Short abstract
The development and maintenance of chronic neuropathic pain involves distorted neuroglial interactions, which result in prolonged perturbations of immune and inflammatory response, as well as disrupted synapses and cellular interactions. Spinal cord stimulation (SCS) has proven effective and safe for more than 40 years, but comprehensive understanding of its mode of action remains elusive. Previous work in our laboratory provided evidence that conventional SCS parameters modulate biological processes associated with neuropathic pain in animals. This inspired the development of differential target multiplexed programming (DTMP) in which multiple electrical signals are used for modulating glial cells and neurons in order to rebalance their interactions. This work compares DTMP with both low rate and high rate programming using an animal model of neuropathic pain. The spared nerve injury model was implemented in 48 rats equally randomized into four experimental groups: No-SCS, DTMP, low rate, and high rate. Naive animals (N = 7) served as a reference control. SCS was applied continuously for 48 h and pain-related behavior assessed before and after SCS. RNA from the spinal cord exposed to SCS was sequenced to determine changes in gene expression as a result of injury (No-SCS vs. naïve) and as a result of SCS (SCS vs. No-SCS). Bioinformatics tools (Weighted Gene Co-expression Network Analysis and Gene Ontology Enrichment Analysis) were used to evaluate the significance of the results. All three therapies significantly reduced mechanical hypersensitivity, although DTMP provided statistically better results overall. DTMP also reduced thermal hypersensitivity significantly. RNA-sequencing corroborated the complex effects of nerve injury on the transcriptome. In addition, DTMP provided significantly more effective modulation of genes associated with pain-related processes in returning their expression toward levels observed in naïve, noninjured animals. DTMP provides a more effective way of modulating the expression of genes involved in pain-relevant biological processes associated with neuroglial interactions.
Keywords: Spinal cord stimulation, differential target multiplexed programming, chronic neuropathic pain, neuroglial interactions, transcriptomics
Introduction
Since its inception in the late 1960s, electrical stimulation of the dorsal aspect of the spinal cord (SCS) has been effectively and safely utilized to provide relief and improve quality of life for many patients suffering from intractable chronic neuropathic pain.1 In recent years, the field has advanced from the traditional paresthesia-based tonic modality to other modalities that make use of larger pulse rates, smaller pulse widths (PWs), lower intensities, or the addition of bursting.2 Such a shift in the paradigm has revolutionized the SCS space from both clinical and commercial perspectives. It has also sparked the scientific interest to investigate and formulate a mode of action that goes beyond the conventional one based on the Gate Control Theory.3–6
Traditionally, the mechanism of action, based on the Gate Control Theory, has been viewed in an electrophysiological fashion, centered on the effect of applying a pulsed tonic electrical field on neuronal transmission. Within this framework, an optimized low rate pulsed electric field applied at the appropriate vertebral segment of the spine would activate mechano-sensitive A-beta fibers in the network, thus gating out the nociceptive information carried out by the smaller sensory A-delta and C-fibers. As a result, the brain perceives a tingling sensation rather than pain.
Our efforts have been placed on investigating the effects of continuous SCS in neural tissue at the molecular level. In 2016, our group published the first transcriptomic-based analysis of traditional SCS on an animal model of neuropathic pain based on microarray analysis.7 This study demonstrated that SCS modulates gene expression in the adjacent spinal cord tissue, and the dorsal root ganglion corresponding with the largest innervation of the injured peripheral nerve. More recently, Guan and coworkers published a similar study using RNA sequencing.8 Both studies reaffirm the molecular complexity of chronic pain. From a transcriptomics perspective, pain involves a multitude of genes that are affected by the nociceptive input and can be grouped into categories based on their known involvement in certain biological processes. Among the various biological processes involved, glial cell activation stands out in both studies. Glia are abundant cells in neural tissue, including astrocytes, oligodendrocytes, satellite cells, Schwann cells, microglia, and ependymal cells. Herculano-Houzel and coworkers have determined that the whole human brain contains about the same number of glia and neurons, although the glia-neuron distribution is variable between different brain structures.9 For instance, glia are more abundant than neurons (3.8 to 1) in the cerebral cortex, but less abundant (1 to 4.3) in the cerebellum. Interestingly, glia largely outnumber neurons in the thalamus, hypothalamus, pons, and medulla (11.4 to 1)10 and the entire spinal cord (5:1).11 Recently, our group measured the glia to neuron ratio in the posterior aspect of the human spinal cord, from segments T8 to T11, where SCS is typically applied. We found that glia outnumbered neurons (20 to 1) in this section of the cord.12 These results imply that when an electrical field is applied to the dorsal aspect of the cord, it affects both neuron and glial cells and thus the ability to modulate the homeostatic milieu resulting from neuroglial interactions.
It is well known that glial cells are electrically excitable and have a resting membrane potential larger than that of neurons. Roitbak and Fanardjian showed that the glial membrane could be depolarized by an external electrical stimulus dependent on applied voltage and frequency.13 Furthermore, Lee and coworkers demonstrated that astrocytes in the thalamus release glutamate, an excitatory neurotransmitter, in a manner dependent on typical adjustable parameters of pulsed signals (intensity, PW, and frequency) as well as the charge balance.14 In addition, Lee et al. confirmed that the release of glutamate by glial cells is associated with the abolishment of thalamic network oscillators.15 Another study by Yamazaki et al. revealed that electrically induced depolarization of oligodendrocytes in the rat hippocampus increases the conduction velocity of action potentials transmitted through the axon being myelinated by the stimulated oligodendrocyte.16
This body of evidence allows us to hypothesize that SCS parameters can be customized to differentially target neurons and glial cells in order to balance neuroglial interactions and optimize the beneficial outcomes of the therapy. This work describes the use of differential target multiplexed programming (DTMP) in an animal model of neuropathic pain in comparison to either conventional low-frequency or high-frequency programming.
Materials and methods
The study was approved by the Institutional Animal Care and Use Committee at Illinois Wesleyan University. A total of 55 male adult Sprague-Dawley rats (Envigo, Indianapolis, IN) weighing in the 275–315 g range were randomized into five groups (Table 1). The control Naïve group had N = 7, while the treatment groups started with N = 12. If the animal did not meet the minimum required 30% decrease in withdrawal threshold (WT) post-spared nerve injury (SNI), it was considered a nonresponder to the injury model and was not used in the study. Animals were housed individually in a temperature and humidity control room and subjected to a 12-h light/dark cycle. Food and water were supplied ad libitum.
Table 1.
Experimental programming groups of the study.
| Experimental group | SNI | SCS |
|---|---|---|
| Naïve | N/A | N/A |
| No-SCS | Yes | N/A |
| DTMP | Yes | Yes |
| LR | Yes | Yes |
| HR | Yes | Yes |
DTMP: differential target multiplexed programming; LR: low rate; HR: high rate; SCS: spinal cord stimulation; SNI: spared nerve injury; N/A: not applicable.
