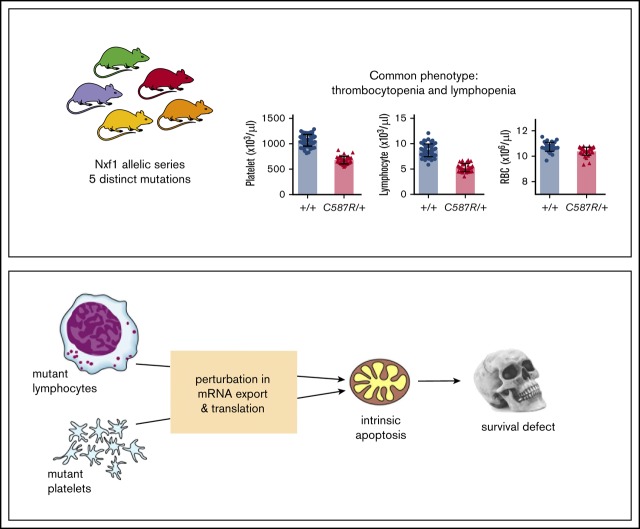
Key Points
Heterozygous Nxf1 mutations sensitize the lymphoid and platelet (but not erythroid) lineages to apoptosis.
Impaired Nxf1 function triggers RNA-metabolism perturbations in blood cells.
Abstract
In eukaryotic cells, messenger RNA (mRNA) molecules are exported from the nucleus to the cytoplasm, where they are translated. The highly conserved protein nuclear RNA export factor1 (Nxf1) is an important mediator of this process. Although studies in yeast and in human cell lines have shed light on the biochemical mechanisms of Nxf1 function, its contribution to mammalian physiology is less clear. Several groups have identified recurrent NXF1 mutations in chronic lymphocytic leukemia (CLL), placing it alongside several RNA-metabolism factors (including SF3B1, XPO, RPS15) whose dysregulation is thought to contribute to CLL pathogenesis. We report here an allelic series of germline point mutations in murine Nxf1. Mice heterozygous for these loss-of-function Nxf1 mutations exhibit thrombocytopenia and lymphopenia, together with milder hematological defects. This is primarily caused by cell-intrinsic defects in the survival of platelets and peripheral lymphocytes, which are sensitized to intrinsic apoptosis. In contrast, Nxf1 mutations have almost no effect on red blood cell homeostasis. Comparative transcriptome analysis of platelets, lymphocytes, and erythrocytes from Nxf1-mutant mice shows that, in response to impaired Nxf1 function, the cytoplasmic representation of transcripts encoding regulators of RNA metabolism is altered in a unique, lineage-specific way. Thus, blood cell lineages exhibit differential requirements for Nxf1-mediated global mRNA export.
Visual Abstract
Introduction
In eukaryotic cells, RNA is transcribed within the nucleus and exported to the cytoplasm through channels called nuclear pore complexes (NPCs). Different classes of RNAs are exported by specific transport molecules capable of interacting with nucleoporins, the proteins that form the NPC.1 The export of ribosomal RNA, transfer RNA, and microRNA is dependent on distinct export mechanisms that are all dependent on the small GTPase Ran. In contrast, the export of messenger RNA (mRNA) is independent of Ran, and is thought to be primarily mediated by the cargo protein known as nuclear RNA export factor1 (Nxf1).
Nxf1 is the vertebrate homolog of Mexp67, a protein identified as an mRNA exporter in yeast.2 The central role of Nxf1 in mRNA export has been established in higher eukaryotes,3-6 where its activity is reportedly dependent on nuclear transport factor 2 like export factor 1 (Nxt1), a binding partner with which it heterodimerizes.5,7 The genetic knockdown of either Nxf1 or Nxt1 inhibits mRNA export, halts protein synthesis, and induces cell death.5 Remarkably, this functional unit is conserved through evolution as demonstrated by the fact that the human Nxf1-Nxt1 heterodimer can functionally replace its homolog in yeast cells.8
Nxf1 has a modular domain organization that is conserved in higher eukaryotes.9 Its N terminus contains a RNA-binding domain and 4 leucine-rich repeats. The C terminus of Nxf1 contains an NTF2 homology domain through which it interacts with Nxt1.7 It also contains a ubiquitin-associated–like domain, which is necessary for nucleocytoplasmic shuttling10 likely via direct interaction with the nucleoporins7,9,10
In eukaryotic cells, nascent mRNA transcripts undergo processing including capping of the 5′ end, removal of introns through splicing, and cleavage of the 3′ end followed by polyadenylation. Processed transcripts are then assembled together with adaptor and RNA-binding factors into ribonucleoprotein particles before being exported to the cytoplasm for translation. These adaptors play crucial roles in enabling Nxf1-mediated export. For example, some adaptors bind and assist in the transfer of mRNAs to Nxf1, whereas others alter Nxf1’s conformation itself, thereby increasing its intrinsic ability to bind and export mRNAs.11,12 A logistical challenge for eukaryotic cells is to specifically export mature mRNAs from the nucleus while retaining incompletely processed transcripts. This issue is partly addressed by the fact that Nxf1 interacts with adaptors and complexes that only bind mature mRNAs. For instance, Nxf1 directly binds to splicing SR proteins13 and to the transcription-export complex, which is present only on spliced and capped transcripts.14-16
Until recently, much of the research that has identified the key regulators, and mechanistic underpinnings of, RNA metabolism has been conducted in cell lines, with comparatively little done to understand the impact of perturbations in RNA metabolism on mammalian physiology. This has begun to change in the last decade, 1 major impetus being the discovery that somatic mutations in mRNA-processing factors occur at high frequency in a range of hematological malignancies including myelodysplastic syndromes, acute myeloid leukemia, chronic lymphocytic leukemia (CLL), and chronic myelomonocytic leukemia.17-20 The subsequent intensive research effort has elucidated novel therapeutic opportunities and fueled several clinical trials,21 but we are still far from a complete understanding of RNA metabolism at an organismal level. Several groups have reported recurrent mutations in NXF1 in CLL.22-24 Detected at frequencies ranging from 1.3% to 6.5%, these mutations affected coding sequences (missense, nonsense, frameshift) as well as splice sites. Clonal analysis pre- and posttherapy suggests that these mutations may be drivers in CLL pathogenesis. By what mechanism, and how Nxf1-mediated RNA metabolism contributes more broadly to hematopoietic biology, remains unclear.
In the current study, we report an allelic series of point mutations in murine Nxf1, and show that they result in dominant lymphopenia and thrombocytopenia, and several additional, more subtle hematological defects. Nxf1 mutations impair platelet and lymphocyte survival in a cell-intrinsic manner apparently by sensitizing them to apoptosis. In contrast, erythrocyte survival is unaltered. Comparative transcriptome analysis of platelets, lymphocytes, and erythrocytes shows that, in response to the heterozygous Nxf1 mutation, hematopoietic cells alter the cytoplasmic representation of transcripts encoding regulators of RNA metabolism in unique, lineage-specific ways. Collectively, our results demonstrate that Nxf1 plays an essential role in hematopoietic cell biology, and illustrate the hematopoietic system’s intrinsic sensitivity to reductions in Nxf1 function relative to other tissues.
