Abstract
Background and Purpose
Patients with locally advanced breast cancer usually receive third‐generation neoadjuvant chemotherapy (NAC). Although NAC treatment improved the overall survival, patients' response varies, some acquire resistance and others exhibit a conversion in their breast cancer molecular subtype. We aimed to identify the molecular changes involved in NAC resistance attempting to find new therapeutic targets in different breast cancer subtypes.
Experimental Approach
We modelled NAC treatments used in clinical practice and generated resistant cell lines in vitro. The resistant cells were generated by consecutive treatment with four cycles of doxorubicin (adriamycin)/cyclophosphamide (4xAC) followed by an additional four cycles of paclitaxel (4xAC + 4xPAC).
Key Results
Our data revealed distinct mechanisms of resistance depending on breast cancer subtype and drugs used. MDA‐MB‐231 cells resistant to 4xAC + 4xPAC activated neuropilin‐1/TNC/integrin β3/FAK/NF‐κBp65 axis and displayed a decrease in breast cancer resistance protein (BCRP/ABCB2). However, MCF7 cells resistant to 4xAC treatments induced HER2 expression, which converted MCF7 subtype from luminal A to luminal B HER2 type, up‐regulated neuropilin‐1, oestrogen receptor‐α, and EGFR, and activated PI3K/Akt/NF‐κBp65 axis. However, MCF7 cells resistant to 4xAC + 4xPAC exhibited down‐regulation of the survival axis and up‐regulated BCRP/ABCG2. Co‐immunoprecipitation demonstrated a novel interaction between HER2 and neuropilin‐1 driving the resistance features.
Conclusions and Implications
The concurrent increase in neuropilin‐1 and HER2 upon resistance and the inverse relationship between neuropilin‐1 and BCRP/ABCG2 suggest that, in addition to HER2, neuropilin‐1 status should be assessed in patients undergoing NAC, and as a potential drug target for refractory breast cancer.
Abbreviations
- AKT
Protein kinase B (PKB)
- DNMT
DNA methyltransferases
- EGFR
Epidermal Growth Factor
- EMT
Epithelial to Mesenchymal Transition
- FAK
Focal Adhesion Kinase
- HER‐2
Human Epidermal Receptor 2
- HDAC
Histone Deacetylase
- ITGB3
Integrin β3
- NF‐κB
Nuclear Factor kappa‐light‐chain‐enhancer ofactivated B cells
- PI3K
Phosphatidylinositol‐4,5‐bisphosphate 3‐kinase
- PTEN
Phosphatase and tensin homolog
- ZEB1
Zinc finger E‐box‐binding homeobox 1
- ABCG2
ATP‐binding cassette transporter G2
- BCRP
breast cancer resistance protein
- ECD
extracellular domain
- ICD
intracellular domain
- NAC
neoadjuvant chemotherapy
- NRP‐1
https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2998
- TNC
tenascin C
What is already known
Clinically, neoadjuvant chemotherapy (NAC) could change breast cancer molecular subtype.
Neuropilin‐1 promotes resistance to anti‐angiogenic agents through TGF‐β signalling.
What this study adds
Distinct mechanisms of neoadjuvant chemotherapy resistance were identified in different subtypes of breast cancer cells.
Neuropilin‐1 binds HER2 and has an inverse expression pattern to ABCG2, conferring chemotherapy resistance.
What is the clinical significance
Neuropilin‐1 could be a potential diagnostic and drug target biomarker for refractory breast cancer.
1. INTRODUCTION
Drug resistance and breast cancer recurrence are still the main hurdles for oncologists. Breast cancer patients with locally advanced tumours are receiving neoadjuvant chemotherapy (NAC) using the metronomic administration of chemotherapies, which refers to the chronic, equally spaced administration of low doses of various chemotherapeutic drugs without extended rest periods (Masood, 2016; Munzone & Colleoni, 2015; Scharovsky, Mainetti, & Rozados, 2009). An example of the combination chemotherapies used in clinical management of breast cancer is anthracycline‐based poly‐chemotherapy consisting of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4789, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7069 (adriamycin), and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7154. Taxanes were also used in combination with anthracycline‐based regimens and improved patient outcomes (Early Breast Cancer Trialists' Collaborative Group, 2005; Laurentiis et al., 2008). Breast tumours recur due to the presence of a certain population of cells, which either acquired resistance during NAC treatment or from new cells evolved from what is known to be a tumourigenic breast cancer cell population (Al‐Hajj, Wicha, Benito‐Hernandez, Morrison, & Clarke, 2003).
In this study, we aimed to understand why the “one‐size‐fits‐all” approach in treating breast cancer is not effective. The response to NAC differs among patients with different breast cancer subtypes receiving the same NAC regimen (Rouzier et al., 2005), which brings up the most important question, what determines response to treatment? Is it the patient's genetics, epigenetics, or molecular signalling that determine the outcome? To answer these questions and understand the molecular differences between different subtypes of breast cancer cells and their response to NAC, we performed a translational in vitro model representing the commonly used NAC setting in clinical practice. We generated resistant variants from the breast cancer cell lines, MDA‐MB‐231 representing the triple negative and MCF7 cells representing the luminal A subtypes (Subik et al., 2010).
Several previous studies have shown the contribution of different molecules in conferring drug resistance. The transmembrane receptor, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2998 (NRP‐1) mediates angiogenesis through https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5085 and enhances https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5060 signalling via Smad and hence promotes resistance to anti‐angiogenic agents (Kwiatkowski et al., 2017). Previously, we reported a significant increase in NRP‐1 levels in the basal subtype of breast cancer, compared with other subtypes (Naik et al., 2017). The proto‐oncogene tyrosine kinase https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2019 is expressed in 20–25% of primary breast cancer cases and is correlated with disease progression and resistance (Modi et al., 2005; Yan, Yu, Zhang, Liu, & Li, 2017). MCF7 cells do not express HER2 (Lee, Oesterreich, & Davidson, 2015) but the recombinant overexpression of HER2 in MCF7 cells leads to intracellular signalling through activation of https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=673‐p110α and thus promoting the phosphorylation of https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=285, which consequently induced resistance to several chemotherapy drugs (Knuefermann et al., 2003). Several reports have shown that HER2 status in breast tumours can be reversed through the course of NAC treatment (Dieci et al., 2012; Niikura et al., 2015; Yoshida et al., 2017). Change of HER2 status from positive to negative in patients with recurring breast cancer was shown to have a poor outcome and low overall survival (Dieci et al., 2012). However, gaining HER2 did not show correlation with disease‐free survival, probably due to the introduction of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5082 in the treatment modalities that improves the treatment strategy (Yoshida et al., 2017). Therefore, it is crucial to study the molecular changes taking place during NAC, which leads to tumour subtype conversion in order to, eventually, find more precise targets for each subtype.
Our results indicated different mechanisms of resistance in the two cell lines. NAC induced resistance in MCF7 cancer cells through the up‐regulation of HER‐2 and NRP‐1 with evidence of their interaction. Our results identified an important role of NRP‐1 up‐regulation in resistance to NAC.
2. METHODS
2.1. Cell culture
Three breast cancer cell lines: MDA‐MB‐231 (CLS, Cat# 300275/NA, RRID:CVCL_0062), MCF7 (CLS, Cat# 300273/p2720_MCF‐7, RRID:CVCL_0031), and BT474 (CLS, Cat# 300131/p705_BT‐474, RRID:CVCL_0179), were purchased from CLS Cell Lines Service, Germany, in 2015. The cells were maintained in monolayer cultures in 5% CO2 incubator at 37°C. The MDA‐MB‐231 cells were grown in DMEM (Sigma, USA) supplemented with 5 mM of sodium pyruvate (Sigma, USA), whereas MCF7 cells were cultured in Opti‐MEM + 1× GlutaMAX (Gibco®, USA) and BT474 cells were grown in RPMI (Gibco, USA) with 5 mM of sodium pyruvate (Sigma, USA). All cells were supplemented with 10% FBS (Gibco, USA) and 2 mg·L−1 of gentamicin (Gibco, USA).
2.2. Establishment of resistant cell lines
The treatment modality of the cell lines was designed to mimic the NAC treatment of breast cancer patients and consisted of a combination of four cycles of 200 nM of doxorubicin; (Adriamycin, Pharmacia, Italy) and 600 nM of cyclophosphamide (4xAC; Cytoxan, Baxter, Germany) followed by an additional four cycles of 50 nM of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2770 (4xAC + 4xPAC; Taxol, EBEWE Pharma, Austria). Resistant breast cancer cells were generated in vitro by consequential treatment similar to a previous study by Kopp, Oak, Wagner, and Roidl (2012). Each treatment cycle was 72 hr long, and the remaining attached cells after each cycle were left to proliferate until confluency, and the following cycle of treatment was initiated immediately after. The resistant cells were collected after four cycles of AC (4xAC), and part of the generated 4xAC cells was further treated with four cycles of paclitaxel (4xPAC), which results in having cells resistant to a total of eight cycles (4xAC + 4xPAC). The generated resistant cells to 4xAC were obtained after 4 months since the start of the treatment, and the resistant cells to 4xAC + 4xPAC were obtained after additional 4 months of subsequent treatment.
2.3. Protein extraction and Western blot
The antibody‐based procedures used in this study comply with the recommendations made by the British Journal of Pharmacology. Protein lysates were extracted from the cells using a 1× lysis buffer (Cell Signaling Technology, USA), which contains 0.1% protease inhibitor (PMFS; Sigma, Germany). Protein concentrations were determined using DC™ protein assay kit (Bio‐Rad). A total of 100 μg of the protein lysate was loaded in 7.5% or 12% acrylamide gel depending on the MW of the protein assayed. Proteins were transferred onto the PVDF membrane (Bio‐Rad) using a semi‐dry Trans‐Blot® Turbo™ Transfer System (Bio‐Rad). Different primary antibodies with their dilutions and corresponding research resource identifier RRID are listed in Table S1. The primary antibodies were diluted in 5% milk and incubated overnight at 4°C. It is important to note that HER2 protein was detected using two different primary antibodies from Cell Signaling Technology that recognize two specific sequences, the extracellular domain HER2‐ECD (Cell Signaling Technology, Cat# 4290, RRID:AB_10557104) and the intracellular domain HER2‐ICD (Cell Signaling Technology, Cat# 2165, RRID:AB_10692490). Depending on the species where the primary antibody was raised, the secondary anti‐rabbit antibody conjugated with HRP (Cell Signaling Technology, Cat# 7074, RRID:AB_2099233) and the secondary anti‐mouse HRP‐conjugated antibody (Cell Signaling Technology, Cat# 7076, RRID:AB_330924) were used to visualize protein expression using the ChemiDoc™ Touch Imaging System (Bio‐Rad). GAPDH was used for normalization. In the ChemiDoc™ Touch Imaging System, there are two modes of exposure, manual and auto exposure. Therefore, on some occasions, the band intensity of the same protein may vary between blots depending on the exposure mode and the adjacent loaded samples, bearing in mind that the loaded quantity is always the same (100 μg).
2.4. RNA extraction and quantitative RT‐PCR
RNA was extracted from adherent control and resistant cells using TRI reagent (Ambion, USA) according to the TRIzol protocol specifications. Prior to cDNA synthesis, RNA concentration and purity were measured by NanoDrop™ 2000c spectrophotometer (Thermo Scientific, USA). Using high‐capacity reverse transcription kit (Applied Biosystems, USA), cDNA was synthesized from 1 μg of RNA and diluted at 5 ng·μl−1 in DEPC‐treated water and stored at −20°C prior to gene quantification. Reaction mixture for real‐time PCR was prepared using Fast SYBR Supermix (Applied Biosystems, USA); 15 ng of cDNA and 100 μM of specific primers were designed using Primer Express Software (Primer Express, RRID:SCR_014326; Applied Biosystems, USA). Primers for tenascin C (TNC) amplification and the reference genes, actin and GUSB, were listed in the additional file of our previous publication (Naik et al., 2018). The 7500 Fast Real‐Time PCR System (Applied Biosystems, USA) was used to quantify the expression of TNC under the following optimized conditions: enzyme activation at 95°C for 20 s followed by 40 cycles of denaturing at 95°C for 3 s and annealing/extension at 63.4°C for 30 s (primers concentration and annealing/extension were optimized before targets quantification). Melting curves were plotted for each reaction. The Ct results generated from the system were analysed using the 2−ΔΔCt method to get relative expression values.
