Summary
Given the critical role that the immune system plays in a multitude of diseases, having a clear understanding of the pharmacology of the immune system is crucial to new drug discovery and development. Here we describe the International Union of Basic and Clinical Pharmacology (IUPHAR) Guide to Immunopharmacology (GtoImmuPdb), which connects expert‐curated pharmacology with key immunological concepts and aims to put pharmacological data into the hands of immunologists. In the pursuit of new therapeutics, pharmacological databases are a vital resource to researchers through providing accurate information on the fundamental science underlying drug action. This extension to the existing IUPHAR/British Pharmacological Society Guide to Pharmacology supports research into the development of drugs targeted at modulating immune, inflammatory or infectious components of disease. To provide a deeper context for how the resource can support research we show data in GtoImmuPdb relating to a case study on the targeting of vascular inflammation.
Keywords: database, immune‐therapeutics, immunopharmacology, pharmacology
The International Union of Basic and Clinical Pharmacology (IUPHAR) Guide to Immunopharmacology (GtoImmuPdb) connects expert‐curated pharmacology with key immunological concepts and aims to put pharmacological data into the hands of immunologists. Pharmacological databases are a vital resource to researchers through providing accurate information on the fundamental science underlying drug action. This extension to the existing IUPHAR/BPS Guide to Pharmacology supports research into the development of drugs targeted at modulating immune, inflammatory or infectious components of disease.

Abbreviations
- BPS
British Pharmacological Society
- CANTOS
Canakinumab Anti‐inflammatory Thrombosis Outcomes Study
- CD
cluster of differentiation
- CIRT
Cardiovascular Inflammation Reduction Trial
- CLL
chronic lymphocytic leukaemia
- COX
cyclooxygenase
- CRP
C‐reactive protein
- CVD
cardiovascular disease
- FDA
US Food and Drug Administration
- GO
Gene Ontology
- GtoImmuPdb
Guide to Immunopharmacology
- GtoPdb
Guide to Pharmacology
- HLA‐E
human leucocyte antigen E
- IL
interleukin
- INN
International Nonproprietary Names
- IRAK
interleukin‐1 receptor‐associated kinase
- IUIS
International Union of Immunological Science
- IUPHAR
International Union of Basic and Clinical Pharmacology
- MAS
macrophage activation syndrome
- MI
myocardial infarction
- NAR
Nucleic Acids Research
- NC‐IUPHAR
Nomenclature Committee of the International Union of Basic and Clinical Pharmacology
- NIH
National Institutes of Health
- NLRP3
NOD‐like receptor family 3
- NOD
nucleotide‐binding and oligomerization domain
- NSAIDs
non‐steroidal anti‐inflammatory drugs
- OMIM
Online Mendelian Inheritance in Man
- PCSK9
proprotein convertase subtilisin/kexin type 9
- SID
PubChem substance record ID
- WHO
World Health Organization
Introduction
The immune system has become a major target for new therapeutics, with approximately 20% of new drug approvals in the last 5 years targeting elements of the immune system (http://www.fda.gov/drugs/development-approval-process-drugs/new-drugs-fda-cders-new-molecular-entities-and-new-therapeutic-biological-products). A high proportion of diseases are also associated with an immune or inflammatory component or process. In particular, chronic age‐related diseases such as Alzheimer's disease, atherosclerosis and diabetes have inflammatory components.1, 2, 3, 4, 5 The significant roles that inflammation and immune mechanisms play in cardiovascular disease (CVD) have also made them potential therapeutic targets in its treatment.6 Autoimmunity is a serious problem, for example in multiple sclerosis,7, 8 Sjögren's syndrome,9 inflammatory bowel disease10, 11 and rheumatoid arthritis.12 Such conditions may coexist with depressive disorders.13 There is also much interest in the use of immune therapies, such as the potential exploitation of dendritic cells, to treat cancer.14, 15
The International Union of Basic and Clinical Pharmacology (IUPHAR) and the British Pharmacological Society (BPS) collaborate on the development and maintenance of the Guide to Pharmacology (GtoPdb, http://www.guidetopharmacology.org). This database is an expert‐curated resource of ligand‐activity‐target relationships, selected from high‐quality pharmacological and medicinal chemistry literature. It has its origins in IUPHAR‐DB and the BPS Guide to Receptors and Channels, both of which focused on receptors and channels.16, 17, 18 The scope of GtoPdb has expanded over the years19, 20, 21, 22 and a Wellcome Trust‐funded project has allowed us to address the priority area of immunity, inflammation and infection.23, 24, 25, 26 In the course of that project, the database has expanded into the field of immunopharmacology.21
Well‐curated pharmacological databases are an important foundation for research on new therapeutics. In the context of immunopharmacology, although there are good internet resources that support purely immunological research, for example Immunopaedia (http://www.immunopaedia.org.za), ImmPort (http://www.immport.org), ImmGen (http://www.immgen.org), InnateBD (http://www.innatedb.com) and IMGT (http://www.imgt.org), none cover the pharmacology of the immune system. The IUPHAR Guide to Immunopharmacology (GtoImmuPdb; http://www.guidetoimmunopharmacology.org) has been developed to deliver a knowledge‐base that, for the first time, connects immunology with pharmacology.27 It expands the data associated with targets and ligands to cover immunological data types and enhances access to the pharmacological data through a user interface tailored to the immunologist. GtoImmuPdb puts valuable pharmacological data into an immunological context and is a resource that enables researchers to easily identify pharmacological agents that can be used experimentally to modulate immune system mechanisms.
The IUPHAR/BPS Guide to Pharmacology
The Guide to Pharmacology holds data on nearly 3000 human proteins, with over 1700 of these ‘targets’ having curated pharmacological interaction data. In total, the database has information on over 9700 ligands, and it contains quantitative data on over 14 000 ligand–target interactions. The selection of content is supported through the expertise of 96 target family subcommittees of the Nomenclature Committee of IUPHAR (NC‐IUPHAR), comprising over 500 scientists worldwide. GtoPdb uses expert human judgement at all stages of curation, in contrast to more automated data and text mining approaches. Curation is not though limited to only the NC‐IUPHAR subcommittees; we also encourage users to make suggestions about content, which when checked often results in appropriate additions or qualifications.
