Abstract
Viruses are very small and most of them can be seen only by TEM (transmission electron microscopy). TEM has therefore made a major contribution to virology, including the discovery of many viruses, the diagnosis of various viral infections and fundamental investigations of virus—host cell interactions. However, TEM has gradually been replaced by more sensitive methods, such as the PCR. In research, new imaging techniques for fluorescence light microscopy have supplanted TEM, making it possible to study live cells and dynamic interactions between viruses and the cellular machinery. Nevertheless, TEM remains essential for certain aspects of virology. It is very useful for the initial identification of unknown viral agents in particular outbreaks, and is recommended by regulatory agencies for investigation of the viral safety of biological products and/or the cells used to produce them. In research, only TEM has a resolution sufficiently high for discrimination between aggregated viral proteins and structured viral particles. Recent examples of different viral assembly models illustrate the value of TEM for improving our understanding of virus—cell interactions.
Keywords: diagnosis, electron microscopy, viral assembly, virus detection, viral morphogenesis
Abbreviations used:
- ELISA
enzyme‐linked immunosorbent assay
- EM
electron microscopy
- ER
endoplasmic reticulum
- ERGIC
ER—Golgi intermediate compartment
- GFP
green fluorescent protein
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- TEM
transmission electron microscopy
Introduction
Most viruses are small enough to be at the limit of resolution of even the best light microscopes, and can be visualized in liquid samples or infected cells only by EM (electron microscopy). However, there has been debate about whether it is useful or useless for medical virology (Curry et al., 1999, 2000a, 2000b; Madeley 2000; McCaughey et al., 2000). In the present article, I will analyse and discuss the benefits of viral detection by EM in various aspects of current and past virology.
The discovery of many viruses and a role in routine diagnosis: the ‘glory days’ of the past
EM was first developed in the 1930s, by physicists in various countries, including Germany, in particular (reviewed in Haguenau et al., 2003). The first microscope for TEM (transmission electron microscopy), which was also known as a ‘supermicroscope’, was initially described by Max Knoll and Ernst Ruska in 1932 (Knoll and Ruska 1932; Ruska 1987). This microscope had a much higher resolution than the light microscopes of the time, and promised to revolutionize many aspects of cell biology and virology. Helmut Ruska, a medical doctor and brother of the physicist Ernst Ruska (Ruska et al., 1939) rapidly recognized the potential of ‘ultramicroscopy’ for investigating the nature of viruses. Despite the lack of appropriate methods of sample preparation for TEM at the time, several viruses were characterized morphologically and an attempt was made to develop a viral classification on the basis of the fundamental science (Ruska, 1943). The first use of TEM in clinical virology concerned the differential diagnosis of smallpox (caused by the variola virus of the poxvirus family) and chicken pox (caused by the varicella‐zoster virus of the herpes family) using fluid from the vesicles on the patients' skin (Nagler and Rake, 1948). Commercially available electron microscopes became widely available from several manufacturers during the 1960s and 1970s. Medical publications from this time featured large numbers of ultrastuctural investigations in thin sections of many embedded cells and organs (infected or uninfected; Figure 1A). The introduction of negative staining, making it possible to detect viruses from liquid samples deposited on carbon‐coated grids and stained with heavy‐metal salts (such as phosphotungstic acid or uranyl acetate), led to the widespread use of TEM in basic virology and rapid viral diagnosis (Brenner and Horne, 1959; Figures 2A and 2B). Negative staining not only makes the virus stand out from the background, but also provides morphological information about symmetry and capsomer arrangement; for example, making the specific identification of viruses or their classification into morphologically similar groups possible. Thus the use of TEM for the study of viruses peaked during the 1970s and 1980s when it contributed to the discovery of many clinically important viruses, such as adeno‐, entero‐, paramyxo‐ and reo‐viruses, which were isolated from diagnostic cell cultures. Differences in virus size and fine structure were used as criteria for classification (Tyrrell and Almeida, 1967). However, TEM failed to detect agents for other diseases, such as hepatitis and gastroenteritis, because susceptible cell cultures were not available for virus isolation or because the virus could not be cultured. However, a major breakthrough was made for these viruses in the 1970s, when TEM was applied to ‘dirty’ clinical samples, such as plasma, urine and faeces (Madeley, 1979). The aetiological agents of hepatitis B (Dane et al., 1970) and hepatitis A (Feinstone et al., 1973) were detected in plasma and stool samples respectively. Parvovirus B19 was discovered during a search for hepatitis B virus in a serum sample from a patient (Cossart et al., 1975). The BK virus, a polyomavirus, was first identified in the urine of patients undergoing organ transplantation (Gardner et al., 1971) (Figure 1B). Rotaviruses were also identified as the main cause of epidemic gastroenteritis in humans and animals by this technique (Bishop et al., 1973; Flewett et al., 1973). However, other viruses were found to be responsible for many outbreaks of gastroenteritis. The first of these viruses was the Norwalk virus, identified during a community outbreak of gastroenteritis in Norwalk, OH, U.S.A. (Kapikian et al., 1972; Kapikian, 2000). Viruses with a similar morphology were subsequently discovered elsewhere and called ‘Norwalk‐like’ or ‘small round‐structured’ viruses, to reflect the similarity of their appearance on TEM (Caul and Appleton, 1982), before being officially renamed noroviruses (Mayo, 2002). Other viruses from the adenovirus (Morris et al., 1975), astrovirus (Appleton and Higgins, 1975; Madeley and Cosgrove, 1975) and calicivirus (Madeley and Cosgrove, 1976) families were also identified in the stool samples of children suffering from gastroenteritis. This large diversity of viruses potentially involved in human gastroenteritis contributed to the use of TEM on negatively stained samples for routine diagnosis by this rapid ‘catch‐all’ method in clinical virology (Figures 2C and 2D).
Figure 1.
