Key Points
Single-cell RNA sequencing reveals an unexpected diversity of healthy human blood NK cells, each population with a distinct transcriptome.
For the first time, we describe a population of NK cells with low ribosomal expression, which may be linked to cellular activation.
Abstract
Human natural killer (NK) cells in peripheral blood perform many functions, and classification of specific subsets has been a longstanding goal. We report single-cell RNA sequencing of NK cells, comparing gene expression in unstimulated and interleukin (IL)-2–activated cells from healthy cytomegalovirus (CMV)-negative donors. Three NK cell subsets resembled well-described populations; CD56brightCD16−, CD56dimCD16+CD57−, and CD56dimCD16+CD57+. CD56dimCD16+CD57− cells subdivided to include a population with higher chemokine mRNA and increased frequency of killer-cell immunoglobulin-like receptor expression. Three novel human blood NK cell populations were identified: a population of type I interferon–responding NK cells that were CD56neg; a population exhibiting a cytokine-induced memory-like phenotype, including increased granzyme B mRNA in response to IL-2; and finally, a small population, with low ribosomal expression, downregulation of oxidative phosphorylation, and high levels of immediate early response genes indicative of cellular activation. Analysis of CMV+ donors established that CMV altered the proportion of NK cells in each subset, especially an increase in adaptive NK cells, as well as gene regulation within each subset. Together, these data establish an unexpected diversity in blood NK cells and provide a new framework for analyzing NK cell responses in health and disease.
Visual Abstract
Introduction
Natural killer (NK) cells are cytotoxic lymphocytes that are crucial for viral defense1 as well as detecting and killing cancer cells. This cell type is therefore of great interest for cancer immunotherapy.2 NK cells have also been implicated in a wide range of diseases: those caused by microbial pathogens,3-5 autoimmune diseases,6-9 reproductive complications,10 and transplantation.11 Understanding how NK cells are involved in these diseases will be informed by better understanding of their different subsets.
Human peripheral blood NK cells, based on surface expression of CD56 and lack of CD3, have long been classified into 2 developmentally related but functionally distinct subsets: CD56dim and CD56bright. CD56dim cells, the largest circulating population, express high levels of activating receptor CD16, lytic granules, and killer-cell immunoglobulin-like receptors (KIRs), called cytolytic cells. CD56bright cells, sometimes termed cytokine-producing cells because they secrete high levels of cytokines and undergo robust proliferation after cytokine stimulation,12 express high levels of NKG2A, CD2, CD62L, and CCR7. An intermediate phenotype between CD56bright and CD56dim has also been proposed; these CD56dimCD94high cells express intermediate levels of CD62L, CD2, and KIR.13,14
Several other phenotypically and functionally distinct subsets have been described. CD56neg cells, found at low frequencies within healthy individuals, are expanded in chronic15,16 and acute viral infections.17 These cells display severely impaired natural cytotoxicity and antibody-dependent cellular cytotoxicity.18 Human cytomegalovirus (CMV) induces the expression of NKG2C19 and CD5720-22 on NK cells, thereby skewing the NK cell repertoire and contributing toward age-associated changes.23,24 These cells express a high frequency of self-KIR and exhibit a highly differentiated phenotype (CD56dimCD16+LILRB1+KIR+NKG2A−) with hallmarks of adaptive immunity; they are therefore commonly termed adaptive or memory NK cells.25,26 In the absence of CMV, CD57 expression marks “mature” NK cells in most human studies, because historical antigenic stimuli are rarely identified. Cytokine-induced memory-like (CIML) NK cells are a recently described subset. First identified in mice27 and later in humans,28,29 these cells show enhanced responsiveness to restimulation, including increased interferon-γ (IFN-γ) and granzyme B production after preactivation with cytokines (interleukin [IL]-12, -15, and -18). CIML NK cells are now being tested in preclinical trials,30 because of their antileukemic properties and in vivo persistence after adoptive transfer.
Thus, several subsets of blood NK cells have been identified in health and disease. By focusing on blood NK cells from healthy human donors, we set out to determine which subsets could be identified by single-cell RNA sequencing (scRNA-seq). Our analysis identified unexpected diversity, encompassing both known and novel types of blood NK cell.
Materials and methods
A full description of experimental and analytical methods can be found in the supplemental Methods.
Sample collection and single-cell preparation
Peripheral blood was obtained with informed consent from 2 healthy donors of Caucasian ancestry (one 60-year-old man and one 60-year-old woman) who were recruited to the National Repository of Healthy Volunteers, The University of Manchester. Ethical approval was obtained from North West Centre for Research Ethics Committee (REC: 99/8/84). Before donation, CMV status was determined with an enzyme-linked immunosorbent assay (human anti-cytomegalovirus IgG ELISA kit, ab108724; Abcam) according to the manufacturer’s instructions. Peripheral blood was similarly acquired by venipuncture from a CMV− female 40-year-old donor from Texas Children’s Hospital (Protocol H-30487) and from 2 anonymous CMV seropositive donors (New York Blood Center, New York, NY).
Peripheral blood mononuclear cells were isolated from fresh blood collected into EDTA-coated tubes (Fisher Scientific) by density gradient centrifugation (Ficoll-Paque Plus; Amersham Pharmacia Biotech) or from leukocyte reduction system chambers through Ficoll density centrifugation. Leukocyte reduction system chambers were purchased from the New York Blood Center. Primary human NK cells were isolated by negative magnetic bead selection (Miltenyi Biotec or Stemcell Technologies), into medium alone (Dulbecco’s modified Eagle medium, 30% F12 Ham, 10% human serum, 1% nonessential amino acids, 1 mM sodium pyruvate [Sigma], 2 mM l-glutamine, 50 U/mL penicillin streptomycin, and 50 µM 2-mercaptoethanol [Gibco]), or into medium supplemented with 200 U/mL recombinant human IL-2 (Roche) for 4 hours at 37°C in 5% CO2.
To assess purity, freshly isolated NK cells were washed in 1% fetal bovine serum/phosphate-buffered saline (PBS) and blocked with 1% human AB serum (Sigma-Aldrich) for 15 minutes at 4°C before staining. NK cells were stained for 20 minutes at 4°C with viability dye (Zombie NIR; Biolegend), phycoerythrin-labeled anti-CD3 monoclonal antibody (mAb; RRID:AB_314062), fluorescein isothiocyanate–labeled anti-CD14 mAb (RRID:AB_314186), AF647-labeled anti-CD19 mAb (RRID:AB_389335), and BV421-labeled anti-CD56 mAb (RRID:AB_11218798) or isotype-matched control mAbs (phycoerythrin-labeled IgG1k (RRID:AB_326435), fluorescein isothiocyanate–labeled IgG2a (RRID:AB_326458), AF647-labeled IgG1k (MOPC-21; Biolegend), and BV421-labeled IgG1k (MOPC-21; Biolegend). Finally, samples were washed in 1% fetal bovine serum/PBS, fixed in 2% paraformaldehyde/PBS for 10 min, assessed by flow cytometry (BD FACS Canto II flow cytometer, BD Biosciences), and then analyzed (FlowJo v 10 software).
Library construction and sequencing
Individually barcoded scRNA-seq libraries were prepared using the Chromium Single Cell Controller and Chromium Single Cell 3′ Library and Gel Bead Kit v2 or v1 kit (10X Genomics, Pleasanton, CA). All steps were performed according to the manufacturers’ instructions. Final libraries were sequenced on 1 flow cell of an Illumina HiSeq 4000 system (Illumina, San Diego, CA) with a read length of 26 bp for read 1 (cell barcode and unique molecule identifier [UMI]), 8-bp i7 index read (sample barcode), and 98 bp for read 2 (RNA read), to yield ∼1.25 billion reads per sample (208 000 reads per cell), or on the Illumina NextSeq500.