The GtoPdb is a well‐used and highly cited resource. Our analytics show that the database is accessed by over 22 000 users worldwide each month and they generate a total of more than 118 000 page views. We produce two main biennial publications. The most prominent of these is the Concise GtoPdb,28 which provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology. The last two editions (2015/1629 and 2017/1830) combined have over 2600 citations. We also produce a biennial publication in Nucleic Acids Research (NAR) Database Issue, which documents database and curatorial updates. Our 201620 and 201821 papers have been cited over 1460 times.
The IUPHAR Guide to Immunopharmacology: development and curation
In establishing the GtoImmuPdb, NC‐IUPHAR expert subcommittees identified targets relevant to immunopharmacology, and they provided detailed curatorial comments on the reasons for their inclusion in the resource. In the 2019.5 database release, 614 targets and 1232 ligands were tagged as relevant to immunopharmacology (http://www.guidetoimmunopharmacology.org//immuno/immunoHelpPage.jsp#gtoimmupdb_content).
Curating GtoImmuPdb data
The first phase of curation involved assessing protein targets and ligands that were already in the GtoPdb for inclusion in the GtoImmuPdb. To extend coverage beyond what was already in GtoPdb, we made use of the Gene Ontology (GO) ‘biological process’ annotations to prioritize targets for curation. We produced a draft list of targets for inclusion in GtoImmuPdb on the basis of both direct involvement in inflammation/immunity and based on involvement in processes known to be important in inflammation/immunity. Ligands for targets that qualified for GtoImmuPdb were then reviewed and included if there was evidence that their activity has a modulatory effect on the inflammation/immune system (e.g. drugs approved to treat inflammatory conditions, or tool compounds used to investigate GtoImmuPdb targets). The selection of content for curation was supported by the NC‐IUPHAR subcommittees, who identified key papers and literature reviews. Examples of inclusions identified at this stage are histamine receptors (http://www.guidetoimmunopharmacology.org/GRAC/FamilyDisplayForward?familyId=33)31 and anti‐histamine drugs, glucocorticoid receptor (http://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=625)32 and anti‐inflammatory glucocorticoid drugs, cyclooxygenase enzymes (http://www.guidetoimmunopharmacology.org/GRAC/FamilyDisplayForward?familyId=269)33 and non‐steroidal anti‐inflammatory drugs, and pattern recognition, cytokine and chemokine receptor families. Examples of new content added during this phase include additional families of pattern recognition receptors34 and immune checkpoint proteins, and ligands and immune checkpoint inhibitors (clinical and investigational) used in immuno‐oncology.
Targets and ligands continue to be added to the GtoImmuPdb as new evidence emerges. Ongoing updates are driven by systematic searches of current literature covering immunology and inflammation to identify lead compounds, their molecular targets and pharmacological data. Other useful resources include pharmaceutical companies' declared development programmes and selective patent analysis that can be used to identify pharmacological data in the absence of peer‐reviewed publications. Review of clinical trial registries, applications to the World Health Organization for new International Nonproprietary Names (INN) (which provide an indication of developments in the immunity/inflammation/immuno‐oncology fields), and monitoring new drug approvals can all identify novel ligands, protein targets and molecular mechanisms of action.
Immunological processes and cell types
The data on targets and ligands have been extended by annotating these with immunological data. This means we have made clear connections between immunological processes, cell types and disease, and the targets and ligands already in the database. We have made use of biological ontologies because they provide an organized, hierarchical and controlled vocabulary against which to annotate data. Ontologies also provide unique accession numbers that identify a particular term, and these are valuable in supporting interoperability between data resources. In the context of GtoImmuPdb, they are also useful in curating protein targets to different categories and in enabling inferred searching. We have used biological processes from the GO35, 36 (http://geneontology.org) and cell types from the Cell Ontology37 (http://obofoundry.org/ontology/cl.html).
The GO is a hierarchical ontology that describes biological processes, including processes that operate in the immune and inflammatory systems.38, 39 GtoImmuPdb uses top‐level process categories, such as T cell (activation) or Cytokine production and signalling, underpinned by GO immune and inflammatory process terms. In the case of T‐cell activation, this includes terms such as ‘T‐cell‐mediated immunity (GO:0002456)’ and ‘regulation of T‐cell differentiation (GO:00045580)’.
The Cell Ontology is designed as a structured vocabulary for cell types, from prokaryotes to mammals. In a similar way, GtoImmuPdb uses top‐level cell type categories, such as Mast cells, because of their relevance in anti‐allergic therapies,40 and Innate lymphoid cells, reflecting the growing understanding of their role within the innate immune system in the control of tissue homeostasis, infection, inflammation, metabolic disease and cancer.41, 42 The top‐level categories are underpinned by Cell Ontology terms, which in the case of Mast cells includes the terms ‘mast cell (CL_0000097)’ and its children, ‘mucosal type mast cell (CL_0000485)’ and ‘connective tissue type mast cell (CL_0000484)’.
Table 1 shows associations between the top‐level processes and the number of human immunopharmacological target proteins. The table also shows the number of human target proteins relevant to immunopharmacology associated with the top‐level cell types. More details of how data have been curated can be found in our recent publication.21
Table 1.
GtoImmuPdb Process and Cell Type categories and the number of human proteins associated with each group
| Process | Annotated human targets | Cell type | Annotated human targets |
|---|---|---|---|
| Barrier integrity | 49 | B cells | 51 |
| Inflammation | 633 | Dendritic cells | 41 |
| Antigen presentation | 142 | Granulocytes | 46 |
| T cell (activation) | 196 | Innate lymphoid cells | 6 |
| B cell (activation) | 161 | Macrophages | 56 |
| Immune regulation | 503 | Mast cells | 39 |
| Tissue repair | 19 | Natural killer cells | 26 |
| Immune system development | 251 | Other T cells | 3 |
| Cytokine production and signalling | 504 | Stromal cells | 1 |
| Chemotaxis and migration | 256 | T cells | 76 |
| Cellular signalling | 476 |
The IUPHAR Guide to Immunopharmacology: accessing the data
The GtoImmuPdb portal allows researchers with a primarily immunological background to find pathways, drugs and targets using an interface built around an immunological perspective. Immunological processes, cell types, pathways and diseases are centre‐stage and connect to search functions that prioritize immunologically relevant pharmacological data. This provides rapid access to lists of targets and ligands relevant to immunopharmacology or allows the viewing of lists of targets and ligands associated with immunological processes, cell types and diseases. In this way, GtoImmuPdb equips immunologists with a means to discover pharmacological agents that are useful in their research and provides a foundation for developing research into therapeutic modifiers of the immune system.
