Summary
We previously discovered in mouse adipocytes an lncRNA (the homolog of human LINC00116) regulating adipogenesis that contains a highly conserved coding region. Here, we show human protein expression of a peptide within LINC00116, and demonstrate that this peptide modulates triglyceride clearance in human adipocytes by regulating lipolysis and mitochondrial β-oxidation. This gene has previously been identified as mitoregulin (MTLN). We conclude that MTLN has a regulatory role in adipocyte metabolism as demonstrated by systemic lipid phenotypes in knockout mice. We also assert its adipocyte-autonomous phenotypes in both isolated murine adipocytes as well as human stem cell-derived adipocytes. MTLN directly interacts with the β subunit of the mitochondrial trifunctional protein, an enzyme critical in the β-oxidation of long-chain fatty acids. Our human and murine models contend that MTLN could be an avenue for further therapeutic research, albeit not without caveats, for example, by promoting white adipocyte triglyceride clearance in obese subjects.
Keywords: adipocyte, metabolism, MTLN, human stem cells, metabolic disease, mitochondrial metabolism
Highlights
-
•
MTLN is expressed in human stem cell-derived adipocytes and murine adipose tissue
-
•
MTLN localizes to mitochondria and associates with mitochondrial trifunctional enzyme
-
•
Adipocytes display decreased fatty acid oxidation upon MTLN knockout
-
•
MTLN KO affects murine serum lipid levels and adipocyte triglyceride accumulation
Cowan and colleagues have demonstrated the expression of mitoregulin (MTLN) in murine adipose tissue as well as human stem cell-derived adipocytes. MTLN affects triglyceride accumulation and lipolysis in adipocytes. It does so through mitochondrial localization and interaction with complexes involved in fatty acid oxidation.
Introduction
Computational annotation of coding genes in the human genome is complicated by myriad non-functional small open reading frames (smORFs) created by random in-frame linking of start and stop codons (Collins and Mansoura, 2001, Venter et al., 2001). For this reason, algorithms predicting coding genes use a minimum of 100 codons, because existing tools cannot discern translated from non-protein-encoding smORFs, resulting in the misclassification of some small coding genes as long non-coding RNAs (lncRNAs). More recently, codon substitution frequency has been used to predict protein-encoding transcripts within lncRNA catalogs (Cabili et al., 2011, Guttman and Rinn, 2012). These analyses suggest the putative existence of thousands of smORF-encoded peptides (SEPs) in the human proteome, many with functional significance in human biology (Anderson et al., 2015, D'Lima et al., 2017, Lee et al., 2015, Matsumoto et al., 2017). Proteomics data suggest that hundreds of these SEPs may be expressed by human cells (Slavoff et al., 2013). Here, we characterized MTLN, an adipocyte-expressed SEP encoded by LINC00116. MTLN has twice previously been described as a mitochondrial regulator of fatty acid oxidation through the mitochondrial trifunctional protein (Makarewich et al., 2018, Stein et al., 2018). Both groups focused on murine muscle, showing changes in mitochondrial respiration upon MTLN knockout (KO). MTLN also regulates C2C12 myoblast differentiation through enhancing mitochondrial respiration (Lin et al., 2019). One of the proteins MTLN interacts with is Cyb5r3, which is one potential way it is regulating lipid metabolism (Chugunova et al., 2019).
However, MTLN is also present in adipose tissue, an important locus of energy homeostasis with high rates of fatty acid metabolism. Adipose tissue comprises an endocrine organ responsible for the storage of excess nutrients in the form of triglycerides (TGs) and mobilization of these energy stores as free fatty acids (FFAs) (Pope et al., 2016). Adipocytes liberate FFAs and glycerol from intracellular TG stores in response to starvation by a mechanism known as lipolysis and secrete these products into the bloodstream for use as fuel by other tissues. Mitochondria catabolize FFAs to produce ATP in a process known as β-oxidation (Roberts et al., 2014). It is postulated that induction of uncoupled mitochondrial respiration in adipocytes (a process referred to as adipocyte browning) could serve as a therapeutic approach to obesity (Moisan et al., 2015). An orthogonal approach would be to augment clearance of fatty acids by mitochondrial β-oxidation. We have explored MTLN's function in lipolysis and β-oxidation using human pluripotent stem cell (hPSC)-derived adipocytes (Lee and Cowan, 2014) as well as murine models.
This approach is very powerful due to the ease with which one can differentiate hPSCs into any desired tissue, and the possibility of genetic manipulation. In addition, any study into human disease or biology is well served by using a human model system. We have exploited these features to study MTLN in human adipocytes with normal, heightened, or abrogated MTLN expression. This approach revealed the mechanism by which MTLN regulates β-oxidation through its physical presence in mitochondria.
Results
Identification of MTLN in Human Adipocytes
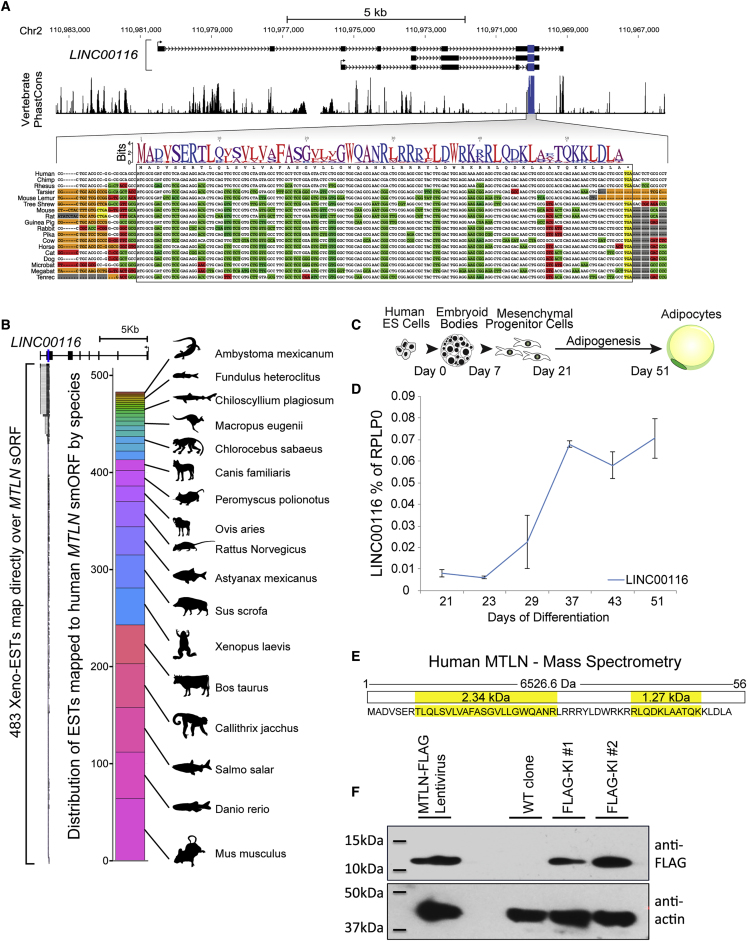
The mouse non-coding RNA 1500011K16Rik, Lnc-RAP5 in the original study, exerts control over lipid accumulation in differentiating mouse pre-adipocytes (Sun et al., 2013). This lncRNA harbors the putative 56-codon MTLN smORF, raising the possibility that the effect on adipocyte function could be driven by either the lncRNA or the encoded SEP. This smORF is conserved in the human homolog of 1500011K15Rik, known as LINC00116 (Figure 1A). Nucleotide and amino acid pairwise identity of this smORF among placental mammals (89.8% and 94.8%, respectively) together with a strong PhyloCSF score (562.72) suggested that the smORF encodes a protein (Lin et al., 2011). This high degree of codon-level conservation is not observed in the immediate flanking regions or elsewhere within the host lncRNA. Evolutionary conservation of the smORF is demonstrated further by 483 annotated expressed sequence tags from 33 vertebrate species that map directly over the human smORF (Figure 1B).
