Abstract
The article summarises the results of more than 30 years of research on palmitoylation (S‐acylation) of viral proteins, the post‐translational attachment of fatty acids to cysteine residues of integral and peripheral membrane proteins. Analysing viral proteins is not only important to characterise the cellular pathogens but also instrumental to decipher the palmitoylation machinery of cells. This comprehensive review describes methods to identify S‐acylated proteins and covers the fundamental biochemistry of palmitoylation: the location of palmitoylation sites in viral proteins, the fatty acid species found in S‐acylated proteins, the intracellular site of palmitoylation and the enzymology of the reaction. Finally, the functional consequences of palmitoylation are discussed regarding binding of proteins to membranes or membrane rafts, entry of enveloped viruses into target cells by spike‐mediated membrane fusion as well as assembly and release of virus particles from infected cells. The topics are described mainly for palmitoylated proteins of influenza virus, but proteins of other important pathogens, such as the causative agents of AIDS and severe acute respiratory syndrome, and of model viruses are discussed.
Keywords: Endoplasmic reticulum/Golgi, Membrane protein, Post‐translational modification, Protein sorting/trafficking/targeting, Virology/viruses
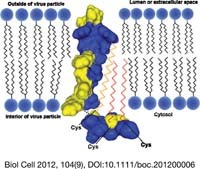
The article covers the biochemistry, cell biology and functional role of palmitoylation of viral proteins, especially of spike proteins. The transmembrane and cytoplasmic domains of influenza virus hemagglutinin are shown, which contains three fatty acids covalently linked to conserved cysteines. The cysteine at the beginning of the transmembrane region is acylated with stearic acid (yellow zigzag line), whereas the two cytoplasmic cysteines contain palmitic acid (red zigzag line). The model was created by Oliver Ernst (Biochemiezentrum, Heidelberg) using BallView 1.3.
Abbreviations used:
APT, acyl‐protein thioesterase; CoV, coronavirus; CRAC, cholesterol recognition/interaction amino acid consensus; DRMs, detergent‐resistant membranes; EM, electron microscopy; ER, endoplasmic reticulum; ESCRT, endosomal sorting complex required for transport; FAST, fusion‐associated small transmembrane; FRET, fluorescence resonance energy transfer; GFP, green fluorescent protein; HA, influenza A virus hemagglutinin; HCV, hepatitis C virus; HEF, hemagglutinin esterase fusion; MHV, mouse hepatitis virus; MS, mass spectrometry; TGN, trans‐Golgi network; SARS, sudden acute respiratory syndrome; SFV, Semliki Forest virus; SIV, simian immunodeficiency virus; TF, trans‐frame; TMR, transmembrane region; VLPs, virus‐like particles; VSV, vesicular stomatitis virus
Introduction
Palmitoylation or S‐acylation is the post‐translational attachment of fatty acids (usually palmitic acid) to cysteine residues by thioester linkage. It is a common modification of integral and peripheral membrane proteins. Protein palmitoylation was originally discovered with viruses (Schmidt and Schlesinger, 1979), but palmitoylated proteins have subsequently been identified in every eukaryotic cell type examined (Bijlmakers and Marsh, 2003; Fukata and Fukata, 2010; Linder and Deschenes, 2007). Bacteria, however, lack the machinery for protein palmitoylation and therefore proteins are devoid of hydrophobic modifications when expressed in Escherichia coli. The only known exceptions are the eukaryotic protein Bet3 and a bacterial virulence factor, which possess an unusual self‐palmitoylating activity (Kummel et al., 2006; Quevillon‐Cheruel et al., 2009).
On the basis of their membrane topology and their function, palmitoylated proteins of viruses can be classified into three groups, two types of transmembrane proteins having different membrane topology and function and peripheral membrane proteins (see Figure 1). The best characterised group are viral spike glycoproteins, most of them with a type I membrane topology. They are composed of an N‐terminal cleavable signal peptide, a long lumenal (extraviral) ectodomain, (at least) one membrane‐spanning region and a cytoplasmic (intraviral) tail, which is typically short. Prominent examples present in human pathogens are the hemagglutinin (HA) of influenza virus, the fusion (F) protein of measles virus, the glycoproteins of filoviruses and retroviruses (including HIV) and the S‐protein of sudden acute respiratory syndrome (SARS)‐coronavirus (CoV) and of other CoVs. In addition, spike proteins of model viruses, such as E1 and E2 of togaviruses (Semliki Forest and Sindbis virus) as well as G of vesicular stomatitis virus (VSV), are palmitoylated. The palmitoylated spike proteins are the major or the only transmembrane protein component inserted into the viral envelope. They are involved in virus entry by catalysing receptor binding and/or membrane fusion. See the glossary for a short description of viruses and their palmitoylated proteins.
Figure 1.
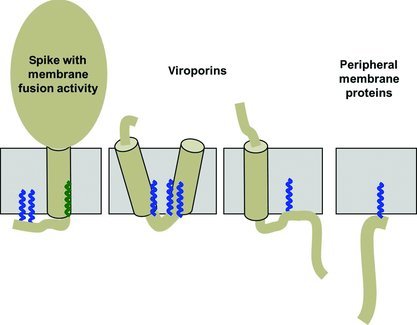
Classification of S‐acylated proteins from viruses
The figure shows the membrane topology of S‐acylated viral proteins. The (presumably) α‐helical transmembrane region is depicted as cylinder embedded within a membrane (grey). Fatty acids linked to cysteine residues are shown as zigzag line. Acylation sites located at the end of the cytoplasmic part of the transmembrane region of viral spike proteins often contain stearic acid (green), acylation sites within the cytoplasmic tail contain palmitic acid (blue).
The second group of palmitoylated viral proteins are viroporins. They are abundantly expressed in infected cells, but only a few copies are integrated into virus particles. They either possess a type III membrane topology (a small lumenal portion, one membrane‐spanning region that also functions as an internal signal peptide and a longer cytoplasmic tail) or a hairpin topology (two membrane‐spanning regions connected by a very short loop). Prominent examples are M2 of influenza A virus and E of CoV; the only example of the second subcategory is the 6K protein of togaviruses. These proteins serve as hydrophilic pores in membranes and are hence termed ‘viroporins’. In addition, they also play a prominent role during assembly and budding of virus particles (Gonzalez and Carrasco, 2003; Wang et al., 2011).
The third group of palmitoylated proteins from viruses are peripheral membrane proteins. In contrast to spike glycoproteins and viroporins, they lack a hydrophobic amino acid sequence for membrane anchoring. Instead, fatty acids anchor the modified protein to membranes or target the protein to subdomains of membranes. These proteins are functionally rather diverse. Notable examples are the non‐structural protein nsP1 of togavirus, UL11 and UL51 of herpesvirus and the core protein of hepatitis C virus (HCV).
Methods to identify S‐acylated proteins
Most palmitoylated proteins were identified by metabolic labelling of virus‐infected cells with [3H]‐palmitic acid, followed by analysis of virus particles or immunoprecipitated viral proteins for incorporation of tritiated fatty acids. However, since the vast majority (>99%) of [3H]‐palmitate is incorporated into lipids, only a tiny amount remains for the labelling of acylated proteins. As a consequence, after SDS‐PAGE of samples, the resulting fluorograms usually have to be exposed for weeks to detect a signal, especially if the acylated protein is not abundantly expressed in the cell under study (Veit et al., 2008).
Recently, a method called acyl–biotin exchange has been developed that allows identifying palmitoylated proteins within a few days of experimental work. The method is based on the exchange of the cysteine‐bound fatty acid with a derivative of biotin that specifically reacts with –SH groups. In order to label only acylation sites, disulphide bonds must first be reduced and free –SH groups need to be blocked chemically. Protein‐bound fatty acids are then removed with hydroxylamine, an agent that selectively cleaves thioester‐linked compounds (thus, fatty acids from S‐acylated proteins). The newly exposed free –SH groups can then react with the biotin derivative, which can then be used to precipitate the acylated proteins using streptavidin beads. Samples not treated with hydroxylamine serve as negative control for the specificity of the reaction. Identification of precipitated proteins may be performed by high‐throughput tandem mass spectrometry (MS)‐based proteomics (Wan et al., 2007). The biotin–streptavidin pull down can be replaced with the use of direct conjugation to thiol‐reactive sepharose resins (Forrester et al., 2011).
Mass spectrometry can be used to quantitatively analyse the fatty acid species linked to individual acylation sites. The increase in the mass of an acylated protein compared with the unmodified protein is rather small (238 Da for palmitate and 266 Da for stearate); therefore, only short proteins can be accurately measured. In the case of viral spike proteins, only the transmembrane and cytoplasmic domains are subjected to the analysis. Purified virus particles are digested with specific proteases, which remove the ectodomain from the spike proteins, and the hydrophobic membrane‐anchoring fragments are then extracted from ‘shaved’ virus particles with chloroform/methanol and directly analysed by MS. Tandem MS sequencing of peptides can be used to identify the acylated cysteine residues (Kordyukova et al., 2004, 2008; Serebryakova et al., 2006).
Location of palmitoylation sites in viral proteins
In viral transmembrane proteins, the acylated cysteine(s) are mostly located in the border region between the transmembrane region (TMR) and the cytoplasmic tail, usually within 20 residues from the TMR boundary (see Table 1). Cysteine residues with a similar intramolecular location have also been determined as palmitoylation sites for numerous cellular transmembrane proteins, mostly receptors and downstream effector proteins involved in important signal transduction pathways at the cell surface (Charollais and Van Der Goot, 2009; Smotrys and Linder, 2004). If a cysteine is too deeply embedded in the membrane bilayer, such as a transmembrane cysteine present in H7‐subype HA, it is not acylated (Veit et al., 1991), probably because it cannot be reached by a putative acyltransferase. An exception might be fusion of measles virus, where a cysteine present in the middle of the transmembrane segment is apparently more strongly acylated than cytoplasmic cysteines (Caballero et al., 1998). Likewise, attachment of palmitate is abolished if a cysteine is moved too far away from the TMR into the cytoplasmic tail (Yang and Compans, 1996, Yik and Weigel, 2002). However, such unusual palmitoylation sites are present in Env of HIV and simian immunodeficiency virus (SIV) (but not in the spike glycoproteins of other retroviruses). In Env, the two acylated cysteines are located more than 50 and 130 amino acids downstream from the TMR. Since the acylation sites are in the vicinity of amphipathic regions, it was proposed that this part of the cytoplasmic tail folds back to the membrane and is anchored by insertion of the fatty acid into the membrane bilayer (Yang et al., 1995).
Table 1.
Acylation sites identified in viral proteins

The (proposed) transmembrane regions are underlined. See Figure 1 for the membrane topology of the respective category of proteins. Identified acylation sites are highlighted in red. Asterisks following a letter indicate that the fatty acid was identified as (mainly) stearic acid (C 18:0), whereas bold C's indicate the presence of predominantly palmitic acid (C 16:0) at the respective cysteine residue. Three dots (…) denote the presence of additional amino acids. For Env glycoproteins of HIV and SIV, the number of amino acids (aa) between the transmembrane region and acylation sites is specified. For abbreviation of viruses and proteins, see the glossary. Sequences were taken from viral zone (http://viralzone.expasy.org/).
The number of cysteines in the palmitoylation region of viral acylproteins varies from one to nine (in the case of CoV S‐protein). However, only a few studies have shown (by MS) that each potentially palmitoylated cysteine residue is indeed stoichiometrically acylated (Kordyukova et al., 2008, 2010). However, for most proteins where palmitoylation was assessed by metabolic labelling, all (or most) cysteines had to be deleted to obtain a completely non‐acylated molecule (Liao et al., 2006; McBride and Machamer, 2010; Steinhauer et al., 1991; Veit et al., 1991). This indicates that all (or most) potential sites are used. Assuming that all nine cysteines of the S‐protein are acylated, the trimeric spike of CoVs contains 27 fatty acid molecules in its cytoplasmic tail. This substantial amount of hydrophobic moieties attached to a small part of the molecule is very likely to profoundly alter the protein's conformation and biophysical properties, and hence its ability to interact with membranes.
Inspection of the amino acids in the vicinity of the acylated cysteine residues of viral (and also cellular) transmembrane proteins reveals no obvious consensus signal for S‐acylation. This is in contrast to other hydrophobic modifications, such as N‐terminal myristoylation and isoprenylation (Resh, 2004a). In the neighbourhood of acylated cysteines, there are often hydrophobic and basic amino acids, but such residues are typical components of this region of membrane proteins. It was initially hypothesized that a transmembrane protein is acylated whenever a cysteine is present in the inner leaflet of the TMR or in the cytoplasmic tail (Veit et al., 1991). However, introducing a cysteine at various positions into the non‐acylated fusion protein of Sendai virus does not automatically lead to palmitoylation, indicating that other constraints exist, such as masking of potential acylation sites within the three‐dimensional protein structure (Ponimaskin and Schmidt, 1998). Nevertheless, a viral transmembrane protein with one or several cysteine residues located between the TMR and the cytoplasmic tail is a likely substrate for palmitoylation.
