Summary
Osteoporosis and osteoporotic fractures lead to decreased life quality and high healthcare costs. Current treatments prevent losses in bone mass and fractures to some extent but have side effects. Therefore, better therapies are needed. This study investigated whether the transcription factor Jun has a specific pro-osteogenic potency and whether modulating Jun could serve as a novel treatment for osteoporosis-associated fractures. We demonstrate that ectopically transplanted whole bones and distinct osteoprogenitors increase bone formation. Perinatal Jun induction disturbs growth plate architecture, causing a striking phenotype with shortened and thickened bones. Molecularly, Jun induces hedgehog signaling in skeletal stem cells. Therapeutically, Jun accelerates bone growth and healing in a drilling-defect model. Altogether, these results demonstrate that Jun drives bone formation by expanding osteoprogenitor populations and forcing them into the bone fate, providing a rationale for future clinical applications.
Keywords: osteoporosis, stem cell therapy, fractures, JUN, bone precursors, bone precursor expansion, osteoprogenitors, skeletal stem cells
Graphical Abstract
Highlights
-
•
Jun drives the formation of bone at the expense of cartilage and stroma
-
•
Jun disturbs the proper differentiation of the developing growth plate
-
•
Jun stimulates hedgehog signaling in skeletal stem cells
-
•
Jun accelerates fracture healing in a drilling-defect model
Wernig and colleagues demonstrate the striking potency of the transcription factor Jun to expand bone precursors, drive bone formation, and accelerate fracture healing. Boosting the regenerative potential of dormant bone precursor cells, they show the potential of stem cell therapy for widespread degenerative disease.
Introduction
Bone formation and resorption is a continuous and normally well-regulated process that meets physiological demands. In healthy individuals both processes should exist in equilibrium, thus maintaining the individual's steady bone mass (Sims and Gooi, 2008). However, in several pathological conditions this balance is disturbed, leading to increased or decreased bone mass and density as can be seen in osteopetrosis and osteoporosis (Cline-Smith et al., 2016, Sobacchi et al., 2013). Decreased bone density is associated with an increased risk of fracture and resulting complications such as immobility, permanent need of care, and even death (Briot and Roux, 2016, Haentjens et al., 2010, Klotzbuecher et al., 2000, LeBlanc et al., 2011). It was previously demonstrated that bone with its osseous, cartilaginous, and stromal tissue components, potentially including bone marrow adipocytes, derive from a hierarchical lineage (Robey et al., 2007). Skeletal stem cells (SSCs) differentiate into bone-cartilage-stromal-progenitors (BCSPs), which then differentiate into Thy+ osteoprogenitors (Chan et al., 2015, Chan et al., 2018). While SSCs and BCSPs are still able to build all components of the bone, differentiation capacity shrinks thereafter (Chan et al., 2015). During development, bones are formed either by desmal or by endochondral ossification. Within the developing bone, the cartilaginous growth plate that comprises different zones (reserve, proliferation, hypertrophy, and resorption) is crucial to maintain length growth, and changes within the cartilage can predict final skeletal proportions (Lui et al., 2018). Thereby, the hedgehog pathway and its effectors Gli2 and Gli3 play an essential role (Hui and Joyner, 1993, Mo et al., 1997, Park et al., 2000). Ihh and PthrP form a feedback loop to regulate growth and development of the growth plate (van den Heuvel and Ingham, 1996). The hedgehog pathway has been demonstrated to be linked to Jun in closing the optic fissure (Weston et al., 2003). Jun belongs to the AP-1 family of transcription factors otherwise including c-Fos, Fra1, Fra2, JunB, and JunD (Shea et al., 1989). As we recently demonstrated, Jun contributes to fibrotic diseases and regulates critical cellular processes such as the cell cycle (Wernig et al., 2017). While Fra2 und JunD have been shown to be important for regular bone development and maintenance, no study has yet investigated the role of Jun in bone formation (Bozec et al., 2010, Kawamata et al., 2008). To study the effect of Jun, we generated two different Jun-inducible mouse models; one with ubiquitous Jun expression under the doxycycline-dependent Rosa26 promoter and the other with bone-restricted Jun expression under the Osterix promoter (Wernig et al., 2017). Therefore, we sought to determine whether and how Jun stimulates bone formation and whether the induction of Jun can be used for therapeutic purposes.
