Abstract
Background
Lactobacillus species are used as probiotics and play an important role in fermented food production. However, use of 16S rRNA gene sequences as standard markers for the differentiation of Lactobacillus species offers a very limited scope, as several species of Lactobacillus share similar 16S rRNA gene sequences. In this study, we developed a rapid and accurate method based on comparative genomic analysis for the identification of 37 Lactobacillus species that are commonly used in probiotics and fermented foods.
Results
To select species-specific sequences or genes, a total of 180 Lactobacillus genome sequences were compared using Python scripts. In 14 out of 37 species, species-specific sequences could not be found due to the similarity of the 16S–23S rRNA gene. Selected unique genes were obtained using comparative genomic analysis and all genes were confirmed to be specific for 52,478,804 genomes via in silico analysis; they were found not to be strain-specific, but to exist in all strains of the same species. Species-specific primer pairs were designed from the selected 16S–23S rRNA gene sequences or unique genes of species. The specificity of the species-specific primer pairs was confirmed using reference strains, and the accuracy and efficiency of the polymerase chain reaction (PCR) with the standard curve were confirmed. The PCR method developed in this study is able to accurately differentiate species that were not distinguishable using the 16S rRNA gene alone. This PCR assays were designed to detect and identify 37 Lactobacillus species. The developed method was then applied in the monitoring of 19 probiotics and 12 dairy products. The applied tests confirmed that the species detected in 17 products matched those indicated on their labels, whereas the remaining products contained species other than those appearing on the label.
Conclusions
The method developed in this study is able to rapidly and accurately distinguish different species of Lactobacillus, and can be used to monitor specific Lactobacillus species in foods such as probiotics and dairy products.
Keywords: Lactobacillus, PCR, Comparative genomics, Probiotic product, 16S rRNA gene, Species-specific primer
Background
Lactobacillus is a Gram-positive, non-spore-forming, rod-shaped, catalase-negative genus of bacteria that often grows best under microaerophilic conditions. Lactobacillus belongs to the family Lactobacillaceae and consists of 170 species and 17 subspecies [1]. Human and animal gastrointestinal tracts harbor a variety of Lactobacillus species, including L. plantarum, L. rhamnosus, L. fermentum, and L. casei [1], while species such as L. gasseri, L. vaginalis, L. crispatus, L. iners, and L. jensenii are known to exist in the vagina [2]. They have a high tolerance to acidic environments and are typically used as starter cultures for fermented foods such as kimchi, yogurt, and cheese [1]. Bifidobacterium and Lactobacillus species are among the most commercially used lactic acid bacteria (LAB) in probiotic products [3]. In particular, L. acidophilus, L. casei, L. rhamnosus, L. plantarum, and L. paracasei are often used in probiotic products in combination with other Lactobacillus species.
Probiotics are human and animal health-promoting bacteria that are generally recognized as safe (GRAS) and known to provide beneficial effects, positively affecting the intestinal microbiota, preventing urogenital infections, decreasing the effect of allergens, reducing the growth of pathogens, on the host such as gut, skin, vagina, and other sites of body [4, 5]. In recent years, the probiotic product market has expanded proportionately with an increased interest in gut health [6, 7]. Despite the widespread use of probiotic products to improve human health, there is increasing concern among consumers regarding the quality and the label claims of commercial probiotic products [3]. In terms of functionality and safety, it is very important that probiotic products contain well-documented probiotic strains that are accurately displayed on the label. However, reports have shown that the LAB species present in some commercial probiotic products do not match those represented on the label [8–10].
The traditional methods used to study microbial communities, such as morphological and physiological characteristics, protein profiling, carbohydrate fermentation patterns, and counts on selective media, are time-consuming and often produce ambiguous outcomes [11, 12]. To achieve the reliable and rapid identification of bacterial species, molecular methods such as 16S rRNA gene sequencing, metagenome sequencing, and denaturing gradient gel electrophoresis (DGGE) have been increasingly applied. 16S rRNA sequencing is commonly used for bacterial identification, including the identification of Lactobacillus species [13–15]. Metagenome sequencing and DGGE based on 16S rRNA gene sequences are useful analytical methods for investigating complex microbial communities without previous isolation of individual bacteria [16–18]. However, 16S rRNA gene sequences in many Lactobacillus species are too similar to be readily distinguished. In particular, closely related species within the L. acidophilus group (L. acidophilus, L. gallinarum, and L. helveticus), the L. casei group (L. casei, L. paracasei, and L. rhamnosus), the L. plantarum group (L. plantarum, L. paraplantarum, and L. pentosus), and the L. sakei group (L. sakei, L. curvatus, and L. graminis) are notoriously difficult to distinguish by 16S rRNA gene sequences [19, 20]. For example, the 16S rRNA gene sequence of the L. casei group and that of the L. sakei group have more than 98.7% similarity between species [19, 20].
In this study, we designed species-specific primer pairs targeting the 16S–23S rRNA gene and species-unique genes, and developed detection and identification methods for 37 Lactobacillus species, which are mainly used in probiotics and difficult to distinguish by conventional identification methods, using single 96 well plate of PCR assays. The developed PCR assays were applied to commercial probiotics and dairy products to distinguish Lactobacillus present in the product to the species level. We have also confirmed that this assay has the ability to determine the composition of Lactobacillus species present in a product, as well as the presence of species not stated on the label.
Results
Selection of species-specific sequences and primer designs
The species-specific primer pairs of 37 Lactobacillus were designed from unique genes or the 16S–23S rRNA region (Table 1). The similarities of the 16S–23S rRNA regions among Lactobacillus species were verified in silico and 23 Lactobacillus species were distinguished with each primer pair designed in the 16S–23S region. Some Lactobacillus species are difficult to distinguish using the 16S–23S rRNA region alone due to the small number of single-nucleotide polymorphisms. Therefore, unique genes of 14 Lactobacillus species were obtained using comparative genomics (Table 2). A membrane protein was found in 4 L. acidipiscis genomes, but was not present in other species of Lactobacillus. Adenylosuccinate lyase and leucine-rich repeat protein were detected as the specific genes in L. amylovorus and L. parabuchneri, respectively. In L. paraplantarum, L. plantarum, L. pentosus, and L. helveticus, MFS (Major Facilitator Superfamily)-type transporter YcnB, LPXTG-motif cell wall anchor domain protein, GHKL domain-containing protein, and decarboxylate/amino acid:cation Na+/H+ symporter family protein were detected as the specific genes to each respective species. We also confirmed the specificity of unique genes using BLAST. The unique genes did not match any of the 52,478,804 sequences found in the NCBI database outside of the target species (Table 3). The selected unique genes confirmed to be present in the genome sequences of the reference strains with 100% identity. However, some genomes of L. casei contained unique genes of L. paracasei. The presence of unique genes in some, but not all, L. casei strains suggests that the genome information given for the strains is incorrect. These L. casei strains were found to be more similar in the 16S rRNA gene to L. paracasei than to the L. casei described in a previous study [21]. Also, one genome of L. gallinarum contained a unique gene of L. helveticus. To clarify the problem of L. gallinarum strain, we further performed a genomic analysis of L. helveticus and L. gallinarum. The result showed that a L. gallinarum strain containing a unique gene of L. helveticus was more similar to other strains of L. helveticus (Fig. 1).
Table 1.
