Summary
Hematopoietic stem cells (HSCs) and skeletal stem cells (SSCs) cohabit in the bone marrow. KITL (C-KIT ligand) from LEPR+ adult bone marrow stromal cells is pivotal for HSC maintenance. In contrast, it remains unclear whether KITL/C-KIT signaling also regulates SSCs. Here, we lineage traced C-KIT+ cells and found that C-KIT was expressed by fetal, but not postnatal skeletal progenitors. Fetal C-KIT+ cells gave rise to 20% of LEPR+ stromal cells in adult bone marrow, forming nearly half of all osteoblasts. Disruption of mTOR signaling in fetal C-KIT+ cells impaired bone formation. Notably, conditional deletion of Kitl from PRX1+ fetal bone marrow stromal cells, but not LEPR+ adult bone marrow stromal cells, significantly increased bone formation. Thus, our work identified C-KIT+ skeletal progenitors as an important source of bones formed during development.
Keywords: skeletal stem cells, hematopoietic stem cells, bone, C-KIT, skeletal progenitors, KITL, SCF
Highlights
-
•
C-KIT was expressed by fetal, but not postnatal skeletal progenitors
-
•
Fetal C-KIT+ cells formed half of all osteoblasts in adult bone marrow.
-
•
Disruption of mTOR signaling in fetal C-KIT+ cells impaired bone formation
-
•
Conditional deletion of Kitl from Prx1+ cells significantly increased bone formation
In this article, Zhou and colleagues identified fetal C-KIT+ stromal cells as an important source of bones formed during development. These cells gave rise to 20% of LEPR+ stromal cells in adult bone marrow, forming nearly half of all osteoblasts. This work highlights the heterogeneity of skeletal progenitors during development.
Introduction
Skeletal stem cells (SSCs), also known as bone marrow mesenchymal stromal cells (BM-MSCs), reside in postnatal bone marrow and give rise to cartilage, bone, marrow adipocytes, and hematopoiesis-supportive stroma (Bianco et al., 2006, Bianco and Robey, 2015, Sacchetti et al., 2007). In vitro, SSCs are colony-forming units-fibroblasts (CFU-F) that are able to undergo osteogenic, adipogenic, and chondrogenic differentiation (Friedenstein et al., 1970, Pittenger et al., 1999). In vivo, most SSCs express PDGFRα (Morikawa et al., 2009), LEPR (Mizoguchi et al., 2014, Zhou et al., 2014a), PRX1 (Greenbaum et al., 2013), MX-1 (Park et al., 2012), Nestin-GFP transgene (Mendez-Ferrer et al., 2010), CXCL12 (Greenbaum et al., 2013), and SCF (Zhou et al., 2014a). For bones formed by endochondral ossification, fetal chondrocytes on the growing cartilage form SSC-enriching bone marrow stromal cells (Maes et al., 2010, Ono et al., 2014). Recent studies have also revealed the contribution of postnatal hypertrophic chondrocytes to SSCs and their lineages (Ono et al., 2014, Zhou et al., 2014b). Thus, chondrocytes are precursors of most SSCs in the bone marrow (Ono et al., 2014). Interestingly, in adult bone marrow, PTHRP+ chondrocytes of the growth plate are multipotent progenitors that give rise to hypertrophic chondrocytes, trabecular bones, as well as SSCs (Mizuhashi et al., 2018). These studies raised a previously unrecognized question concerning the heterogeneity and/or hierarchy of chondrocytes during development and aging.
SSCs are a heterogeneous cell population. Sca-1 expression has been shown to enrich CFU-Fs, which are capable of trilineage differentiation in vitro (Morikawa et al., 2009). GREMLIN-1 is expressed by a small fraction of SSCs that form osteoblasts but not marrow adipocytes (Worthley et al., 2015). Adiponectin is expressed by a subset of SSCs that form most marrow adipocytes but few osteoblasts (Zhou et al., 2017). GLI1-expressing SSCs form bones and are the primary source of myofibroblasts during bone marrow fibrosis (Shi et al., 2017). NG2+ peri-arteriolar SSCs express higher levels of Nestin-GFP transgene and CXCL12 than peri-sinusoidal SSCs (Kunisaki et al., 2013). Neonatal CTSK+ SSCs localize at the periosteal region and contribute to intramembranous bone formation (Debnath et al., 2018). An important yet largely unaddressed question concerns whether the self-renewal and differentiation of different SSC subpopulations are controlled by common or different mechanisms.
C-KIT is a well-known cell surface marker of many adult stem cells, such as hematopoietic stem cells (HSCs). KITL (C-KIT ligand)/C-KIT signaling maintains HSCs in the bone marrow (Ding et al., 2012). Whether C-KIT is expressed by SSCs was a topic with controversy (Deng, 2010, Deng et al., 2006, Hatzistergos et al., 2010). In this study, we assessed the expression of C-KIT in SSCs during development and investigated their potential contribution to SSC lineages, especially osteoblasts. Taking advantage of genetic lineage-tracing techniques, we identified an SSC subpopulation that was derived from fetal C-KIT+ cells. These SSCs formed chondrocytes, osteoblasts, and adipocytes in adult bone marrow. Furthermore, by conditional knockout techniques, we found that the osteogenic activity of C-KIT+ cells was intrinsically regulated by mammalian target of rapamycin (mTOR) and extrinsically regulated by KITL from fetal PRX1+ stromal cells.
Results
Fate-Mapping Postnatal C-KIT+ Cells Did Not Label Any Osteoblasts or Bone Marrow Adipocytes
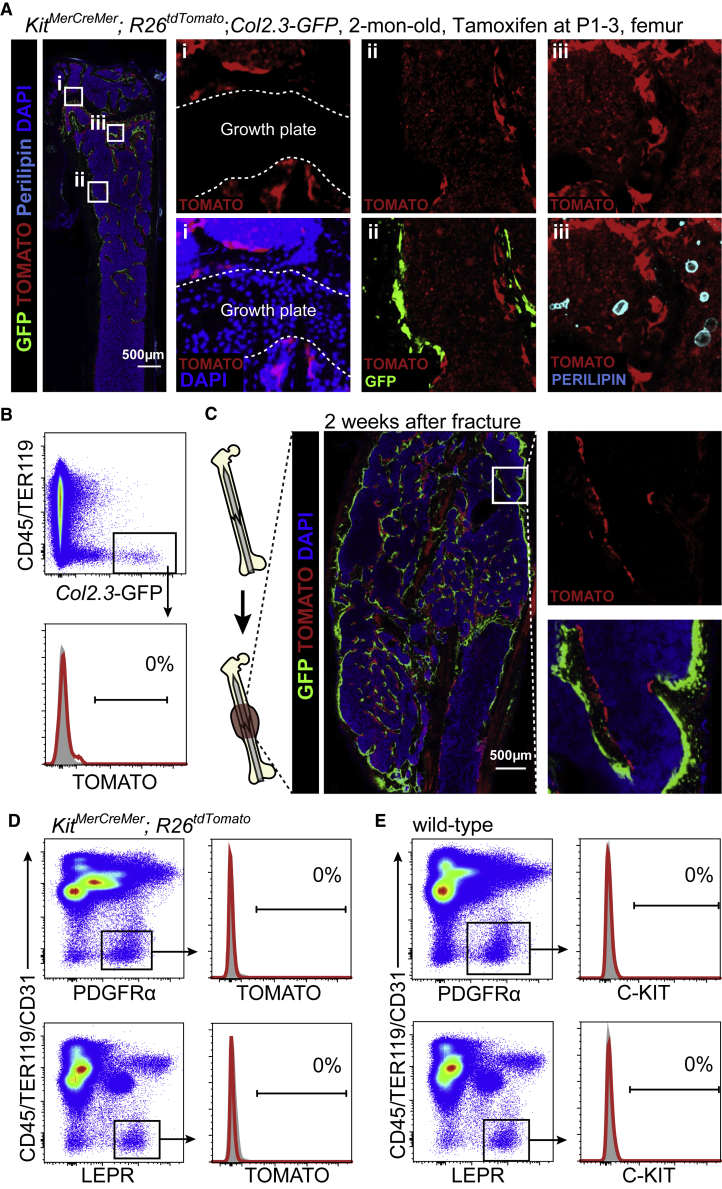
To address whether C-KIT is expressed by postnatal SSCs, we crossed the KitMerCreMer knockin mice (van Berlo et al., 2014) successively with the R26tdTomato knockin mice (Madisen et al., 2010) and the Col1a1∗2.3-GFP (Col2.3-GFP) transgenic mice (Kalajzic et al., 2002) to generate the KitMerCreMer; R26tdTomato; Col2.3-GFP mice. Kit promoter-driven Cre expression did not show any leakiness in the bone marrow without tamoxifen treatment (Figure S1A). By administering tamoxifen to these mice from postnatal days 1–3 (P1–P3) we were able to label and trace the fate of postnatal C-KIT+ cells. In these mice, C-KIT+ cells and their progeny were labeled by TOMATO; osteoblasts were labeled by Col2.3-GFP. Using confocal imaging of thin femur sections from these mice at 2 months of age, we did not detect TOMATO expression in any chondrocytes at the growth plate, Col2.3-GFP+ osteoblasts lining the bone surface, or perilipin+ adipocytes in the bone marrow (Figure 1A). The stromal cell-like cells lining the bone surface that displayed bright TOMATO fluorescence (Figure 1A) were actually tartrate-resistant acid phosphatase+ (TRAP+) osteoclasts, a hematopoietic lineage (Figure S1B). Flow cytometric analysis of enzymatically dissociated bone shafts did not detect any Col2.3-GFP+ osteoblasts that were TOMATO+ in 2-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice (Figure 1B). These data suggested that postnatal C-KIT+ cells do not generate any skeletal lineages in the bone marrow under homeostasis.
