Abstract
Non-centrosomal microtubule-organizing centres (ncMTOCs) have a variety of roles presumed to serve the diverse functions of the range of cell types in which they are found. ncMTOCs are diverse in their composition, subcellular localization, and function. Here we report a perinuclear MTOC in Drosophila fat body cells that is anchored by Msp300/Nesprin at the cytoplasmic surface of the nucleus. Msp300 recruits the MT minus-end protein Patronin/CAMSAP, which functions redundantly with Ninein to further recruit the MT polymerase Msps/XMAP215 to assemble non-centrosomal MTs and does so independently of the widespread MT nucleation factor γ-tubulin. Functionally, the fat body ncMTOC and the radial MT arrays it organizes is essential for nuclear positioning and for secretion of basement membrane components via retrograde dynein-dependent endosomal trafficking that restricts plasma membrane growth. Together, this study identifies a perinuclear ncMTOC with unique architecture and MT regulation properties that serves vital functions.
Introduction
Microtubule (MT) organization supports critical cellular functions in cell division, cell polarity and intracellular trafficking. The best-known microtubule-organizing centre (MTOC) in animal cells is the centrosome. However, various cell types across species, after exit from the cell cycle and upon differentiation, lack a functional centrosome. In these cases, non-centrosomal MTOCs (ncMTOCs) function as alternative sites to accommodate the organization of MT networks specialized for differentiated cell types1–4. Whereas knowledge of the centrosome is extensive, the roles ncMTOCs play in cells, their molecular compositions, and how they are anchored to specific subcellular sites remains largely unknown.
Generation of an ncMTOC requires the nucleation, stabilization, and anchoring of MT minus-ends, generally achieved by MT minus-end-associated proteins. Compared to the relatively large number of MT plus-end proteins, few MT minus-end proteins have been identified. γ-tubulin, a conserved and essential MT minus-end protein that nucleates centrosomal MTs5–7, also nucleates and anchors non-centrosomal MTs in many differentiated cell types1–3, 8. XMAP215/Msps/Stu2/ Dis1/Alp14/ZYG-9/ch-TOG/MOR1 is an ancient family of MT polymerases that was recently identified as a MT nucleator at centrosomes and spindle pole bodies through its association with γ-tubulin9–11; a role for the MT polymerase at nucleating MTs at ncMTOCs has not been reported. Additionally, CAMSAP/Patronin family of MT minus-end proteins has emerged as a critical player at ncMTOCs via unclear mechanisms12–14. Ninein is another MT minus-end anchoring protein, but little is known of its mechanisms of action. To understand the diversity of ncMTOCs and how they serve the unique needs of diverse cell types, it is essential to determine how ncMTOCs are assembled and what the key effectors of MT assembly are.
Here we report the discovery of an ncMTOC that is assembled on the surface of nuclei in Drosophila larval fat body cells, a differentiated cell type that has critical secretory functions and serves the metabolic needs of the organism. This study identifies a perinuclear ncMTOC with unique MT assembly mechanisms that controls physiological roles of fat body cells by supporting nuclear positioning and vital secretory function.
Results
A perinuclear MTOC is assembled in fat body cells
Postmitotic polyploid cell types in Drosophila, such as salivary gland, midgut, and malpighian tubule cells, lack centrosomes15. We found that centrosomes are also lost in polyploid fat body cells (Fig. 1a), which are functionally similar to vertebrate liver and adipose cells having high metabolic and secretory activities16–18.
Figure 1. Fat body cells assemble a perinuclear ncMTOC.
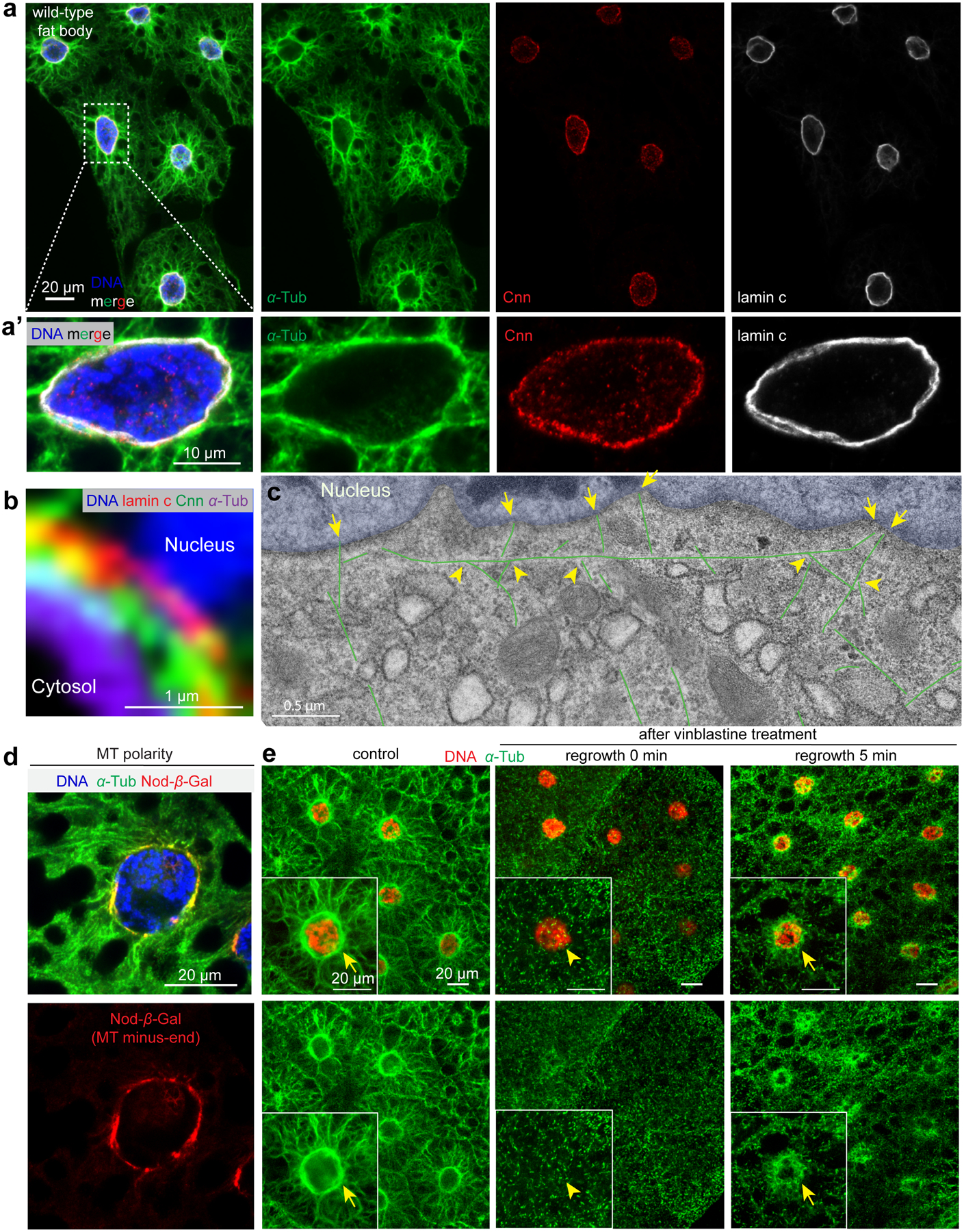
(a, a’) Immunofluorescent (IF) staining of fixed fat body tissues from third instar larvae labelled to reveal MTs (α-tubulin), the centrosomal protein Cnn, nuclear lamin C and DNA (DAPI). (a’) magnified view of a fat body cell. (b) A close-up view of nuclear surface showing MTs, Cnn, lamin and nuclear DNA. (c) EM image of fat body MTs (three stitched images). MTs are coloured in green and nuclei are tinted blue. Two general populations of radial MTs are evident; those anchored at the nuclear surface, presumably by their minus ends (arrows) and another group that appear to branch from the circumferential MTs (arrowheads). See also Extended Fig. 1a. (d) Localization of the Nod-β-gal fusion protein at the nuclear surface marks the MT minus ends. (e) MT regrowth in fat body cells 5 min after recovery from vinblastine treatment shows MT assembly enriched at the perinuclear ncMTOC. Insets show magnification of a single fat body cell; arrows point to sites of MT regrowth. Genotype details are in Supplementary Tables 2 and 3. Experiments were performed 10 times in (a, a’, b), once in (c), twice in (d, e). Scale bars: 20 μm (a, d, e), 10 μm (a’), 1.0 μm (b), 0.5 μm (c).
Fat body cells have a prominent perinuclear organization of MTs. MTs are highly enriched circumferentially at the nuclear surface and also radiate outward toward the plasma membrane (Fig. 1a,a’). Consistent with this being an MTOC, the centrosomal protein Centrosomin (Cnn) and MTs are positioned on the cytoplasmic face of the nuclear envelope (Fig. 1b). Electron microscopy imaging confirmed the presence of MTs oriented circumferentially and perpendicularly (radial) to the nuclear surface (Fig. 1c, Extended Data Fig. 1a). Additionally, localization of a Nod-β-gal fusion protein19 shows that MT minus ends are enriched at the MTOC on the nuclear surface (Fig. 1d). Importantly, MT regrowth experiments show that the nuclear surface is the primary site for MT assembly (Fig. 1e).
Cold treatment, which typically causes MT polymers to disassemble, did not overtly impact the organization of the MT array at the nuclear surface (Extended Data Fig. 1b), indicating that these MTs are highly stable. Consistent with this, fat body MTs are acetylated and polyglutamylated (Extended Data Fig. 1c, d), two tubulin post-translational modifications associated with stabilized MTs20. Together, these data demonstrate an ncMTOC at the nuclear surface of fat body cells that organizes stable MTs.
Fat body microtubules, but not actin, are essential for nuclear positioning
To examine a role for the MT array in fat body cells we disrupted MTs directly by either knocking down the MT subunits (α-tubulin or β-tubulin), or overexpressing MT-severing enzymes Spastin or Katanin-60. Knockdown of either tubulin subunit significantly disrupted MTs (Fig. 2a, b) and also the expression/stability of the other subunit; i.e. knockdown of α-tubulin resulted in reduced β-tubulin expression and vice versa (Fig. 2a’, b’). Overexpression of Spastin or Katanin-60 also disrupted fat body MTs (Fig. 2c–d’).
Figure 2. Microtubule disruption, but not actin disruption impairs nuclear positioning.
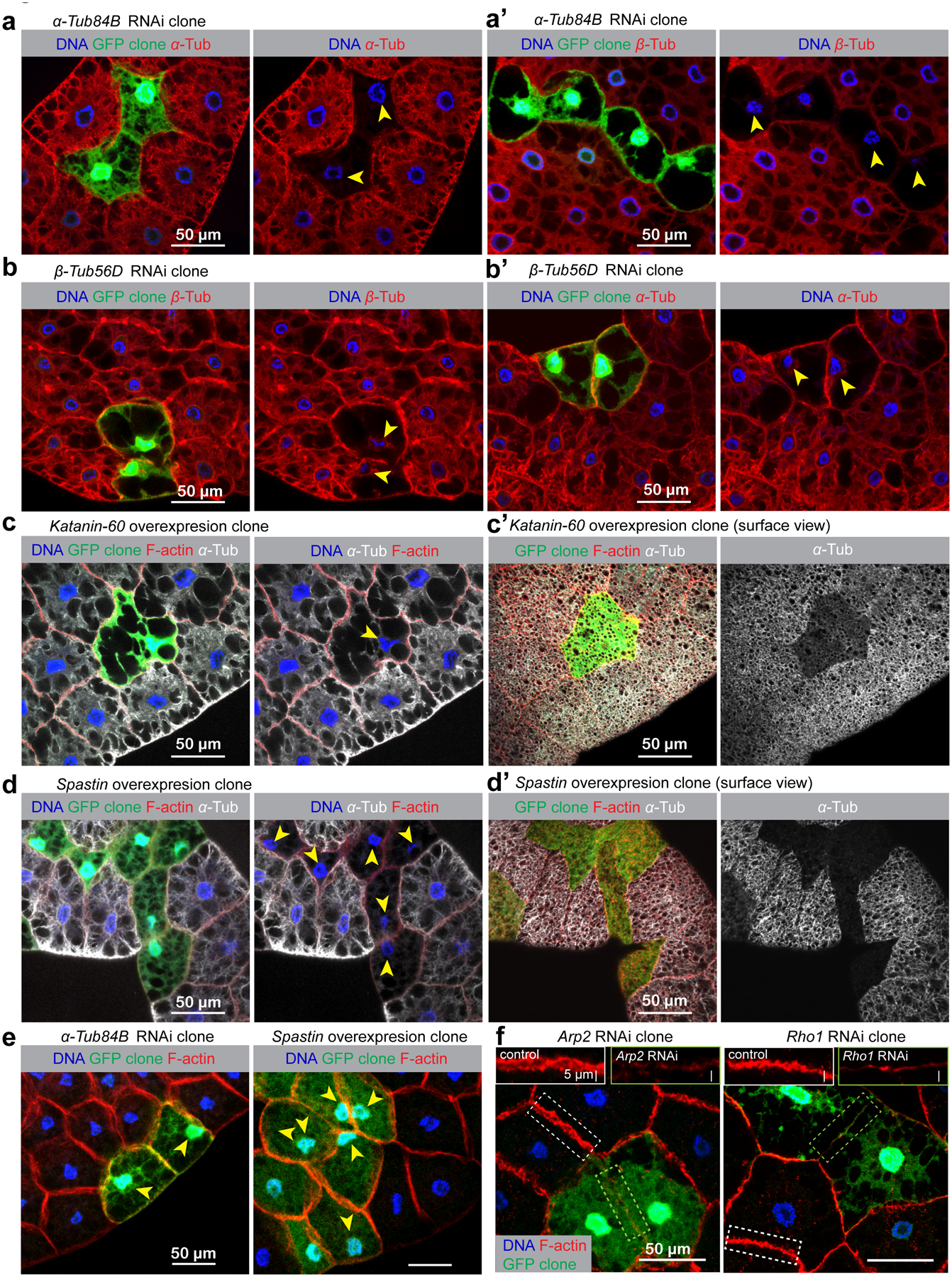
Fat body MTs were disrupted using two approaches, both involving the generation of genetically mosaic “flipout” clones. In the first approach, expression of the genes encoding the MT subunits, α-tubulin and β-tubulin, were knocked down by RNAi. In the second approach, the MT-severing enzymes Spastin or Katanin-60 were overexpressed to disrupt MTs. The flipout clones were marked with GFP expression adjacent to the control cells, enabling comparison of knockdown/overexpression cell phenotypes side-by-side with control cells. Arrowheads indicate nuclei that have lost centricity. (a, a’) knockdown of α-Tub84B significantly reduces expression of α-tubulin (a) and also β-tubulin (a’). Likewise, knockdown of β-Tub56D blocks expression/stability of both tubulin subunits (b, b’). Overexpression of MT severing enzymes Katanin-60 (c, c’) or Spastin (d, d’) in GFP-marked clones also results in MT disruption. (e) All of these MT disruptions result in a loss of nuclear centricity in fat body cells, indicated with arrowheads in (a-d’). (f) Disruption of actin assembly by knockdown of Arp2, an actin nucleator, or Rho1, a GTPase critical for actin assembly did not impair nuclear positioning. Insets show the impaired assembly of cortical F-actin in Arp2 or Rho1 RNAi cell compared to the control cell. Genotype details are in Supplementary Table 3. Fat body clonal analysis in (a-f) were performed twice with similar results. Quantification of nuclear positioning is shown in Fig. 3a. Scale bar, 50 μm.
In each fat body cell the nucleus resides in the geometric centre (centroid) (Figs. 2e, 3a). When fat body MTs were disrupted by either approach, nuclear positioning was significantly affected (Fig. 2e). In contrast to MT disruption, blocking actin nucleation by Arp2 or Rho1 knockdown reduced F-actin signal but did not disrupt nuclear positioning (Fig. 2f). These data show that MTs, but not actin, are necessary for the proper positioning of nuclei in fat body cells.
Figure 3. Msp300, Shot, Msps, Patronin and Nin but not γ-tubulin are required for the fat body ncMTOC.
