Abstract
Objectives
Develop a CT-based radiomics model and combine it with frozen section (FS) and clinical data to distinguish invasive adenocarcinomas (IA) from preinvasive lesions/minimally invasive adenocarcinomas (PM).
Methods
This multicenter study cohort of 623 lung adenocarcinomas was split into training (n = 331), testing (n = 143), and external validation dataset (n = 149). Random forest models were built using selected radiomics features, results from FS, lesion volume, clinical and semantic features, and combinations thereof. The area under the receiver operator characteristic curves (AUC) was used to evaluate model performances. The diagnosis accuracy, calibration, and decision curves of models were tested.
Results
The radiomics-based model shows good predictive performance and diagnostic accuracy for distinguishing IA from PM, with AUCs of 0.89, 0.89, and 0.88, in the training, testing, and validation datasets, respectively, and with corresponding accuracies of 0.82, 0.79, and 0.85. Adding lesion volume and FS significantly increases the performance of the model with AUCs of 0.96, 0.97, and 0.96, and with accuracies of 0.91, 0.94, and 0.93 in the three datasets. There is no significant difference in AUC between the FS model enriched with radiomics and volume against an FS model enriched with volume alone, while the former has higher accuracy. The model combining all available information shows minor non-significant improvements in AUC and accuracy compared with an FS model enriched with radiomics and volume.
Conclusions
Radiomics signatures are potential biomarkers for the risk of IA, especially in combination with FS, and could help guide surgical strategy for pulmonary nodules patients.
Key Points
• A CT-based radiomics model may be a valuable tool for preoperative prediction of invasive adenocarcinoma for patients with pulmonary nodules.
• Radiomics combined with frozen sections could help in guiding surgery strategy for patients with pulmonary nodules.
Electronic supplementary material
The online version of this article (10.1007/s00330-019-06597-8) contains supplementary material, which is available to authorized users.
Keywords: Carcinoma, non-small-cell lung; Machine learning; Frozen sections; Adenocarcinoma of lung; Tomography, spiral computed
Introduction
Lung cancer ranks first in cancer mortality around the world [1]. With the popularization of computed tomography (CT) and the application of low-dose CT for lung cancer screening, substantial early-stage lung cancers have been detected [2]. Most malignant pulmonary nodules are confirmed as adenocarcinoma by pathology [3]. Patients with different types of adenocarcinoma differ in 5-year survival probabilities; e.g., patients with a diagnosis of invasive adenocarcinoma (IA) have a significantly poorer survival probability than those with adenocarcinoma in situ (AIS) or minimally invasive adenocarcinoma (MIA), who have a nearly 100% survival probability [4, 5]. Currently, lobectomy may be a better choice than sublobar resection for patients with IA, and patients with preinvasive lesions (atypical adenomatous hyperplasia (AAH) and AIS) and MIA (collectively PM) are candidates for limited resections [6].
Three methods are most commonly used to perform intraoperative or preoperative diagnosis in clinical practice, namely chest CT scan, biopsy, and intraoperative frozen section (FS). Many radiological studies rely on morphological (semantic) features such as spiculation or lobulation to generate a differential diagnosis. However, qualitative interpretation of the image is hampered by the strong subjectivity introduced by atypical radiology signs, especially in small and in ground-glass nodules [7–10]. Moreover, transbronchial and percutaneous biopsies are limited by the difficulties of sampling and localization [11]. FS has the potential to guide surgical strategy for peripheral small-sized pulmonary nodules by intra-operatively assessing adenocarcinoma type [6]. However, the coincidence rate of pathological diagnosis between frozen and paraffin specimens in early adenocarcinoma is hampered by sampling and interpretation errors, and by suboptimal specimen quality, due to the histologic heterogeneity of lung adenocarcinomas; e.g., it is difficult to identify whether the adenoid structure trapped around the scar is an invasive component or not [11–13]. The International Association for the Study of Lung Cancer emphasized that the diagnosis of adnocarcinoma cannot be firmly established without histologic sampling of the entire tumor. Review of CT images is recommended to add insights to the gross pathologic findings, which motivated the development of a CT-based radiomics model which combined with FS could further help distinguish IA from AIS or MIA [4].
Radiomics is the process that allows quantitative imaging features to be extracted in bulk, creating unique fingerprints for images (or regions of interest (ROI) therein) which can be correlated with clinical data using machine learning approaches [14, 15]. Recently, CT-based radiomics have shown excellent predictive performance to differentiate IA from PM of the lung [16–19]. Therefore, this study aimed to develop and validate a multicenter multifactorial radiomics model combined with FS results and clinical parameters to distinguish IA from PM in pulmonary nodules.
Materials and methods
Patients
The institutional review boards approved this retrospective study registered in http://clinicaltrials.gov (identifier: NCT03872362), and the requirement for informed consent was waived. A total of 582 patients with 623 nodules underwent lung operation and non-contrast-enhanced CT scans between January 2013 and October 2017. The patient cohorts from three centers were identified according to the established inclusion and exclusion criteria. The inclusion criteria were (1) primary lung adenocarcinoma, confirmed by pathology and (2) pulmonary nodules without a visible cavity, which would either alter feature values or otherwise complicate the delineation process. The exclusion criteria were (1) previous history of radiation therapy, chemotherapy, or biopsy before baseline CT scan; (2) the time interval between the CT examination and surgery was more than 2 weeks; and (3) insufficient CT or pathology quality to make a diagnosis. Clinical information was procured from medical records.
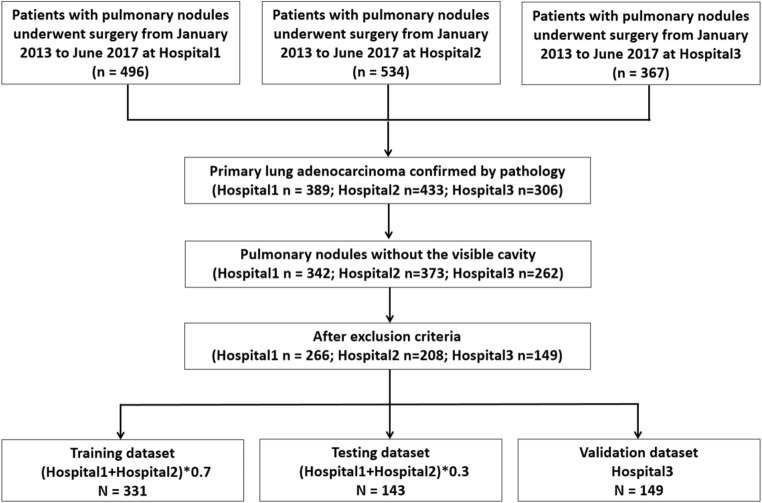
All nodules from Hospital1 and Hospital2 were aggregated and randomly divided into two datasets, 70% for the training dataset and 30% for the testing dataset, while attempting to maintain the original class balance in the sub-cohorts. All nodules from Hospital3 were allocated to the external validation dataset (Fig. 1).
