Summary
Yes-associated protein (YAP) is known to promote the stemness of multiple stem cell types, including pluripotent stem cells, while also antagonizing pluripotency during early embryogenesis. How YAP accomplishes these distinct functions remains unclear. Here, we report that, depending on the specific cells in which it is expressed, YAP could exhibit opposing effects on pluripotency induction from mouse somatic cells. Specifically, YAP inhibits pluripotency induction cell-autonomously but promotes it non-cell-autonomously. For its non-cell-autonomous role, YAP alters the expression of many secreted and matricellular proteins, including CYR61. YAP's non-cell-autonomous promoting effect could be recapitulated by recombinant CYR61 and abrogated by CYR61 depletion. Thus, we define a YAP-driven effect on enhancing pluripotency induction largely mediated by CYR61. Our work highlights the importance of considering the distinct contributions from heterologous cell types in deciphering cell fate control mechanisms and calls for careful re-examination of the co-existing bystander cells in complex cultures and tissues.
Keywords: yes-associated protein, YAP, pluripotency, Yamanaka reprogramming, iPSCs, stem Cell, CYR61, non-cell-autonomous
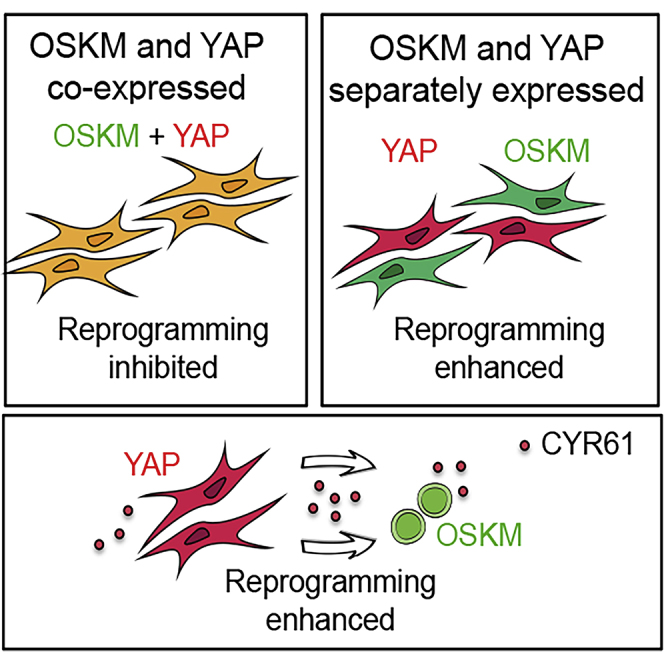
Graphical Abstract
Highlights
-
•
YAP inhibits pluripotency induction when expressed cell-autonomously
-
•
YAP promotes pluripotency induction when expressed non-cell-autonomously
-
•
YAP expression alters the expression of genes that encode extracellular components
-
•
CYR61 is secreted by YAP-expressing cells to promote nearby reprogramming
In this article, Hartman and colleagues show that, depending on the specific cells in which it is expressed, Yes-associated protein (YAP) exhibits opposing effects on pluripotency induction from mouse somatic cells. Specifically, YAP inhibits pluripotency induction cell-autonomously but promotes it non-cell-autonomously. Its non-autonomous function is accomplished by altering the expression of many secreted and matricellular proteins, including CYR61, a known YAP target.
Introduction
Cell fate decisions are instructed by the microenvironment, for which the transcriptional co-activator Yes-associated protein (YAP) is a major signal mediator. Diverse upstream inputs, including cell culture density, extracellular soluble factors, and local extracellular matrix composition converge to regulate YAP's nuclear entry and transcriptional activity (Azzolin et al., 2014, Dupont et al., 2011, Halder et al., 2012, Park et al., 2015, Piccolo et al., 2013, Piccolo et al., 2014, Yu et al., 2012, Zhao et al., 2007, Zhao et al., 2010). Microenvironmental cues also profoundly influence cell fate (Chen et al., 1997, Dupont et al., 2011, Engler et al., 2006, Gilbert et al., 2010, Mammoto and Ingber, 2010, McBeath et al., 2004, Schwartz, 2010, Swift et al., 2013, Vogel and Sheetz, 2006). Accordingly, many have examined YAP's role in cell fate decisions in various biological contexts (Barry et al., 2013, Camargo et al., 2007, Panciera et al., 2016, Qin et al., 2016, Schlegelmilch et al., 2011, Su et al., 2015, Totaro et al., 2017, Zanconato et al., 2016). However, since many YAP target gene products localize outside of the cell (Katsube et al., 2009, Zhang et al., 2009, Zuo et al., 2010), it is possible that YAP could regulate cell fate non-cell-autonomously, a scenario that is underexplored.
In the regulation of pluripotency, YAP's role appears complex and sometimes controversial. YAP functionally antagonizes pluripotency during early mouse embryonic development when the trophectoderm fate is specified versus the pluripotent inner cell mass (Nishioka et al., 2008, Nishioka et al., 2009). These findings contrast those supporting YAP's role in promoting pluripotency, either during pluripotency induction from fibroblasts or in pluripotency maintenance (Lian et al., 2010, Qin et al., 2016, Tamm et al., 2011). Meanwhile, YAP was determined to be dispensable for pluripotency by two other studies (Azzolin et al., 2014, Chung et al., 2016). These apparently conflicting behaviors could potentially be related to YAP's versatile interaction with components of the β-catenin or SMAD signaling pathways (Beyer et al., 2013, Papaspyropoulos et al., 2018, Zhou et al., 2017). Overall, these studies have all focused on YAP's cell-autonomous role in pluripotency. Heretofore, it has been unclear whether YAP functions non-cell-autonomously in pluripotency.
Pluripotency is subject to non-cell-autonomous regulation, prominently exemplified by the use of mitotically inactivated “feeder” cells, which secrete leukemia inhibitory factor (LIF) (Smith and Hooper, 1983, Smith and Hooper, 1987), among other potentially unidentified signals. The somatic-to-pluripotent reprogramming culture comprises a minority of cells that successfully acquire pluripotency and a majority of cells that fail to do so. The potential contribution by the latter cells to the emerging pluripotent fate has been overlooked. Here, we report that, while YAP potently inhibits the emergence of pluripotency cell-autonomously, it promotes pluripotency induction non-cell-autonomously, by increasing expression of the matricellular protein CYR61. This unexpected mode of action could reconcile the apparent discrepancies regarding YAP's role in pluripotency and calls for careful evaluation of YAP's role in other cellular systems, such as malignancy.
Results
YAP Cell-Autonomously Inhibits Pluripotency Induction
An important aspect of YAP regulation is its sensitivity to actin dynamics, with actin polymerization and stress fiber formation known to activate YAP (Reddy et al., 2013, Wada et al., 2011, Zhao et al., 2012). We recently identified a condition in which increased actin polymerization, driven by the transcriptional co-activator MKL1, potently inhibits the induction of pluripotency (Hu et al., 2019). As MKL1 and YAP are often activated by similar upstream signals and transactivate many overlapping target genes (1, 49–52), it was initially puzzling how YAP activity could promote pluripotency (Lian et al., 2010, Qin et al., 2016, Tamm et al., 2011). Previous reports of YAP promoting pluripotency induction utilized co-transduction of viral constructs encoding YAP and the reprogramming factors on separate vectors (Lian et al., 2010, Qin et al., 2016), invariably yielding mixtures of cells expressing either YAP or reprogramming factors, together with cells that express both or neither, potentially confounding the interpretation of the specific mode of YAP's action.
