Diagnostics of antibiotic resistance
P501 Screening of ampC β-lactamases producing Escherichia coli in stool samples
T.D. Huang, C. Bauraing, P. Bogaerts, C. Berhin, Y. Glupczynski (Yvoir, BE)
Objectives: AmpC β-lactamases hyperproducing (ABLH) Enterobacteriaceae have been reported worldwide, but few data are available about their prevalence in human clinical specimens. We aimed to investigate the prevalence of ABLH (chromosomal or plasmid-borne) E. coli in the faecal flora of hospitalised patients.
Material and Methods: 253 consecutive faecal samples from 17 patients collected between April and July 2006 were screened for the presence of ABLH E. coli. Samples were incubated overnight in BHI broth containing vancomycin (20 mg/L) and cefoxitin (12 mg/L) then subcultured onto CHROMagar Orientation agar (BD) plates on which a 30 μg cefoxitin disk and a 30 μg ceftazidim disk (I2A) were placed. E. coli colonies growing close to cefoxitin and/or ceftazidim disks were tested for antimicrobial susceptibility by disk diffusion. Strains with reduced susceptibility to ampicillin, amoxi-clavulanate, cefazolin and cefoxitin (compatible with a ABLH phenotype) were further characterised by AmpC disk test, isoelectric focusing gel (IEF), plasmid-borne ampC PCR and sequence analysis of the chromosomal ampC promoter regions. An ABLH was defined on the basis of molecular tests (presence of a plasmidic ampC gene, a mutated ampC promoter and/or corresponding band by IEF).
Results: 23 E. coli strains (9% of the total faecal samples) isolated on the CHROMagar plates displayed a cephalosporinase resistance phenotype, but only 12 were confirmed as ABLH (prevalence of 6.7% of all patients); 11 strains were chromosomal ampC hyperproducers (prevalence of 6.2%) including 7 showing mutations at position −42 of the promoter region; only 1 strain harboured a plasmidic CMY-2 gene. The other 11 isolates were considered as putative penicillinase producers associated with porin deficiency. Two extended-spectrum β-lactamases (ESBL) producing E. coli strains (including one having both ampC and ESBL enzymes) were recovered by our screening method.
Conclusion: Our screening technique appeared promising to detect ABLH E. coli. In our study, chromosomal ampC hyperproducers E. coli represented the major proportion (85%) of faecal carriage of ABLH E. coli whereas plasmid-mediated ampC was encountered in only 1 of the 12 ABLH isolates. Further studies of larger scale are needed to characterise their prevalence.
P502 Ability of Iranian microbiology laboratories for detection and susceptibility testing of unknown micro-organisms: survey results of 2149 laboratories
M. Rahbar, M. Saremi, R. Sabourian, S. Hekmat yazdi (Tehran, IR)
Objective: The aim of this study was to determine ability of Iranian microbiology laboratories for identification and susceptibility testing of three unknown microorganisms.
Methods: In January 17th run of proficiency testing of Iranian microbiology laboratories carried out by research centre and reference laboratories of Iran. In this survey three microorganisms including Salmonella Para A, Stenotrophomonas maltophilia (ATCC 13637) and Listeria monocytogenes (ATCC 7644) were submitted to 2149, 640 and 1509 laboratories respectively. All laboratories included both hospital and non-hospital microbiology laboratories. S. para A and S. maltophilia were sent to hospital microbiology laboratories and S. para A and Listeria monocytogenes were sent to non-hospital microbiology laboratories. We asked all laboratories to identify each microorganism and susceptibility testing just for S. para A against Ampicillin, Trimethoprim–sulfamethoxazole, Cefotaxime, Ciprofloxacin and Chloramphenicol. Scoring of results performed according criteria recommended by WHO.
Results: Of 2149 laboratories only 1491 (70%) participated in our survey and 30% did not participated in this study. Of 1491 laboratories 513 laboratories identified S. para A correctly and obtained maximum 3 score of points and 317 (21.2%) laboratories misidentified this microorganism. Many laboratories had difficulty in identification of S. maltophilia and only 11% of laboratories were able in identification of S. maltophilia and 54% of laboratories obtained zero score of points. The Third organism, (L. monocytogenes) identified correctly by 24.6% of laboratories and obtained the maximum score and 40.6% of laboratories were not capable in detection of this organism. The other laboratories sent intermediate correctly response. The results of susceptibility testing of S. para A were relatively satisfied and many laboratories obtained maximum five score.
Conclusion: Our study reveals that unfortunately many microbiology laboratories in our country are not capable in detection of microorganism such as S. maltophilia. This problem may be due to lack of some materials and reagents for detection of such microorganism in many laboratories.
P503 Discrepant antimicrobial susceptibility data: an important quality tool for the microbiological laboratory?
A. De Bel, D. Piérard, J. Bellen, A. Van Zeebroeck, S. Lauwers (Brussels, BE)
Objectives: To investigate the usefulness of listings of antimicrobial susceptibility patterns that differ in patients with several isolates of the same species as a tool for improving the quality of antimicrobial susceptibility tests.
Methods: A locally developed data management system was used to detect major differences (S-R) in the antimicrobial susceptibility data of patients with several isolates belonging to the same species. Minor differences (S-I or I-R) were considered as not significant and were not taken into account. The following data were extracted from the laboratory information system: patient name, ID number, organism ID, specimen numbers and the results of antimicrobial susceptibility tests.
Using the pictures of disc diffusion tests stored in the SirScan®, a semi automated image analyser coupled with an expert system, as well as manual notes on the work lists, explanations were searched for the discrepancies.
Results: Between 1 September 2005 and 31 August 2006, a total of 148 sets of discrepant antimicrobial susceptibility data were reviewed on a monthly basis. In 24 cases (16.2%) discrepancies were due to a laboratory error. An explanation was found for the discrepant patterns in 119 cases (2 different strains, etc.) while no conclusion could be drawn in 5 cases.
The following errors were found: erroneous reading of inhibition diameter (n = 9), interpretation of ESBL screenings and confirmation tests (n = 5), interpretation of the D-test for the detection of inducible resistance to clindamycin of staphylococci (n = 3), false sensitive results to clindamycin of anaerobes when the prescribed incubation time is not observed (n = 2), discrepancies between SirScan® results and LIS results (n = 2), not reporting MecA PCR result (n = 1) and incorrect expert rules n = 2).
Although the results of these surveys became available too late to lead to useful corrections for individual patients, they enabled correction of systematic errors. In addition, they were discussed during technical meetings, as an educative tool for all technologists.
Conclusion: Once a local data management system is developed, the generation of monthly listings of discrepant antimicrobial susceptibility data is very effortless. Systematic review of these data increases workload for the microbiologist but is a very informative and useful tool to improve quality. Results are used to implement corrective actions whenever possible and as a training tool.
P504 The frequency of production of metallo-β-lactamases by hospital carbapenem resistant Pseudomonas aeruginosa strains
A. Targosz, P. Nowak, A. Budak, M. Skalkowska, E. Tokarska, D. Wlodarczyk, G. Geza (Cracow, PL)
Objectives: Pseudomonas aeruginosa is the common Gram-negative rod associated with hospital infections. This bacterium is often multidrug resistant. Carbapenem resistance has been observed frequently in P. aeruginosa strains. The one form of carbapenem resistance is mediated by metallo-β-lactamases (MBL). MBLs are Ambler class B enzymes which hydrolyze penicillins, cephalosporins and carbapenems and are not inhibited by site-directed β-lactam inhibitors. The aim of the study was detection of carbapenem resistant P. aeruginosa strains producing MBL.
Methods: Our study concerned 98 P. aeruginosa strains resistant to imipenem and meropenem. Bacteria were isolated from clinical specimens from patients hospitalised in different wards of Rydygier's Hospital in Krakow from 1st January of 2005 till 31st June 2006. P. aeruginosa strains were identified on the basis of typical morphology confirmed by Gram-staining microscopy and biochemical tests – ID 32 GN strips using ATB system (bioMérieux, France). The in vitro antimicrobial susceptibility of P. aeruginosa clinical isolates was routinely determined using disc diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines. P. aeruginosa strains resistant to carbapenems were tested for MBL production. The screening was carried out by two variants of the synergy disk test with EDTA and 2-mercaptopropionic acid as MBL inhibitors and with Etest MBL strips (AB Biodisk, Sweden).
Results: Among 98 strains resistant to imipenem and meropenem 20 produced MBL. P. aeruginosa MBL positive strains were obtained from 9 patients hospitalised at Intensive Care Unit (19 strains) and General Surgery Unit (1 strain). We isolated 4 strains from each of two different patients, 3 strains from one patient, 2 strains from each of three patients and 1 strain from each of three patients. A total of 20 MBL positive strains were collected from: respiratory track (14 strains), urine (3 strains), wound (2 strains) and blood (1 strain). In the group of 78 P. aeruginosa strains the resistance to carbapenems was associated with different various mechanisms.
Conclusions: Detection of carbapenem resistant P. aeruginosa strains producing MBL is the relevant information for clinicists as well as for Infection Control Management
P505 Screening of multidrug-resistant bacteria: clinical evaluation of a novel chromogenic medium chromID” ESBL for the screening of ESBL-producing Enterobacteriaceae
M. Husson, S. Ghiradi, M. Gerard, A. Dediste (Brussels, BE; Craponne, FR)
Objectives: Prevention of healthcare associated infections relies on the development and follow up of wise infection controls programmes. Screening of extended-spectrum β-lactamase producing Enterobacteriaceae (ESBLE) is one of the methods to eradicate the Multi Drug Resistant (MDR) bacteria. The aim of the study was to evaluate the new chromogenic medium chromID” ESBL (bioMérieux, Craponne, France) for the rapid isolation and identification of ESBLE.
Methods: chromID” ESBL was compared to a home made method combining a Mac Conkey agar plate with a disk of ceftazidime (McC). A total of 173 at risk hospitalised patients were tested; 73 rectal swabs, 52 respiratory secretions, 48 urines were directly inoculated on the 2 media which were incubated during 18–24hrs and 48 hrs at 35°C-37°C. Typical colonies growing on chromID” ESBL and on McC were identified. Confirmation of the presence of ESBLs was performed using disk diffusion methods (according to CA-SFM and/or CLSI standards).
Results: A total of 19 specimens were positive for ESBLE, corresponding to 20 ESBLE strains: Escherichia coli (n = 13), Klebsiella spp. (n = 3), Enterobacter aerogenes (n = 2), Enterobacter cloacae (n = 2). After 24hrs incubation, the 19 specimens were positive on chromID” ESBL, whereas only 13 were positive on McC. Negative predictive value for chromID” ESBL and McC was respectively 100% [97.32%; 100%] and 96% [91.37%; 98.17%], corresponding to 6 false negative on McC. After 24hrs, 9 specimens on chromID” ESBL and 17 on McC showed a characteristic growth but were not confirmed ESBL positive (false positives). The specificity was superior for chromID” ESBL than for McC (respectively 94% and 89%), with no statistical significant difference. Selectivity towards non ESBLE was higher for chromID” ESBL.
Conclusion: The new chromID” ESBL, combining two chromogenic substrates, one natural substrate and a selective agent of ESBL character, is well positioned as a screening test in order to exclude ESBLE carriage within 24 hrs and to participate to the development and follow up of infection control programmes.
P506 Infections in pregnant women caused by Streptococcus agalactiae and problems with growing MLSB-type resistance
M. Strus, M. Brzychczy-Wloch, T. Gosiewski, A. Drzewiecki, P. Kochan, D. Pawlik, P. Heczko (Cracow, PL)
Objectives: Over the recent years in Poland one could observe the growing numbers of pregnant women colonised with group B streptococci (GBS). Epidemiologically speaking it raises concern, because of severe consequences, i.e. higher number of infections and deaths in newborns. Therefore, the main aims of this study were to:
-
–
compare the effectiveness of GBS detection in pregnant women using the classical culture method and a modified culture method recommended by the CDC;
-
–
show the frequency of GBS carriage among the populations of healthy pregnant women versus women with high-risk pregnancy;
-
–
examine the frequency of resistance markers to macrolides and lincosamides (MLSB) among the GBS strains isolated from Polish women.
Methods: The study was performed on a group of 629 women between the II and III trimester of pregnancy. 156 women were classified as high-risk pregnancy. The materials collected for microbiological tests were vaginal and anal swabs. The materials were cultured on selective media and evaluated for GBS growth. The bacteria were identified by phenotypic methods and their identification was confirmed by genotyping, using the fluorescent in situ hybridisation (FISH) with the Saga 67 a/b probe selective for Streptococcus agalactiae species. MLSB-type resistance markers were also determined, their presence was verified by looking for the erm a, b, c genes with multiplex PCR.
Results: Carriage of GBS was confirmed in 13.5% of pregnant women by using the classical culture method. Using the CDC recommended method improved the detection to 20.8%. When using the CDC method, it was observed that the high-risk pregnancy had higher rates of carriage of GBS, up to 22.4%. MLSB-type resistance markers were detected in strains isolated with the CDC recommended method.
Conclusion:
-
1
When comparing the effectiveness of GBS detection in pregnant women using the classical culture method versus the CDC recommend method, we showed improved detection (∼5%) using the CDC method.
-
2
We showed that the frequency of GBS carriage among women with high-risk pregnancy is 3% higher than that for healthy pregnant women.
-
3
When analysing the phenotypic resistance of GBS strains to macrolides and lincosamides, we showed an 8% more frequent presence of the MLSB-type resistance among S. agalactiae in women with high-risk pregnancy (n = 156), than in women with normal pregnancy (n = 250).
P507 A novel method for the detection of extended-spectrum β-lactamases, metallo-β-lactamases and AmpC producing Gram-negative bacilli in a single test: the CICA beta-test
F. M'Zali, C. Arpin, V. Dubois, C. Andre, C. Quentin (Bordeaux, FR)
Objectives: Multidrug resistant Gram-negative bacilli are emerging pathogens. Infections caused by these organisms represent a clinical challenge. Currently, evidence related to infectious diseases with Gram-negative rods with extended spectrum β-lactamases (ESBLs) or metallo-β-lactamases (MBLs) or AmpC is accumulating. A rapid, sensitive and reliable method for their early detection is critical. The Cica beta-Test (Kanto Chemical, Japan; Mast, UK) is a new method which uses strips impregnated with a chromogenic cephalosporin (HMRZ-86) coupled with each of clavulanic acid for ESBL detection, boronic acid for AmpC detection and mercapto-acetic-acid and EDTA for detection of MBLs. The aim of this study was to evaluate the Cica beta-Test against a large panel of clinical strains known to harbour these enzymes singly or in combination.
Methods: A collection of 100 epidemiologically distinct clinical isolates from French hospitals and nursing homes were examined. The collection includes strains of the Enterobacteriaecae family, Pseudomonads and Acinetobacter spp. The clinical strains produce previously fully characterised β-lactamases of TEM, SHV, CTX-M, VEB, PER, CMY, GES, VIM, IMP and OXA families. All isolates were re-tested using the Cica beta-Test. The test consists on paper strips each specific for the detection of ESBLs, MBLs, AmpC and penicillinase producing organisms. Only ESBLs and MBLs are able to hydrolyze the β-lactam ring in HMRZ-86, resulting in a colour change from yellow to red. The test organism is grown overnight on Muller Hinton plate. One drop of HMRZ-86 is dispensed on each strip, then one to two colonies of the test organism is spread on the filter pad of the Cica-Beta-Test strip. The test is read after 2 to 15 min.
Results: All mechanisms of resistance in the clinical strains were correctly identified. Presence of combination of mechanisms was also picked up notably (CTX-M +AmpC), (VIM-2 + SHV-5), (VEB-1 +AmpC). In addition, the Cica beta-Test provides us with rapid information on the resistance mechanism(s) (maximum 15 min).
Conclusion: We describe here a technically simple method for the detection of ESBL, MBL and AmpC producing Gram-negative bacilli. The test proved of high sensitivity and specificity and provides useful information for targeted therapy. We recommend this test as a routine screening method for the detection and identification of mechanisms of resistance in Gram-negative bacteria.
P508 Preliminary feasibility study of MIC results obtained with Staphylococcus aureus and cefoxitin using an Investigational Use Only MicroScan Dried Overnight Panel
K. Sei, H. Bains, B. Zimmer (West Sacramento, US)
Objectives: Accurate confirmation of methicillin-resistant S. aureus is important for clinical laboratories. The CLSI recently introduced disk diffusion testing with cefoxitin as an alternate method to testing with oxacillin to determine MRSA; interpretive criteria using cefoxitin broth microdilution have not yet been established. This preliminary feasibility study evaluated cefoxitin microdilution with S. aureus on an Investigational Use Only (IUO) MicroScan Dried Overnight Gram Positive panel. MIC results obtained were compared to results obtained with cefoxitin disk diffusion, cefoxitin and oxacillin MICs using frozen broth microdilution panels, and mecA analysis.
Methods: Thirty-nine S. aureus comprising 17 mecA positive and 22 mecA negative isolates were tested concurrently on a MicroScan IUO Dried Overnight Gram Positive Panel and a frozen reference panel. Both panels contained cefoxitin in doubling dilutions from 2–64 ug/mL and linear dilutions between 4–8 ug/mL. The frozen reference panel contained oxacillin in doubling dilutions from 0.12–64 ug/mL. Panels were inoculated using the turbidity standard method and read visually at 24h. Cefoxitin disk diffusion was performed as described and interpreted by CLSI criteria. All data were analysed using mecA results as the reference method.
Results: All mecA positive isolates gave resistant results using the cefoxitin disk test. Reference oxacillin MICs ranged from 1–>64 ug/mL with 2 isolates giving oxacillin MICs less than or equal to 2 ug/mL. Cefoxitin MICs on frozen panels ranged from 16–64 ug/mL; corresponding results with MicroScan panels ranged from 7–32 ug/mL. All 22 mecA negative isolates gave susceptible results using the cefoxitin disk test. Reference oxacillin MICs ranged from 0.25–4 ug/mL. A total of 20/22 cefoxitin MICs on frozen panels were 4 or 5 ug/mL; the other 2 MICs were 7 and 16 ug/mL. Corresponding results with MicroScan panels gave 21/22 results at 4 ug/mL and 1 result at 5 ug/mL.
Conclusions: This preliminary feasibility study shows the MicroScan IUO Dried Overnight panel correlates well with a CLSI microdilution broth reference panel, cefoxitin disk diffusion testing, and mecA testing for detection of MRSA.
Epidemiology of resistant gastro-intestinal pathogens
P509 Genotypic analysis of human Salmonella typhimurium strains from western Kenya
D. Onyango, E.N. Waindi, R. Kakai, W. Rabsch, E. Tietye, B. Ghebremedhin, W. Konig, B. Konig (Maseno, KE; Berlin, Magdeburg, DE)
Salmonella spp. are recognized as some of the most common pathogens causing enteritis worldwide. In this study we emphasized on molecular characterisation of 20 human S. typhimurium strains from two health centres in Western Kenya.
Identification of S. typhimurium strains was performed using serotyping, biochemical and distinct molecular tests including 16S rRNA sequencing. Antimicrobial screening was done using agar-disc-diffusion, E-test and automated VITEK R 2. Molecular epidemiology and mechanisms of resistance genes to tetracycline, ampicillin, streptomycin, gentamicin, sulfamethoxazole and kanamycin was studied. Strain diversity was analysed using Pulse Field Gel Electrophoresis (PFGE), fluorescence Amplified Fragment Length Polymorphism (fAFLP) and Multi-Locus-Variable-Number-Tandem regions (MLVNTR).
All the 20 S. typhimurium isolates were resistant to ampicillin (MIC > 256 mg/L)and streptomycin (MIC, 48–256 mg/L), sulfamethoxazole (MIC > 32 mg/L), chloramphenicol (MIC > 256 mg/L), 14 to gentamicin, 13 to ceferclor. 3 Salmonella strains (one S. typhimurium, one S. enteritidis and one mixed colony) were tetracycline (MIC, 32 > 256 mg/L) resistant and possessed tetracyline resistance gene tetA responsible for drug efflux. Ciprofloxacin resistance was not detected. The molecular antimicrobial mechanism indicated resistance gene to Grm, aadB, blaPSE1, bla TEM, aadA, and strB. The microrestriction analysis by XbaI gave patterns A, B, C, D and E. Of the 20 typeable isolates, 72.7% were type A, 4.5% were type B, 4.5% were type C, 4.5% were type D, and 4.5% were type E. Phage profile analysis could be categorised into three phage types (type 1, 2, and 3) of which 3 isolates were type 1, 8 were type 2 and 5 were type 3. 3 strains could not be typed. Phage DT 104 was not detected. Nine isolates were positive for class I integron (6 had 1.2 Kb and 3 had 0.8Kb fragments).
Four plasmid profiles of 70, 55, 1.8, 1.4 Kb; 70, 1.8, 1.4 Kb; 70, 4.6 Kb and 60 Kb were observed. Salmonella plasmid virulence factor (spv) and invasive gene (inv) was amplified for all strains. Strains were further subdivided Using FAFLP analysis and selected VNTR loci.
Heterogenicity among S. typhimurium strains from Western Kenya and their difference from strains originating from other parts of Kenya and around the world was observed. S. typhimurium isolates were resistant to various antimicrobials. However the resistance was fairly low as compared to the world resistance patterns.
P510 Phenotypic and genotypic characterisation of antimicrobial resistance in Turkish Salmonella infantis isolates from chicken and minced meat
D. Avsaroglu, E. Junker, R. Helmuth, A. Schroeter, M. Akcelik, F. Bozoglu, K. Noeckler, B. Guerra (Ankara, TR; Berlin, DE)
Objectives: Characterisation of the resistance (R) phenotypes and underlying molecular mechanisms in Salmonella (S.) Infantis strains isolated from Turkish foods.
Methods: 100 Salmonella isolates were isolated from foods bought in free markets in Ankara (2005–2006). One of the most prevalent serotypes was Infantis (13 isolates). Nine of these S. Infantis isolates were considered as epidemiological unrelated strains (different isolation date or place). The nine strains (7 from chickens and 2 from minced meat) were tested for susceptibility to 17 antimicrobial agents by broth microdilution. Resistant strains were screened for 16 R-genes, class 1 and 2 integrons and mutations in the quinolone-R determining regions. Strains were typed by XbaI-PFGE and plasmid profile.
Results: The strains showed two similar XbaI-PFGE-patterns (differences affecting two bands). Six strains showed PFP1 and 3 PFP2. One big plasmid (>200 kb) was present in all of them. All strains were multiresistant, with resistances to 7–9 antimicrobials (6–7 R-determinants). Two phenotypic R-patterns were found: [kanamycin–neomycin–nalidixic acid–streptomycin–spectimomycin–sulfamethoxazole–tetracycline–trimethoprim–sulfamethoxazole/trimethoprim] in eight strains, and the same R-pattern without [kanamycin–neomycin] in one. One R-determinant was responsible for each resistance: aphA1 for kanamycin, aadA1-like for streptomycin-spectinomycin (no strA or strB were found); sul1 for sulfamethoxazole (no sul2 or sul3 were present); tet(A) for tetracycline (no tet(G) or tet(B)); and dfrA14 for trimethoprim (no dfrA1, A12, A7 or A17 were present). All strains harboured a class 1 integron carrying an aadA1 gene. No class 2 integrons were detected. All strains were resistant to nalidixic acid and showed reduced susceptibility to ciprofloxacin (0.25–0.5 microg/mL) conferred by mutations in the gyrA (Ser83 to Tyr83) and parC (Thr57 to Ser57) genes. No quinolone-R genes qnrA, qnrB or qnrS were found.
Conclusions: S. Infantis isolated from foods in Turkey exhibit a wide repertoire of genetic elements to survive under antimicrobial pressure. One specific PFGE-type carrying a big plasmid (>200 kb), and with the antimicrobial multi-R pheno/genotype [KAN-NEO]-[STR-SPE]-SUL-TET-[TMP-SXT]-NAL/aphA1-aadA1-sul1-tet(A)-dfrA14-[gyrA∧Tyr83-parC∧Ser57) is widespread. Since S. Infantis frequently causes human infections, the wide spread of such a multiresistant clone within foods should be considered as a public concern.
P511 Salmonella enterica serotype Typhimurium DT104 as a cause of infantile bacteraemia in Southern Mozambique
J. Ruiz, I. Mandomando, E. Macete, L. Puyol, A. Echeita, P. Alonso (Barcelona, ES; Manhiça, MZ)
Objectives: To report the presence of Salmonella enterica serotype Typhimurium DT104 as a cause of infantile bacteraemia in Southern Mozambique.
Methods: Bacterial infections surveillance has been carried out at Manhiça Health Research Centre since 1998, Maputo, Mozambique. One hundred seventy Salmonella typhimurium isolated from under 15 children admitted at Manhiça District Hospital (MDH) were analysed. Microbial identification was performed by biochemical methods, while serotype and phagotype were established following conventional methodologies. Antimicrobial susceptibility levels to ampicillin (Amp), amoxicillin plus clavulanic acid (AMC), ceftazidime (Caz) gentamicin (Gm), streptomycin (Str) cotrimoxazole (Sxt), tetracycline (Tc), chloramphenicol (Chl), nalidixic acid (Nal) and ciprofloxacin (Cip) were established by the method of Kirby-Bauer. Molecular mechanisms of resistance to β-lactams, chloramphenicol and tetracycline as well as the presence of type 1 integrons were detected by PCR and sequencing.
Results: Two isolates were identified as S. typhimurium DT104, which were resistant to Amp, Str, Chl and Tc, being susceptible to Sxt, Caz, Nal, Cip and Gm. One of the isolates was susceptible to AMC, while the other one was intermediate. The presence of the genes carb2, floR and tetG, as well as two integrons, that carry on the aforementioned carb2 gene and an aadA2 respectively were detected. Presence of the tetA, tetB, cmlA, tem-like, shv-like, oxa-1 like, oxa-2 like and oxa-5 like genes was not detected.
Conclusion: This is the first report of multi-drug resistant Salmonella enterica serotype Typhimurium DT104, as responsible for bacteraemia in rural Mozambican children. Bacterial surveillance studies are need in the country to monitor the emergence of multi-drug resistant Salmonella including this phagotype.
P512 Dissemination of antibiotic multi-resistant Salmonella isolates in Portuguese piggeries
P. Antunes, N. Pestana, C. Réu, L. Peixe (Porto, PT)
Objectives: To study susceptibility to different antibiotics and to characterise the genetic determinants of antimicrobial resistance in Salmonella isolated from Portuguese piggery in order to assess the contribution of this type of animal exploration to the burden of mobile resistance genes and multidrug resistant clones previously observed in human and food products.
Methods: Dry faeces samples collected in 2006 from 2 pig farms in the Center and South of Portugal were positive for the presence of Salmonella (reference method ISO 6579:2002). Antibiotic susceptibility was study by disk diffusion method (CLSI) to 10 antimicrobial agents. Different antibiotic resistant phenotypes were selected for further studies. Detection and characterisation of class 1 integrons was performed by PCR, RFLP (TaqI) and sequencing. Resistance genes were searched by PCR. Conjugation assays and clonality analysis (PFGE-XbaI) were performed.
Results: All the Salmonella isolates recovered were resistant to one (tetracycline) or more antimicrobial agents (streptomycin, gentamicin, ampicillin, nalidixic acid, chloramphenicol, tetracycline, sulfonamides or trimethoprim). Characterisation of class 1 integrons revealed the presence of an array of gene cassettes (dfrA12, aadA) in isolates of S. typhimurium which also carried sul1, sul2 and sul3 genes and in isolates of S. Rissen, carrying sul1. The isolates of both serotypes were clonally related to strains previously observed in human and foodborne isolates widely disseminated in Portugal. Interestingly, MDR isolates of the S. Rissen clone were recovered from both piggeries studied. Other resistance genes (blaTEM, aac(3)-IV, tetA) were identified in the MDR isolates, but not integrated as gene cassettes.
Conclusion: Piggeries are in our country a source of MDR Salmonella isolates. Intensive use of several antimicrobial agents in this type of animal production seems to contribute to the selection of widely disseminated MDR clones.
P513 Dissemination of a new gene cluster comprising sul3 (tnp-sul3-tnp) linked to class 1 integrons with an unusual 3′CS region (qacH) among Salmonella isolates
P. Antunes, J. Machado, L. Peixe (Porto, Lisbon, PT)
Objectives: The objective of this study was to characterise the genetic background of sul3 gene, including their association to integron structures, in non-typhoid Salmonella isolates, in order to explain the dissemination of this sulfonamide resistance gene.
Methods: Forty-seven sul3-carrying Salmonella isolates from different sources (human, food products and environment) and serotypes were studied. Characterisation of class 1 integrons was done by PCR, RFLP (TaqI) and sequencing. Clonality analysis (PFGE-XbaI) and location of the sul3 gene by conjugation assays, plasmid analysis and Southern blot hibrydisation (S1-PFGE) were performed.
Results: A gene cluster comprising sul3 and transposase-like sequences (tnpA-sul3-orf1-IS26) was linked to class 1 integrons with an unusual 3′CS region (qacH). Three types of elements differing in the gene cassette array were observed: type I) 5′CS-dfrA12-orfF-aada2-cmlA1-aadA1-qacH-tnpA-sul3 (ca. 7000bp), located in twelve MDR isolates (four serotypes corresponding to five clones) on large plasmids of different sizes (≥100 Kb, conjugation achieved in five); type II) 5′CS-dfrA12-orfF-aadA2/1-qacH-tnpA-sul3 (ca. 4500bp), only described in the MDR S. Rissen clone on identical conjugative plasmids of ca. 70Kb; type III) 5′CS-estX-psp-aadA2-cmlA1-aadA1-qacH-tnpA-sul3 (ca. 7300 bp), located in thirty-two MDR S. typhimurium isolates (corresponding to three clones) on high molecular weight plasmids of different sizes (between 150 and 240 Kb).
Conclusion: We describe the dissemination of sul3 associated with plasmid-borne class 1 integrons containing an unusual 3′CS site. The presence of similar sul3-integron platforms containing different gene cassettes arrays or hybrid genes suggests evolution of the genetic background by different recombinatorial events. The association with epidemic plasmids and particular MDR clones of Salmonella might contribute to the maintenance and further spread of modular antibiotic resistance elements from food animals to hospitalised humans as reported for other Salmonella genetic elements.
P514 Class 1 integron mediates antibiotic resistance in Aeromonas spp. from rainbow trout farms in Australia
O. Akinbowale, H. Peng, M. Barton (Adelaide, AU)
Objectives: As part of the work carried out in response to the Joint Expert Technical Advisory Committee on Antibiotic Resistance (JETACAR) recommendation on antibiotic resistance surveillance, the presence of integrons and other resistance determinants was investigated in 90 Aeromonas isolates derived from nine freshwater trout farms in Victoria (Australia).
Methods: Polymerase chain reaction (PCR) was carried out for the detection of integrase genes Int1, Int2, Int3, integron variable region, integron associated aadA gene with primers specific for the detection of the aadA1a gene cassette and other closely related aadA gene cassettes (with the exception of aadA4 and aadA5), streptomycin resistance genes strA-strB, sulphonamide resistance gene sul1, quaternary ammonium compound resistance gene qac1, beta lactamase resistance genes blaTEM, blaSHV, and tetracycline resistance gene tetA-E and tetM. Clonality analysis was performed by Pulse field gel electrophoresis (PFGE).
Results: Class 1 integrons were detected in 28 of the 90 (31%) strains investigated. Class 2 and 3 integrons were not detected. Using primers specific for the aadA gene, aadA gene was detected in 19 of the 27 (70%) streptomycin resistant strains. However, when the variable region of the integron was amplified, four strains with streptomycin MIC 128mg/L and one with MIC >128mg/L gave amplicon sizes of 1000 bp each. Other strains having MIC 16mg/L did not give any amplicon possibly due to lack of the 3′conserved segment. Sequence analysis of the products reveals the presence of aadA2 gene. None of the strains harboured the strA-strB genes. PFGE analysis of the five strains reveals genetic relatedness with all five having the same banding pattern even though they came from different farms. Sul1 gene was detected in 13 of the 15 sulphonamide resistant strains and qac1 gene detected in 8 of the 28 integron bearing strains. TetC was detected in all and tetA in 9 of the 18 tetracycline resistant strains. No blaTEM and blaSHV was detected.
Conclusion: Although no antibiotics are licenced for use in Australian aquaculture and there has been no information on resistance determinants, our data suggests that Aeromonas carrying resistance genes as well as integrons are present in farm raised fish and sediments and different fish farms might share a common pool for the aadA2 gene.
P515 Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates in the United Arab Emirates
M. Alfaresi, A. Elkouch, A. Abdulsalam (Abudhabi, AE)
Objectives: The gastric pathogen Helicobacter (H.) pylori produces virulence factors such as CagA and VacA that are associated with symptom severity in infected individuals. In this study, we asked whether there is a correlation between the clarithromycin resistance status of H. pylori clinical isolates and vacA and cagA status, as well as whether these characteristics correlated with the clinical symptoms of gastric disease.
Methods: DNA was extracted from antral gastric biopsy samples from 91 dyspeptic patients in the United Arab Emirates (UAE). Real-time PCR and melting curve analysis was used to identify patients infected with H. pylori and to further identify strains containing the A(2142/43)G or the A(2142)C mutations that are associated with clarithromycin resistance. PCR was also used to identify cagA- and vacA-positive strains. Clinical examination and patient histories were used to classify the clinical symptoms of H. pylori-infected patients.
Results: Real-time PCR analysis detected the presence of H. pylori in 55 samples (60%); further PCR analysis found that 36 of the pathogen-positive samples (65.5%) contained at least one of three point mutations associated with clarithromycin resistance. Patients from the UAE had the same mutation incidence as non-UAE patients. The vacA gene was present in 72.7% and cagA was present in 75.5% of the positive samples. The 55 H. pylori-positive patients were clinically categorised as having non-ulcer dyspepsia (19 patients), peptic ulcers (20 patients) or gastroesophageal reflux disease (16 patients).
Conclusions: The presence of each clarithromycin-resistance inducing mutation was largely independent of the others. The A(2142/43)G mutations were strongly associated with the presence of both the vacA gene and the cagA gene, and there was a strong association of the presence of both the vacA and the cagA genes. Both genes were more likely to be present than absent in samples from all patients, regardless of the symptoms exhibited, but there was no correlation in the incidence of any of the point mutations with any of the categories of clinical symptoms.
Taken together, these results may help physicians identify patients likely to be responsive to standard clarithromycin-based therapy, and also underscore the value of using real-time PCR methods for rapid identification of clarithromycin-resistant H. pylori strains.
P516 Distribution of clarithromycin-resistant strains of Helicobacter pylori in north-west Russia
E. Tarasova, K. Osipov, M. Suvorova (Saint-Petersburg, RU)
Objectives: Recent studies have shown that susceptibility of Helicobacter pylori strains to clarithromycin strongly depends on mutations in the gene of 23SrRNA. The level of clarithromycin resistance is constantly increasing in the population due to accumulation of specific mutations in the epidemic H. pylori strains. In this study we tried to assess the distribution of clarithromycin resistant strains of Helicobacter pylori in north-west Russia.
Material and Methods: 65 patients with H. pylori infection were included in the study. 51 of them were children under 16 years old. Helicobacter pylori infection was analysed by histology, rapid urease tests and PCR employing the primers to urease C gene. From some of the patients both antral and body of stomach biopsies were examined. Clarithromycin resistance was assessed by polymerase chain reaction to identify the presence of point mutations in the peptidyltransferase region of the 23S rRNA gene previously associated with resistance to clarithromycin. PCR products were digested with restriction enzymes MboII, HhaI, and BsaI to detect mutations A2142G, T2717C and A2143G
Results: Results Primary clarithromycin resistance was detected in 13 (20%) patients. The A2143G point mutation was detected in 6 (46.1%) patients, A2142G in 3 (23.%), A2117C in 4 (30%). 12 patients had the same mutations in antral and body stomach sections, but 1 patient had a strain of H. pylori without any mutations in antral section and A2142G mutation in body of stomach.
Conclusions: Our study found that clarithromycin resistance is highly prevalent and that A2143G is the most frequent point mutation involved in north-west Russia region. It is worth examining both antral and body srctions of the stomach. These results suggest that the PCR is a valid tool for rapid assessment of clarithromycin resistance in H. pylori and that in the future it could be used directly on biopsy specimens, avoiding the need for culture-based methods.
P517 Toxigenic status of Korean Clostridium difficile isolates
H. Kim, M. Kim, C-K. Kim, D. Yong, K. Lee, Y. Chong, J-W. Park, T.V. Riley (Seoul, Daejeon, KR; Perth, AU)
Objectives: Toxigenic strains of Clostridium difficile usually produce both toxin A and B, although toxin A variant strains (toxin A-negative, toxin B-positive) have been responsible for C. difficile associated disease (CDAD). Some strains of C. difficile also produce a binary toxin. Recently, epidemics of CDAD with high morbidity and mortality have been reported in Canada, the USA and Europe due to a new strain of C. difficile (PCR ribotype 027). This strain produces more toxin A and B than other strains due to partial deletion of tcdC gene (down regulator of production of toxin A and B) and carried binary toxin gene. The aims of this work were to evaluate the toxigenic status of strains of C. difficile circulating in Korea and to determine whether this new strain had arrived.
Methods: Four hundred and seven randomly selected isolates of C. difficile recovered from patients with diarrhoea in a tertiary teaching hospital in Korea from 1980 to August 2006 were analysed. PCR was used to amplify genes for toxin A (tcdA), toxin B (tcdB), the repeating sequence of toxin A (tcdA rep) and binary toxin (cdtA, cdtB). PCR amplification of tcdC gene and PCR ribotyping were performed in binary toxin-producing strains. The patients' medical records were reviewed in order to evaluate the severity of CDAD.
Results: Of the 407 strains tested, 319 (78%) were toxigenic strains. The proportion of toxin A variant strains increased during the study period: 1980 and 1990 (0%), 1995 (4.2%), 2002 (12.5%), 2003 (15.2%), 2004 (39.6%), 2005 (30.0%), and 2006 (28.6%). The proportion of binary toxin-producing strains was also increased: 1980, 1990, 2002 and 2003 (0%), 1995 (2.1%), 2004 (1.9%), 2005 (2.6%) and 2006 (4.1%). All binary toxin-producing strains were toxin A and B positive, and no partial deletion of the tcdC genes was detected. PCR ribotyping patterns showed that binary toxin-positive strains were unrelated. According to medical records in binary toxin-positive cases, there was no evidence of severe, recurrent or fatal cases.
Conclusion: Toxin A variant strains of C. difficile were very prevalent and binary toxin-producing strains started to emerge in Korea. PCR ribotype 027 strain of C. difficile was not detected in our study. Further multicentre surveys are needed to ensure this new strain is present in Korea.
P518 Mandatory strain sampling programme for Clostridium difficile in English NHS acute hospitals: experience from surveillance in 2005 and options for future implementation
A. Charlett, M. Murray, B. Patel, J. Brazier, K. Wagner, A. Jones, L. Saker, A. Pearson (London, Cardiff, UK)
Objectives: The Department of Health in England (DH) requested that the Health Protection Agency (HPA) introduce various programmes to examine epidemiologic trends on Clostridium difficile associated disease (CDAD). Data from the DH surveillance system suggests increasing incidence of CDAD, with 51,690 cases reported in 2005 compared with 44,107 in 2004, representing a 17.2% increase. In addition to the national mandatory programme for C. difficile, in 2005 the HPA implemented a random sampling scheme to monitor the frequency of C. difficile strains and examine regional variations in types of strains. This paper describes the results of the first year of the C. difficile random sampling programme.
Methods: 173 acute Trusts in England submitted C. difficile samples to the Anaerobe Reference Unit (ARU) in Cardiff as part of a national sampling programme. Each acute trust was randomly allocated one week for sampling during which they provided consecutive positive C. difficile samples (excluding multiple specimens from an outbreak) up to a maximum of ten. C. difficile isolates were identified using PCR Ribotyping.
Results: A total of 1,004 cultures of C. difficile positive stools from 143 trusts were obtained and C. difficile isolates were recovered from 881 of the 1,004 samples. Results indicate wide regional variations in PCR Ribotypes, with C. difficile isolate 106 found predominantly in London (41% of cases) and the West Midlands (40% of cases); epidemic strain 027 identified mainly in the South East (41% of cases) followed by the West Midlands and South West (40% of cases); and isolate 001 found in Yorkshire and the Humber and the North West (48% of cases). Results indicate that there appears to be a major shift from the observed predominance of Ribotype 001, although previous sampling routines focused on analysis of strains submitted from outbreaks. This study indicates a shift in the frequency and distribution of strain types.
Conclusions: The sampling programme underscores the importance of collecting strains for analysis by PCR Ribotying to establish whether there has been a concomitant change in the epidemiology of C. difficile. Options for future sampling schemes for C. difficile include, broadening the sample to include patients under 65 (as these represent 22% of the cases), over-sampling high-prevalence hospitals, instituting specific programmes for examining hyper-endemnicity and emergent outbreaks, and examining temporal trends.
P519 The effect of antibiotic withdrawal on the incidence of antibiotic-resistant Campylobacter spp. in the pig gut
V.I. Enne, A.A. Delsol, P.M. Bennett (Bristol, UK)
Objectives: The fate of antibiotic-resistant bacteria in the absence of antibiotics is poorly understood. We studied the effect of antibiotic withdrawal on the presence of antibiotic-resistant Campylobacter spp. in the pig gut.
Methods: After treatment with tetracycline and penicillin, 6 pigs were placed in a bio-secure unit and antibiotics withdrawn. Faecal samples were collected at withdrawal (t0), and once a week for 8 weeks (t1-t8); Campylobacter spp. were isolated by plating faeces onto CCDA medium followed by microscopic identification. 496 Campylobacter spp. isolates were collected, comprising approximately equal numbers from each time point and pig. 62 of the isolates were speciated by a multiplex PCR assay. The susceptibilities of the isolates to ampicillin, chloramphenicol, ciprofloxacin, streptomycin, sulphamethoxazole and tetracycline were determined by disc diffusion. PCR for the resistance genes aadA, blaOXA-61 and tetO was carried out on a proportion of isolates. RAPD PCR was used to determine relationships between selected isolates.
Results: All 62 isolates speciated by multiplex PCR were C. coli. The incidence of antibiotic resistance was variable from week to week, but antibiotic withdrawal did result in some reductions in the overall prevalence of resistance. Streptomycin resistance decreased significantly from 79% at t0 to 46% at t8. Ampicillin resistance also decreased significantly from 12% at t0 to 0% at t8, and tetracycline resistance decreased from 90% at t0 to 44% at t8. Chloramphenicol resistance remained low throughout the study, while ciprofloxacin and sulphonamide resistances fluctuated, averaging at 31% and 23% resistance respectively. PCR indicated that approximately 75% of streptomycin resistant isolates carried the aadA gene, approximately 90% of tetracycline resistant isolates carried the tetO gene and approximately 60% of ampicillin resistant isolates carried the blaOXA-61 gene. RAPD PCR revealed that at any one time the Campylobacter population was composed of a variety of several different strains, and that the reductions in resistance observed were due to displacement of resistant strains by sensitive ones.
Conclusion: Antibiotic withdrawal resulted in a reduction in the incidence of antibiotic resistance among Campylobacter spp. from the pig gut, although this reduction only occurred several weeks after antibiotic withdrawal. Resistance to agents that both had and had not been administered to the animals was reduced.
P520 Antibiotic resistance conferred by a class 1 integron and SXT element in different Vibrio cholerae O-serotype isolated in Algeria
H. Ammari, R. Ruimy, D. Skurnik, A. Andremont, D. Mazel, A. Guerout, F. Assaous, K. Rahal (Algiers, DZ; Paris, FR)
Objectives: We investigated retrospectively antibiotic resistance, presence of integrons and SXT integrating conjugative element (SXT-ICE) in Vibrio cholerae isolates from the environment and from patients in several cities of Algeria, during the period from 1980 to 2003.
Methods: The strain collection was selected from a large number of V. cholerae studied previously for various phenotypic and genotypic characteristics. Single isolates differing by year, place of isolation and phenotypic pattern of antibiotic resistance were included in this study. Eighteen isolates (8 V. cholerae O1 and 10 V. cholerae non-O1, non-O139) were examined for the presence of: (i) class 1, 2 or 3 integrases by a triplex real-time PCR, (ii) SXT-ICE by PCR detection of the integrase gene. The resistance gene cassettes content within the different class 1 integrons were determined by DNA sequencing of the variable region located between the 5′ and 3′ conserved sequences.
Results: All V. cholerae O1 carried class 1 integron except two isolates of 1980 and one of 1997. One isolate of 1980, and two isolates of 1981, were found to contain two class 1 integrons carrying different cassettes (aacC and aadA1, dfr2 and aadA6). The 1986 isolate carried also 2 integrons which respectively contained dfr15 and aadA1. A single class 1 integron was found in the isolate of 1994, which carried aadA1 only. The SXT-ICE was only found in the isolates of 1994 and 1997.
Concerning V. cholerae non-O1, non-O139, class 1, 2 and 3 integrons were not detected for the isolates of 1985, 1987, 1996, 1997, 1999 and 2003, of which 5 of them were only resistant to ampicillin and one to sulfonamide. Two class 1 integrons were found in an isolate of 2001, which respectively contained aadA7, dfr1 and orfC sequence. Single class 1 integrons were found in three isolates, which contained respectively dfr7, aadA1 and aadA5-dfr17. SXT-ICE was not found in any isolate.
Conclusion: Our findings show that class 1 integrons are widespread among different clinical and environmental V. cholerae O1 and non-O1, non-O139 serotypes in Algeria with a high diversity of resistance cassettes, even in a 1980 strain which appears to be the oldest available V. cholerae O1 from Africa. The SXT-ICE was only found in V. cholerae O1 after 1986.
Parasitology
P521 Prevalence of intestinal parasitism in an urban public hospital of Madrid
D. Monclús, A. Pérez de Ayala, J.M. Azcona, M.C. Del Rey, M. López-Brea (Madrid, ES)
Objective: To determine the prevalence of intestinal parasitism in adults in a 550 beds hospital of Madrid, in a period of four years.
Material and Methods: During the years 2002–2005 a total of 13.765 stool samples from patients of the hospital and area health centres were analysed. The average age was 44.8 years. Microscopic and macroscopic tests were made using methods such as: fresh test with physiological saline solution and lugol's solution, concentration techniques with formaldehyde-eter (Ritchie's test), trichrome stain, Ziehl-Neelsen stain, and Graham test for detection of oxyuros.
Results: The prevalence of the pathogens and commensal parasites was 23.51%. Of the 13,765 samples, 3,237 were positives (23.51%.), and in 686 (21% of the positives samples) we found more than one parasite, with a total of 3,847 parasites. Blastocystis hominis was the main enteroparasitic pathogen found (54.6%), followed by Endolimax nana (16.2%), and Giardia intestinalis (9.5%). The Table 1 shows the parasites detected.
Table 1.
Parasites detected in faeces
| Year | GIA LAM | BLA HOM | END NAN | ENT HI8/DI8 | Other protozoa | Cestodes | Nematodes | Total |
|---|---|---|---|---|---|---|---|---|
| 2002 | 92 | 432 | 47 | 0 | 138 | 3 | 2 | 714 |
| 2003 | 72 | 494 | 142 | 17 | 191 | 5 | 2 | 923 |
| 2004 | 103 | 651 | 222 | 16 | 166 | 1 | 4 | 1,163 |
| 2006 | 100 | 524 | 214 | 15 | 182 | 6 | 6 | 1,047 |
| Total | 367 | 2,101 | 625 | 48 | 677 | 15 | 14 | 3,847 |
GIA LAM: Giardia lamblia; BLA HOM: Blastocystis hominis; END NAN: Endolimax nana; ENT HIS/DIS; Entamoeba histolytica/dispar.
Conclusions: We have found low helminth prevalence compared with protozoa probably due the geographic area and the population group. Most of parasites were commensal or potentially pathogenic. The prevalence of intestinal parasites in our hospital is similar to other areas with similar characteristics.
P522 Use of solar radiation in disinfecting contaminated drinking water with cysts of Giardia lamblia
S. Zinyowera, N. Midzi, C. Muchaneta-Kubara, T. Simbini, T. Mduluza, V. Robertson (Harare, ZW)
Objectives:
-
1
To determine whether solar radiation is capable of inactivating cysts of Giardia lamblia.
-
2
To analyse use of different types of containers.
Methods: This is an experimental study. One hundred stool specimens for the isolation of G. lamblia cysts were collected from pupils of grades one to five from a rural primary school in Zimbabwe. Permission to conduct the study and collect specimens from students was granted including consent from the participants themselves. Specimens were screened for the presence of G. lamblia. They were concentrated using sheaters solution and purified using the discontinuos percoll gradient centrifugation method. Protozoan parasites were then preserved in distilled water, quantified using a hemocytometer, then stored at 4°C. Empty 2L polyethelyne terephthalate (PET) plastic containers were painted black on one side including a 2L high density polypropelene plastic containers. One thousand eight hundred millilitres of tap water were placed in the 2L containers. Initial temperature of each container was measured. One milliliter of the parasite suspension was added to each bottle. The containers were shaken to aerate the water. These were then placed outside on a black surface with the clear side facing the sun. Temperatures were recorded hourly and at each hour, water in a PET bottle painted black on one side would be spun at 500 g for 3 minutes using 50 ml VWR centrifuge tubes. One ml of the pooled sediment was stained with the vital dyes 0.4% trypan blue, 0.3% Congo red and the flourogenic dyes propidium iodide (PI) and 4,6 diamidino-2-phenylindole dihydrochloride (DAPI) were also used.
Results: When there was full sunshine 95% of parasites would be dead after 3 hours at temperatures of 46°C, and after 4 hours when the temperature was above 50°C, 100% of G. lamblia cysts were non-viable. During cloudy conditions when the water temperature was 38°C, 26% of parasites were non-viable. PET bottles painted black on one side absorbed more heat than those not painted including the high density polypropelene plastic containers. The open PET bottles did not kill parasites completely as the water temperature did not rise rapidly.
Conclusion: Solar radiation is effective in killing G. lamblia cysts. More assays need to be carried out to find out if it is the UVA produced by the sun or the heat from the sun that is capable of disinfecting these parasites.
P523 Evaluation of three commercial assays for the detection of Giardia and Cryptosporidium organisms in stool specimens
N. Dauby, C. Moens, S. Mohamed, S. Van den Wijngaert, O. Vandenberg, A. Dediste (Brussels, BE)
Objectives: Giardia is known as the most common pathogenic intestinal protozoa in Belgium, yet Cryptosporidium remains frequently under diagnosed. We compared three commercial assays for the detection of Giardia and Cryptosporidium.
Methods: Stool specimens were collected from 101 children attending a day care centre located in Brussels. Most of those children presented abdominal complaints and/or diarrhoea since two weeks. Therefore, we wanted to investigate this outbreak in order to exclude a parasitic etiology. Stool specimens were examined according to our specific Triple-Feces-Test (TFT) protocol which consists in 3 samples collected on 3 consecutive days (2 with SAF preservative and one fresh specimen) examined with and without concentration techniques and a permanent staining. In addition, a Rhodamine-auramine O staining for Cryptosporidium is performed on the fresh specimen, with confirmation with Kinyoun carbolfuchsin acid-fast staining. Results obtained with our protocol were compared with those from Prospect Crypto/Giardia® (ELISA), ImmunoCard STAT® (IC) and Merifluor® direct fluorescent-antibody (DFA) commercial tests, each of them performed on the unpreserved stool specimen.
Results: Among the 101 children included in the study, 5 and 17 were infected with G. lamblia and Cryptosporidium respectively. The sensitivity of the TFT-protocol was 100% for Giardia which was similar to the sensitivity of the IC and ELISA tests. The specificity of all tested methods was never lower than 97%. The sensitivity of the IC and the ELISA for the detection of Cryptosporidium were 100% and 95.3% respectively compared to 52.9% for the microscopic examination. When we used DFA as screening methods for the diagnosis of Giardia or Cryptosporidium, we found a sensitivity of 80% and 82% respectively. Specificity of the DFA was 100% and 97.6% for Giardia and Cryptosporidium respectively.
Conclusion: Microscopy with the TFT protocol is very sensitive for detection of Giardia but less sensitive for detection of Cryptosporidium than the three commercial kits tested. Regarding to our results, we should consider including ELISA or IC tests in our TFT-protocol for a more reliable detection of Crytosporidium.
P524 The prevalence of Dientamoeba fragilis infection in patients with suspected enteroparasitic disease in Denmark
C.R. Stensvold, M.C. Arendrup, K. Mølbak, H.V. Nielsen (Copenhagen, DK)
A study was taken on to describe the prevalence of D. fragilis in patients with suspected enteroparasitic disease in Denmark and the diagnostic relevance of examining preserved rather than unpreserved stool samples. Parasitological examination of paired stool samples from 103 patients was completed using two different techniques: A formol ethyl-acetate concentration technique (FECT) on unpreserved faeces and a permanent staining technique (PST) on faeces preserved with sodium acetate-acetic acid-formalin (SAF). Using SAF-PST and FECT, 25% and 15% of the specimens were parasite-positive, respectively. D. fragilis was detected only in SAF-preserved stools, and 12/103 (12%) patients were shown to harbour the parasite, only two of which were shown also to host other traditionally acknowledged pathogenic parasites. The present study shows that D. fragilis is remarkably prevalent in Denmark. It confirms the relevance of examining SAF-fixed stools of patients with suspected intestinal parasitosis in settings where the examination of freshly passed, warm stools is not an option, since at least 8/10 cases of potentially symptomatic intestinal parasitosis go undetected using conventional concentration and microscopy of unpreserved stools. D. fragilis was seen mainly in patients aged 0–30 and was not associated with travel activity less than 3 months prior to the submission of stools, suggesting endemic occurrence in Denmark.
P525 Opportunistic properties of insect microsporidia
C. Franzen, S. Fischer, J. Schölmerich, S. Schneuwly, B. Salzberger (Regensburg, DE)
Objectives: Tubulinosema ratisbonensis is a microsporidian pathogen of the fruit fly Drosophila melanogaster belonging to the family Tubulinosematidae. The microsporidia in this family mainly cause infections in invertebrate hosts, but two members of this family, Anncaliia vesicularum and Anncaliia algerae, have been found as cause of infections in humans as well. Moreover, A. algerae could be transmitted to immunodeficient mice and grows in mammalian cell cultures. Thus, the examination of the opportunistic properties of other members of the family Tubulinosematidae is mandatory.
Methods: Spores of T. ratisbonensis, isolated from infected fruit flies, were inoculated on mammalian and insect cell cultures and in immunodeficient mice [NMRI (nu/nu)] (n = 24) at different locations (tail, neck, i.p., eye, leg, oral). On day 60 post infection all mice were euthanised and necropsied. Cultures and mice were examined by light microscopy, scanning and transmission electron microscopy, and by PCR and subsequent DNA sequencing.
Results: In cell cultures parasite growth was only seen in human lung fibroblasts whereas no growth was seen in Vero cells or insect cell cultures. Transmission electron microscopy showed the typical ultrastructure of T. ratisbonensis and scanning electron microscopy showed oval or slightly pyriform spores with some spores having extruded their polar tubes. Sequencing of PCR-fragments, amplified from infected cell cultures, reveals DNA sequences that were 100% identical with the original T. ratisbonensis rRNA sequence. All 24 mice survived the 60 days study period without clear signs of disease. All examined internal organs were free from microsporidia but limited growth was seen at sites with lower body temperature (tail, leg).
Conclusion: As T. ratisbonensis is able to proliferate in mammalian cells and in immunodeficient mice at sites with lower body temperature, it might have opportunistic properties like other members of the family Tubulinosematidae.
P526 Clinical significance and frequency of Blastocystis hominis infection in primary school children in Ardabil, Iran (2003)
A. Daryani, G.H. Ettehad, N. Barmaki, M. Sharif, A. Abedi, H. Ziaei, H. Alimohammadi (Sari, Ardabil, IR)
Objective: There is a dramatic increase in the frequency of Blastocystis hominis infection in association with diarrhoea and distinct clinical symptoms, especially in AIDS patients. The aim of this study was to evaluate the frequency of this parasite, and relate personal data and the presence of signs with the frequency of B. hominis among primary school children.
Methods: This cross-sectional study was performed on 1070 school children between 7–13 years old in Ardabil, Iran, 2003. A questionnaire was completed for each child. Stool specimens were collected by stratified random sampling and were examined for presence of B. hominis using direct wet mount and formalin-ether concentration methods.
Results: The positive rate of B. hominis was 28.2% (302/1070). A total of 109 cases (10.2%) showed more than five parasites per field at a magnification of 400x. The most common symptoms in children who showed only B. hominis were abdominal pain (49.4%), inappetance (35.8%), and nausea (33%).
Conclusion: As B. hominis is quite common among school children, contaminated drinking water is suspected to be the source of infection.
P527 Analysis of IgG and IgM in patients pre-diagnosed with toxoplasmosis
G. Sönmez Tamer, B. Mutlu, A. Willke (Kocaeli, TR)
Objectives: The aim of this study was to compare Toxoplasma seropositivity among the regions of Turkey. For this purpose, we evaluated toxoplasma IgG and IgM seroprevalans retrorespectively from patients who applied to gynecology, neonatalogy, paediatric and other clinics. Serum which were obtained from patients who are prediagnosed with Toxoplasma were included to this study. These serum samples were collected between 01.2003–10.2006.
Methods: Serums tested with Enzyme Immune Assay (Axsym, Abbot®) for IgG and IgM. Data were collected and documented for a period of 3 years.
Results: The results obtained were evaluated in four groups: group 1 gynecology, group 2 neonatal, group 3 paediatric and group 4 others. In 1828 serum samples, we found 32.67 IgG seropositive, 1.25% IgM seropositive and 5.47% IgG+IgM seropositive samples. In 256 serum samples from gynecology 47.65% were IgG seropositive, 1.95% were IgM seropositive and 5.85% were IgG+IgM seropositive. In 239 serum samples from paediatric clinic 50.20% were IgG seropositive, and 1.23% were IgG+IgM seropositive, but fail to find IgM seropositive. In 280 serum samples from neonatal clinic we found 43.57% were IgG seropositive but fail to find IgM and IgM+IgG seropositive samples. In 1053 serum samples from other clinics 22.13% were IgG seropositive, 1.70% were IgM seropositive and 7.79% were IgG+IgM seropositive.
Discussions: Toxoplasma IgG seroprevalance in Kocaeli region were found to be lower than eastern and middle Anatolian parts of Turkey where animal farming is a common practice. The results obtained from this study may indicate that the observed differences may be related to the high socioeconomic status of people better hygiene rules and sanitation in Kocaeli region than other regions.
P528 Prevalence of antibodies to Trypanosoma cruzi in Latin American immigrants in Spain
N. Portocarrero, M. Baquero, M. Gutierrez (Madrid, ES)
Introduction: Chagas' disease is a chronic parasitic infection caused by Trypanosoma cruzi and is endemic in Central and South America. Many immigrants from that region now reside in Spain.
Objective: The aim of this study was to assess the prevalence of antibodies to Trypanosoma cruzi among immigrants from Latin American in Madrid.
Methods: A total of 320 serum samples from Latin American individuals attending the tropical clinic at Carlos III hospital Madrid, between January 2005 and October 2006 were analysed. T. cruzi antibodies were identified using a commercial screening ELISA method (Biokit, Spain).
Results: Geographic distribution of the patients was: 213 Ecuador (66.5%), 52 Colombia (16.2%), 20 Bolivia (6.2%), 9 Brazil (2.8%), 7 Peru (2.1%) and other countries of Latin American (6.2%). Eight patients were considered reactive for antibodies to T. cruzi, most of them from Bolivia 7/20 (35%) and one from Ecuador.
Conclusions: In this study it is worthy to note the high prevalence among Bolivian population (35%) and our results underscore the importance of screening of Latin American immigrants for T. cruzi to prevent transmission possibility through blood transfusion or organ transplantation.
P529 Clinical epidemiology and surveillance of leishmaniasis in the Yungas of Bolivia, 2002–2005
C. Gomez, A. Rodriguez-Morales (La Paz, BO; Trujillo, VE)
Introduction: Leishmaniasis is an endemic disease in many countries in South America, one of them Bolivia. Unfortunately is a neglected disease with few advances in the research and development of drugs against Leishmania spp., the etiological agent of this wide-clinical-spectrum disease.
Objectives: For these reasons surveillance, control and prevention in resource-limited settings where disease is present is continuously needed.
Methods: In this report we describe the clinical epidemiology and surveillance of cases of leishmaniasis seen between 2002 and 2005, in a programme applied in the Yungas of Bolivia. This region includes the Departments of La Paz, Beni, Cochabamba and Santa Cruz. The accomplishments of the programme are describe in terms of number of communities visited for preventive activities, suspected cases find in the active screening, cases reported in healthcare centres, diagnostic tests applied, confirmed cases, clinical distribution (forms of leishmaniasis), and treatments with N-metil glucamine, among others.
Results: For this period 2,606 communities were visited (mean 652 per year), detecting by active screening 2,452 suspected cases (mean 613 per year), and 334 passively (mean 83.5 per year) (p < 0.01); 5,323 tests (mean 1331 per year) were applied finding 1,845 cases of confirmed leishmaniasis (mean 461 per year) (35%, ranging 25%-39.6%), additionally to this 12,596 diagnosed cases were referred for treatment. From this total (14,441), 88.3% corresponded to Visceral Leishmaniasis (AVL), 9.2% to Cutaneous Leishmaniasis (ACL) and 2.5% to Mucosal Leishmaniasis (AMCL). During this 4-year period 69,790 blisters of N-metil glucamine were applied (mean 17,448 per year).
Conclusion: Leishmaniasis remains a major world health problem that continues to increase in incidence. This neglected tropical disease has strong but complex links with poverty. The burden of leishmaniasis falls disproportionately on the poorest segments of the global population, such as most populations in Bolivia. Public investment in treatment, prevention and control would decrease the leishmaniasis disease burden and help to alleviate poverty in such countries.
P530 Impact of climate variability in the occurrence of leishmaniasis in Southern departments of Colombia
R. Cardenas, C. Sandoval, A. Rodriguez-Morales (Cucuta, Pamplona, CO; Trujillo, VE)
Leishmaniasis are transmitted in Americas by Lutzomyia spp., mostly in endemic zones. Previous asian, european and southamerican studies indicated potential changes in vectors climate-related-distribution, but impact outcomes are still need to be furtherly studied. For this reason we report possible climatic impacts and El Niño events during 1985–2002 on leishmaniasis (ATL/AVL) in 11 departments, in the southern region of Colombia, South America. Departments included were: Amazonas (Az), Caquetá (Cq), Cauca (Ca), Huila (Hu), Meta (Mt), Nariño (Na), Putumayo (Py), Tolima (To), Valle (Va), Vaupes (Vp), Vichada (Vi). Climatic data was satellitally obtained; and epidemiological from Health Ministry. NOAA climatic classification and SOI/ONI indexes were used as global climate variability indicators. Yearly variations comparisons and medians trends deviations for disease incidence and climatic variability were made. Statistical analysis used SPSS (conf. 95%). During this period a considerable climatic variability was present, strong El Niño during 6 years and strong La Niña for 8. In this period, 19,212 leishmaniasis cases were registered in these departments (Na 18%, Cq 18%, To 16%, Va 16%, Hu 11%, Mt 8%), mean 4756.83 cases/year (ranging in these departments from 6.89 to 192.5 cases/year). During El Niño years disease increase in a mean of 4.98% (for the whole region) in comparison to La Niña years, but this was spatially heterogeneous with 2 departments evidencing increases during El Niño (Mt 6.95% and Vp 4.84%), but the rest with increase during La Niña (ranging from 1.61% to 64.41%). These differences were significant in Va (p = 0.0092), Py (p = 0.0001), Ca (p = 0.0313), and for the whole region (p = 0.0023) but not in in the rest of departments (p > 0.05). Climate is changing at an unprecedented registered-rate. Shifts in insect distribution indicate their importance. Climate is a relevant temporospatial vectors distribution determinant. These data and other previous presented by our group (Am J Trop Med & Hyg 2006;75:273–7) reflected climate importance on leishmaniasis incidence in different areas of Colombia, given the heterogeneity of impacts more temporal and spatial specific research is needed. This opens further investigations in the area related to forecasting and monitoring systems in public health systems to prevent and control earlierly this emergent infectious disease.
P531 Visceral leishmaniasis – does antimony loose its efficiency?
G. Stevanovic, M. Pelemis, M. Pavlovic, J. Poluga, L. Lavadinovic (Belgrade, RS)
Visceral leishmaniasis is parasitic diseases caused by Leishmania donovani (L. infantum, L. chagasi). Reservoirs of the parasites in our region are mostly dogs and rodents. Vector of transmission is sandflies. Illness was sporadically occurred in the southern regions of our country.
Objective: The objective of this paper was to present our experience in treatment of visceral leishmaniasis and problems in unresponsiveness to antimony therapy.
Method: During four years period in our department we treated 22 patients safer from visceral leishmaniasis. All the patients were citizens of Serbia and Montenegro. In endemic regions of these countries, live 18 patients, others were been during summer period in these regions. No one was traveled out of Europe.
Results: All the patients were adults, average age of 40.24 (range from 22–78) years, 15 of them was mails and 7 were females. Medium duration of the illness before treatment was longer then 4 mounts. Most of them had fever, anaemia or pancytopenia and enlargement of liver and spleen. Diagnosis was established by serological methods and definitive diagnosis was done by microscopic examination of bone marrow smears. As a primary therapy we used antimony (Glukantime®) in the doses of 20 mg/kg during 21–28 days. In one patient we used Pentostam®. Good outcome we have in 17 patients. But in 5 patients in spite of therapy, clinical findings were present. Spelnohepatomegaly was persisted, with pancytopenia. In patients with persistent findings of parasites we repeated therapy with antimony compounds. One of patient had good outcome, but other 4 were needed Amphothericin B. All of them were treated during 15–28 days, given intravenously for a total dose of 20 mg/kg. After two courses of Amphothericin B therapy, only two patients had persisted clinical findings longer then 6 months. These two patients were treated with liposomal amphotericin B (Ambisome®) in daily dose of 2 mg/kg during 5 days. Resolution of the symptoms was achieved during first month after the therapy.
Conclusions: Unresponsiveness to antimony therapy is becoming problem in Asia. In former Yugoslavia, we did not have such problems until now. That was first cases of visceral leishmaniasis that was unresponsive to antimony therapy in Serbia. Favourite outcome was achieved by use of liposomal amphothericin B.
P532 Cases of visceral leishmaniasis in a tertiary hospital in Athens, Greece
P. Karabogia, K. Ziva, M. Orfanidou, E. Sideris, M. Karanika, H. Malamou-Lada (Athens, GR)
Objectives: The aim of this study was to report cases of visceral leishmaniasis in adults with emphasis to epidemiological features and response to treatment in a tertiary hospital in Athens, Greece, during the period 1/2005–10/2006.
Methods: During the study period, 89 patients with high fever of unknown origin were admitted to the hospital. All of them underwent serology testing for CMV, EBV, Rickettsiae, Coxiella burnetii, Legionella, Brucella and Leishmania. Antibodies to Leishmania were detected from serum by an indirect immunofluorescence method (IFA, bioMérieux) and the parasites were detected by direct microscopy of bone marrow smears.
Results: Five out of 89 patients presented high titers of antibodies to Leishmania infantum (1/640–1/1280). Direct Giemsa stain of bone marrow smears indicated Leishmania parasites in four out of five of these patients. The patients median age was 55 years (range 16–80 years, two female-five male). One male patient was immigrant from Albania and four were native Greeks. Three of them were living in Attica area and two in urban areas of central Greece. All patients experienced symptoms for more than one month. Fever and hepatosplenomegaly were observed in all of the patients. The most frequent haematological finding was thrombocytopenia, leukopenia and anaemia. One patient found to have an underlying autoimmune disease. All patients were treated with liposomal amphotericin B and responded well. No adverse effects were detected.
Conclusions: Visceral leishmaniasis should be included in the differential diagnosis of fever of unknown origin, especially in endemic regions like Greece. Early diagnosis leads to more effective treatment, preventing relapses and adverse effects.
P533 Visceral leishmaniasis cases in Romania
S. Florescu, C. Popescu, M. Cotiga, L. Raduta, R. Botgros, C. Voinea, S. Erscoiu, E. Ceausu, P. Calistru (Bucharest, RO)
Background: Tropical diseases are no longer found only between 15 degrees northern and 15 degrees southern latitude. Increasing travelling all around the globe made possible the import of the tropical diseases in the temperate and cold weather regions. In such regions, the diagnosis of tropical diseases takes longer and is more difficult, especially in the visceral leishmaniasis.
Objectives: We analysed the clinical features, the diagnosis tools and the treatment of the imported visceral leishmaniasis in Romania.
Methods: retrospective study of 5 cases of visceral leishmaniasis admitted between 1999–2006 in the Clinic of Infectious and Tropical Diseases “Dr. V. Babes”.
Results: All patients were males, with age limits between 22–35 years. They aquired the disease working in open spaces, in agriculture and building during 3 to 12 months; 2 of them worked in Spain, 2 in Italy and 1 in Greece. The period between the clinical onset and the positive diagnosis ranged from 2 to 14 months, wereas the period between the first medical consult and the positive diagnosis ranged from 2 weeks to 12 months. All patients had fever, chills, malaise, loss of apetite, weight loss from 6 to 20 kgs, liver and splenic enlargement and pancytopenia. Parasitological exam of the medular aspirate showed amastigote forms of Leishmania spp. and was the standard for positive diagnosis. Ethiological treatment consisted of Amfotericin (4 patients) and Pentamidin (1 patient). All patients survived.
Conclusions: Visceral leishmaniasis is a reemergent disease in Romania, due to the masive immigration of the romanian workers, especially in the mediteraneen region. The positive diagnosis is usually delayed because the clinical aspect in nonspecific and aparrently sugests a haematological malignancy; thus, clinician must be aware of the epidemiological data, and mainly about the previous travels of the patient.
P534 Recurrent cutaneous leishmaniasis due to Leishmania (Viannia) guyanensis after treatment with pentamidine: a study of 7 cases and analysis of this nosological entity
J.P. Gangneux, S. Sauzet, S. Donnard, N. Meyer, A. Cornillet, F. Pratlong, C. Guiguen (Rennes, Saint-Aubin du Cormier, Montpellier, FR)
Introduction: Localised cutaneous leishmaniasis (CL) is the most common clinical expression of Leishmania infection in the New World. However, various clinical presentations with therapeutic difficulties and poorer prognosis exist: mucocutaneous leishmaniasis mainly due to L. braziliensis, and diffuse cutaneous leishmaniasis due to L. mexicana and L. amazonensis. Leishmaniasis recidivans is an unusual clinical Old World pattern mostly associated with L. tropica. Recurrence of CL lesions previously cured is also found in the New World CL, for which a few authors identified a specific nosological form, known as leishmaniasis recidiva cutis (LRC) and less than 30 cases have been reported.
Objectives: Here, we report 7 cases of recurrent CL from French Guiana after treatment with pentamidine, and discuss this atypical clinical presentation.
Results: For 15 days, French military personnels spent 3 months in French Guiana and took part in a training programme in the rainforest (Régina and Saint-Georges). 21 of them developed a CL and were treated with Pentacarinat (3 iv or 2 im injections of 4mg pentamidine isothionate/kg on alternate days). All lesions were cured 1 to 3 months after the treatment had ended. For 7 patients (33%), recurrence of the CL lesion was observed after a disease-free interval of 3 to 6 months. For each patient, new lesions appeared on the edge of a healed scar.
Discussion: Physicians should be aware of the risk of early treatment failure but also of recurrent form after Leishmania (Viannia) guyanensis infections evocative of LRC. This clinical presentation stresses the need for long-term follow-up after treatment of American CL and prompt evaluation of specific therapeutic protocols. In our series, all recurrent leishmaniasis were observed after treatment with Pentacarinat, and retreatment with an intensified protocol of 4 iv injections of pentamidine cured the disease. Less than 30 cases of LRC have been reported from Brazil, Colombia, Peru and Ecuador, mainly caused by L. braziliensis, L. amazonensis and L. panamensis. The mechanism of late recurring leishmaniasis is still poorly understood. Immunological data based on skin hypersensitivity, histopathological and immunohistochemical findings support the concept that LRC is a late-onset reactivation after persistence of living parasites around or in “cured” leishmaniasis by as yet unknown stimuli and after an incomplete host immune response to an earlier episode.
P535 Rapid epidemiological assessment of Leishmania donovani infection in eastern Sudan: immune surveillance and application of GIS
A. Sharief, E. Khalil, T. Theander, A. Kharazmi, S. Omer, M. Ibrahim (Khartoum, SD; Copengahegn, DK)
This survey was based on simple in vivo and in vitro immunological techniques combined with clinical history to obtain data about the spectrum of L. donovani infection in communities at risk of developing Visceral Leishmaniasis (VL). Clinical history and immunological tests were conducted in volunteers randomly selected from villages in an endemic area in eastern Sudan. The leishmanin skin reactivity of ≥5 mm was 33.3%. Children <15 years had higher leishmanin non-reactivity (00 mm) of 47.6% compared to 27% in adults. DAT results showed that 19.3% had reciprocal titers of >200 compared to 8.1% with reciprocal titers of >200 and <3200. Titers of ≥6400 were seen in 9% of volunteers. Eight parasite isolates were cultured, characterised as L. donovani using Heteroduplex analysis (HDA) and RFLP. These parasites were tested for their in vitro sensitivities to pentostam and amphotericin B with marked linear reduction in H3-thymidine incorporation. In the J 744-macrophage system, the parasite survival index (PSI) was similar for both drugs.

Effect of Amphotericin B on the survival index of four L. donovani amastigotes infecting a J774-cell line.
The use of clinical interview combined with simple immunological tests can give valuable information about the pattern of L. donovani infection and predict future prevalence of VL in a short time. Leishmanin non-reactive individuals are a useful piece of data to plan for future vaccine efficacy studies. The interactive dynamic map that was produced in the GIS can act as a nidus for development of a Leishmania network in Sudan and the surrounding countries that are endemic for VL.
P536 Concerns on nephrotoxicity and administration schedule of liposomal amphotericin B during treatment of a HIV-related visceral leishmaniasis
R. Manfredi, S. Sabbatani (Bologna, IT)
Introduction: Liposomal amphotericin B (lAB) is the first-line therapy of HIV-related visceral leishmaniasis (VL), based on favourable experiences, although often presented small patient (p) series, and no agreement exists about is schedule of administration.
Case report: Despite HAART, a HIV-infected p had a limited immune recovery (106 CD4+ cells/μL), and VL was diagnosed after a two-month history of fever-fatigue-hepatosplenomegaly An increasing leukopenia-lymphopenia (3660–620 cells/μL, respectively), and anaemia, developed subsequently. A short (6-day) course of lAB was administered at 3 mg/Kg/day. Immediately after the end of therapy, moderate signs of both kidney and haematological toxicity became apparent: increase of creatinine and urate levels, associated with worsening anaemia-leukopenia. Increased fluid administration allowed the control of kidney impairment, and repeated single administrations of lAB (3 mg/Kg), were performed after two and four weeks. A bone marrow biopsy repeated 5 weeks after disclosed a complete disappearance of Leishmania parasites, but a significantly reduced marrow cellularity was shown (40% versus prior 85–90%; p < 0.001).
Discussion: 1AB therapy of HIV-associated VL is a real advance. However, our report raises different problems:
-
1
During HIV disease, VL may mimick opportunistic infections, and diagnosis may be hampered by a negative serology. An elevated clinical suspicion should prompt bone marrow biopsy.
-
2
Kidney-haematological toxicity even after short-term (6-day) administration of lAB recommends frequent monitoring. Nephrotoxicity must be recognized early, while bone marrow toxicity may occur later.
-
3
Our treatment schedule treatment included a 6-day attack, followed by two single doses at 14th and 28th day, but other authors delivered 3–5 further doses according to varied schedules, since no uniformly recognized consensus exist. A standardisation of therapeutic strategies of VL is therefore needed.
P537 Fasciolosis in Spain. Review of ten years
A. Rodríguez Guardado, A. Sempere, M. Rodríguez Pérez, E. Gómez, J. Ordás, P. Suárez Leiva, J. Cartón Sánchez (Oviedo, ES)
Objective: Human infections due to Fasciola hepatica (liver fluke) are common in underdeveloped countries but are rare in Europe. We describe our experience in the diagnosis and management of fasciolosis in Asturias, a province in the north of Spain.
Methods: We have carried out retrospective study of all the cases diagnosed in the Hospital Universitario Central de Asturias (HUCA), during a ten year period (1995–2005). The infection diagnosis was made on the basis of positive serological results (titre :≥ 1/320) using indirect haemaglutination tests and clinical manifestations pointing to fasciolosis.
Results: We found and reviewed five cases. Only one patient was female, the mean age was 40 years (range, 34–45). The mean time from the onset of symptoms until diagnosis was 38 days (range, 30–60). All of them lived in a rural area and they used to eat fresh vegetables. None presented significant underlying diseases. Clinical manifestations in all of them were abdominal pain, fever, malaise and weight loss. One patient presented urticaria. Blood test of all of them showed leukocytosis, mean 14,350 leucocytes/mm3 (range, 11,200–17,400), eosinophilia mean 32% (range, 24%-60%). Hepatic focal injury was seen by computerised tomography in two patients. Serological results were positive in all five, with titres in between 1/5210–1/8920. Direct microscopy examination of stools for presence of parasite eggs was carried out in four of them, with negative results and the duodenal aspirate examination made in one, it was also negative. Four patients received bithionol (40 mgr/Kg every two days during 30 days) and one patient with triclabedazole 10 mg/Kg in alone doses. One had diarrhoea after bithiolol administration but it was not necessary to withdraw medication. Every patient recovered. During follow up serology become negative and eosinophilia disappeared.
Conclusion: The most frequent clinical manifestations of fasciolosis are abdominal pain, fever and weight loss. Eosinophilia is the most frequent laboratory data. Although diagnosis may be established by observation of parasite eggs in faeces, most of the cases can be diagnosed by serology. Biothiolol was a very effective and safe treatment against Fasciola hepatica.
P538 Investigation of Toxocara canis antibodies in patients with eosinophilia and comparison of two methods: ELISA and Western Blot
Y.A. Öner, E. Artinyan (Istanbul, TR)
Objectives: Eosinophylic cells, which are only present in gastrointestinal mucosa and form a very little part of peripheric leucosites, the blood levels are controlled districtly in healthy individuals. However, in many diseases (allergic diseases, parasitic infections and cancer), the count of eosinophylic cells increase meaningfully. Especially in parasitic infections the blood counts of these cells can reach 50,000 cells/mL. Larva migrans (visceral, ocular, cutaneous), in which eosinophilia is a constant finding and is transmitted by pets to humans, is described as the migration of Toxocara spp. larvas to the internal organs, eye or dermal tissues with pathological damage. Almost in all articles and books the primary factor of the disease is mentioned to be Toxocara canis.
Methods: The most prefered method in serological diagnosis is to detect specific anti-Toxocara antibodies. If these antibodies are found to be positive then it is suggested to use Western Blot technique in the distinctive diagnosis of the illness. In our study, in a study group patients with eosinophilia, we compared ELISA and Western Blot methods which is found to be more specific and sensitive than ELISA.
Results: In 92 sera, 25 of them were evaluated as positive (27.2%), five of them were evaluated as equivocal (5.4%) and finally 62 (65.3%) of them were found to be negative with ELISA method. On the other hand, same 92 sera were studied with Western Blot technique; 45 of them were found to be positive with the molecular weight bands of 24, 28, 30 and 35 kDa and 47 of them were evaluated as negative while no anti-Toxocara antibody specific molecular weight bands were formed on the strips.
Conclusion: According to these data; when eosinophilia having blood counts are encountered, the diagnosis probability of Toxocariasis is very high. Moreover, our study indicated once more that in serological diagnosis of Toxocariasis, Western Blot method is a more valuable method with its high specificity and sensitivity than ELISA method.
| Test results | Serum numbers (n) | % |
|---|---|---|
| ELISA(+) WB(+) | 23 | 25 |
| ELISA(−) WB(+) | 17 | 18.5 |
| ELISA(?) WB(+) | 5 | 5.4 |
| ELISA(+) WB(−) | 2 | 2.2 |
| ELISA(−) WB(−) | 45 | 48.9 |
| Total | 92 | 100 |
P539 Acanthamoeba castellanii is a model for eukaryote-prokaryote interaction and a human pathogen
H. Abd, A. Saeed, B. Advinsson, G. Sandström (Stockholm, SE)
Objectives: A. castellanii is a free-living amoeba inhabiting aquatic environments and isolated from different environments. The aim is to study its features, its interaction with pathogenic bacteria and its detection from samples.
Methods: Cultivation, viable count, PCR and microscopy.
Results: A. castellanii grew tenfold and survived for more than 30 days showing a long survival time compared to survival of macrophages. We found that trophozoites as well as cysts emitted autofluorescence helping in the diagnosis of Acanthamoeba and its viability. It is well known that trophozoites feed on different cells by phagocytosis. We observed that A. castellanii could also take up trophozoite or cyst from its cell cultures. The importance of Acanthamoeba species is their ability to be predators or hosts to different bacteria. Our studies showed that the facultative intracellular bacterium Francisella tularensis multiplied inside vacuoles of A. castellanii while the extracellular bacterium Pseudomonas aeruginosa killed this amoeba by type III secretion system effector's proteins. Surprisingly, Vibrio cholerae that is considered as an extracellular bacterium multiplied in the cytoplasm of A. castellanii and behaved same behaviour of the facultative intracellular bacteria Shigella sonnei and S. dysenteriae.
Several Acanthamoeba species are human pathogens. The trophozoites enter human body through respiratory tract, injured skin, invade the central nervous system to cause granulomatous amoebic encephalitis and colonise the cornea causing amoebic keratitis. As evidence to the increasing importance of Acanthamoeba infections, we diagnosed the first Nordic case of fatal meningoencephalitis and two cases of amoebic keratitis caused by A. castellanii.
Conclusions: Characteristics of A. castellanii such as long life because of encystation as well as excystation, phagocytosis, autofluorescence, resistance to many antibiotics, predator, host and victim to different bacteria, make it an ideal unicellular organism to study the interaction between eukaryotes and prokaryotes and to be used as a powerful tool for the culture of some intracellular bacteria in addition to its increased role as human pathogen.
P540 Cysticercosis: correlation between serological and radiological diagnosis
A. Rodríguez Guardado, E. Gómez, M. Rodríguez Pérez, A. Sempere, R. López-Roger Roger, P. Suárez Leiva, V. Asensi Álvarez, J.A. Carton Sánchez (Oviedo, ES)
Objective: Human infections due to Taenia solium are common in underdeveloped countries but they are not frequent in Europe. We describe our experience in the diagnosis and management of cystecercosis in Asturias, a province in northern Spain.
Methods: We have retrospectively reviewed each patient with cisticercosis positive serological result in the Hospital Universitario Central of Asturias (HUCA), during a period of six years (2000–2006). Serum samples of each patient were tested for Taenia solium antigen, antibodies and vesicular fluid using an ELISA test. Computerised tomography or nuclear magnetic resonance were also done on all patients.
Results: We have studied ten cases. None presented significant underlying diseases. Four of them were Spanish and there were six immigrants, four from Ecuador and two from Brasil. The mean age was 45 years (range of age 34–59). Six were female. Cephalea (three cases), epilepsy and subcutaneous nodules (two cases) were the most frequent symptoms. Two patients remained asymptomatic. Eosinophilia in blood tests was not found in any of them. Faeces of each one were examined looking for parasite eggs and were negative. Nine patients presented Taenia solium antibodies, six were positive for vesicular fluid and one for Taenia solium antigen. Three patients were diagnosed of neurocistycercosis, only one of them was positive to T. solium antigen. In two cases of them the computerised tomography was negative. The nuclear magnetic resonance was positive in the three cases Two patients with neurocysticercosis were treated with albendazol during 30 days and steroids in the first seven days. One patient was treated with praziquantel three doses during one day. All the patients cured. The rest of patients received one doses of praziquantel (5 mg/kg). All patients recovered.
Conclusions: Presence of Taenia solium antibodies in serum seems to be useful for cysticercosis diagnosis. However, a positive antigen result in CSF does not seem to indicate a central nervous system infection. Nuclear magnetic resonance is more useful than computerised tomography in neurocysticercosis diagnosis. Albendazol is a very effective and safe treatment. Clinical and epidemiological results consistent with neurocysticercosis are needed in order to ask for antibody and antigen testing
P541 Clinical forms of neurocysticercosis – Our experience
S. Nikolic, G. Stevanovic, V. Peric, T. Stosic-Opincal (Belgrade, RS)
Cysticercosis is one of the commonest parasitic diseases of the central nervous system and frequent cause of convulsions in endemic regions, where incidence rate is about 4% of population.
Objective: The objective of our study was to analyse the clinical forms of neurocysticercosis in patients treated at our department.
Method: The analysis of results of neuroradiological examinations (CT scanning, MR imaging), serological tests, neurological disorders and treatment of patients with neurocysticercosis.
Results: In the period 2000–2005, a total of 65 patients with neurocysticercosis were treated. All patients underwent neuroradiological examinations: endocranial CT in 40 (62%) cases, endocranial MRI in 17 (26), and both CT and MRI of the endocranium in 8 (12%) patients. Parenchymal disease was found in 50 (78%) patients, ventricular in 8 (12%), and parenchymal and subarachnoid in 6 (9%) cases. Only one female patient had spinal form associated with parenchymal, ventricular and subarachnoid, and she had VP shunt implanted due to hydrocephalus. Out of 50 patients with parenchymal disease, cystic changes were verified in 27 (54%) cases, concurrent cysts and calcifications in 15 (30%), and only calcifications in 8 (16%) cases.
Serological test results were positive in all patients with ventricular and subarachnoid forms as well as in the female patient with spinal cysticercosis. Out of 50 patients with parenchymal disease, 22 (44%) had positive results of serological tests. Clinical manifestation of disease correlated with number, extent and localisation of pathological changes. Medicamentous treatment (Albendazole, Praziquantel) was applied in 46 (81%) out of 57 patients with active disease, while 11 (19%) cases underwent surgical interventions (cyst extirpation, V-P shunt), followed by medicamentous therapy. Patients with calcifications were not treated. The outcome was favourable (decrease of a number of cysts, reduction of cyst size, cyst disappearance) in 45 patients (79%), and unchanged findings were observed in 12 (21%) treated patients. There was no lethal outcome.
Conclusion: Parenchymal neurocysticercosis was most prevalent in our patients (77%), while other forms of disease were significantly more infrequent. Active disease (parenchymal, ventricular, subarachnoid, spinal) was detected in 57 (88%) patients. Outcome was favourable in 45 (79%), and unchanged findings were found in 12 (21%) treated patients. No lethal outcome was reported.
P542 Strongyloidiasis hyperinfection: diagnosis problems and management in an intensive care unit
C. Kummerlen, I. Yalaoui, S. Barbar, H. Rahmani, P. Sauder, M. Hasselmann (Strasbourg, FR)
Introduction: Strongyloidiasis hyperinfection (StHI) results from a fulminant dissemination of strongyloides stercoralis larvae in the organism. It occurs essentially in immunocompromised subjects coming from endemic areas. Acute respiratory failure or severe neurologic disorders may require admission in ICU and population migrations may increase the number of StHI also in countries where S stercoralis is usually not present. A delayed or missed diagnosis may then rise different problems, as illustrated by this case report.
Methods: A 80 years old bosnian man, living in France for 13 years, was admitted in the ICU for acute respiratory failure. He had a chronic obstructive pulmonary disease (COPD) with few exacerbations in the past years, with 2 of them accompanied by hypereosinophilia but no specific etiology was found. He developed StHI as he was treated with steroids. Larvae were fortuitously isolated in tracheal secretion and also found in stool and blood. Despite adequate antihelminthic treatment (ivermectine) and respiratory improvement, the patient died after 4 months from irreversible hypertonia probably due to a neurologic localisation of StHI.
Results: This observation exemplifies 4 key points
-
1
StHI has to be evoked in patients coming from endemic areas, even years ago, specially before immunosuppressive treatment. In Europe, South Balkan, eastern and mediterranean countries are concerned. Early symptoms, as dysphagia and bronchospasm in our patient, related to parasitic migration are evocative. Eosinophilia is an alarming clue when present, but was not found.
-
2
Gram negative pneumonia or septicaemia are often associated and may delay the diagnosis of StHI.
-
3
Microscopic identification of larvae in the stool confirms the diagnosis, but a single examination may be insufficient. A positive serology test in a patient with a compatible history or a stay in an endemic area are sufficient grounds for empirical treatment.
-
4
StHI requires contact isolation and in ICU the risk of transmission is theorically high. In this observation, despite an ignored diagnosis, none transmission to healthcare workers was found. However, one other patient developed eosinophilia with a positive serology.
Conclusion: In temperate non-endemic areas, StHI may be underappreciated. Clinical presentation and risk factors should raise awareness of this curable but potentially fatal parasitic infection and so facilitate early diagnosis and treatment.
P543 Primary muscle hydatidosis of the thigh in a pregnant woman
M. Degioanni, A. Maccabruni, C. Bolla, G. Montrucchio, A. Biglino (Asti, Pavia, IT)
Introduction: Primary skeletal muscle hydatidosis is a rare manifestation of cystic hydatid disease. We report a case of muscle hydatidosis of the thigh observed in a pregnant woman.
Methods and Results: Our patient is a 30 years old moroccan woman who has been immigrated to Italy for seven years ago. At the fourth month of her third pregnancy she observed a painful swelling of the right thigh, without any other symptoms; an ultrasonographic scan of the thigh showed a noncalcified cystic mass with little round cystic inside in the posterior muscle compartment (diameter 13 cm). After delivery contrast CT and MRI of right thigh were performed showing that the mass had reached 15 cm in diameter. No additional cysts were found on CT of the thorax, abdomen and pelvis. As hydatidosis was strongly suspected, a specific serology was performed with positive result (IHA 1:320). Treatment with albendazole at 400 mg b.i.d. was immediately started; two months after the ultrasonographic scan showed that the primary cyst was smaller while all but one “daughter” cysts became smaller and oval, with increased echogenicity. The specific antibody titer (IHA) was 1:80. Surgical resection to remove the cyst “in toto” will be the next step of the treatment.
Conclusion: Skeletal muscle hydatid cysts are generally misdiagnosed as either a pyogenic infection or a malignancy. Hydatid disease should be considered in the differential diagnosis of cystic muscular lesions and specific serology has to be performed because routine diagnostic procedures may not always be helpful. The correct diagnosis at the right time allows a radical cure of the disease
P544 Fifteen-year experience with pulmonary hydatidosis: clinical manifestation, diagnosis and treatment
B. Sharifi-Mood, A. Fazaeli, S. Izadi, S. Mokhtari (Tehran, IR)
Background and Objective: Hydatid disease is a major world health problem and pulmonary hydatidosis is a widespread disease. It is presented with different clinical manifestations. In order to determine the most clinical manifestation, diagnostic tools and clinical outcome in our patients, we conducted this study.
Patients and Methods: Forty-nine patients with pulmonary hydatid cysts who were admitted to our hospital in Zahedan (Southeat of Iran) between 1990 and 2005, evaluated. We retrospectively reviewed the patients' symptomatology, diagnostic studies, treatment options, and morbidity and mortality rate.
Results: The ages of the patients ranged from 16 to 68 years (mean 43 years). Seventy-five percent of patients were from male gender. Hemoptysis was the most common clinical presentation in our patients. Radiological studies were the main diagnostic tool. The correct preoperative diagnosis was made in 92% of the patients by chest roentgenogram plus chest CT-Scan. Eighty nine percent of patients were treated by surgical route. Only one patient was expired during surgery.
Conclusion: Upon the results emerged from this study, hemoptysis is the most prevalent clinical manifestation in patients with pulmonary hydatidosis and it can mimick pulmonary tuberculosis in endemic area.
P545 Praziquantel in prevention of complicated echinococcosis relapses
S. Dautovic, N. Koluder, M. Ferhatovic, N. Mostarac, S. Kapisazovic, H. Tanovic, A. Mesic, R. Gojak (Sarajevo, BA)
Human echinococcosis is a large therapeutical problem in all farmer regions of the world. It is an antropozoonosis usually coincidentally diagnosed by radiological methods. It can affect all organs, but most frequently it affects liver. Even today this disease can cause serious disability, when it is lately diagnosed, disseminated or when relapses occur. When the cyst is small and accessible, total cyst and pericyst excision is the only proper therapeutical procedure. Solitaire cyst can be resolved by PAIR method. In complicated cases combined conservative (albendazol)-surgical treatment is recommended (WHO). Relapses rates are usually between 4 and 37%.
Objectives: To investigate the frequency of postoperative relapses in our material and to investigate praziquantel efficiency in combined conservative-surgical treatment.
Methods: This was an open prospective-retrospective study, started in June, 1996. All patients with complicated or large echinococcus cysts, treated at The Clinic for Infectious Diseases and at The Clinic for Abdominal Surgery, Clinical Centre University of Sarajevo, in the period of 1st June, 1996 to 1st June, 2006, were included. Praziquantel was used in doses of 25–50 mg/kg for 14 days with corticosteroids as an adjuvant therapy. All patients were monitored clinically, so as by radiological and serological tests after one, three, six and than every 12 months.
Results: In this period of time 70 patients were treated by combined conservative-surgical treatment, mean age of 34.7 (4–65). There were 42 patients suffered from primary and 28 patients with echinococcosis relapses. In 53 cases evacuation of large cysts was done and 17 patients had cyst rupture. Our results signify that there were 12.5% of recidives in the group of patients which was treated only by surgical treatment in this period. In the group of patients treated by combined treatment there were no relapses, if praziquantel antihelmintic treatment was used in pre and postoperative period. Using praziquantel 10 days after cyst rupture was not effective.
Conclusions: Human echinococcosis is a large public-health problem in Bosnia and Herzegovina. Postoperative relapses in complicated cases occur at rate of 12.5%. After combined treatment, using praziquantel, there were no cases of relapses. Praziquantel can be an antihelmintic of choice in prevention of complicated echinococcosis relapses in combined treatment.
P546 Surveillance Programme in Echinococcosis and importance in the prophylaxis and therapy of human infection
L. Junie, N. Constantea, P. Ciobanca (Cluj Napoca, RO)
Background: The human Echinococcosis is still a serious problem for the public health, in Romania, despite the measures taken for the prophylaxis of the disease.
Methods: There were assessed patients hospitalised in surgical clinics between 2004–2006. The radiographic examinations, ultrasonography, IDR Cassoni and serological test ELISA, have established the diagnosis.
Results: Most of the hydatid cysts (71.5%) were uncomplicated, but 28.5% are diagnosed due to the complications. The evolution of uncomplicated hepatic hydatid cyst is like a biliary dyspeptic syndrome (58%), with tumour-like signs (80%), a biliary colic (6.3%) and portal hypertension (2.1%). Allergic reactions can occur, especially in children (6.2%). The complications of the hydatid cyst are cholecystitis (8.4%), jaundice (4.9%), and infectious syndrome (4.9%) and rupture (18.8%). The rupture occurs in biliary tracts, in pleura, and in peritoneal cavity, with the occurrence of anaphylactic shock (2.5%). In adults, hydatid biliary tract obstruction (2.8%) and cirrhotic signs (3.8%) may occur. Cysts in the lungs ordinarily are uncomplicated (77%) and give rise to cough (63.6%), dyspnea (54.5%) and chest pains (36.4%). The complications of the pulmonary hydatid cyst are fissure and secondary infection of the cyst with the occurrence of an infectious syndrome with fever (36.4%), thoracic twinge (13.6%), sweats (4.5%), anorexia (18.2%), asthenia (9.1%). Either surgical or drug therapy may conclude in a favourable evolution in 90.5% of cases. The unfavourable evolutions after surgical (9.5%) are due to the relapses (4.5%), the secondary location (1%) and the bacterial complications (3.5%). The echinococcosis is present in adults (46.5%) and children (53.5%), with an alarming high incidence in children of 7 to 14 years old (70.1%). In adults, the most common locations are the liver (83.9%), the lungs (9.7%), and the spleen (1%). In children the most common locations are the liver (67.7%), the lungs (27.6%), the spleen (1.9%), the kidneys (0.9%).
Conclusions: Hepatic and pulmonary locations are frequent, leading by their chronic evolution to tumour-like signs, severe complications (1.5%), reserved prognosis (9.5%), and even to death (0.5%). There is an urge for the early diagnosis of the infected patients, by the development of the screening methods and a close collaboration among clinicians and and laboratory doctors, for therapy and prevention of the complications.
P547 Diagnostic E. granulosus particles in hepatic cystis punctate
B. Matica, J. Granic, A. Beus, B. Desnica, K. Granic (Zagreb, HR)
Objective: Incidence of E. granulosus in Croatia is 0.9/100,000 inhabitants in year 2004, 0.46 in 2005 and 0.46 in 2006 (till 1st of November 2006). This high incidence is a challenge for rapid and good algorithm of diagnostic.
Material and Method: We observed 61 patients with hepatic cysts from 1.1.1999 till 1.11.2006, Patients were from age 13–63 years, female 40, male 21, ratio female:male is 1.9:1. Contents of hydatid cysts obtained after punction under echosonographic visualisation, searched for E. granulosus particles and vitality of protoscolices. Liquid samples after aspiration was divided in two portions: first part was examined under microscopy as native specimen and after centrifugation. In second portion 0.1 ml of canine bile or 0.2% solution of sodium taurocholate was added, incubated 48 hours at 37°C, microscopic examined after 24 and 48 hours.
Results: In 61 contents of hepatic cysts patients serological positive on echinococcosis (ELISA and ITF method) 37 had particles of E. granulosus (60.66%), and 24 were negative (39.34%).
Conclusion: Examination of hepatic hydatid cysts contents evacuated after percutaneous punction, demostrate rapid and valid diagnostic tool that enable clinician to establish correct etiological diagnosis and/or to evaluate results of the various therapeutic algorythm.
PAIR method (punction, aspiration, installation, repunction) is a good diagnostic method. Evacuation of hepatic cysts contents is also a good therapeutic procedure.
P548 Current trends in echinococcosis in Latvia
J. Keiss, V. Sondore, A. Cernusenko, L. Viksna, B. Rozentale (Riga, LV)
The objective of study was to analyse echinococcosis cases treated in the Infectology Center of Latvia (ICL) in 1999–2005, to evaluate echinococcosis tendencies in Latvia during this period and at present and to define arising problems to be solved.
Patients and Methods: Retrospective surveys of hospital records covering years 1999–2005 have established the 32 patients (3 – men, 29 – women) with echinococcosis: in 1999 – 1, 2000 – 3, 2001 – 0, 2002 – 7, 2003 – 7, 2004 – 6, 2005 – 8 patients. All patients were rural population, all had dogs and domestic animals and they had not left their residence during the last 20 years. Exclusion is one patient who lived in Kazakhstan and recently moved to Latvia. Diagnosis was based on Echinococcus granulosus and Echinococcus sp. antibodies detection, in all cases liver ultrasonography and computer tomography (CT) was performed, in some cases morphological investigation of biopsy or surgically obtained material from space occupying lesions in the liver also was carried out.
Results: Latvia belongs to countries with a medium echinococcosis rate, predominantly with local source of infection. Before the year 2001 a very few cases were registered, but after the year 2001 there is a considerable increase in the number of Echinococcus infection cases, especially during January-October, 2006, when new 14 echinococcosis patients were treated at the ICL. We suppose the number of invaded persons is more large. Echinococcosis shows a tendency to distribute from the western and central to the eastern regions of Latvia. In 3 of 32 patients alveolar echinococcosis was diagnosed.
Clinically, most of patients had a discomfort in epigastrium, in 5 patients the first clinical symptom was itch with the following jaundice. In 6 patients the focal liver lesions were found accidentally during the abdominal ultrasonography. In dependence on family doctors' understanding of disease the further way of patients was different: 7 patients were forwarded to the surgical clinic, 3 – to oncological clinic and others – to the ICL. All patients were treated with albendazole. One patient died.
Conclusion: 1) Echinococcosis has a tendency to increase in Latvia. 2) Family doctors are often not ready to early diagnostics of echinococcosis. 3) Cooperation with veterinarians and veterinary care need remarkable improvement. 4) In all cases of focal affection in the liver the serological and/or morphological tests for echinococcosis should be performed.
P549 Epidemiological considerations about hydatid disease in Constanta county, Romania
I. Dumitru, S. Rugina, E. Dumitru (Constanta, RO)
Introduction: Hydated disease represent a public health problem in Constanta county because of its high prevalence and multiple health and social implications. The high number of domestic animals (sheep, cattles, goats) and communitarian dogs in this region are the most important causes of this situation.
Objective: To analyse the epidemiology of hydated disease in Constanta county.
Materials and Methods: We performed a study on 340 patients admitted in our department of parasitology in the last 5 years. The diagnosis was based on ELISA reaction for Echinococcus granulosus, chest x-ray, ultrasonography and computed tomography.
Results: The prevalence of hydated disease in our region was high, more than 50 cases/100.000 inhabitants, mainly in urban area (72%). The prevalence was higher in women (57%), the most affected group age was between 50 and 59 years with the highest age at diagnosis beeing 82 years. According to race, the highest prevalence was among romanians, followed by aromanians, turks, tatars and rroms. The epidemiolgical conditions were positive in the majority of cases, most of the patients documented the contact with animals. There were nine familial clusters. Echinococcosis had different locations, involving liver (261), lung (87), spleen (7), kidney (6), brain (2), pericardial (2), mediastinal (1), peritoneum (11) and retro peritoneum (3), muscles (5), bone (2), and parotid glands (1). Secondary hydatidosis was reported in 22.18% cases and multiple cysts were found in 35% cases with liver, pulmonary, muscular, bone, peritoneal and parotid glands hidatidosis and multiple locations in 17% cases. Regarding the treatment, 70% of cases were treated with albendazole, 29% with surgery followed by albendazole and 1% with percutaneous ultrasonography-quided puncture (PAIR).
Conclusions: The prevalence of hydated disease is high in our region. This disease require a particularly attention and health programmes for a better management
P550 A comparative study for the determination of IgG avidity during a toxoplasmosis infection in pregnancy
U. Krispenz, D. Wassenberg, W. Becker, K. Pfrepper (Neuried, Hamburg, DE)
Objectives: The comparison was made to find out if there is a significant advantage for the interpretation of the time of infection during a toxoplasmosis in pregnancy. An antigen specific line assay was used to go further into the question if a conventional EIA using an avidity index is sufficient for the determination of the time of infection or is it possible to obtain a significant improvement by using an antigen specific avidity pattern.
Methods: 180 routine samples from pregnant women were tested positive with Abbott screening Toxo IgG and IgM and confirmed by bioMérieux (BM) IgG and IgM plus avidity. The samples were tested additionally with MIKROGEN recomWell Toxoplasmosis IgG and IgM as well as with recomLine Toxoplasmosis IgG (plus avidity) and IgM (LTG). According to individual band and avidity patterns, the recombinant line assay allows the differentiation into four phases: phase 1 (acute infection, 0–3 months p.i.), phase 2 (acute infecition, 3–6 m.p.i.), phase 3 (subacute infection, 6–12 m.p.i.), and in phase 4 (latent infection, >12 m.p.i.).
Results: In the BM avidity test 120 samples were tested with low avidity. Running these samples with the antigen specific avidity (LTG) the differentiation between an acute infection or a past infection can be made more precisely: 75/120 samples were in the phase 1, 14/120 in phase 2, 19/120 in phase 3, 1/120 sample was in phase 4 and for 9/120 samples it was not possible to determine the time of infection due to an abnormal serological pattern. With the BM assay 17 samples got an intermediate avidity. In contrast to these 17 indeterminable samples only one sample could not be determined with LTG due to an abnormal serological pattern; 6/17 samples were in the acute phase 1, 7/17 in phase 2, 2/17 in phase 3 and 1/17 sample in phase 4. A high avidity was obtained for 43 samples with the BM avidity test.
Running these samples with the LTG 2/43 were in phase 1, 7/43 in phase 2, 14/43 in phase 3, 16/43 in phase 4 and for 4/43 it was not possible to determine the time of infection due to an abnormal serological pattern.
Conclusion: The BM assay turns the user's attention only to a low and high avidity index. In contrast the data revealed that the recomLine Toxoplasma enables a more precise differentiation of a Toxoplasma infection into four specific phases. This shows definitely the advantages of a confirmation assay using recombinant antigens in a line format especially for the avidity testing.
P551 Delayed maturation of toxoplasma immunoglobulin G avidity in pregnant women: impact of spiramycin treatment and gestational age
F. Peyron, M. Lefevre-Pettazzoni, M. Wallon, F. Cozon, A. Bissery, M. Rabilloud (Lyon, FR)
Objectives: The low avidity of immunoglobulin G (IgG) has been reported to be a useful marker of recent infection with Toxoplasma. Nevertheless, discrepant results on the maturation of avidity over time have been reported. The aim of this study was to investigate the maturation of IgG avidity after Toxoplasma seroconversion during pregnancy and to determine factors that could influence its evolution over time.
Methods: IgG avidity was studied retrospectively in our department data base and in 309 sera from 117 pregnant women who seroconverted during pregnancy. Inter-patient variations in the evolution of IgG avidity and factors that potentially influence maturation, such as gestational age at infection and treatment with spiramycin, were investigated.
Results: Persistent low avidity was found in some patients even after a median follow-up period of 6 years. Avidity index of IgG was significantly heterogeneous and ranged from 7.8 to 35.3% for 95% of the women 12 weeks after infection (p < 0.05). When plotted against time after logarithmic transformation, evolution of the avidity index displayed heterogeneous patterns with slopes between −0.017 and 0.051 for 95% of the women (p = 0.011). Maturation of avidity decreased when gestational age at infection increased (p = 0.03) and increased when the delay between infection and onset of treatment increased (p = 0.0003). In conclusion, we demonstrated that maturation of Toxoplasma IgG avidity in pregnant women is highly variable and that persistent low avidity can be observed. Avidity evolution over time is influenced by gestational age at maternal infection and delay in the onset of treatment. The results of this study clearly demonstrate that, in a pregnant woman, an acute toxoplasmic infection cannot be reliably diagnosed solely on the basis of low avidity of immunoglobulin G.
P552 Improved performance of the automated toxoplasmosis IgG, IgM & IgG avidity assays on the Abbott ARCHITECT Instrument
J. Schultess, E. Sickinger, J. Dhein, M. Hausmann, D. Smith, E. Frias, M. Palafox, J. Prostko, D. Pucci, Reno Stricker, Reto Stricker, P. Thulliez, H. Braun (Wiesbaden, DE; Abbott Park, US; Geneva, CH; Paris, FR)
Objectives: Optimisation of IgG, IgM & IgG Avidity immunoassays as a complete screening panel for the acute infection and immunity status to Toxoplasma gondii.
Methods: The Toxo-IgG assay uses recombinant Toxoplasma antigens (P30 and P35) coupled on the solid phase and a labeled anti-human IgG antibody. The Toxo-IgM assay employs a μ-capture format with an anti-human IgM mouse monoclonal antibody bound to the solid phase and tachyzoite lysate complexed with a labeled Anti-P30 antibody as tracer. The Toxo IgG Avidity assay applies a comparative assay format suppressing high avidity IgG antibodies with soluble P30 antigen in an indirect anti-human IgG format. The avidity assay is run with automated, sample specific dilution into a defined concentration range.
The specificity of the Toxo assay panel to detect primary Toxoplasma infections and to detect the correct immune status was determined by testing 988 samples from different populations (518 blood donors, 220 hospitalised patients and 250 pregnant women. 165 seroconversion panels were used to explore sensitivity. For resolution of discordant results, a set of additional Toxo assays was tested, including the respective AxSYM assays.
Results: In early seroconversion, the Architect Toxo IgG ranked equivalent to AxSYM and more sensitive than other comparator assays. It showed an excellent ability to detect past infection low level IgG's, equivalent or better than comparator assays, paired with high specificity to detect immunity even on challenging specimens. The Toxo IgM assay was found to have a relative specificity ranging from 99.7 to 100%, for populations of random blood donors, hospital patients or pregnant females. Its sensitivity to detect seroconversion in acute infection was equivalent to AxSYM Toxo-M. The avidity assay found no specimen drawn less than 4 months after infection as high avidity and showed faster kinetics to higher avidity.
Conclusion: The fully automated Toxoplasmosis panel for the Architect instrument is, in terms of sensitivity and specificity values, comparable to the reference assays. In addition, a fully automated algorithm allows the evaluation of Toxo G and Toxo IgM positive results by Toxo IgG Avidity, to rule out acute infection with Toxoplasma in pregnant women.
P553 Evaluation of the new Vidia® toxoplasmosis IgG and IgM assays in women of childbearing age
J. Zufferey, C. Di Mito, R. Auckenthaler (Lausanne, Geneva, CH)
Objectives: The aim of the present study was to evaluate the performance of the new VIDIA® Toxoplasmosis IgG and IgM assays (bioMérieux, France), using the VIDIA® system easy to use with a high level of traceability, with clinical specimens prospectively collected in women of childbearing age.
Methods: A total of 1000 fresh serum samples consecutively obtained from 1000 women aged 18–44 years (median 30) during a period of 4 weeks were tested with ADVIA® Centaur” Toxoplasma IgG and IgM assays according to routine conditions. Serum aliquots stored at + 4°C were blindly rechecked within 24 hours with the VIDIA system for the same parameters. Discrepancies of results between both methods were resolved considering avidity test, direct agglutination (Toxo-Screen DA, bioMérieux), ISAGA (bioMérieux) and when available, the analysis of previous drawn serum samples.
Results: Among the 1000 women screened with VIDIA, 89.4% had non detectable IgG and IgM (T. gondii seronegative) and 8.2% had a pattern of past acquired infection (positive IgG and no detectable IgM). Positive IgM were detected in 1.1% of them with VIDIA system versus 2% with ADVIA Centaur. Equivocal rate was 0.9% for VIDIA TOXO IgG and 0.5% for VIDIA TOXO IgM (versus 0.9% and 1.1%, respectively for the compared method).
For VIDIA TOXO IgG the relative sensitivity and specificity were 96.7% and 99.7%, respectively. After the resolution of discrepancies, the sensitivity as well as the specificity was 100%.
For VIDIA TOXO IgM the initial relative sensitivity and specificity were 64.7% and 100%, respectively. In fact, 4 of the 6 negative samples with VIDIA and positive with the compared method were found with high avidity index and the remaining two samples were negative with the reference test (ISAGA-IgM). Taking this into account, the absolute sensitivity was found 100%.
Conclusion: The two evaluated assays VIDIA TOXO IgG and TOXO IgM have shown an excellent sensitivity and specificity and are well adapted to the routine screening of toxoplasmosis in pregnant women.
P554 IgG IgM Western Blot in early diagnosis of congenital toxoplasmosis
V. Meroni, F. Genco, L. Piccoli, A. Grosso, P. Lanzarini, L. Bollani, M. Stronati (Pavia, IT)
Introduction: Toxoplasmosis is one of the most frequent congenital infections. Because congenital Toxoplasma infection does not usually produce recognizable signs of infection at birth, most infected newborns are undetected by routine clinical examinations and remain untreated. But serious clinical sequelae such as chorioretinitis can develope later. So the infected children must be identified and treated as early as possible. The aim of this study was to evaluate diagnostic accuracy of IgG IgM Western-blot (IgG IgM-WB LDBIO Lyon France) on 224 newborns at risk of congenital toxoplasmosis.
Methods: 224 neonates born from mother with suspected or certain infection in pregnancy were evaluated retrospectively with IgG IgM WB (LDBIO Lyon France). Serum obtained from all the newborns at birth was compared with maternal sample and then with sample obtained monthly during their first three months of life. Furthermore all the sample were analysed with routine assays: ELISA IgG IgM, IgA(Diasorin Saluggia Italy), IgG ELFA, IgM ISAGA (bioMérieux Marcy L'Etoile France). The patients were tested with all these routine assays monthly until seronegative and then at one year of age.
Results: At the end of the study 40 newborns were found infected. Thirty were diagnosed at birth by the presence of IgM and/or IgA, in the other 10 diagnosis was made by antibody rebound or by IgG positivity at one year of age.
Conclusions: IgG IgM Western blot showed a specificity (96.7) almost superimposable to the traditional tests (98.9). Sensitivity (95) was higher than the traditional tests (75) and the difference was statistically significative (P = 0.028 Yates Corrected χ squared). WB let us find out 8 infected newborns not detectable with traditional tests that could undergone an early treatment, while 178 not infected newborns avoided unnecessary therapy.
Table 1.
| IgM ISAGA+IgA ELISA |
IgG IGM Western blot |
|||||
|---|---|---|---|---|---|---|
| Pos | Neg | Tot | Pos | Neg | Tot | |
| Infected | 30 | 2 | 32 | 38 | 6 | 44 |
| Not infected | 10 | 182 | 192 | 2 | 178 | 180 |
| Tot | 40 | 184 | 224 | 40 | 184 | 224 |
| Sens. 75.0 | Sens. 95.0 | |||||
| (95% CI: 58.8–87.3) | (95% CI: 83.1–99.4)* | |||||
| Spec. 98.9 | Spec. 96.7 | |||||
| (95% CI: 96.1–99.3) | (95% CI: 93.0–98.8) | |||||
Sens., sensitivity; Spec., specificity.
P555 Prevalence of Chagas' disease in pregnant women from Southamerica in Valencia (Spain)
A. Gil Brusola, M.J. Gimenez, Y. García, M.D. Gomez, M. Gobernado (Valencia, ES)
Objectives: Chagas' disease causes high morbidity in some countries in Southamerica. In Europe, where the triatomine vector is not found, its way of transmission can be either mother-to-child or through organ transplant or blood transfusion. The aim of this study was to determine the seroprevalence of this infection in immigrants from Southamerica to our city.
Methods: 354 sera of pregnant women from Southamerica who attended our hospital between September 2003 and September 2005 were tested for anti-Trypanosoma cruzi antibodies (IgG) using an enzyme-linked immunosorbent assay (ELISA). Positive sera were then confirmed with either another ELISA, a particle gel immunoassay or an immunofluorescent assay (IFA).
Results: The mean age of this group was 28. Most of them came from Ecuador (51%), Colombia (20%) or Bolivia (17%). Infection was confirmed in nine women (2.5%). Their mean age was 29. All of them were from Bolivia, resulting in a prevalence of 15% for women from Bolivia. Analysis of the vertical transmission couldn't be conducted due to either miscarriage or lack of sera from the newborns.
Conclusions: Prevalence of Chagas' infection is high in pregnant women from Bolivia. Screening should be carried out in this group of patients before delivery, for follow-up of their children in order to determine the relevance of the disease, and to reduce the risk of transmission in case of organ transplant or blood transfusion.
P556 Optimisation of a flow cytometry protocol for detection of Cryptosporidium parvum in hospital tap water and human stools
J. Barbosa, S. Costa-de-Oliveira, A. Gonçalves Rodrigues, C. Pina-Vaz (Porto, PT)
Cryptosporidium parvum is a waterborne agent, causing diarrhoea in immunocompromised patients. Its diagnosis is based upon microscopic detection of oocysts in faeces following alcohol-acid staining. This conventional procedure presents low sensibility and specificity, being highly dependent of the observe expertise. Our main objective was to optimise a specific Flow Cytometric (FC) protocol for detection of C. parvum in water and stools and to establish its sensibility limit. C. parvum oocysts (Waterborne Inc, USA) were used for protocol optimisation. FC analysis was performed using oocyst suspensions stained with different concentrations of a specific monoclonal antibody conjugated with R-phycoerytrin (Crypt-a-Glo, Waterbone). Serial concentrations (2×105, 2×104, 2×103, 2×102 oocysts/mL) were later stained with an optimised antibody concentration and analysed by FC. Specificity and the sensibility limit of the method were established using both prokaryotic (Escherichia coli, Sthaphylococcus aureus) and eukaryotic microorganisms (Candida albicans, Giardia lamblia cysts). FC analysis was repeated according of the optimised conditions, using human stools and hospital tap water, simulating clinical and environmental settings. Several procedures were also assayed to reduce the loss of oocysts and improve its detection: different filters (gauze, paper filters), centrifugation time and velocity, and distinct flotation solutions (NaCl, ZnSO4.7H2O). As the antibody concentration decreased, a decline of peak intensity was registered. The optimal concentration of antibody was 3.0 μg/mL for 105 oocysts/mL. We established a threshold of detection of 2×103 oocysts/mL. The staining procedure was shown to be specific, no cross-reaction occurring with bacteria, fungi or parasites. However, a decrease in staining was seen especially when Giardia was present, but it did not interfere with the result. Bellow threshold limit, fluorescence was not enough to allow the discrimination of oocysts. Interference of debris was more frequently observed in faeces than in water samples. A prolonged incubation of faeces in a ZnSO4 solution followed by centrifugation for 10 minutes allowed a clear separation of oocysts from debris. With the use of specific antibodies, a distinct cellular population corresponding to oocysts could be represented in the FC histogram. This study describes the first optimised FC protocol for detection of C. parvum in hospital tap water and human faeces.
P557 A multiplex microsphere-based assay for the simultaneous detection of C. parvum, E. histolytica and G. lamblia antigen in human faecal sample
A. Huwe, X. Su, S. Cox, H. Truong, P. Nguyen, E. Laderman, J. Groen (Cypress, US)
Background: Cryptosporidium, Entamoeba, and Giardia are protozoan parasites that infect the gastrointestinal tract of animals and humans by oral-faecal transmission or through contaminated water sources. Infections may be asymptomatic or may cause a range of symptoms including diarrhoea, fever, and vomiting. Immunocompromised individuals are often unable to clear the parasites from their systems and ultimately suffer severe illness and possible death. Current methods of diagnosis include microscopy (O&P), and ELISA; these “single-plex” methods prove to be labour intensive, and require special skills in addition to other limitations. A high performance, multiplexed assay with the ability to simultaneously detect the presence of all three parasites in a single faecal sample is highly desirable. This study describes the development of the Plexus” Parasitic Multi-Analyte Diagnostics assay based on the Luminex xMAP” system (Austin, TX).
Methods: Samples: A retrospective panel of 248 human stool samples submitted for parasite testing was collected. In addition, stool samples, and culture supernatants of common enteric pathogens were collected for cross-reactivity testing. PLEXUS” Parasitic Multi-Analyte Multiplex Assay: Monoclonal antibodies specific for each parasite were covalently linked to microspheres. The capture microspheres were mixed with extracted human faecal samples, washed, and incubated with polyclonal detection antibodies specific for each parasite antigen. Finally, the microspheres were incubated with fluorescent Phycoerythrin (PE) conjugate. The median fluorescence intensity of PE measured by the Luminex 100 System indicates the amount of antigen captured. Reference Assay: Samples were tested by microscopy or with the TechLab” Cryptosporidium II, E. histolytica II, and Giardia II ELISA systems per the manufacturer's suggested protocol.
Results: The sensitivity of the PLEXUS Parasitic Panel compared to ELISA was 100% (62/62) for C. parvum, 95.5% (21/22) for E. histolytica, and 98.4% for G. lamblia. The specificity was determined to be 100% (176/176) for all three parasites. In addition, the system does not display cross-reactivity with any of the common enteric pathogens, bacteria, or viruses.
Conclusion: The multiplex capability of the PLEXUS” Parasitic Multi-Analyte Diagnostics system offers a high performance, time-saving alternative to microscopy and ELISA for the qualitative detection of C. parvum, E. histolytica and G. lamblia.
P558 Evaluation of five commercial screening assays and two commercial immunoblots for the serological diagnosis of Lyme borreliosis
C. Matthys, E. Petrens, E. Padalko, K. Lagrou, L. Van Renterghem (Ghent, Leuven, BE)
Objectives: Lyme borreliosis (LB) caused by Borrelia burgorferi sensu lato is the most common vector-born disease in North America and Europe. According to guidelines serological diagnosis of LB should follow a two-step procedure: a screeningtest and if reactive followed by an immunoblot. A large variety of commercial assay is available for the serological diagnosis of LB. We evaluated the performance of five commercial screening assays for the detection of both IgM and IgG antibodies to B. burgdorferi as well as of two commercial immunoblots used as confirmation of serological LB diagnosis in clinical cases of LB.
Methods: The assays tested were Quick ELISA C6 (Immunetics), VIDAS Lyme (BioMérieux), Enzygnost Borreliosis IgM and IgG (Dade Behring), Enzygnost Lyme link VlsE/IgG (Dade Behring) and Euroimmun Anti-Borrelia IgM and IgG (Euroimmun). The testpanel (n = 73) included sera of patients with a documented clinical history of LB and was divided into 4 groups according to the symptoms: erythema migrans (EM), facialis parese (FP), neuroborreliosis (NB) and other symptoms (OS). All LB patients' sera were also tested by two different immunoblot (WB) assays: the Mikrogen recomblot Borrelia (Mikrogen) and the Euroline-Western Blot (Euroimmun). Additionally, specificities of the screening assays were evaluated by using 50 possible cross-reactive sera.
Results: The sensitivities obtained showed that all screening assays (total antibodies or combination of IgM and IgG assay), provided comparable results for all 4 clinical groups ranging from 94.5% (VIDAS Lyme) to 98.6% (Quick ELISA C6). The lowest sensitivities were observed for the group EM. The specificities ranged from 66% (VIDAS Lyme) to 96% (Quick ELISA C6). The lack of specificity seen with the VIDAS assay was mainly caused by cross-reactivity with samples from patients with active syphilis. The Quick ELISA C6 provided both the best positive (97.3%) and negative (98.0%) predictive value. Considering the confirmatory tests, the Euroline WB IgG achieved the highest sensitivities for all groups, while the Mikrogen recomblot IgM showed a better sensitivity than the Euroline WB IgM. When combining WB IgG and WB IgM, however, both WB had similar sensitivities.
Conclusion: The results obtained for all groups of clinical LB indicate that the Quick ELISA assay performs better than the other 4 screening assays. Sensitivities of Mikrogen and Euroimmun immunoblots were similar when combining both IgM and IgG results.
Extended-spectrum β-lactamases/metallo-β-lactamases
P559 Epidemiology of infections caused by extended-spectrum β-lactamases producing Escherichia coli for a three-year period
MM. Gallardo, M.V. García, R. Rodríguez, F. Ropero, E. Granados, A. Gutierrez, A. Pinedo (Málaga, ES)
Objectives: For the last years we are being witness of an increase of ESBLs productive strain isolations, making more difficult managing carrier patients. Our aim is to get to know the clinical and epidemiologic features of the ESBLs producing E. coli in the population attending our hospital.
Methods: A retrospective study was carried out from January 2003 to December 2005, where a total of 8623 E. coli strains were isolated. Antimicrobial suceptibilities were carried out by the MicroScan Walkaway® automised system and by disk difussion test and E-test. Double-disk synergy test and E-test were used for screening ESBLs. The results interpretation were according NCCLS guidelines.
Results: Out of the total number of isolations obtained, the percentage of ESBLs strains was 4.5% (391) of which 59 (15.1%) were isolated during the year 2003, 81 (20.7%) during 2004 and 351 (64.2%) during 2005. In 71.1% of the cases the origin was extrahospitalarian. Urine was the sample where it was isolated more frequently in 67.5% of the cases followed by exudates (18.2%)and blood (8.4%). The 391 ESBL E. coli strains belonged to 391 patients 247 (63.1%) women and 144 (36.9%) men with an average age of 61.8 years old (range: 1–98). Hospitalised cases were 28.9% (113) and most of them in the internal medicine department (45.5%), followed by the surgical department (30%) and Intensive Care Units (24.5%). Progress of the hospitalised patients went to resolution in 71.4% of the cases being exitus in 28.5% of them. In reference to sensitivity, co-resistance was detected with quinolones, fosfomycin, trimethoprim-sulfamethoxazole (TSX) and aminoglycosides, observing that the co-resistance more frequently found was with quinolones 71.1% followed by TSX 53.2%, aminoglycosides 20.7% and fosfomycin 4.9%. Only 76 (19.4%) did not show co-resistance with other groups of antibiotics. The co-resistance combination more frequently found was ESBLs more resistant to quinolones and TSX in 46% of the cases.
Conclusions: An important increase of ESBLs isolations have been proven from the year 2003–2004 to the year 2005. Most of the strains were of extrahospitalarian origin. A high level of co-resistance has been detected among ESBLs strains, where quinolones are the antibiotic family mostly affected.
P560 A pseudo-outbreak of a Citrobacter with an extended-spectrum β-lactamase in a haemato-oncology ward
L. Jansen, J. Bakkers, M. Wulf (Nijmegen, NL)
Objective: To investigate a sudden increase of Citrobacter species, which were reported to have an extended spectrum β-lactamase (ESBL) on a haemato-oncology ward.
Design: A case-series of 16 patients colonised with a Citrobacter ESBL between January and December 2005 were investigated. Strains were typed by pulsed field gel electrophoresis. On the 22 strains identified by the automated Phoenix system as a possible ESBL, a disk diffusion test with cefpodoxim, ceftazidim, cefotaxim combined with clavulanic acid was performed and ESBL Etest strips were used to confirm the ESBL phenotype. In addition molecular detection of TEM, SHV, GES and CTX-M was done.
Results: Of two 22 strains the Phoenix System indicated the resistance pattern as ‘possible ESBL', 9 strains had a phenotype on disk diffusion and E-tests consistent with this result. Of these 9 strains 4 (44%) could be confirmed by PCR as ESBL positive: 2 strains with SHV, 2 with TEM-26. 3 strains with PCR positive did had a negative disk-diffusion test: 2 strains from one patient carried a GES, one a CTX-M.
Conclusions: The increase of reported Citrobacter ESBL was partly due to a change in laboratory practice where all strains indicated by the Phoenix to have a resistance pattern indicative of ESBL were reported as such. The ‘outbreak’ was not due to a single strain and different genes coding for ESBL were found. 4 of 9 (44%) strains with a susceptibility pattern indicative of ESBL could be confirmed by molecular techniques, whereas 3 strains were found to contain an ESBL that did not have a typical resistance pattern. Detection of ESBL in Gram-negative bacteria other than E. coli and Klebsiella species remains difficult and results should be interpreted with caution.
P561 Identification of a Pseudomonas putida isolate harbouring blaVIM metallo-beta lactamase gene on the hands of an intensive care unit healthcare worker
A. Bisiklis, K. Manolopoulos, K. Tomos, E. Kazakos, I. Isticoglou, S. Alexiou-Daniel (Thessaloniki, GR)
Objectives: We sought to investigate the microbial flora of the hands of healthcare personnel working in Intensive Care Units in order to identify potential dissemination of carbapenemase producing-Pseudomonas spp. during routine care.
Materials and Methods: Cotton swabs were used to obtain surveillance cultures from the hands of 50 healthcare workers attached to Intensive Care Units. Each culture event was performed twice – at the beginning and at the end of their shift. Swabs were placed directly into Mueller–Hinton broth, incubated for 24 hours at 37°C in ambient air and subsequently subcultured onto blood and Mc Conkey agars followed by 18 to 24 hour-incubation under similar conditions. Initial evaluation of growth was performed by means of Gram stain for each unique colony morphotype present on cultures. Definite bacterial identification and antimicrobial susceptibility testing was achieved with the VITEK 2 identification system. Resistance to imipenem and meropenem was further confirmed by the Etest assay according to the manufacturers' instructions and data were interpreted by applying the CLSI criteria. Metallo-β-lactamase (MBL) phenotypes among resistant strains were detected with the use of Etest MBL strips. Carbapenemase-resistant isolates were subjected to PCR analysis to confirm the presence of blaVIM and blaIMP genes.
Results: During the study, two P. aeruginosa, one P. oryzihabitans and four P. putida non-duplicate isolates were recovered. Among these, one Ps. putida strain was characterised phenotypically as metallo-β-lactamase producer. PCR analysis of the latter with primers for blaVIM genes yielded a 261 bp amplicon, indicating the presence of a blaVIM allele. Interestingly, antimicrobial susceptibility testing revealed one P. aeruginosa strain that showed resistance to imipenem (MIC >32 ug/mL) and intermediate susceptibility to meropenem (MIC = 8 ug/mL), however MBL production could not be established by both conventional and molecular assays.
Conclusion: Previous reports have demonstrated the existence of blaVIM genes in Gram-negative bacilli isolated from various contaminated sites – inanimate objects or intact patient skin surfaces – within hospital facilities. Our study provides further evidence regarding the role of healthcare personnel in multidrug-resistant strains transmission and underlines the need for continuous epidemiological surveillance, in order to assess potential reservoirs, and implementation of preventive policies.
P562 Characterisation of an E. coli strain producer of metallo-β-lactamase in a paediatric hospital
R. Gómez-Gil, A. Barrios, J. Mingorance, A. Gutierrez, C. Fernández (La Paz, ES)
Objective: Characterisation of an E. coli strain producer of metallo-β-lactamase (MLBs) isolated in hemocultive and urocultives from a patient with acute pielonefritis.
Material and Methods: Study of 3 isolates of E. coli from a patient of 20 days of age with acute pielonefritis, obtained first in a hemocultive and urocultive and the third in a new urocultive 4 months later. The identification and sensitivity analyses were done using Wider panels MIC/ID Gram negatives (Soria Melguizo, S.A.®) and VITEK2 cards GN and AST-No22 (bioMérieux®). Sensitivity to aztreonam was determined by Etest.
The MLBs phenotype was settled down with EDTA-imipenem/imipenem Etest and diffusion in Mueler-Hinton agar with an imipenem and EDTA containing disks and with Macfarland 5 inocules. The presence of extended-spectrum β-lactamase (ESBL) was descarded by Etest of cefotaxime/cefotaxime-clavulánic, ceftazidime/ceftazidime-clavulánic and cefepime/cefepime-clavulanic. Finally the molecular characterisation was made by ERIC-PCR, and PCR using specific primers (blaVIM, blaIMP)
Results: The 3 isolates corresponded to a same strain of E. coli and had an identical antibiotype. The strain presented resistance to all the β-lactamics, except aztreonam with CMI of 0.12 μg/mL. The CMI of imipenem was of 2 μg/mL. The Etest of imipenem/imipenem-EDTA presented a clear inhibition of the metaloenzime over more than 4 dilutions. In addition, there was resistance to gentamicin, tobramicin and trimethoprim-sulfametoxazole. Analyses by PCR showed the presence of a β-lactamase of the VIM family.
Conclusions: In all the isolates of enterobacteriaceae with CMI to imipenem greater of 1 μg/mL, the MLBs should be discarded even if they seem sensible in antibiogram with CLSI cut offs
-
–
This is the first description in Spain of metaloenzyme of E. coli in children.
