Abstract
Liquiritigenin (LG), isoliquiritigenin (Iso-LG), together with their respective glycoside derivatives liquiritin (LN) and isoliquiritin (Iso-LN), are the main active flavonoids of Glycyrrhiza uralensis, which is arguably the most widely used medicinal plant with enormous demand on the market, including Chinese medicine prescriptions, preparations, health care products and even food. Pharmacological studies have shown that these ingredients have broad medicinal value, including anti-cancer and anti-inflammatory effects. Although the biosynthetic pathway of glycyrrhizin, a triterpenoid component from G. uralensis, has been fully analyzed, little attention has been paid to the biosynthesis of the flavonoids of this plant. To obtain the enzyme-coding genes responsible for the biosynthesis of LN, analysis and screening were carried out by combining genome and comparative transcriptome database searches of G. uralensis and homologous genes of known flavonoid biosynthesis pathways. The catalytic functions of candidate genes were determined by in vitro or in vivo characterization. This work characterized the complete biosynthetic pathway of LN and achieved the de novo biosynthesis of liquiritin in Saccharomyces cerevisiae using endogenous yeast metabolites as precursors and cofactors for the first time, which provides a possibility for the economical and sustainable production and application of G. uralensis flavonoids through synthetic biology.
KEY WORDS: Glycyrrhiza uralensis, Liquiritin, Isoliquiritin, Liquiritigenin, Isoliquiritigenin, Heterologous synthesis, Saccharomyces cerevisiae
Abbreviations: 4CL, 4-coumarate CoA ligase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; CiA, cinnamic acid; F7GT, flavone 7-O-glucosyltransferase; Iso-LG, isoliquiritigenin; Iso-LN, isoliquiritin; LG, liquiritigenin; LN, liquiritin; MeJA, methyl jasmonate; PAL, phenylalanine ammonia-lyase; p-CA, p-coumaric acid; Phe, phenylalanine; UGT, UDP-glucosyltransferase
Graphical abstract
Liquiritigenin, isoliquiritigenin and their glycoside compounds liquiritin and isoliquiritin, the main active flavonoids of Glycyrrhiza uralensis, were de novo heterologously synthesized in Saccharomyces cerevisiae by using raw materials and cofactors from the endogenous metabolic system of yeast.
1. Introduction
As an ancient botanical drug with thousands of years of history, licorice root (Glycyrrhizae Radix et Rhizoma, Gan Cao in Chinese) is a widely used medicine mainly derived from the rare and endangered herb Glycyrrhiza uralensis Fisch. It shows preventive and therapeutic effects against a variety of diseases with high safety, and appears in almost all Chinese medicine prescriptions, additional health care products, and even food1, 2. In addition to triterpenoids such as glycyrrhizin, G. uralensis also contains a large group of flavonoids with strong biological activity3. The flavonoids include liquiritigenin (LG), and isoliquiritigenin (Iso-LG), as well as their respective glycoside derivatives liquiritin (LN) and isoliquiritin (Iso-LN). Iso-LG is a chalcone with high content in G. uralensis, and also a common natural pigment. In spite of its simple structure, Iso-LG has many significant pharmacological activities such as antiinflammatory4, anticancer5, 6, antihistamine7, antioxidation8, antiplatelet agglutination9, antiallergy10, antivirus11 and estrogen-like activities12. Its glycoside derivative Iso-LN is capable of inhibiting tumor angiogenesis13, as well as showing antidepressant effects14. LG is a dihydroflavone compound formed by the isomerization of Iso-LG, which can inhibit the proliferation of certain cancer cells and induce apoptosis15, 16, 17. Its glycoside derivative LN is the quality indicator component of licorice root as prescribed in Chinese Pharmacopoeia. In addition to inducing apoptosis and autophagy in gastric cancer cells18, LN can also fight depression14. These observations indicate that G. uralensis has good development and application prospects in the treatment of cancer and other diseases. Along with the attention of the cosmetics industry in search of additives from natural sources, it was found that flavonoids in G. uralensis, especially the above-mentioned components, can be used for the removal of reactive oxygen species (ROS), as well as the safe and gentle whitening of human skin, and as such has been used by Nivea, Shiseido, YUE-SAI cosmetics, etc19.
Due to the enormous market demand, wild G. uralensis has been overharvested. Once damaged, the G. uralensis population is difficult to recover, and the amount of wild G. uralensis in China had been reduced to less than 500,000 tons as early as 200920. In addition to the large reduction of G. uralensis production, unsustainable development also led to environmental damage and desertification in the growing area21, 22. Coupled with the limitations of chemical synthesis and plant cell culture, sourcing the main flavonoids of G. uralensis through heterologous biosynthesis has become an effective strategy for the sustainable development of G. uralensis resources. In recent years, engineered strains have been used to produce various natural products of plant origin, including artemisinic acid23, 24, ginsenoside25, 26, paclitaxel27, tanshinone28, etoposide aglycone29 and opioids30. As a medicinal plant of great concern, the genome of G. uralensis has been sequenced and published31, and the biosynthesis pathway of the triterpenoid glycyrrhizin has been fully analyzed32, 33, 34, 35. However, the biosynthesis of LN has rarely been investigated, even though it is the main active flavonoid component in G. uralensis.
In this study, the key enzyme-coding genes responsible for LN biosynthesis, starting from phenylalanine ammonia-lyase, were screened out by combined genome and transcriptome analysis of G. uralensis and known homologous genes in the flavonoid biosynthesis pathway. The obtained LN pathway was reconstructed in yeast to realize the heterologous synthesis of the main flavonoids of G. uralensis, including LG, Iso-LG, LN, and Iso-LN, which provides a new method for the production and sustainable utilization of G. uralensis flavonoids.
2. Materials and methods
2.1. Plant materials and stress treatment
Seeds of G. uralensis Fisch., collected in Gansu province, were identified by Prof. Chunsheng Liu (Beijing University of Chinese Medicine, Beijing, China). The seeds were immersed in concentrated sulfuric acid for 70 min, then washed with deionized water and soaked at room temperature for 24 h. The treated seeds were sown in vermiculite and grown for 30 days in an artificial climate box (25 °C, cycles of 16 h light:8 h dark). After treatment with 0.5% NaCl or 100 mmol/L methyl jasmonate (MeJA) for 7 days, the treated plants were washed and frozen in liquid nitrogen and stored at −80 °C.
