ABSTRACT
Sialic acids (Sias) are a group of α‐keto acids with a nine‐carbon backbone, which display many types of modifications in nature. The diversity of natural Sia presentations is magnified by a variety of glycosidic linkages to underlying glycans, the sequences and classes of such glycans, as well as the spatial organization of Sias with their surroundings. This diversity is closely linked to the numerous and varied biological functions of Sias. Relatively large libraries of natural and unnatural Sias have recently been chemically/chemoenzymatically synthesized and/or isolated from natural sources. The resulting sialoglycan microarrays have proved to be valuable tools for the exploration of diversity and biology of Sias. Here we provide an overview of Sia diversity in nature, the approaches used to generate sialoglycan microarrays, and the achievements and challenges arising. © 2013 Wiley Periodicals, Inc. Biopolymers 99: 650–665, 2013.
Keywords: sialic acids, diversity, sialoglycan microarrays
INTRODUCTION
DNA, RNA, proteins, and glycans are the four biopolymers essential to all known life forms.1, 2 Glycans differ from the other types of biopolymers in several ways. Glycan biosynthesis is not template‐driven and cannot be accurately predicted by any known method. In addition, glycans form branched structures, display numerous modifications, and have far more diversity in overall structure. These and other differences partly account for the historical lag in chemical and biological studies of glycans, compared to the other classes of biopolymers. The development of technologies to elucidate the structures and functions of these complex molecules have recently drawn increasing scientific interest and it is now well appreciated that glycans play vital roles in numerous complex biological processes and systems,3, 4 a fact recently recognized by a special report from the US National Academies, urging greater investments in this area.5
Among the various monosaccharide building blocks of glycans, sialic acids (Sias) are rather unusual. They are typically found as terminal residues on the glycan chains of vertebrate glycoconjugates. In contrast to most other common monosaccharides, which are aldoses or ketoses with five or six carbons, Sias are α‐keto acids with a nine‐carbon backbone (Figure 1). Furthermore, the types of natural modifications found on Sias far exceed that of any other monosaccharides (Figure 1).6, 7, 8, 9, 10, 11, 12, 13 Additional differences include the limited occurrence of Sias (primarily in the deuterostome lineage of animals and certain types of bacteria), as well as its unusual form of nucleoside monophosphate‐activated sugar donor (CMP‐Sia).12 These and other distinct features of Sias contribute to higher structural complexity and the potential for more unique and varied biological functions, in comparison to other monosaccharides.
Figure 1.
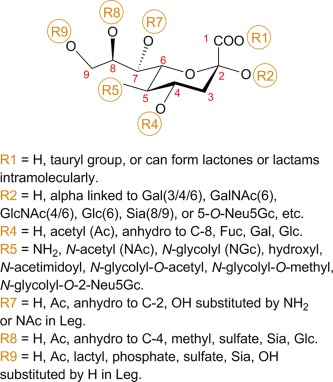
Sialic acid diversity. The nine‐carbon Sia backbone is shown in α configuration. Most known Sia modifications, at positions C‐1, C‐2, C‐4, C‐5, C‐7, C‐8, and C‐9, around the backbone are indicated. Figure modified with permission, from Ref. 13.
Accumulating evidence also indicates that evolutionary selection forces play a critical role in the diversification of glycans, which are intrinsically linked to their biological functions.14, 15, 16 Among various glycan classes, Sias appear to be the most rapidly evolving. In keeping with the recommended style for this contributed article, we will not attempt to cover the whole field; rather we will review our own work within the context of the field, including our personal insights, and speculate on where research is headed in the future. We will begin by briefly discussing the diversity and biology of Sias in nature, with examples from our own labs in the context of work of others. Recent advances in the synthesis of sialosides and the application of sialoglycan microarrays will then be reviewed, from our perspective.
DIVERSITY AND BIOLOGY OF SIALIC ACIDS IN NATURE
Natural Diversity and Biology of Sialic Acid forms and Modifications
The two core Sia forms are neuraminic acid (Neu) and 2‐keto‐3‐deoxy‐d‐glycero‐d‐galacto‐nononic‐acid (Kdn) (Figures 2a and 2b). More than 50 different types of naturally occurring Sia variants (c.f., Table 1 in Ref. 12) have been found based on modifications to these two forms, including 5‐N‐acetylation, hydroxylation of 5‐N‐acetyl group, O‐acetylation, O‐methylation, O‐lactylation, O‐sulfation, O‐phosphorylation, and intramolecular lactam or lactone formation.6, 7, 8, 9, 10, 11, 12, 13 Among the diverse naturally occurring Sias, N‐acetylneuraminic acid (Neu5Ac) and N‐glycolylneuraminic acid (Neu5Gc) are the two most abundant ones in mammals (Figures 2c and d). The nine‐carbon Sia backbone is also shared by other nonulosonic acids (NulOs) that are found in prokaryotes, such as legionaminic acid (Leg) and pseudaminic acid (Pse) (Figures 2e and 2f).17, 18, 19, 20 This review will focus on the more common Sias, some of which are shared by animals and pathogenic bacteria.
Figure 2.
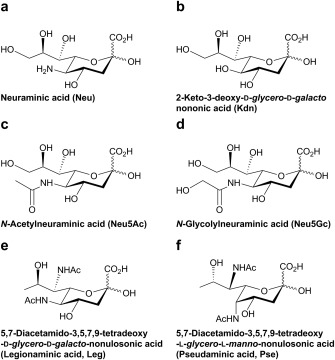
Examples of naturally occurring sialic acid and nonulosonic acid structures. Neu (a) and Kdn (b) are the two most basic Sia forms. Neu5Ac (c) and Neu5Gc (d) are derived from Neu and are the two most abundant Sia structures found in mammals. Leg (e) and Pse (f) are two representatives of the nonulosonic acid family that used to be known as the “bacterial Sias.”
The diversity of Sia forms and modifications are closely associated with biological functions in both intracellular and intercellular environments, from early discovered effects on the physical properties of glycoproteins,21 to their protective and masking roles,22 and to more recently established mediation and modulation of numerous biological processes including signaling, fertilization, immunity, growth and differentiation.6, 7, 8, 9, 10, 11, 12, 23 Among the many Sia O‐modifications, 9‐O‐acetylation is best studied. When modified by O‐acetyl group(s), Sias can be blocked from recognition by intrinsic lectins such as Siglecs (Sia‐binding immunoglobulin‐like lectins).24, 25 O‐Acetylation can also modulate interactions between Sias and microbial proteins, in a manner either benefiting or harming the host.23, 26, 27, 28, 29, 30 O‐Methylation has been found to make Sias resistant to sialidases, indicating a mechanism of biological significance.31
One variation at the C‐5 position has generated some uniquely interesting questions related to human evolution and disease.32 Specifically, hydroxylation of Neu5Ac at the 5‐N‐acetyl group forms Neu5Gc, a molecule that differs from Neu5Ac by a single oxygen atom (Figures 2c and 2d). Humans lost the ability to synthesize Neu5Gc due to a 92 bp exon deletion in the CMAH gene encoding CMP‐Neu5Ac hydroxylase,33, 34 which occurred ∼3 million years ago.35 However, Neu5Gc can be metabolically incorporated into human cells and tissues, via human consumption of Neu5Gc‐rich foods such as red meat,36, 37 even in the face of circulating anti‐Neu5Gc antibodies (Abs) in human blood.38, 39 The interactions between these “xeno‐autoantigens” and “xeno‐autoantibodies” are currently postulated to have numerous effects on human diseases, ranging from induction of chronic inflammation, tumor progression/suppression, effects on biotherapeutic molecules and cells,40, 41, 42, 43, 44, 45, 46 and even to xenotransplantation rejections.47, 48, 49, 50, 51, 52 Moreover, certain bacterial toxins preferentially recognize glycan epitopes containing metabolically incorporated Neu5Gc.53
Natural Diversity and Biology of Sia Linkages and Underlying Glycans
Sias are glycosidically linked via C‐2 to underlying glycans (Figure 1). They can be attached to an underlying galactose (Gal) residue via α2‐3, α2‐4, or α2‐6 linkage, to N‐acetylgalactosamine (GalNAc) via α2‐6 linkage, to N‐acetylglucosamine (GlcNAc) via α2‐4 or α2‐6 linkage, or to another Sia via α2‐8 or α2‐9 linkages.13 The structural diversity of the underlying glycan structures and modifications further increase the complexity of Sia‐containing molecules. Combinations of the multiple forms of Sias, different sialyl linkages, and diverse underlying glycans generate many thousands of possible Sia presentations.
This diversity of Sia linkages and underlying glycans is known to be biologically relevant. One well‐known example is the recognition of different Sia linkages by human and avian adapted influenza A viruses.54 Although the human adapted ones bind Siaα2‐6Gal preferentially, the avian adapted ones prefer Siaα2‐3Gal.27, 55, 56, 57 The relative expression level and distribution of Siaα2‐6Gal and Siaα2‐3Gal in host respiratory tract are believed to play an important role in the infection and transmission of these viruses.58, 59 Endogenous lectins such as Siglecs also have distinct Sia ligand preferences. Human Siglec 2 (CD22), for example, selectively recognizes α2‐6‐linked Sias,60 but human Siglec‐1 (sialoadhesin) prefers α2‐3‐linked Sias.61 In addition, Sia linkage to the underlying glycans plays a role in the progression and spread of human malignancies. For example, carcinoma behavior can be modulated by α2‐6‐sialylation on N‐glycans.62, 63, 64, 65, 66 Moreover, underlying glycans can also influence or even determine Sia binding events. Sias with an underlying Lewis x or Lewis a structure are preferentially recognized by selectins, and sulfation at the C‐6 position of an underlying Gal or GalNAc residue further enhances the binding affinity.67, 68, 69, 70 Additionally, different underlying glycans capped with the same α2‐6 linked Sia were found to show markedly different binding affinities to CD22.71 Numerous other such examples could be cited.
Natural Diversity and Biology of Polysialic Acids
Sia‐containing homopolysaccharides and heteropolysaccharides have been found in both animals and bacteria.7, 17, 72, 73, 74, 75, 76 Homo‐PolySias are found in a few animal glycoproteins, such as α2‐8‐linked PolySia on the N‐glycans of the neural cell adhesion molecule (NCAM) and on O‐glycans of some fish egg glycoproteins. The capsular polysaccharides of certain pathogenic bacteria can express α2‐8‐, α2‐9, or alternating α2‐8‐ and α2‐9‐linked Sias polymers. Heteropolysaccharides containing [‐4Siaα2‐6Galα1‐] and [‐4Siaα2‐6Glcα1‐] repeats have also been found on the capsular polysaccharides of N. meningitidis serotypes W135 and Y, respectively.77 PolySia chains based on Neu5Ac, Neu5Gc, Kdn, or Leg building blocks have been reported, and the inter‐residue linkages can vary from α2‐8, α2‐9, to α2‐4, and to α2‐5‐O glycolylNeu5Gc.75 The degree of polymerization and O‐acetylation,78, 79 homo‐ or hetero‐polymerization, and the dynamic equilibrium between polylactones and polycarboxylates further add to the complexity.80, 81 Altogether, there is a considerable range of diversity in polySia chains.
Natural Diversity and Biology of Sialoglycans in Clustered Saccharide Patches
The concept of “clustered saccharide patches” was first suggested in the context of understanding selectin ligands,67 and was recently updated to take new evidence into account.82 Cell surface glycans can be imagined as the outermost aspects of a tropical rainforest canopy, or more accurately, in a more dynamic fashion as a kelp bed or coral reef in the ocean (analogy suggested by P. Gagneux). Viewed in this manner, one can appreciate the importance of considering the glycome as a whole when studying glycan‐based interactions. Such clustered saccharide patches are proposed to involve multiple glycan components and their surroundings. Thus multiple copies of the same glycan or multiple copies of different glycans, or glycans and nonglycan structures including adjacent sulfates or peptide sequences on the same scaffold/carrier or on different ones, can participate to make up clustered saccharide patches. Indeed, clustered saccharide patches comprised of Sia and adjacent sulfates or peptide sequences were reported as ligands for selectins.70, 83, 84, 85
A similar analogy can be applied to the sialome, as a subset of the glycome (Figure 3).86 Five hierarchical levels of Sia structural and spatial complexities can be conceived of, in analogy to the leaves/flowers, stems, branches, trees and the forest, respectively. This theoretical concept is supported by accumulating experimental evidence.82 For example studies of Ab binding to Sia‐containing ligands revealed that in certain cases, a complex of several sialylated glycans would generate specific binding, despite the fact that a single glycan from the complex was not adequate for recognition.87, 88 Another example involves the “glycosynapse,” where Sia epitopes of the glycans in the cellular surface microdomains play an important functional role. The Sia epitopes cluster with other components in the microdomains and together they mediate cell adhesion and signaling to influence the cellular phenotypes.89 Sia‐containing clustered saccharide patches on RBCs of different blood types can also explain the differential binding of Sia‐recognizing proteins in relation to ABO blood group polymorphisms in humans.90 Furthermore, evidence for Siglec or Ab recognition of bacterial capsular polysaccharides exists, indicating the presence of Sia‐containing clustered saccharide patches on pathogens.91, 92
Figure 3.