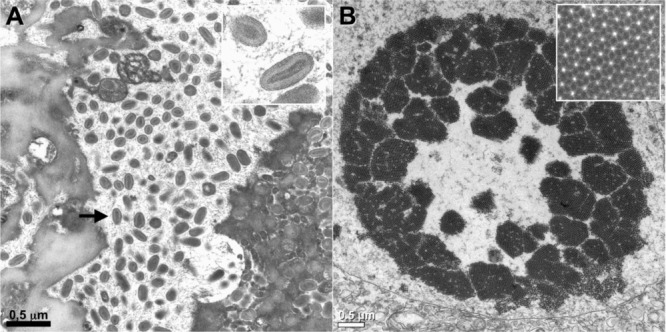
Diagnosis of virus infections by examination of ultrathin sections of human tissues or cells
(A) Parapoxvirus (Orf virus) infection on a human skin biopsy specimen. Multiple oval viral particles (arrow), comprising a dense core surrounded by an envelope (inset, high magnification), are observed in an infected cell. The Orf virus is a parapoxivirus that causes a common skin disease of sheep and goats, and is occasionally transmitted to human. (B) Polyomavirus (BK virus) infection in cells pelleted from a urine sample taken from an organ‐transplant patient. The presence of a large number of viral particles leads to their arrangement into a crystal‐like structure (inset, high magnification).
Figure 2.
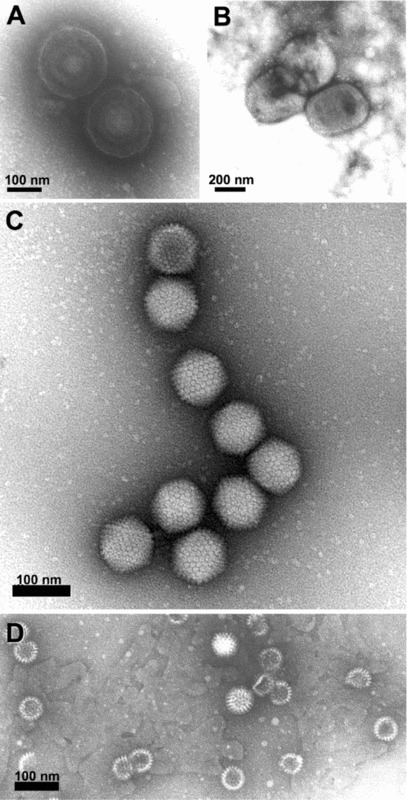
Direct negative staining of virus in fluid recovered from human skin vesicles (A and B) or from stool samples (C and D)
(A and B) Rapid morphological diagnosis and differential diagnosis from a herpesvirus (Varicella, A) and a parapoxvirus (Orf virus, B). The penetration of the negative stain into the herpesvirus particle may reveal the presence of the viral capsid within the envelope. (C and D) Negative staining of viruses involved in gastroenteritis reveals the surface detail of the subunit arrangement of the adenovirus core particle (C), showing its icosahedral form clearly, whereas the rotavirus displays its typical ‘wheel‐like’ appearance (D).
However, by the 1990s, the increasing development of other techniques, such as ELISAs (enzyme‐linked immunosorbent assays) and PCR, had contributed to a gradual decline in the use of TEM for viral diagnosis in cases of gastroenteritis (McCaughey et al., 2000; Biel and Madeley 2001). Indeed, these antigenic and molecular techniques are much more sensitive than TEM, which has a detection limit of 105–106 particles/ml. These new techniques are also more appropriate for the screening of large numbers of samples, and can now be used to detect most of the virus families involved in human gastroenteritis (Medici et al., 2005; Logan et al., 2006, 2007; Oka et al., 2006). A similar change has also been observed in veterinary medicine, in which ELISAs and PCR have progressively replaced TEM for routine viral diagnosis (Guo et al., 2001; van der Poel et al., 2003; Rodak et al., 2005; Tang et al., 2005). In human medicine, EM viral diagnosis for the differentiation of smallpox virus from other viruses present in the vesicle fluids of skin lesions is no longer required due to an intensive worldwide vaccination programme, leading to the successful eradication of the variola virus in 1980 (Henderson, 2002). It has been argued that TEM remains potentially useful for viral diagnosis, because the variola virus might be used for bioterrorism (Miller 2003; Curry et al., 2006). However, the risk of smallpox reappearing is very small, and, even in the unlikely event of smallpox re‐emerging, molecular techniques would certainly surpass TEM for its diagnosis.
Identifying emerging or ‘re‐emerging’ agents and the control of viral biosafety
The benefits of TEM for resolving diagnostic problems in clinical virology have nonetheless been clearly illustrated on several occasions in the last 15 years. TEM proved essential for the identification of a new morbillivirus (Hendra virus, belonging to the Paramyxoviridae) in horses and humans suffering from fatal respiratory infections in 1995 in Australia (Murray et al., 1995). A related virus, the Nipah virus, mostly affecting pig farmers in Malaysia, was discovered more recently (Chua et al., 1999). The aetiology of the SARS (severe acute respiratory syndrome) pandemic in Hong Kong and Southern China in 2003 was first identified as a coronavirus by TEM, leading to subsequent laboratory and epidemiological investigations (Drosten et al., 2003; Ksiazek et al., 2003; Goldsmith et al., 2004). A human monkeypox outbreak in the U.S.A. in 2003 was also diagnosed only once TEM had been used (Reed et al., 2004). TEM is occasionally useful for the identification of new subtypes of viruses involved in human gastroenteritis, such as adenovirus (Jones et al., 2007a) or picornavirus (Jones et al., 2007b). The role of TEM in clinical virology in recent years has thus changed from that of a routine technique to supporting the identification of unknown infectious agents in particular outbreaks. In such investigations, the underlying ‘catch‐all’ principle of this technique is essential for the recognition of an unknown agent. There are also many recent similar examples of the usefulness of TEM for identifying the virus involved in particular outbreaks in veterinary medicine (Coyne et al., 2006; Literak et al., 2006; Matz‐Rensing et al., 2006; Prukner‐Radovcic et al., 2006; Chan et al., 2007; Gruber et al., 2007; Maeda et al., 2007).