Figure 1.
Lineage-Tracing of Neonatal C-KIT+ Cells Did Not Label SSCs in the Bone Marrow
All KitMerCreMer; R26tdTomato; Col2.3-GFP and KitMerCreMer; R26tdTomato mice were tamoxifen treated at P1–3 and analyzed at 2 months of age.
(A) Representative confocal images of femur sections from KitMerCreMer; R26tdTomato; Col2.3-GFP mice. Osteoblasts were revealed by Col2.3-GFP fluorescence; adipocytes were revealed by anti-perilipin staining (n = 3 mice from three independent experiments).
(B) Representative flow cytometry plots of enzymatically dissociated femoral bone shafts showed no TOMATO expression in any Col2.3-GFP+ osteoblasts in KitMerCreMer; R26tdTomato; Col2.3-GFP mice (n = 3 mice from three independent experiments).
(C) Representative confocal images of the callus region of KitMerCreMer; R26tdTomato; Col2.3-GFP mice at 2 weeks after femur fracture. Note that TOMATO was also expressed by hematopoietic cells in these mice but levels of TOMATO expression in stromal cells were ∼10- to 100-fold higher than in hematopoietic cells. Therefore, short-exposure images showed mainly TOMATO fluorescence in stromal cells (n = 3 mice from three independent experiments).
(D) Representative flow cytometry plots of enzymatically dissociated bone marrow cells showed that neither CD45−TER119−CD31−PDGFRα+ nor CD45−TER119−CD31−LEPR+ bone marrow stromal cells expressed TOMATO in KitMerCreMer; R26tdTomato mice. Bone marrow stromal cells from wild-type mice were set as negative controls (gray) (n = 3 mice from three independent experiments).
(E) Neither CD45−TER119−CD31−PDGFRα+ nor CD45−TER119−CD31−LEPR+ bone marrow stromal cells expressed C-KIT in 2-month-old wild-type mice. Isotype controls are shown in gray (n = 3 mice from three independent experiments).
Next, we investigated if postnatal C-KIT+ cells contribute to new osteoblast formation during fracture healing. Two-month-old tamoxifen-administered KitMerCreMer; R26tdTomato; Col2.3-GFP mice were fractured on their right femurs (Figure 1C). At 2 weeks after the fracture, the bone callus was filled with newly formed cancellous bones (Figure 1C). We did not detect any Col2.3-GFP+ osteoblasts that co-expressed TOMATO at the callus region (Figure 1C), suggesting that postnatal C-KIT+ cells do not generate new osteoblasts during bone regeneration.
Postnatal SSCs Did Not Express C-KIT
SSCs express PDGFRα (Morikawa et al., 2009) and LEPR (Mizoguchi et al., 2014, Zhou et al., 2014a). We found by flow cytometry that neither CD45−TER119−CD31−PDGFRα+ nor CD45−TER119−CD31−LEPR+ bone marrow stromal cells expressed TOMATO in 2-month-old KitMerCreMer; R26tdTomato mice (Figure 1D). Consistent with this, C-KIT protein expression was not detected on CD45−TER119−CD31−PDGFRα+ or CD45−TER119−CD31−LEPR+ bone marrow stromal cells in 2-month-old wild-type mice (Figure 1E). Taken together, these data suggested that C-KIT is not expressed by postnatal SSCs.
Fate-Mapping Fetal C-KIT+ Cells Labeled Chondrocytes, Preosteoblasts, and Bone Marrow Stromal Cells at E16.5
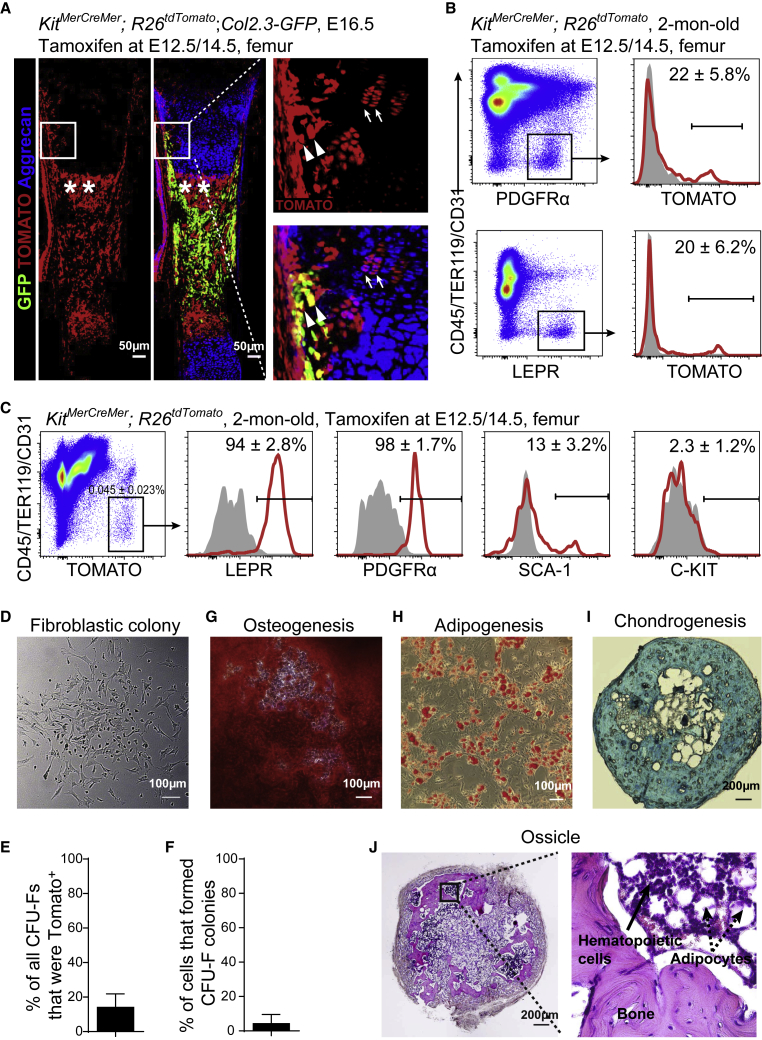
Next, we investigated the expression of C-KIT in fetal bone marrow. KitMerCreMer; R26tdTomato embryos that were treated with tamoxifen at embryonic day 12.5 (E12.5) displayed TOMATO fluorescence only in a few stromal cells in the growth cartilage at E13.5 (Figure S2A). We then treated KitMerCreMer; R26tdTomato; Col2.3-GFP embryos at both E12.5 and 14.5 (E12.5/14.5). Using confocal imaging of femur sections at E16.5 we detected TOMATO expression in a subset of aggrecan+ chondrocytes at the growth cartilage (Figure 2A, arrows). In the osteogenic perichondrium, many Col2.3-GFP+ preosteoblasts that were close to the TOMATO+ chondrocytes also expressed TOMATO (Figure 2A, arrow heads). Moreover, the nascent primary ossification center in the bone marrow was occupied by TOMATO+ stromal cells (Figure 2A, asterisks).
Figure 2.
Lineage-Tracing of Fetal C-KIT+ Cells Labeled a Few Chondrocytes in Fetus and a Subpopulation of SSCs in Adult Bone Marrow
All KitMerCreMer; R26tdTomato; Col2.3-GFP and KitMerCreMer; R26tdTomato mice were tamoxifen treated at E12.5 and 14.5.
(A) Representative confocal images of femur sections from KitMerCreMer; R26tdTomato; Col2.3-GFP mice at E16.5. Arrows indicated TOMATO+ chondrocytes at the growth cartilage; arrowheads indicated TOMATO+Col2.3-GFP+ preosteoblasts at the osteogenic perichondrium; asterisks indicated TOMATO+ stromal cells in the nascent primary ossification center in the bone marrow (n = 3 mice from three independent experiments).
