Abstract
Long noncoding RNA molecules (lncRNAs) are estimated to account for the majority of eukaryotic genomic transcripts, and have been associated with multiple diseases in humans. However, our understanding of their structure–function relationships is scarce, with structural evidence coming mostly from indirect biochemical approaches or computational predictions. Here we describe direct visualization of the lncRNA HOTAIR (HOx Transcript AntIsense RNA) using atomic force microscopy (AFM) in nucleus-like conditions at 37°. Our observations reveal that HOTAIR has a discernible, although flexible, shape. Fast AFM scanning enabled the quantification of the motion of HOTAIR, and provided visual evidence of physical interactions with genomic DNA segments. Our report provides a biologically plausible description of the anatomy and intrinsic properties of HOTAIR, and presents a framework for studying the structural biology of lncRNAs.
Keywords: atomic force microscopy, HOTAIR, long noncoding RNA
INTRODUCTION
Long noncoding RNA (lncRNA) molecules are defined as RNA transcripts longer than 200 nt, which lack an evident ORF and are transcribed by RNA polymerase II. These large transcripts are often also polyadenylated and spliced. Thousands of lncRNAs are annotated to date, but only a few have been studied and defined as functional. Functional lncRNAs are involved in almost every stage of gene expression (Amaral and Mattick 2008; Dinger et al. 2008; Hung and Chang 2010; Fatica and Bozzoni 2014) and have been implicated in a variety of diseases such as cancer and neurodegenerative disorders.
Despite their abundance and emerging importance, our knowledge concerning lncRNA structure is poor. Existing information on the structure of large RNA molecules in general is scarce, with <7% of all RNA structures in the Protein Data Bank being in the size range between 200 and 5000 nt, most of these being ribosomal RNA subunits (Ban et al. 2000) studied by X-ray crystallography. The structure of MALAT1 has been resolved by crystallography (Brown et al. 2014), but available information on other lncRNA structures derives mostly from indirect methods. For example, the structure of SRA (Novikova et al. 2012) and HOTAIR (Somarowthu et al. 2015) were depicted through biochemical methods. Another direction taken to further study lncRNAs such as XIST (Wutz et al. 2002) and GAS5 (Kino et al. 2010) was domain analysis. On the other hand, it has recently been suggested, using a statistical approach, that lncRNAs may not have a structure at all (Rivas et al. 2017). This discrepancy may result from a lack of solid structural information, and its resolution could shed light on the biology of this important class of molecules.
In this work we aimed to obtain such information using one of the most studied lncRNAs, HOTAIR, as a test case. HOTAIR has been shown to bind PRC2 and LSD1 (Tsai et al. 2010) to drive chromatin modification at specific genomic sites (Chu et al. 2011), thus playing a key role in genome silencing. HOTAIR DNA binding sites are focal, specific and numerous, implying HOTAIR as a silencing selector element pinpointing genomic locations to modification (Chu et al. 2011). HOTAIR was shown to be required for the epithelial-to-mesenchymal transition (Pádua Alves et al. 2013; Battistelli et al. 2016), thereby defined as an oncogene and a negative prognostic marker in various cancers (Liu et al. 2013; Wan 2013; Wu et al. 2014).
Our central tool in this study was atomic force microscopy (AFM). AFM has been used to study nucleic acid structures in both fluid and air (Lyubchenko et al. 2011), including the genomic RNA of human immunodeficiency virus (HIV)-1 (Andersen et al. 2004). While AFM is limited in resolution compared with X-ray crystallography or NMR, it enables direct visualization by physically probing native, large molecules under biological conditions. AFM also allows statistical analysis of structurally diverse molecules, as previously suggested to be the case for lncRNAs (Novikova et al. 2013).
RESULTS
HOTAIR has a distinct anatomy
We first generated HOTAIR molecules by in vitro transcription (IVT), using multiple templates and multiple IVT systems in order to avoid method-biased observations. The resulting transcripts were analyzed by gel electrophoresis and RNA-seq and found to be intact and to fully map to the HOTAIR gene sequence in a human reference genome (Supplemental Notes 1, 2). HOTAIR molecules were then scanned by AFM in fluid, under conditions that mimic the chemical environment of the nucleus as reliably as possible (Alberts et al. 2002; Cowan 2002; J Cowan, pers. comm.) and at 37°C (Supplemental Note 3).