TEM currently plays an important role in the control of the biosafety of biological products. Rodent cell lines are widely used as substrates for producing biological therapeutic molecules, such as monoclonal antibodies, recombinant proteins, vaccines and viral vectors for gene therapy. These cell lines have long been known to contain retroviral elements, because the rodent genome contains many copies of endogenous retrovirus‐like sequences (Weiss, 1982). Most of the particles produced in cell culture, such as the intracisternal A‐type and R‐type particles, are defective and are non‐infectious. However, other particles, such as C‐type particles, bud at the cell surface and may infect non‐rodent cells (Lueders, 1991). Some murine retroviruses have been shown to be tumorigenic in primates (Donahue et al., 1992). Murine retroviral vectors have been shown to cause leukaemia in children with severe combined immunodeficiency treated by gene therapy with these vectors (Hacein‐Bey‐Abina et al., 2003). Regulatory agencies therefore recommend the use of a wide safety margin for biological products derived from rodent cells. They also recommend the use of several complementary techniques: reverse transcriptase assays (reverse transcriptase being specific to retroviruses), assays for infectivity in co‐culture and TEM of ultrathin sections (U.S. Food and Drug Administration, http://www.fda.gov/cder/Guidance/Q5A-fnl.pdf; European Medicines Agency, http://www.emea.europa.eu/pdfs/human/ich/029595en.pdf) for the detection and characterization of retroviruses in the manufacturers' master and end‐of‐production cells (Figure 3). These agencies also recommend testing for viruses in unprocessed bulks (one or multiple pools of harvested cells and culture medium). Reverse transcriptase assays with such bulks are hampered by high background levels due to cell‐derived DNA polymerases and are no more sensitive overall than TEM with negative staining for retrovirus detection (Brorson et al., 2002). PCR assays are difficult to set up, because investigators are faced with a multitude of murine viruses, many of which remain uncharacterized. Thus TEM with negative staining appears to be a useful technique for ensuring the biosafety of biological products in these conditions.
Figure 3.
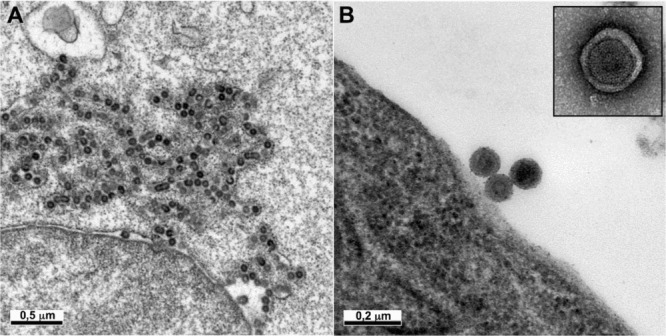
Detection of retroviruses in rodent hybridoma cells used for the production of biological products
Ultrathin sections of cells of different origins may show intracisternal A‐type retroviral particles (A) or C‐type retroviral particles budding at the cell surface (B). The C‐type particles released by the cells can be detected by negative staining in the cell supernatant (B, inset).
Study of virus—host cell interactions: a link between the past, the present and the future
TEM is the only technique able to deliver clear images of viruses due to their small size. Moreover, the fine detail of viral structure may become visible if viral preparations are rapidly frozen and the vitrified specimens examined by cryo‐EM. When combined with data from X‐ray diffraction studies, or with electron tomography or single‐particle analyses of isolated virions, highly detailed structures can be obtained at near atomic resolution (Grunewald et al., 2003; Cyrklaff et al., 2005; Forster et al., 2005; Briggs et al., 2006; Harris et al., 2006; Roux and Taylor, 2007). However, these approaches, which are based on purified viral preparations, are beyond the scope of the present article that focuses on the detection of viruses in fluids or infected cells or tissues. As obligate intracellular parasites, viruses depend on living cells for their replication. Interactions with host cells begin with the binding of the virus to specific receptors on the cell surface (Marsh and Helenius, 2006). Some viruses release their genomes directly into the cell by rupturing the plasma membrane, but most enter cells by endocytosis. Following their internalization, viral particles are sequestered in endocytotic organelles until appropriate conditions for viral genome release into the cytoplasm occur. Two main sequestration strategies are used: enveloped viruses fuse with a cell membrane, whereas non‐enveloped viruses partially disrupt cellular membranes. Further uncoating reactions and/or transport of the core may also be required. The viral genome is then translocated to specific sites in the cytoplasm or nucleus for replication and expression. The formation of ‘factories’ has been reported for many viruses. These factories consist of perinuclear or cytoplasmic foci—mostly excluding host proteins and organelles, but sometimes recruiting specific cell organelles—that form a unique structure characterized by a number of complex interactions and signalling events between cellular and viral factors (Wileman, 2006). The newly synthesized viral proteins and genetic material are then assembled into progeny viruses. Enveloped viruses acquire a lipid membrane by budding through a cellular membrane. Virus assembly may occur at the plasma membrane, but many viruses begin their assembly process in intracellular organelles, such as the ER (endoplasmic reticulum), Golgi apparatus or endosomes (Griffiths and Rottier, 1982). Our understanding of the various stages of the viral life cycle has therefore been greatly enhanced by TEM studies, some of which have included immunolabelling protocols. For example, virus entry pathways via the use of clathrin‐coated pits or caveolae have been shown by TEM analysis for various viruses (Helenius et al., 1980; Kartenbeck et al., 1989; Bousarghin et al., 2003). TEM has been used to study the generation of new virions, providing particularly striking images of viral factories for several virus families (Novoa et al., 2005) (Figure 4). TEM has also proved important for the characterization of morphological features at various stages in the assembly of viral particles, including the acquisition of lipid membranes by enveloped viruses, and for distinguishing between immature and mature particles (Hourioux et al., 2000; Pelchen‐Matthews and Marsh, 2007) (Figure 5).
Figure 4.
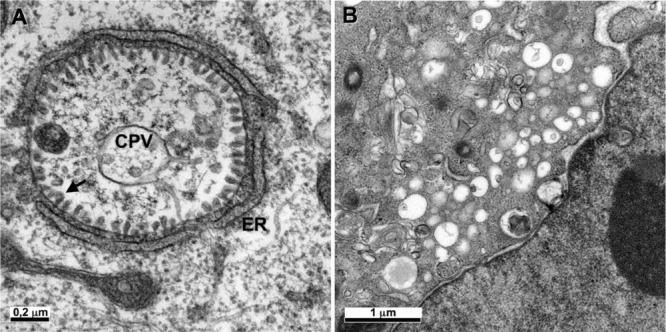
Ultrastructural changes associated with viral replication or viral factories
(A) The Semliki forest virus, an alphavirus, induces the formation of a cytopathic vacuole (CPV), surrounded by the ER. Numerous viral replication complexes (arrow) are anchored in the internal membrane of the CPV. (B) The non‐structural proteins of HCV, a flavivirus, induce the formation of a membranous web in the perinuclear area.
