Abstract
The involvement of host immunity in the gut microbiota-mediated colonization resistance to Clostridioides difficile infection (CDI) is incompletely understood. Here, we show that interleukin (IL)-22, induced by colonization of the gut microbiota, is crucial for the prevention of CDI in human microbiota-associated (HMA) mice. IL-22 signaling in HMA mice regulated host glycosylation, which enabled the growth of succinate-consuming bacteria Phascolarctobacterium spp. within the gut microbiome. Phascolarctobacterium reduced the availability of luminal succinate, a crucial metabolite for the growth of C. difficile, and therefore prevented the growth of C. difficile. IL-22-mediated host N-glycosylation is likely impaired in patients with ulcerative colitis (UC), and renders UC-HMA mice more susceptible to CDI. Transplantation of healthy human-derived microbiotas or Phascolarctobacterium reduced luminal succinate levels and restored colonization resistance in UC-HMA mice. IL-22-mediated host glycosylation thus fosters the growth of commensal bacteria that compete with C. difficile for the nutritional niche.
Introduction
Clostridioidies difficile infection (CDI) causes a severe, potentially life-threatening intestinal inflammation in hospitalized patients 1. The gut microbiota plays a central role in the prevention of CDI, as antibiotic treatment promotes the growth of C. difficile, resulting in the development of CDI 1. However, the precise mechanism by which constituents of healthy microbiotas outcompete C. difficile is not fully understood. Healthy gut microbiotas directly prevent C. difficile from blooming in the gut via increasing the abundance of metabolites that interfere with the growth of C. difficile or decreasing the availability of germination/growth-promoting luminal metabolites required by C. difficile 2, 3. Besides their metabolic functions, the gut microbiotas are also known to activate host antimicrobial immunity, which, in turn, indirectly prevents the colonization and/or growth of many enteric pathogens 4. Despite this fact, the role of host immunity-mediated colonization resistance in CDI, conferred by the gut microbiota, remains underappreciated.
Accumulating evidence has suggested that IL-22 plays a pivotal role in mucosal defense mechanisms in the gastrointestinal tract through promotion of epithelial barrier integrity, secretion of antimicrobial peptides, and induction of iron scavengers 5, 6, 7. Moreover, IL-22 modulates host epithelial glycosylation, which, in turn, prevents the growth of enteric pathogens, such as Citrobacter rodentium and Salmonella 8, 9, 10. In the gut, IL-22 production is regulated by the resident microbiota 11, 12, indicating that IL-22 induction is a mechanism utilized by the gut microbiota to mediate colonization resistance against enteric pathogens. Previous studies have demonstrated that IL-22 is not required for the prevention of C. difficile growth in the intestine 13, 14, 15. However, in these studies, the intestinal microbiota was depleted by treatment with antibiotics prior to CDI, and therefore, microbiota-dependent induction of IL-22 may not have been detected in their models. Thus, the involvement of a microbiota-induced IL-22 pathway and the contribution of its defects to the increased risk of CDI remain largely unknown. Herein, we report that mucosal IL-22 signaling, activated by the colonization of the gut microbiota, modulated glycosylation of host N-linked glycans. Host-derive glycans, in turn, promote the growth of succinate-consuming bacteria Phascolarctobacterium spp.. Phascolarctobacterium spp. consumed succinate, a metabolite that fuels C. difficile, thereby preventing the growth of C. difficile in the intestine. Of note, The IL-22-mediated regulation of host N-glycans is likely impaired in patients with ulcerative colitis (UC), and thereby leading to gut dysbiosis with altered microbial metabolic activities, including succinate metabolism. Hence, gnotobiotic mice that harbor the gut microbiota from UC patients were susceptible to CDI. Thus, IL-22-mediated host glycosylation fosters the growth of commensal bacteria that compete with C. difficile for the nutritional niche, and the defects in this pathway might increase the risk for CDI.
Results
Human microbiotas protect germ-free mice from CDI
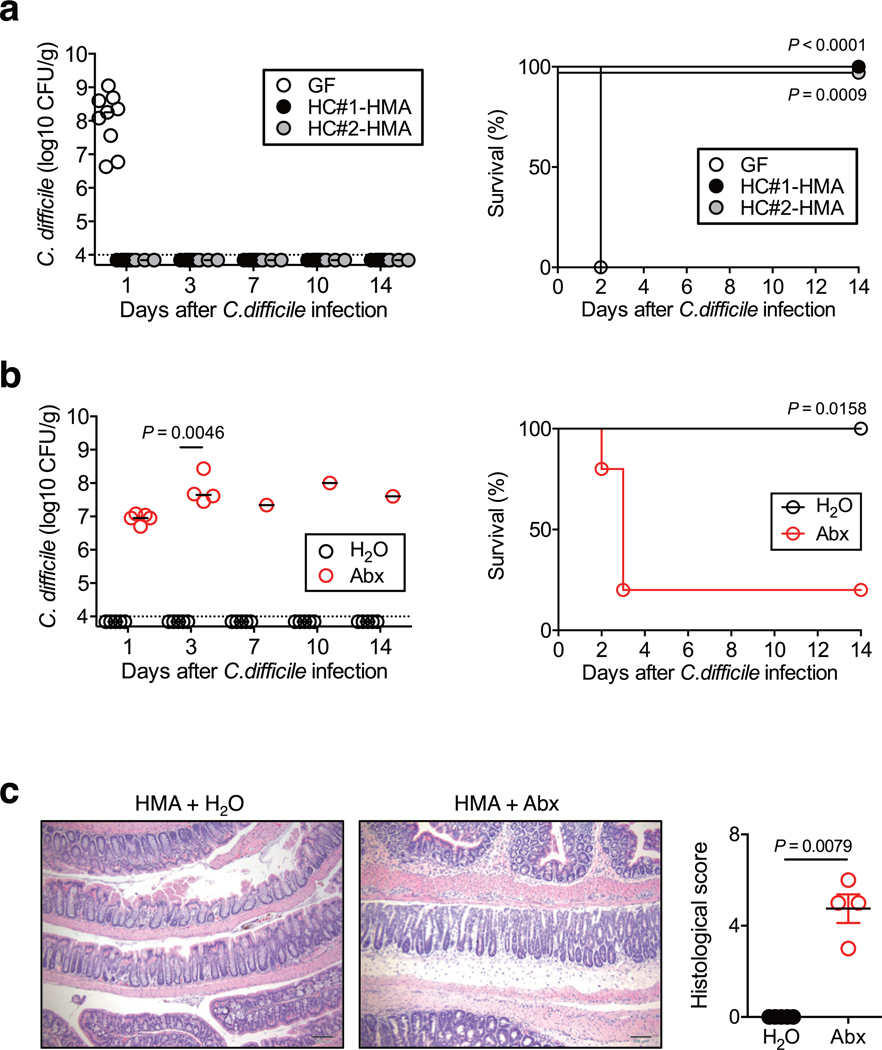
We examined the protective role of human-derived microbiotas against CDI. GF mice were colonized with the gut microbiotas isolated from two healthy controls (HC). HC human microbiota-associated (HC-HMA) mice 16 were infected with C. difficile strain VPI 10463. As a control, GF mice without microbiota reconstitution were also infected with C. difficile. C. difficile was able to colonize control GF mice 1 day after infection and the following day all mice succumbed to CDI (Extended Data Fig. 1a). Consistent with a previous report 17, HC-HMA mice were completely protected against CDI and C. difficile was unable to colonize these mice (Extended Data Fig. 1a). In order to confirm the importance of the gut microbiota, we next treated HC-HMA mice with a broad-spectrum antibiotic cocktail 17. As expected, marked C. difficile colonization (>107–108 CFU/g feces) was observed in antibiotic-treated HC-HMA mice starting on day 1 post-CDI and the majority of mice succumbed to CDI (Extended Data Fig. 1b). Consistent with fecal pathogen burden data, antibiotic-treated HC-HMA mice developed inflammatory pathology in the colon, while no overt inflammation was observed in control HC-HMA mice (Extended Data Fig. 1c). These data suggest that human-derived microbiotas confer colonization resistance against C. difficile in mice.
Microbiota-induced IL-22 prevents CDI
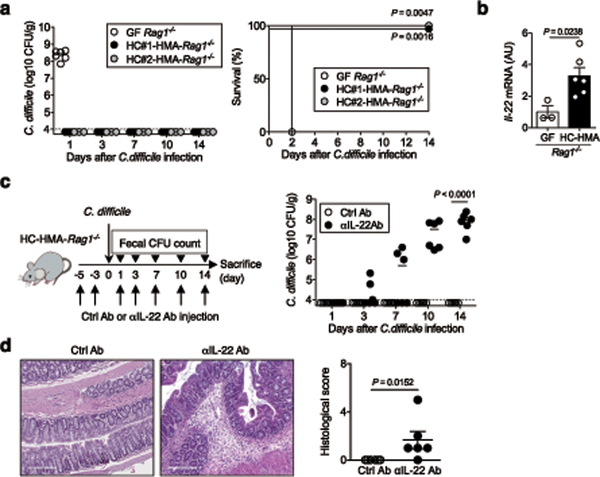
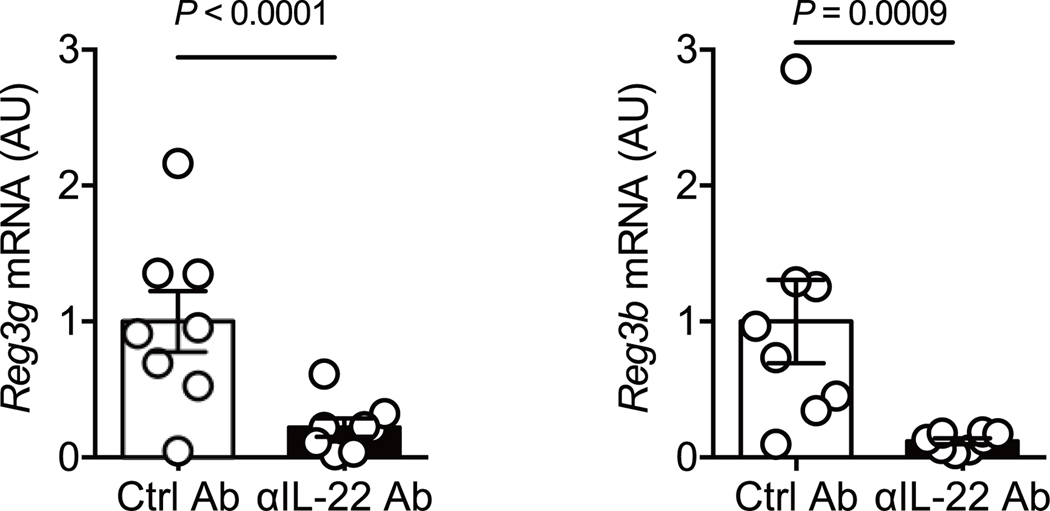
Rag1−/− mice, which lack both T and B cells were used to address the role of host immunity in the human microbiota-conferred colonization resistance against C. difficile. GF Rag1−/− mice were colonized by HC microbiotas to generate HC-HMA-Rag1−/− mice. HC-HMA-Rag1−/− mice and control GF Rag1−/− mice were infected with C. difficile. All GF Rag1−/− mice succumbed to CDI within 2 days of infection. In contrast, C. difficile was unable to grow in HC-HMA-Rag1−/− mice, as was seen in HC-HMA-WT mice (Fig. 1a). This result indicated that host T and B cell immunity is not required for the human microbiota to confer resistance against C. difficile. Rather, the host’s innate antimicrobial response, elicited by the microbiota, plays a role in the prevention of CDI. In this regard, we found that colonization of GF Rag1−/− mice with human microbiotas induced mucosal IL-22 expression (Fig. 1b). This evidence hinted at the involvement of IL-22 in a mechanism underlying microbiota-mediated protection against CDI. To test this hypothesis, we examined the impact of IL-22 signaling blockade in HC-HMA-Rag1−/− mice on CDI susceptibility (Fig. 1c). IL-22 signaling was efficiently inhibited by an αIL-22 antibody in vivo. The antibody treatment also suppressed the expression of Reg3b and Reg3g, two antimicrobial proteins that are strongly induced in response to IL-22 18 (Extended Data Fig. 2). In this setting, colonization resistance of HC-HMA-Rag1−/− mice was abolished and C. difficile was able to colonize the IL-22–neutralized HC-HMA-Rag1−/− mice (Fig. 1c). In accordance with its colonization potential, C. difficile induced massive inflammation in HC-HMA-Rag1−/− mice when IL-22 signaling was blocked (Fig. 1d). Thus, the microbial induction of IL-22 is critical for the prevention of C. difficile blooms in HMA mice.