Information of primer pairs designed for this study
| Species | Target gene | Primer name | Sequence (5′–3′) | Product size (bp) |
Primer conc.b (μM) |
|---|---|---|---|---|---|
| IPCa | 16S–23S region | IPC-F | CAA CGC GAA GAA CCT TAC CAG | 111 | 0.4 |
| IPC-R | CCA ACA TCT CAA CGA CAC GAG C | ||||
| L. gasseri | 16S–23S region | Gasseri-F | TCA AGA GCT GTT AAG GCT GT | 175 | 0.04 |
| Gasseri-R | CTA TCG CTT CAA GTG CTT TC | ||||
| L. rhamnosus | 16S–23S region | Rhamnosus-F | GCC GAT CGT TGA CGT TAG TTG G | 137 | 0.04 |
| Rhamnosus-R | CAG CGG TTA TGC GAT GCG AAT | ||||
| L. brevis | 16S–23S region | Brevis-F | GGG CAA CGA AGC AAG ATC GC | 260 | 0.08 |
| Brevis-R | TTC CAA TCG TGT GCA CAC CA | ||||
| L. sakei | 16S–23S region | Sakei-F | TCG AAC GCA CTC TCG TTT AG | 182 | 0.08 |
| Sakei-R | CGA AAC CAT CTT TCA ACC CT | ||||
| L. johnsonii | 16S–23S region | Johnsonii-F | AGA GAG AAA CTC AAC TTG AAA TA | 195 | 0.4 |
| Johnsonii-R | CCT TCA TTA ACC TTA ACA GTT AA | ||||
| L. jensenii | 16S–23S region | Jensenii-F | AGT TCT TCG GAA TGG ACA TAG | 148 | 0.4 |
| Jensenii-R | GCC GCC TTT TAA ACT TCT T | ||||
| L. fermentum | Unique gene | Fermentum-F | GAC CAG CGC ACC AAG TGA TA | 129 | 0.08 |
| Fermentum-R | AGC GTA GCG TTC GTG GTA AT | ||||
| L. plantarum | Unique gene | Plantarum-F | GCT GGC AAT GCC ATC GTG CT | 147 | 0.12 |
| Plantarum-R | TCT CAA CGG TTG CTG TAT CG | ||||
| L. paracasei | Unique gene | Paracasei-F | CAA TGC CGT GGT TGT TGG AA | 106 | 0.4 |
| Paracasei-R | GCC AAT CAC CGC ATT AAT CG | ||||
| L. paraplantarum | Unique gene | Paraplantarum-F | TTA TTC AAG CCG TCG GAG TG | 128 | 0.4 |
| Paraplantarum-R | TCG CTG GTG CTA ATG CAA TG | ||||
| L. casei | Unique gene | Casei-F | CCA CAA TCC TTG GCT GTT CT | 115 | 0.4 |
| Casei-R | GCT TGA GGC GAT TGT AAT CC | ||||
| L. curvatus | 16S–23S region | Curvatus-F | ACT CTC ATT GAA TTA GGA CGT T | 132 | 0.4 |
| Curvatus-R | CCC GTG TTG GTA CTA TTT AAT | ||||
| L. acidophilus | 16S–23S region | Acidophilus-F | CCT TTC TAA GGA AGC GAA GGA T | 129 | 0.4 |
| Acidophilus-R | ACG CTT GGT ATT CCA AAT CGC | ||||
| L. salivarius | 16S–23S region | Salivarius-F | TAC ACC GAA TGC TTG CAT TCA | 138 | 0.08 |
| Salivarius-R | AGG ATC ATG CGA TCC TTA GAG A | ||||
| L. reuteri | 16S–23S region | Reuteri-F | GAT TGA CGA TGG ATC ACC AGT | 161 | 0.2 |
| Reuteri-R | CAT CCC AGA GTG ATA GCC AA | ||||
| L. coryniformis | 16S–23S region | Coryniformis-F | CAA GTC GAA CGC ACT GAC G | 165 | 0.4 |
| Coryniformis-R | ACA TTC AGG CCA TGT GGT CT | ||||
| L. farciminis | Unique gene | Farciminis-F | ACG AAT CCG GCA GTC AAG AA | 152 | 0.08 |
| Farciminis-R | AAG AAT CGC CAA GCT CTA GG | ||||
| L. zymae | 16S–23S region | Zymae-F | GCT AAA GCA AGC GCA CGA TT | 132 | 0.08 |
| Zymae-R | TCG GCA GTG TGA CAT GGA G | ||||
| L. pentosus | Unique gene | Pentosus-F | GCG GTA TCG ATT CGA TTG GT | 145 | 0.08 |
| Pentosus-R | TGA TGT CAA TCG CCT CTT GG | ||||
| L. crustorum | 16S–23S region | Crustorum-F | GGA ATA GCC CAA ACC AGA G | 145 | 0.2 |
| Crustorum-R | ACT GAA TGG AGT GGG TCA GA | ||||
| L. mucosae | 16S–23S region | Mucosae-F | ACG GAC TTG ACG TTG GTT TAC | 156 | 0.4 |
| Mucosae-R | GTG ATA GCC GAA ACC ACC TT | ||||
| L. buchneri | 16S–23S region | Buchneri-F | CAA GTC GAA CGC GTC TCC AT | 189 | 0.08 |
| Buchneri-R | CCG AAG CCG TCT TTT AAA CC | ||||
| L. helveticus | Unique gene | Helveticus-F | CTA CTT CGC AGG CGT TAA CT | 132 | 0.08 |
| Helveticus-R | GTA CTT GAT GCT CGC ATA CC | ||||
| L. amylovorus | Unique gene | Amylovorus-F | CAA GCA CGA TTG GCA AGA TG | 126 | 0.4 |
| Amylovorus-R | ATT GGA TTC CGC TTC TGT GG | ||||
| L. heilongjiangensis | 16S–23S region | Heilongjiangensis-F | GCT TCA TGA ATC GGA TCT AA | 133 | 0.4 |
| Heilongjiangensis-R | TAA ACT ACG ATC ATG TGA AAG TA | ||||
| L. parabuchneri | Unique gene | Parabuchneri-F | AGC GTC GTG ATT CCT GAT AC | 137 | 0.08 |
| Parabuchneri-R | CGA CTC TCC GAT CGT TGT TA | ||||
| L. acidipiscis | Unique gene | Acidipiscis-F | AGC GGT TCG ATG GCT TAT AC | 125 | 0.08 |
| Acidipiscis-R | TCC AAG TCC GAC ACC AGT CA | ||||
| L. sanfranciscensis | Unique gene | Sanfranciscensis-F | TGG AAC TGA TAC GCG GAT GT | 130 | 0.08 |
| Sanfranciscensis-R | GGC CAA TTC CTC CAA TAA CG | ||||
| L. ruminis | 16S–23S region | Ruminis-F | TTG CAT TCA CCG AAA GAA GC | 129 | 0.4 |
| Ruminis-R | CAT AAA CAT CAT GCG GTG TTC | ||||
| L. agilis | 16S–23S region | Agilis-F | TCG TAG CTT GCT ACA CCG ATT G | 137 | 0.4 |
| Agilis-R | CAT AAT GAC CAT GCG ATC ATC A | ||||
| L. delbrueckii | 16S–23S region | Delbrueckii-F | CAT GTG CAG ACA TGC TAT CCT T | 192 | 0.4 |
| Delbrueckii-R | CTC TGA AGT GCC ATG TCT CAG T | ||||
| L. amylophilus | 16S–23S region | Amylophilus-F | CGA GTT CTG GTT AAG AGT AGC G | 174 | 0.4 |
| Amylophilus-R | CGC CAT CTT TCA AAC ATC TAT C | ||||
| L. kunkeei | 16S–23S region | Kunkeei-F | GAA CGA GCT CTC CCA AAT TGA | 161 | 0.4 |
| Kunkeei-R | GAA CCA TGC GGT TCC AAC TA | ||||
| L. acetotolerans | 16S–23S region | Acetotolerans-F | GAT TAC CTT CGG GTA TGA AGT T | 131 | 0.2 |
| Acetotolerans-R | TCA TGT GAT CTC TCC TTT TAT CC | ||||
| L. lindneri | Unique gene | Lindneri-F | CGG CGT TCT CGA GGA CCA TA | 170 | 0.4 |
| Lindneri-R | CAT CCG GCG TCC TTC ATA GC | ||||
| L. gallinarum | Unique gene | Gallinarum-F | AAC TGG CGG TTA TCG TAG AC | 118 | 0.2 |
| Gallinarum-R | CAC AGC AGG AAC CAT TTT AG | ||||
| L. amylolyticus | 16S–23S region | Amylolyticus-F | TTC GGT AGT GAC GTT TCG GA | 134 | 0.2 |
| Amylolyticus-R | TCA AGC AAG TGC CAT GCA G |
aIPC, internal positive control
bconc., concentration
Table 2.
