Abstract
The lincosamide family antibiotic lincomycin is a widely used antibacterial pharmaceutical generated by Streptomyces lincolnensis, and the high-yield strain B48 produces 2.5 g/L lincomycin, approximately 30-fold as the wild-type strain NRRL 2936. Here, the genome of S. lincolnensis B48 was completely sequenced, revealing a ~10.0 Mb single chromosome with 71.03% G + C content. Based on the genomic information, lincomycin-related primary metabolism network was constructed and the secondary metabolic potential was analyzed. In order to dissect the overproduction mechanism, a comparative genomic analysis with NRRL 2936 was performed. Three large deletions (LDI-III), one large inverted duplication (LID), one long inversion and 80 small variations (including 50 single nucleotide variations, 13 insertions and 17 deletions) were found in B48 genome. Then several crucial mutants contributing to higher production phenotype were validated. Deleting of a MarR-type regulator-encoding gene slinc377 from LDI, and the whole 24.7 kb LDII in NRRL 2936 enhanced lincomycin titer by 244% and 284%, respectively. Besides, lincomycin production of NRRL 2936 was increased to 7.7-fold when a 71 kb supercluster BGC33 from LDIII was eliminated. As for the duplication region, overexpression of the cluster situated genes lmbB2 and lmbU, as well as two novel transcriptional regulator-encoding genes (slinc191 and slinc348) elevated lincomycin titer by 77%, 75%, 114% and 702%, respectively. Furthermore, three negative correlation genes (slinc6156, slinc4481 and slinc6011) on lincomycin biosynthesis, participating in regulation were found out. And surprisingly, inactivation of RNase J-encoding gene slinc6156 and TPR (tetratricopeptide repeat) domain-containing protein-encoding gene slinc4481 achieved lincomycin titer equivalent to 83% and 68% of B48, respectively, to 22.4 and 18.4-fold compared to NRRL 2936. Therefore, the comparative genomics approach combined with confirmatory experiments identified that large fragment deletion, long sequence duplication, along with several mutations of genes, especially regulator genes, are crucial for lincomycin overproduction.
Keywords: Lincomycin, Comparative genomics, Regulation, Overproduction
1. Introduction
Actinobacteria, especially the largest genus Streptomycetes, are high G + C, soil-dwelling Gram-positive filamentous bacteria that are well known for their capacity to produce a prodigious amount of bioactive secondary metabolites. Many of these metabolites have already been commercially used as a source of anti-bacterial, anti-fungal, anti-inflammatory, anti-cancer, anti-parasitic and immunosuppressive agents [1,2]. However, yields of desired metabolites in original producing strains are generally low, which builds a wall hindering their further research, industrialization and commercialization. Traditional efforts to solve the dilemmas focused mainly on fermentation process optimization and mutation breeding methods [3]. Benefitting from massive and easier-to-obtain genomics information about Streptomyces, rational transformations at metabolic or/and regulatory levels are widely adopted in strain improvement currently [4].
Lincomycin A is a member of lincosamide family antibiotics produced by Streptomyces lincolnensis. Lincomycin A and its derivative clindamycin are widely used in clinical practice, for their activities against Gram-positive anaerobic bacteria and protozoans [5]. The structure of lincomycin A consists of an unusual amino acid propylhygric acid (PPL) and an octose methylthio-lincosamide (MTL). Molecular mechanism of lincomycin biosynthesis becomes clearer based on the experimental efforts made in the past 20 years. The PPL originates from l-tyrosine, while the MTL is derived from a transaldol reaction using three sugars from the pentose-phosphate pathway. Then the PPL and MTL are condensed by two small molecule thiols EGT and MSH, and further modified to generate lincomycin A [5]. Studies on the regulation mechanism of lincomycin A biosynthesis have started lately, and only several regulators have been reported. Among them, LmbU acts as a positive cluster-situated regulator for lincomycin biosynthesis, and is assigned as a representative of a novel regulator family [6]. All three famous global regulators, GlnR, AdpA and BldD function as the activators in lincomycin generation [[7], [8], [9]], while the TetR-type regulator SLCG_2919 represses lincomycin synthesis [10]. Besides transcriptional regulation, tRNA BldA promotes lincomycin production through translational modulation [11]. Despite the above insights, what we have known about the complex regulatory network underlying lincomycin biosynthesis is just the tip of the iceberg, which remains an obstacle to be overcome in constructing lincomycin higher-yield strains using genetic manipulation. Traditionally, mutation and screening methods have been widely utilized in enhancement of lincomycin production, and a number of high-yield strains have been obtained. But the intracellular metabolic and regulatory mechanism hidden behind hyperproduction phenotype remains elusive without the aid of comparative genomic analysis.
To date, several comparative genomics studies have been conducted to unravel the high-yield mechanism of antibiotics production. With regard to erythromycin, the overproducing strain Saccharopolyspora erythraea Px contains numerous mutations altering central carbon and nitrogen metabolism, secondary metabolites biosynthesis, as well as basic transcription and translation, in comparison with the wild-type strain [12]. Peano et al. also found 250 variations in rifamycin B overproducer Amycolatopsis mediterranei HP-130, and validated the contributions of two genes encoding methylmalonyl-CoA mutase large subunit and arginyl tRNA synthetase to rifamycin overproduction [13]. Compared with the wild-type strain Streptomyces albus DSM 41398, deletions of a secondary metabolites gene cluster and several regulator-encoding genes led to salinomycin overproduction in the high-yield strain BK 3- 25 [14]. Recently, in a comparative genomics study between acarbose high-yield strain Actinoplanes sp. SE50/110 and wild-type strain SE50, mutations perturbing metabolism and transcription of the gene cluster were illuminated [15]. These cases authenticated the bona fide advantages of comparative genomic analysis, which can elucidate the masked mechanism underlying antibiotic overproduction, in a quick and systematical manner. And the identified key genomics differences can also be targeted for further production enhancement.
Herein, the complete genome of lincomycin high-producing strain B48 was sequenced. Profiles of primary metabolism related to lincomycin biosynthesis and secondary metabolism potential in B48 were thoroughly scanned. After comparison with the wild-type strain NRRL 2936, several mutations that likely rendered B48 a high-yield strain were picked and validated. And the deleted large fragments, duplicated lincomycin biosynthesis cluster and two novel transcriptional regulators, along with mutations of three negative regulators, were all demonstrated to be closely related. This work provides potential targets in high-yield strains construction, for lincomycin and other antibiotics.
2. Materials and methods
2.1. Bacterial strains, plasmids and culture media
Bacterial strains and plasmids used in this study are all listed in Table 1. Escherichia coli DH5α was used for molecular cloning. E. coli S17-1 was used for interspecies conjugal transfer. E. coli strains were cultivated on solid Luria-Bertani (LB) agar medium or in LB liquid medium in the shaker (180 rpm) at 37 °C (or at 28 °C when containing CRISPR/Cas9 editing plasmids).
Table 1.
Strains and plasmids used or constructed in this work.
| Strains or plasmids | Descriptions | Sources or references |
|---|---|---|
| Strains | ||
| S. lincolnensis | ||
| NRRL 2936 | Wild type strain, lincomycin producer | NRRL, USA |
| B48 | Lincomycin high producer | Laboratory stock |
| Δ377 | NRRL 2936 with in-frame deletion of slinc377 | This work |
| Δ4481 | NRRL 2936 with in-frame deletion of slinc4481 | This work |
| Δ6011 | NRRL 2936 with in-frame deletion of slinc6011 | This work |
| Δ6156 | NRRL 2936 with in-frame deletion of slinc6156 | This work |
| ΔBGC29 | NRRL 2936 with deletion of LDII region of BGC29 | This work |
| ΔBGC33 | NRRL 2936 with deletion of BGC33 | This work |
| OlmbB2 | NRRL 2936 ɸC31 attB::pIBlmbB2 | This work |
| OlmbU | NRRL 2936 ɸC31 attB::pIBlmbU | This work |
| O191 | NRRL 2936 ɸC31 attB::pIB191 | This work |
| O348 | NRRL 2936 ɸC31 attB::pIB348 | This work |
| E. coli | ||
| DH5α | F-ϕ80lacZΔM15Δ(lacZYA-argF)U169recA 1endA1hsdR17(rk-mk+)phoAsupE44λ-thi-1gyrA96relA1 |
Laboratory stock |
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | Laboratory stock |
| M. luteus 28001 | Indicator for bioassay analysis of lincomycin | CGMCC |
| Plasmids | ||
| pKCcas9dO | A CRISPR/Cas9 editing plasmid harboring an actII-ORF4-specific gRNA and two homologous arms for in-frame deletion, aac(3)IV, pSG5 | [21] |
| pKCcas9d377 | A CRISPR/Cas9 editing plasmid harboring an slinc377-specific gRNA and two homologous arms for in-frame deletion, aac(3)IV, pSG5 | This work |
| pKCcas9d4481 | A CRISPR/Cas9 editing plasmid harboring an slinc4481-specific gRNA and two homologous arms for in-frame deletion, aac(3)IV, pSG5 | This work |
| pKCcas9d6011 | A CRISPR/Cas9 editing plasmid harboring an slinc6011-specific gRNA and two homologous arms for in-frame deletion, aac(3)IV, pSG5 | This work |
| pKCcas9d6156 | A CRISPR/Cas9 editing plasmid harboring an slinc6156-specific gRNA and two homologous arms for in-frame deletion, aac(3)IV, pSG5 | This work |
| pKCcas9dBGC29 | A CRISPR/Cas9 editing plasmid harboring an BGC29-specific gRNA and two homologous arms for deletion, aac(3)IV, pSG5 | This work |
| pKCcas9dBGC33 | A CRISPR/Cas9 editing plasmid harboring an BGC33-specific gRNA and two homologous arms for deletion, aac(3)IV, pSG5 | This work |
| pIB139 | Integrative vector based on ɸC31 int/attP, aac(3)IV, oriT RK2, PermE* | [57] |
| pIBlmbB2 | pIB139 harboring lmbB2 under control of PermE* | This work |
| pIBlmbU | pIB139 harboring lmbU under control of PermE* | This work |
| pIB191 | pIB139 harboring slinc191 under control of PermE* | This work |
| pIB348 | pIB139 harboring slinc348 under control of PermE* | This work |
Micrococcus luteus 28001 was used for lincomycin bioassay analysis and was cultivated on solid medium III (5 g/L polypepton (Nihon Pharmaceutical, Japan), 3.68 g/L K2HPO4 (Lingfeng, China), 3.5 g/L NaCl (Titan, China), 3 g/L yeast extract (OXOID, USA), 1.5 g/L beef extract (SCRC, China), 1.32 g/L KH2PO4 (Lingfeng, China), 1 g/L glucose (Lingfeng, China), and 18 g/L agar (Shize, China)) at 37 °C.