(B) Flow cytometric analysis showed that ∼22% of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells (upper plots) and ∼20% of CD45−TER119−CD31−LEPR+ bone marrow stromal cells (lower plots) were TOMATO+ in 2-month-old KitMerCreMer; R26tdTomato mice. The corresponding bone marrow stromal cells from wild-type mice were set as negative controls (gray) (n = 3 mice from three independent experiments).
(C) Representative flow cytometry plots of CD45−TER119−CD31−TOMATO+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice. Isotype controls are shown in gray (n = 4 mice from four independent experiments).
(D) Representative bright-field image of colonies formed by TOMATO+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice (n = 3 mice from three independent experiments).
(E) Percentage of all colonies from KitMerCreMer; R26tdTomato mice that were TOMATO+. Macrophage colonies were excluded by staining with anti-CD45 antibody (n = 3 mice from three independent experiments).
(F) Percentage of TOMATO+ bone marrow stromal cells from KitMerCreMer; R26tdTomato mice that formed colonies in culture (n = 384 individual cells from 3 mice in three independent experiments).
(G–I) TOMATO+ CFU-Fs from 2-month-old KitMerCreMer; R26tdTomato mice were able to differentiate into alizarin red S+ osteoblastic cells (G), oil red O+ adipocytes (H), and alcian blue+ chondrocytes (I). ∼500 TOMATO+ bone marrow stromal cells from KitMerCreMer; R26tdTomato mice were cultured in colony-forming medium for 14 days. All colonies were digested and aliquots were cultured in osteogenic, adipogenic, and chondrogenic differentiation medium, respectively, before the staining. Adipogenic and osteogenic differentiation was performed in six-well plates while chondrogenic differentiation was performed as pellets in 15-mL tubes (n = 3 mice from three independent experiments).
(J) Cultured TOMATO+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice were able to form bone ossicles upon subcutaneous transplantation (n = 3 mice from three independent experiments).
C-KIT+ Cell-Derived Bone Marrow Stromal Cells Contained SSCs
To address whether fetal C-KIT+ cells generate SSCs in adult bone marrow, we analyzed 2-month-old KitMerCreMer; R26tdTomato mice that had been treated with tamoxifen at E12.5/14.5. Flow cytometric analysis of enzymatically dissociated bone marrow cells from these mice showed that TOMATO was expressed by approximately 22% of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells and approximately 20% of CD45−TER119−CD31−LEPR+ bone marrow stromal cells (Figure 2B). The CD45−TER119−CD31−TOMATO+ cells from KitMerCreMer; R26tdTomato mice accounted for 0.045% of whole bone marrow cells (Figure 2C). They were uniformly positive for the expression of LEPR and PDGFRα but negative for the expression of C-KIT (Figure 2C). Approximately 13% of them expressed Sca-1 (Figure 2C).
SSCs are characterized as CFU-Fs that are capable of osteogenic, adipogenic, and chondrogenic differentiation in vitro (Friedenstein et al., 1970, Pittenger et al., 1999). To assess CFU-F activity we enzymatically dissociated bone marrow cells from KitMerCreMer; R26tdTomato mice and added them to adherent cultures at clonal density (Figures 2D and S2B). Figure 2E shows that ∼14% of all bone marrow CFU-Fs were TOMATO+. Figure 2F shows that ∼4.2% of TOMATO+CD45−TER119−CD31– bone marrow stromal cells formed colonies. When the nutrient medium was replaced with osteogenic, adipogenic, or chondrogenic medium, these colonies gradually formed alizarin red S+ osteoblastic cells (Figure 2G), oil red O+ adipocytes (Figure 2H), and alcian blue+ chondrocytes (Figure 2I), respectively. Consistent with this, GFP fluorescence and anti-perilipin staining were robustly detected in the TOMATO+ colonies from KitMerCreMer; R26tdTomato; Col2.3-GFP mice when cultured in osteogenic and adipogenic differentiation media, respectively (Figures S2C and S2D).
Ossicle formation capacity distinguished SSCs from other bone marrow stromal cells and non-skeletal stromal cells (Chan et al., 2015, Sacchetti et al., 2007). We cultured ∼500 TOMATO+ bone marrow stromal cells from KitMerCreMer; R26tdTomato mice, allowed them to form colonies, then expanded the colonies by one passage, implanted the cells into denatured collagen sponges, transplanted them subcutaneously, and assessed the development of bony ossicle 8 weeks later (Figure S2E). By hematoxylin and eosin staining, we observed bones, adipocytes, and hematopoietic cells within each ossicle (Figure 2J). Thus, the data above suggested that fate-mapping fetal C-KIT+ cells traces a subset of SSCs in adult bone marrow.
C-KIT+ Cell-Derived Bone Marrow Stromal Cells Showed a Classical Transcriptomic Signature of SSCs
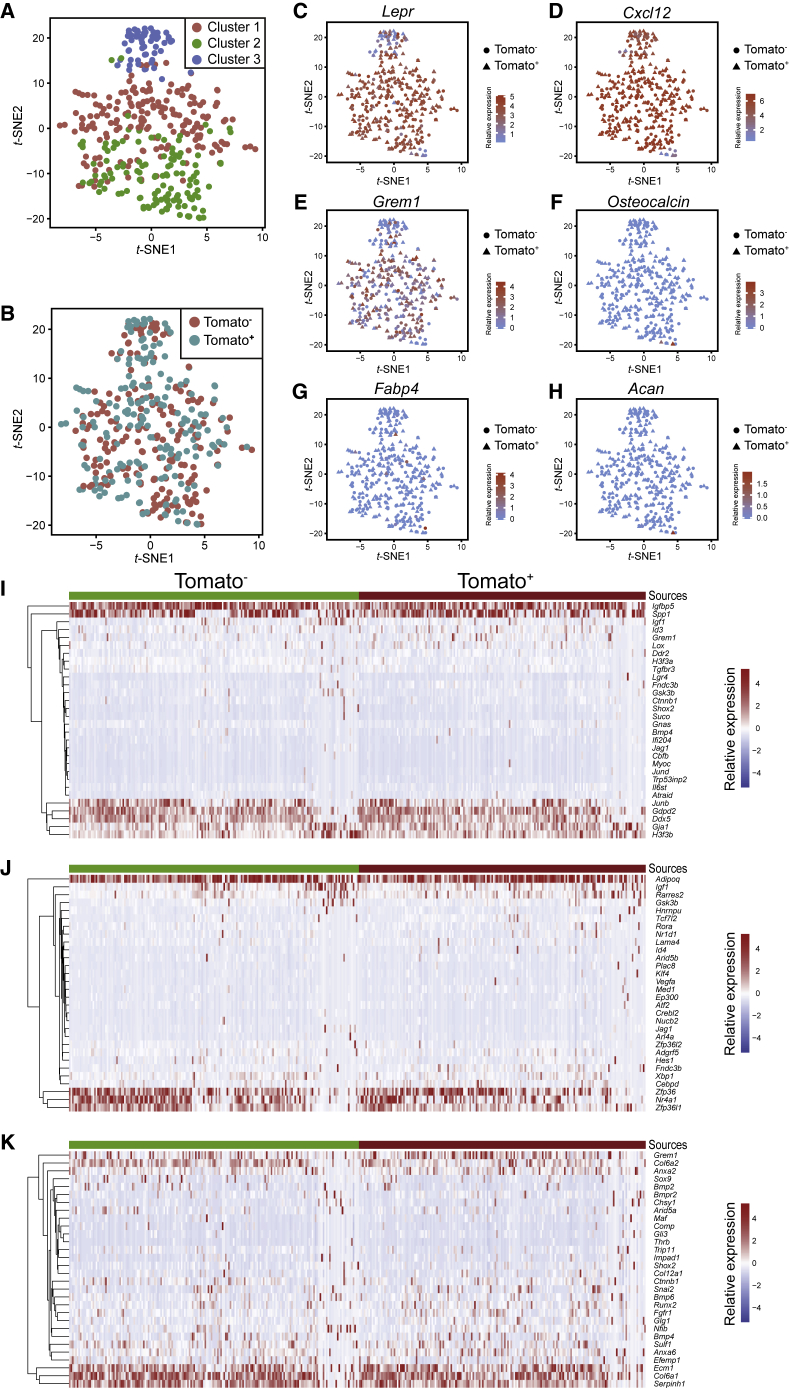
To further evaluate the SSC characteristics of fetal C-KIT+ cell-derived bone marrow stromal cells, we performed single-cell RNA sequencing (scRNA-seq) of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice that had been treated with tamoxifen at E12.5/14.5. A total of 182 TOMATO+ cells and 184 TOMATO− cells from three mice were subjected to Smart-seq2-based sequencing. Clustering analysis showed that the isolated cells grouped into three clusters (Figure 3A). Cluster 1 and 2 were positive for Lepr expression, whereas cluster 3 showed lower and heterogeneous Lepr expression (Figure 3C). TOMATO+ and TOMATO− cells were distributed in all three clusters and were indistinguishable from each other (Figures 3A and 3B).