Data analysis
Raw FASTQ files were processed using CellRanger (v 2.0.1) to map reads against human genome 38 as a reference, filter out unexpressed genes, and count barcodes and UMIs. After alignment, more in-depth quality control, normalization, and analyses were conducted with Seurat (v 2.3.0)31 in R (v 3.3). Only data from high-quality cells were used for further analysis.
For comparative analysis, we used the Seurat alignment strategy31 to identify common cell types across the different donors and conditions. To do this, we calculated highly variable genes within each data set (supplemental Table 2), took the top 2000 per data set and then further selected 1501 genes, which were highly variable in at least 2 data sets; these 1501 genes were used in the canonical correlation analysis (CCA), to identify shared structures across data sets. Next, cells with expression profiles that were not explained by low-dimensional CCA vs low-dimensional principal components analysis (greater than twofold difference) were excluded from the data set and outputted for further investigation. The CCA subspace was then aligned and the cells were clustered by using a shared nearest neighbor modularity, optimization-based clustering algorithm32 and visualized with t-distributed stochastic neighbor embedding (tSNE). Clustering was achieved by using 25 components from the CCA dimensionality reduction, identified by means of the shared correlation strength and using a resolution of 0.8.
To identify cluster-specific markers conserved across stimulation conditions, we determined differential expression for each cluster vs all remaining cells and within each cluster, and the P values were combined by the Wilkinson’s method from the metap R package. Differential expression within a cluster, determined by the Wilcoxon rank sum test, was used to identify markers between stimulatory conditions.
For comparison with published data, each cell was scored based on the expression of genes associated with known NK cell populations or signatures. In brief, mean expression values for each gene of interest within a signature were calculated for each cell, and the aggregated expression of control genes was subtracted, with the result indicating whether a gene’s expression was upregulated or downregulated vs the average. Genes within the signature were binned according to average expression levels, and the control genes were randomly selected from each bin.
Enrichment analysis was conducted by using GeneOntology (GO; http://www.geneontology.org/).33,34 All conserved markers, either upregulated or downregulated within a cluster, with an adjusted P < .05 (in unstimulated cells) were selected for analysis. Enrichment scores (P values) for the selected GO terms were plotted as −log10(P).
Results
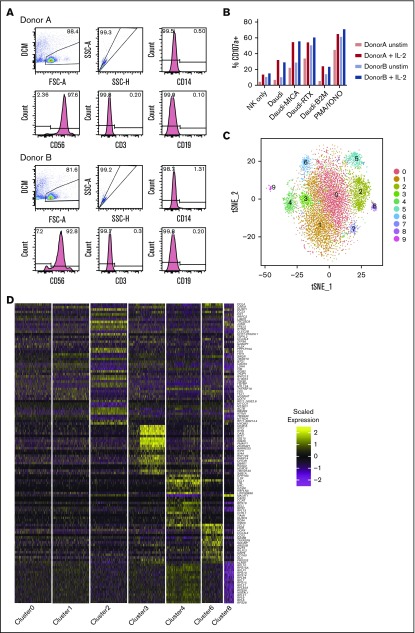
Transcriptional heterogeneity was investigated in blood NK cells from 2 CMV− healthy donors (Figure 1A). NK cells from both donors were treated with IL-2 for 4 hours or left untreated, to gain insight into how different subsets respond to this commonly used cytokine stimulation. Confirming their expected cytolytic potential and cytokine production, cells degranulated in response to HLA-deficient target cells and less to HLA+ cells (Figure 1B) and produced IFN-γ (supplemental Figure 1).
Figure 1.
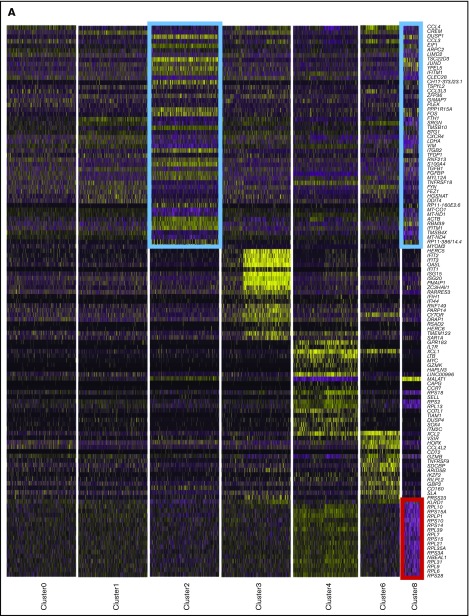
Single-cell sequencing of healthy peripheral blood NK cells. (A) Flow cytometry plots of negatively isolated human blood NK cells from both donors, showing the gating of live singlet cells and then showing the expression of CD14 (monocyte marker). On CD14− cells, expression of CD56 (NK cell marker), CD3 (T-cell marker), and CD19 (B-cell marker) is shown. (B) Total percentage of CD107a+ cells on negatively isolated bulk NK cells after a 5-hour culture, alone (NK only) or with the B lymphoblast cell line Daudi; Daudi cells transfected to express major histocompatibility (MHC) class I–related chain A (Daudi-MICA) or β2m (Daudi-β2m); or rituximab-coated Daudi (Daudi-RTX) or NK cells alone stimulated with phorbol myristate acetate/ionomycin (PMA/IONO). NK cells from both donors are shown separately, with and without IL-2 treatment. (C) tSNE, 2-dimensional plot of 8462 individual NK cells, with clusters (0-9) identified using unsupervised hierarchical clustering. (D) Heat map of top 20 markers distinguishing each NK cell cluster (n = 120 unique genes), identified by differential expression analysis and showing a maximum of 500 genes per cluster, excluding clusters 5, 7, and 9. Cells are plotted in columns, and genes are shown in rows. Gene expression is color coded, using a scale based on z-score distribution. (E) Same tSNE plot as shown in panel C (but excluding clusters 5, 7, and 9), showing the expression of the top 2 markers which distinguish each cluster. Expression is color coded from blue (low) to red (high) and cells positively expressing a marker are brought toward the front of the plot.
In total, we sequenced 8905 cells (4992 unstimulated and 3913 IL-2 stimulated). On average, a median of 935 genes and 305 823 reads per unstimulated cell and 932 genes and 388 361 reads per stimulated cell were detected. Using CCA resulted in a loss of 257 cells that had a lower median gene count compared with the remaining cells (897 vs 940). This left 8462 individual cells (4747 unstimulated and 3715 stimulated) for further analysis.
Seven subsets of NK cells identified by using unsupervised clustering
Unsupervised clustering identified 10 subsets in total (Figure 1C). Cells from all 4 conditions contributed to each cluster (supplemental Figure 2A-B), suggesting that clustering was independent of donor-to-donor variability and stimulatory condition. Furthermore, clustering was not significantly influenced by the cell cycle (supplemental Figure 2C).
Differential expression analysis identified conserved markers per cluster, irrespective of stimulatory condition (supplemental Table 3). A small cluster, representing 1% of the total (cluster 9; Figure 1C), uniquely expressed IFI30, TNFAIP2, CLEC7A, and CD86 (supplemental Table 3), identifying them as antigen-presenting cells (APCs). Another small population (1.97% of the total) expressed CD3E, CD3D, and CD3G and major histocompatibility complex class II molecules (HLA-DPB1, HLA-DQB1, and HLA-DRB1), indicative of T cells or NKT cells (cluster 7; Figure 1C). Of note, CD3 staining by flow cytometry was negligible (Figure 1A), suggesting that this population may represent activated NKT cells that have downregulated the T-cell receptor complex.35 In addition, a population (cluster 5; Figure 1C) representing 5.1% of the total expressed a disproportionate number of heat shock proteins and DNA repair genes, including HSPA1B, HSPA1A, and DNAJB1 (supplemental Table 3). These stressed cells are perhaps a result of the isolation process. These 3 populations (clusters 5, 7, and 9) were excluded from further analysis, leaving 7 NK cell clusters for investigation.
