Abstract
Campylobacter jejuni, a foodborne pathogen, is one of the most common bacterial causes of gastroenteritis in the world. Undercooked poultry, raw (unpasteurized) dairy products, untreated water, and contaminated produce are the most common sources associated with infection. C. jejuni establishes a niche in the gut by adhering to and invading epithelial cells, which results in diarrhea with blood and mucus in the stool. The process of colonization is mediated, in part, by surface-exposed molecules (adhesins) that bind directly to host cell ligands or the extracellular matrix (ECM) surrounding cells. In this review, we introduce the known and putative adhesins of the foodborne pathogen C. jejuni. We then focus our discussion on two C. jejuni Microbial Surface Components Recognizing Adhesive Matrix Molecule(s) (MSCRAMMs), termed CadF and FlpA, which have been demonstrated to contribute to C. jejuni colonization and pathogenesis. In vitro studies have determined that these two surface-exposed proteins bind to the ECM glycoprotein fibronectin (FN). In vivo studies have shown that cadF and flpA mutants exhibit impaired colonization of chickens compared to the wild-type strain. Additional studies have revealed that CadF and FlpA stimulate epithelial cell signaling pathways necessary for cell invasion. Interestingly, CadF and FlpA have distinct FN-binding domains, suggesting that the functions of these proteins are non-redundant. In summary, the binding of FN by C. jejuni CadF and FlpA adhesins has been demonstrated to contribute to adherence, invasion, and cell signaling.
Keywords: pathogenesis, bacteria-host cell interactions, adhesin, MSCRAMM, fibronectin
Recognition of C. jejuni as a Significant Foodborne Pathogen
Campylobacter jejuni has emerged from obscurity to become a leading bacterial cause of diarrheal disease over the course of five decades (Kaakoush et al., 2015). It was not until 1963 that Sebald and Véron proposed the term campylobacter (in Greek, a ‘curved rod’) to distinguish theses microaerophilic vibrios from the vibrios associated with cholera and other halophiles (Sebald and Véron, 1963). Although veterinarians were the first to recognize that this organism caused mild dysentery in cattle and sheep, a major breakthrough occurred in 1968 when C. jejuni was isolated from the diarrheal stool of a young adult using a special filtration technique (Dekeyser et al., 1972). This led to the development of a selective medium for Campylobacter isolation from diarrheal stools of both animals and humans, and a more accurate assessment of the public health burden of C. jejuni infections (Butzler and Skirrow, 1979). Presently, there are an estimated 1.3 million illnesses each year from C. jejuni in the United States and an estimated 96 million cases worldwide annually (Asuming-Bediako et al., 2019). In high-income countries, acute campylobacteriosis is characterized by fever, severe abdominal cramps, and diarrhea containing blood and leukocytes (Blaser et al., 1979; Karmali and Fleming, 1979; Svedhem and Kaijser, 1980). The most common source of C. jejuni infection is the handling or consumption of raw or undercooked poultry products, as chickens are the natural reservoir of this bacterium (Friedman et al., 2004). However, unpasteurized milk, eggs, untreated water, contaminated produce, and contact with animals colonized with C. jejuni have also been implicated as sources of infection (Horrocks et al., 2009; Bronowski et al., 2014; Huang et al., 2015). The economic impact of C. jejuni infections extends beyond the treatment of acute diarrheal illness, as infection with certain C. jejuni strains is correlated with a higher incidence of Guillain-Barré syndrome (GBS). GBS, an autoimmune syndrome, is the leading cause of flaccid paralysis in the post-polio era (Schwerer, 2002). Also, reports of antibiotic-resistant Campylobacter have continued to increase over time for multiple classes of antibiotics (Engberg et al., 2001; Coker et al., 2002; Bae et al., 2005; Gibreel and Taylor, 2006; Ruiz-Palacios, 2007). Recently, the Centers for Disease Control and Prevention listed drug-resistant C. jejuni as a ‘serious threat’1. Overall, C. jejuni has emerged as a pathogen of significance in human health due to the number of infections worldwide, the emergence of antibiotic-resistant isolates, and the bacterium’s association with post-infection sequelae. Given these factors, current efforts to determine the underlying mechanisms by which C. jejuni coordinates virulence during its interactions with host tissues should be expanded to develop new intervention strategies to reduce the global impact of campylobacteriosis. Among the most intensely studied C. jejuni virulence factors to date are host cell-binding proteins. Here we introduce the C. jejuni cell-binding proteins and then focus on the CadF and FlpA proteins, as these are the best-characterized adhesins.
Adhesins Are Key Players at the Bacteria-Host Cell Interface: C. jejuni Adhesive Molecules
Bacteria have evolved an abundance of mechanisms to engage and alter the behavior of host cells. Some of these events are facilitated by hydrophobic interactions resulting in non-specific adhesion, while others are highly specific and dependent upon the binding of a bacterial molecule to a host surface receptor and/or component of the extracellular matrix (ECM) (Stones and Krachler, 2016). A common theme shared among pathogenic bacteria is the presentation of surface-exposed molecules known as adhesins. In this review, the term adhesin is defined as a bacterial molecule(s) that facilitates a specific interaction between an individual bacterium and a eukaryotic cell protein, glycoprotein, or glycolipid that is surface-exposed. Adhesins are highly specialized surface adhesive structures, which are comprised of single monomeric proteins as well as intricate multimeric macromolecules, that can play a crucial role in allowing bacteria to colonize and persist in a host (Pizarro-Cerda and Cossart, 2006). The advantage of a gut bacterium adhering to the host cells is that it can aid in establishing intestinal persistence, as the gastric fluids that bathe the surfaces of tissues, combined with the involuntary rhythmic contractions of peristalsis, may wash away bacteria. The specificity of adhesin:receptor interactions can take on many forms and may change over the course of an infection to enable the pathogen to target different host cells and tissues (tissue tropism). An adhesin may target one host cell molecule or may have several domains that permit binding to multiple surface receptors. Alternatively, multiple adhesins may work in an additive or cooperative manner to enable the bacterium to bind to one surface receptor. Adhesins may also be synthesized at different phases during infection in response to physicochemical properties. The colonization of host tissues can affect the gene expression of virulence-specific genes in bacteria as well as alter the bacterium’s metabolism and respiration. In several instances, bacterial cellular adherence is also used as a platform for type III, type IV, or type VI contact-dependent secretion systems to deliver effectors (virulence factors) that rewire host cell signaling pathways and behavior in dramatic fashion (Tsang et al., 2010; Stones and Krachler, 2016). Similar to other intensely studied bacteria, such as Staphylococcus aureus and Salmonella spp. (Vaca et al., 2019), the foodborne pathogen C. jejuni synthesizes several adhesins to promote binding to host cells. These adhesins are among the first molecules to make physical contact with a host cell. In addition, the FlpA adhesin, and presumably the CadF adhesin, permit the delivery of effector proteins into the cytosol of a host target cell (Larson et al., 2013).
Campylobacter jejuni binding to the cells lining the intestinal epithelium is dependent on multiple factors, including motility, bacterial cell surface charge, and multiple adhesins. Some 30 years ago, C. jejuni was thought not to synthesize specific adhesive proteins or specifically adhere to any tissues, but that host factors such as the O2/CO2 concentration and nutrient availability dictated the site of C. jejuni colonization in a host. However, in the late 1980’s and early 1990’s, Campylobacter researchers identified several proteins and molecules that could bind to host cells or host cell ligands, providing renewed energy for researchers to search for additional adhesins. While this age of discovery for new virulence determinants was an exciting time for Campylobacter researchers, additional studies raised questions regarding whether some of the identified proteins truly act as bacterial adhesins. For example, Cj0268c, Cj0289c (PEB3), Cj0596 (PEB4), Cj0921c (PEB1) are localized in the bacterial periplasmic space, and Cj1349c appears to be localized in the bacterial cytosol (Supplementary Table S1). Moreover, a few of these proteins were determined to have other functions (Leon-Kempis Mdel et al., 2006; Min et al., 2009; Kale et al., 2011). These facts highlight the need for more data to clearly demonstrate that an individual protein is surface exposed and binds to a host cell receptor/ligand. Additional research involving the use of a deletion mutant and a complemented isolate is also necessary to determine if a protein facilitates binding to a host cell ligand. The bacterial flagellum, capsular polysaccharide, and lipopolysaccharide have been reported to contribute to C. jejuni binding to cells, but the precise role of these bacterial structures in cellular adherence remains to be elucidated (Supplementary Table S2). Moreover, generating a deletion of one component of the structure often affects a cellular function (e.g., motility) or influences another bacterial property (e.g., surface charge). For example, deletion in the gene encoding the FlaA filament protein renders the bacteria non-motile. In the past forty years of research on Campylobacter virulence factors, a cellular target has been identified for three C. jejuni proteins; Cj0983 (JlpA) binds to HSP90, Cj1279c (FlpA) binds to fibronectin (FN), and Cj1478c (CadF) binds to FN (Supplementary Table S1). To gain further insight into other C. jejuni adhesins and their potential role in colonization and disease, we refer the reader to two previously published review articles (Rubinchik et al., 2012; Lugert et al., 2015).
Microbial Surface Components Recognizing Adhesive Matrix Molecule(s) (MSCRAMMs), which are synthesized by many pathogenic Gram-positive and Gram-negative bacteria, have been demonstrated to contribute to the disease process. Fibronectin-binding proteins (FNBPs) are members of the MSCRAMM family. This article focuses on the C. jejuni CadF and FlpA FNBPs, two adhesins that facilitate bacterial colonization and contribute to illness in a disease model (Figure 1, and Table 1). Continued investigation of the C. jejuni FNBPs is needed to fully understand the specificity of the CadF and FlpA proteins in mediating host cell interactions and to better compare these two proteins to the FNBPs of Gram-positive pathogens, Gram-negative pathogens, and commensal organisms.
