Abstract
Experimentally measured residual dipolar couplings (RDCs) are highly valuable for atomic-resolution structural and dynamic studies of molecular systems ranging from small molecules to large proteins by solution NMR spectroscopy. Here we demonstrate the first use of magnetic-alignment behavior of lyotropic liquid crystalline polymer macro-nanodiscs (> 20 nm in diameter) as a novel alignment medium for the measurement of RDCs using high resolution NMR. The easy preparation of macro-nanodiscs, high stability against pH and in presence of divalent metal ions, and high homogeneity will render a robust approach to investigate a wide range of molecular systems including natural products, proteins and RNA.
Keywords: Polymer nanodisc, Cytochrome c, Residual dipolar couplings, NMR, Magnetic alignment
Graphical Abstract
Polymer macro-nanodiscs are used as novel alignment medium for the measurement of RDCs using high resolution NMR spectroscopy.
Polymer nanodiscs are being successfully used in structural and functional studies of membrane proteins.[1–6] A polymer nanodisc is composed of a planar lipid bilayer surrounded by an amphiphilic polymer belt.[7] A variety of polymers have been used to form nanodiscs using synthetic lipids or by directly extracting membrane proteins with their native lipid environment.[2, 8–9] Starting with styrene maleic acid,[10] there has been several different polymers[11–13] and their derivatives[14–19] that have been used to overcome the limitations such as size control, metal ion and low pH instability.[14–15, 20] Furthermore the size control enabled the use of macro-nanodiscs (> 20 nm) in solid-state NMR due to their magnetic alignment properties in the presence of an external magnetic field.[14–15, 21] Herein we demonstrate for the first time the use of the magnetic-alignment property of macro-nanodiscs for high-resolution NMR studies of water-soluble biomolecules using residual dipolar couplings (RDCs).
One of the powerful techniques used in the structural characterization of biomolecules is RDCs. RDCs have been shown to provide valuable global orientation constraints to determine and refine high-resolution structures of biomolecules.[22–24] RDCs are typically measured using an anisotropic environment created by magnetically oriented liquid crystalline molecules or mechanically compressed/stretched gels.[23, 25] Previous studies have shown that the degeneracy of bond orientations obtained from RDC values can be minimized using orthogonal tensors by measuring RDCs in different alignment media or modulating the interaction between the alignment medium and protein (or molecular system under investigation).[26] Bicelles have been used to achieve the measurements of orthogonal alignment tensors by changing the net charge via doping with negatively charged lipid[26] or surfactants.[27] However, bicelles are not usable for many biomolecular systems due to the presence of a denaturing detergent. In this study we show that polymer nanodiscs can be used as an alignment medium for measuring RDCs. Our results demonstrate that polymer nanodiscs exhibit a highly ordered homogeneous alignment in the presence of a magnetic field, and the degree of alignment can be scaled by the concentration as well as temperature of the sample. Using cytochrome c (cyt c) as a model system, we show that the experimentally measured RDCs are in good agreement with calculated values from reported structures.
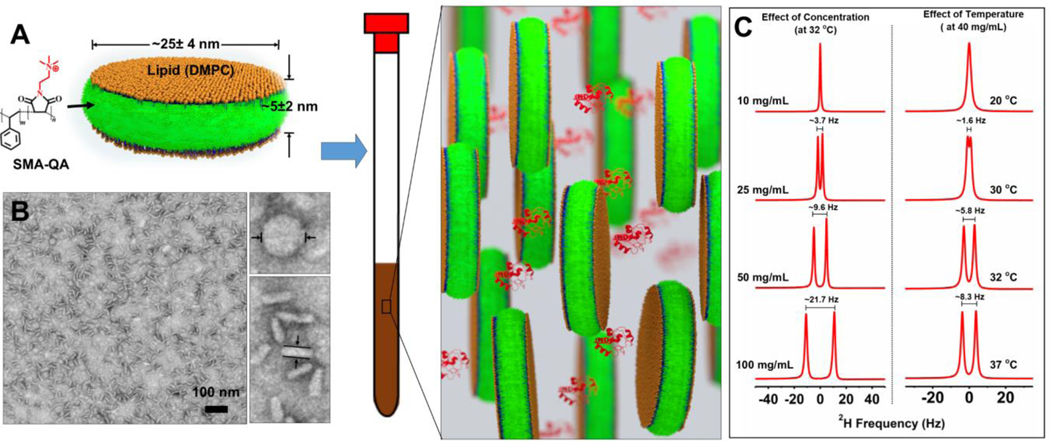
Styrene maleic acid quaternary ammonium (SMA-QA) polymer was synthesized as described previously.[15] Polymer macro-nanodiscs were prepared using 1:0.5 w/w ratio of DMPC and SMA-QA (Figure 1A). The resulting nanodiscs were purified using size exclusion chromatography (SEC), and characterized by dynamic light scattering (Figures S1 and S2) and transmission electron microscopy (TEM). The TEM images showed the presence of macro-nanodiscs that were ~25±4 nm in diameter (Figures 1B and S3).
