Abstract
Modified oligonucleotides represent a promising avenue for drug development, with small interfering RNAs (siRNA) and microRNAs gaining traction in the therapeutic market. Mass spectrometry (MS)-based analysis offers many benefits for characterizing modified nucleic acids. Negative electron transfer dissociation (NETD) has proven valuable in sequencing oligonucleotide anions, particularly because it can retain modifications while generating sequence-informative fragments. We show that NETD can be successfully implemented on a widely-available quadrupole-Orbitrap-linear ion trap mass spectrometer that uses a front-end glow discharge source to generate radical fluoranthene reagent cations. We characterize both unmodified and modified ribonucleic acids and present the first application of activated-ion negative electron transfer dissociation (AI-NETD) to nucleic acids. AI-NETD achieved 100% sequence coverage for both a 6-mer (5’-rGmUrArCmUrG-3’) with 2’-O-methyl modifications and a 21-mer (5’-rCrArUrCrCrUrCrUrArGrArGrGrArUrArGrArArUrG-3’), the luciferase antisense siRNA. Both NETD and AI-NETD afforded complete sequence coverage of these molecules while maintaining a relatively low degree of undesired base-loss products and internal products relative to collision-based methods.
Graphical Abstract
INTRODUCTION
Nucleic acids play vital roles in the storage, processing, and expression of genetic information. Ribonucleic acids (RNAs) exhibit limited primary sequence information, a phenomenon remedied through introduction of post-transcriptional modifications (PTrMs).1–3 Modified RNAs are exceptional in their ability to maintain efficacy in the presence of cellular ribonucleases and confer cellular uptake for proper delivery to their target site.4,5 Consequently, researchers have envisioned an RNA therapeutic for nearly two decades, with the first RNA-based drug finally commercially available in 2018.6 This landmark achievement, which utilizes small interfering RNA (siRNA), represents an inflection point in the burgeoning field of targeted RNA therapeutics.7
Small interfering RNAs are double-stranded RNA molecules that facilitate potent and sequence-specific gene suppression via the mechanism of RNA interference (RNAi).8 Introducing various modifications, such as exchanging certain phosphorous atoms for sulfur atoms and optimizing the position of 2’-fluoro and 2’-O-methyl ribose modifications, allows researchers to increase stability, longevity, and efficacy of biologically active therapeutics in vivo by conferring nuclease resistance or by blocking innate immune stimulation.5 In-depth characterization of both sequence and modification status of RNA therapeutics, of course, is required to understand pharmacological activity. With over 100 diverse chemical modifications known to be introduced enzymatically during the process of RNA maturation, the usually powerful methods of oligonucleotide sequencing, such as RNA-Seq and biochemical labeling strategies, are not optimal for screening oligonucleotide PTrMs with high-throughput.9,10 The high degree of interest in epitranscriptomics, and the current inflection point of the oligonucleotide therapeutic market, require an efficient analysis platform for RNA characterization – mass spectrometry (MS) has emerged as a suitable candidate.11–16
Despite this need, the development of MS-based methods for RNA is limited compared to MS analysis of proteins. Now nearly three decades ago, McLuckey and others successfully electrosprayed various oligonucleotides and characterized fragmentation patterns using collision-based activation methods. Unfortunately, RNA characterization by MS still presents considerable challenges. One reason is that the acidic nature of RNA molecules makes them most suitable for generation of multiply deprotonated analyte anions.17–20 Thus, tandem MS fragmentation methods must be compatible with precursor anions. Traditional collision-based fragmentation, i.e., collision-activated dissociation via resonant excitation (CAD) and beam-type CAD (a.k.a., HCD), are among the most studied tandem MS strategies because of their accessibility and ease of application to deprotonated precursors.21 For RNA anion dissociation, these methods direct cleavage across numerous channels, often leading to highly complex spectra and the generation of internal fragments and neutral losses (which can include the loss of RNA nucleobases themselves).11,22,23 McLuckey and others demonstrated the dependence of charge-state and activation energy on product ion distributions and, for CAD, these drawbacks can be reduced for precursors having low charge states by minimizing collisional energy.24–26
The ideal method for dissociating RNA anions would be indifferent to precursor length, charge, and the presence of modifications. For years a variety of labs have explored alternative fragmentation methods employing electrons or photons to address these shortcomings. Pioneering efforts in the field of ion-ion and ion-molecule reactions of oligonucleotides by McLuckey and co-workers over 20 years ago demonstrated thermodynamic and mechanistic underpinnings of such reactions and highlighted their great potential.27–30 Recent work by McLuckey, Breuker, and others have established negative electron transfer dissociation (NETD) and electron detachment dissociation (EDD) – the negative-mode analogues of electron transfer dissociation (ETD) and electron capture dissociation (ECD), respectively – as promising analytical methods for RNA characterization.31–35 Ultraviolet photodissociation (UVPD), infrared photodissociation (IRMPD), and electron photodetachment dissociation (EPD) have been also explored.36–41 NETD and EDD generally produce d- and w-type product ions (in contrast to c- and y-type ion fragments generated in collisional dissociation), while also retaining labile modifications.29,31 A major challenge of these electron-based methods, however, is their low precursor-to-product-ion conversion efficiency.29 This low efficiency can be somewhat mitigated by the use of supplemental activation following the reaction.31–33,35,40
Whereas EDD is largely reliant on FT-ICR instrumentation, NETD can occur on any instrument with a RF-trapping device.32,36,42 NETD has been implemented on a number of instrument platforms using several different reagent cations, including fluoranthene, sulfur hexafluoride, xenon, metal adducts, and others.28,34,35,43–49 Fluoranthene is the most widely used reagent for characterization of peptides and oligosaccharides.50–53 Using a front-end glow discharge source to generate radical fluoranthene cations, we report here the first implementation of NETD on the widely available quadrupole-Orbitrap-ion trap MS system (Orbitrap Fusion Lumos). And, to our knowledge, this is the first application of fluoranthene cations for dissociation of RNA anions, a 6-mer with two 2’-O-methyl modifications and the 21-mer luciferase antisense siRNA. Finally we explore the use of concurrent infrared photoactivation (AI-NETD) to disrupt higher-order gas-phase structure and improve efficiency of the NETD fragmentation for RNA sequence analysis. 54,55 In all, we provide a new MS platform for RNA characterization that has the ability to rapidly sequence RNA and localize its modifications.