Navigating the database from a starting point of immunological process of cell type
The database contains nearly 200 targets associated with T‐cell activation (Table 1). These can be easily accessed via the processes panel on the GtoImmuPdb portal (Fig. 1a). The targets are organized into sections, one for each target class. Figure 2(b) shows how some cluster of differentiation (CD) molecule targets are displayed in the Other Protein Targets section. The GO terms annotated to a target are shown in the third column of Fig. 1(b); summarized curatorial comments are also displayed. In the example of CD28, its role in the activation, proliferation and survival of T cells is indicated. By clicking on the target name, users can view the detailed targets page, which contains the expanded curators' comments and full pharmacological information on the target.
Figure 1.

Browsing for targets associated with an immunological process. The GtoImmuPdb portal is shown in (a), with the Processes panel linking to lists of targets associated with T‐cell activation (b). Under the Other Proteins section (c) cluster of differentiation (CD) molecule targets are listed, and in the example of CD28, curatorial comments indicate its role in the activation, proliferation and survival of T cells.
Figure 2.

Pharmacological data associated with an immunological cell type. The example shows linking from the portal via the cell type category of Natural killer (NK) cells. The resulting list of targets associated with NK cells includes CD159a. Selecting the link through to the detailed view page shows CD159a interaction with the antibody monalizumab, an anti‐NKG2A clinical lead for haematological cancer.
The annotation of targets to cell types helps to highlight useful pharmacological data relevant to immunopharmacology. For example, the role of natural killer cells in anti‐tumour immunity is well established,43, 44 and the heterodimer CD94/NKG2A is known to have a role in recognition of the main type of human leucocyte antigen class I molecules and functions as a true checkpoint in natural killer cell activation.45 NKG2A (GtoPdb Target 2849; CD159a) is annotated in GtoImmuPdb as being expressed by cells in the natural killer cells category, with the immunopharmacology commentary highlighting its role as an inhibitory checkpoint receptor for human leucocyte antigen E. The detailed view for CD159a (http://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=2849#Antibodies) shows interaction data for the antibody monalizumab (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=summary%26ligandId=8323), an anti‐NKG2A clinical lead molecule that is being developed for solid and haematological cancers (Fig. 2).
Ligand summaries
For ligands, the database contains key information on the biological activity, clinical use, molecular properties, structure and immunopharmacology. These data are displayed on the ligand summary pages, which are easily accessed, either from the Ligands menu bar item, or the Ligands panel on the home page (Fig. 3a). Different categories of ligand can be selected from tabs at the top of the page. When navigating from the GtoImmuPdb portal, the lists contain ligands tagged in the database as relevant to immunopharmacology. Selecting a ligand links through to the ligand summary page where data are organized under several tabs (Fig. 3b). The Immunopharmacology tab contains curator comments on a compound's relevance to immunopharmacology, as well as listing any disease associations. The Summary tab gives general information about the compound, including if the drug is approved for clinical use, and provides a list of trade names (when used clinically), synonyms (such as preclinical names) and INN so that the drug can be identified and tracked in the literature. The ‘Biological Activity’ tab as well as displaying tables of the ligand selectivity at targets in the database also provides access to the Ligand Activity Visualization Tool (Fig. 3c). This tool provides box plots summarizing all the activity data for a ligand taken from ChEMBL46 and GtoPdb across multiple species. The Clinical data tab provides information about molecular mechanisms of action and clinical trials together with trial identifying numbers.
Figure 3.
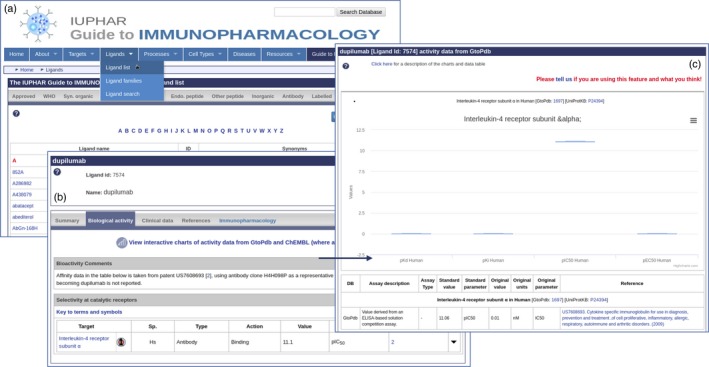
Ligand summary pages. (a) List of ligands is accessed from the menu bar. (b) clicking on a ligand name links to the ligand summary page, here showing dupilumab. Data are presented under several tabs, including one specific to immunopharmacology. Users can link through to the ligand activity visualization tool (c), to compare activities across species.
Disease summaries
The extension for immunopharmacology has also prioritized the development of pages that give consolidated pharmacological summaries for different diseases. In all, there are over 1000 diseases in the GtoPdb that have curated associations with targets and/or ligands. The disease lists, accessed from the portal or menu bar, summarize these (http://www.guidetoimmunopharmacology.org/GRAC/DiseaseListForward?type=Immuno). As a consequence of our recent curatorial focus on immunological data, diseases with significant immunological aspects, such as asthma, rheumatoid arthritis, inflammatory bowel and psoriasis, show the greatest number of associations with targets and/or ligands.
The disease summary pages show targets and ligands associated with a disease and include links to Online Mendelian Inheritance in Man47, 48 (http://omim.org), Orphanet (http://orpha.net) and the Disease Ontology49 (http://disease-ontology.org), providing cross‐references between the diseases in GtoImmuPdb and other resources (http://www.guidetoimmunopharmacology.org/GRAC/DiseaseListForward?type=Immuno). The pages also detail the bioactivities and clinical uses of relevant ligands. For example, chronic lymphocytic leukaemia (http://www.guidetopharmacology.org/GRAC/DiseaseDisplayForward?diseaseId=218; Fig. 4) highlights CD20 as being the molecular target of four antibodies: ofatumumab, veltuzumab, rituximab and obinutuzumab. The summarized view shows these antibodies listed against their molecular target and combines this with detailed disease, clinical use and bioactivity comments (Fig. 4). In the case of rituximab, the pages not only explain its role in treating CD20‐positive non‐Hodgkin's lymphoma and chronic lymphocytic leukaemia, but highlight its role in several other autoimmune conditions and in the suppression of antibody‐mediated organ rejection.50, 51
Figure 4.