Figure 3.
scRNA-Sequencing of Tomato+ and Tomato− Stromal Cells from KitMerCreMer; R26tdTomato Mice
(A and B) tSNE plots of single CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice that had been treated with tamoxifen at E12.5/14.5. TOMATO+ and TOMATO− sub-fractions were subjected to Smart-seq2-based sequencing. Cells were colored by clusters (A) or sources (B). Cells were purified from 3 mice in three independent experiments.
(C–H) tSNE plots of scRNA-seq profiles (points) shaped by sources and colored by expression (Log10(FPKM+1)) of Lepr (C), Cxcl12 (D), Gremlin1 (E), Osteocalcin (F), Fabp4 (G), and Acan (H).
(I–K) Heatmap of genes involved in osteogenic (I), adipogenic (J), and chondrogenic differentiation (K).
Both TOMATO+ and TOMATO− stromal cells from KitMerCreMer; R26tdTomato mice showed typical SSC transcriptomic signatures. They were uniformly positive for Cxcl12 expression (Figure 3D), mostly positive for Lepr expression (Figure 3C), heterogeneously positive for Gremlin1 expression (Figure 3E), and uniformly negative for Osteocalcin (osteoblast marker, Figure 3F), Fabp4 (adipocyte marker, Figure 3G), and Acan expression (chondrocyte marker, Figure 3H). Genes involved in osteogenic differentiation (Figure 3I), adipogenic differentiation (Figure 3J), and chondrogenic differentiation (Figure 3K) showed similar expression patterns in TOMATO+ and TOMATO− stromal cells.
C-KIT+ Cell-Derived SSCs Formed Osteoblasts and Marrow Adipocytes throughout Adulthood
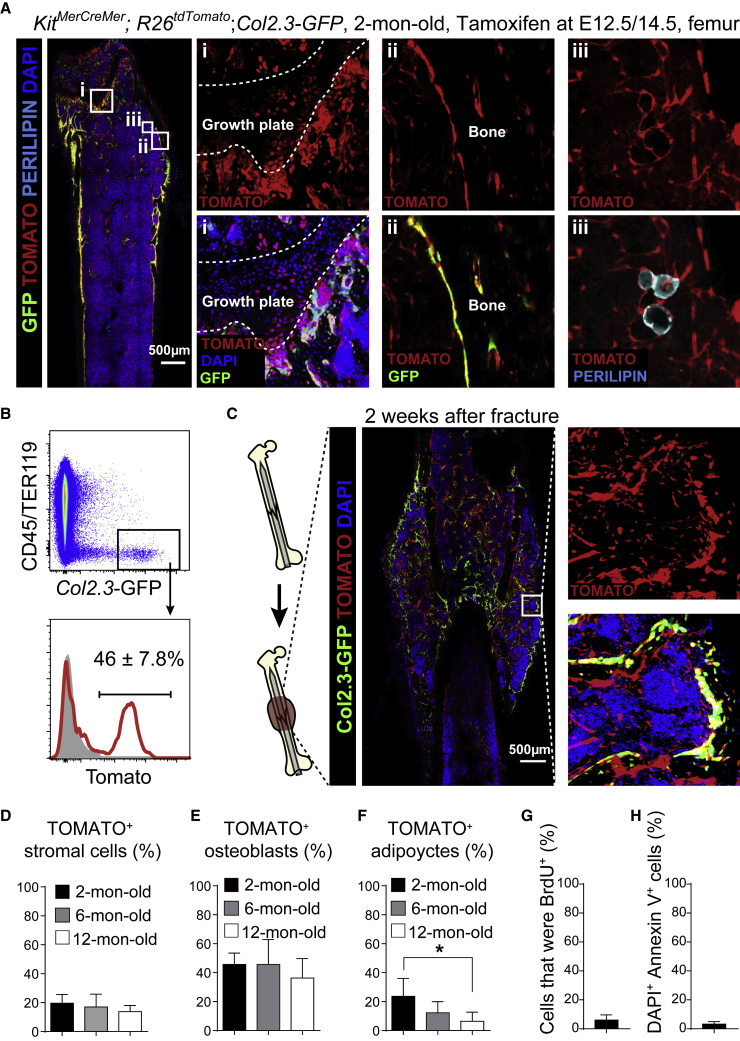
We fate-mapped the fetal C-KIT+ cells to investigate how these cells contribute to skeletal development in vivo. KitMerCreMer; R26tdTomato; Col2.3-GFP mice were treated with tamoxifen at E12.5/14.5 and euthanized at 2 months of age. We found that TOMATO+ chondrocytes were sparsely distributed at the growth plate of the femur from these mice (Figure 4A). We also observed co-labeling of many bone-lining cells by both GFP and TOMATO (Figure 4A), suggesting a contribution of C-KIT+ cells to osteoblasts. By flow cytometry, approximately 46% of all Col2.3-GFP+ osteoblasts were TOMATO+ in 2-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice (Figure 4B).
Figure 4.
Fetal C-KIT+ Cells Contributed to Bone Formation and Regeneration in Adult Bone Marrow
All KitMerCreMer; R26tdTomato; Col2.3-GFP and KitMerCreMer; R26tdTomato mice were tamoxifen treated at E12.5 and 14.5.
(A) Representative confocal images of femur sections from 2-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice. Osteoblasts were revealed by Col2.3-GFP fluorescence; adipocytes were revealed by anti-perilipin staining. Note that TOMATO was also expressed by hematopoietic cells in these mice but levels of TOMATO expression in stromal cells were ∼10- to 100-fold higher than in hematopoietic cells. Therefore, short-exposure images showed mainly TOMATO fluorescence in stromal cells (n = 3 mice from three independent experiments).
(B) Representative flow cytometry plots of enzymatically dissociated femurs showed that 46% of Col2.3-GFP+ osteoblasts were derived from C-KIT+ cells in KitMerCreMer; R26tdTomato; Col2.3-GFP mice (n = 3 mice from three independent experiments).
(C) Representative confocal images of the callus region of KitMerCreMer; R26tdTomato; Col2.3-GFP mice at 2 weeks after femur fracture (n = 3 mice from three independent experiments).
(D–F) Frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells (D), Col2.3-GFP+ osteoblasts (E), and perilipin+ adipocytes (F) that expressed TOMATO in 2-, 6-, and 12-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice. All data represent mean ± SD of 3 mice per age from three independent experiments. The statistical significance of differences among different ages was measured by repeated measures one-way ANOVAs with Greenhouse-Geisser correction and Tukey's multiple comparison tests (∗p < 0.05).
(G) BrdU incorporation (14-day pulse) by TOMATO+ bone marrow stromal cell from 2-month-old KitMerCreMer; R26tdTomato mice (n = 3 mice from three independent experiments).
(H) Percentage of TOMATO+ bone marrow stromal cells from 2-month-old KitMerCreMer; R26tdTomato mice that were DAPI+Annexin V+ (n = 3 mice from three independent experiments).
We performed open fracture surgery on the right femurs of 2-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice. At 2 weeks after the fracture, we detected a significant amount of Col2.3-GFP+ TOMATO+ osteoblasts in the callus (Figure 4C). Thus, the C-KIT+ cell-derived SSCs contribute to bone formation during development and after bone injury.
We also aged the KitMerCreMer; R26tdTomato; Col2.3-GFP mice that had been treated with tamoxifen at E12.5/14.5. We did not observe significant difference in the frequency of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells that expressed TOMATO among 2-, 6-, and 12-month-old mice (Figure 4D). The frequencies of Col2.3-GFP+ osteoblasts that expressed TOMATO were also comparable among different ages (Figure 4E), suggesting that C-KIT+ cell-derived SSCs continuously produce osteoblasts throughout adulthood.
In the bone marrow from 2-month-old KitMerCreMer; R26tdTomato; Col2.3-GFP mice that had been treated with tamoxifen at E12.5/14.5, we also detected TOMATO expression in some perilipin+ marrow adipocytes (Figure 4A). The frequency of TOMATO+ adipocytes gradually reduced from 23% in 2-month-old mice to 6.3% in 12-month-old mice (Figure 4F).