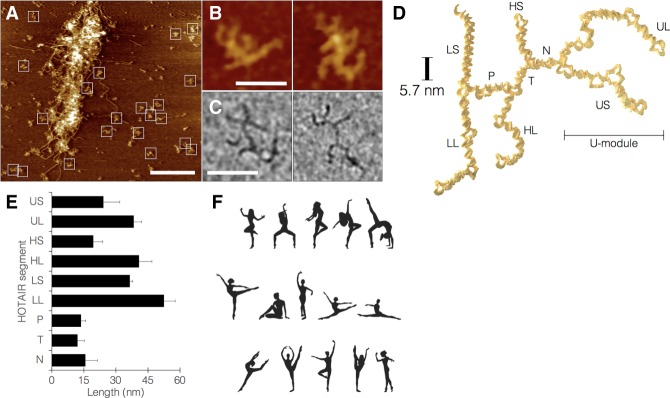
AFM scanning demonstrated that under these conditions, HOTAIR molecules assume a distinct anatomy (Fig. 1A,B), based on a four-limbed body which ends in a branched U-shaped motif, which we termed the U-module. Visualization of HOTAIR by Cryo-EM showed the same anatomy and flexibility observed in AFM (Fig. 1C; Supplemental Note 4). This anatomy was reproduced in seven independent synthesis and scanning repeats. In contrast, a random RNA transcript formed by scrambling HOTAIR sequence formed indistinct shapes and aggregates, with some even not folding (Supplemental Note 5). The archetypal HOTAIR anatomy could be reliably assigned to ∼66% of the observed objects that were intact based on size, with excluded ones being either clear but eccentric or too vague to be reliably assigned. The presence of eccentric forms could reflect that not all HOTAIR products fold properly in the nucleus and are subsequently dysfunctional; however, the folding of HOTAIR in the nucleus could be facilitated by yet unidentified cellular factors. In addition to the whole molecule, the functional HOTAIR modules suggested in previous studies (Tsai et al. 2010; Chu et al. 2011; Wu et al. 2013; Kalwa et al. 2016) were also scanned, showing distinct structures (Supplemental Note 6, Movie S1–S3).
FIGURE 1.
Visualization and modeling of the shape and motion of HOTAIR by AFM. (A) Field imaged at lower magnification showing free HOTAIR molecules, squares mark objects reliably assigned as properly folded HOTAIR. Large mass in the center of the field is an aggregate of dsDNA and HOTAIR RNA (bar = 500 nm). (B) Representative higher magnification images of HOTAIR molecules showing their distinct anatomy (bar = 50 nm). (C) Representative cryo-EM images of HOTAIR (bar = 50 nm). (D) Proposed molecular model of HOTAIR, divided into nine structural segments: (UL) U long, (US) U short, (N) neck, (HS) hand short, (HL) hand long, (T) torso, (P) pelvis, (LS) leg short, (LL) leg long. (E) Structural flexibility of HOTAIR expressed as variation in segment lengths. (F) The human figures frozen in various configurations are analogous to the concept of HOTAIR as a molecule with a distinct anatomy but also high flexibility.
Our HOTAIR model divides the molecule into nine structural segments, which we name as follows: neck (N), torso (T), pelvis (P), leg long (LL), leg short (LS), hand long (HL), hand short (HS), U-module long (UL), U-module short (US) (Fig. 1D). Total length of all segments across multiple samples was constrained to 252 ± 9.4 nm, but the observed variance within segments was 5% to 50% of the mean segment length (shorter segments exhibited higher variance) (Fig. 1E). The high degree of flexibility in our proposed model of HOTAIR may seem contradictory to the conventional meaning of a molecule having a structure; however, this notion is contained within a set of objects that are well-defined anatomically but are globally flexible. A useful analogy is shown here by human dancers frozen in various configurations (Fig. 1F), who also combine these two properties.