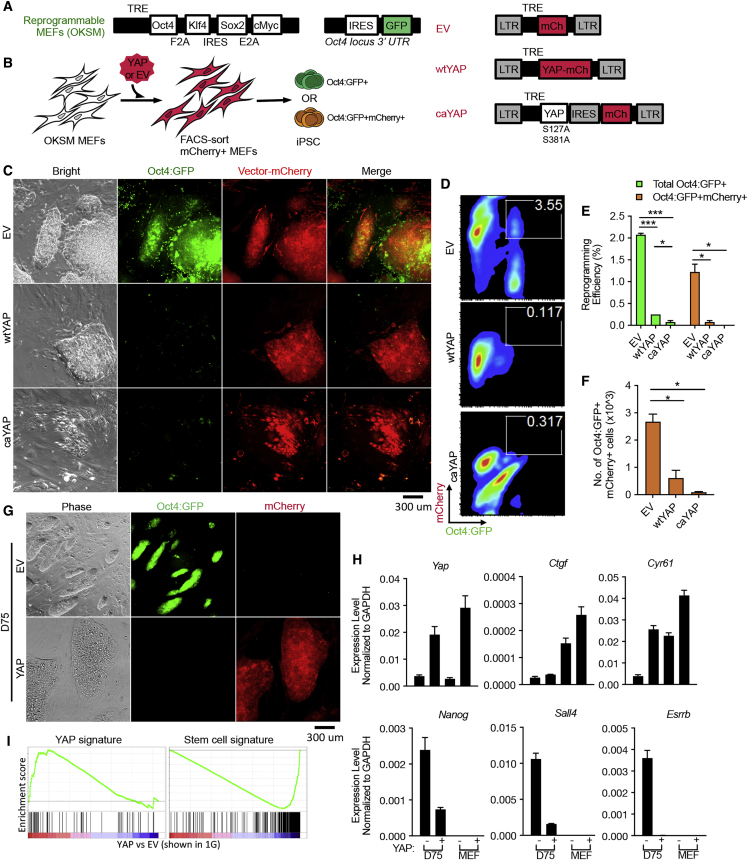
To test the cell-autonomous effect of YAP in pluripotency induction, we transduced either wild-type (wtYAP), constitutively active YAP (caYAP, in which two inhibitory phosphorylation sites are mutated [Zhao et al., 2010, Zhao et al., 2007]), or a control empty vector (EV), into reprogrammable mouse embryonic fibroblasts (MEFs) which express a polycistronic cassette encoding Oct4, Klf4, Sox2, and c-Myc (OKSM) upon doxycycline (Dox) treatment (Stadtfeld et al., 2010) (Figures 1A and 1B). Successfully transduced cells were sorted using fluorescence-activated cell sorting (FACS) based on expression of mCherry, which is encoded by the vectors (Figure 1A), and replated onto mitotically inactivated feeder MEFs for further reprogramming (Figure 1B). This approach ensures a uniform population of cells co-expressing OKSM and YAP. Mature induced pluripotent stem cells (iPSCs) were identified by their expression of GFP from the endogenous Oct4 locus (Stadtfeld et al., 2010) (Figures 1C and 1D), and confirmed by transcriptomic changes, including activation of key pluripotency genes, such as Nanog, Esrrb, Sall4, Dppa2/4, and Lin28a (Figures S1A–S1C). Reprogrammable MEFs co-expressing wtYAP or caYAP produced significantly fewer Oct4:GFP+ iPSCs compared with EV control (Figures 1E and 1F), although mCherry+ cells were abundant in the YAP-expressing cultures (Figures 1C and 1D). The YAP-expressing cells remained negative for Oct4:GFP even after prolonged exposure to OKSM (75 days) (Figure 1G), or further cultured in the 2i medium (Figures S1E and S1F) (Ying et al., 2008). Furthermore, rather than compact dome-shaped colonies characteristic of mouse pluripotency, wtYAP-expressing cells produced large colonies with flat morphology (Figure 1G) and expressed YAP transcriptional signature (Figures 1H and 1I) (Cordenonsi et al., 2011). Importantly, they lacked endogenous pluripotency gene expression (Figures 1H–1I and S1C). Following long-term culture in Dox, a small subset of these cells could emerge as mCherry+ Oct4:GFP+ (Figures S1A and S1B), which no longer displayed increased YAP or its target genes (Figure S1D). These results suggest that stochastic YAP activity dampening may have allowed pluripotency maturation, or that cells with low YAP activity were advantageous during prolonged culture. Of note, the failure of cells with excessive YAP activity to enter pluripotency was not due to impeded Wnt/β-catenin pathway (Figures S2A–S2C) or lack of Tead2 expression (Figure S2D), both of which have been shown to be important for YAP to support pluripotency (Azzolin et al., 2014, Tamm et al., 2011). In conclusion, MEFs co-expressing YAP and OKSM fail to establish pluripotency.
Figure 1.
YAP Inhibits Pluripotency Induction Cell-Autonomously
(A) Top: transgenic reprogrammable system for OKSM expression under the control of a tetracycline-responsive element (TRE). These cells also express GFP from the endogenous Oct4 locus. Right: lentiviral vectors encoding mCherry (EV), wild-type YAP fused to mCherry (wtYAP), or constitutively active YAP followed by an internal ribosome entry site (IRES) and mCherry (caYAP). LTR, long terminal repeats.
(B–D) Experimental scheme illustrating primary OKSM-expressing MEFs transduced with viral vectors in (A), FACS sorted on day 3 of Dox treatment based on mCherry-positivity and replated to allow further reprogramming (B). Oct4:GFP status was determined in the resulting cells in relation to their expression of mCherry, shown in (C and D). (C) Representative images of reprogramming cultures after 15 days of Dox treatment. (D) Representative FACS plots of reprogramming cultures after 20 days of Dox treatment. Gated population denotes Oct4:GFP and mCherry double-positive cells.
(E) Reprogramming efficiency quantified based on the number of Oct4:GFP+ colonies (green) and the number of Oct4:GFP and mCherry double-positive colonies (orange).
(F) Absolute numbers of Oct4:GFP and mCherry double-positive cells in each culture condition of (D).
(G) Representative mature iPSC (top) and YAP-mCherry+ colony (bottom) morphology after long-term culture (day 75) in mESC conditions derived from OKSM MEFs expressing control (EV) or wild-type YAP, respectively.
(H) qRT-PCR analysis of gene expression in cells shown in (G). MEFs expressing EV or wtYAP (denoted as YAP– and YAP+ respectively) are included as controls, which expressed YAP and its target genes (top panel) but not the pluripotency genes (bottom panel).
(I) Differentially expressed genes between cells shown in (G) by gene set enrichment analysis. YAP signature is from Cordenonsi et al. (2011) and stem cell signature is from cluster III Polo et al. (2012). Data from three biological replicates are displayed in (E) and (F), repeated over at least three independent experiments; while data displayed in (H) from three technical replicates, representative of at least three independent experiments.
To examine YAP's cell-autonomous effect on pluripotency induction from other somatic cell types, we expressed YAP in reprogrammable granulocyte-monocyte progenitors (GMPs) (Figure S3A). Similar to MEFs, YAP co-expression inhibited GMP reprogramming (Figures S3B and S3C). Of the Oct4:GFP+ cells that initially arose from YAP-transduced cultures, the percentage of Oct4:GFP+ cells decreased upon further culture, contrasting the EV-transduced cultures (Figures S3D and S3E). Furthermore, the fluorescence intensity of Oct4:GFP was lower in YAP co-expressing cells (Figure S3F), suggesting partial activation of the endogenous Oct4 locus. Taken together, these results further support that YAP compromises pluripotency induction from multiple somatic cell types when co-expressed with the reprogramming factors. Therefore, the two actin polymerization-sensitive transcriptional co-activators, YAP and MKL1 (Hu et al., 2019), both inhibit pluripotency activation (Hu et al., 2019).
