Abstract
TEA Domain Transcription Factors (TEADs) are important in development and serve essential roles in tumorigenesis; however, the role of TEAD2 expression in hepatocellular carcinoma (HCC) has not been widely examined. The present study was conducted to investigate the expression status of TEAD2 in HCC and to evaluate whether the expression of TEAD2 is associated with the prognosis of patients with HCC. mRNA expression data was retrieved for Hippo pathway genes of 50 normal control and 377 HCC samples from The Cancer Genome Atlas data portal. Gene set enrichment, GeneNeighbors, ClassNeighbors and survival analyses were then performed based on the gene expression levels. The mRNA expression of TEAD2 and VGLL4 was significantly higher in HCC compared with the normal control samples, and the mRNA expression of TEAD2 was higher in advanced stages than in early stages. Specifically, survival analysis revealed that higher mRNA expression of TEAD2 was significantly associated with a less favorable overall survival rate (P=0.0067) and there was a trend towards significance between higher mRNA expression of VGLL4 and poor overall survival rate (P=0.051). According to the gene set enrichment analysis, patients with higher mRNA expression of TEAD2 and VGLL4 had strongly enhanced epithelial-mesenchymal transition and angiogenesis, which are associated with tumor progression. In conclusion, increased mRNA expression of TEAD2 is associated with a poor prognosis in patients with HCC. TEAD2 may serve as a prognostic factor for HCC and a novel therapeutic target.
Keywords: TEAD2, hepatocellular carcinoma, The Cancer Genome Atlas data, prognosis, survival analysis
Introduction
Hepatocellular carcinoma (HCC) was the sixth most common cancer type and fourth most common cause of cancer-associated mortalities worldwide in 2018 (1). Although the various therapeutic modalities for HCC have improved significantly in recent years, patients with HCC exhibit a poor survival rate, with a five-year survival rate of ~30% (2). Two leading causes of the unfavorable prognosis are delays in the diagnosis of HCC and a lack of appropriate treatment for advanced HCC (3). Although researchers have evaluated the treatment of advanced HCC with immunotherapy or molecular-targeted therapy, the survival rate of patients with HCC has not significantly improved. The occurrence of HCC is thought to be associated with disturbances in the relationships between various genes (4). Therefore, identifying the genes and proteins that regulate HCC development is important for developing novel therapeutic targets (4).
The Hippo signaling pathway antagonizes the oncogenic transcriptional co-activators yes-associated protein (YAP) and Tafazzin (TAZ) and serves an important role in limiting organ size during developmental processes (5,6). Without the activation of Hippo signaling, non-phosphorylated YAP and TAZ enter the nucleus and activate the expression of antiapoptotic and proliferative genes with TEA domain transcription factors (TEADs) (5,6). Mammals harbor four TEAD genes (TEAD1-4). VGLL is a hippo-independent coactivator that regulates gene expression via interaction with TEAD, and there are four VGLL genes (VGLL1-4). Notably, VGLL4 inhibits tumor growth by binding to TEAD and VGLL1 inhibits YAP/TAZ target genes by competitively binding to TEAD (7–10). Several previous studies revealed that TEADs are involved in human cancer (11–16). However, these studies were primarily focused on TEAD1 and TEAD4, while no clinical studies have examined the association between TEAD2 expression and prognosis in HCC. The purpose of the present study was to investigate the mRNA expression status of TEAD2 in HCC tumors, as well as the prognostic clinical significance of TEAD2 in HCC. Therefore, the role of TEAD2 as a possible prognostic marker and therapeutic target for treating HCC was evaluated.
Materials and methods
Gene expression profiling
Level 3 mRNA expression aggregated data and clinical data from 50 normal and 377 HCC samples were acquired from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) data portal (https://portal.gdc.cancer.gov/) and RNAseqV2-RSEM_genes were obtained from Firebrowse v.1.1.39. (http://firebrowse.org/) for gene expression analysis. All 377 dataset samples were included for profiling of mRNA expression.
Analysis of mRNA expression
R software (v.3.6.1; http://www.r-project.org) was used to analyze the raw data. mRNA expression levels were analyzed with Firebrowse (http://firebrowse.org) and Gene Expression Profiling Interactive Analysis software (GEPIA; http://gepia.cancer-pku.cn/). GeneNeighbors and ClassNeighbors, which are modules in GenePattern (http://broadinstitute.org/cancer/software/genepattern), were used to calculate the nearest gene neighbors and identify genes most significantly correlated with a class template for a specific Hippo pathway gene, respectively. The dataset, which is Illuminahiseq_maseqv2-RSEM_genes_normalized RNA-Seq data obtained from Firebrowse (http://firebrowse.org/?cohort=LIHC&download_dialog=true), was processed using R software (v.3.6.1; http://www.r-project.org) in GeneNeighbors and ClassNeighbors analyses. The 100 genes most strongly associated with TEAD2 and VGLL4 were selected for classification according to Gene Ontology Enrichment Analysis (GO terms) using Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov). Differentially expressed genes (DEGs) were classified by GO terms on the basis of their molecular function, biological process, or cellular component. DAVID provided not only an overview of extensive pathways (www.biocarta.com) with various gene interactions but also the number of DEGs per pathway with a gene enrichment P-value. Gene enrichment score with P<0.05 considered to indicate a strong association rather than random probability (17).
Functional enrichment analysis
Gene set enrichment analysis (GSEA) was performed to identify enriched genes predicted to be associated with pathways in the hallmark and curated gene sets derived from Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) information. A P-value <0.05 and false discovery rate <0.1 were considered to indicate a statistically significant gene.