Results
Jun Drives Bone Growth of Whole Bone Grafts and Osteoprogenitors
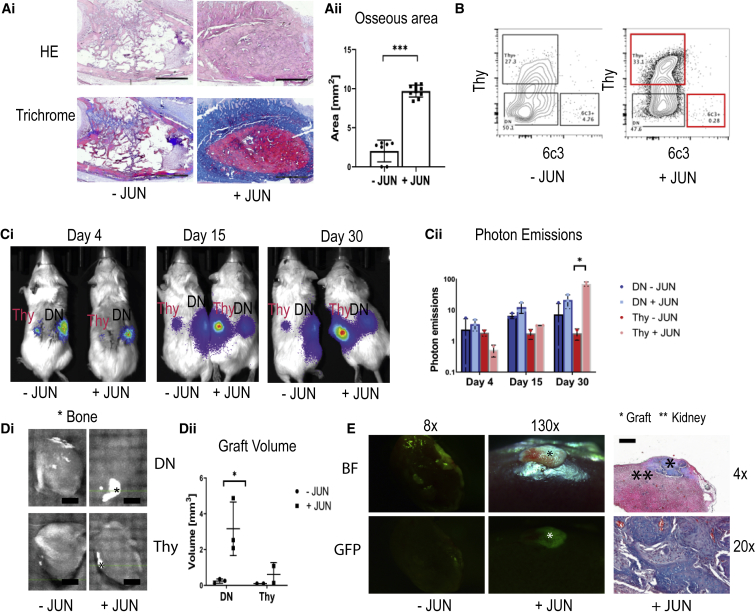
We first determined whether Jun influences bone growth. We transplanted adult long bones from Jun-inducible mice subcutaneously into immunocompromised NOD/Scid/gamma (NSG) mice. We found that with Jun induction (+JUN), osseous areas were significantly increased after 4 weeks (Figure 1A). Thereafter, we investigated the effects of Jun on distinct osteoprogenitor subsets. We classified live CD45+ Tie2− CD51+ cells from bones according to their expression of Thy and 6c3, also known as Ly-51/BP-1 (Figure 1B). We sorted Thy−/6c3− DN (double-negative) osteoprogenitors and Thy+/6c3− (Thy+) osteoprogenitors from Jun-inducible mice, transduced them with a GFP/luciferase virus, and transplanted them as separate groups under the kidney capsule. At day 3, we started to induce JUN in one group (+JUN) while the non-induced group served as a control (−JUN). We then repeatedly measured photon emissions on days 4, 15, and 30. While Jun induction did not lead to increased photon emissions in DN osteoprogenitors, Jun induction caused significantly increased photon emissions in Thy+ osteoprogenitors (Figure 1Cii). At the endpoint, we quantified ectopic bone tissue with micro-computed tomography (microCT). In the absence of Jun induction, we could detect bone in neither DN osteoprogenitors nor Thy+ osteoprogenitors. In contrast, Jun induction caused significant bone formation in DN osteoprogenitors (Figure 1Di). Visualizing the grafts under a dissection microscope, GFP positivity proved that they were originating from the transplanted cells (Figure 1E). Histologically, the grafts demonstrated mature osseous tissue with small amounts of cartilaginous tissue (Figure 1E).
Figure 1.
Jun Drives Ectopic Bone Formation in Osteoprogenitors
Further statistical analyses and raw data are listed in the Data S1. Two-sided t tests were used to determine statistical significances between −JUN (without Jun induction) and +JUN (with Jun induction). ∗p < 0.05, ∗∗∗p < 0.001.
(A) Representative H&E stainings and trichrome stainings of whole bone grafts 4 weeks after subcutaneous transplantations (Ai) and corresponding quantification of the bone area (Aii). Scale bars, 1 mm (n = 7–10). Each data point represents an individual measurement from two independent experiments.
(B) Representative FACS plots of osteoprogenitors with and without Jun.
(C) Optical images (Ci) of transplanted DN (labeled with DN) and Thy+ osteoprogenitors (labeled with Thy) after 4, 15, and 30 days. Corresponding quantification (Cii) of photon emissions normalized to the value at day 4 (n = 3 for DN osteoprogenitors, n = 2 for Thy+ osteoprogenitors). DN − JUN, double-negative osteoprogenitors without Jun induction; DN + JUN, double-negative osteoprogenitors with Jun induction; Thy − JUN = Thy+ osteoprogenitors without Jun induction; Thy + JUN, Thy+ osteoprogenitors with Jun induction. While Jun induction did not to change in photon emissions in double-negative osteoprogenitors, it significantly increased bone formation in Thy+ osteoprogenitors. Each data point represents an independent experiment.
(D) MicroCT of extracted kidneys (Di). Bone is marked with an Asterisk. Scale bar, 2 mm. Corresponding bone volumes (Dii) (n = 3 for DN osteoprogenitors, n = 2 for Thy+ osteoprogenitors). Each data point represents an independent experiment.
(E) Graft visualization under the dissection microscope (BF, brightfield; GFP, fluorescence) and corresponding trichrome stains of a double-negative osteoprogenitor graft under Jun induction.
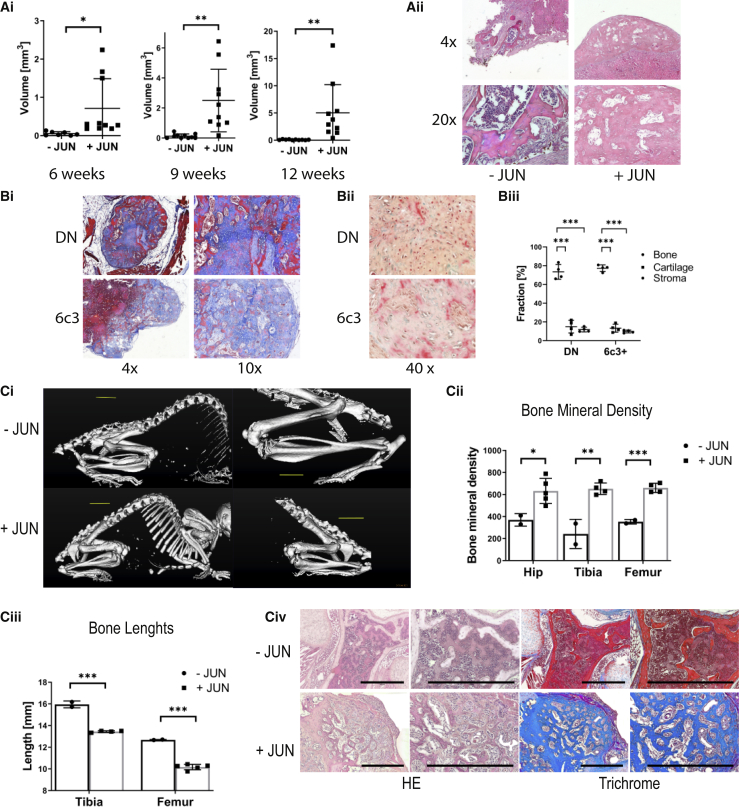
We then sought to determine whether the new bone formation was dependent on the mouse model. For this purpose, we generated a bone-restricted Jun-inducible mouse model under the Osterix promoter. We then isolated osteoprogenitors from this bone-restricted Jun-inducible mouse model and separately transplanted SSCs, BCSPs, and Thy+ osteoprogenitors ectopically under the renal capsule of immunocompromised mice. Subsequently, we tracked new bone formation via microCT over 12 weeks. In accordance with our previous results, Jun led to significant bone formation under induction but only little bone formation occurred without Jun induction (Figure 2Ai). Interestingly, this time all osteoprogenitor subsets were able to form bone, including Thy+ osteoprogenitors, suggesting that Jun-mediated bone formation is time dependent. Histologically, grafts consisted mainly of bone tissue with minimal cartilage, stromal tissue, or bone marrow cavity. In contrast, the small grafts without Jun primarily consisted of stromal parts and bone marrow (Figure 2Aii). In accordance, ectopic grafts from DN and 6c3+ osteoprogenitors mainly consisted of bone after six months of JUN induction (Figure 2B). Altogether, we demonstrate in this section that Jun increases cell-autonomous bone formation in three different bone precursor subsets.