Dimensions of HOTAIR
The molecular dimensions of HOTAIR were measured by an algorithm that combined the absolute size calculated by the AFM with a dsDNA molecule as an internal size reference. We chose to use a dsDNA molecule termed HOTAIR-binding DNA 1 (HBD1) (Liu et al. 2013), which we synthesized for this study (Supplemental Note 7). HBD1 is a 433 bp molecule existing preferentially at the B-DNA geometry (helical rise of 3.5 Å per base with 10.5 bp/turn, yielding a longitudinal density of 3 bp/nm) under the study conditions, thus mapping to 144 nm in length. In contrast, HOTAIR, a 2158 (Rinn et al. 2007) nt RNA molecule which is mostly dsRNA, preferentially exists at the A-DNA geometry (helical rise of 2.6 Å per base with 11 bp/turn, yielding a longitudinal density of 4.23 bp/nm). Our measurements (Supplemental Note 8) showed a mean length of 142 nm for HBD1, and 252 nm for HOTAIR, the latter translating to 1066 bp of dsRNA, which theoretically unfold to 2132 nt of ssRNA. Taken together, this calculation represents a ∼1.5% error in molecular measurements by AFM under the study conditions.
Interaction between HOTAIR and DNA
It is critical to note that our aim in this study was to report intrinsic properties of HOTAIR, particularly its ability to interact with genomic DNA, dissociated from the suggested role of auxiliary proteins in this function, which is still unclear. For example, a recent study reported that genome targeting by HOTAIR is independent of at least one specific protein it interacts with, EZH2, although it did not rule out other proteins such as those that are part of the complex LSD1 (Chu et al. 2011). With that in mind, our aim here was first to describe the structural “baseline” of HOTAIR, on top of which future investigations of the functional complexes it forms inside the cell could be carried out.
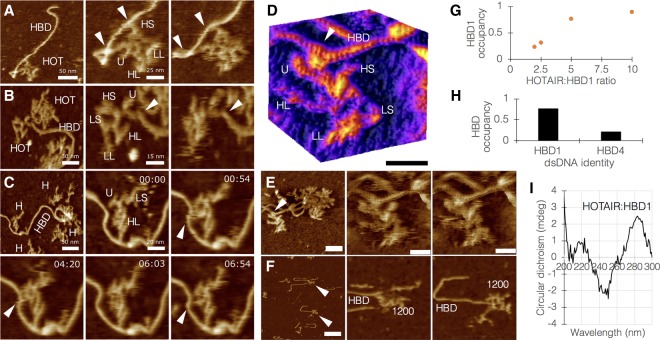
To this end, two dsDNA sequences from a previous study (Chu et al. 2011) were used, one that was found to highly associate with HOTAIR and one that was found not to, termed HOTAIR-binding DNA 1 (HBD1) and HBD4, respectively (Supplemental Note 7). HBD1 and HBD4 were allowed to interact with HOTAIR for short times, up to 30 min at various HOTAIR:HBD ratios, and fast AFM scanning was used to count temporally and positionally defined interactions (remaining bound at the same position along several scan frames). AFM scans demonstrated clear physical interaction between HOTAIR and DNA (Fig. 2A–C; Supplemental Movies S4–S6). Several interesting features of these interactions were observed. First, assignments based on plausible configurations combined with length measurements revealed that HOTAIR:DNA interactions appear to be mediated by the U-module (Fig. 2D,E). In some cases, a second interaction is seen mediated by an H segment (Fig. 2A,C). Second, the 1200 nt domain of HOTAIR, previously shown to associate with DNA (Kalwa et al. 2016), exhibited this ability under the study conditions, suggesting that the U-module maps to this specific domain (Fig. 2F; Supplemental Movie S7). Third, the occupancy of HBD1 by HOTAIR was ratio-dependent (Fig. 2G), suggesting a real biological phenomenon. Finally, the occupancy of HBD1 by HOTAIR was 3.2-fold higher than that of HBD4 (46 occupied out of 60 total HBD1 molecules counted vs. 16 occupied out of 66 total HBD4 counted) (Fig. 2H). Interestingly, HBD1 occupancy by the 1200 nt domain was 18 out of 32 total HBD1 counted, suggesting that an additional domain, such as an H segment as observed here, contributes to the higher binding of the whole molecule. The HOTAIR:HBD interactions observed by AFM were corroborated by flow cytometry, showing that HBD1, but not HBD4, binds HOTAIR (Supplemental Fig. S6).
