Abstract
Background: Higher tumor expression of CD44, a marker of cancer stem cells (CSCs), is associated with poor overall survival (OS) in various cancers. However, the association between CD44 and poor OS remains inconsistent in glioma. We aimed to evaluate the potential predictive role of CD44 for prognosis of glioma patients in a meta-analysis.
Methods: Observational studies comparing OS of glioma patients according to the level of CD44 were identified through searching PubMed, Embase, and Cochrane’s Library databases. Meta-analyses were performed with a random- or fixed-effect model according to the heterogeneity. Subgroup analyses were performed to evaluate the influences of study characteristics.
Results: Eleven retrospective cohort studies were included. Results showed that increased CD44 expression in tumor predicted poor OS in glioma patients (hazard ratio [HR]: 1.42, 95% confidence interval [CI]: 1.02–1.97, P=0.04). Subgroup analyses showed that higher tumor CD44 expression significantly predicted poor OS in patients with World Health Organization (WHO) stages II–III glioma (HR: 2.99, 95% CI: 1.53–5.89, P=0.002), but not in patients with glioblastoma (HR: 1.26, 95% CI: 0.76–2.08, P=0.47; P for subgroup difference = 0.03). Results were not statistically different between subgroups according to patient ethnicity, sample size, CD44 detection method, CD44 cutoff, HR estimation, univariate or multivariate analysis, or median follow-up durations (P-values for subgroup difference all >0.10).
Conclusion: Higher tumor expression of CD44 may predict poor survival in patients with glioma, particularly in those with WHO stage II–III glioma.
Keywords: CD44, Glioblastoma, Glioma, Meta-analysis, Survival
Introduction
Glioma is a common malignant tumor of the central nervous system. According to previous studies, glioma accounts for 50–80% of brain tumors [1]. Currently, glioma can be staged to I–IV in accordance with the World Health Organization (WHO) 2016 classification, and higher stages indicate increased malignancies [2,3]. Among which, the WHO stage IV glioma, also known as glioblastoma, is the most malignant glioma. The median survival of patients with glioblastoma is <15 months [4,5]. Clinically, the WHO stage system is commonly used for prognostic prediction in glioma patients. However, it has been suggested that prognostic prediction in glioma patients may be complicated, and the prognosis in patients with same WHO stage glioma can vary dramatically [6,7]. Therefore, uncovering novel prognostic factors remains important for improving the quality of care for glioma patients.
Cancer stem cells (CSCs) refer to a small proportion of cancer cells which have the capacities of self-renewal and multidifferentiation [8,9]. Current evidence indicates that CSCs play important roles in tumor progression, metastasis, recurrence, resistance to chemotherapy or radiotherapy, and an overall poor prognosis [10,11]. Previous studies showed that CSCs are characterized by higher expressions of certain molecules, which are collectively termed as CSC marker molecules [12,13]. Increasing evidence indicates that CD44 family is one of the most common CSC markers. Pathophysiologically, CD44 is involved in the processes of cell growth, survival, differentiation, motility, tumor growth, proliferation, and metastasis [14]. Moreover, higher expression of tumor CD44 has been associated with poor prognosis in patients with various cancers, such as non-small cell lung cancer [15], gastric cancer [16], hepatocellular carcinoma [17], colorectal cancer [18], renal cell carcinoma [19], breast cancer [20], ovarian cancer [21], head and neck cancer [22], and osteosarcoma [23]. However, the association between higher expression of CD44 and the overall survival (OS) in patients with glioma showed inconsistent results [24]. Some studies demonstrated that higher tumor expression of CD44 was associated with poor OS in patients with glioma [25–27], while others did not show a significant association [28–34]. Moreover, one of the studies even showed that higher tumor expression of CD44 may be associated with improved OS in glioma patients [35]. Since the sample sizes of such studies are limited, they may be too underpowered to show a significant association between tumor CD44 levels and the prognosis in glioma patients. Therefore, we performed a meta-analysis to quantitatively summarize the predictive power of tumor CD44 expression for OS in patients with glioma.
Methods
This meta-analysis was designed, performed, and reported in accordance with the Meta-analysis of Observational Studies in Epidemiology [36] and Cochrane’s Handbook [37] guidelines.
Literature search
The PubMed, Embase, and the Cochrane’s Library electronic databases were queried for relevant studies using the terms ‘CD44’, combined with ‘glioma’, ‘glioblastoma’, ‘glial cell tumor’, or ‘astrocytoma’ from inception to 26 December 2019. The search was limited to human studies published in English or Chinese. The references of the original and review articles were also manually analyzed for possible studies.