Figure 2.
Jun Causes a Striking Phenotype with Shortened and Thickened Bones
Further statistical analyses and raw data are listed in Data S2. Two-sided t tests were used to determine statistical significance between −JUN and +JUN. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(A) CT-based volumes of bone grafts of osteoprogenitors from the bone-restricted mouse model after 6, 9, and 12 weeks (n = 7–10) (Ai). Populations were individually transplanted. The graph represents pooled populations (SSC, BCSP, and Thy). Each data point represents an independent transplantation from three independent experiments. Corresponding H&E stainings (Aii) of BCSP osteoprogenitor grafts.
(Bi) Representative trichrome stains of ectopic grafts six months after transplantation of double-negative and 6c3+ osteoprogenitors and continuous JUN induction (n = 4). (Bii) Corresponding pentachrome stains. (Biii) Corresponding percentages of bone, cartilage and stroma in the grafts. (Ci) Whole body CT of mice after 8 weeks of Jun induction (Cii) Cooresponding CT based measurements of bone mineral density (n = 2–4). Each data point represents an individual bone measurement. Each mouse contributed each type of bone. (Ciii) Corresponding CT based measurements of bone lengths (n = 2–4). Each data point represents an individual bone measurement. Each mouse contributed each type of bone. (Civ) Corresponding H&E and trichrome stains.
Perinatal Jun Induction Disturbs Proper Growth Plate Differentiation and Causes a Striking Phenotype with Shortened and Thickened Bones
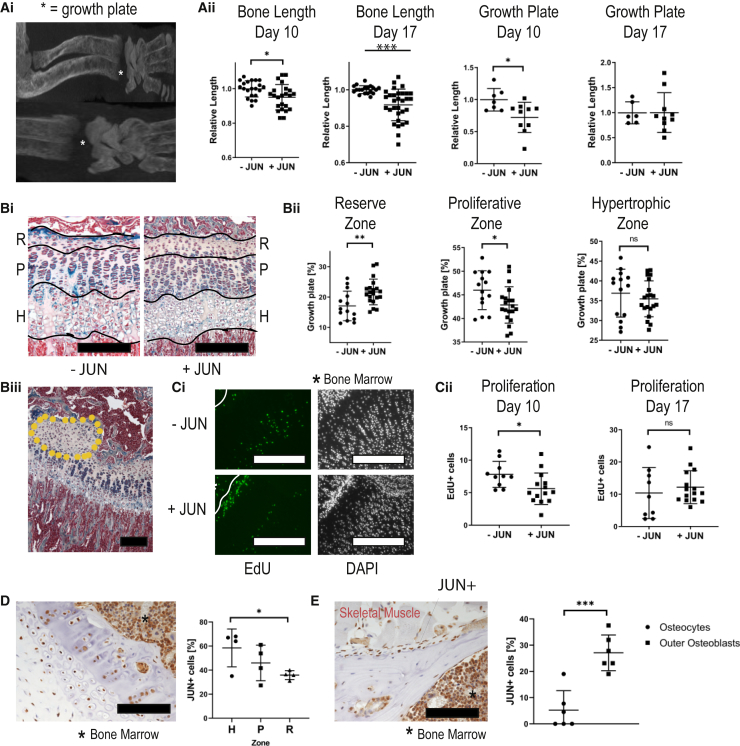
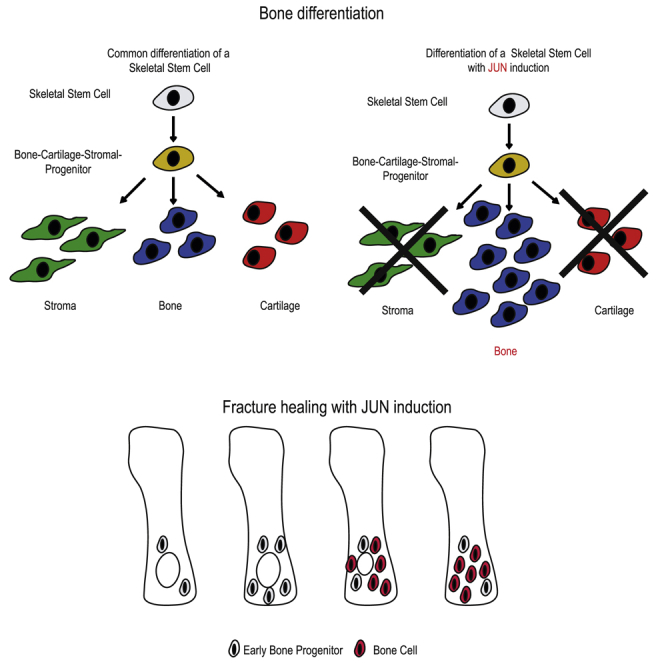
After investigating the cell-autonomous effect of Jun on ectopically transplanted osteoprogenitors, we then determined how Jun affects the skeletal development of perinatal mice. For this purpose, we induced Jun for 8 weeks after birth in the bone-restricted mice before euthanizing them. Visualizing the skeleton at endpoint, mice exhibited a striking phenotype with shortened and thickened bones (Figure 2Bi). Accordingly, bone mineral density was increased and bone length shortened with Jun induction (Figures 2Bii and 2Biii). Histologically, bones showed woven bone formation with only minimal residual bone marrow (Figure 2Biv). We hypothesized that underlying disturbances in the growth plate were the reason for the striking morphological abnormalities. To study our hypothesis, we induced Jun at birth and for 3, 10, and 17 days. At the endpoints, we ran CT scans to measure the length of bones and growth plates and injected 5-ethynyl-2′-deoxyuridine (EdU) 4 h prior to euthanasia to perform cell-cycle analyses (Figure 3Ai). Jun induction caused a decrease in bone length at as early as 10 days, which further increased until day 17. The growth plate length was significantly shortened at day 10 (Figure 3Aii). With respect to the different zones of the growth plate, Jun increased overall cell number and caused an enlargement of the reserve zone (R) at the expense of the proliferative zone (P) (Figures 3Bi and 3Bii). Interestingly, Jun also caused osseous outgrowth in 20% of the growth plates (Figure 3Biii). We detected significantly fewer proliferating cells within the growth plate under Jun induction at day 10, suggesting a previous increase in cell proliferation (Figures 3Ci and 3Cii). We then performed immunohistochemistry to determine which bone and cartilage cells expressed Jun under Jun induction. All zones within the growth plate expressed Jun. Regarding the rest of the bone, osteoblasts on the bone surface expressed significantly more Jun than mature osteocytes within the bone (Figures 3Di and 3Dii). In conclusion, Jun induces the premature differentiation of cartilaginous progenitors in the growth plate, thus disturbing proper skeletal development and causing shortened and thickened bones.
