Summary
ADP-ribosylation factor-like 4aa (Arl4aa) is a member of the ADP-ribosylation factor family. It is expressed in hematopoietic tissue during embryonic development, but its function was unknown. Zebrafish arl4aa is preferentially expressed in the ventral wall of the dorsal aorta (VDA) at 24 and 36 hpf and in caudal hematopoietic tissue at 48 hpf. Morpholino knockdown and transcription activator-like effector nuclease (TALEN) knockout of arl4aa significantly reduced expression of genes associated with definitive hematopoietic stem cells (HSCs). Golgi complex integrity in VDA was disrupted as shown by transmission electron microscopy and immunostaining of Golgi membrane Giantin. Mechanistically, arl4aa knockdown reduced Notch signaling in the VDA and its target gene expression. Protein expression of NICD was also reduced. Effects of arl4aa knockdown on definitive hematopoiesis could be restored by NICD expression. This study identified arl4aa as a factor regulating initiation of definitive HSCs by maintaining the integrity of Golgi complex and, secondarily, maturation of the Notch receptor.
Keywords: Arl4aa, Golgi complex, Notch signaling, definitive hematopoietic stem cells, zebrafish
Highlights
-
•
arl4aa expressed in ventral wall of dorsal aorta and caudal hematopoietic tissue
-
•
arl4aa maintained Golgi complex integrity and Notch signaling in definitive HSCs
-
•
Deletion of arl4aa perturbed initiation of definitive HSCs
In this study, Anskar Y.H. Leung and colleagues show that arl4aa was expressed in the ventral wall of the dorsal aorta and caudal hematopoietic tissue during zebrafish embryo development. Deletion of arl4aa disrupted the integrity of the Golgi complex and Notch signaling and perturbed derivation of definitive hematopoietic stem cells from hemogenic endothelium. arl4aa might emerge as a regulator of embryonic hematopoiesis.
Introduction
Embryonic hematopoiesis occurs in three successive waves characterized by a transient primitive wave that is primarily erythroid and myeloid in lineage, an intermediate wave that generates erythromyeloid progenitors, and the definitive wave arising from hematopoietic stem cells (HSCs) that are capable of self-renewal and differentiation into various hematopoietic lineages (Dzierzak and Bigas, 2018, Shi et al., 2018). In mammals, HSCs arise from the aorta-gonad-mesonephros (AGM) and migrate to fetal liver, thence bone marrow (BM), where lifelong hematopoiesis takes place. In zebrafish, HSCs in definitive hematopoiesis arise from the hemogenic endothelium lining the ventral wall of the dorsal aorta (VDA) via endothelial-to-hematopoietic transition (EHT) (Wattrus and Zon, 2018). HSCs migrate into the cardinal vein and are carried to the caudal hematopoietic tissue (CHT) where they expand and subsequently colonize the kidney marrow, the equivalent of mammalian BM.
The Notch pathway plays essential roles at different stages of hematopoietic development. During early embryognesis, somitic expression of Notch ligands, induced by non-canonical Wnt ligand and fibroblast growth factor, has been shown to play a pivotal role in hematopoietic specification (Butko et al., 2016). Direct interaction between the somite and migratory posteral lateral plate mesoderm induced hemogenic endothelial programming of the latter. Subsequently, proinflammatory signals from primitive neutrophils induced expression of Notch ligands in the endothelial cells conducive to the development of HSCs (Butko et al., 2016, Henrique and Schweisguth, 2019). In embryonic tissues, maturation of the Notch receptor occurs via a series of proteolytic cleavages. The first cleavage occurs in the Golgi complex where the nascent receptor is glycosylated and cleaved by furin-like convertases to a Notch receptor precursor. The latter comprises an N-terminal extracellular and a C-terminal intracellular domain and is transported to cell surface. Once bound and activated, the ligand-bound Notch extracellular domain is cleaved by the ADAM family metalloproteases and endocytosed by adjacent signal-sending cells. The rest of the receptor is cleaved by γ-secretase to release Notch intracellular domain (NICD). NICD translocates into the nucleus where it associates with DNA binding protein CSL (CBF1/Su(H)/Lag-1)/RBPjκ to activate transcription of target genes, including Hes1, Hes5, Hey2, and Gata2 (Butko et al., 2016, Guiu et al., 2013, Kanz et al., 2016). gata2b activation is more restricted to the hemogenic endothelium and accounts for HSC development, whereas gata2a activation occurs more widely in the vascular system (Butko et al., 2015).
ADP-ribosylation factor-like protein 4A (ARL4A) is a member of the ADP-ribosylation factor (ARF) family (D'Souza-Schorey and Chavrier, 2006, Donaldson and Jackson, 2011, Stenmark, 2009). The family comprises 6 ARF (ARF1-6) and over 20 ARL proteins. They are low-molecular-weight guanine-nucleotide binding proteins (G proteins) that control membrane traffic and organelle structure and each member is regulated through activation by GTP binding and inactivation by GTP hydrolysis. In particular, ARL4 encompasses three isoforms (ARL4A, ARL4C, and ARL4D). ARL4A has been shown to express in the cell membrane, cytoplasm, and nuclei (Hofmann et al., 2007, Lin et al., 2000) and participate in Golgi complex organization (Lin et al., 2011), cytoskeleton remodeling (Patel et al., 2011), and membrane protein trafficking (Hofmann et al., 2007). Gene expression profiling by microarray in zebrafish has demonstrated expression of arl4a in hematopoietic tissues during zebrafish development (Leung et al., 2005). However, its role in embryonic hematopoiesis is currently unclear.
In this study, we demonstrate expression of arl4aa, a member of the arl4a subfamily, in hematopoietic tissues during embryonic development and reveal a hitherto undescribed function of arl4aa in the initiation of definitive hematopoiesis by maintaining Golgi complex integrity and the maturation of the Notch receptor in hematopoietic stem cells. The observations provide important insights into the regulation of definitive hematopoiesis during embryonic development.
Results
ARL4 Family Members Are Highly Conserved in Human and Zebrafish Species
The zebrafish Arl4 subfamily comprises five isoforms: Arl4aa, Arl4ab, Arl4ca, Arl4cb, and Arl4d. They share in common an N-terminal helix domain, and switch1, switch2, and inter-switch domains as well as a C-terminal nuclear localization signal domain (Figure S1A). The unique short basic extension in the latter differentiates Arl4 from other family members (Pasqualato et al., 2002). Zebrafish Arl4aa and Arl4ab share 89.5% and 87.6% consensus amino acid sequence with human ARL4A and mouse ARL4A (Figure S1B). Phylogenetic analysis demonstrated that zebrafish Arl4aa and Arl4ab cluster with human, mouse, and Xenopus Arl4a (Figure S1C).
Zebrafish arl4aa Is Specifically Expressed in the AGM Region
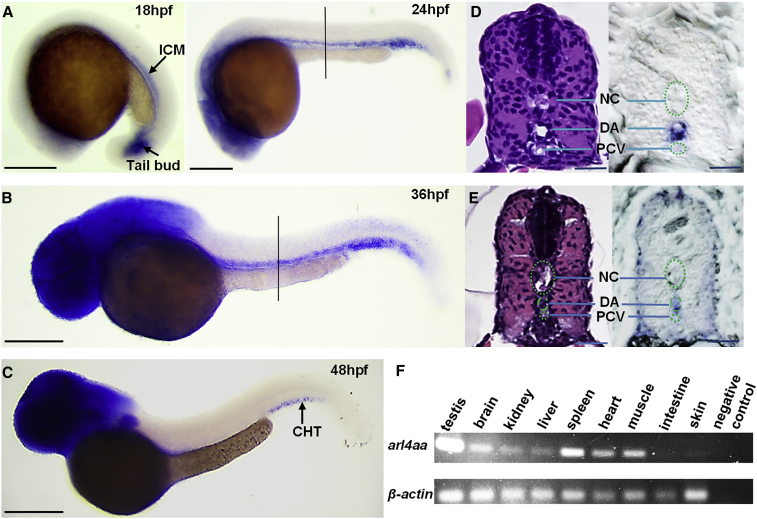
arl4aa is expressed in the intermediate cell mass and tail bud at 18 h postfertilization (hpf), along the DA at 24 and 36 hpf, and in the CHT at 48 hpf. Diffuse expression is also found in the head at 24, 36, and 48 hpf (Figures 1A–1C). Histological sectioning of the trunk showed that arl4aa was expressed almost exclusively in the VDA (Figures 1D and 1E). In the adult, arl4aa is expressed in the kidney, brain, liver, heart, and muscle, and highly expressed in the testis and spleen (Figure 1F).