We administered bromodeoxyuridine (BrdU) to 2-month-old KitMerCreMer; R26tdTomato mice for 14 days. We found that ∼6% of TOMATO+ stromal cells were BrdU+ (Figure 4G). We also examined the frequency of apoptotic cells in C-KIT+ cell-derived bone marrow stromal cells. Using flow cytometry, we found only ∼3% of TOMATO+ stromal cells from 6-month-old KitMerCreMer; R26tdTomato mice that were DAPI+Annexin V+ (Figure 4H). Thus, the descendants of C-KIT+ cells are generally stable and slowly dividing.
C-KIT+ Cells Were Important for Bone Development
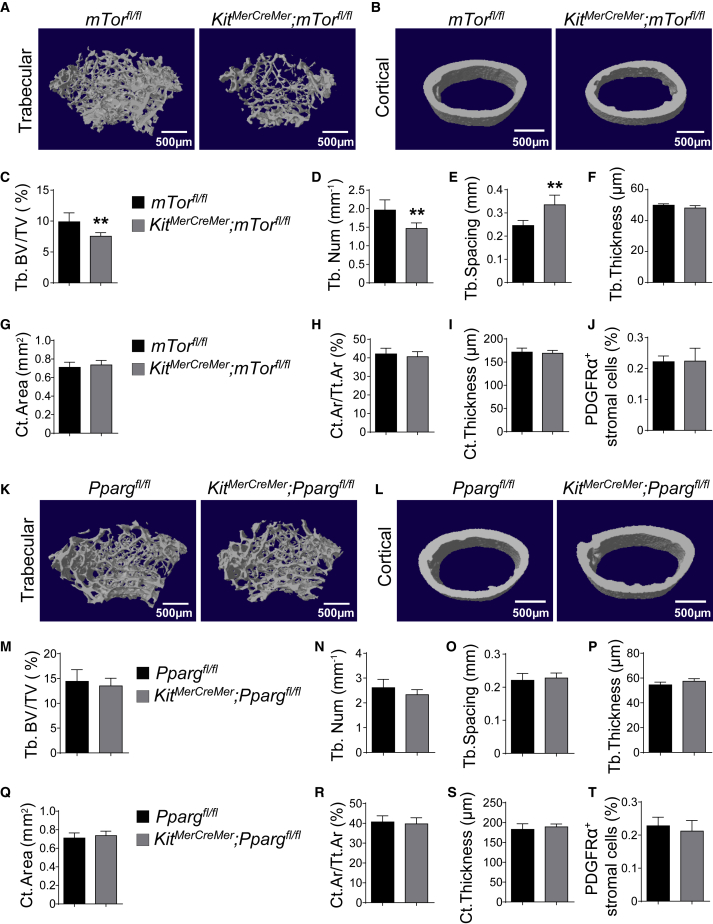
To determine the physiological importance of C-KIT+ cells in bone formation, a standard strategy is to block the osteogenic activity of these cells and see how this affected osteogenesis. mTOR is a well-known regulator of skeletogenesis (Chen and Long, 2014, Dai et al., 2017). We deleted mTor from C-KIT+ cells at E12.5/14.5 and performed micro-computed tomography (microCT) analysis of femurs at 2 months of age. The results showed that KitMerCreMer; mTorfl/fl mice had significantly lower trabecular bone volume (Figures 5A and 5C) and trabecular number (Figure 5D), and significantly larger trabecular spacing (Figure 5E), than their littermate controls. In contrast, the cortical bone parameters of KitMerCreMer; mTorfl/fl mice were generally normal (Figures 5B and 5G–5I). The frequency of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells that expressed TOMATO was normal in these mice (Figure 5J). Thus, disruption of mTOR signaling from fetal C-KIT+ cells and their descendants impairs bone formation in young adults.
Figure 5.
Deletion of mTor, but Not Pparg, from C-KIT+ Cells Impaired Bone Development
All data (mean ± SD) reflected analyses of femurs from 2-month-old mice that had been tamoxifen treated at E12.5/14.5. Different sexes were examined in different experiments such that most data reflected both male and female mice. Two-tailed Student's t tests were used to assess the statistical significance of differences between sex-matched littermates (∗∗p < 0.01).
(A and B) Representative microCT images showed trabecular (A) and cortical bones (B) from mTorfl/fl and KitMerCreMer; mTorfl/fl mice (n = 5 mice per genotype from three independent experiments).
(C–I) MicroCT analyses of the trabecular bone volume/total bone volume (C), trabecular number (D), spacing (E), thickness (F), cortical area (G), cortical area/total area (H), and thickness (I) of femurs from mTorfl/fl and KitMerCreMer; mTorfl/fl mice (n = 5 mice per genotype from three independent experiments).
(J) Frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells in Ppargfl/fl and KitMerCreMer; Ppargfl/fl mice (n = 5 mice from three independent experiments).
(K and L) Representative microCT images showed trabecular bone (K) and cortical bone (L) from Ppargfl/fl and KitMerCreMer; Ppargfl/fl mice (n = 5 mice per genotype from three independent experiments).
(M–S) MicroCT analyses of the trabecular bone volume/total bone volume (M), trabecular number (N), spacing (O), thickness (P), cortical area (Q), cortical area/total area (R), and thickness (S) of femurs from Ppargfl/fl and KitMerCreMer; Ppargfl/fl mice (n = 5 mice per genotype from three independent experiments).
(T) Frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells in Ppargfl/fl and KitMerCreMer; Ppargfl/fl mice (n = 5 mice per genotype from three independent experiments).
PPARG is another well-known regulator of bone homeostasis (Wan, 2010). We generated KitMerCreMer; Ppargfl/fl mice and administered tamoxifen to them at E12.5/14.5. At 2 months of age, we found that KitMerCreMer; Ppargfl/fl, and Ppargfl/fl mice had similar trabecular volume (Figures 5K and 5M), trabecular number (Figure 5N), trabecular spacing (Figure 5O), trabecular thickness (Figure 5P), cortical area (Figures 5L, 5Q, and 5R), and cortical thickness (Figure 5S). The frequency of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells that expressed TOMATO was also similar between control and Pparg-deficient mice (Figure 5T). In contrast, we did detect a small but significant reduction of the density of perilipin+ cells in the bone marrow of 2-month-old KitMerCreMer; Ppargfl/fl mice (Figures S3A–S3C). Bone marrow adipocytes were completely absent from Prx1-cre; Ppargfl/fl mice, in which Pparg was efficiently deleted from early skeletal progenitor (Figures S3D–S3F), excluding the possibility that the Ppargfl allele does not work. These data suggested that conditional deletion of Pparg from fetal C-KIT+ cells does not affect bone development.
KITL/C-KIT Signaling in Fetal, but Not Adult Bone Marrow, Regulated Osteogenesis
C-KIT regulates stem cell functions through its interaction with KITL. Our data above suggested that C-KIT is expressed by skeletal progenitors in the fetus, but not by SSCs in adults. This raises the possibility that KITL/C-KIT signaling regulates fetal, but not adult SSCs. To test this hypothesis, we took genetic approaches to delete Kitl from adult and fetal bone marrow, respectively.
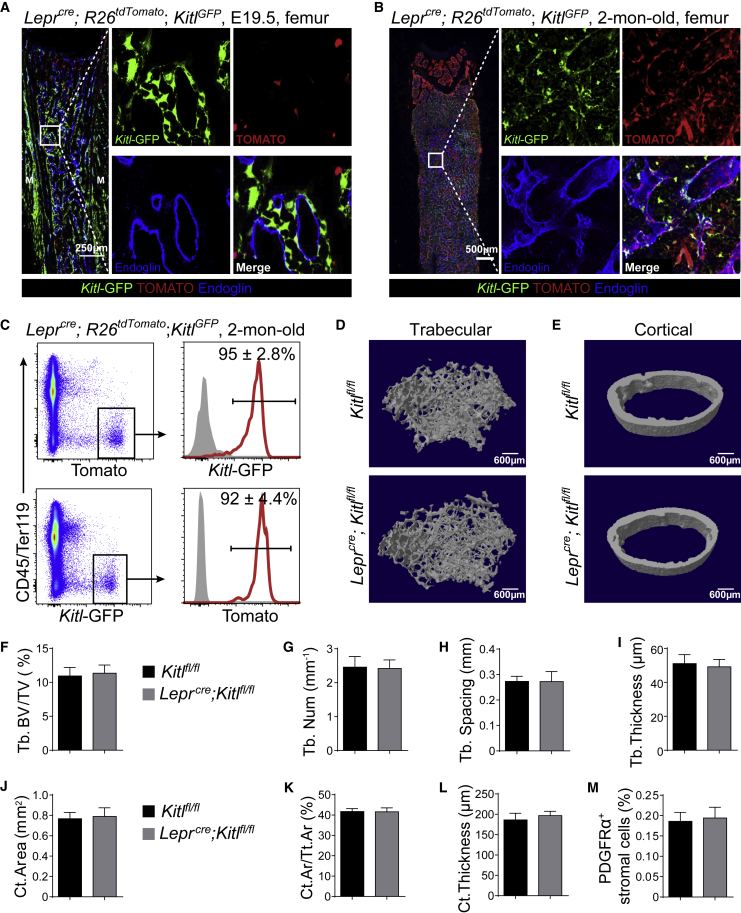
We used Lepr-Cre to delete Kitl from young adult bone marrow. In the bone marrow from Leprcre; R26tdTomato; KitlGFP embryos at E19.5, TOMATO+ cells were rarely observed (Figure 6A). Although TOMATO+ cells usually co-expressed Kitl-GFP, only a minor fraction of all Kitl-GFP+ cells were TOMATO+ (Figure 6A). In contrast, in the bone marrow from Leprcre; R26tdTomato; KitlGFP mice at 2 month of age, TOMATO+ stromal cells distributed throughout the bone marrow (Figure 6B). Most Kitl-GFP+ cells co-expressed TOMATO and vice versa (Figures 6B, 6C, S4A, and S4B). Moreover, RNA from bone marrow stromal cells had much less Kitl mRNA in 2-month-old Leprcre; Kitlfl/fl mice than in Kitlfl/fl mice (Figure S4C). Therefore, Lepr-Cre recombines efficiently in young adult Kitl-expressing cells but poorly in fetal Kitl-expressing cells in the bone marrow.