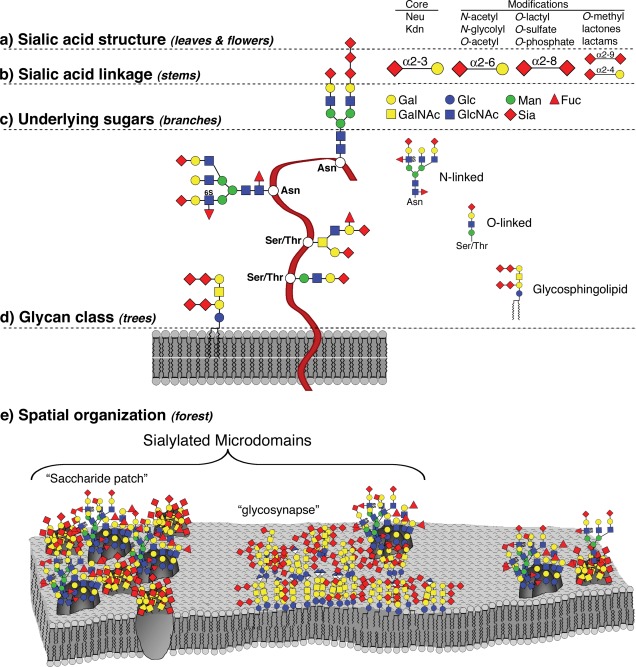
Analogy of the sialome to the canopy of a forest. Five hierarchical levels of structural and organizational complexities are compared. Sia structures are assimilated with tree leaves and flowers (a); Sia linkages are compared to tree stems (b); Underlying glycans are like branches of trees (c); Glycan classes are compared with the tree trunks (d); and finally the spatial organization of glycans and their surroundings are assimilated to the entire forest (e). Figure reproduced with permission.86
SIALIC ACID RECOGNIZING PROTEINS
Many functional roles of glycans are realized or modulated by interactions with glycan‐binding proteins (GBPs).93, 94, 95 Glycan–glycan interactions also play a critical role in biological processes such as cell–cell recognition and membrane organization, but that aspect will not be discussed here. Interested readers are referred to relevant reviews.96, 97
The variety and importance of Sias as ligands in glycan–protein recognition phenomena were extensively reviewed as well.98, 99, 100, 101, 102 A large number of Sia‐binding proteins have already been identified in viruses, bacteria, plants, invertebrates and vertebrates, and this number continues to grow. Besides exploring their occurrence and identifying their cognate ligands, special emphasis has been placed on studying Sia‐binding proteins from an evolutionary point of view, together with their biological significance in human health and disease, especially in immunity, infectious disease and cancer biology.14, 23, 103, 104 In addition to Sia‐binding lectins, Abs that recognize Sias have attracted considerable research interest, and the discovery of cancer biomarkers involving Sia and Sia‐recognizing Abs has become an active research area.46, 105, 106 For example, co‐existence of Neu5Gc and anti‐Neu5Gc Abs in human body represents a unique human phenomenon.38, 39, 40, 41, 43, 44, 46 Another class of Sia‐recognizing proteins are Sia‐recognizing enzymes including sialyltransferases and sialidases. These enzymes are essential to the chemistry and biology of Sias.102 The extensive literature on this subject is not reviewed here. However, examples of their use in sialochemistry will be presented in the following section.
SYNTHESIS OF SIALOGLYCANS BEARING NATURAL AND UNNATURAL SIAS
In nature, the majority of modifications on Sias are added after the sialoglycosidic bond formation (so called carbohydrate postglycosylational modifications or PGMs).107 One clear exception is the formation of Neu5Gc from Neu5Ac, which takes place at the CMP‐Sia level.108, 109, 110 Another exception is the formation of O‐acetylated Neu5Ac‐containing capsular polysaccharides, possibly by direct transfer of O‐acetylated Neu5Ac from O‐acetylated CMP‐Neu5Ac.111 It is also likely that one form of mammalian O‐acetylation takes place at the CMP‐Sia level.112 The gene sequences for most of the enzymes responsible for adding modifications to the Sia residues in sialoglycans have not been identified to date. The few exceptions include an O‐acetyltransferase from Campylobacter jejuni,113 an O‐acetyltransferase for the capsular polysaccharide of Neisseria meningitidis serogroups C, W‐135, and Y,79, 114 and a candidate for a vertebrate enzyme.115 Therefore, it is impractical to synthesize the majority of sialosides containing naturally occurring postglycosylational modifications in vitro, by totally following natural biosynthetic pathways. Among various chemical and chemoenzymatic methods developed for the synthesis of sialoglycans bearing natural and unnatural Sias, the one‐pot multienzyme (OPME) chemoenzymatic sialylation method116 is the most efficient for generating libraries of sialosides102 that can be used for sialoglycan microarray studies. In this approach, naturally occurring and non‐natural Sia forms, or their six‐carbon precursors such as derivatives of N‐acetylmannosamine (ManNAc) or mannose, can be synthesized chemically or enzymatically. These can then be used as substrates, by a CMP‐Sia synthetase and a suitable sialyltransferase in the absence or the presence of a Sia aldolase, for the synthesis of desired sialoglycans. Diverse sialosides can be synthesized due to unusual promiscuities of sialyltransferases and other related sialoside biosynthetic enzymes, including Sia aldolases and CMP‐Sia synthetases, toward substrate modifications. Obtaining a broad array of sialyltransferases,117, 118, 119, 120 CMP‐Sia synthetases,121, 122 and Sia aldolases,122, 123 with high expression levels, good solubility and high activities from bacterial species has made the preparative‐scale and even large‐scale preparation of comprehensive libraries of sialoglycans a reality. Designing enzyme mutants based on protein crystal structures also enables the expansion of the available sialoglycan products and renders the synthetic and purification schemes more efficient.124, 125, 126
STUDIES OF SIA DIVERSITY AND BIOLOGY USING SIALOGLYCAN MICROARRAYS
As discussed above, interactions between glycans and GBPs underlie much of the biological significance of glycans. The advent of glycan microarrays has revolutionized the screening of GBP specificities and fueled the discovery of new GBPs, providing invaluable information in a high‐throughput manner. During the years after its introduction in 2002,127, 128 glycan microarray studies have extended by development of glycan libraries and immobilization chemistries, proof‐of‐principle demonstrations, discovery of novel GBPs and their binding specificities, as well as applications in vaccine/inhibitor identification and biomarker discoveries.127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137 Here, we focus on discussing the development of a subclass of such arrays, the sialoglycan microarrays, and their versatile utility in studying Sia diversity and biology.
Natural and Synthetic Sialoglycan Libraries
To date, one of the most widely used glycan libraries is the one developed by the Consortium for Functional Glycomics (CFG). It consists of over 760 glycan structures (610 mammalian glycans and 153 microbial glycans) of various types, including a total of around 170 Sia‐containing structures (Table 1) (http://www.functionalglycomics.org). However, quite a few Sia‐containing structures in this sialoglycan library have an overlapping or identical terminal di‐/tri‐/tetrasaccharide, some varying merely in spacer structures. Nevertheless, sialoglycan microarrays comprised of ∼50–90 of these structures have been readily produced and applied in various studies.138, 139, 140, 141, 142, 143 During the past few years, Paulson's group has been developing chemical and chemoenzymatic methods to create a large library of sialoside analogues (Table 1).144, 145, 146 In 2008, Blixt et al.144 utilized 9‐azido‐Neu5Ac intermediates and synthesized a library of 44 sialoside analogues bearing un‐natural acyl substituents at the C‐9 position, among which 16 were α2‐3‐linked and 28 α2‐6‐linked. In 2012, Rillahan et al. took advantage of click chemistry and further developed the idea of using a minimum quantity of synthetic sialoglycan analogues on microarrays to screen for high affinity Siglec‐binding ligands.145 This time, a considerably larger library was generated thanks to the facile copper catalyzed azide‐alkyne cycloaddition (CuAAC) chemistry. Besides modifications at the C‐9 position, C‐5 modified Sias were also produced. In total, 224 Sia‐analogues with either α2‐3 or α2‐6 linkages, and with lactose as underlying glycan, were produced. It is noteworthy that these compounds were printed on glycan microarrays for screening without column purifications, resulting in higher throughput than the previous effort.144 However, most of the Sias on these arrays are unnatural ones. Another sialoglycan library comes from the Feizi group, which consists of more than 700 naturally derived and then chemically tagged neoglycolipids.147 Among these, there are about 120 sialoglycans with different Sia forms and linkages, and underlying glycans (Table 1).148, 149, 150, 151 This library of sialoglycans differs from most others by the nature of glycan source, that is, they are derived from natural sources instead of chemically built as in most other cases. It also contains a few α2‐9 linked Sias that other major sialoglycan libraries lack.150 Our own labs have been studying the diversity and biology of naturally occurring Sias and are especially interested in the chemistry and biology of the nonhuman Sia molecule, Neu5Gc, as well as various Sia O‐acetylations (c.f., sections and relevant references above, and additional Refs. 30, 78, 152, 153, 154, 155, 156). A library of over 70 synthetic sialoglycans was thus produced, which is unique in that it contains various pairs of Neu5Ac and Neu5Gc counterparts bearing the same glycosidic linkages, underlying glycans and spacers, that is, matched structures differing only by a single oxygen atom. Moreover, many pairs were further diversified by 9‐O‐acetylation on the core Sia structures (Table 1).46, 157, 158, 159, 160 This library is thus well suited for comparing protein recognition properties between Neu5Ac‐ and Neu5Gc‐containing glycans, as well as for exploring the effects of Sia O‐acetylation on binding. Furthermore, the number of sialoglycans in this library is actively increasing. The Cummings and Smith group also prepared a sialoglycan library in collaboration with one of us. The library has 77 sialoglycans with 16 different Sia modifications based on three core Sia structures (Neu5Ac, Neu5Gc, and Kdn), either α2‐3 or α2‐6 linked, and four different underlying glycan structures (Table 1).158, 161, 162 Among the 16 modified Sias, 13 are naturally occurring and three have not yet been found in nature. This sialoglycan library expanded the Sia diversity formerly covered in glycan microarray systems by adding the methylated and lactylated Sia structures. The Bovin lab and Wong lab have two other libraries of sialoglycans, containing about 40 and 30 different structures, respectively. Both are synthetically produced, and most of them are Neu5Ac‐bearing structures (Table 1).163, 164, 165, 166 Besides monovalent sialoglycans, the Bovin lab also produced sialoglycans on PAA polymer backbones and with additional functionalities such as biotin or fluorescein labels. The Institute for Glycomics in Griffiths University also has a library of ∼20 sialoglycans.167
Table 1.