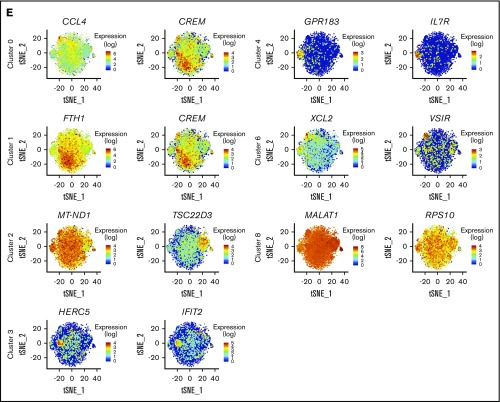
For each remaining cluster, the top 20 defining markers were visualized (Figure 1D). Apart from the 2 largest, denoted 0 and 1 (each having a similar core profile with some distinguishing features), the remaining clusters are clearly distinct. The top 2 markers associated with each cluster were further visualized (Figure 1E). The majority were expressed in a cluster-specific manner, with a few examples of markers shared between clusters. CREM, for example, was highly expressed in cluster 0, but expression was even higher in cluster 1. Together, these data identified 7 NK cell subsets.
Of the conventional CD56dimCD16+ NK cells, only a subset of terminally differentiated NK cells has a distinctive transcriptional signature
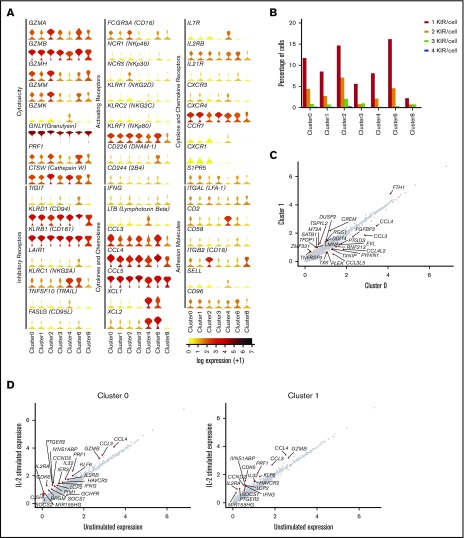
Next, we related these clusters to known NK cell subsets, using canonical markers related to NK cell activation, inhibition, cytotoxicity, and cytokine secretion (Figure 2A). Clusters 0, 1, and 2 represent the largest populations, comprising 34.6%, 28.7%, and 12.2% of the total, respectively. They all share relatively high expression of FCGR3A (CD16) and cytolytic molecules, variable expression of activating/inhibitory receptors, and high levels of chemokines (Figure 2A). Of the assigned KIRs (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, KIR3DL3, and KIR2DL4), clusters 0, 1, and 2 possessed a high proportion of KIR+ cells (Figure 2B), which suggests that those clusters comprise the well-established bulk population of CD56dimCD16+ NK cells.
Figure 2.
Among CD56dim CD16+ NK cells is a distinct subset of CD57+ NK cells. (A) Expression distribution of each cluster (violin plots) using unstimulated cells only, looking at canonical human NK cell markers, grouped by cytotoxicity, inhibitory and activating receptors, cytokines and chemokines, cytokine and chemokine receptors, and adhesion molecules. The shape represents the distribution of cells based on their log(+1) expression values. The color scale represents the mean expression. (B) Percentage of individual NK cells expressing 1, 2, 3, or 4 different KIRs across each cluster, calculated using the full data set of 8462 cells. (C) Comparison of average gene expression values for cluster 0 and cluster 1 in unstimulated cells only. Genes with fold change >0.4 are highlighted. (D) Comparison of average gene expression values for clusters 0 and 1 between unstimulated and IL-2–stimulated cells. Genes with a fold change >0.5 and Bonferroni-corrected P < .05 are highlighted. (E) Selected GO terms using all conserved markers upregulated or downregulated within the individual clusters with an adjusted P < .05. (F) Dot plot (left) of selected markers of interest within unstimulated cells only across the different clusters. The size of the dot represents the percentage of cells expressing the markers, and the color encodes the average scaled expression values. Heat map (right) of the same markers of interest within unstimulated cells only. Clusters are plotted in columns, and genes are shown in rows. Gene expression is color coded using average scaled expression values per cluster, based on a z-score distribution, ranging from low expression (purple) to high expression (yellow). (G) Comparison of average gene expression values for cluster 2 between unstimulated and IL-2–stimulated cells. Genes with a fold change >0.5 and Bonferroni-corrected P < .05 are highlighted. (H) Selected GO terms using all conserved markers upregulated or downregulated within this cluster with an adjusted P < .05.
Clusters 0 and 1 were closely related, evidenced by the high correlation of 0.99 (supplemental Figure 3). In a direct comparison of unstimulated cells (Figure 2C), cluster 0 was found to have significantly higher expression of chemokines (CCL4, CCL3, CCL4L2, and CCL3L3), whereas cluster 1 expressed slightly more TNFRSF9 (4-1BB). Thus, these 2 clusters of CD56dimCD16+ NK cells most likely represent populations of subtly different maturity, rather than distinct cellular subsets.
There was also little to differentiate how these 2 clusters responded to IL-2 stimulation (Figure 2D). Many genes were upregulated, which would be expected with cytokine treatment, including cell-cycle markers (CCND1 and CDK6), IFNG and IL2RA, and regulators of cytokine signaling (SOCS1). In terms of cluster-specific changes, cluster 0 showed increased expression of several genes. Cluster 1 also displayed upregulation, but the fold-change was just below our labeling threshold. GO analysis highlighted enrichment in chemotaxis for cluster 0 and cell activation; whereas cluster 1 was enriched for a broader range of biological processes (Figure 2E). Altogether the data support that clusters 0 and 1 were very similar, each with a subtly distinct CD56dimCD16+ phenotype, such that cluster 1 exhibited a more activated phenotype.
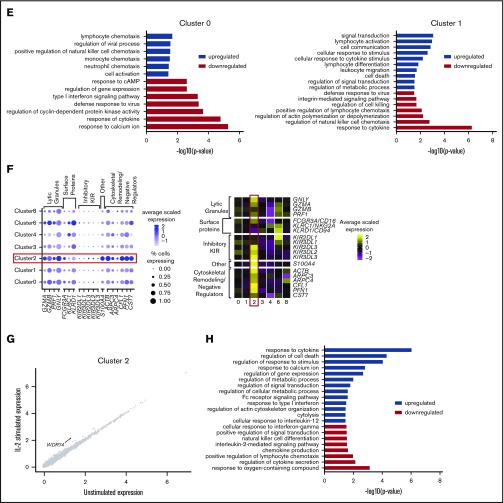
Compared with clusters 0 and 1, cells in cluster 2 expressed more lytic granule genes, FCGR3A (CD16; Figure 2F) and a higher proportion of KIR+ cells (Figure 2B), which suggests that these cells are terminally differentiated CD57+ NK cells that are highly cytotoxic, express CD57, frequently express inhibitory KIR, and restrict proliferative activity to cytokines.36,37 Another defining marker is S100A4 (supplemental Table 3), which increases with NK cell maturation, with highest expression seen on CD57+ NK cells.38 This cluster also highly expressed genes associated with cytoskeletal remodeling, such as ACTB, ARPC3/4, and CFL1, and negative cytolytic regulators, such as CST7 and PFN1, which are needed for optimal cytotoxicity39 (Figure 2F). Although CD57 is underrepresented in our data set, strong evidence that these cells are terminally differentiated is that these cells do not respond to IL-2 stimulation36,37 (Figure 2G). GO analysis further supported these observations, with enrichment for cytolysis and Fc receptor signaling, whereas IL-2–mediated signaling and NK cell differentiation were downregulated (Figure 2H). Thus, cluster 2 represents terminally differentiated CD56dimCD16+CD57+ NK cells.