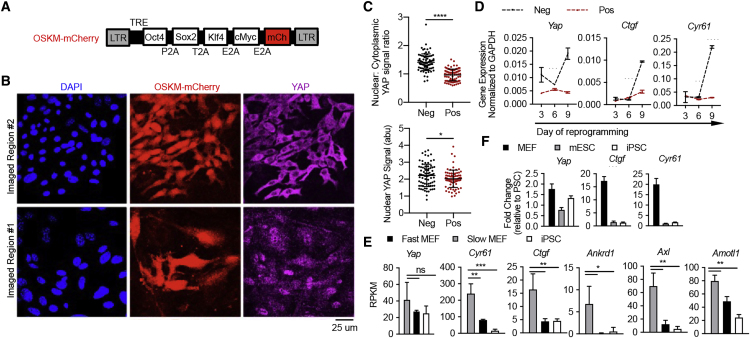
The inhibition of pluripotency induction by co-expressed YAP prompted us to examine the behavior of endogenous YAP during reprogramming. We assessed the subcellular localization of endogenous YAP in MEFs transduced with polycistronic expression of OSKM and mCherry (Figure 2A). By comparing OSKM-expressing (mCherry+) and wild-type (mCherry–) cells within the same heterogeneous culture, the effect of cell density or other culture-related variables are minimized, allowing for examination of YAP subcellular localization largely consequent to reprogramming factor expression. On day 4, the OSKM-expressing (mCherry+) cells displayed significantly lower nuclear YAP signal than the wild-type (mCherry–) cells (Figures 2B and 2C). Consistently, the expression of YAP target genes (Ctgf and Cyr61) remained low in mCherry+ cells as reprogramming proceeded (Figure 2D). Furthermore, reduced YAP target gene expression was evident in cells previously shown to possess enhanced reprogramming capacity, isolated irrespective of YAP activity by us (Figure 2E; re-plotted data from Guo et al., 2014) or by others (Polo et al., 2012). Finally, the expression of YAP target genes is low in iPSCs and embryonic stem cells (ESCs) as compared with MEFs (Figure 2F). Since YAP activity is sensitive to cell size/morphology, low YAP activity in pluripotent cells could be related to the drastic cell size/morphology change accompanying reprogramming (Hu et al., 2019). Indeed, c-Myc expression alone significantly decreased cell size (Figure S3G), with concomitant reduction in YAP target genes (Figure S3H). Taken together, pluripotency coincides with low endogenous YAP target gene expression, supporting YAP's inhibitory role in pluripotency induction cell-autonomously.
Figure 2.
Reduced Endogenous YAP Target Gene Expression Accompanies the Induction of Pluripotency
(A) Schema of polycistronic lentiviral vector encoding OSKM-mCherry.
(B) Primary MEFs transduced with OSKM-mCherry, treated with Dox for 4 days, and stained for YAP. Two representative regions in the same culture of distinct local cell density are shown.
(C) Quantification of endogenous YAP signal within mCherry– (Neg) and mCherry+ (Pos) cells, presented as a ratio of nuclear to cytoplasmic signal (top) or absolute nuclear signal (bottom; a.u., arbitrary unit). Nuclear area is defined as the DAPI+ region; cytoplasm is defined by pan-cellular GFP minus the DAPI+ region. Each dot denotes a single cell (n = 100 each).
(D) RT-qPCR analyses for endogenous Yap and YAP target genes Cyr61 and Ctgf in mCherry– (Neg) and mCherry+ (Pos) cells sorted after 3, 6, or 9 days of Dox treatment.
(E) Expression of endogenous Yap and YAP target genes Cyr61, Ctgf, Ankrd1, Axl, and Amot1 by mRNA sequencing in fast-cycling cells that exhibit enhanced reprogramming efficiency compared with slow-cycling cells and mature iPSCs. Data are re-plotted from (Guo et al., 2014) and are from three biological replicates. RPKM, reads per kilobase of transcript per million mapped reads.
(F) qRT-PCR of endogenous Yap, Ctgf, and Cyr61 in primary MEFs, mESCs, and iPSCs, displayed as the fold change relative to the average level in PSCs normalized to Gapdh. Data from three technical replicates are displayed in (D–F), representative of three independent experiments.
Pluripotency Maintenance Is Not Promoted by YAP Cell-Autonomously
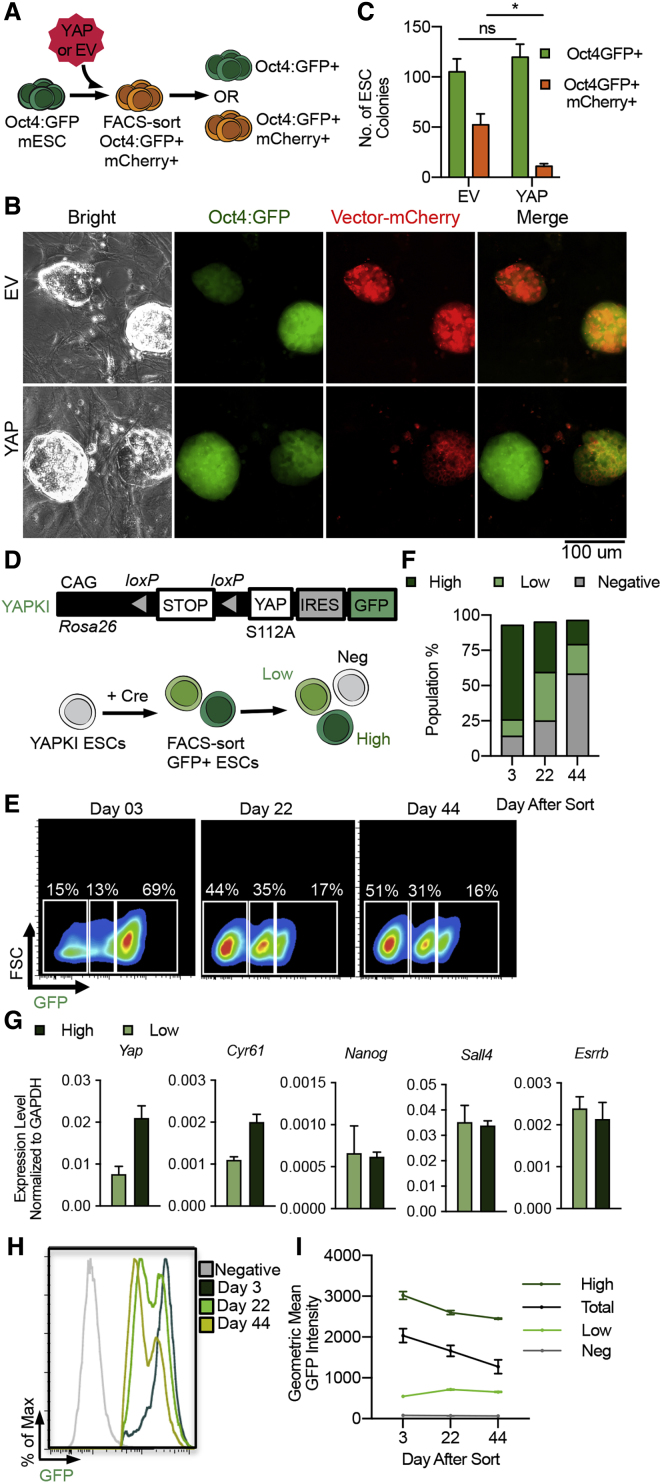
We next considered the possibility that YAP promotes the maintenance of established pluripotency (Lian et al., 2010, Qin et al., 2016), even though it did not promote the somatic-to-pluripotency transition. To examine YAP's cell-autonomous effect on established pluripotency, we transduced wtYAP into mouse ESCs (mESCs) harboring the Oct4:GFP reporter (Figure 3A). Transduced cells (mCherry+) were FACS sorted and replated in mESC maintenance conditions (Figure 3B). Although the total number of Oct4:GFP+ colonies was similar between EV and YAP-transduced cultures, the YAP-transduced ESC cultures had significantly fewer colonies that were mCherry+ (Figure 3C). Therefore, YAP expressed by a viral vector did not favor the maintenance of established pluripotency.