Figure 1.
Identification of a Conserved SEP Expressed by Adipocytes
(A) Alignment of LINC00116's conserved smORF and its homologs from 18 placental mammals, including translation of the SEP.
(B) A total of 483 expressed sequence tags (ESTs) from 33 distinct vertebrate species map over the human MTLN smORF. The distribution of the number of ESTs by species is shown. Xeno-ESTs are derived from translated BLAT alignments of ESTs in GenBank from non-human vertebrates (UCSC “xenoEst” track).
(C) Protocol for differentiation of hPSC-adipocytes. Adipogenesis begins with MPCs on day 21 of the protocol.
(D) qRT-PCR analysis of LINC00116 mRNA levels throughout adipogenesis. n = 3 independent biological replicate experiments.
(E) Primary sequence of the MTLN peptide. Peptides identified by mass spectrometry analysis of hPSC-adipocytes with their respective molecular weights in yellow.
(F) Immunoblot of whole-cell lysate from MTLN-FLAG knockin hPSC-adipocytes and lentivirus-delivered MTLN-FLAG-overexpressing hPSC-adipocytes. β-Actin was used as the loading control.
Data are means ± SD. Related to Figure S1.
To evaluate expression of this smORF in a human adipocyte model, we differentiated hPSC-derived mesenchymal progenitor cells (MPCs) to white adipocytes (hPSC-adipocytes) (Figure 1C). LINC00116 mRNA expression continually increases over 30 days of adipogenesis (Figure 1D). After confirming expression of the LINC00116 mRNA in hPSC-adipocytes, we sought to verify translation of the MTLN peptide. Mass spectrometry of whole-cell lysate from hPSC-adipocytes identified two tryptic peptides (2.34 and 1.27 kDa) derived from MTLN (Figures 1E, S1C, and S1D). In an orthogonal approach, we used CRISPR/Cas9-mediated genome editing to knock in a FLAG tag at the C terminus of the endogenous MTLN gene in the human embryonic stem cell line HUES9 (Figures S1A and S1B). Immunoblotting for the endogenous MTLN-FLAG or an exogenous lentivirus-delivered MTLN-FLAG identified a peptide of >10 kDa, which is not present in parental wild-type (WT) control cells (Figures 1F, S1E, and S1F). Together, these data verify that MTLN is a bona fide SEP encoded by the conserved smORF in LINC00116.
MLTN Knockout Regulates Blood Lipid Levels in Mice
To directly address the physiological role of MTLN in metabolic regulation, we generated MTLN loss-of-function mice by targeting the mouse homolog of MTLN harbored by 1500011K16Rik using CRISPR/Cas9-mediated genome editing (Figure S2A). The resultant MTLN−/− mouse strain carries a 5-base pair deletion at the N terminus of the MTLN smORF (Figure S2B). This mutation does not affect expression of the lncRNA transcript 1500011K16Rik in liver, brown adipose tissue, or gonadal white adipose tissue (Figure 2A).
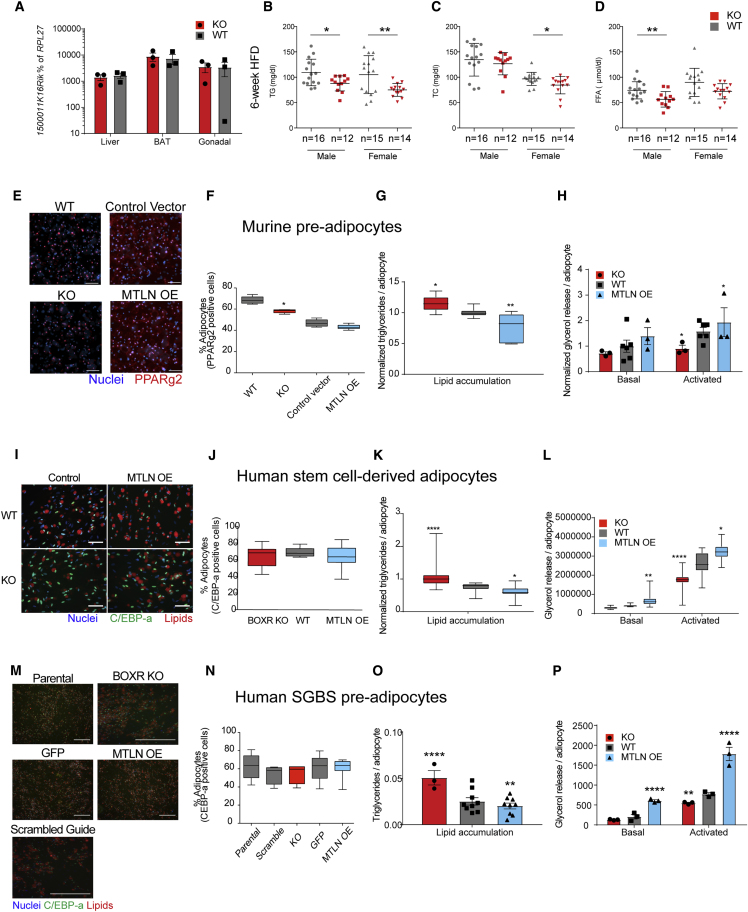
Figure 2.
MTLN Regulates Blood Lipid Levels and Adipocyte Lipid Metabolism
(A) qRT-PCR analysis of 1500011K16Rik in primary mouse liver, brown adipose tissue (BAT), and gonadal adipose tissue (gWAT). n = 3 litter-matched mice, 3 technical replicates each.
(B–D) Serum triglycerides (B), total cholesterol (C), and free fatty acids (D) in mice fed a high-fat diet for 6 weeks, one matched cohort.
(E) Immunofluorescence imaging of murine pre-adipocytes stained for PPARγ2 (red) and nuclei (blue). Scale bars, 100 μm.