2.5. Dual immunofluorescent staining
Prior to immunofluorescent staining, the cells (control, resistant, and HER2 transfected) were grown on positively charged slides. The cells were washed with PBS and fixed in 3.7% paraformaldehyde solution and washed in 0.05% Triton X‐100 for 5 min followed by three washes in PBS. The cells were blocked using 5% goat serum solution (Dako, Denmark) for 30 min, washed in PBS, and then incubated overnight at 4°C with both primary antibodies, rabbit anti‐neuropilin‐1 conjugated with Alexa Fluor® 647 (Abcam, Cat# ab198323, UK) and mouse anti‐ErbB 2 (Abcam, Cat# ab16899, RRID:AB_443534); the antibodies were diluted (1:100) in antibody diluent (Dako REAL Antibody Diluent, Cat# 2022, Denmark). The slides were then washed in PBS and incubated with an anti‐mouse IgG secondary antibody conjugated with Alexa Fluor® 488 (Cell Signaling Technology, Cat# 4408, RRID:AB_10694704) for 1 hr at room temperature. The slides were washed in PBS and counterstained with DAPI. Fluorescent mounting medium was added (Dako, Denmark) and observed blindly under the fluorescent microscope (Nikon H600L, Japan). Staining negative controls was done by incubating the cells in the antibody diluent without primary antibodies. The experiments were repeated five independent times, and the fluorescence intensity of at least 15 cells per replica was measured using ImageJ software (ImageJ, RRID:SCR_003070).
2.6. Co‐immunoprecipitation assay
The interaction between NRP‐1/HER2 and NRP‐1/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1797 was assessed by co‐immunoprecipitation (Co‐IP) assay. The Dynabeads M‐280 Sheep Anti‐Rabbit IgG (Invitrogen, USA) were washed with 1× protein lysis buffer (Cell Signaling Technology, USA) and incubated with either HER2‐ICD or EGFR primary antibodies overnight shaking at 4°C. The next day, the incubated antibody beads were washed three times with 1× cold lysis buffer and re‐incubated with 1,000 μg of protein lysates at 4°C shaking overnight. The following day, the beads were washed three times and the immunoprecipitated proteins were eluted using denaturing sample buffer (0.01% bromophenol blue, 20% SDS, 10% glycerol, 12% 0.5 M of Tris—pH 6.8, and 5% β‐mercaptoethanol) at 95°C for 5 min. The eluted proteins were run on SDS‐PAGE, and Western blot was performed; the blots were probed for NRP‐1 antibody in the first day and re‐probed for the antibody used for IP on the second day confirming the success of the IP procedure. Single and multichannel images were captured using the ChemiDoc™ Touch Imaging System (Bio‐Rad, USA).
2.7. Overexpression of HER2 (Gateway cloning)
HER2 cDNA (3,700 bp, NCBI Reference Sequence: NM_004448) was obtained in an entry Gateway‐compatible plasmid pANT7_cGST from DNASU Plasmid Repository AZ, USA. HER2 cDNA was amplified using NAP150 forward primer 5′‐CCCATTGTATGGGATCTGATC‐3′ and NAP138R reverse primer 5′‐TGTTTCGCCATTTATCACCTTC‐3′ transferred through recombination into the P‐Donr vector using BP Clonase (Invitrogen, USA) and transformed into Escherichia coli cells by electroporation. Transformed cells containing the cloned HER2 in p‐Donr plasmid were selected on LB kanamycin (0.05 mg·ml−1). The purified plasmid containing HER2 cDNA was recombined with the destination vector pcDNA™‐DEST40 (Thermo Scientific, USA) using LR Clonase (Invitrogen, USA). The destination vector containing HER2 cDNA was then transformed to TOP10 competent E. coli cells. The transformed clones were selected on LB ampicillin (0.1 mg·ml−1). HER2 fragment in the expression vector was further confirmed by PCR and sequencing using the two primers present in the destination vector; T7 5′‐TAATACGACTCACTATAG‐3′ and V5 5′‐ACCGAGGAGAGGGTTAGGGAT‐3′ (Macrogen, Seoul, South Korea; Macrogen, RRID:SCR_014454). MDA‐MB‐231 and MCF7 cells were transfected with HER2 expression vector using Lipofectamine 2000 (Thermo Fisher, USA) in opti‐MEM media without FBS (Gibco, USA) and incubated for 48 hr in CO2 incubator at 37°C. Consequently, the transfected cells were selected using 600 μg·ml−1 of geneticin G418. HER2 level was detected using two HER2 antibodies from Cell Signaling: D8F12 detecting the extracellular domain and 29D8 detecting the intracellular domain.
2.8. Spheroid assay
Spheroids were generated by blocking the surface of 96‐well plate with 1.5% agarose to decrease the attachments of the cells to the surface and enhance the adhesion between the cells. The 96‐well plate was incubated for 30 min to solidify before the cells were seeded. Ten thousand cells per well were seeded from MDA‐MB‐231 and MCF7 control, resistant variants and from HER2‐transfected cells. The formed mammospheres were randomized and blindly quantified from five different images and five different independent experiments (n = 5).
2.9. Wound healing assay
To determine the migration ability (motility), MDA‐MB‐231 and MCF7 control and resistant variant cells were seeded in 10‐cm2 plates without FBS and monitored using the phase‐contrast microscope (Zeiss, Germany). After the cells reached confluency, a scratch was made at the centre of the plate. The images were randomized and blindly snapped at days 1, 2, and 3. At least five images per sample and per day were captured, and the width of the gap was measured in micrometres using the microscope software built in ruler. Line graph was plotted to show the migration distance in micrometres. The data were obtained from five independent experiments (n = 5).
2.10. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology. The densitometry of Western blot images listed in the manuscript (main and supplementary figures) was measured using Image Lab Software Version 5.2.1 (Bio‐Rad, USA). The western blots were repeated in five independent experiments (n = 5). The mean density values were normalized to the corresponding housekeeping protein bands of GAPDH and plotted as mean relative protein band intensity (arbitrary unit [a.u.] ± SD). Statistically significant differences among the resistant cells and their respective controls were determined using ANOVA and Tukey's post hoc test. A two‐tailed Sudent's t test was used to compare between HER2‐transfected cells and the corresponding control; the cut‐off value for significance was determined as P < .05. The quantitative RT‐PCR was performed using two independent mRNA isolations (n = 2) from the generated resistant cell lines (each experiment is 8 months long). All other experimental results were from five independent experiments (n ≥ 5) including five in‐experiment replicates.
2.11. Materials
All antibodies used in the study were purchased from either Abcam, UK, or Cell Signaling Technology, USA. All the used antibodies and their concentrations and sources were listed in Table S1.
2.12. Nomenclature of targets and ligands
Key protein targets and ligands used in this article are hyperlinked to corresponding entries in http://www.http://guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2017), and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Cidlowski et al., 2019; Alexander, Fabbro et al., 2019a, b; Alexander, Kelly et al., 2019a, b).
3. RESULTS
3.1. Chemotherapy resistance in MDA‐MB‐231 cells up‐regulated NRP‐1 and integrin β3, which activated FAK/NF‐κBp65 survival axis
MDA‐MB‐231 cells representing the triple negative breast cancer subtype were selected in order to understand the role of NRP‐1 and associated signalling molecules in conferring drug resistance. Resistance to 4xAC alone did not alter the level of NRP‐1 expression. However, resistance to 4xAC + 4xPAC caused a significant increase in NRP‐1 levels (Figure 1). Similarly, TNC transcriptional levels were concomitantly increased (Figure S1A). Both resistance to 4xAC and 4xAC + 4xPAC caused a significant down‐regulation in BCRP/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=792 levels. Similarly, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1871 levels were significantly lower in response to 4xAC and decreased markedly in resistant cells to 4xAC + 4xPAC. http://familydisplayforward/?familyId=673 levels were significantly increased in cells resistant to 4xAC and further increased in cells resistant to 4xAC + 4xPAC. https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2497 levels were significantly increased only in cells resistant to 4xAC + 4xPAC. On the other hand, the https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=285 expression pattern was only down‐regulated significantly in cells resistant to 4xAC + 4xPAC (Figure 1). There was a gradual significant increase in the levels of https://www.guidetopharmacology.org/GRAC/DatabaseSearchForward?searchString=%092457&searchCategories=all&species=none&type=all&comments=includeComments&order=rank in resistant cells to 4xAC and to a higher extent upon resistance to 4xAC + 4xPAC. The p‐FAK levels were increased significantly in cells resistant to 4xAC and 4xAC + 4xPAC. Similarly, the survival‐related transcription factor http://objectdisplayforward/?objectId=2039 was significantly increased in cells resistant to 4xAC and further increased in cells resistant to 4xAC + 4xPAC. https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2050 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2605 proteins showed significant down‐regulation in cells resistant to 4xAC + 4xPAC but not in cells resistant to 4xAC (Figure 1).
Figure 1.
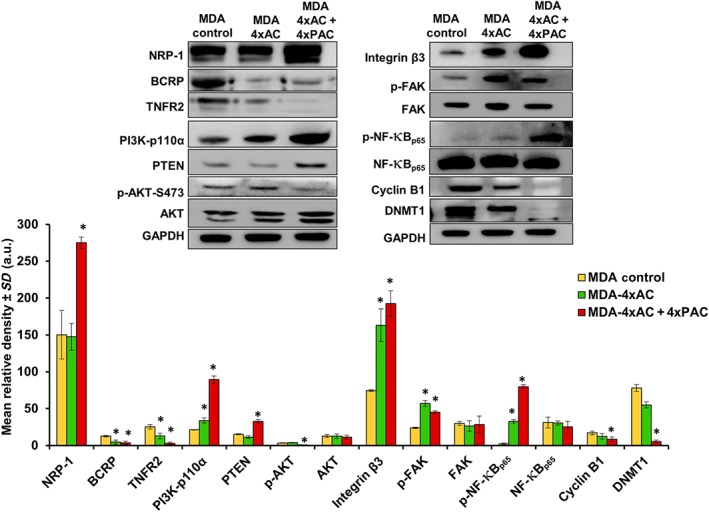
NRP‐1 up‐regulation in resistant MDA‐MB‐231 cells down‐regulates BCRP/ABCG2 and activates the integrin β3/FAK/NF‐κBp65 survival pathway. Representative Western blots and densitometry quantification of five independent experiments (n = 5) for each tested protein indicated that MDA‐MB‐231 cells treated with 4xAC + 4xPAC caused a significant increase in NRP‐1 levels. However, there was a significant down‐regulation of BCRP/ABCG2 and TNFR2 in resistant cells to 4xAC and 4xAC + 4xPAC. NRP‐1 up‐regulation occurs synchronously with a significant increase in PI3K‐p110α in both 4xAC and 4xAC + 4xPAC. PTEN expression was significantly increased in 4xAC + 4xPAC resistant cells, which consequently caused a decrease in phosphorylated AKT‐S473. Resistance to either 4xAC or 4xAC + 4xPAC significantly increased integrin β3, p‐FAK, and p‐NF‐κBp65. Cyclin B1 was significantly down‐regulated in resistant cells to 4xAC + 4xPAC, and DNMT1 was significantly down‐regulated in both 4xAC‐ and 4xAC + 4xPAC resistant cells. Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). *P < .05, significantly different from control cells; ANOVA and Tukey's post hoc test
3.2. Resistant MCF7 cells exhibited conversion from luminal A to luminal B HER2 subtype
In order to compare the mechanism of resistance between different breast cancer subtypes, MCF7 cell line representing the luminal A subtype was used. Resistance to 4xAC but not to 4xAC + 4xPAC increased the levels of NRP‐1 significantly (Figure 2a). Similarly, TNC transcriptional levels concomitantly increased (Figure S1A). Expression of BCRP/ABCG2 was significantly increased in MCF7 cells resistant to 4xAC + 4xPAC. Resistance to 4xAC and 4xAC + 4xPAC in MCF7 cells significantly up‐regulated the proliferation mediator TNFR2. However, resistance to 4xAC caused a significant increase in the PI3K/p‐Akt‐S473 axis but no significant changes in the levels of PTEN (Figure 2a). Although the level of integrin β3 was down‐regulated in cells resistant to 4xAC, levels of FAK, NF‐κBp65, and their phosphorylated forms were significantly up‐regulated (Figure 2a). Cyclin B1 protein was significantly up‐regulated only in cells resistant to 4xAC + 4xPAC (Figure 2a). HDAC3 was up‐regulated in the resistant cells, and HDAC4 was only up‐regulated in cells resistant to 4xAC + 4xPAC, along with a gradual decrease in DNMT1 (Figure 2a). Although HER2 and EGFR proteins were not detected in the untreated control MCF7 cells, the cells resistant to 4xAC and 4xAC + 4xPAC exhibited increased expression of both proteins and their phosphorylated forms (Figure 2b). Similarly, the levels of oestrogen receptor (ER‐α) were increased in cells resistant to both 4xAC and 4xAC + 4xPAC (Figure 2b).