-
–
The association of sensitivity and the expression of the phenotype using imipenem/imipenemEDTA with high inocules allows the phenotypical characterisation of the enzyme
P563 Beta-lactam resistance mechanisms in clinical isolates of Proteus spp. in Portugal: plasmid-mediated inhibitor resistant TEM and extended-spectrum β-lactamases
V. Manageiro, N. Mendonça, D. Louro, E. Ferreira, M. Caniç a on behalf of the Antimicrobial Resistance Surveillance Program in Portugal (ARSIP)
Objectives: The aim of this study was to evaluate β-lactamase mechanisms responsible for β-lactam resistance in clinical strains of Proteus spp. isolated from hospitals covering several regions of Portugal. The clonal diversity of amoxicillin-resistant (AML-R) isolates o f P. mirabilis was also analysed.
Methods: During a 6 months period (January to June 1999), 460 consecutive nonduplicate clinical strains of Proteus spp. (429 P. mirabilis, 29 P. vulgaris and 2 P. penneri) were isolated from patients in 16 Portuguese Hospitals and Public Health Institutions. Antibiotic susceptibility tests were performed according to NCCLS and French Society of Microbiology guidelines. ESBL Etest confirmed ESBL production. Isoelectric points of β-lactamases were estimated by isoelectrofocusing. Beta-lactamase resistant genes were searched by PCR-multiplex method and blaTEM genes were identified by nucleotide sequencing, using specific primers. The genetic relatedness among P. mirabilis TEM-producers (n = 42) was investigated by pulsed-field gel electrophoresis (PFGE).
Results: AML resistance was observed in 35% of isolates (135 P. mirabilis, 27 P. vulgaris and 1 P. penneri). AML-R P. mirabilis strains were divided in 4 groups: non-TEM producer strains (11%), and penicillinase (86%), IRT-type (2%) and ESBL (1%) producer strains. The coding region of blaTEM genes identified in P. mirabilis revealed that 76% were parental genes (blaTEM-1A, blaTEM-1B, blaTEM-1C, blaTEM-1F or blaTEM-1G), 1% was blaESBL gene (blaTEM-10B), 3% blaIRT-19 genes (blaTEM-74F) and 18% were non-ESBL blaTEM genes. A new gene, named blaTEM-156A, was detected in 3% of P. mirabilis strains. Five different promoter regions were identified among blaTEM genes. Thirty-two PFGE profile types were identified: 26 included clones genetically unrelated and the remaining 6 included clones related/closely related or undistinguishable (with >80% and 100% similarity, respectively).
Conclusion: TEM-1 was the main β-lactamase produced by Proteus spp. associated with the weak promoter P3 in the respective coding gene. TEM-10 enzyme was responsible for the ESBL phenotype and TEM-74 (IRT-19) for the IRT phenotype in P. mirabilis. Antimicrobial resistance to cefuroxime and narrow-spectrum cephalosporins in P. vulgaris and P. penneri were associated with the hyperproduction of cefuroximases. This study emphasizes the need to carefully prescribe β-lactams in infections due to P. mirabilis.
P564 Prevalence of confirmed ESBL-production among European Enterobacteriaceae: a ten-year report from the SENTRY Antimicrobial Surveillance Program
L. Deshpande, R. Jones, H. Sader, T. Fritsche (North Liberty, US)
Objectives: To describe the 10-year trend among European (EUR) Enterobacteriaceae (ENT) isolates displaying phenotypic characteristics of extended-spectrum β-lactamases (ESBL). The global emergence of ESBLs has compromised the activity of penicillins, third- and fourth-generation cephems and aztreonam (ATM) as empiric agents when treating infections caused by ENT. Understanding of recommended ESBL detection criteria is critical for the assessment of isolate susceptibility (S) and initiation of necessary infection control measures.
Methods: A total of 30,137 ENT isolates collected from 47 medical centres in 18 EUR countries including Russia, were S tested as part of the SENTRY Antimicrobial Surveillance Program (1997–2006). E. coli (EC), K. pneumoniae (KPN), K. oxytoca (KOX) and P. mirabilis (PM) isolates meeting ESBL-screening criteria (CLSI; MIC values ≥2 mg/L for ATM or ceftriaxone or ceftazidime) were confirmed using ESBL Etests (AB BIODISK) or the clavulanate (CLA) disk approximation method. A cefepime (FEP) MIC at ≥4 mg/L was the ESBL screening criterion for Enterobacter spp. (ESP), Citrobacter spp. (CSP) and Serratia spp. (SER) with confirmation performed using FEP with CLA.
Results: Among the 26,858 ENT isolates examined for ESBL-production, 2,954 (11.0%) qualified as potential ESBL producers. A subset of 1,796 were tested using a confirmatory method with 1,359 (75.7%) being positive. Confirmation of screening criteria occurred most frequently (>70%) in KPN, KOX, EC, CSP and least often (<70%) when testing ESP, SER and PM. Among EC and KPN, ESBL-screen positivity rates have increased from 1997–1999 to 2004–2006 (5.5 to 8.0% and 30.0 to 35.1%, respectively). Occurrence of ESBL-screen positive isolates that did not confirm with CLA inhibition may be attributable to other recognized R mechanisms. Confirmed ESBL-production was often associated with R to fluoroquinolones and aminoglycosides. S to carbapenems, tigecycline, and polymyxin B was retained among ESBL-confirmed isolates.
| Organism (no. tested) | ESBL screening criteria results |
ESBL confirmatory test results |
||
|---|---|---|---|---|
| No. Detected | % Positive | No. Tested | No. (%) Positive | |
| EC (15,026) | 1,022 | 6.8 | 654 | 485 (74.2) |
| KPN (3,881) | 1,034 | 26.6 | 728 | 648 (89.0) |
| KOX (1,121) | 209 | 18.6 | 96 | 80 (83.3) |
| PM (1,590) | 171 | 10.8 | 70 | 40 (57.1) |
| ESP (3,369) | 422 | 12.5* | 190 | 68 (35.8) |
| CSP (683) | 65 | 9.5* | 15 | 11 (73.3) |
| SER (1,188) | 31 | 2.6* | 43 | 27 (62.8) |
| Total (26,858) | 2,954 | 11.0 | 1,796 | 1,359 (75.5) |
ESBL phenotype screening criterion of FEP ≥4 mg/L.
Conclusions: Among tested EUR ENT, ESBL-screening criteria were most often confirmed among KPN (89.0%) and KOX (83.3%) and less so for EC (74.2%) and other species. ESBL prevalence has increased during this 10-year study, primarily in KPN and EC. Plasmidic movement of ESBLs between ENT species is a likely product of the high prevalence within predominant species, a worrisome development warranting continued longitudinal monitoring.
P565 Antimicrobial resistance comparisons amongst extended-spectrum β-lactamase producing Klebsiella spp. phenotypes collected from Brazilian hospitals in 2003 and 2006
C. Mendes, P. Koga, E. Sakagami, P. Turner, C. Kiffer on behalf of the MYSTIC Group Brazil
Objective: To compare the antimicrobial resistance rates of extended-spectrum β-lactamase (ESBL) producing Klebsiella spp. phenotypes responsible for hospital infections in two MYSTIC editions (2003 and 2006) in Brazil.
Methods: All hospitals participating in both surveillance years were included in the analysis. To be included, the isolates had to be consistent with the definition of an ESBL phenotype, established as having a minimal inhibitory concentration (MIC) to cefotaxime, ceftazidime, and/or cefepime ≥1.5 μg/mL determined by E-test method. Additionally, MICs were also determined to piperacillin/tazobactam, imipenem, meropenem, and ciprofloxacin. Interpretations followed the respective year NCCLS/CLSI documents. A chi-square test was applied to identify differences in each antimicrobial susceptibility rate among the two studied years. P values below 0.05 were considered significant.
Results: The same centres participating in both years (n = 19) contributed with the selected ESBL phenotypes and the total Klebsiella spp. isolates for the study (143/279 from 2003 and 93/170 from 2006). Then, 236 Klebsiella spp. ESBL phenotypes clinical isolates entered the analysis. The table presents susceptibility pattern of the Klebsiella spp. ESBL phenotypes according to the year of isolation, the chi-square and the p value for each comparison. Both carbapenems presented elevated susceptibility rates at both years, although a few intermediate and resistant strains occurred. The susceptibility rate for ciprofloxacin did not demonstrate any significant change among the two years. The susceptibility rate for piperacillin/tazobactam in 2003 was 86% and in 2006 61.3%, while the chi-square test yielded a p value <0.0001 for this comparison.
Susceptibility pattern of the Klebsiella spp. ESBL phenotypes according to the year of isolation, the chi-square and the p value for each comparison
| % Susceptibility/year |
P value | χ 2 | ||
|---|---|---|---|---|
| 2003 | 2006 | |||
| n = 143 | n = 93 | |||
| Ciprofloxacin | 73.4 | 80.6 | 0.3 | 1.3 |
| Piperacillin/tazo | 86 | 61.3 | <0.0001 | 17.7 |
| Imipenem | 99.2 | 100 | 0.7 | 0.18 |
| Meropenem | 98.3 | 99 | 0.7 | 0.14 |
Conclusions: The prevalence of isolation of ESBL-producing Klebsiella spp. phenotypes has been stable over the two years in the hospitals evaluated. Stable susceptibility rates were observed among most antimicrobials evaluated in each year against this phenotype, with only the carbapenems presenting virtually full and stable activity. However, susceptibility to piperacillin/tazobactam has decreased significantly over the years, casting doubts on the role of this antimicrobial in empirical therapy for the ESBL-producing phenotypes.
P566 Antibiotic resistance of community-acquired urinary tract infections in south-east Austria. Emergence of E. coli producing extended-spectrum β-lactamase
A. Badura, G. Feierl, A. Grisold, L. Masoud, U. Wagner-Eibel, E. Marth (Graz, AT)
Objective: To determine antimicrobial resistance rates of the most prevalent Gram-negative pathogens responsible for urinary tract infections in community patients in the region of south-east Austria during the last 5 years (2002 to 2006).
Methods: From January 2002 to October 2006, a total of 45.597 urine samples derived from community patients suffering from urinary tract infections from the region of south-east Austria were analysed. Species identification and resistance testing of nonduplicate isolates were done in the routine microbiology laboratory of the Medical University of Graz using conventional methods. Ambiguous results were confirmed with a VITEK2 system using the ID-GNB or ID-GN test cards (bioMérieux) for identification and the VITEK 2 AST-N020 test card (bioMérieux) for resistance testing. Antimicrobial susceptibility results were interpreted using the criteria recommended by CLSI. Production of extended-spectrum β-lactamases (ESBL) was confirmed by the Etest ESBL screen method using strips with cefotaxime, ceftazidime, and cefepime (AB Biodisk, Solna, Sweden).
Results: E. coli was most frequently isolated (70%) followed by Proteus spp. (14%), Klebsiella spp. (8%) and Pseudomonas aeruginosa (7%). The proportion of ESBL-producing E. coli increased from 1 isolate (0.03%) in 2002, 5 (0.2%) in 2003, 9 (0.3) in 2004, 27 (0.9%) in 2005 to 52 (2.1%) in 2006. ForE. coli, a significant increase in resistance could also be observed for ciprofloxacin (5% in 2002 to 12.9% in 2006), cotrimoxazole (15.6% in 2002 to 22% in 2006) and gentamicin (1% in 2002 to 4% in 2006). In total, 11 ESBL-producing Klebsiella spp. were isolated, but no significant change could be observed over the last 5 years. Among Klebsiella isolates, resistance rates to other agents remained stable: ciprofloxacin (mean 2.2%), cotrimoxazole (mean 8.8%) and gentamicin (mean 0.7%).
Generally, resistance patterns of Proteus spp. and Pseudomonas aeruginosa remained constant over the study period with high proportions of ciprofloxacin-resistant isolates (mean 11.3% and 14.8%, respectively).
Conclusion: Community-acquired urinary tract infections due to ESBL-producing Enterobacteriaceae have become a serious problem worldwide because of limited treatment options. This study confirms the rapid emergence of ESBL-producing E. coli among outpatients in the region of south-east Austria. Further analyses concerning the spread of these strains are urgently needed.
P567 Prevalence and antimicrobial susceptibility of extended-spectrum β-lactamases-producing Escherichia coli isolates from community-acquired urinary tract infections in Madrid, Spain
J. Tamayo, B. Orden, J. Cacho, J. Cuadros, J.L. Gómez-Garcés, J.I. Alós (Móstoles, Madrid, Getafe, Alcalá de Henares, ES)
Objectives: To assess the prevalence and monitor antimicrobial susceptibility trends of extended-spectrum β-lactamases (ESBL)-producing Escherichia coli isolates causing community-acquired urinary tract infections in 4 Health Care Areas of Madrid, Spain: Getafe, Argüelles, Alcalá de Henares and Móstoles.
Materials and Methods: A total of 6,721 E. coli isolates were recovered from November 2005 to June 2006. Of these, 279 unique ESBL-producing E. coli strains were studied. Antimicrobial susceptibility testing was performed by the agar dilution method according to guidelines of the CLSI. Thirteen antimicrobial agents were tested (see Table ). Differences in antimicrobial susceptibility related to age distribution and gender were investigated.
| Antibiotic | MIC (mg/L) |
%S | %I | %R | ||
|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||||
| Ampicillin | >16 | >16 | >16 | 0 | 0 | 100 |
| Cefazolin | >16 | >16 | >16 | 0 | 0 | 100 |
| Cefuroxime | >16 | >16 | >16 | 0 | 0 | 100 |
| Cefotaxime | 1–>16 | >16 | >16 | − | − | 100 |
| Amox/clav | 4/2–>32/16 | 8/4 | 32/16 | 56.3 | 29.6 | 14 |
| Piper/taz | ≤1/4–>64/4 | 4/4 | 32/4 | 77.1 | 17.9 | 5.0 |
| Imipenem | ≤0.06–0.12 | 0.25 | 0.5 | 100 | 0 | 0 |
| Ertapenem | ≤0.008–0.12 | 0.03 | 0.06 | 100 | 0 | 0 |
| Gentamycin | ≤0.5–>8 | ≤0.5 | >8 | 81.3 | 2.2 | 16.5 |
| Amikacin | ≤1–>16 | 2 | 8 | 99.3 | 0 | 9.7 |
| Fosfomycin | ≤1–>64 | 2 | 16 | 93.6 | 0 | 6.4 |
| Ciprofloxacin | ≤0.06–>4 | >4 | >4 | 15.5 | 2.5 | 82.0 |
| Co-trimoxazole | ≤0.5/9.5–>2/38 | >2/38 | >2/38 | 37.3 | − | 62.7 |
Results: The table displays the susceptibility of ESBL-producing E. coli isolates to the 13 antimicrobial agents tested. The point prevalence of ESBL-producing E. coli was 4.15% (279/6,721) overall, 5.97% (56/938) in Getafe, 3.94% (75/1,903) in Argüelles, 3.60% (74/2,057) in Alcalá de Henares, and 4.06% (74/1,823) in Móstoles.
No difference was found in antibiotic susceptibility patterns with patients' gender. However, pathogens resistant to co-trimoxazole or ciprofloxacin were detected less frequently in patients aged <50 years than in patients aged ≥50 years (for co-trimoxazole, OR = 0.43, 95%CI: 0.20–0.93, p = 0.018; for ciprofloxacin, OR = 0.32, 95%CI: 0.14–0.74, p = 0.0027).
Conclusions: ESBL-producing E. coli isolates were highly prevalent among patients with community-acquired UTI. All strains tested were susceptible to carbapenems. Ertapenem was the most active agent tested, with an MIC90 of 0.06 mg/L, followed by imipenem (MIC90, 0.5 mg/L), amikacin (MIC90, 8 mg/L), and fosfomycin (MIC90, 16 mg/L). Ciprofloxacin and co-trimoxazole exhibited the lowest activity; isolates from patients aged ≥50 years were more often resistant than those isolated from younger patients.
P568 Virulence genes and pathogenicity islands in an extended-spectrum β-lactamase producing Escherichia coli strain collection from Slovenia
K. Zerjavic, T. Orazem, M. Grabnar, J. Ambrozic Avgustin (Maribor, Ljubljana, SI)
Urinary tract infections (UTIs) belong to most common infectious diseases, being mainly caused by uropathogenic Escherichia coli (UPEC). UPEC strains possess several virulence factors (VFs), including capsules, invasins, siderophores, toxins, proteases, and various types of adhesins, that are involved in UTI pathogenesis. Some of these VFs are encoded by specific regions of bacterial chromosome, called pathogenicity islands (PAIs). Recent data revealed that quinolone resistance is often associated with extended-spectrum β-lactamase (ESBL) producing E. coli strains, which mainly belong to the non-B2 phylogenetic group and that quinolone resistant strains have fewer VFs than their quinolone-susceptible counterparts. The aim of this study was to elucidate the relationship between E. coli phylogenetic groups, virulence determinants, ciprofloxacin resistance and ESBL production in E. coli strains collected between the years 2000 and 2005 at the Institute of public health in Ljubljana. 27 strains from various sources were studied. Twelve of them were ciprofloxacin-susceptible whereas fifteen were ciprofloxacin resistant according to the CLSI guidelines. The phylogenetic group and the prevalence of twenty-four virulence factor genes and gene markers for six PAIs were determined by PCR amplification. Fifteen strains were assigned to phylogenetic group D, five to group B1, four to group B2 and three to group A. Gene markers for PAI I536 and PAI III536 were not detected. Genes associated with PAI II536, PAI IV536, PAI IIJ96 and PAI ICFT073 were detected in 11, 33, 4 and 15 percent of the strains, respectively. The most prevalent virulence genes (occurrence between 37 and 78%) were sfa, iroN, iucD, papG, kpsM, iha, iss and usp. Outstanding was the high prevalence of the picU gene (100%) and the VFs associated with E. coli translocation across the blood-brain barrier i.e. ibeA (41%) and and asl (67%) among ciprofloxacin resistant strains, while some VFs, i.e. papGII, afa, hlyA, cnfI and vat, were found exclusively among ciprofloxacin susceptible strains. Our results are generally in accordance with previous reports and indicate a distribution of UTI strains in two groups i.e. highly virulent and low resistant, and low virulent and highly resistant, thus raising at least two questions: (i) what are the mechanisms that allow strains from the first group to survive and (ii) which VFs do strains from the second group use to cause an infection.
Molecular features of β-lactamases
P569 TEM-142: a novel extended-spectrum β-lactamase derived from TEM-111 via an Asp238Asn substitution
B. Timmerbeil, S. Scherpe, E. Stuerenburg, P. Heisig (Hamburg, DE)
Objectives: Extended spectrum β-lactamases (ESBLs) are a group of β-lactam hydrolysing enzymes which are able to inactivate most β-lactam antibiotics including aztreonam and third generation cephalosporins. The TEM family is the largest and most common ESBL family. Within this family the subtypes differ by a few amino acid substitutions.
Four ESBL-producing Escherichia coli strains were collected from one patient under β-lactam therapy in the period from 2001 to 2004. The β-lactamases were determined and the strains were analysed for clonality.
Methods: Beta-lactamases were identified using a specific blaTEM and blaSHV duplex PCR. Using Sanger sequencing the blaTEM subtype was identified. Clonality analysis was performed by pulsed-field gel electrophoresis (PFGE) and three different fingerprint PCR techniques (RAPD-, ERIC-, RepU1b-PCR). The blaTEM-bearing plasmids were analysed by restriction enzyme mapping. The susceptibilities of the four strains were determined as minimal inhibitory concentrations (MICs) according to CLSI guidelines for AMP, AMP-SUL, AMX-CLA, PIP-SUL, PIP-TZB, CXM, CTX, CRO, CAZ, FEP, FOX, IPM, MEM, ERM, GEN, TOB, AMK, CIP, MXF, DOX, and SXT.
Results: Four ESBL-producing Escherichia coli isolates were collected from the same patient: E. coli Ur7883/2001, E. coli Ur6845/2003, E. coli Ur8093/2003 and E. coli Ur11057/2004.
E. coli Ur7883/2001 was identified to produce the β-lactamase TEM-111. The remaining three isolates are producers of TEM-142. Both β-lactamases differ in just one amino acid position: TEM-111 (Asp238), TEM-142 (Asn238). Results of clonality analysis provide evidence for clonal relationship of all four strains. The blaTEM-carrying plasmids are identical. The MICs of cefepime for the four strains has shown a difference of two serial dilution steps (TEM-111 isolate: 2mg/L, TEM-142 isolates: 8mg/L).
Conclusion: A novel blaTEM subtype was identified with a unique Gly238-Asn amino acid substitution called TEM-142. The β-lactamase is highly related to TEM-111 which was isolated from the same patient three years earlier. All strains are clonally related and the blaTEM containing plasmids are identical. These data suggest that TEM-111 has acquired a point mutation to yield TEM-142. A possible driving force for this evolution might be the selective pressure by antibiotics, like cefepime (differences in the MIC values of cefepime were determined) or enzyme stability.
P570 Molecular epidemiology and typing of CTX-M extended-spectrum
L. Xu, D. Morris, K. Biernacka, N. Woodford, P. Hawkey, K. Nye (Birmingham, UK; Galway, IE; London, UK)
Objectives: The aims of this study were to investigate the molecular epidemiology of CTX-M Extended-Spectrum B-Lactamase-Producing Enterobacteriaceae in both hospital and community patients in the area served by the Heart of England NHS Foundation Trust, Birmingham.
Methods: Between January and April 2006, a total of 2529 consecutive non-duplicate isolates of Enterobacteriaceae were processed by the clinical service. All isolates were screened for susceptibility to cefpodoxime by the BSAC disc diffusion method and ESBL production confirmed using cefpodoxime, cefpirome and ceftazidime with and without clavulanate. Multiplex PCR was used to detect all blaCTX-M genes. The blaCTX-M positive isolates were characterised by dHPLC and results further confirmed by DNA sequencing. Clonal relatedness was identified by PFGE analysis.
Results: A total of 82 isolates (3.2%) were confirmed to produce ESBL, including 53 isolates (64.6%) from hospital and 29 isolates (35.4%) from the community. These isolates included 64 E. coli, 15 Klebsiella spp., 2 E. cloacae and 1 Citrobacter freundii. Multiplex PCR screening identified a blaCTX-M gene in 76 isolates (92.7%). Multiplex PCR screening identified a blaCTX-M gene in 76 isolates. Of these, 71 belonged to a blaCTX-M group 1, and 5 harboured a blaCTX-M group 9 gene. The dHPLC genotyping of group 1 isolates showed that all had blaCTX-M-15 profiles. The 5 group 9 isolates depicted a blaCTX-M-14 chromatogram signature. DNA sequencing confirmed blaCTX-M-15 in 8 representative strains of the 71 dHPLC typed CTX-M-15 isolates and all 5 blaCTX-M-14 producers. PFGE analysis showed the 60 blaCTX-M positive E. coli isolates to represent multiple strains among which 7 clusters of 2 or more isolates and 16 distinct strains (>85% similarity) were observed, eight were strain A.
Conclusion: This study confirms that blactx-m-15 is the commonest cause of ESBL production in this locality. blaCTX-M-14 accounted for 6.6% of all our CTX-M producing isolates, which has not formally reported as causing disease in the UK, it may displace blaCTX-M-15. There was evidence of dissemination of clonally related isolates among patients both in the hospital and the community. Strain A has been reported to comprise 38% UK isolates, but only 13% in our study.
P571 The fate of antibiotic resistance genes in cooked meat
R. Koncan, R. García-Albiach, D. Bravo, R. Cantón, F. Baquero, G. Cornaglia, R. del Campo (Verona, IT; Madrid, ES)
Background: The possibility of transfer of resistant bacteria from animals to humans by the food chain has been demonstrated in several works. Safe food processing implies complete microbial pathogens elimination, but the fate of DNA encoding antibiotic resistance has not been explored. The aim of this work was to evaluate the possibility of detecting the aac(6')-aph(2') codifying aminoglycoside resistance gene in meat after conventional cooking procedures.
Methods: 25 g of chicken, pork and beef meat were contaminated (needle injection) with different dilutions of Enterococcus faecalis (Delaware strain), carrying the bi-functional gene aac(6')-aph(2”) which confers resistance to aminoglycosides. Contaminated meat samples were either boiled (20 min), grilled (10 min), or cooked in an owen microwave (5 min), mimicking conventional cooking procedures. Furthermore, samples were submitted to conventional autoclave sterilisation and high-pressure treatment to eradicate the viable bacteria. No bacterial growth was obtained in plates seeded with tissue samples. Total DNA was extracted using QiaAMP extraction kit for tissues (QiaGen), and all samples were PCR-tested by using selective primers for the aac(6')-aph(2') gene. Quantitative PCR experiments were applied to determine the bacterial DNA concentration recovered after cooking methods. A transformation assay was performed with DNA obtained from meat and E. faecalis JH2–2 as recipient strain.
Results: Growth of the E. faecalis Delaware strain was not detected in any meat type after the different cooking procedures or autoclave sterilisation, but after high-pressure treatment counts ranged from 103 to 106 CFU/mL. Positive PCR results for the bi-functional gene were observed in all experiments, except after the autoclave treatment. Quantitative-PCR results indicated a direct correlation between the density of bacterial inoculum and the amplified DNA. At identical inoculum density, higher amounts of the bifunctional gene were recovered in the beef samples, than in the pork or chicken ones. Transformation experiments with total DNA from samples negative in all cases.
Conclusion: Positive amplification for the antibiotic resistance gene aac(6')-aph(2”) harboured in a E. faecalis strain was obtained after cooking contaminated meat. PCR-amplification procedures might be suitable to retrospectively analyse the contamination of meat samples by antibiotic-resistant bacteria.
P572 Identification of CTX-M-type extended-spectrum β-lactamases in urine based on real-time PCR
C. Oxacelay, A. Özcan, T. Naas, P. Nordmann (Kremlin-Bicetre, FR)
Background: CTX-M extended-spectrum β-lactamases (ESBLs) are increasingly prevalent worldwide mostly in Escherichia coli from community-acquired urinary tract infections. Thus, availability of a fast and reliable technique for identification of CTX-M-enzymes is becoming a challenge for the microbiology laboratory. We have recently developed a powerful tool combining real-time PCR with pyrosequencing for epidemiological studies of CTX-M-positive strains (Naas et al. AAC, 2007, in press). However, this tool required a pyrosequencing machine. Here we report use of an alternative technique based only on real-time PCR.
Method: A fast real-time PCR amplification based on a LightCycler 2.0 amplification system (Roche Diagnostic), using degenerated primers specific for the blaCTX-M alleles coupled to a CTX-M-type-specific detection using hybridisation probes was developed. Five well-characterised CTX-M producers, representing each of the five groups were used as controls. Urine samples were collected at the Bicětre Hospital (France) over a three months period. DNAs were extracted using QiAmp Viral RNA extraction kit (Qiagen). The presence of ESBLs were confirmed by standard microbiological techniques (disk diffusion antibiogram, synergy testing) and by standard PCR followed by sequencing.
Results: 810 urines were collected over the studied period and among them 36 isolates from 29 different patients had an ESBL phenotype (prevalence rate of 3.6%). The bacterial species were: E. coli (22/29: 77%), Klebsiella pneumoniae (2/29: 7%), Citrobacter freundii (2/29: 7%), Providencia stuartii (1/29: 3%), Enterobacter cloacae (1/29: 3%) and Enterobacter aerogenes (1/29: 3%). The patients were mostly from the Nephrology (27%), Emergency (24%), Gerontology (17%) and Urology (10%) wards. Using this PCR approach, 20 out of 29 ESBLs were identified as CTXMs. Five belonged to the CTXM-9-and 15 to the CTXM-1-group as revealed by melting curve analysis. Standard PCR and sequencing identified 5 CTXM-9 and 15 CTXM-15 and the remaining 9 ESBLs were mostly of TEM-type. Patients, with more than one ESBL positive urine were reproducibly identified as CTX-M positive.
Conclusions: The real-time PCR is a powerful technique for a fast identification of CTX-M-positive isolates from urines. It has the potential to be used in a diagnostic microbiology laboratory since up to fifteen clinical samples may be processed in less than 3 h, thus allowing a rapid implementation of isolation measures.
P573 Evolution of bla genes from SHV family
N. Mendonça, M. Caniça (Lisbon, PT)
Objectives: SHV (sulfhydryl variable) is the most prevalent family of β-lactamases produced by Klebsiella pneumoniae strains, an important mechanism of resistance to β-lactam antibiotics. The aim of the study was to evaluate the diversity and molecular evolution of blaSHV genes family.
Methods: PCR specific for blaSHV gene and nucleotide sequencing were performed in 212 clinical K. pneumoniae strains isolated in Portugal (years 1998–2005). In addition 59 blaSHV sequences were downloaded from NCBI GenBank database. All sequences were analysed by Bionumerics software and a specific blaSHV gene fragment of 825bp was used to align all sequences. Phylogenetic and molecular evolutionary analysis was conducted using MEGA software.
Results: Among the 271 sequences analysed we detected 64 different blaSHV gene sequence frameworks (SFWs), from which 40 were here firstly identified. The 64 SFWs emerged from the combination of silent mutations at 38 different nucleotide positions. Silent mutations A402→G, G705→A and C786→G may be considered hot spots, as they appeared in a higher frequency (92%, 62% and 49%, respectively) than others. Phylogenetic analysis showed that the SFW (named ad) presenting those 3 mutations is a possible common ancestral.
Among the 212 blaSHV genes sequenced by us, 58 (27%) were identified as blaSHV-11. This gene showed the highest diversity as it presented 17 different SFWs, followed by 54 (26%) blaSHV-1 genes detected with 13 SFWs, and by 30 (14%) blaSHV-28 genes with only 2 SFWs associated (“a” and “r”). The 111 different sequences of blaSHV genes studied in the phylogenetic approach, presented a total of 53 non-synonymous mutations and 38 synonymous mutations, which allowed to the construction of an unrooted tree.
Overall, the majority of blaSHV-1 and blaSHV-11 sequence genes had the higher number of different SFWs (18 and 20, respectively) and were on opposite sides of the evolutionary tree, which may imply a divergent evolution. Furthermore, the majority of extended-spectrum β-lactamase (ESBL) coding genes (63%) could be detected within the same branch of the unrooted tree, which may indicate a common ancestral or origin.
Conclusions: This study demonstrated that blaSHV genes descend from a yet unidentified common ancestor. The high diversity of blaSHV genes suggests their contribution to the rapid evolution toward blaESBL-SHV genes coding to ESBL enzymes and thus to the emergence of resistance to third generation cephalosporins.
P574 Emergence and dissemination of metallo-β-lactamases producing strains in Europe: report from the SENTRY Antimicrobial Surveillance Program
L. Deshpande, H. Sader, T. Fritsche, R. Jones (North Liberty, US)
Objective: To evaluate the emergence and dissemination of metallo-β-lactamase (MBL) producing strains in European medical centres participating in the SENTRY Program.
Methods: Beginning in 2000 all Gram-negative bacilli submitted to SENTRY with decreased susceptibility to imipenem (IPM), meropenem and ceftazidime were routinely screened for production of MBL by disk approximation tests and/or MBL Etest (AB BIODISK) strips. Isolates with screen-positive results were evaluated by PCR using generic primers for IMP, VIM, SPM and GIM enzyme types. MBL gene sequencing and molecular typing (ribotyping, PFGE) were additionally performed to characterise MBL and to evaluate clonality.
| Organism (no.) | MBL (no. of strains) | Countries (no. of strains) | Detection year |
|---|---|---|---|
| P. aeruginosa (46) | VIM-1 (32) | Germany (4)/Greece (7)/Italy (21) | 2001–2004 |
| VIM-2 (2) | France (1)/Poland (1) | 2001 and 2003 | |
| IMP-13 (6) | Italy (6) | 2002–2002 | |
| GIM-1 (6) | Germany (6) | 2002 | |
| K. pneumoniae (33) | VIM-1 (33) | Germany (1)/Greece (16)/Italy (12)/Spain (3)/Turkey (1) | 2005–2006 |
| E. cloacae (19) | VIM-1 (7) | Germany (3)/Italy (2)/Spain (2) | 2004–2006 |
| IMP-1 (12) | Turkey (12) | 2003–2004 | |
| E. aerogenes (2) | VIM-1 (2) | Greece (2) | 2005 |
| Acinetobacter spp. (2) | IMP-2 (1) | Italy (1) | 2003 |
| VIM-1 (1) | Greece (1) | 2003 | |
| C. koseri (1) | VIM-1 (1) | Italy (1) | 2005 |
| K. ozaenae (1) | VIM-1 (1) | Italy (1) | 2005 |
| P. mirabilis (1) | VIM-1 (1) | Greece (1) | 2005 |
Results: Since 2000, 4,935 P. aeruginosa (PSA), 1,460 Acinetobacter spp. (ASP) and 22,950 Enterobacteriaceae (ENT) have been collected from European centres and tested for susceptibility (S) by reference broth microdilution methods (CLSI, 2006). S rates to IMP remained stable among PSA (75.5% in 2000 and 76.1% in 2006), but varied from 78.5% in 2000 to 51.9% in 2006 among ASP. IMP remained very active against ENT (99.8% S in 2006), but the occurrence of strains with MIC ≥2mg/L increased from 0.4% in 2000 to 1.5% in 2006. A total of 105 MBL-producing strains were identified and characterised since 2000. The most common MBL-producing species were PSA with 46 strains from 5 countries and producing 4 different MBLs; followed by K. pneumoniae (KPN; 33 VIM-1-producing strains from 5 countries) and E. cloacae (ECL; 19 strains, VIM-1 and IMP-1-producers from 4 countries). MBL-producing strains were usually resistant to most antimicrobials tested. Polymyxin B was very active against PSA (100% S) and KPN (94% S); while tigecycline was highly active (100% S) against ASP, KPN and ECL. Molecular typing results indicated clonal dissemination of PSA producing VIM-1 in Germany, Greece and Italy, IMP-13 in Italy and GIM-1 in Germany; KPN producing VIM-1 in Greece, Italy and Spain; and IMP-1-producing ECL in Turkey. In addition, clonal diversity was observed among IMP-13-producing PSA from Rome, Italy; VIM-producing KPN from Athens, Greece and ECL from Leipzig, Germany and Madrid, Spain; and IMP-1 producing ECL from Istanbul, Turkey.
Conclusions: The emergence and dissemination of MBL-producing strains was documented in several European countries and it is of great concern since these enzymes were usually codified by genes located on integrons with great mobility.
Molecular characterisation of gastrointestinal pathogens
P575 Comparison of four real-time PCR-based methods for the detection of food-borne Salmonella enterica
K. Krascsenicsova, E. Kaclíkova, T. Kuchta (Bratislava, SK)
Objectives: The aim of the study was to compare four real-time PCR-based methods – our in-house method and three commercial kits – for the detection of food-borne Salmonella enterica regarding sensitivity and reliability of the results interpretation.
Methods: The next-day method for Salmonella enterica detection developed in our laboratory utilised a two-step enrichment and our original duplex FAM/VIC TaqMan real-time PCR. Sample preparation prescribed for TaqMan Salmonella enterica (Applied Biosystems, Foster City, California, USA), iQ-Check” Salmonella (BioRad, Marnes-la-Coquette, France) and BACIdent Salmonella spp. (GeneScan, Freiburg, Germany) involves a single-step enrichment in non-selective medium. PCR detection of ABI and BacIdent kits is based on duplex FAM/VIC TaqMan real-time PCR. The iQ-Check kit employes duplex FAM/TexasRed Molecular Beacon-based real-time PCR. Detection limits of PCR, as well as the detection limits of whole methods for live and dead Salmonella cells, were determined for each method. One hundred different food samples were analysed and 40 selected food samples were spiked at three contamination levels (100, 101 and 102 CFU per sample) and analysed in parallel by four methods.
Results: Detection limits of 102 CFU per ml for PCR and of 100 CFU per sample were estimated for all presented methods. However, dead cells were detected at a level of 103 – 104 CFU per sample by the commercial kits utilising the single-step enrichment, which may be a potential cause of false positive results. False negative results probably caused by food-borne PCR inhibitors were obtained with some food matrices.
Conclusion: Methods for the detection of Salmonella enterica utilising an enrichment shorter than 24h, followed by duplex real-time PCR including an internal positive control, represent a suitable tool for fast and reliable identification of this food-borne pathogen. The use of a single-step enrichment may be a disadvantage, while a two-step enrichment facilitates a reduction in the concentration of sample-borne PCR inhibitors as well as dead cells.
P576 Comparison of commercially available methods and in-house real-time PCR-based method for the detection of food-borne pathogen Listeria monocytogenes
K. Oravcova, E. Kaclikova, T. Trncikova (Bratislava, SK)
Objectives: Food-borne Listeria monocytogenes causes in susceptible individuals serious illness listeriosis with high mortality rate. Thus methods for its rapid identification in foods are of importance to determine the source and level of contamination. We compared an in-house real-time PCR-based method and three commercially available methods for the detection of L. monocytogenes in food regarding rapid and unambiguous results acquisition.
Methods: All four methods tested were culture-based with the final identification by real-time PCR or on chromogenic agar. Selected 40 food samples artificially contaminated with L. monocytogenes and 12 naturally contaminated samples were analysed using in-house method, TaqMan Listeria monocytogenes Detection Kit (Applied Biosystems, Foster City, CA, USA), iQ-Check Listeria monocytogenes Kit (Bio-Rad, Marnes-la-Coquette, France) and short enrichment procedure (Oxoid, Basingstoke, UK). Method for the next-day identification of L. monocytogenes developed in our laboratory consisted of two-step selective enrichment followed by duplex real-time PCR employing TaqMan probes. Using kits, samples were undergoing one-step selective enrichment and analysed by duplex quantitative real-time PCR employing either dual-labeled TaqMan DNA probes in ABI's kit or Molecular Beacon probes in Bio-Rad's kit. According to Oxoid's enrichment procedure strains were identified on OCLA agar. Detection limits for live as well as for dead cells of L. monocytogenes were set.
Results: Detection limits of all methods for all samples were 100 CFU per sample with the exception of 6 samples giving false negative results by the Oxoid's method due to L. innocua present in the samples and overgrowing the target L. monocytogenes. Detection limits for dead L. monocytogenes cells were 106 CFU per sample for in-house method, 104 – 105 CFU per sample for kits.
Conclusion: Methods for L. monocytogenes identification in foods consisting of short enrichment and real-time PCR detection provide a powerful tool for fast and precise epidemiological studies when compared to only culture-based techniques. If the price of analysis is a decisive factor our in-hause method is of preferable use. Commercially available methods, easy to handle and data interpretation, may suffer from possibility of false positive results due to detection of dead cells potentially present in the food sample.
P577 Evaluation of partial rpoB and 16S rDNA sequencing for identification of Enterococcal species and the comparison to phenotypic methods
G. Haase, R. Lütticken, J. Weber-Heynemann, I. Ramminger, A. Mellmann, D. Harmsen (Aachen, Münster, DE)
Objectives: Enterococci can cause a variety of serious infections in humans and animals. As of spring 2005 this genus compromise 32 species whereas many of the newer species have not yet been included in the database of commercially available identification systems. Therefore, in this study a reference databases for partial RNA polymerase B (rpoB) and the 16S rDNA gene sequences comprising all type strains of the genus Enterococcus was established. Subsequently, the performance of sequence-based identification in comparison to two commercially available, phenotypic based identification systems was evaluated using 44 clinical isolates.
Methods: In addition to the 32 type strains, a panel of 44 strains (mainly human isolates compromising 9 different species) was analysed in order to reveal the relative performance of the different identification methods tested. Direct sequencing was performed using ABI Prism 3100 Analyzer (Applied Biosystems). For quality assured sequence analysis (rpoB: 551 bp, position 2491 to 3041, AF535187; 16S rDNA: 446 bp, position 54 to 510 of E. coli) the Ridom TraceEditPro version 1.0 software (Ridom GmbH, Würzburg, Germany) was used. For phenotypic identification the rapid ID 32 Strep (bioMérieux, Nürtingen, Germany) and the Gram positive panel of the Phoenix” system (BD, Heidelberg, Germany) were used according to the instructions of the manufactures. ID 32 panels were examined using the MiniAPI reader.
Results: Of the 32 type species, 28 exhibit a unique sequence in the respective 16S rDNA/rpoB reference database whereas only the two pairs E. porcinus/E. villorum and E. italicus/E. saccharominimus had identical sequences. Only 7 type strains were correctly identified by ID 32 Strep or the Phoenix system. Analyzing the 44 clinical isolates, 36 (81.8%) could be identified univocally by sequencing (≥99% similarity) using the 16S rDNA/rpoB reference database. Using the rpoB based identification as reference, only 25 (56.8%) and 32 (72.7%) of these strains were correctly identified by the ID 32 Strep or Phoenix test system, respectively.
Conclusion: Sequence based identification, in particular rpoB sequencing turned out as the most discriminatory identification procedure for achieving reliable species identification of Enterococci.
P578 Updated qnr multiplex PCR-based technique: epidemiological survey of Qnr determinants in a collection of ESBL-producing enterobacterial isolates from Kuwait
V. Cattoir, L. Poirel, V. Rotimi, C.-J. Soussy, P. Nordmann (Le Kremlin Bicetre, FR; Kuwait City, KW; Créteil, FR)
Objective: The goal of this study was to develop a single-tube based-PCR technique for detecting simultaneously qnrA, B and S-like genes. This strategy was applied for screening a collection of ESBL(+) enterobacterial isolates from Kuwait.
Material and Methods: A multiplex PCR-based technique was developped for detection of the three known qnr genes using specific primers. Primers were carefully designed for adaptation of variants of these genes (qnrA1 to A5, qnrB1 to B6 and qnrS1 to S2). PCR products were identified as qnrA, B or S according to their sizes in ethidium bromide-stained agarose gels and thus were submitted by direct sequencing. After its optimisation, this technique was used to screen a collection of 63 ESBL-producing enterobacterial isolates obtained in Kuwait from 2002 to 2004.
Results: Multiplex-PCR peformed with specific primers gave three bands of amplification products easily separated: 579 bp, 264 bp and 426 bp for qnrA, -B and -S genes, respectively. All positive and negative controls were according to expected results. A qnr gene was present in 4 (6.3%) of the sixty-three enterobacterial isolates. Whereas qnrA and qnrS were absent, qnrB was present in these four isolates corresponding to 3 Enterobacter cloacae (strains K34, K35 and K37) and 1 Citrobacter freundii (strain K70). After sequencing, amplification products were identical to qnrB2 (441/441) for K34 and K35, similar to qnrB1 (97%) for K70, and similar to qnrB6 (89%) for K70. Analysis of deduced protein structure of Qnr determinants from K37 and K70 strains showed 3 and 5 substitutions compared to QnrB2 and QnrB6, respectively. These novel variants may be designated as QnrB7 and QrB8, respectively.
Conclusion: This multiplex PCR-based technique detected all variants of the three qnr genes. Using this technique, two novel QnrB determinants were discovered. Interestingly, no QnrA-like determinant was identified whereas they have been reported to be associated frequently to ESBL genes.
P579 Analysis of plasmids from multi-resistant and multi-plasmid clinical E. coli strain CZD1527
R. Wolinowska, K. Strzezek, P. Zaleski, J. Chodnicka, A. Lakomy, A. Demianczuk, A. Plucienniczak (Warsaw, PL)
Objectives: Clinical strain of E. coli CZD1527 is resistant to β-lactams, aminoglycosides and tetracycline. Spontaneous changes in level of resistance to antibiotics have been observed for this strain. Analysis of the plasmids present in the strain has been performed in regard to their possible involvement in that changeability of the strain antibiotic resistance.
Methods: Transformation of laboratory E. coli strain NM522 by electroporation. Transformation preceded by insertion of kanamycin transposon. Antimicrobial susceptibility tested by disc-diffusion method. PCR analysis.
Results: Total plasmid preparation obtained from E. coli CZD1527 strain has been analysed on agarose gel and at least 8 bands can be observed. 4 of them migrate like plasmids of 4–5 kb, two bands contain very big molecules and are localised on the top of the gel. The other bands are less visible and placed between these two groups.
Plasmid preparation has been used to direct transformation of laboratory strain E. coli NM522, then selection on plates with antibiotics has been performed. Two kinds of transformants carrying big plasmids have been obtained. Plasmids are above 100 kb and have been named pIGT-15 and pIGAM-1, both encode resistance to β-lactams, pIGT-15 to aminoglycosides and tetracycline as well. Partial sequence of pIGT-15 indicates that resistance to aminoglycosides and β-lactams is bound up with class 1 integron. It shows high homology to plasmid p1658/97 from E. coli (GeneBank Acc. No NC004998). The gene of resistance to tetracycline is located in structure of transposon T1721.
In the second step total plasmid preparation of CZD1527 strain has been subjected to kanamycin transposon insertion and electroporation. Two different plasmids, pIGRW12 and pIGWZ12, have been identified in transformants. 90 colonies of 126 have been tested by PCR, no additional plasmid has been found.
Complete nucleotide sequence of both plasmids is determined, both are cryptic plasmids. Plasmid pIGWZ12 is 4072 bp long (GenBank Acc. No DQ311641, Plasmid 2006, 56, 228–232), pIGRW12 is 4995 bp long (sequence already sent to GenBank). Detailed analyses of two smallest bands from agarose gel reveal that they are single stranded forms of plasmids pIGWZ12 and pIGRW12.
Conclusion:
-
–
Four plasmids have been found in clinical strain E. coli CZD1527.
-
–
Complete sequence of two cryptic plasmids (pIGWZ12 and pIGRW12) has been obtained.
-
–
Partial sequence of pIGT-15 with resistance genes is being determined.
P580 Isolation EPEC strains in the region of Thrace, Greece
E. Alepopoulou, M. Panopoulou, V. Dala, A. Chatzimichail, K. Ritis, S. Kartali (Alexandroupoli, GR)
Objective: Enteropathogenic E. coli (EPEC) is an important category of diarrhoeagenic E. coli which links to infant diarrhoea in the developing world and sporadic cases of diarrhoea in industrial countries. EPEC damage the bowel mucosa with characteristic mechanism (attaching and effacing lesion) mediated by a protein encoded by the eae chromosomal gene.
Methods: Two hundred fifteen stool specimens were examined for EPEC. One hundred sixty five samples proceeded from patients with diarrhoea (group A) and 50 from healthy individuals (group B). Stool specimens or rectal swabs were inoculated on MC agar and incubated at 37°C overnight. Five colonies of E. coli isolated on MC agar were picked. The biochemical identification of the strains was performed by automated system VITEK 2 (bioMérieux) and serotyping by slide-agglutination methods for serotypes O111, O55, O26, O86, O119, O127, O125, O126, O128, O124, O114, O142 (BioRad). The presence of eae gene was detected by PCR that contains primers for eae E. coli gene. The PCR products 384 bp were analysed by electrophoresis in 2% agar gel.
Results: From 215 stools 1207 strains E. coli were examined. EPEC were recovered from 22 (10.2%) stool specimens, 11 (5.1%) of them were from children and 8 (3.7%) from adults (group A), though 2 and 1 strain respectively from group B (control). The EPEC strains encountered in this study belonged to 5 different serotypes of E. coli: O127–8 (36%), O26–5 (23%), O126–4 (18%), O125–4 (18%) and O55–1 (4.5%). These strains did not have the eae gene. However, one strain (0.6%) from group A that yielded gene eae, did not ferment sorbitol and has been isolated from stool of an adult patient with diarrhoea.
Conclusion: The 5.1% EPEC strains were isolated from children and isolation rate is in agreement with those of other investigations. However, according to the decision of the Second International Symposium on EPEC in 1995: strains with A/E histopathology and absence of Shiga toxin should be called EPEC. Consequently, the rate of isolation of EPEC in this study was much lower (0.6%), as only one from the 165 samples has the eae gene. The detection of pathogenic genes altered in high degree the rates of isolation of EPEC and modifies the epidemiologic data of various regions.
P581 Isolation of E. coli O157:H7 and non-O157 in the region of Thrace, Greece
E. Alepopoulou, M. Panopoulou, E. Maltezos, G. Kartalis, S. Kartali (Alexandroupoli, GR)
Aim: The morbidity and mortality associated with several recent large outbreaks of gastrointestinal disease caused by Shiga toxin-producing E. coli has highlighted the threat these organisms pose to public health.
Methods: Two hundred fifteen stool specimens were examined for EHEC. One hundred five samples proceeded from patients with diarrhoea (group A) and 50 from healthy (group B). Stool specimens or rectal swabs were inoculated on MC agar, CT-SMAC agar and incubated at 37°C overnight in aerobic conditions. For the detection of enterohemolysin, five colonies of E. coli from MC agar and sorbitol negative colonies from CT-SMAC were inoculated in WBA (Washed Blood Agar) with CaCl2. For the detection of MUG activity Tryptone X-glucuronide Agar (OXOID) was used. The biochemical identification of the strains was performed by automated system VITEK 2 (bioMérieux) and serologic identification by slide-agglutination methods (Wellcolex * E. coli 157: H7, ABBOTT). Presence of eae, ehx, stx1, stx2 genes was detected by multiplex PCR. The PCR products 384 bp (eae), 534 bp (ehx), 181 bp (stx 1) and 255 bp (stx 2) were analysed by electrophoresis in 2% agar gel.
Results: From 1207 strains isolated, 38 did not ferment sorbitol and two of them produced small, turbid haemolytic zone on WBA with CaCl2 after overnight incubation. One strain (0.46%) was isolated from stool of adult patient (group A), belonged to serotype O157:H7 and it did not yield the eae, ehx, stx1, stx2 genes. From the remainder 37 strains, two (1.2%) have stx toxin genes. These strains were serotype non – O157:H7 E. coli and were isolated from two patients with diarrhoea. One of them has stx 1 and stx 2 toxins genes, and the other has stx 1 and ehx genes.
Conclusion: The isolation rate of E. coli O157:H7 in the region of Thrace, Greece is low. However, the percentage of E. coli non-O157:H7 isolated from stool of patients with diarrhoea is similar with that other of investigations in European countries.
P582 Highly efficient extraction of pathogen DNA from stool using the NucliSENS easyMAG” specific A 1.0.2 protocol
T. Schuurman, R.F. de Boer, P.B.H. van Deursen, P. de Bie, A.M.D. Kooistra-Smid (Groningen, Boxtel, NL)
Objectives: The NucliSENS easyMAG is a fully automated system for extraction of nucleic acids from a wide variety of clinical specimens. However, the extraction of DNA from stool with the standard protocol (Generic) has been shown to be less efficient compared to other clinical specimens. Preliminary data showed that this problem was in part related to impaired elution of the DNA. To overcome this problem, a new protocol, Specific A (bioMérieux) has been developed and validated for use with DNA rich specimen types, such as stool.
Methods: Clinical stools (n = 94), including 43 stools positive for 1 or more intestinal pathogens, were used to challenge both extraction protocols. Relative recovery of DNA was assessed by spiking a known amount of HindIII-digested phage lambda DNA, and comparing the recoveries after gel electrophoresis. Impaired elution for Generic was assessed by a secondary elution of the DNA from the retrieved magnetic silica. Downstream performance with the extracted DNA was assessed with 2 internally controlled (IC) multiplex real-time PCRs each targeting 2–3 intestinal pathogens.
Results: Of the 94 stools tested, 61 showed nearly identical DNA recoveries without impaired elution, although the DNA yield was slightly higher with Specific A in all specimens. For the remaining 33 samples, 19 showed low DNA recovery with Generic, whereas Specific A showed variable but improved recovery. The other 14 specimens could be classified in three groups based on the ratio between the primary (1st) and secondary (2nd) elution for Generic. This ratio was 1st > 2nd, 1st < 2nd, and 1st = 2nd for 4, 4, and 6 specimens, respectively. Results of the real-time PCRs confirmed the DNA recovery results. Ct values for Specific A were on average 0.9 and 1.1 cycles lower for IC and pathogen DNA, respectively, compared to Generic. The distribution of all the paired Ct values also showed a significant difference (paired 2-sided student T-test, p < 0.00002). When the Ct values were addressed to the DNA recovery, the 61 specimens with nearly identical recovery showed on average 0.61 and 0.62 cycles lower Ct values with Specific A for IC and pathogen DNA, respectively, whereas the other specimens showed on average 1.5 and 1.9 cycles lower Ct values. Inhibition was not significantly different between both protocols (6–7%).
Conclusion: Based on the presented data, Specific A, results in a significant improvement in the performance of the NucliSENS easyMAG with stool specimens.
P583 Rapid and sensitive detection of 5 gastro-intestinal pathogens using 2 internally controlled Multiplex real-time PCRs
T. Schuurman, R.F. de Boer, A.M.D. Kooistra-Smid (Groningen, NL)
Objectives: Traditional methods to detect gastro-intestinal pathogens are slow, and/or lack sensitivity. Molecular detection of gastro-intestinal pathogens has proven to be rapid and sensitive. Stool screening requires a large throughput; however, the use of monoplex PCRs greatly limits the capacity. Therefore, a multiplex approach is mandatory.
Methods: Real-time PCR assays for Salmonella enterica (SE), Campylobacter jejuni (CJ), Giardia lamblia (GL), shiga toxin-producing Escherichia coli (STEC), and Shigella spp./enteroinvasive E. coli (SH/EIEC) were developed and subsequently multiplexed in 2 assays combining SE/CJ/GL and STEC/SH/EIEC. Both assays also incorporated an internal control (phocin herpes virus [PhHV]). Stool DNA was extracted with miniMAG or easyMAG (bioMérieux). Assays were validated with regard to selectivity (127 strains), analytical sensitivity (spiked faecal specimens), and clinical performance (851 stool specimens).
Results: Both assays showed 100% selectivity with the tested panel of strains. Analytical sensitivity was in the range of 102–4 CFU/g of stool in both mono- and multiplex approaches. In 281 of the 851 clinical stools, a pathogen targeted by 1 of the multiplex assays was detected by conventional methods (culture or microscopy). Overall, the multiplex assays showed 98.2% concordance in these 281 specimens. In the 570 stools negative for the targeted pathogens by conventional methods, an additional 83 positive results were detected. Furthermore, 13 double infections were detected by the multiplex assays, compared to only 3 by conventional methods. Inhibition of the multiplex PCRs was observed in only 4.85% and 5.43% for the SE/CJ/GL and STEC/SH/EIEC assays, respectively.
Conclusion: Multiplex real-time PCR offers a rapid and sensitive method for the detection of gastro-intestinal pathogens. Multiplexing does not harm the analytical sensitivity if the assay is set-up properly. These multiplex assays will introduce a whole new strategy in screening stool specimens for gastro-intestinal pathogens.
P584 Molecular characterisation of Salmonella typhimurium and Salmonella enteritidis by plasmid analysis and pulsed-field gel electrophoresis
Z. Aktas, M. Day, C. Bal, S. Diren, E. Threlfall (Istanbul, TR; London, UK)
Aims: The aim of this study was to investigate clonal diversity of S. enteritidis and S. typhimurium isolates from various clinical samples in Turkey.
Materials and Methods: Forty-one strains of Salmonella spp. isolates from paediatric patients in Cerrahpasa Faculty of Medicine, Istanbul, Turkey, from 2001 to 2004 were examined for their susceptibility to various antibiotics and the presence of antibiotic resistance genes. Pulsed-field gel electrophoresis and plasmid profiling were used to characterise and determine possible genetic relationships between Salmonella enterica ssp. enterica isolates of clinical isolates.
Results: All the S. enteritidis strains (n = 26) were susceptible to antimicrobial agents tested, whereas one S. typhimurium (n = 15) was resistant to ampicilline, four strains were resistant to ampicilline, chloramphenicol, streptomycin, spectinomycin, sulphonamides and tetracyclines (R-type ACSSpSuT) and the remaining other isolates were susceptible to antimicrobial agents tested. R-type strains were positive for the intI gene and had only one plasmid of 60Mda. Plasmid pattern analysis permitted further differentiation of the S. enteritidis strains into six groups. A serovar-specific virulence plasmid of 38MDa was detected in all of S. enteritidis strains (except one strain). Plasmids, with molecular masses varying between 2.5 and 89MDa, were found in 12 of the 15 of the S. typhimurium strains and five different plasmid patterns were determined. After the XbaI macrorestriction profiles, we observed 23 subtypes which were grouped into five main patterns for S. enteritidis and 15 PFGE profiles were observed among the S. typhimurium strains and four patterns (I, II, III, IV) were found. Plasmids from resistant strains were not transferred by conjugation recipient Escherichia coli cells. Pulsed-field gel electrophoresis and restriction enzyme digestion analysis of DNA revealed different restriction profiles and sizes, indicating these strains usually were not clonaly related whereas MDR- S. typhimurium isolates were clonaly related.
Conclusion: Our study demonstrated the emergence of multiresistant S. typhimurium DT104 infections in our hospital. Therefore, investigation of the antimicrobial susceptibilities, the characteristics of resistant strains and the molecular epidemiology of the strains is more significant. PFGE is more discriminatory and can be used as a confirmatory method.
P585 Comparative study for conjugative plasmids carrying CTX-M genes in Escherichia coli nosocomial isolates
N. Fursova, I. Abaev, O. Korobova, E. Pecherskikh, N. Shishkova, S. Pryamchuk, A. Kruglov, D. Ivanov, L. Weigel, J. Rasheed (Obolensk, Moscow, RU; Atlanta, US)
Objectives: In previous studies we have shown that CTX-M is the most prevalent type of extended-spectrum β-lactamase (ESBL) among recent clinical isolates of Enterobacteriaceae (2003 to 2005 isolates, 14 hospitals across Russian Federation). The blaCTX-M enzymes accounted for 75% of ESBLs. Major groups included CTX-M-1-related enzymes (91%) (in most cases CTX-M-15), CTX-M-9-like (7%), CTX-M-2-like (1%), and a combination of CTX-M-1 and −9-related genes (1%). To determine the potential for spread of CTX-M genes from isolates of Escherichia coli, CTX-M-positive strains were analysed for plasmids, location of CTX-M genes, and the ability to transfer plasmids with CTX-M by conjugation.
Methods: The presence of blaCTX-M-1, blaCTX-M-2, and blaCTX-M-9 genes, as well as blaTEM and blaSHV genes, in ESBL-producing E. coli (n = 161) isolates was analysed by PCR. Conjugations were performed in broth using E. coli C600 (RifRAzR) recipient strain. Antimicrobial resistance phenotypes of donors and transconjugants were determined by disc diffusion and broth microdilution methods. Plasmids were extracted by the alkaline hydrolysis method. blaCTX-M genes were localised to plasmids by DNA-DNA hybridisation using high-sensitive kit Alk-Phos (Amersham). Probes have been generated by amplification of CTX-M genes using universal primers.
Results: Transconjugants were identified for 35 (approx. 22%) of the E. coli isolates including: blaCTX-M-1 gene (27), blaCTX-M-9 gene (7), and blaCTX-M-2 gene (1). Transfer of blaCTX-M-1 was usually observed on the same plasmid with blaTEM. In contrast, the blaCTX-M-9 gene was frequently on a separate replicon from blaTEM (Table 1). In addition to the ESBL genes, transconjugants acquired resistance to doxycycline, gentamicin, amikacin, ciprofloxacin, and sulfonamides (in different combinations). Clinical isolates had 1–12 plasmids of different molecular weights. blaCTX-M genes were consistently transferred on large plasmids (more than 100 Kb).
Donor and transconjugant strains
| Total strains | Successful donors | Transconjugants |
|---|---|---|
| CTX-M1 (n = 128) | CTX-M1 (n = 3) | CTX-M1 (n = 3) |
| CTX-M1+TEM (n = 23) | CTX-M1+TEM (n = 19); CTX-M1 (n = 4) | |
| CTX-M1+TEM+SHV (n = 1) | TEM+SHV (n = 1) | |
| CTX-M9 (n = 29) | CTX-M9 (n = 1) | CTX-M9 (n = 1) |
| CTX-M9+TEM (n = 6) | CTX-M9+TEM (n = 2); CTX-M9 (n = 4) | |
| CTX-M2 (n = 4) | CTX-M2 (n = 1) | CTX-M2 (n = 1) |
Conclusion: Clinical isolates of E. coli isolated in the Russian Federation harbour CTX-M-type ESBLs that are located on high molecular weight conjugative plasmids. In many cases genes encoding resistance mechanisms to several groups of antimicrobial agents were located on the same plasmid capable of horizontal transfer between bacterial strains. These data suggest that the CTX-M-type enzymes, along with multidrug resistance, have the potential to become a widespread problem in this region.
P586 Phylogenetic groups, antibiotic susceptibility, biofilm formation and PFGE in Escherichia coli from community-acquired cystitis
K. Ejrnaes, A. Reisner, B. Lundgren, S. Ferry, S. Holm, T. Monsen, R. Lundholm, N. Frimodt-Moller (Copenhagen, Lyngby, Hvidovre, DK; Umeå, SE)
Objectives: To study Escherichia coli (EC) isolates from community-acquired cystitis with respect to phylogenetic groups, antibiotic susceptibility, biofilm formation and PFGE.
Methods: A subgroup of 243 out of 1162 women with community-acquired cystitis from a placebo-controlled comparative study of three different dosing regimes of mecillinam was studied. We stratified patients into two groups: 1) The mecillinam group (MG), all treated with mecillinam and all with EC at inclusion: a group of all those having EC at follow-up, plus a randomly selected group having negative culture at follow-up (N = 160); and 2) the placebo group (PG), all treated with placebo and all with EC at inclusion: a randomly selected group having EC at follow-up and a randomly selected group having negative culture at follow-up (N = 83). The primary infecting ECs were studied. Susceptibility to mecillinam was tested by agar dilution; MICs of ampicillin, cefpodoxime, chloramphenicol, ciprofloxacin, gentamicin, streptomycin, sulfamethoxazole, trimethoprim and tetracycline were tested with Sensititre. Phylogenetic grouping was determined by a triplex PCR assay. Biofilm formation was measured in 3 media (static growth, 48 hours, crystal violet staining). PFGE was done with XbaI to the primary infecting EC and EC from follow-up.
Results: Only 20% of the strains were resistant to one or more antimicrobials. Susceptibility was significantly associated with phylogenetic group B2 for ampicillin (P = 0.048), chloramphenicol (P = 0.002), streptomycin (P = 0.001), sulfamethizole (P = 0.006), trimethoprim (P = 0.013) and tetracycline (P = 0.003). Resistance to 3 or more antimicrobials was significantly associated with phylogenetic group A (P = 0.033) and D (P = 0.014). Resistance to less than 3 antimicrobials was significantly associated with B2 (P < 0.0001). EC causing relapse/persistence had a higher biofilm formation than those causing reinfection/cure in 2 of 3 media (P = 0.002 and P = 0.011) in the PG, but there was no significant difference in the MG.
Conclusion: Although representing a low-antibiotic-consumption area (Sweden) with low resistance rates, still, antimicrobial susceptibility was significantly associated with phylogenetic group B2 and resistance to 3 or more antimicrobials with A and D. EC causing relapse/persistence had a higher biofilm formation capacity than those causing reinfection/cure in the PG.
P587 Thermophilic helicase-dependent isothermal DNA amplification for molecular detection of Helicobacter pylori
P. Gill, H. Abdul-Tehrani, A. Ghaemi, T. Hashempour, A. Alvandi, M. Noori-Daloii (Tehran, IR)
Objectives: Helicobacter pylori is a Gram-negative, pathogenic bacterium, which specifically colonises in the human gastric mucosa. The infection with this microorganism is one of the most prevalent infections in humans and about 50% of the adults in the industry and more than 90% of the population in developing countries are infected. Several PCR-based methods have been described for molecular diagnosis of this bacterium in biological specimens. However, in this study, we designed and developed a novel procedure for detection of ureC gene of Helicobacter pylori, so called thermophilic helicase-dependent isothermal DNA amplification, (tHDA).
Methods: Like PCR, the tHDA reaction selectively amplifies a target sequence defined by two primers. However, unlike PCR, tHDA uses an additional enzyme called thermophilic helicase to separate DNA rather than heat. Since, this DNA amplification is an isothermal technique, as an advantage it does not need any thermocycler. The accuracy of the technique was checked on DNA extracted from pure culture of Helicobacter pylori.
Results: We obtained same results when several experiments were performed on specimens prepared from infected gastric biopsies. All the results were shown the equal specificity and sensitivity for this technique in comparison to PCR.
Conclusion: THDA can be used for molecular detection of Helicobacter pylori more cost-effectively than PCR in developing countries. Further studies are under taken to develop this technique using ELISA and real-time formats.
P588 Phenotypic versus genotypic identification of bacteria in the clinical microbiology laboratory: the possible impact on patient care
A. Adler, V. Temper, A.E. Moses, O. Nahor, O. Zimchoni, C. Block (Jerusalem, Rehovot, IL)
Objective: Phenotypic bacterial identification methods have numerous strengths but often fail because the phenotype may be variable and subject to biases of interpretation. Sequencing of 16S rRNA and other genes is a more accurate and objective method of identification of microorganisms. We report the study of the causes for phenotypic misidentification in four cases of severe bacterial infections and the possible impact of these errors on patients' care in these cases.
Methods: Phenotypic identification was performed using conventional manual methods and commercial identification kits. Genotypic identification was performed by comparing an 800bp amplicon of the 16S rRNA gene to the GeneBank database. The impact of the phenotypic misidentification was assessed by reviewing the patients' files and questioning the Infectious Diseases specialists that were involved in these cases.
Results: The cases summaries, the phenotypic methods and results, the genotypic results and the possible causes for the phenotypic misidentification are presented in the table . The initial misidentification had no apparent effect on the patients' management and outcome in the first two cases. In the third case, it might have led to missed diagnosis of infectious endocarditis, to inappropriate duration of antibacterial therapy and eventually to valvular replacement surgery. In the forth case, the misidentification of Brucella melitensis led to a suboptimal choice of antibacterial therapy that might have led to persistent infection. Also, it led to inadequate safety measures in the laboratory that resulted in laboratory-associated brucellosis in a technician.
| Case summary and Source of isolates | Phenotypic identification | Phenotypic Methods | Genotypic identification | Possible causes for phenotypic misidentification |
|---|---|---|---|---|
| Pus from a brain abscess in a two-year old girl with congenital cyanotic heart defect | Gemella species | Manual methods | Streptococcus intermedius | Human error |
| Blood culture (2 sets) from a 68-year old female patient with metastatic cancer | Listeria grayii | Manual methods and API CORYNE (bioMérieux) | Lactobacillus plantarum | Pseudocatalase activity resembling catalase led to misidentification by commercial kit. |
| Blood and valvular tissue from a 59-year old male patient with infective endocarditis | Pasteurella haemolytica (isolated from blood culture; valvular tissue culture – no growth) | Manual methods, API 20E (bioMérieux) and RapID NF plus (Remel) | Agregatibacter (Haemophilus) aphrophilus (blood culture isolate and valvular tissue) | Erroneous characterisation of the isolate as “glucose non-fermenters” that led to selection of inappropiate identification systems. |
| Blood culture from a 24-year old female patient with fever and persistent bacteraemia | Actinobacillus ureae | VITEK 1 (the isolate was transferred from another hospital) | Brucella melitensis | Inappropriate reliance on automated indentification system. |
Conclusion: Phenotypic misidentification is multifactorial. It may be caused by an unusual phenotype or by erroneous selection or interpretation of commercial identification systems. Since the consequence of misidentification might be crucial, we believe that genotypic identification of microorganisms should be considered in any case of significant infection, when the results obtained by phenotypic methods are ambiguous or a rare organism is identified.
P589 Occurrence of AmpC chromosomally β-lactamase ACT-1 and extended spectrum β-lactamases type TEM, SHV and CTX-M in clinical isolates of Enterobacter cloacae
M. Biendo, C. Manoliu, B. Canarelli, D. Thomas, F. Hamdad-Daoudi, F. Rousseau, G. Laurans, F. Eb (Amiens, FR)
Objectives: The primary purpose of this investigation was, (i) to study the main drug-resistant mechanism, (ii) to determine the genetic relatedness of strains recovered from different separated sites using pulsed-field gel electrophoresis (PFGE) and, (iii) to describe the molecular epidemiology of the outbreaks.
Methods and Results: We tested 70 consecutive nosocomial E. cloacae isolates recovered from patients admitted in Amiens University Hospital (61 patients), Abbeville General Hospital Center (7 patients), and Iasi Pediatric University Hospital [(Romania) (2 patients)]. Based on phenotypic methods, 100% of isolates produced AmpC β-lactamases, 90% produced ESBLs and 10% were ESBL-nonproducers. 100% of AmpC producers carried AmpD enzyme and blaACT-1 while 7.2% harboured blaFOX-5. PCR amplified, 395 pb and 1,1184 pb segments respectively corresponding to β-lactamases with a pI of 9.0 (ACT-1), and 7.5 (FOX-5) respectively according to IEF and sequencing results. 90% of ESBL-producers carried bla TEM-24, bla SHV-4, blaCTX-M-1 and bla CTX-M-9. PCR amplified 972 pb, 785 pb, and 900 pb segments respectively corresponding to β-lactamases with a pI of 6.5 (TEM-24), 7.8 (SHV-4), 8.4 (CTX-M-1) and 8.0 (CTX-M-9). The 7 ESBL-nonproducers harboured blaTEM-1 and blaSHV-1 encoding β-lactamases with a pI of 5.4 (TEM-1) and 7.6 (SHV-1). Four isolates for which the minimum inhibitory concentrations (MICs) of cefoxitin (FOX), cefotaxime (CTX), ceftazidime (CAZ) and aztreonam (ATM) were of >256 ug/mL and those of imipenem (IPM) were between 12–48 ug/mL showed a diminished level or no expression of a 37- and 38 kDa outer membrane proteins.
Conclusion: It is concluded that the high level of resistance to FOX, CTX, CAZ and ATM and the increase of the MICs of IPM for AmpC β-lactamase- and the ESBL-producing E. cloacae isolates studied are associated with porin deficiency. PFGE analysis showed that these isolates exhibited high genetic diversity (39 different pulsotypes). There was great phenotypic heterogeneity (5 different biotypes), 6 β-lactam R-patterns, and 3 aminoglycosides R-patterns.
P590 Internalin gene in natural atypically haemolytic Listeria innocua strains suggests descent from L. monocytogenes
D. Volokhov, S. Duperrier, A. Neverov, J. George, C. Buchrieser, A. Hitchins (Rockville, College Park US; Paris, FR)
The atypical haemolytic Listeria innocua strain, PRL/NW 15B95, was previously shown to contain a gene cluster analogous to the pathogenicity island 1 (LIPI 1) present in the related food-borne Gram-positive facultative intracellular pathogen Listeria monocytogenes, which causes human and animal listeriosis. LIPI 1 includes the hemolysin gene thus explaining the haemolytic activity of PRL/NW 15B95. However, no other L. monocytogenes specific virulence genes were found to be present. In order to investigate whether any other specific L. monocytogenes genes could be identified, a global approach using a Listeria biodiversity DNA array was applied. According to the hybridisation results the isolate was defined as L. innocua strain of serotype 6a containing LIPI 1. Surprisingly, evidence for the presence of the L. monocytogenes specific inlA gene, previously thought absent, was obtained. The inlA gene codes for the InlA protein, which enables bacterial entry into some non-professional phagocytic cells. In depth PCR and sequence analysis of this region revealed that the flanking background of the inlA gene was identical to that of L. monocytogenes serotype 4b isolates. Sequence analysis of the inlA region identified a small stretch reminiscent of the inlB gene of L. monocytogenes. The presence of more than one cluster of L. monocytogenes specific genes makes it less likely that PRL/NW 15B95 is simply a L. innocua strain altered by horizontal gene transfer. More likely the atypical isolate is a relic of the evolution of L. innocua from an ancestral L. monocytogenes, a process already postulated by others.
P591 Denaturing high pressure liquid chromatography to detect changes in faecal flora of chickens
L.P. Randall, N.G. Coldham, M.J. Woodward (New Haw, UK)
Objectives: Recent studies have shown that it is possible to use Denaturing High Pressure Liquid Chromatography (D-HPLC) to monitor population dynamics of bacteria in environments such as seawater and human faeces. Identification of bacteria relies on separation of amplified 16S rDNA fragments by D-HPLC. The aim of this study therefore was to determine if standard HPLC equipment (Agilent A1100 HPLC system) fitted with a Varian Helix DNA column could be used to perform D-HPLC to monitor changes in the population dynamics of bacteria in chicken faeces.
Methods: Methods were based on those of two previously published papers in which D-HPLC was used to analyse communities of bacteria in human faeces or seawater. Each method utilised different primers for amplification of 16S rRNA, but both previous methods used a dedicated D-HPLC machine. In the present study, fragments of 16S rDNA were amplified by PCR from bacterial strains (n = 16) representative of some that would be found in faeces, to determine if the D-HPLC methods were capable of differentiating between strains. This was coupled with construction of distance maps for sequences of the PCR products. Samples of healthy chicken faeces were then spiked with 0, 104 to 109 Salmonella enterica serovar Typhimurium. Bacterial DNA from faeces was extracted using a QIAamp DNA mini stool kit and this DNA was then used as template for amplification of 16S rDNA prior to D-HPLC.
Results: Phylogenetic trees showed that even for the smaller of the two PCR products (194 bp), there were changes in the 16S rDNA sequences between genera, and in most cases, between different species of the same genera for the strains examined. Fragments from most individual bacterial species gave rise to D-HPLC peaks with different retention times, but when amplicons from different bacterial species were mixed, insufficient resolution was obatined. Faecal samples from chicken gave rise to only two or three D-HPLC peaks. However, when these samples were spiked with Salmonella Typhimurium, a new peak was detected with a detection limit of c. 106 cfu per gram of faeces (Figure 1 ).
Figure 1.

D-HPLC profiles of 16S products from chicken faeces.
Conclusion: Standard HPLC equipment and a readily available column can be used to perform D-HPLC to detect changes in bacterial communities in faeces and it was possible to detect the presence of Salmonella in the chicken faecal samples examined. Further work is needed to determine if it is possible to optimise current methods to improve D-HPLC resolution.
P592 Nosocomial outbreak of Sphingomonas paucimobilis bacteraemia in an oncology and haematology unit
A. Kilic, Z. Senses, A. Kurekci, H. Aydogan, A.C. Basustaoglu (Ankara, TR)
Objectives: The genus Sphingomonas is strictly aerobic, Gram-negative, rod-shaped, usually yellow-pigmented, and motile bacterium with polar flagellum. Sphingomonas paucimobilis strains have been isolated from hospital water systems, respiratory therapy equipment, and miscellaneous clinical specimens. Within a period of one month, we isolated six S. paucimobilis strains, including four from blood cultures of four patients' and two from hospital environment specimens from tap water and bathtub in an oncology and haematology unit.
Methods: All strains were identified by conventional methods and further by the API ID32GN system (bioMérieux, Marcy L'Etoile, France) as S. paucimobilis. We described here these strains' molecular epidemiological analyses by pulsed field gel electrophoresis and antibiotic susceptibilities by E-test.
Results: Despite clinical and environmental isolates yielded three different antibiotic resistances and pulsed field gel electrophoresis patterns, all clinical strains had identical by the both methods. We did not isolate clinical strain clone in healthcare workers and environmental samples as a source of infection.
Conclusion: It was concluded that S. paucimobilis strains could cause a potential outbreak in oncology and haematology units. Genotyping by PFGE is a useful identification technique for epidemiological investigation of outbreak caused by S. paucimobilis in oncology and haematology units.
P593 Diversity in the content and arrangement of CTX genetic element among toxigenic Vibrio cholerae strains isolated during 2004–2006 in Iran
B. Bakhshi, M.R. Pourshafie, F. Navabakbar, A. Tavakoli (Tehran, Isfahan, IR)
Objectives: CTXphi is a filamentous, lysogenic bacteriophage whose genome encodes cholera toxin, the primary virulence factor produced by Vibrio cholerae. In regard to the diversity of the organisation of CTXphi, two different organisational patterns have been reported. However, the distribution and temporal changes in the content of CTX genome of the epidemic strains are still under investigation. In this study, we performed a molecular analysis of the CTX prophages in different toxigenic V. cholerae O1 strains of clinical origin which were isolated during 2004–2006 in Iran.
Methods: To assess the diversity of CTXphi, Long-PCR assay was performed for amplification of an approximately 6.8 kb region using primers specific for conserved region of ig-1 which flanks 3′ end of the rstR, and the intergenic region between CTX and RTX (attB2). Amplified fragments were subjected to RFLP analysis.
Results: The organisation of the CTXphi in the 59 V. cholerae O1 strains revealed 3 patterns, with 6.8, 5.5, 2.7kb in 72.9%, 27.1%, and 10.2% of isolates, respectively. RFLP analysis of the 5.5 kb PCR products with EcoRV, ClaI, DraI, XbaI, and BglI revealed that the diversity was in a region between zot and attB2. PCR assays were also performed with the primers specific for ace/attB2, zot/attB2, and ctx/attB2. In all of the samples an approximately 1.3 kb deletion was detected in this region. Diversity in 2.7 kb PCR products was due to different RS1-CTX arrangement which was confirmed by PCR assays using primers specific for rstR/attB2 and ctxA/attB2. Despite of a defect in organisation of CTXphi in 27.1% of isolates, the PCR assays revealed that these isolates also carried intact copies of the CTXphi.
Conclusion: The results obtained by this study may suggest that the acquisition of CTXphi by V. cholerae may have occurred multiple times and have involved several CTXphi genotypes.
P594 Comparison of real-time PCR and direct culture for the detection of Campylobacter spp. from human faecal samples
K. Eastwood, H. Schuster, D. Gascoyne-Binzi (Leeds, UK)
Objectives: Campylobacter spp. are one of the major causes of food-borne illness worldwide. Current culture techniques are slow and labour intensive and the selective nature of media commonly used for isolation in routine diagnostic laboratories, means that several species of Campylobacter could be overlooked. Real-time PCR offers a specific and rapid method for detection of Campylobacter spp. Newer, commercially available extraction kits, can remove inhibition caused by constituents of faecal samples effectively, but inclusion of an internal control removes the risk of false negative results. A real-time PCR method, with an internal control has been developed for use on chicken faecal samples. This study was designed to evaluate the PCR assay on human faecal samples, for use in the routine diagnostic laboratory.
Method: Human faecal samples were selected for testing using the new real-time PCR method, following routine culture in the Leeds Teaching Hospitals diagnostic laboratory. Briefly, this involved extracting DNA from the samples using the QIAamp DNA stool mini kit (QIAgen, UK) before performing the PCR assay on the Mx3000p QPCR system (Stratagene, Europe) using Brilliant R QPCR mastermix (Stratagene, Europe). Taqman probes for Campylobacter spp. and the internal control organism, Yersinia ruckeri, were used for detection.
Results: One hundred eighty seven faecal samples were collected following routine culture. There were 42 Campylobacter culture positive samples and 145 samples negative for Campylobacter on culture. Fifty eight samples were positive for Campylobacter on PCR, and 129 were negative. All of the culture positive samples were also PCR positive. The sensitivity and specificity of the PCR assay in comparison to culture is 100% and 89% respectively, with a negative predictive value of 100%.
Conclusion: The real-time PCR assay is comparable with routine culture methods for detecting Campylobacter spp. from human faecal specimens. The assay may be more sensitive than culture, and would make a useful screening tool.
P595 Evaluation of a new LightCycler approach for detection of diarrhoeagenic Escherichia coli
R. Ljung, J. Eysturskard, B. Olesen, B. Bruun, D.S. Hansen (Hillerød, DK)
Objectives: Modern laboratory diagnosis of infectious gastroenteritis comprises the classical diarrhoeagenic E. coli: verotoxin-producing E. coli (VTEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC) and enteroinvasive E. coli (EIEC).
Our objective was to compare a novel LightCycler (LC) real-time PCR approach to our standard identification performed by a reference laboratory (ref. lab.).
Methods: During 2 two-months periods, October-November 2005 and May-June 2006, stool samples examined for diarrhoeagenic E. coli were investigated by two different approaches: 1) the standard procedure which was sending the sample to a reference laboratory for analysis by conventional multiplex PCR, and 2) a new LC real-time PCR approach, detecting the eae, vtx1, vtx2, LTI, STIa, STIb, ipaH, and 16S rDNA (internal amplification control) genes from over night cultures. We used the 1.5 LC with 4.0 software, Master Mix from Roche Diagnostics, and primers and hydrolysis probes from TIBMolbiol, Berlin. All protocols used were previously described in the literature except for the ipaH and 16S rDNA genes which were developed in our laboratory.
Results: A total of 371 stool samples were analysed. The number of positive samples and patients in our laboratory and at the ref. lab. were 49 and 34, and 35 and 22, respectively. From 9 of the 14 samples initially not found positive at the ref. lab., 8 E. coli strains were isolated that consequently confirmed our results when examined by the reference laboratory. For one patient our results could be verified, as another sample from the patient was found positive at the ref. lab. Four samples could not be verified as we were unable to isolate the bacteria resulting in the positive PCR result. The number of diarrhoeagenic E. coli found in our laboratory and at the ref. lab. (in brackets), respectively, was: 5 (3) VTEC, 6 (5) EPEC, 11 (9) ETEC, 3 (2) EIEC and 24 (16) intimin producing E. coli (eae positive, but not belonging to the classical EPEC O-groups).
Conclusions: A positive rate of 7% shows that detection of the classical diarrhoeagenic E. coli is diagnostically important. Additionally 6% of samples were positive for intimin producing E. coli, whose clinical importens is disputed. Our results demonstrate that the new LC real-time PCR approach is at least as sensitive (and specific) as standard identification in a reference laboratory, identifying totally 40% more positive samples and 55% more patients.
P596 Detection of enteroaggregative Escherichia coli in faecal samples from patients in the community with diarrhoea
C. Jenkins, M. Tembo, H. Chart, T. Cheasty, A.D. Phillips, D. Tompkins, H. Smith (London, Leeds, UK)
Objectives: A PCR assay targeting the aat, aaiA and astA genes, was used to detect Enteroaggregative Escherichia coli (EAEC) in faecal samples from patients with community-acquired diarrhoea. Strains harbouring one or more of these three genes were assessed for their ability to adhere to HEp-2 cell adhesion assay to confirm their EAEC status. The aim of the study was to assess the usefulness of this PCR for detecting typical and atypical EAEC.
Methods: Five hundred faecal samples were analysed for the presence of EAEC, in addition to routine enteric pathogens. Nutrient broths were inoculated with a sweep of mixed colonies from the MacConkey plates of faecal cultures, and examined for aat, aaiA and astA genes by PCR. In samples of mixed colonies with positive PCR result, a pure colony pick containing either the aat, aaiA or astA genes was obtained from the original MacConkey plate. All single colony isolates were examined by the HEp-2 cell adherence assay, biochemically confirmed as E. coli and serotyped.
Results: The aat, aai or astA gene was found in E. coli isolates faecal from 39 (7.8%) of 500 patients and 20 of these strains adhered to HEp-2 cells in a pattern characteristic of EAEC. Eight isolates carrying the aai or astA gene but not the aat gene were shown to be HEp-2 cell test-positive although 12 strains with this genotype were HEp-2 cell test-negative. Using the HEp-2 adhesion assay as the gold standard, the addition of primers detecting aaiA and astA to the aat PCR increased the number of EAEC isolates detected but identified strains of E. coli that were not EAEC.
Conclusions: The variety of genotypes exhibiting aggregative adherence highlights the problems associated with developing a molecular diagnostic test for EAEC. Our PCR assay detects a variety of strains exhibiting characteristics of the EAEC group making it a useful tool for identifying both typical and atypical EAEC.
Molecular diagnosis of respiratory viruses
P597 Mixed viral infections in hospitalised children with RSV bronchiolitis
V. Poga, K. Kallergi, A. Kossivakis, A. Pangalis, V. Syriopoulou, M. Theodoridou, A. Mentis, M. Giannaki (Athens, GR)
Aim: The detection of mixed infections by Human Metapneumovirus (HMPV), Rhinovirus (RV), Adenovirus (AdV) and Parainfluenza Virus (PIV 1, 2, 3) in children with RSV bronchiolitis.
Material and Methods: The study group included 304 children (7 days – 2 years old) with RSV bronchiolitis, hospitalised in two Athens children's hospitals during the winter period of 2005–2006. RSV antigen was detected by two commercial kits, an immunochromatographic assay (BD) and a direct immunofluorescent assay (Meridian), in nasal aspirates collected in the two first days after admission in the hospital. The same samples were tested for detection of RNA of RSV, HMPV, RV and PIV and DNA of AdV. QIAmp Viral RNA Mini Kit and QIAmp DNA Mini Kit were used for viral RNA and DNA extraction, respectively. Previously published simple and multiplex-nested RT-PCR protocols were used for subtyping RSV A and B viruses and for the detection of HMPV, RV, AdV and PIV 1, 2 and 3 viruses. For the detection of AdV, a PCR protocol was used. All PCR protocols had been optimised before their use.
Results: Two hundred eight samples out of 304 RSV-positive samples were typed as RSVA and 96 as RSVB. Forty nine of the 304 samples, were also positive for at least one of the other viruses checked. HMPV was detected in 19 samples (6.3%), RV in 13 (4.3%), AdV in 10 (3.3%), PIV1 in 5 (1.6%), PIV2 in 1 and PIV3 in 1 (0.3%). Four mixed infections were observed in December, 8 in January, 17 in February, 13 in March, 5 in April and 5 in May.
Conclusions: Apart from RSV, another virus was also detected in 16.1% of the children with bronchiolitis. HMPV was present in most cases of these mixed infections (6.3%). The peak of these mixed infections occurred in February. The clinical importance of the second virus needs further evaluation.
P598 H5N1 diagnostics: experiences with the NucliSens EasyQ® Influenza H5 and N1 reagents based on a real-time NASBA assay on non-human derived specimens
G. Camenish, R. Hoop (Zurich, CH)
Objectives: The bioMérieux Nuclisens EasyQ® Influenza H5 and N1 reagents allow the parallel amplification and detection of gene sequences of haemagglutinin subtype H5 and neuraminidase subtype N1 of Influenza A viruses from human specimens in a real-time NASBA assay. This evaluation consists of analysis of serial dilutions of a non-human clinical sample of H5N1 as well as a H5 transcript, other Influenza A subtypes and avian paramyxovirus strains.
Methods: Influenza A/Duck/Switzerland/2006/V540 strain was used for serial dilution. RNA was extracted using the RNeasy Mini Kit (Qiagen). The extracted RNA was then submitted to H5 and N1 real-time NASBA amplification and detection on the bioMérieux Nuclisens EasyQ® instrument using the Nuclisens EasyQ® H5 and N1 reagents in combination with the Nuclisens EasyQ® Basic Kit v2 following the manufacturer's instructions (bioMérieux). In every run a negative control and a H5N1 positive control (provided with the kit) were included. Obtained data were analysed with the Nuclisens EasyQ® Analysis software which is part of The Nuclisens EasyQ® instrument.
Results: The H5N1 strain was detected down to dilution 10−4 for H5 or 10−6 for N1 genes, respectively. HA and NA subtypes other than H5N1 (H6, H7, H9, N2, N6, N7, N9) were not amplified. However, two H5N9 strains could not be detected. Finally, two avian paramyxovirus-1 strains did not show amplification.
Conclusion: This study reveals a high sensitivity of the Nuclisens EasyQ® Influenza H5 and N1 reagents on avian derived specimens. The assay proves to be highly specific for the actually circulating asian clade of H5N1. The NASBA EasyQ® H5N1 assay provides to be a suitable platform for diagnosis of H5N1 not only for human derived probes.
P599 Use of avian influenza genodiagnostics in the Republic of Kazakhstan
S.U. Mizanbayeva, K.S. Ospanov, S.L. Yingst, A. García-Sastre, M.O. Favorov, S.V. Kazakov, V. V. Zeman (Almaty, KZ; Cairo, EG; New York, US)
A case of mass wild bird die-off was recorded in March 2006 at the Caspian seacoast close to Aktau, the Mangustau Oblast seat, West Kazakhstan. As a disease control measure, a number of samples were collected to investigate the presence of influenza A virus H5 subtype.
Objectives: Determine the possible presence of influenza A virus H5 subtype in:
-
1
Samples obtained from patients who could have had physical contact with wild birds and were hospitalised at the Aktau infectious disease hospital with the flu, pneumonia or acute respiratory viral infection symptoms; and
-
2
Samples obtained from dead wild birds found in the vicinity of Aktau.
Methods: An RT-PCR method was used for the detection of the influenza A virus H5 subtype RNA. The reaction protocol and Flu H5+ 1456 and Flu H5–1685 primers used in the study were provided by NAMRU-3 laboratory, Egypt, Cairo. For verification and detection of the influenza A virus RNA, and concurrent identification of the H5 subtype, we used the AmpliSens Influenza Virus A-H5/H7 test system provided by the Epidemiological Research Institute, Moscow.
Samples Studied:
-
–
12 samples obtained from the tracheas, lungs, intestines, and brains of 3 dead swans;
-
–
1 blood sample obtained from a live swan;
-
–
4 nasopharyngeal swabs obtained from patients presenting symptoms of acute respiratory viral infection or flu;
-
–
16 blood serum samples obtained from patients hospitalised in the pulmînology unit of the Aktau infectious-disease hospital.
Results: The RT-PCR method (based on the use of the NAMRU-3 lab, Cairo, Egypt primers) was negative for influenza A virus H5 subtype in the 16 blood serum samples and 4 nasopharyngeal swabs obtained from patients. RNA from influenza A virus H5 subtype was found in the samples obtained from tracheas, lungs, and intestines of all three birds (swans). Identical results were obtained using the NAMRU-3 lab, Cairo, Egypt primers and the test system provided by the Epidemiological Research Institute, Moscow.
Conclusion: Owing to the RT-PCR method we were able to detect the presence of the influenza A virus H5 subtype among the wild birds in the Mangustau Oblast, the Republic of Kazakhstan.
The results obtained using the NAMRU-3 lab, Cairo, Egypt primers and the AmpliSens Influenza Virus A-H5/H7 test system provided by the Epidemiological Research Institute, Moscow were 100% identical. (This project was supported by the US Department of Defense, Biological Threat Reduction Program, Project #KZ-27.)
P600 Typing and distribution of HCV strains in South Italy (Apulia region) by a cost-effective direct sequencing test
L. Tagliaferro, P. Menegazzi, E. Reho, O. Varnier (Lecce, IT)
Objectives: Standardisation of a rapid, direct and cost-effective test for typing of HCV strains, by sequencing from real time PCR amplicons. Evaluation of epidemiological distribution of HCV isolates in the population of a southern region of Italy (Apulia).
Methods: HCV strains typing is performed by direct sequencing of purified amplicons, using the OpenGene System (Visible Genetics, Bayer diagn.) after removal of FRET probes.
We amplified the HCV 5′ UTR region by an “in house” rapid, single tube LightCycler Real Time PCR with FRET technology. The accuracy and the reproducibility of this test have been confirmed by the very low crossing point coefficient of variation and standard deviation values (3.55 and 1.03, respectively) of the used international WHO HCV human reference standard plasma (HCV Accurun series, BBI Inc.), calculated on more than 4,500 samples tested in 359 runs.
Results: A total of 490 HCV LightCycler amplicons were sequenced using OpenGene System from October 2002 to November 2006. We observed 5 of the 6 wide-world known principal HCV genotypes, identifying 16 types and subtypes, with a homology average of 99.9%. Along with genotypes 1–4, HCV 5 has been found in Italy as well. The sequencer has been able to distinguish 60 different HCV isolates; 186 of 490 samples (38%) have been typed but not subtyped, because of the presence of some undistinguishable sequences, common to several strains.
Conclusions: This HCV sequencing test revealed an interesting cost effectiveness, saving a lot of time and money, and was able to subtype the 62% of observed HCV genotype, with a very high percentage of sequence homology (99.9%). In Apulia region (southern Italy) the most frequently observed genotypes have been 1b (35%) and 2 (28%), with a prevalence of the K0014, HCJ5 and S83 isolates. A different subtypable region and an External Quality Assessment (EQA) programme for HCV typing would be a needed addition.
Biofilms
P601 Ethanol lock therapy: preliminary trial results and future research agenda
J. Broom (Brisbane, AU)
Objectives: Pilot study: Primary aim: To evaluate the safety and efficacy of ethanol lock therapy (in conjunction with intravenous antibiotic therapy) in the treatment of infected tunneled central venous catheters. Prophylaxis study: Develop a clinical trial protocol to assess the efficacy of 70% ethanol lock therapy in prevention of tunneled central venous catheter-associated blood stream infections.
Methods: Pilot study: A prospective non-randomised trial of 70% ethanol locks to treat tunneled central venous catheter-associated blood stream infections was performed in nineteen patients.
Proposed prophylaxis study: We have designed a prospective, randomised, controlled trial of the weekly instillation of a 70% ethanol lock vs sterile heparin saline lock in patients requiring long-term haemodialysis via a tunneled central venous catheterm, to assess the efficacy of ethanol in preventing development of catheter-associted blood stream infections. 56 patients will be recruited to each arm.
Pilot study: These patients had a broad range of pathogens isolated. 12/17 patients completing ethanol lock therapy retained their catheter for greater than 14 days after the initiation of ethanol lock therapy (70% cure rate). 5/17 patients did not retain their catheter for 14 days, but 3 of these patients had their central venous catheter removed for reasons not attributable to recurrence of infection.
Proposed prophylaxis study: Results from the prophylactic study outlined above will demonstrate whether ethanol lock therapy is a safe and effective prophylactic intervention in patients requiring long-term haemodialysis.
Conclusions: Ethanol lock therapy seems effective in treatment of infected central venous catheters but it remains to be demonstrated to be effective in prophylaxis. Patients requiring long-term central venous catheterisation are a diverse group with a broad range of medical conditions. Further larger studies need to be designed to tease out the benefits of ethanol lock therapy these diverse specialty areas.
Results from this proposed prophylaxis trial will determine whether larger studies in more diverse patient groups are warranted to assess ethanol lock therapy.
P602 Analysis of the effect of selected antiseptics and antibiotics on the survival of planktonic celles and biofilm cells
M. Bartoszewicz, A. Rygiel, A. Przondo-Mordarska (Wroclaw, PL)
Objectives: In the clinical practice, infections of biomaterials are still a growing medical and economic problem. The infections are caused both by endogenous bacterial flora and by nosocomial bacteria. The planktonic cells initiate contact with the surface of a catheter or implant. After adhering to the surface, the bacteria start the production of an extracellular slime forming the biofilm. The biofilm structure is not homogenous and the phenotypic and biochemical properties of the organisms forming it are different than those of their planktonic counterparts. The above is the reason why biofilm is very resistant to antibacterial agents. The objective of the paper was to analyse the effect of selected antibiotics and antiseptics on the degree of formation and reduction of biofilm on polystyrene plates.
Methods: A collection of strains from the Department and Institute of Microbiology at the Wrocław Medical University was used isolated from hospitalised patients suffering from generalised catheter-related infections and orthopaedic implant infections as well as a model Staphylococcus epidermidis ATCC 35984 strain. Bacterial survival in the biofilm following the application of an antiseptic or an antibiotic was tested using the microdilution method in microtiter plates (CLSI) and was read as MIC (Minimal Inhibitory Concentration). The following solutions were used for antiseptics testing: 0.1% octenidine dihydrochloride, 7.5% iodine complexed with polyvinylpyrrolidone. Antibiotics were tested using vancomycin, clindamycin, ampicyline, gentamicin and erytromicin in substantia
Results: In the case of antibiotics, despite the strains' sensitivity in planktonic cultures, after transformation into a biofilm structure, the bacteria demonstrated resistance to the tested antibiotics and the minimal inhibitory concentrations were as much as 100 times higher than the therapeutic dose possible to be given to the patient.
As for antiseptics, the lowest minimal inhibitory concentrations in the biofilm, in comparison with the planktonic culture, were observed with respect to the tested strains for octenidine dihydrochloride, while the highest ones for the analysed complexed iodine.
Conclusion: in the tested scope, octenidine dihydrochloride proved to be the most effective agent with respect to biofilm-forming bacteria, while the tested antibiotics were not able to penetrate the biofilm structure to the extent comparable with any of the antiseptics.
P603 Detection, quantifiation and investigation of virulence potential of dental-plaque formers by optic and scanning electron microscopy and microbiological assessment tools
V. Lazar, C. Balotescu, M. Bucur, C. Dragomir, M. Burlibasa, B. Savu, T. Traistaru (Bucharest, RO)
The aim of the present study was to investigate the dental plaque formed on natural teeth surfaces by optic and scanning electron microscopy (SEM), to quantify the microbial density by viable cell counts, to identify the microbial strains recovered after culture and their pathogenicity features.
Material and Methods: Dental plaque specimens were collected from 40 patients in duplicates. One set was fixed on coverslips for SEM, and the second was suspended in phosphate buffered saline and used for further qualitative and quantitative microbiological tests (viable cell counts, microbiological automatic identification and antibiotic susceptibility testing by VITEK system, adherence and invasion capacity on HeLa cells by Cravioto adapted method, adherence on prostetic substrata used in oral medicine by original experimental models for in vitro biofilm development and by slime test, production of extracellular enzymes and exotoxins (haemolysins and other pore-forming toxins, amylase, mucinase, gelatinase, caseinase, aesculin hydrolysis).
Results and Discussion: The scanning electron microscopy revealed a very complex and highly organised architecture of dental plaque (nse masses of microorganisms embedded in a microbial mathrix, biofilm thickness from 50 to 133 micrometers, presence of columns and canalicular system). The qualitative analysis of microorganisms in dental plaque by direct optic examination of Gram-stained smears showed a great diversity of morphological types in 82.5% of cases, with the constant presence of micellian hyphae, the rest of 17.5% being monomorphous (Actinomycetae/Gram-negative cocobacilli). Two non-cultivable spirochetae were present in 12.5% of cases. The quantitative analysis of the dental plaque revealed comparative levels of microbial densities (from 2.4×102 to 6.8×103 CFU/mL). Despite the great diversity of the morphological types observed at direct microscopic examination of the specimens, a maximum of 3 different strains/specimen were recovered after cultivation in aerobic conditions, aspect that is accounting for the great value of direct examination in the investigation of the dental plaque. Out of the total number of 50 microbial strains recovered from the analysed specimens, 50% exhibited ability to adhere to three different polimeric inert substrata used in oral medicine. In exchange, they showed reduced adherence and invasion capacity of HeLa cells, as well as scared expression of soluble enzymatic factors.
P604 Investigation of the antimicrobial activity of different antibiotics on monospecific biofilms developed in vitro by microbial strains isolated from cardiovascular devices associated infections
V. Lazar, C. Balotescu, M. Bucur, O. Banu, G. Dobrin, C. Bleotu, B. Savu, I. Sandulescu, I. Stanciu, R. Cernat (Bucharest, RO)
The aim of this study was to investigate the structure of natural biofilms developed on cardiovascular devices by optic and scanning electron microscopy (SEM), to isolate de biofilm former strains and to reproduce the development of artificial monospecific biofilms on correspondent sterile devices using three in vitro experimental models in order to select the most effective type of treatment in controlling biofilm formation on these devices.
Material and Methods: 31 cardiovascular devices (24 central venous catheters, 3 aortic valves, 3 draining tubes, 1 arterial catheter) taken from patients submitted to cardiovascular surgery were examined by optic microscopy and SEM and seeded on sheep blood agar and nutrient broth. The antibiotic susceptibility of planktonic cells recovered after cultivation was determined by disk diffusion and MICs were established by broth microdillution method. The antibioresistance of adherent cells was tested by three experimental models for in vitro biofilm development: adapted disk diffusion method with bacterial cells embedded in the agar mathrix, development of biofilm on small, sterile device pieces immersed in nutrient agar and inclusion of bacterial cells simultaneously with different antibiotic concentrations in agar mathrix allowing the assessment of cel viability.
Result: The direct optic examination exhibited low predictibility of samples positivity. The isolated microorganisms were: Staphylococcus (S.) epidermidis (3), Acinetobacter baumannii (2) S. aureus (1), Proteus mirabilis (1) and Klebsiella penumoniae (1). All strains proved to be very resistant to all tested antibiotics in planktonic state, excepting colistin, exhibiting low MICs from 0.25 to 2 μg/mL. We further tested the efficiency of colistin on biofilm growing bacteria.
The results demonstrated that adherent bacteria exhibited a higher resistance to colistin (reduced diameters of inhibition zones and MICs 4 to 8 times higher than their planktonic counterparts). However, colistin proved a good penetration into biofilm as demonstrated by the altered structure and reduced thickness of the biofilm revealed by SEM and by loss of cells viability. Our results demonstrate the utility of the biofilm development experimental models in the prediction of the effective antimicrobial agent against biofilm growing bacteria and the utility of colistin in the treatment of Gram-positive as well as Gram-negative biofilm associated infections.
P605 Bactericidal effect and diminution of biofilm using Endox endodontic system
C. Cassanelli, S. Roveta, A. Marchese, E. Debbia (Genoa, IT)
Background: Endox is an instrument used in the endodontic treatment; its action is based on the formation of electromagnetic field created by high frequency alternated current. The aim of this study is to evaluate the role cetrimide 125 mg/l when bacteria are treated with Endox.
Materials and Methods: 0.1 ml of cetrimide solutions 125 ã in NaCl 0.08 M is added to 0.1 ml of suspension Enterococcus faecalis ATCC 29212 biofilm producers cultured in the micro-plates for 48 hours all the cultures were treated with Endox in the presence or not of cetrimide. Quantitative evaluation of biofilm was carried out by measuring the absorbance of the solution (A600). Survivors were determinated by CFU/mL.
Results: The decrease of bacterial population with cetrimide was 92%, while with cetrimide and Endox was 99.99%, The consolidated biofilm was reduced 21.2% using cetrimide in conjunction with Endox, 16.2%with Endox and 9.59% using cetrimide alone.
Conclusions: Present finding indicated that cetrimide might play a role in reducing the consolidated biofilm in culture exposed with Endox.
P606 In vitro biofilm-forming ability and antimicrobial resistance of enterococci from intensive and extensive farming boilers
V. Santos, M. Oliveira, A. Fernandes, C. Carneiro, S.F. Nunes, F. Bernardo, C.L. Vilela (Lisbon, PT; Cambridge, UK)
Objective: Enterococci remain one of the major broiler intestinal colonisers. Antimicrobial exposure may select for resistance that may be spread in and outside the farm environment. Biofilm is a recognized virulence factor that facilitates persistence in the host, immune evasion and bacterial survival at high drug concentrations. This work investigated the relation between biofilm-forming ability and antimicrobial resistance from enterococci field isolates from broilers.
Methods: Biofilm production and antimicrobial resistance of 23 isolates from boiler faecal samples from intensive (n = 6) and extensive (n = 17) farming were evaluated. Isolates were identified as Enterococcus faecalis (n = 6), E. faecium (n = 15), E. durans (n = 1) and E. gallinarum (n = 1). Direct observation of biofilm in bacterial suspensions was performed by Fluorescent In Situ Hybridisation (FISH), using two 16S rRNA oligonucleotide probes (Jansen et al., 2000; Martins da Costa et al., 2006). Minimum Inhibitory Concentrations of vancomycin (VAN), enrofloxacin (ENR), oxytetracyclin (TET) and gentamicin (GEN) were determined by broth microdilution (Clinical Laboratory Standard Institute guidelines). Associations between biofilm production and antimicrobial resistance and between the farming system and biofilm production or antimicrobial resistance were evaluated (Friedman and Mann-Witney Tests).
Results: According to the FISH method, 34.78% of the enterococci isolates could produce biofilm (26.08% from intensive and 8.69% from extensive farming). None of the isolates was resistant to VAN. In extensive farming, resistance to ENR, TET and GEN was found in 4.34%, 21.73%, and 13.04% of the isolates, respectively, while in intensive farming a higher level of resistance was observed (4.35%, 39.13%, and 60.87%, respectively). No significant association was found between biofilm production and antimicrobial resistance nor between farming type and biofilm production or antimicrobial resistance (P > 0.05).
Conclusion: Results suggest that poultry may be colonised by biofilm-producing and antimicrobial resistant enterococci, independently of the farming system. In vitro drug resistance could not be related to biofilm production, re-enforcing the genetic basis of resistance. Nevertheless, biofilm production should be further investigated, since it may hamper therapy, requiring higher antimicrobial concentrations and increasing horizontal gene transfer for resistance.
P607 Bacterial colonisation of original synthesized biomaterials in in vivo examinations
A. Reinis, J. Kroica, J. Vetra, A. Skagers, V. Kuznecova, R. Cimdins, L. Berzina, D. Rostoka (Riga, LV)
Objective: To examine the minimal infective dose of S. epidermidis on different biomaterials in laboratory animal models.
Materials and Methods: Originally synthezised biomaterials – 4N bioactive glass and 4NK bioceramics were contaminated with S.epidermidis strain ATCC 12228 in concentrations 100 CFU/mL and 1000 CFU/mL and incubated for 2 hours. After incubation period non-attached bacteria were removed and contaminated biomaterial discs were inoculated in animal models subdermali on left subscapular region for 2 weeks. After 2 weeks incubation in rabbit model, discs were removed, sonicated (1 min) and vortexed (1 min). Samples were cultivated on TSA to estimate the number of CFU per 1 mm2 on the surface of both biomaterial discs.
Results: Non-contaminated biomaterial discs (4N, 4NK) as well as contaminated with S. epidermidis in concentration 100 CFU/mL remained sterile. The same as bioactive glass disc 4N with S. epidermidis in concentration 1000 CFU/mL remained sterile. The colonisation intensity of the bioceramic glass (4NK) discs in concentration 1000 CFU/mL was 0.04 CFU per mm2.
Conclusions: Different biomaterials have variable attachments of S. epidermidis in laboratory animal models. The minimal infective dose for bioactive glass 4N is 1000 CFU/mL, for bioceramic glass 4NK more than 1000 CFU/mL.
P608 Effect of antibiotics at subMIC concentration on biofilm formation by Streptococcus pyogenes
L. Baldassarri, S. Recchia, R. Creti, M. Imperi, M. Pataracchia, G. Orefici (Rome, IT)
Objectives: To determine the effect of subMIC concentrations of antibiotics on biofilm formation by Streptococcus pyogenes.
Methods: A small collection of strains were chosen on the basis of the genetic determinants for antibiotic resistance they carried (i.e. susceptible, mefA+, or erm(A)-erm(B)+ strains). Four strains for each category were selected. SubMIC concentrations were determined for penicillin, clyndamycin and erythromycin by the microbroth dilution methods. Ninety six wells microtiter plates were filled with serial dilutions (1:2) of a given antibiotic; each strain was inoculated in triplicate for each antibiotic concentration, and allowed to grow for 24 hrs. At the end of the incubation period, absorbance at 630 nm was determined along with CFU counts. Plates were then emptied, dried, and the biofilm deposited on the well's bottom stained with crystal violet. Optical density at 570nm was measured and a biofilm index generated in consideration of different growth rates.
Results: We had already determined that a majority of S. pyogenes strains from a variety of sources are able to form biofilm. In particular, susceptible strains formed thicker biofilm compared to macrolide-resistant strains. In this study we found that subMIC concentration of penicillin, but not erythromycin or clindamycin, were able to stimulate biofilm formation only in susceptible strains. Increase in biofilm index in three of the four susceptible strains examined reached up to 150%, 176% and 585% of the control, respectively. Biofilm formation was not affected in those strains carrying either genetic determinants of antibiotic resistance.
Conclusion: Data obtained in this study confirm that biofilm may represent a way for S. pyogenes to escape antimicrobial treatment even when strains lack the genetic determinants for antibiotic resistance. Penicillin treatment represents the drug of choice for eradication of S. pyogenes in patients with recurrent infections and/or carriers. It should however be considered that a percentage of treatment failures might still be possible in view of this secondary effect of penicillin.
P609 Negative correlation between biofilm formation and antibiotic sensitivity in clinical isolates of clonally related Acinetobacter baumannii
G. Fugazza, P. Landini, R. Migliavacca, M. Spalla, E. Nucleo, A. Navarra, R. Daturi, L. Pagani (Milan, Pavia, IT)
Objectives: Acinetobacter baumannii has emerged worldwide as an important nosocomial pathogen and an increasing number of outbreaks, mainly in ICUs, caused by multidrug resistant (MDR) strains, has been reported over the last years. Since biofilm formation by pathogenic bacteria might increase resistance to antimicrobial agents, we investigated the possibility that biofilm formation might be a resistance factor in A. baumannii.
Methods: A. baumannii analysed in this study included 35 clinical isolates, collected from 2 different hospitals in Northern Italy. Identification and susceptibility testing were carried out following standard procedures. Genotyping was performed by REP-PCR and PFGE analysis. Biofilm formation was tested in two different growth media: M9GSup, a defined growth medium with glucose as main carbon source, and LB, a rich, peptone-based medium. In addition, two different growth temperatures were tested: 37°C (host temperature) and 30°C (sub-optimal growth temperature).
Results: All isolates belonged to the same DNA group and showed either identical or highly similar profiles. Consistent with the identification results, all strains displayed the same MDR phenotype, but were sensitive to both tetracycline and imipenem. Four strains, chosen as representative, were tested for biofilm production and found capable of efficient biofilm formation, thus suggesting that production of adhesion factor is well conserved in this clone of A. baumannii. However, biofilm formation was greatly favoured when bacteria were grown in M9GSup; in contrast, little biofilm formation was observed in LB, possibly suggesting that adhesion factor production might be stimulated by growth on glucose. Growth at 30°C resulted in slight stimulation of biofilm formation compared to growth at 37°C in both media. Interestingly, MICs were roughly 4-fold lower in M9GSup medium for both tetracycline and imipenem, suggesting that sensitivity to antibiotics was actually increased in conditions favouring biofilm formation.
Conclusions: Our results suggest that biofilm formation by A. baumannii does not play a major role in antibiotic resistance. Increased sensitivity to imipenem in M9GSup medium is consistent with the reported bactericidal effect of this antibiotic on slow-growing bacterial cells. Future experiments will allow us to assess sensitivity of A. baumannii biofilm cells to imipenem, in order to evaluate its therapeutic potential against biofilm-related infections.
P610 Biofilm formation in nosocomial pathogens of respiratory tract
V. Hola, F. Ruzicka, R. Tejkalova, M. Votava (Brno, CZ)
Chronic infections caused by biofilm-forming bacteria represent serious medical problem nowadays. Their higher prevalence is associated with more frequent use of artificial implants and medical devices, more frequent invasive manipulation and higher number of immunocompromised patients. The problem is much bigger on the ICUs, where the number of immunocompromised patients is even higher. The biofilm-positive bacteria take advantage of the suppressed immunity. The artificial surface of used implants facilitates adhesion of bacteria, which than form biofilm.
The aim of this study was to compare the ability to form biofilm in two groups of bacteria – bacteria colonising respiratory tract and bacteria causing nosocomial infection of respiratory tract. We collected 448 strains of bacteria (ICU patients, collected from January 2006 to October 2006) which we divided into above-mentioned groups and determined to the species level. Most of them were Gram-negative non-fermentative bacteria, especially Pseudomonas aeruginosa and Acinetobacter baumannii-calcoaceticus group.
In all bacteria we assessed the ability to form biofilm by the modification of Christensen microtiter-plate method. The biofilm was grown on tissue culture microtiter plates. Each strain was cultivated simultaneously in 4 wells and the average optical density was assessed. The results were assessed statistically by the Two-sample analysis (programme R 2.1.1) and General Linear Models (programme Canoco 4.5). In the group of bacteria causing nosocomial infections the biofilm-forming ability was present significantly more often (p < 0.05).
There were differences in spatial arrangement of the ability to form biofilm as the upper and lower respiratory tract is concerned. The differences the biofilm-forming ability in particular species was not statistically significant.
Since the biofilm-forming bacteria are difficult to eradicate with antibiotics and often cause chronic infections, their higher prevalence in bacteria causing nosocomial respiratory tract infections may be very problematic and the regular exchange of tracheostomic tubes and catheters is recommended.
This work was partially supported by the grant MPO – Tandem FT-TA3/098.
P611 Bacterial biofilms in patients with chronic rhinosinusitis
E. Dworniczek, M. Fraczek, J. Kassner, R. Adamski, A. Seniuk, I. Choroszy-Król (Wroclaw, PL)
Objectives: Microbial biofilms that are formed on human tissue surfaces play a role in many chronic diseases. Existing in a biofilm phenotype microorganisms evade host defences and are resistant to systemic and local antibiotic therapy. Biofilms have been implicated in dental and periodontal diseases, chronic tonsilitis, otitis media. We demonstrated that bacterial biofilms are present in patients with chronic rhinosinusitis (CRS). Although many etiological factors contributing to CRS have been described, the role of bacteria is not well definied.
Methods: We reviewed 9 cases of CRS patients using culturing methods, scanning (SEM) and transmission (SEM) electron microscopes. The patients were undergoing functional endoscopic sinus surgery (FESS) or radical antrostomy performed because of failure of past medical therapy. Mucosal specimens and sinus lavage were taken from diseased maxillary sinuses and cultured on media for aerobic and anaerobic bacteria and fungi.
Microrganisms were identified by conventional biochemical tests. The samples were prepared using standard methods for SEM and TEM. Areas of interest were photographed.
Results: Bacterial biofilms observed under electron microscope were detected in 3 patients. Cultures of these specimens contained: Streptococcus spp., Escherichia coli, Klebsiella pneumoniae, Streptococcus agalactiae, Proteus mirabilis. In 2 patients neither bacteria nor fungi were present (negative culture and microscopy). In the samples of 4 patients Gram-negative rods, alpha-haemolytic streptococci, Propionibacterium spp. and Corynebacterium spp. were identified. However no biofilm-like structures were observed under electron microscope.
Conclusion: Biofilms were demonstrated to be present in patients undergoing surgery for CRS. This is one of not numerous documentations of biofilms in association to chronic rhinosinusitis.
P612 Characterisation of bacterial isolates colonising urinary tract catheters
X. Wang, I. Ehren, H. Lunsdorf, A. Fruth, U. Römling, A. Brauner (Stockholm, SE; Braunschweig, Wernigerode, DE)
Objectives: To characterise biofilm and related behaviours of bacteria colonising urine catheters and urine.
Methods: Bacterial isolates were recovered and species identified from urine and urinary catheters samples of 45 patients. All isolates were characterised for biofilm formation and related behaviour such as expression of extracellular matrix components. Serotype was assayed. Catheter samples were investigated by electron microscopy to analyse biofilm formation.
Results: In total 179 bacterial isolates was recovered from urine catheter samples. Most commonly recovered species were coagulase negative staphylococci (CNS), Enterococcus faecalis, Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae and Pseudomonas aeruginosa. Catheter recovered E. coli isolates were significantly more prevalent in patients with prostatic cancer than patients suffering prostatic hyperplasia. Catheter recovered CNS isolates were significantly more prevalent in patients with short-term catheterisation than those with long-term and more prevalence than S. aureus, E. faecalis, Streptococci, E. cloacae and Enterobacteriaceae in patients with short-term catheterisation. 96% of isolates showed biofilm formation in vitro. Isolates of P. aeruginosa demonstrated the highest biofilm formation and adherence capacity among investigated species. E. coli isolates recovered from patients with catheter associated urinary tract infection formed significantly more biofilm and adherence than E. coli isolates causing asymptomatic colonisation. E. coli isolates with UTI-related O-antigen formed significantly more biofilm and adherences than remaining isolates. E. coli isolates with H antigen adhered more and expressed more extracellular matrix than bacteria without H antigen.
Conclusions: Most isolates from catheter can form biofilm in vitro. The capacities to form biofilms contribute to the virulence properties o f P. aeruginosa and E. coli by enabling colonisation of the catheter. Although CNS is commonly found on catheter surfaces it is mainly retrieved during short time catheterisation, which could be due to its inability to persist urine flow and over growth of other bacterial species. The correlation found between E. coli and prostate cancer merits further investigation.
P613 Influence of sub-inhibitory concentrations of antibiotics on biofilm formation by Salmonella typhimurium
J. Majtán, L. Majtánová, V. Majtán (Bratislava, SK)
Objectives: Salmonella typhimurium strains are important food-borne pathogens. Numerous studies have documented the ability of Salmonella spp. to adhere and form biofilms on different surfaces. The aim of this study was to investigate and compare the effect of sub-inhibitory concentrations (sub-MICs) of antibiotics on biofilm formation by clinical S. typhimurium strains.
Methods: The antibiotics used in this study were gentamicin, ciprofloxacin and cefotaxime. Biofilm-forming abilities of three clinical isolates of S. typhimurium (No. 18/06, 41/06, 53/06) in the presence of sub-MICs (1/2, 1/4, 1/8, 1/16 of the MIC value) of these antibiotics were assessed by absorbance at 570 nm of crystal violet-bound cells recovered from 96-well tissue culture plates after growth in TSB growth medium.
Results: The effect of sub-MIC concetrations of antibiotics tested in biofilm formation was determined by the percentage of inhibition of biofilm formation. Each antibiotic had a different effect on biofilm inhibition according to the strain targeted. The most effective in all three strains were sub-MICs of gentamicin in whole concentration range. Sub-MICs of ciprofloxacin expressively inhibited biofilm formation by two strains (41/06 and 53/06). On the other hand cefotaxime markedly stimulated biofilm formation by strain 18/06 in whole concentration range, and by 41/06 and 53/06 strains at 1/2 of the MIC.
Conclusion: These findings showed that gentamicin, ciprofloxacin and cefotaxime influenced biofilm formation by clinical S. typhimurium strains at sub-MIC concentrations. This effect was dependent on the strain and on the type of antibiotic.
P614 Slime production in blood isolates of Staphylococcus aureus under various growth conditions
O. Aslan, B. Aksu, F. Babacan (Istanbul, TR)
Objectives: The production of biofilm represents an important virulence factor of certain strains of Staphylococcus aureus. We aimed to investigate biofilm production of blood-borne S. aureus isolates under different growth conditions.
Methods: Total of 100 blood isolates of S. aureus were included to the study. All the isolates were identified as S. aureus by colony morphology, catalase and tube coagulase reactivity. Slime production of the isolates was detected by cultivation on Congo Red Agar (CRA) plates and quantitative microplate test. Four different Tryptic Soy Broth (TSB) formulas were used in microplates for testing their influence on biofilm production; standard TSB, TSB supplemented with 1% glucose, iron limited TSB (50% less than usual) and iron supplemented TSB (50% more than usual).
Results: Only one isolate was detected as positive with three methods; CRA and quantitative microplate method with standard TSB and iron limited TSB. Of one hundred isolates, 8 were found positive (8%) in glucose supplemented TSB medium which were also found positive in iron limited TSB medium. In addition to these 8 isolates, 37 isolates (45%) were also determined as positive in iron limited medium, but only one isolate (1%) was detected as positive in iron supplemented TSB.
Conclusion: Iron limited TSB medium might be the best approach to evaluate biofilm production of clinical S. aureus isolates.
P615 Adhesion of Staphylococcus epidermidis to a modified cellulose triacetate membrane
C.I. Extremina, P.L. Granja, A. Freitas da Fonseca, A.P. Fonseca (Porto, PT)
Objectives: There is an increase in the use of biomedical materials in modern medicine, despite some problems resulting from this practice. A major drawback are the biomaterial-centred infections. Adhesion of micoorganisms begins when they reach the biomaterial surface. This initial step involves specific interactions between bacteria and surfaces. A high number of the infections affecting medical devices are associated to Staphylococcus epidermidis, which is therefore called an opportunistic pathogen of foreign bodies. The aim of this study was to assess the anti adhesive properties of cellulose triacetate (CTA) membranes incorporating the antibiotic Imipenem in order to prevent Staphylococcus epidermidis adhesion.
Methods: The materials studied were characterised in terms of surface free energy of interaction by contact angle measurements (quantitative measure of hydrophobicity). The antibacterial activity of the materials was assessed in vitro by a modified Kirby Bauer test. The in vitro adhesion of S. epidermidis RP62A expressing capsular polysaccharide/adhesin (PS/A), the most common etiological agent of colonisation of implantable medical devices, to CTA and to CTA with entrapped antibiotic (CTA-Imipenem) was investigated.
Results: The thermodynamic approach showed a good correlation between free energy of interaction between cells, materials' surface and water and bacterial adhesion values. CTA-Imipenem membranes showed an anti-proliferative character, being therefore bacteriostatic. Bacterial adhesion tests showed a statistically significant decrease in the adhesion of S. epidermidis to CTA-Imipenem when compared to its adhesion to CTA alone.
Conclusion: By using the present approach it seems possible to obtain an adequate medical device surface coated with CTA-IMP with anti-adhesive and anti-proliferative properties.
P616 Influence of mutations in genes of the sigma B operon on Staphylococcus epidermidis in vitro and in vivo biofilm formation
V. Pintens, C. Massonet, R. Merckx, J. Van Eldere (Leuven, BE)
Objective: To study the effect of inactivation of the regulatory gene rsbU, the entire regulatory cascade rsbUVW and the entire sigma B operon on Staphylococcus epidermidis biofilm formation in vitro and in vivo.
Methods: Strains used were 8400 (wild type), 8400rsbU (rsbU::erm), 8400rsbUVW (rsbUVW::erm), 8400rsbUVWsigB (rsbUVWsigB::erm), 1457 (wild type), 1457rsbU (rsbU::erm), 1457rsbUVW (rsbUVW::erm) and 1457rsbUVWsigB (rsbUVWsigB::erm) (Mack et al. Infection. Immun. 1992; 60: 2048; Knobloch et al. Infection. Immun. 2004; 72:3838). In vivo, catheter fragments inoculated with the different S. epidermidis strains were implanted subcutaneously in rats as described (S. Vandecasteele et al. Biochem Biophys Res Commun. 2002; 291: 528). Biofilm formation by the different strains was observed after one day in vitro and in vivo. The amount of sessile bacteria in vitro was determined at 0, 2, 6, 24, 48 and 96 hours after inoculation and in vivo catheters were explanted 0, 4, 24, 48, 96, 336 and 504 hours after implantation. The amount of bacteria was determined by Taqman PCR of the housekeeping gene gmk.
Results: After one day as well in vitro as in vivo fewer catheters were infected with rsbU and rsbUVWsigB mutants (78.87%) compared to the wild types and rsbUVW mutants (91.42%). The amount of sessile bacteria was also lower in the rsbU or the rsbUVWsigB mutants. In vitro the difference was significant from 6 hours after inoculation onwards and in vivo from 2 days after implantation onwards. No significant differences were observed between rsbUVW mutants and wild type strains.
Conclusion: Mutation of rsbUVW, a negative regulator of sigma B activity does not increase biofilm formation, relative to the wild type strain. Similar results with regards to number of catheters colonised and amount of sessile bacteria were obtained for wild-type strains and the mutants with inactivation of rsbUVW.
Mutation of rsbUVWsigB reduces catheter colonisation and the number of sessile bacteria, and the same effect was obtained with the rsbU mutants. However, although biofilm formation is reduced, there are still adherent bacteria, suggesting alternative pathways for biofilm formation.
P617 Linezolid inhibits alpha-toxin and biofilm formation in Staphylococcus aureus
K. LaPlante (Providence, US)
Objective: Biofilms are important virulence factors for Staphylococcus aureus. Alpha-toxin (hemolysin) production has been related to the development of biofilms via quorum sensing. By inhibiting alpha-toxin production with antimicrobial agents that target various sites of protein synthesis, we should therefore be able to effect biofilm production.
Methods: We quantified biofilm formation and alpha toxin production in 5 randomly selected biofilm and alpha toxin producing clinical S. aureus strains. We used prototype high-level producers of alpha-toxin and biofilm (ATCC 10832 and ATCC 35556 respectively) as controls. Each isolate was evaluated alone and in the presence of clindamycin, gentamicin, linezolid, tigecycline and vancomycin at ½ he MIC. MIC testing was determined using CLSI methodology. Antimicrobial compounds were evaluated for their ability to influence alpha-toxin and then biofilm production. Alpha-toxin production was quantified by collecting the supernatant of an overnight inoculated broth and assaying the supernatant in the presence of a 2% concentrated sheep blood assay. Biofilm formation was evaluated and quantified using 1 2 the respective MIC of each agent and read using the Calgary Biofilm assay.
Results: Linezolid was the only agent that did not haemolyze sheep blood erythrocytes. There was extensive haemolysis of the erythrocytes in the presence of toxin producing S. aureus and other agents evaluated at 1 2 their respective agents MIC. This may be due to linezolid's ability to inhibit protein synthesis at an earlier point then the other ribosomal targeting agents. We also observed that at ½ the MIC linezolid demonstrated significant inhibitory effects on the production of biofilm (P = 0.002) in S. aureus.
Conclusion: Based upon these data, linezolid has merit to be investigated further as a possible alpha toxin and biofilm deterrent. Not only did this work further validate a link between alpha-toxin and biofilm formation, but at sub-inhibitory concentrations linezolid can significantly reduce alpha toxin and biofilm production when compared to other agents.
P618 The effect of Staphylococcus epidermidis culture supernatants on the biofilm density of other S. epidermidis strains
J.S. Cargill, M. Upton (Cambridge, Manchester, UK)
Objectives: It has been shown that staphylococcal biofilm density can be regulated by the presence of small diffusible molecules, often protein in nature. The aim of the present study was to screen Staphylococcus epidermidis culture supernatants for the ability to affect the biofilm density of other strains of the same organism.
Methods: Fourteen clinical strains of S. epidermidis and the control strain RP62A were characterised as biofilm-positive (optical density (OD) at 450nm above 0.12) or negative. Overnight cultures of the strains were suspended at a dilution of 1:100 in fresh tryptone-soya broth, and 100 microlitres added to the test wells of a sterile, flat-bottomed 96-well plate. Filtered supernatants (100 microlitres, including that from the same strain) were then added to the wells (eight replicates), and the plates incubated for 20 hours. The biofilm density was expressed as a ratio of the OD with that formed by 200 microlitres of the 1:100 dilution (without supernatants). Each combination was repeated twice. Box plots were then used to identify outlying results (greater than one interquartile range above or below the median) which were assumed to show possible supernatant effects. These results were then examined.
Results: All biofilm-positive strains cultured with supernatants (including that of their own strain) showed lower biofilm densities than the control 1:100 dilution. No supernatant was shown to prevent a biofilm-positive S. epidermidis strain from forming a biofilm. No outlying results were seen on the box plots to suggest a significant reduction in biofilm density in any sample. Four outliers were seen with increased biofilm density. One remained a weak biofilm former only. The remaining samples were weakly biofilm-positive in otherwise biofilm-negative strains, although the standard deviation of the eight replicates extended below an OD of 0.12. The results were not reproduced in both tests.
Conclusion: This simple experiment does not suggest that S. epidermidis biofilms are significantly modified by culture supernatants produced by other S. epidermidis strains. Lower biofilm densities are produced by biofilm-positive strains when incubated with any supernatant; this may represent an altered balance of nutrients in the supernatant compared with fresh TSB, or may represent the presence of a general inhibitory effect, such as acid production by S. epidermidis.
P619 Activity of daptomycin on biofilms produced on plastic support by Staphylococcus species
S. Roveta, A. Marchese, G. Schito (Genoa, IT)
Objectives: Antibiotics capable of disrupting or inhibiting the synthesis of biofilms formed by bacterial pathogens may offer therapeutic advantages over molecules without these properties. This study assessed the in vitro activity of the novel lipopeptide, daptomycin, against biofilms produced by staphylococcal species.
Methods: Three recently isolated slime producing strains each of methicillin susceptible (MET-S), methicillin resistant (MET-R) S. aureus and S. epidermidis and two biofilm producing vancomycin intermediate S. aureus (VISA) (Marchese et al., 2000) were tested. Slime formation in 96-well tissue culture plates was quantified spectrophotometrically using a method based on that of Cramton et al. (2001).
Results: Daptomycin at concentrations achievable during therapy (2–64 mg/l) inhibited slime synthesis (>60%) in all strains. Reduction of biofilm production was >80% in both MET-S S. aureus and S. epidermidis, and ranged from 60 to 80% in MET-R S. aureus and from 70 to 95% in MET-R S. epidermidis. At 64 mg/l, biofilm synthesis decreased by 80% in the 2 VISA isolates. Daptomycin also disrupted the biofilm both in initial (5h) and mature (48 h) slimes. Over 50% breakdown of preformed initial biofilm was observed in all strains. Disruption of mature biofilms, however, was more variable (range 20–70%) and was both concentration- and strain-dependent.
Conclusions: Daptomycin promoted a sizable inhibition of slime synthesis and slime disruption in both initial and mature biofilms produced on plastic support by all Staphylococcal strains studied. Biofilm present on tissues during infections is known to be thinner and less organised. If daptomycin interacts positively with the immune defences, its ability to interfere with the physiology of slime in vivo may be further enhanced.
P620 Effect of linezolid and quinupristin/dalfopristin on formation and disruption of Staphylococcus haemolyticus biofilm in vitro
M. Juda, P. Helon, A. Malm (Lublin, PL)
Objectives: Staphylococcus haemolyticus belonging to coagulase negative staphylococci is the common commensal of the skin. This strain may cause bacteraemia, especially in hospitalised patients in the presence of indwelling medical devices. The aim of this study was to assess the effect of linezolid and quinupristin/dalfopristin on formation and disruption of S. haemolyticus biofilm structure on polychloride vinyl (PCV) catheter in vitro.
Methods: Four S. haemolyticus strains were isolated from patients with lung cancer during hospitalisation (three from nasopharynx, one from pleural drain). The routine microbiological methods were used for their isolation and identification. Minimal inhibitory concentration (MIC) of linezolid and quinupristin/dalfopristin was determined by E-test according to criteria of Clinical and Laboratory Standards (CLSI). Modified Richard's et al. method was used to assess of ability for adhesion proccess and biofilm formation on the PCV urological Nelaton's catheter in vitro. For evaluation of linezolid and quinupristin/dalfopristin effect on formation and disruption of S. haemolyticus biofilm structure in vitro following antibiotic concentrations were used: 0.125; 0.25; 0.5; 1.0; 2.0; 4.0; 8.0; 16.0×MIC.
Results: All strains possessed strong ability to adhesion and biofilm formation on the PCV catheter in vitro. Linezolid and quinupristin/dalfopristin inhibited adhesion process in concentrations 0.19–0.375 mg/l (0.25–0.5×MIC) and 0.125–0.75 mg/l (1.0–2.0×MIC), respectively. Linezolid effected formation of biofilm in concentrations between 0.5–0.75 mg/l (0.5–1.0×MIC), while quinupristin/dalfopristin – in concentrations between 0.5–3.0 mg/l (2.0–4.0×MIC). Drug concentrations disrupting the mature biofilm structure was higher: 1.5–2.0 mg/l (2.0×MIC) for linezolid and 1.0–6.0 mg/l (8.0–16.0×MIC) for quinupristin/dalfopristin.
