Abstract
The infectious bronchitis virus is a causative agent of avian infectious bronchitis (AIB), and is is an important disease that produces severe economic losses to the poultry industry worldwide. Recent AIB outbreaks in India have been associated with poor growth in broilers, drop in egg production, and thin egg shells in layers. The complete spike gene of Indian AIB vaccine strain was amplified and sequenced using a conventional reverse transcription polymerase chain reaction and is submitted to the GenBank (accession no KF188436). Phylogenetic analysis revealed that the vaccine strain currently used belongs to H120 genotype, an attenuated strain of Massachusetts (Mass) serotype. Nucleotide and amino acid sequence comparisons have shown that the reported spike gene from Indian isolates have 71.8%–99% and 71.4%–96.9% genetic similarity with the sequenced H120 strain. The study identifies live attenuated IBV vaccine strain, which is routinely used for vaccination, for the first time. Based on nucleotide and amino acid relatedness studies of the vaccine strain with reported IBV sequences from India, it is shown that the current vaccine strain is efficient in controlling the IBV infection. Continuous monitoring of IBV outbreaks by sequencing for genotyping and in vivo cross protection studies for serotyping is not only important for epidemiological investigation but also for evaluation of efficacy of the current vaccine.
Keywords: infectious bronchitis, spike, H120, phylogeny
Abbreviations
- BLAST
Basic Local Alignment and Search Tool
- cDNA
complementary DNA
- CDS
coding sequence
- IBV
infectious bronchitis virus
- LRR
leucine‐rich repeat region
- ORF
open reading frame
- RT‐PCR
reverse transcriptase PCR
1. Introduction
Avian infectious bronchitis (AIB) is an acute respiratory disease mainly found in young chickens in commercial poultry farms, causing significant economic losses. The prevalence of infectious bronchitis virus (IBV) is pandemic in a number of countries including India and assumes a variety of clinical forms, ranging from a respiratory disease to infection of the oviduct leading to permanent damage to immature birds and in hens, leading to reduced egg production and a nephropathogenic form of IBV causing acute nephritis, urolithiasis, and mortality 1. IBV, a causative agent of AIB disease, belongs to the genus Gammacoronavirus, subfamily Coronavirinae, family Coronaviridae, and order Nidovirales (International Committee on Taxonomy of Viruses (ICTV) 2011). The genome of IBV is a positive‐sense single‐stranded RNA of about 27.6 kb with the following genomic organization: 5′‐Pol‐S‐3a‐3b‐E‐M‐5a‐5b‐N‐untranslated region‐3′ 2.
All coronaviruses maintain a set of essential genes, including those that encode the polymerase (Pol), spike (S), small membrane (E), membrane (M), and nucleocapsid (N) proteins 3. The spike gene is translated as a precursor spike So, which is posttranslationally cleaved by host serine protease into S1 and S2 at cleavage sequence motif 4. The S1 protein is the primary cause of antigenic variations in IBV and contains the serotype‐specific neutralization epitopes, whereas the envelope and membrane proteins are conserved. Antigenic variation among IBV strains is common 1, 5, 6, 7, 8; the major prevalent form of IBV in the Indian subcontinent is respiratory and nephropathogenic forms 9.
The IBV classification of different serotypes is based largely on virus‐neutralization tests, a gold standard test for serodiagnosis of infectious bronchitis in infected birds. Serotype determination is of trivial importance as different serotypes do not always cross protect. Genotyping of IBV strains can also be done by genetic characterization of the spike glycoprotein gene by reverse transcription polymerase chain reaction (RT‐PCR), restriction fragment length polymorphism, and nucleotide sequencing, which for the most part correlates with the viral serotype 10, 11. Nucleotide sequencing of the S1 gene is the foremost technique used for differentiation of IBV strains into various genotypes. The emergence of antigenic variants is mostly attributed to variation in the spike gene by recombination 12, 13 and is investigated by nucleotide sequencing of the S1 portion of the S gene coding for the S1 subunit 14, 15, where most of the epitopes to which neutralizing antibodies bind are found 16. The impact of regular emergence of antigenic variants of the disease and subsequent vaccines used to control varies depending on different geographical locations. Genetic assessment of field isolates of viruses from outbreaks is essential for evaluation of vaccine efficiency on antigenic variants that arise. A critical advantage of sequencing S1 is for comparison and analysis of sequences of unknown field isolates and variants with reference vaccine strains for establishing potential relatedness. IBV vaccines currently used are either live attenuated or killed showing varying degrees of efficacy. At present, many countries only permit live vaccines of the Massachusetts type, such as the H120 strain. Some countries have also licensed other live strain vaccine such as Connecticut, Arkansas, or Delaware 072 in the United States or the 4/91 strain in the United Kingdom. In India, Verma, 17 for the first time reported the prevalence of IBV infections in chickens. The emergence of a nephropathogenic IBV with a novel genotype in India has also been reported 9. Recently, an outbreak of nephropathogenic AIB in broiler flocks was reported in the Chhattisgarh region of India 18.