S. lincolnensis NRRL 2936 and its derivatives, as well as B48 were incubated on solid mannitol soya flour (MS) medium [16] at 28 °C for 6 days for routine culture, phenotype observation, and spore preservation, and then were inoculated into liquid yeast extract-malt extract (YEME) medium [17] in the shaker (200 rpm) at 28 °C for DNA extraction and at 37 °C for plasmid curation. Solid ISP4 medium [18] was used for conjugation. Apramycin of 50 μg/mL (Sangon Biotech, China) was supplemented when needed.
2.2. Fermentation and lincomycin bioassay analysis
Spores of S. lincolnensis NRRL 2936, B48 and derivatives of NRRL 2936 were spread on MS medium at 28 °C for sporulation and then inoculated into SFPI medium (30 g/L corn steep liquor (Aladdin, China), 20 g/L soluble starch (Lingfeng, China), 10 g/L glucose, 1.5 g/L (NH4)2SO4 (Lingfeng, China), 1 g/L soya flour, and 4 g/L CaCO3, dissolved in dH2O, pH 7.2) in the shaker (200 rpm) at 28 °C for 4 days. Finally, 1 mL cultures from SFPI were inoculated into SFPII medium (105 g/L glucose, 20 g/L soya flour, 8 g/L NaNO3 (Lingfeng, China), 6 g/L (NH4)2SO4, 5 g/L NaCl, 1.5 g/L corn steep liquor, 0.025 g/L K2HPO4, and 8 g/L CaCO3, dissolved in dH2O, pH 7.8) and fermented in the shaker (200 rpm) at 28 °C for 6 days.
To measure the bioassay of lincomycin produced in fermentation, fermentation broth supernatants of each sample with three replicates were harvested by centrifugation, and then an aforementioned detection method was employed [6,19]. The indicator M. luteus was cultured on solid medium III at 37 °C for 16 h and the lawn was washed off with 0.9% NaCl and suspended for following usage. Lincomycin standard solutions (2, 4, 6, 8, 10, and 12 μg/mL) were used for the standard curve and internal control. Diameters of inhibition zone were linearized with the natural logarithmic of the concentrations of the lincomycin standard solutions. Then, lincomycin concentration of each sample was calculated based on the standard curves. All assays were performed in three replicates and standard deviation of the mean were calculated. To draw the histogram of lincomycin bioassay, the software GraphPad Prism 8.0 was utilized.
2.3. Genome sequencing, assembly, annotation, and analysis of S. lincolnensis B48
The genome sequencing of S. lincolnensis B48 was performed by a combination of PacBio Sequel system and Illumina NovaSeq PE150 at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China). One scaffold with 100% coverage and no gap was generated. Protein-coding and RNA genes were predicted and annotated by NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Mauve and BLAST programs were used to identify the single nucleotide variations (SNVs) and InDels. AntiSMASH (antibiotics & Secondary Metabolite Analysis Shell, http://antismash.secondarymetabolites.org/) was utilized to analyze secondary metabolite gene clusters in S. lincolnensis [20]. To draw the map of genomic comparison, R package circlize (http://cran.r-project.org/web/packages/circlize/index.html) was adopted [21]. The whole genome sequence of B48 can be found at GenBank through the accession number CP046024.
2.4. Construction of gene/cluster deletion and overexpression mutant strains
To delete the LDII region of BGC29 in NRRL 2936, a CRISPR/Cas9-based genetic editing method reported previously was adopted [22]. The BGC29-specific sgRNA was amplified from pKCcas9dO using the primer pair sg29/sgdn. Upstream and downstream homologous arms (approximate 1 kb) of LDII were amplified by PCR with two primer pairs u29-F/R and d29-F/R, respectively, using the genomic DNA of NRRL 2936 as the template. Three products were joined together by overlapping PCRs to generate the LDII deletion cassette. The cassette was then cloned into pKCcas9dO between SpeI and HindIII (Takara Bio, Japan). Then E. coli S17-1 was used to transfer the recombinant plasmid into NRRL 2936 by conjugation. The desired apramycin-resistant conjugants were checked by PCR with primers JD29-F/R. The pKCcas9dBGC29 plasmid was eliminated through two or three rounds of shake flask culture in YEME medium (200 rpm) at 37 °C. BGC33 of NRRL 2936 was knocked out using the similar procedure.
To construct the deletion mutant strain of slinc6156 (named Δ6156), the same CRISPR/Cas9-mediated method was performed but with some modification in plasmid construction process. In brief, after amplification of upstream and downstream homologous arms of slinc6156 (with two primer pairs u6156-F/R and d6156-F/R, respectively), the PCR product of u6156-F/R was amplified again by PCR using the primers sg6156 and u6156-R to add sequence-specific sgRNA sequence. Both sgRNA-containing upstream arm and downstream arm fragments were inserted into pKCcas9dO between SpeI and HindIII to generate pKCcas9d6156 using Super Efficiency Fast Seamless Cloning kit (DoGene, China). Similar procedures were performed to construct other single gene deletion mutants.
To construct lmbB2 overexpression strain OlmbB2, a DNA fragment covering the whole ORF of lmbB2 was amplified by PCR with primers lmbB2-F/R and subsequently trimmed with NdeI/EcoRI (Thermo Fisher Scientific, USA). Then the digested fragment was ligated into the corresponding sites of the integrative vector pIB139. The resulting plasmid pIBlmbB2 was introduced into NRRL 2936 by conjugal transfer mediated by E. coli S17-1 and integrated into the ɸC31 attB site in the chromosome. The desired apramycin-resistant conjugants were checked by PCR with primers 824-F/R. Similar procedures were performed to construct the other overexpression mutants (For slinc191 and slinc348, the restriction enzymes are XbaI/EcoRI instead of NdeI/EcoRI).
All primers used in this study are listed in Table S1, and synthesized by Genewiz (Suzhou, China).