Conclusion: Our data indicate that linezolid was more effecive agent, then quinupristin/dalfopristin in prevention of biofilm formation by S. haemolyticus strains on the PCV catheter and disruption of mature biofilm in vitro.
P621 Prevention of Pseudomonas aeruginosa biofilm formation with antibiotics used in cystic fibrosis patients during early broncopulmonary colonisation
A. Fernandez-Olmos, M. García-Castillo, L. Maiz, A. Lamas, F. Baquero, R. Cantón (Madrid, ES)
Objectives: Early P. aeruginosa isolates colonising cystic fibrosis (CF) airway appear more favourable for eradication with antibiotic therapy than those in chronically colonised patients. We study the antibiotic susceptibilities of non-mucoid P. aeruginosa isolates recovered in early colonisation stages of CF patients and their ability to prevent biofilm formation in these isolates.
Methods: Ciprofloxacin (CIP), tobramycin (TOB), ceftazidime (CAZ) and imipenem (IMP) susceptibility of 27 non-mucoid P. aeruginosa isolates recovered from 18 CF patients with early colonisation were study both using a polystyrene microplate biofilm susceptibility assay (Moskowitz et al. J Clin Microbiol 2004; 42:1915–22) and the standard microdilution method (CLSI). Biofilm was formed by immersing the pegs of a modified microtiter lid into a growth microplate, followed by incubation. Biofilm inhibitory concentration (BIC) was determined by placing the peg lids with the biofilm formed onto microplates containing twofold diluted antibiotics. Biofilm prevention concentration (BPC) was determined after biofilm was formed directly into antibiotic contact. Optical density was measured after 6 and 24 hours of incubation.
Results: CIP, TOB, CAZ and IMP showed, respectively, the following geometric mean values: MIC 1.3, 3.6, 12.1 and 4.9 mg/L; BIC-6-h 2.7, 7.2, 149.3 and 21.8 mg/L; BIC-24-h, 17.3, 37.3, 512.0 and 128.0 mg/L; BPC-6-h, 2.4, 2.7, 42.4 and 5.9 mg/L; BPC-24-h, 10.3, 11.2, 339.5 and 29.6 mg/L. These values showed CIP and TOB similar inhibitory activity when P. aeruginosa growth was either sessile or planktonic at 6-h incubation (BIC/MIC and BPC/MIC ratios of 2x). Higher concentrations were required for all antibiotics to reach BIC and BPC after a 24-h incubation period, with BIC/MIC ratios of 13x, 10x, 42x and 26x for CIP, TOB, CAZ and IMP, respectively, and BPC/MIC ratios of 8x, 3x, 28x and 6x, respectively. In all cases, inhibitory effects required higher concentrations than prevention ones.
Conclusion: Early antibiotic challenged of P. aeruginosa in CF patients might benefit of prevention of biofilm formation, particularly with TOB and CIP. Antibiotic concentration required to prevent biofilm formation was lower than that to inhibit formed biofilm. The BIC and BPC parameters have been postulated in the attempt to correlate in vitro measurements with therapeutic outcomes in CF patients with early P. aeruginosa colonisation biofilm treatment.
P622 Penetration and activity of fosfomycin, ciprofloxacin, amoxicillin plus clavulanic acid and cotrimoxazol in Escherichia coli and Pseudomonas aeruginosa biofilms
J. Rodriguez-Martinez, S. Ballesta, A. Pascual (Seville, ES)
Objectives: The aim of this study was to evaluate the penetration and activity of four oral antimicrobial agents, commonly used to treat uncomplicated UTI, in E. coli and P. aeruginosa biofilms on siliconised latex urinary catheters (SL).
Methods: Strains: E. coli ATCC 25922, ESBL-producing E. coli (urinary clinical isolate) and P. aeruginosa ATCC 27853. Bacterial biofilms: 1 cm in length segments of SL were incubated for 24 h in broth containing 105 cfu/mL of each strain. Segments were washed 3 times with PBS to remove non-adherent bacteria. Segments containing 24 h biofilms were further incubated in broth containing 10×MIC of each antimicrobial for 24 hours. Bacterial survival was determined by sonication of segments, dilution and colony counting. The penetration of antimicrobials into biofilms was also analysed. 24 h biofilms were created on polycarbonate membrane filters and cover with another polycarbonate filter. A disk of known concentration for each antimicrobial was placed on top and removed after 1, 3 and 6 hours. Drug concentrations left in disks were measured using a bioassay method.
Results: No antimicrobial alone was able to completely sterilise the catheter surface. For all of them, the bactericidal activity against 24 h-biofilms was higher than 96%. No differences were observed between ESBL-producing and non-producing E. coli. The kinetics of penetration of the four antimicrobial agents into the bacterial biofilm was similar for E. coli and P. aeruginosa. The penetration of fosfomycin and ciprofloxacin was slightly slower than that of amoxicillin plus clavulanic acid, but still more than 50% of both antimicrobials have accumulated in bacterial biofilms after 6 h incubation.
Conclusions: Fosfomycin, ciprofloxacin, amoxicillin plus clavulanic acid and cotrimoxazole showed high in vitro activity against bacterial biofims. This activity could be partially due to the high accumulation into biofilms.
P624 Production of biofilm and response to oxidative stress in Pseudomonas aeruginosa in dependence on culture media
A. Hostacka, I. Ciznar (Bratislava, SK)
Objectives: P. aeruginosa as one of the most medically relevant biofilm forming bacterium is an important causative agent of many human infections. Effect of six culture media (5 complex, 1 mineral) on formation of biofilm and response to oxidative stress in three clinical P. aeruginosa isolates (pus, ulcer, urine) was evaluated.
Methods: Potential pathogenicity traits of clinical isolates were tested in vitro using assay for biofilm formation (microtiter plate assay with the crystal violet) and for response to oxidative stress evoked by hydrogen peroxide.
Results: The largest biofilm forming ability of the tested strains was observed after pre-incubation of bacteria in tryptone soya broth (TSM) (A550 = 0.374–0.400) or in TSM supplemented with 8% glucose (TSM+GL) (A550 = 0.358–0.384), the lowest level in mineral medium (MM) (A550 = 0.071–0.104). The highest resistance of the tested isolates to oxidative stress was found after cultivation in peptone water (average of the zone of bacterial growth inhibition was 7.5 mm), on the other hand, the highest sensitive cells were observed after incubation in TSM+GL (15.7 mm) and in MM (15 mm). Bacterial cells with the highest as well as with the lowest capacity to form biofilm were in average more sensitive to hydrogen peroxide in comparison with cells growing in other tested complex media (brain-heart infusion, Mueller-Hinton broth, peptone water).
Conclusion: Biofilm forming ability of P. aeruginosa as well as response to oxidative stress was affected by the composition of culture media. This work was supported by Ministry of Health of the Slovak Republic under the project: Analysis of biofilm production in nosocomial strains as a basis to prevent infections in health institutions.
P625 Biofilm formation in non-C. albicans yeasts isolated from blood cultures
F. Ruzicka, V. Hola, P. Hamal, R. Tejkalova, I. Kocmanova, M. Votava (Brno, Olomouc, CZ)
Although Candida albicans remains the most common fungal blood stream pathogen, the incidence of non-albicans yeasts has been approached to C. albicans incidence during last few decades. One of the most important factors of their virulence is the ability to growth in the biofilm form which facilitates colonisation of indwelling implants and defends the microbial cells against effect of the immunity system and antifungal agents. The proof of the ability to form biofilm enables to evaluate the clinical importance and can helps to the physician to choose appropriate strategy of therapy (e.g. removing of the focus).
The seventy-seven strains of non-C. albicans yeasts (C. parapsilosis, 32; C. tropicalis, 21; C. glabrata, 12; C. krusei, 4; C. lusitaniae, 2; C. zeylanoides, 1; C. pelliculosa, 1; Yarrowia lipolytica, 1; Trichosporon asahii, 3) were isolated from blood cultures during last two years (Oct. 2003 – Oct. 2006) and these strains were examined for biofilm formation in this study. The biofilm formation was tested by modification of microtiter plate method. The yeasts were incubated in wells of microtiter plate to form biofilm. The biofilm in wells was stained by means of crystal violet. Quantity of biofilm layer on the wall of each well was evaluated spectrophotometrically. The sensitivity of some biofilm-positive strains to antifungal agents (amphotericin and voriconazole) was evaluated by microdilution assay with colorimetric medium indicating the fungal viability.
The thirty-seven strains (C. parapsilosis, 19; C. tropicalis, 11; C. krusei, 2; C. glabrata, 2; T. asahii, 1; C. zeylanoides, 1 and C. pellicuclosa, 1) were positive for biofilm formation. The relatively high percentage of biofilm-positive strains, especially in C. parapsilosis, and C. tropicalis, indicates the important role of biofilm formation as important virulence factor, because these pathogens are frequently associated with indwelling medical devices infections. Increasing incidence of these yeasts in blood stream can be explained by high frequency of using of indwelling medical devices in the few last decades. All tested strains growing in the biofilm showed higher resistance to all antifungal agents in comparison with their planktonic form.
This work was supported by the grant agency IGA MZ NR/7980–3.
P626 Antifungal activity of human monocytes alone and combined with caspofungin against Candida albicans biofilm and planktonic cells
A. Katragkou, M. Simitsopoulou, M. Dalakiouridou, E. Diza-Mataftsi, C. Tsantali, E. Roilides (Thessaloniki, GR)
Objectives: Candida albicans (CA) forms biofilms (BF) on implanted medical devices, and most commonly on intravascular catheters. BF constitute a niche where CA is protected from innate immunity, an important component of which are monocytes (MNCs), and antifungal agents. Moreover, CA BF have shown resistance to a variety of antifungal agents but susceptibility to caspofungin (CAS). Because little is known about the effects of host defences against Candida BF, we aimed to study the antifungal activity of human MNCs alone or combined with CAS against CA BF in comparison to their planktonic (PL) counterparts.
Methods: CA-M61, a documented biofilm-producing CA intravascular catheter isolate, was used. PL cells were grown in YNB at 37°C overnight. BF were grown on silicone elastomer disks in 96-well plates at 37°C with shaking for 48 h. THP1 monocytic cell line was used as MNC source. MNCs (MNC/target ratios 1:1 and 5:1) and CAS (0.0625 and 0.015 mg/l) alone or in combination were incubated with mature BF and PL cells in RPMI-1640 with 10% fetal bovine serum at 37°C, 5% CO2 for 20h. Plain BF and PL cells served as controls. Percent damage of BF and PL cells was then assessed by XTT colorimetric micro assay as reduction in the metabolic activity after incubation for 30 min at 450 nm with reference wavelength at 690 nm. Statistical analysis (n = 7) included ANOVA and post hoc analysis (P < 0.05).
Results: MNC-induced BF damage was significantly decreased compared to PL cells at 5:1 ratio (mean±SE, 31.5±3.5% vs 43.6±3%, p = 0.01) but not at 1:1 ratio. CAS-induced damage to BF was lower than that to PL cells at 0.0625 mg/l (57.7±4.1% vs 75±11%, p < 0.001). The damage induced by MNCs at 5:1 combined with CAS, at both concentrations, on BF was lower than that on PL cells (71.3±5.2% vs 85±6%, p = 0.001; 65.5±9.3% vs 82.3±6.5%, p = 0.008, respectively). While an additive effect between MNCs at 5:1 ratio and CAS at 0.015 mg/l was noted against PL cells (82.2±6.5% vs 43.6±3%, p < 0.001 and 82.2±6.5% vs 62.8±11%, p < 0.05), no significant collaboration between MNCs and CAS was observed against BF at any MNC/target ratio and any CAS concentration used.
Conclusions: C. albicans BF are more resistant than PL cells of CA to MNCs, to CAS and to the combination of MNCs with CAS. While MNCs and CAS exhibit an additive effect against PL, no significant collaboration between MNCs and CAS exists against BF. The mechanism(s) behind resistance of CA BF to host defences need to be determined.
P627 Effect of disinfectants and caspofungin against planktonic and sessile cells of Candida spp
S. Tobudic, C. Kratzer, W. Graninger, A. Georgopoulos (Vienna, AT)
Objectives: The aim of this study was to evaluate the activity of 3 different disinfectants chlorhexidine digluconate, Akacid plus® and hydrogen peroxide compared to the antifungal caspofungin against planktonic and sessile cells of Candida spp.
Methods: As test strains 40 clinical isolates of Candida spp. including C. albicans, C. krusei and C. tropicalis were used. The activity of active substances was determined against planktonic cells according to CLSI guidelines for antifungals using broth microdilution method. For antifungal susceptibility testing of sessile cells, isolates were incubated overnight in yeast peptone dextrose resuspended in RPMI 1640 to a cellular density equivalent to 1.0×106 CFU/mL. Cells were grown for 48 h in 96-well-microtiter plates, and then treated with 100 μl of Akacid plus®, chlorhexidine and hydrogen peroxide at a final concentration of 0.25, 0.5, 1, 2 and 4% compared to caspofungin at a concentration of 64, 128, 256 and 512 mg/l for 48°C at 35°C. The cells were fixed and stained with crystal violet. The mean optical density was used for quantification using a routine microtiter-plate-reader at 490 nm. Additionally, fungal growth following antimicrobial treatment was examined.
Results: MICs of Akacid plus® and caspofungin against planktonic cells of Candida spp. were comparable and reached MIC values of 0.03–8 mg/l, whereas MICs of chlorhexidine and hydrogen peroxide ranged from 16 to 32 mg/l and from 128 to 256 mg/l. Low concentrations of caspofungin at 64 mg/l caused a 62% reduction of the sessile cells of Candida spp. Treatment with 0.25% chlorhexidine and Akacid plus® led to reduction of the sessile cells in 59 and 74%, whereas hydrogen peroxide showed no effect.. No viable cells of Candida spp. were detected in biofilms treated with 0.25% Akacid plus® and chlorhexidine or 0.1% caspofungin.
Conclusion: Caspofungin and cationic antimicrobials showed high activity against sessile and planktonic cells of Candida spp., whereas hydrogen peroxide was found to be ineffective.
P628 Response of Candida albicans biofilms to bacterial factors
L. Rossi, S. Cagnacci, E. Tavella, O. Muratore, R. Corvò, A. Marchese, E. Debbia (Genoa, IT)
Introduction: Candidoses affecting immunocompromised patients are likely to be associated with certain commensal bacteria attracted by the inflammatory microenvironment. Although the precise outcome of such a cohabitation is not known, a plausible scenario would credit bacteria for influencing the survival of Candida spp. To gain more insights into this aspect of fungal infection, we are looking at the response of Candida biofilms to colonisation by bacteria normally present in the human microflora.
Methods: Candida albicans isolated from an immunocompromised patient were grown in collagen gel to produce multiple biofilms. Conditioned media from Pseudomonas aeruginosa (CMPa), Staphylococcus aureus (CMSa) and Lactobacillus casei (CMLc) were filtrated and freezed as stock solutions. After 5 hrs in collagen gel, nascent Candida biofilms were treated once with the several conditioned media for approximately 10 hrs. Relevant morphological parameters were detected and cell survival/gel was determined by the CFU test.
Results: The value of CFU in the control group was 1×107 cells/mL or above. Conditioned media affected differently the survival of C. albicans. Specifically, 90% of Candida cells died out following treatment with CMPa. CMLc was somewhat less effective in that approximately 50% of the cells survived the treatment. However, CMSa did not damage Candida cells, that displayed a value of CFU comparable to that of the control group. CMPa implemented filamentation of Candida biofilms, resulting in what it appeared a selective inhibition of yeast forms. CMLc and CMSa did not interfere with this process.
Conclusions: The present results indicate that P. aeruginosa releases factors capable of inhibiting Candida biofilms in collagen gel. While the content of CMPa was not analysed, most reports agree that aryl homoserine lactones, quorum sensing (QS) factors regulating the homeostasis of P. aeruginosa biofilms, are mainly responsible for the biological activity of this Gram-negative bacteria. This suggests that certain bacterial QS factors could help to keep under control fungal biofilms, an observation worthy of further investigation. The finding that Lactobacillus casei, a probiotic, can inhibit the growth of Candida albicans has clinical relevance, in that the association of probiotics with existing antimycotic drugs may provide a most effective procedure to cure candidoses. (Supported by Pfizer Italia).
P629 In vitro activity of essential oils and their major components against Candida albicans yeasts growing planktonically and as biofilms
S. Dalleau, E. Cateau, T. Berges, J. Berjeaud, C. Iimbert (Poitiers, FR)
Objectives: Candidiasis can be associated with the formation of biofilms on bioprosthetic surfaces and the intrinsic resistance of C. albicans biofilms to the most commonly used antifungal agents has been demonstrated. In this study, we report on the antifungal activity of 13 terpenes and essential oils on C. albicans growing planktonically or as biofilms.
Methods: The strain ATCC 3153 of C. albicans was used. Nine terpenes (carvacrol, citral, eucalyptol, eugenol, farnesol, geraniol, linalool, menthol and thymol) and 4 essential oils (tea tree, palmarosa, oregano and rosemary) were tested. The anti-biofilm activity of the tested compounds was evaluated using an in vitro model of C. albicans biofilm associated with polystyrene surfaces and the metabolic activity of yeasts within the biofilm was assessed with XTT method.
Results: The majority of the tested compounds showed a significant antifungal activity (MIC < 0.4 mg/mL). Two essential oils exhibited an “intermediate” antifungal activity – tea tree (MIC = 2.25 mg/mL) and rosemary (MIC=1.10 mg/mL) – and two terpenes (farnesol and eucalyptol) were not efficient against planktonic C. albicans (MIC > 74 mg/mL).
Citral, eugenol, palmarosa and rosemary induced a significant inhibition of the metabolic activity of the yeasts included in the C. albicans biofilm (p < 0.001) when added at a concentration <2.25 mg/mL during the early step of the fungal biofilm growth. The concentration needed for carvacrol, geraniol, linalool, oregano and thymol to achieve a significant reduction of the biofilm development was <5 mg/mL. The higher efficient concentrations were obtained for farnesol and menthol and corresponded to 35.5 mg/mL and 17.8 mg/mL respectively.
Conclusion: This study demonstrated the efficiency of almost all the tested terpenes and essential oils to inhibit the biofilm growth which could therefore represent good candidates in the prevention of biofilms associated with implanted medical devices.
P630 Aspergillus fumigatus biofilms are refractory to antifungal challenge
E. Mowat, J. Butcher, C. Williams, G. Ramage (Glasgow, UK)
Background: Aspergillus fumigatus may cause infections in immunocompromised patients including patients with cystic fibrosis. A. fumigatus conidia are readily inhaled and are not cleared by the innate immune system. Respiratory infections are typified by the presence of dense filamentous networks of hyphae in the pulmonary cavity and airways. These fungal mucus plugs are inherently difficult to treat, and have characteristics which resemble biofilms.
Objectives: To develop an in vitro A. fumigatus biofilm model to assess the ability of antifungal agents to inhibit and/or kill these structures.
Methods: Spores were collected from AF293 and standardised at various densities in RPMI medium and grown in 96-well polystyrene plates. Biofilm growth kinetics were then observed over 48 h (1, 2, 4, 6, 24 and 48 h) microscopically and by both metabolic (XTT) and a biomass (crystal violet) assays. Developing biofilms were visualised using confocal laser scanning microscopy (CLSM) and electron scanning microscopy (SEM). The specific effects of voriconazole (0.125–256 μg/mL) was also examined to measure its effectiveness pre- and post challenge.
Results: Optimal spore concentration for confluent biofilms after 24 h was determined to be 1×106 spores/mL. The initial biofilm growth kinetics involved an adherence stage (0–4 h); development of a monolayer of cells (4–8 h); and formation of a three dimensional biofilm structure after 24h. Voriconazole and caspofungin were ineffective against mature biofilms. Amphotericin B was effective between 0.5 – 1.0 μg/mL, however 90% killing was never achieved. When voriconazole was added at the initial stages of adhesion, a dose dependant effects was observed at therapeutic concentrations.
Conclusions: We have developed a robust reproducible in vitro A. fumigatus biofilm model was developed. Early exposure of spores to voriconazole prevented filamentation and biofilm formation. Overall voriconazole potentially offers excellent prophylactic properties against invasive aspergillosis.
Viral infections in the immunocompromised host
P631 Community respiratory virus infections in patients with haematological malignancies
V. Chebotkevich, A. Volkov (St.-Petersburg, RU)
Objectives: Viral infections are the important cause of morbidity and mortality in immunocompromised patients (pts) with haemotological malignancies. With regard to these agents the focus has been on herpes viruses, particularly human Cytomegalovirus (CMV). The studying of last decade showed that community respiratory viruses also play the important role in serious respiratory illnesses in pts with haemoblastosis. The current study was designed to determine the frequency and clinical features of respiratory viral infections in pts with different forms leukaemia.
Methods: Our study included 91 pts with different forms haemoblastosis. The patients were studied during the episodes of respiratory illnesses. Material – blood and nasal swabs were studied by means of polymerase chain reaction (PCR) with primers to the batteries of viral genomes: Adenoviruses, Respiratory syncytial virus (RSV), Influenza type A and B, Parainfluenza type 3 and Coronaviruses.
Results: The signal of Adenoviruses was detected in nasal swabs in 12 (13.1%) cases of respiratory illnesses, RSV in five (5.5%) cases, Influenza A in five (5.5%) cases, Influenza B in two (2.2%), Parainfluenza type 3 in eight (8/8%) and Coronaviruses in 13 (14.3%) cases. In blood the signal of Adenoviruses was detected in 5.4% cases. It is interesting that in all these cases the signal in nasal swab was not founded. In one case Influenza B virus was founded simultaneously in blood and nasal swabs. The main clinical symptoms of respiratory illnesses were the chill (87.5%), fever (87.5%), mostly higher then 38°C, lymphocytosis – 43.7%. In the 50% patients respiratory illnesses were complicated by pneumonia. We did not find any clinical peculiarities of the community respiratory illnesses of different viral etiology.
Conclusions: The community respiratory viral infections are serious illnesses in pts with haemoblastosis. These infections must be controlled as well as CMV and other herpes infections. PCR is adequate method of monitoring viral infection in this group of patients.
P632 Viral infection is responsible for acute renal dysfunction and chronic allograft lesions in paediatric renal-transplant recipients: a prospective study
L. Barzon, M. Pacenti, M. De Pieri, L. Murer, G. Palu' (Padua, IT)
Objectives: Follow-up evaluation of paediatric renal-transplant recipients in order to assess the contribution of viral infection to acute and chronic nephropathy and allograft rejection.
Methods: The presence of viral DNA (i.e., EBV, CMV, HHV6, HHV7, HHV8, VZV, BKV, JCV, SV40 and parvovirus B19) and viral load was prospectively investigated by quantitative real-time PCR methods in the peripheral blood, urine, and in allograft biopsies obtained from a series of 77 consecutive children (31F, 46 M, R/D mean age: 11.9±7.6/11.9±5.3 years) undergoing kidney transplantation in the period 2000–2004. Immunosuppressive therapy included basiliximab, steroids, FK506 or cyclosporinA ± mycophenolate mofetil. Follow-up allograft biopsies were performed at the time of transplantation and at 6, 12, and 24 months post-transplantation; diagnostic biopsies were performed in 11 patients because of acute renal dysfunction. Virological findings were compared with histological analysis according to Banff 97 criteria.
Results: At the time of transplantation, the allografts were positive for parvovirus B19 in 33% of cases, HHV6 23%, BKV 5%, SV40 3%. The cumulative incidence of chronic lesions was 29%, 52%, and 83% at 6, 12, and 24 months post-transplantation, whereas the cumulative incidence of viral DNA detection in biopsies was 63%, 69% and 71%, respectively (coinfections in 25%, 22%, 24%, respectively; the most frequent: EBV, HHV6, BKV, B19). The prevalence of viral genomes was higher in biopsies showing acute (Banff III, IV) or chronic (Banff V) lesions than in normal histology cases, but viral infection or histological damage did not correlate with renal function tests. Moreover, children who developed chronic lesions generally had early and persistent kidney infection (especially from BKV, B19, EBV). Viral-genomes were isolated in 7/11 biopsies performed for acute renal dysfunction: 2 tubulo-intersitial nephropathy (BKV), 3 thrombotic microangiopathy and 1 acute vascular rejection (parvovirus B19), and 2 acute rejection (EBV). A correlation between virological findings in biopsies and viral DNA detection in blood and urine was observed, although the rate of positive tests was higher in biopsies. Evalution of test predictivity is ongoing in a long-term follow-up study.
Conclusions: Viruses not only are responsible for acute renal dysfunction in kidney transplanted children, but also contribute to the development of chronic allograft lesions due to persistent infection.
P633 Whole blood real-time PCR for cytomegalovirus DNA quantification: analysis of PCR data and pp65 antigen test in a cohort of solid-organ transplant recipients
T. Allice, F. Cerutti, F. Pittaluga, S. Varetto, S. Gabella, A. Franchello, M. Rinaldi, V. Ghisetti (Turin, IT)
Cytomegalovirus (CMV) is a major opportunistic agent in solid organ transplantation (SOT). Pre-emptive therapy and a strict infection monitoring with highly sensitive methods have significantly decreased CMV morbidity and mortality. Recently introduced real-time PCR tests for routine CMV DNA quantification require correlation studies with pp65-antigen test as the gold standard. Moreover, there is no consensus as the most appropriate blood compartment (i.e. whole blood, leukocytes, plasma) for PCR test.
Aims: 1) to study the correlation between pp65-antigen test and CMV DNA as quantified by real-time PCR in whole blood (WB) in a cohort of SOT recipients and 2) the identify a CMV DNA cut-off level for pre-emptive anti-CMV therapy.
Methods: WB samples (n = 397) from 41 asymptomatically infected patients (18/41 undergoing pre-emptive therapy with ganciclovir) were monitored the first year after SOT by pp65 antigen test and real-time PCR for the UL123 gene, IE1 exon 4 (Nanogen, I). Extraction was carried out from 220 ul of WB with a fully automated system based on nucleic acid silica-gen affinity (BioRobot 9604, Qiagen, I).
Results: Pp65 and CMV DNA were highly correlated (r = 0.750, 78% concordant samples). CMV DNA level was much higher in pre-emptive treated patients than in not treated ones (mean+SD: 5.8+0.8 log10 copies/mL vs. 4.5+0.6, p < 0.0001). Pre-emptive therapy was started for pp65 values >90 positive cells, corresponding to a median CMV DNA level of 5.6 log10 copies/mL (mean+SD: 5.4+0.8, range 3.6–6.8). According to pp65 positive cells/200,000 examined cells of 0, 1–10, 11–50, 51–100 and >100, median levels of CMV DNA in WB were 3.8, 3.9, 4.6, 5.2 and 5.7 log10 copies/mL, respectively. A plasmid carring the UL123 viral amplified gene was used to probe the couppled extraction and real-time PCR platform sensitivity (100% at 3 copies/ul from the extraction step, 100% at 2 copies/ul from the amplification step) and reproducibility (between assay CV% <19).
Conclusion: CMV DNA evaluation in WB by real-time PCR significantly simplifies and accelerates the production of a reliable quantification for clinical purposes with a good correlation with pp65 antigen tests, excellent sensitivity and reproducibility. Levels of DNA >5.4 log10 copies/mL that is >250,000 copies/mL are highly suggestive of pre-emptive anti-CMV therapy.
P634 Relevance of respiratory viruses among adult immunocompromised patients with community-acquired pneumonia
M. Camps, T. Pumarola, A. Moreno, C. Cervera, A. Torres, M.T. Jiménez de Anta, M.A. Marcos (Barcelona, ES)
Background: Community-acquired pneumonia (CAP) is a serious lower respiratory tract infection associated with significant morbidity and mortality in immunocompromised patients. The present study evaluated the clinical spectrum of CAP in immunocompromised hosts, the role of respiratory viruses (RVs), as well as the profitability of viral diagnostic methods.
Methods: Over a one-year period 92 immunocompromised patients with CAP were prospectively evaluated for the presence of viral, bacterial and other pathogens. A nasopharyngeal swab was collected to study RVs which were processed by indirect immunofluorescence assay (IFA), cell culture and PCR. We defined 4 groups according to aetiology of CAP: group 1 (bacterial or Pneumocystis jiroveci), group 2 (viral), group 3 (mixed) and group 4 (unknown aetiology).
Results: In 61 (66%) of the 92 patients the aetiological diagnosis was achieved. Respiratory infection was due to bacteria or P. jiroveci in 38 (41%) cases of CAP, 12 (13%) were due to a virus and 11 (12%) were mixed infections. The most frequent pathogen detected was Streptococcus pneumoniae in 29 (48%) followed by rhinovirus (RNV) detected in 11 (18%) cases. PCR identified 95% of RVs. Medical records did not show significant differences among aetiological groups. Four patients (4%) required mechanical ventilation and finally died, two of whom had a RNV as the sole aetiological agent.
Conclusion: The high incidence of RV detected among the immunocompromised population studied emphasizes the need for further research on RVs as an important pathogen involved in CAP, with PCR being the most sensitive and rapid technique available to detect these viruses.
P635 HHV-7 primary infection in solid organ transplant recipients
A. Anton, C. Esteva, C. Cervera, T. Pumarola, A. Moreno, M.T. Jiménez de Anta, M.A. Marcos (Barcelona, ES)
Objectives: We aimed to study the seroprevalence of HHV-7 infection in solid organ transplant recipients (SOTR). The incidence of HHV-7, HHV-6, and CMV viraemia after solid organ transplantation among HHV-7 seronegative patients who had HHV-7 seroconversion during the first six months of follow-up was also studied.
Methods: Ninety three SOTR (5 heart, 1 kidney-heart, 28 liver, 2 pancreas, 6 kidney-pancreas and 51 kidney) were included. Plasma and whole blood samples were collected immediately prior to transplantation and at 7, 14, 21, 28, 45, 60, 75, 90 and 180 days post-transplantation. Anti-HHV-7 IgG antibodies in plasma were assessed by indirect inmunoflorescence assays (Advanced Biotechnologies Inc) immediately prior to transplantation, three and six months post-transplantation. Viral DNA was extracted from 200 microL plasma and from 50 microL whole blood using affigene® DNA extraction kit (Sangtec, Bromma, Sweden). An in-house SYBR Green I real-time quantitative PCR assay was performed to detect HHV-7 DNA (detection limit: 5–10 copies/reaction) in blood on the Mx3000P instrument (Stratagene, La Jolla, USA). The PCR primers amplify a specific region from HHV-7 U37 gene sequence. A standard curve was constructed from the threshold cycle values obtained from serial diluted positive control plasmids, cloned into pGEM-T Easy Vector. CMV viral load was determined in plasma by the PCR kit affigene® CMV VL which utilises an ELISA based detection. HHV-6 load in whole blood was detected using the real-time PCR system affigene® HHV-6 trender on the Mx3000P instrument.
Results: Seventy-one (76%) patients had IgG antibodies against HHV-7 detected prior to transplantation. Of 22 (23%) HHV-7-seronegative patients, 5 seroconverted, four at 3rd month and one at 6th month of follow-up. HHV-7 DNA was detected (3rd week of follow-up) in only one of the 5 patients who had HHV-7 seroconversion. In this patient, HHV-6 was simultaneously detected. Of the 4 remaining patients with HHV-7 seroconversion, HHV-6 DNA was also detected in one and CMV DNA was detected in two, while no human beta-herpesvirus DNA was identified in one patient.
Conclusions: While the seroprevalence of HHV-7 infection in SOTR was high, the incidence of HHV-7 among seronegative patients who had HHV-7 seroconverted during the first six months of follow-up was very low. Seroconversion may not only indicate HHV-7 replication but also other human beta-herpesviruses viraemia. Further analysis are necessary.
P636 Stratified treatment of respiratory syncycital virus in haematopoetic stem cell transplanted patients
N. Khanna, A. Widmer, M. Decker, I. Steffen, D. Heim, M. Weisser, A. Gratwohl, U. Flueckiger, H.H. Hirsch (Basle, CH)
Objectives: Respiratory Syncycital Virus (RSV) causes significant mortality in patients after stem cell transplantation (HSCT) when upper respiratory tract infections (RTI) progress to lower RTI and pneumonia. Higher immunodeficiency (ID) is at high risk for lower RTI. Current guidelines recommend treatment of RSV pneumonia with ribavirin (RBV) aerosol inhalation, but impact on mortality has been limited. We evaluated based on standardised protocol treatment using oral or intravenous RBV (10 mg/kg tid), IVIG (0.5 g/kg 3x/week) and pavilizumab (PAB; 15 mg/kg single dose i.v.) according to low or high risk of ID.
Methods: Adult HSCT patients at the University Hospital of Basel between February 2002 and March 2006 were included if RSV was detected by antigen (AG), PCR or culture (Cult) in nasopharyngeal aspirate or bronchioalveolar lavage. High ID was defined as HSCT within the last 6 months, T-cell depletion, B-cell depletion, GvHD > grade 2, leukopenia <2000/uL, lymphopenia <100/uL, or hypogammaglobulinaemia. Patients were stratified to Group-A: low ID or Group-B: high ID.
Results: In the study period, 16 patients (2 autologous, 14 allogeneic HSCT) were identified. 9 (56%) presented as outpatients. RSV infection was detected as positive with AG+Cult+PCR in 4/16; AG+Cult 4/16; AG+PCR 1/16; Cult+PCR 3/16; Cult 2/16; PCR 2/16. 8 (50%) had an upper RTI, 8 (50%) lower RTI. Overall mortality of RSV was 5 (31%), all with lower RTI. Patients in Group-A: 2 (13%); both with upper RTI, received no treatment, and survived without developing lower RTI. Group-B: 14 (87%); 6 with upper RTI, 1/6 received IVIG, 2/6 IVIG+RBV, and 3/6 IVIG+RBV+PAB. No lower RTI developed and all 6 patients survived. Lower RTI was found in 8/14 patients. Treatment (survival) was as follows: 3/8 IVIG+RBV (1/3); 1/8 IVIG+PAB (1/1); 4/8 IVIG+RBV+PAB (1/4). Haemolytic anaemia resulting from oral RBV was found in 1 patient. In group B patients, there was only a trend for better survival using PAB, but these data are limited by small sample size and strong impact of lower RTI.
Conclusion: RSV infection limited to the upper RTI has a better outcome. In patients with high ID treatment may prevent progression to lower RTI and death. In patients with high ID and established lower RTI intervention may be less efficient. The results of PAB treatment should be confirmed in a larger trial.
P637 Quantitative CMV PCR in allogenic stem-cell transplant patients
N. Portocarrero, A. Moriente, D. Monclus, E. Lomas, P. Sanchez, C. Sanchez, L. Cardeñoso (Madrid, ES)
Objective: To assess the clinical value of a commercial quantitative plasma CMV-PCR assay (COBAS AMPLICOR CMV MONITOR test, Roche Molecular System) in allogeneic SCT patients by comparing the results obtained with the PCR and the antigenaemia.
Methods: All patients were monitored weekly antigenaemia and PCR. A total of 1877 blood samples from 94 patients were tested prospectively. CMV seropositive patients (or negative with a seropositive donor < 9 received high dose of acyclovir as prophylaxis. PCR was considered positive when more or equal 400 DNA copies/mL were detected. Antigenaemia was considered positive when one or more positive cells were detected.
Result: All CMV seronegative patients with a seronegative donor were antigenaemia and PCR negative The total patients in study (94), 39 had a positive antigenaemia and/or PCR, with a total of 71CMV infection episodes. None developed CMV disease. PCR detected 49 out of 71 CMV episodes (69%). Overall there were 115 positive antigenaemia (belonging to 35 patients): 65 were PCR positive, 49 PCR negative and 1 PCR inhibited. Four patients (6 episodes) had a positive PCR with a negative antigenaemia. For samples with less than 600 copies/mL a manual calculation of the number of copies was retrospectively done (using a new cut-off). With this lower cut-off, PCR detected 62 out of 71 episodes (87, 4%).
Conclusion: Quantitative CMV PCR assay showed a lower sensibility for de detection of CMV infection in allogeneic SCT compared with antigenaemia. The PCR sensibility was increased without a decrease in the specifity, lowering the cut-off.
P638 Possible BK virus outbreak in haematopoietic stem cell transplant recipients
M. Sonmezoglu, S. Yirmibescik, C. Obek, D. Colak, T. Kocagoz, Y. Koc (Istanbul, Antalya, TR)
BK virus (BKV) is a human polyomavirus that has been implicated as a common cause of late-onset haemorrhagic cystitis (HC) in patients who have undergone bone marrow transplantation. BKV has also been shown to be a significant cause of tubulointerstitial nephritis, vasculopathy, and allograft dysfunction in kidney transplant recipients.
Here we describe five cases of BKV-associated HC in hematopoietic stem cell transplant recipients in a short period. The patients (2 chronic myeloid leukaemia with blastic transformation, 1 Hodgkin lymphoma, 1 acute lymphocytic leukaemia and 1 thalassaemia intermedia) who underwent allogeneic stem cell transplantation (one of them unrelated) developed severe bladder spasms and gross hematuria 46–146 days after receiving immunosuppressant therapy (4 days Busulfan). Physical examination revealed marked suprapubic tenderness, continuous and refractory haemorrhagic cystitis, which required analgesics, continuous bladder irrigation, and blood transfusions. Weekly plasma and urine samples were tested for BK virus DNA by polymerase chain reaction (PCR). PCR of plasma and urine samples yielded results positive for BKV. We administered weekly infusions of 5 mg/kg cidofovir patient in an attempt to clear viruria. Leflunomid were added to improve the condition. Nephrotoxicity occurred in one patient after receiving second dose of cidofovir and later developed renal failure and required daily dialysis. The level of viruria persisted for 4–8 weeks after administration of cidofovir. BKV-associated haemorrhagic cystitis is common in patients following bone marrow transplantation and immunosuppressant therapy. We report a possible HC outbreak due to BK virus in a bone marrow transplant unit seen only in 2 and a half month period and ceased thereafter.
P639 CMV monitoring of transplant patients a ten-year experience
M. Baier, J. Rödel, U. Lang, K. Boden, E. Straube (Jena, DE)
Background: To prevent CMV infection in immuno-compromised patients, in particular after solid organ or bone marrow transplantation regular monitoring is recommended using pp65 antigen detection for screening and quantitative CMV PCR for confirmation or in cases of low cell count.
Methods: In our now 1375-bed-university hospital pp65 detection has been performed since 1996. Quantitative CMV PCR was introduced in 1999. Patient data were extracted from lab database, combined with clinical data and subsequently analysed.
Results: Till November 2006 a total of 35.741 pp65 antigen tests were performed with a share of kidney transplant patients (NTX) of 32.7%, respectively of liver (LTX): 22.2%, bone marrow (KMT): 18.1%, heart transplantation (HTX): 16.4% and of other patients: 10.5%. A total of 4.424 quantitative CMV PCR tests were performed (NTX: 57.8%, LTX: 2.5%, KMT: 15.2%, HTX: 5.8%, others: 18.7%). The number of pp65 tests increased on average 13.4% per year (total: 8 in 1996, 1008 in 1997, 5200 in 2006, mean 1997–2006: 3250/year). The number of quantitative PCR tests peaked in 2000 with 1119 tests but stabilised in the last 6 years on 500 test/year (mean 2001–2006).
Over the 10 year observation period on average 7.9% of all pp65 tests were positive (13.6% in 1997 and 3.64% in 2006), showing a mean decrease in positivity rate of 12.8% per year. From NTX patients 7.9% of samples were positive, respectively 9.3% from LTX, 7.7% from KMT, 6.9% from HTX patients and 7.6% from others. Quantitative CMV PCR was positive on average in 10.9% of tests ranging from 4.6% in 1999 to 15.4% in 2003 with no trend in time. NTX patients were in 11.5% PCR positive, LTX in 10.0%, KMT in 7.6%, HTX in 14.4% and others in 9.9% respectively. In 3982 cases results obtained with both methods were available (239 (6%) positive and 3475 (87.3%) negative in both, 190 (4.8%) were pp65 negative and PCR positive and 78 (2.0%) vice versa). In 183 cases with ≤1 pp65 positive cells/100.000 leucocytes only 60% (n = 110) were PCR positive compared to 134 cases with >1/100.000 cells with 96% (n = 129) PCR positive results. In 40 cases with ≤500 CMV copies/mL plasma 32% (n = 13) were pp65 positive compared to 389 cases with >500 copies/mL with 58% (n = 226) positive pp65 results.
Conclusion: In our setting CMV antigen detection and PCR was well established as useful tools for post transplant monitoring with almost similar sensitivity. Clinical impacts and economical aspects will be discussed.
P640 Fatal viral opportunistic infections and Epstein-Barr virus positive large B-cell lymphoma after alemtuzumab treatment for a refractory Sezary syndrome
N. Roch, D. Salameire, S. Courby, F. Orsini-Piocelle, B. Pegourie-Bandelier, R. Gressin, C. Dumontet, N. Sturm, F. Berger, A. Toffart, J. Guellerin, G. Gereige, V. Hincky, O. Epaulard, J. Brion, P. Pavese, S. Larrat, P. Morand, J. Stahl (Grenoble, Lyon, FR)
Objectives: Alemtuzumab (Campath-1H) is a humanised monoclonal antibody binding to CD52, an antigen expressed on normal and malignant B-lymphocytes and T-lymphocytes. This immunotherapy is effective in the treatment of relapsed or refractory B-cell chronic lymphocytic leukaemia. It has been used in others lymphoproliferative disorders like Sezary syndrome, in autoimmune cytopenias, and in bone marrow and kidney transplantations. The depletion of lymphocytes induced by alemtuzumab can be profound within the first month of therapy and the reconstitution of the lymphoid cell line can be delayed for up to two years. Infectious complications are frequent. Viral, bacterial, fungal and parasitic infections have been described. Cytomegalovirus reactivation is the most common opportunistic infection and recommendations exist to prevent this complication. Others infections were rarely reported.
Methods: We report a case of multiple opportunistic infections after alemtuzumab treatment in a patient with refractory Sezary syndrome. To our knowledge these have been rarely, if ever, reported before.
Results: After three weeks of treatment with alemtuzumab, cytomegalovirus disease developed, successfully treated with ganciclovir. Campylobacter septicaemia, Epstein-Barr virus positive large B-cell lymphoma, adenoviral infection and enteroviral disease appeared respectively at seven, eight, thirteen and eighteen weeks of treatment. In spite of treatment with cidofovir and rituximab evolution of adenoviral disease and Epstein-Barr virus positive large B-cell lymphoma was fatal. The patient died four months after the start of alemtuzumab. Lymphopenia was profound during follow-ups. To our knowledge only two adenoviral diseases during alemtuzumab monotherapy were reported and both were fatal. Epstein-Barr virus positive large B-cell lymphoma after alemtuzumab treatment has been described previously in four observations but never in patients with Sezary syndrome. Campylobacter septicaemia and enteroviral infection have never been reported before.
Conclusion: This confirms the large panel of possible infections during treatment with alemtuzumab. This draws attention to the fact that also viruses other than cytomegalovirus need to be considered in patients with fever of unknown origin while on alemtuzumab treatment. Attention will be focused on these serious complications. Report cases are necessary for a better knowledge of possible opportunistic infections during alemtuzumab treatment.
Clinical complications in HIV infection
P641 Invasive fungal infection as the most common cause of cavitary lung lesions in HIV-infected patients in Taiwan
H-Y. Sun, C-C. Hung, M-Y. Chen, S-M. Hsieh, W-H. Sheng, S-C. Chang (Taipei, TW)
Objectives: Cavitary lung lesions have not been systemically investigated in patients with HIV infection before. This study aimed to analyse the etiology of cavitary lung lesions in HIV-infected patients enrolled in a prospective cohort study.
Methods: Medical records, radiologic and microbiologic data of cases of cavitary lung lesions diagnosed between June 1994 and June 2006 were reviewed using a standardised case record form. During the study period, stepwise investigations were performed to identify the aetiology of cavitary lung lesions, which included microbiologic cultures, chest sonography or computed tomograpgy-guided aspiration and biopsy. Clinical samples were cultured for bacteria, fungi and mycobacteria.
Results: During the 12-year study period, 61 of 1372 (4.4%) HIV-infected patients, 61.7% men having sex with men, 27.3% heterosexuals, and 5.3% injecting drug users, were diagnosed with cavitary lung lesions: 7 of 175 (4.0%) in the pre-HAART era and 54 of 1197 (4.5%) in the HAART era (p = 0.95). Of the 61 patients (56 males) with a median age of 34 years (range, 23–81 years), CD4 count was 25×106 cells/L (range, 1–543×106 cells/L) at the diagnosis of cavitay lung lesions. Fungi caused 31 of the 61 (50.8%) cavitary lung lesions, followed by bacteria (11; 18.0%), tuberculosis (7; 11.5%), and Mycobacterium avium complex (7; 11.5%). Of the 31 cases of fungal pneumonia, 16 (51.6%) were caused by Penicillium marneffei, 11 (35.5%) by Cryptococcus neoformans, 3 (9.7%) by Aspergillus spp., and 1 (3.2%) by unidentified fungus. Compared with 7 (3.5%) of 199 patients with tuberculosis diagnosed during the study period, 16 of 31 (51.6%) patients with P. marneffei infection and 11 of 64 (17.2%) patients with cryptococcosis developed cavitary lung lesions (p < 0.0001). Median CD4 count of the 7 patients with cavitary lung lesions due to tuberculosis (110×106 cells/L) was significantly higher than that of 16 patients with P. marneffei infection (7.5×106 cells/L) and 11 with cryptococcosis (35×106 cells/L).
Conclusion: Our findings suggest that invasive infections due to endemic fungi were the most common cause of cavitary lung lesions in patients at late stage of HIV infection in Taiwan.
P642 Risk of recurrent non-typhoid Salmonella bacteraemia in HIV-infected patients in the era of highly active antiretroviral therapy and increasing trends of quinolone resistance
C.C. Hung, M-N. Hung, P-R. Hsueh, M-Y. Chen, S-M. Hsieh, W-H. Sheng, H-Y. Sun, S-C. Chang (Taipei, TW)
Objectives: This study aimed to assess the trends of quinolone resistance and the risk of recurrent non-typhoid Salmonella (NTS) bacteraemia in HIV-infected patients receiving highly active antiretroviral therapy (HAART).
Methods: Medical records and microbiologic data of cases of NTS bacteraemia between June 1994 and June 2006 were reviewed using a standardised case record form. During the study period, treatment of NTS bacteraemia was ceftriaxone for 7–14 days followed by ciprofloxacin as secondary prophylaxis. While secondary ciprofloxacin prophylaxis for NTS bacteraemia would not be discontinued in the pre-HAART era, the duration of ciprofloxacin prophylaxis in patients receiving HAART was at the discretion of treating physicians. End of follow-up was the date of last clinic contact, date of death, or on 31 October, 2006, whichever occurred first.
Results: During the 12-year study period, 94 of 1372 (6.9%) HIV-infected patients developed 106 episodes of NTS bacteraemia: 16 of 175 (9.1%) in the pre-HAART era and 78 of 1197 (6.5%) in the HAART era (p = 0.27). In the pre-HAART era, 4 of 16 (25%) patients (median CD4 count at NTS bacteraemia, 8×106 cells/L) had 7 recurrent episodes, compared with 3 of 78 (3.8%) (median CD4, 21×106 cells/L) who had 5 recurrent episodes in the HAART era (odds ratio, 8.33 [95% CI, 1.656, 41.95]; p = 0.03). 6 patients in the HAART era died within 30 days of NTS bacteraemia (median, 10 days; range 2–21 days). 5 recurrent episodes occurred in 3 patients in the HAART era: 1 with primary CNS lymphoma receiving chemotherapy and 2 without adherence to HAART. 6 of 23 NTS isolates were resistant to trimethoprim–sulfamethoxazole [TMP-SMX] in the pre-HAART era compared with 16 of 66 isolates in the HAART era (p = 0.99). Although none of the isolates was resistant to ceftriaxone, 14 of 65 (21.5%) isolates in the HAART era were resistant to ciprofloxacin (including 9 isolates resistant to ampicillin, chloramphenicol, TMP-SMX) compared with 0 of 23 isolates in the pre-HAART era (p = 0.02). Eight patients with NTS isolates resistant to ampicillin, chloramphenicol, TMP-SMX, and quinolones who survived NTS bacteraemia and did not receive secondary prophylaxis had no recurrences during HAART.
Conclusion: Our findings suggest that risk of recurrent NTS bacteraemia decreased significantly in the HAART era, although the resistance rate to quinolones was increasing and appropriate oral agents as secondary prophylaxis might not be available.
P643 Causes of liver related death in HIV-infected patients in France: mortality 2005 Survey
D. Salmon, C. Lewden, F. Bonnet, E. Rosenthal, P. Morlat, T. May, C. Burty, D. Costagliola, E. Jougla, C. Semaille, G. Chěne, P. Cacoub on behalf of Mortality 2005 Study group
Objective: To describe frequency and characteristics of liver related death in France in HIV-infected adults and compare the results to those of the same survey performed in 2000.
Method: Physicians involved in the management of HIV infection and those from societies of reanimation, pneumology, hepatology and penitentiary medicine were asked to notify deaths that occurred in 2005. The causes of death were documented using a standardised questionnaire.
Results: The 338 participating wards notified 979 deaths (versus 964 in 2000) and documented 831 deaths with a known status for hepatitis B and C viruses. Median age was 46 years and 75% were men. The main underlying causes of death were AIDS-related (37%, vs 47 in 2000), liver-related including viral hepatitis (15%, vs 13), cancer not related to AIDS or hepatitis (17%, vs 11), cardiovascular disease (9%, vs 7), other infections (5%, vs 7), suicide (5%, vs 4). Among the 125 deaths were related to liver disease, 50% had an alcohol consumption >30 g/day.
Conclusion: The proportion of liver-related deaths and particularly hepatocellular carcinoma increased between 2000 and 2005 in HIV-infected adults, while the proportion of AIDS related death has decreased. This justifies a more aggressive treatment of viral hepatitis C and B, the prevention of alcohol consumption, and an early and regular screening for hepatocellular carcinoma in case of cirrhosis.
| 2000 | 2005 | |
|---|---|---|
| No. of deaths with known status for HBV and HCV | 822 | 831 |
| Death from liver disease among which Hepatocellular carcinoma | 110 (13%) | 125 (15%) |
| 17 (15%) | 31 (25%) | |
| Main cause of liver disease | ||
| HCV | 72 (65%) | 90 (72%) |
| HBV | 14 (13%) | 16 (13%) |
| HCV + HBV | 16 | 6 |
| Alcohol | 8 | 8 |
| Other | − | 5 |
| Median age | 41 | 46 |
| Median CD4 cell count/mm3 | 90 | 165 |
P644 Longitudinal evaluation of the prevalence of insulin resistance in a cohort of HIV vertically-infected children and adolescents
R. Rosso, A. Parodi, E. Repetto, C. Torrisi, C. Bernardini, F. Ginocchio, M.P. Sormani, M. Vignolo, C. Viscoli (Genoa, IT)
Background: To assess the rate of progression insulin resistance (IR) and the associated metabolic disturbances over a 1-year period in HIV vertically-infected children and adolescent and to assess risk factors associated.
Methods: 48 children (age range 6–22 years; 21 F, 27 males) were followed over a mean period of 14 months (range 5–27). Fasting lipids and glucose profile were measured in all children. Therapy and disease history, presence of lipodystrophy, possible co-infections, axiological features were recorded at the baseline and at the end of the follow-up.
Results: At baseline, fasting insulin and HOMA-IR were significantly higher in HIV-infected patients than in healthy children and adolescents (p < 0.001). Therapy duration (r = 0.281, p < 0.05), triglycerides (r = 0.286, p <0.05), age (r = 0.299, p <0.05), BMI (r = 0.485, p <0.001) were significant predictor variables of IR, expressed as HOMA-IR >3. At baseline and at the end of follow-up, 36% vs. 29% of pts showed IR; and 32% vs. 33% showed dyslipidaemia (high cholesterol, or triglycerides or both), respectively. The proportion of patients presenting lipodystrophy remained stable (16.6%). Prospective follow-up showed no progression at all over 1 years, in particular the number of patients with IR remained the same but the level of mean HOMA-IR increased slightly (2.6±1.6 vs 3.8±8.1), showing no correlation with therapy duration as a whole and particularly with PI-based HAART HAART, defined as association of ≥3 antiretroviral drugs, seems not to be related with a worsening in IR. Sex, birth weight, HCV-infection have no relation with IR, while lipodistrophy, Tanner stage IV-V of puberty, hyper triglyceridaemia seem to be predictor variables of IR both at baseline and at the end of follow-up (p <0.05).
Conclusions: In HIV-infected children IR and other metabolic complications, such lipodystrophy are more likely to develop after mid-puberty Progression of disorders of glucose metabolism seems to be slow but, regarding their enlarged life expectancy, efforts to prevent the development of such complications should be targeted toward HIV-infected children with an at least annual (using fasting glycaemia and insulin) follow-up.
P645 Diverse dermatologic manifestations in HIV-infected patients after highly active antiretroviral therapy
K. Jutivorakool, P. Plussind, H. Choengwiwatkit, J. Waiwarawut (Chonburi, TH)
Objective: Highly active antiretroviral therapy (HAART) has increased survival in HIV patients. However, there are few studies on skin manifestations in HIV-infected patients after HAART with somewhat conflicting data. The aim of this study is to evaluate the influence of HAART on the prevalence and spectra of dermatologic disorders in HIV-infected patients
Method: We collected data from 113 HIV-infected patients on whom HAART was initiated in our HIV clinic between 1 June 2005 – 31 May 2006 and thus have received HAART for at least 6 months. The primary outcomes were dermatologicmanifestations after HAART. All diagnoses were based on patient history, physical examination, dermatologic consultation and laboratory investigations.
Results: Dermatologic manifestations occurred in 40 patients (35.4%) after HAART. Mean HAART duration was 55+61 days (range 7–241 days). Eosinophilic folliculitis and drug eruptions were the most common findings (N = 10, 8.8%). The next most common conditions were mycobacterial infections (N = 4, 3.5%), non-specific dermatitis (N = 4, 3.5%), herpes zoster infection (N = 3, 2.7%), idiopathic pruritus (N = 3, 2.7%), lipodystrophy (N = 2, 1.8%), herpes simplex infection (N = 2, 1.8%), histoplasmosis (N = 1, 0.9%) and psoriasis (N = 1, 0.9%). Independent risk factors for skin manifestations after HAART by univariate analysis were female (60% vs 30%, p = 0.003), lower nadir CD4 counts (64.3 vs 107.8, p = 0.024) and higher percentage of eosinophils at the time of dermatologic diagnosis (9.1% vs 5.2%, p = 0.004). Different HAART regimens did not seem to affect the incidence. Female sex and peripheral eosinophilia were confined to the groups of eosinophilic folliculitis and drug eruptions. There was no significant difference in the number of CD4 increase between patients with or without skin manifestations. We observed five cases of skin disorders due to opportunistic infections: four of mycobacterium infections (infiltrative erythematous plaque 1, subcutaneous abscess 1, painful erythematous nodules 1, and visible lymph node protrusion 1 and one of histoplasma lymphadenitis.
| Skin manifestation group |
P-value | ||
|---|---|---|---|
| Yes (N = 40) | No (N = 73) | ||
| Age (years) | 36.98±9.475 | 39.18±9.3 | 0.23 |
| Gender (M:F) | 16:24 | 51:22 | 0.003* |
| HAART regimens (N) | |||
| 1. NVP-based | 14 | 20 | 0.92 |
| 2. EFV-based | 14 | 35 | 0.29 |
| 3. PI-based | 12 | 18 | 0.53 |
| Nadir CD4 count (cell/μl) | 64.35±72.84 | 107.84±129.45 | 0.024* |
| Number of CD4 increase (cell/μl) | 73.8±68.25 | 88.7±125.64 | 0.419 |
| WBC (cell/mm3) | 5951±2552 | 6312±3085 | 0.511 |
| Percentage of eosinophils (%) | 9.13±7.52 | 5.25±3.71 | 0.04* |
| AST (IU/dl) | 92.7±235.9 | 39.38±21.63 | 0.16 |
| ALT (IU/dl) | 65.85±134.17 | 32.44±23.95 | 0.13 |
Conclusion: In the HAART era, eosinophilic folliculitis and drug eruptions are the most prevalent dermatological manifestations in our referral hospital. These are closely related to peripheral eosinophilia in this population. Physicians initiating HAART on a female especially with a low CD4 count should be aware of this consequence.
P646 A prospective study of bloodstream infection in HIV-positive patients during a 15-year period
J. García Rodríguez, H. Álvarez, B. Buño, A. Mariño, M. Rodríguez, P. Sesma (Ferrol, ES)
Objective: To know the evolution of incidence, etiology and prognostic factors for bacteraemia infection in HIV positive patients.
Methods: Prospective study of cases of bacteraemia in our Hospital between 1991 and 2005. Blood cultures were processed using automatic technique and antibiotic susceptibility was determined by microdilution according to the current recommendations from the NCCLS. In patients with HIV positive, demographics as well as a complete set of tests (including clinical features, risk factors to HIV infection, laboratory studies, CD4, treatment and illness evolution) were collected in each case. Statistical analyses were carried out with SPSS.
Results: We studied 1848 episodes of bacteraemia in adult patients, 103 (5.6%) of them in 84 HIV positive patients (incidence by 1000 hospitalised patients-year: 0.62 in pre-HAART and 0.61 in HAART-era). The HIV transmisión were: intravenous drug use 77.7%, heterosexual contact 9.7%, male homosexual contact 2.9%, blood transfusion 1% and unknown 8.7%. Male 87 (84.5%). Mean age 34.2±7 years (range 20–59), 30.4±4.7 in the pre-HAART vs 35.8±7.3 in the HAART-era, p <0.05. The bacteraemia was community acquired in 82 (79.6%) episodes. 98 (95.1%) episodes were due to monomicrobial infection. The more frequently isolated germs were: Staphylococcus aureus 27 and Streptococcus pneumoniae 14; and the origins: endocarditis 18.4% and pneumonia 30.1%. The CD4 cell count was <50 in 31.4% of the patients, 50–200 in 28.6%, 201–500 in 31.4% and >500 in 8.6%. Among 103 episodes, 17 (16.5%) patients died due to the sepsis and 8 (7.8%) cases were not linked to sepsis. Patients in the HAART-era had more comorbidity (41/72 vs 4/31), more frecuently neutropenia (13/72 vs 1/29), pneumococcal vaccine administration (38/63 vs 4/31)and prophylaxis with TMT-SMX (18/66 vs 2/31) than patients in the pre-HAART era. No statistically significant differences were observed between the pre-HAART and the HAART-era in the isolated germs, origins of bacteraemia, mean CD4 cells count, global mortality (7/31 vs 18/72) nor related to bacteraemia mortality (4/31 vs 13/72).
Conclusions: The incidence, etiology, origin and mortality of bacteraemia in HIV patients have remained stable. Patients in the HAART-era had more frecuently associated comorbility, pneumococcal vaccine administration an prophylaxis with TMT-SMX than those in the pre-HAART era.
P647 Frequency of proteinuria among HIV-infected patients
A. Ramezani, M. Mohraz, M. Banifazl, L. Gachkar, F. Yaghmaie, A. Aghakhani, N. Kalantar, K. Nemati, M. Haghighi, M. Rezaie, A. Velayati (Tehran, IR)
Background: Nephropathies associated with human immunodeficiency syndrome (HIVAN) are increasingly prevalent and associated with proteinuria and rapid progression to end-stage renal failure. Some authors reported, proteinuria may be an early marker of HIVAN, and screening for its presence may be beneficial. In this study we aimed to determine the frequency of proteinuria and related risk factors in Iranian HIV positive patients.
Methods: A total of 105 HIV positive patients were enrolled in this study. All the patients filled out a questioner about demographic characteristics and their high risk behaviours. Systolic and diastolic blood pressure was examined by a physician. Patient's blood samples were tested for creatinine, albumin, hemoglobin, HCV Ab and HBs Ag. In all patients CD4 counting were done by flow cytometry. Urine samples were collected and examined for detection of proteinuria. Proteinuria was defined as ≥1 plus in urine exam on 2 consecutive dipstick urine analyses. Personal and lab data's among study groups were compared with the Chi-square using SPSS 11.5 package programme.
Results: Out of 105 HIV positive patients, 20%(n = 21) had proteinuria. Mean age of patients with proteinuria was 35.2±9.19. There was not any significant difference between patients with and without proteinuria, regarding age, sex, race, weight, risk behaviours for HIV acquisition, disease history, staging (HIV/AIDS), concurrent drug therapy (non-nucleoside reverse transcriptase inhibitors or protease inhibitors), systolic and diastolic blood pressure, hemoglobin, albumin, presence of HCV Ab and HBs Ag. Patients with proteinuria had a lower CD4 count (322.47 vs. 439.16 cells/mm3 p < 0.051) and slightly higher creatinine (1.20 vs. 0.96 mg/L p = 0.054) than those without proteinuria.
Conclusion: This study showed a relatively high prevalence (20%) of proteinuria in the HIV-seropositive population. Our study also confirmed the correlation between decreasing CD4 count and the presence of proteinuria. But it was not demonstrated an association between proteinuria and a positive hepatitis C antibody as reported in previous studies.
P648 Infective endocarditis: descriptive analysis of 102 consecutive cases. Influences of human immunodeficiency virus infection in clinical evolution
E. Valencia, V. Moreno, A. Enríquez, L. Martín Carbonero, J. González Lahoz (Madrid, ES)
Objective: 1) To describe the characteristics of infective endocarditis (IE) in a group of inpatients attended in an Infectious Disease Service during the last 14 years and 2) To analyse the influence of different factors, HIV infection and HAART included, in the evolution of these patients
Patients and Methods: We analysed 102 cases of IE diagnosed between 1993 and 2006. Data were collected with regard to the clinical, laboratory and demographics characteristics of patients as well as results of blood cultures and data on clinical outcome. We used the statistical programme SPSS 13.0.
Results: Only four were not intravenous drug addicts (IVDA), most of them (80%) were men and 29 (28.4%) had had a previous episode. The onset was acute in 66 cases (64.7%), blood culture were negative in 56 patients (54.9%) and Staphylococcus aureus was isolated in 25 episodes (24.5%). All patients were febrile and thorax radiography showed septic emboli in 45 cases (44.1%). Affected valves were: tricuspid 86 cases (84.3%), pulmonary 2 (2%), mitral 13 (12.7%) and aortic 11 (10.8%). The most frequent complications were cardiac failure (15 patients, 14.7%), renal insufficiency (8 patients, 7.8%) and arterial emboli (5 patients, 4.9%). Seventy three subjects had a favourable outcome (71.6%), 13 died (12.7%) and 16 (all of them IVDA) left the hospital without finishing the treatment.
Seventy-eight cases had HIV infection (86.7%) and mean CD4+ was 240 cells per mm3 (range: 4–1000). Twenty two patients (21.6%) had a concomitant opportunistic infection and only 14 (13.7%) were receiving highly activity anti-retroviral therapy (HAART). Most of them (87.3%) were active IVDA and 60 patients (58.8%) had hepatitis C virus (HCV) co-infection.
The symptoms and the clinical evolution were independent from the presence of HIV infection. Patients with left sided IE had significantly more cardiac failure and ventricular dysfunction than those with right sided IE regardless HIV infection.
Conclusions: IE continue being a frequent and important problem in IVDA with or without HIV infection. Affected valve is mainly tricuspid and S. aureus is responsible in most of cases. In this group, HIV infection did not influence in clinical manifestations and outcome.
P649 Cardiovascular risk evaluation in HIV-infected adults
S. Sousa, E. Valadas, J.P. Lopes Cruz, M. Doroana, L. Tavares, L. Caldeira, N. Duarte, F. Antunes (Lisbon, PT)
Objectives: Metabolic changes caused by antiretroviral therapy may increase risk of cardiovascular disease. The aim of this study is to quantify cardiovascular risk (CVR) and to determine the prevalence of CVR factors in HIV-infected adults, as well as, to suggest strategies for decreasing CVR.
Methods: In a cross-sectional study, HIV-infected individuals were evaluated during a six-month period, at Hospital de Santa Maria, Lisbon. Data collected includes: demographic variables, antiretroviral therapy, duration of HIV infection, smoking habit, diabetes mellitus and family history of cardiovascular disease. Height, weight, blood pressure, total cholesterol, HDL-cholesterol, glucose and triglycerides levels were determined. Estimates of 10-year CVR were based on Framingham's equation.
Results: In the 1,340 patients included: the average age was 42.1 years, 66.7% were men, 49.1% were smokers, 36% had hypertension, 4.4% had diabetes, 15% had elevated cholesterol levels (>6.2 mmol/l) and 13.5% had low HDL-cholesterol levels (<0.9 mmol/l). Estimated 10-year CVR average was 5.2%. CVR average for patients on antiretroviral treatment (5.86%) is almost twice than for naïve patients (3.2%). An elevated CVR (>20%) was found in 4.4% of the patients, most of them were on non-nucleoside reverse transcriptase inhibitors or on protease inhibitors based treatments. To stop smoking was the measure with the highest impact on CVR decrease, followed by normalising lipid levels.
Conclusion: HIV-infected patients show several cardiovascular risk factors. Patients should be encouraged to change their lifestyle (smoking cessation, diet and exercise) and in some cases of important lipidaemia changes in antiretroviral treatment should be considered.
P650 Human immunodeficiency virus infected patients with community-acquired pneumonia: implication of respiratory viruses
R. Perello, A. Moreno, C. Cervera, A. Smithson, J. Miro, L. Linares, C. Agusti, M. Camps, M. Marcos (Barcelona, ES)
Background: Community acquired pneumonia (CAP), is an important source of morbidity/mortality in human immunodeficiency virus (HIV) infected patients. The aim of the present study is to evaluate the implication of respiratoty viruses (RV) in CAP in HIV-infected patients.
Methods: From June 2003 to December 2005, 67 adult HIV patients (mean age 41.39 years) with CAP were prospectively included. RV were identified by polymerase chain reaction, cellular cultures and immunofluorescence techniques from samples obtained by nasopharyngeal smears.
Results: A microbiological diagnostic was achieved in 48 patients. Forty CAPs had an isolated bacterial ethiology being S. pneumoniae the most common pathogen (85%). RV were implicated in 14 cases; in 8 as the only pathogen and in 6 in combination with bacteria's, being adenovirus plus S. pneumoniae the most frequent combination. Rhinovirus was the most common RV implicated followed by adenovirus. No statistical significant differences were found in CAP in which VR were implicated compared to CAP without presence of RV regarding: male gender (50 vs 70%; P = 0.21), mean age (40.6 vs 41 years; P = 0.92), median CD4 cell count (229.5 vs 228.5; P = 0.75), mean logarithmic viral charge (4.04 vs 3.68; P = 0.408), HAART (36 vs 41.5%; P = 0.694), positive hemoculture (21.4 vs 28.3; P = 0.85), admission in ICU (7 vs 15%; P = 0.67), mechanical ventilation (7 vs 2%; P = 0.37), APACHE II score (10.5 vs 11; P = 0.38), death (7 vs 0%; P = 0.29) and median of hospitalisation (5 vs 7 days; P = 0.076).
Conclusions: The implication of RV in CAP of HIV infected patients is not associated to a worst outcome.
P652 Incidence, reasons, and risk factors for hospital admissions in patients starting their clinical management in the era of combination antiretroviral therapy
A. Tone, N. Viget, P. Choisy, V. Baclet, F. Ajana, Y. Gerard, H. Melliez, Y. Mouton, Y. Yazdanpanah (Tourcoing, FR)
Objective: To assess the incidence and reasons for hospital admissions and to determine factors associated with hospitalisation in a French clinical cohort of HIV-infected patients
Methods: We conducted our study on HIV-infected patients from the Tourcoing AIDS Reference Center who started their clinical management from January, 1997 to August, 2004 and followed through December, 2004. We categorised diagnoses into seven disease groups. Time to hospital admission was calculated using the Kaplan-Meier method, from enrolment to the first admission. A multivariate Cox model was used to determine independent risk factors for hospital admission among demographic, clinical and laboratory characterises of patients at the inclusion.
Results: Among 781 patients followed for a median period of 2.9 years (IQ25–75, 1.3–5.3), 325 patients (41.6%) experienced at least one hospital admission, 179 (22.9%) at least two admissions, and 112 (14.3%) at least three admissions. The risk for the hospital admission was estimated at 33.2%, 39.3%, and 43.9% at one, two, and three years after enrolment, respectively. The most common reasons for hospitalisation were AIDS defining illness (26.3%), non AIDS-defining infections (16.7%), neoplasms (14.3%)and toxic events related to treatments (10.7). The less frequents reasons for hospitalisation were the cardiovascular diseases (2.2%). In the multivariate analysis, age >50 years (vs. <30, hazard ratios [HR], 1.7; 95% Confidence Interval [95%CI], 1.1–2.5), initial AIDS stage (HR, 8.2; 95%CI, 6.0–11.1), HIV viral load >5 log copies/mL (vs. <4.5; HR, 1.4; 95%CI, 1.1–1.9), viral hepatitis infection (HR, 2.0; 95%CI [1.4–2.8), and a history of mental illness at enrollment (HR, 1.7; 95%CI, 1.2–2.4) were predictive of hospital admission.
Conclusion: Our data indicates that in the era of combination antiretroviral therapy, hospital admission remain substantial among HIV-infected patients. Specific interventions should target subgroups of patients at risk of hospital admission to reduce the occurrence of events for which hospitalisations are required. Given the high cost of hospital admissions, these interventions might be particularly efficient and should be prioritised.
P653 Mycobacterium tuberculosis disease in HIV-infected patients in HAART era
J. Guardiola, L. Matas, A. Mauri, S. Herrera, M. Mateo, M. Fuster, M. Sambeat, J. Cadafalch, P. Domingo (Barcelona, ES)
Background: The incidence and prevalence of M. tuberculosis in HIV-infected patients remains variable. The aim of this study was to evaluate the presence of M. tuberculosis diseases during the HAART era in our HIV population.
Methods: We retrospectively studied all HIV-AIDS records from 1997–2004 at a teaching-urban hospital in Barcelona (Spain). All positive microbiologic cultures of M. tuberculosis from our data-base and medical records from 1997 to 2004 were reviewed.
Results: We analysed 1502 HIV-infected patients. During this period, a total of 623 (41%) patients showed one or more microbiologically documented infections. A total of 71/1502 (4.7%) patients showed one or more positive cultures of M. Tuberculosis, with 132 positive microbiological samples (30 patients had more than one site infection), representing 11% of total prevalence of documented infections during this period (71/623). 68.8% were men. Mean age was 40 + 9 (range: 18–79). 89% of patients were on highly-active antiretroviral therapy. The most frequent samples isolates were: sputum 63 samples, bronchial lavage 18 samples, urine 12 samples, adenopathy 12 samples, and hemoculture 11 samples. 31/71 (43%) patients showed disseminated infection.
Conclusions:
-
1
Almost 5% of patients had a M. tuberculosis infection.
-
2
M. tuberculosis constituted 11% of all infections.
-
3
Respiratory tract infections were the most prevalent site of infection.
-
4
43% patients showed disseminated infection.
P654 Neoplasia in patients with HIV infection: effect of HAART
A.A. Garcia-Egido, M.A. Escobar, S.P. Romero, J.A. Bernal, C. Asencio, J.L. Puerto, J. Gonzalez-Outon, F. Gomez (El Puerto Santa Maria, ES)
Surveillance data indicate that, the incidence and type of HIV related neoplasias has changed after the introduction of HAART.
Objective: To determine the incidence and types of cancers in the pre-HAART and post-HAART eras, and the differences between women and men.
Methods: Retrospective record review of HIV infected patients with cancer from January 1991 to December 2003 at a teaching hospital.
Results: A total of 215 HIV patients with cancer were identified, 173 men (age: 38±11 years, CD4: 123±39/μL) and 42 women (age: 44±16 years, CD4: 68±27/μL), of them 90 were detected in the pre-HAART (47%) and 102 in the post-HAART era (53%, p = n/s). AIDS defining cancers (Total: 215, 72.4%; of them 108, 77.7% in men vs. 31, 22.3% in women) were more frequent pre-HAART (Total: 76, 54.7%; of them 60, 43% in men vs. 16, 11.5% in women) than post-HAART (Total: 63, 45.3%; of them 48, 34.5% in men vs. 15, 10.8% in women). Non AIDS defining cancers (Total: 53, 27.6%; of them 34, 64% in men vs. 19, 36% in women) were less frequent pre-HAART (Total: 14, 26.4%; of them 9, 17% in men vs. 5, 5.3% in women) than post-HAART (Total: 39, 73.6%; of them 25, 47.2% in men vs. 14, 26.4% in women). Total cancer related mortality was higher pre-HAART (61% vs 52%, p <0.05) and in women (60% vs 55%, p = n/s) than post-HAART (Total: 53, 27.6%; of them 36, 18.8% in men vs. 17, 8.9% in women). AIDS defining cancer related mortality was higher pre-HAART (65.8% vs 36.5%, p <0.01) in both sexes (p <0.01), while non AIDS defining cancer related mortality was higher post-HAART (76.9% vs 35.7%, p <0.01) in both sexes (p <0.01).
Conclusions: The incidence of non AIDS defining cancer increased, although the total incidence of HIV related neoplasia has not changed after HAART Total cancer related mortality is higher pre-HAART and in women. Both sexes have a higher mortality by AIDS defining cancer pre-HAART and, by non AIDS defining cancer post-HAART.
P655 Incidence and risk factor of major opportunistic infections after initiation of antiretroviral therapy between HIV-infected patients with baseline CD4 cell counts ≤50 cells/mm3 and >50 cells/mm3
W. Manosuthi, A. Chaovavanich, S. Tansuphaswadikul, Y. Inthong, S. Chottanapund, C. Sittibusaya, V. Moolasart, S. Sungkanuparph (Nonthaburi, TH)
Objectives: To study incidence and risk factors for new episodes of major opportunistic infections (OIs) after antiretroviral therapy (ART) among HIV-infected patients.
Methods: A retrospective cohort study was conducted among naïve HIV-infected patients who were initiated ART between January 2001 and December 2003. Patients were categorised into two groups based on baseline CD4 cell counts: group A (≤50 cells/mm3) and group B (>50 cells/mm3). All patients were followed until 15 months after ART initiation.
Results: There were 793 patients with a mean age of 35.1±7.4 years and 56% male. Of 793 patients, 531 (67.0%) were in group A and 262 (33.0%) were in group B. Median (IQR) CD4 cell count was 13 (6–26) cells/mm3 in group A and 116 (78–167) cells/mm3 in group B. Median (IQR) baseline plasma HIV RNA was 300,500 (138,500–556,000) copies/mL and 185,000 (68,500–577,500) copies/mL in the corresponding groups (P < 0.05). Group A had a higher proportion of male, previous OIs, body weight, transaminase enzymes (P < 0.05). Incidence of major OIs (i.e., tuberculosis, cryptococcosis, pneumocystis pneumonia, histoplamosis and CMV disease) at 1, 3, 6, and 12 months after ART were 2.8%, 6.6%, 8.3% and 9.8% in group A; and 1.5%, 1.9%, 3.1% and 3.5% in group B (log rank test, P = 0.002). Cox's proportional hazard model showed that the patients with baseline CD4 cell count ≤50 copies/mL was associated with high incidence of OIs after initiation of ART (P = 0.018, HR = 4.292, 95%CI = 1.289–14.286).
Conclusions: HIV-infected patients who had baseline CD4 ≤50 cells/mm3 had a higher probability of the occurrence of major OIs after initiation of ART than those who had baseline CD4 >50 cells/mm3. Closed monitoring of clinical condition is strongly recommended after ART initiation in patients with low baseline CD4 cell counts.
P656 GB virus C infection among HIV-positive patients in Estonia
K. Denks, K. Huik, R. Avi, M. Sadam, T. Krispin, T. Karki, I. Lutsar (Tartu, EE)
Objectives: GB virus C (GBV-C) infection is not associated with any human disease. Some studies have shown that HIV-1 and GBV-C coinfected patients may have improved AIDS-free survival and higher CD4+ T-cell counts than GBV-C negative patients. The aim of current study was to determine the GBV-C infection frequency in HIV infected subjects in Estonia and to analyse if GBV-C infection associates with any other parameter of the study population.
Methods: Blood samples were collected from 95 HIV positive subjects (68 male and mean age of 28 years), between October 2005 and September 2006. 38 subjects were prisoners. All study subjects were infected with HIV between 2000 and 2005. 70 (74%) patients were or had been intravenous drug users (IDU), the remaining 25 were infected heterosexually Among IDUs there were significantly more men than women; 61 (90%) vs 9 (33%) (OR = 17.4; p < 0.0001). 65 (68%) subjects were coinfected with hepatitis C (HCV) and/or hepatitis B virus (HBV). At the time of sampling, 14 study subjects were treated with antiretrovirals. GBV-C infection was detected using PCR of NS5A region [1]. The positive result was confirmed and genotyped using type-specific PCR targeting GBV-C 5′ nontranslated region 2]. PCR products were analysed in agarose gel electrophoresis or by sequencing.
Results: GBV-C infection was detected in 33/95 (35%) subjects; the genotype 2 was the only one found. GBV-C infection was associated with IDU – 28 (41%) of past or current IDUs carried GBV-C infection versus only 5 (33%) of non-IDUs (OR = 6.7; p < 0.0001). Of men, 27 (40%) were GBV-C positive as compared with six (22%) women, but this difference arose from IDU rather from sex. The GBV-C carriage was not associated with HCV and/or HBV infection nor with the age. The CD4+ T-cell number and HIV load were measured six months prior the sampling in 55 and 43 patients, respectively. The average CD4+ T-cell count was similar in GBV-C positive and negative subjects; 289 vs 412 cells/mL, respectively. The average viral load was 50,580 copies/mL in GBV-C negative patients vs 207,754 copies/mL in GBV-C positive patients (p = 0.04).
Conclusion: The GBV-C infection was detected in one third of HIV-positive patients and its presence was associated with higher HIV loads. The relationship between GBV-C infection and IDU suggests that GBV-C could mainly be transmitted via intravenous route.
P657 HPV lesions in both genitalia and mouth of HIV sero-positive male patients
E. Giovani, R. Müller, F. Aguiar, J. Melo, P. Armonia, R. Andia-Merlin (São Paulo, BR)
Objectives: HIV has become an important risk factor for HPV infection and the development of HPV associated lesions. So, the aim of this study was to correlate HPV lesions in male genitalia and in mouth of HIV sero-positive patients.
Methods: 179 male patients were selected from the Attendance Center of Special Patients with Venereal Diseases and HIV/Aids – SP/Brazil from April/03 to March/06. These patients had the HPV lesions confirmed in genitalia after clinical examination, peniscopy and biopsy performed by an Urologist. All the patients were submitted to serologic exams (HIV/Aids), and CD4 T-lymphocyte count was carried out for those sero-positives for HIV The age, skin colour and exposure category were also analysed for all patients. The whole group was sent to a dentist for HPV lesions detection and diagnosis in oral cavity. When lesions were diagnosed, the results were confirmed by anatomy-pathological exam. After the results, the prevalence of the lesions was tabulated and a linear correlation was performed. The treatment protocol used for oral lesions was the topic administration of 75% trichloracetic acid.
Results: The mean age of the patients was 34 years, 136 (58%) leucoderm and 43 (18%) melanoderm. 46 (26%) were HIV– (32 homosexuals and 14 heterosexuals) and 133 (74%) were HIV+ (101 homo and 32 hetero). 27 patients (15%) showed concomitants lesions in genitalia and oral cavity and all they were HIV+ (20 homo and 7 hetero). 8 patients (30%) presented T-CD4 cells <200 for mm3 of blood, 15 (55%) presented 200 to 500 T-CD4, 4 (15%) presented T-CD4>500. All patients had total remission of the lesion after treatment, but for those that showed T-CD4 cells <200, more sessions of treatment were necessary (>4 sessions).
Conclusion: There is a positive linear correlation between HPV lesions in male genitalia and in mouth. HIV is an important risk factor for HPV infection and the development of HPV lesions in mouth.
P658 Clinical and paraclinical manifestations of thyroid dysfunction among patients with HIV/AIDS Tehran, Iran (2004–2005)
M. Rasoolinejad, S. Afhami, M. Izadi, M. Hajabdolbaghi, P. Khairandish (Tehran, IR)
Objective: The aim of this study was to determine the prevalence of thyroid dysfunction and associated risk factors in human immunodeficiency virus infected and acquired immunodeficiency syndrome patients.
Materials and Methods: In this cross sectional study, HIV/AIDS patients referring to the AIDS Clinic at Imam Khomeini Hospital in Tehran were included. All patients underwent complete history taking and physical examination regarding signs and symptoms of thyroid dysfunction and a sample of venous blood was drown in order to test for free T4, free T3, and thyroid-stimulating hormone (TSH) levels. Data on age, sex, weight variation, duration of HIV infection, CD4 cell count, HIV–HCV co-infection, and antiretroviral treatment were also collected.
Results: Between March 2004 to March 2005, 88 patients (mean age: 35.2±6.9 years) with HIV/AIDS, consisting of 73 males and 12 females, were included. Eighteen patients (20.5%) had abnormal thyroid function tests at different levels: one patient (1.1%) had clinical hypothyroidism, 2 patients (2.2%) had sub-clinical hypothyroidism, 12 patients (13.6%) had low free T4 and 3 patients (3.4%)had low free T3 levels. After exclusion of the last 3 patients, a case-control study was conducted which compared hypothyroid (15) with euthyroid (70) patients with respect to other influencing factors. Univariate and also multivariate analysis did not show any significant relationship between the studied parameters of age, sex, weight, CD4 count, HCV co-infection, stage of the disease and antiretroviral therapy of HIV/AIDS patients and presence of thyroid dysfunction.
Conclusion: According to the findings of this study hypothyroidism is prevalent in HIV/AIDS patients therefore, it is recommended that thyroid function should be evaluated all HIV/AIDS patients during routine follow-ups in order to detect any abnormality and initiate appropriate management at an earlier stage.
Resistance surveillance of Gram-negatives
P659 Susceptibility of ESBL, AmpC and K1 β-lactamase producing Enterobacteriaceae to carbapenems in Copenhagen, Denmark
H. Fjeldsøe-Nielsen, C. Schlegel, K. Schønning, A. Friis-Møller (Hvidovre, DK)
Objective: Carbapenems are often chosen for treatment of serious infections with cephalosporin resistant Enterobacteriaceae. In the present study we investigated the susceptibility of isolates of Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca producing ESBL (extended-spectrum β-lactamase) and/or AmpC or chromosomal K1 β-lactamase to three different carbapenems (meropenem, ertapenem and imipenem).
Methods: During the first 6 months of 2006 145 isolates of cephalosporin resistant E. coli (106), K. pneumoniae (30), K. oxytoca (9) collected in our routine laboratory were tested by E-test for meropenem (SRGA breakpoint 0.12/8 mg/l), ertapenem (0.25/2 mg/l) and imipenem (1/8 mg/l) according to Oxoid E-test technical manual. Isolates had already been characterised as being ESBL positive, AmpC positive, ESBL and AmpC positive, or K. oxytoca hyperproducing chromosomal K1 β-lactamase.
Results: The isolates collected were classified as follows: 65 ESBL, 40 AmpC, 38 ESBL and AmpC, 2 K1. Meropenem had the lowest MIC90 (minimal inhibitory concentration) values for ESBL positive and AmpC positive E. coli and K. pneumoniae. Among these isolates the difference between ertapenem and imipenem was minimal. However, among isolates possessing both ESBL and AmpC β-lactamases MIC90 was highest for ertapenem compared to both meropenem and imipenem and also compared to isolates possessing either ESBL or AmpC. When tested for ertapenem 15 of 145 isolates had MIC values >0.25 mg/l. Of these 11 were both ESBL and AmpC positive. Among the remaining isolates 130/145 had MIC values ≤0.25 mg/l for ertapenem. 5/145 isolates were intermediate to meropenem all of which were both ESBL and AmpC positive.
Conclusion: Most cephalosporin resistant Enterobacteriaceae remain susceptible to carbapenems. However, in our study isolates harbouring both ESBL and AmpC β-lactamases seem to be less sensitive to ertapenem and meropenem than isolates containing either ESBL or AmpC β-lactamases.
P660 Pseudomonas aeruginosa in German intensive care units
E. Meyer, F. Schwab, P. Gastmeier, H. Rueden, D. Jonas (Freiburg, Berlin, Hannover, DE)
Objective: To analyse resistance data of Pseudomonas aeruginosa (PAE) in German ICUs participating in the project SARI (Surveillance of Antimicrobial use and antimicrobial Resistance in German Intensive Care Units.)
Methods: From 2000–2005 resistance rates in 45 ICUs were calculated and correlated with antibiotic use and structure parameters. Temporal changes were tested by Wilcoxon test for paired samples.
Results: A total of 7187 PAE were included. The mean resistance rate to imipenem was 22.9% (range 0–50.4), to piperacillin-tazobactam 21 (range 0–50.4), to ceftazidime 16.1 (0–55.6), to ciprofloxacin 17.1 (range 0–50.4), to meropenem 13.8 (range 0–100). At 7% resistance to amicacin was lowest (range 0–68.6). Mean ceftazidime resistance increased significantly from 2000–2003 to 2004–2005 (from 14.6 to 18.8%) whereas amicacin resistance decreased from 8.2 to 4.5%. Ceftazidime resistance was significantly higher in hospitals with >1000 beds and resistance to amicacin was significantly higher in interdisciplinary ICUs. Carbapenem use correlated significantly and with a correlation coefficient >0.5 with imipenem, meropenem and ceftazidime resistance of PAE.
Conclusion: Over 20% of PAE in German ICUs are resistant to imipenem and piperacillin-tazobactam. This has not changed over the last 5 years. Carbapenem use and resistance correlate significantly.
P661 ICU antimicrobial susceptibility rates among Gram-negative bacilli isolated from infections in the USA: results from ICU surveillance study 2005
G. Gallagher, H. Wilson, M. Abramson (West Point, US)
Background: ISS (ICU Surveillance Study) is an ongoing US antimicrobial surveillance programme that has focused on infections from Intensive Care Units (ICU). The objective of this analysis was to assess antimicrobial susceptibility patterns among aerobic & facultative Gram-negative bacilli recovered at participating sites in the USA during 2005.
Methods: 39 centres in the USA each tested the in vitro activity of 15 antimicrobial agents. Consecutive unique Gram-negative bacilli from all body sites were tested using microdilution techniques according to CLSI guidelines and breakpoints. Production of extended-spectrum β-lactamases (ESBL) was confirmed in isolates with a MIC of ceftriaxone, ceftazidime, or cefepime ≥2 μg/mL by comparing ceftazidime, cefotaxime, and cefepime MICs with and without clavulanate.
Results: A total of 4304 isolates were recovered from 3665 patients. The bacteria species with a prevalence of >5.0% were P. aeruginosa (1030 isolates; 24%), E. coli (747 isolates; 17%), K. pneumoniae (658 isolates; 15%), E. cloacae (376 isolates; 9%), A. baumannii (302 isolates; 7%), and S. marcescens (244 isolates; 6%). The percent susceptible are reported in the table . Most prevalent body sites were respiratory isolates (2,224; 52%), blood isolates (812; 19%), and urine isolates (574; 13%).
| Pseudomonas aeruginosa | Escherichia coli | Klebsiella pneumoniae | Enterobacter cloacae | Acinetobacter baumannii | Serratia marcescens | |
|---|---|---|---|---|---|---|
| Ertapenem | N/A | 98 | 89 | 98 | N/A | 96 |
| Imipenem | 76 | 99 | 93 | 99 | 78 | 97 |
| Cefepime | 62 | 91 | 73 | 70 | 33 | 91 |
| Cefotaxime | 8 | 95 | 78 | 62 | 22 | 78 |
| Cefoxitin | N/A | 90 | 76 | 7 | N/A | 29 |
| Ceftazidime | 71 | 89 | 71 | 54 | 38 | 83 |
| Ceftriaxone | 18 | 89 | 71 | 54 | 29 | 86 |
| Amp/Sulbactam | 2 | 52 | 60 | 16 | 50 | 4 |
| Pip/Tazobactam | 81 | 94 | 78 | 75 | 38 | 83 |
| Amikacin | 84 | 98 | 83 | 96 | 73 | 96 |
| Gentamicin | 57 | 88 | 81 | 86 | 39 | 90 |
| Tobramycin | 73 | 87 | 74 | 85 | 63 | 85 |
| Ciprofloxacin | 60 | 76 | 76 | 86 | 37 | 89 |
| Lexofloxacin | 60 | 77 | 79 | 90 | 38 | 95 |
| Aztreonam | 59 | 89 | 71 | 57 | 12 | 87 |
Conclusion: In this study, the most common Enterobacteriaceae organisms were E. coli and K. pneumoniae. P. aeruginosa was the predominant Non-Enterobacteriaceae. Amikacin was the most active agent in vitro for P. aeruginosa. Ertapenem, imipenem, and amikacin were the most reliably active drugs in vitro against Enterobacteriaceae.
P662 Affect of length of hospitalisation on susceptibility patterns of Gram-negative bacilli isolated from intra-abdominal infections: SMART 2005
G. Gallagher, F. Rossi, P.R. Hsueh, J. Báez-Villaseñor, H.M. Wilson, M. Abramson (West Point, US; Taipei, TW; São Paulo, BR)
Background: SMART (Study for Monitoring Antimicrobial Resistance Trends) is an ongoing global antimicrobial surveillance programme focused on clinical isolates from intra-abdominal infections (IAI). Isolates identified after 48 hours (h) of hospitalisation have been shown to have less susceptibility than those taken within the first 48h. This 2005 sub-analysis assessed susceptibility patterns among Gram-negative bacilli from 5 regions of the world.
Methods: 50 major medical centres in North America, Latin America, Europe, Middle East/Africa, & Asia/Pacific tested the in vitro activity of 12 antimicrobial agents. Microdilution techniques followed CLSI guidelines. Enterobacteriaceae susceptibility rates were compared between isolates recovered <48 h & >48 h after hospitalisation.
Results: Enterobacteriaceae were recovered from 3226 pts (3422 isolates) worldwide. 648 isolates (19%) were Inducible; 2774 (81%) were non-Inducible Enterobacteriaceae. 238 inducible (7%) & 1423 (42%) non-Inducible Enterobacteriaceae were recovered <48 h after hospitalisation. The susceptibility rates are presented in the table .
| Susceptibility rate (%) |
||||
|---|---|---|---|---|
| Inducible Enterobacteriaceae* |
Non-inducibleEnterobacteriaceae# |
|||
| <48 h | ≥48 h | <48 h | ≥48 h | |
| Entrapenem | 96 | 94 | 97 | 97 |
| Imipenem | 97 | 96 | 98 | 98 |
| Cefepime | 94 | 87 | 93 | 81 |
| Cefotaxime | 79 | 59 | 93 | 81 |
| Cefoxitin | 27 | 24 | 93 | 81 |
| Ceftazidime | 77 | 57 | 92 | 79 |
| Ampicillin/Sulbactam | 34 | 21 | 62 | 43 |
| Piperacillin/Tazobactam | 34 | 21 | 62 | 43 |
| Amikacin | 99 | 93 | 97 | 95 |
| Ciprofloxacin | 91 | 83 | 84 | 70 |
| Levofloxacin | 93 | 88 | 86 | 72 |
Length of hospitalisation not reported for 6* & 33# isolates.
Conclusion: Susceptibility rates for most Enterobacteriaceae were slightly higher for isolates recovered <48 h after hospitalisation. Ertapenem, imipenem, & amikacin were the most active drugs in vitro regardless of time of hospitalisation.
P663 Worldwide antimicrobial susceptibility patterns among E. coli isolated from intra-abdominal infections (IAI): results from SMART 2005
G. Gallagher, P.R. Hsueh, F. Baquero, D. Paterson, J. Báez-Villaseñor, H.M. Wilson, M. Abramson (West Point, Pittsburgh, US; Taipei, TW; Madrid, ES)
Background: SMART (Study for Monitoring Antimicrobial Resistance Trends) is an ongoing global antimicrobial surveillance programme focused on clinical isolates from intra-abdominal infections (IAI). The aim of the 2005 analysis was to assess antimicrobial susceptibility patterns among E. coli from 5 different regions of the world.
Methods: 48 centres in the North America (NA), Latin America (LA), Europe (EU), Middle East/Africa (ME/A), & Asia/Pacific (A/P) tested the in vitro activity of 12 antimicrobial agents Microdilution techniques followed CLSI guidelines. E. coli were screened for extended-spectrum β-lactamase (ESBL) production by ceftriaxone, cefotaxime, cefepime, or ceftazidime MIC greater than or equal to 2 ug/mL & confirmed by comparing ceftazidime, cefotaxime, and cefepime MICs ± clavulanate.
Results: E. coli was recovered from 1852 patients of the study's 3553 patients (3805 isolates) worldwide. % susceptibilities are reported by region in the table . ESBL-producing E. coli constituted 1%, 9%, 4%, 9%, & 21% of isolates from NA, LA, EU, ME/A, & A/P, respectively. 53% (979/1852) were recovered <48 hrs of hospitalisation. Of those, 3% (32/979) were ESBL producers.
| NA N = 156 | LA N = 190 | EU N = 1036 | M E/A N = 32 | A/P N = 438 | |
|---|---|---|---|---|---|
| Ertapenem | 100 | >99 | >99 | 97 | 90 |
| Imipenem | 100 | 100 | >99 | 100 | 90 |
| Ceftriaxone | 97 | 89 | 93 | 91 | 68 |
| Ceftazidime | 97 | 90 | 92 | 91 | 68 |
| Cefotaxime | 97 | 91 | 94 | 91 | 71 |
| Cefoxitin | 94 | 95 | 94 | 84 | 77 |
| Cefepime | 99 | 91 | 94 | 91 | 71 |
| Ampicillin-Sulbactam | 60 | 43 | 48 | 34 | 36 |
| Piperacillin-Tazobactam | 96 | 96 | 95 | 88 | 82 |
| Amikacin | 99 | 98 | >99 | 97 | 87 |
| Ciprofloxacin | 84 | 75 | 82 | 75 | 56 |
| Levofloxacin | 84 | 77 | 83 | 75 | 58 |
Conclusion: Prevalence of ESBL-producing strains may affect choice of empirical therapy. Many ESBL-producing E. coli may have been hospital acquired. Overall, ertapenem, imipenem, and amikacin were the most reliably active drugs in vitro against E. coli.
P664 Antimicrobial susceptibility patterns of inducible Enterobacteriaceae isolated from intraabdominal infections in Europe: results from SMART 2005
G. Gallagher, F. Baquero, F. Rossi, J. Báez-Villaseñor, H.M. Wilson, M. Abramson (West Point, US; Madrid, ES; São Paulo, BR)
Background: SMART (Study for Monitoring Antimicrobial Resistance Trends) is an ongoing global antimicrobial surveillance programme focused on clinical isolates from intraabdominal infections (IAI). The 2005 analysis assessed antimicrobial susceptibility patterns among inducible Enterobacteriaceae from Europe.
Methods: 25 European sites tested the in vitro activity of 12 antimicrobial agents. Microdilution techniques followed CLSI guidelines. All Enterobacter, Serratia, Citrobacter, Providencia spp., Morganella morganii, Hafnia alvei, & Proteus vulgaris were considered to have inducible β-lactamases for this study.
Results: Inducible Enterobacteriaceae were recovered from 19% of pts (347/1820); 18% (350/1964) of the total isolates. 42% (145) of these isolates were recovered <48 hrs of hospitalisation. Enterobacter spp. (46%), Citrobacter spp. (25%), M. morganii (12%) & Serratia spp. (6%) were the most common isolates. Susceptibility rates are listed in the table .
| Susceptibility (%) |
||||
|---|---|---|---|---|
| Enterobacter spp. N = 162 | Citrobacter spp. N = 86 | Morganella morganii N = 43 | Serratia spp. N = 20 | |
| N = 162 | N = 86 | N = 43 | N = 20 | |
| Ertapenem | 96 | 99 | 100 | 100 |
| Imipenem | 98 | 99 | 100 | 100 |
| Cefoxitin | 5 | 20 | 70 | 35 |
| Ceftriaxone | 59 | 64 | 95 | 90 |
| Ceftazidime | 57 | 61 | 86 | 100 |
| Cefotaxime | 61 | 69 | 88 | 90 |
| Cefepime | 91 | 98 | 100 | 100 |
| Ampicillin-Sulbactam | 20 | 48 | 9 | 10 |
| Piperacillin-Tazobactam | 79 | 84 | 100 | 90 |
| Amikacin | 98 | 99 | 100 | 100 |
| Ciprofloxacin | 90 | 88 | 88 | 95 |
| Levofloxacin | 93 | 94 | 93 | 95 |
Conclusion: The majority of isolates were recorded >48 hrs of hospitalisation. Enterobacter species were the most common isolates. Cefoxitin & ampicillin-sulbactam were the least active agents; ertapenem, imipenem & amikacin were the most active agents in vitro.
P665 Multicentre surveillance of Pseudomonas aeruginosa susceptibility patterns in nosocomial infections
J. Van Eldere, I. Devenijns (Leuven, BE)
Objectives: To determine susceptibility rates and patterns of nosocomially acquired Pseudomonas aeruginosa from Belgian hospitals against commonly used antibiotics and compare these data with data from a similar surveillance study performed 5 years earlier.
Methods: 1250 strains from 40 Belgian hospitals were collected in 2005. Only clinically significant non-duplicated isolates from blood, deep respiratory samples, sterile body fluids and urine samples taken >48 hrs after admission were included. All strains were centralised in a single lab, re-identified and MIC values were determined with an agar-dilution method according to CLSI guidelines against the following antibiotics: piperacillin-tazobactam (P-T), aztreonam (AZ), ceftazidime (CT), cefepime (CP), meropenem (ME), amikacin (AK), tobramycin (TB), gentamicin (GN), ciprofloxacin (CF), levofloxacin (LF).
Results: Applying CLSI breakpoints, susceptibility rates in decreasing order were 91% for P-T, 90% for AK, 88% for ME, 84% for CT and TB, 80% for CP, 77% for GN and CF, 72% for LF and 65% for AZ. Corresponding MIC50 and MIC90 values were respectively 8/64 for P-T, 4/16 for AK, 1/8 for ME, 2/32 for CT, 1/128 for TB, 4/16 for CP, 2/64 for GN, 0.25/32 for CF, 1/32 for LF and 8/32 for AZ. Significant differences were observed between hospitals but not according to sample origin. Compared to a similar 1999 survey, there was a significantly increased susceptibility to all β-lactam antibiotics and fluoroquinolones, whereas susceptibility to aminoglycosides remained stable. Beta-lactam and fluoroquinolone MIC distribution curves showed a clear shift to the left between 1999 and 2005.
Conclusion: Resistance to β-lactams and fluoroquinolones has decreased compared to 5 years ago. In spite of reduced overall resistance, no single anti-pseudomonas antibiotic has sufficiently high susceptibility levels to warrant monotherapy for nosocomial P. aeruginosa infections.
P666 Comparative in vitro activity of tigecycline against ICU- and non-ICU isolates of five clinically important Gram-negative pathogens: results of the German T.E.S.T. Surveillance Programme 2005
M. Kresken, J. Brauers, H. Geiss, E. Halle, E. Leitner, G. Peters, H. Seifert on behalf of the German T.E.S.T. Surveillance Programme
Objectives: Tigecycline (TGC), the first glycylcycline antibacterial agent, has been shown to be highly effective against a wide range of bacteria including multiple resistant Gram-negative pathogens such as extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. G.-T.E.S.T. is a surveillance programme comprising 15 German laboratories which monitors the susceptibility of bacterial pathogens to TGC. The objective of this study was to evaluate the in vitro activity of TGC against both ICU- and non-ICU isolates of Acinetobacter baumannii (Ab), Stenotrophomonas maltophilia (Sm) and three major members of the family Enterobacteriaceae.
Methods: A total of 410 ICU isolates (36 Ab, 120 Enterobacter spp. [En], 93 Escherichia coli [Ec], 87 Klebsiella spp. [Kl], 74 Sm) and 679 non-ICU isolates (93 Ab, 140 En, 187 Ec, 181 Kl, 78 Sm) were tested against TGC, doxycycline (DOX), ciprofloxacin (CIP), cefotaxime (CTX), imipenem (IMP), piperacillin-tazobactam (P/T) and other drugs. MICs were determined by broth microdilution according to German DIN guidelines in a central laboratory. The MICs of TGC were interpreted by both EUCAST and FDA criteria. DIN breakpoints were applied to the other drugs.
Results: TCG exhibited comparable activity against ICU- and non-ICU isolates of all five organisms tested. MIC50/90 values for ICU- vs non-ICU isolates were 0.25/0.5 vs 0.25/0.5 for Ab, 0.5/1 vs 0.5/2 for En, ≤0.125/0.25 vs ≤0.125/0.25 for Ec, 0.5/4 vs 0.5/2 for Kl, and 0.5/2 vs 0.5/1 for Sm, respectively. In contrast, resistance to CTX and/or P/T in En and Kl was more frequently distributed among ICU- than non-ICU isolates. Overall, susceptibility rates for ICU/non-ICU isolates (%) were as follows: Ab – TGC (no breakpoints available, n. b. a.), DOX 97/90, CIP 69/71, CTX 0/5, IMP 94/100, P/T 64/69, En – TGC EUCAST:93/88 FDA:93/94, DOX 33/29, CIP 94/94, CTX 53/74, IMP 100/100, P/T 58/69, Ec – TGC EUCAST:100/100 FDA:100/100, DOX 47/47, CIP 76/79, CTX 95/94, IMP 100/100, P/T 88/94, Kl – TGC EUCAST:82/88 FDA:90/91, DOX 59/66, CIP 89/93, CTX 93/97, IMP 99/100, P/T 74/96, and Sm – TGC (n. b. a.), DOX 34/38, CIP 82/79, CTX 0/1, IMP 0/1, P/T 0/1.
Conclusion: TGC demonstrated excellent in vitro activity against major Gram-negative pathogens, with almost equivalent activity against strains from ICU- and non-ICU patients. TGC seems to be a useful drug for the treatment of infections caused by clinically important Gram-negative bacteria, even in patients on intensive care units.
P667 Assessment of the polymyxin B antimicrobial activity tested against 26,921 clinical strains of Gram-negative bacilli collected in Europe: report from 10 years of the SENTRY Antimicrobial Surveillance Program
H. Sader, T. Fritsche, M. Janechek, R. Jones (North Liberty, US)
Objective: To evaluate the in vitro activity of polymyxin B tested against Gram-negative organisms isolated from patients hospitalised in European medical centres. Emergence of multidrug-resistant (MDR) P. aeruginosa, Acinetobacter spp. and K. pneumoniae isolates causing life-threatening infections has restored the potential therapeutic indication for the parenteral use of the polymyxin class (polymyxin B or colistin).
Methods: A total of 26,921 Gram-negative bacilli isolated from diverse sites of infection were tested for susceptibility (S) against polymyxin B by reference broth microdilution method and the results were interpreted according to the S breakpoint established by the CLSI (2007) for Acinetobacter spp. and P. aeruginosa (≤2 mg/L). The isolates were collected through the SENTRY Antimicrobial Surveillance Program between 2001 and 2006 in 34 medical centres located in 12 European countries, Turkey and Israel. Concurrent quality control was obtained and all results were within CLSI ranges.
Results: Polymyxin B showed excellent potency and spectrum against 4,137 P. aeruginosa (MIC90, 2mg/L; 99.5% S) and 1,191 Acinetobacter spp. strains (MIC90, ≤1 mg/L; 97.9% S). Among other non-fermentative Gram-negative bacilli, polymyxin B S rates were 92.9% for other Pseudomonas spp., 88.5% for Aeromonas spp., 78.8% for S. maltophilia, but only 25.0% for Burkholderia cepacia. Against Enterobacteriaceae, polymyxin B showed excellent activity overall (MIC90, ≤1mg/L; >98% S) against Citrobacter spp., E. coli and Klebsiella spp.; but inconsistent activity against Enterobacter spp. (MIC50, ≤1mg/L; 83.3% susceptible) and Salmonella spp. (MIC50, 2 mg/L, 65.5% S). Very limited activity (MIC50, >8mg/L) against Serratia spp. (6.2% susceptible), indole-positive Proteus (1.5% susceptible) and Proteus mirabilis (0.8% susceptible) was documented for polymyxin B.
Conclusions: Polymyxin B was highly active against Acinetobacter spp., P. aeruginosa and Klebsiella spp., including MDR strains. The emergence of acquired resistance to polymyxin B (also colistin) is of great concern since these agents are typically regarded as agents of last resort when no therapeutic options remain. Prudent use of this class is recommended guided by recently developed in vitro testing guidelines (M7-A7, M100-S17).
P668 Antimicrobial resistance and prevailing resistance mechanisms among problematic clinical isolates of Pseudomonas aeruginosa from the university hospitals in Sofia, Bulgaria
T. Strateva, V. Ouzounova-Raykova, B. Markova, Y. Marteva-Proevska, I. Mitov (Sofia, BG)
Objectives: To assess the current levels of antimicrobial susceptibility and to evaluate the resistance mechanisms to antipseudomonal antimicrobial agents among Bulgarian nosocomial isolates of Pseudomonas aeruginosa.
Methods: A total of 203 clinical isolates of P. aeruginosa, resistant to one or more of the following groups of antimicrobials – extended-spectrum cephalosporins, carbapenems, aminoglycosides, fluoroquinolones, were collected from 5 University hospitals in Sofia during 2001–2006. Antimicrobial susceptibilities were detected by the disk diffusion method and Etest (AB Biodisk, Solna, Sweden). The resistance mechanisms were studied with phenotypic methods as previously described by Jarlier V. et al., Lee K. et al. and Miller G. et al. Polymerase chain reaction and sequencing of genes, encoding Ambler class A, B and D β-lactamases, were performed.
Results: The antibiotic resistance rates were: to carbenicillin – 93.1%, azlocillin – 91.6%, piperacillin – 86.2%, piperacillin/tazobactam – 56.8%, ceftazidime – 45.8%, cefoperazone – 86.2%, cefepime – 48.9%, cefpirome – 58.2%, aztreonam – 49.8%, imipenem – 42.3%, meropenem – 45.5%, amikacin – 59.1%, gentamicin – 79.7%, tobramycin – 89.6%, netilmicin – 69.6%, and ciprofloxacin – 80.3%. 101 of the studied P. aeruginosa isolates (49.8%) were multidrug-resistant. Structural genes encoding Ambler class A and class D β-lactamases showed the following frequency: blaVEB-1 – 33.1%, blaPSE-1 – 22.5%, blaPER-1 – 0%, blaOXA-groupI – 41.3%, and blaOXA-groupII – 8.8%. IMP- and VIM-type carbapenemases from molecular class B were not detected.
Conclusions: The studied clinical stains of P. aeruginosa were problematic nosocomial pathogens. VEB-1 ESBLs appear to have a significant presence among the clinical P. aeruginosa isolates from Sofia. The carbapenem resistance was related to non-enzymatic mechanisms such as OprD deficiency and active efflux.
Beta-lactamases in epidemic Gram-negatives
P669 Epidemiology and genotypes of plasmid-mediated AmpC β-lactamase produced by clinical isolates of Klebsiella pneumoniae in Korea
W. Song, C.H. Lee, J.S. Kim, H.S. Kim, Y. Uh, J. Lee, K. Lee (Seoul, Choongju, Wonju, Daegeon, KR)
Objectives: Plasmid-mediated AmpC β-lactamases (pAmpCs) are cephalosporinases that confer resistance to a wide variety of β-lactam drugs and that may thereby create serious therapeutic problems. The pAmpC-producing organisms are a major concern in nosocomial infections and should therefore be monitored in surveillance studies. The present study was conducted to determine the epidemiology and genotypic distributions of pAmpCs among Klebsiella pneumoniae isolates in Korea.
Methods: During the period May to July 2004, 60 cefoxitin non-susceptible isolates of 735 consecutive, nonrepeat isolates of K. pneumoniae at five Korean university hospitals were tested for antimicrobial susceptibility by broth microdilution methodology. The cefoxitin non-susceptible isolates were further investigated by AmpC disk tests, double disk synergy and antagonism tests, isoelectric focusing, multiplex AmpC PCR, allele-specific PCR, DNA sequencing, and pulsed-field gel electrophoresis (PFGE)
Results: pAmpC producers were found at all the 5 sites in 48/735 K. pneumoniae (6.5%). Thirty-one of 48 pAmpC producers (64.6%) also positive tested by double disk synergy tests for extended-spectrum β-lactamases. Susceptibilities of the pAmpC producers were as follows: ceftazidime 2%, aztreonam 19%, cefepime 49%, and imipenem 96%. Among the 48 K. pneumoniae isolates, there were 47 DHA-1 and 1 CMY-1 β-lactamase. Ten PFGE patterns were shown by the DHA-1-producing K. pneumoniae isolates.
Conclusion: pAmpC producers widespread among Korean medical institutions. A DHA-1 type in K. pneumoniae was the predominant enzyme detected. Overall, despite many different PFGE patterns of the pAmpC producers, some outbreak and epidemic clones appear to be prevalent according to the hospitals in Korea. Prevention of the spread of pAmpC producers requires clinical laboratories test for this resistance mechanism.
P670 Molecular epidemiology of VIM-1 producing Klebsiella pneumoniae isolates
E. Kraniotaki, E. Platsouka, E. Belesiotou, M. Nepka, Z. Psaroudaki, A. Argyropoulou, O. Paniara (Athens, GR)
Objectives: This study was performed to investigate the molecular epidemiology of MBL producing blood isolates of Klebsiella pneumoniae, collected in our tertiary care hospital in the two-year period 2005–2006.
Methods: All consecutive K. pneumoniae isolates from blood cultures of 140 inpatients (49 in medical wards, 17 in surgical wards, 74 in ICUs) were tested. They were identified by standard methods and MICs were determined by the broth microdilution method, according to CLSI guidelines. MBL production was screened by Etest MBL. blaVIM-1 alleles were detected by PCR. The clonality of the VIM-1 positive isolates was examined by PFGE, using the restriction enzyme XbaI.
Results: Seventy out of one hundred forty isolates were found to produce an MBL activity by the Etest. The MICs of imipenem(IMI)and meropenem (MER) of the above MBL(+) isolates ranged from 0.5 to ≥16 μg/mL. The same seventy isolates, 50% of the total K. pneumoniae blood isolates, were found positive for the presence of the blaVIM-1 gene. PFGE results revealed four distinct genotypes among the MBL(+)isolates.
Conclusion: The presence of the blaVIM-1 gene in 50% of our K. pneumoniae blood isolates is high. The fact that four distinct genotypes were involved in the nosocomial spread of the MBL resistance, indicates horizontal transfer of the blaVIM-1 gene. Continuous surveillance and control measures, comprising molecular investigation methods, are necessary in order to eliminate the MBLs.
P671 Emergence of Klebsiella pneumoniae with an AmpC and blaSHV-11 in a Belgian hospital
T. Vanwynsberghe, A. Boel, R. Cartuyvels, M. Raymaekers, H. De Beenhouwer (Aalst, Hasselt, BE)
Objectives: From August to October 2006 we observed 12 unrelated clinical isolates of Klebsiella pneumoniae expressing an unusual antibiotic susceptibility pattern. Characterisation of these strains was performed.
Methods: Isolates were found in routine because of flagging by Phoenix® (BD) as “possible extended-spectrum bèta-lactamases (ESBL) positive”. Identification was confirmed by 16S rDNA sequencing. Control testing for ESBL was done with a modified double-disk synergy method also including cefoxitin. In order to verify the presence of an AmpC β-lactamase, the AmpC disk test presented by Black et al (2005), was performed. The clinical isolates were examined for the presence of blaTEM, blaTEM-24, blaSHV and blaCTX-M by polymerase chain reaction (PCR), using consensus primers for the different genes. Furthermore typing with pulsed field gel electrophoresis was performed.
Results: Strains were confirmed as Klebsiella pneumoniae. All were cefoxitin-resistant. A resistance-induction phenomenon potentiated by amoxicillin-clavulanic acid was observed with cefotaxime and aztreonam. Besides that, scattered colonies were found in the inhibition zones of ceftazidime, ceftriaxone, aztreonam, cefotaxime, and amoxicillin-clavulanic acid, but not for cefepime. On the other hand the cefepime inhibition zone showed a phantom zone in the neighbourhood of amoxicillin-clavulanic acid.
The cefoxitin-resistance leads to only a few possible causes of the expressed pattern: porin loss, AmpC β-lactamase production, or carbapenemase production (metallo-β-lactamase) are all described. In these strains the AmpC disk test was positive pointing to the presence of an AmpC β-lactamase. Furthermore the molecular investigation showed the presence of the blaSHV-11 gene, a gene which is not classified as an ESBL-producing gene.
Strains of Klebsiella pneumoniae containing SHV-11 and an AmpC β-lactamase are rare and have been described mainly in Taiwan. Until now strains producing these enzymes have been found rarely on other continents. However in the Far East the prevalence of AmpC β-lactamases in Klebsiella pneumoniae is rising.
Characteristics of the first strain discovered
| Ref. nr. | AMC |
AT M |
CAZ |
CRO |
CTX |
FEP |
FOX |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | Ø | MIC | Ø | MIC | Ø | MIC | Ø | MIC | Ø | MIC | Ø | MIC | Ø | |
| 67224 | >16 | 11a | ≤2 | 23a | >16 | 20a | ≤2 | 26 | − | 23a,b | ≤2 | 29c | − | 6 |
MIC in μg/mL; inhibition zone (Ø) in mm. AMC: amoxicillin-clavulanic acid; ATM: aztreonam; CAZ: ceftazidim; CRO: ceftriaxone; CTX: cefotaxime; FEP: cefepime; FOX: cefoxitin.
Scattered colonies in the inhibition zone
flattening of the inhibition zone towards AMC
phantom zone present towards AMC.
Conclusion: Based on the phenotype and the molecular findings an AmpC β-lactamase together with SHV-11 is very suggestive. Strains of Klebsiella pneumoniae harbouring the combination of the blaSHV-11 gene and an AmpC gene are infrequently found in Europe.
P672 Spread of extended-spectrum β-lactamases among clinical Klebsiella pneumoniae isolates from an Algerian hospital
N. Ramdani-Bouguessa, J. Leitão, E. Ferreira, M. Tazir, M. Caniça (Algiers, DZ; Lisbon, PT)
Objectives: Extended-spectrum β-lactamases (ESBLs) are a cause of resistance to third-generation cephalosporins and aztreonam in Klebsiella pneumoniae pathogens, which have an important role in nosocomial infections. The aim of this study was the characterisation of resistance mechanisms in clinical K. pneumoniae strains from an Algerian hospital.
Methods: Thirty six clinical K. pneumoniae isolates were selected for this study among a total of 131 Klebsiella spp. ESBL producer strains, collected among January and June 2005 from different specimens. MICs were determined by microdilution broth method. PCR and sequencing were used to screen and identify blaTEM, blaSHV, blaOXA, ampC and blaCTX-M genes. Environmental context of blaCTX-M genes was also analysed by PCR, searching for ISEcp1, IS26 and IS903 insertion sequences (IS). Biochemical characterisation was performed by isoelectric focusing (IEF). Genetic relatedness among strains was analysed by pulsed-field gel electrophoresis (PFGE).
Results: Of a total of 36 isolates, 32 (89%) were multidrug-resistant (MDR): 30 (94%) were resistant to β-lactam, aminoglycoside and trimethopim-sulfamethoxazole families, and 2 (6%) also to fluoroquinolones. Molecular characterisation revealed the presence of blaTEM-type (n = 33), blaSHV-1 (n = 7), blaSHV-11 (n = 11), blaSHV-28 (n = 1), blaSHV-33 (n = 1), blaSHV-type with G794 → T mutation (n = 3) or with A299 → G mutation (n = 1), other blaSHV-type (n = 11), blaCTX-M-3 (n = 10), blaCTX-M-14 (n = 1) and blaCTX-M-15 (n = 20) genes. The novel blaSHV-100 gene was also identified. ISEcp1 (n = 29) elements were detected upstream of blaCTX-M genes and IS26 (n = 14) were also downstream. IS903 (n = 2) were detected downstream of those genes. IEF confirmed strains as ESBL producers. PFGE analysis revealed a high clonal heterogeneity, with 16 unique profile types and genetically related or closely related (>80% similarity) strains forming clusters I to VII.
Conclusions: K. pneumoniae strains from Algeria showed to be ESBL producers mainly due to CTX-M (83%) enzymes, however, 19% were simultaneous ESBL producers from SHV family. The presence of IS elements in ESBL (CTX-M)-producing K. pneumoniae strains suggests their important role in the dissemination of these enzymes. Overall, the presence of IS and the first report of CTX-M-14 enzyme in this country, as well as the new SHV-100 enzyme and the concomitant MDR phenotypes constitute a microbial threat, as they contribute to the reducing therapeutic choices.
P673 Description of blaTEM-48 ESBL gene carried in addition to blaSHV-5 gene within the same megaplasmid of nosocomial Klebsiella pneumoniae strains
M. Alcántar-Curiel, J. Ortíz, R. Morfin, C. Gayosso, S. Esparza, A. Carlos, E. Rodríguez-Noriega, J. Santos, C. Alpuche-Aranda (Mexico, Jalisco, MX)
Background: Prevalence of ESBL-producing Klebsiella pneumoniae (Kpn) and other Gram-negatives in the United States of America (USA) is relatively lower compare to Mexico. The most common ESBLs (85%) described in México are SHV-5 and TLA-1 and in contrast to US there was no description of TEM ESBLs in Mexico before.
Methods: We collected nosocomial Kpn strains isolated over a 45 month period in a Mexican Hosp. Antimicrobial susceptibility patterns were determined by diffusion and dilution methods. Genotyping was determined by PFGE. E-test, isoelectrical point focusing (pI) and kinetic studies by spectrophotometric assay were used to define ESBL production. Conjugation, restriction endonuclease and Southern blot were used to characterise plasmids. PCR and sequencing analysis were used to molecular characterise ESBLs.
Results: Sixty two per cent of the 168 Kpn strains collected were ESBL producers and they were part of 23 different clones. ESBL in addition to other antibacterial resistance phenotype were transferred by conjugation of one plasmid. Most of the clones produced ESBLs with pI 7.5 and 8.2 and only two (G18 and G56) produced ESBL pI 6.0, 7.5 and 8.2. Those with pI 6.0 and 8.2 were transferred in the same identical megaplasmid in both clones. ESBL pI 8.2 corresponded to SHV-5. ESBL pI 6.0 preferentially hydrolyzed cefotaxime and it was PCR amplified by blaTEM specific primers. The deduced protein sequences showed to be 100% identical to blaTEM-48.
Conclusion: Prevalence of ESBL-producing Kpn strains in Mexico is very high mostly related to blaSHV-5 gene carried in a plasmid. However blaTEM genes can also be present in these plasmids. This is the first report of an ESBL-TEM family produced in Mexico.
P674 ESBL evolution in Italy: rapid regional spread of a multidrug-resistant Klebsiella pneumoniae strain producing CTX-M-15
C. Mugnaioli, E. Manso, G. Rossolini (Siena, Ancona, IT)
Objectives: Extended-spectrum β-lactamases (ESBLs) play a crucial role as emerging resistance determinants to expanded-spectrum cephalosporins in Enterobacteriaceae. The CTX-M-type ESBLs have recently undergone a massive spread in several settings, showing a remarkable potential for dissemination. In Italy, we have recently observed a remarkable countrywide dissemination of these enzymes in Escherichia coli (54.8% of ESBL producers) while their prevalence in Klebsiella pneumoniae was found to be much more limited (12.3% of ESBL producers). Here we describe the occurrence of a rapid regional spread of a multidrug resistant (MDR) K. pneumoniae strain producing CTX-M-15.
Methods: susceptibility testing was carried out as recommended by CLSI. Genomic relatedness among the isolates was investigated by RAPD profiling. ESBL determinants were detected by PCR, and the DNA sequence was determined by direct amplicon sequencing. Transfer of resistance genes by conjugation was assayed by mating experiments. Plasmids were characterised by RFLP analysis.
Results: during the period October 2005 – June 2006, 65 consecutive nonreplicate ESBL-positive isolates of K. pneumoniae showing an MDR phenotype including β-lactams (except carbapenems), aminoglycosides and fluoroquinolones were referred to the regional Clinical Microbiology Laboratory of the Marche region (central Italy) from 8 different hospitals. Most isolates were from ICUs, but some were from medical and surgical wards. RAPD analyses revealed clonal relatedness among isolates, all of which were found to carry the blaCTX-M-15 ESBL gene. The gene was carried on a conjugative plasmid that could be efficiently transferred to E. coli (conjugation frequencies in the order of 10−4 transconjugants per recipient). Plasmid analysis revealed apparent identity among CTX-M-15-encoding plasmids from all the isolates.
Conclusion: Results of this study underscore the ability for rapid multifocal spreading of MDR clones of K. pneumoniae that have acquired the CTX-M-15 determinant.
P675 High frequency of CTX-M genes among ESBL-producing Klebsiella pneumoniae in a university hospital in Belgium
H. Rodriguez-Villalobos, C. Laurent, R. Castany-Prado, A. Deplano, B. Byl, R. de Mendonça, M.J. Struelens (Brussels, BE)
Objectives: CTX-M enzymes are emerging in Europe and have become prevalent in many institutions among E. cloacae and E. coli but less frequently in K. pneumoniae. We investigated the molecular epidemiology of ESBL-producing K. pneumoniae (ESBL-KP) isolates over a 6 year period in a university hospital in Brussels, Belgium.
Methods: In 2000–2005, ESBL production was screened by double-disk synergy test. ESBLs were characterised by multiplex PCR for bla genes of the SHV, TEM and CTX-M family and DNA sequencing. MIC of 12 antimicrobials was tested by agar dilution. ESBL-producing strains were screened for class I and II integrase by PCR. Strains from a cluster in 2005 were genotyped by ERIC2-PCR. The clinical charts of patients were retrospectively reviewed.
Results: Strains (n = 137) were isolated from 84 males and 53 females with a mean age of 56 years (1–96). Charts (n = 96) patients showed that acquisition was either nosocomial (45% of them originated from ICU) or from previous contact with our institution or from other care centres. Incidence density (cases/10.000 pts-day) since 2000 to 2005 were 0.87, 0.80, 0.16, 0.61, 0.84 and 2.85 respectively. Isolates included screening isolates from rectal swabs (40%), clinical isolates from urinary tract (24%), respiratory tract (15%), wounds (7%), blood (4%) and other sites (10%). Sixty-five percent of isolates harboured CTX-M enzymes: CTX-M group 1 in 83%, CTX-M group 2 in 5% and CTX-M group 9 in 13%. The mean proportion of ESBL-KP harbouring bla CTX-M genes by year was 44% (0% in 2002 to 73% in 2005). SHV and TEM ESBLs were harboured by 25% and 4% of strains respectively. Class I integrase was detected in 91% of strains. ERIC typing showed 3 major types (11 to 21 pts) sharing close spatio-temporal linkage (2 harbouring CTX-M-15 enzymes) in 2005. The proportion of non-susceptible isolates were: temocillin 9%, ceftazidime 79%, cefepime 45%, piperacillin-tazobactam 35%, cotrimoxazole 82%, ciprofloxacin 43%, gentamicin 48%, tobramycin 61%, and amikacin 22%. Meropenem showed good activity with MIC50 of 0.06 and MIC90 of 0.25 mg/L.
Conclusion: Since 2000 ESBL-KP isolates in our institution frequently harboured bla CTX-M genes with predominance of group 1. Integrons of class I were present in the majority of these strains. Two CTX-M-15 clones were associated with one outbreak in ICU during 2005. Co-resistance to aminoglycosides and quinolones was frequent but no resistance to meropenem was observed.
P676 Epidemiology of hospital-acquired Klebsiella pneumoniae producing CTX-M β-lactamases in Slovenia
K. Meško Meglic, S. Koren, M.F.I. Palepou, E. Karisik, D.M. Livermore, A. Andlovic, S. Jeverica, V. Križan-Hergouth, M. Mueller-Premru, K. Seme, N. Woodford on behalf of the Slovenian ESBL Study Group
Objectives: The epidemiology of extended-spectrum β-lactamases (ESBLs) has changed dramatically, with the emergence of CTX-M enzymes. They have accumulated rapidly in Escherichia coli, including those from the community. Less has been reported on the spread of CTX-M ESBLs in K. pneumoniae though evidence suggests they are replacing TEM/SHV ESBLs, e.g. in the UK. We investigate the emergence and epidemiology of CTX-M ESBLs in K. pneumoniae in Slovenian hospitals.
Methods: From January 2005 to May 2006, isolates of ESBL-producing K. pneumoniae causing nosocomial infections and/or colonisation were collected at 11 hospitals in Slovenia (one isolate per patient). ESBL production was detected by double-disc synergy tests and ESBL Etests. MICs were determined and interpreted according to CLSI criteria. Isolates were screened for blaCTX-M genes using multiplex PCR and were compared by PFGE of XbaI-digested genomic DNA. Data were analysed using BioNumerics software.
Results: A total of 177 ESBL-producing K. pneumoniae isolates were collected. Of these, 60 (33.9%), from 8 hospitals, were positive for blaCTX-M, all with group-1 enzymes. Clonal relationships among these 60 isolates were studied in comparison with 27 CTX-M-negative strains from the same hospitals. The isolates represented multiple strains, but several clusters of related isolates (>80% similarity) were observed, some of them including isolates from more than one centre. The largest cluster consisted of 26 clonally-related isolates with group-1 CTX-M enzymes, 24 of them from one hospital. This outbreak needs further investigation. Two isolates of this outbreak strain were identified in further hospitals, suggesting inter-site transmission. Most isolates had substantial resistance to both cefotaxime (>128mg/L) and ceftazidime (>16mg/L), possibly indicating CTX-M-15.
Conclusion: K. pneumoniae with CTX-M enzymes are an emerging problem in Slovenian hospitals, but currently represent a minority of ESBL producers of this species. This contrasts with the dominance of CTX-M enzymes among ESBL-producing E. coli in most European countries. The epidemiology of K. pneumoniae with CTX-M enzymes was complex, with the spread of several strains within and between hospitals. Since K. pneumoniae is an important hospital pathogen this worrying development merits closer monitoring.
P677 Characterisation of PER-1 extended-spectrum β-lactamase producing P. aeruginosa clinical isolates from Hungary and Serbia
B. Libisch, Z. Lepsanovic, B. Krucso, M. Muzslay, B. Tomanovic, Z. Nonkovic, V. Mirovic, G. Szabo, B. Balogh, M. Füzi (Budapest, HU; Belgrade, RS)
Objectives: The aim of our study was to assess the contribution of ESBLs to the MDR phenotype among P. aeruginosa clinical isolates from Hungary and Serbia and to examine their molecular epidemiology.
Methods: The double-disk synergy test was performed with cefepime, ceftazidime and amoxicillin-clavulanic acid disks on MH agar plates for phenotypic ESBL screening. The positive isolates were characterised by serotyping, RAPD and PFGE analysis and submitted to PCR reactions using blaPER, blaVEB, blaGES and class 1 integron specific primers. The full length of the coding region of the identified blaPER genes were sequenced. Mating out assays were carried out on MH agar plates and transconjugants were selected on plates containing 16 mg/l ceftazidime and 300 mg/l rifampicin using the P. putida strain UWC1 as recipient.
Results: A strain collection of MDR P. aeruginosa representing all geographical regions of Hungary and one Belgrade hospital was screened by phenotypic methods. This work yielded 4 positive P. aeruginosa isolates from the Belgrade hospital in Serbia and 2 isolates from two different hospitals in Budapest, Hungary. PCR experiments revealed the presence of blaPER genes in all six isolates and indicated that these genes are not located on class 1 integrons. Sequencing of their coding region identified the PER-1 allele in every case. All isolates belonged to serotype O11. The four positive isolates from the same Belgrade hospital are clonally related by both PFGE and RAPD thus represent a cluster of PER-1 positive infections. While macrorestriction profiling could not establish genetic relatedness between the isolates from Budapest and Belgrade using an 80% cut off value, RAPD indicated a potential clonal relationship between them with the Dice coefficients ≥ 89%. Interestingly, one of the PER-1 isolates from Budapest was cultured from the nasal swab sample of a Romanian citizen on admission to the hospital. This observation raises the possibility that this strain was imported to Hungary from abroad. The PER-1 gene proved to be transferable by in vitro conjugation experiments for both strains from Hungary.
Conclusions: This is the first report of ESBL-producing Pseudomonas spp. from Serbia and Hungary. Our results indicate that ESBL producers occur only sporadically among P. aeruginosa clinical isolates in Hungary while a cluster of PER-1 positive infections was identified in the Belgrade hospital.
P678 Characterisation VIM-1-producing Pseudomonas aeruginosa, Enterobacter cloacae and Klebsiella pneumoniae strains from Germany: report from SENTRY Antimicrobial Surveillance Program
L. Deshpande, A. Rodloff, G. Just-Nuebling, R. Jones, H. Sader (North Liberty, US; Frankfurt, Leipzig, DE)
Objective: To characterise carbapenem (CARB)-resistant P. aeruginosa (PSA) and Enterobacteriaceae strains isolated in German medical centres participating in the SENTRY Program. The GIM class of metallo-β-lactamases (MBL) was originally described by our programme in clinical isolates from Germany and has not been observed again or in other nations. In Germany, MBL-producing strains are still rare in contrast to other European countries.
Methods: From 2000 to 2006, a total of 8,846 isolates were submitted to the SENTRY Program monitor from six German centres, including 596 PSA, 256 E. cloacae (ECL) and 348 K. pneumoniae (KPN). The isolates were tested for susceptibility (S) by reference (CLSI) broth microdilution methods and those with decreased S to imipenem (IPM), meropenem and ceftazidime were routinely screened for MBL production by disk approximation tests and/or MBL Etest (AB BIODISK) strips. Isolates with screen-positive results were confirmed by PCR using generic primers for IMP, VIM, SPM and GIM enzyme types. MBL gene sequencing and molecular typing (automated ribotyping, PFGE) were additionally performed to characterise MBL and to evaluate clonality.