Figure 1. Healthy human microbiota-driven IL-22 prevents C. difficile blooms in the gut.
(a) (Left) GF-Rag1−/− mice were colonized with healthy microbiotas for 2 weeks (HC-HMA-Rag1−/− mice). GF or HC-HMA-Rag1−/− mice (GF: n=6, HC#1:n=5 and HC#2:n=3, biologically independent animals) were infected with C. difficile VPI 10463 spores (103 spores/mouse). C. difficile load in feces was determined on indicated days post-infection. Dots represent individual mice. Bars represent median. (Right) The mortality of C. difficile infected mice. Statistical significance was assessed by Log-rank test (two-sided). Data were pooled from two independent experiments. (b) Il22 mRNA levels in GF-Rag1−/− (n=3) or HC-HMA-Rag1−/− mice (n=6) were measured by qPCR. Expression was normalized to that of the murine Actb gene. Data are presented as mean ± s.e.m.. Data were pooled from two independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (c) C. difficile infected HC-HMA-Rag1−/− mice were treated with control or αIL-22 antibody twice before C. difficile inoculation (3 and 5 days prior to infection) and 3 times a week post-inoculation. C. difficile load in feces was determined on indicated days post-inoculation (n=6, biologically independent animals). Dots represent individual mice. Bars represent median. Data were pooled from two independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (d) Representative histological images and associated histological scores. Scale bar is 200 μm. Data are presented as mean ± s.e.m.. Statistical significance was assessed by Mann-Whitney U test (two-sided).
IL-22 shapes the gut microbial community and its metabolic function
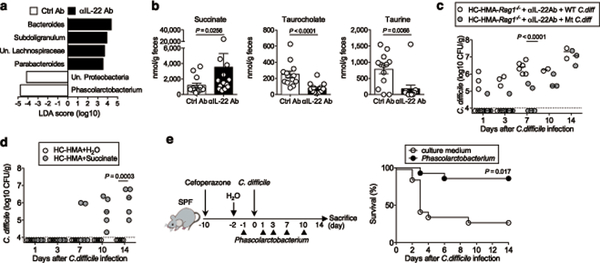
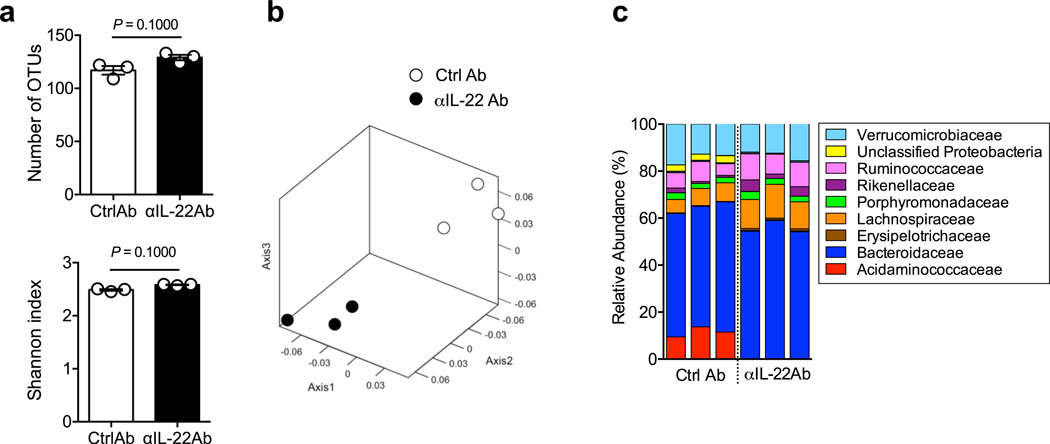
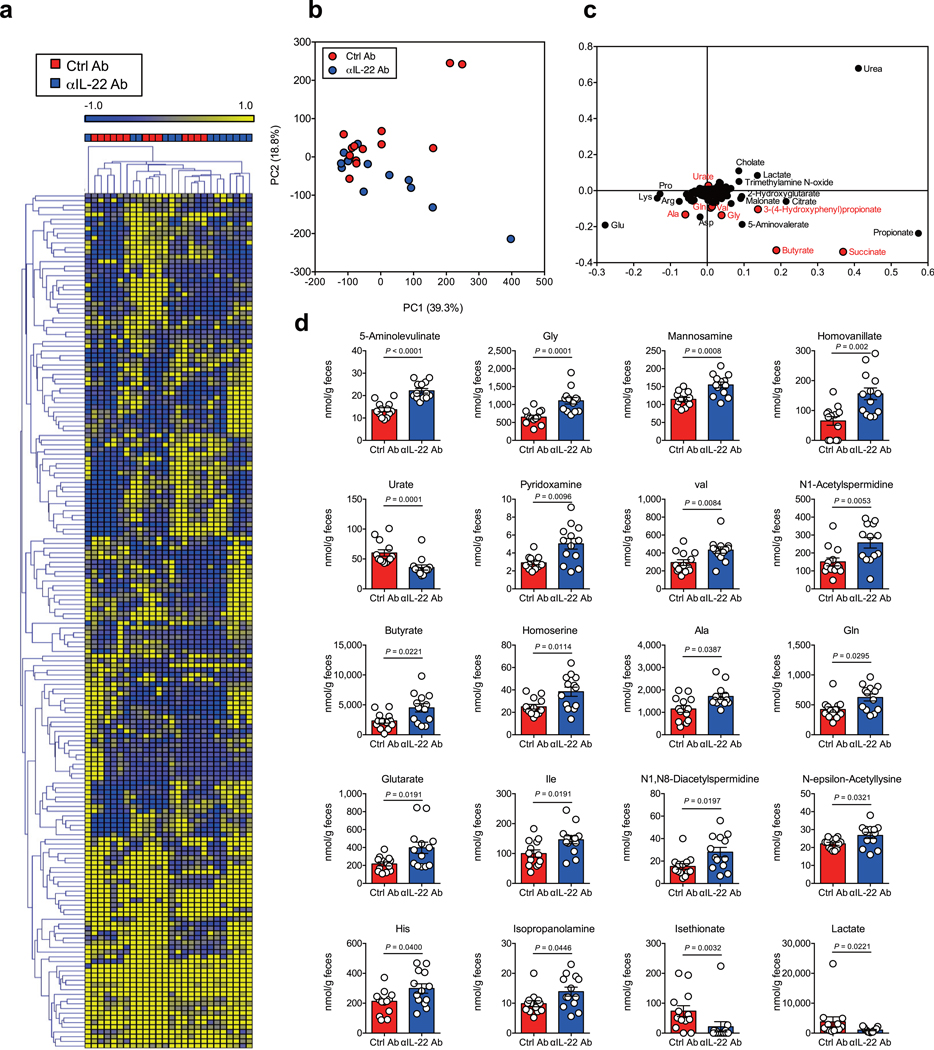
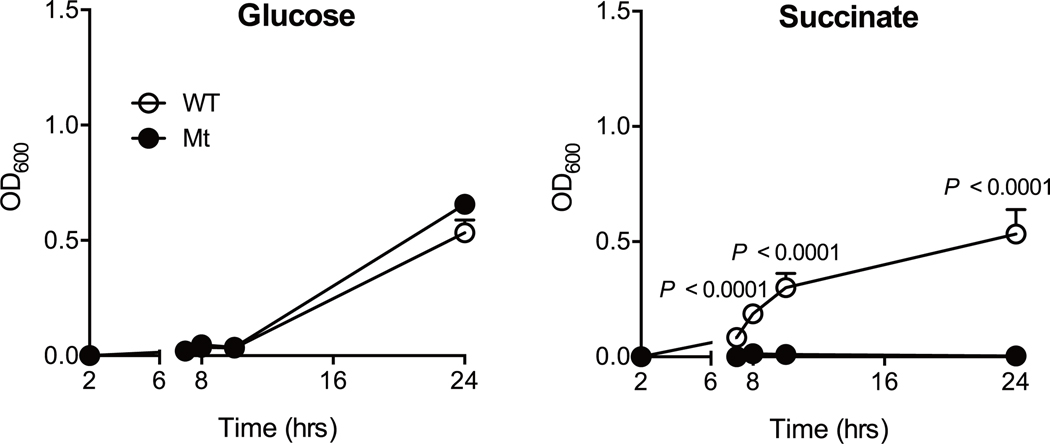
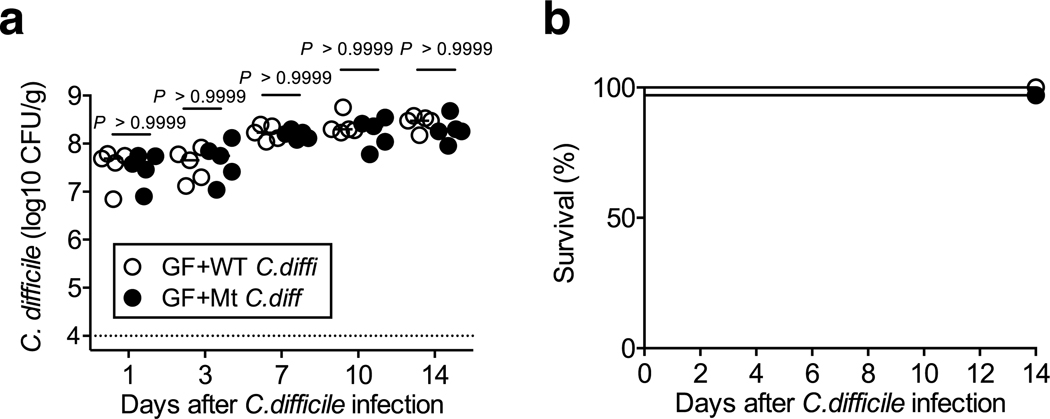
We next sought to study the mechanism by which IL-22 prevents CDI and examine the effect of IL-22 on the composition of the gut microbial community. We analyzed the gut microbiome of HC-HMA-Rag1−/− mice treated with the αIL-22 antibody. IL-22 neutralization did not change the diversity and richness of the microbial communities (Extended Data Fig. 3a), but did change the composition of the gut microbiota (Extended Data Fig. 3b). In particular, it resulted in a decrease in the abundance of Acidaminococcaceae following IL-22 neutralization (Extended Data Fig. 3c). LEfSe analysis further identified bacterial genera that became over- and under-represented following IL-22 neutralization (Fig. 2a). Among them, we focused on genus Phascolarctobacterium (family Acidaminococcaceae), which was significantly decreased as a result of IL-22 blockade (Fig. 2a). Phascolarctobacterium spp. are the major consumers of succinate generated by other bacterial species, such as Bacteroides spp. 19, 20. Thus, the decreased abundance of Phascolarctobacterium may lead to abnormal succinate metabolism in the gut. Consistent with this hypothesis, the luminal levels of succinate were significantly elevated in IL-22-neutralized HC-HMA-Rag1−/− mice (Fig. 2b, Extended Data Fig. 4, and Supplementary Table 1). Importantly, luminal succinate is known to aid the growth of C. difficile in the intestine 21. Hence, an imbalance in succinate metabolism due to gut dysbiosis, induced by a blockade of IL-22 signaling, may affect the host’s susceptibility to CDI. To explore this notion, we utilized a mutant strain of C. difficile that lacks the succinate utilization gene operon CD2344 21. The CD2344 mutant of C. difficile (Cd-CD2344−), unlike its isogenic WT strain JIR8094, is unable to grow on succinate (Extended Data Fig. 5) 21. IL-22-neutralized HC-HMA-Rag1−/− mice, which are susceptible to CDI, were infected with either WT JIR8094 or Cd-CD2344− (Fig. 2c). As seen in Fig. 1, WT C. difficile was able to colonize IL-22-neutralized HC-HMA-Rag1−/− mice (Fig. 2c). In contrast, early colonization by Cd-CD2344− was markedly impaired (Fig. 2c). To address the extent to which succinate plays a key role in the increased susceptibility to CDI, we administrated succinate to HC-HMA mice. We observed that the administration of succinate promoted the growth of C. difficile in HC-HMA mice (Fig. 2d), indicating that succinate is a key metabolite whose abundance controls the colonization of C. difficile in the gut. Interestingly, Cd-CD2344− was able to colonize in GF mice (Extended Data Fig. 6), suggesting that C. difficile can utilize alternate nutritional sources for its growth in the absence of commensal microbiota. Next, we asked if colonization by the succinate-consuming bacteria Phascolarctobacterium spp. can prevent CDI. Specific pathogen-free (SPF) C57BL/6 mice were pre-treated with cefoperazone to breach the gut colonization resistance 3. As previously reported, the cefoperazone-treated mice succumbed to CDI (Fig. 2e). Colonization by two strains of Phascolarctobacterium – P. faecium JCM 30894 and P. succinatutens JCM 16074 – significantly improved mortality of C. difficile–infected mice (Fig. 2e). These results suggest that IL-22, induced by the microbiota, modulates the abundance of specific commensal bacteria, such as Phascolarctobacterium, and can lead to the accumulation of luminal metabolites, including succinate, which may foster the growth of C. difficile.