(F) High-content imaging quantification of the percentage of PPARγ2-positive nuclei in differentiated murine pre-adipocytes. n = 6.
(G) Quantification of triglyceride accumulation in differentiated murine pre-adipocytes. n = 16 for WT, 8 for KO, 7 for OE biological replicates from one differentiation.
(H) Concentration of glycerol released into the medium of differentiated murine pre-adipocytes under basal or forskolin-stimulated conditions. n = 6 for WT, 3 for KO, 3 for OE.
(I) Immunofluorescence imaging of genome-edited hPSC-adipocytes stained for C/EBP-α (CCAAT/enhancer binding protein alpha) (green), neutral lipids (red), and nuclei (blue). Scale bars, 100 μm.
(J) High-content imaging quantification of the percentage of C/EBP-α-positive nuclei in differentiated hPSC-adipocytes. n = 20 for WT, 40 for KO and OE.
(K) Quantification of triglyceride accumulation in hPSC-adipocytes. n = 33 for WT, 54 for KO, 75 for OE.
(L) Concentration of glycerol released into the medium of hPSC-adipocytes under basal or forskolin-stimulated conditions. n = 30/21 for WT, 42/42 for KO, 66/90 for OE basal/stimulated.
(M) Immunofluorescence imaging of genome-edited SGBS adipocytes stained for C/EBP-α (green), neutral lipids (red), and nuclei (blue). Scale bars, 500 μm.
(N) High-content imaging quantification of the percentage of C/EBP-α-positive nuclei in differentiated SGBS adipocytes. n = 9 for parental, GFP, and MTLN OE, 12 for MTLN KO and scrambled guide RNA control.
(O) Quantification of triglyceride accumulation in SGBS adipocytes. n = 9 for WT, 3 for KO, 12 for OE.
(P) Concentration of glycerol released into the medium of SGBS adipocytes under basal or forskolin-stimulated conditions. n = 3 for WT, KO, and OE.
Data are means ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; one- or two-way ANOVA. Unless specified, “WT” groups include both untransduced and control vector overexpression cells. All n values are independent biological replicates, with 25 fields per replicate averaged for the imaging, and all lipid accumulation and glycerol release done in technical duplicate. Related to Figure S2.
High-fat diets (HFDs) are used to assess metabolic dysfunction in mice, and adipose phenotypes are often uncovered and exacerbated by this feeding regimen. MTLN−/− mice were challenged with HFD to assess the effects of MTLN depletion on lipid handling. During HFD, circulating TG levels were decreased in both male and female MTLN−/− mice compared with WT littermates (Figure 2B). Serum total cholesterol and FFAs also were decreased in female and male MTLN−/− mice, respectively (Figures 2C and 2D). Despite these differences in blood lipid concentrations, there was no change in body weight between MTLN−/− and WT littermates (Figure S2C). On normal chow diet, global depletion of MTLN reduced serum total cholesterol level in female mice (Figure S2E), but had no effect on circulating TG (Figure S2D) or FFA (Figure S2F).
MTLN Regulates TG Accumulation and Metabolism in Murine and Human Adipocytes
HFD experiments using MTLN−/− mice demonstrated that circulating lipid levels are affected by global MTLN depletion. To investigate the adipocyte-autonomous effects of MTLN, we isolated murine pre-adipocytes from WT and MTLN−/− mice. Loss of MTLN led to a mild decrease in differentiation efficiency as measured by PPARg2, although overexpression of MTLN had no effect (Figures 2E and 2F). MTLN did have a significant effect on TG accumulation, leading to increased lipid in the KO, and reduced TGs in the MTLN-overexpressing (OE) cells (Figure 2G). Consistent with this phenotype MTLN also affected forskolin-activated lipolysis in murine adipocytes, which was lower in the KO and higher in the OE cells (Figures 2H and S2K). These results suggest that MTLN has an adipocyte-autonomous effect on lipid handling.
To investigate the human-specific effects of MTLN, we created MTLN KO hPSCs by leveraging CRISPR/Cas9 genome editing (Figure S2G). The resulting four KO hPSC clones contain homozygous frameshift mutations in the MTLN smORF (Figure S2H). Overexpression of MTLN (Figure S1F) persisted after lentiviral transduction of MPCs and differentiation to adipocytes. MTLN KO did not affect transcription of the endogenous LINC00116 transcript (Figure S2I). We used MTLN KO and MTLN OE cells to study the effect of MTLN on lipid metabolism in differentiated human adipocytes.
We assessed human adipocyte differentiation efficiency by measuring the percentage of nuclei positive for the mature adipocyte marker CCAAT/enhancer binding protein alpha (Figure 2I). There was no significant difference in human adipocyte differentiation efficiency between WT, KO, or MTLN OE cells (Figure 2J). In contrast, MTLN had a significant effect on TG accumulation (Figure 2K) and lipolysis (Figures 2L and S2L). MTLN KO caused increased accumulation of TG, whereas MTLN OE resulted in a decrease in intracellular TG. In accord with this phenotype, basal lipolysis was lower in MTLN KO hPSC-adipocytes and increased in MTLN OE hPSC-adipocytes. During stimulation of lipolysis with forskolin, MTLN KO hPSC-adipocytes liberate fewer FFAs into the medium than WT or MTLN OE hPSC-adipocytes. The lipid accumulation and lipolysis phenotypes are strikingly similar between murine and human adipocytes.
Cumulatively, these results suggest that MTLN does not affect human adipogenesis but affects a lipolytic phenotype in human white adipocytes. To validate these findings, we replicated this set of experiments in an immortalized human pre-adipocyte cell line that readily differentiates into mature adipocytes: Simpson-Golabi-Behmel syndrome (SGBS) cells. We targeted the MTLN ORF using CRISPR/Cas9 to make MTLN KO SGBS cells (Figure S2J). In agreement with the hPSC-adipocyte phenotypes, MTLN KO or MTLN OE had no effect on SGBS differentiation efficiency (Figures 2M and 2N). TG accumulation in SGBS adipocytes was reduced by MTLN OE and increased by MTLN KO (Figure 2O). Lipolytic phenotypes of MTLN KO and MTLN OE SGBS adipocytes were also consistent with hPSC-adipocytes (Figure 2P). The consistent phenotypes of MTLN KO and OE adipocytes in both murine and human models implied that the MTLN peptide might localize to the lipid droplet or mitochondria, the sites of lipolysis and fatty acid oxidation.