Figure 1.
Expression Pattern of arl4aa in Zebrafish Embryos and Adults
(A–C) Whole-mount in situ hybridization (WISH) of arl4aa at (A) 18 and 24 hpf, (B) 36 hpf, and (C) 48 hpf.
(D and E) Cross-sections of the trunk at 24 (D) and 36 hpf (E) as shown by H&E staining (left panels) and WISH (right panels), at the level shown by the black lines in (A and B). H&E, hematoxylin and eosin; hpf, hours postfertilization; NC, notochord; DA, dorsal aorta; PCV, posterior cardinal vein. (F) Expression of arl4aa in different adult zebrafish tissues detected by semi-quantitative RT-PCR. Scale bars, 250 μm (A–C) and 50 μm (D and E).
Knockdown and Knockout of arl4aa Reduced Definitive Hematopoiesis
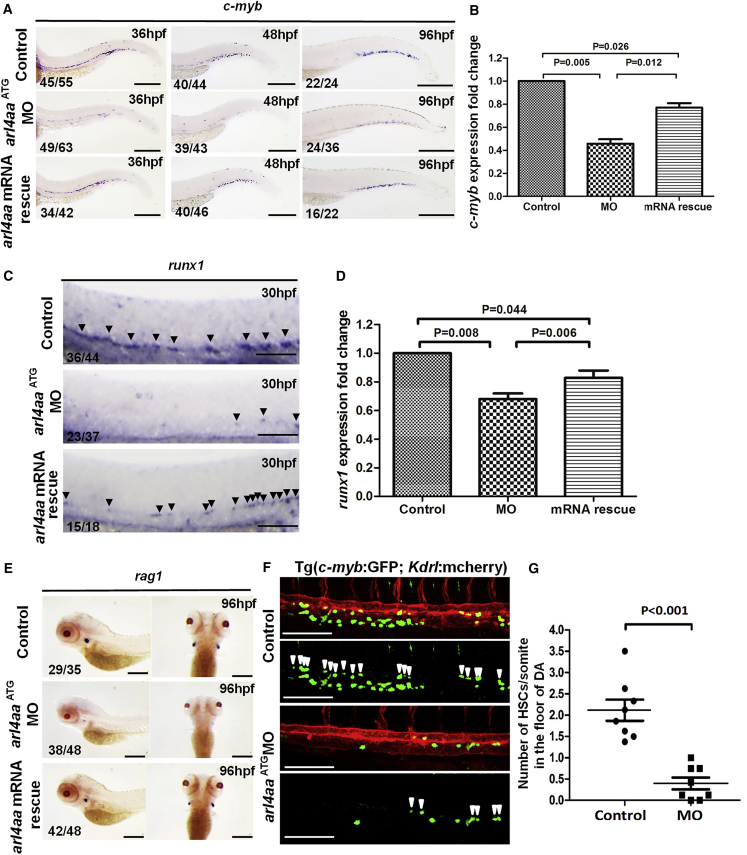
Embryos at 1- to 4-cell stage were injected with arl4aa morpholino (MO) targeting translation start site codon. Dose-finding experiments showed that 1 ng of MO perturbed hematopoiesis with minimal effects on embryonic development and this dose was used throughout the study (Figure S2A). Molecular targeting was confirmed by the quenching of mosaic fluorescence in arl4aa 5′ UTR-GFP injected embryos by arl4aa MO (Figure S2B). Vasculogenesis as well as arterial or venous specification of axial vasculature were unaffected (Figure S2C). c-myb expression in DA at 36 hpf and in CHT at 48 and 96 hpf (Figures 2A and 2B) was significantly reduced. Similar observations of runx1 expression were seen (Figures 2C and 2D). rag1 expression in the thymus at 96 hpf was also reduced (Figure 2E). Defective hematopoiesis could be rescued by MO-resistant arl4aa mRNA that carried 4-bp mismatch in the MO binding sequence but without amino acid changes in the coding region, proving the specificity of the hematopoietic phenotype (Figures 2A–2E). To examine the effects on EHT, a double transgenic kdrl:mCherry; c-myb:GFP fish line was used. The number of kdrl + cmyb + HSCs in the VDA per somite was significantly reduced in the morphant embryos (control embryos: 2.12 ± 0.25, n = 8; morphants: 0.40 ± 0.14, n = 8, p < 0.001, Figures 2F and 2G).
Figure 2.
Knockdown of Zebrafish arl4aa Perturbed Definitive Hematopoiesis
(A) c-myb expression in un-injected control (top), arl4aa morphant (MO) (middle), and morphant rescued by arl4aa mRNA (bottom) at 36 (left), 48 (middle), and 96 hpf (right).
(B) qRT-PCR of c-myb expression in control, MO, and arl4aa mRNA rescue groups (mRNA rescue) in embryos at 48 hpf. Data are representative of three independent experiments with error bars representing the mean ± SEM.
(C) runx1 expression at 30 hpf in un-injected control (top), arl4aa ( middle), and morphant rescued by arl4aa mRNA (bottom). Arrowheads indicate runx1-expressing cells in the ventral wall of the dorsal aorta (DA).
(D) qRT-PCR of runx1 expression in control, MO, and arl4aa mRNA rescue groups (mRNA rescue) in embryos at 30 hpf. Data are representative of three independent experiments with error bars representing the mean ± SEM.
(E) rag1 expression at 96 hpf between control (top), MO (middle), and arl4aa mRNA rescue groups (bottom).
(F) Confocal microscope imaging of double transgenic (cmyb:GFP; kdrl:mCherry) embryos injected with arl4aa MO. Arrowheads denote cmyb + kdrl + HSCs along the DA. The embryos were examined at 36 hpf.
(G) Number of HSCs at the floor of the DA per somite in Tg (cmyb:GFP; kdrl:mcherry) embryos at 36 hpf. The results are presented as mean ± SEM (p < 0.001; n = 8 embryos per group in three independent experiments). Scale bars, 250 μm (A) and 100 μm (C and E).
See also Figure S2.