CD56bright NK cells respond most potently to IL-2 stimulation
Cluster 4 comprised 5.5% of the total and had a distinct transcriptional profile at an extreme end of the spectrum, revealed in diffusion map analysis (supplemental Figure 4). This cluster exclusively expressed GPR183, IL7R, LTB, GZMK, CD62L, and CCR7 (supplemental Table 3). Furthermore, these cells expressed high levels of CD2 and KLRC1 (NKG2A), low levels of FCGR3A (CD16), and little or no PRDM1 (which decreases cytokine production and limits proliferation in CD56dim cells)40,41 (Figure 3A). This cluster also displayed a diminished cytolytic signature (Figure 2A). Together, these markers identify cluster 4 as CD56bright NK cells.42
Figure 3.
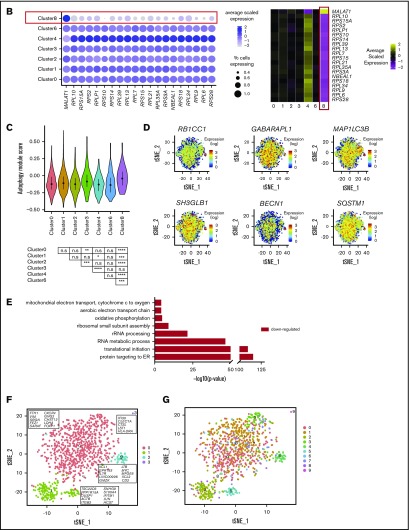
CD56bright NK cells respond most potently to IL-2 stimulation. (A) Dot plot (left) of selected markers of interest within unstimulated cells only (columns) across the different clusters (rows). The size of the dot represents the percentage of cells expressing the markers, and the color encodes the average scaled expression values. Heat map (right) of the same markers of interest within unstimulated cells only. Clusters are plotted in columns, and genes are shown in rows. Gene expression is color coded using average scaled expression values per cluster, based on a z-score distribution, ranging from low expression (purple) to high expression (yellow). (B) Comparison of average gene expression values for cluster 4 between unstimulated and IL-2–stimulated cells. Genes with a fold change >0.5 and Bonferroni-corrected P < .05 are highlighted. (C) Selected GO terms using all conserved markers upregulated or downregulated within this cluster with an adjusted P < .05. (D) Module scores for each NK cell cluster at the single-cell level, defined using the top 100 markers from bulk expression profiles of sorted CD56dimCD16+ and CD56brightCD16− NK cells.47 Module scores were calculated for unstimulated cells only. CD56dim module score (left), CD56bright module score (middle), and custom CD56bright module score, excluding CTSW, DUSP1, JUN, FOS and CD69 (right). Violin plots represent the distribution of the module scores for each cluster, and the error bars represent the median and interquartile range. One-way analysis of variance with Bonferroni’s multiple comparison. Nonsignificant (n.s) P > .05; *P < .03; **P < .02; ***P < .0002; ****P < .0001.
Human CD56bright NK cells are transcriptionally similar to innate lymphoid cells (ILCs).43 To determine whether ILCs contributed to cluster 4, negatively isolated NK cells were stained for CD56, CD127, and perforin in separate healthy donors. ILCs in healthy blood express CD127 (IL7R) and generally lack CD56 and perforin.44 In contrast, here, most of the CD127+ cells stained positively for both CD56 and perforin (supplemental Figure 5A). Furthermore, CD127+ non-NK cell ILC markers reported by scRNA-seq45 were expressed in a very small fraction of cells and were expressed across all clusters and at low levels of expression. Thus, this subset represents a specific subset of bona fide NK cells, rather than other ILCs (supplemental Figure 5B-C).
This cluster responded the most potently to IL-2 stimulation. Some of the genes influenced were those also effected in the 2 CD56dimCD16+ populations (clusters 0 and 1) (Figure 3B). In addition, however, upregulation of β-actin (ACTB), lymphotoxin α/β (LTA/LTB), and TNFSF10 (TRAIL) was also observed. Transcripts for IL7R decreased (as previously shown46), as well as CXCR4, ZFP36L2, and ATM. Consistent with the observation that cluster 4 represents CD56bright NK cells, GO analysis indicated enrichment for response to cytokine and cytokine-mediated signaling pathway terms, while concurrently displaying a diminished cytolytic profile (Figure 3C).
Module scores calculated at a single-cell level using bulk expression profiles of sorted CD56dim and CD56bright NK cells47 clearly showed that cluster 4 most resembled CD56bright NK cells, whereas the remaining clusters resembled CD56dim NK cells (Figure 3D: left and middle plots). Surprisingly, terminally differentiated NK cells (cluster 2) displayed features of CD56bright NK cells (Figure 3D; middle plot), pinpointed to 5 markers (CTSW, DUSP1, FOS, JUN, and CD69), which were upregulated in a proportion of cells in cluster 2 (supplemental Figure 6). CD69, DUSP1, FOS, and JUN are either early activation markers or immediate early response genes (IEGs), and this analysis indicates that these markers overlap for activated versions of both CD56dim and CD56bright NK cells.
A fraction of peripheral blood NK cells is a population of type I IFN–responding cells
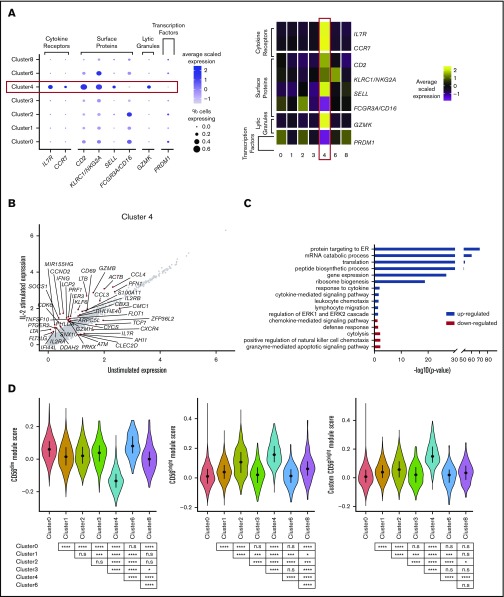
So far, this analysis has identified the more commonly described NK cell subsets: 2 populations of CD56dimCD16+ cells, a population of terminally differentiated NK cells, and a CD56brightCD16− population, leaving 3 clusters (clusters 3, 6, and 8). Of these, cluster 3 was the largest, representing 5.9% of the total. These cells expressed less FCGR3A (CD16) and cytolytic markers than did the CD56dim cells (clusters 0, 1, and 2; Figure 4A), but more than the CD56bright cells (cluster 4), suggesting a diminished cytolytic capacity. Such a phenotype is attributed to nonclassic CD56negCD16+CD7+ NK cells, which also lack SIGLEC7 expression, display reduced expression of KLRC1 (NKG2A) and natural cytotoxicity receptors, but still release substantial amounts of chemokines.18 In our data, cluster 3 expressed substantial amounts of CCL3 and CCL4 and significantly more CCL5 than all other clusters (Figure 4A).
Figure 4.
A fraction of peripheral blood NK cells is a population of type I IFN-responding cells. (A) Dot plot (left) of selected markers of interest within unstimulated cells only (columns) across the different clusters (rows). The size of the dot represents the percentage of cells expressing the markers, and the color encodes the average scaled expression values. Heat map (right) of same markers of interest within unstimulated cells only. Clusters are plotted in columns, and genes are shown in rows. Gene expression is color coded using average scaled expression values per cluster, based on a z-score distribution, ranging from low expression (purple) to high expression (yellow). (B) Comparison of average gene expression values (calculated in unstimulated cells only) for cluster 3 and grouped CD56dim clusters (clusters 0, 1, and 2). The markers shown differentiate CD56neg and CD56dim blood NK cells by flow cytometry.48 *Markers that are not consistent with flow cytometry. (C) Comparison of average gene expression values for cluster 3 between unstimulated and IL-2–stimulated cells. Genes with a fold change >0.5 and Bonferroni-corrected P < .05 are highlighted. (D) Heat map (right) of top 20 markers which define cluster 3 within unstimulated cells only. Format and expression scale are as described for the heat map in panel A. (E) Selected GO terms using all conserved markers upregulated or downregulated within this cluster with an adjusted P < .05.