Figure 6.
Deletion of Kitl After Birth Did Not Affect Osteogenesis
(A and B) Representative confocal images of femur sections from Leprcre; R26tdTomato; KitlGFP mice at E19.5 (A) or 2 months of age (B). Bone marrow vessels were revealed by anti-ENDOGLIN staining (n = 3 mice/age from three independent experiments).
(C) Representative flow cytometry plots of enzymatically dissociated bone marrow cells from 2-month-old Leprcre; R26tdTomato; KitlGFP mice showed that most TOMATO+ cells were GFP+ and vice versa (n = 3 mice from three independent experiments).
(D and E) Representative microCT images showed trabecular bone (D) and cortical bone (E) from Kitlfl/fl and Leprcre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments).
(F–L) MicroCT analyses of the trabecular bone volume/total bone volume (F), trabecular number (G), spacing (H), thickness (I), cortical area (J), cortical area/total area (K), and thickness (L) of femurs from Kitlfl/fl and Leprcre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments).
(M) Frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells in Kitlfl/fl and Leprcre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments). Two-tailed Student's t tests were used to assess the statistical significance of differences between sex-matched littermates in (F–M).
MicroCT analysis of femurs from 2-month-old paired Kitlfl/fl and Leprcre; Kitlfl/fl mice showed that Leprcre; Kitlfl/fl mice did not differ from Kitlfl/fl mice in any bone parameters, including trabecular volume (Figures 6D and 6F), trabecular number (Figure 6G), trabecular spacing (Figure 6H), trabecular thickness (Figure 6I), cortical area (Figures 6E, 6J and 6K), and cortical thickness (Figure 6L). The frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells that expressed TOMATO were similar between Kitlfl/fl and Leprcre; Kitlfl/fl mice (Figure 6M). These data suggested that deletion of Kitl from young adult bone marrow does not affect bone formation.
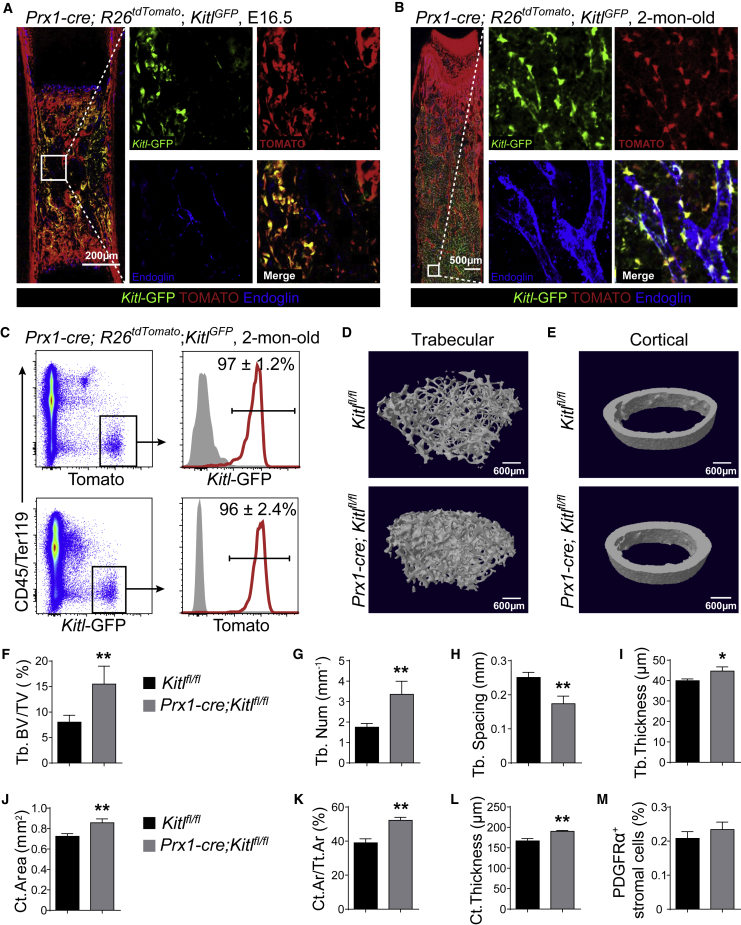
We used Prx1-Cre to delete Kitl from fetal bone marrow. In Prx1-cre; R26tdTomato; KitlGFP embryos at E16.5, TOMATO+ stromal cells spread throughout the bone marrow (Figure 7A). Virtually all Kitl-GFP+ cells co-expressed TOMATO (Figure 7A). Similarly, in 2-month-old Prx1-cre; R26tdTomato; KitlGFP mice, virtually all Kitl-GFP+ bone marrow cells were TOMATO+, as revealed by confocal imaging (Figure 7B) and flow cytometry (Figures 7C, S5A, and S5B). Moreover, RNA from bone marrow stromal cells had much less Kitl mRNA in 2-month-old Prx1-cre; Kitlfl/fl mice than in Kitlfl/fl mice (Figure S5C). Therefore, Prx1-Cre recombines efficiently in Kitl-expressing cells in both fetal and young adult bone marrow stromal cells.
Figure 7.
Deletion of Kitl in Fetus Increased Osteogenesis During Development
(A and B) Representative confocal images of femur sections from Prx1-cre; R26tdTomato; KitlGFP mice at E19.5 (A) or 2 months of age (B). Bone marrow vessels were revealed by anti-ENDOGLIN staining (n = 3 mice/age from three independent experiments).
(C) Representative flow cytometry plots of enzymatically dissociated bone marrow cells from 2-month-old Prx1-cre; R26tdTomato; KitlGFP mice showed that most TOMATO+ cells were GFP+ and vice versa (n = 3 mice from three independent experiments).
(D and E) Representative microCT images showed trabecular bone (D) and cortical bone (E) from Kitlfl/fl and Prx1-cre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments).
(F–L) MicroCT analyses of the trabecular bone volume/total bone volume (F), trabecular number (G), spacing (H), thickness (I), cortical area (J), cortical area/total area (K), and thickness (L) of femurs from Kitlfl/fl and Prx1-cre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments).
(M) Frequencies of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells in Kitlfl/fl and Prx1-cre; Kitlfl/fl mice (n = 5 mice per genotype from three independent experiments).
Two-tailed Student's t tests were used to assess the statistical significance of differences between sex-matched littermates in (F–M) (∗p < 0.05, ∗∗p < 0.01).
MicroCT analysis showed that 2-month-old Prx1-cre; Kitlfl/fl mice had significantly increased trabecular bone volume (Figures 7D and 7F), trabecular number (Figure 7G), and trabecular thickness (Figure 7I) and significantly decreased trabecular spacing (Figure 7H) as compared with Kitlfl/fl mice. All cortical bone parameters, including cortical area (Figures 7E, 7J and 7K) and cortical thickness (Figure 7L), were also significantly increased in Prx1-cre; Kitlfl/fl mice. The frequency of CD45−TER119−CD31−PDGFRα+ bone marrow stromal cells that expressed TOMATO was normal in Prx1-cre; Kitlfl/fl mice (Figure 7M). The density of TRAP+ osteoblasts in the bone marrow was comparable between Prx1-cre; Kitlfl/fl and Kitlfl/fl mice (Figures S5D and S5E). These data suggested that deletion of Kitl from fetal bone marrow augments osteogenic differentiation by acting on skeletal progenitors during development.