Characteristics of unique genes to each species
| Species | Gene name | Accession no. |
|---|---|---|
| L. sanfranciscensis | Acetyltransferase | KRM80157.1 |
| L. acidipiscis | Membrane protein | KRM26780.1 |
| L. fermentum | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase | EEI21326.1 |
| L. amylovorus | Adenylosuccinate lyase | KRK41078.1 |
| L. pentosus | GHKL domain-containing protein | AYJ41677.1 |
| L. plantarum | LPXTG-motif cell wall anchor domain protein | EFK29584.1 |
| L. helveticus | Dicarboxylate/amino acid:cation Na+/H+ symporter family protein | EEW67281.1 |
| L. farciminis | DUF262 domain-containing protein | ATO45673.1 |
| L. parabuchneri | Leucine-rich repeat protein | KRM47288.1 |
| L. paraplantarum | MFS-type transporter YcnB | KRL48501.1 |
| L. gallinarum | LacI family transcriptional regulator | KRL21687.1 |
| L. casei | Putative truncated melibiose symporter | BAN74848.1 |
| L. paracasei | Cation transport ATPase | ABJ68989.1 |
| L. lindneri | Accessory Sec system protein Asp2 | ANZ57695.1 |
Table 3.
The BLASTN results of unique genes
| Species | Description | Identity (%) | Target species match | Non-target species match | ||||
|---|---|---|---|---|---|---|---|---|
| Species | No. of strains | Identify (%) | Species | No. of strains | Identity (%) | |||
| L. sanfranciscensis | L. sanfranciscensis TMW 1.1304 | 99 | L. sanfranciscensis | 20/20 | 100 ~ 98.94 | – | – | – |
| L. acidipiscis | L. acidipiscis strain ACA-DC 1533 | 99.58 | L. acidipiscis | 5/5 | 100 ~ 99.17 | – | – | – |
| L. fermentum | L. fermentum strain B1 28 | 100 | L. fermentum | 63/63 | 100 ~ 98.57 | – | – | – |
| L. amylovorus | L. amylovorus DSM 20531 | 100 | L. amylovorus | 14/15 | 100 ~ 98.84 | – | – | – |
| L. pentosus | L. pentosus strain DSM 20314 | 100 | L. pentosus | 22/22 | 100 ~ 98.35 | – | – | – |
| L. plantarum | L. plantarum strain IDCC3501 | 100 | L. plantarum | 449/453 | 100 ~ 97.14 | – | – | – |
| L. helveticus | L. helveticus isolate NWC_2_3 | 100 | L. helveticus | 56/57 | 100 ~ 98.70 | L. gallinarum | 1/7 | 99.64 |
| L. farciminis | L. farciminis KCTC 3681 | 100 | L. farciminis | 7/7 | 100 | – | – | – |
| L. parabuchneri | L. parabuchneri strain FAM21731 | 99.97 | L. parabuchneri | 25/25 | 100 ~ 96.57 | – | – | – |
| L. paraplantarum | L. paraplantarum strain DSM 10667 | 100 | L. paraplantarum | 10/11 | 100 ~ 98.78 | – | – | – |
| L. gallinarum | L. gallinarum DSM 10532 | 100 | L. gallinarum | 6/7 | 100 ~ 99.39 | – | – | – |
| L. casei | L. casei subsp. casei ATCC 393 | 100 | L. casei | 14/25 | 100 ~ 96.41 | – | – | – |
| L. paracasei | L. paracasei ATCC 334 | 100 | L. paracasei | 109/164 | 100 ~ 98.51 | L. casei | 3/25 | 98.39 ~ 98.14 |
| L. lindneri | L. lindneri strain TMW 1.481 | 100 | L. lindneri | 12/12 | 100 | – | – | – |
Fig. 1.
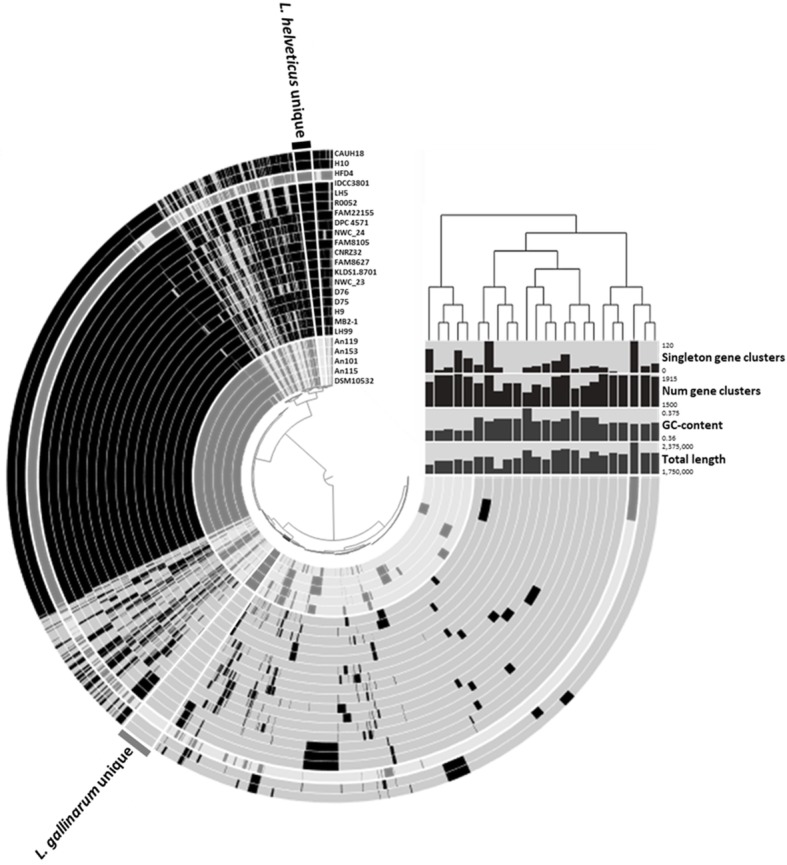
Pan-genome distribution across Lactobacillus gallinarum and L. helveticus. Each ring represents L. gallinarum and L. helveticus strain and each layer displays the pan-genome distribution. The gray and black rings represent the genomes of L. gallinarum and L. helveticus, respectively
Specificity of designed primer pairs
To confirm whether primer pairs were species-specific for the identification of each Lactobacillus species, conventional PCR assays were performed with 37 Lactobacillus reference strains. For each of the primer pairs, the amplification product was exclusive to each target strain with a high specificity. The results of the conventional PCR assays confirmed 100% specificity for all Lactobacillus species.
Specificity and accuracy of the developed PCR assays
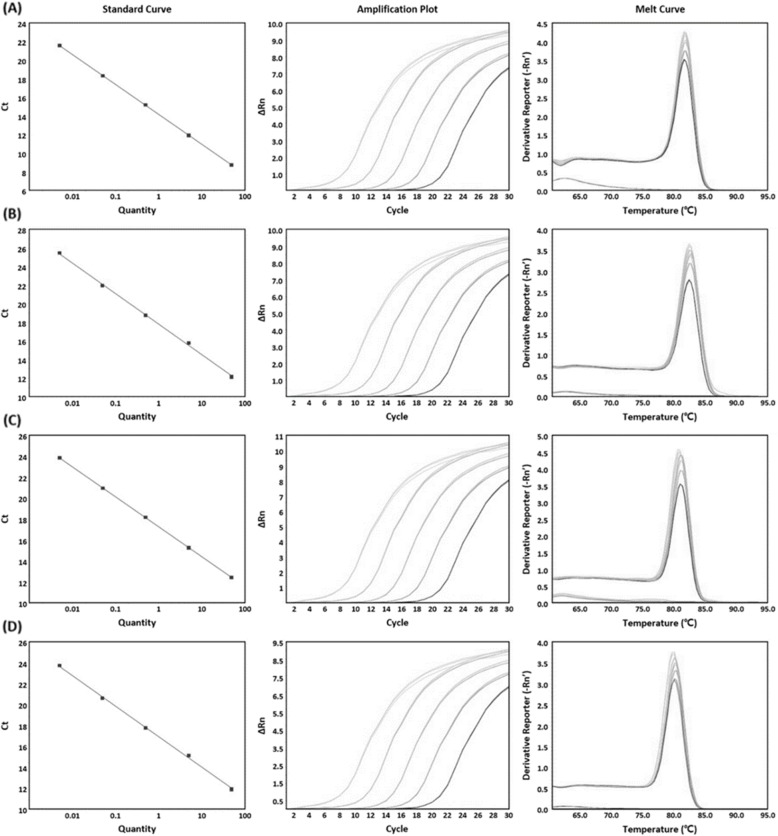
The accuracy and efficiency of the PCR assays were validated using the template DNA of the Lactobacillus reference species. All primer pairs exhibited a linear relationship over the range of 0.005 to 50 ng. The slopes for the specific primer pairs of L. acetotolerans, L. casei, L. parabuchneri, and L. lindneri were − 3.209, − 3.284, − 3.207, and − 3.595, respectively, and the R2 values were 1, 0.999, 1, and 0.985, respectively (Fig. 2). The R2 and slope values of the remaining primer pairs are shown in Table 4.