FIGURE 2.
Characterization of the physical interaction between HOTAIR and dsDNA. (A–C) Fast scanning AFM images of HOTAIR interacting with HOTAIR-binding DNA 1 (HBD1). White arrowheads point at sites of interaction. (B) Center panel is at preinteraction; right panel is at interaction. (C) Time series showing a live interaction. HOT, HOTAIR. HBD, HOTAIR-binding DNA 1. Reliably assigned HOTAIR segments are denoted by their abbreviation. HOT (ag) shows HOTAIR molecules in aggregate. (D) 3D surface plot of HOTAIR:HBD1 interaction; white arrowhead points at interaction site. The light–dark stripe pattern on HBD and HOTAIR is the actual major/minor grooves (respectively) that build the double helical structure of dsDNA and dsRNA (bar = 20 nm). Image processing details can be found in Supplemental Note 3. (E) Raw specimen used for D, HOTAIR and HBD1 currently at interaction. White arrowhead points to the interaction site (left bar = 35 nm; center and right bars = 15 nm). (F) Binding of the 1200 nt domain of HOTAIR to HBD1 (bar = 70 nm). (G) Occupancy of HBD1 by various HOTAIR:HBD1 ratios shows ratio dependence (n from 2 to 10 = 73, 240, 60, and 18). (H) Occupancy of HBD1 versus HBD4 by HOTAIR at a 5:1 ratio (n = 60 vs. 66). (I) Circular dichroism spectrum of HOTAIR:HBD1, showing evidence of a physical triplex between ssRNA and dsDNA (negative peak at 210 nm, shifted positive peak at 280 nm).
Evidence for a triplex helix mediating HOTAIR:DNA interaction
HOTAIR:DNA interactions have been proposed to be mediated through a triple helix structure (Chu et al. 2011). A more recent study (Kalwa et al. 2016) showed by electrophoretic mobility shift assay that HOTAIR segments may form RNA–DNA–DNA triplexes, however in a nonbiological system (e.g., boiling to 60°C and cooling). Hoping to shed light on this mechanism, we initially used the Triplexator (Buske et al. 2012) package to perform sequence-based predictions of potential sites for triplex formation between HOTAIR and HBD1. Triplexator retrieved three potential triplex scenarios, all within the HBD1-binding, 1200 nt domain of HOTAIR. Out of these, two were biologically probable, that is, sequences with sufficient guanine residues to support Hoogsteen and reverse-Hoogsteen base-pairing, and antiparallel configuration. Examination of the highest-scoring combination of oligonucleotides by circular dichroism (CD) spectroscopy showed evidence of a formed triplex involving ssRNA and dsDNA, namely a negative peak at 210 nm and a shifted positive peak at 280 nm (Fig. 2I; Supplemental Note 9).
Motion of HOTAIR using fast-scan AFM
Our observations of HOTAIR revealed a highly flexible molecule, which exhibited a striking diversity of configurations, albeit converging to the same anatomy. A central question is thus whether the known mechanics of RNA allows for such flexibility. Previous works have reported that the flexibility of dsRNA is lower than that of dsDNA, and measurements by orthogonal techniques have yielded persistence length values around 62 nm (Hagerman 1997; Abels et al. 2005), which is longer than the observed discrete segments of HOTAIR. However, these estimations may not properly reflect the behavior of biological RNA molecules. Computational predictions, as well as recent experimental works that include biochemical methods and direct imaging (Andersen et al. 2004; Somarowthu et al. 2015), show that these molecules are rich in unpaired loops of varying sizes, which can be thought of as mechanical joints that allow for the observed flexibility. In order to quantitate this freedom of motion within our experimental system, we measured the range of movement of a single RNA joint in the AFM. We scanned 12-nt RNA joints connected at the edges of DNA origami rectangles used as AFM imaging guides. Segments connected by these joints were able to pivot up to approximately ±100° in the study conditions (Supplemental Note 10). Given the fact that, based on biochemical analysis as well as computational predictions applied locally (Supplemental Note 11), HOTAIR is rich in such joints, its actual flexibility is likely significantly larger than that predicted for an idealized dsRNA shaft.