Behavioral testing for mechanical and thermal allodynia
All rats were assessed for hind paw sensitivity in response to mechanical and thermal stimulus prior to surgery as well as before and during SCS. Mechanical sensitization was assessed by measuring the WT using standard von Frey filaments (DanMic Global, San Jose, CA) varying in diameter and tensile strength to provide a gradual increase in force applied with each subsequent fiber. Animals were placed in specialized cages on a stand with a wire mesh floor and were allowed to acclimate for approximately 20 min or until exploratory behavior had ceased. Testing was performed by a researcher, blinded to treatment groups, throughout the study. The von Frey up-down method was used as previously described.17 Briefly, the starting von Frey filament was pressed against the rat’s hind paw at a perpendicular angle and advanced until either the animal withdrew its paw or a bend in the filament was observed over a period of six rounds of testing. If a withdrawal occurs, the next weaker filament is applied. If the bend occurred prior to paw-withdrawal then the next sturdiest filament would be used. The WT was then calculated based on the logarithmic trend line generated from plotting the filament strengths (2–180 g) against filament numbers (1–10).
Hot and cold allodynia was assessed by dynamic hot/cold (IITC Life Science, Woodland Hills, CA) testing based on established methods.18,19 For hot testing, the plate was set to an initial acclimation temperature of 30°C and the temperature was set to increase to 46°C at a rate of 6°C/min. As the temperature increased, the rat was observed for nociceptive behavior. If the rat retracted its paw for more than 2 s, the temperature change was stopped and the temperature at the point of withdrawal was recorded. If no response was elicited, the final temperature was recorded. The experiment was repeated with 2–4 min in between trials. Cold testing was performed in the same format, with the initial acclimation temperature of 20°C, decreasing to 4°C at a rate of 6°C/min.
Lead implantation and SNI induction
Following presurgical behavioral testing, animals were anesthetized and epidurally implanted with a custom-made, small diameter, four-electrode SCS cylindrical lead (0.62 mm diameter, 1 mm contact length; Heraeus Medical, Minneapolis, MN). The lead was advanced to the L1 vertebral level where the L5 nerve root synapses to the cord, and properly anchored to prevent migration. Electrode cables were terminated in a connector block, attached to a customized harness, which connected the lead to an external neurostimulator (ENS, 37022-MRS; Medtronic, Minneapolis, MN). Motor response was induced to verify lead placement and determine which limb was ipsilateral to the stimulation. For animals receiving the SNI model,20 the ipsilateral tibial and common peroneal nerves were transected with 1–2 mm of nerve tissue removed, leaving the sural branch intact.
Five days postsurgery, mechanical sensitivity was tested to verify the establishment of the SNI model. A 30% reduction in paw WT relative to baseline scores was used as a cutoff to determine nonresponse to injury. Those that did not meet or exceed the cutoff were not included in the study. Rats receiving stimulation were connected to the ENS, which was mounted on a counter-balancing swivel system that allowed the animal to roam freely in the cage during stimulation.
The DTMP approach utilizes multiplexed charge-balanced pulsed signals with frequencies in the 20–1200 Hz range and a maximum PW of 500 μs. In this particular study, signals were multiplexed in order to provide stimulation with components at frequencies of 50 Hz (150 µs PW) and 1200 Hz (50 µs PW), distributed over the four contacts of the lead. Low rate (LR) programming was set to 50 Hz and 150 μs PW, while high rate (HR) programming was set at 1200 Hz and 50 μs PW. The same set of SCS parameters were used for each animal in a treatment group. Signal intensities were set to ∼70% of the motor threshold (MT) tested under a given stimulation program and were in the 0.02–0.10 mA range for HR, 0.03–0.09 mA range for LR, and 0.03–0.10 mA range for DTMP signals. None of the stimulations were duty-cycled, and the intensities were kept constant throughout the 48 h of SCS.
RNA isolation
Animals were euthanized via CO2 inhalation following behavioral testing after 48 h of SCS. The overlaid region of the spinal cord, ipsilateral dorsal quadrant of the L1–L2 segment, was harvested and stored in RNAlater® (Thermo Fisher Scientific, Waltham, MA) at –20°C. Tissue was homogenized in 1 ml Tri Reagent and RNA was extracted using TRIzol® according to the manufacturer’s instructions (Molecular Research Center, Cincinnati, OH). RNA was treated with DNase I (Thermo Fisher Scientific, Waltham, MA) in the presence of RNase inhibitor and then column-purified (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. RNA was sent to the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for RNA sequencing and bioinformatics analyses.
RNA sequencing
Construction of libraries and sequencing on the Illumina HiSeq® 4000 were performed at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. Total RNAs were DNAse-treated with the RNAse-free DNase set from Qiagen (Germantown, MD) and run on a Fragment Analyzer (Advanced Analytical, Ankeny, IA) to evaluate RNA integrity. RNAseq libraries were constructed with the TruSeq® Stranded mRNA Sample Prep kit (Illumina, San Diego, CA). Briefly, polyadenylated messenger RNAs (mRNAs) were enriched from 1 μg of high-quality DNA-free total RNA with oligo dT beads. The mRNAs were chemically fragmented, annealed with a random hexamer and converted to double stranded cDNAs, which were subsequently blunt-ended, 3’-end A-tailed and ligated to indexed adaptors. Each library was ligated to a unique dual indexed adaptor (unique dual indexes) to prevent index switching. The adaptor-ligated double-stranded cDNAs were amplified by Polymerase Chain Reaction for eight cycles with the high fidelity (HiFi) polymerase (Kapa Biosystems, Wilmington, MA) to reduce the likeliness of multiple identical reads due to preferential amplification. The final libraries were quantitated with Qubit™ (ThermoFisher, Waltham, MA) and the average library fragment length was determined on a Fragment Analyzer. The libraries were diluted to 10 nM and further quantitated Polymerase Chain Reaction (qPCR) on a CFX Connect™ Real-Time qPCR system (Biorad, Hercules, CA) for accurate pooling of the barcoded libraries and maximization of the number of clusters in the flow cell.
The pooled barcoded libraries were loaded on three lanes of an eight-lane flow cell for cluster formation and sequenced on an Illumina HiSeq® 4000. The libraries were sequenced from one end of the cDNA fragments for a total of 100 base pairs (bp). The fastq read files were generated and demultiplexed with the bcl2fastq v 2.17.1.14 Conversion Software (Illumina, San Diego, CA), which also trims adaptor sequences and removes reads less than 35 bp after trimming. The quality of the resulting fastq files was evaluated with the FastQC software (Brabaham Bioinformatics, Cambridge, UK), which generates reports with the quality scores, base composition, k-mer, sequence guanine-cytosine (GC) and per base N contents, sequence duplication levels, and overrepresented sequences.