Figure 5.
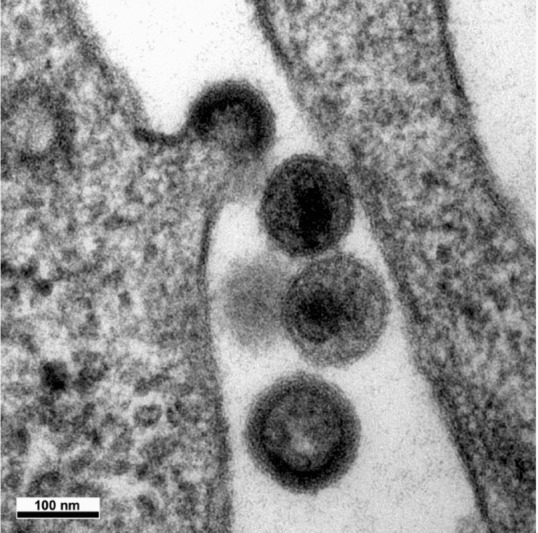
Budding of HIV
The viral particle at the top shows virus formation with distortion of a cellular membrane away from the cytoplasm. The budding particle and the particle at the bottom are immature viral particles, whereas the two particles in the centre are mature and have a truncated cone‐shaped core. Thus maturation of the core by the viral protease occurs shortly after the release of the particle from the host cell membrane.
However, the recent development of new imaging techniques for light microscopy has progressively limited the use of TEM for studies of virus—cell interactions. Viruses labelled with small fluorescent proteins or small dye molecules are currently the most powerful tools for studying dynamic interactions between viruses and the cellular machinery (reviewed in Brandenburg and Zhuang, 2007). It is now possible to follow the trafficking of a single virus in a single cell. One of the chief disadvantages of TEM is the need to use dead and fixed samples. The new techniques, which can be used on living cells, make it possible to follow the fate of individual viral particles and to monitor dynamic interactions between viruses and cellular structures, making it possible to study steps in infection that could previously not be observed. For example, the long‐standing debate concerning whether poliovirus breaches the plasma membrane barrier or relies on endocytosis to deliver its genome into cells was settled recently using this approach (Brandenburg et al. 2007). This study (Brandenburg et al. 2007) demonstrated that poliovirus enters cells via clathrin‐, caveolin‐ and microtubule‐independent, but tyrosine kinase‐ and actin‐dependent, endocytosis. Many studies have also demonstrated that various viruses hijack the microtubule‐ and actin filament‐based transport machinery for their transport within living cells (Brandenburg and Zhuang, 2007).
Nevertheless, these fluorescence methods do not reveal detailed information about the structure of viruses. Viruses can be visualized as small spots on fluorescence microscopy, but the resolution of this technique is too low to determine whether these fluorescent spots correspond to assembled virions or aggregated viral proteins. Thus TEM remains an essential technique for visualizing structured virions in infected cells, as shown by the following examples. HCV (hepatitis C virus) is unusual in that it was first identified by the cloning of its genome, before TEM visualization (Roingeard et al., 2004). Even today, little is known about the morphogenesis of this virus. TEM of a VLP (virus‐like particle) model obtained by expressing genes encoding the HCV structural proteins has demonstrated that viral budding occurs at the ER membrane and that the HCV core protein drives this process (Blanchard et al., 2003). Fluorescence microscopy has shown that most of the HCV core protein is associated with the surface of lipid droplets (McLauchlan et al., 2002), and TEM has shown that viral budding occurs at the ER membrane in the close vicinity of these lipid droplets (Ait‐Goughoulte et al., 2006; Hourioux et al., 2007) (Figure 6). TEM has also shed light on the morphogenesis and intracellular trafficking of subviral envelope particles associated with HBV (hepatitis B virus). According to an initial model based on biochemical and fluorescence microscopy studies, the major HBV envelope protein forms dimers in the ER membrane before it is transported, as transmembrane dimers in vesicles, to the ERGIC (ER—Golgi intermediate compartment) where its self‐assembly leads to the morphogenesis of subviral envelope particles (Huovila et al., 1992). However, recent TEM studies have shown that HBV subviral particles self‐assemble into filaments within the ER lumen (Patient et al., 2007). TEM has also shown that these filaments are packed into crystal‐like structures for transport by ER‐derived vesicles to the ERGIC, where they are unpacked and relaxed (Figure 7). HIV morphogenesis also provides a remarkable example of intensive research into virus—host cell interactions, for which the debate concerning the site of virus assembly remains unresolved. Studies in this field make use of TEM to determine the site of virus assembly, which may occur at the plasma membrane or in late endosomes, depending on the virus—host cell system (Grigorov et al., 2006; Jouvenet et al., 2006; Pelchen‐Matthews and Marsh, 2007; Welsch et al., 2007). Thus light microscopy and TEM are not exclusive and rather complementary. One particularly interesting development is CLEM (correlative light electron microscopy), which combines ultrastructural and fluorescence visualization (Frishknecht et al., 2006). The use of quantum dots (Giepmans et al., 2005) or small genetic tags, such as tetra‐cysteine motifs (Gaietta et al., 2002; Lanman et al., 2007), which are readily visible on both TEM and light microscopy, is useful for such approaches. Other methods, such as GRAB [GFP (green fluorescent protein) recognition after bleaching], use oxygen radicals generated during the GFP bleaching process to photo‐oxidize diaminobenzidine into an electron‐dense precipitate that can be visualized by EM (Grabenbauer et al., 2005).
Figure 6.