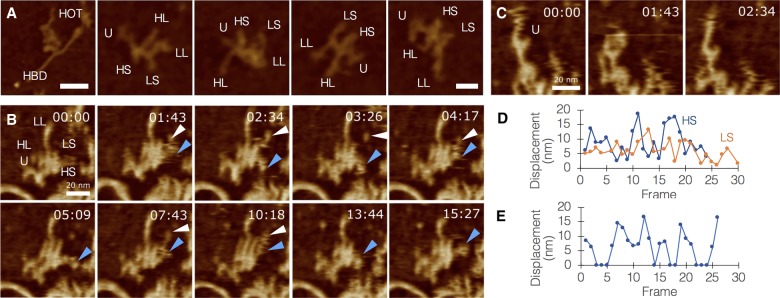
We utilized the fast scanning capability of our AFM system in order to quantitate the dynamics of minimally constrained HOTAIR molecules under biological conditions. The mica surface was covered with poly-L-ornithine in order to pin down, as quickly as possible, the DNA–RNA complexes that formed during the short incubation time. Once the complex is captured by the mica, DNA is no longer moving, whereas RNA appears as softer, smaller, shows a complex 3D structure with multiple nonbinding points allowing it to move more freely and adopt a range of possible configurations. Our initial observations revealed a very diverse range of HOTAIR morphologies (Fig. 3A), and further investigation into the dynamics of these molecules demonstrated that this diversity most likely derives from movement, and not degradation or misfolding (Supplemental Movies S8–S10). All limbs of HOTAIR are capable of pivoting around joints, extending, or retracting (Fig. 3B; Supplemental Movie S8), including the U-module, which exhibited pincer-like motion and at least one clear joint in segment UL (Fig. 3C). These movements occupied a radius of up to 20 nm from body (Fig. 3D,E).
FIGURE 3.
Quantitating the flexibility of HOTAIR. (A) A range of HOTAIR morphologies, segment names are abbreviated. Left panel shows HOTAIR interacting with HBD1 through the U-module (left panel bar = 50 nm; other panels bar = 20 nm). (B) Fast AFM scan of HOTAIR, tracking the movement of segments HS (blue arrowheads) and LS (white arrowheads; bar = 20 nm). (C) Fast AFM scan of HOTAIR focusing on movements of the U-module, exhibiting pincer-like opening/closure and a joint in segment UL (bar = 25 nm). (D) Quantitative analysis of the displacement (nm) of HL/HS movements. Numbers were corrected to account for center-mass drift. (E) Quantitative analysis of U-module behavior (0 nm represents closed state; other numbers represent varying levels of opening).
Visualizing RNA:DNA structures by AFM
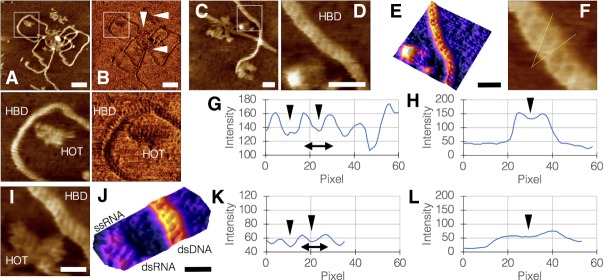
Our AFM scans enabled direct observations of structural configurations of DNA and RNA under the study conditions, as well as the identification of differences between their material properties using phase imaging. In this imaging mode, the phase difference between the cantilever and the drive signal is measured, a measurement sensitive to the stiffness or softness of the sample, providing an additional layer of information in samples where height may be equal throughout, but that are made of various materials. Phase imaging of our samples was able to discriminate between DNA and RNA based on the fact that the DNA is more rigid than RNA (Fig. 4A,B). For this reason, we frequently used phase imaging in complex samples to be able to reliably determine molecular identity of sample objects.
