Abstract
Human gut is home to a diverse and complex microbial ecosystem encompassing bacteria, viruses, parasites, fungi, and other microorganisms that have an undisputable role in maintaining good health for the host. Studies on the interplay between microbiota in the gut and various human diseases remain the key focus among many researchers. Nevertheless, advances in sequencing technologies and computational biology have helped us to identify a diversity of fungal community that reside in the gut known as the mycobiome. Although studies on gut mycobiome are still in its infancy, numerous sources have reported its potential role in host homeostasis and disease development. Nonetheless, the actual mechanism of its involvement remains largely unknown and underexplored. Thus, in this review, we attempt to discuss the recent advances in gut mycobiome research from multiple perspectives. This includes understanding the composition of fungal communities in the gut and the involvement of gut mycobiome in host immunity and gut-brain axis. Further, we also discuss on multibiome interactions in the gut with emphasis on fungi-bacteria interaction and the influence of diet in shaping gut mycobiome composition. This review also highlights the relation between fungal metabolites and gut mycobiota in human homeostasis and the role of gut mycobiome in various human diseases. This multiperspective review on gut mycobiome could perhaps shed new light for future studies in the mycobiome research area.
1. Introduction
Human gut is a complex ecosystem inhabited by a myriad of microorganisms including bacteria, fungi, archae, and viruses [1]. Thus far, studies on gut bacteria or gut “microbiome” have received the most attention due to the abundance of bacterial flora present in the gut. For example, large-scale projects including Metagenomics of the Human Intestinal Tract (MetaHIT) and National Institutes of Health (NIH) Human Microbiome Project (HMP) were among the projects that were initiated to study the composition of bacterial flora and their impacts on human health [1, 2]. Nevertheless, the shift in focus towards the “rare biosphere” [3] in the gut, particularly on fungi, has gathered increasing traction nowadays. Studies on “mycobiome,” a term used to describe the fungal community of the microbiome, have escalated despite still being in its infancy stage as it may be significant in the context of human health and diseases [4].
The studies on human gut mycobiome have received little attention for the past decades because fungal presence is relatively insignificant in the gut compared to the bacterial communities. In addition to that, fungi have been traditionally studied by culture-dependent methods [5, 6], which limits the in-depth understanding of the fungal microbiota. Nonetheless, recent advances in deep-sequencing technologies and bioinformatics analysis have shed light on the complexity of the fungal communities that reside on both mucosal and luminal surfaces in the gut and further highlighted our current understanding on this poorly understood compartment that reside in the gut. Increasing evidences have demonstrated the undisputable role of fungal components in driving the pathogenesis of various gut-associated and metabolic diseases [7–9]. In addition to that, specific fungi possess the ability to modulate host immune response and could be a risk factor for immunological disorders seen in genetically susceptible individuals [10]. More profoundly, gut mycobiome could be the reservoir for opportunistic pathogens in immunocompromised hosts [11–13]. These clearly indicate that gut mycobiome is crucial in host homeostasis and disease development. Thus, in this review, we seek to summarize the role of gut mycobiota from multiple perspectives and to highlight the latest research on gut mycobiome on disease development, in order to provide new insights and productive direction for future studies in this newly emerged research area.
2. Fungal Communities in Gut
In general, fungi constitute a minor component of the entire gut microbiome. Recent shotgun metagenomics sequencing analysis has revealed that fungi consist of nearly 0.1% of the total microbes in the gut [1, 14]. On the other hand, despite numerous published data on gut mycobiome, the fungal communities in the gut remain poorly understood. There is no consensus in defining a healthy gut mycobiome due to a variety of factors such as low abundance and diversity of fungi in the gut, temporal instability of gut mycobiota throughout the development periods, and high intervolunteer and intravolunteer variability of the gut mycobiome [15, 16]. Nevertheless, the fungal communities in the gut have been highlighted in many studies [8, 16, 17]. In terms of phyla, thus far, most studies have suggested that Ascomycota is the most predominant phylum found in the gut, followed by Zygomycota and Basidiomycota phyla [15, 18–20]. Meanwhile, in the context of genus, a recent review by Hallen-Adams and coworkers had identified potential fungal species that inhabit the intestinal niche, belonging to the genera Candida, Cryptococcus, Malassezia, Aspergillus, Saccharomyces, Galactomyces, Trichosporon, and Cladosporium [21]. Recently, through the Human Microbiome Project (HMP), Nash and coworkers (2017) sequenced 317 stool samples in a healthy cohort via Internal Transcribed Spacer 2 (ITS2) region and 18S rRNA gene. The authors reported that gut mycobiota is mainly dominated by Malassezia, Candida, and Saccharomyces, with S. cerevisiae, M. restricta, and C. albicans identified in 96.8%, 88.3%, and 80.8% of the samples, respectively [16]. Reports from earlier studies using culture-dependent analysis showed that less than 30% of fungal species are present in the human gut [22, 23]. Collectively, these studies suggest that gut mycobiome is relatively low in diversity as compared to bacterial communities, and the high prevalence of several fungal species identified across samples further suggested that a core mycobiota may exist in the gut.
In the context of fungal development in infants, Schloss and coworkers performed analysis on gut microbiome profile between family members and unrelated subjects and discovered that similar gut microbiome profile are identified in people sharing same life history and environment [24]. A study by Palmer et al. demonstrated that variable levels of fungi were detected from the stool samples of healthy full-term infants after birth and also at defined intervals throughout the first year of life using microarray and PCR analysis. Also, the authors reported the absence of fungal species from the initial stool sample in one of the healthy infants, where the mother of the infant showed detectable fungal levels in the vagina, raising the possibility that vertical transmission (vaginal microbiota) has minimal impact on shaping the fungal colonization in the infant [25]. Bliss et al. had demonstrated vertical and horizontal transmission of C. albicans from mother to infant through DNA fingerprinting techniques [26]. On the other hand, diet could affect the composition of the infant's mycobiota as well. For instance, Candida species have been identified in breast-feeding women with symptoms of mammary candidiasis [27]. A study by LaTuga et al. identified several fungal species in extremely low birth weight infants in their first postnatal month. These fungal species include S. cerevisiae followed by Candida spp., Cladosporium spp., and Cryptococcus spp. [28]. Another study also reported a lack of fungal diversity and richness in eleven infants using combined NGS technology (targeted on fungal ITS2 amplicons) with qPCR analysis. The most predominant fungi identified in this study were C. albicans, C. parapsilosis, and Leptosphaerulina, with C. albicans being detected in all infants. The authors speculate that the relatively low diversity of fungi in infants as compared to adults could be due to age factor where these infants may not yet been fully colonized by a myriad of different fungal species. Further, the authors highlighted the challenge of detecting low abundance fungi, where low levels of Candida parapsilosis and Candida krusei sequences were detected in a few samples; however, their presence was only detected in one sample via species-specific qPCR [29]. On the other hand, a study by Strati et al. demonstrated the influence of age and gender on the gut fungal composition. In this study, the authors reported that infants and children have higher fungal richness than in adults via culture-independent analysis. Meanwhile, the diversity and fungal richness was also higher in female than male healthy subjects [30]. Together, the diversity of fungal composition, the timing and the mechanism involved in assembling fungi into gut microbiome, the influence of genetic and environmental factors in shaping the infant mycobiota, and the contribution of fungi to the microbiome phenotype remain to be explored further.
3. Gut Mycobiome and Host Interactions
As discussed previously, gut ecosystem remains a complex environment harboured by various “microscopic” and “macroscopic” organisms. It is believed that the ecosystem in the gut is maintained via multiple interactions among gut members (bacteria, fungi, and virus) as suggested by Filyk and Osborne [31]. On the other hand, accumulated evidences also suggest significant involvement of gut fungi in maintaining host dynamic and the potential of utilizing fungal metabolites in clinical applications. Thus, in this section, we will discuss interaction between fungi with host, deliberate on fungal-bacteria interaction and gut-brain axis, examine the impact of gut fungi on diet and host immunity, and also highlight the influence of gut fungi on health and diseases (Figure 1).
Figure 1.
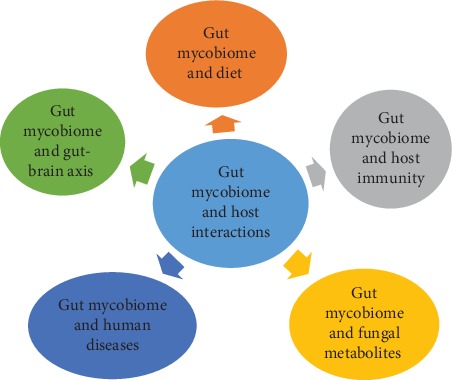
The interactions between gut mycobiome and host in various areas.
3.1. Fungal-Bacteria Interactions
Generally, the dialogue between fungi and bacteria is studied through the induction of dysbiosis in the gut followed by treatment with either antifungal or antibacterial drugs. It is well known that antibiotics specific to anaerobic bacteria or broad-spectrum antibiotics can have differential impacts on fungal susceptibility, in particular C. albicans [32, 33]. Some studies even suggested that the administration of C. albicans to mice after antibiotic exposure could significantly change the gut microbiome composition, from phyla to family level, and the changes are irreversible in the long run [34, 35]. Further, mycobiome equilibrium is known to have impact on microbiome stability. Such impact is demonstrated in mice model of dextran sulphate sodium- (DSS-) induced colitis. In this model, it was found that the prescription of antifungal drug in the mice had significantly reduced the fungal diversity along with the increase of pathogenic bacterial diversity, which in turn exacerbated the severity of colitis inflammation [36]. On the other hand, a study by Jiang et al. demonstrated that commensal fungi such as C. albicans or S. cerevisiae can functionally replace intestinal bacteria in the event of bacterial dysbiosis after antibiotic exposition. Moreover, these fungal species confer protection against colitis and influenza A virus infection through alleviation of mucosal tissue injuries and host immune modulation [37]. Another robust example of fungal-bacteria interaction is secretion of extracellular enzymes such as phosphatases and proteases by S. boulardii, which aids in deactivating the toxins produced by C. difficile and E. coli [38, 39]. Furthermore, studies had shown that R. gnavus and C. albicans can cause lesion formation in the gut by degrading the protective mucin layer through the action of mucolytic enzymes [40, 41]. Meanwhile, some fatty acid metabolites secreted by bacterial flora also seem to modulate C. albicans germination [42]. A more recent study by García et al. demonstrated that gut microbial metabolites inhibit the invasion of human enterocytes and C. albicans hyphal growth through the target of rapamycin (TOR) signaling pathway [43]. Other than that, interaction between fungal and bacterial cells can cause adverse effects on host via proinflammatory cytokines secretion, which result in apoptotic cell death and oxidative damage on host [44].