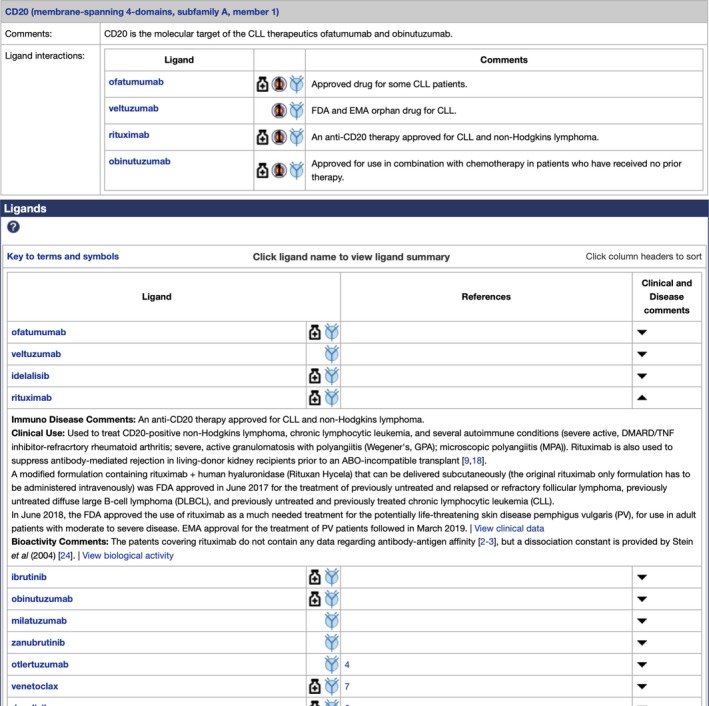
An example disease summary page illustrating chronic lymphocytic leukaemia (CLL; http://www.guidetopharmacology.org/GRAC/DiseaseDisplayForward?diseaseId=218). Four antibodies are highlighted, all of which are therapeutics for CLL, that target CD20. The ligands section provides extended curatorial commentary on the clinical use and bioactivity of the compounds.
Immunopaedia
Through the partnership between IUPHAR and the International Union of Immunological Sciences (IUIS) to create standard tools and nomenclature (https://iuis.org/news/2018-iuis-council-meeting-summary/), GtoImmuPdb has been working in collaboration with the IUIS resource, Immunopaedia (http://www.immunopaedia.org.za). Immunopaedia provides materials for teaching and learning immunology, from the basic immune system to advanced immunology and specialized focus areas. They are an official provider for online resources for the IUIS, creating and hosting online courses to educate and support participants before and after immunology conferences worldwide. We have undertaken to provide links from key ligands in GtoImmuPdb to the rich and detailed clinical case studies hosted by Immunopaedia (Fig. 5).
Figure 5.

Example of ligand summary page links to relevant Immunopaedia clinical case studies. The illustrated link shows that the antibody rituximab was used in the chemotherapy treatment of a case of lymphadenopathy.
Searching, web services and PubChem
The search mechanisms across the website have been extended, such that the new immunological data types are incorporated. The search algorithm itself has been tailored so that when using the GtoImmuPdb URL, results of immunological relevance are upweighted. The immunological relevance of a target or ligand is determined by the amount of immunological data associated with it in the database. Our application programming interface has also been extended to incorporate parameters to retrieve immunopharmacology tagged data.
The GtoPdb maintains strong connectivity with PubChem, the open chemistry database at the National Institutes of Health.52 On each database release of GtoPdb, we submit our chemical structures to PubChem. As part of this process, we include Depositor Comments in the substance records that we submit to PubChem. These comments, among other things, indicate if a structure is part of GtoImmuPdb and contains any immunopharmacology curatorial comments. Described in more detail in our most recent NAR paper,22 the inclusion of these comments in our PubChem submissions make it possible to run domain‐specific queries related to immunopharmacology when searching via PubChem.
Case study: targeting vascular inflammation
The best way to illustrate the potential usefulness of GtoImmuPdb is through a case study. We have chosen vascular inflammation, because in the last three decades, experimental data have clearly shown the causal role played by immune and inflammatory responses in the initiation and development of atherosclerosis, and in the regulation of plaque instability.6 Epidemiological studies have also called attention to vascular inflammation. To date, however, there is no immunomodulatory treatment in routine use for prevention of atherosclerosis.53 How might GtoImmuPdb help to change this?
The Canakinumab Anti‐inflammatory Thrombosis Outcomes Study (CANTOS; http://clinicaltrials.gov/ct2/show/NCT01327846)53, 54 has been the first large (>10 000), randomized, double‐blind, placebo‐controlled trial to target the inflammatory cytokine interleukin, interleukin‐1β (IL‐1β) (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=biology%26ligandId=4974) for secondary prevention (to reduce the number of new or severe cases of the disease) of atherosclerosis. In CANTOS, the human monoclonal antibody canakinumab (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=immuno%26ligandId=6773) significantly reduced the rate of a composite end‐point of major cardiovascular events in patients previously affected by myocardial infarction (MI) and who had high levels of C‐reactive protein (CRP).53 The CANTOS trial represents the first clinical evidence that targeting inflammation may be a viable approach in atherosclerosis and it started an important discussion on how to target vascular immune‐inflammatory responses in the most efficient way (which might not be by targeting IL‐1β).
The CANTOS was followed by the Cardiovascular Inflammation Reduction Trial (CIRT; http://clinicaltrials.gov/ct2/show/NCT01594333).54, 55 In CIRT, treatment with low‐dose methotrexate failed to reduce cardiovascular event rates in patients with previous multi‐vessel coronary artery disease or MI also affected by metabolic syndrome or type 2 diabetes.55 It should be noted that despite co‐morbidities, CIRT patients had normal CRP levels and therefore had not been selected on the basis of residual inflammatory risk. Given that high levels of CRP are associated with an increased risk of cardiovascular events, this may help to explain the difference between the CANTOS and CIRT results. In fact, in post hoc observations within CANTOS, patients with the largest reduction in IL‐6 and CRP in response to IL‐1β inhibition56 showed the greatest reduction in cardiovascular mortality, whereas methotrexate had no effect on circulating inflammatory mediators in CIRT. It is worth noting that typing the name of a clinical trial into the main search box on GtoImmuPdb returns a list of ligands involved in the trial, where these data have been curated.