Discussion
KITL is a well-known growth factor that keeps stem cells, such as HSCs, in a quiescent state. When Kitl is deleted from their niche, stem cells lose quiescence and differentiate. In the new scenario of SSCs, we observed significantly increased bone formation upon Kitl deletion from fetal bone marrow stromal cells (Figures 7D–7M). It is likely that C-KIT+ cells enter into cell cycle and are more prone to differentiate when KITL/C-KIT signaling is blocked. We speculate that the overall number of C-KIT+ cell-derived SSCs decreases and the number of C-KIT+ cell-derived osteoblasts increases in Prx1-cre; Kitlfl/fl mice. This hypothesis can be tested by genetic tracing C-KIT+ cells under the background of Prx1-cre; Kitlfl/fl. However, it will require the generation a new allele with a different recombinase, such as kit-DreER.
It had been shown that Pparg deletion from Osx+ osteogenic progenitors significantly increased bone formation (Sun et al., 2013). However, we did not observe any changes to any bone parameters after Pparg deletion from C-KIT+ cells (Figures 5K–5T), although we did observe a slight reduction of bone marrow adiposity in these mutant mice (Figures S3A–S3C). A possible explanation is that Pparg is required for osteogenesis by SSCs that are not derived from C-KIT+ cells. There may also be some compensation mechanisms between different SSC subpopulations. When osteogenesis by C-KIT+ cell-derived SSCs was elevated by Pparg deletion, the activity of other SSCs decreases proportionally.
At the current stage, we have not determined the cellular identity of C-KIT+ skeletal progenitors. Based on the short-term lineage-tracing experiment, C-KIT was expressed by stromal cells at the growth cartilage (Figure S2A). Whether they represent proliferating chondrocytes, hypertrophic chondrocytes, or emerging Osx+ stromal cells during development is not clear yet. Although new evidence has highlighted the importance of fetal and neonatal chondrocytes in bone formation (Mizuhashi et al., 2018, Ono et al., 2014, Zhou et al., 2014b), the bone marrow stromal cells are still believed to be the main source of SSCs for bone formation and regeneration in adult bone marrow (Mizoguchi et al., 2014, Zhou et al., 2014a). Future studies need to address the division-of-labor between different SSC subpopulations in bone formation and regeneration.
Our data showed that tracing C-KIT+ cells labeled osteoclasts at endosteal regions (Figure S1B). This is expected as osteoclasts are one of the myeloid lineages that are derived from C-KIT+ HSCs. Although conditional deletion of Scf from bone marrow stromal cells reduced HSC frequency (Ding et al., 2012), it did not affect the frequency of osteoclasts in the bone marrow (Figures S5D and S5E). Unlike Prx1-cre; Scffl/fl mice, which showed increased bone formation (Figures 7D–7L), mice carrying Kit loss-of-function mutation exhibited an osteopenia phenotype, likely due to elevated osteoclast differentiation (Lotinun and Krishnamra, 2016). Although we cannot fully rule out the possibility that conditional Scf deletion by Prx1-Cre affects bone resorption, the different phenotypes of conditional Scf mutants and germline Kit mutants imply that they attribute to distinct mechanisms.
Experimental Procedures
Mice
KitMerCreMer (van Berlo et al., 2014), R26tdTomato (Madisen et al., 2010), Col2.3-GFP (Kalajzic et al., 2002), mTorfl/fl (Risson et al., 2009), Ppargfl/fl (He et al., 2003), KitlGFP (Ding et al., 2012), Kitlfl/fl (Ding et al., 2012), Leprcre (Cohen et al., 2001), and Prx1-cre (Logan et al., 2002) mice were housed in the Animal Facility at Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Chinese Academy of Sciences. To induce CreER activity in neonatal mice, breeder mice were gavaged with 2 mg tamoxifen (Sigma) dissolved in corn oil daily for 3 consecutive days. To induce CreER activity in embryos, pregnant mice were gavaged with 1 mg tamoxifen, together with 0.5 mg progesterone (to avoid abortion, Sigma), dissolved in corn oil. All procedures were approved by the Institutional Animal Care and Use Committees of SIBCB.
Flow Cytometry
Whole bone marrow cells were flushed using Hank’s balanced salt solution (HBSS) (Mg2+ and Ca2+ plus) and digested with 200 U/mL DNase I and 250 μg/mL Collagenase I and IV (Worthington) at 37°C for 30 min. Cell suspension in 100 μL HBSS with 2% bovine serum was stained with antibodies, including anti-LEPR-biotin (1:100, R&D Systems, BAF497), anti-CD45 (1:200, eBioscience, 30F-11), anti-CD31 (1:200, eBioscience, 390), anti-TER119 (1:200, eBioscience, TER-119), anti-SCA-1 (1:200, eBioscience, E13–161.7), and/or anti-PDGFRα-biotin (1:100, BioLegend, APA5), and then brilliant violet 421 streptavidin (1:500, BioLegend). Samples were analyzed using a FACSCelesta flow cytometer (BD Biosciences). Data were analyzed by FlowJo software.
For BrdU incorporation assays, mice were given an intraperitoneal injection of 1 mg BrdU (Sigma) per 6 g of body mass in PBS and maintained on 1 mg/mL of BrdU in the drinking water for 14 days. The frequency of BrdU+ cells was then analyzed by flow cytometry using the APC BrdU Flow Kit (BD Biosciences).
For apoptotic cell staining, cells were stained by anti-Annexin V antibody before flow cytometry following the manufacture's introductions (Thermo Fisher).
Bone Sectioning, Immunostaining, and Confocal Imaging
Freshly dissected femurs were fixed in 4% paraformaldehyde overnight followed by 3-day decalcification in 10% EDTA. O.C.T. (SciGen)-embedded bones were sectioned using the CryoJane Tape-Transfer system (Leica Biosystems). Sections were blocked in PBS with 5% donkey serum for 1 h and then stained overnight with rabbit-anti-perilipin (Sigma, 1:800, P1873), goat-anti-ENDOGLIN (R&D, 1:500, AF1320), rabbit-anti-TRAP (Abcam, 1:200, ab185716) or rabbit-anti-AGGRECAN (Millipore, 1:400, AB1031). Donkey anti-rabbit Alexa Fluor 647 or donkey anti-goat Alexa Fluor 647 were used as secondary antibody (both from Invitrogen, 1:500). Non-immune immunoglobulins of the same isotype as the primary antibodies were used as negative controls. Slides were mounted with anti-fade solution (Invitrogen). Images were acquired with a Zeiss LSM880 or a Leica SP8 confocal microscope.
Colony-Forming Efficiency Assay and In Vitro Differentiation
Whole bone marrow or sorted cells were cultured in six-well plates at densities ensuring that colonies would form at clonal density. The cultures were incubated at 37○C in a gas-tight chamber (Billups-Rothenberg) with 5% O2 and 10% CO2 for 7–14 days. For in vitro differentiation assays, Colonies were digested with 0.25% Trypsin/EDTA, pooled together, and then separated into three aliquots permissive for adipocyte, chondrocyte, and osteoblastic differentiation, respectively. Adipogenic and osteogenic differentiation was performed in 6-well plates while chondrogenic differentiation was performed as pellets in 15-mL tubes. Adipocyte, chondrocyte, and osteoblastic differentiation was induced with StemPro differentiation kits (Invitrogen) and detected by staining with oil red O (Sigma), alcian blue (Sigma) and alizarin red S (Sigma), respectively.
Subcutaneous Ossicle Formation Assay
Colonies from ∼500 flow cytometry-purified bone marrow stromal cells were passaged to generate ∼106 cells, then transferred into 6 × 6 × 6 mm sterile collagen sponges (Gelfoam; Pfizer) and incubated at 37○C for 2 h. Cell-loaded sponges were then transplanted subcutaneously into recipient mice. The sponges were harvested 8 weeks after transplantation and examined by hematoxylin and eosin staining.
Single-Cell RNA-Seq
Single cells were sorted into 96-well plates (Bio-Rad, HSP9601) on a 4○C holder and subjected to a modified full-length single-cell RNA-seq protocol (Smart-seq2) (Chen et al., 2017). In brief, cells were lysed in single-cell RNA lysis buffer containing 0.45% NP-40, then subjected to reverse transcription with SuperScript II reverse transcriptase (Invitrogen) and whole transcription amplification with a KAPA HiFi HotStart ReadyMix (2×; KAPA Biosystems). PCR products were purified with AMPure XP beads (Agencourt) and quantified with a Qubit dsDNA HS Assay Kit (Thermo Fisher). cDNA libraries were constructed with a Nextera XT DNA Library Preparation Kit (Illumina) and were sequenced on an Illumina HiSeq X Ten instrument in 150-bp paired-end-read mode by Berry Genomics. The amplification, cDNA purification, and library construction were performed on the Agilent Bravo automatic liquid-handling platform in the core facility for molecular biology at SIBCB.