Materials and methods
Animals
Mice carrying the CD23-Cre,25 the Ubiquitin-GFP Tg,26 the germline deletion of Bak1,27 Ifnar,28 and Ifnγ29 and the floxed Bax allele30 have been previously described. For bone marrow (BM) chimera experiments, recipients were lethally irradiated (11 Gy) and reconstituted with 5 × 106 BM cell suspension from either a single donor or from 2 distinct congenic donors (1:1 ratio). All animal experiments complied with the regulatory standards of, and were approved by, the Walter and Eliza Hall Institute Animal Ethics Committee.
Flow cytometry
Flow cytometric acquisition was performed with BD Biosciences LSR Fortessa and FACSCalibur analyzers; Flow Jo software was used for analysis. Flow cytometry antibodies are listed in supplemental Methods. Organ cellularity was determined by flow cytometry by counting cell suspensions with CaliBRITE beads (BD Biosciences) in the presence of 20 µg/mL propidium iodide (Sigma-Aldrich). Enumeration of reticulated platelets and red blood cells (RBCs) was performed as previously described.31 Cell sorting was done on Influx (BD Biosciences) and MoFlo (Beckman Coulter) cell sorters with >99.5% purity.
Statistical analysis
Analyses were performed using Prism software (GraphPad). P values were calculated by the Student unpaired t test with Welch correction.
Results
Nxf1 mutations cause lymphopenia and thrombocytopenia in mice
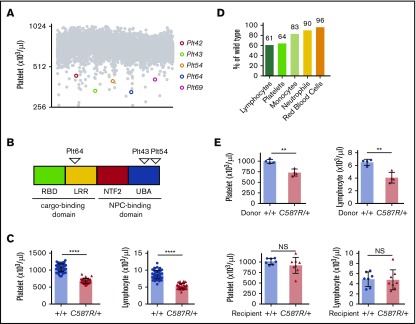
We conducted an N-ethyl-N-nitrosurea forward genetic screen to identify regulators of blood cell homeostasis in mice (described in Masters et al32). Five pedigrees were isolated in which affected animals exhibited dominant heritable thrombocytopenia and lymphopenia, and for which the causative mutation mapped genetically to the same 3-Mb interval (6 580 813-9 028 410) on chromosome 19 (Figure 1A). These pedigrees were initially designated Plt42, Plt43, Plt54, Plt64, and Plt69. Using a combination of custom genomic capture arrays and high-throughput sequencing, we identified a distinct heterozygous point mutation in the Nxf1 gene in each pedigree (Table 1). No other variants common to all 5 pedigrees were identified. The Plt42 and Plt69 mutations were both located within splice donor sites, whereas the Plt64, Plt43, and Plt54 mutations were located within exons 9, 19, and 20, respectively (Figure 1B).
Figure 1.
Mutations in the Nxf1 gene cause lymphopenia and thrombocytopenia. (A) Peripheral blood platelets count from 7-week-old G1 offspring of N-ethyl-N-nitrosurea–mutagenised BALB/c males. (B) Position of amino acid substitutions on the Nxf1 protein. (C) Automated analysis of platelet and lymphocyte counts in Nxf1C587R/+ mice and WT littermate controls. (D) Average decrease in the hematopoietic population. (E) The C587R phenotype is BM-derived. Platelet and lymphocyte counts in lethally irradiated WT recipients reconstituted with C587R/+ or littermate control BM (top). Cell counts in lethally irradiated C587R/+ and control recipients reconstituted with WT BM (bottom). Circles and triangles represent cell counts for an individual mouse. Data show mean with standard deviation (SD). **P < .01; ****P < .0001. LRR, leucine-rich repeat; NS, nonsignificant; NTF2, nuclear transport factor 2–like domain; Plt, platelet; RBD, RNA-binding domain; UBA, ubiquitin-associated–like domain.
Table 1.
Nxf1 mutations causing lymphopenia and thrombocytopenia
| Mutation | Location of mutation | Nucleotide change | Type of mutation | Predicted effect |
|---|---|---|---|---|
| Plt42 | 19:8 757 261 | T>G | N/A | Splice donor site |
| Plt43 | 19:8 769 146 | T>C | S574P | Missense |
| Plt54 | 19:8 770 261 | T>C | C587R | Missense |
| Plt64 | 19:8 764 557 | T>C | I285T | Missense |
| Plt69 | 19:8 768 666 | T>G | N/A | Splice donor site |
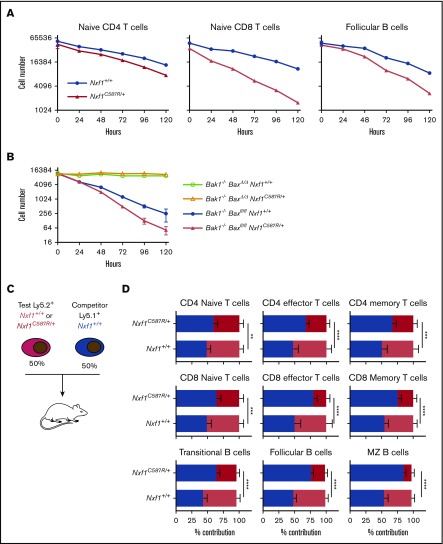
Initial characterization of each pedigree indicated that all 5 exhibited the same phenotype, with varying degrees of severity (Figure 1A-D; supplemental Figure 1A-D). We therefore elected to focus our analyses on one of them, the Plt54 pedigree. The Plt54 mutation is predicted to encode a cysteine to arginine substitution at position 587 (Table 1). This cysteine residue is highly conserved across species (supplemental Figure 1E), and is reportedly critical for binding to nucleoporins.33 Adult mice heterozygous for the Nxf1C587R allele exhibited peripheral blood platelet and lymphocyte counts ∼60% those of wild-type (WT) littermates (Figure 1C-D). Although Nxf1 is expressed in all blood lineages34 (supplemental Figure 2A), the impact on other mature blood cell lineages was subtle, in some cases reaching statistical significance (supplemental Figure 2B). In reciprocal BM transplantation experiments, thrombocytopenia and lymphopenia were observed in recipients reconstituted with cells from Nxf1+/C587R, but not Nxf1+/+, donors (Figure 1E), demonstrating that these defects are intrinsic to the hematopoietic compartment.
To investigate the effects of Nxf1C587R homozygosity, we intercrossed Nxf1+/C587R breeders. No homozygous mutants were observed at birth, suggesting that Nxf1C587R homozygosity results in embryonic lethality. We subsequently intercrossed Nxf1+/C587R animals and recovered 35 embryos at embryonic day 12. Of these, none were homozygous mutant (the ratio of wild type [WT] to heterozygous to homozygous was 15:20:0 against an expected mendelian ratio of 9:17:9), consistent with Nxf1C587R homozygosity being incompatible with normal development.
