Abstract
Study on the 2‐phenylchroman‐4‐one derivatives and their anti‐MERS‐CoVactivities.
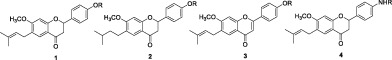
Keywords: MERS‐CoV, anti‐MERS, 2‐phenylchroman‐4‐one, Bavachin
Middle East respiratory syndrome coronavirus (MERS‐CoV) has been one of the most fearful diseases by its mortality and possibility of global outbreak. Until 23 January 2019, 2279 laboratory‐confirmed cases of MERS‐CoV, including 806 associated deaths (case‐fatality rate: 35.5%) were reported globally.1 In Korea on May 2015, 186 confirmed cases including 38 associated deaths (20.4%) were reported by an outbreak caused by only one infected person from Saudi Arabia.2 At present, no effective vaccine or therapeutics are available for the prevention or treatment of MERS‐CoV infection.3, 4, 5 However, various basic and clinical research are on‐going.6
We screened a variety of natural products against MERS‐CoV as an attempt of developing anti‐MERS‐CoV drugs, and many flavonoids showed anti‐MERS activities. Among them, 2‐phenylchroman‐4‐one derivatives e.g., bavachin and bavachnin separated from dry seed of Psoralea corylifolia L., Korean medicinal herb exhibited comparatively good activities. Bavachin derivatives have been reported as their diverse biological activities, including anti‐cancer,7 PPAR agonist,8 anti‐inflammatory,9 anti‐Alzheimer,10 immunomodulatory,11 anti‐osteoporosis,12 and anti‐viral (against carp virus13 and influenza A14) activities.
The synthesis of racemic mixtures of bavachin derivatives were reported15, 16 and these compounds have been proven to be PPAR agonists16 and anticancer reagents.17 However, exchanges of phenol group to aniline group have not been tried.
Bavachin and bavachinin showed good anti‐MERS‐CoV activities of 2.9 and 7.9 μM respectively by phenotypic cellular screening with vero cell. As the small structural difference between two compounds (‐OH vs. –OMe) could change the anti‐viral activity, we tried to synthesize the bavachin derivatives for structure–activity‐relationship study (SAR study).
First, we synthesized racemic bavachinin (1a) according to the procedure reported by Du et al.15 The double bond in the side chain of 1a was reduced by hydrogenation with Pd/C to prepare 2a, and oxidation of chromane ring with I2 was performed to produce 3a (Scheme 1). 6a was synthesized by the modified procedure of that of 1a.15, 16 4 was reacted with N‐Boc‐protected 4‐aminobenzaldehyde by aldol condensation to give 5, which was cyclized by KF to produce 6a (Scheme 3). The further derivatizations of 1a, 2a, 3a, and 6a were performed by the substitutions of phenolic OH (1a, 2a, 3a) and NH2 (6a) groups.
Scheme 1.
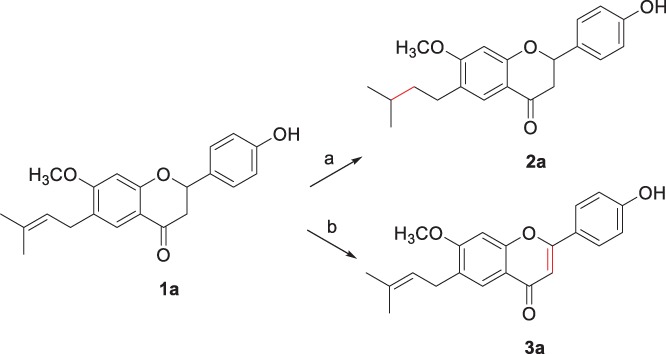
Synthesis of 2a and 3a from 1a. Reagents and conditions: (a) H2, 10% Pd/C, ethanol, rt., 90%; (b) I2, pyridine, 100 °C, 45%.
For example, racemic bavachnin, 1a was reacted with iodomethane, 2‐bromopropane, and benzyl bromide in the presence of potassium carbonate to afford 1b, 1c, and 1d, respectively. The derivatizations of phenol group of 2a and 3a were performed by the similar procedures as shown in Scheme 2 to prepare 2b–2d and 3b. 6b and 6c were prepared by acetylation and propargylation of the amino group of 6a as shown in Scheme 3.