Inclusion and exclusion criteria
Studies were included if the following criteria were fulfilled: (1) full-length article in English or Chinese; (2) included patients with histopathologically diagnosed glioma; (3) observed the association between tumor CD44 expression and OS of the patients;reported hazard ratios (HRs) and 95% confidence intervals (CIs) for poor OS during a follow-up in patients with higher vs. lower CD44 level in tumor, or the data were efficient to estimate the HR and 95% CIs from the survival analysis. Reviews, preclinical studies, and duplicate reports were excluded.
Data extraction and quality assessment
Database search, data extraction, and quality evaluation were independently performed by two authors. Discrepancies were resolved by discussion with the corresponding author. Data containing the study characteristics (name of the first author, study design, publication year, and location), patient characteristics (age, gender of the patients, and WHO grade of glioma), CD44 measurements (methods and cut-off values), characteristics of survival analysis (HR reported in the context or estimated via the survival curve, univariate or multivariate analyses), median follow-up durations, and variables adjusted for when presenting the results were extracted. Some of the published articles did not provide HR and 95% CI directly. In such cases, two reviewers independently digitized and extracted the data through the Kaplan–Meier survival curves using GetData Graph Digitizer 2.24 (http://getdata-graph-digitizer.com), and then reconstructed the HR and its variance (GraphPad Software, Inc.). We assessed the quality of the included studies using the Newcastle–Ottawa Scale [38], which judges each study according to three domains: selection of the study groups; the comparability of the groups, and the ascertainment of the primary outcome.
Statistical analyses
Meta-analyses were performed to summarize the association between tumor miR-210 expression and the risk of poor OS outcome. HR and the corresponding standard errors (SEs) were estimated from 95% CIs or P-values, and were logarithmically transformed to obtain a normal distribution [37]. The Cochrane’s Q test and I2 test were performed to evaluate the heterogeneity [39]; an I2 > 50% indicates significant heterogeneity. A random-effect model was applied if significant heterogeneity was detected, otherwise, a fixed-effect model was applied [37]. A predefined subgroup analysis was performed to evaluate the influence of study characteristics on the outcome, including patient ethnicity, sample size, WHO grade, CD44 detection method, CD44 cutoff, HR estimation, univariate or multivariate analysis, and median follow-up durations. Medians were used as cutoffs for grouping according to continuous variables. Potential publication bias was assessed by the visual examination of funnel plots for symmetry, as well as the Egger regression asymmetry test [40]. The RevMan (Version 5.1; Cochrane Collaboration, Oxford, U.K.) and STATA software (Version 12.0; Stata Corporation, College Station, TX) were used for the statistical analyses.
Results
Results of literature search
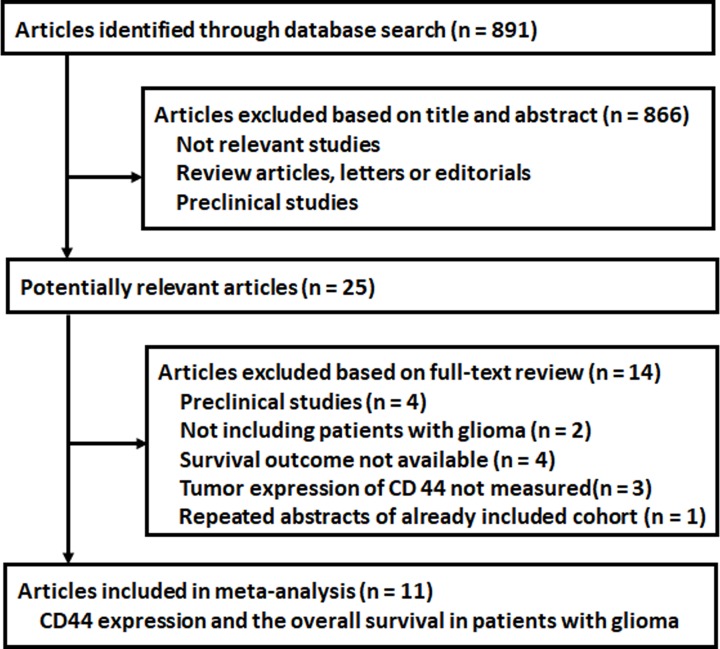
As shown in Figure 1, 891 studies were initially identified by the database search, and 866 were subsequently excluded based on the content of the titles and/or abstracts, mainly because they were not relevant to the purpose of this meta-analysis. The remaining 25 studies underwent full-text review. Of them, 14 studies were further excluded; four were preclinical studies, two did not include patients with glioma, four did not report the outcome of OS, three did not measure tumor expression of CD44, and one was a repeated report of an included study. Finally, 11 studies [25–35] were included for subsequent meta-analysis.