Fig. 2.
Examples of PCR standard curves, amplification curves and melting curves: aL. acetotolerans standard curve between 50 and 0.005 ng (y = − 3.209x + 14.197, R2 = 1, left), amplification plot (middle) and melt curve (right); bL. casei standard curve (y = − 3.284x + 17.817, R2 = 0.999, left), amplification plot (middle), melt curve (right); cL. parabuchneri standard curve (y = − 3.207x + 17.19, R2 = 1, left), amplification plot (middle) and melt curve (right); and (d) L. lindneri standard curve (y = − 3.595x + 16.261, R2 = 0.982, left), amplification plot (middle) and melt curve (right)
Table 4.
Slope, R2, and efficiency of Lactobacillus reference strain in the PCR assay
| Species | Slope | R2 | Efficiency (%) |
|---|---|---|---|
| L. gasseri | −3.214 | 0.999 | 104.701 |
| L. rhamnosus | − 3.362 | 0.998 | 98.35 |
| L. brevis | − 3.444 | 1 | 95.158 |
| L. sakei | − 3.212 | 1 | 104.797 |
| L. johnsonii | −3.214 | 0.999 | 104.701 |
| L. jensenii | −3.328 | 0.996 | 99.764 |
| L. fermentum | −3.56 | 0.995 | 90.955 |
| L. plantarum | −3.221 | 0.995 | 104.396 |
| L. paracasei | −3.305 | 0.98 | 100.694 |
| L. paraplantarum | −3.256 | 0.998 | 102.822 |
| L. casei | −3.284 | 0.999 | 101.612 |
| L. curvatus | −3.485 | 0.999 | 93.617 |
| L. acidophilus | −3.506 | 1 | 92.845 |
| L. salivarius | −3.564 | 1 | 90.809 |
| L. reuteri | −3.342 | 0.999 | 99.161 |
| L. coryniformis | −3.217 | 0.989 | 104.578 |
| L. farciminis | −3.386 | 0.991 | 97.39 |
| L. zymae | −3.5 | 0.997 | 93.073 |
| L. pentosus | −3.292 | 0.999 | 101.251 |
| L. crustorum | −3.438 | 0.999 | 95.366 |
| L. mucosae | −3.478 | 0.986 | 93.886 |
| L. buchneri | −3.411 | 0.993 | 96.424 |
| L. helveticus | −3.230 | 0.998 | 103.98 |
| L. amylovorus | −3.582 | 0.993 | 90.167 |
| L. heilongjiangensis | −3.462 | 1 | 94.458 |
| L. parabuchneri | −3.207 | 1 | 105.049 |
| L. acidipiscis | −3.528 | 0.984 | 92.075 |
| L. sanfranciscensis | −3.229 | 0.999 | 104.034 |
| L. ruminis | −3.295 | 1 | 101.153 |
| L. agilis | −3.508 | 1 | 92.795 |
| L. delbrueckii | −3.31 | 0.999 | 100.479 |
| L. amylophilus | −3.481 | 0.984 | 93.768 |
| L. kunkeei | −3.571 | 0.998 | 90.568 |
| L. acetotolerans | −3.209 | 1 | 104.92 |
| L. lindneri | −3.559 | 0.982 | 90.972 |
| L. gallinarum | −3.346 | 0.999 | 98.989 |
| L. amylolyticus | −3.552 | 0.996 | 91.209 |
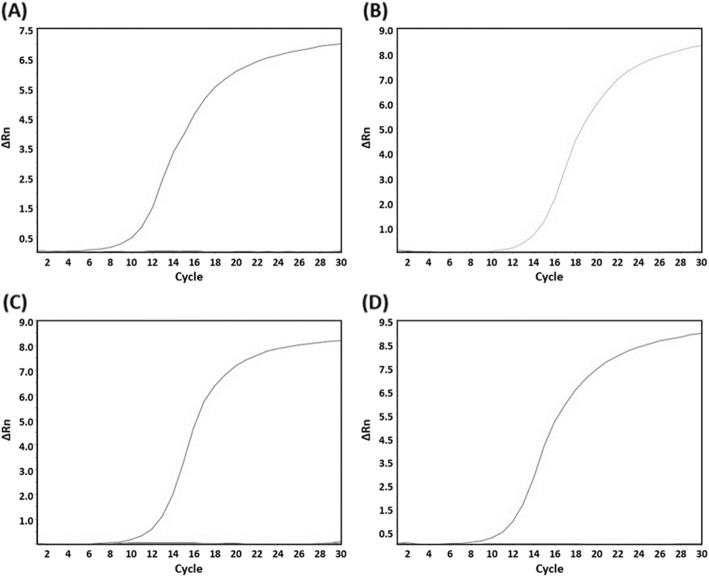
The specificities of all 37 Lactobacillus reference strains were evaluated for each species-specific primer pair. A non-template was used as a negative control, and the template DNA of 37 Lactobacillus reference stains was used as a positive control for each primer pair. All genomic DNA from Lactobacillus species yielded detectable amplicon signals in the well containing each primer pair, whereas none of the non-target Lactobacillus species generated any signals at all (Fig. 3). The Ct ranges were 9.0 to 15.0 for each Lactobacillus species (Table 5). Thus, all primer pairs were considered specific for the detection of an individual Lactobacillus species. To verify the accuracy of the assay, a primer pair targeting the 16S rRNA gene was used as an IPC; the amplification of the target region was observed within the Ct value range of 5.7 to 9.1 for all tested Lactobacillus species.
Fig. 3.
Specificities of species-specific primer pairs against 37 Lactobacillus species: a specificity of L. acetotolerans specific primer pair, amplification curve: L. acetotolerans KACC 12447; b specificity of L. casei specific primer pair, amplification curve: L. casei KACC 12413; c specificity of L. parabuchneri specific primer pair, amplification curve: L. parabuchneri KACC 12363; and (d) specificity of L. lindneri specific primer pair, amplification curve: L. lindneri KACC 12445
Table 5.