2.2. Chemicals
All of the chemical reference substances, including liquiritigenin (CAS: 578-86-9), isoliquiritigenin (CAS: 961-29-5), isoliquiritin (CAS: 5041-81-6), liquiritin (CAS: 551-15-5), phenylalanine (CAS: 63-91-2), cinnamic acid (CAS: 140-10-3), p-coumaric (CAS: 501-98-4), malonyl-CoA (CAS: 108347-84-8), coumaroyl-CoA (CAS: 119785-99-8), naringenin chalcone (CAS: 73692-50-9), UDP-glucose (CAS: 28053-08-9), and NADPH (CAS: 2646-71-1), had purity >98% and were commercially available (Sigma–Aldrich, Saint Louis, MO, USA; Yuanye, Shanghai, China).
2.3. Deep Illumina sequencing and transcriptome analysis
Total RNA was extracted from three biological replicates of G. uralensis roots using the Plant Easy Spin RNA Miniprep Kit (BIOMIGA, San Diego, CA, USA). A cDNA library was constructed after the RNA samples were qualified by Novogene (Beijing, China), and then sequenced on an HiSeq 4000 platform (Illumina, San Diego, CA, USA). The reference genome of G. uralensis (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl.)31 was used for bioinformatic transcriptome analysis, and supplemented gene functions were comprehensively annotated based on the following databases: Nr (NCBI nonredundant protein sequences), Nt (NCBI nonredundant nucleotide sequences), Pfam (protein families), KOG/COG (clusters of orthologous groups of proteins), SwissProt (a manually annotated and reviewed protein sequence database), KEGG (Kyoto encyclopedia of genes and genomes database), GO (gene ontology) and KO (KEGG Orthology). The abundance of unigenes was normalized using the FPKM (Fragments Per Kilobase of exon per Million mapped fragments) values.
According to the annotation, the sequences of seven candidate key functional genes in the flavonoid biosynthesis pathway, including the sequences which encoding phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), chalcone synthase (CHS), chalcone reductase (CHR), chalcone isomerase (CHI) and isoflavonoid 7-O-glycosyltransferase (F7GT/UGT), were selected. The homologous sequences of candidate genes were searched against the NCBI database and evolutionary analyses were conducted in MEGA 6.0 (Phoenix, USA)36 by sampling 1000 bootstrap replicates. Differential gene expression analysis was performed using HemI 1.0 (Wuhan, China)37, the abundance of candidate genes was represented by log2 values of FPKM, and sequences with log2 FPKM≤−1 were assigned as low expression in the samples which were excluded from the statistical data. The full-length cDNAs of the target genes were amplified by PCR using Phusion High-Fidelity PCR Master Mix (BioLabs, Ipswich, USA), and the primers designed based on the transcriptomic data (Supporting Information Table S1).
2.4. Bacterial expression and in vitro characterization
The ORFs of PAL, CHS, CHR, CHI and UGT were individually inserted between the KpnI and XhoI restriction sites of pET-32a(+) using the EasyGeno Assembly Cloning kit (Tiangen, Beijing, China), and transferred into Escherichia coli BL21 (DE3). All primers used in vector construction are listed in Supporting Information Table S2. Transformants were screened on Luria–Bertani (LB) solid culture medium containing 100 mg/mL ampicillin and single clones were picked for sequencing verification. The recombinant cells were cultured in 200 mL of LB medium containing 100 mg/mL ampicillin at 37 °C to an OD600 = 0.6–1.0, after which expression was induced by isopropyl β-d-thiogalactoside (IPTG) at a final concentration of 0.2 mmol/L and continued at 16 °C for 10 h. The expressing cells were harvested by centrifugation at 8000×g and 4 °C, resuspended in 3 mL phosphate buffered solution (pH = 8.0), and disrupted by ultrasonication in an ice-bath. The crude lysate was cleared from cell debris by centrifugation at 12,000×g and 4 °C and the supernatant was collected for purification. Recombinant protein was purified using the His-Tagged Protein Purification Kit (soluble protein, CWBIO, Beijing, China), and concentrations were measured using the Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China) with BSA as the standard.
The 1 mL enzymatic reaction systems contained about 0.5 mmol/L recombinant protein (10 mmol/L phosphate buffer solution, pH = 8.0), 1 mmol/L d,l-dithiothreitol (DTT), and 1 mmol/L substrate. For CHR, an equal amount of recombinant CHS protein with known function and 1 mmol/L NADPH were added to the enzymatic system. 25 mmol/L UDP-glucose was added to the enzymatic reaction system of UGT. The reaction was incubated at 30 °C for 10 h and stopped by the addition of 200 μL methanol. The catalytic products were analyzed by HPLC‒Q-TOF-MS.
2.5. Yeast expression and in vivo characterization
Cytochrome P450 gene C4H was expressed in Saccharomyces cerevisiae WAT11 using pESC-HIS as expression vector. While the catalytic product of 4CL is unstable, the candidate 4CL and the CHS with known function were co-expressed in WAT11 using the binary vector pESC-LEU as expression vector. The yeast expression vector was constructed using the EasyGeno Assembly Cloning kit (Tiangen, Beijing, China). C4H and CHS were individually inserted between the SpeI and NotI sites, and 4CL was inserted between the NheI and BamHI sites of the CHS recombinant vector. The recombinant plasmid was transferred into WAT11 using the Frozen-EZ Yeast Transformation Kit II (Zymo Research, Los Angeles, USA), and transformants were grown on corresponding auxotrophy synthetic medium (SC-His or -Leu) with 2% glucose and 2% agar at 30 °C for 4 days. Positive clones were cultivated in the corresponding liquid auxotrophy medium (2% glucose) and shaken at 30 °C to an OD600 of about 0.8. The 2% glucose medium was exchanged for induction medium containing 2% galactose, and after induction at 30 °C and 220 rpm (Honour, Tianjing, China) for 6 h, 20 μmol/L cinnamic acid or coumaric acid was added and the cultivation continued for another 12 h. The fermentation broth was extracted with an equal volume of ethyl acetate three times, and after evaporation of the solvent, the products re-dissolved in methanol and analyzed by HPLC–Q-TOF-MS after passing through a 0.22 μm polytetrafluoroethylene (PTFE) filter.