Figure 1. Flowchart of the database search and study identification.
Study characteristics and quality evaluation
The characteristics of the included studies are displayed in Table 1. Since one study reported datasets in patients with stage II–III glioma and glioblastoma independently, we included these datasets separately [26]. Overall, this meta-analysis included 967 patients with glioma from 11 retrospective cohort studies (12 datasets) performed in China [27,35], Japan [25,26], Russia [32], Spain [34], Germany [33], Sweden [29], Italy [30], France [31], and Argentina [28]. All the studies were published in English. The amount of included patients varied from 13 to 280, with mean patient ages varying from 52 to 67 years. Eight datasets included patients with glioma [25,26,30–35], while two datasets included patients with WHO grades II–III glioma [26,27], and the remaining two included patients with WHO grades II–IV glioma [28,29]. For seven studies, glioma tissue CD44 expression was measured by immunohistochemistry [28–32,34,35], while for the rest studies, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was applied [25–27,33]. The median of CD44 expression values were used as cutoff for patient grouping in seven studies [25–27,31–33,35], while for the remaining four studies, CD44 expression of >70–75% was used as the cut-off value [28–30,34]. Data of HRs and 95% CI were reported in seven studies [26,27,31–35] and were estimated from survival curves in four studies [25,28–30]. Multivariate analyses adjusted for age, gender, and treatments were applied in six studies when reporting the association between CD44 level and OS [26,27,31–34], while the other five studies used univariate analyses [25,28–30,35]. The median follow-up durations varied from 15 to 40 months. The Newcastle–Ottawa scale varied from 5 to 8 in the included studies (Table 2), indicating moderate overall quality.
Table 1. Characteristics of the included studies.
| Study | Country | Study design | Sample size | Mean age | Male | WHO grade | CD44 detection method | CD44 cutoff | HR estimation | Survival analysis | Median follow-up duration | Variables adjusted |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| years | % | Months | ||||||||||
| Rauuncolo, 2002 | Argentina | RC | 84 | 52.1 | 61.9 | II–IV | IHC | >70% | Survival curve | Univariate | 40 | NA |
| Wei, 2010 | China | RC | 42 | NR | 78.6 | IV | IHC | Median | Reported | Univariate | 18 | NA |
| Sooman, 2014 | Sweden | RC | 97 | 59.5 | 56.7 | III–IV | IHC | >75% | Survival curve | Univariate | 15 | NA |
| Guadagno, 2016 | Italy | RC | 25 | 60.3 | 72 | IV | IHC | >70% | Survival curve | Univariate | 30 | NA |
| Pinel, 2017 | France | RC | 122 | 60.3 | 66.3 | IV | IHC | Median | Reported | Multivariate | 16 | Age, gender, and treatments |
| Tsidulko, 2017 | Russia | RC | 74 | 52.0 | 52 | IV | IHC | Median | Reported | Multivariate | 20 | Age, gender, Ki67, and treatments |
| Nishikawa, 2018 | Japan | RC | 13 | NR | NR | IV | qRT-PCR | Median | Survival curve | Univariate | 18 | NA |
| Bien-Möller, 2018 | Germany | RC | 77 | 67.0 | 64.9 | IV | qRT-PCR | Median | Reported | Multivariate | 26 | Age, gender, and treatments |
| Alameda, 2019 | Spain | RC | 280 | 59.0 | 58.6 | IV | IHC | >70% | Reported | Multivariate | 16 | Age, gender, KPS, and treatments |
| Hou, 2019 | Japan | RC | 112 | NR | 51.8 | II–III | qRT-PCR | Median | Reported | Multivariate | 32 | Age, gender, KPS, IDH, and treatments |
| Dong, 2019a | China | RC | 16 | NR | 42.7 | II–III | qRT-PCR | Median | Reported | Multivariate | 36 | Age, gender, and KPS |
| Dong, 2019b | China | RC | 38 | NR | 61.1 | IV | qRT-PCR | Median | Reported | Multivariate | 36 | Age, gender, and KPS |
The study Dong 2019 included stratified datasets in patients with WHO grade II–III glioma and glioblastoma, which were included in the meta-analysis separately.
Abbreviations: IDH, isocitrate dehydrogenase; IHC, immunohistochemistry; KPS, Karnofsky Performance Scale; NA, not applicable; NR, not reported; RC, retrospective cohort.