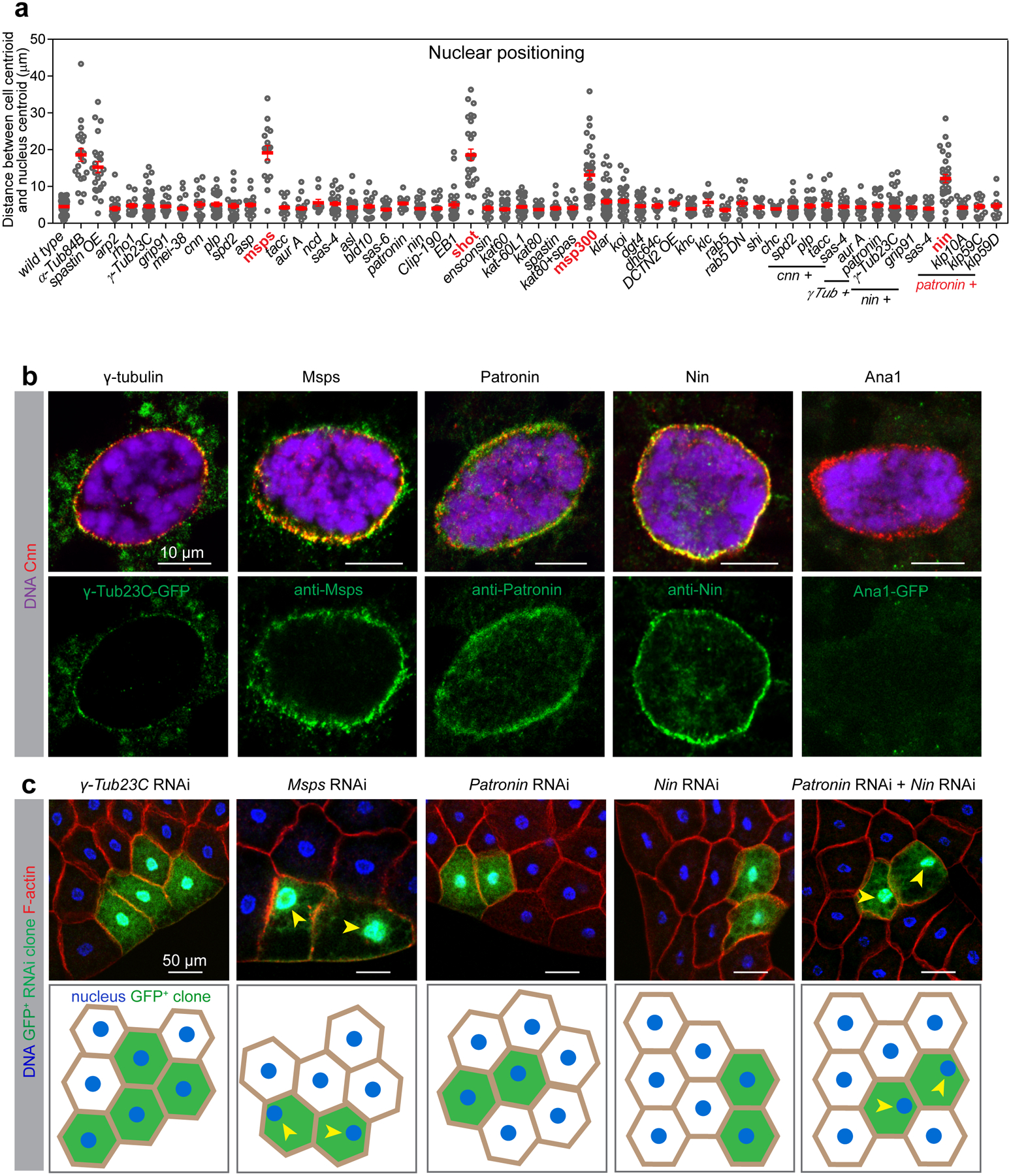
(a) The indicated mutants or RNAi-mediated knockdowns were surveyed for their requirement to position the nucleus at the centroid relative to cortical F-actin or myr-RFP (myristoylation domain of Src fused to mRFP), both of which mark the cell boundary. Nuclear positioning in the indicated genotypes is quantified in the graph. Data are shown as mean ± s.e.m. Statistics by two-tailed Student’s t-test. Number of independent clones or cells (n) analyzed and other statistical details are shown in Source Data Fig. 3_Statistical Source Data. (b) Images of fat body cells stained for Cnn in red and counterstained in green for the indicated proteins using either antibody staining or expression from a fluorescent protein-tagged transgene as indicated. Anti-GFP was applied for GFP-tagged transgenes to enhance signals. See also Extended Data Fig. 2. (c) The indicated genes were knocked down by RNAi in GFP-marked clones. The top panel: fat bodies containing control and GFP-marked knockdown clones were co-stained for DNA (Hoechst) and actin (CF568-Phalloidin) to assess nuclear centricity. Bottom panel: illustrations depicting the nuclear positioning in the indicated RNAi clones. Arrowheads indicate a loss of nuclear centricity. RNAi lines used here are mspsHMS01906, γTub23CGL01171, PatroninHMS01547, NinHMJ23837 (see RNAi validation in Extended Data Fig. 4b, c, 5a, 6a, g, h, see phenotype verification with other independent RNAi lines in Supplementary Table 4). Genotype details are in Supplementary Table 3. Staining experiments in b, c were performed three times with similar results. Scale bar, 10 μm (b), 50 μm (c).
Centrosomal proteins are localized at the fat body ncMTOC but are not required for MT assembly
We next determined the requirement of centrosomal proteins and MT regulatory proteins for fat body MTOC functions using nuclear positioning as a methodological readout (Supplementary Table 1, Movie 1). We also surveyed the localization of centrosomal proteins and MT regulators at the perinuclear MTOC (Supplementary Table 1). This survey showed that the major core PCM components were present at the fat body MTOC (with the exception of Spd-2), but core centriolar proteins were absent (Extended Data Fig. 2, Supplementary Table 1).
However, no single centrosomal mutant that we tested disrupted fat body nuclear centricity or ncMTOC assembly (Supplementary Table 1, Fig. 3a, Extended Data Fig. 3). Reasoning that there might be functional redundancy in MT assembly, as is the case with Cnn and Spd-2 at centrosomes21, we tested pairwise combinations of mutants or RNAi knockdowns, but still found no combination of centrosomal proteins that was required for the fat body ncMTOC to assemble MTs or affect nuclear positioning (Fig. 3a, Extended Data Fig. 3). These data indicate that although the fat body ncMTOC shares a majority of the core centrosome PCM components, it may not require them for MTOC function.
γ-tubulin is not required for MT assembly at the fat body ncMTOC
γ-tubulin, partnering with γ-tubulin complex proteins (GCPs) to form the γ-tubulin ring complex (γ-TuRC), is widely employed as a MT nucleator or anchor at centrosomes and other MTOCs1–3, 5–8, 22, 23. Compared to larval brains, fat bodies express about 120-fold lower levels of γ-tubulin (Extended Data Fig. 4a). Nevertheless, its localization is detected at the ncMTOC throughout larval stages (Fig 3b, Extended Data Fig. 4d), together with other γ-TuRC components including GCP2/3 (Extended Data Fig. 2).
To investigate the requirement for γ-tubulin at the fat body ncMTOC, we examined a null mutant and three independent RNAi lines, all of which showed depletion by western blotting and/or no detectable γ-tubulin at the nuclear surface in the larval fat body throughout developmental stages (Extended Data Fig. 4a–d’). The lack of detectable signal in 1st instar larvae indicates that the mutant effectively eliminated expression, and that there is no significant residual maternal supply in fat body cells prior to the third instar larval stage where most of our experiments were performed. Surprisingly, we found that fat body cells lacking γ-tubulin (γTub23C) or GCP3 (grip91) had normal nuclear positioning (Fig. 3a, c) and MT assembly (Fig. 4a–d, Extended Data Fig. 4e, e’). Since fat body MTs are highly stabilized, a requirement for γ-tubulin during early stages of fat body MTOC, when some residual maternal supply might persist, could be masked by MT stabilization in the γ-tubulin mutant. To address this possibility, we performed MT regrowth and found that MT regrowth was indistinguishable between wild-type and γ-tubulin-depleted fat body cells (Extended Data Fig. 4f), demonstrating that γ-tubulin has no significant role in MT assembly at the fat body ncMTOC.
Figure 4. Patronin plus Ninein cooperatively assemble perinuclear MTs and recruit Msps for MT elongation independently of γ-tubulin.
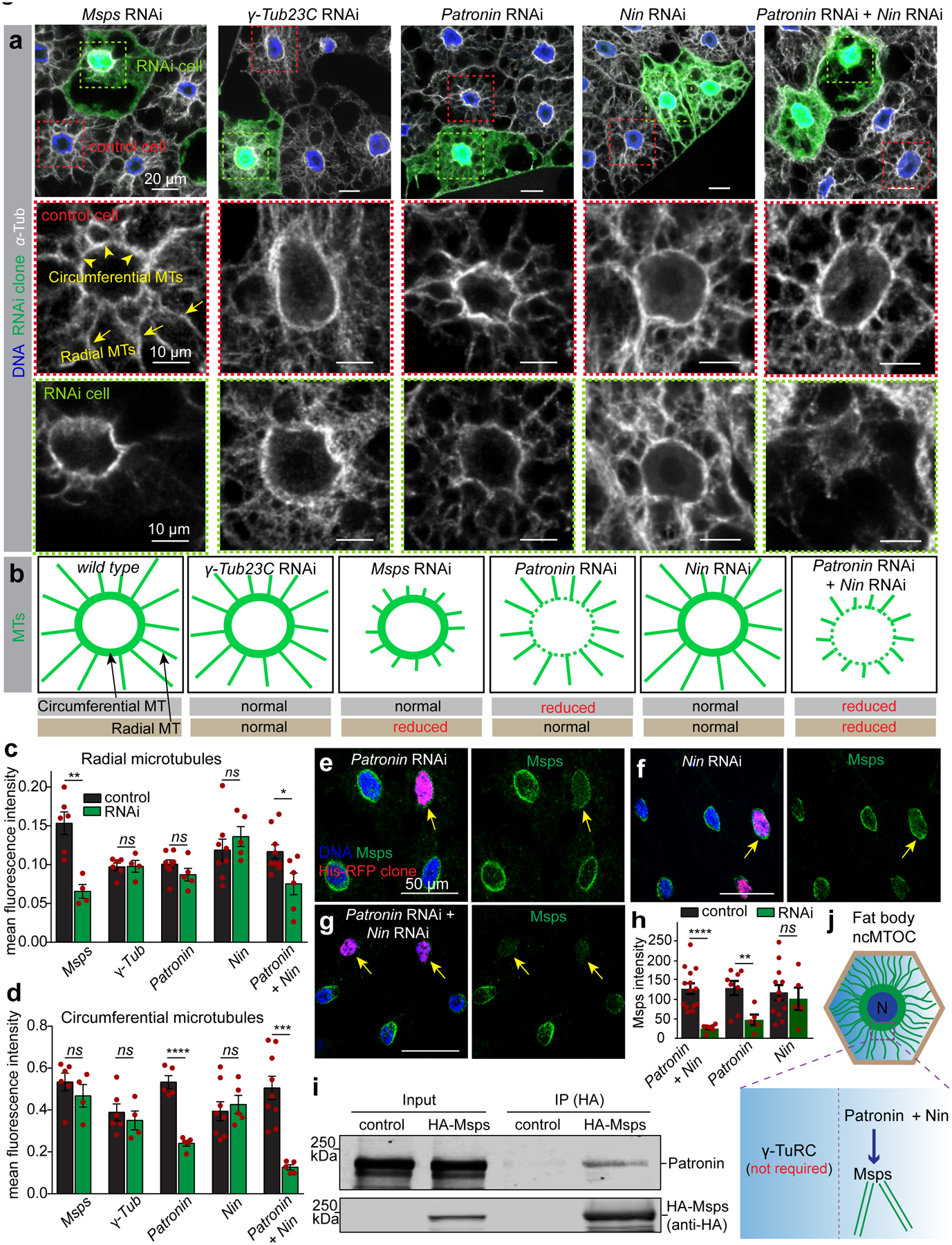
(a) Mosaic fat bodies were stained for anti-α-tubulin to assess MT assembly defects at the ncMTOC in control and GFP-marked RNAi cells. Middle and bottom panels show close-ups of the MT staining in the control (red box) and RNAi cells (green box). Circumferential MTs (arrowheads) and radial MTs (arrows) are indicated in a control cell. This experiment was performed three times with similar results. (b) Diagrams depicting circumferential and radial MT assembly phenotypes in fat body cells from wild type and the indicated RNAi. (c, d) Quantification of mean fluorescent intensity of radial microtubules (c) and circumferential microtubules (d) in the indicated RNAi clones compared to control cells. msps RNAi vs control, p=0.002 (**, radial, n=6 control, n=4 RNAi), p=0.3572 (ns, circumferential, n=6 control, n=4 RNAi); γ-Tub RNAi vs control, p=0.9316 (ns, radial, n=6 control, n=4 RNAi), p=0.5535 (ns, circumferential, n=6 control, n=4 RNAi); Patronin RNAi vs control, p=0.1845 (ns, radial, n=8 control, n=5 RNAi), p=0.00002 (****, circumferential, n=5 control, n=5 RNAi); Nin vs control, p=0.4156 (ns, radial, n=8 control, n=5 RNAi), p=0.6417 (ns, circumferential, n=8 control, n=5 RNAi); Patronin RNAi + Nin RNAi vs control, p=0.0213 (*, radial, n=9 control, n=6 RNAi), p=0.0004 (***, circumferential, n=8 control, n=5 RNAi). (e, f, g) Msps localization at the nuclear surface (arrows) in the indicated RNAi clones marked with His-RFP. (h) Quantification of Msps perinuclear intensity shown in e, f, g. Patronin + Nin RNAi vs control, p=0.00007 (****, n=14 control, n=7 RNAi), Patronin RNAi vs control, p=0.00867 (**, n=8 control, n=5 RNAi), Nin RNAi vs control, p=0.6572 (ns, n=12 control, n=5 RNAi). (i) HA-Msps (anti-HA) coimmunoprecipitation with endogenous Patronin (anti-Patronin) in Drosophila S2 cells. Input was 2% of total lysate. Full blot is shown in Source Data Fig. 4_Uncropped Western Blots. CoIP experiment was repeated twice with consistent Co-IP. (j) Working mechanism in fat body ncMTOC. N: nucleus. Data in c, d, h are the means ± s.e.m by two-tailed Student’s t-test, n=number of independent clones analyzed, statistical details in Source Data Fig. 4_Statistical Source Data. Scale bar, 20 μm (a, top), 10 μm (a, middle and bottom), 50 μm (e-g).
At centrosomes, γ-tubulin is the major regulator of MTs while AurA independently regulates the remainder24–26. At the fat body ncMTOC, however, co-depletion of γTub23C and aurA had no effects on nuclear positioning and MT assembly (Fig. 3a), consistent with a similar lack of requirement for these MT regulators at the apical ncMTOC in C. elegans embryonic intestine23. Therefore, the widespread MT nucleator γ-tubulin is not required for MT assembly at the fat body ncMTOC (Fig.4j).
Msps controls fat body radial MT assembly independent of conventional partners
Among the 39 candidates tested in our survey (Fig. 3a), minispindles (msps) knockdown (Extended Data Fig. 5a) singularly impaired nuclear centricity (Fig. 3a, c). Msps functions at the growing MT plus end as a processive MT polymerase27. Msps localizes to the fat body ncMTOC (Fig. 3b) and msps knockdown significantly impaired radial MT assembly while the dense circumferential MTs at the nuclear surface appeared unaffected (Fig. 4a–d), indicating that MT elongation is specifically impaired.
TACC is a partner of Msps, and its localization at centrosomes is regulated by Aurora A kinase28–30. In oocytes, Msps is transported to spindle poles by the Kinesin-14 motor Ncd28–30. However, these components are not required for ncMTOC assembly or nuclear centricity in the fat body (Extended Data Fig. 5b), indicating a distinct mechanism for Msps deployment at the fat body ncMTOC.
It was recently shown that Msps homologs function as MT nucleators by directly binding to and cooperating with γ-tubulin complexes9–11. However, the role for Msps at the fat body ncMTOC is clearly distinct from these other contexts because the γ-TuRC is not required for the fat body ncMTOC. Previous work supports a role for Msps in MT growth independent of γ-tubulin in interphase S2 cells31.