Proteomic analysis of CD56neg and CD56dim blood NK cells from healthy donors48 found marked similarities between the 2 (principal components analysis correlation 0.92), as also observed at the transcript level (supplemental Figure 3). Across 14 protein markers reported to have decreased expression on CD56neg cells, we observed that 12 of these also showed a decrease in transcript abundance (Figure 4B). These CD56neg cells respond to IL-2, similarly to how CD56dim populations respond (Figure 4C).
Strikingly, the top 20 defining markers for CD56neg cells (cluster 3), are almost all exclusively type I IFN-stimulated genes (Figure 4D). A strong IFN signature has been reported in another single-cell analysis of NK cells published recently.49 Those authors termed these NK cells “inflamed” and found them in the bone marrow. Our data imply that such NK cells are also present in blood. Consistent with this finding, GO analysis indicates enrichment for pathways involved in defense response to viruses, response to type I IFN, and diminished cytolysis (Figure 4E). A subunit of the type I IFN receptor, IFNAR2, is also more highly expressed on a subset of cluster 3 compared with all other cells (supplemental Table 3). Upregulation of IFNAR can occur as result of IL-12 and polyIC stimulation.50 Since CD56neg cells (cluster 3) express IFNAR2 and large amounts of transcripts for CCL3, CCL4, and CCL5, it is also possible that these cells are particularly important for cross-talk with plasmacytoid dendritic cells, which express the ligand for CCL3, CCL4, and CCL5.
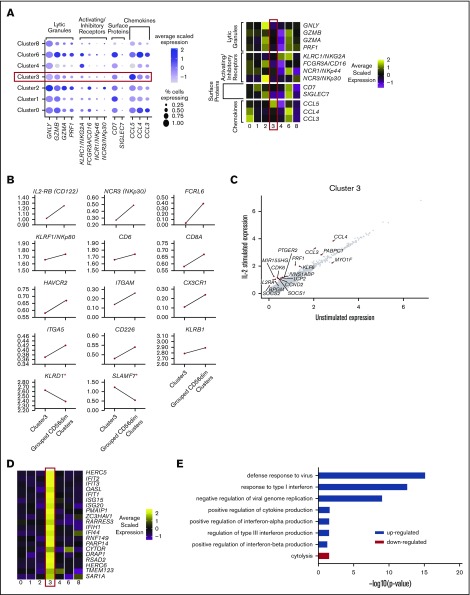
A small fraction of peripheral blood NK cells displays a cytokine-induced memorylike phenotype
Cluster 6, representing 3.7% of the total, displays a phenotype that is something of a hybrid between CD56dim and CD56bright NK cells. Akin to CD56bright cells, these cells express high levels of KLRC1 (NKG2A), IL2RA, XCL1, and XCL2 (Figure 5A). However, they also express FCGR3A (CD16) (to a lesser extent than CD56dim cells), GZMA, PRF1, and GNLY (at levels similar to those expressed by CD56dim cells); with even higher GZMB. This cluster also contains a high proportion of KIR+ cells (Figure 2B), which is reminiscent of CD56dimCD94high intermediary NK cells.14 However, cluster 6 did not express transcripts at a level to match previously identified protein markers14 (supplemental Figure 7).
Figure 5.
A small fraction of peripheral blood NK cells displays a cytokine-induced memory-like phenotype. (A) Dot plot (left) of selected markers of interest within unstimulated cells only (columns) across the different clusters (rows). The size of the dot represents the percentage of cells expressing the markers, and the color encodes the average scaled expression values. Heat map (right) of the same markers of interest within unstimulated cells only. Clusters are plotted in columns, and genes are shown in rows. Gene expression is color coded, using average scaled expression values per cluster, based on a z-score distribution, ranging from low expression (purple) to high expression (yellow). (B) Expression distribution of each cluster and stimulation condition (violin plots) specifically of granzyme B (GZMB) expression. The shape represents the distribution of cells based on their log(+1) expression values. The color scale represents the mean expression. (C) Module score for each NK cell cluster at the single-cell level, defined using CIML markers.28 Module scores were calculated for unstimulated cells only. Violin plots represent the distribution of the module scores for each cluster, and the error bars represent median and interquartile range. One-way analysis of variance with Bonferroni’s multiple comparison. Nonsignificant (n.s) P > .05; *P < .03; **P < .02; ***P < .0002; ****P < .0001. (D) Comparison of average gene expression values for cluster 6 between unstimulated and IL-2–stimulated cells. Genes with a fold change >0.5 and Bonferroni-corrected P < .05 are highlighted. (E) Selected GO terms using all conserved markers upregulated within this cluster with an adjusted P < .05. No markers were downregulated within this cluster at this significance threshold.
The most prominent feature of this cluster is significant upregulation of GZMB after IL-2 treatment (Figure 5B). This is an important feature of enhanced-recall responses observed with in vitro differentiated CIML NK cells. Module score analysis using markers reported by time of flight cytometry, comparing CIML to control cells maintained in IL-15,28 showed that cluster 6 most resembles CIML NK cells (Figure 5C). Upon IL-2 stimulation, several markers are upregulated that are common to other NK cell subsets (Figure 5D). Those specifically upregulated include CMTM6 (which positively regulates PD-L1 expression51,52), RAB11FIB1, and SAMSN1. Although CIML NK cells are not new to the NK cell field, it is new to suggest that cells with features of CIML NK cells are constitutively present in circulating human blood.
GO analysis indicates enrichment of markers within a variety of pathways, including regulation of cell activation and cell communication and regulation of IL-10 and -12 production and granzyme B production (Figure 5E). Overall, this analysis demonstrated that cells within cluster 6 have a strongly activated phenotype and are primed to respond to IL-2 stimulation.
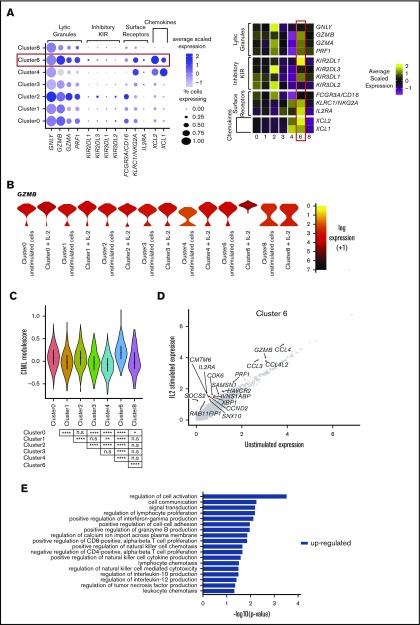
A small, novel population of blood NK cells exhibits loss of ribosomal expression
Cluster 8 makes up 1.6% of the total population and displays a phenotype not documented previously. It resembles terminally differentiated cells (cluster 2; Figure 6A; blue boxes) and both are remarkably similar according to correlation analysis (supplemental Figure 3). However, the definitive feature of this cluster is a striking loss of ribosomal expression (Figure 6A; red box, Figure 6B). Under nutrient starvation, ribosomes are targeted for autophagy-mediated degradation, as they are a major source of amino acids and nucleotides.53,54 Here, we observed modest enrichment for autophagy-related markers within cluster 8 (Figure 6C). In particular, a proportion of cells showed increased expression of RB1CC1 (ATG17/FIP200), GABARAPL11 (ATG8), MAP1LC3b (LC3B), SH3GLB1 (BIF-1), BECN1 (BECLIN 1), and SQSTM1 (p62) (Figure 6D). The number of genes expressed (nGene) and number of unique molecular identifiers (nUMIs) were significantly lower in cluster 8 (supplemental Figure 8A). This is reminiscent of cells undergoing senescence or quiescence, but we did not observe any enrichment for genes specifically defining these processes (supplemental Figure 8B). The relative percentage of mitochondrial RNA did not differ across any of the subsets (supplemental Figure 8C; left) and module score analysis using proapoptotic markers were similar across all NK cell subsets, showing that these cells were not simply dying (supplemental Figure 8C; right). There were also no significant changes in this population upon IL-2 stimulation, but this may be related to insufficient power, given the small number of cells (supplemental Figure 8D). GO analysis indicated downregulation of oxidative phosphorylation (Figure 6E). Upon lymphocyte activation, cells switch from oxidative phosphorylation to glycolysis.55 Interestingly, a proportion of cells in cluster 8 overexpressed the same IEG markers as seen in cluster 2, indicating that cells had been recently activated (supplemental Figure 6), which suggests that low ribosomal expression is linked to increased energy demands because of recent cellular activation.