Results: Decreased S to CARB (IMP MIC, ≥2 mg/L) was observed in 3 ECL (1.2%) and 1 KPN (0.3%) strains, while 77 PSA (13.1%) were R to IPM (MIC, ≥16 mg/L). Among IPM-R PSA, 10 strains had a positive MBL screen test and were PCR-positive for blaGIM-1 (6 strains from Düsseldorf described in 2002) or blaVIM-1 (4 strains from Frankfurt isolated in 2005–2006). The ECL and KPN strains were from Leipzig (2005 and 2006) and PCR-positive for blaVIM-1. The blaVIM-1 was located within a class 1 integron for all VIM-1-producing strains. VIM-1-producing PSA showed identical/similar PFGE patterns, while the ECL strains each had a distinct molecular typing pattern.
Distribution of MBL types in German samples (2000–2006)
| MBL-type | Location (no.) | Species (no.) | No. of PFGE patterns |
|---|---|---|---|
| GIM-1 | Düsseldurf (6) | P. aeruginosa (6) | 1 |
| VIM-1 | Frankfurt (4) | P. aeruginosa (4) | 1 |
| Leipzig (4) | E. cloacae (3) | 3 | |
| K. pneumoniae (1) | 1 |
Conclusions: blaVIM-1 has emerged and is rapidly disseminating as clones and also among clonally unrelated strains in diverse areas of Germany. Long-term surveillance and continued screening for MBL genes among isolates with decreased S to CARBs will be essential to control of dissemination by this important R mechanism.
P679 Dissemination of a CMY-16 producing clone of P. mirabilis in long-term care and rehabilitation facilities of northern Italy
E. Nucleo, R. Migliavacca, M. Spalla, C. Terulla, M. Debiaggi, M. Balzaretti, A. Migliavacca, A. Navarra, L. Pagani (Pavia, Milan, IT)
Background: The use of cephamycins and β-lactam-inhibitor combinations to counter the threat of extended-spectrum β-lactamases (ESBLs) mediated resistance determined a shift toward non-ESBL phenotypes in species without inducible chromosomal AmpCs. The CMY-LAT-type enzymes are a group of molecular acquired class C β-lactamases (CBLs) that exhibit a broader spectrum of resistance than classical ESBLs.
Methods: 204 non-repetitive P. mirabilis isolates intermediate/resistant to cefotaxime, collected from May 2003 to March 2006 from inpatients in three Long Term Care and Rehabilitation facilities of Northern Italy (ASP S. Margherita, IRCCS S. Maugeri and ASP P. Redaelli), were included in the study. The isolates were recovered from urinary tract. The production of an ESBL activity was screened by the CLSI diffusion test; IEF of crude bacterial lysates was performed to detect the pIs of β-lactamase bands. The nature of the resistance genes and the clonal relationships between the strains, were studied by molecular techniques such as amplification, sequencing and PFGE (SfiI).
Results: 18/204 (8.8%) strains showed an AmpC phenotype. Analytical IEF revealed the presence of 2 β-lactamase bands, of pI 5.4 and >8.4 respectively, in all the isolates. The pI 5.4 band was unable to hydrolyze extended-spectrum cephalosporins (CTX, CAZ, FEP), monobactams (ATM) and cefoxitin (FOX) and was likely contributed by a TEM-type enzyme. The alkaline pI band was active against both FOX, CTX and CAZ, suggesting the presence of an acquired CBL. PCR and sequencing revealed the occurrence of the CMY-16 enzyme, a variant of the CMY/LAT lineage. All the 18 CMY-16 producers were clonally related. The incidence of CMY genes within the three hospitals was: 10/64 (15.6%) at ASP S. Margherita; 2/15 (13.3%) at IRCCS S. Maugeri; 6/125 (4.8%) at ASP Redaelli respectively.
Conclusion: Resistance of P. mirabilis to expanded-spectrum cephalosporins is an increasing problem in several settings. Acquired AmpC-type β-lactamases are overall less common than class A ESBLs, but emergence of these enzymes in P. mirabilis has been reported in some areas. This report focus on the increasing diffusion of an AmpC-type variant in P. mirabilis; the clinical strains investigated in this work were all clonally related, and shared a common structure of genetic environment of CMY-16 determinant, suggesting a worrisome vertical spread of diffusion.
P680 Occurrence of metallo-β-lactamases in Pseudomonas aeruginosa clinical isolates resistant to carbapenems
D. Vitti, E. Protonotariou, D. Sofianou (Thessaloniki, GR)
Objectives: The main mechanism of resistance to carbapenems is the production of metallo-β-lactamases (MBLs). These enzymes are plasmid-mediated and their spread in P. aeruginosa has become of great concern. The aim of this study was to determine the occurrence of MBLs in clinical isolates of P. aeruginosa resistant to carbapenems in our hospital.
Methods: A total of 36 nonrepetitive strains of P. aeruginosa resistant to carbapenems were obtained from clinical specimens between February 2003 to November 2005. The identification and susceptibility testing were performed by the VITEK-2 automated system (bioMérieux, France). Imipenem (IMP) and Meropenem (MER) MICs were determined by E-test. EDTA-IMP double disc synergy test was carried out for screening of MBLs production. All isolates were subjected to PCR assay with specific primers and DNA sequencing. Pulsed-field gel electrophoresis (PFGE) was used to delineate clonal relationship between strains.
Results: Among the 36 carbapenem-resistant isolates of P. aeruginosa, 18 were positive by PCR for the presence of blaVIM. All these isolates had MIC of IMP and MER >32 μg/mL and showed a synergy between EDTA-IMP. Sequence analysis of PCR product revealed the presence of VIM-2 gene. Eight VIM producing strains harboured also a blaSHV gene and gave a synergy using disks of amoxicillin+clavulanate and 3rd generation cefalosporins. Phylogenetic tree using UPGMA (pairwise-unweighted) algorithm with QuantityOne Software (BIORAD), revealed different PFGE patterns with predominance of type A that comprised 5 isolates.
Conclusions: This study illustrates the spread of genes encoding MBLs i n P. aeruginosa. High percentage of MBLs-producing strains were, in addition, SHV-producers. Appropriate control measures including introduction of MBL screening in regular basis, are necessary in order to prevent wider dissemination of these genes.
Epidemiology of vancomycin-resistant enterococci
P681 Emergence of worldwide epidemic clones of vancomycin-resistant Enterococcus faecium in a Northern Spain hospital
M. Romo, H. Tomita, Y. Ike, L. Martínez-Martínez, M. Francia (Santander, ES; Maebashi, JP)
Objectives: Vancomycin-resistant enterococcal outbreaks are unusual in Spain. However, between October 2002 and April 2004, 31 clinical isolates of VanA E. faecium were reported in our hospital. These isolates showed high-level resistance to ampicillin, erythromycin, ciprofloxacin and aminoglycosides. Preliminary PFGE typing indicated the presence of two different patterns. The objective of this study was to determine the genetic lineages from which our isolates were derived along with the conjugative elements involved in the vancomycin resistance dissemination in our institution.
Methods: E. faecium isolates were genotyped by MultiLocus Sequence Typing (MLST). The clonal relationship between the isolates was analysed by the allelic profiles of the sequence types (STs) through the web site (http://www.mlst.net). Southern Hybridisation methods using a digoxigenin-based nonradioisotope system (Boehringer GmbH, Mannheim, Germany) and standard protocols were performed to determine the presence of pMG1-like plasmids.
Results: Among the 31 E. faecium isolates, two STs were identified, ST132 (representing 85% of the isolates) and ST18 (15%), being coincidental with the two PFGE patterns previously shown. Analysis of the allelic profile of the STs suggested all the isolates be assigned to clonal complex 17, with each ST being a double locus variant of ST17, a well-known world wide epidemic strain. ST132 strains were shown to harbour the highly conjugative pMG1-like plasmids, frequently found in vancomycin resistant E. faecium clinical isolates in Japan and USA. pMG1 was absent from ST18 isolates.
Conclusions: Our data suggested the outbreak occurred in our hospital was caused by two hospital adapted multi-resistant E. faecium clones forming part of the major circulating epidemic lineage in the world. pMG1-like plasmids appear to be involved in the vancomycin dissemination, at least in our predominant clone.
P682 Increase of VRE in a German university hospital
A. Kola, F. Schwab, I. Chaberny, S. Suerbaum, P. Gastmeier (Hannover, Berlin, DE)
Objective: To analyse the epidemiology of vancomycin-resistant E. faecium (VREfm) at Hannover Medical School (MHH), a 1400 bed university hospital with 40000 admitted patients per year. Starting in 2004, an increase of VREfm was observed: The proportion of VREfm raised from 1.2% (first half-year of 2004) to 31.5% (first half-year of 2006) in spite of isolation precautions.
Methods: VREfm were typed by PFGE (SmaI-restriction). PFGE profiles yielding a similarity of >80% using the Dice coefficient (<4 different fragments) were considered as clonally related. The simple diversity index SD (different genotypes//N isolates * 100) was calculated. Results were compared to those obtained for 239 isolates of Vancomycin-susceptible E. faecium (VSEfm). In addition, multiple-locus variable-number tandem repeat analysis (MLVA) was performed. Typing results and conventional epidemiology were applied for transmission analysis.
Results: Excluding copy-strains, 171 isolates of 166 patients hospitalised on 30 wards were analysed: PFGE revealed 57 different genotypes, of which 36 were unique and 21 appeared in clusters of 2 – 31 isolates belonging to the same type. The most frequent genotype was present on 14 wards. With the exception of three clusters of 2, 3 and 5 isolates, there was no genotype that was related to a specific ward. Sixty-one patients (37%) with VREfm were in the general surgery unit and 38 patients (23%) in the haematological oncology unit. In these units, 30% of VREfm were due to patient-to-patient transmissions. SD was significantly different between VREfm and VSEfm (33.3 vs. 44.8, RR VREfm/VSEfm = 0.745, CI95: 0.58–0.96, p = 0.024), as were the cluster sizes (number of isolates belonging to one genotype) of VREfm and of VSEfm (mean: 6.47 vs. 4.77, median: 3 vs. 2, p = 0.028). MLVA-analysis of the most prominent PFGE-cluster revealed the involvement of the epidemic clonal complex-17 (MT 1, MT 3 and MT 7).
Conclusion: The lower SD and the greater median cluster size of VREfm express a higher genetic relationship of VREfm in comparison to VSEfm. Considering the 36 unique genotypes and the occurrence of identical genotypes on different wards without epidemiological linkage, this is not explained by simple patient-to-patient transmission of VREfm, which accounted for approximately 30% of VREfm. Further analysis has to be done to clarify the causes of the increase in VREfm at MHH.
P683 Molecular characterisation of vancomycin-resistant Enterococcus isolates in clinical samples from Chile
G. Saavedra, J. Hormazábal, A. Maldonado, J. Mota, F. Baquero, J. Silva, R. del Campo (Panamá, PA; Santiago de Chile, CL; Madrid, ES; Antofagasta, CL)
Objectives: The aim of this work was to characterise a collection of vancomycin-resistant isolates recovered from clinical samples in Chile.
Methods: A total of 70 vancomycin resistant enterococal strains recovered during 2003–2005 was included (10 E. faecalis and 60 E. faecium). Strains were recovered from different samples (34 faecal swabs, 15 urine, 6 exudates, 3 blood and 12 others) corresponding to unrelated patients attended in 16 different hospitals corresponding to four regions of Chile. All strains presented vancomycin-resistance and were sent to the National Reference Institute. Susceptibility to several antimicrobial agents was tested using the microdilution test. Clonal relationships were studied by PFGE-SmaI assays, and presence of the vanA or vanB genes were demonstrated by PCR experiments. Amplicons were purified and sequenced. Presence of ace, agg, cylA, esp, efaA and gelE genes was also investigated in the different clones detected.
Results: All strains were resolved into 7 different clones (2 E. faecalis and 5 E. faecium) showing resistance to vancomycin, and susceptibility to teicoplanin. Resistance to macrolides, tetracycline, quinolones, β-lactams (only in E. faecium clones), and high level resistance to aminoglycosides was observed in all enterococcal clones. Positive amplification with the generic vanB primers was observed, and the sequences correspond to the previously described vanB2 variant. Presence of ace, agg, cylA, esp, efaA and gelE genes was only detected in the E. faecalis clones.
Conclusions: vanB2-containing E. faecium and E. faecalis clones have been isolates dispersed in 16 different hospitals in Chile.
P684 Molecular epidemiology of vancomycin-resistant enterococci isolated in 2003–2005 in a large teaching hospital in Warsaw
W. Grzybowska, G. Mlynarczyk, A. Mlynarczyk, A. Mrowka, S. Tyski, M. Luczak (Warsaw, PL)
Objectives: Enterococcal infections caused by multiresistant bacteria might constitute a severe problem for patients with serious diseases. Glycopeptides are the most frequently used antibiotics in such cases. In the investigated hospital, vancomycin resistant enterococci (VRE) did not occur frequently up to the 2003. A significant increase in the number of VRE isolated recently has suggested a possibility of an outbreak. The aim of this study was to perform epidemiological analysis of VRE strains isolated from clinical materials of hospitalised patients.
Methods: All VRE strains originated from patients of the investigated hospital and were isolated in the years 2003–2005. Strains were initially identified in the Diagnostic Laboratory of Microbiology Department. Identification was performed by API ID32 Strep and the susceptibility was tested by ATB Entero. Vancomycin resistant or intermediate isolates were checked by E-tests. All resistant strains were analysed for the presence of vanA, van B, vanD or vanG ligase genes by PCR. The identification of the strains was proven by PCR with application of the ddl primers. Only one VRE isolate from one patient was taken to epidemiological analysis. Strains were compared on the basis of differences in patterns obtained by PFGE of SmaI-restricted whole genomes analysis.
Results: Although the frequency of enterococci isolation in our laboratory was usually about 1000 strains per year, until the 2003, VRE occurred sporadically. In 2003 they appeared in one of internal wards, and were isolated from urine (9 patients) and from blood (1 patient). In 2004 VRE were isolated from 9 patients of the same ward and appeared in The Intensive Therapy Unit (OIOT) (2 patients) and in surgery (2 patients). In 2005 VRE were isolated from 11 patients of internal ward, from one patient of OIOT and from 21 patients of surgery. All together 53 VRE isolates were available for PFGE examination (12 E. faecalis, 41 E. faecium). All examined strains possessed vanA determinant of resistance. PFGE analysis revealed that most E. faecium strains belonged to one of the 3 clusters. The same strains occurred in three wards of the hospital. The PFGE relationship analysis between isolated E. faecium and between E. faecalis strains showed their high similarity: 75% and 82%, respectively.
Conclusion: The PFGE and SmaI restrictase turned out to be very useful for epidemiological analysis of enterococci.
P685 Molecular characterisation of vancomycin-resistant Enterococcus faecium isolated from German hospitals between 1991 and 2006 reveals differences in emergence and spread
G. Werner, I. Klare, W. Witte (Wernigerode, DE)
Objectives: To investigate the molecular background of vanA-type glycopeptide resistance in 57 clinical vancomycin-resistant Enterococcus faecium (VREF) from 29 German hospitals of three separate periods (group I, 1991–1995; II, 1996–1999; III, 2004–2006).
Methods: Isolates were characterised by multi-locus sequence typing (MLST), SmaI macrorestriction analysis in pulsed-field gel electrophoresis (PFGE), and multiple-locus variable-number tandem repeat analysis (MLVA). Phylogenetic relatedness between strains was identified using BioNumerics and eBURST software. Distribution of virulence genes esp and hylEfm was investigated by PCR. The structure of the vanA gene clusters was investigated by PCR, long PCR, sequencing and Southern hybridisations.
Results: Group I isolates (n = 6) are quite diverse by different typing methods, lack mostly any of the investigated virulence genes, and possess all an identical vanA cluster type. Two and all group II and III isolates belong to the clonal complex of hospital-adapted strain types (MLST CC17, MLVA C1). Isolates of group II (n = 8) are identical by MLST (ST-117), PFGE and MLVA (MT-12), harbour all the esp but not the hylEfm gene and possess an identical or slightly modified vanA cluster type (insertion of ISEf1). Group III VREF (n = 43) are rather diverse constituting different strain types, different virulence markers and vanA clusters. Within this diversity we found supportive data for a dissemination of related VREF among various hospitals and spread of identical vanA gene clusters into different strain types within single hospitals.
Conclusions: Our data suggest that VREF from the three periods investigated here have evolved differently. The increase in the rates of VREF among German hospital patients within the last two years might be rather complex and due to different molecular events and scenarios.
P686 Nosocomial outbreak of vancomycin-resistant Enterococcus faecium at a Turkish University Hospital
A. Ozcan, B. Ozhak Baysan, D. Ogunc, D. Inan, T. Naas, D. Colak, P. Nordmann (Antalya, TR; Kremlin-Bicetre, FR)
Objectives: Vancomycin-resistant enterococci (VRE) are widespread worldwide. Despite growing concern about VRE as nosocomial pathogens, especially in the USA, they are rarely isolated in Turkish hospitals. In 2001, unrelated VRE isolates were first described in Turkey (Colak D, JAC, 2002, 50:397–401.). Here we report the emergence of a novel VRE clone responsible of a nosocomial outbreak at a Turkish University Hospital.
Methods: Strains correspond to either clinical samples or rectal swab specimens for analysing carriage. Identification, susceptibility testing, molecular characterisation and molecular comparison of the strains (PFGE and MLST) were performed according to standard techniques. Virulence genes were searched for by PCR.
Results: Thirty six VRE were isolated from ten patients between June 2005 and January 2006, of whom nine were hospitalised at the Department of Pediatrics and one of them at the Department of Haematology. Six patients were carriers, two had urinary tract infections and two patients had bloodstream infections. All isolates were E. faecium and expressed high level glycopeptide resistance (MIC for vancomycin: >256 mg/L and for teicoplanin: >48 mg/L). These strains were also resistant to ampicillin, clindamycin, erythromycin and displayed high-level resistance to gentamicin, kanamycin and streptomycin whereas they remained susceptible to levofloxacin, linezolid, and tigecycline.
PFGE analysis revealed that these 36 E. faecium isolates were clonally related, except for four isolates (three from patient 9 and one from patient 10). The epidemic clone was found in all 10 patients. They were all of vanA genotype, and esp positive. The vanA gene was part of a vanA-type operon for expression of resistance located on a transposon similar to Tn1546 in the VanA prototype strain E. faecium BM4147, except for the presence of Insertion Sequences.
Conclusion: This work further reports emergence of VRE in Turkey as observed now in many European countries. Implementation of stringent hand disinfection and environmental disinfection policies, as well as measures for patient isolation contained this outbreak of VRE, and since February 2006 no new VanA E. faecium strains was isolated from these units.
P687 Vancomycin-resistant Enterococcus faecium emergence during a nosocomial outbreak of Clostridium difficile diarrhoea
A. Moreno, A. del Rio, D. Pereda, A. Cangussu, M. Tuset, M. Castellá, C. Cervera, L. Linares, J. Martinez, C. Mestres, J. Vila (Barcelona, ES)
Objectives: Vancomycin-resistant Enterococcus faecium (VRE) have been recognized as microorganisms capable of causing epidemics in critically ill patients; however, in our institution are very infrequent. For this reason, we describe three cases involving VRE infection during a nosocomial outbreak of Clostridium difficile diarrhoea in patients with vascular diseases.
Methods; From January to April 2006, 45 patients admitted from the vascular surgery department developed diarrhoea. Faeces samples were taken from these patients in order to determine cultures, parasites and toxins of C. difficile. Data related to the diarrhoea outbreak and the appearance of Vancomycin-resistant Enterococcus faecium strains in this population were recorded.
Results: We included 45 patients with diarrhoea, 31 of whom were men (70%), with a mean age of 70 years (SD 10). Median length of stay was 16 days (IQR 8;31). Thirty patients (66%) received antibiotics due to surgical wound infection prior to the diarrhoea, 15 received vancomycin during 3 days (IQR 1; 10) and 26 received quinolones during 7 days (IQR 2.7; 10.2). Six patients died (13%) due to other causes not related to the diarrhoea. All bacterial cultures and parasites from faeces were negative. Nineteen cases (42%) had a positive C. difficile toxin. Thirteen patients received oral vancomycin (29%) and 32 (71%) oral metronidazole. Median length of stay of patients with C. difficile diarrhoea was 18 days (IQ 9; 27) vs. 12 (IQ 7; 38) days of patients without a C. difficile diagnosis (p=NS). In 54% of patients that received quinolones prior to the diarrhoea, a positive C. difficile toxin was found (p = 0.07). Three patients had a VRE surgical infection developed during the diarrhoea treatment. Three out of 23 patients that received vancomycin developed VRE (13%), one before diarrhoea and two as a result of C. difficile diarrhoea.
Conclusions: During a nosocomial outbreak of C. difficile diarrhoea, three Vancomycin-resistant Enterococcus faecium strains were isolated in patients with surgical wound infection. These three patients had been treated with vancomycin. Therefore we suggest not using this drug as initial treatment for nosocomial C. difficile diarrhoea.
P688 Interhospital dissemination of glycopeptide resistance among enterococcal clinical isolates from Portugal is associated to spread of epidemic pMG1-like plasmids carrying diverse Tn1546 variants
A. Freitas, C. Novais, T.M. Coque, L. Peixe (Porto, PT; Madrid, ES)
Objectives: Vancomycin resistant enterococci (VRE) are increasing in European hospitals, Portugal having one of the highest VRE rates. Our objective was to characterise the genetic context of representative clinical strains in order to understand the role of horizontal gene transfer in the dramatic recent spread of VRE in our area.
Methods: We analysed 43 E. faecium and 21 E. faecalis isolates (37 PFGE types) collected from 3 Portuguese hospitals in different regions (1996–2003). Susceptibility to 13 antibiotics was studied by standard agar dilution method. Clonality was established by PFGE and transfer of vancomycin resistance was achieved by broth and filter matings. Tn1546 backbone was determined by overlapping PCR and Tn1546 location was identified by Southern hybridisation of I-CeuI and/or S1 nuclease-digested genomic DNA. Plasmids characterisation included comparison of EcoRI-RFLP patterns and hybridisation with probes for vanA and pMG1, pAD1 and pAMβ1 plasmids.
Results: Fourteen Tn1546 types, mostly containing ISEf1 or IS1216V within vanX-vanY region, and located on conjugative plasmids of variable size (50–225 kb) were identified. Two Tn1546 types represent 56% of the cases (PP4 and PP5). Plasmids of 80–95 kb showing similar EcoRI-RFLP patterns and related 4 ISEf1-Tn1546 variants (types PP4, PP5, PP9 and PP24) were collected from 21 strains of 3 hospitals during 7 years. Plasmids of 45–110kb corresponding to 7 unrelated patterns harbouring IS1216V-Tn1546 variants (4 Tn1546 types) were collected from 12 strains of 3 hospitals during 2 years. All of them showed homology with known pMG1 enterococcal plasmid by hybridisation.
Conclusions: Persistent epidemic plasmids highly related to the wordlwide spread pMG1 have a significantly role in intra and interhospital dissemination of VRE in Portugal. The Tn1546 diversity found among related plasmids suggests evolution of specific elements by different genetic events.
P689 Genotypic characterisation by PFGE and ribotyping of clinical isolates of vancomycin-resistant enterococci in Iran
S. Eshraghi, M. Pourshafie (Tehran, IR)
Objectives: Vancomycin resistant enterococci (VRE) is becoming a major concern in the clinical settings as the rate of occurrence of Enterococcus spp. is increasing. Epidemiological knowledge of the clonal dissemination of vancomycin resistant. enterococci (VRE) spp. is fundamental for the implementation of control measures. This study conceived to provide information on the diversity of vancomycin resistant enterococci in Iran.
Methods: The clinical enterococcal isolates which were collected sporadically from the hospitalised patients and outpatients in three major hospitals in Tehran. A single specimen was obtained from each patient. The susceptibility tests of the isolates were performed and interpreted according to the guidelines from the Clinical and Laboratory Standards Institute (CLSI). The structures of Tn1546 like elements were analysed by long PCR-RFLP using a single primer targeting the inverted repeat sequence of Tn1546. All of isolates were typed by Pulsed-field Gel Electrophoresis (PFGE) using SmaI and ribotyping using EcoRI restriction enzymes.
Results: Fifty (5.7%) VRE isolates were obtained out of nine hundred enterococci spp. samples were collected. All VRE isolates showed a high level vancomycin resistance and harboured vanA gene. Antibiotic susceptibility tests showed that the isolates were resistant to ampicillin (98%), ciprofloxacin (92%), gentamicin (96%), erythromycin (92%), tetrracyclin (28%), and chloramphenicol (4%). L-PCR-RFLP revealed the presence of two groups among the 50 human strains tested. Genotyping by PFGE using SmaI and ribotyping using EcoRI restriction enzyme grouped the isolates into 18 and 12 types, respectively.
Conclusion: The results indicated that the clinical VRE strains are highly diverse in term of bacterial clonality. But it was shown that different VRE isolated from patients' samples carried a similar Tn1546 with same genomic structure.
P690 Clonal diversity of vancomycin-resistant Enterococcus faecium isolated from three university hospitals in Daegu, Korea
D.T. Cho, J.Y. Oh (Daegu, KR)
Objectives: To determine the evidence for molecular epidemiology by genetic characterisation of related resistant genes and virulence associated genes in high-level glycopeptide and aminoglycoside resistant clinical isolates of Enterococus faecium.
Methods: Species identification of a total of 58 vancomycin-resistant E. faecium (VREFM) was confirmed by ddl gene PCR. Conjugal transfer of resistance was done by filter mating. Vancomycin resistance gene (van), aminoglycoside resistance genes and five virulence genes were detected by multiplex PCR. Mapping of transposon Tn1546 structure by PCR, pulse-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST) were attempted for clustering of isolates to study clonal diversity and genetic relatedness.
Results: All isolates carried vanA gene. The aac6-aph2 gene was detected in 53 (91.4%) of isolates and 27 (46.6%) showed aadE gene. Macrolide resistance gene ermA was detected in 3 (5.2%) VREFM isolates while 55 (94.8%) showed ermB. Virulence associated genes, esp and hyl were detected in all 58 VREFM and in 34 (58.6%) respectively. Other virulence genes asa1, gelE and cylA were not detected. Virulence gene hyl was transferred by conjugation with vanA and gentamicin-resistance gene. Eleven types of (A to H) transposon Tn1546-like element were obtained by PCR mapping. Insertion sequence IS1216 in the vanX-vanY intergenic region in Tn1546 structure were confirmed in 49 strains (A to F). Six clusters could be estimated by PFGE analysis. MLSTs of 58 VREFM isolates showed 11 different sequence types (ST). Thirty-eight (65.5%) of 58 strains belonged to clonal complex CC78 and then these were subdivided into three types; ST192 (32.8%), ST203 (19.0%) and ST78 (13.8%). ST203 was detected in all three university hospitals but ST192 and ST78 were present only in KNU-hospital.
Conclusion: Vancomycin resistance gene, vanA and aminoglycoside resistance gene were located in conjugally transferable plasmid which carried Tn1546-like element and virulence gene hyl. Clustering based on PFGE analysis showed relatively more close relatedness with Tn1546 type and MLST than those relatedness between Tn1546 type and MLST. Genomic evolution of nosocomial isolates of VREFM could be confirmed through molecular characterisation, including PFGE, transposon typing and MLST.
P691 Vancomycin-resistant enterococci still persist in slaughtered poultry in Hungary 8 years after the ban on avoparcin
A. Ghidan, O. Dobay, E. Kaszanyitzky, P. Samu, S. Amyes, K. Nagy, F. Rozgonyi (Budapest, HU; Edinburgh, UK)
Objectives: Vancomycin-resistant enterococci (VRE) are one of the major problems in the field of antibiotic resistance. Fortunately, the proportion of human VRE isolates remains very low in Hungary; however, VREs persist in animal samples. In this report we examined the glycopeptide susceptibility of enterococci, isolated in 2005, from slaughtered animals, within the confines of Hungarian Antibiotic Resistance Monitoring System. We determined the presence of the van genes and their genetic relatedness in enterococci from poultry.
Methods: Enterococcus sp. (n = 175) were collected from intestinal samples of slaughtered poultry in 2005. The origin of the samples was registered at county level. After screening the strains with 30 microgram vancomycin disc 19 (86%) intermediate resistant and 4 (3%) resistant strains were found. The MICs of vancomycin and teicoplanin were determined by agar dilution. The presence of the van genes was detected by PCR. The identity of the van gene carrying strains was identified by PCR using genus-specific and species-specific primers. The potential similarities of these strains was determined by PFGE (digesting with SmaI).
Results: The distribution of MICs among 23 enterococcus strains which were intermediate or resistant to vancomycin were 0.25 mg/L (4.4%), 2 mg/L (8.6%), 4mg/L (8.6%), 8mg/L (61%), 16mg/L (8.6%) and 256 mg/L (8.6%). The MICs of teicoplanin were 0.25 mg/L (4.3%), 1 mg/L (8.6%), 4 mg/L (78.3%), 16mg/L (4.3%), and 256 mg/L (4.3%). The two most vancomycin-resistant strains were vanA carriers (1 E. faecalis and 1 E. faecium). No strains carried vanB, vanC1 or vanC2. The E. faecalis strain had high resistance to both glycopeptides (MICs =256 mg/L) and the E. faecium had high resistance to vancomycin (MIC=256 mg/L) and intermediate resistance to teicoplanin (MIC = 16 mg/L). These strains originated from two different counties of Hungary. These 23 strains were not closely related to one another.
Conclusion: Avoparcin was used as a growth promoter for broiler chickens since 1989 but was banned in 1998. Despite this prohibition, the VRE strains only disappeared in 2003. However, in 2004 strains only with higher MICs to vancomycin were detected and, in 2005, two strains were vanA positive. The farms that produced these strains can be reservoirs of VRE and the affected farms should change the technology of disinfection and breeding in order to prevent the emergence of high numbers of human VRE isolates in Hungary.
P692 A two-year study of antibiotic resistance in Enterococci isolated from food, abattoirs and farms in Scotland and its link to hospital-acquired infections
L. Vali, A. Hamouda, A. Mateus, D.J. Walker, J. Dave, A. Gibb, S.G.B. Amyes (Edinburgh, Glasgow, UK)
Objective: To establish whether there is any significant link between enterococci originated from animals and the clinical human strains, indicating animals infecting human populations in hospitals.
Methods: From 2004–2006 a total of 55 different food stuff including dairy and meat products from commercial outlets, 100 faecal samples from cattle on Scottish beef cattle farms, and from abattoirs; 100 faecal, 460 nostril, skin, and ear swab samples collected from pigs, cattle and sheep. Fifty clinical E. faecium and E. faecalis were isolated from hospitalised patients from two hospitals in Edinburgh. Selective media was used for isolation followed by phenotypic and genotypic species characterisations. The minimum inhibitory concentrations of antibiotics were determined by the agar dilution method. PCR and sequencing determined the van genes responsible for vancomycin resistance. Pulsedfield gel electrophoresis (PFGE) was used for molecular typing of the strains, and the presence of esp virulence gene was determined by PCR.
Results: Enterococci were isolated from 20 food, 50 faecal samples, and 27 swab samples. In total 14 strains were resistant to vancomycin, of which 5 were highly resistant to vancomycin (MIC > 32) and teicoplanin (MIC > 16). vanA and vanC1 genes were detected in 10 and 7 strains respectively. PFGE demonstrated that there is diversity within enterococci population isolated from animals and humans; suggesting the host specificity. However closely related clones were identified from outlet food and the clinical strains; suggesting the possibility of contamination by food handlers. esp gene was not detected in enterococci obtained from animal sources.
Conclusion: Host specificity of enterococci suggests that animal strains do not generally impose a serious threat to nosocomial infections in clinical settings. In hospitalised patients the threat of infections is caused by bacteria that largely spread clonally. Moreover the presence of esp gene that identifies the successful nosocomial clones was not detected in any of the strains from the animal origin. Although the presence of van genes causes concern regarding the spread and transfer of vancomycin resistance, it is unlikely that resistance in hospital acquired infections is caused by animal strains. Enterococci may spread from food to humans only if the foodstuff is contaminated through human handling. Safe food handling is therefore crucial especially to high risk patients in hospitals.
P693 Epidemiology of multiresistant Enterococcus faecalis and Enterococcus faecium collected during 2004 to 2006 in a Lisbon hospital
R. Pires, M. Pereira, P. Rodrigues, S. Carvalho, T. Ferreira, R. Mato (Oeiras, Lisbon, PT)
Objectives: Determination of the prevalence, evolution of antimicrobial resistance, and clonality of high-level gentamicin (HLGR) and glycopeptide resistant (GR) E. faecalis (E.fl) and E. faecium (E.fm) recovered from infection products during 2004 to 2006 in a public community (350-beds) Lisbon hospital.
Methods: Microbial identification and antimicrobial susceptibility testing were performed using the VITEK2 system. HLGR and GR E.fl and E.fm were confirmed by PCR detection of aac(6')-aph(2”) and vanA/B/C1/C2 genes and clones were assessed by SmaI-PFGE and carriage of the esp gene. Multiplex-PCR was used to detect the cylA, asaI, gelE and hyl virulence genes among GR isolates.
Results: A total of 403 isolates were recovered during 2004–2006. The prevalence of E.fl and other enterococcal species (E. durans, E. casseliflavus, E. avium by decreasing order of frequency) increased from 68 to 81% and from 0.6 to 6%, respectively, while E.fm decreased from 30 to 11%. HLGR-E.fl increased from 31 to 41% and resistance to ampicillin-Amp (4%), ciprofloxacin-Cip (40%), erythromycin-E (89%), and vancomycin-Va (1%) remained stable. Most (90%) of the E.fm were resistant to Amp, Cip and E, and resistance to Va decreased from 14 to 8%. HLGR-E.fm increased from 62 to 77%.
All HLGR isolates (n = 162) carried the aac(6')-aph(2”) genes. GR-E.fl (n = 3) and GR-E.fm (n = 5) were vanA-positive. HLGR-E.fl (n = 107) were of 27 PFGE patterns. PFGE-AO was prevalent (n = 53) and increased in prevalence during the study period (37.5% to 61.5%). esp-positive HLGR-E.fl increased from 25 to 82%, in parallel with the increment of esp-positive HLGR-E.fl PFGE-AO from 8 to 92%. PFGE-AO also included GR-E.fl isolates positive for cylA, asaI, gelE and one isolate was also esp-positive. HLGR-E.fm (n = 55) were of 11 PFGE types. Three major clones (PFGE-a, -o and -c) coexisted in 2004. PFGE-a decreased from 2005 (76%) to 2006 (20%) while PFGE-c emerged as dominant (50%). The esp gene increased from 68 to 90%. GR-E.fm were of PFGE-a, -o (two esp-positive isolates), -c and -d (all esp-negative, HLG susceptible). PFGE-o and -c isolates carried only gelE.
Conclusions: Enterococcal infections mainly by HLGR-E.fl and E.fm seem to be increasing. The prevalence of GR strains was lower that the published data. One major E.fl nosocomial clone is acquiring virulence traits over time. To our best knowledge this is the first molecular epidemiological study of enterococcal infections from a Portuguese hospital.
P694 Nationwide increase of invasive ampicillin resistant Enterococcus faecium in the Netherlands
J. Top, R. Willems, S. van der Velden, M. Asbroek, M. Bonten (Utrecht, NL)
Objectives: We recently described an increase in the proportion of invasive enterococcal infections caused by ampicillin resistant Enterococcus faecium (AREfm) from 2% in 1994 to 32% in 2005 and a partially replacement of ampicillin-susceptible (ampS) E. faecalis by E. faecium (75% AREfm) among enterococcal bloodstream infections in our hospital (Top et al., CMI in press). In our hospital all AREfm belonged to CC17 E. faecium, a clonal complex associated with nosocomial outbreaks worldwide. A nationwide study was initiated to determine whether these ecological changes had also occurred in other hospitals in the Netherlands.
Methods: All 66 microbiology laboratories serving all hospitals of the country were asked to provide data on annual numbers of all invasive ampicillin resistant (ampR) enterococcal isolates and the first 30 enterococcal blood stream isolates (1 per patient) from 1994 until 2006. Multiplex PCR based on the ddl gene was performed to distinguish E. faecium and E. faecalis and susceptibility to ampicillin was determined.
Results: Thirty labs (45.5%) provided data, and 8 (12%) labs also provided isolates. The mean number of invasive ampR enterococcal isolates increased from 5 in 1994 to 25 in 2005. This increase was more pronounced in academic and large non-academic hospitals (>500 beds); from 5 in 1994 to 45 in 2005 in academic hospitals and from 4 in 1994 to 19 in 2005 in non-academic hospitals. Among enterococcal blood isolates proportions AREfm increased from 13% in 1993 to 40% in 2006; from 13% in 1993 to 51% in 2006 in academic hospitals and from 13% in 1999 to 26% in 2006 in non-academic hospitals. All E. faecalis isolates were ampicillin susceptible, while 75% of the E. faecium isolates were ampicillin resistant.
Conclusions: In the Netherlands invasive AREfm increased nationwide and have partially replaced ampS E. faecalis. The difference in prevalence among academic and non-academic hospitals probably reflects differences in patient population with haematology and transplant patients having the highest risks for AREfm bacteraemia being over represented in academic hospitals. All E. faecium isolates will be genotyped to determine the molecular epidemiology of AREfm in the Netherlands.
In vitro susceptibility of Gram-positives
P695 Susceptibility of streptococci to antibiotics and biocides isolated from Saudi Arabia and UK
A.M. Al-Qorashi (Dammam, SA)
The assessment of the in vitro activities of 16 antibiotics, 10 biocides and 6 metallic compounds against clinical isolates of Streptococcus pyogenes (group A) from UK and Saudi Arabia and its control strains from UK. 16 antibiotics (benzypenicillin, erythromycin, lincomycin, methicillin, rifampicine, trimethoprim, vancomycin, ampicillin, chloramphenicol, fucidic acid, gentamicion, kanamycin, nalaedixic acid, novobiocin and streptomycin), 10 biocides (3 phenolics: chlorocresol, cresol, phenol; 4 parabens: methyl, ethyl, butyl and propyl esters of para-4-hydroxybenzoic acid); a bisbiguanide: chlorhexidine diacetatate; 2 quaternary ammonium compounds: cetylpyridinium chloride and benzethonium chloride) and 6 metallic compounds (mercuric chloride, phenylmercuric nitrate, cadmium chloride, cupric chloride, zinc chloride and silver nitrate) were tested against controls and clinical isolates of Streptococcus pyogenes (group A) from the university hospital of Wales UK as well as clinical isolates from King Fahd University Hospital, Al-Khobar and King Abdulaziz University Hospital, Jaddah, Saudi Arabia by the disc diffusion method and the determination of minimal inhibitory concentration (MIC).
All strains of Streptococcus pyogenes (group A) were β-lactamase-negative. All British strains were sensitive to all of the antibiotics tested, except gentamicin and streptomycin. Some of the Saudi strains were resistant to gentamicin and erythromycin and all were resistant to lincomycin. The British and Saudi Arabian strains showed the same order of response to phenols and parabens. Similarly, the British and Saudi Arabian strains were equally sensitivity to phenylmercuric nitrate, however, response to other metallic compounds were variable.
P696 In vitro activity of fosfomycin tromethamine and linezolid against clinical vancomycin-resistant Enterococcus faecium isolates
F. Cilli, H. Pullukcu, S. Aydemir, O. Sipahi, M. Tasbakan, A. Turhan, S. Ulusoy (Izmir, TR)
Objectives: Infections with vancomycin resistant enterococci are emerging problems. The aim of this study was investigate the in vitro effect of fosfomycin tromathanole and linezolid against 117 vancomycin-resistant isolates of Enterococcus faecium.
Method: MIC of fosfomycin and linezolid were determined by E test (Biodisk, Sweden). A 0.5 McFarland suspension of the microorganism in 0.9% saline was inoculated into Mueller-Hinton agar (Oxoid, England). E test strips were placed on the culture plates and the MIC read after 24 h. Since E test strips for fosfomycin contained glucose-6-phosphate, extra supplementation of the compound in the culture medium was not done. The readings were tabulated and the MIC50 and MIC90 values determined. The breakpoint criteria to determine susceptibility were based on the CLSI. All isolates were Enterococcus faecium (89 from rectal swabs, 24 from hemoculture, 3 from tissue biopsy culture, 1 from urine culture).
Results: The MIC90 and MIC50 of fosfomycin were 192mg/L and 512 mg/L and for linezolid 1mg/L and 2 mg/L, respectively. Overall MIC for linezolid ranged between 0.5–3 mg/dl.
Conclusion: Linezolid but not fosfomycin tromethanol had good in vitro activity against 117 isolates of vancomycin resistant E. faecium.
P697 In vitro activity of daptomycin and linezolid against vancomycin-resistant Gram-positive pathogens from cancer patients
R. Prince, E. Coyle, K. Rolston, M. Kapadia, A. Kaur, S. McCauley (Houston, US)
Objectives: To compare the in-vitro activity of daptomycin (DAP) and linezolid (LIN) to vancomycin (VAN) against Van-resistant Gram-positive (GP) organisms isolated from cancer patients.
Background: GP organisms cause 40–70% of documented bacterial infections in cancer patients. VAN has been the drug of choice for the treatment of GP infections in such patients. Although uncommon, VAN-resistant organisms are being isolated with increasing frequency and alternative agents need to be evaluated in this setting.
Methods: The organisms were isolated between January 2004 and September 2006 and >90% were from blood cultures. E-test” strips (AB Biodisk, Solna, Sweden) were used for susceptibility testing. S. aureus ATC-29213 was used as the control strain.
Results: MIC50, MIC90 values (if >10 strains were tested) and range of activity (in μg/mL) are shown in the Table below.
| Agent | MIC (μg/mL) |
|||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| VAN-resistant enterococci (34) | DAP | 1.0 | 3.0 | 0.25–30 |
| LIN | 1.5 | 1.5 | 0.38–2.0 | |
| VAN | >256 | >256 | >256 | |
| Leuconostoc spp. (7) | DAP | 0.064–0.38 | ||
| LIN | 2.0–8.0 | |||
| VAN | >256 | >256 | >256 | |
| Pediococcus spp. (5) | DAP | 0.25–1.0 | ||
| LIN | 4.0–8.0 | |||
| VAN | >256 | >256 | >256 | |
Conclusions: Daptomycin appears to have excellent in-vitro activity against most VAN-resistant organisms. It also has better bactericidal activity than VAN and LIN (data not shown). LIN has excellent activity against VRE but appears to be less potent against Leuconostoc and Pediococcus spp. The clinical potential of these agents needs to be evaluated, although infections caused by some of these pathogens are rare.
P698 VITEK diagnostics of Enterococcus species in urine samples
V. Vuksanovic (Podgorica, ME)
Objectives: Urinary infections (UTI) are common in patients with anatomically normal urinary tract, manifested as a no complicated infections, where Escherichia coli is the most frequent aetiological cause. In UTI, with structional or functional abnormality of urinary tract, the resistant enterobacteria, enterococci and Candida are more common aetiological cause. Enterococci as a opportunistic pathogens can cause severe urinary infection, infections of surgery wounds, bacteraemia and bacterial endocarditis. As the high resistance of micro organisms to antibiotics has been registered the aim of this work is to examine sensitivity of enterococci originated from patients' urine.
Methods: After isolation in blood agar, identification and sensitivity of 67 enterococci was done by using VITEK 2 system (bioMérieux). Out of these patients 42 were hospitalised in Clinical Centre Podgorica, and 25 samples were taken from patients visited outpatient healthcare services.
Results: Out of 62 isolated enterococci, 63% (42) were from hospitalised patients, and E. faecium was identified in 50% (21) cases. This was not case with urine samples taken from patients who visited outpatient healthcare services, where E. faecium was isolated in only 4% (1) of cases. All types of E. faecalis showed sensitivity in over 95% of cases to beta lactamic antibiotics: ampicilin 95%, ampicilin sulbactam 98% and imipenem 95%, as well as to linezolid 95% and glycopeptides (teicoplanin and vancomycin 98%). Lower percentage of sensitivity was found in quinolones (73%). The highest resistance of E. faecalis was to tetracycline 72% (37), while half of the types were resistant to erythromycin 51% and gentamycin 53%. Although E. faecium is normally resistant to beta lactamic antibiotics, one urine sample had wild phenotype sensitive to ampicilin, ampicilin sulbactam and imipenem. E. faecium showed resistance to ciprofloxacin in over of 80% cases (86%), erythromycin 86% and gentamycin 82%, while sensitivity to linezolid, teicoplanin and vancomycin was registred in 90% of cases.
Conclusion: This study shows that E. faecalis is the most sensitive to penicillin preparations and it is still drug of first choice in our region. Drugs of choice for E. faecium are linezolid, teicoplanin and vancomycin, in infections of hospitalised patients caused by enterococci. The final identification of enterococci and antibiogram is mandatory procedure, if it is available, in treatment of urinary infections
P699 Antimicrobial effects of non-antibiotics on resistant Gram-positive bacteria
O. Hendricks, T. Butterworth, J. Christensen, H. Sahly, R. Podschun, J. Kristiansen (Sønderborg, Copenhagen, DK; Kiel, DE)
Objectives: Psychotropic therapeutics, especially the phenothiazines are employed for the treatment of psychosis though they exhibit the additional property of an anti-microbial activity. We defined MICs for selected compounds on clinically relevante Gram-positive bacteria. The evaluation of the combined effect of these compounds and conventional antibiotics of clinical relevance showed a significant synergistic effect. As intracellular survival of Gram positive species is another possible explanation for the insufficiency of antibiotic treatment, we investigated the influence of phenothiazines on bacterial invasion in human epithelial cell lines. The project investigates the anti-microbial activity of Non-antibiotics against clinically relevant Gram-positiv bacteria, e.g. staphylococci, streptococci and enterococci.
Methods: Agardilution, Microdilution, Human ephitelcell-models A-549, HCT-8, T-24.
Results: All Gram-positive bacterial strains, regardless of their susceptibility to regularly used antibiotics, were inhibited by the testsubstances at concentrations of 8–64 g/L. Combination of the antibiotics and the choosen testcompounds at subinhibitory concentration demonstrated a restored activity for the investigated antibiotics. We documented interference of our test-compounds with efflux based multidrug resistance. Furthermore, we recorded a significant reduction of the mean bacterial invasion ability in the investigated cell lines in the presence of selected agents. Overall, these results indicated a significant reduction of the mean invasion ability of the Gram-positive bacteria in all epithelial cell lines (18.9%±1.8) as compared to the invasion in absence of the substance (52.1%±4.4) (p < 0.0001).
Conclusions: The present experiment shows that phenothiazine derivates, especially thioridazine, have an antimicrobial effect against the investigated strains. Our studies offer new information on the effect of phenothiazines on Resistant Gram-positive bacteria. The anti-microbial activity of these compounds may have a place in the treatment of infections where the possibilities for current antibiotic treatment are limited.
P700 Comparison of antibiotics susceptibility of different vancomycin-resistant Enterococcus faecium clones
A. Mironova, G. Kliasova, A. Brilliantova (Moscow, RU)
Background: The aim of this study was to compare the antibiotic resistance between different clones of vancomycin-resistant Enterococcus faecium (VREF).
Methods: A total of 123 isolates were collected from patients with haematological malignancies during May 2003 to September 2006. Minimal Inhibitory Concentrations (MICs) were determined by broth microdilution method (CLSI). Resistance genes were detected by PCR. Macrorestriction analysis of SmaI digests was performed by pulsed-field gel electrophoresis.
Results: PFGE typing revealed 21 strain types of VREF. 78 (69%) isolates belonged to type A and 12 (11%) – to type F. The remaining 24 VanA isolates were distributed among 17 different PFGE types. All strains were resistant to ampicillin, penicillin, erythromycin, levofloxacin, streptomycin and gentamicin (high-level resistance). Different resistance levels to chloramphenicol and tetracycline were exhibited among PFGE types. Tetracycline susceptibility was 82% (64/78) for type A isolates, 50% (6/12) for type F and 83% (20/24) for the strains, belonged to the other 17 different PFGE types. Chloramphenicol susceptibility was 45% (35/78) for type A isolates, 25% (3/12) for type F and 54% (13/24) for the other strains. During the period of investigation from 2004 to 2006 the number of clone A isolates with intermediate susceptibility to teicoplanin increased from 18% (5/28) in 2005 to 82% (36/44) in 2006, with susceptibility to chloramphenicol increased from 32% (9/28) in 2005 to 59% (26/44) in 2006. Linesolide was highly active against all investigated VREF strains. The distribution of linesolide MICs in all VREF was unchanged during 2004 to 2006 (MIC90=2mg/L).
Conclusion: VREF presented high resistance rates to the antibiotics tested. Linesolide is uniformly active against all VREF regardless of phenotype and PFGE type. Differences between PFGE types were discovered to compare resistant to chloramphenicol and tetracycline.
P701 Comparison of baseline profiles of RO4908463 (CS-023) against recent isolates of target Gram-positive pathogens exhibiting key resistance phenotypes obtained from Europe and USA 2003–2006
D. Sahm, N. Brown, M. Jones, D. Draghi, B. Dannemann (Herndon, US; Breda, NL; Nutley, US)
Objectives: RO4908463 (CS-023) is a carbapenem that has activity against a broad spectrum of bacterial pathogens including MRSA and P. aeruginosa and is under development for lower respiratory tract infections and complicated skin and skin structure infections. Because susceptibility profiles of target organisms obtained from different geographic areas can vary, this study was done to compare the current activity of RO49 against key Gram-positive pathogens isolated from Europe (EU) and from the United States of America (USA).
Methods: RO4908463 and other agents were tested by broth microdilution (CLSI; M7-A7, 2006) against a total of 1,231 Gram-positive bacteria, isolated from the EU (n = 599) and USA (n = 632), that included S. aureus (SA), S. epidermidis (SE), E. faecalis (EF), E. faecium (EM), S. pneumoniae (SP), S. pyogenes (SY), and S. agalactiae (SG). The EU isolates were from 14 different countries and the US isolates were collected from across all 9 US Census Bureau Regions collected during 2003–2006 (11 isolates dating pre-2003).
Results: See the table .
| Organism | Phenotypea | RO4908463 MIC parameter (mg/L) |
|||||
|---|---|---|---|---|---|---|---|
| EU |
USA |
||||||
| Total n | Range | MIC90 | Total n | Range | MIC90 | ||
| S. aureus | All | 312 | 0.06–>32 | 16 | 333 | 0.06–32 | 4 |
| MRSA | 161 | 0.5–>32 | 16 | 172 | 0.25–32 | 4 | |
| MSSA | 151 | 0.06–2 | 0.25 | 161 | 0.06–2 | 0.25 | |
| S. epidermidis | All | 56 | 0.06–16 | 1 | 56 | 0.03–8 | 4 |
| MRSE | 23 | 0.06–16 | 4 | 29 | 0.06–8 | 4 | |
| MSSE | 33 | 0.06–0.06 | 0.06 | 27 | 0.03–0.12 | 0.12 | |
| E. faecalis | 51 | 0.06–>32 | 4 | 54 | 0.03–4 | 4 | |
| E. faecium | 30 | 4–>32 | >32 | 30 | 0.06–>32 | >32 | |
| S. pneumoniae | All | 52 | ≤0.008–1 | 0.5 | 55 | ≤0.008–1 | 1 |
| PEN-S | 32 | ≤0.008–0.03 | ≤0.008 | 32 | ≤0.008–0.03 | 0.015 | |
| PEN-NS | 20 | 0.03–1 | 0.5 | 23 | 0.25–1 | 1 | |
| S. pyogenes | 47 | ≤0.008–0.015 | 0.015 | 51 | ≤0.008–0.06 | 0.015 | |
| S. agalactiae | 51 | 0.015–0.06 | 0.06 | 53 | 0.03–0.06 | 0.06 | |
MR, methicillin-resistant; MS, methicillin-susceptible; PEN, penicillin; S, susceptible; NS, non-susceptible.
Conclusion: RO4908463 demonstrated good in vitro activity against most Gram-positive pathogens, including MRSA. The level of RO4908463 activity was comparable regardless of the geographic source of the strains, although for MRSA MIC90 and range was higher among the EU strains than those from the USA. The reason for this difference is currently unknown, but studies are underway to confirm the higher MIC phenotype and determine if there may be a clonal relationship among the EU strains that exhibited the higher MIC. These data obtained with strains from widely distributed geographic areas indicate that RO4908463 has potent anti-Gram-positive activity and provides an important baseline for detecting any changes in the activity of RO4908463 as clinical development continues.
In vitro susceptibility of staphylococci
P702 Synergistic effect of lactoferricin/amoxicillin associations against “two canine” amoxicillin-resistant Staphylococcus intermedius strains
P. Butty, B. Manco, L. Florent (Libourne, FR)
Objectives: The bacteriostatic and bactericidal effects of lactoferricin/amoxicillin combinations against two S. intermedius strains (00-A519 and 00–5191) isolated from a dog infected by pyoderma and resistant to amoxicillin were compared to the effect of lactoferrin/amoxicillin combinations. The synergistic, addition or antagonistic activity was determined by calculating the fractional inhibitory concentration (FIC) index. The FIC-index value related to synergism must be 0.5 or less when the concentration for each antimicrobial and for the most favourable point is at least 4 times below the MIC obtained for each antimicrobial used alone.
Methods: Checkerboard array technique in liquid medium was used to assess the bacteriostatic activity of these combinations. The bactericidal activity was tested by sub-cultures on agar free-drug medium from the combination inhibitory concentrations. The concentrations tested for each antimicrobial were ranged from five dilutions below the minimal inhibitory concentration (MIC) to twice the MIC for amoxicillin and to the maximum possible concentration for lactoferricin and lactoferrin depending of their respective solubility.
Results: The tested strains showed a lactoferricin MIC of 1,280 μg/mL, a lactoferrin MIC equal to 51,200 μg/mL and an amoxicillin MIC ranging from 64–512 μg/mL. Lactoferricin/amoxicillin combinations showed synergic effect with FIC-index between 0.16 and 0.27 and amoxicillin MIC was reduced to 8 μg/mL. Concerning the lactoferrin/amoxicillin combinations, the synergistic effect was observed with FIC-index equal to 0.25. The respective concentrations of the lactoferricin/amoxicillin and lactoferrin/amoxicillin combinations showing the bacteriostatic effect correspond also to a bactericidal effect.
Conclusion: Considering that lactoferricin represents the purified active fraction of the lactoferrin in our approach, lactoferricin could be a good alternative to maximise the antibacterial activity of amoxicillin against emerging antibioresistant strains of canine S. intermedius.
P703 Comparative activity of moxifloxacin vs. trimethoprim-sulfamethoxazole, cloxacillin, linezolid, clindamycin, and ciprofloxacin against intracellular methicillin-sensitive and community-acquired methicillin-resistant S. aureus
S. Lemaire, F. Van Bambeke, P. Tulkens, Y. Glupczynski (Brussels, BE)
Objectives: Recurrence and persistence of S. aureus infection is often ascribed to intracellular bacterial persistence, which may also be a cause of emergence of resistance. We showed that the activity of antibiotics is always weaker when tested against intracellular bacteria in comparison to those growing in broth (AAC 2006, 50:841–851). In this context, we have examined the intraphagocytic activity of moxifloxacin (MXF; recently approved for skin and soft-tissues infections) against fully sensitive S. aureus (MSSA) and community-acquired methicillin-resistant S. aureus (CA-MRSA), in comparison with (i) antibiotics commonly used for the treatment of CA-MRSA infections (trimethoprim-sulfamethoxazole [TMP-SMX], cloxacillin [CLX], linezolid [LNZ], clindamycin [CLI]), and (ii) ciprofloxacin (CIP).
Methods: MSSA (ATCC 25923) and CA-MRSA (NRS 192) were used. MICs were determined by microdilution in MH broth according to CLSI guidelines with a 10e5 cfu/mL original inoculum. Intracellular activity was assessed on infected THP-1 macrophages (as described in details in AAC 2006, 50:841–851) after 24 h exposure to drug concentrations corresponding to their respective human Cmax (see Table ). Controls cells (no antibiotic added) were incubated with gentamicin [0.5×MIC] to prevent extracellular growth (see validation in AAC 2006, 50: 841–851).
| Drugs (Cmax*) | ATCC 25923 (MSSA) |
NRS 192 (CA-MRSA) |
||
|---|---|---|---|---|
| MIC (mg/L) | intracell. activity (Δ log cfu) | MIC (mg/L) | intracell. activity (Δ log cfu) | |
| TMP-SMX (25) | 1 | +0.6±0.1 | 1–2 | +0.7±0.0 |
| CLX (8) | 0.125 | −0.8±0.1 | 0.5–1 | −0.07±0.1 |
| LNZ (21) | 1 | −0.7±0.1 | 2 | −0.7±0.1 |
| CLI (12) | 0.06 | −1.0±0.1 | 0.125 | −0.9±0.1 |
| CIP (4) | 0.125 | −1.3±0.1 | 0.5 | −1.4±0.1 |
| MXF (4) | 0.03 | −2.0±0.0 | 0.03 | −1.8±0.1 |
As commonly observed in humans after conventional administration and used as extracellular concentration (total drug) to assess intracellular activity.
Results: The table shows the MICs of each drug, together with their corresponding intracellular activity (change in cfu from post-phagocytosis inoculum).
Conclusions: All antibiotics tested, with the exception TPM-SMX, allowed for a reduction of the intracellular inoculum, but MXF demonstrated the largest effect (∼2 log decrease), but this could be due, partly, to its low MIC towards the strains examined.
P704 Susceptibility of Staphylococcus aureus biofilms to vancomycin and gentamicin
D. Kotulova, L. Slobodnikova, A. Longauerova (Bratislava, SK)
Objectives: To determine changes in MBIC (microbial biofilm inhibitory concentration) and MBEC (microbial biofilm eradicating concentration) to vancomycin and gentamicin in comparison with MIC (minimum inhibitory concentration) of a planktonic form of S. aureus strains, isolated from indwelling medical devices of patients of the Bratislava Teaching Hospital.
Patients and Methods: Fifteen S. aureus strains isolated from central venous catheters, intratracheal cannulae and surgical drains from patients of the Teaching Hospital were investigated. The strains were identified by standard microbiological methods. Antimicrobial susceptibility was tested by the disc diffusion method according to NCCLS 2004 and 2005, respectively. MIC values to vancomycin and gentamicin were established by broth dilution according to NCCLS 2004, 2005 and, to vancomycin by E test, as well. Biofilm production was detected by the Christensen and microtitre biofilm methods. MBIC and MBEC were detected in microtitre plates using different concentrations of the investigated drugs, equal or higher than MIC detected in a planktonic form.
Results: Of the 15 investigated strains, 4 were methicillin resistant, but susceptible to vancomycin and gentamicin. When using Christensen method, a weaker biofilm production was detected in all of the tested strains in comparison with the microtitre method. No differences were found between vancomycin MIC (median 2mg/L) and MBIC (median 2 mg/L). However, the concentration required for biofilm eradication was higher in all tested strains – MBEC values were 4 mg/L (S) in two strains, 8–16 mg/L (I) in ten strains, and 32 mg/L (R) in two strains.
While investigating the effects of gentamicin, it was found that in comparison with MIC (median 1mg/L), the MBICs were increased in 14 strains (median 2.7 mg/L). MBEC values were increased only in two strains – to 4 mg/L.
Conclusions: Our preliminary results validated the fact that determining bacterial susceptibility in a planktonic form is insufficient in efforts to successfully eradicate S. aureus biofilms on biomaterials: MBIC and MBEC values should be determined. Such results may help physicians in administration of drugs in correct doses to individual patients. Standardisation of this susceptibility testing method will be necessary in the future, due to the increasing incidence of indwelling and implanted medical devices infections.
P705 Activity of moxifloxacin and cefuroxime against MSSA in time-kill and in vitro PK/PD studies
R. Endermann, G. Weskott, A. Pickbrenner, J. Schuhmacher, I. Kneževic (Wuppertal, DE)
Objectives: Methicillin-susceptible Staphylococcus aureus (MSSA) is frequently involved in complicated skin and skin structure infections (cSSSI). Moxifloxacin (MXF) and cefuroxime (CXM) have been approved for this indication. This study compares bactericidal activity of MXF and CXM against three MSSA strains. Both agents were compared in time-kill experiments and in a hollow fibre pharmacokinetic (PK) model.
Methods: MIC values were determined by the broth microdilution method. MIC and time-kill experiments were performed according to CLSI guidelines. The MSSA strain DSM 11823 and a recent clinical isolate, HF 1012717 were used for the time kill experiments at concentrations of 0.25–16 times the MIC. Using the hollow fibre PK model, the strains ATCC 29213 and HF 1012717 were exposed to free drug serum concentrations according to human PK after a dose of MXF 400mg IV (Cmax 2.2mg/L, T1/2 13h) or CXM 1500mg t.i.d. (Cmax 70 mg/L, T1/2 1.5 h). For the PK model an initial inoculum of 108 CFU/mL was used. Antibiotic concentrations were confirmed by bioassay and HPLC. Antibacterial effect was measured over time and specifically by time to 3-log reduction (bactericidal kill) in viable counts.
Results: The MIC for CXM was 2 mg/L for all three test strains and for MXF was 0.06 mg/L for ATCC 29213 or 0.03mg/L for HF 1012717 and DSM 11823. In time-kill experiments, exposure to MXF and CXM at 1–16 times the MIC led to bactericidal kill of strain DSM 11823. With MXF, 3-log kill was achieved earlier than with CXM (1 vs 3 h at 8 times the MIC). Against the clinical isolate HF 1012717, MXF showed a bactericidal kill at 4–16 times the MIC, while CXM achieved at best a 2-log reduction at 16 times the MIC. In the hollow fibre PK model 3-log kill was achieved for both strains (HF 1012717 and ATCC 29213) at 1.5–2 h for MXF. When CXM PK was simulated, 3-log kill was noted at 4.5–6 h for ATCC 29213 and at 7–10 h for HF 1012717. There was no regrowth for any strain or regimen after 24h, and the MICs of the isolated colonies at 24 h were not elevated.
Conclusions: MXF and CXM were bactericidal against all three MSSA strains when tested in time-kill experiments and in a hollow fibre PK model. With regard to time to 3-log reduction, MXF was more effective than CXM in both models.
P706 The evaluation of the bactericidal activity of daptomycin, vancomycin and linezolid and determination of the interactions of these antimicrobials with gentamicin or rifampin against S. aureus
H. Sader, T. Fritsche, R. Jones (North Liberty, US)
Objective: To determine the bactericidal activities of daptomycin (DAP), vancomycin (VAN) and linezolid (LZD), and the drug interaction categories for these 3 antimicrobials combined with each other or tested with gentamicin (GEN) or rifampin (RIF) against S. aureus strains.
Methods: Minimum inhibitory concentrations (MIC) were determined by CLSI broth microdilution methods. Bactericidal activity (MBC) was evaluated on 207 S. aureus (101 wild-type [WT] oxacillin-resistant [MRSA], 64 VAN-heteroresistant [hVISA], 37 VAN-intermediate [VISA] and 5 VAN-resistant [VRSA]) by plating the entire broth content from the well onto appropriate growth media. Quantitative colony counts were performed and compared to the initial inoculum. Checkerboard synergy tests were performed on 18 S. aureus (15 MRSA) using broth microdilution trays containing 2 agents in 2-fold dilutions dispensed in a checkerboard format. DAP, VAN and LZD were combined with each other and with GEN or RIF, resulting in 9 combinations. The fractional inhibitory concentration (FIC) was calculated for each agent and the summation of both FICs was used to classify the combined activity of antimicrobials as synergistic (SYN; ≤0.5), partially synergistic (PSYN; >0.5 and <1), additive (ADD; 1), indifferent (IND; >1 and <4) and antagonistic (ANT; ≥4).
Results: DAP was bactericidal against all S. aureus groups while VAN MBC/MIC ratios consistent with tolerance were observed at a rate of 14.9; 68.8; and 86.5% for WT-MRSA, hVISA and VISA respectively, and LZD MBC/MIC ratios consistent with tolerance were observed at a rate of 99.0%; 100.0%; 94.6% and 100.0% for WT-MRSA, hVISA, VISA and VRSA respectively. Checkerboard results showed no ANT or SYN with any of the antimicrobial combinations evaluated. The majority of strains (77.8%) showed PSYN (50.0%) or ADD (28.8%) interactions when DAP was combined with GEN; while ADD and IND effects predominated when DAP was combined with LZD (94.4%), RIF (88.9%) or VAN (72.2%). All isolates showed IND effect when LZD was combined with GEN. VAN combinations exhibited predominant ADD (16.7–38.9%) or IND interactions (44.4–77.8%).
MBC results for daptomycin linezolid and vancomycin tested against S. aureus
| Antimicrobial agent | No. of isolates (cumulative %) with MBC at: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | |
| Daptomycin | |||||||||
| MRSA-WT (101) | 0 (0) | 29 (29) | 64 (92) | 7 (99) | 1 (100) | − | − | − | − |
| hVISA (64)c | 0 (0) | 1 (2) | 32 (52) | 30 (98) | 1 (100) | − | − | − | − |
| VISA (37) | 0 (0) | 0 (0) | 2 (5) | 23 (65) | 9 (92) | 3 (100) | − | − | − |
| VRSA (5) | 0 (0) | 0 (0) | 4 (80) | 1 (100) | − | − | − | − | − |
| Linezolid | |||||||||
| MRSA-WT (101)a | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) | 0 (1) | 100 (100) |
| hVISA (64)c | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 64 (100) |
| VISA (37) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (5) | 0 (5) | 0 (5) | 0 (5) | 35 (100) |
| VRSA (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (100) |
| Vancomycinb | |||||||||
| MRSA-WT (101) | NTc | NT | 2 (2) | 38 (40) | 23 (62) | 9 (71) | 14 (85) | 3 (88) | 12 (100) |
| hVISA (64)c | NT | NT | 0 (0) | 3 (3.0) | 9 (12) | 3 (17) | 2 (20) | 4 (24) | 43 (100) |
| VISA (37) | NT | NT | 0 (0) | 0 (0) | 0 (0) | 2 (5) | 1 (8) | 0 (8) | 34 (100) |
Clinical MRSA isolates with vancomycin MIC ≤ 2 mg/L (homogeneous populations) collected from medical centres worldwide in 2003.
MBC was not evaluated on VRSA strains (five strains with vancomycin MIC >32mg/L)
NT not tested
Conclusions: DAP was the only agent highly bactericidal against S. aureus strains independent of their susceptibility to VAN. The combinations of DAP with any other agent were never antagonistic. DAP with GEN and LZD with RIF generally exhibited a PSYN or ADD interaction; while IND and ADD interactions predominated among all other combinations.
P707 Reduced efficacy of chlorhexidine against clinical MRSA residues compared to standard strains
S. Davies, L. Vali, S. Amyes (Edinburgh, UK)
Objectives: To determine whether sub-inhibitory concentrations of chlorhexidine acetate (CHX) are effective against surface dried MRSA and whether there are differences in the susceptibility of clinical strains compared to control EMRSA 16 and Staphylococcus aureus strains.
Methods: A surface disinfectant test was performed as follows: Washed bacterial suspensions (overnight growth in nutrient broth, washed three times in saline) of control strains (EMRSA 16 and S. aureus NCTC 6571) and clinical MRSA were dried onto surfaces for 2 and 24 hours. After drying, a sub-inhibitory concentration of chlorhexidine diacetate (CHX) was added. Following a 5 minute contact time, neutraliser was added and the cells resuspended for 5 minutes using magnetic followers. Enumeration was performed using the drop-counting method (serial dilution in SDW followed by 3×10 ul drops of each dilution on nutrient agar plates). Microbiocidal effect (log reduction in CFU/mL following CHX exposure) was determined by evaluating triplicate counts against a control without CHX for each isolate, enabling the results from different isolates to be compared.
Results: CHX was more effective after longer bacterial drying times (figure ).

Mean microbiocidal effect of CHX on bacterial residues.
This difference was more pronounced in the control EMRSA 16 (C) and NCTC 6571 (S) strains than in the clinical isolates (LF26, LF67 and LF93), in which the effectiveness of CHX after 24 hours bacterial drying time was not greatly increased. At both drying times, CHX was less effective on the clinical isolates than the control strains. Even after 24 hours bacterial drying, there was less than a 3-log CFU/mL reduction in the clinical isolates following exposure to CHX.
Conclusions: These results suggest that sub-inhibitory concentrations of CHX, as might occur during misuse of biocides in hospitals, are minimally effective against clinical isolates of MRSA, and may therefore lead to selection of reduced susceptibility MRSA isolates in the clinical environment. The difference between CHX efficiency against control strains compared to clinical isolates has implications for the tested efficacy of biocides, as it may be that clinical strains are less susceptible than efficacy testing on standard strains would imply.
P708 Bactericidal activity of moxifloxacin against Staphylococcus aureus and S. epidermidis at concentrations simulating bone penetration
J. LeBlanc, S. Campbell, R.J. Davidson (Halifax, CA)
Objectives: Staphylococcal osteomyelitis is frequently managed with a combination of ciprofloxacin and rifampin, the combination preventing the emergence of resistance to either agent. Moxifloxacin has significantly enhanced activity against Gram-positive bacteria including Staphylococcus spp. We describe a series of pharmacodynamic experiments designed to investigate the bactericidal activity of moxifloxacin against S. aureus and S. epidermidis at concentrations simulating bone penetration.
Methods: Eight fluoroquinolone susceptible isolates (4 S. aureus and 4 S. epidermidis) were challenged with moxifloxacin in a pharmacodynamic model utilising Mueller-Hinton broth. Cultures were inoculated at a density of 1×107 cfu/mL, incubated at 35°C, and examined for viable growth at 0, 1, 2, 4, 6, 12, and 24 h after exposure to moxifloxacin at concentrations simulating bone penetration (1.8 μg/mL). The model also simulated the elimination of moxifloxacin over a 24 h dosing interval. The moxifloxacin MICs of S. aureus and S. epidermidis ranged between 0.03 and 0.12 μg/mL for both species. All isolates were screened by PCR and parC/gyrA DNA sequencing to ensure the absence of target site mutations prior to initiating the experiments.
Results: Moxifloxacin was rapidly bactericidal against all 8 staphylococcal isolates. For S. aureus, a 4 log decrease in the viable colony count was observed after 1 h exposure in all 4 strains and complete eradication was achieved in 4h. For S. epidermidis, a 3 log decrease in the viable colony count was observed after 1 h exposure in 2 strains and a 4 log decrease in the remaining 2 strains. Complete eradication was achieved in 3 strains after 4 h and 6 h in the remaining strain. Despite simulating elimination of moxifloxacin over a 24 h dosing interval, we failed to detect re-growth in any of the isolates.
Conclusions: Moxifloxacin was very rapidly bactericidal against both S. aureus and S. epidermidis at concentrations achievable in bone. Complete eradication was achieved after 4 h in the majority of isolates and re-growth was not detected in any isolate even after 24 h. Our findings suggest that moxifloxacin may be an excellent choice in the management of osteomyelitis.
P709 Differences in antibiotic resistance between MRSA and MSSA strains isolated in Hungary, Austria and Macedonia
A. Horváth, G. Malmos, N. Pesti, K. Kristóf, K. Nagy, Z. Cekovska, G. Kotolácsi, R. Gattringer, W. Graninger, F. Rozgonyi (Budapest, HU; Skopje, MK; Vienna, AT)
Objectives: The aim of the study was to compare the quantitative susceptibility of methicillin-resistant (MRSA) and methicillin-sensitive (MSSA) strains of Staphylococcus aureus to antistaphylococcal agents. Antimicrobial sensitivity of 123 MSSA and 158 MRSA strains isolated in Hungary, 115 MSSA and 40 MRSA strains isolated in Austria and 72 MRSA strains isolated in Macedonia were tested.
Methods: Identification of S. aureus strains was performed by classical and molecular methods (presence of catalase, clumping factor, nucA and 23S rDNA genes). The mecA gene was detected by polymerase chain reaction (PCR). Minimum inhibitory concentrations (MICs) of antibiotics were determined by broth microdilution method according to NCCLS/CLSI recommendations. PFGE analysis of the strains is in process.
Results: All tested strains were sensitive to vancomycin. Majority of Hungarian and Austrian MRSA strains were sensitive to amikacin, while 70.8% of Macedonian strains were resistant. Resistance of Austrian and Macedonian MRSA strains to gentamicin exceeded 90%, Hungarian MRSA strains were gentamicin resistant in 73.7%. None of the MRSA strains were sensitive to clindamycin. To clarithromycin, ciprofloxacin, levofloxacin and moxifloxacin more than 90% of MRSA strains were resistant.
All tested MRSA strains were multidrug resistant. The most frequent resistance phenotype of Hungarian and Austrian strains was the resistance to gentamicin, clindamycin, clarithromycin and to fluoroquinolones. The most common phenotype of Macedonian strains was the resistance to these antibiotics and amikacin as well.
Both Hungarian and Austrian MSSA strains were mainly sensitive to aminoglycosides. While MSSA strains were mostly sensitive to clarithromycin, resistance of Hungarian and Austrian MSSA strains to clindamycin were 69% and 77%, respectively. The vast majority of MSSA strains were sensitive to all 3 tested fluoroquinolones, Austrian strains were less sensitive than Hungarians.
Conclusion: Resistance rates and degrees of MRSA strains to a variety of antimicrobials were significantly higher than those of the MSSA strains. Therapeutic options differ according to countries. In MSSA infections all antistaphylococcal drugs except for penicillin and clindamycin can be used, while in MRSA infections for empiric therapy only vancomycin and teicoplanin is recommended.
Supported by OTKA No.: T 46186 and ÖAAD-TéT grant No. A-19/02
P710 Baseline activity of ceftobiprole against methicillin-susceptible and -resistant staphylococcal clinical isolates from Europe collected in 2005–2006
T. Davies, D. Sahm, R. Flamm, K. Bush (Raritan, Herndon, US)
Objectives: Ceftobiprole is an investigational parenteral cephalosporin that is distinguished from currently marketed cephalosporins by its antimicrobial activity against methicillin-resistant (MR) staphylococcal isolates. It is currently under clinical development for pneumonia and complicated skin and skin structure infections. The activity of ceftobiprole and comparators was tested against methicillin-susceptible (MS) and -resistant (MR) staphylococcal isolates from Europe to establish a baseline for longitudinal tracking.
Methods: A total of 1203 S. aureus (SA), 214 S. epidermidis (SE), and 125 other coagulase-negative staphylococci (CoNS), primarily S. haemolyticus, S. sciuri, S. saprophyticus, and S. simulans, were collected from 31 cites in 13 European countries during 2005–2006. Ceftobiprole (BPR), clindamycin (CLI), erythromycin (ERY), gentamicin (GEN), linezolid (LZD), oxacillin, trimethoprim-sulfamethoxazole (SXT), and vancomycin (VAN) MICs were determined by broth microdilution according to CLSI methods.
Results: Against MSSA, SXT had the lowest MIC90 (≤0.25 mg/L) followed by BPR and GEN (0.5 mg/L). The SXT MIC90 (≤0.25 mg/L) was also the lowest for MRSA. BPR, LZD, and VAN had MIC90s of 2 mg/L for MRSA. SXT was significantly less active against SE and other CoNS. BPR and GEN were the most active agents tested against MSSE and MSCoNS with MIC90s of 0.12 to 0.25 mg/L. For MRSE and MRCoNS BPR, VAN, and LZD had the lowest MIC90s of 1 to 2 mg/L.
| Organism | N | MIC50/MIC90 (mg/L) |
|||||
|---|---|---|---|---|---|---|---|
| BPR | VAN | LZD | GEN | ERY | SXT | ||
| All SA | 1203 | 0.5/2 | 1/1 | 2/2 | 0.25/>16 | >8/>8 | ≤0.25/≤0.25 |
| MSSA | 404 | 0.25/0.5 | 1/1 | 2/2 | 0.25/0.5 | 0.5/>8 | ≤0.25/≤0.25 |
| MRSA | 799 | 1/2 | 1/2 | 2/2 | 0.5/>16 | >8/>8 | ≤0.25/≤0.25 |
| All SE | 214 | 1/1 | 2/2 | 1/2 | ≤0.06/>16 | >8/>8 | ≤0.25 >4 |
| MSSE | 69 | ≤0.12/0.25 | 2/2 | 1/1 | ≤0.06/0.12 | 0.25/>8 | ≤0.25/4 |
| MRSE | 115 | 1/2 | 2/2 | 1/1 | 16/>16 | >8/>8 | 2/>4 |
| All CoNSa | 125 | 0.5/2 | 2/2 | 1/2 | ≤0.06/>16 | >8/>8 | ≤0.25/>4 |
| MSCoNS | 32 | ≤0.12/0.25 | 2/1 | 1/1 | ≤0.06/0.12 | 0.25/>8 | ≤0.25/>4 |
| MRCoNS | 93 | 1/2 | 2/2 | 1/2 | 2/>16 | >8/>8 | 0.5/>4 |
Excluding SE.
Conclusion: Ceftobiprole had excellent activity against methicillin-susceptible and -resistant staphylococci, similar to or slightly better than linezolid and vancomycin. Ceftobiprole MICs were lower in methicillin-susceptible isolates compared to MR strains; however, the ceftobiprole MIC rarely exceeded 2mg/L.
Surveillance and epidemiology of bacterial infections in immunocompromised patients
P711 Legionnaires' disease in solid-organ transplant recipients revisited
C. Garcia-Vidal, C. Gudiol, R. Verdaguer, N. Fernández-Sabé, J. Carratalà (Barcelona, ES)
Objectives: We aimed to ascertain the incidence, characteristics, and outcome of Legionnaires' disease (LD) in solid-organ transplant (SOT) recipients. We also looked at differences on time to diagnosis between cases identified by urine antigen and cases documented when this test was not available.
Methods: We reviewed medical charts of all cases of LD occurring in SOT recipients from 1985 to 2006. Since 1997 urine antigen testing was routinely performed whenever a SOT recipient presented with pneumonia. Information concerning clinical characteristics, diagnosis, treatment, and outcome was collected.
Results: Among 2,789 SOT recipients, 12 (0.4%) cases of LD were diagnosed. The incidence according to the type of allograft received was as follows: 0.8% (2 of 246) for heart recipients, 0.5% (8 of 1560) for kidney recipients, and 0.2% (2 of 983) for liver recipients. Ten patients were males (83%), with a mean age of 47.8 years (range, 26–65 years). All patients were receiving more than one immunosuppressive drug by the time of diagnosis: corticoesteroids (10), cyclosporine (9), mycophenolate mofetil (5), rapamycin (2), tacrolimus (2), and other agents (2). Nine cases were community-acquired. The mean time to the development of LD was 1046 days (range, 31–2920 days). Among all patients, 25% had early-onset LD (≤3 months after SOT) and 75% had late-onset LD (>3 months). LD occurred in the setting of transplant rejection in 50% of cases. Patients frequently had high fever (62%), chills (70%), and multilobar pneumonia (44%). Pleural effusion and hyponatraemia were also relatively common. Microbiological diagnosis was established by bronchoalveolar lavage (4), transthoracic needle aspiration (3), tracheal aspirate (2), and/or urine antigen test (4). Legionella pneumophila was the species identified in all cases. Two patients had dual infection (cytomegalovirus viraemia 1, and nocardiosis 1). The mean time to diagnosis was shorter for cases identified by urine antigen testing than for the remaining cases (1.3 days vs. 8.5 days; p = 0.005). Eight patients were given erythromycin (plus rifampin in 4 cases) and 4 patients received levofloxacin. Overall case fatality-rate was 17%.
Conclusion: The incidence of LD among SOT recipients is relatively low but it causes significant mortality. LD occurs mainly as a late complication of transplantation involving patients with allograft rejection. Urinary antigen testing has improved early recognition of cases.
P712 Risk factors for Nocardia infection in organ transplant recipients: a matched case-control study over an 11-year period
A. Peleg, S. Husain, Z. Qureshi, F. Silveira, M. Sarumi, K. Shutt, E. Kwak, D. Paterson (Boston, Pittsburgh, US)
Objectives: Risk factors for Nocardia infection in organ transplant recipients have not been formally assessed in the current era of transplantation. Our objectives were to systematically assess these risk factors and to describe the clinical, radiological and microbiological characteristics of the largest number of Nocardia infections across the broadest range of organ transplant recipients yet reported from a single institution.
Methods: We performed a matched case-control study (1:2 ratio) from January 1995 through December 2005. Controls were matched for transplant type and timing. Univariate matched odds ratios and conditional logistic regression were performed to identify independent risk factors. Clinical and microbiological characteristics of all cases were reviewed.
Results: Thirty-five cases (0.6%) of Nocardia infection were identified from 5126 organ transplant recipients. The highest frequency was in lung recipients (18/521 [3.5%]), followed by heart (10/392 [2.5%]), intestinal (2/155 [1.3%]), kidney (3/1717 [0.2%]) and liver (2/1840 [0.1%]) recipients. Compared to 70 matched controls, high dose steroids (OR 27 [3.2–235], P = 0.003) and CMV disease (OR 6.9 [1.02–46], P = 0.047) in the preceding 6 months, and a high median calcineurin inhibitor level in the preceding 30 days (OR 5.8 [1.5–22], P = 0.012), were independent risk factors for Nocardia infection. The majority of cases had pulmonary disease only (27 [77%]). Seven recipients (20%) had disseminated disease. Nocardia nova was the most common species (17 isolates [49%]), followed by N. farcinica (9 isolates [28%]), N. asteroides (8 isolates [23%]) and N. brasiliensis (1 isolate [3%]). Of the 35 cases, 24 (69%) were receiving trimethoprim-sulfamethoxazole for PCP prophylaxis. Thirty-one cases (89%) were cured of their Nocardia infection.
Conclusions: High dose steroids, CMV disease and high levels of calcineurin inhibitors are independent risk factors for Nocardia infection in organ transplantation. Our study provides insights into the epidemiology of Nocardia infection over the current era, a time where immunosuppressive and prophylactic regimens have greatly evolved.
P713 Risk factor analysis of blood stream infection and pneumonia in neutropenic patients after peripheral blood stem-cell transplantation
E. Meyer, J. Beyersmann, H. Bertz, S. Wenzler-Roettele, R. Babikir, M. Schumacher, F. Daschner, H. Rueden, M. Dettenkofer and the ONKO-KISS study group
Objectives: To analyse risk factors for blood stream infection (BSI) and pneumonia in neutropenic patients who have undergone peripheral blood stem cell transplantation (PBSCT). Data taken from the ONKO-KISS (Hospital Infection Surveillance System for Patients with Hematologic/Oncologic Malignancies) multicentre surveillance project.
Methods: Infections were identified using CDC definitions (laboratory-confirmed BSI) and modified criteria for pneumonia in neutropenic patients. The multivariate analysis was performed using the Fine-Gray regression model for the cumulative incidences of the competing events ‘infection', ‘death’ and ‘end of neutropenia'. The risk factors investigated were: sex, age, underlying disease (early and advanced disease), and type of transplant (autologous or allogeneic, related donor or unrelated donor).
Results: From 1/2000 to 6/2004, a total of 1699 patients in 20 hospitals were investigated. In the multivariate analysis, male patients had a significantly higher risk of acquiring BSI than female patients (p = 0.002). The risk of acquiring BSI is highest in patients with advanced acute myeloid leukaemia (AML). In the univariate and multivariate analysis, unrelated donor allogeneic transplantation constituted a risk factor for pneumonia (p = 0.012).
Conclusion: ONKO-KISS provides reference data on the incidence of pneumonia and BSI. The increased risk for BSI in males and patients with advanced AML, and the increased risk for pneumonia in unrelated donor allogeneic PBSCT patients should be targeted to prevent infections in these higher risk groups.
P714 Surveillance of nosocomial sepsis and pneumonia in patients with haematologic stem-cell transplantation: five years of ‘ONKO-KISS’
M. Dettenkofer, R. Babikir, H. Bertz, E. Meyer, H. Rüden, F. Daschner and the ONKO-KISS Study Group
For surveillance of nosocomial bloodstream infections (BSI) and pneumonia during neutropenia in adult patients undergoing bone marrow transplantation (BMT) or peripheral blood-stem cell transplantation (PBSCT), in the year 2000 an ongoing multicentre surveillance project was initiated by the German National Reference Centre for Surveillance of Nosocomial Infections (ONKO-KISS).
Methods: Nosocomial Infections are identified using CDC definitions for laboratory-confirmed BSI and modified criteria for pneumonia in neutropenic patients [for detailed information in German language see http://www.nrz-hygiene.de/surveillance/onko.htm].
Results: Over the 60-month period up to June 2006, 4,203 patients with 62,338 neutropenic days were investigated (26 centres in German-speaking countries). Of this number, 2,492 (59%) had undergone allogeneic and 1,711 (41%) autologous BMT or PBSCT. The mean length of neutropenia was 14.8 days (9.4d after autologous and 18.6 d after allogeneic transplantation). In total, 776 bloodstream infections and 353 cases of pneumonia were identified. The site-specific incidence densities (pooled mean) were: 12.4 BSIs per 1,000 neutropenic days (10.6 for allogeneic vs. 17.8 for autologous transplantations) and 5.7 cases of pneumonia per 1,000 neutropenic days (5.8 for allogeneic vs. 5.3 for autologous transplantations). There was a trend toward lower incidence densities over the five years. Following allogeneic transplantation, 19.7 BSI/100 patients and 10.8 cases of pneumonia/100 patients occurred, whereas following autologous transplantation 16.7 cases of BSI/100 patients and 5.0 cases of pneumonia/100 patients were observed. The main pathogens associated with BSI were coagulase-negative staphylococci (52%).
Conclusions: The ongoing ONKO-KISS project adds to improving the quality of care in HSCT-patients. Since 2006, surveillance has been extended to neutropenic patients with acute leukaemia to allow centres that do not perform HSCT to participate.
P715 Trends in bacteraemia due to Pseudomonas aeruginosa: comparison between oncohaematologic and non-oncohaematologic patients
P. Martin-Davila, G. Ayala, E. Fernandez-Cofrades, J. Fortun, E. Loza, J. Lopez-Martin, J. Lopez-Jimenez, R. Canton, F. Baquero, S. Moreno (Madrid, ES)
Objectives: To review epidemiological aspects of P. aeruginosa. (PA) bacteraemia, prognostic factors, and changes in their antimicrobial (Ab) susceptibility patterns in oncohaematologic (OH) and non-OH patients.
Methods: A retrospective review during a 10-year period (1996–2005) in our institution was performed including all episodes with at least one positive blood culture yielding PA from patients with diagnosis of active cancer, receiving or not chemotherapy at bacteriemic episode (OH group) and non-OH patients. Clinical, microbiological and epidemiological data were obtained and compared. Bacteraemia was considered nosocomial if the diagnosis was done ≥72 h after hospital admission and no evidence of bacteraemia was present at the time of admission. Also patients diagnosed after discharge within 60 days of a previous hospital admission.
Results: From Jan-96 to Dec-05 a total of 354 episodes of PA bacteraemia were identified, 342 for full-analysis (125 episodes occurring in OH patients). Total PA bacteraemia episodes per year remains stable but time trends analysis showed an increased rate of episodes occurring in OH patients. Mean ages were similar in both groups (61 vs 60 years). Mean time from hospital admission to bacteraemia was 16 days in OH vs. 24 days in non-OH (p = 0.04). Bacteraemia was considered nosocomial-acquired in 86% of OH and 83% of non-OH groups. Patients staying at ICU during bacteraemia were 4% in OH and 45.5% in non-OH patients (p < 0.001). Ab susceptibility profiles are shown in the table .
Ab susceptibility profilesa
| 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % CAZ | OH | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 9 | 6 | 10 |
| Non-OH | 12 | 11 | 0 | 0 | 4.5 | 11.5 | 11.5 | 18 | 12 | 5 | |
| % FQ | OH | 0 | 0 | 0 | 0 | 0 | 0 | 12.5 | 9 | 33 | 34.5 |
| Non-OH | 4 | 11 | 0 | 0 | 14 | 4 | 19 | 18 | 32 | 35 | |
| % AG | OH | 0 | 17 | 9 | 0 | 20 | 0 | 0 | 9 | 28 | 31 |
| Non-OH | 4 | 9 | 0 | 0 | 4.5 | 0 | 8 | 0 | 16 | 32.5 | |
| % Carbapenems | OH | 0 | 0 | 0 | 0 | 0 | 11 | 12.5 | 4 | 11 | 14 |
| Non-OH | 4 | 11 | 0 | 0 | 9 | 19 | 38.5 | 18 | 28 | 15 | |
| % Colistin | OH | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | ||
| % Non-resistance | OH | 100 | 83 | 82 | 100 | 80 | 89 | 87.5 | 74 | 56 | 55 |
| Non-OH | 76 | 57 | 0 | 0 | 82 | 73 | 50 | 59 | 44 | 55 | |
| % R to ≥3 groups | OH | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 10 |
| Non-OH | 0 | 3 | 0 | 0 | 4.5 | 0 | 11.5 | 4.5 | 12 | 10 |
No. of isolates: 125 OH, 217 Non-OH.
No significant differences were found when compared Ab susceptibilities between both groups, but a general decrease in susceptibility was observed. In 2005, 10% of isolates were multidrug-resistant (≥3 Ab groups) in OH and non-OH patients Mortality at 30 days after bacteraemia was similar in both groups (38% vs 31%, p = NS).
Conclusions: Bacteraemia due to PA increased in OH patients. Analysis of Ab susceptibility patterns showed an increase in resistance to ciprofloxacin, aminoglycosides and, to a lesser degree, to carbapenems in both studied groups. Mortality remains high in both groups.
P716 Febrile neutropenia in children with cancer; a retrospective Norwegian multicentre study
N. Stabell, E. Nordal, K. Wiger, B. Lund, E. Stensvold, A. Taxt, F. Buhring, M. Greve-Isdahl, H. Fornebo, G. Simonsen, C. Klingenberg (Tromsø, Oslo, Trondheim, Bergen, Stavanger, Tønsberg, NO)
Objectives: Febrile neutropenia (FN) is a frequent and potentially life-threatening complication in children with cancer under chemotherapy. Empirical antibiotic therapy of FN in Norway differs from international guidelines with ampicillin or benzyl penicillin combined with an aminoglycoside commonly being used as first line treatment.
Methods: During a 2-year period we retrospectively reviewed clinical and laboratory data on FN episodes in children with cancer below 16 years of age at seven hospitals in Norway. Empirical antibiotic therapy was aggregated into two groups; (i) benzyl penicillin or ampicillin in combination with an aminoglycoside (Regimen A) and (ii) monotherapy with a third generation cephalosporin or a combination of a third generation cephalosporin with other agents (Regimen B).
Results: 95 patients (52.6% boys) with a total of 236 FN episodes were included. Baseline clinical parameters including degree of neutropenia were similar in both treatment groups. Time to defervescence was shorter in patients receiving Regimen A (136 episodes, median 1 day) than in Regimen B (100 episodes, median 2 days) (p = 0.026). There were no statistically significant differences regarding duration of neutropenia, duration of antibiotic therapy, change of first-line empirical antibiotic therapy, subsequent number of changes of therapy, and frequency of positive blood cultures. Thirty-nine of 236 episodes (17%) revealed positive blood cultures. Children with leukaemia had higher rates of blood culture positive FN episodes (30/141, 21%) compared to children with other cancer diagnoses (9/95, 9%) (p = 0.017). Twenty-eight (72%) bacterial isolates were Gram-positive species. High rates of resistance to penicillin and ampicillin were found in both Gram-positive and Gram-negative species (19/39, 49%). All Gram-negative isolates were susceptible to aminoglycosides. Only coagulase negative staphylococci were frequently resistant to both β-lactams and aminoglycosides. One infection-related death occurred during the study period, a child who died of candida sepsis.
Conclusion: Our data indicate that ampicillin or benzyl penicillin combined with an aminoglycoside is not inferior to a regimen based on third generation cephalosporins for the empirical treatment of paediatric febrile neutropenia in Norway.
P717 Bacteraemia and fungaemia in haematologic patients
G. Kliasova, L. Speranskaya, E. Trushina, A. Mironova, M. Maschan, S. Vereschagina, L. Kaporskaja, N. Yuritcina, T. Pospelova, L. Krainova, A. Brilliantova, O. Markina (Moscow, Irkutsk, Samara, Novosibirsk, Ekaterinburg, RU)
Objectives: To evaluate the frequency of pathogens isolated from bloodstream of haematologic patients hospitalised at seven medical centres of Russia.
Methods: During the period from 01 January 2003 to 10 November 2006 we performed a prospective study of all clinically significant bloodstream pathogens. Coagulase-negative staphylococci, Corynebacterium spp. were evaluated in the cases of isolation from two separate blood cultures.
Results: A total of 882 bloodstream isolates were recovered from 654 patients. Age of patients ranged from 5 months to 88 years old, most patients (49%) had leukaemia. 440 (50%) episodes were caused by Gram-negative bacilli, 390 (44%) by Gram-positive bacteria, 50 (5.8%) by fungi and 2 (0.2%) by anaerobic bacteria. In 5% of patients, the first episode of bacteraemia was polymicrobial. The most frequent pathogens were E. coli (20%), coagulase negative staphylococci (18%, CoNS), Enterococcus spp. (10%), P. aeruginosa (7.5%), K. pneumoniae (7%), Viridans streptococci (7%). The prevalence of ESBL-producing strains ranged from 34% (59/175) for E. coli to 58.5% (38/65) for K. pneumoniae. Incidence of Acinetobacter spp. was 4% (n = 33), Stenotrophomonas maltophilia – 3% (n = 24). Among Enterococcus spp. isolates, E. faecium (69%) was predominant. One isolate of E. faecium was resistant to vancomycin (vancomycin MICs – 512 mg/mL, teicoplanin MICs – 32 mg/mL). Oxacillin-resistance rates were 23% and 79% among S. aureus and CoNS, respectively, isolates. 50 episodes of fungaemia included Paecilomyces javanicus (2), Fusarium spp. (1), C. neoformans (1), G. capitatum (1), C. albicans (11), Candida non-albicans (34).
Conclusions: Gram-negative bacilli were predominant in bloodstream infections among haematologic patients. Fungi were isolated from blood rarely but the broad spectrum of pathogens was the cause of fungaemia.
P718 Bloodstream infections complicating orthotopic liver transplant: comparison between the recipients from cadaver and living donors
A. Bedini, C. Venturelli, M. Codeluppi, S. Cocchi, F. Prati, F. Di Benedetto, M. Masetti, C. Mussini, G. Guaraldi, V. Borghi, F. Rumpianesi, G.E. Gerunda, R. Esposito (Modena, IT)
Objective: We evaluated the incidence, the prevalence of the microorganisms isolated and the impact on the survival of the bloodstream infections (BSIs) in two groups of patients: recipients of orthotopic liver transplant (OLT) from cadaveric donor (group A) and from living donor (group B).
Methods: Between October 2001 and September 2006, all the episodes of BSIs that occurred in the first year after OLT in 205 patients were evaluated. One hundred seventy (82.9%) patients received OLT from cadaveric donor and 32 (15.6%) from living donor.
Results: During the study period, 91 episodes of BSI occurred in 205 patients with an incidence rate of 0.44 episodes per patients. The patients of group B had a higher incidence of BSIs than the patients of group A (40.0% and 19.4%, respectively; p = 0.008). No differences in the prevalence of the microorganisms isolated were observed between the 2 groups (group A: Gram positives 49%; Gram negatives 42%; fungi: 9%; group B: Gram positives 41%; Gram negatives 50%; fungi: 9%). The episodes of BSI occurred within the first 2 months after transplant in the 50.8% in the group A and 42.4% in the group B (p = n.s.). The crude 30-day mortality rate was 21.2% in the group A and 14.2% in the group B (p = n.s). Survival rate 1 year after OLT was 80.0% in the patients from both groups. Considering the patients all together, we observed that 1-year survival of the patients with BSIs was lower than those without BSIs (p = 0.0001).
Conclusions: In our population, the patients receiving OLT from living donor had a higher risk of BSIs than recipients of OLT from cadaveric donor. These data could be due to the higher complexity of surgical interventions used in OLT from living donor. Notwithstanding of this, the 30-day mortality rate resulted lower in the group B and the survival at 1 year after OLT resulted the same in both groups. The results of this study confirm that BSIs affect the survival of OLT patients, particularly during the first 1 year after transplantation.
P719 First reported case of human infection with Psychrobacter submarinus
S. Musaad, N.D. Smith, A. English, D. Gascoyne-Binzi, A. Glaser, R. Phillips, N. Young (Leeds, UK)
We present the first reported human infection involving Psychrobacter submarinus. This was from a case of suspected line infection in a 7 year old, male, Caucasian child who was diagnosed 3 months prior to this episode with widespread Burkitts lymphoma in association with high Epstein Barr virus titres. He made a good symptomatic response to standard chemotherapy. Following his fifth course of intensive myelosuppressive therapy, he was admitted on day 20 with fever, abdominal pain and tenderness. These symptoms settled with observation and antibiotic therapy as detailed below.
Luminal blood cultures revealed mixed streptococci and Gram-negative coccobacilli (GNCB).
The child clinically improved on iv cefuroxime and the focus of the infection was thought to be his line. Further luminal blood cultures revealed only enterococci after a repeat fever spike 3 days later. The child was switched and responded well to vancomycin, tobramycin and piperacillin/tazobactam treatment.
The GNCB grew on blood agar after 6 days in CO2 at 37°C, was oxidase positive and was sensitive to most anti Gram-negative antibiotics including amoxicillin, cefuroxime and tobramycin. It failed to identify by conventional methods so was submitted for PCR and sequencing (in both directions) of the 16S ribosomal gene. Using the GenBank database, it was found to have 98.5% similarity with Psychrobacter submarinus. The genus Psychrobacter was first described in 1986 and belongs to the family Moraxellaeceae. Members of this genus have been isolated from sea water, skins of fish and other marine life. P. submarinus is a new species described in 2002 as a marine organism. It is a GNCB and is catalase and oxidase positive. Some isolated case reports have been described as being caused by Psychrobacter spp. (in an AIDS patient in 1994, meningitis in a neonate in 1991 and an ocular infection in 1990). Recently (2006), a surgical wound infection due to Psychrobacter spp. and a bacteraemia in a cirrhotic patient due to P. phenylpyruvicus were described.
This is the first reported case of P. submarinus in humans and involved a line infection in an immunocompromised patient.
P720 Successful treatment of a linezolid- and vancomycin-resistant Enterococcus faecium sepsis with daptomycin plus doxycycline in an allogenic stem cell transplant recipient
J. Dahlke, I. Sobottka, H. Rhode, G. Franke, T. Zabelina, H. Lellek, A. Muth, C. Wolschke, A. Zander, N. Kröger (Hamburg, DE)
Background: Nosocomial bacteraemia is a major cause of morbidity in patients with cancer. Of major concern is the growing resistance of Enterococci such as E. faecium to multiple antibiotic agents. While vancomycin-resistant E. faecium (VRE) remains a problem in treatment, the linezolid- and vancomycin-resistant E. faecium (LRVRE) comes into focus.
Case: During induction chemotherapy a 41-year-old woman with CML developed fever and inflammation of the colon with severe diarrhoea. Neutropenic fever was treated with vancomycin and imipenem but fever persisted. Blood- and urine culture yielded Enterococci: VRE with additional resistance to ampicillin-, erythromycin-, clindamycin and flourquinolones but susceptibility to linezolid and tetracycline. Treatment with linezolid was successful with recovery of the intestinal mucosa. However the patient remained in neutropenia and bone marrow showed persistent leukaemia. The patient was transferred to BMT-department for allogeneic stem cell transplantation (SCT). During pretransplant conditioning the patient remained neutropenic and developed fever and an increase of CRP. Neutropenic fever persisted under antibiotic therapy of linezolid in combination with ceftazidim and tobramycin. The CT of the abdomen detected a perianal abscess. The further clinical courses showed a clinical picture deteriorated with critically systemic inflammatory response. Blood cultures obtained at day 5 and 1 before SCT as well as genital and anal smears were positive for VRE again that however since then emerged as resistant to linezolid additionally. So far the LRVRE isolates were susceptible to tetracycline only. In view of the critical state of the patient we decided to combine tetracycline with daptomycin (4 mg/kg) that subsequently was found to be effective also. Within the next three days the patient became apyrexial and the CRP fell rapidly. Blood- and stool cultures obtained regularly after starting daptomycin/tetracycline did not yield LRVRE. The perianal abscess resolved completely.
Conclusions: This is the first report of a LRVRE strain isolated in Germany that successfully could be treated with the combination of daptomycin and doxycycline. The case presented emphasizes that daptomycin should be considered as therapeutic option for severe infections with multiresistant organisms such as LRVRE.
P721 Pulmonary nocardiosis in a tertiary care hospital: 8-year experience (1999–2006)
E. Papaefstathiou, M. Kanellopoulou, N. Skarmoutsou, M. Martsoukou, P. Katiska, E. Papafrangas, A. Tsakris (Athens, GR)
Objectives: Pulmonary nocardiosis (PN) is a subacute or indolent pneumonia caused by aerobic actinomycetes of the genus Nocardia. The disease is difficult to diagnose and therefore its incidence is not well established. We report our experience on PN in a case study conducted in our hospital (108 pulmonary beds) during a period of eight years (1999–2006).
Methods: Sputa of all admitted patients were smeared, stained by Gram and ZN and plated in appropriate media. The identification of Nocardia species was performed by drug susceptibility patterns (MIC, E-test, Disk diffusion method) in conjunction with the profiles of conventional biochemical assays. Information was collected on demographic data, clinical details, underlying diseases and immunosuppressive therapy.
Results: Thenty-nine patients with PN were studied (17 males and 12 females; mean age 52 y). Pathogens were N. asteroides I, II, VI (22/29), N. farcinica (6/29) and N. transvalensis complex (1/29). Underlying diseases included haematological and other malignancies (11 cases), cystic fibrosis (10 cases), COPD (3 cases), multiple sclerosis (2 cases), diabetes mellitus (3 cases). Diabetes mellitus was concomitant underlying disease in cystic fibrosis (2 cases) and haematological malignancies (3 cases). Seven patients had received steroid treatment. Therapy with TMP/SMX (20/29) or with the combination of imipenem and amikacin (9/29) was given to the patients for 2 to 6 months. In all but seven cystic fibrosis patients, treatment resulted in microbiological eradication of the pathogen.
Conclusion: PN is an infrequent infection that mainly affects immunocompromised patients. In our study, several Nocardia species were identified among patients with various predisposing diseases. To our experience direct Gram-stain in sputum smears was found useful to drive the diagnosis of PN, particularly in patients with underlying diseases and a clinical suspicious of the disease.
P722 Clonal dissemination of Gram-negative bacteria causing bloodstream infections in patients with haematological malignancies
A. Brilliantova, G. Kliasova, A. Mironova, S. Sidorenko (Moscow, RU)
Objectives: The purpose of the study was to investigate the clonal dissemination of Gram-negative strains causing bloodstream infections in one haematological centre.
Methods: A total of 141 Gram-negative strains, which included E. coli (n = 80), Enterobacter spp. (n = 15), K. pneumonia (n = 27) and P. aeruginosa (n = 19) isolated from blood of haematological patients form January 2003 to December 2005, were analysed by pulsed-field gel electrophoresis (PFGE). Genomic DNA of E. cloacae, and P. aeruginosa isolates was digested with SpeI enzyme, K. pneumonia and E. coli DNA – with XbaI and NotI enzymes respectively. Interpretative criteria used were those described by F. Tenover et al.
Results: Genotyping of P. aeruginosa strains revealed 15 different PFGE types, 37% (7/19) of P. aeruginosa isolates belonged to three closely related clonal types. Enterobacter spp. isolates represented 12 PFGE types, among them 33% (5/15) were genetically related and arouse from two clones. Isolates of K. pneumonia were grouped into 23 PFGE types, 26% (7/27) of isolates belonged to three closely related clonal types. E. coli isolates belonged to 79 different PFGE types and showed the highest level of clonal diversity. Only 3% of E. coli (2/80) isolates were clonally related, which differed significantly from the amount of clonally related isolates of P. aeruginosa (37%), Enterobacter spp.(33%), K. pneumonia (26%), p < 0.001.
Conclusion: E. coli isolates demonstrated significantly higher clonal diversity than other Gram-negative bacteria isolated from blood. We suppose that the majority of E. coli bloodstream infections in haematological patients are of endogenous origin. Infections caused by other Enterobacteriaceae and P. aeruginosa are more likely to be acquired from hospital environment.
P723 Diagnostic relevance of interleukin-6 and tumour necrosis factor alpha in discriminating high-risk and low-risk groups in febrile
M. Parsa, S. Najjar najafi, N. Jonaidi jafari, M. Mohraz, A. Ghavam zadeh, B. Bahar, M. Izadi, H. Radfar, H. Ghofrani (Tehran, IR)
Objectives: Early diagnosis of serious infection in an important issue in feverish patients with neutropenia. Identifying serum markers of immunologic response may be useful for distinguish patients with high risk or low risk. Serum IL-6 and TNF-alpha have shown to increase in response to sepsis and infection in non-neutropenic patients. The present study was designed to determine diagnostic value of IL-6 and TNF-alpha in patients with fever and neutropenia.
Methods: This is a prospective study of 133 patients admitted to two university hospital in Tehran, Iran with fever and neutropenia. Patients were divided two groups as low risk and high-risk groups. Cytokines level compared with Mann-Whitney test in study groups of patients and ROC curves used to determine best cut-off points level for cytokines discriminating risk groups.
Results: Mean age of patients was 26.8±2.5 years and 7.5% of patients allocated in low risk group. The mean IL-6 and TNF-alpha serum level below 17 pg/mL was defined as best cut-off point determining low risk group patients with sensitivity and specificity of 70% and 67.5% respectively. However, we cannot define a statistically significant cut-off point for TNF-alpha to use as a diagnostic test.13.5% of patients of our study have positive blood cultures (6% Gram-negative, 6% Gram-positive, 1.5% fungi), but no statistical difference had found in serum IL-6 and TNF-alpha levels in blood culture groups.
Conclusion: Despite our findings about IL-6 diagnostic value in neutropenic patients with fever and its advantages in discriminating risk groups of patients it seem necessary to design a randomised controlled trial before use of this marker.
P724 Serum levels of IL-6, IL-8 and IL-10 at fever onset in neutropenic patients: a rapid test for the prediction of Gram-negative bacteraemia? Results of an EORTC Infectious Disease Group multicentre study
H. Akan, M. Paesmans, O. Marchetti, W. V. Kern on behalf of the EORTC Infectious Disease Group
Objectives: Previous studies have suggested that serum or plasma levels of cytokines such as IL-6 or TNF-alpha may have prognostic value in patients with severe sepsis. These and other cytokines have also been studied in febrile neutropenic patients, but the test characteristics for prediction of outcomes in this setting have been variable. We wondered whether the levels of IL-6, IL-8 or IL-10 in samples obtained at fever onset were helpful in predicting bacteraemia in general or, more specifically, Gram-negative bacteraemia (GNB).
Methods: Blood samples were obtained from 573 patients with fever and neutropenia who were included in a multicentre therapeutic trial of the EORTC (EORTC-IDG Trial 46971). The samples were collected at fever onset, centrifuged within 1 hour, frozen and shipped to a central facility where they were stored at −70°C until testing. The cytokine concentrations were measured in duplicate by an immunoluminiscence assay (Immulite) allowing single serum sample measurements within ∼45 min.
Results: Most patients had acute leukaemia. Their median age was 46 years (range, 2–86). Fifteen percent of the patients had GNB (anaerobes excluded), and eight percent died. For all three cytokines, there was a significant correlation between serum concentrations and length of fever. Levels of the three cytokines were higher in patients who failed initial empirical therapy because of clinical deterioration. They were highest in the subgroup of patients with GNB. The areas under the receiver operating characteristic curve (AUROCs) for the prediction of bloodstream infection (any organism) was best for IL-10 (0.73; 95% CI, 0.68–0.78) but the differences between IL-10 and IL-6 or IL-8 AUROCs were small and statistically nonsignificant. Similarly, the AUROC for the prediction of GNB was better for IL-10 (0.82; 95% CI, 0.77–0.87) than for IL-8 (0.75; 95% CI, 0.69–0.81) and IL-6 (0.73; 95% CI, 0.67–0.80). At specificity levels of 95%, sensitivities of the three cytokine assays, however, were <50% for the prediction of bacteraemia or GNB.
Conclusion: We conclude that a single serum sample analysis for measurement of one of these interleukins at the onset of febrile neutropenia currently has limited predictive value.
P725 European survey on the use of antibacterial prophylaxis in neutropenic cancer patients: a joint project of the EORTC and the EBMT
H. Akan, O. Marchetti, C. Cordonnier, T. Calandra for the Infectious Diseases Group of the European Organization for Research and Treatment of Cancer (EORTC-IDG) and the Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation (EBMT-IDWP)
Background: Bacterial infections are frequently observed in neutropenic cancer patients. The use of antibacterial prophylaxis (Px) for prevention of these complications may differ among centres due to the emergence of bacterial resistance and the uncertain impact on infectious morbidity and mortality. Few data are available on the use of antibacterial Px in onco-haematological patients in Europe.
Objective: To conduct an European survey on the use of antibacterial Px in neutropenic cancer patients.
Methods: A questionnaire on antibacterial prophylaxis was distributed in 2004–2005 to all members of the EORTC-IDG and EBMT-IDWP. Four clinical settings were studied: short-duration neutropenia (<10d) in solid tumours/haematological malignancies, long-lasting neutropenia (>10 d) in acute leukaemia, autologous hematopoietic stem cell transplant (HSCT) and allogeneic HSCT.
Results: 105 out of 586 EBMT centres reached by web page and 32 EORTC-IDG members reached by e-mail (70% university, 16% university-affiliated and 14% community or private hospitals) from 25 European countries participated in the survey. Specialties of investigators were: 86% haematology–oncology, 9% infectious diseases, and 5% other. 62% of responding investigators were involved in patients care as primary physicians, 26% as consultants, while 12% were not in charge of this type of patients. Assessment of the risk of bacterial infections and use of antibacterial prophylaxis in the 4 patient populations. Median (range) daily dose of ciprofloxacin: 1000mg (500–1500). Px was given orally in 100% of the patients with neutropenia < or > 10 days and in autologous HSCT, while it was given i.v. in 6.5% of allogeneic HSCT.
| Neutropenia |
HSCT |
|||
|---|---|---|---|---|
| <10d (n = 97) | >10d (n = 101) | Autologous (n = 101) | Allogeneic (n = 100) | |
| Proportion of investigators estimating that patients are at high risk of bacterial infections | 9% | 78% | 51% | 80% |
| Proportion of patients receiving antibacterial Px | 19% | 51% | 50% | 76% |
| Antibiotics used for Px: | ||||
| Ciprofloxacin | 55% | 52% | 56% | 53% |
| Other fluoroquinolone | 19% | 20% | 18% | 22% |
| Cotrimoxazole | 23% | 15% | 18% | 22% |
| Selective digestive decontamination | 3% | 10% | 7% | 11% |
Conclusions: In Europe, antibacterial prophylaxis is used routinely in 50% to 76% of neutropenic cancer patients with long-duration neutropenia. Oral fluoroquinolones, mostly ciprofloxacin, are the agents used most commonly.
P726 Usefulness of a general chemotherapy myelotoxicity score to predict febrile neutropenia in haematological cancers
A. Georgala, M. Paesmans, A. Schwarzbold, F. Muanza, M. Aoun, A. Ferrant, D. Bron, J. Klastersky, M. Moreau (Brussels, BE)
Introduction: Chemotherapy-induced neutropenia is the most common adverse effect of chemotherapy and is often complicated by febrile neutropenia (FN). In many institutions, the decision to give granulocyte colony-stimulating factors as prophylaxis is mainly based on the myelosuppressive potential of the chemotherapy regimen (CR) but clear regimen-specific risks are not defined. The objective of this study is to validate a classification of aggressiveness of a CR and to evaluate its usefulness in a risk prediction model of FN in patients with haematological cancers (HC) at the beginning of a chemotherapy cycle.
Methodology: Patients aged above 16 years, diagnosed with HC of any type were prospectively enrolled and followed in the “Institut Jules Bordet” and the “Université Catholique de Louvain” between 2001 and 2005. Out of the 266 enrolled patients, 22.9%, 43.6% and 33.5% were followed respectively during one cycle, 2 to 4 cycles and more than 4 cycles, totalising 1053 cycles. Relevant patient's information were collected at the beginning of the first cycle (sex, age, diagnosis and concomitant diseases). At the beginning of each cycle, the CR score was computed, a blood examination performed and the temperature measured. In the follow-up, the number of days of FN (neutrophils <500/μl and fever) were counted. This outcome was dichotomised (no FN vs ≥1 day of FN). Generalised Estimating Equations (GEE) was used for the analysis as it takes into account the correlation structure between the outcome as well as the covariates within the same patient.
Results: In 35.3% of the cycles, patients experienced FN. In the final model, aggressive CR is the major predictor of FN (OR 4.4 [2.7–7.0]), compared to those not receiving an aggressive CR. The other independent predictors are a diagnosis of “myeloid tumour” (2.9 [1.7–5.1], a baseline monocyte count <150/μl (2.3 [1.4–3.8]), an involvement of bone marrow (2.1 [1.4–3.2]), the first cycle in the same treatment line (2.2 [1.5–3.2]) and a baseline hemoglobin dosage <12 gr/l (1.7 [1.0–2.7]). Using the estimates of the regression coefficients, a rule of prediction of FN was computed (sensitivity: 82.2%, specificity: 78.1%, positive predictive value: 68.5% and negative predictive value: 88.4%).
Conclusions: Further studies are needed to validate this score as well as investigating new factors in order to be able to better predict FN.
P727 The use of cepfoperazone/sulbactam in oesophageal cancer patients after Lewis-Tanner oesophagectomy via an abdominal-right thoracotomy approach
Z. Volkova (Moscow, RU)
Objective: The aim of the study was the comparison of two regimens of cefoperazone/sulbactam (CS) in oesophageal cancer patients (pts) undergoing Lewis-Tanner oesophagectomy via an abdominal-right thoracotomy approach for prevention of postoperative infectious complications.
Materials and Methods: 64 oesophageal cancer pts were studied. Group (gr) A (31 pts) received preventive therapy with CS 4 g/day for 5 days. Gr B (33 pts) received preventive therapy with CS 8 g/day for 5 days. First dose of CS started 1 hour before operation. Both groups were comparable (p > 0.05) for the main parameters (underlying diseases, age, weight, cancer diseases, ã-therapy, chemotherapy). Bacteriological analyses were made with automatic systems: “ATB-Expression” and “VITEK2” (“BioMerioux”, France).
Results: 11 (35.5%) pts of gr A had infectious complications during first 7 days after operation: sepsis, septic shock – in 2/31 (6.5%) pts; pneumonia in – 11/31 (35.5%) pts; supperative tracheobronchitis in – 7/31 (22.6%), pleural abscess in 1/31 (3.2%) pts. 9 (29.0%) pts had two or more infections: pneumonia and bronchitis in – 6/9 (66.7%) pts; sepsis, septic shock and pneumonia in – 2/9 (22.2%) pts; pneumonia, abscess and pleural empyema in – 1/9 (11.1%). Only 4/33 (12.1%) pts of gr B (p < 0.05) had infections complications during first 7 days after operation. 4/33 (12.1%) pts – pneumonia; 1/33 (3.0%) pts – pneumonia and bronchitis.
67.9% of strains were Gram-negative rods: 28.4% – P. aeruginosa, 13.3% – S. malthophilia, 9.8% – K. pneumoniae, 7.4% – E. coli. 59% strains of P. aeruginosa was multiresistant. Nosocomial microflora contaminated pts after 13–14 days postoperation. The pneumonia cases was found 2.8 times more often in gr A vs gr B (p < 0.05) and it was assiciated with sepsis, septic chock in 2/13 (15.4%) pts vs 0% pts in group A and B, respectively.
Conclusion: Preventive therapy with cefoperazone/sulbactam 8 g per day, in oesophageal cancer patients after Lewis-Tanner oesophagectomy via an abdominal-right thoracotomy approach reduced the rate of infectious complications in early postoperative period.
P728 Initial experience with ertapenem in clinical practice: treatment of twenty patients with chemotherapy-induced low-risk febrile neutropenia in an outpatient setting
F. Krasniqi, A. Ho, G. Egerer (Heidelberg, DE)
Objectives: Ertapenem is a novel, once-a-day parenteral carbapenem with a broad spectrum, including extended-spectrum β-lactamases (ESBL) producers and AmpC producing enterobacteriaceae. An observational study was performed in low-risk (defined by the AGIHO) cancer patients with chemotherapy-induced febrile neutropenia to determine the safety and efficacy of ertapenem given in an outpatient setting.
Material and Methods: In a prospective observational study carried out between December 2005 and September 2006 20 febrile episodes were recorded. Inclusion criteria were neutropenia (neutrophil count <500/μL or <1000/μL with predicted decline to 500/μL within the next 2 days) and fever (≥38.3°C). Patients with acute leukaemia (n = 7), multiple myeloma (n = 5), high-grade Non-Hodgkin-Lymphoma (n = 4), low-grade Non-Hodgkin-Lymphoma (n = 3) and Hodgkin's disease (n = 1) were enrolled with a median age of 58 years (range 25–74) and a median Karnofsky-performance-score of 8.0 (range 7.0–10.0).
The causes of fever were: FUO in 10 patients, clinically defined infection (CDI) in 6 patients and 4 patients with MDI and CDI. All patients were initially treated with an empirical treatment with ertapenem 1 g per day, either alone (n = 8) or in combination with other agents (n = 12).
Results: The initial treatment with ertapenem was successful in 14 patients, who were treated in an outpatient setting. However admission to hospital was necessary for another 6 patients, two of whom died (one of mykoplasmapneumonia, another one of septicaemia with Enterococcus faecium).
Conclusion: Ertapenem is effective and safe in patients with febrile neutropenia. In terms of increasing Gram-negative multi-drug resistance ertapenem is an effective, safe and economic option for the treatment of low-risk febrile neutropenia. However, further studies are necessary.
P729 Tigecycline usage in cancer patients with refractory pneumonia: a report on 38 cases. A single-institution study
R. Chemaly, R. Hachem, S. Hanmod, J. Adachi, H. Hogan, V. Mulanovich, D. Kontoyiannis, A. Safdar, I. Raad, K. Rolston (Houston, US)
Background: Tigecycline (TG), first in a new class of Glycylclines, is a novel agent approved for treatment of complicated soft tissue and intraabdominal infections in adults. Clinical data on the safety and efficacy of TG in cancer pts with pneumonia is lacking.
Methods: We reviewed records of cancer pts with pneumonia treated with TG for >72 h between Sept'05 and Sept'06. Data collection included demographics, cancer type, indication for TG, side effects and outcome.
Results: Thirty-eight pts with pneumonia were identified, 4 (10%) of them had ventilator-associated pneumonia. Median age was 56 years (23–79 y). Most pts (28, 74%) had haematologic malignancies, including 14 allogeneic HSCT pts; 13 pts (34%) were neutropenic (ANC < 500/mm3). Twenty-six pts (68%) were in the ICU of whom 18 (69%) required ventilator support after development of pneumonia. Thirty-six (95%) pts received TG as second line agent (after failure of other broad-spectrum antibiotics) and in combination with an anti-pseudomonal drug(s) and the remaining 2 pts received it as a first line agent because of allergy to vancomycin and/or β-lactams. Median duration of therapy was 11d (4–35 d). Twenty-eight pts (74%) received TG for refractory pneumonia of unknown etiology and 10 pts (26%) for microbiologically documented pneumonia with multi-drug resistant organisms (MDRO) [MRSA, S. maltophilia, E. coli, VRE, and Acinetobacter]. Clinical response was noted in 24 pts (63%); including 4 of the 5 pts who had associated bacteraemia and 2 pts with associated intra-abdominal infections. Eight of the 10 (80%) patients who had microbiologically documented pneumonia responded to the treatment. The remainder 14 pts died (37%). The cause of death was multi-organ failure and pneumonia of unknown etiology in 10 pts; MDR P. Aeuroginosa bacteraemia, aspergillosis, S. maltophilia and VRE pneumonia in 1 each. Of the 9 pts who were not on anti-emetics and were not intubated, only 1 developed mild nausea. Diarrhoea was noted in 1 pt.
Conclusions: Combination of TG with anti-pseudomonal drug(s) appears to be an appropriate choice for treatment of refractory pneumonia secondary or not to MDRO (excluding P. aeuroginosa) in cancer patients, including HSCT recipients.
Antibiotic susceptibility of respiratory bacterial isolates
P730 Resistance of Streptococcus pneumoniae to widely used antibiotics in a training hospital in Ankara, Turkey
N. Balaban, I. Mumcuoglu, N. Hayirlioglu, Z. Karahan, N. Sultan, H. Bodur (Ankara, TR)
Objectives: The worldwide increase of resistance to widely used antibiotics in S. pneumoniae treatment has become a serious problem within the last 20 years. The aim of this study was to determine the antimicrobial susceptibility of clinical S. pneumoniae isolates to penicillin and other widely used antibiotics.
Methods: Seventyeight S. pneumoniae strains were isolated from clinical samples between June 2004 and May 2005 at the Department of Microbiology and Clinical Microbiology in Ankara Numune Training and Research Hospital and Gazi University Hospital. 57 (73.1%) of the strains were isolated from sputum and 21 (26.9%) from sterile body sites. Identification of the S. pneumoniae strains was performed by Gram stain, colony morphology, optochin sensitivity and bile solubility tests. Susceptibility of the strains to penicillin, erythromycin, azithromycin, trimethoprim-sulfamethoxazole, cefotaxime, cefuroxime, doxycycline, ofloxacin and levofloxacin were determined by the E test method (AB-Biodisk, Sweeden). Results were evaluated according to the CLSI standards. S. pneumoniae ATCC 49619 was used as the quality control strain.
Results: The rates of penicilin resistance and intermediate resistance were 7.7% and 21.8% respectively. All resistant strains were isolated from sputum. The resistance rates for trimethoprim-sulfametaxazole, azithromycin and erithromycin were 28.2%, 16.7% and 14.1%. 98.7% of the pneumococcal isolates were susceptible to both levofloxacin and cefotaxime. All penicillin resistant isolates and 82.4% of the intermediate resistant isolates were resistant to at least two antimicrobial agents.
Conclusion: Increasing rates of penicillin resistant S. pneumoniae (PRSP) has been shown in Turkey in recent studies. Knowing penicillin resistance rates of the isolates is important for planning empirical antimicrobial therapy in pneumococcal infections such as community acquired pneumonia and meningitidis. It is worth reminding the clinicians about the fact that, PRSP may cause treatment failure in respiratory tract infections as they are usually resistant to many commonly used antimicrobial agents such as macrolides and trimethoprim-sulfametaxazole.
P731 S. pneumoniae resistance patterns in a chest diseases hospital, for the decade 1997–2006
M. Makarona, H. Moraitou, N. Tsagarakis, S. Karabela, A. Gioga, I. Kouseris, V. Spyropoulos, S. Triantafyllou, A. Pefanis, S. Kanavaki (Athens, GR)
Objectives: The aim of this study was to investigate the resistant patterns of S. pneumoniae strains isolated in ‘Sotiria’ Chest Diseases Hospital from 1997 to 2006, a period covering the last decade.
Material and Methods: A total of 480 S. pneumoniae strains were isolated in our laboratory during the last decade. Most of them (373/480, 77.7%) derived from sputum samples, while 107/480 (22.3%) derived from different invasive infection sites, such as blood (61/480, 12.7%), pleural effusion (35/480, 7.29%) and CSF (5/480, 1.04%).
Culture and susceptibility tests were performed according to NCCLS 2004 guidelines. All strains were tested by Kirby-Bauer disk diffusion method for penicillin, erythromycin, tetracycline, co-trimoxazole, ciprofloxacin and cefotaxime susceptibility. Oxacillin 1 μg/mL disks checked resistance to penicillin. Zone diameter ≥20 mm indicated Penicillin susceptible strains (PSSP) and ≤19mm penicillin non-susceptible (PNSP). Penicillin MIC for PNSP strains was determined by E-test, according to NCCLS 2004 guidelines.
Results: Regarding penicillin, 366/480 (76.25%) were PSSP strains and 114/480 (23.75%) were PNSP. A number of 92/114 (80.7%) showed intermediate resistance and 22/114 (19.3%) resistance. Most of the PNSPs (90/114, 78.9%) derived from sputum cultures. It is worthy of remark that 81/114 (71.0%) of PNSPs and 76/366 (20.7%) of PSSPs conferred resistance also to Erythromycin. Erythromycin resistance rates remained stable around 30% throughout the decade. Erythromycin MIC was performed on 108 strains and 48/108 (44.4%) showed high levels of resistance (MIC ≥128 μg/mL). Tetracycline resistance rate was at 14.6%. Cefotaxime resistance rate was at 1.0% and Ciprofloxacin at 2.0%. 96/480 (20.0%) strains proved multi-drug resistant (MDR), while 76/96 (79.0%) were PNSPs.
Discussion: All medical practitioners should be aware of current S. pneumoniae resistance rates, before empirical treatment is offered to community-acquired respiratory system infections.
P732 Comparison between respiratory and blood isolates of community-acquired Streptococcus pneumoniae from the UK and Ireland: resistance and serotypes
R. Reynolds, D. Felmingham, R. Hope for the BSAC Working Parties on Resistance Surveillance
Objective: Results from the BSAC Respiratory and Bacteraemia Resistance Surveillance Programmes were compared to identify differences between respiratory and blood isolates of S. pneumoniae.
Methods: 31 centres collected 5083 community-acquired lower respiratory S. pneumoniae from 1999/2000 to 2005/06; 29 centres collected 1157 isolates from blood from 2001 to 2005. Ten of these centres contributed to both programmes. MICs were measured by BSAC methods in two central laboratories, one for each programme. Respiratory isolates from 2005/06 and all blood isolates were serotyped. The 285 blood isolates taken >48 hours after hospital admission differed from presumed community-acquired blood isolates in penicillin non-susceptibility (11% vs. 5%), patient age and sex, and were excluded from the results below. Logistic and multinomial logit models used robust errors to account for clustering of effects by centre.
Results: The top ten serotypes in blood were 14, 1, 9V, 23F, 8, 4, 3, 19F, 6B, 22F (total 70%); this distribution did not vary significantly over 5 years. The distribution was different in respiratory isolates: the top ten were 19F, 23F, 6B, 3, 6A, 9V, 14, 11, 15, 19A (total 63%). Serotype distributions varied with age group but not with sex or care setting (community vs. hospital), and prevalence of penicillin non-susceptibility varied between serotypes.
Penicillin non-susceptibility was more prevalent in respiratory than in blood isolates; this difference was apparent in each age group, sex and care setting, and within some common serotypes. Care setting and isolate source (respiratory vs. blood) were significant independent predictors of penicillin non-susceptibility; sex and age group were not. The role of serotype distribution is uncertain, as serotypes were too numerous to include satisfactorily in models, but it is unlikely to explain these effects completely since differences could be seen within some major serotypes.
Conclusion: Penicillin non-susceptibility is uncommon in community-acquired S. pneumoniae in the UK and Ireland. It is more common in isolates from lower respiratory sources than those from blood, but some caution in interpretation is required as few centres contributed both blood and respiratory isolates. The serotype distribution of blood isolates was stable over 5 years. These results provide a baseline for comparison should serotype distributions and associated resistance change with future use of the 7-valent conjugate vaccine.
S. pneumoniae penicillin non-susceptibility
| Non-susceptible |
||||
|---|---|---|---|---|
| Respiratory |
Blood |
|||
| N | %PEN-NS | N | %PEN-NS | |
| All isolates | 5065 | 8.3 | 843 | 4.7 |
| Care setting | ||||
| community | 2023 | 6.4 | 162 | 4.3 |
| hospital (<48 hours) | 3042 | 9.5 | 681 | 4.8 |
| Age | ||||
| 0–19 | 481 | 8.9 | 114 | 6.1 |
| 20–59 | 1837 | 6.8 | 275 | 4.0 |
| ≥60 | 2742 | 9.2 | 447 | 4.9 |
| Gender | ||||
| female | 2074 | 7.0 | 406 | 4.9 |
| male | 2989 | 9.1 | 434 | 4.6 |
| Serotype | ||||
| 14 | 38 | 34 | 135 | 3 |
| 23F | 72 | 6 | 60 | 3 |
| 19F | 83 | 10 | 43 | 2 |
| 9V | 39 | 31 | 67 | 34 |
| 3 | 51 | 0 | 46 | 0 |
| 1 | 11 | 0 | 74 | 0 |
| 6B | 53 | 9 | 32 | 6 |
P733 The first nationwide surveillance of bacterial respiratory pathogens conducted by the Japanese Society of Chemotherapy
Y. Niki, S. Kohno, N. Aoki, A. Watanabe, M. Yagisawa, J. Sato, H. Hanaki on behalf of the JSC Surveillance Committee & The Kitasato Institute
Objectives: The main approach in the control of antibiotic-resistant infections is through the precise usage of specific antimicrobial agents. Comprehensive data on susceptibilities of the major pathogens to currently available agents is currently lacking. In 2006 the Japanese Society of Chemotherapy (JSC) initiated a nationwide study of major bacterial RTI pathogens (Staphylococcus aureus, Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae, Moraxella catarrhalis, Klebsiella pneumoniae and Pseudomonas aeruginosa) in Japan.
Methods: A total of 924 clinical isolates from well-diagnosed adult RTI patients were obtained from 34 hospitals throughout Japan between January and April 2006. Susceptibility of these strains to 40 antimicrobial agents was tested at the central laboratory according to CLSI standards for broth microdilution method. Beta-lactamases were detected by the Nitrocefin disc method. Extended-spectrum β-lactamase (ESBL) and metallo-β-lactamase (MBL) were detected by the Cica-Beta Test (Kanto Chemical, Tokyo).
Results: See the table . One MBL-producing multi-drug resistant P. aeruginosa was found among 143 strains, and two ESBL-producing K. pneumoniae were found among 74 strains.
| Strain | No. of strains | Antimicrobial agent | % resistance |
|---|---|---|---|
| S. aureus (MRSA) | 205 | methicillin | 63.4* |
| vancomycin | 0 | ||
| teicoplanin | 0 | ||
| linezolid | 0 | ||
| S. pneumoniae | 200 | penicillin | 39a |
| erythromycin | 73 | ||
| H. influenzae | 165 | ampicillin (also | 29.1 |
| β-lactamase negative) |
4% resistant, 35% intermediate.
Conclusion: In the first nationwide surveillance of bacterial respiratory pathogens, valuable data on resistance patterns to currently available antimicrobial agents were elucidated. JSC intend to update this data annually to promote precise usage of antimicrobial agents. We also intend to extend the study to include uropathogens.