We have learned to a great extent from both trials, but we still have a long way to go before anti‐inflammatory therapies become standard care in the treatment of CVD.57
Canakinumab is an expensive agent and it is very unlikely that it will be used in CVD prevention. Several further directions may be investigated, and the first clear opportunity is represented by the targeting of mediators that sit either just above or below IL‐1β. More recent analysis from the CANTOS trial revealed that there remains substantial residual inflammatory risk related to both IL‐18 and IL‐6 after IL‐1β inhibition.58 Therefore, targeting IL‐18 or IL‐6 signalling59 could be a way forward. These present us with several questions – can we find a good way to target IL‐18 or IL‐6? Can GtoImmuPdb help in finding a good way to modulate either of these molecules?
Accessing ligand summaries for IL‐6 and IL‐18
To access information about IL‐6 in GtoImmuPdb, go to the portal and type IL‐6 into the database search at the top of any page. IL‐6 is the top‐hit from this search and clicking on the ligand name links through to its ligand summary page. Ligand summary pages can also be accessed by browsing via the Ligand menu bar item, either via Ligand List (alphabetical; Fig. 1), or Ligand Families, which has several groupings of ligands, including one for interleukins, where IL‐6 can be found (Fig. 6).
Figure 6.
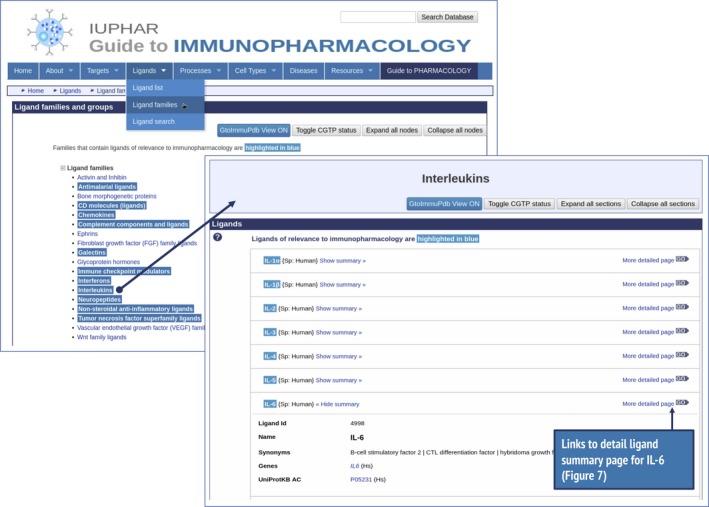
Illustrates accessing ligand summary data using interleukin‐6 (IL‐6) as an example. Browsing via the menu bar for ligand families, selecting the Interleukins group, opens the link through to the Interleukins group. Users can then link through from these points to the IL‐6 ligand summary page (Fig. 7).
Information on IL‐6 is contained under several tabs on the ligand summary page (Fig. 7). Figure 7(a) shows information under the immunopharmacology tab, highlighting its pro‐inflammatory and anti‐inflammatory effects and indicating its role in the treatment of rheumatoid arthritis. Figure 7(b) shows biological activity data, which list ligands with which IL‐6 interacts, including binding affinity data and indications of whether the ligands are approved drugs, as is the case for siltuximab. As a starting point when considering a way to potentially target IL‐6, these pharmacological data and immunological context are helpful. They show that IL‐6 is already a validated drug target and a primary target of three ligands including the approved drug siltuximab.
Figure 7.
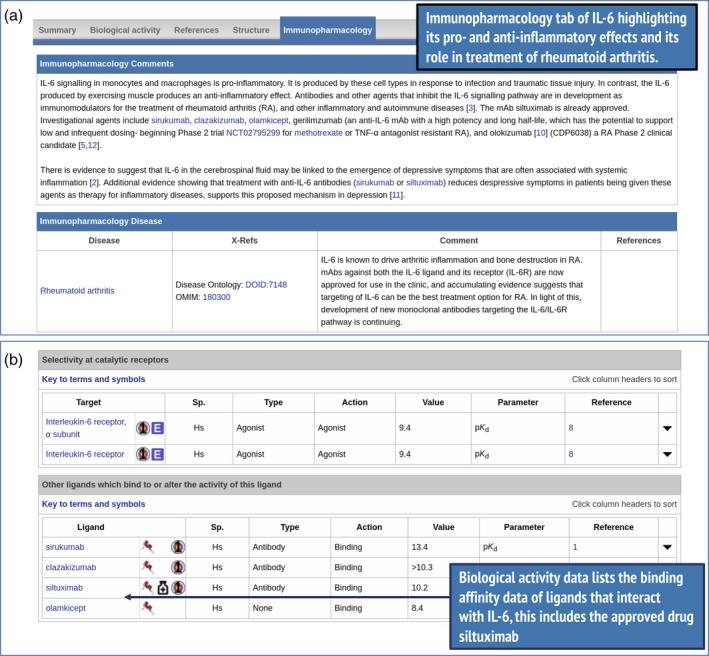
Highlights from the interleukin‐6 (IL‐6) ligand summary page (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?ligandId=4998). Curator's comments (a) are shown under the immunopharmacology tab and indicate links with rheumatoid arthritis. Parts of the biological activity tab (b), show ligands that interact with IL‐6, including the approved drug siltuximab.
Similarly, information on IL‐18 can be accessed in the same way. Figure 8 shows some of the highlights from the IL‐18 ligand summary page, including tadekinig α, a peptide ligand that binds to and inhibits the pro‐inflammatory activity of IL‐18 and has US Food and Drug Administration orphan drug designation for the treatment of macrophage activation syndrome. This is useful pharmacological information and context for further investigation of targeting IL‐18.
Figure 8.