MTLN Localizes to the Mitochondria and Regulates Mitochondrial Respiration
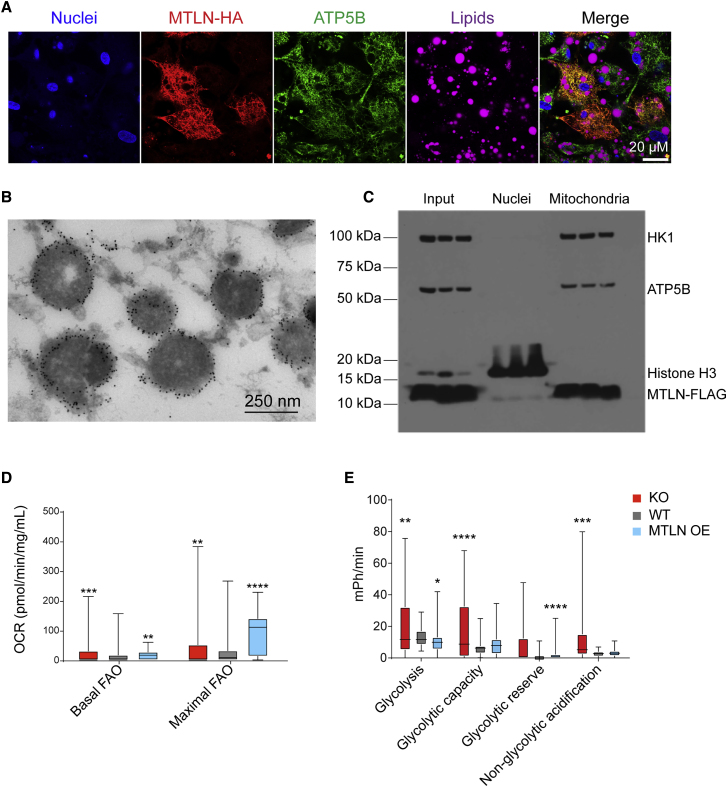
We determined the localization of MTLN using MTLN-HA and MTLN-FLAG OE hPSC-adipocytes by immunofluorescent confocal microscopy as well as transmission electron microscopy with immunogold labeling. Immunofluorescent labeling indicated that MTLN co-localizes with the mitochondrial marker hexokinase-1 (HK1) and the mitochondrial ATP synthase subunit beta (ATP5B) (Figures 3A and S3A–S3C). The immunogold-labeled MTLN was found at the mitochondrial membrane (Figures 3B and S3D–S3F). To verify these measures of subcellular localization, we performed cell fractionation to purify mitochondria from the nuclear and cytoplasmic fractions. HK1, ATP5B, and MTLN-FLAG all localized together in the mitochondrial fraction, whereas only histone H3 was present in the nuclear fraction (Figure 3C). We have been unable to generate an antibody to MTLN, and therefore, have only visualized the overexpressed MTLN constructs. Although neither the FLAG- nor HA-tag have been reported to localize to the mitochondria, we cannot fully exclude an effect of the tag on MTLN localization. However, our findings are consistent with previous publications localizing MTLN to the inner mitochondrial membrane (Makarewich et al., 2018, Stein et al., 2018).
Figure 3.
MTLN Localizes to the Mitochondrion and Affects Energetics Regulation
(A) Confocal microscopy showing MTLN-HA (red), ATP5B (green), neutral lipids (purple), and nuclei (blue) in differentiated hPSC-adipocytes. Scale bar, 20 μm.
(B) Immunogold labeling of MTLN-HA in differentiated hPSC-adipocytes.
(C) Subcellular fractionation of differentiated hPSC-adipocytes. Whole-cell lysate is blotted as input material.
(D) Basal and maximal fatty acid oxidation of hPSC-adipocytes. n = 35.
(E) Comparison of glycolysis, glycolytic capacity, and reserve in hPSC-adipocytes. n = 30. Bars represent mean values ±SD.
∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; two-way ANOVA. All n values are independent biological replicates. Related to Figure S3.
The mitochondrial localization of MTLN and its effect on lipolysis and TG accumulation suggest that the peptide might affect cellular energetic pathways. To test this hypothesis, we assayed glycolysis, glucose-driven mitochondrial respiration, and fatty acid oxidation (FAO) in hPSC-adipocytes. Consistent with our previous lipolysis assays, both basal and maximal FAO were decreased in the KO and increased in the MTLN OE cells (Figures 3D and S3I). To compensate for this energetic imbalance, the KO displayed increased glycolysis, while the OE cells showed reduced glycolysis (Figures 3E and S3G). The KO also showed higher glycolytic capacity, while the OE showed increased glycolytic reserve. Non-FAO mitochondrial oxidative respiration was decreased in MTLN OE hPSC-adipocytes and increased in the KO (Figures S3H and S3J). Despite these dramatic changes in mitochondrial metabolism, we saw no evidence of adipocyte browning or increased mitochondrial content (Figures S3K and S3L). These results dovetail well with previous reports, but add a layer of complexity due to the specific regulation of fatty acid metabolism in adipocytes (Makarewich et al., 2018, Stein et al., 2018). The suppressive effect of MTLN on glycolysis and oxidative respiration and the increase in FAO suggests that MTLN causes adipocytes to favor fatty acid β-oxidation for energy production. The protein constituents of the mitochondrial β-oxidation pathway are well known, so we hypothesized that MTLN interacts with known β-oxidation pathway proteins.
