Abstract
Background
Papillary thyroid carcinoma (PTC), which is the most common endocrine malignancy, has been steadily increasing worldwide in incidence over the years, while mechanisms underlying the pathogenesis and diagnostic for PTC are incomplete. The purpose of this study is to identify potential biomarkers for diagnosis of PTC, and provide new insights into pathogenesis of PTC.
Methods
Based on weighted gene co-expression network analysis, Robust Rank Aggregation, functional annotation, GSEA and DNA methylation, were employed for investigating potential biomarkers for diagnosis of PTC.
Results
Black and turquoise modules were identified in the gene co-expression network constructed by 1807 DEGs that from 6 eligible gene expression profiles of Gene Expression Omnibus database based on Robust Rank Aggregation and weighted gene co-expression network analysis. Hub genes were significantly down-regulated and the expression levels of the hub genes were different in different stages in hub gene verification. ROC curves indicated all hub genes had good diagnostic value for PTC (except for ABCA6 AUC = 89.5%, the 15 genes with AUC > 90%). Methylation analysis showed that hub gene verification ABCA6, ACACB, RMDN1 and TFPI were identified as differentially methylated genes, and the decreased expression level of these genes may relate to abnormal DNA methylation. Moreover, the expression levels of 8 top hub genes were correlated with tumor purity and tumor-infiltrating immune cells. These findings, including functional annotations and GSEA provide new insights into pathogenesis of PTC.
Conclusions
The hub genes and methylation of hub genes may as potential biomarkers provide new insights for diagnosis of PTC, and all these findings may be the direction to study the mechanisms underlying of PTC in the future.
Keywords: Papillary thyroid carcinoma, Biomarkers, DNA Methylation, Robust rank aggregation, Weighted gene co-expression network analysis
Background
Thyroid carcinoma is the most common endocrine cancer, and papillary thyroid cancer (PTC) accounts for the highest proportion of thyroid carcinoma. In recent years, the incidence of PTC has been steadily increasing worldwide, which may be due to a real increase, or may be due to the improvement and widespread use of screening techniques [1]. Increased TSH, autoimmune diseases and inflammation were considered risk factors for thyroid cancer [2]. In recent years, researchers believe that the major carcinogenic event of PTC is the activation of mitogen-activated protein kinase (MAPK) [3]. However, the mechanisms underlying the pathogenesis of PTC have not yet been elucidated. Surgical resection, TSH inhibition therapy and radioactive iodine therapy are the conventional treatment methods for PTC [4]. With these therapeutic approaches, most patients with PTC have a good prognosis, but there were some challenges for both patients and clinicians. The consequence of surgical excision that is one of the important treatments for thyroid cancer may be the increased incidence of hypothyroidism [5]; PTC is often difficult to diagnose because of similarities between malignant and benign nodules, which results in some benign patients having their thyroid removed [6]; treatment of refractory radioiodine differentiated thyroid cancer still faces challenges and some slow-moving tumors were overdiagnosed and overtreated. In terms of molecular therapy, the combined use of immune checkpoint inhibitors and BRAF (especially BRAFV600E) inhibitors is aussichtsreich in the treatment of thyroid cancer and some progress has been made [7]. However, not all tumors have mutations in BRAF [8–10], and other biomarkers are needed. In addition, immunotherapy is associated with immune-related adverse events (autoimmune toxicities) [11]. For example, the use of mAbs anti-cytotoxic T lymphocyte antigen 4 (anti-CTLA-4) and anti-programmed cell death protein-1/programmed cell death ligand-1 (anti-PD-1/PD-L1) causes thyroid dysfunction (including painless thyroiditis and so on) [12] in 10 percent of cancer patients [7]. DNA methylation, which belongs to epigenetics [13], can affect gene expression by affecting the structural stability of chromosomes and the interaction between DNA and proteins [14]. Abnormal levels of DNA methylation had been reported in almost all cancers. The study of DNA methylation is very important for the pathogenesis, early diagnosis and prognosis prediction of tumors and methylation drugs are the darlings of recent targeted therapies for cancer for DNA methylation itself is reversible. The combination of genetic alterations and DNA methylation alterations may improve their clinical value. However, the specific gene map and methylation map of PTC are not complete. Therefore, it is necessary to further understand the underlying biological mechanisms underlying the onset and development of PTC and to identify potential biomarkers for diagnosis. More accurate diagnosis leads to more personalized treatment, which is valuable in further improving patient survival.
As a bioinformatics method that can integrate gene lists of different technology platforms, Robust Rank Aggregation (RRA) is widely used in cancer research [15–17]. This method can reduce noise increase signal while integrating data information of different platforms, which makes the research results more reliable [16]. The weighted gene co-expression network analysis (WGCNA) [18] method is widely used in cancer research, but there is still room for the research of establishing gene co-expression network to identify the hub genes closely related to PTC. In order to better explain the biological function genes, gene set enrichment analysis (GSEA) on hub genes was performed. GSEA can assess whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states [19]. Furthermore, we analyzed the relationship between hub genes and tumor immune infiltrating cells since microenvironment composed of tumor immune infiltrating cells can play an important role in the occurrence and progression of tumor by promoting tumor and anti-tumor [20], and the influence of different infiltrating immune cells on tumor is different [21]. Our study might provide some new insights into current diagnosis and pathogenesis for PTC.
Methods
Thyroid cancer gene expression datasets collection and identification of robust DEGs
The data sets enrolled in this study needs to meet the main conditions [1]: The data set must include the gene expression profile of PTC and normal thyroid tissue [2]. The genes in the platform need to be above 5000. The 6 unprocessed gene expression profiles that met the inclusion criteria of this study were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database: GSE6004 [22], GSE58545 [23], GSE27155 [24, 25], GSE53157 [26], GSE60542 [27] and GSE33630 [28, 29]. The relevant data are shown in Table 1. For more detailed sample information, please see Additional file 2: Table S1. After data normalization using robust multi-array averaging algorithm [30], “limma” [31] package were used to assess statistical significance (P-value) for each gene based on a linear model implemented. Then, gene lists in six datasets were integrated using the RRA method to identify differentially expressed genes (DEGs) according to P-value in “RobustRankAggreg” package of R software, and P-value < 0.05 was set as the threshold for DEGs.