Figure 3.
Jun Disturbs the Regular Architecture of the Growth Plate
Further statistical analyses and raw data are listed in the Data S3. Two-sided t tests were used to determine statistical significances between −JUN and +JUN. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(A) Representative CT images of growth plates (Ai). CT-based measurements of bone lengths and growth plate lengths in all long bones, normalized to the values without Jun (Aii). Each of four animals contributed multiple long bones to the analysis (n = 13–22 for bone lengths, n = 6–10 for growth plates). Each data point represents individual bone measurements from two independent experiments.
(B) Representative pentachrome stainings of growth plates (Bi) indicating the reserve zone (R), proliferative zone (P), and hypertrophic zone (H). Scale bar,100 μm. Lengths of the different zones without and with Jun induction (n = 14–20) (Bii). Each data point represents an individual bone measurement. Each mouse contributed each type of bone. Representative osseous outgrowth (encircled area) under JUN induction (Biii). Scale bar, 100 μm.
(C) Representative EdU stainings without and with Jun induction (Ci). Scale bar, 100 μm. Counting of EdU+ cells per 100 μm growth plate width (n = 9–15) (Cii). Each data point represents an individual bone measurement. Each mouse contributed each type of bone.
(D) Representative immunostaining against Jun of the growth plate and corresponding percentage of Jun+ cells in the growth plate, showing that all zones of the growth plate express. Scale bar, 100 μm.
(E) Representative immunostaining against Jun of the mature bone and corresponding percentage of Jun+ cells in the mature bone (n = 4–6). Data points represent individual measurements from two independent experiments. Scale bar, 100 μm.
Ns, not significant.
Jun Activates Hedgehog Signaling in Skeletal Stem Cells
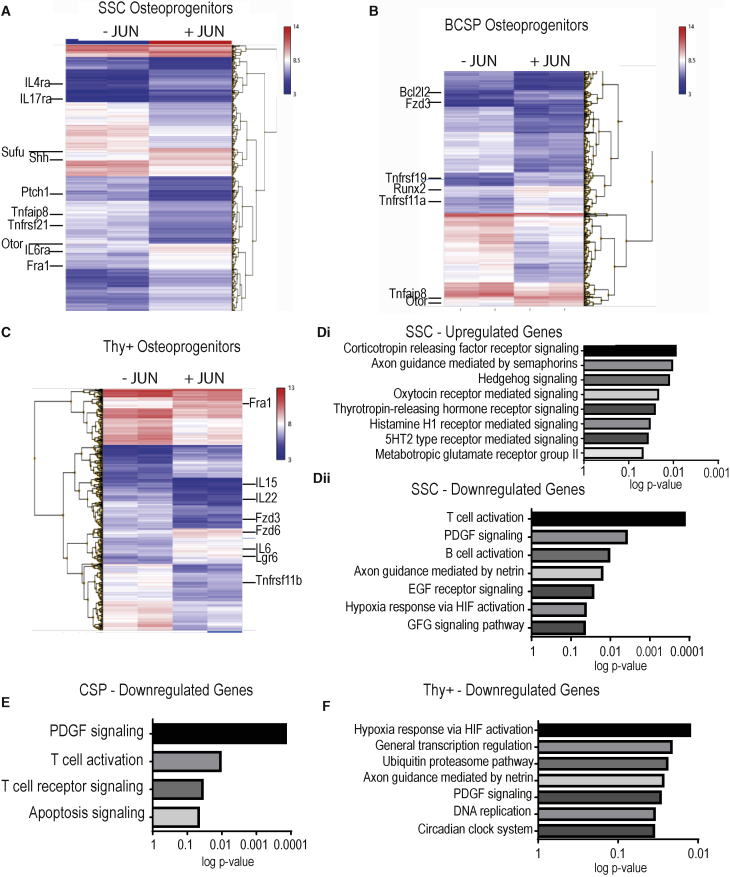
Next, we aimed at identifying the underlying molecular mechanism driving bone formation under Jun. For this purpose, we evaluated the effects of Jun on gene expression in fluorescence-activated cell sorting (FACS)-purified osteoprogenitors after 2 days of Jun induction via microarray. In all osteoprogenitor subsets, distinct gene clusters occurred. Jun induction upregulated hedgehog-associated genes such as Sonic Hedgehog (Shh) and Suppressor of Fused Homolog (Sufu) in SSC osteoprogenitors. Additionally, we found that Otor as an important inducer for the development of cartilage and Fra1 as another AP-1 transcription family member were upregulated in SSC osteoprogenitors under Jun induction (Figure 4A). BCSP osteoprogenitors increased the expression of the important osteogenic factor Runx2 (Figure 4B). In Thy+ osteoprogenitors, Fra1 gene expression was again enhanced in addition to the increased expression of interleukin-6 and the Wnt-associated gene Lgr6 (Figure 4C). Performing pathway analyses, we found that the gene expression signature under Jun induction significantly correlated with the hedgehog pathway, T cell activation, and platelet-derived growth factor receptor (PDGFR) signaling in SSC osteoprogenitors (Figure 4D). In BCSP osteoprogenitors, expression patterns significantly correlated with PDGFR signaling, T cell activation, and apoptosis (Figure 4E). In Thy+ osteoprogenitors, Jun again correlated with PDGFR signaling (Figure 4F).