3. Results
3.1. Phenotypic differences between the high-yield strain S. lincolnensis B48 and wild-type strain NRRL 2936
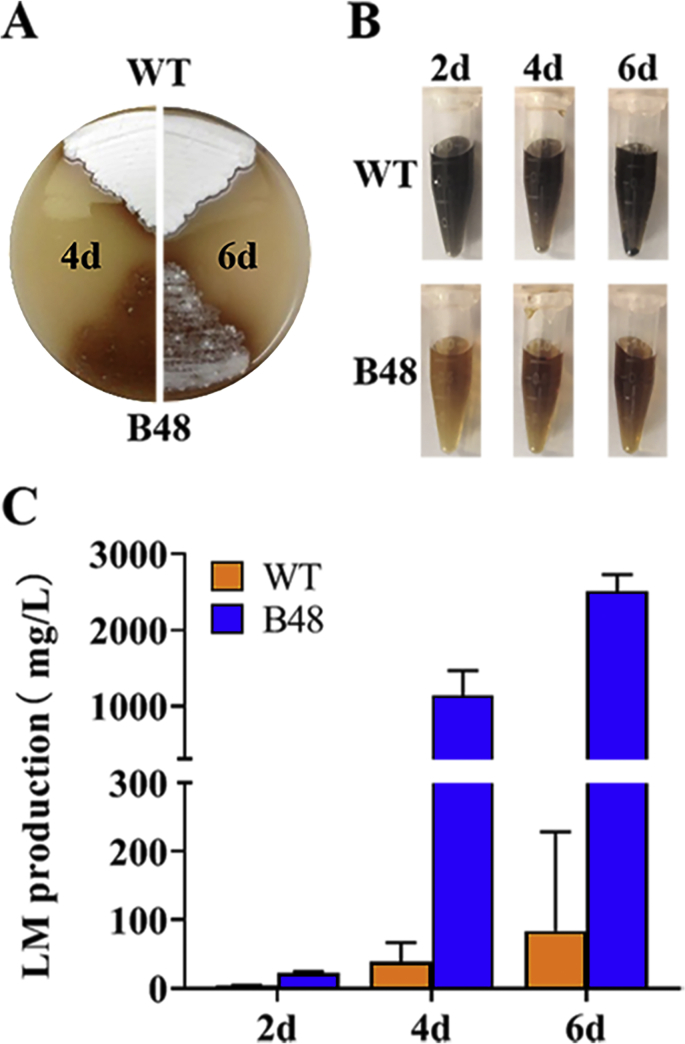
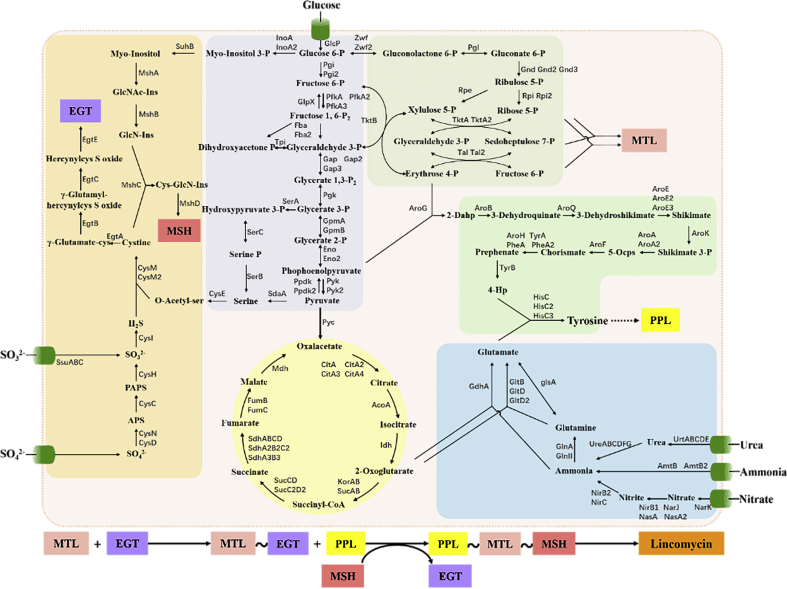
S. lincolnensis B48, possessing significant phenotypic variations, is a high-yield strain gained by traditional mutation and screening method from the wild-type strain NRRL 2936. Compared with NRRL 2936, B48 shows an obviously lagged and defective sporulation process when growing on MS medium (Fig. 1A). When fermented simultaneously, lincomycin generation of B48 was apparently earlier and higher than that of NRRL 2936. The final lincomycin titer of B48 can reach 2.5 g/L in culture supernatant, approximately 30-fold higher than that of NRRL 2936, whose titer was only 87.3 mg/L (Fig. 1C). Furthermore, the color of the fermentation broth of B48 was light, similar to uninoculated medium, while that of NRRL 2936 was darker (Fig. 1B).
Fig. 1.
Phenotype differences between the wild-type strain S. lincolnensis NRRL 2936 and the high-yield strain B48. (A) Colonies of WT (NRRL 2936) and B48 cultured on MS plates at 4 and 6 days. (B) Lincomycin production of WT and B48. Error bars represent SD of three replicates. (C) Fermentation broths of WT and B48.
The genetic divergences hidden behind the distinct phenotype differences remained elusive, thus requiring a systematical interpretation through genomic analysis.
3.2. Genome sequencing and sequence analysis of B48
General features of B48 genome. Sequencing of B48 genome was performed by Illumina and PacBio sequencing technologies. After sequence assembly, a single linear chromosome of 10,008,966 bp with an average 71.03% G + C content was obtained. The genome of B48 contains 8558 protein-encoding genes, 70 tRNA genes and 6 copies of rDNAs.
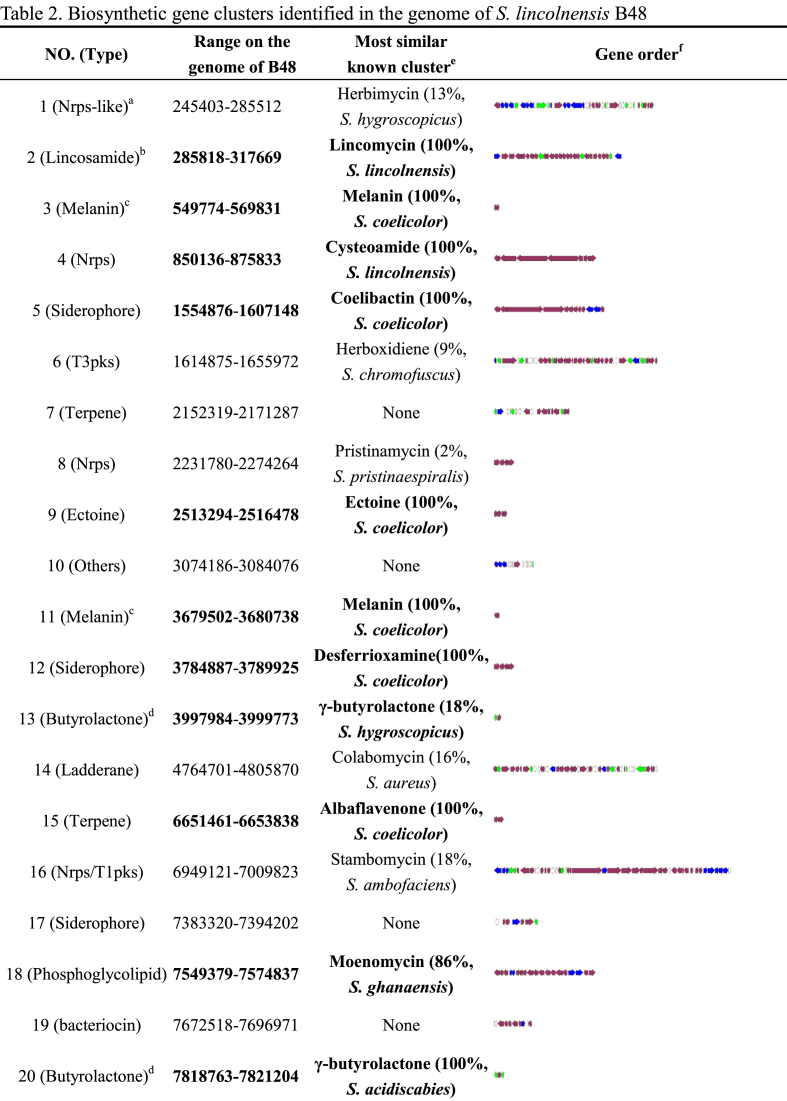
Lincomycin-related metabolic network. Essential metabolism pathways providing building precursors of lincomycin were selected based on KEGG analysis and manual correction via alignments with Streptomyces coelicolor, as depicted in Fig. 2. For convenience sake, lincomycin-related metabolism network was divided into three modules: Central carbon metabolism, l-Tyrosine synthesis and Sulfur metabolism. (1) Central carbon metabolism, consisting of glycolysis (Embden-Meyerhof-Parnas pathway, EMP), citrate cycle (TCA) and pentose phosphate pathway (PPP), can furnish not only major energy for consumption in anabolism, but also precursors to directly support lincomycin generation [5]. B48 contains complete pathways in central carbon metabolism, of which most steps are performed by more than one isozyme. (2) l-Tyrosine that derives the PPL moiety is composed of 4-Hp (Hydroxyphenylpyruvate) from the shikimate pathway and the amino group provided by l-Glutamate. The shikimate pathway initiates from condensation of d-erythrose 4-phosphate and phosphoenolpyruvate from central carbon metabolism. The major organic nitrogen donor l-Glutamate is generated via two different ways depending on ammonia availability. Intracellular ammonia of B48 can be obtained by direct assimilation, reduction of nitrate, and hydrolysis of urea. (3) Sulfur metabolism genes that encode enzymes related to generation of mycothiol (MSH) and ergothioneine (EGT) are all found in B48 genome. As for sulfur-containing amino acids, B48 has all cys genes encoding enzymes for sulfur assimilation and cysteine synthesis.
Fig. 2.
Schematic illustration of lincomycin-related primary metabolic network in B48. Primary metabolic pathways that generate precursors (l-tyrosine, d-fructose 6-phosphate, d-sedoheptulose 7-phosphate, d-ribose 5-phosphate, MSH and EGT) are displayed. Black texts represent substances and gray texts represent enzymes.