Figure 2.
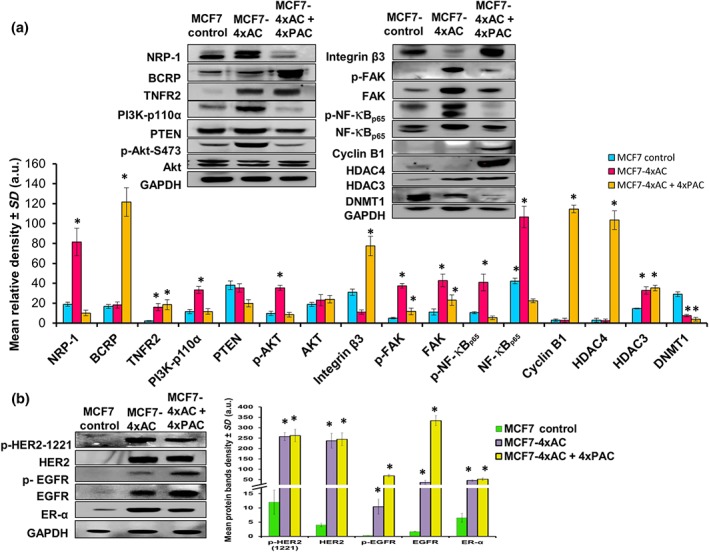
Resistant MCF7 cells responded differently to the different NAC treatmenst and up‐regulated HER2 and EGFR signalling and changed their subtype from luminal A to luminal B HER2 type. (a) Representative western blots illustrating the comparison between protein levels in control and resistant MCF7 variants. The MCF7 cells resistant to 4xAC showed significantly increased NRP‐1, TNFR2, PI3K‐p110α, PTEN (trend of increase), p‐AKT‐S473, FAK, and its phosphorylated form (p‐FAK and p‐NF‐κBp65) and HDAC3 but down‐regulated integrin β3 significantly. Unlike cells resistant to 4xAC, those resistant to 4xAC + 4xPAC showed a significant down‐regulation in NRP‐1 and an up‐regulation in BCRP levels. On the other hand, resistance to 4xAC + 4xPAC down‐regulated PI3K‐p110α and the phosphorylated forms of Akt‐S473, FAK, and NF‐κBp65. Additionally, cells resistant to 4xAC + 4xPAC overexpressed TNFR2, integrin β3, cyclin B1, and HDAC4. DNMT1 was down‐regulated in the cells resistant to 4xAC and to a higher extent in cells resistant to 4xAC + 4xPAC. (b) Although HER2 and EGFR were not detected in the control cells, their levels and the corresponding phosphorylated forms were induced in the resistant variant cells. ER‐α was also overexpressed in resistant cells. Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). *P < .05, significantly different from control cells; ANOVA and Tukey's post hoc test
3.3. Immunofluorescence staining and single‐drug treatment confirmed dual overexpression of NRP‐1 and HER2 in MCF7 cells
In order to confirm the western blot results, the up‐regulation of HER2 and NRP‐1 in MCF7 cells was assayed by immunofluorescent staining. HER2 (green) expression levels were significantly increased in resistant cells. NRP‐1 (red) levels were significantly increased in resistant cells to 4xAC compared with the control and cells resistant to 4xAC + 4xPAC (Figure 3a). To determine which drug converted the MCF7 subtype from HER2 negative to HER2 positive, MCF7 cells were treated with cyclophosphamide, doxorubicin or paclitaxel as single agents for short periods ranging from 2 to 48 hr (Figure 3b). The treatment with either one of the three drugs increased the expression of NRP‐1 with a concomitant induction of HER2 (Figures 3b and S2–S4). Only paclitaxel caused a decrease in HER2 and NRP‐1 expression starting at 8 hr and decreased further after 48 hr of treatment (Figures 3b and S4). HER2 phosphorylation at site 1221 was induced after 48 hr as a response to cyclophosphamide and doxorubicin and after 2 and 4 hr of paclitaxel treatment, which declined after 8 and 24 hr and disappeared after 48 hr (Figures 3b and S2–S4). Epigenetic enzymes https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2659 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2617 were affected by the different cytotoxic drugs in the same expression pattern as HER2 and NRP‐1 (Figures 3b and S2–S4). However, the methylation‐related enzyme DNMT1 showed a different expression pattern (Figures 3b and S2–S4). The blots were repeated independently five times (n = 5) and the mean value ± SD of the band density was calculated.
Figure 3.
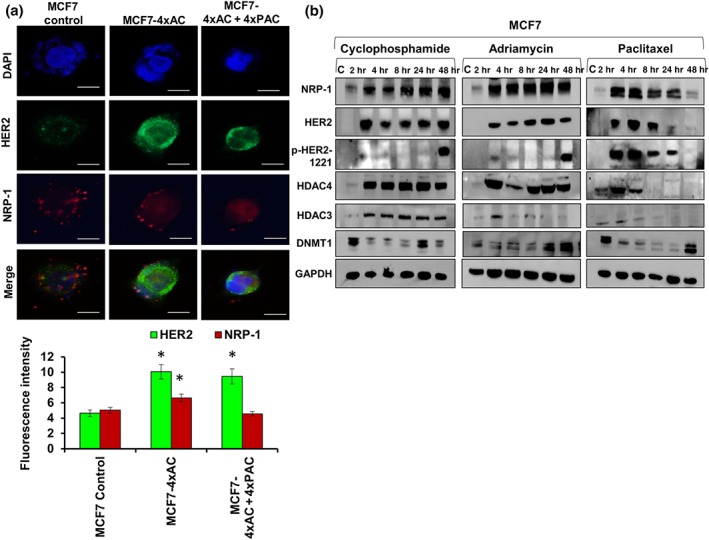
Immunofluorescent staining in resistant cells and after short period treatments with individual drugs in MCF7 cells confirmed the up‐regulation of HER2 and NRP‐1. (a) Fluorescence intensity of at least 15 cells showed a significant increase of NRP‐1 in resistant cells to 4xAC only. HER2 overexpression was detected in resistant cells to both 4xAC and 4xAC + 4xPAC. Fluorescence intensity was measured by ImageJ software, and the data are means ± SD, * P < .05, significantly different from control cells; ANOVA and Tukey's post hoc test. Images were captured at the same exposure using the 40× objective, scale bar = 100 μm. (b) Short period treatments with either of the three drugs cyclophosphamide, doxorubicin (adriamycin) or paclitaxel showed a rapid induction of NRP‐1, HER2, HDAC3, and HDAC4. Phosphorylated HER2 at site 1221 was up‐regulated after 48 hr of cyclophosphamide or adriamycin treatment. Paclitaxel increased the level of p‐HER2 (1221) after 2 and 4 hr of treatment but decreased after 8 and 24 hr and further reduced after 48 hr. Finally, DNMT1 had a different expression pattern in response to either of the three used drugs. The densitometry quantification of the protein bands from five independent experiments (n = 5) can be found in Figures S2–S4
3.4. Recombinant HER2 overexpression in MCF7 and MDA‐MB‐231 cells activated the survival axis and triggered NRP‐1 overexpression
To understand the role of HER2 overexpression in resistant cells, Gateway cloning of HER2 was used to induce the expression of HER2 in the parental MCF7 and MDA‐MB‐231 cells. HER2 overexpression and associated NRP‐1 up‐regulation was confirmed by western blot in both transfected cell lines (Figure 4a–c). MCF7 cells overexpressing HER2 (MCF7‐HER2) exhibited a significant increase in the phosphorylated HER2 at site 1221, in addition to overexpression of HER2‐ECD and HER2‐ICD, p‐EGFR, ER‐α, NRP‐1, p‐NF‐κBp65, and p‐https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=574 (Figure 4a,b). In MDA‐HER2 cells, although the overexpression of p‐HER2 (site 1221) was not as high as in MCF7 cells, there were significantly higher levels of HER2‐ICD with a significant increase in NRP‐1, integrin β3, p‐NF‐κBp65, and p‐FAK (Figure 4a,c). Dual immunofluorescence staining assays confirmed the recombinant overexpression of HER2 in the cells and the consequent overexpression of NRP‐1. In MCF7‐HER2 cells, the HER2 levels were so high (green), compared with the levels of NRP‐1 (red), that they masked the expression of NRP‐1 in the overlaid image (Figure 5a). Additionally, MCF7‐HER2 and MDA‐HER2 cells formed larger spheroids and a greater number of mammospheres respectively (Figure 5b).
Figure 4.
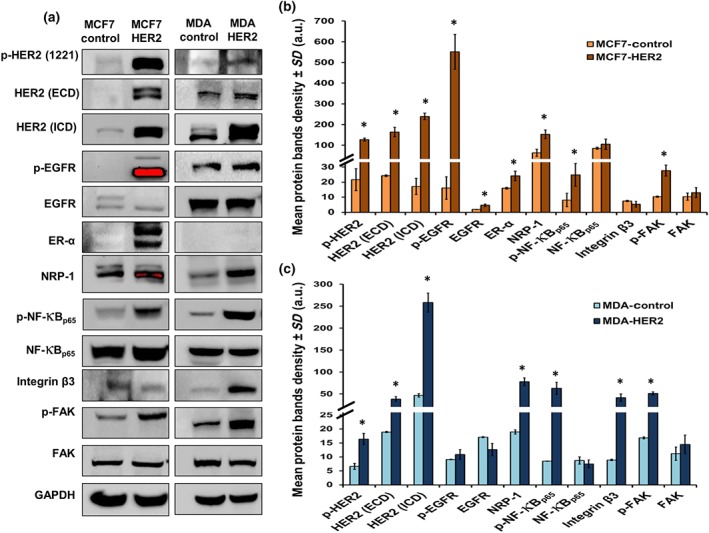
HER2 transfection triggered the features observed in resistant MCF7 cells. (a, b) MCF7‐HER2 cells exhibited a significant up‐regulation of phosphorylated HER2, HER2‐ICD, HER2‐ECD and EGFR, ER‐α, NRP‐1, p‐NF‐κBp65, and p‐FAK with a decrease in the levels of integrin β3. On the other hand, (a, c) MDA‐HER2 cells exhibited significant increase in phosphorylated HER2, HER2‐ICD, HER2‐ECD, NRP‐1, p‐NF‐κBp65, integrin β3, and p‐FAK. EGFR and p‐EGFR were not changed in the transfected cells. There were no bands of ER‐α detected in MDA control nor in the MDA‐HER2, and a positive control of this white blot can be found in Figure S6. Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). *P < .05 significantly different from control cells; two‐tailed Student's t test
Figure 5.