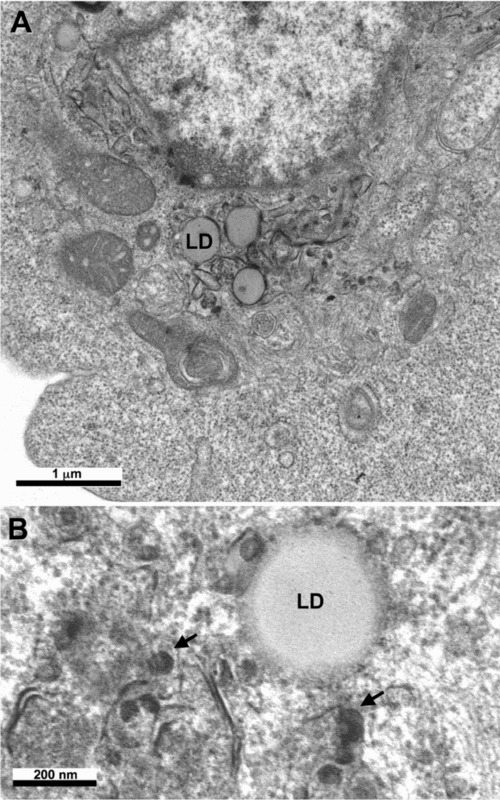
Budding of HCV
Ultrastructural analysis of cells producing the HCV core protein shows that this protein self‐assembles into HCV‐like particles (arrows) at convoluted and electron‐dense ER membranes surrounding the lipid droplets (LD) present in the perinuclear area.
Figure 7.
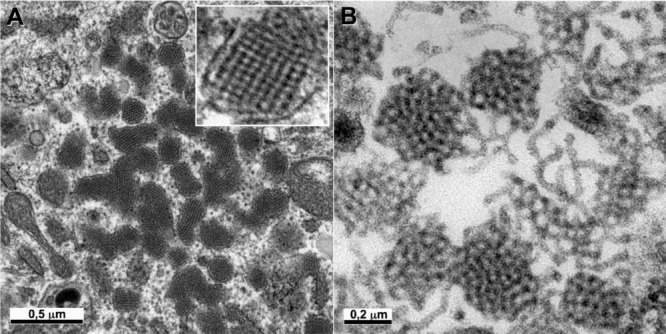
Hepatitis B subviral envelope particle morphogenesis and intracellular trafficking
Ultrastructural analysis of cells producing HBV major envelope protein shows that this protein self‐assembles in the ER into filaments packed into crystal‐like structures (A, see also a high magnification of these packed filaments in the inset). These filaments are transported to the ERGIC, where they are unpacked and relaxed (B).
In conclusion, TEM is clearly less important than it once was in the field of diagnostic virology, but this technique remains useful for the occasional identification of unknown agents during particular outbreaks. In this respect, it is a valuable technique for controlling the viral safety of biological products. For research, TEM is complementary to other investigative techniques for elucidating many aspects of the life‐cycle of the virus in an infected cell, including viral assembly, in particular, as ultrastructural analyses may be remarkably informative.
Acknowledgments
I thank Brigitte Arbeille, Elodie Beaumont, Emmanuelle Blanchard, Denys Brand, Christophe Hourioux, Romuald Patient, Pierre‐Yves Sizaret (Université François Rabelais, Tours, France), Delphine Muriaux, Jean‐Luc Darlix (Ecole Normale Supèrieure de Lyon, Lyon, France), Jean Dubuisson, Yves Rouillé (Institut de Biologie de Lille, Lille, France), Gérard Prensier (Université Blaise Pascal, Clermont‐Ferrand, France) and Ali Salib (Université de Paris VII, Paris, France) for sharing data concerning our ongoing projects on virus—host cell interactions, and Fabienne Arcanger and Sylvie Trassard (Université François Ralelais et CHRU, Touss, France) for their invaluable help with TEM techniques. INSERM ERI 19 is supported by grants 06038 and 06199 from the ANRS (Agence Nationale de la Recherche sur le SIDA et les hépatites virales), a Virodynamics grant from the ANR (Agence Nationale de la Recherche) and the Centre Region (ESPRI). The Electron Microscopy Facility, Université François Rabelais, is part of the French RIO/IBISA (Réunion Inter Organismes/Infrastructures en Biologie Sante et Agronomie) network.
References
- Ait‐Goughoulte M. Hourioux C. Patient R. Trassard S. Brand D. Roingeard P. Core protein cleavage by signal peptide peptidase is required for hepatitis virus‐like particle assembly J. Gen. Virol. 2006. 87 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton H. Higgins P.G. Viruses and gastroenteritis in infants. Lancet. 1975;i:1297. doi: 10.1016/s0140-6736(75)92581-7. [DOI] [PubMed] [Google Scholar]
- Biel S.S. Madeley D. Diagnostic virology—the need for electron microscopy: a discussion paper J. Clin. Virol. 2001. 22 1–9 [DOI] [PubMed] [Google Scholar]
- Bishop R.F. Davidson G.P. Holmes I.H. Ruck B.J. Virus particles in epithelial cells of duodenal mucosa from children with acute non‐bacterial gastroenteritis Lancet 1973. ii 1281–1283 [DOI] [PubMed] [Google Scholar]
- Blanchard E. Hourioux C. Brand D. Ait‐Goughoulte M. Moreau A. Trassard S. Sizaret P.‐Y. Dubois F. Roingeard P. Hepatitis C virus‐like particle budding: role of the core protein and importance of its Asp111 J. Virol. 2003. 77 10131–10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousarghin L. Touze A. Sizaret P.‐Y. Coursaget P. Human papillomavirus types16, 31, and 58 use different endocytosis pathways to enter cells J. Virol. 2003. 77 3846–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B. Zhuang X. Virus trafficking—learning from single‐virus tracking Nat. Rev. Microbiol. 2007. 5 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B. Lee L.Y. Lakadamyali M. Rust M.J. Zhuang X. Hogle J.M. Imaging poliovirus entry in live cells. PloS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. Horne R.W. A negative staining method for high resolution electron microscopy of viruses Biochim. Biophys. Acta 1959. 34 103–110 [DOI] [PubMed] [Google Scholar]
- Briggs J.A. Grunewald K. Glass B. Forster F. Krausslich H.G. Fuller S.D. The mechanisms of HIV‐1 core assembly: insights from three‐dimensional reconstructions of authentic virions Structure 2006. 14 15–20 [DOI] [PubMed] [Google Scholar]
- Brorson K. Xu Y. Swann P.G. Hamilton E. Mustafa M. de Wit C. Norling L.A. Stein K.E. Evaluation of a quantitative product‐enhanced reverse transcriptase assay to monitor retrovirus in mAb cell‐culture Biologicals 2002. 30 15–26 [DOI] [PubMed] [Google Scholar]
- Caul E.O. Appleton H. The electron microscopical and physical characteristics of small round human fecal viruses: an interim scheme for classification J. Med. Virol. 1982. 9 257–265 [DOI] [PubMed] [Google Scholar]
- Chan K.W. Lin J.W. Lee S.H. Liao C.J. Tsai M.C. Hsu W.L. Wong M.L. Shih H.C. Identification and phylogenetic analysis of orf virus from goats in Taiwan Virus Genes 2007. 35 705–712 [DOI] [PubMed] [Google Scholar]
- Chua K.B. Goh K.J. Wong K.T. Kamarulzaman A. Tan P.S. Ksiazek T.G. Zaki S.R. Paul G. Lam S.K. Tan C.T. Fatal encephalitis due to Nipah virus among pig‐farmers in Malaysia Lancet 1999. 354 1257–1259 [DOI] [PubMed] [Google Scholar]