Fungi and bacterial cells are also believed to interact with each other within a biofilm habitat, a so called “mixed species biofilm.” Such a habitat can help them to persistently colonize and survive in specific microenvironments such as gut, skin, and oral cavity. Additionally, this “mixed species biofilm” can give extra protection against antimicrobial agents and host immune evasion [45]. Notably, this “mixed species biofilm” provides mutual benefits for both bacterial and fungal cells. Fungi can strengthen their virulence determinants within the biofilm habitat, such as increasing the ability to invade host through hyphal induction and production of extracellular enzymes such as aspartic proteinases. Meanwhile, bacteria may benefit from this habitat via the increase in their resistance towards antimicrobial treatment [45]. Kalan et al. had demonstrated a rapid formation of mixed species biofilms between Trichosporon asahii and Staphylococcus simulans or C. albicans and Citrobacter freundii in vitro, revealing a close relation between fungal and bacterial cells [46]. Similarly, Hoarau et al. reported formation of robust biofilm between C. tropicalis, S. marcescens, and E. coli in in vitro biofilm model [47]. Other studies also demonstrated that production of lipopolysaccharides by S. marcescens and E. coli enhanced fungal biofilm maturation [48, 49]. Clearly, there is a strong connection between fungal and bacteria cells. A more in-depth analysis of this interkingdom interaction can provide us with more information on the gut-associated pathogenesis driven by this interaction.
3.2. Mycobiome and Gut-Brain Axis
It is interesting that besides gut microbiome, fungi are involved in the gut-brain axis (GBA). Growing evidences from both clinical and experimental studies suggest that fungi are involved in the bidirectional communication between brain and gut through neuro-immuno-endocrine mediators, which is comparable to microbiome-gut-brain axis [50]. An excellent example of this mycobiome-gut-brain communication is via a study by Botschuijver et al., where the authors investigated the association of intestinal fungal components in patients with inflammatory bowel syndrome (IBS) and in a rat model of visceral hypersensitivity. In the study, the authors observed a mycobiome dysbiosis in the rat model of visceral hypersensitivity, where administration of fungicide, cecal mycobiomes, and soluble β-glucans improved hypersensitized rats [51].
3.3. Gut Mycobiome and Immunity
The crosstalk between fungi and host immunity had been extensively reviewed [7, 44]. Of all, Dectin-1 appears as one of the most crucial pattern recognition receptors (PRRs) in shaping fungal immunity [10, 52]. Dectin-1 interacts with β-1,3 glucan motif found on the fungal cell walls and elicits host immune response against them. The importance of Dectin-1 has been implicated in the mouse model of DSS-induced colitis where Dectin-1 knockout mice experienced more severe colitis when compared to wild-type mice. In addition, expansion in Candida and Trichosporon genera and a decrease in Saccharomyces genus, together with increased of proinflammatory cytokines, IFN-γ, IL-17, and TNF-α, also aggravated inflammation in Dectin-1 knockout mice [10]. Further, clinical study has pointed out that polymorphisms in Dectin-1 gene are likely to contribute towards disease exacerbation in patients with ulcerative colitis (UC) [10]. In addition, other evidences also showed that deficiency in Dectin-1 is also linked with increased gastrointestinal colonization with Candida species in patients who received transplants [53, 54]. Taken together, these studies provide evidence on the protective role Dectin-1 in fungal infection and the importance of Dectin-1 to keep fungal in check.
Caspase recruitment domain-containing protein 9 (CARD9), a crucial downstream molecule for antifungal receptors including C-lectin receptors, is also implicated in defense against fungi. In the mice model of colitis, CARD9 knockout mice had increased antifungal antibodies, and the severity of colitis was mitigated upon antifungal treatment, suggesting the protective role of CARD9 signaling against fungi [55]. Meanwhile, IL-17, an effector cytokine for Th-17 helper cells, is involved in the mucosal immune response against fungi. The roles of IL-17 during mucosal fungal infections have been documented in both clinical and animal experimental studies [56–58]. Nevertheless, it remains obscure on the effect of IL-17 on gut mycobiota. One clinical study has reported that higher incidence of fungal infections accompanied with severe intestinal pathology were observed in patients with Crohn's disease upon IL-17A blockade, indicating the possible role of IL-17 pathway in regulating fungal communities in the gut [59]. A review by Conti et al. also suggested the pertinent role of IL-17 against opportunistic C. albicans [56]. On the other hand, IL-22, a cytokine that is similar to IL-17, is also closely linked with mucosal immunity against fungi. It has been demonstrated that IL-22 regulates gastrointestinal fungi, where mice lacking IL-22 are more prone to gastrointestinal candidiasis upon intragastric challenge with C. albicans [60]. Furthermore, both IL-17 and IL-22 are potent inducers of antimicrobial peptides (AMPs) by epithelial cells, which have indisputable role in clearing mucosal Candida spp. and Aspergillus spp. infections [61–64].
Crosstalk between gut mycobiome and host immune system can modulate the disease outcome. For example, Qamar et al. reported protective effect of Saccharomyces boulardii against Clostridium difficile colitis-induced mice model. In this study, the authors showed that administration of S. boulardii stimulates production of intestinal immunoglobulin A (IgA) against Clostridium difficile toxin A in mice [65]. In another study, Thomas and coworkers reported the anti-inflammatory effects of S. boulardii in patients with inflammatory bowel diseases, mainly via inhibition of T and dendritic cells activation, reduced levels of proinflammatory cytokines including tumor necrosis factor-α and interleukin- (IL-) 6 and increase the production of IL-10, which consequently promotes epithelial restitution relevant in IBD [66]. On the other hand, crosstalk between immune system and fungi may influence bacteria in the gut and vice versa. A study by Tang et al. demonstrated that the colon of Clec7a−/− mice was protected by Lactobacillus murinus-induced regulatory T cell expansion in the absence of Candida species. Nevertheless, the presence of C. tropicalis seems to cancel this protective effect and exacerbates intestinal inflammation [67]. Meanwhile, colonization of Candida species as seen in Card9−/− mice also reduced the populations of tryptophan-metabolizing bacteria, including lactobacilli, which intensify the severity of colitis in Card9−/− mice. The reduction of lactobacilli is accompanied with reduced levels of aryl hydrocarbon receptor (AHR) ligands, Reg3g, Reg3b, and Il22 expression in the colons of Card9−/− mice [68]. In addition to that, fungal microbiota also helped in shaping the host immune system in regards to the development of colorectal cancer. A recent study by Wang et al. demonstrated the adaptor protein CARD9 confers protection against colon cancer through restriction of Mycobiota-Mediated Expansion of Myeloid-Derived Suppressor Cells (MDSCs) in mice model [69].
Fungal species such as S. cerevisiae and C. albicans are capable of modifying immune response in a significant way. For example, chitin from S. cerevisiae is able to induce “trained immunity (a de facto immune memory of the innate immunity)” in monocytes in a strain dependent manner by increasing cytokine productions such as TNF-α and IL-6 and through direct antimicrobial activity upon stimulation with bacterial, fungal, and TLR ligands [70]. Similarly, C. albicans can induce “trained immunity” along with functional reprogramming of monocytes, which confers protection against reinfection [71]. Taken together, these studies suggest that “trained immunity” is important in maintaining gut immune homeostasis and confer protection on host against invading pathogens.
During gut-brain communication, the effect of intestinal inflammation on the central nervous system is often associated with anxiety, depression, or “sickness behavior” [72]. These psychological behavioral changes are always considered as comorbidities in patients with persistent intestinal inflammation, including inflammatory bowel diseases and irritable bowel diseases [73]. The underlying mechanism could be due to imbalances in serotonergic activity and hyperreactivity of hypothalamic-pituitary-adrenal axis (HPPA) [74, 75]. It is believed that this systemic effect is mediated by host immune factors, especially cytokines including interleukin-6 (IL-6), interleukin-1β (IL-1β), and tumor necrosis factor (TNF) [76]. Studies have shown that fungi such as C. albicans, S. cerevisiae, and A. fumigatus are able to modulate cytokine expression, particularly IL-6 [70, 77]. Thus, it is speculated that gut mycobiota may elicit local immune response at the gut site, where these immune mediators, especially cytokines will cross the blood-brain barrier (BBB), and reaching the brain to stimulate specific brain areas, in particular the hypothalamus and circumventricular organs [78].
Meanwhile, there is increasing evidences showing the influence of gut mycobiota on extragastrointestinal organs immune responses. A typical example is through a study by Wheeler et al., where the authors demonstrated an aggravation of allergic airway disease accompanied with altered gut mycobiota prior to fluconazole administration in mice with DSS-induced colitis. In this study, an expansion of Wallemia sebi, Aspergillus amstelodami, and Epicoccum nigrum and a reduction of Penicillium brevicompactum and C. tropicalis were observed [79]. Meanwhile, McAleer et al. demonstrated the ability of gut microbiota in shaping the pulmonary immune response upon A. fumigatus oropharyngeal challenge. In this study, mice treated with vancomycin for a month had reduced levels of IL-17 and IL-22 and increased the level of IL-4 in the lungs, which indicates the involvement of Th-17 and Th-2 responses against fungi in the lungs [80]. A recent study by Skalski et al. demonstrated that an expansion of Wallemia spp. (W. mellicola) in the gut modified the pulmonary immune response and intensified the airway inflammation in the intratracheal house dust mite (HDM) mice model of allergic airways disease. Additionally, Wallemia was not detected in the lung, suggesting the ability of gastrointestinal colonization of Wallemia to modulate lung immune response remotely. Further analysis of the gut mycobiota revealed that the expansion of W. mellicola in the gut was associated with perturbation in both fungal and bacteria communities [81]. Another study by Li et al. reported that gut fungal dysbiosis induced by fluconazole persistently exacerbates allergic airway disease (AAD) in mice, where this effect is mainly via fungal sensing by gut-resident CX3CR1+ MNPs on peripheral immunity [82]. Overall, these studies revealed a clear link between gut mycobiota, pulmonary immune responses, and lung diseases, which further consolidate the idea of the existence of mycobiota-gut-lung axis.