Fig. 1.
Flowchart for patient selection from three hospitals
Image acquisition, semantic features, and pathological evaluation
CT scans were performed on a 64- or 128-detector row CT system (Somatom Definition, Siemens Medical Solutions) with the following acquisition and reconstruction parameters: tube voltage 100 kV or 120 kV; the tube current is automatically calculated; pitch 0.75–1.5; collimation 0.6 mm; matrix 512 × 512. The reconstruction algorithm of “bone plus” for thin-section helical scans was used with a thickness of 1.0–1.5 mm. Further detailed acquisition parameters are provided in Supplementary S1.
Two chest radiologists each with 8 years of experience were blinded to evaluate the images in the lung window setting (window, − 600 HU; width, 1200 HU) and the mediastinal window setting (window, 40 HU; width, 350 HU). The type of ground-glass nodule (GGN) and solid nodule was defined as well as the lesion diameter was reported according to the guidelines from Fleischner Society [20, 21]. FS and final pathology results were a blind assessment by two pathologists according to the International classification of lung adenocarcinoma [4]. Atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), and MIA were categorized as PM. Cases of disagreement for semantic features and pathological assessments were resolved through consultation.
Segmentation
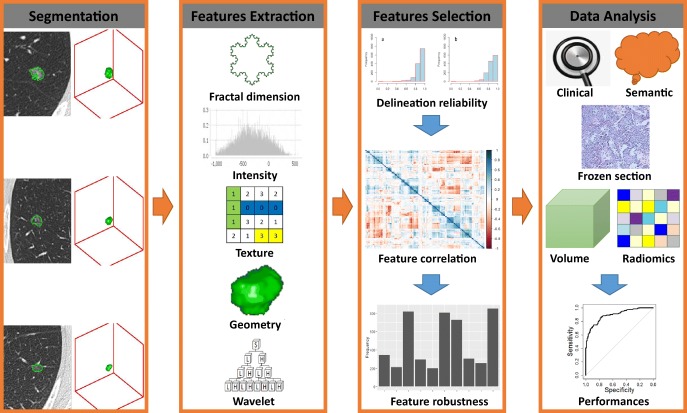
The workflow of radiomics from segmentation to data analysis is shown in Fig. 2. The ROI was manually contoured slice-wise on the axial projection to arrive at a 3D-segmentation using MIM (MIM Software Inc.). One radiologist with 8 years of experience performed segmentation for all cases. To assess intra-reader agreement, 50 randomly selected cases were segmented again by the same radiologist, as well as by another medical doctor with 3 years of contouring experience. Apart from the volume feature computed by the radiomics software, tumor volume was also computed by MIM after contouring.
Fig. 2.
Flowchart showing the process of radiomics
Image processing and feature extraction
All images were resampled to an in-plane pixel spacing of 0.75 mm and a slice thickness of 1.5 mm using linear interpolation to partially counter the heterogeneous reconstruction settings found in the database [22]. For non-filtered features, excluding first-order statistics features, voxels values, represented in Hounsfield units (HU), were aggregated into bins of 25 HU wide in order to reduce noise and inter-scanner variability. Filtered features used a fixed number of bins equal to the number calculated for non-filtered features. Feature extraction was performed using the RadiomiX Discovery Toolbox (OncoRadiomics SA). The features extracted describe fractal dimension, intensity histogram, first-order statistics, texture, local intensity, shape, and features extracted from wavelet-filtered images. Descriptions and mathematical definitions of the features have been described in detail previously [23].
Feature selection
The intra-/inter-class correlation coefficient (ICC) was used to assess the robustness of features between the individual radiologist contours. Features with ICC values < 0.8 were removed from further analysis. Features with little variance across the cohort (the ratio of the frequency of the most common value to the frequency of second most common is greater than 95/5) have little explanatory value and hence were removed. Likewise, highly correlated features needlessly inflate the dimensionality of feature space. For feature pairs with a high Spearman correlation (r > 0.8) in the data from Hospital1 and Hospital2, the feature with the highest mean correlation with all remaining features was removed. The cases from Hospital1 and Hospital2 were randomly spilt into training (70%) and testing (30%) datasets 1000 times. For each iteration, the top ten features were ranked and selected in the training dataset using recursive feature elimination with the treebag method and a cross-validation technique (tenfold, 10 times), and a random forest model built in the training dataset using the top 5 features and evaluated in the testing dataset. The features with the highest selection frequencies in the 1000 iterations were retained. Finally, features with high Spearman correlation with volume (r > 0.8) were removed since volume is examined as an independent feature in univariate and multivariate analyses.
Model training and validation
Random forest binary classification models were trained using an increasing number of features from the previous step, starting with the highest ranked feature, and their performance was tested on the testing dataset until the area under the curve (AUC) of the receiver operator characteristic (ROC) increased < 0.02 in order to strike a balance between good performance and possible overfitting. The final model (with a set number of features) is trained on the combined training and testing dataset and validated on the external dataset. The radiomics model was developed using CT-based radiomics features, the clinical model was created based on clinical variables (age, gender, smoking status, and family history of lung cancer), and the semantic model was built with semantic features (location, diameter, and nodule type). Lesion volume values were used to build the volume model. The radiomics model combined with volume was named RV, which was further combined with clinical and semantic information (CSRV). FS results were combined with the radiomics and volume to build a multifactorial model (FSRV), and likewise, volume was added to the FS model (FSV). Finally, all data including clinical variables, semantic features, FS, radiomics features, and tumor volume were used to build a combined model called CSFSRV. In order to examine the entire pipeline for the ability to find spurious correlations, the outcomes were randomized and the process repeated, including feature selection and model building.
TRIPOD guidelines, radiomics quality score, and statistical analysis
This study was followed by the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guidelines [24]. The radiomics quality score (RQS) was used to evaluate the radiomics workflow [25].