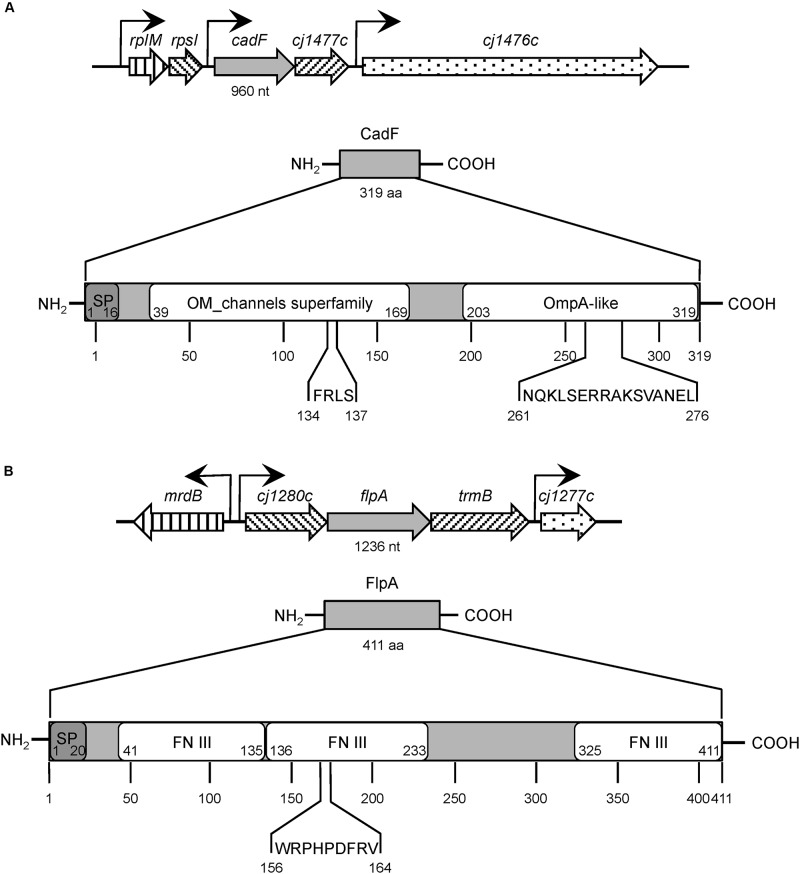
FIGURE 1.
Structural organization of Campylobacter jejuni CadF and FlpA fibronectin-binding proteins. Panel (A) C. jejuni CadF, a 319 amino-acid protein, is encoded by the cadF gene, 960 nucleotides in size, expressed as the first gene of a bicistronic operon. CadF has two dominant protein domains: an outer membrane (OM) channel superfamily and an OmpA-like domain. The four amino-acid residues (FRLS) of fibronectin (FN)-binding motif are located in the OM channel superfamily domain at position 134 through 137. The C-terminal region of CadF also contains the 16-residue peptidoglycan-binding motif (NQKLSERRAKSVANEL) located in the OmpA-like domain at position 261 through 276. Panel (B) The C. jejuni FlpA protein is comprised of 411 amino-acid residues and is encoded by the flpA gene. The flpA gene is 1,236 nucleotides in size and is expressed as the second gene of a tri-cistronic operon. The FlpA protein has three individual FN type III-like (FN III) domains. The FN-binding motif of FlpA is nine amino-acid residues long (WRPHPDFRV) and located in the second FN III domain at position 156 through 164. The signal peptide sequences, which were identified using SignalP 5.0, are indicated with the ‘SP’ for both (A and B). The abbreviations ‘nt’ and ‘aa’ refers to nucleotides and amino acids, respectively. The gene information was obtained from the NCBI GenBank database; Accession No: AL111168.1. The protein structure domains were obtained from the UniProt database; CadF UniProtKB Accession No: Q0P8D9, FlpA UniProtKB Accession No: Q0P8X7.
TABLE 1.
Campylobacter jejuni CadF and FlpA fibronectin (FN)-binding proteins (FNBPs).
| Property or characteristic | CadF (Campylobacter adhesion to Fibronectin) | FlpA (Fibronectin-like protein A) |
| Gene organization | Predicted to be the first gene of a bicistronic operon (cadF, Cj1477c) | Predicted to be the second gene in an operon containing three genes (Cj1280c, flpA, and Cj1278c) |
| No. of nucleotides, residues, Mr (strain NCTC 11168) | 960 nts, 319 aa, 37 kDa | 1,236 nts, 411 aa, 46 kDa |
| Prominent features | Signal peptide of 16 residues in length (Konkel et al., 1997) and a consensus peptidoglycan-binding motif NX2LSX2RAX2VX3l (von Heijne, 1985) Fractionates in the outer membrane preparations and has been demonstrated to be surface-exposed using a rabbit CadF-specific serum (Konkel et al., 1997) | Lipoprotein signal sequence of 20 residues comprised of a tripartite structure and an invariant cysteine after the carboxy-terminus of the signal (Konkel et al., 2010) Deduced amino acid sequence contains three FN type III-like domains (Konkel et al., 2010) Fractionates in the outer membrane preparations and has been demonstrated to be surface-exposed using a rabbit FlpA-specific serum (Konkel et al., 2010) |
| Host cell target | FN | FN |
| FN-binding domain | Purified CadF displays dose-dependent and saturable FN-binding activity (Konkel et al., 2010) FN-binding domain has been localized to four amino acids [AA 134–137, Phe-Arg-Leu-Ser (FRLS)] (Konkel et al., 2005) FN-binding to an FRLS containing peptide is saturable (Konkel et al., 2005) FRLS domain determined to be surface-exposed using a mouse α-CadF peptide polyclonal antibody (Konkel et al., 2005) A rCadF protein containing the Ala-Ala-Gly-Ser residues at AA 134–137 exhibited a decrease in FN-binding (Konkel et al., 2005) | Purified FlpA protein displays dose-dependent and saturable FN-binding activity (Konkel et al., 2010; Larson et al., 2013) FN-binding domain has been localized to nine amino acids [AA 156–164, Trp-Arg-Pro-His-Pro-Asp-Phe-Arg-Val (WRPHPDFRV)] (Larson et al., 2013) |
| Site of binding to FN | The site of CadF binding on FN is not known | FlpA exhibits dose-dependent and saturable binding to the 40 kDa gelatin-binding domain of FN (Larson et al., 2013) |
| Adhesion to cells | A cadF mutant demonstrates reduced adhesion to human INT 407 epithelial cells (Monteville et al., 2003) The binding of C. jejuni to INT 407 cells is reduced by peptides containing the FRLS residues (Konkel et al., 2005) Competitive inhibition assays revealed that a cadF mutant inefficiently competes against the C. jejuni wild-type strain for binding to INT 407 cells compared to another wild-type strain (Monteville et al., 2003) Phenotypic changes were initially confirmed by studies performed with a Cj1477c mutant (Monteville et al., 2003) | A flpA deletion mutant demonstrates reduced adhesion to human INT 407 epithelial cells (Konkel et al., 2010) and chicken LMH hepatocellular carcinoma epithelial cells (Flanagan et al., 2009) Phenotypic changes were confirmed by in trans complementation studies (Konkel et al., 2010) |
| Additional cell assays | FN-facilitated invasion of T84 eukaryotic cells by C. jejuni occurs preferentially at the basolateral cell surface (Monteville and Konkel, 2002) | A polyclonal serum against FlpA blocks C. jejuni adherence to INT 407 cells in a concentration-dependent manner (Konkel et al., 2010) |
| Host cell signaling pathways | Phosphorylation of paxillin is reduced with a C. jejuni cadF insertional mutant (Monteville and Konkel, 2002) Rac1 and Cdc42 GTPase dependent cell-signaling events are blunted with a C. jejuni cadF mutant (Krause-Gruszczynska et al., 2007) A C. jejuni cadF flpA double mutant is impaired in the activation of the epidermal growth factor receptor and Rho GTPase Rac1 (Larson et al., 2013) | FlpA is required for phosphorylation of Erk1/2 during C. jejuni infection (Larson et al., 2013) A C. jejuni cadF flpA double mutant is impaired in the activation of epidermal growth factor receptor and Rho GTPase Rac1 (Larson et al., 2013) |
| Infection of chickens | A cadF insertional mutant is impaired in chicken colonization (Ziprin et al., 1999; Flanagan et al., 2009) | A flpA deletion mutant is impaired in chicken colonization – 2 of 10 chickens were colonized (Flanagan et al., 2009) |
| Vaccination of chickens | Vaccination results in a reduction in the median level of C. jejuni cecal colonization when compared to the C. jejuni-inoculated, non-vaccinated control group (Neal-McKinney et al., 2014) | Vaccination results in a reduction in the median level of C. jejuni cecal colonization when compared to the C. jejuni-inoculated, non-vaccinated control group (Neal-McKinney et al., 2014) |
| Human antibody response | Individuals infected with C. jejuni generate antibodies against CadF (Konkel et al., 1997) | Not yet tested |
| Other information related to potential disease | Abiotic IL-10–/– mice show a reduced median pathology score for the cadF mutant and lower levels of TNF-α and IFN-γ compared to the wild-type strain (Schmidt et al., 2019) Undergoes post-translational processing to form smaller proteins of 24 kDa (CadF24) and 22 kDa (CadF22) that retain FN binding but that loses immunogenicity (Scott et al., 2010) | FlpA is required for C. jejuni dissemination to the spleen in IL-10–/– germ-free mice (Larson et al., 2013) |
Fibronectin: a Multidomain Glycoprotein
Many bacterial adhesins target and bind to FN, which is ubiquitously present in the ECM of a variety of human tissues and organs, including intestinal epithelial cells (Frantz et al., 2010). Mature FN is a large glycoprotein (220 kDa) and can exist as two forms. Plasma FN is synthesized by hepatocytes and secreted into the blood plasma, saliva, and other body fluids, where it circulates as a compact dimer (To and Midwood, 2011). In contrast, cellular FN is synthesized and secreted by many cell types, including fibroblasts and endothelial cells, and becomes incorporated into the ECM to form a fibrillar-type (insoluble) matrix (Sottile and Hocking, 2002; To and Midwood, 2011). One of the stages of matrix assembly is FN unfolding, where cryptic binding sites are exposed (Smith et al., 2007; Weinberg et al., 2017). The unfolding of FN molecules enhances its binding to host cells, especially involving the β1 integrin subunit (Henderson et al., 2011). In mammals, there are 18 α and 8 β integrin subunits that can associate to form 24 integrin heterodimers, which are transmembrane ECM-binding proteins. The binding of FN to the α5β1 integrin heterodimer induces integrin clustering and promotes fibrillar assembly. Moreover, a tripartite linkage is established between FN, the integrins, and the actin cytoskeleton, allowing the cytoplasmic domains of the integrins to trigger intracellular signaling (Singh et al., 2010). FN incorporation into the cellular matrix has been demonstrated to play an important role in cell adhesion, cell migration, cell signaling, ECM remodeling, and tissue regeneration (To and Midwood, 2011).