2.6. HPLC‒Q-TOF-MS analysis of catalytic product
The catalytic products were analyzed using an Agilent 1200 HPLC system coupled with an Agilent Q-TOF 6520 mass spectrometer (Agilent, Santa Clara, CA, USA) equipped with an electrospray ionization (ESI) device. Gradient elution was performed on an Agilent XDB-C18 column (150 mm × 2.1 mm, 3.5 μm) at room temperature with a flow rate of 0.3 mL/min using a linear gradient with water containing 0.1% formic acid (A) and acetonitrile (B) as the mobile phases as follows: 0–3 min, 5% B; 3–9 min, 25% B; 9–11 min, 25%–55% B; 11–14 min, 55%–95% B; 14–27 min, 95% B; 27–30 min, 95%–5% B. The injection volume was 20 μL.
2.7. Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from roots, stems and leaves of G. uralensis seedlings, and reverse transcription was done used the PrimeScript™ 1st-Strand cDNA Synthesis Kit (Takara, Dalian, China) according to the manufacturer's instructions. The qRT-PCR was performed using the KAPA SYBR FAST Universal qPCR Kit (Kapa Biosystems, Boston, MA, USA) with gene-specific primer pairs (Supporting Information Table S3). Three technical replicates and three biological replicates were analyzed for each sample. The relative amounts of the target genes were evaluated based on the relative expression index of mRNA using the 2–△△Ct method38, with Guβ-actin as the reference gene34.
2.8. Extraction of main flavonoids from G. uralensis
After fully pulverizing in liquid nitrogen, samples comprising about 100 mg of freeze-dried roots, stems or leaves of G. uralensis seedlings that were grown for one month were accurately weighed, after which 1 mL 70% ethanol was added and the samples ultrasonically extracted at 30 °C for 30 min. The product was centrifuged, and the supernatant was passed through a 0.22 μm PTFE filter for quantitative UPLC‒MS analysis.
2.9. Reconstitution of the liquiritin biosynthesis pathway in yeast and product analysis
The genes of the G. uralensis flavonoid pathway with known function (GuPAL1, GuC4H1, Gu4CL1, GuCHS1, GuCHR1, GuCHI1, and GuUGT1) were inserted in different combinations into the binary vector pESC downstream of the GAL1 or GAL10 promoters. The 3ʹ end of GuCHS1 was fused to the 5ʹ end of GuCHR1 via the linker sequence GGTGGTGGTTCT (GuCHS1::GuCHR1). The constructs were verified by bi-directional sequencing, and the resulting plasmids were used to transform the yeast strain WAT11 using the Frozen-EZ Yeast Transformation kit II (Zymo Research, Los Angeles, CA, USA). The transformants were selected on synthetic dropout plates (SC-His, -Leu, -Trp or -Ura) according to the vector label and the positive yeast clones were cultured in SC medium with 2% glucose at 30 °C and 220 rpm (Honour, Tianjing, China), and then harvested and resuspended in SC medium with 2% galactose followed by expression for 36 h. All recombinant yeast strains used in this study are listed in Supporting Information Table S4. 500 μL fermentation broth was taken and mixed with an equal volume of methanol, ultrasonicated for 60 min and centrifuged at 13,400×g for 10 min. The supernatant was analyzed by UPLC–MS after passing through a 0.22 μm PTFE filter.
2.10. Fermentation
Strain WM4-3 was used for the production of liquiritin in fed-batch fermentation. Synthetic dropout medium (SC-His-Leu-Trp-Ura) was used for both seed preparation and the fermentation. The seed culture was prepared by inoculating a 250 mL flask containing 100 mL culture medium with 2% glucose. The cells were grown at 30 °C and 250 rpm (Honour, Tianjing, China) for 48 h, and then transferred into 1 L of fresh seed medium and incubated at 30 °C and 220 rpm (Honour, Tianjing, China) for 36 h. The resulting seed was harvested and used to inoculate an 11-L New Brunswick™ BioFlo®/CelliGen® 115 fermenter (Eppendorf, Hamburg, Germany) containing 6 L of induction medium (SC-His-Leu-Trp-Ura with 2% galactose) to an initial OD600 = 1.5–2.0. The fermentation was carried out at 30 °C. The dissolved oxygen concentration (DOC) was kept above 40% and the pH was controlled at 5.0 using automatic addition of ammonium hydroxide. Concentrated synthetic dropout medium (SC-His-Leu-Trp-Ura; 80 g/L total solids) and 40% galactose were automatically fed to the fermenter separately at a rate of 6.25 mL/h from the second day. The fermentation broth was sampled at intervals of 24 h to measure the OD600, and an aliquot of 500 μL volume was mixed with an equal volume of methanol, ultrasonicated for 60 min, centrifuged at 13,400×g for 10 min, and the supernatant was stored at −20 °C until UPLC–MS analysis.
2.11. Quantitative UPLC–MS analysis
The separation of 1 μL filtrate was performed using an Agilent ZORBAX RRHD SB-C18 column (100 mm × 2.1 mm, 1.8 μm) on an Agilent 1290 Infinity UPLC system (Agilent, Santa Clara, CA, USA) by gradient elution with a mobile phase comprising 0.1% (v/v) formic acid aqueous solution (A) and acetonitrile (B) at a flow rate of 0.3 mL/min. The gradient program was as follows: 0–1.0 min, 25% B; 1.0–4.0 min, 25%–90% B; 4.0–4.5 min, 90% B; 4.5–4.51 min, 90%–25% B; 4.51–5.5 min, 25%. The column temperature was set at 25 °C.
The analyte was quantified using an AB Sciex LC–MS/MS Qtrap 6500 mass spectrometer equipped with an electro-spray ionization (ESI) source (AB Sciex, Singapore). The multiple reaction monitoring (MRM) scan type was used in the negative scan mode to increase the specificity of the analysis. The mass parameters are listed in Table 1. The software MultiQuant 3.0.1 (AB Sciex, Singapore) was used to perform the data analysis.
Table 1.