Scheme 2.
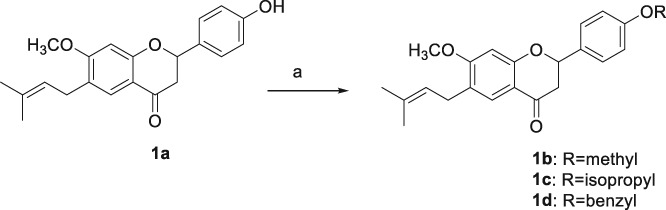
O‐Alkylations of 1a. Reagents and conditions: (a) Iodomethane, K2CO3, DMF, rt., overnight, 68% (1b); 2‐bromopropane, K2CO3, DMF, 60 °C, overnight, 41% (1c); benzyl bromide, K2CO3, DMF, rt., overnight, 58% (1d).
Scheme 3.
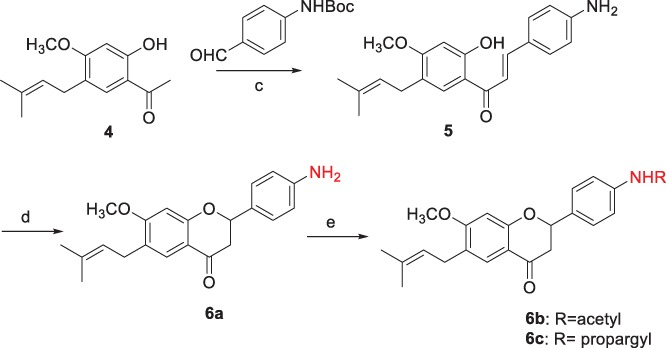
Synthesis of 6a, 6b, and 6c. Reagents and conditions: (c) Potassium trimethylsilanolate, ethanol, reflux, 15 h, 57%; (d) KF, methanol, reflux, 15 h, 60%; (e) Acetic anhydride, trimethylamine, DCM, rt., 2 h, 89% (6b); propargyl chloride, trimethylamine, DCM, rt., overnight, 45% (6c).
Total 12 compounds of bavachinin derivatives in four core structures were evaluated to figure out their anti‐MERS‐CoV activity and cell‐cytotoxicity by cellular phenotypic screening method as shown in Table 1. The activity of synthesized racemic bavachin (1a) was around half of the natural bavachin but O‐methylated 1a, racemic bavachnin (1b) showed similar activity of natural bavachinin (Entry 1 and 2 in Table 1). The further alkylations of phenolic OH of 1a with isopropyl and benzyl group decreased the anti‐MERS activities (Entry 3 and 4 in Table 1).
Table 1.
Anti‐MERS‐CoV activities of 2‐phenylchroman‐4‐one derivatives
| ||||
|---|---|---|---|---|
| Entry | Compound | R | IC50 (μM)a | CC50 (μM)b |
| 1 | 1a | H | 6.6 | 13.0 |
| 2 | 1b | Methyl | 6.3 | >25 |
| 3 | 1c | Isopropyl | 13.2 | >25 |
| 1d | 1d | Benzyl | 10.0 | >25 |
| 4 | 2a | H | 3.3 | 4.0 |
| 5 | 2b | Methyl | 11.1 | 15.2 |
| 6 | 2c | Isopropyl | 5.6 | 12.8 |
| 7 | 2d | Benzyl | 6.7 | 15.0 |
| 8 | 3a | H | 4.8 | >25 |
| 9 | 3b | Methyl | 14.4 | >25 |
| 10 | 6a | H | 11.6 | >25 |
| 11 | 6b | Acetyl | 8.4 | >25 |
| 12 | 6c | Propagyl | 12.0 | >25 |
| 13 | Bavachin | 2.9 | 8.2 | |
| 14 | Bavachinin | 7.9 | 11.1 | |
| 15 | Chloroquinec | 26.2 | 200 | |
Inhibition concentration.
Cytotoxicity concentration.
Positive control.
