ABSTRACT
Trauma, burn injury, sepsis, and ischemia lead to acute and chronic loss of skeletal muscle mass and function. Healthy muscle is essential for eating, posture, respiration, reproduction, and mobility, as well as for appropriate function of the senses including taste, vision, and hearing. Beyond providing support and contraction, skeletal muscle also exerts essential roles in temperature regulation, metabolism, and overall health. As the primary reservoir for amino acids, skeletal muscle regulates whole-body protein and glucose metabolism by providing substrate for protein synthesis and supporting hepatic gluconeogenesis during illness and starvation. Overall, greater muscle mass is linked to greater insulin sensitivity and glucose disposal, strength, power, and longevity. In contrast, low muscle mass correlates with dysmetabolism, dysmobility, and poor survival. Muscle mass is highly plastic, appropriate to its role as reservoir, and subject to striking genetic control. Defining mechanisms of muscle growth regulation holds significant promise to find interventions that promote health and diminish morbidity and mortality after trauma, sepsis, inflammation, and other systemic insults. In this invited review, we summarize techniques and methods to assess and manipulate muscle size and muscle mass in experimental systems, including cell culture and rodent models. These approaches have utility for studies of myopenia, sarcopenia, cachexia, and acute muscle growth or atrophy in the setting of health or injury.
Keywords: C2C12, gene transfer, genotype, methods, muscle atrophy, muscle hypertrophy, myoblasts, myogenesis, phenotyping, Skeletal muscle
What follows is an invited review summarizing a presentation by Teresa Zimmers in the Master Class Session of the 2018 Annual Meeting of the Shock Society.
INTRODUCTION
Skeletal muscle is a dynamic tissue comprising 40% of the total mass of a human body. In addition to providing force production for respiration, eating and locomotion, skeletal muscle acts as an endocrine organ regulating whole body metabolism, glucose and lipid metabolism, inflammation, and even sleep and circadian rhythm (1–3). Muscle-derived mechanical signals, secreted factors (known as “myokines”) (4), and exosomes (5) all exert substantial influence over adjacent tissues such as bone as well as distant tissues including liver, adipose and the pancreas. Muscle also represents an energy repository and storehouse of amino acids and nitrogen which is liberated during states of acute infection, injury and starvation (6–8). Increased muscle mass and muscle quality are associated with better outcomes and prognoses across disease conditions, while muscle loss is associated with poor outcomes and increased mortality (9–13). Moreover, products of muscle destruction can precipitate coagulation and kidney function and injury (14).
Skeletal muscle mass and quality is a powerful modulator of morbidity and mortality in health and disease, including sepsis, shock, burns and trauma. Myopenia at injury is associated with poor outcomes, demonstrating the need for adequate skeletal muscle at baseline. Furthermore, injury induces muscle wasting, often both in the acute injury period and in the longer term. This muscle loss and accompanying muscle dysfunction often lead to dysmobility, asthenia, disability, and ultimately, death (Fig. 1). Thus, assessment of skeletal muscle mass, quality, and regulation of muscle growth and wasting are important for understanding systemic disease conditions and for predicting patient outcomes.
Fig. 1.
Overview of this invited review.
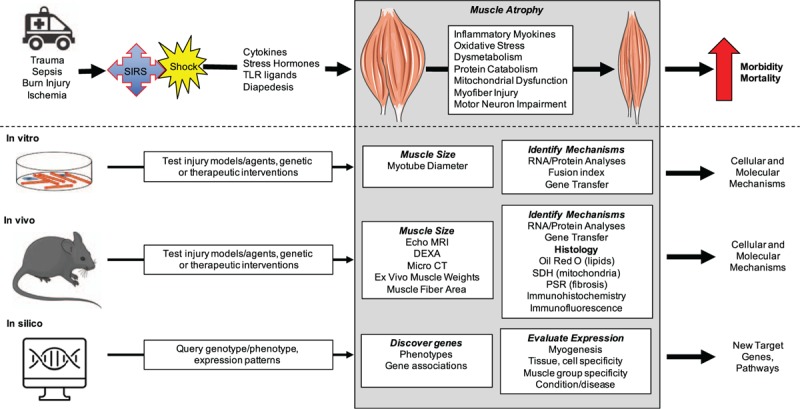
Trauma leads to a systemic inflammatory response syndrome (SIRS) and shock, which produces mediators that act on muscle to produce atrophy through effects on multiple processes affecting the myofibers but also the motor neurons. This leads to morbidity and mortality. We review methods to interrogate the mechanisms underlying this injury-induced atrophy using cell culture, mouse models, and available datasets. CT indicates computed tomography; DXA, dual energy x-ray absorptiometry, PSR, picrosirius red; SDH, succinate dehydrogenase; TLR, toll-like receptor.
Discovery of mediators of muscle growth regulation requires responsive experimental systems and methods to quantify muscle size and mass. Given the conservation of many growth regulatory pathways and the food production value of muscle, multiple model organisms and a variety of species have been studied, ranging from worms, flies, and fish (zebrafish as well as salmon), to rodents, ruminants, and pigs. Here, however, we will focus on cell culture of mouse and human muscle cells, as well as on the use of mouse models and available cell, murine, and human data given their predominance in the biomedical literature. In this invited review, we provide an overview of in vitro, in vivo, and in silico methods to probe muscle growth and wasting in the setting of normal biology or in the presence of trauma, burn, sepsis, inflammation, or other injury relevant to the readers of Shock (Fig. 1).
IN VITRO ASSESSMENT OF MUSCLE SIZE REGULATION
C2C12 myogenesis
Skeletal muscle myogenesis, muscle growth, and muscle atrophy can be investigated in culture using myoblast cell lines, primary cells, and isolated myofibers. The C2C12 murine myoblast cell line is a well-established, readily accessible in vitro model for assaying potential regulators of muscle differentiation and muscle growth (15). These immortalized myoblasts can be passaged as mononuclear cells in conditions of growth factor abundance (growth medium or GM: DMEM with 10–20% fetal calf serum), but when washed and placed in restricted growth factor conditions (differentiation medium or DM: DMEM with 2% donor horse serum), the cells express myogenin, withdraw from the cell cycle and express Cdkn1a/p21, then progressively fuse and express contractile proteins, resulting in long, multi-nucleated myotubes (Fig. 2A). Detailed protocols for generating and visualizing myotubes have been described previously (16).
Fig. 2.
Assessing effects on muscle size using C2C12 myotube cultures.