Figure 2. IL-22–mediated succinate pathway drives the colonization of C. difficile in the healthy microbiota.
(a) Fecal microbiome from HC-HMA-Rag1−/− mice treated with control or αIL-22 antibody (n=3, biologically independent animals) was analyzed. Significantly enriched bacterial taxa in αIL-22 antibody-treated mice (black bars) and control mice (white bars) were identified by LEfSe analysis. Data are representative of 2 independent experiments. (b) Luminal metabolites were analyzed by CE-TOF/MS. Data are presented as mean ± s.e.m. (n=13, biologically independent animals ). Dots represent individual mice. Data were pooled from 3 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (c) αIL-22 antibody-treated HC-HMA-Rag1−/− mice were infected with WT JIR8094 or Cd-CD2344− mutant C. difficile (WT: n=7,Mut: n=8, biologically independent animals). C. difficile load in feces was determined on the indicated days. Dots represent individual mice. Bars represent median. Data were pooled from two independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (d) HC-HMA-WT C57BL/6 mice were given regular water or 1% succinate. C. difficile load in feces was determined on the indicated days, post-inoculation. Dots represent individual mice (n=5, biologically independent animals). Bars represent median. Data were pooled from two independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (e) Cefoperazone pre-treated SPF C57BL/6 mice were infected with C. difficile VPI 10463 spores (103 spores/mouse) by oral gavage. Mice were inoculated orally with P. faecium JCM 30894 and P. succinatutens JCM 16074 (106 CFU each strain) or its culture medium (n=14, biologically independent animals). The mortality of C. difficile–infected mice was assessed. Data were pooled from 3 independent experiments. Statistical significance was assessed by Log-rank test (two-sided).
Host mucus glycosylation regulates gut microbiota
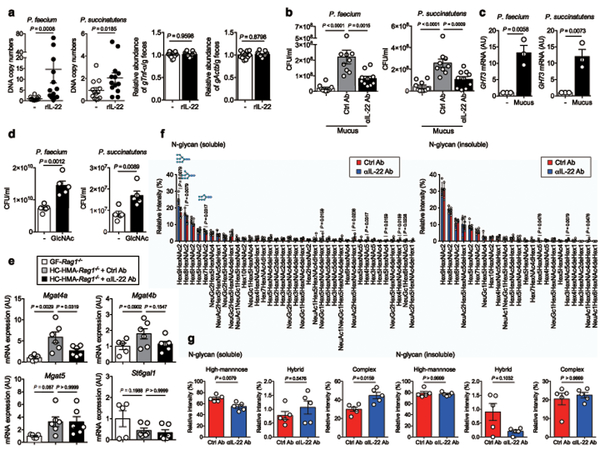
Given the decreased abundance of succinate-consuming bacteria Phascolarctobacterium in the gut significantly decreased when IL-22 signaling was blocked, it is conceivable that IL-22 signaling promotes the growth of Phascolarctobacterium in the gut. To test this hypothesis, GF mice were colonized with two strains of Phascolarctobacterium, P. faecium JCM 30894, and P. succinatutens JCM 16074, followed by treatment with an IL-22-Fc fusion protein. The amount of host genomic DNA in feces remained unchanged as a result of rIL-22 treatment (Fig. 3a). The gut colonization of both strains (gene copies normalized to host genomic DNA) was significantly enhanced by IL-22-Fc, suggesting that IL-22 directly regulates the growth of Phascolarctobacterium (Fig. 3a). The growth of Phascolarctobacterium is primarily limited by the availability of succinate, while carbohydrates and/or SCFAs do not appear to be of major importance 19. However, succinate is not available in GF mice since succinate-producing commensal bacteria are absent 21. This indicates that Phascolarctobacterium can utilize other nutritional sources for its growth in gnotobiotic mice treated with IL-22-Fc. We found that P. faecium JCM 30894 and P. succinatutens JCM 16074 harbors enzymes that belong to glycoside hydrolase (GH) families, GH73 (GenBank/EMBL/DDBJ accession# AB490811). Given that GH73 is a member of the host glycan-related enzyme families (N-acetylglucosamine (GlcNAc)), it is plausible that Phascolarctobacterium has become adapted to consume host-derived glycans in the gut. To examine this, we isolated mucus from SPF mice treated with the αIL-22 antibody or the isotype control antibody. Isolated mucus was added to the Phascolarctobacterium growth medium in vitro. Supplementation with mucus isolated from control mice promoted the growth of P. faecium and P. succinatutens (Fig. 3b). In contrast, the growth of P. faecium and P. succinatutens was significantly weakened when mucus isolated from IL-22–neutralized mice was used (Fig. 3b). Consistent with this result, P. faecium and P. succinatutens expressed GH73 when cultured in mucus (Fig. 3c). Moreover, supplementation with GlcNAc enhanced the growth of Phascolarctobacterium (Fig. 3d). These results suggest that IL-22–induced host mucus glycosylation likely promotes the growth of Phascolarctobacterium. We also assessed the expression of host glycosyltransferases in HC-HMA mice, and whether or not these enzymes are regulated via IL-22 signaling. As shown in Fig. 3e, host glycosyltransferases that are related to N-glycosylation, such as Mgat4a, were induced in GF mice upon colonization with a human microbiota. The microbiota-induced IL-22 was responsible for the induction of some of these enzymes, as their expression levels were significantly reduced when IL-22 signaling was blocked (Fig. 3e). We next analyzed the host glycosylation status of HMA-Rag1−/− mice with or without IL-22 signal blockade. We found that N-glycosylation of the host proteins was impaired in IL-22-neutralized mice both in the insoluble fractions (mainly mucus proteins, such as Muc2) and in the soluble fractions (intestinal cells, such as epithelial cells). Notably, high-mannose glycans were reduced in the soluble fraction and hybrid glycans tended to be decreased in the insoluble fraction in IL-22–neutralized mice (Fig. 3f, g, and Supplementary Table 2). In contrast, O-glycisylation was unchanged in IL-22-neutralized mice (Supplementary Table 3). These findings suggest that IL-22–mediated host N-glycosylation may foster the growth of some commensal microbiota, such as Phascolarctobacterium.
Figure 3. Host glycosylation regulated by IL-22 influences the growth of Phascolarctobacterium species.
(a) GF mice were inoculated orally with P. faecium JCM 30894 and P. succinatutens JCM 16074 (106 CFU each strain). Mice were then injected with either a mouse IL-22Fc fusion protein or a control Fc protein. (Left) The DNA copy numbers of P. faecium and P. succinatutens in the feces were quantified by qPCR. Each copy number was normalized to that of the host genomic DNA (murine gTnf). Relative abundance to the control group is shown. (Right) The DNA copy numbers of host genomic DNAs (murine Tnf and Actb) in the 1g of feces were quantified by qPCR. Relative abundance to the control group is shown. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=13, biologically independent animals). Data were pooled from 3 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (b) In vitro growth of P. faecium and P. succinatutens in PYS medium supplemented with colonic mucus derived from control (n=10) or αIL-22 antibody–treated mice (n=9, independent experiments). Data are presented as mean ± s.e.m.. Dots represent independent experiments. Statistical significance was assessed by 1-way ANOVA with Bonferroni post-hoc test (two-sided). (c) The GH73 mRNA expression in P. faecium and P. succinatutens during the growth on mucus was analyzed by qPCR. Expression was normalized to Eubacteria 16s rRNA. Data are presented as mean ± s.e.m. (n=3 independent experiments). Statistical significance was assessed by Student t-test (two-sided). (d) P. faecium and P. succinatutens growth on GlcNAc. Data are presented as mean ± s.e.m. (n=5 independent experiments). Statistical significance was assessed by Student t-test (two-sided). (e) The expression of glycosyltransferase genes in the colonic mucosa of GF Rag1−/− (n=6, biologically independent animals) and HC-HMA-Rag1−/− mice injected with control or αIL-22 antibody was determined by qPCR. Expression was normalized to that of the murine Actb gene. Data are presented as mean ± s.e.m.. Data were pooled from 2 independent experiments. Statistical significance was assessed by 1-way ANOVA with Bonferroni post-hoc test (two-sided). (f) Colonic mucus derived from control or αIL-22 antibody–treated HMA-Rag1−/− mice were separated into GuHCl soluble and insoluble fractions. N-glycans were analyzed by liquid chromatography–electrospray ionization tandem mass spectrometry. (g) The relative intensity of high-mannose, hybrid, and complex N-glycans in soluble and insoluble fractions. Data are presented as mean ± s.e.m. (n=5, biologically independent animals). Dots represent individual mice. Statistical significance was assessed by Mann-Whitney U test (two-sided) (f, g).
