Abstract
Background & Aims
Claudin-7 (Cldn7) is a tight junction (TJ) membrane protein located at the apical TJ and basolateral side of intestinal epithelial cells. Deletion of Cldn7 by gene targeting leads to the inflammatory bowel disease–like phenotype in mice, which includes weight loss, diarrhea, mucosa ulceration, and severe intestinal epithelial damage. In this study, we test our hypothesis that Cldn7 plays a critical role in regulating intestinal crypt stem cell functions.
Methods
Gene expression microarray, quantitative reverse-transcription polymerase chain reaction, in situ hybridization, histologic examinations, immunoblotting, 3-dimensional organoid culture, and various treatments to rescue Cldn7-deficient organoid defects were conducted using global Cldn7 knockout mice and inducible, conditional Cldn7 knockout mice.
Results
Gene deletion of Cldn7 in intestines showed significant alteration of expression profiles with striking down-regulation of intestinal crypt stem cell markers such as Olfm4, dislocated proliferative cells, and disrupted epithelial cell differentiation. In addition, the isolated Cldn7-deficient crypts where the stem cells reside were either unable to survive at all or formed defective spheroids, highlighting the functional impairment of crypt stem cells in the absence of Cldn7. Remarkably, the Cldn7-expressing organoids with buddings underwent rapid cell degeneration within days after turning off Cldn7 expression in the culture. We identified that activation of Wnt/β-catenin signaling rescued the organoid defects caused by Cldn7 deletion.
Conclusions
In this study, we show that Cldn7 is indispensable in controlling Wnt/β-catenin signaling–dependent intestinal epithelial stem cell survival, self-renewal, and cell differentiation. This study could open a door to study roles of TJ proteins in stem cell regulations in other tissues and organs.
Keywords: Crypt Stem Cells, Organoid Culture, Wnt/β-Catenin Signaling
Abbreviations used in this paper: 4OH-TAM, 4-hydroxytamoxifen; cCldn7fl/fl-T, inducible, conditional Cldn7 knockout mice; Cldn, claudin; cKO, tamoxifen-injected cCldn7fl/fl-T mice with inducible, conditional Cldn7 knockout; DMSO, dimethyl sulfoxide; EE, enteroendocrine; FABP-1, Fatty Acid-Binding Protein 1; FISH, fluorescence in situ hybridization; gKO, global claudin-7 knockout; GSK3β, glycogen synthase kinase 3 beta; IESC, intestinal epithelial stem cell; mRNA, messenger RNA; PCNA, proliferating cell nuclear antigen; PN, postnatal day; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SI, small intestine; TJ, tight junction; TUNEL, terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling; WT, wild-type
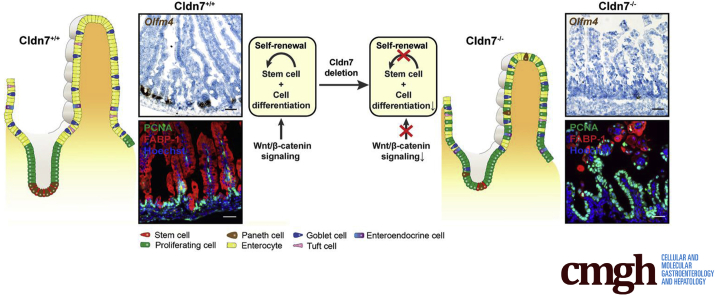
Graphical abstract
Summary.
We report a previously unidentified role of tight junction protein claudin 7 in intestinal epithelial stem cell function and regulation by using 2 independent claudin 7 knockout mouse models. Our data show that claudin 7 controls intestinal crypt stem cell survival, self-renewal, and epithelial differentiation through Wnt/β-catenin signaling.
The intestinal epithelial sheet is dynamically maintained by its self-renewal ability. The matured epithelial cells at the tip of villi are constantly replaced by newly differentiated cells derived from multipotent intestinal epithelial stem cells (IESCs) located in the crypt regions. IESCs give rise to proliferating progenitor cells, which subsequently differentiate into the nutrient-absorbing enterocytes, mucus-secreting goblet cells, neuropeptide-secreting enteroendocrine (EE) cells, antimicrobial peptide-secreting Paneth cells, and immune-sensing tuft cells.1 IESCs are destined to become active crypt stem cells or quiescent stem cells located adjacent to the Paneth cells.1,2 Lgr5,3 along with co-expressed genes such as Olfm4,4 Ascl2,5 and Sox9 low,6 are defined as the markers for active crypt stem cells that rapidly generate all epithelial cell types of the small intestine (SI). Quiescent stem cells that play an essential role in repairing epithelial cells after injury are marked by Bmi1,7 Hopx,8 and Lrig1.9
The IESC self-renewal process is critical for intestinal injury repair and regeneration. However, the factors and underlying molecular mechanisms regulating this process are still not well understood. Studies have shown that the Wnt/β-catenin signaling pathway is a key regulator of stem cell fate. In the gastrointestinal tract, Wnt signaling activation drives homeostasis and damage-induced repair. When the Wnt ligand is present, it binds to its receptor and inhibits the activity of GSK3β. The key pathway component β-catenin then is free and forms a complex with transcriptional factor T cell factor/lymphoid enhancer factor and induces target gene transcription. When Wingless/integrated (Wnt) ligand is absent, glycogen synthase kinase 3 beta (GSK3β) phosphorylates β-catenin at serines 33 and 34, and threonine 41, triggering the destabilization and degradation of β-catenin in the cytosol, leading to the suppression of Wnt signaling.10,11
Tight junctions (TJs) form a paracellular barrier restricting the free diffusion of ions and small molecules between cells. Claudins are a family of TJ integral membrane proteins. Deletion of TJ membrane protein claudin-7 (Cldn7) in mice leads to mucosa ulceration and severe intestinal epithelial damage.12,13 Cldn18 deletion promotes the proliferation of pulmonary epithelial progenitors and develops intraepithelial neoplasia in the stomach.14,15 Interestingly, when intestinal epithelial differentiation is induced in intestinal organoids, claudins are found to distribute heterogeneously among the various cell types including intestinal stem cells, Paneth cells, and enterocytes.16 These studies suggest that, in addition to their traditional roles in regulating epithelial barrier function and polarity, claudins also may regulate cell functions, such as proliferation, that could contribute to intestinal epithelial self-renewal. However, it is unknown whether claudins are involved in stem cell functions and regulations.
In this study, we show that Cldn7 is essential in maintaining IESC functions and intestinal epithelial self-renewal. We showed that deletion of Cldn7 reduces the number of IESCs and disrupts epithelial differentiation and proliferation in both global Cldn7 knockout mice (gKO) and inducible, conditional Cldn7 knockout mice (cCldn7fl/fl-T) model systems. By using a genome-wide gene expression approach and subsequent verifications by cell and molecular analyses, as well as a 3-dimensional intestinal organoid culture system, we discovered that Cldn7 deletion-mediated intestinal defects are caused by the disruption of IESC functions through the Wnt/β-catenin–dependent signaling pathway. This study thus establishes an unexpected and novel role for TJ proteins in regulating IESC functions and epithelial self-renewal and differentiation.