In peripheral membrane proteins of viruses, acylation sites are located in the middle or (more often) at either the N‐ or C‐terminus of the protein. The latter regions are likely to be flexible and/or exposed at the surface of the molecule such that cysteine residues are accessible for fatty acid transfer. In cellular proteins, the N‐ and C‐terminus of palmitoylated proteins often bear other hydrophobic modifications, for example myristate at an N‐terminal glycine residue or isoprenoids at a C‐terminal CAAX‐box. Surprisingly, only one viral protein, UL 11, of herpes virus contains another hydrophobic moiety (myristate) in addition to palmitate, but a search for viral sequences with a cysteine residue in the vicinity of either an N‐terminal myristoylation signal (MGXXXS/T) or a C‐terminal CAAX‐box might reveal further peripheral membrane proteins likely to be palmitoylated.
Intracellular site for acylation of virus proteins
At what stage during biosynthesis and intracellular transport of a protein is the fatty acid attached? Such questions have been studied with influenza virus HA, which is considered as a model system for glycoproteins in general. The polypeptide chain of the transmembrane protein HA is synthesised on membrane‐bound ribosomes and translocated into the lumen of the endoplasmic reticulum (ER), where signal peptide cleavage, N‐linked glycosylation, folding and disulphide‐bond formation occur. Post‐translationally but still in the ER, three HA molecules assemble to a trimer, which is the rate‐limiting step for transport out of the ER. Trimming of HA‐linked carbohydrates occurs in medial Golgi cisternae, and terminal glycosylation in the trans‐Golgi network (TGN). Also in the TGN, HA proteins with polybasic cleavage sites are split into the membrane‐spanning HA2 and the large N‐terminal HA1 subunits by the cellular endoprotease furin. HA is then transported to the plasma membrane and incorporated into virus particles during the budding process (Whittaker, 2001). Most other viral spike proteins discussed in this review follow the same pathway of processing and transport to the cell surface.
We have analysed timing of palmitoylation of HA relative to its other modifications by pulse–chase experiments (Veit and Schmidt, 1993). Freshly acylated HA (i.e. labelled with a short pulse of [3H]‐palmitic acid), is completely trimerised but has not undergone proteolytical cleavage or trimming of its N‐linked carbohydrates, suggesting that palmitoylation of HA localises to the ‘late’ ER, to the ER–Golgi intermediate compartment or to the cis‐Golgi cisternae.
Likewise, the S‐protein of CoV, E1/E2 of togaviruses and G of VSV are also acylated prior to the acquisition of Endo‐H‐resistant carbohydrates (Bos et al., 1995; McBride and Machamer, 2010; Berger and Schmidt, 1985; Bonatti et al., 1989; Schmidt and Schlesinger, 1980). For other viral glycoproteins, which are cleaved at a polybasic site by enzymes in the TGN, it has been described that it is the uncleaved precursor molecule that is the substrate for palmitoylation (Yang et al., 1995). For the core protein of HCV, an early compartment of the exocytic pathway can be inferred as acylation site from the fact that the protein is retained at specific locations of the ER connected to lipid droplets, the budding sites of these viruses (Majeau et al., 2009). The virus particle travels through the exocytic pathway in order to be secreted by the cell, and the glycoproteins are subject to post‐translational modification of their ectodomains, but palmitoylation of the endodomain is unlikely to occur at this stage, since it is shielded inside the virus particle. In summary, acylation of viral transmembrane proteins typically occurs along the secretory pathway at late ER and/or early Golgi.
The site for palmitoylation of peripheral membrane proteins from viruses has not been analysed. Palmitoylation is often considered as a signal that targets intrinsically hydrophilic proteins to membranes. However, since palmitoylation is a post‐translational event that occurs on membranes of the vesicular pathway, initial targeting of a protein destined to be palmitoylated has to be achieved by other means. It is known from the study of various cellular proteins involved in signal transduction that targeting can occur by (i) co‐translational attachment of myristate to an N‐terminal glycine residues, (ii) attachment of isoprenoids to a C‐terminal CAAX‐box or (iii) by stretch of basic amino acids located in the vicinity of the acylation site. These signals provide initial, but weak membrane affinity and palmitoylation is then utilised to permanently anchor the protein to the membrane (Resh, 2004a).
Fatty acid species bound to S‐acylated proteins
It has been shown early that [3H]‐palmitic acid, which is used to label acylated proteins, can be converted into other fatty acid species of different chain length and saturation before it is attached to the acylprotein. Thus, if radioactivity derived from labelling with [3H]‐palmitic acid is eventually detected in a given protein, this does not necessarily mean that the protein‐bound acyl chain species is exclusively palmitic acid (C16:0). Indeed, varying amounts of other fatty acid species including myristic acid (C14:0), stearic acid (C18:0) and traces of oleic acid (C18:1) are detected (Schmidt, 1984). The hemagglutinin esterase fusion (HEF) glycoprotein of influenza C virus and the E1 protein of Semliki Forest virus (SFV) are unique in this respect because they contain mainly stearic acid as protein‐bound fatty acid (Veit et al., 1990, 1996).
This analysis relied on chromatographic determination of the protein‐bound, [3H]‐labelled fatty acids. This allows for a rough estimation of a protein's fatty acid pattern. Furthermore, such bulk analysis cannot tell how the different fatty acid species are distributed within an acylprotein with multiple acylation sites.
Advancements in MS have recently allowed to precisely quantify the amount and the type of a fatty acid linked to an acylprotein or even to a single acylation site. These studies revealed that influenza B virus HA possessing two cytoplasmic cysteines contains palmitate, whereas HEF of influenza C virus having one transmembrane cysteine is stearoylated. HAs of influenza A virus having one transmembrane and two cytoplasmic cysteines contain both palmitate and stearate, but the latter is exclusively attached to the cysteine positioned in the TMR of HA (Kordyukova et al., 2008). We also report that G of VSV contains palmitate at a cytoplasmic cysteine, whereas fusion of Newcastle Disease virus and E1 of SFV are acylated with stearate at a transmembrane cysteine (Kordyukova et al., 2010). Thus, site‐specific attachment of palmitate or stearate is a common feature of viral spike proteins and the location of the acylated cysteine relative to the transmembrane span might be the main molecular signal controlling attachment of specific carbon chains. However, individual amino acids in the vicinity of the transmembrane cysteine or other structural factors might affect fatty acid selection, since different influenza virus HA subtypes containing acylated cysteines at the same position differ moderately in the amount of attached stearate (12–35%) (Kordyukova et al., 2008, 2011).
Palmitoylation is not limited to mammalian cells. Insect cells are also able to acylate viral proteins, for example the baculovirus spike protein Gp64 (Zhang et al., 2003) as well as proteins expressed with the baculovirus expression system, for example HA of influenza virus and Gp of filoviruses (Funke et al., 1995; Kuroda et al., 1991). Interestingly, transfer of stearic acid to HA was not observed (Reverey et al., 1996), indicating that differential acylation of proteins is apparently an achievement of higher cells. This might be important to consider if recombinant proteins purified from insect cells are intended for use with therapeutic purpose.
The biological function of heterogenous protein acylation of viral spikes is unclear. However, fatty acids which differ by only two methylene groups, such as myristate (C14) and palmitate (C16), show a significant difference in their hydrophobicity, which has a profound effect on the affinity of the acylated peptide for lipids. Although myristate barely contributes enough hydrophobicity for membrane partitioning of a protein, a single palmitoyl chain promotes efficient membrane binding (Peitzsch and McLaughlin, 1993; Shahinian and Silvius, 1995). In addition, the length of a covalently attached acyl chain might also influence the strength of protein–protein interactions. The α‐subunit of the photoreceptor G‐protein transducin is heterogenously acylated at its N‐terminus with laurate (C12) or myristate (C14), and the α‐subunits containing the latter interact more strongly with the β/γ subunit (Kokame et al., 1992). However, it is difficult to imagine how stearate attached right to the beginning of the TMR might affect interactions of the cytoplasmic tail with either lipids or other proteins, for example internal components of virus particles. Figure 2 and the final paragraph describes a speculation how stearate might associate with amino acids in the TMR of HA, an interaction that might be important for the raft localisation of HA.
Figure 2.
The transmembrane region of influenza virus HA contains a groove that might accommodate a fatty acid
Hydrophobic/hydrophilic properties (left panel) and landscape (right panel) of the solvent‐accessible surface of the transmembrane region helix of influenza A virus hemagglutinin (HA). Surfaces are projected onto a cylinder and shown as 2D maps (angle of rotation along the helical axis and rise along it). Hydrophobic/hydrophilic properties are mapped on the surface according to molecular hydrophobicity potential (MHP) approach and expressed in water–octanol logP units. Maps are coloured according to the scales given below. The hydrophilic groove composed of Ala 20, Gly 16 and Ser 9 could accommodate the acyl chain of a fatty acid attached to Cys 26. The peptide NH2‐ILAIYSTVSSSLVLVGLIIAVGLWMC‐COOH, corresponding to the TMR of H6‐subtype HA, was used for the analysis, which was performed by Anton Polyansky (Shemyakin‐Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow).
The publisher did not receive permission from the copyright owner to include this object in this version of this product. Please refer either to the publisher's own online version of this product or the printed product where one exists. Wiley Periodicals, Inc.
Enzymology of acylation and de‐acylation of proteins
As described in more detail below, palmitoylation is essential for the function of several viral proteins and thus for the replicative cycle of the respective virus. Thus, it will be helpful to identify palmitoylating enzymes and to elucidate their mechanism of action and their three‐dimensional structure in order to develop specific inhibitors of protein palmitoylation as promising candidates for anti‐viral drugs. Unfortunately, enzymes catalysing acylation of viral proteins have not been identified so far, and even the reaction mechanism (enzymatic versus non‐enzymatic) is controversial, as described in the following paragraph.
Palmitoyl acyltransferases
It was demonstrated early that cellular membranes contain an acylating activity of proteinaceous nature operating on viral glycoproteins (Berger and Schmidt, 1984). However, attempts of many laboratories have failed to purify a palmitoyl acyltransferase (PAT) with biochemical methods. Moreover, some viral and cellular proteins can, in the test tube, even be palmitoylated at authentic sites in the absence of any enzyme source with palmitoyl‐coenzyme A (CoA) as lipid donor (Berger and Schmidt, 1984; Duncan and Gilman, 1996; Kummel et al., 2006; Veit, 2000). Therefore, it was questioned whether an enzyme is necessary for protein palmitoylation.
Initially, using a genetic approach with yeast cells, a member of the DHHC protein family was identified as PAT. DHHC proteins are polytopic membrane proteins containing an Asp–His–His–Cys (DHHC) motif as the catalytic centre in one of their cytoplasmic domains (Lobo et al., 2002; Roth et al., 2002). The alteration in the palmitoylation level of putative substrate proteins upon co‐over‐expression with DHHC proteins or upon small interfering RNA‐mediated inhibition of DHHC expression could be linked to DHHC proteins and established their role in protein palmitoylation in both yeast and mammalian cells. Using such assays, it was shown that most substrate proteins can be palmitoylated by several, but not every of the various DHHC proteins, indicating that the seven yeast and 23 mammalian enzymes show distinct, but overlapping substrate specificities (Fukata and Fukata, 2010; Hou et al., 2009; Mitchell et al., 2006; Roth et al., 2006).
The enzymatic nature of protein palmitoylation was recently challenged again by a chemical–biological approach to directly monitor dynamic palmitoylation in single cells. Microinjected fluorescent peptides representing the palmitoylation site of various peripheral membrane proteins accumulate rapidly (T 1/2 < 1 min) at the Golgi, presumably due to membrane anchoring by palmitoylation. Importantly, peptides composed of either β‐amino acids (that contain an additional carbon atom between the amino and carboxy groups) or d‐amino acids (stereoisomeric counterparts of natural amino acids) accumulate at the Golgi with the same kinetics as natural peptides. Apparently, the sequence context flanking the target cysteine does not contribute to substrate recognition. Thus, protein palmitoylation does not have the typical features of classical enzyme–substrate recognition and substrate specificity. Rather, any cysteine exposed at a protein's surface with transient access to the cytosolic face of Golgi membranes seems to become palmitoylated (Rocks et al., 2010).