Together, these data show that Msps is required for MT assembly at the fat body ncMTOC, specifically for radial MT elongation, independently of previously described mechanisms.
Patronin controls circumferential MT assembly at the ncMTOC
Patronin/CAMSAP family proteins are MT minus-end proteins that stabilize MTs and have emerged as critical factors at ncMTOCs in a variety of organisms and cell types32. Patronin localized at the fat body ncMTOC (Fig.3b), however, its knockdown (Extended Data Fig. 6a) had no significant impact on nuclear centricity (Fig. 3a, c). Interestingly, Patronin knockdown significantly reduced the circumferential MTs but not the radial MTs (Fig. 4a–d), indicating that Patronin is involved in stabilization or assembly of the MTs most proximal to the MTOC.
Patronin/CAMSAP stabilizes MT minus ends and antagonizes the activity of Kinesin-13 family depolymerases12–14. However, this antagonism does not prevail at the fat body ncMTOC because Kinesin-13 knockdown (Klp10A, Klp59C or Klp59D), did not suppress the reduced circumferential MTs in Patronin knockdown cells (Extended Data Fig. 6b).
Patronin/CAMSAP associates with the MT severing enzyme Katanin33–35, an association proposed to remodel MTs and perhaps amplify MT minus ends36, 37. However, in fat body cells, loss of katanin p60 (Kat60), katanin p60-like 1(Kat-60L1), the regulatory subunit p80 (Kat80) or another severing enzyme Spastin did not overtly impact MT assembly or nuclear positioning, and neither did the co-depletion of Kat80 and Spastin (Fig.3a, Extended Data Fig. 6c, d). Disruption of another MT regulator, the augmin/HAUS complex38, 39, 40, 41, with a dgt4 mutant also had no effect on the centroid nucleus or MT assembly in fat body cells (Extended Data Fig. 6e). These two potential MT amplification mechanisms appear not required for MT assembly at the fat body ncMTOC.
Taken together, these results show that the ncMTOC regulator Patronin/CAMSAP is partially responsible for MT assembly or stability at the fat body MTOC, but does not function via the known mechanisms established in other contexts.
Patronin cooperates with Ninein to organize the fat body ncMTOC by recruiting Msps
We reasoned that Patronin might work in conjunction with other minus-end proteins to assemble the fat body ncMTOC. We investigated γ-tubulin and Ninein (Nin), two other MT minus-end proteins. CAMSAP and γ-tubulin are proposed to act sequentially in the generation of non-centrosomal MTs in neurons where γ-tubulin initiates MT nucleation and CAMSAP stabilizes MTs42. In C. elegans larval epidermis, Patronin works in parallel with γ-tubulin at ncMTOCs43. In Drosophila fat body cells, however, co-depletion of Patronin and γTub23C did not enhance the MT-organization phenotype of Patronin single knockdown (Extended Data Fig. 6f), indicating that γ-tubulin does not function redundantly or cooperatively with Patronin at the fat body ncMTOC.
Nin is a MT binding protein with anchoring function at the centriole subdistal appendages in mammals44, 45, but Nin also has ncMTOC roles in mammals, Drosophila and C. elegans43, 45–48. Nin localizes to Drosophila muscle and wing epithelial ncMTOCs46 and to the oocyte ncMTOC48. Nin also localized to the fat body ncMTOC (Fig. 3b). Mutations in ninein are viable in fly and mouse46, 48–50, and Nin mutants or knockdowns (Extended Data Fig. 6g) showed no overt effects on fat body nuclear positioning (Fig. 3a, c) or MT assembly (Fig. 4a–d). However, double knockdown of Patronin and Nin significantly disrupted nuclear positioning (Fig. 3a, c). Moreover, Patronin Nin double knockdown impaired both radial and circumferential MT assembly, while Patronin knockdown mostly affected circumferential MTs and Nin knockdown had no overt effects on its own (Fig. 4a–d). These data demonstrate that Patronin and Nin function redundantly in MT assembly at the fat body ncMTOC and indicates that each can effectively compensate for the other to control assembly of radial MTs.
Nin associates with γ-tubulin44, 46. In C. elegans, γ-tubulin recruits NOCA-1 (Nin homolog) to the ncMTOC in epidermal cells, where they function together and in parallel to Patronin to regulate non-centrosomal MT assembly43. However, at the fat body ncMTOC, γ-tubulin did not recruit Nin (Extended Data Fig. 6i), and γTub23C knockdown or null mutant did not enhance Patronin knockdown phenotypes (Extended Data Fig. 6f). Moreover, Nin knockdown together with γTub23C or gcp3 had no effect on nuclear positioning in fat body cells (Extended Data Fig. 6j). Altogether, these findings show that Nin and Patronin cooperate in MT assembly at the ncMTOC without functional involvement of γ-tubulin.
We then examined whether Patronin and Ninein are required to recruit Msps to the ncMTOC. Knockdown of Patronin reduced Msps recruitment to the nuclear surface to 37.0% of control, while Nin knockdown had no obvious impact (Fig. 4e, f, h). Patronin Nin double knockdown, however, significantly diminished Msps localization to 19.6% relative to control (Fig. 4g, h). We used CoIP assays and show that Patronin associates with Msps (Fig. 4i). These combined data point to a mechanism for MT assembly at the ncMTOC where Patronin, and perhaps also Nin, cooperate to recruit Msps to promote elongation of radial MTs independently of γ-tubulin (Fig. 4j).
The Nesprin Msp300 anchors the fat body ncMTOC at the nuclear surface and recruits Shot and Patronin
We investigated how the fat body ncMTOC is anchored to the nuclear surface and reasoned that a likely candidate is the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex, comprising KASH domain-containing Nesprins and SUN domain-containing proteins that span both nuclear membranes51. Drosophila has two Nesprins: Klarsicht (Klar) and Msp300, and one major SUN protein Klaroid (Koi), all of which are localized to the nuclear envelope in fat body cells (Fig. 5a). Null alleles of klar or koi had little or moderate effect on nuclear centricity (Fig. 3a, Supplementary Table 1). However, knockdown or mutation of Msp300 significantly disrupted nuclear centricity (Fig. 5b, c) and MT assembly (Fig. 5d, d’), demonstrating its requirement for a functional ncMTOC.
Figure 5. Msp300/Nesprin is required for MTOC assembly at the fat body nuclear surface by recruiting Shot and Patronin.
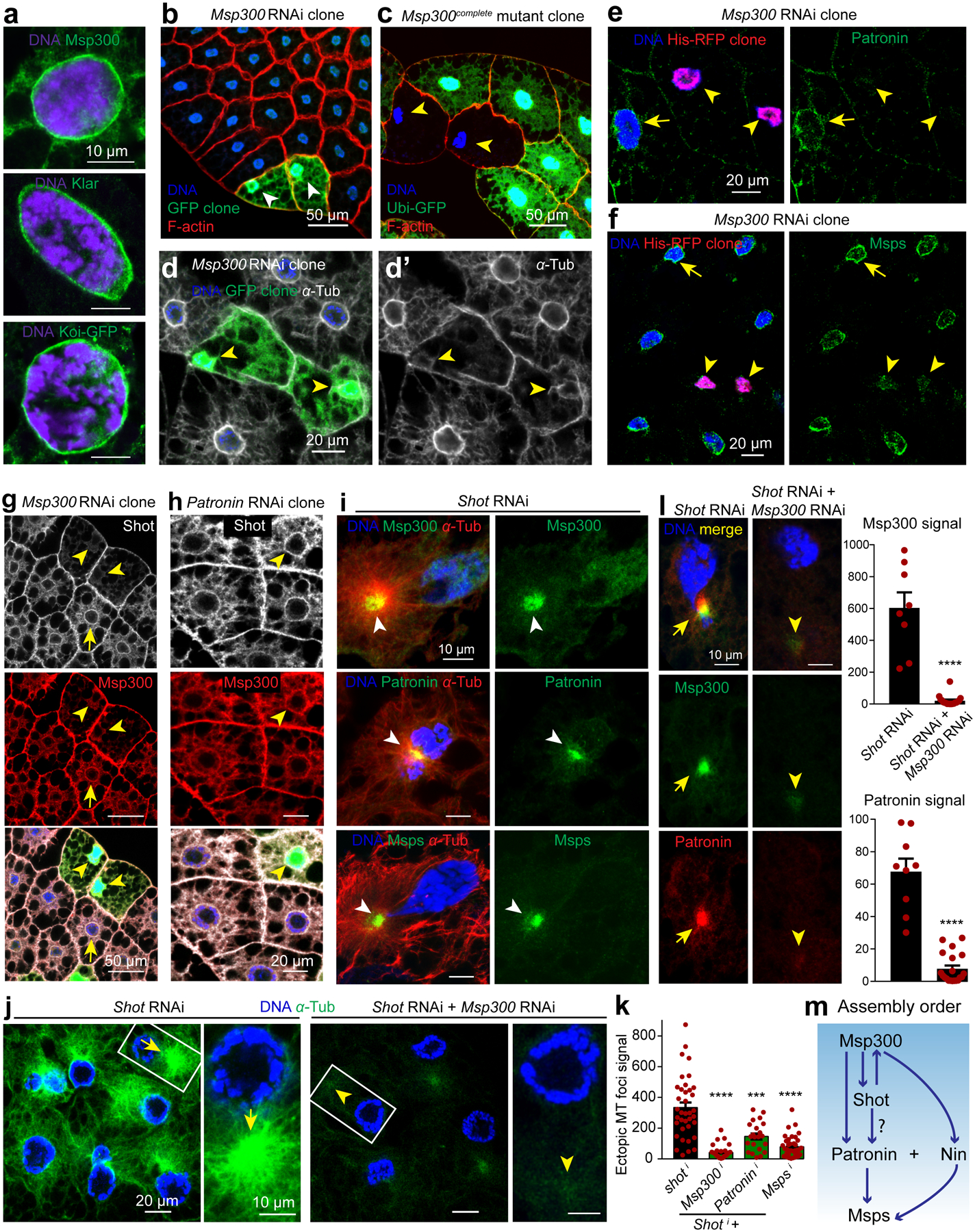
RNAi/mutant clones are marked with arrowheads and controls with arrows throughout figure. (a) LINC complex components localized to the fat body nuclear periphery. (b, c) Msp300 maintains nuclear centricity. Msp300 RNAi clones marked with GFP (b) and mutant clones by loss of GFP expression (c). (d) Circumferential and radial MTs are diminished with Msp300 knockdown. (e) Patronin localization to the nuclear surface is reduced in Msp300 RNAi clones (His-RFP). Quantification of Patronin perinuclear signal: 66.48±10.21 (n=10 control) vs 2.983±0.7863 (n=9 Msp300 RNAi clone), p=0.000019(****). (f) Msps localization to the nuclear surface is reduced in Msp300 RNAi clones marked with His-RFP. Quantification of Msps perinuclear signal: 132.8±11.13 (n=22 control) vs 30.5±10.18 (n=7 Msp300 RNAi clone), p=0.000036(****). (g) Shot localization to the perinuclear MTOC (arrow) is disrupted in Msp300 RNAi clones. (h) Shot and Msp300 localization (antibody staining) to the perinuclear MTOC do not require Patronin. (i) Msp300, Patronin and Msps (antibody staining) are delocalized to an ectopic MTOC in shot knockdown cells. (j) Double knockdown of Msp300 and shot with two combined fat body drivers (SPARC-Gal4+Cg-Gal4) significantly diminished ectopic MT foci compared to shot knockdown alone. (k) Quantification of ectopic MT foci in j and with two other combinations. From left to right, p=8.87e-12(****), p=0.0001(***), p=1.62e-10(****), n=37, 33, 22, 38 independent cells analyzed respectively. (l) Msp300 and Patronin delocalized to ectopic foci by shot RNAi; shot+Msp300 RNAi significantly reduced them. Msp300 signal: p=3.44e-09(****, n=8 shot RNAi, n=18 double RNAi), Patronin signal: p=1.69e-10(****, n=9 shot RNAi, n=20 double RNAi). (m) Epistasis for key components. Experiments were performed three times in a, twice in g, h, five times in i. Quantification for b is in Figure 3a. Images in c, d are representative of 30 and 14 independent clones, respectively. Data in e, f, k, l are presented as the means±s.e.m by two-tailed Student’s t-test; n=number of independent clones (e,f) or cells (k,l) analyzed. Statistical details in Source Data Fig. 5_Statistical Source Data. Scale bar, 10 μm (a, i, l, right panels in j), 50 μm (b, c, g), 20 μm (d, e, f, h, left panels in j).
Consistent with Msp300 controlling assembly of the ncMTOC, Patronin and Msps localization at the nuclear surface required Msp300 (Fig. 5e, f). Additionally, Short stop (Shot), the only spectraplakin in Drosophila, was localized to the nuclear surface in an Msp300-dependent manner (Fig. 5g). Shot associates with Patronin and is required to localize Patronin to ncMTOCs in oocyte and ovarian follicle cells35, 52. Knockdown of Patronin did not impact Msp300 or Shot localization (Fig. 5h), consistent with a dependence of Patronin on Msp300 and/or Shot for perinuclear localization. These combined analyses suggest that Msp300 is the primary organizer of the perinuclear ncMTOC by recruiting Patronin with possible involvement of Shot. Patronin, together with Nin, generates circumferential MTs, and they also recruit Msps to assemble radial MTs (Fig. 5m).
Shot depletion shifts the perinuclear ncMTOC to an ectopic MTOC
While Shot localization to the perinuclear ncMTOC depends on Msp300 (Fig. 5g), Msp300 localization also depends on Shot. In contrast to Msp300 knockdown, knockdown of shot delocalized the perinuclear MTOC components, including Msp300, Patronin and Msps to a centrosome-like MTOC focus in the cytoplasm (Fig. 5i, Extended Data Fig. 7a), whereas proteins not required for MTOC function, such as Cnn and γ-tubulin, were not (Extended Data Fig. 7b–d). To test the idea that Msp300 may be the primary organizer of the ectopic MTOC induced by shot RNAi, we co-depleted Msp300 and shot. We found that Msp300 was critical for the shot RNAi-induced ectopic MTOC (Fig. 5j, k), and also for the recruitment of Patronin (Fig.5i), suggesting that Msp300 may directly recruit Patronin. Co-depletion of shot with Patronin or msps also attenuated the ectopic MTOC (Fig. 5k), further indicating that Msp300, Patronin and Msps are key for MTOC function in the fat body. Together, these data indicate that Shot and Msp300 are co-dependent for their assembly into the perinuclear ncMTOC, and that Msp300 may recruit Patronin and Msps independently of Shot (Fig. 5l).
Shot disruption generates additional pleiotropic phenotypes besides perturbing the perinuclear MTOC. Knockdown of shot reorganized F-actin into large aggregates that accumulate near the displaced MTOC (Extended Data Fig. 7e); deformed the nucleus (Extended Data Fig. 7f); reorganized organelles including ER, Golgi and mitochondria (Extended Data Fig. 7g); and blocked secretion of basement membrane components (Extended Data Fig. 7h, i, i’), which accumulated near the ectopic MTOC (Extended Data Fig. 7h). These severe disruptions involve more than Shot’s role at the ncMTOC, because Msp300 knockdown blocked Shot localization to the ncMTOC but did not result in the severe and pleiotropic effects seen with shot knockdown.
The fat body ncMTOC is essential for basement membrane (BM) secretion
The fat body is the major source of synthesis and secretion of the collagen IV trimer53, whose subunits are encoded by Collagen IV α1 (Cg25c) and α2 (vkg) (Fig. 6a). A small fraction of secreted collagen IV is incorporated into fat body cell junctions54, but the majority is secreted, transported via the hemolymph, and deposited along with other BM components, like Perlecan (Trol), LanB1 and Nidogen (Ndg) at destination organs like imaginal discs and brain (Fig. 6a)53.
Figure 6. The fat body ncMTOC is essential for secretion of basement membrane proteins.