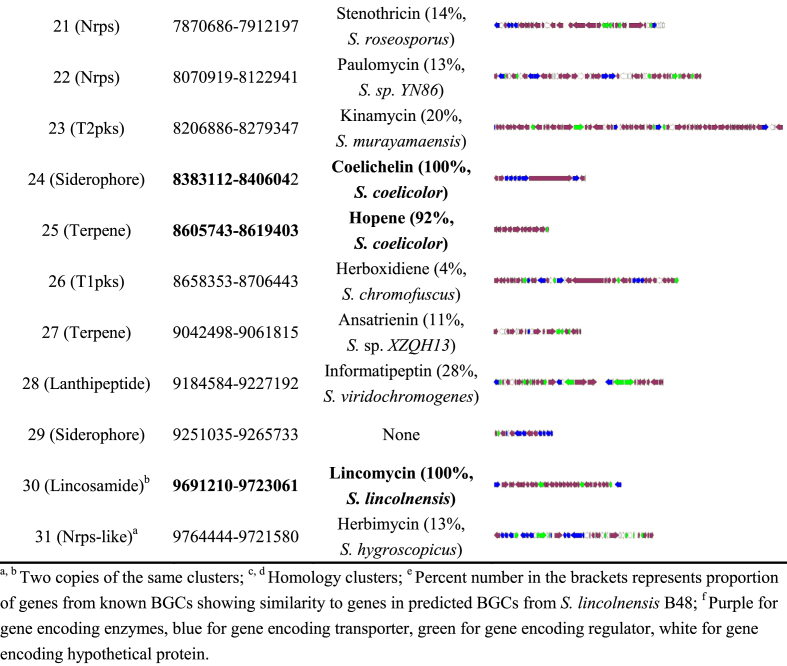
Secondary metabolic gene clusters. Given the huge potential of secondary metabolism stored in Streptomyces genome, complete sequence of B48 was submitted to antiSMASH, and 31 putative BGCs (biosynthesis gene clusters) were found (shown in Table 2). Precise boundaries of BGCs that are homologous to known clusters were manually corrected to remedy the inevitable errors generated by in silico prediction, as shown in bold. Among them, BGC2 and BGC30 are two copies of lincomycin biosynthesis clusters, the same as another high-yield strain 78-11 [23]. BGC4 was previously identified to direct the biosynthesis of cysteoamide [24]. BGC18 is supposed to synthesize a kind of phosphoglycolipid antibiotic structurally similar to moenomycin. Among all 19 genes in BGC18, 18 members are high homologues of genes in the moenomycin BGC from S. ghanaensis [25] (Table S3). Furthermore, 10 BGCs show high identities to studied clusters. BGC5, BGC12, and BGC24 are homologues of three widely studied siderophore synthesis clusters in S. coelicolor, coelibactin, desferrioxamine and coelichelin, respectively [26]. Both BGC3 and BGC11 contain gene pairs homology to tyrosinase/tyrosinase cofactor-encoding genes [27], thus being assigned as melanin synthesis clusters. As important auto-regulators in Streptomycetes [28], two kinds of γ-butyrolactones (GBLs) can be likely produced by BGC13 and BGC20, for the presence of GBL synthetase/GBL receptor-encoding gene pairs. Salt and heat stress protectant ectoine may be generated by BGC9 [29]. BGC15 encodes two homology proteins that synthesize sesquiterpene-type antibiotic albaflavenone [30]. BGC25 was deduced to be involved in biosynthesis process of membrane-distribution lipid hopanoid that is related to morphological differentiation [31].
Table 2.
Biosynthetic gene clusters identified in the genome of S. lincolnensis B48.
3.3. Comparative genomic analysis between B48 and NRRL 2936
To gain insights into the mechanism about lincomycin overproduction, a comparison between genome of B48 and that of NRRL 2936 sequenced previously (GenBank accession number CP016438.1) was performed.
Three large fragment deletions in B48. Genome of B48 is 310 kb smaller than that of NRRL 2936, mainly resulting from three large fragment deletions (Fig. 3). One large deletion (LDI) with 27.3 kb is near the left chromosome terminus, at 455,105-482,421 bp, coincidently taking up most of a putative ladderane BGC in NRRL 2936. The 24.7 kb LDII is situated on the right arm of chromosome, a region that BGC28 occupies, spanning from 9,243,876 to 9,268,615 bp. The last but largest deletion (LDIII) is located on the right terminus, with a length up to 777 kb. As a result of 3 large deletions, 483 genes are absent from the genome of B48, including 5 putative secondary metabolic gene clusters (Table S2).
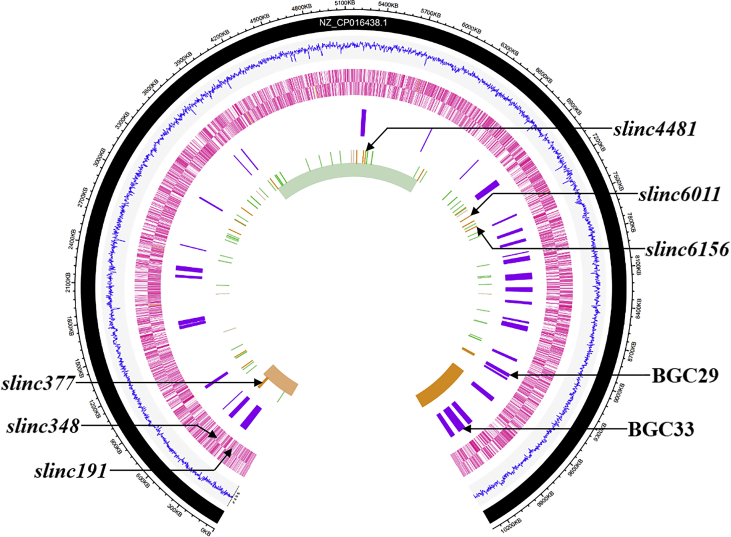
Fig. 3.
Chromosome map of genetic variations between B48 and NRRL 2936. From the outside in, circle 1: the chromosomal regions of NRRL 2936 (black); circle 2: G + C content of NRRL 2936 (blue); circles 3 and 4: (forward and reverse strands), the predicted protein coding genes (pink); circle 5: distribution of putative secondary gene clusters (purple); circle 6: positions of Indels (green for inserts and orange for deletions) and SNVs (plum for transition and pale green for transversion) between B48 and NRRL 2936; circle 7: positions of inversion (aquamarine) and duplication (sandy brown). Validated mutations that contributed to lincomycin overproduction are marked with arrows. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
A large inverted duplication in B48. A 523,875 bp sequence of the left chromosome terminus doubles and occupies the location where LDIII is. The duplication sequence, named as LID, has no difference with the original sequence and comprises 509 putative genes. As a ripple effect, the original short terminal inverted repeat (TIR) in the chromosome of NRRL 2936 (24.3 kb) is replaced by a longer one (523 kb) in the genome of B48. In view of this point, genome stability of B48 is supposed to be poorer than that of NRRL 2936. Lincomycin BGC is doubled due to the duplication, and the similar cases were found in producers of kirromycin and pristinamycin [32,33].
An inversion in B48. In terms of chromosome structural mutations, besides deletions and duplication, there is also an inversion in the middle region, from 3,875,155 to 6,213,573 bp. The left inversion site is located in a galactose oxidase-encoding gene, accompanied by a 159 bp deletion. The right inversion site is situated in a tryptophan tRNA ligase-encoding gene, along with a 205 bp deletion.
Small mutations in B48. Apart from large mutations mentioned above, 50 single nucleotide variations (SNV, include 42 transversions and 8 transitions), 13 insertions (INS), and 17 deletions (DEL, include 2 deletions on wings of inversion) were found in genomic comparison. Coding sequences of 41 genes are predicted to be affected, including 8 regulators, 5 transporters, 22 enzymes and 6 hypothetical proteins (Table 3).
Table 3.
Mutated proteins in S. lincolnensis B48.
| Gene NO. in NRRL 2936 | Putative function | Variationsa | Effects on protein |
|---|---|---|---|
| 0098 | terminal protein | SNV | T15A |
| 0933 | hypothetical protein | SNV | G209R |
| 1635 | MFS transporter | SNV | Nonsense |
| 1746 | ABC transporter substrate-binding protein | SNV | I263T |
| 2047 | cell division protein | SNV | A14T |
| 2468 | hypothetical protein | DEL | Frameshift |
| 2672 | Xre family transcriptional regulator | SNV | G210D |
| 2953 | hybrid sensor histidine kinase/response regulator | INS | Amino acid insertion |
| 3088 | sugar-binding protein | SNV | G9D |
| 3257 | glycosyl transferase | DEL | Frameshift |
| 3258 | galactose oxidase | DEL | Deletion |
| 3400 | TetR family transcriptional regulator | INS | Frameshift |
| 3901 | Two-component system sensor kinase | SNV | L65P |
| 4040 | cation:proton antiporter | SNV | Q237E |
| 4158 | hypothetical protein | SNV | G132V |
| 4453 | GntR family transcriptional regulator | DEL | Frameshift |
| 4481 | ATPase and TPR domain-containing protein | DEL | Amino acid insertion |
| 5157 | hypothetical protein | SNV | Nonsense |
| 5216 | tryptophan--tRNA ligase | DEL | Deletion |
| 5259 | polysaccharide pyruvyl transferase | SNV | G343D |
| 5644 | polyprenyl synthetase | INS | Frameshift |
| 5823 | ABC transporter permease | SNV | R104 M |
| 5869 | ABC transporter ATP-binding protein | SNV | I2R |
| 5912 | esterase | SNV | W221Y |
| 5941 | phosphodiesterase | SNV | L163V |
| 6011 | 16S rRNA (guanine(966)-N(2))-methyltransferase | DEL | Frameshift |
| 6096 | γ-aminobutyric acid aminotransferase | DEL | Deletion |
| 6097 | ATP-binding protein | DEL | Deletion |
| 6125 | hypothetical protein | SNV | P27L |
| 6160 | ribonuclease J | DEL | Frameshift |
| 6222 | TetR family transcriptional regulator | SNV | V166I |
| 6601 | 2,3-diaminopropionate biosynthesis protein | SNV | V118I |
| 6731 | AMP-binding protein | SNV | I159L |
| 6835 | hypothetical protein | SNV | F45L |
| 6965 | xyloglucanase | SNV | N284K |
| 7161 | carnitine dehydratase | SNV | P127L |
| 7316 | cytochrome P450 | SNV | L374P |
| 7507 | TetR family transcriptional regulator | SNV | F160S |
| 7967 | Zn-dependent hydrolase | DEL | Deletion |
SNV stands for single-nucleotide variant, DEL stands for deletion, INS stands for insertion.