Impaired N-glycosylation–related gene expression in UC patients
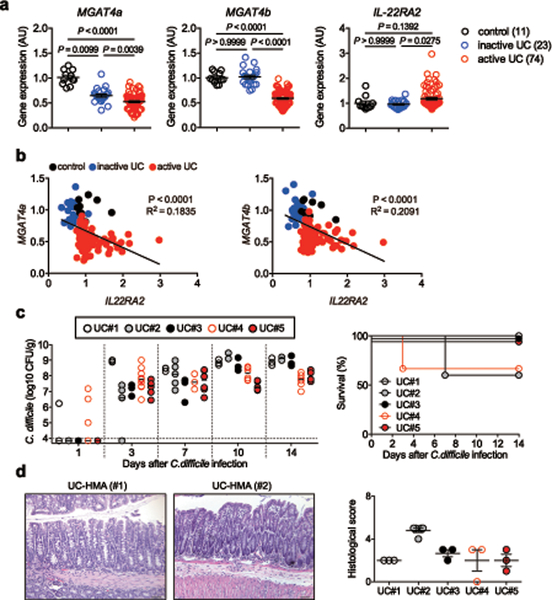
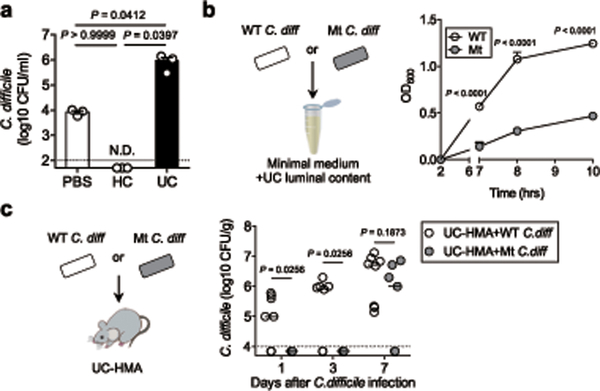
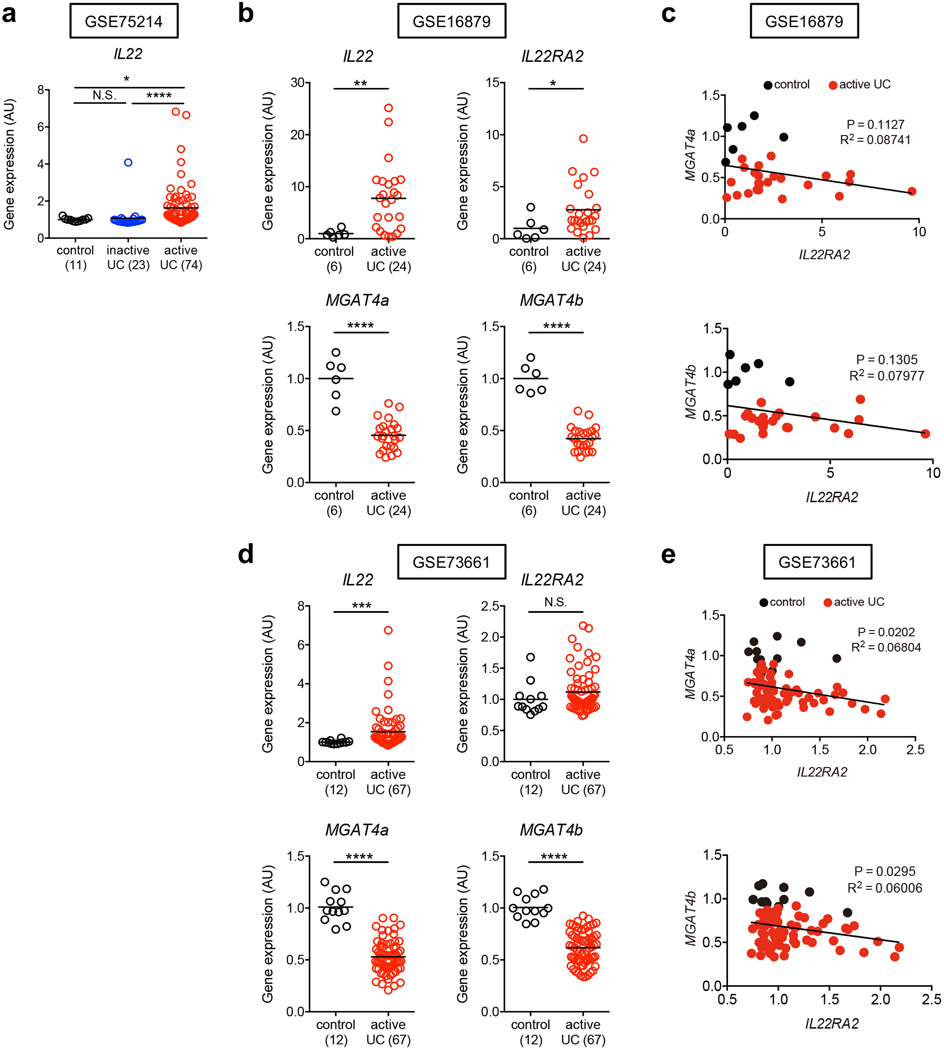
We next examined the link between IL-22–mediated host glycosylation and increased host susceptibility to CDI resulting from underlying disease. To this end, we focused on patients with ulcerative colitis (UC), who are at higher risk for CDI compared to normal individuals 22, 23, 24. Using a publically available database (GSE75214) 25, we found that the expression of host glycosyltransferases, such as MGTA4A and MGAT4B, was significantly impaired in UC patients compared to normal controls (Fig. 4a). The expression of IL22RA2 (also known as IL-22 binding protein (IL-22BP), a soluble IL-22 receptor that blocks IL-22 signaling, was markedly up-regulated in patients with UC, while IL-22 expression was not affected (Fig. 4a and Extended Data Fig. 7a). Notably, IL22RA2 expression was inversely correlated with the expression of these glycosyltransferases, indicating that impaired IL-22 signaling, resulting from the overexpression of IL-22BP, led to decreased host glycosylation in UC (Fig. 4b). An analysis of two other deposited cohorts (GSE16879 26 and GSE73661 27) revealed similar expression patterns, and inverse correlation between IL22RA2 and host glycosylation enzymes (Extended Data Fig. 7b-e). Consistent with these results, HMA mice-colonized with the microbiotas derived from UC patients (UC-HMA mice) were unable to resist C. difficile colonization (Fig. 4c, 4d). Consistent with the in vivo data, inoculated C. difficile failed to proliferate ex vivo using luminal contents isolated from HC-HMA mice (Fig. 5a). On the other hand, C. difficile grew from luminal contents isolated from UC-HMA mice (Fig. 5a). As expected, the growth of C. difficile was dependent on succinate, as the Cd-CD2344− mutant exhibited impaired growth in UC luminal contents in vitro and in vivo (Fig. 5b, c).
Figure 4. N-glycan related glycosyltransferase gene expression and CDI risk in UC patients.
(a) The mRNA expression of glycosyltransferase-related genes in the colonic tissue from control subjects (n=11), patients with inactive UC (n=23) and patients with active UC (n=74). Data were derived from Gene Expression Omnibus (GEO) data sets GSE75214. Dots represent biologically independent subjects. Data are presented as mean ± s.e.m.. Statistical significance was assessed by Kruskal-Wallis test with Dunn’s post-hoc test (two-sided). (b) The correlation between MGAT4A/MGAT4B and IL22RA2 mRNA expression in 3 groups. Statistical significance was measured by Pearson correlation test (two-sided). (c) GF WT C57BL/6 mice were colonized with the gut microbiotas obtained from UC patients (UC-HMA mice). UC-HMA mice were then infected with C. difficile VPI 10463 spores (UC#1: n=3, UC#2: n=5, UC#3: n=3, UC#4: n=9, and UC#5: n=6, biologically independent animals). C. difficile load in feces was determined on indicated days, post-infection. Dots represent individual mice. Bars represent median. Data were pooled from 3 independent experiments. (Right) The mortality of C. difficile infected UC-HMA mice. (d) Representative histological images and associated histological scores. Scale bar is 100 μm. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=3, biologically independent animals) from pooled 3 independent experiments.
Figure 5. C. difficile utilizes luminal succinate for its growth in UC patients.
(a) Ex vivo germination and growth of C. difficile strain VPI 10463 in cecum contents isolated from HC-HMA and UC-HMA mice (n=3 biological independent animals). Data are presented as mean ± s.e.m. from pooled 3 independent experiments. Statistical significance was assessed by 1-way ANOVA with Bonferroni post-hoc test (two-sided). (b) Ex vivo germination and growth of WT or Cd-CD2344− mutant C. difficile in UC luminal contents. Data are presented as mean ± s.d. (n=3 technical replicates). Data are representative of 3 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (c) UC-HMA mice were infected with WT or Cd-CD2344− mutant C. difficile (WT: n=8, Mut: n=7, biologically independent animals). C. difficile load in feces was determined on indicated days post-infection. Dots represent individual mice. Bars indicate median. Data were pooled from 3 independent experiments Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided).
Restoration of microbial metabolism reduces the risk for CDI
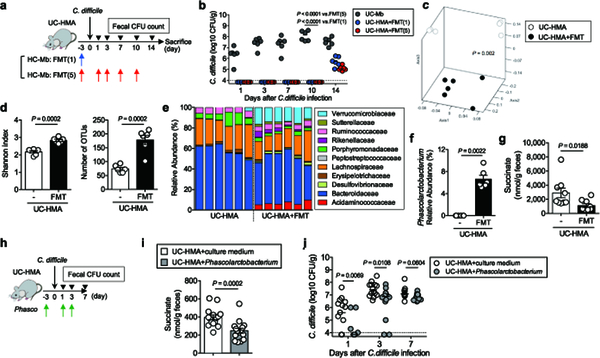
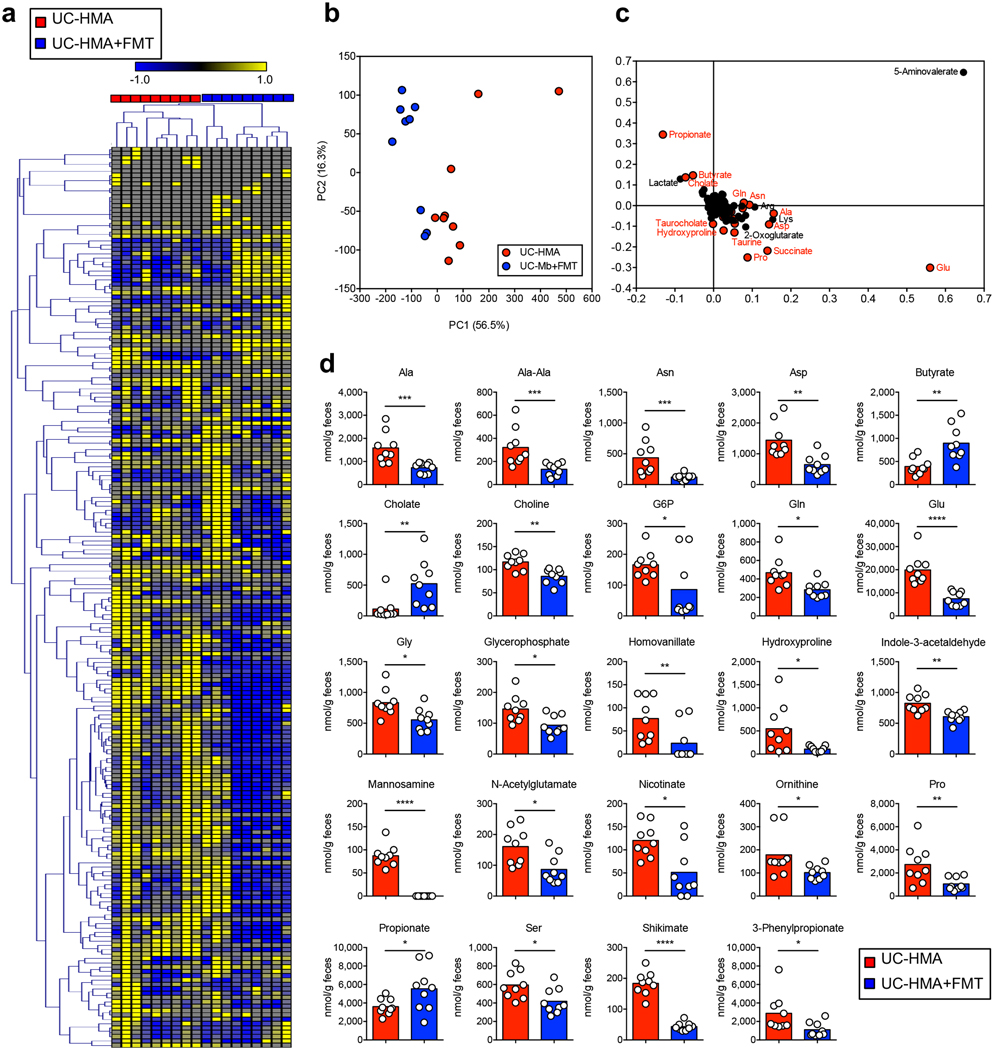
Finally, we tested whether fecal microbiota transplantation (FMT) could restore normal microbial metabolic activity and colonization resistance in UC-HMA mice. Healthy human-derived microbiotas were inoculated into UC-HMA mice, which were then infected with C. difficile (Fig. 6a). As a result, FMT almost completely prevented the growth of C. difficile in the gut (Fig. 6b). We found that a single FMT was equally effective at preventing CDI as multiple FMTs (Fig. 6b). Gut dysbiosis seen in UC-HMA mice improved significantly as a result of FMT (Fig. 6c–6e), which restored the abundance of Phascolarctobacterium (Fig. 6f and Extended Data Fig. 8). Consistent with this, the levels of luminal succinate were significantly reduced following FMT (Fig. 6g, Extended Data Fig. 9, and Supplementary Table 4). To validate the importance of Phascolarctobacterium in the prevention of CDI, we inoculated UC-HMA mice with succinate consumers, P. faecium and P. succinatutens, instead of a full FMT (Fig. 6h). As expected, colonization of P. faecium and P. succinatutens significantly reduced the concentration of luminal succinate (Fig. 6i). According to the reduction of luminal succinate availability, Inoculation of Phascolarctobacterium significantly inhibited the colonization of C. difficile, particularly in early stages of infection (Fig. 6j).