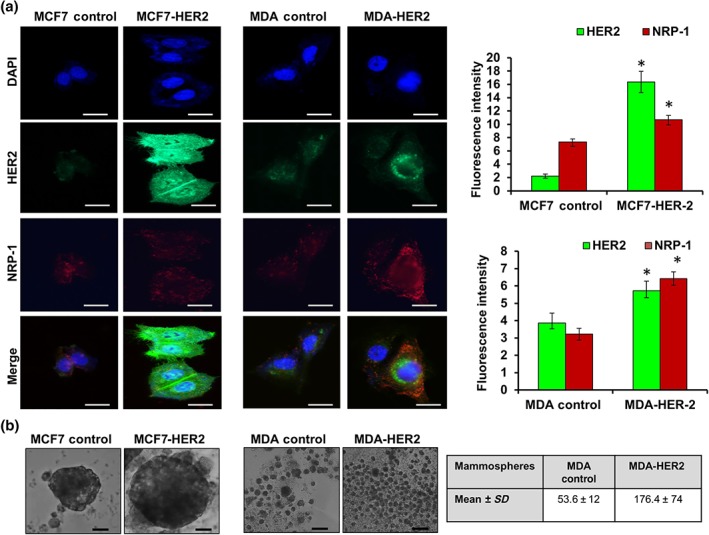
Dual immunofluorescence staining confirmed the up‐regulation of HER2 (green) and NRP‐1 (red) in HER2‐transfected cells. (a) Notably, in MCF7‐HER2 cells, HER2 was significantly overexpressed and showed a cytoplasmic and nuclear staining, which was high enough to mask NRP‐1 staining in the overlaid image. In MDA‐HER2 cells, NRP‐1 was significantly increased and detected in the cell membrane and cytoplasm; however, HER2 surrounded the nucleus. Images were captured at the same exposure using the 40× objective, scale bar = 100 μm. Fluorescence intensity was measured by ImageJ software, and the data were presented as mean value ± SD, two‐tailed t test was performed, and * P < .05 was the cut‐off for significance. (b) MCF7‐HER2 formed larger spheroids compared with MCF7 control. MDA‐HER2 formed greater numbers of mammospheres (mean ± SD, 176.4 ± 74) relative to MDA‐MB‐231 control cells (mean ± SD, 53.6 ± 12), scale bar = 500 μm. All data are from five independent experiments (n = 5). * P < .05 significantly different from control cells; two‐tailed Student's t test
3.5. HER2 has higher affinity to bind NRP‐1 in MCF7 cells compared with either MDA‐MB‐231 or BT‐474 cells
In order to determine the association between NRP‐1 and HER2 in breast cancer cells, three cell lines with various levels of NRP‐1 and HER2 were used. The baseline levels of NRP‐1 from the total cell lysates were detected by western blot analysis in MDA‐MB‐231, MCF7, and BT‐474 cells. MDA‐MB‐231 cells expressed the highest levels of NRP‐1, MCF7 expressed moderate levels relative to MDA‐MB‐231 cells, and BT‐474 cells expressed the lowest levels of NRP‐1 protein (Figure 6a).
Figure 6.
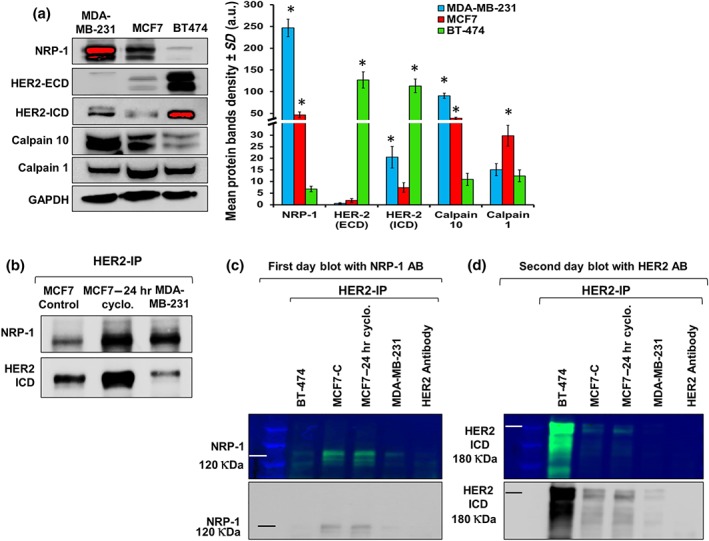
Co‐immunoprecipitation revealed that HER2 binds NRP‐1 in a pattern differing between breast cancer cell lines. (a) Baseline levels of NRP‐1, HER2‐ECD, HER2‐ICD, calpain 10, and calpain 1 were determined in the three breast cancer cell lines (MDA‐MB‐231, MCF7, and BT‐474). MDA‐MB‐231 cells expressed the highest levels of NRP‐1, MCF7 cells expressed moderate levels, and BT‐474 cells expressed the least levels of the protein. The levels of HER2‐ECD exhibited an opposite expression pattern to the levels of NRP‐1. MDA‐MB‐231 cells expressed the least levels of HER2‐ECD, MCF7 cells expressed very low levels, and BT‐474 cells expressed the highest levels of the protein. However, HER2‐ICD was expressed in higher levels in MDA‐MB‐231 cells compared with the levels in MCF7 and were less than the levels detected in BT‐474 cells. The different HER2 band size and pattern in MCF7 and MDA‐MB‐231 cells was explained by the levels of the cysteine proteases calpain 10 and calpain 1 levels. Calpain 10 was expressed in significantly higher levels in MDA‐MB‐231 cells compared with the levels in MCF7 and BT‐474 cells, while calpain 1 had the opposite pattern of expression in the three cell lines. Densitometry quantification (n = 5) of the proteins is presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). * P < .05, significantly different from control cells; ANOVA and Tukey's post hoc test. (b) Western blot analysis of MCF7, MCF7 treated with cyclophosphamide for 24 hr, and MDA‐MB‐231 cells showed NRP‐1 band of 120 kDa in the immunoprecipitated (IP) samples using HER2‐ICD antibody. Re‐blotting the same membrane with HER2 confirmed the binding between the two proteins. (c, d) Western blot analysis containing IP eluted proteins using HER2‐ICD antibody that were blotted in the first day with NRP‐1 and HER2 in the second day. The multichannel coloured western blot images were displayed to show the protein molecular marker. (c) MCF7 cells and MCF7 cells treated with cyclophosphamide for 24 hr showed NRP‐1 band when protein lysate was immunoprecipitated using HER2‐ICD, and very faint NRP‐1 band was detected in BT‐474 and MDA‐MB‐231 cells. (d) Second day blot re‐incubated with HER2 antibody showed that HER2 was highly expressed in BT‐474, moderately expressed in MCF7 and MCF7‐treated cyclophosphamide cells, and very low levels were detected in MDA‐MB‐231 cells. The experimental data are from five independent experiments (n = 5)
MDA‐MB‐231 cells expressed a very faint band of HER2‐ECD with larger size, MCF7 cells expressed very low levels, and BT‐474 cells expressed the highest levels of HER2‐ECD protein. In terms of HER2‐ICD expression, MDA‐MB‐231 cells expressed higher levels than in MCF7 cells but lower than those in BT‐474 cells.
It has been reported that HER2‐ECD gets cleaved by the cysteine protease calpain 10 (Panis et al., 2015) and the HER2‐ICD is cleaved by https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2336&familyId=731&familyType=ENZYME (Kulkarni et al., 2010). The calpain 10 exhibited an opposite expression pattern to HER2‐ECD, and calpain 1 was only increased in MCF7 cells that had the least HER2‐ICD (Figure 6a).
To explain the concurrent increase in both NRP‐1 and HER2 upon NAC resistance in MCF7 cells, the association between HER2 and NRP‐1 was confirmed by Co‐IP experiments. As shown in Figure 6b, the eluted proteins using HER2‐ICD antibody were first blotted with NRP‐1 antibody, which showed that NRP‐1 was bound to HER2 in control MCF7, MCF7 treated with cyclophosphamide for 24 hr, and in MDA‐MB‐231 cells (Figure 6b).
Additional Co‐IP experiments were performed to show the association of NRP‐1 and HER2 in the three cells lines, simultaneously. Proteins precipitated, using the HER2‐ICD antibody, showed a clear band of NRP‐1 in MCF7 and MCF7 treated with cyclophosphamide; however, there were faint bands in the lanes loaded with eluted proteins from BT‐474 and MDA‐MB‐231 cells (Figure 6c). The same blot was re‐blotted with HER2 antibody to confirm the HER2 IP. The results showed that HER2 was precipitated in very high amounts using HER2 IP from the BT‐474 cells, medium levels from MCF7 and MCF7 cyclophosphamide‐treated cells, and very low levels from MDA‐MB‐231 cells (Figure 6d). Both antibodies HER2 and NRP‐1 incubated with beads alone without protein lysates were used as negative controls for the expressed proteins (Figure 6c,d). Other confirmatory blots for this result can be found in Figure S5A.
3.6. Co‐IP in MDA‐HER2 and MCF7‐HER2 cells confirmed the binding between HER2 and NRP‐1
In order to further confirm the binding between HER2 and NRP‐1, Co‐IP was performed in HER2‐transfected MDA‐MB‐231 (MDA‐HER2) and MCF7 (MCF7‐HER2) cells. As shown in Figure 7a, immunoprecipitation of HER2 pulled higher levels of NRP‐1 and HER2 (ECD + ICD) in MDA‐HER2 cells, compared with parental control MDA‐MB‐231 cells (Figure 7a). Unexpectedly, immunoprecipitation using HER2 bound more NRP‐1 in control MCF7 cells, compared with MCF7‐HER2 cells, which still show successful elution of both forms of HER2 (Figure 7a). To confirm this result, the same cell lysates used for Co‐IP in Figure 7a were also used to detect the basal levels of both NRP‐1 and HER2 (Figure 7b). Finally, we performed immunoprecipitation with EGFR and detected NRP‐1 by western blot. The Co‐IP results showed that NRP‐1 in MDA control and MDA‐HER2 binds to EGFR, similar to a previous report (Rizzolio et al., 2012), and strong bands were observed (Figure 7c). However, NRP‐1 was barely detected in either control or MCF7‐HER2 cells (Figure 7c). The baseline levels of EGFR and NRP‐1 proteins in MDA‐MB‐231 cells were higher than those levels in MCF7 cells as shown in Figure 7d. Another confirmatory blot of HER2 IP in transfected cells with HER2 can be found in Figure S5B,C.
Figure 7.