Figure 1:
A) Schematic of a polymer lipid bilayer nanodisc and a representation of its use as an alignment medium. B) Transmission electron microscope image of macro-nanodiscs prepared using 1:0.5 w/w ratio of DMPC:SMA-QA. C) Deuterium NMR spectra of macro-nanodiscs with a varying lipid concentration (at 32 °C) and at different temperatures (at 40 mg/mL). All spectra were acquired using a 500 MHz NMR spectrometer.
Macro-nanodiscs with a varying lipid concentration (10–100 mg/mL) were prepared in a 10% D2O containing phosphate buffer. 2H NMR was used to examine the anisotropic property of macro-nanodiscs. An isotropic peak was observed at a low lipid concentration (10 mg/mL) at 32° C, whereas by increasing the concentration to 20 mg/mL a doublet was observed with ~3.7 Hz 2H quadrupolar splitting (ΔνQ) from HOD (Figure 1C). Further increase in the lipid concentration to 100 mg/mL increased the observed 2H quadrupole splitting to ~21.7 Hz. These observations suggest that the degree of magnetic-alignment of macro-nanodiscs is scalable by altering the lipid concentration; and the observed symmetric, narrow peaks (line width <2 Hz) indicate a highly ordered homogeneous alignment of nanodiscs. The effect of temperature on the alignment was examined using 40 mg/mL lipid concentration of nanodiscs. An isotropic peak was observed at 20 °C (which is below the gel-to-liquid crystalline phase transition temperature of DMPC), while a doublet with a quadrupole splitting of ~1.6 Hz appeared as the sample temperature increased to 30 °C. Further increase in temperature increased the observed quadrupole splitting as shown in Figure 1C suggesting that the macro-nanodiscs exhibit a lyotropic liquid crystal behavior, in which the liquid crystalline phase depends on the temperature and concertation, and are suitable for RDC measurements.[22]
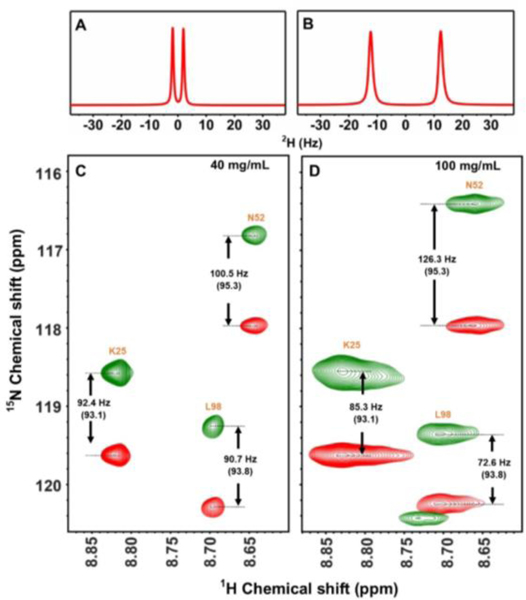
To demonstrate the feasibility of using nanodiscs as an alignment medium for NMR structural studies, uniformly 15N labeled cyt c was used as a model protein in this study. cyt c is a positively charged, water-soluble protein with pI of 9.6.[28] We chose positively charged SMA-QA macro-nanodiscs to avoid any charge-charge interaction with cyt c.[29] 15N labeled cyt c was added to a solution of macro-nanodiscs (40 mg/mL) and used in 15N-1H In-Phase, Anti-Phase Heteronuclear Single-Quantum Coherence (IPAP-HSQC)[30] experiments to measure RDCs. 2D IPAP-HSQC spectra acquired at 20 °C showed the 1JNH-couplings measured in the presence and absence of nanodisc are comparable (Figure S4), suggesting that the nanodiscs medium is isotropic at 20 °C. This is also confirmed from the appearance of an isotropic 2H peak for the same sample (Figure S5). An anisotropic nanodiscs medium was obtained by increasing the temperature to 32 °C, for which the alignment was confirmed from the appearance of a doublet in deuterium NMR spectra (Figure 2A). Selected regions of 2D IPAP-HSQC spectra are shown in Figure 2C. RDCs were obtained by subtracting coupling (1JNH or 1JNH + DNH) values measured from 2D IPAP-HSQC spectra acquired at isotropic (20 °C) and anisotropic (32 °C) conditions. Our experimental results show that the RDC values can be scaled by simply increasing the nanodiscs concertation or the temperature as evident from the increased RDCs obtained at higher concentration: ±6 Hz for 40 mg/mL and ±40 Hz for 100 mg/mL (see Figures 2(C and D), 3(A and B) and Figure S6).