Table 2. Details of quality evaluation by the Newcastle–Ottawa Scale.
| Studies | Representativeness of the exposed cohort | Selection of the non-exposed cohort | Ascertainment of exposure | Outcome of interest not present at baseline | Adjustment of age and gender | Adjustment of other confounding factors | Assessment of outcome | Follow-up long enough | Adequacy of follow-up of cohorts | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Rauuncolo, 2002 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 6 |
| Wei, 2010 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 5 |
| Sooman, 2014 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 6 |
| Guadagno, 2016 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 6 |
| Pinel, 2017 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Tsidulko, 2017 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Nishikawa, 2018 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 5 |
| Bien-Möller, 2018 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Alameda, 2019 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Hou, 2019 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Dong, 2019a | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| Dong, 2019b | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
The study Dong 2019 included stratified datasets in patients with WHO grade II–III glioma and glioblastoma, which were included in the meta-analysis separately.
Expression of CD44 and OS in glioma patients
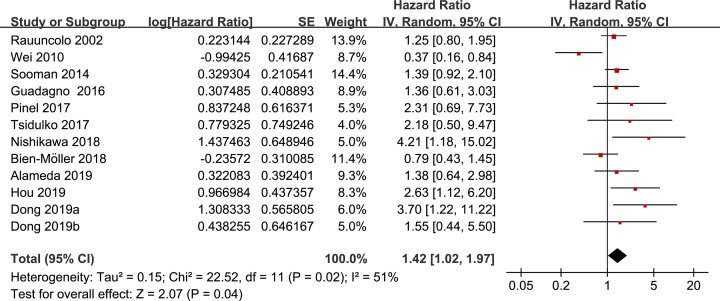
The pooled results of the 12 datasets from the 11 studies with a random-effect model showed that higher tumor CD44 expression significantly predicted the poor OS in patients with glioma during follow-up (HR: 1.42, 95% CI: 1.02–1.97, P=0.04; Figure 2) with significant heterogeneity (P for Cochrane’s Q test = 0.02, I2 = 51%). Subgroup analyses showed that the association between tumor CD44 expression and poor OS may be different according to the WHO stages of the tumor. Specifically, higher tumor CD44 expression significantly predicted poor OS in patients with WHO stages II–III glioma (HR: 2.99, 95% CI: 1.53–5.89, P=0.002; I2=0%), but not in patients with glioblastoma (HR: 1.26, 95% CI: 0.76–2.08, P=0.47; I2=55%). The difference between the subgroups was significant (P=0.03; Table 3). Results were not statistically different between subgroups according to study characteristics such as patient ethnicity, sample size, CD44 detection method, CD44 cutoff, HR estimation, univariate or multivariate analysis, or median follow-up durations (P-values for subgroup difference all >0.10; Table 3).
Figure 2. Forest plots for the meta-analysis of the prognostic efficacy of tumor CD44 for OS in patients with glioma.
Table 3. Subgroup analyses.
| OS | |||||
|---|---|---|---|---|---|
| Study characteristics | Datasets number | HR (95% CI) | I2 | P for subgroup effect | P for subgroup difference |
| Ethnicity | |||||
| Non-Asian | 7 | 1.28 [1.01, 1.61] | 0% | 0.04 | |
| Asian | 5 | 1.80 [0.67, 4.81] | 78% | 0.24 | 0.50 |
| Sample size | |||||
| >80 | 5 | 1.45 [1.12, 1.89] | 0% | 0.005 | |
| ≤80 | 7 | 1.39 [0.73, 2.64] | 67% | 0.32 | 0.89 |
| WHO grade | |||||
| II–III | 2 | 2.99 [1.52, 5.89] | 0% | 0.002 | |
| IV | 8 | 1.26 [0.76, 2.08] | 55% | 0.37 | 0.03 |
| CD44 detection method | |||||
| IHC | 6 | 1.21 [0.75, 1.95] | 52% | 0.45 | |
| qRT-PCR | 6 | 1.72 [1.03, 2.89] | 57% | 0.04 | 0.32 |
| CD44 cutoff | |||||
| IHC | 8 | 1.63 [0.86, 3.08] | 69% | 0.14 | |
| qRT-PCR | 4 | 1.33 [1.02, 1.74] | 0% | 0.03 | 0.57 |
| HR estimation | |||||
| Survival curve | 4 | 1.41 [1.06, 1.87] | 4% | 0.02 | |
| Reported | 8 | 1.41 [0.81, 2.46] | 63% | 0.22 | 0.99 |
| Survival analysis | |||||
| Multivariate | 7 | 1.68 [1.06, 2.64] | 35% | 0.03 | |
| Univariate | 5 | 1.20 [0.72, 2.01] | 67% | 0.49 | 0.34 |
| Median follow-up duration | |||||
| >20 months | 6 | 1.46 [0.97, 2.20] | 42% | 0.07 | |
| ≤20 months | 6 | 1.40 [0.77, 2.54] | 64% | 0.28 | 0.90 |
Publication bias
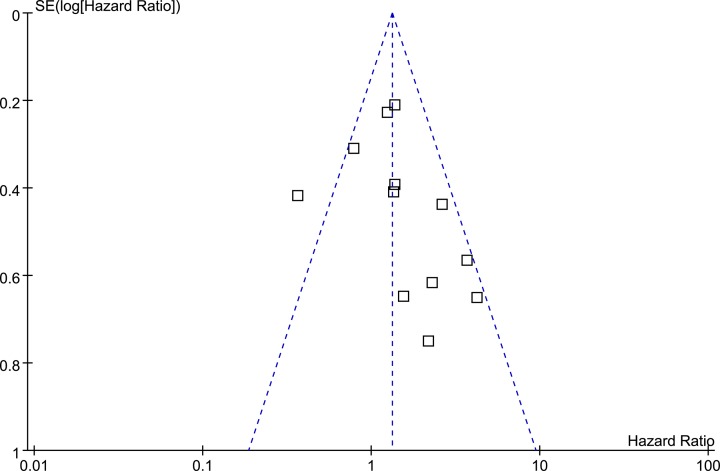