Table 1.
Characteristics of the data sets enrolled in the study
| Dataset ID | Number of normal samples | Number of tumor samples | Country | GPL ID | Number of rows per platform |
|---|---|---|---|---|---|
| GSE6004 | 4 | 7 | USA | GPL570 | 54,675 |
| GSE58545 | 18 | 27 | Poland | GPL96 | 22,283 |
| GSE27155 | 4 | 51 | USA | GPL96 | 22,283 |
| GSE53157 | 2 | 7 | Portugal | GPL570 | 54,675 |
| GSE60542 | 26 | 28 | Belgium | GPL570 | 54,675 |
| GSE33630 | 45 | 49 | Belgium | GPL570 | 54,675 |
Function enrichment analysis
The top 300 DEGs were uploaded to Database for Annotation, Visualization, and Integrated Discovery (DAVID) [32] for Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The top terms were visualized in “GOplot” [33] of R software.
Weighted co-expression network construction and key modules identification
1807 DEGs with P < 0.05 obtained from RRA to construct gene co-expression networks with expression data retrieved from The Cancer Genome Atlas (TCGA). Specifically, 1807 genes were selected from the TCGA gene list, and then the 1807 genes with their gene expression values and TCGA sample Numbers formed a correlation matrix. The threshold value of outliers was Z.K value < − 2.5. The correlation matrix (Sij) was converted to an adjacency matrix (Aij) based on soft threshold β that can approximate the scale-free distribution (R2 > 0.8). This transformation allows us to build networks with higher biological signals, which is the focus of the WGCNA approach. Topological overlap matrix (TOM) realizes the visualization of network, which is a simplified network diagram for identifying modules. The hierarchical clustering tree formed by average linkage hierarchical clustering also participates in the formation of modules. By the way, some genes without characteristics were assigned to the gray module. In order to identify the key modules closely related to PTC, module eigengene (ME), gene significance (GS), module membership (MM) and other parameters were calculated [34]. The genes in the key modules were uploaded to DAVID [32] for GO functional annotation and KEGG enrichment analysis to explore the biological functions of the key modules.
Identification, validation and efficacy evaluation for hub genes
The hub gene is defined as the gene with the highest degree of connectivity in the key module. Specifically, the genes with geneModuleMembership > 0.9 and geneTraitSignificance > 0.5 were determined as the hub genes in the study. Samples in the Cancer Genome Atlas-Thyroid carcinoma (TCGA-THCA) and a separate data set GSE29265 was used to verify that hub genes can distinguish between non-tumor and PTC. If the P-value < 0.01, the selection of the hub gene is considered statistically significant. In addition, we conducted a study to understand the expression patterns of hub genes between different stages of PTC based on GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/) [35], which was a web server for cancer and normal gene expression profiling and interactive analyses. To assess diagnostic values of hub genes, receiver operating characteristic (ROC) curve was plotted and area under the ROC curve (AUC) was calculated with “pROC” of R package [36] to evaluate the capability of distinguishing tumor and normal tissue.
Methylation analysis of hub genes
Methylation is one of the earliest and most widely studied epigenetics to be included in our study. We conducted methylation analysis of hub genes based on methylation data obtained from the human disease methylation database version 2.0 [37, 38] (DiseaseMeth 2.0, http://bioinfo.hrbmu.edu.cn/diseasemeth/), which collects and annotates the abnormal methylation of various cancers, is a useful resource platform for further understanding the molecular mechanisms of human disease. Data in the platform based on huge international disease projects including TCGA and public genome databases including GEO [37]. Furthermore, the relationship between hub genes expression and their DNA methylation status were investigated based on MEXPRESS (http://mexpress.be) [39].
Hub genes and tumor-infiltrating immune cells
Due to tumor immune infiltrating cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) are closely related to prognosis, thus it is necessary to investigate the correlation between the expression of identified hub genes and tumor infiltrating immune cells, which might provide new ideas for immunotherapy. This analysis for immune infiltrating cells and their interactions with tumor cells [40, 41] was performed on the basis of TIMER, which use to comprehensively investigate molecular characterization of tumor-immune interactions.
Gene set enrichment analysis
GSEA, which is widely used to predict the biological functions of hub genes, were performed in GSEA 4.0.3 and the top terms were visualized in R. The samples in TCGA were divided into two groups (high expression vs. low expression) with median expression values of each hub gene.
Results
Identification of robust DEGs
The P-value of each gene in 6 datasets was calculated based on the “limma” package. Gene lists of six datasets were integrated by RRA method and 1807 DEGs including 796 up-regulated genes and 1011 down-regulated genes were identified to the threshold of P-value < 0.05. The top 20 down-regulated and up-regulated genes were listed based on heat maps (Additional file 1: Figure S1).