Plasma (soluble) and cellular (insoluble) FN arise from alternative splicing of a single pre-mRNA (a single gene) (Pankov and Yamada, 2002). While there are variations in the composition of cellular and plasma FN, the proceeding discussion focuses primarily on cellular FN found in the ECM of tissues. The gene encoding cellular FN harbors Extra Domains A and B (EDA and EDB, also termed EIIIA and EIIIB) and a segment connecting two type III repeats (see below) called the type III connecting segment (IIICS, also termed the variable, or V, region). The alternative splicing of the EDA, EDB, and IIICS elements during transcription allows for the expression of different cellular FN isoforms (Schwarzbauer et al., 1983). In contrast to cellular FN, plasma FN does not harbor the EDA and EDB domains, and only one FN subunit possesses a IIICS segment. Typically, both plasma and cellular FN consist of three types of repetitive domains: 12 FN type I repeats (FN I), 2 FN type II repeats (FN II), and 15-18 FN type III repeats (FN III) (Pankov and Yamada, 2002). FN I and FN II are structurally rigid. In contrast, the FN III repeats are flexible due to the absence of disulfide bonds and are required for FN polymerization (Potts and Campbell, 1994). The multimodular structure and intermodular regions of FN provide the molecule with the ability to interact with other host proteins, including collagen, laminin, integrins, and fibrin. Moreover, the complexity of the FN molecule provides multiple targets for FNBPs to bind to this molecule (White et al., 2008).
Bacterial Binding to Fibronectin
Pathogenic bacteria that have FNBPs can use them to promote tissue adherence and interface with host cell signaling complexes (Stones and Krachler, 2015). More specifically, a three-component bridge is formed between the FNBPs, host cell-associated FN, and transmembrane integrins (Hymes and Klaenhammer, 2016). The linkage between the bacteria and FN stimulates integrin occupancy and clustering, triggering the recruitment of cell signaling molecules and rearrangement of the host cell actin cytoskeleton (Wu, 2007; Zaidel-Bar et al., 2007; Deakin and Turner, 2008). In some instances, it is the intimate binding mediated by FNBPs that enables host cell invasion and intracellular multiplication, resulting in acute disease (Hymes and Klaenhammer, 2016).
In general, FNBPs contain an N-terminal signal sequence, a central binding region, and a C-terminal cell wall anchor (LPXTG motif). The very first observation that a bacterium, S. aureus, was capable of binding to FN was reported in 1978 (Kuusela, 1978). More specifically, the FNBPA and FNBPB proteins from S. aureus have been reported to bind to the heparin-binding domain of FN (Bae and Schneewind, 2003; Schwarz-Linek et al., 2003) and that the deletion of both genes is required to abolish the organism’s ability to bind FN (Greene et al., 1995). Since the initial discovery of an FNBP in S. aureus, at least 100 other FNBPs have been identified in both Gram-positive and Gram-negative bacteria (Henderson et al., 2011). Although many of these proteins have been found to possess distinct FN-binding domains, even more interesting is the fact that they bind to different regions within the FN molecule. For example, S. aureus FNBPA and FNBPB only bind to the N-terminal five-module region (FN I1–5, heparin-binding domain), whereas the Streptococcus pyogenes F1 and Sfb1 bind to the FN I1–5 region as well as the FN I6–9 gelatin-binding domain (Sela et al., 1993; Bae and Schneewind, 2003; Schwarz-Linek et al., 2003; Schwarz-Linek et al., 2004). Other FNBPs target different FN regions. For example, Borrelia burgdorferi BBK32 binds to multiple sites, including FN I2–3 of the heparin-binding domain, FN I8–9 of the gelatin-binding domain, and FN III1–3 (Kim et al., 2004; Harris et al., 2014). The binding of these FNBPs to different sites within the FN molecule likely has different biological consequences.
CadF Is a C. jejuni Outer Membrane Protein Containing Fibronectin-Binding Residues
In the 1980-90s, studies were conducted to determine if C. jejuni were able to bind to various components of the ECM, including FN, collagen, laminin, and vitronectin (Kuusela et al., 1989; Konkel et al., 1997; Moser et al., 1997). Collagen is the most abundant protein found in the ECM and provides structural support to resident cells. Laminin is a major component of the basal lamina, influencing cell differentiation, migration, and adhesion. Vitronectin binds to the α5β3 integrin, promoting cell adhesion and spreading. Kuusela et al. (1989) were the first to report that C. jejuni isolates were able to bind to FN, and also reported that some isolates were able to bind to Type I, III, and V collagens. Several years later, the observation was made that C. jejuni bound to host cell retraction fibers, which are fingerlike projections enriched in ECM components. This prompted researchers to further investigate C. jejuni binding to various ECM components. In contrast to laminin and vitronectin, C. jejuni bound to coverslips that were treated with FN (FN-coated coverslips). Radioactive FN ([125I]-FN) was then used to probe blots of C. jejuni outer membrane protein (OMP) extracts and was found to specifically bind to a 37 kDa OMP (CadF) (Konkel et al., 1997). A rabbit anti-serum was generated against the 37 kDa protein and used to screen Campylobacter genomic phage expression libraries. This led to the identification of Cj1478c, designated cadF for Campylobacter adhesion to Fibronectin (Konkel et al., 1997). The cadF gene encodes a protein of 326 amino acids, with a calculated molecular mass of 36,872 Da (Figure 1).
The C. jejuni CadF protein is a surface-exposed OMP that binds to soluble and insoluble FN (Table 1). Key features of the deduced amino acid sequence included a signal peptide of 16 residues (von Heijne, 1985) and a consensus peptidoglycan-binding motif, NX2LSX2RAX2VX3l. Sequence analysis showed that the protein consists of an N-terminal transmembrane domain that forms a β-barrel pore and a C-terminal domain forming a mixed α/β fold. Additional evidence confirmed that the 37 kDa protein bound to FN. First, biotinylated FN bound to a protein with an apparent molecular mass of 37 kDa (CadF) in the C. jejuni OMP extracts as judged by ligand-binding blots (Konkel et al., 1997). Second, the FN-binding domain within CadF was localized to four amino acids [AA 134–137, Phe-Arg-Leu-Ser (FRLS)] using overlapping 30-mer and 16-mer peptides coupled with enzyme-linked immunosorbent assays (ELISA) (Konkel et al., 2005). Third, C. jejuni mutants containing insertions in cadF, which disrupted the coding sequence, demonstrated a significant reduction in binding to FN. Moreover, the FRLS domain of CadF was determined to be surface-exposed using a mouse polyclonal α-CadF peptide antibody coupled with laser scanning confocal microscopy (Konkel et al., 2005). Finally, a recombinant CadF protein containing mutated FRLS residues (AA 134-137, FRLS > AAGS) exhibited a 91% decrease in FN-binding activity compared to the unmodified/native CadF protein. Although the FN-binding residues within CadF have been identified, researchers have yet to identify the CadF-binding site in FN.
FlpA Is a C. jejuni Outer Membrane Lipoprotein Containing Fibronectin-Binding Residues
A second FNBP was identified in 2009 and was designated FlpA for Fibronectin-like protein A (Flanagan et al., 2009). The C. jejuni flpA gene is 1,236 nucleotides (411 residues) and is capable of encoding a protein with a calculated molecular mass of 46,124 Da (Figure 1, Table 1). The flpA gene is located in an operon containing three genes (Cj1280c, flpA, and Cj1278c). Sequence analysis of FlpA revealed the presence of three domains with similarity to the FN type III domain. Based on the presence of the FN type III domains, assays were performed to determine if FlpA binds to FN and the FN-binding phenotype of a C. jejuni flpA mutant. Purified FlpA protein displays dose-dependent and saturable FN-binding activity, as judged by ELISA using purified FlpA-GST protein (Konkel et al., 2010). The FN-binding site within FlpA was localized to a span of nine amino acids: Trp-Arg-Pro-His-Pro-Asp-Phe-Arg-Val (Larson et al., 2013). Assays were also performed to determine where FlpA binds within the FN molecule; proteolysis of FN with thermolysin (protease, type X) is a well-documented method for generating fragments of 29, 40–45, 65, and 130 kDa. Interestingly, thermolysin-digested FN fragments retain their biological activity. FlpA was determined to bind to the 40–45 kDa fragment (gelatin-binding domain) composed of four FN I repeats (FN I6–9) and two FN II repeats (FN II1,2). FlpA is likely a surface-exposed lipoprotein based on the data obtained from analysis using SignalP 5.0 (Almagro Armenteros et al., 2019) coupled with the application of indirect immunofluorescence microscopy of C. jejuni incubated with the FlpA-specific serum (Konkel et al., 2010).
Role of CadF AND FlpA in C. jejuni Colonization of Chickens
Campylobacteriosis often results from the handling and consumption of foods cross-contaminated with raw poultry products. The linkage between human infection and the handling of fresh poultry is mainly due to the fact that C. jejuni endemically colonizes commercial chicken flocks. Given that the antibodies passed from hens to chicks are partially protective against Campylobacter colonization, research has been performed to identify C. jejuni membrane-associated proteins recognized by maternal antibodies (Sahin et al., 2003). Immunoblots coupled with tandem mass spectrometry revealed a list of C. jejuni proteins recognized by maternal antibodies that included CadF and FlpA (Shoaf-Sweeney et al., 2008). This finding is consistent with previous findings that a C. jejuni cadF mutant and a C. jejuni flpA mutant are impaired in colonizing chickens (Ziprin et al., 1999; Flanagan et al., 2009). Based on the premise that specific adhesins are pivotal to colonization, disruption of C. jejuni adherence by anti-adhesin antibodies seems to be an obvious way to incapacitate the bacterium. In this regard, vaccination of chickens with a combination of CadF and FlpA peptides together with the full-length CadF and FlpA proteins resulted in an antibody response and a reduction in the median level of C. jejuni cecal colonization when compared to the C. jejuni-inoculated, non-vaccinated control group (Neal-McKinney et al., 2014). These results support the proposal that CadF and FlpA significantly contribute to C. jejuni chicken colonization.
Additional work is required to understand the interaction of C. jejuni within the gut of chickens and the biological consequences of C. jejuni infection in poultry (Awad et al., 2018). For decades, C. jejuni has been considered a commensal organism of poultry, as chickens, in contrast to humans, do not develop disease symptoms. More specifically, C. jejuni, which are principally found in the ceca (mucosal crypts) of chickens at very high CFUs, have been reported to stimulate a poor or inefficient inflammatory response, leading to tolerance and persistent cecal colonization (Hermans et al., 2012). In contrast, it has also been reported that certain Campylobacter strains can adversely affect the health and welfare of chickens and that the clinical signs of C. jejuni infection of poultry are not obvious (Humphrey et al., 2014). As our understanding of the bacteria and host cell factors that influence C. jejuni infection in chickens results in a better understanding of disease outcome, studies are warranted to determine if CadF and FlpA contribute to steps beyond the colonization of chickens.