Survival analysis
Cutoff Finder (http://molpath.charite.de/cutoff) was used to determine the cut-off values for LIHC mRNA expression. Illuminahiseq_maseqv2-RSEM_genes_normalised RNA-seq data of Hippo pathway genes were acquired from a tab-separated files, and the columns represented variables and the rows represented patients (http://molpath.charite.de/cutoff/load.jsp). The determination of optimal cut-off point in Cutoff Finder was based on overall survival rate for significance on the basis of the log-rank test for outcome of patient (http://molpath.charite.de/cutoff/assign.jsp). The Kaplan-Meier method was performed to estimate the cumulative event (death) rate from the date of operation until death used as the outcome variable. Survival curves stratified by high and low expression groups on each Hippo pathway genes were compared using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Human liver samples
In total, 79 patients with HCC were retrospectively screened who were treated by surgical resection between 1999 and 2016 at Chungnam National University Hospital (Daejeon, South Korea). The median age of patients was 59 years (range, 18–78 years). All tissue samples were obtained from specimens removed during lobectomy or segmentectomy. Clinical data was obtained by reviewing the medical records of all patients. Clinicopathological review of all cases performed was by JSJ and HSE. The histologic grade of HCC was determined according to the Edmondson and Steiner grading system and the Tumor-Node-Metastasis (TNM) staging system for HCC was determined according to the 8th edition of the American Joint Committee on Cancer TNM staging system at the time of surgery (18,19).
Of the 79 HCC cases, fresh-frozen 79 primary HCC and paired 79 non-tumor liver tissues were acquired for quantitative (q)PCR from the National Biobank of Korea, Chungnam National University Hospital, a member of the Korea Biobank Network. In total, one vial (100 mg) of tumor sample and one vial (100 mg) of non-tumor sample was obtained from the Biobank of Chungnam National University Hospital. The present research was approved by the Institutional Review Board of Chungnam National University Hospital (approval no. CNUH 2017-05-013).
Reverse transcription (RT)-qPCR
Total RNA was extracted from liver tissues with the RNeasy Mini kit (Qiagen, Inc.; cat. no. 74106) or TRIzol reagent (Thermo Fisher Scientific, Inc.) in accordance with the manufacturers' instructions. We extracted RNA from all liver tissues used in this experiment using RNeasy Mini kit. Reverse transcription was performed using the same quantity of total RNA to generate cDNA using amfiRivert cDNA synthesis master mix (GenDEPOT). The following temperature protocol were used for RT: Annealing, 25°C for 5 min and extension at 50°C for 60 min. After that, Reverse Transcriptase inactivation was performed at 70°C for 15 min. Finally, the samples were held at 4°C. qPCR was performed using SYBR Green Real-time PCR Master mix (Toyobo Life Science). The qPCR thermocycling condition were as follows: First, pre-denaturation step is performed at 95°C for 10 min. This is followed by 40 cycles of denaturation at 95°C for 45 sec, annealing at 60°C for 45 sec and extension for 60 sec at 72°C. After that, the final denaturation reaction is performed for 15 sec at 95°C followed by a final extension for 5 sec at 65°C. To quantify transcription, the mRNA expression levels of the target genes were normalized to those of β-actin. Table SI shows the primers used in this study. All samples were evaluated in duplicate, and the relative fold-changes in gene expression levels were calculated as 2−Δ∆Cq (20).
Experimental and recurrence, survival analyses
SPSS 13.0 (SPSS, Inc.) and Prism v.5.0 (GraphPad Software, Inc.) were used for data analysis. In our experimental analysis, complement DNA (cDNA) was synthesized from paired liver tissues (tumor and paired non-tumor tissue) and gene expression was analyzed by amplifying TEAD2 and β-actin on the cDNA. Τhe mRNA expression data of TEAD2 was analyzed in tumors and normalized to β-actin. Arbitrary values were then obtained by defining (Tumor TEAD2/Tumor β-actin)/(Non-tumor TEAD2/Non-tumor β-actin). Groups stratified into high and low expression according to the median values and these groups were converted to categorical variables based on this. Survival analysis was performed for recurrence and overall survival related with these two groups. Overall survival (OS) was defined as the duration between the date of diagnosis and the date of death/final follow up, from any cause. Recurrence-free survival (RFS) was defined as the interval between the date of surgery and the date of first recurrence or the date of the last follow-up. In the experiment, the gene expression value obtained from duplicated results for each gene and set the expression value of the patient using the mean value of the duplicated results. The experiment was performed once using the paired tissue of the patients. Comparison of the distributions between two groups were analyzed using the χ2 test (or Fisher's exact test when the expected frequency in any group was <5) for categorical variables and by paired Student's t-test (or by Kolmogorov-Smirnov test when the expected frequency in any group was <5) for continuous variables. One-way ANOVA followed by Student Newman-Keuls post-hoc comparisons were used to compare ≥3 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