Figure 6.
A small, novel population of blood NK cells exhibits loss of ribosomal expression. (A) Same heat map of the top 20 markers distinguishing each NK cell cluster as shown in Figure 1D, highlighting the similarities (blue boxes) and dissimilarity (red box) between clusters 2 and 8. Cells are plotted in columns, and genes are shown in rows. Gene expression is color coded using a scale based on z-score distribution, ranging from low expression (purple) to high expression (yellow). (B) Dot plot (left) of 20 top markers associated with cluster 8 within unstimulated cells only (columns) across the different clusters (rows). The size of the dot represents the percentage of cells expressing the markers, whereas the color encodes the average scaled expression values. Heat map (right) of the same markers of interest within unstimulated cells only. Clusters are plotted in columns, and genes are shown in rows. The gene expression scale is as in panel A. (C) Module score analysis for each NK cell cluster at the single-cell level, defined using markers of mammalian autophagy.63 Module scores were calculated for unstimulated cells only. Violin plots represent the distribution of the module scores for each cluster and the error bars represent median and interquartile range. One-way analysis of variance with Bonferroni’s multiple comparison. Nonsignificant (n.s) P > .05; *P < .03; **P < .02; ***P < .0002; ****P < .0001. (D) tSNE plot (excluding clusters 5, 7, and 9), showing the expression of selected autophagy markers within the full data set. Expression is color coded from blue (low) to red (high) and cells positively expressing a marker were brought toward the front of the plot. (E) Selected GO terms using all conserved markers downregulated within this cluster with an adjusted P < .05. No markers were upregulated within this cluster at this significance threshold. (F) tSNE, 2-dimensional plot of 1000 NK cells from the unstimulated condition (500 randomly selected from each donor). The analysis was performed using the first 15 principle components and a resolution of 0.4. The top markers positively associated with each cluster are highlighted. (G) The same tSNE plot as in panel F) but cells are color coded according to the cluster identities assigned using the full data set of 8462 cells.
Down-sampling results in 3 clusters of NK cells
A recent scRNA-seq analysis of human blood NK cells identified only 2 clusters.56 In that study, analysis was performed per ∼1000 cells. Although there are differences in read depths between the previous study and this one, the number of cells has been shown to affect clustering to a greater degree.57 We therefore sought to determine whether the extra clusters observed in this study were related to an increase in the number of cells. To achieve this, the analysis was limited to 1000 random cells. Following unsupervised clustering using 15 principal components and a low resolution of 0.4 (as used in the previous study56), we observed 4 clusters (Figure 6F). The smallest, cluster 3, comprised the contaminating APCs, leaving 3 NK cell clusters. We overlaid the original cluster identities assigned using the full data set and found that the contaminating NKT cells (cluster 7), the cells that resemble CIML cells (cluster 6), and the type I IFN–responding cells (cluster 3) clustered with the 2 CD56dimCD16+CD57− populations (clusters 0 and 1) (Figure 6G), whereas the cells highly expressing heat shock proteins (cluster 5) clustered with the terminally differentiated CD57+ population (cluster 2). Overall, this analysis shows that the number of cells profiled is an important consideration of single-cell studies and establishes that our data are consistent with those of a previous work.56 This study revealed additional NK cell subsets because a greater number of cells were analyzed.
Peripheral blood NK cells from CMV+ individuals have several altered features
Having defined 7 populations of NK cells in healthy human blood, we sought to use their transcriptional signatures to understand NK cell diversity in the context of latent CMV infection. Data obtained on blood NK cells isolated from 2 CMV+ but otherwise healthy donors and 3 CMV− healthy donors were combined, looking for populations common across donors and limiting the study to 8000 random cells for comparability to the previous analyses. After unsupervised clustering, using 20 components from the CCA dimensionality reduction and a resolution of 0.7 resulted in 11 clusters (Figure 7A) that included a small population of APCs and a cluster of stressed cells that were removed, leaving 9 NK cell subsets for analysis.
Figure 7.
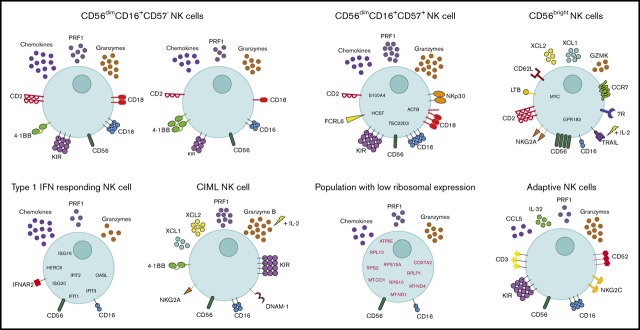
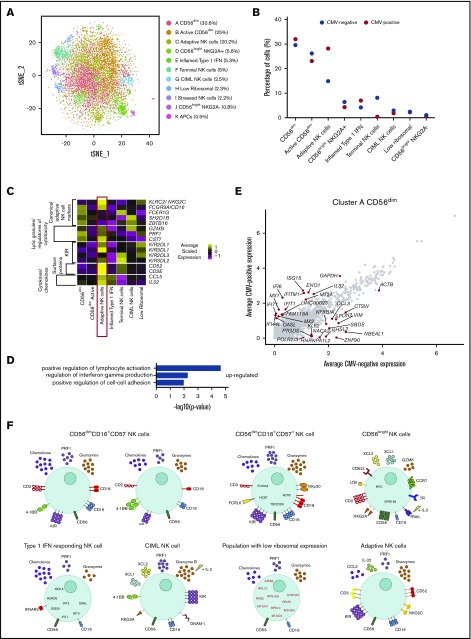
Peripheral blood NK cells from CMV+ individuals have several altered features. (A) tSNE, 2-dimensional plot of 8000 individual NK cells, with clusters (A-K) identified using unsupervised hierarchical clustering. (B) Mean percentage of CMV− and CMV+ cells contributing toward each cluster. (C) Heat map of canonical adaptive NK cell markers and markers that distinguish this cluster from other cells. Clusters are plotted in columns, and genes are shown in rows. Gene expression is color coded using average scaled expression values per cluster, based on a z-score distribution, ranging from low expression (purple) to high expression (yellow). (D) Selected GO terms using all markers upregulated within cluster C, adaptive NK cells, with an adjusted P < .05. (E) Within cluster A, CD56dim NK cells, comparison of average gene expression values for CMV− cells and CMV+ cells. Genes with a fold-change >1 are highlighted. (F) Summary schematic of NK cell populations identified by scRNA-seq. Markers highlighted in red represent downregulation within that population. Of note, the schematic is based on transcript levels that may not entirely correlate with protein expression.
