Abstract
Precisely identifying and killing tumor cells are diligent pursuits in oncotherapy. Synthesized gene circuits have emerged as an intelligent weapon to solve these problems. Gene circuits based on post-transcriptional regulation enable a faster response than systems based on transcriptional regulation, which requires transcription and translation, showing superior safety. In this study, synthetic-promoter-free gene circuits possessing two control layers were constructed to improve the specific recognition of tumor cells. Using split-TEV, we designed and verified the basic control layer of protein-protein interaction (PPI) sensing. Another orthogonal control layer was built to sense specific proteins. Two layers were integrated to generate gene circuits sensing both PPI and specific proteins, forming 10 logic gates. To demonstrate the utility of this system, the circuit was engineered to sense alpha-fetoprotein (AFP) expression and the PPI between YAP and 14-3-3σ, the matching profile of hepatocellular carcinoma (HCC). Gene-circuit-loaded cells distinguished HCC from other cells and released therapeutic antibodies, exhibiting in vitro and in vivo therapeutic effects.
Keywords: gene circuit, logic gate, post-transcriptional-regulation, HCC
Graphical Abstract

This research used split TEV to design synthetic-promoter-free gene circuits for PPI sensing and specific protein sensing. Ten logic gates were formed to integrate the two sensing units. The circuit was utilized to distinguish HCC and release therapeutic antibodies in vitro and in vivo through sensing AFP expression and YAP/14-3-3σ interaction.
Introduction
Eliminating malignant cells without harming healthy tissue has been one of the main challenges in cancer therapy. After years of exploration, scientists developed an intelligent weapon called gene circuit that could automatically detect and kill cancer cells. Those synthesized gene circuits are composed of interconnected gene switches that could not only recognize endogenous cancer biomarkers but also execute subsequent suicide programs or express reporter genes as previously designed. As one of the most promising advances achieved recently, these kinds of gene circuits are also named synthetic cancer-cell-specific kill switches or cancer computers.1 Several cancer computers have been designed to treat or diagnose breast cancer, ovarian cancer, cervical cancer, squamous cancer of the head and neck, colorectal carcinoma, hepatocarcinoma, bladder cancer, relapsed acute lymphoblastic leukemia, lung carcinoma, prostate cancer, and liver metastases.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14
Most of the established gene circuits utilized synthetic promoters and are based on transcriptional regulation, which is inherently slower than systems based on post-transcriptional regulation like riboswitch and protease.15, 16, 17 This time-consuming feature may impair the safety of the designed cellular logic system because of delayed information processing. For example, in tumor-killing gene circuits, if the promoter of the toxic gene is designed to be inhibited by a synthetic-promoter-controlled protein, the expression of this toxic gene will continue until the inhibiting protein was synthesized. Those outputs released in the time window are surely toxic to the cell, causing safety issues. Gene circuits regulated in the post-transcription level allow a faster response to signals, but without synthetic promoters that possess natural advantages in gathering cell information and integrating multiple signals, combining and integrating synthetic-promoter-free modules to perform similar functions has been an urgent task.
In naturally evolved pathways, cellular signaling is predominantly achieved by the alteration of protein abundance in certain subcellular regions and protein-protein interactions (PPIs), both of which can be used as faster alternative solutions to gather cell information. Specific cytosolic PPIs can be monitored by split-TEV, a technology to split tobacco etch virus protease (TEVp) into two complementary fragments that are fused to the paired domains of different chosen proteins.18 If those proteins bind to each other, the two fragments will be spatially close enough to form a complete TEVp protein and exhibit proteolytic activity to cleave the TEVp cleavage site (tevS). Thus, this technology can transform PPI into the tevS-cleavage activity of TEVp. The protein abundance in the cytoplasm can be sensed by the specific riboswitch, which is composed of an aptamer domain that binds to the protein and an expression platform that regulates the gene expression of its own mRNA. Many aptamers with high affinity for cancer-related proteins have been screened and verified, such as beta-catenin, AFP, bFGF, NF-kappa B, c-Met, p50, p53, and hTERT, making riboswitch a promising tool in sensing cancer-related features.19, 20, 21, 22, 23, 24, 25, 26, 27
Most malignant cells possess more than one distinguishable feature, the single member of which can also be found in other tissues, so targeting tumor cells using a single feature could result in misidentification. This might hamper the safety of cancer computers in therapeutic usage. To increase the safety and reliability of the circuits, one promising option is introducing more sophisticated control layers into the circuits to regulate suicide genes more precisely by logic calculation on the basis of more than one selected cancer feature. For example, in most hepatocellular carcinoma (HCC) cells, AFP is significantly increased and the Hippo pathway is distinctly altered.28, 29, 30 Designing a gene circuit recognizing both the AFP and Hippo pathway changes will greatly improve the specificity and safety for further applications.
In the design of synthetic-promoter-free circuits to improve targeting safety and specificity in mammalian cells, three main characters were set to be achieved: (1) sensing both PPI and protein expression to form the matching profile; (2) logic processing (AND, OR, NAND, NOR, XOR, XNOR, I1 IMPLY I2, I1 NIMPLY I2, I2 IMPLY I1, and I2 NIMPLY I1); and (3) secreting output proteins such as NanoLuc or anti-vascular endothelial growth factor (anti-VEGF) single-chain variable fragment (scFV) (Figure 1A). Kinetic results showed that this system responded faster than the transcription-based control. Additionally, after the engineering, gene-circuit-loaded cells distinguished HCC from other cells and released therapeutic antibodies, exhibiting in vitro and in vivo therapeutic effects.