The funnel plots for the meta-analysis of the prognostic efficacy of tumor CD44 level for poor OS are shown in Figure 3. The funnel plots were determined to be symmetrical based on visual inspection, suggesting low risk of publication bias. The result of the Egger’s regression test for the meta-analysis also demonstrated similar result (P=0.37).
Figure 3. Funnel plots for the meta-analysis of the prognostic efficacy of tumor CD44 for OS in patients with glioma.
Discussion
In the present study, by pooling the results of available cohort studies, we found that higher tumor expression of CD44 is a prognostic factor for poor survival in patients with glioma. Subsequent subgroup analyses showed that disease stages may affect the potential prognostic efficacy of tumor CD44 expression for the survival in patients with glioma. Specifically, the predictive efficacy of higher CD44 tumor expression for poor OS may be significant in patients with WHO stage II–III glioma, but not for those with glioblastoma (WHO stage IV). In addition, the association between higher expression of CD44 and poor survival in patients with glioma does not seem to affect by study characteristics such as patient ethnicity, sample size, CD44 detection method, CD44 cutoff, HR estimation strategy, univariate or multivariate analysis, or median follow-up durations. Taken together, these results demonstrated that higher tumor expression of CD44 may predict poor survival in patients with glioma, particularly in those with WHO stage II–III glioma. These findings should be validated in large-scale prospective cohort studies, and the clinical importance of tumor CD44 for the risk stratification and treatment of glioma deserve further investigation.
To the best of our knowledge, this is the first meta-analysis to evaluate the predictive value of higher tumor CD44 expression for the prognosis in patients with glioma. The potential association between higher tumor expression of CD44 and overall poor prognosis in patients with glioma could be explained by the roles of CD44 in the proliferation, invasion, metastasis of glioma, and resistance to chemotherapy and radiation therapy [24]. CD44, as a cell membrane glycoprotein, exerts diverse cellular processes including cell motility, proliferation, apoptosis, and angiogenesis, via binding extracellular ligands, principally hyaluronic acid (HA). Increased expression of CD44 has been early confirmed in human glioma cells as compared with normal brain tissue, and suppression of CD44 expression decreases migration and invasion of human glioma cells [41]. In another study, depletion of CD44 was shown to block glioblastoma growth and sensitizes glioblastoma cells to cytotoxic drugs in vivo, and CD44 functions upstream of the mammalian Hippo signaling pathway in glioblastoma cells [42]. Moreover, tumor expression of CD44 is shown to be up-regulated during glioma progression in the brain, which may aid in tracing and targeting the invading glioma cells [43]. Importantly, a subsequent study showed that CD44 transduces HA-based stiffness cues, temporally precedes integrin-based adhesion maturation, and facilitates invasion, demonstrating that the HA-CD44 axis is critical for cell adhesion, invasion, and mechanosensing of an HA-based matrix in glioma [44]. Stemness of cancer cells has been associated with poor survival [45]. It was shown that CD44 modulates the hypoxic response of glioma cells and that the pseudo-hypoxic phenotype of stem-like glioma cells is achieved by stabilization of hypoxia-inducible factors-2α through interaction with CD44, thereby contributing to the stemness of glioma cells [46]. These studies highlight the multiple roles of CD44 in the pathogenesis and invasion of glioma, and the potential therapeutic significance of CD44 depletion for patients with glioma. Further studies are needed to determine the key signaling pathways underlying the role of CD44 in glioma, which may be helpful to develop the CD44-based target therapy for patients with glioma.