Zebrafish arl4aa Was Also Knocked Out by TALEN
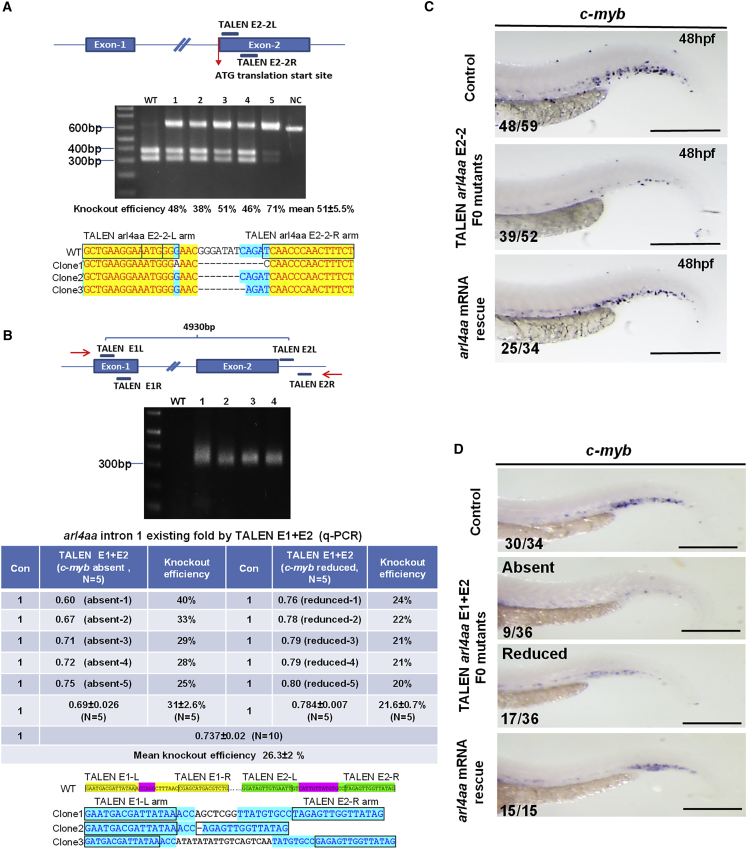
A TALEN pair that induced small insertions and deletions (Indel) at the translation start site (TALEN E2-2 pair) (Figure 3A), as well as two TALEN pairs (TE1+TE2) that induced a large gene deletion between exon 1 and 2, were designed (Figure 3B). TALEN E2-2 (60 pg each arm) showed knockout efficiency of 51% ± 5.5% (Figure 3A) and significantly reduced c-myb expression in CHT at 48 hpf (Figure 3C). Co-injection of TE1 + TE2 (E1 + E2 mRNA 22.5 + 30 pg per arm) resulted in a 4,611-bp deletion with knockout efficiency of 26.3% ± 2.0% (Figure 3B). To examine definitive hematopoiesis, c-myb expression in CHT at 48 hpf was abolished in 25% (9 out of 36) and reduced in 47% (17 out of 36) of F0 embryos (Figure 3D). Genotyping of individual embryos after evaluation of hematopoietic gene expression by whole-mount in situ hybridization (WISH) showed that knockout efficiency was correlated with hematopoietic phenotype, being 31% ± 2.6% and 21.6% ± 0.7% (Figure 3B), respectively, for embryos whose c-myb expression was completely absent and reduced. For both TALEN knockout models, the hematopoietic defects could be rescued by wild-type (WT) arl4aa mRNA (Figures 3C and 3D).
Figure 3.
Knockout of Zebrafish arl4aa Perturbed Definitive HSC Development in F0 Somatic Mutants
(A) Knockout of arl4aa by a TALEN pair (E2-2) targeting the translation start site. Upper panel: the TALEN pair; middle panel: genotyping of single embryos (numbers 1–5) injected with TALEN E2-2. Mean knockout efficiency was 51% ± 5.5% (n = 5); lower panel: DNA sequence of three mutant clones from F0 embryos.
(B) Knockout of arl4aa by two TALEN pairs targeting exons 1 and 2 (E1 + E2). Upper panel: the TALEN pairs, primer positions indicated by red arrows. Middle panel (top): genotyping of single embryos (numbers 1–4) injected with E1 + E2, showing an amplicon of 319 bp. Middle panel (bottom): knockout efficiency was based on qPCR amplifying arl4aa intron 1 existing fold, which was deleted by E1 + E2. Mean knockout efficiency was 26.3% ± 2% (n = 10). Knockout efficiency was correlated with hematopoietic phenotype, being 31% ± 2.6% (n = 5) and 21.6% ± 0.7% (n = 5) for embryos whose c-myb expression was completely absent and reduced. Lower panel: DNA sequencing of three mutant clones from F0 embryos.
(C) c-myb expression in control (left arm TALEN E2-2L) (upper), E2-2 injected (middle), and E2-2 and arl4aa mRNA co-injected (lower) embryos at 48 hpf.
(D) c-myb expression in control (top), E1 + E2 injected embryos showing absent (second) and reduced (third) c-myb expression, and E1 + E2 and arl4aa mRNA co-injected (bottom) embryos. Data are shown as means ± SEM. Scale bars, 250 μm (in WISH). The numbers in each WISH showed the numbers of embryos with typical gene expression as shown over the total number of embryos injected in each group.
To address the potential role of arl4aa in early HSC specification, we examined the effects of arl4aa on somite formation at 12 and 18 hpf, which are sources of Notch signaling required for HSC specification. We demonstrated that arl4aa knockout perturbed early somite formation (Figures S3A and S3B). Furthermore, Notch signaling was reduced, as shown by reduced tp1 expression (Figures S3C–S3E). The observations suggest that arl4aa might play a role in Notch signaling and possibly HSC specification during early embryonic development. A potential link between arl4aa knockout and Notch signaling perturbation is investigated in subsequent sections.
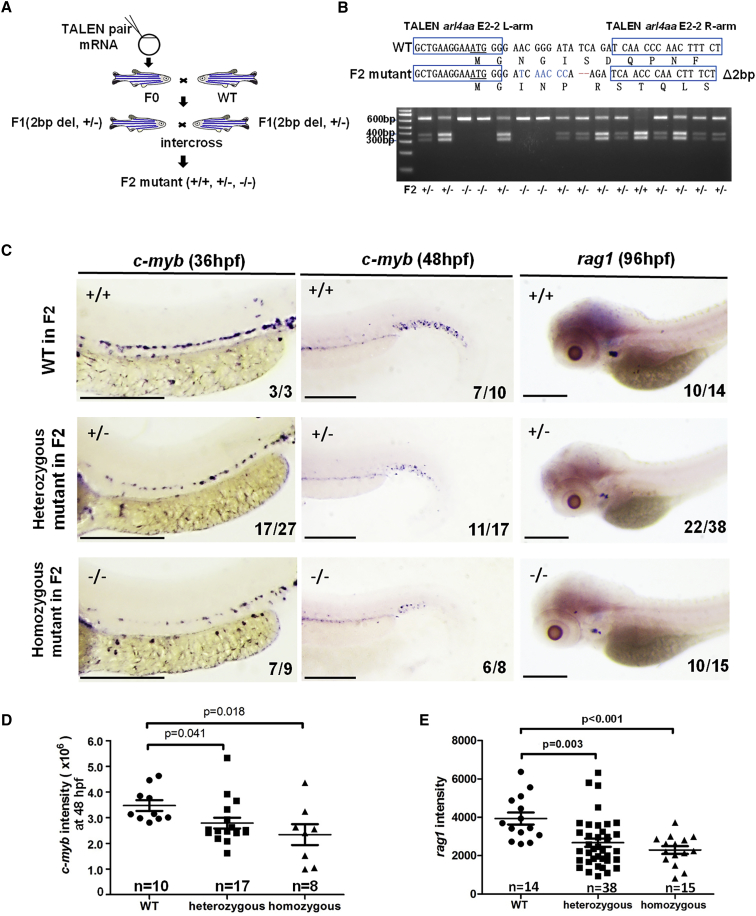
F0 embryos generated by TALEN E2-2 pair were raised and outcrossed with WT fish to generate F1 embryos. The latter were raised and genotyped by tailfin clipping. F1 fish that carried a 2-bp deletion and 6-bp substitutions, with a frameshift in arl4aa, were intercrossed to generate F2 embryos (Figures 4A and 4B). At 36 hpf, 7 out of 9 (78%) homozygous and 17 out of 27 (63%) heterozygous embryos showed a decrease in c-myb expression along the DA. At 48 hpf, 6 out of 8 (75%) homozygous and 11 out of 17 (65%) heterozygous embryos showed reduced c-myb expression in CHT (Figure 4C). rag1 expression in the thymus was also reduced. Quantification of gene expression was performed by measuring the intensities of c-myb expression at 48 hpf and rag1 expression at 96 hpf in WISH using ImageJ software. Both heterozygous and homozygous mutants showed reduced c-myb and rag1 expression compared with their WT siblings (Figures 4D and 4E). The number of embryos in each subgroup was limited, as the F1 adult fish showed lower than expected fecundity. However, the observations arising from multiple gene expression and time points collectively supported the proposition that arl4aa knockout significantly perturbed definitive hematopoiesis.
Figure 4.