Results
Cldn7 Is Essential for Epithelial Cell Differentiation
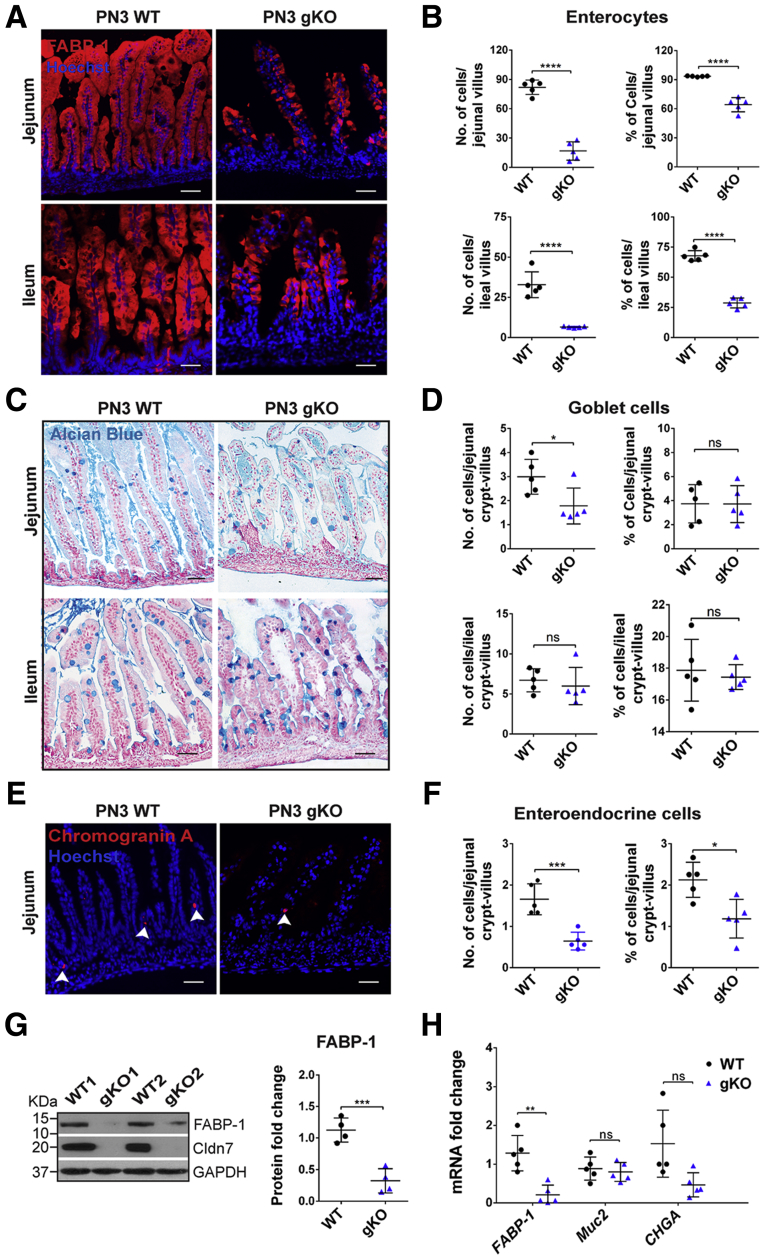
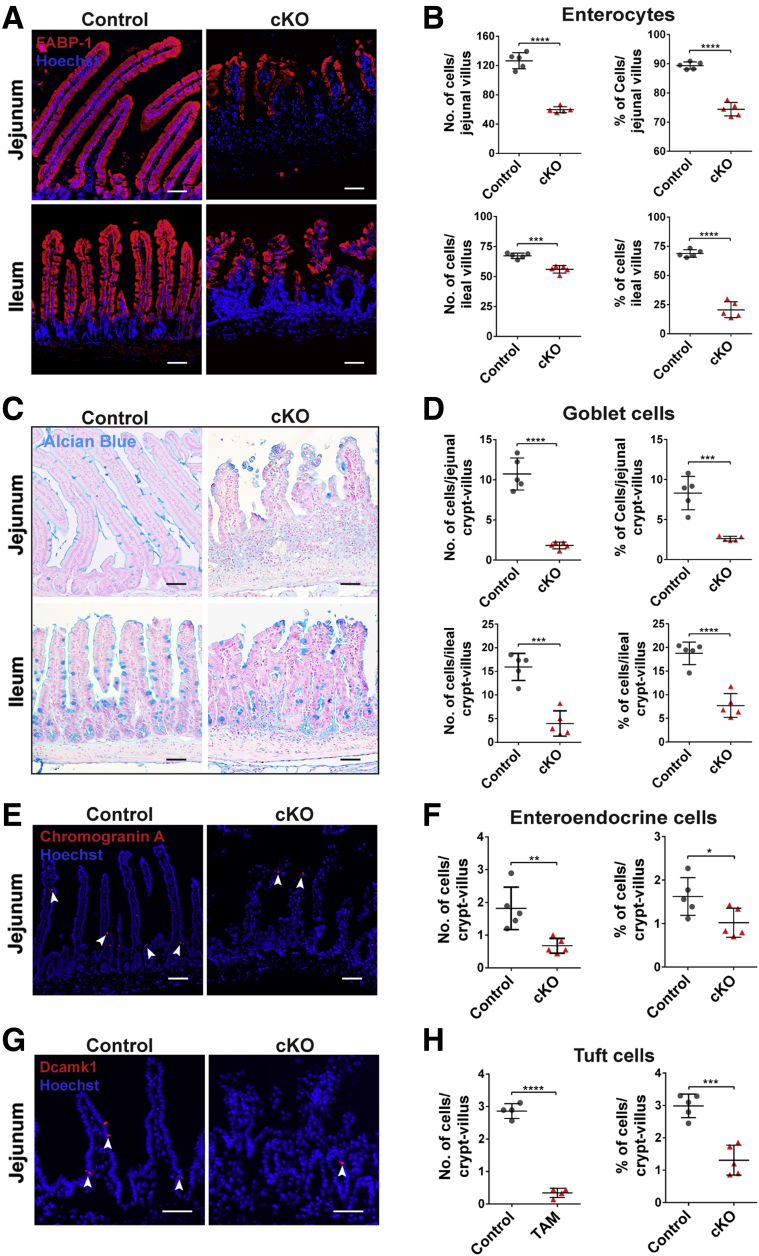
We previously reported that the intestines of gKO mice showed disrupted mucosal architecture, epithelial cell sloughing, and inflammation.12 To determine whether the deletion of Cldn7 affected epithelial differentiation, we used specific cellular markers on the SI of postnatal day (PN)3 wild-type (WT) and gKO mice. We found that the total number of enterocytes (Fatty Acid-Binding Protein 1 [FABP-1]) (Table 1) and the percentage of FABP-1–positive cells per villus were decreased significantly in both the jejunum and ileum of gKO mice compared with those of WT SI (Figure 1A and B). On the other hand, the total number of goblet cells (Alcian blue staining) was decreased significantly, although the percentage of goblet cells per crypt–villus axis showed no significant changes between WT and gKO SI (Figure 1C and D). This indicates that the reduced number of goblet cells is the result of the loss of villi. For EE cells (chromogranin A), both the total number and percentage were decreased in the jejunum of gKO compared with that of WT (Figure 1E and F). Paneth cells and tuft cells are not detectable at this age because they are not differentiated until 2 weeks after birth.17,18 These observations were consistent with the reduced protein level of FABP-1 detected by immunoblotting (Figure 1G) and reduced FABP-1 and chromogranin A (CHGA) messenger RNA (mRNA) levels detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using WT and gKO SIs (Figure 1H). The mRNA expression level of Muc2 showed no significant difference between WT and gKO SIs. These data show that deletion of Cldn7 impairs the enterocyte and EE cell differentiation, but not goblet cells in gKO mice.
Table 1.
List of Antibodies
| Antibody | Source (vendor/catalog number) | Dilution | Use |
|---|---|---|---|
| FABP-1 | Cell Signaling (Danvers, MA)/13368 | 1:200 | IF |
| FABP-1 | Cell Signaling/13368 | 1:1000 | WB |
| Cleaved caspase-3 | Cell Signaling/9664 | 1:1000 | WB |
| Cleaved PARP | Cell Signaling/94885 | 1:1000 | WB |
| Chromogranin A | Santa Cruz Biotechnology (Dallas, TX)/sc-393941 | 1:100 | IF |
| Claudin 1 | Zymed (Waltham, MA)/71-7800 | 1:500 | WB |
| Claudin 4 | Invitrogen (Waltham, MA)/364800 | 1:1000 | WB |
| Claudin 7 | IBL (Gunma, Japan) Japan/18875 | 1:1000 | WB |
| Claudin 7 | IBL Japan/18875 | 1:200 | IF |
| Claudin 15 | IBL Japan/18805 | 1:500 | WB |
| E-cadherin | BD Transduction Labs (San Jose, CA)/610181 | 1:1000 | WB |
| Dcamkl1 | Abcam (Cambridge, MA)/ab37994 | 1:100 | IF |
| GAPDH | Cell Signaling/2118 | 1:1000 | WB |
| Hopx | Santa Cruz Biotechnology/sc-30216 | 1:500 | WB |
| Lysozyme | Abcam/ab36362 | 1:200 | IF/IHC |
| Muc2 | Santa Cruz Biotechnology/sc-7314 | 1:200 | IHC |
| Nonphospho(active)β-catenin | Cell Signaling/8814 | 1:1000 | WB |
| Olfm4 | Novus Biologicals (Centennial, CO)/NBP2-24535 | 1:800 | WB |
| Olfm4 | Cell Signaling/39141 | 1:200 | IF |
| Occudin | Zymed/71-1500 | 1:2000 | WB |
| PCNA | Cell Signaling/2586 | 1:200 | IF |
| Phopho-GSK3β(S9) | Cell Signaling/9336 | 1:1000 | WB |
| Phospho–β-catenin (S33/37/T41) | Cell Signaling/9561 | 1:1000 | WB |
| Sox9 | Millipore (Bedford, MA)/AB5535 | 1:100 | IF |
| Tcf4 | Santa Cruz Biotechnology/sc-166699 | 1:1000 | WB |
| Total GSK3β | BD Transduction Labs/610202 | 1:1000 | WB |
| Total β-catenin | Cell Signaling/8480 | 1:1000 | WB |
| Wnt3a | Santa Cruz Biotechnology/sc-74537 | 1:1000 | WB |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IF, immunofluorescence; IHC, immunohistochemistry; Muc2, mucin 2; S33/37/T41 serines 33 and 37, and threonine 41; WB, Western blot.
Figure 1.