Interaction of MTLN with Key Regulators of Mitochondrial Metabolism
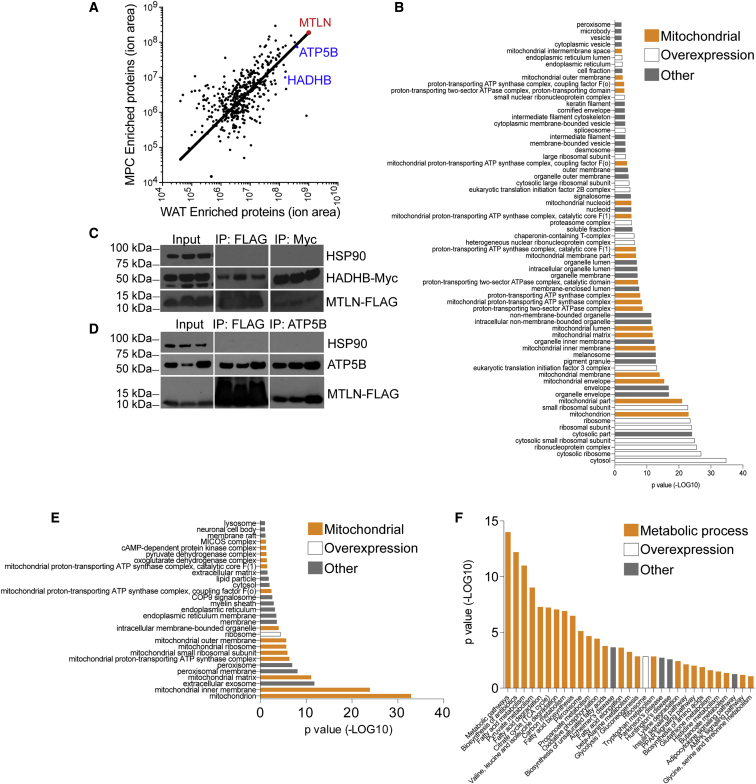
We affinity-purified MTLN-FLAG and interacting proteins from MPCs and hPSC-adipocytes to identify protein binding partners interacting with the peptide. Mass spectrometry analysis suggested that MTLN itself was the most highly enriched protein in both cell types, confirming the specificity of our purification strategy (Figure 4A). Gene ontology analysis of the eluted proteins' subcellular compartmentalization identified the mitochondrion as the most highly enriched cellular compartment associated with MTLN, secondary to translation machinery inherent to our overexpression strategy (Figure 4B). Biological processes significantly associated with MTLN-interacting proteins included fatty acid metabolism and oxidative phosphorylation, among other lipid and metabolite handling processes (Figure S4A). We verified the co-immunoprecipitation of exogenously expressed mitochondrial trifunctional enzyme, subunit beta (HADHB) and the endogenous ATP synthase beta subunit (ATP5B) using immunoblotting (Figures 4B, 4C, and S4B–S4D). The mitochondrial trifunctional enzyme interaction had been described earlier, but the found interaction with ATP5B seems to be a novel or adipocyte-specific process (Makarewich et al., 2018, Stein et al., 2018). As with our visualization experiments, our approach does have the drawback of only using MTLN-FLAG overexpression, not the endogenous gene. We compared these with samples not expressing MTLN-FLAG. Therefore, we followed up by restricting our analysis to adipocytes, where endogenous MTLN is more highly expressed, we see an even stronger bias toward mitochondrial localization and processes. This approach also removed the background inherent to our overexpression strategy. On the other hand, MTLN-protein interactions specific to adipocytes in our dataset almost solely localize to mitochondrial compartments (Figure 4E). Similarly, all the proteins found this way are directly involved in mitochondrial or metabolic processes (Figure 4F). The involvement of MTLN in mitochondrial compartments and metabolic processes, and the direct interaction between MTLN and ATP5B, as well as other lipid-handling proteins, such as the β subunit of the mitochondrial trifunctional enzyme, strengthen our conclusion that MTLN regulates lipid handling in adipocytes.
Figure 4.
MTLN Interacts with the Mitochondrial Energetics Machinery
(A) Mass spectrometric profiling of co-immunoprecipitated proteins bound to MTLN-FLAG in undifferentiated MPCs and hPSC-adipocytes. Red, MTLN; blue, ATP5B and HADHB. Trendline represents the linear regression of these data using a non-linear model. Data based on two independent biological replicates each of MPCs and WAT. Data in Supplemental Table 2.
(B) DAVID gene ontology analysis of the cellular compartment categorization of co-immunoprecipitated proteins bound to MTLN-Flag. Orange, mitochondrial localization; white, protein translation machinery; gray, other. Modified Fisher exact p value, EASE score. Data in Supplemental Table 3.
(C) Immunoblot of HADHB co-immunoprecipitated with MTLN-FLAG. Whole-cell lysate is shown as input material.
(D) Immunoblot of ATP5B co-immunoprecipitated with MTLN-FLAG. Whole-cell lysate is shown as input material.
(E) DAVID gene ontology analysis of the cellular compartment categorization of co-immunoprecipitated proteins bound to MTLN-Flag in hPSC-adipocytes but not MPCs. Orange, mitochondrial localization; white, protein translation machinery; gray, other. Modified Fisher exact p value, EASE score.
(F) Gene ontology analysis describing the biological pathways enriched in the co-immunoprecipitated proteins bound to MTLN-Flag in hPSC-adipocytes but not in MPCs. Orange, mitochondrial processes; white, protein translation and overexpression; gray, other. Pathways and processes are colored through personal interpretation of DAVID terms. Modified Fisher exact p value, EASE score.
Related to Figure S4.
Discussion
Our experiments point toward the MTLN SEP of LINC00116 as an important functional element in β-oxidation of fatty acids in human adipocytes. This situates MTLN among the rapidly expanding class of SEPs encoded by lncRNAs, supporting the hypothesis that there are many functional small peptides yet to be discovered. MTLN localizes to the mitochondria and affects energetics and lipid metabolism in human adipocytes, mice, and isolated murine adipocytes. Interaction with HADHB and accumulation of TGs in MTLN KO hPSC-adipocytes implicates MTLN in long-chain fatty acid catabolism. HADHB catalyzes the final step in long-chain FAO, thought to be the rate-limiting step (Li and Schulz, 1988, Olowe and Schulz, 1982). Our data suggest MTLN augments the rate of β-oxidation of long-chain fatty acids, possibly through direct interaction with HADHB. It does so at the expense of other energy-producing pathways, including glycolysis and non-FAO mitochondrial respiration. In this model, decreased intracellular fatty acids would decrease feedback inhibition of lipolysis (Fain and Shepherd, 1975), resulting in the lipolytic phenotype observed in our in vitro systems. Ongoing research will determine the mechanism by which MTLN modulates the rate of β-oxidation, how the peptide is trafficked to the mitochondria, and how MTLN overexpression affects systemic phenotypes in mice.
Our data suggest that there is regulation of MTLN's function through certain environmental conditions. MTLN does not have an effect on browning adipocytes from our data; therefore, we speculate it is not regulated by cold exposure. However, it appears that nutritional status has a profound effect on MTLN's function. Our Seahorse data show that oxidative phosphorylation is specifically enhanced under nutrient starvation conditions, when the adipocytes are utilizing endogenous lipids for FAO. This phenomenon may be very interesting for further study into the fuel source preference of MTLN activity.
Aside from the endogenous expression, most of our experiments have been performed with tagged overexpression of MTLN. We were not able to generate an antibody to the endogenous peptide; hence, this was our only viable approach. It is hoped that future work can follow up on our data and confirm the peptide interactions using the endogenous MTLN.
The pool of undiscovered functional SEPs remains fertile ground for drug discovery and development. hPSCs are a perfect model to study the role of these novel genes in a human system as they are amenable to functional screening, genetic manipulation, as well as differentiation into any desired tissue to mechanistically understand a SEP’s purpose. This study can serve as a blueprint for functional discovery of uncharacterized SEPs, of which there are now predicted to be a plethora.
The small size of SEPs lends them to repurposing as peptide therapies, particularly if they are secreted and function in an endocrine fashion to modulate systemic phenotypes (Lee et al., 2015). MTLN's apparent function recommends its application as a gene therapy for clearance of excess lipids in adipose tissue. Obesity is on the rise in developed and developing countries, with 500 million cases worldwide (Malik et al., 2013). Gene therapies delivering MTLN to white adipose could combat this global trend, serving as a biological trigger for the catabolism of FFAs by β-oxidation. However, further research is warranted as care needs to be taken that this does not lead to the unfavorable metabolic state of deleterious increased plasma TG levels.