MS parameters for the quantification of analytes.
| Component | Q1 Mass (Da) | Q3 Mass (Da) | Declustering potential (V) | Collision energy (V) |
|---|---|---|---|---|
| p-Coumaric acid | 163 | 119 | −60 | −23 |
| 93 | −60 | −40 | ||
| Isoliquiritigenin | 255 | 135 | −80 | −21 |
| 119 | −80 | −30 | ||
| Isoliquiritin | 417 | 255 | −180 | −25 |
| 135 | −180 | −30 | ||
| Liquiritigenin | 255 | 119 | −100 | −25 |
| 135 | −100 | −20 | ||
| Liquiritin | 417 | 255 | −70 | −27 |
| 135 | −70 | −40 |
2.12. Accession numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: GuPAL1 (MK341789), GuC4H1 (MK341785), Gu4CL1 (MK341782), GuCHS1 (MK341787), GuCHR1 (MK341786), GuCHI1 (MK348532), GuUGT1 (MK341792); the transcriptome datasets: Gu-CK, SRR8400027; Gu-NaCl, SRR8400026; Gu-MeJA, SRR8468083.
3. Results
3.1. Screening of liquiritin pathway genes
Water deficiency can increase the yield of LN in the roots of G. uralensis39, and MeJA is also believed to increase the production of flavonoids40. Consequently, deep sequencing of the G. uralensis root transcriptome was performed after NaCl- and MeJA-treatment, respectively (Supporting Information Tables S5 and S6). The reference genome of G. uralensis was used in combination with the Nr, Nt, Pfam, KOG/COG, SwissProt, KO and GO databases for bioinformatics analysis, and gene functions were comprehensively annotated, and a total of 61 genes that were annotated to encode the seven enzymes required for the biosynthesis of LN were screened from the transcriptome database (Supporting Information Table S7). Hierarchical cluster analysis of the candidate genes revealed that GuPAL1, GuC4H1, GuCHS1, GuCHR1, GuCHR4, GuCHI1, GuCHI5 and GuUGT1 had relatively high expression levels in both NaCl- or MeJA-treated and control G. uralensis. Furthermore, the treatment, and especially NaCl stress, increased the expression levels of these genes to some extent. The expression level of Gu4CL1 under NaCl stress was significantly higher than under MeJA stress or in the CK group (Fig. 1). In addition, multiple sequence alignment results showed that GuPAL1, GuC4H1, Gu4CL1, and GuCHS1 were respectively 78%, 92%, 72%, and 85% identical to Populus hybrid (P. trichocarpa × P. deltoides) PAL41, Glycine max C4H41, Petroslinum crispum 4CL-242,43, and P. hybrid (P. trichocarpa × P. deltoides) CHS142, 43, 44, which were commonly used for heterologous synthesis of plant-specific flavones.
Figure 1.
Hierarchical clustering and corresponding heatmaps of the differentially expressed unigenes across the flavonoid biosynthesis pathway. CK, control; NaCl, root sample treated with 0.5% NaCl; MeJA, root sample treated with 100 mmol/L MeJA. The heatmap was drawn using HemI 1.0 with log2 values of FPKM (Fragments Per Kilobase of exon per Million mapped fragments) of the candidate genes, and the sequences with log2FKPM≤−1 were assigned as low expression in the samples which were excluded from the statistical data. Depths of color in the red and green rectangles indicate higher and lower Z-scores (log2) of the corresponding RNA expression levels. The red font indicates the sequences used for liquiritin pathway reconstruction in yeast.
Subsequently, to screen for the unique CHR and CHI, BLASTX analysis was performed using the NCBI database with the nucleic acid sequences that were annotated as encoding chalcone reductase (or NAD(P)H-dependent 6ʹ-deoxychalcone synthase) and chalcone isomerase, respectively. The sequences with higher identity were used for the neighbor-joining tree construction. Four CHR candidate genes were divided into three subgroups in the neighbor-joining tree (Fig. 2A) with GuCHR1, GuCHR2, GuCHR3 belonging to subgroup A, wherein GuCHR1 shares 87% identity to G. max CHR5, which had the closest correlation with the accumulation of abundant 5-deoxyisoflavonoids in soybean root45. GuCHR4, which is also highly expressed in G. uralensis, was clustered in subgroup B with G. max CHR4, which previous studies suggested to have no CHR activity for the production of Iso-LG45. The eight CHI homologs from G. uralensis were classified into four CHI types, whereby GuCHI1 belonged to type II CHIs, which were thought to have the activity of isomerizing Iso-LG into LG46, 47, whereas GuCHI5 fell into the type IV subfamily (Fig. 2B).
Figure 2.
Phylogenetic analysis of the amino acid sequences of CHR and CHI based on the G. uralensis transcriptome. The evolutionary analyses were conducted in MEGA6.0 using the neighbor-joining method, and the percentage of replicate trees in which the associated taxa clustered together among the 1000 replicates in the bootstrap test are shown next to the branches. Sequences from the transcriptome were marked with red triangles. Red font indicates the sequences used for liquiritin pathway reconstruction in yeast.
3.2. Functional characterization of liquiritin pathway enzymes
In order to confirm the catalytic function of the enzymes encoded by the candidate genes, GuPAL1, GuCHS1, GuCHR1, GuCHI1 and GuUGT1 were expressed in E. coli and the recombinant proteins were extracted and purified (Supporting Information Fig. S1) for in vitro enzymatic experiments, while the cytochrome P450 GuC4H1 was expressed in S. cerevisiae WAT11. As indicated by LC–MS, incubation of the staring substrate phenylalanine with GuPAL1 yielded cinnamic acid (Fig. 3B). Similarly, the recombinant yeast harboring GuC4H1 was able to produce p-coumaric acid upon feeding cinnamic acid and galactose (Fig. 3C). In an in vitro enzymatic system with coumaroyl-CoA and malonyl-CoA as substrates at a molar ratio of 1:3, naringenin chalcone was produced when only GuCHS1 was added. When the same amount of recombinant GuCHS1 and GuCHR1 proteins and a high concentration of NADPH (1 mmol/L) was added, the production of Iso-LG was also detected in addition to naringenin chalcone (Fig. 3E). As the catalytic product of 4CL was unstable, Gu4CL1 and GuCHS1 were co-expressed in WAT11 using the binary plasmid pESC-LEU as yeast expression vector. Upon feeding with p-coumaric acid, naringenin chalcone could also be detected in the culture extracts (Fig. 3D). GuCHI1, which belongs to type II CHIs, was able to isomerize Iso-LG into LG in in vitro enzymatic experiments, demonstrating its CHI activity (Fig. 3F). Thus, GuPAL1, GuC4H1, Gu4CL1, GuCHS1, GuCHR1 and GuCHI1 formed a complete biosynthetic pathway of liquiritigenin with phenylalanine as the precursor (Fig. 3A).