Deletion of Cldn7 affected epithelial cell differentiation in gKO SIs. Detection of (A) enterocytes by FABP-1 immunofluorescent staining, (C) goblet cells by Alcian blue staining, and (E) EE cells by immunofluorescent staining of chromogranin A in PN3 WT and gKO SIs. Images are representative of more than 3 independent experiments. Scale bars: 100 μm. Quantification of the total number and percentage per villus (or crypt–villus) of (B) enterocyte, (D) goblet cells, and (F) EE cells. Each point represents the calculation from 50 crypt–villus in each mouse. n = 5; 2-tailed, unpaired t test; *P < .05, ***P < .001, ****P < .0001, P > .05. (G) Protein expression levels of FABP-1 in 2 representative PN3 WT and gKO SIs were determined by immunoblotting (left) and then quantified (right). n = 4; 2-tailed, unpaired t test, ***P < .001. (H) The qRT-PCR analysis of FABP1, Muc2, and CHGA mRNA in PN3 WT and gKO SIs. n = 5; 2-tailed, paired t test; P < .05, **P < .01, P > .05. Data are represented as means ± SEM. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Deletion of Cldn7 Changes the Landscape of Gene Expression Profiles in gKO
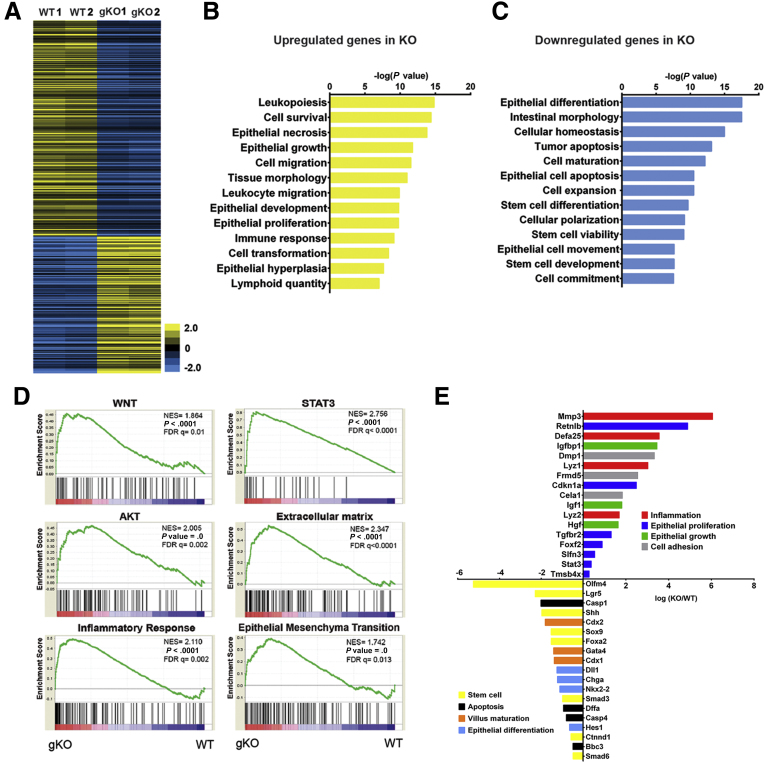
To unravel the molecular mechanism underlying the Cldn7 deletion-mediated intestinal defects in a nonbiased approach, we performed genome-wide microarray analysis using RNA samples from PN3 WT and global claudin-7 knockout mice littermates. More than 6000 gene expression patterns were analyzed and grouped into up-regulation and down-regulation clusters in relation to those of WT SI. Among 29,990 annotated transcripts, 1238 transcripts (4.1%) were down-regulated and 276 transcripts (0.09%) were up-regulated in gKO compared with WT (P < .01) as shown in the heatmap (Figure 2A). Using the ingenuity pathway analysis tool, our data showed that the up-regulated genes were related to leukopoiesis, epithelial growth, proliferation, and hyperplasia (Figure 2B). Importantly, those down-regulated genes were related to epithelial differentiation and maturation, stem cell differentiation, stem cell development, and cell commitment (Figure 2C). Gene set enrichment analysis showed that 77 gene sets were enriched in global claudin-7 knockout mice SI (P < .01 and false-positive values; false-discovery rate, < 25%). These enriched gene sets include signaling pathways of WNT, signal transducer and activator of transcription 3, AKT, extracellular matrix, inflammatory response, and epithelial–mesenchymal transition (Figure 2D). Moreover, genes related to inflammation, epithelial proliferation, epithelial growth, and cell adhesion all were up-regulated, whereas genes related to stem cells, apoptosis, villus maturation, and epithelial differentiation all were down-regulated (P < .01; q < 0.05) (Figure 2E). These consistent data suggest that Cldn7 deletion-induced gene expression changes were related especially to epithelial cell proliferation and differentiation, as well as IESCs. One of the most striking findings from the microarray data was that several IESC markers all were down-regulated in gKO SI, especially Olfm4, which was the top one reduced gene expression (Figure 2E). This is significant because although more than 12 claudins are expressed along the gastrointestinal tract, none of them have been reported to be involved in IESC functions and regulation.
Figure 2.
Genome-wide microarray analysis for WT and gKO SI. (A) The heatmap shows the gene expression profile of 2 independent sets of PN3 WT and gKO SIs by microarray analysis. Data are color-coded to reflect the gene expression level. Genes that have false-positive values of q < 0.05 are shown. (B and C) Ingenuity analysis of gene expression related to differential gastrointestinal functions between WT and gKO SIs. Genes with P < .001 and q < 0.05 were analyzed. (D) Gene Set Enrichment Analysis of enriched pathways activated by Cldn7 deletion. P < .01, false-discovery rate (FDR) q < 25%. (E) Key up-regulated and down-regulated genes related to essential intestinal epithelial functions are shown. AKT, a serine/threonine-specific protein kinase; NES, nestin; STAT3, signal transducer and activator of transcription 3.
Deletion of Cldn7 Results in the Loss of Active Crypt Stem Cells and Hyperproliferation of Epithelial Cells in gKO
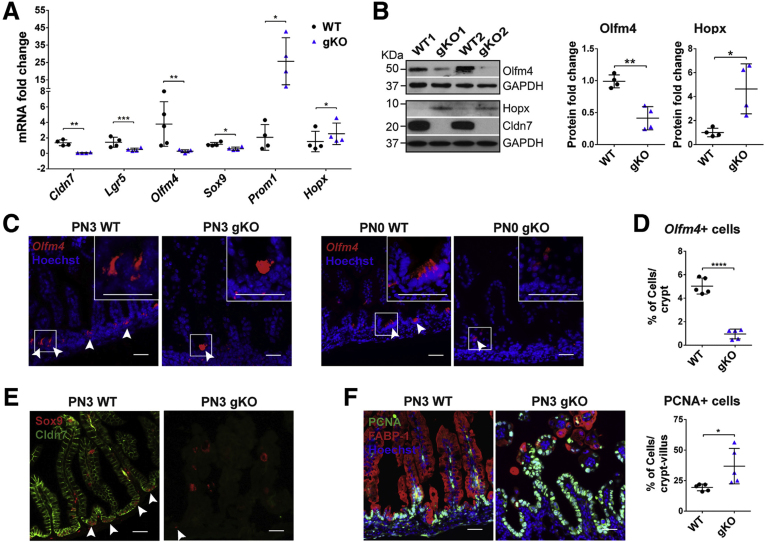
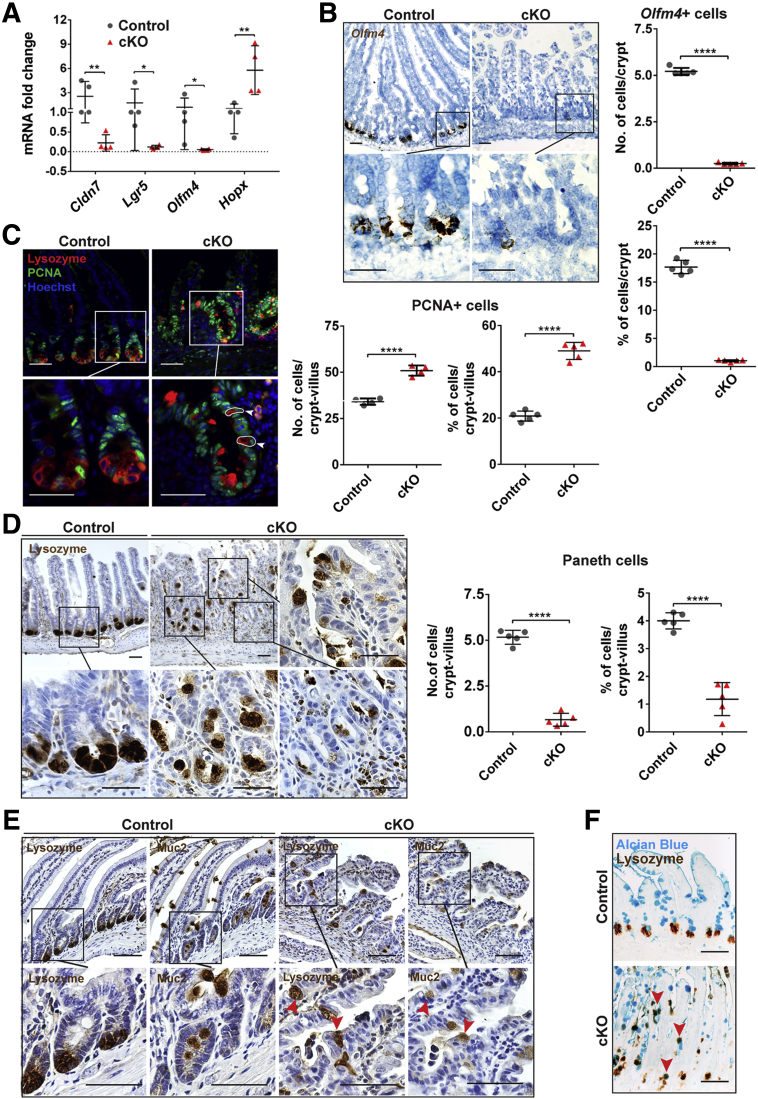
We performed a series of experiments to verify the data obtained from the microarray and to test whether Cldn7 has any effect on IESCs and transit-amplifying cells. Because intestinal crypt cells are grouped into active stem cells, quiescent stem cells, and transit-amplifying cells, we first examined the mRNA levels of representative cell marker genes by qRT-PCR. Lgr5, Olfm4, and Sox9 mark active stem cells. Hopx and Prom1 represent quiescent IESCs and transit-amplifying cells, respectively.19,20 Surprisingly, Cldn7 deletion resulted in significantly reduced mRNA levels of Lgr5, Olfm4, and Sox9, but increased the mRNA levels of Prom1 and Hopx (Figure 3A). To test whether the mRNA level changes translated to changes in protein levels, we probed Olfm4 and Hopx using Western blot. The protein level of Olfm4 was decreased significantly, while the protein level of Hopx was increased significantly, suggesting that the quiescent stem cells and/or transit-amplifying cells were expanding, while active stem cells were decreased when Cldn7 was absent (Figure 3B).
Figure 3.
Deletion of Cldn7 resulted in the loss of active crypt stem cells and an increase in epithelial cell proliferation in gKO SIs. (A) The qRT-PCR analysis of IESC marker genes in PN3 WT and gKO SIs. Lgr5, Olfm4, and Sox9 are active intestinal stem cell marker genes. Hopx is a quiescent stem cell marker gene and Prom1 is a transit-amplifying cell marker gene. n = 4–5; 2-tailed, paired t test; *P < .05, **P < .01, ***P < .001. (B) The protein levels of Olfm4 and Hopx were detected by immunoblotting (left) and quantified (right). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. n = 4; 2-tailed, unpaired t test; *P < .05, **P < .01. (C) The IESC maker Olfm4 gene was detected by FISH in PN3 (left) and PN0 (right) WT and gKO SIs. Arrowheads indicate the Olfm4+cells. (D) Quantification of Olfm4+cells in panel C (PN3). n = 5; 2-tailed, unpaired t test; ****P < .0001. (E) Double-immunofluorescent labeling of Sox9 (red) and claudin-7 (green). Arrowheads indicate the Sox9+ cells. (F) Co-immunolabeling of proliferating cell marker PCNA (green) with enterocyte marker FABP-1 (red) (left). The quantification of PCNA-positive cells is shown (right). n = 5; 2-tailed, unpaired t test; *P < .05. Each point represents the calculation from 50 crypt–villus in each mouse. Data are represented as means ± SEM. Images are representative of more than 3 independent experiments. (C, E, and F) Scale bars: 100 μm.