Mutations in Nxf1 impair platelet survival and life span
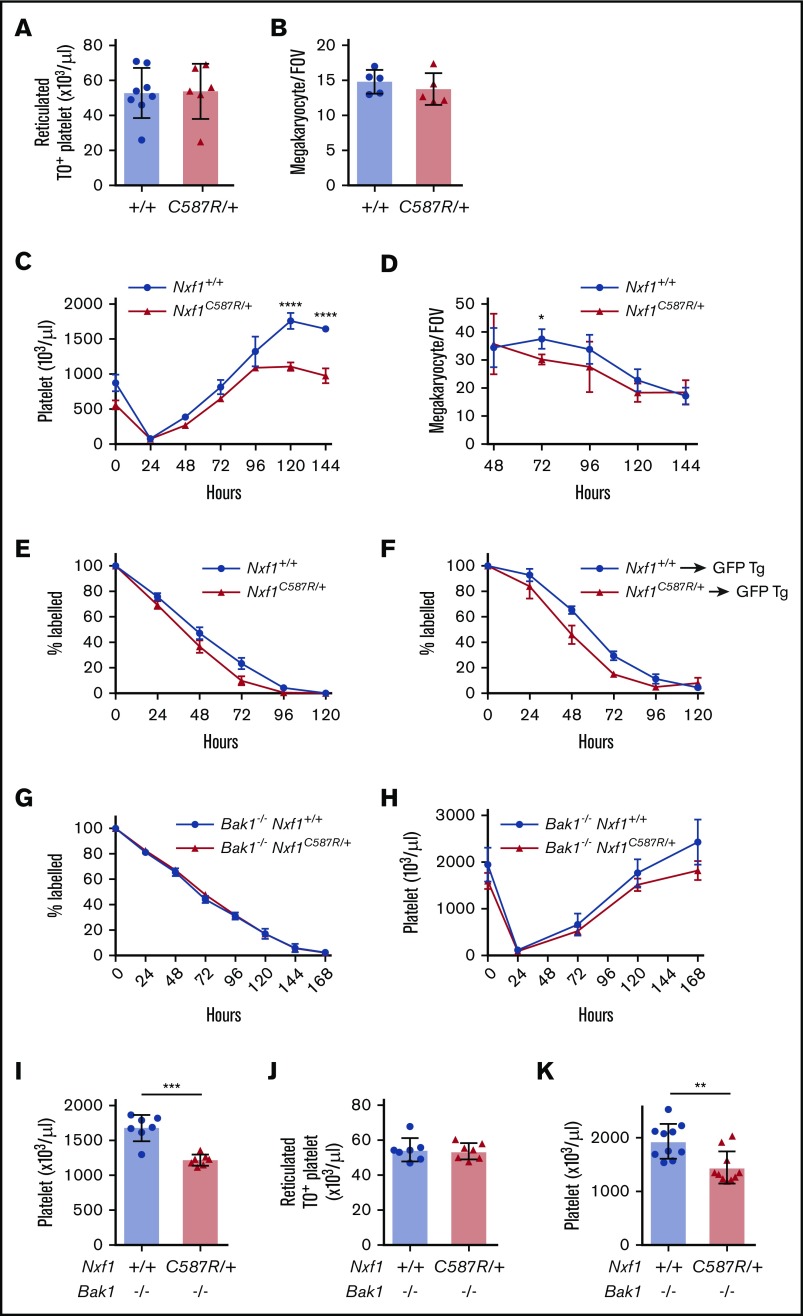
To understand the basis of thrombocytopenia in Nxf1C587R heterozygous mice, we conducted a detailed analysis of the megakaryocyte lineage. Enumeration of megakaryocytes in sternal sections revealed no differences between Nxf1+/C587R mice and WT littermates (Figure 2B). Similarly, the number of RNA-containing reticulated platelets in the blood was unchanged, indicating that platelet biogenesis at steady state was not impaired (supplemental Figure 3A; Figure 2A). We induced acute thrombocytopenia by injecting mice with antiplatelet serum (APS) and monitored platelet counts over a period of 6 days. Platelet counts in mutant mice rebounded with WT kinetics over the first 96 hours, but plateaued from 120 hours (Figure 2C). No decrease in megakaryocyte number was observed, suggesting the possibility of a defect in platelet life span. (Figure 2D). We therefore examined platelet survival in vivo. Platelets in Nxf1+/C587R animals disappeared a day earlier than those of WT littermates (Figure 2E). This survival defect was cell-intrinsic as it was still observed when platelets from Nxf1+/C587R donors were transferred into WT hosts (Figure 2F).
Figure 2.
The C587R mutation sensitizes platelet to apoptosis without compromising megakaryopoiesis. (A) Absolute numbers of reticulated platelets were determined by thiazole orange staining. (B) Average megakaryocyte number per field of view (average of 6 fields per individual mice; n = 5 mice per group). (C) Average platelet count in response to APS-induced thrombocytopenia in C587R/+ mutant and littermate controls; n = 5 mice per group. (D) Average megakaryocyte number per field of view (average of 5 fields per individual mice; n = 3 mice per group) upon APS-induced thrombocytopenia. (E) C587R/+ platelets have a survival defect. Platelet survival curve in C587R/+ mutant and control mice. Platelets were labeled via IV injection of DyLight 488–conjugated anti-GPIbβ antibody. Data represent mean ± SD; n = 6 mice per group. (F) Survival defect is cell-intrinsic. Platelets purified from C587R/+ mice and littermate controls were injected IV into GFP-transgenic recipients to assess their survival; n = 5 mice per group. (G) Loss of proapoptotic gene Bak1 restores survival defect. Platelet survival curve in Bak1−/−Nxf1C587R/+ and Bak1−/− control mice. (H) Platelet counts in response to APS-induced thrombocytopenia in Bak1−/−Nxf1C587R/+ and Bak1−/− littermate controls; n = 3-4 mice per group. (I) Platelets count in splenectomized Bak1−/−Nxf1C587R/+ and Bak1−/− control mice. (J) Absolute numbers of TO+ reticulated platelets in Bak1−/−Nxf1C587R/+ and Bak1−/− control mice. (K) Platelet counts in splenectomized Bak1−/−Nxf1C587R/+ and Bak1−/− control mice. (A-B,I-K) Circles and triangles represent cell counts for an individual mouse. Data show mean with SD. *P < .05; **P < .01; ***P < .001; and ****P < .0001. FOV, field of view; GFP, green fluorescent protein; Tg, transgenic; TO, thiazole orange.