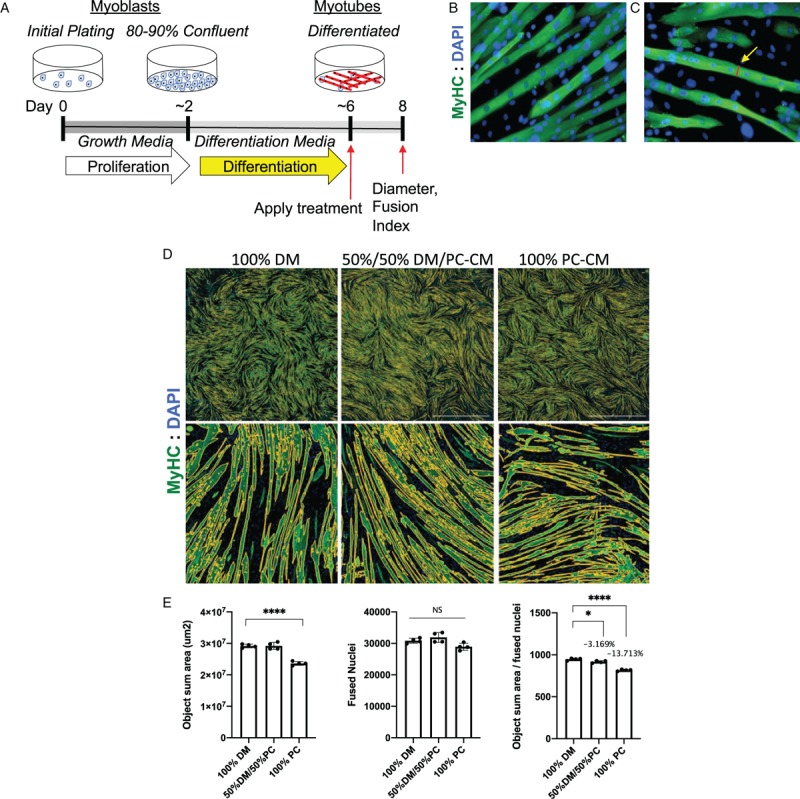
A, Schematic of a typical C2C12 differentiation study in which myoblasts are grown in fetal calf serum-containing growth media, then washed and switched to low growth factor containing differentiation media (DM) for 4 days, then incubated with test factors for 48 h, then washed, fixed, and measured on day 8. B, Myotubes are visualized by immunofluorescence with antimyosin heavy chain antibody (green) and DAPI to visualize nuclei (blue). C, Manual measurements on calibrated micrografts can be done manually using ImageJ. D, Alternatively, wells can be scanned on digital scanning microscopes (Lionheart was used here), and E, automated image analysis used to determine total area covered by myotubes (object sum area), the number of nuclei within myotubes (fused nuclei), and the ratio. Here 4-day-old myotubes were incubated for 48 h in 100% DM or 50% DM with 50% pancreatic cancer cell conditioned media (PC-CM) or 100% PC-CM. Automated imaging demonstrates a reduction in overall green area, a non-significant reduction in fused nuclei, and reductions in the ratio, indicating smaller myotube cell body size per nucleus or atrophy. ∗P < 0.05; ∗∗∗∗P < 0.0001. These studies were done using four replicate wells per condition.
Under ideal conditions, myoblasts will fully fuse into multinucleated myotubes 4 to 5 days after transfer into DM, producing striking patterns of striated and often visibly contracting myotubes (Fig. 2B). Growth of these myotubes, as growth of myofibers, is the result of both accretion of nuclei from newly fused and differentiating myoblasts and of protein synthesis and resulting cellular hypertrophy. Effects of nuclear accretion can be eliminated after a point by treating cultures with antiproliferative agents such as arabinosylcytosine (AraC; Sigma) to kill myoblasts, leaving a purer culture of myotubes. Often this is done by transferring myotubes from DM to GM with AraC for 48 h and then back to DM for 24 h for recovery. Whether myoblasts are depleted or not, once myotubes are well differentiated, agents specific to an hypothesis (e.g., drugs, cytokines, lipids, exosomes, tumor cell conditioned media, mouse or human serum/plasma, etc.) can be added to the differentiation media and effects on myotube diameter can be assessed. Furthermore, therapeutic interventions in combination with atrophy inducing agents can be used to treat myotubes to evaluate the efficacy of an intervention to maintain or increase myotube size.
Myotube morphometry
Myotubes can be visualized via phase contrast or bright field microscopy without staining, or by staining with Ponceau S (17) or Cresyl violet. To readily distinguish myotubes from unfused cells however, observers often use immunofluorescence for myosin heavy chain (MyHC) to highlight cell bodies and DAPI to visualize nuclei (18). Fluorescent myotubes and nuclei are then imaged using a camera-equipped fluorescence microscope or plate scanner in such a manner as to ensure equivalent sampling of wells. This can be done by photographing the entire well or by sampling the same regions within each well across experimental conditions. Note that the center and edges of the wells will have greater and lesser density of myotubes, respectively and this should be accounted for while sampling images.
Appropriate constraints should be specified a priori and could include measurement of only myotubes that stain positive for MyHC or other muscle markers such as dystrophin, that are fully in frame, and that contain a minimum number of nuclei (variously specified in the literature as 2 to 10). Myotube measurements should be done away from nuclear accretions and should be standardized across conditions. Myotube diameter and fusion index can be measured either manually from micrographs using opensource software programs such as Image J (Fig. 2C) or automatically using scanning digital microscopes such as the Incucyte, Lionheart (Biotek), or similar product. While segmentation and measurement of individual myotubes is difficult with automated systems, generally proprietary software can estimate differences across wells by measuring total green area as a marker of total MyHC presence/total myotube area, and fusion index by determining the number of DAPI-positive nuclei within the green area (Figs. 2D, E). The automated assessment of myotubes across wells is useful with higher throughput experiments (e.g., screening compound libraries) to detect differences across wells in up to 48-well plates. Effects detected by automated analysis can subsequently be confirmed independently via manual measurements.
Experimental considerations
Controls are useful when assaying novel conditions for effects on myotube size. Positive controls for hypertrophy could include a hypertrophic stimulus leading to enhanced protein synthesis, such as addition of recombinant IGF-1 (long IGF) (19, 20), which ultimately activates AKT, or transfection/infection with constructs to express constitutively activated AKT. Positive controls for atrophy could include use of glucose-depleted differentiation medium, or addition of glucocorticoids (20, 21) or other atrophy-inducing stimuli including LPS plus INF-g (17), myostatin, TNF, or Interleukin-6 (22). Myotube experiments are also useful for investigating molecular changes associated with muscle atrophy and hypertrophy. Using the same culture approach as described, alongside wells meant for imaging, wells of myotubes can processed to collect media, exosomes, and cellular DNA, RNA and protein for various downstream analyses. In this fashion, markers of hypertrophy could be quantified using as puromycin incorporation into nascent proteins and detection by Western blotting analysis, or by total protein or total muscle specific protein. Similarly, one could assay markers of muscle proteolysis such as total protein ubiquitylation by Western blotting analysis or RNA expression of muscle-specific ubiquitin ligases MAFbx/Atrogin-1/Fbxo32, Murf-1/Trim63 (23), or others such as MUSA1 and SMART (24) by quantitative real-time reverse transcription and PCR (qPCR).