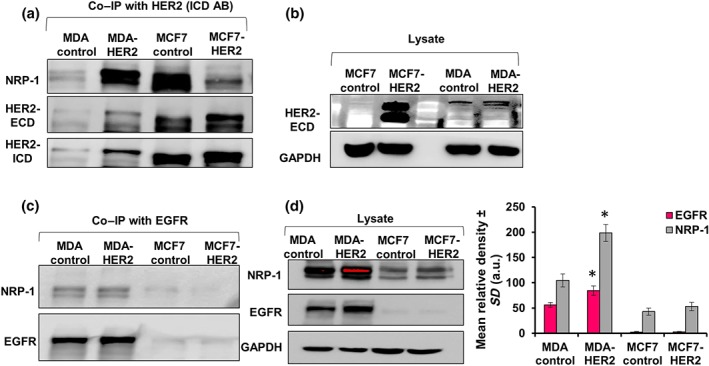
HER2 overexpression in MDA‐MB‐231 (MDA‐HER2) and MCF7 (MCF7‐HER2) cells confirmed the binding between HER2 and NRP‐1. (a) HER2 IP and NRP‐1 blotting did not show a strong NRP‐1 band in MDA‐MB‐231 control cells; however, in MDA‐HER2 cells, a strong NRP‐1 band was detected. Conversely, MCF7 control cells showed a strong NRP‐1 band, and MCF7‐HER2 cells did not show a strong NRP‐1 band. Re‐blotting the same membrane showed the expression levels of immunoprecipitated HER2 detected using the antibodies recognizing the ECD and ICD. Clearly, the immunoprecipitated levels of HER2 (ECD and ICD) were higher in MCF7 cells compared with MDA‐MB‐231 cells. (b) Baseline HER2 (ECD) levels in the cell's lysate used in blot A extracted from MCF7 control and MCF7‐HER2 cells as well as MDA‐MB‐231 control and MDA‐HER2 cells showed different band expression pattern and size of HER2 protein between the two cells and their transfected forms. (c) Immunoprecipitating EGFR and blotting with NRP‐1 followed by re‐blotting with EGFR showed that EGFR was able to pull NRP‐1 in MDA‐MB‐231 control and MDA‐HER2 cells in a higher capacity compared with both MCF7 control and MCF7‐HER2 cells. (d) Baseline NRP‐1 and EGFR levels in the cell's lysate used in blot C showed that both NRP‐1 and EGFR proteins were significantly higher in MDA‐MB‐231 cells compared with MCF7 cells. Expressed levels of NRP‐1 and EGFR were significantly higher in MDA‐HER2 compared with MDA‐MB‐231 control cells. The experiments were performed independently for five times (n = 5). Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). *P < .05 significantly different from control cells; two‐tailed Student's t test
3.7. Resistant MDA‐MB‐231 cells to 4xAC + 4xPAC exhibited slow migration and a decrease in mammosphere formation
To translate the molecular findings, we checked cellular responses, following the development of resistance. Wound healing assays showed faster migration in control MDA‐MB‐231 cells relative to the resistant cells, and the migration distance (in μm) is shown in Figure 8a (images and bar graph). Although control untreated MDA‐MB‐231 cells formed one clumped mammosphere, the resistant MDA‐MB‐231 cells to 4xAC formed larger numbers of mammospheres, compared with the cells resistant to full treatment (4xAC + 4xPAC; Figure 8b). MDA‐MB‐231 cells as a mesenchymal type of cells do not express E‐cadherin, either in the untreated control or in the generated resistant variants (Figure 8c). However, the MDA‐MB‐231 cells resistant to 4xAC + 4xPAC up‐regulated the epithelial to mesenchymal transition (EMT) markers, vimentin and ZEB1 (Figure 8c).
Figure 8.
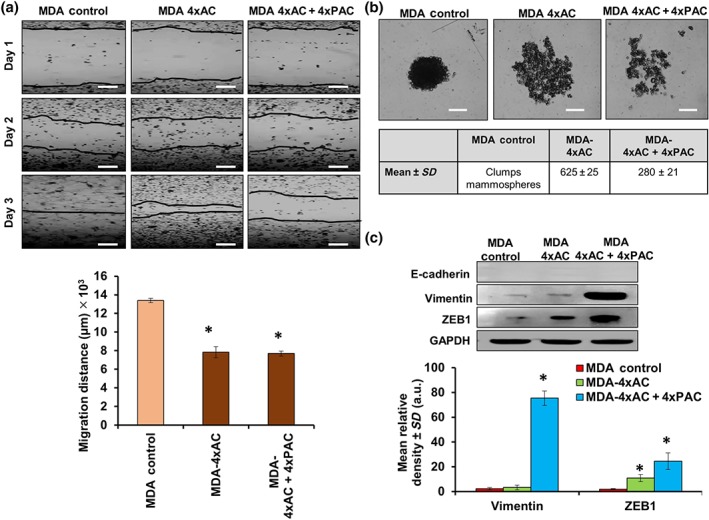
Resistant MDA‐MB‐231 cells exhibited less migration and up‐regulated EMT‐related proteins. (a) Phase‐contrast images show that the wound healing process was slower in MDA‐MB‐231 cells resistant to 4xAC and 4xAC + 4xPAC. The graph represents the migration ability of the cells. Subtracting the distance of the scratch on day 3 from the distance on day 1 showed a significant decrease in migration of both resistant MDA‐MB‐231 cells to 4xAC and 4xAC + 4xPAC. (b) Images represent mammosphere formation in control MDA‐MB‐231 cells, which formed clumped mammospheres, while MDA‐MB‐231 cells resistant to 4xAC formed more mammospheres (mean ± SD, 625 ± 25) relative to the cells resistant to 4xAC + 4xPAC (mean ± SD, 280 ± 21), scale bar = 2,000 μm. (c) Resistant or control MDA‐MB‐231 cells do not express E‐cadherin (positive E‐cadherin control blot can be found in Figure S6); however, cells resistant to 4xAC + 4xPAC overexpressed vimentin and ZEB1 to a greater extent. The graph represents the quantification of protein bands from five independent western blot experiments showing a significant increase in vimentin and ZEB1 in MDA‐MB‐231 cells resistant to 4xAC + 4xPAC. The experimental data are from five independent experiments (n = 5). *P < .05 significantly different from control cells; ANOVA and Tukey's post hoc test
3.8. Resistant MCF7 cell migration and spheroid formation was drug‐type dependent
The migration ability of the MCF7 cells resistant to 4xAC + 4xPAC was less than that of the cells resistant to 4xAC or untreated control cells (Figure 9a). Control MCF7 cells formed compact spheroids, whereas MCF7 cells resistant to 4xAC formed multiple compacted spheroids and cells resistant to 4xAC + 4xPAC formed clumped spheroids (Figure 9b). The cellular adhesion molecules, E‐cadherin, ZO1, N‐cadherin, and claudin‐1, were overexpressed in MCF7 cells resistant to 4xAC. However, resistance to 4xAC + 4xPAC caused even a higher level of E‐cadherin compared with 4xAC resistance and slight increase in the other adhesion molecules compared with control untreated cells (Figure 9c).
Figure 9.
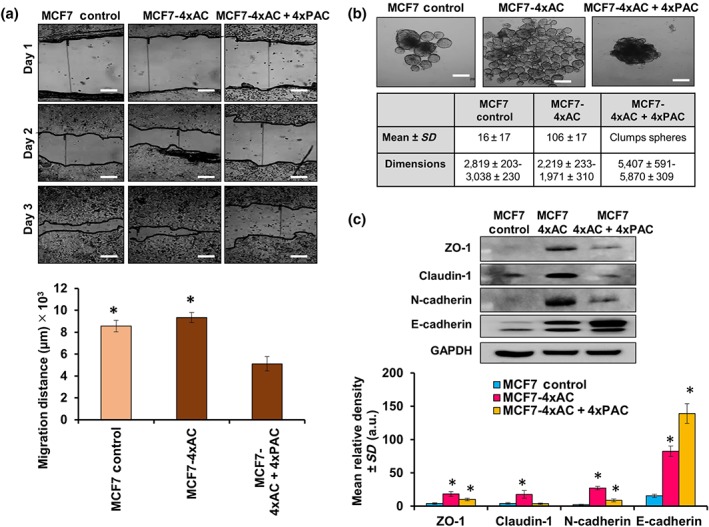
MCF7 cells resistant to 4xAC migrated faster, formed larger number of spheroids, and overexpressed adhesion molecules when compared with resistant variants to 4xAC + 4xPAC. (a) Wound healing assay showed a significant reduction in the migration ability of cells resistant to 4xAC + 4xPAC when compared with control (n = 5) and cells resistant to 4xAC (n = 5). The graph represents the migration distance measured in micrometres, scale bar = 2,000 μm. (b) Spheroids formed by MCF7 control and resistant cells have different pattern in terms of density and number formed. MCF7 resistant to 4xAC + 4xPAC formed the densest spheroid, scale bar = 2,000 μm. (c) Resistant MCF7 cells exhibited increased adhesion mediators such as ZO1, claudin‐1, N‐cadherin, and E‐cadherin that were significantly increased in MCF7 cells resistant to 4xAC and 4xAC + 4xPAC. All experimental data are from five independent experiments (n = 5). *P < .05 significantly different from control cells; ANOVA and Tukey's post hoc test
4. DISCUSSION
NAC used in breast cancer treatment was linked with the development of a small population of resistant cells that is responsible for the recurrence and relapse of the disease (Tsuruo et al., 2003). The response to NAC depends on the subtype of breast cancer (Rouzier et al., 2005) and has important consequences for the surgery required (Spanheimer et al., 2013). In addition, NAC changed the molecular subtype of breast cancer (Lim et al., 2016). Therefore, there is a need to analyse the mechanisms of resistance and response to NAC in different subtypes of breast cancer cells and to identify the precise molecular targets for each subtype.
The resistance is typically driven by the up‐regulation of cell survival molecules (Abdullah & Chow, 2013; Chow, 2013). In this study, we identified distinct mechanisms of resistance in the two different subtypes of breast cancer cells, as illustrated in Figure 10. We have already reported that recombinant overexpression of NRP‐1 in BT‐474 (BT‐474 NRP‐1) breast cancer cells induced the TNC/integrin β3/FAK/NF‐κBp65 survival axis (Naik et al., 2018). In this study, we showed that resistance to NAC in MDA‐MB‐231 cells caused an increase in NRP‐1 levels and hence increased the same survival axis, indicating that NRP‐1 up‐regulation is involved in MDA‐MB‐231 cell survival and resistance to NAC. The EMT is a mechanism contributing to drug resistance (Arumugam et al., 2009; Guo et al., 2017). The overexpression of NRP‐1 triggered the up‐regulation of EMT‐related molecules vimentin and ZEB1, which is consistent with our previous finding that NRP‐1 is positively correlated with EMT (Adham et al., 2014). Recently, we showed that higher NRP‐1 levels in the plasma and tissue of breast cancer patients after NAC were correlated with reduced patient response and overall survival, respectively, and NRP‐1 knockdown in MDA‐MB‐231 cells improved the sensitivity of the cells to chemotherapy (Al‐Zeheimi et al., 2019). Although the transporter protein BCRP/ABCG2 is a known marker of multidrug resistance (Li, Chua, Kunnath, & Chowdhury, 2012), its expression in resistant MDA‐MB‐231 model was decreased while NRP‐1 and the downstream survival molecules were increased, indicating that BCRP/ABCG2 does not necessarily drive NAC resistance in the context of breast cancer. Instead, NRP‐1 plays a key role in promoting resistance in these cells. The same inverse relationship between NRP‐1 and BCRP/ABCG2 expression was observed in resistant MCF7 cells and the previous resistance model in BT‐474 cells (Naik et al., 2018). Down‐regulation of BCRP/ABCG2 expression in resistant MDA‐MB‐231 cells can be a consequence of PTEN overexpression and down‐regulation of p‐AKT similar to that reported in chronic myeloid leukaemia cells (Huang et al., 2014). However, the PTEN levels in MCF7 cells were not affected in the resistant variants and did not seem to affect the BCRP/ABCG2 levels. The increased BCRP/ABCG2 levels in MCF7 cells resistant to 4xAC + 4xPAC appears rather to be due to the down‐regulation of NRP‐1 levels. Unlike MCF7 cells resistant to 4xAC, resistance to 4xAC + 4xPAC down‐regulated NRP‐1. This finding could be due to paclitaxel inhibiting the crosstalk between NRP‐1 and FAK (Ellison et al., 2015), leading to inhibition of the FAK pathway and NRP‐1 simultaneously. This would be consistent with earlier findings showing NRP‐1 inhibition suppressing the FAK/p130cas signalling pathway in MCF7 cells (Zeng et al., 2014). The integrin β3 levels in MCF7 cells resistant to 4xAC, were decreased, with a concurrent increase in NRP‐1 and p‐FAK, a correlation consistent with a previous report showing that integrin β3 suppression induced a neuropilin‐1‐dependent change in FAK remodelling (Ellison et al., 2015). Taken together, our data showed that different drugs had different actions on molecular signalling pathways in MCF7 cells.