mRNA expression of Hippo pathway genes in HCC
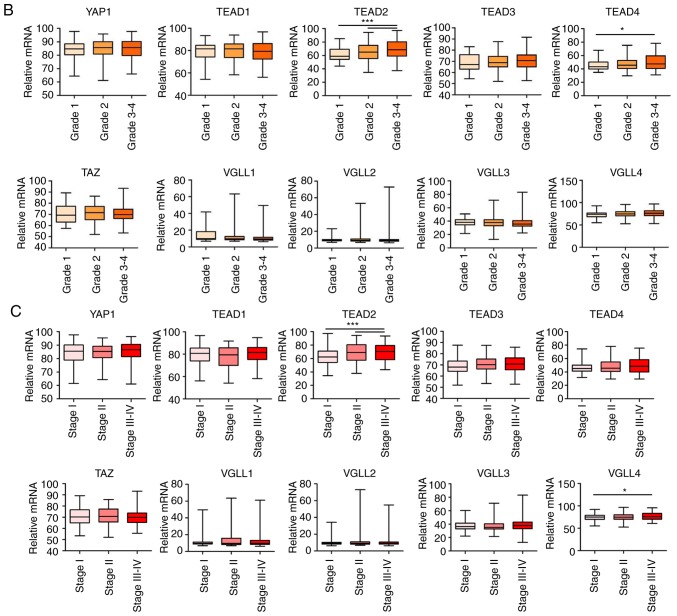
Table I exhibits the clinicopathological characteristics of patients included in the present study. The expression of Hippo pathway genes was examined in patients with HCC using the GEPIA database (Fig. 1A and Table II). Comparison of mRNA expression levels between HCC and normal control samples revealed that TEAD2 expression was significantly higher in HCC tumor tissue compared with normal control samples. The mRNA expression levels of YAP, TEAD4 and VGLL4 were higher in HCC tissue compared with normal control tissue, although the differences were not significant (Fig. 1A and Table II). Specifically, the mRNA expression of TEAD2 was significantly increased in histologic grades 3–4 samples compared with histologic grades 1–2 samples and the mRNA expression of TEAD4 was significantly increased in histologic grades 3–4 samples compared with histologic grades 1 samples (Fig. 1B). The mRNA expression of TEAD2 was significantly increased in patients with TNM stage III and IV compared with TNM stage I and II, and the mRNA expression of VGLL4 was also significantly increased in TNM stage III and IV compared with in TNM stage I (Fig. 1C). Among the ten genes evaluated, TEAD2 exhibited higher expression in HCC tumor tissue compared with normal control tissue, and this was significantly associated with histological grade or stage of HCC (Fig. 1B and C).
Table I.
Clinicopathologic information of patients with hepatocellular carcinoma derived from TCGA data portal.
| Characteristics | Total, n (%) |
|---|---|
| Patients | 377 (100.0) |
| Sex | |
| Female | 122 (32.3) |
| Male | 255 (67.7) |
| Age, years | |
| ≤60 | 180 (47.7) |
| >60 | 196 (52.0) |
| NA | 1 (0.3) |
| TNM stage (8th AJCC staging system) | |
| Stage I | 175 (46.4) |
| Stage II | 87 (23.1) |
| Stage III | 86 (22.8) |
| Stage IV | 5 (1.3) |
| NA | 24 (6.4) |
| Histological grade (Edmondson-Steiner grade) | |
| Grade 1 | 55 (14.6) |
| Grade 2 | 180 (47.7) |
| Grade 3 | 124 (33.0) |
| Grade 4 | 13 (3.4) |
| NA | 5 (1.3) |
| Vital status | |
| Alive | 245 (65.0) |
| Dead | 132 (35.0) |
| Child-Pugh classification | |
| A | 223 (59.1) |
| B | 21 (5.6) |
| C | 1 (0.3) |
| NA | 132 (35.0) |
| Histological type | |
| Hepatocholangiocarcinoma | 7 (1.9) |
| Hepatocellular carcinoma | 367 (97.3) |
| Fibrolamellar carcinoma | 3 (0.9) |
| Adjacent hepatic tissue inflammation extent type | |
| Mild | 101 (26.8) |
| Severe | 19 (5.0) |
| None | 119 (31.6) |
| NA | 138 (36.6) |
| Ishak fibrosis score | |
| 0-no fibrosis | 76 (20.1) |
| 1,2-portal fibrosis | 31 (8.2) |
| 3,4-fibrous septa | 30 (8.0) |
| 5-nodular formation and incomplete cirrhosis | 9 (2.4) |
| 6-established cirrhosis | 72 (19.1) |
| NA | 159 (42.2) |
| Thrombocytopenia (<150×109/l) | |
| Yes | 76 (20.1) |
| No | 234 (62.1) |
| NA | 67 (17.8) |
| Albumin level, g/dl | |
| >3.5 | 217 (57.6) |
| ≤3.5 | 86 (22.8) |
| NA | 74 (19.6) |
| AFP, ng/ml | |
| ≤20 | 152 (40.3) |
| >20 | 132 (35.0) |
| NA | 93 (24.7) |
| History of hepatocellular carcinoma risk | |
| Hepatitis B | 105 (27.9) |
| Hepatitis C | 51 (13.5) |
| Hepatitis B + C | 7 (1.9) |
| Alcohol consumption | 118 (31.3) |
| Non-alcoholic fatty liver disease | 18 (4.8) |
| NA | 78 (20.6) |
TNM, Tumor-Node-Metastasis; AJCC, American Joint Committee on Cancer; AFP, alpha-fetoprotein; NA, not applicable.
Figure 1.
Gene expression of Hippo pathway in HCC. (A) Gene expression of the Hippo pathway in HCC, based on the Gene Expression Profiling Interactive Analysis database was compared between normal and tumor samples and is given as Log2 (Transcripts per kilobase million + 1). *P<0.01. (B) Relative mRNA expression differences of Hippo pathway genes in HCC according to histological grade (Edmondson-Steiner grade). (C) Relative mRNA expression differences of Hippo pathway genes in HCC according to Tumor-Node-Metastasis stage. One-way ANOVA was conducted in more than two groups. *P<0.05; **P<0.01; ***P<0.001. HCC, hepatocellular carcinoma; TEAD, transcriptional enhancer associated domain; VGLL, vestigial like family; TAZ, tafazzin; YAP, yes-associate protein; TNM, Tumor-Node-Metastasis.