Figure 3.
Ectopic YAP Expression Does Not Promote Pluripotency Maintenance
(A–I) Experimental scheme illustrating Oct4:GFP-expressing mESCs transduced with EV or wtYAP (same as in Figure 1A) and FACS sorted for Oct4:GFP and mCherry double-positive cells (A). The expression status of mCherry was determined in the resulting mESCs, shown in (B and C). (B) Representative colony images after 7 days of culture. (C) Number of total Oct4:GFP+ colonies and those co-expressing mCherry, from three biological replicates, representative of three independent experiments (D) Top: schema of YAPKI allele in the Rosa26 locus, containing a coding sequence for caYAP and IRES-GFP preceded by a loxP-flanked STOP signal. Bottom: mESCs harboring this allele undergo Cre-mediated recombination, yielding two populations of GFP+ cells, which were FACS sorted and cultured in standard mESC maintenance condition. The relative abundance of the GFP-negative, GFP-low, and GFP-high populations was analyzed over time, shown in (E–I). (E) Representative FACS plots of the GFP-negative, GFP-low, and GFP-high populations on days 3, 22, and 44 after sorting. (F) Quantification of data shown in €. (G) qRT-PCR analyses of endogenous Yap, Cyr61, and pluripotency genes Nanog, Sall4, and Esrrb in the GFP-high (High) and GFP-low (Low) mESCs 3 days after sorting. (H) Representative FACS plot showing the evolution of fluorescence intensity of the GFP+ population over time. (I) Quantification of GFP fluorescence intensity (geometric mean) in the total GFP+ population (Total), GFP-low (Low), GFP-high (High), and GFP-negative (Negative) populations over time (n = 103 cells). Data in (F–G) from three technical replicates, representative of three independent experiments.
To circumvent potential silencing of the virally expressed YAP, we induced ectopic YAP expression in mESCs containing a loxP-STOP-loxP-caYAP(S112A)-IRES-GFP cassette (YAPKI mESCs) in the Rosa26 locus (Su et al., 2015). We transduced the YAPKI mESCs with a lentiviral Cre, which permanently activated the YAPKI-IRES-GFP allele (Figure 3D). Shortly after Cre transduction, recombined cells were FACS sorted and replated in maintenance conditions (Figure 3D). Consistent with the original report describing the YAPKI allele (Su et al., 2015), two populations of GFP+ cells with distinct intensities emerged (Figures 3E and 3F), with only the GFP-high cells expressing increased YAP target genes (Su et al., 2015). We confirmed that the GFP-high mESCs indeed exhibited increased Yap and target gene expression, while expressing comparable levels of pluripotency genes to the control GFP-low cells (Figure 3G). The percentage of GFP-high cells decreased relative to the GFP-low cells in the same culture over time (Figures 3E, 3F, 3H, S4A, and S4B). A substantial portion of the culture became GFP–, likely due to expansion of the few GFP– cells from the original sorting (Figures 3E and 3F). Furthermore, even among cells that remained within the GFP-high gate, their GFP intensity decreased over time (Figures 3I and S4C). Thus, YAP expressed by a knockin allele did not support the maintenance of established pluripotency either.
To gain insights why YAP-expressing pluripotent stem cells did not persist, we compared the transcriptome of control Oct4:GFP+ iPSCs (from EV), or those emerged Oct4:GFP+ cells carrying YAP-mCherry (Figures S1A and S1B). Although they expressed pluripotency genes similarly (Figure S1D), gene set enrichment analysis (GSEA) revealed a prominent apoptotic signature in the YAP-mCherry+ cells (Figures S4D and S4E). To test whether YAP activity indeed induces apoptosis in mESCs, we performed annexin V staining in the YAPKI mESCs 3 days after lentiviral Cre transduction (Figure 3D). Consistent with the GSEA results, the GFP-high YAPKI mESCs were more apoptotic (Figures S4F–S4H), illustrating a cellular outcome for the mESCs with excessive YAP activity. Taken together, these data suggest that ectopic YAP expression does not promote, but rather is competitively unfavorable for, pluripotency maintenance when expressed cell-autonomously.
YAP Promotes Pluripotency Induction in a Non-cell-Autonomous Manner
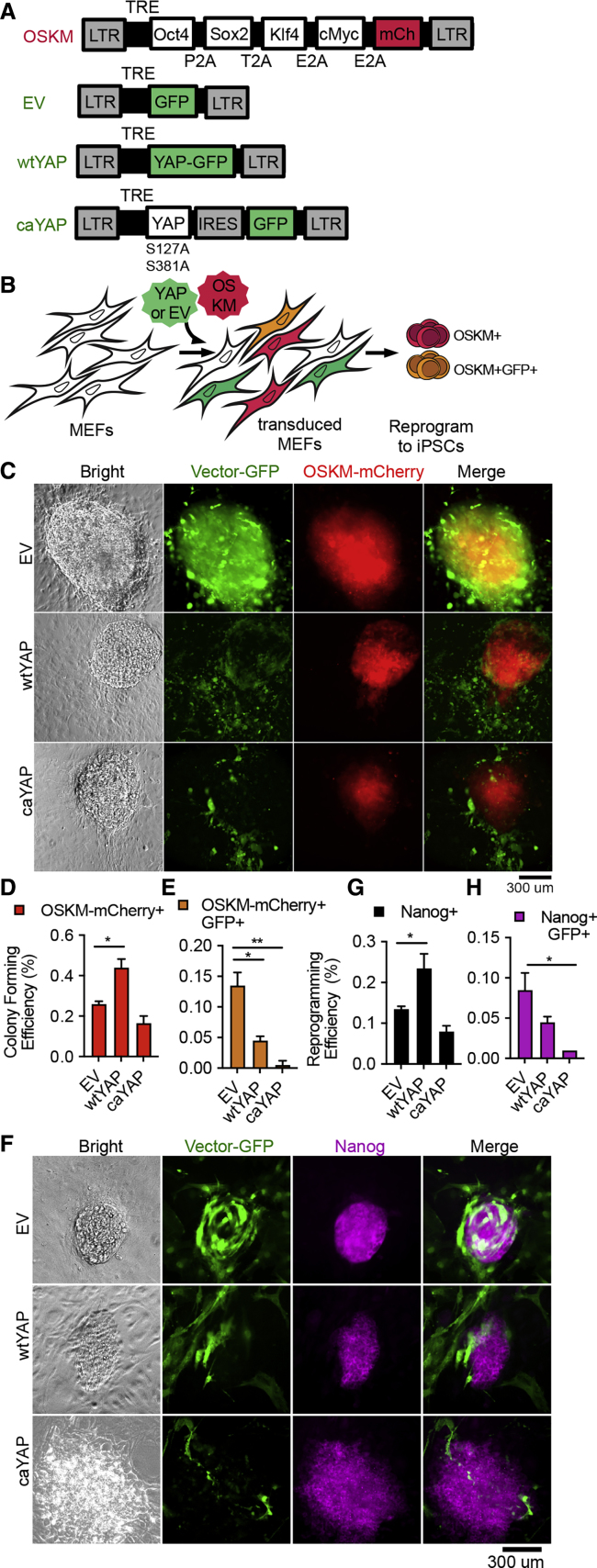
Having ruled out a cell-autonomous promoting effect by YAP in pluripotency induction and maintenance, we examined whether YAP promotes pluripotency non-cell-autonomously. We delivered the reprogramming factors and YAP via separate lentiviral constructs into MEFs to intentionally establish a heterogeneous cell culture comprised of cells expressing OSKM, YAP, both, or neither (Figures 4A and 4B), similar to the previous studies (Lian et al., 2010, Qin et al., 2016). Importantly, we tagged YAP- and OSKM-expressing vectors with GFP and mCherry, respectively, to track the contribution of heterogeneous cell types co-existing in the co-transduced cultures (Figures 4B and 4C). Consistent with the previous report (Lian et al., 2010), we observed a 2-fold increase in the total number of colonies when wtYAP was co-transduced (Figure 4D). While more colonies were present in the wtYAP co-transduced cultures (Figure 4D), few of them co-expressed YAP as indicated by their lack of GFP (i.e., YAP) expression (Figures 4C–4E). caYAP co-transduction also failed to increase the number of total colonies (Figure 4D). The absence of YAP+ colonies could not be simply accounted for by enhanced vector silencing, as GFP+ cells were abundant even though they did not appear as colonies (Figure 4C). Assessing the activation of pluripotency more stringently by immunofluorescence staining of endogenous Nanog confirmed these results (Figures 4F–4H). Overall, these data demonstrate that, while co-transduced wtYAP promotes the emergence of iPSC colonies, the colonies themselves do not co-express YAP. The presence of non-pluripotent YAP-expressing fibroblasts strongly suggests a non-cell-autonomous effect mediated by YAP. The fact that promotion was only observed with co-transduced wtYAP, but not caYAP (Figures S4I–S4J), suggests that the non-cell-autonomous effect is limited to wtYAP, while cell-autonomous inhibition is shared by both forms of YAP.