Figure 10.
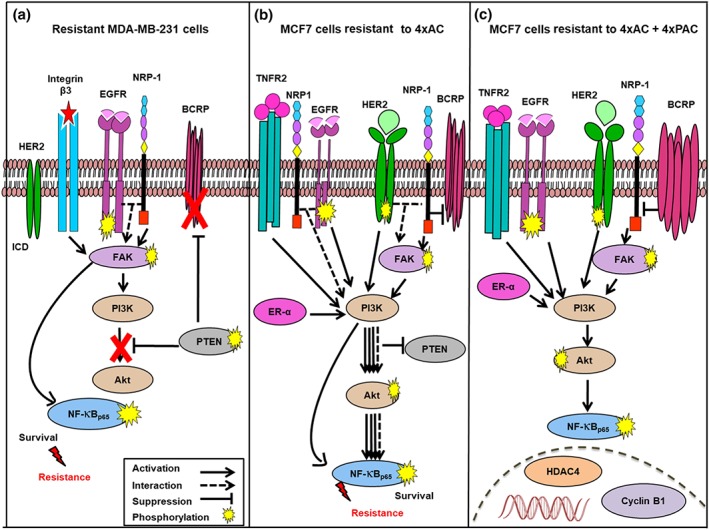
In vitro modelling of neoadjuvant chemotherapy (NAC) resistance revealed different pathways of resistance in MDA‐MB‐231 and MCF7 cells. (a) Resistant MDA‐MB‐231 cells activated NRP‐1/integrin β3/FAK/NF‐κBp65 but down‐regulated BCRP/ABCG2. On the other hand, (b) MCF7 cells resistant to 4xAC overexpressed NRP‐1 and ER‐α and induced EGFR expression and phosphorylation, which consequently activated the FAK/PI3K/AKT/NF‐κBp65 pathway, in addition to increasing ER‐α, EGFR2, HER‐2, and TNFR2. (c) Additional paclitaxel cycles in MCF7 cells (4xAC + 4xPAC0 up‐regulated BCRP/ABCG2 in response to the increase in cyclin B1 and deactivated the FAK/PI3K/AKT/NF‐κBp65 signalling pathway
The mechanism of resistance in MCF7 cells was different from the mechanism found in MDA‐MB‐231 cells, probably due to the induction and overexpression of HER2 in resistant MCF7 cells changing the cell subtype. As HDAC3 and HDAC4 enzymes were shown to promote HER2 regulation and epigenetic alterations are common in breast cancer (Garee, Chien, Li, Wellstein, & Riegel, 2014), the overexpression of both enzymes in resistant MCF7 cells might be the reason behind HER2 overexpression in MCF7 cells but not in resistant MDA‐MB‐231 cells in which both HDAC3 and HDAC4 levels were not affected (Figure S1A). This feature of acquired resistance in MCF7 cells was further confirmed in HER2 transfected MCF7 and MDA‐MB‐231 cells (MCF7‐HER2 and MDA‐HER2), which triggered the up‐regulation of NRP‐1 and FAK/NF‐κBp65 survival axis. Similarly, it has been shown that the co‐operation between HER2 and NF‐κB enhanced tumour resistance to radiotherapy (Ahmed, Cao, & Li, 2006). Therefore, we postulate that the overexpression of HER2 in MCF7 cells confers resistance to NAC, supporting the current regulations in re‐checking HER2 status after NAC (Yoshida et al., 2017).
Although NRP‐1 is well‐known as a co‐receptor for https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9902 and VEGF in angiogenesis (Geretti, Shimizu, & Klagsbrun, 2008), NRP‐1 also binds to EGFR and regulates its signalling, independently of effects on tumour angiogenesis (Rizzolio et al., 2012). In this study, we identified a new association between NRP‐1 and HER2 in breast cancer cells and their resistance to NAC. The binding between HER2 and NRP‐1 was more obvious in MCF7 cells, which up‐regulated HER2 protein. Although immunoprecipitation is not a precise quantitative method, the levels of NRP‐1 bound by HER2 in MCF7 cells were higher than the levels bound in either MDA‐MB‐231 or BT‐474 cells. This might be due to the very low baseline levels of HER2 in MDA‐MB‐231 cells and very low baseline levels of NRP‐1 in BT‐474. MDA‐MB‐231 and MCF7 cells have been classified as HER2 negative cell lines (Dai, Cheng, Bai, & Li, 2017), even though both forms of HER2 ECD and ICD were detected in the cells, which can be explained by the different processing of HER2 protein in the two cell lines by the action of calpains 1 and 10 (Panis et al., 2015; Figure 6a).
The interaction between HER2 and NRP‐1 was further confirmed in MCF7‐HER2 and MDA‐HER2‐transfected cells. Although MDA‐MB‐231 cells are known to represent the triple negative subtype, the HER2‐ICD was still detectable (Figures 4a and 6a), and this is consistent with previous reports that indicated that some basal‐like tumours exhibit the expression of HER2‐ICD (Moasser, Basso, Averbuch, & Rosen, 2001; Panis et al., 2015). Unlike MDA‐HER2 cells, the binding between HER2 and NRP‐1 was greater in control MCF7 cells compared with MCF7‐HER2 cells. Immunoprecipitating HER2 from MCF7‐HER2 cells showed a faint NRP‐1 band, despite the high levels of HER2, an observation that might indicate that the transfected HER2 protein into MCF7 cells was processed differently by these cells which prevented its interaction with NRP‐1, which was not the case of MDA‐HER2 cells. EGFR binding to NRP‐1 was greater in MDA‐HER2 cells because EGFR baseline levels in MDA‐MB‐231 cells are higher than the levels in MCF7 cells (Figure 7c,d; Rizzolio et al., 2012). Previous studies (Ahmed et al., 2006; Nahta, Yuan, Zhang, Kobayashi, & Esteva, 2005; Tanner et al., 2004) showed that inhibition of HER2 alone was not enough to target resistant cells. A role for neuropilin‐1 in acquired resistance for oncogene targeted therapy was reported recently (Rizzolio et al., 2018). Therefore, we postulate that targeting both HER2 and NRP‐1 in HER2/NRP‐1 positive breast cancer patient might improve the response to the treatment.
The observed molecular changes caused by NAC resistance in the two cell lines were translated in phenotypic assays. Thus, resistant MDA‐MB‐231 cells exhibited a decrease in the migration ability that might be due to the down‐regulation in BCRP/ABCG2 and a profound decrease in cyclin B1, as the inhibition of cyclin B1 in breast cancer cells reduced the cellular proliferation and sensitized the cancer cells to paclitaxel treatment (Androic et al., 2008). Absence of E‐cadherin expression in control or resistant MDA‐MB‐231 cells prevents them from forming big compact spheroids. However, NRP‐1 and NF‐κB overexpression enhanced the formation of mammospheres, similar to a previous report that showed that NRP‐1 and active NF‐κB in cancer stem cells were responsible for mammosphere formation (Glinka, Mohammed, Subramaniam, Jothy, & Prud'homme, 2012).
The overexpression of both HER2 and EGFR in MCF7 cells resistant to doxorubicin enhanced cell migration (Jin et al., 2016). However, the migration of resistant MCF7 cells in our model was drug‐dependent. For instance, MCF7 cells resistant to 4xAC exhibited an increase in their migration ability, which was confirmed by the overexpression of the adhesion molecules ZO1, N‐cadherin, and claudin‐1 known to enhance breast cancer progression (Yu et al., 2009). Inversely, the migration (cell motility) in MCF7 cells resistant to 4xAC + 4xPAC was decreased as the level of E‐cadherin increased (thus increasing the cell‐to‐cell adhesion; Frixen et al., 1991), with a simultaneous decrease in ZO1, N‐cadherin, and claudin‐1. These changes also explain the formation of dense and compacted spheroids as these cells up‐regulated BCRP/ABCG2, integrin β3, and cyclin B1 known to be involved in the formation of compact spheroids (Dai et al., 2013; Sodek, Ringuette, & Brown, 2009; Zhang, Rahbari, He, & Kebebew, 2011).
In conclusion, we have identified a new role for NRP‐1 in acquired resistance to multi‐drug chemotherapy, which could be translated to recommend the clinical assessment of NRP‐1 expression as a new diagnostic marker and a new drug target.
AUTHOR CONTRIBUTIONS
S.A.A. contributed in the conceptualization, supervision, investigation, writing the original draft and review, and editing of the manuscript. N.A.‐Z. conducted the experiments, performed the data analysis, and wrote the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for https://bpspubs.onlinelibrary.wiley.com/doi/abs/10.1111/bph.14207, and https://bpspubs.onlinelibrary.wiley.com/doi/abs/10.1111/bph.14208 (Alexander et al., 2018), as recommended by funding agencies, publishers, and other organizations engaged with supporting research.
Supporting information
Figure S1: MDA‐MB‐231 control and resistant variant cells to either 4xAC or 4xAC + 4xPAC lack the expression of p‐HER2–1221, HER2 (ICD) and ER‐α proteins (blots containing positive control samples are presented in supplementary figure 6 (Figure S6). There were no significant differences between MDA‐MB‐231 and resistant variants in the expression levels of the following proteins: p‐EGFR, EGFR, HDAC3, HDAC4 (n = 5). (B) Quantifying the transcriptional level of TNC using qRT‐PCR showed an increase in the level of the gene in resistant MDA‐MB‐231 cells to 4xAC + 4xPAC compared with either control untreated or resistant cells to 4xAC cells. (C) Transcriptional levels of TNC were upregulated only in MCF7 cells resistant to 4xAC (the experiment was done in duplicate considering that the generation of the resistant cells takes 8 months to prepare).
Figure S2: Cyclophosphamide treatment for short periods ranging from 2 hours to 48 hours caused a significant increase (compared to untreated control cells) in NRP‐1, HER2 (ECD) HDAC3, and HDAC4. However, p‐HER2‐1221 was only upregulated after 48 hours of the treatment. DNMT1 was significantly downregulated after 2 h, 4 h, 8 h and 48 h of cyclophosphamide treatment. Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells).
Figure S3: Adriamycin treatment for short periods ranging from 2 hours to 48 hours caused a significant increase in NRP‐1, HER2 (ECD), HER‐3 and HDAC4). p‐HER2 (1221) were only significantly overexpressed after 48 h of Adriamycin treatment. HDAC3 was significantly increased after 2 h of Adriamycin treatment. DNMT1 was increased significantly after 24 h and 48 h of Adriamycin treatment respectively. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells).
Figure S4: Paclitaxel treatment had a distinct effect on protein expression compared with cyclophosphamide and Adriamycin. Paclitaxel caused a significant sharp increase within the first 2‐4 hours in the following proteins; p‐HER2‐1221, HER2, NRP‐1 and HDAC4 however, a significant decrease in the levels of the proteins were observed after 8 hours of treatment. HDAC3 levels were proportionally decreased with Paclitaxel treatment after 24 and 48 hours, the DNMT1 level decreased significantly after 2, 4, 8 and 24 hours of the treatment and got back to the levels found in the untreated cells after 48 hours. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells). semaphorins
Figure S5: Five independent Co‐IP repeated experiments confirmed the binding between NRP‐1 and HER2 in breast cancer cell lines. (A) First experiment, Immunoprecipitation using HER2 antibody and blotting using NRP‐1 confirmed the binding between the two proteins more notably in MCF7 and MCF7 treated with cyclophosphamide compared with BT‐474 and MDA‐MB‐231 cells. The same blot was re‐incubated with HER2 confirming the immunoprecipitated HER2 protein showed that BT‐474 cells expressed the highest levels of the protein and MDA‐MB‐231 cells expressed the lowest levels. (B) Second experiment, Co‐IP using HER2 antibody confirmed the attachment with NRP‐1 in MCF7 control and HER2 transfected cells (MCF7‐HER2). Blotting the membrane with NRP‐1 antibody in the first day showed thicker NRP‐1 band in MCF7‐HER2 cells compared with control cells, the flow through was loaded as well to show the basal level of the protein. Re‐blotting the membrane with HER2 antibody on the following day showed HER2 protein bands in the IP samples but not on the flow through indicating that the total HER2 was captured by the used beads. (C) Third experiment confirming the binding between HER2 and NRP‐1 was performed on MDA‐MB‐231 cells known to express high levels of NRP‐1 and un‐detectable levels of HER2 (ICD). The first day the blot incubated with NRP‐1 antibody which showed thick NRP‐1 band in MDA‐HER2 cells only since HER2 (ICD) is not detected in the control MDA‐MB‐231 cells. Re‐blotting the membrane with HER2 showed that HER2 was successfully immunoprecipitated from the control and MDA‐HER2 cells and the flow through does not have HER2 protein since it is expressed in very low levels in these cells. This experiment was repeated independently five times n = 5.