One important regulator of platelet survival is the intrinsic apoptosis pathway, which culminates with the activation of the prodeath Bak.31 Loss of Bak results in a doubling of circulating platelet life span. Crossing the Nxf1+/C587R mutation onto a Bak1-deficient background restored platelet survival kinetics to those observed in WT littermates (Figure 2G). Strikingly, the response to acute thrombocytopenia induced by APS was also normalized (Figure 2H). Surprisingly, however, although platelet counts in Bak1−/−Nxf1+/C587R animals were equivalent to those observed in WT counterparts (mean, 1219 × 109/L), they did not reach the levels exhibited by Bak1−/− mice (mean, 1679 × 109/L) (Figure 2I). Splenectomy of Bak1−/− Nxf1+/C587R animals did not restore platelet counts in mutant mice (Figure 2H), indicating that the defect was not due to abnormal splenic sequestration. Consistent with results obtained on the WT background (Figure 2A), platelet biogenesis was normal in Bak1−/−Nxf1+/C587R mice (Figure 2J). Thus, the exact basis of the thrombocytopenia observed in Nxf1+/C587R animals remains somewhat enigmatic. Cell-intrinsic defects in platelet survival and life span are clearly responsible, but the inability of Bak deletion to restore platelets counts to Bak1−/− levels (despite rescuing platelet survival kinetics) suggests a complex interplay between the intrinsic apoptosis pathway and some as-yet-unidentified mechanism of platelet homeostasis.
Mutations in Nxf1 reduced peripheral mature lymphocyte numbers
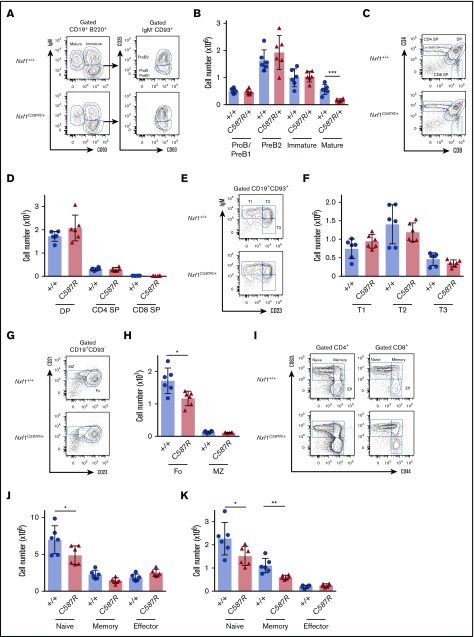
We next examined the basis of lymphopenia. Flow cytometric analyses revealed that, in the BM, hematopoietic stem cell and progenitor populations were present at WT frequency and number in Nxf1+/C587R mice (supplemental Figure 3B-I). Although developing B-cell populations were unaltered, there was a threefold reduction in the numbers of mature B cells (Figure 3B). Thymocyte populations were present in normal numbers (Figure 3C-D).
Figure 3.
Peripheral lymphocytes but not progenitors are decreased in C587R/+ animals. (A) Representative FACS plots of B-cell compartments in the BM of Nxf1C587R/+ and WT control animals. Gates indicate CD93+IgM− progenitor B cells, CD93+ IgM+ immature B cells, and CD93−IgM+ mature B cells. CD93+IgM−CD25+ are pro-B/pre-B1 and CD93+IgM−CD25− are pre-B2. (B) Absolute cell numbers in C587R/+ mutant and control animals. (C) Representative FACS plots of developing T cells in the thymus of Nxf1C587R/+ and WT control animals. (D) Absolute cell numbers. (E) Representative plots of transitional B cells in the spleen of Nxf1C587R/+ and WT control animals. Gates indicate IgMhighCD23low T1, IgMhighCD23+ T2, and IgMlowCD23+ T3 transitional B cells. (F) Absolute cell numbers in C587R/+ mutant and control animals. (G) Representative plots of Fo CD21+CD23+ B cells and CD21highCD23low MZ B cells in the spleen of Nxf1C587R/+ and WT control animals. (H) Fo B cells are decreased mutant animals. Absolute splenic B-cell numbers. (I) Representative plots of CD4 and CD8 T-cell compartments in spleen of Nxf1C587R/+ and WT control animals. Gates indicate CD62L+CD44low naive, CD62L+CD44high memory and CD62L−CD44high effector T cells. (J) Naive T cells are decreased in C587R/+ mutant animals. Absolute T cells number within the CD4 (J) and CD8 (K) compartments. Circles and triangles represent cell numbers for an individual mouse. Data show mean with SD. *P < .05; **P < .01; and ***P < .001. DP, double positive; IgM, immunoglobulin M; SP, single positive.
Upon exiting the BM, immature B cells migrate to the spleen where they go through a transitional stage before entering the pool of mature cells. Transitional B cells are characterized by the coexpression of CD19 and CD93,35 and can be further subdivided between T1, T2, and T3 stages.36 These 3 populations were present in normal numbers in Nxf1+/C587R mice (Figure 3E-F). Among CD93− mature B cells, the follicular (Fo) B cells were significantly decreased, whereas marginal zone (MZ) B cells were present in normal numbers (Figure 3G-H). CD4- and CD8-naive T cells, as well as memory CD8 T cells, were present in reduced numbers (Figure 3I-K). Taken together, these results indicate that Fo B cells and peripheral T lymphocytes are haploinsufficient for Nxf1, whereas the lineage-committed progenitors from which they originate are not.
Nxf1+/C587R peripheral lymphocyte survival is compromised
We next examined lymphocyte survival in an in vitro growth factor deprivation assay. We found that naive CD4 and CD8 T and Fo B cells flow-sorted from Nxf1+/C587R mice died more rapidly than those from WT littermate mice (Figure 4A). To establish whether the increased death was the result of intrinsic apoptosis, we deleted Bak and Bax, the 2 essential mediators of this pathway.30 Loss of both rescued survival in Nxf1+/C587R Fo B cells (Figure 4B). Lymphocytes with an increased propensity to undergo intrinsic apoptosis exhibit a competitive defect.37 We therefore generated chimeras in which Ly5.2+ Nxf1+/C587R (test) BM cells were mixed with equal numbers of Ly5.1+ WT (competitor) cells before being transferred into lethally irradiated Ly5.1+ Ly5.2+ recipients (Figure 4C). Twelve weeks postreconstitution, the proportion of test-derived cells in hematopoietic stem cell, progenitor, and myeloid populations were identical, irrespective of genotype (supplemental Figure 4A), demonstrating that the Nxf1+/C587R mutation neither compromises the repopulating capacity of hematopoietic stem cells, nor appreciably affects the competitiveness of progenitor and myeloid compartments.
Figure 4.
C587R mutant peripheral lymphocytes are sensitized to apoptosis and underrepresented under competitive condition (A) Peripheral lymphocytes carrying the C587R mutation are sensitized to apoptosis. Growth factor deprivation assay of FACS-sorted naive CD4, CD8 T cells and Fo B cells from either C587R/+ or WT animals. Representative experiment of 3. Each point is the average of 3 technical replicates and bar represents standard deviation. (B) The deleterious effect of the C587R mutation on B-cell survival is masked by the deletion of Bax and Bak1. Representative experiment of 2. Each point is the average of 3 technical replicates and bar represents standard deviation. (C) BM competitive transplant assay. (D) Peripheral B- and T-cell compartments are underrepresented in competitive BM chimera. Twelve weeks posttransplant, the relative representation of “test” and “competitor” cells among splenic lymphocytes was assessed. Data show mean with SD. **P < .01; ***P < .001; and ****P < .0001.