3.4. Mutations contributing to lincomycin overproduction in B48
To verify the contributions of the aforementioned genomic mutations to lincomycin overproduction, a series of gene/cluster knock-out strains and gene overexpression strains were constructed based on NRRL 2936. The capacities of these mutants for lincomycin production were examined to identify the fundamental mutations.
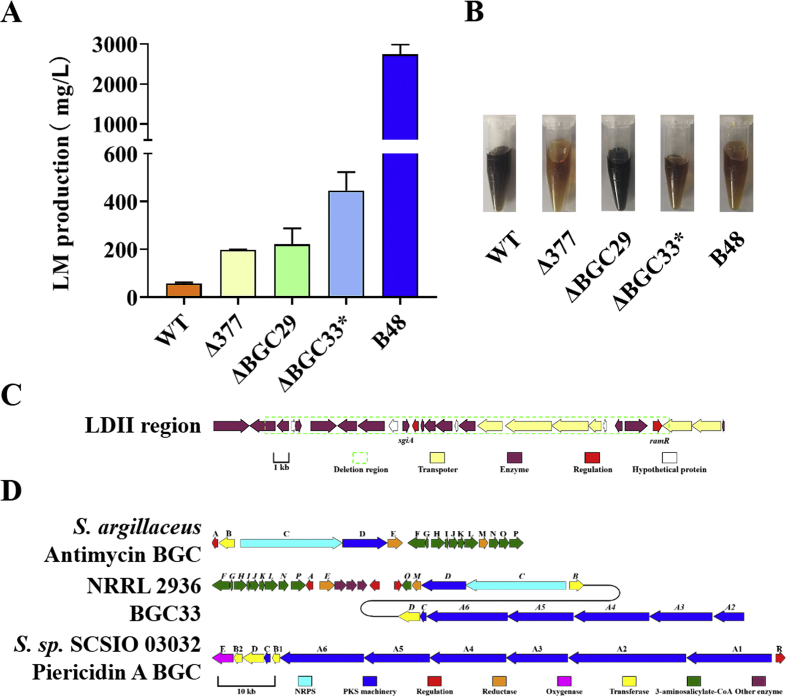
LDs prompting lincomycin production. Deletions of large chromosomal fragments have already been found to facilitate antibiotics production [14,34]. In view of this, the possible promotional effects of three large deletions was validated. Since the attempt on deletion of the whole LDI region in NRRL 2936 was unfortunately failed, as an alternative, the gene slinc377 from LDI that encodes a MarR family transcriptional regulator was knocked out. Lincomycin titer in Δ377 was 3.4-fold as that of NRRL 2936 (Fig. 4A), proving that LDI contributed to lincomycin high production. Deletion of LDII region (Fig. 4C) in NRRL 2936 (named ΔBGC29, for it takes over part of BGC29 in NRRL 2936) showed an enhancement phenotype in lincomycin production, with a titer as 3.8-fold as NRRL 2936 (Fig. 4A). The color of the fermentation broth of ΔBGC29 was darker than that of NRRL 2936 (Fig. 4B). BGC33 in LDIII is a supercluster that contains two subunits homologous to reported BGCs for biosynthesis of antimycin [35] and piericidin A [36] (Fig. 4D). Elimination of a 71 kb fragment containing BGC33 conspicuously improved lincomycin titer by 673% (the plasmid for knockout remained in the cell). All these data demonstrated the contributions of these three large deletions to lincomycin hyperproduction.
Fig. 4.
Effects of LDs on lincomycin production. (A) Lincomycin production of WT, B48 and knockout mutants based on WT, after fermentation for 6 days. Error bars represent SD of three replicates. (B) Fermentation broths of WT, B48 and mutants, after fermentation for 6 days. (C) Gene organization of LDII region. (D) Gene organization of BGC33 and homology clusters (antimycin and piericidin A). Letters above the arrows stands for the names of ant and pie series, and italics letters above the arrows stands for homologues of corresponding ant or pie genes.
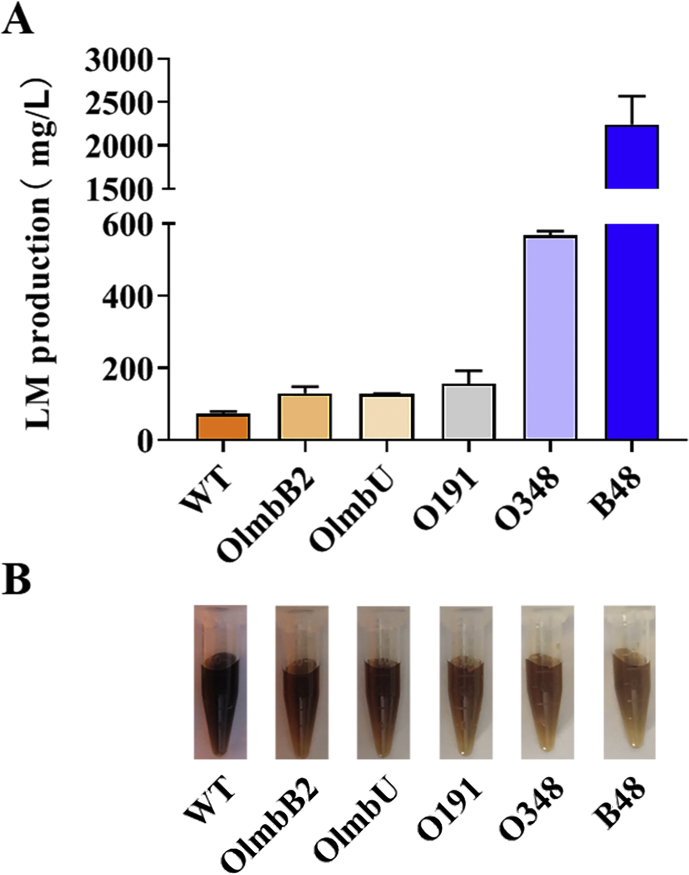
LID prompting lincomycin production. It was essential to explore the possibility that the duplication event, especially the amplification of lincomycin BGC, made great contributions to lincomycin titer enhancement. Several genes from the lincomycin biosynthesis cluster were selected to be integrated into the chromosome of NRRL 2936, under the control of PermE* promoter. Among them, overexpressions of lmbB2 (encodes l-tyrosine 3-hydroxylase that initiates PPL biosynthesis) and lmbU (encodes cluster-situated positive regulator) showed 77% and 75% increase on lincomycin titer, respectively (Fig. 5A). These results provide an evidence that duplication of the cluster, at least some of genes, promoted lincomycin production in B48.
Fig. 5.
Effects of genes from LID on lincomycin production. (A) Lincomycin production of WT, B48 and overexpression mutants based on WT, after fermentation for 6 days. Error bars represent SD of three replicates. (B) Fermentation broths of WT, B48 and mutants, after fermentation for 6 days.
Besides, putative transcriptional regulator-encoding genes that are located at the duplication region but out of the lincomycin BGC were also selected to verify whether it is related to positive regulation on lincomycin biosynthesis. Two novel transcriptional regulators (SLINC191 and SLINC348, gene loci are marked in Fig. 3) were identified as positive regulators on lincomycin biosynthesis. Overexpression of slinc191 resulted in a 114% improvement on lincomycin titer, while O348 strain presented a more apparent enhancement, equivalent to 7.0-fold of NRRL 2936 (Fig. 5A).
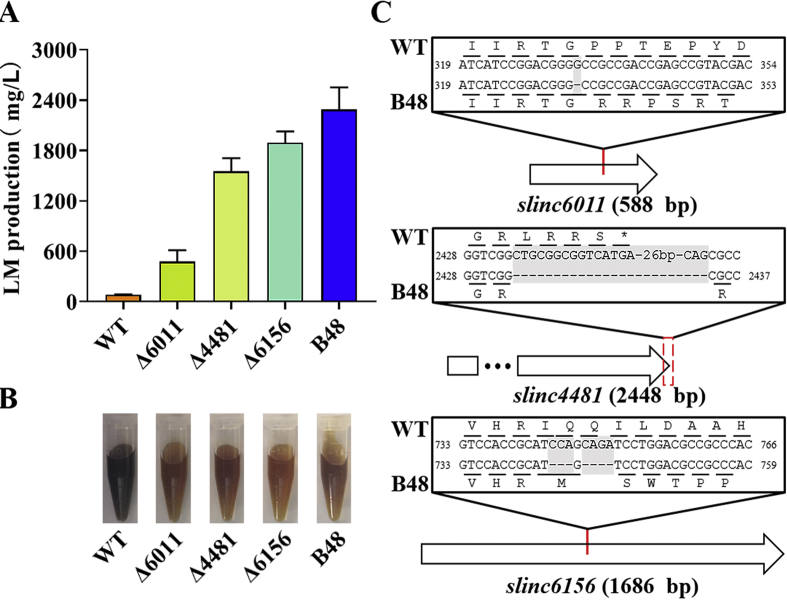
Crucial single gene mutations prompting lincomycin production. To verify the contributions of single gene destructions, mutated genes with potential regulatory functions were knocked out from genome of NRRL 2936. After fermentation, three mutants showed an increase more than 5.0-fold. As for the color of fermentation broths, they were all similar to B48 but obviously different to NRRL 2936 (Fig. 6A and B). The locations of the three genes in the chromosome are marked in Fig. 3.