Figure 6. Restoration of luminal metabolites reduces the risk of C. difficile infection.
(a-g) UC-HMA mice were infected with C. difficile VPI 10463 spores (103 spores/mouse). Mice were transplanted with a healthy, human-derived fecal microbiota (FMT) (n=6, biologically independent animals). (b) C. difficile load in feces. Dots represent individual mice. Bars indicate median. Data were pooled from 2 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (c-f) Fecal microbiota before CDI (day 0) was harvested and bacterial 16S rRNA sequences were analyzed. (c) Microbial community structures were analyzed using the Yue and Clayton dissimilarity distance metric (θYC). Statistical significance was assessed by AMOVA test (two-sided) (d) Shannon index (α-diversity) and number of OTUs (richness). Data are presented as mean ± s.d. (n=6, biologically independent animals). Statistical significance was assessed by Mann-Whitney U test (two-sided). (e) Bacterial taxonomy at the family level in the feces. (f) The relative abundance of Phascolarctobacterium species in the feces. Data are presented as mean ± s.d. (n=6 each). Statistical significance was assessed by Mann-Whitney U test (two-sided). (g) Fecal succinate concentration at day 0 was analyzed by CE-TOF/MS. Data are presented as mean ± s.d. (n=9, biologically independent animals). Dots represent individual mice. Data were pooled from 2 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (h-j) UC-HMA mice were infected with C. difficile VPI 10463 spores (103 spores/mouse). Mice were inoculated with P. faecium JCM 30894 and P. succinatutens JCM 16074 (106 CFU each strain). (i) Luminal succinate concentration was analyzed by LC-MS. Data are presented as mean ± s.d. (n=15, biologically independent animals) from pooled 3 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (j) C. difficile load in feces. Data are presented as mean ± s.d. (n=15, biologically independent animals) from pooled 3 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided).
Discussion
By utilizing the HMA mouse model, we demonstrated that IL-22, induced by colonization of the gut microbiota, shapes the composition of the gut resident microbiota. IL-22 signaling promotes the expression of host glycosyltransferases, such as Mgat4a and Mgat4b, which encode enzymes that catalyze the transfer of N-acetylglucosamine (GlcNAc) to the core mannose residues28, 29. The overall abundance of GlcNAc seemed unchanged, although N-glycosylation in both soluble and insoluble fraction of mucus was compromised by the impairment of IL-22 signaling. This suggests that the transfer of GlcNAc to N-linked glycans may be impaired only in subsets of host-derived proteins. Host glycosylation plays an important role in maintaining specific populations of gut commensal bacteria, such as Phascolarctobacterium spp., which utilize host-derived GlcNAc for their growth19, 30. The dysbiotic microbiota, as a result of the impaired IL-22 signaling, has a compromised metabolic function, thereby increasing the opportunistic growth of potential pathogens, such as C. difficile.
It has been reported that the abundance of Phascolarctobacterium is markedly reduced in CDI patients compared to healthy control subjects 31. Likewise, the abundance of Phascolarctobacterium was restored in CDI patients following FMT, resulting in the amelioration of their clinical symptoms 32. Thus, the abundance of Phascolarctobacterium is likely involved in host protection against CDI. However, the mechanisms by which Phascolarctobacterium spp. are regulated within the gut microbiota remain poorly understood. In this study, we found that Phascolarctobacterium spp. harbor glycoside hydrolases associated with host glycan degradation, suggesting that Phascolarctobacterium spp. have the ability to forage host glycans in addition to consuming succinate. Therefore, IL-22-mediated host N-glycosylation likely promotes the growth of this bacterial genus in the gut. We speculate that the increased abundance of Phascolarctobacterium spp. leads to a more efficient consumption of succinate in the gut lumen, which limits the opportunistic growth of C. difficile whose proliferation is aided by succinate 21. Interestingly, the administration of Phascolarctobacterium spp. sufficiently protects antibiotic-treated mice from CDI. Further, Phascolarctobacterium improves the mortality of infected animals, even if they are inoculated after the colonization by C. difficile (Extended Data Fig. 10). These data suggest that Phascolarctobacterium might be useful for treating patients who are already infected with C. difficile. However, it is noteworthy that single-dose, pre-treatment with Phascolarctobacterium was not sufficient to prevent C. difficile colonization and subsequent lethal colitis (Extended Data Fig. 10), suggesting that Phascolarctobacterium must retain an optimal number in the intestine to maximize their protective effect against CDI.
Phascolarctobacterium spp. are not the only bacterial species regulated by IL-22–mediated gut conditioning. LEfSe analysis showed that unclassified Proteobacteria were also underrepresented in IL-22–neutralized HMA mice. It remains possible that these not-yet-identified bacteria also play an important role in the protection against CDI. In addition to succinate, several other luminal metabolites were differentially abundant in control and IL-22–neutralized HMA mice, including taurocholate and taurine. Taurocholate is known to promote the germination of C. difficile spores 3, while microbiota-derived taurine is known to control microbial–host mutualism by modulating NLRP6-mediated anti-microbial peptide production by the intestinal epithelial cells 33. Moreover, the concentration of various luminal amino acids was significantly increased in IL-22–neutralized HMA mice. As amino acid availability is closely associated with the risk for CDI 34, they may also contribute to the susceptible phenotype we observed in those mice. Consistent with these observations, the Cd-CD2344− strain, which cannot utilize succinate, was still able to proliferate in IL-22–neutralized HMA mice, although the early colonization of this strain was dramatically impaired compared to the WT strain. This result indicates that succinate is an important metabolite that promotes the proliferation of C. difficile in the gut early after its colonization. In later colonization, C. difficile may be able to use alternative nutritional sources, such as amino acids, for its growth. Thus, targeting different metabolic pathways, such as bile acid metabolism, succinate metabolism, and amino acid metabolism, by using multiple strains of commensal bacteria may be a more rational, and effective, strategy to prevent CDI.
Recent epidemiologic studies have shown that inflammatory bowel disease (IBD) patients, particularly UC, are at higher risk for CDI compared with non-IBD individuals 22, 23, 24. Since CDI induces IBD flares and worsens disease outcome, CDI in IBD patients is recognized as a major clinical complication 22, 23, 24. In the current study, we recapitulated increased susceptibility to CDI seen in UC patients using our HMA mouse model. Since it is known that the abundance of Phascolarctobacterium is decreased in IBD patients, both in UC and CD 35, the gut microbiotas derived from UC patients might have similar defects as those observed in IL-22–neutralized HMA mice, namely decreased abundance of Phascolarctobacterium and elevated levels of luminal succinate. Consistently, although the expression of IL-22 is up-regulated in the intestine of IBD patients 36, 37, there is evidence of possible defective IL-22 signaling in IBD, particularly UC, patients, such including the up-regulation of IL-22BP, which blocks IL-22 bioactivity 38, 39, 40, 41, 42, 43, 44. Collectively, IL-22 signaling is likely compromised in UC patients. In this study, we discovered that the expression of IL-22BP (IL22RA2) is significantly up-regulated in UC patients, and that it inversely correlates with the expression of host glycosyltransferases, MGAT4A and MGAT4B. Reduced host N-glycosylation has been reported in active UC patients 45. Our findings suggest that impaired IL-22 signaling in UC patients may cause abnormal host glycosylation, which triggers gut dysbiosis, including a reduction in the abundance of Phascolarctobacterium spp. Further study will be required to link IL-22 signaling, host glycosylation, and gut dysbiosis in patients with UC.
The increased luminal succinate level is likely only one facet of the complex process that increases the susceptibility of IBD patients to CDI. Indeed, FMT is much more effective than the administration of Phascolarctobacterium spp. to prevent CDI in UC-HMA mice, although Phascolarctobacterium spp. treatment significantly reduces the availability of luminal succinate. Consistently, FMT reduces the levels of various metabolites, such as amino acids, which are known to foster the growth of C. difficile 34. It suggests that Phascolarctobacterium spp. may be used with other bacterial strains that target distinct metabolic pathways related to C. difficile germination and growth (e.g., amino acid consumption, bile acid conversion) to improve clinical effectiveness further. In this regard, we are comparing the prevention efficacy of Phascolarctobacterium with that of a previously reported protective bacteria, such as Clostridium scindens, which resist to C. difficile in a secondary bile acid–dependent fashion 2. We speculate that the combination of Phascolarctobacterium and C. scindens might have a synergistic effect in preventing CDI. However, further study is needed for developing a recipe of the effective cocktail of metabolic competitors that resists CDI. Although succinate may not be the only target for prevention of CDI in IBD patients, it remains possible that these parameters (i.e., the abundance of Phascolarctobacterium, the concentration of luminal succinate) could be used to screen and identify IBD patients who are at high risk for CDI. These findings also raise the question of whether modulation of IL-22 by current therapies for IBD could affect the incidence of C. difficile colitis.
Compared to CDI, clinical response to FMT in IBD is variable 46, 47. This unresponsiveness of IBD patients to FMT might be, at least in part, due to insufficiencies of the nutritional niche caused by impaired host glycosylation. In this case, the combination of FMT and recombinant IL-22 (or co-inoculation with IL-22-inducing bacteria 48) could maximize the colonization potential of transplanted beneficial bacteria, such as Phascolarctobacterium in the gut.