P734 Nosocomial outbreak of telithromycin- and fluoroquinolone-resistant Streptococcus pneumoniae in a Japanese hospital
T. Muratani, T. Kobayashi, T. Matsumoto on behalf of the Hibiki Research Group for Clinical Microbiology
Background: Streptococcus pneumoniae is a major pathogen causing community-acquired pneumonia and acute bronchitis. Amoxicillin, macrolides, and fluoroquinolones have been used in clinically, because of their potent activity against S. pneumoniae. Recently, the resistant isolates to β-lactams including amoxicillin and macrolides, such as erythromycin, clarithromycin, rokitamycin, azithromycin have accounted for majority in clinical isolates. But telithromycin and fluoroquinolone have kept potent activity. We isolated high resistant isolates to telithromycin and fluoroquinolones in 2004 and 2006. The aim of the this study is to analyse these resistant isolates.
Materials and Methods: The resistant isolates to telithromycin and levofloxacin were isolated from 2 patients in 2004, and 9 patients in 2006 from a same hospital. The MICs of various antimicrobials against the isolates were determined by the two-fold serial agar dilution method. PCR and DNA sequencing technique were used to analyse mechanism of resistance to telithromycin. PFGE technique was performed to analyse clonal spread.
Results: The MICs of levofloxacin and telithromycin were 8–16 and 16–32 mg/L, respectively. According to PCR results these isolates had ermB, but didn't have mefA, mefE, ermTR. The DNA sequence of 23S rRNA, rplD and rplV of these resistant isolates were the same as the susceptible strain. The ermB of resistant isolates had an amino acid change (Asn100Ser), and 1 or 2 mutations in upstream base sequence. PFGE technique using SmaI digested DNA of resistants was performed against 7 isolates. The results revealed the 7 resistant isolates were the same clone.
Conclusions: We isolated 11 telithromycin and fluoroquinolone high resistant S. pneumoniae. The mechanisms of resistance to telithromycin were considered mutated ermB with changed upstream base sequence. It was considered that the 11 resistant isolates were spread by nosocomial infection. We must minimise the spread of such resistant S. pneumoniae.
P735 Prevalence and antimicrobial susceptibility profiles of leading nosocomial pneumonia pathogens: the ten-year report from the European SENTRY Antimicrobial Surveillance Program
T. Fritsche, H. Sader, P. Strabala, R. Jones (North Liberty, US)
Objectives: To present a 10-year summary of bacterial pathogens (prevalence and antimicrobial susceptibility [S] trends; SENTRY Antimicrobial Surveillance Program) recovered from European patients hospitalised with pneumonia (HAP). The emergence of resistance (R) among pathogens responsible for HAP is changing approaches to empiric therapy, with increasing dependence on carbapenems (CARB), fluoroquinolones (FQ), and β-lactamase inhibitor combinations.
Methods: Non-duplicate, clinically-significant pneumonia isolates (10,780) were collected from 25 medical centres in Europe participating in the SENTRY Program from 1997–2006. Identifications were confirmed by the central monitoring laboratory and all isolates were S tested using CLSI methods and interpretive criteria (M100-S17) against commonly used antimicrobial agents used for the empiric or directed therapy of HAP.
Results: Ranking European HAP pathogens between the years 1997–2006 included S. aureus (SA) > P. aeruginosa (PSA) > Klebsiella spp. (KSP) > E. coli (EC) > Enterobacter (ESP) > Acinetobacter (ASP) > Serratia spp. > S. maltophilia. MRSA rates have remained essentially unchanged during the study, although dramatic differences were noted between nations. R-emergence is most notable among Gram-negative bacilli, especially ASP, where R increases have been seen in all sampling periods for CARB, cephalosporins, FQ and aminoglycosides. Modest increases in R have also been detected with PSA (imipenem [IPM], FQ), KSP (ceftazidime [CAZ], FQ, amikacin [AMK]), EC (FQ), and ESP (CAZ, FQ). ESBL-phenotype rates for KSP have more than doubled (>27%) since the start of the Program and are also of concern among EC (9.7%). IPM-R isolates are sporadically detected among KSP and ESP, usually due to the presence of metallo-carbapenemases (VIM-1; Italy, Greece and Turkey).
| Organism (R pattern) | % Inhibited at CLSI Breakpoints |
||
|---|---|---|---|
| 1997–1999 | 2000–2002 | 2004–2006 | |
| S. aureus (SA; 2,456) | |||
| Methicillin-R (MRSA) | 37.3 | 39.6 | 38.3 |
| P. aeruginosa (PSA; 2,377) | |||
| IPM-NSa | 23.6 | 25.1 | 29.3 |
| CAZ-R | 15.2 | 21.1 | 19.3 |
| LEV-R | 21.2 | 25.1 | 28.6 |
| AMK-R | 8.7 | 8.2 | 7.5 |
| Klebsiella spp. (KSP; 956) | |||
| IPM-NSa | 0.0 | 0.0 | 0.4 |
| CAZ-R | 10.2 (13.6)b | 12.4 (19.4)b | 17.0 (27.1)b |
| LEV-R | 2.4 | 2.3 | 12.6 |
| AMK-R | 1.0 | 2.6 | 5.4 |
| E. coli (EC; 847) | |||
| CAZ-R | 1.7 (6.8) | 3.1 (8.2) | 3.6 (9.7) |
| LEV-R | 4.0 | 7.4 | 17.9 |
| Enterobacter spp. (ESP; 748) | |||
| IPM-NSa | 0.5 | 0.3 | 1.1 |
| CAZ-R | 21.9 | 24.5 | 30.7 |
| LEV-R | 7.1 | 7.8 | 17.0 |
| Acinetobacter (ASP; 568) | |||
| IMP-NSa | 33.3 | 20.9 | 47.5 |
| CAZ-R | 37.9 | 59.6 | 69.1 |
| LEV-R | 45.2 | 51.7 | 68.3 |
| AMK-R | 45.9 | 53.1 | 62.2 |
NS, non-susceptible
Number in parentheses reflects the ESBL-phenotype rate (MIC values ≥2 mg/L).
Conclusions: The SENTRY Program has documented emerging HAP pathogens and changing susceptibility profiles within European medical canters for 10 years. During this time, dramatic changes have been noted with declining S among widely used classes including the cephalosporins (Enterobacteriaceae [ENT]), CARB (PSA), and FQ (ENT, PSA and ASP); marked declines in S to all tested agents were noted with ASP. Changing patient demographics, antimicrobial usage and recognition of R genotypes with highly mobile genetic elements (class 1 integrons) within hospital environments have altered antibiograms, resulting in continued R emergence among HAP pathogens.
P736 In vitro synergism between rokitamycin and cotrimoxazole against S. pyogenes and S. pneumoniae
S. Roveta, A. Marchese, E.A. Debbia, R. Bandettini (Genoa, IT)
Objectives: Using standard susceptibility tests (CLSI, 2005), synergism between cotrimoxazole (SXT) and rokitamycin (ROK) was observed on S. pyogenes and S. pneumoniae. The aim of this study was to confirm this phenomenon on a large number of isolates, displaying different macrolide resistance phenotypes, employing time-kill tests.
Methods: Synergism between SXT and ROK on 100 S. pyogenes and 100 S. pneumoniae recently isolated was detected by a double-disk screening test. Time kill experiments were performed on representative strains adopting standard procedures (CLSI 2005).
Results: The combination of SXT plus ROK reacted synergistically against 93% S. pyogenes strains and 51% S. pneumoniae strains. On pneumococci SXT-S this percentage arise to 64%, while, on SXT-R it was 29%. In no instances antagonism was demonstrated. Synergism was not observed against S. pyogenes strains showing cMLSB phenotype. In S. pneumoniae no relationship between different mechanisms of macrolide resistance and the results of interactions was found. Results of time-kill experiments confirmed those obtained with double-disk assay in all the strains tested.
Conclusion: Synergism between SXT and ROK was more frequently encountered among S. pyogenes than S. pneumoniae strains. Different macrolide-resistance mechanisms (reduced binding due to modification of the 50S subunit or efflux pump) among the various bacteria may eplain the observed differences
P737 Current susceptibility patterns for Streptococcus pneumoniae and Haemophilus influenzae isolates from Europe: findings of the 2005–2006 GLOBAL Surveillance Program
M. Jones, N. Brown, D. Draghi, C. Thornsberry, D. Sahm (Breda, NL; Herndon, US)
Objective: Multi-drug resistant (MDR) has become problematic for clinicians treating infections caused by S. pneumoniae (SP). Monitoring susceptibility patterns along with geographic trends for MDR prevalence can be useful. H. influenzae (HI) commonly associated with community-acquired respiratory tract infections can become resistant (R) to commonly prescribed agents and these resistance rates can vary according to regional distributions. The GLOBAL Surveillance initiative was undertaken to track resistance patterns among common respiratory pathogens.
Methods: During ‘05-'06, 1542 SP and 1579 HI were collected from 5 countries in Europe (EU; France [FR], Germany [GE], Italy [IT], Spain [SP], and the United Kingdom [UK]. All isolates were centrally tested by broth microdilution (CLSI M7-A6, 2003) and results were interpreted according to CLSI M100-S15, 2005. For SP, MDR was defined as R to ≥2 agents of the following agents: penicillin (PEN), cefuroxime (CFX), azithromycin (AZI), and trimethoprim-sulfamethoxazole (SXT).
Results: For all countries combined the prevalence of MDR SP was 18.7%; the most common MDR phenotype was R to PEN, AZI, CFX, and SXT. LFX-R was not commonly associated with MDR; 98.6% of MDR were LFX-susceptible (S). By country, the MDR rates (%) were as follows: 33.2 FR, 28.5 SP, 17.6 GE, 13.4 IT, and 4.3 UK. In each of those countries, LFX S rates among MDR SP remained high ranging from 96.6% (GE) to 100% (IT and UK). Overall, there were 61% of isolate pan-S; 20.4% 1-drug R; 5.3% 2-drug R; 7.3% 3-drug R; and 6% 4-drug R. For all countries combined 219 HI isolates were β-lactamase (BL) positive (13.9%) and 1360 isolates were BL-negative (86.1%). Overall MIC90s (mg/L), were >8 for ampicillin (AMP), 0.015 for LFX, and >4 for SXT. By country LFX retained potent activity against HI, based on MIC90s (0.015 mg/L for all) and %S (100% for all). Only 1 isolate was encountered that was BL-negative AMP-resistant (BLNAR), which came from Italy.
Conclusions: The MDR rates among SP varied by country studied; however, most frequently involved resistance to three classes of agents including β-lactams, macrolides, and SXT. Susceptibility to LFX remains high among SP. Among HI, resistance to SXT and AMP (β-lactamasemediated) is prevalent throughout EU. In contrast, current susceptibility to LFX remains high throughout EU. Continued surveillance is warranted to monitor any changes that may occur among the bacterial landscape.
P738 Respiratory tract isolates of Streptococcus pneumoniae – susceptibility to antimicrobial agents in Slovenia, 2002 and 2004
I. Štrumbelj, T. Oražem, T. Franko-Kancler, V. Božanic, I. Grmek Košnik, M. Kavcic, L. Sarjanovic, T. Harlander (Murska Sobota, Ljubljana, Maribor, Celje, Kranj, Koper, Nova Gorica, Novo Mesto, SI)
Objectives: To determine and compare rates of susceptibility to selected antimicrobial agents in respiratory tract isolates of Streptococcus pneumoniae in Slovenia in 2002 and 2004.
Methods: Non-duplicated isolates of Streptococcus pneumoniae from respiratory tract from 2002 and 2004 were identified by standard methods in eight Slovenian laboratories. Oxacillin zones and susceptibility to seven non-betalactam antimicrobials were determined by NCCLS disc-diffusion procedure. Isolates with oxacillin inhibition zone less than 20mm were tested by Etest: MICs for penicillin and cefotaxime were interpreted according to NCCLS criteria for isolates from respiratory tract. Results from 2002 and 2004 were compared (chi-square test, statistical significance: p < 0.05).
Results: Number of isolates in 2002 and 2004 was 850 and 976, respectively. All isolates were susceptible to vancomycin. Rates of susceptibility in years 2002 and 2004, respectively, were: to penicillin 81% and 76%, to cefotaxime 99% and 100%, to erythromycin 87% and 80%, to clindamycin 90% and 84%, to trimethoprim-sulfamethoxazole 70% and 63%, to tetracycline 85% and 82%, to levofloxacin and moxifloxacin 99.8% and 99.8%. Differences between 2002 and 2004 were statistically significant for penicillin, erythromycin, clindamycin, trimethoprim-sulfamethoxazole and tetracycline.
Conclusion: Susceptibility to five antimicrobial agents decreased from 2002 to 2004 in Slovenian S. pneumoniae respiratory tract isolates. Surveillance and actions to decrease resistance are necessary.
P739 Reversibility of antimicrobial resistance in respiratory isolates in HIV-positive Cambodian children after 36 months of HAART
E. Kalavsky, P. Beno, A. Shahum, J. Benca, L. Seng Duong, A. Augustinova, Z. Havlikova, A. Liskova, V. Krcmery (Bratislava, Trnava, SK; Phnom Penh, KH)
Objective: Aim of this study was to assess, if restauration of the immune system after 36 months treatment with HAART in cambodian children has an impact on antibiotic resistance and its reversibility.
Methods: Study participants were HIV positive cambodian children treated with HAART stavudine, lamivudine and nevirapine or efavirenz. Respiratory tract isolates (nose, pharyngeal, ear svabs) from 32 cambodian previously ART naive children 3–11 years old were assessed every 3 months within 36 months of HAART.
Results: Analysing relationship between duration of HAART, and colonisation with any specific resistance pattern, MRSA appeared to emerge after 6–12 months of HAART in comparison to pre-HAART period (was 90–93% after 9–12 months vs 50% in HAART naive, P < 0.01). Presence of multiresistant Klebsiella, and Enterobacter spp. was high already at baseline and in first months of HAART and the proportion of multiresistant Gram-negative bacteria (MR GNB) decreased later to 0 and 20% (P < 0.02). Susceptibility of both Gram-negative and Gram-positive bacteria showed biphasic but increasing tendency. Proportion of MR GNB decreased from 21/23 (90%) in the first 6 months of HAART, to 0–11% in those receiving HAART for 15–18 months and to 20–50% after 33 months of HAART. Reversibility of MR in GNB took 15–18 months. However, the baseline of resistance in GNB were relatively high. Proportion of MRSA increased from 50–55% in first 6 months to 93–85.7% after 9–18 months but than decreased to 20–33% after 36 months of HAART. Emergence of MRSA was slower. Reversibility of MR in Staphylococcus aureus was longer and took approximately 24–30 months. Ratio of Gram-positive to Gram-negative decreased from 1:3.9 (HAART naive) to 1:1.1 (30–36 month of HAART).
Conclusion: Reversibility of resistance among isolates from respiratory system was probably due to the reconstitution of their immune system due to the HAART and therefore less exposition with therapeutic ATB. In MRSA, the reversibility of resistance took 15–21 months and was slower than in MR Gram-negative bacilli (Klebsiella, Enterobacter) where the increase of susceptibility (and the decrease of resistance) took 9–12 months. Prophylactic administration of cotrimoxazol 3x weekly did not affect the reversibility of resistance and seems to be less promotive for antibiotic resistance.
P740 Changes in the catchment population – changes in the rate of penicillin-resistant Streptococcus pneumoniae strains?
E. Hajdu, M. Matuz, R. Benko, A. Ordas, E. Nagy (Szeged, HU)
Objectives: The catchment population of our laboratory has changed during the years. We analysed whether this change had any impact on penicillin resistance rates of S. pneumoniae strains.
Methods: We included in our analysis the S. pneumoniae isolates if the symptoms or the source of the isolate proved of its clinical relevance. According to the changes in the catchment population (between 1998 and 2001 we provided our service exclusively for the in- and outpatient departments of the university hospital while from the year 2002 increasing number of general practitioners sent samples to our laboratory) we set up two time periods: Period I (1998–2001) and Period II (2002–2005). Isolation and identification were carried out by conventional methods.
All resistance data were collected from our laboratory database system. The same methodology was used to detect penicillin resistance throughout the study. The susceptibility of isolates was determined by using the breakpoints recommended in the NCCLS/CLSI guidelines. MIC determination was carried out by using the E test. Repeat isolates from individual patients were excluded. Differences in the distribution of resistance patterns between the two time periods were analysed by chi-square test and Fisher's exact test, using the SPSS programme package (version 13.0). A P value <0.05 was considered to be statistically significant.
Results: Our results are summarised in the table .
| Age group |
S. pneumoniae isolates |
P value | |||||
|---|---|---|---|---|---|---|---|
| Total number |
Number of penicillin-resistant isolates |
||||||
| HR* |
LR** |
||||||
| Period I | Period II | Period I | Period II | Period I | Period II | ||
| In-patients | |||||||
| 0–2 | 158 | 335 | 32 (20) | 13 (4) | 58 (37) | 180 (54) | <0.001 |
| 3–14 | 193 | 297 | 26 (13) | 7 (2) | 68 (35) | 129 (43) | <0.001 |
| 15–65 | 85 | 103 | 7 (8) | 3 (3) | 22 (26) | 28 (27) | 0.270 |
| >65 | 41 | 55 | 4 (10) | 1 (2) | 12 (29) | 11 (20) | 0.097 |
| All | 477 | 790 | 69 (14) | 24 (3) | 160 (34) | 348 (44) | <0.001 |
| Outpatients | |||||||
| 0–2 | 115 | 373 | 24 (21) | 9 (2) | 36 (31) | 208 (56) | <0.001 |
| 3–14 | 181 | 545 | 12 (7) | 9 (2) | 55 (30) | 297 (53) | 0.001 |
| 15–65 | 84 | 92 | 1 (1) | 1 (1) | 16 (19) | 25 (27) | 0.444 |
| >65 | 4 | 9 | 0 (0) | 0 (0) | 0 (0) | 2 (0) | 0.304 |
| All | 384 | 1019 | 37 (10) | 19 (2) | 107 (28) | 522 (51) | <0.001 |
HR: high level of resistance
LR: low level of resistance including resistant (R) and intermediate resistant (IR) strains.
Conclusion: Contradictory publications exist about the rate of penicillin resistance among S. pneumoniae isolates in Hungary [1, 2]. In our study the rate of penicillin resistance was found to be low and further decreasing was observed with the change of the catchment population. The importance of taking into account the used methodology for detecting resistance and the origin of the isolates (inpatient vs. outpatient) is highlighted by our study.
P741 Prevalence and mechanisms of Streptococcus pyogenes resistance to macrolides in Moscow, Russian Federation
S.A. Grudinina, O.Y. Filimonova, E.N. Ilina, S.V. Sidorenko (Moscow, RU)
Objectives: Streptococcus pyogenes (Spy) is a leading cause of bacterial pharyngitis in humans it is also implicated in some life-threatening infections in last years. Although Spy isolates are fully susceptible penicillin G, macrolides are recommended for some groups of patients. However Spy resistance to macrolides due to ribosomal methylation or efflux is increasing. The objective of the study was to determine the prevalence and mechanisms of resistance among Spy to macrolides in Moscow region.
Methods: Clinically significant Spy isolates were prospectively collected in Moscow region from adult and paediatric patients with upper respiratory tract and skin and soft tissues infections during 2002–2005. The species identity and purity of all isolates was confirmed in central laboratory. MICs of erythromycin, clindamycin and telithromycin were determined using a broth microdilution method as recommended by CLSI. The erm and mef genes were amplified using specific primers. Discrimination of mef genes subclasses was carried out in thermocyclic primer extension reaction, followed by mass-spectrometric detection of the products. Sequencing of selected mef genes was performed using ABI Prism 3100 Genetic Analyzer (Applied Biosystems, USA).
Results: Trends in resistance prevalence (% of nonsusceptible isolates) among Spy during the study period are presented in the Table .
| Antibiotic | Year (number of isolates) |
|||
|---|---|---|---|---|
| 2002 (n = 194) | 2003 (n = 175) | 2004 (n = 189) | 2005 (n = 158) | |
| Erythromycin | 6.7% | 4.6% | 7.4% | 11.4% |
| Clindamycin | 0.5% | 0% | 1.1% | 1.3% |
| Telithromycin | 0 | 0 | 0 | 0 |
Thirty erythromycin-nonsusceptible isolates were available for molecular studies. ermA genes were detected in three isolates, and ermB in one. All these isolates demonstrated MLS phenotype. Thirteen isolates demonstrating M phenotype were mef positive in PCR with specific primers. All mef genes were recognized as mef(I) subclass in primer extension reaction, and in 10 isolates presence of mef genes was confirmed by sequencing. No resistant determinants were detected in 13 isolates, demonstrating M phenotype.
Conclusions: Prevalence of macrolide resistance among Spy isolates in Russia is similar to other regions of Central and Eastern Europe. Efflux mediated by mef genes which belong to recently described mef(I) subclass is predominant mechanism of resistance.
P742 Microbial and antimicrobial susceptibility patterns from patients with chronic otitis media in Ardebil, Iran
G.H. Ettehad, S. Refahi, A. Nemmati, A. Pirzadeh, Z. Tazakori (Ardebil, IR)
Objective of this study is to identify the commonest microorganisms associated with chronic suppurative otitis media (CSOM) and their antimicrobial sensitivities. This study was carried out from 2003 – 2004 at the Department of ear and nose and throat of Ardebil University of medical sciences. Sixty one patients with chronic suppurative otitis media were prospectively studied. They had chronic ear discharge and had not received antibiotics for the previous five days. Also they had no cholesteatoma. Swabs were taken, and cultured for bacteria. Bacteriological specimens were processed and identified with standard cultures. Antimicrobial susceptibility of these bacterial isolates was assessed by an agar disc diffusion method. Isolates were tested against 10 antibiotics: The most frequently isolated organism in chronic suppurative otitis media was Staphylococcus aureus 19 (31.15%), followed by Pseudomonas aeruginosa 16 (26.23%) and Proteus sp. 12 (19.67). Fungi accounted for 4 (6.56%) of the isolates. Sensitivity results showed majority of isolates were susceptible to Ciprofloxacin (85.71%), and resistant to Penicillin (84.97%). In conclusion, the in vitro susceptibility results indicate that Ciprofloxacin can be an effective antibiotic in the treatment of active chronic suppurative otitis media.
P743 Antimicrobial resistance of Streptococcus pneumoniae strains to penicillin and ceftriaxone, isolated in the Niš district, Romania during 1999–2006
S. Mladenovic-Antic, B. Kocic, P. Stojanovic, S. Ivic, V. Mladenovic (Nis, RS)
Streptococcus pneumoniae has shown an increase in resistance to anitimicrobial drugs which significantly hinders therapy.
Objectives: To determine the level of resistance to penicillin and ceftriaxon, and to study it in relation to its resistance in other European countries.
Methods: In the period between January 1999-December 2003 and January 2005-September 2006, 523 isolates of S. pneumoniae of various origins were studied. They were identified by means of morphological, cultural and antigen characteristics, and tested by the agar-dilution method, to indicate their response to penicillin and ceftriaxon according to the recommendations of the Clinical Laboratory Standards Institute.
Results: In the first period, from the 320 S. pneumoniae obtained from various clinical material, the greatest percentage was found in the population of children under the age of 15 (84, 07%). The nose smear (81, 4%) and aspirate (7, 51%) were the most frequent. Testing their sensitivity gave the following results: 68, 2% of the isolates showed a reduced sensitivity to penicillin (21, 3% were resistant and 46, 9% intermediate), and 19, 4% showed a reduced sensitivity to ceftriaxon – I (9, 4%) and R (10%). In the second period of investigation, 203 isolates of S. pneumoniae of various origins were studied. The greatest percentage was found in the population of children under the age of 15 (68, 9%). The nose smear (68, 9%) and aspirate (12, 3%) were the most frequent. Testing their sensitivity gave the following results: 79, 4% of the isolates showed a reduced sensitivity to penicillin (27% resistant and 52,4% intermediate). Among this isolates, 6, 9% had MIC ≥ 4 μg/mL – HLR isolates. 18, 67% showed a reduced sensitivity to ceftriaxon – I (10, 3%) and R (8, 37, 0%).
Conclusion: The level of resistance and percentage of highly resistant isolates (MIC ≥4 μg/mL) 6, 9% rank us among those European countries with the highest rate of resistance to penicillin. The level of resistance to penicillin is the highest among the isolates obtained from children under the age of 15 (28%). In the case of ceftriaxon, the level of resistance is highest among hospital isolates (8%), while the percentage of isolates which are resistant (8, 37%) and intermediary (10, 3%) to ceftriaxon. The level of resistance to penicillin is significantly higher among the isolates from the second period of investigation than among the isolates obtained from children, p ≤ 0.001).
P744 Changes in United States regional variations in penicillin-resistant rates of Streptococcus pneumoniae, 1999 to 2006
S. Bouchillon, B. Johnson, R. Badal, M. Hackel, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: The percentage rates of penicillin-resistant (PenR) S. pneumoniae (SPN) vary by country and region. Earlier studies have documented US regional variations in PenR SPN. The purpose of this study was to determine changes in regional variations, if any, of PenR and PenNS strains of SPN, and the current activity of tigecycline (TIG), amoxicillin-clavulanic acid (AC), ceftriaxone (CFX), levofloxacin (LEV), linezolid (LNZ), and vancomycin (VAN) to PenR isolates.
Methods: 2,071 clinically relevant isolates of SPN were collected from patients in 172 hospitals from 2004–2006. MIC's to all agents tested were determined by broth microdilution and interpreted following CLSI guidelines. Regions are defined by the CDC.
Results: PenNS rate was 42.2% for all regions varying from a high of 62.5% (East South Central) to a low of 31.2% (Pacific). PenR decreased in all regions but one (New England) with a corresponding increase in PenI rates in all regions. Regional changes from 1999–2000 to 2004–2006 are noted. Tigecycline and vancomycin had the lowest MIC90s (μg/mL) against PenR SPN at 0.25 and 0.5, respectively, followed by LEV and LNZ at 1, and CFX at 2.
| Regions | Pen I+R (%) |
Net Gain/(Loss) (%) | |
|---|---|---|---|
| 1999–2000 n = 1948/4751 | 2004–2006 n = 873/2071 | ||
| All regions | 41.0 | 42.2 | 1.2 |
| East North Central | 38.7 | 41.7 | 3.0 |
| East South Central | 53.3 | 62.5 | 9.2 |
| Middle Atlantic | 36.9 | 37.6 | 0.7 |
| Mountain | 41.1 | 37.1 | (4.0) |
| New England | 26.1 | 35.1 | 9.0 |
| Pacific | 34.6 | 31.2 | (3.4) |
| South Atlantic | 47.9 | 45.3 | (2.6) |
| West North Central | 37.1 | 45.3 | 8.2 |
| West South Central | 47.5 | 45.1 | (2.4) |
Conclusions: PenNS for SPN has increased slightly since 1999, with large increases in levels of PenI in almost all regions, but significant decreases in PenR levels. VAN, LNZ, LEV and TIG MIC90 values remain unaffected by pen phenotypes.
P745 Antimicrobial susceptibility among clinical isolates of Haemophilus influenzae in a tertiary hospital in Greece
D. Kofteridis, G. Notas, S. Maraki, G. Samonis (Heraklion, GR)
Objectives: Haemophilus influenzae (HI) is an important human pathogen as well as a commensal of the respiratory tract of children and adults. The increasing antimicrobial resistance of this organism is a matter of concern. The antimicrobial resistance of HI strains isolated from patients of the University Hospital of Heraklion, Crete, Greece during a 10-year period was investigated.
Materials and Methods: A total of 930 clinical strains of HI were isolated from various clinical specimens, between January 1996 and December 2005. The strains were characterised according to the production of β-lactamase, and their in vitro susceptibilities to 24 antimicrobial agents on the basis of the current Clinical and Laboratory Standards Institute guidelines.
Results: Overall, 9.5% of the isolates were producing β-lactamase. An increase in penicillin resistance from 31% in 1996 to 77% in 2005 was observed with an overall resistance of 50%. No increase in amoxicillin and amoxicillin-clavulanate resistance was observed with overall resistance of 11% and 0.6%, respectively. A high percentage of β-lactamase producing strains were isolated from ophthalmic specimens (36.5%), while most penicillin resistant ones from ear, ophthalmic and bronchoalveolar lavage (71, 77 and 64%, respectively). Some uncommon strains such as 2 β-lactamase negative-ampicillin resistant (BLNAR), and 4 β-lactamase positive-amoxicillin-clavulanate resistant (BLPACR) were identified. BLNAR represented 0.22% and BLPACR 0.44% of all studied isolates. A significant increase in tetracycline resistance from 1.6% in 1996 to 38% in 2005 was observed, being most prominent during the last years. Resistance to erythromycin was 99% and remained steady and high during all the study period. Newer macrolides were not included in the investigation. Resistance to quinolones was extremely low during all years (0.8% for ofloxacin, and 0.2% for ciprofloxacin). Resistance to other non-β-lactam agents varied from 37.5% for trimethoprim-sulfamethoxazole to 2.6% for rifampin and 0.4% for chloramphenicol.
Conclusions: An increase in penicillin resistance during the study period was observed among HI isolates, as opposed to amoxicillin and amoxicillin-clavulanate resistance that remained unchanged. Among non β-lactams increased resistance was observed to tetracycline and trimethoprim-sulfamethoxazole.
P746 Bacteriological efficacy of azithromycin on non-typeable Haemophilus influenzae adhered to and entered cultured human epithelial cells
N. Yamanaka, M. Hotomi, K. Fujihara, S. Tamura, T. Tomita, S. Saito (Wakayama, JP)
Objectives: Nontypeable Haemophilus influenzae (NTHi) is a respiratory tract pathogen that is not traditionally regarded as an intracellular bacterium. However, NTHi was shown to reside and replicate intracellulary in human macrophage-like cells found subepithelially in human adenoid tissue. The possibility that NTHi might be shielded from the local immune response and antibiotics by entering macrophages and epithelial cells may explain the persistence of NTHi in prolonged otitis media, sinusitis and bronchitis. The aim of the present study was to study bacteriological efficacy of azithromycin on NTHi adhered to and internalised by cultured human epithelial cells.
Methods: Clinical isolates of NTHi and the human carcinoma epithelial cell lines, Hep2 and Detroit562, were used in this study. Bacteriological efficacies of azithromycin(AZM) and ceftriaxon(CTRX) were studied in adherence and internalisation assay. In adherence assay for NTHi, after separating the extracellular nonadhered bacteria in supernatant, 1MBC of AZM and CTRX were added to the well with NTHi adhered to Hep2 cells. The number of adhered and intracellular NTHi was determined the following day. To examin internalisation and penetration activity of NTHi, we used 24 well multiplate with transwell polyester membrane filters cultured with Detroit562 cells. Bacterial suspension of NTHi was added to monolayer cells pre-treated with various concentrations of AZM, followed by overnight incubation, and internalised bacteria after the mechanical lysis and penetrated bacteria in the lower chamber were counted.
Results: CTRX did not show bacteriocidal effect on adhered and intracellular NTHi even after 12 h incubation. On the other hand, AZM showed marked bacteriocidal effect on adhered and intracellular NTHi as well as extracellular bacteria after 4 h incubation. AZM higher than 10 ug/mL incubated with monolayer Detroid562 cells showed marked bacteriocidal effects on both internalised and penetrated NTHi and no viable bacterium was determined.
Conclusion: In the present study, we observed a wide spectrum in the level of NTHi adherence, internalisation and penetration. AZM showed marked bacteriological efficacy on NTHi adhered to, internalised by, and penetrated with cultured human epithelial cells, in contrast with CTRX which was bacteriocidal only to extracellular NTHi. These data suggest that AZM may be of high therapeutic significance on prolonged respiratory tract infections due to NTHi.
P747 Comparative minimum inhibitory concentration and mutant prevention concentration of azithromycin, cefuroxime, gemifloxacin, moxifloxacin and telithromycin against clinical isolates of Haemophilus influenzae
J. Blondeau, S. Borsos (Saskatoon, CA)
Objective: Haemophilus influenzae (HI) is an important and prevalent respiratory pathogen; it is the most prevalent bacterial cause of acute exacerbations of chronic bronchitis (AECB). Beta-lactam, macrolide and quinolone compounds are used for AECB therapy, however, little is known about the resistance selection potential of these compounds with HI. We tested azithromycin (AZI), cefuroxime (CFX), gemifloxacin (GEM), moxifloxacin (MXF) and telithromycin (TEL) by minimum inhibitory concentration (MIC) and mutant prevention concentration (MPC) against clinical isolates of HI.
Methods: For MIC testing, the recommended procedure of the Clinical and Laboratory Standards Institute (CLSI) were followed using 10 CFU/mL of organism in brain heart infusion broth and 5% Fildes exposed to doubling-drug concentrations and incubation in optimal atmosphere and temperature. For MPC testing, ≥109 CFU were exposed to doubling-drug concentrations on Haemophilus test medium agar plates and incubated for 24–48 h. The concentration preventing growth was either the MIC or MPC depending on the method used.
Results: MIC and MPC data for the antimicrobials vs HI (n = 42–45) are shown in the Table . For GEM and MXF, MIC and MPC values for all strains were below the susceptibility breakpoint as compared to 100% and 0% for AZI, 100% and 0% for CFX and 96.3% and 20% for TEL.
| MIC (mg/L) |
MPC (mg/L) |
|||||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MPC50 | MPC90 | Range | |
| AZI | 1 | 2 | 0.5–2 | 16 | 32 | 8–64 |
| CFX | 0.5 | 0.5 | 0.25–2 | >16 | >16 | >16 |
| GEM | 0.004 | 0.008 | 0.002–0.016 | 0.063 | 0.125 | 0.016-≥0.125 |
| MXF | 0.016 | 0.031 | 0.008–0.031 | 0.125 | 0.25 | ≤0.031–0.5 |
| TEL | 2 | 2 | 1–8 | 8 | 16 | 2–32 |
Conclusions: MPC values were low for GEM and MXF and below susceptibility breakpoints for 100% of isolates tested suggesting a low propensity of these compounds to select for resistance from high density inocula. MPC values for AZI and CFX were elevated beyond susceptibility breakpoints, however, the mechanism of this observation remains unknown. For TEL, MPC values were above the susceptibility breakpoint for 80% of isolates tested.
Epidemiology and resistance of streptococci
P748 EARSS results: S. pneumoniae resistance related to serogroups?
M.E.A. de Kraker, G. Kahlmeter, N. van de Sande-Bruinsma, E.W. Tiemersma, J.C.M. Monen, H. Grundmann and EARSS participants
Objective: Since three years the European Antimicrobial Resistance Surveillance System (EARSS) has, next to resistance data, been collecting serotype information for S. pneumoniae isolates (SPN). We analysed the relationship between penicillin and erythromycin resistance and serotype among SPN isolates.
Methods: Since 1999, EARSS collects routine antimicrobial susceptibility test data amongst others from invasive SPN isolates. According to standard protocols, macrolide (ery) and β-lactam (pen) susceptibility are reported. Serotype data is reported according to the Danish Kauffman-Lund scheme. We chose two countries that represented a low and a high endemic situation and that reported serotype information for most SPN isolates in the database. We compared resistance rates among the most common serogroups between the high and low endemic country by chi-square test.
Results: The total number of SPN isolates with serogroup information over those tested for both antimicobial groups in 2005 was 1536/1539 for the high endemic country and 1182/1373 for the low endemic country. Pen resistance was reported 4% for the low endemic country and 12% for the high endemic country, ery resistance 11% versus 31% and dual resistance 1 versus 9%. The six most common serogroups in both countries were 1, 6, 9, 14, 19 and 23. Ery resistance was higher in the high endemic country for all six serogroups (p < 0.01), pen resistance was significantly higher for serogroups 14, 19, 23 compared to the low endemic country.
Conclusion: Irrespective of serogroup, pen and ery resistance were higher for the high endemic country. This indicates that resistance in SPN is not based on the expansion of particular clones, but rather influenced by antimicrobial consumption.
P749 Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates from ocular infections
S. Maraki, E. Nioti, A. Georgiladakis, Z. Gitti, I. Neonakis, M. Katrinaki, Y. Tselentis (Heraklion, Crete, GR)
Objective: To determine the antimicrobial susceptibilities and serotypes of Streptococcus pneumoniae recovered from ocular and periocular infections from 1997 through 2006 in the University Hospital of Crete, Greece.
Material and Methods: A total of ninety Streptococcus pneumoniae isolates were studied. Pneumococci were identified by using standard techniques, including Gram stain characteristics, colonial morphology, optochin susceptibility, and bile solubility. Susceptibility tests were done by the E-test method according to manufacturer's recommendations and were interpreted according to the CLSI guidelines. The following antibiotics were tested: penicillin G, cefuroxime, cefotaxime, ceftriaxone, cefepime, imipenem, meropenem, erythromycin, clarithromycin, clindamycin, ciprofloxacin, levofloxacin, sparfloxacin, moxifloxacin, chloramphenicol, tetracycline, trimethoprim/sulfamethoxazole, and vancomycin. Serotyping was performed by the capsular swelling method with specific antisera.
Results: Fourteen isolates (15.5%) showed intermediate resistance and 7 (7.8%) high-level resistance to penicillin. Erythromycin, clarithromycin, clindamycin, chloramphenicol, tetracycline, and trimethoprim/sulfamethoxazole resistance rates were 32.2, 32.2, 14.4, 1.1, 25.6 and 15.6%, respectively. All isolates were sensitive to vancomycin and to all 4 quinolones tested. The most prevalent serotype was 19, followed by 6, 9 and 14.
Conclusion: The increased resistance rates of S. pneumoniae to penicillin, macrolides and other antibiotics indicate the importance of performing antimicrobial susceptibility testing in order to determine the appropriate therapy. Fluoroquinolones are highly active in vitro against ocular pneumococcal isolates including penicillin resistant strains and may offer enhanced coverage for these organisms.
P750 Regional differences in pneumococcal serotype distribution in Germany
I. Seegmüller, M. van der Linden, C. Heeg, R. Reinert (Aachen, DE)
Objectives: The German National Reference Centre for Streptococci collects S. pneumoniae isolates from allover the country. Together with the isolates we receive epidemiological data concerning the residence of the patient, the federal state the patient lives in and the federal state of the centre which sent the isolate. Taking into account the longstanding coexistence of two german states we wanted to know whether this separation has a still lasting effect on serotype distribution. We wanted to know whether there is a difference in the serotype distribution between the eastern federal states and the western federal states and if so whether it changes with time.
Methods: Between 2000 and 2005 we received 3678 S. pneumoniae isolates of which we have information on serotype and the federal state of patients residence and/or on the federal state of the centre which sent the isolates.
First we determined the five most common serotypes of all samples sent. Afterwards we divided the German federal states into belonging to the western or eastern part of the country, resulting in 10 western states and 6 eastern states (including the city and state of Berlin). Thereafter we split the time period between 2000 and 2005 into two separate periods (2000–2002, 2003–2005) and calculated whether there was a difference in the relative amounts of the five most common serotypes changing with time.
Results: The five most common serotypes among the 3678 isolates taken into account are 14, 23F, 6B, 19F and 3 of which serotype 14 (21.6%) is by far the most common one. Between 2000–2002 (2003–2005) we received 1643 (2143) samples of which 184 (241) were sent from the eastern part of Germany and 1459 (1902) from the western part of Germany. The relative amount of serotype 14 was stable in western Germany (20.2% vs 20.1%) but changed dramatically in eastern parts of Germany (37.5% vs 24.9%).
Conclusions: Serotype distribution is different in eastern and western parts of Germany and it is dramatically changing.
P751 Epidemiology of invasive Streptococcus pneumoniae in Taiwan, 2001–2003
C-J. Chen, Y-C. Huang, L-H. Su, T-Y. Lin (Taoyuan, TW)
Objectives: To characterise the microbiological features of Streptococcus pneumoniae invasive isolates from a tertiary care hospital in Taiwan before the introduction of 7-valent pneumococcal conjugate vaccine (PCV).
Methods: Between January 2001 and December 2003, a total of 272 invasive isolates (115 from children and 157 from adults) were collected from the clinical microbiologic laboratory of Chang Gung Memorial Hospital. The susceptibilities to various antibiotics, the serotypes and pulsed field gel electrophoresis (PFGE) typing were determined in all isolates.
Results: Of 272 isolates, 12 serogroups (18 serotypes) and 78 PFGE types were identified. The most frequent serotypes were types 14 (30.1%), followed by 23F (20.6%), 6B (16.2), 19F (7.4%), 3 (5.9%) and 9V (4.0%). The serotype distributions were similar in children and adults, except for serotype 9V, which was identified exclusively in adult isolates. The genotypes of isolates were diverse and a variety of PFGE types were identified in isolates with same serotype. However, it was not uncommon to identify predominant clones of isolates in each of the six most common serotypes. The non-susceptible rate to penicillin was 73.9% and the incidence of high-level resistance (MIC ≥ 2 ug/mL) was 43.4%. The isolates also demonstrated high rate of resistance to erythromycin (91.2%), cefuroxime (55.5%) and ceftriaxone (29.0%). The isolates expressing PCV-serotypes comprised 95.0% of the penicillin-non-susceptible strains. When compared to the isolates from adults, the isolates from children had a significant higher rate of resistance to penicillin (80.9% vs. 68.8%, p = 0.025) and erythromycin (95.7% vs. 87.9%, p = 0.0259) but a lower resistant rate to ceftriaxone (21.7% vs. 37.4%, p = 0.0231). The coverage of 23-valent polysaccharide vaccine and 7-valent PCV of the serotypes was 91.5% and 83.0%, respectively, in children; and 85.7% and 78.6%, respectively, in adults.
Conclusion: In Taiwan, the invasive isolates of S. pneumoniae harboured high-level resistances to macrolide and β-lactams and exhibited distinct resistant profile between strains from children and adults. The current commercial pneumococcal vaccines had adequate coverage of the serotypes among the pneumococcal invasive diseases.
P752 5-year study of resistance, serotype and genetic diversity of resistant Streptococcus pneumoniae strains isolated in West Pomerania region of Poland
M. Nowosiad, S. Giedrys-Kalemba (Szczecin, PL)
Objectives: Streptococcus pneumoniae is responsible for the majority of upper respiratory tract infection incidents and some invasive diseases. Abuse of antibiotics in empirical ambulatory practice caused an abrupt increase in antibiotics resistance as well as clonal spread of resistant strains in recent years. This explains the importance of epidemiological studies and searching for new antipneumococcal agents of which vaccination seems to be the most effective. The aim of this study was to analyse resistance patterns, serotypes and genetic diversity of resistant S.p. strains isolated in our region during a 5-year period (2001–2005).
Methods: Using E-test method and the CLSI criteria for benzylpenicillin(P), erythromycin(E), clindamycin(L), tetracycline(T), cotrimoxazole(S), ceftriaxone(C), chloramphenicol(H), vankomycin(V), imipenem(I), 158 strains resistant or intermediate to at least one drug were obtained. Strains were serotyped with the reference antisera kit from the Statens Serum Insitut (Copenhagen). Molecular typing was performed using PFGE method with SmaI restriction enzyme and analysed with Molecular Analyst (BioRad) software. The resistance pattern, serotype and PFGE profile was determined for each strain.
Results: Resistance to 8 out of the 9 antibiotics (except vancomycin) has been observed and resistance to cotrimoxazole was the most frequent (86.7%). Strains showed 32 different resistance patterns. The dominant four: TSH (15.2% strains), S, ELTS, PSI concerned 48% of strains. Resistance degree reached 8 drugs (0.6% strains) and 53.8% of strains were MDR. Strains belonged to 13 serotypes and 11 of them are found in the 23-valent vaccine and cover 83.5% of strains. The most frequent serotype was 19F (22.2%). We have found 70 PFGE profiles: 44 unique and 26 clusters (A-Z) consisting of 2–30 strains with similarity exceeding 87%. The most numerous cluster I consists of 30 strains that were isolated over the 5 years of the study and showed the same or very similar resistant patterns: TSH (70%), SH, TH, T, H, S and serotype 19F.
Conclusions: Population of resistant S.p. strains in our region presents high genetic diversity and numerous different resistance patterns. They do not show many serotypes and majority of resistant strains are still covered by the 23-valent vaccine. The I cluster presents the most clonal spread and doesn't show similarity to any of the international clones.
P753 Antimicrobial resistance within Streptococcus pneumoniae isolates from eastern and southern Mediterranean countries
M.A. Borg, E.A. Scicluna, E.W. Tiemersma, M. de Kraker, N. van de Sande-Bruinsma, J. Monen, H. Grundmann and ARMed Project Collaborators
Objective: Streptococcus pneumoniae is a common cause of invasive disease, such as meningitis, sepsis and pneumonia. Treatment of these serious conditions is compromised by antimicrobial resistance which has been identified to be particularly prevalent some Northern Mediterranean countries, including France and Spain. Information about the prevalence of antimicrobial resistance in the countries of the southern and eastern Mediterranean has, however, been sparse. The Antibiotic Resistance Surveillance and Control in the Mediterranean Region (ARMed) project [www.slh.gov.mt/armed] provided a first time opportunity for a longitudinal multi-year study of trends of antimicrobial resistance amongst this species within countries of the southern and eastern Mediterranean.
Methods: ARMed used almost identical protocols to those adopted and validated by the European Antimicrobial Resistance Surveillance System (EARSS) and collected susceptibility test results from invasive isolates of S. pneumoniae routinely isolated from clinical samples of blood and cerebrospinal fluid in the participating laboratories situated in Algeria, Cyprus, Egypt, Jordan, Lebanon, Malta, Morocco, Tunisia and Turkey.
Results: In total, 1298 S. pneumoniae invasive isolates were reported to ARMed from 2003 to 2005. Overall 27% of these isolates were reported non-susceptible (intermediate or resistant) to penicillin (PNSP). In 2005, the lowest proportions of PNSP were found in Malta (15%, n = 13) and Morocco (17%, n = 42). The highest proportions of PNSP were reported from Algeria (44%, n = 71). In 2005, the lowest proportions of erythromycin non-susceptibility (ENSP) were found in Turkey (10%, n = 98) and Morocco (12%, n = 41). The highest proportions of ENSP were reported from Malta (46%, n = 13) and Tunisia (39%, n = 33). In 2005, the highest evidence of dual resistance to both penicillin and erythromycin was seen from Tunisia (24.2%, n = 33) with the lowest proportion reported from Egypt (2.5%, n = 121) and Malta (0%, n = 13). A significant increase was seen in Turkish laboratories (3% in 2003 to 10% in 2005).
Conclusion: ARMed countries showed a greater prevalence of penicillin resistance in S. pneumoniae than their northern European Mediterranean counterparts. However, with the exception of Tunisia, erythromycin susceptibility was greater and less instances of multiple resistance were identified in the southern and eastern Mediterranean centres.
P754 Analysis of in vitro Streptococcus pneumoniae non-susceptibility to several antimicrobial agents in a children's hospital in Turkey (1997–2005)
D. Gur, B. Guciz Dogan (Ankara, TR)
Objectives: The trends in resistance to nine antimicrobial agents in Streptococcus pneumoniae (SP) isolated in a nine-year period in a university hospital were evaluated.
Methods: A total number of 1406 clinical isolates of SP isolated between january 1997 and december 2005 were included for evaluation. Susceptibility tests were performed following the CLSI guidelines. Susceptibilities to penicillin (PEN) and cefotaxime (CTX) were determined with E-test and disk diffusion method was used for erythromycin (ERY), chloramphenicol (CHL), ofloxacin (OFX), levofloxacin (LEV), trimethoprim/sulfamethoxazole (T/S), tetracycline (TET) and vancomycin (VAN). Rates of resistance to antimicrobial agents were evaluated by year for each group of specimens (respiratory tract, CSF and blood). Annual mean MIC and inhibition zone diameter values were compared for each antimicrobial agent in each group of specimens.
Results: PEN non-susceptible isolates were 36.9% in 1997 and increased to 40.9% in 2005 (p = 0.052). Overall 2.4% of the isolates were highly and 30.0% were intermediately resistant. There was a significant increase in the mean MIC's of PEN over the years (p = 0.031). ERY resistance increased from 5.0% to 27.6% in (p = 0.000). Dual resistance to PEN+ERY increased from 5.0% to 20.5% (p = 0.000). Resistance to T/S increased from 51.9% to 62.6% and to TET increased from 8.0% to 25.7%. Overall two isolates were intermediately resistant to LEV and all were susceptible to VAN.
Conclusion: Resistance rates to PEN has not increased over the years, however, mean MIC's for penicillin and resistance rates for ERY, TET, CHL, T/S have increased significantly in nine years.
P755 Rapid variations in the macrolide resistance frequency, phenotypes and clones of group A streptococci from pharyngeal colonisation and infections, 2000 to 2006
D. Rolo, S. Custódio, A. Nunes, R. Cabral, M. Rato, V. Oliveira, D.A. Tavares, R. Pires, A. Morais, M.C. Faria, R.M. Barros, I. Peres, G. Trigueiro, C. Cardoso, J.G. Marques, I. Santos-Sanches (Caparica, Oeiras, Covilhã, Lisbon, Miraflores, PT)
Objectives: To compare the macrolide resistance frequencies, phenotypes, clones and population structure among isolates of Group A streptococci (GAS) from pharyngeal colonisation and infections over time.
Methods: A total of 1,425 GAS collected during 2000–2006 in Portugal from asymptomatic pharyngeal carriage (CA, n = 938) and tonsillitis/pharyngitis (TS, n = 487) of children and adults were tested for macrolide resistance frequency and phenotypes (M or MLSB) by disk diffusion. The macrolide-resistant (MR) isolates from CA (n = 153) and TS (n = 101) were characterised for serologic T-types and for SmaI or SfiI DNA-band Pulsed-Field Gel Electrophoresis (PFGE) patterns. Isolates of major PFGE patterns were tested by PCR and sequencing for emm-types and for sequence types (ST) by Multi Locus Sequence Typing (MLST).
Results: Resistance to macrolides gradually increased since 2000 (12%) to 2003 (43%) and decreased since 2004 (21%) to 2006 (13%) in TS isolates and the same trend was observed in CA isolates: an increase since 2000 (9%) to 2003 (28%) and a decrease until 2006 (14%). The M phenotype among CA isolates increased rapidly from 40% in 2000 to 88% in 2001, became stable (>80%) during 2000/04 and was undetected in 2005/06, while among TS isolates was almost constant from 2000/05 (>50%) and decreased to 17% in 2006. In parallel, the MLSB phenotype became dominant in 2005/06. Most (76%) of the MR isolates were of three M phenotype international lineages that emerged among isolates of both origins in different years: ST39 (PFGE AA-emm4,emm75-T8.25Imp19,T4,others), ST36 (PFGE R-emm12-T12,others) and ST28 (PFGE Z-emm1-T1). ST28 emerged in 2000/01, was dominant in 2001 (71%) and disappeared in 2003. ST39 and ST36 emerged in 2001/02, were dominant in 2003 (c.a.40%) and disappeared in 2005/06. One MLSB phenotype lineage, ST52 (PFGE F or PFGE AK-emm28-T28), resistant to bacitracin and also seen as epidemic worldwide emerged in 2000/02 and in 2006 included all the MLSB isolates from CA and TS.
Conclusions: The epidemiology of GAS from throat colonisation and infection underwent major shifts during 2000–2006, which included: (i) decrease in macrolide resistance since 2003 to 2006; (ii) inversions of macrolide resistance phenotypes in 2000 and 2005; (iii) fluctuation and replacement of major macrolide-resistant clones. In this study, lineage ST52 of MLSB phenotype recognized as capable of causing a broad range of streptococcal infections was first detected in carriers.
P756 Epidemiology of invasive Streptococcus pyogenes infections in Germany
R. Wahl, R. Lütticken, S. Stanzel, M. van der Linden, R. Reinert (Aachen, DE)
Objectives: This study sought to identify epidemiological markers of invasive S. pyogenes disease in Germany.
Methods: A nationwide laboratory-based surveillance study of invasive S. pyogenes infections was conducted in Germany from 1996 until 2002. Demographic and clinical information on the invasive cases were obtained from medical files. 464 isolates from 475 patients were available for emm typing and characterisation of pathogenicity factors. Isolates were identified by their haemolysis on sheep blood agar, Lancefield grouping, using a commercially available agglutination technique and standard biochemical procedures. The presence of emm genes was determined by PCR using ‘all M’ primers following a previously published protocol. The presence of the genes speA, speC, speF, or ssa was determined by PCR. Multiple logistic regression analysis was performed to determine risk factors for fatal outcome.
Results: Invasive isolates were obtained from 475 patients, with 251 (52.8%) isolates cultured from blood. The most frequent emm types were emm 1 (36.4%), emm 28 (8.8%) and emm 3 (8%). The genes speA and speC and ssa were present at variable frequencies in different emm types. The highest rate of speA and speC were found in emm 1 (93.6%) and emm 4 (94.7%), respectively. The number of the annual estimated incidence of invasive GAS disease was at least 0.1 cases per 100,000 persons. Complete clinical information was available in 165 cases. The overall case fatality rate was 40.6% and highest in the age group 60–69 years (65.2%). Shock, age ≥30 years and adult respiratory distress syndrome (ARDS) were predictors of fatal outcome in a multiple logistic regression analysis. 6.7% of the cases were categorised as having been nosocomially acquired. Nine cases of puerperal sepsis were observed.
Conclusions: The study underscores the importance of invasive S. pyogenes disease in Germany. Chemoprophylaxis of selected contacts of patients with invasive infection, infection control, and prompt investigation of outbreaks in hospitals and nursing homes are possible measures to reduce the high burden of S. pyogenes disease.
P757 Antimicrobial resistance of S. pyogenes in Russia: results of prospective multicentre study PEHASus
R. Kozlov, O. Sivaja (Smolensk, RU)
Objective: S. pyogenes (GAS) is the one of the common bacterial pathogens casing community-acquired infections (tonsillopharyngitis, skin and skin structure infections). In addition it might cause streptococcal toxic shock syndrome and also necrotising fasciitis, incidence of which is increasing in Russia. Therefore it is significant to determine the most active antimicrobials against S. pyogenes.
Methods: The study was conducted in 16 centres in 2001–2003 and 13 centres in 2004–2005 of Russia. Identification of the strains were done on the basis of colony morphology, Gram stain, bacitracin (0.02 IU) susceptibility and latex agglutination tests. Susceptibility to 13 antimicrobials was performed in central laboratory by broth microdilution method. Breakpoints were those of NCCLS (2005) except for SPI (<1; >4 mg/L) and MID (<1; >4 mg/L).
Results: A total of 1,057 in non-duplicate clinical isolates of S. pyogenes were included in this study.
| 2001–2003, n = 683 |
2004–2005, n = 374 |
|||
|---|---|---|---|---|
| I/R (%) | MIC90 (mg/L) | I/R (%) | MIC90 (mg/L) | |
| Penicillin G | 0 | 0.06 | 0 | 0.008 |
| Erythromycin | 2.9/5.3 | 5.3 | 4.0/4.8 | 0.25 |
| Clarithromycin | 3.1/3.8 | 1.2 | 1.6/2.9 | 0.125 |
| Azithromycin | 0.4/8.3 | 8.3 | 4.4/5.3 | 0.06 |
| Midecamycin | 2.0/0 | 0.5 | 0/0.3 | 0.25 |
| Spiramycin | 2.0/0 | 0.5 | 0/0.3 | 0.5 |
| Clindamycin | 0/0.7 | 0.03 | 0.3/0.3 | 0.03 |
| Levofloxacin | 0 | 0.5 | 0 | 1 |
| Moxifloxacin | 0 | 0.25 | 0 | 0.25 |
| Linezolid | 0 | 1 | 0 | 0.5 |
| Chloramphenicol | 1.0/13.0 | 16 | 0.8/12.6 | 16 |
| Tetracycline | 1.6/44.5 | 32 | 4.0/43.1 | 32 |
Conclusions: Expectedly, there were no non-susceptible strains to penicillin G, levofloxacin, moxifloxacin and linezolid. Percentage of non-susceptibility to erythromycin, clarithromycin, azithromycin and clindamycin was less than 10% and relatively stable during the study period. Midecamycin and spiramycin possessed the higher in vitro activity in comparison with 14- and 15-membered macrolides suggesting efflux as main mechanism of resistance. The highest non-susceptibility was observed to chloramphenicol and tetracycline that compromises their potential for usage for treatment of streptococcal infections.
P758 Differences in the epidemiology of paediatric and adult Streptococcus pyogenes invasive infections in Greece, 2003–2005
A. Stathi, J. Papaparaskevas, L. Zachariadou, A. Pangalis, P. Tassios, A. Tseleni-Kotsowili, N. Legakis for The Hellenic Strep-EURO Study Group
Objectives: To investigate possible clinical and epidemiological differences in Streptococcus pyogenes invasive infections (iGAS) among the adult and paediatric populations in Greece.
Materials and Methods: A total of 101 cases of iGAS infections during 2003–2005 were studied. Epidemiological and clinical information collected included: age, gender, specimen type, date of isolation, clinical manifestation, treatment, outcome, and risk factors. All isolates were typed by emm gene sequencing.
Results: Children represented 68 (67.3%) of the cases, and adults 33 (32.7%). The male to female distribution was 37 (54%) to 31 (46%) in children, and 24 (73%) to 9 (27%) in adults. Age range was 0.5–14 years (mean 5.3) in children, and 22–90 years (mean 51.3) in adults. Annual incidence was 0.8 cases/100,000 inhabitants/year. Blood and deep abscesses were the most frequent sites of isolation (41.6 and 35.6%, respectively). A second isolation site was found in 21% of the cases. Bactaeraemia was a more frequent clinical manifestation among adults (p = 0.054). Varicella infection was detected as a risk factor only among children (23%), though a marked decrease was recorded during 2005 (7%, compared to 20% and 30% for 2003 and 2004, respectively), when trauma emerged as the predominant risk factor in children (39% – compared to 11% and 6% in 2003 and 2004, respectively). Trauma was overall the main risk factor among adults (11%). Immunosupression was more frequent among adults than children (5 and 0.8%, respectively), whilst surgery was required more frequently among children (p = 0.003). Only 2 cases were fatal, both paediatric, presented with meningitis and STSS (paediatric case fatality rate: 3%). The most frequent (>5%) emm types were 1 (26.7%) and 12 (8.9%). Some emm types were exclusively observed in adult (11, 19, 50, 80, 101, 108, 113, and 117), or paediatric isolates (22, 75, 77, 78, 83, and 115).
Conclusions: In addition to distinct emm-types affecting either the adult or the paediatric population, there were also differences in clinical manifestations, and predisposing factors between iGAS in adult and paediatric infections in Greece.
Acknowledgments: Strep-EURO project is funded by the European Commission (QLK2-CT-2002–01398).
M. Foustoukou, A. Avlamis, H. Malamou-Ladas, E. Papafrangas, A. Bethimouti, G. Kouppari, B. Gizaris, S. Levidiotou-Stefanou, O. Paniara, A. Halakatevaki, A. Perogambros, A. Vogiatzi, V. Petroxeilou-Pashou and M. Iordanidou.
P759 High-level multidrug-resistance among viridians group streptococci isolated from Turkey: report from the SENTRY Antimicrobial Surveillance Program
D. Biedenbach, D. Gur, V. Korten, G. Soyletir, R. Jones (North Liberty, US; Ankara, Istanbul, TR)
Objectives: Viridans group streptococci (VGS) are composed of numerous species which are usually considered as harmless commensal of the oral cavity, gastrointestinal and female genital tract. However, VGS can cause invasive disease including endocarditis, deep abscesses, and bacteraemia, and are especially problematic among neutropenic patients. The SENTRY Program has monitored VGS in Europe (EUR) since 1997 including centres in Turkey. After ten years of monitoring these sites, it was observed that VGS from Turkey were markedly more MDR compared to other EUR sites and this study documents the MDR patterns detected.
Methods: A total of 1,361 isolates of VGS were collected from EUR sites (1997–2006) of which 119 strains were from Turkey. Isolates were identified to species level by participants and referred to a central laboratory for confirmation using bile solubility, colony morphology and commercial kits when needed. The majority of VGS from Turkey were included in the S. mitis group. The susceptibility (S) profile was determined using broth microdilution methods in lysed-horse blood supplemented CAMHB according to CLSI recommendations/interpretations (2006). A subset of 18 strains that were high-level resistant (R) to penicillin (PEN) and mupirocin (MUP) were tested for clonality using PFGE.
Results: The table showing R-profiles among 10 antimicrobials for strains collected in Turkey compared to centres in EUR. PEN non-S was 69.8% in Turkey (approx. 60% of these were non-S to ceftriaxone [CRO] or cefepime [FEP]) compared to VGS isolated in EUR at 25% PEN non-S; 30% non-S to CRO and FEP. Erythromycin (ERY)-R was also greater in Turkey (53.8%) compared to EUR (28.3%) with a constitutive clindamycin (CC)-R rate of 56 and 35%, respectively. R to levofloxacin (LEV) was two-fold higher, tetracycline (TET)-R was nearly double, and strains R to quin/dalfo (Q/D) and MUP were rarely isolated outside Turkey. PFGE revealed clonally related strains within this population of unusually R patterns.
MDR patterns of Turkish viridans-group streptococci
| VGS/Antimicrobial agent | Turkey (n = 119) |
Europe and Israel (n = 1,242) |
||
|---|---|---|---|---|
| % Intermediatea | %Ra | % Intermediatea | % Ra | |
| PEN | 32.8 | 37.0 | 19.6 | 5.0 |
| CRO | 5.8 | 36.5 | 3.9 | 3.3 |
| FEP | 10.9 | 30.3 | 4.6 | 3.6 |
| ERY | 5.9 | 53.8 | 4.0 | 28.3 |
| CC | 0.0 | 30.3 | 1.0 | 9.8 |
| LEV | 0.0 | 3.4 | 0.6 | 1.5 |
| TET | 3.4 | 51.3 | 2.0 | 28.9 |
| Q/D | 5.9 | 1.7 | 0.9 | 0.0 |
| Imipenem | − | 35.6b | − | 3.1b |
| MUP | − | 18.0c | − | 1.0c |
% based upon published CLSI S ranges (M100-S16)
% of isolates ≥1mg/L
% of isolates ≥16 mg/L.
Conclusions: This study documents significant variability among the S profile of Turkish VGS compared to strains from adjacent EUR nations. The percentage of strains from Turkey that were R to commonly prescribed antimicrobial agents such as β-lactams and macrolides was alarming as was the R rate to fluoroquinolones, TET, Q/D and MUP. Continued monitoring of VGS will be necessary because of the geographic variability in R for this increasingly important pathogen.
Infection control: Clostridium difficile
P760 Emergence of PCR-ribotype 027 Clostridium difficile-associated disease, Northern France, 2006
B. Coignard, F. Barbut, K. Blanckaert, J. Thiolet, A. Carbonne, P. Astagneau, J. Petit, M. Popoff, J. Desenclos (Saint-Maurice, Paris, Lille, FR)
Background: Clostridium difficile (CD)-associated disease (CDAD) has been recognized as an increasing cause of nosocomial infections (NI) since 2003 when a 027 epidemic strain emerged in Northern America and Europe. To timely detect and control CDAD clusters in France, the Institut de Veille Sanitaire (InVS) and regional infection control coordinating centres (CClin) strengthened the surveillance of NI and set up with the Anaerobe national reference centre (NRC) a network of regional laboratories to characterise CD isolates. We describe the introduction and spread of 027 CDAD in northern France in 2006.
Methods: Using ECDC case definitions, CDAD were notified by healthcare facilities (HCF) to CClin and district health departments through the mandatory national NI early warning and response system. CD strains were sent to the regional laboratories and NRC for confirmatory testing and PCR-ribotyping. CClin assisted HCF in the investigation and implementation of control measures. InVS coordinated the investigation and centralised epidemiological and microbiological data.
Results: The first cluster of 027 CDAD occurred in a HCF of the Nord – Pas de Calais region in April 2006 and accounted for 41 cases. Until November 2006, 30 HCF and 3 nursing homes (NH) notified 400 CDAD cases: 23 facilities had clusters of which 7 with 10 cases or more. Among 387 cases diagnosed in HCF, 328 (85%) were healthcare-associated and 54 (14%) community-acquired. Cases occurred mostly among elderly patients in geriatric or rehabilitation wards; 105 (27%) patients died; 22 (6%) deaths were attributed to CDAD. Of 236 C. difficile strains obtained from stool, 167 (71%) (31 HCF and 2 NH) belonged to the 027 epidemic strain. As of November 7th, 6 clusters were still considered as active. Intensive control measures were needed and included contact precautions, reinforcement of handwashing, glove use, environmental cleaning, and patients' isolation/cohorting. In other French regions, 49 HCF notified 138 cases with 60 strains sent for typing: none belonged to the 027 epidemic strain.
Conclusion: Our data confirm the emergence and spread of the 027 epidemic strain in France, which is presently clustered in the Northern region. This is probably related to close relationships with Northern Europe countries previously affected by this strain (United Kingdom, Belgium and The Netherlands). Control of 027 CDAD needs timely, organised and intensive surveillance and infection control resources.
P761 Succesful control of Clostridium difficile type 027 outbreak by cohort isolation of infected patients
A.A. Bentohami, C.C.M. Nolte, C.M.F. Verberg, M. Tensen, E. F. P. IJzerman, G. Heuff, E.J. Kuijper (Hoofddorp, Leiden, NL)
Objectives: Since 2005, outbreaks due the hypervirulent C. difficile type 027 have been recognized in the western part of the Netherlands. The outbreaks are difficult to control and advises are contradictory with respect to isolation protocols. We performed a prospective comparative study of cohort isolation with isolation of a patient in a single room to the incidence of CDAD.
Methods: From January until December 2005, an outbreak of CDAD occurred in a 400 beds general hospital encompassing 129 patients in total. The mortality attributable to CDAD was 3.1%. Of 17 availabe strains, 12 (71%) belonged to PCR ribotype 027. The departments of Internal Medicine (81 beds) and Surgery (76 beds) were the two most affected departments with 42 and 60 patients, respectively. At week 18, cohort isolation was introduced at the Surgery department, whereas isolation on a single room was continued at the Internal Medicine. Patients were isolated on a clinical suspicion of CDAD and/or a positive toxin test of a faeces sample by enzyme immunoassay. Data on the antibiotic use were obtained from the pharmacy database and calculated as daily defined doses (DDD). Patients characteristics were obtained using a home made standardised questionnaire.
Results: Before week 18 of the outbreak, the incidence of CDAD was 38.5 per 1000 admissions at the Department of Internal medicine and 35.4 per 1000 admission at the Surgery department. The construction of both departments was identical. Patients did not differ in mean age, gender, classification of American Society of Anesthesiology characteristics, previous antibiotic use and days of admission before CDAD developed. After introduction of cohort isolation, the incidence decreased in the following 20 weeks to 4.4 per 1000 admissions whereas the incidence remained unchanged to 39.8 per 1000 admission at the Internal Medicine. Patients with CDAD after week 18 did not differ from patients with CDAD before week 18. Environmental disinfection with hypochlorite was similar at the two departments, as were handhygiene with water and soap and the use of protective clothing. DDD of cephalosporines and fluoroquinolones were 463 and 1281 at the Internal Medicine per 12 months and 463 and 1134 at the Surgery department.
Conclusion: In contrast with isolation on a single room, cohort isolation resulted in a rapid decrease of the incidence of CDAD during an outbreak of the hypervirulent type 027.
P762 Clinical features of Clostridium difficile-associated disease and molecular characterisation of the isolated strains in a cohort of Danish hospitalised patients
L. Søes, I. Brock, S. Persson, K.E.P. Olsen, M. Kemp (Copenhagen, DK)
Objective: Recently outbreaks of severe cases of Clostridium difficile associated disease (CDAD) due to bacteria with increased virulence have been reported in hospitals in North America and in parts of Europe. These virulent strains have been associated with increased production of toxin A (TcdA) and toxin B (TcdB) and production of binary toxin (CDT). Furthermore a deletion in the toxinregulating gene tcdC has been reported in these virulent strains. These findings have emphasized the importance of an ongoing surveillance of CDAD. At Statens Serum Institut a multiplex-PCR method for the simultaneously detection of C. difficile toxin genes has been developed for this purpose. The aim of this study was to determine the toxin profile of the strains involved in CDAD in a Danish cohort and to compare the toxin profiles to clinical features.
Methods: A questionnaire was devised to reveal clinical features such as severity of disease, symptoms, predisposing illnesses and prior use of antibiotics. During a period of 6 months stool samples positive in culture for C. difficile were recorded and the local hospital ward contacted to fill out the questionnaire by an interview. The isolates were then analysed by PCR for detection of the toxin genes: tcdA, tcdB and cdtA/cdtB. Furthermore PCR was applied for detection of deletions in the tcdA and a sequencing based method was applied to reveal deletions in the tcdC regulatory gene.
Results: 104 new cases of CDAD were diagnosed by stool culture, and at present 89 clinical charts could be reviewed. Age ranged from 1 month to 93 years with 8 patients less than 3 years old. Excluding these, mean age was 66 years. Male/female ratio was 44/56%. Contributing factors, such as haematological malignancies, preexisting renal insufficiency, malignant solid tumours or inflammatory bowel disease, were seen in 52% of the patients. Immunosuppressive agents were used in one third of the patients. Bloody stools were seen in about one fifth of the patients and endoscopy was performed in 12%. According to preliminary results three quarters of the patients received antibiotics prior to symptoms and positive culture. Clinical isolates were studied and the toxin profile investigated as well as deletion studies.
Conclusion: Among other characteristics, these clinical features will be compared to toxin profiles of the isolates and observed deletions in tcdA and tcdC in order to evaluate the pathogenic potential of the isolated C. difficile.
P763 Epidemiological investigation of Clostridium difficile associated diarrhoea in a tertiary care hospital
V. Mela, K. Dimarogona, A. Pantazatou, D. Rebelou, A. Katsandri, J. Papaparaskevas, A. Avlamis (Athens, GR)
Objective: To investigate for epidemiological differences among patients with various underline disease severity, that were diagnosed and treated for Clostridium difficile associated diarrhoea (CDAD).
Materials and Methods: During the period March 2005-May 2006, out of a total of 640 specimens submitted for investigation of CDAD, 65 were culture-positive for C. difficile, of which 33 were also positive for toxin A and B. Culture for C. difficile was performed on cefoxitine-cycloserine-fructose agar after alcohol shock treatment of the specimen. Toxin A and B production was investigated by a commercially available enzyme immunoassay (Kytolone A+B kit, Meridian). Epidemiological and clinical information collected included: age, gender, type of ward, nosocomial acquisition of diarrhoea or not, duration of symptoms, classification of underlying disease according to Mac Cabe score (cases with score A were designated as group A, and those with scores B+C as group B), prior antimicrobial therapy or hospitalisation, CDAD treatment, and outcome. Statistical analysis was performed with the SPSS 13.0 programme using the x2 procedure.
Results: Among the 65 patients, 34 (52%) were women. The majority (45/65, 69%) were admitted in internal medicine wards, and previous hospitalisation was identified in 44 patients (68%). CDAD treatment received 40 patients (62%). Improvement was detected in 50 patients (77%). Nosocomial acquisition (>48 h from admission) was identified in 55 patients (85%) and was associated with group B cases (p = 0.001). Previous hospitalisation among group B patients was associated with positive toxin production (p = 0.036). Group B patients who received treatment for CDAD were more frequently improved than patients without treatment (p = 0.024), an association that was not detected among group A patients. Increased length of diarrhoea (>7 days) affected negatively the outcome of CDAD among group B patients (p = 0.009), but not among group A patients. Prior quinolone and prior 4th generation cephalosporin therapy was associated with increased length of diarrhoea (>7 days) among group B (p = 0.040 and 0.019, respectively) but not among group A patients.
Conclusions: Differences were detected between the two groups of patients regarding nosocomial acquisition, prior hospitalisation, prior antimicrobial therapy, length of diarrhoea, and final outcome. These differences are considered useful for optimisation of measures for prevention, control and treatment of CDAD.
P764 Mortality of patients with antibiotic-associated diarrhoea – the impact of Clostridium difficile
J. Bishara, S. Pitlik, Z. Samra (Petah-Tiqva, IL)
Background: Clostridium difficile infection is implicated in 20 to 30% of cases of antibiotic-associated diarrhoea. Previous studies have shown conflicting results relating to mortality attributable to C. difficile infection. The objective of this study was to determine the impact of C. difficile infection on short- and long-term mortality in hospitalised patients with antibiotic-associated diarrhoea.
Methods: All patients hospitalised from October 2003 to January 2004 who had antibiotic-associated diarrhoea and underwent stool enzyme immunoassay for C. difficile TOX A/B were followed prospectively. For univariate survival analysis the Kaplan-Meier and the log-rank test were used. The Cox regression model was used for multivariate analysis of 28-day and long-term mortality.
Results: Fifty-two (24%) of the 217 patients who met the study criteria were positive for C. difficile TOX A/B. The crude 28-day and long-term mortality rates of the entire cohort were 12.4% and 56%, respectively. On Cox regression analysis, hypoalbuminaemia, impaired functional capacity, and elevated serum urea levels were found to be the only independent and significant variables associated with long-term mortality. C. difficile toxin positivity per se was not associated with increased short- or long-term mortality rates.
Conclusions: Antibiotic-associated diarrhoea is associated with high rates of short- and long term mortality. Hypoalbuminaemia, renal failure, and impaired function capacity predict mortality. C. difficile involvement by itself does not further increase the risk of death in these patients.
P765 Co-existence of Clostridium difficile and vancomycin-resistant enterococci in stool samples, in a tertiary hospital in Greece, during a one-year period
M. Orfanidou, E. Vagiakou, P. Karabogia, M. Karanika, A. Strouza, H. Malamou-Lada (Athens, GR)
Objectives: The surveillance of vancomycin resistant enterococci (VRE) from stool specimens submitted for Clostridium difficile (Cd) testing, in a tertiary Hospital in Athens, Greece, during one year period (6/2005 – 6/2006)
Methods: During the study period 553 stool samples from patients with hospital acquired diarrhoea were examined for both Cd and VRE using cycloserine cefoxitin blood agar (BD) and esculin azide agar with 6 mg/L vancomycin (VA), respectively. Cd strains were identified by rapid ANA II (Remel, Lenexa) and latex test (Culturette, BD). VRE strains were identified by VITEK 2 (bioMérieux). Toxin A was detected from Cd strains by an ELISA (Vidas, bioMérieux) and a chromatographic assay (ColorPak, BD). Both toxins A&B were detected by an EIA (Premier Toxins A&B, Meridian).
Results: Cd strains were isolated in 67/553 (12%) and VRE in 125/553 (22.6%) faecal specimens. Co-existence of Cd and VRE was observed in 17/67 (25.4%) specimens. Toxin A+B+ producers were 9/17, A-B+ 5/17 and A-B– 3/17 Cd strains. These 17 specimens were obtained from patients of Internal Medicine, Gastroenterology, Surgical, Hematology and Nephrology department. The mean age was 66.7 years (range 44–83 years). The underlying diseases were: diabetes mellitus, cancer, Crohn's disease and operation during the past six months. All of the patients were under antibiotic therapy, mainly with β-lactams and glycopeptides, during their hospitalisation. All of Cd-positive patients were treated with metronidazole (MTZ), with the exception of one patient who presented pseudomembranous colitis and to whom MTZ was given simultaneously with VA. None of the 17 patients presented VRE bacteraemia. Interestingly, 7/67 Cd-positive patients who were treated successfully for their intestinal infection (five with MTZ and two with VA) presented VRE in their stool samples after their treatment.
Conclusions: In our hospital co-existence of Cd and VRE is high. This may result in the emergence of vancomycin resistant Cd posing new challenges in the management of hospital associated diarrhoea. So appropriate antibiotic policy and strict enteric precautions should be implemented to break the vicious circle between these two bacteria.
P766 Clostridium difficile in Australia
B. Elliott, B. Chang, T. Riley (Perth, AU)
Background: Clostridium difficile is an important nosocomial pathogen. Toxigenic strains usually produce toxins A and B, which are primary virulence factors of C. difficile. Some strains produce an additional toxin, an adenosine-diphosphate ribosyltransferase known as binary toxin, the role of which in pathogenicity is unknown. There has been concern about the recent emergence of a hypervirulent fluoroquinolone-resistant strain (PCR ribotype 027) in North America and Europe.
Objectives: To determine the circulating molecular types of C. difficile in Australia, and the prevalence of different toxin genotypes including binary-toxin positive strains. The extent of antimicrobial resistance was also investigated.
Methods: A total of 115 recent Western Australian (WA) clinical isolates was examined and compared to 34 clinical isolates from the Eastern States (ES) of Australia and a PCR ribotype 027 isolate. PCR was used to detect C. difficile toxin genes and PCR ribotyping to type isolates.
Results: Of the WA isolates, 10 of 103 (10%) were toxin A-negative, B-positive while 21 of 103 (20%) were non-toxigenic. Only 1 of 33 (3%) ES isolates was toxin A-negative, B-positive, while 4 of 33 (12%) were non-toxigenic. These differences were not statistically significant. Binary toxin genes were detected in 17 of 103 (17%) WA isolates, but only 3 of 33 (9%) ES isolates. Two toxin A-negative, B-negative strains possessing binary toxin genes were detected in WA. Computer analysis of ribotyping patterns divided 95 Australian isolates into 51 PCR ribotypes. No isolate with a ribotyping pattern matching that of PCR ribotype 027 was found. Little antimicrobial resistance was found in WA C. difficile isolates. Fluoroquinolone resistance was detected in one WA and one ES isolate. Clindamycin resistance was detected in 6 WA isolates (6%). No metronidazole or vancomycin resistance was detected.
Conclusions: Clinical isolates of C. difficile in WA are diverse with respect to both their toxin genotype and PCR ribotype. Antimicrobial resistance, including fluoroquinolone resistance, is low in WA, possibly as a reflection of restricted antimicrobial usage. The PCR ribotype 027 strain was not detected.
P767 Quantification of Clostridium difficile by real-time PCR in hospital environmental samples
C. Nonnenmacher, R. Kropatsch, S. Schumacher, R. Mutters (Marburg, DE)
Objective: The aim of this study was to detect and quantify Clostridium difficile in environmental samples obtained in hospital units where Clostridium difficile symptomatic patients were hospitalysed.
Methods: A real-time PCR assay was established for the detection and quantification of C. difficile in environmental samples. Quantification was performed with specific 16S rRNA target sequences using double fluorescence labeled probes. 221 samples were collected from environmental sites considered to be commonly exposed to patients and healthcare staff from the hospital settings. Sampling was performed with sterile cotton wool swabs moistened with 0.25% Ringer's solution. The samples were placed immediately in a Schaedler boullion and incubated under anerobic conditions for 3 days, and subsequently DNA extraction was performed.
Results: The sites sampled comprised bed frames, commodes, toilet environment, patient side room, floors, staff and patient hands. 86 isolates (40.6%) recovered from the hospital environment were positive for the presence of Clostridium difficile. Quantification of the positive samples ranged from 6.7×104 cells/mL to 1.0×103 cells/mL with the higher numbers of of C. difficile being found in the hands of patients and staff, staff gloves and in the toilets.
Conclusion: Considering the importance of staff and the inanimate hospital environment as a potential source of C. difficile, close attention should be paid to the hygiene of the clinical settings.
Infection control: surveillance and networking
P768 WHO first Global Patient Safety Challenge: current achievements
B. Allegranzi, A. Leotsakos, J. Storr, G. Dziekan, L. Donaldson, D. Pittet (Geneva, CH)
Objective: The WHO World Alliance for Patient Safety identified healthcare-associated infection (HAI) as the topic of the first Global Patient Safety Challenge (GPSC). The GPSC aims at reducing HAI worldwide by strengthening integrated actions in the areas of blood safety, injection safety, clinical procedure safety, and water, sanitation and waste management safety, with the promotion of hand hygiene (HH) in healthcare as the cornerstone.
Methods: The GPSC team, supported by a core group of renowned international experts and WHO collaborating partners, identified the following key success factors: 1) raise global awareness about the importance of HAI as a priority patient safety issue; 2) catalyze country commitment to actually face this problem; 3) prepare evidence-based guidelines for HH improvement in healthcare; 4) design and pilot test a strategy to translate into practice the GPSC, in particular the newly produced guidelines.
Results: Over the past 12 months, a formal statement has been signed by 35 Ministries of Health as a pledge of their support to implement actions to reduce HAI. Twenty additional countries have planned to sign the pledge by 2007, leading to 75% coverage of the world population. Following the pledge, 13 countries have recently documented their actual progress, including establishment of new policies, resources, national campaigns and guidelines, training programmes and surveillance systems. A dedicated web page has been constructed as well as a database of more than 2,000 contacts in the field of patient safety and infection control acting as stakeholders worldwide. A multimodal strategy is proposed by WHO to improve HH in healthcare settings together with other infection control interventions. The implementation is supported by a range of practical tools of different types to address different targets: operational, advocacy and information, monitoring, HH product procurement, education, impact evaluation. A test phase is currently ongoing in several sites.
Conclusions: In only one year of work, the GPSC has attained several achievements related to pre-established key success factors. The aim of the ongoing testing of the GPSC implementation strategy is to obtain feedback about feasibility, acceptability and sustainability in healthcare settings worldwide. The combined efforts expected under the GPSC have the potential to save millions of lives and engender major cost savings by improvement of basic procedures such as HH.
P769 National Resource for Infection Control (www.nric.org.uk): meeting a need?
S. Wiseman, P. Kostkova, F. Hansraj (London, UK)
The National Resource for Infection Control (NRIC) was launched in May 2005 in response to National Audit Office (2000/04) recommendations for a national infection control manual. The project funded by the Department of Health (UK) and endorsed by the UK National electronic Library of Infection (www.neli.org.uk) has also been timely in assisting infection control professionals in reviewing/updating their local evidence based infection control policies, now a statutory requirement under the Health Act: Code of Practice (2006) National policy and guidance documents are available on NRIC, with level of evidence for each resource clearly noted, documents are organised by settings, clinical practice tasks, modes of transmission and diseases, organisms. Eighteen months on are we succeeding? A preliminary evaluation study investigating the NRIC web server logs, search keywords used and user feedback survey analysis demonstrates over 2000 distinct users and thousands of hits per month. Geographical distribution demonstrates 32% of users are from the UK, and 68% users from non-UK countries (see Table 1).

On average, users spent 2.32 sec on NRIC and visit 3.44 pages; 25.0% visit NRIC page from other NRIC page, 21.3% come from Google, 6.4% from NeLI, 2.3% from Department for Health, UK website, 5.3% from other search engines. User feedback has been positive indicating increasing need for evidence-based knowledge accessed online. NRIC has now received a ‘face lift', adding several useful tools for easier access to other information such as News, upcoming events and over 1000 infection control professionals have signed up for the monthly eNewsletter. Whilst valid for research the impact of NRIC/NeLI on public/professionals knowledge, attitudes and subsequent patient care also needs to be evaluated. Preliminary research has indicated the potential of these resources to impact on patient care by changing knowledge & attitudes. Further research is needed to see if these resources are influencing clinical outcomes, policy writing and the rise in shared internet-based infection control manuals by UK healthcare trusts or whether NRIC needs to develop a wider remit. This presentation will describe the progress made, the detailed results of the user evaluation and outline a research protocol for a review of NRIC users, changes in attitudes/knowledge following use and cost effectiveness in terms of impact on decision making in policy development and subsequent practice. Ref-NRIC
P770 Surveillance of surgical site infections after ambulatory surgery
A. Blaich, R. Babikir, E. Meyer, P. Gastmeier, M. Dettenkofer (Freiburg, Hannover, DE)
Objective: For patients ambulatory surgery must not be associated with an higher risk for surgical site infections (SSI) than in hospital settings. Several studies have demonstrated that SSI result in considerable morbidity and excess healthcare costs, mostly from extended duration of hospitalisation and antibiotic use. Surveillance programmes have been shown to reduce the risk of SSI in surgical patients and are strongly recommended for prevention.
Methods: In 2002 the German National Reference Center for Surveillance of Nosocomial Infections established a surveillance module in order to provide sound data for prevention and control of SSI in ambulatory surgery (AMBU-KISS). Until today the project centre (Institute of Enviromental Medicine and Hospital Epidemiology at the University Medical Center Freiburg, Germany) has analysed data of 101,584 procedures for a total of 8 operative indicator procedures from 135 participating institutions. Three of these indicator procedures were compared with corresponding procedures of the OP-KISS modul (SSI Surveillance in hospital settings). Included in the comparison were SSI of the OP-KISS modul in patients with arthroscopic surgery of the knee, inguinal hernias (risk group 0) and in patients with vein – stripping (risk group 0, 1, 2 and 3).
Results: Results obtained for three indicator procedures show a significant difference in SSI rates for ambulatory surgery institutions and hospital settings (OP – KISS). The arithmetic mean values of SSI rates in arthroscopic surgery of the knee are 0.09% in AMBU KISS and 0.18% in OP–KISS. For inguinal hernias, the respective rates are 0.35% versus 0.68%, and for vein-stripping 0.27% versus 0.84%. The arithmetic mean values of SSI rates in ambulatory surgery are 0.14% for orchidotomy and 0% for lumbar intervertebral disc operations, breast excisions, breast enlargements and nasal septum operations.
Conclusion: The results display that AMBU – KISS is suitable to assess the frequency of SSI in outpatient institutions and that there is a lower risk to contract SSI than in hospital settings. However, these data have to be subject of validation. In ambulatory surgery a highly experienced team is active. The resulting shorter duration of the operative procedures and a more gentle technique may be important reasons for the lower rates in ambulatory surgery.
P771 Are our mobile phones clean?
F. Ulger, S. Esen, A. Dilek, K. Yanik, M. Gunaydin, H. Leblebicioglu (Samsun, TR)
Objective: Inanimate objects can be contaminated with epidemiologically important nosocomial pathogens. Cellular phones are widely used non medical devices by healthcare workers. In this study the contamination rates of health care worker's (HCW) mobile phones were evaluated and the relationship with the contaminated hands of healthcare workers were investigated.
Material and Methods: A total of 200 HCWs included in the study. Microbial samples were collected from mobile phones and dominant hands of HCWs. For each HCW, a sterile swab moistened with sterile water was rotated over the surface of both sides of his/her phone, a second swab for the sampling of the dominant hand and both swabs were immediately streaked on to two plates that consist of blood agar supplemented with 5% defibrinated sheep blood and eosin methylene blue agar. Plates were incubated aerobically at 37°C for 24 h. Oxacillin sensitivity of the staphylococci and ceftazidime sensitivity of the Gram-negative isolates were investigated by disk diffusion method.
Results: In total, 94.5% of phones demonstrated evidence of bacterial contamination. The distribution of the isolated microorganisms was similar to hand isolates. Some of the mobile phones sampled grew bacteria that are known to cause nosocomial infections (Table ). It was found that 49.0% of phones grew one bacterial species, 34.0% grew two different species and 11.5% grew three or more different species. 52.0% of the S. aureus strains that were isolated from mobile phones and 37.7% of the strains from the hands were resistant to methicillin. 31.3% of the Gram-negative strains that were isolated from mobile phones and 39.5% of the strains from the hands were resistant to ceftazidime.
Types of bacteria isolated from phones and hands of HCWs
| Bacteria | Mobile phones (n = 200) | Dominant hands of HCWs (n = 200) |
|---|---|---|
| CoNSa | 181 | 193 |
| Staphylococcus aureus | 50 | 53 |
| Moulds | 20 | 19 |
| Nonenteric Gram-negatives | 19 | 26 |
| Coliforms | 15 | 12 |
| Streptococci | 12 | 18 |
| Enterococcus spp. | 7 | 9 |
| Yeasts | 3 | 3 |
CoNS: Coagulase-negative staphylococci.
Conclusions: This study demonstrates that mobile phones may be contaminated by the hands of HCWs. In this study, some of the contaminated microorganisms were epidemiologically important nosocomial drug resistant pathogens. Development of effective preventive strategies such as regular decontamination of mobile phones with alcohol disinfectant wipes can reduce cross-infection related with this frequently used devices in healthcare settings.
Malaria
P772 Sociodemographic and spatial influences on the malaria incidence of infants in the Ashanti Region/Ghana
B. Kreuels, R. Kobbe, S. Adjei, O. Adjei on behalf of the Agona IPTi Trial Team
Introduction: Malaria incidence rates have been shown to vary greatly over short distances and periods of time in areas of low to medium transmission. In areas of high, perennial transmission, due to early acquired immunity malaria incidence variation is considered to be far less various. However in small children who have not yet developed immunity this is not necessarily true.
Methods: We recruited a group of 535 children from 9 villages in an area of high transmission in Ghana at the age of 3 months and followed them monthly over a period of 21 months. Malaria was defined as a new fever episode together with a positive thick smear of at least 500 parasites/μl. Socio-economic characteristics were evaluated with the help of a questionnaire on recruitment. Geographical characteristics of the villages and distances of households to the forest fringe were analysed on satellite images with a Geographic Information System (GIS). Ecological analyses and multivariate Poisson regressions were performed to determine the most important factors responsible for the spatial variation of malaria incidence.
Results: Malaria incidence in the study area was highly heterogeneous between the 9 villages with a variation of 0.83–2.20 episodes per person year at risk. Incidence rates were strongly correlated to village area (r2 = 0.74) and village population (r2 = 0.68) while altitude variation in the study area was low and did not correlate to incidence rates. We found significant lower malaria incidences in children born in May (p < 0.001), September (p < 0.01) or October (p < 0.01), in children with literate mothers (p < 0.02), in those with use of bednets (p < 0.001) and window-screens (p < 0.001), and those born into families with a higher financial status (p < 0.01). On the other hand malaria incidence was higher in children of Northern Ghanaian ethnicity (p < 0.001) and children whose mothers worked as farmers (p < 0.01). An independent and strongly significant influence could be seen for the distance of the household from the forest fringe with a linear rate reduction of 0.2 episodes every 50 meters further away from the forest (p < 0.001).
Conclusion: Our results indicate that the variation of malaria incidence rates in children from rural areas of high transmission may have been underestimated in the past. Taking this variation into account when designing new and implementing current intervention measures could possibly increase their efficiency and cost-effectiveness.
P773 Trend in malaria vector resistance or susceptibility to insecticides in Cameroon
J. Etang, P. Nwane, M. Chouaibou, L. Manga, P. Awono-Ambene, C. Antonio-Nkondjio, E. Fondjo, F. Simard (Yaounde, Brazzaville, CM)
Objectives: The National Strategic Plan for Malaria Prevention in Cameroon mainly releases on vector control by Insecticide treated nets and indoor residual spraying. Vector resistance to insecticides is therefore seen as a threat for interventions.
The objectives of the study were: (1) to evaluate the susceptibility of malaria vectors to insecticides used in vector control, (2) to identify involved resistance mechanisms and their distribution.
Methodology: A large scale programme was conducted in Cameroon from 2002 to 2006, using WHO protocol for adult bioassays as well as molecular and biochemical analyses, to evaluate susceptibility to DDT and pyrethroid insecticides in 45 field populations of Anopheles gambiae Giles and An. arabiensis Patton, 3 populations of An. funestus Giles, 2 populations of An. nili Theobald and 2 populations of An. moucheti Evans. These anopheline species are the major malaria vectors in Cameroon (300 infected bites/man/year).
Results: An. funestus, An. nili and An. moucheti were found susceptible to all insecticides, with almost 100% mortality rates. However, different patterns of resistance were seen in An. gambiae M and S molecular forms, and An. arabiensis. In the southern equatorial region, An. gambiae displayed resistance to DDT (30–60% mortality rate), sometimes coupled with resistance to pyrethroids (60–80% mortality rates). This resistance was mainly due to target site insensitivity resulting from a single nucleotide polymorphism in the gene encoding subunit 2 of the sodium channel. This mutation, known as kdr mutation, leads to substitution of Leucine (TTA) amino acid to Phenylalanine (TTT) or Serine (TCA). Both mutations were broadly distributed in the S molecular form, but slightly in the M form. A polymorphism was seen in discriminative nucleotides between M and S, at positions 702 and 703 of the subunit 1 of the sodium channel, but not at position 896, suggesting 3 independent mutational events and introgession between M and S forms. In the northern part of the country, both An. gambiae S form and An. arabiensis displayed low level resistance to pyrethroids and sometimes to DDT (80–100% mortality rates). This resistance was attributed to elevated activity of esterases or oxidases rather than kdr mutations.
Conclusion: This is the first report of the distribution of kdr mutations in Cameroon. Current data will enable the National Malaria Control Programme to elaborate strategies for effective malaria prevention in Cameroon.
P774 Malaria cases in Turkey: do climatic changes play any role?
Ö. Ergönül, A. Azap, S. Akgündüz (Istanbul, Ankara, TR)
Introduction: Malaria is one of the vector borne infectious diseases that was affected by climatic changes.
Objective: To demonstrate the effects of temperature and rainfall on malaria cases in Turkey.
Methods: The data was obtained from two sources: Number of the malaria cases in the last 30 years from the Ministry of Health (MOH) of Turkey and the changes in the temperature and rainfall in the last 70 years from The Research Unit in Turkish State Meteorological Service. Temperature and rainfall variations and trends for Turkey were analysed by using a data set including monthly averages of daily mean, and minimum temperatures. Non-parametric Kruskal-Wallis (K-W) test was performed in order to detect homogeneity in mean annual series. The non-parametric Mann-Kendall (M-K) rank correlation test was used to detect any possible trend in temperature series, and to test whether or not such trends are statistically significant. The Cramer test was used to detect the difference in temperature and rainfall between given time periods and the longer period.
Results: In recent 35 years, there were two important peaks of malaria cases in Turkey, one is at the 1977–1984 period, and the other is at the 1993–1999 period. The mean temperature in 1977–1987 period was significantly higher than mean temperature between 1930 and 2004 in some provinces where malaria incidence was high. Malaria cases increased in parallel to the increase in mean temperature within certain time intervals in certain provinces. Although no significant decrease in temperature was observed, the incidence of malaria has declined significantly after year 2000. Extensive control efforts implemented by local and governmental healthcare authorities might contribute to the decline of malaria cases. No association was found between rainfall and the incidence of malaria in any region for any time period.
Conclusion: Although climatic changes may play some role on the incidence of malaria, preventive efforts for controlling malaria have a substantial impact.
P775 Effects of intermittent preventive anti-malarial treatment of infants in a holoendemic area
R. Kobbe, S. Adjei, O. Adjei, J. May on behalf of the Agona IPTi Trial Team
Background: Intermittent preventive antimalarial treatment of infants (IPTi) with sulfadoxine-pyrimethamine reduces falciparum malaria and anaemia in areas of moderate and seasonal transmission. To date, IPTi has not been assessed in areas of intense perennial malaria transmission. We investigated the protective efficacy of IPTi in a holoendemic area of perennial high transmission and analysed the time-dependency as well as the therapeutic and prophylactic fraction of the effects.
Methods: A randomised, double-blinded, placebo-controlled trial on IPTi with sulfadoxine-pyrimethamine at three, nine and fifteen months of age was conducted with 1070 children in an area holoendemic for malaria inthe Ashanti Region, Ghana. Participants were monitored for 21 months after recruitment by active follow-ups and passive case detection. Primary endpoint was the reduction of malaria incidence. Additional outcome measures were incidence of anaemia, number of outpatient visits, frequency of hospital admissions, and mortality.
Results: Overall protective efficacy against malaria episodes was 20% (95% CI: 11%–29%; p < 0.001). The frequency of malaria episodes was reduced after the first two sulfadoxine-pyrimethamine applications (protective efficacy 23%; 95% CI: 6%–36%; p = 0.01 and 17%; 95% CI: 1%–30%; p = 0.04, respectively). After the third sulfadoxine-pyrimethamine administration at month 15, however, no further protection was achieved. Stratified analyses for six-months periods after each treatment were performed. Protection against the first or single episode of anaemia was only significant after the first IPTi dose (30%; 95% CI: 5%–49%; p = 0.02) and the frequency of anaemia episodes increased during the rebound period (–24%; 95% CI: −50%–2%; p = 0.03). For all IPTi applications the prophylactic effects clearly exceeded the therapeutic effects.
Conclusion: In an area of intense perennial malaria transmission, the protective efficacy of sulfadoxine-pyrimethamine based IPTi is age-dependent and based on the prophylactic effects of the antimalarial drug.
P776 Clinical symptoms, treatment and outcome of Highlands malaria in Eldoret (2,400 m a.s.l.) and comparison to malaria in hyper-immune population in endemic region of South Sudan
A. Farkasova, S. Seckova, Z. Nagyova, A. Kolenova, P. Olejcekova, I. Sabo, L. Gabrhelova, V. Krcmery, P. Kisac, E. Kalavsky, M. Kiwou, M. Taziarova, P. Bukovinova, M. Ocenas, J. Kralova, K. Kutna, J. Mykyta, M. Velganova, A. Augustinova, M. Pivarnik, M. Duris, L. Uhercik, J. Benca, T. Pastekova (Trnava, Bratislava, SK; Eldored, KE)
Aim of the study and Introduction: Malaria should not be present in altitudes more than 1,800 m a.s.l. However due to global warming, so called Highlands malaria (HM) sporadically occurs up to 2,000 m a.s.l. The purpose of this study is compare of clinical picture and prognosis of HM and compare it to malaria in endemic region of South Sudan (endemic malaria – EM) among hyper-immune population.
Patients and Methods: Analysis of HM from November 2005 to November 2006 were reviewed (64 cases), symptoms, therapy and outcome were compared to 215 cases of EM occurring in Gordhim, South Sudan, where malaria is endemic. Imported cases from Rift Valley to Eldoret were excluded.
Results: Analysis of 64 cases of sporadic HM showed mild clinical picture – fever 100%, chills up to 80%, headache 15%, gastrointestinal symptoms were present in 35.6%, respiratory symptoms in 64.4%. Only 1 case (1.6%) of cerebral malaria occurred. Amodiaquine in 81% and amodiaquine/arthesunate in 19% were used for therapy with only 1 failure.
Analysis of 215 cases of EM showed more severe symptoms: 58 (27%) had severe clinical course, 10 patients (4.7%) cerebral, 1 patient (0.5%) lung edema, severe anaemia with hemoglobinuria 21.8%, seizures 8 patients (3.7%), severe hypoglycaemia 12 (5.75%). Amodiaquine plus artesunate was used for therapy, in some cases, arthemeter or/and quinine were prescribed. 17 patients in Sudan (4%) died, mostly on CNS and respiratory failure due to cerebral malaria and/or severe anaemia.
Conclusion: Sporadic malaria occurs also in heights up to 2,400 m a.s.l. We documented 64 cases of non-imported malaria in population living above 2,000 m a.s.l., probably due to global warming in Equatorial Africa. Clinical symptoms despite of high immunity among population were much severe in endemic area of South Sudan (27%) including 17 deaths (4%) in comparison to no deaths and 1.6% occurrence of severe malaria in semi-immune population of Eldoret (Highlands malaria).
P777 The risks of reintroduction of malaria into Belgrade area, Serbia
Z. Dakic, N. Stajkovic, Z. Kulisic, M. Pelemis, J. Poluga, M. Cobeljic, I. Ofori-Belic, D. Tomic, M. Pavlovic (Belgrade, RS)
Objectives: Since 1974, when the WHO officially declared Serbia malaria-free, only imported cases of malaria have occurred. The population of mosquitoes has generally been reduced by the campaign of malaria eradication and frequent insecticide use. During the 1990s, severe financial constraints contributed to the suspension of the vector-control. Most patients with imported malaria are treated in Belgrade.
Methods: The pattern of imported malaria was monitored during the period 1994 to 2005 in Belgrade, as a whole and available data on the presence of the potential vectors for malaria are presented for comparison.
Results: The mosquitoes present in Belgrade include 20 species of mosquitoes family of Culicidae. In centre of city the molestants are distributed. Before eradication, the main vectors of malaria in Belgrade's area were Anopheles maculipennis s. s., and secondary vectors were An. messeae and An. atroparvus. Mainly in the areas of the river Danube and Sava basin, they have recolonised their previous habitats. But, the relation between the species of An. maculipennis complex has changed. An. maculipennis s. s. is very rare now, while there is a constant increase of An. atroparvus.
From 1994 to 2005, a total of 166 imported cases of malaria were treated in Belgrade. Most patients were treated in the Institute for Infectious and Tropical Diseases (96%), and others in Military Medical Academy. The vast majority of malaria infections in our patients were due to P. falciparum alone or in mixed infections (67%). Of all imported cases during the period theoretically favourable to transmission (n = 63), only one third of them (n = 18) were gametocyte carriers. Of these 18 gametocyte carriers, 67% were of P. falciparum, and 33% of P. vivax. Most of them were imported from Africa, only 1 from Asia. The length of potential exposure of gametocyte carriers to mosquitoes is very low. All patients are treated in 2 hospitals in the centre of city, and that limits mosquito-human contact.
Conclusion: From 2000, when travel restrictions stopped, there has not been a significant increase in the total number of imported malaria cases. Belgrade's vulnerability is low because of the low presence of gametocyte carriers during June-September. The three species of the Anopheles maculipennis complex, particularly An. messeae and An. atroparvus, are considered as potential vectors of malaria in Belgrade. Their roles as vectors requires additional investigation.
New antimicrobials I
P778 A novel not β-lactam inhibitor of DD-peptidase 64–575
J. Solecka, A. Rajnisz, A. Laudy, W. Kurzatkowski (Warsaw, PL)
Objectives: The increasing bacterial resistance to antibiotics is at present a great therapeutic problem. Multi-resistant pathogenic bacteria occur more and more frequently. The solution of this problem involves screening for microorganisms producing novel antimicrobial drugs. Streptomycetes are very well known producers of antibiotic. The purpose of this work was search for novel antimicrobial agents (inhibitors of DD-peptidases).
Methods: 1) Screening for Streptomyces producing exocellular inhibitors of DD-carboxypeptidase/transpeptidase 64–575 (DD-peptidase 64–575). After cultivation in liquid medium, supernatants of 110 Streptomyces strains (strain collections: IAUR, ISP, IMAN) were tested for DD-peptidase 64–575 inhibition (mode of action of β-lactams antibiotics) (Solecka J et al., Acta Pol Pharm, 2003; 60: 115–8). 2) Assay the supernatant stability of DD-peptidase 64–575 inhibitors after class A β-lactamase incubation. 3) Purification of inhibitors: acetone protein precipitation from the culture supernatant, use of Strongly Basic Anion Exchanging Resin column (acetic acid pH modification) and RP HPLC with C18 modified columns (Atlantis, Waters)(eluent: TFA and acetonitrile; gradient methods). 4) Structural elucidation was conducted using the following methods: HPLC-MS, MS/MS, LR-MS, IR, 1H, 13C and 2D NMR. 5) Antimicrobial activity assay: dilution method was used.
Results: After 48 h of cultivation the supernatant of the Streptomyces sp. 1 strain showed 90% of the DD-peptidase 64–575 inhibition. This strain was selected to further experimentation. The most inhibiting compound – AR1 (not degraded by penicillinase) was purified to one peak. Molecular weight of AR1 is 207.2 Da. It is an aromatic not β-lactam compound. The AR1 DD-peptidase 64–575 inhibition is expressed by the value of ID50(M) = 0.12×10−2. AR1 showed antibacterial activity against Proteus vulgaris NCTC 4635 and Staphylococcus aureus NCTC 4163.
Conclusions: A novel non β-lactam inhibitor of DD-peptidase 64–575 with antimicrobial activity was isolated. Further experiments should be done to synthesize the AR1 compound and to continue the chemical characterisation.
Grant no. 3P0 5F 033 24, Ministry of Education and Science, Warsaw, Poland.
P779 Pharmacokinetics of ceftobiprole following single and multiple intravenous infusions administered to healthy subjects
B. Murthy, D. Skee, D. Wexler, D. Balis, I. Chang, D. Desai-Kreiger, G. Noel (Raritan, US)
Objectives: Ceftobiprole, an investigational broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci, is currently in late stage clinical development. The purpose of this study was to evaluate the single- and multiple-dose pharmacokinetics (PK) of ceftobiprole 500 mg q8h given as a 2-h infusion.
Methods: This was a single-centre, open-label study in 28 healthy adult males and females. Subjects received a single infusion on Day 1 and multiple infusions on Days 3 through 5. Serial blood and urine samples were collected through 24 h after the start of infusion on Days 1 and 5 for estimation of ceftobiprole medocaril (prodrug), ceftobiprole, and open-ring metabolite (M1) via an LC-MS/MS method. PK parameters were estimated using model-independent methods.
Results: PK parameter estimates for ceftobiprole and M1 are shown in the table . Plasma concentrations of the prodrug were measurable only during the infusion, indicating instantaneous conversion to ceftobiprole. Systemic exposure of ceftobiprole was comparable between Days 1 and 5 with minimal accumulation. Clearance (CL) remained unchanged from Day 1 to Day 5. Ceftobiprole was primarily excreted unchanged in the urine (≥83%). Systemic exposure of M1 increased from Day 1 to 5 with an accumulation index of 1.5. Elimination half-life of M1 was 5–6 h. Less than 7% of ceftobiprole is excreted in the urine as the open-ring metabolite. Systemic exposure of ceftobiprole and M1 was higher in females compared to males, which normalised upon correction for body weight. On Day 5, approximately 100% of the dose was excreted in the urine as the prodrug, ceftobiprole, and M1. No subject had a serious adverse event. No subjects were withdrawn from the study due to an adverse event.
| Parameter | Ceftobiprole |
M1 |
||
|---|---|---|---|---|
| Day 1 | Day 5 | Day 1 | Day 5 | |
| Cmax (mg/L) | 29.2 (5.52) | 33.0 (4.83) | 0.804 (0.214) | 1.07 (0.254) |
| AUC0–8h (mg·h/L) | 90.0 (12.4) | 102 (11.9) | 3.68 (0.882) | 5.45 (1.35) |
| t1/2 (h) | 3.1 (0.3) | 3.3 (0.3) | 4.7 (0.8) | 5.8 (0.9) |
| CL (L/h) | 4.89 (0.687) | 4.98 (0.582) | − | − |
Conclusion: The pharmacokinetics of the ceftobiprole 500mg q8h regimen given as a 2-h infusion are similar to results previously reported with no accumulation observed. The majority of the administered dose was recovered in the urine. This regimen was safe and well tolerated.
P780 Activity of ceftobiprole tested against contemporary European Enterobacteriaceae and Pseudomonas aeruginosa (2005–2006)
T. Fritsche, H. Sader, P. Strabala, R. Jones (North Liberty, US)
Objectives: To present results assessing in vitro potency of ceftobiprole (BPR) against the most commonly occurring Enterobacteriaceae (ENT) and non-fermentative Gram-negative bacilli isolates in Europe. BPR, an investigational parenteral cephalosporin, is currently in clinical trials for complicated skin and skin structure infections and nosocomial pneumonia. This agent is unique amongst its class, being active against methicillin-resistant S. aureus (MRSA) as well as other Gram-positive and -negative pathogens, making it an attractive candidate for broad-spectrum therapy.
Methods: Non-duplicate clinically-significant isolates of ENT (3,399), P. aeruginosa (PSA; 666) and Acinetobacter spp. (ASP; 230) were collected from 25 medical centres in Europe participating in a BPR surveillance programme during 2005–2006. Identifications were confirmed by the central monitoring laboratory and all isolates were susceptibility (S) tested using CLSI methods against BPR and comparators including ceftazidime (CAZ) and cefepime (FEP).
Results: BPR, CAZ and FEP results are listed in the Table . BPR was similar in potency to the third- and fourth-generation cephems (MIC50 values, ≤1mg/L) for all tested ENT. Coverage against EC was nearly identical for the three agents (Table; 94–95% inhibited at ≤4 mg/L). Whereas FEP provided enhanced coverage against KSP (88% at ≤8 mg/L vs. 76–81% for BPR and CAZ), BPR and FEP were superior to CAZ against ESP and CIT All were equally active against PM, SER and Salmonella spp. Against PSA, BPR was equal in potency to CAZ (MIC50, 2mg/L) and two-fold more potent than FEP, although % inhibited for these agents at ≤2/4/8 mg/L was similar (49–54/65–70/76–80%, respectively). None of these agents inhibited >45% of ASP at 8 mg/L.
| Species (no. tested) | MIC90 (% at ≤2/4/8 mg/L) |
||
|---|---|---|---|
| BPR | CAZ | FEP | |
| E. coli (EC; 1889) | ≤0.06 (94/94/94) | ≤1 (94/95/96) | 0.25 (95/95/96) |
| Klebsiella spp. (KSP; 624) | >8 (75/75/76) | >16 (78/79/81) | 16 (82/85/88) |
| Enterobacter spp. (ESP; 381) | >8 (81/84/88) | >16 (66/68/69) | 4 (89/94/96) |
| Citrobacter spp. (CIT; 79) | 1 (99/99/99) | >16 (72/73/76) | 1 (99/99/99) |
| P. mirabilis (PM; 143) | ≤0.06 (96/96/96) | ≤1 (96/97/98) | ≤0.12 (97/97/97) |
| Serratia spp. (SER; 142) | 0.5 (96/96/96) | <1 (96/96/97) | 0.5 (98/98/98) |
| PSA (666) | >8 (54/65/79) | >16 (57/70/76) | 16 (49/65/80) |
| ASP (230) | >8 (37/39/40) | >16 (12/31/37) | >16 (22/33/45) |
Conclusions: BPR is a new anti-MRSA β-lactam with recognized activity against the most commonly occurring ENT and PSA, similar to that of extended-spectrum cephems. These characteristics warrant continued evaluation of BPR as empiric therapy for severe pneumonia, especially in those European institutions/regions where MRSA and PSA may be prevalent.
P781 Activity of ceftobiprole tested against staphylococcal and streptococcal isolates recovered from patients in European medical centres (2005–2006)
T. Fritsche, H. Sader, P. Strabala, R. Jones (North Liberty, US)
Objectives: To present in vitro potency of ceftobiprole (BPR) against staphylococci and streptococci originating from European (EUR) patients in 2005 and 2006. BPR, an investigational parenteral cephalosporin with a broad spectrum against Gram-negative and -positive pathogens, including MRSA, is currently in clinical trials targeting complicated skin and skin structure infections (cSSSI) and nosocomial pneumonia (NP).
Methods: Non-duplicate clinically-significant isolates (4,957 isolates) of S. aureus (SA; oxacillin-susceptible [OXA-S] and – resistant [R]), coagulase-negative staphylococci (CoNS; OX-S And OX-R), S. pneumoniae (SPN; penicillin (PEN)-S and PEN-R), viridans group streptococci (VGS), beta-haemolytic streptococci (BHS) were submitted from 25 medical centres in EUR participating in BPR surveillance (2005–2006). Central laboratory processing included identification confirmation and susceptibility (S) testing to BPR and other comparative β-lactams using CLSI reference methods and interpretive criteria.
Results: BPR inhibited 100 and >99% of tested S. aureus and CoNS at ≤4 mg/L, respectively, and all SPN and BHS at ≤0.5 mg/L. While MIC90 values for OXA-R strains were four-and eight-fold higher for SA and CoNS, respectively, published PK/PD characteristics suggest that target attainment for both OXA-S and -R populations would be achievable. BPR potency against OXA-S and OXA-R SA from North America and Latin America were nearly identical to those presented here for EUR (MIC50/90, 0.25/0.5 and 1–2/2 mg/L, respectively). All streptococci were readily inhibited by BPR (MIC90 values ≤0.5 mg/L); only VGS included strains with elevated MIC values (6% at >0.5 mg/L). BPR potency against SPN was equivalent to that of imipenem (MIC90, 0.25 mg/L). While BPR is generally inactive against E. faecium, the majority of E. faecalis strains (93% of 720 isolates) were inhibited at ≤4 mg/L (data not shown).
| Organism (no. tested) | BPR MIC (mg/L) |
Cum. % inhibited at MIC (mg/L) |
||||||
|---|---|---|---|---|---|---|---|---|
| 50% | 90% | ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | |
| S. aureus (SA) | ||||||||
| OXA-S (1,197) | 0.25 | 0.5 | 3 | 74 | >99 | >99 | 100 | |
| OXA-R (781) | 1 | 2 | <1 | <1 | 16 | 65 | 96 | 100 |
| CoNS | ||||||||
| OXA-S (324) | 0.12 | 0.25 | 68 | 97 | 100 | |||
| OXA-R (826) | 1 | 2 | 1 | 7 | 42 | 81 | 93 | >99 |
| S. pneumoniae (SPN) | ||||||||
| PEN-S (141) | ≤0.06 | ≤0.06 | 100 | |||||
| PEN-R (169) | 0.25 | 0.5 | 1 | 55 | 100 | |||
| VGS (289) | ≤0.06 | 0.25 | 89 | 92 | 94 | 95 | 96 | 97 |
| BSH (510) | ≤0.06 | ≤0.06 | >99 | 100 | ||||
Conclusions: BPR displayed a, antibacterial spectrum of activity against EUR pathogens responsible for cSSSI and NP, including OXA-R SA and OXA-R CoNS. BPR was also among the most active β-lactams tested against SPN, equivalent in potency to the carbapenems.
P782 Dalbavancin activity tested against rarely isolated Gram-positive organism species from Europe
R. Jones, H. Sader, T. Fritsche, M. Stilwell (North Liberty, US)
Objectives: To determine the in vitro dalbavancin activity against European isolates of Gram-positive species that may rarely be pathogens in significant infections. A large surveillance platform was utilised (SENTRY Antimicrobial Surveillance Program) to evaluate over 1,300 strains (≥10 isolates/species) tested by reference (CLSI) methods against dalbavancin and selected comparators.
Methods: A total of 1,314 strains were available for dalbavancin testing, all isolated since 2003. Only those species (26) with at least 10 isolates were considered representative. A total of 30 locations contributed strains from 13 countries, all tested by broth microdilution method (CLSI) with appropriate solvents and polysorbate-80 (0.002%). Quality control results with S. aureus ATCC 29213, E.faecalis ATCC 29212 and S. pneumoniae ATCC 49619 were within published ranges.
Results: The distribution of tested strains was: Bacillus spp. (19), Corynebacterium spp. (31), enterococci other than E. faecalis/faecium (82; 4 species), Listeria spp. (24), CoNS (699, 8 species), Micrococcus spp. (21) and streptococci not pneumococci or beta-haemolytic (439; 10 species). The overall dalbavancin MIC50 was ≤0.03 mg/L and the MIC90 results ranged from ≤0.03 (streptococci, micrococci) to 0.25 mg/L (S. haemolyticus and S. warnerii). These results were comparable to the MIC90 results for the most studied dalbavancin-indicated species (beta-haemolytic streptococci and S. aureus; MICs, 0.03–0.06 mg/L). Examples of specific species (≥50 strains) and their MIC90 (in mg/L) are: Staphylococcus capitis (0.06), S. hominis (0.06), Streptococcus anginosus (≤0.03), S. bovis (0.06), S. mitis (≤0.03), S. oralis (0.06) and S. parasanguis (≤0.03). All four enterococcal species (avium, casseliflavus, durans, and gallinarum) had the same MIC90 at 0.12 mg/L.
Activity of dalbavancin against 1,314 uncommonly isolated Gram-positive species (Europe 2002–2006)
| Organism (no. tested) | MIC (mg/L) |
||
|---|---|---|---|
| 50% | 90% | Range | |
| B. cereus (19) | ≤0.03 | 0.12 | ≤0.03–0.12 |
| Corynebacterium spp. (31) | 0.06 | 0.12 | ≤0.03–0.25 |
| Enterococci (82; 4 spp.) | 0.06 | 0.12 | ≤0.03–0.25 |
| L. monocytogenes (24) | 0.06 | 0.06 | ≤0.03–0.06 |
| M. luteus (21) | ≤0.03 | ≤0.03 | ≤0.03–0.06 |
| CoNS (699; 8 spp.) | ≤0.03 | 0.12 | ≤0.03–0.5 |
| Streptococci (439; 10 spp.) | ≤0.03 | ≤0.03 | ≤0.03–0.06 |
Conclusions: Dalbavancin exhibited a wide and potent spectrum of activity (MIC90 range, ≤0.03–0.25 mg/L) against European strains of 26 rarely isolated Gram-positive organisms other than the usual indicated species (e.g. S. aureus or S. pyogenes). Generally, the most dalbavancin-susceptible organisms were streptococci species (viridians group) and M. luteus; while S. haemolyticus showed the most elevated MIC values (0.5 mg/L). Dalbavancin demonstrated a wide potential application as therapy for Gram-positive infections encountered in European medical centres.
P783 QT/QTc assessment of ceftobiprole in a single-dose study
D. Balis, R. Strauss, B. Murthy, H. Tiian, G. Noel (Raritan, US)
Objectives: Delayed cardiac repolarisation is a potential side effect of some non-antiarrhythmic drugs. Because delayed cardiac repolarisation has been associated with the increased risk of serious and potentially life-threatening arrhythmia, rigorous characterisation of a drug's effect on the QT/QTc interval is warranted. The current study was conducted to evaluate the effects of ceftobiprole on QT/QTc interval.
Methods: This was a randomised, double-blind, placebo- and positive-controlled, double-dummy, 4-way crossover, single-centre study of ceftobiprole medocaril at therapeutic (500 mg) and supratherapeutic (1000 mg) doses intravenously infused over 2 h in healthy adult volunteers. Sixty subjects received either a single dose of ceftobiprole medocaril (500 mg or 1000 mg) or matching placebo intravenous (i.v.) infusion on Day 1. In addition, the subjects received either a single dose of moxifloxacin 400 mg as a positive control or a matching placebo as oral administration prior to i.v. infusion on Day 1. Serial 12-lead ECGs were measured on Day −1 (baseline) and Day 1 (post-dose). Fridericia correction was used as the primary correction method for statistical evaluation of serial ECG data.
Results: The upper limits of the 90% confidence intervals for difference in mean change in QTcF (QTc interval using Fridericia correction formula) between ceftobiprole 1000mg and placebo (change in QTcF) were below 10ms at all time points. The same result was demonstrated for change in QTcF between ceftobiprole 500 mg and placebo. Therefore, this analysis establishes the noninferiority of ceftobiprole to placebo with respect to QT/QTc interval duration. The lower limit of 90% confidence intervals for the difference in mean change in QTcF between moxifloxacin and placebo was above zero for all time points between 1.25 and 24 h, with a mean difference of 5.5ms to 13.1 ms. Results from this analysis establish assay sensitivity.
Conclusion: In this QT/QTc study conducted in healthy adult subjects, the effect of single i.v. administrations of ceftobiprole at therapeutic (500 mg) and supratherapeutic (1000 mg) doses on QT/QTc prolongation was comparable to placebo.
P784 In vitro activity of ceftobiprole, dalbavancin and tigecycline against methicillin-resistant Staphylococcus aureus strains from hospitalised patients in Belgium
O. Denis, C. Nonhoff, A. Deplano, M. Hallin, R. De Ryck, S. Rottiers, S. Crevecoeur, M.J. Struelens (Brussels, BE)
Background: The in-vitro activity of 20 antimicrobial agents including ceftobiprole, dalbavancin, tigecycline was tested against methicillin-resistant Staphylococcus aureus (MRSA) strains collected during a national survey conducted in 2005 in Belgian hospitals.
Methods: 335 MRSA strains from 116 hospitals were identified by PCR for 16S rRNA, nuc and mecA genes. Strains were genotyped by spa typing, SCCmec type. MICs of 20 antimicrobials were determined by agar dilution method according to CLSI. Resistance genes for aminoglycosides and macrolides-lincosamides-streptogramins were tested by PCR.
Results: By molecular typing, 80% of MRSA strains belonged to 5 epidemic genotypes: spa CC38-SCCmec IV (45%), spa CC8-SCCmec IV (21%), spa CC2-SCCmec II (9%), spa CC2-SCCmec IV (4%), spa CC790-SCCmec IV (4%). The proportion of MRSA strains resistant to aminoglycosides ranged from 2% for gentamicin to 41% for tobramycin; 54% were resistant to erythromycin and 39% to clindamycin. 90% of isolates were susceptible to minocycline and 85% to tetracycline. More than 95% of strains were susceptible to fusidic acid, rifampin and cotrimoxazole. 93% of isolates were resistant to ciprofloxacin and moxifloxacin. 2.7% of strains were high level resistant (MIC > 524 mg/l) to mupirocin. No strains resistant to glycopeptides, dalbavancin, linezolid, tigecycline or ceftobiprole were detected. Resistance to aminoglycosides was mainly encoded by ant4 gene and to MLS by ermA and ermC genes. The table shows MIC distributions for the new agents.
| Antimicrobial agent | MIC (mg/L) |
||
|---|---|---|---|
| MIC50 | MIC90 | Range | |
| Dalbavancin | 0.12 | 0.12 | 0.06–1 |
| Tigecycline | 0.25 | 0.25 | 0.06–0.5 |
| Ceftobiprole | 1 | 2 | 0.5–4 |
| Vancomycin | 0.5 | 1 | 0.25–1 |
Conclusion: Ceftobiprole, dalbavancin and tigecycline showed excellent antibacterial activity against recent isolates of sporadic and epidemic multi-drug resistant MRSA clones from hospitalised patients in Belgium.
P785 In vitro activitiy of dalbavancin against clinical vancomycin-resistant enterococci isolates
B. Saager, M. Horstkotte, H. Rohde, I. Sobottka, P. Heisig (Hamburg, DE)
Objectives: Dalbavancin is a lipoglycopeptide holding bactericidal antimicrobial activity against Gram-positive bacteria bringing forth noscomial infections, e.g. methicillin-resistant Staphylococcus aureus (MRSA). It is currently being filed for FDA approval for complicated skin and skin structure infections. The mechanism of action of lipoglycopeptides is similiar to other glycopeptides (e.g. teicoplanin or vancomycin) by inhibiton of cell-wall synthesis through binding to the terminal D-alanyl-D-alanine of nascent peptidoglycan chains. A collection of genotypically characterised vancomycin-resistant enterococci (VRE) was tested towards their susceptibility with a view to compare the activity of dalbavancin with recently approved and other clinical well proven antibiotics.
Methods: A number of 61 strains previously described as VRE isolated at the University Medical Center Hamburg – Eppendorf (Germany) between 1995 and 2006 was chosen for susceptibility testing. MICs of dalbavancin, daptomycin, linezolid, teicoplanin, vancomycin, ampicillin, and gentamicin were determined by broth microdilution according to CLSI guidelines. Characterisation of clinically relevant glycopeptide genotypes (vanA, vanB, vanC1, vanC2/3) and species identification for enterococci were done by polymerase chain reaction.
Results: A number of 57 isolates were resistant to vancomycin, while 4 four isolates showed susceptibility or intermediate susceptibility to vancomycin. MICs ranged from 4 to >256 for vancomycin and from 16 to 256 for teicoplanin. Corresponding MIC90 values were >256 and 256 for vancoymcin and teicoplanin, respectively. Determination of MICs for dalbavancin showed values ranging from ≤0.03 to >32 with a MIC90 of 16. The MIC90 values for daptomycin and linezolid were 4 and 2 respectively, MICs were ranging from ≤0.016 to 4 for daptomycin and from 1 to 16 for linezolid.
Conclusion: In comparison to the well established antibiotics vancomycin and teicoplanin, dalbavancin shows improved in vitro activity against the tested isolates. With regard to the newer antimicrobial agents introducing new mechanism of actions, daptomycin and linezolid, the in vitro activity of dalbavancin was comparable. Thus, dalbavancin is a potential drug for the treatment of VRE, with its awaiting approval hopefully supplementing the arsenal of antimicrobial drugs for the treatment of nosocomial infections.
P786 Susceptibility of Staphylococcus aureus, Enterococcus faecium and Enterococcus faecalis to daptomycin at a university medical centre Hamburg-Eppendorf, Germany
B. Saager, M. Horstkotte, H. Rohde, I. Sobottka, P. Heisig (Hamburg, DE)
Objectives: Daptomycin (Cubicin®) is the first lipopeptide bactericidal antimicrobial agent and introduces a new mechanism of action against Gram-positive bacteria. By binding to and integrating into bacterial membranes it causes a rapid depolarisation of membrane potential resulting in the inhibition of metabolism and ultimately in cell death. Being approved for the treatment of complicated skin and skin structure infections due to S. aureus, including methicillin resistant S. aureus (MRSA) and E. faecalis, daptomycin presents a promising therapeutical option for treatment of noscomial Gram-positive pathogens.
To investigate the susceptibility of daptomycin facing isolates with multiple resistance phenotype in vitro, clinical isolates of S. aureus, E. faecium and E. faecalis were chosen for susceptibility testing against daptomycin and established clinically important antibiotics.
Methods: 285 strains being isolated at the University Medical Center Hamburg – Eppendorf (Germany) between 2004 and 2006 were chosen for susceptibilty testing. This collective comprised 221 isolates of S. aureus including 15.4% MRSA, 41 isolates of E. faecalis and 23 isolates of E. faecium with two isolate of E. faecium being resistant to vancomycin. MICs of daptomycin, penicillin, oxacillin, erythromycin, ciprofloxacin, gatifloxacin, moxifloxacin, sparfloxacin gentamicin, linezolid, teicoplanin, and vancomycin were determined for S. aureus and those of daptomycin, ampicillin, linezolid, teicoplanin, vancomycin and gentamicin for E. faecalis and E. faecium by broth microdilution according to CLSI guidelines.
Results: All strains tested of S. aureus were susceptible to daptomycin (MICs ≤ 1 μg/mL) as well as all strains of E. faecium and E. faecalis (MICs ≤4 μg/mL). MICs were ranging between 0.03–1 for S. aureus and 0.25–4 and 0.5–4 for E. faecalis and E. faecium, respectively. MIC90 values were 0.5 (S. aureus), 2 (E. faecalis) and 4 (E. faecium). A total number of 25 S. aureus (11.3%) isolates with multiple resistant phenoytpe were found showing resistance to ciprofloxacin, erythromycin and oxacillin.
Conclusion: Our data show that daptomycin offers good in vitro activity against S. aureus, E. faecalis and E. faecium, including isolates with multiple-resistant phenotype.
P787 Comparative in vitro activity of dalbavancin against clinical isolates of vanA, vanB, and vanC enterococci
E. Cercenado, O. Cuevas, M. Marin, M.J. Goyanes, E. Bouza (Madrid, ES)
Objectives: Dalbavancin (DAL) is a new lipoglycopeptide with activity against Gram-positive microorganisms. Its half-life of 8.5 days enables once-weekly dosing. In this study we compared the activity of DAL with other agents against clinical isolates of vancomycin-resistant enterococci (VRE).
Methods: We tested a total of 238 well characterised non-duplicate clinical isolates of VRE recovered from patients at our institution. Origin of isolates were: wounds (69 isolates), urine (45), blood (38), peritoneal fluid (24), abscesses (17), other sterile fluids (12), intravascular catheter (12), and others (21). Multiplex PCR was used to detect vanA, vanB, vanC-1, and vanC-2 genes. The isolates were 83 E. faecalis (64 vanA, 19 vanB), 80 E. faecium (37 vanA, 43 vanB), 50 E. gallinarum (vanC1), and 25 E. casseliflavus (vanC2). Susceptibility testing was performed by the broth microdilution method in cation-adjusted Mueller-Hinton broth following CLSI guidelines. The antimicrobial agents evaluated were: dalbavancin (DAL), vancomycin (VAN), teicoplanin (TEI), and linezolid (LZD). E. faecalis ATCC 29212 and S. aureus ATCC 29213 were used as control strains.
Results: The MIC50, MIC90, and MIC range values (mg/L) are summarised in the table .
| Organisms and type (No.) | Antimicrobials | MIC (mg/L) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| VanA VRE (101) | Dalbavancin | 8 | 64 | ≤0.12–64 |
| Vancomycin | 512 | 1,024 | 256–1,024 | |
| Teicoplanin | 64 | 128 | 32–128 | |
| Linezolid | 1 | 2 | 1–2 | |
| VanB VRE (62) | Dalbavancin | ≤0.12 | 1 | ≤0.12–2 |
| Vancomycin | 32 | 512 | 16–1,024 | |
| Teicoplanin | 0.5 | 0.5 | ≤0.12–1 | |
| Linezolid | 1 | 2 | 1–2 | |
| VanC VRE (75) | Dalbavancin | ≤0.12 | ≤0.12 | ≤0.12–0.5 |
| Vancomycin | 8 | 16 | 4–16 | |
| Teicoplanin | 0.25 | 0.5 | ≤0.12–0.5 | |
| Linezolid | 1 | 2 | 1–2 | |
Conclusions: Dalbavancin was more active than comparator glycopeptides and presented good activity against vanB, and vanC enterococci, however, it was inactive against vanA enterococci.
P788 Efficacy and safety of Uro-vaxom treatment for patients with recurrent cystitis: open multicentre study
U-S. Ha, S-G.C. Chang, H.K. Park, S.J. Yoon, Y-H. Cho (Suwon, Seoul, Incheon, KR)
Objectives: To investigate the efficacy and safety of the immunotherapeutic Uro-vaxom in uncomplicated recurrent cystitis in female patients only.
Methods: Adult female patients could enroll in this multicentre, open-label study if they had acute cystitis at the enrollment visit and positive results of urine culture (≥103 CFU/mL). Patients were treated for 3 months with one capsule daily of Uro-vaxom after antibiotic therapy and observed for further 3 months. Primary efficacy criteria were cystitis recurrence rates over 6 months, distribution of cystitis and proportion of patients of cystitis.
Results: A total of 50 patients were evaluated. During the 6-month trial the number of recurrences from cystitis was significantly reduced in comparison with the 6-month pretrial period (on the average 0.64 as compared to 3.0 recurrences, p < 0.001). The case for the incidence of frequency, urgency and dysuria was remained low until the end of the trial. Uro-vaxom was well tolerated: side-effects were mentioned in 8% of the 50 patients, in the absence of case leading to treatment withdrawal.
Conclusions: Uro-vaxom significantly reduced the incidence of cystitis during the 6 months of the study including 3 months of treatment. These results demonstrate that Uro-vaxom is a valuable product for the prophylaxis of recurrent cystitis.
P789 A comparison of the CLSI (formerly NCCLS), EUCAST and various EU country standard methods for the susceptibility testing of retapamulin, a novel pleuromutilin
L. Williams, A. Colclough, D. Felmingham (London, UK)
Objective and Methods: Retapamulin[RE] is the first semi-synthetic pleuromutilin formulated as a topical antibacterial for treating skin infections. In this study, minimum inhibitory concentrations (MIC) and inhibition zone diameters of RE were determined for selected populations of Staphylococcus aureus [SA] (1000 isolates) and Streptococcus pyogenes [SP] (503) using CLSI standard methods and compared with various methods used in EU countries including those defined by EUCAST, DIN, SRGA, BSAC and the CA-SFM.