Major Sialoglycan Libraries Used in Sialoglycan Microarrays
| Research | Number of | Sia | Underlying | Immobilization | |||
|---|---|---|---|---|---|---|---|
| Group | Sialoglycans | Sources | Form | Linkage | Glycan | Chemistry | Refs |
| CFG | ∼170 natural Sia structures | Mainly synthetic | Mainly Neu5Ac, a few Neu5Gc and Kdn | α2‐3, α2‐6, α2‐8 | Multiple types | Mainly amine + NHS‐ester | http://www.functionalglycomics.org |
| Paulson | >260 sialoside analogues | Synthetic | 5/9‐substituted Neu analogues | α2‐3, α2‐6 | LacNAc/Lactose | Amine + NHS‐ester | 144–146 |
| Feizi | ∼120 natural Sia structures | Mainly naturally derived | Mainly Neu5Ac, a few Neu5Gc and Kdn | α2‐3, α2‐6, α2‐8, α2‐9 | Multiple types | Non‐covalent hydrophobic attachment | 148–151 |
| Varki and Chen | ∼70 natural Sia structures | Synthetic (Chen) | Pairs of Neu5Ac & Neu5Gc, w/ or w/o 9‐OAc, and Kdn | α2‐3, α2‐6, α2‐8 | Multiple types | Amine + epoxide | 46,157–160 |
| Cummings, Smith and Chen | ∼70 natural Sia structures | Synthetic (Chen) | Methylated, acetylated, and lactylated Neu5Ac/Gc, Kdn | α2‐3, α2‐6 | LacNAc/Lactose/NA2/LNnT | Amine + NHS‐ester | 158, 161, 162 |
| Bovin | ∼40 natural Sia structures | Synthetic | Mainly Neu5Ac, two Neu5Gc | α2‐3, α2‐6, α2‐8 | Multiple types | Amine + NHS‐ester | 163 |
| Wong | ∼30 natural Sia strucutres | Synthetic | Mainly Neu5Ac, three Neu5Gc | α2‐3, α2‐6, α2‐8 | Multiple types | Amine + NHS‐ester | 164–166 |
Current Platforms for Sialoglycan Microarrays and the Immobilization Chemistries
Based on the libraries discussed above, standard sialoglycan microarray platforms have been developed. Typically, functional group modified glass substrates are used to accommodate robotically printed microarrays that contain hundreds to thousands of tiny sialoglycan solution spots. After incubation to ensure proper attachment, the glycan microarrays are blocked and are then ready for high‐throughput screening studies. Many immobilization chemistries have been developed in the past decade.134, 135 Although elegant proof‐of‐principle studies regarding each chemistry have been extensively demonstrated, most of them have not yet been tested broadly for more advanced applications. In the case of sialoglycan microarrays, only three immobilization methods are widely used and applied for various studies (Table 1). These are chemistries involving reactions of an amine with NHS‐ester, or with epoxide, and noncovalent hydrophobic attachment. Occasionally, the high affinity noncovalent biotin and streptavidin interaction was utilized for immobilizing sialoglycans, but mostly in a multiwell plate format.157, 168, 169
Besides the standard sialoglycan microarrays, a few mini‐sialoglycan array platforms appeared to couple with other attractive techniques/technologies and provide interesting potential. In 2008, the Cheng group in collaboration with one of us reported a sialoglycan biosensor platform based on surface plasmon resonance (SPR) spectroscopy, to study Sia‐lectin interactions in a label‐free format and in real‐time.170 The biotin‐neutravidin interaction was used to immobilize four biotinylated Sias, Neu5Acα2‐3Lac, Kdnα2‐3Lac, Neu5Acα2‐6Lac, and Kdnα2‐6Lac on the sensor chip and the system was interrogated with a number of lectins. Interaction kinetics and affinity data were obtained, and the effects of Sia structure and linkage variations on lectin binding proved to be detectable by the system. In 2009, a follow‐up SPR imaging study was carried out for more detailed characterizations of the sensor surface chemistry and lectin interactions with additional techniques including fluorescence microscopy and atomic force microscopy (AFM).171 In 2011, the Flitsch group presented a platform to generate sialoglycans in situ on surfaces in an array format and the synthesis was monitored by MALDI‐TOF (matrix‐assisted laser desorption ionization time‐of‐flight) mass spectrometry (MS). After completion of the in situ chemoenzymatic reactions, the sialoglycan surfaces were directly used for studying interactions by cells bearing recombinant Siglecs.172 The authors demonstrated that a self‐assembled monolayer (SAM) surface was suitable for studying whole mammalian cell interactions with limited nonspecific interactions. Combination of glycan microarray and MALDI‐TOF techniques was also exploited to assess enzymatic activities and specificities of influenza neuraminidases from whole viruses.173 However, this study used different surfaces and immobilization chemistry. Their sialoglycan array took advantage of a DNA/DNA hybridization method and glass slides bearing microreactors were produced to anchor the surface DNA strand. In 2012, the Sun group reported two interesting developments involving sialoglycan microarrays.174, 175 In one case, two Neu5Ac‐containing glycopolymers were chemoenzymatically synthesized. The Sias were α2‐3‐ or α2‐6‐linked to underlying lactose, and the polymer scaffold was functionalized with an O‐cyanate group at the chain‐end for surface immobilization. The oligo‐Sia macroligands were immobilized to amine‐functionalized glass slides via amine‐O‐cyanate chemistry, leading to an oriented presentation of the Sia‐bearing polymer.174 The system was interrogated by a range of lectins, in the presence or absence of free Sia competitors. An SPR biosensor system was also developed based on the same immobilization chemistry to study lectin and influenza hemagglutinin (HA) binding to the attached sialoglycan polymers. The authors claim this system better mimics the 3D nature of cell surface Sia presentation and should provide considerable advantages for relevant studies. In the other study, Ma et al.175 fabricated a liposomal sialoglycan microarray by the Staudinger ligation method. Gangliosides GM1 and GM3 inserted liposomes were immobilized on glass slides and remained intact, after which lectin and bacterial toxin bindings were evaluated. This system was claimed to present sialoglycans on a surface mimicking native cell membranes, and could thus be deemed an important tool for various applications.
All of the above mentioned mini‐sialoglycan array platforms provide interesting additional functionality to the standard sialoglycan microarrays and show considerable promise. However, most of these examples included only a few sialoglycans, and some systems were designed in a well‐based array format. Thus, expansion of the sialoglycan libraries and miniaturization of the array spots in these systems are yet to be explored, so that truly high‐throughput evaluation of sialoglycan‐involved interactions can be realized.
Studying Sia‐Recognizing Proteins by Sialoglycan Microarrays
It is evident that Sia binding lectins can be found in many microbes, plants and animals.98, 99, 100, 101, 102 Studies of these Sia‐recognizing proteins by sialoglycan microarrays are rapidly growing. In part due to a joint effort between the CFG and the Centers for Disease Control (CDC), and partly because of the global public health concern/high importance of the topic, the proteins from influenza viruses of various origins, subtypes, clades, and strains have been extensively studied by sialoglycan microarrays. These include influenza virus HAs from various subtypes such as H5N1,138, 139, 141 H2N2,140, 143 H1N1,164, 165, 176, 177 and H3N2.164 As influenza virus HAs bind to Sias, a central aim for these screenings has been to find out whether the specificity could act as a pathogenic pandemic risk factor. The molecular basis for difference of HA ligand specificity between zoonotic influenza viruses and corresponding human‐adapted ones is being actively explored, however, clearer relevance between the HA ligand specificity and the risk of pathogenic pandemic is yet to be neatly demonstrated.
Wang et al.166 took a different approach to study the influenza HAs using sialoglycan microarrays. They examined the effects of HA glycosylation on its host receptor binding and found that removal of certain glycan structures on viral surface glycoproteins could result in higher‐affinity Abs elicited and better neutralization activities of these Abs. More recently, both the influenza HA homologue from bat influenza virus (H17N10) and the N10 neuraminidase‐like protein from the same origin were found to show no binding to ligands on the CFG sialoglycan microarrays.178, 179 In contrast, a neuraminidase mutant from human H3N2 influenza virus avidly bound to Sia ligands on the sialoglycan microarrays. The observed neuraminidase binding was of much higher affinity (μM range) compared to HA bindings (usually low mM).180 Other Sia‐binding proteins from different types of viruses have been studied by sialoglycan microarrays, including human JC polyomavirus, serotype 1 reovirus, bovine coronavirus and canine adenovirus,142, 151, 158, 181 as well as parasites like Toxoplasma gondii and Eimeria tenella.150, 182
The sialoglycan microarrays derived from more extensive glycan libraries (e.g., CFG and the Feizi group) have been used to study specific proteins or microbes directly. However, the two more recently established sialoglycan microarrays were tested extensively against a wide range of proteins and microbes, from common plant lectins to viruses.158, 161
Because of their important roles in human immunity,24, 103, 183, 184 Siglecs are another type of favored candidates for sialoglycan microarray studies.144, 145, 149, 157, 158 The Paulson lab and the Chen lab utilized different chemical and chemoenzymatic methods to generate relatively large libraries of unnatural and natural Sias in a high‐throughput manner. These libraries of Sias could be directly used for array fabrication and Siglec ligand identification.144, 145, 149, 157 A few of the unnatural Sia analogues generated by Rillahan et al.145 have proved to be of high affinity to a range of Siglecs. Besides the C‐5 and C‐9 position modifications on the Sia core structure, the Kelm lab very recently showed that modifications at the Sia C‐4 position could act synergistically with C‐9 modification and also enhance Siglec binding.185 Higher affinity Sia analogues could be potentially used as Siglec inhibitors or for targeted therapeutic delivery to treat, for example, B cell leukemia.145 However, Siglecs like CD22 are recently noted to be expressed not only on B cells, but also on dendritic cells and gastrointestinal eosinophils.186, 187 Thus, precautions and extra investigations are needed to successfully pursue this strategy.
Sialoglycan microarrays have also been used to identify cancer biomarkers. Taking advantage of its unique feature, the paired Neu5Gc/Neu5Ac sialoglycan microarray platform was utilized in our labs to study the unusual anti‐Neu5Gc Abs in humans.46 Sera from cancer and noncancer patients were characterized, and Abs against Neu5Gcα2‐6GalNAcα1‐O‐Ser/Thr (GcSTn) were found to be more prominently present in patients with carcinomas than with other diseases. Furthermore, the corresponding patient sera and purified polyclonal Abs which showed strong anti‐GcSTn reactivity both proved capable of killing human tumors expressing GcSTn, via either complement‐dependent cytotoxicity or Ab dependent cellular cytotoxicity.46 This study was followed up by a detailed LC‐MS analysis of the polyclonal human anti‐Neu5Gc Abs, and all four IgG subclasses of Abs were confirmed to present after the human immune response to the xeno‐autoantigen Neu5Gc.159 The same sialoglycan microarray platform was also used to detect anti‐Neu5Gc Ab responses in patients with Kawasaki Disease.160 Other glycan microarray platforms (including microarrays with various non‐Sia glycan epitopes), have also been used to study complex samples like human sera and for biomarker discoveries.106, 163, 188, 189, 190, 191 Aside from studying Sia‐recognizing lectins and Abs, sialoglycan microarrays can also be applied to measure enzymatic activities of various neuraminidases/sialidases.173, 192 In addition, sialyltransferase reactions have been monitored directly on glycan microarrays.172, 193
Studying Viruses, Bacteria, and Whole Mammalian Cells by Sialoglycan Microarrays
No matter what one finds using purified glycan‐binding proteins, there is always the possibility that the binding specificity will be different when studying the intact organism that expresses the same protein. Thus there is a need to study interactions of intact organisms with arrays. To date, the most studied examples are influenza viruses. Receptor specificities of human, avian, and porcine influenza viruses have been examined and comparisons among these strains have yielded insights into the molecular basis for their receptor specificity, transmissibility, as well as virulence.139, 141, 148, 161, 165, 194, 195, 196, 197, 198 In most of these studies, intact or biotinylated viruses were applied to the sialoglycan microarrays and detected by fluorochrome‐labeled virus‐binding Abs or streptavidin‐fluorochromes, respectively. However, fluorochrome‐conjugated viruses could be directly used for glycan microarray studies.199 Other types of viruses were also studied by sialoglycan microarrays. In 2007, a neoglycolipid microarray was used to study receptor specificity of simian virus 40 (SV40), finding that the N‐glycolyl GM1 ganglioside was a preferred receptor.200 Cell studies and molecular modeling further supported the finding. Preferential binding to Neu5Gc over Neu5Ac was also observed for some other viruses and bacterial toxins.53, 201, 202, 203 There are many more such proteins and microbes that await exploration. The potential impact of this type of preferential binding in host–pathogen interactions on human health and disease warrants further research, and sialoglycan microarrays containing pairs of Neu5Gc and Neu5Ac glycans will serve as a novel platform for such investigations.
Surprisingly, there are very few reports to date on studying intact bacteria using sialoglycan microarrays. In 2004, Seeberger and coworkers tested Escherichia coli binding on a different type of glycan microarray comprised of mannose (Man), glucose (Glc), N‐acetyl‐glucosamine (GlcNAc), galactose (Gal), or fucose (Fuc) structures. Unexpectedly, the printed glycan microarrays with oligomannose structures showed no increase in bacterial binding capacity compared to mono‐mannose arrays.204 The authors attributed this phenomenon to the possibility that the tested bacterial stain only required a single mannose residue for recognition and the multivalency of mannose and stereochemistry of the intermannose linkages played little role. However, a more recent study from the same group showed that trimannose and nonamannose structures attached to a cantilever array sensor tip did show differential binding affinities to the same E. coli strain tested earlier, indicating an increased multisite and multivalent binding for the nonamannose structure compared to less complex mannose structures.205 Taken together, these studies showed a need to improve the glycan microarray platform for studying intact bacteria. In this regard we have recently embarked on using sialoglycan microarrays to investigate a group of Gram‐positive bacteria (Deng and Varki, et al., unpublished observations). Bacterial adhesins/mutants and corresponding intact isogenic strains are being tested on printed sialoglycan microarrays, and after careful optimization, the data show interesting correlations between ligand spectra/affinity and virulence. The effect of shear force on bacterial binding also needs investigation.