CadF AND FlpA Regulation in Response to Intestinal Conditions
The intestinal life cycle of C. jejuni requires transition to an animal’s gut, where it responds to changes in physiological conditions. The abundance of the CadF and FlpA proteins has been demonstrated to be responsive to host conditions, including temperature, oxygen levels, oxidative stress, and mucin (Tu et al., 2008; Hong et al., 2014; Koolman et al., 2016; Guccione et al., 2017; de Oliveira et al., 2019). For example, Hong and colleagues reported that culturing C. jejuni with porcine mucin resulted in an increase in 32 proteins and a decrease in 20 proteins compared to bacteria grown in the absence of mucin for 24 h using a label-free LC-MS/MS technique (Hong et al., 2014). In this study, more than a 3-fold increase was detected in CadF abundance after growth in medium supplemented with porcine mucin. In contrast, Tu et al. (2008) reported a 1.9-fold decrease in the expression of cadF when C. jejuni were cultured in medium with a component of human mucin (MUC2) for 12 h using quantitative RT-PCR. Based on this finding, the investigators concluded that the CadF protein is not required for the bacteria to penetrate the mucus barrier. Although the two investigations used different methodologies (Tu et al., 2008; Hong et al., 2014), it is possible that CadF levels change over the course of an infection. Furthermore, in a study conducted by Guccione et al., a 2.4-fold increase in CadF abundance and a 1.7-fold increase in FlpA abundance was reported following an oxygen-tension downshift (from high to low oxygen tension), as determined by label-free proteomic analysis (Guccione et al., 2017). Noteworthy is that the abundance of the Peb1A protein was also reduced in response to the shift from high to low oxygen-tension. The investigators rationalized their findings based on the primary functions of the proteins. Peb1A, originally proposed to be an adhesin, was subsequently found to be an ABC-transporter involved in aspartate and glutamate uptake whereas CadF and FlpA are dedicated adhesins. Although much work needs to be done to determine how C. jejuni responds to the host environment, the studies conducted to date suggest that this bacterium responds to the gut, in part, by increasing CadF and FlpA protein levels.
In Vitro Evidence of Fibronectin Recognition by C. jejuni in Promoting Host Cell Adherence
Molecular biologists have utilized mutational studies to assess the function of a particular gene product. Likewise, a genetic approach has been taken to assess the individual role of CadF and FlpA in C. jejuni-host cell interactions by generating single-gene mutations. A cadF mutant was created by disrupting the cadF gene by homologous recombination via a single crossover event between the cadF gene on the chromosome and an internal fragment of the cadF gene on a suicide vector (Konkel et al., 1997). As mentioned above, the C. jejuni cadF knockouts were deficient in FN-binding. At the time, one of the obstacles facing Campylobacter researchers for the generation of a cadF complemented isolate was the fact that repeated attempts to clone the entire cadF gene and its endogenous promoter in E. coli failed, perhaps due to toxicity (Monteville et al., 2003; Mamelli et al., 2006). Because cadF and Cj1477c are the first and second genes of a bicistronic operon, the possibility was raised that Cj1477c, which encodes a putative hydrolase, might contribute to the observed reduction in host cell adherence. To address this concern, a knockout was generated in Cj1477c. While the C. jejuni cadF mutant was found to be deficient in binding to INT 407 cells (Monteville et al., 2003), no reduction was noted in the binding of the Cj1477c knockout to INT 407 cells when compared to a C. jejuni wild-type isolate. To address the contribution of the CadF adhesin in C. jejuni-host cell attachment in the context of other adhesive proteins, competitive inhibition adherence assays were performed with a C. jejuni cadF mutant and a C. jejuni wild-type strain and compared with a competition between two different C. jejuni wild-type strains. The adherence assay performed with the two wild-type strains revealed a dose-dependent decrease in the adherence of one strain when the inoculum of the competing wild-type strain was increased (Monteville et al., 2003). However, the C. jejuni cadF mutant was unable to competitively inhibit the binding of a C. jejuni wild-type strain to INT 407 cells (Monteville et al., 2003). The application of a polarized cell model revealed that C. jejuni translocate a cell monolayer via a paracellular route, and then bind to FN localized on the basolateral surface of cells via CadF (Monteville and Konkel, 2002). These findings were in accordance with earlier studies, indicating that CadF binds to host cell-associated FN and promotes C. jejuni-host cell adherence.
The generation of a flpA knockout and complemented isolate was more straightforward than for cadF, as flpA containing its endogenous promoter can readily be cloned in E. coli. The flpA deletion mutant was generated using standard molecular techniques, and the mutant isolate was complemented by the introduction of a shuttle vector harboring flpA driven by a constitutive promoter (in trans complementation). Adherence assays revealed that the binding of the C. jejuni flpA mutant to INT 407 epithelial cells was significantly reduced compared with that of a wild-type strain. The reduction in binding of the C. jejuni flpA mutant was judged to be specific since complementation of the mutant in trans with a wild-type copy of the gene restored the organism’s binding to INT 407 cells (Konkel et al., 2010; Larson et al., 2013). Moreover, rabbit polyclonal serum generated against FlpA blocked C. jejuni adherence to INT 407 cells in a concentration-dependent manner.
C. jejuni CadF and FlpA Stimulate Host Cell Signaling Pathways Associated With Host Cell Invasion
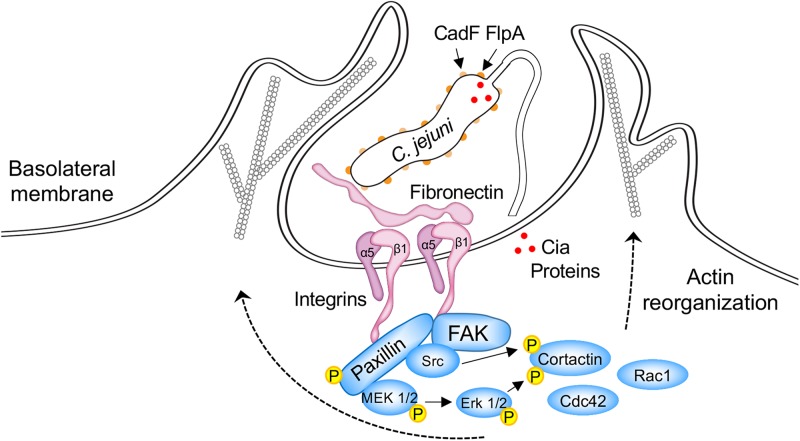
A model of the C. jejuni-host cell interface is presented in Figure 2. This figure shows the interaction of the CadF and FlpA proteins with host cell-associated FN and α5β1 integrins, and the cell signaling pathways activated upon C. jejuni binding FN. C. jejuni manipulates focal adhesions (FAs) to stimulate host cell signaling and promote cell invasion (Eucker and Konkel, 2012; Konkel et al., 2013; Larson et al., 2013; Samuelson and Konkel, 2013). FAs are dynamic cellular structures that link ECM components, including FN, to intracellular cytoskeletal structures. They are comprised of integrin receptors, adaptor proteins, signaling proteins, and actin. Central to signaling through the FA is paxillin, which serves to integrate and disseminate signals from integrins to associated kinases that regulate membrane trafficking and cytoskeletal rearrangement (Schaller, 2001; Zaidel-Bar et al., 2007; Deakin and Turner, 2008). Although paxillin does not exhibit enzymatic activity, when phosphorylated, it serves as a central hub for other proteins. More specifically, phosphorylated paxillin acts as a scaffold for both signaling [kinases such as focal adhesion kinase (FAK) and Src] and adaptor proteins (vinculin and p130Cas). It is the establishment of the mature FA that ultimately results in signals being transduced from the integrins to the actin cytoskeleton (outside-in signaling).
FIGURE 2.
Schematic overview of host cell signaling events triggered by C. jejuni CadF and FlpA binding to fibronectin and engagement of the α5β1 integrin heterodimer. Focal adhesions (FAs) are dynamic protein complexes that connect the extracellular matrix to intracellular actin bundles. FAs contain over 100 proteins, including paxillin, focal adhesion kinase (FAK), and Src. Paxillin is a multi-domain adaptor protein that localizes to FAs. During FA assembly, paxillin becomes indirectly associated with the tails of β integrin subunits, is phosphorylated on multiple tyrosine, serine, and threonine residues, and serves as a central hub for other proteins. In particular, the phosphorylation of paxillin provides a binding platform for FAK and Src. The FAK-Src dual-activated signaling complex is responsible for the recruitment and activation of additional signaling and adaptor proteins that result in the activation of the Cdc42 and Rac1 Rho GTPases. Cortactin is a monomeric cytoplasmic protein that is also involved in polymerization and rearrangement of the actin cytoskeleton. The coordinated effort of key signaling proteins, scaffold proteins, and cytoskeletal components stimulate the formation of actin-based membrane protrusions and bacterial invasion. C. jejuni cadF flpA mutants are impaired in the ability to stimulate the activation of paxillin, Erk1/2, and Cdc42/Rac1 Rho GTPases (see the text for additional detail). The delivery of the Cia effector proteins (red dots) to the cytosol of host cells is impaired in a C. jejuni flpA mutant. Not known is whether CadF participates in the delivery of the effector proteins to host cells. The reader is referred to other papers for detailed models of C. jejuni-host cell interactions (Krause-Gruszczynska et al., 2007, 2011; Boehm et al., 2011; Eucker and Konkel, 2012; Konkel et al., 2013; Samuelson and Konkel, 2013).
CadF and FlpA appear to set the stage for cell invasion by promoting C. jejuni binding to the FN present on the basolateral surfaces of cells (i.e., C. jejuni bound to FN bound to the α5β1 integrin) and by initiating host cell signaling events. More specifically, C. jejuni infection of INT 407 epithelial cells results in the phosphorylation of paxillin, and the phosphorylation is partially dependent on CadF (Monteville et al., 2003). This is based on the findings that a lower Multiplicity of Infection (MOI) of the C. jejuni cadF mutant (100 to 1) did not induce phosphorylation of paxillin. However, paxillin phosphorylation was observed in cells infected with a higher MOI of the C. jejuni cadF mutant (2000 to 1). This implies that CadF is not solely responsible for the host cell signaling. Consistent with paxillin activation playing a role in C. jejuni invasion, treatment of epithelial cells with TAE 226 and PP2, selective inhibitors of FAK and Src kinase activity, results in a significant decrease in C. jejuni internalization as judged by the gentamicin-protection assay (Eucker and Konkel, 2012). C. jejuni internalization is also significantly reduced in the presence of both PD168393 and erlotinib, which are specific inhibitors of epidermal growth factor (EGF) receptor tyrosine phosphorylation (Eucker and Konkel, 2012). This is relevant because the EGF receptor can be stimulated in the absence of an extracellular ligand via integrin signaling, and the activation of this receptor can alter components of the cytoskeleton involved with actin organization, FA formation and resolution, and cell-cell adhesion (Thelemann et al., 2005). Researchers have reported that C. jejuni induces EGF receptor activation in a CadF and FlpA dependent manner; however, whether one or both proteins are required for EGF receptor activation is currently not known (Eucker and Konkel, 2012).