Excellent technique and attention to culture conditions is important. High-quality studies with C2C12 myotubes require consistency between experiments, maintaining plating density, depth of media, regular changes of media, and timepoints of analysis. The system should be optimized and validated within each laboratory and by each operator. Morphologic changes can be benchmarked against myogenic gene expression. Points of reference are available in multiple published datasets, including those deposited into the Gene Expression Omnibus as GSE11415 (6 timepoints from confluence to 5 days (25)), GSE20059 (6 conditions of differing confluence and differentiation stage (26)), and GSE17039 (26 conditions in early myogenesis (27)). These can be readily queried using a paid or free version of Illumina BaseSpace Correlation Engine (28) or GEO2R (29), the NCBI R-based web application to query genes of interest along the myogenic course.
During growth conditions, myoblasts must be kept at low confluence to avoid selecting against cells with differentiation potential. Well-cared for cell lines should elicit robust differentiation with most of the surface covered with myotubes (Fig. 2D). Timing is important; effects observed after 4-day of differentiation might be absent in 7-day cultures, due to changes in gene expression. The presence or absence of myoblasts could also influence outcomes and interpretation. Substantial variation in this system across laboratories is evident in the literature, a pitfall of this approach. Discrepancies could be due to specifics of culture conditions, including serum type (30, 31) and subclone, given the well-recognized variable differentiation potential of lines shared among laboratories. As well, lower passage clones tend to differentiate better than higher passage lines. Thus, reviewers should demand representative images from authors and reviewers and readers both should be cautious of studies that draw conclusions from cultures of few or sparse myotubes.
Rigor
Rigor is increased with distribution of plating variables across different plates rather than between plates, with use of multiple technical replicates, with blinding of the observer to experimental conditions, and with sampling of several hundred to a thousand myotubes per condition, determining the average myotube diameter of each well, and calculating standard deviations and statistical differences based upon the number of wells and not on thousands of myotubes. Care should also be taken to compare equivalent sections of each well, given that cells can sometimes pool in the center of the well, generating gradients of cell density and myogenesis. Results should be repeated at least once in a separate culture experiment.
Alternative cell lines
In addition to the C2C12 line and the similar rat L6 myoblast cell line, investigators can isolate or purchase primary satellite cells/myoblasts/muscle progenitor cells for similar myogenesis studies. Abundant resources describe multiple protocols for the isolation and culture of primary myoblasts, ranging from explanting muscle and harvesting of migrating cells to automated tissue digestion and magnetic selection or flow cytometry approaches. Use of primary satellite cells or myoblasts can leverage genetically modified mice. Human primary myoblasts can be purchased from several vendors including Lonza and Cook Myosite; however, these cell preparations are expensive, have limited replication potential prior to onset of senescence, and display variable myogenic potential. Recently, hTERT/cdk4 immortalized human myogenic cell lines have been developed, which could greatly facilitate studies of human myogenesis and muscle growth regulation (32).
In vitro genetic manipulation of muscle size
Muscle mass is remarkably plastic and research over the past decades has identified key cell autonomous and non-cell autonomous pathways regulating hypertrophy and atrophy of skeletal muscle (33–35). Targeting these pathways has been an area of interest for the treatment of muscle wasting associated with various pathologies including cancer, muscular dystrophy, burn injury, sepsis, and aging (8, 36, 37). Using cultured myotubes to genetically manipulate muscle size provides an excellent first screen prior to embarking upon more costly and time-consuming in vivo methods. Specific genes can be expressed, knocked down, or knocked out by transfecting myoblasts with cDNA- or shRNA-expressing plasmids, with RNAs such as siRNA, lnRNA, miRNA, or with CRISPR constructs using plasmids or gRNAs, then differentiating the myoblasts into myotubes. (It should be noted that the potential polyploidy and limited differentiation potential of C2C12 myoblasts make CRISPR-based approaches challenging.) Genetic manipulation is generally accomplished by transfecting myoblasts with plasmids or siRNA using Lipofectamine 3000 or other transfection reagents or by infecting myoblasts with lentivirus to manipulate genes of interest (38, 39). Transfection of myoblasts can affect their differentiation potential; however, so proper controls are essential. In theory, myoblasts could be transfected with an inducible construct, permitting expression only after differentiation into myotubes; in practice, such a system is not widely reported.
Differentiated myotubes are notoriously resistant to transfection via conventional means, regardless of transfection reagent. Physical transfection approaches including particle-based magnetic and biolistic approaches are generally inefficient (data not shown and (40)). Electroporation of myotubes seeded on coverslips has been reported although not widely adopted (41). Electroporation with an immersion electrode is efficient but results in copious fusion events, grossly altering myotube morphology (TAZ, data not shown). Genetic manipulation in myotubes is most efficient and straightforward through use of viral vectors. Adenovirus readily infects C2C12 myotubes (22), but for safety of personnel involved in the preparation and handling of vectors, adeno-associated viruses are preferred and also highly effective (42). Recombinant adeno-associated viruses readily transduce both dividing and non-dividing cells and persist as episomal chromatin in muscle (43), allowing for long-term expression in differentiated myotubes in vitro and muscle fibers in vivo. A recent study using AAV6 documents infection of 5-day-old myotubes with an anti-atrophy gene resulting in resistance to challenge with a atrophy-inducing stimulus 48 h later (44), demonstrating the power and utility of this approach.
IN VIVO AND EX VIVO ASSESSMENT OF MUSCLE MASS AND SIZE REGULATION
In vivo measurement of lean mass
Estimation of muscle mass in living mice and rats begins with whole body weights and is refined by quantitative assessments of whole-body lean mass. Quantitative time domain nuclear magnetic resonance (qNMR) and dual energy X-ray absorptiometry (DXA) can also be used to estimate lean body mass (45). Assessments by qNMR (EchoMRI and Bruker instruments) can be done on awake animals, while DXA requires general anesthesia. Although the literature is littered with reports equating lean mass to muscle mass, neither of these approaches quantifies skeletal muscle specifically, which requires quantitative imaging approaches such as micro X-ray computed tomography (microCT) (46). MicroCT can be used to segment and measure individual muscle volumes in mice and rats longitudinally across an experiment; however, the long times under anesthesia, the complexity of quantitative image analysis, and the expense and relative rarity of such systems usually render them impractical for simple assessments of mass. More typical is the use of euthanasia, dissection and direct measurement of muscle mass and size in tissues.
Ex vivo measurement of muscle mass
Destructive analysis of muscle mass begins with identification, excision, and weighing of individual muscles immediately post-mortem to determine changes to muscle mass. To account for size differences between animals, ex vivo muscle weight can be normalized to body weight at the beginning of a longitudinal experiment or to tibia length in most any experiment. Anatomic maps of mouse skeletal muscles are available (47–49) and a video demonstrating muscle identification, excision, and collection is published and available via open access (50).