3.4. Gut Mycobiome and Diet
Diet could be one of the determinants in driving the changes in gut fungal mycobiota composition between individuals. An investigation on the link between diet and fungal mycobiota has been carried out by Hoffmann and his coworkers using culture-independent analysis. In the study, the authors had identified 66 fungal genera, with Candida, Cladosporium, and Saccharomyces being the most common genera identified. The authors speculate that the high prevalence of Saccharomyces could be due to consumption of yeast-containing foods such as beer and bread while high level of Candida was strongly correlated with the recent consumption of carbohydrates. Although this study has identified various fungal communities in the gut, one of the concerns raised in this study is whether these fungal mycobiota identified are permanent residents in the gut or just transient species [19]. Meanwhile, another study had profiled the short-term effect of plant and animal diet on the gut microbiome. In this study, David et al. showed that short-term diet consisting of both animal and plant products alters the microbial community structure. In addition to that, diversified fungal genera including Scopulariopsis, Penicillium, Debaryomyces, and Candida were identified in this study [83]. Moreover, diet therapy and antibiotics seem to reduce the abundance of fungal species in patients with Crohn's disease, as implicated by Lewis et al. [84]. Collectively, studies on the impact of diet on gut mycobiome are still in its infancy. Nonetheless, more studies are warranted to evaluate and to associate the effect of diet and food-associated fungi in shaping the diversity, composition, and functionality of human microbiome as well as the effect on diet on gut-associated diseases.
3.5. Gut Mycobiome and Fungal Metabolites
Since the invention of first antibiotic-Penicillin G from Penicillium notatum by Sir Alexander Fleming in 1928, the era of using fungi for medicinal purposes has started. Fungi have shown indispensable role in various areas, including food, biotechnology, and pharmaceutical industries. Fungi also possess great potential in producing a broad spectrum of metabolites, which can be applied for medication or therapeutic purposes. For example, griseofulvin isolated from Penicillium griseofulvum served as antimycotic drug [85], while fusidic acid from Fusidium coccineum [86] and cephalosporins from Acremonium chrysogenum act as antibacterial agents, and lovastatin isolated from Aspergillus terreus and mevastatin from Penicillium citrinum [87] can be used as lipid lowering agents.
Besides this, similar like bacteria, fungi can express metabolites which can influence host homeostasis and exert biological effects on host, as part of fungi-host interactions [17]. Fungal species such as Saccharomyces boulardii, C. albicans, and Saccharomyces cerevisiae may secrete molecules like farnesol, fusel alcohols, tyrosol, and fatty acids, which are autoregulatory molecules of growth. These molecules enable fungal cells to regulate adhesion, yeast-to-hyphae transition and biofilm formation themselves, which in turn facilitate the colonization, invasion, and dissemination in host [88]. On the other hand, polysacharide β1, 3-glucan is a fungal-derived molecule found in the inner cell wall of C. albicans. β1, 3-glucan has strong link with host immunity. A recent study suggested that β1, 3-glucan can trigger “trained immunity” prior to exposure to monocytes, generating more robust immune response upon fungal reinfection. Additionally, β1, 3-glucan can induce epigenetic changes including histone methylation upon exposure to monocytes [71].
Meanwhile, the probiotic yeast Saccharomyces boulardii is able to produce a low molecular weight, water soluble anti-inflammatory factor, which is capable to mediate signal transduction pathway in host cells, such as NF-κB. It also helps to conserve the tight junction integrity between enterocytes in the small intestine and modulates signal transduction pathway during enteropathogenic E. coli infection [89, 90]. Other than that, Saccharomyces boulardii has been applied in treating various gastrointestinal disorders, as reviewed by Kelesidis. Nonetheless, safety issues concerned with using fungi as probiotics need to be considered, especially in immunocompromised or critically ill patients. These concerns include the possibility of developing fungemia after fungi treatment, gastrointestinal allergic reaction, and the environment risk prior to exposure of fungi in the air [91]. Saccharomyces boulardii may also exert trophic effect on the intestinal enterocytes via endoluminal release of polyamines. A study by Buts et al. reported that daily administration of lyophilized S. boulardii had significantly increased sucrase and maltase activities in rats' intestines [92]. The actual mechanism remains unknown, but it is likely through the release of spermine and spermidine. Meanwhile, S. boulardii produces a 54 kDa serine protease which can directly inhibit Clostridium difficile toxin A in rat ileum and Clostridium difficile toxins A and B in human colonic mucosa [38, 93]. Additionally, it also produces a 63-kDa phosphatase, which degrades Escherichia coli endotoxin by dephosphorylation [39]. Another study also documented the effect of Capric acid produced by S. boulardii in inducing adhesion, yeast-to-hyphae transition, and biofilm formation [94]. Moreover, administration of S. boulardii increases short-chain fatty acids (SCFAs), particularly butyrate, which may confer protective effect in total enteral nutrition- (TEN-) induced diarrhea [95]. Similarly, SCFAs produced by Malassezia species through enzymatic reaction (lipases and phospholipases) on host triglycerides found abundantly on skin could serve as metabolic sources for fungi [96, 97].
Some of the fungal-derived molecules are also known to mediate the interaction between fungal and bacteria. For example, ethanol from Saccharomyces cerevisiae is able to trigger the growth of Acinetobacter (A. baumannii, A. johnsonii, A. haemolyticus, and A. radioresistens) in vitro [98]. Farnesol, a small molecule produced by C. albicans, can also modify quorum sensing regulation in Pseudomonas aeruginosa [99, 100]. This showed that fungi are able to produce assorted secondary metabolites, and more studies can be conducted to shed light on the contribution of fungal metabolites produced in the gut on health and disease.
3.6. Gut Mycobiome and Disease Susceptibility
The significant involvement of fungi in the development and progression of human diseases have been reviewed extensively recently. Often, fungal dysbiosis contributes towards development and progression of several human diseases. A summary of the significant involvement of fungi in human diseases is depicted in Table 1.
Table 1.
Significant involvement of fungi in human disease development.
| Disease | Disease subtypes | Significant findings that involved fungi | Reference |
|---|---|---|---|
| Inflammatory bowel disease | Crohn's disease (CD) | (i) Fungal dysbiosis is closely related to CD in most of the conducted studies | [9, 47, 84, 101–103] |
| (ii) Interkingdom interaction between fungal and bacteria was observed | [9, 47] | ||
|
| |||
| Inflammatory bowel syndrome (IBS) | — | (i) Fungal dybiosis, predominant by Saccharomyces cerevisiae and Candida albicans in IBS patients | [51] |
|
| |||
| Cancers | Colorectal cancer | (i) Fungal dysbiosis is observed in most of the reported studies | [109–112] |
|
| |||
| Infectious diseases | Hepatitis B | (i) High levels of Aspergillus, Candida, Galactomyces, Saccharomyces, and Chaetomium were identified | [11] |
| (ii) Richness and diversity of fungal species is associated with chronic HBV infection | |||
| HIV | (i) C. parvum, C. difficile, and C. albicans are significantly present in HIV-seropositive patients | [114] | |
| (ii) C. albicans, C. krusei, and C. tropicalis were associated with diarrhea in HIV patients | [115] | ||
| (iii) Fungal dysbiosis and high prevalence of Candida species were observed in HIV patients | [116] | ||
| (iv) Prevalence of Candida in HIV patients without antiretroviral treatment was higher than HIV patients with active antiretroviral treatment | [117] | ||
|
| |||
| Noncommunicable diseases | Obesity | (i) Candida, Nakaseomyces, and Penicillium genera were commonly identified in obese subjects | [119] |
| (ii) Mucor racemosus and M. fuscus were identified in nonobese patients. | |||
| (iii) Specific fungal composition could be potentially used to distinguish between obese and nonobese patients | |||
| Diabetes | (i) C. albicans is more prevalence in type 1 diabetes | [123–125] | |
| (ii) C. albicans is more prevalence in type 1 and type 2 diabetes | |||
| (iii) No difference is found between C. albicans colonization in type 1 and type 2 diabetes. | [124] | ||
| (iv) Isolated fungal species from type 1 diabetes patient is more resistant towards antifungal treatment | [125] | ||
|
| |||
| Atherosclerosis | — | (i) Phylum Zygomycota, which consists of family Mucoraceae and genus Mucor, was negatively correlated with the risk of cardiovascular disease development through carotid intima-media thickness (cIMT) method | [129] |
|
| |||
| Alcoholic liver disease | — | (i) Decreased in fungal diversity along with Candida overgrowth in alcohol-dependent patients | [130] |
|
| |||
| Central nervous system diseases | Rett syndrome | (i) High abundance of Candida genus were detected | [134] |
| Autism Spectrum disorder | (i) Candida, Malassezia, Aspergillus, and Penicilliun genera were identified | [135] | |
| Schizophrenia | (i) Increased levels of S. cerevisiae and C. albicans species | [136] | |
| (ii) Close association of gastrointestinal tract disturbance with elevation of C. albicans species and lower cognitive score | [137] | ||
3.6.1. Inflammatory Bowel Disease (IBD)
Inflammatory bowel disease (IBD) is a group of intestinal diseases characterized by persistent inflammation of the digestive tract. The two most common diseases under IBD include Crohn's disease (CD) and ulcerative colitis (UC). A number of sources have proven a causal relation between gut mycobiome and IBD. For example, Ott et al. highlighted the differences between IBD with mucosal and fecal microbiota. The authors surmised that there is an alteration in the diversity and composition of fungal microbiota between patients with IBD and controls. However, the authors reported that there are no significant changes between CD and UC [101]. Subsequently, another study by Li et al. demonstrated that fungal dysbiosis in CD are associated with mucosal inflammation in patients with CD. In this study, the authors reported that the feces of patients with CD were characterized by the abundance of Aspergillus clavatus, C. neoformans, and Candida albicans, while Alternaria brassicicola, Gibberella moniliformis, Cryptococcus neoformans, and Candida spp. were reported in the inflamed mucosa [102]. Additionally, the diversity and richness of fungal species identified in this study were associated with the expression of, IFN-γ, IL-10, or TNF-α. Meanwhile, a study by Lewis et al. had identified fungal dysbiosis in patients with CD with an increase in Saccharomyces cerevisiae, Cyberlindnera jadinii, Clavispora lusitaniae, Kluyveromyces marxianus, and Candida albicans, accompanied with a change in bacteria composition [84]. A study by Hoarau et al. reported that an expansion of C. tropicalis is found in patients with Crohn's disease as compared to their healthy relatives. Furthermore, the authors reported a positive correlation between C. tropicalis, S. marcescens, and E. coli, suggesting that an interkingdom microbial interaction could be one of the key determinants in CD development [47]. Meanwhile, Liguori and collaborators showed that global fungus load increased in CD flare, predominantly by Basidiomycota and Ascomycota phyla. Meanwhile, Filobasidium uniguttulatum and Saccharomyces cerevisiae species were correlated with noninflamed mucosa, while Xylariales order was linked with inflamed mucosa [103]. A more recent study by Sokol et al. also revealed the distinctive mycobiome profile between IBD and healthy subjects. The authors reported a spike in Basidiomycota/Ascomycota ratio, accompanied with an expansion of Malassezia sympodialis and C. albicans and a decreased proportion of Saccharomyces cerevisiae in IBD in comparison with healthy controls. This study also highlighted the significant changes in fungal microbiota between remission and relapse stage within IBD cohort. For instance, C. albicans were remarkably increased during relapse when compared to remission stage. Lastly, based on the concomitant analysis of bacterial and fungal microbiota, the authors also suggest that an interkingdom interaction between fungi and bacteria could contribute towards IBD pathogenesis [9]. Hence, modulation of gut mycobiota could be a potential approach in the treatment of IBD [104].