To further investigate whether Cldn7 deletion reduced the signal of Olfm4, a robust IESC marker, we detected its mRNA using fluorescence in situ hybridization (FISH).4 Olfm4+ cells were decreased significantly in gKO SI compared with those in WT (Figure 3C, left panel). Olfm4 is a glycoprotein and is associated closely with many inflammatory digestive diseases.21 Significant inflammation was shown in the SI of PN3 gKO mice, whereas little inflammation was detected in newborn mice.12 To test whether the decrease of Olfm4+ IESCs was the result of intestinal inflammation, we examined the Olfm4 mRNA in PN0 mice. We found that the Olfm4+ IESCs also were decreased significantly in PN0 gKO SI, suggesting that the decrease of IESCs was not caused by inflammation, but rather the loss of Cldn7 (Figure 3C, right panel). Figure 3D shows the statistical data comparing Olfm4+ IESCs between PN3 WT and gKO SI. Sox9 also was used to label active IESCs.6 We labeled Sox9 in PN3 WT and gKO SI and found that the positive signal in the gKO crypt area was reduced, suggesting a decrease in the active stem cell population (Figure 3E). Sox9 also labels some of the differentiated epithelial cells in the villi, such as a subset of enteroendocrine cells.6
Previously, we showed that the bromodeoxyuridine incorporation was increased significantly in gKO.12 In this study, we found that the proliferating cell nuclear antigen (PCNA)-positive proliferating cells in gKO SI not only were increased significantly, but also, in some areas, were no longer restricted in the crypt region as those in WT (Figure 3F). These proliferating cells were not enterocytes, as indicated by FABP-1 signal (Figure 3F, left panel). PCNA-positive cells also were not co-localized with goblet cells or EE cells (data not shown).
The earlier-described data show that Cldn7 deletion results in a hyperproliferative intestinal epithelium and a decrease in IESCs that is not owing to inflammation.
Cldn7 Deletion in Adult Mice Leads to the Loss of Active Crypt Stem Cells and Alters the Number and Position of Paneth Cells
To determine whether Cldn7 deletion in adult mice also affects IESCs, the mRNA levels of robust IESC marker genes Lgr5, Olfm4, and quiescent IESC marker gene Hopx were detected by qRT-PCR in the tamoxifen-induced Cldn7 knockout mice (cKO); the inducible, intestinal-specific Cldn7 knockout (cCldn7fl/fl-T) mice were generated by crossing Cre mice (Villin-CreERT2) with Cldn7-flox mice. Similar to gKO mice, Cldn7 deletion significantly reduced the mRNA levels of Lgr5 and Olfm4 but increased the mRNA level of Hopx (Figure 4A). In addition, the number of Olfm4+ cells in cKO SIs, determined by FISH, was significantly lower compared with control mice (Figure 4B). Approximately 50% of the crypts in cKO SIs contained no Olfm4+ cells, suggesting the significant loss of IESCs.
Figure 4.
Cldn7 deletion in adult cKO SIs led to the loss of active crypt stem cells, increased epithelial cell proliferation, and aberrant positioning of Paneth cells. (A) The qRT-PCR analysis of Cldn7, Lgr5, Olfm4, and Hopx mRNA in 3-month-old control and cKO SIs. n = 4–5; 2-tailed, paired t test; *P < .05, **P < .01. (B) Active IESCs were labeled using FISH (left) and quantified (right) in 3-month-old control and cKO SIs. Each point represents the calculation from 50 crypts in each mouse. n = 5; 2-tailed, unpaired t test, ****P < .0001. (C) The control and cKO SIs were triple-labeled with lysozyme/PCNA/Hoechst to show the positions of Paneth and proliferating cells (left). Arrowheads and circles highlight the epithelial cells with positive PCNA and lysozyme signals. The statistical analysis of PCNA-positive cells is shown (right). Each point represents the calculation from 50 crypt–villus in each mouse. n = 5; 2-tailed, unpaired t test, ****P < .0001. (D) Paneth cells were labeled using antilysozyme antibody by immunohistochemistry to compare the position of the Paneth cells between control and cKO SIs (left, images in the square were enlarged as indicated by corresponding lines). The statistical analysis of Paneth cells is shown (right). Each point represents the calculation from 50 crypt–villus in each mouse. n = 5; 2-tailed, unpaired t test; ****P < .0001. Data are represented as means ± SEM. (E) Parallel labeling of lysozyme and mucin 2 (muc2) on the neighboring control and cKO SI sections. Arrowheads highlight the epithelial cells with positive lysozyme and muc2 signals. The images in the square were enlarged as indicated by corresponding lines. (F) Co-labeling of Alcian blue (blue) and antilysozyme antibody by immunohistochemistry (brown). Arrowheads indicate the epithelial cells with positive Alcian blue and lysozyme signals (n = 4). Scale bars: 100 μm.
To investigate whether Cldn7 deletion in adult mice affects Paneth cells and cell proliferation, triple staining of lysozyme, PCNA, and Hoechst was performed using control and cKO SIs. Paneth cells were located at the base of crypts, and proliferating cells were in the crypt region, as expected, in the control mice. Surprisingly, both Paneth cells and proliferating cells in cKO mice were no longer restricted to the crypt region, they also appeared along the villi (Figure 4C, cKO in the top panel). The number of proliferating cells in cKO SIs was increased significantly, as observed in gKO SIs (Figure 4C, right panel). The Paneth cells in cKO SIs showed remarkable aberrant localization and were distributed randomly throughout the villi and crypts as visualized by immunohistochemistry, while Paneth cells in the control mice were located only at the base of crypts (Figure 4D, left panel). Statistical analysis showed that the Paneth cells were reduced significantly in cKO SIs (Figure 4D, right panel).
We observed that many aberrantly positioned Paneth cells were cup-shaped and resembled goblet cells (Figure 4D), therefore, we compared the locations of lysozyme-positive (lysozyme+) Paneth cells and mucin 2–positive goblet cells in the neighboring intestinal slides from the same intestinal specimens. We found that some lysozyme+ cells also expressed mucin 2 (Figure 4E). This result was confirmed by the fact that some of the Alcian blue–positive cells also were labeled with lysozymes, suggesting the existence of hybrid cells (Figure 4F) and indicating that these Paneth and goblet cells were not fully differentiated into their destinations.
To test whether Cldn7 deletion affects the epithelial differentiation in adult SIs, enterocytes were labeled by FABP-1, goblet cells by Alcian blue, EE cells by chromogranin A, and tuft cells by Dcamk1, respectively. The number of enterocytes (Figure 5A and B), goblet cells (Figure 5C and D), EE cells (Figure 5E and F), and tuft cells (Figure 5G and H) all were decreased significantly in cKO SIs, showing that Cldn7 deletion also impeded the epithelial cell differentiation in mature SIs. As shown in Figure 5E and 5G, EE cells and tuft cells were located at villi and near the crypt region.
Figure 5.
Disruption of epithelial differentiation in cKO SIs. (A) Enterocytes were labeled by anti–FABP-1 antibody, (C) goblet cells were stained by Alcian blue staining, (E) EEs were labelled by anti–chromogranin A antibody (Arrowheads indicate EE+ or tuft+ cells, respectively), and (G) tuft cells were labelled by anti-Dcamk1 antibody in 3-month-old control and cKO SIs. Images are representative of more than 3 independent experiments. Quantification of the total number and percentage per villus (or crypt–villus) of (B) enterocytes, (D) goblet cells, (F) EE cells, and (H) tuft cells. Each point represents the calculation from 50 crypt–villus in each mouse. n = 5; 2-tailed, unpaired t test; *P < .05, **P < .01, ***P < .001, ****P < .0001. Data are represented as means ± SEM. Scale bars: 100 μm.
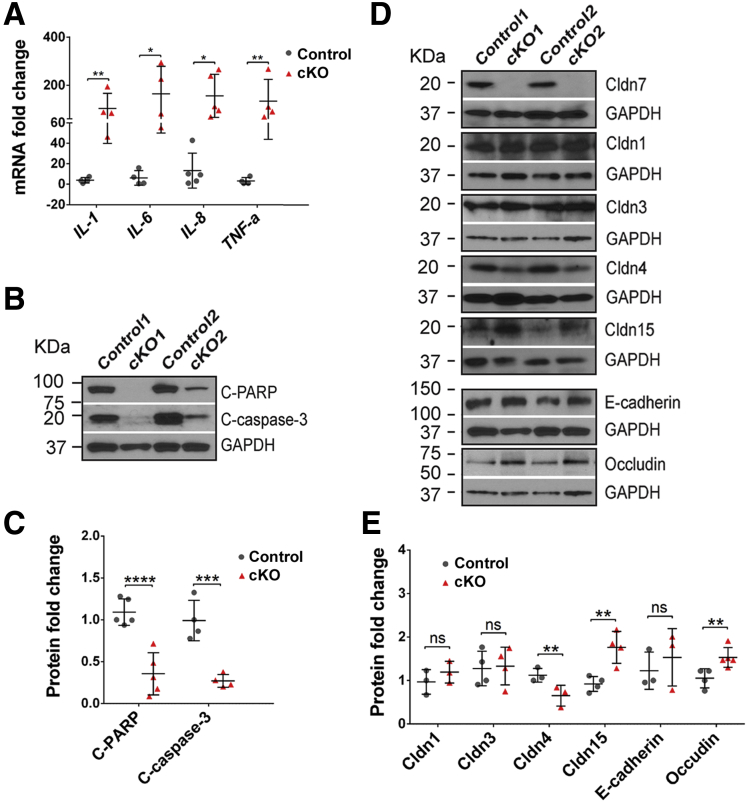
Histologic examinations of small intestines from cKO mice showed that the lamina propria contained an increased number of leukocytes compared with that in control mice, indicating an immune response after Cldn7 deletion.22 This observation was confirmed by the significantly increased mRNA levels of IL1, IL6, IL8, and TNF-α in cKO SI using qRT-PCR (Figure 6A). Apoptotic marker proteins, cleaved poly ADP-ribose polymerase and cleaved caspase-3, were used to examine the cell apoptosis status. The result showed that both cleaved poly ADP-ribose polymerase and cleaved caspase-3 were reduced significantly in the cKO SIs (Figure 6B and C), consistent with the microarray analysis showing that most cell apoptosis-related genes were down-regulated (Figure 2C and E). In addition, a panel of junctional proteins was assayed by Western blot. The results showed that there were no significant changes in expression levels of Cldn1, Cldn3, and E-cadherin, although Cldn15 and occludin expression levels were increased significantly and the expression level of Cldn4 was decreased significantly in cKO SIs (Figure 6D and E). The significance of these protein expression level changes needs to be investigated further.