Fixation of excised muscles
Muscle mass in normal mice is absolutely linked to myofiber number and myofiber size. In pathological conditions, muscle mass could be affected by infiltrating immune cells, fatty change, fibrosis and extracellular matrix expansion, and water content. Histomorphometry is used to distinguish these. Skeletal muscle is high in water content and this must be considered during the fixation process. Muscle is commonly fixed using two methods, either with formalin or by freezing. Choice of fixation is determined by the downstream analyses. Excellent protocols for formalin fixed, paraffin embedded (FFPE) muscle preparation, and analysis are widely available, including here (51). To prevent shrinkage of muscle, isolated muscles or biopsy specimens are gently pulled to their normal length using a clamp or by pinning to a wax substrate (e.g., dental wax sheets). Fixation is performed by submerging tissue (∼50–100 mg) in formalin for 24 to 72 h, depending on the volume of the specimen. The tissue is then processed, paraffin embedded as usual, and sectioned on a microtome. Formalin fixation cross-links proteins within the tissue, which needs to be reversed prior to immunohistochemistry using a chemical or heat-mediated antigen retrieval process, which can be quite harsh and alter the structure of antigens. Deparaffinization removes almost all lipids from the sample, leaving empty space behind. FFPE sections are typically stained with H&E and provide visualization of invading inflammatory cells, fibrosis, cell body size, and presence of centrally located nuclei.
Snap freezing and cryosectioning of muscle can be preferable to paraffin fixation because it eliminates an antigen retrieval step during immunohistochemistry (IHC), and because freezing preserves both lipids and protein/enzyme activity. Frozen sections can be stained with Oil Red O or other lipophilic stain to reveal extent of myosteatosis (Fig. 3A) and fatty infiltrates, both of which are frequent markers and potential mediators of muscle atrophy (52–54). Enzymatic assays including succinate dehydrogenase (SDH) and nicotinamide adenine dinucleotide (NADH) reactions can help localize mitochondria and identify muscle fiber oxidative capacity (Fig. 3B), key mediators of muscle mass, function, and metabolism (55, 56). Snap freezing muscle is not trivial and requires some practice to produce useable specimens. A video demonstrating snap freezing and sectioning is available online (50). When snap freezing the muscle, the fascicles must be perpendicular to the disc to obtain true cross-sectional slices or parallel to the disc to obtain longitudinal sections. For measurements of muscle fiber size, muscles must be cryosectioned to the mid-belly, the region of greatest cross-sectional area. Frozen sections can be stored in slide boxes wrapped in plastic at −80°C until use. Alternatively, for visualizing transgenic GFP or other highly soluble proteins, frozen sections can be stored overnight in a box containing formaldehyde-soaked filter paper for vapor phase fixation (57), storing serial sections without fixation to preserve enzyme activity. Properly stored frozen sections can also be scraped from slides and used for preparation of DNA, RNA, or protein.
Fig. 3.
Histological assessment of murine tibialis anterior muscle sections.
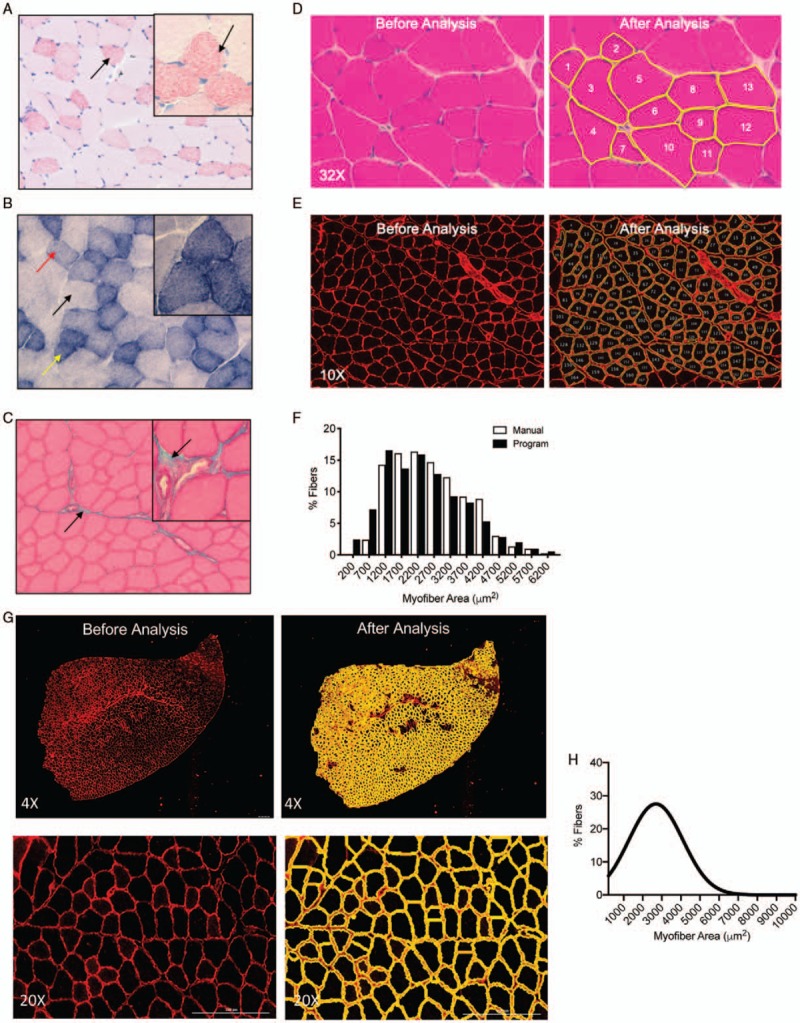
A, Oil Red O staining of frozen tibialis anterior reveals increased lipid accumulation in small oxidative fibers versus larger glycolytic fibers. B, Enzymatic staining for succinate dehydrogenase shows light, medium, and strong staining reflective of the increasing oxidative capacity of each fiber. C, Trichrome staining reveals blue-stained collagen outside fibers. D, Manual measurements of individual fibers on H&E stained sections using ImageJ. E, Immunofluorescence using antilaminin antibody results in high contrast images suitable for automated segmentation using an ImageJ macro. F, Results from either manual (white) or automated (black) measurements are highly concordant. G, Immunofluorescence visualized by Lionheart LX microscope of antidystrophin antibody (red) on tibialis anterior sections. Segmentation (yellow) and quantification in a histogram using Gen5 software, H. The mice from which the muscles were taken in (D–F) were much younger than those in (G, H), hence the difference in mean fiber size.
Morphometric analysis of muscle fiber cross-sectional area (CSA)
Increase or decrease in the average CSA of muscle fibers is associated with muscle hypertrophy and atrophy respectively. Analysis of muscle fiber CSA can be performed on either FFPE or frozen sections and generally involves staining the tissue with H&E (Fig. 3D) or performing immunofluorescence using antidystrophin or antilaminin antibodies (Fig. 3E) to outline the myofibers. If H&E is used, the resulting images are low contrast and fiber CSA must be quantified manually using programs such as Image J (Fig. 3D, right). High contrast images produced by immunofluorescence permit automated analysis of fiber CSA using well-described macros and open source software such as ImageJ (58) (Fig. 3E, right) or CellProfiler (59). Cell Profiler has also been used to automatically segment and measure specific fiber types in images obtained by multiplex IHC. Digital scanning microscopes such as the Lionheart LX with embedded analysis programs such as Gen5 (Fig. 3G, H) can also be used to both image and measure myofibers. These automated methods are an order of magnitude faster than manual measurements and allow for a large increase in sample size (both in fibers and sections) while maintaining a high level of accuracy (Fig. 3, F–H).