Fig. 6.
Single gene mutations that prompted lincomycin production. (A) Lincomycin production of WT, B48 and knockout mutants based on WT, after fermentation for 6 days. Error bars represent SD of three replicates. (B) Fermentation broths of WT, B48 and mutants, after fermentation for 6 days. (C) DNA and deduced protein variations between WT and B48 of 3 genes. Mutated sites are marked using red lines/dotted boxes, and mutated base pairs are marked in gray background. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Firstly, inactivation of a ribonuclease RNase J-encoding gene slinc6156 enhanced lincomycin production drastically. Surprisingly, the titer of the mutant strain Δ6156 achieved 1.89 g/L, equivalent to 22.4-fold of NRRL 2936 and 82.7% of B48. As shown in Fig. 6C, CCA in position 744–746 and CAGA in position 748–751 were lost in B48, resulting in a frameshift mutation from I248. As a chain effect, its translation was stopped prematurely, with its amino acid sequence shortened from 561 AA to 280 AA (Fig. 6C and Fig. S1A). Secondly, knockout of slinc4481 increased lincomycin titer to 1.55 g/L, equivalent to 18.4-fold of NRRL 2936 and 67.9% of B48. Gene slinc4481 encodes an 815 AA unknown protein, which consists of an AAA ATPase domain near the N-terminal and a C-terminal TPR domain. Due to a 44 bp sequence deleting event occurred in the region covering the stop codon (Fig. 6C), its coding product in B48 was lengthened to 1119 AA. Finally, deletion of 16S rRNA (guanine(966)-N(2))-methyltransferase-encoding gene slinc6011 in NRRL 2936 raised lincomycin titer to 0.48 g/L, equivalent to 5.6-fold of NRRL 2936 and 20.7% of B48. As for the mutation type of slinc6011 in B48, guanine in position 333 missed, leading to a frameshift mutation, with a peptide elongation from 195 AA to 377 AA (Fig. 6C and Fig. S3A). All the results from these mutants broadened regulation network of lincomycin to post-transcriptional, translational and other mechanisms.
4. Discussion
The proposal of the study was based on the apparent phenotypic discrepancies in lincomycin titer, as well as the sporulation process between the wild-type strain S. lincolnensis NRRL 2936 and the high-yield strain B48 (Fig. 1). Although several rational genetic engineering attempts have been carried out on lincomycin-producers [7,37,38], most of them were low-in-pertinence and time-consuming, owing to lack of understanding on genetic mechanisms hidden in the genome of overproducers. High-yield strains obtained in the past half century provided valuable materials for lincomycin overproduction mechanisms elucidation, and the faster-and-cheaper DNA sequence technique facilitates the implement.
Primary metabolism, such as carbon metabolism, nitrogen metabolism, phosphate metabolism and sulfur metabolism, provides precursors or intermediates for secondary metabolites [39], serving as building blocks (e.g. amino acids and sugars), donors for structural modification (e.g. glycosylation, methylation and amination), and enzyme cofactors (e.g. NADPH, NADH and ATP). Therefore, secondary metabolism is tightly tied to and seriously limited by primary metabolism. Thus primary metabolic network related to lincomycin biosynthesis was analyzed. No change in the metabolic network at the DNA level was found in B48. So more “omics” (transcriptomics, proteomics and metabolomics) researches might need to be executed to elucidate the intracellular differences on primary metabolism. But there are also some appealing discoveries when compared with other Streptomyces. The first one originated from nitrogen metabolism. It should be noted that S. lincolnensis (both NRRL 2936 and B48) lacks 8 of 9 constituents for 3 respiratory nitrate reductases (Nar) that participate in nitrate reduction [40], except for a homologue of NarJ1 (69% identity). In light of the significance of Nar in spore survival and mycelium germination, how S. lincolnensis completes this vital life process under oxygen-limiting conditions remains to be solved. Additionally, because two thiols (MSH and EGT) play a key role in condensation steps for lincomycin synthesis, sulfur metabolism is also noteworthy. Although synthesis pathways of cysteine seemed complete, MetXY mediated sulfhydrylation pathway for methionine synthesis [41] does not exist in S. lincolnensis, leaving a problem to be solved in the future.
Furthermore, for secondary metabolism, in view of the conspicuous difference exhibited in pigment of fermentation broth between the two strains, melanin synthesis clusters were sought from the genome sequence. Sequences of the two putative melanin clusters (BGC3 and BGC11) had no difference between NRRL 2936 and B48. In BGC3, slinc492 and slinc493 encode analogues of tyrosinase cofactor (MelC1, 58% identity) and tyrosinase (MelC2, 53% identity) from S. coelicolor. As for BGC11, there exists another copy of presumptive melC1/C2 gene pair (slinc3117/3116), whose coding products show 81% and 51% identities with MelC1 and MelC2, respectively. The difference in color of fermentation broth may arise from differential gene expression, which needs further validation. Stemmed from the interest on mining novel natural products, BGC18 has attracted more attentions. BGC18 is identical to biosynthesis cluster of phosphoglycolipid antibiotic moenomycin from S. ghanaensis [25], for homologues of 18 genes in the latter can be found in BGC18, with 65%–90% identities in deduced protein sequences (Table S3). However, homologues of genes encoding a hexose-4,6-dehydratase, a hexose-4-ketoreductase and an amide synthetase are absent from BGC18. There is an additional gene encoding putative cyclase in BGC18. These similarities and discrepancies indicate that B48 might potentially produce a moenomycin-like novel antibiotic.
Loss of two BGCs in B48 was proved to be involved with lincomycin high-production. The LDII region covers 22 complete ORFs and 2 partial ORFs that belong to a putative lanthipeptide BGC (numbered 29 in NRRL 2936 whereas 28 in B48). Besides elevation in lincomycin titers, the spores of ΔBGC29 were slightly sparser than that of WT strain (Data not shown). BLAST analysis was conducted to find the homologies of these 24 genes. Among them, SLINC7721 is a protease inhibitor showing 72% identity with SgiA from S. coelicolor. SgiA was reported to control actinorhodin production and morphological differentiation [42]. But inactivation of slinc7721 made no difference to morphological differentiation, and lincomycin production was abated instead when fermented in 24 deep-well plates (Data not shown). Therefore, it might be not involved in phenotypic variations in B48. SLINC_RS46710 is a homologue of two-component system response regulator RamR (49% identity), an important morphogenesis regulator in S. coelicolor [43]. Further research is needed to validate the possible role of SLINC_RS46710. Among four putative BGCs in LDIII (Table S2), three of them have no highly homologous known clusters. BGC33 is a ‘supercluster’ consisting of two clusters highly homologous to antimycin and piericidin A clusters. As shown in Fig. 4D and Table S4, slinc8366-8376 and slinc8382-8385 are homologues of all 15 genes from antimycin cluster of S. argillaceus [35]. Biosynthesis of antimycin requires L-Tryptophan as a substrate, whose generation is a competitive approach for l-Tyrosine biosynthesis, for both of them originate from chorismate in shikimate pathway (Fig. 2). The increase in supply of precursors, if experimentally validated, might explain the enhanced lincomycin titer in ΔBGC33. Besides, slinc8377-8379 encode ketoacyl-ACP synthetase, 3-hydroxybutyryl-CoA dehydrogenase and thioesterase, which are common in PKS-type clusters, but not found in antimycin cluster. These data indicate that NRRL 2936 has the potential to produce an antimycin-like metabolite with the novel structure. Other genes (slinc8387-8393) in BGC33 are homology to 7 of 11 genes in piericidin A cluster from Streptomyces sp. SCSIO 03032 (Fig. 4D and Table S4). Genes encoding the cluster-situated regulator and enzymes for tailoring steps of hydroxylation and methylation are absent in BGC33 [36], suggesting that BGC33 can direct the biosynthesis of a metabolite similar to piericidin A. Deletion of more BGCs from the genome of B48 is worthy to be done, due to the above results and similar cases [14,34].
Meanwhile, subtle and precise regulatory systems have been evolved to force secondary metabolic processes under environmental and developmental control [2,39,44,45]. Detailed mechanism dissection of the regulatory networks will undoubtedly promote works on strain improvement for lincomycin production. But the previous mechanism study mainly focused on single strains, and differences in regulation between the overproducing strains and the lower one remained mysterious. Here two positive (SLINC191 and SLINC348) and four negative regulators (SLINC377, SLINC4481, SLINC6011 and SLINC6156) that participate in lincomycin synthesis were found out.