Figure 4.
YAP Promotes Pluripotency Induction in Mixed Cultures
(A) Schema of lentiviral vectors used to generate a mixture of cells that express OSKM, YAP, both, or neither.
(B–H) Experimental scheme illustrating the heterogeneous cell types co-existing in culture (B). The expression status of GFP from EV- or YAP-expressing vectors was determined in the resulting colonies, shown in (C–H). (C) Representative OSKM-mCherry+ colony images after 18 days of reprogramming from MEFs co-transduced with EV, wtYAP, or caYAP, each expressing a GFP reporter. (D) Quantification of total OSKM-mCherry+ colonies in (C). (E) Quantification of the OSKM-mCherry+ colonies also positive for GFP in (C). (F) Representative iPSC colonies after 18 days of reprogramming immunostained for the endogenous Nanog protein. (G) Quantification of total Nanog+ iPSC colonies in (F). (H) Quantification of Nanog+ iPSC colonies also positive for GFP in (F). Data from three biological replicates are displayed in (D–E, G, and H), repeated over at least three independent experiments.
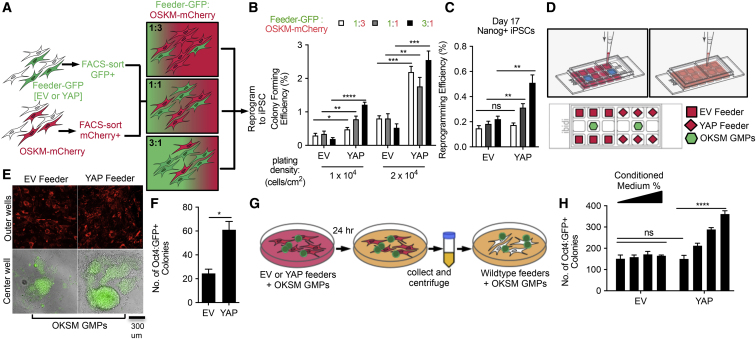
To directly test the possibility of a non-cell-autonomous role, we mixed, in a controlled manner, two types of cells: one expressing wtYAP-GFP (YAP) or a control GFP (EV) and the other expressing OSKM-mCherry, with GFP+ cells serving as the feeder cells (Figure 5A). After FACS sorting each population, cells were replated together at varying ratios of feeder-to-reprogramming cells, while maintaining identical overall cell-plating density. Strikingly, all conditions in which reprogramming cells were co-cultured with YAP feeders produced more colonies (Figures 5B and 5C). The promotional effect became more pronounced as the ratio or number of YAP feeders increased (Figures 5B and 5C). These data demonstrate that YAP-expressing fibroblasts promote pluripotency induction non-cell-autonomously.
Figure 5.
YAP Promotes Pluripotency Induction Non-cell-Autonomously
(A) Experimental scheme illustrating the mixing of two populations of primary MEFs: one expressing GFP (EV or YAP) corresponding to the “feeder” cells and the other expressing OSKM-mCherry. The cells were FACS sorted and replated at varying ratios as indicated, at two plating densities (1 × 104 or 2 × 104 cells/cm2). Resulting iPSC colonies were scored on days 15–17 as shown in (B and C).
(B) Colony-forming efficiency (mCherry+) on day 15 of reprogramming.
(C) Nanog+ iPSC colonies on day 17, from cultures plated at 1 × 104 cells/cm2.
(D) Top: schema illustrating a co-culture device that allows physically separated cells to share medium. Each of two major wells has nine minor wells. After seeding cells in the individual minor wells, the major well was filled with medium so that it is shared among cells across the nine-minor-well unit. Bottom: schema illustrating the co-culture of mitotically inactivated feeders expressing either control (EV) or wtYAP fused to mCherry (YAP) in outer minor wells and reprogrammable GMPs in the central minor well.
(E) Representative images of Oct4:GFP+ iPSCs (bottom) formed in the center well, fed by either EV feeders or YAP feeders (top), on day 5 of reprogramming.
(F) Quantification of data shown in (E).
(G) Experimental scheme illustrating the preparation of conditioned medium.
(H) Quantification of Oct4:GFP+ iPSC colonies cultured with varying proportion of conditioned medium to fresh medium (in order from left to right: 10%, 25%, 50%, and 75%) on day 5 of reprogramming. Data from three biological replicates are displayed in (B, C, F, and H), repeated over at least three independent experiments.
To determine if the promoting effect requires direct cell-cell contact, we utilized a co-culture device in which different cell types are kept physically separate, while sharing the same medium (Figure 5D). Reprogrammable GMPs were plated in the center minor well to be “fed” by mitotically inactivated feeder MEFs expressing either EV (mCherry) or YAP (YAP-mCherry) in the surrounding outer minor wells (Figure 5D). For experiments using this co-culture device, we tested GMPs in the center well as the wells had limited growth area and were not amenable to accommodating the extensive cell proliferation required for MEF reprogramming. The OKSM GMPs sharing medium with YAP feeders yielded more Oct4:GFP+ colonies compared with those sharing medium with EV feeders (Figures 5E and 5F). Thus, direct cell-cell contact is not necessary for YAP feeders to promote pluripotency induction. To directly test whether conditioned medium is sufficient to mediate the YAP feeder effect, we compared the reprogramming efficiency of reprogrammable GMPs fed by medium conditioned by YAP feeders or EV feeders (Figure 5G). More Oct4:GFP+ iPSC colonies arose when cultured in YAP feeder-conditioned medium, depending on the proportion of such conditioned medium to fresh medium (Figure 5H). As the promoting effect could be recapitulated by physically separated, mitotically inactivated feeder cells, or even medium conditioned by such feeder cells, the non-cell-autonomous promotion by YAP on pluripotency induction is at least partly mediated by components that exist in the shared medium.
YAP Target CYR61 Promotes Pluripotency Induction
For the conditioned medium to be effective, it could either contain increased levels of factor(s) that promote pluripotency induction or reduced levels of inhibitory factor(s). To uncover the identity of potential secreted factor(s), we first performed cytokine/growth factor detection arrays using medium conditioned by YAP feeders or EV feeders (Figures S5A and S5B). Only 3 out of 111 probed proteins were differentially present (at least 2-fold difference) in 2 independent experiments: Pentraxin-3 (PTX-3), CCL6/C10, and CCL11 (Figures S5A and S5B), with PTX-3 being the only protein increased in the YAP feeder-conditioned medium (Figures S5A and S5B).