GEO accession number for RNA-seq data: GSE146002.
Preparing Total RNA for qRT-PCR
Cells were sorted directly into TRIzol. Total RNA was extracted according to the manufacturer's instructions (Invitrogen). Total RNA was subjected to reverse transcription and then qRT-PCR using SYBR green on a LightCycler 96 system (Roche). Primers used in this study were: β-actin: 5′-GCTCTTTTCCAGCCTTCCTT-3′ and 5′-CTTCTGCATCCTGTCAGCAA-3′; Kitl: 5′-GCCAGAAACTAGATCCTTTACTCCTGA-3′ and 5′-CATAAATGGTTTTGTGACACTGACTCTG-3′.
Single-Cell RNA-Seq Data Analysis
Sequencing quality of raw data was evaluated by FASTQC. Reads were aligned to the GRCm38 genome using HISAT (version 2-2.1.0) (Pertea et al., 2016), and gene counts were generated using HTseq (version 0.6.0) with default parameters. All mapped reads were processed by StringTie (version 1.3.3b) to evaluate the gene expression levels (measured by FPKM [fragments per kilobase per million]) with default parameters. After that, gene expression values for each cell were log10 (FPKM+1) transformed. Highly variable genes were used for principal-component analysis, and then the top 500 PC loading genes for each of PC1-5 (using FactoMineR in R) were chosen to perform clustering analysis. Seurat package (version 3.0) (Butler et al., 2018) was used and UMAP was applied to visualize the results. Differential expressed genes were identified by using DEseq2 with log2 (fold change) > 1.5 and at a false discovery rate < 0.01 (Love et al., 2014).
Analysis of Bone Volume by MicroCT
Femurs were fixed in 70% ethanol and scanned at an isotropic voxel size of 10 μm (80 kV, 450 μA, and 2,000 ms integration time) with a SkyScan1176 MicroCT scanner (Bruker). Three-dimensional images were reconstructed using a fixed threshold. The bone microstructure was then analyzed according to the literature (Bouxsein et al., 2010).
Bone Fracture
After mouse anesthesia using an isoflurane tank, one of the mouse legs was shaved, scrubbed with alcohol and then betadine. To stabilize impending fractures a 0.01-inch-diameter, stainless-steel wire was retrogradely inserted into the intramedullary canal of the tibia. Fracture was then produced by 3-point bending and the wound was sealed by non-absorbable sutures. Buprenorphine was then used. The mouse would not be transferred into the cage until it was fully recovered. The mouse was then housed in an independent cage and checked twice a day. The steel wire was moved out of the marrow 12 days after the fracture.
Statistical Methods
Data in all figures represent multiple independent experiments performed on different days with different mice. Sample sizes were not based on power calculations. No randomization or blinding was performed. Variation is always indicated using standard deviation. For analysis of the statistical significance of differences between the two groups, we performed two-tailed Student's t tests. For analysis of the statistical significance of differences among three or more groups, we performed repeated measures one-way ANOVAs with Greenhouse-Geisser correction (if variances between groups were not equal) and Tukey's multiple comparison tests with individual variances computed for each comparison.
Author Contributions
D.D.H. and B.O.Z. conceived and initiated the project. D.D.H. analyzed mTor, Pparg, and Kitl conditional mutant mice. X.T.T. conducted most flow cytometry experiments. G.C., X.T.T., G.P., and N.J. performed scRNA-seq analyses. W.D. performed most immunostaining and confocal imaging experiments. X.Y. performed all in vitro experiments. Y.C. assisted in confocal microscopy. B.O.Z. and X.T.T. wrote the manuscript.
Acknowledgments
We thank X. Fang from Beijing Institute of Genomics for technical support in bioinformatic and statistical analysis. This work was supported by the National Key Program on Stem Cell and Translational Research (2017YFA0106400 and 2018YFA0107201), the Strategic Priority Research Program on Stem Cell and Translational Research (2017YFA0106400), the National Natural Science Foundation of China (31771637 and 81730006), and a SKLEH-Pilot Research Grant (ZK17-04). None of the authors of this work has a financial interest related to this work.
Published: March 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.03.001.
Supplemental Information
References
- Bianco P., Kuznetsov S.A., Riminucci M., Gehron Robey P. Postnatal skeletal stem cells. Methods Enzymol. 2006;419:117–148. doi: 10.1016/S0076-6879(06)19006-0. [DOI] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K., Seo E.Y., Chen J.Y., Lo D., McArdle A., Sinha R., Tevlin R., Seita J., Vincent-Tompkins J., Wearda T. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Long F. mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development. 2014;141:2848–2854. doi: 10.1242/dev.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Suo S., Tam P.P., Han J.J., Peng G., Jing N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- Cohen P., Zhao C., Cai X., Montez J.M., Rohani S.C., Feinstein P., Mombaerts P., Friedman J.M. Selective deletion of leptin receptor in neurons leads to obesity. J. Clin. Invest. 2001;108:1113–1121. doi: 10.1172/JCI13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Xu Z., Ma X., Niu N., Zhou S., Xie F., Jiang L., Wang J., Zou W. mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ. 2017;24:1886–1899. doi: 10.1038/cdd.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath S., Yallowitz A.R., McCormick J., Lalani S., Zhang T., Xu R., Li N., Liu Y., Yang Y.S., Eiseman M. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562:133–139. doi: 10.1038/s41586-018-0554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. Mesenchymal stem cells express C-KIT. Circ. Res. 2010;107:e17. doi: 10.1161/CIRCRESAHA.110.230961. author reply e18. [DOI] [PubMed] [Google Scholar]
- Deng W., St Hilaire R.C., Chattergoon N.N., Jeter J.R., Jr., Kadowitz P.J. Inhibition of vascular smooth muscle cell proliferation in vitro by genetically engineered marrow stromal cells secreting calcitonin gene-related peptide. Life Sci. 2006;78:1830–1838. doi: 10.1016/j.lfs.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedenstein A.J., Chailakhjan R.K., Lalykina K.S. The development of fibroblast colonies in monolayer cultures of Guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzistergos K.E., Quevedo H., Oskouei B.N., Hu Q., Feigenbaum G.S., Margitich I.S., Mazhari R., Boyle A.J., Zambrano J.P., Rodriguez J.E. Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circ. Res. 2010;107:913–922. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic Z., Liu P., Kalajzic I., Du Z., Braut A., Mina M., Canalis E., Rowe D.W. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–660. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Lotinun S., Krishnamra N. Disruption of c-kit signaling in kit(W-sh/W-sh) growing mice increases bone turnover. Sci. Rep. 2016;6:31515. doi: 10.1038/srep31515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C., Kobayashi T., Selig M.K., Torrekens S., Roth S.I., Mackem S., Carmeliet G., Kronenberg H.M. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Pinho S., Ahmed J., Kunisaki Y., Hanoun M., Mendelson A., Ono N., Kronenberg H.M., Frenette P.S. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuhashi K., Ono W., Matsushita Y., Sakagami N., Takahashi A., Saunders T.L., Nagasawa T., Kronenberg H.M., Ono N. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563:254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Mabuchi Y., Kubota Y., Nagai Y., Niibe K., Hiratsu E., Suzuki S., Miyauchi-Hara C., Nagoshi N., Sunabori T. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Ono W., Nagasawa T., Kronenberg H.M. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Spencer J.A., Koh B.I., Kobayashi T., Fujisaki J., Clemens T.L., Lin C.P., Kronenberg H.M., Scadden D.T. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Shi Y., He G., Lee W.C., McKenzie J.A., Silva M.J., Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017;8:2043. doi: 10.1038/s41467-017-02171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Kim J.K., Mortensen R., Mutyaba L.P., Hankenson K.D., Krebsbach P.H. Osteoblast-targeted suppression of PPARgamma increases osteogenesis through activation of mTOR signaling. Stem Cells. 2013;31:2183–2192. doi: 10.1002/stem.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo J.H., Kanisicak O., Maillet M., Vagnozzi R.J., Karch J., Lin S.C., Middleton R.C., Marban E., Molkentin J.D. C-KIT+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y. PPARgamma in bone homeostasis. Trends Endocrinol. Metab. 2010;21:722–728. doi: 10.1016/j.tem.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Worthley D.L., Churchill M., Compton J.T., Tailor Y., Rao M., Si Y., Levin D., Schwartz M.G., Uygur A., Hayakawa Y. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yu H., Yue R., Zhao Z., Rios J.J., Naveiras O., Morrison S.J. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 2017;19:891–903. doi: 10.1038/ncb3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:1042–1055. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., von der Mark K., Henry S., Norton W., Adams H., de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10:e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.