Within the B lineage, Nxf1+/C587R cells were slightly underrepresented in the pro-B/pre-B1 and immature compartments (supplemental Figure 4A), and were greatly reduced within the transitional, Fo, and MZ compartments (Figure 4D). Nxf1+/C587R cells were present in normal frequency among all thymocyte subsets (supplemental Figure 4B), but were severely underrepresented within all peripheral T-cell populations (Figure 4D). Collectively, these experiments demonstrate that Nxf1 heterozygosity does not compromise early hematopoietic progenitors, myeloid cells, or the developing lymphoid compartments, but potently undermines the competitiveness of peripheral lymphocytes.
Impaired Nxf1 function results in altered cytoplasmic representation of RNA-metabolism transcripts
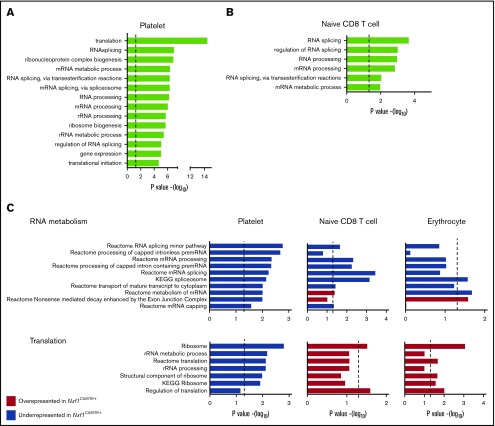
Our results indicated that mutations in Nxf1 affect both platelets and peripheral lymphocytes, but whether this was linked to the role of Nxf1 in mRNA export remained unclear. We therefore conducted RNA-sequencing (RNAseq) expression profiling on platelets and cytoplasmic fractions from naive CD8 T cells (see supplemental Methods). Platelets are anuclear cells containing a repertoire of mRNA exported from the megakaryocyte nucleus, and therefore represent a naturally occurring cytoplasmic fraction. The 14 most significant gene sets identified by gene ontology (GO) analysis of the transcripts underrepresented in Nxf1+/C587R platelets compared with WT were all specific to RNA metabolism and translation regulation (Figure 5A). Analysis of the transcripts underrepresented in the cytoplasmic fraction of Nxf1+/C587R-naive CD8 T cells identified 6 gene sets that were common with those found with the platelet data set and all 6 were specific to RNA metabolism (Figure 5B).
Figure 5.
C587R-mutant hematopoietic cells have altered representation of transcripts encoding RNA metabolism and translation regulators. (A) 14 most significant gene sets identified by Gene Ontology (GO) analysis done on genes underrepresented in C587R/+ platelets compared with +/+ control cells. The dotted line represents the statistical significance at P = .02. (B) Six gene sets identified by GO analysis performed on underrepresented genes in cytoplasmic fraction of C587R/+ naive CD8 T cells. These genes sets were also among the 14 most significant in platelets. The dotted line represents the statistical significance at P = .02. (C) Rotation gene set testing (ROAST) analysis using independent gene sets specific for RNA metabolism, mRNA splicing, and translation on platelets, naive CD8 T cells, and RBCs. The dotted line represents the statistical significance at P = .02. KEGG, Kyoto Encyclopedia of Genes and Genomes; rRNA, ribosomal RNA.
As part of this experiment, we also included erythrocytes, a cell type that exhibited no defects, and also represent a naturally occurring cytoplasmic fraction (Figure 1D; supplemental Figure 5). We were curious to understand whether heterozygosity for the Nxf1C587R mutation had any impact on gene expression in these cells. We ran a rotation gene set testing analysis38 using independent gene sets specific for genetic information processing. Confirming and extending the GO analysis, this analysis revealed that genes regulating RNA metabolism were less abundant in mutant cells from all 3 lineages, and independently confirmed that translation genes were underrepresented in platelets from Nxf1+/C587R mice (Figure 5C). Although they possess less statistical power, these results further suggested that transcripts encoding proteins that regulate translation were overrepresented in mutant lymphocytes and erythrocytes (Figure 5C). These results indicate that decreased Nxf1 function leads to alterations of RNA metabolism in 3 distinct hematopoietic lineages, and illustrate the ability of cells to detect defects in the Nxf1 pathway, and in response, modulate the representation of specific RNA transcripts immediately upstream and downstream of mRNA export.
Nxf1 mutations trigger an IFNγ signature in lymphocytes
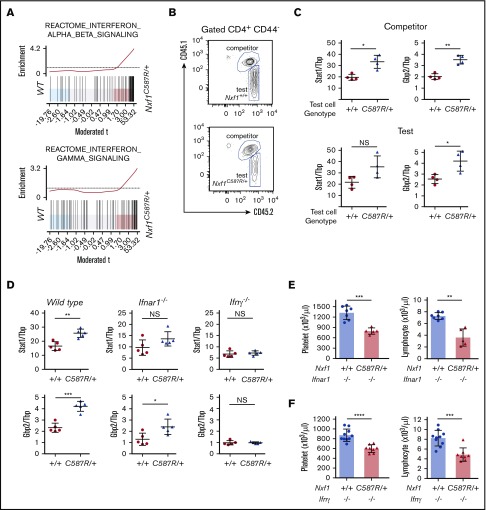
Consistent with our results showing that Nxf1 mutations sensitize lymphocytes to apoptosis, GO analysis (biological process) on mRNAs overrepresented in mutant CD8-naive T cells showed that many apoptosis gene sets were highly enriched (supplemental Figure 6A). Gene-set enrichment analysis revealed that gene sets specific for interferon (IFN) responses were also enriched among transcripts overrepresented in mutant cells (Figure 6A), a result independently confirmed via GO analysis (supplemental Figure 6B). To test whether the IFN signature was hematopoietic in origin and intrinsic to mutant lymphocytes, we generated mixed BM chimeras in WT hosts and characterized naive CD4 T WT competitor and Nxf1+/C587R test cells (Figure 6B). Nxf1+/C587R test cells exhibited increased expression of the IFN-stimulated genes (ISGs) Stat1 and Gbp2, relative to WT test cells (Figure 6C top panel), confirming that the IFN signature was hematopoietic in origin. Increased expression of these 2 ISGs was also observed in WT competitor cells that coexisted with mutant test cells (Figure 6C bottom panel), demonstrating that the IFN signature was not intrinsic to T cells, but rather, resulted from paracrine signaling. To establish whether this signature arose in response to the secretion of type I or type II IFN, we backcrossed the Nxf1C587R allele onto Ifnar1- and Ifnγ-deficient backgrounds. Naive Nxf1+/C587R CD4+ T cells displayed increased Stat1 and Gbp2 expression levels in the absence of the Ifnar gene (Figure 6D), ruling out a major role for type I IFN signaling. In contrast, deletion of Ifnγ restored these ISG expression to WT levels (Figure 6D), demonstrating that Ifnγ signaling caused the IFN signature observed in peripheral lymphocytes. Despite this, loss of Ifnγ did not ameliorate either lymphopenia or thrombocytopenia in Nxf1+/C587R mice (Figure 6E-F).