- Cossart Y.E. Field A.M. Cant B. Widdows D. Parvovirus‐like particles in human sera Lancet 1975. i 72–73 [DOI] [PubMed] [Google Scholar]
- Coyne K.P. Jones B.R. Kipar A. Chantrey J. Porter C.J. Barber P.J. Dawson S. Gaskell R.M. Radford A.D. Lethal outbreak of disease associated with feline calicivirus infection in cats Vet. Rec. 2006. 158 544–550 [DOI] [PubMed] [Google Scholar]
- Curry A. Bryden A. Morgan‐Capner P. Fox A. Guiver M. Martin L. Mutton K. Wright P. Mannion P. Westwell A. Cheesbrought J. Aston I. Blackey A. A rationalised virological electron microscope specimen testing policy: PHLS north west viral gastroenteritis and electron miscroscopy subcommittee J. Clin. Pathol. 1999. 52 471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. Bryden A. Morgan‐Capner P. Caul E.O. Rationalised virological electron microscope specimen testing policy. J. Clin. Pathol. 2000a;53:163. doi: 10.1136/jcp.53.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. Caul E.O. Morgan‐Capner P. Rationalised virological electron microscope specimen testing policy. J Clin. Pathol. 2000b. 53 722–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. Appleton H. Dowsett B. Application of transmission electron microscopy to the clinical study of viral and bacterial infections: present and future Micron 2006. 37 91–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff M. Risco C. Fernandez J.J. Jimenez M.V. Esteban M. Baumeister W. Carrascosa J.L. Cryo‐electron tomography of vaccinia virus Proc. Natl. Acad. Sci. U.S.A. 2005. 102 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dane D.S. Cameron C.H. Briggs M. Virus‐like particles in serum of patients with Australia antigen‐associated hepatitis Lancet 1970. i 695–698 [DOI] [PubMed] [Google Scholar]
- Donahue R.E. Kessler S.W. Bodine D. McDonagh K. Dunbar C. Goodman S. Agricola B. Byrne E. Raffeld M. Moen R. Bacher J. Zsebo K.M. Niehia A.W. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer J. Exp. Med. 1992. 4 1125–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C. Gunther S. Preiser W. van der Werf S. Brodt H.R. Becker S. Rabenau H. Panning M. Kolesnikova L. Fouchier R.A. Berger A. Burguiere A.M. Cinatl J. Eickmann M. Escriou N. Grywna K. Kramme S. Manuguerra J.C. Muller S. Rickerts V. Sturmer M. Vieth S. Klenk H.D. Osterhaus A.D. Schmitz H. Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome New Engl. J. Med. 2003. 348 967–976 [DOI] [PubMed] [Google Scholar]
- Feinstone S.M. Kapikian A.Z. Purcell R.H. Hepatitis A: detection by immune electron microscopy of a virus‐like antigen associated with acute illness Science 1973. 182 1026–1028 [DOI] [PubMed] [Google Scholar]
- Flewett T.H. Bryden A.S. Davies H. Virus particles in gastroenteritis. Lancet. 1973;i:1497. doi: 10.1016/s0140-6736(73)92760-8. [DOI] [PubMed] [Google Scholar]
- Forster F. Medella O. Zauberman N. Baumeister W. Fass D. Retrovirus envelope protein complex structure in situ studied by cryo‐electron tomography Proc. Natl. Acad. Sci. U.S.A. 2005. 102 4729–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht F. Renaud O. Shorte S.L. Imaging today's infectious animalcules Curr. Opin. Microbiol. 2006. 9 297–306 [DOI] [PubMed] [Google Scholar]
- Gaietta G. Deerinck T.J. Adams S.R. Bouwer J. Tour O. Laird D.W. Sosinsky G.E. Tsien R.Y. Ellisman M.H. Multicolor and electron microscopic imaging of connexin trafficking Science 2002. 296 503–507 [DOI] [PubMed] [Google Scholar]
- Gardner S.D. Field A.M. Coleman D.V. Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation Lancet 1971. i 1253–1257 [DOI] [PubMed] [Google Scholar]
- Giepmans B. Deerinck T.J. Smarr B.L. Jones Y.Z. Ellisman M.H. Correlated light and electron microscopic imaging of multiple endogenous protein using quantum dots Nat. Methods 2005. 2 743–749 [DOI] [PubMed] [Google Scholar]
- Goldsmith C.S. Tatti K.M. Ksiazek T.G. Rollin P.E. Comer J.A. Lee W.W. Rota P.A. Bankamp B. Bellini W.J. Zaki S.R. Ultrastructural characterization of SARS coronavirus Emerging Infect. Dis. 2004. 10 320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenbauer M. Geerts W.J. Fernandez‐Rodriguez J. Hoenger A. Koster A.J. Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation Nat. Methods 2005. 2 857–862 [DOI] [PubMed] [Google Scholar]
- Griffiths G. Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway Semin. Cell Biol. 1982. 3 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B. Arcanger F. Roingeard P. Darlix J.L. Muriaux D. Assembly of infectious HIV‐1 in human epithelial and T‐lymphoblastic cell lines J. Mol. Biol. 2006. 359 848–862 [DOI] [PubMed] [Google Scholar]
- Gruber A. Grabensteiner E. Kolodziejek J. Nowotny N. Loupal G. Poxvirus infection in a great tit (Parus major) Avian Dis. 2007. 51 623–625 [DOI] [PubMed] [Google Scholar]
- Grunewald K. Desai P. Winckler D.C. Heymann J.B. Belnap D.M. Baumeister W. Steven A.C. Three‐dimensional structure of herpes simplex virus from cryo‐electron tomography Science 2003. 302 1396–1398 [DOI] [PubMed] [Google Scholar]