How do the published data on palmitoylation of viral proteins fit to these different concepts for protein palmitoylation? On the one hand, the apparent lack of a consensus sequence for palmitoylation in viral transmembrane proteins implies a non‐enzymatic (or unusual) mode of acylation. Furthermore, the few studies addressing recognition of cellular substrates by DHHC proteins indicate that regions far distant from the palmitoylation site are required for efficient acylation (Nadolski and Linder, 2009; Roth et al., 2011). However, viral spike proteins contain only one TMR and a (usually very short) cytoplasmic tail and hence do not expose any region outside the acylation site to a putative DHHC protein.
On the other hand, the site‐specific attachment of stearate versus palmitate observed for viral spike proteins (Kordyukova et al., 2008) argues against a purely non‐enzymatic reaction mechanism. Acylation without an enzyme would not show any preference for a particular fatty acid, but should reflect the concentration of individual acyl‐CoAs present in the membrane where acylation occurs. It is thus more likely that individual enzymes exist that differ in their acyl‐CoA specificities, as recently demonstrated for DHHC 2 and 3 (Jennings and Linder, 2012). Note that most DHHC proteins are located at the ER and the Golgi apparatus (Ohno et al., 2006), the intracellular site of viral spike protein acylation. One can imagine that the active site of a DHHC protein with a preference for stearoyl‐CoA might penetrate deeper into the membrane to attach stearate to a transmembrane cysteine compared with an enzyme with specificity for palmitoyl‐CoA. Such an enzyme would not recognise amino acids in the vicinity of the acylation site, but rather its position in the interior of a lipid bilayer. However, this notion has not been confirmed experimentally for viral glycoproteins. Hence, much work remains to be done to fully delineate the mechanism of palmitoylation of viral proteins.
Acyl‐protein thioesterase
In contrast to other hydrophobic modifications, palmitoylation of several cellular acyl‐proteins is reversible with cycles of acylation and de‐acylation. Global profiling of protein palmitoylation revealed that palmitoylation is a stable event for the majority of cellular proteins and that only a subset of proteins is dynamically palmitoylated, especially those that are involved in cell growth, migration and cancer (Martin et al., 2012). Depalmitoylation, which has a kinetic range from seconds to several minutes, occurs on all membranes of the vesicular pathway (Rocks et al., 2010) and is mediated by a cytosolic enzyme, acyl‐protein thioesterase (APT) (Duncan and Gilman, 1998). Acylation cycles may mediate shuttling of peripheral membrane proteins between compartments. The activity of APT releases these proteins from membranes (Dekker et al., 2010). Depalmitoylated proteins then rapidly diffuse to the Golgi where they are again palmitoylated and distributed by the vesicular pathway to their site of action (Rocks et al., 2010). Furthermore, the activity of some transmembrane receptors has been shown to be regulated by the acylation status (Linder and Deschenes, 2007). In contrast to the diversity of DHHC proteins, there are only two APT isoforms (Tomatis et al., 2010), which are expressed in most mammalian cell type examined. Substrate recognition features of APT and its regulation have not been established and hence the likelihood of depalmitoylation of a protein cannot be predicted.
Surprisingly, very few experiments have been performed to analyse whether palmitoylation of viral proteins is reversible. We did not observe cleavage of [3H]‐palmitic acid from HA of influenza virus during its transport to the plasma membrane (Veit and Schmidt, 1993). In accordance, no proteolytic fragments from virus particles representing non‐acylated or under‐acylated peptides were detected by MS, neither for influenza virus HA nor for G of VSV, E1/E2 of SFV and fusion of NDV (Kordyukova et al., 2008, 2010). Thus, during their transit along intracellular membranes, these viral spike proteins are stoichiometrically acylated and the fatty acids remain stably attached during virus budding. Likewise, palmitoylation of the S‐protein of SARS‐CoV is a stable modification (McBride and Machamer, 2010).
Nevertheless, purified APT, when incubated with detergent‐extracted virus particles, rapidly and almost completely cleaves fatty acids from HA proteins of influenza A and C virus, the G‐protein of VSV and E2 of SFV. In contrast, E1 of SFV is largely resistant to APT activity (Veit and Schmidt, 2001). Thus, APT1 can in principle recognise viral spike proteins; hence, their acylation sites in the cytoplasmic tails must be somehow shielded from the enzyme inside cells to maintain spikes completely acylated.
Function of palmitoylation of virus proteins
Attachment of peripheral proteins to membranes and membrane domains
The proteins UL11 and UL51 of herpes simplex virus are palmitoylated at one (UL51) or several (UL11) cysteine residues in the N‐terminal part of the respective molecule. UL11 is additionally modified with myristate at the N‐terminal glycine. The hydrophobic modifications are required to anchor the intrinsically hydrophilic proteins to the cytoplasmic site of the Golgi (Baird et al., 2010; Nozawa et al., 2003). These proteins are assumed to play a role in the secondary envelopment of the viral nucleocapsid at the TGN.
The non‐structural protein nsP1, which is part of the replication complex of togaviruses, is palmitoylated at one (Sindbis virus) or three consecutive (SFV) cysteine residues located in the C‐terminal half of the protein (Ahola et al., 2000). Acylation strengthens the association of the protein with the cytoplasmic surface of the plasma membrane without being absolutely essential. However, only palmitoylated nsP1 associates with filopodia, which are induced during SFV replication. This suggests that palmitoylation localises the protein to specialised membranes (Laakkonen et al., 1996). Mutations preventing nsP1 palmitoylation caused reduced synthesis of virus‐encoded RNA and attenuated virus replication (Kiiver et al., 2008) or led to the emergence of second‐site compensatory mutations that enhance viral infection (Karo‐Astover et al., 2010; Zusinaite et al., 2007).
The hydrophilic core protein of HCV, which forms the capsid of the virus, contains palmitate at a C‐terminal cysteine residue (Majeau et al., 2009). The strategy of HCV to synthesise his structural proteins as a polyprotein precursor ensures that the core protein is delivered to the ER. After synthesis of the core on cytosolic ribosomes, the following signal peptide of the glycoprotein E1 protein targets the nascent polyprotein to the ER. During translocation of E1 into the lumen of the ER, the membrane‐integrated signal peptide is cleaved off from both E1 and from the core protein. Palmitate is then transferred to the core protein to stabilise its interaction with the membrane (Moradpour et al., 2007). Palmitoylation is not essential for membrane binding per se but controls association of the core protein with lipid droplets and associated parts of the ER, the assembly site of HCV particles and this association is essential for virion production (Majeau et al., 2009).
Palmitoylation as raft‐targeting signal in viral proteins
Membrane rafts are heterogenous lipid assemblies enriched in cholesterol and sphingolipids, which are highly dynamic and short lived, but can be fused to larger, more stable platforms (Simons and Gerl, 2010). The property of rafts to exclude most proteins and selectively incorporate others might be utilised by certain enveloped viruses to concentrate their components prior to budding (Simons and Toomre, 2000; Suomalainen, 2002). Lipid analysis of virus particles indicated that budding of influenza virus and HIV occurs primarily through rafts (Brugger et al., 2006; Gerl et al., 2012), whereas SFV and VSV bud through the bulk phase of the membrane (Kalvodova et al., 2009).
Dual acylation and in particular palmitoylation is one of the best characterised signals for association of proteins with rafts (Levental et al., 2010a, 2010b). It is thought that the straight chain of a saturated fatty acid associates with tightly packed inner leaflet rafts, which presumably contain cholesterol and phospholipids with saturated acyl chains. Indeed, modification with unsaturated (‘kinked’) fatty acids and with isoprenoids (branched, unsaturated hydrocarbon chains) is not compatible with raft association of the proteins (Liang et al., 2001; Melkonian et al., 1999; Zacharias et al., 2002).
Raft association of a protein is often deduced from the partitioning of a protein into detergent‐resistant membranes (DRMs). Using such assays, it has been shown in many studies that deletion of palmitoylation sites from influenza HA of various subtypes abolishes or reduces their partition into DRMs (Chen et al., 2005; Melkonian et al., 1999; Wagner et al., 2005). Likewise, blocking palmitoylation of the S‐protein of mouse hepatitis virus (MHV) and removal of acylation sites from the Env glycoprotein of retroviruses (HIV [Bhattacharya et al., 2004; Rousso et al., 2000] or MuLV [Li et al., 2002]) abolished incorporations of these proteins into DRMs. However, others have reported that Env does not require palmitoylation for association with DRMs (Chan et al., 2005; Vzorov et al., 2007).
Conflicting results are not uncommon if partition into DRMs is used to determine raft association of a protein (Munro, 2003). Despite these reservations, the cold extraction method has been surprisingly successful as starting point to identify raft proteins, but more sophisticated methods should be used to confirm and characterise the raft localisation of a protein.
Förster's (or fluorescence) resonance energy transfer (FRET) is exceptionally well suited to demonstrate very close association between two molecules, for example if they populate the same small raft domain. FRET relies on the transfer of energy from an excited donor fluorophore, such as the cyan fluorescent protein (CFP), to an acceptor fluorophore, for example the yellow fluorescent protein (YFP), if they are in very close vicinity to each other (<10 nm). Influenza virus HA, fused at its cytoplasmic tail to CFP, clusters with an established marker for inner leaflet rafts, double‐acylated YFP (Engel et al., 2010; Zacharias et al., 2002). Furthermore, an artificial HA‐derived FRET probe, consisting of a signal peptide, a fluorescent protein and the transmembrane as well as cytoplasmic domain of HA (Scolari et al., 2009), clusters with a glycolipid‐anchored protein, an established marker for rafts of the outer leaflet. For both HA constructs, clustering was significantly reduced when rafts were disintegrated by cholesterol extraction and when the palmitoylation sites were removed from HA. Removing palmitoylation sites from HA increased its mobility in the plasma membrane (Engel et al., 2010), but only slightly indicating that HA does not stay in slowly diffusing rafts for long time periods. In general, the type of membrane anchorage (TMR versus lipid moieties) and not the association with rafts is the determining factor for mobility of proteins within membranes (Kenworthy et al., 2004).
In the case of the ion channel protein M2 of influenza virus, it was hypothesized that it is localised to the edge of the viral budding site, which is considered a large, coalesced raft phase (see Figure 3). S‐acylation in concert with binding of the raft lipid cholesterol is proposed to target M2 to rafts, but complete immersion of the protein in the more ordered, hence thicker raft domains is thought to be prevented by the relatively short TMR of M2 (Schroeder et al., 2005). Thereby, M2 becomes located in an ideal position to mediate the scission of virus particles, supposedly triggered by an amphiphilic helix in M2's cytoplasmic tail, which is inserted into the membrane in a wedge‐like manner to induce curvature (see below) (Rossman et al., 2010b). Several overlapping cholesterol recognition/interaction amino acid consensus (CRAC) motifs, which mediate the interaction with cholesterol (Rossman et al., 2010a; Schroeder et al., 2005; Thaa et al., 2011), and the single acylation site are both located within this amphiphilic helix.
Figure 3.
Role of palmitoylation of viral membrane proteins during the life cycle of influenza virus
The left part of the figure shows the individual steps of the replicative cycle of the virus: attachment to a receptor, endocytosis of virus particles, acidification of the endosome, fusion of viral and cellular membrane, release of the viral genome, synthesis of genomic and mRNA in the nucleus, protein synthesis on soluble and membrane‐bound ribosomes, vesicular trafficking of membrane proteins, assembly and budding of virus particles at membrane rafts in the plasma membrane. The steps where palmitoylation of HA and/or M2 plays a role are depicted in blue. Details of these events are pictured in the right part of the figure. The red arrows in the lower part show the location of the fatty acids during membrane fusion. Driven by the conformational change of HA, they might perturb the organisation of the membrane lipids to allow for completion of membrane fusion. Fatty acids mediate localisation of HA to membrane rafts and might be required for the interaction of the cytoplasmic tail of HA with M1, an internal component of virus particles. The fatty acid attached to M2 (in concert with a cholesterol‐binding site) is believed to target the protein to the edge of rafts, where M2 causes scission of assembled virus particles. Similar functions are envisaged for acylated membrane proteins from other virus families.
The publisher did not receive permission from the copyright owner to include this object in this version of this product. Please refer either to the publisher's own online version of this product or the printed product where one exists. Wiley Periodicals, Inc.
Mixed results were obtained when raft localisation of M2 was tested experimentally. M2 expressed in transfected cells is not associated with DRMs (Zhang et al., 2000) and does not interact with the double‐acylated marker for inner leaflet rafts in FRET experiments (Thaa et al., 2010). However, in similar FRET experiments, M2 associates with HA (Thaa et al., 2010), which supposedly organises the viral budding site. Likewise, using immuno electron microscopy (EM), M2 has been localised to the base of budding filamentous virus particles (Rossman et al., 2010b). In addition, preparation of giant plasma membrane vesicles from cells expressing M2–GFP (green fluorescent protein) showed that the protein is partly present in the co‐alesced raft phase. Raft targeting of M2 does not require the CRAC motifs, but is dependent on palmitoylation, similar to cellular proteins that were tested with this model system (Levental et al., 2010b; Thaa et al., 2011). Although in none of these experiments localisation of M2 to the edge of a raft could be directly demonstrated, the experiments might be taken as an indication that M2 has features of both a raft‐associated and a non‐raft‐associated protein.