Pearson’s chi-squared test was used for the statistical analysis of essential demographic characteristics. The performances of the random forest binary classification models were evaluated with receiver operating characteristic (ROC) curve to calculate the area under the curve (AUC), and the 95% confidence interval (CI) was derived leave-one-out cross-validation. Other diagnosis values (e.g., accuracy, sensitivity, specificity, negative predictive values (NPV), and positive predictive values (PPV)) were measured. The Hosmer-Lemeshow test was used to estimate the goodness-of-fit of models and the calibration plots were performed to test the consistency of models between predicted risk and observed risk in the validation dataset. The decision curve analysis was performed to assess the net benefits based on different threshold probabilities in models. Two-sided p values less than 0.05 were considered as a statistical significance. All statistical analysis, model building, and model evaluation were performed in R (version 3.5.2; http://www.r-project.org). Detailed statistical process, R packages, and R functions are described in Supplementary S2.
Results
Demographic characteristics
The demographic characteristics of the three datasets are summarized in Table 1. There were no significant differences in terms of clinical record (age, p = 0.34; gender, p = 0.14; smoking, p = 0.16; family history, p = 0.49), radiology semantic information (diameter, p = 0.54; location, p = 0.37; nodules type, p = 0.09), and volume values (p = 0.60) among three datasets. Additionally, there were no significant differences in final pathological type and surgical type (p = 0.08 and p = 0.17, respectively) among all datasets.
Table 1.
Demographic and clinical characteristics of patients on different datasets
| Variable | Training (n = 331) | Testing (n = 143) | Validation (n = 149) | p value |
|---|---|---|---|---|
| Age, n (%) | ||||
| ≤ 60 | 172 (52.0) | 74 (51.7) | 67 (45.0) | 0.34 |
| > 60 | 159 (48.0) | 69 (48.3) | 82 (55.0) | |
| Gender, n (%) | ||||
| Male | 111 (33.5) | 45 (31.5) | 62 (41.6) | 0.14 |
| Female | 220 (66.5) | 98 (68.5) | 87 (58.4) | |
| Smoking, n (%) | ||||
| Yes | 46 (13.9) | 19 (13.3) | 30 (20.1) | 0.16 |
| No | 285 (86.1) | 124 (86.7) | 119 (79.9) | |
| Family history, n (%) | ||||
| Yes | 9 (2.7) | 6 (4.2) | 7 (4.7) | 0.49 |
| No | 322 (97.3) | 137 (95.8) | 142 (95.3) | |
| Final pathology, n (%) | ||||
| IA | 200 (60.4) | 86 (60.1) | 105 (70.5) | 0.08 |
| PM | 131 (39.6) | 57 (39.9) | 44 (29.5) | |
| Diameter (cm), n (%) | ||||
| ≤ 1 | 132 (39.9) | 54 (37.8) | 52 (34.9) | 0.54 |
| 1.1–2.0 | 131 (39.6) | 51 (35.7) | 63 (42.3) | |
| 2.1–3.0 | 68 (20.5) | 38 (26.6) | 34 (22.8) | |
| Location, n (%) | ||||
| LUL | 78 (23.6) | 31 (21.7) | 26 (17.4) | 0.37 |
| LLL | 49 (14.8) | 28 (19.6) | 22 (14.8) | |
| RUL | 127 (38.4) | 51 (35.7) | 51 (34.2) | |
| RML | 23 (6.9) | 10 (7.0) | 14 (9.4) | |
| RLL | 54 (16.3) | 23 (16.1) | 36 (24.2) | |
| Nodule type, n (%) | ||||
| pGGN | 124 (37.5) | 49 (34.3) | 37 (24.8) | 0.09 |
| PSN | 150 (45.3) | 71 (49.7) | 79 (53.0) | |
| Solid | 57 (17.2) | 23 (16.1) | 33 (22.1) | |
| Volume (mm3), n (%) | ||||
| < 500 | 115 (34.7) | 47 (32.9) | 43 (28.9) | 0.60 |
| 500–1000 | 67 (20.2) | 29 (20.3) | 27 (18.1) | |
| > 1000 | 149 (45.0) | 67 (46.9) | 79 (53.0) | |
| Surgical type, n (%) | ||||
| Lobectomy | 256 (77.3) | 103 (72.0) | 104 (69.8) | 0.17 |
| Limited resection | 75 (22.7) | 40 (28.0) | 45 (30.2) | |
IA, invasive adenocarcinoma; PM, preinvasive lesions/minimally invasive adenocarcinomas; LUL, left upper lobe; LLL, left lower lobe; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; pGGN, pure ground-glass nodule; PSN, part-solid nodule. p values calculated using Pearson’s chi-squared test
Table 2 summarizes cohort differences between patients with final diagnosis of IA and PM without correction for multiple testing. There were significant differences in age, gender, diameter, nodule type, volume, and surgical type (p < 0.01 for all), while smoking, family history, and location have no significant differences between two groups. The predictive and diagnosis performance of individual clinical, semantic, and radiomics features are shown in Table 3.
Table 2.
Demographic and clinical characteristics of patients on IA and PM groups
| Variable | IA (n = 391) | PM (n = 232) | p value |
|---|---|---|---|
| Age, n (%) | < 0.01* | ||
| ≤ 60 | 168 (43.0) | 145 (62.5) | |
| > 60 | 223 (57.0) | 87 (37.5) | |
| Gender, n (%) | < 0.01* | ||
| Male | 155 (39.6) | 63 (27.2) | |
| Female | 236 (60.4) | 169 (72.8) | |
| Smoking, n (%) | 0.089 | ||
| Yes | 67 (17.1) | 28 (12.1) | |
| No | 324 (82.9) | 204 (87.9) | |
| Family history, n (%) | 0.06 | ||
| Yes | 18 (4.6) | 4 (1.7) | |
| No | 373 (95.4) | 228 (98.3) | |
| Diameter (cm), n (%) | < 0.01* | ||
| ≤ 1 | 57 (14.6) | 181 (78.0) | |
| 1.1–2.0 | 211 (54.0) | 34 (14.7) | |
| 2.1–3.0 | 123 (31.5) | 17 (7.3) | |
| Location, n (%) | 0.27 | ||
| LUL | 89 (22.8) | 46 (19.8) | |
| LLL | 57 (14.6) | 42 (18.1) | |
| RUL | 139 (35.5) | 90 (38.8) | |
| RML | 27 (6.9) | 20 (8.6) | |
| RLL | 79 (20.2) | 34 (14.7) | |
| Nodule type, n (%) | < 0.01* | ||
| pGGNs | 62 (15.9) | 148 (63.8) | |
| PSN | 217 (55.5) | 83 (35.8) | |
| Solid | 112 (28.6) | 1 (0.4) | |
| Volume (mm3), n (%) | < 0.01* | ||
| < 500 | 47 (12.0) | 158 (68.1) | |
| 500–1000 | 81 (20.7) | 42 (18.1) | |
| > 1000 | 263 (67.3) | 32 (13.8) | |
| Surgical type, n (%) | < 0.01* | ||
| Lobectomy | 377 (96.4) | 86 (37.1) | |
| Limited resection | 14 (3.6) | 146 (62.9) |
IA, invasive adenocarcinoma; PM, preinvasive lesions/minimally invasive adenocarcinomas; LUL, left upper lobe; LLL, left lower lobe; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; pGGN, pure ground-glass nodule; PSN, part-solid nodule. *p < 0.05. p values calculated using Pearson’s chi-squared test
Table 3.
