humans40. There are also ethical, cost, and clinical relevance concerns for studies in non-human primates currently used to evaluate radiation countermeasure drug efficacy and safety, as well as political pressures to ban their use. Thus the human BM Chip could provide a badly needed alternative for development of these types of therapeutics.
A system that can recapitulate marrow recovery in vitro would represent a valuable tool for therapeutic discovery by providing a means to test potential methods to enhance restoration of marrow function, and this is often difficult to study using clinically relevant drug exposure profiles in animal models. To address this possibility, we took advantage of the fact that clinical trial data show the hematologic side effects of barasertib were readily reversed by withholding drug or stimulating with growth factors35,37, implying that efficient marrow recovery occurs upon cessation of AZD2811 treatment. This is in keeping with the observation that most CD34+ cells survived the highest dose of AZD2811 (2 μM.h) regardless of infusion regimen (Fig. 4c). When we cultured the human BM Chip in drug-free medium for 7 additional days after administering this dose of AZD2811, we observed robust neutrophil recovery with both the 2 and 48-hour dosing regimens, as well as erythroid recovery with the 2-hour regimen (Fig. 4c). In contrast, neutrophil recovery was relatively impaired in plate-based suspension cultures even though CD34+ cell numbers were unaffected by AZD2811 treatment (Supplementary Fig. 5c). These results suggest that the BM Chip could be used to study a full cycle of BM injury and recovery in response to therapeutics using human-relevant PK profiles during preclinical drug evaluation, development, and regulatory assessment, which cannot be done using animal models.
Recapitulation of hematopoietic abnormalities in patients with a genetic disorder
Shwachman-Diamond Syndrome (SDS) is a genetic BM failure syndrome and the majority of patients harbor biallelic mutations in the SBDS gene41–43. The marrow is typically hypocellular for age and neutropenia is observed in almost all patients44. Hematologic dysfunction may involve other lineages (e.g., thrombocytopenia or anemia) and there is a predisposition to developing myeloid malignancies as well41,42. The study of hematopoiesis in the context of SDS has been limited because animal models do not faithfully recapitulate the human disease45–49. Thus, to assess the ability of the BM Chip to phenocopy the hematopoietic defects of SDS, we cultured CD34+ cells from two SDS patients (Supplementary Fig. 6a) together with normal BMSCs and endothelial cells in the 2-channel microfluidic device.
After 2 weeks of culture, these SDS patient-derived BM Chips displayed both broad defects in hematopoiesis and cell-type specific responses. Overall, significantly fewer cells developed in the SDS BM Chips compared to control BM Chips (Fig. 5a, Supplementary Fig. 6b), analogous to the hypoplastic phenotype observed in SDS patient marrow biopsies41,44. This decrease in cell numbers was present in both the neutrophil and erythroid lineages (Fig. 5b). Maintenance of CD34+ cell numbers in the SDS BM Chip was also impaired (Fig. 5c), consistent with an intrinsic defect in CD34+ progenitor function in SDS patients and past demonstration of their impaired colony forming ability50,51. In these studies, we noticed that neutrophils in the SDS BM Chip exhibited impaired maturation and developed a smaller percentage of mature CD16hi cells (Fig. 5d, Supplementary Fig. 6c). In contrast, no maturation defect was observed in the erythroid lineage, evidenced by similar numbers of mature E3 erythroid cells in SDS and control BM Chips (Fig. 5e). Instead, there was an unexpected loss of early E1 erythroid progenitors (Fig. 5e).
Fig. 5 |. Human BM Chip recapitulates hematopoietic abnormalities observed in Shwachman-Diamond Syndrome patients.
a, Photographs of BM Chips seeded with CD34+ cells from normal donors versus SDS patients at 2 weeks of culture (n=4 chips per condition; representative of 2 independent experiments). Scale bar, 5mm. Neutrophil (b, left), erythroid (b, right), and CD34+ (c) cell numbers were quantified by flow cytometry. d, Percentages of neutrophils with a mature CD16hi surface phenotype in control versus SDS BM Chips were quantified by flow cytometry. e, Graph showing the number of erythroid cells at different maturation states (left) and representative flow plots (right) depicting the percentages of the erythroid subpopulations (E1: immature, E3: mature), as quantified by flow cytometry. f, Representative flow plots depicting the abnormal CD13 vs CD16 neutrophil maturation pattern observed in SDS BM Chips (top) and SDS patient bone marrow aspirates (bottom) as compared to normal controls (SDS = blue, normal = gray). After gating on CD16hi neutrophils in BM Chips and patient marrow samples, CD13 expression for SDS vs control samples was assessed as shown by the histograms (middle, each histogram represents a different patient) and quantified by comparing median fluorescence intensities (right, square and triangle data points belong to the 2 SDS patients tested in BM Chip cultures). (b-f, BM Chips: n=8, error bars represent s.d., data pooled from 2 independent experiments, each using cells from a different normal and SDS patient with 4 chips per experiment; Patient BM aspirates: n=4 normal and n=8 SDS patients; numbers indicate P values for normal versus SDS samples using two-tailed Student’s t-test except CD13 MFI levels for SDS patients fell into 2 distinct groups and were compared with that of control patients by Mann-Whitney U test; ***P < 0.001).
Interestingly, the impaired neutrophil maturation was further discernable as an aberrantly “blunted” CD13/CD16 expression pattern, where mature CD16hi neutrophils in SDS BM Chips failed to upregulate CD13 to the same degree as their normal BM Chip counterparts (Fig. 5f top). This has not been previously reported in the BM of SDS patients, although mild morphologic dysplasia in the neutrophil lineage has been observed41,44. To assess whether this finding represents an in vitro artifact of the BM Chip, we retrospectively reviewed clinical BM aspirate flow cytometry results from Boston Children’s Hospital and compared the CD13/CD16 expression pattern during neutrophil development in 8 SDS patients with those of control patients without a BM disorder. Interestingly, 50% (4/8) of these patients displayed the same aberrant CD13/CD16 pattern we observed in the SDS BM Chips (Fig. 5f bottom), while the other half exhibited a normal pattern. Notably, both patients from whom CD34+ cells were available for BM Chip cultures belonged to the group that exhibited aberrantly blunted maturation (Fig. 5f bottom), confirming that the human patient-specific BM Chip results replicate their in vivo BM phenotype with high fidelity. Further study is needed to determine the functional implications of these observations, but these data demonstrate that patient-specific SDS BM Chips recapitulate the clinical manifestations of this disease, including neutropenia, anemia, and BM failure.