FIGURE 4.
High-resolution imaging of DNA and RNA by AFM. (A,B) HOTAIR molecules interacting with HBD1, imaged by height (A) and phase (B), the latter showing differences between HBD1 and HOTAIR molecules based on the higher rigidity of DNA compared with the softer RNA. Bottom panels are magnifications of white squares in A and B (bar = 50 nm). (C,D) Segment from a HOTAIR:HBD1 sample focusing on the dsDNA structure, D is a magnification of white square in C, showing the right-handed double helix (bar in C = 20 nm; bar in D = 10 nm). (E) 3D surface plot of D (bar = 10 nm). (F) Helical twist angle measurement, yielding 31.89° relative to helix plane. (G) Plot profile along the axis showing the major grooves (black arrowheads) and helical pitch (double-sided arrow) of 3.76 nm. (H) Plot profile orthogonal to the axis. Black arrowhead pointing at oblique segment of a major groove. (I) Segment of a HOTAIR:HBD1 sample showing dsDNA, dsRNA, and ssRNA together in a single frame (bar = 10 nm). (J) 3D surface plot of I, emphasizing the different structures in the sample (bar = 10 nm). (K) Plot profile along the axis showing the major grooves (black arrowheads) and helical pitch (double-sided arrow) of 2.89 nm. (L) Plot profile orthogonal to the axis. Black arrowhead pointing at oblique segment of a major groove. Note the lower profile of dsRNA compared with dsDNA (H) due to RNA being softer than DNA.
Multiple scans have repeatedly shown the right-handed, double helical structure of DNA under the study conditions in exquisite detail (Fig. 4C–E). Our measurements yielded a dsDNA helical twist angle of 31.89° relative to helix plane, which is within the observed range (Dickerson and Klug 1983) of 27.7°–42.0° (Fig. 4F). Measured mean helical pitch was 3.76 nm, within 3 Å of the accepted value of 3.4 nm for B-DNA (Fig. 4G,H). Measurements of dsRNA (Fig. 4I,J) yielded a helical twist angle of 18.86° relative to helix plane, within the observed range of 16.1°–44.1°, and a helical pitch of 2.89 nm, with the accepted value being 2.82 nm for A-DNA (Fig. 4K,L). Interestingly, dsRNA exhibited a lower profile than dsDNA due to its relative softness.
DISCUSSION
LncRNA molecules are emerging as abundant and important players at multiple levels of regulation over gene expression. Our ability to study structure-function relationships in this new group of molecules would be critical to our understanding of their biology, their roles in health and disease, and the potential ways to correct their malfunction.
Our conclusions regarding structure-function relationship of HOTAIR need to be taken carefully for several reasons. First, our model does not take into account HOTAIR-protein interactions, which may be indispensable to its cellular state and functionality. Such interactions have been previously described, for example, with the proteins PRC2 and LSD1 (Tsai et al. 2010), which are involved in specific chromatin modifications. While our observations indicate that HOTAIR has an inherent structure, this structure could be very well modulated in the cell by such interactions. Future studies could build on the approach we describe here, and incorporate these proteins in the study systems.
Second, we chose to visualize HOTAIR in conditions that mimic, to the best of our knowledge and ability, the chemical conditions inside the nucleus. Particularly, divalent and monovalent ions are expected to play important roles for lncRNA molecules, that is, stabilizing long-range interactions. Our study conditions included, for example, a low magnesium concentration; a higher magnesium concentration could theoretically produce different anatomies and configurations than the ones observed here.