Testing whole mammalian cells on sialoglycan microarrays is also an emerging endeavor. Although whole cell bindings were demonstrated in a millimeter‐spot‐sized microarray system as early as in 2004,206 robotically printed micrometer‐sized glycan arrays have not been used to study whole mammalian cells until very recently.145, 167 Rillahan et al.145 demonstrated binding of Siglec‐bearing Chinese hamster ovary (CHO) cells and CD22‐expressing human B‐cells on their Sia analogue microarrays. In another report, Arndt et al.167 tested a range of human cancer cell lines directly on their glycan microarrays (various types of glycans, not completely sialoglycans) and on a lectin microarray, and they found an inverse relationship between how many glycans the tested cells could recognize and how many types of intrinsic glycans were expressed on those cells. This study also pointed to the importance of characterizing/evaluating cell surface glycosylation status when using those common laboratory cell lines for cell‐based assays. Well‐plate based glycan arrays have also been developed for studies of glycan‐cell interactions recently.172, 207 Initial efforts to pair the screening of binding specificity of secreted proteins with corresponding protein‐bearing cells are currently undertaken as well.208
Decoding Clustered Saccharide Patches by Sialoglycan Microarrays
Current glycan microarray platforms are usually composed of individual spots each displaying a single glycan structure. However, cell surfaces present glycans in a highly heterogeneous fashion. Thus there is also a need to try to explore glycan–protein interactions in a manner more similar to the natural state. Although difficult to prove conclusively, the hypothesis of clustered saccharide patches is worth testing.67, 82, 86, 209 The information gained could further enhance our understanding of glycan–protein interactions and improve drug design strategies. This concept can be to some extent tested using glycan microarrays.
In 2010, Wu and coworkers210 published a glycan microarray design of such a nature. They spotted the glass slides with mixed glycans, for example, SSEA4/Gb5, Globo H/Gb5, Gb4/Gb5, Gb3/Gb5, Gb2/Gb5, Bb2/Gb5, in 1:1 molar ratio and interrogated the slides with anti‐Gb5 Ab. Interestingly, the SSEA4/Gb5 mixed glycan spot consistently showed higher binding than the Gb5‐alone glycan spot. On the other hand, neighboring glycans could also exert a negative effect on the Gb5 and anti‐Gb5 interaction, possibly via steric hindrance, as evidenced by lower binding of the other mixed glycan spots compared to the Gb5‐alone spot. Further tests were done by mixing glycans in varied concentrations and by using synthetic oligomannose dendrimers containing different ratios of Man4 and Man9 structures. Both experiments demonstrated Ab binding could be affected by the density and structures of neighboring glycans. In parallel, the Willison lab combined a few techniques and recently produced polyvinylidene difluoride membranes (PVDM) affixed glass slides for the generation of sialoglycan (ganglioside) microarrays. They called this setup a “combinatorial glycoarray,” and used it primarily to study neuropathy‐associated anti‐glycolipid autoantibodies.211, 212 These Abs specifically recognize complexes of glycolipid pairs, but fail to interact with either of the glycolipids alone. Other exciting developments include that relatively stable lipid bilayer coated glycan microarray slides have been readily made. This is promising due to its better mimicry of the cell surface features.213, 214 It can be envisaged that this type of surface may serve as an ideal platform for decoding the effects of clustered saccharide patches on protein and cell bindings.
COMPARISONS OF SIALOGLYCAN MICROARRAY PLATFORMS
In the DNA microarray field, the microarray quality control (MAQC) consortium has been established and major efforts toward standardization of processing and reporting microarray data have been made.215, 216 As the number of new glycan microarray platforms continues to grow and applications using these platforms expand, the need for cross‐comparison among platforms and established guidelines for glycan microarray experiments has become increasingly relevant and urgent.
In 2012, two labs took the initiative and cross‐compared their newly developed sialoglycan microarray platforms, both developed using the same chemistries.158 These two glycan microarrays presented comparable sialoglycans produced by the same synthetic strategy from the same group. However, the immobilization chemistries used for attaching the sialoglycans on the two platforms were different, and the linkage monosaccharide ring was opened in one of the two arrays. These microarrays were reciprocally tested against various Sia‐binding proteins and analyzed in the two labs.158 This comparison yielded a lot of useful information but also identified major challenges in the standardization of glycan microarray experiments. For instance, immobilization methods appeared to strongly influence some binding results. Also, the close‐ring immobilization method was evidently critical for some glycan‐recognition events, and an open‐ring immobilization could sometimes lead to the absence of detectable binding. This is in keeping with another intragroup comparison of glycan microarrays differing only in the immobilization method, where it was found that the ring‐closed immobilization method was critical for many glycan‐recognition events.217 Lower signal‐to‐noise ratio bindings based on open‐ring immobilization method was also observed in another independent array comparison study.162 Besides these, binding data with various glycans from the same source but arrayed at different locations were also compared and reported.218 More recently, a larger scale glycan microarray comparison involving five different research groups has been carried out (personal communication from Dr. Lara K. Mahal, NYU).
CONCLUSIONS AND PERSPECTIVES
The diversity and biology of Sias represent part of the vast information and knowledge gained in the field of glycosciences. It is evident that sialoglycan microarrays are playing an increasingly important role in elucidating this diversity and biology. During the past decade, the advancement of glycan microarray development and applications is indeed highly laudable and exemplary; however, there is still plenty of room for further developments and improvements.
Larger libraries of sialoglycans, consisting of either natural structures or synthetic analogues, are still greatly desired. Just by comparing currently available sialoglycan libraries and the diversity of Sia structures occurring in nature12 one can realize how big the gap is, let alone the unlimited possibilities of synthetic Sia analogues. For example, it will be interesting to see sialoglycan microarrays made of 4‐O‐acetylated Sias or polySias. These structures are largely un‐explored by sialoglycan microarrays. In addition, current efforts are mainly focused on elucidating binding of terminal glycan sequences. However, binding effects resulting from intact natural sialoglycan classes and underlying peptides/lipids have to be further evaluated.
Sialoglycan microarrays have shown to be a suitable platform to demonstrate the concept of clustered saccharide patches. Further studies such as mixing more components in a single glycan spot, controlling spacing between glycans in the spot, and testing their effects on protein binding, are worthwhile.
Ligand specificity of many more Sia‐recognizing proteins, viruses, bacteria and whole mammalian cells are worthy of investigation in detail.98, 99, 100, 101, 102 This would require more extensive collaborations between glycoscientists (chemists and biologists) and researchers from other fields, such as microbiologists. The joint effort will certainly benefit all parties involved.
To date, more than 20 different types of immobilization methods for glycan microarray fabrication have been developed.134, 135 For example, the immobilization of glycans by photochemistry is a highly versatile and efficient method.219 This can be realized by producing photoprobe‐derivatized glycans,220 direct attachment of underivatized glycans,221 or a “photo‐click” hybrid method.222 Photogenerated glycan microarrays have also been successfully applied to identify immunogenic glycan antigens specific to certain bacterium.223 However, most of the immobilization methods developed to date were only demonstrated in proof‐of‐concept type of studies. The exceptional versatility and efficiency of the photochemical immobilization methods warrant further development and broader applications. Cross‐comparison of these immobilization methods and insight into their merits and disadvantages are needed for further microarray standardization and applications.
Technical issues such as Sia contamination of the most widely used glycan microarray blocking reagent bovine serum albumin (BSA) (our observations, unpublished result), and sample dilution effects,224 and so forth, are also important considerations in conducting glycan microarray experiments and analyzing glycan microarray data.
Initial efforts have been undertaken, but the cross‐comparison and standardization of sialoglycan microarray experiments and microarray data processing has a long way to go. Guiding rules for the interpretation of glycan microarray data are also to be established. Moreover, building common glycan microarray databases and further enabling relevant glycobioinformatics would be very helpful and beneficial to the entire glycoscience community.225
Combination of glycan microarrays with other methods and techniques will, on the one hand, help validate the glycan microarray data, and on the other hand, often add another dimension to the findings. For example, real‐time monitoring of the binding events and additional binding kinetics data can be obtained with biosensors like SPR220, 221 and quartz crystal microbalance (QCM).222, 226 Isothermal titration calorimetry (ITC) can provide thermodynamic information,227 and detailed single molecular interaction data can be acquired by using AFM.228 Combination of microarrays with nanotechnology can greatly enhance the sensitivity of detection.229, 230 Furthermore, comparisons with other assays like enzyme‐linked immunosorbent assay (ELISA) and dot blots,231, 232 and combination with MS, computational modeling, X‐ray crystallography, etc., have been and will continue to play major roles in the elucidation of glycan–protein interactions.
After glycan microarray screening of glycan–protein interactions and complementary methods to validate the bindings, follow‐up biomedical and/or biological studies are critical. These will ultimately move the initial findings into more clinically and/or biologically relevant and more profound discoveries/applications to combat diseases and benefit human health.
This review focused on studies of Sia diversity and biology by using sialoglycan microarrays, but the concepts, trends, and ideas discussed in this incomplete survey are readily applicable to other types of glycans and glycan microarrays.
ACKNOWLEDGMENTS
The authors thank Miriam Cohen, Jerry Fong, and Stevan Springer for helpful comments.