3.6.2. Inflammatory Bowel Syndromes (IBS)
Inflammatory bowel syndrome is a functional gastrointestinal disorder associated with altered bowel habits. Few studies have documented the link between IBS and fungal microbiota in the gut. Overall, the representation of fungi in healthy individuals is an insignificant proportion (~0.1%) of the entire microbiome as reported by various studies, with the three major fungal genera being Saccharomycyes, Candida, and Cladosporium [105]. An earlier study by Levine et al. concluded that overgrowth of Candida species is associated with diarrhea symptom in patients receiving antibacterial therapy [106]. Subsequently, another study documented that patients with antibiotic-associated diarrhea have Candida overgrowth in the gastrointestinal tract [107]. Furthermore, Santelmann and Howard reported that “IBS associated symptoms” or “Candida syndrome” could be triggered by Candida products, antigens and cross-antigens [108]. A more recent study by Botschuijver et al. demonstrated fungal dybiosis, predominant by Saccharomyces cerevisiae and Candida albicans in patients with IBS [51]. Taken together, the possible role of fungal microbiota in IBS remains much to explore. Additionally, whether a disruption in bacteria microbiota causes a skewed commensal mycobiota profile in IBS remains a question.
3.6.3. Cancers
The gut mycobiome has been linked with the pathogenesis of colorectal adenoma, an inducer of colorectal cancer (CRC). A recent study by Luan et al. has discerned the role of gut fungi in different stages of colorectal adenoma. In the study, the authors characterized mucosal mycobiota in twenty-seven paired samples of adenomas and adjacent tissues, where Glomeromycota and Basidiomycota phyla dominated both adenomas and adjacent tissues from all subjects. In terms of genera, opportunistic pathogens Phoma and Candida were frequently identified (45%). Also, a decrease in fungal diversity in colorectal adenomas based on operational taxonomic unit (OTU) analysis was recorded [109]. Although the study illustrates the possible role of fungal microbiota in colorectal adenoma development, more studies are warranted to elucidate the cause-effect relationship between fungal microbiota and colorectal cancer. On the other hand, fungal dysbiosis is observed in patients with colorectal cancers and polyps. Additionally, the authors demonstrated that an increase in the Ascomycota/Basidiomycota ratio accompanied with the expansion of opportunistic fungi Trichosporon and Malassezia populations may facilitate the progression of colorectal cancer [110]. Similarly, Coker et al. reported fungal dysbiosis in patients with colorectal cancer. In addition to that, the authors also demonstrated an increase in the Basidiomycota : Ascomycota ratio with enrichment of fungal class Malasseziomycetes and depletion of classes Pneumocystidomycetes and Saccharomycetes were detected [111]. A recent study by Chin et al. has identified the presence of Schizosaccharomyces pombe in the guts of colorectal cancer patients and healthy subjects. The authors discovered that proteins secreted by Schizosaccharomyces pombe were present in high intensity in colorectal cancer patients, with four secreted proteins being closely related with the late stage of colorectal cancer [112]. Collectively, these studies suggest the undisputable role of fungi in colorectal cancer development and provide a new paradigm in utilizing these specific fungal signatures in disease diagnosis or therapeutic approaches.
3.6.4. Hepatitis B and HIV Infections
Infectious viruses such as hepatitis B virus and HIV virus, which can affect host immunity, pose the potential to modify the role of other microbes in the gut including fungi, in the context of disease progression and exacerbation. One study characterized the association of fungi in the gut with varying degrees of chronic hepatitis B virus infection via culture-independent and culture-dependent analysis. In this study, Chen et al. reported high prevalence of Aspergillus, Candida, Galactomyces, Saccharomyces, and Chaetomium in patients with hepatitis B. Moreover, the authors surmised that richness and diversity of fungal species in hepatitis B patients are positively correlated with the disease progression in patients with chronic HBV infection [11]. Nevertheless, the positive correlation does not differentiate between cause and effect, which have yet to explored in recent studies.
On the other hand, considerable efforts investigating the relationship between gut mycobiota and HIV are also receiving much attention, as fungal infections often result in diarrhea wasting syndrome, as seen in HIV/AIDS [113]. Jha et al. assessed the clinical and microbiological profile of HIV/AIDS cases in association with diarrhea. The authors discovered that the prevalence of fungi was higher in HIV-seropositive patients in comparison with HIV-negative controls. The authors also discovered that C. parvum, C. difficile, and C. albicans are significantly present in HIV-seropositive patients [114]. Similarly, Awoyeni et al. reported an association between candidiasis and HIV patients with diarrhea. The most common species identified in this study were C. albicans, C. krusei, and C. tropicalis [115]. Meanwhile, Gouba and Drancourt also reported a reduction in fungal diversity in HIV-positive patients accompanied with higher prevalence of Candida species [116]. In another study, Esebelahie et al. reported that the prevalence of Candida in HIV patients without antiretroviral treatment was higher than HIV patients with active antiretroviral treatment. The Candida species recovered from the study were C. albicans, C. glabrata, C. krusei, C. tropicalis, and C. parapsilosis [117]. Collectively, these studies suggest that fungal microbiota especially Candida species are associated with the secondary immunodeficiency in HIV-positive patients, in the case of diarrhea and antibiotic treatment.
3.6.5. Obesity
The association between fungi and obesity has been implicated in an obese subject with BMI of 48.9. In this study, an increase in fungal diversity is observed. Additionally, numerous fungi species identified in this study apparently originated from food sources (11 out of 16 fungal species). Nevertheless, the findings in this study may be overrepresented due to only one subject was involved in the study [118]. A recent study involving a higher number of obese subjects (n = 52) and controls was conducted. Based on the findings, there is a significant difference in the fungal composition between obese subjects and controls. Nonetheless, Candida, Nakaseomyces, and Penicillium were the most predominant genera identified in obese subjects, whereas Mucor racemosus and M. fuscus were the most represented in nonobese patients. Additionally, the authors found that Mucor genus was relatively increased in obese subjects upon weight loss. The authors surmise that targeting fungal communities in the gut could be a novel strategy in handling obesity [119].
3.6.6. Diabetes
Micobiome analysis has been performed in a few studies to elucidate the role of microbiota in driving the pathogenesis of diabetes mellitus [120–122]. In the context of fungal involvement, Soyucen and collaborators found that besides Echerichia coli, C. albicans and Enterobacteriaceae colonization were increased in the patients with type 1 diabetes mellitus (T1DM), whereas Bifidobacterium colonization was reduced [123]. Apart from that, Gosiewski et al. measured the quantitative changes of Candida species in both type 1 (T1DM) and type 2 diabetes mellitus (T2DM) patients via real-time PCR (qPCR). From the findings, C. albicans were predominant in the feces of patients with T1DM and T2DM. Nonetheless, there are no significant differences between T1DM and T2DM in terms of C. albicans colonization. Furthermore, the authors observed that the quantity of Candida is negatively correlated with serum lipids in T2DM patients [124]. Likewise, higher fungal species diversity was observed in T1DM with C. albicans being apparently less significant as compared to controls. The discrepancy could be due to greater recovery of fungal diversity in the T1DM cohort rather than a true reduction of C. albicans levels. Moreover, fungal species isolated from this study were resistant towards antifungal treatment [125]. Collectively, these studies showed that Candida species are more prevalent in patients with diabetes, particularly T1DM. This warrants further studies to elucidate the actual role of Candida in the pathogenesis of T1DM. Meanwhile, the role of gut fungi in T2DM remains much to be explored.
3.6.7. Atherosclerosis
Recently, numerous studies have suggested the association of gut microbiota with the development of atherosclerosis and cardiovascular diseases [126–128]. Indeed, a recent preliminary study based on cardiovascular risk has documented the possible involvement of gut mycobiota in carotid atherosclerosis. This study recruits thirty-three obese subjects (men and women) where the risk of developing cardiovascular disease was determined through Framingham risk score (FRS) and the carotid intima-media thickness (cIMT). From the study, the authors identified the abundance of the phylum Zygomycota, which consists of family Mucoraceae and genus Mucor showing negative correlation with cIMT. Additionally, M. racemosus was the most prevalent species found in subjects with a low cardiovascular risk profile, and this species was negatively associated with FRS and cIMT. The authors suggest that M. racemosus could be a relevant biomarker for cardiovascular risk [129]. Thus, it seems that mycobiota could have its role in cardiovascular diseases and more studies are needed to unravel the beneficial effects of mycobiota in cardiometabolic diseases.
3.6.8. Alcoholic Liver Disease
The role of gut mycobiota alcohol liver disease has been unraveled in a study involving animal model. In this study, Yang et al. observed an increased in fungal communities and β-glucan translocation into systemic circulation in mice after chronic alcohol administration. The authors reported that the effect of β-glucan-induced liver inflammation is mainly via C-type lectin-like receptor (CLEC7A) on Kupffer cells, which increases IL-1β expression, promotes hepatocyte damage, and consequently leads to the development of ethanol-induced liver disease. The authors further demonstrated a reduction in intestinal fungal growth and β-glucan translocation, followed by alleviation of ethanol-induced liver disease upon antifungal treatments. Meanwhile, the authors observed a decrease in fungal diversity along with Candida overgrowth in alcohol-dependent patients. Further, elevation in immune response against mycobiota was detected in alcoholic cirrhosis patients in comparison with patients with non–alcohol-related cirrhosis and healthy individuals. The authors surmised that chronic alcohol consumption is closely related with changes in gut mycobiota and translocation of fungal products. Thus, the authors suggest that manipulation of gut mycobiota could be an effective intervention for relieving alcohol-related liver disease [130]. A recent review by Szabo commented on the influence of fungal mycobiome on the complexity of alcohol-induced gut-liver axis from multiple aspects and some new insights to unravel the contribution of fungal microbiota between gut and liver in alcoholic liver disease [131].