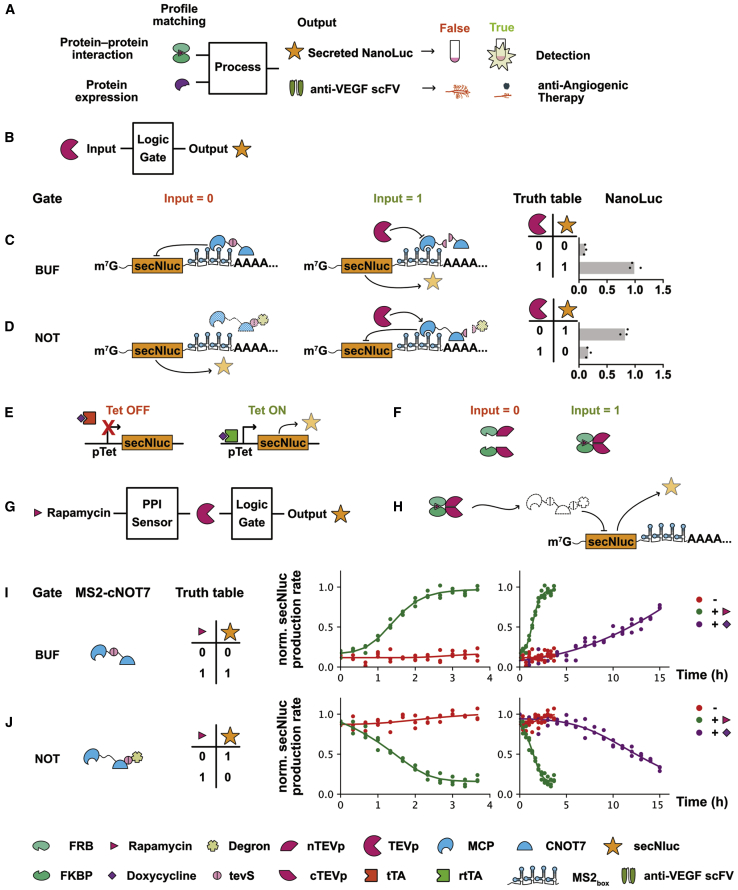
Figure 1.
Design of PPI-Sensing Components Using Split-TEV and an FRB/FKBP Heterodimer
(A) Circuit blueprints of the dual-control layers. (B) Schematic of the logic gate sensing TEVp as the input signal and output NanoLuc. (C) The schematic and verification of the BUF gate. (D) The schematic and verification of the NOT gate. (E) Schematic of the Tet-on and Tet-off system, which were used as a transcription-based control. (F) Schematic of rapamycin-induced FRB-FKBP interaction, which could restore the protease activity of TEVp. (G) Schematic of gene circuit, which integrated the rapamycin-induced PPI sensor, TEVp, logic gate, and output. (H) A more detailed schematic of the gene circuit. (I) The verification of the BUF gate. Normalized secNluc production rate was continuously monitored. Compared with the transcription-based control, the designed circuit showed faster response. (J) The verification of the NOT gate. Normalized secNluc production rate was continuously monitored. Compared with the transcription-based control, the designed circuit showed faster response.
Results
The Basic Control Layer of PPI Sensing Was Designed Using the Split-TEV
We first constructed the basic control layer to sense PPI. As explained earlier, PPIs can be sensed by the split-TEV and transformed into the cleavage activity of TEVp. The logic gate should sense TEVp activity as the input signal and, after logic calculation, control the expression of a secreting protein as the output signal (Figure 1B). Gao et al.16 reported that the target protein can be regulated by TEVp by adding tevS and degron to it. But this strategy failed to produce sufficient regulation of secreting protein in our previous study, possibly because the secreting protein shifted from the cytosol to lumen soon after translation.31 We then tried the protease-responsive MCP-cNOT7 repressor devices reported by Wroblewska et al.7 MCP-cNOT7 protein was constructed by combining two functional domains: MS2 bacteriophage coat protein (MCP) and CCR4-NOT transcription complex subunit 7 (cNOT7).32,33 The MCP endows MCP-cNOT7 protein with the ability to target the 3′ UTR of mRNAs containing the MS2 box, and after this targeting, the target mRNA can then be degraded by cNOT7, which possesses deadenylation activity. A plasmid transcribing secNluc_MS2 mRNA was constructed by inserting MS2 box to the 3′ end of secNluc coding sequence. This mRNA can be targeted and degraded by MCP-cNOT7. Without the degradation of MCP-cNOT7, secNluc_MS2 mRNA will be translated into secNluc and detected as an output. Using TEVp as the input and secNluc as the output, the regulation of TEVp to MCP-cNOT7 containing tevS can be validated by detecting the activity of secNluc.
Inspired by the circuit reported by Cella et al., MCP-tevS-cNOT7 was constructed by inserting tevS between MCP and cNOT7.34 A BUF logic gate controlling the expression of a secreting protein as the output was formed: in cells containing MCP-tevS-cNOT7 but without TEVp (input = 0), secNluc_MS2 mRNA was degraded by MCP-tevS-cNOT7 (output = 0); in cells containing both MCP-tevS-cNOT7 and TEVp (input = 1), tevS was cleaved by TEVp, cNOT7 was departed form MCP, mRNA containing MS2 box was spared from degradation, and secNluc_MS2 mRNA was translated to secreted NanoLuc (output = 1) (Figure 1C).
The NOT gate reported by Cella et al. was formed by a protease-protease system in which two orthogonal proteases were needed.34 We refined the design by introducing the degron strategy. MCP-cNOT7-tevS-degron was constructed by fusing a dihydrofolate reductase (DHFR) degron to MCP-cNOT7 via tevS. So, only one protease was needed to form a NOT gate: in cells containing MCP-cNOT7-tevS-degron but without TEVp (input = 0), degron mediated the degradation of MCP-cNOT7-tevS-degron through the proteasome pathway, so secNluc_MS2 mRNA was spared and translated to secreted NanoLuc (output = 1); in cells containing both MCP-cNOT7-tevS-degron and TEVp (input = 1), degron was removed by TEVp-mediated cleavage of tevS, and then MCP-cNOT7 was stabilized, which then degraded secNluc_MS2 mRNA, resulting in no secreted NanoLuc (output = 0) (Figure 1D).