In this study, we amplified and sequenced the spike gene of the live infectious bronchitis attenuated vaccine virus, which is routinely used in India. Bioinformatics was applied to evaluation of the spike gene for characterization and spike gene's relatedness with previously reported Indian IBV isolates.
2. Materials and Methods
2.1. Virus
Viral RNA was isolated from the AIB vaccine, living bp vet (Mass type strain; Ventri Biologicals, Pune, India). A virus sample was obtained in a freeze‐dried form that was dissolved in 1 mL normal saline; 0.1 mL of the inoculum was inoculated in five 10‐day‐old specific pathogen free chicken eggs (Venky's Hatcheries, Pune, India), and the eggs were candled daily for 96 H. The allantoic fluid was collected and centrifuged at 1700 g for 10 Min at room temperature, and a supernatant was stored at –20°C until use.
2.2. RNA isolation and RT‐PCR
Total RNA from the infected allantoic fluid was extracted by a Trizol reagent (Sigma, St. Louis, MO, USA) as per the manufacturer's protocol. The extracted RNA was first reverse‐transcribed with gene‐specific primers and a thermoscript RT kit (Invitrogen, Carlsbad, CA, USA) to synthesize the first‐strand cDNA. Synthesized cDNA was screened for the presence of IBV genome by a polymerase chain reaction (PCR) with a set of primers specific to the spike gene of IBV. Primers were used to amplify the complete coding sequence (CDS) of the S gene including forward 5′‐CCCGAATTCATGTTGGTAACACCTCTTTTACTAG‐3 (EcoRI) (nucleotide position 20374‐20398) and reverse primer 5′‐GCGGAGCTCTTAAACAGACTTTTTAGGTC‐3′ (SacI) (nucleotide position 20843–23862), designed from Massachusetts M41 serotype (FJ904723). PCR amplification was carried out using Pfu DNA polymerase (Thermo Scientific, Waltham, MA, USA) with the following cycling conditions: 94°C for 5 Min followed by 30 cycles of 94°C for 1 Min, 60°C for 1 Min, 72°C for 1.5 Min, with a final extension of 72°C for 10 Min. The PCR‐amplified gene fragment was cloned with a cloning vector CloneJETTM (Thermo Scientific). The size of amplicon was 3489 bp. The PCR product was visualized by agar gel electrophoresis and confirmed by restriction enzyme digestion and sequencing.
2.3. Sequence and phylogenetic analysis
Sequences were analyzed using Chromas Lite 2.1.1. The nucleotide sequence of the S gene of Indian IBV vaccine strain was assembled and aligned by clustalW 19 after Basic Local Alignment and Search Tool (BLAST) search with published IBV sequences deposited to the GenBank database. Sequence identities by BLAST analysis were included in the alignment and phylogenetic tree construction. The multiple sequence alignments and phylogenetic tree construction with the neighbor joining method were performed using MEGA version 5 20. The bootstrap values were determined from 1,000 replicates of the original data. Phylogenetic analysis of nucleic acid and deduced amino acid sequences was carried out with the neighbor joining method using the Jukes–Cantor model and pairwise deletion. The S gene sequences from the GenBank database, which were used for comparison or phylogenetic analysis in this study, are enlisted in Tables 1 and 2.
Table 1.