EXPERIMENTAL SECTION
Materials
The short, synthetically modified ribonucleic acid 5’-rGmUrArCmUrG-3’ with 2’-O-methyl modifications on the second and fifth nucleotides and the luciferase antisense siRNA with sequence 5’-rCrArUrCrCrUrCrUrArGrArGrGrArUrArGrArArUrG-3’ were obtained from Integrated DNA Technologies (Coralville, IA) on the 1.0 μmole scale and used without further purification. Both oligonucleotides contained 3’- and 5’-terminal hydroxyl groups. Oligonucleotide single strand concentrations were determined spectrophotometrically by Beer’s Law using the extinction coefficients provided by the manufacturer. For electrospray mass spectrometry analysis, solutions were diluted to 5 pmol/μL of oligonucleotide by preparing in 50:50 (v/v) H2O:CH3OH with 10 mM ammonium acetate. Methanol (HPLC grade), H2O (HPLC grade), ammonium acetate (>97%), and 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) (>99%) were obtained from Sigma-Aldrich (St. Louis, MO).
Mass Spectrometry
All MS and MS/MS experiments were performed on a quadrupole-Orbitrap-quadrupole linear ion trap (q-OT-QLT) hybrid mass spectrometer system (Orbitrap Fusion Lumos, Thermo Fisher Scientific, San Jose, CA) which was modified to perform negative-mode electron transfer dissociation. Additionally, the mass spectrometer was modified to include a Firestar T-100 Synrad 60 W CO2 continuous wave IR laser (10.6 μm) (Mukilteo, WA) to allow for the excitation of precursor ions within the quadrupole linear ion trap during ion-ion reactions, as previously reported.56 Oligonucleotide samples were directly infused at a flow rate of 3–5 μL/min using a Hamilton GasTight Valco syringe (Reno, NV) and a Chemyx Fusion 101 syringe pump (Stafford, TX), and anions were generated via electrospray in the negative ion mode with a heated electrospray voltage of −2.7 kV relative to ground and a transfer tube temperature of 275 °C. The ion funnel RF was held at 30%. To aid in desolvation, nitrogen sheath and auxiliary gas were applied at 15 and 10 arbitrary units, respectively. Data was collected in profile mode. For increased statistics, MS/MS spectra are the sum of 20 individual scans, with averaging performed in the vendor’s post-acquisition software (Xcalibur Qual Browser, version 2.2). No microscans were performed.
For NETD experiments, precursor anions were selected in the quadrupole with a 4 Th isolation window and were accumulated in the middle section of the high-pressure trap of the quadrupole linear ion trap, followed by accumulation of fluoranthene reagent cation (202 m/z) within the front section of the trap, and subsequent charge sign independent trapping for the NETD reaction. The fluoranthene radical cation reagent is generated by a chemical ionization source at the front of the instrument.57 All product ions and unreacted precursor ions were subsequently sent to the Orbitrap for mass analysis at a resolving power of 120k at 200 m/z. During AI-NETD experiments, the 60 W continuous wave CO2 laser irradiated the trapping volume of the high-pressure cell (i.e., the reaction chamber) for the duration of the NETD reaction only. The IR laser power output was manipulated through an external controller and varied from 6 W (10% maximal output) to 30 W (50% maximal output) as noted in the text.
For CAD and HCD experiments, various normalized collision energies and precursor charge states were tested to optimize dissociation efficiency. For CAD, an activation time of 10 ms was used with a q value of 0.25. Quadrupole isolation and Orbitrap mass analysis was performed the same as in the NETD experiments described above.
Data Analysis
A C# script, based on the design framework of the C# Mass Spectrometry Library (CSMSL, https://github.com/dbaileychess/CSMSL), was written to model RNA fragmentation patterns illustrated in Figure 1. While directly reading in Thermo Xcalibur .raw files, the C# script allowed for extraction of fragment ions and their intensities, as well as that of base-loss fragments, neutral loss fragments, and internal fragments. All matches were made within a 10 ppm tolerance. Note, due to the slight palindromic nature of the nucleic acids used and product types possible, sporadic indistinguishable overlaps in coverage likely occurred. The script was written to incorporate potential chemical modifications at any site along the phosphodiester bond, ribose sugar, or nucleobase, accounting for the diverse array of enzymatic modifications referenced in RNA modification databases.58,59
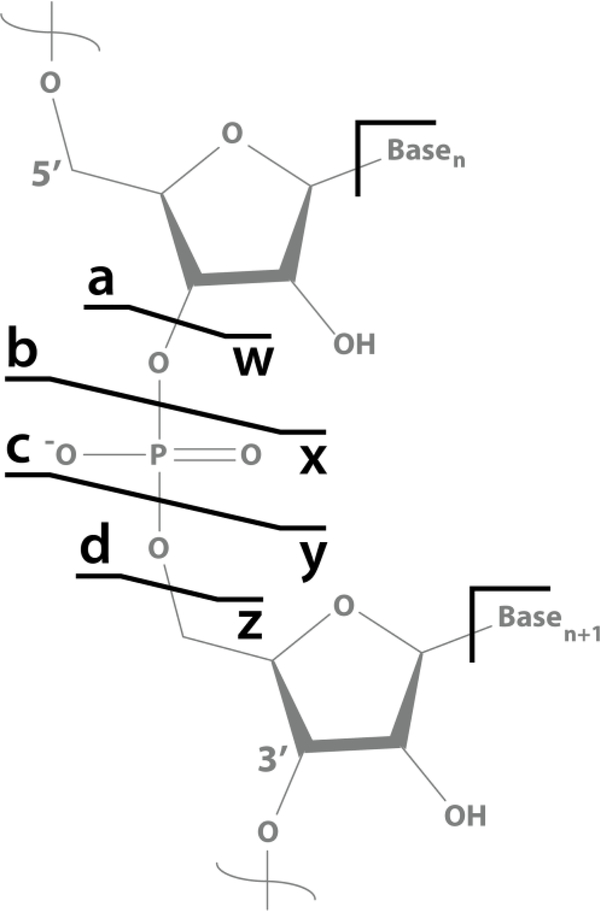
Figure 1. Schematic of potential product ion fragments obtained by tandem mass spectrometry of oligonucleotide polymers.
Base losses may also occur at a nucleotide, leaving a product ion or intact oligonucleotide without a base.
RESULTS AND DISCUSSION
Introduction to nucleic acid anion dissociation
Figure 1 provides a schematic of RNA backbone dissociation schemes as defined by McLuckey et al.11 Collision-based fragmentation methods typically produce complementary c- and y-type ions along with numerous base-loss and internal fragments.60 Conversely, NETD generates mainly non-complementary d- and w-type product ions with considerably less base-loss and internal fragmentation. One challenge with NETD is that precursors with low charge density can undergo an electron transfer event that fails to dissociate the two product ions due to higher-order gas-phase structure. This sequence uninformative event, termed nondissociative negative electron transfer dissociation (NETnoD), arises from product ions being noncovalently bound together in secondary complexes.61–63 To circumvent this issue, supplemental activation via collisions or photons can be introduced, providing vibrational excitation to facilitate dissociation. NETcaD uses resonant excitation after the ion-ion reaction to collisionally activate the NETnoD products, whereas NEThcD shuttles all NETD products and remaining precursor to the Ion Routing Multipole for subsequent beam-type collisional activation. AI-NETD uses photoactivation of the ion trap trapping volume concurrent with the NETD ion-ion reaction, which disrupts noncovalent interactions and promotes more efficient fragmentation and better depth of coverage than NETD alone.55 Here we compare NETD and AI-NETD to collision-based fragmentation for RNA dissociation.