Based on split-TEV technology, fusion proteins named nTEVp-FRB and FKBP-cTEVp were used to construct a PPI sensor, which could transfer FRB-FKBP PPI to the protease activity of TEVp.18 Rapamycin was used to control this PPI (Figure 1F). Next, a gene circuit containing a split-TEV-based PPI sensing unit and a TEVp-centered logic processing unit was constructed to validate the composability of two units (Figures 1G and 1H). A 293T cell line was established in which nTEVp-FRB, FKBP-cTEVp, MCP-tevS-cNOT7, and secNluc_MS2 mRNA coexisted. A BUF gate was formed: when PPI did not happen between FKBP and FRB (input = 0), secNluc_MS2 mRNA was degraded by MCP-tevS-cNOT7 (output = 0); when PPI happened between FKBP and FRB (input = 1), tevS was cleaved by the restored proteolytic activity of TEVp, cNOT7 was departed from MCP, mRNA containing MS2 box was spared from degradation, and secNluc_MS2 mRNA was translated to secreted NanoLuc (output = 1) (Figure 1I). In the same way, another 293T cell line was established in which nTEVp-FRB, FKBP-cTEVp, MCP-cNOT7-tevS-degron, and secNluc_MS2 mRNA coexisted. A NOT gate was formed: when PPI did not occur between FKBP and FRB (input = 0); the degradation of the MCP-cNOT7-tevS-degron was mediated by degron through the proteasome pathway, while secNluc_MS2 mRNA was translated to secreted NanoLuc (output = 1); when PPI occurred between FKBP and FRB (input = 1), tevS was cleaved by the restored proteolytic activity of TEVp, degron was removed, and secNluc mRNA was degraded by the remaining MCP-cNOT7 (output = 0) (Figure 1J). A doxycycline (Dox) induced Tetracycline-on/Tetracycline-off (Tet-on/Tet-off) system was used as a transcription-based control (Figure 1E). By comparing the kinetics of the designed PPI sensing circuit and the Tet-on or Tet-off system, we showed that the designed PPI system reacted faster than control (Figures 1I and 1J).
An Extra Control Layer Was Designed to Sense Specific Proteins
Introducing an extra control layer endows the gene circuit with superior safety by adding another defined signature of input signal. Gene circuits with multiple inputs could regulate suicide genes more precisely by introducing more sophisticated control layers on the premise that there is no crosstalk between the multiple layers. The orthogonality of biological parts transferring and processing with different inputs is quite critical to guarantee the insulation. To serve in the same system, the logic gate for another control layer was designed to sense a different protease, tobacco vein mottling virus protease (TVMVp), as the input signal and control the expression of NanoLuc as the output signal (Figure 2A). Similar to MCP-tevS-cNOT7 and MCP-cNOT7-tevS-degron, MCP-tvmvS-cNOT7 and degron-tvmvS-MCP-cNOT7 were designed. Figures 2B and 2C show the BUF gate and NOT gate for this control layer. To avoid the crosstalk between different signal layers, the orthogonality between the logic gate of two control layers was checked, showing fine orthogonality (Figure 2D).
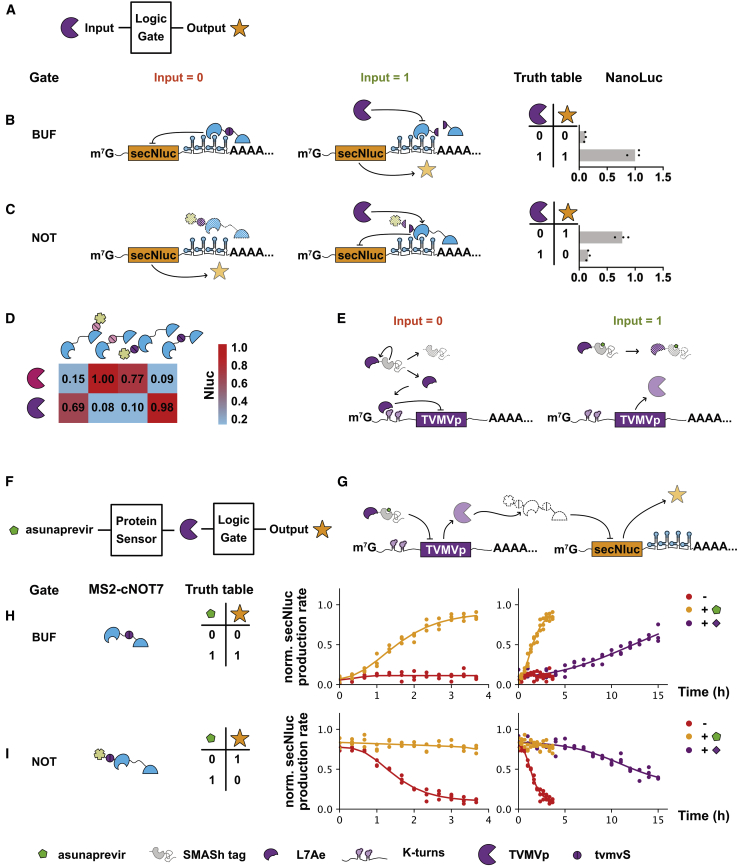
Figure 2.
Design of Components Able to Sense Specific Proteins Using L7Ae Riboswitches and TVMVp
(A) Schematic of the logic gate sensing TVMVp as the input signal and output NanoLuc. (B) The schematic and verification of the BUF gate. (C) The schematic and verification of the NOT gate. (D) The orthogonality between the logic gate of two control layers was verified. (E) Schematic of asunaprevir-induced regulation of TVMVp through the interaction between L7Ae-SMASh and K-turn. (F) Schematic of gene circuit integrated asunaprevir-controlled protein sensor, TVMVp, logic gate, and output. (G) A more detailed schematic of the gene circuit. (H) The verification of the BUF gate. Normalized secNluc production rate was continuously monitored every 20 min. Compared with the transcription-based control, the designed circuit showed faster response. (I) The verification of the NOT gate. Normalized secNluc production rate was continuously monitored. Compared with the transcription-based control, the designed circuit showed faster response.
The protein-sensing unit was constructed by adding a riboswitch to the 5′ UTR of the mRNA-expressing TVMVp, which could transfer the abundance of certain cytoplasmic protein to the protease activity of TVMVp. L7Ae was chosen to study this layer.35 L7Ae was fused to a small molecule-assisted shutoff (SMASh) tag to shut off further production of L7Ae in response to asunaprevir.36 Because kink-turns (K-turns) can specifically bind to L7Ae, the riboswitch containing K-turns was chosen to construct the L7Ae-sensing unit (Figure 2E). Next, a gene circuit containing the riboswitch-based protein-sensing unit and the TVMVp-centered logic processing unit was constructed to validate the composability of two units (Figures 2F and 2G). A 293T cell line was established in which L7Ae-SMASh, MCP-tvmvS-cNOT7, K-turns_TVMVp mRNA, and secNluc_MS2 mRNA coexisted. A BUF gate was formed. Without asunaprevir (input = 0), the SMASh tag was decreased by self-cleavage, leaving untagged L7Ae, which then combined with K-turns to suppress the translation of TVMVp. Spared from the cleavage of TVMVp, MCP-tvmvS-cNOT7 was stabilized and suppressed the translation of secNluc (output = 0). While in the presence of asunaprevir (input = 1), the self-cleavage of the SMASh tag was inhibited, and then the tagged L7Ae was degraded along with the tag. Without L7Ae, K-turns was no longer able to suppress the translation of TVMVp, resulting in cleaved MCP-tvmvS-cNOT7 and the translation of secNluc (output = 1) (Figure 2H). Similarly, another 293T cell line was established in which L7Ae-SMASh, degron-tvmvS-MCP-cNOT7, K-turns_TVMVp mRNA, and secNluc_MS2 mRNA coexisted. A NOT gate was formed (Figure 2I). Compared with the response of transcriptional regulation executed by the Dox-induced Tet-on or Tet-off system, the designed gene circuits showed a faster feature after the administration of asunaprevir.