Selected IBV strains used in this study
| Strain | Year | Country | NCBI accession number |
|---|---|---|---|
| H120 | 1989 | The Netherlands | M21970 |
| H120 | 1960 | The Netherlands | GU393335 |
| H120 | 2009 | Taiwan | EU822341 |
| H120 | 1960 | The Netherlands | FJ88835 |
| M41 | 2009 | India | GQ219712 |
| M41 | 2006 | United States | FJ904713 |
| M41 | 1972 | United States | FJ904721 |
| M41 | 2006 | United States | DQ834384 |
| M41 | 1965 | United States | FJ904720 |
| Beaudette | 2005 | Singapore | DQ001334 |
| Beaudette | 2005 | Singapore | DQ001336 |
| Beaudette | 2005 | Singapore | DQ001340 |
| Conn46 | 1991 | United States | FJ904719 |
| Conn46 | 1983 | United States | FJ904718 |
| Conn46 | 1972 | United States | FJ904717 |
| Ark‐dpi | 2009 | United States | EU418976 |
| JMK | 1964 | United States | GU393338 |
| Holte | 1954 | United States | GU393336 |
| 3071/03 | 2004 | Taiwan | AY606319 |
| 6/82 | 1993 | United Kingdom | X04723 |
| CK/CH/LSD/05I | 2008 | China | EU637854 |
| CK/CH/LDL971/97 | 2004 | China | DQ068701 |
| CK/CH/LGD/120724 | 2012 | China | KC119407 |
| CK/CH/LZJ/111113 | 2011 | China | JX195176 |
| 4/91 | 1998 | United Kingdom | AF093794 |
| 4/91 | 2011 | United Kingdom | JN192154 |
| Egypt/F/03 | 2003 | Egypt | DQ487085 |
| Nephropathogenic HBC | 2006 | China | DQ973112 |
| Nephropathogenic | 2006 | China | DQ973114 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 2.
Selected Indian IBV isolates used in this study
| Isolate | Year | Region/strain | NCBI accession number |
|---|---|---|---|
| IBS1_UP_09_15 | 2009 | Uttar Pradesh, India | GU967401 |
| IBS1_AP_09_15 | 2008 | Andhra Pradesh, India | GU967405 |
| IBS1_UKND_08_14 | 2008 | Massachusetts | GU967392 |
| India‐744‐AD‐04 | 2004 | Massachusetts | HQ291840 |
| India‐764‐AD‐04 | 2004 | Massachusetts | HQ291841 |
| India‐627‐AD‐02 | 2002 | Massachusetts | HQ291842 |
| India‐16‐V‐AD‐07 | 2007 | Massachusetts | HM179146 |
| India/LKW/IVRI/56/08 | 2008 | Massachusetts | HM163471 |
| PDRC/PUNE/India/9/99 | 2004 | Nephropathogenic | AY091551 |
| India/NMK/72/IVRI/10 | 2010 | Namkkal | HM748585 |
| India M41 | 2009 | Massachusetts | GQ219712 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2.4. Bioinformatics analysis
N‐Glycosylation sites were predicted by using the facility available at http://www.cbs.dtu.dk/services/NetNGlyc. Leucine‐rich repeat regions (LRR) were determined by LRRfinder, which is available at http://www.lrrfinder.com/result.php.
3. Results
3.1. PCR amplification and sequencing of the spike gene
The spike gene was amplified using a cDNA template prepared from isolated RNA of commercial IBV vaccine available in India. On agarose gel electrophoresis, a positive amplification of 3.4‐kb band was seen (Fig. 1), which was gel eluted and cloned. The resulting full‐length CDS of spike gene was submitted to the GenBank (accession number KF188436).
Figure 1.