Function enrichment analysis
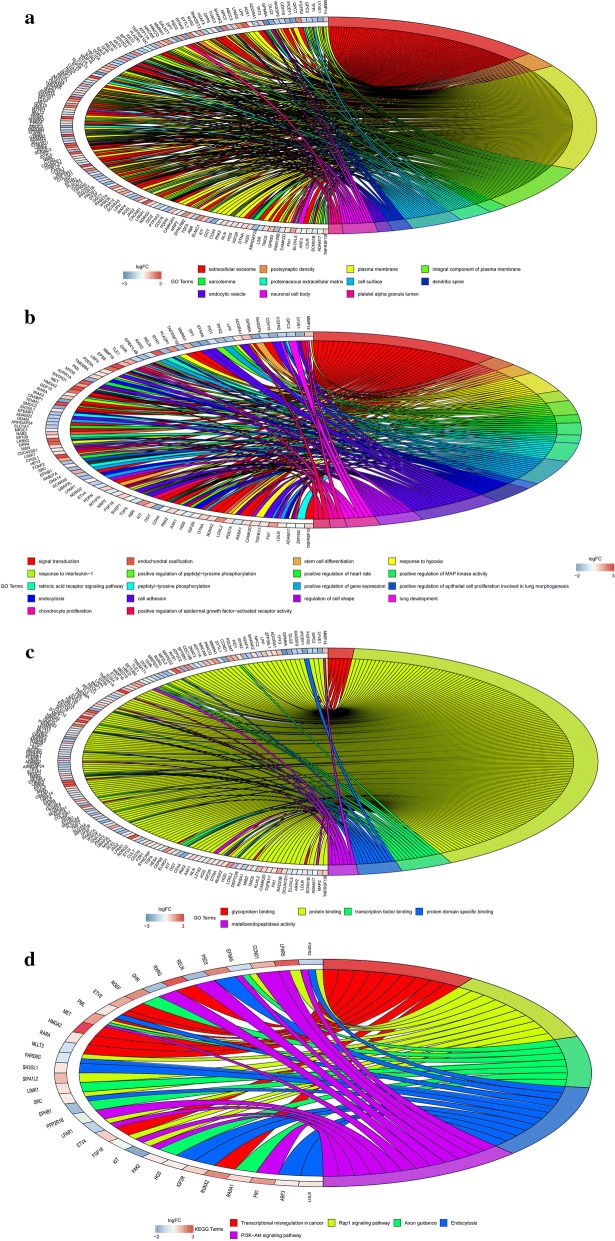
Chord diagrams showed the biological pathways in which the top 300 genes were involved. The top terms were illustrated in Fig. 1. Figure 1a shows that these genes are enriched into extracellular exosome, plasma membrane, integral component of plasma membrane etc. based on GO cellular components. Figure 1b shows that these genes are enriched into signal transduction, response to interleukin − 1, stem cell differentiation and positive regulation of MAP kinase activity etc. based on GO biological processes. Figure 1c shows that these genes are enriched to glycoprotein binding, protein binding and metalloendopeptidase activity etc. based on GO molecular function. These genes were enriched into the transcriptional misregulation in cancer, Rap1 signaling pathway and PI3K − Akt signaling pathway etc. based on KEGG (Fig. 1d).
Fig. 1.
Chord diagrams for GO and KEGG analysis of top 300 DEGs. The link between genes and pathways was described grounded on GO cellular components. a The link between genes and pathways was described grounded on GO biological processes. b The link between genes and pathways was described grounded on GO molecular function. c The link between genes and pathways was described grounded on KEGG. d Different genes and pathways are color-coded according to the catalog
Weighted co-expression network construction and key modules identification
Since there were 5 samples with Z.K value < − 2.5 (TCGA.KS.A41F, TCGA.EM.A3O3, TCGA.DJ.A2Q1, TCGA.DE.A4MA and TCGA.BJ.A3F0), these 5 samples were considered as outliers and excluded from the subsequent analysis (Additional file 1: Figure S2). Matrix transformation is based on soft threshold β = 7 (scale free R2 = 0.88) (Additional file 1: Figure S3), which is selected according to scale-free topological criteria. 10 gene modules were identified based on TOM and average linkage hierarchical clustering (Additional file 1: Figure S4). In order to identify the modules associated with clinical characteristics, the ME that represents the gene expression profile of each module was calculated. The MEblack (r = − 0.64, P = 5e − 38) and MEturquoise (r = − 0.65, P = 3e − 39) were considered to be the key modules most associated with thyroid cancer (Additional file 1: Figure S5). At the same time, the module membership vs. gene significance showed that the black and turquoise modules were closely related to the disease (Additional file 1: Figure S6). GO and KEGG indicated that black module (Fig. 2a) was mainly enriched to extracellular region, extracellular matrix and extracellular space etc. GO and KEGG indicated that turquoise module was (Fig. 2b) mainly enriched to retinoic acid receptor signaling pathway, oxidoreductase activity and cellular response to zinc ion etc.
Fig. 2.
Enrichment analysis of black and turquoise modules based on GO and KEGG. a Indicates enrichment analysis of black module, and b indicates enrichment analysis of turquoise module. The pathways (vertical axis) and rich factor (horizontal axis) were shown, and the size and color of nodes that represent genes were depicted according to the legends
Identification, validation and efficacy evaluation for hub genes
16 genes with geneModuleMembership > 0.9 and geneTraitSignificance > 0.5 were determined as the hub genes in Table 2. Figure 3 (TCGA-THCA) and Fig. 4 (GSE29265) intuitively showed that 16 hub genes are significantly down-regulated. Figure 5 ploted in GEPIA illustrated that there were some differences in expression patterns in different stages of PTC. The main difference was between stage I, II, and III: the expression patterns of ABCA6, PID1and TFPI were down in stage I–II–up in stage II–III, ACACB, BCL2, CASC2, ITPR1, MPPED2, MRO, PRKCQ, RMDN1, RNF150, RPS6KA6,SLC26A7, SLC4A4 and TTC30A were up in stage I–II–down in stage II–III (Table 2). Furthermore, ROC curves showed high diagnostic value of 16 hub genes for PTC (Additional file 1: Figure S7A, B): except for ABCA6 AUC = 89.5%, the 15 genes with AUC > 90%. To our knowledge, the eight hub genes (ABCA6, ACACB, TTC30A, RMDN1, RNF150, RPS6KA6, PID1 and TFPI) in Fig. 3 have been poorly studied. Therefore, we focused on these 8 hub genes in this study.
Table 2.