Thus, in many cases where an S‐acylated viral protein is present in rafts, the fatty acids are required for or at least contribute to raft association. Conversely, S‐acylation per se, either with palmitate or with stearate, is not sufficient to cause raft localisation of a viral glycoprotein. Examples for S‐acylated, but not DRM‐associated proteins are the mono‐acylated glycoprotein HEF of influenza C virus and G of VSV. Even the attachment of several fatty acids does not necessarily cause raft localisation of a spike protein. The E1/E2 heterotrimer of SFV contains 15 covalently linked fatty acids, six of them being stearate, but the spike protein is nevertheless completely extractable with cold Triton‐X 100 from the viral membrane. SFV is considered not to bud through raft domains (Kalvodova et al., 2009; Kordyukova et al., 2010).
Since rafts are believed to be minor domains in most membranes (an exception might be the apical membrane in polarised cells [Meder et al., 2006]), association of proteins with rafts is thought to concentrate the viral components (Suomalainen, 2002; Veit and Thaa, 2011). This concept can explain how protein palmitoylation as a raft‐targeting signal might influence assembly and budding of virus particles as well as the membrane fusion activity of a viral spike protein. Enrichment of spikes in rafts is thought to provide a platform for virus assembly, concentrating the viral components to facilitate their interactions. Likewise, the membrane fusion activity of a spike protein critically depends on its surface density (Bentz and Mittal, 2003). Hence, clustering of spikes in rafts might yield a concentration high enough to support fusion.
Involvement of protein palmitoylation in virus assembly and budding
To create a functional virus particle, all its structural proteins and the viral genome must be transported to the assembly site. Assembly is achieved by interactions between the membrane‐embedded spike proteins and internal components of virus particles, such as matrix or (nucleo)capsid proteins, which in turn encase the genome. The subsequent budding process needs at least one protein which physically drives the curvature of membranes to generate a bud and another (or the same) protein that mediates the scission of the enveloped virus particle from the membrane. Membrane bending and scission are often achieved by cellular endosomal sorting complex required for transport (ESCRT) proteins, which are recruited by a viral protein to the budding site. The contribution of ESCRT is however limited to some virus families, most prominently retroviruses (such as HIV). Conversely, influenza virus buds independently of ESCRT proteins, and no involvement of ESCRT proteins has been reported for CoVs and togaviruses, the two other virus families where protein palmitoylation is known to affect virus budding (Chen and Lamb, 2008). In that case, viral proteins might take over the function of the ESCRT proteins, matrix proteins or capsid proteins, the viral spike protein itself and/or small and hydrophobic proteins, often with ion channel activity (viroporins).
Small and hydrophobic proteins with ion channel activity
M2 of influenza A virus is an integral membrane protein that is abundantly expressed at the cell surface, but only poorly incorporated into virus particles. It forms a disulphide‐linked tetramer which conducts protons from the acidic endosome into the interior of virus particles during virus entry. The protons are required for release of the viral genome from M1 and thus mediate disassembly of the particle (Pinto and Lamb, 2006). Additionally, M2 has also been implicated in the ultimate step in virus budding, the scission of the virus particle from the plasma membrane. Since peptides representing the amphiphilic helix of M2 induced the budding of vesicles from giant unilamellar vesicles, it was concluded that this amphiphilic helix, situated in the cytoplasmic tail of M2, inserts into the membrane in a wedge‐like manner to induce curvature (Rossman et al., 2010a, 2010b).
Does palmitoylation of the amphiphilic helix affect the various activities of M2? Loss of the palmitoylation site does not influence the ion channel activity of M2 (Holsinger et al., 1995). More surprisingly, acylation does not affect the production of virus particles either, although it affects targeting of M2 in the absence of other viral proteins, as described above. In cell culture, recombinant viruses where the acylated cysteine in M2 (or in CM2 of influenza C virus) has been replaced grow similarly well as the corresponding wild‐type virus (Castrucci et al., 1997; Muraki et al., 2011), even if acylation is deleted simultaneously with the second raft‐targeting signal, the cholesterol‐binding CRAC motif (Thaa et al., 2012). Moreover, there are natural virus strains that lack the acylation site in M2 (Thaa et al., 2012). Thus, acylation of M2 is neither required nor beneficial for virus replication in cell culture. Intriguingly, however, attenuation of virus infectivity is observed upon infection of mice with virus containing non‐acylated (Grantham et al., 2009) or CRAC‐disrupted (Stewart et al., 2010) M2. This indicates that there is a more complex influence of M2 in the context of the infected host that is not accounted for in cell culture experiments. Moreover, it is likely that budding of virus particles is a redundant process, that is other proteins can take over the function of a partially defective M2 protein. It is still not unambiguously defined which of the influenza virus proteins (if any) is the driving force for budding, that is executes membrane bending and scission (Chen and Lamb, 2008).
The small envelope protein E of CoVs shares several structural and functional features with M2 of influenza virus. It is likely to have a type III membrane topology, might function as a cation‐selective channel and is involved in budding of virus particles (Liu et al., 2007). E from murine hepatitis virus, infectious bronchitis virus and SARS are palmitoylated at three cytoplasmic cysteine residues, two of which are present in all members of the CoV family (Boscarino et al., 2008; Corse and Machamer, 2001; Liao et al., 2006; Lopez et al., 2008). Budding of virus‐like particles (VLPs) and replication of MHV virions is impaired if at least two of the palmitoylation sites in E are deleted. Since virus entry is not affected in those mutants, it was concluded that palmitoylation affects virus budding (Boscarino et al., 2008; Lopez et al., 2008). However, enhanced degradation of non‐palmitoylated E might also contribute to the observed decrease in virus yield (Boscarino et al., 2008). Interestingly, the palmitoylated cysteine residues are embedded in a hydrophobic surrounding that might run perpendicular to the plane of the membrane, similarly to the amphiphilic helix of M2 (Boscarino et al., 2008).
The 6K protein of togaviruses is another small and hydrophobic viroporin, which, in contrast to E and M2, is assumed to have a hairpin topology. 6K is heavily S‐acylated at five cysteine residues, most of which are located in the short cytoplasmic region that connects both membrane‐spanning domains (Gaedigk‐Nitschko and Schlesinger, 1990). Removal of palmitoylation sites decreased release of Sindbis virus particles from infected cells. Electron microscopy of isolated virions showed that the 6K mutations led to large numbers of aberrant particles containing multiple nucleocapsids within one membrane structure. This ‘daisy chain structure’ suggests that mutated virus particles are defective in release from the cellular plasma membrane and that palmitoylation of 6K is therefore involved in scission of virus particles (Gaedigk‐Nitschko et al., 1990).
However, new data on the translation of the togavirus genome might lead to a (partial) re‐evaluation of the mentioned experiments. It was reported recently that ribosomal −1 frameshifting, with an estimated efficiency of ∼10–18%, occurs in the C‐terminal region of the 6K protein. This results in the synthesis of an additional protein, termed TransFrame (TF) protein, in which the C‐terminal amino acids are encoded by the −1 frame (Firth et al., 2008). However, since both 6K and TF contain all the palmitoylation sites, this new observation does not touch the general conclusion that palmitoylation of either 6K or the TF protein affects budding of togaviruses.
Taken together, viroporins are apparently involved in budding of influenza, corona and togaviruses, with a more or less prominent functional contribution of palmitoylation.
Viral spike glycoproteins
In the case of influenza virus, the main spike protein HA seems to be a key player for virus assembly and budding. The release of VLP from transfected cells depends on the expression of HA (Chen and Lamb, 2008). Yet other viral factors also contribute to budding, which is a redundantly organised process for influenza virus.
The cytoplasmic tail of HA plays a critical role for influenza virus budding. Viruses lacking this part of the protein were found to have severe defects in assembly and genome packaging as well as irregular morphology (Jin et al., 1994, 1997). Direct evidence that palmitoylation of HA is involved in virus release was obtained for HA of subtype H3 (Chen et al., 2005). Virus particles containing HA without either one of the two‐palmitoylated cytoplasmic cysteines revealed defects in virus release and incorporated reduced amounts of the internal components NP and M1. Importantly, exchange of the M1 protein by that of a different influenza virus restored assembly of viruses with non‐palmitoylated HA. This observation links palmitoylation of HA to the matrix protein M1, but amino acids in the latter that might be required for binding to the cytoplasmic tail of HA could not be identified. However, similar experiments with H7‐subtype HA did not reveal a defect in virus budding, but in virus entry by membrane fusion (see below) (Wagner et al., 2005). Nevertheless, the cumulative evidence just described indicates that the cytoplasmic tail of HA plays an important role in virus budding, and it is hard to imagine that the fatty acids do not affect the conformation of the very short cytoplasmic tail (11 amino acids) and hence its interaction with internal components of virus particles (see Figure 3).
Similar data were reported for the function of palmitoylation of the S‐protein of CoVs. S of murine hepatitis virus (MHV) and of SARS are heavily acylated at up to nine cysteine residues located in the cytoplasmic tail; at least seven of them are present in the S‐proteins of other CoVs suggesting an important role of acylation for the function of the protein (Bos et al., 1995; Petit et al., 2007). Blocking palmitoylation of S of MHV using a pharmacological approach reduced incorporation of S into virus particles and abolished its interaction with the M protein (Thorp et al., 2006). Interestingly, removal of only some palmitoylated cysteine residues had no effect, suggesting that not every fatty acid binding site is required for association with M (Thorp et al., 2006). In contrast, removal of palmitoylation sites from S of SARS by genetic means does not affect its interaction with the M protein (McBride and Machamer, 2010), but incorporation of S into VLPs (Ujike et al., 2012).
There is strong evidence that togaviruses require palmitoylation of their hetero‐oligomeric E1/E2 spike protein. Viruses with a deletion of the single acylation sites in E1 or with deletion of transmembrane or cytoplasmic acylation sites in E2 grew slower in cell culture, and less infectious virus particles were released from infected cells. In addition, aberrant particles containing multiple nucleocapsids were found, similar to that described for the Sindbis mutants with under‐acylated 6K protein, indicating that the mutations caused a defect in virus scission (Ivanova and Schlesinger, 1993; Ryan et al., 1998).
The published data on the requirements of palmitoylation for incorporation of the Env glycoprotein into budding HIV and SIV particles are not completely consistent. Exchange of one (SIV) or two (HIV) palmitoylation sites present in the long cytoplasmic tail by short amino acids (serine, alanine) reduced viral infectivity and incorporation of Env into virus particles (Bhattacharya et al., 2004; Rousso et al., 2000; Vzorov et al., 2007). Interestingly, exchange of cysteine with bulky and hydrophobic amino acids having a large membrane insertion capacity, especially phenylalanine and tyrosine residues, can partly substitute for an acylated cysteine (Bhattacharya et al., 2004), indicating that palmitoylated cysteines attach the cytoplasmic tail of Env to the membrane. However, others have not seen any effect of palmitoylation on incorporation of Env into virus particles and on virus infectivity (Chan et al., 2005).
Rous sarcoma virus containing a deletion of one of the two cysteines in the TMR of the Env protein is only poorly infectious and contains less Env protein in the envelope. However, it was also observed that Env with a deletion of the second acylation site is rapidly internalised from the cell surface and degraded. Therefore, poor incorporation of Env into virus particles is at least partly due to mistargeting of the protein (Ochsenbauer‐Jambor et al., 2001).
Deletion of both palmitoylation sites adjacent to the transmembrane domain of the glycoprotein N (gN) of human cytomegalovirus does not affect its oligomerisation with gM and transport of the complex to the TGN, but the secondary envelopment of the viral capsid by viral glycoprotein‐containing membranes and hence virus replication is inhibited (Mach et al., 2007).
In summary, the incorporation of a palmitoylated spike protein into virus particles upon assembly and budding often depends on the presence of the fatty acid(s).
Involvement of palmitoylation in spike‐catalysed membrane fusion
In most cases, the main function of the viral spike glycoproteins is to catalyse the fusion of viral and cellular membranes upon virus entry, which allows for uncoating of the virus and release of its genome. The fusion of viral and cellular membranes assumingly proceeds via a ‘hemifusion’ stage, where only the outer leaflets of the two fusing membranes are connected; then, a fusion pore in the membrane opens, flickers and ultimately dilates (Chernomordik and Kozlov, 2005; Nikolaus et al., 2011). The hemifusion stage is characterised by mixing of lipids between the two membranes, solutes of the two compartments are not exchanged before fusion is complete.