Dissociation of a short, modified nucleic acid anion
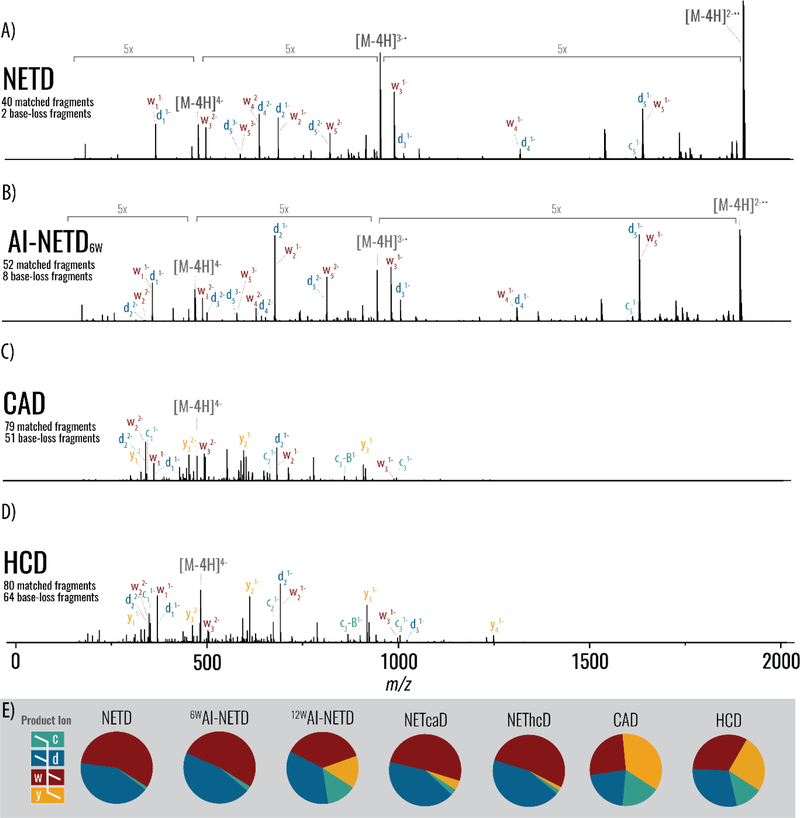
First, we examined NETD and AI-NETD for analysis of a modified hexanucleotide, GmUACmUG, which includes 2’-O-methyl modifications. Hexanucleotide precursors are useful models as many studies employ endoribonuclease digestion, leaving digestion products 4 – 6 nucleotides in length for LC-MS/MS analysis.64 Figure 2 presents annotated tandem mass spectra following dissociation of the quadruply deprotonated anion at m/z 475 using either NETD, AI-NETD, ion trap CAD, or HCD. NETD generated 40 unique fragments and two unique base-loss fragments (Figure 2a), whereas AI-NETD (6 W) produced 52 unique fragments and eight unique base-loss fragments (Figure 2b). Both methods elicited complete phosphodiester backbone fragmentation of the hexanucleotide. AI-NETD at 12 W laser power further increased product ion generation to 67 unique fragments; however, 35 unique base-loss fragments were additionally produced. When the concurrent IR irradiation was performed, charge-reduced precursor peak intensities were significantly diminished, while overall spectral quality increased. Specifically, nearly two thirds of the total ion current (64%) was occupied by charge-reduced precursors following NETD; that amount was reduced by half (31%) following AI-NETD. Supplemental activation with resonant excitation (NETcaD) and beam-type collisional activation (NEThcD) offer predominantly d- and w-ions while generating more unique fragments for greater depth of coverage than NETD alone, prompting detection of 55 and 50 unique product ions, respectively.
Figure 2. Tandem mass spectra and summary fragmentation profiles for a short, modified ribonucleic acid with various activation methods.
(A) MS/MS spectra of the 6-nucleotide modified RNA after 50ms of NETD, (B) 50ms of AI-NETD at 6 W, (C) CAD, and (D) HCD. Note each panel presents a spectrum that results from averaging 20 single-scan spectra and are shown on the same intensity scale. Charge-reduced intact species’ peaks are indicated by brackets in grey text. (E) Summary of product ion distributions, based on relative intensity, for the various activation methods.
Here NETD performs well on its own, even in the presence of the sequence-uninformative, charge transfer events without dissociation. Previous electron transfer studies on oligonucleotides have generated similar NETnoD species but did not yield high sequence coverage until a secondary activation technique was implemented.34,35 IR photoirradiation during the NETD reaction unfolds the RNA to release product ions and limit the presence of nondissociative NETnoD species. Additional efficiencies likely result from the robust front-end reagent source, ion transmission, and ion trapping of the linear ion trap arrangement used here. Figure 2c and 2d display the spectra resulting from collision-activated dissociation (ion trap CAD and HCD, respectively). Both methods achieve backbone fragmentation for this short oligonucleotide; however, they produce many undesired base-loss products. HCD generated 80 unique product ions with 64 base-loss fragments, whereas ion trap CAD generated 79 unique product ions with 51 additional base-loss fragments. Minimizing formation of sequence-uninformative side-products is key to both simplifying spectra and achieving good sensitivity.
For RNA anion fragmentation, c- and y-type ions are the main products following collision-activated dissociation, whereas d- and w-ions are the main products of electron-based activation methods.65 Our data confirm these previous reports that NETD predominantly generates d- and w-ions (Figure 2e). CAD and HCD create a nearly even assortment of c-, d-, w-, and y-type ions with a notable absence of intermediate-to-high m/z peaks. For AI-NETD, multiple laser power levels were evaluated. We supposed that as the power was increased, dissociation through vibrational channels could occur (i.e., IRMPD). Indeed, when higher IR power levels are reached in AI-NETD, the relative ratio of c-, d-, w-, and y-product ions approaches unity. At higher laser power levels (12–30 W), the relative intensity of d- and w-type ion products diminishes, and the spectra more closely resemble those generated via ion trap CAD or HCD methods.
While most previous reports have combined electron-based with collision-based fragmentation data, AI-NETD at higher laser-power levels establishes an activation type capable of accessing both electronic and vibrational dissociation pathways. Notably, here AI-NETD also improves depth of coverage, but is concurrent with the NETD reaction, reducing the instrumental duty cycle. This will play an important role in future LC-MS/MS experiments, where fast scan speed is vital to maximal identification and quantification. Attaining 100% sequence and modification characterization via NETD and AI-NETD, while generating d- and w-fragment types complementary to traditional methods (CAD, IRMPD) reveals the potential of NETD-based methods for large-scale analysis of modified and unmodified oligonucleotides.