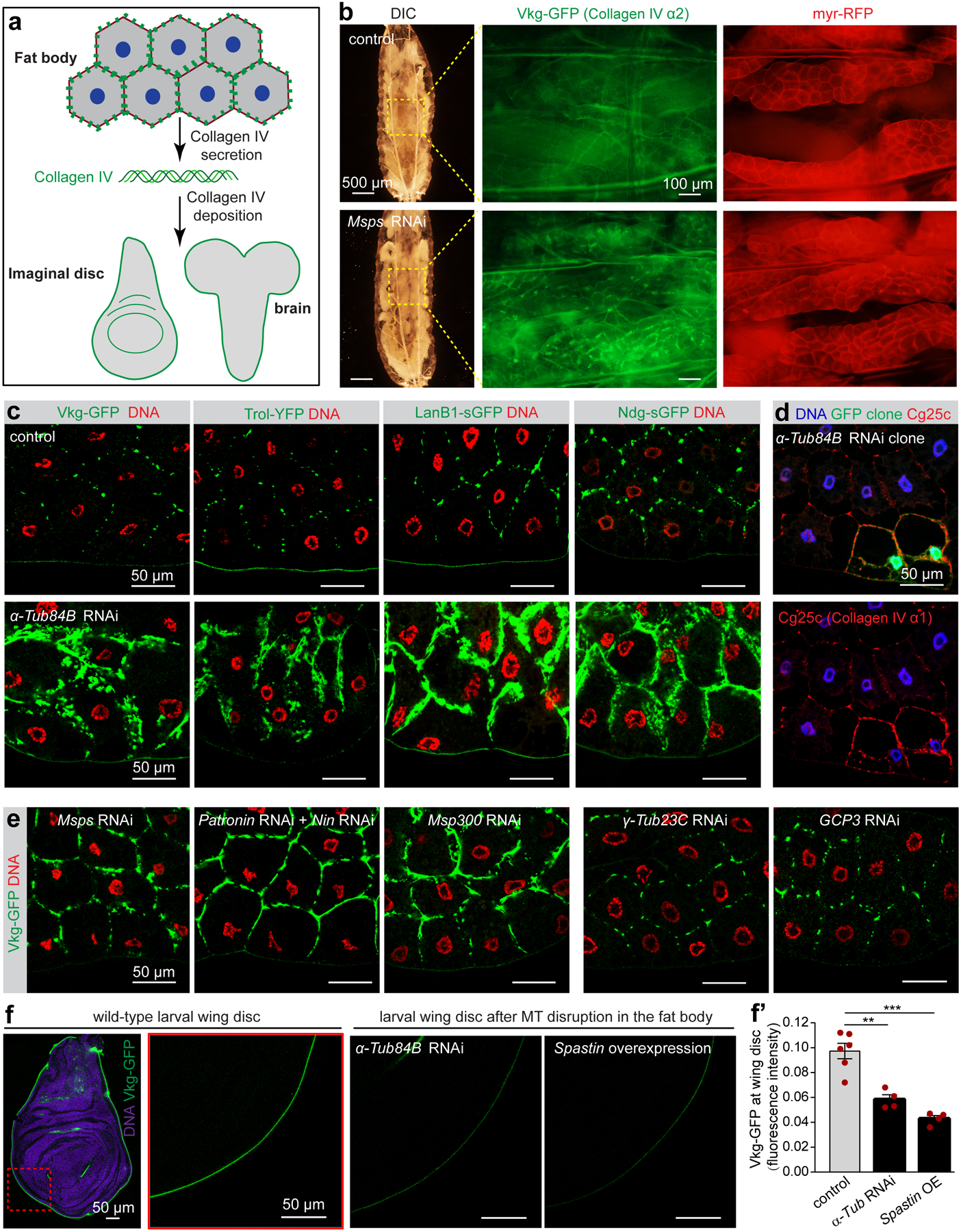
(a) Diagram of basement membrane component collagen IV production in fat bodies and its secretion and deposition to destination tissues as basement membrane. (b) DIC and fluorescent imaging of collagen IV α2 (Vkg-GFP) in live fat bodies labelled with the membrane marker myr-RFP from control larvae and larvae with fat body-specific knockdown of Msps. Images are representatives of 12 larvae. (c) Images showing fat body accumulation of indicated basement membrane components after MT disruption in fat body cells by α-Tub84B RNAi. Vkg-GFP (collagen IV α2) and Trol (Perlecan)-YFP are shown by GFP or YFP autofluorescence. LanB1-sGFP and Ndg-sGFP are stained with anti-GFP antibody. (d) Images showing fat body accumulation of Cg25c (collagen IV α1) in α-Tub84B RNAi clones marked with GFP. (e) Images showing fat body accumulation of Vkg-GFP after the indicated RNAi in fat body. (f) Images showing reduced wing disc deposition of Vkg-GFP after MT disruptions in fat body cells by α-Tub84B RNAi or spastin overexpression. (f’) Quantification of Vkg-GFP fluorescent intensity in f. Data are the means ± s.e.m (n=6 control, and n=4 independent experiments for α-Tub84B RNAi and spastin overexpression). Significance by two-tailed Student’s t-test. α-Tub84B RNAi vs control, p= 0.0016 (**); spastin overexpression vs control, P=0.0001 (***). Statistical details are shown in Source Data Fig. 6_Statistical Source Data. The experiments in c, d, e were repeated three times with similar results. Genotype details are in Supplementary Table 3. Scale bar, 500 μm (b, left), 100 μm (b, right), 50 μm (c, d, e, f).
When fat body MTs were disrupted or if the MTOC was impaired, collagen IV and other BM proteins accumulated to significantly higher levels at the plasma membrane of fat body cells and were visible in intact larvae (Fig. 6b) or dissected fat bodies (Fig. 6c–e, Extended Data Fig. 8a). Disruption of actin, on the other hand, did not cause accumulation of BM proteins (Extended Data Fig. 8b). As a consequence of fat body MT disruption, deposition of BM components at the distant wing imaginal disc (Fig. 6f, f’) were significantly reduced. Depletion of γ-TuRC components did not cause accumulation of BM components on the plasma membrane (Fig. 6e), in accord with no involvement of the γ-TuRC in fat body MT organization.
The fat body ncMTOC is essential to restrict plasma membrane growth and avert extracellular entrapment of basement membrane components
The accumulation of BM proteins at the plasma membrane following disruption of the fat body ncMTOC is extracellular, as demonstrated by antibody binding to Vkg-GFP on fixed but non-permeabilized fat body cells (Fig. 7a). Thus, BM proteins are secreted but trapped on the outside of the plasma membrane. This phenotype of extracellular BM entrapment occurs in fat body cells when endocytosis is disrupted55. When endocytosis is impeded, the plasma membrane overgrows (Fig. 7b,c) and becomes highly convoluted, thus traping secreted collagen within the folds of excess plasma membrane55. Disruption of the fat body MTOC caused similar overgrowth of the plasma membrane as seen with endocytosis block (Fig. 7b,c). Knockdown of Msp300 or msps, but not γTub23C, resulted in plasma membrane overgrowth (Fig. 7b) and entrapment of collagen IV (Fig. 6e).
Figure 7. The fat body ncMTOC is essential for retrograde dynein-dependent endosome trafficking to restrict plasma membrane growth.
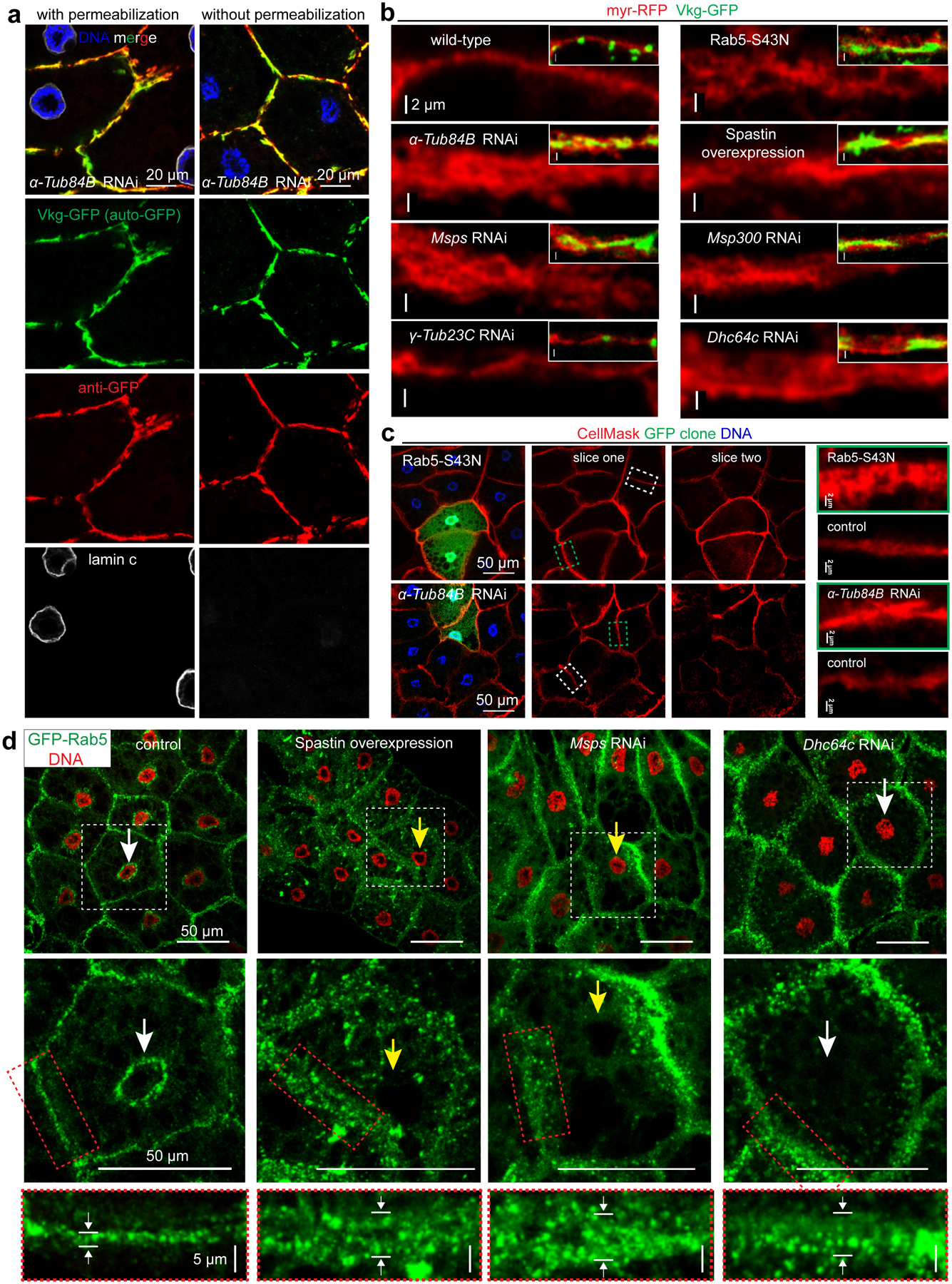
(a) Vkg-GFP accumulation at the plasma membrane is extracellular in α-Tub84B RNAi fat body cells by immunostaining with or without detergent permeabilization. Vkg-GFP autofluorescence is shown as green and GFP antibody staining is shown as red. LaminC staining is included as control for cell permeabilization. (b) Overgrowth of plasma membrane (marked by myr-RFP) in fat body cells after disruption of MT directly (α-Tub84B RNAi or spastin overexpression) or by impairing the ncMTOC (msps or Msp300 RNAi), inactivation of dynein motor (Dhc64c RNAi) or endocytic machinery (Rab5–S43N, dominant negative Rab5, a positive control). Insets show merged images with Vkg-GFP at the plasma membrane. (c) Overgrowth of plasma membrane using the membrane dye CellMask Orange in GFP-marked fat body cells that express dominant negative Rab5 or α-Tub84B RNAi. Slice one and two are separate image sections from the same confocal image z-stack. The dashed boxes for control (white) and GFP clone (green) plasma membrane are shown in the panels on the right. (d) Images showing endosome distribution (GFP-Rab5) in the indicated fat bodies. GFP-Rab5 is distributed at two main sites that are proximal to the plasma membrane and nuclear surface (control), but shifts to localizing predominantly near the plasma membrane after MT disruption (spastin overexpression or msps RNAi), or inactivation of dynein motor by Dhc64c RNAi. The middle panels are zoom-in views of cells from the white boxed region in the top panel to highlight the membrane and perinuclear Rab5-GFP localizations. The bottom panels are magnified views of membrane GFP-Rab5 from the middle panels (red rectangle). Note nucleus positioning is impaired (yellow arrows) in spastin overexpression or msps RNAi cells, but normal (white arrows) in Dhc64c RNAi and wild type. Experiments in a, b, c, d were repeated twice with similar results. Genotype details are in Supplementary Table 3. Scale bar, 20 μm (a), 2 μm (b, right panel in c), 50 μm (left panel in c, top and middle panels in d), 5 μm (bottom panels in d).
The fat body ncMTOC supports retrograde dynein-dependent endosome trafficking to maintain proper plasma membrane growth
Using GFP-Rab5 as an endosomal vesicle marker, we showed that in normal fat body cells GFP-Rab5 vesicles were enriched at the plasma membrane and also encircled the nucleus where the ncMTOC resides (Fig. 7d). Following disruption of the fat body ncMTOC, the perinuclear pool of GFP-Rab5 vesicles was diminished and instead became elevated at the plasma membrane (Fig. 7d). Thus, fat body MT arrays are required for retrograde trafficking of endocytic vesicles from the plasma membrane to perinuclear sites.
Retrograde trafficking of endosomes requires the minus-end directed MT motor dynein in other systems56. Loss of retrograde dynein motor function, by dynein RNAi or overexpression of dynamitin, shifted the perinuclear pool of GFP-Rab5 to the plasma membrane (Fig. 7d, Extended Data Fig. 8c). This blockage of endosomal trafficking by dynein inhibition phenocopied the effect of MT disruption on plasma membrane overgrowth (Fig. 7b, d) and entrapment of collagen at the plasma membrane (Extended Data Fig. 8d), but not nuclear mispositioning (Fig. 3a, 7d, Extended Data Fig. 8d), consistent with dynein being required for trafficking along MTs but not the organization of the MT arrays. When the Kinesin-1 motor was knocked down to block anterograde MT trafficking, there was no effect on BM secretion (Extended Data Fig. 8d) or nuclear positioning (Extended Data Fig. 8d). Therefore, a primary function for the fat body MTOC is to support the retrograde endosomal trafficking by dynein.
Taken together, we show two key functions for the fat body MTOC: nuclear positioning and retrograde endosomal trafficking. These two functions are separable because blockade of endosomal machinery or dynein motor activity caused membrane thickening and BM protein accumulation without affecting nuclear positioning.
Discussion
The rich diversity of non-centrosomal MT arrays and ncMTOC sites has long been recognized, yet the molecular architecture of ncMTOCs and whether their makeup and assembly mechanisms are similar to the centrosome have been underexplored. Here we show that an ncMTOC assembles on the nuclear surface in Drosophila fat body cells (Extended Data Fig. 9a). A Nesprin-Shot complex anchors the ncMTOC at the nuclear surface to recruit the MT minus-end regulators Patronin and Nin for assembly, anchoring and/or stabilization of MT seeds that are elongated by Msps (Extended Data Fig. 9b). γ-tubulin and known Msps cofactors, however, are notably not required. Neither is MT amplification by severing enzymes or the augmin complex. This work thus reveals unconventional paradigms for ncMTOC structure and for non-centrosomal MT assembly.
While sharing some properties in common with the muscle perinuclear MTOC57–64 like stable MTs and the requirement of Msp300/Nesprin and Shot, the fat body perinuclear ncMTOC is unique in its molecular architecture and the mechanisms by which MT assembly is regulated. The fat body perinuclear MTOC differs from the muscle perinuclear MTOC in that the muscle requires ensconsin/MAP765, CLIP-19066, EB164, Kinesin-165, and Dynein66 for myonuclear positioning or morphology, but these proteins are not required for fat body MTOC organization or nuclear positioning.
Functionally, the fat body ncMTOC maintains nuclear centricity, and also serves a critical function in providing a trafficking conduit for dynein motor-based retrograde transport of endocytic vesicles, whose trafficking maintains the balance of plasma membrane growth (Extended Data Fig. 9c). Maintaining proper balance of membrane growth is an important function of the fat body MTOC to support the secretion of large molecules beyond the fat body. Large protein assemblies such as collagen IV and other BM components become trapped in the membrane folds resulting from plasma membrane overgrowth when the MTOC is impaired.