2a was prepared by hydrogenations of external double bonds of 1a, which showed the best anti‐MERS activities regardless of their O‐substituents. Interestingly, O‐isopropyl and O‐benzyl derivatives (2c and 2d, respectively) showed similar activity with 2a (non‐substituted) better than 2b (O‐methylated), but the cytotoxicity for vero cell also increased. 3a showed good activity similar to that of 2a, however its cytotoxicity was much less than that of 2a. Considering activity and cytotoxicity, 3a was thought to be the best compounds for anti‐MERS drug.
The pharmacokinetic study of 1a was performed on rats by IV and PO. 1a could not be detected in plasma after 30 min in both IV and PO study by its rapid clearance, but its liver microsomal phase I stability (% of remaining after 30 min) in rat was 45%. We decided to change the phenolic OH group to amino group (6a) based on the assumption that phenolic OH group was concerned with its instability in plasma. Though bioavailability (4%) was not satisfactory and liver microsomal phase I stability was not improved (36%), the results from pharmacokinetic study of 6a afforded acceptable curves in both IV and PO study as shown in Figure 1. The anti‐MERS activity of 6a was a little decreased compared to 1a, however, the anti‐MERS activities of its derivatives were conserved.
Figure 1.
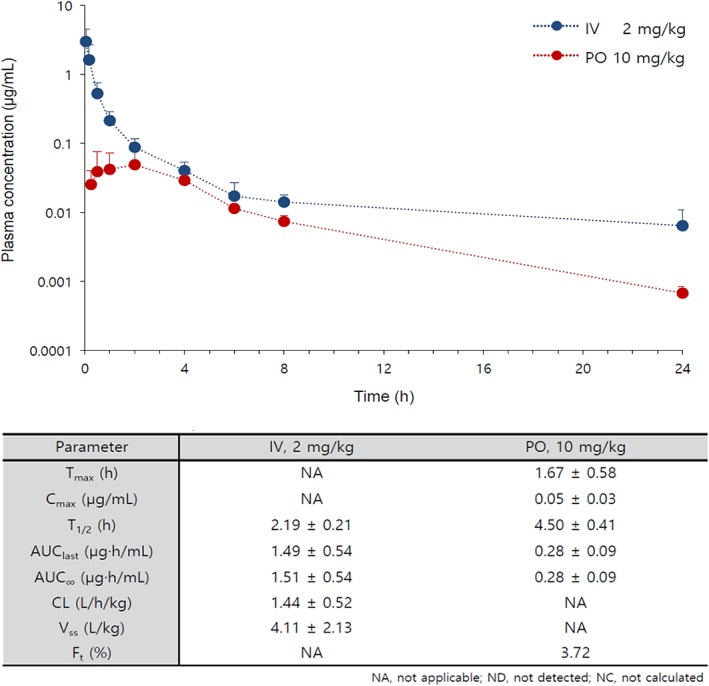
Preliminary pharmacokinetic profiles of 6a in rats.
1a also exhibited anti‐viral activity against SARS‐CoV (10.2 μM) in vero cell.
As a conclusion, a series of 2‐phenylchroman‐4‐one derivatives were synthesized for the chemical modifications of bavachin, and they exhibited anti‐MERS activities in vero cell. 2a showed the best activity, but 3a was thought to be the best compounds for anti‐MERS drug in consideration of cytotoxicity. The modification of phenol group (1a) to amino group (6a) could much improve pharmacokinetic properties. We expect the study on bavachin derivatives can contribute to the development of anti‐MERS drug.
Experimental
General
1H and 13C NMR spectra were recorded on a Varian Gemini 300 spectrometer (Palo Alto, CA, USA) using chloroform‑d and DMSO‑d 6 as a solvent with TMS internal standard. HRMS analysis was performed on a JEOL JMS‐700 (Tokyo, Japan) with EI mode.