Dissociation of an intact, ribonucleic acid anion
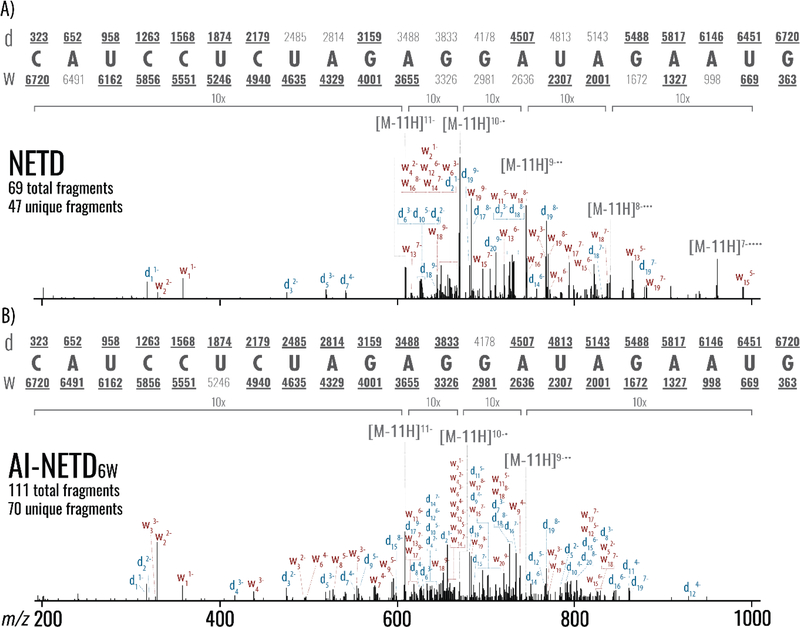
The ability to dissociate longer, intact RNAs is especially important – i.e., native siRNA and tRNA molecules. Obtaining high sequence coverage using collisional-activation of such anionic precursors is challenging.19 To determine the potential utility of NETD we next analyzed an intact, medium-length oligoribonucleotide (5’-rCrArUrCrCrUrCrUrArGrArGrGrArUrArGrArArUrG-3’, the luciferase siRNA antisense strand). Figure 3 illustrates a comparison of NETD and AI-NETD tandem MS spectra for the z = −11 precursor anion of the 6.72 kDa ribooligonucleotide. Evinced by the high intensity charge-reduced intact species, NETD has relatively low efficiency. Still NETD generated 69 total fragments and 47 unique fragments to give 80% phosphodiester bonds broken considering only d- and w-ions and complete coverage when all eight product ion types are used (Figure 3a). From these data we conclude NETD can effectively dissociate intact nucleic acids without supplemental activation. Figure 3b presents dissociation of the same anionic precursor using AI-NETD experiments. Note the charge-reduced peaks are greatly diminished, having been converted into sequence-informative product ion signal. This spectrum is characterized by d- and w-type ions and the lack of the NETnoD products. The AI-NETD spectrum demonstrated a greater depth of coverage, producing 111 total fragments – a 160% boost over NETD. This greater depth of coverage affords 100% phosphodiester bond cleavage, even when only d- and w-type product ions are searched.
Figure 3. Comparison of NETD and AI-NETD MS/MS of the 21-nucleotide luciferase antisense siRNA strand.
NETD (A) or AI-NETD (B) MS/MS of the −11 charge state RNA precursor. Note both spectra result from the average of 20 single-scan spectra and are presented on the same intensity scale. Predicted d- and w-ion mass values are shown above or below the RNA sequence, respectively.
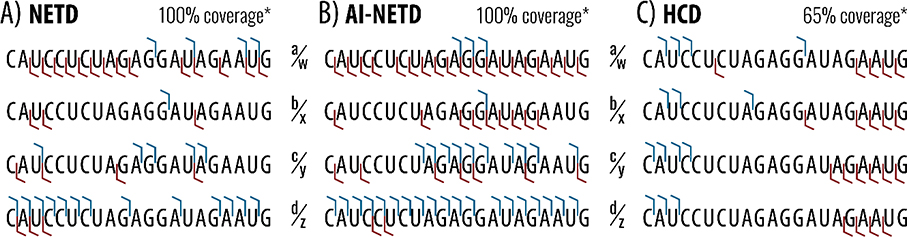
When characterizing nucleic acids, whether DNA, RNA, or modified sequences, the ability to attain 100% sequence coverage is paramount. Figure 4 illustrates the relative sequence coverage of this precursor from each of the eight product ion types (a, b, c, d, w, x, y, and z), which are shown in complementary pairs. With NETD/AI-NETD dissociation of the 21-nucleotide luciferase antisense siRNA strand, good sequence coverage is obtained by both d- and w-type ions independently. Interestingly, d- and w-type ions are non-complementary, as depicted in Figure 1. Others have proposed mechanisms for the rearrangement of radical -type ions into wn-ions after electron detachment or electron transfer. Further, an equally likely pathway for d-type ion derivation from radical -type ions has been proposed.31,34 The NETD work exhibited here supports these concepts, as one may otherwise expect detection of complementary product ions. All other product ions, typical of collision-based methods, are here only formed in a limited capacity.
Figure 4. Fragmentation profiles for NETD, AI-NETD, and HCD of the 21-nucleotide luciferase siRNA antisense strand.
The four pairs of complementary fragment types are displayed for (A) NETD, (B) AI-NETD, and (C) HCD tandem mass spectrometry dissociation. Sequence coverage denotes the percentage of phosphodiester bond cleavage using all eight product types.
AI-NETD models a very similar fragmentation pattern as NETD but enables more comprehensive coverage (Figure 4b). Better breadth of coverage by d-type ions is especially noticeable in the direct comparison of AI-NETD to NETD. Better depth of coverage is also noticeable in Figure S4, where multiple charge-states are present for many AI-NETD product ions. As depicted in Figure 4a, NETD captures many short and long d- and w-ion products but produces fewer medium length fragments. This may be due to secondary structure interactions within the interior of the RNA strand, preventing dissociation. Concurrent IR laser irradiation with the NETD reaction significantly reduces the coverage gaps of NETD.