Materials and Methods
Drosophila stocks
Information for the source, identifier, and original reference for all fly strains used, including mutants, RNAi lines, transgenic lines and drivers were listed in Supplementary Table 2. Detailed experimental genotypes and their associations with each figure are provided in Supplementary Table 3. Flies were maintained on standard food and incubated at 25°C, except that RNAi-mediated knockdowns were crossed at 29°C.
Plasmid constructs and generation of UASp-CFP-Msps Drosophila strain
The coding sequences for Msps were amplified by PCR from cDNA clone LP04448 (Acc# BT023496), cloned into pENTR-DTopo (ThermoFisher), and sequenced before shuttling into pPCW-attB (gift from Michael Buszczak) to generate UASp-CFP-Msps, and into pPHW-attB to generate UASp-HA-Msps. UASp-CFP-Msps plasmid was injected into embryos for targeted insertion on chromosome 3 at ZH-96E by Bestgene, Inc.
Generation of mutant and FLP-out expression clones in fat bodies
Gain-of-function (Flp-out) clones were generated by crossing virgin females: hsFlp; UAS-Dcr-2; Act>CD2>Gal4, UAS-GFP /TM6B or hsFlp; Act>CD2>Gal4, UAS-His-RFP/TM6B to UAS-driven RNAi lines or other transgenes. FLP-mediated excision of the CD2 insert induces gene knockdown or overexpression driven by Act-Gal4 with UAS-Dcr-2 at 29°C in GFP-marked cells. These clones were generated in early embryos by shift to 29˚C, and fat body precursor cells as evidenced by clones that typically include multiple cells due to mitotic expansion of the clone prior to differentiation.
hsFLP/Flippase Recognition Target (FRT)-mediated Msp300 loss-of-function clones in larval fat body were induced in 0–6 h embryos by a 1-h heat shock at 37°C using an FRT-linked Msp300complete mutant allele. Mutant clones were marked by loss of GFP.
Antibodies
All antibodies used are described in Supplementary Table 5.
Immunostaining
Most stainings were done in fat bodies from late third instar wandering larvae, except that early stages of larvae were included in Extended Data Fig. 4d, d’, e, e’.
MTOC components staining in larval fat bodies was performed after methanol fixation as previously described with minor modification46. Fat bodies were dissected from late third instar wandering larvae in 1× phosphate-buffered saline (PBS). The dissected fat bodies from one larvae were transferred to 8 μl of PEM solution (100 mM PIPES, pH 6.9, 1 mM EGTA, and 2 mM MgSO4) on a slide. A 22 × 22 mm siliconized coverslip containing 1 μl of 18.5% formaldehyde in PEM was placed on the slide, allowing the weight of the coverslip to flatten the fat bodies for 10 s, followed by snap freezing of the slide in liquid nitrogen. The coverslip was pried off using a razor blade and the slide with attached fat bodies was immediately fixed in −20°C methanol for 10 min, followed by PBS rinse. A hydrophobic ring was drawn around the fixed fat bodies using a Super PAP Pen (Immunotech, Monrovia, CA). The fat bodies were incubated with 50 μl of antibody solution (1×PBS, 5% BSA and 0.1% saponin) in a humid chamber overnight at 4°C for primary antibody and 2 hr at room temperature for secondary antibody.
Fat body co-staining of microtubules and filamentous actin (F-actin) was performed after fixation with paraformaldehyde (PFA). Briefly, dissected fat bodies were fixed for 10 min in 50 μl of 4% PFA within a hydrophobic ring drawn on a slide. The fixed fat bodies were washed twice with PBS, followed by blocking with PBSBT (1×PBS, 0.1% Triton X-100, and 1% BSA) for 1 h and incubated with FITC-conjugated DM1A (1:200, Sigma) and CF568-conjugated Phalloidin (1:200, Cat#00044, Biotium) in PBSBT for 2 h in the hydrophobic ring.
For plasma membrane staining with CellMask Orange (C10045, Invitrogen), dissected fat bodies were directly incubated with CellMask Orange (1:1000) for 5 min without inclusion of detergents. After washing three times with PBS, the stained fat bodies were then fixed with 4% PFA for 10 min and washed three times again after fixation. Mounted samples were immediately imaged.
Non-permeabilization anti-GFP staining of larval fat bodies expressing Vkg-GFP was performed similarly in 4% PFA-fixed fat bodies as above described, except that no detergent was used during the whole wash, block and antibody incubation.
Transmission electron microscopy (TEM)
TEM analysis of the MTs at the fat body ncMTOC was performed essentially as described67, except that fat bodies from 30 third instar larve were fixed for 72 hrs in 1 ml Karnovsky’s fixative (EM Sciences Cat#15720). Embedding, staining, sectioning and preparation of grids were performed by the Core Facility at UT Southwestern Medical Center. After three rinses with 0.1 M sodium cacodylate buffer, samples were embedded in 3% agarose and sliced into small blocks (1mm3), rinsed with the same buffer three times and post-fixed with 1% osmium tetroxide and 0.8 % potassium ferricyanide in 0.1 M sodium cacodylate buffer for 1.5 h at room temperature. Samples were rinsed with water and en bloc stained with 4% uranyl acetate in 50% ethanol for 2 h. They were then dehydrated with increasing concentration of ethanol, transitioned into propylene oxide, infiltrated with Embed-812 resin and polymerized in a 60°C oven overnight. Blocks were sectioned with a diamond knife (Diatome) on a Leica Ultracut 7 ultramicrotome (Leica Microsystems) and collected onto copper grids, post stained with 2% aqueous Uranyl acetate and lead citrate. Images were acquired on a Phillips CM120 Biotwin transmission electron microscope at the Florida State University Biological Science Imaging Resource (BSIR).
Coimmuniprecipitation (CoIP)
Co-IP of HA-Msps and Patronin was performed with Drosophila S2 cells. S2 cells were cultured in Shields and Sangs M3 medium (Sigma Cat# S3652) with 10% fetal bovine serum (Gibco cat#10437–010) at room temperature. pMT-GAL4 (DGRC Cat#1042) was used to induce expression of pUASp-HA-Msps. Plasmids (pMT-GAL4 plus or minus pUASp-HA-Msps) were transfected using Lipofectamine 3000 (Thermo Fisher) in 6-well dishes. After 48 hrs of culture, expression was induced by addition of 1mM CuSO4 for 24 hr. Cells were harvested and lysed in 500 μL lysis buffer: 50mM Tris pH 7.5, 150mM NaCL, 1mM EDTA, 05% NP-40 plus protease inhibitors (0.5mM PMSF (VWR Cat#EM-7110), 1mM Phenanthroline (Sigma Cat#P3975), 1mM Benzamidine (Sigma Cat#B6506), 10mM N-Ethylmaleimide (Sigma Cat#E3876), 1X PI cocktail (Sigma Cat#P8340)) for 10 min on ice. Immunoprecipitation was performed using anti-HA-Agarose (Sigma Cat#2095) following the manufacturer’s recommended protocol. For western blots, input lanes represent 2.1% of the total lysates and IP lanes represent 62.5% of the total immunoprecipitate. Mouse anti-HA (HA-7, Sigma Cat#H9658, 1:40,000) and rabbit anti-Patronin (1:500)13 were used to detect HA-Msps and endogenous Patronin, respectively. Western blotting was performed as described below.
Western blot
Fat bodies or brains dissected from five wandering larvae were lysed respectively in 50 μL or 10 μL of 2×SDS-PAGE buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 0.02% bromophenol blue, 20% glycerol, 5% β-mercaptoethanol). 10 whole larvae at wandering stage were lysed with motorized pestle in 200 μL of 2×SDS-PAGE loading buffer. After boiling for 5 min, larval lysates were centrifuged at 13,200g for 5 min to clear pellets. 10 μL of larval lysates were loaded for SDS-PAGE gel electrophoresis, followed by semi-dry transfer to nitrocellulose membrane. The membranes were blocked with 5% nonfat milk in 1×TBS for 1 h at room temperature and then probed with primary antibodies diluted in 1×TBS containing 0.1% Tween (1×TBST) overnight at 4°C. After washing with 1×TBST three times, the membranes were incubated with secondary antibodies conjugated with IRDye-800CW or IRDye-680LT (1:20,000) for 1 h at room temperature. Blots were scanned on an Odyssey Infrared Imaging system (LI-COR Bioscience, Lincoln, NE). Signals were quantified using Li-Cor Image Studio software. Images were processed in Adobe Photoshop CS4 and presented in monochrome.
Microtubule regrowth assay
Wild-type fat bodies were dissected and placed in 15 μl of 10 mg/ml fibrinogen (EMD#341573) in D-PBS (Dulbecco’s PBS, Invitrogen) on a clean slide within a hydrophobic ring drawn with a super PAP pen. 2 μl of thrombin solution (Sigma T9549–50UN; 100 U/ml in D-PBS) was pipetted to the fibrinogen solution to induce a fibrin clot. The fat bodies were treated with 30 μM vinblastine in Shields and Sangs M3 medium (Sigma # S3652) for 1–2 h to induce MT disassembly at room temperature. The vinblastine was washed out in ice-cold D-PBS (0–4˚C, or at a constant 4˚C) for 1 h, followed by a time-course recovery at 25˚C. The fat bodies were fixed at the respective time points in −20°C methanol for 10 min and stained with anti-α-tubulin and DAPI to label MTs and nuclei, respectively. Microtubule regrowth in the γ-Tub23C RNAi clone experiment (Extended Data Fig. 4f) was fixed with 4% PFA to enable staining with CF568-phalloidin.
Image acquisition
Fixed fat body samples were imaged using a Nikon A1 laser scanning confocal microscope (Nikon, Japan) using a 60×/1.49 NA oil immersion objective. All confocal images were captured with a spacing of 0.25 μm or 0.5 μm between z-sections using Nikon NIS-Elements AR software (version 4.6) and are presented as maximum intensity projections of z stacks. Images of larval wing discs and brains were acquired by stitching together images acquired with 25% overlap using in Elements using a 60×/1.49NA oil immersion objective. Gamma-correction was applied to images.
Live imaging of intact fat body in whole larvae expressing vkg-GFP and myr-RFP was performed by a Macro Zoom microscope (MVX10, Olympus, Japan) equipped with DP72 camera. DIC view of whole larvae was imaged at 32-fold magnification. Fluorescent imaging of Vkg-GFP and myr-RFP expressed in larval fat bodies at single-cell resolution was performed at 130-fold magnification. Representative still frames were shown in Figure 6b.
Live imaging of nuclear positioning/movement in intact fat body expressing myr-RFP and Histone-GFP was performed on whole live third instar wandering stage larvae restrained between a 22×22 mm and a 24×40 mm 1.5 coverslip using clear 1/4-inch wide office tape. Images were captured every 3.0 s using 488 nm and 568 nm laser excitation with a 20×/0.75 NA Plan Apo objective on a Nikon A1 confocal microscope with the pinhole open to 8.0. The time-lapse image sequence was converted to.avi video at 20 fps.
Quantification analysis
Nuclear positioning, or centricity analysis, in fat bodies was measured as the distance between the cell’s geometric centre and nucleus’ geometric centre using Nikon NIS-Elements AR software (version 4.6). Cell and nuclear geometric centres were measured using the “centroid” tool after auto- or manual thresholding of cell boundary (cortical F-actin by Phalloidin staining) and nucleus (Hoechst staining). The distance between cell and nuclear geometric centres was measured using the length tool in NIS-Elements.
Fluorescent intensity of circumferential and radial MTs, Msps or Patronin in GFP or His-RFP-marked clones was analyzed in Image J. Integrated density and area for circumferential or radial MTs was measured in 8-bit inverted monochrome images. Mean fluorescence intensity was measured as integrated density per unit area using the measure tool in ImageJ on 8-bit monochrome inverted and thresholded images. For each RNAi knockdown, analysis was performed in control and GFP-marked clones for side-by-side comparison.
Vkg-GFP fluorescent intensity in larval wing discs was quantified in Image J. Mean fluorescent intensity of GFP auto-fluorescence was measured after GFP signal thresholding in 8-bit inverted images.
Statistics and reproducibility
Sample size was chosen according to standard practices in the field to ensure adequate statistical power. No data were excluded from our studies. For experiments where image parameters were quantified, a minimum of 4 independent images were quantified and in most experiments it was 8 or more. Almost all of the images were examined as clones: experimentally manipulated cells generated within an otherwise wild-type tissue. This approach adds notable and significant rigor to the experimental design and the measurements taken, as the control cells are measured side-by-side with the experimental cells. The numbers (n) measured were significant as indicated by the low p values (<0.01) generated from the data. Data are presented as the mean ± s.e.m. The number of replicates and sample sizes are indicated in the figure legends. The statistical significance was determined by unpaired two-tailed Student’s t-test using Prism 7 (Graphpad, San Diego, CA). Differences were considered statistically significant when p<0.05. * denotes p<0.05, ** for p<0.01, *** for p<0.001 and **** for p<0.0001 as shown in figures and figure legends.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Data not included are available from the corresponding authors upon reasonable request.
Code Availability
There are no custom coding or algorithms reported in the manuscript or used to generate any of the results.
Extended Data
Extended Data Fig. 1. Fat body MTs are stabilized.
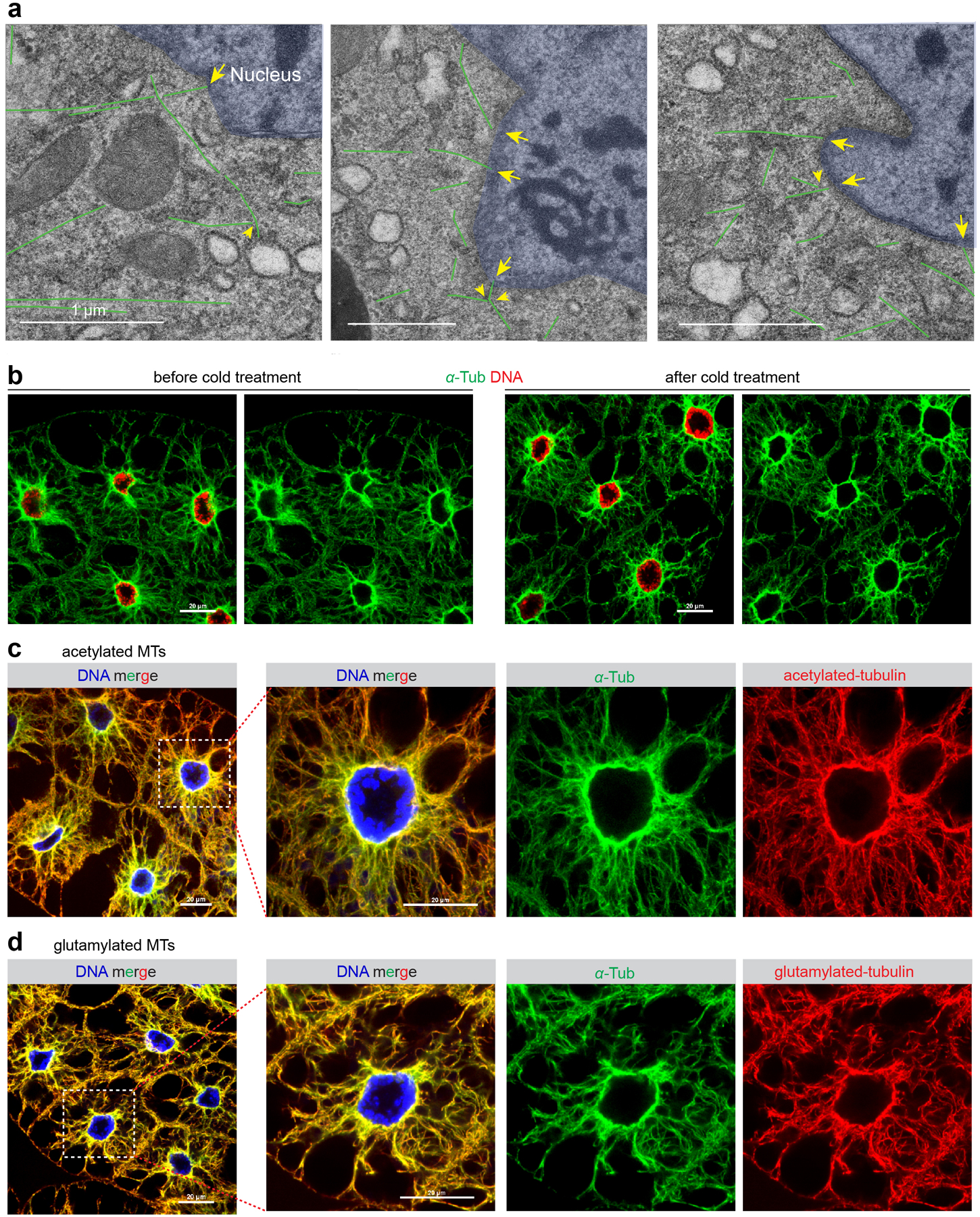
(a) Additional EM images, related to Figure 1c. (b) Images of IF stained fat body cells with or without cold treatment for 1 hr, which did not cause significant reduction of the MT array. (c, d) IF staining shows that the fat body contains MTs that are acetylated (c) and polyglutamylated (d), two post-translational modifications attributed to stabilized MTs. Genotype details are in Supplementary Table 3. Experiments were performed once in (a), twice in (b, c, d) with similar results. Scale bar, 1 μm (a), 20 μm (b, c, d).