Experimental Procedures
Mouse Models
All procedures used in animal experiments were in accordance and with approval from the pertinent Institutional Animal Care and Use Committees at Harvard University. MTLN KO mice were generated via CRISPR/Cas9 technology in the Genome Modification Facility at Harvard University. Both male and female MTLN global KO mice and WT littermates were used in this study. They were housed three to five mice per cage with free access to water and normal chow diet or HFD (60/FAT, TD.06414). Blood was collected from tail tips after 16-h fasting, and plasma was further isolated.
Cell Lines
HUES9 embryonic stem cells were cultured as described previously (Cowan et al., 2004) in mTeSR growth medium (STEMCELL Technologies). Adipocyte differentiation of embryonic stem cells was performed as described previously (Lee and Cowan, 2014). Genome editing in the HUES9 cell line was performed following a published protocol (Peters et al., 2013) using the guide RNA and screening primer sequences in Supplemental Table 1. Cell lines were tested for mycoplasma contamination and were not contaminated during this study.
The SGBS cell line was a gift from Dr. Martin Wabitsch's lab and cultured as described previously (Wabitsch et al., 2001). Fully confluent SGBS cells were differentiated to adipocytes using adipogenic medium: DMEM (Corning) supplemented with 7.5% Knockout Serum Replacement, 1% penicillin/streptomycin, 0.5% non-essential amino acids, 1 μM dexamethasone, 10 μg/mL insulin, and 0.5 μM rosiglitazone for 21 days. To generate Cas9-positive SGBS cells, lentivirus was packaged using the LentiCas9-Blast plasmid available Addgene. After application of lentivirus, Cas9-positive SGBS cells were selected using 10 μg/mL blasticidin for 5 days. Guide RNAs were cloned into the LentiGuide-Puro plasmid and lentivirus was packaged according to a published protocol (Lee and Cowan, 2014). Three guide RNA lentiviruses were pooled and applied to SGBS-Cas9 cells. These cells were then selected with puromycin (2 μg/mL) for 4 days. Selected cells were then differentiated according to the above protocol.
Isolation and Differentiation of Pre-adipocytes from Mouse White Adipose Tissue
Inguinal fat pads from 6- to 8-week-old mice were carefully dissected, cut into small pieces, and digested with 1 mg/mL collagenase A at 37°C for 45 min. The cell suspension was then filtered through a 100-μm cell strainer followed by centrifugation at 150 × g for 5 min. The cell pellet was resuspended with complete medium (DMEM/F12 with 10% fetal bovine serum [FBS], 1% penicillin/streptomycin) and plated in gelatin-coated 10-cm plates. For differentiation 50,000 cells were seeded per well of a 24-well plate. The next day, induction medium (complete medium with 1 μM dexamethasone, 5 μg/mL insulin, 0.5 mM IBMX, and 5 μM rosiglitazone) was added. On day 4, maintenance medium (complete medium with 1 μg/mL insulin and 5 μM rosiglitazone) was added and the cells further differentiated for 11 days before performing the assays.
RNA Isolation, cDNA Synthesis, and qPCR
RNA was extracted from adherent cells using TRIzol reagent reagent according to the manufacturer's instructions (Thermo Scientific). cDNA synthesis was performed using qScript cDNA SuperMix according to the manufacturer's instructions (Quanta Biosciences). qPCR of LINC00116, MTLN, UCP1, CPT1, RPLP0, 1500011K16Rik, and RPL27 was performed using primer sets listed in Table 1 in conjunction with Fast SYBR Green Master Mix (Thermo Scientific). Expression data are presented after calculating the relative expression compared with the housekeeping gene RPLP0 or Rpl27, using the equation relative quantification (RQ) = 100/(2ˆ(Target Gene Ct – RPLP0 Ct)).
Immunofluorescence
Cells were washed in phosphate-buffered saline (PBS) and then fixed in ice-cold methanol at −20°C for 15 min. Cells were then washed three times with PBS, and blocked for 1 h at room temperature with 5% (w/v) bovine serum albumin (BSA), Fraction V (Millipore), 0.1% Triton X-100 (Sigma) in PBS. Primary antibodies were incubated overnight at 4°C. Cells were then washed three times in PBS, and incubated with secondary antibodies conjugated to fluorophores for 1 h at room temperature. Nuclei were labeled with Hoechst (Thermo Scientific) and neutral lipids were stained with the LipidTOX Red or LipidTOX Deep Red (Thermo Scientific) fluorescent dye for 30 min at room temperature. Antibodies and dyes with working dilutions are listed in Table 1.
TG Quantification
Cells were washed in PBS, scraped in PBS supplemented with 5% Nonidet P40 Substitute (Sigma). Cells were lysed by two 30-s cycles of sonication on high power using a BioRuptor sonicator (Diagenode). Debris was pelleted at 14,000 × g for 10 min at 4°C, then soluble protein concentration was measured using the bicinchoninic acid assay following the manufacturer's instructions (Thermo Scientific). In both a commercial standard (Abcam) and experimental samples, TGs were solubilized by two cycles of heating to 95°C for 2 min followed by vortexing. TG concentrations were measured using the Infinity Triglycerides Reagent (Thermo Scientific) as per the manufacturer's instructions. Concentrations were normalized to protein concentration and differentiation efficiency, and plotted as TGs/adipocyte.
Lipolysis Assay
Differentiated adipocytes were starved for 4 h in serum-free no-glucose DMEM, and treated with or without forskolin for an additional 4 h at 37°C. Medium was collected, and cells were harvested as described above for quantification of TG content. Concentration of FFAs and glycerol in conditioned medium was quantified using the Lipolysis Assay Kit (ZenBio, Durham, NC) according to the manufacturer's instructions. Concentrations were normalized to TG and protein concentration of whole-cell lysate and differentiation efficiency, and plotted as FFA release/adipocyte or glycerol release/adipocyte.
Expression Plasmid Construction
The evolutionarily conserved MTLN peptide sequence was downloaded from NCBI and synthesized in vitro (Eurofins Genomics) for cloning with a multi-site gateway cloning strategy (Thermo Scientific). A lentiviral packaging vector, a donor plasmid carrying the EF1 alpha promoter, and a donor plasmid carrying the MTLN peptide tagged with a C-terminal 3× FLAG or 3× HA peptide were recombined in a three-way cloning reaction.
Cell Fractionation
Nuclei were isolated from five million cells using a 2% Nonidet P40 Substitute (Sigma Aldrich) solution, otherwise following a published protocol (Nabbi and Riabowol, 2015). Mitochondria were then pelleted by centrifugation at 17,000 × g for 15 min at 4°C.