Figure 6.
Cldn7 deletion increased inflammatory cytokines and reduced apoptotic signals in cKO SIs. (A) The qRT-PCR analysis of common inflammatory cytokine genes, IL-1, IL-6, IL-8, and TNF-a, in 3-month-old control and cKO SIs. n = 4–5; 2-tailed, paired t test; *P < .05; **P < .01. (B) Protein levels of cleaved poly ADP-ribose polymerase and cleaved caspase-3 from 3-month-old control and cKO SIs were detected by immunoblotting. (C) The quantification of protein expression levels shown in panel B. n = 4; 2-tailed, paired t test; ***P < .001; ****P < .0001. (D) Protein expression levels of claudin 1, 3, 4, 7, 15, E-cadherin, and occludin from 3-month-old control and cKO SIs were detected by immunoblotting. (E) The quantification of protein expression levels shown in panel D. n = 3–4; 2-tailed, paired t test; **P < .01, P > .05. C-PARP, poly ADP-ribose polymerase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Deletion of Cldn7 Impairs Intestinal Organoid Survival and Growth Owing to Disruption of the Wnt/β-Catenin Signaling Pathway
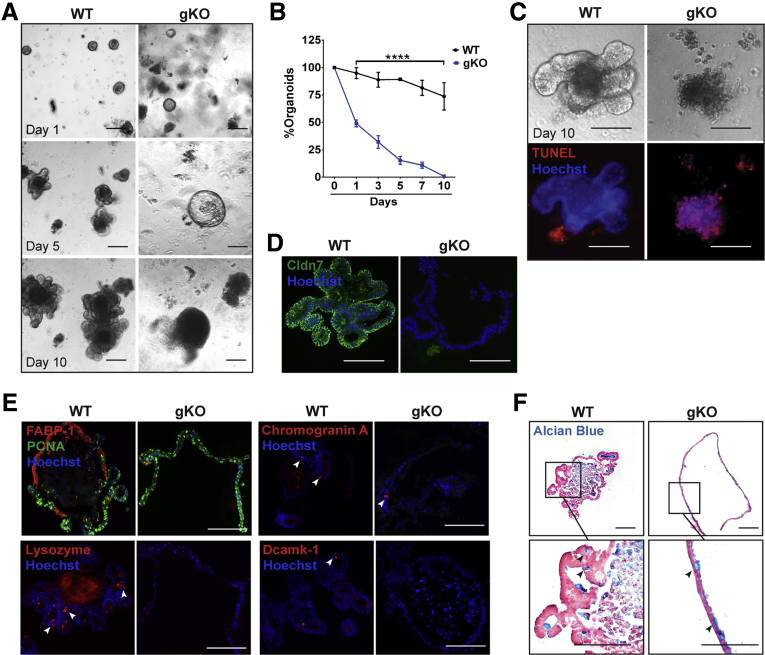
To further examine the effect of Cldn7 deletion on IESCs, 100 crypts were isolated from WT and gKO SIs and plated for 10 days. Organoids were counted on days 1, 3, 5, 7, and 10. From days 1 through 10, the number of gKO organoids decreased to 50%, 30%, 15%, 10%, and 0.8%, respectively (Figure 7A and B). On day 5, the crypts from gKO SIs formed spheroids without any budding, while typical organoids with finger-like buddings were observed in WT groups (Figure 7A). On day 10, the clustered cells in gKO cultures were identified as apoptotic cells via terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining, while organoids from WT SIs were larger, had numerous buddings, and tested negative for TUNEL staining (Figure 7C). The absence of Cldn7 in gKO spheroids was confirmed by immunofluorescence (Figure 7D). These results show that loss of Cldn7 impairs the intestinal organoid survival and growth.
Figure 7.
Cldn7 is essential for in vitro gKO organoid survival and growth. (A) Representative bright-field images of in vitro organoid growth of PN3 WT and gKO SI crypts. Images were captured 1, 5, and 10 days after plating. (B) Comparison of organoid numbers on different days between WT and gKO SI crypts. n = 6; percentage of organoids were compared by 1-way analysis of variance, followed by the Dunnett multiple comparison test; ****P < .0001. Data are represented as means ± SEM. (C) Representative images of bright-field and whole-mount TUNEL staining from WT and gKO organoids after 10 days of culture (n = 3). (D) The 5-day WT and gKO organoids were double-stained with anti–claudin-7 antibody and Hoechst. (E) Enterocytes, EE cells, Paneth cells, and tuft cells were labeled by anti–FABP-1, anti–chromogranin A, antilysozyme, and anti–Dcamk-1 antibodies, respectively, on WT and gKO SI organoids after 5 days of culture. PCNA-positive proliferating cells were co-labeled with FABP-1. Arrowheads point to the corresponding cells labeled by specific antibodies. Images are representative of 3 independent experiments. (F) Goblet cells were stained by Alcian blue on WT and gKO SI organoids after a 5-day culture. Images are representative of 3 independent experiments. Scale bars: 100 μm.
Organoids and spheroids were collected from 5-day cultures and processed for histologic evaluation. Strikingly, the numbers of FABP-1–positive enterocytes, chromogranin A-positive EE cells, lysozyme-positive Paneth cells, and Dcamk1-positive tuft cells had almost diminished in gKO spheroids, whereas these cells in WT organoids were abundant or present after 5 days of in vitro growth, suggesting that Cldn7 is required for organoid epithelial differentiation (Figure 7E). Interestingly, the number of Alcian blue–positive goblet cells did not change much between organoids and spheroids, suggesting that the loss of Cldn7 did not affect goblet cell differentiation (Figure 7F).
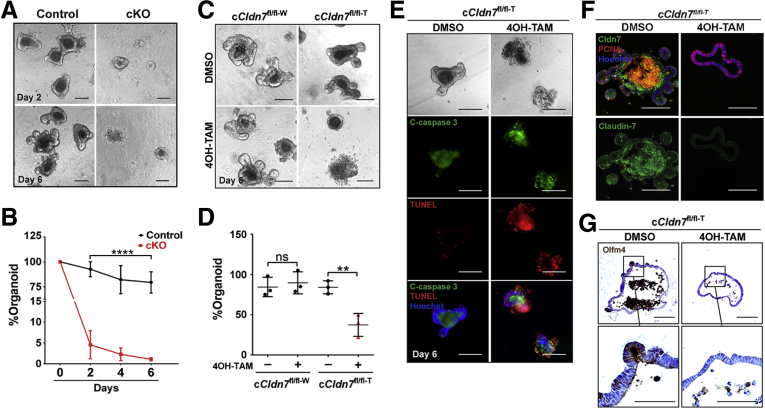
To investigate whether the IESCs from cKO have survival and self-renewal defects in vitro, the crypts were isolated from 3-month-old control and cKO SIs, from which Cldn7 had been deleted by tamoxifen injections. Figure 8A shows a striking loss of surviving cKO spheroids compared with control organoids with buddings. On day 2 after plating, only 5% of spheroids survived, although fewer than 2% were found on day 6, suggesting a critical role of Cldn7 in maintaining in vitro survival of adult IESCs (Figure 8B).
Figure 8.
Cldn7 is essential for in vitro organoid survival and growth of adult cKO SIs. (A) Representative bright-field images of control and cKO organoids isolated from 3-month-old control and cKO SI crypts treated with tamoxifen in vivo. Images were captured on days 2 and 6 after plating. (B) Quantification of organoid numbers. The percentage of organoids were compared by 1-way analysis of variance, followed by the Dunnett multiple comparison test; n = 3; ****P < .0001, P > .05. Scale bars: 100 μm. (C) Representative bright-field images of organoids isolated from 3-month-old cCldn7fl/fl-W and cCldn7fl/fl-T SI crypts. Then these organoids were treated with either DMSO or 4OH-TAM (3 μmol/L) in vitro. All images were captured on day 6 after plating. (D) Quantification of organoid numbers. The percentage of organoids were compared by 1-way analysis of variance, followed by the Dunnett multiple comparison test; n = 3; **P < .01, P > .05. (E) Representative images of bright-field (top), whole-mount cleaved caspase-3 and TUNEL staining of cCldn7fl/fl-T organoids treated with either DMSO or 4OH-TAM after a 6-day culture. Images are representative of 3 independent experiments. (F) Detection of Cldn7 (green) and proliferating cells (red) by immunofluorescent staining in DMSO- or 4OH-TAM–treated cCldn7fl/fl-T organoids after a 6-day culture (n = 3). (G) Detection of Olfm4+ stem cells by immunohistochemistry in DMSO- or 4OH-TAM–treated cCldn7fl/fl-T organoids after a 6-day culture (n = 3).