β1 integrins, FAK, Src, and paxillin also contribute to the activation of the MAP kinase Erk1/2 in C. jejuni infected cells (Eucker et al., 2014). Moreover, the phosphorylation of Erk1/2 is associated with FlpA-mediated activation of β1 integrins and EGF receptor signaling (Larson et al., 2013). Consistent with this finding, inhibition of Erk1/2 activation with PD98059 results in a significant reduction in C. jejuni internalization (Zaidel-Bar et al., 2007; Deakin and Turner, 2008). Finally, FAs can also activate the Rac1 and Cdc42 Rho GTPases, thereby stimulating the formation of actin-based membrane protrusions and bacterial invasion (von Heijne, 1985; Mitra et al., 2005; Hu et al., 2006; Krause-Gruszczynska et al., 2007; Eucker and Konkel, 2012; Neal-McKinney and Konkel, 2012; Samuelson and Konkel, 2013; Figure 2). Noteworthy is that GTPase dependent cell-signaling events are blunted with a C. jejuni cadF mutant compared to a wild-type strain (Krause-Gruszczynska et al., 2011; Boehm et al., 2012). Collectively, these findings demonstrate that C. jejuni binding to host cell-associated FN, via CadF and FlpA, is necessary to stimulate signaling pathways associated with cellular invasion. A potential caveat to these findings is that the binding of the C. jejuni cadF and flpA mutants to cells is reduced when compared to a wild-type strain, which could disrupt the delivery of secreted effector proteins.
Delivery of C. jejuni Effector Proteins to Cells
Researchers have reported that the treatment of C. jejuni with chloramphenicol, a specific inhibitor of bacterial protein synthesis, significantly reduced host cell internalization, as compared to untreated controls (Konkel and Cieplak, 1992; Negretti et al., 2019). However, chloramphenicol treatment has little effect on C. jejuni adherence to host cells. Together, these data demonstrate that C. jejuni binding to host cell ligands is facilitated by adhesins, which are not sufficient to promote host cell internalization, but rather that internalization requires a second signal to stimulate host cell signaling pathways. The simplest view is that the C. jejuni FNBPs promote the bacterium’s binding to host cells, whereas C. jejuni invasion of epithelial cells requires de novo bacterial protein synthesis (McSweegan and Walker, 1986; Karlyshev and Wren, 2005). The question then arises of whether the C. jejuni CadF or FlpA proteins set the stage for cell invasion by merely binding to FN or whether they prime/stimulate host cell signaling pathways (albeit not to the level that would promote cell invasion).
The translocation of bacterial Type III secretion effectors to the cytosol of a host cell has been demonstrated using the adenylate cyclase domain (ACD) of Bordetella pertussis CyaA as a reporter (Sory and Cornelis, 1994; Desvaux et al., 2006; Ono et al., 2006; Schechter et al., 2006; Neal-McKinney and Konkel, 2012). The basis of the ACD assay is that adenylate cyclase (adenylyl cyclase) only catalyzes the production of cAMP when bound by calmodulin in the eukaryotic cell cytosol (Ladant and Ullmann, 1999; Dautin et al., 2002). C. jejuni was first reported to secrete proteins in 1999 (Konkel et al., 1999b) and later found to export proteins via the flagellar Type III Secretion System (T3SS) (Konkel et al., 2004). These secreted proteins were termed the Campylobacter invasion antigens (Cia), as they were found to contribute to cell invasion (Konkel et al., 1999b).
The delivery of most Type III secreted effectors requires the bacteria to contact the host cells (Schlumberger et al., 2005; Winnen et al., 2008). Different Cia proteins have been studied in several different C. jejuni strain backgrounds, including 81-176, NCTC 11168, and F38011. The Cia proteins have been found to play a role in C. jejuni cell invasion and survival but not in cell adherence (Christensen et al., 2009; Novik et al., 2010; Buelow et al., 2011; Samuelson et al., 2013; Naz, 2014). The most studied Cia proteins to date are CiaC and CiaD, which are necessary for maximal C. jejuni cellular invasion (Christensen et al., 2009; Neal-McKinney and Konkel, 2012; Samuelson and Konkel, 2013; Samuelson et al., 2013). Cj1242 (ciaC) and Cj0788 (ciaD) were initially identified to harbor a T3SS export signal using a genetic screen in a heterologous system, and Cj0788 (ciaD) was also identified to play a role in C. jejuni cell invasion using a non-targeted transposon screen (Christensen et al., 2009; Novik et al., 2010). Relevant to the role of C. jejuni adhesins in Cia delivery to host cells was the discovery that the CiaC effector protein was delivered to the cytosol of host target cells using the ACD assay (Neal-McKinney and Konkel, 2012). Two additional experiments support the proposal that CiaC delivery requires bacteria-host cell contact. First, no increase was observed in intracellular cAMP levels when a bacterial supernatant containing the CiaC-ACD fusion protein was added to INT 407 cells versus non-infected cells. Second, no cAMP production was observed when a 0.2 μm pore filter was used to block the physical contact of C. jejuni with host cells (a two-chamber system: the apical chamber contained a C. jejuni strain harboring a ciaC-ACD construct and the bottom chamber contained the epithelial cells) (Neal-McKinney and Konkel, 2012). Similar methods were then used to test whether CiaC could be delivered to the host cell cytosol using a C. jejuni flpA mutant (Larson et al., 2013). The assay was performed in parallel with a C. jejuni wild-type strain transformed with the ciaC-ACD construct. Significantly less cAMP was detected in the cytosol of host cells infected with the C. jejuni flpA mutant when compared to cells inoculated with the C. jejuni wild-type strain transformed with the ciaC-ACD construct. Experiments have yet to be performed to determine if a Cia effector can be delivered to host cells from a C. jejuni cadF mutant. Taken together, it seems reasonable that the C. jejuni adhesins work in conjunction with the effector proteins to alter host cell behavior, including promoting host cell internalization.
Potential Role of CadF and FlpA in Human Disease
Presumably, the ability of C. jejuni to bind to the cells lining the human gastrointestinal tract is necessary for disease. In one study, C. jejuni isolates recovered from individuals with fever and diarrhea were found to adhere to cultured cells at a greater efficiency than those strains isolated from individuals without diarrhea or fever (Fauchere et al., 1989). Whether the adherence capacity and disease severity are correlated for C. jejuni isolates needs to be further studied. Based on the data generated with C. jejuni infection of cultured human intestinal cells, it is probable that CadF and FlpA mediate C. jejuni binding to the epithelial cells lining the human intestinal tract. This binding is likely to set the stage for host cell invasion and intracellular multiplication, resulting in acute disease (van Spreeuwel et al., 1985; Black et al., 1988). Several options exist for how C. jejuni may gain access to FN as a substrate; three possibilities are discussed below. Each possibility is predicated on the belief that the bacteria must be able to penetrate the mucus overlying the intestinal epithelial cells and that FN is primarily found at the basolateral (internal) surface or between cells within the gut epithelium. First, C. jejuni could target sites where the host intestinal cells are shed by extrusion (at the tips of the intestinal villi), similar to that proposed for Listeria monocytogenes (Pentecost et al., 2006). Data supporting this possibility has been observed in C. jejuni-infected piglets, where the infection site is evident from villus blunting and cell necrosis (Babakhani et al., 1993). Piglets infected with different strains of C. jejuni develop a range of clinical symptoms similar to humans, ranging from watery stools to bloody diarrhea. The dysenteric-like illness (blood in the stool) is a severe form of the disease and is illustrative of cell adherence/invasion. Second, C. jejuni could reach the basolateral surfaces of the intestinal cells by cellular translocation. Laboratory studies with polarized intestinal cells have demonstrated that C. jejuni can translocate across an intact cell barrier by migrating between cells (i.e., a paracellular route of translocation) (Monteville and Konkel, 2002; Boehm et al., 2012). Third, C. jejuni may breach the intestinal barrier by transcytosis across Microfold (M) cells in Peyer’s patches of the intestine (Walker et al., 1988). Together, it is possible for the bacteria to either initially colonize at sites where the epithelium is disrupted and FN is readily accessible and/or bind to FN on the basolateral surface of epithelial cells following cellular translocation or via passage across Peyer’s patches.
Two animal studies have been done to date in order to specifically address the role of CadF and FlpA in disease (Larson et al., 2013; Schmidt et al., 2019). Recently, the contribution of CadF in clinical disease was assessed using an abiotic (gnotobiotic) IL-10–/– mouse model (Schmidt et al., 2019). In this model, the mice were inoculated with 109 CFU of a C. jejuni wild-type isolate and cadF mutant, on two consecutive days, and disease parameters were assessed daily for six days. While the authors concluded that CadF is not required for campylobacteriosis in mice, the pro-inflammatory responses, such as TNF-α and IFN-γ, were lower for the cadF mutant compared to the wild-type isolate. In addition, the median pathology score for the cadF mutant was approximately 8 at day six, whereas a median pathology score of 12 was recorded for the mice infected with the wild-type strain. Disease severity was determined based on the presence of blood in the stool, diarrhea, and animal behavior. Thus, the mice inoculated with the cadF mutant develop less severe disease symptoms when compared to mice inoculated with the wild-type strain. These data demonstrate that CadF, while not being essential for disease, contributes to the severity of disease in a mouse model. Because C. jejuni-cell adherence is multifactorial, we speculate that it is possible to overcome the involvement of CadF and FlpA in a disease model by using high or repeated doses of mutant bacteria. Regarding the role of the FlpA protein in disease, a study with IL-10–/– germ-free mice reported a reduced number of the C. jejuni flpA mutant in spleen when compared to mice inoculated with the C. jejuni wild-type strain (Larson et al., 2013). Studies are required to determine whether CadF and FlpA are necessary for human disease.