Figure 3.
Analysis of catalytic products of the enzymes encoded by the candidate genes. (A) The liquiritin biosynthesis pathway of G. uralensis, the histogram next to each pathway enzyme name shows relative expression levels of the encoding gene (GuPAL1, GuC4H1, Gu4CL1, GuCHS1, GuCHR1, GuCHI1 and GuUGT1) in roots, stems and leaves of G. uralensis determined by qRT-PCR; (B, E-G) HPLC–Q-TOF-MS profiles of the in vitro enzymatic products of recombinant GuPAL1 (B, phenylalanine (Phe) as substrate), GuCHS1 and GuCHR1 (E, coumaroyl-CoA as substrate), GuCHI1 (F, isoliquiritigenin (Iso-LG) as a substrate), GuUGT1 (G, liquiritigenin (LG) as a substrate) expressed in E. coli using pET-32a(+) as expression vector; (C, D) HPLC–Q-TOF-MS profiles of the fermentation products of yeast harboring the recombinant vector pESC-GuC4H1 (C, cinnamic acid (CiA) as a substrate) or pESC-Gu4CL1-GuCHS1 (D, p-coumaric acid (p-CA) as substrate) with external precursor addition. The peaks and mass spectrum of the product indicated by the asterisk, are shown (electron ionization in negative-ion mode, [M-H]–). Extracted-ion chromatogram (EIC) of the analyte, as indicated, at m/z 147, 164 (B), m/z 147, 163 (C), m/z 271 (D), m/z 255, 271 (E), m/z 255 (F), m/z 255, 417 (G). All the reactions were performed with empty vector as control.
In the final step of LN biosynthesis, GuUGT1 was able to produce LN from LG in vitro with UDP-glucose as sugar donor (Fig. 3G). The kinetic parameters of GuUGT1, GuUGT2 and GuUGT3, respectively belong to UGT88E, UGT88H and UGT88A subfamilies within the UGT88 family (Supporting Information Fig. S2), were subsequently determined. Not surprisingly, GuUGT1 (Km 68.17 μmol/L and Vmax 0.87 μmol/(min·mg)) exhibited a higher maximum activity, and its catalytic efficiency (Vmax/Km) was also clearly higher (Supporting Information Section 1, Fig. S3 and Table S8). To investigate the structural basis for the binding of LG and UDP-glucose, a homology model was generated for GuUGT1 using the crystal structure of Arabidopsis thaliana UGT (PDB ID: 2VCE) with 34% identity as template (Supporting Information Section 2 and Fig. S4)48. The key residues of GuUGT1 for liquiritigenin glycosylation (Gly-14 and Gly-277) predicted by molecular docking (Supporting Information Section 3, Tables S9, S10 and Fig. S5) and residue scanning based on simulated mutations (Supporting Information Section 4 and Table S11) were different from those of the known flavonoid glucosyltransferase VvGT1 (His-20 and Asp-119) or isoflavonoid 7-O-glycosyltransferase GmIF7GT (Glu-392), which also belong to the UGT88E subfamily.
A comparison was made between the expression profiles of the enzymes whose catalytic functions and flavonoid accumulation in different tissues of G. uralensis were characterized. A pattern emerged that indicated that most LN pathway genes were mainly expressed in leaves, except for the last UGT responsible for glycosylation, which had the highest relative expression level in the roots (Fig. 3A). However, LG and Iso-LG, as well as their respective glycosylation products LN and Iso-LN are mainly accumulated in the roots (Supporting Information Fig. S6). We therefore inferred that the upstream step of LN synthesis in G. uralensis is mainly carried out in the leaves, and aglycones are transferred from the leaves to the roots, followed by glycosylation to form glycosides and accumulation in the roots.
3.3. Heterologous production of G. uralensis flavonoids in yeast
Having demonstrated the catalytic abilities of all the key enzymes in the G. uralensis flavonoid biosynthesis pathway in vitro or in vivo, we attempted to reconstruct the main flavonoid pathway in yeast (Fig. 4A). When GuPAL1 and GuC4H1 were simultaneously introduced into S. cerevisiae WAT11, harboring the A. thaliana NADPH-cytochrome P450 reductase ATR149, which provides the reducing equivalents essential for the activity of plant CYP450s such as C4H, the obtained recombinant yeast WM1 was able to produce p-coumaric acid under galactose induction (Fig. 4B), and the content of coumaric acid reached 7.59 μmol/L after cultivation for 36 h (Fig. 4C). When the vector carrying Gu4CL1 and GuCHS1::GuCHR1 was transferred into WM1, a small amount of Iso-LG was detected in the fermentation broth (Fig. 4B, WM2-1). Interestingly, Iso-LG was not detected when GuCHS1 and GuCHR1 were co-expressed in yeast that was fed with the substrate. After GuCHI1 and GuUGT1 were successively transferred to WM2-1, strains WM3-1 and WM4-1 with the capacity to produce LG and LN were respectively obtained (Fig. 4B). Compared with LN, more Iso-LG in WM4-1 was glycosylated before isomerization, resulting in 4.7 times higher Iso-LN production than that of LN in shake flasks (Fig. 4E).
Figure 4.
Reconstitution of the biosynthesis pathway of liquiritin in yeast (A) Schematic of the recombinant yeast strain WM4-1. (B) Chromatogram of selected ions with m/z 163, 255, 271 and 417 from the fermentation products of the recombinant yeasts using HPLC−Q-TOF-MS; (C) Production of p-CA and Iso-LG by yeast stains WM1 (harboring GuPAL1, GuC4H1), WM2-1 (harboring GuPAL1, GuC4H1, Gu4CL1, GuCHS1::GuCHR1) and WM2-2 (GuCHS1::GuCHR1 overexpressed in WM2-1); (D) Production of p-CA, Iso-LG and LG by yeast stains WM3-1 (harboring GuPAL1, GuC4H1, Gu4CL1, GuCHS1::GuCHR1 and GuCHI1), WM3-2 (GuCHI1 overexpressed in WM3-1) and WM3-3 (GuCHS1::GuCHR1 overexpressed in MW3-2) after induction with galactose for 36 h; (E) Fermentation products of yeast stains WM4-1 (harboring GuPAL1, GuC4H1, Gu4CL1, GuCHS1::GuCHR1, GuCHI1 and GuUGT1), WM4-2 (GuCHI1 overexpressed in WM4-1) and WM4-3 (GuCHS1::GuCHR1 overexpressed in WM4-2) after induction with galactose in shake flasks. GuCHS1::GuCHR1 represents a construct in which the 3ʹend of GuCHS1 was fused to the 5ʹ end of GuCHR1 via the linker sequence GGTGGTGGTTCT.