Because Cldn7-deficient crypts cannot form organoids, we wondered whether this was caused by Cldn7 deletion-induced impairment in the closure of open crypts at the beginning of culture. To test this possibility, we established organoid cultures from 3-month-old cCldn7fl/fl-T mice, allowed the crypts to close and grow for 1 day, followed by the addition of 4-hydroxytamoxifen (4OH-TAM), the active form of tamoxifen, for 5 days to turn off the Cldn7 expression. Crypts from control mice were included to test the toxicity of 4OH-TAM. On day 6, approximately 90 of 100 plated control crypts (cCldn7fl/fl-W) treated with dimethyl sulfoxide (DMSO) or 4OH-TAM formed organoids with buddings (Figure 8C and D, cCldn7fl/fl-W). The crypts isolated from cCldn7fl/fl-T mice formed normal organoids when treated with DMSO, but the number of organoids was reduced to 40% after treatment with 4OH-TAM (Figure 8C and D, cCldn7fl/fl-T). In addition, Cldn7-deficient crypts failed to form budding organoids. Instead, they formed spheroids without buds (Figures 7A, middle panel, and 8C, bottom right). Similar to the gKO organoid culture, the clustered cells in 4OH-TAM–treated cultures were identified as apoptotic cells via cleaved caspase-3 and TUNEL staining, while the control organoids (treated with DMSO) grew normal with buddings and the apoptotic signals were in the lumen (Figure 8E). Cldn7 signal was highly expressed in the cCldn7fl/fl-T culture treated with DMSO and was completely absent after treatment with 4OH-TAM, indicating the successful induction of Cldn7 deletion by 4OH-TAM (Figure 8F). Moreover, the Olfm4-positive cells were barely found in the Cldn7-deficient spheroids, although they were clearly present in the control organoids (Figure 8G, DMSO). All of the earlier-described data strongly support our hypothesis that Cldn7 deletion reduces the number of IESCs and impairs intestinal epithelia self-renewal and that Cldn7 is essential in maintaining organoid survival and growth.
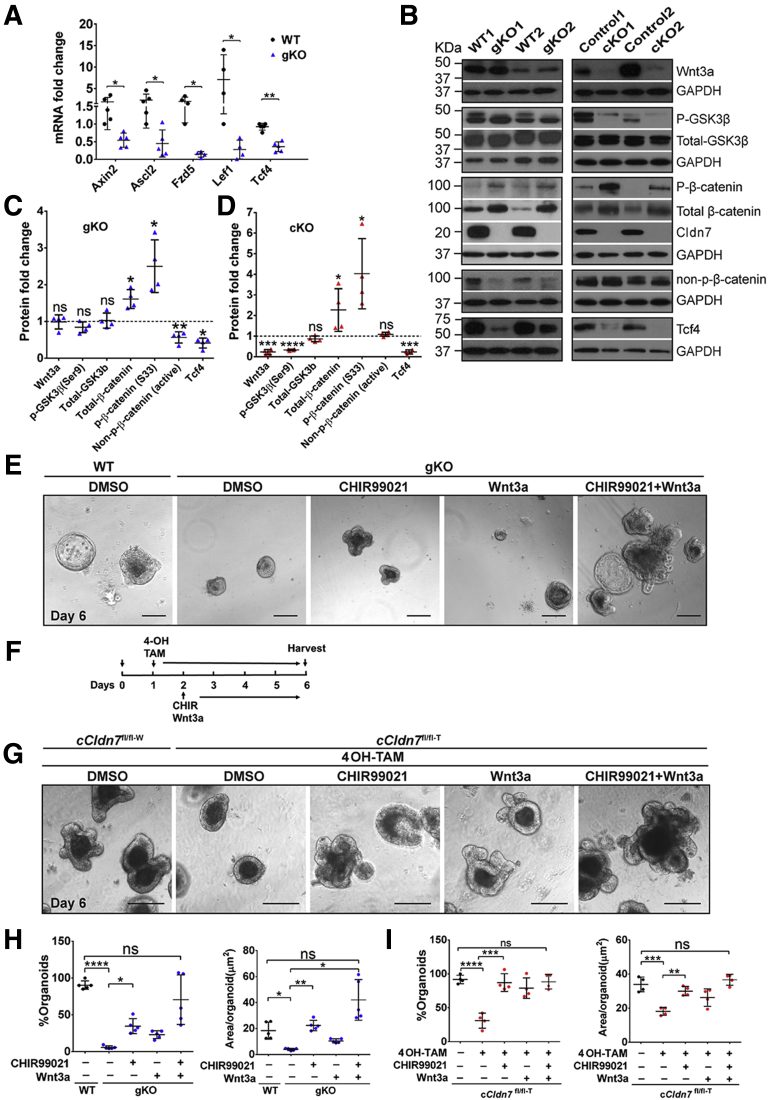
The gene enrichment analysis (Figure 2D) showed that the Wnt signaling pathway, a critical pathway for IESC survival, is one of the most affected pathways.23 Of note, IESC marker genes Lgr5, Olfm4, and Sox9, related to Wnt signaling, all were down-regulated significantly in gKO SIs (Figure 3A)24, 25, 26; therefore, we hypothesized that deletion of Cldn7 disrupts the Wnt signaling pathway. We examined the Wnt target genes Axin2, Ascl2, Fzd5, Lef1, and Tcf4.10,11 The mRNA levels of these genes all were down-regulated significantly in gKO SIs, suggesting that Wnt signaling was suppressed (Figure 9A). Then, we tested the effect of Cldn7 deletion on different components of the Wnt signaling pathway. Interestingly, the protein level of Wnt3a did not change in PN3 gKO SIs, but was reduced significantly in adult cKO SIs. In addition, phosphorylated β-catenin at serines 33 and 34, and threonine 41, was significantly higher in both gKO and cKO SIs, whereas the ratio of nonphosphorylated β-catenin/total-β-catenin was decreased, suggesting that β-catenin was destabilized without Cldn7 (Figure 9B). Furthermore, Tcf4, a Wnt target gene and a transcriptional factor, also was down-regulated, further confirming the suppression of Wnt signaling in the absence of Cldn7. Figure 9C and D show statistical data from 5 independent experiments. These results show that in Cldn7-deficient SIs, the Wnt/β-catenin signaling pathway was suppressed in both PN3 (gKO) and adult (cKO) mice.
Figure 9.
Activation of Wnt/β-catenin signaling rescued the organoid defects resulting from Cldn7 deletion. (A) The qRT-PCR analysis of Wnt/β-catenin signaling target genes in PN3 WT and gKO SIs. n = 4–5; 2-tailed, paired t test; *P < .05; **P < .01. (B–D) Protein levels of Wnt/β-catenin signaling pathway from PN3 WT and gKO SIs (B, left column) and 3-month-old control and cKO SI crypts (B, right column) were detected by immunoblotting and quantified (C, gKO; D, cKO). n = 4; 2-tailed, paired t test; *P < .05, **P < .01, ***P < .001, ****P < .0001, P > .05. (E) Organoids from PN3 gKO SIs were treated with DMSO, CHIR99021 (2 μmol/L), wnt3a (200 ng/mL), and a combination of CHIR99021 (2 μmol/L) and wnt3a (200 ng/mL). Organoids from PN3 WT SIs were treated with DMSO as a control. (F) The experimental time schedules of organoid treatments in in vitro Cldn7-deleted organoids by 4OH-TAM. (G) Organoids from 3-month-old cCldn7fl/fl-T SIs were treated with 4OH-TAM for 5 days. On the next day after adding 4OH-TAM to induce claudin-7 deletion, claudin-7–deficient organoids were treated with DMSO, CHIR99021 (2 μmol/L), Wnt3a (200 ng/mL), and a combination of CHIR99021 (2 μmol/L) and wnt3a (200 ng/mL) for 4 days. Organoids from 3-month-old control cCldn7fl/fl-W SIs were treated with 4OH-TAM (3 μmol/L) and DMSO as a control. All images are representative of more than 3 independent experiments. Scale bars: 100 μm. Quantification of organoid number per well and area (μm2) per organoid for (H) PN3 WT vs gKO and (I) 3-month-old cCldn7fl/fl-T treated with or without 4OH-TAM. n = 4; 1-way analysis of variance, followed by the Dunnett multiple comparison test; *P < .05; **P < .01; ***P < .001; ****P < .0001, P > .05. Data are represented as means ± SEM. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Because Wnt/β-catenin signaling is essential for maintaining IESCs,23 we investigated whether poor survival and growth of Cldn7-deficient crypts were the result of suppression of Wnt/β-catenin signaling. Exogenous Wnt3a and GSK3β inhibitor CHIR99021 were added into the PN3 crypt culture.11 We found that the number of Cldn7-deficient organoids was increased 6-fold by CHIR99021 and 4-fold by Wnt3a and was close to the control level when combined (Figure 9E and H, left). The organoids grew to their normal size by CHIR99021, 2-fold by Wnt3a, and even larger than control organoids by combining the 2 (Figure 9E and H, right). Importantly, the organoid buddings also were stimulated synergistically, especially when they were treated with both Wnt3a and CHIR99021. Next, we tested whether activation of Wnt/β-catenin signaling could rescue the control organoids with Cldn7 deletion induced in culture. Crypts were isolated from the cCldn7fl/fl-T mice without tamoxifen injection and cultured for 1 day. Cldn7 was deleted in the cCldn7fl/fl-T organoids using 4OH-TAM and then supplemented with CHIR99021 or Wnt3a or both on day 2 (Figure 9F). On day 6, the number of Cldn7-deficient organoids had increased to the control level by CHIR99021, by Wnt3a, or by both of them, while the size of organoids returned to control level by CHIR99021 or by combined treatment (Figure 9G and I).
Our data from both PN3 gKO mice and adult cKO mice collectively show that Cldn7 deletion disrupts IESC self-renewal, and that stimulating the suppressed Wnt/β-catenin signaling in Cldn7-deficient crypts could rescue organoid survival and growth.
Discussion
In this study, we provide in vivo and in vitro evidence that Cldn7 plays an essential role in IESC functions. By sustaining the stable pools of IESCs for epithelial self-renewal and by promoting differentiation into mature epithelial cells, Cldn7 is vital in the process to replenish daily epithelial cell loss at the villi tips.