Average quality scores were above 40 for all samples through the end of the reads, so no quality-based trimming was performed. Salmon v 0.8.221 was used to quasi-map read to the NCBI’s Rnor_6.0 transcriptome based on Annotation Release 106 and to quantify the abundance of each transcript. The transcriptome was first indexed, then quasi-mapping was performed to map reads to the transcriptome with additional arguments –seqBias and –gcBias to correct sequence-specific and GC content biases and –numBootstraps = 30 to compute bootstrap transcript abundance estimates. Gene-level counts were then estimated based on transcript-level counts using the “bias-corrected counts without an offset” method from tximport v 1.6.0; this method provides more accurate gene-level counts estimates and keeps multimapped reads in the analysis compared to traditional genome alignment methods.22
Weighted Gene Co-expression Network Analysis
Gene-level counts were imported into R v 3.4.3 and genes without at least 0.5 counts per million after trimmed-mean of M values (TMM) normalization23 in at least four samples were filtered out. TMM normalization factors were recalculated after filtering and log2-based count per million values (logCPM) were calculated using the cpm() function from edgeR v 3.20.524 with prior.count = 3 to help stabilize fold-changes of extremely low expression genes. Multidimensional scaling clustering of the top 5000 most variable genes indicated two samples were slight outliers and a slight bit of unexplained variation among all samples. To help correct for these issues, we used surrogate variables analysis25,26 to estimate six surrogate variables to add to the statistical model as quantitative co-variates. Differential gene expression analysis was performed using the limma-trend method27,28 on the logCPM values and a one-way analysis of variance (ANOVA) across 10 experimental groups was computed. The top 50% of genes with lowest p-values were selected for Weighted Gene Co-expression Network Analysis (WGCNA)29,30 using WGCNA v1.61 in order to reduce the dimensionality of the data set from thousands of genes to dozens of modules containing genes with similar expression patterns. The effects of the six surrogate variables were removed from the logCPM values so they would not influence the module detection, and a soft-thresholding power β = 4 was picked to fit a scale-free topology. The blockwiseModules() function was run with default parameters except for power = 4, maxBlockSize = 20,000, networkType = “signed hybrid,” TOMType = “signed,” deepSplit = 2, minModuleSize = 20, and mergeCutHeight = 0.2. The overall expression pattern of each module was summarized using module eigengene values for each sample, and these values were tested for differential expression again using the limma-trend method, but without the surrogate variables added because their effects had already been removed. Specific pairwise comparisons were pulled from the model to identify which modules showed the effects of specific therapies.
Gene Ontology Enrichment Analysis
Gene ontology analysis was performed on individual modules using the Gene Ontology Enrichment Analysis (GOEA) and Visualization tool (GOrilla) open-source software (http://cbl-gorilla.cs.technion.ac.il/).31 All genes in a particular module were input as a query set and run through the Rattus norvegicus database as a single ranked list of genes. Results provided all ontologies related to the query set. Within each ontology, the associated genes were listed along with p-values, false discovery rate (FDR) q-values, and enrichment values. The process was performed for biological process ontologies and then repeated for molecular functions.
Statistical analysis
A one-way repeated measurements ANOVA was performed to evaluate behavioral efficacy at 48 h of therapy relative to prestimulation scores. A post-hoc multiple comparison analysis was carried out to compare differences in behavioral scores between the different therapies, including the No-SCS control after 48 h. Analyses were done using R-Studio statistical software.32 Statistical significance was established when p < 0.05.
Results
Effects on mechanical and thermal hypersensitivity
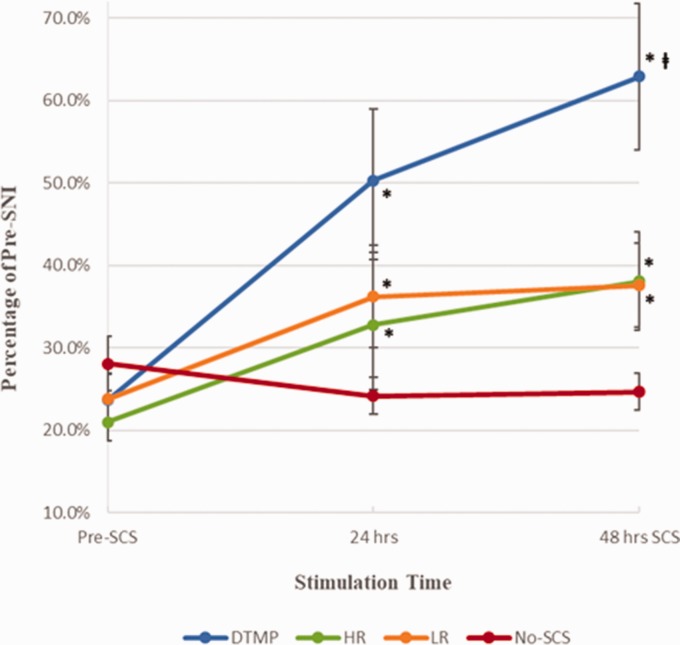
A total of six rats did not meet the minimum requirement of at least a 30% WT reduction post-SNI and were therefore not included in the study. This equated to a 13% nonresponder rate to the injury model. Figure 1 shows mean values (±standard error) for mechanical WTs (WT as a percentage relative to presurgery scores) for DTMP, LR, HR, and No-SCS groups. All therapies significantly improved mechanical sensitivity in injured animals relative to the pre-SCS measurements, both after 24 and 48 h of SCS. DTMP (62.9 ± 8.9%, N = 9) is significantly more effective than LR (37.6 ± 6.0%, N = 10) and HR (38.3 ± 5.5%, N = 11) after 48 h. All were significantly better than No-SCS (24.6 ± 2.6%, N = 10).
Figure 1.
Mean mechanical withdrawal thresholds as percentage of pre-SNI for DTMP, HR, LR, and No-SCS (Sham for Stim). Error bars are SEM values. * represents p < 0.05 vs. pre-SCS; represents p < 0.05 vs. LR or HR. SNI: spared nerve injury; SCS: spinal cord stimulation; DTMP: Differential target multiplexed programming; LR: low rate; HR: high rate.
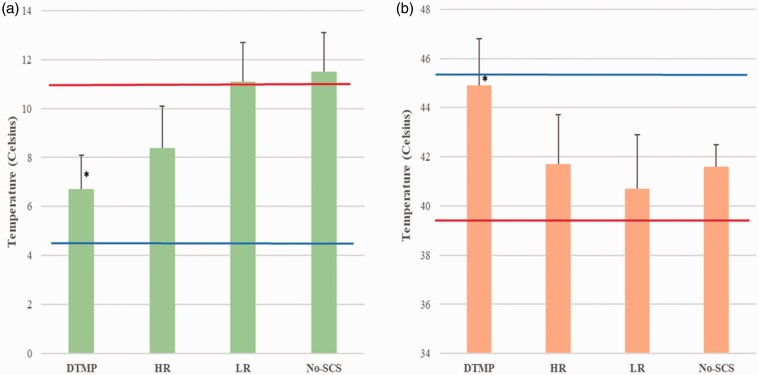
Figure 2 shows mean threshold temperatures (±standard error) for thermal hypersensitivity to touching a cold (TC) or hot (TH) surface. In terms of cold sensitivity (Figure 2(a)), the mean TC presurgery was 4.2 ± 0.1°C, which increased to a mean value of 11.0 ± 0.4°C in injured animals. DTMP significantly reduced TC to 6.7 ± 1.4°C relative to untreated animals. HR decreased TC to 8.4 ± 1.7°C, although not significantly relative to untreated animals, while LR had no effect yielding a TC of 11.1 ± 1.6°C. In terms of hot sensitivity (Figure 2(b)), the mean TH presurgery was 45.5 ± 0.1°C, which decreased to a mean value of 39.4 ± 0.4°C upon injury. DTMP significantly reduced heat hypersensitivity as the mean value of TH increased to 44.9 ± 1.9°C relative to untreated animals. Both HR and LR did not increase TH significantly (41.7 ± 2.0°C and 40.7 ± 2.2°C, respectively).
Figure 2.