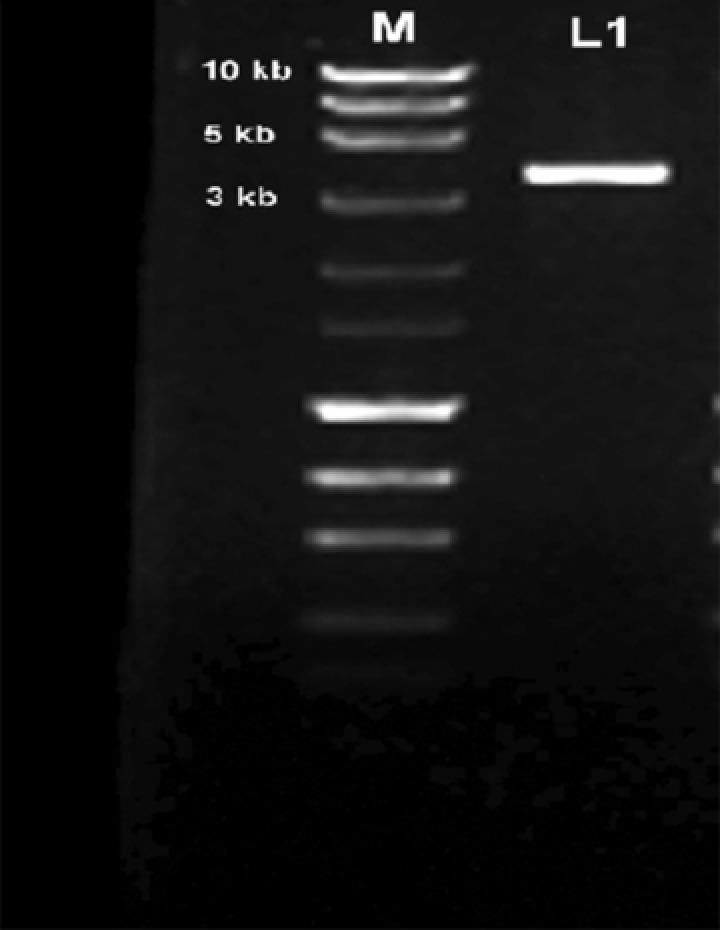
PCR amplification product of the spike gene in 1.2% agarose.
3.2. Phylogenetic analysis and percent identity of spike gene sequences
The partial CDS of the S gene was compared with relevant available IBV sequences in the GenBank (Tables 1 and 2). Phylogenetic analyses of the Indian vaccine strain of IBV were carried out based on the partial CDS of the spike gene. Published IBV sequences of Massachusetts type, M41, H120, Baudette, 4/91, Conn46, Ark‐dpi, JMK, Holte, 3071/03, 6/82, CK/CH, 4/91, Egypt/F/03, and nephropathogenic and Indian IBV isolates available in the GenBank were used to determine phylogenetic characteristics and molecular epidemiology of the Indian vaccine strain. No complete spike gene sequences of the Indian IBV isolates genotype were available in the GenBank; therefore partial CDS of KF188436 and other previously reported Indian isolates were analyzed. The phylogenetic analysis based on the partial S1 gene sequence of the Indian vaccine strain showed that it belongs to the H120 strain of IBV, which is a type of Massachusetts strain routinely used as a vaccine strain worldwide (Fig. 2). Some Indian isolates were clustered together to form a distinct cluster, whereas AY091551 and HM748585 were clustered with the Chinese nephropathological strain (DQ973112 and DQ973114) and 6/41 of UK (AF093794 and JN192154), respectively. Interestingly, one Indian isolate HM163471 was clustered with the Egyptian isolate DQ487085, which is a nephropathogenic strain of AIB virus found in Egypt.
Figure 2.
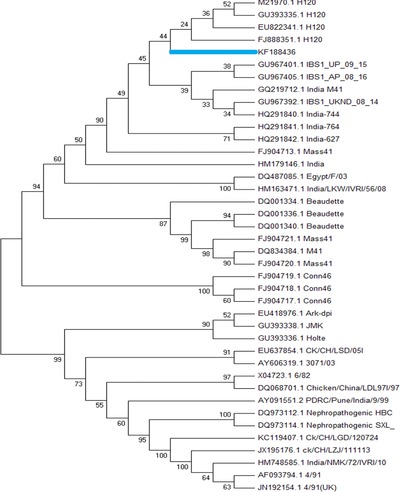
Phylogenetic analysis of the partial CDS of the S gene of IBV obtained from the Indian vaccine strain at the nucleotide level.
To determine the relatedness among Indian isolates, a comparative analysis of the nucleotide sequences of the spike gene (KF188436.1) with previously reported sequences of the S gene from Indian isolates was carried out. A higher nucleotide sequence homology was shown by Indian isolates GU967405, GQ219712, and GU967392 with KF188436, which is around 98.4%, 99.0%, and 99.0%, respectively. The isolates GU967387, HQ291841, HQ291842, and HM179146 showed around 90.6%–93.7% nucleotide sequence homology to KF188436. The lowest sequence homology to KF188436 was shown by isolates HM748585 and HQ291840, which was about 71.8% and 87.7%, respectively. The nucleotide sequence divergence of reported Indian isolates from KF188438 was 0.1%–3.5% with the exception of HM748585. The Spike gene of infectious bronchitis harbors recombinational hotspots; no crossover was observed between the vaccine strain and field isolates.