The predictive performance of individual feature on clinical, semantic, and radiomics model on the training dataset
| Individual features | ACC (95% CI) | Sensitivity | Specificity | AUC (95% CI) |
|---|---|---|---|---|
| Clinical | ||||
| Age | 0.57 (0.51–0.62) | 0.28 | 0.76 | 0.63 (0.57–0.69) |
| Gender | 0.60 (0.55–0.66) | 0 | 1 | 0.56 (0.51–0.61) |
| Smoking | 0.60 (0.55–0.66) | 0 | 1 | 0.55 (0.51–0.58) |
| Family history | 0.60 (0.55–0.66) | 0 | 1 | 0.50 (0.49–0.52) |
| Semantic | ||||
| Diameter | 0.71 (0.66–0.76) | 0.60 | 0.78 | 0.81 (0.77–0.86) |
| Location | 0.57 (0.52–0.63) | 0.02 | 0.94 | 0.51 (0.44–0.57) |
| Nodule type | 0.77 (0.72–0.82) | 0.69 | 0.83 | 0.80 (0.76–0.84) |
| Radiomics | ||||
| LocInt_peakLocal | 0.68 (0.64–0.73) | 0.60 | 0.74 | 0.83 (0.80–0.87) |
| Wavelet_HLL_Stats_max | 0.68 (0.63–0.72) | 0.59 | 0.73 | 0.85 (0.82–0.89) |
| GLRLM_LGRE | 0.76 (0.72–0.80) | 0.68 | 0.81 | 0.90 (0.87–0.92) |
| Wavelet_LLL_Stats_cov | 0.73 (0.68–0.77) | 0.63 | 0.79 | 0.87 (0.84–0.90) |
ACC, accuracy; AUC, area under curve; CI, confidence interval
Feature selection
A total of 1322 radiomics feature were extracted. After analysis of features stability to ROI variations, 325 features with ICC values less than 0.8 were removed (Supplementary S3). Ten features were removed due to little variance and 875 features (including volume) were removed due to high correlation with other features, leaving 112 features. The frequency of the top ten selected features and the distribution of the AUC values of the generated 10-feature models for the 1000 iterations are shown in Supplementary S4. When applying the pipeline to data with randomized outcomes, the mean AUC upon 1000 iterations for 5 feature models was 0.53 (Supplementary S4).
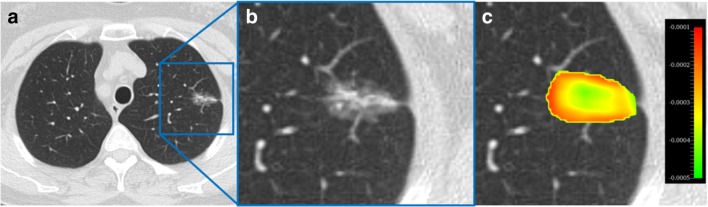
Of the features selected with high frequency, the top 5 features (with frequencies 850, 817, 805, 727, and 343 per 1000 iterations) were pre-selected using our methodology. One of these features (Wavelet_LHH_GLDZM_LIE) was removed due to a high correlation with tumor volume (r = − 0.84), arriving at a model with four robust radiomics features. The feature map is shown in Fig. 3.
Fig. 3.
a CT axial view of a pulmonary nodule; b zoomed in view; c feature map overlaid on the zoomed in CT
AUC of radiomics vs. clinical, semantic, volume, and FS
The radiomics model performed well when classifying between IA and PM with an AUC of 0.89 (95% CI, 0.86–0.93), 0.89 (95% CI, 0.83–0.94), and 0.88 (95% CI, 0.81–0.94) on the training, testing, and validation datasets, respectively. There was no significant difference between the AUC values of the radiomics and FS models on the three datasets (training: AUC = 0.90 (95% CI, 0.87–0.93), p = 0.82; testing: AUC = 0.93 (95% CI, 0.88–0.97), p = 0.20; validation: AUC = 0.92 (95% CI, 0.87–0.96), p = 0.29). The radiomics models performed better than the clinical model on all datasets, and outperformed semantic and volume models only in the training dataset, while no significant differences were seen on the testing and validation dataset (Table 4; Supplementary S5).
Table 4.