Table II.
Genes regulating the Hippo pathway hepatocellular carcinoma. The gene alteration contains gene amplification, deep deletion, missense mutation (unknown significance), mRNA upregulation and truncating mutation (unknown significance) of each analysed gene on HCC tissues.
| Symbol | Gene name | Chromosome location | Gene alteration (%) |
|---|---|---|---|
| YAP1 | Yes-Associated Protein 1 | 11q22.1 | 7 |
| TAZ | Tafazzin | Xq28 | 9 |
| TEAD1 | TEA Domain Transcription Factor 1 | 11p15.3 | 4 |
| TEAD2 | TEA Domain Transcription Factor 2 | 19q13.33 | 6 |
| TEAD3 | TEA Domain Transcription Factor 3 | 6p21.31 | 13 |
| TEAD4 | TEA Domain Transcription Factor 4 | 12p13.33 | 8 |
| VGLL1 | Vestigial Like Family Member 1 | Xq26.3 | 3 |
| VGLL2 | Vestigial Like Family Member 2 | 6q22.1 | 3 |
| VGLL3 | Vestigial Like Family Member 3 | 3p12.1 | 4 |
| VGLL4 | Vestigial Like Family Member 4 | 3p25.2 | 6 |
Poor prognosis of patients with HCC with high expression of TEAD2 and VGLL4
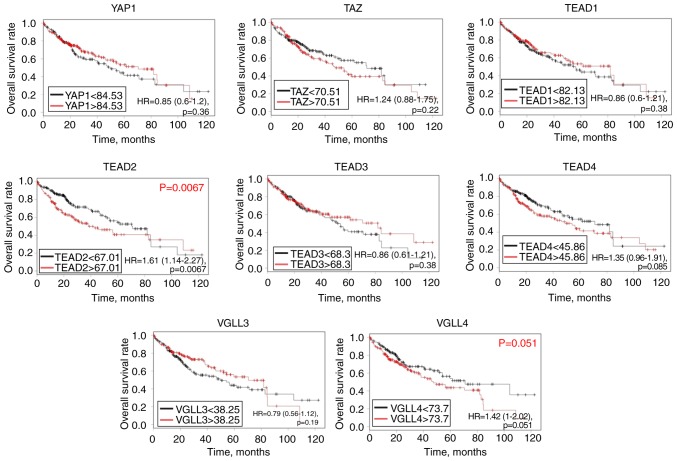
Based on the log-rank test and TCGA database, abundant mRNA expression of TEAD2 [hazard ratio (HR), 1.61; 95% confidence interval (CI), 1.14–2.27; P=0.0067] was significantly associated with a poor OS rate in patients with HCC, and there was a tendency towards significance between TEAD4 (HR, 1.35; 95% CI, 0.95–1.91; P=0.085) and VGLL4 mRNA expression (HR, 1.42; 95% CI, 1–2.02; P=0.051), and a poor prognosis (Fig. 2). However, YAP1, TAZ, TEAD1, TEAD3 and VGLL3 mRNA levels were not significantly associated with the prognosis of patients with HCC. To validate the association between the mRNA expression of TEAD2 and prognosis, RT-qPCR was performed and a log-rank test was conducted on 79 clinical HCC tissues samples. The clinicopathological characteristics of the patients with HCC are summarized in Table SII. Although the results were not significant, there was a tendency for a less favorable prognosis in patients with increased TEAD2 mRNA expression compared with those with decreased TEAD2 mRNA expression, affecting both in recurrence-free and overall survival rate (P=0.257, P=0.217, respectively; Fig. 3).
Figure 2.
Survival analysis of Hippo pathway genes in hepatocellular carcinoma. Kaplan-Meier analysis of the association between mRNA expression of Hippo pathway genes and overall survival rate of patients. TEAD, transcriptional enhancer associated domain; VGLL, vestigial like family; TAZ, tafazzin; YAP, yes-associated protein.
Figure 3.
OS rate and RFS rate analysis of patients with HCC based on TEAD2 mRNA expression. Groups were stratified according to median expression level and were converted to categorical variables accordingly. Survival curves were compared by the log-rank test. (A) Kaplan-Meier analysis of RFS rate. Patients who did not exhibit recurrence at either the date of death or the date of last follow-up were censored. (B) Kaplan-Meier analysis of OS rate. Patients who were still alive at the end of the follow-up period were censored. HCC, hepatocellular carcinoma; OS, overall survival; RFS, recurrence-free survival; TEAD, transcriptional enhancer associated domain.
GeneNeighbors analysis of TEAD2 and VGLL4
The present data revealed that TEAD2 and VGLL4 are closely associated with a poor prognosis in overall survival rate analysis using TCGA database, and that TEAD2 is associated with a poor recurrence-free survival rate according to the current clinical sample-based analyses (Figs. 2 and 3). Therefore, GeneNeighbors analysis was performed to determine the effect of TEAD2 and VGLL4 on the survival of patients with HCC. The 100 genes most strongly associated with TEAD2 and VGLL4 were identified via GeneNeighbors analysis (Fig. 4A and B) and classified using the DAVID (https://david.ncifcrf.gov/). The selected genes were divided into three categories of biological processes, molecular functions and cellular components according to gene ontology (GO) terms. In GeneNeighbors analysis of TEAD2, highly expressed genes in HCC were primarily associated with ‘positive regulation of small GTPase’, ‘vasculogenesis’, ‘positive regulation of transcript’, ‘regulation of cell migration’, ‘regulation of VEGF signaling’, ‘actin cytoskeleton organization’, ‘peptidyl-arginine methylation’, ‘B cell apoptotic process’ and ‘positive regulation of vascular permeability’ in biological processes (Fig. 4A). For cellular components, highly expressed genes in HCC were strongly associated with ‘focal adhesion’, ‘cytoplasm’, ‘cytosol’ and ‘RNA polymerase II transcription factor’. For molecular functions, highly expressed genes in HCC were mainly associated with ‘transcription factor binding’, ‘protein heterodimerization activity’, ‘protein-arginine methyltransferase activity’, ‘profilin binding’ and ‘protein binding’ (Fig. 4A).