Mean temperature threshold for hypersensitivity to (a) cold (TC) and (b) heat (TH) for DTMP, HR, LR, and No-SCS (sham for stim). Error bars represent SEM values. * represents p < 0.05 vs. pre-SCS. Pre-SNI (blue horizontal line) represents the mean value for naïve animals, while Pre-SCS (red horizontal line) represents the mean value for naïve injured animals before SCS. DTMP: differential target multiplexed programming; LR: low rate; HR: high rate; SCS: spinal cord stimulation.
WGCNA and GOEA
High-quality reads and alignment rates were obtained across all samples and they all passed quality control (QC) procedures. At least 70% of reads were aligned to the Rattus norvegicus transcriptome. Library sizes range from 15 million to 38 million reads which were sufficient for the subsequent analysis. Transcript counts were summed to the gene level using the “lengthScaledTPM” method of the tximport package, which adjusts for transcript lengths and scales the counts to the original library sizes so that the sum of the gene-level counts is the same as the sum of the transcript-level counts. Of the 31,060 genes listed in the genome, those that did not have at least 0.5 counts per million (CPM; or ∼9 to 16 counts per sample) were filtered out after TMM normalization and kept 19,944 genes for the analysis. The expression of these genes was compared relative to that in the naïve group in terms of a fold-change for every experimental group (DTMP, HR, LR, and No-SCS). In addition, the expression of genes in every group in which there was active stimulation was compared to the no-stimulation group (No-SCS).
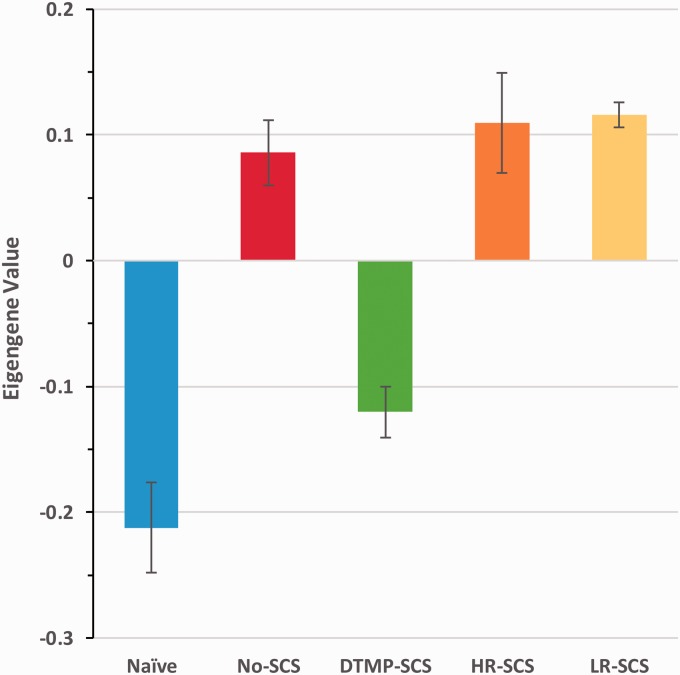
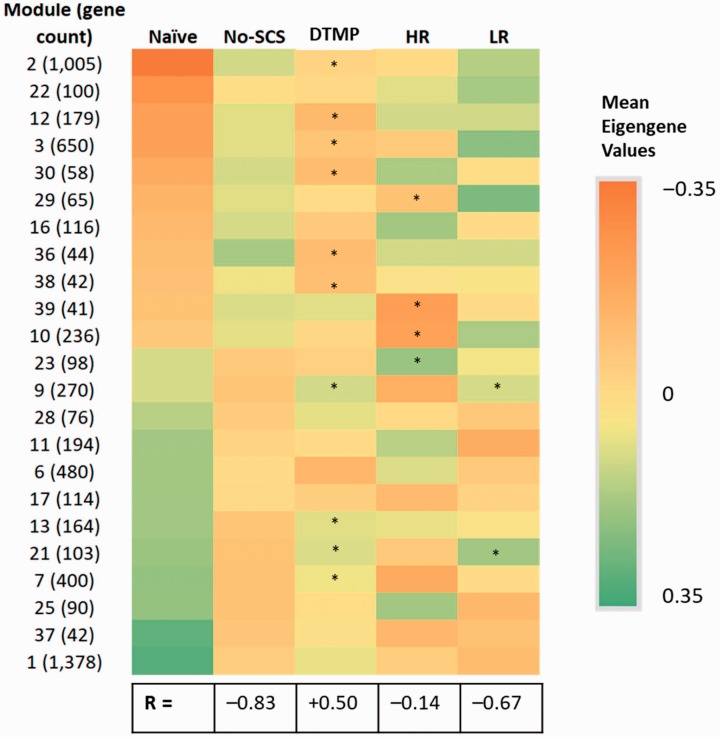
Given the large number of genes and multiple comparisons, a WGCNA allows the analysis in terms of the hierarchical clustering of genes (i.e., modules) based on expression patterns. The WGCNA allows for a comparison of experimental groups by analyzing the eigengene value, which is the first principal component of the expression profiles of a particular experimental group, in a given module. Figure 3 shows a representative bar graph with mean eigengene values for a given module. The WGCNA yielded 41 modules accounting for 9972 transcripts. Out of these, 24 modules containing 7436 genes transcripts are relevant to this work since expression patterns of the genes in these modules were significantly affected by the injury. Figure 4 contains a heat map comparing the mean eigengene values of 23 of these 24 WGCNA modules (module 0 is not included) for naïve animals, injured animals (No-SCS), and treated animals with SCS programs. Those modules in which either therapy also has a significant change in the mean eigengene value relative to the No-SCS animals are marked. DTMP significantly modulated the expression patterns of 11 out of these 23 modules (48%) in the direction of expression patterns shown by naïve animals, indicating that DTMP provides a path to normalization of expression patterns shown by animals before injury. In contrast, HR only reversed expression patterns significantly in 5 of these modules (22%), while LR reversed significantly 2 out of the 23 (9%). Furthermore, Pearson correlations comparing No-SCS and the SCS programs to Naïve for these modules provide a way of comparing the general effect of SCS. The Pearson correlation coefficient (R) for No-SCS vs. Naïve is negative and high (–0.83) as expected since it illustrates the opposite trend in the gene expression due to injury. The correlation coefficients between HR or LR vs. Naïve are also negative (–0.14 and –0.67, respectively) and of smaller magnitude indicating a small to moderate effect on the SNI towards Naïve. In contrast, the correlation coefficient between DTMP and Naïve is positive indicating that modulation of gene expression by DTMP trends largely towards the naïve state.
Figure 3.
A representative graph illustrating mean eigengene values for WGCNA module 12. Error bars are standard deviation. DTMP: differential target multiplexed programming; LR: low rate; HR: high rate; SCS: spinal cord stimulation.
Figure 4.
Heat map of mean module eigengene values for modules with significantly different comparisons (FDR-p < 0.2) between injured untreated animals (No-SCS) and Naïve animals. A total of 23 modules out of the total 39 are affected. Asterisks (*) indicate significantly different module eigengene values when comparing either SCS treatment to untreated animals (No-SCS). R is the Pearson coefficient for the correlation between eigengene values for naïve and each of the other groups. A negative value indicates an opposite trend. DTMP: differential target multiplexed programming; LR: low rate; HR: high rate; SCS: spinal cord stimulation.