Gene loci of slinc191 and slinc348, as marked in Fig. 3, are 43 kb away from the left end and 104 kb far from the right terminal of the lincomycin BGC, respectively. Gene slinc191 encodes a putative ArsR/SmtB family transcriptional regulator, several members of which were reported to manage intracellular metal-ion concentration and antibiotic biosynthesis in response to metal excess [39]. Deduced product of slinc348 belongs to Xre family, and few studies have been done in characterizing the members of this family, except the well-known global regulator BldD [9,39].
Gene slinc6156 encodes a widely distributed ribonuclease RNase J, with 97% and 88% identical to homologues from S. coelicolor and S. venezuelae, respectively (Fig. S1A). RNase J has both endonuclease and 5’ exonuclease activities, and participates in forming functional ribosomes in Bacillus subtilis, Streptococcus pyogenes and Mycobacterium smegmatis [[46], [47], [48]]. Deletion of RNase J-encoding genes resulted in reduced antibiotics production in S. coelicolor and S. venezuelae [49,50]. A structural analysis revealed the presence of the essential β-lactamase core and β-CASP domain in SLINC6156. A frameshift mutation event from I248 in B48 deleted motif B and C of β-CASP domain (Fig. S1B). It is most likely that function loss of RNase J affected ribosome processing and/or mRNA degradation in B48, thus enhancing lincomycin production through changes in translational and/or posttranscriptional regulation. After protein BLAST of SLINC6156 using NCBI, approx 560 homologues from Streptomycetes with more than 95% coverage and higher than 90% identity were found out. Among them, 19 homologous sequences from known antibiotic-producers were selected to draw the evolutionary tree with SLINC6156 (Fig. S1C). The high conservation of SLINC6156 homologues among the genus implies widely regulatory functions in different species. It can be inferred that modification of RNase J-encoding genes can be a general strategy for antibiotics yield enhancement.
TPR domain-containing proteins, such as NsdA [51] and NsdB [52], are reported to play important roles on regulation of secondary metabolism. Due to the 44 bp sequence deletion, the stop codon of native SLINC4481 was erased and the length of mutated protein was prolonged to 1119 AA. Based on the domain architecture analysis using CDD (Conserved Domain Database), the approximately 300 AA extra sequence has low similarity with PHA03307 super family domain, which is originally found in virus immediate-early regulatory protein ICP4 (Fig. S2A). There is a great possibility that a structural domain formed by the extended sequence blocks the functional TPR region (Fig. S2B), thereby inactivating SLINC4481. Based on protein BLAST of SLINC4481, only 19 homologues with more than 90% coverage and higher than 60% identity had been deposited in NCBI (Fig. S2C). How SLINC4481 represses lincomycin production requires further study.
Gene slinc6011 encodes a 195 AA SAM-dependent methyltransferase showing 39% identity with 16S rRNA (guanine(966)-N(2))-methyltransferase RsmD from E. coli, and 49% identity with RsmD homologue (Rv2966c) in M. tuberculosis (Fig. S3A). The methylation modification role that RsmD plays on the loop of 16S rRNA helix 31 was first reported in E. coli, and a conserved DPPY/F motif essential for catalytic activity was identified [53]. For the only case in Actinomycetes, Rv2966c from M. tuberculosis can complement rsmD-deleted E. coli cells [54]. In slinc6011 ORF of B48, guanine in position 333 was deleted, leading to a frameshift mutation from P112, a foremost position of catalytic region. The subsequent delay of translation stop has prolonged SLINC6011 to 377 AA (Fig. S3B). m2G966 was structurally revealed to be in direct contact with P-site-bound tRNA [55]. It can be inferred that catalytic function deficient of SLINC6011 altered modification state of ribosome, thereby changing protein expression profile. Mutations in another methyltransferase RsmG (methylate G527 of 16S rRNA) have been widely found in antibiotics overproducers and applied in ribosome engineering for strain improvement [56]. It can be indicated that the mutation of RsmD promoted lincomycin production in a mechanism similar to the RsmG mutations, and can be exploited in strain improvement for more antibiotics.
In summary, complete genome sequence of the high-yield lincomycin-producing strain B48 was determined. Primary metabolism network related to lincomycin biosynthesis was analyzed based on gene annotation. Secondary metabolic potential was also predicted. More importantly, a comparative genomic analysis with the wild type strain NRRL 2936 was carried out, and crucial mutations contributing to lincomycin overproduction were found out. Findings in this study broadened our horizons in regulatory mechanism of lincomycin production, and laid a foundation for strain improvement of antibiotic producers.
CRediT authorship contribution statement
Ruida Wang: Methodology, Investigation, Software, Writing - original draft. Fanjing Kong: Investigation, Formal analysis. Haizhen Wu: Methodology, Writing - review & editing. Bingbing Hou: Validation, Writing - review & editing. Yajing Kang: Investigation. Yuan Cao: Validation. Shiwei Duan: Software. Jiang Ye: Conceptualization, Methodology. Huizhan Zhang: Supervision, Methodology.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (NSFC) (31900059), the China Postdoctoral Science Foundation (2019M650079), and the Research Program of State Key Laboratory of Bioreactor Engineering. We thank Dr. Weihong Jiang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for kindly providing the plasmid pKCcas9dO.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2020.03.001.
Contributor Information
Jiang Ye, Email: yyjj413@163.com.
Huizhan Zhang, Email: huizhzh@ecust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Barka E.A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev. 2006;80:1–43. doi: 10.1128/mmbr.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Heul H.U., Bilyk B.L., McDowall K.J., Seipke R.F., van Wezel G.P. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep. 2018;35:575–604. doi: 10.1039/c8np00012c. [DOI] [PubMed] [Google Scholar]
- 3.Bilyk O., Luzhetskyy A. Metabolic engineering of natural product biosynthesis in actinobacteria. Curr Opin Biotechnol. 2016;42:98–107. doi: 10.1016/j.copbio.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Hopwood D.A. Soil to genomics: the Streptomyces chromosome. Annu Rev Genet. 2006;40:1–23. doi: 10.1146/annurev.genet.40.110405.090639. [DOI] [PubMed] [Google Scholar]
- 5.Spížek J., Řezanka T. Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem Pharmacol. 2017;133:20–28. doi: 10.1016/j.bcp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Hou B., Lin Y., Wu H., Guo M., Petkovic H., Tao L. The novel transcriptional regulator LmbU promotes lincomycin biosynthesis through regulating expression of its target genes in Streptomyces lincolnensis. J Bacteriol. 2018;200 doi: 10.1128/jb.00447-17. e00447-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng S., Wu H., Wang L., Zhang B., Bai L. Enhancement of antibiotic productions by engineered nitrate utilization in actinomycetes. Appl Microbiol Biotechnol. 2017;101:5341–5352. doi: 10.1007/s00253-017-8292-7. [DOI] [PubMed] [Google Scholar]
- 8.Kang Y., Wang Y., Hou B., Wang R., Ye J., Zhu X. AdpAlin, a pleiotropic transcriptional regulator, is involved in the cascade regulation of lincomycin biosynthesis in Streptomyces lincolnensis. Front Microbiol. 2019;10:2428. doi: 10.3389/fmicb.2019.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Wang N., Tang Y., Cai X., Xu Y., Liu R. Developmental regulator BldD directly regulates lincomycin biosynthesis in Streptomyces lincolnensis. Biochem Biophys Res Commun. 2019;518:548–553. doi: 10.1016/j.bbrc.2019.08.079. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y., Ke M., Li J., Tang Y., Wang N., Tan G. TetR-type regulator SLCG_2919 is a negative regulator of lincomycin biosynthesis in Streptomyces lincolnensis. Appl Environ Microbiol. 2019;85 doi: 10.1128/aem.02091-18. e02091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou B., Tao L., Zhu X., Wu W., Guo M., Ye J. Global regulator BldA regulates morphological differentiation and lincomycin production in Streptomyces lincolnensis. Appl Microbiol Biotechnol. 2018;102:4101–4115. doi: 10.1007/s00253-018-8900-1. [DOI] [PubMed] [Google Scholar]
- 12.Peano C., Talà A., Corti G., Pasanisi D., Durante M., Mita G. Comparative genomics and transcriptional profiles of Saccharopolyspora erythraea NRRL 2338 and a classically improved erythromycin over-producing strain. Microb Cell Factories. 2012;11:32. doi: 10.1186/1475-2859-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peano C., Damiano F., Forcato M., Pietrelli A., Palumbo C., Corti G. Comparative genomics revealed key molecular targets to rapidly convert a reference rifamycin-producing bacterial strain into an overproducer by genetic engineering. Metab Eng. 2014;26:1–16. doi: 10.1016/j.ymben.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Lu C., Bai L. Mechanism of salinomycin overproduction in Streptomyces albus as revealed by comparative functional genomics. Appl Microbiol Biotechnol. 2017;101:4635–4644. doi: 10.1007/s00253-017-8278-5. [DOI] [PubMed] [Google Scholar]