In vivo genetic manipulation of muscle size
Genetic manipulations to assay effects on muscle size are possible in mice without generation of germline transgenic and knockout mice. Use of in vivo electroporation permits the uptake of plasmids and siRNAs, resulting in long-term expression/repression of the genes of interest (60, 61). To mark transfected fibers, genes of interests are often co-expressed with reporter genes, marked with reporter tags, or are co-electroporated at a 10:1 ratio with reporter plasmids. Common reporters include TdTomato, mCherry, EGFP and YFP. Marked fibers expressing genes of interest are measured against similarly transfected control fibers in the opposite limb. Note that while electroporation produces little overt injury, gene expression analyses of electroporated muscle reveal evidence of cytokine release and local inflammation and thus results should not be overinterpreted as evidence of muscle growth regulation in health.
Adeno-associated virus (AAV) is also widely used to express/repress specific genes in vivo, a technology driven by gene therapy efforts in congenital myopathies. Previous studies have identified specific AAV serotypes with high specificity for skeletal muscle infection (62). Skeletal muscle specificity is enhanced by using synthetic high expressing, muscle-specific promoters. While local infection in adult muscle groups can permit either assessment of effects on infected fibers marked with reporter proteins, or effects on distant muscles in the case of expression of secreted proteins, infection of neonates can result in long-term and wide-spread expression, the effects of which can be assayed in adults. Detailed protocols for such studies are available (63–66).
Germline transgenic and knockout mouse models are available for the study of atrophy and hypertrophy in skeletal muscle. However, the use of conditional knockouts enables attribution of phenotype to muscle-specific mechanisms. Genetic recombination for skeletal muscle-specific transgenics or knockouts typically use the muscle progenitor cell/satellite cell-specific Pax7 promoters or other muscle-specific promoters including myogenic factor 5 (Myf5), myogenic differentiation 1 (Myod1), muscle creatine kinase (MCK), myosin light chain (MLC), and human alpha-skeletal actin (HSA)/actin alpha 1 (ACTA1), among others, that are activated in progressively more committed cells during myogenesis (67–74). To induce mutations or genes in adult myofibers, many investigators choose the HSA-Cre transgene, in which the cre-recombinase is activated by a synthetic estrogen receptor modulator, tamoxifen, given to the mouse either by injection or through food and water.
In vivo effects on skeletal muscle size are assessed using gross muscle weights at euthanasia and at the myofiber level, by measuring the mean fiber CSA and minimum Feret's diameter to eliminate artefact from off-center sections or fibers. Rigor is enhanced in electroporation and AAV experiments with the use of empty vectors or scrambled siRNAs in the contralateral limb. Rigor in all muscle size studies is increased by use of both sexes, by separating results by sex, by use of multiple individual mice per condition/genotype, and by co-housing mice of different genotypes and treatments to account for cage effects. Our experience demonstrates that a sample size of 10 mice at 10 weeks of age is sufficient to reliably detect a difference in muscle mass as small as 10%—the rule of 10 s. Appropriate statistical analysis of myofiber size requires quantifying the average myofiber diameter of each individual mouse and SD of the experimental group and calculating statistics based upon mouse number rather than myofiber number, a common mistake in the literature.
IN SILICO ASSESSMENT OF MUSCLE SIZE REGULATION
Phenotype-genotype analysis
Abundant data relating to muscle growth regulation are available in online repositories, permitting much in silico investigation prior to embarking upon wet lab experiments. Primary phenome data from genetically diverse laboratory mice measured in multiple laboratories using standardized protocols, including body weights, lean body mass, and skeletal muscle morphology traits, are collated in the Mouse Phenome Database, available at https://phenome.jax.org(75, 76). Published instructions and guidance for genetic analysis of specific phenotypes are available for life span and health span, and these can be adapted for other phenotype queries (77). Such data across strains can be the starting point for discovering novel modulators of muscle mass. However, specific genes and loci regulating body size and thus potentially muscle growth/wasting can be discovered by querying the International Mouse Phenotyping Consortium (IMPC) website, available at https://www.mousephenotype.org. At this writing, IMPC had tested 3,126 genes; none thus far demonstrate abnormal skeletal muscle mass, although 484 demonstrate abnormal body size. As well, 19 genetic alterations demonstrate abnormal muscle contractility and 382 abnormal physical strength in mice. For characterized genes, one can access lists of significant phenotypes, primary measurements at single mouse resolution, expression data and images, and more. Mouse Genome Informatics (MGI) provides integrated genetic, genomic, and biological data on known mouse genes, largely culled from the published studies (78). Searching the Mammalian Phenotype Browser tool within MGI for abnormal muscle morphology returns 2,811 genotypes and 5,529 annotations, including 80 genotypes and annotations related to abnormal muscle weight, 10 of which show increased muscle weight (79).
Transcriptomics
Phenotype data can be followed up by probing gene expression. The Gene eXpression Database (GXD) leverages existing datasets and is freely available at www.informatics.jax.org/expression.shtml. Data localizing or quantifying endogenous expression across developmental stages, between and within tissues, and between the sexes can be visualized using GXD. Detailed explanations and instructions are available at the website and are frequently updated (80). Similar queries can be done for single or multiple gene expression, tissue-level gene expression, and histology of human tissues using Genotype-Tissue Expression (GTEx), a resource to study tissue-specific gene expression available at www.gtexportal.org. Samples of 54 non-diseased tissues across nearly 1,000 persons were characterized by whole genome sequencing, whole exome sequencing, and RNAseq. Sex stratification of expression data is included and biospecimens are available for request.
All existing cataloged information for any gene or protein can be readily determined by querying all databases at the National Center for Biotechnology Information portal at www.ncbi.nlm.nih.gov. Datasets exhibiting differential expression of a gene can be identified at the Gene Expression Omnibus (GEO) Profiles portal at www.ncbi.nlm.nih.gov/geoprofiles, narrowing the search by including keywords such as “muscle.” These datasets can be further analyzed using additional GEO tools, including Profile neighbors to find similarly regulated genes within a bioset.
Another powerful tool for learning about a particular gene is Illumina BaseSpace Correlation Engine's (28) QuickView function, which reveals all available information about a gene of interest present in this curated searchable dataset. The most correlated tissues, diseases, pharmacological compounds, gene perturbations, and omics studies are shown and readily accessible, providing abundant unbiased data about a gene of interest. Links to literature and relevant clinical trials are also posted. Such data can immediately associate a gene with particular disease conditions, other genes or compounds, enabling the planning of wet lab and mouse studies.
Narrowing analysis to skeletal muscle specifically, the resource MuscleDB, available at www.muscledb.org, can be used to determine expression of specific genes of interest in a wide array of murine skeletal muscle groups, from tongue and eye to masseter, diaphragm, limb muscles and more, versus cardiac, or smooth muscle (aorta) (81). The murine datasets were generated using male C57BL/6J mice; however, MuscleDB also provides searchable RNAseq data for male and female soleus and extensor digitoris longum muscles from rats, as well as miRNA from mice and rats (in beta testing as of this writing).