The list of sixteen hub genes identified in gene expression network
| Gene symbol | GeneModuleMembership | GeneTraitsignificance | Expression pattern |
|---|---|---|---|
| PID1 | 0.915896493 | 0.66001985 | Down in stage I–II–up in stage II–III |
| ABCA6 | 0.911773582 | 0.524366279 | Down in stage I–II–up in stage II–III |
| TFPI | 0.903220611 | 0.599708264 | Down in stage I–II–up in stage II–III |
| MPPED2 | 0.945621212 | 0.637198678 | Up in stage I–II–down in stage II–III |
| RPS6KA6 | 0.932048783 | 0.611335416 | Up in stage I–II–down in stage II–III |
| MRO | 0.931567859 | 0.632361438 | Up in stage I–II–down in stage II–III |
| RMDN1 | 0.923154211 | 0.610969324 | Up in stage I–II–down in stage II–III |
| ACACB | 0.919880602 | 0.610967988 | Up in stage I–II–down in stage II–III |
| SLC4A4 | 0.913670371 | 0.575022712 | Up in stage I–II–down in stage II–III |
| TTC30A | 0.912368213 | 0.563286233 | Up in stage I–II–down in stage II–III |
| RNF150 | 0.909497328 | 0.625793803 | Up in stage I–II–down in stage II–III |
| BCL2 | 0.909385573 | 0.588759014 | Up in stage I–II–down in stage II–III |
| CASC2 | 0.908891219 | 0.567289908 | Up in stage I–II–down in stage II–III |
| PRKCQ | 0.907179413 | 0.615140746 | Up in stage I–II–down in stage II–III |
| SLC26A7 | 0.903842658 | 0.648569813 | Up in stage I–II–down in stage II–III |
| ITPR1 | 0.900631876 | 0.587725656 | Up in stage I–II–down in stage II–III |
GeneModuleMembership, GeneTraitSignificance and expression pattern of hub genes were showed. The cut-off thresholds of hub genes were geneModuleMembership > 0.9 and geneTraitSignificance > 0.5, and all hub genes are significantly down-regulated
Fig. 3.
Sixteen hub genes were verified based on TCGA. The 16 hub genes were significantly down-regulated
Fig. 4.
Sixteen hub genes were verified based on GSE29265. The 16 hub genes were significantly down-regulated
Fig. 5.
Pathological stage plot of THCA from GEPIA
Methylation analysis of hub genes
As far as we know, DNA methylation shuts down the activity of some genes and demethylation induces gene expression. Additional file 1: Figure S8 shown that ABCA6, ACACB, RMDN1 and TFPI were significantly different (P < 0.05), so ABCA6, ACACB, RMDN1 and TFPI were defined as differentially methylated genes (DMGs). Moreover, the Additional file 1: Figure S9A–D illustrated that the expression levels of 4 DMGs are negatively correlated with DNA methylation in MEXPRESS. That indicate the abnormal down-regulation of DMGs likely resulted from hypermethylation.
Hub genes and tumor-infiltrating immune cells
Immune cells play an important role in the development and progression of tumors. Immunotherapy for cancer is becoming more and more important, so it is necessary to study the relationship between hub genes and immune cell infiltration. We studied the relationship between 8 hub genes and different immune cells based on TIMER. However, RMDN1was not involved in this analysis since hub genes RMDN1 is not recognized by TIMER. The seven hub genes were all correlated with the immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) and tumor purity in the study (Additional file 1: Figure S10A–G).
Gene set enrichment analysis
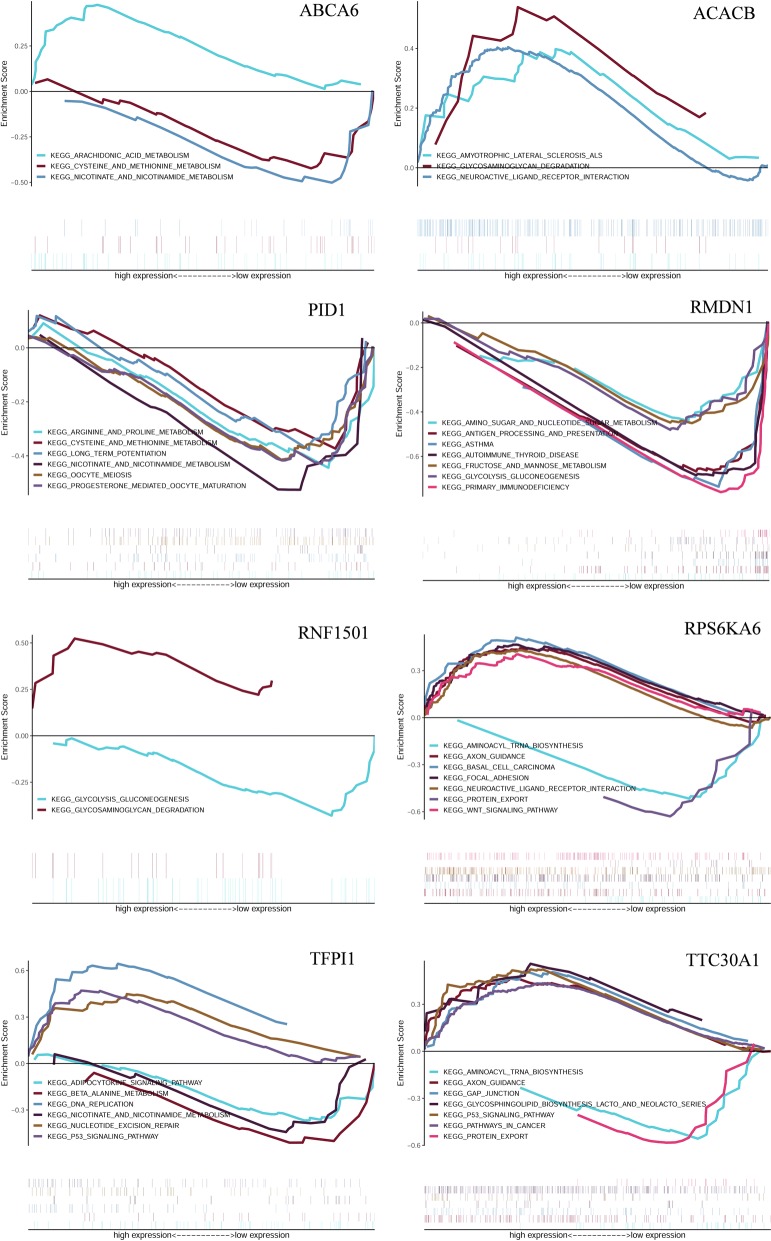
In order to further explore the expression pathways of 8 hub genes, we conducted GSEA, which is widely used to predict the biological functions of hub genes. These results suggest that these genes are associated with PTC (Fig. 6). ABCA6, PID1, RMDN1, RPS6KA6, TTC30A and TFPI were involved in amino acid metabolism. ACACB, RMDN1, RNF150, TTC30A have been shown to be involved in carbohydrate anabolism. PID1, TFPI and ABCA6 may be involved in the niacin and niacinamide metabolism.