Knockout of Zebrafish arl4aa Perturbed Definitive Hematopoiesis in F2 Mutants
(A) Strategy of raising F2 mutant embryos.
(B) DNA sequencing and genotyping of F2 mutant embryos containing 2-bp deletion and 6-bp substitutions. The amino acids are labeled under the DNA sequence.
(C) Expression of c-myb in F2 embryos (upper, WT; middle, heterozygous; lower, homozygous) at 36 (left panel) and 48 hpf (middle panel) and of rag1 at 96 hpf (right panel).
(D and E) Intensity of c-myb expression in CHT at 48 hpf (D) and rag1 expression in thymus at 96 hpf (E) as measured by ImageJ software. Data are shown by means ± SEM; n and p values are indicated in (D) and (E). Scale bars, 250 μm (in WISH). The numbers in each WISH showed the numbers of embryos with typical gene expression as shown over the total number of embryos injected in each group.
arl4aa Is Required for the Maintenance of the Integrity of the Golgi Complex
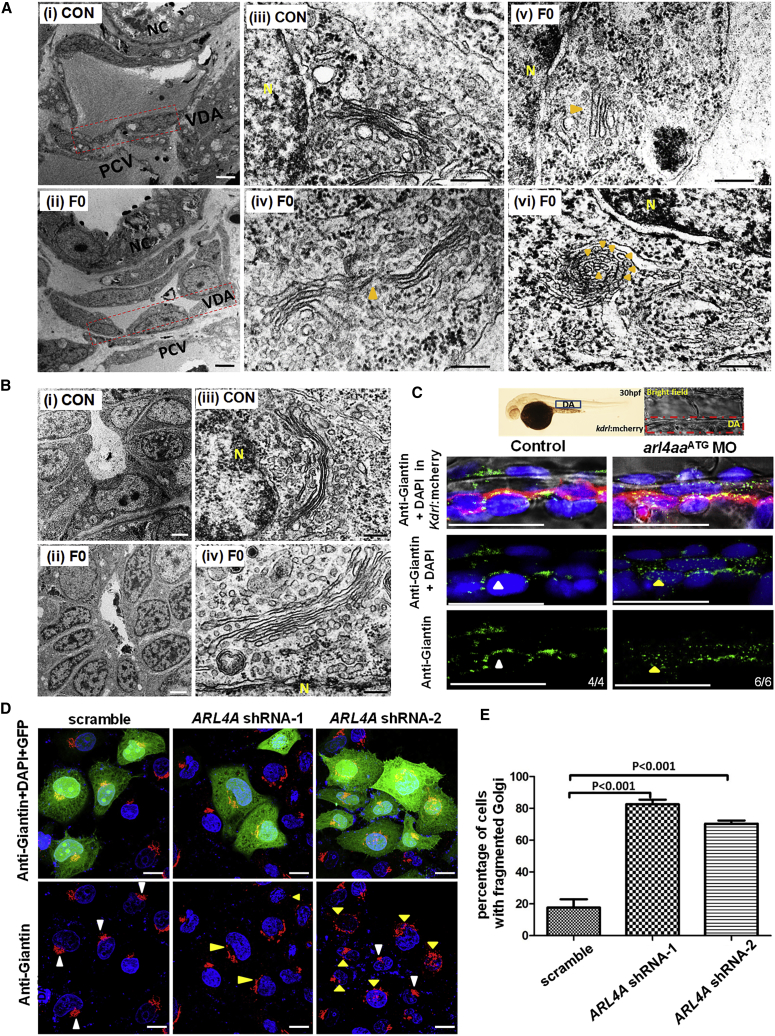
As human ARL4A has been shown to maintain the structural integrity of the Golgi complex, we examined the effects of arl4aa knockout on the Golgi complex in VDA, where arl4aa was intensely expressed. At 30 hpf, subcellular structures of VDA endothelial cells were examined by transmission electronic microscope (TEM) (Figure 5Ai-ii). Confirmation of cell types in each section was based on their anatomical location and neighboring structures. Ten sections each from WT and F0 embryos that contained VDA with identifiable Golgi complex were obtained. Knockout of arl4aa by the TALEN E2-2 pair resulted in morphologic changes of the Golgi complex in F0 embryos. Compared with WT embryos (Figure 5Aiii), the Golgi complexes in mutant embryos were elongated and disrupted (Figure 5Aiv). In others, the Golgi complex was under-developed (Figure 5Av) and arranged into “onion-skin” configuration (Figure 5Avi). Electron microscope photographs of all WT and mutant embryos are shown in Figures S5A and S5B. Overall, abnormal Golgi complex morphology was shown in 9 out of 10 mutant cells and in only 1 out of 10 WT cells (Figures S4A–S4C). Importantly, morphology of mitochondria and endoplasmic reticulum (ER) in the VDA were unaffected in mutant embryos (Figures S5A–S5D). Furthermore, the Golgi complexes in the neural tubes of mutant embryos were unaffected (Figures 5B, S5E, and S5F). These observations suggested that arl4aa functions were specific to the maintenance of the Golgi integrity of the VDA. The integrity of the Golgi complex at the VDA was also evaluated by immunostaining for Golgi membrane protein Giantin. Giantin expression in WT embryos was more localized and condensed in the perinuclear regions, whereas in arl4aa MOs Giantin was more dispersed (Figure 5C). Similar dispersion of Giantin expression was seen in HeLa cells upon ARL4A knockdown by two short hairpin RNAs (shRNAs) (Figures 5D, 5E, S6A, and S6B).
Figure 5.
Deletion of arl4aa and ARL4A Caused Fragmentation of Golgi Complex in VDA Endothelial Cells in Zebrafish and Human HeLa Cells
(A) Electron microscopy of endothelial cells of VDA of un-injected (control [CON]) and F0 embryos injected with TALEN E2-2 at 30 hpf. (i–ii) Endothelial cells of VDA are shown in the rectangles. (iii–vi) High power magnification of control (iii) and F0 embryos (iv–vi). Golgi complex shows typical configuration in control embryos (9/10 cells from 4 control embryos) but aberrant morphology in F0 embryos being unusually elongated with disruption (iv, 6/10 cells from 6 embryos), under-developed (v, 1/10 cells from 6 embryos), and “onion-skin” appearance (vi, 2/10 cells from 6 embryos). NC, notochord; VDA, ventral wall of dorsal aorta; PCV, posterior cardinal vein; N, nucleus.
(B) Electron microscopy of neural tube in CON (i) and F0 embryos (ii) injected with TALEN E2-2, showing normal Golgi complex configuration (iii and iv). Scale bars, 2 μm (Ai,ii and Bi,ii) and 200 nm (Aiii–vi and Biii, iv). Orange arrowheads: abnormal morphology of the Golgi complex.
(C) Immunostaining of Golgi membrane Giantin in control and arl4aa morphant (MO) embryos at 30 hpf. The insert on top shows the morphology of 30-hpf embryos with the rectangle highlighting the region of the DA under detailed examination. The dotted red line in the bright field defines the DA. Immunostaining: Giantin was stained with anti-Giantin followed by Alexa Fluor 488-conjugated antibody and the nuclear outline is shown by DAPI staining. The yellow arrowhead denotes the fragmentation of the Golgi complex in endothelial cells from the VDA. The white arrowhead denotes normal expression of Giantin at the Golgi complex. Scale bar, 100 μm.
(D) Immunostaining of Giantin followed by Alexa Fluor 594-conjugated antibody in scramble sequence control, ARL4A shRNA-1, and ARL4A shRNA-2 transfected HeLa cells. GFP+ labeled the successfully transfected cells. White arrowheads indicate normal Golgi complex and yellow arrowheads denote the fragmented and dispersed Giantin staining. Scale bar, 20 μm.
(E) Both ARL4A shRNA-1 and shRNA-2 significantly increased the proportion of cells with fragmented Golgi complex as shown by Giantin expression, data are shown as means ± SEM (n = 3 experiments, p < 0.001).