Although CadF is highly immunogenic, Scott and colleagues published an article revealing the possibility that CadF could retain its adhesive property and escape immune recognition in a host by post-translational processing (Scott et al., 2010). More specifically, the investigators demonstrated that CadF undergoes post-translational processing (proteolytic cleavage) to form smaller proteins of 24 kDa (CadF24) and 22 kDa (CadF22). Interestingly, the CadF24 and CadF22 variant forms were fully capable of binding to FN but were not recognized by patient sera. In addition, the processing of CadF appeared to be less abundant in the NCTC 11168 laboratory-adapted or avirulent strain (“GS” genome sequenced strain) when compared to the NCTC 11168 virulent strain (“O” original strain). The investigators concluded that the processing of CadF to the CadF24 and CadF22 variants provided antigenic variation that enabled evasion from the host immune response while enabling the protein to retain its adhesin-like function. Based on these findings, studies are warranted to determine whether the post-translational processing of CadF is common in other C. jejuni strains. Taken together, the data support the proposal that CadF may play a role in C. jejuni pathogenesis in a human host.
Presence of CadF and FlpA in C. jejuni Isolates
Previous survey studies have suggested that the cadF and flpA genes are conserved amongst C. jejuni isolates (Konkel et al., 1999a; Ripabelli et al., 2010; Ghorbanalizadgan et al., 2014; Levican et al., 2019; Wei et al., 2019). To explore this proposition, we assessed 20,218 full C. jejuni genome sequences that were available on the GenBank FTP server as of August 1st, 2019. After the removal of misidentified isolates (Campylobacter coli, Campylobacter upsaliensis, and Campylobacter lari), the remaining 20,166 sequences were searched for the presence of the cadF or flpA genes using the blastn command line tool with the default parameters. The cadF gene was absent in the genomes deposited for eight C. jejuni isolates, and the flpA gene was absent in the genomes for another seven isolates (Supplementary Figure S1). No genomes were identified in which both the cadF and flpA genes were absent. The caveat of this analysis is that the genomes reported for these 15 isolates may not be complete; it is not possible to determine the accuracy/completeness of the deposited sequences. At the very least, this analysis indicates that at a minimum, 99.93% of the sequenced C. jejuni isolates have both cadF and flpA. Based on this bioinformatic analysis, we conclude that cadF and flpA are ubiquitous and highly conserved amongst C. jejuni isolates.
Molecular-based diagnostics are being used more and more frequently in epidemiological studies. As opposed to conventional diagnostic methods utilizing a combination of culture and biochemical testing, molecular methods (e.g., polymerase chain reaction, PCR) offer the advantage of being fast. Relevant to this review is that cadF is a common target gene for the testing of both human and animal samples for Campylobacter spp. More specifically, epidemiologists have utilized cadF as a C. jejuni/C. coli-specific diagnostic marker. In contrast to other potential virulence-associated genes [e.g., the CDT subunit genes (cdtA, cdtB, and cdtC), iamA, iamB, and virB11], cadF is present in nearly 100% of C. jejuni and C. coli isolates (Supplementary Figure S1) (Ghorbanalizadgan et al., 2014; Levican et al., 2019; Wei et al., 2019). The cadF and flpA genes do not appear to be present in other pathogenic bacteria. Given that molecular diagnostics is becoming a more common approach, we have cited only a few articles where researchers have used PCR amplification of the cadF gene for diagnostic purposes (Ghorbanalizadgan et al., 2014; Francois et al., 2018; Levican et al., 2019; Wei et al., 2019).
Perspective (Summary and Future Directions)
MSCRAMMs are fascinating in that they help establish an infection for many Gram-positive and Gram-negative bacteria. While many pathogenic bacteria harbor MSCRAMMs, it is evident from several decades of research that C. jejuni is a unique pathogen. The identification of CadF and FlpA in C. jejuni, two distinctive MSCRAMMs, is a perfect illustration of this fact, as the FN-binding domains within CadF and FlpA are unique among FNBPs. Moreover, CadF and FlpA are two proteins that have drawn the attention of researchers due to their adhesive properties, as well as their potential role in stimulating host cell signaling pathways and cytoskeletal rearrangement (via a mechanism that is either direct or indirect).
The CadF and FlpA FNBPs from C. jejuni are currently recognized as important virulence factors, as they are involved in the early interactions occurring at the bacteria-host cell interface. Both CadF and FlpA bind to FN, facilitate C. jejuni adherence to host cells, and enable C. jejuni colonization of chickens at a high level. Also known are the FN-binding domains within both the CadF and FlpA proteins, as well as the FN fragment to which the FlpA protein binds. Experimental evidence suggests that C. jejuni binding to FN allows the microorganism to communicate with the cell cytoskeleton via outside-in signaling through integrins, leading to bacterial internalization into a host cell. Internalization by an intestinal cell allows the bacterium to avoid the powerful killing mechanisms of a phagocyte. Several studies suggest that C. jejuni invasion of the cells lining the intestinal tract is a primary mechanism of colonic damage (van Spreeuwel et al., 1985; Babakhani et al., 1993; Russell et al., 1993). While CadF and FlpA are required for maximal host cell invasion (Eucker and Konkel, 2012), the contribution of these two proteins to invasion is, in part, due to their role in triggering host cell signaling pathways, possibly by aiding the delivery of effector proteins. Relevant to this point is that CadF and FlpA are involved in the activation of Cdc42 and Rac1 in human cells (Krause-Gruszczynska et al., 2011; Boehm et al., 2012; Eucker and Konkel, 2012; Larson et al., 2013); these are the two primary host cell Rho GTPases involved in C. jejuni invasion. These findings are likely due to the fact that C. jejuni adhesion mutants, including a flpA mutant, are impaired in the delivery of effector proteins to host cells (Larson et al., 2013). In addition, we propose that CadF and FlpA are both involved in the activation of the β1 integrin, which is required for C. jejuni invasion (Krause-Gruszczynska et al., 2011; Konkel et al., 2013). Studies are needed to address outstanding questions (e.g., Where does CadF bind within the FN molecule?; Do CadF and FlpA bind to FN simultaneously or is binding sequential?; Do CadF and FlpA bind to other substrates or ECM components?; What role do CadF and FlpA play individually in binding to FN and initiating downstream host cell signaling events?; Do other C. jejuni adhesins work cooperatively with CadF and FlpA?; Are CadF and/or FlpA necessary for biofilm formation?, etc.).
In spite of CadF and FlpA having unique FN-binding domains, the mechanistic basis for how they modify FN function will likely have broader implications given the number of pathogenic microbes that have FNBPs. As mentioned earlier, the FN molecule is a complex glycoprotein composed of multiple repeating domains. The different domains in FN allow this glycoprotein to bind to cells and to molecules within the surrounding matrix simultaneously. Moreover, pathogen binding to specific domains may afford different functional outcomes (e.g., cell binding, cell invasion, etc.). While many FNBPs target the 29 kDa N-terminal fragment of FN that is composed of five FN I repeats (FN I1–5), other FNBPs bind to the 40–45 kDa FN fragment that is comprised of four FN I and two FN II repeats (FN I6 - FN II1–2 - FN I7–9). While FlpA has been determined to bind to the 40–45 kDa fragment, it is not known where CadF binds to the FN molecule. It is possible that the binding of a pathogen to a distinct FN domain can alter various FN functions, including the modulation of integrin function. It will be of interest to compare the functional attributes of the CadF and FlpA proteins from C. jejuni with the FNBPs from other more intensely studied bacterial pathogens, as this additional information may provide further insight into pathogenic mechanisms. Finally, continued investigation of FNBPs is necessary to provide a greater understanding of their diversity and specificity in mediating bacteria-host cell interactions. Please note that following the submission of this article, we submitted a research article on the cooperative interaction of CadF and FlpA in binding to FN and host cells that has been published (Talukdar et al., 2020).
Author Contributions
MK worked on the original draft preparation, Table 1 preparation, project design and management. PT reviewed the manuscript, prepared Figure 1 and Supplementary Tables S1, S2. NN reviewed the manuscript, prepared Figure 2 and Supplementary Figure S1. CK reviewed the manuscript and prepared Figure 2.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Kerry Cooper (University of Arizona), Dr. Christopher R. Gourley, and Kyrah L. Turner for proofreading this manuscript.