Fig. 6.
Gene set enrichment analysis for hub gene
Discussion
The information of 6 data sets was integrated based on RRA, and 1807 DEGs were screened as the threshold of P-value < 0.05. In order to explore the specific biological role of these genes, the top 300 DEGs were analyzed by GO functional annotation and KEGG enrichment based on DAVID. Functional annotation of the top 300 DEGs may be helpful to the underlying mechanism of PTC. GO terms were obtained such as signal transduction, response to interleukin-1, positive regulation of MAP kinase activity, extracellular exosome, plasma membrane, integral component of plasma membrane and metalloendopeptidase activity. Signal transduction is very important in the occurrence of tumor [42]. Interleukin-1 plays a role in regulating innate immunity and adaptive immunity [43]. In particular, interleukin-1 has dual roles of anti-tumor and pro-tumor [43]. Positive regulation of MAP kinase activity and metalloendopeptidase activity are thought to be associated with thyroid cancer [44, 45]. This is a validation of the major carcinogenic event of PTC is the activation of MAPK. Meanwhile, KEGG terms such as transcriptional misregulation in cancer, Rap1 signaling pathway and PI3K-Akt signaling pathway were obtianed. PI3K-Akt signaling pathway had been reported to play an important role in the development of thyroid cancer [46, 47], breast cancer [48], colorectal cancer [49], non-small cell lung cancer [50] and gastric cancer [51] etc.
In order to identify the hub genes in the 1807 DEGs, 1807 genes were used to construct a gene co-expression network. MEblack and MEturquoise showed high correlations with PTC. Enrichment analysis showed that the genes in the black module were mainly related to extracellular matrix, and the genes in the turquoise module were enriched to retinoic acid receptor signaling pathway, oxidoreductase activity and cellular response to zinc ion etc. As the main bioactive metabolite of vitamin A, retinoic acid plays a regulatory role in proliferation and differentiation [52], and it has a profound impact on adipogenesis by activating retinoic acid receptors [53]. It is important to note that retinoic acid receptor signaling pathway may play a role in immune suppression. Previous studies have shown that abnormalities in the retinoic acid receptor signaling pathway are associated with various malignancies [54], but the specific mechanism between this pathway and PTC remains unclear. We identified 16 hub genes based on our selection criteria and ROC curve indicated that 16 hub genes had good diagnostic value, 8 of which were focused on. Not only did these genes differ significantly between tumours and non-tumours, but their expression patterns differed at different stages based on validation for hub genes. Methylation analysis showed that ABCA6, ACACB, RMDN1 and TFPI were identified as DMGs, and significant down-regulation of DMGs in patients with PTC might be realized by the hypermethylation. DNA methylation is reversible, so targeted therapies are attractive in cancer. The combination of genetic alterations and DNA methylation alterations may improve their clinical value. Therefore, 16 hub genes and DNA methylation of DMGs as potential biomarkers may be used to diagnose state (inert or invasive) of PTC for selecting the most appropriate treatment plan at present, so as to avoid overtreatment, improve the diagnosis of invasive tumor. This may provide new insights into refractory radioiodine differentiated tumors. The expression levels of 8 hub genes were significantly correlated with tumor purity, and these genes were moderately correlated with tumor-infiltrating immune cells. Furthermore, GSEA for hub genes predicted biological functions of these genes. ABCA6 belongs to the superfamily of ATP-binding cassette transporters and ABCA6 may be related to lipid homeostasis in macrophages [55]. Although some members of the superfamily of ATP-binding cassette transporters play important roles in tumor-generating mechanisms and drug resistance [56], the specific biological role of ABCA6 is not clearly understood. ACACB has been reported in laryngeal squamous cell carcinoma [57], nasopharyngeal carcinoma [58] and hepatocellular carcinoma [59]. PID1, a gene that regulates the sensitivity of fat and muscle cells to insulin signals, has been reported as a feature gene in several cancers, including thyroid cancer [60]. RPS6KA6, also known as RSK4, is considered as a tumor suppressor gene due to its resistance to invasion and metastasis, which may be related with inhibition of the MAPK pathway [61]. The activation of MAPK likely resulted from down-regulated of RPS6KA6. RPS6KA6 has been significantly down-regulated in colorectal cancer [62], ovarian cancer [63], non-small cell lung cancer [64], breast cancer, acute myeloid leukemia [65], etc., while this study is the first to mention that RPS6KA6 is significantly down-regulated in PTC. By the way, RPS6KA6 is also considered a DMG in esophageal cancer [66]. It is worth mentioning that RPS6KA6 was considered as a drug resistance marker for the treatment of cancer by protein kinase inhibitors in a study in 2012 [67]. In general, there has been little research on these 8 hub genes, especially RMDN1, TTC30A and RNF150, thus, it is imperative to fully uncover the specific relationship between hub genes and PTC.
Although WGCNA is widely used as a powerful data-driven tool for various diseases including various solid malignancies and hematologic malignancies, there is little research on establishing gene co-expression networks to identify genes that play a pivotal role in PTC. A new study using the WGCNA method to identify hub genes suggests that 11 hub genes may be involved in the recurrence of papillary thyroid cancer [68]. A total of 16 hub genes were identified in this study, and these 16 genes did not overlap with the above 11 genes. In particular, our study is the first to study PTC using a combination of RRA and WGCNA. However, we cannot deny that there may be bias due to the small sample sizes, more studies are needed to validate our results.