Our subgroup analysis showed that the predictive efficacy of higher CD44 tumor expression for poor OS may be significant in patients with WHO stage II–III glioma, but not for those with glioblastoma. The potential mechanisms underlying these findings remain unclear. However, from a clinical perspective, since the median survival in patients with WHO stage II–III glioma is generally longer than those with glioblastoma, introducing additional risk stratification factors besides WHO grade system may be more important in those with stage II–III glioma as compared with patients with glioblastoma. A previous study showed that the expression of CD44 in glioblastoma may be heterogeneous, and the tumorigenicity of primary GBM differs between CD44low/CD133high and CD44high/CD133low for gene expression profiles [47]. Subsequently, the molecular heterogeneity among tumors may affect the prognostic value of CD44 in glioblastoma [25]. The exact mechanisms underlying the potential difference between the prognostic power of higher tumor CD44 expression for survival in patients with WHO stage II–III glioma and in those with glioblastoma deserve further investigation.
The strengths of our study include extensive literature search, rigorous meta-analysis according to the MOOSE and Cochrane’s guidelines, and comprehensive subgroup analyses. However, our study has the following limitations which should be noted when interpreting the results. First, only 11 studies with 12 datasets were included. The number of available studies remains small. Therefore, results of subgroup analyses should be interpreted with caution. Large-scale cohort studies are needed to confirm our results. Second, only retrospective cohort studies were included in the meta-analysis, which may be confounded by limitations such as recall bias. Therefore, prospective studies are needed to validate these findings. Third, significant heterogeneity existed among the included studies. Although subgroup analyses showed that WHO stages of the diseases might contribute to the heterogeneity, results of subgroup analyses should be treated cautiously since the limited studies available for each stratum. Besides, other factors, such as comorbidities of the patients, cancer treatments, and cut-off value to determine the increased tumor CD44 expression may also contribute to the heterogeneity. However, since these factors were rarely reported in each study, we were unable to evaluate the potential prognostic role of tumor CD44 expression for survival according to these characteristics. Future studies are warranted. In addition, since the cut-off values for grouping patients with higher or lower tumor CD44 varied among the studies, it may lead to heterogeneity. The optimal tumor CD44 cut-off values for the prognosis prediction in patients with glioma should be determined in future studies. Finally, the potential therapeutic significance of CD44 as a treatment target for glioma also should be determined in the future.
In conclusion, results of meta-analysis showed that higher tumor expression of CD44 may predict poor survival in patients with glioma, particularly in those with WHO stage II–III glioma. These findings should be validated in large-scale prospective cohort studies, and the clinical role of CD44 for glioma deserves further investigation.
Abbreviations
- CI
confidence interval
- CSC
cancer stem cell
- HA
hyaluronic acid
- HR
hazard ratio
- OS
overall survival
- WHO
World Health Organization
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
Gang Wu and Yuhong Chen designed the study. Gang Wu and Xinghui Song performed literature search and data extraction. Wang Wu, Jun Liu, Shize Li, Weiqin Gao, Mingxing Qiu, Changjin Yang, and Yiming Ma performed statistical analyses and contributed to data interpretation. Gang Wu, Xinghui Song, and Yuhong Chen drafted the manuscript. All authors critically reviewed the manuscript and approved its submission.