Third, because of the observed flexibility of HOTAIR, the molecule orientates freely and displays variable anatomy. This phenotype highlighted the need to rely on multiple parameters, such as measured lengths and position of modules relative to each other, while eliminating shapes that cannot be assigned beyond a reasonable likelihood. Despite that, image-based assignment of molecular identities is an inherent limitation of molecular imaging in AFM, and our conclusions in this report should be taken with this limitation in mind. Better assignment could be achieved by adding information to the system, for example, by chemical modification of the molecule under observation. A simple way to do that is by hybridizing a nanoparticle-labeled DNA or RNA probe to a specific part of the molecule; the nanoparticle could be relatively easily identified in the AFM, indicating with high probability the identity of the part it binds to. However, such methods constantly face the risk of the modification perturbing the native conformation of the molecule, in a molecular analog of Heisenberg's uncertainty principle. In this study we chose to do careful assignment as natively as possible. Further studies using additional methods could further improve our understanding of the complex structure and behavior of molecules such as HOTAIR.
With that said, our observations of HOTAIR produce a biologically plausible model of its anatomy, quantitate itsmotility, and confirm it can intrinsically target genomic DNA. Moreover, our findings demonstrate that structural study of lncRNAs can be done using AFM as a tool of choice, owing to its ability to enable direct, high-resolution, and dynamic visualization of nucleic acids in a liquid, cell-like environment, and at physiological temperature. Although it was introduced more than three decades ago (Binnig et al. 1986), AFM is still not a mainstream technique in molecular, cellular, and structural biology. Our findings make a convincing case in favor of adding this versatile tool to X-ray crystallography, NMR, and cryo-EM, in order to enable new forms of understanding of the behaviors of biological molecules.
MATERIALS AND METHODS
In vitro transcription (IVT) and RNA-seq
LZRS-HOTAIR and pCDNA3-HOTAIR were a kind gift from Prof. Howard Chang. pJ-HOTAIR was purchased from DNA2.0. IVT templates were either plasmids linearized with EcoRI restriction enzyme (NEB), or PCR amplicons. IVT was carried out using two separate kits (from New England Biolabs and Megascript). IVT was carried out for 3 h at 37°C and followed by DNA template digestion (using DNase included in the kits). Samples were analyzed by gel electrophoresis using formamide as denaturing agent to verify purity from prematurely terminated transcripts. Purity was confirmed by HPLC using an Agilent 1100 series instrument. RNA was purified using MegaClear kit. RNA-seq was performed at the Nancy and Stephen Grand Israel National Center for Personalized Medicine (G-INCPM) at the Weizmann Institute of Science. Library preparation was done using in-house protocols.
AFM and Cryo-EM
Samples were analyzed using a NanoWizard ULTRA Speed AFM (JPK Instruments) mounted on an inverted optical microscope (Nikon Eclipse TE2000-U or Zeiss AxioObserver.A1), or equipped with a JPK TopViewOptics. Samples were imaged in buffer at ambient temperature in amplitude-modulation or phase-modulation AC mode. Fast-scanning high-resonant ultra-short cantilevers (USC-F0.3-k0.3, NanoWorld) with a nominal resonance frequency of 300 kHz in air, spring constant of 0.3 N/m, reflective chromium/gold-coated silicon chip, and high-density carbon tips with a radius of curvature of 10 nm were used. Prior to deposition on substrate, RNA and HBDs molecules were incubated in filtered nuclear-like buffer (NLB; 5 mM NaCl, 140 mM K+, 0.5 mM Mg2+, 10−4 mM Ca2+, pH = 7.2) for 30 min at 37°C. For cryo-EM, HOTAIR-bearing grids were plunge-frozen in liquid ethane cooled by liquid nitrogen, using a Leica EM-GP plunger (4 sec blotting time, 80% humidity), and imaged at liquid nitrogen temperature on an FEI Tecnai TF20 electron microscope operated at 200 kV with a Gatan side entry 626 cryo-holder. Images were recorded on a K2 Summit direct detector (Gatan) mounted at the end of a GIF Quantum energy filter (Gatan). Images were collected in counting mode, at a calibrated magnification of 16,218 yielding a pixel size of 3.083 Å. Additional detailed methods can be found in the Supplemental Notes.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank M. Gershovits, S. Gilad, Y. Spector, and all the staff at the Nancy & Stephen Grand Israel National Center for Personalized Medicine (Weizmann Institute) for discussions and technical assistance, N. Elad (Weizmann Institute) for discussions and technical assistance, F. Buske for kind discussions and technical assistance, S. Dori (Tel-Aviv University) and O. Girshevitz (Bar-Ilan University) for technical assistance, A. Munitz (Tel-Aviv University) for valuable discussions, G. Church (Harvard Medical School) for endless inspiration and valuable discussions, and all the team at Augmanity for valuable assistance and discussions.