Synthesis of the Compounds
1a: 1H NMR (300 MHz, chloroform‑d) δ 7.68 (s, 1H), 7.35 (d, J = 8.4 Hz, 2H), 6.89 (d, J = 8.4 Hz, 2H), 6.44 (s, 1H), 5.38 (dd, J = 13.2, 2.7 Hz, 1H), 5.27 (t, J = 7.5 Hz, 1H), 5.20 (s, 1H), 3.84 (s, 3H), 3.24 (d, J = 7.2 Hz, 2H), 3.08–2.98 (m, 1H), 2.77 (dd, J = 17.1, 3.0 Hz, 1H), 1.71 (d, J = 13.5 Hz, 3H). HRMS (EI+): m/z calcd for C21H23NO3 [M]+ 338.1518; found 338.1517.
1b: 1H NMR (300 MHz, Chloroform‑d) δ 7.68 (s, 1H), 7.46–7.33 (m, 7H), 7.03 (d, J = 8.7 Hz, 2H), 6.44 (s, 1H), 5.39 (dd, J = 13.2, 3.0 Hz, 1H), 5.30–5.24 (m, 1H), 5.10 (s, 2H), 3.84 (s, 3H), 3.24 (d, J = 7.2 Hz, 2H), 3.09–2.99 (m, 1H), 2.77 (dd, J = 17.1, 3.0 Hz, 1H), 1.72 (d, J = 13.5 Hz, 6H).
1c: 1H NMR (300 MHz, Chloroform‑d) δ 7.68 (s, 1H), 7.38 (d, J = 9.0 Hz, 2H), 6.93 (d, J = 6.0 Hz, 2H), 6.45 (s, 1H), 3.38 (dd, J = 15.0, 3.0 Hz, 1H), 5.30–5.25 (m, 1H), 4.62–4.53 (m, 1H), 3.84 (s, 3H), 3.24 (d, J = 6.0 Hz, 2H), 3.10–2.99 (m, 1H), 2.77 (dd, J = 12.0, 3.0 Hz, 1H), 1.72 (d, J = 12.0 Hz. 6H), 1.35 (d, J = 6.0 Hz, 6H).
1d: 1H NMR (300 MHz, chloroform‑d) δ 7.68 (s, 1H), 7.46–7.33 (m, 7H), 7.03 (d, J = 8.7 Hz, 2H), 6.44 (s, 1H), 5.39 (dd, J = 13.2, 3.0 Hz, 1H), 5.30–5.24 (m, 1H), 5.10 (s, 2H), 3.84 (s, 3H), 3.24 (d, J = 7.2 Hz, 2H), 3.09–2.99 (m, 1H), 2.77 (dd, J = 17.1, 3.0 Hz, 1H), 1.72 (d, J = 13.5 Hz, 6H).
2a: 1H NMR (300 MHz, chloroform‑d) δ 7.05–7.08 (d, 2H), 6.82 (s, 1H), 6.76 (d, J = 8.4 Hz, 2H), 6.34 (s, 1H) 4.63 (s, 1H) 4.47 (s, 1H) 3.76 (s, 3H) 2.47–2.63 (m, 4H), 1.83–1.93 (m, 2H), 1.36–1.44 (m, 2H) 0.92 (d,, J = 6.3 Hz, 6H).
2b: 1H NMR (300 MHz, chloroform‑d) δ 7.12 (d, J = 8.7 Hz, 1H), 7.07 (d, J = 8.4 Hz, 1H), 6.84 (d, J = 7.8 Hz, 2H), 6.74 (d, J = 8.4 Hz, 1H), 6.42 (s, 1H), 6.34 (s, 1H), 4.45 (s, 1H), 3.18 (d, J = 3.0 Hz, 3H), 3.78 (d, J = 10.2 Hz, 3H), 2.64–2.48 (m, 2H), 1.94–1.81 (m, 2H), 1.62–1.53 (m, 2H), 1.45–1.37 (m, 2H).
2c: 1H NMR (300 MHz, Chloroform‑d) δ 7.08 (t, J = 8.4 Hz, 2H), 6.83 (t, J = 5.7 Hz, 2H), 6.74 (d, J = 8.4 Hz, 1H), 6.34 (s, 1H), 4.57–4.43 (m, 2H), 3.77 (d, J = 7.8 Hz, 3H), 2.63–2.47 (m, 2H), 1.94–1.79 (m, 2H), 1.59–1.53 (m, 2H), 1.44–1.67 (m, 2H), 1.33 (d, J = 6.0 Hz, 6H), 0.93 (d, J = 6.6 Hz, 6H).