Results: For isolates of SA, there was very good correlation between MICs of RE determined by broth microdilution using CLSI, with those by EUCAST, DIN and SRGA methods (median MIC 0.06 mg/L for all methods; >90% of results within ±1 doubling dilution of each other). Similar results were observed when MICs of RE determined by agar dilution using CLSI, were compared to those determined by EUCAST, DIN, SRGA, BSAC and CA-SFM methods. Close agreement was observed between inhibition zone diameters (median diameters within 1 mm) using CLSI compared to DIN, SRGA, BSAC and CA-SFM methods. For isolates of SP, MICs of RE determined using the broth microdilution methods of the CLSI, EUCAST, DIN and SRGA were all in close agreement (median MIC 0.015–0.03 mg/L; >95% of results within ±2 doubling dilutions). For agar dilution, there was also good correlation between MICs determined using CLSI compared to EUCAST, SRGA, BSAC and CA-SFM (median MIC 0.06 mg/L for all methods; >95% of results within ±1 doubling dilution). However, there was poor agreement between MICs obtained by the CLSI and DIN agar dilution methods. It is possible that the difference in the recommended amount of lysed horse blood (2% for DIN and 5% for CLSI) had an impact, because the median MIC by the DIN method (0.015 mg/L) was four-fold lower than by CLSI and only 44.73% of results within ±2 doubling dilutions of each other. Consistent with these results, inhibition zone diameters obtained using CLSI, SRGA, BSAC and CA-SFM correlated well (median values 20–22 mm) but those produced using the DIN method were appreciably larger (median 29 mm).
Conclusion: Minor differences were seen for both SA and SP, in MICs and inhibition zone diameters determined by the various methods examined in this study and such differences are unlikely to affect qualitative interpretation of MICs. The differences noted between CLSI and DIN methods warrant further investigation.
P790 In vitro activity of retapamulin, a novel pleuromutilin, against Staphylococcus spp. (n = 1413) and Streptococcus pyogenes (n = 503) from 26 European centres
L. Williams, J. Northwood, N. Crowhurst, D. Felmingham (London, UK)
Background and Objectives: Retapamulin (RE) is the first agent of the novel pleuromutilin class, formulated as a semi-synthetic topical antibacterial for treating skin infections. It has a unique mode of action, a low potential for the development of resistance and no target specific cross-resistance to other antibacterial classes. The in vitro activity of RE against Staphylococcus spp. (n = 1413) and Streptococcus pyogenes (SP) (n = 503) collected from 26 centres in 14 European countries (January 2003 – March 2005) was compared with that of erythromycin (ERY), fusidic acid (FUS), gentamicin (GEN) and mupirocin (MUP).
Methods: All testing was conducted using CLSI methods.
Results: MIC90 of RE, ERY, FUS, GEN and MUP for isolates of S. aureus (SA), S. epidermidis (SE), other coagulase-negative staphylococci (CNS) and SP are shown in the table . 99.9% of the isolates of Staphylococcus spp. were inhibited by ≤2 mg/L RE this activity being unaffected by resistance to methicillin, ERY, FUS or MUP (MIC90 range 0.06–0.5 mg/L). All isolates of SP were inhibited by ≤0.06 mg/L (MIC90 0.03 mg/L).
| MIC90 (mg/L) |
||||||
|---|---|---|---|---|---|---|
| Species | Phenotype (n) | RE | ERY | FUS | GEN | MUP |
| SA | All isolates (1048) | 0.12 | ≥128 | 4 | ≥64 | 0.5 |
| MRSA (318) | 0.12 | ≥128 | ≥64 | ≥64 | 16 | |
| ERYR (369) | 0.12 | ≥128 | 8 | ≥64 | 16 | |
| FUSR (144) | 0.12 | ≥128 | ≥64 | ≥64 | 0.25 | |
| MUPR (76) | 0.12 | ≥128 | 0.25 | ≥65 | ≥0.25 | |
| SE | All isolates (256) | 0.06 | ≥128 | 16 | ≥64 | ≥128 |
| MRSE (178) | 0.12 | ≥128 | 16 | ≥64 | ≥128 | |
| ERYR (159) | 0.06 | ≥128 | 16 | ≥64 | ≥128 | |
| FUSR (89) | 0.12 | ≥128 | 32 | ≥64 | ≥128 | |
| MUPR (73) | 0.06 | ≥128 | 16 | ≥64 | ≥128 | |
| Other CNS | All isolates (109) | 0.25 | ≥128 | 8 | 4 | ≥128 |
| MRCNS (75) | 0.25 | ≥128 | 8 | 4 | ≥128 | |
| ERYR (49) | 0.12 | ≥128 | 8 | 16 | ≥128 | |
| FUSR (56) | 0.5 | ≥128 | 16 | 2 | 4 | |
| MUPR (12) | 0.25 | ≥128 | 16 | 4 | ≥128 | |
| SP | All isolates (503) | 0.03 | 16 | 8 | 8 | 0.12 |
| ERYR (94) | 0.03 | ≥128 | 8 | 8 | 0.25 | |
Conclusions: RE was the most potent agent tested in vitro and has excellent activity against Staphylococcus spp. (including those isolates resistant to either MET, ERY, FUS or MUP) and Streptococcus pyogenes (including ERY-resistant strains). It has potential for the topical therapy of uncomplicated skin infections involving these organisms.
P791 Novel small-molecule inhibitors of gyrase B: antibacterial activity in vitro and in murine models of infection
D. Haydon, P. Lancett, J. Bennett, L. Czaplewski (Oxford, UK)
Objectives: The bacterial type II topoisomerases, DNA gyrase and topoisomerase IV, are well-validated targets for antibacterial therapy. A series of novel compounds have been synthesized that inhibit the ATPase activity of these enzymes. The objective of this work was to explore the structure activity relationships (SAR) of the series and to progress the compounds through preclinical development towards candidate nomination.
Methods: The biochemical activity of compounds was determined using an in vitro ATPase assay. Minimal inhibitory concentrations (MIC) were determined using the CLSI broth microdilution method. Resistance frequency, time-kill and post-antibiotic effect were investigated using standard microbiological methods. The ADMET profile of various compounds was evaluated using in vitro methodology. Compounds from the series were evaluated in acute systemic infection models in mice.
Results: A medicinal chemistry programme to explore the SAR of the compound series resulted in the synthesis of potent derivatives that have on-target activity against a range of species. Compounds within the series have nanomolar IC50s against gyrase and topo IV ATPase and single-digit, or better, microgram/mL MICs against S. aureus, E. faecalis, S. pneumoniae, M. catarrhalis and H. influenzae. The spontaneous resistance frequency observed for the series is low, consistent with dual targeting of DNA gyrase and topo IV. The compounds have a cidal mode of action with a time, but not concentration, dependant kill and no post-antibiotic effect. The protein binding, cytotoxicity, chemical, plasma and microsomal stability, Caco-2 permeability, cytochrome P450 and hERG ion channel activities of the series have been explored.
Selected derivatives have been characterised in vivo. The compounds were well tolerated and there was good bioavailability following IP or oral administration. Selected compounds demonstrated efficacy in murine septicaemia models of staphylococcal infection.
Conclusion: The properties of the inhibitors are consistent with a compound series capable of optimisation into an antibiotic targeting both respiratory tract and drug-resistant Gram-positive infections.
P792 Comparative activity of tigecycline, cefepime and imipenem against clinical isolates of Klebsiella pneumoniae producing extended spectrum β-lactamases, plasmid-mediated AmpC-type β-lactamases or both, associated or not with porin deficiency
M. Conejo, J. Hernández, A. Pasual (Seville, ES)
Objective: To study the in-vitro activity of tigecycline against clinical isolates of Klebsiella pneumoniae producing extended spectrum β-lactamases (ESBL), plasmid-mediated AmpC-type β-lactamases (pACBL) or both, associated or not with porin (POR) deficiency.
Methods: Fifty K. pneumoniae clinical strains were studied, including 2 ESBL-/pACBL-/POR-, 10 ESBL+/POR+, 12 ESBL+/pACBL-/POR-, 18 ESBL-/pACBL+/POR+, 1 ESBL-/pACBL+/POR-, 4 ESBL+/pACBL+/POR+, and 3 ESBL+/pACBL+/POR-. Antimicrobial susceptibilities to tigecycline, cefepime and imipenem were determined by broth microdilution (CLSI guidelines) with standard and 10-fold higher inocula. The inoculum effect was defined as an eight-fold or greater MIC increase on testing with the highest inoculum.
Results: The MIC50 of tigecycline, cefepime and imipenem were 0.25, 2 and 0.5 mg/L respectively, the MIC90 were 1, 128 and 1mg/L, respectively and the percentages of susceptible isolates were 98, 80 and 94%, respectively. Tigecycline was the most active drug, with no resistance detected and only one isolate with a MIC of 4 mg/L (1 isolate ESBL-/pACBL-/POR-). No inoculum effect was observed with this antimicrobial agent. Imipenem showed excellent activity, except for the group ESBL+/pACBL+/POR-, and inoculum effect was only observed in pACBL+ strains. Cefepime was the least active drug, and inoculum effect was very frequently observed among ESBL+ and pACBL+ isolates.
Conclusions: Tigecycline showed excellent activity against clinical isolates of Klebsiella pneumoniae producing ESBL, pACBL or both, associated or not with POR deficiency. Its activity was comparable to that of imipenem against isolates producing ESBL, and higher than imipenem against pACBL producers.
P793 Pharmacokinetic/pharmacodynamic modelling of the plasma bactericidal activity of NXL103 against Streptococcus pneumoniae and Staphylococcus aureus in phase I studies
H. Merdjan, J. Lowther, A-M. Girard, C. Delachaume, M. Rangaraju (Romainville, FR)
Objective: NXL103 is a novel oral streptogramin antibiotic associating 2 components RPR202868 (PI) and RPR132552 (PII). The objective of this study was to characterise the relationship between the plasma bactericidal activity against S. pneumoniae and S. aureus strains, and PI and PII plasma concentrations measured in samples collected in 2 phase I studies.
Methods: The dataset for building the PK/PD model was composed of 1067 plasma samples from a phase I repeated (10-day) oral administration study in healthy male volunteers. Four cohorts of 10 subjects (8 received NXL103 and 2 placebo) received 0.5 g bid, 0.75 g bid, 1.5 g oad, or 1 g bid. Blood samples were taken on 7 occasions from day 1 to day 11. PI and PII concentrations were measured in plasma by LC-MS/MS. Separate aliquots of the same samples were serially diluted into a growth medium/control plasma mixture, and incubated with S. pneumoniae (030MV1 strain) and S. aureus (011HT18 strain) to measure bactericidal activity. For the purpose of PK/PD modelling, cidality data were recoded as 0 (no activity) or 1 (cidal activity), irrespective of the intensity of effect. Fits were performed using WinNonlin®.
Results: The model predicted bactericidal activity on S. pneumoniae if both PI and PII were more than 0.175 mg/L and 0.523 mg/L, respectively. Breakpoints for S. aureus were 0.123 mg/L and 0.081 mg/L, respectively. The descriptive performance of the model was 93.5% and 83.9% accurate against S. pneumoniae and S. aureus, respectively. The predictive performance of the model was assessed on a separate validation dataset composed of 416 human plasma samples collected in a previous study, where 32 healthy male volunteers received a single oral dose of either 0.5 g, 1g, 1.5 g or 2 g of NXL103. Plasma concentrations of PI and PII, and bactericidal activity were measured as above. Model-predictions were 91.8% and 93.5% accurate against S. pneumoniae and S. aureus, respectively, the remainder being evenly distributed between false positive and false negative predictions.
Conclusion: A PK/PD model was built that predicts the plasma bactericidal activity against two pathogens of therapeutic interest. These results should be taken into account in defining dose(s) for phase II trial(s).
P794 The efficacy of ceftazidime combined with NXL104, a novel β-lactamase inhibitor, in a mouse model of kidney infections induced by β-lactamase producing Enterobacteriaceae
M. Borgonovi, C. Miossec, J. Lowther (Romainville, FR)
Objectives: NXL104 is a novel β-lactamase inhibitor that has been shown in vitro and in vivo to inhibit both class A and class C β-lactamases. The occurrence of Enterobacteriaceae producing extended-spectrum β-lactamases and AmpC enzymes needs to be considered in the therapy of complicated urinary tract infections (UTI). The aim of the study was to demonstrate in a murine model that the combination of ceftazidime (CAZ) with NXL104 restored the bactericidal efficacy of CAZ against strains refractive to CAZ alone due to β-lactamases.
Methods: Kidney infections were induced in immunodepressed, anaesthetised male CD1 mice by direct injection of 104 cells in 0.02 ml of exponentially growing culture by 25 gauge needle. Typically the kidney bacterial burden increased by 1.5log10 within 48 hours. Therapy commenced subcutaneously 4 hours after infection bid for 2 days. Bacteria were enumerated in the kidneys of treated and control mice 48 hours post infection.
Results: CAZ alone was ineffective against all 6 strains tested compared to the non-treated control group. The combination CAZ/NXL104 (4:1) was effective in a dose range 10–25 mg/kg in reducing the inoculum and preventing proliferation of Escherichia coli (one ClassA and one AmpC), Enterobacter cloacae (AmpC), Klebsiella pneumoniae (ClassA + AmpC), Morganella morganii (AmpC) and Citrobacter freundii (AmpC). In each case CAZ/NXL104 was significantly effective, reducing bacterial kidney burden by 2.6 to 4.5log10 compared to the CAZ treated group (p < 0.05, Bonferroni).
Conclusion: The combination CAZ/NXL104 (4:1) was effective against representative strains of CAZ-resistant Enterobacteriaceae species in a murine kidney infection model. This combination could represent a useful therapeutic option for the treatment of infections due to β-lactamase producing Enterobacteriaceae species, which are increasing in frequency in complicated UTI.
P795 Tolevamer, a novel toxin-binding polymer, neutralises the large clostridial toxins from the B1/027 epidemic strain of Clostridium difficile
R. Barker, P. Hinkson, J. Klinger, D. Davidson (Waltham, US)
The incidence of C. difficile-associated diarrhoea (CDAD) has more than doubled over the past decade, accompanied by an increase in the proportion of isolates associated with the epidemic strain BI/027. The BI/027 epidemic is associated with greater morbidity and mortality than previously seen with CDAD, and this strain has been shown to produce high levels of the large clostridial toxins in vitro, perhaps due to a mutation in the tcdC gene, a putative negative regulator of toxin production. Tolevamer is a non-antibiotic polymer that is being developed as a novel therapy for CDAD. Previous studies have shown that tolevamer binds toxins A and B (CdtA and CdtB) in vitro, preventing cell-rounding in the Vero cell cytotoxicity assay. Furthermore, tolevamer prevented mortality in a hamster model, and most importantly, was not inferior to vancomycin with respect to resolving diarrhoea in a phase II clinical study.
Objectives: Given their virulence and rising incidence, the capacity of tolevamer to neutralise CdtA and CdtB from BI/027 isolates obtained from different geographic areas was assessed.
Methods: Cultures were grown in broth culture for 48hr, the supernatant was removed, centrifuged, and sterile filtered. Supernatants were diluted onto confluent Vero cell monolayers and toxin titre was determined by microscopic examination of cell morphology after 24hr. The titre used for tolevamer neutralisation studies was defined as 2-fold greater than the minimum concentration required for 100% cell rounding. For toxin neutralisation studies, dilutions of tolevamer were added to Vero cell monolayers in combination with tittered supernatants; cells were incubated 24 hr and then were assessed microscopically. Neutralisation was defined as that concentration of tolevamer that prevented >90% cell-rounding.
Results: Titred supernatants from all strains tested were neutralised by the same concentration of tolevamer.
Conclusions: Tolevamer neutralises the large clostridial toxins from the epidemic BI/027 strain as well as it does toxins from other C. difficile strains.
P796 In vitro and in vivo pharmacokinetics of 2-methylamino-benzodiazepines to treat Helicobacter pylori infections
J. Newman, M. Rooney, B. de Jonge, A. Kutschke, C. Cederberg (Waltham, US; Molndal, SE)
Objectives: 2-Methylamino-benzodiazepines were identified as selective H. pylori inhibitors that suppress growth through MurI, an enzyme essential for peptidoglycan synthesis. We describe the pharmacokinetics of these analogs and the pharmacodynamic parameters required for gastric H. pylori monotherapy.
Methods: In vitro kinetics were determined upon exposure to cells to various concentrations of compound. In vivo pharmacokinetics were determined after intravenous and oral dosing in rats and mice. Drug levels were measured in plasma and gastric juice by LC-MS/MS. In vivo efficacy against H. pylori was assessed in a 4-day mouse model. Pharmacodynamics for amoxicillin were determined in this model as a comparator, since it also blocks cell-wall biosynthesis with slow killing kinetics.
Results: 2-Methylamino-benzodiazepines were determined to be cidal agents in vitro with slow killing kinetics. They exhibited moderate clearance in rats (∼20 ml/min/kg) and moderate bioavailability (∼40%). In addition, these compounds were detected in rat gastric juice after i.v. dosing, indicating penetration to the site of infection. Analysis of amoxicillin efficacy in mice, and correlation to plasma drug levels, indicated that T>MIC for 24 hours was the index associated with suppression of infection. Therefore, benzodiazepines were administered to mice in various dosing schedules designed to exceed the MIC for the full 4-day course of therapy. Multiple daily dosing, and use of a metabolism inhibitor, produced drug levels above the MIC for most of day 1, but levels were lower on days 2–4. Consequently, no suppression of infection was observed.
Conclusions: Analogs in this series exhibit suitable in vitro killing kinetics with good PK properties in rodents, but efficacy against H. pylori was hampered by sub-optimal exposure.
Host parameters of infection
P797 Correlation of C-reactive protein to complications and outcome during the intensive care unit course of sepsis patients
O. Alici, N. Bavbek, A. Kargili, A. Kaya, F. Karakurt, A. Kosar (Ankara, TR)
Purpose: Elevated concentrations of CRP have been reported to be in correlation with increased risk of death in different patient groups. There are conflicting results about the prognostic role of CRP in sepsis patients. To determine the prognostic informative value of CRP in patients admitted to the ICU with sepsis, 213 patients hospitalised in our unit were retrospectively studied.
Materials and Methods: Demographic data, comorbid diseases, all clinical, microbiological, and laboratory data were registered retrospectively. Plasma levels of CRP on days 1, 3, 5 and 7 were registered.
Results: The overall mortality rate was 37%. Nonsurvivors had significantly higher CRP levels than survivors on the 3rd day (150.8±21.9 mg/L vs 96.4±6.5 mg/L, p = 0.002). Survivors had significantly lower CRP levels on the 3rd day than the 1st day (96.4±6.5 mg/L vs 160.2±11.8 mg/L, p = 0.000) while nonsurvivors had significantly higher CRP levels on the 3rd day than the 1st day 150.8±21.9 mg/L vs 130.9±17.3 mg/L, p = 0.000). In patients with sepsis, age and diabetes mellitus were the important prognostic variables for intensive care unit mortality.
Conclusions: In sepsis, progressively increasing concentrations of CRP are correlated with increased risk of organ failure and death. Serial measurements are helpful to identify these patients who require more intensive treatment to prevent complications.
P798 The investigation of correlation between serum cytokine levels, C-reactive protein and erythrocyte sedimentation rate in acute brucellosis
N. Ates Arica, F.S. Erdinc, G. Tuncer Ertem, C. Bulut, M.A. Yetkin, U. Onde, B. Oral, A.P. Demiröz (Ankara, TR)
Objectives: Cytokines are thought to have an important role in the pathogenesis of brucellosis and the T helper 1/T helper 2 balance may be involved in the susceptibility or resistance to the disease. Effective immune response is mediated by T helper 1 cells in brucellosis. The aim of this study was to detect serum interferon gamma (IFN-γ), interleukin-6 (IL-6) and interleukin-12 (IL-12) levels in patients with acute brucellosis and to investigate the correlation between those cytokines, C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR).
Methods: Fifty patients with acute brucellosis followed between August 2002 and August 2006 in Ankara Research and Training Hospital Infectious Diseases and Clinical Microbiology Department were included in to the study. The control group included 31 healthy persons. Serum cytokine levels were measured by ELISA method. ESR values were determined by Westergren method and CRP levels were determined by nephelometric method. Data were analysed with SPSS 13.0 for Windows.
Results: The mean serum levels in patients with acute brucellosis and control group were as follows; IFN-γ; 14.2 pg/mL and 0.4 pg/mL (p = 0.021), IL-6; 44.7 pg/mL and 2.4 pg/mL (p = 0.012) and IL-12; 176.0 pg/mL and 76.9 pg/mL (p = 0.001), respectively. The difference between two groups was stastically significant (p < 0.05).
In acute brucellosis group, for CRP, we found positive correlation with IFN-γ, IL-12 and ESR (r = 0.31, p < 0.05; r = 0.31, p < 0.05; r = 0.53, p < 0.01, respectively). Also there was a positive correlation between IL-12 and IFN-γ (r = 0.62, p < 0.01). There was no significant correlation between ESR and the cytokines. For IL-6, there was not any significant correlation with the other parameters.
Conclusion: In conclusion, our study is in accordance with the results of the previous studies suggesting that immunity against brucellosis is mediated by T helper 1 cells. CRP and ESR are proved parameters to be related with acute brucellosis. Significantly high IFN-γ, IL-6 and IL-12 levels in acute brucellosis might imply the role of the cytokines in the pathogenesis of brucellosis.
P799 Phagocyte Fcg receptors expression in bacteraemia
A.A. Garcia-Egido, M.A. Escobar, F.J. Fernandez, F.M. Gomez-Soto, P. Ruiz, M. Rosety, C. Rivera, F. Gomez (El Puerto Santa Maria, ES)
Early markers of bacteraemia are useful for prognosis and, in decision making for i.v. antibiotic therapy.
Objectives: To assess the diagnostic power of the surface expression of Fc receptors for IgG (FcgRs) for the prediction of bacteraemia in febrile patients.
Methods: We performed a 4 year prospective case-control study on 166 consecutive patients (pts) with an episode of bacteraemia as compared to 174 randomly selected concurrent febrile pts with negative blood cultures (control). Demographic and clinical data were collected by chart review and/or questioning their attending physicians. Plasma levels of C-reactive protein (CRP), TNFg, IL-1a, IL-6, IL-8 and IL-10 were determined. The surface expression of Fc receptors for IgG (FcgRs): FcgRI, FcgRII and FcgRIII on peripheral blood monocytes (M) and granulocytes (G) was assessed by flow cytometry. These studies were done concomitantly with blood cultures.
Results: Both groups were not different for age, gender, previous administration of immunosupressants or antibiotics, clinical severity index or comorbid conditions. In univariate analysis, cases had significantly higher levels of CRP (p < 0.001), TNFg (p < 0.001), IL-1a (p < 0.001) and IL-6 (p < 0.01) than controls. The expression of FcgRIIA and FcgRIII by M and, that of FcgRI on G was significantly enhanced (p < 0.001) in bacteriemic patients as compared to culture-negative febrile pts; while the expression of FcgRIIB by either M or G was significantly decreased (p < 0.03). Setting a cut-off value = 25% of the mean fluorescence intensity over controls for FcgRs surface expression and, assuming a prevalence of bacteraemia of 5–10% among hospitalised patients undergoing blood cultures, results in a sensitivity, specificity, positive and, negative predictive values of: 77%, 97%, 74%, and 98%, respectively for M-FcgRIIA, 73%, 96%, 74% and 97%, respectively for M-FcgRIII, 58%, 93%, 49% and 95%, respectively for G-FcgRI and, 71%, 91%, 57% and 83%, respectively for G-FcgRIIB.
Conclusions: Our results suggest that the surface expression of Fc receptors for IgG on peripheral blood monocytes and granulocytes may help clinicians to rule out bacteraemia in febrile patients.
P800 Modification of a haemolytic assay for the measurement of functional human mannose-binding lectin by classical pathway inhibition
B.A.W. de Jong, B.L. Herpers, B. Dekker, P.C. Aerts, H. van Dijk, G.T. Rijkers, H. van Velzen-Blad (Nieuwegein, Utrecht, NL)
Objective: Low levels of functional mannose-binding lectin (MBL), impairing early complement activation via the lectin pathway, have been associated with disease. Therefore, diagnostic procedures for MBL levels and lectin pathway activity are widely performed. In a previously described assay MBL levels are measured by bystander-haemolysis after binding mannan on Saccharomyces cerevisiae (Kuipers et al, 2002). All down-stream complement components are provided by MBL-deficient serum. As both the ligand-binding and complement-activating properties are assessed simultaneously without interference of downstream deficiencies, this is the only assay to quantitatively measure the level of functional MBL. Interference by classical pathway activity has been postulated, for example by C1q-binding immune complex formation of antibodies to S. cerevisiae (ASCA). Here we describe the classical pathway interference in the haemolytic MBL assay and the modification of this assay to prevent this artifact.
Methods: To determine the minimal inhibitory concentration, classical pathway activity was inhibited in a CH50 assay by preincubation of three 1:10 diluted ASCA-positive test sera with 0–150 ug/mL anti-C1q monoclonal antibodies. To examine the interference of classical pathway activity in the functional MBL assay, haemolytic activity was assessed in the MBL assay in absence and presence of 50 ug/mL anti-C1q in the three ASCA-positive sera and in 35 selected sera of donors with known genotypes.
Results: Anti-C1q inhibited all classical pathway activity dose-dependently, with full inhibition at 50 ug/mL. In three selected ASCA-positive sera, anti-C1q decreased haemolysis in the MBL assay in two samples (figure 1a ).
Figure 1.

Effect of anti-C1q on haemolysis in the haemolytic MBL assay in (a) three selected ASCA-positive sera and (b) 35 sera of known genotypes.
Most of the 35 selected sera did not show a significant difference with or without anti-C1q (figure 1b). A striking effect was found in some sera from donors with deficient genotypes XA/0 and 0/0, where anti-C1q decreased haemolytic activity completely.
Conclusion: The haemolytic MBL assay is designed to measure functional MBL levels by its ability to bind its ligand and activating complement, without interference of down-stream deficiencies. However, in some samples classical pathway activity can interfere with this functional MBL measurement, either via ASCA or other C1q-binding immune complex formation. This interference can and should be inhibited by anti-C1q to prevent that MBL deficiency is overlooked and patients are misdiagnosed.
Antimicrobial effects and mechanisms
P801 Eradication of induced mouse fibrosarcoma with combination of verotoxin1 and monophosphoryl lipid A
H. Hosain Zadegan, M. Sattari, Z. Mohammad Hasan, A. Allameh (Khorram Abad, Tehran, IR)
Objectives: Because of disadvantages of routine cancer therapies such as chemotherapy and radiotherapy, four essential approaches of different microbial metabolites have been used recently in treatment of cancers. These are enterotoxins, bacterial proteins, recombinant toxins, and combination therapies. Antitumour effects of verotoxin1 have known from several years ago, that produced from some kinds of Escherichia coli. Our purpose was to study the synergistic cytotoxicity of verotoxin1 and monophosphoryl lipidA (MPL) a nontoxic derivative of lipopolysaccharide on induced fibrosarcoma of Balb/c.
Methods: For determining treatment doses of Verotoxin1 and MPL on animals, lethal dose 50 percent of them calculated with Reed & Munch method. Then intradermal tumours (mean diameter of 0/7–1 centimeter) induced with WEHI-164 cells on back of animals, and divided in four groups. Group 1 used as control, and groups 2, 3, 4 injected with MPL, verotoxin1, and combinations of MPL and verotoxin1 respectively. After treatment, eradication or decrease in volume of tumours monitored daily and compared in 4 groups.
Results: verotoxin1 like standard vero cell line indicated cytotoxicity on WEHI-164 line in In vitro experiments. Simultaneous treatment of tumours with verotoxin1 & MPL eradicated malignant fibrosarcoms of balb/c. Animals of control group died after severe weight loss in comparison.
Conclusion: Our results indicate that verotoxin1 have synergistic effects with MPL in eradication of malignant fibrosarcomas of Balb/c. And after further studies, it can be used for treatment of different tumours. Application of tow microbial metabolite as a treatment of tumours proposed and studies firstly in this study. On the other hand results indicate that tow factors have synergy in pathogenesis of verotoxigenic E. coli infections.
P802 Effect of Berberis aetnensis C. Presl extract on tissue transglutaminase expression in primary astroglial cell cultures exposed to glutamate
A. Campisi, A. Speciale, R. Acquaviva, G. Raciti, I. Barbagallo, S. Puglisi, L. Iauk (Catania, IT)
Berberis aetnensis C. Presl (Berberidaceae) is a bushy spiny shrub presents on Mount Etna (Sicily). It contains several alkaloids showing several pharmacological properties (febrifugal, hypotensive, immunostimulating, antinflammatory). It is well known that excitotoxicity is a common feature in various neurological disorders. The excitotoxicity, increasing the intracellular calcium levels and activating specific genes, results in synthesis of several enzymes involved in cellular stress response, including tissue transglutaminase (TG-2). This is a multifunctional protein implicated in numerous cellular processes, as well as cellular differentiation, signal transduction, cell survival, and apoptosis. In previous researches, we demonstrated that the alterations of cellular redox state, evoked by glutamate in primary astrocyte cultures, are associated with the increase of TG-2 expression, and its effects were reduced by the antioxidant treatments.
In this study, we investigated the protective effects of B. aetnensis extract in glutamate-evoked TG-2 up-regulation in primary rat astroglial cell cultures.
Primary rat astroglial cell cultures were prepared from newborn albino cerebral cortex (1–2-day-old Wistar strain), maintained in cultures for two weeks, and exposed to 500 μM of glutamate for 24 hrs. Some cultures were also treated with the methanolic extract of plant roots at different concentrations and then exposed to the neurotransmitter.
The exposure of the astroglial cell cultures to glutamate caused a dose-dependent depletion of the glutathione levels, increased the reactive oxygen species (ROS) production and induced DNA fragmentation. The treatment of the cells with B. aetnensis extracts recovered the oxidative status and reduced the glutamate-increased of TG-2 expression.
These data suggest that B. aetnensis, preventing TG-2 up-regulation and reducing the oxidative stress induced by glutamate in primary astroglial cell cultures, may represent a new strategies in the neuropathological conditions associated to excitotoxicity.
P803 Different antimicrobial activity of beta defensins and colonic biopsy extracts against aerobic and anaerobic bacterial strains of the gastrointestinal tract
S. Nuding, L. Zabel, K. Fellermann, J. Wehkamp, H.A.G. Mueller, E.F. Stange (Stuttgart, Goppingen, DE)
Objectives: The human gastrointestinal tract harbours a variety of aerobic and anaerobic microorganisms. To prevent colonisation and invasion of bacteria, the intestinal mucosa synthesizes antimicrobial peptides such as the cationic defensins. Based on RT-PCR and western blot, we determined a high induction of beta defensins in ulcerative colitis, whereas beta defensins in Crohn's disease were less induced. To elucidate possible functional consequences for the intestinal barrier, we investigated the antimicrobial activity of defensins and cationic protein extracts, taken from the colon of patients with Crohn's disease, ulcerative colitis or controls, against bacteria of the intestinal flora.
Methods: To quantitate the antimicrobial activity, we established a flow cytometric assay with the membrane potential sensitive dye bis-(1,3-dibutylbarbituric acid) trimethine oxonol. Depolarisation of the bacterial cell membrane caused by antimicrobial peptides leads to an uptake of the dye followed by an increasing green fluorescence. We tested the antibacterial activity of the constitutive human beta defensin HBD-1, the inducible defensin HBD-3 and cationic biopsy extracts of 22 patients with colonic Crohn's disease, 32 with ulcerative colitis and 13 controls against ATCC strains of Escherichia coli and Staphylococcus aureus and a clinical isolate of Bacteroides vulgatus.
Results: In populations of the aerobic strains E. coli and S. aureus as well as the anaerobic strain B. vulgatus 80% of the bacteria were killed by 2.5–5 μg HBD-3/mL. In contrast, HBD-1 in concentrations up to 15 μg/mL, which kill E. coli and S. aureus, exerted no bactericidal effect against B. vulgatus. The antimicrobial activity of cationic biopsy extracts was significantly diminished in extracts of patients with Crohn's disease against E. coli and B. vulgatus compared to ulcerative colitis. The activity in extracts of Crohn's disease against S. aureus was also reduced, but the differences versus controls and ulcerative colitis were less pronounced.
Conclusion: The viability of B. vulgatus is not influenced by the constitutive HBD-1, this is in concordance with its augmented presence in the normal intestinal flora. The inducible defensin HBD-3 is a potent antimicrobial peptide against all 3 strains tested. The lower antimicrobial activity in extracts of Crohn's disease corresponds well to the deficient beta defensin induction. The impact of other antimicrobial peptides must still be clarified.
P804 Inhibitory effect of Melaleuca alternifolia (tea tree oil) on influenza A/PR/8 virus replication
R. Timpanaro, A. Garozzo, B. Bisignano, A. Stivala, P.M. Furneri, G. Tempera, A. Castro (Catania, IT)
Objectives: The Melaleuca alternifolia (Tea Tree) is a coniferous tree found in tropical regions, whose needles contain an essential oil that is used in medical and cosmetic products. The Tea Tree Oil (TTO) consists of about 50% oxygenated monoterpenes and 50% terpene hydrocarbons, terpinen-4-ol is the main active component. TTO has a wide spectrum of antimicrobial activity against bacteria, yeasts and fungi. The antimicrobial activity has been principally attributed to terpinen-4-ol. The aim of this study was to investigate the antiviral activity and the mechanism of action of Tea Tree Oil and its components, terpinen-4-ol, γ-terpinene, p-cymene, α-terpinene, terpinolene and α-terpineol, against Influenza A/PR/8 virus subtype H1N1 in MDCK cells.
Methods: The inhibitory effect was studied by measuring hemoagglutinin units (HAU) and 50% cytopathic effect (CPE50). In order to study the mode of action of the TTO, we carried out a series of experiments, including virucidal assay, pre-treatment assay, inhibition of attachment assay and time of addition assay.
Results: The TTO, the terpinen-4-ol, the terpinolene, the α-terpineol were found to have an inhibitory effect on influenza virus replication at doses below the cytotoxic dose. No of the tested compounds showed virucidal activity nor any protective action for the MDCK cells.
The effect of compound on different steps of the replicative cycle of influenza virus in MDCK cells was studied by adding compound at various times after infection (0, 1, 2, 4, 6 and 9 h). Viral replication, assessed as HAU/mL and CPE50 24 h after infection, revealed that this was significantly inhibited only if TTO was added within 2 h of infection, indicating an interference with an early step of the viral replicative cycle of Influenza virus. The influence of the compound on the virus adsorption step, studied by the infective centre assays, indicated that TTO did not interfere with cellular attachment of virus.
Conclusion: These data suggest that TTO interferes with an early events of Influenza virus replication, after viral adsorption. Further studies are necessary to understand the precise mode of action of this compound.
P805 Essential oil composition and antibacterial effects of Ziziphora clinopodioides
M. Chitsaz, M. Barton, M. Bazargan, M. Kamalinejad (Tehran, IR; Adelaide, AU)
Background: The plant of Ziziphora clinopodioides (LAM), which is habitant of Iran, has been used in Iranian Traditional Medicine for treatment of some infectious conditions. In this survey, its antibacterial effects were examined against some bacterial species.
Methods: Hydro-alcoholic extract of plant obtained by solvent (methanol 70%) and essential oil obtained by hydro-distillation procedure. The methanolic extract and essential oil were tested against Staphylococcus aureus (ATCC25923), Streptococcus pyogenes (PTCC1470), Escherichia coli (ATCC25922), Salmonella typhimurium (PTCC1609), Klebsiella pneumoniae (PTCC1053), and Pseudomonas aeruginosa (ATCC27853). Standard agar diffusion method was used for antimicrobial assay of methanolic extract and essential oil. MIC and MBC were determined by using macrobroth dilution method. Compounds of essential oil separated and identified by GC and GC/MS analysis.
Results: The growth of Gram-positive organisms (S. aureus and S. pyogenes) were inhibited by methanolic extract at a concentration of 25 mg/well, but it did not inhibit any of the tested Gram-negative species. Essential oil halted the growth of all tested Gram-positive and Gram-negative organisms, with highest effect on S. typhimurium (with MIC and MBC of 225 micg/mL). Gas chromatographic analysis revealed 22 different compounds in essential oil which five of them comprise more than 73% of oil and pulegon is the highest one.
Conclusion: The results suggest that Ziziphora clinopodioides has potent antibacterial effects on some Gram-positive and Gram-negative bacterial species especially on Salmonella typhimurium.
Pathogenesis of Gram-negative infections
P806 Assessment of tryptophan metabolism and cytokine profile in cerebrospinal fluid samples from patients with bacterial meningitis
L.G. Coutinho, C.L. Bellac, D. Grandgirard, M. Wittwer, R.S. Coimbra, S. Christen, L.F. Agnez-Lima, L.A. Marinho, S.L. Leib (Berne, CH)
Objectives: Activation of the kynurenine (KYN) pathway (KP) has been observed in experimental bacterial meningitis (BM). Here we assessed the association of chemo-/cytokine levels with the concentration of KP metabolites in CSF and plasma samples from patients with BM.
Methods: CSF samples were collected from 22 hospitalised patients. Nine patients were diagnosed with bacterial meningitis, 6 patients with viral meningitis and 7 patients with non-infectious neurological disorders. Microsphere-based multiplex assays (Lincoplex®, Linco Research Inc., St Charles, MA, USA) was used to assess the concentrations of 14 chemo-/cytokines separately in CSF and serum. The CSF and serum concentration of metabolites from the kynurenine pathway was assessed by high pressure liquid chromatography.
Results: The concentration of TNF-α, IL-6, IL-1β, IFN-γ, IL-10, IL-1 receptor antagonist, MIP-1α, MIP-1β, MCP-1 and G-CSF were 100-fold higher in CSF from patients with BM compared to the two other groups. In all CSF samples the concentration of IL-2, IL-12(p70), IL-4 and GM-CSF was below the detection limit.
In plasma samples the concentrations of IL-6, IL-10, IL-1 receptor antagonist, MCP-1 and G-CSF were significantly increased in patients with BM. The concentrations of the KP metabolites kynurenine, anthranilic acid and kynurenic acid were 10-fold higher in CSF of patients with BM compared to the other two groups. In contrast to what was found in CSF, the concentrations of KP metabolites in the plasma were not significantly different between the three groups. Tryptophan levels in plasma samples were higher than in CSF samples and were significantly decreased in patients with BM.
Conclusion: BM is associated with increased levels of pro-inflammatory cytokines and KP metabolites. This increase in KP metabolites is most likely due to activation of KP by IFN-γ and TNF-alpha. Based on the comparison of tryptophan and KP metabolite concentrations between plasma and CSF samples we conclude that the activation of the tryptophan pathway upon BM occurs within the brain.
P807 Characterisation of the soluble domain of nitrite reductase from Neisseria meningitidis strains
P. Stefanelli, G. Colotti, A. Neri, M.L. Salucci, R. Miccoli, L. Di Leandro, R. Ippoliti (Rome, L'Aquila, IT)
Objectives: The objective of the study was to characterise the molecular and biochemical properties of the soluble domain of the AniA protein of N. meningitidis serogroup B MC58, and thereafter evaluate genomic conservation and protein expression in a panel of N. meningitidis clinical isolates.
Methods: The soluble domain of the aniA gene of N. meningitidis MC58 was cloned and expressed in E. coli following standard procedures. Nitrite consumption was evaluated by the Griess method at different time points. For the pH dependence, the experiments were performed using different buffers 20 mM acetate buffer in the range 4.5–5.5, 20 mM Hepes buffer in the range 5.5–6.6, and 20 mM phosphate buffer in the range 6.5–7.5. Direct Nitrogen Oxide production was recorded by a specific electrode (WPI, UK). A total of 11 N. meningitidis strains, 7 disease-associated and 4 carriers, were cultured with 5% CO2 at 37°C on Thayer-Martin plates. The aniA sequences were analyzed with the BLAST programme. The antisera was prepared with 1.5 mg of purified protein to immunise New Zealand rabbits, Charles River and used to perform western-blot analysis.
Results: The biochemical results show an ordered mechanism for the catalysis, since the binding of nitrite needed to induce electron transfer from the type I to the type II Cu-site. Furthermore sAniA shows a dependence of the catalytic activity upon acidification. Sequence analysis of the coding region of the gene in 11 N. meningitidis strains showed that, notwithstanding a high degree of gene sequence similarity among them, two gene variants were found due to insertions or deletions. In 1 disease associated strain a 9-residue insertion was located close to the catalytic site. No amplification was obtained in 2 carried strains due to the presence of a gene for a transposase protein. The AniA was expressed in all the strains, except for two with no amplification reaction.
Conclusion: These preliminary results describe the basic biochemical properties of the sAniA from N. meningitidis MC58, in particular its pH dependence. The conservation of the gene among the majority of the examined strains suggests that preservation of its integrity is important for bacterial survival. However, further investigation will be performed to understand how ani A gene variations in the two carrier strains could affect the enzymatic activity of the protein.
P808 Distinct Neisseria meningitidis isolates display different cytokine induction capacities in an ex vivo whole-blood model
A. Fiserova, O. Beran, J. Kalmusova, M. Musilek, P. Kriz, M. Holub (Prague, CZ)
Objectives: The aim of the present study was to examine the cytokine induction capacities of heat-inactivated Neisseria meningitidis isolates from patients with distinct courses of invasive meningococcal disease (IMD) in an ex vivo whole-blood model.
Methods: Three healthy adult volunteers with negative antimeningococcal anti-B and anti-C antibodies (as detected by serum bactericidal assay) were selected for whole blood-model. Refludan®-anticoagulated whole blood (0.05 mg/mL) and heat-inactivated bacteria at a final concentration of 106 CFU/mL in blood samples were incubated for 4 hr at 37°C under constant shaking. Flow cytometric quantification of tumour necrosis factor (TNF)-α, interleukin(IL)-1β, IL-6, IL-8, IL-10, and IL-12p70 levels in supernatants was performed using cytometric bead array.
Results are summarised in the table , n = 6 (each donor in two independent experiments). There are statistically significant differences between strains 137/05 vs. (versus) 285/05, 137/05 vs. 113/04, 137/05 vs. 171/05, 285/05 vs. 109/05, 113/04 vs. 109/05, and 109/05 vs. 171/05 in TNF-α, IL-1β, and IL-6 production (P < 0.01) except for strains 113/04 vs. 109/5 in IL-1β production (P < 0.05).
| Neisseria meningitidis strains and clinical parameters of IMD | TNF-α (ng/mL) | IL-1β (ng/mL) | IL-6 (ng/mL) |
|---|---|---|---|
| 137/05 group C strain | 3,724 | 5,844 | 8,071 |
| (C:NT:P1.2,5; ST-11 HC) M+S, APACHE 14, SOFA 3, moderate course | ±1,326 | ±3,556 | ±7,138 |
| 285/05 group B strain | 13,540 | 18,773 | 45,319 |
| (B:1:NST; ST-41/44 complex) M+S, APACHE 10, SOFA 4, mild course | ±4,038 | ±4,223 | ±14,563 |
| 113/04 group B strain | 10,908 | 15,661 | 37,123 |
| (B:22:P1.14; ST-18 HC) SS+WFS, APACHE 25, SOFA 12, severe course | ±3,043 | ±6,283 | ±14,575 |
| 109/05 group B strain | 3,399 | 6,664 | 10,493 |
| (B:NT:NST; ST-919, not HC) M+S, APACHE 17, SOFA 6, mild course | ±1,999 | ±4,290 | ±7,686 |
| 171/05 group B strain | 14,043 | 19,862 | 46,854 |
| (B:15:P1.4; ST-5126, not HC) M, APACHE 11, SOFA 3, mild course | ±4,103 | ±9,660 | ±14,629 |
HC: hypervirulent complex; IMD: invasive meningococcal disease; M: meningitis;
S: sepsis; SS: septic shock; WFS: Waterhouse–Friderichsen syndrome.
Data are expressed as mean±SD.
Conclusion: N. meningitidis isolates display different cytokinestimulating potential in whole-blood model. The cause might be in differences in the quantity and quality of bacteria structural components and remains to be solved.
Acknowledgement: The study is supported by grants MSM 0021620806 and IGA NR/8014–3.
P809 Roles of uropathogenic Escherichia coli pili in pathogenesis of urinary tract infection
S. Najar Peerayeh, N. Nooritalab, M. Sattari (Tehran, IR)
Introduction: Urinary tract infections (UTI) are one of the most common infectious diseases in man with a prevalence strongly influenced by gender and age. Uropathogenic E. coli (UPEC) strains account for 90% of all UTI and up to 50% of all nosocomial UTI. Infection is initiated when UPEC binds to the superficial epithelial cells by type 1 pili. In addition to attachment, the presence of type 1 pili can lead to bacterial invasion to bladder epithelial cells. However, P piliation of UPEC is characteristic of strains causing upper urinary tract infection as well as pyelonephritis leading to urosepsis. In this study we determine the roles of type 1 and P pili in interaction of UPEC with human polymorphonuclear leukocyts (PMNS).
Materials and Methods: Type 1 and P piliated and unpiliated strains (obtained by growth at a pilus-restrictive temperature) of UPEC were used for determining the effects of these adhesins on migration of neutrophils towards bacteria in Boyden chamber. The lectinophagocytosis and intracellular killing of bacteria with purified human neutrophils were estimated by counting of the number of viable bacteria in 45 min.
Results: Type 1 piliated UPEC stimulated significantly greater chemotaxis than did P piliated, unpiliated bacteria and bacteria in which the piliation was suppressed. Phagocytosis of type 1 piliated UPEC occurred in the direct and opsonin-independent manner. In contrast, P piliated and unpiliated bacteria failed to bind to PMNS.
Conclusion: The results indicated that type 1 pili have a chemotatic effect, and there was a positive correlation between type 1 piliation and bacterial killing by PMNS. In contrast, PMNS did not chemotaxis to UPEC with type P pili, and unable to react with these bacteria. Therefore the expression of type P pili is critical to UPEC establishment in upper urinary tract.
P810 Salmonella ability to use B-lymphocytes as a reservoir is due to overcoming bactericidal mechanisms, in addition to inability to produce inflammatory cytokines
R. Rosales-Reyes, A. Pérez-López, M. Ramírez-Aguilar, V. Ortiz-Navarrete, M. Urbán-Reyes, C. Alpuche-Aranda (Mexico City, MX)
The ability of Salmonella to survive and replicate within macrophages and dendritic cells is a relevant pathogenic factor to develop disseminated disease. It has also been shown that Salmonella is able to infect B cells in vitro and in vivo. In the mouse model, Salmonella is localised inside B cells (CD19+, B220+) and macrophages (Mac1+) of spleen and bone marrow after 60 days of initial infection.
Objectives: To identify B-Lymphocytes ability to produce bactericidal mechanisms and inflammatory cytokines profile against Salmonella infection.
Methods: Intracellular Salmonella were detected by FACS and UFCs. The nitrogen intermediates reactive (NIR) were quantified by Griess method. The cytotoxicity (ctx) was assay by LDH quantification. TNF-α and IL-1β were detected by ELISA.
Results: The efficiency of Salmonella uptake by B cells increased 10 times when the bacterium was opsonised with specific IgG anti-Salmonella Ab. In addition, the intracellular bacteria were not ctx on infected B cells (even MOI of 1:50) as macrophages (10% vs 38% respectability). Salmonella survives more efficiently in B cells than macrophages at 24 hours post-infection. On the other hand, these Salmonella infected B cells were able to produce NIRs (14.05±3.61 microM without infection, 29.33±3.33 microM with MOI = 50, 75.72±1.39 microM with MOI = 100) in response to magnitude of infection as macrophages (9.33±1.67 microM without infection, 89.33±1.11 microM with MOI = 50 and 116.27±1.39 microM with MOI = 100). Infected B cells are not able to produce IL-1β by inflamosome complex as macrophages (non detected vs 20000 pg/mL respectively).
Conclusions: Salmonella internalisation by B cells is favoured by Fc receptor phagocytosis and is able to survive and evade the bactericidal effect of NIRs. The infected B cells are not able to produce inflammatory cytokines by inflamosome complex (such as IL-1beta) involved in recruitment of effectors immune cells. These data suggest that B cells are a “safe” intracellular reservoir for Salmonella and perhaps they are also a mechanism of transportation and evading effectors immune cells during in vivo infection.
P811 The role of typical and atypical enteropathogenic Escherichia coli in diarrhoea
M.Y. Alikhani, M.M. Aslani, A. Mirsalehian, B. Fatollahzadeh (Hamadan, Tehran, IR)
Objectives: Infectious diarrhoea is one of the world's leading causes of morbidity and mortality, resulting in about two million deaths per year. Enteropathogenic Escherichia coli (EPEC) is a major cause of infantile diarrhoea in developing countries. The present study was performed to investigate the contribution of typical and atypical enteropathogenic Escherichia coli (EPEC) as a cause of infectious diarrhoea in Iranian children less than 10 years of age.
Methods: A total of 1355 children were chosen for the presence of EPEC in their faeces. The samples were cultured for enteric pathogen by standard methods. Strains biochemically identified as Escherichia coli were selected and Serogroups of lactose positive and lactose negative E. coli strains were determined by slide agglutination tests. EPEC isolates was examined by PCR with the specific primers for the following virulence genes: eaeA, bfpA and stx.
Results: During the summer months, 247 specimens from children with diarrhoea, and 1108 from asymptomatic cases were analyzed for the presence of typical and atypical EPEC. Potential enteric pathogens were identified in 140 patients (56.7%). EPEC was the most frequently identified agent (111 patients). EPEC isolates were examined for the presence of the eaeA, bfpA and stx genes by PCR. EPEC isolates were classified as typical (eaeA+, bfpA+, stx-) or atypical (eaeA+, stx- or bfpA+, stx-). The results obtained indicated that typical EPEC was diagnosed in 35 patient (11.8%) compared to 8 (0.4%) of asymptomatic persons (P < 0.05). Atypical EPEC strains were isolated in 23 patients (9.3%) compared to 13 (1.2%) of the healthy cases (P < 0.05).
Conclusion: Detection of the eaeA and bfpA genes is a useful method for identification and differentiation between typical and atypical EPEC strains. Typical and atypical EPEC were prevalent in children with diarrhoea than healthy cases. Our data suggest that typical and atypical EPEC are important causes of diarrhoea in Iranian children.
P812 Haemoglobin-binding activity and haemoglobin-binding protein of Prevotella nigrescens
S. Fujimura (Shiojiri, JP)
Objectives: Prevotella nigrescens, a Gram-negative, black-pigmented, anaerobic rod is implicated as an etiological agent of periodontitis as Porphyromonas gingivalis or Prevotella intermedia. These periodontopathogens, however, lack iron-chelating systems (siderophores) which overcome a deficiency of iron. Based on the assumption that P. nigrescens acquires iron from haemoglobin derived from erythrocytes in periodontal lesions, we examined the binding activity of the envelope of P. nigrescens to haemoglobin and efforts were made to isolate haemoglobin-binding protein (HbBP).
Methods: P. nigrescens ATCC33563 was cultured anaerobically in a glove box in Trypticase-base medium containing haemin and menadione. The envelope was prepared by ultrasonic treatment of cells and centrifugation at 120,000 g and solubilised using a detergent (CHAPS). Insoluble material of the envelope was used as the outer membrane. Binding of haemoproteins including haemoglobin to the envelope was tested by photometrically. Dot blot assay was employed to confirm the binding of HbBP and haemoglobin using peroxidase-labelled haemoglobin on nitrocellulose membranes.
Results: Binding of the envelope to haemoglobin occurred in a strictly pH-dependent manner; the binding was observed at pH 4.5 to 5.5, but was not in the neutral and alkaline buffers. Furthermore, haemoglobin bound to the envelope dissociated in the high pH buffers. The same level of haemoglobin binding activity was observed in the whole cell, but the outer membrane demonstrated only one third the activity of the envelope. Other haemoproteins such as myoglobin, cytochrome c and catalase also were found to bind to the envelope. We hypothesised the existence of haemoglobin-binding protein (HbBP) in the envelope responsible for this binding, and proved its presence by dot blot method. HbBP was purified from the solubilised material of the envelope by the combination of ion-exchange chromatography, affinity chromatography and isoelectric focusing.
Conclusion: Binding of the envelope of P. nigrescens to haemoproteins including haemoglobin was confirmed and haemoglobin-binding protein in the envelope was isolated.
P813 Alterations of serum lipoproteins in patients with acute brucellosis
F. Apostolou, A. Kostoula, E. Liberopoulos, I. Gazi, A. Spyrou, E. Tsianos, E. Bairaktari, A. Tselepis, M. Elisaf (Ioannina, GR)
Objectives: Changes in serum lipid parameters are observed during inflammation. These changes may be associated with the pathogenesis of atherosclerosis. There are no data on the possible effect of acute infection with Brucella on serum lipoproteins.
Methods: Serum lipid parameters were determined in 28 patients (18 male and 10 female, aged 52.9±14.6 years) with acute brucellosis on admission and 4 months after the initiation of appropriate treatment. Furthermore, 24 matched controls were studied. Fasting levels of total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), triglycerides, apolipoproteins (Apo) AÉ, B, E and lipoprotein (a) [Lp(a)] were measured. Low-density lipoprotein (LDL) subclass analysis was performed by use of the Lipoprint LDL System.
Results: On admission, patients exhibited significantly lower levels of TC, HDL-C, LDL-C and ApoAI compared with controls, while there were no significant differences in TG, ApoB, ApoÅ and Lp(a) levels. Furthermore, mean LDL particle diameter was significantly lower in patients compared with controls. Of interest, small dense LDL-C (sdLDL-C) levels as well as the sdLDL-C/LDL-C, LDL-C/HDL-C and ApoB/ApoAI ratios were significantly higher in patients compared with controls.
Four months after treatment initiation, a significant increase in levels of TC (by 23%), HDL-C (by 37%), LDL-C (by 23%), and ApoAI (by 39%) was noticed. On the other hand, levels of TG, ApoB, ApoE and Lp(a) were not significantly altered. Furthermore, sdLDL-C levels, the sdLDL-C/LDL-C ratio, the LDL-C/HDL-C ratio and the mean LDL particle diameter were not significantly changed. However, the ApoB/ApoAI ratio was significantly decreased.
Conclusions: Significant quantitative and qualitative alterations of serum lipoproteins were seen during acute infection with Brucella. These changes were not fully restored 4 months after treatment initiation.
P814 Association of FCgamma RIIA polymorphism with brucellosis
A. Rafiei, R. Ghasemian, Z. Hosseini khah, A. Ajami (Mazandaran, IR)
Background and Purpose: Brucellosis is a major bacterial zoonosis of global importance. Effective host defence against brucella depends on immunoglobulin G-mediated phagocytosis of the bacteria and it has been shown that the FC RIIA polymorphism (FCgamma RIIA-R131 vs FCgamma RIIA –H131) determines the capacity of immunoglobulin G-mediated phagocytosis via this receptor. The aim of the study was evaluation of FCgamma RIIA polymorphism in patients with brucellosis and reveals any relation between this polymorphism and disease progression.
Materials and Methods: In this study we evaluated FCgamma RIIA polymorphisms (R/R131, R/H131, H/H131) in 67 patients with serologically proven brucellosis and 67 age, gender and geographical healthy volunteers. FCgamma RIIA polymorphism was determined using a polymerase chain reaction (SSCP-PCR).
Results: The frequency of FCgamma RIIA-R/R131 genotype was higher in patients with brucellosis compared Controls (47.8% vs 28.4%). This genotype has an OR = 2.1, 95%CI = 1.3–4.2, P = 0.039) Significant correlation with brucellosis. But we did not find a significant difference between patients with brucellosis and controls (P = 0.2).
Although the frequency of FCgamma RIIA-R/R131 was higher in patients with brucellosis compared controls, we did not find any statistically differences (53.8% vs46.3%, P = 0.2). As a result there was no significant association between FCgamma RIIA genotype and severity of brucellosis.
P815 Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles
T.T. Tan, M. Mörgelin, A. Forsgren, K. Riesbeck (Malmö, Lund, SE)
Objectives: Moraxella catarrhalis causes respiratory tract infections in children and adults with COPD. It is often isolated as a co-pathogen with Haemophilus influenzae. The underlying mechanism for this cohabitation is unclear. The aim of this study was to investigate if M. catarrhalis is able to contribute to the survival of H. influenzae in human serum.
Methods: The M. catarrhalis ubiquitous surface proteins (Usp) A1 and A2 are known to contribute to M. catarrhalis serum resistance by interactions with various complement proteins. M. catarrhalis also secretes outer membrane vesicles (blebs) in vitro. Hence, we examined nasopharyngeal samples of a 9-year old child who had grown pure cultures of M. catarrhalis during an episode of sinusitis. Both electron microscopy and gold labeled anti-UspA1 and A2 antibodies were used to analyse the samples. The effect of M. catarrhalis blebs on the bacteriolytic activity of human serum against H. influenzae was examined by incubating blebs with serum before performing bactericidal assays. The interaction of these blebs with the complement system was further documented by dot-blot assays.
Results: Blebs carrying Usp A1/A2 are secreted both in vivo and in vitro. These blebs do carry UspA1 and A2 and they absorb the third component of complement system (C3), counteracting the complement cascade directed against H. influenzae. An improved survival was evident in all clinical strains (n = 5) of H. influenzae tested. In contrast, UspA1/2 deficient blebs are much weaker inhibitors of the complement-dependent killing of H. influenzae.
Conclusion: Our results suggest a novel strategy in which pathogens collaborate to conquer innate immunity and that the M. catarrhalis vaccine candidates, UspA1/A2 play a major role in this interaction.
P816 Does Pseudomonas aeruginosa need free living amoebae in environment?
C. Roger, S. Bouyer, Y. Hechard, C. Imbert, M. Rodier (Poitiers, FR)
Objectives: According to most authors, amoebae of genus Acanthamoeba do not play an important role as reservoirs for Pseudomonas aeruginosa, frequently isolated from hospital water. In our hospital, beside Acanthamoeba, amoebae of genus Hartmanella are also often recovered from water taps. We investigated in this work the ability for H. vermiformis to allow growth and/or survival of P. aeruginosa.
Methods: Trophozoites of H. vermiformis ATCC 50256 (5.105/mL) were incubated with 5.104/mL bacteria (MOI = 0.1) of three P. aeruginosa strains (ATCC 1111, one strain from a patient and one environmental strain from hospital water). The mixture was incubated in PBS at 27°C during 24, 48 and 72 h. Each P. aeruginosa strain was also cocultivated with trophozoites of A. castellanii (ATCC 30234), A. polyphaga (ATCC 30461) and A. culbertsoni (ATCC 30171) in the same conditions. Controls were realised by incubating bacteria in PBS without amoebae. After incubation and before plating on Mueller Hinton medium, the mixture was passed through a 27 gauges needle five times to ensure lysis of amoebae. Dilutions of the lysate were plated to determine CFU. All the experiments have been made three times, each time in triplicate. To visualise the potential interactions between bacteria and amoebae, transmission electron microscopy was used.
Results: In this study, we have shown that there was neither growth nor survival of P. aeruginosa in the free living amoebae used. No internalisation of the bacteria was shown by transmission electron microscopy.
Conclusion: In our conditions, neither the H. vermiformis strain nor the Acanthamoeba strains were able to allow the survival and the growth of the three strains of P. aeruginosa used. Further studies are nevertheless necessary to confirm that most of the P. aeruginosa strains do not need reservoirs to survive in water.
P817 Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa
Y. Hirakata, A. Kondo, K. Hoshino, T. Miyazaki, S. Kurihara, K. Izumikawa, M. Seki, K. Yanagihara, Y. Miyazaki, S. Kamihira, S. Kohno (Nagasaki, Tokyo, JP)
Objectives: We have recently shown a mexAB-oprM deletion strain (delta-ABM) derived from P. aeruginosa PAO1 was compromised in its capacity to invade or transmigrate into MDCK cells (Hirakata Y, et al. J Exp Med 2002; 196: 109). These data suggest that P. aeruginosa exports invasion determinants predominantly using MexAB-OprM efflux system. On the other hand, multi-drug efflux systems are thought to contribute to the intrinsic resistance of a number of the antimicrobials. MexAB-OprM is known to be constitutively expressed in P. aeruginosa PAO1. We studied the influence of efflux pump inhibitors (EPIs), broad-spectrum efflux inhibitor MC-207,110 and MexAB-OprM specific efflux pump inhibitor (ABS-EPI) D13–9001, on the P. aeruginosa invasion using the Madin-Darby canine kidney (MDCK) epithelial cell monolayer penetration assay model.
Methods: P. aeruginosa PAO1, its delta-ABM mutant, nalB (MexAB-OprM overproducing strain) mutant, and nfxB (MexCD-OprJ overproducing strain) mutant were used in this study. Broad-spectrum EPI, Phe-Arg β-naphthylamide (MC-207,110) and MexAB-OprM-specific efflux pump inhibitor (ABS-EPI) D13–9001 were used. MDCK cell monolayer penetration assay was performed as reported previously (Hirakata Y, et al. J Infect Dis 2000; 181: 765). Bacterial strains were cultured together with several concentrations of EPIs in LB broth overnight at 37°C with vigorous shaking. The bacteria in the baso-lateral medium were counted by plating at timed intervals.
Results: The invasiveness of PAO1 wild-type (WT) and nalB (MexAB-OprM overproducing strain) was concentration-dependently inhibited in the presence of EPIs. However, the invasiveness of nfxB (MexCD-OprJ overproducing strain) was not reduced in the presence of EPIs.
Conclusion: The findings support that invasion determinant(s) are predominantly exported by P. aeruginosa via MexAB-OprM, and that the EPIs are useful to reduce not only antimicrobial resistance but also the invasiveness of P. aeruginosa.
P818 Qnr prevalence and their association with orf513 in Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, and Serratia marcescens: a multicentre study from Korea
Y.J. Park, J.K. Yu, E.J. Oh, J.J. Park, G.J. Woo (Seoul, KR)
Objectives: In this study, we investigated the prevalence of all known qnr determinants (qnrA, qnrB and qnrS) in Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, and Serratia marcescens collected from various parts of Korea, their distribution of quinolone (ciprofloxacin and nalidixic acid) MICs and association with orf 513 and orf 1005.
Methods: A total of 644 consecutive, non-duplicate isolates, including E. cloacae (186), E. aerogenes (154), C. freundii (138), and S. marcescens (166), were collected from clinical specimens at 12 clinical laboratories. The MICs of ciprofloxacin (CIP) (0.062–1 mg/L) and nalidixic acid (NAL) (4–32 mg/L) were determined by an agar dilution method as described in CLSI guideline (2003). For isolates with cinprofloxacin MICs of ≥0.25 mg/L or nalidixic acid MICs of ≥16 mg/L, qnr determinants were screened by PCR using primers reported previously:
for qnrA;
qnrA-F, atttctcacgccaggatttg-3′,
qnrA-R, 5′-gatcggcaaaggttaggtca-3′,
for qnrB;
qnrB-F, 5′-gatcgtgaaagccagaaagg-3′,
qnrB-R, 5′-acgatgcctggtagttgtcc-3′, for qnrS;
qnrS-F, 5′-acgacattcgtcaactgcaa-3′,
qnrS-R, 5′-taaattggcaccctgtaggc-3′, for orf513;
orf513-F, 5′-aag gaacgccacggcgagtcaa-3′,
orf513-R, 5′-tgcaaagacgccgtggaagc-3′, for orf1005;
orf1005-F, 5′-ctt ggtattcgaagctggtc-3′,
orf1005-R, 5′-ctaccgtttgcaacagtaag-3′.
Strains positive for qnrA were generous gifts from Professor Nordmann and Professor Kim and positive control strains for qnrB, qnrS were from Professor Jacoby (Lahey clinic).
Results: Of the total of 644 isolates, 47.9% (89/186) of E. cloacae, 16.9% (26/154) of E. aerogenes, 47.1% (65/138) of C. freundii, and 66.3% (110/166) of S. marcescens showed cinprofloxacin MICs of ≥0.25 mg/L or nalidixic acid MICs of ≥16 mg/L. Of them, 54 E. cloacae, 8 E. aerogenes, 42 C. freundii and two S. marcescens isolates harboured qnr determinants. One E. cloacae harboured both qnrA and qnrB. Almost all (93.5%) of the qnrA producers and about half (47.8%) of the qnrB producers harboured orf513.
None of the qnrB producers harboured orf1005. The occurrences of qnr determinants did not show relationship with quinolone MICs in either E. cloacae or C. freundii (Fig. 1 ).

% of qnr in ECL
Conclusion: The prevalence of qnr determinants was very high among E. cloacae and C. freundii, but very low in S. marcescens. In E. cloacae and E. aerogenes, proportion of qnrA and qnrB was similar, but qnrB was predominant in C. freundii.
P819 Characteristics of phylogenetic group D Escherichia coli isolates from bacteraemia
S. Soto, S. Martí, J. Sánchez-Céspedes, J. Martínez, J. Mensa, J. Vila (Barcelona, ES)
Objective: The most frequent cause of Gram-negative extraintestinal bacterial infections, such as bacteraemia, is Escherichia coli. In our hospital 50% of E. coli strains most frequently isolated as a cause of bacteraemia belonged to phylogenetic group D, in contrast to the data found by other research groups. The aim of our study was to characterise phylogenetic group D bacteraemic E. coli isolates by analysing the epidemiological relationship, the virulence profiles, and the capacity to form “in vitro” biofilm and compare bacteraemic group D E. coli from a urinary focus with those from another focus.
Methods: Ninety-nine E. coli blood isolates belonged to phylogenetic group D collected from patients with bacteraemia in the Hospital Clinic of Barcelona were included in the study. The epidemiological relationship was analysed by REP-PCR. The presence of VFs was determined by PCR using specific primers. Biofilm assay was carried out using minimal glucose medium (M63).
Results: Yersiniobactin, type 1 fimbriae, and the Ag43 were the VFs most frequently found (78%, 73%, and 71%, respectively). When the comparison of blood isolates from a urinary focus with blood isolates from an other focus (intra-abdominal, pneumonia, venous catheter, skin and soft tissue infections, and unknown source) was made, several observations are of note: (i) the isolates from a non-urinary focus tended to be more resistant to nalidixic acid (p = 0.09); (ii) there were significant differences in the presence of several VFs between the two groups of isolates. In fact, E. coli isolates from a urine focus frequently presented: hlyA, cnf1, malX, and P-fimbriae-related genes (papA, papC, papG, and papEF) (p = 0.01, 0.02, 0.006, 0.03, 0.001, 0.02, and 0.00001, respectively).
Conclusion: The phylogenetic group D was the most predominant among the E. coli isolates causing bacteraemia in our region. The virulence capacity was related to the original foci of the isolates, with the blood isolates from a urinary focus being more virulent probably because the UPEC isolates causing bacteraemia need to invade the kidney and then disseminate to the bloodstream.
P820 Escherichia coli virulence factors in chronic bacterial prostatitis
J. Vranes, A. Antolovic-Pozgain, M. Senjug, B. Bedenic (Zagreb, Osijek, HR)
Objectives: Urinary tract infection (UTI) is the principal extraintestinal syndrome caused by Escherichia coli. Although most common among women, it is also significant problem among men, who can experience cystitis, pyelonephritis and prostatitis. Better enderstanding of the pathogenesis of UTI and prostatitis in men is necessary to guide the development of effective preventive measures. The aim of this study was to determine the prevalence of various virulence factors among E. coli strains causing chronic prostatitis (CPR, n = 25) and compare them with the strains isolated in acute (AP, n = 23) and chronic pyelonephritis (CP, n = 22), acute cystitis (AC, n = 22), and asymptomatic bacteriuria (ABU, n = 18).
Methods: Total of 110 E. coli strains were isolated from 110 patients who presented with one of five clinical syndromes. Bacterial susceptibility to serum killing was measured by assessing bacterial regrowth after incubation in normal human serum. The expression of adhesins was defined by hemagglutination and inhibition of hemagglutination in microtiter plates. Serotyping was performed on glass slides and the production of hemolysin was tested on human blood agar plates.
Results: The most virulent strains were isolated from patients with AP, followed by strains isolated from patients with CPR. Strains isolated from patients with CPR were significantly more often resistant to the bactericidal activity of serum (BAS), significantly more often produced hemolysin and significantly more often expressed adhesins (p < 0.01) than did ABU and CP strains. P-fimbriated strains were most commonly detected in the AP and CPR groups, although AP strains expressed P-fimbriae significantly more often than did CPR strains (p < 0.05). CPR and AP groups of strains were significantly more often resistant to BAS than strains isolated in other diagnostic groups (p < 0.01). There were significant correlations between resistance of E. coli strains to BAS and expression of mannose-resistant adhesins (C = 0.365). The difference in antigenic structure did not influence on serum sensitivity of the strains.
Conclusion: E. coli virulence factors such as adhesins, hemolysin and serum resistance may have important roles in the pathogenesis of chronic prostatitis.
P821 Urease-like IgG activity in H. pylori-positive patients with duodenal ulcer
M. Konorev, I. Generalov, V. Okulich (Vitebsk, BY)
Objectives: To investigate the urease-like IgG activity in Hp-positive patients with duodenal ulcer.
Methods: Randomly selected 37 Hp-positive duodenal ulcer patients and 40 Hp-negative patients with gastritis in the age of range 21–61 years were examined. Identification of H. pylori (Hp) was performed by rapid urease test, morphological method, ELISA. Serum IgG were purified from serum of patients according to Paul et al. Confirmation of IgG purity was made by size-exclusion Toyopearl HW 55 chromatography or Diasorb Diol 400 HPLC in acid glycine-HCl buffer, 0.05 M, pH 2.8 and by SDS-polyacrylamide gradient gel electrophoresis. For detection of urease IgG activity 0.1 ml IgG (1.5 mg/mL) and 0.1 ml substrate solution (20 g/L urea (Serva) with 2 g/L NaN3 in 0.1 ml FBB 0.1 M, pH 8.2) were mixed. Control tests include 0.1 ml albumin (1.5 mg/mL) or 0.1 ml NaCl (9.0 mg/mL) and 0.1 ml substrate solution. The samples were incubated at least for 120 hours (t = 37°C) to attain reliable differences between experimental and control tests. The control of sterility of IgG preparations, albumin, substrate, experimental and control tests did't reveal any bacterial contamination (estimated in 72, 120 and 148 hours). The fall of urea concentration (in mM) was assessed by enzymatic urea reagent (BUN Reagent, ECO-MED-POLL, Austria) with spectrophotometer “Spectrum II” detection (Abbott, USA; λ = 340 nm). Conversion of results into international units of activity (pikoCatals) was carried out according to regression equation Y = 4.6X (in pCat), where X is the quantity of hydrolyzed urea (mM).
Results: All Hp-negative patients didn't have Hp in the stomach and demonstrated the negative rapid urease test. IgG fractions of all Hp-negative patients were lack of urease activity (P > 0.05); at the same time they didn't contain IgG antibodies to Hp by the ELISA test. All Hp-positive patients had Hp in the stomach, serum IgG antibodies to Hp, and demonstrated the positive rapid urease test. IgG fraction of 28 patients with Hp infection and duodenal ulcer showed the high incidence and levels of urease-like IgG activity (75.7%; 4.6±1.4 pCat; P < 0.01) and possessed IgG antibodies to Hp, determined by the ELISA test (2.4±0.2 ISR value).
Conclusion: Serum IgG fractions of Hp-positive patients with duodenal ulcer demonstrate the discernible levels of urease-like Ig activity. The latter is expected to be an intrinsic property of IgGs, as far as the enzyme urease is not produced by mammalian cells.
P822 Evidence for the role of Haemophilus influenzae in the pathogenesis of recurrent tonsillitis
C. Skoulakis, E. Tigiroglou, E. Malli, D. Klapsa, C. Papadakis, E. Petinaki (Volos, Larissa, GR)
Objectives: Frequently, cultures of tonsils from patients with recurrent tonsillitis did not reveal the presence of pathogenic bacteria such as S. pyogenes. The bacteria probably enter into the core and conventional methods failed to detect them. In this study, the tonsils of 60 patients undergoing tonsillectomy were studied in order to determine the correlation between bacteria isolated from tonsillar surface and core using conventional and molecular methods.
Methods: Sixty tonsillar tissues, from patients with recurrent tonsillitis (n: 29), as well from individuals undergoing tonsillectomy for snoring or sleep apnea (control group, n: 31), were collected. Samples from both surface and core were cultured, while the identification of bacteria isolated was done using conventional methods. In addition, DNA was extracted from specimens obtained exclusively from tonsil core using a commercial kit (QIAGEN), and then it was PCR amplified with a pair of primers targeting the 16SrRNA gene, followed by sequence analysis. The viability of microorganisms were detected by RT-PCR, after RNA extraction, by using specific primers for bacteria whom DNAs were detected.
Results: Among patients with recurrent tonsillitis, 25% of the surface cultures did not grow any bacteria, while 75% showed the presence of Streptococcus mitis and oralis; in some cases we observed the co-existence of Staphylococcus aureus (17%) and Haemophilus parainfluenzae (20%). On the other hand, the core cultures were negative for bacteria. However, into the core of nineteen tonsils, molecular methods revealed the presence of mRNA of S. pyogenes in three cases (10.3%) and H. influenzae in sixteen cases (55%) confirming their viability. Among the control group, surface cultures showed the presence of Streptococcus mitis and S. oralis (72%), with the co-existence of Staphylococcus aureus (5%) and Haemophilus parainfluenzae (20%), while the core cultures remained negative. No bacterial DNAs were detected in the core of the control group.
Conclusions: The high incidence of H. influenzae isolated in patients with recurrent tonsillitis, comparing to the control group, suggests the potential pathogenic role of this bacterium in this type of infection.
In vitro susceptibility of glycolipopeptides
P823 Europe surveillance initiative profiling the activity of telavancin against streptococci by specimen source from 2004 to 2005
D. Draghi, B. Benton, M. Jones, K. Krause, C. Thornsberry, D. Sahm (Herndon, South San Francisco, US; Breda, NL)
Objectives: Telavancin (TLV) is a novel, rapidly bactericidal lipoglycopeptide with potent activity against a wide spectrum of Gram-positive bacteria. TLV has recently completed two Phase 3 clinical trials for the treatment of complicated skin and skin structure infections. Ongoing trials are assessing TLV for the treatment of hospital-acquired pneumonia. The current study was performed to establish a baseline of TLV activity versus recent isolates of various streptococcal species that may be encountered in the clinical setting.
Methods: During 2004–2005, 168 Streptococcus pneumoniae (SP), 67 viridans streptococci (VGS) and 106 β-haemolytic streptococci (BS; including 46 S. pyogenes, 42 S. agalactiae and 18 Group C, G and F streptococci) were collected from Europe (33 hospital sites in 14 countries). Isolates were centrally tested by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) methodology (M7-A7) against TLV and comparators. Isolates were analysed in groups by specimen source (SPEC), which included blood, lower respiratory tract, upper respiratory tract and skin structure.
Results: The table shows the activity of TLV against the different streptococcal isolates.
Activity of telavancin against Streptococcus pneumoniae (SP), viridans streptococci (VGS) and β-haemolytic streptococci (BS) isolates
| Organism | Specimen sourcea | Phenotypeb | No. of isolates | MIC (mg/L) |
|
|---|---|---|---|---|---|
| Range | MIC90 | ||||
| SP | Blood | All | 52 | ≤0.001–0.06 | 0.03 |
| PEN-S | 44 | ≤0.001–0.06 | 0.03 | ||
| PEN-NS | 8 | 0.015–0.03 | NAc | ||
| LRT | All | 67 | 0.002–0.03 | 0.03 | |
| PEN-S | 42 | 0.002–0.03 | 0.03 | ||
| PEN-NS | 25 | 0.015–0.03 | 0.03 | ||
| URT | All | 49 | 0.008–0.03 | 0.03 | |
| PEN-S | 35 | 0.008–0.03 | 0.03 | ||
| PEN-NS | 14 | 0.015–0.03 | 0.03 | ||
| VGS | Blood | All | 40 | 0.03–0.5 | 0.12 |
| ERY-S | 21 | 0.03–0.5 | 0.12 | ||
| ERY-NS | 19 | 0.03–0.12 | 0.12 | ||
| SST | All | 27 | 0.015–0.5 | 0.06 | |
| ERY-S | 17 | 0.03– | |||
| 0.06–0.06 | |||||
| ERY-NS | 10 | 0.015–0.5 | 0.06 | ||
| BS | Blood | All | 26 | ≤0.001–0.25 | 0.06 |
| ERY-S | 21 | 0.015–0.25 | 0.06 | ||
| ERY-NS | 5 | ≤0.001–0.06 | Nac | ||
| SST | All | 80 | 0.002–0.12 | 0.06 | |
| ERY-S | 59 | 0.002–0.12 | 0.06 | ||
| ERY-NS | 21 | 0.015–0.12 | 0.06 | ||
LRT, lower respiratory tract; URT, upper respiratory tract; SST, skin structure.
PEN-S/PEN-NS, penicillin-susceptible/nonsusceptible; ERY-S/ERY-NS, erythromycin-susceptible/nonsusceptible.
NA, not available; MIC90 values were not determined for <10 isolates.
Conclusion: TLV demonstrated potent activity against all streptococcal species and groups encountered in this surveillance initiative, regardless of resistance to other antimicrobial classes. TLV MIC90 values for each streptococcal group were equivalent or within one doubling dilution when studied by SPEC. This baseline evaluation provides a useful benchmark of TLV activity as clinical development and use progresses.
P824 Europe surveillance initiative profiling the anti-staphylococcal activity of telavancin by specimen source from 2004 to 2005
D. Draghi, B. Benton, M. Jones, K. Krause, C. Thornsberry, D. Sahm (Herndon, South San Francisco, US; Breda, NL)
Objectives: The increase and spread of methicillin (oxacillin)-resistant staphylococci present challenges for clinicians. Telavancin (TLV), a novel, rapidly bactericidal lipoglycopeptide, has recently completed two Phase 3 clinical trials for the treatment of complicated skin and skin structure infections. Ongoing trials are assessing TLV for the treatment of hospital-acquired pneumonia. Surveillance was initiated to establish a baseline for the activity of TLV against recent staphylococcal isolates, according to specimen source (SPEC).
Methods: Overall, 1721 Staphylococcus aureus (SA) and 226 coagulase-negative staphylococcus (CoNS) isolates were collected from Europe (33 hospital sites in 14 countries) during 2004–2005. Isolates were centrally tested by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) methodology (M7-A7) against TLV and comparators. Isolates were analysed in groups by SPEC, which included blood, skin structure, upper respiratory tract (SA only) and lower respiratory tract (SA only).
Results: The oxacillin-resistance (OX-R) rate for all SA was 30.5% and the OX-R rate for CoNS was 73.5%. The activity of TLV against the SA and CoNS isolates is shown in the Table . Based on MIC90 values, TLV (0.5 mg/L) was 16 times more potent than teicoplanin (TEI; 8 mg/L) against CoNS. Additionally, MIC ranges for TLV (0.06–2 mg/L) were lower compared with TEI (0.25–32 mg/L).
Activity of telavancin against Staphylococcus aureus (SA) and coagulase-negative staphylococci (CoNS) isolates
| Organism | Specimen sourcea | Phenotypeb | No. of isolates | MIC (mg/L) |
|
|---|---|---|---|---|---|
| Range | MIC90 | ||||
| SA | SST | All | 951 | ≤0.015–1 | 0.25 |
| OX-S | 709 | ≤0.015–1 | 0.25 | ||
| OX-R | 242 | 0.06–1 | 0.5 | ||
| Blood | All | 365 | 0.06–1 | 0.25 | |
| OX-S | 235 | 0.06–1 | 0.25 | ||
| OX-R | 130 | 0.12–1 | 0.5 | ||
| LRT | All | 349 | ≤0.015–1 | 0.5 | |
| OX-S | 209 | ≤0.015–1 | 0.25 | ||
| OX-R | 140 | 0.12–1 | 0.5 | ||
| URT | All | 17 | 0.12–0.5 | 0.5 | |
| OX-S | 16 | 0.12–0.5 | 0.25 | ||
| OX-R | 1 | 0.5–0.5 | NAd | ||
| CoNS | SST | All | 25 | 0.06–0.25 | 0.25 |
| OX-S | 10 | 0.06–0.25 | 0.25 | ||
| OX-R | 15 | 0.12–0.25 | 0.25 | ||
| Blood | All | 190 | 0.06–2 | 0.5 | |
| OX-S | 48 | 0.12–2 | 0.5 | ||
| OX-R | 142 | 0.06–2 | 0.5 | ||
c 39 SA isolates and 11 CoNS isolates were from an unknown or other source.
SST, skin structure; LRT, lower respiratory tract; URT, upper respiratory tract.
OX-R, oxacillin-resistant; OX-S, oxacillin-susceptible.
Not available; MIC90 values were not determined for <10 isolates.
Conclusion: TLV demonstrated potent activity against all SA and CoNS isolates, regardless of SPEC or resistance to oxacillin or TEI. This evaluation of TLV activity will provide a baseline for comparison of in vitro activity as clinical development and use progresses.
P825 Comparative activity of telavancin against Gram-positive pathogens using European and CLSI susceptibility testing methods
D. Farrell, A. Colclough, L. Williams (London, UK)
Objectives: Telavancin (TLV) is a novel lipoglycopeptide antibiotic that is in late stage development for the treatment of serious infections in humans caused by Gram-positive bacterial pathogens. In this study, the in vitro activity of TLV was determined using the Clinical and Laboratory Standards Institute (CLSI) reference methodology and 5 European guidelines (Deutsches Institut für Normung [DIN], European Committee on Antimicrobial Susceptibility Testing [EUCAST], Swedish Reference Group for Antibiotics [SRGA], British Society for Antimicrobial Chemotherapy [BSAC] and Comité de l'Antibiogramme de la Société Française de Microbiologie [CASFM]).
Methods: MICs for TLV versus 478 bacterial isolates (150 staphylococci [STA], 206 streptococci [STR], 102 enterococci [ENT] and 20 Corynebacterium jeikeium [CJ]) were determined under 6 experimental conditions representing 4 microbroth dilution (MD) and 6 agar dilution (AD) MIC methods.
Results: The MIC range and mode for each of the 6 experimental methods are shown in the Table . Against staphylococci, consistent results were obtained among all methodologies used. The MIC distribution for ENT was bimodal, with VanA phenotype Enterococcus faecalis being responsible for the higher mode. The greatest inter-method variability was found for STR, with 2 of the AD methods having modal values 4 to 8 times higher than the other methods.
MIC range and mode for telavancin Gram-positive pathogens using 6 CLSI and European susceptibility testing methods
| Telavancin MIC range (mode) mg/L |
||||
|---|---|---|---|---|
| Methoda | STA | ENT | STR | CJ |
| 1 (MD) | 0.015–1 | 0.015–8 | 0.004–0.25 | 0.008–0.03 |
| (0.12) | (0.25 and 4b) | (0.015) | (0.015) | |
| 2 (MD) | 0.03–1 | 0.03–8 | 0.008–0.25 | 0.015–0.06 |
| (0.12) | (0.06 and 4b) | (0.03) | (0.03) | |
| 3 (AD) | 0.06–2 | 0.03–4 | 0.015–0.12 | 0.015–0.12 |
| (0.12) | (0.06 and 2b) | (0.12) | (0.06) | |
| 4 (AD) | 0.03–1 | 0.03–4 | 0.015–0.12 | 0.015–0.03 |
| (0.12) | (0.06 and 4b) | (0.03) | (0.03) | |
| 5 (AD) | 0.03–1 | 0.03–4 | 0.015–0.06 | 0.015–0.03 |
| (0.06) | (0.06 and 2b) | (0.03) | (0.015) | |
| 6 (AD) | 0.06–2 | 0.03–4 | 0.015–0.12 | 0.015–0.06 |
| (0.12) | (0.06 and 2b) | (0.12) | (0.03) | |
NL>
Methods: 1. CLSI-DIN-EUCAST (MD); 2. SRGA (MD); 3. DIN (AD); 4. BSAC (AD); 5, SRGA (AD); 6. CLSI-CASFM-EUCAST (AD).
E. faecalis with VanA phenotype.
Conclusion: There was very good correlation between all methods and between MD and AD. These results will be useful when establishing TLV breakpoints in countries using these susceptibility testing methods.
P826 Europe surveillance initiative profiling the anti-enterococcal activity of telavancin by specimen source from 2004 to 2005
D. Sahm, D. Draghi, B. Benton, M. Jones, K. Krause, C. Thornsberry (Herndon, South San Francisco, US; Breda, NL)
Objectives: Enterococci are frequently associated with hospital-based infections, including bacteraemia. Telavancin (TLV), a novel, rapidly bactericidal lipoglycopeptide, has recently completed two Phase 3 clinical trials for the treatment of complicated skin and skin structure infections. Ongoing trials are assessing TLV for the treatment of hospital-acquired pneumonia. Because TLV will be used in the hospital setting, profiling its activity against enterococci is important.
Methods: A total of 434 Enterococcus faecalis (EF) and 348 E. faecium (EM) isolates were collected from Europe (35 hospital sites in 14 countries) during 2004–2005. Isolates were centrally tested by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) methodology (M7-A7) against TLV and comparators. Results were analysed according to blood and non-blood specimen sources (SPECs).
Results: The vancomycin-nonsusceptible (VAN-NS) rate for all EF was 5.5% and the VAN-NS rate for EM was 22.7%. The activity of TLV against the EF and EM isolates is shown in the Table .
Activity of telavancin against Enterococcus faecalis (EF) and E. faecium (EM) isolates
| Organism | Specimen source | Phenotypea | No. of isolates | MIC (mg/L) |
|
|---|---|---|---|---|---|
| Range | MIC90 | ||||
| EF | Blood | All | 181 | 0.06–8 | 1 |
| VAN-S | 164 | 0.06–2 | 0.5 | ||
| VAN-NS | 17 | 4–8 | 8 | ||
| Non-blood | All | 253 | 0.06–8 | 0.5 | |
| VAN-S | 246 | 0.06–8 | 0.5 | ||
| VAN-NS | 7 | 0.5–8 | 8 | ||
| EM | Blood | All | 144 | 0.03–8 | 4 |
| VAN-S | 100 | 0.03–2 | 0.25 | ||
| VAN-NS | 44 | 0.12–8 | 4 | ||
| Non-blood | All | 204 | 0.03–8 | 2 | |
| VAN-S | 169 | 0.03–2 | 0.25 | ||
| VAN-NS | 35 | 0.12–8 | 4 | ||
VAN-S/VAN-NS, vancomycin-susceptible/nonsusceptible.
Conclusion: TLV demonstrated potent activity against all EF and EM isolates, regardless of SPEC. Based on MIC90 values, TLV was 16-fold more potent against vancomycin-susceptible (VAN-S) isolates compared with VAN-NS isolates among EF and EM. This evaluation of TLV activity will provide a baseline for comparison of in vitro activity as clinical development and use continues.
P827 Influence of polysorbate 80 on susceptibility of Gram-positive bacteria to oritavancin
F. F. Arhin, I. Sarmiento, A. Belley, G. McKay, D. Draghi, P. Grover, D. Sahm, T.J. Parr Jr., G. Moeck (Saint Laurent, CA; Herndon, US)
Objectives: Oritavancin (ORI) is a lipoglycopeptide (LG) with activity against Gram-positive bacteria. Broth microdilution (BMD) assays with dalbavancin, another LG, require 0.002% polysorbate 80 (P80). Following initial observations that P80 reduced ORI BMD minimum inhibitory concentrations (MICs) for reference strains of Staphylococcus aureus (Sa) ATCC 29213 and Enterococcus faecalis (Ef) ATCC 29212 but not for Streptococcus pneumoniae (Sp) ATCC 49619, we examined the effect of P80 on ORI MICs for 301 clinical isolates of these three genera.
Methods: BMD assays were conducted on clinical isolates of Sa (n = 76), coagulase-negative staphylococci (CNS; n = 26), Ef (n = 70), E. faecium (Em; n = 30), Sp (n = 19), S. agalactiae (n = 29), S. pyogenes (n = 29), Groups C and G streptococci (n = 8), and Viridans Group streptococci (n = 14), either following CLSI guidelines (no P80) or with P80, in parallel. When tested in the presence of P80, ORI, vancomycin (VAN) and teicoplanin (TEI) were dissolved in 0.002% P80 and maintained in 0.002% P80 thereafter. Reference strains of Sa, Ef and Sp were tested concurrently.
| MIC range (MIC 90a), mg/L |
||||||
|---|---|---|---|---|---|---|
| ORI | ORI+P80 | TEI | TEI+P80 | VAN | VAN+P80 | |
| Enterococci | ||||||
| E. faecalis (n = 70) | 0.12–4 (2) | 0.008–0.5 (0.12) | 0.06–64 (0.5) | 0.06–64 (0.12) | 0.12–>256 (2) | 0.5–>256 (2) |
| E. faecium (n = 30) | 0.12–4 (4) | 0.004–0.5 (0.25) | 0.12–256 (64) | 0.06–128 (128) | 0.25–>256 (>256) | 0.25–>256 (>256) |
| Staphylococci | ||||||
| S. aureus (n = 76) | 1–8 (4) | 0.015–0.25 (0.12) | 0.12–1 (0.5) | 0.12–2 (0.5) | 0.5–1 (1) | 0.5–16 (1) |
| CNS (n = 26) | 0.12–8 (4) | 0.008–0.5 (0.25) | 0.12–4 (2) | 0.06–4 (2) | 0.5–2 (1) | 0.5–2 (2) |
| Streptococci | ||||||
| S. agalactiae (n = 29) | 0.03–0.5 (0.25) | 0.03–0.5 (0.25) | 0.03–0.12 (0.06) | 0.03–0.12 (0.06) | 0.25–0.5 (0.5) | 0.25–0.5 (0.5) |
| S. pneumoniae (n = 19) | 0.00025–0.004 (0.004) | 0.0005–0.004 (0.004) | 0.015–0.06 (0.06) | 0.015–0.06 (0.06) | 0.12–0.5 (0.25) | 0.12–0.5 (0.25) |
| S. pyogenes (n = 29) | 0.015–0.5 (0.25) | 0.015–0.5 (0.25) | ≤0.001–0.25 (0.06) | 0.015–0.06 (0.03) | 0.25–0.5 (0.25) | 0.25–0.5 (0.25) |
| Group C and G (n = 8) | 0.004–1 (na) | 0.004–1 (na) | 0.03–0.06 (na) | 0.03–0.06 (na) | 0.25–0.5 (na) | 0.25–0.5 (na) |
| Viridans group (n = 14) | 0.004–2 (1) | 0.004–2 (1) | 0.002–0.12 (0.06) | 0.002–0.25 (0.12) | 0.25–1 (0.5) | 0.25–0.5 (0.5) |
na, MIC 90 was not calculated for groups containing less than 10 isolates; P80: polysorbate 80.
Results: P80 reduced ORI MIC90s by 16- to 32-fold for enterococci and staphylococci. In contrast, TEI and VAN MIC90s were identical (the same or within one doubling dilution) with and without P80 with the exception of a 4-fold reduced TEI MIC90 for Ef with P80. P80 did not change MIC90s for any agent against streptococci grown in cation-adjusted Mueller-Hinton broth containing 2% lysed horse blood (LHB).
Conclusions: Significant (16- to 32-fold) reductions in ORI MIC90s were observed for enterococci and staphylococci with P80. A modest (4-fold) reduction in TEI MIC90 was observed for Ef +P80. ORI MICs ± P80 for streptococci were identical in the CLSI-recommended medium which contains LHB. Companion binding studies promote the idea that LHB may substitute for P80, which may help to explain the observed lack of shift in ORI MICs with P80 for streptococci.
P828 Activity of daptomycin against coryneform bacteria isolated from clinical samples
C. Salas, J. Calvo, L. Martínez-Martínez (Santander, ES)
Objectives: To evaluate the activity of Daptomycin (DAP) and six other agents against coryneform bacteria of clinical origin.
Methods: We evaluated 179 clinical isolates (1 per patient) including Corynebacterium striatum [Cstr (58)], C. amycolatum [Camy (25)], C. jeikeium [Cjei (25)], C. pseudodiphtheriticum [Cpse (21)], Listeria monocytogenes [Lmon (24)] and Arcanobacterium haemolyticum [Ahae (26)]. A microdilution assay, according to CLSI guidelines (M45-P), was used. The following antimicrobials were included: Clindamycin (CLI), Vancomycin (VAN), Teicoplanin (TEC), Linezolid (LNZ) and Quinupristin-dalfopristin (QDA). For DAP, Mueller-Hinton broth with 3% laked horse blood was supplemented with 50 mg/L of Ca++. S. pneumoniae ATCC 49619, S. aureus ATCC 29213 and E. faecalis ATCC 29212 were used as reference control strains.
Results: MICs 50/90 (mg/L) values are presented in table 1 .
Table 1.
| Antimicrobial | MIC50/MIC90 |
|||||
|---|---|---|---|---|---|---|
| Cstr | Camy | Cjei | Cpse | Lmon | Ahae | |
| CLI | 32/>32 | >32/>32 | >32/>32 | >32/>32 | 0.5/1 | ≤0.015/≤0.015 |
| VAN | 0.25/0.5 | 0.25/0.5 | ≤0.06/0.25 | 0.25/0.25 | 0.5/1 | 0.25/0.5 |
| TEC | ≤0.06/0.12 | 0.12/0.25 | ≤0.06/0.12 | ≤0.06/0.25 | ≤0.06/0.25 | ≤0.06/≤0.06 |
| LNZ | 0.25/0.5 | 0.12/0.25 | 0.25/0.5 | 0.25/0.5 | 2/4 | 0.25/0.5 |
| QDA | 0.12/0.5 | ≤0.06/0.25 | 0.25/1 | ≤0.06/≤0.06 | 0.5/1 | ≤0.06/≤0.06 |
| DP | ≤0.06/0.5 | ≤0.06/≤0.06 | 0.25/0.25 | ≤0.06/≤0.06 | 2/4 | 0.5/0.5 |
Conclusions: Daptomycin has a good in vitro activity against C. striatum, C. amycolatum, C. jeikeium, C. pseudodiphtheriticum and A. haemolyticum, and presented reduced activity against L. monocytogenes. Teicoplanin, Vancomycin, Linezolid and Quinupristin-dalfopristin are also active against these organisms. Clindamycin was poorly active, in vitro, except against A. haemolyticum and L. monocytogenes.
P829 Intracellular activity of daptomycin against methicillin-sensitive, methicillin-resistant and vancomycin-intermediate S. aureus
S. Lemaire, F. Van Bambeke, P. Tulkens (Brussels, BE)
Objectives: Relapsing and chronic S. aureus infections have been associated with intracellular bacterial persistence. The activity of antibiotics may differ markedly between the extracellular and the intracellular milieus, and intracellular activity needs, therefore, to be assessed in specific models. Daptomycin (DAP) is a cyclic lipopeptide with fast and extensive bactericidal activity against extracellular forms of S. aureus. Our aim was to examine its intracellular activity in a newly developed model of human macrophages allowing for extended exposure periods (AAC 2006; 50:841–851), and using strains with different resistance phenotypes of clinical significance.
Methods: MSSA (ATCC 25923), MRSA (ATCC 33591) and VISA (NRS 23) were used. MICs were determined in MH broth (supplemented with 50 mg/L of Ca2+) by microdilution method. Infection of THP-1 macrophages was performed as described previously (AAC 2006; 50:841–851). Intracellular localisation of the bacteria was examined by confocal and electron microscopy. Activity was measured after 24 h exposure to a DAP concentration corresponding to its reported human Cmax (77 mg/L), in comparison with controls (cells incubated with gentamin [0.5×MIC] to prevent extracellular growth; see AAC 2006, 50:841–851 for validation).
Results: In controls, phagocytised bacteria were seen sojourning and multiplying in membrane-bounded vacuoles (phagolysosomes). MICs (broth) and intracellular changes in cfu are shown in the table .
| Strains | MIC (mg/L) | log10 intracell. change in cfu (±SD)a |
|
|---|---|---|---|
| controls | + DAP (77 mg/L) | ||
| MSSA | 0.125 | +1.30±0.05 | −1.66±0.07 |
| MRSAb | 0.125 | +1.63±0.13 | −1.46±0.07 |
| VISA | 0.5 | +1.39±0.05 | −1.30±0.05 |
compared to the original inoculum (approx. 106 cfu/mg cell protein).
Hospital-acquired.
Conclusions: DAP shows intracellular activity against MSSA, MRSA and VISA.
P830 Assessing the in vitro activity of daptomycin against well-characterised methicillin-resistant Staphylococcus aureus strains representing different spa types
K. Becker, B. Grünastel, A. Mellmann, G. Peters, C. von Eiff (Münster, DE)
Objectives: Daptomycin is a recently introduced cyclic lipopeptide antibiotic that is rapidly bactericidal in vitro against a broad spectrum of Gram-positive bacteria including multi-resistant strains. The purpose of the present study was to assess the in vitro activity of daptomycin against a well-characterised collection of methicillin-resistant Staphylococcus aureus (MRSA) isolates collected in multi-centre studies.
Methods: The MicroScan WalkAway-96 system for Gram-positive (GP) organisms (Dade Behring) was used to assess the in vitro activity of daptomycin against MRSA strains. For this purpose, clinical MRSA strains of different geographic origin and different clonality were included. To determine the clonal lineages of MRSA strains, the repeat region of the spa gene was amplified by PCR. spa types were determined using the Ridom StaphType software version 1.3.
Results: All MRSA strains tested were susceptible to daptomycin. The MIC for all strains tested was ≤1 mg/l (MIC90 ≤1 mg/l). These MRSA strains representing 100 different spa types comprised the most common European spa types, such as t003, t032, t008, t002, t044, t001, t004, and t037, as well as a broad range of rarely occurring types. Daptomycin was also active towards those strains harbouring the Panton Valentine leukocidin (PVL) encoding genes.
Conclusion: Daptomycin was shown to be potent against all MRSA strains tested representing the most common European MRSA lineages.
P831 In vitro activities of daptomycin against isolates of Gram-positive organisms causing skin and soft tissue infections, compared to vancomycin, linezolid, and teicoplanin
J. Fairest (London, UK)
Objectives: Gram-positive pathogens are a major cause of skin and soft tissue infection (SSTI), and daptomycin is a new novel drug developed primarily for the treatment of SSTIs.
The aim of this study was to compare daptomycin with the current conventional antimicrobials used to treat SSTIs (vancomycin, linezolid, and teicoplanin).
Methods: In total 260 isolates were collected comprising of 20 Staphylococcus aureus, 50 Staphylococcus epidermidis, 50 MRSA, 50 Enterococci, 10 Vancomycin-Resistant Enterococci (VRE), and 50 group A Streptococci (S. pyogenes), over a period of one month. Minimum inhibitory concentrations (MICs) were obtained using two different methods, the E-test method and the broth microdilution method. Data analysis was carried out using the paired t-test to compare mean MICs obtained from both tests.
Results: All isolates proved to be highly susceptible to daptomycin. Low mean MICs were obtained with daptomycin against the staphylococci (0.24 ul/mL for S. aureus, 0.32 ul/mL for S. epidermidis, and 0.18 ul/mL for MRSA). Higher mean MICs (1.13 ul/mL) were found with daptomycin against the enterococci. Daptomycin was found to be highly active against VRE, with a mean MIC of 1.78 ul/mL. However, linezolid had a lower mean MIC of 0.54 ul/mL. For the group A streptococci (S. pyogenes) daptomycin had the lowest mean MIC (0.07 ul/mL) compared to the other antimicrobials.
Conclusion: Overall, daptomycin has proved to be highly effective agent against most Gram-positive organisms responsible for SSTIs. Daptomycin was found to be comparable to linezolid, however superior to vancomycin and teicoplanin in most cases. Daptomycin appears to be a useful alternative in the treatment of SSTIs, especially when multi-drug resistant organisms such as MRSA and VRE are encountered.
Clinical trials of antibiotics
P832 Temporal trends in the methodological quality of randomised controlled trials of antimicrobial agents
M.E. Falagas, E. Pitsouni, I. Bliziotis (Athens, GR)
Objective: The aim of this study was to investigate the potential improvement of the methodological quality of RCTs of antimicrobial agents published during the last 30 years.
Methods: We randomly selected from the Cochrane Central Register of Controlled Trials database 70 RCTs of antibacterial agents that were published during a 30-year study period (1975–2005); specifically we randomly selected 10 RCTs published during each of the following years: 1975, 1980, 1985, 1990, 1995, 2000, and 2005. In each of the selected RCTs we searched for information regarding quality of randomisation, double blinding, quality of the blinding, description of withdrawals or dropouts, eligibility of the study group, interventions, outcomes, baseline data, allocation concealment, adverse events, statistical significance, and reporting of conflicts of interest and funding of the research. We graded the methodological quality of the reviewed RCTs to evaluate trends for possible improvement.
Results: No improvement was noted in most of the analyzed methodological aspects of the RCTs during the 30-year study period. Quality of randomisation, double blinding, quality of the blinding, and allocation concealment were rather scarce among the reviewed RCTs, without observing a trend for improvement during the study period. We noted improvement in reporting and analyzing baseline data as well as in reporting the presence or not of statistical significance and the statistical cut off of significance. In only 2 of the reviewed 70 RCTs all 12 of the examined methodological quality aspects were met.
Conclusion: We did not observe considerable overall improvement in the quality of the methodology of RCTs on antibacterial agents during the last 30 years. The methodological quality aspects that need the most improvement are these that ensure (when present) minimisation of various types of biases.
P833 Intravenous therapy with doripenem versus levofloxacin with an option for oral step-down therapy in the treatment of complicated urinary tract infections and pyelonephritis
K. Naber, R. Redman, P. Kotey, L. Llorens, K. Kaniga (Munich, DE; Mountain View, Raritan, US)
Objective: Doripenem (DOR) is an investigational carbapenem with broad-spectrum activity against common pathogens of complicated urinary tract infections (cUTIs). This study evaluated the efficacy and safety of DOR for the treatment of complicated lower UTIs and pyelonephritis (complicated and uncomplicated) in a large, multinational phase 3 study.
Methods: This randomised, double-blind, double-dummy, multicentre study compared DOR (500 mg q8h) with levofloxacin (LVX) (250 mg q24h) in the treatment of cUTIs in adults. Both study drugs were administered as 60-min IV infusions. After 9 or more doses of IV drug therapy, subjects could be switched to oral therapy with LVX (250 mg q24h). Total duration of study therapy was 10 days. Non-inferiority of DOR to LVX was evaluated with respect to microbiologic response in patients who were microbiologically evaluable at the test of cure (ME at TOC) visit (6–9 days after completion of study drug therapy). Non-inferiority was defined as a lower limit of the 95% confidence interval (CI) for the difference in response rates greater than −10%. The primary end point was the microbiological cure rate at the TOC visit; this was evaluated in subjects who were ME at TOC, and as a co-primary analysis in the microbiological modified intent-to-treat (mMITT) population. A key secondary end point was clinical cure in the clinically evaluable (CE) at TOC analysis set.
Results: A total of 753 patients were randomised; 376 received DOR and 372 received LVX. The microbiologic cure rate in the ME at TOC population was 82.1% (230/280) for DOR and 83.4% (221/265) for LVX (95% CI: −8.0, 5.5). The microbiologic cure rates in the mMITT population were 79.2% (259/327) and 78.2% (251/321), respectively (95% CI: −5.6, 7.6). The clinical cure in the CE at TOC population was 95.1% (272/286) for DOR and 90.2% (240/266) for LVX (95% CI: 0.2, 9.6). DOR was effective against major causative organisms of cUTIs, including Escherichia coli, Proteus mirabilis, and Klebsiella pneumoniae. Gastrointestinal disorders were most commonly reported as adverse events, occurring in 25% of DOR- and 27% of LVX-treated patients. Headache was also frequently reported (16%, DOR; 15%, LVX). No seizures were reported in patients receiving DOR.
Conclusion: DOR was microbiologically and clinically effective and therapeutically non-inferior to LVX in this study for the treatment of cUTIs and was generally safe and well-tolerated, with an AE profile similar to that of LVX.
P834 Treatment of complicated intra-abdominal infections: doripenem versus meropenem
C. Lucasti, A. Jasovich, O. Umeh, J. Jiang, K. Kaniga (Somers Point, US; Buenos Aires, AR; Mountain View, Raritan, US)
Objective: Doripenem (DOR) is an investigational carbapenem with broad-spectrum activity against common causes of complicated intraabdominal infections (cIAIs), such as Enterobacteriaceae and anaerobes. This study evaluated the efficacy and safety of DOR for the treatment of cIAIs in a large phase 3 study (DORI 07).
Methods: This phase 3, randomised, double-blind, double-dummy, multicentre study compared IV DOR (500 mg q8h over 60 min) with IV meropenem (MER) (1 g q8h administered as a bolus over 3–5 min) in the treatment of adults with cIAIs. Subjects could be switched to oral amoxicillin/clavulanate after 9 or more doses of DOR or MER. Total duration of study therapy was 5–14 days. Non-inferiority of DOR to MER was inferred if the lower limit of the 2-sided 95% confidence interval (CI) for the difference in clinical cure rates (doripenem minus meropenem) was greater than or equal to −15%. The primary efficacy end points included clinical cure rate at the test of cure (TOC) visit (21–60 days post therapy) in subjects who were microbiologically evaluable (ME) at TOC, and the clinical cure rate occurring up to 60 days after the last dose of study drug therapy in the microbiological modified intent-to-treat (mMITT) population.
Results: A total of 476 patients were randomised; 237 received DOR and 239 received MER. The clinical cure rate at the TOC visit in the ME at TOC population was 85.9% (140/163) for DOR and 85.3% (133/156) for MER (95% CI: −7.7, 9.0). The clinical cure rates in the mMITT population were 77.9% (152/195) and 78.9% (150/190), respectively (95% CI: −9.7, 7.7). DOR was microbiologically effective against major causative organisms of cIAIs, including Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Streptococcus intermedius, Enterococcus faecalis, Bacteroides caccae, Bacteroides thetaiotaomicron, Bacteroides fragilis, and Bacteroides uniformis. The most common study drug-related adverse events reported were nausea (DOR, 6.8%; MER, 1.3%), diarrhoea (DOR, 6.4%; MER, 4.7%), and phlebitis (DOR, 3.4%; MER, 2.1%). No seizures were reported.
Conclusion: For the treatment of cIAIs, DOR was generally well-tolerated and therapeutically non-inferior to MER at the doses studied.
P835 Adequacy and efficacy of initial empiric antibiotic treatments in severe nosocomial infections in ICU departments: results of multicentre randomised study
S. Yakovlev, V. Beloborodov, S. Sidorenko, A. Ovechkin, V. Yakovlev (Moscow, RU)
Objectives: The incidence of nosocomial infections due to multi-drug resistant pathogens especially ESBL-producing microorganisms has increased markedly in recent years in most ICU departments. The resistant pathogens are the main reason of empiric antimicrobial treatment failure. In this open randomised multicentre study, the adequacy and effectiveness of meropenem monotherapy was compared with traditional cephalosporins or fluoroquinolones plus aminoglycoside combination for initial empiric antibiotic therapy of severe infections in ICU departments.
Methods: This multicentre study was conducted in 18 ICU departments in 15 cities of Russia. The adult patients with confirmed nosocomial infection (pneumonia or intra-abdominal) were randomly allocated to one of two treatment arms: meropenem (MER) IV 0.5–1 g TID as monotherapy or conventional combination treatment (CT) with 3rd or 4th generation cephalosporin or fluoroquinolone (ciprofloxacin, ofloxacin or levofloxacin) plus aminoglycoside and ± metronidazole. Only the patients with severe infection (clinical signs of severe sepsis and APACHE II score 15–25) were included. The initial therapy was empiric provided the adequate samples for bacteriological investigation.
Results: Of a total of 166 patients enrolled, 135 were clinically evaluable: 62 in the MER arm and 73 in CT arm; 102 patients were microbiology evaluable. Baseline demographic and disease characteristics as well as the severity of infection were similar for both treatment groups. At the TOC assessment, 50 of 62 (80.6%) clinically evaluable patients in the MER group and 34 of 73 (46.6%) in the CT group had a favourable clinical response (P < 0.01). The bacteriological efficacy was also better after MER therapy – 89.6 and 48.1%, respectively (P < 0.001). The microbiological eradication rates was significantly higher in the MER group for P. aeruginosa (88 and 40%, P = 0.007), E. coli (100 and 46.7%, P = 0.003) and Acinetobacter spp. (90.9 and 40.0%, P = 0.02). The adequacy of empiric therapy was 2.7 times higher in MER group (P < 0.001). The cost-effectiveness ratio for MER therapy was significantly lower than for CT (2031 and 4483 per cured patient taking into consideration only direct medical cost).
Conclusion: Empiric meropenem monotherapy of severe infections in ICU appears to be more adequate and more cost-effective than conventional combination therapy with broad-spectrum antibiotics.
P836 Efficacy and safety of colistin
A. Levcovich, Y. Lischenski, P. Leon, S. Lev, R. Hazzan, M. Chowers, J. Bishara, L. Leibovici, M. Paul (Petah Tikva, Kfar Saba, IL)
Objectives: The use of colistin decreased substantially in the 1970s following reports of renal failure and neuropathy. The appearance of highly resistant Gram-negative bacteria in hospitals, such as Acinetobacter baumannii, Pseudomonas spp. and Klebsiella spp., rendered colistin a drug of last resort for patients infected with these bacteria. The objectives of this study were to establish the safety and efficacy profile of colistin, in comparison to carbapenems and ampicillin-sulbactam.
Methods: Prospective observational cohort study including all adult patients treated with colistin, imipenem, meropenem or ampicillin/sulbactam with bacteraemia or fulfilling CDC diagnostic criteria for pneumonia, urinary tract infection, meningitis, catheter-related or catheter-associated infections. We compared outcomes for colistin vs. comparator antibiotics. Data were collected at the time of infection presentation with follow-up of 30 days. Mortality was defined as 30-day all-cause mortality. Renal failure was defined as ≥50% increase of creatinine levels from baseline and above 1.4 mg/dL at 2 weeks follow-up.
Results: Data were collected between March to October 2006 in 2 medical centres in Israel; 31 patients were treated with colistin and 44 with comparator antibiotics (meropenem 12, imipenem 15, ampicillin-sulbactam 17). All infections were healthcare associated (median time in hospital before infection 17 days, 0–34). Prior antibiotic treatment was administered to 27 patients with colistin vs. 38 patients with comparator antibiotics. Treatment was administered for microbiologically documented infections in all but 2 patients. Acinetobacter, Klebsiella and Pseudomonas spp. were implicated in 56%, 56% and 31% of infections, respectively. Colistin was administered as single drug to 27/31 colistin-treated patients; bacteria were susceptible only to colistin in 23/3. Baseline patient characteristics and outcomes are shown in the table . Mortality was significantly higher with colistin vs. comparators, 48% vs. 20%, p = 0.01. A multivariate analysis including 75 patients and the variables age, McCabe score, independent functional status and treatment arm as covariates, revealed that colistin remained significantly associated with 30-day mortality (p = 0.03).
| Baseline patient characteristics | Colistin | Comparators | P-value |
|---|---|---|---|
| No patients | 31 | 44 | |
| Female gender | 14 (45.2%) | 19 (43.2%) | 0.86 |
| Age (mean, SD) | 68 (±16) | 57 (±20) | 0.029 |
| Service | |||
| Internal medicine | 12 (38.7%) | 14 (31.8%) | 0.29a |
| Surgery | 4 (12.9%) | 14 (31.8%) | |
| ICU | 15 (48.4%) | 16 (36.4%) | |
| Baseline functional status (full | 17 (54.8%) | 31 (70.5%) | 0.17 |
| activity) | |||
| Central line | 10 (32.3%) | 19 (43.2%) | 0.34 |
| Mechanical ventilation | 18 (58.1%) | 25 (56.8%) | 0.91 |
| Recent invasive procedure | 18 (58.1%) | 29 (65.9%) | 0.49 |
| Charison score (median, range) | 2 (1–6) | 1 (1–7) | 0.31 |
| McCabe score | |||
| 1 | 17 (54.8%) | 32 (72.7%) | 0.11b |
| 2 | 7 | 8 | |
| 3 | 7 | 4 | |
| SOFA score (median, range) | 4 (1–14) | 4 (1–14) | 0.68 |
| Chronic renal failure | 10 (32.3%) | 11 (25%) | 0.49 |
| Albumin g/dL (mean, SD) | 2.2 (±0.46) | 2.4 (±0.66) | 0.33 |
| WBC K/micl (mean, SD) | 16.587 (±9.6) | 14.294 (±8.4) | 0.28 |
| Bacteraemia | 12 (38.7%) | 13 (29.5%) | 0.41 |
For ICU vs. others
for 1 vs. others.
Conclusions: Treatment with colistin was associated with a significantly higher mortality rate at 30 days and a non-significantly higher rate of renal failure development. Higher-risk patients were treated with colistin.
P837 Clinical factors associated with daptomycin outcomes in skin and soft-tissue infections
K. Lamp, L. Friedrich, K. Lindfield (Lexington, US)
Objectives: To examine the impact of multiple clinical factors on daptomycin (DAP) clinical outcomes in patients (pts) with skin and soft-tissue infections (SSTI).
Methods: The Cubicin® Outcomes Registry and Experience (CORE∧sm 2005) is a multicentre (52 US institutions), retrospective study evaluating DAP outcomes. Pts unevaluable for outcome were excluded. Factors that may impact clinical outcomes (initial creatinine clearance [CrCl] <30 mL/min, dialysis, complicated SSTI [cSSTI], bacteraemia, methicillin-resistant Staphylococcus aureus [MRSA], sepsis, ICU stay, diabetes, cancer history, chronic kidney disease, age ≥66 yrs, concomitant antibiotic (abx) use, community location, prior abx category) were investigated in a multivariate logistic regression analysis (MV). Prior abx categories were defined as: no prior abx (DAP first line, FL), DAP after vancomycin (VAN) failure (VF), after VAN without VAN failure (VNF), after non-VAN failure (NVF), and after non-VAN without non-VAN failure (NVNF). Prior abx failure was determined by the investigator.
Results: There were 486 evaluable pts with SSTI including noncatheter-related bacteraemia. The number of pts in prior abx categories were FL (n = 127, 26%), VF (n = 56, 12%), VNF (n = 121, 25%), NVF (n = 86, 18%), NVNF (n = 96, 20%). Pts were 51% female, 21% ≥66 yrs, 12% with CrCl <30 mL/min or dialysis, 57% were in the community 2 days prior to DAP. The population consisted of 70% cSSTI, 30% uncomplicated SSTI and 5% of all pts had bacteraemia. The most common pathogen cultured was MRSA, 52%. The median DAP dose (4 mg/kg) and duration (12 days) did not differ by prior abx category. Overall, 94% of pts achieved clinical success after DAP therapy. By logistic regression DAP success rates were higher for FL than VF (98% vs. 88%, P = 0.018). The presence of an initial low CrCl (P = 0.015), ICU stay (P = 0.007) and sepsis (P = 0.009) were significant predictors of lower DAP success in MV There was a trend that VF (P = 0.07) or NVF (P = 0.07) were associated with lower DAP success.
Conclusion: In almost 500 SSTI pts treated with DAP, clinical success rates exceeded 90%. In the MV, a low initial CrCl, ICU stay or presence of sepsis were associated with lower DAP success which is consistent for markers of severity of illness. Prior abx category, in the presence of other patient factors, in MV resulted in the observation that DAP success was independent of the prior abx. Further studies are needed to confirm these findings.
P838 Clinical experience trends with daptomycin in the first two years: report from a registry
K. Lamp, L. Friedrich (Lexington, US)
Objectives: To describe trends in the use of daptomycin (DAP) in the United States during the first two years on the market.
Methods: The Cubicin® Outcomes Registry and Experience (CORESM) collected data in 2004 and 2005 at 45 and 52 US institutions, respectively. CORESM is a retrospective, observational chart review evaluating outcomes of DAP patients (pts). Investigators collected demographic, disease state, clinical and microbiological data; outcomes were assessed using standard definitions.
Results: There were 1160 and 1172 pts enrolled in CORESM 2004 and 2005, respectively. Pts with more than one infection type were stratified by severity of infection (endocarditis > osteomyelitis > bacteraemia > other [foreign body, septic arthritis, pyelonephritis/UTI, CNS, necrotising infection] > complicated skin and soft-tissue, cSSTI > uncomplicated skin and soft-tissue, uSSTI). The patients in 2004/2005 (%/%) by infection type were: endocarditis (4/3), osteomyelitis (12/12), bacteraemia (19/25, P < 0.01), other (15/15), cSSSI (32/33), uSSSI (18/12, P < 0.01). These findings were consistent even if restricted to sites participating in both years. Pt gender, age ranges and location 48 hours prior to starting DAP were similar for both years. The percentage of pts with an initial creatinine clearance <30 mL/min was higher in 2004 (30% vs. 18%, P < 0.001). Overall, DAP was used as initial therapy in 24% of pts. Staphylococcus aureus was the most common pathogen, isolated in approximately one-half of patients (methicillin-resistant > methicillin-susceptible), followed by coagulase-negative staphylococci and vancomycin-resistant enterococci. The mean DAP dose was higher for all infection types in 2005; however, the median dose differed only for bacteraemia and other infections (P < 0.05). The median DAP duration was 13 days for both years. Clinical outcomes for 2004/2005 (%/%) were success, cure plus improved, (80/75), failure (5/6), and nonevaluable (15/19).
Conclusion: DAP was primarily used to treat SSTI. In 2005, DAP was used in more pts with bacteraemia and at higher doses, which may reflect the release of data from a phase 3 study in S. aureus bacteraemia and endocarditis. In a population of pts with a high rate of previous antibiotic use (76%), DAP was associated with good clinical outcomes. Further studies are needed to describe the effectiveness of DAP.
P839 Daptomycin is clinically more rapidly effective than comparator for treatment of complicated skin and skin-structure infections
L. Friedrich, J. Krige, K. Lindfield, C. Otradovec, W. Martone, D. Katz, F. Tally (Mt. Pleasant, US; Cape Town, ZA; Lexington, US)
Objective: To compare rapidity of clinical response between daptomycin (DAP) and comparator (COMP) (vancomycin, semi-synthetic penicillin) in South African (SA) patients from 2 large randomised, blinded studies.
Methods: A subset of clinically evaluable SA patients comparing DAP to COMP for complicated skin and skin-structure infections (cSSSI) treatment with either 0 or 1 clinical comorbidity were studied. Patients were evaluated at baseline, on therapy (day 3–4), end of therapy (EOT), and test of cure (TOC). Clinical symptom (edema, erythema, fluctuance, induration, necrotic tissue, purulent drainage, tenderness, ulceration) severity was assessed as none, mild, moderate, or severe by a blinded investigator at each visit. These assessments were combined into 2 categories (none/mild and moderate/severe) and tallied for each visit. The Mantel-Haenszel chi-square was used to assess overall differences between DAP and COMP symptom severity at any visit with chi-square tests to determine the source(s) of the overall effect for each symptom at each visit. Nonparametric tests were used for all other comparisons. A P < 0.05 was statistically significant and 0.05 ≤ P < 0.10 marginal.
Results: Of 356 clinically evaluable patients, 326 (92%) had 0 or 1 clinical comorbidity and were eligible for analysis (DAP = 174, COMP = 152). Common comorbidities included SIRS (∼30%), diabetes (7%-9%), and peripheral vascular disease (2%-3%); there were no demographic differences between groups. Wound infection (∼50%) and major abscess (∼20%) were most common. Diabetic ulcer occurred in 3% of each group. Staphylococci and Streptococci were the most common pathogens. Only 4% of COMP patients received vancomycin. Clinical success was 93% for DAP and 95% for COMP. DAP patients had a shorter median duration of therapy (7 [3–15] vs. 8 [3–17] days, P < 0.0001). Overall, DAP patients improved more quickly (P = 0.038). No differences in symptom severity were noted between DAP and COMP at baseline. DAP symptom severity significantly decreased by day 3–4 for induration (P = 0.027), erythema (P = 0.048) and marginally for edema (P = 0.096) and necrotic tissue (P = 0.097). At EOT, DAP showed a significant decrease in severity for induration (P = 0.043) and a marginal decrease in tenderness (P = 0.056). At TOC, the decrease in severity for DAP for induration was still evident (P = 0.031).
Conclusion: DAP length of therapy was significantly shorter and produced a more rapid clinical response than COMP as evidenced by a significant decrease in the severity of clinical symptoms.
P840 Hospital length of stay in patients with methicillin-resistant Staphylococcus aureus infections: retrospective analysis of a clinical study comparing tigecycline and vancomycin
R. Mallick, S. Sun (Collegeville, US)
Objective: Using data from a recent clinical study, to investigate which presenting characteristics were risk factors for prolonged length of stay (LOS) in hospitalised patients treated for serious infections involving MRSA.
Method: For this analysis, data from patients with MRSA in a multinational, double-blind clinical study conducted in patients with confirmed serious infections involving resistant Gram-positive pathogens (MRSA and vancomycin-resistant enterococci [VRE]) was extracted. In the study, patients with MRSA were randomly assigned in a 3:1 ratio to receive IV tigecycline or vancomycin. Treatment duration was to be 7–28 days, depending on site and severity of the infection. Hospital length of stay was based on additional systemic evaluation. We (a) summarised baseline characteristics and (b) estimated multiple regression models – adjusting for hospital death and study discontinuation as necessary – to identify risk factors for prolonged LOS.
Results: Among patients with MRSA infections who met minimal disease criteria (n = 133) and had complete hospitalisation data (n = 131), diabetes (20.7%) and peripheral vascular disease (PVD) (17.4%) were leading co-morbidities. Complicated skin and skin structure infections (69.5%), complicated intra-abdominal infections (16.0%) and bacteraemia – including catheter-related bacteraemia – (9.9%) constituted the most common infection types. Mean APACHE severity score was 7.9; 9.9% of patients had an APACHE score >15. Mean duration of IV antibiotic therapy was 12.1 days, mean LOS was 15.0 days, re-hospitalisation rate was 9.2%, mortality rate was 3.8%. Bacteraemia (+4.8 days; p = 0.0014) and peripheral vascular disease (+2.7 days; p = 0.029) were notably associated with prolonged IV treatment duration. An APACHE score >15 was associated with a substantial impact (+15.8 days; p < 0.0001) on hospital LOS. There was no significant difference between the tigecycline and vancomycin groups in IV treatment duration, LOS, re-hospitalisation rate or mortality rate.
Conclusions: In this retrospective analysis of a clinical study in patients with selected serious infections involving MRSA, bacteraemia and peripheral vascular disease emerged as presenting risk factors for prolonged IV antibiotic treatment, and an APACHE score >15 for prolonged hospitalisation; there were no significant differences between tigecycline and vancomycin.
P841 Duration of hospital length of stay in patients with community-acquired pneumonia treated in European centres: retrospective analysis of a clinical study comparing tigecycline and levofloxacin
R. Mallick, S. Sun (Collegeville, US)
Objective: To investigate risk factors for prolonged length of stay (LOS) in hospitalised CAP patients (pts) in European centres.
Method: Data from European centres in a double-blind clinical study among hospitalised pts with CAP who were stratified by the FINE Pneumonia Severity Index and randomised to receive intravenous (IV) tigecycline or IV levofloxacin, was retrospectively analyzed. Treatment termination was based on resolution of infection symptoms and hospital discharge on additional systemic evaluation. We (a) summarised baseline characteristics and (b) estimated multiple regression models – adjusting for hospital death and study discontinuation as necessary – to identify risk factors for prolonged LOS.
Results: Among 345 pts (23.2% age >65 yrs) hospitalised for CAP in European centres, diabetes (10.1%) and COPD (8.1%) were leading co-morbidities; 57.1% of pts were past or current smokers. Median FINE category and CURB-65 severity index scores were 3 (range 1–5) and 1 (range 0–4) respectively. Blood urea nitrogen (BUN) concentration >7 mmol/L (22.0% of pts) and respiratory rate >30/minute (18.0% of pts) were most common severity components. Among 246 pts with known microbiology, Streptococcus pneumoniae (41.5%) and Mycoplasma pneumoniae (28.1%) were leading pathogens; 42.7% of pts had a poly-microbial infection. Among all pts, mean duration of IV antibiotic therapy was 10.0 days; mean LOS was 13.8 days. Alcohol abuse (hazard ratio [HR] = 0.47; p = 0.0039), co-morbid liver disease (HR = 0.60; p = 0.0485) and BUN >7 mmol/L (HR = 0.74; p = 0.0282) were associated with slower hospital discharge rates (prolonged LOS). Among the 246 pts with known microbiology, presence of a polymicrobial infection (HR = 0.69; p = 0.007), Legionella pneumoniae (HR = 0.41; p = 0.005), or Haemophilus influenzae (HR = 0.57; p = 0.013) was associated with prolonged LOS, as was absence of cure (HR = 0.28; p < 0.0001). There was no significant difference between the tigecycline and levofloxacin groups in LOS, however, tigecycline was associated with a slight trend towards a lower re-hospitalisation rate (tigecycline 0.6%, levofloxacin 4.1%; p = 0.067).
Conclusions: Co-existing liver disease, alcohol abuse, abnormal blood urea nitrogen, and among pts with known microbiology, presence of polymicrobial infections, Legionella pneumoniae or Haemophilus influenzae emerged as risk factors for prolonged hospitalisation; there were no differences between tigecycline and levofloxacin.
P842 An open-label, randomised, comparative study of the patient compliance, efficacy and safety of treatment with clarithromycin extended-release versus clarithromycin immediate-release for the treatment of acute bacterial exacerbation of chronic bronchitis
P. Kardas (Lodz, PL)
Objectives: The aim of this study was to compare patient compliance as well as the efficacy and safety of clarithromycin extended-release (ER) tablets administered once daily (QD) and immediate-release (IR) tablets administered twice daily (BID).
Methods: This was an open-label, randomised, comparative study in adult outpatients (age ≤ 60 years) with a diagnosis of acute bacterial exacerbation of chronic bronchitis (ABECB). Eligible patients were randomised 1:1 to receive either 1 clarithromycin ER 500-mg tablet QD for 7 days or 1 clarithromycin IR 250-mg tablet BID for 7 days. Studied drugs were given to the patients in excessive number in MEMS 6 containers, which enabled precise electronic measurement of compliance parameters. Clinical response was assessed at day 7 and at the test-of-cure visit (study day 21). For the assessment of patient compliance, the overall compliance (defined with the ratio of the number of container openings to the number of prescribed doses), as well as days with correct number of doses taken, were employed. Safety was assessed based on the incidence of study drug-related adverse events.
Results: A total of 122 patients were randomised (60 to ER and 62 to IR group). Clinical cure rates were similar for evaluable patients treated with ER (94.5%, 52/55) and those treated with IR (88.1%, 52/59) (P > 0.05). Mean overall compliance was 94.3% in the ER group and 82.3% in the IR group, and the mean days with correct number of doses taken was 80.2% and 70.3% for the respective treatment groups (P < 0.001). The incidence of adverse events was 8% (5/60) in the ER group and 8% (5/62) in the IR group.
Conclusion: Clarithromycin ER 500-mg tablets QD for 7 days were equally effective and well tolerated as clarithromycin IR 250-mg tablets BID for 7 days in treating adults with ABECB, and were associated with significantly higher rates of patient compliance.
P843 Effectiveness of linezolid in nosocomial and severe community-acquired pneumonia: meta-analysis
O. Yeniova, S. Ascioglu (Ankara, TR; Boston, US)
Objective: To review the evidence for the effectiveness of linezolid treatment in patients with nosocomial or severe community acquired pneumonia (CAP).
Methods: We searched PubMed and Cochrane databases (January 1990 to 31 October 2006) using linezolid and pneumonia as text words, also hand searched the bibliographies of published studies. We tried to be inclusive in our inclusion criteria due to scarcity of studies in this area. Studies met our inclusion criteria if they were randomised controlled trials or observational studies comparing linezolid with a suitable comparator in adult patients with pneumonia. Pooled estimates of the risk ratio (RR) for successful outcome were obtained using a fixed effects model. Test of heterogeneity was not significant (MH test, p = 0.950).
Results: Our search yielded a total of 117 references. After we examined all abstracts, we identified 6 randomised controlled trials. There weren't any observational studies or non-randomised trials which met our inclusion criteria. Only 2 of the included trials had a primary objective of evaluating effectiveness in nosocomial pneumonia patients, 3 trials had subgroups of nosocomial pneumonia patients and 1 trial included hospitalised patients with severe CAP. In all but 1 of the studies comparator drug was a glycopeptide (2 studies teicoplanin, 3 vancomycin), in the severe CAP study only, comparator drug was ceftriaxone. The pooled analysis of all trials showed that linezolid was not associated with a significantly better outcome than the comparator treatments (RR: 1.05, 95% CI: 0.98–1.12) (Figure ). We did a sensitivity analysis including only nosocomial pneumonia studies; omission of the CAP study which appeared to be the one that dominated the other trials attenuated the effect estimate (RR: 1.02, 95% CI: 0.92–1.13). Our sample size was too small to evaluate publication bias.

Conclusions: Linezolid had a very small favourable effect on the outcome of patients with severe CAP and nosocomial pneumonia and this result wasn't statistically significant. Future trials addressing linezolid treatment in these patient groups are urgently needed.
P844 The ATLAS studies: double-blind, randomised, active controlled, multinational Phase 3 trials comparing telavancin with vancomycin for the treatment of complicated skin and skin structure infections
G.R. Corey, M.E. Stryjewski, V.G. Fowler Jr, V. Skerk, A. Hopkins, M.M. Kitt, S.L. Barriere for the ATLAS Study Group
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) is a common pathogen isolated from complicated skin and skin structure infections (cSSSI). Telavancin is a novel, rapidly bactericidal lipoglycopeptide with a multifunctional mechanism of action against Gram-positive pathogens, including MRSA. The ATLAS trials compared the efficacy and safety of telavancin with vancomycin for the treatment of cSSSI, especially in patients with infections due to MRSA.
Methods: ATLAS 1 and 2 were identical double-blind, randomised, active controlled, multinational Phase 3 trials. Patients were 18 years of age or older with suspected or confirmed MRSA resulting in cSSSI (major abscess, infected burn, deep/extensive cellulitis, infected ulcer or wound infection) requiring ≥7 days of IV antibiotic therapy. Patients received telavancin 10 mg/kg administered IV once daily or vancomycin 1 g IV once every 12 h for 7–14 days. Efficacy was assessed as the clinical cure at the test-of-cure visit (7–14 days after administration of the last dose of study drug) in the combined population. Safety was also assessed.