Using these resources enables the harvesting of considerable knowledge about a new gene, which could then be functionally tested in C2C12 cultures, by electroporation or gene transfer into mouse models, or through genetically modified mice. Resources for testing a new gene can be found by searching GeneCards, www.genecards.org, which is focused on human genes but contains links for products related to murine orthologs. Availability of ES cells, embryos, mice, or sperm carrying a mutation or transgene for a particular gene of interest can be determined by searching the International Mouse Strain Resource (IMSR) at www.findmice.org(82).
An example of in silico analysis
A search of the IMPC database for genotype-phenotype relationships related to grip strength identified female-specific increased grip strength in 1700007K13Rik (human ortholog C90rf116) homozygous null mice (Fig. 4). MGI identified multiple targeted alleles and phenotypes from two alleles, including a lethal phenotype due to severe laterality defects, suggesting strain effects or effects of specific targeting events. The NCBI all database portal returned no literature focused on 1700007K13Rik, although the gene is mentioned in the full text of 13 PubMed Central articles. One Gene link and two Protein links were provided, along with 1,068 Gene Expression Omnibus (GEO) profiles. Querying GEO Profiles on “1700007K13Rik AND muscle” revealed 61 profiles, with Nebulin deficiency identified as the top profile for subgroup effect in profile GDS5880. Using “Profile neighbors,” the genes Dhrs7, Usp11, Atat1, and others were found to be highly co-expressed. BaseSpace Correlation Engine identified highest expression in expression in skeletal muscle psoas and Fallopian tube. Searching the most correlated studies for keyword “skeletal muscle.” Correlation Engine further revealed that 1700007K13Rik expression increased 4.83-fold in the setting of a titin-related muscular dystrophy model (dataset GSE33157) and in denervated muscle (dataset GSE49826), among other results. In the denervation dataset, mice with SMAD4 deletion showed 20.8-fold increased 1700007K13Rik over innervated, while wild-type mice with denervation showed a 12.5-fold increase. MuscleDB showed highest expression of 1700007K13Rik in tongue, while GTEx demonstrated low expression in human skeletal muscle compared with all other tissues except blood. These results suggest that 1700007K13Rik/C90rf116 could play a role in skeletal muscle mass or function. GeneCards revealed many commercially available products including siRNAs, vectors, and antibodies for future wet lab studies. Moreover, IMSR revealed 15 available strains or lines, with 11 bearing mutations exclusively in 1700007K13Rik, including three lines with available sperm and/or embryos. While the functions of 1700007K13Rik/C90rf116 remain to be determined, this exercise demonstrates the utility of in silico analyses preparatory to other testing.
Fig. 4.
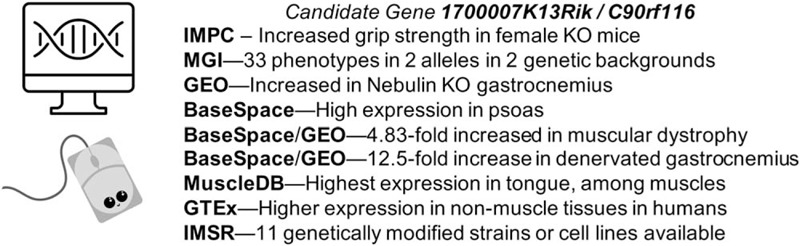
Example of in silico discovery of a novel gene modulating muscle function, 1700007K13Rik.
SUMMARY
Skeletal muscle mass and quality is a powerful modulator of morbidity and mortality in health and disease, including sepsis, shock, burns, and trauma. Robust model systems are available for the interrogation of molecular mechanisms of skeletal muscle growth regulation. These systems span in vitro to in vivo studies that are accessible to most biomedical research laboratories. In silico studies can provide powerful insights to plan and interpret gene level data. Ultimately, such studies will lead to means of promoting muscle growth and muscle health, improving outcomes and survival in patients.
Acknowledgments
Antibodies to myosin heavy chain and dystrophin were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA.
Footnotes
This work was funded in part by grants to TAZ from the National Institutes of Health (Grants R01CA122596 and R01CA194593), the VA (Grant I01BX004177), the IU Simon Cancer Center, and the Lustgarten Foundation. JER was funded in part through an NIH training grant to Hal Broxmeyer (Grant T32 HL007910). DHAJ was funded in part through the Adam Herbert Fellowship and the IU Simon Cancer Center Cancer Biology Training Program.
The authors report no conflicts of interest.
REFERENCES
- 1.Argiles JM, Campos N, Lopez-Pedrosa JM, Rueda R, Rodriguez-Manas L. Skeletal muscle regulates metabolism via interorgan crosstalk: roles in health and disease. J Am Med Dir Assoc 17 (9):789–796, 2016. [DOI] [PubMed] [Google Scholar]
- 2.Ehlen JC, Brager AJ, Baggs J, Pinckney L, Gray CL, DeBruyne JP, Esser KA, Takahashi JS, Paul KN. Bmal1 function in skeletal muscle regulates sleep. eLife 2017; 6: pii: e26557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukund K, Subramaniam S. Skeletal muscle: a review of molecular structure and function, in health and disease. Wiley Interdiscip Rev Syst Biol Med 12:e1462, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das DK, Graham ZA, Cardozo CP. Myokines in skeletal muscle physiology and metabolism: recent advances and future perspectives. Acta Physiol (Oxf) 2019. e13367. [DOI] [PubMed] [Google Scholar]
- 5.Rome S, Forterre A, Mizgier ML, Bouzakri K. Skeletal muscle-released extracellular vesicles: state of the art. Front Physiol 10:929, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneki M. Metabolic inflammatory complex in sepsis: septic cachexia as a novel potential Therapeutic Target. Shock 48 (6):600–609, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao T, Herndon DN, Porter C, Chondronikola M, Chaidemenou A, Abdelrahman DR, Bohanon FJ, Andersen C, Sidossis LS. Skeletal muscle protein breakdown remains elevated in pediatric burn survivors up to one-year post-injury. Shock 44 (5):397–401, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murton A, Bohanon FJ, Ogunbileje JO, Capek KD, Tran EA, Chao T, Sidossis LS, Porter C, Herndon DN. Sepsis increases muscle proteolysis in severely burned adults, but does not impact whole-body lipid or carbohydrate kinetics. Shock 52 (3):353–361, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chargi N, Bril SI, Emmelot-Vonk MH, de Bree R. Sarcopenia is a prognostic factor for overall survival in elderly patients with head-and-neck cancer. Eur Arch Otorhinolaryngol 276 (5):1475–1486, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha YS, Kim SW, Kwon TG, Chung SK, Yoo. ES. Decrease in skeletal muscle index 1 year after radical cystectomy as a prognostic indicator in patients with urothelial bladder cancer. Int Braz J Urol 45 (4):686–694, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroenke CH, Prado CM, Meyerhardt JA, Weltzien EK, Xiao J, Cespedes Feliciano EM, Caan BJ. Muscle radiodensity and mortality in patients with colorectal cancer. Cancer 124 (14):3008–3015, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Xia J, Zhang XI, Gathirua-Mwangi WG, Guo J, Li Y, McKenzie S, Song Y. Associations of muscle mass and strength with all-cause mortality among US older adults. Med Sci Sports Exerc 50 (3):458–467, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sierzega M, Chrzan R, Wiktorowicz M, Kolodziejczyk P, Richter P. Prognostic and predictive implications of sarcopenia in Western patients undergoing gastric resections for carcinoma of the stomach. J Surg Oncol 120 (3):473–482, 2019. [DOI] [PubMed] [Google Scholar]