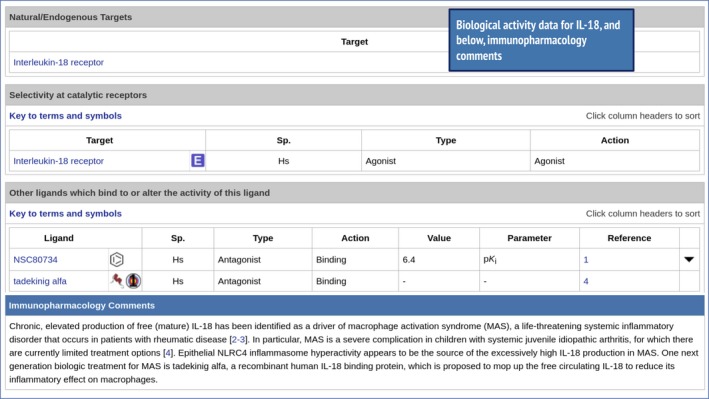
Showing the biological activity and immunopharmacology data from the interleukin‐18 (IL‐18) ligand summary page (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?ligandId=4983). Interaction with the tadekinig α peptide is highlighted, which plays a role in reducing the inflammatory effect of IL‐18.
Accessing immunopharmacology data for NLRP3 and PCSK9
Targeting nucleotide‐binding and oligomerization domain (NOD) ‐like receptor family 3 (NLRP3) inflammasome inhibitors that can inhibit both IL‐1β and IL‐1860 may also present a viable way forward. In this regard, using GtoImmuPdb to view the detailed target page for NLRP3 may be helpful. It is possible to use the direct search to find NLRP3, but it can also be found by browsing through the Catalytic Receptors targets, where NLRP334 is found under the pattern recognition receptors and NOD‐like receptor subfamilies. Figure 9 shows inhibitors and immunopharmacology comments from the NLRP3 detailed target page. Two of the three ligands, CY‐09 and MCC950, have quantitative interaction data for NLRP3 and all three are indicated as having relevance to immunopharmacology. CY‐09 and MCC950 (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=immuno%26ligandId=10057#immuno) are shown to have significant therapeutic effects in NLRP3‐driven diseases,61 MCC950 (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=immuno%26ligandId=8228#immuno) has the potential to block NLRP3‐induced events and there is evidence that dapansutrile (http://www.guidetoimmunopharmacology.org/GRAC/LigandDisplayForward?tab=immuno%26ligandId=10056) is a clinical lead for autoinflammatory disease and heart failure.
Figure 9.
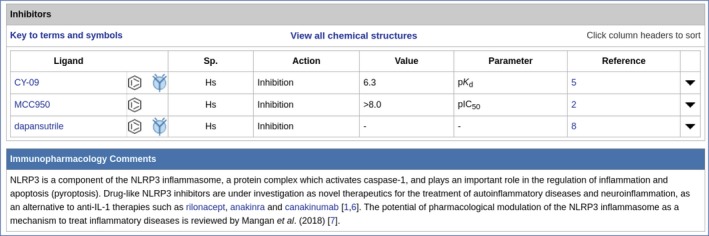
Inhibitors and immunopharmacology data from the detailed target page for NRLP3 (http://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=1770#Inhibitors). Immunopharmacology comments highlight its role in the regulation of inflammation. Both CY‐09 and MCC950 have quantitative interaction data and these are marked with the immuno‐icon, showing that they have relevance to immunopharmacology.
A further translational direction may be the development of a novel combination of lipid‐lowering and anti‐inflammatory treatments by design of monoclonal antibodies that could simultaneously inhibit proprotein convertase subtilisin/kexin type 9 (PCSK9; Fig. 10) and either IL‐1β or IL‐6. Figure 10 shows inhibitor data from the GtoImmuPdb for PCSK9, showing three monoclonal antibodies with quantitative interaction data for PCSK9. Both evolocumab and alirocumab are approved drugs, and bococizumab is being evaluated in Phase III clinical trials.
Figure 10.
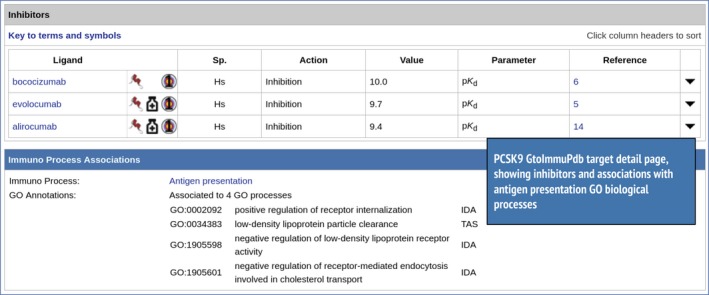
Inhibitors and immunopharmacology data from the detailed target page for PCSK9 (http://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=2388#Inhibitors).
Another alternative in targeting the IL‐1β pathway could be targeting the IL‐1 receptor itself, or modulating signal transduction downstream of the activated receptors such as members of the IL‐1 receptor‐associated kinase (IRAK) family. In GtoImmuPdb, details for the IL‐1 receptor (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1905) show that it is already targeted by the antagonist peptide mimic anakira (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?tab=clinical%26ligandId=6972). For IRAK4, the target detail page shows 11 inhibitors (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2045%26familyId=579%26familyType=ENZYME#Inhibitors), six of which are selective, including the Pfizer compound (PF‐06650833; http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9667) which is a clinical lead for rheumatoid arthritis, demonstrating that IRAK4 is a druggable target in the pathway.
The testing of new drugs should move in parallel with the identification of better biomarkers for patient stratification, the development of novel molecular imaging modalities for diagnosis and monitoring of vascular inflammation, as well as novel drug‐delivery systems for selective in situ targeting of vascular immune pathways and consequent reduced risk of systemic immunosuppression.62
Concluding remarks
The recent appreciation that most chronic diseases include immune aspects, and that modulation of immunity can have a profound effect on disease progression or resolution, makes the immune system a critical target for new therapies. The historically small overlap between immunological and pharmacological research communities has probably hindered the rapid development of immunologically relevant therapeutics. The IUPHAR GtoImmuPdb database and search tools provide a partial solution to this problem, allowing researchers with immunological training to use search terms framed in the concepts of immunology to find pharmacological information and tools relevant to them. In this way, the database should accelerate discovery and development of new strategies against chronic disease.