2d: 1H NMR (300 MHz, Chloroform‑d) δ 7.45–7.32 (m, 5H), 7.11 (d, J = 8.4 Hz, 1H), 7.03 (d, J = 8.4 Hz, 1H), 6.90 (d, J = 12.8 Hz, 1H), 6.87 (m, 2H), 6.71 (d, J = 8.4 Hz, 1H), 6.58 (s, 1H), 5.04 (s, 2H), 4.50 (s, 1H), 3.77 (d, J = 3.6 Hz, 3H), 2.64–2.47 (m, 2H), 1.94–1.85 (m, 2H), 1.61–1.51 (m, 2H), 1.44–1.38 (m, 2H), 0.93 (d, J = 6.3 Hz, 6H).
3a: 1H NMR (300 MHz, DMSO‑d 6) δ 10.25 (s, 1H), 7.92 (d, J = 8.4 Hz, 2H), 7.68 (s, 1H), 7.26 (s, 1H) 6.92 (d, J = 8.4 Hz, 2H), 6.75 (s, 1H), 5.28 (t, J = 7.5 Hz, 1H), 3.94 (s, 3H), 3.26 (d, J = 9.0 Hz, 2H), 1.68 (d, J = 15.0 Hz, 6H). HRMS (EI+): m/z calcd for C21H23NO3 [M]+ 336.1362; found 336.1368.
3b: 1H NMR (300 MHz, Chloroform‑d) δ 7.67 (s, 1H), 7.32 (d, J = 8.4 Hz, 2H), 6.73 (d, J = 8.4 Hz, 2H), 6.44 (s, 1H), 5.37–5.24 (m, 2H), 3.96 (d, J = 2.4 Hz, 2H), 3.83 (s, 1H), 3.24 (d, J = 7.5 Hz, 2H), 3.11–2.72 (m, 2H), 1.72 (d, J = 13.0 Hz, 6H).
5: To a mixture of 1‐(4‐methoxy‐2‐((3‐methylbut‐2‐en‐1‐yl)oxy)phenyl)ethan‐1‐one (4) (2.0 g, 8.5 mmol) and N‐Boc‐(4‐aminobenzaldehyde) (1.9 g, 8.5 mmol) in ethanol (25 mL) was added potassium trimethylsilanolate (4.3 g, 30 mmol) and the reaction mixture was heated under reflux overnight. The reaction was quenched by the addition of saturated NH4Cl solution. The organic layer was extracted with ethyl ether, dried over MgSO4, and concentrated in vacuum. The residue was purified by silica gel column chromatography (hexane/EtOAc) to afford (E)‐3‐(4‐((tert‐butoxymethyl)amino)phenyl)‐1‐(2‐hydroxy‐4‐methoxy‐5‐(3‐methylbut‐2‐en‐1‐yl)phenyl)prop‐2‐en‐1‐one (5) (1.6 g, 57%) as yellow solid.
1H NMR (300 MHz, Chloroform‑d) δ 13.61 (s, 1H), 7.82 (d, J = 15.0 Hz, 1H), 7.59 (s, 1H), 7.49 (d, J = 8.4 Hz, 2H), 7.41 J = 7.2 Hz, 2H), 1.77 (d, J = 11.0 Hz, 6H). HRMS (EI+): m/z calcd for C21H23NO3 [M]+ 337.1678; found 337.1663.
6a: Amixture of 6 (47 mg, 0.14 mmol) and potassium fluoride (28 mg, 0.49 mmol) in methanol (1 mL) was heated underreflux overnight. The mixture was extracted with EtOAc, dried over MgSO4, and filtered. The filtrate was separated by silica gel column chromatography (hexane/EtOAc) to afford 6a (28 mg, 60%) as yellow solid.