The Integration of Two Control Layers by Logic Calculation
To integrate and process those two layers, a series of modules composed of MCP-cNOT7 was constructed to respond to the cleavage of TEVp and TVMVp (Figures 3A and 3B). Because all logic circuits shared the same sensing unit and actuation unit, we stably expressed the components of those units, including L7Ae-SMASh tag, FRB-nTEVp, FKBP-cTEVp, K-turns_TVMVp mRNA, and secNluc_MS2 mRNA, in 293T cells. For ease of description, we defined rapamycin as input 1 (I1) (I1 = 0 represents no rapamycin, while I1 = 1 represents the existence of rapamycin) and the asunaprevir as input 2 (I2) (I2 = 0 represents no asunaprevir while I2 = 1 represents the existence of asunaprevir). Matrix [I1, I2] was used to describe input status. The Tet-on or Tet-off system was also used as a transcription-based control. The kinetics of all circuits was studied by detecting the production rate of secNluc, showing that the gene circuits designed in this research responded faster.
Figure 3.
The Integration of Two Control Layers by Logic Calculation
(A) Schematic of the integration of two control layers by two-input logic gates. (B) A more detailed schematic of the integration. (C–G) The verification of the AND (C), OR (D), NAND (E), NOR (F), and XOR (G) logic gates. Normalized secNluc production rate was continuously monitored. Compared with the transcription-based control, the designed circuit showed faster response.
The verifications of 10 logic calculations (AND, OR, NAND, NOR, XOR, XNOR, I1 IMPLY I2, I1 NIMPLY I2, I2 IMPLY I1, and I2 NIMPLY I1) were shown in Figures 3 and 4. Taking the AND logic gate as an example (Figure 3C), MCP-tevS-cNOT7 and MCP-tvmvS-cNOT7 was used to construct this gate. Because secNluc_MS2 mRNA can be degraded by both kinds of MCP-cNOT7, only at [1, 1] input status when MCP-tevS-cNOT7 and MCP-tvmvS-cNOT7 were destroyed by TEVp and TVMVp could this mRNA containing the MS2 box be spared from degradation (output = 1). Otherwise, at [0, 0], [0, 1], or [1, 0] status, at least one kind of MCP-cNOT7 remained to degrade secNluc_MS2 mRNA (output = 0). Similarly, MCP-tevS-tvmvS-cNOT7 was used to construct the OR gate (Figure 3D); degron-tvmvS-MCP-tevS-degron was used to construct the NAND gate (Figure 3E); MCP-cNOT7-tevS-degron and degron-tvmvS-MCP-cNOT7 were used to construct the NOR gate (Figure 3F); degron-tvmvS-MCP-cNOT7-tevS-degron and MCP-tevS-tvmvS-cNOT7 were used to construct the XOR gate (Figure 3G); degron-tvmvS-MCP-tevS-cNOT7 and MCP-tvmvS-cNOT7-tevS-degron were used to construct the XONR gate (Figure 4A); MCP-tvmvS-NOT7-tevS-degron was used to construct the I1 IMPLY I2 gate (Figure 4B); MCP-tevS-cNOT7 and degron-tvmvS-MCP-cNOT7 were used to construct the I1 NIMPLY I2 gate (Figure 4C); degron-tvmvS-MCP-tevS-cNOT7 was used to construct the I2 IMPLY I1 gate (Figure 4D); MCP-cNOT7-tevS-degron and MCP-tvmvS-cNOT7 were used to construct the I2 NIMPLY I1 gate (Figure 4E).
Figure 4.
The Integration of Two Control Layers by Logic Calculation
(A–E) The verification of the XNOR (A), I1 IMPLY I2 (B), I1 NIMPLY I2 (C), I2 IMPLY I1 (D), and I2 NIMPLY I1 (E) logic gates. Normalized secNluc production rate was continuously monitored. Compared with the transcription-based control, the designed circuit showed faster response.
The Antiangiogenic Activity of Full Circuits Was Observed in HCC
Next, a gene circuit was designed to distinguish HCC cells from other cells. As a typical oncogenic transcription coactivator, yes associated protein (YAP) translocates from the cytosol to the nucleus after dephosphorylation (Hippo-off). After the phosphorylation of Ser127 (Hippo-on), YAP interacts with 14-3-3σ proteins and is located in the cytoplasm.37 This mechanism is critical for nucleocytoplasmic trafficking, by which the localization of YAP can be controlled. Thus, the PPI between the 14-3-3σ proteins and YAP was used as input 1 (Figure 5A). AFP is a tumor biomarker successfully used in the clinic for general investigation, diagnosis, evaluation therapeutic effects, and prediction of relapse in HCC.38 Thus, AFP was chosen as input 2 (Figure 5F). Because AFP expressed in HCC cells is relatively high and the interaction level between YAP and 14-3-3σ proteins is relatively low, we designed MCP-cNOT7-tevS-degron and degron-tvmvS-MCP-cNOT7 to integrate two different control layers. Based on split-TEV technology, fusion proteins named nTEVp-YAP and 14-3-3σ-cTEVp were designed to restore the proteolytic activity of TEVp when PPI occurred between YAP and 14-3-3σ, forming heterodimers. To verify the design of the split-TEV, we established a 293T cell line in which nTEVp-YAP, 14-3-3σ-cTEVp, MCP-cNOT7-tevS-degron, and secNluc_MS2 mRNA coexisted (Figure 5B). The truth table was presented in Figure 5C. The readout was decreased by overexpressing STK3 and LATS1, the upstream gene of YAP, either in combination or alone (Figure 5D). Conversely, the NanoLuc signal got increased when YAP1, LATS1, and MOB1B were depleted by the RNA interference (RNAi) (Figure 5E).
Figure 5.