References
- 1.Lapointe S., Perry A. and Butowski N.A. (2018) Primary brain tumours in adults. Lancet 392, 432–446 10.1016/S0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- 2.Bates A., Gonzalez-Viana E., Cruickshank G. and Roques T. (2018) Primary and metastatic brain tumours in adults: summary of NICE guidance. BMJ 362, k2924 10.1136/bmj.k2924 [DOI] [PubMed] [Google Scholar]
- 3.Brandner S. and Jaunmuktane Z. (2018) Neurological update: gliomas and other primary brain tumours in adults. J. Neurol. 265, 717–727 10.1007/s00415-017-8652-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A. and Dwivedi T. (2017) A Simplified Overview of World Health Organization Classification Update of Central Nervous System Tumors 2016. J. Neurosci. Rural Pract. 8, 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis D.N., Perry A., Reifenberger G. et al. (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 6.Quan G., Zheng Y., Chen J. et al. (2018) Prediction value of unmeasurable MR enhancement at early stage after gross-total resection on the survival state of patients with high-grade glioma. J. Neuro Oncol. 140, 359–366 10.1007/s11060-018-2961-y [DOI] [PubMed] [Google Scholar]
- 7.Wang Z.L., Zhang C.B., Liu Y.Q., Wang Z. and Jiang T. (2019) Peripheral blood test provides a practical method for glioma evaluation and prognosis prediction. CNS Neurosci.Ther. 25, 876–883 10.1111/cns.13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Shi P., Zhao G. et al. (2020) Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 5, 8 10.1038/s41392-020-0110-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budillon A., Curley S., Fusco R. and Mancini R. (2019) Identification and targeting of stem cell-activated pathways in cancer therapy. Stem Cells Int. 2019, 8549020 10.1155/2019/8549020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen L., de Sousa e Melo F., Richel D.J. and Medema J.P. (2012) The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 13, e83–89 10.1016/S1470-2045(11)70257-1 [DOI] [PubMed] [Google Scholar]
- 11.Najafi M., Mortezaee K. and Majidpoor J. (2019) Cancer stem cell (CSC) resistance drivers. Life Sci. 234, 116781 10.1016/j.lfs.2019.116781 [DOI] [PubMed] [Google Scholar]
- 12.Murar M. and Vaidya A. (2015) Cancer stem cell markers: premises and prospects. Biomark. Med. 9, 1331–1342 10.2217/bmm.15.85 [DOI] [PubMed] [Google Scholar]
- 13.Kim W.T. and Ryu C.J. (2017) Cancer stem cell surface markers on normal stem cells. BMB Rep. 50, 285–298 10.5483/BMBRep.2017.50.6.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skandalis S.S., Karalis T.T., Chatzopoulos A. and Karamanos N.K. (2019) Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell. Signal. 63, 109377 10.1016/j.cellsig.2019.109377 [DOI] [PubMed] [Google Scholar]
- 15.Zhao S., He J.L., Qiu Z.X. et al. (2014) Prognostic value of CD44 variant exon 6 expression in non-small cell lung cancer: a meta-analysis. Asian Pac. J. Cancer Prev. 15, 6761–6766 10.7314/APJCP.2014.15.16.6761 [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Fu Z., Xu S., Xu Y. and Xu P. (2014) The prognostic value of CD44 expression in gastric cancer: a meta-analysis. Biomed. Pharmacother. 68, 693–697 10.1016/j.biopha.2014.08.001 [DOI] [PubMed] [Google Scholar]
- 17.Luo Y. and Tan Y. (2016) Prognostic value of CD44 expression in patients with hepatocellular carcinoma: meta-analysis. Cancer Cell Int. 16, 47 10.1186/s12935-016-0325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z., Tang Y., Xie L. et al. (2019) The prognostic and clinical value of CD44 in colorectal cancer: a meta-analysis. Front. Oncol. 9, 309 10.3389/fonc.2019.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Ma X., Chen L. et al. (2015) Prognostic value of CD44 expression in renal cell carcinoma: a systematic review and meta-analysis. Sci. Rep. 5, 13157 10.1038/srep13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Wang Q., Wang Y. and Chen J. (2017) Prognostic significance of CD24 and CD44 in breast cancer: a meta-analysis. Int. J. Biol. Markers 32, e75–e82 10.5301/jbm.5000224 [DOI] [PubMed] [Google Scholar]