Extended Data Fig. 2. Survey of localization of centrosomal proteins and MT regulators at the fat body ncMTOC.
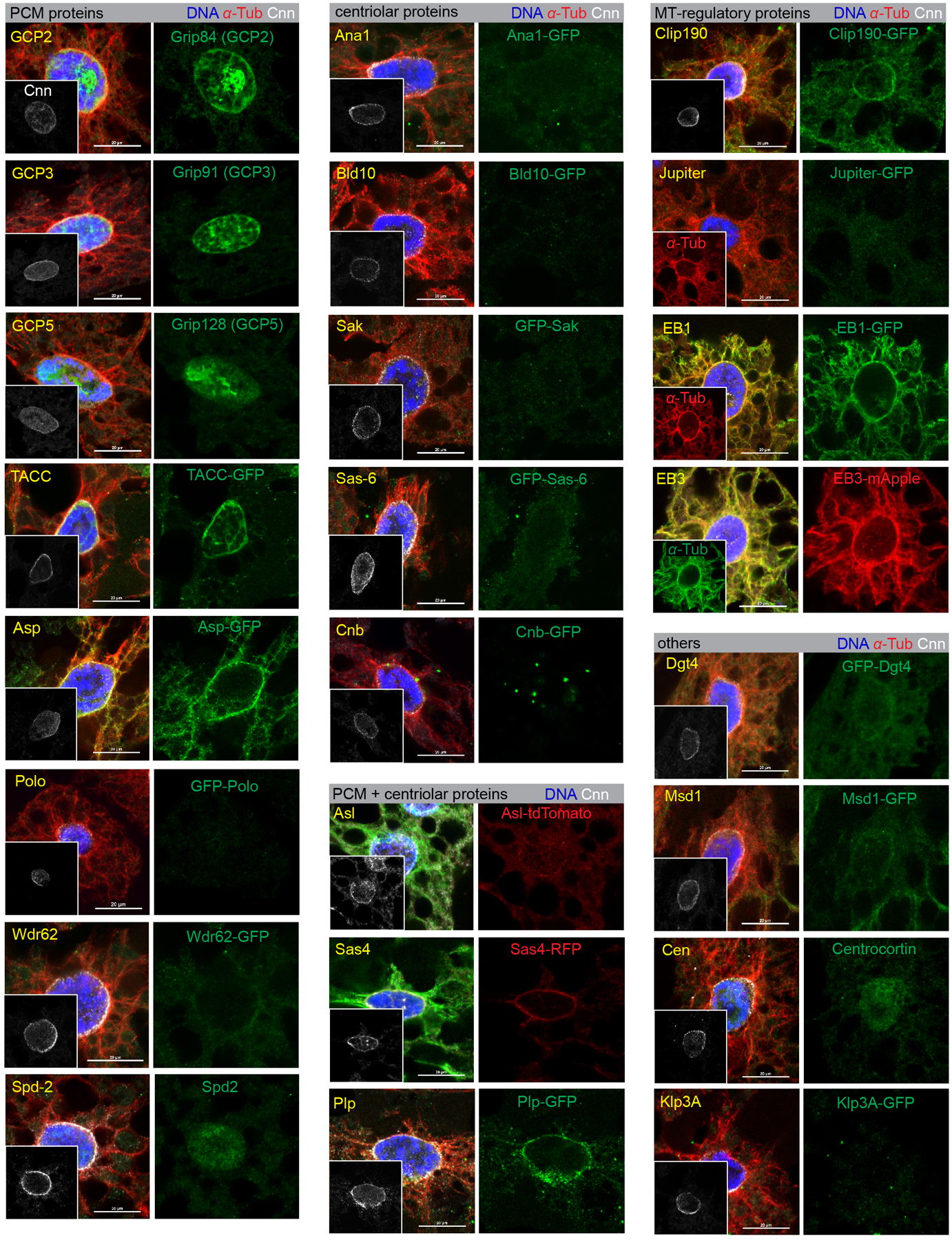
Images of fat body cells stained for Cnn and counterstained in green for the indicated proteins using either antibody staining or expression from a fluorescent protein-tagged transgene as indicated. The insets are Cnn staining as positive control. See Supplementary Table 1 for the complete list of proteins assayed and the summary of their localization at the MTOC as well as the information about promoters used to drive transgenes. Anti-GFP or anti-RFP staining was applied for GFP-tagged or RFP-tagged transgenes. The centriolar proteins are core centriolar proteins. The “PCM + centriolar” proteins are proteins that reside in both compartments or straddle both compartments and are known to function in PCM organization. The “MT regulatory” are effector proteins with established roles in regulating MT assembly or anchoring. Since some of the transgenic proteins were expressed ectopically, it is possible that some of the positive localizations determined in this manner do not reflect endogenous protein localization. Collectively, the fat body MTOC contains some proteins in common with the centrosome PCM, but also some distinct components including Patronin. Note that Ana1-GFP image in Fig. 3b is reproduced from this figure. Genotype details are in Supplementary Table 3. Staining experiments were performed twice with similar results. Scale bar, 20 μm.
Extended Data Fig. 3. Single or double mutant combinations of centrosomal protein genes do not overtly impair MT assembly at the fat body ncMTOC.

Fat body cells were stained for DNA (Hoechst) and filamentous actin (CF568-Phalloidin) to assay for nuclear centricity (top panels). MTs (anti-α-tubulin) and DNA (DAPI) were stained to assay MT organization with respect to nuclei (bottom panels). None of the single or double mutants tested shows defects in nuclear positioning or MT assembly at the MTOC. Nuclear positioning (top panel) are quantified in Fig. 3a. Genotype details are in Supplementary Table 3. Experiments were performed twice with similar results. Scale bar, 50 μm (top panels), 20 μm (bottom panels).
Extended Data Fig. 4. γ-tubulin is not required for MT assembly at the fat body ncMTOC.
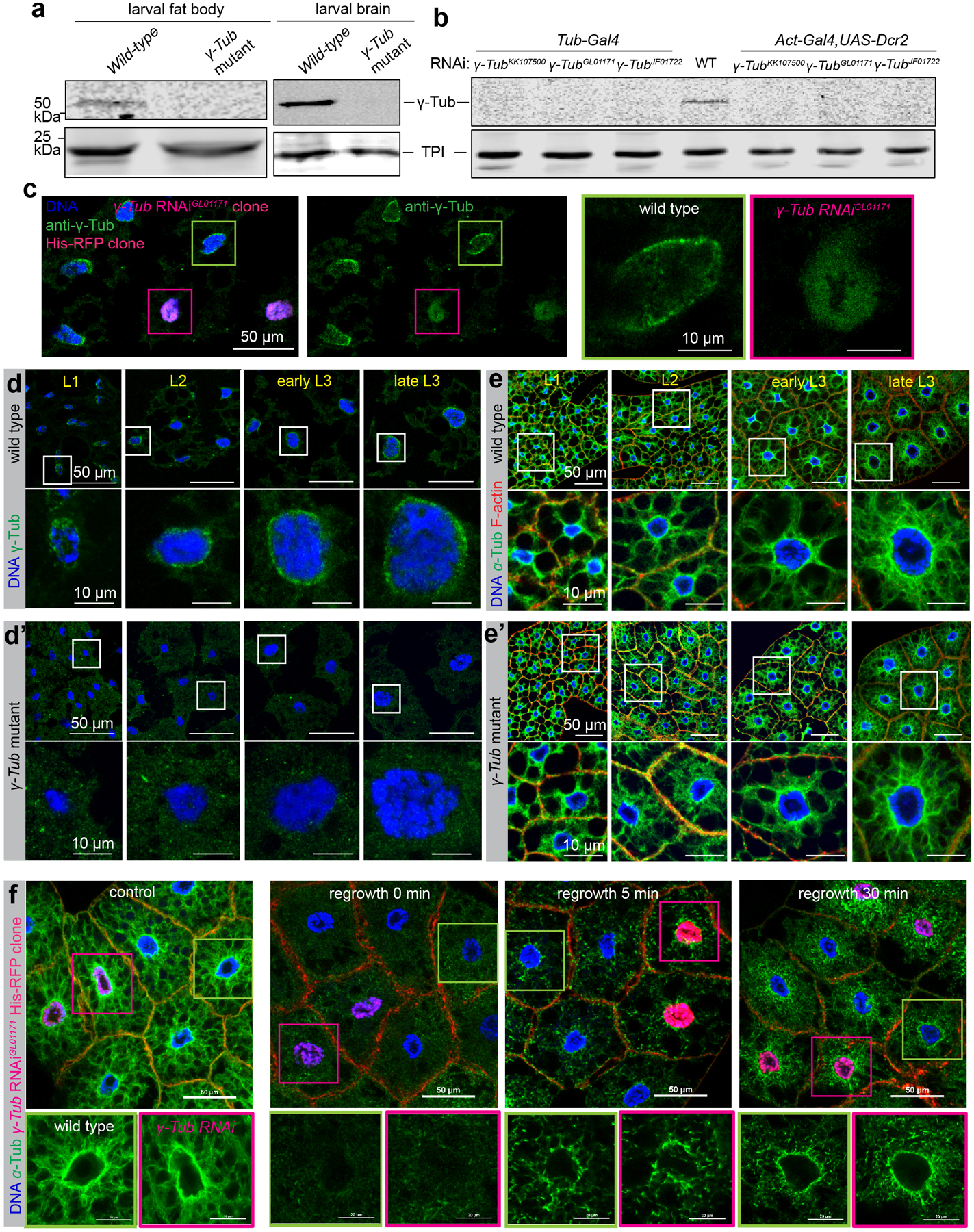
(a) Western blot detection of γ-tubulin in wild-type and γTub23C mutant larval fat body and brain lysates. Wild type is w1118, γTub mutant is γ-Tub23CA15−2/Df (2L)JS17. Triose-phosphate isomerase (TPI) is the loading control. γ-tubulin levels, as measured from the shown blot, were about 120 fold lower in wild type larval fat bodies compared to brains when normalized against Tpi. (b) Western blot detection of γ-tubulin in whole larval lysates from wild-type and three independent γTub23C RNAi lines driven by two ubiquitous strong promoters: Tub-Gal4 and with Act5c-Gal4 plus UAS-Dicer-2. (c) Images showing γ-tubulin staining in γ-Tub23CGL01171 RNAi fat body clone marked by His-RFP. (d, d’) Images showing γ-tubulin staining in wild-type (d) and γ-Tub23C mutant (d’) fat body at different developmental stages as indicated. (e, e’) Images showing nuclear positioning and MT assembly in wild-type (e) and γ-Tub23C mutant (e’) fat body cells. (f) Images showing MT regrowth in wild-type and γ-Tub23C depleted fat body cells from γ-Tub23CGL01171 RNAi clones marked by His-RFP. Control: no vinblastine treatment; the other three groups were treated with vinblastine and underwent MT regrowth for 0, 5, 30 min as indicated. The western blots in a and b, staining in c, d, d’, e, e’, f were repeated twice with similar results. Full blots for a, b are shown in Source Data Extended Data Fig. 4_Uncropped Western Blots. Scale bar, 50 μm (left panels in c, top panels in d-e’, top panels in f), 10 μm (right panels in c, bottom panels in d-e’), 20 μm (bottom panels in f).
Extended Data Fig. 5. Msps localization at the fat body ncMTOC does not require TACC, AurA or Ncd.
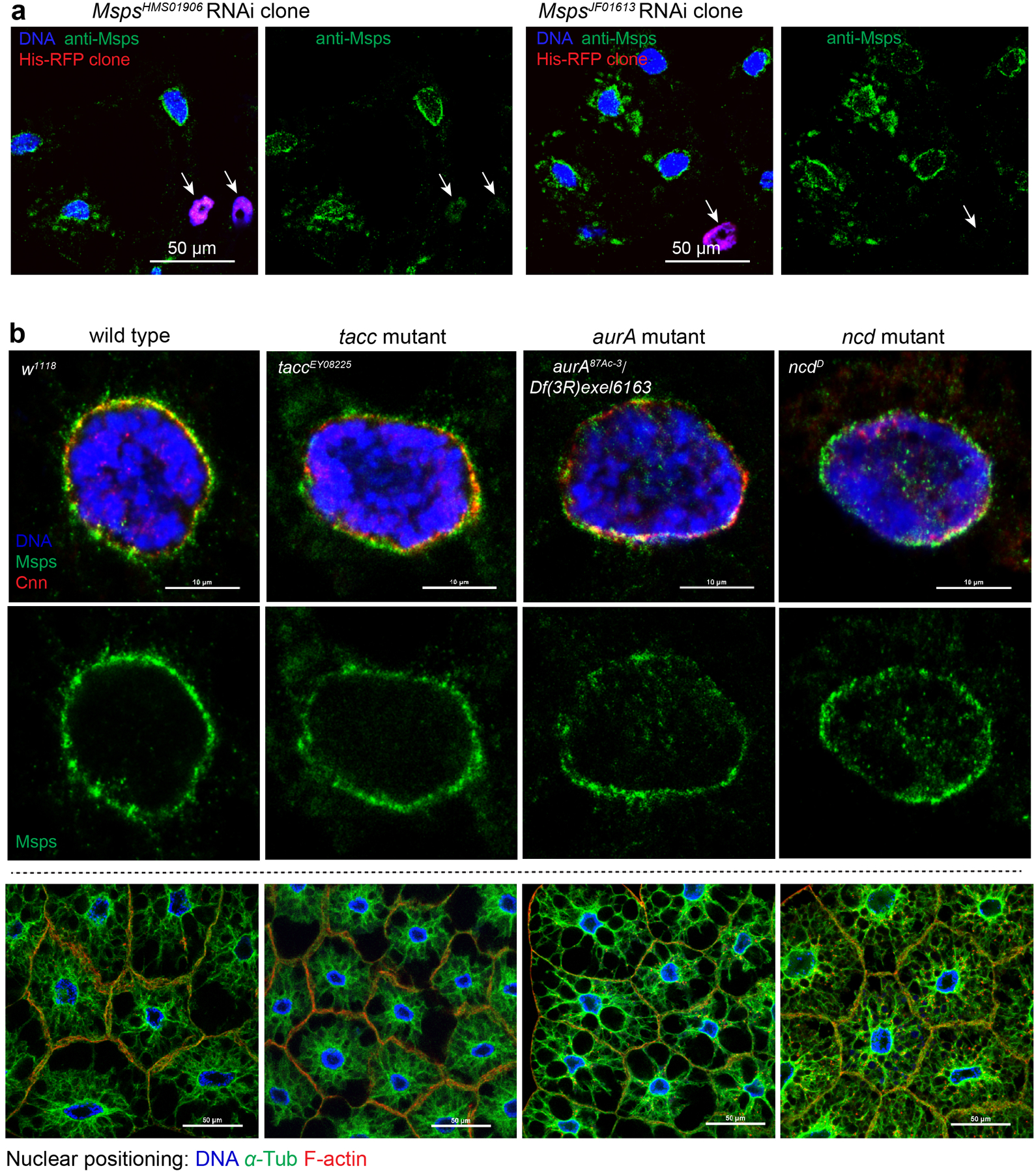
(a) Staining for Msps in msps RNAi fat body clones marked by His-RFP. Arrow indicates the loss of Msps in an msps RNAi cell. (b) Msps localization at centrosomes or spindle poles relies on TACC, Aurora A kinase phosphorylation of TACC, and the Kinesin-14 motor Ncd. None of these components are required for Msps localization at the fat body ncMTOC (top and middle panels), or for fat body nuclear positioning (bottom panels). Fat bodies were stained with antibodies against Msps and Cnn, or stained for DNA (DAPI), MTs using a FITC-conjugated DM1A antibody, and actin with CF568-conjugated phalloidin. Staining was repeated twice in a, three times in b with similar results. Nuclear positioning in b was quantified in Figure 3a. Genotype details provided in Supplementary Table 3. Scale bar, 50 μm (a, bottom panels in b), 10 μm (top panels in b).