Funding. Research in the Konkel Lab is supported, in part, by a grant from the National Institutes of Health to MK (Award Number R01AI125356). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00564/full#supplementary-material
References
- Almagro Armenteros J. J., Tsirigos K. D., Sonderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37 420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Asuming-Bediako N., Parry-Hanson Kunadu A., Abraham S., Habib I. (2019). Campylobacter at the human-food interface: the African perspective. Pathogens 8:E87. 10.3390/pathogens8020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad W. A., Hess C., Hess M. (2018). Re-thinking the chicken-Campylobacter jejuni interaction: a review. Avian Pathol. 47 352–363. 10.1080/03079457.2018.1475724 [DOI] [PubMed] [Google Scholar]
- Babakhani F. K., Bradley G. A., Joens L. A. (1993). Newborn piglet model for campylobacteriosis. Infect. Immun. 61 3466–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2003). The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185 2910–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Kaya K. N., Hancock D. D., Call D. R., Park Y. H., Besser T. E. (2005). Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl. Environ. Microbiol. 71 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R. E., Levine M. M., Clements M. L., Hughes T. P., Blaser M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157 472–479. [DOI] [PubMed] [Google Scholar]
- Blaser M. J., Berkowitz I. D., Laforce F. M., Cravens J., Reller L. B., Wang W. L. (1979). Campylobacter enteritis: clinical and epidemiologic features. Ann. Intern. Med. 91 179–185. [DOI] [PubMed] [Google Scholar]
- Boehm M., Hoy B., Rohde M., Tegtmeyer N., Baek K. T., Oyarzabal O. A., et al. (2012). Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog. 4:3. 10.1186/1757-4749-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Krause-Gruszczynska M., Rohde M., Tegtmeyer N., Takahashi S., Oyarzabal O. A., et al. (2011). Major host factors involved in epithelial cell invasion of Campylobacter jejuni: role of fibronectin, integrin beta1, FAK, Tiam-1, and DOCK180 in activating Rho GTPase Rac1. Front. Cell. Infect. Microbiol. 1:17. 10.3389/fcimb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronowski C., James C. E., Winstanley C. (2014). Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 356 8–19. 10.1111/1574-6968.12488 [DOI] [PubMed] [Google Scholar]
- Buelow D. R., Christensen J. E., Neal-McKinney J. M., Konkel M. E. (2011). Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol. Microbiol. 80 1296–1312. 10.1111/j.1365-2958.2011.07645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzler J. P., Skirrow M. B. (1979). Campylobacter enteritis. Clin. Gastroenterol. 8 737–765. [PubMed] [Google Scholar]
- Christensen J. E., Pacheco S. A., Konkel M. E. (2009). Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol. Microbiol. 73 650–662. 10.1111/j.1365-2958.2009.06797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker A. O., Isokpehi R. D., Thomas B. N., Amisu K. O., Obi C. L. (2002). Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N., Karimova G., Ladant D. (2002). Bordetella pertussis adenylate cyclase toxin: a versatile screening tool. Toxicon 40 1383–1387. [DOI] [PubMed] [Google Scholar]
- de Oliveira M. G., Rizzi C., Galli V., Lopes G. V., Haubert L., Dellagostin O. A., et al. (2019). Presence of genes associated with adhesion, invasion, and toxin production in Campylobacter jejuni isolates and effect of temperature on their expression. Can. J. Microbiol. 65 253–260. 10.1139/cjm-2018-0539 [DOI] [PubMed] [Google Scholar]
- Deakin N. O., Turner C. E. (2008). Paxillin comes of age. J. Cell Sci. 121 2435–2444. 10.1242/jcs.018044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekeyser P., Gossuin-Detrain M., Butzler J. P., Sternon J. (1972). Acute enteritis due to related vibrio: first positive stool cultures. J. Infect. Dis. 125 390–392. [DOI] [PubMed] [Google Scholar]
- Desvaux M., Hebraud M., Henderson I. R., Pallen M. J. (2006). Type III secretion: what’s in a name? Trends Microbiol. 14 157–160. [DOI] [PubMed] [Google Scholar]
- Engberg J., Aarestrup F. M., Taylor D. E., Gerner-Smidt P., Nachamkin I. (2001). Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eucker T. P., Konkel M. E. (2012). The cooperative action of bacterial fibronectin-binding proteins and secreted proteins promote maximal Campylobacter jejuni invasion of host cells by stimulating membrane ruffling. Cell Microbiol. 14 226–238. 10.1111/j.1462-5822.2011.01714.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eucker T. P., Samuelson D. R., Hunzicker-Dunn M., Konkel M. E. (2014). The focal complex of epithelial cells provides a signalling platform for interleukin-8 induction in response to bacterial pathogens. Cell Microbiol. 16 1441–1455. 10.1111/cmi.12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchere J. L., Kervella M., Rosenau A., Mohanna K., Veron M. (1989). Adhesion to HeLa cells of Campylobacter jejuni and C. coli outer membrane components. Res. Microbiol. 140 379–392. [DOI] [PubMed] [Google Scholar]
- Flanagan R. C., Neal-McKinney J. M., Dhillon A. S., Miller W. G., Konkel M. E. (2009). Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 77 2399–2407. 10.1128/IAI.01266-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois R., Yori P. P., Rouhani S., Siguas Salas M., Paredes Olortegui M., Rengifo Trigoso D., et al. (2018). The other Campylobacters: not innocent bystanders in endemic diarrhea and dysentery in children in low-income settings. PLoS Negl. Trop. Dis. 12:e0006200. 10.1371/journal.pntd.0006200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Stewart K. M., Weaver V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C. R., Hoekstra R. M., Samuel M., Marcus R., Bender J., Shiferaw B., et al. (2004). Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3) S285–S296. [DOI] [PubMed] [Google Scholar]
- Ghorbanalizadgan M., Bakhshi B., Kazemnejad Lili A., Najar-Peerayeh S., Nikmanesh B. (2014). A molecular survey of Campylobacter jejuni and Campylobacter coli virulence and diversity. Iran Biomed. J. 18 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibreel A., Taylor D. E. (2006). Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58 243–255. [DOI] [PubMed] [Google Scholar]
- Greene C., Mcdevitt D., Francois P., Vaudaux P. E., Lew D. P., Foster T. J. (1995). Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17 1143–1152. [DOI] [PubMed] [Google Scholar]
- Guccione E. J., Kendall J. J., Hitchcock A., Garg N., White M. A., Mulholland F., et al. (2017). Transcriptome and proteome dynamics in chemostat culture reveal how Campylobacter jejuni modulates metabolism, stress responses and virulence factors upon changes in oxygen availability. Environ. Microbiol. 19 4326–4348. 10.1111/1462-2920.13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G., Ma W., Maurer L. M., Potts J. R., Mosher D. F. (2014). Borrelia burgdorferi protein BBK32 binds to soluble fibronectin via the N-terminal 70-kDa region, causing fibronectin to undergo conformational extension. J. Biol. Chem. 289 22490–22499. 10.1074/jbc.M114.578419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B., Nair S., Pallas J., Williams M. A. (2011). Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 35 147–200. 10.1111/j.1574-6976.2010.00243.x [DOI] [PubMed] [Google Scholar]
- Hermans D., Pasmans F., Heyndrickx M., Van Immerseel F., Martel A., Van Deun K., et al. (2012). A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut. Crit. Rev. Microbiol. 38 17–29. 10.3109/1040841X.2011.615298 [DOI] [PubMed] [Google Scholar]
- Hong S., Cha I., Kim N. O., Seo J. B., Kim S. Y., Kim J. H., et al. (2014). Comparative proteomic label-free analysis of Campylobacter jejuni NCTC 11168 cultured with porcine mucin. Foodborne Pathog. Dis. 11 240–247. 10.1089/fpd.2013.1596 [DOI] [PubMed] [Google Scholar]
- Horrocks S. M., Anderson R. C., Nisbet D. J., Ricke S. C. (2009). Incidence and ecology of Campylobacter jejuni and coli in animals. Anaerobe 15 18–25. 10.1016/j.anaerobe.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Hu L., Mcdaniel J. P., Kopecko D. J. (2006). Signal transduction events involved in human epithelial cell invasion by Campylobacter jejuni 81-176. Microb. Pathog. 40 91–100. [DOI] [PubMed] [Google Scholar]
- Huang H., Brooks B. W., Lowman R., Carrillo C. D. (2015). Campylobacter species in animal, food, and environmental sources, and relevant testing programs in Canada. Can. J. Microbiol. 61 701–721. 10.1139/cjm-2014-0770 [DOI] [PubMed] [Google Scholar]
- Humphrey S., Chaloner G., Kemmett K., Davidson N., Williams N., Kipar A., et al. (2014). Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio 5:e01364-14. 10.1128/mBio.01364-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymes J. P., Klaenhammer T. R. (2016). Stuck in the Middle: fibronectin-binding proteins in Gram-positive bacteria. Front. Microbiol. 7:1504. 10.3389/fmicb.2016.01504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A., Phansopa C., Suwannachart C., Craven C. J., Rafferty J. B., Kelly D. J. (2011). The virulence factor PEB4 (Cj0596) and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni. J. Biol. Chem. 286 21254–21265. 10.1074/jbc.M111.220442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev A. V., Wren B. W. (2005). Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl. Environ. Microbiol. 71 4004–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmali M. A., Fleming P. C. (1979). Campylobacter enteritis in children. J. Pediatr. 94 527–533. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Singvall J., Schwarz-Linek U., Johnson B. J., Potts J. R., Hook M. (2004). BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J. Biol. Chem. 279 41706–41714. [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Christensen J. E., Keech A. M., Monteville M. R., Klena J. D., Garvis S. G. (2005). Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol. Microbiol. 57 1022–1035. [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Cieplak W., Jr. (1992). Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect. Immun. 60 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Garvis S. G., Tipton S. L., Anderson D. E., Jr., Cieplak W., Jr. (1997). Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24 953–963. [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Gray S. A., Kim B. J., Garvis S. G., Yoon J. (1999a). Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J. Clin. Microbiol. 37 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Kim B. J., Rivera-Amill V., Garvis S. G. (1999b). Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32 691–701. [DOI] [PubMed] [Google Scholar]
- Konkel M. E., Klena J. D., Rivera-Amill V., Monteville M. R., Biswas D., Raphael B., et al. (2004). Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186 3296–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Larson C. L., Flanagan R. C. (2010). Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 192 68–76. 10.1128/JB.00969-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Samuelson D. R., Eucker T. P., Shelden E. A., O’loughlin J. L. (2013). Invasion of epithelial cells by Campylobacter jejuni is independent of caveolae. Cell Commun. Signal. 11:100. 10.1186/1478-811X-11-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolman L., Whyte P., Burgess C., Bolton D. (2016). Virulence gene expression, adhesion and invasion of Campylobacter jejuni exposed to oxidative stress (H2O2). Int. J. Food Microbiol. 220 33–38. 10.1016/j.ijfoodmicro.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Krause-Gruszczynska M., Boehm M., Rohde M., Tegtmeyer N., Takahashi S., Buday L., et al. (2011). The signaling pathway of Campylobacter jejuni-induced Cdc42 activation: role of fibronectin, integrin beta1, tyrosine kinases and guanine exchange factor Vav2. Cell Commun. Signal. 9:32. 10.1186/1478-811X-9-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause-Gruszczynska M., Rohde M., Hartig R., Genth H., Schmidt G., Keo T., et al. (2007). Role of the small Rho GTPases Rac1 and Cdc42 in host cell invasion of Campylobacter jejuni. Cell Microbiol. 9 2431–2444. [DOI] [PubMed] [Google Scholar]
- Kuusela P. (1978). Fibronectin binds to Staphylococcus aureus. Nature 276 718–720. [DOI] [PubMed] [Google Scholar]
- Kuusela P., Moran A. P., Vartio T., Kosunen T. U. (1989). Interaction of Campylobacter jejuni with extracellular matrix components. Biochim. Biophys. Acta 993 297–300. [DOI] [PubMed] [Google Scholar]
- Ladant D., Ullmann A. (1999). Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol. 7 172–176. [DOI] [PubMed] [Google Scholar]