Discussion
Taken together, our data show the microfluidic human BM Chip faithfully recapitulates key aspects of human hematopoiesis, as well as marrow dysfunction induced by drugs, radiation, and genetic mutation. It is worth noting that this study does not address issues of hematopoietic stem cell (HSC) presence or maintenance, though methods to effectively support, expand, and even create HSCs in vitro are active areas of investigation by others1,2,10,52–54. Instead, our work was directed towards developing an enabling technology with near-term translational potential as an effective preclinical model of human marrow pathophysiology that would allow analysis of human responses to clinically relevant drug PK profiles and radiation dose exposures. As the primary function of the BM in vivo is the production of blood cells, we engineered a system that balances proliferation, multilineage differentiation, and continued progenitor survival to achieve extended myeloerythroid development while also improving bulk CD34+ cell maintenance. Our data show that the human BM Chip supports erythroid differentiation as well as myeloid development and mobilization (intravasation of mature neutrophils) over 4 weeks of culture while improving maintenance of CD34+ progenitors over traditional culture methods. More importantly, in comparison to static suspension or 3D gel cultures, the BM Chip was better able to recapitulate the toxicity responses of human marrow to clinically relevant dose exposures of AZD2811 and 5-FU and also demonstrated an improved ability to recover after injury. Thus, the BM Chip could be used to assess both drug toxicity and recovery without requiring the usual animal studies, which has significant ethical implications, in addition to expediting the drug development process. Finally, BM Chips seeded with SDS patient-derived CD34+ cells demonstrated hematopoietic dysfunctions that parallel key hematologic abnormalities observed in SDS patients and led to the discovery of a neutrophil maturation abnormality that we confirmed exists in a significant subset of these patients in vivo. Thus, this in vitro human marrow model could facilitate drug development, basic research, and translational studies for a range of hematopoietic disorders and toxicities, and potentially offer a human-specific alternative to animal testing for regulatory assessment.
Over 60 studies have reported on the development of 3D or dynamic human hematopoietic culture systems (Supplementary Table 1). These include static cultures of hematopoietic cells embedded within various gel formulations, or seeded into soft or rigid porous scaffolds, as well as dynamic cultures varying from automated medium dilution setups to spinner flask cultures to both microfluidic and macroscale perfused devices. Of these, only 2 studies modeled drug-induced marrow injury and these studies were not focused on characterizing the effects on hematopoiesis8,9. In contrast, we show that by recreating patient-relevant drug exposure PK profiles in the BM Chip, we also recapitulate the regimen-dependent side effect profiles seen in human patients in Phase I clinical studies of a small molecule drug in active clinical development. This shows that the BM Chip has strong translational potential and could be used for human risk assessments with other new drug candidates and differs from past studies with microphysiological systems and Organ Chips that mostly used only model reference compounds. The ability to capture this type of nuanced, but clinically significant, hematotoxicity could benefit the later stages of drug development, particularly when designing clinical trials and exploring mode of toxicity during regulatory assessment. While the biologic mechanisms underlying how different regimens of Aurora B kinase inhibition cause different lineage-specific effects are not understood, the BM Chip provides an experimental tool to gain insight into this unexpected behavior.
As biologic variability is an important determinant of a model system’s utility, we note that our radiation and 5-FU toxicity results were highly reproducible across 5 and 6 different CD34+ donors, respectively. We found that the BM Chip is intrinsically reproducible but have further minimized variability by taking advantage of the ability to benchmark toxicity responses against control conditions that utilize the same CD34+ donor cells. Additionally, these experiments were performed with BMSCs from one donor as well as one commercial lot of HUVECs. By extension, one could systematically alter the source (e.g., normal vs. diseased, male vs. female) of the CD34+, BMSC, or endothelial cell compartments, independently or combination, to examine how changes in those cells affect drug responses or contribute to disorders of myeloerythroid development. The reproducibility we obtained in our studies support the potential utility of the human BM Chip for addressing questions that require robust human-relevant models of drug and radiation-induced myelosuppression, as well as modeling of rare disease phenotypes.
While sustaining hematopoiesis for 1 month in vitro is sufficient to successfully study many types of BM disorders, certain types of experiments may require a longer timeframe. In these situations, combining the BM Chip with methods that improve hematopoietic stem and progenitor maintenance in vitro, such as addition of small molecules SR1 and UM17111,12, might significantly increase the time period over which the BM Chip can approximate human marrow behavior. It is important to note, however, that these compounds expand CD34+ cells at the expense of inhibiting hematopoietic differentiation11,12, which may be a limitation of any experimental manipulation designed to optimize hematopoietic stem and progenitor numbers in future studies.
The specific configuration of the BM Chip we used in the present study was effective for modeling clinically relevant dysfunctions of myeloerythroid development, but the technology could be further optimized for other applications by altering the types of cytokines and cells included in the model. For example, adding different growth factors or using lymphoid-supporting stromal cells might enable the study of lymphoid development, as has been described in co-culture systems with stromal monolayers55,56. Alternatively, decreasing exogenous cytokine supplementation could help determine which factors might be produced by the endothelium and BMSCs, and thus facilitate dissection of cellular crosstalk between the endothelial cells, BMSCs, and hematopoietic cells. These types of studies would be difficult to perform outside of the in vitro setting. Another point of customizability follows from the observation that the BM Chip is capable of maintaining stable oxygen levels in the context of a densely cellular and proliferative culture system. In a static culture, cells are increasingly subject to nutrient scarcity and higher concentrations of metabolic waste products as they proliferate. Our system can maintain an improved equilibrium state due to a continuous supply of new medium and removal of accumulated soluble waste products through the vascular channel. And while our experiments were performed under atmospheric oxygen conditions, experimental oxygen tensions could be matched to those found in in vivo microenvironments in future studies as we have recently demonstrated this capability using our 2-channel Organ Chip27; these types of studies could yield additional insight into the important role that oxygen plays in hematopoietic cell biology57–60.