Figure S6. Full size blots with positive controls were added to prove the absence of bands (white blots) in some blots presented in figure 4A ER‐α, figure 8C E‐cadherin, supplementary figure 1A p‐HER2‐1221, HER2 ICD and ER‐α.
Table S1 Lists of primary antibodies used for the western blots
ACKNOWLEDGEMENTS
We thank Mr Dawood Al‐Salhi, senior pharmacist in Sultan Qaboos University Hospital, for providing the chemotherapy drugs. We thank Prof. Mansour S. Al Moundhri, Dr Adviti Naik and Miss Maiya Al Maawali for advice and help.
This work was supported by The Research Council of Oman (TRC) under Grant Agreement No. ORG/HSS/14/006 (TRC#137) SQU# RC/SCI/BIOL/15/02 and Sultan Qaboos University, College of Science, Internal Grant Fund No. IG/SCI/BIOL/18/03 given to S.A.A.
Al‐Zeheimi N, Adham SA. Modeling Neoadjuvant chemotherapy resistance in vitro increased NRP‐1 and HER2 expression and converted MCF7 breast cancer subtype. Br J Pharmacol. 2020;177:2024–2041. 10.1111/bph.14966
REFERENCES
- Abdullah, L. N. , & Chow, E. K.‐H. (2013). Mechanisms of chemoresistance in cancer stem cells. Clinical and Translational Medicine, 2(1), 3‐9. 10.1186/2001-1326-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adham, S. A. , Al Harrasi, I. , Al Haddabi, I. , Al Rashdi, A. , Al Sinawi, S. , Al Maniri, A. , … Coomber, B. (2014). Immunohistological insight into the correlation between neuropilin‐1 and epithelial–mesenchymal transition markers in epithelial ovarian cancer. The Journal of Histochemistry and Cytochemistry, 62(9), 619–631. 10.1369/0022155414538821 [DOI] [PubMed] [Google Scholar]
- Ahmed, K. M. , Cao, N. , & Li, J. J. (2006). HER‐2 and NF‐κB as the targets for therapy‐resistant breast cancer. Anticancer Research, 26(6B), 4235–4243. [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Cidlowski, J. A. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators (2019). The Concise Guide to PHARMACOLOGY 2019/20: Nuclear hormone receptors. British Journal of Pharmacology, 176, S229–S246. 10.1111/bph.14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators (2019a). The Concise Guide to PHARMACOLOGY 2019/20: Catalytic receptors. British Journal of Pharmacology, 176, S247–S296. 10.1111/bph.14751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators (2019b). The Concise Guide to PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Faccenda, E. , … CGTP Collaborators (2019a). The Concise Guide to PHARMACOLOGY 2019/20: Introduction and Other Protein Targets. British Journal of Pharmacology, 176, S1–S20. 10.1111/bph.14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , … CGTP Collaborators (2019b). The Concise Guide to PHARMACOLOGY 2019/20: Transporters.
- Alexander, S. P. , Roberts, R. E. , Broughton, B. R. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology . British Journal of Pharmacology, 175(3), 407–411. Wiley‐Blackwell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Hajj, M. , Wicha, M. S. , Benito‐Hernandez, A. , Morrison, S. J. , & Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences, 100(7), 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Zeheimi, N. , Naik, A. , Bakheit, C. S. , Al Riyami, M. , Al Ajarrah, A. , Al Badi, S. , … Al‐Moundhari, M. S. (2019). Neoadjuvant chemotherapy alters neuropilin‐1, PlGF, and SNAI1 expression levels and predicts breast cancer patients response. Frontiers in Oncology, 9, 323–335. 10.3389/fonc.2019.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androic, I. , Krämer, A. , Yan, R. , Rödel, F. , Gätje, R. , Kaufmann, M. , … Yuan, J. (2008). Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer, 8(1), 391–402. 10.1186/1471-2407-8-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam, T. , Ramachandran, V. , Fournier, K. F. , Wang, H. , Marquis, L. , Abbruzzese, J. L. , … Choi, W. (2009). Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Research, 69(14), 5820–5828. 10.1158/0008-5472.CAN-08-2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, E. K.‐H. (2013). Implication of cancer stem cells in cancer drug development and drug delivery. Journal of Laboratory Automation, 18(1), 6–11. 10.1177/2211068212454739 [DOI] [PubMed] [Google Scholar]
- Dai, L. , Guinea, M. C. , Slomiany, M. G. , Bratoeva, M. , Grass, G. D. , Tolliver, L. B. , … Toole, B. P. (2013). CD147‐dependent heterogeneity in malignant and chemoresistant properties of cancer cells. The American Journal of Pathology, 182(2), 577–585. 10.1016/j.ajpath.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , Cheng, H. , Bai, Z. , & Li, J. (2017). Breast cancer cell line classification and its relevance with breast tumor subtyping. Journal of Cancer, 8(16), 3131–3141. 10.7150/jca.18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci, M. , Barbieri, E. , Piacentini, F. , Ficarra, G. , Bettelli, S. , Dominici, M. , … Guarneri, V. (2012). Discordance in receptor status between primary and recurrent breast cancer has a prognostic impact: A single‐institution analysis. Annals of Oncology, 24(1), 101–108. 10.1093/annonc/mds248 [DOI] [PubMed] [Google Scholar]
- Ellison, T. S. , Atkinson, S. J. , Steri, V. , Kirkup, B. M. , Preedy, M. E. , Johnson, R. T. , … Robinson, S. D. (2015). Suppression of β3‐integrin in mice triggers a neuropilin‐1‐dependent change in focal adhesion remodelling that can be targeted to block pathological angiogenesis. Disease Models & Mechanisms, 8(9), 1105–1119. 10.1242/dmm.019927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frixen, U. H. , Behrens, J. , Sachs, M. , Eberle, G. , Voss, B. , Warda, A. , … Birchmeier, W. (1991). E‐cadherin‐mediated cell‐cell adhesion prevents invasiveness of human carcinoma cells. The Journal of Cell Biology, 113(1), 173–185. 10.1083/jcb.113.1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garee, J. P. , Chien, C. D. , Li, J. V. , Wellstein, A. , & Riegel, A. T. (2014). Regulation of HER2 oncogene transcription by a multifunctional coactivator/corepressor complex. Molecular Endocrinology, 28(6), 846–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti, E. , Shimizu, A. , & Klagsbrun, M. (2008). Neuropilin structure governs VEGF and semaphorin binding and regulates angiogenesis. Angiogenesis, 11(1), 31–39. 10.1007/s10456-008-9097-1 [DOI] [PubMed] [Google Scholar]
- Glinka, Y. , Mohammed, N. , Subramaniam, V. , Jothy, S. , & Prud'homme, G. J. (2012). Neuropilin‐1 is expressed by breast cancer stem‐like cells and is linked to NF‐κB activation and tumor sphere formation. Biochemical and Biophysical Research Communications, 425(4), 775–780. 10.1016/j.bbrc.2012.07.151 [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group (2005). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15‐year survival: An overview of the randomised trials. The Lancet, 365(9472), 1687–1717. [DOI] [PubMed] [Google Scholar]
- Guo, C. , Ma, J. , Deng, G. , Qu, Y. , Yin, L. , Li, Y. , … Zeng, S. (2017). ZEB1 promotes oxaliplatin resistance through the induction of epithelial–mesenchymal transition in colon cancer cells. Journal of Cancer, 8(17), 3555–3566. 10.7150/jca.20952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … Bryant, C. (2017). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46(D1), D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F.‐F. , Zhang, L. , Wu, D.‐S. , Yuan, X.‐Y. , Chen, F.‐P. , Zeng, H. , … Zhao, X. L. (2014). PTEN regulates BCRP/ABCG2 and the side population through the PI3K/Akt pathway in chronic myeloid leukemia. PLoS ONE, 9(3), 1–10, e88298 10.1371/journal.pone.0088298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. , Zhang, W. , Wang, H. , Zhang, Z. , Chu, C. , Liu, X. , & Zou, Q. (2016). EGFR/HER2 inhibitors effectively reduce the malignant potential of MDR breast cancer evoked by P‐gp substrates in vitro and in vivo. Oncology Reports, 35(2), 771–778. 10.3892/or.2015.4444 [DOI] [PubMed] [Google Scholar]
- Knuefermann, C. , Lu, Y. , Liu, B. , Jin, W. , Liang, K. , Wu, L. , … Fan, Z. (2003). HER2/PI‐3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene, 22(21), 3205–3212. 10.1038/sj.onc.1206394 [DOI] [PubMed] [Google Scholar]
- Kopp, F. , Oak, P. S. , Wagner, E. , & Roidl, A. (2012). miR‐200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression. PLoS ONE, 7(11), e50469 10.1371/journal.pone.0050469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. , Reddy, K. , Esteva, F. J. , Moore, H. , Budd, G. , & Tubbs, R. (2010). Calpain regulates sensitivity to trastuzumab and survival in HER2‐positive breast cancer. Oncogene, 29(9), 1339–1350. 10.1038/onc.2009.422 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski, S. C. , Guerrero, P. A. , Hirota, S. , Chen, Z. , Morales, J. E. , Aghi, M. , & McCarty, J. (2017). Neuropilin‐1 modulates TGFβ signaling to drive glioblastoma growth and recurrence after anti‐angiogenic therapy. PLoS ONE, 12(9), e0185065 10.1371/journal.pone.0185065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurentiis, M. D. , Cancello, G. , D'Agostino, D. , Giuliano, M. , Giordano, A. , Montagna, E. , … Pennacchio, R. (2008). Taxane‐based combinations as adjuvant chemotherapy of early breast cancer: A meta‐analysis of randomized trials. Journal of Clinical Oncology, 26(1), 44–53. 10.1200/jco.2007.11.3787 [DOI] [PubMed] [Google Scholar]
- Lee, A. V. , Oesterreich, S. , & Davidson, N. E. (2015). MCF‐7 cells—Changing the course of breast cancer research and care for 45 years. JNCI: Journal of the National Cancer Institute, (7), 107–111, djv073. 10.1093/jnci/djv073 [DOI] [PubMed] [Google Scholar]
- Li, Y. T. , Chua, M. J. , Kunnath, A. P. , & Chowdhury, E. H. (2012). Reversing multidrug resistance in breast cancer cells by silencing ABC transporter genes with nanoparticle‐facilitated delivery of target siRNAs. International Journal of Nanomedicine, 7, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S. K. , Lee, M. H. , Park, I. H. , You, J. Y. , Nam, B.‐H. , Kim, B. N. , … Lee, E. S. (2016). Impact of molecular subtype conversion of breast cancers after neoadjuvant chemotherapy on clinical outcome. Cancer Research and Treatment: Official Journal of Korean Cancer Association, 48(1), 133–141. 10.4143/crt.2014.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masood, S. (2016). Neoadjuvant chemotherapy in breast cancers. Women's Health, 12(5), 480–491. 10.1177/1745505716677139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moasser, M. M. , Basso, A. , Averbuch, S. D. , & Rosen, N. (2001). The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2‐driven signaling and suppresses the growth of HER2‐overexpressing tumor cells. Cancer Research, 61(19), 7184–7188. [PubMed] [Google Scholar]
- Modi, S. , DiGiovanna, M. P. , Lu, Z. , Moskowitz, C. , Panageas, K. S. , Van Poznak, C. , … Carter, D. (2005). Phosphorylated/activated HER2 as a marker of clinical resistance to single agent taxane chemotherapy for metastatic breast cancer. Cancer Investigation, 23(6), 483–487. 10.1080/07357900500201301 [DOI] [PubMed] [Google Scholar]