- Larson C. L., Samuelson D. R., Eucker T. P., O’loughlin J. L., Konkel M. E. (2013). The fibronectin-binding motif within FlpA facilitates Campylobacter jejuni adherence to host cell and activation of host cell signaling. Emerg. Microbes Infect. 2:e65. 10.1038/emi.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kempis Mdel R., Guccione E., Mulholland F., Williamson M. P., Kelly D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60 1262–1275. [DOI] [PubMed] [Google Scholar]
- Levican A., Ramos-Tapia I., Briceno I., Guerra F., Mena B., Varela C., et al. (2019). Genomic analysis of Chilean strains of Campylobacter jejuni from human faeces. Biomed. Res. Int. 2019:1902732. 10.1155/2019/1902732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert R., Gross U., Zautner A. E. (2015). Campylobacter jejuni: components for adherence to and invasion of eukaryotic cells. Berl. Munch. Tierarztl. Wochenschr. 128 90–97. [PubMed] [Google Scholar]
- Mamelli L., Pages J. M., Konkel M. E., Bolla J. M. (2006). Expression and purification of native and truncated forms of CadF, an outer membrane protein of Campylobacter. Int. J. Biol. Macromol. 39 135–140. [DOI] [PubMed] [Google Scholar]
- McSweegan E., Walker R. I. (1986). Identification and characterization of two Campylobacter jejuni adhesins for cellular and mucous substrates. Infect. Immun. 53 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min T., Vedadi M., Watson D. C., Wasney G. A., Munger C., Cygler M., et al. (2009). Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes. Biochemistry 48 3057–3067. 10.1021/bi802195d [DOI] [PubMed] [Google Scholar]
- Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005). Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6 56–68. [DOI] [PubMed] [Google Scholar]
- Monteville M. R., Konkel M. E. (2002). Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 70 6665–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteville M. R., Yoon J. E., Konkel M. E. (2003). Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149 153–165. [DOI] [PubMed] [Google Scholar]
- Moser I., Schroeder W., Salnikow J. (1997). Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol. Lett. 157 233–238. [DOI] [PubMed] [Google Scholar]
- Naz N. (2014). Reinvestigation into the Mechanism of Campylobacter jejuni Invasion of Intestinal Epithelial Cells. Doctoral Dissertation, London School of Hygiene and Tropical Medicine, London. [Google Scholar]
- Neal-McKinney J. M., Konkel M. E. (2012). The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front. Cell Infect. Microbiol. 2:31. 10.3389/fcimb.2012.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal-McKinney J. M., Samuelson D. R., Eucker T. P., Nissen M. S., Crespo R., Konkel M. E. (2014). Reducing Campylobacter jejuni colonization of poultry via vaccination. PLoS One 9:e114254. 10.1371/journal.pone.0114254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negretti N. M., Clair G., Talukdar P. K., Gourley C. R., Huynh S., Adkins J. N., et al. (2019). Campylobacter jejuni demonstrates conserved proteomic and transcriptomic responses when co-cultured with human INT 407 and Caco-2 epithelial cells. Front. Microbiol. 10:755. 10.3389/fmicb.2019.00755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik V., Hofreuter D., Galan J. E. (2010). Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 78 3540–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Park K. S., Ueta M., Iida T., Honda T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74 1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Yamada K. M. (2002). Fibronectin at a glance. J. Cell Sci. 115 3861–3863. [DOI] [PubMed] [Google Scholar]
- Pentecost M., Otto G., Theriot J. A., Amieva M. R. (2006). Listeria monocytogenes invades the epithelial junctions at sites of cell extrusion. PLoS Pathog. 2:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerda J., Cossart P. (2006). Bacterial adhesion and entry into host cells. Cell 124 715–727. [DOI] [PubMed] [Google Scholar]
- Potts J. R., Campbell I. D. (1994). Fibronectin structure and assembly. Curr. Opin. Cell Biol. 6 648–655. [DOI] [PubMed] [Google Scholar]
- Ripabelli G., Tamburro M., Minelli F., Leone A., Sammarco M. L. (2010). Prevalence of virulence-associated genes and cytolethal distending toxin production in Campylobacter spp. isolated in Italy. Comp. Immunol. Microbiol. Infect. Dis. 33 355–364. 10.1016/j.cimid.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Rubinchik S., Seddon A., Karlyshev A. V. (2012). Molecular mechanisms and biological role of Campylobacter jejuni attachment to host cells. Eur. J. Microbiol. Immunol. (Bp) 2 32–40. 10.1556/EuJMI.2.2012.1.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M. (2007). The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44 701–703. [DOI] [PubMed] [Google Scholar]
- Russell R. G., O’donnoghue M., Blake D. C., Jr., Zulty J., Detolla L. J. (1993). Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168 210–215. [DOI] [PubMed] [Google Scholar]
- Sahin O., Luo N., Huang S., Zhang Q. (2003). Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Appl. Environ. Microbiol. 69 5372–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson D. R., Eucker T. P., Bell J. A., Dybas L., Mansfield L. S., Konkel M. E. (2013). The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease. Cell Commun. Signal. 11:79. 10.1186/1478-811X-11-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson D. R., Konkel M. E. (2013). Serine phosphorylation of cortactin is required for maximal host cell invasion by Campylobacter jejuni. Cell Commun. Signal. 11:82. 10.1186/1478-811X-11-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M. D. (2001). Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20 6459–6472. [DOI] [PubMed] [Google Scholar]
- Schechter L. M., Vencato M., Jordan K. L., Schneider S. E., Schneider D. J., Collmer A. (2006). Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 19 1180–1192. [DOI] [PubMed] [Google Scholar]
- Schlumberger M. C., Muller A. J., Ehrbar K., Winnen B., Duss I., Stecher B., et al. (2005). Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc. Natl. Acad. Sci. U.S.A. 102 12548–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. M., Escher U., Mousavi S., Tegtmeyer N., Boehm M., Backert S., et al. (2019). Immunopathological properties of the Campylobacter jejuni flagellins and the adhesin CadF as assessed in a clinical murine infection model. Gut Pathog. 11:24. 10.1186/s13099-019-0306-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. (1983). Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell 35 421–431. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Hook M., Campbell I. D., et al. (2004). High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J. Biol. Chem. 279 39017–39025. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., et al. (2003). Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423 177–181. [DOI] [PubMed] [Google Scholar]
- Schwerer B. (2002). Antibodies against gangliosides: a link between preceding infection and immunopathogenesis of Guillain-Barré syndrome. Microbes Infect. 4 373–384. [DOI] [PubMed] [Google Scholar]
- Scott N. E., Marzook N. B., Deutscher A., Falconer L., Crossett B., Djordjevic S. P., et al. (2010). Mass spectrometric characterization of the Campylobacter jejuni adherence factor CadF reveals post-translational processing that removes immunogenicity while retaining fibronectin binding. Proteomics 10 277–288. 10.1002/pmic.200900440 [DOI] [PubMed] [Google Scholar]
- Sebald M., Véron M. (1963). Base DNA content and classification of vibrios. Ann. Inst. Pasteur (Paris) 105 897–910. [PubMed] [Google Scholar]
- Sela S., Aviv A., Tovi A., Burstein I., Caparon M. G., Hanski E. (1993). Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10 1049–1055. [DOI] [PubMed] [Google Scholar]
- Shoaf-Sweeney K. D., Larson C. L., Tang X., Konkel M. E. (2008). Identification of Campylobacter jejuni proteins recognized by maternal antibodies of chickens. Appl. Environ. Microbiol. 74 6867–6875. 10.1128/AEM.01097-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Carraher C., Schwarzbauer J. E. (2010). Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 26 397–419. 10.1146/annurev-cellbio-100109-104020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. L., Gourdon D., Little W. C., Kubow K. E., Eguiluz R. A., Luna-Morris S., et al. (2007). Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 5:e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory M. P., Cornelis G. R. (1994). Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14 583–594. [DOI] [PubMed] [Google Scholar]
- Sottile J., Hocking D. C. (2002). Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol. Biol. Cell 13 3546–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stones D. H., Krachler A. M. (2015). Fatal attraction: how bacterial adhesins affect host signaling and what we can learn from them. Int. J. Mol. Sci. 16 2626–2640. 10.3390/ijms16022626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stones D. H., Krachler A. M. (2016). Against the tide: the role of bacterial adhesion in host colonization. Biochem. Soc. Trans. 44 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedhem A., Kaijser B. (1980). Campylobacter fetus subspecies jejuni: a common cause of diarrhea in Sweden. J. Infect. Dis. 142 353–359. [DOI] [PubMed] [Google Scholar]
- Talukdar P. K., Negretti N. M., Turner K. L., Konkel M. E. (2020). Molecular dissection of the Campylobacter jejuni CadF and FlpA virulence proteins in binding to host cell fibronectin. Microorganisms 8:389. 10.3390/microorganisms8030389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelemann A., Petti F., Griffin G., Iwata K., Hunt T., Settinari T., et al. (2005). Phosphotyrosine signaling networks in epidermal growth factor receptor overexpressing squamous carcinoma cells. Mol. Cell. Proteomics 4 356–376. [DOI] [PubMed] [Google Scholar]
- To W. S., Midwood K. S. (2011). Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair 4:21. 10.1186/1755-1536-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang T. M., Felek S., Krukonis E. S. (2010). Ail binding to fibronectin facilitates Yersinia pestis binding to host cells and Yop delivery. Infect. Immun. 78 3358–3368. 10.1128/IAI.00238-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q. V., Mcguckin M. A., Mendz G. L. (2008). Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 57 795–802. 10.1099/jmm.0.47752-0 [DOI] [PubMed] [Google Scholar]
- Vaca D. J., Thibau A., Schutz M., Kraiczy P., Happonen L., Malmstrom J., et al. (2019). Interaction with the host: the role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med. Microbiol. Immunol. 10.1007/s00430-019-00644-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spreeuwel J. P., Duursma G. C., Meijer C. J., Bax R., Rosekrans P. C., Lindeman J. (1985). Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 26 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1985). Signal sequences. The limits of variation. J. Mol. Biol. 184 99–105. [DOI] [PubMed] [Google Scholar]
- Walker R. I., Schmauder-Chock E. A., Parker J. L., Burr D. (1988). Selective association and transport of Campylobacter jejuni through M cells of rabbit Peyer’s patches. Can. J. Microbiol. 34 1142–1147. [DOI] [PubMed] [Google Scholar]
- Wei B., Kang M., Jang H. K. (2019). Genetic characterization and epidemiological implications of Campylobacter isolates from wild birds in South Korea. Transbound. Emerg. Dis. 66 56–65. 10.1111/tbed.12931 [DOI] [PubMed] [Google Scholar]
- Weinberg S. H., Mair D. B., Lemmon C. A. (2017). Mechanotransduction dynamics at the cell-matrix interface. Biophys. J. 112 1962–1974. 10.1016/j.bpj.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. S., Baralle F. E., Muro A. F. (2008). New insights into form and function of fibronectin splice variants. J. Pathol. 216 1–14. 10.1002/path.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnen B., Schlumberger M. C., Sturm A., Schupbach K., Siebenmann S., Jenny P., et al. (2008). Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS One 3:e2178. 10.1371/journal.pone.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (2007). Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh. Migr. 1 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. (2007). A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120 137–148. [DOI] [PubMed] [Google Scholar]
- Ziprin R. L., Young C. R., Stanker L. H., Hume M. E., Konkel M. E. (1999). The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 43 586–589. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.