Figure 4.
Jun Hedgehog Signaling in SSC Osteoprogenitors
Top up- and downregulated genes in the individual osteoprogenitor populations are shown in Data S4. n = 2 for both groups.
(A) Microarray analysis in SSC progenitors after 2 days of Jun induction.
(B) Microarray analysis in BCSP progenitors after 2 days of Jun induction.
(C) Microarray analysis in Thy+ progenitors after 2 days of Jun induction.
(D) Pathway analysis of upregulated genes under Jun induction in SSC progenitors (Di) and pathway analysis of downregulated genes under Jun induction in SSC progenitors (Dii).
(E) Pathway analysis of downregulated genes under Jun induction in BCSP progenitors.
(F) Pathway analysis of downregulated genes under Jun induction in SSC progenitors.
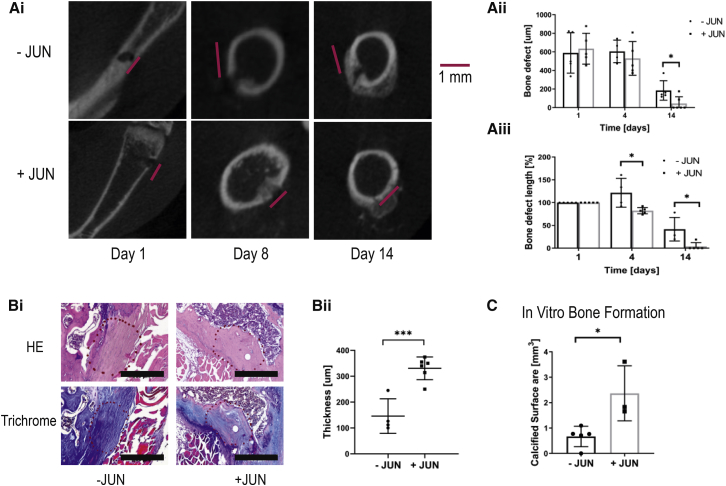
Jun Accelerates Healing in a Fracture Model
Next, we wanted to test whether the induction of Jun could be used for therapeutic purposes. Therefore, we studied a drilling-defect model in which we drilled holes into the femur of the mice with bone-restricted Jun expression. We induced Jun in one group and performed microCT imaging over a time course of 2 weeks. We used the CT scans to quantify the bone defects. Initial bone defects were similar between both groups. Strikingly, whereas the control mice still exhibited a significant bone defect after 2 weeks, the bone defect was almost completely healed with Jun induction (Figure 5A). Additionally, after 1 week the fracture site was already filled with denser tissue under Jun induction (Figure 5A). Histologically, the fracture site without Jun induction was mainly filled with connective tissue (Figure 5B). In contrast, Jun induction led to a repair with osseous tissue (Figure 5B). Finally, we studied the bone-forming potential of bone cells from fracture sites. Measuring calcified areas by alizarin red, Jun resulted in increased formation of calcified tissue (Figure 5C). In conclusion, Jun induction compensated for decreased bone mass and accelerated fracture healing.
Figure 5.
Jun Accelerates Fracture Healing in a Drilling-Defect Model
Further statistical analyses and raw data are listed in Data S5. Two-sided t tests were used to determine statistical significances between −JUN and +JUN. ∗p < 0.05, ∗∗∗p < 0.001.
(A) Representative CT images of fracture sites over time (Ai) and corresponding absolute and relative (compared with day 0) bone defects (Aii and Aiii) (n = 4–6). Each data point represents a fracture site from an independent mouse.
(B) Representative H&E and trichrome stains from the previous fracture site (encircled area) (Bi). The fracture site is filled with connective tissue without Jun and with bone tissue with Jun induction. Scale bar, 500 μm. Quantification of the bone thickness at the previous fracture site (Bii). Each data point represents a fracture site from an independent mouse.
(C) Calcified surface produced by cells from fracture sites without and with Jun induction (n = 3). Data points represent individual measurements from three independent experiments.
Discussion
Bone homeostasis results from the balance between new bone formation by osteoblasts and bone resorption by osteoclasts, and proper skeletal development requires both coordinated proliferation and differentiation. In this study, we investigated the effect of the transcription factor and AP-1 family member Jun on bone formation, ectopically, systemically, orthotopically, and on the molecular level.