Specificity results of the PCR assay
| Primer name | Detected species | Ct value | Tm (°C) |
|---|---|---|---|
| Gasseri-F,R | L. gasseri KCTC 3163 | 5.366 | 83.749 |
| Rhamnosus-F,R | L. rhamnosus KCTC 3237 | 11.258 | 79.329 |
| Brevis-F,R | L. brevis KCTC 3498 | 5.762 | 85.319 |
| Sakei-F,R | L. sakei KCTC 3603 | 11.139 | 82.441 |
| Johnsonii-F,R | L. johnsonii KCTC 3801 | 6.450 | 84.193 |
| Jensenii-F,R | L. jensenii KCTC 5194 | 6.583 | 81.378 |
| Fermentum-F,R | L. fermentum KACC 11441 | 4.260 | 88.582 |
| Plantarum-F,R | L. plantarum KACC 11451 | 10.715 | 82.027 |
| Paracasei-F,R | L. paracasei KACC 12361 | 12.012 | 80.746 |
| Paraplantarum-F,R | L. paraplantarum KACC 12373 | 10.884 | 82.306 |
| Casei-F,R | L. casei KACC 12413 | 10.739 | 82.513 |
| Curvatus-F,R | L. curvatus KACC 12415 | 13.832 | 82.686 |
| Acidophilus-F,R | L. acidophilus KACC 12419 | 12.383 | 79.308 |
| Salivarius-F,R | L. salivarius KCTC 3600 | 14.905 | 81.806 |
| Reuteri-F,R | L. reuteri KCTC 3594 | 9.142 | 83.439 |
| Coryniformis-F,R | L. coryniformis KACC 12411 | 13.638 | 84.793 |
| Farciminis-F,R | L. farciminis KACC 12423 | 10.678 | 80.465 |
| Zymae-F,R | L. zymae KACC 16349 | 7.546 | 82.568 |
| Pentosus-F,R | L. pentosus KACC 12428 | 11.603 | 84.268 |
| Crustorum-F,R | L. crustorum KACC 16344 | 12.467 | 82.012 |
| Mucosae-F,R | L. mucosae KACC 12381 | 11.598 | 83.109 |
| Buchneri-F,R | L. buchneri KACC 12416 | 11.606 | 82.206 |
| Helveticus-F,R | L. helveticus KACC 12418 | 12.087 | 79.059 |
| Amylovorus-F,R | L. amylovorus KACC 12435 | 11.256 | 82.037 |
| Heilongjiangensis-F,R | L. heilongjiangensis KACC 18741 | 11.922 | 81.205 |
| Parabuchneri-F,R | L. parabuchneri KACC 12363 | 9.377 | 81.604 |
| Acidipiscis-F,R | L. acidipiscis KACC 12394 | 10.743 | 81.566 |
| Sanfranciscensis-F,R | L. sanfranciscensis KACC 12431 | 10.273 | 79.814 |
| Ruminis-F,R | L. ruminis KACC 12429 | 9.724 | 82.341 |
| Agilis-F,R | L. agilis KACC 12433 | 11.758 | 82.095 |
| Delbrueckii-F,R | L. delbrueckii KACC 12420 | 8.621 | 83.114 |
| Amylophilus-F,R | L. amylophilus KACC 11430 | 10.943 | 82.733 |
| Kunkeei-F,R | L. kunkeei KACC 19371 | 8.542 | 83.217 |
| Acetotolerans-F,R | L. acetotolerans KACC 12447 | 11.912 | 82.031 |
| Lindneri-F,R | L. lindneri KACC 12445 | 12.910 | 79.917 |
| Gallinarum-F,R | L. gallinarum KACC 12370 | 10.132 | 78.138 |
| Amylolyticus-F,R | L. amylolyticus KACC 12374 | 11.694 | 83.460 |
Application of the developed PCR assays in probiotics and dairy products
The PCR assays was applied to identify Lactobacillus species from commercial probiotics and dairy products. A total of 31 products were evaluated using the PCR assays we have developed, and the assay results were compared with the probiotic label claims. Probiotic products were tagged as P1 to P19, whereas dairy products were designated as D1 to D12. As a result of the validation process, 17 products were confirmed to match their label claims (Table 6). However, the label claims of four products (P14, P15, P17, and P18) identified L. helveticus but contained L. acidophilus, and three products (P14, P15, and P17) contained L. paracasei instead of the L. casei indicated on the label. In one product (P16), we detected additional Lactobacillus species that were not listed on the label. We were also able to identify the Lactobacillus species from products labeled with the compound LAB. Our PCR results confirmed that these products contained either L. acidophilus and L. delbrueckii or L. paracasei and L. helveticus.
Table 6.
Results of application test of the developed PCR assay to commercial probiotic and dairy products
| Name | Country | Label claim | Detected species |
|---|---|---|---|
| P1 | Korea | L. plantarum | L. plantarum |
| P2 | USA | L. rhamnosus | L. rhamnosus |
| P3 | Korea | L. acidophilus | L. acidophilus |
| P4 | Korea | L. delbrueckii, L. paracasei | L. delbrueckii, L. paracasei |
| P5 | Korea | L. acidophilus, L. rhamnosus | L. acidophilus, L. rhamnosus |
| P6 | Korea | L. acidophilus, L. rhamnosus | L. acidophilus, L. rhamnosus |
| P7 | Korea | L. acidophilus, L. delbrueckii | L. acidophilus, L. delbrueckii |
| P8 | Korea | L. acidophilus, L. plantarum, L. reuteri | L. acidophilus, L. plantarum, L. reuteri |
| P9 | Korea | L. acidophilus, L. plantarum, L. reuteri | L. acidophilus, L. plantarum, L. reuteri |
| P10 | Korea | L. acidophilus, L. fermentum, L. plantarum | L. acidophilus, L. fermentum, L. plantarum |
| P11 | USA | L. acidophilus, L. brevis, L. casei, L. delbrueckii, L. paracasei, L. plantarum, L. salivarius | L. acidophilus, L. brevis, L. casei, L. delbrueckii, L. paracasei, L. plantarum, L. salivarius |
| P12 | Canada | L. acidophilus, L. casei, L. gasseri, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus | L. acidophilus, L. casei, L. gasseri, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus |
| P13 | Korea | L. rhamnosus | L. rhamnosus |
| P14 | Canada | L. acidophilus, L. casei, L. rhamnosus | L. helveticus, L. paracasei, L. rhamnosus |
| P15 | Canada | L. acidophilus, L. casei, L. rhamnosus | L. helveticus, L. paracasei, L. rhamnosus |
| P16 | Korea | L. rhamnosus | L. rhamnosus, L. helveticus, L. reuteri |
| P17 | Canada | L. acidophilus, L. casei, L. plantarum, L. rhamnosus | L. helveticus, L. paracasei, L. plantarum, L. rhamnosus |
| P18 | Canada | L. acidophilus, L. paracasei, L. rhamnosus, L. salivarius | L. helveticus, L. paracasei, L. rhamnosus, L. salivarius |
| P19 | Korea | L. delbrueckii, L. plantarum, LAB mixed powder | L. delbrueckii, L. plantarum, L. amylovorus, L. helveticus, L. paracasei, L. rhamnosus |
| D1 | Korea | L. acidophilus, L. casei | L. acidophilus, L. casei |
| D2 | Korea | L. delbrueckii, L. rhamnosus | L. delbrueckii, L. rhamnosus |
| D3 | Korea | L. delbrueckii, L. rhamnosus | L. delbrueckii, L. rhamnosus |
| D4 | Korea | L. delbrueckii, L. rhamnosus | L. delbrueckii, L. rhamnosus |
| D5 | Korea | L. rhamnosus, LAB | L. rhamnosus, L. helveticus, L. paracasei |
| D6 | Korea | LAB, probiotic LAB | L. acidophilus, L. delbrueckii, L. paracasei |
| D7 | Korea | Compound LAB | L. acidophilus, L. delbrueckii, L. fermentum |
| D8 | Korea | LAB | L. acidophilus, L. delbrueckii |
| D9 | Korea | LAB | L. helveticus, L. paracasei |
| D10 | Korea | LAB | L. helveticus, L. paracasei |
| D11 | Korea | LAB | L. helveticus, L. paracasei |
| D12 | Korea | LAB | L. helveticus, L. paracasei |
LAB lactic acid bacteria
Discussion
A variety of methods have been used to identify LAB in foods or in the environment. The most representative method is a conventional method consisting of phenotypic and biochemical tests, which have limitations in accuracy among isolates possessing similar physiological specificities and fermentation profiles at the species level [22, 23]. To overcome these difficulties, several genotype-based methods such as DGGE and metagenome sequencing have been developed [23]. In addition, metagenome sequencing based on the 16S rRNA gene is a common approach in investigating microbial communities but is limited to distinguishing similar species [24]. Because metagenome sequencing remains a time-consuming process and requires specialized equipment and techniques, it is unsuitable for analyzing a large number of samples. To combat this, we have developed PCR assays that can rapidly and easily analyze Lactobacillus communities in fermented foods and potentially environmental samples.