To look for secreted protein factors beyond those on the growth factor array, we carried out mRNA sequencing comparing MEFs transduced with EV or wtYAP (Figure S5C). Upregulated Cyr61 and Ctgf in the YAP-overexpressing samples (Figure S5D) confirmed increased YAP transcriptional activity. The overall gene expression was similar between EV- and YAP-expressing cells (Figures S5E and S5F). Of note, however, many of the differentially expressed genes belonged to the “extracellular region,” “extracellular exosome,” or “extracellular matrix” (Figures S5G–S5J). This was the case for both the upregulated and downregulated genes. These data suggest that YAP expression potentially altered the molecular compositions of the microenvironment, providing a molecular basis for YAP's non-cell-autonomous function.
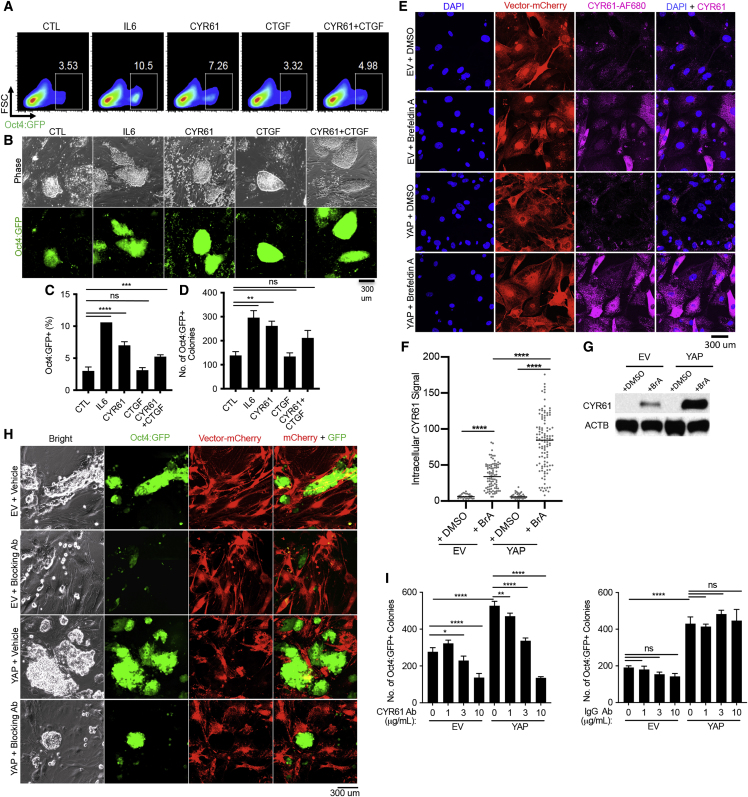
Given the recent discoveries of interleukin 6 (IL-6) in promoting pluripotency induction (Brady et al., 2013, Mosteiro et al., 2018), we focused on upregulated genes in YAP-expressing cells, revealing 42 genes that encode putative secreted proteins (Figure S5K). Two of these candidates (CYR61 and CTGF) and one from the cytokine array (PTX3) were selected for further testing; specifically, whether their recombinant forms could promote reprogramming. As a positive control, we included recombinant IL-6 since its promoting effect, as well as its effective concentration (10 ng/mL), have been described (Brady et al., 2013, Mosteiro et al., 2018). Recombinant IL-6 increased the number of Oct4:GFP+ colonies, as expected (Figure S6A). Recombinant CTGF and PTX3 had no consistent effect on reprogramming over the concentration range tested, even though they were biologically active as determined in an independent assay (Figures S6B–S6E). In contrast, a promoting effect was readily detected at the lowest concentration for recombinant CYR61 (Figure S6A). This low concentration was used in all subsequent experiments. Furthermore, recombinant CYR61, but not PTX3 (Figures S6D–S6E), also promoted MEF reprogramming assessed by the percentage of Oct4:GFP+ cells (Figures 6A and 6C) or number of Oct4:GFP+ iPSC colonies (Figures 6B and 6D). The extent of promotion by recombinant CYR61 was similar to that reported for recombinant IL-6 (Brady et al., 2013).
Figure 6.
CYR61 Promotes Pluripotency Induction
(A) Representative FACS plots for Oct4:GFP+ cells from reprogrammable MEFs in the presence of recombinant proteins IL-6, CYR61, CTGF, or both CYR61 and CTGF (10 ng/mL) on day 15 of reprogramming.
(B) Representative images of the resulting iPSCs colonies, following the same treatment in (A).
(C) Quantification of the percentage of Oct4:GFP+ cells in (A).
(D) Quantification of the number of Oct4:GFP+ iPSCs in (B).
(E) Immunofluorescence for CYR61 in EV- and YAP-expressing feeders, treated overnight with DMSO or 0.5 ng/mL Brefeldin A in DMSO (BrA).
(F) Quantification of endogenous CYR61 signal in cells shown in (E). Each dot denotes the CYR61 signal of a single cell (n = 100 each).
(G) Immunoblot for CYR61 in EV- and YAP-expressing feeders, treated overnight with DMSO or 0.5 ng/mL Brefeldin A (BrA), with β-actin (ACTB) as a loading control.
(H) Representative Oct4:GFP+ iPSCs formed from reprogrammable GMPs at day 5 when plated on EV- or YAP-expressing fibroblasts ± CYR61 blocking antibody (10 μg/mL).
(I) Quantification of the total number of Oct4:GFP+ iPSCs formed at day 5 of reprogramming when plated on EV- or YAP-expressing fibroblasts ± CYR61 blocking antibody (left) or control IgG (right) at the indicated concentrations. Data from three biological replicates are displayed in (C, D, and I), repeated over at least three independent experiments.
To test whether CYR61 is indeed produced by feeder cells, we directly probed for the endogenous CYR61 protein in EV- and YAP-expressing feeder MEFs. Anticipating CYR61 is being secreted, we treated EV- and YAP-expressing feeder MEFs with Brefeldin A, an inhibitor of the protein secretory pathway (Sciaky et al., 1997). While CYR61 was barely detectible in vehicle DMSO-treated cells, Brefeldin A treatment increased CYR61 in both EV- and YAP-expressing feeders (Figures 6E and 6F). These results suggest that CYR61 protein is indeed being actively secreted, even in the EV control feeder cells, such that detection of the intracellular protein requires inhibition of its secretory pathway. Importantly, the intracellular accumulation of CYR61 became exaggerated in the YAP-expressing feeders (Figures 6E and 6F). In addition to immunofluorescence staining, similar results were obtained by western blot (Figure 6G). Thus, the YAP-expressing feeder cells supply higher levels of CYR61 to the shared culture medium.
To assess the extent of the contribution to YAP's non-cell-autonomous effect by CYR61, we performed reprogramming in the presence of a CYR61 blocking antibody (Haseley et al., 2012, Imhof et al., 2016, Kim et al., 2013, Kim et al., 2015). Although YAP-expressing feeder cells supported higher reprogramming efficiency than control feeders, as expected, this promoting effect diminished in the presence of the CYR61 blocking antibody in a dose-dependent manner (Figures 6H and 6I). A control immunoglobulin G (IgG) had no effect at all equivalent concentrations (Figure 6I). At higher blocking antibody concentration (10 μg/mL), reprogramming efficiency supported by the control feeders also decreased (Figure 6I), suggesting that the control feeder cells produce CYR61 at a baseline level. Taken together, CYR61 plays an important role in mediating YAP's non-cell-autonomous promotion of pluripotency induction, summarized in the graphic abstract.