The deduced amino acid sequences of the spike glycoprotein from the Indian IBV vaccine strain and previously reported Indian isolates exhibited 71.4%–96.9% homology. Indian isolates GQ219712 (96.9%), GU967392 (96.6%), GU967401 (96%), and GU967405 (94.9%) showed the highest percent homology with KF188436, whereas isolates HQ291841 (93.1%) and HQ291842 (93.9%) showed intermediate homology. The lowest percent homology to the vaccine strain was shown by Indian isolates HM163471 (89.7%), HM179146 (89.7%), HM748585 (71.4%), and HQ291840 (71.4%).
3.3. Analysis of the spike protein cleavage site
A deduced amino acid sequence of Indian isolates has shown a cleavage sequence motif Arg–Arg–Phe–Arg–Arg in a sequenced KF188436 H120 vaccine strain and other reported Indian isolates at the amino acid position 533 21 (Table 3).
Table 3.
Cleavage site motif in Indian isolates of IBV
| Indian isolate | Type | Cleavage site sequence motif |
|---|---|---|
| KF188436 | H120 | 533 Arg–Arg–Phe–Arg–Arg 537 |
| GQ219712 | Massachusetts 41 | 533 Arg–Arg–Phe–Arg–Arg 537 |
| GU967392 | Massachusetts 41 | 533 Arg–Arg–Phe–Arg–Arg 537 |
| GU967401 | Massachusetts 41 | 533 Arg–Arg–Phe–Arg–Arg 537 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.4. Prediction of potential N‐glycosylation sites
N‐Glycosylation sites were predicted based on the presence of conserved motif Asn–Xaa–Ser/Thr in a sequence where an Asn residue is N‐glycosylated. Twenty‐six N‐glycosylation sites were found in the KF188436 strain. In other Indian isolates, most of the sites were found to be conserved with some exception (Table 4)
Table 4.
Distribution of N‐glycosylation in the S1 sequence of the spike gene (up to cleavage motif)
| Position of N‐glycosylation sites (amino acid position) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indian isolate | 51 | 77 | 103 | 144 | 163 | 178 | 212 | 237 | 247 | 264 | 271 | 276 | 283 | 306 | 425 | 447 | 530 |
| KF188436 | ++ | + | ++ | +++ | ++ | + | ++ | + | + | + | + | + | + | + | + | + | ++ |
| GU967392 | × | ||||||||||||||||
| HM163471 | × | ||||||||||||||||
| HM179146 | × | ||||||||||||||||
| HQ291841 | × | × | × | ||||||||||||||
| HQ291842 | × | ||||||||||||||||
Cleavage site at the amino acid position = 53. + indicates the presence of N‐glycosylation.
× indicates the absence N‐glycosylation.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.5. Determination of leucine‐rich repeat regions and signal peptide
The LRR region of 84 amino acids in length was found in the infectious bronchitis vaccine strain KF188436 starting at position 1079 (LEKLSILKTYI). A signal peptide MLVTPLLLVTLLCALCSA was found in the vaccine strain H120 with a signal peptide probability of 0.729.
4. Discussion
In this study, we carried out propagation, amplification, sequencing, and bioinformatic analysis of the complete spike gene from the Indian vaccine strain, routinely used for vaccination of poultry birds. Sequencing result showed that the open reading frame (ORF) of the Indian vaccine strain was 3,489 bp with a cleavage site for spike located at 1,596 bp, which is equivalent to 532 amino acids. This is the first report on a complete CDS spike gene of an Indian IBV vaccine strain. Universal primers are available for cloning and sequencing of the spike gene; self‐designed primers were used in this study. In phylogenetic analysis, the spike sequence was clustered together with the H120 vaccine strain of IBV. The H120 strain is a live attenuated vaccine strain of Massachusetts (Mass) serotype, originally prepared after 120th serial passage of strain H, isolated from embryonated chicken eggs in the year 1956 in the Netherlands 22. The H120 strain is considered to be one of the safest vaccine strains and is used worldwide as a primary vaccine in broilers, breeders, and future layers.