3.4. Promoting the production of flavonoids by gene overexpression
To promote the transformation of the upstream metabolites to the downstream flavonoid structure along the liquiritin pathway, we tried to overexpresses the key genes for Iso-LG biosynthesis in the described recombinant strains. The results showed that overexpression of GuCHS1::GuCHR1 significantly increased the synthesis of Iso-LG, and the production of Iso-LG by WM2-2 was increased 18 times compared with WM2-1 (Fig. 4C). To solve the problem of the much lower production of the target product LN than Iso-LN in WM4-1, GuCHI1 was overexpressed in WM3-1 and WM4-1, after which the yield of LG and LN increased 1.3-fold (Fig. 4D, WM3-2) and 2.1-fold (Fig. 4E, WM4-2), respectively. Moreover, simultaneous overexpression of GuCHS1::GuCHR1 and GuCHI1 increased the LG production of WM3-3 5.3-fold compared with WM3-1 (Fig. 4D), and the accumulation of LN in WM4-3 was 6.4 times higher than in WM4-1 (Fig. 4E). In addition, it was found that overexpression of both GuCHI1 and GuCHS1::GuCHR1 in this yeast system not only increased the production of the downstream products of these enzymes, but also significantly increased the accumulation of the upstream precursor compound p-coumaric acid. Additionally, more glycosides (Iso-LN: 89.3%, LN: 90.3%) were secreted into the culture medium, whereas more than 30% of the Iso-LG and LG remained inside the yeast cells (Supporting Information Fig. S7).
3.5. Metabolite accumulation in the fermenter varies with the amount of cells
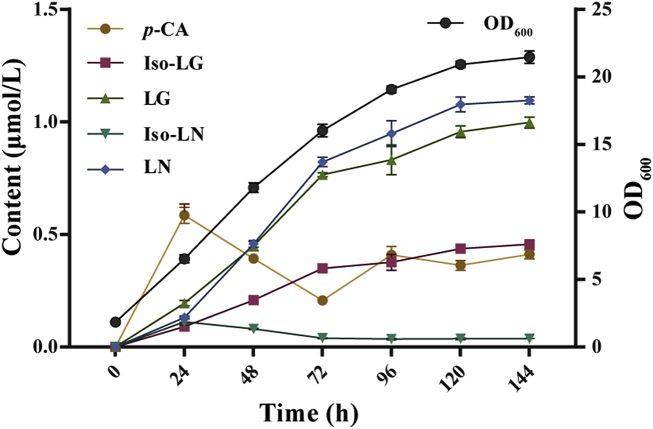
The changes of metabolites in the recombinant yeast strain WM4-3 with increasing induction time and biomass were also investigated in the 11 L benchtop fermenter with controlled DOC (>40%) and pH (5.0) (Fig. 5). In the early stage of fermentation, the upstream pathway accumulated a large amount of p-coumaric acid. Subsequently, the metabolites rapidly flowed to the final product after 24 h of fermentation, which was different from the production of a large amount of Iso-LN instead of LN observed in shake flasks (Fig. 4E). At 144 h, the accumulation of LG and LN were respectively reached 1.0 and 1.1 μmol/L. It can be seen that the fermentation conditions can greatly affect the metabolic flow direction, and the potential of this yeast fermentation system can be fully developed in the future via metabolic engineering and fermentation optimization.
Figure 5.
Cell growth and fermentation products of strain WM4-3 in fed-batch fermentation. The dissolved oxygen concentration (DOC) was kept above 40% and the pH was maintained at 5.0 using automatic addition of ammonium hydroxide. Three replicates were performed for each analysis and the error bars represented the standard deviation (SD).
4. Discussion
Flavonoids are an important class of compounds widely found in nature. In addition to their contribution to the gorgeous colors of plants, their enormous medicinal potential has become the most active area of flavonoid research in recent years50. With the rapid development of genetic and metabolic engineering, great progress has been made in the analysis of biosynthetic pathways of flavonoids51. The complete pathways of plant-specific flavonoids such as quercetin, resveratrol, kaempferol41, scutellarin52, baicalein and scutellarein43 have been reconstructed in S. cerevisiae or E. coli. Some researchers have also expressed a part of the genes encoding the enzymes in the LG biosynthetic pathway from plasmids in E. coli or S. cerevisiae, and attempted to convert exogenously fed phenylpropanoid acids into 5-deoxyflavonoids such as Iso-LG and LG, but the main fermentation product they obtained was naringenin42. In this study, the complete biosynthetic pathway of the main flavonoid of G. uralensis, LN, was characterized and reconstructed in S. cerevisiae, and achieved the de novo biosynthesis of LN using raw materials and cofactors from the endogenous yeast metabolism. This provides a possibility of economically and sustainably producing G. uralensis flavonoids through synthetic biology.
The biosynthesis of LN is initiated via the general phenylpropanoid metabolism, but the downstream steps of its synthetic pathway leads to the production of 5-deoxyflavonoids (Iso-LG, LG) due to the role of CHR, which is different from the synthetic pathways of quercetin and other 5-hydroxyisoflavonoids. CHR, also known as NADPH-dependent 6′-deoxychalcone synthase, can only work with CHS at high concentrations of NADPH (≥ 0.1 mmol/L) to convert malonyl-CoA and coumaroyl-CoA to synthesize Iso-LG. Otherwise, only naringenin chalcone, the catalytic product of CHS, can be generated53. Therefore, in the in vitro enzymatic reaction system of CHR, 0.1 mmol/L NADPH was added. Nevertheless, naringenin chalcone still accounted for the main part of the catalytic products in spite of the production of Iso-LG (Fig. 3E). In addition, we also used S. cerevisiae WAT11 as a natural NADPH supplier, in which GuCHS1 and GuCHR1 were co-expressed, and the precursor compound was fed during fermentation, but no Iso-LG was detected in the extract of the fermentation broth. Although studies have shown that CHR has no obvious interaction with CHS or even other cytoplasmic enzymes in the isoflavone pathway except for IFS45, interestingly, the recombinant yeast expressing the fusion of GuCHS1 and GuCHR1 was able to produce Iso-LG instead of naringenin chalcone (Fig. 4B). This is in contrast to the LG-producing strain constructed by the Mattheos research team42, which produces more by-product, naringenin chalcone, than the target product Iso-LG. The fusion of GuCHS1 and GuCHR1 may play a role in shortening the time and distance that the unstable intermediates must traverse from CHS to CHR, which is similar to the immobilization of CHS and CHR recombinant proteins on Ni2+-coated beads, which was reported to significantly increase the proportion of deoxygenated products45.