Elucidation of the X‐ray structure of the ectodomain of influenza virus HA at neutral and mildly acidic pH (which are equivalent to the HA structure before and after membrane fusion) has led to a model on how conformational changes of HA execute membrane fusion. The hydrophobic fusion peptide at the N‐terminus of HA2, which is buried inside the trimeric structure at neutral pH, becomes exposed on the molecule's surface after acidification and is then able to interact with the cellular membrane. A second conformational change then bends the HA molecule, thereby drawing the fusion peptide towards the TMR. This leads to close apposition of both lipid bilayers. Since many other viral fusion proteins, for example S of CoVs, the fusion protein of paramyxoviruses and Env of HIV, have a similar structure as influenza HA in their post‐fusion conformation, it was suggested that they cause apposition of membranes by a similar mechanism (Earp et al., 2005; Skehel and Wiley, 2000).
Lipid mixing was not impaired in cells expressing non‐acylated HA from all influenza virus strains examined, suggesting that palmitoylation is not required for hemifusion. In accordance with this, it was observed that the HA ectodomain (without transmembrane domain and cytoplasmic tail, but anchored to the outer membrane leaflet by a glycolipid anchor) catalyses hemifusion, but not full fusion. This indicates that the TMR and the cytoplasmic tail are required for this process (Kemble et al., 1994).
Inconsistent data have been published concerning the effect of acylation site removal on the ability of HA to cause full fusion. It was reported that non‐acylated H2 and H3 subtype HA mediate cell–cell fusion or show unperturbed transfer of aqueous dyes into HA‐expressing cells (Chen et al., 2007; Naim et al., 1992; Steinhauer et al., 1991). In contrast, non‐acylated HA mutants from the H1, H7, another HA from H2 subtype and from influenza B virus show impaired fusion pore or syncytium formation (Naeve and Williams, 1990; Sakai et al., 2002; Ujike et al., 2004; Wagner et al., 2005). These data might be seen as an indication that the function of the fatty acids depends on the HA subtype or variant to which they are attached. It might be that different HAs execute the final merger of membranes differentially and that acylation contributes only to one of these fusion mechanisms. Alternatively, the discrepancies might be due to the experimental setup, since the methods used to measure fusion are not ideally suited for this purpose. The fusion activity of HA depends on its density on the cell surface (Bentz and Mittal, 2003). This can hardly be measured accurately and barely controlled in expression systems; it is hence conceivable that a defect in fusion might be masked by over‐expression of HA. An experimental fusion system composed of closely controlled amounts of purified viral fusion protein (with or without fatty acids) reconstituted into lipid vesicles with the authentic composition of the viral envelope and fluorescently labelled liposomes as the target membrane would be helpful to quantitatively analyse the contribution of protein‐linked fatty acids to fusion pore formation and its widening.
HA is an enormously variable molecule with very low amino acid conservation (≈20%) through all subtypes (Nobusawa et al., 1991). However, comparison of all HA sequences from viruses isolated from humans or from various animals present in the NCBI database showed that each HA molecule contains three, some even four cysteine residues located in the cytoplasmic domain and at the cytosol‐facing end of the TMR. The position of the most C‐terminal cysteine is strictly conserved; the positions of the other two or three cysteines are sometimes shifted by one or more amino acids, but are strictly conserved within an HA subtype ([Veit and Schmidt, 2006] and Andrei Alexeevski, Lomonosov Moscow State University, personal communication). Thus, assuming these cysteine residues (and the attached fatty acids) would not play an essential role for the life cycle of influenza viruses, it is hard to conceive why they have not been exchanged by similar amino acids during evolution of influenza viruses. Indeed, it was not possible to generate influenza virus mutants with more than one HA palmitoylation site removed, which implies that this modification is essential for virus growth (Wagner et al., 2005; Zurcher et al., 1994).
Palmitoylation is also required for the S‐protein of CoVs (SARS, MHV) to perform its fusion activity. Exchange of all nine palmitoylated cysteine residues completely abolishes fusion activity of the SARS S‐protein (McBride and Machamer, 2010). The cysteines located at the beginning of the cytoplasmic tail of S from both SARS and MHV have a stronger effect than those located in the distal part of the cytoplasmic tail (Petit et al., 2007; Thorp et al., 2006). A more detailed study on the various steps of membrane fusion was reported for S of MHV. S with deleted acylation sites is especially sensitive to inhibitory peptides that bind only to folding intermediates of the fusion process, implying that refolding of S to the post‐fusion conformation is delayed (Shulla and Gallagher, 2009). This interesting observation suggests that palmitoylation affects the conformational change of S. Hence, functional coupling between the palmitoylated endodomain and the fusion‐mediating ectodomain of S must occur.
However, this conclusion cannot be extended to all palmitoylated viral fusion proteins. In the case of VSV, the New Jersey strain naturally contains a non‐acylated G‐protein (lacking a cysteine in its TMR). This would argue in favour of a non‐essential role of palmitoylation for the function of G. Indeed, no effect on virus replication was found when the palmitoylation site was removed from the G‐protein of the Indiana strain of VSV (Whitt and Rose, 1991). Likewise, no function could be attributed to palmitoylation of gp64 of baculovirus (Zhang et al., 2003) and of the filovirus glycoprotein (Funke et al., 1995; Ito et al., 2001).
The fusion‐protein of many paramyxoviruses is acylated at one or several cysteines located in the inner part of the TMR. An exception is fusion of Sendai virus, a very well characterised viral fusogen, indicating that viral membrane fusion per se does not require protein‐bound fatty acids (Veit et al., 1989). Likewise, in the case of acylated fusion from Newcastle disease and human respiratory syncytial virus, removal of palmitoylation sites does not affect cell–cell fusion (Branigan et al., 2006; Sergel and Morrison, 1995). In contrast, removal of certain (but not each) acylated cysteine residue from the fusion of measles virus reduced its cell‐cell fusion activity (Caballero et al., 1998).
Deletion of the acylation site from Env of the retrovirus MulV revealed a slower kinetics of syncytium formation, but also reduced surface expression (Li et al., 2002).
Noteworthy is also the non‐structural protein p10 from the family of non‐enveloped orthoreoviruses. P10 is an fusion‐associated small transmembrane (FAST) protein that mediates fusion of cells that subsequently lyse, thereby facilitating virus release. FAST proteins are typical type I transmembrane protein, but do not contain an fusion peptide or coiled‐coil domains indicating that they catalyse membrane fusion by a different mechanism compared with viral spike proteins (Boutilier and Duncan, 2011). However, p10 is palmitoylated at two conserved cysteine residues adjacent to the membrane span and the fatty acids are essential for syncytium formation (Shmulevitz et al., 2003).
In a nutshell, many viral spike proteins require palmitoylation for a late step in the fusion process, the opening or widening of the fusion pore. Membrane fusion is believed to proceed via an fusion stalk, where lipids connect the viral envelope with the cellular membrane such that lipid exchange occurs between the outer leaflets of both membranes occurs (Chernomordik and Kozlov, 2005). It is conceivable that protein‐bound fatty acids might perturb the organisation of the membrane lipids at this stage of the fusion process, which would then allow membrane fusion to proceed to completion (see Figure 3). Alternatively, protein‐bound fatty acids might not work directly during fusion, but attract cholesterol (or another lipid) to the viral envelope, which could serve a specific function during membrane fusion (Biswas et al., 2008).
Conclusions and Outlook
Generalised conclusions can hardly be drawn from the multitude of studies described above. Yet, it is evident that palmitoylation of viral proteins often plays similar roles for the function of individual proteins as those usually considered for cellular proteins. The fatty acids are often required for attachment of proteins, especially intrinsically hydrophobic ones, to membranes or for their targeting to membrane subdomains, such as rafts. Many studies on palmitoylated spike proteins from different virus families conclude that the fatty acids play a role during virus entry by membrane fusion and/or during assembly and budding of new virus particles. During membrane fusion, the fatty acids might perturb the organisation of the lipid layers during the late stage of the fusion process, as exemplarily shown for HA of influenza virus in Figure 3. Acylation of the cytoplasmic tail of the spike might be required for interactions with internal components of virus particles and/or the incorporation of the spike into membrane rafts, the budding sites of many enveloped viruses. The fatty acid attached to M2 (in concert with a cholesterol‐binding site) is believed to target the protein to the edge of rafts, where the protein causes scission of newly assembled virus particles. Fatty acids linked to other viroporins also affect the budding process in an unknown manner. Some viral proteins might be acylated simply because they (accidentally) contain a cysteine residue at the right position, without any essential or even beneficial role for the function of the modified protein.
One important feature of protein palmitoylation has hitherto not been observed for viral proteins, namely regulatory cycles of de‐ and re‐acylation (Bijlmakers and Marsh, 2003). Why are many cellular proteins depalmitoylated, but viral spike proteins apparently not? After performing their function, cellular proteins are re‐used several times, and acylation cycles regulate the localisation (and hence the function) of the modified protein. In contrast, the viral spike proteins perform their function during virus assembly and cell entry only once, and thus acylation cycles to ‘reset’ the protein into its original condition are not necessary.
Furthermore, some cellular proteins require palmitoylation for proper transport along the exocytic pathway (Greaves and Chamberlain, 2007); the transport of acylated viral transmembrane proteins, however, has not been described to need the presence of the fatty acid modification. In the case of influenza virus HA, the kinetics of folding, trimerisation and transport through the Golgi are the same in the case of palmitoylated HA and HA with deleted acylation sites (Engel et al., 2012; Naim et al., 1992; Veit et al., 1991). Likewise, many other palmitoylated viral spike proteins do not mis‐localise if they lack the fatty acids (Mach et al., 2007; McBride and Machamer, 2010; Yang and Compans, 1996), but enhanced degradation resulting in decreased surface expression was observed for some proteins (Lopez et al., 2008; Ochsenbauer‐Jambor et al., 2001).
To analyse the role of palmitoylation not only for a single protein, but for complete cycles of virus replication, acylation sites were exchanged in the context of the viral genome. This allowed determining whether (and to what extent) acylation affects the infectivity of the respective virus and whether virus entry or virus budding is disturbed. Most studies concluded that palmitoylation of transmembrane proteins (spike proteins and viroporins) affects budding of virus particles and/or their subsequent cell entry by membrane fusion. Although very useful from a virological point of view, the method of reverse genetics is in principle not suitable to provide a mechanistic understanding of the role of protein palmitoylation on a molecular level.
Such knowledge relies on information on the protein structure in the vicinity of the acylation site. The molecular structure of the transmembrane and cytoplasmic domain of a given protein needs to be resolved, preferentially embedded within a membrane. Recent advancements in cryo‐EM might be suitable for that purpose. Images taken from virus particles allowed to partially visualise one of the fatty acids linked to E2 of Barmah Forest Virus, a togavirus related to SFV (Kostyuchenko et al., 2011). However, for most palmitoylated proteins, similarly detailed information is not available, mainly because the various crystal structures of viral spike proteins were gathered from their ectodomains only. The lack of structural information of the acylation site's vicinity also hinders the identification of signals that might direct S‐acylation in general or the preferential attachment of palmitate versus stearate. If such conformational signals exist, they are not obvious from the primary structure of the protein. For most spike proteins, it is even uncertain at which amino acid exactly a TMR ends and the cytoplasmic domain starts. Hence, it is not known whether a cysteine destined to become acylated is exposed to the hydrophobic lipid environment of the bilayer, the transition region containing the head groups of the lipids or to the hydrophilic milieu of the cytosol. Thus, presently one can only speculate how a fatty acid interacts with its surroundings, lipids or proteins.
Stearic acid attached to a cysteine located right at the border of the TMR might interact with amino acid side chains of the transmembrane domain. A projection of HA's transmembrane α‐helix surface with mapped polar and surface properties onto a cylinder shows that there are small and slightly hydrophilic residues (glycine, alanine and serine) above the acylated cysteine, which are surrounded by large and hydrophobic amino acids. This surface morphology of the TMR forms a groove into which a saturated (therefore flexible) acyl chain might fit into (see Figure 2). The larger acyl chain of stearate would fill the groove more completly compared with the shorter chain of palmitate. The accommodation of the acyl chain into this cavity converts an irregular and rough TMR surface into a smooth one which is more compatible with raft association of HA. Most (non‐acylated) transmembrane proteins are not associated with rafts since their rough TMR surface would disrupt the tight packing of lipids characteristic of raft domains (Kusumi et al., 2011). The accommodation of the acyl chain is also assumed to ‘lubricate’ the TMR of HA for subsequent interaction with the planar sterol ring of cholesterol to allow for raft association. In addition, the fatty acids might attract specific lipids, such as cholesterol. This has been suggested in a recent crystallographic study of the palmitoylated β‐adrenergic receptor, in which cholesterol molecules were resolved in the vicinity of covalently bound fatty acids (Hanson et al., 2008).