We observed a very small, novel population of cells (Figure 7A; cluster J) which, upon further investigation, came primarily from 1 CMV− donor. These cells resembled CD56bright NK cells (supplemental Figure 9A-C), but lacked expression of KLRC1 (NKG2A) and some other CD56bright NK cell markers, suggesting that they were transitioning or intermediate CD56bright NK cells.14 Strikingly, we identified a population of adaptive NK cells found at substantially increased frequency in CMV+ individuals (Figure 7B; cluster C). A proportion of this cluster displayed higher expression of adaptive NK cell markers: KLRC2 (NKG2C) and KIR genes (Figure 7C). This population also expressed lower levels of FCER1G (FсεR1γ), SH2D1B (EAT-2), and ZBTB16 (PLZF), as shown previously,58 and high expression of cytolytic molecules (GZMB and PRF1) and CST7 (a negative cytolytic regulators), in line with their potent cytotoxic potential. In addition, we observed increased expression of CD3, CD52, and IL-32, which have also been recently observed in adaptive NK cells49 (Figure 7C). GO highlighted enrichment for the regulation of IFN-γ production, again supporting the strong functionality attributed to adaptive NK cells (Figure 7D).
For other NK cell subsets, frequencies altered slightly between the viral states (particularly for the larger populations), most likely to account for the change in adaptive NK cells. Terminally differentiated NK cells were less frequent in this combined analysis, perhaps because of a proportional shift caused by the adaptive NK cells. Inflamed type I IFN NK cells, CIML NK cells, and the low-ribosomal cells (supplemental Figure 9D-F) were found in similar proportions in both CMV− and CMV+ individuals (Figure 7B). Interestingly, across all clusters, there was an upregulation of type I IFN genes and IL-32 in the CMV+ donors; as shown for cluster A (Figure 7E). Of note, IFIT1, OAS1, IFI6, IFITM1, MX2, and IL-32 were increased in expression across all NK cell subsets in CMV+ donors compared with CMV− donors. Thus, CMV impacts both the frequencies of different NK cell subsets and their transcriptional profiles.
Discussion
scRNA-seq is a powerful technique for discovering transcriptional diversity. Toward unraveling NK cell diversity, scRNA-seq studies have been performed in human tonsil NK cells, in comparison with developmentally related ILCs,45 and on matched spleen and blood human NK cells56 or NK cells from human bone marrow and blood.49 However, these studies either did not resolve subsets, only resolved the CD56dim and CD56bright subsets, or were focused on unhealthy individuals or were performed with a relatively small number of blood NK cells. Thus, our objective was to define baseline blood NK cell heterogeneity from healthy individuals in a large number of individual cells, including an assessment of how these populations differentially respond to IL-2 stimulation. By analyzing ∼8000 individual cells, we identified an additional 4 subsets to the commonly identified CD56dim and CD56bright populations, while also observing subdivision within the CD56dimCD16+CD57− compartment.
CD56bright cells have a distinctive phenotype, with their robust capacity to respond to cytokines. Closely associated with this subset was another cytokine-sensitive population: CD56dimNKG2Ahigh cells, which showed enhanced cytotoxicity to IL-2, reminiscent of CIML NK cells. Cytokine-activated responses generated in vivo are functionally distinct from in vitro CIML cells generated with exogenous cytokines.59 Our study highlighted similarities and differences from in vitro generated CIML cells. For example, NKp80 was more highly expressed in our study, but is downregulated on in vitro CIML cells,28 whereas very little NKp30 was observed. Nevertheless, our data support the existence of CIML NK cells, or closely related cells, in peripheral human blood.
At the other end of the spectrum were terminally differentiated CD57+ NK cells, distinguished by their minimal response to IL-2, expressing many transcripts relating to cytolysis and several surface receptors for membrane-bound ligands, such as NKp30 and KIR. This subset seems to be specialized in recognition of pathogens via specific antigens. Multiple subpopulations of CD57+ have been distinguished using time of flight cytometry.60 Our results suggest that, whereas individual CD57+ NK cells each have a specific repertoire of activating and inhibitory receptors, transcriptionally these cells form 1 population.
A subset closely related to CD56dim cells was defined, by its response, not to IL-2, but to type I IFNs. This subset may be specialized for communication with plasmacytoid dendritic cells and T cells through their secretion of type I IFNs and CCL3, CCL4, and CCL5. Recent work in cancer has shown that NK cell–mediated recruitment of dendritic cells to the tumor microenvironment, through the chemokine CCL5 among others, is associated with patient survival.61 It remains to be determined whether these IFN-activated cells are the key to switching the immune response from a tolerogenic response to a cytotoxic one. Overall, this subset looks to have a specialized role in immune responses.
Cluster 8 represents a type of NK cell not previously characterized, resembling terminally differentiated CD57+ cells, but with significantly reduced ribosomal expression. Reduced ribosomal expression is a feature of a certain type of autophagy. First described in yeast cells53 and more recently in mammalian cells,54 under nutrient starvation, ribosomes are targeted for degradation and recycling in a process termed “ribophagy.” GO analysis highlighted a reduction of oxidative phosphorylation, which, when coupled with expression of IEGs, suggests a switch in metabolic states to support cellular activation. This would be consistent with the notion that these cells degrade ribosomes as a source of biosynthetic material to support cellular activation and function.
We also report subdivision within the CD56dimCD16+CD57− population. There were higher levels of 4-1BB on one subset and greater amounts of chemokines and a higher proportion of KIR+ cells on the other. However, their responses to IL-2 were very similar, suggesting that these represent subtle subdivisions. Education may play a role in setting the transcriptome of blood NK cells, but in a recent study, Goodridge et al.62 concluded that education was controlled at the protein level and that transcriptional differences between educated and noneducated cells could not be observed using sorted populations and microarray technology.
Our data showed that CMV altered the frequencies of particular NK cell subsets, as well as gene regulation within each subset. The most striking difference was an increased frequency of adaptive NK cells in CMV+ donors. A small proportion of adaptive NK cells were still found in CMV− individuals. A lower proportion of the adaptive cells within CMV− individuals were NKG2C+ and, on average, expressed less NKG2C compared with CMV+ individuals, consistent with adaptive NK cells arising from infections other than CMV, but with CMV being a particularly strong driver for NK cell adaptive immunity.
In summary, in this study NK cells within healthy individuals displayed an exquisite division of labor (summarized in Figure 7F). The expansion, contraction, or alteration, of these subsets in pathological settings will give insight into NK cell responses during disease progression.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the Genomic Technologies Core Facility, Faculty of Biology, Medicine, and Health (FMBH), The University of Manchester for performing scRNA-seq; Gareth Howell and the Manchester Collaborative Centre for Inflammation Research (MCCIR) Flow Cytometry Facility; and Clair Gardiner and Matt Hepworth for advice. The authors also acknowledge help from IT Services and the use of the Computational Shared Facility at The University of Manchester.
This work was supported by the Medical Research Council (MC_PC_15072) a Wellcome Trust investigator Award (110091), Versus Arthritis fellowship (21745), the NIHR Manchester Biomedical Research Centre, Versus Arthritis grant 20385, the Manchester Academic Health Sciences Centre, and the MCCIR (funded by a precompetitive open-innovation award from GSK, AstraZeneca, and The University of Manchester), National Institutes of Health, National Institute on Aging grant AI130760, and an American Society for Hematology Junior Scholar Award (E.M.M.).
Footnotes
Authorship
Contributions: All authors conceived the project and designed the experiments; S.L.S., P.R.K., J.D.W., K.B.S., E.M.M., B.M., S.S.C., A.H., R.S., M.L.S., and K.A. performed the experiments; S.L.S., P.R.K., P.M., E.M.M., S.S., E.H.S., A.H., R.S., M.L.S., K.A., and Y.-C.W. analyzed the data; S.L.S., P.R.K., and D.M.D. wrote the manuscript; and all authors critically appraised and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.S.C. is Qiagen, Houston, TX.
Correspondence: Daniel M. Davis, Manchester Collaborative Centre for Inflammation Research, University of Manchester, 46 Grafton St, Manchester M13 9NT, United Kingdom; e-mail: daniel.davis@manchester.ac.uk.