Discussion
We report that YAP inhibits pluripotency induction cell-autonomously, consistent with studies of early embryogenesis (Nishioka et al., 2008, Nishioka et al., 2009), but promotes it in a non-cell-autonomous manner by reprogramming the microenvironment. This dual mode of action could potentially contribute to YAP's seemingly conflicting roles in pluripotency regulation. Specifically, one of YAP's targets, the matricellular protein CYR61, mediates this promotional effect. It remains possible that additional mechanisms mediate YAP's non-cell-autonomous effect (Chen et al., 2008, Kosaka et al., 2010, Vickers et al., 2011, Wang et al., 2010). We noted that, while caYAP is competent for the cell-autonomous inhibition of pluripotency induction, it failed the non-cell-autonomous promotion. The inability of caYAP-expressing cells to promote pluripotency induction may be related to the production of other yet to be identified factors that could counter the effect of CYR61. Alternatively, one of the inhibitory phosphorylation sites on YAP might mediate some critical protein-protein interactions required for executing its non-autonomous action. Although the molecular basis of the distinction between wtYAP and caYAP remains unclear, it is consistent with the previous report where cell density beyond an optimal range failed to promote pluripotency induction (Stadtfeld et al., 2010). We have demonstrated YAP's non-cell-autonomous effect in pluripotency induction from embryonic fibroblasts and adult hematopoietic progenitors, both of which are of mesoderm origin. It remains possible that different cell types could respond differently to this YAP-CYR61-mediated mechanism. Our study does not exclude the possibility that human pluripotent stem cells may follow distinct regulation regarding YAP, a scenario to be further tested.
Our work demonstrates that, within a heterogeneous reprogramming culture, the non-reprogramming cells are not merely passive bystanders. Instead, they could actively participate in nearby cell fate conversion by reprogramming the microenvironment they share. Since their isolation, mESCs have been traditionally cultured on mitotically inactivated feeder MEFs, as they inhibit spontaneous differentiation by secreting LIF (Smith et al., 1988, Smith et al., 1992, Smith and Hooper, 1983, Smith and Hooper, 1987). Besides LIF, the contribution by feeders has been largely overlooked. Our work unveils another secreted protein, CYR61, under the control of YAP in supporting pluripotency. CYR61 modulates inflammation and senescence (Jun and Lau, 2010), both of which have been implicated in pluripotency induction via non-cell-autonomous mechanisms. Specifically, activation of innate immunity was shown to increase reprogramming in the process of “transflammation” (Lee et al., 2012). A recent study reveals that reprogramming induced in live animals triggers senescence in some cells and reprogramming in others (Mosteiro et al., 2016, Mosteiro et al., 2018). Whether YAP plays a similar non-cell-autonomous role in in vivo reprogramming and whether CYR61's mechanism of action involves inflammation or senescence awaits further investigation.
Outside of pluripotency, YAP is well-known for its role in controlling organ size (Camargo et al., 2007, Lee et al., 2016, Richardson and Portela, 2017, Yimlamai et al., 2014). YAP deregulation not only results in overgrown tissues and organs, but also has a well-documented role in cancer (Harvey et al., 2013). Although this has been traditionally attributed to YAP's cell-autonomous role, our work suggests the possibility that deregulated YAP promotes malignancy in part by altering the local secretory microenvironment or the tumor niche. Cancer-associated fibroblasts and tumor-associated macrophages are reasonable candidate cell types in this context, as their involvement in cancer by modulating the secretory microenvironment is well documented (Aras and Zaidi, 2017, Kalluri, 2016). YAP's non-cell-autonomous role in tumor development should be more extensively examined (Mugahid et al., 2020).
Experimental Procedures
Mice and Constructs
All mouse work was approved by the Institutional Animal Care and Use Committee of Yale University. All research animals were maintained in facilities of Yale Animal Resource Center. The polycistronic OSKM-2A-mCherry cassette is cloned into the pFUW backbone. Expression constructs for wild-type YAP was subcloned from Addgene no. 21126, and caYAP from Addgene no. 33069. Lentiviral Cre was a gift from the Valentina Greco lab. Various mouse strains have been described previously (Guo et al., 2014, Perl et al., 2002, Stadtfeld et al., 2010, Su et al., 2015). ESCs were derived from E3.5 blastocysts at Yale Animal Genomic Services.
Cell Culture
Isolation and maintenance of primary MEFs and GMPs were described previously (Guo et al., 2010, Takahashi and Yamanaka, 2006). Reprogramming cells and mESCs were cultured on feeder MEFs at 37°C/5% CO2 (Hu et al., 2019). Primary MEFs were transduced with lentiviral constructs encoding EV or YAP, sorted on mCherry and further expanded, followed by irradiation at 8,000 rads. Using the ibidi co-culture device, EV and YAP feeders were plated in the outer minor wells at 4.5 × 104 cells/cm2, with 25 reprogrammable GMPs plated per central minor well. The following morning, medium was added to the entire major well and cultured without further medium change for 4–5 days. Recombinant CYR61, CTGF, PTX-3, and IL-6 (see Supplemental Experimental Procedures) were reconstituted following the manufacturer’ instructions and used at 10, 50, and 200 ng/mL. Fresh recombinant protein and medium were replenished every 48 h for MEF reprogramming. CYR61 blocking antibody or control IgG (Novus Biologicals) was added in similar manner.
Conditioned Medium Collection and Cytokine/Growth Factor Identification Assay
Medium was conditioned for 24 h by EV or YAP feeders plated at 4.5 × 104 cells/cm2 in ESC medium with Dox and reprogrammable GMPs. When indicated, conditioned medium was diluted with fresh medium. For cytokine/growth factor array (Proteome Profiler Mouse XL Cytokine Array Kit, R&D Systems), medium conditioned for 5 days within the ibidi device was used. Spot densitometry was quantified using the QuickSpots software (H&L Image).
Author Contributions
A.A.H. designed, performed, analyzed most of the experiments and wrote the manuscript. S.G. supervised the project, designed experiments and wrote the manuscript. S.M.S. performed some of the cell culture, Q-PCR and apoptosis assays. J.Z. analyzed RNA-seq data. X.H., X.C. A.E.E. C.Y. and S.G. performed various intellectual and technical support. All the authors reviewed the manuscript.
Acknowledgments
This research is supported by Connecticut Innovations 15-RMB-YALE-03 (A.A.H, S.G.) and NIH 1DP2GM123507 (S.G.).
Published: April 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.03.006.