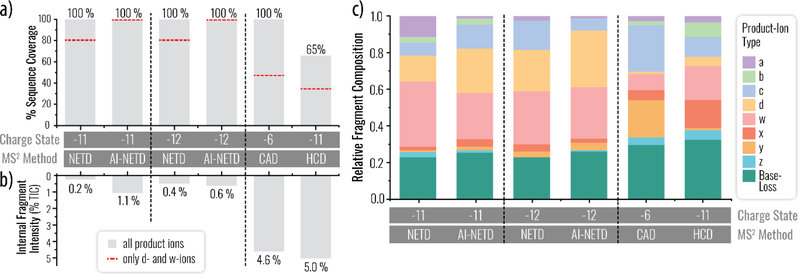
Collisional activation of this precursor at the most abundant precursor charge states provides limited sequence information, especially when compared to NETD and AI-NETD spectra. Specifically, CAD and HCD of the of these most abundant, high charge-density precursor populations produce predominantly short 5’- and 3’-terminal fragment ions, pathways that seems to become more prominent for longer oligonucleotides (Figure S5). Notably, this issue is somewhat mitigated when low charge state precursors are utilized. Here, NETD and AI-NETD produce d- and w-type product ions throughout the siRNA sequence, which outperforms the fragmentation profile of collisional activation methods traditionally employed in the field. The complementary nature of NETD and AI-NETD relative to collisional methods is further illustrated in Figure 5a by the d- and w-type product ion generation. While NETD of a z = −11 or −12 precursor gives 80% phosphodiester bonds broken through d- and w-type ions (and complete sequence coverage when all eight product types are searched), CAD produces only 45% coverage through d- and w-type ions, while coverage from HCD is further reduced to 35%. When precursor charge state and normalized collision energy of collisional dissociation reactions are optimized, 100% coverage at z = −6 was realized primarily through the CAD-induced generation of c- and y-type ions. The complete sequence coverage attained by these NETD-based methods is noteworthy.
Figure 5. Sequence coverage and product ion distributions following dissociation of the 21-nucleotide RNA strand via HCD, CAD, NETD, and AI-NETD.
(A) The percent sequence coverage is shown for electron- and collision-based dissociation methods. Grey bars indicate when all eight fragment types were considered, whereas a dashed orange line indicates integration of only d- and w-type ion data, the main products of NETD approaches. (B) The undesirable internal fragments are monitored as a function of a percentage of the total ion current (% TIC) for each individual experiment. (C) The overall product ion distribution, based on relative intensity, for all eight fragment types and base-loss fragments.
Another challenge of nucleic acid dissociation is the production of internal fragments – products that complicate spectra and divert informative fragment ion signal. We sought to quantify the abundance of these products that contain neither termini. Figure 5b illustrates the degree of internal fragment generation across the methods at the optimized precursor charge state. The vibrational methods, CAD and HCD, produced numerous internal fragments that in total accounted for 4.6% and 5.0% of the spectral ion current from the z = −6 and z = −11 precursor populations, respectively. NETD of the z = −11 precursor does produce internal fragments, but their signal is ~ 25-fold reduced (0.2% of total ion current). Dissociation of the z = −12 precursor by NETD results in more internal fragmentation (0.4%). AI-NETD produced higher internal fragment intensities than NETD (1.1% of total ion current) for the precursor with z = −11. At a slightly higher and relatively abundant −12 charge state, the internal fragment production is reduced two-fold (0.6% of total ion current). Previously, Håkansson and coworkers reported IRMPD-based methods for the dissociation of nucleic acids, noting fragmentation is similar to CAD-based methods.37,66 Brodbelt and coworkers have reported on alternative photodissociation methods for oligonucleotides, including 260 nm and 193 nm UVPD, revealing that UVPD produces fewer nucleobase-loss products than IRMPD; however, internal products appear numerous in UVPD and electron photo-detachment (EPD) spectra, but their abundance has not been quantified.38,40,67,68
Dissociative base-loss along the phosphodiester backbone, from either the intact precursor or product ions, is another fragmentation channel that can hinder nucleotide anion characterization. Similar to reducing the extent of internal fragmentation, the ideal dissociation method will minimize production of base-loss products. We note low energy collisional activation may preferentially cleave nitrogenous bases while leaving the sequence fully intact.12 That said, for most experiments, base-loss dissociation is undesired as it adds additional spectral complexity. In Figure 5c we quantify the relative product ion type intensities for NETD, AI-NETD, CAD, and HCD spectra, including base-loss fragments. As discussed previously, d- and w-type ions are the main products of NETD and AI-NETD, whereas HCD offers a more even sampling of the entire product ion landscape and CAD offers primarily c- and y-type ions. Noticeably, NETD and AI-NETD produce lower levels of base-loss fragments (dark green) relative to collision-based approaches, even when precursor charge state and collisional energy are optimized for each method. Figure S6 demonstrates a direct comparison of the relevant undesired products from NETD and CAD methods across charge state. Specifically, base-loss fragments account for 22.3% of all fragment (a, b, c, d, w, x, y, z, and base-loss) intensities for the NETD spectrum in Figure 3a, whereas the AI-NETD base-loss fragments explain 24.5% of the total fragment intensity. In contrast, HCD and CAD produced base-loss fragments accounting for 32.0% and 29.5% of the total spectral fragment intensity, respectively. For a breakdown of the relative base-loss fragment types generated via each method, see Figure S7. Reducing the unintended side-products within a tandem mass spectrum simplifies manual and database-dependent spectral interpretation and reduces the number of false-positive discoveries.
CONCLUSION
Efforts in oligonucleotide sequencing have begun to reveal the complex molecular network underlying genetic regulation and expression; however, powerful chemical labelling approaches often fail to reveal necessary site-specificity, while traditional methods in mass spectrometry are difficult to implement or can be limited by sequence length. Here, we outline the utility of NETD and AI-NETD tandem mass spectrometry methods for characterization of modified and unmodified nucleic acids. These methods produce interpretable spectra that provide complete sequence coverage and offer spectra with much lower complexity as compared to dissociation with traditional collision-based methods. NETD and AI-NETD each obtain complete sequence coverage for both a short, modified RNA and a longer siRNA strand via predominantly d- and w-type product ion generation. These MS/MS methods produce ions complementary to CAD and HCD experiments and show promise for top-down analyses of larger, intact nucleic acids such as heavily modified translational RNAs.
Perhaps the largest detriment to common collision-based MS methods is their non-specific cleavage of the weakest bonds within a precursor anion. Complete structural analyses of intact tRNA sequences, which are 74 – 95 nucleotides in length and often heavily modified with some secondary interactions, are especially challenging.31,32 Here we demonstrate that NETD permits retention of modifications for confident localization and does not require secondary activation. Isomeric modifications, including monomethylated nucleobases, represent future challenges, although solutions to such hurdles have been reported previously.69 NETD and AI-NETD can directly and informatively dissociate intact modified tRNAs or mRNA transcripts, which may expose yet undiscovered pathways within the epitranscriptome and toward proteoform generation.70
Many modern strategies to sequence oligonucleotides feature digestion followed by an on-line liquid chromatography separation of digestion products and subsequent mass spectrometry analysis (LC/MS).64,71,72 The ability to account for only two product types in a future large database search, as is common with LC/MS/MS methods, will reduce the number of false-positive spectral matches, quicken analysis time, and provide a more robust experimental method. Future work will include the analysis of such complex mixtures using either NETD or AI-NETD for dissociation of cellular and synthesized ribonucleic acids, deoxyribonucleic acids, and modified oligonucleotide sequences with greater throughput.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health [P41GM108538 to J.J.C.] and the National Human Genome Research Institution through a training grant to the Genomic Sciences Training Program [5T32HG002760 to T.PC.]. The authors acknowledge invaluable support from John E. P. Syka, Nicholas Riley, Christopher Mullen, Jae Schwartz, Jean Lodge and other members of the Coon Lab.