Figure 6.
The C587R mutation in the hematopoietic lineage causes an interferon-signature in lymphocytes. (A) Representative barcode enrichment plots of IFN signatures in C587R/+ CD8-naive T cells. (B) Mixed BM chimeras were set up as shown for Figure 4C. Representative FACS plot showing chimerism in the CD4-naive compartment: WT CD45.1+CD45.2+ “competitor cells” coexisting with either WT (top panel) or C587R-mutant CD45.2+ “test cells” (bottom panel). (C) The IFN signature is hematopoietic in origin and the result of paracrine signaling. Quantitative PCR results show that the IFN-stimulated genes Stat1 and Gbp2 are upregulated in “competitor cells” when in presence of C587R/+ mutant “test cells.” Each point represents the value obtained from CD4 T cells sorted an individual mouse. (D) The IFN signature is dependent on IFNγ signaling. The IFN signature was maintained in C587R mutant CD4-naive T cells purified from mice lacking the Ifnar1 gene but not the Ifnγ gene. Each point represents the value obtained from CD4 T cell sorted an individual mouse. The loss of Ifnar1 (E) or Ifnγ (F) did not restore neither lymphopenia nor thrombocytopenia. (E) Circles and triangles represent cell numbers for an individual mouse. Data show mean with SD. *P < .05; **P < .01; ***P < .001; and ****P < .0001.
Nxf1+/C587R lymphocytes are sensitized to translation inhibition
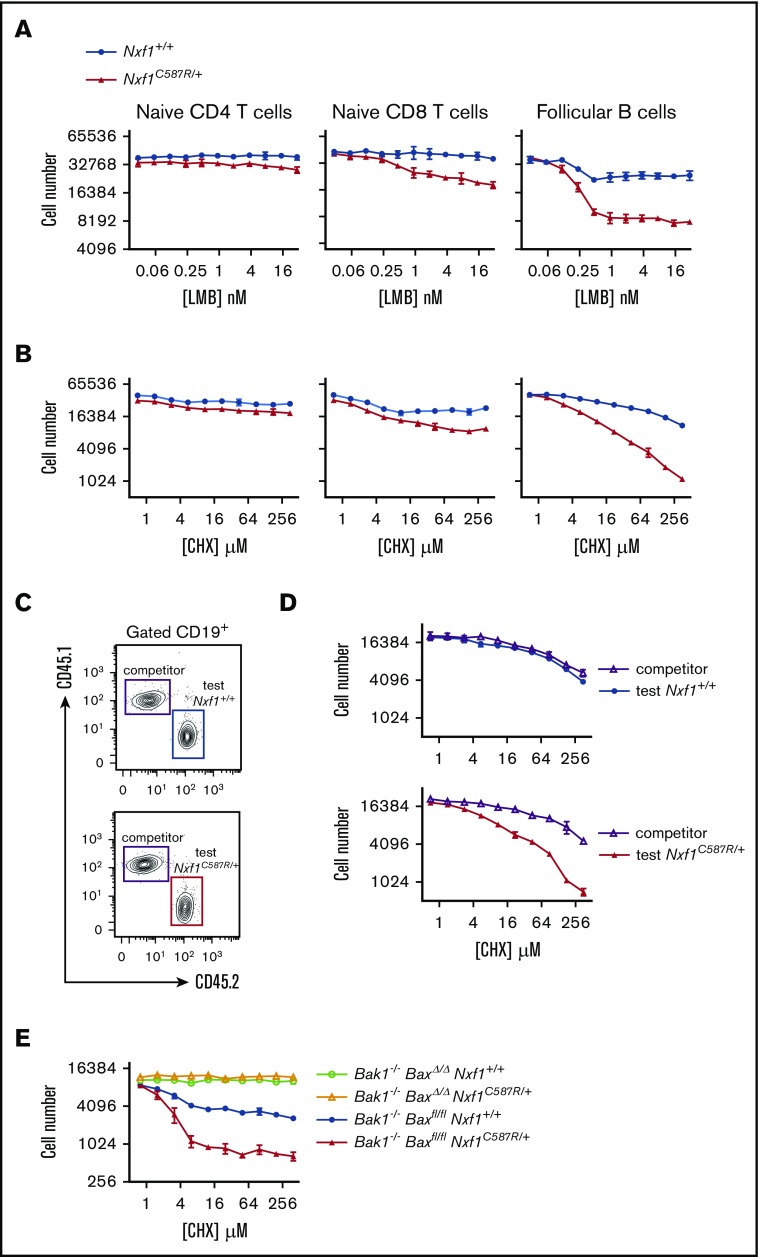
Our RNAseq data were consistent with Nxf1 playing a role in global mRNA export. We reasoned that cells bearing Nxf1 mutations may be more sensitive to the inhibition of other nuclear exporters such as the ribosomal RNA nuclear exporter CRM1.39,40 To test this, we treated lymphocytes with leptomycin B (LMB), a highly specific inhibitor of CRM1.41 Both naive CD8 T and Fo B cells exhibited greater sensitivity to LMB than WT counterparts (Figure 7A). They were also more sensitive to the translation inhibitor cycloheximide (CHX) (Figure 7B). In contrast, naive Nxf1+/C587R CD4 T cells were normosensitive to both LMB and CHX (Figure 7A-B), highlighting the differential requirement for Nxf1 function among these lymphocytes.
Figure 7.
The C587R mutation sensitizes lymphocytes to translation inhibition. (A) FACS-sorted lymphocytes were submitted to various concentrations of LMB (A) or CHX (B) for 24 hours. Representative experiment of 2. Each point is the average of 3 technical replicates and bar represents standard deviation. (C) Mixed BM chimeras were set up as shown for Figure 4C. Representative FACS plot showing chimerism in the B-cell compartment: WT CD45.1+ “competitor cells” together with either WT (top panel) or C587R/+ mutant CD45.2+ “test cells” (bottom panel). (D) Increased sensitivity to translation inhibition is intrinsic to C587R-mutant Fo B cells. Representative experiment of 2. Each point is the average of 3 technical replicates and bar represents standard deviation. (E) Loss of Bax and Bak1 in B cells masks the deleterious effect of the C587R mutation in response to translation inhibition. Representative experiment of 2. Each point is the average of 3 technical replicates and bar represents standard deviation.
To establish whether this was cell-intrinsic or caused by extrinsic factors such as IFNγ, we generated mixed BM chimeras, this time isolating test and competitor cells from the Fo B cell compartment (Figure 7C). As expected, Nxf1+/C587R test cells were sensitized to CHX translation-inhibition whereas WT competitor cells from the same recipient were normosensitive, showing the defect was cell intrinsic (Figure 7D). Nxf1+/C587R Fo B cells lacking both the Bax and Bak1 genes were entirely resistant to CHX (Figure 7E), further supporting the link between apoptosis and perturbations in Nxf1 function. These results altogether suggest that impaired Nxf1 function renders lymphocytes more susceptible to apoptosis in response to translation inhibition.