Conclusions
In summary, we identified 1807 DEGs from 6 datasets of PTC data based on RRA methods. From the gene co-expression network, we identified 16 hub genes, which were shown to be significantly down-regulated and expression patterns differed at different stages according to hub gene verification. ROC curve indicated that 16 hub genes had good diagnostic value. For methylation analysis, ABCA6, ACACB, RMDN1 and TFPI were identified as DMGs, and MEXPRESS indicated that decreased expression level of these genes may relate to abnormal DNA methylation. Hub genes and methylation of DMGs may as potential biomarkers provide new insights for diagnosis of PTC. The expression levels of 8 hub genes were correlated with tumor purity as well as tumor-infiltrating immune cells. Although these hub genes were found in PTC, the specific role of these hub genes in the underlying mechanism of PTC is not clear. Therefore, GSEA provides insights into the biological functions of hub genes, which might be the direction of future work. In addition, functional annotations for the top300 DEGs and key modules provide insight into the underlying mechanism of PTC. To further understand the pathogenesis of PTC is the key to adjust the current diagnosis, which can further improve the prognosis of PTC patients.
Supplementary information
Additional file 1: Figure S1. Heat map for top 20 down-regulated and top 20 up-regulated genes. Figure S2. Sample dendrogram and trait heatmap. Figure S3. Analysis of network topology for various soft-thresholding powers. Figure S4. Cluster dendrogram. Modules identified by WGCNA based on a dissimilarity measure (1-TOM). Figure S5. Heat map for the correlation between ME and clinical traits of thyroid cancer. Each gene module was colored according to legend. Figure S6. Scatter diagram for black and turquoise modules. Figure S7A-B. ROC curves for hub genes. Figure S8. Boxplot for methylation analysis of 8 hub genes. Figure S9A-D. Association of Methylation sites with expression of 4 differentially methylated genes (DMGs). Figure S10. The relationship between hub gene expression and tumor purity and tumor infiltrating immune cells.
Additional file 2: Table S1. Details of the six databases.
Acknowledgements
None.
Abbreviations
- PTC
Papillary thyroid cancer,
- MAPK
Mitogen-activated protein kinase,
- anti-CTLA-4
Anti-cytotoxic T lymphocyte antigen 4,
- anti-PD-1/PD-L1
Anti-programmed cell death protein-1/programmed cell death ligand-1
- BRAF
B-Raf proto-oncogene, serine/threonine kinase
- DAVID
Database for annotation, visualization, and integrated discovery
- DEGs
Differentially expressed genes
- ROC
Receiver operating characteristic
- AUC
Area under the ROC curve
- DMGs
Differentially methylated genes,
- GEPIA
Gene expression profiling interactive analysis
- GEO
Gene expression omnibus
- GO
Gene ontology
- GS
Gene significance
- GSEA
Gene set enrichment analysis
- KEGG
Kyoto encyclopedia of genes and genomes
- ME
Module eigengene
- MM
Module membership
- RRA
Robust rank aggregation
- TCGA
The cancer genome atlas
- TCGA-THCA
The cancer genome atlas-thyroid carcinoma
- TOM
Topological overlap matrix
- WGCNA
Weighted gene co-expression network analysis
Authors’ contributions
QW and CZ conceived and designed this study. YL, ZYY and TYC carried out the analysis procedure, TYC and YL analyzed the results, QW, CZ and WF contributed analysis tools, TYC and YL participated in the manuscript writing. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The data that support the findings of this study are openly available in GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structuralgenomics/tcga).
Ethics approval and consent to participate
Not required.
Consent for publication
That the article is original, has not already been published in a journal, and is not currently under consideration by another journal.
Conflicts of interest
All authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Liu and Ting-Yu Chen contributed equally to this study
Contributor Information
Qian Wu, Email: wuqian@xjtu.edu.cn.
Chao Zhang, Email: zhangchao0803@126.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12967-020-02327-7.
References
- 1.Kitahara CM, Devesa SS, Sosa JA. Increases in thyroid cancer incidence and mortality-reply. JAMA. 2017;318(4):390–391. doi: 10.1001/jama.2017.7910. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari SM, Fallahi P, Elia G, Ragusa F, Ruffilli I, Paparo SR, et al. Thyroid autoimmune disorders and cancer. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Riesco-Eizaguirre G, Santisteban P. ENDOCRINE TUMOURS: advances in the molecular pathogenesis of thyroid cancer: lessons from the cancer genome. Eur J Endocrinol. 2016;175(5):R203–R217. doi: 10.1530/EJE-16-0202. [DOI] [PubMed] [Google Scholar]
- 4.Zhang K, Li C, Liu J, Tang X, Li Z. DNA methylation alterations as therapeutic prospects in thyroid cancer. J Endocrinol Invest. 2019;42(4):363–370. doi: 10.1007/s40618-018-0922-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang LX, Wu J, Guo ML, Zhang Y, Ma SG. Suppression of long non-coding RNA TNRC6C-AS1 protects against thyroid carcinoma through DNA demethylation of STK4 via the Hippo signalling pathway. Cell Prolif. 2019;52(3):e12564. doi: 10.1111/cpr.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yim JH, Choi AH, Li AX, Qin H, Chang S, Tong ST, et al. Identification of tissue-specific DNA methylation signatures for thyroid nodule diagnostics. Clin Cancer Res. 2019;25(2):544–551. doi: 10.1158/1078-0432.CCR-18-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varricchi G, Loffredo S, Marone G, Modestino L, Fallahi P, Ferrari SM, et al. The immune landscape of thyroid cancer in the context of immune checkpoint inhibition. Int J Mol Sci. 2019;20(16):3934. doi: 10.3390/ijms20163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almubarak H, Qassem E, Alghofaili L, Alzahrani AS, Karakas B. Non-invasive molecular detection of minimal residual disease in papillary thyroid cancer patients. Front Oncol. 2019;9:1510. doi: 10.3389/fonc.2019.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celik M, Bulbul BY, Ayturk S, Durmus Y, Gurkan H, Can N, et al. The relation between BRAFV600E mutation and clinicopathological characteristics of papillary thyroid cancer. Medicinski Glasnik. 2020;17(1). [DOI] [PubMed]