PCR is generally considered to be a rapid, sensitive, and time-saving method for the detection of bacterial species [25–27]. The accuracy of PCR is determined by the specificity of the primer pairs used. The 16S rRNA gene is considered a marker gene for bacterial genotypic analysis and is useful for the accurate identification of bacteria [12, 28]. Studies focusing on the identification of Lactobacillus have mainly used PCR-based molecular analysis by primer pair targeting variable regions of the 16S rRNA gene sequences [23, 29]. However, for closely related species such as the members of the L. casei, L. sakei, L. plantarum, and L. acidophilus groups, each of which has a 16S rRNA gene similarity of more than 98% [30–32], only species-specific PCR primer pairs could sufficiently differentiate species.
To overcome the limitations of the 16S rRNA gene, we developed 37 Lactobacillus species-specific primer pairs based on 16S–23S rRNA gene analysis and comparative genome analysis. Species-specific primer pairs were designed to have a small amplicon size (~ 260 bp) to increase amplification efficiency and detect Lactobacillus species present in processed foods. The specificities of the species-specific primer pairs were confirmed using the 37 Lactobacillus species, and amplification was observed only in the target species DNA without any cross-reactivity. Also, it was confirmed that species such as the L. casei group, L. acidophilus group, and L. plantarum group, which are not distinguished by the conventional identification method, were differentiable using the species-specific primer pairs. According to the CODEX guidelines, the slope values of − 3.1 to − 3.6 are considered to indicate a high PCR efficiency. The coefficient value of determination should be at least 0.98 to be considered viable data [33]. Therefore, these results demonstrate that the developed PCR assays provides high accuracy and efficiency.
The developed PCR assays was used to assess probiotics and dairy products. Using this assays, 17 products were determined to contain the Lactobacillus species advertised on the label. In the remaining products, the species indicated on the labels were either replaced with or contaminated by another species. For example, L. acidophilus was replaced by L. helveticus and L. casei was replaced by L. paracasei in four probiotic products. Though these products were produced by different companies, the same strains were identified. As described above, L. acidophilus belongs to the same group as L. helveticus, and L. casei belongs to the same group as L. paracasei. The likely reason a label names species other than the one detected is misidentification [20, 34]. In one product, additional Lactobacillus species that were not indicated on the label were detected by PCR. These were detected at much higher Ct values than the Lactobacillus species indicated on the label, suggesting that such strains were only present in low concentrations [35]. We were also able to accurately identify the species contained in products labeled compound LAB. In all of these products, we detected L. acidophilus and L. delbrueckii or L. helveticus and L. paracasei. These results confirm that our PCR assays can detect all species of Lactobacillus contained in these products.
Many researchers have provided evidence that the advertised contents of commercial probiotic products containing LAB are significantly different from the actual contents [25, 34]. Lewis et al. (2016) reported that only one of the 16 commercial probiotic products corresponded exactly with the Bifidobacterium species claimed on the label [36]. In addition, some products are inconsistent from one lot to another. These results indicate inadequate quality control for these products.
Conclusion
In this study, we developed specific primer pairs using comparative genomics to identify Lactobacillus accurately and rapidly at the species level, then applied this technology in the PCR assays that can detect and identify 37 Lactobacillus species in a single 96 well plate. The developed PCR assays were able to accurately discriminate species that were not distinguishable by the conventional identification method. To verify the developed PCR assays, we compared the label claims of probiotics and dairy products with the Lactobacillus species detected using the PCR method. The PCR assays that we have developed were successfully applied to commercial probiotic and dairy products, and showed that some products did not accurately match the Lactobacillus species listed on their labels. Thus, this assays will be helpful for monitoring the reliability of commercial probiotic and dairy product labels. In addition to its application in probiotic products, the assays can be applied to identify Lactobacillus communities in various food or environmental samples.
Methods
Bacterial strains and probiotic and dairy products
The Lactobacillus reference strains were obtained from the Korean Collection for Type Cultures (KCTC; Daejeon, South Korea; https://kctc.kribb.re.kr/) and the Korean Agricultural Culture Collection (KACC; Jeonju, South Korea; http://genebank.rda.go.kr/) (Table 7). All reference strains were cultured in Lactobacilli MRS Broth (Difco, Becton & Dickinson, Sparks, MD, USA) at 30 °C for 48 h under anaerobic conditions. The probiotic and dairy products tested in this study were obtained from various markets around the world (South Korea, United States, and Canada). The samples used in this study included 19 probiotic products (10 capsule-form pharmaceuticals and 9 powder-form food supplements) and 12 dairy products manufactured by 19 different companies. All products were labeled with bacterial species or LAB compounds.
Table 7.
Lactobacillus reference strains used in this study
| Species | Strain no. |
|---|---|
| L. gasseri | KCTCa 3163 |
| L. rhamnosus | KCTC 3237 |
| L. brevis | KCTC 3498 |
| L. sakei | KCTC 3603 |
| L. johnsonii | KCTC 3801 |
| L. jensenii | KCTC 5194 |
| L. fermentum | KACCb 11,441 |
| L. plantarum | KACC 11451 |
| L. paracasei | KACC 12361 |
| L. paraplantarum | KACC 12373 |
| L. casei | KACC 12413 |
| L. curvatus | KACC 12415 |
| L. acidophilus | KACC 12419 |
| L. salivarius | KCTC 3600 |
| L. reuteri | KCTC 3594 |
| L. coryniformis | KACC 12411 |
| L. farciminis | KACC 12423 |
| L. zymae | KACC 16349 |
| L. pentosus | KACC 12428 |
| L. crustorum | KACC 16344 |
| L. mucosae | KACC 12381 |
| L. buchneri | KACC 12416 |
| L. helveticus | KACC 12418 |
| L. amylovorus | KACC 12435 |
| L. heilongjiangensis | KACC 18741 |
| L. parabuchneri | KACC 12363 |
| L. acidipiscis | KACC 12394 |
| L. sanfranciscensis | KACC 12431 |
| L. ruminis | KACC 12429 |
| L. agilis | KACC 12433 |
| L. delbrueckii | KACC 12420 |
| L. amylophilus | KACC 11430 |
| L. kunkeei | KACC 19371 |
| L. acetotolerans | KACC 12447 |
| L. lindneri | KACC 12445 |
| L. gallinarum | KACC 12370 |
| L. amylolyticus | KACC 12374 |
aKCTC Korean Collection for Type Cultures
bKACC Korean Agricultural Culture Collection
DNA extraction
All Lactobacillus reference strains were grown in MRS broth at 30 °C for 48 h under anaerobic conditions. The cultured cells were harvested by centrifugation at 13,600×g for 5 min, after which the supernatant was removed. Genomic DNA was extracted using a bacterial genomic DNA extraction kit (Intron Biotechnology, Seongnam, South Korea) according to the manufacturer’s instructions. Total genomic DNA from the probiotic and dairy products was extracted using a DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the method described in a previous study [37]. DNA concentration and purity were determined by absorbance using a MaestroNano® spectrophotometer (Maestrogen, Las Vegas, NV, USA).
Identification of Lactobacillus species-specific regions and primer designs
In total, 180 genome sequences, which contain 37 Lactobacillus species, were obtained from the National Center for Biotechnology Information (NCBI; ftp://ftp.ncbi.nlm.nih.gov/genomes/) database (Additional file 1: Table S1). The 16S–23S rRNA regions, including the intergenic spacer regions, of 180 strains were extracted from the Lactobacillus genomes using a script written in the Python language, and the extracted regions were aligned using the Geneious program ver. 11.1.2 (Biomatters Limited, Auckland, New Zealand). According to the alignment results, primer pairs were designed on the basis of species-specific sequences in the 16S–23S rRNA gene. Some Lactobacillus species are difficult to distinguish at the species level because of the high degree of similarity in their 16S–23S rRNA gene sequences. For these species, we have developed species-specific primer pairs from unique genes that exist only in the target species obtained through comparative genomic analysis.