Among bone precursors, we studied SSC, BCSP, and Thy+ osteoprogenitors. They build a hierarchy with SSCs as the most immature and multipotent osteoprogenitors. As osteoprogenitors differentiate, their capacity to form the three components of mature bone (bone, cartilage, and stroma) shrinks. In this study, we show the dramatic bone-forming potential of osteoprogenitors with Jun induction, whereby not only the amount of formed bone after ectopic transplantations under the renal capsule is striking, but also the observation that all three tested osteoprogenitor types are able to build equal amounts of bone. Histologically, the grafts mainly comprised bone and only small amounts of cartilage and stroma. This indicates that Jun promotes the differentiation of osteoprogenitors away from cartilage and stroma into the direction of bone tissue. Importantly, the potential to form bone tissue does not decrease from SSC osteoprogenitors to Thy+ osteoprogenitors.
At ectopic sites, bone cells are deprived of their innate cytokines and neighbors. To investigate how Jun effects niche-dependent bone development, we induced Jun over several weeks after birth in a bone-restricted Jun-inducible mouse model. In this model, mice developed strikingly shortened and thickened bones. In accordance with this, Jun-induced mice exhibited abnormal growth plate architectures with increased osseous components and reduced proliferation. The proper development of the growth plate is crucial for the normal growth of the skeleton. Within the growth plate, proliferating residual cells that develop into cartilage give rise to the continuous maintenance of the growth plate. If cells prematurely differentiate into bone cells, the growth plate closes earlier and overall skeleton length is reduced. Therefore, the observation of the shortened bones confirmed our hypothesis from the previous experiment, namely that Jun promotes osteoprogenitors to differentiate into bone cells at the expense of cartilage and stroma. Gene expression studies then indicated the involvement of the hedgehog pathway in SSC osteoprogenitors and the Wnt pathway in Thy+ osteoprogenitors. Interestingly, the pro-osteogenic factor Runx2 was highly expressed in BCSP osteoprogenitors (Komori, 2009). This suggests that BCSP osteoprogenitors represent the hierarchical level at which Jun determines the bone fate of bone precursors.
Finally, we addressed the question of whether the pro-osteogenic potential of Jun can be used for therapeutic purposes by using a drilling-defect model. Jun induction led to a strikingly accelerated closure of the bone defect, an observation that was supported by the increased capacity of cells in the injury site to build calcified tissue under Jun.
Altogether, our study shows the striking potential of Jun to induce bone formation in osteoprogenitors, primarily by steering them toward the osseous fate on the BCSP level. Importantly, the number of osteoprogenitors even in adult mice is sufficient to stimulate overall bone growth in a fracture setting. Osteoporosis and osteoporotic fractures pose a major health burden. Our study shows the benefits that the limited induction of a single transcription factor can have under these conditions. Therefore, further studies are warranted to investigate how Jun can be safely manipulated in bones offering novel therapeutic approaches to severe diseases.
Experimental Procedures
Subcutaneous Whole Bone Transplantations
Whole, long bones were harvested from adult Jun-inducible mice and subcutaneously transplanted into immunocompromised mice between the shoulders. Jun was then induced by giving doxycycline (2 mg/mL) (MilliporeSigma, MA, USA) through the drinking water. Mice were euthanized after 4 weeks and the long bones were extracted for subsequent histological analysis.
Cell Transplantation under the Kidney Capsule
After anesthetizing the mice, areas over the right and/or left flank were shaved and disinfected. A flank cut was made and the subcutaneous tissue was bluntly removed from the underlying soft tissue. An incision was cut into the abdominal wall and the kidney was luxated out of the abdominal cavity. The renal capsule was pierced and bluntly detached from the renal tissue. Two thousand to 10,000 cells suspended in 5 μL of Matrigel (ECM Gel from Engelbreth-Holm-Swarm murine sarcoma, MilliporeSigma) were injected under the kidney capsule. The kidney was replaced into the abdominal cavity, and the abdominal cavity and skin were closed using sutures.
Intrafemoral Cell Transplantation
After anesthetizing mice, the knee was shaved and disinfected. The knee was bent and a cut was made over the knee. The soft tissue over the knee was moved to the side so that the femur near the skin became visible. The knee was bent again and a cortical hole made with a 25-gauge needle. A 21-gauge needle was then used to widen the hole, followed by a 19-gauge needle. Two thousand to 5,000 cells were resuspended in 5 μL of FACS buffer (PBS + 2% FCS + 25 mM HEPES + 1 mM EDTA + 1% penicillin/streptomycin) and pipetted through the cortical hole into the femur. Finally, the skin was closed using sutures.
Drilling-Defect Model
Mice were anesthetized and knees were shaved and disinfected. The knee was bent and a cut was made over the knee. The soft tissue over the knee was moved to the side so that the femur became visible. Thereafter, holes were drilled into the femur using a 20-gauge syringe causing a complete cortical defect on one side of 0.5–1 mm in length. Finally, the skin was closed using sutures.
Luciferase-Based Optical Imaging
One hundred microliters of luciferin substrate (15 mg/mL) (Biosynth) were intraperitoneally injected. Fifteen minutes later, optical imaging was performed using the Lago optical imaging system (Spectral Imaging Instruments). Analysis was conducted with the Aura Software from the same manufacturer.
CT Imaging
For CT Imaging, the Bruker SkyScan 1276 (Bruker Micro-CT) was used. CT imaging was carried out under anesthesia and analyses were performed using the Bruker software. The settings were 1,024 × 1,024 (resolution) and 40 μm (thickness).