Figure 2.
(A and B) Deuterium NMR spectra of SMA-QA:DMPC macro-nanodiscs containing 200 μM 15N-cytC in 10% D2O and phosphate buffer at 32 °C with a lipid concentration of 40 mg/mL (A) and 100 mg/mL (B). Selected region of 2D 1H-15N IPAP-HSQC spectra obtained for a lipid concentration of 40 mg/mL (C) and 100 mg/mL (D) at 32 °C in a 800 MHz NMR spectrometer. Isotropic 1JNH values were obtained at 20 °C are shown in parenthesis (See Figure S5). NMR spectra (Figs. S10–14) and SEC profiles (Fig.15) are included in the supplementary information.
Figure 3.
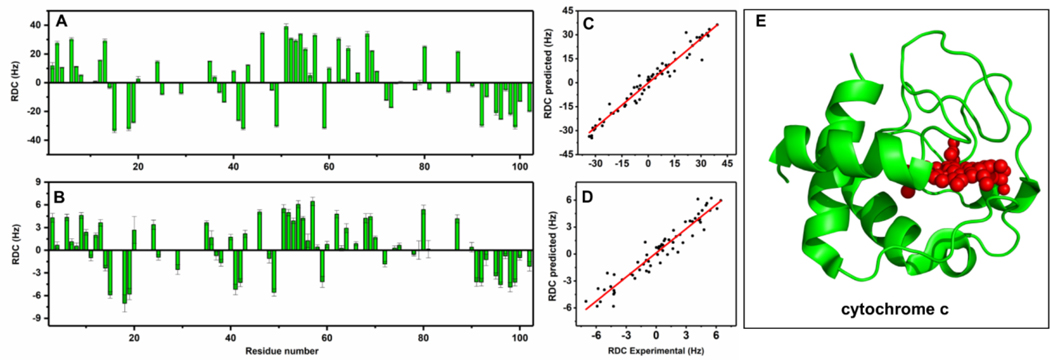
A) Experimentally measured residual dipolar couplings for 15N cytochrome c for a lipid concentration of 100 mg/mL (A) and 40 mg/mL (B). Correlation of experimental and calculated RDCs at 100 mg/mL (C) and 40 mg/mL (D). (E) X-ray crystal structure of cyt c (PDB:6FF5).
To validate the accuracy of experimentally measured RDC values, RDCs were calculated using reported crystal structures of cyt c. Figure 3(C and D) shows the correlation between experimental and calculated RDCs with R values 0.958 for 40 mg/mL and 0.979 for 100 mg/mL. We further compared the experimental RDCs with values calculated from different high-resolution structures reported for cyt c (Figures S7–S9). These comparisons can be used to confirm the correct structure of the protein used in the NMR sample.