This article was originally published online as an accepted preprint. The “Published Online” date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com
References
- 1. Marth, J. D. Nat Cell Biol 2008, 10, 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Varki, A. Cold Spring Harb Perspect Biol 2011, 3, 1–14; doi:pii: a005462. 10.1101/cshperspect.a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varki, A. ; Cummings, R. D. ; Esko, J. D. ; Freeze, H. H. ; Stanley, P. ; Bertozzi, C. R. ; Hart, G. W. ; Etzler, M. E. Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: Plainview, NY, 2009. [PubMed] [Google Scholar]
- 4. Hart, G. W. ; Copeland, R. J. Cell 2010, 143, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. National Research Council , Transforming Glycoscience: A Roadmap for the Future; The National Academies Press: Washington, DC, 2012. [PubMed] [Google Scholar]
- 6. Schauer, R. Adv Carbohydr Chem Biochem 1982, 40, 131–234. [DOI] [PubMed] [Google Scholar]
- 7. Troy, F. A. Glycobiology 1992, 2, 5–23. [DOI] [PubMed] [Google Scholar]
- 8. Varki, A. Glycobiology 1992, 2, 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kelm, S. ; Schauer, R. Int Rev Cytol 1997, 175, 137–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zanetta, J. P. ; Pons, A. ; Iwersen, M. ; Mariller, C. ; Leroy, Y. ; Timmerman, P. ; Schauer, R. Glycobiology 2001, 11, 663–676. [DOI] [PubMed] [Google Scholar]
- 11. Schauer, R. Glycoconjugate J 2000, 17, 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angata, T. ; Varki, A. Chem Rev 2002, 102, 439–469. [DOI] [PubMed] [Google Scholar]
- 13. Varki, A. ; Schauer, R. Essentials of Glycobiology, 2009; pp 199–218. [Google Scholar]
- 14. Varki, A. Cell 2006, 126, 841–845. [DOI] [PubMed] [Google Scholar]
- 15. Bishop, J. R. ; Gagneux, P. Glycobiology 2007, 17, 23R–34R. [DOI] [PubMed] [Google Scholar]
- 16. Springer, S. A. ; Gagneux, P. J Biol Chem 2013, 288, 6904–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vimr, E. R. ; Kalivoda, K. A. ; Deszo, E. L. ; Steenbergen, S. M. Microbiol Mol Biol Rev 2004, 68, 132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knirel, Y. A. ; Shashkov, A. S. ; Tsvetkov, Y. E. ; Jansson, P. E. ; Zahringer, U. Adv Carbohydr Chem Biochem 2003, 58, 371–417. [DOI] [PubMed] [Google Scholar]
- 19. Lewis, A. L. ; Desa, N. ; Hansen, E. E. ; Knirel, Y. A. ; Gordon, J. I. ; Gagneux, P. ; Nizet, V. ; Varki, A. Proc Natl Acad Sci USA 2009, 106, 13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watson, D. C. ; Leclerc, S. ; Wakarchuk, W. W. ; Young, N. M. Glycobiology 2011, 21, 99–108. [DOI] [PubMed] [Google Scholar]
- 21. Lukowsky, W. A. ; Painter, R. H. Can J Biochem 1972, 50, 909–917. [DOI] [PubMed] [Google Scholar]
- 22. Schauer, R. Trends Biochem Sci 1985, 10, 357–360. [Google Scholar]
- 23. Varki, A. ; Gagneux, P. Ann N Y Acad Sci 2012, 1253, 16–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varki, A. ; Angata, T. Glycobiology 2006, 16, 1R–27R. [DOI] [PubMed] [Google Scholar]
- 25. Schauer, R. ; Srinivasan, G. V. ; Wipfler, D. ; Kniep, B. ; Schwartz‐Albiez, R. Adv Exp Med Biol 2011, 705, 525–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schauer, R. Curr Opin Struct Biol 2009, 19, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suzuki, Y. Biol Pharm Bull 2005, 28, 399–408. [DOI] [PubMed] [Google Scholar]
- 28. de Groot, R. J. Glycoconj J 2006, 23, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weiman, S. ; Dahesh, S. ; Carlin, A. F. ; Varki, A. ; Nizet, V. ; Lewis, A. L. Glycobiology 2009, 19, 1204–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weiman, S. ; Uchiyama, S. ; Lin, F. Y. ; Chaffin, D. ; Varki, A. ; Nizet, V. ; Lewis, A. L. Biochem J 2010, 428, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelm, A. ; Shaw, L. ; Schauer, R. ; Reuter, G. Eur J Biochem 1998, 251, 874–884. [DOI] [PubMed] [Google Scholar]
- 32. Varki, A. Proc Natl Acad Sci USA 2010, 107 (Suppl 2), 8939–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Irie, A. ; Koyama, S. ; Kozutsumi, Y. ; Kawasaki, T. ; Suzuki, A. J Biol Chem 1998, 273, 15866–15871. [DOI] [PubMed] [Google Scholar]
- 34. Chou, H. H. ; Takematsu, H. ; Diaz, S. ; Iber, J. ; Nickerson, E. ; Wright, K. L. ; Muchmore, E. A. ; Nelson, D. L. ; Warren, S. T. ; Varki, A. Proc Natl Acad Sci USA 1998, 95, 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chou, H. H. ; Hayakawa, T. ; Diaz, S. ; Krings, M. ; Indriati, E. ; Leakey, M. ; Paabo, S. ; Satta, Y. ; Takahata, N. ; Varki, A. Proc Natl Acad Sci USA 2002, 99, 11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tangvoranuntakul, P. ; Gagneux, P. ; Diaz, S. ; Bardor, M. ; Varki, N. ; Varki, A. ; Muchmore, E. Proc Natl Acad Sci USA 2003, 100, 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Banda, K. ; Gregg, C. J. ; Chow, R. ; Varki, N. M. ; Varki, A. J Biol Chem 2012, 287, 28852–28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Padler‐Karavani, V. ; Yu, H. ; Cao, H. ; Chokhawala, H. ; Karp, F. ; Varki, N. ; Chen, X. ; Varki, A. Glycobiology 2008, 18, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor, R. E. ; Gregg, C. J. ; Padler‐Karavani, V. ; Ghaderi, D. ; Yu, H. ; Huang, S. ; Sorensen, R. U. ; Chen, X. ; Inostroza, J. ; Nizet, V. ; Varki, A. J Exp Med 2010, 207, 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malykh, Y. N. ; Schauer, R. ; Shaw, L. Biochimie 2001, 83, 623–634. [DOI] [PubMed] [Google Scholar]
- 41. Nguyen, D. H. ; Tangvoranuntakul, P. ; Varki, A. J Immunol 2005, 175, 228–236. [DOI] [PubMed] [Google Scholar]
- 42. Hedlund, M. ; Tangvoranuntakul, P. ; Takematsu, H. ; Long, J. M. ; Housley, G. D. ; Kozutsumi, Y. ; Suzuki, A. ; Wynshaw‐Boris, A. ; Ryan, A. F. ; Gallo, R. L. ; Varki, N. ; Varki, A. Mol Cell Biol 2007, 27, 4340–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hedlund, M. ; Padler‐Karavani, V. ; Varki, N. M. ; Varki, A. Proc Natl Acad Sci USA 2008, 105, 18936–18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pham, T. ; Gregg, C. J. ; Karp, F. ; Chow, R. ; Padler‐Karavani, V. ; Cao, H. ; Chen, X. ; Witztum, J. L. ; Varki, N. M. ; Varki, A. Blood 2009, 114, 5225–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghaderi, D. ; Taylor, R. E. ; Padler‐Karavani, V. ; Diaz, S. ; Varki, A. Nat Biotechnol 2010, 28, 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Padler‐Karavani, V. ; Hurtado‐Ziola, N. ; Pu, M. ; Yu, H. ; Huang, S. ; Muthana, S. ; Chokhawala, H. A. ; Cao, H. ; Secrest, P. ; Friedmann‐Morvinski, D. ; Singer, O. ; Ghaderi, D. ; Verma, I. M. ; Liu, Y. T. ; Messer, K. ; Chen, X. ; Varki, A. ; Schwab, R. Cancer Res 2011, 71, 3352–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park, J. Y. ; Park, M. R. ; Bui, H. T. ; Kwon, D. N. ; Kang, M. H. ; Oh, M. ; Han, J. W. ; Cho, S. G. ; Park, C. ; Shim, H. ; Kim, H. M. ; Kang, M. J. ; Park, J. K. ; Lee, J. W. ; Lee, K. K. ; Kim, J. H. Cell Reprogram 2012, 14, 353–363. [DOI] [PubMed] [Google Scholar]
- 48. Padler‐Karavani, V. ; Varki, A. Xenotransplantation 2011, 18, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yeh, P. ; Ezzelarab, M. ; Bovin, N. ; Hara, H. ; Long, C. ; Tomiyama, K. ; Sun, F. ; Ayares, D. ; Awwad, M. ; Cooper, D. K. Xenotransplantation 2010, 17, 197–206. [DOI] [PubMed] [Google Scholar]
- 50. Basnet, N. B. ; Ide, K. ; Tahara, H. ; Tanaka, Y. ; Ohdan, H. Xenotransplantation 2010, 17, 440–448. [DOI] [PubMed] [Google Scholar]
- 51. Ezzelarab, M. ; Ayares, D. ; Cooper, D. K. Immunol Cell Biol 2005, 83, 396–404. [DOI] [PubMed] [Google Scholar]
- 52. Miwa, Y. ; Kobayashi, T. ; Nagasaka, T. ; Liu, D. ; Yu, M. ; Yokoyama, I. ; Suzuki, A. ; Nakao, A. Xenotransplantation 2004, 11, 247–253. [DOI] [PubMed] [Google Scholar]
- 53. Byres, E. ; Paton, A. W. ; Paton, J. C. ; Lofling, J. C. ; Smith, D. F. ; Wilce, M. C. ; Talbot, U. M. ; Chong, D. C. ; Yu, H. ; Huang, S. ; Chen, X. ; Varki, N. M. ; Varki, A. ; Rossjohn, J. ; Beddoe, T. Nature 2008, 456, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paulson, J. C. ; Sadler, J. E. ; Hill, R. L. J Biol Chem 1979, 254, 2120–2124. [PubMed] [Google Scholar]
- 55. Daniels, P. S. ; Jeffries, S. ; Yates, P. ; Schild, G. C. ; Rogers, G. N. ; Paulson, J. C. ; Wharton, S. A. ; Douglas, A. R. ; Skehel, J. J. ; Wiley, D. C. EMBO J 1987, 6, 1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chandrasekaran, A. ; Srinivasan, A. ; Raman, R. ; Viswanathan, K. ; Raguram, S. ; Tumpey, T. M. ; Sasisekharan, V. ; Sasisekharan, R. Nat Biotechnol 2008, 26, 107–113. [DOI] [PubMed] [Google Scholar]
- 57. Nicholls, J. M. ; Chan, R. W. ; Russell, R. J. ; Air, G. M. ; Peiris, J. S. Trends Microbiol 2008, 16, 149–157. [DOI] [PubMed] [Google Scholar]
- 58. Imai, M. ; Kawaoka, Y. Curr Opin Virol 2012, 2, 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gagneux, P. ; Cheriyan, M. ; Hurtado‐Ziola, N. ; Brinkman Van Der Linden, E. C. ; Anderson, D. ; McClure, H. ; Varki, A. ; Varki, N. M. J Biol Chem 2003, 278, 48245–48250. [DOI] [PubMed] [Google Scholar]
- 60. Powell, L. D. ; Sgroi, D. ; Sjoberg, E. R. ; Stamenkovic, I. ; Varki, A. J Biol Chem 1993, 268, 7019–7027. [PubMed] [Google Scholar]
- 61. Kelm, S. ; Schauer, R. ; Manuguerra, J.-C. ; Gross, H.-J. ; Crocker, P. R. Glycoconjugate J 1994, 11, 576–585. [DOI] [PubMed] [Google Scholar]
- 62. Seales, E. C. ; Shaikh, F. M. ; Woodard‐Grice, A. V. ; Aggarwal, P. ; McBrayer, A. C. ; Hennessy, K. M. ; Bellis, S. L. J Biol Chem 2005, 280, 37610–37615. [DOI] [PubMed] [Google Scholar]
- 63. Seales, E. C. ; Jurado, G. A. ; Brunson, B. A. ; Wakefield, J. K. ; Frost, A. R. ; Bellis, S. L. Cancer Res 2005, 65, 4645–4652. [DOI] [PubMed] [Google Scholar]
- 64. Zhuo, Y. ; Chammas, R. ; Bellis, S. L. J Biol Chem 2008, 283, 22177–22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hedlund, M. ; Ng, E. ; Varki, A. ; Varki, N. M. Cancer Res 2008, 68, 388–394. [DOI] [PubMed] [Google Scholar]