Design of Complete Circuits to Identify HCC
(A) Schematic of the gene circuit sensing PPI between YAP and 14-3-3σ and output secNluc. (B) A more detailed schematic of the gene circuit. (C) The truth table of the circuit. (D) STK3 and LATS1, alone or together, were overexpressed in the 293T cell line loaded with the gene circuit. Nluc/Firefly was detected in each group. (E) YAP1, LATS1, and MOB1B were knocked down by siRNAs in the 293T cell line loaded with the gene circuit. Nluc/Firefly was detected in each group. (F) Schematic of the gene circuit sensing AFP and output secNluc. (G) A more detailed schematic of the gene circuit. (H) The truth table of the circuit. (I) AFP was overexpressed in the PLC/PRF/5 cell line loaded with the gene circuit. Nluc/Firefly was detected in each group. (J) AFP was knocked down by an siRNA in the PLC/PRF/5 cell line loaded with the gene circuit. Nluc/Firefly was detected in each group. Each experiment was independently performed three times in triplicate.
Plasmids transcribing AFP-riboswitch_TVMVp mRNA were constructed by replacing the two copies of K-turn structures in K-turns_TVMVp mRNA with two copies of aptamers specifically binding AFP.26 We then designed a gene circuit sensing AFP as the input (Figure 5F). A 293T cell line was established in which degron-tvmvS-MCP-cNOT7, AFP-riboswitch_TVMVp mRNA, and secNluc_MS2 mRNA coexisted (Figure 5G). The truth table was presented in Figure 5H. The readout was increased after the overexpression of AFP (Figure 5I). We also constructed a PLC/PRF/5 cell line in which degron-tvmvS-MCP-cNOT7, AFP-riboswitch_TVMVp mRNA, and secNluc_MS2 mRNA coexisted. The NanoLuc signal got decreased after the RNAi-mediated-depletion of AFP in PLC/PRF/5 cells (Figure 5J).
As one of the hallmarks of cancer, angiogenesis is critical for the growth, progression, invasiveness, and metastasis of tumors.39 So, the VEGF was chosen as the target, and anti-VEGF scFV was designed as the output of the therapeutic circuits.40 Plasmids transcribing anti-VEGF_MS2 mRNA were constructed by replacing secNluc with anti-VEGF-scFV.41 The treatment of human umbilical vein endothelial cells (HUVECs) with supernatant from PLC/PRF/5 cells promoted the formation of capillary-like tubes, whereas supernatant from PLC/PRF/5 cells transfected with anti-VEGF_MS2 mRNA impaired the vascular structure formation observed in control cells (Figure 6A). Two kinds of PLC/PRF/5 cells were established by overexpressing anti-VEGF and control vectors. Those cells were implanted subcutaneously in the left and right flanks of immunocompromised NOD-PrkdcscidIl2rgnull (NPG) mice to establish two groups of solid tumor xenograft models. The inhibition of tumor growth was observed in anti-VEGF group compared with the control (Figures 6B and 6C). These data demonstrated that the actuator unit can effectively inhibit angiogenesis in vitro and in vivo.
Figure 6.
The Antiangiogenic Activity of Full Circuits in HCC
(A) For the tube formation assay, HUVECs were treated with PLC/PRF/5 vector supernatant (left) and PLC/PRF/5 anti-VEGF supernatant (right). Scale bars, 200 μm, ∗∗p < 0.01, determined with a t test. (B) PLC/PRF/5 cells overexpressing anti-VEGF_MS2 or control vectors were implanted subcutaneously in the left and right flanks of immunocompromised NPG mice to establish two groups of solid tumor xenograft models (n = 3). (C) Tumor volume was monitored. Graphs show average tumor volumes ± SD of three mice. ∗∗p < 0.01, determined with a pairwise comparison test. (D) Schematic of the gene circuit sensing PPI between YAP and 14-3-3σ and AFP as two inputs and outputting anti-VEGF. (E) A more detailed schematic of the gene circuit. (F) The truth table of the gene circuit. (G) Four kinds of PLC/PRF/5 cells were established and implanted subcutaneously in the left and right flanks of immunocompromised NPG mice: AFP knockdown cells were used to establish group 1 xenograft models (n = 3); wild-type (WT) cells transfected with control vector were used to establish group 2 xenograft models (n = 3); PPP2CA knockdown cells were used to establish group 3 xenograft models (n = 3); AFP and PPP2CA knockdown cells were used to establish group 4 xenograft models (n = 3). The scale bars represent 1 cm. (H) Tumor volume was monitored. Graphs show average tumor volumes ± SD of three mice. ∗∗p < 0.01, determined with a pairwise comparison test. (I) Immunohistochemistry of CD31 shows blood vessels. Full circuits group showed significantly decreased blood vessels. (J) Tumor weights were measured at the endpoint using an electronic scale. ∗∗p < 0.01, determined with a t test.
Gene circuits sensing YAP-14-3-3σ interaction and AFP as two inputs were designed, in which anti-VEGF scFV was used as the output (Figures 6D and 6E). Next, a series of experiments were performed to verify whether such gene circuits could selectively inhibit the proliferation of HCC cells expressing AFP and Hippo-off. The truth table was showed in Figure 6F. First, four kinds of PLC/PRF/5 cells were established: (1) AFP knockdown cells, which then were used to establish group 1 xenograft models; (2) wild-type (WT) group transfected with control vector, which then were used to establish group 2 xenograft models; (3) PPP2CA knockdown group, which then were used to establish group 3 xenograft models; and (4) AFP and PPP2CA knockdown group, which then were used to establish group 4 xenograft models. These cells were transfected with control circuits (YAP-nTEVp, 14-3-3σ-cTEVp, MCP-cNOT7-tevS-degron, degron-tvmvS-MCP-cNOT7, AFP-riboswitch_TVMVp mRNA, and secNluc_MS2 mRNA) and full circuits (YAP-nTEVp, 14-3-3σ-cTEVp, MCP-cNOT7-tevS-degron, degron-tvmvS-MCP-cNOT7, AFP-riboswitch_TVMVp mRNA, and anti-VEGF_MS2 mRNA) to generate four pairs of PLC/PRF/5 cells that were then implanted subcutaneously in the left and right flanks of immunocompromised NPG mice to establish solid tumor xenograft models. In the four groups transfected with gene circuits, group 2 (WT PLC/PRF/5 cells with control vector) showed the inhibition of tumor growth (Figures 6G, 6H, and 6J). Immunostaining of CD31 shows significant reduction of the formation in new blood vessels (Figure 6I). Only the full circuits side of group 2 showed tumor inhibition effect, while the other three groups showed no significant difference. This result validated the effectiveness and safety of the designed gene circuits.