Western Blotting
Bis-Tris polyacrylamide gels (4%–12%) were run using LDS sample buffer with MOPS running buffer (Thermo Scientific). Cells were lysed using RIPA buffer, cell debris was pelleted at 14,000 × g, and soluble protein concentrations were measured as described above. Protein samples were supplemented with a final concentration of 5 mM dithiothreitol before heating at 95°C for 5 min. After polyacrylamide gel electrophoresis using a Mini-Cell apparatus (Thermo Scientific), proteins were transferred to a nitrocellulose membrane in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol) for 1 h by applying 100 V of constant voltage at 4°C. Primary and secondary antibodies and working dilutions are detailed in Table 1.
Transmission Electron Microscopy
MTLN OE or WT hPSC-adipocytes were detached from the dish with a cell scraper. The cell suspension was layered on top of a 200-μL cushion of 4% paraformaldehyde (PFA)/0.1% glutaraldehyde (in 0.1 M sodium phosphate buffer [pH 7.4]) in an Eppendorf tube and pelleted for 3 min at 3,000 rpm. The supernatant was removed carefully, fresh 4% PFA/0.1% glutaraldehyde mixture was added without resuspending the pellet. After 2 h of fixation at room temperature the fixative was replaced with PBS without disturbing the pellet. PBS then was replaced with 2.3 M sucrose in PBS (containing 0.2 M glycine to quench free aldehyde groups) for 15 min. The pellet then was flash frozen in liquid nitrogen. Samples were sectioned at −120°C, and the sections were transferred to formvar- and carbon-coated copper grids. Grids were floated on PBS until immunogold labeling was carried out. Gold labeling was carried out at room temperature on a piece of parafilm. All antibodies and protein A-gold were diluted in 1% BSA. The diluted antibody solution was centrifuged for 1 min at 14,000 rpm before labeling to avoid possible aggregates. Grids were floated on drops of 1% BSA in PBS for 10 min to block for non-specific labeling, transferred to 5-μL drops of primary antibody, and incubated for 30 min. The grids were then washed in 4 drops of PBS for a total of 15 min, transferred to 5 μL drops of protein A-gold for 20 min, washed in 4 drops of PBS for 15 min and 6 drops of double distilled water. Contrasting/embedding of the labeled grids was carried out on ice in 0.3% uranyl acetete in 2% methyl cellulose for 10 min. Grids were picked up and the excess liquid was removed by streaking on filter paper (Whatman grade 1), leaving a thin coat of methyl cellulose. The grids were examined in a JEOL 1200EX and images were recorded with an AMT 2k CCD camera.
Extracellular Flux Analysis
A Seahorse XF24 Analyzer (Seahorse Bioscience) was used to assess oxygen consumption and extracellular acidification rates. MPCs (40,000) were plated in each well 2 days before induction of adipogenesis by the above protocol. The mitochondrial stress test, glycolysis stress test, and the XF Palmitate-BSA FAO Substrate kits were used according to the manufacturer's protocol. Carbonyl cyanide m-chlorophenylhydrazone was used at 0.5 μM final concentration in the mitochondrial stress test, and at 2 μM final concentration in the FAO assay. Rate measurement cycles consisted of 2 min of mixing, 1 min of waiting, and 5 min of measurement.
Co-immunoprecipitation
Differentiated adipocytes were lysed in Pierce IP Buffer (Thermo Scientific) supplemented with protease and phosphatase inhibitors (Roche Life Science). Cell debris was pelleted at 20,000 × g for 15 min at 4°C. After removal of the lipid supernatant, 10 mg of protein from the infranatant was incubated overnight on an end-over-end rotator at 4°C with antibodies directed against the FLAG-tag (conjugated to magnetic beads, Sigma Aldrich, 20 μL of packed gel volume), ATP synthase, beta subunit (10 μg, Abcam, ab128743), or the C-terminal Myc tag of HADHB (10 μg, Abcam, ab9106) (Table 1). Pre-immune immunoglobulin immunoprecipitations (IPs) were performed as controls with equal quantities of protein, using magnetic bead-conjugated mouse immunoglobulin G (IgG) (Cell Signaling Technology) or rabbit IgG (Santa Cruz). After overnight incubation, ATP synthase and HADHB IP reactions and rabbit IgG control reactions were incubated with protein A-conjugated magnetic beads (25 μL, Thermo Scientific) for 1 h at room temperature on an end-over-end rotator. Bead-protein complexes were pelleted on a magnetic stand and washed with Tris-buffered saline ten times. Protein was eluted from the beads using 100 μL of 0.1 M glycine (pH 3.0). Samples were equilibrated to neutral pH by addition of 1 M triethylammonium bicarbonate (TEAB) buffer (pH 8.0) to a final concentration of 100 mM TEAB.
Co-immunoprecipitation Proteomics
Each sample was submitted for a single liquid chromatography-tandem mass spectrometry experiment that was performed on an LTQ Orbitrap Elite (Thermo Fischer) equipped with a Waters nanoACQUITY HPLC pump. Peptides were separated onto a 100-μm inner diameter microcapillary trapping column packed first with approximately 5 cm of C18 ReproSil resin (5 μm, 100 Å, Dr. Maisch, Germany) followed by an analytical column ∼20 cm of ReproSil resin (1.9 μm, 200 Å, Dr. Maisch). Separation was achieved by applying a gradient from 5% to 27% acetonitrile in 0.1% formic acid over 90 min at 200 nL min−1. Electrospray ionization was enabled by applying a voltage of 1.8 kV using a homemade electrode junction at the end of the microcapillary column and sprayed from fused silica pico tips (New Objective). The LTQ Orbitrap Elite was operated in the data-dependent mode for the mass spectrometry methods. The mass spectrometry survey scan was performed in the Orbitrap in the range of 395–1,800 m/z at a resolution of 6 × 104, followed by the selection of the 20 most intense ions (TOP20) for collision-induced dissociation (CID)-tandem mass spectrometry fragmentation in the ion trap using a precursor isolation width window of 2 m/z, automatic gain control (AGC) setting of 10,000, and a maximum ion accumulation of 200 ms. Singly charged ion species were not subjected to CID fragmentation. Normalized collision energy was set to 35 V and an activation time of 10 ms, AGC was set to 50,000, and the maximum ion time was 200 ms. Ions in a 10-ppm m/z window around ions selected for tandem mass spectrometry were excluded from further selection for fragmentation for 60 s.
Statistical Analysis of qPCR, Mouse Cohort, Differentiation Efficiency, TG Assay, Lipolysis Assay, and Extracellular Flux Analyzer Data
Prism 5 (GraphPad) was used to create charts and perform statistical analyses in all figures. Student's t test (paired), one- (three-group) or two-way ANOVA (three or more groups, multiple conditions) was used to compare KO and OE genotypes or treatment conditions to the WT group.
Gene Ontology Analyses
Co-immunoprecipitated proteins were analyzed using DAVID (Huang da et al., 2009). Ion area values of background control IP samples were subtracted from MTLN-Flag IP sample values to determine MTLN-Flag-specific enrichment of protein interactants.