FACS Sorting
For the isolation of osteoprogenitors, perinatal mice from Jun-inducible mice (both systemic and bone-specific) were euthanized. After removing the skin, their bones (tibia, fibula, femur, spine, rib cage, humerus, radius, ulna, and skull) were harvested, dissected with scissors, smashed with a pestle, and digested in α-minimal essential medium (Gibco) substituted with 2.2 mg of collagenase I/mL (Thermo Fisher Scientific, MA, USA) for up to three rounds of 25 min under constant agitation at 37°C in the cell incubator. Thereafter, debris and red blood cells were removed by gradient centrifugation with Histopaque-1119 (MilliporeSigma). After washing, cells were stained with primary antibodies for 45 min and with the secondary antibody for 20 min (Table S1). Cells were then resuspended in FACS buffer (PBS + 2% FCS + 1% penicillin/streptomycin + 1 mM EDTA + 25 mM HEPES) and sorted for different subsets of osteoprogenitors using the BD FACSAria III sorter (Becton Dickinson). To mark negative and positive gates, we used FMO (full-minus-one) controls. Populations were double-sorted to ensure purity. We used FlowJo (FlowJo LLC) in its newest version for the analysis.
RNA Extraction
FACS-purified cells were sorted into TRIzol (Thermo Fisher Scientific). To extract RNA, we added chloroform and centrifuged the tubes. We transferred the upper phase to a new tube and added 70% EtOH. Thereafter, the complete volume was added to the columns of the RNEasy MiniElute Cleanup Kit (Qiagen). Between the next three centrifugation steps, we washed the columns with 80% EtOH (+H2O), 80% EtOH (+RPE), and 70% EtOH. After letting the membranes slightly dry, we added water onto the membranes, centrifuged the columns, and measured the RNA quantity and quality of the flow-through with a NanoDrop 2000 (Thermo Fisher Scientific).
Jun Staining
Tissue was fixed overnight at 4°Cin 4% paraformaldehyde (Thermo Fisher Scientific), then stored in 70% EtOH and submitted to the Stanford Human Pathology/Histology Service Center for embedding. Paraffin-embedded sections were first deparaffinized and rehydrated, followed by heat-induced antigen retrieval in sodium citrate (pH 6). Slides were then incubated in 0.3% H2O2 for 15 min and blocked in 10% normal serum for 1 h. Sections were incubated with the primary antibody (anti-c-Jun antibody, Abcam ab31419) overnight at 4°C, washed with PBST (PBS with Tween 20), and incubated with the HRP-conjugated secondary antibody (Goat Anti-Rabbit IgG H&L, Abcam, ab205718) for 30 min. After washing with PBST, 3,3′-diaminobenzidine was added for 10 min, followed by washing under running water for 5 min, counterstaining with hematoxylin for 4 min, dehydration, and mounting with Permount (Thermo Fisher Scientific).
Hematoxylin Staining
Tissue was incubated in hematoxylin for 4 min, followed by incubation in Bluing Reagent for 2 min and in eosin for 2 min. Slides were then dehydrated with ethanol and xylene and covered using Permount (Thermo Fisher Scientific).
Trichrome Staining
Trichrome stainings were done with a trichrome stain kit (American MasterTech, CA, USA). After deparaffinization and rehydration, tissue was incubated in Bouin's fluid overnight, followed by modified Mayer's hematoxylin for 7 min and One Step Trichrome Stain for 5 min. Slides were dehydrated with ethanol and xylene and covered with Permount (Thermo Fisher Scientific).
Pentachrome Staining
After deparaffinizing and rehydrating paraffin-embedded sections, sections were first washed with PBS for 10 min and ddH2O for 2 min. Thereafter, sections were incubated in Alcian blue (MilliporeSigma) for 15 min, followed by a wash step with running water for 15 min and 60-min incubation in alkaline alcohol (25 mL ammonium hydroxide + 225 mL 96% EtOH). After washing the sections with running water for 15 min, sections were incubated in brilliant crocein (MilliporeSigma) for 8 min, followed by three drips into 0.5% acetic acid, 20 min of incubation in phosphotungstic acid (MilliporeSigma) and 1 min of incubation in 0.5% acetic acid. Sections were then incubated in 100% ethanol for 5 min three times, followed by an incubation for 60 min in saffron (MilliporeSigma) and three incubations for 5 min each in 100% ethanol. Finally, sections were incubated in xylene for 3 min and covered with coverslips using Permount (Thermo Fisher Scientific).
Statistics
GraphPad Prism (GraphPad Software, CA, USA) was used for the statistical analysis of all experiments except the microarrays. Statistical significances were determined via two-sided t tests. p values below 0.05 were regarded as significant. Every experiment was run at least twice. Technical replicates were not used. Data represent mean ± SD. Gene expression data from the microarrays were uploaded onto the Gene Expression Omnibus (GEO: GSE144829). For analysis, the filter criteria were: fold change ≤2 or ≥2 + p < 0.05. Data were corrected for badge effects. For correlation with different pathways and signatures, commonly available databanks were used (Panther, WikiPathway, KEGG, and BioCarta).
Mouse Husbandry and Jun Induction
Jun mice were kept in the facilities of the Veterinary Service Center at Stanford University. Both ubiquitously and bone-restricted Jun-inducible mice had a B6/129 background. To generate the bone-restricted model, we crossed the ubiquitously Jun-inducible mouse with the Osx1-GFP::Cre mouse purchased from the Jackson Laboratory. Nod.Scid.Gamma (strain NOD/ShiLtj) mice were also purchased from the Jackson Laboratory. As published previously, genotyping in ubiquitously inducible Jun was run using primers for the transgene Jun and the Rosa26 promoter, and genotyping in bone-restricted inducible Jun mice was performed using primers for the transgene Jun and Cre (Wernig et al., 2017). Mice were not backcrossed and generally between 1 and 6 months old, except neonatal mice (3–5 days old) or older female NSG mice (>6 months). Under the Rosa26 promoter, Jun was induced by adding doxycycline (2 mg/mL) (MilliporeSigma) to the drinking water, and under the Osterix promoter, Jun was induced by removing doxycycline from the drinking water. Both male and female neonatal mice were used.
Study Approval
Animal trials were performed in accordance with preapproved protocols by the institutional review board at Stanford.