See also Figures S4–S6.
arl4aa Is Required for the Maturation of Notch Receptor
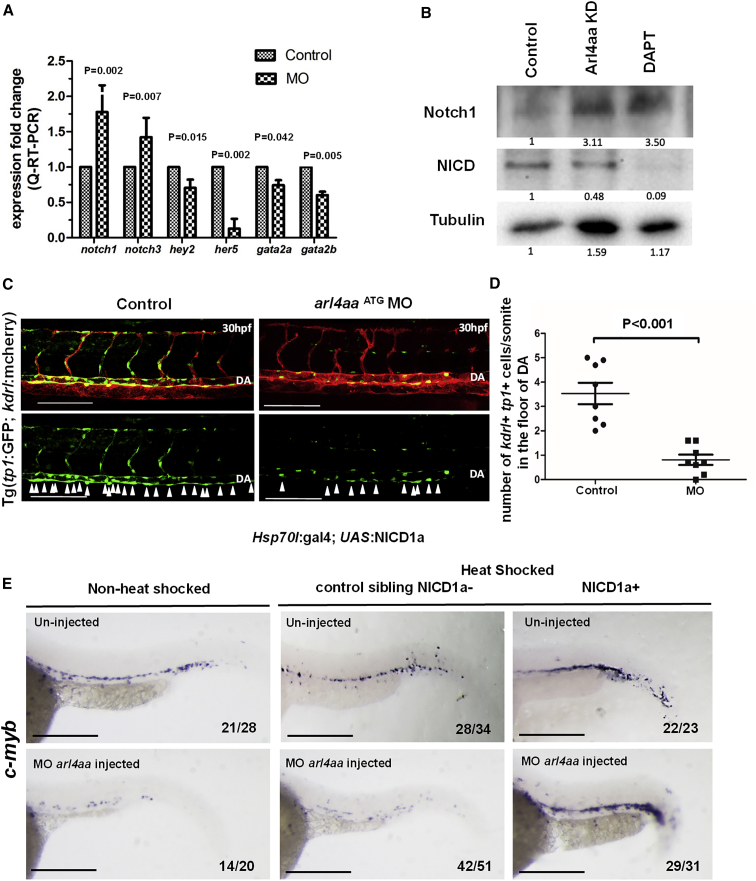
Notch signaling plays a pivotal role in the specification of the hemogenic endothelium (Butko et al., 2016, Clements and Traver, 2013, Kanz et al., 2016, Kim et al., 2014). As arl4aa was important for maintaining the integrity of the Golgi complex and the latter is crucial for Notch receptor maturation, the mechanistic link between arl4aa and Notch signaling was examined. arl4aa MO significantly downregulated expression of Notch target genes hey2, her5, gata2a, and gata2b but increased that of notch1 and notch3 receptors (Figure 6A). Notch1 protein expression was increased but that of NICD was reduced (Figure 6B). arl4aa MO significantly reduced Notch signaling in VDA at 30 hpf as shown by the decrease in tp1 + kdrl + cells in a double transgenic line tp1:GFP; kdrl:mcherry (Figures 6C and 6D). To demonstrate the relationship between arl4aa and Notch signaling, arl4aa was knocked down in a double transgenic line (hsp70l:gal4; UAS:NICD1a-myc) with inducible NICD expression. Heat shock induction of NICD1a expression at the 14–15 somite stage restored c-myb + HSC population along the DA in the arl4aa morphant at 36 hpf (Figure 6E).
Figure 6.
Knockdown of arl4aa Blocked Cleavage of Notch Receptor Precursor and Downregulated Notch Signaling Pathway along DA Floor
(A) Expression of Notch signaling genes in control and arl4aa morphant groups at 42 hpf (notch1, notch3, hey2, and her5) and 33hpf (gata2a and gata2b). Data are representative of three independent experiments with error bars representing the mean ± SEM.
(B) Western blot showing Notch1 and NICD protein expression in control, arl4aa morphant embryos, and DAPT-treated embryos at 42 hpf. The numbers indicate the image intensities of immunoblotting relative to those of tubulin in each embryo group. The intensity in the controls was set arbitrarily as 1. The results are representative of at least three experiments. DAPT, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester.
(C) Confocal microscope imaging of double transgenic tp1:eGFP; kdrl:mCherry embryos injected with arl4aa MO and examined at 30 hpf. Arrowheads indicate cells in the floor of the DA with active Notch signaling. Scale bar, 100 μm.
(D) Enumeration of tp1 + kdrl + HSCs in the floor of DA (n = 8 embryos per group, p < 0.001). Data are representative of three independent experiments with error bars representing the mean ± SEM.
(E) hsp70l:Gal4; UAS:NICD1a embryos injected with arl4aa MO were heat shocked at 14–15 somites and c-myb expression was performed at 36 hpf. Sibling embryos without NICD1a were similarly heat shocked as control. Scale bar, 250 μm. The number of embryos with typical c-myb expression as shown over the total number of embryos examined is shown in each panel.
To provide additional evidence of the proposed mechanistic link, we performed immunofluorescence in HeLa cells upon ARL4A knockdown, with particular reference to the abundance and subcellular localization of the Golgi complex, NOTCH1, and NICD. shRNAs targeting ARL4A resulted in significant reduction in ARL4A protein, attesting to a high knockdown efficiency (Figure S6C). Immunostaining for the Golgi complex was dispersed (Figures S6D and S6E), a finding consistent with our previous observations. Furthermore, there was significant increase in immunostaining for NOTCH1, some of which was located in the peri-nuclear zone (Figure S6D, arrows). There was also a significant decrease in NICD expression (Figure S6E). The observations consolidated our findings in zebrafish and underscored the mechanistic link between ARL4A and maintenance of Golgi complex integrity, where NOTCH receptors are processed (Figure 7). Furthermore, knockdown and knockout of arl4aa induced significant increase in gene expression associated with primitive macrophages (Figures S7A–S7E) but not erythropoiesis or primitive progenitors (Figure S7F). The mechanistic link between Golgi complex integrity and primary macrophage is being investigated.
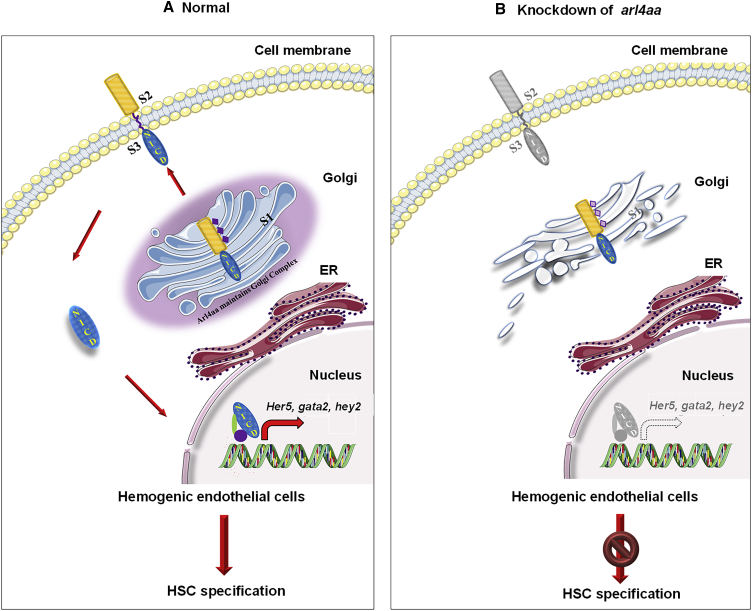
Figure 7.
Proposed arl4aa Function in Zebrafish Definitive HSC Development
(A) Notch signaling in normal hemogenic endothelium (HE) cells. Sequential cleavages of Notch receptors occur in the Golgi complex (S1) and cell membranes (S2, S3). NICD is released and translocated to the nucleus where it activates downstream gene expression.
(B) Deletion of arl4aa disrupted Golgi complex and the first cleavage of Notch precursor, hence production of NICD and subsequent Notch signaling.