Results: A total of 928 patients received telavancin (n = 745 clinically evaluable; n = 278 clinically evaluable with MRSA) and 939 received vancomycin (n = 744 clinically evaluable; n = 301 clinically evaluable with MRSA). Clinical cure at the test-of-cure visit was similar for the two drugs in the combined clinically evaluable population (telavancin, 88.3%; vancomycin, 87.1%; difference, 1.2% [95% CI, −2.1, 4.6]). Clinical cure was also comparable in clinically evaluable patients with MRSA infections (telavancin, 90.6%; vancomycin 86.4%; difference, 4.1% [95% CI, −1.1, 9.3]), regardless of diabetes status, gender, race, type of cSSSI or body mass index. Serious adverse events and discontinuations due to adverse events were slightly greater in the telavancin group. The most common adverse events associated with telavancin were generally mild and transient taste disturbance, nausea, vomiting and foamy urine. Renal adverse events were infrequent, but were greater with telavancin than vancomycin (3% vs 1%).
Clinical cure and microbiological eradication at test-of-cure clinic visit
| Endpoint | Cure rate |
||
|---|---|---|---|
| Telavancin, n (%) | Vancomycin, n (%) | Difference, % (95%CI) | |
| Clinical cure | |||
| All-treated population | 710/928 (76.5) | 697/939 (74.2) | 2.3 (−1.6, 6.2) |
| Clinically evaluable population (MRSA) | 658/745 (88.3) | 648/744 (87.1) | 1.2 (−2.1, 4.6) |
| 252/278 (90.6) | 260/301 (86.4) | 4.1 (−1.1, 9.3) | |
| Microbiological eradication | |||
| Modified all-treated population | 527/680 (77.5) | 526/703 (74.8) | 2.7 (−1.8, 7.2) |
| Microbiologically evaluable population (MRSA) | 473/527 (89.8) | 468/536 (87.3) | 2.4 (−1.4, 6.2) |
| 250/278 (89.9) | 257/301 (85.4) | 4.4 (−0.9, 9.8) | |
| Overall therapeutic response (cure + eradication) | |||
| Modified all-treated population | 516/680 (75.9) | 515/703 (73.3) | 2.6 (−2.0, 7.2) |
| Microbiologically evaluable population (MRSA) | 467/527 (88.6) | 462/536 (86.2) | 2.4 (−1.6, 6.4) |
| 250/278 (89.8) | 255/301 (84.7) | 5.1 (−0.3, 10.5) | |
Conclusion: Combined data from ATLAS 1 and 2 have consistently shown that telavancin is non-inferior to vancomycin for the treatment of patients with a cSSSI, including those caused by MRSA. Telavancin had an acceptable adverse event profile for the treatment of serious infections due to resistant bacteria.
P845 ATLAS 1: the first Phase 3 study evaluating the new lipoglycopeptide, telavancin, for the treatment of patients with complicated skin and skin structure infections
G.R. Corey, M.E. Stryjewski, W.D. O'Riordan, V.G. Fowler Jr., D.P. Ross, A. Hopkins, M.M. Kitt, S.L. Barriere for the ATLAS Study Group
Objectives: Recent publications have highlighted the critical shortage of new antimicrobial agents currently in Phase 2/3 development [1]. Telavancin, a novel, rapidly bactericidal lipoglycopeptide with a multifunctional mechanism of action against Gram-positive pathogens, was recently evaluated in two parallel Phase 3 studies involving patients with complicated skin and skin structure infections (cSSSIs). Here we report results from the first of these trials, ATLAS 1.
Methods: ATLAS 1 was a double-blind, randomised, active controlled, multinational study comparing the efficacy and safety of telavancin (10 mg/kg IV q 24 h) with vancomycin (1 g IV q 12 h). Eligible patients included men and women, aged 18 years or older, with a cSSSI (major abscess, infected burn, deep/extensive cellulitis, infected ulcer or wound infection) requiring 7 to 14 days of IV antibiotic therapy. The primary endpoint, clinical response at the test-of-cure visit (TOC; 7–14 days after administration of the last dose of study medication), was evaluated using non-inferiority (delta defined as 10%). Secondary objectives included cure rates by pathogen and microbiological eradication. Safety was also evaluated.
Results: Among all patients treated with telavancin (n = 426) or vancomycin (n = 429), 81% were clinically evaluable (CE; 346 and 349, respectively) and over 70% of the CE population was also microbiologically evaluable (237 and 255, respectively). Clinical cure at the TOC visit was similar for the two treatment groups (Table ), thus demonstrating non-inferiority for telavancin relative to vancomycin in this study. The safety profile of telavancin was acceptable for the treatment of serious infections due to resistant bacteria. Discontinuations due to adverse events were reported for 7% of telavancin-treated patients and 5% of vancomycin-treated patients.
Clinical cure and microbiological eradication at test-of-cure clinic visit
| Endpoint | Cure rate |
||
|---|---|---|---|
| Telavancin, n (%) | Vancomycin, n (%) | Difference, % (95%CI) | |
| Clinical cure | |||
| All-treated population | 323/426 (75.8) | 321/429 (74.8) | 1.0 (−4.8, 6.8) |
| Clinically evaluable population | 304/346 (87.9) | 302/349 (86.5) | 1.3 (−3.6, 6.3) |
| Clinically evaluable population (MRSA) | 101/116 (87.1) | 118/138 (85.5) | 1.6 (−6.9, 10.0) |
| Microbiological eradication | |||
| Modified all-treated population | 240/307 (78.2) | 241/322 (74.8) | 3.3 (−3.3, 10.0) |
| Microbiologically evaluable population | 212/237 (89.5) | 219/255 (85.9) | 3.6 (−2.2, 9.4) |
| Microbiologically evaluable population (MRSA) | 101/116 (87.1) | 117/138 (84.8) | 2.3 (−6.3, 10.8) |
| Overall therapeutic response (cure+ eradication) | |||
| Modified all-treated population | 235/307 (76.5) | 239/322 (74.2) | 2.3 (−4.4, 9.1) |
| Microbiologically evaluable population | 210/237 (88.6) | 218/255 (85.5) | 3.1 (−2.8, 9.0) |
| Microbiologically evaluable population (MRSA) | 101/116 (87.1) | 117/138 (84.8) | 2.3 (−6.3, 10.8) |
Conclusion: Telavancin is a novel lipoglycopeptide that acts by inhibiting bacterial cell wall synthesis and disrupting the functional integrity of the bacterial plasma membrane. The results of ATLAS 1, the first of two parallel, Phase 3 studies evaluating telavancin for the treatment of serious skin infections, consistently favoured telavancin compared with vancomycin.
P846 ATLAS 2: a double-blind, randomised, active controlled, multinational Phase 3 study comparing telavancin with vancomycin for the treatment of patients with complicated skin and skin structure infections
G.R. Corey, M.E. Stryjewski, V.G. Fowler Jr., O. Teglia, A. Hopkins, M.M. Kitt, S.L. Barriere for the ATLAS Study Group
Objectives: Telavancin is a novel, rapidly bactericidal lipoglycopeptide with activity against Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA). It has a multifunctional mechanism of action, which includes inhibiting cell wall synthesis and disrupting plasma membrane functional integrity. The efficacy and safety of telavancin vs vancomycin have been compared in patients with complicated skin and skin structure infections (cSSSIs) in two Phase 3 studies. Here we report results from the second of these trials, ATLAS 2.
Methods: ATLAS 2 was a double-blind, randomised, active controlled, multinational study involving men and women, aged 18 years or older. Patients had a cSSSI (major abscess, infected burn, deep/extensive cellulitis, infected ulcer or wound infection) requiring ≥7 days of IV antibiotic therapy. Patients were randomised to IV infusions of telavancin 10 mg/kg once daily or vancomycin 1 g once every 12 h for 7–14 days. The primary efficacy endpoint was the clinical response at the test-of-cure visit (7–14 days after the last dose of study drug), evaluated using non-inferiority criteria (with a non-inferiority delta of 10%). Secondary endpoints included clinical cure in patients with MRSA infections, clinical cure by pathogen and microbiological eradication. Safety was also evaluated.
Results: A total of 502 patients received telavancin (n = 399 clinically evaluable; n = 290 microbiologically evaluable; n = 204 with MRSA) and 510 received vancomycin (n = 395 clinically evaluable; n = 281 microbiologically evaluable; n = 202 with MRSA). Clinical cure and eradication at the test-of-cure visit were similar for the two treatments (Table ), demonstrating the non-inferiority of telavancin vs vancomycin, since the lower bound of the 95% confidence interval was greater than −10%. Telavancin achieved a higher overall therapeutic response (cure + eradication) than vancomycin. Adverse events were comparable in the two treatment arms. Overall, 43 (8.6%) telavancin-treated patients and 28 (5.5%) vancomycin-treated patients discontinued due to adverse events.
Clinical cure and microbiological eradication at test-of-cure clinic visit
| Endpoint | Cure rate |
||
|---|---|---|---|
| Telavancin, n (%) | Vancomycin, n (%) | Difference, % (95%CI) | |
| Clinical cure | |||
| All-treated population | 387/502 (77.1) | 376/510 (73.7) | 3.4 (−1.9, 8.7) |
| Clinically evaluable population | 354/399 (88.7) | 346/395 (87.6) | 1.1 (−3.4, 5.6) |
| Clinically evaluable population (MRSA) | 151/162 (93.2) | 142/163 (87.1) | 6.1 (−0.3, 12.5) |
| Microbiological eradication | |||
| Modified all-treated population | 287/373 (76.9) | 285/38 (74.8) | 2.1 (−4.0, 8.2) |
| Microbiologically evaluable population | 261/290 (90.0) | 249/281 (88.6) | 1.4 (−3.7, 6.5) |
| Microbiologically evaluable population (MRSA) | 149/162 (92.0) | 140/163 (85.9) | 6.1 (−0.7, 12.9) |
| Overall therapeutic response (cure +eradication) | |||
| Modified all-treated population | 281/373 (75.3) | 276/381 (72.4) | 2.9 (−3.4, 9.2) |
| Microbiologically evaluable population | 257/290 (88.6) | 244/281 (86.8) | 1.8 (−3.6, 7.2) |
| Microbiologically evaluable population (MRSA) | 149/162 (92.0) | 138/163 (84.7) | 7.3 (0.4, 14.2) |
Conclusion: In the ATLAS 2 study, telavancin was found to be effective for the treatment of patients with cSSSIs, including those caused by MRSA. The results, including overall therapeutic response, consistently favoured telavancin compared with vancomycin. Telavancin also had an acceptable adverse event profile for the treatment of serious infections due to resistant bacteria.
P847 Activity of telavancin against Staphylococcus aureus isolates carrying the Panton-Valentine leukocidin gene in the ATLAS studies
V.G. Fowler Jr., T. Rude, C. Nelson, M.E. Stryjewski, M.M. Kitt, S.L. Barriere, D. Friedland, G.R. Corey (Durham, South San Francisco, US)
Objectives: Telavancin (TLV) is a novel, rapidly bactericidal lipoglycopeptide antibiotic with a unique, multifunctional mechanism of action against a broad range of Gram-positive pathogens. TLV was recently evaluated in two large, international, Phase 3 studies (ATLAS 1 and 2) involving patients with complicated skin and skin structure infections (cSSSI). Panton-Valentine leukocidin (PVL) is a highly potent cytotoxin which is increasingly associated with community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infections. This analysis examined PVL gene prevalence in S. aureus isolates from patients with cSSSI enrolled in ATLAS 1 and 2.
Methods: ATLAS 1 and 2 were identical, double-blind, randomised, active-controlled, multinational, Phase 3 studies. Patients were ≥ 18 years of age with a cSSSI (major abscess, infected burn, deep/extensive cellulitis, infected ulcer or wound infection) caused by a Gram-positive pathogen and required ≥7 days of IV antibiotic therapy. Patients were randomised to TLV (10 mg/kg administered IV q 24 h) or vancomycin (1 g IV q 12 h). Clinical cure was determined at the test-of-cure (TOC) visit (7–14 days after administration of the last dose of study medication). S. aureus isolates were screened for PVL genes (lukS-PV and lukF-PV) by PCR methods.
Results: Isolates from a total of 870 clinically evaluable patients in the two studies were available for analysis (543 MRSA; 327 methicillin-sensitive S. aureus [MSSA]). The PVL gene was detected in isolates from 557/870 (64%) patients. Rates of PVL-positivity were higher in patients with MRSA isolates (459/543 [85%]) than MSSA isolates (98/327 [30%]). Clinical cure at the TOC visit was similar for TLV and vancomycin in the combined ATLAS 1 and 2 microbiologically evaluable populations, regardless of PVL status (Figure ).

Overall treatment difference in clinical response at test-of-cure by pathogen PVL characteristics; ATLAS 1 and ATLAS 2 combined, CE population. *Weighting for point difference was bazed on the size of ATLAS 1 and ATLAS 2 trials. Percentages shown to the right were not weighted.
Conclusion: The PVL gene was present in a large proportion of S. aureus isolates from cSSSI patients in the ATLAS studies. PVL status did not seem to influence the outcome of treatment. In these studies, TLV was effective in the treatment of cSSSI caused by S. aureus, including strains carrying the PVL gene.
P848 Efficacy of tigecycline (TGC) compared with levofloxacin (LEV) for treating Streptococcus pneumoniae bacteraemia in patients (pts) hospitalised with community-acquired pneumonia (CAP)
N. Dartois, G. Dukart, C.A. Cooper, N. Castaing, H. Gandjini on behalf of the 308 and 313 Study Groups
Objective: Tigecycline (TGC), a first-in-class glycylcycline approved for treating complicated skin/skin structure and intra-abdominal infections, has an expanded spectrum of activity against Gram-positive, Gram-negative and atypical bacteria including some resistant strains. CAP pts with Streptococcus pneumoniae bacteraemia frequently have more severe disease and increased mortality than pts without bacteraemia. We evaluated the efficacy and safety of TGC vs. levofloxacin (LEV) in a subset of hospitalised CAP pts with pneumococcal bacteraemia.
Method: Two Phase 3, multicentre, double blind studies were conducted in hospitalised CAP pts. Pts were randomised to receive IV TGC (100 mg then 50 mg ql2h) or IV LEV (500 mg q24h or q12h). In 1 study, pts could be switched to oral LEV after ≥3 days of IV dosing. Clinical response was evaluated at test-of-cure (TOC). Results are presented for the microbiologically evaluable (ME) and microbiologic modified intent-to-treat (m-mITT) populations.
Results: 846 pts received at least 1 dose of study drug in these 2 trials. Of the 345 ME and 457 m-mITT pts, 40 (11.6%) and 50 (10.9%) pts, respectively, had S. pneumoniae (sp.) bacteraemia. At TOC (ME population), TGC cured 20/22 (90.9%)and LEV cured 13/18 (72.2%) – absolute difference TGC–LEV, 18.7% (95%CI −8.8,45.6). The cure rate for TGC in sp. bacteraemic pts was similar to that of TGC in ME pts without sp. bacteraemia (127/138, 92.0%). The cure rate for LEV in bacteraemic pts (ME) was numerically lower than that for LEV pts without bacteraemia (146/159, 91.8%). In the m-mITT population, TGC cured 22/27 (81.5%) and LEV cured 15/23 pts (65.2%) – absolute difference TGC–LEV 16.3% (95%CI −10.5,41.4). Again, the cure rate for TGC in sp. bacteraemic pts was similar to that achieved by TGC in pts without sp. bacteraemia (162/189, 85.7%), whereas the cure rate for LEV was lower in bacteraemic vs. non-bacteraemic pts (177/203, 87.2%). The minimum inhibitory concentration (MIC) of TGC for S. pneumoniae blood isolates ranged from 0.03–0.12 μg/mL, whereas the MIC of LEV for these isolates ranged from 0.5–1.0 μg/mL.
Conclusions: In these 2 studies, TGC appeared safe and achieved cure rates similar to LEV in hospitalised pts with CAP. Further, in a subset of pts with S. pneumoniae bacteraemia, TGC achieved cure rates similar to those in TGC-treated CAP pts without bacteraemia. Finally, TGC achieved a numerically higher cure rate than LEV, although the difference was not statistically significant.
P849 Efficacy of tigecycline versus levofloxacin in patients hospitalised with community-acquired pneumonia: analysis of risk factors
N. Dartois, G. Dukart, C. Cooper, N. Castaing, I. Gandjini, D. Sarkozy on behalf of the 308 and 313 Study Groups
Objective: The initial severity of community-acquired pneumonia (CAP) and eventual clinical outcome may be influenced by a number of risk factors. Tigecycline (TGC), a first-in-class glycylcycline approved for treating complicated skin/skin structure and intra-abdominal infections, exhibits expanded activity against Gram-positive, Gram-negative and atypical bacteria and some resistant strains. The efficacy of TGC vs levofloxacin (LEV) was studied in hospitalised pts with CAP. In addition to overall response, exploratory analyses were performed evaluating response to treatment for a variety of risk factors.
Method: Two Phase 3, multicentre, double blind studies were conducted in hospitalised pts with CAP who were randomised to receive IV TGC (100 mg initially then 50 mg ql2h) or IV LEV (500 mg q24h or q12h). In 1 study, pts could be switched to oral LEV after ≥3 days of IV dosing. At randomisation, pts were stratified by: geographic location in 1 study and by Fine Pneumonia Severity Index in both studies. Clinical response was evaluated at test-of-cure. Risk factors, such as age, Fine Pneumonia Severity Index and estimated CURB-65 scores, and co-morbidities including diabetes (DM), chronic obstructive pulmonary disease, congestive heart failure (CHF) and cerebrovascular disease (CVD) were evaluated. Results will be presented for the clinically evaluable (CE) population.
Results: 846 pts received at least 1 dose of study drug in these 2 trials and the CE population consisted of 574 pts (TGC = 282, LEV = 292). Overall, the cure rates (CE) for TGC (253/282 pts, 89.7%) were similar to LEV (252/292, 86.3%). Although the number of pts was small for some subgroups, a statistically significant subgroup by treatment interaction was observed for some factors, such as age <55 vs. ≥55 and presence or absence of CVD, CHF, and DM. The cure rates for TGC pts were generally consistent across the subgroups, while those for LEV were more variable. Failure analysis revealed no significant risk factors for TGC pts, whereas multilobar disease, prior antibiotic failures, creatinine clearance (≤70 mL/min), and geographic region (Western Europe and US/Canada) were significant risk factors for LEV.
Conclusions: In both studies, TGC appeared safe and achieved cure rates similar to LEV in hospitalised pts with CAP. The cure rates for TGC pts were generally consistent across the subgroups. Further, in an analysis of risk factors, which contributed to failure of the study regimen, no prognostic factors were identified for TGC treated subjects.
P850 Comparison of clinical outcomes in patients with Staphylococcus aureus bacteraemia and endocarditis presenting with or without systemic inflammatory response syndrome
Z. Kanafani, G. Vigliani, H. Boucher, H. Chambers, M. Rupp, S. Nasraway, S. Rehm, M. Campion, E. Abrutyn, A. Karchmer, D. Levine, G. Fatkenheuer, H. Brodt, T. Wolf, R. Corey (Durham, Lexington, Boston, San Francisco, Omaha, Cleveland, Philadelphia, Detroit, US; Cologne, Frankfurt, DE)
Objectives: S. aureus is a unique pathogen with an ability to invade normal tissues including normal heart valves. Determining the optimal length of therapy (LOT) at onset of S. aureus bacteraemia (SAB) is therefore difficult. In addition, the effect of the systemic inflammatory response syndrome (SIRS) on the outcome of patients with documented SAB (SIRS + SAB = sepsis) has not been determined. We evaluated the final diagnosis and outcomes in patients with documented SAB enrolled in the recently reported SAB/S. aureus infective endocarditis (SAIE) daptomycin trial who presented with and without SIRS (sepsis) at baseline.
Methods: The SAB/SAIE daptomycin trial was a randomised study conducted between August 2002 and February 2005 in 44 sites in the United States and Europe. Eligible patients had ≥1 positive blood cultures for S. aureus within two days of enrolment. Patients were randomised to daptomycin 6 mg/kg/d or combination of either vancomycin 1 g every 12 hours or antistaphylococcal penicillin 2 g every 4 hours, both with gentamicin 1 mg/kg every 8 hours for 4 days. Investigators determined the duration of treatment based on the working diagnosis. Final diagnosis and outcome at the end of therapy (EOT) and test of cure (TOC) were determined by an adjudication committee blinded to treatment group assignment. SIRS was considered present at baseline if patients met 2 or more of the following criteria: temperature <36°C or >38°C, heart rate >90 bpm, respiratory rate >20 breaths/min, WBC <4,000 mm3 or 12,000/mm3 or bands >10%.
Results: One hundred and seventy-six of the 235 patients enrolled with SAB (75%) met SIRS criteria. The proportion with MRSA was similar in 38.6% patients with and 35.6% without SIRS. Complicated SAB or SAIE was the final diagnosis in 133/176 (75.6%) with SIRS and 41/59 (69.5%) without SIRS. Patients with and without SIRS had similar success rates at EOT (78.4 vs 71.2%) and TOC (71.0 vs 64.4%) and similar mortality by 42 days after treatment (11.4 vs 11.9%).
Conclusion: SAB is associated with a high rate of complicated disease and SAIE irrespective of the presence of SIRS at baseline. The absence of SIRS is not a useful predictor of outcome and should not be used to limit duration of therapy in patients with SAB.
| Final diagnosis | SIRS (N = 176) n (%) | No SIRS (N = 59) n (%) |
|---|---|---|
| R-sided IE | 29 (16.5) | 6 (10.2) |
| L-sided IE | 14 (8.0) | 4 (6.8) |
| Complicated SAB | 90 (51.1) | 31 (52.5) |
| Uncomplicated SAB | 43 (24.4) | 18 (30.5) |
P851 The eradication of MRSA nasal carrier status by mupirocin pomadeand the effect of eradication on the prevelance of MRSA infection
H. Irmak, S. Cesur, F. Yildiz, C. Bulut, S. Kinikli, Z. Aygün, A.P. Demiröz (Ankara, TR)
Objectives: The aim of this study is to compare the rate of eradication of MRSA carrier status in hospital staff with mupirocin with placebo and to establish the effect of eradication on the prevalance of MRSA infection.
Methods: This study was carried out in Infectious Diseases and Clinical Microbiology Department of Ankara Training and Research Hospital.
Totally eigty-eight hospital staff, 44 of them in mupirocin grup (20 MRSA nasal carriers, 24 MSSA nasal carriers) and further 44 hospital staff, who are MSSA carriers and administered placebo as control group, were included in the study. Informed consent of the patients and approval of ethics committee was obtained. Control group received only placebo.
MRSA prevalance in the three months before administration of mupirocin and the prevalance in the three months following administration were calculated.
Mupirocin pomad was applied three times a day for 5 days to the nose. 6th day, 1 month and 2 months after application, nasal cultures from obtained form the staff applied muciporin and those receiving placebo and eradication rates established. and compared statistically. In statistical analysis Chi-square and Fisher's Exact tests were used.
Results: Eradication rates were found to be respectively 91.7%, 66.6% and 58.3% after 6 days, 1 month and 2 months in mupirocin group while they were 4.5%, 6.8% and 11.4% respecively in the control group. In the mupirocin group, resistance developed against mupirocin in 2 (4.5%) hospital staff. In another 6 (13.6%) hospital staff receiving mupirocin, side effects such as feeling of burning developed in the nose. After treatment ended, these complaints resolved spontaneously.
When the eradication rates at 6 days, 1 month and 2 months were compared between two groups, statistically significant difference was found (p < 0.001). While MRSA infection prevalence was 9.6% (12/125) in the three months before application of mupirocin, it was 6.8% (8/118) in the the three months after application., with no statistically significant difference (p > 0.05).
Conclusion: In conclusion, although the eradication of nasal carrier status of MRSA is higher with mupirocin compared with placebo, mupirocin per se is not adequate in the reductionof the prevalence of MRSA infection. In order to reduce the prevalance of MRSA infection, all infection control measures for MRSA infection (isolation, contact isolation, hand washing, laboratory based surveillance) and training of the staff are required.
P852 A multicentre, double-blind, randomised clinical trial of parenteral cefepime in the treatment of acute bacterial infections
X.J. Lu (Chengdu, CN)
Objective: To evaluate the clinical efficacy and safety of domestic injectable cefepime for the treatment of acute moderate and severe bacterial infections.
Methods: A multicentre double-blind randomised clinical trial was conducted comparing the efficacy and safety of domestic injectable cefepime with imported one (Maxipime). Cefepime was administered intravenously at a dose of 1–2 g twice daily for 7 to 14 days.
Results: A total of 211 patients were enrolled in the study. There were 109 patients who enrolled in FAS analysis and 104 in PPS analysis in treatment group, while 108 patients were enrolled in FAS analysis and 107 patients were eligible for PPS analysis in control group. At the end of treatment, the cure rate and effective rate were 42.30% and 84.61% in trial group; while 48.59% and 79.43% in control group. The bacterial eradication rates were 91.3% and 86.7%, respectively. There was no statistical difference between the two groups (P > 0.05). Adverse reactions were observed in 8.93% and 5.41% of patients in the two groups, dominated by skin rash (1/112), temporary acidophilia (2/112; 2/111) and mild elevated transaminases (4/112; 2/111).
Conclusion: Domestic injectable cefepime is effective and safe for the treatment of acute moderate and severe bacterial infections.
P853 Efficacy and tolerance of linezolid-rifampicin combination prolonged oral therapy for bone and joint infections
E. Senneville, L. Legout, X. Lemaire, C. Dehecq, E. Beltrand, C. Loiez, H. Migaud, Y. Yazdanpanah (Tourcoing, Lille, FR)
Objectives: To evaluate the efficacy and safety of linezolid-rifampicin combination (LRC) in the treatment of bone and joint infections.
Methods: The records for patients treated with LRC for more than 4 weeks because of chronic bone and joint infections were reviewed for clinical outcome and tolerance. Primary end points were the clinical outcome at follow-up after the end of treatment (F-EOT), and the occurrence of adverse effects.
Results: Between June 1999 and July 2005, 29 adult patients with 18 infected orthopaedic devices including 11 prosthetic joints, and 11 chronic osteomyelitis, were eligible for the study. Pathogens were predominantly methicillin-resistant staphylococci (16/30 strains, 53.3%). The mean treatment duration was 17.8±7.5 weeks. Patients were given intravenous therapy for 6–8 days as inpatients, and then, as outpatients, were changed to oral therapy with weekly haematological monitoring. Reversible anaemia (haemoglobin <90 g/L) was reported in 6 patients (20.7%), of whom 4 had to receive blood transfusions. Mean time from treatment initiation to anaemia onset was 7.5 weeks (range 4–11). No other LRC-related haematological toxicity episodes (i.e. leukopenia and thrombocytopenia) were recorded. Five patients (17.2%) had to stop the antibiotic treatment because of the occurrence of serious adverse effects (anaemia = 4, and peripheral neuropathy = 1). In the 10 patients who had two-stage exchange of prosthetic device, no linezolid-resistant strains grew from the samples taken during reimplantation of the prosthesis. At the end of treatment, 26 patients (89.6%) were cured. During the post-treatment follow-up (median duration: 17 months, range: 12–36), 1 patient experienced relapses of infection, resulting in an overall success rate of 25/29 (86.2%).
Conclusions: LRC prolonged oral therapy seems effective for treating patients with resistant Gram-positive cocci bone and joint infections, including infected orthopaedic devices. However, close haematological and neurological monitoring of patients receiving LRC prolonged therapy is needed in order to reduce the occurrence of severe adverse effects.
P854 Efficacy of gentamicin for 5 days plus doxycycline for eight weeks versus streptomycin for 2 weeks plus doxycycline for 45 days in the treatment of human brucellosis
M.R. Hasanjani Roushan, M.J. Soleimani Amiri, N. Janmohammadi, S. Soleimani Amiri (Babol, IR)
Objectives: Regimen of choice and duration of therapy in human brucellosis is unknown. The purpose of this study was to evaluate the efficacy of gentamicin for 5 days plus doxycycline for 8 weeks versus streptomycin for 2 weeks plus doxycycline for 45 days in the treatment of human brucellosis
Methods: From April 2005 to October 2006, this comparative clinical study was conducted to compare the efficacy of gentamicin for 5 days plus doxycycline 100 mg twice daily for 8 weeks (regimen GD) versus streptomycin 1 gr for 2 weeks plus the same dose of doxycycline (regimen SD) for 45 days at the department of infectious diseases, in Babol, Iran. All cases were followed for 12 months after cessation of therapy.
Results: Forty-nine cases with the mean age of 34±14 years and 44 cases with the mean age of 36.5±15.3 years were treated by GD and SD regimens, respectively. The clinical manifestations and laboratory test results in these two treated groups were similar. Relapse was seen in 2 (4.1%) cases treated by regimen GD and in 2 (4.5%) cases treated by regimen SD (p > 0.05). Cure rate with regimen GD was 95.9% and with regimen SD was 95.5% (p > 0.05). Both regimens of therapy were well tolerated.
Conclusion: The result of this study shows that gentamicin for 5 days plus doxycycline for 8 weeks is as effective as standard regimen for therapy of human brucellosis.
P855 Effectiveness and tolerability of amoxycillin and azithromycin in treatment of patients with mild-to-moderate community-acquired pneumonias in outpatient settings
A. Kondratenko, I. Bereznyakov, V. Pozhar (Kharkiv, UA)
Background: Although amoxycillin and azithromycin are recommended in Ukraine as first-line antibiotics to treat adult outpatients with community-acquired pneumonias (CAP), there are no direct comparisons between these antibiotics.
Objective: to compare an effectiveness and tolerability of amoxycillin and azithromycin in outpatients with CAP confirmed by chest X-ray with no co-morbidities and other modifying factors and no bacterial sample requirements for patient (pt) inclusion.
Methods: We performed a randomised open-label study in outpatients with mild-to-moderate CAP aged 18–60 years. 56 pts with CAP confirmed clinically and by chest X-ray were randomly assigned for treatment with amoxycillin (0.5 g tid po for 7–10 days), 45 pts were assigned for treatment with azithromycin (0.5 g qd po for 3 days or 0.5 g po x 1, than 0.25 g/d for 4 days). Dynamics of clinical, objective and laboratory data was estimated in 48–72 hours, 7±1 and 12±2 days after the beginning of treatment.
Results: Adverse reactions were registered in 1 pt (2.2%) from azithromycin group and 2 pts (3.6%) from amoxycillin group; initial antibiotic was changed in 1 case in the last group. Clinical recovery with chest X-ray confirmation (i.e. excellent results) were registered in 20 pts (35.7%) from amoxycillin group and 15 pts (33.3%) from azithromycin group, clinical recovery with no chest X-ray confirmation (i.e. good results) were determined in 11 (19.6%) and 7 pts (15.6%) respectively. Satisfactory results (i.e. clinical improvement) were defined in 13 (23.2%) and 17 pts (37.8%) correspondingly. In 4 cases (7.1%) from amoxycillin group and 3 cases (6.7%) from azithromycin group it was not possible to estimate results. Unsatisfactory results of therapy were documented in 2 pts (4.4%) from azithromycin group and 8 pts (14.3%) from amoxycillin group.
Conclusion: in 12±2 days of outpatient treatment positive results were reached in 78.5% cases in amoxycillin group and 86.7% cases in azithromycin group. There was a similar tolerability of both regimens of treatment.
P856 Comparison of ornidazole and metronidazole in the treatment of trichomoniasis in men
A. Khryanin, O. Reshetnikov (Novosibirsk, RU)
Objective: Trichomonas vaginalis is a globally common sexually transmitted human parasite. Many strains of T. vaginalis from around the world have been described to be resistant to the current drug of choice, metronidazole. The aim of the study was to compare efficacy of metronidazole and ornidazole in the treatment of urogenital trichomoniasis in men.
Methods: A series of consecutive outpatients attending our STD clinic in 2000–2004 were included in the study. Four hundred twenty seven men aged from 20 to 48 years were randomly assigned to receive either 250 mg metronidazole t.i.d. over 10 days (210 subjects) or 500 mg ornidazole (Tiberal®) b.i.d. over 10 days (217 subjects). Clinical efficacy was assessed 1, 2 and 3 weeks after end of treatment regimen as well as microbiological efficacy with microscopy and culture.
Results: Clinical efficacy of metronidazole or ornidazole after 3 weeks was 57.6% and 94.5%, microbiological efficacy – 77.1% and 98.2%, respectively. Side effects were reported by 59.0% metronidazole arm versus only 3.7% in ornidazole arm.
Conclusion: Thus, ornidazole is more effective and safe medication than metronidazole in the treatment of T. vaginalis infection in males.
P857 Clindamycin/aminoglycoside versus β-lactam monotherapy for the treatment of intra-abdominal infections: a meta-analysis of randomised controlled trials
M.E. Falagas, D.K. Matthaiou, E.A. Karveli, G. Peppas (Athens, GR)
Objective: We sought to compare the effectiveness and safety of clindamycin/aminoglycoside combination therapy with broad-spectrum β-lactam monotherapy in patients with intra-abdominal infections by performing a meta-analysis of the available relevant randomised controlled trials (RCTs).
Methods: The studies for our meta-analysis were retrieved from searches of the PubMed database and reviewed by two independent reviewers. We considered a study eligible if: (1) it was a RCT comparing clindamycin/aminoglycoside combination therapy with β-lactam monotherapy for the treatment of patients with intra-abdominal infections, (2) reported data regarding the effectiveness of the treatment, mortality, and/or adverse effects, (3) included at least 10 evaluable patients, and (4) at least 70% of the study population had an intra-abdominal infection.
Results: A total of 28 RCTs were included in the analysis. Beta-lactam monotherapy was more effective regarding cure of the infection than clindamycin/aminoglycoside combination (3,177 clinically evaluable patients, fixed effects model, OR = 0.67, 95% CI 0.55 to 0.81, p < 0.001). The same result was found in several subset sensitivity analyses. There was no difference in all-cause mortality and attributable to infection mortality [2,382 intention-to-treat (ITT) patients, fixed effects model, OR = 1.25, 95% CI 0.74 to 2.11 and 1,976 ITT patients, fixed effects model, OR = 1.19, 95%CI 0.59 to 2.41, respectively]. There was no difference regarding overall adverse effects and ototoxicity (1,460 ITT patients, fixed effects model, OR = 1.05, 95%CI 0.80 to 1.37, and 1,404 ITT patients, fixed effects model, OR = 3.22, 95% CI 0.72 to 14.45, respectively). However, treatment with clindamycin/aminoglycoside was more likely to be associated with nephrotoxicity compared to β-lactam (3,065 ITT patients, fixed effects model, OR = 3.7, 95%CI 2.09 to 6.57, p<0.001). On the other hand, clindamycin/aminoglycoside was less likely to be associated with antibiotic associated diarrhoea compared to β-lactam (3,050 ITT patients, fixed effects model, OR = 0.68, 95% CI 0.46 to 1, p = 0.051).
Conclusion: The results of our meta-analysis suggest that β-lactams are more effective in the treatment of intra-abdominal infections compared to clindamycin/aminoglycoside combination.
P858 Beta-lactam alone compared to β-lactam-macrolide combination therapy for community-acquired pneumonia: prospective, observational study using a propensity score
M. Paul, A. Nielsen, A. Gafter-Gvili, E. Tacconelli, S. Andreassen, N. Almanasreh, E. Goldberg, R. Cauda, U. Frank, L. Leibovici for the TREAT Study Group
Objectives: Observational studies have shown improved outcomes for patients treated with β-lactam (BL)-macrolide combinations compared to single BL treatment for CAP. However patients treated for an atypical pathogen are probably a-priori different from patients treated with a BL drug alone. Physicians are likely to reflect in their choice of treatment common wisdom as to the presentation of “atypical” pathogens. We used a propensity score to adjust for these differences and compared treatment groups.
Methods: Patients were enrolled as part a multicentre trial assessing a decision support system for antibiotic treatment in Israel, Germany and Italy. We compared all 30-day mortality for patients with radiologically proven CAP treated empirically with BL alone vs. combination therapy. Baseline characteristic of the two treatment groups were used to develop a propensity score for combination therapy and comparisons were matched by patients' propensity score. In addition, we used the propensity score as a covariate in a logistic model for mortality.
Results: Patients treated with BLs alone (N = 169) were older (mean age 70.6±17.3 vs. 65.0±19.6 years), more often residents of nursing homes (9% vs 4%), bed-ridden (60% vs. 40%), had a higher chronic diseases score and a different clinical presentation compared to patients given combination therapy (N = 282). Accordingly, the propensity scores differed markedly, 0.179±0.139 vs. 0.074±0.103, p <0.001, respectively. Unadjusted fatality was significantly higher with BL monotherapy 22% vs. 7% (p<0.001) and remained significantly higher on multivariate analysis without adjusting for the propensity score. Only 27 patients in the BL group could be matched to (27) patients in the BL-macrolide group using the propensity score with a precision of 3 figures after the decimal point. Among these patients, mortality was identical in the two study groups, 3 demises each (11%, p = 1.0, OR = 1.0, 95% CI 0.2–5.5). A multivariable analysis for mortality, adjusting for the propensity score showed no significant difference between the study groups (OR = 0.84, 95%CI 0.27–2.51).
Conclusions: Patients given single BL treatment for CAP are markedly different from patients given combination therapy. Only a small percentage of patients could be matched using a propensity score and when matched mortality is identical. Classical multivariable techniques may not adjust correctly for these differences.
P859 Comparison of 2 options for antibacterial treatment of tularaemia type B
A. Stepanyan (Yerevan, AM)
Objectives: To compare the effect of antibacterial treatment with streptomycin or tetracycline on lymphadenitis during the Francisella tularensis type B infection.
Methods: This was a randomised clinical trial using indicated antibiotics for this infection. Thirty male military personnel with tularaemia type B, aged 18 to 20 years, were randomly prescribed to get either streptomycin 1.0×2 i.m. (14 patients) or tetracycline hydrochloride 0.5×4 p.o. (16 patients) for 10 days. All the cases were mild to moderate and presented as pharyngeal-bubonic or ocular-bubonic forms during the first week of disease. No antibiotic has been taken before admission to the hospital. Patients stayed at the hospital until the resolution or sclerosis of lymph node or healing of fistula after suppuration. In the case of suppuration (determined by fluctuation of node), early surgical drainage was performed, after test puncture. Sclerosis of lymph node was defined as no suppuration occurred during the first 3 weeks in hospital. From previous own experience it was observed that fluctuation usually occurs within the first 4 weeks of disease and if not so it will develop to sclerosis. The outcomes of study were the duration of stay in the hospital and the resolution/suppuration or sclerosis of lymph node. The t test was used for significance of difference for hospital stay and chi squared test for lymphadenitis outcomes.
Results: Patients in the tetracycline group stayed in the hospital 20±2.1 days, while patients receiving streptomycin 28±3.4 days (P < 0.01). In 14 patients of tetracycline group (87.5%) and 12 patients of streptomycin group (85.7%) resolution or suppuration of lymph nodes were observed (P > 0.05). All the suppurations occurred in the 2nd or 3rd weeks of hospital stay. In other cases sclerotic lymph node remained.
Discussion: Tularaemia type B infection is mild to moderate infection, which causes significant lymphadenitis. The outcomes of bubo are resolution, suppuration or sclerosis. First two outcomes are favourable, because simple surgery or no surgery is required for complete recovery. Unlike it, sclerotic lymph node remains in the subcutaneous tissue in the third case, which needs cosmetic surgery. Treatment by tetracycline leaded to shorter hospital stay but there was no significant difference for bubo outcomes.
It can be concluded that using tetracycline in the case of mild to moderate tularaemia type B reduces the time necessary for hospital stay or medical care.
P860 Efficacy and safety of gatifloxacin for chronic prostatitis (NIH category II or IIIa) in Korea
S. Lee, M. Kim, C. Kim, B. Shim, D. Kim, C. Han, Y. Ha, Y. Cho (Busan, Seoul, Gwangju, Daegu, KR)
Objectives: The objective of this study was to investigate the efficacy and safety of gatifloxacin for patients with chronic prostatitis (NIH category II or IIIa) in Korean urologic practice.
Materials and Methods: A total of 16 outpatient urology clinics at tertiary care medical centres in Korea participated. Gatifloxacin (400 mg/day) treatment (S.D.) of 149 patients (20 patients with category II and 129 patients with category IIIa) with prostatitis, mean age 45.8 (13.3) years, was carried out for 41.7 (33.1) days. A 4-glass test according to Meares and Stamey or two glass test was carried out at study entry and one month after the end of treatment. Clinical response, safety and bacteriological response were assessed before treatment (within 48 hours of initiation of the study medication) and at one month after treatment completion.
Results: In a total of 149 patients, the total NIH-CPSI score was significantly reduced from 20.5 to 10.0 (response rate 86.7%; 95% CI 80.2–93.2%) (p< 0.05). Sub-scores of pain, urinary symptoms and impact on the quality of life were also significantly reduced from 8.9 to 3.8 (response rate 83.8%; 95%CI 76.8–90.9%), from 4.2 to 2.0 (response rate 73.3%; 95% CI 64.9–81.8%) and from 7.4 to 4.2 (response rate 79.0%; 95% CI 71.3–86.8%), respectively (p <0.05). In terms of the overall clinical efficacy assessment by investigators, out of 149 patients with prostatitis, 71.2% were assessed to be responders. Bacteriological studies in expressed prostatic secretion (EPS) or post prostate massage urine (VB3) at 1 month after treatment completion demonstrated that the overall eradication rates of pathogens was 85% and the pyuria (more than 10 WBC/HPF) rates in the NIH category II and IIIa were 35% and 18.6%, respectively (overall rate 20.8%). There were 16.1% of patients that presented with some adverse events considered by investigators to be related to the drug. The majority of adverse events were considered to be of mild (87.5%) or moderate (8.3%) intensity.
Conclusions: These results suggest that gatifloxacin in Korean urologic practice is well tolerated and improves the clinical outcomes in the patients with chronic prostatitis (NIH category II or IIIa).
Antibiotic resistance mechanisms
P861 ECL-1, a novel plasmid-mediated class A β-lactamase with carbenicillinase characteristics from Escherichia coli
C. Papagiannitsis, A. Loli, L.S. Tzouvelekis, E. Tzelepi, G. Arlet, V. Miriagou (Athens, GR; Paris, FR)
Objectives: Characterisation of a novel class A β-lactamase (ECL-1) encoded by an 80-kb self-transferable plasmid from Escherichia coli.
Methods: E. coli EC-3521r was isolated in 2002 from a urine sample of a patient treated in a general hospital. MICs of β-lactam antibiotics were determined by agar dilution. Isoelectric points of the produced β-lactamases were determined by IEF of cell extracts. Hydrolysis rates of β-lactams and inhibitory activity of clavulanic acid, tazobactam, and sulbactam, were determined by UV spectrophotometry. Bla-genes were identified by PCR. Mating experiments were performed in mixed broth cultures. Bla gene encoding fragments were cloned into the pBCSK(+) vector and the nucleotide sequences of inserts were determined.
Results: By PCR screening, the E. coli EC-3521r isolate was found to be positive for blaACC and blaTEM. Analysis of the β-lactamase content by IEF, indicated production of three main β-lactamase species with isoelectric points (pIs) of 7.8 corresponding to ACC-1, 5.4 (TEM-1) and 5.8. Identities of the former two enzymes were confirmed by sequencing of PCR products. Beta-lactam resistance was transferred by an 80-kb plasmid (pR3521) that encoded all three β-lactamases.
A cloned fragment of pR3521 encoded production of the β-lactamase with a pI of 5.8 and mediated resistance to penicillins but not cephalosporins. The respective recombinant plasmid carried a 3,833-bp Sau3A fragment containing an ORF of 867-bp homologous to the orf1 observed in ACC-1-encoding plasmids. This orf did not exhibit significant homology with any known sequence. However, the deduced polypeptide (288aa) possessed the typical motifs of a class A β-lactamase and was designated blaECL-1. ECL-1 exhibited 51% amino acid sequence identity with the chromosomal carbenicillinases of the RTG. The enzyme was effective against ampicillin and carbenicillin while the relative hydrolysis rates of oxacillin, cephalothin and cephaloridine were low. Tazobactam was the most potent inhibitor of ECL-1 followed by clavulanic acid and sulbactam.
Conclusion: The ECL-1 is a novel class A β-lactamase of unknown origin, that is functionally and phylogenetically related to the RTG subgroup of the CARB β-lactamases.
P862 Prevalence and molecular epidemiology of CTX-M β-lactamases in Croatia
B. Bedenic, M. Tonkic, I. Jajic-Bencic, S. Kalenic, A. Baraniak, J. Fiett, M. Gniadkowski (Zagreb, Split, HR; Warshaw, PL)
Objectives: The aim of this study was to characterise β-lactamases produced by these isolates.
Methods: Eleven E. coli strains were isolated from two hospitals in Zagreb and one in Split. Extended-spectrum β-lactamases (ESBL) were detected by double-disk synergy test and CLSI combined disk test. Susceptibility to a wide range of antibiotics was determined by the broth microdilution method according to CLSI. ESBLs were characterised by isoelectric focusing, substrate profile determination, PCR and sequencing of blaCTX-M genes. The two latter approaches were also used to study the genetic environment of the genes. Genetic relatedness between the strains was tested by PFGE.
Results: Despite some similarities, susceptibility testing revealed significant differences between the groups of isolates identified in the three hospitals.
All of the isolates produced β-lactamases with high pI values, either 8.9 in the case of the isolates from the University Hospital Center Zagreb or 8.4 in the case of the all remaining isolates. PCR for blaCTX-M genes was positive and sequencing these identified the pI 8.9 enzymes as CTX-M-15, whereas those with a pI of 8.4 as CTX-M-3 β-lactamase. An insertion sequence ISFcp1 was identified 48 bp upstream of all blaCTX-M-15 producers and 128 bp upstream of one blaCTX-M-3 gene, whereas the remaining blaCTX-M-3 genes were accompanied by the IS26 element. The five CTX-M-15-producing isolates were indistinguishable in the PFGE analysis, and different from the all remaining isolates. The group of CTX-M-3 producers from Split was much more diverse with only two isolates that were clonally related to each other. The isolate from the Sisters of Mercy Hospital in Zagreb was unrelated to any other study isolate.
Conclusions: This study reported the appearance of CTX-M-3 and CTX-M-15 in clinical isolates of E. coli from Croatia. Despite the high similarity of these enzymes, differences in the genetic context of their genes indicated that they emerged independently on each other. Moreover, it is possible that there were also two different origins of CTX-M-3, which altogether is striking considering the relative small geographic region of the isolates identification. CTX-M-15 producers disseminated in the hospital in Zagreb by clonal spread, whereas in Split the blaCTX-M-3 gene was probably horizontally transmitted between non-related strains. CTX-M β-lactamases are still rare in Croatia.
P863 A single-tube real-time PCR and melting-curve analysis for detection and characterisation of TEM-type extended-spectrum β-lactamases
A. Nikulin, Y. Alexeev, M. Edelstein (Smolensk, Moscow, RU)
Objectives: ESBLs of the TEM-type are among the most common plasmid-mediated enzymes conferring resistance to oxyimino-β-lactams in Enterobacteriaceae. Despite the overall diversity, all the known TEMs capable of hydrolising oxyimino-β-lactams differ from TEM-1 and TEM-2 penicillinases in at least one of the three aa positions: 104, 164, and 238, suggesting their importance for ESBL activity. Here we describe a simple and rapid method for identifying mutations at these positions in a single real-time PCR reaction.
Methods: The proposed method utilises asymmetric amplification of three ultrashort (50–63 nt) fragments of blaTEM genes encompassing the key mutation sites. Each fragment is amplified using one excess primer internally labeled with the BHQ quencher in the presence of 3'-fluorophore-labeled probe that covers the mutation site and anneals to the extension product of the BHQ-labeled primer just 1 to 3 nucleotides apart from its 3′-end. Thus, binding of the probe to its target brings the fluorophore and quencher into close contact and results in fluorescence quenching. Owing to this design, several nucleotide polymorphisms (SNPs) in each site can be discerned using post-PCR melting curve analysis. When the probe is fully complementary to the product, it melts at higher temperature, if an SNP is present, the melting temperature (Tm) specifically decreases depending on the type and position of mutation. In this study, three probes were designed to match the blaTEM-1 sequences at codons 104, 164, and 237–240, respectively, and one probe was designed to match the mutant codon (AAG) for Lys240. The probes were labeled with different dyes to allow multiplex detection in the same reaction.
Results: The applicability of the real-time PCR method described herein for detection of SNPs associated with ESBL activity was evaluated using a collection of 20 laboratory and 100 clinical strains producing TEM-1, TEM-2 (non-ESBL controls), and various TEM-type ESBLs carrying the following mutations: E104K, R164C, R164H, R164S, A237T, G238S and G240K. All of these mutations were successfully detected and discriminated from the wild-type sequences according to specific Tm-s. Most notably, the proposed design of primers and probes made possible an unambiguous identification of key mutations regardless of the presence of closely spaced silent SNPs.
Conclusion: The simplicity and rapidity of the described method make it useful for epidemiological studies on TEM ESBLs.
P864 Complete nucleotide sequence of pEK499, a multidrug-resistance plasmid from the UK's most prevalent Escherichia coli strain with CTX-M-15 β-lactamase
E. Karisik, A. Underwood, M.J. Ellington, D.M. Livermore, N. Woodford (London, UK)
Objectives: Multi-resistant E. coli with CTX-M (mostly CTX-M-15) β-lactamases are prevalent in the UK, with several epidemic strains and many unrelated producers. Strain A is the most widespread producer clone, recorded from >45 centres. We report the complete nucleotide sequence of pEK499, a multi-resistance plasmid encoding CTX-M-15 enzyme in epidemic strain A.
Method: pEK499 was randomly sheared and the 2–3 kb fraction was cloned into the pGEM-Teasy vector, prior to transformation into E. coli DH10b. Inserts were sequenced by dye terminator chemistry. Sequences were assembled using the Staden Package. Combinatorial PCRs, directed PCRs, and walking reads on selected clones were used to assemble the sequences and to fill-in gaps.
Results: pEK499 was found to be a circular molecule of 117,536 bp belonging to incompatibility group FII. It harboured up to 185 predicted genes and encoded multi-resistance and virulence factors. With the exception of blaTEM-1, all antibiotic resistance genes were clustered in a 25-kb region. They included blaCTX-M-15 and blaOXA-1 as well as genes conferring resistance to aminoglycosides and ciprofloxacin (aac6'-Ib-cr), macrolides [mph(A)], chloramphenicol (catB3) and tetracycline [tet(A)]. A 1.8-kb class I integron was present within the multi-resistance region; this carried dfrA17 and aadA5, encoding trimethoprim and streptomycin resistance respectively, also sulI encoding sulphonamide resistance. Virulence-associated genes present on pEK499 included the serum survival gene traT, as well as vagC/D. pEK499 also encoded the F-plasmid-derived CcdA/B toxin/antitoxin addiction system.
Conclusion: Plasmid pEK499 harboured 10 antibiotic resistance genes affecting 8 antibiotic classes, along with different virulence determinants. It may contribute towards the epidemiological success of E. coli strain A in the UK. The toxin/antitoxin addiction system will ensure its maintenance in the absence of antibiotic pressure.
P865 A new metallo-β-lactamase gene (blaIMP-22) harboured in class 1 integron from Pseudomonas fluorescens
C. Pellegrini, G. Celenza, P. Mercuri, M. Galleni, B. Segatore, G. Amicosante, M. Perilli (L'Aquila, IT; Liège, BE)
Objectives: Intensive use of β-lactams in human and animals practice promoted spreading and evolution of resistance determinants also among environmental bacteria. These bacteria represent an important reservoir of mobile metallo-β-lactamase determinants.
Methods: Pseudomonas fluorescens was isolated from environment samples derived from effluent of urban sewage. PCR experiments using specific primers for blaIMP and blaVIM genes were performed on genomic DNA of P. fluorescens. The presence of mobile elements such as integrons was investigated by PCR using the specific primers for 5′CS and 3′CS region. The nucleotide sequence was submitted to the EMBL/GenBank whit accession number DQ361087. The blaIMP-22 gene was cloned in pBC-SK vector to yield the recombinant plasmid pBC-IMP22. In vitro susceptibility of E. coli HB101 harbouring the pBC-IMP-22 plasmid versus different antibiotics was investigated.
Results: P. fluorescens was highly resistant to imipenem (32 μg/mL), meropenem (32 μg/mL), cefotaxime (>128 μg/mL) and ceftazidime (>64 μg/mL) and susceptible to penicillins (piperacillin, MIC = 16 μg/mL; amoxicillin, MIC = 8 μg/mL). The blaIMP gene-isolated by PCR revealed an ORF of 741 bp that encode for a pre-protein of 246 amino-acid, named IMP-22. Compared to other known IMP β-lactamases, the IMP-22 enzyme is quite divergent from other IMP variants: its closest relative is IMP-16 (94% sequence identity) and IMP-1 is 85% identical to IMP-22. The IMP-22 shows three unique substitutions at the following positions: E104D, K108Q and E174V (EBL numbering). Sequence mapping obtained from PCR products, carried out using 5′CS for and 3′CS rev primers and P. fluorescens genomic DNA, demonstrated that blaIMP-22 is located in a class 1 integron. The genetic context in which was located blaIMP-22 is upstream represented by typical structure of 5′CS region whereas the 3′CS region is represented by qaEDelta and sul. Downstream the blaIMP-22 gene cassette an ORF of 327 bp was found. The amino acid translation of this last region corresponds to a putative unknown protein.
Conclusion: Detection of a new metallo-β-lactamse in environmental strain is very interesting but not rare. In this case blaIMP-22 gene was harboured by a class 1 integron whose structure is typical of most of integrons found in clinical isolates, but not in environmental bacteria.
P866 Mutators are not responsible for the association between β-lactam and quinolone resistance in Escherichia coli with AmpC hyperproduction phenotype
M. Conejo, G. Amblar, E. Perea, L. Martínez-Martínez (Seville, ES)
Objective: Clinical isolates of Esherichia coli (Eco) resistant to quinolones displaying an AmpC hyperproduction phenotype (resistance to both cefoxitin and amoxicillin-clavulanate in the absence of extended-spectrum β-lactamases) have been increasingly observed in our institution. The aim of this study was to determine if the frequency of hypermutable strains is higher in this group of organisms than in isolates without combined resistance to both quinolones and β-lactams.
Method: Rifampin (Rif) resistance mutation frequency (MF) was studied in 97 Eco isolates from the Univ. Hosp. V. Macarena, Seville, Spain, including 60 isolates showing an AmpC hyperproduction phenotype resulting from several point mutations in the promoter and or the attenuator of the chromosomal ampC gene (ACBL+) and co-resistance to ciprofloxacin and nalidixic acid (QUINR), 9 isolates ACBL+ and susceptible to both quinolones (QUINS), and 28 isolates susceptible to both cefoxitin and amoxicillin-clavulanate (ACBL-) and either QUINR or QUINS (15 and 13 strains respectively). Three Luria-Bertani (LB) tubes for each strain were inoculated with an independent colony from a blood agar plate. After 24 h of incubation, appropriate dilutions were seeded onto LB agar plates with and without Rif (100 mg/L), and colony counts were performed after 48 h. MFs were reported as a proportion of the number of Rif-resistant colonies to the total viable count. The results were the mean value obtained in three independent experiments that were repeated in cases of discrepancies. Eco AB1157 mutS::Tn10, with a defective mutS gene, was used as a strong mutator control, and Eco ATCC25922 as a non hypermutable control. Statistical differences were determined by the chi-square test (p <0.05: significant).
Results: Strains were considered non hypermutable when the MF was <4× 10−8, weak mutators if 4 × 10−8 ≤ MF < 4 × 10−7 and strong mutators if MF ≥4×10−7. Of the 97 strains, 65 were non hypermutable, 29 were weak mutators and 3 were strong mutators. Hypermutable strains were detected in higher proportions in the isolates with ACBL+QUINS and ACBL-QUINR (54.5% and 40%, respectively) than in isolates with ACBL+QUINR (29.7%). Among ACBL-QUINS isolates only 15% were hypermutable. No significant differences were observed among groups.
Conclusion: Hypermutability could not explain the association between β-lactam and quinolone resistance in clinical isolates of Eco with AmpC hyperproduction phenotype.
P867 Carbapenem resistance mechanisms in Norwegian clinical isolates of Pseudomonas aeruginosa
Ø. Samuelsen, L. Buarø, B. Aasnæs, C.G. Giske, B. Haldorsen, G.S. Simonsen, T.M. Leegaard, A. Sundsfjord (Tromsø, NO; Stockholm, SE; Oslo, NO)
Objectives: The objective of the study was to investigate resistance mechanisms to carbapenems and evaluate phenotypic selection criteria for metallo-β-lactamase (MBL) testing in carbapenem resistant clinical isolates of P. aeruginosa in a low-prevalence country like Norway.
Methods: 31/138 imipenem resistant P. aeruginosa clinical isolates from Norwegian hospitals (year 2004 to 2006) with a positive MBL Etest (IP/IPI ratio of ≥8 and/or ellipse/phantom zone) were retested at the Reference Centre. A broad spectrum β-lactam MIC screen by Etest was also done. Verification of MBL presence was done by PCR (blaIMP and blaVIM) and by spectrophotometric analysis of imipenem hydrolysis by crude cell extracts. As part of an ongoing study a subset of the strains (n = 10) was evaluated for changes in the transcription of oprD and mexB by quantitative RT-PCR.
Results: Of the 31 MBL Etest positive isolates 7 were positive upon retesting. Interestingly, the IP MIC was reproducible while the IPI MIC was consistently increased, often by more than 2-fold dilutions. MBL production was verified in two isolates by PCR and hydrolysis of imipenem, resulting in 29 and 5 false-positive isolates in the initial and retesting respectively. Analysis of the resistance profile of the isolates showed that additional selection criteria for MBL testing such as resistance to meropenem and/or ceftazidime using EUCAST breakpoints would reduce the number of false positives further. qRT-PCR on the subset of the isolates (n = 10) revealed a significant downregulation of oprD (n = 6) and/or upregulation of mexB (n = 4) in the tested isolates.
Conclusions: (i) In this study MBL Etest results were difficult to reproduce and overestimated the presence of MBL in a low prevalence country like Norway. (ii) Additional selection criteria such as meropenem and/or ceftazidime resistance will reduce false-positive results and should be considered before more labour intensive analysis are performed. (iii) Gene transcription analyses indicate that decreased permeability and efflux are more prevalent mechanisms than MBL production in carbapenem resistant Norwegian P. aeruginosa isolates. (iv) False positive test results are probably due to the permeabilising effect of EDTA.
P868 Occurrence of OXA-58 and an OXA-58 variant in Acinetobacter baumannii isolates from blood cultures in a university hospital in Athens, Greece
I. Galani, V. Sakka, L. Galani, M. Souli, Z. Chryssouli, H. Giamarellou (Athens, GR)
Objectives: Imipenem (IMP)-resistant A. baumannii isolates, collected from blood cultures of hospitalised patients in the University General Hospital “Attikon”, between September 2004 – May 2005, were studied for their clonality and carbapenemase content.
Methods: Susceptibility to antimicrobials was determined using a broth microdilution method and E-test. Bacterial clones were identified by PFGE with ApaI. Metallo-β-lactamases were detected by the IMP-EDTA disc synergy test and by PCR with primers specific for blaVIM. Multiplex PCR with primers amplifying blaOXA-23-like, blaOXA-24-like, blaOXA-51-like and blaOXA-58-like, was performed according to Woodford et al (Int J Antimicrob Agents 2006; 27:351–353). Sequencing of cloned PCR products was performed by MWG-THE Genomic Company. Sequence similarity searches were carried out with the BLAST programme found at the website of the NCBI.
Results: Twenty-seven non-repetitive IMP-resistant isolates, 70.2% of all A. baumannii strains isolated from blood cultures, were tested. All isolates presented a multi-resistance pattern, with all being susceptible to colistin (MICs: 0.125–0.75) and 33% being susceptible also to ampicillin/sulbactam. A. baumannii isolates were distributed into seven genotypes by PFGE profiles, with 12, 4, 4, 3, 2, 1, and 1 strains in each group. The three genotype-groups of 12 and 4 isolates were further divided into 2 subgroups. None of the isolates was positive for metallo-β-lactamase production by IMP-EDTA synergy test or PCR. Multiplex PCR for OXA-type carbapenemases revealed the presence of blaOXA-51-like, which is believed to be intrinsic to A. baumannii and blaOXA-58-like in all isolates. Sequencing of blaOXA-58 PCR amplicon revealed the presence of the blaOXA-58 in most of the isolates but also the existence of a novel variant of OXA-58 in which an A to G substitution revealed a Thr to Ala amino acid change.
Conclusions: Several clones of IMP-resistant A. baumannii producing both blaOXA-58 and naturally occurring blaOXA-51-like have emerged as important bloodstream pathogens in our hospital. This observation emphasizes the importance of both the restriction of carbapenem usage as well as of the strict implementation of hand hygiene techniques for the containment of this emerging threat.
P869 Spread of clinical extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli in Portugal
J. Leitão, N. Mendonça, V. Manageiro, D. Louro, E. Ferreira, M. Caniça on behalf of the Antimicrobial Resistance Surveillance Program in Portugal (ARSIP)
Objectives: CTX-M extended-spectrum β-lactamases, which had a rapid growing and geographic dissemination, are the main recent cause of resistance to third-generation cephalosporins. The aim of this study was to characterise the resistance mechanisms to β-lactam antibiotics of clinical Escherichia coli strains, using phenotypic, biochemical and molecular approaches.
Methods: During a 2-year period, started on March 2004, 181 nonrepetitive E. coli strains were isolated as ESBL producers from different clinical specimens at 9 hospitals from 3 different Portuguese regions. In this work, were only investigated the ESBL (CTX-M)-producing strains. Antimicrobial susceptibility was performed by broth-microdilution method. PCR and sequencing were used to screen and identify blaTEM, blaSHV, blaOXA, ampC and blaCTX-M genes. Biochemical characterisation was performed by isoelectric focusing. The genetic environment of blaCTX-M was characterised by PCR, regarding for ISEcp1, IS26 and IS903 elements. Strains were subtyped by using PFGE.
Results: Of the 181 strains 119 (66%) were CTX-M producers. Susceptibility towards β-lactams confirmed all isolates as ESBL producers, also suggesting CTX-M enzymes expression, which was corroborated by pI values. More than 98% of strains producing CTX-M group-1 were resistant to cefotaxime and ceftazidime, while all CTX-M group-9 producers were resistance to cefotaxime and 11% resistant to ceftazidime. Overall, multidrug-resistant (92%) strains were predominant in outpatients (50%) than inpatients (37%). PCR-sequence analysis confirmed the presence of blaCTX-M-15 (n = 110) and blaCTX-M-32 (n = 1) genes from CTX-M-1 group, blaCTX-M-14 (n = 9) genes from CTX-M-9 group, blaTEM-1 (n = 104) and blaOXA-30 (n = 101). No strain carried the blaSHV gene. ISEcp1 elements were found upstream of all blaCTX-M genes and IS903 was detected downstream of one blaCTX-M-14 gene. Genetic relatedness analysis revealed 5 clusters and indicated that 76% of all isolates (from cluster IV) corresponded to a single epidemic strain.
Conclusions: Our work confirms the geographic spread in Portugal of CTX-M-type β-lactamases, mainly CTX-M-15, and suggests that the horizontal transfer of blaCTX-M genes, mediated by plasmids and/or mobile elements, contribute to the dissemination of those enzymes to community and hospital environments. Given the potent extended-spectrum activity of these enzymes, their continuous spread would have a distressing development.
P870 GES-1 and TEM-1-like β-lactamases of Pseudomonas aeruginosa strains isolated from patients in two Warsaw hospitals
A.E. Laudy, A. Cieloch, D. Dzierzanowska, S. Tyski, J. Patzer (Warsaw, PL)
Objectives: Ambler class A extended-spectrum β-lactamases are produced not only by Enterobacteriaceae family rods but also by P. aeruginosa strains isolated from clinical materials of hospitalised patients world wide. Recently, the presence of PER-1 enzymes produced b y P. aeruginosa isolated in Poland has been described.
The aim of the study was to identify strains producing TEM-, SHV-, GES- and VEB-type ESBLs, among P. aeruginosa isolated in Warsaw hospitals.
Methods: The analysed 35 P. aeruginosa strains were isolated from clinical materials of patients in two Warsaw hospitals. The presence of ESBL in the strains was detected by a double-discs synergy test with inhibitors: clavulanic acid, sulbactam, tazobactam and imipenem. The antibiotics MICs value were determined by agar dilution methods, according to CLSI recommendation. The presence of class 1 integron and genes coding ESBL-type enzymes among P. aeruginosa was detected by PCR. Gene cassettes were identified by sequencing of the obtained amplicons.
Results: The ESBL-type enzymes were detected among 12 strains, by double-discs synergy test with inhibitors of serine β-lactamases. However, this test showed to be not useful for detection of ESBLs in the remaining 23 isolates. Three out of 23 strains were resistant to all β-lactams. In 2 isolates the gene coding class A β-lactamases was identified – blaGES-1 in a strain from hospital A and blaTEM-1-like in a strain received from hospital B. The clinical isolate producing GES-1 exhibited the inhibitor-sensitive phenotype. The synergy pattern between inhibitors (clavulanic acid or imipenem) and ceftazidime, cefepime or aztreonam was observed, as well as resistance to gentamicin and all β-lactams except imipenem. The gene blaGES-1 is located on the variable region of class 1 integron. The other strain, producing TEM-1-like enzyme, was resistant to all β-lactams and no synergy between inhibitors and β-lactams was observed. Additionally in case of this strain no VIM-type metallo-enzymes were found.
Conclusion: GES-1 and TEM-1-like β-lactamases producing P. aeruginosa strains were identified the first time in Poland. The double-discs synergy test was not useful for detection of all Ambler class A ESBLs among P. aeruginosa isolates.
P871 Phenotypic detection of AmpC in E. coli: comparison of cloxacillin, boronic acid and EDTA disk synergy assays
C. Giske, B. Haldorsen, E. Lundblad, B. Aasnæs, G. Simonsen, A. Sundsfjord (Stockholm, SE; Tromsø, NO)
Objectives: Disk synergy tests using cephalosporins ± boronic acid or cloxacillin have been found to be both sensitive and specific for phenotypic detection of chromosomal hyperproduction of AmpC and plasmid-mediated AmpC in E. coli. Also, an assay based on EDTA permeabilisation followed by analysis of distortion of cefoxitin inhibiton zone has been described. The purpose of this study was to compare the three methods against a previously characterised strain collection of E. coli either hyperproducing chromosomal AmpC or featuring plasmid-mediated AmpC.
Materials: A collection of 23 isolates from 12 different Norwegian laboratories were characterised with respect to hyperproduction of chromosomal AmpC (mediated by promotor mutations) and the presence of plasmid-mediated AmpC β-lactamases. Control strains of Enterobacteriaceae with defined plasmid-mediated AmpC or hyperproducing chromosomal AmpC as well as negative controls (ESBL-producers and wild-type E. coli) were included. Thirteen isolates were AmpC hyperproducers, whereas CMY β-lactamases were detected in 10 isolates. All isolates were subjected to disk synergy testing using cefoxitin, cefotaxime and ceftazidime ± boronic acid (400 ug) or cloxacillin (750 ug). A zone diameter difference of ≥5 mm was considered indicative of AmpC production.
Results: Synergy between cefoxitin and cloxacillin was found in all isolates, while synergy between cefoxitin and boronic acid could be demonstrated in all but two isolates. However, the latter two isolates were positive when results obtained with cefotaxime and ceftazidime ± boronic acid were taken into consideration. The EDTA test was negative in two isolates hyperproducing chromosomal AmpC.
Conclusions: The cefoxitin ± cloxacillin assay correctly identified all AmpC hyperproducers and CMY-producers. The cefoxitin ± boronic acid assay failed to identify two isolates, but both of these were positive with cefotaxime and ceftazidime ± boronic acid. This is in agreement with earlier observations that synergy between cefoxitin and boronic acid can sometimes be masked in strains featuring porin defects. Although the strain collection is relatively small the results indicate that cefoxitin ± cloxacillin may be promising as a stand alone test for phenotypic detection of AmpC production in E. coli. Further studies are warranted to determine whether cefotaxime and ceftazidime should be included in the cloxacillin assay.
P872 Extended-spectrum β-lactamase among non-typhoidic Salmonella in paediatric unit of Algerian hospital
M. Ouar-Korichi, F. Assaous, L. Gilly, G. Paul, K. Rahal (Algiers, DZ; Paris, FR)
Objectives: The purpose of this work is to characterise the different types of extended-spectrum β-lactamases (ESBL's) produced by non typhoidic Salmonella and to study their evolution from 1989 to 2005.
Methods: From 1989 to 2005, 240 of non typhoidic Salmonella producing ESBL's were recovered from patient of multiple outbreaks (n = 28) in paediatric unit from 13 hospitals. ESBL's were detected by disc diffusion method and confirmed with MIC s of cefotaxime (CTX) and ceftazidime (CAZ) plus clavulanate, sulbactam and tazobactam by microdilution broth method. Isoelectric focusing was used to characterise Pi of β-lactamases. PCR was performed with specific primers to type β-lactamase genes like bla TEM, bla SHV, bla CTX-M. The sequencing of the gene was used to identify the specific type of the β-lactamase.
Results: 240 strains were confirmed as ESBL's producers with the disc diffusion methods and the MIC's. The serotypes of the Salmonella enterica were: m'bandaka (n = 136), brunei (n = 40), kedougou (n = 29), seftemberg (n = 28), typhimurium (n = 7). These Salmonella were isolated from following biological products: stool (n = 217), blood (n = 15), urine (n = 5), tear fluid (n = 1), pus (n = 1), physiological water (n = 1).
The strains of 18 outbreaks had the gene bla TEM. The sequencing of the gene was made for 9 outbreaks and the results revealed that the strains encoded for different enzymes TEM-1 (Pi 5.4), TEM 25 (Pi 5.4) and TEM 110 (Pi 5.4) in the outbreak's in 1989–1990, TEM 25 (Pi 5.4) in 1991, TEM 47 (Pi 6) in the years 1993, 1994, 1995, 1999 and TEM 19 (Pi 5.4) in 1994. But since 2001 we characterised strains with CTX-M15 (Pi 8.8) extended-spectrum β-lactamase and CTX-M 3 (Pi 8.4) in 3 outbreaks in 2004–2005. In our study no strains produced SHV extended β-lactamase.
Conclusion: Our study showed an evolution with different TEM ESBL's before 2001 and after that we found only CTX-M like CTX-M3 and CTX-M 15.
P873 Prevalence of plasmid-mediated quinolone resistance determinants: qnrA, qnrB and qnrS in extended-spectrum β-lactamase producing Enterobacteriaceae strains in Hungary
D. Szabó, N. Pesti, K. Kristóf, L. Rókusz, J. Szentandrássy, K. Katona, K. Nagy (Budapest, HU)
Objectives: To determine the prevalence of the plasmid-mediated quinolone resistance genes: qnrA, qnrB and qnrS in a selected collection of extended-spectrum β-lactamase(ESBL)-producing Enterobacteriaceae strains.
Methods: A total of 92 non-repetitive ESBL-producing Klebsiella spp., 58 ESBL-producing Enterobacter spp., and 20 ESBL-producing E. coli isolates were collected from seven different hospitals of Budapest, Hungary. The minimum inhibitory concentrations (MICs) of ciprofloxacin, levofloxacin and moxifloxacin were determined by microdilution technique according to the Clinical and Laboratory Standards Institute (CLSI). All the isolates were screened for the presence of the qnrA, qnrB and qnrS genes by PCR.
Results: 75% (15/20) of the ESBL-producing E. coli isolates were resistant to all the three fluoroquinolones. QnrA gene was detected only in one strain and none of them harboured qnrB gene. QnrS was present also in one strain (5%). 84% (49/58) of the ESBL-producing Enterobacter. isolates were resistant to all the three fluoroquinolones. Three Enterobacter strains were qnrA positive (5%), 19 strains were qnrB positive (33%), and two strains were qnrS positive (3%). Among the Klebsiella isolates 55/97 (56%) were resistant to all the three fluroquinolones. Five Klebsiella strains were qnrA positive (5%), and two strains were qnrB positive (2%) and one strain were qnrS positive (1%). One qnrA positive Enterobacter strain, one qnrA positive Klebsiella strain and two qnrB positive Enterobacter strains were susceptible to the quinolones based on their MIC values. One qnrB positive Enterobacter strain and the qnrS positive Klebsiella strain's MIC values belonged to the intermediate range.
Conclusion: This is the first report of qnr-positive ESBL-producing Enterobacteriaceae strains in Hungary. The prevalance of qnrA and qnrS gene was similar to the international studies, but the prevalence of the qnrB gene was high among Enterobacter. suggesting widespread distribution of plasmids conferring resistance to extended-spectrum cephalosporins and quinolones in clinical isolates.
P874 Quinolone-resistant Brucella selection by treatment with fluoroquinolones in a macrophage infection experimental model
R. Rodríguez Tarazona, J. Muñoz Bellido, J. García Rodríguez (Salamanca, ES)
Background: Brucellosis remains an important public health problem in wide areas around the world. Classical treatments, though quite effective, are not exempt of side effects. Fluoroquinolones (FQ) were proposed years ago as a possible alternative, but the treatment failures rate was unacceptable. A reduced activity of FQ due to the low pH in the phagolysosome has been proposed, but not definitively demonstrated. The possibility of failures as a consequence of resistant mutants selection has not been explored. We have studied the emergence of FQ-resistant Brucellae after experimental infection of macrophages cultures and treatment with FQs.
Methods: We infected dog macrophages cultures with a B. melitensis type strain, according previously described methods. Macrophage cultures were infected with B. melitensis, opsonised with specific antibodies. After 1 hour incubation to allow Brucellae to penetrate into the macrophages, extracellular microorganisms were killed by adding gentamicin. Then, ciprofloxacin (CIP), levofloxacin (LEV) or moxifloxacin (MOX) were added. After 2 hours, cultures were lysed and spread onto Brucella agar with CIP, LEV or MOX at 4x wild type B. melitensis MIC. MICs of CIP, LEV and MOXI were determined for the colonies grown on plates. When a significant increase of resistance was confirmed, the gyrA and parC regions similar to regions containing the QRDR in E. coli were amplified and sequenced.
Results: MICS of CIP, LEV and MOX for the original strain were 0.2, 0.1 and 0.2 μg/mL respectively. MOX did not selected any resistant strain. CIP and LEV selected resistant isolates with a high frequency, around 1×10–4. Most of these resistant isolates showed increased MICs for all the fluoroquinolones tested, MICs ranging between 2–4 μg/mL for CIP, 1–2 μg/mL for LEV and 2–8 μg/mL for MOX. One isolate showed significant MICs increases only for MOX (2 μg/mL). gyrA and parC regions sequencing did not show significant changes, and MICs in presence of reserpine did not show differences as compared to antibiotics alone.
Conclusions: The treatment with FQs in macrophage cultures infected with Brucella select resistant strains with a high frequency. This fact might play an important role in treatment failures of brucellosis with FQs.
P875 Development of multidrug efflux mediated fluoroquinolone resistance in Salmonella hadar
S. Schmidt, P. Heisig (Hamburg, DE)
Objectives: Fluoroquinolones (FQs) are often the antibiotics of choice to combat gastrointestinal, respiratory and urinary tract infections as well as infections of the skin and soft tissue. However, bacterial resistance is becoming an increasing problem. Resistance to FQs is acquired by alterations of the target (mutations in type II topoisomerase genes gyrAB, parC) as well as by decreased uptake and increased efflux. Multi-drug efflux in E. coli is known to be due to elevated expression of several RND-type efflux pumps like AcrAB-TolC. Enhanced expression of efflux systems can be a result of mutations in local repressor genes or an altered expression of global regulators like marA.
For investigating the development of multi-drug efflux mediated FQ resistance in S. hadar a selection experiment has been performed and the resulting first and second step mutants have been analysed by sequencing of target genes, performing susceptibility testings as well as quantitative real-time PCR to determine putative expression changes of efflux pumps and global regulators.
Methods: For in vitro selection of FQ resistant mutants S. hadar was plated on ciprofloxacin and sparfloxacin (2–8 × MIC). Susceptibility testing of representative mutants was done according to CLSI-guidelines using phe-arg-γ-naphthylamide as efflux pump inhibitor (EPI). For qrt-PCR total RNA was isolated, reverse transcription was done using gene specific primers and expression changes were determined using SYBR Green.
Results: First step selection mutants show elevated MICs (4–6 dilution steps) to several FQs which are due to mutations in gyrA. Second step mutants are even more resistant (8 dilution steps) without having additional mutations in the target genes. An MIC-decrease of 3 dilution steps in the presence of an EPI indicates that elevated efflux is responsible for the increase in resistance in the second step mutants. qrt-PCR experiments show that expression of efflux-pumps acrAB-tolC, acrEF-tolC, mdtABC-tolC and ydhE as well as global regulators marA and rma is elevated.
Conclusions: In S. hadar elevated expression of several efflux pumps as well as global regulators add to the resistance phenotype. Still unknown is the complex interplay between regulators and efflux pumps. However insight is essential for the development of new chemotherapeutics acting on the level of bacterial efflux.
P876 Fluoroquinolone resistance in Clostridium difficile Italian clinical isolates
P. Spigaglia, F. Barbanti, P. Mastrantonio on behalf of ESCMID Study Group on Clostridium difficile
Objectives: Decreased susceptibility to fluoroquinolones in Clostridium difficile has been recently described. In particular, the epidemic strain NAP1/027 shows to be resistant to moxifloxacin and levofloxacin. Since no data are available in our Country, the aim of this study was to evaluate the minimal inhibitory concentrations (MICs) to ciprofloxacin and moxifloxacin of 60 C difficile clinical isolates and to investigate the mechanism of resistance.
Methods: Sixty C difficile isolates, collected between 1987 and 2005 from different Italian hospitals, were examined. MICs were determined using both the agar dilution technique performed on Muller Hilton plates and the E-test. E-test was performed on pre-reduced brucella agar plates containing vitamin K1, hemin, and 5% defibrinated sheep red blood cells. The breakpoints for ciprofloxacin and moxifloxacin were ≥16 mg/L and ≥4 mg/L, respectively. The quinolone resistance-determining region of gyrA was amplified using the primers 5-AA TGAGTGTTATAGCTGGACG-3 and 5-TCTTTTAACGACTCATCAAAGT T-3, whereas that of gyrB was amplified using the primers 5-AGTTGATGAACTGGGGTCTT-3 and 5-TCAAAATCTT CTCCAATACCA-3. Both strands of the amplified fragments were sequenced.
Results: 16 of the 60 isolates (27%) were highly resistant to both ciprofloxacin and moxifloxacin by both methods, except for two strains showing MICs = 16 mg/L for ciprofloxacin by agar dilution method and one strain showing a MIC = 4 mg/L for moxifloxacin by E-test. The majority of resistant strains were isolated after 1999. All resistant strains, except one, showed amino acid substitutions in gyrA or/and gyrB. Two amino acid substitutions were found in gyrA: from Thr82 to Ile and from Gly113 to Glu. Four different amino acid substitutions were found in gyrB: from Ser416 to Ala, from Asp426 to Asn, from Arg447 to Lys and Asp481 to Asn. The amino acid substitutions regarding Gly113 in gyrA, Ser416, Arg447 and Asp481 in gyrB have been not yet described in C difficile. In gyrA, the substitutions of Gly113 and Thr82 were always observed together. Substitutions of Ser416 and Asp481 in gyrB were found only in strains showing amino acid substitutions in gyrA.
Conclusions: This study shows an increase of C. difficile isolates resistant to ciprofloxacin and moxifloxacin in Italy since 1999 and indicates the necessity of further studies to investigate the possible association of the different amino acid substitutions with resistance patterns to other fluoroquinolones.
P877 The impact of a drastic change in selection pressure on trimethoprim resistance genes in clinical Escherichia coli isolates
A. Motakefi, M. Sundqvist, G. Kahlmeter, M. Grape (Stockholm, Växjö, SE)
Objectives: A rapid increase in resistance against trimethoprim (TRI) has been observed during the last ten years. The main mechanism of TRI-resistance is the integron-carried dihydrofolate reductase genes (dfr-genes). During 2 years (from Oct 1, 2004) an intervention project was carried out in the county of Kronoberg, Sweden, the TRICK study, where the use of trimethoprim and cotrimoxazole was reduced by 85%. By monitoring the effect of the intervention on the distribution of the five most common dfr-genes (dfrA1, dfrA5, dfrA7, dfrA12 and dfrA17) we describe changes in the mechanisms of resistance development and reversibility.
Methods: Before, during and after the intervention 100 consecutive clinical urinary E. coli isolates resistant to TRI were collected. Two multiplex PCR (mPCR) protocols using similar primers were developed, one conventional mPCR and one real-time mPCR respectively. The first ten PCR-products of each dfr-gene were sequenzed to validate the PCR methods. Both protocols were run on each isolate and the results were compared. The frequency of the five different dfr genes, were then determined as well as the distribution of these genes at each timepoint.
Results: The five selected dfr-genes were responsible for 86% of TRI resistance prior to and at mid-point of the intervention. dfrA1 was the most prevalent gene amongst the selected five genes. The distribution in the isolates collected prior to the intervention was; 39% dfrA1, 21% dfrA5, 2% dfrA7, 7% dfrA12 and 31% dfrA17. At mid-point only small shifts in the proportions of the five selected genes were seen 37% dfrA1, 15% dfrA5, 6% dfrA7, 2% dfrA12 and 27% dfrA17.
Conclusion: A high proportion of the isolates carried the five selected genes indicating that the selection of genes was relevant. 12 months of substantially reduced TRI consumption did not affect the distribution of dfr-genes to any significant extent. This implies that other factors than use of trimethoprim are of importance for determining the distribution of genetic elements encoding TRI resistance. The modest changes observed in the distribution of the five dfr-genes are being investigated further as are the hundred strains collected at the end of the intervention.
P878 First detection of invasive non-typeable Haemophilus influenzae strains with heterogeneous resistance to imipenem in Italy
M. Cerquetti, R. Cardines, M. Giufrè, P. Mastrantonio (Rome, IT)
Objectives: Carbapenems are considered an useful alternative to third-generation cephalosporins in treating invasive infections caused by ampicillin-resistant Haemophilus influenzae isolates. Here we report the detection of two nontypeable H. influenzae (NTHi) strains with heterogeneous resistance to imipenem. The resistance mechanisms were investigated.
Methods: NTHi strains 183 and 184 were isolated from elderly patients with meningitis. MICs were determined by E-test. Genetic relatedness between the isolates was examined by PFGE. The sequences of the ftsI gene encoding the penicillin-binding protein (PBP) 3 and of the dacB gene encoding PBP 4 were determined. Rd strain was transformed with the PCR-amplified ftsI gene from strain 183. The imipenem susceptibilities of the Rd-transformant and of strains Rd and 183 were compared by population analysis. To investigate the presence of additional mechanisms, the acrR regulatory gene of the AcrAB efflux pump was sequenced.
Results: By E-test, strains 183 and 184 showed MICs ranging from 4 mg/L to 6 mg/L, respectively, but colonies grew inside the inhibition zone up to 32 mg/L in both strains. By PFGE, the two isolates appeared closely related. Analysis of the deduced amino acid sequences of PBP3 transpeptidase region revealed the presence of 6 common amino acid substitutions in both strains plus an additional one unique for strain 183, compared to the Rd strain. This pattern of amino acid changes was not found in a group of imipenem-susceptible strains. Conversely, no relationship between amino acid substitution patterns in PBP4 and imipenem susceptibility was observed. Replacement of the ftsI gene in the Rd strain (MIC of imipenem = 0.094 mg/L) with the ftsI from 183 resulted in a transformant with a imipenem MIC ranging from 1.00 mg/L to 1.50 mg/L. When strains 183 and Rd-transformant were examined by the quantitative method of population analysis, both were heterogeneously resistant to imipenem and contained subpopulations growing up to 32 mg/L, at frequency of 10–5 and 10–8, respectively. No subpopulations were detected in Rd strain above 2 mg/L. No change in sequence of the acrR regulatory gene was found.
Conclusion: To our knowledge, this is the first report of H. influenzae strains showing heterogeneous resistance to imipenem in a clinical setting in Italy. Although the resistant phenotype seems to partly depend on PBP3 mutations, additional mechanisms could be involved.
P879 Cloning of a mdfA-like gene, encoding an efflux pump of an Acinetobacter baumannii clinical isolate
S. Marti, J. Sanchez-Cespedes, J.M. Sierra, M. Ruiz, S.M. Soto, J. Vila (Barcelona, ES)
Objectives: Acinetobacter baumannii is an important cause of nosocomial infection over the world; the increasing multiresistance of this microorganism is becoming a problem. An important way of acquiring this multiresistance is by overexpressing antimicrobial efflux pumps. The objective was to clone and characterise a mdfA-like gene encoding an efflux pump.
Methods: The search for efflux pumps was done by designing primers homologous to well characterised efflux pumps in other microorganisms. We compared the homology between the gene mdfA in Escherichia coli and the ORF57 of Acinetobacter calcoaceticus (presumably mdfA). A set of primers was designed in the areas of higher homology between these two genes. A small portion of the mdfA gene was amplified and sequenced. In order to obtain the whole gene, the genomic DNA of A. baumannii strain RUH134 was digested with MspI, a high frequency cutting restriction enzyme. The fragments obtained were auto ligated and the piece of DNA containing the mdfA gene was detected by PCR. Afterwards, this fragment of DNA was sequenced to obtain the complete gene sequence.
Results: The whole mdfA gene of A. baumannii strain RUH134 was sequenced. This gene had 1,230 nucleotides and it showed a 51.6% homology with the gene in Escherichia coli, and a 71.1% homology with Acinetobacter calcoaceticus ADP1. As regard to amino acids, the gene had a 42.7% homology with E. coli and a 76.5% homology with A. calcoaceticus ADP1. The prevalence of this gene was analysed in a group of sixty-four epidemiologically unrelated strains collected from Spanish hospitals in the year 2000. All the strains were positive for the gene mdfA and therefore, this suggests that the gene mdfA is present in the bacterial genome and not associated to a plasmid.
Conclusion: The mdfA gene codifying a putative multidrug efflux pump was identified and completely sequenced in one strain of A. baumannii. This gene also appeared in all the strains from a collection of A. baumannii obtained from Spanish hospitals suggesting a genomic implication. Further analyses will be necessary to determine the function of this gene and the involvement in acquiring antimicrobial resistance.
P880 Single-step selection of double mutations leading to high antibiotic-resistance in hyper-mutable Pseudomonas aeruginosa
M. García-Castillo, M.I. Morosini, A. Oliver, F. Baquero, J.L. Martínez, R. Cantón (Madrid, Palma de Mallorca, ES)
Objective: To determine the resistance mechanisms in single-step variants from PAO1deltamutS P. aeruginosa and those derived from the isogenic wild-type strain.
Methods: Both the PAO1deltamutS and the wild-type PAO1 P. aeruginosa strains (5×109–1×1010 cfu/mL) were exposed to agar plates containing different antibiotic concentrations (range 0.25–256 mg/L) of tobramycin (TOB), ceftazidime (CAZ), imipenem (IMP), ciprofloxacin (CIP), and levofloxacin (LEV). Sixty five mutants recovered from plates immediately below the mutant prevention concentration were selected and susceptibility (standard CLSI agar dilution) to different antibiotics was determined. Topoisomerase (II and IV) mutations, expression of efflux pumps (MexAB-OprM, MexCD-OprJ, and MexXY) and AmpC β-lactamase, and OprD deficiency were analyzed in selected mutants.
Results: The original PAO1deltamutS strain was less susceptible than its parental wild-type PAO1 strain for CAZ (8 and 1 mg/L, respectively) and TOB (2 and 0.5 mg/L). Mutants derived form the PAO1deltamutS strain were obtained in plates with higher CAZ, IMP, or TOB antibiotic concentrations (1–2 log dilutions) than those permitting the growth of mutants originated in normo-mutable PAO1 strain. Indeed mutants obtained from PAO1deltamutS strain were regularly less susceptible (1–4 log dilutions) than those selected form the wild-type PAO1 strain. Mutants selected on CAZ and IMP have reduced susceptibility restricted to β-lactam antibiotics. In these mutants, the MICs and enzymatic activity were compatible with a higher production of chromosomal AmpC enzyme in the case of CAZ selected mutants, whereas AmpC hyper-production superimposed with OprD reduction was observed in the case of IMP. In mutants selected with CIP or LEV, overproduction of efflux pumps was mainly observed with PAO1 strain and overproduction of pumps superimposed with gyrA mutations with PAO1deltamutS strain.
Conclusions: The higher antibiotic resistance levels of mutants derived from dense populations of PAO1deltamutS P. aeruginosa when compared with those from the corresponding normo-mutable PAO1 wild-type strain might result from the selection of cells carrying double mutations. Hyper-mutable strains contribute to the single-step selection of variants with higher levels of antibiotic resistance.
P881 Altered outer membrane protein profiles of Pseudomonas aeruginosa highly susceptible to the efflux pump inhibitor Phe-Arg-β-naphthylamide
A.B. Campo-Esquisabel, S. Martí, J.M. Sierra, L. Martinez-Martinez, J. Vila (Santander, Barcelona, ES)
Objective: Multidrug effllux systems play an important role in resistance o f P. aeruginosa (Pae) to several antibiotics. Phe-Arg-β-naphthylamide (PAN) is usually able to inhibit these systems. When evaluating the relevance of efflux in some clinical isolates of Pae we noticed that some of them were inhibited by low concentration of PAN alone. We have evaluated OMP expression in Pae highly susceptible to PAN.
Methods: Three Pae clinical isolates (HUMV1, HUMV2 and HUMV3) for which MIC of FEP was at least 2 times higher than that of CAZ were selected among a collection of 20 isolates with a similar phenotype. MICs of PAN and of FEP in the absence and in the presence of 20 mg/l of PAN were determined by reference microdilution (CLSI guidelines). The study of clonal relation between the isolates was performed by pulsed-field gel electrophoresis (PFGE) digested with SpeI. Fractions of enriched cell envelope proteins from the three organisms grown in media without or with subMIC of PAN profiles were characterised by SDS-PAGE. Proteins were further analysed by MALDI-TOF mass spectometry after in-gel trypsin digestion.
Results: MICs (mg/l) of FEP for HUMV1, HUMV2 and HUMV3 were 64, 32 and 16 mg/l, respectively. MIC of FEP+PAN for HUMV3 was 4 mg/l, but HUMV1 and HUMV2 did not grow in the presence of 20 mg/l of PAN. In fact, MICs of PAN alone for HUMV1 and HUMV2 were 4 and 2 mg/l, respectively. The PFGE profile for HUMV1 and HUMV3 was identical, and their corresponding protein profiles differed in three proteins identified as: major outer membrane lipoprotein I (present in HUMV3, absent in HUMV1), 30S ribosomal protein S16 (reduced expression in HUMV1) and lisozyme (present in HUMV1, absent in HUMV3). Major outer membrane lipoprotein I was also absent in the PAN-susceptible HUMV2 isolate.
Conclusions: Hypersusceptibility of two P. aeruginosa strains to the efflux pump inhibitor Phe-Arg-β-naphthylamide was linked to loss of major outer membrane lipoprotein I.
P882 Biological cost of resistance to fosfomycin in Escherichia coli isolates
J.I. Alós, P. García-Peña, J. Tamayo (Móstoles, ES)
Fosfomycin is a bactericidal antibiotic that acts by inhibiting the cell wall, and which is used mainly in the treatment of uncomplicated urinary tract infections (UTI). Resistance to fosfomycin develops rapidly in experimental conditions, although despite its frequent use in UTI, resistance in E. coli, the main uropathogen, is very low (1–3%), and has remained so for many years. The objective of this study was to ascertain whether E. coli fosfomycin-resistant strains have less fitness than those that are fosfomycin-sensitive in competing, and would therefore tend to disappear in their competition with fosfomycin-sensitive strains in the absence of antibiotics.
Methods: Fosfomycin-resistant strains (n = 8) with different phenotypes of resistance to other antibiotics. All but one were lactose (+). Fosfomycin-resistant strains (n = 13) that had same phenotypes of resistance to other antibiotics as the resistant strains and which furthermore had the opposite pattern of lactose fermentation.
Thirty-three (33) competition experiments by pairs of strains were conducted (Fosfomycin-R versus Fosfomycin-S with the rest of resistance determinants being equal and with a different lactose fermentation capacity to perform the differential counts in MacConkey agar). The experiments were performed in nutrient broth (NB). Equal amounts of the strains were challenged (approx. 50% and approx. 50%) for 4 days, with a daily change to a new medium. Five differential counts were performed (days 0, 1,2, 3, and 4). The 33 experiments were performed in duplicate, and the mean of both results are presented below.
Results: In 20 experiments (60.6%) there was a relative increase in the fosfomycin-sensitive strain that translated into a count of >60% (between 65%-99%) on the fourth day. In 6 experiments (18.2%) there was a relative increase in the fosfomycin-resistant strain that translated into a count of >60% (between 65%-75%) on the fourth day. In 7 experiments (21.2%), on the fourth day none of the strains reached 60%. When the data of the 26 (20+6) experiments in which there were changes were analysed by the Chi2 test there was an statistically significant difference (P = 0.044).
Conclusions:
-
•
Resistance to fosfomycin entails a biological cost (less fitness) for the majority of the E. coli strains assayed.
-
•
This biological cost hinders their competition with fosfomycin-sensitive strains in the normal intestinal flora, which would make them less likely to cause UTI.
P883 Antibiotic resistance of enterococci isolated from blood cultures during 2003–2006
U. Arslan, B. Feyzioglu, E.B. Uysal, E.I. Tuncer, D. Findik (Konya, TR)
Objectives: Enterecocci have become a significant problem due to their aetiologic role in bacterial infections. An alarming problem is the increasing rate of enterococci resistant to vancomycin (VRE) and therefore the source and spreading of these strains are very important epidemiological problems. The aim of the study was to evaluate antibiotic resistance and monitoring of VRE isolated from blood cultures during July 2003 – October 2006 in our hospital.
Methods: A total of 138 strains of enterococci were isolated from blood cultures. The identification of the isolated bacteria was performed by conventional methods and bioMérieux API 20 STREP test (Marcy I'Etoile, France). The susceptibility testing was carried out by disc-diffusion method and Etest. Determination of glycopeptide resistance genotypes (van A, van B, van C1, van C2/3) of VRE was performed by GenoType Enterococcus assay (Hain Lifescience GmbH, Nehren, Germany).
Results: Of the 138 enterococci strains isolated 72 strains were E. faecium, 59 E. faecalis, five E. gallinarum, two E. casseliflavus. 123 (89.1%) enterococ strains were isolated from inpatients. 53 (38.4%) of them were isolated from intensive care units, 40 (29%) from medical units, 28 (20.3%) from paediatric units, 17 (12.3%) from surgery units. A total of 108 strains were resistant to ampicillin (78.3%), 58 to high level gentamycin (42.0%) and 9 to vancomycin and teicoplanin (6.5%). All of VRE strains was identified as E. faecium. All of the VRE strains were susceptible only to linezolide. VRE showing high resistance to vancomycin and teicoplanin suggest the presence of Van A phenotype. In all of the VRE strains with glycopeptide resistance genotypes we detected Van A genotype.
Conclusion: This study indicates that in our hospital Enterecoccus spp. shows high resistance to high level aminoglycosides and to aminopenicillins and VRE strains carry Van A gene.
P884 Effect of multiple passages of S. typhimurium in the presence of disinfectants on susceptibility to antimicrobials, on persistence in the one-day-old chick model and efflux systems
M. Bagnall, L. Randall, N. Coldham, A. Karatzas, T. Humphrey, L. Piddock, M. Woodward (Addlestone, Bristol, Birmingham, UK)
Objectives: Reduced susceptibility to certain disinfectants has been linked with reduced susceptibility to some antibiotics. If persistence and accumulation of such mutants persist within the food chain this would be of concern. In this study, mutants derived from Salmonella typhimurium following multiple exposures to two different farm disinfectants (either a combination formaldehyde, glutaraldehyde and quaternary ammonium compound aldehyde-based disinfectant (F/G/QAC) or an oxidising compound disinfectant (OXC) disinfectant) were analysed for their susceptibility to antimicrobials and fitness in the day old chick model. The proteomes were also compared and molecular flux investigated using the fluorescent dye bisbenzimide (H33342).
Methods: Minimum Inhibitory Concentrations (MIC) of various disinfectants and antibiotics were determined by the BSAC agar dilution method. In competitive index experiments one-day-old chicks were infected by oral gavage (c.104 cfu per bird, c.1:1 mix of mutant and parent strain). Infection was monitored by cloacal swabbing at 3, 10, 17, 24, 31 and 35 days post-infection. Infection was calculated as cfu/g of faecal matter. In conjunction with the cloacal swabs 5 birds/group were sacrificed at 7, 14, 21 and 28 days post-infection and the caecal contents enumerated as previously described. Proteomes were determined by 2D-LC-MS and uptake of bisbenzimide (2.5 mM) using a fluorescent plate reader in a 96 well format at 350 and 460 nm respectively.
Results: The mutant strains were significantly less able to persist in the one-day-old chick model than their parent strains. Both mutants showed reduced susceptibility (circa x 4) to some clinical antibiotics (cip, chlor, tet and amp) and were also cyclohexane resistant. This suggests that the mechanism of resistance is due to up-regulated efflux and/or reduced porins. The proteomics confirmed increased expression of the AcrAB/TolC efflux pump. Reduced uptake of the dye bisbenzimide was consistent with these changes in protein expression.
Conclusions: Repeated exposure of Salmonella Typhimurium to disinfectants gave rise to mutants with reduced susceptibility to disinfectants and/or antibiotics. This was coupled with increased efflux, as measured by bisbenzimide uptake assays, and increased differences in key efflux proteins.
P885 Investigation of autolysin and autolysis of glycopeptide-intermediate Staphylococcus aureus induced from methicillin-resistant Staphylococcus aureus
J.I. Yoo, E.J. Park, J.S. Yoo, K.M. Lee, G.T. Chung, Y.S. Lee (Seoul, KR)
Background: Methicillin-resistant Staphylococcus aureus are the most important pathogens causing nosocomial infections. The glycopeptide antibiotics has been used successfully for the treatment of serious infections caused by MRSA for several decades. However, clinical isolates with reduced susceptibility to glycopeptide antibiotics have arisen during long-term vancomycin therapy in various contries around the world since 1997. The efficacy of vancomycin has been limited by the emergence of both GISA and VRSA in clinical isolates. In this study, we induced GISA phenotype from vancomycin susceptible strains and investigated characteristics of autolysis and autolysin.
Methods: The 6 MRSA clinical strains showing prevalent PFGE types in Korea were selected to induce resistance. The agr types were determined with agr group specific primers by multiplex PCR. Strains were grown in TSB in the presence of vancomycin at 1 to 2 ug/mL or were above the MIC for a given strain and induced to 4 ug/mL and 8 ug/mL MICs, continuously. The change of autolysis was quantitated as percentage of the initial OD600 remaining at each sampling time point. The autolysin was analyzed with intracellular protein and extracellular protein from GISA strain. The function of the agr operon in GISA strains was measured by δ-hemolysin production experiment.
Results: All of 6 strains showed 3 different agr types (I, II, III) and all strains were induced to MICs 4–8 ug/mL. All induced GISA strains showed a slower rate of autolysis than did their parents. And two isolates among GISA strains showed decreased δ-hemolysin expression (the other 4 strains had no δ-hemolysin activity in their parents). The expression of major intracellular autolysin (53 kDa) was decreased in GISA strains and the other 3 or 4 autolysin bands were shown. In extracellular proteins, the expression of 53 kDa autolysin was also decreased but that of 37 kDa autolysin was increased in 2 GISA strains.
Conclusion: The reduction of autolytic activity and δ-hemolysin activity in GISA strains is supposed that the agr operon has relation with the activity of autolysin. Among the several autolysin, 53 kDa and 37 kDa autolysins are play a role in GISA strains. And also, the autolysin activity is reduced against vancomycin to tolerate the antibiotic stress.
P886 Macrolide resistance in Streptococcus spp. and Enterococcus spp. clinical isolates: phenotypic and genotypic analysis
J. Perez, E. Culebras, M. Gomez, J. Picazo (Madrid, ES)
Objectives: The growing number of macrolide resistant strains of Enterococcus spp. and Streptococcus spp. is an increasing problem worldwide. In this study we evaluated 44 clinical isolates of both genera obtained at Microbiology Department of Hospital Clínico San Carlos. All of these macrolide resistant strains were tested for their antimicrobial susceptibility, resistance phenotypes and genotypes.
Methods: A total of 25 Streptococcus spp. (10 S. pyogenes, 10 S. agalactiae, 5 group G Streptococcus and 1 S. pneumoniae) and 19 Enterococcus spp. (10 E. faecalis, 7 E. faecium, 1 E. gallinarum and 1 E. avium) clinical isolates resistant to erythromycin and/or clindamicin were included. Organisms were collected at the Hospital Clínico San Carlos from January to June of 2006. All strains were identified by standard methods and by the Rapid ID32 Strep system. Susceptibility to penicillin (P), erythromycin (ER), clindamycin (CC), clarithromycin (CL), azithromycin (AZ), josamycin (J), telithromycin (TL), Quinupristin/Dalfopristin (Q/D), gentamicin (G), tetracycline (T), minocycline (M) and tigecycline (TG) was determined by the agar dilution methods. The presence of erm(A), erm(B) and mefA/E genes was determined by PCR amplification with specific primers.
Results: In general, Enterococcus spp. were more resistant than Streptococcus spp. Of the 25 Streptococcus spp. studied, 68%, 36%, 8% and 8% were resistant to CC, T, Q/D and TL respectively. The Enterococcus spp. clinical isolates show TL MICs ranged between ≤ 0.006 and 4 mg/L, and Q/D ranged between ≤0.006 and >256 mg/L. 68% and 58% of Enterococcus spp. were resistant to ER and T respectively. All strains remain susceptible to TG. There was 22 strains with erm(B) gene alone (5 S. agalactiae, 5 S. pyogenes, 1 S. pneumoniae, 6 E. faecalis, 4 E. faecium and 1 E. gallinarum). erm(A) alone was present in 5 strains (4 group G Streptococcus and 1 S. agalactiae), in 3 of these strains MIC to CC was higher than MIC to ER. The mefA/E gene was amplified in all strains with M phenotype. Three strains harboured erm(A) and erm(B) both genes. We were no able to detected any of the studied gene in 13 strains.
Conclusions: 1) erm(B) was the most prevalent gene in our clinical isolates; 2) A bimodal distribution of ER MICs was noted in erm(A) isolales. 3) All clinical isolates resistant to TL harboured erm(B) gene. 4) Resistance to Q/D is common in our E. faecium.
P887 Linezolid resistance in clinical isolates of Staphylococcus haemolyticus
J. Calvo, M. Cano, M. Marín, E. Cercenado, L. Martinez-Martinez (Santander, Madrid, ES)
Objectives: To characterise four isolates of linezolid-resistant S. haemolyticus obtained from different patients.
Methods: Identification and preliminary susceptibility testing of organisms were performed with the WalkAway system. Linezolid MICs were determined by reference microdilution and Etest assays, according to CLSI guidelines and manufacturer's instructions, respectively. Detection of the G2576T mutation was performed by PCR amplification and sequencing of the domain V of 23S rRNA gene (Tsiodras et al, Lancet 2001). Clonal relationship of the isolates was examined by PFGE of SmaI macrorestricted genomic fragments.
Results: Four linezolid-resistant S. haemolyticus isolated in pure cultures from blood samples (n = 3) or bile (n = 1) were identified among 17 organisms of this species obtained in our laboratory in the period March-April 2006. All four isolates were from different patients admitted to ICUs of our hospital. Reference MICs of linezolid were 64 mg/L for 3 isolates and 128 for the other one. Etest yielded one twofold dilution lower in all isolates. The G2576T mutation was detected in all four isolates and all of them were homozygous. Similar resistance phenotype (WalkAway) was observed for all 4 isolates, being resistant to oxacillin, teicoplanin, ciprofloxacin, clindamycin, erythromycin, gentamicin, tobramycin, amikacin, rifampin, trimethoprim-sulfamethoxazol and susceptible to vancomycin, tetracycline and quinupristin-dalfopristin. Two closely related PFGE-patterns were observed for the 4 isolates, each one corresponding to two isolates.
Conclusions: We have identified the emergence of multiresistant S. haemolyticus isolates presenting homozygous resistance to linezolid.
P888 Macrolide efflux pumps mef(E) and mef(I) detected in clinical isolates of S. pyogenes isolated from PROTEKT 1999–2005 and PROTEKT US 1999–2004
J. Northwood, L. Cossins, M. Coley, A. Pantosti, M. Del Grosso, D. Farrell (London, UK; Rome, IT)
Objectives: To determine the presence of the macrolide efflux mechanism of resistance, mef(A) subclass mef(E), in community-acquired lower respiratory tract infections of Streptococcus pyogenes, from a global collection of clinical isolates (n = 28,892) 1999–2005.
Methods: Isolates of S. pyogenes with MIC values demonstrating the M phenotype of resistance to macrolides were screened for the mef(E) gene using a novel TaqMan™ assay designed for this project. Isolates which were positive for the presence of mef(E) using the TaqMan™ assay were confirmed by di-deoxy sequencing of a 300 bp segment of the mef(E) gene.
Results: Twenty-Two isolates (0.07%) were considered to be mef(E) although 3 isolates had 1 amino acid substitution from mef(E): M30I and 1 isolate had 3 amino acid substitutions from mef(E): C97V, M104I, and I105V (5 SNPs). These isolates may have additional substitutions outside of the 300 bp region examined, and further sequence analysis of the mef gene is required. Five isolates (0.02%) of S. pyogenes were consider to have the mef(A) subclass mef(I) gene present, all of these isolates had 1 amino acid substitution from mef(I): K39R. One isolate was considered to be a novel subclass of mef(A) due to the following variations: mef(A) 4 amino acid substitutions L52V, S89A, T92A and A125S; mef(E) 5 amino acid substitutions I59V, L60F, C97Y, I105V and A125S; mef(I) 4 amino acid substitutions L96F, V107I, V114I and A125S and mef(O) 3 amino acid substitutions L96F, V107I, A125S (14 SNPs). There was a global distribution of mef(E) in S. pyogenes whereas 4 out of the 5 isolates positive for mef(I) came from Belgium.
Conclusions: This study definitively shows the presence of mef(E) in S. pyogenes and its global distribution, the previously un-described presence of mef(I) in S. pyogenes and the presence of a novel subclass of mef(A).
P889 Emergence of linezolid-resistant Enterococcus faecalis in Spain and rapid characterisation by real-time PCR
E. Cercenado, M. Marin, O. Cuevas, E. Bouza (Madrid, ES)
Objectives: Linezolid was introduced in our institution in 2004. From May 2005 to September 2006 we determined the in vitro activity of linezolid against all staphylococci and enterococci isolated in our laboratory. We characterise the first linezolid-resistant (LNZ-R) clinical isolates of Enterococcus faecalis in Spain.
Methods: MICs were determined by the broth microdilution method using commercialised MicroScan panels (Dade-Behring). CLSI guidelines (2006) were followed for determination of breakpoints. LNZ-R isolates were confirmed by E-test. Detection of the G2576T mutation was performed by PCR and sequenciation of the 23S rRNA gene, as well as by real-time PCR (Woodford et al; JCM 2002; 40: 4298–4300) using a fluorescent dye-labeled detection probe.
Results: Over the period of study, we tested 6,214 staphylococci (3,817 S. aureus; 2,407 coagulase-negative staphylococci), and 1,850 enterococci (1,439 E. faecalis; 340 E. faecium; 71 other species). All staphylococci were susceptible to linezolid (MIC ≤ 4 mg/L). Three E. faecalis isolates (0.16%) were LNZ-R (MICs 64, and 128 mg/L; 2 and 1 isolate, respectively. The isolates were recovered in 2006 and corresponded to 3 patients hospitalised in different wards that received linezolid for >15 days previous to the isolation of the LNZ-R isolates. PCR and sequenciation detected the G2576T mutation only in 2 isolates (MICs 64 mg/L). Real-time PCR was positive in all 3 isolates, being one strain (MIC = 128 mg/L) heterozygous, and the other 2 homozygous. Results were obtained in less than 2 h. The G2576T mutation in the heterozygous strain was not detected by PCR amplification and sequencing of the 23S rRNA gene.
Conclusions: We characterise for the first time in Spain the emergence of LNZ-R E. faecalis isolates in patients previously treated with linezolid. Since the G2576T mutation was not detected in the heterozygous isolate by PCR and sequencing, the real-time PCR must be the method of choice for the characterisation of this emergent resistance mechanism.
P890 Linezolid resistance in a Staphylococcus haemolyticus strain isolated in an intensive care unit
A. Mazzariol, G. Lo Cascio, R. Koncan, L. Maccacaro, R. Fontana, G. Cornaglia (Verona, IT)
Objectives: Although even recent reports confirm resistance to linezolid to be minimal among coagulase-negative staphylococci, and virtually confined to a few cases of Staphylococcus epidermidis encountered in the USA, a number of documented cases have occurred in South America and Europe, too. In a recent report from intensive care units (ICUs) in Madrid, Spain, variable resistance percentages were found in Staphylococcus epidermidis (0.81%), Staphylococcus hominis (0.71%), and most noticeably Staphylococcus haemolyticus (6.84%).
A 65-year-old male patient was admitted to the ICU of the Verona University Hospital on 1 August 2006, after extensive surgery for acute pancreatitis. Empirical therapy with teicoplanin (600 mg) was started on admission. On the 16th day of therapy, a strain of S. haemolyticus was isolated from the blood culture. The strain proved intermediate to teicoplanin (MIC, 16 mg/L), which was immediately suspended and replaced with linezolid (MIC, 0.25 mg/L). After a further 8 days, a new blood culture showed that the same strain had become resistant to linezolid (MIC, 32 mg/L).
Methods: Antimicrobial susceptibility testing was routinely performed by diffusion test according to the latest CLSI documents. The MICs to all antibiotics were determined by means of the E-test and confirmed by dilution tests. PFGE was carried out by standard procedures.
Results: MICs for recently released anti-staphylococcal compounds were as follows: daptomycin, 0.12 mg/L; quinopristin/dalfopristin, 0.5 mg/L; telavancin, 0.12 mg/L; tigecycline, 1 mg/L.
After serial passage on antibiotic-free medium, the isolate's resistance to high concentrations of linezolid was unvaried. Compared with two linezolid-susceptible S. haemolyticus strains, isolated from the same ICU in the same time period, the linezolid-resistant isolate demonstrated a significant difference in its in-vitro growth characteristics. PFGE of the two linezolid-susceptible strains would suggest a clonal relationship with the linezolid-resistant one. Further resistance characterisation and clonal analysis are ongoing.
Conclusions: After three years of linezolid use, multifocal emergence of linezolid resistance in coagulase-negative staphylococci has become an important matter of concern and involves species other than S. epidermidis. Because of the paucity of newer effective antimicrobial agents, this fact mandates stricter control over the use of this antibiotic to preserve its clinical utility.
P891 Mycobacterium leprae DNA gyrase: expression, purification, inhibition by quinolones and functional analysis of two mutant enzymes
S. Matrat, S. Petrella, E. Cambau, W. Sougakoff, V. Jarlier, A. Aubry (Paris, FR)
Objectives: M. leprae is still uncultivable in vitro and evaluation of antibiotic activity against this bacteria relies mainly on the mouse foot pad model that requires 8 to 12 months experiments. It is therefore important to establish an in vitro approach allowing to test rapidly the efficiency of quinolones against wild type and quinolone resistant mutants of M. leprae. Mutations in the DNA gyrase (the sole target of quinolones in M. leprae) was previously described. The aim of the present study was to purify the DNA gyrase of M. leprae in order (i) to test the ability of quinolones to inhibit this enzyme and (ii) to study the implication of gyrase mutations in the resistance of M. leprae clinical isolates to quinolones.
Methods: We separately overexpressed in Escherichia coli the M. leprae GyrA and GyrB subunits as His-Tagged proteins using pET plasmids harbouring the gyrA and gyrB genes. Two mutant enzymes (A91V in GyrA and D205N in GyrB, corresponding to amino acids GyrA 83 and GyrB 183 in E. coli numbering system), representing mutations found in M. leprae clinical isolates (Cambau et al., Lancet, 1997; Kim et al., FEMS, 2003), were introduced by site directed mutagenesis. Wild type and mutant subunits were purified by nickel chelate chromatography. The drug concentrations that induced 25% of DNA cleavage (CC25) were measured for 3 quinolones (MXF, OFX and LVX).
Results: The soluble 97.5-kDa GyrA and 74.5-kDa GyrB wild type and mutant subunits obtained after purification were mixed to reconstitute enzymes showing a DNA supercoiling activity. The CC25s of the GyrB mutant D205N were similar to those found with the wild type subunit. In contrast, the CC25s of OFX, LVX and MXF for the GyrA mutant A91V were 4-, 6- and 20-fold higher than the wild type ones respectively.
Conclusion: Our results show that mutation D205N in GyrB is not involved in acquired resistance of M. leprae to quinolones whereas A91V in GyrA is involved in quinolone resistance of M. leprae. In addition, we showed that drug-induced DNA cleavage assays from purified wild type and mutant M. leprae DNA gyrases represent a safe, quick and useful screening test for identifying quinolones with potential activity against M. leprae and related multidrug resistant strains.
P892 Genetic basis of macrolide and lincosamide resistance among staphylococcal clinical isolates in a cancer centre
V. Kolliou, M. Lymperopoulou, M. Tsaparli, A. Markogiannakis, E.N. Manolis, N.J. Legakis, A. Tsakris (Athens, Piraeus, GR)
Objectives: To investigate the incidence of macrolide and lincosamide resistance phenotypes and the distribution of the related resistance genes among staphylococcal isolates collected in a cancer hospital in Greece.
Materials and Methods: The study included 148 Staphylococcus aureus and 24 cogulase-negative staphylococci (CNS) clinical isolates recovered during 2002–2004 from separate patients in the Metaxa Cancer Hospital. MICs of erythromycin, clindamycin and oxacillin were tested by an agar dilution method. The macrolide-lincosamide-streptogramin B (MLSB), macrolide-streptogramin B (MS) and lincosamide (L) phenotypes were screened by the disk approximation test with erythromycin and clindamycin. Detection of the genes encoding resistance to macrolides and lincosamides was performed by PCR, while clonal strains were identified by PFGE analysis of SmaI-digested DNA.
Results: One hundred-one (68.2%) of the S. aureus isolates were characterised as MRSA and 19 (79.2%) of the CNS as MRCNS. Seventy-six (44.2%) staphylococcal isolates (56 S. aureus and 20 CNS isolates) exhibited resistance or intermediate level of resistance to at least one of erythromycin and clindamycin. The MLSB constitutive phenotype was detected in as many as 69 (90.8%) isolates, followed by the MLSB inducible phenotype (4 isolates; 5.3%), the MSB phenotype (2 isolates; 2.6%) and the L phenotype (one isolate; 1.3%). The ermA gene was identified in 69.7% of the isolates, while less frequently were identified the ermC gene (17.1% of isolates), the combination of ermC and msrA/B genes (6.6% of isolates), the msrA/B gene (1.3% of isolates) and the lnuA/A′ gene (1.3% of isolates). In 3 (3.9%) isolates resistance genes were not detected. PFGE showed the dissemination of two major clonal types among S. aureus isolates.
Conclusions: The data indicate that in the cancer centre of this study the overall macrolide and lincosamide resistance is driven by the epidemic spread of the MLSB constitutive phenotype among clonal staphylococcal isolates containing either ermA or ermC gene.
P893 Prevalence and genetic background of fluoroquinolone resistance in clinical isolates of Proteus mirabilis
R. Nakano, M. Abe, M. Inoue, R. Okamoto (Sagamihara, Kanagawa, JP)
Objectives: Fluoroquinolone (FQ) resistance among Enterobacteriaceae was reported increasingly. It occurs primarily through mutation in the QRDRs of the DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE). In this study, we carried out a survey to assess the prevalence of the FQ resistant P. mirabilis and investigate the mechanism of the resistance.
Methods: 1855 isolates of P. mirabilis were collected from 7 different hospitals in Kanagawa Prefecture, Japan in 2000–2005. The susceptibility of levofloxacin (LVFX) was determined by the agar dilution method according to the CLSI guidelines. DNA sequences of the QRDRs of gyrA, gyrB, parC, and parE in 50 clinical isolates with susceptibility or resistance to LVFX were determined by PCR and sequencing.
Results: Of the 297 strains were found to resistant to LVFX (MIC > 2 μg/mL). It was gradually increase the incidence in 2000–2004 and was significantly increased 45.9% in 2005. The DNA sequences of the QRDRs of P. mirabilis were identified. Susceptibility strains (MIC, 0.125 to 2 μg/mL) and MIC 4 μg/mL strains showed change in gyrA (S83I) and parC (S80I). Low-level resistant strains (MIC, 8 to 16 μg/mL) showed change in gyrA (S83I), gyrB (S464Y or E467D), and parC (S80I). High-level resistant strains (MIC, 32 to 256 μg/mL) showed change in gyrA (S83I+E87K), parC (S80I), and parE (D420N). Highest-level resistant strains (MIC, 512 μg/mL) strains showed change in gyrA (S83I+E87G), gyrB (S464F), parC (S80I), and parE (D420N).
Conclusion: The prevalence of LVFX resistant P. mirabilis was steadily increasing in Japan. Susceptibility profiles for LVFX were correlated with amino acid changes in QRDRs. Low-level resistant strains (MIC, 8 to 16 μg/mL) were associated with mutation in gyrB (S464Y or E467D). High-level resistant strains (MIC, 32 to 256 μg/mL) were associated with mutations in gyrA (E87K) and parE (D420N). Highest-level resistant strains (MIC, 512 μg/mL) were associated with mutations in gyrA (E87G), gyrB (S464F), and parE (D420N).
P894 Characterisation of insertion sequence, conjugative transferase gene and mupA gene of mupR plasmid from MuH methicillin-resistant staphylococci
J.I. Yoo, G.T. Chung, E.S. Shin, Y.S. Lee (Seoul, KR)
Objectives: Mupirocin has been used to eliminate the carriage of S. aureus and prevent staphylococcal infections. However, mupirocin-resistant S. aureus has been increased and the high level mupirocin resistance (MuH) is associated with an isoleucyl tRNA synthetase that is encoded by a novel plasmid-encoded gene, mupA. In this study, we investigated transferable mupirocin resistance element containing insertion sequence (IS), trs and mupA gene of mupR plasmid from MuH methicillin resistant staphylococci isolates.
Methods: Against 680 staphylococci isolates from tertiary hospitals, antimicrobial susceptibility test including mupirocin was done by disk diffusion and MIC of mupirocin was determined by E-test method. The plasmid-encoded mupA gene was detected by PCR. The mupA-IS and trsLM-mupA specific primers were used to identify the presence and genetic location of IS element, trs and mupA gene.
Results: 12 MuH-staphylococci isolates with different PFGE patterns were selected from hospitals (680 isolates) and long-term care facilities (407 isolates) in Korea. MuH MRSA (8 isolates), MuH S. haemolyticus (2 isolates), MuH S. hominis (1 isolates) and MuH S. epidermidis (1 isolates) were identified. The trsLM-mupA PCR showed 4 different products such as 7,033 bp (6 isolates), 5,024 bp (3 isolates), 4,530 bp (2 isolates) and 6,105 bp (1 isolates). IS sequence of all 12 isolates identified to have IS257 sequences except 2 nucleotide substitution (A174G, A536G) and one thymine nucleotide deletion at 644 nucleotide. The mupA was flanked by two directly repeated IS257 like sequences. The sequences of trsLM were also different in 68 nucleotide site compared to previously reported trsLM sequences.
Conclusion: The high-level mupirocin resistant staphylococci isolated from Korea hospitals had different IS257 sequences and trsLM sequences. Because sequences of all MuH isolates showed the same sequences, the IS257 like sequences and trsLM sequences is typical in Korean isolates. According to the trsLM-mupA sequence analysis, there was no specific difference between MRSA and MRCNS. It indicated the possibility of mupA transfer from MRSA to MRCNS and vice versa. And also the sequence relation of trs, IS and mupA against MRCNS is the first report in the world.
Molecular diagnostics for staphylococci
P895 Evaluation of a real-time PCR assay and an multiplex-reverse hybridisation system for the detection of methicillin-resistant Staphylococcus aureus
M. Ieven, M. Michiels, H. Jansens, H. Goossens (Edegem, BE)
Objective: To evaluate the performance of a commercial multiplex PCR – reverse hybridisation system, Hyplex StaphyloResist® (BAG Germany distributed in the Benelux by AlphaOmega instruments), and a real-time PCR assay for the detection of MRSA from 24 hours enrichment broths compared to the routine culture method.
Methods: A total of 500 enrichment broths from MRSA screenings were included in the study. On arrival in the lab, swabs were cultured by direct plating and subsequently put in MRSA enrichment broth. After 24 hours incubation the broth was subcultured on routine media and MRSA was identified by conventional methods. A broth sample was taken for DNA extraction and two PCR assays. The Hyplex StaphyloResist® consists of a multiplex PCR amplifying simultaneously mecA gene, coagulase gene and a “housekeeping” gene. Specimens were processed according to the manufacturer's instructions. For the real-time PCR assay specimens were extracted using the Qiagen blood mini kit with consecutive detection of mecA, nuc and SCCmec genes, according to a modified protocol by Huletsky et al (JCM 2004; 42: 1875–84). PCR results were considered positive if confirmed by culture or the second PCR.
Results: In total 78 (16%) of the 500 MRSA enrichment broths tested were culture and both PCRs positive, 6 were culture negative and both PCRs positive, 1 was culture positive and both PCRs negative. 6 broths were culture positive and one PCR negative (5 real-time PCR assay, 1 Hyplex StaphyloResist® assay) and 28 broths were culture negative and one PCR negative (14 real-time PCR assay, 14 Hyplex StaphyloResist® assay). The sensitivity, specificity and negative and positive preditive values of the Hyplex StaphyloResist® assay for the detection of MRSA from broth were 98%, 96%, 99% and 83% and 93%, 96%, 98% and 83% for the real-time PCR assay, respectively. Both PCR assays provided same day results on 24 hours enrichment broths compared to 48 to 96 hours for routine culture method. The time needed to complete the Hyplex StaphyloResist® and real-time PCR assays were 3 hrs 25 min and 2 hrs 30 min with a hands-on time of 1 hrs 30 min and 1 hrs 15 min, respectively.
Conclusion: There is no significant difference in sensitivity and specificity between the Hyplex StaphyloResist® assay and the real-time assay for the detection of MRSA from enrichment broth. More than 90% of MRSA positives can be identified after one day by either PCR if performed on MRSA enrichment broth.
P896 Comparison of NucliSens easyMAG and Qiagen nucleic acid extraction using screening broths for the detection of MRSA
M. Ieven, M. Michiels, K. Bergs, D. Ursi, H. Goossens (Edegem, BE)
Background: The NucliSens easyMAG platform (bioMérieux) is a second generation system for automated isolation of nucleic acids (NA) from clinical samples, based upon silica extraction technology. Currently, no data are available on the NucliSens easyMAG for the extraction of MRSA DNA from screening samples.
Objectives: To evaluate the performance and user convenience of the NucliSens easyMAG platform for NA extraction from MRSA enrichment broths compared to manual Qiagen extraction.
Materials and Methods: 500 enrichment broths from MRSA screenings were included in the study: on arrival in the lab, swabs were cultured by direct plating and enrichment cultures. 200 μl broth was used for NA extraction using the Qiagen blood mini kit and 1 ml broth was frozen. NAs extracts were analysed by real-time PCR targeting the mecA, nuc and SCCmec genes for the detection of MRSA DNA. 60 broths proven positive by culture and/or PCR were retrospectively extracted using the NucliSens easyMAG protocol on 200 μl of the frozen aliquots. An equal number of negative samples were analysed by the same extraction and PCR protocol.
Results: The real-time PCR detected MRSA DNA in 60 enrichment broths as well after Qiagen as after easyMAG extraction. In 49, 48 and 48 of the extracted MRSA positive broths Icycler Ct values for detection of SCCmec, mecA and nuc genes respectively were lower after NucliSens easyMAG extraction compared to the values after Qiagen extraction with a mean Ct difference of 3.02, 1.79 and 2.04. If the Ct values after Qiagen extraction were lower than after NucliSens easyMAG extraction, which was the case in less than 10 of the extracted broths, the mean Ct difference was minimal: 1.2, 1.3 and 1.2 for the detection of SCCmec, mecA and nuc genes, respectively. For all 60 negative broths, both NucliSens easyMAG and Qiagen produced clear cut negative results. The time needed for the Nuclisens easyMAG extraction procedure was 40 minutes for 24 samples, with a hands-on time of less than 20 min, compared to 85 minutes for Qiagen extraction.
Conclusion: In this study the Nuclisens easyMAG extracted more efficiently the MRSA DNA of the enrichment broths by showing on average lower and higher Ct values for respectively MRSA positive and negative samples in the real-time PCR assays. The instrument features user-friendly, intuitive software, and delivers high throughput capabilities with 40 minutes turn-around-times.
P897 Expression of icaA and icaD genes by quantitative real-time PCR in correlation to biofilm synthesis among methicillin-resistant coagulase-negative staphylococci
A. Foka, V. Chini, E. Petinaki, G. Dimitracopoulos, I. Spiliopoulou (Patras, GR)
Objectives: Among pathogens causing hospital infections methicillin-resistant coagulase–negative staphylococci (MR-CNS) have become predominant, especially in neonatal intensive care units (nICU). This results their ability to form biofilm, especially among patients with invasive procedures. The purpose of the present study was to investigate slime production in correlation to the presence and expression of ica operon among MR-CNS isolated from different patients hospitalised in a nICU.
Methods: During a two-year period 132 (MR-CNS) were isolated from different inpatients at the nICU. Biofilm production was detected by the qualitative and quantitative methods. PCR was performed for the detection of the four genes of the ica operon, (icaA, icaD, icaB, icaC) and of the insertion sequence element IS256. Fourteen representative strains, 11 slime-positive and three slime-negative, were selected for reverse transcription quantitative relative Real time PCR (qRT-PCR) for the detection of the expression levels of icaA and icaD genes. Total RNA was isolated by the Trizol method, and a part of the 23S rRNA was used as the reference gene in the relative quantification of ica genes using the SYBR® Green I chemistry; results were calculated by the Pfaffl method.
Results: Both methods for bioflilm formation revealed that 117 (89%) isolates were biofilm-positive. PCR showed a variation of possession of ica genes. The selected 14 strains were positive for icaA and icaD by PCR screening. Seven out of the 11 slime-positive strains carried also IS256. Among them, two showed high, two medium and three strains low expression levels of both ica genes by qRT-PCR. In the remaining four slime-positive strains that did not carry IS256, two showed medium and two low expression levels of both ica genes. Two out of the three slime-negative MR-CNS possessed IS256 and all three had low expression levels of ica genes.
Conclusions: The majority of MR-CNS associated with infections in the nICU produced biofilm. The insertion sequence element IS256 does not always inactivate the expression of ica operon. Other factors are involved in biofilm formation.
P898 StaphPlex System for rapid and simultaneous identification, antibiotic resistance determination and Panton-Valentine leukocidin detection of staphylococci from positive blood cultures
Y.W. Tang, A. Kilic, Q. Yang, H.J. Li, R.S. Miller, M. McCormac, K.D. Tracy, C.W. Stratton, J. Han (Nashville, Huntsville, US)
Objectives: Current phenotypic methods take several days for identification and antimicrobial susceptibility testing of staphylococcal isolates after Gram-positive cocci in clusters (GPCC) are seen in positive blood cultures. We developed and validated a StaphPlex system that amplifies and detects 18 gene targets simultaneously in one reaction for identification, Panton-Valentine leukocidin (PVL) gene detection, and antimicrobial resistance determination of staphylococci.
Methods: We collected positive blood culture specimens in which GPCC were seen and compared the results of the StaphPlex System with staphylococcal identification and antimicrobial resistance determination obtained by phenotypic methods.
Results: Among a total of 360 GPCC specimens, 265 (73.6%), 41 (11.4%), 32 (8.9%), 11 (3.1%), and 11 (3.1%) were identified as coagulase-negative Staphylococci (CoNS), methicillin-resistant Staphylococcus aureus (MRSA), methicillin-susceptible S. aureus (MSSA), mixed infections of CoNS and MRSA, and non-staphylococci, respectively. The total agreement rate was 91.9% in comparison with those identified by conventional phenotypic methods. The 277 CoNS specimens were further speciated to 205 (74.0%) Staphylococcus epidermidis, 9 (3.3%) Staphylococcus haemolyticus, 25 (9.0%) Staphylococcus hominis, 1 (0.4%) Staphylococcus lugdunensis, and 37 (13.4%) other CoNS with an accordance rate of 84.6% when compared to an API STAPH identification. High specificity and low sensitivity were noticed when aacA, ermA, ermC, tetM and tetK genes were detected to predict in vitro antimicrobial susceptibilities. The StaphPlex system presented a sensitivity of 100.0% and specificity ranged from 95.5% to 100.0% when used for staphylococcal cassette chromosome mec typing and PVL gene detection.
Conclusion: StaphPlex provides accurate and relevant staphylococcal identification, PVL gene detection, and antimicrobial resistance determination within five hours, which significantly shortens the time usually needed for phenotypic identification and antimicrobial susceptibility testing.
P899 Comparison of enterotoxin genes in Staphylococcus aureus from nasal carriage and bacteraemia
J.Y. Baek, K.S. Ko, K.R. Peck, J.H. Song (Seoul, KR)
Background and Objective: The prevalence of enterotoxin genes within S. aureus isolated in Korea including both in community-setting and in hospital-setting has not been reported.
Material and Methods: We analyzed 95 S. aureus colonisers (18 MRSA and 77 MSSA) isolated from children attending outpatient clinics and 70 S. aureus isolates (36 MRSA and 34 MSSA) from patients with bacteraemia. spa typing, multilocus sequence typing, and SCCmec typing were performed to characterise S. aureus isolates. Enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sek, and tst) and pvl gene were assayed by method of PCR.