Figure 4.
GeneNeighbors analysis and ClassNeighbors analysis of TEAD2 and VGLL4 in 377 HCC samples. Class A comprised the highest 10% TEAD2 and VGLL4-upregulated HCC samples and class B included the 10% of HCC samples with the lowest expression levels of TEAD2 and VGLL4. (A) GeneNeighbors analysis of TEAD2. (B) GeneNeighbors analysis of VGLL4. (C) ClassNeighbors analysis of TEAD2. (D) ClassNeighbors analysis of VGLL4. HCC, hepatocellular carcinoma; TEAD2, transcriptional enhancer associated domain 2; VGLL4, vestigial like family member 4.
In GeneNeighbors of VGLL4, highly expressed genes in HCC were primarily associated with ‘integrin-mediated signaling’, ‘cell adhesion mediated by integrin’, ‘extracellular matrix (ECM) organization’, ‘cell-matrix adhesion’, ‘cell-cell adhesion’, ‘mesodermal cell differentiation’, ‘cell motility’, ‘focal adhesion assembly’, ‘cell adhesion’ and ‘regulation of cell migration in biological processes’ (Fig. 4B).
For cellular components, highly expressed genes in HCC were associated with ‘focal adhesion’, ‘cell-cell adherent junction’, ‘integrin complex’, ‘extracellular exosome’ and ‘ruffle’. For molecular functions, highly expressed genes in HCC were mainly associated with ‘cadherin binding involved in cell-cell adhesion’, ‘integrin binding’, ‘laminin binding’, ‘protein binding’ and ‘protein kinase C binding’ (Fig. 4B).
ClassNeighbors analysis of TEAD2 and VGLL4
Analysis by ClassNeighbors yielded two classes of HCC samples: Class A included the highest 10% expressed TEAD2 and VGLL4-upregulated HCC samples and class B included the lowest 10% expressed TEAD2 and VGLL4-downregulated HCC samples (Fig. 4C and D). The 150 most highly expressed genes in classes A and B were identified from 20,502 probe sets. These genes were divided into three categories; biological processes, molecular functions and cellular components based on gene ontology (GO) terms (Fig. 4C and D). In ClassNeighbors of TEAD2, highly expressed genes in class A were primarily associated with ‘drug transmembrane transport’, ‘regulation of cell morphogenesis’, ‘signal transduction’, ‘cell migration’, ‘regulation of transforming growth factor beta receptor’, ‘cell growth’, ‘ECM disassembly’, ‘cellular component organization’, ‘vasculature development’ and ‘L-alpha-amino acid transmembrane transport’ in biological processes; ‘integral component of plasma membrane’, ‘neuron part’ and ‘ruffle’ in cellular components; and ‘drug transmembrane transporter activity’ and ‘amino acid transmembrane transporter activity’ in molecular functions. Highly expressed genes in class B were closely related to ‘oxidation-reduction’, ‘alpha-amino acid catabolic process’, ‘ammonium ion metabolic process’, ‘alcohol metabolic process’, ‘glutamate metabolic process’, ‘amino-acid betaine catabolic process’, ‘acyl-CoA metabolic process’, ‘alpha-amino acid biosynthetic process’, ‘lipid transport’ and ‘organic hydroxyl compound transport’ in biological process; oxidoreductase activity and coenzyme binding in molecular function; mitochondrial matrix and apical part of cell in cellular components (Fig. 4C).
In ClassNeighbors of VGLL4, highly expressed genes in class A were closely associated with ‘cell adhesion mediated by integrin’, ‘cell migration’, ‘epithelial cell migration’, ‘tyrosine kinase’, ‘cell adhesion’, ‘cellular compartment organization’, ‘cell-matrix adhesion’, ‘cellular response to endogenous stimulus’, ‘integrin-mediated signaling pathway’ and ‘angiogenesis’ in biological processes; ‘collagen-containing ECM’, ‘cell surface’ and ‘integrin complex’ in cellular components; and ‘integrin binding’ and ‘signaling receptor binding’ in molecular functions. Highly expressed genes in class B were closely associated with ‘oxidation-reduction’, ‘xenobiotic metabolic process’, ‘ammonium ion metabolic process’, ‘glutamate metabolic process’, ‘carboxylic acid biosynthetic process’, ‘protein targeting to peroxisome’, ‘chemokine-mediated signaling’, ‘monocarboxylic acid catabolic process’, ‘exogenous drug catabolic process’ and ‘alpha-amino acid catabolic process’ in biological process; ‘arachidonic acid epoxygenase activity’, ‘iron ion binding’ and ‘aromatase activity’ in molecular function; and ‘peroxisomal matrix’ and ‘intracellular non-membrane organelle’ in cellular components (Fig. 4D).