In summary, we have successfully demonstrated the use of polymer macro-nanodiscs as an effective alignment medium for the measurement of RDC values in the structural studies using NMR experiments. Our results showed that the degree of alignment can be scaled by varying either the lipid-nanodiscs concentration or temperature. Using cyt c as a model system, in this study we showed that the experimentally measured RDCs are in good agreement with the calculated values from known structures of the protein. It is remarkable that the pH resistant and divalent metal ion tolerant SMA-QA nanodiscs can be further exploited to study a variety of proteins and other biomolecules for RDCs based NMR structural studies under various conditions. The alignment tensor of the macro-nanodiscs alignment medium can be modulated by altering the interaction between the protein and the alignment medium via changing the lipid composition, and thereby enabling the measurement of RDCs with orthogonal tensors to restrict the determined bond orientations. Another advantage of polymer nanodiscs is that the protein (or any biomolecule) under study can be recovered from the sample using SEC after NMR experiments. Due to these unique properties, we believe that the use of polymer nanodiscs as an alignment medium will be valuable for high-resolution structural studies on water-soluble biomolecules as well as to probe the interaction of molecules (such as peptides, proteins, ligands, RNA) with lipid membrane.
Supplementary Material
Acknowledgements
This study was supported by the funds from NIH (GM084018 to A.R.). We thank Dr. Nagao for help with the production of cyto-chrome c.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB, Nature 2018, 557, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR, Nat. Protoc. 2016, 11, 1149–1162. [DOI] [PubMed] [Google Scholar]
- [3].Stroud Z, Hall SCL, Dafforn TR, Methods 2018. [DOI] [PubMed] [Google Scholar]
- [4].Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA, Eur Biophys J 2016, 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, Silvestro D, Hatzakis NS, Wenk MR, Dafforn TR, Olsen CE, Motawia MS, Hamberger B, Møller BL, Bassard J-E, Science 2016, 354, 890. [DOI] [PubMed] [Google Scholar]
- [6].Lee SC, Pollock NL, Biochem Soc Trans 2016, 44, 1011–1018. [DOI] [PubMed] [Google Scholar]
- [7].Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Grobner G, Watts A, Angew Chem Int Ed Engl 2012, 51, 4653–4657. [DOI] [PubMed] [Google Scholar]
- [8].Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A, Nano Lett. 2012, 12, 4687–4692. [DOI] [PubMed] [Google Scholar]
- [10].Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M, J Am Chem Soc 2009, 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- [11].Oluwole AO, Danielczak B, Meister A, Babalola JO, Vargas C, Keller S, Angew Chem Int Ed Engl 2017, 56, 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi JI, Ramamoorthy A, J Am Chem Soc 2017, 139, 18657–18663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hardin NZ, Ravula T, Mauro GD, Ramamoorthy A, Small 2019, 15, e1804813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, Angew Chem Int Ed Engl 2017, 56, 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [15].Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A, Angew Chem Int Ed Engl 2018, 57, 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [16].Lindhoud S, Carvalho V, Pronk JW, Aubin-Tam ME, Biomacromolecules 2016, 17, 1516–1522. [DOI] [PubMed] [Google Scholar]
- [17].Fiori MC, Jiang Y, Altenberg GA, Liang H, Sci Rep 2017, 7, 7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A, Langmuir 2017, 33, 10655–10662. [DOI] [PubMed] [Google Scholar]
- [19].Hall SCL, Tognoloni C, Charlton J, Bragginton EC, Rothnie AJ, Sridhar P, Wheatley M, Knowles TJ, Arnold T, Edler KJ, Dafforn TR, Nanoscale 2018, 10, 10609–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang R, Sahu ID, Liu L, Osatuke A, Comer RG, Dabney-Smith C, Lorigan GA, Biochim Biophys Acta 2015, 1848, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Radoicic J, Park SH, Opella SJ, Biophys J 2018, 115, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tjandra N, Bax A, Science 1997, 278, 1111–1114. [DOI] [PubMed] [Google Scholar]
- [23].Prestegard JH, Bougault CM, Kishore AI, Chemical Reviews 2004, 104, 3519–3540. [DOI] [PubMed] [Google Scholar]
- [24].Bax A, Protein Science 2003, 12, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lipsitz RS, Tjandra N, Annu Rev Biophys Biomol Struct 2004, 33, 387–413. [DOI] [PubMed] [Google Scholar]
- [26].Lorieau JL, Maltsev AS, Louis JM, Bax A, J Biomol NMR 2013, 55, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ramirez BE, Bax A, J Am Chem Soc 1998, 120, 9106–9107. [Google Scholar]
- [28].Hüttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I, Mitochondrion 2011, 11, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ravula T, Hardin NZ, Bai J, Im SC, Waskell L, Ramamoorthy A, Chem Commun (Camb) 2018, 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ottiger M, Delaglio F, Bax A, J Magn Reson 1998, 131, 373–378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.