The detailed AUC values and p values among models on the three datasets
| Dataset | Model | AUC | 95% CI | P1 | P2 | P3 |
|---|---|---|---|---|---|---|
| Training | Clinical | 0.58 | 0.52–0.65 | < 0.01* | ||
| Semantic | 0.85 | 0.81–0.89 | 0.01* | |||
| Volume | 0.84 | 0.80–0.88 | 0.01* | |||
| FS | 0.90 | 0.87–0.93 | 0.82 | 0.58 | < 0.01* | |
| Radiomics | 0.89 | 0.86–0.93 | ||||
| RV | 0.90 | 0.87–0.94 | 0.18 | < 0.01* | ||
| CSRV | 0.91 | 0.88–0.94 | ||||
| FSV | 0.94 | 0.91–0.96 | 0.06 | |||
| FSRV | 0.96 | 0.94–0.98 | ||||
| CSFSRV | 0.96 | 0.94–0.98 | < 0.01* | 0.74 | ||
| Testing | Clinical | 0.55 | 0.45–0.65 | < 0.01* | ||
| Semantic | 0.85 | 0.78–0.92 | 0.28 | |||
| Volume | 0.87 | 0.81–0.93 | 0.54 | |||
| FS | 0.93 | 0.88–0.97 | 0.20 | 0.21 | 0.01* | |
| Radiomics | 0.89 | 0.83–0.94 | ||||
| RV | 0.88 | 0.82–0.93 | 0.10 | < 0.01* | ||
| CSRV | 0.89 | 0.84–0.94 | 0.21 | |||
| FSV | 0.98 | 0.96–1 | 0.50 | |||
| FSRV | 0.97 | 0.94–1 | ||||
| CSFSRV | 0.97 | 0.94–0.99 | < 0.01* | 0.25 | ||
| Validation | Clinical | 0.61 | 0.51–0.72 | < 0.01* | ||
| Semantic | 0.87 | 0.81–0.92 | 0.75 | |||
| Volume | 0.93 | 0.88–0.98 | 0.16 | |||
| FS | 0.92 | 0.87–0.96 | 0.29 | 0.97 | 0.01* | |
| Radiomics | 0.88 | 0.81–0.94 | ||||
| RV | 0.91 | 0.86–0.96 | 0.71 | 0.03* | ||
| CSRV | 0.92 | 0.87–0.96 | ||||
| FSV | 0.97 | 0.94–0.99 | 0.62 | |||
| FSRV | 0.96 | 0.93–0.99 | ||||
| CSFSRV | 0.96 | 0.94–0.99 | 0.01* | 0.30 |
FS, frozen section; RV, radiomics combining with volume; CSRV, radiomics combing with clinical, semantic, and volume; FSV, frozen section combining with volume; FSRV, frozen section combining with radiomics and volume; CSFSRV, radiomics combining with clinical, semantic, volume, and frozen section; AUC, the area under the curve; CI, confidence interval. *p < 0.05; P1 = p values between radiomics and other models; P2 = p values between CSRV and other models; P3 = p values between FSRV and other models. p values calculated using roc test by Delong method
AUC of CSRV vs. FS, RV, and CSFSRV
Compared with the FS and RV model, the CSRV model did not show improved AUC value (training: AUC = 0.91, 95% CI 0.88–0.94; testing: AUC = 0.89, 95% CI 0.84–0.94; validation AUC = 0.92, 95% CI 0.87–0.96; p > 0.05), while it was significantly worse than the CSFSRV model (training: AUC = 0.96, 95% CI 0.94–0.98, p < 0.01; testing AUC = 0.97, 95% CI 0.94–0.99, p < 0.01; validation: AUC = 0.96, 95% CI 0.94–0.99, p = 0.01).
AUC of FSRV vs. RV, FS, FSRV, and CSFSRV
Adding FS into the radiomics and volume (FSRV) model improved the classification performance between IA and PM (training: AUC = 0.96, 95% CI 0.94–0.98; testing: AUC = 0.97, 95% CI 0.94–1; validation: AUC = 0.96, 95% CI 0.93–0.99), which significantly outperforms both RV and FS alone on all three datasets. There were no significantly differences between FSRV and both FSV and CSFSRV models on all datasets (Table 4; Supplementary S6).
Accuracy of models
The FS model was able to discriminate between IA and PM on the training, testing, and validation dataset with an accuracy of 0.90, 0.92, and 0.90, respectively. The radiomics model had a higher accuracy than other single-factor models including clinical, semantic, and volume on all three datasets (0.82, 0.79, and 0.85). When FS combined with RV (0.83, 0.80, and 0.87), the resulting FSRV model showed an improved accuracy with values of 0.91, 0.94, and 0.93 on the three datasets, outperforming FSV (0.89, 0.92, and 0.91). When the FS model is enriched with CSRV (0.83, 0.83, and 0.87), the resulting CSFSRV had lower accuracy than the FSRV model in the testing and validation (0.92 and 0.91) and same accuracy in the training (0.91). The detailed accuracy, sensitivity, specificity, PPV, and NPV are summarized in Table 5.
Table 5.
The detailed diagnosis values of models on three datasets
| Dataset | Model | Accuracy (95% CI) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| Training | Clinical | 0.63 (0.58–0.68) | 0.31 | 0.84 | 0.56 | 0.65 |
| Semantic | 0.79 (0.74–0.83) | 0.79 | 0.79 | 0.71 | 0.85 | |
| Volume | 0.66 (0.61–0.71) | 0.61 | 0.69 | 0.56 | 0.73 | |
| FS | 0.90 (0.86–0.93) | 0.91 | 0.89 | 0.84 | 0.94 | |
| Radiomics | 0.82 (0.78–0.86) | 0.79 | 0.85 | 0.77 | 0.86 | |
| RV | 0.83 (0.79–0.87) | 0.79 | 0.86 | 0.78 | 0.86 | |
| CSRV | 0.83 (0.78–0.87) | 0.80 | 0.85 | 0.77 | 0.87 | |
| FSV | 0.89 (0.85–0.92) | 0.89 | 0.89 | 0.84 | 0.93 | |
| FSRV | 0.91 (0.88–0.94) | 0.90 | 0.92 | 0.88 | 0.93 | |
| CSFSRV | 0.91 (0.87–0.94) | 0.89 | 0.92 | 0.88 | 0.93 | |
| Testing | Clinical | 0.63 (0.54–0.71) | 0.23 | 0.90 | 0.59 | 0.64 |
| Semantic | 0.78 (0.71–0.85) | 0.68 | 0.85 | 0.75 | 0.80 | |
| Volume | 0.69 (0.60–0.76) | 0.61 | 0.73 | 0.60 | 0.74 | |
| FS | 0.92 (0.87–0.96) | 0.95 | 0.91 | 0.87 | 0.96 | |
| Radiomics | 0.79 (0.71–0.85) | 0.70 | 0.85 | 0.75 | 0.81 | |
| RV | 0.80 (0.73–0.87) | 0.74 | 0.85 | 0.76 | 0.83 | |
| CSRV | 0.83 (0.75–0.88) | 0.77 | 0.86 | 0.79 | 0.85 | |
| FSV | 0.92 (0.87–0.96) | 0.95 | 0.91 | 0.87 | 0.96 | |
| FSRV | 0.94 (0.88–0.97) | 0.86 | 0.99 | 0.98 | 0.91 | |
| CSFSRV | 0.92 (0.87–0.96) | 0.84 | 0.98 | 0.96 | 0.90 | |