GSEA for TEAD2 and VGLL4
Using GSEA of mRNAs associated with hallmark pathways and KEGG pathways (Fig. 5), the top 10% of HCC samples with upregulated TEAD2 and VGLL4 and bottom 10% of samples with downregulated TEAD2 and VGLL4 were investigated. In the hallmarks pathways, high mRNA expression of TEAD2 and VGLL4 was strongly associated with genes related to ‘epithelial-mesenchymal transition (EMT)’ and ‘angiogenesis’. In KEGG pathway analysis, increased mRNA expression of TEAD2 and VGLL4 was closely associated with genes involved in cancer pathways (Fig. 5A and B).
Figure 5.
Gene Set Enrichment Analysis (GSEA) results for HCC with high TEAD2 and VGLL4 expression. Representative GSEA data with P-values are exhibited for (A) TEAD2 and (B) VGLL4. HCC, hepatocellular carcinoma; TEAD2, transcriptional enhancer associated domain 2; VGLL4, vestigial like family member 4; EMT, epithelial-mesenchymal transition; NOM, nominal; ES, enrichment score; NES, normalized enrichment score.
Discussion
TEAD protein serves an essential role in the early stage of the developmental process and cellular ageing (21–23). It has been demonstrated that specific TEAD proteins influence cancer as activators of transcription of pro-growth genes (24). In previous studies, TEAD was revealed to be upregulated in gastric cancer (16), breast cancer (11,25) and renal cell carcinoma (15). In prostate cancer, an increased TEAD1 level was correlated with unfavorable clinical outcomes (12), and in colorectal cancer and gastric cancer an increased TEAD4 expression level was associated with poor outcomes (13,14,26). Regarding HCC, expression of YAP-TEAD was reported to be upregulated, with most studies examining the expression of TEAD1 and TEAD4 (26–28). According to previous cellular-based assays, inhibition of LATS2, an upstream regulator of YAP/TAZ in the Hippo signaling pathway, induces nuclear localization of YAP1 and YAP1/TEAD2 interactions, which in turn promotes HCC progression (26). However, this study only explained the molecular mechanism underlying the action of YAP1/TEAD2 via LATS2 but did not reveal the clinical significance of TEAD2 in HCC. In the present study, based on analyses of TCGA database and clinical samples, as the HCC stage and histologic grade of tumor increased, TEAD2 expression adversely affected overall survival rate and recurrence-free survival rate.
In TCGA data analysis, increased mRNA expression of TEAD2 was significantly associated with a less favorable prognosis in patients with HCC in overall survival rate, and the potential of TEAD2 as a prognostic marker was predicted. Moreover, clinical data analysis revealed a similar tendency as the results of TCGA data analysis. However, additional studies are needed to confirm the association between mRNA expression of TEAD2 and prognosis.
To investigate the function and mechanism of TEAD2 in HCC, bioinformatics analysis was performed. GeneNeighbors analyses demonstrated that ‘angiogenesis-associated genes’ (VEGF, vasculogenesis and vascular permeability), ‘regulation of cell migration’ and ‘actin cytoskeleton organization’ were highly correlated with TEAD2 in HCC. ClassNeighbors analysis classified TEAD2-expressing HCC into Class A, which expresses genes associated with ‘signal transduction’, ‘cell migration’, ‘ECM disassembly’, ‘cell morphogenesis’, ‘transforming growth factor β signaling’ and ‘vasculature development’. It was also classified into Class B, which expresses genes associated with metabolic pathways. Class A genes enhance EMT and angiogenesis. GSEA was performed on TEAD2, which was significantly associated with the prognosis of patients with HCC. EMT, angiogenesis and cancer pathway-related genes were strongly associated with high mRNA expression of TEAD2. During EMT, cancer cells lose their apical basal polarity and cell-cell adhesions and develop a more invasive phenotype (29). Diepenbruck et al (30) reported that increased TEAD2 transcriptional activity promotes the induction of EMT in breast cancer cells and mammary gland epithelial cells. Moreover, angiogenesis also serves an important role in tumor growth and metastasis (31,32). For a tumor to grow above a certain size the development of new blood vessels is required, and tumors induce angiogenesis by secreting various growth factors including VEGF (33,34).
In the current study, the mRNA expression of VGLL4 was higher in HCC compared with normal control samples although the difference was not significant, and there was a tendency towards significance between VGLL4 expression and a poor prognosis (P=0.051). GeneNeighbors analyses demonstrated that ‘ECM organization’, ‘integrin-mediated signaling pathway’ and ‘cell adhesion’ associated genes were highly correlated with VGLL4 expression in HCC. ClassNeighbors analysis classified VGLL4-expressing HCC into Class A, which includes genes associated with ‘integrin’, ‘cell adhesion’ and ‘angiogenesis’, and Class B, which includes genes associated with ‘metabolic pathways’. Class A genes enhance EMT and angiogenesis. Remodeling of the ECM and changes in cell-ECM interactions are necessary for the induction and progression of EMT (35). Integrin complexes allow cells to receive signals from ECM proteins via interactions with signal transduction mediators. Some integrins play essential roles in EMT progression (35). GSEA revealed that EMT-, angiogenesis- and cancer pathway-associated genes were strongly associated with high mRNA expression of VGLL4. Previous studies have reported that VGLL4 suppresses multiple cancer types, such as gastric, lung, colon and breast carcinoma by suppressing the WNT and Hippo signaling pathways (36–40). Xie et al (41) demonstrated that VGLL4 causes G2/M phase arrest in HCC cells, and Shu et al (42) reported that the YAP/VGLL4 ratio is higher in patients with HCC, which is associate with tumor progression and a poor prognosis (41,42). By contrast to previous studies, the present study reported tendency towards significance between increased VGLL4 mRNA expression and the poor prognosis of patients with HCC. Unlike other cancer types, unknown mechanisms specific to HCC may have contributed to this discrepancy, and further studies are needed.