Author Contributions
T.L., experimental conception, collection and assembly of data, data analysis and interpretation, writing manuscript. L.C., collection and assembly of data, data analysis and interpretation. C.M., writing manuscript. A.S., data analysis and interpretation. C.v.N., data analysis and interpretation. P.D., data analysis and interpretation. C.C., experimental conception, data analysis and interpretation. G.W., experimental conception, collection and assembly of data, financial support, administrative support, data analysis and interpretation, writing manuscript.
Acknowledgments
The following funding sources made this work possible. Funding institutions include the National Heart, Lung, and Blood Institute (NHLBI) and Desmoid Tumor Research Foundation (DTRF). The DFG granted a research fellowship to T.L. We obtained FACS data on Aria Sorter purchased by the FACS Core at the Stanford Institute for Stem Cell Biology and Regenerative Medicine and ran CT scans on a Bruker Skyscan 1276 purchased by the Stanford Center for Innovation in In-Vivo Imaging with an NIH S10 Shared Instrumentation Grant (1S10OD02349701, PI Timothy C. Doyle) We thank Pauline Chen and Shirley Kwok for their help and elaborate expertise in embedding and cutting various forms of tissue.
Published: March 19, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.02.009.
Supplemental Information
References
- Bozec A., Bakiri L., Jimenez M., Schinke T., Amling M., Wagner E.F. Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 2010;190:1093. doi: 10.1083/jcb.201002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briot K., Roux C. Actualités du traitement de l’ostéoporose post-ménopausique. Rev. Med. Interne. 2016;37:195–200. doi: 10.1016/j.revmed.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Chan C.K.F., Seo E.Y., Chen J.Y., Lo D., McArdle A., Sinha R., Tevlin R., Seita J., Vincent-Tompkins J., Wearda T. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K.F., Gulati G.S., Sinha R., Tompkins J.V., Lopez M., Carter A.C., Ransom R.C., Reinisch A., Wearda T., Murphy M. Identification of the human skeletal stem cell. Cell. 2018;175:43–56.e21. doi: 10.1016/j.cell.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline-Smith A., Gibbs J., Shashkova E., Buchwald Z.S., Aurora R. Pulsed low-dose RANKL as a potential therapeutic for postmenopausal osteoporosis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.88839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haentjens P., Magaziner J., Colón-Emeric C.S., Vanderschueren D., Milisen K., Velkeniers B., Boonen S. Meta-analysis: excess mortality after hip fracture among older women and men. Ann. Intern. Med. 2010;152:380–390. doi: 10.1059/0003-4819-152-6-201003160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C.-c., Joyner A.L. A mouse model of Greig cephalo-polysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Kawamata A., Izu Y., Yokoyama H., Amagasa T., Wagner E.F., Nakashima K., Ezura Y., Hayata T., Noda M. JunD suppresses bone formation and contributes to low bone mass induced by estrogen depletion. J. Cell. Biochem. 2008;103:1037–1045. doi: 10.1002/jcb.21660. [DOI] [PubMed] [Google Scholar]
- Klotzbuecher C.M., Ross P.D., Landsman P.B., Abbott T.A., Iii, Berger M. Patients with prior fractures have an increased risk of future fractures: a summary of the literature and statistical synthesis. J. Bone Miner. Res. 2000;15:721–739. doi: 10.1359/jbmr.2000.15.4.721. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2009;339:189. doi: 10.1007/s00441-009-0832-8. [DOI] [PubMed] [Google Scholar]
- LeBlanc E.S., Hillier T.A., Pedula K.L., Rizzo J.H., Cawthon P.M., Fink H.A., Cauley J.A., Bauer D.C., Black D.M., Cummings S.R. Hip fracture and increased short-term but not long-term mortality in healthy older women. Arch. Intern. Med. 2011;171:1831–1837. doi: 10.1001/archinternmed.2011.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J.C., Jee Y.H., Garrison P., Iben J.R., Yue S., Ad M., Nguyen Q., Kikani B., Wakabayashi Y., Baron J. Differential aging of growth plate cartilage underlies differences in bone length and thus helps determine skeletal proportions. PLoS Biol. 2018;16:e2005263. doi: 10.1371/journal.pbio.2005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R., Freer A.M., Zinyk D.L., Crackower M.A., Michaud J., Heng H.H., Chik K.W., Shi X.M., Tsui L.C., Cheng S.H. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Park H.L., Bai C., Platt K.A., Matise M.P., Beeghly A., Hui C.C., Nakashima M., Joyner A.L. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- Robey P.G., Kuznetsov S.A., Riminucci M., Bianco P. Skeletal (“mesenchymal”) stem cells for tissue engineering. In: Hauser H., Fussenegger M., editors. Tissue Engineering. Humana Press; 2007. pp. 83–99. [DOI] [PubMed] [Google Scholar]
- Shea E.K., Rutkowski R., Stafford W.F., Kim P.S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- Sims N.A., Gooi J.H. Bone remodeling: multiple cellular interactions required for coupling of bone formation and resorption. Semin. Cell Dev. Biol. 2008;19:444–451. doi: 10.1016/j.semcdb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Sobacchi C., Schulz A., Coxon F.P., Villa A., Helfrich M.H. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat. Rev. Endocrinol. 2013;9:522. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M., Ingham P.W. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Wernig G., Chen S.-Y., Cui L., Van Neste C., Tsai J.M., Kambham N., Vogel H., Natkunam Y., Gilliland D.G., Nolan G. Unifying mechanism for different fibrotic diseases. Proc. Natl. Acad. Sci. U S A. 2017;114:4757–4762. doi: 10.1073/pnas.1621375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C.R., Wong A., Hall J.P., Goad M.E.P., Flavell R.A., Davis R.J. JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev. 2003;17:1271–1280. doi: 10.1101/gad.1087303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.