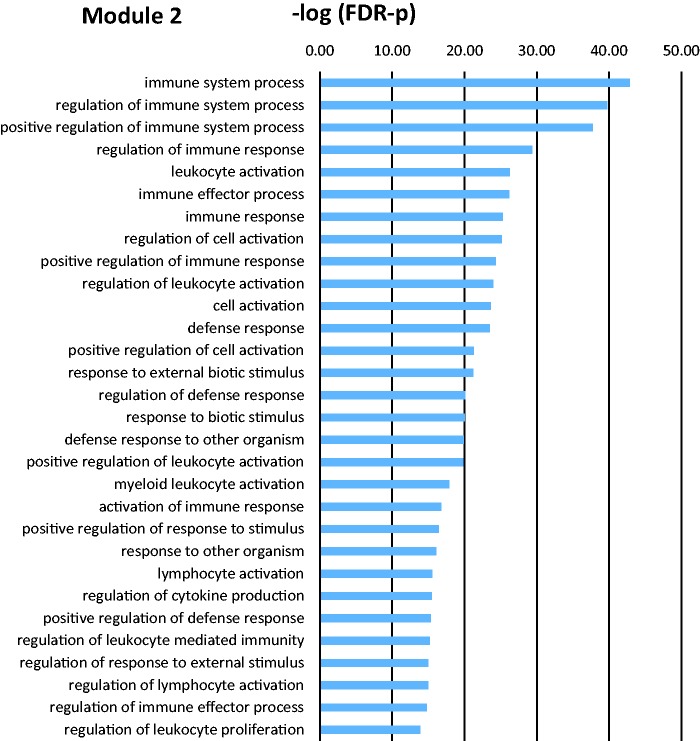
GOEA for each module yielded relevant biological processes. Figure 5 shows a representative bar graph with significantly enriched biological processes obtained from the GOEA for a given module. Table 2 shows the most relevant biological processes gene ontology terms for significant modules. Some modules did not render any enriched processes because of the low number of genes in them.
Figure 5.
Bar graph illustrating the 30 most significant biological processes GO terms for module 2, which are significantly affected by the injury model relative to naïve animals. There are 451 GO terms with FDR-p value below 0.01 in this module. FDR: false discovery rate.
Table 2.
Relevant significantly enriched biological process gene ontology terms for modules with expression patterns significantly affected by SNI (vs. Naïve) and reversed significantly by SCS.
| Module | Biological process gene ontology terms | SCS therapy |
|---|---|---|
| 1 | Regulation of transport, signaling, trans-synaptic signaling, cell–cell signaling, G-protein-coupled receptor signaling pathway, cellular calcium ion homeostasis | N.A. |
| 2 | Response to stress, regulation of immune system process, regulation of response to stimulus, signal transduction, regulation of transport | DTMP |
| 3 | Translation, peptide biosynthetic process, RNA metabolic process | DTMP |
| 6 | Response to oxygen levels, cellular developmental process, tissue development | N.A. |
| 7 | Cell–cell signaling, synaptic signaling, trans-synaptic signaling, regulation of protein phosphorylation, regulation of MAPK cascade, regulation of nervous system development, regulation of ion transport, regulation of nervous system development | DTMP |
| 9 | Regulation of cell-cycle process, DNA repair, cellular response to stress | DTMP, LR |
| 10 | Cellular response to stress, regulation of mRNA processing, primary metabolic process | HR |
| 11 | Signal transduction | N.A. |
| 12 | Signal transduction by trans-phosphorylation, response to chemokine, regulation of metallopeptidase activity | DTMP |
| 13 | Neurotransmitter receptor transport, regulation of postsynaptic membrane neurotransmitter receptor levels, protein transport within plasma membrane | DTMP |
| 16 | Sphingosine-1-phosphate receptor signaling pathway | DTMP |
| 17 | Regulation of neurogenesis, regulation of BMP signaling pathway, regulation of signal transduction | N.A. |
| 21 | N.A. | DTMP, LR |
| 22 | Axon ensheathment, myelin assembly, chemical homeostasis | N.A. |
| 23 | Cellular metabolic process, adenosine receptor signaling pathway, aerobic respiration | HR |
| 25 | Immune system process, regulation of cell-cell adhesion, cytokine-mediated signaling pathway, regulation of response to stress | HR |
| 28 | Ion transport, G-protein-coupled receptor signaling pathway, transmembrane transport | N.A. |
| 29 | rRNA metabolic process, glycosyl compound biosynthetic process | HR |
| 30 | Regulation of reactive oxygen species metabolic process, regulation of cytokine production, regulation of immune system process, inflammatory response | DTMP |
| 36 | Regulation of myelination, regulation of gliogenesis | DTMP |
| 37 | Regulation of amyloid precursor protein biosynthetic process, neuropeptide signaling pathway | N.A. |
| 38 | N.A. | DTMP |
| 39 | Lipid homeostasis | HR |
N.A.: did not render any enrichment; MAPK = Mitogen-Activated Protein Kinase; BMP = Bone Morphogenetic Protein.
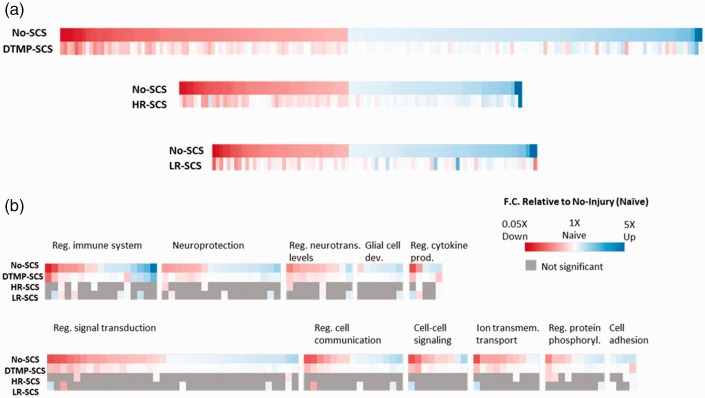
Figure 6 shows heat maps for fold changes relative to gene expression in naïve animals for the three therapies. The maps only contain those genes that have significant expression changes upon injury (i.e., represent a pain state) and that had been significantly modulated by the therapy. DTMP affects the expression of genes affected by injury to a larger extent than HR and LR. For instance, DTMP significantly modulated 252 genes significantly affected by injury towards naïve, while HR modulated 130 genes and LR modulated 120 genes. Furthermore, DTMP returned 167 of these genes within 10% of their expression in naïve animals. In contrast, HR and LR returned 69 and 91 genes to within 10% of the expression in naïve animals, respectively.
Figure 6.
(a) Heat maps illustrating the most significantly changed gene expressions due to injury (No-SCS) and the effect of treatments (DTMP, HR, or LR) relative to expression in naïve animals. Each colored column is a gene. Note that DTMP modulates a larger number of genes towards the naïve state (represented by white columns) than HR and LR. (b) Heat map of genes regrouped in terms of involvement in relevant biological processes. Gray boxes indicate that therapy did not produce a significant change. The intensity of red color is proportional to the extent to down regulation relative to expression in naïve; the intensity of blue color is proportional to the extent of upregulation relative to expression in naïve. White indicates that therapy modulated expression back to levels of naïve animals. DTMP: differential target multiplexed programming; LR: low rate; HR: high rate; SCS: spinal cord stimulation.