| Validation | Clinical | 0.70 (0.62–0.77) | 0.18 | 0.91 | 0.47 | 0.73 |
| Semantic | 0.79 (0.72–0.85) | 0.68 | 0.84 | 0.64 | 0.86 | |
| Volume | 0.79 (0.72–0.85) | 0.66 | 0.85 | 0.64 | 0.86 | |
| FS | 0.90 (0.84–0.94) | 0.95 | 0.88 | 0.76 | 0.98 | |
| Radiomics | 0.85 (0.78–0.90) | 0.61 | 0.94 | 0.82 | 0.85 | |
| RV | 0.87 (0.81–0.92) | 0.68 | 0.95 | 0.86 | 0.88 | |
| CSRV | 0.87 (0.81–0.92) | 0.73 | 0.93 | 0.82 | 0.89 | |
| FSV | 0.91 (0.85–0.95) | 0.95 | 0.89 | 0.78 | 0.98 | |
| FSRV | 0.93 (0.87–0.96) | 0.93 | 0.92 | 0.84 | 0.97 | |
| CSFSRV | 0.91 (0.86–0.95) | 0.89 | 0.92 | 0.83 | 0.95 |
FS, frozen section; RV, radiomics combining with volume; CSRV, radiomics combining with clinical, semantic, and volume; FSV, frozen section combining with volume; FSRV, frozen section combining with radiomics and volume; CSFSRV, radiomics combining with clinical, semantic, volume, and frozen section; PPV, positive predictive values; NPV, negative predictive values; CI, confidence interval
Calibration, decision curve analysis, RQS, and TRIPOD
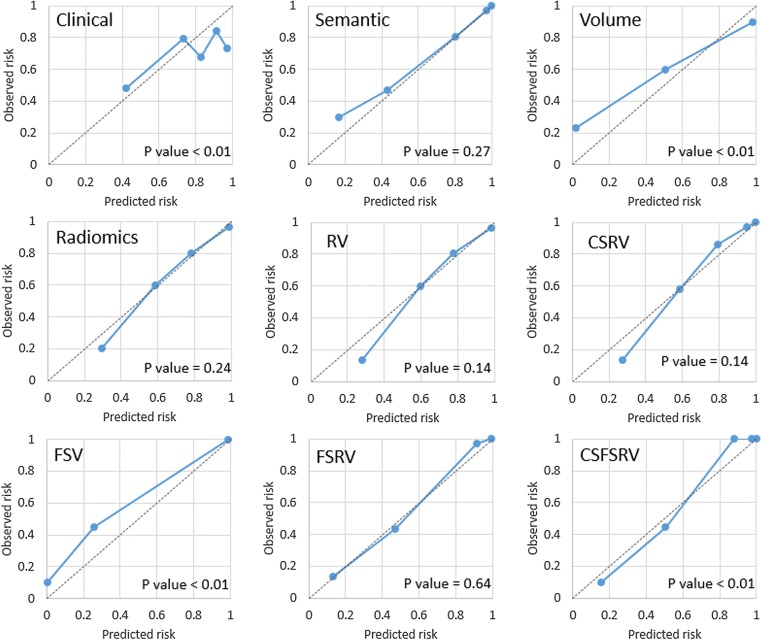
Among all models, the semantic, radiomics, RV, and FSRV models showed good calibrations with p values of 0.27, 0.24, 0.14, and 0.64, respectively, in the validation dataset (calibration plots depicted in Fig. 4). From the decision curve analysis on the validation dataset, we found that the clinical and volume models alone perform worse than semantic, radiomics, RV, and CSRV models. Models combining FS with other modalities (i.e., FSV, FSRV, and CSFSRV) perform the best. However, it seems that there is no obvious difference between the FSV, FSRV, and CSFSRV models (Fig. 5). The RQS points and total score of this study were 17 and 47.2%, respectively. We concluded this signature could be classified as TRIPOD 3 (Supplementary S6).
Fig. 4.
The calibration plots of the single and complex models on the validation dataset
Fig. 5.
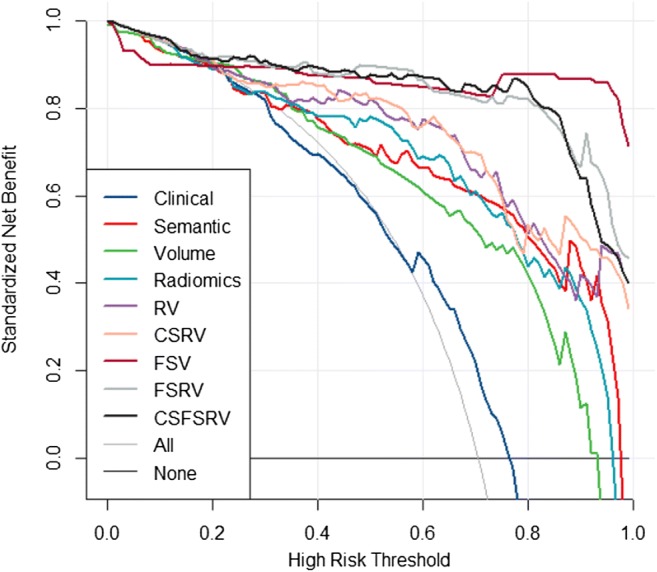
The decision curve of models performed on the validation dataset
Discussion
In this multicenter study, multiple univariate and multivariate binary classification models have been built to distinguish IA from PM using combinations of radiomics features as well as clinical features, semantic features, volume, and frozen section results. A method to select quantitative imaging features that are robust to spurious distributions of patient subgroups within the cohort as well as being mostly independent of the ROI volume is presented. The performance of the radiomics classifier was compared with models informed by clinical or semantic features, volume, and frozen section. The performance of the multifactorial FSRV diagnostic model was also compared with FS, RV, FSV, and CSFSRV models. Our results show that a multifactorial model based on radiomics features combined with FS and volume had excellent classification performance and diagnostic accuracy, suggesting that it can potentially be employed to gauge the risk of invasiveness in malignant pulmonary nodules.
Smoking is one of the most important risk factors for developing lung cancer, which is strongly correlated with the number of years and the amount of tobacco smoked [26]. Moreover, a population-based prospective study indicated that the risk factor for developing lung cancer increases with age and with a family history of lung cancer for female patients [27]. However, in this study, only age and gender significantly differ between cohorts diagnosed with IA and PM, with males older than 60 years having a significantly higher probability to be diagnosed with IA. Age has been reported elsewhere to increase the risk factor of IA diagnosis, while gender differences in the adenocarcinoma spectrum need further study [8–10]. Our results also show that a model informed purely on clinical variables has low sensitivity and relatively high specificity for the identification of IA, which may lead to moderate accuracy for diagnosis and low benefit from decision curve. This result, however, should be interpreted with caution, because clinical variables are varied in different populations.
Another study also looked at semantic features, proposing that pulmonary nodules with a larger diameter, located in the upper lobe, spiculation, and PSN (part-solid nodule) had a higher probability to be malignant [27]. However, it has been shown that semi-automated volume analysis is a more robust method than a simple measurement of the diameter to measure the size of the pulmonary nodule [28], and spiculation is an uncommon feature in early-stage lung cancer [8]. Our study finds that nodule diameter and nodule type are significantly different between cohorts diagnosed with IA and PM, with nodules with smaller diameter and pure GGN types increasing the probability of PM diagnosis. These two semantic features by themselves, as well as the semantic model, show high AUC and accuracy values for prediction and diagnosis of IA. Overall, our results indicate both a semantic feature model and a lesion volume model show similar predictive performance compared with radiomics, while radiomics has higher accuracy than semantic and volume models.