- Munzone, E. , & Colleoni, M. (2015). Clinical overview of metronomic chemotherapy in breast cancer. Nature Reviews. Clinical Oncology, 12(11), 631–644. 10.1038/nrclinonc.2015.131 [DOI] [PubMed] [Google Scholar]
- Nahta, R. , Yuan, L. X. , Zhang, B. , Kobayashi, R. , & Esteva, F. J. (2005). Insulin‐like growth factor‐I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Research, 65(23), 11118–11128. 10.1158/0008-5472.CAN-04-3841 [DOI] [PubMed] [Google Scholar]
- Naik, A. , Al‐Yahyaee, A. , Abdullah, N. , Sam, J.‐E. , Al‐Zeheimi, N. , Yaish, M. W. , & Adham, S. A. (2018). Neuropilin‐1 promotes the oncogenic Tenascin‐C/integrin β3 pathway and modulates chemoresistance in breast cancer cells. BMC Cancer, 18(1), 533–547. 10.1186/s12885-018-4446-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, A. , Al‐Zeheimi, N. , Bakheit, C. S. , Al Riyami, M. , Al Jarrah, A. , Al Moundhri, M. S. , … Adham, S. A. (2017). Neuropilin‐1 associated molecules in the blood distinguish poor prognosis breast cancer: A cross‐sectional study. Scientific Reports, 7(1), 3301–3315. 10.1038/s41598-017-03280-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura, N. , Tomotaki, A. , Miyata, H. , Iwamoto, T. , Kawai, M. , Anan, K. , … Tokuda, Y. (2015). Changes in tumor expression of HER2 and hormone receptors status after neoadjuvant chemotherapy in 21 755 patients from the Japanese breast cancer registry. Annals of Oncology, 27(3), 480–487. 10.1093/annonc/mdv611 [DOI] [PubMed] [Google Scholar]
- Panis, C. , Pizzatti, L. , Corrêa, S. , Binato, R. , Lemos, G. F. , do Amaral, A. C. , … Abdelhay, E. (2015). The positive is inside the negative: HER2‐negative tumors can express the HER2 intracellular domain and present a HER2‐positive phenotype. Cancer Letters, 357(1), 186–195. 10.1016/j.canlet.2014.11.029 [DOI] [PubMed] [Google Scholar]
- Rizzolio, S. , Cagnoni, G. , Battistini, C. , Bonelli, S. , Isella, C. , Van Ginderachter, J. A. , … Tamagnone, L. (2018). Neuropilin‐1 upregulation elicits adaptive resistance to oncogene‐targeted therapies. Journal of Clinical Investigation, 128(9), 3976–3990. 10.1172/JCI99257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolio, S. , Rabinoviz, N. , Rainero, E. , Lanzetti, L. , Serini, G. , Norman, J. C. , … Tamagnone, L. (2012). Neuropilin‐1‐dependent regulation of EGF‐receptor signaling. Cancer Research, 72(22), 5801–5811. [DOI] [PubMed] [Google Scholar]
- Rouzier, R. , Perou, C. M. , Symmans, W. F. , Ibrahim, N. , Cristofanilli, M. , Anderson, K. , … Pusztai, L. (2005). Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clinical Cancer Research, 11(16), 5678–5685. 10.1158/1078-0432.CCR-04-2421 [DOI] [PubMed] [Google Scholar]
- Scharovsky, O. G. , Mainetti, L. E. , & Rozados, V. R. (2009). Metronomic chemotherapy: Changing the paradigm that more is better. Current Oncology, 16(2), 7–15. 10.3747/co.v16i2.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek, K. L. , Ringuette, M. J. , & Brown, T. J. (2009). Compact spheroid formation by ovarian cancer cells is associated with contractile behavior and an invasive phenotype. International Journal of Cancer, 124(9), 2060–2070. 10.1002/ijc.24188 [DOI] [PubMed] [Google Scholar]
- Spanheimer, P. M. , Carr, J. C. , Thomas, A. , Sugg, S. L. , Scott‐Conner, C. E. , Liao, J. , & Weigel, R. J. (2013). The response to neoadjuvant chemotherapy predicts clinical outcome and increases breast conservation in advanced breast cancer. The American Journal of Surgery, 206(1), 2–7. 10.1016/j.amjsurg.2012.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subik, K. , Lee, J.‐F. , Baxter, L. , Strzepek, T. , Costello, D. , Crowley, P. , Xing L, Hung MC, Bonfiglio T, Hicks DG, Tang P (2010). The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki‐67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer: Basic and Clinical Research, 4, 117822341000400004. [PMC free article] [PubMed]
- Tanner, M. , Kapanen, A. I. , Junttila, T. , Raheem, O. , Grenman, S. , Elo, J. , … Isola, J. (2004). Characterization of a novel cell line established from a patient with Herceptin‐resistant breast cancer. Molecular Cancer Therapeutics, 3(12), 1585–1592. [PubMed] [Google Scholar]
- Tsuruo, T. , Naito, M. , Tomida, A. , Fujita, N. , Mashima, T. , Sakamoto, H. , & Haga, N. (2003). Molecular targeting therapy of cancer: Drug resistance, apoptosis and survival signal. Cancer Science, 94(1), 15–21. 10.1111/j.1349-7006.2003.tb01345.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. , Yu, K. , Zhang, K. , Liu, L. , & Li, Y. (2017). Efficacy and safety of trastuzumab emtansine (T‐DM1) in the treatment of HER2‐positive metastatic breast cancer (MBC): A meta‐analysis of randomized controlled trial. Oncotarget, 8(60), 102458–102467. 10.18632/oncotarget.22270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A. , Hayashi, N. , Suzuki, K. , Takimoto, M. , Nakamura, S. , & Yamauchi, H. (2017). Change in HER2 status after neoadjuvant chemotherapy and the prognostic impact in patients with primary breast cancer. Journal of Surgical Oncology, 116(8), 1021–1028. 10.1002/jso.24762 [DOI] [PubMed] [Google Scholar]
- Yu, M. , Smolen, G. A. , Zhang, J. , Wittner, B. , Schott, B. J. , Brachtel, E. , … Haber, D. A. (2009). A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes & Development, 23(15), 1737–1742. 10.1101/gad.1809309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, F. , Luo, F. , Lv, S. , Zhang, H. , Cao, C. , Chen, X. , … Chen, Y. (2014). A monoclonal antibody targeting neuropilin‐1 inhibits adhesion of MCF7 breast cancer cells to fibronectin by suppressing the FAK/p130cas signaling pathway. Anti‐Cancer Drugs, 25(6), 663–672. 10.1097/CAD.0000000000000091 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Rahbari, R. , He, M. , & Kebebew, E. (2011). CDC23 regulates cancer cell phenotype and is overexpressed in papillary thyroid cancer. Endocrine‐Related Cancer, 18(6), 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: MDA‐MB‐231 control and resistant variant cells to either 4xAC or 4xAC + 4xPAC lack the expression of p‐HER2–1221, HER2 (ICD) and ER‐α proteins (blots containing positive control samples are presented in supplementary figure 6 (Figure S6). There were no significant differences between MDA‐MB‐231 and resistant variants in the expression levels of the following proteins: p‐EGFR, EGFR, HDAC3, HDAC4 (n = 5). (B) Quantifying the transcriptional level of TNC using qRT‐PCR showed an increase in the level of the gene in resistant MDA‐MB‐231 cells to 4xAC + 4xPAC compared with either control untreated or resistant cells to 4xAC cells. (C) Transcriptional levels of TNC were upregulated only in MCF7 cells resistant to 4xAC (the experiment was done in duplicate considering that the generation of the resistant cells takes 8 months to prepare).
Figure S2: Cyclophosphamide treatment for short periods ranging from 2 hours to 48 hours caused a significant increase (compared to untreated control cells) in NRP‐1, HER2 (ECD) HDAC3, and HDAC4. However, p‐HER2‐1221 was only upregulated after 48 hours of the treatment. DNMT1 was significantly downregulated after 2 h, 4 h, 8 h and 48 h of cyclophosphamide treatment. Individual protein band density quantification was normalized against their corresponding GAPDH bands. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells).
Figure S3: Adriamycin treatment for short periods ranging from 2 hours to 48 hours caused a significant increase in NRP‐1, HER2 (ECD), HER‐3 and HDAC4). p‐HER2 (1221) were only significantly overexpressed after 48 h of Adriamycin treatment. HDAC3 was significantly increased after 2 h of Adriamycin treatment. DNMT1 was increased significantly after 24 h and 48 h of Adriamycin treatment respectively. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells).
Figure S4: Paclitaxel treatment had a distinct effect on protein expression compared with cyclophosphamide and Adriamycin. Paclitaxel caused a significant sharp increase within the first 2‐4 hours in the following proteins; p‐HER2‐1221, HER2, NRP‐1 and HDAC4 however, a significant decrease in the levels of the proteins were observed after 8 hours of treatment. HDAC3 levels were proportionally decreased with Paclitaxel treatment after 24 and 48 hours, the DNMT1 level decreased significantly after 2, 4, 8 and 24 hours of the treatment and got back to the levels found in the untreated cells after 48 hours. Data are presented as the mean relative density (arbitrary units, a.u.) ± SD (n = 5). The levels of significance were evaluated by ANOVA followed by Tukey as a post hoc test (*P < 0.05 significantly different from control cells). semaphorins
Figure S5: Five independent Co‐IP repeated experiments confirmed the binding between NRP‐1 and HER2 in breast cancer cell lines. (A) First experiment, Immunoprecipitation using HER2 antibody and blotting using NRP‐1 confirmed the binding between the two proteins more notably in MCF7 and MCF7 treated with cyclophosphamide compared with BT‐474 and MDA‐MB‐231 cells. The same blot was re‐incubated with HER2 confirming the immunoprecipitated HER2 protein showed that BT‐474 cells expressed the highest levels of the protein and MDA‐MB‐231 cells expressed the lowest levels. (B) Second experiment, Co‐IP using HER2 antibody confirmed the attachment with NRP‐1 in MCF7 control and HER2 transfected cells (MCF7‐HER2). Blotting the membrane with NRP‐1 antibody in the first day showed thicker NRP‐1 band in MCF7‐HER2 cells compared with control cells, the flow through was loaded as well to show the basal level of the protein. Re‐blotting the membrane with HER2 antibody on the following day showed HER2 protein bands in the IP samples but not on the flow through indicating that the total HER2 was captured by the used beads. (C) Third experiment confirming the binding between HER2 and NRP‐1 was performed on MDA‐MB‐231 cells known to express high levels of NRP‐1 and un‐detectable levels of HER2 (ICD). The first day the blot incubated with NRP‐1 antibody which showed thick NRP‐1 band in MDA‐HER2 cells only since HER2 (ICD) is not detected in the control MDA‐MB‐231 cells. Re‐blotting the membrane with HER2 showed that HER2 was successfully immunoprecipitated from the control and MDA‐HER2 cells and the flow through does not have HER2 protein since it is expressed in very low levels in these cells. This experiment was repeated independently five times n = 5.
Figure S6. Full size blots with positive controls were added to prove the absence of bands (white blots) in some blots presented in figure 4A ER‐α, figure 8C E‐cadherin, supplementary figure 1A p‐HER2‐1221, HER2 ICD and ER‐α.
Table S1 Lists of primary antibodies used for the western blots