Discussion
In this study, we examined the hitherto undescribed expression pattern, functions, and mechanisms of action of arl4aa during zebrafish hematopoiesis. Zebrafish arl4aa shared sequence similarity with human ARL4 and was expressed preferentially in hematopoietic tissues during embryonic development. Both MO knockdown and TALEN knockout demonstrated the pivotal function of arl4aa in the initiation of definitive hematopoiesis. Mechanistically, arl4aa was crucial to Notch signaling by maintaining the integrity of the Golgi complex and hence maturation of the Notch receptor. These observations provided important insights to the regulation of embryonic hematopoiesis.
First, to our knowledge this is the first evidence linking arl4aa, the Golgi complex, and definitive hematopoiesis. Specifically, the process of EHT was perturbed, as shown by the decrease in the kdrl + cmyb + HSC population in the VDA of arl4aa morphant embryos. The hematopoietic effects of perturbing this hitherto ubiquitous gene function might be related to the localized expression of arl4aa at the vicinity of the DA and CHT. Similar expression has been described in a large-scale in situ hybridization screening (Thisse et al., 2004) and a transcriptome profile of blood and endothelial cells in zebrafish embryos (Cannon et al., 2013). Specificity of molecular targeting by arl4aa MO was shown by quenching of EGFP of co-injected arl4aa 5′ UTR-GFP plasmids. Specificity of hematopoietic phenotypes in arl4aa TALEN knockout and MO knockdown embryos could be confirmed by mRNA rescue. Reduction of arl4aa expression was also confirmed in F2 heterozygous and homozygous mutants and was probably due to non-sense-mediated mRNA decay (Lykke-Andersen and Jensen, 2015). The observations that arl4aa knockdown by MO and knockout by TALEN in both F0 and F2 embryos generated the same hematopoietic phenotype also validated these genetic manipulation approaches. Vasculogenesis was unaffected. These observations underscored the specific and fundamental roles of arl4aa in the initiation of EHT. arl4aa was also expressed in the posterior blood island (PBI), although it is presently unclear if arl4aa is involved in the initiation or maintenance of erythromyeloid progenitors.
Second, the observations provided a mechanistic link between arl4aa and Notch signaling. The latter was critically important for HSC specification from hemogenic endothelium. Formation of the aorta was relatively unaffected as shown by kdrl:mcherry transgenic embryos and WISH of efnb2a expression. Knockdown of arl4aa significantly reduced Notch signaling based on reporter transgenic line tp1:GFP, western blot for NICD, and qRT-PCR of Notch target genes. Expression of full-length Notch1 precursor was unaffected; supporting the proposition that arl4aa interfered with the maturation of Notch receptor and subsequent release of NICD. Intriguingly, notch1 and notch3 receptor transcripts were significantly increased. Whether it was mediated by modulation of Notch target genes, of which some were negative regulators of Notch receptor transcription, was unclear (Borggrefe and Oswald, 2009). Drosophila arf1, an ortholog of ARF1, was shown to play an important role in the maintenance of hematopoiesis by providing a nurturing cellular niche in the lymph gland (Khadilkar et al., 2014). Perturbation of arf1 resulted in loss of niche cell and differentiation of prohemocytes (HSCs) and aberrant stalling of NICD in the endosomes of crystal cells. Therefore, the findings of this study might support an evolutionarily conserved mechanism whereby members of the ARF and ARL families regulate hematopoiesis and its niche via their effects on Notch signaling. The impact of Golgi complex integrity on Wnt, VEGF, and Hedgehog pathways that are closely associated with Notch signaling and EHT (Kanz et al., 2016) will need to be investigated.
Third, this study demonstrated a specific role of the Golgi complex in HSC specification. The Golgi complex receives material from the ER, performs post-translational protein modification, and transfers specific cargoes to various cellular destinations, such as endolysosomes and the plasma membrane (Potelle et al., 2015, Zhong, 2011). Perturbation of these processes has been associated with human diseases ranging from neuropsychiatric and musculoskeletal defects to defective erythropoiesis and anemia (Khoriaty et al., 2014, Nieradka et al., 2014). The underlying mechanisms whereby arl4aa maintains the integrity of the Golgi complex are unclear. Human ARL4A has been shown to bind GCC185, one of the coiled-coil proteins known as golgins that are involved in the organization of the Golgi complex and intracellular vesicle transport. GCC185 recruits a family of microtubule-binding proteins, known as cytoplasmic linker-associated proteins, to the Golgi complex that together maintain its structure integrity (Lin et al., 2011). In this study, deletion of arl4aa disrupted the Golgi complex particularly in the VDA of developing embryos. Their ultrastructural changes were correlated with a consistent KO efficiency and high penetrance of hematopoietic defects in the same clutch of embryos, providing a mechanistic link between arl4aa knockout and the morphologic defects of the Golgi complex. Importantly, the morphologies of mitochondria and ER were not affected, suggesting a specific role of arl4aa in the maintenance of Golgi complex integrity. Knockdown of human ARL4A also resulted in disintegration of the Golgi complex in HeLa cells, as shown by the dispersed immunostaining of Giantin. Our results corroborated with those of a previous study showing that deletion of ARL4A, GCC185, and its interacting domain, with ARL4A caused Golgi disruption (Lin et al., 2011). Compared with the VDA, the PBI was less defined under TEM. Therefore, the role of arl4aa in the maintenance of the Golgi complex of the PBI is presently unclear.
Based on the aforementioned observations, we proposed a mechanistic link of arl4aa-Golgi complex-Notch definitive hematopoiesis. Perturbed Notch signaling was central to the phenotype induced by defective arl4aa function as expression of NICD completely restored definitive hematopoiesis. Although disruption of the Golgi complex might induce widespread effects on cellular protein processing, effects of arl4aa on Notch and definitive hematopoiesis were specific, resulting from its unique expression at the VDA where Notch induced initiation of definitive hematopoiesis. Knockdown of methionine aminopeptidase 2 (metap2), a cytosolic metalloproteinase that removes NH2-terminal methionine from newly synthesized peptides and hence essential for protein maturation, also perturbed initiation of definitive hematopoiesis (Ma et al., 2011). When taken together, these observations might suggest an important role of post-translational protein modification in the modulation of signal transduction during hematopoietic specification.
Experimental Procedures
Zebrafish Husbandry, Strains, and Treatment
Embryos and adult fish were raised under standard laboratory conditions at 28°C (Faculty Zebrafish Core Facility, LKS Faculty of Medicine, HKU) and embryos were staged according to Kimmel et al. (1995). The study was approved by the Committee of the Use of Laboratory Animals for Teaching and Research at the University of Hong Kong. The transgenic lines kdrl:mCherry, c-myb:GFP, and mpeg1:GFP were generously provided by Dr Yiyue Zhang (Southern Medical University, Guangzhou, China) and tp1:GFP, hsp70l:gal4 as well as UAS:NICD1a-myc were obtained from Zebrafish International Resources Center (ZIRC). Specifically, heterozygous Tg(hsp70l:gal4) and Tg(UAS:NICD1a-myc) embryos were raised to adulthood, genotyped, and crossed to generate both homozygous and double heterozygous embryos. The specific genotyping primers were provided by ZIRC (Table S1). Embryos of double transgenic line (hsp70l:Gal4; UAS:NICD1a-myc) were placed in E3 medium and incubated in a 37°C water bath for 45 min at the 14–15 somite stage. Specific genotypes of individual embryos after c-myb WISH were also confirmed by PCR.