Careful consideration was given to how to best model the genetic disorder Shwachman Diamond Syndrome given the limited availability of primary patient-derived cells. Ultimately, BM Chips were created using SDS CD34+ cells with normal BMSCs as opposed to also using BMSCs from SDS patients. This limited potential variability and allowed a better comparison to BM Chips seeded with normal CD34+ cells and BMSCs. Moreover, it is known that the hematopoietic cells are the primary drivers of BM dysfunction in this disease as transplants of normal HSCs can cure the marrow phenotype in SDS patients42, and this is consistent with our results that demonstrate the presence of cell-intrinsic defects in CD34+ progenitors from SDS patients. However, future experiments could examine possible contributions of the stromal microenvironment, which is a subject of active investigation in the field46,50,61.
Relatively little is known about how SBDS mutation affects the differentiation of human hematopoietic cells in the marrow. The impaired neutrophil development we observed in the SDS BM Chips aligns with prior work showing impaired function of peripheral blood neutrophils in SDS patients51,62,63, and it is also supported by results from SBDS inhibition and knockout studies in mice47,48. The discovery that the aberrant maturation phenotype we detected with the BM Chip also exists in vivo, in a significant portion (50%) of the 8 SDS patients whose clinical flow cytometry results could be analyzed, illustrates the value that patient-specific BM Chips could have for translational research and precision medicine, especially for patients with rare disorders that would otherwise be difficult to study. Combining the BM Chip with genomic editing tools to facilitate the study of rare marrow diseases could be another exciting application of this technology.
In summary, the vascularized human BM Chip represents a new in vitro preclinical model of human hematopoiesis that recapitulates many clinically relevant features of BM pathophysiology in response to clinically relevant exposures to drugs and ionizing radiation, as well as genetic mutation. Future use of the BM Chip may enable the accurate prediction of myelotoxicity during drug safety testing and thus be used to systematically search for dosing regimens that minimize toxicity in a manner not possible with human subjects, which would then inform clinical trial design. The BM Chip could also facilitate the discovery of therapeutic strategies that enhance marrow recovery and expedite translational research and precision medicine for patients with genetic BM failure disorders, as well as benefit the study of a broader variety of human marrow pathologies for which there are no current effective experimental models.
Methods
Hematopoietic CD34+ cell isolation
Mobilized peripheral blood and leukapheresis product were anonymously collected from donors undergoing stem cell mobilization at the Massachusetts General Hospital (MGH) under Institutional Review Board approved protocol #2015P001859. Mononuclear cells were purified via Histopaque 1077 gradient (Sigma, 10771). CD34+ cells were isolated using positive magnetic bead selection using CD34 MicroBead Kit (Miltenyi Biotec, 130-046-702) and LS columns (Miltenyi Biotec, 130-042-401) according to manufacturer recommendations. CD34+ purity routinely exceeded 85% as assessed by flow cytometry. Aliquots of 3–5 × 105 cells were frozen in RPMI 1640 (Gibco, 12633-012) + 10% DMSO (Sigma, 41640) + 10% FBS (Gibco, 10082-147) using the CoolCell XL (Corning) at −80°C and then transferred to liquid nitrogen cryogenic storage (VWR, CryoPro). Upon thawing, CD34+ cell viability was >90% as assessed by trypan blue (Lonza, 17-942E) and the yield was typically ~60% of frozen cells.
Bone marrow stromal cell isolation
Femoral head specimens were anonymously collected from patients undergoing hip replacement at the MGH under Institutional Review Board approved protocol #2015P001859. Fragments of trabecular bone were digested with 1 mg/mL collagenase type I (Gibco, 17100-017), 25mM HEPES (Thermo Fisher Scientific, 15630-080), and 10% FBS in DMEM (Gibco, 11885-084) for 1 hour at 37°C and then vortexed vigorously to dissociate cells. Cells were then washed 1× with PBS and mononuclear cells were isolated via Histopaque 1077 gradient (Sigma, 10771). Mononuclear cells were then plated in EGM-2 medium (Lonza, CC-3162) containing 10% FBS at high density (>50 million cells per well in 5mL in a 6-well plate). Non-adherent cells were washed away with medium changes on days 1, 3, and 6. Cells were then sequentially passaged and used up to passage 6 with mixed medium containing 10% FBS and composed of 3:1 ratio of DMEM (Gibco, 11885-084) to EGM-2. Immunophenotyping by flow showed that cells expressed typical BMSC markers including CD90 (5E10 clone, Biolegend) CD105 (AD2 clone, Biolegend), and CD73 (43A3 clone, Biolegend) and were negative for hematopoietic (CD45, HI30 clone, BioLegend) and endothelial (CD31, WM59 clone, eBioscience) markers.
Shwachman-Diamond Syndrome CD34+ cells and clinical flow cytometry data
Written informed consent was provided by patients and normal donors in accordance with the Declaration of Helsinki. Primary human-derived bone marrow mononuclear cells were collected and frozen according to research protocols approved by the IRB of Boston Children’s Hospital. Upon thawing, SDS patient or normal donor-derived bone marrow mononuclear cells were allowed to recover in StemSpan SFEM II (STEMCELL Technologies, 09655) supplemented with recombinant human cytokines (all from PeproTech): 100ng/mL Flt3 (AF-300-19-50UG), 100ng/ml SCF (AF-300-07-50UG), 100ng/ml TPO (AF-300-18-50UG), 20ng/ml IL3 (AF-200-03-50UG). After 30–32 hours of recovery, CD34+ cells were sorted using magnetic beads as above and cultured in the BM Chip as described before.