The genome sequences of target species were blasted against the genome of target species using the UBLAST function of USEARCH program ver. 9.0 [38], with 80% cutoff identity to obtain genes with high similarity [39]. The genes that showed a significant match with the genomes of all target species were considered as core genes of target species. Those genes were then blasted against all of the Lactobacillus genomes except the target species using the UBLAST function of USEARCH program with default parameter settings of 50% cutoff identity [38]. Genes that found no match to all genomes of the non-target species were identified as potential unique genes. The identified potential unique genes were verified using the Basic Local Alignment Search Tool (BLAST) for 52,478,804 sequences including Lactobacillus genomes. Also, it was confirmed whether the unique genes exist in the genome sequences of reference strains using USEARCH program. The genes were confirmed to be unique genes in the species level and found all in the target species used in this study. The species-specific primer pairs were designed based on these genes. To verify the presence of genomic DNA from Lactobacillus species, primer pairs were designed from the conserved regions of 37 Lactobacillus species in the 16S rRNA gene sequence and used as an internal positive control (IPC). All primer pairs were designed using Primer Designer (Scientific and Educational Software, Durham, NC, USA) and synthesized by Bionics Co. Ltd. (Seoul, South Korea).
Specificity of species-specific primer pairs
PCR assays were performed to confirm the specificity of the designed species-specific primer pairs. The specificity was evaluated using 37 Lactobacillus reference strains. PCR products were amplified using the following conditions in a thermocycler (Astec, Fukuoka, Japan): 94 °C for 10 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min. The 25 μL reaction mixtures contained 20 ng of template DNA of a Lactobacillus reference strain, 0.5 unit of Taq DNA polymerase (TaKaRa BIO Inc., Tokyo, Japan), and species-specific primer pairs. The optimal concentration of each species-specific primer pair obtained from the experiments is shown in Table 1. The amplification products were confirmed by electrophoresis on a 2% agarose gel, and the product bands were visualized under a UV transilluminator (Vilber Lourmat, Marne La Vallee, France).
Development of PCR assays
In this study, we developed the PCR assays that allows each primer pair to run independently to cover each full assays using one primer pair in each well and 37 wells. The PCR assays were performed on the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the following conditions: 95 °C for 2 min, followed by 30 cycles of 95 °C for 5 s and 60 °C for 30 s. The melting curve data were generated using 1 cycle of 95 °C for 15 s, 60 °C for 1 min, 95 °C for 30 s, and 60 °C for 15 s. The amplification mixture with a final volume of 20 μL for real-time PCR assays included 2X LeGene SB-Green Real-Time PCR Master Mix (LeGene Biosciences, San Diego, CA, USA), template DNA, and species-specific primer pairs at optimal concentrations shown in Table 1. To evaluate the analytical accuracy of the PCR assays, a standard curve was constructed using serial dilutions (50 to 0.005 ng) of genomic DNA from Lactobacillus reference strains in triplicate. The specificities of the species-specific primer pairs were tested using 20 ng of DNA extracted from 37 Lactobacillus reference strains. PCR amplifications of IPC were also confirmed with 37 Lactobacillus reference strains. The results of the PCR were confirmed using 7500 Software V2.3 (Applied Biosystems).
Application of the developed PCR assays in probiotic and dairy products
We designed a validation test to detect 37 Lactobacillus species with PCR in a single 96 well plate using primer pairs. Each well of a reaction plate contained each primer pair and IPC for the detection of 37 Lactobacillus species (Additional file 2: Fig. S1). Briefly, 20 ng of product DNA and 2X Master Mix (LeGene Biosciences) were added to each well of the reaction plate containing species-specific primers. Then, PCR was performed in the 7500 Real-Time PCR system (Applied Biosystems). The real-time PCR conditions were similar to those described in “Development of PCR assays” section. Our method included one primer pair in each well, so 37 wells were used for the full assay of each product sample. Therefore, for all products, including mixed samples, the PCR results determined that the corresponding species was included in the product when amplified in a well containing specific primer pair.
Supplementary information
Additional file 1: Table S1. General genome features of Lactobacillus species.
Additional file 2: Figure S1. Real-time PCR 96-well plate layout for validation of probiotic products. P: Internal positive control, N: no template control, 1: L. gasseri specific primer set, 2: L. rhamnosus specific primer set, 3: L. brevis specific primer set, 4: L. sakei specific primer set, 5: L. johnsonii specific primer set, 6: L. jensenii specific primer set, 7: L. fermentum specific primer set, 8: L. plantarum specific primer set, 9: L. paracasei specific primer set, 10: L. paraplantarum specific primer set, 11: L. casei specific primer set, 12: L. curvatus specific primer set, 13: L. acidophilus specific primer set, 14: L. salivarius specific primer set, 15: L. reuteri specific primer set, 16: L. coryniformis specific primer set, 17: L. farciminis specific primer set, 18: L. zymae specific primer set, 19: L. pentosus specific primer set, 20: L. crustorum specific primer set, 21: L. mucosae specific primer set, 22: L. buchneri specific primer set, 23: L. helveticus specific primer set, 24: L. amylovorus specific primer set, 25: L. heilongjiangensis specific primer set, 26: L. parabuchneri specific primer set, 27: L. acidipiscis specific primer set, 28: L. sanfranciscensis specific primer set, 29: L. ruminis specific primer set, 30: L. agilis specific primer set, 31: L. delbrueckii specific primer set, 32: L. amylophilus specific primer set, 33: L. kunkeei specific primer set, 34: L. acetotolerans specific primer set, 35: L. lindneri specific primer set, 36: L. gallinarum specific primer set, 37: L. amylolyticus specific primer set.
Acknowledgements
Not applicable.
Abbreviations
- PCR
Polymerase Chain Reaction
- GRAS
Generally Recognized As Safe
- 16S rRNA
16S ribosomal Ribonucleic Acid
- LAB
Lactic acid bacteria
- DGGE
Denaturing Gradient Gel Electrophoresis
- IPC
Internal Positive Control
- MFS
Major Facilitator Superfamily
- BLAST
Basic Local Alignment Search Tool
- NCBI
National Center for Biotechnology Information
- GHKL
Gyrase, Hsp90, Histidin Kinase, MutL
Authors’ contributions
EK, SMY, BL, SHP, and HYK designed the experiment. EK, SMY, and BL contributed the experiment. EK performed the bioinformatics analysis. EK and SMY confirmed primer specificity and performed application tests using real-time PCR. EK prepared a draft manuscript. EK, SHP, BR, and HYK reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (grant number 918005–4). None of the funders had any role in designing and conducting of the study.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01781-z.