By using global and inducible, intestinal-specific Cldn7 KO mice, our studies show a unique role of Cldn7 in the intestinal epithelial self-renewal process. Our data show that Cldn7 deletion results in loss of IESCs and impedes intestinal epithelial differentiation (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5). Notably, loss of IESCs is not caused by intestinal inflammation. Instead, it is a direct consequence of Cldn7 deletion, as determined by the following evidence. First, the loss of IESCs was found in PN0 gKO neonatal mice that showed minimal inflammation (Figure 3C). Second, loss of IESCs also was observed in organoid culture after Cldn7 was deleted in vitro (Figure 8). Third, and, most importantly, the ability of intestinal crypts to establish and form budding organoids was impaired in both gKO and cKO mice (Figures 7 and 8).
Interestingly, although Cldn7 deletion led to the loss of IESCs, quiescent stem cell markers were increased significantly in PN3 gKO and adult cKO mice. Studies have reported that intestinal damage or injury can activate the quiescent stem cells to enter the proliferative state for repair and regeneration.6 In addition, proliferative intestinal epithelial cells were seen along the villi in SIs of both PN3 gKO and adult cKO mice. Inflammatory cytokines are known to trigger intestinal repair machinery and stimulate epithelial proliferation.27,28 The intestinal-specific Cldn7 knockout mice reported by Tanaka et al showed inflammation in colons.29 Although inflammatory cytokines were up-regulated in gKO mice12 and cKO (Figure 6A), epithelial hyperproliferation also was observed in the in vitro organoid culture (Figures 7F and 8F), a system that lacks immune cells, suggesting that the hyperproliferative epithelium was a direct consequence of Cldn7 deletion. Our genome-wide microarray analysis showed that the deletion of Cldn7 up-regulated genes related to epithelial hyperplasia and epithelial mesenchymal transition. For example, signal transducer and activator of transcription 3 and AKT signaling pathways were up-regulated in gKO SIs, which could cause epithelial hyperproliferation. Previously, we also reported that knockdown of Cldn7 in human lung cancer cells resulted in increased cell proliferation.30 Future research is needed to unravel the underlying mechanism of how Cldn7 can suppress epithelial overgrowth.
In this study, both gKO and cKO mice showed an impediment to intestinal differentiation. In PN3 mice, Cldn7 deletion suppressed the differentiation of enterocytes and EE cells (Figure 1). Although the total number of goblet cells was decreased, the ratio of goblet cells vs the total epithelial cells in each crypt villus was not affected after loss of Cldn7, indicating that the decrease in total number was owing to the sloughing of villi epithelial cells. This finding was confirmed by the cell types of spheroids derived from Cldn7-deficient crypts. In the adult mice, loss of Cldn7 suppressed all lineage differentiation, including that of Paneth cells and tuft cells, which were not detected until 2 weeks after birth (Figure 5). At least 60% of enterocytes were lost after Cldn7 deletion, which quickly deteriorated the nutrient-absorbing ability of SIs. This is most likely the primary cause of rapid weight loss and early death of Cldn7 KO mice. These data show that Cldn7 is essential for epithelial cell differentiation.
The potential molecular mechanism driving the disruption of intestinal epithelial self-renewal was investigated in the current study. The Wnt signaling pathway was uniquely identified by gene enrichment analysis (Figure 2D), and its target genes all were down-regulated significantly in Cldn7-deficient SIs (Figure 9A–D). The activity of a key molecule, β-catenin, also was down-regulated because phospho-β-catenin was increased significantly in both gKO SIs and cKO crypts (Figure 9B), indicating that more β-catenin was directed for proteasome degradation. The Wnt/β-catenin signaling pathway is essential for IESC survival and its defect will lead to the depletion of IESCs.23 Cldn7 has been linked to the Wnt/β-catenin signaling pathway in the context of colorectal tumorigenicity, in which Cldn7 is suppressed by Tcf4 and Sox9 in aggressive colorectal cancer cells.31 It also has been reported that Cldn7 overexpression enhances β-catenin/Tcf activity and promotes tumor formation in xenograft mice.32 These data suggest that Cldn7 interacts with the Wnt/β-catenin signaling pathway in controlling intestinal epithelial cell differentiation and proliferation.
The strong evidence that the Wnt/β-catenin signaling was the major disrupted pathway came from the in vitro organoid culture. Our data show that activation of Wnt/β-catenin signaling by the GSK-3β inhibitor and Wnt3a effectively can rescue the defect of Cldn7-deficient crypts in that the number and morphology of organoids from Cldn7-deficient crypts were returned to those of the control organoids (Figure 9E and G–I). This finding is significant because it not only confirms that Wnt/β-catenin signaling is the key pathway disrupted in Cldn7 KO SIs, but also identified a vital molecular pathway causing the severe disruption of intestinal epithelial homeostasis in Cldn7 KO mice. Future studies will include the nature of the interaction between Cldn7 and the Wnt/β-catenin signaling pathway and how Cldn7 deletion leads to the suppression of Wnt signaling.
In summary, this study shows that Cldn7 is one of the major factors regulating IESC functions and epithelial self-renewal. By using organoid culture as a powerful in vitro 3-dimensional model, we have identified Wnt/β-catenin signaling as one of the key pathways involved in Cldn7 deletion-mediated intestinal defects. These findings suggest that deletion of a TJ protein can disrupt the function of a biological system and induce a fatal gastrointestinal disorder. The current study provides a therapeutic target for potential intervention and the treatment of intestinal disorders in the future.
Materials and Methods
Global and Conditional Cldn7 Knockout Mice
The generation of global and inducible intestinal-specific Cldn7 knockout mice using a gene-targeting strategy was described previously.12,22 PN0 and PN3 (±1 day) gKO were used in the experiments. The inducible cCldn7fl/fl-T mice were generated by crossing Cre mice (Villin-CreERT2) with Cldn7-flox mice. Three-month-old cCldn7fl/fl-T mice were obtained by injecting 2 mg tamoxifen (catalog number T5648; Sigma-Aldrich, St. Louis, MO) dissolved in sunflower oil (catalog number S5007; Sigma-Aldrich) (10 mg/mL) daily for 4 days (±1 day; cKO). The intestinal tissues then were collected and verified for the successful knockout of Cldn7 before using for various experiments. All animal experiments and procedures were approved by the East Carolina University Animal Care and Use Committee and conducted in compliance with guidelines from the National Institutes of Health and the East Carolina University on laboratory animal care and use.
Intestinal Crypt Isolation and In Vitro Organoid Culture
Small intestines were isolated from PN3 WT and gKO pups, as well as 3-month-old control and cKO mice, and cut open longitudinally. After villi were removed by physical scratching, the intestinal tissues were chopped into 5-mm pieces. After a 30-minute incubation at 4°C in 2 mmol/L EDTA, crypts were freed by continuous handshaking for 2 minutes. The crypt-containing solution was filtered through a 70-μm cell strainer, pelleted at 200 × g for 10 minutes, washed in phosphate-buffered saline, and then resuspended in culture medium. The isolated crypts were mixed with growth factor–reduced Matrigel (catalog number 356230; Corning, Corning, NY), seeded at 10 crypts/μL Matrigel in 48- or 96-well plates. The organoid culture medium was overlaid at a volume of 10:1 to Matrigel in the plates (advanced Dulbecco’s modified Eagle medium/F12 supplemented with 1× penicillin/streptomycin, 10 mmol/L HEPES, 1× GlutaMAX, 1× B27, and 1× N2; all from Life Technologies, Waltham, MA), 50 ng/mL murine recombinant epidermal growth factor (catalog number PMG8041; Life Technologies), 100 ng/mL murine recombinant Noggin (catalog number 250-38; Peprotech, Rocky Hill, NJ), and 500 ng/mL R-spondin 1 (catalog number 3474-RS; R&D system, Minneapolis, MN).
To induce Cldn7 gene deletion in organoid culture, crypts isolated from cCldn7fl/fl-T mice without tamoxifen injection were cultured for 24 hours, followed by the addition of 3 μmol/L 4OH-TAM (catalog number 94873; Sigma), and then cultured for 5 days. The culture medium containing fresh 4OH-TAM was changed every 2 days. For the rescue experiments, the organoid culture was treated with different reagents after the addition of 4OH-TAM for 24 hours. Images of organoids were taken using a Zeiss Axio Imager M2 microscope (Carl Zeiss, Thornwood, NY), equipped with AxioVision imaging software.
Genome-Wide Microarray Data Analysis
For genome-wide gene expression microarray experiments, intestinal fragments (2 cm) from PN3 WT and gKO pups were collected in RNAlater solution (catalog number: AM7020; Thermo Fisher Scientific, Waltham, MA). The total RNA was isolated using the RNeasy mini kit (catalog number: 74104; Qiagen, Valencia, CA) following the manufacturer's instructions. RNA quality was assessed by electrophoresis using Bioanalyzer, and the high-integrity RNA samples were used for microarray hybridization. Microarray profiling was conducted at the Genomics and Bioinformatics Core at the University of North Carolina at Chapel Hill using the Agilent SurePrint GE 8 × 60K mouse whole genome chip (Agilent Technologies, Inc, Santa Clara, CA). Microarray data were background-corrected and normalized using R software (version 3.3.0). Differential expression was tested using an equal variance t test, and P values were transformed to q values using Benjamini and Hochberg’s approach.33 Genes were identified as differentially expressed if the q value was less than 0.05 and absolute fold-change was greater than 2. Heatmaps were created using Treeview software.34 Two group comparisons were performed using Gene Set Enrichment Analysis software (version 3.0.) against the molecular signatures database to identify a differentially expressed gene set (also requiring q < 0.05).35
qRT-PCR
The total RNA was isolated from dissected intestinal tissues from different samples using the RNeasy mini kit (catalog number: 74104; Qiagen), and the concentration was measured using spectrophotometry. The complementary DNA synthesis was performed using high-capacity complementary DNA reverse-transcription kits (catalog number: 4374966; Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Real-time qRT-PCR reactions were performed using the RT2 Profiler PCR Array system (catalog number: PAMM-143Z; Qiagen). Primers used for real-time qRT-PCR are listed in Table 2. Samples with a glyceraldehyde-3-phosphate dehydrogenase value less than 30 cycle threshold (Ct) value were considered acceptable for analysis. The relative changes in gene expression from real-time qRT-PCR were analyzed using the 2−ΔΔCt method.12
Table 2.