Extended Data Fig. 6. Patronin cooperates with Nin but not with γ-tubulin or other known Patronin interactors in fat body.
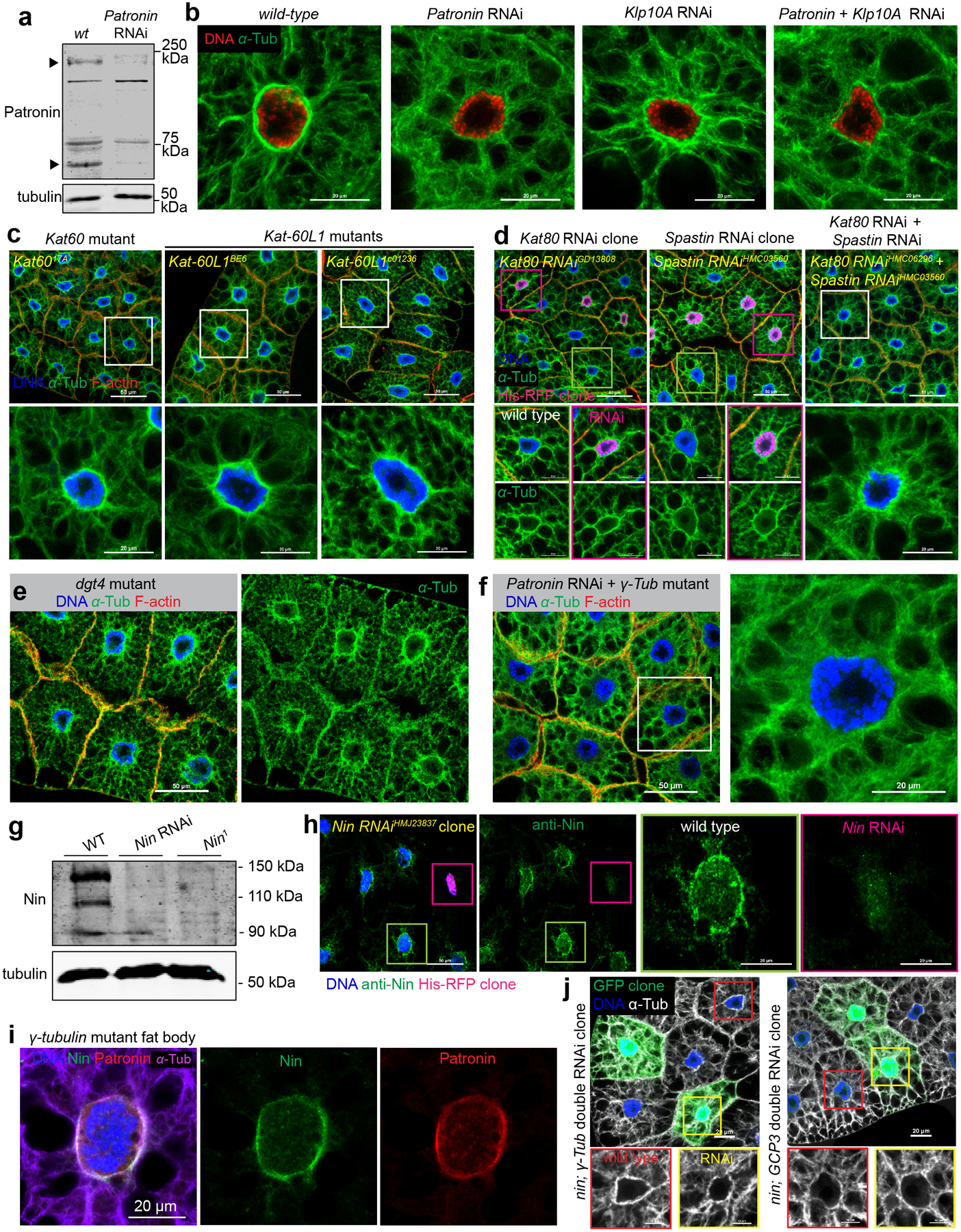
(a) Western blot detection of Patronin in larval fat body lysates from wild-type and Patronin knockdown. Arrows indicate the established Patronin isoform at 180 kDa, and another putative isoform at approximately 65 kDa. (b) MT staining in wild-type, single knockdown of Patronin or Klp10A (Kinesin-13), and double knockdown of Patronin plus Klp10A. (c) Mutants for MT severing enzymes katanin p60 or katanin p60-like 1 do not affect nuclear positioning (top) or MT assembly (bottom). (d) Knockdown of MT severing enzymes kat80 (c) or spastin (d) or double knockdown of kat80 plus spastin does not affect nuclear positioning (top) or MT assembly (bottom). (e) Mutant for dgt4 does not affect nuclear positioning (left) or MT assembly (right). (f) γTub23C mutation together with Patronin RNAi does not significantly affect nuclear positioning (left) or radial MTs (right). (g) Western blot detection of Nin in larval brain lysates from wild-type, Nin RNAi, and Nin1 mutant. (h) Nin staining in Nin RNAi fat body clone marked by His-RFP. (i) Nin, Patronin and MTs in γTub23C mutant fat body. (j) Double knockdown of Nin and γTub23C, or Nin and gcp3 does not affect nuclear positioning and MT assembly in fat body clones. The western blots in a and g were performed twice with similar results. Full blots are shown in Source Data Extended Data Fig. 6_Uncropped Western Blots. The staining were repeated three times in (b, c, e, i), twice in (d, f, h, j) with similar results. Scale bar, 20 μm (b, bottom panel in c, middle and bottom panels in d, right panel in h, i, top panels in j), 50 μm (top panels in c, d, e, left panels in f, h), 10 μm (magnified panels in j).
Extended Data Fig. 7. Shot depletion generates an ectopic MTOC and results in severe and pleiotropic disruption of subcellular compartments.
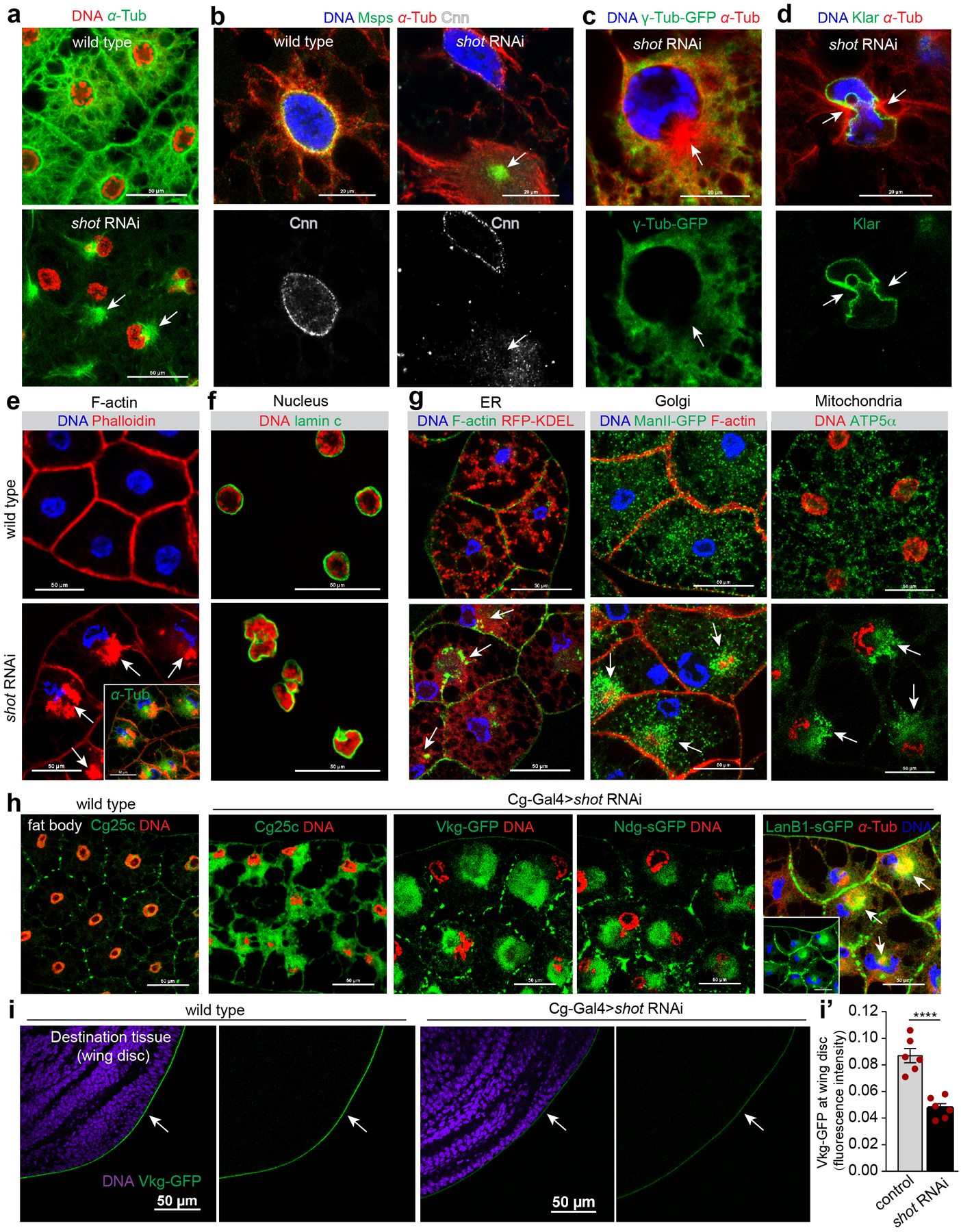
(a) shot knockdown results in a reorganization of the MTOC from the nuclear surface to a centrosome-like focus in the cytoplasm (arrows). (b, c, d) Cnn (b), γTub23C (c) and Klar (d) are not delocalized from the nuclear surface to the ectopic MTOC (arrows) generated by shot knockdown. Arrow in c indicates no γTub23C-GFP enrichment in the ectopic centrosome-like MTOC. (e) Actin ‘collapses’ into an aggregate (arrows) near the MTOC after shot knockdown. Note that nuclear centricity is also affected. Inset shows the merged image with MTs. (f, g) Nuclear morphology (f) and organization of organelles (g) are disrupted in shot knockdown fat bodies. Arrows point to accumulation of ER (RFP-KDEL), Golgi (ManII-GFP) and mitochondria (anti-ATP5α) near actin aggregates. (h) shot knockdown results in accumulation of basement membrane components in the cytoplasm, proximal to the ectopic MTOC (arrows). (i) Deposition of collagen IV α2 (Vkg-GFP, arrows) on the wing disc is decreased when shot is knocked down in fat body cells. (i’) Quantitation of Vkg-GFP in (i). Data are the means ± s.e.m (n=6 independent experiments), p=0.000088 (****) by two-tailed Student’s t-test. Statistical details are shown in Source Data Extended Data Fig. 7_Statistical Source Data. Experiments in a-h were performed three times with similar results. Genotype details are in Supplementary Table 3. Scale bar, 50 μm (a, e, f, g, h, i), 20 μm (b, c, d).
Extended Data Fig. 8. Supporting data for secretion of basement membrane components and retrograde trafficking as well as plasma membrane growth.
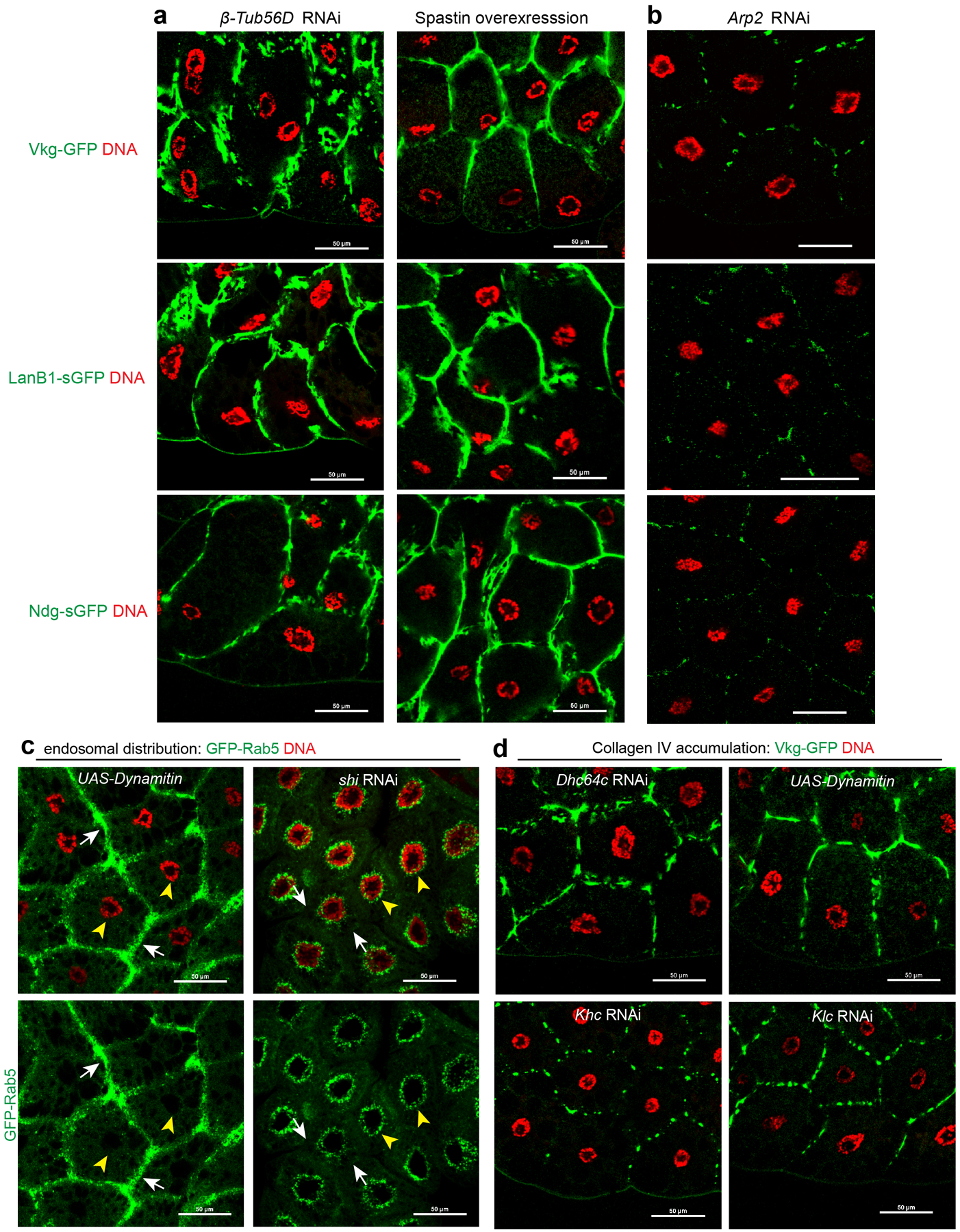
(a) Images showing fat body accumulation of different basement membrane components after disruption of MTs in fat body cells with β-Tub56D RNAi or spastin overexpression. (b) Images showing no accumulation of different basement membrane components after actin disruption in fat body by Arp2 RNAi. (c) Images showing that GFP-Rab5 endosomes shift from their perinuclear location (yellow arrowheads) to the plasma membrane site (white arrows) after inactivation of dynein activity (Dynamitin overexpression). Conversely, dynamin loss (shi RNAi) disrupts endocytic vesicle budding, causing a loss of GFP-Rab5 vesicles at the plasma membrane. (d) Images showing fat body accumulation of Vkg-GFP at the plasma membrane after inactivation of dynein activity either by Dhc64c RNAi or Dynamitin overexpression, but not after inactivation of kinesin-1 activity by Kinesin heavy chain (Khc) or Kinesin light chain (Klc) RNAi. Experiments in a, b, c, d were repeated twice with similar results. Genotype details are in Supplementary Table 3. Scale bar, 50 μm.