3.6.9. Neurological Disorders
On the other hand, gut mycobiota is considered as one of the etiological agents in driving the pathophysiology of central nervous diseases (both psychiatric and nonpsychiatric disorders). It is believed that exposure to infectious pathogens at the critical stages of neurodevelopment may deteriorate central nervous system and elicit behavioral anomalies and psychiatric disorders during adulthood [132, 133]. Strati et al. reported dysbiosis in both gut fungal and bacteria microbiota in subjects with Rett syndrome (RTT), a progressive neurological disorder frequently linked with constipation and gastrointestinal dysfunctions. The authors also reported that high abundance of Candida species was detected [134]. Meanwhile, in autism spectrum disorders (ASD), Strati et al. observed an alteration in both fungal and bacteria gut microbiota of ASD patients, with Candida, Malassezia, Aspergillus, and Penicilliun genera commonly identified. Further, the authors even observed an increase in Candida genus in autistic compared to neurotypical subjects in ASD [135]. Meanwhile, several studies suggested that schizophrenia, a psychiatric disorder, is associated with fungal dysbiosis, as portrayed by elevation of S. cerevisiae and C. albicans species [136]. Moreover, Severance et al. also demonstrated the presence of antibody against C. albicans and a robust link between gastrointestinal disturbances with elevated C. albicans in patients with schizophrenia and bipolar disorder, accompanied with lower cognitive scores in these patients [137]. Subsequently, the authors showed that supplementation with a probiotic Lactobacillus rhamnosus and Bifidobacterium animalis greatly improved psychiatric symptoms and normalized blood levels of C. albicans antibodies in schizophrenia [138]. A recent review by Forbes et al. also describes the recent advancement in knowledge regarding gut mycobiome and the plausible role of gut mycobiota in neurological disorders [139].
4. Challenges and Future Direction
Numerous sources have proven that gut mycobiota have certain weightage in maintaining host homeostasis. Nevertheless, there remain challenges that hindered the progression of fungal research and in-depth understanding on the fungal community in human. Culture-dependent methods utilizing conventional microbiological techniques, such as biochemical assays [140], microscopy [141], and observation of the fungal growth in culture media (Sabouraud dextrose agar and potato dextrose agar) [142], remain the preference for scientists in resolving the complexity of fungal community in microbial ecosystems. This is due to the fact that culture-dependent methods are inexpensive and cost effective in many laboratories, and alternatives to culture-dependent methods are yet to be developed.
On the other hand, advancement in the molecular era has shifted researchers towards nonculture-dependent methods in microbial community analysis. The approaches such as polymerase chain reaction (PCR) [143] and high-throughput next generation sequencing (NGS) technologies [144, 145] diversified fungal identification and analysis without the need of for complex culturomics [146, 147]. Nevertheless, gold standard methods for culture-independent analysis to study the complexity of gut mycobiota are still lacking. Recently published data using different molecular approaches such as denaturing gradient gel electrophoresis (DGGE) and cloning [148], qPCR and NGS [29], and NGS alone [149] in gut mycobiome studies raise the concern on the precision of these studies. There is no comparative analysis being undertaken to analyse and to corroborate the findings using nonculture-dependent methods thus far.
Furthermore, the number and significance of fungi in the gut may be underestimated due to several reasons. First, fungal analysis through sequencing efforts and matched with available annotated reference sequences may be underrepresented. Technical problems such as misspellings, poor annotation of fungal database, incomplete representation, and other factors may arise when assigning taxonomy to sequencing reads [150, 151]. Second, the gut fungal composition is influenced by a number of factors, including age, diet, host immunity, medication, host genetics, and also bacterial microbiome through interkingdom interactions [19, 30]. It remains largely unknown to what extent these factors might affect the diversity and stability of gut mycobiota throughout different developmental stages. Third, fungal cell is a substantial mass of biomaterial with its size approximately 100 times larger as compared to a typical bacterial cell, where simple genome-counting numbers may fail to characterize [7]. Lastly, there remain uncultivable fungi yet to discovered, with unknown function and taxonomy [7]. Hopefully, with the advance of culturomics study, the role of uncultivable fungi can be explored, and a full understanding of the fungal compartment in the gut can be attainable.
Although certain fungal species are commensals of human at various body parts, certain “foodborne fungi” or food contamination by fungi pose a serious threat to human health. “Foodborne fungi” illnesses can be caused by fungi from the genera Aspergillus, Alternaria, Fusarium, Candida, and mucormycetes [152–154] or can be due to secondary metabolites such as mycotoxins. An estimation of 600 million foodborne diseases are reported annually [155], but the disease burden due to mycotoxin and fungus remains largely [156]. Moreover, we still lack a distinctive method to differentiate between foodborne fungi contaminants and commensal fungi that colonize the gut. The only distinguishable characteristic thus far is the visible clinical signs and symptoms often associated with foodborne microbial pathogens whereas the colonizers are often asymptomatic for the individuals that harbor the latter. A review mentioned that surrogate biomarkers such as procalcitonin in addition to the conventional but nonspecific neutrophil count could be used in conjunction with clinical evaluation for identifying infection as opposed to colonization [157]. Frequently, fungal contamination occurs during processing [158, 159], due to improper storage [160, 161], or due to the presence of fungi intrinsically in certain food products [162, 163]. Culturomics and noncultoromics methods are still actively being employed to identify fungi from both sources. Other possible future strategies could exploit the host factors to distinguish between the colonizers and the contaminants, since colonizers often elicit detectable immune responses whereas the colonizers do not cause untoward immune reactions or inflammations. Moreover, an ideal detection method should be developed to identify the presence of foodborne fungi at all stages of food production. As reviewed previously, a good detection tool should satisfy a few criteria such as high specificity and sensitivity, cost-effectiveness, is nonlaborious, and is not time consuming [164, 165].
Clearly, there is a strong connection between gut mycobiota and brain, and the potential of gut mycobiota in driving brain-associated diseases. Indeed, a review by Enaud et al. had proposed possible mechanisms involved in mycobiome-gut-brain axis interaction (GBA) [166]. Further, Candida species appear to be one of the most prevalent species identified in the studies discussed above. This indicates that Candida species could have notable role in the mycobime-GBA. Further studies should focus on the putative role of Candida species in this gut-brain interaction. Additionally, targeting gut mycobiota could be one of the potential intervention strategies to relieve neurological and neuropsychiatric disorders. For example, administration of Saccharomyces boulardii CNCM I-745 had improved intestinal neuromuscular anomalies in IBS-induced mouse model prior to Herpes Simplex Virus type 1 (HSV-1) exposure [167]. A recent study also documented the protective role of mycobiota in reducing inflammation in the central nervous system. In this study, Takata et al. reported that the administration of C. kefyr alleviated the severity of experimental autoimmune encephalomyelitis (EAE), an animal model of multiple sclerosis. The authors surmised that the protection could be due to alteration of bacterial microbiome, accompanied with increase of Tregs and regulatory dendritic cells in mesenteric lymph nodes and diminished production of T-helper 17 cells in the intestinal lamina propria [168].
5. Conclusion
From the accumulated evidences, it is clear that gut mycobiome has an indisputable role in host homeostasis and disease development, despite constituting only a small proportion in the gut. Nevertheless, the journey of uncovering the mystery of gut mycobiome must be continued. More multifaceted and multidisciplinary approaches have to be adopted in such a way to identify those uncultivatable or low abundance fungi in the gut, to characterize the fungal species and strain diversity in the gut, and also to differentiate permanent and transient fungal species that reside in the gut. In addition to that, interaction between fungi and their metabolites with various host players (brain, lungs, and host immune system) and xenobiotic components (diets, environments, and others) has to be emphasized, where this could offer new insight into the role of gut mycobiome on host physiology and disease development. Moreover, interkingdom interaction of various fungal species with other members (bacteria, parasite, and virus) present in the gut and how these interactions could affect us are areas to be further explored.
Acknowledgments
The study was supported by the Fundamental Research Grant Scheme (FRGS/1/2019/SKK11/TAYLOR/01/1) and (FRGS/1/2019/SKK11/PERDANA/03/1) from Ministry of Education, Malaysia.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
CVK, CPP, RB, YVC, SAN, and MA conceptualized the idea. CVK and CPP performed literature search, wrote, and revised the manuscript. CVK, YVC, CPP, SAN, RB, and MA revised the manuscript. All authors agreed to the final version of the manuscript.