In the reconstruction of the LN pathway in yeast, the overexpression of GuCHI1 significantly increased the yield of p-coumaric acid in the upstream pathway even though CHIs have relatively slower kinetics in the conversion of Iso-LG to LG54, and overexpression of GuCHS1::GuCHR1 was able to significantly increase the accumulation of both upstream and downstream products (Fig. 4C–E). This phenomenon suggests that there may be an interaction between the different genes of the pathway, or that the downstream product of the LN pathway exerts a positive feedback effect on the upstream pathway. The possible mechanisms need further study.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81773838), the National Program for Special Support of Eminent Professionals and the Key Project at central government level (No. 2060302, China). We wish to sincerely thank the other members in our laboratory, including Ying Huang, Jing Chen, Zhen Xu, Xiaosong Hu, and Ting Li, for their generous help with plant cultivation, sample collection and critical review of this manuscript. We are also grateful to Wenbin Li (Beijing University of Chinese Medicine, China) for providing the picture of Glycyrrhiza uralensis Fisch.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Author contributions
Wei Gao and Chunsheng Liu designed the research. Yan Yin and Yanpeng Li performed the experiments under the guidance of Wei Gao and Chunsheng Liu. Yan Yin and Yanpeng Li finished the sketch, Dan Jiang and Xianan Zhang analyzed the data and revised the manuscript. All authors contributed a lot to this work and approved the final version of the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.07.005.
Contributor Information
Wei Gao, Email: weigao@ccmu.edu.cn.
Chunsheng Liu, Email: max_liucs@263.net.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gao X., Wang W., Wei S., Li W. Review of pharmacological effects of Glycyrrhiza radix and its bioactive compounds. China J Chin Mater Med. 2009;34:2695–2700. [PubMed] [Google Scholar]
- 2.Li T., Zhuang S., Wang Y., Wang Y., Wang W., Zhang H. Flavonoid profiling of a traditional Chinese medicine formula of Huangqin Tang using high performance liquid chromatography. Acta Pharm Sin B. 2016;2:148–157. doi: 10.1016/j.apsb.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., Yang R., Yuan B., Liu Y., Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B. 2015;4:310–315. doi: 10.1016/j.apsb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Traboulsi H., Cloutier A., Boyapelly K., Bonin M.A., Marsault É., Cantin A.M. The flavonoid isoliquiritigenin reduces lung inflammation and mouse morbidity during influenza virus infection. Antimicrob Agents Chemother. 2015;59:6317–6327. doi: 10.1128/AAC.01098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanazawa M., Satomi Y., Mizutani Y., Ukimura O., Kawauchi A., Sakai T. Isoliquiritigenin inhibits the growth of prostate cancer. Eur Urol. 2003;43:580–586. doi: 10.1016/s0302-2838(03)00090-3. [DOI] [PubMed] [Google Scholar]
- 6.Gao F., Zhang J., Fu C., Xie X., Peng F., You J. iRGD-modified lipid-polymer hybrid nanoparticles loaded with isoliquiritigenin to enhance anti-breast cancer effect and tumor-targeting ability. Int J Nanomed. 2017;12:4147–4162. doi: 10.2147/IJN.S134148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D.C., Choi S.Y., Kim S.H., Yun B.S., Yoo I.D., Reddy N.R. Isoliquiritigenin selectively inhibits H2 histamine receptor signaling. Mol Pharmacol. 2006;70:493–500. doi: 10.1124/mol.106.023226. [DOI] [PubMed] [Google Scholar]
- 8.Chen H., Zhang B., Yuan X., Yao Y., Zhao H., Sun X. Isoliquiritigenin-induced effects on Nrf2 mediated antioxidant defence in the HL-60 cell monocytic differentiation. Cell Biol Int. 2013;37:1215–1224. doi: 10.1002/cbin.10156. [DOI] [PubMed] [Google Scholar]
- 9.Li Y.Y., Yang L., Chai X., Yang J.J., Wang Y.F., Zhu Y. Four major urinary metabolites of liquiritigenin in rats and their anti-platelet aggregation activity. Chem Nat Compd. 2018;54:443–446. [Google Scholar]
- 10.Hou Y., Che D., Ma P., Zhao T., Zeng Y., Wang N. Anti-pseudo-allergy effect of isoliquiritigenin is MRGPRX2-dependent. Immunol Lett. 2018;198:52–59. doi: 10.1016/j.imlet.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Feng Yeh C., Wang K.C., Chiang L.C., Shieh D.E., Yen M.H., San Chang J. Water extract of licorice had anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J Ethnopharmacol. 2013;148:466–473. doi: 10.1016/j.jep.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maggiolini M., Statti G., Vivacqua A., Gabriele S., Rago V., Loizzo M. Estrogenic and antiproliferative activities of isoliquiritigenin in MCF7 breast cancer cells. J Steroid Biochem Mol Biol. 2002;82:315–322. doi: 10.1016/s0960-0760(02)00230-3. [DOI] [PubMed] [Google Scholar]
- 13.Liu J.H., Wei H.B., Yao Y., Zheng Q.S. Anti-tumor angiogenesis of four different active ingredients from Glycyrrhiza. Prog Mod Biomed. 2010;10:2731–2734. [Google Scholar]
- 14.Wang W., Hu X., Zhao Z., Liu P., Hu Y., Zhou J. Antidepressant-like effects of liquiritin and isoliquiritin from Glycyrrhiza uralensis in the forced swimming test and tail suspension test in mice. Prog Neuropsychopharmacol Biol Psychiatr. 2008;32:1179–1184. doi: 10.1016/j.pnpbp.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Xie S.R., Wang Y., Liu C.W., Luo K., Cai Y.Q. Liquiritigenin inhibits serum-induced HIF-1α and VEGF expression via the AKT/mTOR-p70S6K signalling pathway in heLa cells. Phytother Res. 2012;26:1133–1141. doi: 10.1002/ptr.3696. [DOI] [PubMed] [Google Scholar]