Our data clarify the role of IL-22 in the feedback regulation of the gut microbiota. IL-22-induced host glycosylation governs the growth of certain protective members of the gut microbiota, such as Phascolarctobacterium, and IL-22 normalizes the composition of the microbiota and influences its metabolic activities, which may be critical for the prevention of opportunistic colonization/expansion of C. difficile in the gut.
Methods
Mice
Specific pathogen-free (SPF) mice were housed in the Animal Facility at the University of Michigan. GF C57BL/6 and GF Rag1−/− mice were housed in the Germ-Free Animal Facility at the University of Michigan. GF mice were maintained in flexible film isolators and their GF status was checked weekly by aerobic and anaerobic culture. The absence of the microbiota was verified by microscopic analysis of stained cecal contents that detects any unculturable contamination. Histologically and endoscopically diagnosed UC patients and HC subjects were recruited through the University of Michigan IBD Databank (HUM0004184), and stool samples were collected from those donors. Individuals who received any antibiotics in the past 3 months were excluded. Individuals who had previous bacteria infections (e.g., C. difficile) or virus infections (e.g., hepatitis B virus, hepatitis C virus, or human immunodeficiency virus) were also excluded. Patient information was provided in Supplementary Table 5. Stool samples were resuspended in pre-reduced phosphate-buffered saline (in 1:10 ratio) in an anaerobic chamber and then filtrated through 100 μm cell strainer. Stool suspension was then inoculated into GF C57BL/6 mice or GF Rag1−/− mice (100 μl/mouse) 16.
Human microbiota-associated mice
The HMA mice were housed in positive-pressure individually ventilated cages (IVCs) (ISOcage P; Techniplast, West Chester, PA) in order to prevent cross-contamination and maintain their gnotobiotic status 49, 50. All mice were fed a sterilized rodent breeder diet 5013 (LabDiet, St. Louis, MO). 8- to 16-week-old female and male mice were used in all experiments. All animals were handled in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan. To attain robust C. difficile infection in HC-HMA mice, the animals were treated with a cocktail of antibiotics ofr 5 days. The cocktail contains kanamycin (0.4 mg/ml), gentamicin (0.035 mg/ml), colistin (850 U/ml), metronidazole, (0.215 mg/ml), and vancomycin (0.045 mg/ml). Clindamycin (10 mg/kg) was then injected intra-peritoneally at day 5. One day after clindamycin injection, mice were challenged by C. difficile 24 h post-IP injection 17.
Bacterial strain and C. difficile infection of mice
The spores of C. difficile strain VPI 10463 (ATCC 43255) were cultured at 65°C for 20 min at 65°C to kill vegetative bacteria. WT JIR8094 and Cd-CD2344−21 were grown overnight in brain heart infusion medium supplemented with 100 mg/l L-cysteine and 5 mg/mL yeast extract (BHIS). Mice were infected with C. difficile VPI10463 spores (103 spores per mouse) or with an overnight culture of JIR8094/Cd-CD2344− (OD600 = 0.5–1.0, 200 μl per mouse) by oral gavage. Stool samples were collected from C. difficile–infected mice on days 1, 3, 7, 10, and 14 post-challenge. The samples were then serially diluted and plated on TCCFA selective plates to quantify the number of C. difficile spores under anaerobic conditions. For the αIL-22 antibody treatment, HC-HMA Rag1−/− mice were injected intraperitoneally twice before C. difficile inoculation (3 and 5 days prior to infection) and 3 times per week post-infection with the αIL-22 antibody (150 μg/mouse per dose) (Genentech, South San Francisco, CA, USA) or an equivalent amount of the isotype-matched control antibody (Genentech). For the IL-22-Fc fusion protein treatment, GF mice were injected intraperitoneally once before Phascolaectobacterium spp. (P. faecium JCM 30894 and P. succinatutens JCM 16074, 1 × 106 CFU each strain/mouse) inoculation (1 day prior to inoculation) and twice post inoculation (2 and 5 days) with IL-22-Fc (100 μg/mouse per dose) (Genentech) or an equivalent amount of the isotype-matched control protein (Genentech). For the succinate treatment, HC-HMA mice were given 1% succinate (Acros Organics, Belgium, WI) in drinking water 3 days prior to infection until sacrifice. C. difficile–infected UC-HMA mice were transplanted with a healthy human–derived microbiota (fecal microbiota transplantation; FMT) or Phascolarctobacterium (P. faecium JCM 30894 and P. succinatutens JCM 16074, 1 × 106 CFU each strain/mouse) once before C. difficile inoculation (3 days prior to infection) and 4 times post-inoculation (1, 3, 7, 10 days post-infection). Regarding the CDI experiments using SPF C57BL/6 mice, the mice were given cefoperazone (MP Biomedicals, Solon, OH) in drinking water (0.5 mg/mL) for 8 days, followed by regular water for 2 days. Then, the cefoperazone-treated SPF mice were infected with C. difficile VPI 10463 spores. For the prevention study, cefoperazone-treated SPF mice were inoculated with P. faecium JCM 30894 and P. succinatutens JCM 16074 (1 × 106 CFU each strain/mouse) once before C. difficile inoculation (3 days prior to infection) and 4 times post-inoculation (1, 3, 7, 10 days post-infection). HMA mice were sacrificed 7 days or 2 weeks post C. difficile inoculation. Cecum and colon tissues were collected and fixed with 4% paraformaldehyde. Fixed tissues were then processed, embedded, sectioned and stained with hematoxylin and eosin (H&E). Histological inflammation was scored at the University of Michigan Unit for Laboratory Animal Medicine In-Vivo Animal Core. A veterinary pathologist evaluated histological inflammation in a blinded fashion. Histological scores were calculated based on following criteria: [1] Inflammation (0, none; 1, minimal multifocal inflammation [few foci]; 2, moderate multifocal inflammation [numerous foci]; 3, severe multifocal coalescing inflammation; and 4, same as a score of 3 with abscesses or extensive mural involvement); [2] edema (0, none; 1, mild focal or multifocal edema, minimal submucosal expansion (<2); 2, moderate focal or multifocal edema, moderate submucosal expansion (2–3); 3, severe multifocal to coalescing inflammation; and 4, same as a score of 3 with diffuse submucosal expansion); [3] epithelial damage (0, none; 1, mild, multifocal, superficial damage; 2, moderate, multifocal, superficial damage; 3, severe, multifocal to coalescing mucosal damage ± pseudomembrane ± ulcer; 4, same as a score of 3 with significant pseudomembrane or ulcer formation).
Quantitative Real-Time PCR
RNA was extracted with E.Z.N.A. Total RNA Kit (Omega Bio-tek, Norcross, GA) according to the manufacturer’s instructions. RNA was reverse transcribed using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster, CA) and cDNA was then used for quantitative PCR analysis using SYBR Green Gene-Expression Assay on an ABI 7900HT analyzer (ABI 7900HT analyzer). The following primer sets were used for amplification: Actb-F; 5′-AAGTGTGACGTTGACATCCG, Actb-R; 5′-GATCCACATCTGCTGGAAGG, Il22-F; 5′-TTTCCTGACCAAACTCAGCA, Il22-R; 5′-TCTGGATGTTCTGGTCGTCA, Reg3b -F; 5′-CTCTCCTGCCTGATGCTCTT, Reg3b -R; 5′-GTAGGAGCCATAAGCCTGGG, Reg3g-F; 5′-TCAGGTGCAAGGTGAAGTTG, Reg3g-R; 5′-GGCCACTGTTACCACTGCTT, GH73-F; 5′-GCGTTTATGAAGTGCTGGAAAAG, GH73-R; 5′-ATTCCATACGCAGTTTCTCAGGT, Mgat4a-F; 5′-GCGACAGACAGAAGGCAAACC, Mgat4a-R; 5′-CCGACAGAGACGAGTGTAGGC, Mgat4b-F; 5′-AGGTGACGTGGTGGACATTT, Mgat4b-R; 5′-GCTTCAGGCTCTCTTGCTCA, Mgat5-F; 5′-GGAAATGGCCTTGAAAACACA, Mgat5-R; 5′-CAAGCACACCTGGGATCCA, St6gal1-F; 5′-TGCGTGTGGAAGAAAGGGAGC, St6gal1-R; 5′-CTCCTGGCTCTTCGGCATCTG, Eubacteria-F; 5′-ACTCCTACGGGAGGCAGCAGT, Eubacteria-R; 5′-ATTACCGCGGCTGCTGGC, P. faecium-F; 5′-CCTTTAGACGGGGACAACATTC, P. faecium-R; 5′-ATCGCCTTGGTAGTCCGTTACA, P. succinatutens-F; 5′-AGCAATCTCGCATGAGGATGCTGT, P. succinatutens-R; 5′-GCCGTGGCTTATTCGTTTACTACCG, mouse genomic Tnf (gTnf)−F; 5′-CAACCCTTATTCTCGCTCACA, gTnf−R; 5′-CTCCACACTCTCCTCCACCT, mouse genomic Actb (gActb)-F; 5’ -ATGACCCAGATCATGTTTGA. mouse genomic Actb (gActb)-R; 5’ TACGACCAGAGGGCATACAG
Microbiome analysis
Genomic DNA was extracted using a modified Qiagen DNeasy Blood and Tissue kit protocol (Qiagen, Valencia, CA) 16. These modifications included the following steps: (1) UltraClean fecal DNA bead tubes (Mo Bio Laboratories, Inc, West Carlsbad, CA) and a Mini-Beadbeater-16 (BioSpec Products, Inc, Bartlesville, OK) were used to homogenize samples (1.5 min); (2) the amount of buffer ATL used in the initial steps of the protocol was increased (from 180 to 400 μl); (3) the volume of proteinase K was increased (from 20 to 40 μl); and (4) the amount of buffer AE used to elute DNA at the end of the protocol was decreased (from 200 to 75 μl). The V4 region of bacterial 16S rRNA was sequenced using the Illumina MiSeq at the Host Microbiome Initiative University of Michigan. The sequence results were then analyzed by Mothur (v.1.33) 51, 52. Operational taxonomic units (OTUs) (>97% identity) were curated and converted to relative abundance using Mothur. Shannon diversity index (H’) and OUT richness were used to show community diversity. To compare the diversity between communities (β-diversity), we employed the Yue and Clayton (θYC) dissimilarity distance metric. θYC values were shown as non-metric multidimensional scaling (NMDS) plot. Linear discriminant analysis effect size (LEfSe) analysis was performed to identify differentially abundant taxa 53.
Metabolome analysis
The fecal samples for metabolome analysis were collected from HMA mice (2 weeks after inoculation with human stool sample) before CDI (at day 0). The fecal metabolomic profile was analyzed using capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) 54. Lyophilized fecal samples were homogenized by a Shake Master (Biomedical Science, Tokyo, Japan) with 3.0-mm Zirconia Beads (Biomedical Science, Tokyo, Japan). Homogenized fecal samples were then resuspended in methanol:chloroform:water solution to extract metabolites. The Agilent CE System, the Agilent G3250AA LC/MSD TOF System, the Agilent 1100 Series Binary HPLC Pump, the G1603A Agilent CE-MS adapter, and the G1607A Agilent CE-ESI-MS Sprayer Kit (Agilent Technologies, Santa Clara, CA) were used for CE-TOFMS analysis. Data are processed by the MasterHands software. For luminal succinate analysis, frozen stool samples were analyzed using liquid chromatography–mass spectrometry (LC-MS) at the Michigan Regional Comprehensive Metabolomics Resource Core 55. In brief, fecal samples were resuspended in 0.5 ml of extraction solution (methanol: chloroform: water = 8:1:1) containing 13C4-labeled succinate. The samples were then homogenized and incubated at 4°C for 10 min. The extract was harvested by centrifuging suspension at 4°C, 14,000 rpm for 10 min. An A 1260 UPLC module coupled with a 6520 quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies, Santa Clara, CA.) was used for LC-MS analysis. A 150 × 1 mm Luna NH2 hydrophilic interaction chromatography column (Phenomenex, Torrance, CA) was used to separate metabolites. MassHunter Quantitative Analysis Reporting B.07.00 Service Pack (Agilent Technologies) was used for data processing. Succinate was normalized to its isotopically labeled internal standard and quantitated using 2 replicated injections of 5 standards to create a linear calibration curve with accuracy better than 80% for each standard. Other compounds in the analysis were normalized to the nearest internal standard, and the peak areas were used for differential analysis between groups.