1H NMR (300 MHz, chloroform‑d) δ 7.67 (s, 1H), 7.26 (d, J = 8.4 Hz, 2H), 6.72 (d, J = 8.4 Hz, 2H), 6.44 (s, 1H), 5.35–5.25 (m, 1H), 3.83 (s, 3H), 3.24 (d, J = 7.2 Hz, 2H), 3.11–2.71 (m, 2H), 1.72 (d, J = 13.0 Hz, 6H). 13C NMR (500 MHz, CDCl3): δ 191.1, 164.6, 162.2, 139.1, 133.1, 131.6, 127.5, 126.9, 125.4, 123.0, 121.4, 113.5, 98.7, 78.8, 55.8, 43.9, 27.7, 25.7, 17.7. HRMS (EI+): m/z calcd for C21H23NO3 [M]+ 337.1678; found 337.1688.
6b: 1H NMR (300 MHz, chloroform‑d) δ 7.67 (s, 1H), 7.57 (d, J = 8.4 Hz, 2H), 7.43 (d, J = 8.4 Hz, 2H), 6.45 (s, 1H), 5.41 (dd, J = 13.0, 3.0 Hz, 1H), 5.27 (t, J = 9.9 Hz, 1H), 3.85 (s, 3H), 3.24 (d, J = 7.2 Hz, 2H), 3.05–2.75 (m, 2H), 2.20 (s, 3H), 1.71 (d, J = 14 Hz, 6H).
6c: 1H NMR (300 MHz, chloroform‑d) δ 7.67 (s, 1H), 7.32 (d, J = 8.4 Hz, 2H), 6.73 (d, J = 8.4 Hz, 2H), 6.44 (s, 1H), 5.37–5.24 (m, 2H), 3.96 (d, J = 2.4 Hz, 2H), 3.83 (s, 1H), 3.24 (d, J = 7.5 Hz, 2H), 3.11–2.72 (m, 2H), 1.72 (d, J = 13 Hz, 6H).
Screening of Compounds for anti‐MERS‐CoV Activity
A primary screening was performed in vero cells infected with a Korean clinical MERS‐CoV. To evaluate the ability of the molecules to inhibit the replication of MERS‐CoV, viral infectivity was determined by monitoring the cells expressing spike (S) protein using immunofluorescence assay (IFA). Vero cells were seeded in 384‐well μ‐clear plates at a density of 1.2 × 104 cell per well in 30 μL SFM and incubated for 24 h prior to infection. Compounds were transferred from the library into intermediate 384‐well polypropylene plates containing SFM and were mixed using an automated liquid handling. Subsequently, the diluted compounds were added to the cell plates in 10 μL volumes (final DMSO concentration of 0.5% (v/v)). For infection, the plates were transferred into the BSL‐3 containment facility prior to adding 10 μL MERS‐CoV at a multiplicity of infection (MOI) of 0.0625. The cells were fixed at 24 hpi with 4% PFA and the infected cells were identified by IFA as described below.
For immunofluorescence staining, the fixed cells (above) were permeabilized with 0.25% Triton X‐100 (Sigma‐Aldrich, St. Louis, MO, USA) for 20 min. Then the cells were incubated with rabbit anti‐MERS‐CoV Spike antibody for 1 h at 37 °C. After three washes with PBS, the cells were incubated with Alexa 488‐conjugated goat anti‐rabbit IgG (H + L) secondary antibody and Hoechst 33342 (Life Technologies, Waltham, MA, USA) for 1 h at 37 °C. Images were acquired by Perkin Elmer Operetta (20×; Waltham, MA, USA). The acquired images were analyzed with in‐house‐developed Image‐Mining 3.0 (IM 3.0) plug‐in software as previously described (PMID: 24205414). In the analyzed image, the total number of cells and the number of infected cells were determined by counting Hoechst‐stained nuclei and spike protein‐expressing cells, respectively. The infection ratio of each well was normalized using the average infection ratio of mock control as 0% and the average infection ratio of negative control (0.5% DMSO) as 100%, in each assay plate. Cell ratio was determined according to the number of cells of each well versus the average number of cells of mock control, in each assay plate.
All work with MERS‐CoV was performed in an enhanced biosafety level‐3 (BSL‐3) facility at Institut Pasteur Korea.