Footnotes
The authors declare no competing financial interests.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Figure S1, annotated electrospray mass spectra for the modified 6-nt RNA and 21-nt siRNA; Figure S2, overall sequence coverage of luciferase siRNA antisense strand (NETD, AI-NETD, CAD, HCD); Figure S3, Sequence maps of 21nt siRNA following NETD and AI-NETD; Figure S4, table of d- and w-type ions identified in NETD and AI-NETD experiments of 21-nt siRNA; Figure S5, MS/MS raw spectra following HCD and CAD of the 21-nt antisense siRNA strand; Figure S6, undesired base-loss and internal fragments generated via NETD and CAD methods across all precursor charge states; Figure S7, base-loss fragmentation by product ion type.
REFERENCES
- (1).Eddy SR Non-Coding RNA Genes and the Modern RNA World. Nat. Rev. Genet 2001, 2, 919–929. [DOI] [PubMed] [Google Scholar]
- (2).Helm M Post-Transcriptional Nucleotide Modification and Alternative Folding of RNA. Nucleic Acids Res 2006, 34 (2), 721–733. 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chow CS; Lamichhane TN; Mahto SK Expanding the Nucleotide Repertoire of the Ribosome with Post-Transcriptional Modifications. ACS Chem. Biol 2007, 2 (9), 610–619. 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Li S; Mason CE The Pivotal Regulatory Landscape of RNA Modifications. Annu. Rev. Genomics Hum. Genet 2014, 15, 127–150. 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- (5).Wittrup A; Lieberman J Knocking down Disease: A Progress Report on SiRNA Therapeutics. Nat. Rev. Genet 2015, 16, 543–552. 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Morrison C Alnylam Prepares to Land First RNAi Drug Approval. Nat. Rev. Drug Discov 2018, 17 (3), 156–157. 10.1038/nrd.2018.20. [DOI] [PubMed] [Google Scholar]
- (7).Foster DJ; Brown CR; Shaikh S; Trapp C; Schlegel MK; Qian K; Rajeev KG; Jadhav V; Manoharan M; Kuchimanchi S; et al. Advanced SiRNA Designs Further Improve in Vivo Performance of GalNAc-SiRNA Conjugates. Mol. Ther 2018, 26 (3), 708–717. 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Morris K V; Mattick, J. S. The Rise of Regulatory RNA. Nat. Rev. Genet 2014, 15, 423–437. 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mortazavi A; Williams BA; Mccue K; Schaeffer L; Wold B Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5 (7), 621–628. 10.1038/NMETH.1226. [DOI] [PubMed] [Google Scholar]
- (10).Wang Z; Gerstein M; Snyder M RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Drug Discov 2009, 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mcluckey SA; Berkel, Van GJ; Glish GL Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides. J. Am. Soc. Mass Spectrom 1992, 3, 60–70. [DOI] [PubMed] [Google Scholar]
- (12).Mcluckey SA; Habibi-goudarzi S Decompositions of Multiply Charged Oligonucleotide Anions. J. Am. Chem. Soc 1993, 115 (25), 12085–12095. 10.1021/ja00078a054. [DOI] [Google Scholar]
- (13).Mcluckey SA; Habibi-goudarzi S Ion Trap Trandem Mass Spectrometry to Small Multiply Charged Oligonucleotides with a Modified Base. J. Am. Soc. Mass Spectrom 1994, 5, 740–747. https://doi.org/1044-0305/94. [DOI] [PubMed] [Google Scholar]
- (14).Kowalak JA; Pomerantz SC; Crain PF; Mccloskey JA; City SL A Novel Method for the Determination of Post-Transcriptional Modification in RNA by Mass Spectrometry. Nucleic Acids Res. 1993, 21 (19), 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Little DP; Chorush RA; Speir JP; Senko MW; Kelleher NL; Mclafferty FW Rapid Sequencing of Oligonucletides by High-Resolution Mass Spectrometry. J. Am. Chem. Soc 1994, 116 (11), 4893–4897. 10.1021/ja00090a039. [DOI] [Google Scholar]
- (16).Hofstadler SA; Sannes-lowery KA; Hannis JC Analysis of Nucleic Acids by FTICR MS. Mass Spectrom. Rev 2005, 24, 265–285. 10.1002/mas.20016. [DOI] [PubMed] [Google Scholar]
- (17).Gupta R; Kapur A; Beck JL; Sheil MM Positive Ion Electrospray Ionization Mass Spectrometry of Double-Stranded DNA/Drug Complexes. Rapid Commun. Mass Spectrom 2001, 15, 2472–2480. 10.1002/rcm.524. [DOI] [PubMed] [Google Scholar]
- (18).Sannes-lowery KA; Mack DP; Hu P; Mei H; Loo JA Positive Ion Electrospray Ionization Mass Spectrometry of Oligonucleotides. J. Am. Soc. Mass Spectrom 1997, 8, 90–95. [Google Scholar]
- (19).Keller KM; Brodbelt JS Collisionally Activated Dissociation and Infrared Multiphoton Dissociation of Oligonucleotides in a Quadrupole Ion Trap. Anal. Biochem 2004, 326, 200–210. 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- (20).Ganisl B; Taucher M; Riml C; Breuker K Charge as You like! Efficient Manipulation of Negative Ion Net Charge in Electrospray Ionization of Proteins and Nucleic Acids. Eur. J. Mass Spectrom 2011, 17, 333–343. 10.1255/ejms.1140. [DOI] [PubMed] [Google Scholar]
- (21).Mcluckey SA Principles of Collisional Activation in Analytical Mass Spectrometry. J. Am. Soc. Mass Spectrom 1992, 3, 599–614. [DOI] [PubMed] [Google Scholar]
- (22).Marzilli LA; Barry JP; Sells T; Law SJ; Vouros P; Harsch A Oligonucleotide Sequencing Using Guanine-Specific Methylation and Electrospray Ionization Ion Trap Mass Spectrometry. J. Mass Spectrom. 1999, 34 (4), 276–280. . [DOI] [PubMed] [Google Scholar]
- (23).McLuckey SA; Vaidyanathan G; Habibi‐Goudarzi S Charged vs. Neutral Nucleobase Loss from Multiply Charged Oligonucleotide Anions. J. Mass Spectrom 1995, 30 (9), 1222–1229. 10.1002/jms.1190300903. [DOI] [Google Scholar]
- (24).Huang T. yi; Kharlamova A; Liu J; McLuckey SA Ion Trap Collision-Induced Dissociation of Multiply Deprotonated RNA: C/y-Ions versus (a-B)/w-Ions. J. Am. Soc. Mass Spectrom 2008, 19 (12), 1832–1840. 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- (25).Taucher M; Rieder U; Breuker K Minimizing Base Loss and Internal Fragmentation in Collisionally Activated Dissociation of Multiply Deprotonated RNA. J. Am. Soc. Mass Spectrom 2010, 21, 278–285. 10.1016/j.jasms.2009.10.010. [DOI] [PubMed] [Google Scholar]