Ex vivo growth of C. difficile (JIR8094 and Cd-CD2344−)
Overnight cultures of JIR8094 and Cd-CD2344− were diluted 5000-fold with a minimal medium (10 g/l peptone, 10 g/l beef extract, 3 g/l yeast extract, 1.55 g/l NaH2PO4・H2O, 5.95 g/l Na2HPO4) supplemented with 1% glucose, 0.1% succinate, or 100 μg/mL UC luminal content. A handheld spectrophotometer (Biochrom US, Holliston, MA) was used for optical density (OD600) measurements.
Ex vivo germination and growth of C. difficile (VPI10463)
Mouse cecal contents from HC-HMA and UC-HMA mice were quantified inside an anaerobic chamber and diluted 10-fold with PBS. Heat-treated (65°C, 20 min) C. difficile spores (103 spores) were added to the cecal contents, which were then incubated anaerobically at 37°C for 6 h. After incubation, bacterial samples were serially diluted, plated on TCCFA plates in order to quantify germination and growth of C. difficile 3.
In vitro growth of Phascolarctobacterium
P. faecium JCM 30894 and P. succinatutens JCM 16074 (JCM, Ibaragi, Japan) were grown overnight in a peptone yeast extract medium supplemented with 80 mM sodium succinate (PYS) under anaerobic conditions 19. The overnight culture of Phascolarctobacterium spp. was diluted 100-fold with PYS medium. In some experiments, 100 mM GlcNAc (MilliporeSigma, St. Louis, MO) was added. For the growth assay on mucus medium, SPF WT C57BL/6 mice (some, but not all mice were littermates) were co-housed for over 2 weeks, and then randomly separated into two groups. One group was treated with αIL-22 antibody and the other with the control antibody. Colonic mucus was scraped from colonic walls into HEPES-Hank’s buffer (8.0 g/Ll NaCl, 0.4 g/l KCl, 0.05 g/l CaCl2・H2O, 0.35 g/l KH2PO4, 0.2 g/l MgSO4 • 7H2O, and 2.6 g/l HEPES, pH 7.4). Contaminating epithelial cells and membranes were removed by centrifugation, once at 12,000 g for 10 min, and once at 27,000 g for 15 min at 4°C. Colonic mucus in HEPES-Hank’s buffer was stored at −80°C until further use 56. 300 μg of colonic mucus scrape from the control and the αIL-22 antibody–treated mice were added to the PYS medium. Phascolarctobacterium spp. were cultured anaerobically for 24–48 h. Bacterial RNAs were purified and GH family gene expression was quantified by qPCR. The CFU count was determined by culturing samples on PYS agar plates.
Glycan analysis
The mucus samples were scraped from the colonic tissues. The scraped mucus was suspended in GuHCl buffer (6 M GuHCl, 5 mM EDTA, 10 mM NaH2PO4, and 5 mM PMSF), on a rolling wheel, in a cold room, overnight. The samples were separated into a soluble fraction (supernatant) and an insoluble fraction (pellet) by centrifugation at full speed for 10 min at 4°C. The insoluble fraction was solubilized by adding 0.975 mL solubilization buffer (6 M GuHCl, 100 mM Tris, 5 mM DETA) along with 25 mM DTT and 5 mM PMSF. The solution was kept on a rolling wheel overnight in a cold room. After incubation, iodoacetamide was added, and the solution was incubated in the dark for 4 h at room temperature. The pellet was removed by centrifugation at full speed for 10 min at 4°C. Both soluble and insoluble fractions were transferred to a PVDF membrane using dot-blotting. The membrane was stained with Alcian blue, followed by destaining with methanol. N- and O-linked oligosaccharides were released from glycoproteins blotted on PVDF membranes for glycan analysis 57. The N-glycans were first released from the PVDF membrane. The membrane was blocked with 1% PVP solution (polyvinyl pyrrolidone 40,000, w/w) in 50% methanol and then washed 3 times in water and shaken for 5 min. Next, the membrane was added 50 mM NH4Ac (pH 8.4) and PNGase. After overnight incubation at 37°C, the sample was dried using SpeedVac. The left membrane dots were subjected to O-glycans release. The N-glycans were reduced by incubating with 20 mM NaOH and 0.5 M NaBH4 overnight at 50°C. The salt was removed as O-glycans. The O-glycans on the dot blots were released by reducing beta-elimination and desalted. Released O-glycans were resuspended in water and analyzed by liquid chromatography–electrospray ionization tandem mass spectrometry (LC-ESI/MS). A 5 μm porous graphite particles column (Hypercarb, Thermo-Hypersil, Runcorn, UK) was used to separate oligosaccharides. The loaded oligosaccharides were eluted with 0%−45% gradient of Buffer B (10 mM ammonium bicarbonate in 80% acetonitrile) followed by a wash step with 100% Buffer B. The column was then equilibrated with Buffer A (10 mM ammonium bicarbonate). An LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA) in negative ion mode was used to analyze the samples. Xcalibur software (Version 2.0.7) was used for data acquisition and analysis.
The Gene Expression Omnibus (GEO) accession number
Publicly available gene expression data used in this study are available from the NCBI Gene Expression Omnibus under the following accession codes: GSE75214 25, GSE16879 26, and GSE73661 27.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism software version 6.0 (GraphPad Software Inc.). Statistical tests used for the analysis of data are identified in the legend of each figure. Differences of P < 0.05 were considered significant.
Extended Data
Extended Data Fig. 1. Healthy human microbiotas prevent C. difficile infection.
(a) (Left) GF B6 mice were colonized with healthy control (HC) microbiotas for 2 weeks (human microbiota-associated (HMA) mice). GF or HC-HMA mice (GF: n=9, HC#1:n=8 and HC#2:n=3, biologically independent animals) were then infected with C. difficile VPI 10463 spores (103 spores/mouse). C. difficile load in feces was determined on indicated days post-infection. Dots represent individual mice. Bars indicate median. Data were pooled from 2 independent experiments. (Right) The mortality of C. difficile infected GF or HC-HMA mice. Statistical significance was assessed by Log-rank test (two-sided). (b and c) (b) HC-HMA mice were treated with a cocktail of antibiotics or regular water and then infected with C. difficile VPI 10463 spores (n=5, biologically independent animals). (Left) CFU in feces. Dots represent individual mice. Bars indicate median. Data were pooled from 2 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided). (Right) The mortality of mice. Statistical significance was assessed by Log-rank test (two-sided). (c) Representative histological images and associated histological scores. Scale bar is 100 μm. Data are presented as mean ± s.e.m. from pooled two independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided).
Extended Data Fig. 2. IL-22 neutralization inhibits the expression of antimicrobial proteins.
Reg3b and Reg3g mRNA levels in control or αIL-22 antibody-treated HC-HMA Rag1−/− mice were determined by qPCR. Expression was normalized to that of the murine Actb gene. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=8, biologically independent animals) from pooled 2 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided).
Extended Data Fig. 3. IL-22 shapes the gut microbial community.
HC-HMA-Rag1−/− mice were treated with control or αIL-22 antibody twice, on day −5 and day −3, before fecal samples were collected. Bacterial 16S rRNA sequences were analyzed. (a) Shannon index (α-diversity) and number of OTUs (richness) of control and αIL-22 antibody-treated HMA-Rag1−/− mice. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=3, biologically independent animals). Data are representative of 2 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided). (b) Microbial community structures were analyzed using the Yue and Clayton dissimilarity distance metric (θYC) and are shown in a nonmetric, multidimensional scaling plot. (c) Bacterial taxonomy at the family level in the feces.
Extended Data Fig. 4. Luminal metabolomic analysis in IL-22–neutralized HMA mice.
(a) Fecal samples were collected after treatment with αIL-22 antibody, twice and before CDI (day 0). A heat map showing the quantified metabolic profiles of control or αIL-22 antibody-treated HC-HMA-Rag1−/− mice. All concentrations of quantified metabolites were transformed into Z-scores and clustered according to their Euclidean distance. Gray areas in the heat map indicate that respective metabolites were not detected. (b) Principal component analysis of the metabolome data. (C) A loading scatter plot of the principal component analysis. (d) Luminal metabolites were analyzed by CE-TOF/MS. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=13, biologically independent animals) from pooled 3 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided).
Extended Data Fig. 5. C. difficile growth on succinate.
In vitro growth of WT JIR8094 or Cd-CD2344- mutant C. difficile in a minimal medium supplemented with glucose or succinate. Data are presented as mean ± s.d. (n=3 technical replicates). Data are representative of 3 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided).
Extended Data Fig. 6. Succinate is not required for the growth of C. difficile in germ-free mice.
GF C57BL/6 mice were infected with WT JIR8094 or Cd-CD2344− mutant C. difficile strains. C. difficile load in feces was determined on indicated days post-infection (n=5, biologically independent animals). Dots represent individual mice. Bars indicate median. Data were from pooled 2 independent experiments. Statistical significance was assessed by 2-way ANOVA with Bonferroni post-hoc test (two-sided).
Extended Data Fig. 7. Gene expression profiles in UC patient cohorts.
(a) The mRNA expression of IL22 mRNA in the colonic tissue from control subjects (n=11), patients with inactive UC (n=23) and patients with active UC (n=74). Data were derived from GEO data set GSE75214. Dots represent biologically independent subjects. Data are presented as mean ± s.e.m.. Statistical significance was assessed by Kruskal-Wallis test with Dunn posttest (two-sided). (b and d) The mRNA expression of IL22, IL22RA, MGAT4A and MGAT4B mRNA in the colonic tissue from control subjects and patients with active UC. (c and e) Correlation between MGAT4A/MGAT4B and IL22RA mRNA expression in 3 groups. Statistical significance was measured by Pearson correlation test (two-sided). Dots represent biologically independent subjects. Data were derived from GEO data sets GSE16879 (control: n=8 and active UC: n=24) and GSE73661 (control: n=12 and active UC: n=67). Data are presented as mean ± s.e.m.. Statistical significance was assessed by Mann-Whitney U test (two-sided). (b and d). Statistical significance was measured by Pearson correlation test (two-sided) (c and e).