It has been reported that the differences in S1 probably contribute to failure of cross protection. However, common epitopes exist between isolates, which are of importance in cross‐immunity; some of these are likely to be on proteins other than S1. The degree of cross protection induced by UK/6/82 against homologous and heterologous IBV isolates diminished as the similarity of the S1 proteins diminished 12. In a study involving vaccination of birds with single vaccination with 6/82 failed to protect all chicks against isolates with extremely similar (>98% identity) S1 sequences, indicating that a slight change in the S1 sequence may have led to the failure of vaccine 12. The Indian IBV vaccine strain H120 showed 71.4%–96.9% homology at an amino acid level and 71.8–99% homology at a nucleotide level. So the degree of protection may be higher with homologous genotype compared to heterologous ones. It is presumed that the degree of cross protection between different serotypes is less, but some strains of the IBV viruses do induce cross protection against other serotypes and are known to as protectotypes. The H120 strain of IBV is a well‐known protectotype and has been extensively used for vaccination. To overcome the problem associated with different serotypes and genotypes, it has been reported that vaccinating with two different types of IBV vaccine can provide broad protection against different IBV types 5. A different combination of protectotypes can be tested to achieve a high level of cross protection and to overcome a problem of vaccination failure. The study involving the Indian vaccine strain Ma41 with 94.8%–98.8% homology to field isolates showed good cross protection 23. Other than spike, enhancement of humoral immunity against IBV by a bicistronic DNA vaccine plasmid encoding nucleocapsid protein and interleukin‐2 has shown up to 80% of protection 24.
Twenty‐six N‐glycosylation sites were found in the vaccine strain, out of which 17 sites are present in the S1 region, which is an ectodomain of the spike gene. The reported Indian IBV isolates have lost some of the N‐glycosylation sites (Table 4). As N‐glycosylation is a regular event in posttranslational modification of the protein, changes in a glycosylation pattern can modulate antigenic properties of the protein as antibodies raised against glycoprotein may be specific for a carbohydrate moiety of the glycoprotein 25. Diversification of the glycosylation pattern may influence a survival pattern and a transmission property of virus, in lactate dehydrogenase elevating virus, a member of the Coronaviridae family; determinants of changes in virulence and cellular tropism of virus are loss or acquisition of N‐glycosylation sites of the protein 26.
The precursor spike glycoprotein (S) of IBV is posttranslationally cleaved by a host serine protease at the cleavage site motif into two subunits (S1 and S2). The cleavage recognition site consists of five basic amino acid residues, and variation in a sequence and composition does not correlate with serotype or pathogenicity of IBV as different serotypes as well as attenuated and pathogenic isolates contain the same cleavage recognition site that correlate with viruses of different geographic regions 21. The Indian vaccine strain KF188436, which belongs to Massachusetts (Mass) serotype, is shown to have an Arg–Arg–Phe–Arg–Arg cleavage recognition site, which correlates with isolates THA241251, THA280252, THA290252, and THA320352 of Thailand 21; FJ888351, GQ154655, and FJ888351 of China; and GU393335 of the United States. It shows that the H120 strain mostly has a RRFRR cleavage recognition site. LRR is a structural motif of protein like alpha/beta horseshoe fold and is responsible for protein==protein interactions 27, 28. In KF188436, LRR is present at position 1079 that is a part of the S2 protein and may be responsible for an interaction with other proteins of IBV.
In conclusion, the results of the current study show that the current vaccine strain, which is routinely used for field vaccination of poultry birds, belongs to the H120 strain. The percent identity of the spike gene sequence of the H120 strain with the Indian isolates shows varying degrees of similarity. The vaccine strain used is sufficiently protective based on a similarity study but further in vivo cross protection studies should be carried out to check the cross protection potential of the current vaccine against field isolates.
5. Acknowledgments
The authors are thankful to the CSIR‐UGC, New Delhi, India, and the Director, Indian Veterinary Research Institute, Bareilly, India, for providing necessary grants and facilities to carry out this research work. The authors hereby declare that there is no conflict of interest.