- 10.Ge J, Wang J, Wang H, Jiang X, Liao Q, Gong Q, et al. The BRAF V600E mutation is a predictor of the effect of radioiodine therapy in papillary thyroid cancer. J Cancer. 2020;11(4):932–939. doi: 10.7150/jca.33105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ihara K. Immune checkpoint inhibitor therapy for pediatric cancers: a mini review of endocrine adverse events. Clin Pediatr Endocrinol. 2019;28(3):59–68. doi: 10.1297/cpe.28.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari SM, Fallahi P, Elia G, Ragusa F, Ruffilli I, Patrizio A, et al. Autoimmune endocrine dysfunctions associated with cancer immunotherapies. Int J Mol Sci. 2019;20(10):2560. doi: 10.3390/ijms20102560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Wang Z, Jia E, Ouyang T, Pan M, Lu J, et al. Analysis of genome-wide in cell free DNA methylation: progress and prospect. The Analyst. 2019;144(20):5912–5922. doi: 10.1039/C9AN00935C. [DOI] [PubMed] [Google Scholar]
- 14.Chandhok NS, Prebet T. Insights into novel emerging epigenetic drugs in myeloid malignancies. Ther Adv Hematol. 2019;10:2040620719866081. doi: 10.1177/2040620719866081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song ZY, Chao F, Zhuo Z, Ma Z, Li W, Chen G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging. 2019;11(13):4736–4756. doi: 10.18632/aging.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinform. 2012;28(4):573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Lu Q, Shao X, Mo B, Nie X, Liu W, et al. Development of a three-gene prognostic signature for hepatitis B virus associated hepatocellular carcinoma based on integrated transcriptomic analysis. J Cancer. 2018;9(11):1989–2002. doi: 10.7150/jca.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinform. 2007;23(23):3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 20.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–1040. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 22.Vasko V, Espinosa AV, Scouten W, He H, Auer H, Liyanarachchi S, et al. Gene expression and functional evidence of epithelial-to-mesenchymal transition in papillary thyroid carcinoma invasion. Proc Natl Acad Sci USA. 2007;104(8):2803–2808. doi: 10.1073/pnas.0610733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusinek D, Swierniak M, Chmielik E, Kowal M, Kowalska M, Cyplinska R, et al. BRAFV600E-Associated gene expression profile: early changes in the transcriptome, based on a transgenic mouse model of papillary thyroid carcinoma. PLoS ONE. 2015;10(12):e0143688. doi: 10.1371/journal.pone.0143688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, Wilhelm KG, Jr, et al. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12(7 Pt 1):1983–1993. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 25.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, et al. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24(44):6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- 26.Pita JM, Banito A, Cavaco BM, Leite V. Gene expression profiling associated with the progression to poorly differentiated thyroid carcinomas. Br J Cancer. 2009;101(10):1782–1791. doi: 10.1038/sj.bjc.6605340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarabichi M, Saiselet M, Tresallet C, Hoang C, Larsimont D, Andry G, et al. Revisiting the transcriptional analysis of primary tumours and associated nodal metastases with enhanced biological and statistical controls: application to thyroid cancer. Br J Cancer. 2015;112(10):1665–1674. doi: 10.1038/bjc.2014.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dom G, Tarabichi M, Unger K, Thomas G, Oczko-Wojciechowska M, Bogdanova T, et al. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107(6):994–1000. doi: 10.1038/bjc.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomas G, Tarabichi M, Gacquer D, Hebrant A, Dom G, Dumont JE, et al. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene. 2012;31(41):4490–4498. doi: 10.1038/onc.2011.626. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 33.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinform. 2015;31(17):2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 34.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):w98. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Wei Y, Gu Y, Zhang S, Lyu J, Zhang B, et al. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic acids research. 2017;45(D1):D888–d895. doi: 10.1093/nar/gkw1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv J, Liu H, Su J, Wu X, Liu H, Li B, et al. DiseaseMeth: a human disease methylation database. Nucleic acids research. 2012;40(Database issue):D1030–D1035. doi: 10.1093/nar/gkr1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch A, Jeschke J, Van Criekinge W, van Engeland M, De Meyer T. MEXPRESS update 2019. Nucleic Acids Res. 2019;47(W1):W561. doi: 10.1093/nar/gkz445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Cruz Silva E, Dontenwill M, Choulier L, Lehmann M. Role of integrins in resistance to therapies targeting growth factor receptors in cancer. Cancers. 2019;11(5):692. doi: 10.3390/cancers11050692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker KJ, Houston A, Brint E. IL-1 family members in cancer; two sides to every story. Front Immunol. 2019;10:1197. doi: 10.3389/fimmu.2019.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanly EK, Rajoria S, Darzynkiewicz Z, Zhao H, Suriano R, Tuli N, et al. Disruption of mutated BRAF signaling modulates thyroid cancer phenotype. BMC Res Notes. 2014;7:187. doi: 10.1186/1756-0500-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roncevic J, Djoric I, Selemetjev S, Jankovic J, Dencic TI, Bozic V, et al. MMP-9-1562 C/T single nucleotide polymorphism associates with increased MMP-9 level and activity during papillary thyroid carcinoma progression. Pathology. 2019;51(1):55–61. doi: 10.1016/j.pathol.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Li Q, Jin X, Guo H, Li Y. Long non-coding RNA H19 knockdown inhibits the cell viability and promotes apoptosis of thyroid cancer cells through regulating the PI3K/AKT pathway. Exp Ther Med. 2019;18(3):1863–1869. doi: 10.3892/etm.2019.7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Yan X, Zhang H, Zhan X. CircRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med Sci Monit. 