Furthermore, it was suggested that a fatty acid might alter the orientation of the transmembrane span. When the TMR is longer than the thickness of the bilayer, palmitoylation might help the helix to adopt a tilted position. In this way, a hydrophobic mismatch with the surrounding lipid bilayer is avoided. On the other hand, palmitoylation at juxtamembraneous cysteines might increase the effective hydrophobic length of a TMR to adapt a short hydrophobic protein sequence to a thicker hydrophobic bilayer (Charollais and Van Der Goot, 2009). The thickness of membrane bilayers increases along the exocytic pathway, concomitantly with the cholesterol content, and is most pronounced in membrane rafts (van Meer et al., 2008).
A fatty acid attached to cytoplasmic cysteines is likely to insert into the membrane and will thus profoundly affect the conformation of a protein's cytoplasmic tail. This could, in turn, influence its interaction with internal components of virus particle. It is assumed that palmitoylation promotes protein–protein interactions by positioning a binding site in close proximity to a membrane‐localised partner protein (Charollais and Van Der Goot, 2009). However, one can also imagine that the fatty acid directly interacts with a partner protein, for example by insertion into a hydrophobic groove of the molecule. Upon loose interaction of a spike protein with a binding partner, the fatty acid might be extruded from the lipid bilayer and contact the binding partner to stabilise the interaction. This is reminiscent of the well described myristoyl switch of the retroviral Gag protein. There, the fatty acid is hidden within the myristoylated protein and only exposed upon Gag oligomerisation to facilitate membrane binding of the protein (Resh, 2004b).
In conclusion, a combination of structural, biochemical and biophysical methods will be required to decipher the function of protein palmitoylation on a molecular level, and any model derived from such studies needs to be tested by reverse genetics to understand its relevance for virus replication.
Glossary of viruses and their palmitoylated proteins
Baculovirus: double‐stranded, linear DNA genome encoding 150 genes.
Infect only invertebrates, especially insects. Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) was isolated from the alfalfa looper, a Lepidoptera. It is used for the expression of recombinant proteins in insect cells.
Gp64: Major glycoprotein required for budding and mediating cell attachment and membrane fusion.
Coronavirus: single‐stranded, positive‐sense RNA genome containing seven to 10 genes.
Coronaviruses infect a wide range of mammals and birds. SARS‐coronavirus (SARS‐CoV) is the causative agent of the severe acute respiratory syndrome. Mouse hepatitis virus (MHV) causes an epidemic illness with high mortality, especially among colonies of laboratory mice.
S: trimeric spike protein mediating cell attachment and membrane fusion at acidic pH.
E: small and hydrophobic membrane protein involved in virus budding. Viroporin.
Filovirus: single‐stranded, negative‐sense RNA genome containing seven genes.
Marburg and Ebolavirus cause severe disease in humans and certain primates in the form of viral hemorrhagic fevers. Virus particle has filamentous shape.
Gp: trimeric spike glycoprotein mediating cell attachment and membrane fusion at acidic pH.
Flavivirus: positive‐sense, single‐stranded RNA genome encoding one open reading frame.
The polyprotein precursor is proteolytically cleaved into four non‐structural proteins and three structural proteins. HCV is the causative agent of a liver disease.
Core: protein forming the capsid of the virus.
Herpesvirus: double‐stranded, linear DNA genome encoding 100–200 genes.
Large virus family infecting a wide variety of vertebrates. They produce latent, recurring infections. Infection with Human cytomegalovirus (HCMV) is typically unnoticed in healthy people, but can be life threatening for the immunocompromised or newborn infants. Herpes simplex virus (HSV) produces cold soar and genital herpes.
gN: glycoprotein N. Dimer with gM required for budding of viral capsid into the Golgi.
UL11 and UL51: non‐structural proteins. Required for budding of the viral capsid into the Golgi.
Orthomyxovirus: single‐stranded, negative‐sense, segmented RNA genome with 11 genes.
Although influenza B and C virus (Flu B, Flu C) infect mainly humans, influenza A virus (Flu A) infects also pigs, horses and a large variety of birds, especially waterfowl.
HA: hemagglutinin of Flu A and B, HEF: hemagglutinin, esterase, fusion protein of Flu C: trimeric spikes mediating attachment to cellular receptor and membrane fusion. HEF has in addition receptor‐destroying (esterase) activity.
M2 of Flu A and CM2 of Flu C: forms proton channel and M2 is involved in virus scission.
Orthoreovirus: double‐stranded, segmented RNA genome encoding one open reading frame.
The only non‐enveloped virus family for which palmitoylated proteins have been described. These viruses infect vertebrates (including humans), but no disease symptoms are normally seen.
P10: fusion‐associated small transmembrane (FAST) protein. Non‐structural protein, that catalyses membrane fusion by a different mechanism compared with viral spike proteins.
Paramyxovirus: single‐stranded, negative‐sense RNA genome with six to 10 genes.
Large virus family infecting a wide variety of vertebrates. Measles virus causes the childhood disease with the same name, whereas respiratory syncytial virus (hRSV) is an important cause of bronchiolitis and pneumonia in infants and children. Newcastle Disease virus (NDV): infects domestic and wild avian species and produces (depending on the strain) severe nervous and respiratory signs or only mild signs with negligible mortality:
Fusion protein: trimeric surface protein mediating membrane fusion at neutral pH.
Retrovirus: positive‐sense, single‐stranded RNA genome replicating via a DNA intermediate.
Rous sarcoma virus (RSV) causes sarcoma in chickens. Murine leukemia viruses (MuLV) causes cancer in mice. Human immunodeficiency virus (HIV) causes acquired immunodeficiency syndrome (AIDS), a condition in humans in which progressive failure of the immune system allows life‐threatening opportunistic infections and cancers to thrive. Simian immunodeficiency virus (SIV) infects African primates, but often does not cause any disease in these animals.
Env: glycoprotein encoded by the env gene. Required for cell attachment and membrane fusion.
Rhabdovirus: single‐stranded, negative‐sense RNA genome with five genes.
Vesicular stomatis virus (VSV) is an important model virus, whereas Rabies virus causes the disease of the same name, a fatal infection of the central nervous system.
G: trimeric spike mediating cell attachment and membrane fusion at acidic pH.
Togavirus: single‐stranded, positive‐sense RNA genome containing two open reading frames.
The two‐polyprotein precursors are proteolytically cleaved into four non‐structural proteins and five structural proteins. Sindbis and Semliki Forest virus (SFV) have been used extensively in biological research as a model of the viral life cycle.
E1/E2: hetero‐oligomeric spike mediating cell attachment and membrane fusion at acidic pH.
6K: small hydrophobic membrane protein with hairpin topology. Involved in budding. Viroporin.
nsP1: non‐structural protein, component of the viral replication complex.
Funding
The work done in the author's laboratory is funded by the German Research Foundation (DFG) (priority programme 1175, Collaborative Research Centre 740, and individual projects Ve 141/10, Ve 141/11) and by 7th Framework Programme of the European Commission, (Marie Curie Initial Training Network ‘Virus‐Entry’).
Conflict of interest statement
The author has declared no conflict of interest.
Acknowledgements
The authors thank Larisa Kordyukova (Lomonosow University Moscow), Evgeni Ponimaskin (Medizinische Hochschule Hannover), Oliver Rocks (MDC, Berlin), Christian Ungermann (Univ. of Osnabrück) and especially Bastian Thaa (FU Berlin) for suggestions on the manuscript.
References
- Ahola, T. , Kujala, P. , Tuittila, M. , Blom, T. , Laakkonen, P. , Hinkkanen, A. and Auvinen, P. (2000) Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J. Virol. 74, 6725–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, N.L. , Starkey, J.L. , Hughes, D.J. and Wills, J.W. (2010) Myristylation and palmitylation of HSV‐1 UL11 are not essential for its function. Virology 397, 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz, J. and Mittal, A. (2003) Architecture of the influenza hemagglutinin membrane fusion site. Biochim. Biophys. Acta 1614, 24–35 [DOI] [PubMed] [Google Scholar]
- Berger, M. and Schmidt, M.F. (1984) Cell‐free fatty acid acylation of Semliki Forest viral polypeptides with microsomal membranes from eukaryotic cells. J. Biol. Chem. 259, 7245–7252 [PubMed] [Google Scholar]
- Berger, M. and Schmidt, M.F. (1985) Protein fatty acyltransferase is located in the rough endoplasmic reticulum. FEBS Lett. 187, 289–294 [DOI] [PubMed] [Google Scholar]
- Bhattacharya, J. , Peters, P.J. and Clapham, P.R. (2004) Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J. Virol. 78, 5500–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlmakers, M.J. and Marsh, M. (2003) The on–off story of protein palmitoylation. Trends Cell Biol. 13, 32–42 [DOI] [PubMed] [Google Scholar]
- Biswas, S. , Yin, S.R. , Blank, P.S. and Zimmerberg, J. (2008) Cholesterol promotes hemifusion and pore widening in membrane fusion induced by influenza hemagglutinin. J. Gen. Physiol. 131, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatti, S. , Migliaccio, G. and Simons, K. (1989) Palmitylation of viral membrane glycoproteins takes place after exit from the endoplasmic reticulum. J. Biol. Chem. 264, 12590–12595 [PubMed] [Google Scholar]
- Bos, E.C. , Heijnen, L. , Luytjes, W. and Spaan, W.J. (1995) Mutational analysis of the murine coronavirus spike protein: effect on cell‐to‐cell fusion. Virology 214, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino, J.A. , Logan, H.L. , Lacny, J.J. and Gallagher, T.M. (2008) Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 82, 2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier, J. and Duncan, R . (2011) The reovirus fusion‐associated small transmembrane (FAST) proteins: virus‐encoded cellular fusogens. Curr. Top Membr. 68, 107–140 [DOI] [PubMed] [Google Scholar]
- Branigan, P.J. , Day, N.D. , Liu, C. , Gutshall, L.L. , Melero, J.A. , Sarisky, R.T. and del Vecchio, A.M. (2006) The cytoplasmic domain of the F protein of human respiratory syncytial virus is not required for cell fusion. J. Gen. Virol. 87, 395–398 [DOI] [PubMed] [Google Scholar]
- Brugger, B. , Glass, B. , Haberkant, P. , Leibrecht, I. , Wieland, F.T. and Krausslich, H.G. (2006) The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U. S. A. 103, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero, M. , Carabana, J. , Ortego, J. , Fernandez‐Munoz, R. and Celma, M.L. (1998) Measles virus fusion protein is palmitoylated on transmembrane‐intracytoplasmic cysteine residues which participate in cell fusion. J. Virol. 72, 8198–8204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrucci, M.R. , Hughes, M. , Calzoletti, L. , Donatelli, I. , Wells, K. , Takada, A. and Kawaoka, Y. (1997) The cysteine residues of the M2 protein are not required for influenza A virus replication. Virology 238, 128–134 [DOI] [PubMed] [Google Scholar]
- Chan, W.E. , Lin, H.H. and Chen, S.S. (2005) Wild‐type‐like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J. Virol. 79, 8374–8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais, J. and van der Goot, F.G. (2009) Palmitoylation of membrane proteins (Review). Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- Chen, B.J. and Lamb, R.A. (2008) Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372, 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.J. , Leser, G.P. , Morita, E. and Lamb, R.A. (2007) Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid‐derived virus‐like particles. J. Virol. 81, 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.J. , Takeda, M. and Lamb, R.A. (2005) Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79, 13673–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik, L.V. and Kozlov, M.M. (2005) Membrane hemifusion: crossing a chasm in two leaps. Cell 123, 375–382 [DOI] [PubMed] [Google Scholar]
- Corse, E. and Machamer, C.E. (2001) Infectious bronchitis virus envelope protein targeting: implications for virus assembly. Adv. Exp. Med. Biol. 494, 571–576 [DOI] [PubMed] [Google Scholar]
- Dekker, F.J. , Rocks, O. , Vartak, N. , Menninger, S. , Hedberg, C. , Balamurugan, R. , Wetzel, S. , Renner, S. , Gerauer, M. , Scholermann, B. , Rusch, M. , Kramer, J.W. , Rauh, D. , Coates, G.W. , Brunsveld, L. , Bastiaens, P.I. and Waldmann, H. (2010) Small‐molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- Duncan, J.A. and Gilman, A.G. (1996) Autoacylation of G protein alpha subunits. J. Biol. Chem. 271, 23594–23600 [DOI] [PubMed] [Google Scholar]