Review of clinical flow cytometry data was performed at Boston Children’s Hospital under an approved IRB protocol. All available SDS patient bone marrow aspirates were gathered and parsed for cases that allowed analysis of the CD13/CD16 maturation pattern of the neutrophil lineage. Flow data from 8 SDS patients was found that included CD13 FITC and CD16 PerCP-Cy5.5; some patients had multiple aspirate samples at different timepoints and these were aggregated together as the CD13/CD16 patterns were similar. Similarly stained BM aspirates from patients that did not receive a diagnosis of a primary bone marrow disorder served as controls.
Chip fabrication and preparation
The device was fabricated using polydimethylsiloxane (PDMS; SYLGARD® 184 silicone elastomer kit) with previously described soft lithography techniques21,64 or obtained from Emulate, Inc. The chip design, which was similar to previously published devices21, included apical and basal microchannels with dimensions of 1 × 1 × 16.7 mm and 1 × 0.2 × 16.7 mm (w x h x l), respectively, that were separated by a 50 μm thick porous PDMS membrane (7μm diameter holes with 40μm spacing). Both channels were washed with 70% ethanol, filled with 0.5 mg/mL sulfo-SANPAH solution (Thermo Fisher Scientific, A35395) in 50mM HEPES pH 8 and placed under a UV lamp (Nailstar, NS-01-US) for 20 minutes to activate the surface.The microchannels were then rinsed sequentially with 50 mM HEPES buffer and cold PBS, and then filled with a coating solution of PBS containing 100 μg/ml fibronectin (Gibco, 33016-015) and 50 μg/ml collagen (Sigma, C2249) and placed at 37°C for 2 hours. The solution was then aspirated from the chip and it was allowed to air dry overnight.
Organ Chip Culture
The apical channel was first seeded with 1×104 primary human CD34+ cells and 5 ×103 BMSCs in a solution containing 5 mg/ml fibrinogen (Millipore, 341578), 0.2 mg/ml collagen I (Sigma, C2249), 25 μg/ml aprotinin (Sigma, A3428), and 0.5 U/ml thrombin (Millipore, 605195), that was allowed to gel on-chip. To prevent premature crosslinking, medium containing fibrinogen and aprotinin was prepared separately from the medium containing cells, collagen, and thrombin. The two solutions were then carefully mixed at a ratio of 1:3 (vol:vol) to avoid bubbles, immediately pipetted into the apical channel (25uL per chip), and allowed to gel for 20–30 minutes at room temperature. HUVECs (Lonza, CC-2517 or C2519A) were seeded either at this point or at day 8 by adding medium containing the cells (2–3×106/ml; 30uL/chip) into the lower fibronectin- and collagen-coated channel followed by immediate inversion of the device and static incubation at 37°C for 2 hours to promote adhesion to the central membrane. The devices were then placed right-side up and apical channel ports were sealed with sterile tape. For experiments over 21 days in length, the overgrowth of stromal cells was prevented via irradiation of BMSCs at 12Gy before initiation of BM Chip cultures. The entire BM Chip was fed via medium perfused only through the endothelium-lined channel using a 16-channel peristaltic pump (Ismatec, ISM 938D) with 0.25 mm inner diameter (ID) pump tubing (Ismatec, 95723-12) with 19G bent metal pins. Outflow medium was collected using 0.89 mm ID tubing (Cole-Parmer, 95809-26) with 19G bent metal pins into a sterile reservoir. All parts were sterilized using oxygen plasma (Diener ATTO; 0.3 mbar pressure, 50W power and 2 min plasma time). Pump speeds were set at 1.2–1.6μL/min (varied depending on manufacturer’s lot of tubing) with regular pauses calculated to deliver a total flow-through volume of 200 μL/day for the first seven days and 400μL/day thereafter. When drug treatments were initiated, pump speeds remained unchanged but the pauses were shortened such that the total flow-through volume increased to 1.6 mL/day in order to facilitate equilibration of drug into the chip and match human pharmacokinetic profiles.
In all studies, we used SFEM II medium (STEMCELL Technologies, 09655) supplemented with 10% FBS (Gibco, 10082-147), 100 U/l penicillin and 100 mg/ml streptomycin (Pen/Strep; Gibco, 15140-122), 12.5 μg/ml aprotinin (Sigma, A3428), 20 ng/ml EPO (PeproTech, 100-64), 1 ng/ml G-CSF (PeproTech, 300-23), 100 ng/ml Flt3-L (PeproTech, 300-19), 100 ng/ml TPO (PeproTech, 300-18), 50 ng/ml SCF (PeproTech, 300-07), and select EGM-2 BulletKit (Lonza, CC-4176) components (hFGF-B, VEGF, R3-IGF-1, hEGF, ascorbic acid, and heparin) according to the manufacturer’s instructions. Medium was degassed under vacuum for at least 15 minutes before use to minimize bubble formation. Chips were kept humidified with damp tissues (replaced every 2 days; dampened with sterile water containing Lysol No-Rinse Sanitizer diluted 500×) placed on top of the chips and the whole setup was covered with aluminum foil.
Static gel and suspension cultures
For static gel cultures, the same numbers of CD34+ cells and BMSCs as used in the BM Chips were seeded into individual wells of a 96 well plate (Corning, 353072) in 50μL of the same fibrin+collagen gel used in BM Chips in order to cover the entire well surface. After crosslinking, 200μL of the same medium used in BM Chips was added on top of the gel; the medium was fully replaced on day 3 and then daily starting at day 5. For suspension cultures, the same numbers of CD34+ cells were seeded into individual wells of a 96 well plate in 250μL of medium, and 50% of the medium volume was replaced daily.
Oxygen measurements
BM Chips with embedded oxygen sensors were created as recently described27. For static gel culture measurements, an oxygen sensor spot (PreSens Precision Sensing GmbH, SP-PSt3-NAU) was placed at the bottom of a 96 well flat bottom plate before cell seeding. Cells were then cultured as described above. Oxygen measurements were optically measured daily using an OXY-4 mini instrument (PreSens Precision Sensing GmbH).