References
- 1.Mace EM, Orange JS. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev. 2019;287(1):202-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer [published correction appears in Nat Rev Innunol. 2018;18:726]. Nat Rev Immunol. 2018;18(11):671-688. [DOI] [PubMed] [Google Scholar]
- 3.Ogbomo H, Mody CH. Granule-dependent natural killer cell cytotoxicity to fungal pathogens. Front Immunol. 2017;7:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf AS, Sherratt S, Riley EM. NK cells: Uncertain allies against malaria. Front Immunol. 2017;8:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horowitz A, Stegmann KA, Riley EM. Activation of natural killer cells during microbial infections. Front Immunol. 2012;2:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanvillard C, Jacolik RF, Infante-Duarte C, Nayak RC. The role of natural killer cells in multiple sclerosis and their therapeutic implications. Front Immunol. 2013;4:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogel LA, Yokoyama WM, French AR. Natural killer cells in human autoimmune disorders. Arthritis Res Ther. 2013;15(4):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gianchecchi E, Delfino DV, Fierabracci A. NK cells in autoimmune diseases: Linking innate and adaptive immune responses. Autoimmun Rev. 2018;17(2):142-154. [DOI] [PubMed] [Google Scholar]
- 9.Poli A, Michel T, Patil N, Zimmer J. Revisiting the Functional Impact of NK Cells. Trends Immunol. 2018;39(6):460-472. [DOI] [PubMed] [Google Scholar]
- 10.Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest. 2014;124(5):1872-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benichou G, Yamada Y, Aoyama A, Madsen JC. Natural killer cells in rejection and tolerance of solid organ allografts. Curr Opin Organ Transplant. 2011;16(1):47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity. 2017;47(5):820-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Mao HC, Wei M, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115(2):274-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu PF, Hultin LE, Hultin P, et al. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10(3):331-340. [PubMed] [Google Scholar]
- 16.Gonzalez VD, Falconer K, Björkström NK, et al. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. 2009;183(10):6612-6618. [DOI] [PubMed] [Google Scholar]
- 17.Björkström NK, Lindgren T, Stoltz M, et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208(1):13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Björkström NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 2010;31(11):401-406. [DOI] [PubMed] [Google Scholar]
- 19.Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104(12):3664-3671. [DOI] [PubMed] [Google Scholar]
- 20.Foley B, Cooley S, Verneris MR, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012;119(11):2665-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Vergès S, Milush JM, Schwartz BS, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. 2011;108(36):14725-14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Béziat V, Dalgard O, Asselah T, et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol. 2012;42(2):447-457. [DOI] [PubMed] [Google Scholar]
- 23.Goodier MR, White MJ, Darboe A, et al. Rapid NK Cell Differentiation in a Population with Near-Universal Human Cytomegalovirus Infection Is Attenuated by NKG2C Deletions. Blood. 2014;124(14):2213-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solana R, Campos C, Pera A, Tarazona R. Shaping of NK cell subsets by aging. Curr Opin Immunol. 2014;29(1):56-61. [DOI] [PubMed] [Google Scholar]
- 25.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells [published correction appears in Nature. 2009;457(7233):1168]. Nature. 2009;457(7229):557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16(2):112-123. [DOI] [PubMed] [Google Scholar]
- 27.Cooper MA, Elliott JM, Keyel PA, et al. Cytokine-induced memory-like natural killer cells. 2009;106(6):1915-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romee R, Rosario M, Berrien-Elliott MM, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. 2016;8(357):357ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romee R, Schneider SE, Leong JW, et al. Cytokine activation induces human memory-like NK cells. Blood. 2012;120(24):4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baragaño Raneros A, López-Larrea C, Suárez-Álvarez B. Acute myeloid leukemia and NK cells: two warriors confront each other. Oncoimmunology. 2018;8(2):e1539617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waltman L, Van Eck NJ. A smart local moving algorithm for large-scale modularity-based community detection. Eur Phys J B. 2013;86(11):471. [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, et al. ; The Gene Ontology Consortium . Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbon S, Douglass E, Dunn N, et al. ; The Gene Ontology Consortium . The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330-D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson MT, Johansson C, Olivares-Villagómez D, et al. The response of natural killer T cells to glycolipid antigens is characterized by surface receptor down-modulation and expansion. Proc Natl Acad Sci USA. 2003;100(19):10913-10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Vergès S, Milush JM, Pandey S, et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116(19):3865-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Björkström NK, Riese P, Heuts F, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116(19):3853-3864. [DOI] [PubMed] [Google Scholar]
- 38.Urlaub D, Höfer K, Müller M-L, Watzl C. LFA-1 Activation in NK Cells and Their Subsets: Influence of Receptors, Maturation, and Cytokine Stimulation. J Immunol. 2017;198(5):1944-1951. [DOI] [PubMed] [Google Scholar]
- 39.Mace EM, Dongre P, Hsu HT, et al. Cell biological steps and checkpoints in accessing NK cell cytotoxicity. Immunol Cell Biol. 2014;92(3):245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins PL, Cella M, Porter SI, et al. Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell. 2019;176(1-2):348-360.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith MA, Maurin M, Cho HI, et al. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185(10):6058-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michel T, Poli A, Cuapio A, et al. Human CD56bright NK Cells: An Update. J Immunol. 2016;196(7):2923-2931. [DOI] [PubMed] [Google Scholar]
- 43.Allan DSJ, Cerdeira AS, Ranjan A, et al. Transcriptome analysis reveals similarities between human blood CD3- CD56bright cells and mouse CD127+ innate lymphoid cells. Sci Rep. 2017;7(1):3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spits H, Bernink JH, Lanier L. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol. 2016;17(7):758-764. [DOI] [PubMed] [Google Scholar]
- 45.Björklund AK, Forkel M, Picelli S, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing [published correction appears in Nat Immunol. 2016;17:740]. Nat Immunol. 2016;17(4):451-460. [DOI] [PubMed] [Google Scholar]
- 46.Xue H-H, Kovanen PE, Pise-Masison CA, et al. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. Proc Natl Acad Sci USA. 2002;99(21):13759-13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. Novel insights on human NK cells’ immunological modalities revealed by gene expression profiling. J Immunol. 2004;173(11):6547-6563. [DOI] [PubMed] [Google Scholar]
- 48.Voigt J, Malone DFG, Dias J, et al. Proteome analysis of human CD56neg NK cells reveals a homogeneous phenotype surprisingly similar to CD56dim NK cells. Eur J Immunol. 2018;48(9):1456-1469. [DOI] [PubMed] [Google Scholar]
- 49.Yang C, Siebert JR, Burns R, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10(1):3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duluc D, Tan F, Scotet M, et al. PolyI:C plus IL-2 or IL-12 induce IFN-γ production by human NK cells via autocrine IFN-beta. Eur J Immunol. 2009;39(10):2877-2884. [DOI] [PubMed] [Google Scholar]
- 51.Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burr ML, Sparbier CE, Chan YC, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602-610. [DOI] [PubMed] [Google Scholar]
- 54.Wyant GA, Abu-Remaileh M, Frenkel EM, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360(6390):751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donnelly RP, Loftus RM, Keating SE, et al. mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. J Immunol. 2014;193(9):4477-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crinier A, Milpied P, Escalière B, et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity. 2018;49(5):971-986.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CG000148 Rev A, Technical Note. Resolving Cell Types as a Function of Read Depth and Cell Number. Pleasanton, CA: 10X Genomics; 2018. https://support.10xgenomics.com/single-cell-gene-expression/sequencing/doc/technical-note-resolving-cell-types-as-a-function-of-read-depth-and-cell-number.
- 58.Schlums H, Cichocki F, Tesi B, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42(3):443-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nabekura T, Lanier LL. Tracking the fate of antigen-specific versus cytokine-activated natural killer cells after cytomegalovirus infection. J Exp Med. 2016;213(12):2745-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horowitz A, Strauss-Albee DM, Leipold M, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5(208):208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Böttcher JP, Bonavita E, Chakravarty P, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172(5):1022-1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodridge JP, Jacobs B, Saetersmoen ML, et al. Remodeling of secretory lysosomes during education tunes functional potential in NK cells. Nat Commun. 2019;10(1):514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.