- 21.Lin J. and Ding D. (2017) The prognostic role of the cancer stem cell marker CD44 in ovarian cancer: a meta-analysis. Cancer Cell Int. 17, 8 10.1186/s12935-016-0376-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Zhou J., Lu J., Xiong H., Shi X. and Gong L. (2014) Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer 14, 15 10.1186/1471-2407-14-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Ding C., Wang J. et al. (2015) Prognostic significance of CD44V6 expression in osteosarcoma: a meta-analysis. J. Orthop. Surg. Res. 10, 187 10.1186/s13018-015-0328-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney K.L., Choy W., Sidhu S. et al. (2016) The role of CD44 in glioblastoma multiforme. J. Clin. Neurosci. 34, 1–5 10.1016/j.jocn.2016.05.012 [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa M., Inoue A., Ohnishi T. et al. (2018) Significance of glioma stem-like cells in the tumor periphery that express high levels of CD44 in tumor invasion, early progression, and poor prognosis in glioblastoma. Stem Cells Int. 2018, 5387041 10.1155/2018/5387041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Q., Li Q., Wang M. et al. (2019) Elevated CD44 expression predicts poor prognosis in patients with low-grade glioma. Oncol. Lett. 18, 3698–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou C., Ishi Y., Motegi H. et al. (2019) Overexpression of CD44 is associated with a poor prognosis in grade II/III gliomas. J. Neuro Oncol. 145, 201–210 10.1007/s11060-019-03288-8 [DOI] [PubMed] [Google Scholar]
- 28.Ranuncolo S.M., Ladeda V., Specterman S. et al. (2002) CD44 expression in human gliomas. J. Surg. Oncol. 79, 30–35, 10.1002/jso.10045 [DOI] [PubMed] [Google Scholar]
- 29.Sooman L., Freyhult E., Jaiswal A. et al. (2015) FGF2 as a potential prognostic biomarker for proneural glioma patients. Acta Oncol. 54, 385–394 10.3109/0284186X.2014.951492 [DOI] [PubMed] [Google Scholar]
- 30.Guadagno E., Borrelli G., Califano M., Cali G., Solari D. and Del Basso De Caro M. (2016) Immunohistochemical expression of stem cell markers CD44 and nestin in glioblastomas: evaluation of their prognostic significance. Pathol. Res. Pract. 212, 825–832 10.1016/j.prp.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 31.Pinel B., Duchesne M., Godet J. et al. (2017) Mesenchymal subtype of glioblastomas with high DNA-PKcs expression is associated with better response to radiotherapy and temozolomide. J. Neuro Oncol. 132, 287–294 10.1007/s11060-016-2367-7 [DOI] [PubMed] [Google Scholar]
- 32.Tsidulko A.Y., Kazanskaya G.M., Kostromskaya D.V. et al. (2017) Prognostic relevance of NG2/CSPG4, CD44 and Ki-67 in patients with glioblastoma. Tumour Biol. 39, 1010428317724282 10.1177/1010428317724282 [DOI] [PubMed] [Google Scholar]
- 33.Bien-Moller S., Balz E., Herzog S. et al. (2018) Association of glioblastoma multiforme stem cell characteristics, differentiation, and microglia marker genes with patient survival. Stem Cells Int. 2018, 9628289 10.1155/2018/9628289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alameda F., Velarde J.M., Carrato C. et al. (2019) Prognostic value of stem cell markers in glioblastoma. Biomarkers 24, 677–683 10.1080/1354750X.2019.1652345 [DOI] [PubMed] [Google Scholar]
- 35.Wei K.C., Huang C.Y., Chen P.Y. et al. (2010) Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 30, 253–259 [PubMed] [Google Scholar]
- 36.Stroup D.F., Berlin J.A., Morton S.C. et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283, 2008–2012 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 37.Higgins J. and Green S. (2011) Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. The Cochrane Collaboration www.cochranehandbook.org [Google Scholar]
- 38.Wells G.A., Shea B., O’Connell D. et al. (2010) The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp [Google Scholar]
- 39.Higgins J.P. and Thompson S.G. (2002) Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 40.Egger M., Davey Smith G., Schneider M. and Minder C. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada H., Yoshida J., Sokabe M., Wakabayashi T. and Hagiwara M. (1996) Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int. J. Cancer 66, 255–260 [DOI] [PubMed] [Google Scholar]
- 42.Xu Y., Stamenkovic I. and Yu Q. (2010) CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 70, 2455–2464 10.1158/0008-5472.CAN-09-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiranowska M., Ladd S., Smith S.R. and Gottschall P.E. (2006) CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain Cell Biol. 35, 159–172 10.1007/s11068-007-9009-0 [DOI] [PubMed] [Google Scholar]
- 44.Kim Y. and Kumar S. (2014) CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res. 12, 1416–1429 10.1158/1541-7786.MCR-13-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saygin C., Matei D., Majeti R., Reizes O. and Lathia J.D. (2019) Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 24, 25–40 10.1016/j.stem.2018.11.017 [DOI] [PubMed] [Google Scholar]
- 46.Johansson E., Grassi E.S., Pantazopoulou V. et al. (2017) CD44 interacts with HIF-2alpha to modulate the hypoxic phenotype of perinecrotic and perivascular glioma cells. Cell Rep. 20, 1641–1653 10.1016/j.celrep.2017.07.049 [DOI] [PubMed] [Google Scholar]
- 47.Fu J., Yang Q.Y., Sai K. et al. (2013) TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro Oncol. 15, 1353–1365 10.1093/neuonc/not079 [DOI] [PMC free article] [PubMed] [Google Scholar]