Oligonucleotide Primers
| Primer | Forward sequence (5’-3’) | Reverse sequence (5’-3’) |
|---|---|---|
| FABP-1 | ATGAACTTCTCCGGCAAGTACC | CTGACACCCCCTTGATGTCC |
| ChA | CAAGCTAGGAGCCCCCATCTA | CACTCTGGCATTTCCATTACT |
| Cldn7 | TTCATTGTGGCAGGTCTTG | CCGATAAAGATGGCAGGT |
| GAPDH | CTTTGTCAAGCTCATTTCCTG | TCTTGCTCAGTGTCCTTGC |
| Hopx | GAGGACCAGGTGGAGATCCT | TCCGTAACAGATCTGCATTCC |
| Lgr5 | ACCCGCCAGTCTCCTACATC | GCATCTAGGCGCAGGGATTG |
| Muc2 | ATGCCCACCTCCTCAAAGAC | GTAGTTTCCGTTGGAACAGTGAA |
| Olfm4 | TGGCCCTTGGAAGCTGTAGT | ACCTCCTTGGCCATAGCGAA |
| Prom1 | CACTCCTGACTGAAACACCAAAGC | TGCCATCCAGGTCTGAGAATGC |
| Sox9 | CAGACCAGTACCCGCATCTG | CTCCGCTTGTCCGTTCTTCA |
| IL1 | ACGGACCCCAAAAGATGAAG | TTCTCCACAGCCACAATGAG |
| IL6 | CAAAGCCAGAGTCCTTCAGAG | GTCCT AGCCACTCCTTCTG |
| IL8 | TCTTCCAGTTCAACCAGCC | ATCCACCTTGAATTCTCCCATC |
| TNF-α | GACCCTCACACTCAGATCAT | GAGATCCATGCCGTTGG |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Muc2, mucin 2; TNF-α, tumor necrosis factor-α.
FISH
The mRNA localization was detected by the ViewRNA ISH tissue 1-Plex assay kit (catalog number: VB1-10818; Affymetrix, Waltham, MA), or the RNAscope 2.5 HD detection kit (catalog number: 322300; ACD, Newark, CA) according to the manufacturer's instructions. Briefly, intestinal tissue sections prepared for immunohistochemistry were used for FISH. Sections were pretreated in a 94°C water bath for 15 minutes to allow for the unmasking of mRNA and probe accessibility. The endogenous alkaline phosphatase was inactivated using 0.2 mol/L HCl/300 mmol/L NaCl for 15 minutes, followed by its fixation in 4% paraformaldehyde. The target mRNAs were hybridized to specific labeled probes for 3 hours. The probe (Mm-Olfm4, catalog number: 311831; ACD) hybridization and amplification were performed at 40°C. The signal amplification was conducted via a series of sequential hybridization steps to achieve 3000-fold amplification per target mRNA.
TUNEL Assay
Apoptotic cells in the organoids were assessed using the In-Situ Cell Death Detection Kit (catalog number: 12156792910; Roche, Mannheim, Germany). Briefly, organoids grown in Matrigel were fixed with 4% paraformaldehyde for 24 hours at 4°C and then permeabilized with 0.5% Triton X-100 in phosphate-buffered saline before detection using the TUNEL reaction according to the manufacturer’s protocol. DNA strand breaks were labeled by terminal deoxynucleotidyl transferase, which catalyzed the polymerization of labeled nucleotides to free 3’-OH DNA ends in a template-independent manner. The negative control without terminal deoxynucleotidyl transferase and positive control with DNase I recombinant (Qiagen) were included in the experiments. Whole-mount imaging of organoids was performed using a Zeiss Axiovert S100 microscope equipped with MetaMorph software (Molecular Devices, San Jose, CA).
Statistical Analysis
Statistical analysis was performed using either Origin8 (OriginLab, Northampton, MA) or GraphPad Prism (GraphPad Software, San Diego, CA). Differences between the 2 groups were analyzed using the unpaired Student t test. One-way analysis of variance, followed by the Dunnett multiple comparison test, was performed when more than 2 groups were involved. A P value less than .05 was considered significant.
Acknowledgment
The authors would like to thank Dr Scott Magness’s laboratory at the University of North Carolina at Chapel Hill for helping set up the intestinal organoid culture, and Dr Hemant Kelkar from the University of North Carolina Center for Bioinformatics for helping with the ingenuity pathway analysis. The authors also thank Joani Zary Oswald, Rodney Tatum, and Beverly G. Jeansonne for their technical assistance.
Footnotes
Author contributions Tiaosi Xing performed experiments, collected and analyzed data, and wrote the first draft of the manuscript; Lesley Jasmine Benderman, Stephiya Sabu, and Jeffrey Yang performed experiments and collected data; Joel Parker conducted the bioinformatic analysis of microarray data; Qun Lu conceptualized the study and edited the manuscript; Lei Ding designed and supervised the study and edited the manuscript; and Yan-Hua Chen designed and supervised the study, interpreted data, and finalized the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Institutes of Health grant DK103166 (Y.-H.C.) and National Natural Science Foundation of China grant 81772557(L.D.).
Contributor Information
Lei Ding, Email: dinglei1005@126.com.
Yan-Hua Chen, Email: cheny@ecu.edu.
References
- 1.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 4.van der Flier L.G., Haegebarth A., Stange D.E., van de Wetering M., Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., van der Flier L.G., Cuppen E., van Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Formeister E.J., Sionas A.L., Lorance D.K., Barkley C.L., Lee G.H., Magness S.T. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108–G1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., Sangiorgi E., Capecchi M.R., Kuo C.J. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., Myant K., Versteeg R., Sansom O.J., van Es J.H., Barker N., van Oudenaarden A., Mohammed S., Heck A.J., Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Nusse R., Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Ding L., Lu Z., Foreman O., Tatum R., Lu Q., Renegar R., Cao J., Chen Y.H. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology. 2012;142:305–315. doi: 10.1053/j.gastro.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing T., Camacho Salazar R., Chen Y.H. Animal models for studying epithelial barriers in neonatal necrotizing enterocolitis, inflammatory bowel disease and colorectal cancer. Tissue Barriers. 2017;5 doi: 10.1080/21688370.2017.1356901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen S.J., Ang L.H., Zheng Y., Karahan S.N., Wu J., Wang Y.E., Caron T.J., Gad A.P., Muthupalani S., Fox J.G. Loss of tight junction protein claudin 18 promotes progressive neoplasia development in mouse stomach. Gastroenterology. 2018;155:1852–1867. doi: 10.1053/j.gastro.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B., Flodby P., Luo J., Castillo D.R., Liu Y., Yu F.X., McConnell A., Varghese B., Li G., Chimge N.O., Sunohara M., Koss M.N., Elatre W., Conti P., Liebler J.M., Yang C., Marconett C.N., Laird-Offringa I.A., Minoo P., Guan K., Stripp B.R., Crandall E.D., Borok Z. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J Clin Invest. 2018;128:970–984. doi: 10.1172/JCI90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce S.C., Al-Jawadi A., Kishida K., Yu S., Hu M., Fritzky L.F., Edelblum K.L., Gao N., Ferraris R.P. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 2018;16:19. doi: 10.1186/s12915-018-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bry L., Falk P., Huttner K., Ouellette A., Midtvedt T., Gordon J.I. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci U S A. 1994;91:10335–10339. doi: 10.1073/pnas.91.22.10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerbe F., Legraverend C., Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69:2907–2917. doi: 10.1007/s00018-012-0984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tetteh P.W., Farin H.F., Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25:100–108. doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Snippert H.J., van Es J.H., van den Born M., Begthel H., Stange D.E., Barker N., Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–2194 e1. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Anholt R.R. Olfactomedin proteins: central players in development and disease. Front Cell Dev Biol. 2014;2:6. doi: 10.3389/fcell.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W.J., Xu C., Wang K., Li T.Y., Wang X.N., Yang H., Xing T., Li W.X., Chen Y.H., Gao H., Ding L. Severe intestinal inflammation in the small intestine of mice induced by controllable deletion of claudin-7. Dig Dis Sci. 2018;63:1200–1209. doi: 10.1007/s10620-018-4973-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fevr T., Robine S., Louvard D., Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., Stange D.E., van Es J.E., Guardavaccaro D., Schasfoort R.B., Mohri Y., Nishimori K., Mohammed S., Heck A.J., Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 25.Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W., Li H., Hong S.H., Piszczek G.P., Chen W., Rodgers G.P. Olfactomedin 4 deletion induces colon adenocarcinoma in Apc(Min/+) mice. Oncogene. 2016;35:5237–5247. doi: 10.1038/onc.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Hernandez V., Quiros M., Nusrat A. Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci. 2017;1397:66–79. doi: 10.1111/nyas.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka H., Takechi M., Kiyonari H., Shioi G., Tamura A., Tsukita S. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut. 2015;64:1529–1538. doi: 10.1136/gutjnl-2014-308419. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z., Kim do H., Fan J., Lu Q., Verbanac K., Ding L., Renegar R., Chen Y.H. A non-tight junction function of claudin-7-interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol Cancer. 2015;14:120. doi: 10.1186/s12943-015-0387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darido C., Buchert M., Pannequin J., Bastide P., Zalzali H., Mantamadiotis T., Bourgaux J.F., Garambois V., Jay P., Blache P., Joubert D., Hollande F. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68:4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 32.Bhat A.A., Pope J.L., Smith J.J., Ahmad R., Chen X., Washington M.K., Beauchamp R.D., Singh A.B., Dhawan P. Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene. 2015;34:4570–4580. doi: 10.1038/onc.2014.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochberg Y.B. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:12. [Google Scholar]
- 34.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]