- 66. Schultz, M. J. ; Swindall, A. F. ; Bellis, S. L. Cancer Metastasis Rev 2012, 31, 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Varki, A. Proc Natl Acad Sci USA 1994, 91, 7390–7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McEver, R. P. ; Moore, K. L. ; Cummings, R. D. J Biol Chem 1995, 270, 11025–11028. [DOI] [PubMed] [Google Scholar]
- 69. Rosen, S. D. ; Bertozzi, C. R. Curr Biol 1996, 6, 261–264. [DOI] [PubMed] [Google Scholar]
- 70. McEver, R. P. ; Cummings, R. D. J Clin Invest 1997, 100, S97–103. [PubMed] [Google Scholar]
- 71. Powell, L. D. ; Jain, R. K. ; Matta, K. L. ; Sabesan, S. ; Varki, A. J Biol Chem 1995, 270, 7523–7532. [DOI] [PubMed] [Google Scholar]
- 72. Troy, F. Annu Rev Microbiol 1979, 33, 519–560. [DOI] [PubMed] [Google Scholar]
- 73. Finne, J. ; Finne, U. ; Deagostini‐Bazin, H. ; Goridis, C. Biochem Biophys Res Commun 1983, 112, 482–487. [DOI] [PubMed] [Google Scholar]
- 74. Muhlenhoff, M. ; Eckhardt, M. ; Gerardy‐Schahn, R. Curr Opin Struct Biol 1998, 8, 558–564. [DOI] [PubMed] [Google Scholar]
- 75. Inoue, Y. ; Inoue, S. Pure Appl Chem 1999, 71, 789–800. [Google Scholar]
- 76. Rutishauser, U. Nat Rev Neurosci 2008, 9, 26–35. [DOI] [PubMed] [Google Scholar]
- 77. Claus, H. ; Stummeyer, K. ; Batzilla, J. ; Muhlenhoff, M. ; Vogel, U. Mol Microbiol 2009, 71, 960–971. [DOI] [PubMed] [Google Scholar]
- 78. Lewis, A. L. ; Nizet, V. ; Varki, A. Proc Natl Acad Sci USA 2004, 101, 11123–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li, Y. ; Chen, X. Appl Microbiol Biotechnol 2012, 94, 887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kakehi, K. ; Kinoshita, M. ; Kitano, K. ; Morita, M. ; Oda, Y. Electrophoresis 2001, 22, 3466–3470. [DOI] [PubMed] [Google Scholar]
- 81. Zhang, Y. ; Lee, Y. C. J Biol Chem 1999, 274, 6183–6189. [DOI] [PubMed] [Google Scholar]
- 82. Cohen, M. ; Varki, A. Int Rev Cell Mol Biol, in press. [DOI] [PubMed] [Google Scholar]
- 83. Leppanen, A. ; Parviainen, V. ; Ahola‐Iivarinen, E. ; Kalkkinen, N. ; Cummings, R. D. Glycobiology 2010, 20, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang, J. ; Shiratori, I. ; Satoh, T. ; Lanier, L. L. ; Arase, H. J Immunol 2008, 180, 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sun, Y. ; Senger, K. ; Baginski, T. K. ; Mazloom, A. ; Chinn, Y. ; Pantua, H. ; Hamidzadeh, K. ; Ramani, S. R. ; Luis, E. ; Tom, I. ; Sebrell, A. ; Quinones, G. ; Ma, Y. ; Mukhyala, K. ; Sai, T. ; Ding, J. ; Haley, B. ; Shadnia, H. ; Kapadia, S. B. ; Gonzalez, L. C. ; Hass, P. E. ; Zarrin, A. A. J Biol Chem 2012, 287, 15837–15850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cohen, M. ; Varki, A. OMICS 2010, 14, 455–464. [DOI] [PubMed] [Google Scholar]
- 87. Kaida, K. ; Kanzaki, M. ; Morita, D. ; Kamakura, K. ; Motoyoshi, K. ; Hirakawa, M. ; Kusunoki, S. J Neurol Neurosurg Psychiatry 2006, 77, 1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kusunoki, S. ; Kaida, K. J Neurochem 2011, 116, 828–832. [DOI] [PubMed] [Google Scholar]
- 89. Hakomori Si, S. I. Proc Natl Acad Sci USA 2002, 99, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cohen, M. ; Hurtado‐Ziola, N. ; Varki, A. Blood 2009, 114, 3668–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Carlin, A. F. ; Lewis, A. L. ; Varki, A. ; Nizet, V. J Bacteriol 2007, 89, 1231–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pincus, S. H. ; Moran, E. ; Maresh, G. ; Jennings, H. J. ; Pritchard, D. G. ; Egan, M. L. ; Blixt, O. Vaccine 2012, 30, 4849–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Esko, J. D. ; Sharon, N. 2009, Essentials of Glycobiology, 489–500. [Google Scholar]
- 94. Cummings, R. D. ; Esko, J. D. 2009, Essentials of Glycobiology, 387–402. [PubMed] [Google Scholar]
- 95. Varki, A. ; Etzler, M. E. ; Cummings, R. D. ; Esko, J. D. Essentials of Glycobiology, 2009; pp 375–386. [PubMed] [Google Scholar]
- 96. Hakomori, S. Glycoconj J 2004, 21, 125–137. [DOI] [PubMed] [Google Scholar]
- 97. Bucior, I. ; Burger, M. M. Glycoconj J 2004, 21, 111–123. [DOI] [PubMed] [Google Scholar]
- 98. Mandal, C. Experientia 1990, 46, 433–441. [DOI] [PubMed] [Google Scholar]
- 99. Varki, A. FASEB J 1997, 11, 248–255. [DOI] [PubMed] [Google Scholar]
- 100. Lehmann, F. ; Tiralongo, E. ; Tiralongo, J. Cell Mol Life Sci 2006, 63, 1331–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Varki, A. Nature 2007, 446, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 102. Chen, X. ; Varki, A. ACS Chem Biol 2010, 5, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Crocker, P. R. ; Paulson, J. C. ; Varki, A. Nat Rev Immunol 2007, 7, 255–266. [DOI] [PubMed] [Google Scholar]
- 104. Varki, A. Trends Mol Med 2008, 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tian, Y. ; Esteva, F. J. ; Song, J. ; Zhang, H. Mol Cell Proteomics 2012, 11, M111.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Blixt, O. ; Boos, I. ; Mandel, U. , Anticarbohydrate Antibodies. From Molecular Basis to Clinical Application; Springer, Vienna: Wien, 2012; pp 283–306. [Google Scholar]
- 107. Yu, H. ; Chen, X. Org Biomol Chem 2007, 5, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shaw, L. ; Schauer, R. Biol Chem Hoppe Seyler 1988, 369, 477–486. [DOI] [PubMed] [Google Scholar]
- 109. Kozutsumi, Y. ; Kawano, T. ; Yamakawa, T. ; Suzuki, A. J Biochem (Tokyo) 1990, 108, 704–706. [DOI] [PubMed] [Google Scholar]
- 110. Muchmore, E. A. ; Milewski, M. ; Varki, A. ; Diaz, S. J Biol Chem 1989, 264, 20216–20223. [PubMed] [Google Scholar]
- 111. Lewis, A. L. ; Cao, H. ; Patel, S. K. ; Diaz, S. ; Ryan, W. ; Carlin, A. F. ; Thon, V. ; Lewis, W. G. ; Varki, A. ; Chen, X. ; Nizet, V. J Biol Chem 2007, 282, 27562–27571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lrhorfi, L. A. ; Srinivasan, G. V. ; Schauer, R. Biol Chem 2007, 388, 297–306. [DOI] [PubMed] [Google Scholar]
- 113. Houliston, R. S. ; Endtz, H. P. ; Yuki, N. ; Li, J. ; Jarrell, H. C. ; Koga, M. ; van Belkum, A. ; Karwaski, M. F. ; Wakarchuk, W. W. ; Gilbert, M. J Biol Chem 2006, 281, 11480–11486. [DOI] [PubMed] [Google Scholar]
- 114. Claus, H. ; Borrow, R. ; Achtman, M. ; Morelli, G. ; Kantelberg, C. ; Longworth, E. ; Frosch, M. ; Vogel, U. Mol Microbiol 2004, 51, 227–239. [DOI] [PubMed] [Google Scholar]
- 115. Arming, S. ; Wipfler, D. ; Mayr, J. ; Merling, A. ; Vilas, U. ; Schauer, R. ; Schwartz‐Albiez, R. ; Vlasak, R. Glycobiology 2011, 21, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yu, H. ; Chokhawala, H. A. ; Huang, S. ; Chen, X. Nat Protoc 2006, 1, 2485–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yu, H. ; Chokhawala, H. ; Karpel, R. ; Yu, H. ; Wu, B. ; Zhang, J. ; Zhang, Y. ; Jia, Q. ; Chen, X. J Am Chem Soc 2005, 127, 17618–17619. [DOI] [PubMed] [Google Scholar]
- 118. Yu, H. ; Huang, S. ; Chokhawala, H. ; Sun, M. ; Zheng, H. ; Chen, X. Angew Chem Int Ed Engl 2006, 45, 3938–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Yu, H. ; Cheng, J. ; Ding, L. ; Khedri, Z. ; Chen, Y. ; Chin, S. ; Lau, K. ; Tiwari, V. K. ; Chen, X. J Am Chem Soc 2009, 131, 18467–18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ding, L. ; Yu, H. ; Lau, K. ; Li, Y. ; Muthana, S. ; Wang, J. ; Chen, X. Chem Commun (Camb) 2011, 47, 8691–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yu, H. ; Cao, H. ; Tiwari, V. K. ; Li, Y. ; Chen, X. Bioorg Med Chem Lett 2011, 21, 5037–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yu, H. ; Yu, H. ; Karpel, R. ; Chen, X. Bioorg Med Chem 2004, 12, 6427–6435. [DOI] [PubMed] [Google Scholar]
- 123. Li, Y. ; Yu, H. ; Cao, H. ; Lau, K. ; Muthana, S. ; Tiwari, V. K. ; Son, B. ; Chen, X. Appl Microbiol Biotechnol 2008, 79, 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Li, Y. ; Yu, H. ; Cao, H. ; Muthana, S. ; Chen, X. Appl Microbiol Biotechnol 2012, 93, 2411–2423. [DOI] [PubMed] [Google Scholar]
- 125. Sugiarto, G. ; Lau, K. ; Li, Y. ; Khedri, Z. ; Yu, H. ; Le, D. T. ; Chen, X. Mol Biosyst 2011, 7, 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sugiarto, G. ; Lau, K. ; Qu, J. ; Li, Y. ; Lim, S. ; Mu, S. ; Ames, J. B. ; Fisher, A. J. ; Chen, X. ACS Chem Biol 2012, 7, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang, D. ; Liu, S. ; Trummer, B. J. ; Deng, C. ; Wang, A. Nat Biotechnol 2002, 20, 275–281. [DOI] [PubMed] [Google Scholar]
- 128. Fukui, S. ; Feizi, T. ; Galustian, C. ; Lawson, A. M. ; Chai, W. Nat Biotechnol 2002, 20, 1011–1017. [DOI] [PubMed] [Google Scholar]
- 129. Laurent, N. ; Voglmeir, J. ; Flitsch, S. L. Chem Commun (Camb) 2008, 4400–4412. [DOI] [PubMed] [Google Scholar]
- 130. Horlacher, T. ; Seeberger, P. H. Chem Soc Rev 2008, 37, 1414–1422. [DOI] [PubMed] [Google Scholar]
- 131. Wu, C. Y. ; Liang, P. H. ; Wong, C. H. Org Biomol Chem 2009, 7, 2247–2254. [DOI] [PubMed] [Google Scholar]
- 132. Song, E. H. ; Pohl, N. L. Curr Opin Chem Biol 2009, 13, 626–632. [DOI] [PubMed] [Google Scholar]
- 133. Taylor, M. E. ; Drickamer, K. Glycobiology 2009, 19, 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rillahan, C. D. ; Paulson, J. C. Annu Rev Biochem 2011, 80, 797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Park, S. ; Gildersleeve, J. C. ; Blixt, O. ; Shin, I. Chem Soc Rev 2013, 42, 4310–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Smith, D. F. ; Cummings, R. D. Mol Cell Proteomics 2013, 12, 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chevolot, Y. Carbohydrate Microarrays: Methods and Protocols; Humana Press, Springer Science+Business Media, LLC, 2012; pp 1–427. [Google Scholar]
- 138. Stevens, J. ; Blixt, O. ; Tumpey, T. M. ; Taubenberger, J. K. ; Paulson, J. C. ; Wilson, I. A. Science 2006, 312, 404–410. [DOI] [PubMed] [Google Scholar]