- (26).Huang TY; Liu J; Liang X; Hodges BDM; McLuckey SA Collision-Induced Dissociation of Intact Duplex and Single-Stranded SiRNA Anions. Anal. Chem 2008, 80 (22), 8501–8508. 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- (27).Herron WJ; Goeringer DE; Mcluckey SA Gas-Phase Electron Transfer Reactions from Multiply-Charged Anions to Rare Gas Cations. J. Am. Chem. Soc 1995, 117, 11555–11562. [Google Scholar]
- (28).Stephenson JL; Mcluckey SA Charge Reduction of Oligonucleotide Anions via Gas-Phase Electron Transfer to Xenon Cations. Rapid Commun. Mass Spectrom 1997, 11, 875–880. [DOI] [PubMed] [Google Scholar]
- (29).McLuckey SA; Stephenson JL; O’Hair RAJ Decompositions of Odd- and Even-Electron Anions Derived from Deoxy-Polyadenylates. J. Am. Soc. Mass Spectrom 1997, 8 (2), 148–154. 10.1016/S1044-0305(96)00231-0. [DOI] [Google Scholar]
- (30).Herron WJ; Goeringer DE; McLuckey SA Ion-Ion Reactions in the Gas Phase: Proton Transfer Reactions of Protonated Pyridine with Multiply Charged Oligonucleotide Anions. J. Am. Soc. Mass Spectrom 1995, 6 (6), 529–532. 10.1016/1044-0305(95)00199-N. [DOI] [PubMed] [Google Scholar]
- (31).Taucher M; Breuker K Top-down Mass Spectrometry for Sequencing of Larger (up to 61 Nt) RNA by CAD and EDD. J. Am. Soc. Mass Spectrom 2010, 21, 918–929. 10.1016/j.jasms.2010.02.025. [DOI] [PubMed] [Google Scholar]
- (32).Taucher M; Breuker K Characterization of Modified RNA by Top-down Mass Spectrometry. Angew. Chemie 2012, 51, 11289–11292. 10.1002/anie.201206232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Huang T; Mcluckey SA Gas-Phase Ion/Ion Reactions of Rubrene Cations and Multiply Charged DNA and RNA Anions. Int. J. Mass Spectrom 2011, 304, 140–147. 10.1016/j.ijms.2010.06.019. [DOI] [Google Scholar]
- (34).Gao Y; Mcluckey SA Electron Transfer Followed by Collision-Induced Dissociation (NET-CID) for Generating Sequence Information from Backbone-Modified Oligonucleotide Anions. Rapid Commun. Mass Spectrom 2013, 27, 249–257. 10.1002/rcm.6428. [DOI] [PubMed] [Google Scholar]
- (35).Gao Y; Yang J; Cancilla MT; Meng F; Mcluckey SA Top-down Interrogation of Chemically Modified Oligonucleotides by Negative Electron Transfer and Collision Induced Dissociation. Anal. Chem 2013, 85, 4713–4720. 10.1021/ac400448t. [DOI] [PubMed] [Google Scholar]
- (36).Mo J; Håkansson K Characterization of Nucleic Acid Higher Order Structure by High-Resolution Tandem Mass Spectrometry. Anal. Bioanal. Chem 2006, 386, 675–681. 10.1007/s00216-006-0614-z. [DOI] [PubMed] [Google Scholar]
- (37).Yang J; Håkansson K Characterization of Oligodeoxynucleotide Fragmentation Pathways in Infrared Multiphoton Dissociation and - Electron Detachment Dissociation by Fourier. Eur. J. Mass Spectrom 2009, 15, 293–304. 10.1255/ejms.966. [DOI] [PubMed] [Google Scholar]
- (38).Gabelica V; Tabarin T; Antoine R; Compagnon I; Broyer M; Pauw, De E; Dugourd P Electron Photodetachment Dissociation of DNA Polyanions in a Quadrupole Ion Trap Mass Spectrometer. Anal. Chem 2006,78 (18), 6564–6572. 10.1021/ac060753p. [DOI] [PubMed] [Google Scholar]
- (39).Yang J; Håkansson K Fragmentation of Oligoribonucleotides from Gas-Phase Ion-Electron Reactions. J. Am. Soc. Mass Spectrom 2006, 17, 1369–1375. 10.1016/j.jasms.2006.05.006. [DOI] [PubMed] [Google Scholar]
- (40).Smith SI; Brodbelt JS Characterization of Oligodeoxynucleotides and Modifications by 193 Nm Photodissociation and Electron Photodetachment Dissociation. Anal. Chem 2010, 82, 7218–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Little DP; Speir JP; Senko MW; Connor PBO; Mclafferty FW Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing. Anal. Chem 1994, 66 (18), 2809–2815. 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- (42).Wolff JJ; Amster IJ; Chi L; Linhardt RJ Electron Detachment Dissociation of Glycosaminoglycan Tetrasaccharides. J. Am. Soc. Mass Spectrom 2007, 18, 234–244. 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Coon JJ; Shabanowitz J; Hunt DF; Syka JEP Electron Transfer Dissociation of Peptide Anions. J. Am. Soc. Mass Spectrom 2005, 16, 880–882. 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- (44).Huzarska M; Ugalde I; Kaplan DA; Hartmer R; Easterling ML; Polfer NC Negative Electron Transfer Dissociation of Deprotonated Phosphopeptide Anions: Choice of Radical Cation Reagent and Competition between Electron and Proton Transfer. Anal. Chem 2010, 82, 2873–2878. [DOI] [PubMed] [Google Scholar]
- (45).Wolff JJ; Leach III FE; Laremore TN; Kaplan DA; Easterling ML; Linhardt RJ; Amster IJ Negative Electron Transfer Dissociation of Glycosaminoglycans. Anal. Chem 2010, 82, 3460–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wei J; Wu J; Tang Y; Ridgeway ME; Park MA; Costello CE; Zaia J; Lin C Characterization and Quantification of Highly Sulfated Glycosaminoglycan Isomers by Gated-Trapped Ion Mobility Spectrometry Negative Electron Transfer Dissociation MS/MS. Anal. Chem 2019, 91, 2994–3001. 10.1021/acs.analchem.8b05283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wu J; Mcluckey SA Ion/Ion Reactions of Multiply Charged Nucleic Acid Anions: Electron Transfer, Proton Transfer, and Ion Attachment. Int. J. Mass Spectrom 2003, 228, 577–597. 10.1016/S1387-3806(03)00165-9. [DOI] [Google Scholar]