- Guo M. Evermann J.F. Saif L.J. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink Arch. Virol. 2001. 146 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein‐Bey‐Abina S. von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. de Basile G. Saint Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa‐Lyonnet D. Romana S. Radford‐Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana‐Calvo M. LMO2‐associated clonal T cell proliferation in two patients after gene therapy for SCID‐X1 Science 2003. 302 415–419 [DOI] [PubMed] [Google Scholar]
- Haguenau F. Hawkes P.W. Hutchison J.L. Satiat‐Jeunemaitre B. Simon G.T. Williams D.B. Key events in the history of electron microscopy Microsc. Microanal. 2003. 9 96–138 [DOI] [PubMed] [Google Scholar]
- Harris A. Cardone G. Winckler D.C. Heyman J.B. Brecher M. White J.M. Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography Proc. Natl. Acad. Sci. U.S.A. 2006. 103 19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A. Kartenbeck J. Simons K. Fries E. On the entry of Semliki forest virus into BHK‐21 cells J. Cell Biol. 1980. 84 404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D.A. Countering the posteradication threat of smallpox and polio Clin. Infect. Dis. 2002. 34 79–83 [DOI] [PubMed] [Google Scholar]
- Hourioux C. Brand D. Sizaret P.‐Y. Lemiale F. Lebigot S. Barin F. Roingeard P. Identification of the glycoprotein 41TM cytoplasmic tail domains of human immunodeficiency virus type1 that interact with Pr55Gag particles AIDS Res. Hum. Retroviruses 2000. 16 1141–1147 [DOI] [PubMed] [Google Scholar]
- Hourioux C. Ait‐Goughoulte M. Patient R. Fouquenet D. Arcanger‐Doudet F. Brand D. Martin A. Roingeard P. Core protein domain involved in hepatitis C virus‐like particle assembly and budding at the endoplasmic reticulum membrane Cell Microbiol. 2007. 9 1014–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovila A.P. Eder A.M. Fuller S.D. Hepatitis B surface antigen assembles in a post‐ER, pre‐Golgi compartment J. Cell Biol. 1992. 118 1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.S. Harrach B. Ganac R.D. Gozum M.M. de la Cruz W.P. Riedel B. Pan C. Delwart E.L. Schnurr D.P. New adenovirus species found in a patient presenting with gastroenteritis J. Virol. 2007a. 81 5978–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.S. Lukashov V.V. Ganac R.D. Schnurr D.P. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin J. Clin. Microbiol. 2007b. 45 2144–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N. Neil S.J.D. Bess C. Johnson M.C. Virgen C.A. Simon S.M. Bieniasz P.D. Plasma membrane is the site of productive HIV‐1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian A.Z. The discovery of the 27‐nm Norwalk virus: an historic perspective J. Infect. Dis. 2000. 181 S295–S302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian A.Z. Wyatt R.G. Dolin R. Thornhill T.S. Kalica A.R. Chanock R.M. Visualization by immune electron microscopy of a 27‐nm particle associated with acute infectious nonbacterial gastroenteritis J. Virol. 1972. 10 1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J. Stukenbrok H. Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum J. Cell Biol. 1989. 109 2721–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll M. Ruska E. Das elektronenmikroscop Zeitschrift fur Physik 1932. 78 318–329 [Google Scholar]
- Ksiazek T.G. Erdman D. Goldsmith C.S. Zaki S.R. Peret T. Emery S. Tong S. Urbani C. Comer J.A Lim W. Rollin P.E. Dowell S.F. Ling A.E. Humphrey C.D. Shieh W.J. Guarner J. Paddock C.D. Rota P. Fields B. De Risi J. Yang J.Y. Cox N. Hughes J.M. Le Duc J.W. Bellini W.J. Anderson L.J. The SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome New Engl. J. Med. 2003. 348 1953–1966 [DOI] [PubMed] [Google Scholar]
- Lanman J. Crum J. Deerinck T.J. Gaietta G.M. Schneemann A. Sosinsky G.E. Ellisman M.H. Johnson J.E. Visualizing flock house virus infection in Drosophilia cells with correlated fluoresence and electron microscopy Struct. Biol. 2007. 161 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I. Smid B. Dubska L. Bryndza L. Valicek L. An outbreak of the polyomavirus infection in budgerigars and cockatiels in Slovakia, including a genome analysis of an avian polyomavirus isolate Avian Dis. 2006. 50 120–123 [DOI] [PubMed] [Google Scholar]
- Logan C. O'Leary J.J. O'sullivan N. Real‐time reverse transcription‐PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children J. Clin. Microbiol. 2006. 44 3189–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C. O'leary J.J. O'sullivan N. Real‐time reverse transcription PCR detection of norovirus, sapovirus and astrovirus as causative agents of acute viral gastroenteritis J. Virol. Methods 2007. 146 36–44 [DOI] [PubMed] [Google Scholar]
- Lueders K.K. Genomic organization and expression of endogenous retrovirus‐like elements in cultured rodent cells Biologicals 1991. 1 1–7 [DOI] [PubMed] [Google Scholar]
- Madeley C.R. Viruses in the stools J. Clin. Pathol. 1979. 32 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeley C.R. Rationalised virological electron microscope specimen testing policy J. Clin. Pathol. 2000. 53 722–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeley C.R. Cosgrove B.P. 28 nm particles in faeces in infantile gastroenteritis Lancet 1975. ii 451–452 [DOI] [PubMed] [Google Scholar]
- Madeley C.R. Cosgrove B.P. Caliciviruses in man Lancet 1976. i 199–200 [DOI] [PubMed] [Google Scholar]
- Maeda Y. Shibahara T. Wada Y. Kadota K. Kanno T. Uchida I. Hatama S. An outbreak of teat papillomatosis in cattle caused by bovine papilloma virus (BPV) type6 and unclassified BPVs Vet. Microbiol. 2007. 121 242–248 [DOI] [PubMed] [Google Scholar]
