Abstract
DiscoveryDotTM is a novel solution‐phase technology for chemical compound microarrays which has been validated for several targets (e.g. serine proteases, cysteine proteases, metalloproteinases, histone deacetylases, phosphatases and various kinases) of significance for drug discovery. The historical context of microarrays and the advantages of the DiscoveryDotTM technology are highlighted. The success of this chemical microarray technology will provide unprecedented possibility and capability for parallel functional analysis of hundreds of thousands of chemical compounds.
DiscoveryDotTM Technology for Biochemical Microarrays
In retrospect, high‐throughput screening (HTS) with microtiter plates has been the major tool used in pharmaceutical industry to explore the chemical diversity space to find lead compounds and pharmacophores for specific biological targets (including those arising from genomic and proteomic research). To reduce the screening cost and increasing screening efficiency, many new tools have emerged in HTS application, and chemical microarray is one of the new technologies showing great promise. The earlier development of chemical microarray followed DNA array technology by immobilizing chemical compounds on the surface of microarray to probe the biological target with binding assay (1). However, given >1040 potential low molecular weight chemical compounds available in theory (2), many difficulties will be encountered with immobilization, drying or caging compound with other material. To avoid these problems, we have developed a novel solution‐phase chemical compound microarray, DiscoveryDotTM (Reaction Biology Corporation, Malvern, PA, USA). In this format, each compound is individually arrayed on glass surface with reaction buffer plus low concentration of glycerol, and activated with biological targets by delivering analytes with an aerosol deposition technology (Figure 1) (3, 4, 5). As this technology exactly mimics the conventional well‐based screenings, chemical compounds are always in reaction solutions; therefore, any organic compound library can be manufactured. With different detection methods, this microarray can be used for assaying enzymatic reactions, protein–protein interactions, antigen–antibody screening and potentially receptor bindings.
Figure 1.
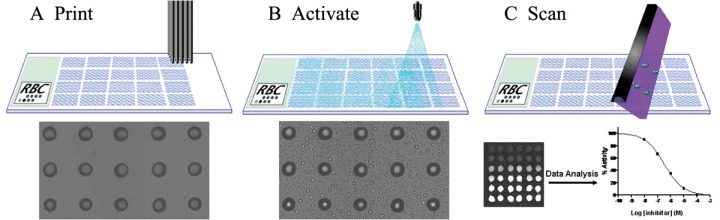
The execution procedure of DiscoveryDot TM platform. A) Compounds are arrayed as solution‐phase individual reaction centers. B) Microarray is activated by a fine aerosol mist of analytes. C) Reactions were detected and analyzed with imaging instruments.
Currently 6600 individual nanoliter reactions are screened on one regular 1′′ × 3′′ microarray chip, which is equal to 17 of 384‐well plates. A single printing run can rapidly create hundred sets of the library; piezo‐aerosol only need microliters analytes to activate all nanoliter reactions at the same time, this technology could reduce the HTS cost over 90%. For example, a 66 000 compound screen required only $10 of Factor Xa (4). In conventional kinases HTS and profiling, peptide substrates are often used instead of native protein substrates because of the high cost of generating pure protein substrates. However, this practice creates large number of inhibitors which are target unspecific. As only microgram of native proteins is needed for DiscoveryDotTM‐based screening, kinase HTS and profiling could be performed with native protein substrate at very low cost (5). For example, less than ¢0.1 per reaction of native protein substrate, dephosphorylated myelin basic protein, is needed for screening p38α MAP kinase against LOPAC library (Figure 2). More recently, DiscoveryDotTM technology has been successfully used in the discovery of a cathepsin L inhibitor, MDL28170, which prevented severe acute respiratory syndrome (SARS) virus from entering targeted cells (6). This study opened a new target class for developing drugs that could be therapeutically very useful to against the SARS virus.
Figure 2.
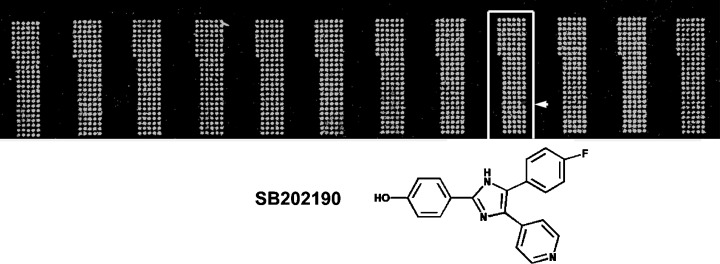
Serine/threonine kinases p38 α screening against LOPAC library. Dephosphorylated myelin basic protein was used as substrate to coat microarray, and LOPAC library (Sigma, St Louis, MO, USA) was then microarrayed and activated by p38α. Compound, SB202190, a potent and cell permeable inhibitor of p38 MAP kinase stood out as strong inhibitor during screening (see arrow at subarray#9, column 1 and row 18), which was then confirmed by well‐based reaction.
References
- 1. Kuruvilla F.G., Shamji A.F., Sternson S.M., Hergenrother P.J., Schreiber S.L. (2002) Dissecting glucose signaling with diversity‐oriented synthesis and small‐molecule microarrays. Nature;416: 653–657. [DOI] [PubMed] [Google Scholar]
- 2. Gribbon P., Sewing A. (2005) High‐throughput drug discovery: what can we expect from HTS? Drug Discov Today;10: 17–22. [DOI] [PubMed] [Google Scholar]
- 3. Gosalia D.N., Diamond S.L. (2003) Printing Chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci USA;100: 8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma H., Horiuchi K.Y., Wang Y., Kucharewicz S.A., Diamond S.L. (2005) Nanoliter homogenous ultra high throughput screening microarray for lead discoveries and IC50 profiling. Assay Drug Dev Technol;3: 177–187. [DOI] [PubMed] [Google Scholar]
- 5. Horiuchi K.Y., Wang Y., Diamond S.L., Ma H. (2005) Microarrays for the functional analysis of the chemical‐kinase interactome.. J Biomol Screening, in press. Epub. Nov. 28. [DOI] [PubMed] [Google Scholar]
- 6. Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA;102: 11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]