WISH and Histological Sectioning
WISH was performed as described previously (Ma et al., 2011) using probes for zebrafish arl4aa, c-myb, rag1, scl, pu1, l-plastin, mpeg1, mpo, gata1, ɑ-hemoglobin, efnb2ɑ, and flt4 and myoD. Probe for runx1 was a gift from Dr Yiyue Zhang (Southern Medical University, Guangzhou, China). Probe for myoD was a gift from Miss Chen Chen (Peking University Shenzhen Graduate School, Shenzhen, China). The embryos were observed using a Nikon SMZ800 stereomicroscope (Nikon Corporation, Kawasaki, Japan) and photographed using a Nikon Digital Sight DS-Fi1 camera (Nikon Corporation, Hong Kong, Hong Kong). To provide objective and quantitative assessments of hematopoiesis in zebrafish embryos, the total area of WISH staining was examined and analyzed by ImageJ software (LOCI, University of Wisconsin, Wisconsin). In brief, by adjusting the detection threshold of images, the software was able to distinguish pixels representing positive signals from those that represented the background. The area of positive staining in individual zebrafish was measured and expressed with reference to a defined area of WISH staining.
For histological sectioning, after WISH embryos were fixed and paraffin embedded. Histological sections (10 μm) were stained with hematoxylin and eosin (Sullivan-Brown et al., 2011). Images were taken using a Nikon Eclipse Ni-U microscope (Nikon Corporation, Tokyo, Japan).
Morpholinos, mRNA Synthesis, and Microinjection
Antisense MO (Gene Tools, Philomath, OR) was designed to target the translation start codon of zebrafish arl4aa. A random sequence MO was used as negative control. To test the molecular specificity of MO, 5′ UTR of arl4aa gene containing the MO binding sequence was cloned into the BamHI-EcoRI site of the pEGFP-N3 vector (Clontech, Palo Alto, CA) and the plasmid (25 pg) was injected into one-cell-stage embryos with or without MO. To test if mRNA can rescue the hematopoietic defects induced by MO, arl4aa complete coding sequence was cloned to pGEMT easy vector (Promega, Madison, WI) and capped mRNA was synthesized using the mMESSAGE mMACHINE SP6/T7 Kit (Ambion, Austin, TX). Silent mutations with 4-bp mismatch were introduced into the MO binding sequence using arl4aa-rescue F primer (Table S1) to generate an arl4aa mRNA that was resistant to MO. MO (1.0 ng) and mRNA (50 pg) were injected into 1- to 4-cell stage embryos.
Generation of arl4aa Zebrafish Mutant
Three TALEN pairs were designed by parameters described previously (Ma et al., 2013); the targeted binding sequences and the unique restriction enzyme (RE) site within the spacer regions are described in Table S2. The genome loci was amplified using the genotyping primers (Table S1) and sequenced to avoid any polymorphism within the binding and spacer sites. In brief, the length of the TAL binding domain should be 15 repeat variable di-residues (RVDs) with a spacer region of between 14 and 18 bp. T nucleotide upstream of both TAL binding domains resulted in high activity. Golden Gate TALEN and TAL Effector Kit 2.0 were used to assemble TALEN pairs. TALEN RVDs were cloned into a GoldyTALEN scaffold. Then the constructed plasmid was linearized by SacI and purified using a QIAquick PCR Purification Kit (QIAGEN, Valencia, CA). Capped mRNA encoding the TALEN pairs was synthesized using a T3 mMESSAGE mMACHINE Transcription Kit (Ambion), and injected into one-cell-stage embryos (TALEN E2-2 pair 60 pg per arm, TALEN E1 pair 22.5 pg each arm, and TALEN E2 pair 30 pg each arm). Optimum doses were determined by dose-response trials. Typically, the highest dose with less than 50% embryos developing morphological abnormality was chosen. Somatic and germline TALEN-induced mutations were evaluated by PCR and restriction fragment length polymorphisms. For somatic F0 analysis, genomic DNA was extracted from 24 hpf injected embryos and used as template to perform PCR. PCR product was subsequently digested using RE. The efficiency of designed TALENs can be further evaluated by the intensity of bands using ImageJ software. The efficiency of the combination of two TALEN pairs (TALEN E1 + E2) was tested by quantitative PCR targeting arl4aa intron using arl4ab intron as internal control. Primers used were described in Table S1. The phenotype of somatic mutants was analyzed by WISH followed by genotyping of a single embryo. The F0 embryos generated by the TALEN E2-2 pair were raised to adulthood and outcrossed with WT fish to generate F1 embryos. F1 fish carrying a 2-bp deletion and 6-bp substitutions were intercrossed to generate F2 embryos. Hematopoietic gene expression was shown by WISH and the embryos were classified into homozygous, heterozygous, and WT groups after genotyping.
qRT-PCR
Total RNA isolated from trunks of 20–30 zebrafish embryos was reverse transcribed using a SuperScript II Reverse Transcriptase Kit (Life Technologies, Rockvile, MD). Primers to detect transcripts are described in Table S1. Relative gene expression was calculated according to the difference in threshold cycle numbers (Ct) (2−ΔΔCt) between control and experimental arms.
Western Blot
Protein was harvested by homogenizing 50 embryos (only the trunk region was included) at 42 hpf with 100 μL of CelLytic MT Cell Lysis Reagent (Sigma-Aldrich, St. Louis, MO) supplemented with a protease and phosphatase inhibitor cocktail (1:100, Thermos Scientific, Rockford, IL). WT and morphant embryos as well as embryos treated with 100 μM of N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) (Sigma-Aldrich) from 24 to 42 hpf were examined. DAPT is a γ-secretase inhibitor that inhibits NICD release from Notch receptor. Protein lysates were centrifuged and boiled for 5 min, separated by electrophoresis on 10% (for anti-NICD incubation) and 6% (for anti-Notch1 incubation) (wt/vol) SDS polyacrylamide gel and electro-transferred to nitrocellulose membranes. The latter were then blocked in 5% milk in Tris-buffered saline with Tween 20 for 1 h, incubated at 4°C overnight with rabbit anti-Notch1 (1:750, 20687-1-AP, Proteintech, Chicago, IL), rabbit anti-Notch1 intracellular domain antibody (anti-NICD, 1:1,000, ab83232, Abcam, Cambridge, MA), or mouse anti-β-actin (1:2,000, A2228, Sigma-Aldrich) antibody diluted in blocking buffer, incubated with secondary horseradish peroxidase (HRP)-linked electrochemiluminescence antibodies (sheep anti-mouse-HRP antibody [1:3,000, NA931V, Amersham Biosciences, Little Chalfont, UK], and goat anti-rabbit-HRP antibody [1:1,000, 32,460, Thermo Fisher Scientific, Rockford, IL]) for 2 h at room temperature. Proteins were detected using Luminate Forte Western HRP substrate (Millipore, Billerica, MA) and signals were developed using ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA).
Immunofluorescence Staining
Embryos were fixed in 4% paraformaldehyde (PFA), permeabilized in cold acetone (10 min), 0.1% trypsin (10 min), and 0.5% hyaluronidase (10 min), blocked in 10% Lamb serum in PBS, and incubated with rabbit anti-Giantin (Abcam, ab24586) at 1:300 at 4°C overnight. The next day, the embryos were incubated in goat anti-rabbit IgG Alexa Fluor 488-conjugated (A11008, Invitrogen, Cambridge, MA) at 1:1,000 dilution and 200 ng/mL DAPI (Life Technologies, Carlsbad, CA) at room temperature for 2 h. Thereafter, the embryos were washed with PBST. Images were acquired with a LSM710 confocal microscope (Carl Zeiss, Carlsbad, CA). Details about human ARL4A shRNA construct, transfection in HeLa cell line and immunofluorescence staining were described in Supplemental Experimental Procedures.
Confocal Microscopy Imaging
Tg(cmyb:GFP; kdrl:mCherry), Tg(tp1:GFP; kdrl:mCherry), and Tg(tp1:GFP) embryos were imaged by confocal microscopy. Z sections of the DA region were captured by an LSM 710 confocal microscope (Carl Zeiss, MicroImaging, Jena, Germany) and images were generated by maximum projections using a ZEN 2010 D (Carl Zeiss, MicroImaging GmbH, Jena, Germany) software.