Supplemental Information
References
- Aras S., Zaidi M.R. TAMeless traitors: macrophages in cancer progression and metastasis. Br. J. Cancer. 2017;117:1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Barry E.R., Morikawa T., Butler B.L., Shrestha K., de la Rosa R., Yan K.S., Fuchs C.S., Magness S.T., Smits R., Ogino S. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer T.A., Weiss A., Khomchuk Y., Huang K., Ogunjimi A.A., Varelas X., Wrana J.L. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Brady J.J., Li M., Suthram S., Jiang H., Wong W.H., Blau H.M. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-seq. Nat. Cell Biol. 2013;15:1244–1252. doi: 10.1038/ncb2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chung H., Lee B.K., Uprety N., Shen W., Lee J., Kim J. Yap1 is dispensable for self-renewal but required for proper differentiation of mouse embryonic stem (ES) cells. EMBO Rep. 2016;17:519–529. doi: 10.15252/embr.201540933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C., Inui M., Montagner M., Parenti A.R., Poletti A. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Gilbert P.M., Havenstrite K.L., Magnusson K.E., Sacco A., Leonardi N.A., Kraft P., Nguyen N.K., Thrun S., Lutolf M.P., Blau H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Lu J., Schlanger R., Zhang H., Wang J.Y., Fox M.C., Purton L.E., Fleming H.H., Cobb B., Merkenschlager M. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. U S A. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Zi X., Schulz V.P., Cheng J., Zhong M., Koochaki S.H., Megyola C.M., Pan X., Heydari K., Weissman S.M. Nonstochastic reprogramming from a privileged somatic cell state. Cell. 2014;156:649–662. doi: 10.1016/j.cell.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G., Dupont S., Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Haseley A., Boone S., Wojton J., Yu L., Yoo J.Y., Yu J., Kurozumi K., Glorioso J.C., Caligiuri M.A., Kaur B. Extracellular matrix protein CCN1 limits oncolytic efficacy in glioma. Cancer Res. 2012;72:1353–1362. doi: 10.1158/0008-5472.CAN-11-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Liu Z.Z., Chen X., Schulz V.P., Kumar A., Hartman A.A., Weinstein J., Johnston J.F., Rodriguez E.C., Eastman A.E. MKL1-actin pathway restricts chromatin accessibility and prevents mature pluripotency activation. Nat. Commun. 2019;10:1695. doi: 10.1038/s41467-019-09636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof B.A., Jemelin S., Ballet R., Vesin C., Schapira M., Karaca M., Emre Y. CCN1/CYR61-mediated meticulous patrolling by Ly6Clow monocytes fuels vascular inflammation. Proc. Natl. Acad. Sci. U S A. 2016;113:E4847–E4856. doi: 10.1073/pnas.1607710113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.I., Lau L.F. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat. Cell Biol. 2010;12:676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- Katsube K., Sakamoto K., Tamamura Y., Yamaguchi A. Role of CCN, a vertebrate specific gene family, in development. Dev. Growth Differ. 2009;51:55–67. doi: 10.1111/j.1440-169X.2009.01077.x. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Chen C.C., Alpini G., Lau L.F. CCN1 induces hepatic ductular reaction through integrin alphavbeta(5)-mediated activation of NF-kappaB. J. Clin. Invest. 2015;125:1886–1900. doi: 10.1172/JCI79327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Chen C.C., Monzon R.I., Lau L.F. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Mol. Cell Biol. 2013;33:2078–2090. doi: 10.1128/MCB.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Park J.O., Kim T.S., Kim S.K., Kim T.H., Kim M.C., Park G.S., Kim J.H., Kuninaka S., Olson E.N. LATS-YAP/TAZ controls lineage specification by regulating TGFbeta signaling and Hnf4alpha expression during liver development. Nat. Commun. 2016;7:11961. doi: 10.1038/ncomms11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Sayed N., Hunter A., Au K.F., Wong W.H., Mocarski E.S., Pera R.R., Yakubov E., Cooke J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I., Kim J., Okazawa H., Zhao J.G., Zhao B., Yu J.D., Chinnaiyan A., Israel M.A., Goldstein L.S.B., Abujarour R. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Gene Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T., Ingber D.E. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., Alcazar N., Marion R.M., Chondronasiou D., Rovira M., Fernandez-Marcos P.J., Munoz-Martin M., Blanco-Aparicio C., Pastor J. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354 doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., de Martino A., Serrano M. Senescence promotes in vivo reprogramming through p16(INK)(4a) and IL-6. Aging Cell. 2018;17 doi: 10.1111/acel.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugahid D., Kalocsay M., Liu X., Gruver J.S., Peshkin L., Kirschner M.W. YAP regulates cell size and growth dynamics via non-cell autonomous mediators. Elife. 2020;9 doi: 10.7554/eLife.53404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N. The hippo signaling pathway components lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Nishioka N., Yamamoto S., Kiyonari H., Sato H., Sawada A., Ota M., Nakao K., Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech. Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Fujimura A., Di Biagio D., Frasson C., Bresolin S., Soligo S., Basso G., Bicciato S., Rosato A. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaspyropoulos A., Bradley L., Thapa A., Leung C.Y., Toskas K., Koennig D., Pefani D.E., Raso C., Grou C., Hamilton G. RASSF1A uncouples Wnt from Hippo signalling and promotes YAP mediated differentiation via p73. Nat. Commun. 2018;9:424. doi: 10.1038/s41467-017-02786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.W., Kim Y.C., Yu B., Moroishi T., Mo J.S., Plouffe S.W., Meng Z.P., Lin K.C., Yu F.X., Alexander C.M. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A.K., Wert S.E., Nagy A., Lobe C.G., Whitsett J.A. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. U S A. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Cordenonsi M., Dupont S. Molecular pathways: YAP and TAZ take center stage in organ growth and tumorigenesis. Clin. Cancer Res. 2013;19:4925–4930. doi: 10.1158/1078-0432.CCR-12-3172. [DOI] [PubMed] [Google Scholar]
- Piccolo S., Dupont S., Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Hejna M., Liu Y., Percharde M., Wossidlo M., Blouin L., Durruthy-Durruthy J., Wong P., Qi Z., Yu J. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Deguchi M., Cheng Y., Hsueh A.J. Actin cytoskeleton regulates Hippo signaling. PLoS One. 2013;8:e73763. doi: 10.1371/journal.pone.0073763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H.E., Portela M. Tissue growth and tumorigenesis in Drosophila: cell polarity and the Hippo pathway. Curr. Opin. Cell Biol. 2017;48:1–9. doi: 10.1016/j.ceb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J.R., Zhou D., Kreger B.T., Vasioukhin V., Avruch J., Brummelkamp T.R. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.A. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb. Perspect. Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky N., Presley J., Smith C., Zaal K.J., Cole N., Moreira J.E., Terasaki M., Siggia E., Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G., Heath J.K., Donaldson D.D., Wong G.G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Smith A.G., Hooper M.L. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev. Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- Smith A.G., Nichols J., Robertson M., Rathjen P.D. Differentiation inhibiting activity (DIA/LIF) and mouse development. Dev. Biol. 1992;151:339–351. doi: 10.1016/0012-1606(92)90174-f. [DOI] [PubMed] [Google Scholar]
- Smith T.A., Hooper M.L. Medium conditioned by feeder cells inhibits the differentiation of embryonal carcinoma cultures. Exp. Cell Res. 1983;145:458–462. doi: 10.1016/0014-4827(83)90025-3. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Borkent M., Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Bondar T., Zhou X., Zhang C., He H., Medzhitov R. Two-signal requirement for growth-promoting function of Yap in hepatocytes. Elife. 2015;4 doi: 10.7554/eLife.02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tamm C., Bower N., Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J. Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- Totaro A., Castellan M., Battilana G., Zanconato F., Azzolin L., Giulitti S., Cordenonsi M., Piccolo S. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 2017;8:15206. doi: 10.1038/ncomms15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Wada K., Itoga K., Okano T., Yonemura S., Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Wang K., Zhang S., Weber J., Baxter D., Galas D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J.G., Yuan H.X., Tumaneng K., Li H.R. Regulation of the hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M., Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.M., Ji J.Y., Yu M., Overholtzer M., Smolen G.A., Wang R., Brugge J.S., Dyson N.J., Haber D.A. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–U1134. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.Y., Guan K.L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF beta-TRCP. Gene Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Wang L., Wang C.Y., Yu J., Guan K.L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Gene Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Chadarevian J.P., Ruiz B., Ying Q.L. Cytoplasmic and nuclear TAZ exert distinct functions in regulating primed pluripotency. Stem Cell Reports. 2017;9:732–741. doi: 10.1016/j.stemcr.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo G.W., Kohls C.D., He B.C., Chen L., Zhang W., Shi Q., Zhang B.Q., Kang Q., Luo J., Luo X. The CCN proteins: important signaling mediators in stem cell differentiation and tumorigenesis. Histol. Histopathol. 2010;25:795–806. doi: 10.14670/hh-25.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.