Flow cytometry analysis
Cells were harvested from BM Chips and static gel cultures after digestion using 2.5 mg/mL nattokinase (Japan Bio Science Laboratory, NSK-SD), 1 mg/mL collagenase type I (Gibco, 17100-017), 25mM HEPES (Thermo Fisher Scientific, 15630-080), and 10% FBS in DMEM (Gibco, 11885-084). To digest BM Chips, one port of the bottom channel was plugged with a 200μL filter tip and 100μL of digestion solution was added via the other port. The tape on top of the apical channel ports was removed and a 200μL filter tip containing 50μL of digestion solution was then plugged in each apical channel port. After incubation for 1 hour at 37°C, the gel was broken up by pipetting the digestion solution in the chip up and down. If necessary, an additional 30–60 min of incubation was allowed. To digest static gel cultures, all medium was removed and 100μL of digestion solution was added on top; after incubation for 1 hour at 37°C, cells were harvested or allowed an additional 30 min of digestion if necessary.
Harvested cells were centrifuged, resuspended in flow staining buffer composed of 1% FBS (Gibco, 10082-147), 25mM HEPES (Thermo Fisher Scientific, 15630-080), 1mM EDTA (Thermo Fisher Scientific, 15575-020), and 0.05% sodium azide (VWR, BDH7465-2) in DPBS (Gibco, 14190-144), and filtered through 105 μm pore nylon mesh (Component Supply Co, U-CMN-105-A). Antibody staining was performed for 30 min in 96 well V-bottom plates (Nunc, 249944) in 100μL of staining buffer using one of 2 panels:
Panel 1: anti-CD235a-Pacific Blue (HI264 clone, BioLegend, 349108, dilution 1:80), anti-CD15-Brilliant Violet 510 (W6D3 clone, BioLegend, 323028, dilution 1:50), anti-CD45-Brilliant Violet 570 (HI30 clone, BioLegend, 304034, dilution 1:50), anti-CD33-PE (WM53 clone, BioLegend, 303404, dilution 1:50), anti-CD16-PE/Dazzle 594 (3G8 clone, BioLegend, 302054, dilution 1:80), anti-CD41-PE/Cy5 (HIP8 clone, BioLegend, 303708, dilution 1:133), anti-CD71-PerCP/Cy5.5 (CY1G4 clone, BioLegend, 334114, dilution 1:50), anti-CD34-PE/Cy7 (581 clone, BioLegend, 343516, dilution 1:50), anti-CD13-APC (WM15 clone, BioLegend, 301706, dilution 1:80). Panel 2: anti-CD45-Brilliant Violet 570 (HI30 clone, BioLegend, 304034, dilution 1:50), anti-CD34-PE/Cy7 (581 clone, BioLegend, 343516, dilution 1:50), anti-CD38-APC (HB-7 clone, BioLegend, 356606, dilution 1:80). Both panels included 20 nM Syto16 (Thermo Fisher Scientific, S7578, dilution: 1:500), Zombie NIR dye (BioLegend, 423106, dilution: 1:500), Fc Block (BioLegend, 422302, dilution 1:20), Monocyte block (BioLegend, 426103, dilution 1:20), and Brilliant stain buffer (BD, 566385, dilution 1:20). Additionally, 5 × 103 counting beads (Spherotech, ACRFP-100-3) were added to each sample to enable quantification of cell numbers. Stained cells were analyzed using the LSRFortessa (BD Biosciences). Results were analyzed using FlowJo V10 software (Flowjo, LLC). A representative gating strategy is illustrated in Supplementary Fig. 7.
Immunofluorescence microscopy
BM Chips were fixed by plugging one bottom channel port with a 200μL filter tip and gently adding 200μL of 2% paraformaldehyde (Electron Microscopy Sciences, 15730), 25mM HEPES (Thermo Fisher Scientific, 15630-080) in DPBS (Gibco, 14190-144) via the other bottom channel port. After 2 hours, the fixation solution was removed and replaced with DPBS. The chips were then kept at 4°C until sectioning at 500μm with a vibratome (Leica, VT1000S). Sections were blocked/permeabilized using 0.1% Triton X-100 (Sigma, T8787), 5% Normal Goat Serum (Jackson Immunoresearch, 005-000-121), 25mM HEPES in DPBS for 30 minutes at room temperature. Samples were then stained overnight at 4°C in the same buffer containing Fc Block (BioLegend, 422302, 1:20), anti-CD41 Alexa Fluor 488 (Biolegend, 303724, 1:20), anti-CD71 purified (Biolegend, 334102, 1:400), anti-CD235 purified (Biolegend, 349102, 1:400), anti-CD45 Alexa Fluor 594 (Biolegend, 304060, 1:20). The next day, after 3 washes of PBS, sections were then stained overnight at 4°C in the same buffer containing goat anti-mouse IgG2a Alexa Fluor 555 (ThermoFisher, A-21137, 1:500), and Hoechst 33342 (Life Technologies, H3570, 1:10,000). Images were taken using a laser scanning confocal immunofluorescence microscopes (Zeiss TIRF/LSM 710) with a 405-nm diode laser, a 489–670 nm white light laser, 488 nm and 496 nm argon laser and coupled to a photomultiplier tube or HyD detectors. Acquired images were analyzed using IMARIS software (Bitplane).
Cell growth assay
Growing cells were detected using the Click-iT™ Plus EdU Alexa Fluor™ 350 Flow Cytometry Assay Kit (Thermo Fisher Scientific, C10645). BM Chips were perfused with medium containing 10μM EdU for 2 hours immediately before cell harvesting as described above. After surface staining with panel 1 antibodies, samples were fixed with 100 μl of 2% paraformaldehyde (Electron Microscopy Sciences, 15730), 25mM HEPES (Thermo Fisher Scientific, 15630-080) in DPBS (Gibco, 14190-144) for 20 min at room temperature. Samples were then processed as per manufacturer’s protocol.