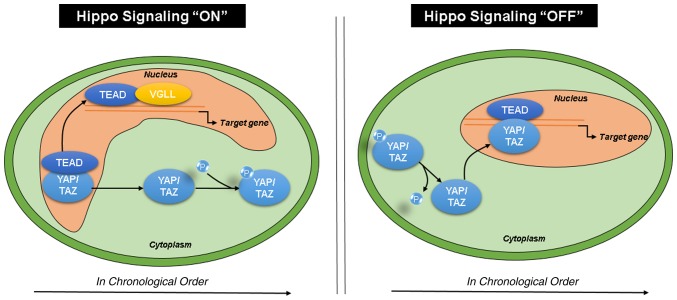
According to previous studies, when Hippo signaling is activated, YAP/TAZ are phosphorylated and sequestered in the cytoplasm, whereas TEAD and VGLL bind in the nucleus. When Hippo signaling is deactivated, YAP/TAZ are dephosphorylated and translocate to the nucleus to bind TEAD, activating the downstream transcription of target genes (Fig. 6) (43,44). In the present study, mRNA expression and poor prognosis on overall survival rate for both TEAD2 and VGLL4 exhibited similar tendencies in patients with HCC. Generally, when Hippo signaling pathway is ‘On’, TEAD and VGLL maintain their binding status; therefore, the similar tendency between TEAD2 and VGLL4 is likely related to our findings. However, further mechanistic studies are needed to validate this prediction. The present analysis did not reveal that TEAD2 and VGLL4 significantly influence each other. However, TEAD2 and VGLL4 expression were significantly correlated in TCGA HCC cohorts (P<0.001) (data not shown). Therefore, further experimental studies are required to determine which factors are the most significant.
Figure 6.
Hippo signaling pathway. Upon activation of Hippo signaling, YAP/TAZ are phosphorylated and sequestered in the cytoplasm, whereas TEAD and VGLL bind in the nucleus. When Hippo signaling is deactivated, YAP/TAZ are dephosphorylated and translocate to the nucleus to bind TEAD, activating the downstream transcription of target genes. TEAD, transcriptional enhancer associated domain; VGLL, vestigial like family; HCC, hepatocellular carcinoma; TEAD2, transcriptional enhancer associated domain 2; VGLL4, vestigial like family member 4; YAP, yes-associated protein.
The present study demonstrated that an increase in expression of TEAD4 was associated with an increase in histological grade; however, survival tended to decrease with increasing TEAD4 expression. Thus, in addition to histological grades, other factors may influence patient survival.
In conclusion, the present study was the first to evaluate the association between TEAD2 and the survival of patients with HCC. The current data revealed that a high expression level of TEAD2 in resected tumor tissue from patients with HCC was positively associated with a poor prognosis. Therefore, TEAD2 may represent a useful prognostic marker and therapeutic target for HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Datasets were both generated and presented in the present study (patient tissue analysis) and retrieved from online databases (TCGA). TCGA-LIHC dataset is available in GDC data portal (https://portal.gdc.cancer.gov/) and Firebrowse (http://firebrowse.org/?cohort=LIHC&download_dialog=true).
Authors' contributions
BSL and HSE designed the study and conducted critical revision of the manuscript. JSJ, SYC, WSR, HSE, JSK, SHKang, ESL, HSM, SHKim and JKS conducted acquisition, analysis and interpretation of the data. BSL carried out supervision of the analysis. JSJ and SYC wrote the manuscript. ISK verified and contributed statistical analysis. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
This research was approved by the Institutional Review Board of Chungnam National University Hospital (approval no. CNUH 2017-05-013).
Patient consent of publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ, Wu F. Solitary large hepatocellular carcinoma: A specific subtype of hepatocellular carcinoma with good outcome after hepatic resection. Ann Surg. 2009;249:118–123. doi: 10.1097/SLA.0b013e3181904988. [DOI] [PubMed] [Google Scholar]
- 3.Aravalli RN, Cressman EN, Steer CJ. Cellular and molecular mechanisms of hepatocellular carcinoma: An update. Arch Toxicol. 2013;87:227–247. doi: 10.1007/s00204-012-0931-2. [DOI] [PubMed] [Google Scholar]
- 4.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HH, Maeda T, Mullett SJ, Stewart AF. Transcription cofactor Vgl-2 is required for skeletal muscle differentiation. Genesis. 2004;39:273–279. doi: 10.1002/gene.20055. [DOI] [PubMed] [Google Scholar]
- 8.Chen HH, Mullett SJ, Stewart AF. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279:30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 9.Günther S, Mielcarek M, Kruger M, Braun T. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 2004;32:791–802. doi: 10.1093/nar/gkh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, Pan D. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci USA. 2012;109:E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight JF, Shepherd CJ, Rizzo S, Brewer D, Jhavar S, Dodson AR, Cooper CS, Eeles R, Falconer A, Kovacs G, et al. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99:1849–1858. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim B, Park JL, Kim HJ, Park YK, Kim JH, Sohn HA, Noh SM, Song KS, Kim WH, Kim YS, et al. Integrative genomics analysis reveals the multilevel dysregulation and oncogenic characteristics of TEAD4 in gastric cancer. Carcinogenesis. 2014;35:1020–1027. doi: 10.1093/carcin/bgt409. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Wang G, Yang Y, Mei Z, Liang Z, Cui A, Wu T, Liu CY, Cui L. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene. 2016;35:2789–2800. doi: 10.1038/onc.2015.342. [DOI] [PubMed] [Google Scholar]