- Duncan, J.A. and Gilman, A.G. (1998) A cytoplasmic acyl‐protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273, 15830–15837 [DOI] [PubMed] [Google Scholar]
- Earp, L.J. , Delos, S.E. , Park, H.E. and White, J.M. (2005) The many mechanisms of viral membrane fusion proteins. Curr. Top Microbiol. Immunol. 285, 25–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, S. , de Vries, M. , Herrmann, A. and Veit, M. (2012) Mutation of a raft‐targeting signal in the transmembrane region retards transport of influenza virus hemagglutinin through the Golgi. FEBS Lett. 586, 277–282 [DOI] [PubMed] [Google Scholar]
- Engel, S. , Scolari, S. , Thaa, B. , Krebs, N. , Korte, T. , Herrmann, A. and Veit, M. (2010) FLIM‐FRET and FRAP reveal association of influenza virus haemagglutinin with membrane rafts. Biochem. J. 425, 567–573 [DOI] [PubMed] [Google Scholar]
- Firth, A.E. , Chung, B.Y. , Fleeton, M.N. and Atkins, J.F. (2008) Discovery of frameshifting in Alphavirus 6K resolves a 20‐year enigma. Virol. J. 5, 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester, M.T. , Hess, D.T. , Thompson, J.W. , Hultman, R. , Moseley, M.A. , Stamler, J.S. and Casey, P.J. (2011) Site‐specific analysis of protein S‐acylation by resin‐assisted capture. J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata, Y. and Fukata, M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- Funke, C. , Becker, S. , Dartsch, H. , Klenk, H.D. and Muhlberger, E. (1995) Acylation of the Marburg virus glycoprotein. Virology 208, 289–297 [DOI] [PubMed] [Google Scholar]
- Gaedigk‐Nitschko, K. , Ding, M.X. , Levy, M.A. and Schlesinger, M.J. (1990) Site‐directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology 175, 282–291 [DOI] [PubMed] [Google Scholar]
- Gaedigk‐Nitschko, K. and Schlesinger, M.J. (1990) The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 175, 274–281 [DOI] [PubMed] [Google Scholar]
- Gerl, M.J. , Sampaio, J.L. , Urban, S. , Kalvodova, L. , Verbavatz, J.M. , Binnington, B. , Lindemann, D. , Lingwood, C.A. , Shevchenko, A. , Schroeder, C. and Simons, K. (2012) Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 196, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, M.E. and Carrasco, L. (2003) Viroporins. FEBS Lett. 552, 28–34 [DOI] [PubMed] [Google Scholar]
- Grantham, M.L. , Wu, W.H. , Lalime, E.N. , Lorenzo, M.E. , Klein, S.L. and Pekosz, A. (2009) Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. J. Virol. 83, 8655–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, J. and Chamberlain, L.H. (2007) Palmitoylation‐dependent protein sorting. J. Cell Biol. 176, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M. A. , Cherezov, V. , Griffith, M.T. , Roth, C.B. , Jaakola, V.P. , Chien, E.Y. , Velasquez, J. , Kuhn, P. and Stevens, R.C. (2008) A specific cholesterol binding site is established by the 2.8 A structure of the human beta2‐adrenergic receptor. Structure 16, 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger, L.J. , Shaughnessy, M.A. , Micko, A. , Pinto, L.H. and Lamb, R.A. (1995) Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 69, 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, H. , John Peter, A.T. , Meiringer, C. , Subramanian, K. and Ungermann, C. (2009) Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two‐step reaction mechanism. Traffic 10, 1061–1073 [DOI] [PubMed] [Google Scholar]
- Ito, H. , Watanabe, S. , Takada, A. and Kawaoka, Y. (2001) Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 75, 1576–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova, L. and Schlesinger, M.J. (1993) Site‐directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 67, 2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings, B.C. and Linder, M.E. (2012) Dhhc protein S‐acyltransferases use a similar ping‐pong kinetic mechanism but display different acyl‐coA specificities. J. Biol. Chem 287, 7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Leser, G.P. and Lamb, R.A. (1994) The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 13, 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Leser, G.P. , Zhang, J. and Lamb, R.A. (1997) Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16, 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova, L. , Sampaio, J.L. , Cordo, S. , Ejsing, C.S. , Shevchenko, A. and Simons, K. (2009) The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 83, 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo‐Astover, L. , Sarova, O. , Merits, A. and Zusinaite, E. (2010) The infection of mammalian and insect cells with SFV bearing nsP1 palmitoylation mutations. Virus Res. 153, 277–287 [DOI] [PubMed] [Google Scholar]
- Kemble, G. W. , Danieli, T. and White, J.M. (1994) Lipid‐anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76, 383–391 [DOI] [PubMed] [Google Scholar]
- Kenworthy, A.K. , Nichols, B.J. , Remmert, C.L. , Hendrix, G.M. , Kumar, M. , Zimmerberg, J. and Lippincott‐Schwartz, J. (2004) Dynamics of putative raft‐associated proteins at the cell surface. J. Cell Biol. 165, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiiver, K. , Tagen, I. , Zusinaite, E. , Tamberg, N. , Fazakerley, J.K. and Merits, A. (2008) Properties of non‐structural protein 1 of Semliki Forest virus and its interference with virus replication. J. Gen. Virol. 89, 1457–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokame, K. , Fukada, Y. , Yoshizawa, T. , Takao, T. and Shimonishi, Y. (1992) Lipid modification at the N terminus of photoreceptor G‐protein alpha‐subunit. Nature 359, 749–752 [DOI] [PubMed] [Google Scholar]
- Kordyukova, L.V. , Ksenofontov, A.L. , Serebryakova, M.V. , Ovchinnikova, T.V. , Fedorova, N.V. , Ivanova, V.T. and Baratova, L.A. (2004) Influenza A hemagglutinin C‐terminal anchoring peptide: identification and mass spectrometric study. Protein Pept. Lett. 11, 385–391 [DOI] [PubMed] [Google Scholar]
- Kordyukova, L.V. , Serebryakova, M.V. , Baratova, L.A. and Veit, M. (2008) S‐acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site‐specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 82, 9288–9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordyukova, L.V. , Serebryakova, M.V. , Baratova, L.A. and Veit, M. (2010) Site‐specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology 398, 49–56 [DOI] [PubMed] [Google Scholar]
- Kordyukova, L.V. , Serebryakova, M.V. , Polyansky, A.A. , Kropotkina, E.A. , Alexeevski, A.V. , Veit, M. , Efremov, R.G. , Filippova, I.Y. and Baratova, L.A. (2011) Linker and/or transmembrane regions of influenza A/Group‐1, A/Group‐2, and type B virus hemagglutinins are packed differently within trimers. Biochim. Biophys. Acta 1808, 1843–1854 [DOI] [PubMed] [Google Scholar]
- Kostyuchenko, V.A. , Jakana, J. , Liu, X. , Haddow, A.D. , Aung, M. , Weaver, S.C. , Chiu, W. and Lok, S.M. (2011) The structure of barmah forest virus as revealed by cryo‐electron microscopy at a 6‐angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 85, 9327–9333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummel, D. , Heinemann, U. and Veit, M. (2006) Unique self‐palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc. Natl. Acad. Sci. U. S. A. 103, 12701–12706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, K. , Veit, M. and Klenk, H.D. (1991) Retarded processing of influenza virus hemagglutinin in insect cells. Virology 180, 159–165 [DOI] [PubMed] [Google Scholar]
- Kusumi, A. , Suzuki, K.G. , Kasai, R.S. , Ritchie, K. and Fujiwara, T.K. (2011) Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 36, 604–615 [DOI] [PubMed] [Google Scholar]
- Laakkonen, P. , Ahola, T. and Kaariainen, L. (1996) The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J. Biol. Chem. 271, 28567–28571 [DOI] [PubMed] [Google Scholar]
- Levental, I. , Grzybek, M. and Simons, K. (2010a) Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49, 6305–6316 [DOI] [PubMed] [Google Scholar]
- Levental, I. , Lingwood, D. , Grzybek, M. , Coskun, U. and Simons, K. (2010b) Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. U.S.A, 107, 22050‐‐22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Yang, C. , Tong, S. , Weidmann, A. and Compans, R.W. (2002) Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J. Virol. 76, 11845–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Nazarian, A. , Erdjument‐Bromage, H. , Bornmann, W. , Tempst, P. and Resh, M.D. (2001) Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J. Biol. Chem. 276, 30987–30994 [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Yuan, Q. , Torres, J. , Tam, J.P. and Liu, D.X. (2006) Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349, 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, M.E. and Deschenes, R.J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- Liu, D.X. , Yuan, Q. and Liao, Y. (2007) Coronavirus envelope protein: a small membrane protein with multiple functions. Cell Mol. Life Sci. 64, 2043–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo, S. , Greentree, W.K. , Linder, M.E. and Deschenes, R.J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae . J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- Lopez, L.A. , Riffle, A.J. , Pike, S.L. , Gardner, D. and Hogue, B.G. (2008) Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 82, 3000–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach, M. , Osinski, K. , Kropff, B. , Schloetzer‐Schrehardt, U. , Krzyzaniak, M. and Britt, W. (2007) The carboxy‐terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J. Virol. 81, 5212–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau, N. , Fromentin, R. , Savard, C. , Duval, M. , Tremblay, M.J. and Leclerc, D. (2009) Palmitoylation of hepatitis C virus core protein is important for virion production. J. Biol. Chem. 284, 33915–33925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, B.R. , Wang, C. , Adibekian, A. , Tully, S.E. and Cravatt, B.F. (2012) Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbride, C.E. and Machamer, C.E. (2010) Palmitoylation of SARS‐CoV S protein is necessary for partitioning into detergent‐resistant membranes and cell–cell fusion but not interaction with M protein. Virology 405, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder, D. , Moreno, M.J. , Verkade, P. , Vaz, W.L. and Simons, K. (2006) Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 103, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian, K.A. , Ostermeyer, A.G. , Chen, J.Z. , Roth, M.G. and Brown, D.A. (1999) Role of lipid modifications in targeting proteins to detergent‐resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274, 3910–3917 [DOI] [PubMed] [Google Scholar]
- Mitchell, D. A. , Vasudevan, A. , Linder, M.E. and Deschenes, R.J. (2006) Protein palmitoylation by a family of DHHC protein S‐acyltransferases. J. Lipid Res. 47, 1118–1127 [DOI] [PubMed] [Google Scholar]
- Moradpour, D. , Penin, F. and Rice, C.M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- Munro, S. (2003) Lipid rafts: elusive or illusive? Cell 115, 377–388 [DOI] [PubMed] [Google Scholar]
- Muraki, Y. , Okuwa, T. , Furukawa, T. , Matsuzaki, Y. , Sugawara, K. , Himeda, T. , Hongo, S. and Ohara, Y. (2011) Palmitoylation of CM2 is dispensable to influenza C virus replication. Virus Res. 157, 99–105 [DOI] [PubMed] [Google Scholar]
- Nadolski, M.J. and Linder, M.E. (2009) Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3. J. Biol. Chem. 284, 17720–17730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeve, C.W. and Williams, D. (1990) Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 9, 3857–3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim, H.Y. , Amarneh, B. , Ktistakis, N.T. and Roth, M.G. (1992) Effects of altering palmitylation sites on biosynthesis and function of the influenza virus hemagglutinin. J. Virol. 66, 7585–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaus, J. , Warner, J.M. , O’Shaughnessy, B. and Herrmann, A. (2011) The pathway to membrane fusion through hemifusion. Curr. Top Membr. 68, 1–32 [DOI] [PubMed] [Google Scholar]
- Nobusawa, E. , Aoyama, T. , Kato, H. , Suzuki, Y. , Tateno, Y. and Nakajima, K. (1991) Comparison of complete amino acid sequences and receptor‐binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182, 475–485 [DOI] [PubMed] [Google Scholar]
- Nozawa, N. , Daikoku, T. , Koshizuka, T. , Yamauchi, Y. , Yoshikawa, T. and Nishiyama, Y. (2003) Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77, 3204–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer‐Jambor, C. , Miller, D.C. , Roberts, C.R. , Rhee, S.S. and Hunter, E. (2001) Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 75, 11544–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, Y. , Kihara, A. , Sano, T. and Igarashi, Y. (2006) Intracellular localization and tissue‐specific distribution of human and yeast DHHC cysteine‐rich domain‐containing proteins. Biochim. Biophys. Acta 1761, 474–483 [DOI] [PubMed] [Google Scholar]