Author contribution
SDH designed and developed the database and wrote the manuscript. CS, EF and AP curated the GtoImmuPdb database. PM made a significant contribution to the writing of the manuscript, in particular the case study. SPHA and APD had input on writing the manuscript and as grant holders had roles in the planning of the project. DF contributed to GtoImmuPdb in curation of protein kinases. FLS contributed to GtoImmuPdb in curation of cellular targets, pathways and monoclonal antibodies. MS as grant holder had roles in the planning of the project. JAD contributed to the writing of the manuscript and is the principal investigator of the database development and curation team at the University of Edinburgh.
Disclosures
There are no conflicts of interest to declare.
Acknowledgements
The authors wish to thank the grant holders and scientific advisors for the Guide to Immunopharmacology not already listed on this manuscript: Prof. Clare Bryant and Prof. Steve Anderton. The authors appreciate the contributions made by Prof. Adriano G. Rossi, Dr Georgia Perona‐Wright and the IUPHAR Immunopharmacology Section (https://iuphar.org/sections-subcoms/immunopharmacology/). We also would like to acknowledge Joanna L. Sharman for database development advice and support. Thanks also to the members of Immunopaedia: Prof. Clive Gray, Prof. Michelle Letarte and Bon Holtak. This work was funded by the Wellcome Trust (108420/Z/15/Z); the International Union of Basic and Clinical Pharmacology; and the British Pharmacological Society (MED791).
References
- 1. Licastro F, Candore G, Lio D, Porcellini E, Colonna‐Romano G, Franceschi C et al Innate immunity and inflammation in ageing: a key for understanding age‐related diseases. Immun Ageing A 2005; 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Candore G, Balistreri CR, Grimaldi MP, Vasto S, Listì F, Chiappelli M et al Age‐related inflammatory diseases: role of genetics and gender in the pathophysiology of Alzheimer's disease. Ann N Y Acad Sci 2006; 1089:472–86. [DOI] [PubMed] [Google Scholar]
- 3. Shadfar S, Hwang CJ, Lim M‐S, Choi D‐Y, Hong JT. Involvement of inflammation in Alzheimer's disease pathogenesis and therapeutic potential of anti‐inflammatory agents. Arch Pharm Res 2015; 38:2106–19. [DOI] [PubMed] [Google Scholar]
- 4. Boe DM, Boule LA, Kovacs EJ. Innate immune responses in the ageing lung. Clin Exp Immunol 2017; 187:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Accardi G, Caruso C. Immune‐inflammatory responses in the elderly: an update. Immun Ageing A 2018; 15:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welsh P, Grassia G, Botha S, Sattar N, Maffia P. Targeting inflammation to reduce cardiovascular disease risk: a realistic clinical prospect? Br J Pharmacol 2017; 174:3898–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams KC, Ulvestad E, Hickey WF. Immunology of multiple sclerosis. Clin Neurosci N Y N 1994; 2:229–45. [PubMed] [Google Scholar]
- 8. Ziemssen T, Ziemssen F. The role of the humoral immune system in multiple sclerosis (MS) and its animal model experimental autoimmune encephalomyelitis (EAE). Autoimmun Rev 2005; 4:460–7. [DOI] [PubMed] [Google Scholar]
- 9. Pontarini E, Lucchesi D, Bombardieri M. Current views on the pathogenesis of Sjögren's syndrome. Curr Opin Rheumatol 2018; 30:215–21. [DOI] [PubMed] [Google Scholar]
- 10. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol 2010; 28:573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silva FAR, Rodrigues BL, Ayrizono M LS, Leal RF. The immunological basis of inflammatory bowel disease. Gastroenterol Res Pract [Internet]. 2016; 2016:2097274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McInnes IB, Schett G. Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet Lond Engl 2017; 389:2328–37. [DOI] [PubMed] [Google Scholar]
- 13. Nerurkar L, Siebert S, McInnes IB, Cavanagh J. Rheumatoid arthritis and depression: an inflammatory perspective. Lancet Psychiatry 2019; 6:164–73. [DOI] [PubMed] [Google Scholar]
- 14. Bryant CE, Sutherland S, Kong B, Papadimitrious MS, Fromm PD, Hart DNJ. Dendritic cells as cancer therapeutics. Semin Cell Dev Biol 2019; 86:77–88. [DOI] [PubMed] [Google Scholar]
- 15. Veglia F, Gabrilovich DI. Dendritic cells in cancer: the role revisited. Curr Opin Immunol 2017; 45:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR et al IUPHAR‐DB: the IUPHAR database of G protein‐coupled receptors and ion channels. Nucleic Acids Res 2009;37(Database issue):D680–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharman JL, Mpamhanga CP, Spedding M, Germain P, Staels B, Dacquet C et al IUPHAR‐DB new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res 2011;39(Database issue):D534–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharman JL, Benson HE, Pawson AJ, Lukito V, Mpamhanga CP, Bombail V et al IUPHAR‐DB: updated database content and new features. Nucleic Acids Res 2013;41(Database issue):D1083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SPH, Buneman OP et al The IUPHAR/BPS Guide to PHARMACOLOGY: an expert‐driven knowledgebase of drug targets and their ligands. Nucleic Acids Res 2014;42(Database issue):D1098–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 2016; 44(D1):D1054–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res 2018; 46(D1):D1091–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Southan C, Sharman JL et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res 2019; 48(D1):D1006–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiligada E, Ishii M, Riccardi C, Spedding M, Simon H‐U, Teixeira MM et al The expanding role of immunopharmacology: IUPHAR Review 16. Br J Pharmacol 2015; 172:4217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carvalho S, Levi‐Schaffer F, Sela M, Yarden Y. Immunotherapy of cancer: from monoclonal to oligoclonal cocktails of anti‐cancer antibodies: IUPHAR Review 18. Br J Pharmacol 2016; 173:1407–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landolina N, Levi‐Schaffer F. Monoclonal antibodies: the new magic bullets for allergy: IUPHAR review 17. Br J Pharmacol 2016; 173:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishii M. Immunology proves a great success for treating systemic autoimmune diseases – a perspective on immunopharmacology: IUPHAR Review 23. Br J Pharmacol 2017; 174:1875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harding SD, Faccenda E, Southan C, Maffia P, Davies JA. A new guide to immunopharmacology. Nat Rev Immunol 2018; 18:729. [DOI] [PubMed] [Google Scholar]