Discussion
The SNI model produces both mechanical and thermal hypersensitivity that are associated with pain-like behavior. The behavioral changes observed are in agreement with previous literature on this model. Furthermore, the transcriptomics indicates that the injury model affects various biological processes such as the activation of immune processes as well as signaling processes that are associated with glial activation in the spinal cord. There is a large body of evidence supporting the importance of glial cells, particularly microglia and astrocytes, in the development and maintenance of a chronic neuropathic state33–35 Glial cells are able to sense the state of neuronal activity and react accordingly to maintain the proper homeostasis within the synapses. The development of a chronic pain state involves the activation of microglia. Peripheral nerve injury and ectopic activity of neurons trigger the transition of microglia to a reactive state that results in the release of inflammatory mediators that also activates astrocytes. Persistent pain is therefore the result of glia-mediated sensitization of the neural tissue in the spinal cord which is conducive to phenotypic changes of the cell.36 Results from the WGCNA are consistent with this. The SNI model affects 58% of the WGCNA modules, in which the expression of 1574 transcripts is significantly changed relative to naïve levels. Furthermore, module 2, which includes transcripts involved in the regulation of the immune system, response to stimulus, signal transduction, and transport, was largely and significantly modulated, with 54% of the genes (540) in this module significantly affected by the peripheral nerve injury. All of these were upregulated relative to the expression in naïve animals.
The beneficial effect of SCS therapy is evident with the three different programming modalities used, but DTMP is statistically better to a significant degree at relieving mechanical and thermal hypersensitivity. The transcriptomics of the spinal cord of animals treated with DTMP seems to correlate with the behavioral results. DTMP modulates more genes than both HR and LR. More importantly, DTMP significantly modulates the expression of 252 genes that were significantly affected by the SNI back toward naïve levels. Ninety percent (90%) of these genes have expression levels that are within 25% of the naïve levels, and 66% are within 10% of naive expression levels. In contrast, HR modulates 132 genes back towards naïve levels, with 83% and 53% of these within 25% and 10% of naïve levels, respectively, while LR modulates 128 genes, the fewest among DTMP and HR back towards naïve levels, with 85% and 71% of these within 25% and 10% of the naïve expression levels.
Previously, we demonstrated that utilization of SCS pulses at 50 Hz (20 μs PW, 70% MT) for 72 h in the SNI model modulated the expression of pain-related genes and emphasized the importance of exploring the molecular mechanism of pain using “omics”-based methods.7 Recently, Stephens et al. used RNA sequencing to describe the transcriptomics of SCS (50 Hz, 200 μs PW, 80% MT) on the chronic constriction injury model of neuropathic pain.8 Both studies found that regardless of the injury model, SCS at 50 Hz upregulated genes involved in immune-related processes and downregulated genes involved in neurotransmission and synaptic signaling. The results of this study are congruent with both studies in that LR increases the expression of genes involved in immune-related processes relative to the expression in injured animals, as shown by the WGCNA for module 2 (Figure 4). The previous studies hypothesized that the upregulation of such genes may be associated with a neuroprotective role of the neural tissue associated with glia activation, although both implied that further investigation is warranted. Interestingly, as can be seen in Figure 4, both DTMP and HR trend opposite than LR in terms of their effect on the enriched genes involved in immune-related processes. Both of these SCS treatments reverse the expression pattern towards the expression state of naïve animals, although only DTMP reversed it back significantly. Our previous work and that done by Stephens et al. imply that SCS modulates the behavior of glial cells. Under the experimental conditions used in these studies, the expression of glia-related genes such as Tlr2, Cxcl16, Cd68, Gfap, Ccl2, and Itgam was associated with the effects of SCS on a neuroinflammatory state induced by injury. The current study, which included naïve animals for comparison, corroborated the fact that SNI increases the expression of various glia-related genes. For instance, the expression of both Cd68 and Itgam, which are associated with microglial activation, are elevated in the injured, untreated animals relative to naïve animals (1.6-fold and 2.1-fold, respectively). Also, the expression of Gfap, an astrocyte marker, is also significantly increased (1.8-fold) by the SNI. The expression of the toll-like receptor genes involved in microglia-related processes, such as Tlr2 and Tlr4, are also increased by the SNI (1.6-fold to 1.7-fold). In agreement with our previous work and that by Stephens et al., we also found that LR increases the expression of Tlr2, Itgam, and Ccl2 by 19% or more relative to untreated animals. However, under our LR programming conditions, there was no increase on the expression of Cd68 and Gfap. In contrast, HR and DTMP significantly decreased the expression of Gfap relative to untreated animals and also decreased the expression of Itgam by 15–20% although not significantly. The role of microglia and astrocytes on the establishment and maintenance of chronic pain is well established.33–35 These and oligodendrocytes maintain a homeostatic balance of the neural tissue, which is perturbed by a state of chronic pain. Balanced neuroglial interactions are key to maintain the proper level of neurotransmitters, membrane receptors, and synaptic proteins. Key biological processes that are associated with the neuroglial interaction involve the regulation of the immune response to injury, signal transduction, and cell to cell communication.37,38 Activation of microglia upon nerve injury leads to the release of cytokines and chemokines, which affect the normal state of signal transduction by neurons at the synapse by setting an unbalanced release of neurotransmitters. The prolonged effects of the neural changes induced by microglia also activate astrocytes, which in turn disturbs their normal interaction with neurons and neighboring astrocytes. For instance, our results indicate that SNI increases the expression of immune-related genes such as C1qa (3.0-fold), which encodes for a complement-system protein involved in synaptic pruning;39 Casp1 (1.5-fold), which encodes for a proteolytic enzyme that catalyzes the formation of proinflammatory cytokines, such as IL1B;40,41 and Tal1 (1.5-fold), which encodes for transcription factor that regulates phenotyping of microglia into a neurotoxic state.40 These genes, which are part of module 2, are involved in the immune response and are significantly modulated back towards naïve levels by DTMP only. The recovery toward naïve expression caused by DTMP is 49% for C1qa, 71% for Casp1, and 84% for Tal1. Besides regulating the immune response, DTMP also regulates signal transduction and cell to cell communications (module 7). SNI downregulates the expression of genes such as Glra2 (1.4-fold), which encodes for the glycine receptor alpha 2 protein, which is part of the glycine receptor, a ligand-gated chloride channel that is important for suppression of neuronal excitability via inhibitory postsynaptic currents.42 SNI also downregulates Sst (2.1-fold), a gene that encodes for somatostatin (SST), a neuropeptide that is released by a particular set of GABAergic interneurons that mediate mechanical pain transmission in the spinal cord.43,44 Astrocytes in the somatosensory cortex of the brains are able to drive inhibitory/excitatory balance and calcium signaling by GABAergic interneurons that release SST. DTMP significantly increases the expression of Sst towards levels shown by naïve animals (44% recovery), while both HR and LR also increase it slightly (16–18%) but not significantly. These genes provide some examples of their relevance in the aforementioned biological processes, which are significantly affected by DTMP. Many other genes are obviously involved and affected (see Figure 5). Ultimately, these genes are modulated in a concerted manner by the applied electric field in a way that provides a robust effect on the neural tissue that drives the balance of the system towards the naïve (i.e., healthy) state as evidenced in Figures 4 and 6.