- Marsh M. Helenius A. Virus entry: open sesame Cell 2006. 124 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz‐Rensing K. Ellerbrok H. Ehlers B. Pauli G. Floto A. Alex M. Czerny C.P. Kaup F.J. Fatal poxvirus outbreak in a colony of New World monkeys Vet. Pathol. 2006. 43 212–218 [DOI] [PubMed] [Google Scholar]
- Mayo M.A. A summary of taxonomic changes recently approved by ICTV Arch. Virol. 2002. 147 1655–1663 [DOI] [PubMed] [Google Scholar]
- McCaughey C. O'Neill H. Wyatt D.E. Coyle P.V. Rationalised virological electron microscope specimen testing policy. J. Clin. Pathol. 2000;53:163. doi: 10.1136/jcp.53.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J. Lemberg M.K. Hope G. Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets EMBO J. 2002. 21 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici M.C. Martinelli M. Ruggeri F.M. Abelli L.A. Bosco S. Arcangeletti M.C. Pinardi F. De Conto F. Calderaro A. Chezzi C. Dettori G. Broadly reactive nested reverse transcription‐PCR using an internal RNA standard control for detection of noroviruses in stool samples J. Clin. Microbiol. 2005. 43 3772–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.E. Bioterrorism and electron microscopic differentiation of poxviruses from herpesviruses: dos and don'ts Ultrastruct. Pathol. 2003. 27 133–140 [DOI] [PubMed] [Google Scholar]
- Morris C.A. Flewett T.H. Bryden A.S. Davies H. Epidemic viral enteritis in a long‐stay children's ward Lancet 1975. i 4–5 [DOI] [PubMed] [Google Scholar]
- Murray K. Selleck P. Hooper P. Hyatt A. Gould A. Gleeson L. Westbury H. Hiley L. Selvey L. Rodwell B. Ketterer P. A morbillivirus that caused fatal disease in horses and humans Science 1995. 268 94–97 [DOI] [PubMed] [Google Scholar]
- Nagler F.P. Rake G. The use of the electron microscope in diagnosis of variola, vaccinia, and varicella J. Bacteriol. 1948. 55 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa R.R Calderita G. Arranz R. Fontana J. Granzow H. Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis Biol. Cell 2005. 97 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T. Katayama K. Hansman G.S. Kageyama T. Ogawa S. Wu F.T. White P.A. Takeda N. Detection of human sapovirus by real‐time reverse transcription‐polymerase chain reaction J. Med. Virol. 2006. 78 1347–1353 [DOI] [PubMed] [Google Scholar]
- Patient R. Hourioux C. Sizaret P‐Y. Trassard S. Sureau C. Roingeard P. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking J. Virol. 2007. 81 3842–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen‐Matthews A. Marsh M. Electron microscopy analysis of viral morphogenesis Methods Cell Biol. 2007. 79 515–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prukner‐Radovcic E. Luschow D. Ciglar‐Grozdanic I. Tisljar M. Mazija H. Vranesic D. Hafez H.M. Isolation and molecular biological investigations of avian poxviruses from chickens, a turkey, and a pigeon in Croatia Avian Dis. 2006. 50 440–444 [DOI] [PubMed] [Google Scholar]
- Reed K.D. Melski J.W. Graham M.B. Regnery R.L. Sotir M.J. Wegner M.V. Kazmierczak J.J. Stratman E.E. Li Y. Fairley J.A. Swain G.R. Olson V.A. Sargent E.K. Kehl S.C. Frace M.A. Kline R. Foldy S.L. Davis J.P. Damon I.K. The detection of monkeypox in humans in the Western Hemisphere New Engl. J. Med. 2004. 350 342–350 [DOI] [PubMed] [Google Scholar]
- Rodak L. Valicek L. Smid B. Nevorankova Z. An ELISA optimized for porcine epidemic diarrhoea virus detection in faeces Vet. Microbiol. 2005. 105 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard P. Hourioux C. Blanchard E. Brand D. Ait‐Goughoulte M. Hepatitis C virus ultrastructure and morphogenesis Biol. Cell 2004. 96 103–108 [DOI] [PubMed] [Google Scholar]
- Roux K.H. Taylor K.A. AIDS virus envelope spike structure Curr. Opin. Struct. Biol. 2007. 17 244–252 [DOI] [PubMed] [Google Scholar]
- Ruska H. Versuch zu einer ordnung der virusarten Arch. Gesamte Virusforsch. 1943. 2 480–498 [Google Scholar]
- Ruska E. Nobel lecture: the development of the electron microscope and of electron microscopy Biosci. Rep. 1987. 7 607–629 [DOI] [PubMed] [Google Scholar]
- Ruska H. von Borries B. Ruska E. Die bedeutung der ubermikroskopie fur die virusforschung Arch. Gesamte Virusforsch. 1939. 1 155–159 [Google Scholar]
- Tang Y. Wang Q. Saif M. Development of a ssRNA internal control template reagent for a multiplex RT‐PCR to detect turkey astroviruses J. Virol. Methods 2005. 126 81–86 [DOI] [PubMed] [Google Scholar]
- Tyrrell D.A. Almeida J.D. Direct electron‐microscopy of organ culture for the detection and characterization of viruses Arch. Gesamte Virusforsch. 1967. 22 417–425 [DOI] [PubMed] [Google Scholar]
- van der Poel W.H. van der Heide R. Verschoor F. Gelderblom H. Vinje J. Koopmans M.P. Epidemiology of Norwalk‐like virus infections in cattle in The Netherlands Vet. Microbiol. 2003. 92 297–309 [DOI] [PubMed] [Google Scholar]
- Weiss R.A. Retroviruses produced by hybridomas. New Engl. J. Med. 1982;307:1587. doi: 10.1056/NEJM198212163072522. [DOI] [PubMed] [Google Scholar]
- Welsch S. Kepper O.T. Haberman A. Allespach I. Krijnse‐Locker J. Krausslich H.G. HIV‐1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication Science 2006. 312 875–878 [DOI] [PubMed] [Google Scholar]