References
- 1.Qin J., Consortium M. H. I. T., Li R., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson J., Garges S., Giovanni M., et al. The NIH human microbiome project. Genome Research. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sogin M. L., Morrison H. G., Huber J. A., et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui L., Morris A., Ghedin E. The human mycobiome in health and disease. Genome Medicine. 2013;5(7):p. 63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage D. C. Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology. 1977;31(1):107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy M. J. Regulation of candida albicans populations in the gastrointestinal tract: mechanisms and significance in GI and systemic candidiasis. In: McGinnis M. R., Borgers M., editors. Current topics in medical mycology. New York, NY, USA: Springer; 1989. pp. 315–402. [DOI] [PubMed] [Google Scholar]
- 7.Underhill D. M., Iliev I. D. The mycobiota: interactions between commensal fungi and the host immune system. Nature Reviews Immunology. 2014;14(6):405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huseyin C. E., O’toole P. W., Cotter P. D., Scanlan P. D. Forgotten fungi—the gut mycobiome in human health and disease. Microbiological Reviews. 2017;41(4):479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 9.Sokol H., Leducq V., Aschard H., et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iliev I. D., Funari V. A., Taylor K. D., et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Chen Z., Guo R., et al. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagnostic Microbiology and Infectious Disease. 2011;70(4):492–498. doi: 10.1016/j.diagmicrobio.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Miceli M. H., Díaz J. A., Lee S. A. Emerging opportunistic yeast infections. The Lancet Infectious Diseases. 2011;11(2):142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 13.Polvi E. J., Li X., O’Meara T. R., Leach M. D., Cowen L. E. Opportunistic yeast pathogens: reservoirs, virulence mechanisms, and therapeutic strategies. Cellular and Molecular Life Sciences. 2015;72(12):2261–2287. doi: 10.1007/s00018-015-1860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam M., Raes J., Pelletier E., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallen-Adams H. E., Suhr M. J. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8(3):352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash A. K., Auchtung T. A., Wong M. C., et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5(1):p. 153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogilnicka I., Ufnal M. Gut mycobiota and fungal metabolites in human homeostasis. Current Drug Targets. 2018;20(2):232–240. doi: 10.2174/1389450119666180724125020. [DOI] [PubMed] [Google Scholar]
- 18.Hibbett D. S., Binder M., Bischoff J. F., et al. A higher-level phylogenetic classification of the fungi. Mycological Research. 2007;111(5):509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann C., Dollive S., Grunberg S., et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8(6, article e66019) doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sam Q., Chang M., Chai L. The fungal mycobiome and its interaction with gut bacteria in the host. International Journal of Molecular Sciences. 2017;18(2):p. 330. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallen-Adams H. E., Kachman S. D., Kim J., Legge R. M., Martínez I. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecology. 2015;15:9–17. doi: 10.1016/j.funeco.2015.01.006. [DOI] [Google Scholar]
- 22.Hamad I., Ranque S., Azhar E. I., et al. Culturomics and amplicon-based metagenomic approaches for the study of fungal population in human gut microbiota. Scientific Reports. 2017;7(1):16788–16788. doi: 10.1038/s41598-017-17132-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Hamad I., Raoult D., Bittar F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: taxonomy and detection methods. Parasite Immunology. 2016;38(1):12–36. doi: 10.1111/pim.12284. [DOI] [PubMed] [Google Scholar]
- 24.Schloss P. D., Iverson K. D., Petrosino J. F., Schloss S. J. The dynamics of a family's gut microbiota reveal variations on a theme. Microbiome. 2014;2(1) doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. Development of the human infant intestinal microbiota. PLoS Biology. 2007;5(7):p. e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bliss J. M., Basavegowda K. P., Watson W. J., Sheikh A. U., Ryan R. M. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. The Pediatric Infectious Disease Journal. 2008;27(3):231–235. doi: 10.1097/INF.0b013e31815bb69d. [DOI] [PubMed] [Google Scholar]
- 27.Mutschlechner W., Karall D., Hartmann C., et al. Mammary candidiasis: molecular-based detection of Candida species in human milk samples. European Journal of Clinical Microbiology & Infectious Diseases. 2016;35(8):1309–1313. doi: 10.1007/s10096-016-2666-0. [DOI] [PubMed] [Google Scholar]
- 28.LaTuga M. S., Ellis J. C., Cotton C. M., et al. Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants. PLoS One. 2011;6(12, article e27858) doi: 10.1371/journal.pone.0027858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heisel T., Podgorski H., Staley C. M., Knights D., Sadowsky M. J., Gale C. A. Complementary amplicon-based genomic approaches for the study of fungal communities in humans. PLoS One. 2015;10(2, article e0116705) doi: 10.1371/journal.pone.0116705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strati F., Di Paola M., Stefanini I., et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filyk H. A., Osborne L. C. The multibiome: the intestinal ecosystem's influence on immune homeostasis, health, and disease. eBioMedicine. 2016;13:46–54. doi: 10.1016/j.ebiom.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark J. D. Influence of antibiotics or certain intestinal bacteria on orally administered Candida albicans in germ-free and conventional mice. Infection and Immunity. 1971;4(6):731–737. doi: 10.1128/IAI.4.6.731-737.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samonis G., Gikas A., Anaissie E. J., et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrobial Agents and Chemotherapy. 1993;37(1):51–53. doi: 10.1128/AAC.37.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason K. L., Erb Downward J. R., Mason K. D., et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infection and Immunity. 2012;80(10):3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erb Downward J. R., Falkowski N. R., Mason K. L., Muraglia R., Huffnagle G. B. Modulation of Post-Antibiotic Bacterial Community Reassembly and Host Response by Candida albicans. Scientific Reports. 2013;3(1, article 2191) doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu X., Zhang F., Yang X., et al. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Scientific Reports. 2015;5(1) doi: 10.1038/srep10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang T. T., Shao T. Y., Ang W. X. G., et al. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host & Microbe. 2017;22(6):809–816.e4. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castagliuolo I., Riegler M. F., Valenick L., LaMont J. T., Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infection and Immunity. 1999;67(1):302–307. doi: 10.1128/IAI.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buts J.-P., Dekeyser N., Stilmant C., Delem E., Smets F., Sokal E. Saccharomyces boulardii Produces in Rat Small Intestine a Novel Protein Phosphatase that Inhibits Escherichia coli Endotoxin by Dephosphorylation. Pediatric Research. 2006;60(1):24–29. doi: 10.1203/01.pdr.0000220322.31940.29. [DOI] [PubMed] [Google Scholar]
- 40.Colina A. R., Aumont F., Deslauriers N., Belhumeur P., de Repentigny L. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infection and Immunity. 1996;64(11):4514–4519. doi: 10.1128/IAI.64.11.4514-4519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Png C. W., Lindén S. K., Gilshenan K. S., et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. The American Journal of Gastroenterology. 2010;105(11):2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 42.Noverr M. C., Huffnagle G. B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infection and Immunity. 2004;72(11):6206–6210. doi: 10.1128/IAI.72.11.6206-6210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García C., Tebbji F., Daigneault M., et al. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere. 2017;2(6) doi: 10.1128/mSphere.00555-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iliev I. D., Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nature Reviews Immunology. 2017;17(10):635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghannoum M. Cooperative evolutionary strategy between the bacteriome and mycobiome. MBio. 2016;7(6) doi: 10.1128/mBio.01951-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalan L., Loesche M., Hodkinson B. P., et al. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. MBio. 2016;7(5) doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoarau G., Mukherjee P. K., Gower-Rousseau C., et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio. 2016;7(5) doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Brucker K., Tan Y., Vints K., et al. Fungal β-1, 3-glucan increases ofloxacin-tolerance of Escherichia coli in a polymicrobial E. coli–Candida albicans biofilm. Antimicrobial Agents and Chemotherapy. 2015;59(6):3052–3058. doi: 10.1128/AAC.04650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong E. F., Tsui C., Kucharíková S., Andes D., Van Dijck P., Jabra-Rizk M. A. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans Biofilm matrix. MBio. 2016;7(5) doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carabotti M., Scirocco A., Maselli M. A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Annals of Gastroenterology. 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- 51.Botschuijver S., Roeselers G., Levin E., et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. 2017;153(4):1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Moyes D. L., Naglik J. R. Mucosal immunity and candida albicans infection. Clinical & Developmental Immunology. 2011;2011, article 346307:9. doi: 10.1155/2011/346307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plantinga T. S., van der Velden W. J. F. M., Ferwerda B., et al. Early stop polymorphism in human DECTIN-1 is associated with increased Candida colonization in hematopoietic stem cell transplant recipients. Clinical Infectious Diseases. 2009;49(5):724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 54.van der Velden W. J. F. M., Netea M. G., de Haan A. F. J., Huls G. A., Donnelly J. P., Blijlevens N. M. A. Role of the mycobiome in human acute graft-versus-host disease. Biology of Blood and Marrow Transplantation. 2013;19(2):329–332. doi: 10.1016/j.bbmt.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Sokol H., Conway K. L., Zhang M., et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145(3):591–601.e3. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conti H. R., Gaffen S. L. IL-17-mediated immunity to the opportunistic fungal pathogen Candida albicans. Journal of Immunology. 2015;195(3):780–788. doi: 10.4049/jimmunol.1500909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Netea M. G., Joosten L. A. B., van der Meer J. W. M., Kullberg B.-J., van de Veerdonk F. L. Immune defence against Candida fungal infections. Nature Reviews Immunology. 2015;15(10):630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 58.Plato A., Hardison S. E., Brown G. D. Pattern recognition receptors in antifungal immunity. Seminars in Immunopathology. 2015;37(2):97–106. doi: 10.1007/s00281-014-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hueber W., Sands B. E., Lewitzky S., et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61(12):1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Luca A., Zelante T., D'Angelo C., et al. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunology. 2010;3(4):361–373. doi: 10.1038/mi.2010.22. [DOI] [PubMed] [Google Scholar]