- 139. Stevens, J. ; Blixt, O. ; Chen, L. M. ; Donis, R. O. ; Paulson, J. C. ; Wilson, I. A. J Mol Biol 2008, 381, 1382–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Xu, R. ; McBride, R. ; Paulson, J. C. ; Basler, C. F. ; Wilson, I. A. J Virol 2010, 84, 1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Maines, T. R. ; Chen, L. M. ; Van Hoeven, N. ; Tumpey, T. M. ; Blixt, O. ; Belser, J. A. ; Gustin, K. M. ; Pearce, M. B. ; Pappas, C. ; Stevens, J. ; Cox, N. J. ; Paulson, J. C. ; Raman, R. ; Sasisekharan, R. ; Katz, J. M. ; Donis, R. O. Virology 2011, 413, 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Rademacher, C. ; Bru, T. ; McBride, R. ; Robison, E. ; Nycholat, C. M. ; Kremer, E. J. ; Paulson, J. C. Glycobiology 2012, 22, 1086–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Xu, R. ; Krause, J. C. ; McBride, R. ; Paulson, J. C. ; Crowe, J. E. J. ; Wilson, I. A. Nat Struct Mol Biol 2013, 20, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Blixt, O. ; Han, S. ; Liao, L. ; Zeng, Y. ; Hoffmann, J. ; Futakawa, S. ; Paulson, J. C. J Am Chem Soc 2008, 130, 6680–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Rillahan, C. D. ; Schwartz, E. ; McBride, R. ; Fokin, V. V. ; Paulson, J. C. Angew Chem Int Ed Engl 2012, 51, 11014–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Rillahan, C. D. ; Schwartz, E. ; Rademacher, C. ; McBride, R. ; Rangarajan, J. ; Fokin, V. V. ; Paulson, J. C. ACS Chem Biol 2013, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Liu, Y. ; Childs, R. A. ; Palma, A. S. ; Campanero‐Rhodes, M. A. ; Stoll, M. S. ; Chai, W. ; Feizi, T. Methods Mol Biol 2012, 808, 117–136. [DOI] [PubMed] [Google Scholar]
- 148. Childs, R. A. ; Palma, A. S. ; Wharton, S. ; Matrosovich, T. ; Liu, Y. ; Chai, W. ; Campanero‐Rhodes, M. A. ; Zhang, Y. ; Eickmann, M. ; Kiso, M. ; Hay, A. ; Matrosovich, M. ; Feizi, T. Nat Biotechnol 2009, 27, 797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Redelinghuys, P. ; Antonopoulos, A. ; Liu, Y. ; Campanero‐Rhodes, M. A. ; McKenzie, E. ; Haslam, S. M. ; Dell, A. ; Feizi, T. ; Crocker, P. R. J Biol Chem 2011, 286, 34522–34532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lai, L. ; Bumstead, J. ; Liu, Y. ; Garnett, J. ; Campanero‐Rhodes, M. A. ; Blake, D. P. ; Palma, A. S. ; Chai, W. ; Ferguson, D. J. ; Simpson, P. ; Feizi, T. ; Tomley, F. M. ; Matthews, S. PLoS Pathog 2011, 7, e1002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Reiss, K. ; Stencel, J. E. ; Liu, Y. ; Blaum, B. S. ; Reiter, D. M. ; Feizi, T. ; Dermody, T. S. ; Stehle, T. PLoS Pathog 2012, 8, e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chen, H. Y. ; Challa, A. K. ; Varki, A. J Biol Chem 2006, 281, 7825–7833. [DOI] [PubMed] [Google Scholar]
- 153. Martin, L. T. ; Marth, J. D. ; Varki, A. ; Varki, N. M. J Biol Chem 2002, 277, 32930–32938. [DOI] [PubMed] [Google Scholar]
- 154. Krishna, M. ; Varki, A. J Exp Med 1997, 185, 1997–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sjoberg, E. R. ; Powell, L. D. ; Klein, A. ; Varki, A. J Cell Biol 1994, 126, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Cheresh, D. A. ; Reisfeld, R. A. ; Varki, A. P. Science 1984, 225, 844–846. [DOI] [PubMed] [Google Scholar]
- 157. Chokhawala, H. A. ; Huang, S. ; Lau, K. ; Yu, H. ; Cheng, J. ; Thon, V. ; Hurtado‐Ziola, N. ; Guerrero, J. A. ; Varki, A. ; Chen, X. ACS Chem Biol 2008, 3, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Padler‐Karavani, V. ; Song, X. ; Yu, H. ; Hurtado‐Ziola, N. ; Huang, S. ; Muthana, S. ; Chokhawala, H. A. ; Cheng, J. ; Verhagen, A. ; Langereis, M. A. ; Kleene, R. ; Schachner, M. ; de Groot, R. J. ; Lasanajak, Y. ; Matsuda, H. ; Schwab, R. ; Chen, X. ; Smith, D. F. ; Cummings, R. D. ; Varki, A. J Biol Chem 2012, 287, 22593–22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lu, Q. ; Padler‐Karavani, V. ; Yu, H. ; Chen, X. ; Wu, S. L. ; Varki, A. ; Hancock, W. S. Anal Chem 2012, 84, 2761–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Padler‐Karavani, V. ; Tremoulet, A. H. ; Yu, H. ; Chen, X. ; Burns, J. C. ; Varki, A. PLoS One 2013, 8, e58443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Song, X. ; Yu, H. ; Chen, X. ; Lasanajak, Y. ; Tappert, M. M. ; Air, G. M. ; Tiwari, V. K. ; Cao, H. ; Chokhawala, H. A. ; Zheng, H. ; Cummings, R. D. ; Smith, D. F. J Biol Chem 2011, 286, 31610–31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bradley, K. C. ; Galloway, S. E. ; Lasanajak, Y. ; Song, X. ; Heimburg‐Molinaro, J. ; Yu, H. ; Chen, X. ; Talekar, G. R. ; Smith, D. F. ; Cummings, R. D. ; Steinhauer, D. A. J Virol 2011, 85, 12387–12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Jacob, F. ; Goldstein, D. R. ; Bovin, N. V. ; Pochechueva, T. ; Spengler, M. ; Caduff, R. ; Fink, D. ; Vuskovic, M. I. ; Huflejt, M. E. ; Heinzelmann‐Schwarz, V. Int J Cancer 2012, 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Liao, H. Y. ; Hsu, C. H. ; Wang, S. C. ; Liang, C. H. ; Yen, H. Y. ; Su, C. Y. ; Chen, C. H. ; Jan, J. T. ; Ren, C. T. ; Chen, C. H. ; Cheng, T. J. ; Wu, C. Y. ; Wong, C. H. J Am Chem Soc 2010, 132, 14849–14856. [DOI] [PubMed] [Google Scholar]
- 165. Yen, H. L. ; Liang, C. H. ; Wu, C. Y. ; Forrest, H. L. ; Ferguson, A. ; Choy, K. T. ; Jones, J. ; Wong, D. D. ; Cheung, P. P. ; Hsu, C. H. ; Li, O. T. ; Yuen, K. M. ; Chan, R. W. ; Poon, L. L. ; Chan, M. C. ; Nicholls, J. M. ; Krauss, S. ; Wong, C. H. ; Guan, Y. ; Webster, R. G. ; Webby, R. J. ; Peiris, M. Proc Natl Acad Sci USA 2011, 108, 14264–14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wang, C. C. ; Chen, J. R. ; Tseng, Y. C. ; Hsu, C. H. ; Hung, Y. F. ; Chen, S. W. ; Chen, C. M. ; Khoo, K. H. ; Cheng, T. J. ; Cheng, Y. S. ; Jan, J. T. ; Wu, C. Y. ; Ma, C. ; Wong, C. H. Proc Natl Acad Sci USA 2009, 106, 18137–18142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Arndt, N. X. ; Tiralongo, J. ; Madge, P. D. ; von Itzstein, M. ; Day, C. J. J Cell Biochem 2011, 112, 2230–2240. [DOI] [PubMed] [Google Scholar]
- 168. Bochner, B. S. ; Alvarez, R. A. ; Mehta, P. ; Bovin, N. V. ; Blixt, O. ; White, J. R. ; Schnaar, R. L. J Biol Chem 2005, 280, 4307–4312. [DOI] [PubMed] [Google Scholar]
- 169. Gambaryan, A. S. ; Matrosovich, T. Y. ; Philipp, J. ; Munster, V. J. ; Fouchier, R. A. ; Cattoli, G. ; Capua, I. ; Krauss, S. L. ; Webster, R. G. ; Banks, J. ; Bovin, N. V. ; Klenk, H. D. ; Matrosovich, M. N. J Virol 2012, 86, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Linman, M. J. ; Taylor, J. D. ; Yu, H. ; Chen, X. ; Cheng, Q. Anal Chem 2008, 80, 4007–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Linman, M. J. ; Yu, H. ; Chen, X. ; Cheng, Q. ACS Appl Mater Interfaces 2009, 1, 1755–1762. [DOI] [PubMed] [Google Scholar]
- 172. Sardzik, R. ; Sharma, R. ; Kaloo, S. ; Voglmeir, J. ; Crocker, P. R. ; Flitsch, S. L. Chem Commun (Camb) 2011, 47, 5425–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Pourceau, G. ; Chevolot, Y. ; Goudot, A. ; Giroux, F. ; Meyer, A. ; Moules, V. ; Lina, B. ; Cecioni, S. ; Vidal, S. ; Yu, H. ; Chen, X. ; Ferraris, O. ; Praly, J. P. ; Souteyrand, E. ; Vasseur, J. J. ; Morvan, F. Chembiochem 2011, 12, 2071–2080. [DOI] [PubMed] [Google Scholar]
- 174. Narla, S. N. ; Sun, X. L. Biomacromolecules 2012, 13, 1675–1682. [DOI] [PubMed] [Google Scholar]
- 175. Ma, Y. ; Sobkiv, I. ; Gruzdys, V. ; Zhang, H. ; Sun, X. L. Anal Bioanal Chem 2012, 404, 51–58. [DOI] [PubMed] [Google Scholar]
- 176. Xu, R. ; Zhu, X. ; McBride, R. ; Nycholat, C. M. ; Yu, W. ; Paulson, J. C. ; Wilson, I. A. J Virol 2012, 86, 9221–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Xu, R. ; McBride, R. ; Nycholat, C. M. ; Paulson, J. C. ; Wilson, I. A. J Virol 2012, 86, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Zhu, X. ; Yu, W. ; McBride, R. ; Li, Y. ; Chen, L. M. ; Donis, R. O. ; Tong, S. ; Paulson, J. C. ; Wilson, I. A. Proc Natl Acad Sci USA 2013, 110, 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhu, X. ; Yang, H. ; Guo, Z. ; Yu, W. ; Carney, P. J. ; Li, Y. ; Chen, L. M. ; Paulson, J. C. ; Donis, R. O. ; Tong, S. ; Stevens, J. ; Wilson, I. A. Proc Natl Acad Sci USA 2012, 109, 18903–18908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhu, X. ; McBride, R. ; Nycholat, C. M. ; Yu, W. ; Paulson, J. C. ; Wilson, I. A. J Virol 2012, 86, 13371–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Neu, U. ; Maginnis, M. S. ; Palma, A. S. ; Stroh, L. J. ; Nelson, C. D. ; Feizi, T. ; Atwood, W. J. ; Stehle, T. Cell Host Microbe 2010, 8, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Blumenschein, T. M. ; Friedrich, N. ; Childs, R. A. ; Saouros, S. ; Carpenter, E. P. ; Campanero‐Rhodes, M. A. ; Simpson, P. ; Chai, W. ; Koutroukides, T. ; Blackman, M. J. ; Feizi, T. ; Soldati‐Favre, D. ; Matthews, S. EMBO J 2007, 26, 2808–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cao, H. ; Crocker, P. R. Immunology 2011, 132, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Varki, A. Glycobiology 2009, 19, 810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Kelm, S. ; Madge, P. ; Islam, T. ; Bennett, R. ; Koliwer‐Brandl, H. ; Waespy, M. ; von Itzstein, M. ; Haselhorst, T. Angew Chem Int Ed Engl 2013, 52, 3616–3620. [DOI] [PubMed] [Google Scholar]
- 186. Edwards, A. D. ; Chaussabel, D. ; Tomlinson, S. ; Schulz, O. ; Sher, A. ; Reis e Sousa, C. J Immunol 2003, 171, 47–60. [DOI] [PubMed] [Google Scholar]
- 187. Wen, T. ; Mingler, M. K. ; Blanchard, C. ; Wahl, B. ; Pabst, O. ; Rothenberg, M. E. J Immunol 2012, 188, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Huflejt, M. E. ; Vuskovic, M. ; Vasiliu, D. ; Xu, H. ; Obukhova, P. ; Shilova, N. ; Tuzikov, A. ; Galanina, O. ; Arun, B. ; Lu, K. ; Bovin, N. Mol Immunol 2009, 46, 3037–3049. [DOI] [PubMed] [Google Scholar]