- 14.Coleman JR, Moore EE, Zilberman-Rudenko J, Samuels JM, Cohen MJ, Silliman CC, Banerjee A, Sauaia A, Griffin JH, Deguchi H. Rapid Communication: Cardiac and Skeletal Muscle Myosin Exert Procoagulant Effects. Shock 52:554–555, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32 (4):1171–1180, 1983. [DOI] [PubMed] [Google Scholar]
- 16.Liang R, Dong W, Shen X, Peng X, Aceves AG, Liu Y. Modeling myotonic dystrophy 1 in C2C12 myoblast cells. J Vis Exp 2016; (113): doi: 10.3791/54078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frost RA, Nystrom GJ, Lang CH. Endotoxin and interferon-gamma inhibit translation in skeletal muscle cells by stimulating nitric oxide synthase activity. Shock 32 (4):416–426, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Au ED, Desai AP, Koniaris LG, Zimmers TA. The MEK-inhibitor selumetinib attenuates tumor growth and reduces IL-6 expression but does not protect against muscle wasting in Lewis lung cancer cachexia. Front Physiol 7:682, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3 (11):1009–1013, 2001. [DOI] [PubMed] [Google Scholar]
- 20.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14 (3):395–403, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Sato AY, Richardson D, Cregor M, Davis HM, Au ED, McAndrews K, Zimmers TA, Organ JM, Peacock M, Plotkin LI, et al. Glucocorticoids Induce bone and muscle atrophy by tissue-specific mechanisms upstream of E3 ubiquitin ligases. Endocrinology 158 (3):664–677, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonetto A, Aydogdu T, Jin X, Zhang Z, Zhan R, Puzis L, Koniaris LG, Zimmers TA. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab 303 (3):E410–E421, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294 (5547):1704–1708, 2001. [DOI] [PubMed] [Google Scholar]
- 24.Milan G, Romanello V, Pescatore F, Armani A, Paik JH, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun 6:6670, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Q, Chirn GW, Szustakowski JD, Bakhtiarova A, Kosinski PA, Kemp D, Nirmala N. Uncovering mechanisms of transcriptional regulations by systematic mining of cis regulatory elements with gene expression profiles. BioData Min 1 (1):4, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18 (4):662–674, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajan S, Chu Pham Dang H, Djambazian H, Zuzan H, Fedyshyn Y, Ketela T, Moffat J, Hudson TJ, Sladek R. Analysis of early C2C12 myogenesis identifies stably and differentially expressed transcriptional regulators whose knock-down inhibits myoblast differentiation. Physiol Genomics 44 (2):183–197, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 2010; 5 (9): e13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41 (Database issue):D991–D995, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini A, Rullman E, Lilja M, Mandic M, Melin M, Olsson K, Gustafsson T. Asymmetric cellular responses in primary human myoblasts using sera of different origin and specification. PLoS One 13 (2):e0192384, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultan KR, Henkel B, Terlou M, Haagsman HP. Quantification of hormone-induced atrophy of large myotubes from C2C12 and L6 cells: atrophy-inducible and atrophy-resistant C2C12 myotubes. Am J Physiol Cell Physiol 290 (2):C650–C659, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Thorley M, Duguez S, Mazza EMC, Valsoni S, Bigot A, Mamchaoui K, Harmon B, Voit T, Mouly V, Duddy W. Skeletal muscle characteristics are preserved in hTERT/cdk4 human myogenic cell lines. Skelet Muscle 6 (1):43, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387 (6628):83–90, 1997. [DOI] [PubMed] [Google Scholar]
- 34.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3 (11):1014–1019, 2001. [DOI] [PubMed] [Google Scholar]
- 35.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol 49 (1):59–68, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashiwagi S, Khan MA, Yasuhara S, Goto T, Kem WR, Tompkins RG, Kaneki M, Martyn JA. Prevention of burn-induced inflammatory responses and muscle wasting by GTS-21, a specific agonist for alpha7 nicotinic acetylcholine receptors. Shock 47 (1):61–69, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov 14 (1):58–74, 2015. [DOI] [PubMed] [Google Scholar]
- 38.Counsell JR, Asgarian Z, Meng J, Ferrer V, Vink CA, Howe SJ, Waddington SN, Thrasher AJ, Muntoni F, Morgan JE, et al. Lentiviral vectors can be used for full-length dystrophin gene therapy. Sci Rep 7:44775, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, Reyes-Ordonez A, Chen J. Lentivirus-mediated RNAi in skeletal myogenesis. Methods Mol Biol 1889:95–110, 2019. [DOI] [PubMed] [Google Scholar]
- 40.Antolik C, De Deyne PG, Bloch RJ. Biolistic transfection of cultured myotubes. Sci STKE 2003 (192):PL11, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Sandri M, Bortoloso E, Nori A, Volpe P. Electrotransfer in differentiated myotubes: a novel, efficient procedure for functional gene transfer. Exp Cell Res 286 (1):87–95, 2003. [DOI] [PubMed] [Google Scholar]
- 42.Naso MF, Tomkowicz B, Perry WL, 3rd, Strohl WR. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31 (4):317–334, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penaud-Budloo M, Le Guiner C, Nowrouzi A, Toromanoff A, Cherel Y, Chenuaud P, Schmidt M, von Kalle C, Rolling F, Moullier P, et al. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol 82 (16):7875–7885, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin Q, Qiao C, Li J, Xiao B, Li J, Xiao X. A GDF11/myostatin inhibitor, GDF11 propeptide-Fc, increases skeletal muscle mass and improves muscle strength in dystrophic mdx mice. Skelet Muscle 9 (1):16, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halldorsdottir S, Carmody J, Boozer CN, Leduc CA, Leibel RL. Reproducibility and accuracy of body composition assessments in mice by dual energy x-ray absorptiometry and time domain nuclear magnetic resonance. Int J Body Compos Res 7 (4):147–154, 2009. [PMC free article] [PubMed] [Google Scholar]
- 46.Badea CT, Hedlund LW, Mackel JF, Mao L, Rockman HA, Johnson GA. Cardiac micro-computed tomography for morphological and functional phenotyping of muscle LIM protein null mice. Mol Imaging 6 (4):261–268, 2007. [PMC free article] [PubMed] [Google Scholar]
- 47.Mathewson MA, Chapman MA, Hentzen ER, Friden J, Lieber RL. Anatomical, architectural, and biochemical diversity of the murine forelimb muscles. J Anat 221 (5):443–451, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charles JP, Cappellari O, Spence AJ, Hutchinson JR, Wells DJ. Musculoskeletal geometry, muscle architecture and functional specialisations of the mouse Hindlimb. PLoS One 11 (4):e0147669, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedrich HJ. The Laboratory Mouse. AP, Elsevier: Amsterdam; 2012. [Google Scholar]