The role of oligodendrocytes in chronic pain is emerging.45 Ablation of oligodendrocytes of the spinal cord in mice triggered sensory changes resembling neuropathic pain, suggesting that their dysfunction can lead to central neuropathic pain, independent of other immune contributions.46 Our results indicate that the SNI upregulates certain genes predominantly expressed by oligodendrocytes. These are Mobp (1.6-fold), Klk6 (1.6-fold), S1pr5 (1.5-fold), Gjc2 (1.3-fold), and Nkx6-2 (1.4-fold). Mobp encodes for myelin oligodendrocyte basic protein, a marker of oligodendrocytes. Shi et al. reported an increase in the expression of this protein, and others related to mature oligodendrocytes and oligodendrocyte precursor cells (OPCs), in patients with neuropathic pain associated with HIV.47 S1pr5 is a gene that encodes for the sphingosine-1-phosphate receptor 5. The ligand, sphingosine-1-phosphate has been linked to signaling processes that modulate pain. Its receptors (S1PRs) can play both anti- and pro-nociceptive roles, depending on their localization in specific cell types.48 Specifically, S1PR5 is only expressed by oligodendrocytes and plays a critical role in the migration of OPCs.49 Nkx6-2, Gjc2, and Klk6 have not been previously associated with pain processes but are known to mediate oligodendrocyte communication and oligodendrocyte-based immunity. Nkx6-2 is a gene that encodes for a transcription factor with repressor activity in the regulation of axon-oligodendrocyte interactions.50 Gjc2 encodes for connexin-47 a protein expressed in oligodendrocytes of white matter in the spinal cord that forms gap junctions that facilitate communication between oligodendrocytes or between oligodendrocytes and astrocytes.51 Klk6 is a gene that encodes for kallikrein 6 (KLK6), a serine protease released by oligodendrocytes that regulates immune response. It is known that suppression of KLK6 reduces the expression levels of inflammatory cytokines, chemokines, and their receptors.52 Interestingly, these oligodendrocyte-only related genes are in module 36, which is significantly changed by DTMP back to the naïve expression pattern. The expression of these genes is significantly reversed towards the naïve state with recoveries larger than 80%. Although both HR and LR result in a decrease of expression levels towards naïve, these are not significant, and in most cases, do not correspond to more than 30% recovery. These results suggest that DTMP also regulates oligodendrocytes and their interactions with neurons and other glial cells.
The results indicate that DTMP modulates immune-related processes as well as neurotransmission and synaptic signaling, biological processes that are key to maintaining balanced neuroglial interactions. They also suggest that its effects are associated with the modulation of activated glial cells engaged in the immune response and the inflammatory and neuroprotective states of the spinal cord as a result of the peripheral injury. Current work addresses whether DTMP reverts injury-induced pathological glial states by investigating cell-specific transcriptomes of such cells and how they are affected by the pain model and ensuing therapies relative to the naïve state.
There are certain limitations of this animal study. This extensive study provides a snapshot of the history of the development of the neuropathic pain model. In this particular case, we intervened at an early stage (5 days after injury), which may be considered largely an acute state transitioning into a chronic state, taking into account that, on average, 16 rat days are equivalent to one human year.53 Further research would be needed to determine the evolution of the transcriptomics of the pain model and the effect of SCS on it. Such research might provide insightful information about whether SCS may provide more beneficial effects if it were to be used earlier in the establishment of chronic pain. This may be the case, as a longer duration of a chronic pain state may imply the development of irreversible phenotypic and epigenetic changes in the cells involved. SCS was also applied for a limited time (48 h), thus it is uncertain if the effects of HR and LR would have eventually leveraged the effects shown by DTMP. Clinical applications are also longer, as the majority of patients are required to keep their SCS systems running most of the time. Our study also did not evaluate if gene expression would return to the levels measured in untreated animals if the SCS system was turned off. It is known that interrupting SCS for a given period of time results in the onset of pain back to baseline levels, suggesting that the transcriptomics of the tissue could reverse to the injured state. Future proteomics studies will complement the results presented here since proteins and their activations and post-translational modifications are of extreme importance in understanding chronic pain and the effect of SCS therapy.
Acknowledgments
Special thanks to Medtronic (Minneapolis, MN) for providing the modified external neurostimulators and programmers and Dr Jenny Drnevich at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for her statistical assistance on RNA-sequencing.
Author Contributions
DLC and RV designed the experiments. CAK performed experiments; DLC, CAK, AV, WJS, AG, and RV analyzed the data. All authors contributed to the drafting and revisions of the manuscript. DLC and RV managed the project.
Declaration of Conflicting Interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: DLC and RV are co-inventors in patents related to DTMP. RV, DLC, AG, and CAK were employed by Millennium Pain Center. RV and DLC had membership interest in Stimgenics. CAK was a consultant of Stimgenics.
Funding
The author(s) disclose receipt of the following financial support for the research, authorship, and/or publication of this article: The study was funded by Millennium Pain Center and Stimgenics.
ORCID iDs
Ashim Gupta https://orcid.org/0000-0003-1224-2755
David L Cedeño https://orcid.org/0000-0001-5421-802X
References
- 1.Shealy CN, Mortimer JT, Reswick JB. Electrical inhibition of pain by stimulation of the dorsal columns: preliminary clinical report. Anesth Analg 1967; 46: 489–491. [PubMed] [Google Scholar]
- 2.Caylor J, Reddy R, Yin S, Cui C, Huang M, Huang C, Rao R, Baker DG, Simmons A, Souza D, Narouze S, Vallejo R, Lerman I. Spinal cord stimulation in chronic pain: evidence and theory for mechanisms of action. Bioelectron Med 2019; 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melzack R, Wall PD. Pain mechanisms: a new theory. Science 1965; 150: 971–979. [DOI] [PubMed] [Google Scholar]
- 4.Vallejo R, Bradley K, Kapural L. Spinal cord stimulation in chronic pain: mode of action. Spine (Phila Pa 1976) 2017; 42: S53–S60. [DOI] [PubMed] [Google Scholar]
- 5.Deer TR, Jain S, Hunter C, Chakravarthy K. Neurostimulation for intractable chronic pain. Brain Sci 2019; 9: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivanesan E, Maher DP, Raja SN, Linderoth B, Guan Y. Supraspinal mechanisms of spinal cord stimulation for modulation of pain: five decades of research and prospects for the future. Anesthesiol J Am Soc Anesthesiol 2019; 130: 651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallejo R, Tilley DM, Cedeño DL, Kelley CA, DeMaegd M, Benyamin R. Genomics of the effect of spinal cord stimulation on an animal model of neuropathic pain. Neuromodulation 2016; 19: 576–586. [DOI] [PubMed] [Google Scholar]