Discussion
Given the fundamental role of RNA in cell biology,42 it is not surprising that mutations in genes regulating the processing, the export, or the translation of mRNA cause disease. Somatic mutations in splicing factors are associated with hematopoietic malignancies,17-20,23,24,43,44 whereas germline mutations in mRNA export factors lead to severe motor neuron disease and intellectual disabilities.45-47 To shed light on the etiology of these diseases, a better understanding of how RNA perturbations impact mammalian tissues and how they alter the biology of different cell lineages is required. We present here an allelic series that represents a novel opportunity to investigate the lineage-specific roles of Nxf1 in mammalian biology. It is to be noted that, for this study, we analyzed heterozygous mutant animals, which were healthy; however, the impossibility of generating homozygous mutants at embryonic day 12.5 demonstrates that, as in Caenorhabditis elegans and Drosophila,6,48 Nxf1 exerts crucial and nonredundant functions in mammalian embryogenesis. Five independent mutations in the Nxf1 gene all caused the same phenotype in mice: thrombocytopenia and lymphopenia accompanied by subtler hematological defects. This strongly suggests that each allele we isolated represents a loss of function (rather than gain or dominant negative) at the Nxf1 locus. This is further supported by the fact that the effects of the C587R mutation on transcript representation in primary blood cells closely resemble those reported in human cell lines upon Nxf1 depletion.49
For reasons that are elusive, genetic lesions in ubiquitously expressed RNA-metabolism genes often have tissue-specific effects. For instance, mice carrying mutations in genes encoding splicing factor or spliceosome components suffer from lineage-specific cytopenias50,51 whereas patients with defective expression of the exon junction complex component RBM8A present with bone malformations and thrombocytopenia.52 Germline Nxf1 mutations in Drosophila lead specifically to neuronal, muscular, and bristle defects,53 suggesting that primary tissues have different sensitivities to Nxf1 perturbation. Although Nxf1 is ubiquitously expressed within the hematopoietic lineage,34 mutations undermining its function do not compromise all blood cell lineages equally. In fact, Nxf1 mutations primarily decrease the numbers and the survival of platelets and peripheral lymphocytes, whereas they have almost no detectable effect on the numbers and life span of RBCs. It is unlikely that this lineage-specific effect results from a different reliance on Nxf1 for mRNA export in these different cell types because our RNAseq analysis revealed that these 3 lineages responded similarly to the Nxf1 mutations. In fact, the nonredundant role of Nxf1 in mRNA export has been observed in all tested tissues in Drosophila48 and there is, to date, no known cell type whose mRNA export is independent of Nxf1.
Although it is well established that the genetic perturbation of Mex67/Nxf1 is lethal both at the cellular2,5 and at the organismal level,6,48 the cause of cell death has not been addressed. Our results establish a functional link between the Nxf1 pathway and the activation of the intrinsic apoptosis pathway. We show that platelets have a decreased life span and that this cell-intrinsic defect can be corrected by deleting Bak1, the primary instigator of apoptosis in these cells.31 Hence, Nxf1 mutations impair platelet survival by actively inducing Bak-mediated apoptosis. Similarly, both our in vitro and in vivo results suggest that Nxf1 mutations cause lymphopenia by sensitizing peripheral lymphocytes to intrinsic apoptosis.
How are mRNA export and intrinsic apoptosis functionally linked? The answer may lie in the relative expression of anti- and proapoptosis genes that will depend on the generation, export, stability, and translation of their corresponding transcripts. Consistently, our RNAseq data show that the representation of many apoptosis genes is altered in mutant lymphocytes. In addition, the perturbations downstream of Nxf1 mutations may be directly sensed by BH3-only proteins, which can trigger apoptosis in response to a range of cellular stresses.54
Our transcriptome analyses suggest that Nxf1 mutations trigger large-scale perturbations in RNA metabolism and protein synthesis in the 3 lineages we investigated. In line with the observation that the transcripts bound by the exporter Mex67 are enriched for RNA processing and translation factors,55 Nxf1 depletion in human cells strongly decreased the cytoplasmic representation of this functional class of transcripts.49 Our results further show that transcripts encoding for RNA processing and translation factors are strongly underrepresented in mutant platelets and suggest that the export of these mRNAs is exquisitely sensitive to Nxf1’s activity. Interestingly, the export of this functional class of transcripts is also dependent on the germinal center–associated protein (GANP),49 whereas the transport of transcripts with a long 3′ untranslated region relies on the concerted activities of Nxf1 with CFI-68.56 Altogether, these results support a model whereby the concerted activities of Nxf1 with different partners ensure the export of a specific class of transcript.55
Our RNAseq analyses on CD8 and RBCs do not possess the same statistical power as the platelet data, but suggest that, in contrast to what is observed in platelets, transcripts encoding for translation factors are overrepresented. Further investigation is necessary to elucidate the underlying mechanisms, but this suggests that these 2 lineages use compensatory transcriptional programs to increase the expression of translation factors. These results further exemplify the diversity of strategies in primary cells facing RNA-metabolism perturbations and suggest that protein synthesis may be impaired. In agreement with this notion, we found that mutant CD8 T and Fo B cells were sensitized to translation inhibition. The phenotype was rescued by deleting Bax and Bak in Fo B cells, suggesting that translation defects sensitize mutant cells to apoptotic cell death and thereby compromise their ability to compete in vivo. This hypothesis is consistent with the observation that both translation efficiency and apoptosis are crucial in determining cell competition outcomes in vivo.57-59 Together with the previous report that Nxf1 depletion blocks protein synthesis in Drosophila cells,5 our results add weight to the hypothesis that impairments in Nxf1 function compromise the protein production machinery. How this might relate to Nxf1 mutations in the pathogenesis of CLL cells is yet to be established. Whether translation inhibition might present synthetic lethal possibilities in CLL cells carrying mutations in RNA-metabolism regulators remains to be explored.
Altogether, our results suggest that the Nxf1 pathway regulates RNA maturation and protein synthesis at the genetic level by altering the cytoplasmic abundance of transcripts encoding RNA processing and translation factors. Given the centrality of these pathways for gene expression at a global level, it is likely that Nxf1 mutations thereby indirectly impact the expression of most cellular mRNAs.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank S. Ross, M. Midson, K. Franks, L. Johnson, T. Carle, K. Stoev, M. Dayton, and Walter and Eliza Hall Institute of Medical Research Bioservices staff for mouse work, and J. Corbin and Y. Liao for technical assistance.
This work was supported by a Swiss National Science Foundation Fellowship for Advanced Researchers (PA00P3_139695) (S.C.), grants from the Australian National Health and Medical Research Council (1113577, 1063008, 1016647, and 1154970), and a Senior Medical Research Fellowship from the Sylvia and Charles Viertel Foundation.