Wright-Giemsa staining
1×105 cells in 200 μl DPBS were spun (300 rpm, 5 minutes, RT) onto Superfrost Plus slides (VWR, 48311-601) using a Cytospin 4 (Thermo Fisher Scientific) cytocentrifuge and air dried. Slides were placed for 4 minutes in full-strength staining solution (Sigma, WG16) and then moved to diluted staining solution (1 part WG16 to 5 parts DPBS) for 30 minutes. Slides were then washed in deionized water, air dried, and cover slipped using Cytoseal 60 (Electron Microscopy Sciences, 18006). Representative images were taken using a Zeiss Axioplan 2 microscope with AmScope MU1830 digital camera.
Colony forming unit (CFU) assay
CFU assays were performed using MethoCult™ medium (STEMCELL Technologies, 04434) in meniscus-free 6 well plates (STEMCELL Technologies, 27370) as per manufacturer’s suggestions. Each well contained total cells sufficient to include 50–70 CD34+ cells as calculated from same-day flow cytometry results. Plates were placed into a secondary humidified chamber within an incubator and analyzed using the STEMvision instrument and software (STEMCELL Technologies) after 14 days.
Drug treatment
AZD2811 (formerly known as AZD1152 hydroxy-quinazoline pyrazole anilide) was provided by AstraZeneca in DMSO and stored at −20°C. Fresh medium with drug at the indicated concentrations was prepared daily and added to BM Chip reservoirs and plate cultures for the indicated times. To stop drug treatment, BM Chips were flushed at maximum pump speed (using the MAX/CAL button) for 30 seconds while suspension cultures were harvested, diluted with additional medium, centrifuged at 300g for 10 min, and replated in fresh medium. Dosing was based on results from a Phase I clinical trial35,37, which showed that cytopenia-inducing doses of drug corresponded to a total Area Under the Curve (AUCTotal) per dose of 4776 – 13520 ng.h/mL in human plasma. Accounting for the molecular weight (507 g/mol) and protein binding of this drug (4% fraction unbound in human plasma; internal AstraZeneca testing), this corresponds to hematologic side effects being induced by free drug AUCTotal amounts of 0.376–1.06 μM.h. Correcting for protein binding of AZD2811 in the BM Chip medium (37.5% fraction unbound as determined by mass spectrometry) results in a corresponding AUCTotal range in BM Chip medium of 1.00 – 2.83 μM.h. Thus, BM Chips were dosed at 0.5, 1, and 2 μM.h to match patient exposure levels.
5-Fluorouracil (Sigma, F6627) was dissolved in DMSO and stored at −20°C. Fresh medium with drug at the indicated concentrations was prepared on the day prior to use and kept overnight at 4°C before being added to BM Chip reservoirs and plate cultures for the indicated times. Dosing was determined based upon the 48 hour infusion used in many 5-FU containing chemotherapy regimens (e.g. FOLFOX, FOLFIRI, FOLFIRINOX, and LV5FU2) and target patient plasma concentrations30. For suspension cultures that required removal of 5-FU, cells were harvested into V-bottom plates, spun down, and washed 4 times with blank SFEM II medium before being put back into the standard culture medium used in all experiments.
Mass spectrometry analysis of AZD2811
Drug concentrations in medium samples were determined by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). Briefly, 50 μL of each sample was dispensed into a 96-well plate followed by addition of 250 μL of protein precipitation solution (80:20 acetonitrile:methanol) containing internal standard (clozapine). Plates were vortexed for 5 min and then centrifuged at 2150 × g for 5 min at 4 °C. Clear supernatant solutions (225 μL) were transferred to clean plates and dried to completeness under nitrogen, reconstituted in 80:20 10mM ammonium formate in water: acetonitrile (200 μL) and analyzed using LC-MS/MS. Liquid chromatography was performed using a Waters XBridge C18 3.5um, 30 × 3mm. The ionization was conducted in positive mode and the m/z transitions were 507.9/130.2 for AZD2811. Mass spectrometry was performed using an AB Sciex 6500 Triple quadrupole mass spectrometer (AB Sciex, Foster City, CA, USA) operated in electrospray ionization mode. Data were acquired and analyzed using Analyst software (v 1.6.2).
The unbound fraction of AZD2811 in culture medium was determined using equilibrium dialysis. Medium samples spiked with 1μM of compound were dialyzed using the Rapid Equilibrium Dialysis device (Thermo Fisher Scientific Inc, Rockford, IL) by transferring 300 μL of spiked medium to the dialysis unit’s medium chamber and 500 uL of PBS to the buffer chamber. The dialysis unit was incubated at 37°C and 400 rpm for 18 hours. At the end of the incubation, 25 μL of each sample from the medium and buffer chambers was transferred to a 96 well plate. Blank medium or buffer (25 μL) was added to the buffer or medium samples, respectively, to yield identical matrices. Samples were then quantified using LC-MS/MS, as described above, and the unbound drug fraction (Funbound) in the medium was calculated as follows: Funbound = [Buffer chamber] / [Medium chamber].
PK modeling and simulation
All model fitting and simulations were performed using WinNonlin Phoenix NLME 6.4A. Clinical pharmacokinetic profiles of AZD2811 plasma concentration versus time for a population average patient were simulated at each Phase 1 study dose level (75, 100 & 150 mg administered by 2 hour infusion; 100, 150 & 225 mg administered by 48 hour infusion) using the 3-compartment population PK model for AZD2811 described by Keizer et al65. The free fraction in human plasma was assumed to be 0.04 (AstraZeneca derived value).