Discussion
The Hippo signaling pathway plays a critical role in the occurrence and development of HCC. The interaction between YAP and the 14-3-3σ protein is the central part of the Hippo signaling pathway.28, 29, 30 Therefore, the Hippo pathway may become an applicable target for HCC identification. Wehr et al.18 performed a split-TEV-based genome-wide RNAi screen for modulators of Hippo signaling based on the interaction between Yki and the 14.3.3 protein (the homologous protein of YAP and 14-3-3σ protein in Drosophila). In their study, YAP-nTEVp and 14-3-3σ-cTEVp were designed as the basic control layer of gene circuits. The split-TEV parts could adjust TEVp activity according to the status of the Hippo pathway in mammalian cells.
AFP is a cancer-associated protein and has long been used as a serum tumor marker to monitor disease progression and judge prognosis. In addition, AFP is strongly relevant to the proliferation of HCC.38 Here, AFP aptamers or K-turns were used as a part of the riboswitches in extra control layers. L7Ae-sensing units could be transformed to AFP sensing units by switching the K-turns to AFP aptamers, indicating that the input protein could be changed by replacing the aptamers of the riboswitches.
In this study, we used a bottom-up approach to design gene circuits (both synthetic promoter free and dual-control layered) that could regulate functional gene expression after sensing the PPI in the cytoplasm, integrate extra control layers detecting special gene expression, and conduct logic calculations to guarantee both safety and specificity. Based on the profile of HCC, gene circuits were engineered to validate the utility of the system. If the cell is in the status of AFP expression and Hippo-off, it will be recognized as an HCC cell and release antiangiogenic scFV; otherwise, the cell will be recognized as a non-HCC cell and spared from killing. The efficiency of the gene circuits was tested by in vivo experiments.
Multi-input gene circuits have been proposed in several studies. Liu et al.1 synthetized gene circuits with an AND gate for the identification of bladder cancer cells using the CRISPR-Cas9 technique. Prochazka et al.6 created highly modular bow-tie gene circuits with programmable dynamic behavior. Therefore, we constructed gene circuits with bilayered structures to sense two inputs to specifically distinguish HCC cells from other cells.
A synthetic RNA-based immunomodulatory gene circuit was designed by Nissim et al.14 for cancer immunotherapy. Similarly, in our research, gene circuits were loaded into tumor cells to inhibit proliferation by secreting therapeutic molecules after matching profiles. The RNA-only single-output AND gate was composed of synthetic intronic microRNAs and miR-binding sites in Morel et al.’s research.3 One of the problems caused by miRNAs is unspecific gene silencing due to off-target effects, which indicates that the status of the logic gates might be affected by other innate microRNAs or that the expression of other molecules might be silenced by the synthetic intronic microRNAs. To solve this problem, we used a logic gate formed by MCP-cNOT7 to avoid microRNAs, providing higher safety.
Two control layers were integrated by inserting protease cleavage sites between the MCP and cNOT7 domain of MCP-cNOT7, similar to the RNA-binding protein reported by Wroblewska et al.7 in 2015. We then designed biological parts fusing MCP-cNOT7 to a DHFR degron via protease cleavage sites, which endowed the protease with the function of regulating MCP-cNOT7 as needed, making it possible to possess all functions of dual input logic gates using fewer biological parts.
Circuits of hacked orthogonal modular proteases (CHOMP) was constructed by Gao et al.16 using the integrability of engineered viral proteases, in which target proteases were inhibited by input proteases by binding and cleavage. In their study, all of the functional proteins were activated/inactivated/stabilized/unstabilized after being cleaved, which confined the number of available functional genes. Hyperactivation might occur when two adjustable functional proteins exist at the same time; for example, the fluorescent intensity of the XOR [1, 0] state was twice the intensity of the [0, 1] state. We solved the output problem of CHOMP by introducing RNA-binding proteins into the protease-based regulation. Because the regulation occurred at the mRNA level, the range of available genes was expanded. Taking our research as an example, a secretory protein containing signal peptides was designed as the output. This single functional protein design also prevented hyperactivation. A design concept that was both synthetic-promoter-free and RNAi-free was introduced into the design; therefore, the gene circuits had the potential to function in an RNA-only delivery mode. For further research, a modified mRNA or self-replicating RNA delivery gene circuit might be designed to adjust the aptamer and therapeutic antibodies for optimization.
Consequently, this multilayer control circuits based on proteases combined with HCC-specific markers may provide a new insight into the diagnosis and treatment of HCC.
Materials and Methods
Cell Culture and Luciferase Assay
PLC/PRF/5 and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. PUMC-HUVEC-T1 cells (National Infrastructure of Cell Line Resource, Beijing, China) were maintained in DMEM supplemented with 10% fetal bovine serum, 40 μU/L insulin (HY-P0035, MCE, Monmouth Junction, NJ, USA), and 1% non-essential amino acids (NEAA) (Gibco, Carlsbad, CA, USA). All cells were examined by mycoplasma and Short Tandem Repeat analysis (STRa).42 Transfection of HEK293T cells for luciferase assays was carried out using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). Luminescence was measured on a Synergy HTX multi-mode reader (BioTek, Winooski, VT, USA) or a microplate luminometer (Centro LB 960; BERTHOLD, Bad Wildbad, Germany) following addition of Nano-Glo luciferase assay substrate (Promega, Madison, WI, USA).
Plasmid Construction
The plasmid construction is detailed in the Supplemental Information.
RNAi for Gene Knockdown
A pool of three small interfering RNAs (siRNAs) was used to knockdown YAP1 (sc-38637, Santa Cruz Biotechnology, Dallas, TX, USA), LATS1 (sc-35797, Santa Cruz Biotechnology, Dallas, TX, USA), MOB1B (sc-88864, Santa Cruz Biotechnology, Dallas, Texas), and AFP (sc-270319, Santa Cruz Biotechnology, Dallas, TX, USA). PLC/PRF/5 cells were transiently transfected with siRNAs mixed with Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols.
Tube Formation Assay
The formation of tube-like structures was assayed in a 24-well plate using growth factor reduced Matrigel (Corning, NY, USA). HUVECs were plated on top of Matrigel and treated with the cultured supernatant of PLC/PRF/5 cells. The images were captured after 24 h, and total tube areas were quantified as the mean pixel density obtained from the analysis of five random microscopic fields using the ImageJ software (https://imagej.nih.gov/nih-image/).