Footnotes
Stéphane Chappaz (stephane.chappaz@monash.edu) can be contacted for data that are not publicly accessible. Transcriptome profiling of blood cells is available at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141161).
Authorship
Contribution: S.C. and B.T.K. designed research; S.C. and R.M.L. performed research; S.C. and M.R.D. collected, analyzed, and interpreted data; C.W.L., G.K.S., and M.E.R. performed bioinformatics analysis; K.T.C., L.H.N., and V.O.W. provided fruitful discussions; and S.C. and B.T.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for B.T.K. is Faculty of Health and Medical Sciences, University of Adelaide, Adelaide, SA, Australia.
Correspondence: Stéphane Chappaz, Monash University, 19 Innovation Walk, Clayton, VIC 3800, Australia; e-mail: stephane.chappaz@monash.edu; or Benjamin T. Kile, Faculty of Health and Medical Sciences, University of Adelaide, Adelaide, SA 5005, Australia: e-mail: benjamin.kile@adelaide.edu.au.
References
- 1.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8(10):761-773. [DOI] [PubMed] [Google Scholar]
- 2.Segref A, Sharma K, Doye V, et al. . Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16(11):3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276(23):20536-20543. [DOI] [PubMed] [Google Scholar]
- 4.Grüter P, Tabernero C, von Kobbe C, et al. . TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1(5):649-659. [DOI] [PubMed] [Google Scholar]
- 5.Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7(12):1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 6.Tan W, Zolotukhin AS, Bear J, Patenaude DJ, Felber BK. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6(12):1762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suyama M, Doerks T, Braun IC, Sattler M, Izaurralde E, Bork P. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 2000;1(1):53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katahira J, Strässer K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18(9):2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herold A, Suyama M, Rodrigues JP, et al. . TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20(23):8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachi A, Braun IC, Rodrigues JP, et al. . The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6(1):136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA. 2008;105(13):5154-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viphakone N, Hautbergue GM, Walsh M, et al. . TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller-McNicoll M, Botti V, de Jesus Domingues AM, et al. . SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30(5):553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127(7):1389-1400. [DOI] [PubMed] [Google Scholar]
- 15.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19(13):1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viphakone N, Sudbery I, Griffith L, Heath CG, Sims D, Wilson SA. Co-transcriptional loading of RNA export factors shapes the human transcriptome. Mol Cell. 2019;75(2):310-323.e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graubert TA, Shen D, Ding L, et al. . Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papaemmanuil E, Cazzola M, Boultwood J, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium . Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Lawrence MS, Wan Y, et al. . SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 21.Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. 2019;9(11):1493-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin NA, Seymour E, Saiya-Cork K, Parkin B, Shedden K, Malek SN. A quantitative analysis of subclonal and clonal gene mutations before and after therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2016;22(17):4525-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landau DA, Tausch E, Taylor-Weiner AN, et al. . Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puente XS, Beà S, Valdés-Mas R, et al. . Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. [DOI] [PubMed] [Google Scholar]
- 25.Kwon K, Hutter C, Sun Q, et al. . Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28(6):751-762. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell Immunol. 2001;214(2):110-122. [DOI] [PubMed] [Google Scholar]
- 27.Lindsten T, Ross AJ, King A, et al. . The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6(6):1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang SY, Hertzog PJ, Holland KA, et al. . A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92(24):11284-11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259(5102):1739-1742. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, Korsmeyer SJ. Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proc Natl Acad Sci USA. 2005;102(32):11272-11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason KD, Carpinelli MR, Fletcher JI, et al. . Programmed anuclear cell death delimits platelet life span. Cell. 2007;128(6):1173-1186. [DOI] [PubMed] [Google Scholar]
- 32.Masters SL, Gerlic M, Metcalf D, et al. . NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 2012;37(6):1009-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grant RP, Neuhaus D, Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol. 2003;326(3):849-858. [DOI] [PubMed] [Google Scholar]
- 34.Choi J, Baldwin TM, Wong M, et al. . Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 2019;47(D1):D780-D785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolink AG, Andersson J, Melchers F. Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol. 1998;28(11):3738-3748. [DOI] [PubMed] [Google Scholar]
- 36.Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167(12):6834-6840. [DOI] [PubMed] [Google Scholar]
- 37.Lee EF, Grabow S, Chappaz S, et al. . Physiological restraint of Bak by Bcl-xL is essential for cell survival. Genes Dev. 2016;30(10):1240-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26(17):2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24(16):2862-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci. 2003;116(Pt 12):2409-2419. [DOI] [PubMed] [Google Scholar]
- 41.Kudo N, Matsumori N, Taoka H, et al. . Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96(16):9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp PA. The centrality of RNA. Cell. 2009;136(4):577-580. [DOI] [PubMed] [Google Scholar]
- 43.Meggendorfer M, Roller A, Haferlach T, et al. . SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012;120(15):3080-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papaemmanuil E, Gerstung M, Bullinger L, et al. . Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nousiainen HO, Kestilä M, Pakkasjärvi N, et al. . Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40(2):155-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ylikallio E, Woldegebriel R, Tumiati M, et al. . MCM3AP in recessive Charcot-Marie-Tooth neuropathy and mild intellectual disability. Brain. 2017;140(8):2093-2103. [DOI] [PubMed] [Google Scholar]
- 47.Kumar R, Corbett MA, van Bon BW, et al. . THOC2 mutations implicate mRNA-export pathway in X-linked intellectual disability. Am J Hum Genet. 2015;97(2):302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkie GS, Zimyanin V, Kirby R, et al. . Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA. 2001;7(12):1781-1792. [PMC free article] [PubMed] [Google Scholar]
- 49.Wickramasinghe VO, Andrews R, Ellis P, et al. . Selective nuclear export of specific classes of mRNA from mammalian nuclei is promoted by GANP. Nucleic Acids Res. 2014;42(8):5059-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirai CL, Ley JN, White BS, et al. . Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27(5):631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang D, Yue T, Choi JH, et al. . Syndromic immune disorder caused by a viable hypomorphic allele of spliceosome component Snrnp40. Nat Immunol. 2019;20(10):1322-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albers CA, Paul DS, Schulze H, et al. . Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet. 2012;44(4):435-439, S431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korey CA, Wilkie G, Davis I, Van Vactor D. Small bristles is required for the morphogenesis of multiple tissues during Drosophila development. Genetics. 2001;159(4):1659-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27(suppl 1):S128-S136. [DOI] [PubMed] [Google Scholar]
- 55.Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33(2):155-161. [DOI] [PubMed] [Google Scholar]
- 56.Chen S, Wang R, Zheng D, et al. . The mRNA export receptor NXF1 coordinates transcriptional dynamics, alternative polyadenylation, and mRNA export. Mol Cell. 2019;74(1):118-131.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42(2):211-221. [DOI] [PubMed] [Google Scholar]
- 58.Clavería C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500(7460):39-44. [DOI] [PubMed] [Google Scholar]
- 59.Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416(6882):755-759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.