It is important also to point out that the ground truth used for diagnosis in this study is fairly unique as resections are not generally considered for pGGNs in guidelines in most countries outside of Asia where pGGNs are followed up until a solid component appears or the tumor progresses [29]. Moreover, pGGN adenocarcinomas are more common in low-risk Asian females than other populations, and the patients more often request surgery. Around 34% of nodules in this study are pGGNs, 30% of which are confirmed as IA, which may reflect doctors’ and patients’ more positive attitudes towards surgery.
In our study, the CT-based radiomics model shows a similar predictive performance with FS in distinguishing IA from PM. Selected features (Wavelet_HLL_Stats_max, Wavelet_LLL_Stats_cov, and LocInt_peakLocal) reflect the distribution of intensity values within the ROI, and another selected feature (GLRLM_LGRE) describes the heterogeneity of the density within the ROI [23]. Lim et al found that the mean density differs between IA and non- or minimally IA [8]. Moreover, a previous study reported that IA tends to appear more heterogeneous on CT images than PM [30]. Therefore, we hypothesize that radiomics features describing density and heterogeneity are related to tumor biology and pathology and are an excellent predictor for identification of IA [25].
CT and positron emission tomography radiomics studies have shown predictive features could be a surrogate of lesion volume and knowledge of which features correlate highly with volume is therefore important [31–33]. Upon volume correlation analysis, we excluded one feature that correlated highly with volume and found no change in model performance. The volume was embedded into the radiomics signature since radiomics is synonymous with quantitative imaging; features that contribute to model performance should not be excluded a priori. In this study, a radiomics plus volume model (RV) showed slight improvement of accuracy compared with the radiomics-alone model, and it had similar AUC and accuracy values as the CSRV model. In addition, we found that our models employing radiomics (i.e., radiomics alone, RV, and CSRV) had similar predictive performance (AUC) as the frozen section models. However, the accuracy of these models was lower than that of FS.
Although the FS can be a precise diagnostic method to guide intraoperative resection procedures for lung adenocarcinoma, it remains difficult to recommend a definitive assessment by FS alone [34]. Borczuk suggested that combining clinical and radiologic information with FS could reduce diagnostic errors [35]. Our results show no significant difference in the AUC values between the FSRV and FSV models, but the former model has better accuracy and calibration. Furthermore, we found that the AUC of the CSFSRV model is not significantly different from that of the FSRV model, did not increase the accuracy, and got bad calibration. In addition, the decision curve indicates that the models containing FS all had better performance than the models without FS. Therefore, we conclude that the addition of radiomics (with volume) to FS analysis potentially creates a substantial biomarker for assessing the risk of invasive adenocarcinomas and could be applied in clinical practice.
Nevertheless, this study has certain limitations. First, because of the retrospective data collection, selection bias is unavoidable. Further prospectively international investigation as a registered clinical trial is paramount. Second, different population cohorts, tumor morphology, and CT parameters are known to influence the results of radiomics features [36]. Further external validation datasets are desired to verify the reliability of our model, especially including diverse cohorts to fully capture phenotype heterogeneity. Third, the ROIs were contoured manually, which is time-consuming and highly prone to error. Therefore, a reliable and robust automatic segmentation tool is necessary to address this issue [37], also taking into account, e.g., peritumoral and normal tissue, to increase the accuracy of quantitative image-based models. Fourth, the accuracy and specificity of the FS analysis in our cohort were lower than the results from previous studies [6, 11]. We speculate that we included more small size and GGN cases, which have lower accuracy than larger tumors as most studies found [6, 11, 12]. Future prospects include prospective validation and deep learning methods for automatic segmentation and in combination with the ones described in this study, novel parametric imaging techniques. While this work focuses on the correlation of radiomics features with the underlying biology (histology), future work will also focus on the prediction of clinical outcomes directly, such as overall survival, progression free survival, or response to therapy.
In conclusion, a radiomics signature can be employed as a preoperative tool to distinguish invasive adenocarcinoma from preinvasive lesions or MIA. Furthermore, a multifactorial model combining radiomics with FS analysis is a potential biomarker for assessing the risk of invasive adenocarcinoma during surgery, and this model could help the therapeutic strategy for patients with pulmonary nodules.
Electronic supplementary material
(DOCX 454 kb)
Abbreviations
- AAH
Atypical adenomatous hyperplasia
- AIS
Adenocarcinoma in situ
- AUC
Area under the curve
- CI
Confidence interval
- CT
Computed tomography
- FS
Frozen section
- GGN
Ground-glass nodule
- IA
Invasive adenocarcinoma
- ICC
Intra-/inter-class correlation coefficients
- MIA
Minimally invasive adenocarcinoma
- NPV
Negative predictive values
- pGGN
Pure ground-glass nodule
- PM
Preinvasive lesions or minimally invasive adenocarcinomas
- PPV
Positive predictive values
- PSN
Part-solid nodule
- ROC
Receiver operating characteristic
- ROI
Regions of interest
- ROV
Radiomics plus volume model
- TRIPOD
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis
Funding information
This study was financially supported by the program of China Scholarships Council (n° 201808210318), ERC advanced grant (ERC-ADG-2015, n° 694812 - Hypoximmuno), and ERC-2018-PoC (n° 81320 – CL-IO). This research is also supported by the Dutch technology Foundation STW (grant n° P14-19 Radiomics STRaTegy), which is the applied science division of NWO, and the Technology Programme of the Ministry of Economic Affairs. This is also financially supported by the SME Phase 2 (RAIL - n°673780), EUROSTARS (DART, DECIDE, COMPACT), the European Program H2020-2015-17 (ImmunoSABR - n° 733008, PREDICT - ITN - n° 766276), TRANSCAN Joint Transnational Call 2016 (JTC2016 “CLEARLY”- n° UM 2017-8295), Interreg V-A Euregio Meuse-Rhine (“Euradiomics”), and Kankeronderzoekfonds Limburg (KOFL) from the Health Foundation Limburg and the Dutch Cancer Society.
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Philippe Lambin.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Two of the authors have significant statistical expertise.
Informed consent
The requirement for informed consent was waived and patient data are anonymized.
Ethical approval
The institutional review boards approved this retrospective study registered in http://clinicaltrials.gov (identifier: NCT03872362).
Methodology
• Retrospective
• Diagnostic or prognostic study
• Multicenter study
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangyao Wu, Email: g.wu@maastrichtuniversity.nl.