- 15.Xie H., Zhao Q., Zhang X., Kang Q., Bai L. Comparative functional genomics of the acarbose producers reveals potential targets for metabolic engineering. Synth Syst Biotechnol. 2019;4:49–56. doi: 10.1016/j.synbio.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbs G., Frazer C.M., Gardner D.C., Cullum J.A., Oliver S.G. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. doi: 10.1007/bf00258408. [DOI] [Google Scholar]
- 17.Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. John Innes Foundation; Norwich: 2000. Practical Streptomyces genetics. [Google Scholar]
- 18.Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 19.Pharmacopoeia Commission of the Ministry of Health of the People’s Republic of China . China Medico-Pharmaceutical Science & Technology Publishing House; Beijing, China: 1990. Pharmacopoeia of the People's Republic of China; pp. 113–116. [Chinese] [Google Scholar]
- 20.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 22.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin. 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 23.Peschke U., Schmidt H., Zhang H., Piepersberg W. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol Microbiol. 1995;16:1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang M., Chen D., Zhao Q., Liu W. Isolation, structure elucidation, and biosynthesis of a cysteate-containing nonribosomal peptide in Streptomyces lincolnensis. J Org Chem. 2018;83:7102–7108. doi: 10.1021/acs.joc.8b00044. [DOI] [PubMed] [Google Scholar]
- 25.Ostash B., Saghatelian A., Walker S.A. A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bentley S.D., Chater K.F., Cerdeño-Tárraga A.M., Challis G.L., Thomson N.R., James K.D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2) Nature. 2002;417:141. doi: 10.1038/news020506-8. [DOI] [PubMed] [Google Scholar]
- 27.Vasanthabharathi V., Lakshminarayanan R., Jayalakshmi S. Melanin production from marine Streptomyces. Afr J Biotechnol. 2011;10:11224–11234. doi: 10.5897/ajb11.296. [DOI] [Google Scholar]
- 28.Biarnes-Carrera M., Breitling R., Takano E. Butyrolactone signalling circuits for synthetic biology. Curr Opin Biotechnol. 2005;28:91–98. doi: 10.1016/j.cbpa.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Bursy J., Kuhlmann A.U., Pittelkow M., Hartmann H., Jebbar M., Pierik A.J. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3 (2) in response to salt and heat stresses. Appl Environ Microbiol. 2008;74:7286–7296. doi: 10.1128/aem.00768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin X., Cane D.E. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor. Mechanism and stereochemistry of the enzymatic formation of epi-isozizaene. J Am Chem Soc. 2009;131:6332–6333. doi: 10.1021/ja901313v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghimire G.P., Koirala N., Sohng J.K. Activation of cryptic hop genes from Streptomyces peucetius ATCC 27952 involved in hopanoid biosynthesis. J Microbiol Biotechnol. 2015;25:658–661. doi: 10.4014/jmb.1408.08058. [DOI] [PubMed] [Google Scholar]
- 32.Rückert C., Szczepanowski R., Albersmeier A., Goesmann A., Iftime D., Musiol E.M. Complete genome sequence of the kirromycin producer Streptomyces collinus Tü 365 consisting of a linear chromosome and two linear plasmids. J Biotechnol. 2013;168:739–740. doi: 10.1016/j.jbiotec.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Tian J., Yang J., Li L., Ruan L., Wei W., Zheng G. The complete genome sequence of a high pristinamycin-producing strain Streptomyces pristinaespiralis HCCB10218. J Biotechnol. 2015;214:45–46. doi: 10.1016/j.jbiotec.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Li L., Zheng G., Chen J., Ge M., Jiang W., Lu Y. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab Eng. 2017;40:80–92. doi: 10.1016/j.ymben.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Becerril A., Álvarez S., Braña A.F., Rico S., Díaz M., Santamaría R.I. Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PloS One. 2018;13 doi: 10.1371/journal.pone.0198145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Zhang W., Zhu Y., Zhang Q., Tian X., Zhang S. Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis. Org Lett. 2014;16:736–739. doi: 10.1021/ol4034176. [DOI] [PubMed] [Google Scholar]
- 37.Pang A., Du L., Lin C., Qiao J., Zhao G. Co-overexpression of lmbW and metK led to increased lincomycin A production and decreased byproduct lincomycin B content in an industrial strain of Streptomyces lincolnensis. J Appl Microbiol. 2015;119:1064–1074. doi: 10.1111/jam.12919. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y., Tan G., Ke M., Li J., Tang Y., Meng S. Enhanced lincomycin production by co-overexpression of metK1 and metK2 in Streptomyces lincolnensis. J Ind Microbiol Biotechnol. 2018;45:345–355. doi: 10.1007/s10295-018-2029-1. [DOI] [PubMed] [Google Scholar]
- 39.Romero-Rodríguez A., Robledo-Casados I., Sánchez S. An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta-Gene Regul Mech. 2015;1849:1017–1039. doi: 10.1016/j.bbagrm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Falke D., Biefel B., Haase A., Franke S., Fischer M., Sawers R.G. Activity of spore-specific respiratory nitrate reductase 1 of Streptomyces coelicolor A3 (2) requires a functional cytochrome bcc-aa3 oxidase supercomplex. J Bacteriol. 2019;201:e00104–e00119. doi: 10.1128/jb.00104-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkarni A., Zeng Y., Zhou W., Lanen S.V., Zhang W., Chen S. A branch point of Streptomyces sulfur amino acid metabolism controls the production of albomycin. Appl Environ Microbiol. 2016;82:467–477. doi: 10.1128/aem.02517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D., Hesketh A., Kim E., Song J., Lee D., Kim I. Complex extracellular interactions of proteases and a protease inhibitor influence multicellular development of Streptomyces coelicolor. Mol Microbiol. 2008;70:1180–1193. doi: 10.1111/j.1365-2958.2008.06471.x. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen K.T., Willey J.M., Nguyen L.D., Nguyen L.T., Viollier P.H., Thompson C.J. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol Microbiol. 2002;46:1223–1238. doi: 10.1046/j.1365-2958.2002.03255.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu G., Chater K.F., Chandra G., Niu G., Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/mmbr.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urem M., Świątek‐Połatyńska M.A., Rigali S., van Wezel G.P. Intertwining nutrient‐sensory networks and the control of antibiotic production in Streptomyces. Mol Microbiol. 2016;102:183–195. doi: 10.1111/mmi.13464. [DOI] [PubMed] [Google Scholar]
- 46.Even S., Pellegrini O., Zig L., Labas V., Vinh J., Brechemmier-Baey D. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bugrysheva J.V., Scott J.R. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol Microbiol. 2010;75:731–743. doi: 10.1111/j.1365-2958.2009.07012.x. [DOI] [PubMed] [Google Scholar]
- 48.Taverniti V., Forti F., Ghisotti D., Putzer H. Mycobacterium smegmatis RNase J is a 5′‐3′ exo‐/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol Microbiol. 2011;82:1260–1276. doi: 10.1111/j.1365-2958.2011.07888.x. [DOI] [PubMed] [Google Scholar]
- 49.Bralley P., Aseem M., Jones G.H. SCO5745, a bifunctional RNase J ortholog, affects antibiotic production in Streptomyces coelicolor. J Bacteriol. 2014;196:1197–1205. doi: 10.1128/jb.01422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones S.E., Leong V., Ortega J., Elliot M.A. Development, antibiotic production, and ribosome assembly in Streptomyces venezuelae are impacted by RNase J and RNase III deletion. J Bacteriol. 2014;196:4253–4267. doi: 10.1128/jb.02205-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Guo S., Guo W., Xi D., Xiang W. Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis. J Antibiot. 2009;62:309. doi: 10.1038/ja.2009.33. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L., Li W., Zhao C., Chater K.F., Tao M. NsdB, a TPR-like-domain-containing protein negatively affecting production of antibiotics in Streptomyces coelicolor A3 (2) Wei sheng wu xue bao= Acta Microbiologica Sinica. 2007;47:849–854. [PubMed] [Google Scholar]
- 53.Sergiev P.V., Bogdanov A.A., Dontsova O.A. Ribosomal RNA guanine-(N2)-methyltransferases and their targets. Nucleic Acids Res. 2007;35:2295–2301. doi: 10.1093/nar/gkm104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar A., Saigal K., Malhotra K., Sinha K.M., Taneja B. Structural and functional characterization of Rv2966c protein reveals an RsmD-like methyltransferase from Mycobacterium tuberculosis and the role of its N-terminal domain in target recognition. J Biol Chem. 2011;286:19652–19661. doi: 10.1074/jbc.m110.200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lesnyak D.V., Osipiuk J., Skarina T., Sergiev P.V., Bogdanov A.A., Edwards A. Methyltransferase that modifies guanine 966 of the 16 S rRNA functional identification and tertiary structure. J Biol Chem. 2007;282:5880–5887. doi: 10.1074/jbc.m608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu S., Duan Y., Huang Y. The application of ribosome engineering to natural product discovery and yield improvement in Streptomyces. Antibiotics. 2019;8:133. doi: 10.3390/antibiotics8030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkinson C.J., Hughes-Thomas Z.A., Martin C.J., Böhm I., Mironenko T., Deacon M. Increasing the efficiency of heterologous promoters in actinomycetes. J Mol Microbiol Biotechnol. 2002;4:417–426. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.