6. References
- 1. Cavanagh D., and Gelb J., Jr. (Eds.). (2008) Diseases of Poultry Location of publisher: Ames, Iowa, USA, pp. 117–135, Wiley‐Blackwell. [Google Scholar]
- 2. Cook, J. K. A. (1984) Avian Pathol. 13, 733–741. [DOI] [PubMed] [Google Scholar]
- 3. Dawson, P. S. , and Gough, R. E. (1971). Arch. Gesamte Virusforsch 34, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hofstad, M. S. (1958) Am. J. Vet. Res. 19, 740–743. [PubMed] [Google Scholar]
- 5. Ignjatovic, J. , and Sapats, S. (2000) Rev. Sci. Tech. Off. Int. Epiz. 19, 493–508. [DOI] [PubMed] [Google Scholar]
- 6. Bayry, J. , Goudar, M. S. , Nighot, P. K. , Kshirsagar, S. G. , Ladman, B. S. , Gelb, J., Jr. , Ghalsasi, G. R. , and Kolte, G. N. (2005) J. Clin. Microbiol. 43, 916–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kulkarni, A. B. , and Resurreccion, R. S. (2010) Avian Dis. 54, 1144–1151. [DOI] [PubMed] [Google Scholar]
- 8. Sumi, V. , Singh, S. D. , Dhama, K. , Gowthaman, V. , Barathidasan, R. , and Sukumar, K. (2012) Trop. Anim. Health Prod. 44, 1791–1795. [DOI] [PubMed] [Google Scholar]
- 9. Cavanagh, D. , Davis, P. J. , Cook, J. K. A. , Li, D. , Kant, A. , and Koch, G. (1992) Avian Pathol. 21, 33–43. [DOI] [PubMed] [Google Scholar]
- 10. Zwaagstra, K. A. , Van Der Zeijst, B. A. M. , and Kusters, J. G. (1992) J. Clin. Microbiol. 30, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavanagh, D. (1991) Rauischhol. Germ 34, 147. [Google Scholar]
- 12. Kusters, J. G. , Niesters, H. G. M. , Lenstra, J. A. , Horzinek, M. C. , and Van Der Zeijst, B. A. M. (1989) Virology 169, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koch, G. , Kant, A. , Cook, J. K. A. , and Cavanagh, D. (1992) J. Gen. Virol. 73, 591–596. [DOI] [PubMed] [Google Scholar]
- 14. Fateh, S. , Sanjay, S. , and Nidhi, R. (2009) Indian J. Poult. Sci. 44, 379–338. [Google Scholar]
- 15. Verma (1964). MVSc thesis, India.
- 16. Chenna, R. , Sugawara, H. , Koike, T. , Lopez, R. , Gibson, T. J. , Higgins, D. G. , and Thompson, J. D. (2003) Nucleic Acid Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. , and Kumar, S. (2011) Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pohuang, T. , Chansiripornchai, N. , Tawatsin, A. , and Sasipreeyajan, J. (2011) Virol. Gen. 43, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bijlenga, G. , Cook, J. K. A. , Gelb, J. , and de Wit, J. J. (2004) Avian Pathol. 33, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar, K. , Dinkar Raj, G. Raja, A. , and Ramdass, P. (2006) Indian J. Biotechol. 6, 41–44. [Google Scholar]
- 21. Tang, M. , Wang, H. , Zhou, S. , and Tian, G. (2008) J. Virol. Methods 149, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lisowska, E. (2002) Cell. Mol. Life Sci. 59, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li, K. , Schuler, T. , Chen, Z. , Glass, G. E. , Childs, J. E. , and Plagemann, P. G. (2000) Virol. Res. 67, 153–162. [DOI] [PubMed] [Google Scholar]
- 24. Jackwood, M. W. , Hilt, D. A. , Callison, S. A. , Lee, C.‐W. , Plaza, H. , and Wade, E. D. (2001) Avian Dis. 45, 366–372. [PubMed] [Google Scholar]
- 25. Gay, N. J. , Packman, L. C. , Weldon, M. A. , and Barna, J. C. (1991) FEBS Lett. 291, 87–91. [DOI] [PubMed] [Google Scholar]
- 26. Kobe, B. , and Kajava, A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- 27. Mardani, K. , Noormohammadi, A. H. , Hooper, P. , Ignjatovic, J. , and Browning, G. F. (2008) J. Virol. 82, 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai, M. M. , and Cavanagh, D. (1997) Adv. Virol. Res 48, 1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]