- 189. Wu, C. S. ; Yen, C. J. ; Chou, R. H. ; Li, S. T. ; Huang, W. C. ; Ren, C. T. ; Wu, C. Y. ; Yu, Y. L. PLoS One 2012, 7, e39466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. von Gunten, S. ; Smith, D. F. ; Cummings, R. D. ; Riedel, S. ; Miescher, S. ; Schaub, A. ; Hamilton, R. G. ; Bochner, B. S. J Allergy Clin Immunol 2009, 123, 1268–76.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Campbell, C. T. ; Gulley, J. L. ; Oyelaran, O. ; Hodge, J. W. ; Schlom, J. ; Gildersleeve, J. C. Clin Cancer Res 2013, 19, 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Parker, R. B. ; McCombs, J. E. ; Kohler, J. J. ACS Chem Biol 2012, 7, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Blixt, O. ; Allin, K. ; Bohorov, O. ; Liu, X. ; Andersson‐Sand, H. ; Hoffmann, J. ; Razi, N. Glycoconj J 2007, 25, 59–68. [DOI] [PubMed] [Google Scholar]
- 194. Belser, J. A. ; Blixt, O. ; Chen, L. M. ; Pappas, C. ; Maines, T. R. ; Van Hoeven, N. ; Donis, R. ; Busch, J. ; McBride, R. ; Paulson, J. C. ; Katz, J. M. ; Tumpey, T. M. Proc Natl Acad Sci USA 2008, 105, 7558–7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wan, H. ; Sorrell, E. M. ; Song, H. ; Hossain, M. J. ; Ramirez‐Nieto, G. ; Monne, I. ; Stevens, J. ; Cattoli, G. ; Capua, I. ; Chen, L. M. ; Donis, R. O. ; Busch, J. ; Paulson, J. C. ; Brockwell, C. ; Webby, R. ; Blanco, J. ; Al‐Natour, M. Q. ; Perez, D. R. PLoS One 2008, 3, e2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Stevens, J. ; Chen, L. M. ; Carney, P. J. ; Garten, R. ; Foust, A. ; Le, J. ; Pokorny, B. A. ; Manojkumar, R. ; Silverman, J. ; Devis, R. ; Rhea, K. ; Xu, X. ; Bucher, D. J. ; Paulson, J. ; Cox, N. J. ; Klimov, A. ; Donis, R. O. J Virol 2010, 84, 8287–8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Chen, L. M. ; Rivailler, P. ; Hossain, J. ; Carney, P. ; Balish, A. ; Perry, I. ; Davis, C. T. ; Garten, R. ; Shu, B. ; Xu, X. ; Klimov, A. ; Paulson, J. C. ; Cox, N. J. ; Swenson, S. ; Stevens, J. ; Vincent, A. ; Gramer, M. ; Donis, R. O. Virology 2011, 412, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Chen, L. M. ; Blixt, O. ; Stevens, J. ; Lipatov, A. S. ; Davis, C. T. ; Collins, B. E. ; Cox, N. J. ; Paulson, J. C. ; Donis, R. O. Virology 2012, 422, 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Heimburg‐Molinaro, J. ; Tappert, M. ; Song, X. ; Lasanajak, Y. ; Air, G. ; Smith, D. F. ; Cummings, R. D. Methods Mol Biol 2012, 808, 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Campanero‐Rhodes, M. A. ; Smith, A. ; Chai, W. ; Sonnino, S. ; Mauri, L. ; Childs, R. A. ; Zhang, Y. ; Ewers, H. ; Helenius, A. ; Imberty, A. ; Feizi, T. J Virol 2007, 81, 12846–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Yu, X. ; Coulson, B. S. ; Fleming, F. E. ; Dyason, J. C. ; von Itzstein, M. ; Blanchard, H. J Mol Biol 2011, 413, 929–939. [DOI] [PubMed] [Google Scholar]
- 202. Yu, X. ; Dang, V. T. ; Fleming, F. E. ; von Itzstein, M. ; Coulson, B. S. ; Blanchard, H. J Virol 2012, 86, 13456–13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Lofling, J. ; Michael Lyi, S. ; Parrish, C. R. ; Varki, A. Virology 2013, 440, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Disney, M. D. ; Seeberger, P. H. Chem Biol 2004, 11, 1701–1707. [DOI] [PubMed] [Google Scholar]
- 205. Mader, A. ; Gruber, K. ; Castelli, R. ; Hermann, B. A. ; Seeberger, P. H. ; Radler, J. O. ; Leisner, M. Nano Lett 2012, 12, 420–423. [DOI] [PubMed] [Google Scholar]
- 206. Nimrichter, L. ; Gargir, A. ; Gortler, M. ; Altstock, R. T. ; Shtevi, A. ; Weisshaus, O. ; Fire, E. ; Dotan, N. ; Schnaar, R. L. Glycobiology 2004, 14, 197–203. [DOI] [PubMed] [Google Scholar]
- 207. Grabosch, C. ; Kolbe, K. ; Lindhorst, T. K. Chembiochem 2012, 13, 1874–1879. [DOI] [PubMed] [Google Scholar]
- 208. Otto, D. M. ; Campanero‐Rhodes, M. A. ; Karamanska, R. ; Powell, A. K. ; Bovin, N. ; Turnbull, J. E. ; Field, R. A. ; Blackburn, J. ; Feizi, T. ; Crocker, P. R. Anal Biochem 2011, 411, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Jimenez Blanco, J. L. ; Ortiz Mellet, C. ; Garcia Fernandez, J. M. Chem Soc Rev 2013, 42, 4518–4531. [DOI] [PubMed] [Google Scholar]
- 210. Liang, C. H. ; Wang, S. K. ; Lin, C. W. ; Wang, C. C. ; Wong, C. H. ; Wu, C. Y. Angew Chem Int Ed Engl 2011, 50, 1608–1612. [DOI] [PubMed] [Google Scholar]
- 211. Rinaldi, S. ; Brennan, K. M. ; Goodyear, C. S. ; O'Leary, C. ; Schiavo, G. ; Crocker, P. R. ; Willison, H. J. Glycobiology 2009, 19, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Rinaldi, S. ; Brennan, K. M. ; Willison, H. J. Methods Mol Biol 2012, 808, 413–423. [DOI] [PubMed] [Google Scholar]
- 213. Deng, Y. ; Wang, Y. ; Holtz, B. ; Li, J. ; Traaseth, N. ; Veglia, G. ; Stottrup, B. J. ; Elde, R. ; Pei, D. ; Guo, A. ; Zhu, X. Y. J Am Chem Soc 2008, 130, 6267–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Zhu, X. Y. ; Holtz, B. ; Wang, Y. ; Wang, L. X. ; Orndorff, P. E. ; Guo, A. J Am Chem Soc 2009, 131, 13646–13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Shi, L. ; Reid, L. H. ; Jones, W. D. ; Shippy, R. ; Warrington, J. A. ; Baker, S. C. ; Collins, P. J. ; de Longueville, F. ; Kawasaki, E. S. ; Lee, K. Y. ; Luo, Y. ; Sun, Y. A. ; Willey, J. C. ; Setterquist, R. A. ; Fischer, G. M. ; Tong, W. ; Dragan, Y. P. ; Dix, D. J. ; Frueh, F. W. ; Goodsaid, F. M. ; Herman, D. ; Jensen, R. V. ; Johnson, C. D. ; Lobenhofer, E. K. ; Puri, R. K. ; Schrf, U. ; Thierry‐Mieg, J. ; Wang, C. ; Wilson, M. ; Wolber, P. K. ; Zhang, L. ; Amur, S. ; Bao, W. ; Barbacioru, C. C. ; Lucas, A. B. ; Bertholet, V. ; Boysen, C. ; Bromley, B. ; Brown, D. ; Brunner, A. ; Canales, R. ; Cao, X. M. ; Cebula, T. A. ; Chen, J. J. ; Cheng, J. ; Chu, T. M. ; Chudin, E. ; Corson, J. ; Corton, J. C. ; Croner, L. J. ; Davies, C. ; Davison, T. S. ; Delenstarr, G. ; Deng, X. ; Dorris, D. ; Eklund, A. C. ; Fan, X. H. ; Fang, H. ; Fulmer‐Smentek, S. ; Fuscoe, J. C. ; Gallagher, K. ; Ge, W. ; Guo, L. ; Guo, X. ; Hager, J. ; Haje, P. K. ; Han, J. ; Han, T. ; Harbottle, H. C. ; Harris, S. C. ; Hatchwell, E. ; Hauser, C. A. ; Hester, S. ; Hong, H. ; Hurban, P. ; Jackson, S. A. ; Ji, H. ; Knight, C. R. ; Kuo, W. P. ; LeClerc, J. E. ; Levy, S. ; Li, Q. Z. ; Liu, C. ; Liu, Y. ; Lombardi, M. J. ; Ma, Y. ; Magnuson, S. R. ; Maqsodi, B. ; McDaniel, T. ; Mei, N. ; Myklebost, O. ; Ning, B. ; Novoradovskaya, N. ; Orr, M. S. ; Osborn, T. W. ; Papallo, A. ; Patterson, T. A. ; Perkins, R. G. ; Peters, E. H. ; Peterson, R. ; Philips, K. L. ; Pine, P. S. ; Pusztai, L. ; Qian, F. ; Ren, H. ; Rosen, M. ; Rosenzweig, B. A. ; Samaha, R. R. ; Schena, M. ; Schroth, G. P. ; Shchegrova, S. ; Smith, D. D. ; Staedtler, F. ; Su, Z. ; Sun, H. ; Szallasi, Z. ; Tezak, Z. ; Thierry‐Mieg, D. ; Thompson, K. L. ; Tikhonova, I. ; Turpaz, Y. ; Vallanat, B. ; Van, C. ; Walker, S. J. ; Wang, S. J. ; Wang, Y. ; Wolfinger, R. ; Wong, A. ; Wu, J. ; Xiao, C. ; Xie, Q. ; Xu, J. ; Yang, W. ; Zhang, L. ; Zhong, S. ; Zong, Y. ; Slikker, W. J. Nat Biotechnol 2006, 24, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Brazma, A. ; Hingamp, P. ; Quackenbush, J. ; Sherlock, G. ; Spellman, P. ; Stoeckert, C. ; Aach, J. ; Ansorge, W. ; Ball, C. A. ; Causton, H. C. ; Gaasterland, T. ; Glenisson, P. ; Holstege, F. C. ; Kim, I. F. ; Markowitz, V. ; Matese, J. C. ; Parkinson, H. ; Robinson, A. ; Sarkans, U. ; Schulze‐Kremer, S. ; Stewart, J. ; Taylor, R. ; Vilo, J. ; Vingron, M. Nat Genet 2001, 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 217. Liu, Y. ; Feizi, T. ; Campanero‐Rhodes, M. A. ; Childs, R. A. ; Zhang, Y. ; Mulloy, B. ; Evans, P. G. ; Osborn, H. M. ; Otto, D. ; Crocker, P. R. ; Chai, W. Chem Biol 2007, 14, 847–859. [DOI] [PubMed] [Google Scholar]
- 218. Bovin, N. ; Obukhova, P. ; Shilova, N. ; Rapoport, E. ; Popova, I. ; Navakouski, M. ; Unverzagt, C. ; Vuskovic, M. ; Huflejt, M. Biochim Biophys Acta 2012, 1820, 1373–1382. [DOI] [PubMed] [Google Scholar]
- 219. Carroll, G. T. ; Wang, D. ; Turro, N. J. ; Koberstein, J. T. Glycoconj J 2008, 25, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Deng, L. ; Norberg, O. ; Uppalapati, S. ; Yan, M. ; Ramstrom, O. Org Biomol Chem 2011, 9, 3188–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Tyagi, A. ; Wang, X. ; Deng, L. ; Ramstrom, O. ; Yan, M. Biosens Bioelectron 2010, 26, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Norberg, O. ; Deng, L. ; Aastrup, T. ; Yan, M. ; Ramstrom, O. Anal Chem 2011, 83, 1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Wang, D. ; Carroll, G. T. ; Turro, N. J. ; Koberstein, J. T. ; Kovac, P. ; Saksena, R. ; Adamo, R. ; Herzenberg, L. A. ; Herzenberg, L. A. ; Steinman, L. Proteomics 2007, 7, 180–184. [DOI] [PubMed] [Google Scholar]
- 224. Shilova, N. ; Navakouski, M. ; Khasbiullina, N. ; Blixt, O. ; Bovin, N. Glycoconj J 2012, 29, 87–91. [DOI] [PubMed] [Google Scholar]
- 225. Kletter, D. ; Singh, S. ; Bern, M. ; Haab, B. B. Mol Cell Proteomics 2013, 12, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Norberg, O. ; Deng, L. ; Yan, M. ; Ramstrom, O. Bioconjug Chem 2009, 20, 2364–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Deng, L. ; Wang, X. ; Uppalapati, S. ; Norberg, O. ; Dong, H. ; Joliton, A. ; Yan, M. ; Ramstrom, O. Pure Appl Chem 2013; ASAP article, published online: http://dx.doi.org/10.1351/PAC-CON-12-08-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Madwar, C. ; Kwan, W. C. ; Deng, L. ; Ramstrom, O. ; Schmidt, R. ; Zou, S. ; Cuccia, L. A. Langmuir 2010, 26, 16677–16680. [DOI] [PubMed] [Google Scholar]
- 229. Wang, X. ; Matei, E. ; Deng, L. ; Koharudin, L. ; Gronenborn, A. M. ; Ramstrom, O. ; Yan, M. Biosens Bioelectron 2013, 47, 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Gao, J. ; Liu, D. ; Wang, Z. Anal Chem 2008, 80, 8822–8827. [DOI] [PubMed] [Google Scholar]
- 231. Pochechueva, T. ; Jacob, F. ; Goldstein, D. R. ; Huflejt, M. E. ; Chinarev, A. ; Caduff, R. ; Fink, D. ; Hacker, N. ; Bovin, N. V. ; Heinzelmann‐Schwarz, V. Glycoconj J 2011, 28, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Walz, A. ; Odenbreit, S. ; Mahdavi, J. ; Boren, T. ; Ruhl, S. Glycobiology 2005, 15, 700–708. [DOI] [PubMed] [Google Scholar]