For the BM Chip, a three-compartment PK model was fitted to the LC-MS/MS quantified AZD2811 concentrations. The central compartment in the model was viewed as representing the vascular channel and therefore the volume was fixed at the physical volume of the channel (20 μL) and the clearance fixed at the flow rate of medium through the channel (72 μL/hr). The first peripheral compartment was viewed as representing the hematopoietic channel and the volume fixed at the physical volume of the channel (25 μL). The rate and extent of distribution into the hematopoietic channel was described using a distribution clearance (Cl2) and a partition coefficient (Kphem) with both parameter value estimates being obtained during the model fitting process. Distribution into a second peripheral compartment (described by parameters V3 and Cl3) was found to be required to describe the second phase in the outlet medium concentration profile during the washout of drug from the system. Model parameter value estimates (Kphem = 4.5; Cl2 = 99 mL/hr; V3 = 163 μL; Cl3 = 5.6 μL/hr) were derived by fitting the PK model to the combined outlet medium concentration data from all AZD2811 dosed BM Chips using a naïve-pooled approach. Residual unexplained variability (between model and observed concentration data) was modelled using a multiplicative error model.
Radiation exposure
For radiation exposure studies, BM Chips were disconnected from peristaltic pumps, transported in sterile containers to an irradiation facility at Boston Children’s Hospital, and exposed to a single selected γ-irradiation dose (Cs-137; Gammacell 40 Exactor) at 0.98 Gy/min. BM Chips were then reconnected to the peristaltic pumps, flushed for 5 seconds at maximum pump speed to remove bubbles, and placed in the incubator.
Statistical Analyses
All graphs depict mean ± standard deviation (s.d.) and tests for differences between two groups were performed using two-tailed unpaired Student’s t-test unless otherwise specified. Prism 7 (GraphPad Software) was used for statistical analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All the data supporting the results in this study are available within the Article and its Supplementary Information. The broad range of raw datasets acquired and analysed (or any subsets of it), which for reuse would require contextual metadata, are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
This research was sponsored by funding from the US Food and Drug Administration grant HHSF223201310079C, Defense Advanced Research Projects Agency under Cooperative Agreement Number W911NF-12-2-0036, AstraZeneca, and the Wyss Institute for Biologically Inspired Engineering (to D.E.I.); US National Institutes of Health (R24 DK099808 and 5U01HL134812 to A.S., R01 DK102165 to C.D.N., and 5T32CA009216-37 training grant to D.B.C.); and the Department of Defense (W81XWH-14-1-0124 to C.D.N.). Additional funding was provided by the Dana-Farber Cancer Center Claudia Adams Barr Award (C.E.J.) and the EPSRC Centre for Innovative Manufacturing in Regenerative Medicine (A.R.). The authors would like to thank S. Sweeney for helpful discussions, P. Machado and J. Caramanica for machining expertise, and M. DeLelys, R. Matthews, J. Houston, J. Patel, D. Kingman, A. Shay, J. Graham, S. Chung, T. Spitzer MD, and F. Preffer PhD at Massachusetts General Hospital as well as M. Fleming MD DPhil and M. Armant PhD at Boston Children’s Hospital for their invaluable help in working with patient data and samples.
Footnotes
Competing interests
D.E.I. is a founder and holds equity in Emulate, Inc., and chairs its scientific advisory board. D.B.C., V.F., Y.M., L.S.M.T., O.L., R.N., and D.E.I. are co-inventors on a patent application describing the BM Chip. R.D., P.P.-D., D.F., A.M., and L.E. are employed by AstraZeneca, which is developing AZD2811.
References
- 1.Doulatov S, Notta F, Laurenti E & Dick JE Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Eaves CJ Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 125, 2605–2613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wognum B, Yuan N, Lai B & Miller CL Colony forming cell assays for human hematopoietic progenitor cells. Methods Mol Biol 946, 267–283 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Dexter TM, Allen TD & Lajtha LG Conditions controlling the proliferation of haemopoietic stem cells in vitro. J Cell Physiol 91, 335–344 (1977). [DOI] [PubMed] [Google Scholar]
- 5.Sieber S et al. Bone marrow-on-a-chip: Long-term culture of human haematopoietic stem cells in a three-dimensional microfluidic environment. J Tissue Eng Regen Med 12, 479–489 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Thon JN et al. Platelet bioreactor-on-a-chip. Blood 124, 1857–1867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Maggio N et al. Toward modeling the bone marrow niche using scaffold-based 3D culture systems. Biomaterials 32, 321–329 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Rodling L et al. 3D models of the hematopoietic stem cell niche under steady-state and active conditions. Sci Rep 7, 4625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourgine PE et al. In vitro biomimetic engineering of a human hematopoietic niche with functional properties. Proc Natl Acad Sci U S A 115, E5688–E5695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SJ & Scadden DT The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boitano AE et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science (80-. ). 329, 1345–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fares I et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science (80-. ). 345, 1509–1512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira MS et al. Cord blood-hematopoietic stem cell expansion in 3D fibrin scaffolds with stromal support. Biomaterials 33, 6987–6997 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Lee HY et al. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature 522, 474–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pineault N & Abu-Khader A Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol 43, 498–513 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Pessina A et al. Application of the CFU-GM assay to predict acute drug-induced neutropenia: an international blind trial to validate a prediction model for the maximum tolerated dose (MTD) of myelosuppressive xenobiotics. Toxicol Sci 75, 355–367 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Boulais PE & Frenette PS Making sense of hematopoietic stem cell niches. Blood 125, 2621–2629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Méndez-Ferrer S et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacchetti B et al. Self-Renewing Osteoprogenitors in Bone Marrow Sinusoids Can Organize a Hematopoietic Microenvironment. Cell 131, 324–336 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Rafii S, Butler JM & Ding BS Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassell BA et al. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep 21, 508–516 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Huh D et al. Reconstituting organ-level lung functions on a chip. Science (80-. ). 328, 1662–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorrell C, Gan OI, Pereira DS, Hawley RG & Dick JE Expansion of human cord blood CD34(+)CD38(−) cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID repopulating cell (SRC) frequency: dissociation of SRC phenotype and function. Blood 95, 102–110 (2000). [PubMed] [Google Scholar]