- 8.Stephens KE, Chen Z, Sivanesan E, Raja SN, Linderoth B, Taverna SD, Guan Y. RNA-seq of spinal cord from nerve-injured rats after spinal cord stimulation. Mol Pain 2018; 14: 174480691881742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Bartheld CS, Bahney J, Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J Comp Neurol 2016; 524: 3865–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci 2009; 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahney J, von Bartheld CS. The cellular composition and glia–neuron ratio in the spinal cord of a human and a nonhuman primate: comparison with other species and brain regions. Anat Rec 2018; 301: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz‐Sauri A, Orduña‐Valls JM, Blasco‐Serra A, Tornero‐Tornero C, Cedeño DL, Bejarano‐Quisoboni D, Valverde-Navarro AA, Benyamin R, Vallejo R. Glia to neuron ratio in the posterior aspect of the human spinal cord at thoracic segments relevant to spinal cord stimulation. J Anat 2019; 235: 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roitbak AI, Fanardjian VV. Depolarization of cortical glial cells in response to electrical stimulation of the cortical surface. Neuroscience 1981; 6: 2529–2537. [DOI] [PubMed] [Google Scholar]
- 14.Tawfik VL, Chang SY, Hitti FL, Roberts DW, Leiter JC, Jovanovic S, Lee KH. Deep brain stimulation results in local glutamate and adenosine release: investigation into the role of astrocytes. Neurosurgery 2010; 67: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KH, Hitti FL, Chang SY, Lee DC, Roberts DW, McIntyre CC, Leiter JC. High frequency stimulation abolishes thalamic network oscillations: an electrophysiological and computational analysis. J Neural Eng 2011; 8: 046001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki Y, Hozumi Y, Kaneko K, Sugihara T, Fujii S, Goto K, Kato H. Modulatory effects of oligodendrocytes on the conduction velocity of action potentials along axons in the alveus of the rat hippocampal CA1 region. Neuron Glia Biol 2007; 3: 325–334. [DOI] [PubMed] [Google Scholar]
- 17.Vallejo R, Gupta A, Kelley CA, Vallejo A, Rink J, Williams JM, Cass CL, Smith WJ, Benyamin R, Cedeño DL. Effects of phase polarity and charge balance spinal cord stimulation on behavior and gene expression in a rat model of neuropathic pain. Neuromodulation 2020; 23: 26–35. [DOI] [PubMed] [Google Scholar]
- 18.Ögren SO, Berge OG. Test-dependent variations in the antinociceptive effect of p-chloroamphetamine-induced release of 5-hydroxytryptamine. Neuropharmacology 1984; 23: 915–924. [DOI] [PubMed] [Google Scholar]
- 19.Yalcin I, Charlet A, Freund-Mercier MJ, Barrot M, Poisbeau P. Differentiating thermal allodynia and hyperalgesia using dynamic hot and cold plate in rodents. J Pain 2009; 10: 767–773. [DOI] [PubMed] [Google Scholar]
- 20.Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain 2000; 87: 149–158. [DOI] [PubMed] [Google Scholar]
- 21.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017; 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015; 4: 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leek JT, Storey JD. A general framework for multiple testing dependence. Proc Natl Acad Sci 2008; 105: 18718–18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Lun ATL, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 2016; 5: 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw 2012; 46: pii: i11. [PMC free article] [PubMed] [Google Scholar]
- 31.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009; 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rstudio Team. RStudio: integrated development for R. Boston, MA: RStudio, 2015. [Google Scholar]
- 33.Ji RR, Berta T, Nedergaard M. Glia and pain: is chronic pain a gliopathy? Pain 2013; 154: S10–S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallejo R, Tilley DM, Vogel L, Benyamin R. The role of glia and the immune system in the development and maintenance of neuropathic pain. Pain Pract 2010; 10: 167–184. [DOI] [PubMed] [Google Scholar]
- 35.Milligan ED, Watkins LR. Pathological and protective roles of glia in chronic pain. Nat Rev Neurosci 2009; 10: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grace PM, Hutchinson MR, Maier SF, Watkins LR. Pathological pain and the neuroimmune interface. Nat Rev Immunol 2014; 14: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benarroch EE. Central neuron-glia interactions and neuropathic pain: overview of recent concepts and clinical implications. Neurology 2010; 75: 273–278. [DOI] [PubMed] [Google Scholar]
- 38.Ji RR, Strichartz G. Cell signaling and the genesis of neuropathic pain. Sci STKE 2004; 2004: reE14. [DOI] [PubMed] [Google Scholar]
- 39.Györffy BA, Kun J, Török G, Bulyáki É, Borhegyi Z, Gulyássy P, Kis V, Szocsics P, Micsonai A, Matkó J, Drahos L, Juhász G, Kékesi KA, Kardos J. Local apoptotic-like mechanisms underlie complement-mediated synaptic pruning. Proc Natl Acad Sci USA 2018; 115: 6303–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Bernhardi R, Eugenín-von Bernhardi L, Eugenín J. Microglial cell dysregulation in brain aging and neurodegeneration. Front Aging Neurosci 2015; 7: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkley KS, Popichak KA, Afzali MF, Legare ME, Tjalkens RB. Microglia amplify inflammatory activation of astrocytes in manganese neurotoxicity. J Neuroinflammation 2017; 14: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutertre S, Becker CM, Betz H. Inhibitory glycine receptors: an update. J Biol Chem 2012; 287: 40216–40223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mederos S, Perea G. GABAergic‐astrocyte signaling: a refinement of inhibitory brain networks. Glia 2019; 67: 1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamessian A, Young M, Qadri Y, Berta T, Ji RR, Van De Ven T. Transcriptional profiling of somatostatin interneurons in the spinal dorsal horn. Sci Rep 2018; 8: 6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borghi SM, Fattori V, Hohmann MSN, Verri WA. Contribution of spinal cord oligodendrocytes to neuroinflammatory diseases and pain. Curr Med Chem 2019; 26: 5781–5810. [DOI] [PubMed] [Google Scholar]
- 46.Gritsch S, Lu J, Thilemann S, Wörtge S, Möbius W, Bruttger J, Karram K, Ruhwedel T, Blanfeld M, Vardeh D, Waisman A, Nave K-A, Kuner R. Oligodendrocyte ablation triggers central pain independently of innate or adaptive immune responses in mice. Nat Commun 2014; 5: 5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Shu J, Liang Z, Yuan S, Tang SJ. Oligodendrocytes in HIV-associated pain pathogenesis. Mol Pain 2016; 12: 174480691665684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim-Selley LJ, Wilkerson JL, Burston JJ, Hauser KF, McLane V, Welch SP, Lichtman AH, Selley DE. Differential tolerance to FTY720-induced antinociception in acute thermal and nerve injury mouse pain models: role of sphingosine-1-phosphate receptor adaptation. J Pharmacol Exp Ther 2018; 366: 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novgorodov AS, El‐Awani M, Bielawski J, Obeid LM, Gudz TI. Activation of sphingosine-1-phosphate receptor S1P5 inhibits oligodendrocyte progenitor migration. FASEB J 2007; 21: 1503–1514. [DOI] [PubMed] [Google Scholar]
- 50.Southwood C. CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure. J Neurosci 2004; 24: 11215–11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orthmann-Murphy JL, Abrams CK, Scherer SS. Gap junctions couple astrocytes and oligodendrocytes. J Mol Neurosci 2008; 35: 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bando Y, Hagiwara Y, Suzuki Y, Yoshida K, Aburakawa Y, Kimura T, Murakami C, Ono M, Tanaka T, Jiang Y-P, Mitrovi B, Bochimoto H, Yahara O, Yoshida S. Kallikrein 6 secreted by oligodendrocytes regulates the progression of experimental autoimmune encephalomyelitis. Glia 2018; 66: 359–378. [DOI] [PubMed] [Google Scholar]
- 53.Sengupta P. The laboratory rat: relating its age with human’s. Int J Prev Med 2013; 4: 624. [PMC free article] [PubMed] [Google Scholar]