TEM
Embryos at 30 hpf were fixed in 2.5% glutaraldehyde at 4°C overnight and post-fixed in 1% osmium tetroxide for 1 h at room temperature, both in cacodylate buffer (0.1 M [pH 7.4]). They were dehydrated in ethanol (50%, 70%, 90%, and 100%), embedded in epoxy resin, sectioned (100 nm) with an ultramicrotome, and stained with 2% aqueous uranyl acetate and Reynold's lead citrate. Images were obtained with a Philips CM100 Transmission Electronic Microscope (Philips/FEI, Eindhoven, Holland). The assessor of Golgi morphology was blinded to the genotype of the embryos.
Flow Cytometry and mpeg1+ Cell Sorting
Twenty embryos of mpeg1:GFP at 24 hpf were dissociated as described (Ma et al., 2011) and the percentage of GFP+ cells was evaluated by flow cytometry (Cytomics FC 500MPL; Beckman Coulter, Brea, CA).
Statistical Analysis
Data are shown as mean ± SEM. Comparisons were evaluated by Student's t test. p value less than 0.05 was considered statistically significant.
Author Contributions
Y.G. and B.Y.L.C. designed and performed the experiments, analyzed the data, and wrote the manuscript. D.W. and A.C.H.M. designed and performed the experiments and analyzed the data. B.-L.H., T.K.M., X.S., and N.K.L.N. performed the cloning, microinjection, and WISH experiments. M.P.L.C. performed the electronic microscopy experiments and analyzed the data. A.Y.H.L. designed the experiments, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank the HKU LKS Faculty of Medicine confocal microscopy core facility and Zebrafish Core Facility. We appreciate Mr. Lee W.S. from the EM unit of HKU for his assistance in transmission electronic microscopy and Professor Yiyue Zhang and Professor Wenqing Zhang from Southern Medical University, Guangzhou, China, for the generous supply of zebrafish transgenic lines and runx1 WISH probe. We thank Miss Chenchen and Professor Shuo Lin from Perking University Shenzhen Graduate School, Shenzhen, China, for the kind supply of zebrafish myoD WISH probe. A.Y.H.L. is the Li Shu Fan Medical Foundation Professor in Haematology and received funding from its endowment. This work was supported by the General Research Fund (77613), the Collaborative Research Fund (CityU9/CRF/13G), and the NSFC (81328004).
Published: March 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.02.012.
Supplemental Information
References
- Borggrefe T., Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko E., Distel M., Pouget C., Weijts B., Kobayashi I., Ng K., Mosimann C., Poulain F.E., McPherson A., Ni C.W. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development. 2015;142:1050–1061. doi: 10.1242/dev.119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko E., Pouget C., Traver D. Complex regulation of HSC emergence by the Notch signaling pathway. Dev. Biol. 2016;409:129–138. doi: 10.1016/j.ydbio.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J.E., Place E.S., Eve A.M., Bradshaw C.R., Sesay A., Morrell N.W., Smith J.C. Global analysis of the haematopoietic and endothelial transcriptome during zebrafish development. Mech. Dev. 2013;130:122–131. doi: 10.1016/j.mod.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements W.K., Traver D. Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat. Rev. Immunol. 2013;13:336–348. doi: 10.1038/nri3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Donaldson J.G., Jackson C.L. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22:639–651. doi: 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Guiu J., Shimizu R., D'Altri T., Fraser S.T., Hatakeyama J., Bresnick E.H., Kageyama R., Dzierzak E., Yamamoto M., Espinosa L. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J. Exp. Med. 2013;210:71–84. doi: 10.1084/jem.20120993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D., Schweisguth F. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development. 2019;2:dev172148. doi: 10.1242/dev.172148. [DOI] [PubMed] [Google Scholar]
- Hofmann I., Thompson A., Sanderson C.M., Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr. Biol. 2007;17:711–716. doi: 10.1016/j.cub.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kanz D., Konantz M., Alghisi E., North T.E., Lengerke C. Endothelial-to-hematopoietic transition: Notch-ing vessels into blood. Ann. N.Y. Acad. Sci. 2016;1370:97–108. doi: 10.1111/nyas.13030. [DOI] [PubMed] [Google Scholar]
- Khadilkar R.J., Rodrigues D., Mote R.D., Sinha A.R., Kulkarni V., Magadi S.S., Inamdar M.S. ARF1-GTP regulates Asrij to provide endocytic control of Drosophila blood cell homeostasis. Proc. Natl. Acad. Sci. U S A. 2014;111:4898–4903. doi: 10.1073/pnas.1303559111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoriaty R., Vasievich M.P., Jones M., Everett L., Chase J., Tao J., Siemieniak D., Zhang B., Maillard I., Ginsburg D. Absence of a red blood cell phenotype in mice with hematopoietic deficiency of SEC23B. Mol. Cell Biol. 2014;34:3721–3734. doi: 10.1128/MCB.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.D., Stachura D.L., Traver D. Cell signaling pathways involved in hematopoietic stem cell specification. Exp. Cell Res. 2014;329:227–233. doi: 10.1016/j.yexcr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Leung A.Y., Mendenhall E.M., Kwan T.T., Liang R., Eckfeldt C., Chen E., Hammerschmidt M., Grindley S., Ekker S.C., Verfaillie C.M. Characterization of expanded intermediate cell mass in zebrafish chordin morphant embryos. Dev. Biol. 2005;277:235–254. doi: 10.1016/j.ydbio.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Huang P.H., Liao W.L., Cheng H.J., Huang C.F., Kuo J.C., Patton W.A., Massenburg D., Moss J., Lee F.J. ARL4, an ARF-like protein that is developmentally regulated and localized to nuclei and nucleoli. J. Biol. Chem. 2000;275:37815–37823. doi: 10.1074/jbc.M002470200. [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Chiang T.C., Liu Y.T., Tsai Y.T., Jang L.T., Lee F.J. ARL4A acts with GCC185 to modulate Golgi complex organization. J. Cell Sci. 2011;124:4014–4026. doi: 10.1242/jcs.086892. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Ma A.C., Fung T.K., Lin R.H., Chung M.I., Yang D., Ekker S.C., Leung A.Y. Methionine aminopeptidase 2 is required for HSC initiation and proliferation. Blood. 2011;118:5448–5457. doi: 10.1182/blood-2011-04-350173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A.C., Lee H.B., Clark K.J., Ekker S.C. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS One. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieradka A., Ufer C., Thiadens K., Grech G., Horos R., van Coevorden-Hameete M., van den Akker E., Sofi S., Kuhn H., von Lindern M. Grsf1-induced translation of the SNARE protein Use1 is required for expansion of the erythroid compartment. PLoS One. 2014;9:e104631. doi: 10.1371/journal.pone.0104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato S., Renault L., Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for 'front-back' communication. EMBO Rep. 2002;3:1035–1041. doi: 10.1093/embo-reports/kvf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Chiang T.C., Tran V., Lee F.J., Cote J.F. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 2011;286:38969–38979. doi: 10.1074/jbc.M111.274191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potelle S., Klein A., Foulquier F. Golgi post-translational modifications and associated diseases. J. Inherit. Metab. Dis. 2015;38:741–751. doi: 10.1007/s10545-015-9851-7. [DOI] [PubMed] [Google Scholar]
- Shi X., He B.L., Ma A.C., Leung A.Y. Fishing the targets of myeloid malignancies in the era of next generation sequencing. Blood Rev. 2018;30:119–130. doi: 10.1016/j.blre.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sullivan-Brown J., Bisher M.E., Burdine R.D. Embedding, serial sectioning and staining of zebrafish embryos using JB-4 resin. Nat. Protoc. 2011;6:46–55. doi: 10.1038/nprot.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., Kirchner J., Parkhill J.P., Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Wattrus S.J., Zon L.I. Stem cell safe harbor: the hematopoietic stem cell niche in zebrafish. Blood Adv. 2018;2:3063–3069. doi: 10.1182/bloodadvances.2018021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W. Golgi during development. Cold Spring Harb. Perspect. Biol. 2011;3:a005363. doi: 10.1101/cshperspect.a005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.