Acknowledgments
This work was supported by the grant of National Research Council of Science & Technology (NST) by the Korean government (MSIP) (No. CRC‐16‐01‐KRICT). The chemical library used in this study was kindly provided by Korea Chemical Bank of Korea Research Institute of Chemical Technology. MERS‐CoV Korea strain MERS‐CoV/KOR/KNIH/002_05_2015 (Genebank accession no. KT029139.1) was kindly provided by Division of Respiratory Viruses, Center for Infectious Diseases, Korea National Institute of Health (KNIH).
References
- 1. https://www.who.int/emergencies/mers-cov/en/
- 2. https://www. http://mers.go.kr/
- 3. Falzarano D., de Wit E., Rasmussen A. L., Feldmann F., Okumura A., Scott D. P., Brining D., Bushmaker T., Martellaro C., Baseler L., A. G. Benecke, M. G. Katze, V. J. Munster, H. Feldmann, Nat. Med. 2013, 19, 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dyall J., Coleman C. M., Hart B. J., Venkataraman T., Holbrook M. R., Kindrachuk J., Johnson R. F., Olinger G. G. Jr., Jahrling P. B., Laidlaw M., Johansen L. M., Lear‐Rooney C. M., Glass P. J., Hensley L. E., Frieman M. B., Antimicrob. Agents Chemother. 2014, 58, 4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu L., Xia S., Ying T., Jiang S., Emerg. Microbes Infect. 2015, 4, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liang R., Wang L., Zhang N., Deng X., Su M., Su Y., Hu L., He C., Ying T., Jiang S., Yu F., Viruses 2018, 10, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Kuntz S., Wenzel U., Daniel H., Eur. J. Nutr. 1999, 38, 133. [DOI] [PubMed] [Google Scholar]; (b) Song P., Yang X. Z., Yuan J. Q., J. Asian Nat. Prod. Res. 2013, 15, 624. [DOI] [PubMed] [Google Scholar]
- 8. Guo, F. ; Huang, C. ; Feng, L. ; Li, Y. U.S. Pat. US10,034,852 B2 (20180731).
- 9. Lee S. W., Yun B. R., Kim M. H., Park C. S., Lee W. S., Oh H. M., Rho M. C., Planta Med. 2012, 78, 903. [DOI] [PubMed] [Google Scholar]
- 10. Chen X., Yang Y., Zhang Y., FEBS Lett. 2013, 587, 2930. [DOI] [PubMed] [Google Scholar]
- 11. Sharma M. L., Singh B., Chandan B. K., Khajuria A., Kaul A., Bani S., Banerjee S. K., Gambhir S. S., Phytomedicine 1996, 3, 191. [DOI] [PubMed] [Google Scholar]
- 12. Weng Z.‐B., Gao Q.‐Q., Zhao G.‐H., Yin F.‐Z., Wang F., Cai B.‐C., Chen Z.‐P., Li W.‐D., Mol. Cell. Endocrinol. 2015, 417, 103. [DOI] [PubMed] [Google Scholar]
- 13. Chen C., Shen Y.‐F., Hu Y., Liu L., Chen W.‐C., Wang G.‐X., Zhu B., Virus Res. 2018, 255, 24.29913251 [Google Scholar]
- 14.(a)Choi J., Jin Y.‐H., Kim J.‐H., Oh T. W., Yim N.‐H., Cho W.‐K., Ma J. Y., Front. Pharmacol. 2016, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhao H., Z. Chen., J. Chromatogr. A 2014, 1340, 139. [DOI] [PubMed] [Google Scholar]
- 15.(a) Du G., Feng L., Yang Z., Shi J., Huang C., Guo F., Li B., Zhu W., Bioorg. Med. Chem. Lett. 2015, 25, 2579. [DOI] [PubMed] [Google Scholar]; (b) Harwood L. M., Loftus G. C., Oxford A., Thomson C., Synth. Commun. 1990, 20, 649. [Google Scholar]
- 16. Du G., Zhuo Y., Feng L., Yang Z., Shi J., Huang C., Guo F., Li B., Zhu W., Li Y., ChemMedChem 2017, 12, 183. [DOI] [PubMed] [Google Scholar]
- 17. Gupta N., Qayum A., Raina A., Shankar R., Gairola S., Singh S., Sangwan P. L. E., J. Med. Chem. 2018, 145, 511. [DOI] [PubMed] [Google Scholar]