Jianlin Wu, Email: cjr.wujianlin@vip.163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.National Lung Screening Trial Research Team. Church TR, Black WC, et al. Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med. 2013;368:1980–1991. doi: 10.1056/NEJMoa1209120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maldonado F, Boland JM, Raghunath S, et al. Noninvasive characterization of the histopathologic features of pulmonary nodules of the lung adenocarcinoma spectrum using computer-aided nodule assessment and risk yield (CANARY)--a pilot study. J Thorac Oncol. 2013;8:452–460. doi: 10.1097/JTO.0b013e3182843721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Wang R, Zhang Y, et al. Precise diagnosis of intraoperative frozen section is an effective method to guide resection strategy for peripheral small-sized lung adenocarcinoma. J Clin Oncol. 2016;34:307–313. doi: 10.1200/JCO.2015.63.4907. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura H, Saji H, Shinmyo T, et al. Close association of IASLC/ATS/ERS lung adenocarcinoma subtypes with glucose-uptake inpositron emission tomography. Lung Cancer. 2015;87:28–33. doi: 10.1016/j.lungcan.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Lim HJ, Ahn S, Lee KS, et al. Persistent pure ground-glass opacity lung nodules /=10 mm in diameter at CT scan: histopathologic comparisons and prognostic implications. Chest. 2013;144:1291–1299. doi: 10.1378/chest.12-2987. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Goo JM, Lee CH, et al. Predictive CT findings of malignancy in ground-glass nodules on thin-section chest CT: the effects on radiologist performance. Eur Radiol. 2009;19:552–560. doi: 10.1007/s00330-008-1188-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee KH, Goo JM, Park SJ, et al. Correlation between the size of the solid component on thin-section CT and the invasive component on pathology in small lung adenocarcinomas manifesting as ground-glass nodules. J Thorac Oncol. 2014;9:74–82. doi: 10.1097/JTO.0000000000000019. [DOI] [PubMed] [Google Scholar]
- 11.Zhu E, Xie H, Dai C, et al. Intraoperatively measured tumor size and frozen section results should be considered jointly to predict the final pathology for lung adenocarcinoma. Mod Pathol. 2018;31:1391–1399. doi: 10.1038/s41379-018-0056-0. [DOI] [PubMed] [Google Scholar]
- 12.Yeh YC, Nitadori J, Kadota K, et al. Using frozen section to identify histological patterns in stage I lung adenocarcinoma of ≤3 cm: accuracy and interobserver agreement. Histopathology. 2015;66:922–938. doi: 10.1111/his.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Group of Respiration Diseases, Chinese Society of Pathology Consensus on early stage non-mucinous lepidic lung adenocarcinoma frozen section diagnosis. Zhonghua Bing Li Xue Za Zhi. 2019;48:3–10. doi: 10.3760/cma.j.issn.0529-5807.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan L, Fang M, Li Z, et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol. 2019;29:889–897. doi: 10.1007/s00330-018-5530-z. [DOI] [PubMed] [Google Scholar]
- 17.She Y, Zhang L, Zhu H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol. 2018;28:5121–5128. doi: 10.1007/s00330-018-5509-9. [DOI] [PubMed] [Google Scholar]
- 18.Chae HD, Park CM, Park SJ, Lee SM, Kim KG, Goo JM. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology. 2014;273:285–293. doi: 10.1148/radiol.14132187. [DOI] [PubMed] [Google Scholar]
- 19.Lee SM, Park CM, Goo JM, Lee HJ, Wi JY, Kang CH. Invasive pulmonary adenocarcinomas versus preinvasive lesions appearing as ground-glass nodules: differentiation by using CT features. Radiology. 2013;268:265–273. doi: 10.1148/radiol.13120949. [DOI] [PubMed] [Google Scholar]
- 20.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246:697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 21.Bankier AA, MacMahon H, Goo JM, Rubin GD, Schaefer-Prokop CM, Naidich DP. Recommendations for measuring pulmonary nodules at CT: a statement from the Fleischner Society. Radiology. 2017;285:584–600. doi: 10.1148/radiol.2017162894. [DOI] [PubMed] [Google Scholar]
- 22.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 24.Moons KG, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 25.Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol. 2018;127:349–360. doi: 10.1016/j.radonc.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Powell HA, Iyen-Omofoman B, Hubbard RB, Baldwin DR, Tata LJ. The association between smoking quantity and lung cancer in men and women. Chest. 2013;143:123–129. doi: 10.1378/chest.12-1068. [DOI] [PubMed] [Google Scholar]
- 27.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369:910–919. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heuvelmans MA, Walter JE, Vliegenthart R, et al. Disagreement of diameter and volume measurements for pulmonary nodule size estimation in CT lung cancer screening. Thorax. 2018;73:779–781. doi: 10.1136/thoraxjnl-2017-210770. [DOI] [PubMed] [Google Scholar]
- 29.Bai C, Choi CM, Chu CM, et al. Evaluation of pulmonary nodules: Clinical Practice Consensus Guidelines for Asia. Chest. 2016;150:877–893. doi: 10.1016/j.chest.2016.02.650. [DOI] [PubMed] [Google Scholar]
- 30.Ost DE, Gould MK. Decision making in patients with pulmonary nodules. Am J Respir Crit Care Med. 2012;185:363–372. doi: 10.1164/rccm.201104-0679CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 32.Orlhac F, Soussan M, Maisonobe JA, Garcia CA, Vanderlinden B, Buvat I. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med. 2014;55:414–422. doi: 10.2967/jnumed.113.129858. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim A, Vallières M, Woodruff H, et al. Radiomics analysis for clinical decision support in nuclear medicine. Semin Nucl Med. 2019;49:438–449. doi: 10.1053/j.semnuclmed.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Chen D, Dai C, Kadeer X, Mao R, Chen Y, Chen C. New horizons in surgical treatment of ground-glass nodules of the lung: experience and controversies. Ther Clin Risk Manag. 2018;14:203–211. doi: 10.2147/TCRM.S152127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borczuk AC. Challenges of frozen section in thoracic pathology: lepidic lesions, limited resections, and margins. Arch Pathol Lab Med. 2017;141:932–939. doi: 10.5858/arpa.2016-0415-RA. [DOI] [PubMed] [Google Scholar]
- 36.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 37.Charbonnier JP, Chung K, Scholten ET, et al. Automatic segmentation of the solid core and enclosed vessels in subsolid pulmonary nodules. Sci Rep. 2018;8:646. doi: 10.1038/s41598-017-19101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 454 kb)