- 50.Bonetto A, Rupert JE, Barreto R, Zimmers TA. The colon-26 carcinoma tumor-bearing mouse as a model for the study of cancer cachexia. J Vis Exp 2016; (117): doi: 10.3791/54893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Joyce NC, Oskarsson B, Jin LW. Muscle biopsy evaluation in neuromuscular disorders. Phys Med Rehabil Clin N Am 23 (3):609–631, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamrick MW, McGee-Lawrence ME, Frechette DM. Fatty infiltration of skeletal muscle: mechanisms and comparisons with bone marrow adiposity. Front Endocrinol (Lausanne) 7:69, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brioche T, Pagano AF, Py G, Chopard A. Muscle wasting and aging: experimental models, fatty infiltrations, and prevention. Mol Aspects Med 50:56–87, 2016. [DOI] [PubMed] [Google Scholar]
- 54.De Carvalho FG, Justice JN, Freitas EC, Kershaw EE, Sparks LM. Adipose tissue quality in aging: how structural and functional aspects of adipose tissue impact skeletal muscle quality. Nutrients 2019; 11 (11): pii: E2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coen PM, Musci RV, Hinkley JM, Miller BF. Mitochondria as a target for mitigating sarcopenia. Front Physiol 9:1883, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rontoyanni VG, Malagaris I, Herndon DN, Rivas E, Capek KD, Delgadillo AD, Bhattarai N, Elizondo A, Voigt CD, Finnerty CC, et al. Skeletal muscle mitochondrial function is determined by burn severity, sex, and sepsis, and is associated with glucose metabolism and functional capacity in burned children. Shock 50 (2):141–148, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jockusch H, Voigt S, Eberhard D. Localization of GFP in frozen sections from unfixed mouse tissues: immobilization of a highly soluble marker protein by formaldehyde vapor. J Histochem Cytochem 51 (3):401–404, 2003. [DOI] [PubMed] [Google Scholar]
- 58.Minamoto VB, Hulst JB, Lim M, Peace WJ, Bremner SN, Ward SR, Lieber RL. Increased efficacy and decreased systemic-effects of botulinum toxin A injection after active or passive muscle manipulation. Dev Med Child Neurol 49 (12):907–914, 2007. [DOI] [PubMed] [Google Scholar]
- 59.Lau YS, Xu L, Gao Y, Han R. Automated muscle histopathology analysis using CellProfiler. Skelet Muscle 8 (1):32, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiFranco M, Quinonez M, Capote J, Vergara J. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J Vis Exp 2009; (32): pii: 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dona M, Sandri M, Rossini K, Dell’Aica I, Podhorska-Okolow M, Carraro U. Functional in vivo gene transfer into the myofibers of adult skeletal muscle. Biochem Biophys Res Commun 312 (4):1132–1138, 2003. [DOI] [PubMed] [Google Scholar]
- 62.Louboutin JP, Wang L, Wilson JM. Gene transfer into skeletal muscle using novel AAV serotypes. J Gene Med 7 (4):442–451, 2005. [DOI] [PubMed] [Google Scholar]
- 63.Kimura T, Ferran B, Tsukahara Y, Shang Q, Desai S, Fedoce A, Pimentel DR, Luptak I, Adachi T, Ido Y, et al. Production of adeno-associated virus vectors for in vitro and in vivo applications. Sci Rep 9 (1):13601, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun 8:14454, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piekarowicz K, Bertrand AT, Azibani F, Beuvin M, Julien L, Machowska M, Bonne G, Rzepecki R. A muscle hybrid promoter as a novel tool for gene therapy. Mol Ther Methods Clin Dev 15:157–169, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pozsgai ER, Griffin DA, Heller KN, Mendell JR, Rodino-Klapac LR. Systemic AAV-mediated beta-sarcoglycan delivery targeting cardiac and skeletal muscle ameliorates histological and functional deficits in LGMD2E mice. Mol Ther 25 (4):855–869, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Batran R, Gopal K, Martin MD, Ho KL, Almutairi M, Aburasayn H, Eaton F, Campbell JE, Ussher JR. Skeletal muscle-specific Cre recombinase expression, controlled by the human alpha-skeletal actin promoter, improves glucose tolerance in mice fed a high-fat diet. Diabetologia 61 (8):1849–1855, 2018. [DOI] [PubMed] [Google Scholar]
- 68.Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev Biol 332 (1):131–141, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JC, Mortimer J, Marley J, Goldhamer DJ. MyoD-cre transgenic mice: a model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis 41 (3):116–121, 2005. [DOI] [PubMed] [Google Scholar]
- 70.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460 (7255):627–631, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138 (2):371–384, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miniou P, Tiziano D, Frugier T, Roblot N, Le Meur M, Melki J. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res 27 (19):e27, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishijo K, Hosoyama T, Bjornson CR, Schaffer BS, Prajapati SI, Bahadur AN, Hansen MS, Blandford MC, McCleish AT, Rubin BP, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J 23 (8):2681–2690, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao P, Monks DA. A tetracycline-inducible and skeletal muscle-specific Cre recombinase transgenic mouse. Dev Neurobiol 69 (6):401–406, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bogue MA, Grubb SC, Walton DO, Philip VM, Kolishovski G, Stearns T, Dunn MH, Skelly DA, Kadakkuzha B, TeHennepe G, et al. Mouse Phenome Database: an integrative database and analysis suite for curated empirical phenotype data from laboratory mice. Nucleic Acids Res 46 (D1):D843–D850, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bogue MA, Churchill GA, Chesler EJ. Collaborative cross and diversity outbred data resources in the mouse phenome database. Mamm Genome 26 (9–10):511–520, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogue MA, Peters LL, Paigen B, Korstanje R, Yuan R, Ackert-Bicknell C, Grubb SC, Churchill GA, Chesler EJ. Accessing data resources in the mouse phenome database for genetic analysis of murine life span and health span. J Gerontol A Biol Sci Med Sci 71 (2):170–177, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE. TMGD Group. Mouse Genome Database (MGD) 2019. Nucleic Acids Res 8 (D1):D801–D806, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mammalian Phenotype Browser. Vol. 2019. [Google Scholar]
- 80.Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, Baldarelli RM, Beal JS, Campbell JC, Corbani LE, et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res 47 (D1):D774–D779, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Terry EE, Zhang X, Hoffmann C, Hughes LD, Lewis SA, Li J, Wallace MJ, Riley LA, Douglas CM, Gutierrez-Monreal MA, et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. eLife 2018; 7: pii: e34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eppig JT, Motenko H, Richardson JE, Richards-Smith B, Smith CL. The International Mouse Strain Resource (IMSR): cataloging worldwide mouse and ES cell line resources. Mamm Genome 26 (9–10):448–455, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]