Author contributions: R.S.S. spearheaded research. R.S.S., D.S., L.A., H.H., and J.G. designed and performed experiments and analyzed data. L.A. and H.J. performed computational modeling. E.S. and R.U. analyzed data. I.B. and A.A.H. oversaw the project and designed experiments. I.B. analyzed data. R.S.S., D.S., and I.B. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.074633.120.
REFERENCES
- Abels JA, Moreno-Herrero F, van der Heijden T, Dekker C, Dekker NH. 2005. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys J 88: 2737–2744. 10.1529/biophysj.104.052811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Walter P, Raff M, Roberts K. 2002. Molecular biology of the cell, 4th ed.: International Student Edition Garland Science, NY. [Google Scholar]
- Amaral PP, Mattick JS. 2008. Noncoding RNA in development. Mamm Genome 19: 454–492. 10.1007/s00335-008-9136-7 [DOI] [PubMed] [Google Scholar]
- Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. 2004. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem 279: 22243–22249. 10.1074/jbc.M314326200 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, de Nonno V, Grassi G, Amicone L, Tripodi M. 2016. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 36: 942–955. 10.1038/onc.2016.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. 1986. Atomic force microscope. Phys Rev Lett 56: 930–933. 10.1103/PhysRevLett.56.930 [DOI] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. 2014. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol 21: 633–640. 10.1038/nsmb.2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske FA, Bauer DC, Mattick JS, Bailey TL. 2012. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res 22: 1372–1381. 10.1101/gr.130237.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan JA. 2002. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals 15: 225–235. 10.1023/A:1016022730880 [DOI] [PubMed] [Google Scholar]
- Dickerson RE, Klug A. 1983. Base sequence and helix structure variation in B and A DNA. J Mol Biol 166: 419–441. 10.1016/S0022-2836(83)80093-X [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Hagerman PJ. 1997. Flexibility of RNA. Annu Rev Biophys Biomol Struct 26: 139–156. 10.1146/annurev.biophys.26.1.139 [DOI] [PubMed] [Google Scholar]
- Hung T, Chang HY. 2010. Long noncoding RNA in genome regulation. RNA Biol 7: 582–585. 10.4161/rna.7.5.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalwa M, Hänzelmann S, Otto S, Kuo C-C, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, et al. 2016. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res 44: 10631–10643. 10.1093/nar/gkw802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. 2010. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-H, Liu Z-L, Sun M, Liu J, Wang Z-X, De W. 2013. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 13: 464 10.1186/1471-2407-13-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS, Ando T. 2011. Imaging of nucleic acids with atomic force microscopy. Methods 54: 274–283. 10.1016/j.ymeth.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res 40: 5034–5051. 10.1093/nar/gks071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2013. Tackling structures of long noncoding RNAs. Int J Mol Sci 14: 23672–23684. 10.3390/ijms141223672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pádua Alves C, Fonseca AS, Muys BR, de Barros E Lima Bueno R, Bürger MC, de Souza JES, Valente V, Zago MA, Silva WA Jr. 2013. Brief report: the lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 31: 2827–2832. 10.1002/stem.1547 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Henry Goodnough L, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 14: 45–48. 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. 2015. HOTAIR forms an intricate and modular secondary structure. Mol Cell 58: 353–361. 10.1016/j.molcel.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X-P. 2013. The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med 33: 325–332. 10.3892/ijmm.2013.1570 [DOI] [PubMed] [Google Scholar]
- Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. 2013. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52: 9519–9527. 10.1021/bi401085h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-H, Wang X-L, Tang H-M, Jiang T, Chen J, Lu S, Qiu G-Q, Peng Z-H, Yan D-W. 2014. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep 32: 395–402. 10.3892/or.2014.3186 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.