- 61.Puel A., Döffinger R., Natividad A., et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. The Journal of Experimental Medicine. 2010;207(2):291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gessner M. A., Werner J. L., Lilly L. M., et al. Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus. Infection and Immunity. 2012;80(1):410–417. doi: 10.1128/IAI.05939-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Luca A., Carvalho A., Cunha C., et al. IL-22 and IDO1 affect immunity and tolerance to murine and human vaginal candidiasis. PLoS Pathogens. 2013;9(7, article e1003486) doi: 10.1371/journal.ppat.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti H. R., Bruno V. M., Childs E. E., et al. IL-17 receptor signaling in oral epithelial cells is critical for protection against oropharyngeal candidiasis. Cell Host & Microbe. 2016;20(5):606–617. doi: 10.1016/j.chom.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qamar A., Aboudola S., Warny M., et al. Saccharomyces boulardii stimulates intestinal immunoglobulin A immune response to Clostridium difficile toxin A in mice. Infection and Immunity. 2001;69(4):2762–2765. doi: 10.1128/IAI.69.4.2762-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas S., Metzke D., Schmitz J., Dörffel Y., Baumgart D. C. Anti-inflammatory effects of Saccharomyces boulardii mediated by myeloid dendritic cells from patients with Crohn’s disease and ulcerative colitis. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2011;301(6):G1083–G1092. doi: 10.1152/ajpgi.00217.2011. [DOI] [PubMed] [Google Scholar]
- 67.Tang C., Kamiya T., Liu Y., et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing Lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host & Microbe. 2015;18(2):183–197. doi: 10.1016/j.chom.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 68.Lamas B., Richard M. L., Leducq V., et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nature Medicine. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T., Fan C., Yao A., et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity. 2018;49(3):504–514.e4. doi: 10.1016/j.immuni.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rizzetto L., Ifrim D. C., Moretti S., et al. Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. The Journal of Biological Chemistry. 2016;291(15):7961–7972. doi: 10.1074/jbc.M115.699645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quintin J., Saeed S., Martens J. H. A., et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host & Microbe. 2012;12(2):223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dantzer R., Konsman J. P., Bluthé R. M., Kelley K. W. Neural and humoral pathways of communication from the immune system to the brain: parallel or convergent? Autonomic Neuroscience. 2000;85(1-3):60–65. doi: 10.1016/S1566-0702(00)00220-4. [DOI] [PubMed] [Google Scholar]
- 73.Graff L. A., Walker J. R., Bernstein C. N. Depression and anxiety in inflammatory bowel disease: a review of comorbidity and management. Inflammatory Bowel Diseases. 2009;15(7):1105–1118. doi: 10.1002/ibd.20873. [DOI] [PubMed] [Google Scholar]
- 74.Clarke G., Quigley E. M. M., Cryan J. F., Dinan T. G. Irritable bowel syndrome: towards biomarker identification. Trends in Molecular Medicine. 2009;15(10):478–489. doi: 10.1016/j.molmed.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 75.El Aidy S., Dinan T. G., Cryan J. F. Immune modulation of the brain-gut microbe axis. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunn A. J. Effects of cytokines and infections on brain neurochemistry. Clinical Neuroscience Research. 2006;6(1-2):52–68. doi: 10.1016/j.cnr.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Czakai K., Leonhardt I., Dix A., et al. Kruppel-like Factor 4 modulates interleukin-6 release in human dendritic cells after in vitro stimulation with Aspergillus fumigatus and Candida albicans. Scientific Reports. 2016;6(1, article 27990) doi: 10.1038/srep27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El Aidy S., Dinan T. G., Cryan J. F. Immune modulation of the brain-gut-microbe axis. Frontiers in Microbiology. 2014;5 doi: 10.3389/fmicb.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wheeler M. L., Limon J. J., Bar A. S., et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host & Microbe. 2016;19(6):865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McAleer J. P., Nguyen N. L. H., Chen K., et al. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. Journal of Immunology. 2016;197(1):97–107. doi: 10.4049/jimmunol.1502566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skalski J. H., Limon J. J., Sharma P., et al. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathogens. 2018;14(9, article e1007260) doi: 10.1371/journal.ppat.1007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X., Leonardi I., Semon A., et al. Response to fungal dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes aggravates allergic airway disease. Cell Host & Microbe. 2018;24(6):847–856.e4. doi: 10.1016/j.chom.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.David L. A., Maurice C. F., Carmody R. N., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis J. D., Chen E. Z., Baldassano R. N., et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host & Microbe. 2015;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banani H., Marcet-Houben M., Ballester A. R., et al. Genome sequencing and secondary metabolism of the postharvest pathogen Penicillium griseofulvum. BMC Genomics. 2016;17(1) doi: 10.1186/s12864-015-2347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Curbete M. M., Salgado H. R. N. A critical review of the properties of fusidic acid and analytical methods for its determination. Critical Reviews in Analytical Chemistry. 2016;46(4):352–360. doi: 10.1080/10408347.2015.1084225. [DOI] [PubMed] [Google Scholar]
- 87.Manzoni M., Rollini M. Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Applied Microbiology and Biotechnology. 2002;58(5):555–564. doi: 10.1007/s00253-002-0932-9. [DOI] [PubMed] [Google Scholar]
- 88.Shareck J., Belhumeur P. Modulation of morphogenesis in Candida albicans by various small molecules. Eukaryotic Cell. 2011;10(8):1004–1012. doi: 10.1128/EC.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Czerucka D., Dahan S., Mograbi B., Rossi B., Rampal P. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infection and Immunity. 2000;68(10):5998–6004. doi: 10.1128/IAI.68.10.5998-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sougioultzis S., Simeonidis S., Bhaskar K. R., et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-κB-mediated IL-8 gene expression. Biochemical and Biophysical Research Communications. 2006;343(1):69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 91.Kelesidis T., Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therapeutic Advances in Gastroenterology. 2012;5(2):111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buts J.-P., Keyser N. D., Raedemaeker L. D. Saccharomyces boulardii Enhances Rat Intestinal Enzyme Expression by Endoluminal Release of Polyamines. Pediatric Research. 1994;36(4):522–527. doi: 10.1203/00006450-199410000-00019. [DOI] [PubMed] [Google Scholar]
- 93.Castagliuolo I., LaMont J. T., Nikulasson S. T., Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infection and Immunity. 1996;64(12):5225–5232. doi: 10.1128/IAI.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murzyn A., Krasowska A., Stefanowicz P., Dziadkowiec D., Łukaszewicz M. Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS One. 2010;5(8, article e12050) doi: 10.1371/journal.pone.0012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schneider S. M., Girard-Pipau F., Filippi J., et al. Effects of Saccharomyces boulardii on fecal short-chain fatty acids and microflora in patients on long-term total enteral nutrition. World Journal of Gastroenterology. 2005;11(39):6165–6169. doi: 10.3748/wjg.v11.i39.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Velegraki A., Cafarchia C., Gaitanis G., Iatta R., Boekhout T. Malassezia infections in humans and animals: pathophysiology, detection, and treatment. PLoS Pathogens. 2015;11(1, article e1004523) doi: 10.1371/journal.ppat.1004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.White T. C., Findley K., Dawson T. L., et al. Fungi on the skin: dermatophytes and Malassezia. Cold Spring Harbor Perspectives in Medicine. 2014;4(8) doi: 10.1101/cshperspect.a019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith M. G., Des Etages S. G., Snyder M. Microbial synergy via an ethanol-triggered pathway. Molecular and Cellular Biology. 2004;24(9):3874–3884. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cugini C., Calfee M. W., Farrow J. M., Morales D. K., Pesci E. C., Hogan D. A. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Molecular Microbiology. 2007;65(4):896–906. doi: 10.1111/j.1365-2958.2007.05840.x. [DOI] [PubMed] [Google Scholar]
- 100.Peleg A. Y., Hogan D. A., Mylonakis E. Medically important bacterial-fungal interactions. Nature Reviews Microbiology. 2010;8(5):340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 101.Ott S. J., Kühbacher T., Musfeldt M., et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scandinavian Journal of Gastroenterology. 2008;43(7):831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 102.Li Q., Wang C., Tang C., He Q., Li N., Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. Journal of Clinical Gastroenterology. 2014;48(6):513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liguori G., Lamas B., Richard M. L., et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. Journal of Crohn's & Colitis. 2016;10(3):296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lam S., Zuo T., Ho M., Chan F. K. L., Chan P. K. S., Ng S. C. Review article: Fungal alterations in inflammatory bowel diseases. Alimentary Pharmacology & Therapeutics. 2019;50(11-12):1159–1171. doi: 10.1111/apt.15523. [DOI] [PubMed] [Google Scholar]
- 105.Chong P. P., Chin V. K., Looi C. Y., Wong W. F., Madhavan P., Yong V. C. The microbiome and irritable bowel syndrome – a review on the pathophysiology, current research and future therapy. Frontiers in Microbiology. 2019;10, article 1136 doi: 10.3389/fmicb.2019.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Levine J., Dykoski R. K., Janoff E. N. Candida-associated diarrhea: a syndrome in search of credibility. Clinical Infectious Diseases. 1995;21(4):881–886. doi: 10.1093/clinids/21.4.881. [DOI] [PubMed] [Google Scholar]
- 107.Krause R., Reisinger E. C. Candida and antibiotic-associated diarrhoea. Clinical Microbiology and Infection. 2005;11(1):1–2. doi: 10.1111/j.1469-0691.2004.00978.x. [DOI] [PubMed] [Google Scholar]
- 108.Santelmann H., Howard J. M. Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. European Journal of Gastroenterology & Hepatology. 2005;17(1):21–26. doi: 10.1097/00042737-200501000-00005. [DOI] [PubMed] [Google Scholar]
- 109.Luan C., Xie L., Yang X., et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Scientific Reports. 2015;5(1, article 7980) doi: 10.1038/srep07980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao R., Kong C., Li H., et al. Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. European Journal of Clinical Microbiology & Infectious Diseases. 2017;36(12):2457–2468. doi: 10.1007/s10096-017-3085-6. [DOI] [PubMed] [Google Scholar]