Animals and Xenografts
All animal procedures were performed according to National Institutes of Health (NIH) guidelines. This study was approved by the the Southern Medical University Institutional Animal Care and Use Committee, and animals were treated humanely and with regard to the alleviation of suffering. In brief, immunodeficient NPG mice were obtained from Vitalstar Biotechnology (Beijing, China). Cultured cells were collected by trypsinization before injection. Approximately 106 cells were suspended in a 1:1 mixture of phosphate-buffered saline (PBS) and Matrigel and implanted subcutaneously into the NPG mice. Each pair of modified cell lines was assigned to a group of three mice; control groups were injected in the right armpits, and experimental groups were injected in the other side. Tumor volume was measured every 3 days after the tumor volume reached 100 mm3. The greatest longitudinal diameter (length) and the greatest transverse diameter (width) were determined. Tumor volume was calculated based on caliper measurements using the modified ellipsoidal formula: tumor volume = 1/2 (length × width2). The animals were humanely sacrificed before the tumor volume exceeded 1,500 mm3. Tumor volume and weight were presented as mean ± SD (n = 3). The quantitative data were analyzed by prism 6 (GraphPad, San Diego, CA, USA). p values were determined by Student’s t test. With a p value < 0.05, the differences between experimental groups and control groups were considered statistically significant.
Immunostaining
Xenograft tumors were routinely fixed in 10% formalin and embedded in paraffin. 4-μm-thick tissue sections were deparaffinized in xylene and rehydrated over a graded ethanol series. Then, sections were immunostained with primary antibody against CD31 (1:50 dilution, ab28364, Abcam, Cambridge, MA, USA).
Author Contributions
X.H., J.Y., F.Z., J.W., Y.Z., Q.P., and K.L. conducted the experiments; X.H., J.Y., K.L., and S.D. designed the experiments and wrote the paper. Y.G. and L.S. supervised the project.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFA0108200 and 2018YFC1106400) and the China Postdoctoral Science Foundation funded project (2017M620376).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.03.008.
Contributor Information
Shigang Ding, Email: dingshigang222@163.com.
Kaiyu Liu, Email: ky_liu@icloud.com.
Yi Gao, Email: drgaoy@126.com.
Supplemental Information
References
- 1.Liu Y., Zeng Y., Liu L., Zhuang C., Fu X., Huang W., Cai Z. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 2014;5:5393. doi: 10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
- 2.Wu H.C., Tsao C.Y., Quan D.N., Cheng Y., Servinsky M.D., Carter K.K., Jee K.J., Terrell J.L., Zargar A., Rubloff G.W. Autonomous bacterial localization and gene expression based on nearby cell receptor density. Mol. Syst. Biol. 2013;9:636. doi: 10.1038/msb.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morel M., Shtrahman R., Rotter V., Nissim L., Bar-Ziv R.H. Cellular heterogeneity mediates inherent sensitivity-specificity tradeoff in cancer targeting by synthetic circuits. Proc. Natl. Acad. Sci. USA. 2016;113:8133–8138. doi: 10.1073/pnas.1604391113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude S.L., Frey N., Shaw P.A., Aplenc R., Barrett D.M., Bunin N.J., Chew A., Gonzalez V.E., Zheng Z., Lacey S.F. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014;371:1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloss C.C., Condomines M., Cartellieri M., Bachmann M., Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 2013;31:71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prochazka L., Angelici B., Haefliger B., Benenson Y. Highly modular bow-tie gene circuits with programmable dynamic behaviour. Nat. Commun. 2014;5:4729. doi: 10.1038/ncomms5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wroblewska L., Kitada T., Endo K., Siciliano V., Stillo B., Saito H., Weiss R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015;33:839–841. doi: 10.1038/nbt.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kojima R., Scheller L., Fussenegger M. Nonimmune cells equipped with T-cell-receptor-like signaling for cancer cell ablation. Nat. Chem. Biol. 2018;14:42–49. doi: 10.1038/nchembio.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roybal K.T., Rupp L.J., Morsut L., Walker W.J., McNally K.A., Park J.S., Lim W.A. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Della Peruta M., Badar A., Rosales C., Chokshi S., Kia A., Nathwani D., Galante E., Yan R., Arstad E., Davidoff A.M. Preferential targeting of disseminated liver tumors using a recombinant adeno-associated viral vector. Hum. Gene Ther. 2015;26:94–103. doi: 10.1089/hum.2014.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danino T., Prindle A., Kwong G.A., Skalak M., Li H., Allen K., Hasty J., Bhatia S.N. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 2015;7:289ra84. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrhardt K., Guinn M.T., Quarton T., Zhang M.Q., Bleris L. Reconfigurable hybrid interface for molecular marker diagnostics and in-situ reporting. Biosens. Bioelectron. 2015;74:744–750. doi: 10.1016/j.bios.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Din M.O., Danino T., Prindle A., Skalak M., Selimkhanov J., Allen K., Julio E., Atolia E., Tsimring L.S., Bhatia S.N., Hasty J. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissim L., Wu M.R., Pery E., Binder-Nissim A., Suzuki H.I., Stupp D., Wehrspaun C., Tabach Y., Sharp P.A., Lu T.K. Synthetic RNA-Based Immunomodulatory Gene Circuits for Cancer Immunotherapy. Cell. 2017;171:1138–1150.e15. doi: 10.1016/j.cell.2017.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fink T., Lonzarić J., Praznik A., Plaper T., Merljak E., Leben K., Jerala N., Lebar T., Strmšek Ž., Lapenta F. Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat. Chem. Biol. 2019;15:115–122. doi: 10.1038/s41589-018-0181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X.J., Chong L.S., Kim M.S., Elowitz M.B. Programmable protein circuits in living cells. Science. 2018;361:1252–1258. doi: 10.1126/science.aat5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suess B., Weigand J.E. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 18.Wehr M.C., Laage R., Bolz U., Fischer T.M., Grünewald S., Scheek S., Bach A., Nave K.A., Rossner M.J. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 19.Piater B., Doerner A., Guenther R., Kolmar H., Hock B. Aptamers Binding to c-Met Inhibiting Tumor Cell Migration. PLoS ONE. 2015;10:e0142412. doi: 10.1371/journal.pone.0142412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang D.B., Vu D., Cassiday L.A., Zimmerman J.M., Maher L.J., 3rd, Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi J., Zhang X., Rabbani Z.N., Liu Y., Su Z., Vujaskovic Z., Kontos C.D., Sullenger B.A., Clary B.M. H1 RNA polymerase III promoter-driven expression of an RNA aptamer leads to high-level inhibition of intracellular protein activity. Nucleic Acids Res. 2006;34:3577–3584. doi: 10.1093/nar/gkl482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jellinek D., Lynott C.K., Rifkin D.B., Janjić N. High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding. Proc. Natl. Acad. Sci. USA. 1993;90:11227–11231. doi: 10.1073/pnas.90.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshney A., Bala J., Santosh B., Bhaskar A., Kumar S., Yadava P.K. Identification of an RNA aptamer binding hTERT-derived peptide and inhibiting telomerase activity in MCF7 cells. Mol. Cell. Biochem. 2017;427:157–167. doi: 10.1007/s11010-016-2907-7. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Rashid F., Shah A., Awan H.M., Wu M., Liu A., Wang J., Zhu T., Luo Z., Shan G. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc. Natl. Acad. Sci. USA. 2015;112:10002–10007. doi: 10.1073/pnas.1502159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H.K., Choi Y.S., Park Y.A., Jeong S. Modulation of oncogenic transcription and alternative splicing by beta-catenin and an RNA aptamer in colon cancer cells. Cancer Res. 2006;66:10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y.J., Lee S.W. Regression of hepatocarcinoma cells using RNA aptamer specific to alpha-fetoprotein. Biochem. Biophys. Res. Commun. 2012;417:521–527. doi: 10.1016/j.bbrc.2011.11.153. [DOI] [PubMed] [Google Scholar]
- 27.Mi J., Zhang X., Rabbani Z.N., Liu Y., Reddy S.K., Su Z., Salahuddin F.K., Viles K., Giangrande P.H., Dewhirst M.W. RNA aptamer-targeted inhibition of NF-kappa B suppresses non-small cell lung cancer resistance to doxorubicin. Mol. Ther. 2008;16:66–73. doi: 10.1038/sj.mt.6300320. [DOI] [PubMed] [Google Scholar]
- 28.Han S.X., Bai E., Jin G.H., He C.C., Guo X.J., Wang L.J., Li M., Ying X., Zhu Q. Expression and clinical significance of YAP, TAZ, and AREG in hepatocellular carcinoma. J. Immunol. Res. 2014;2014:261365. doi: 10.1155/2014/261365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Notarpaolo A., Layese R., Magistri P., Gambato M., Colledan M., Magini G., Miglioresi L., Vitale A., Vennarecci G., Ambrosio C.D. Validation of the AFP model as a predictor of HCC recurrence in patients with viral hepatitis-related cirrhosis who had received a liver transplant for HCC. J. Hepatol. 2017;66:552–559. doi: 10.1016/j.jhep.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Sohn B.H., Shim J.J., Kim S.B., Jang K.Y., Kim S.M., Kim J.H., Hwang J.E., Jang H.J., Lee H.S., Kim S.C. Inactivation of Hippo Pathway Is Significantly Associated with Poor Prognosis in Hepatocellular Carcinoma. Clin. Cancer Res. 2016;22:1256–1264. doi: 10.1158/1078-0432.CCR-15-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelham H.R. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asensio M.A., Morella N.M., Jakobson C.M., Hartman E.C., Glasgow J.E., Sankaran B., Zwart P.H., Tullman-Ercek D. Correction to A Selection for Assembly Reveals That a Single Amino Acid Mutant of the Bacteriophage MS2 Coat Protein Forms a Smaller Virus-like Particle. Nano Lett. 2016;16:8034. doi: 10.1021/acs.nanolett.6b04653. [DOI] [PubMed] [Google Scholar]
- 33.Van Etten J., Schagat T.L., Hrit J., Weidmann C.A., Brumbaugh J., Coon J.J., Goldstrohm A.C. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cella F., Wroblewska L., Weiss R., Siciliano V. Engineering protein-protein devices for multilayered regulation of mRNA translation using orthogonal proteases in mammalian cells. Nat. Commun. 2018;9:4392. doi: 10.1038/s41467-018-06825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito H., Kobayashi T., Hara T., Fujita Y., Hayashi K., Furushima R., Inoue T. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 36.Chung H.K., Jacobs C.L., Huo Y., Yang J., Krumm S.A., Plemper R.K., Tsien R.Y., Lin M.Z. Tunable and reversible drug control of protein production via a self-excising degron. Nat. Chem. Biol. 2015;11:713–720. doi: 10.1038/nchembio.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher B., Skwarczynska M., Rose R., Ottmann C. Structure of a 14-3-3σ-YAP phosphopeptide complex at 1.15 A resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:978–984. doi: 10.1107/S1744309110025479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai Q., Melandro F., Pinheiro R.S., Donfrancesco A., Fadel B.A., Levi Sandri G.B., Rossi M., Berloco P.B., Frattaroli F.M. Alpha-fetoprotein and novel tumor biomarkers as predictors of hepatocellular carcinoma recurrence after surgery: a brilliant star raises again. Int. J. Hepatol. 2012;2012:893103. doi: 10.1155/2012/893103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossenta M., Busato D., Baboci L., Cintio F.D., Toffoli G., Bo M.D. New Insight into Therapies Targeting Angiogenesis in Hepatocellular Carcinoma. Cancers (Basel) 2019;11:11. doi: 10.3390/cancers11081086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilar-Cazares D., Chavez-Dominguez R., Carlos-Reyes A., Lopez-Camarillo C., Hernadez de la Cruz O.N., Lopez-Gonzalez J.S. Contribution of Angiogenesis to Inflammation and Cancer. Front. Oncol. 2019;9:1399. doi: 10.3389/fonc.2019.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang T., Wang H., Chen N.G., Frentzen A., Minev B., Szalay A.A. Expression of anti-VEGF antibody together with anti-EGFR or anti-FAP enhances tumor regression as a result of vaccinia virotherapy. Mol. Ther. Oncolytics. 2015;2:15003. doi: 10.1038/mto.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng J., Li Y., Cai L., Li T., Peng G., Fu C., Han X., Li H., Jiang Z., Zhang Z. Elimination of Mycoplasma Contamination from Infected Human Hepatocyte C3A Cells by Intraperitoneal Injection in BALB/c Mice. Front. Cell. Infect. Microbiol. 2017;7:440. doi: 10.3389/fcimb.2017.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.