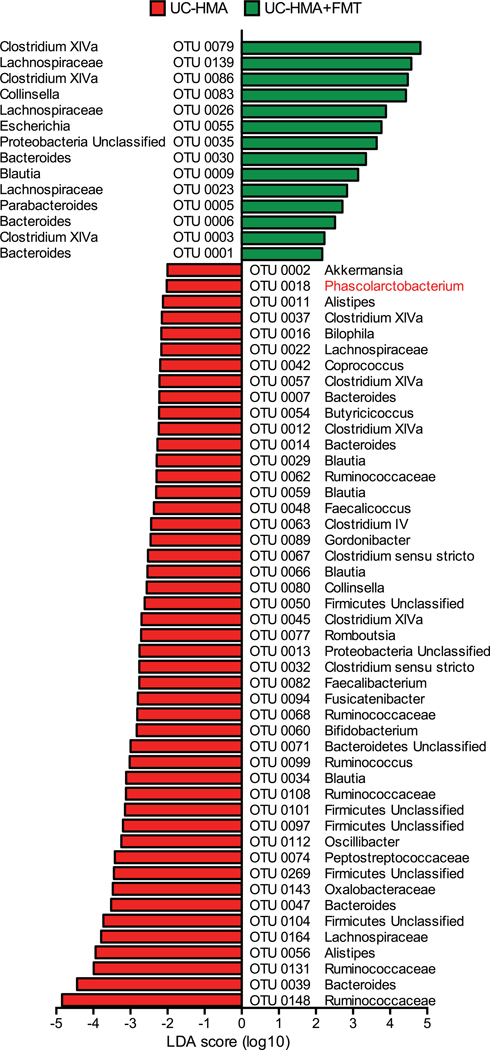
Extended Data Fig. 8. FMT restores the microbial composition in UC-HMA mice.
Significantly altered bacteria in pre–C. difficile infected UC-HMA mice with or without FMT were identified by LEfSe analysis. UC-HMA mice–enriched taxa have a positive LDA score (red, n=6, biologically independent animals), and FMT-treated UC-HMA mice–enriched taxa have a negative LDA score (green, n=6, biologically independent animals). A P value of < 0.05 and a score ≥ 2.0 were considered significant in Kruskal–Wallis and pairwise Wilcoxon tests (two-sided), respectively.
Extended Data Fig. 9. Luminal metabolomic analysis in FMT-treated UC-HMA mice.
(a) Fecal samples were collected after treatment with FMT (day −3) and before CDI (day 0). A heat map showing the quantified metabolic profiles of UC-HMA or FMT-treated UC-HMA mice. All concentrations of quantified metabolites were transformed into Z-scores and clustered according to their Euclidean distance. Gray areas in the heat map indicate that respective metabolites were not detected. (b) The principal component analysis of the metabolome data (n=9, biologically independent animals). (C) A loading scatter plot of the principal component analysis. (d) Luminal metabolites were analyzed by CE-TOF/MS. Dots represent individual mice. Data are presented as mean ± s.e.m. (n=9, biologically independent animals) from pooled 2 independent experiments. Statistical significance was assessed by Mann-Whitney U test (two-sided).
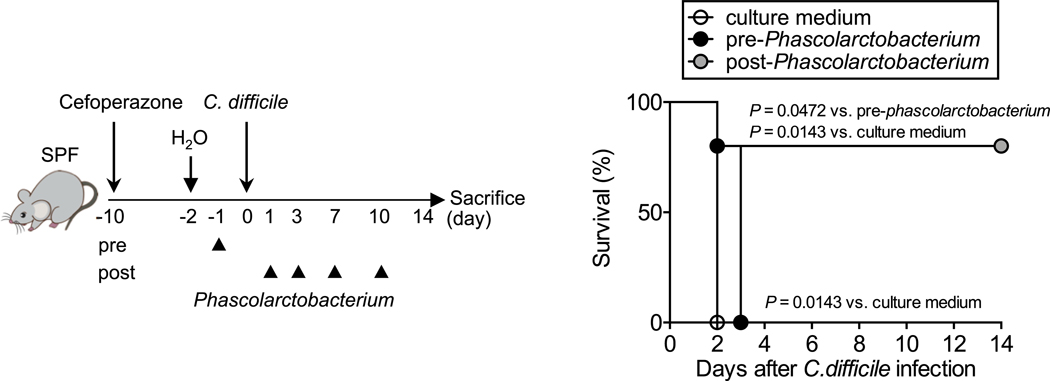
Extended Data Fig. 10. Phascolarctobacterium inoculation protect mice from CDI.
SPF C57BL/6 mice were treated with cefoperazone (0.5 mg/mL in drinking water). After 8 days, the mice were switched to regular water and allowed to recover for 2 days before being infected with C. difficile VPI spores. Mice were treated with P. faecium JCM 30894 and P. succinatutens JCM 16074 (106 CFU each strain) or with its culture medium by oral gavage, once, before C. difficile inoculation (1 day prior to CDI, pre-treatment) or 4 times post-inoculation (1, 3, 7, 10 days post-infection, post-treatment). Dots represent individual mice (n=5, biologically independent animals). The mortality of C. difficile–infected mice was assessed. Statistical significance was assessed by Log-rank test (two-sided) from pooled 2 independent experiments.
Supplementary Material
Acknowledgments
The authors thank Andrew B. Shreiner for technical assistance, the University of Michigan Center for Gastrointestinal Research (UMCGR) (DK034933), Host Microbiome Initiative, the Germ-free Animal Facility, the Michigan Regional Comprehensive Metabolomics Resource Core (MRC2) (DK097153) for core services, and Ingrid L. Bergin from the In-Vivo Animal Core at the University of Michigan Unit for Laboratory Animal Medicine for histological assessment. This work was supported by National Institute of Health DK110146 and DK108901 (N.K.), Crohn’s and Colitis Foundation of America (N. K and H. N.-K.), Young Investigator Grant from the Global Probiotics Council (N.K.), University of Michigan Center for Gastrointestinal Research DK034933 (N. K.), Joint Usage/Research Program of Medical Mycology Research Center Chiba University 18–1 (N.K. and Y.G.), JSPS Postdoctoral Fellowship for Research Abroad (H. N.-K. and S.K.), the Uehara Memorial Foundation Postdoctoral Fellowship Award (S.K.), Clinical and Translational Science Awards (CTSA) Program (S.K.), and Prevent Cancer Foundation (S.K.). JSPS KAKENHI 16H04901, 17H05654, 18H04805 (S.F.), PRESTO JPMJPR1537 (S.F.), JST ERATO JPMJER1902 (S.F.), AMED-CREST JP19gm1010009 (S.F.), the Takeda Science Foundation (S.F.), the Food Science Institute Foundation (S.F.). MS analysis of glycans was performed by the Swedish infrastructure for biological mass spectrometry (BioMS) supported by the Swedish Resarch Council.
Data Availability:
Source data for all Figures and Extended data Figures are provided with the paper. The microbiome data in this study is available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive under BioProject PRJNA594915. The metabolome data is available at the NIH Common Fund’s Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench, http://www.metabolomicsworkbench.org, where it has been assigned Project ID PR000882 (αIL-22 Ab experiment in Fig. 2b and Extended Data Fig. 4), PR000881 (FMT experiment in Fig. 6f and Extended Data Fig. 9), and PR000869 (Phascolarctobacterium administration in Fig. 6i).
Footnotes
Disclosures: The authors declare no competing interests.
References
- 1.Britton RA & Young VB Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146, 1547–1553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theriot CM et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5, 3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N & Nunez G Role of the gut microbiota in the development and function of lymphoid cells. J Immunol 190, 1389–1395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenberg GF et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science 336, 1321–1325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14, 282–289 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto K et al. IL-22 Controls Iron-Dependent Nutritional Immunity Against Systemic Bacterial Infections. Science immunology 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pham TA et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 16, 504–516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickard JM et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto Y et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 345, 1254009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh-Takayama N et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Sanos SL et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol 10, 83–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadighi Akha AA et al. Interleukin-22 and CD160 play additive roles in the host mucosal response to Clostridium difficile infection in mice. Immunology 144, 587–597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa M et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 41, 620–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abt MC et al. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe 18, 27–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagao-Kitamoto H et al. Functional characterization of inflammatory bowel disease-associated gut dydbiosis in gnotobiotic mice. Cellular and Molecular Gastroenterology and Hepatology 2, 468–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins J, Auchtung JM, Schaefer L, Eaton KA & Britton RA Humanized microbiota mice as a model of recurrent Clostridium difficile disease. Microbiome 3, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenberg GF & Artis D Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity 37, 601–610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y, Nagai F & Morotomi M Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol 78, 511–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topping DL & Clifton PM Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiological reviews 81, 1031–1064 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Ferreyra JA et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodemann JF, Dubberke ER, Reske KA, Seo da H & Stone CD Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin Gastroenterol Hepatol 5, 339–344 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Reddy SS & Brandt LJ Clostridium difficile infection and inflammatory bowel disease. J Clin Gastroenterol 47, 666–671 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Berg AM, Kelly CP & Farraye FA Clostridium difficile infection in the inflammatory bowel disease patient. Inflamm Bowel Dis 19, 194–204 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Vancamelbeke M et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm Bowel Dis 23, 1718–1729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arijs I et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 4, e7984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arijs I et al. Effect of vedolizumab (anti-alpha4beta7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut 67, 43–52 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Johnson JL, Jones MB, Ryan SO & Cobb BA The regulatory power of glycans and their binding partners in immunity. Trends Immunol 34, 290–298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley P What Have We Learned from Glycosyltransferase Knockouts in Mice? Journal of molecular biology 428, 3166–3182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F et al. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Experimental and therapeutic medicine 14, 3122–3126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L et al. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe 34, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Hourigan SK et al. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment Pharmacol Ther 42, 741–752 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Levy M et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163, 1428–1443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battaglioli EJ et al. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan XC et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13, R79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolk K et al. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn’s disease. J Immunol 178, 5973–5981 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Schmechel S et al. Linking genetic susceptibility to Crohn’s disease with Th17 cell function: IL-22 serum levels are increased in Crohn’s disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis 14, 204–212 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Mann ER et al. Human gut dendritic cells drive aberrant gut-specific t-cell responses in ulcerative colitis, characterized by increased IL-4 production and loss of IL-22 and IFNgamma. Inflamm Bowel Dis 20, 2299–2307 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Leung JM et al. IL-22-producing CD4+ cells are depleted in actively inflamed colitis tissue. Mucosal Immunol 7, 124–133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamas B et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 22, 598–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin JC et al. IL-22BP is produced by eosinophils in human gut and blocks IL-22 protective actions during colitis. Mucosal Immunol 9, 539–549 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Pelczar P et al. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science 354, 358–362 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Xu AT et al. High suppressor of cytokine signaling-3 expression impairs STAT3-dependent protective effects of interleukin-22 in ulcerative colitis in remission. Inflamm Bowel Dis 21, 241–250 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Li Y et al. Increased suppressor of cytokine signaling-3 expression predicts mucosal relapse in ulcerative colitis. Inflamm Bowel Dis 19, 132–140 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Dias AM et al. Dysregulation of T cell receptor N-glycosylation: a molecular mechanism involved in ulcerative colitis. Hum Mol Genet 23, 2416–2427 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Lopez J & Grinspan A Fecal Microbiota Transplantation for Inflammatory Bowel Disease. Gastroenterology & hepatology 12, 374–379 (2016). [PMC free article] [PubMed] [Google Scholar]
- 47.Paramsothy S et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Atarashi K et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 49.Paik J et al. Potential for using a hermetically-sealed, positive-pressured isocage system for studies involving germ-free mice outside a flexible-film isolator. Gut Microbes 6, 255–265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hecht G et al. A simple cage-autonomous method for the maintenance of the barrier status of germ-free mice during experimentation. Lab Anim 48, 292–297 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Schloss PD et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segata N et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirayama A et al. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Analytical and bioanalytical chemistry 404, 3101–3109 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lorenz MA, Burant CF & Kennedy RT Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical chemistry 83, 3406–3414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen PS & Laux DC Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods in enzymology 253, 309–314 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Schulz BL, Packer NH & Karlsson NG Small-scale analysis of O-linked oligosaccharides from glycoproteins and mucins separated by gel electrophoresis. Analytical chemistry 74, 6088–6097 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.