- 24.von Laer D et al. Loss of CD38 antigen on CD34+CD38+ cells during short-term culture. Leukemia 14, 947–948 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Danet GH, Lee HW, Luongo JL, Simon MC & Bonnet DA Dissociation between stem cell phenotype and NOD/SCID repopulating activity in human peripheral blood CD34(+) cells after ex vivo expansion. Exp Hematol 29, 1465–1473 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Csaszar E et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 10, 218–229 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Jalili-Firoozinezhad S et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. (2019). doi: 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longley DB, Harkin DP & Johnston PG 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3, 330–338 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Lee JJ, Beumer JH & Chu E Therapeutic drug monitoring of 5-fluorouracil. Cancer Chemother Pharmacol 78, 447–464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saif MW, Choma A, Salamone SJ & Chu E Pharmacokinetically guided dose adjustment of 5-fluorouracil: a rational approach to improving therapeutic outcomes. J Natl Cancer Inst 101, 1543–1552 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Santini J et al. 5-FU therapeutic monitoring with dose adjustment leads to an improved therapeutic index in head and neck cancer. Br J Cancer 59, 287–290 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trump DL et al. Pharmacokinetic and pharmacodynamic analysis of fluorouracil during 72-hour continuous infusion with and without dipyridamole. J Clin Oncol 9, 2027–2035 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Malerba I, Casati S, Diodovich C, Parent-Massin D & Gribaldo L Inhibition of CFU-E/BFU-E and CFU-GM colony growth by cyclophosphamide, 5-fluorouracil and taxol: development of a high-throughput in vitro method. Toxicol Vitr. 18, 293–300 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Carmena M, Wheelock M, Funabiki H & Earnshaw WC The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13, 789–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boss DS et al. Clinical evaluation of AZD1152, an i.v. inhibitor of Aurora B kinase, in patients with solid malignant tumors. Ann Oncol 22, 431–437 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Kantarjian HM et al. Stage I of a phase 2 study assessing the efficacy, safety, and tolerability of barasertib (AZD1152) versus low-dose cytosine arabinoside in elderly patients with acute myeloid leukemia. Cancer 119, 2611–2619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz GK et al. Phase I study of barasertib (AZD1152), a selective inhibitor of Aurora B kinase, in patients with advanced solid tumors. Invest New Drugs 31, 370–380 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Yang J et al. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood 110, 2034–2040 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Waselenko JK et al. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med 140, 1037–1051 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Singh VK & Seed TM A review of radiation countermeasures focusing on injury-specific medicinals and regulatory approval status: part I. Radiation sub-syndromes, animal models and FDA-approved countermeasures. Int J Radiat Biol 93, 851–869 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Dror Y et al. Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome. Ann N Y Acad Sci 1242, 40–55 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Myers KC, Davies SM & Shimamura A Clinical and molecular pathophysiology of Shwachman-Diamond syndrome: an update. Hematol Oncol Clin North Am 27, 117–28, ix (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bezzerri V & Cipolli M Shwachman-Diamond Syndrome: Molecular Mechanisms and Current Perspectives. Mol. Diagnosis Ther 23, 281–290 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Myers KC et al. Variable clinical presentation of Shwachman-Diamond syndrome: update from the North American Shwachman-Diamond Syndrome Registry. J Pediatr 164, 866–870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finch AJ et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev 25, 917–929 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raaijmakers MH et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 464, 852–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawls AS, Gregory AD, Woloszynek JR, Liu F & Link DC Lentiviral-mediated RNAi inhibition of Sbds in murine hematopoietic progenitors impairs their hematopoietic potential. Blood 110, 2414–2422 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zambetti NA et al. Deficiency of the ribosome biogenesis gene Sbds in hematopoietic stem and progenitor cells causes neutropenia in mice by attenuating lineage progression in myelocytes. Haematologica 100, 1285–1293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Shi M, Hui CC & Rommens JM Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol 26, 6656–6663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dror Y & Freedman MH Shwachman-Diamond syndrome: An inherited preleukemic bone marrow failure disorder with aberrant hematopoietic progenitors and faulty marrow microenvironment. Blood 94, 3048–3054 (1999). [PubMed] [Google Scholar]
- 51.Kuijpers TW et al. Hematologic abnormalities in Shwachman Diamond syndrome: lack of genotype-phenotype relationship. Blood 106, 356–361 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Walasek MA, van Os R & de Haan G Hematopoietic stem cell expansion: challenges and opportunities. Ann N Y Acad Sci 1266, 138–150 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Lis R et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimura R et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohn LA et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat Immunol 13, 963–971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rawlings DJ, Quan SG, Kato RM & Witte ON Long-term culture system for selective growth of human B-cell progenitors. Proc Natl Acad Sci U S A 92, 1570–1574 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison JS, Rameshwar P, Chang V & Bandari P Oxygen saturation in the bone marrow of healthy volunteers. Blood 99, 394 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Nombela-Arrieta C et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol 15, 533–543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer JA et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broxmeyer HE, O’Leary HA, Huang X & Mantel C The importance of hypoxia and extra physiologic oxygen shock/stress for collection and processing of stem and progenitor cells to understand true physiology/pathology of these cells ex vivo. Curr Opin Hematol 22, 273–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andre V et al. Mesenchymal stem cells from Shwachman-Diamond syndrome patients display normal functions and do not contribute to hematological defects. Blood Cancer J 2, e94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dror Y et al. Immune function in patients with Shwachman-Diamond syndrome. Br J Haematol 114, 712–717 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Orelio C & Kuijpers TW Shwachman-Diamond syndrome neutrophils have altered chemoattractant-induced F-actin polymerization and polarization characteristics. Haematologica 94, 409–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novak R et al. Scalable Fabrication of Stretchable, Dual Channel, Microfluidic Organ Chips. J. Vis. Exp (2018). doi: 10.3791/58151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keizer RJ, Zandvliet AS, Beijnen JH, Schellens JH & Huitema AD Two-stage model-based design of cancer phase I dose escalation trials: evaluation using the phase I program of barasertib (AZD1152). Invest New Drugs 30, 1519–1530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the results in this study are available within the Article and its Supplementary Information. The broad range of raw datasets acquired and analysed (or any subsets of it), which for reuse would require contextual metadata, are available from the corresponding author on reasonable request.