- 28. Alexander SPH, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF et al THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: introduction and other protein targets. Br J Pharmacol 2019; 176(S1):S1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander SP, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E et al The concise Guide to PHARMACOLOGY 2015/16: overview. Br J Pharmacol 2015; 172:5729–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexander SP, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al The concise GUIDE TO PHARMACOLOGY 2017/18: overview. Br J Pharmacol 2017; 174(Suppl 1):S1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chazot P, Cowart M, Fukui H, Ganellin CR, Gutzmer R, Haas HL et al Histamine receptors (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHARBPS Guide Pharmacol CITE [Internet] 2019; 2019 10.2218/gtopdb/F33/2019.4 [DOI] [Google Scholar]
- 32. Cain D, Cidlowski J, Edwards DP, Fuller P, Grimm SL, Hartig S et al 3C. 3‐Ketosteroid receptors (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHARBPS Guide Pharmacol CITE [Internet] 2019; 2019 10.2218/gtopdb/F98/2019.4 [DOI] [Google Scholar]
- 33. Izzo AA, Mitchell JA. Cyclooxygenase (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHARBPS Guide Pharmacol CITE [Internet] 2019; 2019 10.2218/gtopdb/F269/2019.4 [DOI] [Google Scholar]
- 34. Bryant C, Monie TP. Pattern recognition receptors (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHARBPS Guide Pharmacol CITE [Internet] 2019. URL http://journals.ed.ac.uk/gtopdb-cite/article/view/3276 [accessed on 4 November 2019]. [Google Scholar]
- 35. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. The Gene Ontology Consortium . The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019; 47(D1):D330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol 2005; 6:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diehl AD, Lee JA, Scheuermann RH, Blake JA. Ontology development for biological systems: immunology. Bioinformatics 2007; 23:913–5. [DOI] [PubMed] [Google Scholar]
- 39. Lovering RC, Camon EB, Blake JA, Diehl AD. Access to immunology through the Gene Ontology. Immunology 2008; 125:154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gangwar RS, Landolina N, Arpinati L, Levi‐Schaffer F. Mast cell and eosinophil surface receptors as targets for anti‐allergic therapy. Pharmacol Ther 2017; 170:37–63. [DOI] [PubMed] [Google Scholar]
- 41. Artis D, Spits H. The biology of innate lymphoid cells. Nature 2015; 517:293–301. [DOI] [PubMed] [Google Scholar]
- 42. Zook EC, Kee BL. Development of innate lymphoid cells. Nat Immunol 2016; 17:775–82. [DOI] [PubMed] [Google Scholar]
- 43. Kim N, Kim HS. Targeting checkpoint receptors and molecules for therapeutic modulation of natural killer cells. Front Immunol 2018; 9:2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nicholson SE, Keating N, Belz GT. Natural killer cells and anti‐tumor immunity. Mol Immunol 2019; 110:40–7. [DOI] [PubMed] [Google Scholar]
- 45. Mariotti FR, Quatrini L, Munari E, Vacca P, Moretta L. Innate lymphoid cells: expression of PD-1 and other checkpoints in normal and pathological conditions. Front Immunol 2019; 10:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D et al The ChEMBL database in 2017. Nucleic Acids Res 2017; 45(D1):D945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43(Database issue):D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinforma 2017; 58:1.2.1–1.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schriml LM, Mitraka E, Munro J, Tauber B, Schor M, Nickle L et al Human disease ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 2019; 47(D1):D955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fehr T, Stussi G. ABO‐incompatible kidney transplantation. Curr Opin Organ Transplant 2012; 17:376–85. [DOI] [PubMed] [Google Scholar]
- 51. Macklin PS, Morris PJ, Knight SR. A systematic review of the use of rituximab as induction therapy in renal transplantation. Transplant Rev 2015; 29:103–8. [DOI] [PubMed] [Google Scholar]
- 52. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S et al PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 2019; 47(D1):D1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C et al Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017; 377:1119–31. [DOI] [PubMed] [Google Scholar]
- 54. Everett BM, Pradhan AD, Solomon DH, Paynter N, Macfadyen J, Zaharris E et al Rationale and design of the Cardiovascular Inflammation Reduction Trial: a test of the inflammatory hypothesis of atherothrombosis. Am Heart J 2013; 166:199–207.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ridker PM, Everett BM, Pradhan A, MacFadyen JG, Solomon DH, Zaharris E et al Low‐dose methotrexate for the prevention of atherosclerotic events. N Engl J Med 2019; 380:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ridker PM, MacFadyen JG, Everett BM, Libby P, Thuren T, Glynn RJ et al Relationship of C‐reactive protein reduction to cardiovascular event reduction following treatment with canakinumab: a secondary analysis from the CANTOS randomised controlled trial. Lancet 2018; 391:319–28. [DOI] [PubMed] [Google Scholar]
- 57. Maffia P, Guzik TJ. When, where, and how to target vascular inflammation in the post‐CANTOS era? Eur Heart J 2019; 40:2492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ridker PM, MacFadyen JG, Thuren T, Libby P. Residual inflammatory risk associated with interleukin‐18 and interleukin‐6 after successful interleukin‐1β inhibition with canakinumab: further rationale for the development of targeted anti‐cytokine therapies for the treatment of atherothrombosis. Eur Heart J 2019. 10.1093/eurheartj/ehz542 [DOI] [PubMed] [Google Scholar]
- 59. Ridker PM. From C‐reactive protein to interleukin‐6 to interleukin‐1: moving upstream to identify novel targets for atheroprotection. Circ Res 2016; 118:145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Toldo S, Abbate A. The NLRP3 inflammasome in acute myocardial infarction. Nat Rev Cardiol 2018; 15:203–14. [DOI] [PubMed] [Google Scholar]
- 61. Jiang H, He H, Chen Y, Huang W, Cheng J, Ye J et al Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J Exp Med 2017; 214:3219–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cicha I, Chauvierre C, Texier I, Cabella C, Metselaar JM, Szebeni J et al From design to the clinic: practical guidelines for translating cardiovascular nanomedicine. Cardiovasc Res 2018; 114:1714–27. [DOI] [PMC free article] [PubMed] [Google Scholar]