- 111.Coker O. O., Nakatsu G., Dai R. Z., et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68(4):654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chin S. F., Megat Mohd Azlan P. I. H., Mazlan L., Neoh H. M. Identification of Schizosaccharomyces pombe in the guts of healthy individuals and patients with colorectal cancer: preliminary evidence from a gut microbiome secretome study. Gut Pathogens. 2018;10(1):p. 29. doi: 10.1186/s13099-018-0258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kashyap B., Bhalla P., Uppal B. Enteric pathogens in HIV/AIDS from a tertiary care hospital. Indian Journal of Community Medicine. 2009;34(3):237–242. doi: 10.4103/0970-0218.55291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jha A. K., Uppal B., Chadha S., et al. Clinical and microbiological profile of HIV/AIDS cases with diarrhea in North India. Journal of Pathogens. 2012;2012:7. doi: 10.1155/2012/971958.971958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Awoyeni A., Olaniran O., Odetoyin B., et al. Isolation and evaluation of Candida species and their association with CD4+ T cells counts in HIV patients with diarrhoea. African Health Sciences. 2017;17(2):322–329. doi: 10.4314/ahs.v17i2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gouba N., Drancourt M. Digestive tract mycobiota: a source of infection. Médecine et Maladies Infectieuses. 2015;45(1-2):9–16. doi: 10.1016/j.medmal.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 117.Esebelahie N. O., Enweani I. B., Omoregie R. Candida colonisation in asymptomatic HIV patients attending a tertiary hospital in Benin City, Nigeria. Libyan Journal of Medicine. 2013;8(1, article 20322) doi: 10.3402/ljm.v8i0.20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gouba N., Raoult D., Drancourt M. Plant and fungal diversity in gut microbiota as revealed by molecular and culture investigations. PLoS One. 2013;8(3, article e59474) doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mar Rodríguez M., Pérez D., Javier Chaves F., et al. Obesity changes the human gut mycobiome. Scientific Reports. 2015;5(1, article 14600) doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Paun A., Yau C., Danska J. S. The influence of the microbiome on type 1 diabetes. Journal of Immunology. 2017;198(2):590–595. doi: 10.4049/jimmunol.1601519. [DOI] [PubMed] [Google Scholar]
- 121.Han H., Li Y., Fang J., et al. Gut microbiota and type 1 diabetes. International Journal of Molecular Sciences. 2018;19(4):p. 995. doi: 10.3390/ijms19040995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vallianou N. G., Stratigou T., Tsagarakis S. Microbiome and diabetes: where are we now? Diabetes Research and Clinical Practice. 2018;146:111–118. doi: 10.1016/j.diabres.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 123.Soyucen E., Gulcan A., Aktuglu-Zeybek A. C., Onal H., Kiykim E., Aydin A. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatrics International. 2014;56(3):336–343. doi: 10.1111/ped.12243. [DOI] [PubMed] [Google Scholar]
- 124.Gosiewski T., Salamon D., Szopa M., Sroka A., Malecki M. T., Bulanda M. Quantitative evaluation of fungi of the genus Candida in the feces of adult patients with type 1 and 2 diabetes - a pilot study. Gut Pathogens. 2014;6(1):p. 43. doi: 10.1186/s13099-014-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kowalewska B., Zorena K., Szmigiero-Kawko M., Wąż P., Myśliwiec M. Higher diversity in fungal species discriminates children with type 1 diabetes mellitus from healthy control. Patient Preference and Adherence. 2016;10:591–599. doi: 10.2147/PPA.S97852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jie Z., Xia H., Zhong S. L., et al. The gut microbiome in atherosclerotic cardiovascular disease. Nature Communications. 2017;8(1):p. 845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tang W. H. W., Kitai T., Hazen S. L. Gut microbiota in cardiovascular health and disease. Circulation Research. 2017;120(7):1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoshida N., Yamashita T., Hirata K. I. Gut microbiome and cardiovascular diseases. Diseases. 2018;6(3):p. 56. doi: 10.3390/diseases6030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chacón M. R., Lozano-Bartolomé J., Portero-Otín M., et al. The gut mycobiome composition is linked to carotid atherosclerosis. Beneficial Microbes. 2018;9(2):185–198. doi: 10.3920/BM2017.0029. [DOI] [PubMed] [Google Scholar]
- 130.Yang A. M., Inamine T., Hochrath K., et al. Intestinal fungi contribute to development of alcoholic liver disease. The Journal of Clinical Investigation. 2017;127(7):2829–2841. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Szabo G. Gut-liver axis beyond the microbiome: how the fungal mycobiome contributes to alcoholic liver disease. Hepatology. 2018;68(6):2426–2428. doi: 10.1002/hep.30055. [DOI] [PubMed] [Google Scholar]
- 132.Knuesel I., Chicha L., Britschgi M., et al. Maternal immune activation and abnormal brain development across CNS disorders. Nature Reviews Neurology. 2014;10(11):643–660. doi: 10.1038/nrneurol.2014.187. [DOI] [PubMed] [Google Scholar]
- 133.Labouesse M. A., Langhans W., Meyer U. Long-term pathological consequences of prenatal infection: beyond brain disorders. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2015;309(1):R1–R12. doi: 10.1152/ajpregu.00087.2015. [DOI] [PubMed] [Google Scholar]
- 134.Strati F., Cavalieri D., Albanese D., et al. Altered gut microbiota in Rett syndrome. Microbiome. 2016;4(1):p. 41. doi: 10.1186/s40168-016-0185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Strati F., Cavalieri D., Albanese D., et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5(1):p. 24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Severance E. G., Alaedini A., Yang S., et al. Gastrointestinal inflammation and associated immune activation in schizophrenia. Schizophrenia Research. 2012;138(1):48–53. doi: 10.1016/j.schres.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Severance E. G., Gressitt K. L., Stallings C. R., et al. Candida albicans exposures, sex specificity and cognitive deficits in schizophrenia and bipolar disorder. NPJ Schizophrenia. 2016;2(1, article 16018) doi: 10.1038/npjschz.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Severance E. G., Gressitt K. L., Stallings C. R., et al. Probiotic normalization of Candida albicans in schizophrenia: a randomized, placebo-controlled, longitudinal pilot study. Brain, Behavior, and Immunity. 2017;62:41–45. doi: 10.1016/j.bbi.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Forbes J. D., Bernstein C. N., Tremlett H., van Domselaar G., Knox N. C. A fungal world: could the gut mycobiome be involved in neurological disease? Frontiers in Microbiology. 2019;9, article 3249 doi: 10.3389/fmicb.2018.03249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Khatib R., Riederer K. M., Ramanathan J., Baran J. Faecal fungal flora in healthy volunteers and inpatients. Mycoses. 2001;44(5):151–156. doi: 10.1046/j.1439-0507.2001.00639.x. [DOI] [PubMed] [Google Scholar]
- 141.de Repentigny L., Phaneuf M., Mathieu L. G. Gastrointestinal colonization and systemic dissemination by Candida albicans and Candida tropicalis in intact and immunocompromised mice. Infection and Immunity. 1992;60(11):4907–4914. doi: 10.1128/IAI.60.11.4907-4914.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ouanes A., Kouais A., Marouen S., Sahnoun M., Jemli B., Gargouri S. Contribution of the chromogenic medium CHROMagar®Candida in mycological diagnosis of yeasts. Journal de Mycologie Médicale. 2013;23(4):237–241. doi: 10.1016/j.mycmed.2013.07.058. [DOI] [PubMed] [Google Scholar]
- 143.Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mardis E. R. Next-generation sequencing platforms. Annual Review of Analytical Chemistry. 2013;6(1):287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 145.Zoll J., Snelders E., Verweij P. E., Melchers W. J. G. Next-generation sequencing in the mycology lab. Current Fungal Infection Reports. 2016;10(2):37–42. doi: 10.1007/s12281-016-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Landlinger C., Bašková L., Preuner S., Willinger B., Buchta V., Lion T. Identification of fungal species by fragment length analysis of the internally transcribed spacer 2 region. European Journal of Clinical Microbiology & Infectious Diseases. 2009;28(6):613–622. doi: 10.1007/s10096-008-0683-3. [DOI] [PubMed] [Google Scholar]
- 147.Shokralla S., Spall J. L., Gibson J. F., Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Molecular Ecology. 2012;21(8):1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- 148.Scanlan P. D., Marchesi J. R. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. The ISME Journal. 2008;2(12):1183–1193. doi: 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 149.Tang J., Iliev I. D., Brown J., Underhill D. M., Funari V. A. Mycobiome: approaches to analysis of intestinal fungi. Journal of Immunological Methods. 2015;421:112–121. doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Suhr M. J., Hallen-Adams H. E. The human gut mycobiome: pitfalls and potentials—a mycologist’s perspective. Mycologia. 2016;107(6):1057–1073. doi: 10.3852/15-147. [DOI] [PubMed] [Google Scholar]
- 151.Halwachs B., Madhusudhan N., Krause R., et al. Critical issues in mycobiota analysis. Frontiers in Microbiology. 2017;8:p. 180. doi: 10.3389/fmicb.2017.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tomsíková A. Risk of fungal infection from foods, particularly in immunocompromised patients. Epidemiol Mikrobiol Imunol. 2002;51(2):78–81. [PubMed] [Google Scholar]
- 153.Brenier-Pinchart M. P., Faure O., Garban F., et al. Ten-year surveillance of fungal contamination of food within a protected haematological unit. Mycoses. 2006;49(5):421–425. doi: 10.1111/j.1439-0507.2006.01257.x. [DOI] [PubMed] [Google Scholar]
- 154.Pitt J. I., Hocking A. D. Fungi and Food Spoilage. 3. New York: Springer; 2009. [Google Scholar]
- 155.[WHO] World Health Organization. WHO estimates of the global burden of foodborne diseases. 2015, http://apps.who.int/iris/bitstream/10665/199350/1/9789241565165_eng.pdf.
- 156.Marroquin-Cardona A. G., Johnson N. M., Phillips T. D., Hayes A. W. Mycotoxins in a changing global environment – A review. Food and Chemical Toxicology. 2014;69:220–230. doi: 10.1016/j.fct.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 157.Wunderink R. G. Surrogate markers and microbiologic end points. Clinical Infectious Diseases. 2010;51(S1):S126–S130. doi: 10.1086/653061. [DOI] [PubMed] [Google Scholar]
- 158.Lazar S. P., Lukaszewicz J. M., Persad K. A., Reinhardt J. F. Rhinocerebral Mucor circinelloides infection in immunocompromised patient following yogurt ingestion. Delaware Medical Journal. 2014;86(8):245–248. [PubMed] [Google Scholar]
- 159.Vallabhaneni S., Walker T. A., Lockhart S. R., et al. Notes from the field: fatal gastrointestinal mucormycosis in a premature infant associated with a contaminated dietary supplement—Connecticut, 2014. MMWR. 2015;64(6):155–156. [PMC free article] [PubMed] [Google Scholar]
- 160.Aboltins C. A., Pratt W. A. B., Solano T. R. Fungemia secondary to gastrointestinal Mucor indicus infection. Clinical Infectious Diseases. 2006;42(1):154–155. doi: 10.1086/498751. [DOI] [PubMed] [Google Scholar]
- 161.Gurgui M., Sanchez F., March F., et al. Nosocomial outbreak of Blastoschizomyces capitatus associated with contaminated milk in a haematological unit. Journal of Hospital Infection. 2011;78(4):274–278. doi: 10.1016/j.jhin.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 162.Sutherland J. C., Jones T. H. Gastric mucormycosis: report of case in a Swazi. South African Medical Journal. 1960;34:p. 161. [PubMed] [Google Scholar]
- 163.Radosavljevic M., Koenig H.´.`., Letscher-Bru V.´., et al. Candida catenulata fungemia in a cancer patient. Journal of Clinical Microbiology. 1999;37(2):475–477. doi: 10.1128/JCM.37.2.475-477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Benedict K., Chiller T. M., Mody R. K. Invasive fungal infections acquired from contaminated food or nutritional supplements: a review of the literature. Foodborne Pathogens and Disease. 2016;13(7):343–349. doi: 10.1089/fpd.2015.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Priyanka B., Patil R. K., Dwarakanath S. A review on detection methods used for foodborne pathogens. Indian Journal of Medical Research. 2016;144(3):327–338. doi: 10.4103/0971-5916.198677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Enaud R., Vandenborght L. E., Coron N., et al. The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms. 2018;6(1):p. 22. doi: 10.3390/microorganisms6010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Brun P., Scarpa M., Marchiori C., et al. Saccharomyces boulardii CNCM I-745 supplementation reduces gastrointestinal dysfunction in an animal model of IBS. PLoS One. 2017;12(7, article e0181863) doi: 10.1371/journal.pone.0181863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Takata K., Tomita T., Okuno T., et al. Dietary yeasts reduce inflammation in central nerve system via microflora. Annals of Clinical Translational Neurology. 2015;2(1):56–66. doi: 10.1002/acn3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]