- 16.Liu C., Wang Y., Xie S., Zhou Y., Ren X., Li X. Liquiritigenin induces mitochondria-mediated apoptosis via cytochrome c release and caspases activation in heLa Cells. Phytother Res. 2011;25:277–283. doi: 10.1002/ptr.3259. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S.P., Zhou Y.J., Liu Y., Cai Y.Q. Effect of liquiritigenin, a flavanone existed from Radix glycyrrhizae on pro-apoptotic in SMMC-7721 cells. Food Chem Toxicol. 2009;47:693–701. doi: 10.1016/j.fct.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 18.Wei F., Jiang X., Gao H.Y., Gao S.H. Liquiritin induces apoptosis and autophagy in cisplatin (DDP)-resistant gastric cancer cells in vitro and xenograft nude mice in vivo. Int J Oncol. 2017;51:1383–1394. doi: 10.3892/ijo.2017.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J.G., Zhou Z., Liu H.F., Wang J.X. Application of active glycyrrhizic constituents to cosmetics. China Surf Deter Cosmet. 2004;34:249–251. [Google Scholar]
- 20.Huang M., Wang W., Wei S. Investigation on medicinal plant resources of Glycyrrhiza uralensis in China and chemical assessment of its underground part. China J Chin Mater Med. 2010;35:947–952. doi: 10.4268/cjcmm20100802. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q., Li J., Simayi H. Utilization status and protection measures for licorice resources in Xinjiang. Grass-Feed Livest. 2018;2018:52–56. [Google Scholar]
- 22.Yang L., Chen J., Yang T., Li Z., Hu W., Wang Y. Geographic distribution and resource survey of the wild medicinal plants licorice (Glycyrrhiza uralensis) in northwest China. Chin Wild Plant Res. 2013;32:27–31. [Google Scholar]
- 23.Ro D.K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 24.Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 25.Dai Z., Wang B., Liu Y., Shi M., Wang D., Zhang X. Producing aglycons of ginsenosides in bakers' yeast. Sci Rep. 2014;4:3698. doi: 10.1038/srep03698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Ajikumar P.K., Xiao W.H., Tyo K.E., Wang Y., Simeon F., Leonard E. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y.J., Gao W., Rong Q., Jin G., Chu H., Liu W. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 29.Lau W., Sattely E.S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mochida K., Sakurai T., Seki H., Yoshida T., Takahagi K., Sawai S. Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 2017;89:181–194. doi: 10.1111/tpj.13385. [DOI] [PubMed] [Google Scholar]
- 32.Seki H., Ohyama K., Sawai S., Mizutani M., Ohnishi T., Sudo H. Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci U S A. 2008;105:14204–14209. doi: 10.1073/pnas.0803876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seki H., Sawai S., Ohyama K., Mizutani M., Ohnishi T., Sudo H. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell. 2011;23:4112–4123. doi: 10.1105/tpc.110.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu G., Cai W., Gao W., Liu C. A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol. 2016;212:123–135. doi: 10.1111/nph.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Liu Y., Zhang X., Zhan X., Liu C. Cloning and characterization of the gene encoding β-amyrin synthase in the glycyrrhizic acid biosynthetic pathway in Glycyrrhiza uralensis. Acta Pharm Sin B. 2013;6:416–424. [Google Scholar]
- 36.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Li W.D., Hou J.L., Wang W.Q., Tang X.M., Liu C.L., Xing D. Effect of water deficit on biomass production and accumulation of secondary metabolites in roots of Glycyrrhiza uralensis. Russ J Plant Physiol. 2011;58:538–542. doi: 10.1134/S1021443711030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong Y.J., An C.H., Park S.C., Pyun J.W., Lee J., Kim S.W. Methyl jasmonate increases isoflavone production in soybean cell cultures by activating structural genes involved in isoflavonoid biosynthesis. J Agric Food Chem. 2018;66:4099–4105. doi: 10.1021/acs.jafc.8b00350. [DOI] [PubMed] [Google Scholar]
- 41.Trantas E., Panopoulos N., Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng. 2009;11:355–366. doi: 10.1016/j.ymben.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Yan Y., Huang L., Koffas M.A. Biosynthesis of 5-deoxyflavanones in microorganisms. Biotechnol J. 2007;2:1250–1262. doi: 10.1002/biot.200700119. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Tian C., Xia Y., Mutanda I., Wang K., Wang Y. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab Eng. 2019;52:124–133. doi: 10.1016/j.ymben.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Leonard E., Yan Y., Fowler Z.L., Li Z., Lim C.G., Lim K.H. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm. 2008;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- 45.Mameda R., Waki T., Kawai Y., Takahashi S., Nakayama T. Involvement of chalcone reductase in the soybean isoflavone metabolon: identification of GmCHR5, which interacts with 2-hydroxyisoflavanone synthase. Plant J. 2018;96:56–74. doi: 10.1111/tpj.14014. [DOI] [PubMed] [Google Scholar]
- 46.Ralston L., Subramanian S., Matsuno M., Yu O. Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol. 2005;137:1375–1388. doi: 10.1104/pp.104.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ban Z., Qin H., Mitchell A.J., Liu B., Zhang F., Weng J.K. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc Natl Acad Sci U S A. 2018;115:E5223–E5232. doi: 10.1073/pnas.1802223115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brazier-Hicks M., Offen W.A., Gershater M.C., Revett T.J., Lim E.K., Bowles D.J. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci U S A. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban P., Mignotte C., Kazmaier M., Delorme F., Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 50.Harborne J.B., Williams C.A. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 51.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 2018;9:448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayabe S.I., Udagawa A., Furuya T. NAD(P)H-dependent 6'-deoxychalcone synthase activity in Glycyrrhiza echinata cells induced by yeast extract. Arch Biochem Biophys. 1988;261:458–462. doi: 10.1016/0003-9861(88)90362-1. [DOI] [PubMed] [Google Scholar]
- 54.Jez J.M., Noel J.P. Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences. J Biol Chem. 2002;277:1361–1369. doi: 10.1074/jbc.M109224200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.