Analysis of Proteomics Data
Raw data were submitted for analysis in Proteome Discoverer 2.1.0.81 (Thermo Scientific) software. Assignment of tandem mass spectrometry spectra was performed using the Sequest HT algorithm by searching the data against a protein sequence database, including all entries from the Human UniProt database (SwissProt 16,768 and TrEMBL 62,460 [total of 79,228 protein forms], 2015) and other known contaminants, such as human keratins and common lab contaminants. Sequest HT searches were performed using a 20-ppm precursor ion tolerance and requiring each peptide N/C terminus to adhere with trypsin protease specificity while allowing up to two missed cleavages. Cysteine carbamidomethyl (+57.021) was set as static modification while methionine oxidation (+15.99492 Da) was set as variable modification. Tandem mass spectrometry spectra assignment false discovery rate of 1% on protein level was achieved by applying the target-decoy database search. Filtering was performed using a Percolator (64 bit version, reference 6). For quantification, a 0.02-m/z window centered on the theoretical m/z value of each the six reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were exported in result files of Proteome Discoverer 2.1 searches as Excel tables. All fold changes were analyzed after normalization between samples based on total unique peptide ion signals.
Author Contributions
Manuscript Preparation, M.F. and C.R.W.; Study Supervision, C.A.C.; Study Support, J.L.R. and C.A.C.; smORF Conservation, L.A.G. and J.L.R.; Human PSC and Mouse Genome Editing, Q.D.; Mass Spectrometry, M.H.C.F. and C.R.W.; Mouse Experiments, H.Y. and T.T.; Murine Pre-adipocyte Characterization, M.F., H.Y., and T.T.; Human Adipocyte Characterization, M.F. and C.R.W.; SGBS Pre-adipocyte Characterization, M.F. and H.Y.; smORF Localization, M.F. and C.R.W.; Metabolic Assays, M.F.; Expression and Proteomics Analysis, M.F.; Supporting Experiments, M.H.C.F., C.S., and B.D.P. All authors approved the results and manuscript.
Acknowledgments
The authors thank the Harvard University Mass Spectrometry and Proteomics Resource Laboratory for their assistance with proteomics analyses. Thanks to Maria Ericsson and the Harvard Medical School Electron Microscopy core for performing immunogold experiments. Thanks to Feng Zhang for providing us with the LentiCas9-Blast and LentiGuide-Puro plasmids. This study was funded by NIH/NIDDK R01DK097768-01, NIH/NIDDK RO1DK095384, and NIH/NHLBI U01HL107440.
Published: April 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.03.002.
Supplemental Information
References
- Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugunova A., Loseva E., Mazin P., Mitina A., Navalayeu T., Bilan D., Vishnyakova P., Marey M., Golovina A., Serebryakova M. LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. U S A. 2019;116:4940–4945. doi: 10.1073/pnas.1809105116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Mansoura M.K. The Human Genome Project. Revealing the shared inheritance of all humankind. Cancer. 2001;91:221–225. doi: 10.1002/1097-0142(20010101)91:1+<221::aid-cncr8>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P., Wang S., Morton C.C., McMahon A.P., Powers D. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- D'Lima N.G., Ma J., Winkler L., Chu Q., Loh K.H., Corpuz E.O., Budnik B.A., Lykke-Andersen J., Saghatelian A., Slavoff S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017;13:174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain J.N., Shepherd R.E. Free fatty acids as feedback regulators of adenylate cyclase and cyclic 3':5'-AMP accumulation in rat fat cells. J. Biol. Chem. 1975;250:6586–6592. [PubMed] [Google Scholar]
- Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Lee C., Zeng J., Drew B.G., Sallam T., Martin-Montalvo A., Wan J., Kim S.J., Mehta H., Hevener A.L., de Cabo R. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.K., Cowan C.A. Differentiation of white and brown adipocytes from human pluripotent stem cells. Methods Enzymol. 2014;538:35–47. doi: 10.1016/B978-0-12-800280-3.00003-7. [DOI] [PubMed] [Google Scholar]
- Li J.X., Schulz H. 4-Bromo-2-octenoic acid specifically inactivates 3-ketoacyl-CoA thiolase and thereby fatty acid oxidation in rat liver mitochondria. Biochemistry. 1988;27:5995–6000. doi: 10.1021/bi00416a025. [DOI] [PubMed] [Google Scholar]
- Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27 doi: 10.1093/bioinformatics/btr209. i275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.F., Xiao M.H., Chen H.X., Meng Y., Zhao N., Yang L., Tang H., Wang J.L., Liu X., Zhu Y. A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 2019;10:528. doi: 10.1038/s41419-019-1767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarewich C.A., Baskin K.K., Munir A.Z., Bezprozvannaya S., Sharma G., Khemtong C., Shah A.M., McAnally J.R., Malloy C.R., Szweda L.I. MOXI is a mitochondrial micropeptide that enhances fatty acid beta-oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V.S., Willett W.C., Hu F.B. Global obesity: trends, risk factors and policy implications. Nat. Rev. Endocrinol. 2013;9:13–27. doi: 10.1038/nrendo.2012.199. [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- Moisan A., Lee Y.K., Zhang J.D., Hudak C.S., Meyer C.A., Prummer M., Zoffmann S., Truong H.H., Ebeling M., Kiialainen A. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat. Cell Biol. 2015;17:57–67. doi: 10.1038/ncb3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabbi A., Riabowol K. Rapid isolation of nuclei from cells in vitro. Cold Spring Harb. Protoc. 2015;2015:769–772. doi: 10.1101/pdb.prot083733. [DOI] [PubMed] [Google Scholar]
- Olowe Y., Schulz H. 4-Bromocrotonic acid, an effective inhibitor of fatty acid oxidation and ketone body degradation in rat heart mitochondria. On the rate-determining step of beta-oxidation and ketone body degradation in heart. J. Biol. Chem. 1982;257:5408–5413. [PubMed] [Google Scholar]
- Peters D.T., Cowan C.A., Musunuru K. Genome editing in human pluripotent stem cells. In: Girard L., editor. StemBook. Harvard Stem Cell Institute; 2013. [DOI] [PubMed] [Google Scholar]
- Pope B.D., Warren C.R., Parker K.K., Cowan C.A. Microenvironmental control of adipocyte fate and function. Trends Cell Biol. 2016;26:745–755. doi: 10.1016/j.tcb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L.D., Bostrom P., O'Sullivan J.F., Schinzel R.T., Lewis G.D., Dejam A., Lee Y.K., Palma M.J., Calhoun S., Georgiadi A. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C.S., Jadiya P., Zhang X., McLendon J.M., Abouassaly G.M., Witmer N.H., Anderson E.J., Elrod J.W., Boudreau R.L. Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D.R., Hendrickson D.G. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wabitsch M., Brenner R.E., Melzner I., Braun M., Moller P., Heinze E., Debatin K.M., Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.