- Peitzsch, R.M. and Mclaughlin, S. (1993) Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry 32, 10436–10443 [DOI] [PubMed] [Google Scholar]
- Petit, C.M. , Chouljenko, V.N. , Iyer, A. , Colgrove, R. , Farzan, M. , Knipe, D.M. and Kousoulas, K.G. (2007) Palmitoylation of the cysteine‐rich endodomain of the SARS‐coronavirus spike glycoprotein is important for spike‐mediated cell fusion. Virology 360, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, L.H. and Lamb, R.A. (2006) The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 281, 8997–9000 [DOI] [PubMed] [Google Scholar]
- Ponimaskin, E. and Schmidt, M.F. (1998) Domain‐structure of cytoplasmic border region is main determinant for palmitoylation of influenza virus hemagglutinin (H7). Virology 249, 325–335 [DOI] [PubMed] [Google Scholar]
- Quevillon‐Cheruel, S. , Leulliot, N. , Muniz, C.A. , Vincent, M. , Gallay, J. , Argentini, M. , Cornu, D. , Boccard, F. , Lemaitre, B. and van Tilbeurgh, H. (2009) Evf, a virulence factor produced by the Drosophila pathogen Erwinia carotovora, is an S‐palmitoylated protein with a new fold that binds to lipid vesicles. J. Biol. Chem. 284, 3552–3562 [DOI] [PubMed] [Google Scholar]
- Resh, M.D. (2004a) Membrane targeting of lipid modified signal transduction proteins. Subcell. Biochem. 37, 217–232 [DOI] [PubMed] [Google Scholar]
- Resh, M.D. (2004b) A myristoyl switch regulates membrane binding of HIV‐1 Gag. Proc. Natl. Acad. Sci. U. S. A. 101, 417–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverey, H. , Veit, M. , Ponimaskin, E. and Schmidt, M.F. (1996) Differential fatty acid selection during biosynthetic S‐acylation of a transmembrane protein (HEF) and other proteins in insect cells (Sf9) and in mammalian cells (CV1). J. Biol. Chem. 271, 23607–23610 [DOI] [PubMed] [Google Scholar]
- Rocks, O. , Gerauer, M. , Vartak, N. , Koch, S. , Huang, Z.P. , Pechlivanis, M. , Kuhlmann, J. , Brunsveld, L. , Chandra, A. , Ellinger, B. , Waldmann, H. and Bastiaens, P.I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- Rossman, J. S. , Jing, X. , Leser, G.P. , Balannik, V. , Pinto, L.H. and Lamb, R.A. (2010a) Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84, 5078–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman, J.S. , Jing, X. , Leser, G.P. and Lamb, R.A. (2010b) Influenza virus M2 protein mediates ESCRT‐independent membrane scission. Cell 142, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F. , Feng, Y. , Chen, L. and Davis, N.G. (2002) The yeast DHHC cysteine‐rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F. , Papanayotou, I. and Davis, N.G. (2011) The yeast kinase Yck2 has a tripartite palmitoylation signal. Mol. Biol. Cell 22, 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F. , Wan, J. , Bailey, A.O. , Sun, B. , Kuchar, J.A. , Green, W.N. , Phinney, B.S. , Yates, J.R., 3rd and Davis, N.G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso, I. , Mixon, M.B. , Chen, B.K. and Kim, P.S. (2000) Palmitoylation of the HIV‐1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 97, 13523–13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C. , Ivanova, L. and Schlesinger, M.J. (1998) Effects of site‐directed mutations of transmembrane cysteines in sindbis virus E1 and E2 glycoproteins on palmitylation and virus replication. Virology 249, 62–67 [DOI] [PubMed] [Google Scholar]
- Sakai, T. , Ohuchi, R. and Ohuchi, M. (2002) Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J. Virol. 76, 4603–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M.F. (1984) The transfer of myristic and other fatty acids on lipid and viral protein acceptors in cultured cells infected with Semliki Forest and influenza virus. EMBO J. 3, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M.F. and Schlesinger, M.J. (1979) Fatty acid binding to vesicular stomatitis virus glycoprotein: a new type of post‐translational modification of the viral glycoprotein. Cell 17, 813–819 [DOI] [PubMed] [Google Scholar]
- Schmidt, M.F. and Schlesinger, M.J. (1980) Relation of fatty acid attachment to the translation and maturation of vesicular stomatitis and Sindbis virus membrane glycoproteins. J. Biol. Chem. 255, 3334–3339 [PubMed] [Google Scholar]
- Schroeder, C. , Heider, H. , Moncke‐Buchner, E. and Lin, T.I. (2005) The influenza virus ion channel and maturation cofactor M2 is a cholesterol‐binding protein. Eur. Biophys. J. 34, 52–66 [DOI] [PubMed] [Google Scholar]
- Scolari, S. , Engel, S. , Krebs, N. , Plazzo, A.P. , de Almeida, R.F. , Prieto, M. , Veit, M. and Herrmann, A. (2009) Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time‐resolved fluorescence imaging. J. Biol. Chem. 284, 15708–15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryakova, M.V. , Kordyukova, L.V. , Baratova, L.A. and Markushin, S.G. (2006) Mass spectrometric sequencing and acylation character analysis of C‐terminal anchoring segment from influenza A hemagglutinin. Eur. J. Mass Spectrom. (Chichester, Eng.) 12, 51–62 [DOI] [PubMed] [Google Scholar]
- Sergel, T. and Morrison, T.G. (1995) Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology 210, 264–272 [DOI] [PubMed] [Google Scholar]
- Shahinian, S. and Silvius, J.R. (1995) Doubly‐lipid‐modified protein sequence motifs exhibit long‐lived anchorage to lipid bilayer membranes. Biochemistry 34, 3813–3822 [DOI] [PubMed] [Google Scholar]
- Shmulevitz, M. , Salsman, J. and Duncan, R. (2003) Palmitoylation, membrane‐proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 77, 9769–9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla, A. and Gallagher, T. (2009) Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 284, 32725–32734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K. and Gerl, M.J. (2010) Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell. Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- Simons, K. and Toomre, D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- Skehel, J.J. and Wiley, D.C. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E. and Linder, M.E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- Steinhauer, D.A. , Wharton, S.A. , Wiley, D.C. and Skehel, J.J. (1991) Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology 184, 445–448 [DOI] [PubMed] [Google Scholar]
- Stewart, S.M. , Wu, W.H. , Lalime, E.N. and Pekosz, A. (2010) The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence. Virology 405, 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, M. (2002) Lipid rafts and assembly of enveloped viruses. Traffic 3, 705–709 [DOI] [PubMed] [Google Scholar]
- Thaa, B. , Herrmann, A. and Veit, M. (2010) Intrinsic cytoskeleton‐dependent clustering of influenza virus M2 protein with hemagglutinin assessed by FLIM‐FRET. J. Virol. 84, 12445–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaa, B. , Levental, I. , Herrmann, A. and Veit, M. (2011) Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem. J. 437, 389–397 [DOI] [PubMed] [Google Scholar]
- Thaa, B. , Tielesch, C. , Moller, L. , Schmitt, A. O. , Wolff, T. , Bannert, N. , Herrmann, A. and Veit, M. (2012) Growth of influenza A virus is not impeded by simultaneous removal of the cholesterol‐binding and acylation sites in the M2 protein. J. Gen. Virol. 93, 282–292 [DOI] [PubMed] [Google Scholar]
- Thorp, E.B. , Boscarino, J.A. , Logan, H.L. , Goletz, J.T. and Gallagher, T.M. (2006) Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 80, 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatis, V. M. , Trenchi, A. , Gomez, G.A. and Daniotti, J.L. (2010) Acyl‐protein thioesterase 2 catalyzes the deacylation of peripheral membrane‐associated GAP‐43. PLoS One 5, e15045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike, M. , Huang, C. , Shirato, K. , Matsuyama, S. , Makino, S. and Taguchi, F. (2012) Two palmitylated cysteine residues of the severe acute respiratory syndrome coronavirus spike protein are critical for S‐incorporation into virus‐like particles, but not for M‐S co‐localization. J. Gen. Virol. 93, 823‐‐828. [DOI] [PubMed] [Google Scholar]
- Ujike, M. , Nakajima, K. and Nobusawa, E. (2004) Influence of acylation sites of influenza B virus hemagglutinin on fusion pore formation and dilation. J. Virol. 78, 11536–11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer, G. , Voelker, D.R. and Feigenson, G.W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, M. (2000) Palmitoylation of the 25‐kDa synaptosomal protein (SNAP‐25) in vitro occurs in the absence of an enzyme, but is stimulated by binding to syntaxin. Biochem. J. 345, 145–151 [PMC free article] [PubMed] [Google Scholar]
- Veit, M. , Herrler, G. , Schmidt, M.F. , Rott, R. and Klenk, H.D. (1990) The hemagglutinating glycoproteins of influenza B and C viruses are acylated with different fatty acids. Virology 177, 807–811 [DOI] [PubMed] [Google Scholar]
- Veit, M. , Kretzschmar, E. , Kuroda, K. , Garten, W. , Schmidt, M.F. , Klenk, H.D. and Rott, R. (1991) Site‐specific mutagenesis identifies three cysteine residues in the cytoplasmic tail as acylation sites of influenza virus hemagglutinin. J. Virol. 65, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, M. , Ponimaskin, E. and Schmidt, M.F. (2008) Analysis of S‐acylation of proteins. Methods Mol. Biol. 446, 163–182 [DOI] [PubMed] [Google Scholar]
- Veit, M. , Reverey, H. and Schmidt, M.F. (1996) Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem. J. 318, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, M. and Schmidt, M.F. (1993) Timing of palmitoylation of influenza virus hemagglutinin. FEBS Lett. 336, 243–247 [DOI] [PubMed] [Google Scholar]
- Veit, M. and Schmidt, M.F. (2001) Enzymatic depalmitoylation of viral glycoproteins with acyl‐protein thioesterase 1 in vitro . Virology 288, 89–95 [DOI] [PubMed] [Google Scholar]
- Veit, M. and Schmidt, M.F. (2006) Palmitoylation of influenza virus proteins. Berl. Munch. Tierarztl. Wochenschr. 119, 112–122 [PubMed] [Google Scholar]
- Veit, M. , Schmidt, M.F. and Rott, R. (1989) Different palmitoylation of paramyxovirus glycoproteins. Virology 168, 173–176 [DOI] [PubMed] [Google Scholar]
- Veit, M. and Thaa, B. (2011) Association of influenza virus proteins with membrane rafts. Adv. Virol. 2011, 370606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vzorov, A. N. , Weidmann, A. , Kozyr, N.L. , Khaoustov, V. , Yoffe, B. and Compans, R.W. (2007) Role of the long cytoplasmic domain of the SIV Env glycoprotein in early and late stages of infection. Retrovirology 4, 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, R. , Herwig, A. , Azzouz, N. and Klenk, H.D. (2005) Acylation‐mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J. Virol. 79, 6449–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J. , Roth, A. F. , Bailey, A.O. and Davis, N.G. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- Wang, K. , Xie, S. and Sun, B. (2011) Viral proteins function as ion channels. Biochim. Biophys. Acta 1808, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt, M.A. and Rose, J.K. (1991) Fatty acid acylation is not required for membrane fusion activity or glycoprotein assembly into VSV virions. Virology 185, 875–878 [DOI] [PubMed] [Google Scholar]
- Whittaker, G.R. (2001) Intracellular trafficking of influenza virus: clinical implications for molecular medicine. Expert Rev. Mol. Med. 2001, 1–13 [DOI] [PubMed] [Google Scholar]
- Yang, C. and Compans, R.W. (1996) Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology 221, 87–97 [DOI] [PubMed] [Google Scholar]
- Yang, C. , Spies, C.P. and Compans, R.W. (1995) The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. U. S. A. 92, 9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik, J.H. and Weigel, P.H. (2002) The position of cysteine relative to the transmembrane domain is critical for palmitoylation of H1, the major subunit of the human asialoglycoprotein receptor. J. Biol. Chem. 277, 47305–47312 [DOI] [PubMed] [Google Scholar]
- Zacharias, D.A. , Violin, J.D. , Newton, A.C. and Tsien, R.Y. (2002) Partitioning of lipid‐modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Pekosz, A. and Lamb, R.A. (2000) Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74, 4634–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.X. , Han, Y. and Blissard, G.W. (2003) Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts. J. Virol. 77, 6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurcher, T. , Luo, G. and Palese, P. (1994) Mutations at palmitylation sites of the influenza virus hemagglutinin affect virus formation. J. Virol. 68, 5748–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusinaite, E. , Tints, K. , Kiiver, K. , Spuul, P. , Karo‐Astover, L. , Merits, A. and Sarand, I. (2007) Mutations at the palmitoylation site of non‐structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations. J. Gen. Virol. 88, 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]