Extended Data Fig. 9. Summary and model for perinuclear non-centrosomal MTOC assembly and function in fat body cells.
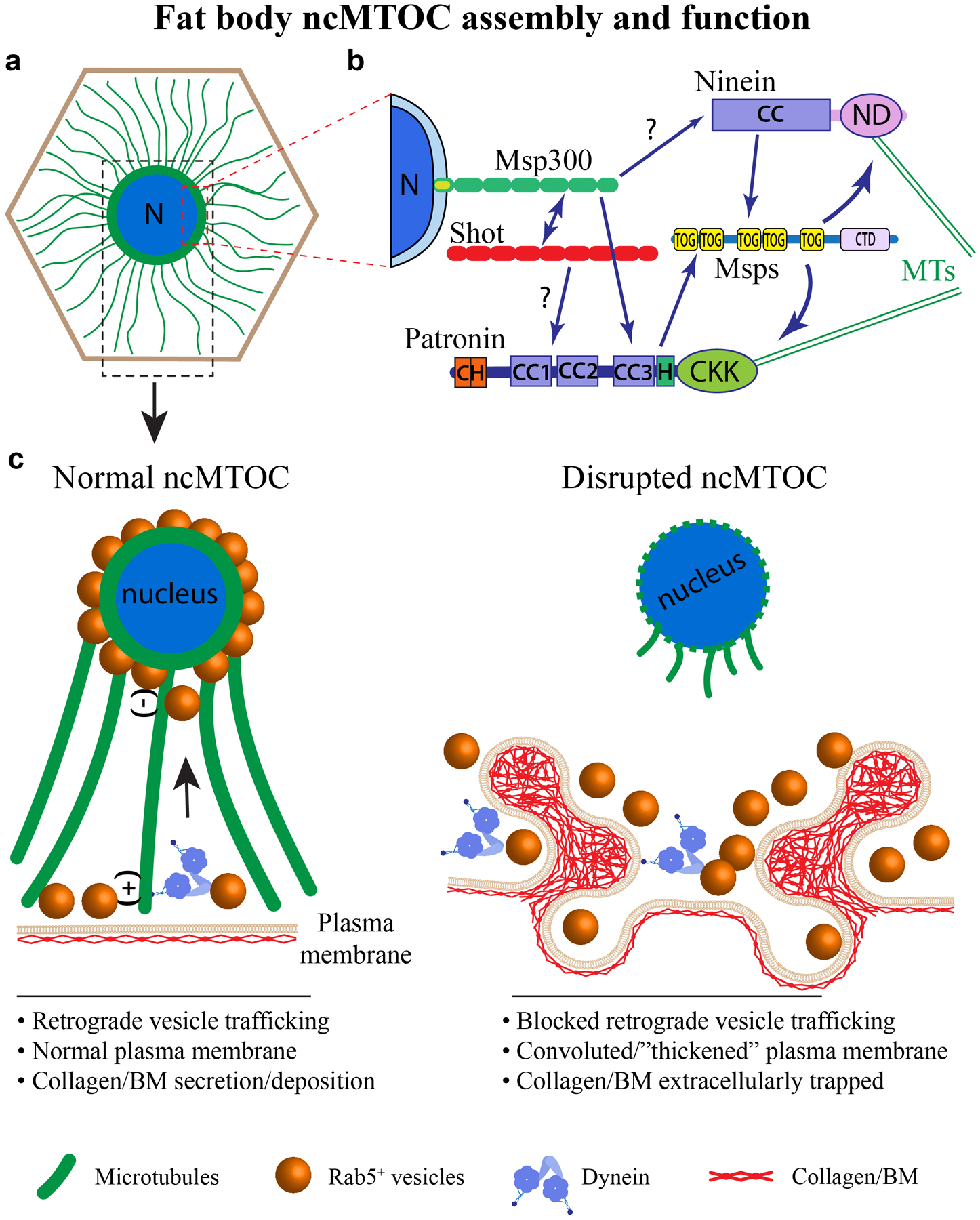
(a) Drosophila fat body cells, a differentiated cell type analogous to human adipocytes and liver, assemble a perinuclear ncMTOC that organizes dense circumferential MTs and radial MTs. The radial MTs are polarized, emanating from the nuclear surface (minus-end) towards the plasma membrane (plus-end). N: nucleus. (b) The perinuclear ncMTOC has unique architecture and MT assembly mechanisms. Msp300/Nesprin anchors the ncMTOC at nuclear surface, requiring and recruiting Shot, and is epistatic to Patronin/CAMSAP. Patronin and Nin cooperate to recruit Msps for radial MT elongation independently of γ-tubulin. Domains are shown for each protein. (c) The perinuclear ncMTOC has two critical physiological functions: nuclear positioning and control of plasma membrane growth via retrograde trafficking. The fat body perinuclear ncMTOC controls endosomal retrograde trafficking in coordination with minus-end directed dynein to restrict plasma membrane overgrowth. Disruption of the ncMTOC or inactivation of dynein blocks retrograde membrane trafficking, leading to excessive plasma membrane growth and convoluted “thickened” plasma membranes. Consequentially, secreted BM components are trapped within the convoluted plasma membrane folds. The entrapment of BM proteins in fat body cells leads to reduced BM deposition in destination tissues, including imaginal discs and brains (not depicted).
Supplementary Material
Supplementary Tables 1 through 5.
1. Survey of fat body ncMTOC components and key assembly regulators.
2. Relevant genotypes of strains and stocks used.
3. Detailed genotypes associated with each figure
4. Summary of mutant and RNAi lines used for each gene and associated phenotypes (Related to Supplementary Table 1)
5. Antibodies used in this work
Movie 1. Nucleus positioning in live wild type larval fat bodies. In wild-type fat bodies, nucleus positioning was recorded in live whole larvae expressing His-GFP (nucleus, green) ubiquitously, and myr-RFP (plasma membrane, red) under the control of the fat body driver SPARC-Gal4. Movie records one hour of time-lapse imaging, sped up 60-fold. Note very little movement and maintenance of centroid positioning of nuclei.
Movie 2. Nucleus positioning defects after disruption of the fat body ncMTOC. In Msp300 RNAi fat bodies, nucleus positioning was recorded in live whole larvae expressing His-GFP (nucleus, green) ubiquitously, and myr-RFP (plasma membrane, red) under the control of the fat body driver SPARC-Gal4. Movie records one hour of time-lapse imaging, sped up 60-fold. Note very little nuclear movement and loss of nuclear centricity in these ncMTOC-disrupted fat body cells.
Acknowledgements
We thank Megraw lab members for numerous discussions and critiques of the manuscript, and three anonymous reviewers for constructive recommendations that improved the work. We thank Melissa Rolls, Wu-Min Deng, Jordan Raff, Ron Vale, Talila Volk, Nina Sherwood, Renata Basto, Nasser Rusan, Eric Lécuyer, Tomer Avidor-Reiss, Maurice Ringuette, Dorothy Lerit, Steve Rogers, James Wakefield, Hiroyuki Nakanishi, and Volodya Gelfand for antibodies and Drosophila stocks. We thank the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, Kyoto Stock Center for Drosophila stocks and the Developmental Studies Hybridoma Bank, University of Iowa, for antibodies. We apologize to colleagues whose work was not cited due to space limitations or oversight on our part. We are grateful for funding from NIH grants R15GM119078 and R15HD099648 (T.L.M).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Tillery MML, Blake-Hedges C, Zheng Y, Buchwalter RA & Megraw TL Centrosomal and Non-Centrosomal Microtubule-Organizing Centers (MTOCs) in Drosophila melanogaster. Cells 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez AD & Feldman JL Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Current opinion in cell biology 44, 93–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muroyama A & Lechler T Microtubule organization, dynamics and functions in differentiated cells. Development 144, 3012–3021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin M & Akhmanova A Coming into Focus: Mechanisms of Microtubule Minus-End Organization. Trends in cell biology (2018). [DOI] [PubMed] [Google Scholar]
- 5.Farache D, Emorine L, Haren L & Merdes A Assembly and regulation of gamma-tubulin complexes. Open biology 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollman JM, Merdes A, Mourey L & Agard DA Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol 12, 709–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oakley BR, Paolillo V & Zheng Y gamma-Tubulin complexes in microtubule nucleation and beyond. Molecular biology of the cell 26, 2957–2962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JV, Buchwalter RA, Kao LR & Megraw TL A Splice Variant of Centrosomin Converts Mitochondria to Microtubule-Organizing Centers. Curr Biol 27, 1928–1940.e1926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flor-Parra I, Iglesias-Romero AB & Chang F The XMAP215 Ortholog Alp14 Promotes Microtubule Nucleation in Fission Yeast. Curr Biol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thawani A, Kadzik RS & Petry S XMAP215 is a microtubule nucleation factor that functions synergistically with the gamma-tubulin ring complex. Nature cell biology 20, 575–585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunzelmann J et al. The microtubule polymerase Stu2 promotes oligomerization of the gamma-TuSC for cytoplasmic microtubule nucleation. eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atherton J et al. A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat Struct Mol Biol 24, 931–943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin SS & Vale RD Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263–274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendershott MC & Vale RD Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc Natl Acad Sci U S A 111, 5860–5865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfelder KP et al. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development (Cambridge, England) 141, 3551–3560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrese EL & Soulages JL Insect fat body: energy, metabolism, and regulation. Annual review of entomology 55, 207–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Droujinine IA & Perrimon N Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annual review of genetics 50, 539–570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshizaki DK et al. Embryonic fat-cell lineage in Drosophila melanogaster. Development 120, 2489–2499 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Clark IE, Jan LY & Jan YN Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development 124, 461–470 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Song Y & Brady ST Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends in cell biology 25, 125–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conduit PT et al. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife 3, e03399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin TC, Neuner A & Schiebel E Targeting of gamma-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends in cell biology 25, 296–307 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Sallee MD, Zonka JC, Skokan TD, Raftrey BC & Feldman JL Tissue-specific degradation of essential centrosome components reveals distinct microtubule populations at microtubule organizing centers. PLoS Biol 16, e2005189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motegi F, Velarde NV, Piano F & Sugimoto A Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Developmental cell 10, 509–520 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Hannak E et al. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. The Journal of cell biology 157, 591–602 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strome S et al. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Molecular biology of the cell 12, 1751–1764 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouhard GJ et al. XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barros TP, Kinoshita K, Hyman AA & Raff JW Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. The Journal of cell biology 170, 1039–1046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cullen CF & Ohkura H Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nature cell biology 3, 637–642 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Lee MJ, Gergely F, Jeffers K, Peak-Chew SY & Raff JW Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nature cell biology 3, 643–649 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Rogers GC, Rusan NM, Peifer M & Rogers SL A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Molecular biology of the cell 19, 3163–3178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J & Akhmanova A Microtubule-Organizing Centers. Annu Rev Cell Dev Biol 33, 51–75 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Jiang K et al. Structural Basis of Formation of the Microtubule Minus-End-Regulating CAMSAP-Katanin Complex. Structure 26, 375–382 e374 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Jiang K et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Developmental cell 28, 295–309 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Nashchekin D, Fernandes AR & St Johnston D Patronin/Shot Cortical Foci Assemble the Noncentrosomal Microtubule Array that Specifies the Drosophila Anterior-Posterior Axis. Developmental cell 38, 61–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roll-Mecak A & Vale RD Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? The Journal of cell biology 175, 849–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp DJ & Ross JL Microtubule-severing enzymes at the cutting edge. Journal of cell science 125, 2561–2569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goshima G, Mayer M, Zhang N, Stuurman N & Vale RD Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. The Journal of cell biology 181, 421–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawo S et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 19, 816–826 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Huertas C et al. Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat Commun 7, 12187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunha-Ferreira I et al. The HAUS Complex Is a Key Regulator of Non-centrosomal Microtubule Organization during Neuronal Development. Cell reports 24, 791–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yau KW et al. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron 82, 1058–1073 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Wang S et al. NOCA-1 functions with γ-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. eLife 4, e08649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delgehyr N, Sillibourne J & Bornens M Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. Journal of cell science 118, 1565–1575 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V & Bornens M Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. Journal of cell science 113 (Pt 17), 3013–3023 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y et al. The Seckel syndrome and centrosomal protein Ninein localizes asymmetrically to stem cell centrosomes but is not required for normal development, behavior, or DNA damage response in Drosophila. Molecular biology of the cell 27, 1740–1752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldspink DA et al. Ninein is essential for apico-basal microtubule formation and CLIP-170 facilitates its redeployment to non-centrosomal microtubule organizing centres. Open biology 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowanda M et al. Loss of function of the Drosophila Ninein-related centrosomal protein Bsg25D causes mitotic defects and impairs embryonic development. Biology open 5, 1040–1051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lecland N, Hsu CY, Chemin C, Merdes A & Bierkamp C Epidermal development requires ninein for spindle orientation and cortical microtubule organization. Life science alliance 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosen JN et al. The Drosophila Ninein homologue Bsg25D cooperates with Ensconsin in myonuclear positioning. The Journal of cell biology 218, 524–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mejat A & Misteli T LINC complexes in health and disease. Nucleus (Austin, Tex.) 1, 40–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khanal I, Elbediwy A, Diaz de la Loza Mdel C, Fletcher GC & Thompson BJ Shot and Patronin polarise microtubules to direct membrane traffic and biogenesis of microvilli in epithelia. Journal of cell science 129, 2651–2659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastor-Pareja JC & Xu T Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Developmental cell 21, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai J, Ma M, Feng Z & Pastor-Pareja JC Inter-adipocyte Adhesion and Signaling by Collagen IV Intercellular Concentrations in Drosophila. Current Biology 27, 2729–2740.e2724 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Zang Y et al. Plasma membrane overgrowth causes fibrotic collagen accumulation and immune activation in Drosophila adipocytes. eLife 4, e07187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granger E, McNee G, Allan V & Woodman P The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol 31, 20–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fant X, Srsen V, Espigat-Georger A & Merdes A Nuclei of Non-Muscle Cells Bind Centrosome Proteins upon Fusion with Differentiating Myoblasts. Plos One 4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srsen V, Fant X, Heald R, Rabouille C & Merdes A Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol 10, 28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gimpel P et al. Nesprin-1alpha-Dependent Microtubule Nucleation from the Nuclear Envelope via Akap450 Is Necessary for Nuclear Positioning in Muscle Cells. Curr Biol 27, 2999–3009 e2999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Espigat-Georger A, Dyachuk V, Chemin C, Emorine L & Merdes A Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. Journal of cell science 129, 4227–4237 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Bugnard E, Zaal KJM & Ralston E Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskel 60, 1–13 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Elhanany-Tamir H et al. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. The Journal of cell biology 198, 833–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao CX, Wen X, Jin S & Zhang YQ Increased acetylation of microtubules rescues human tau-induced microtubule defects and neuromuscular junction abnormalities in Drosophila. Disease models & mechanisms 10, 1245–1252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S, Reuveny A & Volk T Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. The Journal of cell biology 209, 529–538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metzger T et al. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Folker ES, Schulman VK & Baylies MK Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development (Cambridge, England) 139, 3827–3837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen JV et al. Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. The Journal of cell biology 211, 435–453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1 through 5.
1. Survey of fat body ncMTOC components and key assembly regulators.
2. Relevant genotypes of strains and stocks used.
3. Detailed genotypes associated with each figure
4. Summary of mutant and RNAi lines used for each gene and associated phenotypes (Related to Supplementary Table 1)
5. Antibodies used in this work
Movie 1. Nucleus positioning in live wild type larval fat bodies. In wild-type fat bodies, nucleus positioning was recorded in live whole larvae expressing His-GFP (nucleus, green) ubiquitously, and myr-RFP (plasma membrane, red) under the control of the fat body driver SPARC-Gal4. Movie records one hour of time-lapse imaging, sped up 60-fold. Note very little movement and maintenance of centroid positioning of nuclei.
Movie 2. Nucleus positioning defects after disruption of the fat body ncMTOC. In Msp300 RNAi fat bodies, nucleus positioning was recorded in live whole larvae expressing His-GFP (nucleus, green) ubiquitously, and myr-RFP (plasma membrane, red) under the control of the fat body driver SPARC-Gal4. Movie records one hour of time-lapse imaging, sped up 60-fold. Note very little nuclear movement and loss of nuclear centricity in these ncMTOC-disrupted fat body cells.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. Data not included are available from the corresponding authors upon reasonable request.