2019;25:1342–1349. doi: 10.12659/MSM.913463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He H, Wang X, Chen J, Sun L, Sun H, Xie K. High-Mobility group box 1 (HMGB1) promotes angiogenesis and tumor migration by regulating hypoxia-inducible factor 1 (HIF-1alpha) expression via the Phosphatidylinositol 3-Kinase (PI3K)/AKT signaling pathway in breast cancer cells. Med Sci Monit. 2019;25:2352–2360. doi: 10.12659/MSM.915690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Q, Yao W, Yu C, Zou L, Shen Y, Zhu Y, et al. Identification of microRNA-181 as a promising biomarker for predicting the poor survival in colorectal cancer. Cancer Med. 2019;8(13):5995–6009. doi: 10.1002/cam4.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F, Geng J, Xu S, Meng Q, Chen K, Liu F, et al. FAM83A signaling induces epithelial-mesenchymal transition by the PI3K/AKT/Snail pathway in NSCLC. Aging. 2019;11:6069. doi: 10.18632/aging.102163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan HM, Lang WY, Yao LJ, Wang Y, Li XL. shRNA-interfering LSD1 inhibits proliferation and invasion of gastric cancer cells via VEGF-C/PI3K/AKT signaling pathway. World J Gastrointest Oncol. 2019;11(8):622–633. doi: 10.4251/wjgo.v11.i8.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmutzler C, Brtko J, Bienert K, Kohrle J. Effects of retinoids and role of retinoic acid receptors in human thyroid carcinomas and cell lines derived therefrom. Exp Clin Endocrinol Diabetes. 1996;104(4):16–19. doi: 10.1055/s-0029-1211693. [DOI] [PubMed] [Google Scholar]
- 53.Wang B, Yang Q, Harris CL, Nelson ML, Busboom JR, Zhu MJ, et al. Nutrigenomic regulation of adipose tissue development—role of retinoic acid: a review. Meat Sci. 2016;120:100–106. doi: 10.1016/j.meatsci.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.di Masi A, Leboffe L, De Marinis E, Pagano F, Cicconi L, Rochette-Egly C, et al. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol Aspects Med. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Kaminski WE, Wenzel JJ, Piehler A, Langmann T, Schmitz G. ABCA6, a novel a subclass ABC transporter. Biochem Biophys Res Commun. 2001;285(5):1295–1301. doi: 10.1006/bbrc.2001.5326. [DOI] [PubMed] [Google Scholar]
- 56.Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Nat Cancer Instr. 2014 doi: 10.1093/jnci/dju149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding Y, Wu Y, Gao W, Zhang C, Zhao Q, Guo H, et al. Analysis of gene expression profiling variations induced by hsamiR1455poverexpression in laryngeal squamous cell carcinoma cell line Tu177. Mol Med Rep. 2017;16(5):5863–5870. doi: 10.3892/mmr.2017.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge Y, He Z, Xiang Y, Wang D, Yang Y, Qiu J, et al. The identification of key genes in nasopharyngeal carcinoma by bioinformatics analysis of high-throughput data. Mol Biol Rep. 2019;46(3):2829–2840. doi: 10.1007/s11033-019-04729-3. [DOI] [PubMed] [Google Scholar]
- 59.Pan WY, Zeng JH, Wen DY, Wang JY, Wang PP, Chen G, et al. Oncogenic value of microRNA-15b-5p in hepatocellular carcinoma and a bioinformatics investigation. Oncol Lett. 2019;17(2):1695–1713. doi: 10.3892/ol.2018.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen JX, Li XQ, Chang Y. Signature gene identification of cancer occurrence and pattern recognition. J Comput Biol. 2018;25(8):907–916. doi: 10.1089/cmb.2017.0261. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Vicente L, Pons B, Coch L, Teixido C, Hernandez-Losa J, Armengol G, et al. RSK4 inhibition results in bypass of stress-induced and oncogene-induced senescence. Carcinogenesis. 2011;32(4):470–476. doi: 10.1093/carcin/bgr003. [DOI] [PubMed] [Google Scholar]
- 62.Cai J, Ma H, Huang F, Zhu D, Zhao L, Yang Y, et al. Low expression of RSK4 predicts poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2014;7(8):4959–4970. [PMC free article] [PubMed] [Google Scholar]
- 63.Arechavaleta-Velasco F, Zeferino-Toquero M, Estrada-Moscoso I, Imani-Razavi FS, Olivares A, Perez-Juarez CE, et al. Ribosomal S6 kinase 4 (RSK4) expression in ovarian tumors and its regulation by antineoplastic drugs in ovarian cancer cell lines. Med Oncol. 2016;33(2):11. doi: 10.1007/s12032-015-0724-6. [DOI] [PubMed] [Google Scholar]
- 64.Li A, Liu D, Liu Y, Zhou Y, Du Z, Song J. A pilot study of RSK4 expression in patients with human non-small cell lung carcinoma. Ann Clin Lab Sci. 2018;48(4):484–489. [PubMed] [Google Scholar]
- 65.Rafiee M, Keramati MR, Ayatollahi H, Sadeghian MH, Barzegar M, Asgharzadeh A, et al. Down-regulation of ribosomal S6 kinase RPS6KA6 in acute myeloid leukemia patients. Cell J. 2016;18(2):159–164. doi: 10.22074/cellj.2016.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xi T, Zhang G. Epigenetic regulation on the gene expression signature in esophagus adenocarcinoma. Pathol Res Pract. 2017;213(2):83–88. doi: 10.1016/j.prp.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 67.Bender C, Ullrich A. PRKX, TTBK2 and RSK4 expression causes Sunitinib resistance in kidney carcinoma- and melanoma-cell lines. Int J Cancer. 2012;131(2):E45–E55. doi: 10.1002/ijc.26486. [DOI] [PubMed] [Google Scholar]
- 68.Zhai T, Muhanhali D, Jia X, Wu Z, Cai Z, Ling Y. Identification of gene co-expression modules and hub genes associated with lymph node metastasis of papillary thyroid cancer. Endocrine. 2019;66(3):573–584. doi: 10.1007/s12020-019-02021-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Heat map for top 20 down-regulated and top 20 up-regulated genes. Figure S2. Sample dendrogram and trait heatmap. Figure S3. Analysis of network topology for various soft-thresholding powers. Figure S4. Cluster dendrogram. Modules identified by WGCNA based on a dissimilarity measure (1-TOM). Figure S5. Heat map for the correlation between ME and clinical traits of thyroid cancer. Each gene module was colored according to legend. Figure S6. Scatter diagram for black and turquoise modules. Figure S7A-B. ROC curves for hub genes. Figure S8. Boxplot for methylation analysis of 8 hub genes. Figure S9A-D. Association of Methylation sites with expression of 4 differentially methylated genes (DMGs). Figure S10. The relationship between hub gene expression and tumor purity and tumor infiltrating immune cells.
Additional file 2: Table S1. Details of the six databases.
Data Availability Statement
The data that support the findings of this study are openly available in GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structuralgenomics/tcga).