References
- 1.Goldstein EJC, Tyrrell KL, Citron DM. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis. 2015;60:S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 2.Jespers V, Menten J, Smet H, Poradosú S, Abdellati S, Verhelst R, et al. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol. 2012;12:1–10. doi: 10.1186/1471-2180-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masco L, Huys G, De Brandt E, Temmerman R, Swings J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int J Food Microbiol. 2005;102:221–230. doi: 10.1016/j.ijfoodmicro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 4.Patrignani F, Siroli L, Parolin C, Serrazanetti DI, Vitali B, Lanciotti R. Use of Lactobacillus crispatus to produce a probiotic cheese as potential gender food for preventing gynaecological infections. PLoS One. 2019;14(1):e0208906. doi: 10.1371/journal.pone.0208906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 6.Stanton C, Gardiner G, Meehan H, Collins K, Fitzgerald G, Lynch PB. Market potential for probiotics. Am J Clin Nutr. 2001;73:476S–483S. doi: 10.1093/ajcn/73.2.476s. [DOI] [PubMed] [Google Scholar]
- 7.Temmerman R, Scheirlinck I, Huys G, Swings J. Culture-independent analysis of probiotic products by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003;69:220–226. doi: 10.1128/AEM.69.1.220-226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung PSM, Sanders ME, Kitts CL, Cano R, Tong PS. Species-specific identification of commercial probiotic strains. J Dairy Sci. 2002;85:1039–1051. doi: 10.3168/jds.S0022-0302(02)74164-7. [DOI] [PubMed] [Google Scholar]
- 9.Weese JS. Evaluation of deficiencies in labeling of commercial probiotics. Can Vet J. 2003;44:982–983. [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon HS, Yang EH, Yeon SW, Kang BH, Kim TY. Rapid identification of probiotic Lactobacillus species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23S rRNA. FEMS Microbiol Lett. 2004;239:267–275. doi: 10.1016/j.femsle.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 11.Dickson EM, Riggio MP, Macpherson L. A novel species-specific PCR assay for identifying Lactobacillus fermentum. J Med Microbiol. 2005;54:299–303. doi: 10.1099/jmm.0.45770-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee CM, Sieo CC, Wong CMVL, Abdullah N, Ho YW. Sequence analysis of 16S rRNA gene and 16S–23S rRNA gene intergenic spacer region for differentiation of probiotics Lactobacillus strains isolated from the gastrointestinal tract of chicken. Ann Microbiol. 2008;58:133–140. doi: 10.1007/BF03179457. [DOI] [Google Scholar]
- 13.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 14.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/MMBR.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Sullivan DJ. Methods for analysis of the intestinal microflora. Curr Issues Intest Microbiol. 2000;1:39–50. [PubMed] [Google Scholar]
- 16.Fasoli S, Marzotto M, Rizzotti L, Rossi F, Dellaglio F, Torriani S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int J Food Microbiol. 2003;82:59–70. doi: 10.1016/S0168-1605(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 17.Hong Y, Yang HS, Li J, Han SK, Chang HC, Kim HY. Identification of lactic acid bacteria in salted Chinese cabbage by SDS-PAGE and PCR-DGGE. J Sci Food Agric. 2014;94:296–300. doi: 10.1002/jsfa.6257. [DOI] [PubMed] [Google Scholar]
- 18.De Filippis F, Parente E, Zotta T, Ercolini D. A comparison of bioinformatic approaches for 16S rRNA gene profiling of food bacterial microbiota. Int J Food Microbiol. 2018;265:9–17. doi: 10.1016/j.ijfoodmicro.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Huang CH, Liou JS, Huang L, Watanabe K. Developing novel species-specific DNA markers for PCR-based species identification of the Lactobacillus sakei group. Lett Appl Microbiol. 2017;66:138–144. doi: 10.1111/lam.12825. [DOI] [PubMed] [Google Scholar]
- 20.Huang CH, Huang L. Rapid species- and subspecies-specific level classification and identification of Lactobacillus casei group members using MALDI Biotyper combined with ClinProTools. J Dairy Sci. 2018;101:979–991. doi: 10.3168/jds.2017-13642. [DOI] [PubMed] [Google Scholar]
- 21.Kang J, Chung WH, Lim TJ, Whon TW, Lim S, Nam YD. Complete genome sequence of Lactobacillus casei LC5, a potential probiotics for atopic dermatitis. Front Immunol. 2017;8:413. doi: 10.3389/fimmu.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohania D, Nagpal R, Kumar M, Bhardwaj A, Yadav M, Jain S, et al. Molecular approaches for identification and characterization of lactic acid bacteria. J Dig Dis. 2008;9:190–198. doi: 10.1111/j.1751-2980.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- 23.Karapetsas A, Vavoulidis E, Galanis A, Sandaltzopoulos R, Kourkoutas Y. Rapid detection and identification of probiotic Lactobacillus casei ATCC 393 by multiplex PCR. J Mol Microbiol Biotechnol. 2010;18:156–161. doi: 10.1159/000308518. [DOI] [PubMed] [Google Scholar]
- 24.Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. Peer J. 2017;5:e3375. doi: 10.7717/peerj.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youn SY, Ji GE, Han YR, Park MS. Development of strain-specific primers for identification of Bifidobacterium bifidum BGN4. J Microbiol Biotechnol. 2017;27:909–915. doi: 10.4014/jmb.1610.10018. [DOI] [PubMed] [Google Scholar]
- 26.Cho Y, Kim E, Han SK, Yang SM, Kim MJ, Kim HJ, et al. Rapid identification of Vibrio species isolated from the southern coastal regions of Korea by MALDI-TOF mass spectrometry and comparison of MALDI sample preparation methods. J Microbiol Biotechnol. 2017;27:1593–1601. doi: 10.4014/jmb.1704.04056. [DOI] [PubMed] [Google Scholar]
- 27.Kim MJ, Kim HY. Development of a fast duplex real-time PCR assay for simultaneous detection of chicken and pigeon in raw and heat-treated meats. Food Control. 2018;85:1–5. doi: 10.1016/j.foodcont.2017.09.012. [DOI] [Google Scholar]
- 28.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teanpaisan R, Dahlén G. Use of polymerase chain reaction techniques and sodium dodecyl sulfate–polyacrylamide gel electrophoresis for differentiation of oral Lactobacillus species. Oral Microbiol Immunol. 2006;21:79–83. doi: 10.1111/j.1399-302X.2006.00259.x. [DOI] [PubMed] [Google Scholar]
- 30.Koort J, Vandamme P, Schillinger U, Holzapfel W, Björkroth J. Lactobacillus curvatus subsp. melibiosus is a later synonym of Lactobacillus sakei subsp. carnosus. Int J Syst Evol Microbiol. 2004;54:1621–1626. doi: 10.1099/ijs.0.63164-0. [DOI] [PubMed] [Google Scholar]
- 31.Berthier F, Ehrlich SD. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol Lett. 1998;161:97–106. doi: 10.1111/j.1574-6968.1998.tb12934.x. [DOI] [PubMed] [Google Scholar]
- 32.Huang CH, Chang MT, Huang L, Chu WS. The dnaJ gene as a molecular discriminator to differentiate among species and strain within the Lactobacillus casei group. Mol Cell Probes. 2015;29:479–484. doi: 10.1016/j.mcp.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol. 2014;37:115–126. doi: 10.1016/j.tifs.2014.03.008. [DOI] [Google Scholar]
- 34.Patro JN, Ramachandran P, Barnaba T, Mammel MK, Lewis JL, Elkins CA. Culture-independent metagenomic surveillance of commercially available probiotics with high-throughput next-generation sequencing. mSphere. 2016;1:1–12. doi: 10.1128/mSphere.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caraguel CGB, Stryhn H, Gagné N, Dohoo IR, Hammell KL. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. J Vet Diagn Investig. 2011;23:2–15. doi: 10.1177/104063871102300102. [DOI] [PubMed] [Google Scholar]
- 36.Lewis ZT, Shani G, Masarweh CF, Popovic M, Frese SA, Sela DA, et al. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr Res. 2016;79:445–452. doi: 10.1038/pr.2015.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volk H, Piskernik S, Kurinčič M, Klančnik A, Toplak N, Jeršek B. Evaluation of different methods for DNA extraction from milk. J Food Nutr Res. 2014;53:97–104. [Google Scholar]
- 38.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Chaudnari NM, Gautam A, Gupta VK, Kaur G, Dutta C, Paul S. PanGFR-HM: a dynamic web recource for pan-genomic and functional profiling of human microbiome with comparative features. Front Microbiol. 2018;9:2322. doi: 10.3389/fmicb.2018.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. General genome features of Lactobacillus species.
Additional file 2: Figure S1. Real-time PCR 96-well plate layout for validation of probiotic products. P: Internal positive control, N: no template control, 1: L. gasseri specific primer set, 2: L. rhamnosus specific primer set, 3: L. brevis specific primer set, 4: L. sakei specific primer set, 5: L. johnsonii specific primer set, 6: L. jensenii specific primer set, 7: L. fermentum specific primer set, 8: L. plantarum specific primer set, 9: L. paracasei specific primer set, 10: L. paraplantarum specific primer set, 11: L. casei specific primer set, 12: L. curvatus specific primer set, 13: L. acidophilus specific primer set, 14: L. salivarius specific primer set, 15: L. reuteri specific primer set, 16: L. coryniformis specific primer set, 17: L. farciminis specific primer set, 18: L. zymae specific primer set, 19: L. pentosus specific primer set, 20: L. crustorum specific primer set, 21: L. mucosae specific primer set, 22: L. buchneri specific primer set, 23: L. helveticus specific primer set, 24: L. amylovorus specific primer set, 25: L. heilongjiangensis specific primer set, 26: L. parabuchneri specific primer set, 27: L. acidipiscis specific primer set, 28: L. sanfranciscensis specific primer set, 29: L. ruminis specific primer set, 30: L. agilis specific primer set, 31: L. delbrueckii specific primer set, 32: L. amylophilus specific primer set, 33: L. kunkeei specific primer set, 34: L. acetotolerans specific primer set, 35: L. lindneri specific primer set, 36: L. gallinarum specific primer set, 37: L. amylolyticus specific primer set.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.