- (48).Rush MJP; Riley NM; Westphall MS; Syka JEP; Coon JJ Sulfur Pentafluoride Is a Preferred Reagent Cation for Negative Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom 2017, 28, 1324–1332. 10.1007/s13361-017-1600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Smith SI; Brodbelt JS Electron Transfer Dissociation of Oligonucleotide Cations. Int. J. Mass Spectrom 2009, 283, 85–93. 10.1016/j.ijms.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Shaw JB; Madsen JA; Xu H; Brodbelt JS Systematic Comparison of Ultraviolet Photodissociation and Electron Transfer Dissociation for Peptide Anion Characterization. J. Am. Chem. Soc 2012, 23, 1707–1715. 10.1007/s13361-012-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Stickney M; Sanderson P; Leach III FE; Zhang F; Linhardt RJ; Amster IJ Online Capillary Zone Electrophoresis Negative Electron Transfer Dissociation Tandem Mass Spectrometry of Glycosaminoglycan Mixtures. Int. J. Mass Spectrom 2019, 445 10.1016/j.ijms.2019.116209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Leach III FE; Riley NM; Westphall MS; Coon JJ; Amster IJ Negative Electron Transfer Dissociation Sequencing of Increasingly Sulfated Glycosaminoglycan Oligosaccharides on an Orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom 2017, 28, 1844–1854. 10.1007/s13361-017-1709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Huang Y; Yu X; Mao Y; Costello CE; Zaia J; Lin C De Novo Sequencing of Heparan Sulfate Oligosaccharides by Electron-Activated Dissociation. Anal. Chem 2013, 85, 11979–11986. 10.1021/ac402931j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ledvina AR; Mcalister GC; Gardner MW; Smith SI; Madsen JA; Schwartz JC; Stafford GC; Syka JEP; Brodbelt JS; Coon JJ Infrared Photoactivation Reduces Peptide Folding and Hydrogen- Atom Migration Following ETD Tandem Mass Spectrometry. Angew. Chemie Int. Ed 2009, 48, 8526–8528. 10.1002/anie.200903557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Riley NM; Westphall MS; Coon JJ Activated Ion Electron Transfer Dissociation for Improved Fragmentation of Intact Proteins. Anal. Chem 2015, 87, 7109–7116. 10.1021/acs.analchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Riley NM; Westphall MS; Hebert AS; Coon JJ Implementation of Activated Ion Electron Transfer Dissociation on a Quadrupole-Orbitrap-Linear Ion Trap Hybrid Mass Spectrometer. Anal. Chem 2017, 89, 6358–6366. 10.1021/acs.analchem.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. PNAS 2004, 101 (26), 9528–9533. https://doi.org/10.1073兾pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Cantara WA; Crain PF; Rozenski J; Mccloskey JA; Harris KA; Zhang X; Vendeix FAP; Fabris D; Agris PF The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res. 2011, 39, 195–201. 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Machnicka MA; Milanowska K; Oglou OO; Purta E; Kurkowska M; Olchowik A; Januszewski W; Kalinowski S; Dunin-horkawicz S; Rother KM; et al. MODOMICS : A Database of RNA Modification Pathways — 2013 Update. Nucleic Acids Res. 2013, 41, 262–267. 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Schürch S Characterization of Nucleic Acids by Tandem Mass Spectrometry - the Second Decade (2004 – 2013): From DNA to RNA and Modified Sequences. Mass Spectrom. Rev. 2016, 35, 483–523. 10.1002/mas.21442. [DOI] [PubMed] [Google Scholar]
- (61).Pitteri SJ; Chrisman PA; Mcluckey SA Electron-Transfer Ion/Ion Reactions of Doubly Protonated Peptides: Effect of Elevated Bath Gas Temperature. Anal. Chem 2005, 77 (17), 5662–5669. 10.1021/ac050666h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Liu J; Mcluckey SA Electron Transfer Dissociation: Effects of Cation Charge State on Product Partitioning in Ion/Ion Electron Transfer to Multiply Protonated Polypeptides. Int. J. Mass Spectrom 2012, 332, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Xia Y; Han H; McLuckey SA Activation of Intact Electron-Transfer Products of Polypeptides and Proteins in Cation Transmission Mode Ion/Ion Reactions. Anal. Chem 2008, 80 (4), 1111–1117. 10.1021/ac702188q. [DOI] [PubMed] [Google Scholar]
- (64).Ross R; Cao X; Yu N; Limbach PA Sequence Mapping of Transfer RNA Chemical Modifications by Liquid Chromatography Tandem Mass Spectrometry. Methods 2016, 107, 73–78. 10.1016/j.ymeth.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wu J; Mcluckey SA Gas-Phase Fragmentation of Oligonucleotide Ions. Int. J. Mass Spectrom 2004, 237, 197–241. 10.1016/j.ijms.2004.06.014. [DOI] [Google Scholar]
- (66).Håkansson K; Chalmers MJ; Quinn JP; McFarland MA; Hendrickson CL; Marshall AG Combined Electron Capture and Infrared Multiphoton Dissociation for Multistage MS/MS in a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem 2003, 75 (13), 3256–3262. 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- (67).Smith SI; Brodbelt JS Hybrid Activation Methods for Elucidating Nucleic Acid Modifications. Anal. Biochem 2011, 83, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Gabelica V; Rosu F; Tabarin T; Kinet C; Antoine R; Broyer M; De Pauw E; Dugourd P Base-Dependent Electron Photodetachment from Negatively Charged DNA Strands upon 260-Nm Laser Irradiation. J. Am. Chem. Soc 2007, 129 (15), 4706–4713. 10.1021/ja068440z. [DOI] [PubMed] [Google Scholar]
- (69).Nakayama H; Yamauchi Y; Nobe Y; Sato K; Takahashi N; Shalev-Benami M; Isobe T; Taoka M Method for Direct Mass-Spectrometry-Based Identification of Monomethylated RNA Nucleoside Positional Isomers and Its Application to the Analysis of Leishmania RRNA. Anal. Chem 2019, 91, 15634–15643. 10.1021/acs.analchem.9b03735. [DOI] [PubMed] [Google Scholar]
- (70).Smith LM; Kelleher NL Proteoform: A Single Term Describing Protein Complexity. Nat. Methods 2013, 10 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Kellner S; Seidu-larry S; Motorin Y; Poincare H; Universite N A Multifunctional Bioconjugate Module for Versatile Photoaffinity Labeling and Click Chemistry of RNA. Nucleic Acids Res. 2011, 39 (16), 7348–7360. 10.1093/nar/gkr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Zhang N; Shi S; Jia TZ; Ziegler A; Yoo B; Yuan X; Li W; Zhang S A General LC-MS-Based RNA Sequencing Method for Direct Analysis of Multiple-Base Modifications in RNA Mixtures. Nucleic Acids Res. 2019, 47 (20), e125 10.1093/nar/gkz731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.