- 15.Schütte U, Bisht S, Heukamp LC, Kebschull M, Florin A, Haarmann J, Hoffmann P, Bendas G, Buettner R, Brossart P, Feldmann G. Hippo signaling mediates proliferation, invasiveness, and metastatic potential of clear cell renal cell carcinoma. Transl Oncol. 2014;7:309–321. doi: 10.1016/j.tranon.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou GX, Li XY, Zhang Q, Zhao K, Zhang CP, Xue CH, Yang K, Tian ZB. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev. 2013;14:5199–5205. doi: 10.7314/APJCP.2013.14.9.5199. [DOI] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmondson HA, Steiner PE. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 19.Ghassan K A-A, Timothy M. (Eighth ed) Chicago, IL: Springer; 2017. Pawlik, Junichi Shindoh and Jean-Nicolas Vauthey editors: AJCC Cancer Staging Manual; pp. 287–293. [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- 22.Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko KJ, Kohn MJ, Liu C, DePamphilis ML. Transcription factor TEAD2 is involved in neural tube closure. Genesis. 2007;45:577–587. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holden JK, Cunningham CN. Targeting the Hippo pathway and cancer through the TEAD family of transcription factors. Cancers (Basel) 2018;10(pii):E81. doi: 10.3390/cancers10030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Nie Z, Zhou Z, Zhang H, Liu R, Wu J, Qin J, Ma Y, Chen L, Li S, et al. The interplay between TEAD4 and KLF5 promotes breast cancer partially through inhibiting the transcription of p27Kip1. Oncotarget. 2015;6:17685–17697. doi: 10.18632/oncotarget.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo C, Wang X, Liang L. LATS2-mediated YAP1 phosphorylation is involved in HCC tumorigenesis. Int J Clin Exp Pathol. 2015;8:1690–1697. [PMC free article] [PubMed] [Google Scholar]
- 27.Mao B, Hu F, Cheng J, Wang P, Xu M, Yuan F, Meng S, Wang Y, Yuan Z, Bi W. SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene. 2014;33:1468–1474. doi: 10.1038/onc.2013.88. [DOI] [PubMed] [Google Scholar]
- 28.Bai N, Zhang C, Liang N, Zhang Z, Chang A, Yin J, Li Z, Luo N, Tan X, Luo N, et al. Yes-Associated Protein (YAP) increases chemosensitivity of hepatocellular carcinoma cells by modulation of p53. Cancer Biol Ther. 2013;14:511–520. doi: 10.4161/cbt.24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Tilló E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diepenbruck M, Waldmeier L, Ivanek R, Berninger P, Arnold P, van Nimwegen E, Christofori G. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial-mesenchymal transition. J Cell Sci. 2014;127:1523–1536. doi: 10.1242/jcs.139865. [DOI] [PubMed] [Google Scholar]
- 31.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 32.Folkman J. Tumor angiogenesis: Therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 33.McDougall SR, Anderson AR, Chaplain MA. Mathematical modelling of dynamic adaptive tumour-induced angiogenesis: Clinical implications and therapeutic targeting strategies. J Theor Biol. 2006;241:564–859. doi: 10.1016/j.jtbi.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 34.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 35.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F, Han X, Feng Y, Zheng C, Wang Z, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiao S, Li C, Hao Q, Miao H, Zhang L, Li L, Zhou Z. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun. 2017;8:14058. doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin HS, Park HS, Shin JH, Kim DH, Jun SH, Lee CJ, Lee TH. A novel inhibitor of apoptosis protein (IAP)-interacting protein, Vestigial-like (Vgl)-4, counteracts apoptosis-inhibitory function of IAPs by nuclear sequestration. Biochem Biophys Res Commun. 2011;412:454–459. doi: 10.1016/j.bbrc.2011.07.117. [DOI] [PubMed] [Google Scholar]
- 39.Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Shen H, Withers HG, Yang N, Denson KE, Mussell AL, Truskinovsky A, Fan Q, Gelman IH, Frangou C, Zhang J. VGLL4 Selectively represses YAP-dependent gene induction and tumorigenic phenotypes in breast cancer. Sci Rep. 2017;7:6190. doi: 10.1038/s41598-017-06227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu B, Zhai M, Miao X, He C, Deng C, Fang Y, Luo M, Liu L, Liu S. Serotonin and YAP/VGLL4 balance correlated with progression and poor prognosis of hepatocellular carcinoma. Sci Rep. 2018;8:9739. doi: 10.1038/s41598-018-28075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie W, Hao J, Zhang K, Fang X, Liu X. Adenovirus armed with VGLL4 selectively kills hepatocellular carcinoma with G2/M phase arrest and apoptosis promotion. Biochem Biophys Res Commun. 2018;503:2758–2763. doi: 10.1016/j.bbrc.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. The TEAD family and its oncogenic role in promoting tumorigenesis. Int J Mol Sci. 2016;17(pii):E138. doi: 10.3390/ijms17010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin KC, Park HW, Guan KL. Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42:862–872. doi: 10.1016/j.tibs.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets were both generated and presented in the present study (patient tissue analysis) and retrieved from online databases (TCGA). TCGA-LIHC dataset is available in GDC data portal (https://portal.gdc.cancer.gov/) and Firebrowse (http://firebrowse.org/?cohort=LIHC&download_dialog=true).