Results: The most common enterotoxin genes were seg, sei, and tst in both nasal carriage strains and S. aureus from bacteraemia (60.0% and 58.6% for seg, 69.5% and 55.7% for sei, and 52.6% and 42.9% for tst, respectively). Possession rate of enterotoxin genes was not significantly different between isolates from nasal carriage and bacteraemia, except for sec and seh genes. While only two isolates of nasal carriage (2.1%) were carrying sec gene, 34.3% of isolates from bacteraemia were positive. Interestingly, sec gene was found only in MRSA isolates. For seh gene, it was positive in 30.5% of nasal carriage strains, but only 8.6% of bacteraemia strains possessed it. No isolates contain pvl gene. While the most prevalent clone was ST30 (spa motif, WG/FKAOMQ) (34/95 isolates, 35.8%) in nasal carriage, ST5 (spa motif, DMGMK) was the most frequently found clone in bacteraemia strains (24/70 isolates, 34.3%). Bacteraemia strains belonging to the same clone have the similar enterotoxin gene pattern, but it did not in nasal carriage strains.
Conclusion: S. aureus isolates from bacteraemia and nasal carriage showed different genetic backgrounds, that is, no close relation each other. There was no evidence that S. aureus isolates from bacteraemia possessed more enterotoxins genes than nasal carriage.
P900 Performance of the GeneXpert® system for detection of methicillin-resistant Staphylococcus aureus including the non-typeable clone from animal origin
M.W.M. Wassenberg, S. Nijssen, X.W. Huijsdens, A.M.C. Bergmans, C.M.J.E. Vandenbroucke-Grauls, M.J.M. Bonten, J.A.J.W. Kluytmans (Utrecht, Breda, Bilthoven, Amsterdam, NL)
Objectives: The rapid and accurate identification of methicillin-resistant Staphylococcus aureus (MRSA) is of great importance to control the spread of MRSA. Since 2003 a MRSA clone from animal origin is observed with rapidly increasing prevalence in the Netherlands and this isolate is non-typable (NT) with SmaI PFGE. The recently developed GeneXpert® system allows molecular detection of MRSA with a simple procedure in 75 minutes.
The objective of this study was to assess the in vitro sensitivity of the Xpert MRSA Assay performed in the GeneXpert® system to detect MRSA including NT-MRSA isolates.
Methods: A collection consisting of 35 NT-MRSA and 73 typable MRSA strains isolated from humans in the period January 2003 to September 2005 was used. The typable strains were obtained from the same area and time period as the NT-MRSA. All strains were coded during testing. The GeneXpert® was performed on fresh cultures of the isolates and according to the manufacturer's protocol.
Results: 108 MRSA isolates were tested. In 4 cases there was an invalid result or technical error. Of the remaining 104 isolates, 99 (95%) were detected with the GeneXpert®. The sensitivity among NT-MRSA isolates was 31 of 35 (89%) and among the other strains 68/69 (99%). Four of the 5 false negative MRSA isolates had SCCmec type III. Of the 76 strains that were SCCmec typed, 14 had SCCmec type III (18%).
Conclusion: This study demonstrates that the GeneXpert® is a rapid and reliable system for detecting MRSA in vitro. The assay is also able to detect NT-MRSA. Clinical studies are required to determine the reliability of the GeneXpert® to detect MRSA from clinical samples. The majority of the strains that were not detected were of SCCmec type III. This should be studied in more detail.
P901 A new automated magnetic bead based nucleic acid sample preparation directly from nasal swabs for MRSA diagnostics
T.E. Kolpus, H.K. Høidal Berthelsen, M. Lorentzen, U.H. Refseth (Oslo, NO)
Objectives: The term MRSA (methicillin resistant Staphylococcus aureus) is used to describe Staphylococcus aureus that is resistant to commonly used antibiotics. The bacterium is known to mainly colonise the nostrils, but the respiratory tract, open wounds, catheters and urinary tract are also potential sites of infection. It is believed that as many as 53 million people carry MRSA. The traditional method of testing for MRSA is both cumbersome and time consuming, and can take several days before a result can be reported.
We have developed a unique principle for bacterial DNA extraction directly from nasal swabs. Collection and transport systems from alternative manufacturers have been successfully tested, such as rayon and nylon swabs in Liquid Stuart or Amies (liquid and gel) Transport Medium. The dimensions of many types of transport tubes limits full automation because they are unsuitable for insertion into the automation instruments, which necessitates manual transfer of sample into sample preparation tubes. In this study we present results of a fully automated BUGS' n BEADS system for bacterial DNA isolation, using swab and transport devices manufactured by Copan, Brescia, Italy.
Methods: The specimen is collected from the patient using a flocked nylon swab that is submerged into a polypropylene screw-cap tube containing 1 ml of Liquid Amies Transport Medium following transport to the test laboratory, the tube is vortexed, the swab removed and the tube is placed straight into the automation instrument to serve as the primary tube for automated transfer of sample into the sample preparation tube. Microbial contents remaining in the primary tube after withdrawal of sample for BUGS' n BEADS may be used for cultivation.
Results: Automated BUGS' n BEADS extraction of MRSA DNA directly from swab was achieved using automation instrument compatible polypropylene screw-cap tubes containing Liquid Amies Transport Medium, in combination with flocked nylon swabs (Copan, Brescia, Italy). The obtained sensitivity was 150 cfu MRSA per swab.
Conclusions: MRSA is detected directly from swab without prior cultivation, at a sensitivity of about 150 cfu per swab, using Copan swab and transport tubes in fully automated BUGS' n BEADS sample preparation.
P902 Development of a multiplex real-time PCR for detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine leukocidin directly from clinical samples
L. Renwick, A. Hardie, E.K. Girvan, A.P. Gibb, D. Morrison, J. Dave, K.E. Templeton (Edinburgh, Glasgow, UK)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen responsible for significant numbers of healthcare associated infections. Isolates containing Panton-Valentine leukocidin (PVL) that cause severe skin infections are emerging as a problem in the community. Rapid, PCR-based detection of MRSA in screening and wound swabs would be an invaluable tool in a diagnostic laboratory. The aim of this study is to develop a novel multiplex real-time PCR assay for detection of MRSA and PVL directly from clinical samples.
Methods: Individual assays for MRSA (SCCmec) and PVL (LukF and LukS) were optimised and combined and the new multiplex assay evaluated with screening and wound swabs. Swabs (n = 240) from all S. aureus positive patients taken during May and June 2006 were collected and tested by multiplex PCR. All PVL positives were confirmed by second PCR.
Results: MRSA and PVL positive isolates were detected by the assay with an analytical sensitivity of 100 cells/reaction. No other bacterial species were amplified. Five of 240 swabs (2.1%) were positive for PVL. All five were independently confirmed by different PCR in the Scottish MRSA reference Laboratory. Three PVL positives were methicillin-sensitive S. aureus and two were MRSA. Each positive specimen was collected from a patient with a skin infection at an outpatient clinic or within 24 hours of admission to hospital.
Conclusion: This assay is a powerful, sensitive diagnostic tool giving rapid results and could allow timelier treatment and infection control decisions to be taken. It can also provide valuable epidemiological information.
P903 Methicillin-resistant Staphylococcus aureus strains from animal origin are recognized by IDI-MRSA PCR
R. Roosendaal, J.A.J.W. Kluytmans, J.H.C. Woudenberg, X. Huijsdens, C.M.J.E. Vandenbroucke-Grauls (Amsterdam, Bilthoven, NL)
Objectives: The Netherlands is one of few countries in the world that is able to keep the prevalence of MRSA low, grace to a very stringent infection control policy. Recently, however, a new problem has emerged: a high percentage of pig-farmers appears colonised with MRSA strains that are non-typable in pulse field electrophoresis (PFGE). These strains have also been detected in large numbers of pig-farms and pigs. Because these strains represent a new threat to our national policy, we determined whether these strains of animal origin can also be rapidly recognized by the PCR assay described by Huletsky et al [1], and marketed as IDI-MRSA Test by Becton Dickinson.
Methods: MRSA strains collected at the RIVM between January 2003 and September 2005 and defined as “non-typable” by routine PFGE were tested with the IDI-MRSA test. In addition, the presence of the mecA-gene was determined. SCCmec type and spa type were determined in a previous study at the RIVM.
Results: The IDI-MRSA identified correctly 29/33 strains (86.2%). Of these, 28 were of MLST sequence type (ST) 398, and one ST 752 (a single locus variant of ST 398). Furthermore, spa types t034 (5 strains), t108 (14 strains), t011(8 strains), t571 (2 strains) and SCCmec types V (21 strains), IVa (3 strains), and III (4 strains) were detected in the 29 correctly identified strains. In one strain the SCCmec type could not be determined.
The four misidentified strains were MLST ST398 (3 strains) and ST753 (1 strain). The spa types were t034 (1 strain), t567 (2 strains) and t898 (1 strain). The SSCmec types were all type III in these four strains, hence half of the strains with this SSCmec (4/8) were not correctly identified by the IDI-MRSA test.
Conclusion: The IDI-MRSA performs well with MRSA strains of animal origin. We hypothesise that the strains that are not recognized harbour point mutations at critical sites in SCCmec. This is currently under investigation.
P904 Comparison of culture and real-time PCR for point prevalence surveillance of methicillin-resistant Staphylococcus aureus in a large public teaching hospital
D. Fuller, S. Wilson, R. Buckner, M. Commiskey, K. Prugh, C. Miramonti, G. Huffman, H. Cui, T. Davis (Indianapolis, US)
Objective: Controlling MRSA is a primary focus of most hospital infection control programmes. Currently, the standard surveillance method for detecting MRSA is culture, which is very laborious and time intensive. A rapid and more sensitive method to screen for MRSA will represent a definite advantage for infection control programmes. The objective of this pilot study is to evaluate the prevalence of MRSA colonisation in patients at Wishard Health Services (Indianapolis, IN) using conventional culture and real-time PCR (RT-PCR) technology.
Methods: We conducted a prevalence survey of all inpatients at Wishard excluding children and prisoners. A total of 259 subjects were tested to establish MRSA prevalence. Anterior nares specimens were collected from each subject using a double swab set. One swab from each set pair was streaked to conventional agar media for culture and RT-PCR testing was performed on the other swab from each swab set pair. Cultures positive for SA were then tested for oxacillin sensitivity. Discrepant results were resolved by retesting the culture swab using PCR technology and by retesting the RT-PCR swab with an alternative amplification assay (Analyte Specific Reagent). Percent agreement was calculated in which a true positive was defined as (a) culture positive or (b) any 2 positive PCR results.
Results: Of the 259 subjects tested, 183 (71%) were negative for MRSA with both conventional culture and RT-PCR while 57 were positive with both methods. Nineteen subjects had discrepant results (17 were RT-PCR positive/culture negative and 2 were RT-PCR negative/culture positive). All discrepant results were resolved after retesting and showed 100% agreement.
Conclusions: RT-PCR technology proved to be much more sensitive than conventional culture methods for detection of MRSA. This patient population had an overall MRSA prevalence rate of 29% (74/259). Using culture only instead of PCR technology would have missed 23% (17/74) of MRSA colonised patients which has profound implications for infection control.
P905 Low specificity of GenoType® MRSA Direct test when result interpretation is restricted to hybridisation of PCR products
S. Borgmann, A. Kick, M. Schuierer, A. Reber, H. Gruber, K. Beyser, R. Abel, A. Lindauer (Weiden, Bayreuth, DE)
Objectives: Identification of MRSA bearing patients might be accelerated by performing PCR from primary materials (swabs, etc.) circumventing bacterial culture. Recently, Robert-Koch-Institute recommended GenoType® MRSA Direct test (Hain, Nehren, Germany) to reveal MRSA colonisation. In our lab this test has been performed since January 2006 servicing more than 40 hospitals in Northern Bavaria with microbiological analyses. Here we present data covering all PCR examinations collected over a time period between April and October 2006.
Methods: MRSA PCR and hybridisation of PCR products against specific probes linked to nitrocellulose strips were performed according to the manufacturers' recommendations. In addition, all PCR products were examined by agarose gel electrophoresis. All results were documented and compared to those of bacterial cultures performed within the following 7 days.
Results: A total of 861 samples was examined by PCR out of which 457 samples were grown and characterised through microbiological routine within the next 7 days (Table 1 ).
Table 1.
Summary of MRSA direct PCR and bacterial culture resultsa
| Culture | Gel− Hyb− | Gel− Hyb+ | Gel+ Hyb+ |
|---|---|---|---|
| n.d. | 376 | 5 | 23 |
| + | 7 | 2 | 60 |
| − | 351 | 12 | 25 |
Gel: results of agarose gel electrophoresis. Hyb: results of hybridisation reaction; n.d., not done.
Among the cohort of PCR negative MRSA samples only 2% showed bacterial growth (column 2). Hybridisation positive MRSA without detectable PCR products on agarose gels (column 3) could be grown in bacterial cultures in only 14% of the samples, suggesting a high percentage of false positive PCR results. Even with a detectable PCR product on an agarose gel combined with a positive hybridisation signal (column 4) growth of MRSA was observed in only 71% of the samples. Since this result also indicates a low specificity these 25 cases were backtracked in more detail. Eventually there were only 5 cases left for a fair comparison of positive PCR results versus negative bacterial growth results suggesting specificity higher than 90%.
Conclusion: GenoType® MRSA Direct test is a sensitive tool for identifying MRSA bearing patients. However, interpretation of PCR results needs to be based on hybridisation combined with agarose gel electrophoresis.
P906 Evaluation of the IDI-MRSA assay for detection of methicillin-resistant Staphylococcus aureus from clinical specimens
K. Lucke, M. Hombach, G.E. Pfyffer (Lucerne, CH)
Persistent spread of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and other healthcare facilities requires suitable and efficient infection control strategies. Early detection of MRSA carriers and the rapid availability of diagnostic results are paramount for the implementation of efficient control measures. To determine the benefit of a recently developed molecular technique 691 high risk patients were screened for carriage of MRSA by real time PCR using the commercially available IDI-MRSA assay (GenOhm, San Diego, CA). Its efficacy as a screening test was assessed by analysing nasal, inguinal, rectal, and vaginal swabs as well as swabs from axilla, wound and throat. In addition, backup cultures for all specimens were done in parallel. Thirty-two of 691 specimens were both PCR and culture positive, while 633 specimens were negative with both methods. Sixteen specimens were PCR positive but culture negative, whereas 8 specimens were PCR negative but culture positive. For two specimens the PCR reaction was inhibited. Among the 16 specimens with a false susceptible PCR result 10 contained a methicillin-susceptible Staphylococcus aureus strain, but no MRSA. Overall sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the IDI-MRSA assay were 80.0%, 97.5%, 66.6%, and 98.8%, respectively.
In addition, our study demonstrates that the test is also applicable for specimens other than nasal swabs. With its excellent NPV the IDI-MRSA is a perfect screening test for a rapid exclusion of MRSA carriers. However, due to the incidence of false positive results we strongly recommend backup cultures. Taking into account the high cost for patient isolation and other infection control measures and in particular, the prevention of pathogen transmission within the hospital the expenses for the IDI-MRSA assay are more than paid off.
P907 Rapid detection of methicillin-resistant Staphylococcus aureus by real-time PCR from clinical specimens
B. Jovanic, B. Jobst, C. Schmidt, T. Miethke (Munich, DE)
To avoid spread of methicillin-resistant Staphylococcus aureus (MRSA) within the clinic rapid detection of the bacterium is desirable. Culture based methods require up to 5 days to identify MRSA from patient specimens. In order to replace this time consuming method a real time PCR was developed by our group to rapidly identify MRSA within 3 hours (Hagen et al. 2006, Int. J. Med. Microbiol. 295:77). The real time PCR assay targets the 3′ integration site of the staphylococcal cassette chromosome mec (SCCmec) at the orfX locus.
In the present study we improved our real time PCR to detect in addition to SCCmec types I–IV the type V cassette. We demonstrate here that 226 MRSA, 111 methicillin-sensible Staphylococcus aureus (MSSA), 50 methicillin-resistant coagulase-negative Staphylococci and 12 methicillin-sensible coagulase-negative Staphylococci were correctly identified. However, 3 MSSA were false positive and the reason for this observation remains currently unknown.
Furthermore, we evaluated our method with nasal and wound swabs from patients. In our ongoing clinical study we have analyzed 519 swabs by culture and PCR. 489 swabs were negative in both experimental procedures. All together 30 swabs contained MRSA. 15 swabs had positive results in PCR and culture method, 8 swabs were only in PCR positive and 7 only by culture. The swabs, which were only positive in PCR, were confirmed to be MRSA-positive in an independent PCR-assay detecting mecA and a Staphylococcus aureus-specific gene. To rule out the possibility that our SCCmec-PCR was unable to detect the 7 MRSA-strains that were only positive in culture, we repeated the PCR-assay in 5 out the 7 cases (in 2 cases the strains were unfortunately not available) from colony material. In all 5 cases the SCCmec-PCR was positive.
The earlier analysis (3 hours versus at least 5 days) brings an overall cost reduction in the clinic because hygiene measures can be employed quicker and the spread of MRSA thus prevented. Isolation measures of patients with former MRSA-carriage can be finished sooner, if the SCCmec-PCR assay is negative.
In conclusion the SCCmec-PCR is a reliable and time saving detection method for MRSA in the clinic and our studies show that this PCR assay equals the accuracy of culture.
P908 The ability of a rapid PCR-based method to identify surgical patients colonised with methicillin-resistant Staphylococcus aureus
K.J. Hardy, S. Gossain, C. McMurray, S. Shabir, I. Alobwede, A. Williams, N. Athamneh, P.M. Hawkey (Birmingham, UK)
Objectives: To determine the ability of the rapid IDI MRSA PCR based assay to identify surgical patients colonised with MRSA compared to conventional culture methods.
Methods: During a 9 month period all patients admitted to 4 surgical wards had nasal swabs taken for detection of MRSA on admission and every 4 days subsequently. Each swab was plated directly onto MRSA ID agar and then tested using the IDI MRSA assay. Additionally all swabs were placed in brain heart infusion broth incubated overnight and subcultured onto MRSA ID and Baird Parker agar for detection of MRSA and MSSA respectively.
Results: A total of 7545 nasal samples were examined from 3769 patients. Seven hundred and fifty six samples were positive by the IDI MRSA assay, of which MRSA was isolated on direct culture from 374 (49.5%). Of the 382 samples that were negative on direct culture, MRSA was isolated on broth enrichment from 138 samples, and MSSA from 42 samples. Of the 244 samples that were IDI MRSA assay positive but did not have MRSA isolated, 77 (31.6%) came from patients that had previous or subsequent samples from which MRSA was isolated. When individual patients are examined a total of 465 of the 3769 patients (12.3%) sampled had at least one sample that was positive by the IDI MRSA assay, of which 254 (54.6%) were positive by direct culture. An additional 76 patients had MRSA isolated from broth enrichment, but a total of 135 patients (29%) were only ever found to be positive by the IDI MRSA assay and did not have MRSA isolated using culture. Overall the IDI MRSA assay has a sensitivity of 97%, a specificity of 96.1%, NPV of 99.7% and a PPV of 71%.
Conclusion: The IDI MRSA assay provides a rapid and sensitive way of screening surgical patients for MRSA, with a very high negative predictive value. The increased number of patients identified as colonised with MRSA using the IDI MRSA assay compared to conventional culture, may be due to the increased sensitivity of PCR compared to culture. Rapid and sensitive identification of surgical patients colonised with MRSA aids in the prompt decolonisation and isolation of patients.
P909 Genome variability of methicillin-resistant Staphylococcus aureus as revealed by comparative genome hybridisation with a multi-strain microarray
G. Kuhn, T. Koessler, P. Francois, A. Huyghe, J. Schrenzel, P. Francioli, D.S. Blanc (Lausanne, Geneva, CH)
Objectives: Using comparative genome hybridisation (CGH), we firstly aimed to analyse core and accessory genome compartments in a diverse collection of Methicillin resistant S. aureus (MRSA). Secondly, we aimed to know whether genetic elements have specifically been gained or lost during evolution of S. aureus Clonal Complexes (CC's).
Methods: Our collection comprised 21 MRSA isolates of 5 different CC's. It included 1 isolate of CC1, 6 isolates of CC5, 7 isolates of CC8, 1 isolate of CC22, 7 isolates of CC45 and 2 singletons (ST 80 and 15). CGH was performed on a multi-strain oligonucleotide microarray. It comprised probes for S. aureus strains N315, Mu50, COL, and MW2 covering 99% of all genomes with 2535, 2663, 2633, and 2625 genes respectively. Based on genome sequences of all 4 strains, 2019 genes are core. Oligonucleotide probes included common probes targeted at conserved gene regions (based on alignment of orthologs of all 4 strains) and specific probes targeted at variable gene regions. Common probes were designed to avoid absence of signals in CGH due to sequence variation. For analysis of S. aureus CC evolution we subtracted isolates of CC5, CC8, or CC45 from all other isolates.
Results and Discussion: CGH of DNA from 21 strains to common probes showed that the core genome comprised only 1851 genes. Interestingly, we noted that a considerable number of genes, which were located on mobile genetic elements, were present in all strains. In contrast to this, other genes, which were clearly not associated with mobile genetic elements, were variably lost.
A phylogenetic analysis based on presence and absence of accessory genes was performed and showed a weak congruence with CC groups and this was especially true for isolates of CC8. In line with this observation, we did not find genes which were specifically associated with either CC5, CC8, or CC45. This was in contradiction to previous studies using CGH.
Conclusions: By performing CGH analysis on 21 genetically diverse strains using a microarray which comprised a set of common probes we were able to clearly differentiate core and accessory genes. Our results show that accessory genes are not useful for phylogenetic analyses.
P910 Rule-out Staphylococcus aureus, including MRSA, directly from primary swab cultures using S. aureus PNA FISH
A. Voss, M. Wulf, M. Sørum, R. Skov, K.G. Madsen (Nijmegen, NL; Copenhagen, Vedbæk, DK)
Objectives: In July 2006, we performed a study among veterinarians attending the International Pig Veterinary Society Congress (IPVS) in Copenhagen. This study was set-up to screen the attendees for MRSA carriership and at the same time to validate S. aureus PNA FISH (AdvanDx Inc.) as a rapid rule-out test from primary swab cultures. S. aureus PNA FISH is a rapid molecular diagnostic test with a 2.5 hours turn-around time once culture swab culture reach the laboratory.
Methods: 264 swabs from the anterior nares and throat (combined culture) were collected from participants at the IPVS Congress. The swabs were transported to Statens Serum Institut for over-night enrichment in salt-enriched semi-selective nutrient broth. One ml of this primary swab culture was frozen and shipped to Nijmegen, The Netherlands for analysis by S. aureus PNA FISH. Subsequently, the broth was subcultured on blood agar and MRSA-ID agar plates (bioMérieux). Staphylococcus aureus was identified by colony morphology and tube coagulase test. Oxacillin-resistance was determined by cefoxitine-disc-diffusion according to CLSI criteria and confirmed by in-house mecA PCR.
Results: 34 of the 264 samples were MRSA-positive according to the above mentioned method. Using S. aureus PNA FISH, 112 of the samples were positive for S. aureus, including 32 of the 34 MRSA-positive samples. Two MRSA's were found S. aureus PNA FISH false negative. The 80 false positive S. aureus PNA FISH results were presumably caused by the presence of methicillin-susceptible S. aureus. The sensitivity, specificity, positive predicted value and negative predicted value (NPV) of S. aureus PNA FISH for MRSA were 94.1%, 65.2%, 28.5%, and 98.7%, respectively, such that after only 24 hours, 150 out of the 264 samples could be ruled out as being non-S. aureus and therefore also non-MRSA's.
Conclusion: The high NPV (98.7%) of S. aureus PNA FISH suggest that this test could be a rapid method to rule-out MRSA directly from primary surveillance swab cultures. Using this rapid test as a pre-screen, only the minority of samples that yield a positive S. aureus PNA FISH result would require continued isolation and testing using conventional screening and/or other MRSA specific rapid diagnostic tests.
Molecular detection of sexually transmitted infectious agents
P911 The Becton Dickinson ProbeTec ET system detects Chlamydia trachomatis and Neisseria gonorrhoeae from male “self collected” glans/urethral dry swab specimens thus advancing the potential for prevention and control through “home collection”
D.V. Ferrero, D.E. Schultz, N. Burgess, S.A. Willis, G.M. Ferrero (Stockton, Santa Clara, US)
Nucleic Acid Amplification Tests (NAATs) have proven to be useful tools in identifying cases of Chlamydia trachomatis (CT) and Neisseria gonorrhoeae (NG) throughout the world. Unfortunately, infection rates continue to increase. Increasing CT rates and ever-present NG infections in women, have given rise to consideration of male CT/NG screening programmes in order to address the male reservoir. Because, current efforts have not adequately controlled these diseases, Public Health Officials must consider other means. Home collection may be a viable mechanism for reaching certain populations that in the past have not been accessed. Effortlessly collected, non-invasive, self-collected male specimens that are stable and easy to transport would enhance the success of male screening programmes. We designed a study to consider the effectiveness of male dry noninvasive self collected glans/urethral swab (SCS) specimens to detect CT and NG when compared to clinician collected swab (CCS) specimens and first catch urine (FCU) specimens. 284 male patients attending an STD clinic were tested for CT and NG using CCS, FCU and SCS. All patients enrolled signed an approved Investigational Review Board (IRB) consent. The three specimens collected per patient were processed according to the manufacturer's recommended procedure using the Becton Dickinson ProbeTec system. There were a total of 51 specimens detected positive for either CT or NG. The CT prevalence was 12.0%. The NG prevalence was 5.6%. The overall sensitivity of SCS was 91% with a specificity of 99%. Sensitivity and specificity were based on an infected patient status of any two or more results being considered true results. There was an overall SCS agreement of 97.5% with CCS specimens and 98.1% with FCU specimens. Male dry self-collected glans/urethral swab specimens are a viable specimen choice. Dry swab specimens are easy to collect, transport and test using the Becton Dickinson ProbeTec system. The BD ProbeTec system showed high sensitivity and specificity for detection of CT/NG and high agreement when compared to CCS or FCU specimens. Simpler methods of collection and transport that produce reliable test results may increase the potential for screening difficult to test populations. This study is particularly important as the data indicates there is potential for NAATs to adequately detect CT and NG in male SCS in many venues including one's own home.
P912 Molecular genetic-based diagnosis of sexually transmitted Mycoplasma and Ureaplasma using an ELISA-based Multiplex PCR system
R. Raßhofer, F. Schwarzmann, S. Hofstetter, F-W. Tiller (Munich, DE)
Mycoplasmatales are the causative agents of a number of non-chlamydial non-gonococcal infections in the urogenital tract. It was shown further that neonates become infected during delivery resulting in colonisation of the respiratory tract leading to pneumonia and meningitis. Diagnosis of Mycoplasmatales in clinical samples by culture is difficult and time-consuming. Thus, we have evaluated the molecular genetic-based hyplex STD ID system for the rapid and reliable detection of U. urealyticum/U. parvum, M. hominis, and M. genitalium.
Twenty-two vaginal, 3 cervical, 1 urethral and 14 glans swabs as well as 6 ejaculate and 46 urine samples were selected from 46 patients presenting with acute clinical symptoms of inflammation characteristic for chlamydial or mycoplasmal infections. DNA was extracted automatically using MagNA Pure technology and samples were analysed in parallel using the molecular genetic hyplex technology and culture.
Using the hyplex system, the pathogen-specific DNA was amplified by multiplex PCR and subsequently detected by hybridisation to immobilised Mycoplasma/Ureaplasma-specific probes in an ELISA-based format with colour-coded wells.
21.3% (10/46) of the samples were identified positive for Mycoplasma or Ureaplasma by bacterial culture and 32.6% (15/46) by the molecular genetic assay. In 2/46 samples bacterial growth was detected by culture, however, interpretation was not possible. The respective samples scored positive for both Mycoplasma and Ureaplasma by the molecular genetic assay. In 73.9% (34/46) of the samples culture was negative whereas the molecular genetic-approach did detect neither Mycoplasma nor Ureaplasma only in 67.4% (31/46) of the samples.
14/15 positive samples were vaginal or cervical swabs whereas 13/14 glans swabs and 6/6 ejaculate samples were negative. 12/15 patients' positive swabs were confirmed with corresponding positive urine samples. 3/15 positive swabs could not be confirmed in the urine whereas 2/11 negative glans swabs were re-tested positive.
In conclusion, the hyplex STD ID system proved to be a fast and reliable tool for the diagnosis of Mycoplasma or Ureaplasma. In comparison to the standard culture the sensitivity of the molecular genetic hyplex assay was 100%, specificity was 91.2%, NPV was 100%, and PPV was 80%. Nevertheless, urines should be handled with caution keeping in mind that large volumes may result in inappropriate dilution and probably false negative results.
P913 High throughput-screening for Chlamydia trachomatis in urogenital clinical samples using an ELISA-based PCR-system
F. Schwarzmann, S. Fesenmeir, R. Raßhofer, S. Hofstetter, F-W. Tiller (Munich, DE)
Screening of asymptomatic women has been shown to reduce significantly the incidence of severe ascending infections caused by Chlamydia trachomatis. The enzyme immunoassay is used most commonly for diagnosis but lacks the sensitivity required for screening. Thus, we tested the hyplex STD Chlamydia PCR-ELISA-System as a cost-effective and sensitive tool for high throughput applications.
217 urogenital swabs, 22 urine and 11 ejaculate samples were randomly selected and DNA was extracted automatically using MagNA Pure technology. The samples were analysed in parallel using the hyplex technology and the Cobas Amplicor system as a gold standard. Using the hyplex system, the target-DNA was amplified by conventional PCR and subsequently the Chlamydia-specific products were detected by hybridisation to immobilised probes in an ELISA-based format with colour-coded wells.
9.2% (20/217) of the urogenital swabs, 0% (0/22) of the urine samples, and 0% (0/11) of the ejaculate samples were identified positive using the hyplex system. There was an overall concordance of 100% with the gold standard. 88.9% (193/217) of the swabs, 86.4% (19/22) of the urine samples and 100% (11/11) of the ejaculate samples were identified negative both using the hyplex system and the Amplicor system. However, 1.9% (4/217) of the swabs and 13.6% (3/22) of the urine samples, which were identified negative by the Amplicor system, were excluded from the analysis by the hyplex system due to missing internal control (IC), which uses a bacterial house-keeping gene to detect inhibition as well as insufficient quality of the sample. In conclusion, testing urogenital swabs, urine and ejaculate samples the hyplex assay showed 100% concordance in clinical sensitivity compared to the gold standard. Referring to the negative results, there was a concordance of 97.9%, 86.4%, and 100% for swabs, urine and ejaculate samples, respectively. This was exclusively due to “invalid” IC results of the hyplex test, supposedly related to differences in the type of IC. Although there was not sufficient material available for retesting, we suppose that the observed differences were due to efficient detection of insufficient sample quality by the hyplex IC. In conjunction with an ELISA-processor the recently CE-certified hyplex ELISA-based PCR-assay is a cost-effective alternative to well established systems allowing analysis of up to 96 samples for C. trachomatis DNA within four hours of time.
P914 Sequencing of Treponema pallidum ssp. pertenue Samoa D genome
P. Matejkova, M. Strouhal, D. Šmajs, S.J. Norris, G.M. Weinstock (Brno, CZ; Houston, US)
Objectives: Treponema pallidum ssp. pertenue (TPE) causes endemic treponematosis yaws. Genome of this pathogen was shown to be 99% identical to Treponema pallidum ssp. pallidum (TPA), the causative agent of syphilis. Although syphilis and yaws have different clinical manifestation, the pathogens are microscopically and serologically indistinguishable. Genome scale approaches (microarray hybridisations and whole genome fingerprinting – WGF) showed that there are no gene deletions and multiplications and no large indels. Relatively subtle genetic differences responsible for different pathogenesis of syphilis and yaws thus require complete genome sequence of TPE.
Methods: Comparative genome sequencing (CGS) is a technique based on hybridisation of the test and the reference genomic DNA to the oligonucleotide chip and has two stages. In the first stage, heterologous regions of the test genome were mapped and in the second step, positions of single nucleotide polymorfisms (SNPs) in the test genome were revealed. Additional regions containing clustered SNPs and indels were subjected to dideoxyterminator (DDT) sequencing. Physical map of complete genome sequence obtained by combination of CGS and DDT of heterologous regions was confirmed by WGF.
Results: CGS together with DDT sequencing and WGF was employed to obtain complete genome sequence of TPE Samoa D strain. The unfinished complete genome sequence comprised 1 linear contig. CGS revealed 904 SNPs and 84 regions were suggested for DDT sequencing. DDT sequencing of these regions revealed more than additional 500 SNPs and 85 indels in the length range of 1 303 bp. WGF was used to verify the assembly of the genomic sequence and to conclude the exact number of repeats in 2 tandem repeat regions and in additional 4 regions comprising T. pallidum repeat (tpr) genes and hypothetical genes in their vicinity. In silico proteome analysis revealed that more than 300 proteins (out of 1039 proteins predicted in TPA Nichols strain) were affected by sequence changes.
Conclusion: The genome sequence of yaws treponeme was used to define intersubspecies difference between TPE and TPA.
This work was supported by the grants from the Internal Grant Agency of the Ministry of Health of the Czech Republic (NR/8967–4/2006) and from the Grant Agency of the Czech Republic (310/04/0021).
P915 Comparative genomics of closely related treponemal strains
M. Strouhal, P. Matejkova, D. Šmajs, E. Sodergren, S.J. Norris, G.M. Weinstock (Brno, CZ; Houston, US)
Objectives: Genus Treponema includes several pathogenic spirochetes: Treponema pallidum ssp. pallidum (TPA) is the causative agent of sexually transmitted syphilis, Treponema pallidum ssp. pertenue (TPE) causes endemic non-veneral treponemal infections and T. paraluiscuniculi (TPAR) is the etiologic agent of veneral syphilis in rabbits and does not infect humans. These treponemes cannot be continuously cultivated under in vitro conditions and are indistinguishable by morphological and serological methods. In this study, genomes of closely related treponemes were compared.
Methods: Whole genome fingerprinting (WGF) was used to compare four treponemal strains including Nichols (TPA), SS14 (TPA), Samoa D (TPE) and Cuniculi A (TPAR). The genomes of analysed strains were divided into 97 overlapping intervals covering the whole genome and each of these segments was amplified and digested by restriction enzymes BamH I, EcoR I and Hind III. The resulting restiction profiles were used for identification of heterologous regions in chromosomal DNA.
Results: When compared to the Nichols genome the prominent indels and sequence changes were preferentially localised in tpr genes (tprD – TP0131, tprE – TP0313, tprF – TP0316, tprG – TP0317, tprI – TP0620, tprJ – TP0621, tprK – TP0897 and tprL – TP1031) and in the hypothetical genes in the vicinity of tpr genes. Differences in number of tandem repetitions within the gene TP470 and genes TP0433–4 (hypothetical genes) were detected in all analysed strains. In the second case fusion of both genes (TP0433–4) resulted in one acidic repeat protein gene (arp gene). Altogether, 2 deletions and 2 insertions, 7 deletions and 3 insertions, 12 deletions and 7 insertions were found in the strains SS14, Samoa D and Cuniculi A, respectively.
Conclusions: In this study, WGF was shown to be essential approach for verification of the whole genome assembly based on comparative genome sequencing and dideoxyterminator sequencing data. During completion of the SS14 and Samoa D genome sequences, tandem repeat regions and regions comprising paralogous genes were selectively identified by WGF.
This work was supported by the grants from the Internal Grant Agency of the Ministry of Health of the Czech Republic (NR/8967–4/2006) and from the Grant Agency of the Czech Republic (310/04/0021).
P916 Treponema pallidum in the vitreous tap
M. Müller, I. Ewert, F. Hansmann, C. Tiemann, H. Hagedorn, J. Roider, H. Hoerauf, H. Laqua, W. Solbach (Lubeck, Herford, Kiel, DE)
Background: Syphilis of the eye still poses a clinical challenge due the chameleonic behaviour of the disease. As the serodiagnosis has significant limitations, the direct detection of Treponema pallidum (TP) in the vitreous of the eye represents a desirable diagnostic tool.
Methods: Real-time polymerase chain reaction (PCR) for the detection of TP was applied in specimens from diagnostic vitrectomies of 2 patients with acute chorioretinitis. Qualitative verification of TP by real-time PCR and melting point analysis according a modified protocol was ruled out. Patients underwent complete serological workup (TP antibodies, immunoblot, antilipoidal antibodies), ophthalmologic examination with fundus photographs, fluorescein angiography, antibiotic treatment and follow-up.
Results: In 2 cases of acute chorioretinitis of unknown origin, real-time PCR of vitreous specimens of both patients provided evidence of TP and was 100% specific. Initial diagnosis of presumed viral retinitis was ruled out by PCR of vitreous specimen. Patients were treated with systemic antibiotics and showed prompt improvement in visual function and resolution of fundus lesions.
Conclusions: In cases of acute chorioretinitis the use of PCR-based assays of vitreous specimens in the diagnostic evaluation of patients is advisable. With real-time PCR, detection of TP in the vitreous was possible and delivered a sensitive, quick and inexpensive answer in a disease rather difficult to assess. Although syphilitic chorioretinitis is a rare disease, PCR should include search for TP, as diagnostic dilemmas prolong definitive treatment in a sight threatening disease.
P917 A new fast automated magnetic bead based nucleic acid sample preparation for STI diagnostics combined with BD ProbeTec™ ET
T. Engen, M. Anglès d'Auriac, S. Harja, M. Tauvo-Kallio, K. Rantakokko-Jalava, U. Refseth (Oslo, NO; Turku, FI)
Objectives: Chlamydia trachomatis is a leading cause of sexually transmitted disease worldwide. Current NAAT methods on urine samples require substantial cumbersome manual handling, centrifugation and lead to inhibition with a significant number of samples, further requiring costly retesting. Automation of the sample preparation will reduce hands-on time, the risk of cross-contamination, increase reproducibility and sample throughput. Here, we present a new rapid automated magnetic sample preparation method for C. trachomatis from urine samples compared with the manual BD ProbeTec™ ET (Becton Dickinson) sample preparation, both combined with the SDA detection system of BD ProbeTec™ ET. The new rapid method uses the same Bacteria Binding Beads as the validated BUGS'n BEADS™ (BnB, Genpoint AS, Norway) on a customised Tecan MiniPrep 75 instrument. This new method called BUGS'n BEADS STI-fast, includes no active lysis, uses no hazardous or inhibiting reagents and is faster.
Methods: C. trachomatis was isolated in parallel from urine samples using BD ProbeTec™ ET sample preparation and BUGS'n BEADS STI –fast, both combined with the BD ProbeTec™ ET detection system. The amplification control (AC) in the BD ProbeTec™ ET kit was included to reveal any potential inhibition. Discrepant results were resolved by repeating both methods in duplicate.
Results: Based on the 442 specimens processed so far, it seems that sensitivity is improved using this new and faster sample preparation system obtaining 97.4% compared to 84.6% using the BD ProbeTec™ ET sample preparation. Meanwhile, specificity remained excellent of 100% for both methods. The average signal to noise ratio for positive samples is about 20000 MOTA units higher for BUGS'n BEADS STI –fast suggesting that the new method increases the end result robustness for BD ProbeTec™ ET. In addition, inhibition was observed for the BD ProbeTec™ ET sample preparation at the rate of about 1.5%, whereas no inhibition has been observed using the new method. The automated isolation of 48 samples including primary sample transfer to robot tubes and transfer of finished isolated sample to BD priming wells takes close to 1.5 hours.
Conclusions: This new robust and faster DNA preparation method avoids inhibition, increases throughput, reduces hands-on time and uses no hazardous reagents while obtaining comparable sensitivity with the standard BD protocol.
P918 Evaluation of the Abbott M2000 sp/rt real-time PCR on Chlamydia trachomatis and Neisseria gonorrhoeae using urine samples, and comparison with the Roche Cobas Amplicor Ctr/Ngo PCR assays
D. Luijt, J. Schirm (Groningen, NL)
Objectives: Chlamydia trachomatis (Ctr) and Neisseria gonorrhoeae (Ngo) are the most common micro organisms diagnosed in sexually transmitted diseases. Nowadays laboratory diagnosis of Ctr and Ngo is mainly based on molecular methods. The aim of this study was to evaluate the recently introduced ABBOTT M2000 sp/rt real-time Ctr/Ngo PCR assay in urine specimens, and to make a comparison with the ROCHE COBAS AMPLICOR (CA) Ctr/Ngo PCR test.
Methods and Materials: Dilutions of positive control stocks in urine and QCMD quality control panels were used for the analytical sensitivity testing. In addition 846 clinical urine samples were tested prospectively. Positive results of the CA Ngo assay were confirmed with a specific real-time in-house PCR using the opa gene.
Results: For both Ctr and Ngo the dilution series showed a higher sensitivity for the M2000 assay than for the CA assay. In the QCMD panels the M2000 assay had a 100% performance, whereas the CA assay missed the sample with the lowest copy number of both Ctr and Ngo. In the prospective study 5 (0.6%) samples were inhibited in the M2000 assay and 20 (2.4%) samples were inhibited in the CA assay. Of the remaining 821 urine samples 61 were positive and 749 negative for Ctr in both assays. Of the 11 discrepant samples 8 were positive with the Ctr M2000-positive/CA-negative, and 3 were Ctr CA-positive/M2000-negative. The 8 M2000-positive/CA-negative samples were all weak positive. Because of the higher analytical sensitivity of the M2000 assay these samples were presumable true Ctr positive samples. The 3 Ctr CA-positive/M2000-negative samples were all negative after retesting in the CA assay.
For Ngo 5 samples were positive and 799 samples negative in both assays. All 5 positive samples were also positive in the Ngo specific real-time confirmation assay. The other 17 discrepant samples were all positive in the CA assay but negative in the M2000 assay. All these 17 samples were negative in the real-time in-house PCR confirmation assay.
Conclusions: In conclusion, the M2000 assays for both Ctr and Ngo are sensitive and specific assays, with a very low inhibition rate. For Ngo the M2000 the assay is a more specific test than the Ngo CA assay. The ABBOTT M 2000sp/rt system is very suitable for routine diagnostic laboratories because of the automation, the high throughput, and the good specificity.
P919 Specimen pooling for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urogenital specimens
M. Altwegg, O. Süess, K. Jaton, G. Greub, K. Egli (Lucerne, Lausanne, CH)
Objectives: To test whether pooling of 3 clinical specimens in combination with automated DNA extraction and a home-brew duplex test for the detection of Chlamydia trachomatis (CT) and Neisseria gonorrhoeae (NG) performs similarly as a well established commercial system with individual specimens and thus might help reducing costs for analysis.
Methods: 231 clinical specimens (swabs in Roche transport medium, native urines) were analyzed for CT and NG using the CobasAmplicor system (Roche). Inhibited specimens were retested after DNA extraction and NG+ specimens were confirmed using an independent test. The same specimens were further analyzed individually (N = 99) and pooled (N = 231; 3 consecutive specimens per pool). This included DNA extraction using the easyMAG system (bioMérieux) and analysis with a home-brew duplex test for CT and NG (5′exonuclease format) on the LightCycler (LC; Roche). In case of a positive result for a given pool, the 3 specimens were analyzed individually using the same system. The volumes of clinical specimens analyzed per PCR on the Cobas were 50ul and 25ul for urines and swabs, respectively, 29ul for individual and 18ul for pooled specimens in our own assay. Inhibition of PCR was determined using the internal amplification control on the Cobas and an external control on the LC.
Results: Of the 231 specimens, 4 were CT+, 1 NG+ and 2 CT+/NG+. An additional 2 specimens were false positive for NG on the Cobas. All pools gave the expected result when tested by home-brew PCR. Analysis of individual specimens from positive pools correctly identified the positive specimens (which were all in separate pools). The 2 false positive NG specimens did not result in a positive pool signal. Inhibition occurred in 19/231 (8.2%) of the individual specimens tested on the Cobas, in 1/99 (1.0%) individual specimens and in 2/73 (2.7%) pools tested on the LC. No inhibition was detected when the members of inhibitory pools were individually extracted and tested on the LC.
Conclusions: Pooling of urogenital specimens is a promising strategy to reduce costs for CT/NG testing. Because of the small number of positive results the sensitivity of this procedure requires confirmation. Optimal pool sizes depend on the number of specimens to be analyzed and on the prevalence of the two organisms. Amplification controls are recommended for pools, however, this may not be necessary for individual specimens because for those the use of automated DNA extraction results in an inhibition rate of only 1%.
P920 Screening for Chlamydia trachomatis in women attending outpatient clinic in Lithuania
D. Ambrasiene, R. Mikalauskas (Kaunas, Vilnius, LT)
Chlamydia trachomatis (C. trachomatis) is associated with a spectrum of diseases. The most well-known is the sexually transmitted disease (STD) commonly referred to as “chlamydia”. This pathogen causes genital tract infection in both men and women, leading to sometimes serious consequences such as infertility, ectopic pregnancy and chronic pelvic pain. Infection with C. trachomatis is often asymptomatic, delaying its diagnosis and treatment. The present study was initiated to determine the prevalence of sexually transmitted pathogen C. trachomatis in Lithuanian females.
Objectives: The aim of this study was to determinate the incidence of C. trachomatis in asymptomatic women who attended the outpatient clinic, for their routine annual visit.
Material and Methods: The study was carried out in Biomedical Research Centre using highly sensitive nucleic acid amplification technique where this method was successfully introduced in 2001. A total of 827 women were studied for presence of C. trachomatis by PCR method. The vaginal and/or cervical swabs specimens have been collected from each patient. DNA was extracted from clinical samples by using rapid in-house procedures. The presence C. trachomatis was determined by using in house PCR with specific primers. Five age groups were studied: group A – under 20 years (n = 46), group B – 21–30 years (n = 415), group C – 31–40 years (n = 205), group D – 41–50 years (n = 126), group E – 51–60 years (n = 35).
Results: C. trachomatis infection was detected in 60 out of 833 samples (7.2%). The prevalence of C. trachomatis genital infection in the studied groups were: group A – 24.3% (46/9), group B – 8.6% (415/33), group C – 4.6% (205/9), group D – 6.8% (126/8), group E – 2.9% (35/1).
Conclusion: The C. trachomatis infection is common and largely asymptomatic. Therefore, early detection strategies of C. trachomatis with screening programmes among young women are important in prevention and control of target infection.
P921 Utility of molecular methods in the diagnosis of infections related to abnormal vaginal discharge
L. García-Agudo, M. Roman-Enry, J.R. León, I. Jesus de la Calle, M. Rodriguez-Iglesias (Cadiz, ES)
Objective: Abnormal vaginal discharge is among the most common causes women ask for medical advice, resulting in 5 to 10 million visits annually. The principal diseases associated with vaginitis, in order of frequency, are bacterial vaginosis (BV), vulvovaginal candidosis and trichomoniasis. We evaluated a DNA hybridisation test for simultaneous molecular detection of Gardnerella vaginalis, Candida spp. and Trichomonas vaginalis, as an alternative to conventional microbiological methods.
Methods: We analysed 273 vaginal samples from symptomatic women attending the outpatient clinic of Gynecology at our hospital. We obtained two swabs from each woman, one for mycological culture and the other for wet mount, Gram stain and molecular detection. Wet mounts were examined for the presence of motile trichomonads and Gram smears were evaluated according to Nugent criteria, considering intermediate flora as negative for BV. DNA hybridisation method (Affirm VPIII, Becton Dickinson) was performed as described by the manufacturer. The sensitivity cutoff indicates nucleic acid estimation equal or more than 2×105 CFU of G. vaginalis, 1×104 cells of Candida spp. and/or 5×103 of T. vaginalis.
Results: Of the 273 patients, 34 had Gram smear compatible with BV. Ten patients demonstrated intermediate Gram stain. G. vaginalis was detected by hybridisation method in 27 of 34 specimens positive for BV by Gram stain. Compared to the Gram stain, the DNA hybridisation test had a sensitivity of 79%, a specificity of 79%, a positive predictive value of 35% and a negative predictive value of 96%. Seventy-two women presented a positive mycological culture for Candida spp. Fifty-seven of them were positive by hybridisation. Compared to mycological culture, the molecular technique had a sensitivity of 79%, a specificity of 92%, a positive predictive value of 78% and a negative predictive value of 92%. There was only a positive wet mount for T. vaginalis which was confirmed by hybridisation.
Conclusion: Although we count with several laboratory methods to diagnose vaginitis, there is a need for a rapid and highly selective tool to help distinguish them and establish the appropriate treatment. Affirm VPIII test correlates well, resulting more objective than Gram stain and faster than mycological culture, although it is more expensive. Its high negative predictive value compared with any of the other diagnostic methods, permits its use as a screening test.
P922 Evaluation of the role of Chlamydia trachomatis in chronic prostatic infection
V. Ouzounova-Raykova, G. Mizgova, E. Ouzounova, I. Mitov (Sofia, BG)
Objectives: Urinary tract infection (UTI) is one of the most common reason for adults to refer to a medical specialist. Half of all men experience symptoms of prostatitis at some time in their life. The most frequently recognized causes of prostatitis are classical urinary tract pathogens rather than sexually transmitted pathogens. They are the reason for the so called acute or chronic bacterial prostatitis. According to the NIH consensus classification of prostatitis there is established category of chronic prostatitis/chronic pelvic pain syndrome (CPPS). Opinion among researchers is divided as to whether Chlamydia trachomatis, which cannot be cultured on conventional bacteriological culture media, is a causative organism of chronic prostatitis.
Our study was designed to establish the prevalence of C. trachomatis in men with chronic prostatitis and to evaluate the role of the bacterium as possible etiological agent of chronic prostatitis syndrome. From December 2005 to September 2006 a total of 236 men were enrolled in the study and evaluated for the syndrome of chronic prostatitis.
Methods: Diagnosis of chronic prostatitis was made using medical histories, physical examinations, scoring with Prostate Symptom Score Index (PSSI) and NIH Chronic Prostatitis Symptom Index (CPSI), pre-massage and post-massage test (PPMT), culture and microscopic examinations of prostatic fluids and urine. In all cases with suspicion of CPPS culture and PCR of urethral smears were performed.
Results: Based on laboratory findings 195 patients (82.6%) were diagnosed with chronic bacterial prostatitis caused by one or more of the following pathogens – Enterococus spp., S. aureus, E. coli, T. vaginalis, P. mirabilis. 41 men (17.4%) were examined for C. trachomatis infection and 5 (12.2%) of them resulted positive either on PCR and culture.
Conclusion: C. trachomatis is frequent cause for STDs in European countries. Diseases caused by C. trachomatis range from asymptomatic to those with severe sequelae. Especially in prostatitis the exact role is still under debate. The found prevalence of C. trachomatis in men with CPPS in this study resulted comparable or a little higher to that in health population. It is important to continue with the investigation including more patients for a longer period. In our opinion although the role of C. trachomatis in pathogenesis of prostatitis remains speculative, however, testing for infections is highly recommended.
P923 Detection of Ureaplasma urealyticum biovars in semen of infertile men by polymerase chain reaction
H. Zeighami, S. Najar Peerayeh, R. Salman Yazdi (Tehran, IR)
Introduction: Ureaplasma urealyticum can be produce disorders such as non gonococal urethritis, epididymitis, prostatitis, in men and immature neonatal birth, abortion and fallopian infection in female. Ureaplasma urealyticum can be effect in fertility rate in both sexes. Ureaplasma urealyticum have a two biovars and 14 serovars. Biovar 1 (Parvum biovar) has serotypes 1,3,6 and 14 and biovar 2 (Urealyticum) include serovars 2, 4, 5, 6, 7, 8, 9, 10, 11, 12 and 14. These bacteria can be present in healthy and patient people but it is important that specific biovar may be effect on diseases. Therefore in this study, we investigated frequency of Ureaplasma urealyticum in infertile and healthy men by PCR method, and then detect responsible biovars in semen of theirs.
Materials and Methods: In this study semen samples were obtained from 100 infertile and 100 healthy men with age between 22–55 years. After collection of samples, spermogram test were made. Samples DNA extracted by Cadieux method. For all extracted DNA, PCR by genus primer for Ureaplasma urealyticum (U4, U5) that based on Urease gene were done. For all positive PCR samples PCR by biovars specific primers (UMS263, UMA61) for urealyticum biovar and (UMA57, UMS222) for parvum biovar which based on 5′ end of MBA gene were done.
Results: From 100 infertile men in 12% and in healthy men in 3% U urealyticum was detected (p<0.05). From 12 positive samples in infertile men in 9% urealyticum biovar and 3% parvum biovar were seen. In 3% of healthy men in 2% parvum biovar and 1% urealyticum biovar were detected. Spermogram test and most semen parameters decrease in men with positive PCR test and in men with uealyticum biovar disorders such as varicocele, azoospermia, abortion in wife and etc were seen.
Conclusion: Results of this study showed that differences in frequency of this bacteria in two groups was significant (p < 0.05). Furthermore urealyticum biovar in dominant biovar in infertile men (such other studies) and may be urealyticum biovar have a high potency than parvum biovar for produce disease.
P924 Comparison of DiaSorin Liaison® and Architect® immunoassay systems for syphilis screening
L. Grassi, D. Campisi, M.R. Sarcinelli, I. Papagni, R. Frigato (Milan, IT)
Objectives: Syphilis, which is caused by the spirochete Treponema pallidum is a chronic bacterial infection that remains a public health concern worldwide. As the World Health Organization estimated, venereal syphilis is rapidly increasing in USA and in Europe. The laboratory diagnosis of syphilis is a crucial point in the epidemiological and diagnostic evaluation of the diseases.
Aim of the present study was to compare the performance of two automated immunoassay systems for Syphilis immunoassay in blood donors and random clinical specimens sent to the laboratory for Treponema pallidum screening.
Methods: Study group: 419 blood donors serum samples and 458 random specimens sent to the laboratory for Treponema pallidum screening. The following tests were used: TPPA (Fujirebio, Tokio, Japan), Syphilis TP Architect® (Abbott Diagnostics, Weisbaden, Germany), Treponema Screen LIAISON® (DiaSorin S.p.A, Saluggia, Italy) were used on all specimen from blood donors. Random clinical samples were tested with all the kits as above and with RPR (Alfa Wasserman). Samples with discordant results were tested by Western blot (Mikrogen).
Specimens were graded as reactive, non-reactive or equivocal in accordance with the manufacturer's interpretation criteria.
Results: Blood donors serum samples: All results from the 419 blood donors samples showed complete agreement between Syphilis TP Architect® and Treponema Screen LIAISON® on blood donors samples. Random clinical specimens: The agreement between Syphilis TP Architect® and Treponema Screen LIAISON® was 99.6%: two samples positive with Architect® were negative with Liaison. These 2 samples were further analyzed by all the methods and they were scored as negative.
Conclusions: The performance evaluation data demonstrate that both automated immunoassay LIAISON® Treponema Screen and Syphilis TP Architect® have equivalent performance.
Gastro-intestinal pathogens
P925 Evolution of antimicrobial susceptibility of Campylobacter jejuni strains isolated from hospitalised children in Athens, Greece
H. Papavasileiou, K. Papavasileiou, S. Chatzipanagiotou, A. Makri, I Andrianopoulou, A. Voyatzi (Athens, GR)
Objective: To investigate the current epidemiology and the antimicrobial susceptibility of Campylobacter jejuni strains, isolated from children with gastroenteritis.
Material and Methods: 170 strains were isolated from stools of children aged up to 14 years during a two year period (2004–2005). The specimens were plated on charcoal, blood free, selective medium (oxoid, UK) and incubated for 48 h at 42°C in a microaerophilic atmosphere (BBL). The identification was performed by typical growth, microscopic examination, hippurate hydrolysis, positive catalase and oxidase reactions. For the susceptibility of Campylobacter the diffusion disk method was used and the determination of MIC was performed by E-test, (AB, Biodisk, Solna Sweden) for the following antibiotics: erythromycin, amoxicillin + clavulanic acid, ampicillin, gentamicin, tetracycline, ciprofloxacin, clindamycin. As control, the ATCC33560 strain was used. Results were interpreted according to the criteria of the NCCLS for Enterobacteriaceae. The serotyping was performed, based on Penner's heat-stable (HS) serogroups, in the Greek reference laboratory.
Results: From the 170 strains of C. jejuni isolated from gastroenteritic cases, 30% showed resistance to ciprofloxacin (MIC ≥4 mg/l), 55% to tetracycline (MIC > 8 mg/l), 13% to clindamycin (MIC ≥ 8 mg/l), 4% to ampicillin (MIC > 16 mg/l), 6% to erythromycin (MIC ≥ 8 mg/l) and 4% to amoxicillin + clavulanic acid (MIC ≥ 16/8 mg/l). In gentamicin all strains were sensitive.
The MIC50 and MIC90 of the strains are shown in the table .
| Strain | MIC (mg/L) |
|
|---|---|---|
| MIC50 | MIC90 | |
| Erythromycin | 0.50 | 2 |
| Ciprofloxacin | 0.50 | >32 |
| Gentamicin | 0.50 | 0.50 |
| Clindamycin | 0.25 | 32 |
| Tetracycline | 16,256 | |
| Ampicillin | 2 | 4 |
| Co-amoxiclav | 0.50 | 2 |
Most of the strains resistant to ciprofloxacin were also to tetracycline. Only two strains found to be multiresistant. Two serotypes were more predominant: HS: 4, 13, 16, 43, 50 and HS: 1,44, while a significant number of strains was non-typeable (NT).
Conclusion: Resistance of C. jejuni to erythromycin, ampicillin, amoxicillin + clavulanic acid, still remains low, whereas to tetracycline and ciprofloxacin increases. As long as resistance of C. jejuni to quinolones increases, verification of antimicrobial susceptibility is suggested as a routine practice in laboratories.
P926 Prevalence of antimicrobial resistance among Polish Campylobacter jejuni and Campylobacter coli strains isolated from humans and chicken meat
E. Rozynek, K. Dzierzanowska-Fangrat, D. Korsak, S. Wardak, J. Szych, D. Dzierzanowska (Warsaw, PL)
Objectives: Campylobacter jejuni and Campylobacter coli have been recognized as a major cause of foodborne bacterial gastrointestinal infections in humans. Campylobacteriosis is often associated with handling raw poultry or eating undercooked poultry meat. Many studies have shown an increasing antibiotic resistance among poultry and human strains of Campylobacter.
The aim of this study was to compare the prevalence of antimicrobial resistance among C. jejuni and C. coli strains isolated from children with diarrhoea and from chicken meat in central Poland.
Methods: All isolates were collected during 2003–2005. Samples of chicken meat were obtained from selected supermarkets in Warsaw. Isolation of Campylobacter sp. from chicken meat was performed according to the ISO10272 guidelines. Campylobacter strains isolated from children with diarrhoea were identified according to the WHO procedure. Species identification was confirmed by the PCR method. Susceptibility to nalidixic acid, ciprofloxacin, tetracycline, erythromycin, gentamycin and ampicilin was determined by the E-test method.
Results: A total of 203 Campylobacter sp. strains were isolated. Resistance to tetracycline in all strains did not exceed 30%, but the resistance in chicken isolates increased from 0% in 2003 to 17.3% in 2005 (p<0.05). Tetracycline had lower activity against human C. jejuni strains comparing to chicken isolates. Ampicillin and ciprofloxacin showed low activity against both C. jejuni and C. coli strains, regardless of their origin. Resistance to ampicillin rose from 8% in 2003 to 35.4% in 2005 (p<0.05) in human isolates, and from 5.8% to 30.4% in chicken strains (p<0.05). High rates of ciprofloxacin resistance (>40%) were noted in both human and chicken isolates and they did not change significantly in the study period. Only one human C. jejuni strain was resistant to erythromycin and azithromycin. Resistance to gentamycin was detected in three C. jejuni strains isolated from children and in one chicken strain. Double resistance was detected twice as frequently in child isolates comparing to chicken strains (37% versus 16.7%, respectively).
Conclusion: Differences in resistance patterns between human and chicken strains may indicate that chickens are not the main source of Campylobacter infection in Polish children.
P927 Ten years of surveillance on antimicrobial resistance in non-typhoidal Salmonella from humans in a Spanish hospital: 1996–2005
C. Rodriguez-Avial, O. López, I. Rodriguez-Avial, J. Picazo (Madrid, ES)
Objectives: In the present work we studied the evolution of resistance and the serotypes of nontyphoidal Salmonella isolated during ten years in a hospital of Madrid.
Methods: From 1996 thought 2005 a total of 1647 nontyphoidal Salmonella strains were isolated from stools. The identification and susceptibility to 14 antibiotics were performed by a microdilution method (Wyder system). All strains were serotyped using the somatic and flagellar antigens following the Kauffman-White Scheme.
Results: Of the 1647 isolates, 25 different serotypes were identified. The most common were S. Enteritidis (76.5%), S. typhimurium (19.5%) and S. Hadar (2%). Overall, 43, 7% of the strains were susceptible to all antibiotics tested. The more relevant antimicrobial resistance rates were: nalidixic-acid (Na) 30, 5%, amoxicilin (A) 22%, chloranphenicol (CL) 9%, gentamycine (G), tobramycin (T) 4, 7%, and cotrimoxazol (SXT) 4.7%. Resistance to Na increased from 13% in 1996 to 38, 5% in 2005. No resistance to ciprofloxacin (CP) was detected, although decreased susceptibility (CMI > 0.12 mg/L) was observed in 89% of Na resistant strains Resistance to CL decreased from 20% in 1996 to 10.5% in 2005 and was associated with S. typhimurium. In 2004 cefoxatime (CTX), ceftazidime (TAZ) and cefoxitin (CX) resistance was observed in four S. Heidelberg strains, all expressed a β-lactamase of pI 8.9 with amplicons showing a high level of homology with CMY-2. The most frequent resistance association was A+ Na (5.5%). Six strains (6 S. typhimurium) were resistant to A+Na+SXT+CL and eleven strains (8 S. typhimurium) to A+SXT+G+CL.
Conclusions: S. enteritidis was the most frequent serotype isolated, which showed an increase from 63% in 1996 to 76, 5% in 2005. S. typhimurium was associated with multidrug resistance. Resistance to NA with decreased susceptibility to ciprofloxacin was associated with S. Enteritidis and S. Hadar serotypes. In 2004 four S. Heidelbeg strains were CMY-2 β-lactamase producers.
P928 Report of the susceptibility pattern and toxigenicity of Clostridium difficile strains isolated from patients in a tertiary hospital in Greece during four years
M. Orfanidou, H. Kafkoula-Alevizou, E. Vagiakou, H. Tzoumakidou, A. Strouza, H. Malamou-Lada (Athens, GR)
Objective: To report the susceptibility pattern and toxigenicity of Clostridium difficile (Cd) strains isolated from hospitalised patients suffering from Cd associated diarrhoea, in a tertiary hospital in Athens, Greece, during four years period (3/02–3/06).
Methods: During the study period 2035 stool samples were examined for C.d. using cycloserine cefoxitin fructose agar with 5% egg yolk (CCFA) and cycloserine cefoxitin blood agar (BD). The strains were identified by rapid ANA II (Remel, Lenexa) and latex test (Culturette, BD). Toxin A was detected from C.d. strains by an ELISA (Vidas, bioMérieux) and a chromatographic assay (ColorPac, BD). Both toxins A&B were detected by an EIA (Premier Toxins A&B, Meridian) and a chromatographic assay (Immunocard Toxins A&B, Meridian). Antibiotic susceptibility testing was performed by E-test (AB Biodisk, Solna).
Results: C.d. strains were isolated in 171/2035 (8.4%) stool specimens. Toxin A was detected in 133/171 (77.8%) strains, toxin B in 22/171 (12.9%) and A–B in 16/171 (9.3%) strains. The resistance rate of the isolated C.d. strains to penicillin (PEN) was 78.8% (MICs 0.016->32 mg/L), clindamycin (DA) 55.6% (MICs 0.094->256 mg/L), tetracycline (TE) 33.4% (MICs 0.023–96 mg/L), while no resistance was observed to metronidazole (MTZ) (MICs 0.016–0.38 mg/L), vancomycin (VA) (MICs 0.032–4 mg/L) and piperacillin/tazobactam (TZP) (MICs 0.75–16 mg/L), although one strain presented to VA a high level MIC of 4 mg/L. Especially for meropenem (MP) the resistance rate was 7.6% (MICs 0.016->32 mg/L), while 67 strains were tested also to ertapenem (ERT) and were found to be resistant 20% of them (MICs 0.094->32 mg/L). The MICs range of moxifloxacin (MOX) and erythromycin (ERY) for 80 C.d. strains was found to be 0.25->32 mg/L and 0.125->256 mg/L and their resistance rate was 31.2% and 46% respectively. Linezolid (LZ) was tested in 100 strains and all were found susceptible (MICs 0.5–4 mg/L). All strains that produced toxin B were found resistant to PEN, DA and ERY (MICs 1.5->32 mg/L, >256 mg/L, 12->256 mg/L respectively).
Conclusions:
-
–
The most common toxin detected remains toxin A but there is an up going presence of toxin B
-
–
There is a high resistance rate to PEN, DA, ERY, TE and MOX, while there is still no resistance to MTZ and VA
-
–
A new resistance in MP and ERT is being revealed with meropenem being more effective. No resistance was found to LZ.
P929 Primary and secondary resistance of Helicobacter pylori to metronidazole and azithromycin in the northern part of Croatia
Z. Marušic, V. Plecko, M. Katicic, L. Žele-Starcevic, A. Budimir, B. Bedenic, A. Presecki Stanko, Z. Bošnjak, S. Kalenic (Zagreb, HR)
Objectives: The aim of this study was to assess primary and secondary antibiotic resistance rates in Helicobacter pylori isolates over a period of three and a half years (2003–2005 and the first 6 months of 2006).
Methods: A total of 378 H. pylori from 347 patients were isolated from 1211 gastric biopsies taken from patients visiting CHC Zagreb and CHC Merkur in Zagreb, Croatia. Isolates from January 1st 2003 to July 1st 2006 were included in the study. From each patient 4 gastric biopsies were taken for histology and 2 were sent to the Department for Clinical and Molecular Microbiology CHC Zagreb for culture and determination of antibiotic activity against H. pylori by means of agar dilution. Susceptibility to clarithromycin, azithromycin, metronidazole, tetracycline and amoxicillin was determined. Primary resistance was assessed in H. pylori strains isolated before the first eradication therapy. Secondary resistance was assessed in H. pylori strains repeatedly isolated after treatment failure.
Results: Resistance of H. pylori to amoxicillin and tetracycline was not detected. Clarithromycin and azithromycin showed a common resistance pattern in all tested isolates. Among the 342 pre-therapy isolates, 124 (36.25%) were susceptible to all antibiotics tested. Primary resistance to macrolids was detected in 167 (48.83%) strains and 192 (56.14%) strains were resistant to metronidazole. The mean dual resistance rate (resistance to both macrolids and metronidazole) was 37.72% (129/342). Among the 36 isolates cultured after unsuccessful therapy, only 1 was susceptible to all antibiotics tested (2.78%), 2 (5.56%) were resistant only to macrolids and 5 (13.89%) were resistant only to metronidazole. The dual resistance rate was 77.78% (28/36). Among the macrolid resistant strains, therapy-induced resistance was determined in 30.0% (9/30), while the rest (21/30) was attributed to primary resistance. Therapy-induced resistance was determined in 21% (7/33) of the metronidazole resistant strains.
Conclusions: The resistance rates to metronidazole and macrolids and dual resistance rates are high among H. pylori in northern Croatia. There are increased rates of resistance to macrolids and metronidazole among post-therapy isolates which can mainly be attributed to pre-existing primary resistance and in part to therapy-induced resistance.
P930 Primary and secondary resistance to metronidazole and clarithromycin in Spanish Helicobacter pylori clinical isolates obtained from children
L. Cibrelus, T. Alarcon, P. Urruzuno, M. Martinez, A. Perez De Ayala, J. Diaz-Regañon, M. Lopez-Brea (Madrid, ES)
Objective: To determine the primary and secondary resistance to metronidazole and clarithromycin in Spanish Helicobacter pylori (HP) clinical isolates obtained from paediatric patients from January 2002 to June 2006.
Methods: Samples were collected from gastric biopsies of symptomatic paediatric patients and H. pylori cultured as previously described. Resistance was determined by E-test. Strains were considered resistant if MIC ≥ 2 mg/l for amoxicillin, ≥ 4 mg/l for tetracyclin, ≥ 8 mg/l for metronidazole and ≥ 1 mg/l for clarithromycin and intermediate if MIC = 0.5 mg/l for clarithromycin, Medical record were retrospectively reviewed to get histories. Statistical analysis was based on chi2 tests using Stata software. Significance was construed for p <0.05.
Results: 101 patients were included: 38 males and 63 females (gender ratio M/F = 0.6). They were aged 10 years in average (standard deviation 3.3, ranges 4–18 years). Thirty-five patients (34.7%) had history of treatment failure, and were therefore considered as secondary HP-infection, with no statistical difference in distribution of gender (0.5 M/F ratio for patients with history of treatment failure versus 0.7 without, p = 0.61) or class breaks of age (45.7% (n = 16) over 12 among patients with history of treatment failure versus 27.0% (n = 17) without, p = 0.06).
All strains were susceptible to amoxicillin and tetracycline, 35.7% (n = 35) were resistant to metronidazole, 54.6% (n = 54) to clarithromycin and 2.0% (n = 2) were intermediate to clarithromycin. Double resistance to metronidazole and clarithromycin rated to 17.2% (n = 17). Primary and secondary resistance rates to metronidazole and clarithromycin (including double resistance) are detailed in the table .
| Antibiotic | Primary resistancea, n (%) | Secondary resistancea, n (%) | RRb | p |
|---|---|---|---|---|
| Metronidazole | 21 (32.8%) | 14 (41.2%) | 1.3 | 0.41 |
| Clarithromycin | 32 (49.2%) | 24 (70.6%) | 1.4 | 0.04 |
| Double resistance | 10 (15.4%) | 9 (26.5%) | 1.7 | 0.18 |
Intermediate strains to clarithromycin were considered as resistant.
RR of resistance in case of treatment failure.
Conclusion: Resistance to clarithromycin (56.6%) was higher than to metronidzole (35.7%) in the HP strains studied herein. Clarithromycin resistance was very high even in strains from paediatric patients not previously treated for HP infection (49.2%).
Acknowledgement: The study was supported by FIS PI 052452.
P931 Susceptibility of micro-organisms causing bacterial gastroenteritis in a Spanish hospital
M. Quiles, S. Garcia-Bujalance, J. Mingorance, R. Gómez, A. Gutierrez (Madrid, ES)
Objective: To study the antimicrobial resistance of Salmonella sp., Shigella sp., Aeromonas sp., Yersinia sp., and Campylobacter sp., isolated from clinical samples.
Material and Methods: 5504 strains isolated from stool samples between January 2000 to December 2005 were studied. The isolates were cultured on the selective media routinely used for the study of enteric pathogens. Identification was made according to standard microbiological procedures. Antimicrobial susceptibility was performed by the broth microdilution method using an automated system (Wider®). The antimicrobials studied were amoxicillin (Amx), amoxicillin-clavulanate (A/C), cotrimoxazole (Cotr), ciprofloxacin (Cipr) and clarithromycin (Clar) (for Campylobacter sp. only)
Results: The 5504 strains studied accounted for 15.86% of the total of stool samples analyzed. The isolates were as follows: Campylobacter sp (2415 strains), Salmonella sp. (2309 strains), Aeromonas sp. (191 strains), Yersinia enterocolitica 0.3 (89 strains) and Shigella sp. (50 strains). The Campylobacter sp. resistance rate were 86.6% to Cipr, 1.98% to Clar and 0.2% to Amx. The Salmonella enterica ssp. enterica resistance rate were 44.16% to Amx, 9.12% to Cotr, 0.72% to A/C and 0.63% to Cipr. The Aeromonas sp. resistance rate were 86% to Amx, 14.6% to Cotr, 7.3% to A/C and 0% to Cipr. Aeromonas hydrophila and Aeromonas veronii var. sobria were the most resistant species. The Yerisinia entercolitica 0.3 resistance rate were 100% to Amx, 13.5% to Cotr, 1.12% to A/C and 0% to Cipr. The Shigella sp. resistance rate were 74% to Cotr, 26% to Amx, 2% to A/C and 0% to Cipr.
Conclusion: Our results show that all the isolates of Aeromonas, Yersinia and Shigella and most of Salmonella (except two strains) were susceptible to ciprofloxacin confirming that it is the empirical first-line treatment of bacterial gastroenteritis. Campylobacter sp. keeps susceptibility to clarithromycin but there was a trend to increasing levels of resistance to ciprofloxacin, reaching a 92.43% in 2005.
P932 Evolution of susceptibility of non-typhi Salmonella in Spanish hospital: six years of surveillance
M.I. Quiles, S. Garcia-Bujalance, J. Mingorance, R. Gomez-Gil, A. Gutierrez (Madrid, ES)
Objective: To study the evolution of the antimicrobial resistance of non-typhi Salmonella in a Spanish Hospital during the last five years (2000–2005).
Materials and Methods: We studied 2309 Salmonella enterica ssp. enterica (designated ssp. I) isolated from clinical samples. All strains were isolated in the culture media routinely used for the isolation of enteric pathogens. Identification was made according to standard microbiological procedures and was confirmed by serotyping with commercial antisera (Difco Laboratories). Antimicrobial susceptibility was performed by the broth microdilution method using an automated system (Wider®). The antimicrobials studied were amoxicillin (Amx), amoxicillin-clavulanate (A/C), cotrimoxazole (Cotr) and ciprofloxacin (Cipr). The majority of the isolates (90%) belonged to serotypes 0:4 (B) and 0:9 (D1), and therefore only these have been included in this study.
Results: See Table 1, Table 2 .
Table 1.
Evolution of number of isolates by serotype
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
|---|---|---|---|---|---|---|
| 0:4B | 77 | 75 | 78 | 61 | 80 | 79 |
| 0:9D | 236 | 311 | 323 | 274 | 295 | 151 |
Table 2.
Evolution of resistance (%) to different antimicrobial agents by serotype
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | ||
|---|---|---|---|---|---|---|---|
| AMX | 0:4B | 80.52 | 68 | 85.9 | 75.41 | 71.25 | 51.9 |
| 0:9D | 17.37 | 8.7 | 14 | 17.15 | 8.13 | 4 | |
| A/C | 0:4B | 1.3 | 0 | 2.56 | 3.3 | 0 | 0 |
| 0:9D | 0.85 | 0 | 0.31 | 0 | 0.33 | 0 | |
| COTR | 0:4B | 23.37 | 13.33 | 19.23 | 27.9 | 10 | 12.65 |
| 0:9D | 0.42 | 0.64 | 0.31 | 0.36 | 0.7 | 1.32 | |
| CIPRO | 0:4B | 0 | 0 | 0 | 0 | 1.25 | 1.26 |
| 0:9D | 0 | 0 | 0 | 0 | 0 | 0 |
Conclusions: 1) There is a strong relationship between serotype and antimicrobial resistance, being serotype 0:4(B) more resistant than 0:9 (D1). 2) In spite of the low resistance rate of serotype 0:4(B) to A/C there were high level (range 17–31%) of moderately resistant (intermediate) strains. 3) Resistance to Cotr was low in serotype 0:9(D1) and moderate (range 13% to 23%) in serotype 0:4(B). 4) In 2004 and 2005 appeared two strains of serotype 0:4(B) resistant to Cipr, probably in relation with a high frequency of using these antibiotics as first choice drugs in the treatment of acute diarrhoea in adults.
P933 Emergence of resistance to new antimicrobials detected among Shigella isolates in Finland
K. Haukka, A. Siitonen (Helsinki, FI)
Objectives: In Finland most cases of shigellosis are related to traveling abroad. Shigella strains have rapidly evolved resistance to the commonly used antimicrobials. We studied the geographical resistance patterns and emerging new resistances to antimicrobials among Shigella strains isolated from patients in Finland between 1990 and 2005.
Methods: Antimicrobial drug resistance of 1834 Shigella strains isolated from Finnish patients during 1990–2005 was studied by diffusion method using discs for 12 antimicrobial agents. Since 2000, E-test for ciprofloxacin MIC determination has been done to the nalidixic acid resistant isolates (n = 65).
Results: The proportion of strains resistant to the studied antimicrobials increased during the study period. However, the frequencies of resistance differed between S. sonnei and the other Shigella serogroups. The proportion of multiresistant strains (resistant to at least 4 antimicrobials) was highest among the strains imported from China and India. Resistance to nalidixic acid has become common (87% in 2005) among the strains from the Far-East. In general, only one strain resistant to nalidixic acid per year has been obtained from the other geographical areas. In 2000–2001, susceptibility to ciprofloxacin had decreased (MIC ≥0.125 mg/L) in 44% and in 2004–2005 even in 83% of the nalidixic acid resistant strains. First three isolates, fully resistant to ciprofloxasin by the disc diffusion method were obtained during 2004–2005. By mid-November 2006, three more ciprofloxasin resistant isolates have been detected. All these six isolates belonged to the S. flexneri 2a serotype (MICs 3–32 mg/L), and most of them originated from India. Also six gentamicin, two cefotaxime and several mecillinam resistant strains have been encountered during the recent years.
Conclusion: The proportion of multiresistant Shigella strains is growing and, moreover, resistance to several new antimicrobials is emerging. The antimicrobial resistance patterns detected among the isolates from Finnish travelers reflect the resistance situation and usage of antimicrobials in the countries the infections were obtained from. Fluorokinolones, especially ciprofloxasin, are the drug of choice when treatment is needed. Unfortunately, Shigella strains seem to be rapidly evolving resistance to this group of antimicrobials as well. South and East Asia seem to be the areas where the new resistances emerge.
P934 Characterisation of multi-resistant Helicobacter pylori emerging in the UK
S.A. Chisholm, R.J. Owen (London, UK)
Objectives: To identify and characterise multi drug resistant (MDR) Helicobacter pylori in the UK. Only four antibiotics, metronidazole (MTZ), clarithromycin (CLA), tetracycline (TET) and amoxicillin (AMX) are recommended for H. pylori eradication. MTZ and CLA resistance rates account for 33% and 11% of H. pylori recovered pretreatment in the UK, with significantly higher rates observed post-treatment. Resistance to these agents substantially reduces eradication efficacy. In contrast TET and AMX resistance is rarely observed and MDR (resistant to three or more drugs) H. pylori have not been reported in the UK.
Methods: Antibiotic susceptibility data for all isolates referred to our reference unit from 2000 to date were examined for multi-resistance. As fluoroquinolones and rifamycins are used to treat refractive infection, additional susceptibilities to ciprofloxacin (CIP) and rifampicin (RIF) were tested. Antibiotic susceptibilities were re-tested for eight single colony picks (scps) for each MDR isolate. All scps were screened for mutations associated with CLA, TET and CIP resistance by real-time PCR or sequencing.
Results: Since 2004, eight isolates of H. pylori with atypical MDR profiles were identified. All had originated from patients in Southeast England, of whom seven had failed prior eradication therapy. The incidence of MDR strains increased annually, with four examples observed in 2006. Seven resistance profiles were identified: MTZ/CLA/TET/AMX (n = 2), MTZ/CLA/AMX (n = 1), MTZ/CLA/TET (n = 1), CLA/TET/AMX/CIP (n = 1), MTZ/CLA/TET/CIP (n = 1), MTZ/CLA/AMX/CIP (n = 1), MTZ/CLA/TET/AMX/CIP (n = 1). Re-testing scps for each isolate confirmed the MDR profiles in five cases. Repeat testing failed to confirm CIP resistance in two cases and AMX resistance (known to be unstable) in one case. All CLA, TET and CIP resistant isolates contained mutations in the 23S rRNA, 16S rRNA and gyrA genes, respectively.
Conclusions: Multi-resistance in H. pylori is extremely rare in Europe and has not been documented previously in the UK. We suggest that this may be an emerging problem. Further characterisation of scps confirmed that most were genuine MDR strains and not examples of mixed infections. While the clinical impact of these rare MDR strains is unknown, this study highlights the importance of continued surveillance of antibiotic resistance in H. pylori.
Bloodstream infections
P935 Time-to-positivity of aerobic blood cultures by using BacTec 9120 blood culture system
St. Fokas, Sp. Fokas, G. Altouvas, M. Tsironi, S. Kaptanis, M. Kalkani, M. Dionysopoulou (Eghio, Sparta, GR)
Objectives: The aim of this study was to determine the time to detection of positive aerobic blood cultures with the BACTEC 9120 system.
Methods: We retrospectively analyzed the data of 3250 aerobic blood cultures incubated for a period of 7 days in the BACTEC 9120 system from June 2004 to June 2006. Blood samples were drawn from peripheral veins and were inoculated into two aerobic/F bottles for each blood culture. Whenever there was a sign of positive culture the detection time, measured in hours, was documented for each bottle. Negative blood cultures from patients with clinical evidence of brucellosis were subcultured blindly.
Results: A total of 247 (7.6%) positive cultures were detected and 62 (1.9%) of them were clinically insignificant. Gram-negative organisms were recovered more frequently than Gram-positives in true bacteraemias (78%, 144/185 strains versus 22%, 41/185 strains) while CoNS were the most common contaminants. The mean detection time for all microbes (435 isolates) was 24.13 h (range 2.88 h to 122 h) and 72% (313/435) of them were detected within the first 24 h of incubation. The mean detection times for the Gram-negative and Gram-positive bacteria were 31.08 h and 21.9 h respectively (E. coli 9.3 h, Proteus spp. 9.2 h, Klebsiella spp. 9.2 h, P. aeruginosa 14.2 h, Brucella melitensis 65.39 h, S. aureus 15.5 h, CoNS 23.6 h, S. pneumoniae 10 h, Enterococcus spp. 19.8 h, Streptococcus spp. 36.6 h, Candida spp. 15.6 h).
Conclusions: The recovery time of Enterobacteriaceae (9.2 h) was significantly shorter of Gram-positive bacteria and 85% of Enterobacteriaceae were isolated within the first 24 h. All pathogenic microbes, including Brucella spp. were recovered within 5 days of incubation and the detection times of clinically significant isolates were shorter than contaminants. A protocol of 5-days incubation period is possible to apply with the BACTEC 9120.
P936 Is there any relation between time to positivity in blood cultures and source of infection in patients with Enterococcus spp. bacteraemia?
J. Alava, C. Ezpeleta, G. Ezpeleta, V. Cabezas, I. Atutxa, C. Busto, E. Gomez, J. Unzaga, R. Cisterna (Bilbao, ES)
Objective: The aim of this study is to know if, like in other bacteraemias, the time between blood culture incubation and growth detection measured by the time to positivity in a continuously monitored system correlates with the source of infection and the outcome of the patient who suffers and enterococcal bacteraemia.
Methods: We performed a retrospective, observational study involving adult inpatients that had Enterococcus spp. bacteraemia between 1 October 2003 and 30 September 2006 at a University hospital. Measurements included time to positivity in initial blood culture series, duration of bacteraemia episode, gender, age, rate of metastatic infection, and outcome.
Results: A total of 38 Enterococcus spp. bacteraemias (>1 positive blood culture result) were reported for patients with ages between 1 day–94 years (median age, 69 years); 5 (13.15%) bacteraemias were associated with endocarditis. The microbiological documentation of the source of infection was achieved only in half of the cases. The mortality rate was 21.8%.
The duration of bacteraemia was 1–47 days (median duration, 8 days; average duration 11.65 days). The time to positivity ranged from 40 minutes to 1 day (median time to positivity, 8.45 h). There was significantly shorter for patients with an endocarditis or catheter related infection, compared with the other sources of bacteraemia (p = 0.05) but no statistical difference was observed when both endovascular sources of infections were compared. Analysis using logistic regression found that a short time to positivity was an independent predictor of an endovascular source of infection but not the outcome of the patient. In fact, all the deaths recorded in this study were non-infection related.
Conclusions: Time to positivity in Enterococcus spp. bacteraemia may provide useful diagnostic information of the source of infection but not prognostic information. Meanwhile due to the reduced number of cases further studies are needed.
P937 Time to positivity in blood cultures of patients with Staphylococcus aureus bacteraemia: possible correlation with the source of infection
C. Ezpeleta, J.A. Alava, G. Ezpeleta, V. Cabezas, I. Atutxa, C. Busto, E. Gomez, J. Unzaga, R. Cisterna (Bilbao, ES)
Objective: The aim of this study is to asses if the time between blood culture incubation and growth detection in Staphylococcus aureus bacteraemia measured by the time to positivity in a continuously monitored system correlates with the source of infection and the outcome of the patient.
Methods: We performed a retrospective, observational study involving adult inpatients who had S. aureus bacteraemia between 1 October 2002 and 30 September 2006 at a University hospital. Measurements included time to positivity in initial blood culture series, duration of bacteraemia episode, gender, age, rate of metastatic infection, and outcome.
Results: A total of 211 S. aureus bacteraemias (>1 positive blood culture result) were reported for patients with ages between 7–94 years (median age, 69 years); 21 (9.95%) bacteraemias were associated with endocarditis. The microbiological documentation of the source of bacteraemia was achieved only in half of the cases. The mortality rate was 21.8%.
The duration of bacteraemia was 1–60 days (median duration, 8 days; average duration 9.8 days). The time to positivity ranged from 40 minutes to 3 days (median time to positivity, 11.19 h) and was significantly shorter for patients with an endovascular source of infection (endocarditis or catheter related infection), compared with the other sources of bacteraemia (p = 0.001). Analysis of the data using logistic regression revealed that a time to positivity shorter than 10 hours, was an independent predictor of an endovascular source of infection and outcome of the patient.
Conclusions: Time to positivity in S. aureus bacteraemia may provide useful diagnostic and prognostic information. Growth of S. aureus within 10 h after the initiation of incubation may identify patients with a high risk of fatal infection. Meanwhile due to the reduced number of cases further studies are needed.
P938 Time-to-report blood culture Gram stain and tentative susceptibility result
G. Just-Nübling, S. Franck, M. Hahn, V. Rickerts, P.M. Shah (Frankfurt am Main, DE)
Objective: Rapid detection of pathogens in blood culture bottles shortens hospitalisation (Beekmann et al.) and appropriate empirical antimicrobial treatment improves outcome. (Behrendt et al., 1999, Hautala et al., 2005) How long does it take to report Gram stain result, tentative categorisation (TC) “susceptible, intermediate or resistant” from directly inoculated agar plates and how often were very major (VME), major (ME) or minor errors (mE) when TC results were compared to the final susceptibility results?
Method: Bactec bottles (Plus aerobic/anaerobic) were inoculated on the wards as recommended by the manufacturer. All bottles were incubated in the BACTEC 9120/9140. When growth was reported to the laboratory personnel a Gram stain was performed and an aliquot was streaked on blood and chocolate agar plates. Depending on the Gram stain result appropriate antibiotic disks were placed on additional Müller-Hinton or Columbia blood agar plates and TC was judged by using agar diffusion method. The Gram stain result was reported to the clinician. Tentative susceptibility was interpreted after ≥6–8 hours of incubation and TC was compared with the final categorisation obtained from MIC determined using the E-Test method.
Results: 141 blood culture bottles were prospectively studied. Gram stain results were available in 0.6 (standard deviation [SD] 0.6, range 3.6–0.1) hours and it took 0.9 (SD 0.7, range 3.3–0.1) hours to report the result to the attending physician. 2,538 antibiotic interpretations (TC) were available within 22.5 (SD 16.4, range 97.8–5.1) hours. VME were seen in 9 (0.35%), ME in 4 (0.16%) and mE in 27 (1.06%).
Conclusions: Gram stain result and direct susceptibility result are available within 23.1 hours and can be safely used to modify empirical treatment, as very major or major errors were recorded in less than 1%.
P939 Effectiveness of the lysis centrifugation method isolator 10 system compared to BacT/Alert 3D in detecting bacteraemia
K. Kharazmi, C. Kratzer, G. Tucek, M. Klimpfinger, A. Georgopoulos (Vienna, AT)
Objectives: Aim of this study was to compare the effectiveness of the lysis centrifugation method Isolator 10 system to the conventional BacT/Alert 3D system in diagnosis of bacteraemia.
Methods: 300 hospitalised patients with fever of unknown origin were included in the present study. At three different times, especially when patients' blood temperature was rising, one blood sample was taken and divided among the aerobic, the anaerobic BacT/Alert bottle and the Isolator 10 tube.
Results: 129 bacterial and 4 fungal isolates were recovered by using both methods. Isolator 10 System showed a higher detection rate of 31.7% compared to 14% of the BacT/Alert 3D (χ2 test p<0.001). The Isolator 10 system recovered significantly more coagulase-negative staphylococci, viridans streptococci, pneumococci, non-fermenters and fungi, but no obligate anaerobic bacteria were identified. A strong positive correlation (Spearman = 0.76, T-test p< 0.001) between the microbial counts detected by Isolator 10 system and a positive BacT/Alert blood culture was found. All Isolator cultures with a microbial count >12 cfu/mL were also positive using BacT/Alert, whereas microbial counts <5 cfu/mL were not detectable by the conventional blood culture system. Isolator 10 system required 8 to 22 hours to detect staphylococci, Enterobacteriaceae and non-fermenters compared to 21 to 24 hours with BacT Alert. A time range till 48 hours was needed to detect streptococci by using both blood culture systems.
Conclusion: Isolator 10 system showed high effectiveness in detecting pathogens with low microbial counts. Due to high potentiality of contamination and poor recovery of anaerobic bacteria by Isolator 10 system, the combination of both blood culture methods could optimise the diagnosis of bacteraemia.
P940 First notification of positive blood cultures: can we rely on the Gram stain?
M. Søgaard, M. Nørgaard, H. Schønheyder (Aalborg, DK)
Objectives: When blood cultures (BCs) turn positive the attending physicians are usually notified immediately about Gram stain findings. Despite the implications for antibiotic treatment and other therapeutic interventions, information on the accuracy of Gram staining is very limited. In this study we examined the accuracy of the preliminary BC reports based on wet-mount microscopy and Gram stain.
Methods: Observational study in North Jutland County, Denmark including the years 1996, 2000, 2001, and 2003. We retrieved data from the County Bacteraemia Registry, the departmental information system, and technicians' laboratory notes. The study was restricted to BCs with one morphological type; for patients with bacteraemia only the first positive BC was included. We defined 6 morphological groups: Gram-positive cocci in clusters, Gram-positive cocci in chains or diplococci, Gram-positive rods, Gram-negative cocci, Gram-negative rods, and yeasts. The sensitivity, specificity, positive (PPV) and negative (NPV) predictive values were estimated for each group using the species diagnosis as reference. We also evaluated Gram stain and wet-mount findings for the most frequent bacterial species/groups.
Results: A total of 5833 positive BCs were obtained during the four study years. The Table shows the distribution, sensitivity, specificity, PPV and NPV for the defined morphological groups. The sensitivity for the most frequent pathogens was in the range 88.2–100% with non-haemolytic streptococci being one notable exception (sensitivity 88.2%; 95% CI 82.6–92.4%). Wet-mount reports were more inaccurate (sensitivity 30–70% for species with peritrichous motility). Enterobacteriaceae (Salmonella spp. being most prominent) accounted for 25% of the bacteria with a report of polar motility.
| Cocci |
Rods |
Yeasts | ||||
|---|---|---|---|---|---|---|
| Gram(+) |
Gram(−) | Gram(+) | Gram(−) | |||
| clusters | chains/diplococci | |||||
| Number (%) | 2088 (36%) | 826 (14%) | 35 (0.6%) | 612 (10%) | 2180 (37%) | 92 (1.6%) |
| Correct evaluation | 2082/2088 | 794/826 | 33/35 | 566/612 | 2151/2180 | 90/92 |
| Sensitivitya | 99.7 (99.4–99.9) | 96.1 (94.6–97.3) | 94.3 (80.1–99.3) | 92.5 (90.1–94.4) | 98.7 (98.1–99.1) | 97.8 (92.4–99.7) |
| Specificitya | 99.1 (98.7–99.4) | 99.8 (99.6–99.9) | 100 (99.9–100) | 99.7 (99.5–99.8) | 99.1 (98.7–99.4) | 100 (99.9–100) |
| PPVa | 98.4 (97.8–98.9) | 98.6 (97.6–99.3) | 94.3 (80.1–99.3) | 97.1 (95.4–98.3) | 98.4 (97.8–98.9) | 100 (96.0–100) |
| NPVa | 99.8 (99.7–99.9) | 99.4 (99.1–99.6) | 100 (99.9–100) | 99.1 (98.8–99.4) | 99.2 (98.9–99.5) | 100 (99.9–100) |
95% CI in parentheses.
Conclusion: We demonstrated a high accuracy of Gram stain reports. Non-haemolytic streptococci formed an exception, which probably reflected this group's taxonomic heterogeneity. Wet-mount microscopy was generally less accurate.
P941 An evaluation of the rapid transport of BacT/ALERT blood culture bottles via a vacuum sample transport system at a UK regional hospital
L. Joslin (Exeter, UK)
Objectives: Blood cultures should be transported to the laboratory as quickly as possible, preferably within two hours [1]. The location of the laboratory, in relation to hospital wards at the Royal Devon and Exeter Hospital, meant that the deliveries of blood cultures were sometimes delayed by up to 15 hours. The objective of the study was to evaluate the possibility of rapidly transporting BacT/ALERT polycarbonate blood culture bottles to the laboratory via a vacuum sample transport system.
Methods: A two phase evaluation proticol was followed. We investigated the robustness of the polycarbonate bottles both experimentally and in routine use.
Experimental evaluation: Fifty uninoculated BacT/ALERT PF bottles and BacT/ALERT SA and SN bottle pairs were transported from hospital wards to the microbiology laboratory. To apply stress greater than that expected in routine use each culture bottle travelled eight stop/start journeys over a total distance of four miles. Bottles were placed in sample collection bags only, supplemental bottle transport devices or padding were not included. Bottles were visually inspected for leaks and cracks at the end of each run.
Routine use evaluation: The transport of approximately 28,000 blood culture sets via a vacuum transport system was monitored over a two-year period. The incidence of cracks or leaks were monitored and recorded.
Results: Experimental evaluation: On inspection, cracks or leaks were not detected in 49 BacT/ALERT PF bottles and 50 BacT/ALERT SA and SN paired bottles. One PF bottle was ‘lost’ within the vacuum transport system and was not returned to the laboratory for investigation.
Routine use evaluation: Over a two-year period approximately 28,000 blood culture sets have been received by the laboratory the majority if which arrived via vacuum tube. No incidences of cracks or leaks in the polycarbonate blood culture bottles have been reported.
Conclusion: The routine transport of polycarbonate BacT/ALERT blood cultures via rapid vacuum transport systems was demonstrated to be a viable option. Blood cultures can be received to the laboratory within minutes rather than hours potentially decreasing the time to report both positive and negative culture results.
P942 Time to positivity of blood cultures in patients Escherichia coli bacteraemia
G. Peralta, B. Ceballos, L. García-Mauriño, M.P. Roiz, J.C. Garrido, I. De Benito, L. Ansorena, M.B. Sanchez (Torrelavega, ES)
Objectives: Bacterial blood concentration is associated with prognosis of patients with bacteraemia caused by some microorganisms. However quantitative blood cultures are not performed in clinical practice. Time from the starting of incubation to a positive reading of blood cultures has been considered a surrogate marker of bacterial blood concentration and can be associated with prognosis. We analyze time to positivity (ttp) relationships with clinical parameters in patients with E. coli bacteraemia.
Methods: charts of all patients with monomicrobial E. coli bacteraemia attended at our hospital between January 1997 and December 2004 were identified by using the Microbiology laboratory database and reviewed. BacT/ALERT (bioMérieux) microbial detection system, was used for blood cultures and TTP detection. When multiple cultures were positive only the shortest TTP was selected for the analysis. Mann-Whiney U test was used for the comparison of median values, chi square test and Fisher's exact test for the comparison of categorical data.
Results: during the study period we identified 567 cases of E. coli monomicrobial bacteraemia, with a mortality of 4.9%. Median (interquartile range) TTP was: 11.3 h (10.2 h–11.8 h). Significant correlation among TTP and the number of positive blood cultures was found (Spearman's coefficient = −0.5, p<0.0001). No differences in TTP were detected depending on the comorbidities, or nosocomial or postsurgical acquisition. However TTP was longer in patients treated with antibiotics when blood cultures were performed (21.2 h±27.4 h vs 14.01 h±12.06 h, p = 0.01), and shorter in patients with a non urinary origin of the bacteraemia (12.5±10.1 h vs 16±16.8 h, p<0.0001), severe sepsis or shock (9.7±3.3 h vs 15.2±1.3 h, p < 0.0001), or patients who died (10.7±5.4 h vs 14.9±15 h, p = 0.006). ROC analysis revealed that a time to positivity of 10.4 h was associated with the best sensitivity and specificity for predicting dead (70% and 59%, respectively).
Conclusions: in patients with E. coli bacteraemia TTP is influenced by antibiotic treatment and has relationship with relevant clinical parameters as the origin of the bacteraemia, the presence of severe sepsis or shock and with the outcome. Its usefulness as a prognostic marker should be explored.
P943 Time to positivity of BACTEC blood culture bottles
S. Baka, I. Logginidis, V. Efstratiou, E. Panagiotopoulou, G. Kaparos, K. Gerolymatos, E. Kouskouni (Athens, GR)
Objective: The aim of this study was to detect how long it takes for blood cultures to become positive when using the BACTEC system bottles.
Methods: Using a continuously monitoring blood culture system (BACTEC 9050, Becton Dickinson, USA) we determined the time to positivity of blood cultures obtained from the patients of our hospital. We processed a total of 2125 blood culture sets and detected 420 (19.7%) positive blood cultures.
Results: Out of the 420 positive blood cultures we recovered 1 pathogen from 392 (93.3%) and 2 pathogens from 28 (6.7%) of them. Four hundred and forty-eight clinically significant pathogens were identified in the following order: 185 (41.3%) Gram-negative rods, 153 (34.1%) staphylococci [131 (29.2%) coagulase-negative staphylococci and 22 (4.9%) Staphylococcus aureus], 51 (11.4%) streptococci, 49 (10.9%) yeasts, 8 (1.8%) anaerobes and 2 (0.5%) Gram-positive rods. Two hundred and twenty-eight (54.3%) blood cultures were obtained from ICU patients, 183 (43.6%) from surgical patients, 7 (1.7%) from OB-GYN patients and only 2 (0.4%) from neonates. In respect to the time to detection or to positivity 274 (65.2%) blood cultures turned positive during the first 24 h of incubation, 119 (28.3%) up to 48 h, 21 (5.0%) in 72 h, while 4 (1.0%) and 2 (0.5%) were positive after 4 and 5 days of incubation, respectively.
Conclusions: The time to positivity for 65.2% of our positive blood cultures was up to 24 h, while in the first 2 days of incubation a total of 93.5% of the positive cultures were detected. Although we maintain the recommended period (7 days for aerobic and anaerobic bottles and 14 days for mycosis bottles), we could consider reducing the incubation time to 5 days (at least for the aerobic and anaerobic bottles) since all our blood cultures turned positive during the first 5 days.
P944 Bacteraemia caused by high-level gentamicin-resistant Enterococcus: risk factors, clinical features and outcomes
H.C. Jang, W. Park, C-I. Kang, S. Kim, C. Lee, S-W. Park, E. Kim, H-B. Kim, N-J. Kim, E-C. Kim, M-D. Oh, K-W. Choe (Seoul, Jeonju, Cheonan, Kyongju, KR)
Objectives: High-level gentamicin resistance (HLGR) was increased in enterococci since 1980, but there were few studies about the impact of HLGR on clinical features and outcomes. So, we performed this study to determine the risk factors, clinical features and outcomes of bacteraemia due to enterococcus with HLGR.
Methods: All patients with blood cultures positive for E. faecalis and E. faecium during the period of January 1999 – August 2003 (inclusive) were identified from a retrospective review of the Clinical Laboratory Records at Seoul National University Hospital in Korea. HLGR was determined by using disk diffusion method with 120 ug gentamicin disc (Oxoid), following the recommendation of the NCCLS.
Patients who had bacteraemia due to enterococcus without HLGR (Group 1) were compared with patients who had bacteraemia due to enterococcus with HLGR (Group 2). Statistical analysis of the data was performed using SPSS for Windows, version 11.0 (SPSS). Logistic regression analysis was done for multivariate analysis.
Clinical features of 215 patients with enterococcal bacteraemia
| HLGR+ (N = 79) |
HLGR– (N = 136) |
||||
|---|---|---|---|---|---|
| n | % | n | % | P value | |
| Demographic data | |||||
| Male gender | 52 | 66 | 92 | 68 | NS |
| Age | 58.1(±14.2) | 56.2(±16.8) | NS | ||
| Underlying disease | |||||
| Haematologic disease | 4 | 5 | 27 | 20 | 0.002 |
| Neutropenia | 4 | 5 | 39 | 29 | <0.001 |
| Cancer | 42 | 53 | 70 | 51 | NS |
| Kidney disease | 2 | 3 | 8 | 6 | NS |
| Urologic disease | 5 | 6 | 7 | 5 | NS |
| Biliary disease | 9 | 11 | 9 | 7 | NS |
| GI disease | 5 | 6 | 6 | 4 | N |
| Infection site | |||||
| Vascular catheter | 2 | 3 | 12 | 9 | NS |
| Infective endocarditis | 6 | 8 | 3 | 2 | NS |
| Urinary tract | 12 | 15 | 20 | 15 | NS |
| GI (except biliary tract) | 13 | 16 | 35 | 26 | NS |
| Biliary tract | 35 | 44 | 33 | 24 | 0.004 |
| Other status | |||||
| B. faecium infection | 45 | 57 | 105 | 77 | 0.003 |
| Nosocomial infection | 51 | 65 | 115 | 85 | 0.001 |
| Polymicrobial infection | 30 | 38 | 22 | 16 | <0.001 |
| ICU stay at culture | 7 | 9 | 36 | 26 | 0.002 |
| Appropriate treatment | 42 | 53 | 54 | 40 | NS |
| APACHE II scores | 14.4(±7.3) | 20.7(±8.3) | <0.001 | ||
| Outcome | |||||
| 14-day mortality | 12 | 15 | 50 | 37 | 0.001 |
| 30-day mortality | 17 | 22 | 66 | 49 | 0.002 |
Results: 215 cases of clinically significant E. faecalis and E. faecium bacteraemia were identified. 136 (63.3%) were caused by enterococci with HLGR. Hospital acquired infection, monomicrobial bacteraemia, E. faecium bacteraemia, underlying haematologic malignancy and neutropenia were more common in Group 2 than in Group 1 (p< 0.05). Biliary tract infection was less common in Group 2 than in Group 1 (p<0.05). Independent factors for occurrence of HLGR enterococcal bacteraemia were ICU stay at culture, 3rd generation cephalosporin use and monomicrobial infection (p < 0.05). 14-day and 30-day mortality in Group 2 were higher than in Group 1 in univariate analysis (p<0.05). But HLGR was not an independent risk factor for mortality in multivariate analysis.
Conclusion: Independent factors for occurrence of HLGR enterococcal bacteraemia were ICU stay at culture, 3rd generation cephalosporin use and monomicrobial infection. HLGR was not an independent risk factor for mortality.
P945 Occurrence of virulence determinants, antibiotic resistance in Swedish Enterococcus faecium blood-culture isolates during a six-year period
H. Billström, Å. Sullivan, B. Lund (Stockholm, SE)
Background: Recently the isolation ratio between E. faecalis and E. faecium causing infections has shifted from 10:1 to 3:1. Suggested explanation for this has been the increasing antibiotic resistance in E. faecium. However, changes in virulence cannot be excluded. Evidences are now emerging indicating a nosocomial origin of these infections.
Objectives: To detect the presence of virulence determinants in E. faecium infectious isolates and determine changes in frequencies over time. Furthermore, to investigate the levels of antibiotic resistance towards clinically relevant antibiotics and possible genetic relationships between isolates with suspected clonality.
Methods: A total of 278 strains E. faecium isolated from bacteraemia patients during year 2000 to 2006 at the Karolinska University Hospital, Huddinge were used. All isolates were screened for the seven virulence genes, aggregation substance (asa1), cytolysin (cylA), collagen binding protein (ace), E. faecalis endocarditis antigen (efafm), enterococcal surface protein (espfm), gelatinase (gelE), and hyaluronidase (hylfm), using PCR. Minimal inhibitory concentrations were determined towards ampicillin, ciprofloxacin, daptomycin, gentamicin, linezolid and vancomycin using agar dilution. PFGE was used to detection possible identical clones.
Results: A total of 54.3% of the isolates was esp-positive. In 14 of the isolates, hyl was found. 2% of the isolates harboured one or more of ace, asa1, efaAfm and gelE. None of the isolates were cytolysin positive. Ampicillin, ciprofloxacin and imipenem resistance were high, 76%, 90% and 80% respectively, while low frequencies were found for vancomycin (4.7%) and gentamicin (1.8%). Resistance towards linezolid and daptomycin was not found.
Discussion: During the course of this 6-year study, the levels of antibiotic resistance seem to be stable. The occurrence of espfm and hylfm also appear to be constant. The more rare virulence determinants, exclusively found during the last two study years, might indicate that an increase is occurring. The high frequency of espfm and antibiotic resistance suggests that these factors are of importance for infection development. However, if this is the case further unknown traits probably exist since a significant part of the isolates lacks all the investigated determinants.
P946 Recurrent cases of Escherichia coli bacteraemia in the county of Funen, Denmark in 1996–2001
B. Olesen, E.M. Nielsen, P. Søgaard, F. Scheutz, H.J. Kolmos, B.G. Bruun (Hillerød, Copenhagen, Odense, DK)
Objectives: To characterise a cohort of recurrent E. coli bacteraemia bacteriologically and epidemiologically with emphasis on differences between cases of relapse and re-infection.
Methods: The following definitions were used:
-
–
An episode of bacteraemia was defined as the presence of E. coli in a blood culture set.
-
–
Recurrence occurred when an isolate was obtained from an episode taking place more than 4 weeks after the first episode.
-
–
Polymicrobial bacteraemia occurred when more than one clinically relevant species or two different strains of E. coli were present in the same blood culture.
-
–
A recurrent infection with the same strain was defined as a relapse and was assumed when the PFGE profiles were indistinguishable or had <4-fragment difference.
-
–
All other cases were considered to be re-infections.
All isolates were serotyped and typed by Pulsed-Field Gel Electrophoresis (PFGE) with XbaI.
Results: Out of a total of 1,648 patients with E. coli bacteraemia, 106 patients (6.4%) suffered (a) recurrence(s) during the 6-year study period: 191 isolates from 89 patients were available for examination. 79 of these patients had 2 episodes of bacteraemia, 8 patients had 3, 1 patient had 4 and 1 patient had 5 episodes. Three patients were found in both the relapse and re-infection group.
“The relapse group”: 48 patients had 102 recurrent episodes with 48 sets of identical strains:
-
•
The isolates comprised 18 different O groups.
-
•
Thirteen strains (27%) possessed capsular antigen K1 and 12 (25%) capsular antigen K5.
-
•
The median time between episodes was 95 days (range 28–1278).]
-
•
Four episodes (4%) were polymicrobial.
“The re-infection group”: 44 patients had 89 recurrent episodes with 89
different isolates:
-
–
The isolates comprised 30 different O groups.
-
–
Five strains (6%) possessed capsular antigen K5 and two strains (2%) capsular antigen K1.
-
–
The median time between episodes was 222 days (range 40–2011).
-
–
Fourteen episodes (16%) were polymicrobial.
Conclusions: 6.4% of 1,648 patients with E. coli bacteraemia suffered (a) recurrence(s) during the 6-year study period. 54% of 89 patients had a relapse with the same strain. O6:K5:[H1] was the single most common serotype in both groups. Characteristics of cases of relapse compared to re-infections were: significantly higher frequency of possession of K1 or K5 capsular antigen, shorter median time between recurrences, and lower frequency of polymicrobial bacteraemia.
P947 Bacteraemia due to extended-spectrum β-lactamase producing Enterobacteriaceae in a tertiary care hospital in Salford, UK: one-year retrospective study
S. Ambalkar, K. Ryan, P.R. Chadwick, C.P.K. Subudhi (Salford, UK)
Objectives: Extended-spectrum β-lactamase (ESBL) producing strains of Enterobacteriaceae have caused major therapeutic problems worldwide since the majority of these are resistant to multiple antibiotics. This retrospective observational study was conducted to determine the prevalence, antibiotic susceptibility pattern and outcome of treatment of ESBL-producing Enterobacteriaceae isolated from blood culture over a one year period.
Methods: The laboratory database was used to identify patients who had positive blood cultures for Enterobacteriaceae during the period October 2004 to September 2005. Case notes and electronic patient records were reviewed to determine the outcome of episodes of bacteraemia in patients due to ESBL producers. Antibiotic susceptibility pattern of ESBL positive isolates were analysed to determine the prevalence of co-resistance to ciprofloxacin, gentamicin and piperacillin-tazobactam.
Results: There were 220 patients who had positive blood cultures for Enterobacteriaceae during this period. Approximately 10% (23/220) of these patients had ESBL-producing isolates. Bacteraemia due to ESBL-producing organisms in this study was healthcare-associated in all 23 patients (100%). 70% of these patients were aged more than 65 years. E. coli was the most common (70%) isolate amongst the ESBL-producing organisms. All the ESBL-producing isolates were susceptible to carbapenems. Resistance to ciprofloxacin, gentamicin and piperacillin-tazobactam was present in 80% (19/23), 55% (12/23) and 24% (5/23) of these isolates respectively. Majority (72%) of patients had inappropriate empirical antibiotic therapy with ceftriaxone. There was 48% (11/23) 30 day associated mortality amongst patients with bacteraemia due to ESBL-producing organisms.
Conclusions: ESBL-producing strains of Enterobacteriaceae are a significant cause of healthcare-associated blood stream infections. They carry a high risk of mortality. Empirical treatment of sepsis caused by Enterobacteriaceae needs to be reconsidered in areas where there is high prevalence of ESBL producers as there are therapeutic implications due to co-resistance to multiple antibiotics.
P948 Risk factors and outcome of bloodstream infections caused by enterobacteria producing extended-spectrum β-lactamases
A. Endimiani, F. Luzzaro, A. Toniolo (Varese, IT)
Objectives: Enterobacteria-induced bloodstream infections (BSI) represent a common clinical entity. This study was initiated to evaluate risk factors and outcome of BSI episodes caused by members of the Enterobacteriaceae family producing extended-spectrum β-lactamases (ESBLs).
Methods: Two-hundred-eighteen BSI episodes caused by enterobacteria occurring at the Ospedale di Circolo (Varese, Italy) from 1997 to 2005 have been analyzed. ESBL production was assessed by phenotypic and molecular methods. Clinical records of BSI-patients were examined retrospectively: demographic data, underlying diseases (McCabe and Jackson classification scheme and Charlson weighted index), risk factors, antimicrobial therapy, treatment outcome and clinical outcome were investigated. Cases due to ESBL-positive strains were compared to those due to ESBL-negative strains. Student's unpaired t-test and Mann-Whitney U-test were used to compare continuous and not normally distributed continuous variables. The Chi-square test was used to compare the treatment and clinical outcome.
Results: Ninety-eight isolates were shown to produce ESBLs (mainly TEM-52/92, CTX-M-1 and SHV-12). Compared with the ESBL-negative group, ICU stay, use of bladder and intravascular catheters, previous surgery, previous use of antibiotics, and intubation were significant risk factors for BSI due to ESBL-positive isolates (all P < 0.01). BSI cases due to ESBL-positive enterobacteria were more frequently associated to lower respiratory tract infections than those due to ESBL-negative strains (32.7% and 8.3%, respectively; P < 0.01). Compared with cases due to ESBL-negative isolates, those caused by ESBL-positive strains showed higher mean hospital stay (74.4 and 33.4 days, respectively; P < 0.01) and crude mortality rate (26.5% and 8.3%, respectively; P < 0.01). Empirical treatment was more frequently adequate among patients with BSI due to ESBL-negative than in the case of ESBL-positive isolates (76.7% and 49.0%, respectively; P < 0.01). BSI cases due to ESBL-negative isolates responded to therapy more frequently than those due to ESBL-positive strains (81.3% and 62.5%, respectively; P < 0.10).
Conclusion: Therapeutic failure and high mortality rate are a common feature of BSI episodes caused by ESBL-positive enterobacteria. Prompt recognition of ESBL-positive strains appears critical for the clinical management of patients with systemic enterobacterial infections.
P949 High rate of extended-spectrum β-lactamases among Enterobacteriaceae blood-culture isolates from Bulgarian hospitals
E. Keuleyan, R. Markovska, I. Schneider, T. Kantardjiev, M. Sredkova, D. Ivanova, K. Rachkova, B. Markova, E. Dragijeva, I. Mitov, A. Bauernfeind (Sofia, BG; Munich, DE; Pleven, Stara Zagora, BG)
Objectives: (1) To reveal the rate of ESBL producers among bacteraemia isolates. (2) To characterise the spectrum of ESBL genes and their possible spread.
Methods: Surveillance data have been obtained through the national programme BulStar since 1998. Eight hospitals from three towns provided 71 bacteraemia isolates in an ESBLs survey, conducted from 1996 to 2003. The Medical Institute-Ministry of the Interior (MIMI) served as a referral centre. ESBLs were determined and characterised by double disk synergy-, disk diffusion confirmatory- (CLSI, 2005) tests; conjugation was performed on solid medium; Isoelectric focusing and bioassay were followed by ESBL-group specific PCR and nucleic acid sequencing; RAPD was carried out to detect epidemiological relationships.
Results: ESBLs, 3% among Enterobacteriaceae from septicaemia in 1998, augmented up to 15% in E. coli, 35% in K. pneumoniae and 24% in S. marcescens during 2004 (BulStar); their rate at MIMI from 2003 to 2005 was: 6% – E. coli, 50% – K. pneumoniae and 33% – S. marcescens. The survey on ESBLs revealed the incidence of 3 groups: SHV-, TEM- and CTX-M; and 5 particular enzymes: SHV-12 (47%), SHV-2 (4%), TEM-3-like (19%), CTX-M-3 (14%) and CTX-M-15 (9%); 5 strains produced 2 ESBLs. The majority of the pathogens from MIMI and other Sofia hospitals produced SHV-12; TEM-3-like was found at the medical centre-Pleven. K. pneumoniae, the predominant species collected (60% of all blood-culture isolates), produced most frequently SHV-12 (29 out of 42 strains), followed by TEM-3-like (6), CTX-M-15 (3), CTX-M-3 and SHV-2 (by 1). Among 11 E. coli, CTX-M-15 ESBLs were most common (4), followed by TEM-3-like (2) and 5 strains produced combinations of 2 enzymes. S. marcescens (7 strains) produced CTX-M-3 (3), SHV-12 (2) and SHV-2 (2). Two small outbreaks and one larger could be identified. Most of the strains showed multiple resistance, e.g. 74% to aminoglycosides and ciprofloxacin, however they were susceptible to carbapenems.
Conclusion: A high proportion of Enterobacteriaceae strains (3–50%) isolated from patients with bacteraemia were shown to elaborate ESBLs from three groups, five different enzymes, which is worrying and represents one of the highest percentage in Europe. Strong attention should be paid at national and hospital level towards antibiotic use and Infection control.
P950 Quantifying the relationship between Staphylococcus aureus bacteraemia and Staphylococcus aureus bacteriuria
M. Ekkelenkamp, J. Verhoef, M. Bonten (Utrecht, NL)
Objectives: Common belief is that Staphylococcus aureus bacteraemia (SAB) causes S. aureus bacteriuria, especially in the case of endocarditis. Evidence for this theory is however limited. We hypothesised that if SAB truly causes S. aureus bacteriuria this phenomenon would be observed also in patients without urinary tract infection (UTI) and without risk factors for urinary colonisation with S. aureus (specifically the presence of urinary catheters and oliguria).

Methods: We conducted a retrospective cohort study over a 5-year period to assess the frequency of S. aureus bacteriuria in patients with SAB. We analyzed the diagnosis of these patients and determined whether they had risk factors for colonisation or whether the S. aureus bacteriuria could solely be attributed to SAB.
Results: In the five-year period, 515 patients had demonstrated SAB. From 164 patients urine cultures had been taken concomitantly with the blood cultures and of these, 23 patients had S. aureus bacteriuria. In all 23 patients the bacteriuria could be explained either by UTI, the presence of urinary catheters or oliguria.
P951 Trends in nosocomial bacteraemias: results from a university hospital in the Netherlands
H.S.M. Ammerlaan, A. Troelstra, H. Blok, M.J.M. Bonten (Utrecht, NL)
Objective: To evaluate the burden (morbidity and mortality) of nosocomial bacteraemias (NB) in a MRSA-free hospital.
Methods: The UMC Utrecht is a 997-bed tertiary care centre with about 21,100 admissions annually. We analyzed all patients admitted between January 1996 and December 2005, aged ≥18. Day-care admissions and those treated in psychiatry were excluded. The hospitals' microbiological database was linked to the patient-administrative system, thereby providing all blood culture results and data on gender, date of birth, department, length of stay, and mortality at hospital discharge. NB was defined as isolation of bacteria or yeast from ≥1 blood culture set >2 days after hospital admission, with only the first episode analyzed. Skin organisms commonly associated with contamination, were excluded. Annual incidence and mortality rates of NB were calculated, with subgroup analysis for different micro-organisms.
Results: 1,769 episodes of NB were identified, with incidence rates increasing from 89.9 per 100,000 patient-days in 1996 to 112.6 in 2005, due to an increase in both Gram-positive and Gram-negative infections. The RR of male on acquiring NB was 1.6 compared to female. The RR of patients in the ICU was 6.0 compared to the other departments. In this period no trend towards increasing resistance was found, accept for a striking increase of amoxicillin resistance among Enterococcus species in the 2nd half of the study from 12.1% to 46.6% (p = 0.001). The length of stay (LOS) within the hospital decreased from 10.7 (95% CI: 10.6–10.8) in 1995 to 7.9 (95%CI: 7.8–7.9) days in 2005. The average LOS for patients with NB did not change between 1995 and 2004 (50.4 days (95% CI: 48.0–52.7)), but decreased in 2005 to 41.5 days (95% CI: 36.2–46.8). Average LOS for patients with NB ranged from 39.6 days (95%CI: 24.6–54.5) for Morganella morganii to 73.7 days (95% CI: 49.2–98.1) for Serratia species. Overall hospital mortality for patients with NB was 26.4% (ranging from 12.0% for Acinetobacter species to 52.1% for Candida). The RR to die in the hospital for patients with NB was 8.5 when compared to non-affected patients. This did not change significantly from 1996 to 2005.
Conclusion: The burden of nosocomial bacteraemias did not change significantly over the past 10 years within the UMC Utrecht. While the mean LOS within the hospital decreases, the incidence rate of HAB increases, suggesting an increasing severity of illness of hospitalised patients.
P952 Epidemiology and outcome of primary and secondary community-acquired bacteraemia in adult patients
M. Ortega, M. Almela, J. Martínez, F. Marco, A. Soriano, J. López, M. Sánchez, A. Muñoz, J. Mensa (Barcelona, ES)
Objectives: To know the associated factors and outcome of community-acquired primary bacteraemia (PB); to describe the most frequently isolated microorganisms, its antibiotic resistance pattern and to guide the most adequate antibiotic treatment.
Patients and Methods: 1640 community-acquired bacteraemias in non-neutropenic adults were consecutively enrolled from January 2003 to May 2006 and prospectively followed up. Non-conditional logistic regression methods were used with PB and death as dependent variables.
Results: 1440 were secondary bacteraemia (SB) and 200 (12%) were PB. The independent factors associated with PB were: male gender (OR: 1.69, 95%CI: 1.27–2.25, p = 0.001), ultimately or rapidly fatal prognosis of underlying disease (OR: 2.48, 95%CI: 1.84–3.34, p = 0.001) and incorrect empirical antibiotic therapy (OR: 2.33, 95%CI: 1.61–3.33, p = 0.001). The most frequently isolated microorganisms in PB were E. coli and other enterobacterias (26% and 22%) and S. aureus (15%). There were 28% and 37% of ciprofloxacin resistant E. coli strains in SB and PB, respectively (p = 0.2). Mortality was significantly higher in PB cases (13% vs 8%, p = 0.04). The independent factors associated with mortality in PB were: ultimately or rapidly fatal prognosis of underlying disease (OR: 2.1, 95%CI: 1.41–3.13, p = 0.001) lack of fever at the moment of bacteraemia (OR: 2.38, 95%CI: 1.18–4.76, p = 0.02) and incorrect empirical antibiotic therapy (OR: 2.01, 95%CI: 1.22–3.33, p = 0.006).
Conclusions: The initial empiric antibiotic treatment is more frequently incorrect in PB than in SB and this is a predictor factor for mortality in PB. The resistance pattern of E. coli, other enterobacterias and S. aureus in every setting should guide the most appropriated empirical treatment for PB.
P953 A six-year surveillance of the epidemiology and aetiology of the bacteriaemias at a Milan hospital. The clinical relevance of haemoculture
M. Cainarca, C. Tarricone, R. Ranieri, C. Erba, A. D'Arminio Monforte, G. Melzi d'Eril (Milan, IT)
Objectives: In recent years, hospitalised patients are at increasing risk of acquiring a nosocomial infection, mainly due to the widespread use of invasive procedures. Haemocolture is considered as the gold standard for defining and treating a patient at high risk for developing a bacteraemia. Our aim was to perform a six-year surveillance of the infectious events, nosocomial and community bacteraemias and pseudobacteraemias occurring at San Paolo Hospital (about 500 beds for admissions).
Methods: We performed a retrospective collection of infectious events, bacteraemias and pseudobacteraemias during the period between 2000 and 2005. The bacterial strains were identified by the instrument VITEK 2 (bioMérieux).
Results: The number of infectious events remained stable overtime (from 893 in 2000 to 891 in 2005), as well as the number of colture isolates/infectious events (from 133 in 2000 to 133 in 2005). The most frequent isolated bacteria was Staphylococcus aureus (21.1%), followed by Escherichia coli (20.5%), enterococci (9.2%) and negative coagulase staphylococci (8.35%). Nosocomial bacteraemias were mainly due to Staphylococcus aureus (23.2%); on the contrary Escherichia coli was the most frequent agent in community bacteraemias (23%). The mean percentage of the bacteraemias/infectious events in the period 2000–2005 was 14% with a difference between the surgical department (18.7%) and medical department (15.7%). The rate of nosocomial bacteraemias (2.76/1000 admissions) and community bacteraemias (4.08/1000 admissions) did not vary during the period of surveillance.
Conclusion: Our data confirm that Staphylococcus aureus is the main responsible bacteria for nosocomial infections as Escherichia coli is in community bacteraemias. The steadiness of the number of isolates/infectious events and the rate of nosocomial bacteraemias suggest the need of an educational interventions to remark the role of the haemocolture in definining bacteraemias and the establishment of approaches to reduce the incidence of hospital acquired infections.
P954 Predictors of positive blood culture in an emergency service. A prospective study
B. Ceballos, G. Peralta, L. Ansorena, M. Rodriguez-Lera, M. Roiz, M. Sánchez (Torrelavega, ES)
Objectives: The microbiology laboratory is of limited usefulness for bloodstream infection detection, because only a low proportion of blood cultures have positive results, and the final results take at least 24 h to be known. We design a prospective study to develop a prediction model for bacteraemia, based on clinical data of patients assisted in an Emergency Service.
Methods: The study was developed in a 250 bed community teaching hospital placed in the North of Spain with around 65,000 presentations to the emergency service annually. All patients to whom blood cultures were performed from January to June 2006 were recruited for the study. Univariate analyses and stepwise logistic regression were performed to identify factors associated with bacteraemia.
Results: Blood cultures were performed in 390 patients in the period of study. Bacteraemia was detected in 60 (15.4%). Isolated microorganisms were E. coli in 35 (9%), S. aureus in 3 (0.8%), S. agalactiae in 2 (0.5%), P. aeruginosa in 2 (0.5%), Listeria spp. in 2 (0.5%), and others in 16 (4.1%). No significant differences among patients with positive and negative blood cultures in age, gender, proportion of comorbibities (renal chronic failure, cardiac failure, diabetes, dementia, immunosuppression, cirrhosis, neoplasia), Charlson index score, or clinical manifestations (presence of chills, or vomiting, and mean systolic blood pressure) were detected. However patients with positive blood cultures had shorter duration of the acute illness (29.3±32.5 h vs 59.4±85.7 h), more frequently had no previous antibiotic treatment (96.7% vs 84.2%), more frequently had temperature higher than 38°C (60% vs 40.2%), severe sepsis (21.7% vs 7.9%) or altered consciousness (20% vs 6.7%). In a logistic regression model the predictors of bacteraemia were absence of previous antibiotic treatment (OR: 4.87, 95CI: 1.12–21.17), altered consciousness at arrival (OR: 3.85, 95CI: 1.55–9.54) and the presence of fever (OR: 2.18, 95CI: 1.16–3.97).
Conclusions: Several clinical parameters can be useful for predict bacteraemia in an Emergency service. The absence of previous antibiotic administration, altered consciousness at arrival, and the presence of fever are associated with a higher risk of bacteraemia
P955 Epidemiological characteristics of nosocomial blood infections at a clinical centre, Belgrade, Serbia in 2005
B. Jovanovic, V. Mioljevic, S. Jovanovic, N. Mazic, I. Palibrk (Belgrade, RS)
Introduction: Nosocomial bloodstream infections (BSI) represent actual and one of the most severe medical problems due to their frequency, prolonged hospitalisation and increase of medical costs. Their incidence varies depending on the characteristics of the observed patients and department, ranging between 1.3 and 18.4 per 1000 patient days. Major causes of BSI include coagulase-negative staphylocci(CNS), Staphylococcus aureus, Enterococcus spp. and Candida spp.
Purpose: Determining of BSI incidence and distribution of BSI causes in percentages in patients hospitalised for more than 48 hours at the Institute of Digestive Diseases in 2005.
Methods: BSI incidence is calculated using Atlanta CDC methodology. Cultivation, isolation, identification and susceptibility tests of the causative organisms obtained from the patient material was carried out using standard microbiological methods in the Microbiological Laboratory within the Emergency Center of the Clinical Center of Serbia.
Results: BSI incidence at the Institute of Digestive Surgery was 5.76 per 1000 patient days. One third (28%) of all blood infections verified in the laboratory was caused by CNS, followed by Klebsiella spp. (14%), Staphylococcus aureus and Candida spp. (12.5%). Methicillin resistance was evidenced in 80% of CNS isolates and 90% of Staphylococcus aureus isolates. ESBL was produced by 18% of Klebsiella spp. isolates.
Conclusion: The incidence of BSI recorded at the Institute of Digestive Diseases in 2005 was 5.76 per 1000 patient days. One third of all blood infections verified in the laboratory at the Institute during 2005, was caused by CNS, followed by Klebsiella spp., Staphylococcus aureus and Candida spp. All the isolates show high degree of resistance to antimicrobial drugs. Epidemiological surveillance of BSI enables insight into epidemiology of BSI, monitoring of resistance of causative organisms and application of the specific preventive measures against the infections.
P956 Surveillance of nosocomial sepsis and identifcation of relevant risk factors in neutropenic haematology-oncology patients
F. Tabibnia, F. Mattner, A. Ganser, P. Gastmeier, I. Chaberny (Hannover, DE)
Objectives: Implementation of a prospective surveillance and identification of risk factors concerning nosocomial bloodstream infections (BSI) during neutropenia in adult patients of a haematology–oncology unit after hematopoetic stem cell transplantation or high dose chemotherapy (CT).
Methods: BSIs were identified using CDC definitions (laboratory confirmed BSI) in neutropenic patients (NPs) with haematological and solid malignancies (absolute neutrophile counts <1000/mm3). Recorded risk factors (RFs): Age, gender, diagnosis, type of transplantation (autologous, allogeneic sibling donor, matched unrelated donor), type of central access (CVC, Port) or use of peripheral access, fever days. Uni- and multivariate analyses were used to determine RFs.
Results: During the 12-month period of the study 202 patients with 2,843 neutropenic days (NDs) were investigated. 41 patients received autologous transplant (345 NDs), 161 were treated for other reasons (2,498 NDs). The median age was 49.5 years (17–77). The mean number of NDs (range: 2–72) was 14.1 (15.5 and 8.4 after CT and autologous transplantation (AT), respectively). A total of 37 BSIs were diagnosed. The incidence densities were 13.0 BSIs/1,000 NDs (10.2 and 23.2 after CT and AT, respectively) or a rate of 18.3 BSIs/100 NPs (18.0 and 19.5 after CT and AT, respectively). The main pathogens were coagulase-negative staphylococci (36.4%), Gram-negative rods (31.8%) and Gram-positive cocci (20.5%). The shortest mean duration of neutropenia until development of BSI among the pathogens was found for Gram-negative rods counting 5 days. The mean length of fever was 4 days (with BSI 8 days). On univariate analysis, the duration of fever (p<0.001) and of neutropenia (p < 0.001) was significantly associated with development of BSI. On multivariate analysis, the independent RFs for BSI were the number of fever days (Hazards ratio 1.2 increased risk/day) and use of CVC (Odds ratio [OR], 6.4; 95% confidence intervall, 1.5–27.7, Hazards ratio 5.8).
Conclusions: The study shows and confirms that the use of CVC is a major RF for nosocomial BSIs in NPs. The incidence of BSI/1000 NDs was higher among AT recipients of this study than of the participating hospitals of ONKO-KISS (pooled data: 17.8). Compared to the results in former studys (Dettenkofer et al., CID 2005;40:926–31) a higher share of Gram-negative rods (31.8% vs. 15%) was found associated with BSI.
P957 The influence of antimicrobial resistance on mortality in critically ill patients with bacteraemia
J.Y. Choi, Y.K. Kim, M.S. Kim, Y.S. Park, Y.G. Song, D. Yong, K. Lee, J.M. Kim (Seoul, KR)
Background: Among all hospitalised patients, ICU patients are at greatest risk for nosocomial infections. Predominant pathogens include Pseudomonas aeruginosa, Enterobacter cloacae, Staphylococcus aureus, Enterococci, and Candida spp. They are more likely to be resistant to antimicrobial agents than are isolates from elsewhere in the hospital. The isolation of antimicrobial resistant pathogens increase the risk of inadequate antimicrobial therapy. However, clinical importance of antimicrobial resistance, as a risk factor for hospital mortality and other adverse clinical outcomes, has not been systematically evaluated in the ICU setting. The objective of this study was to evaluate the effects of the isolation of antimicrobial resistant pathogens on clinical outcomes among ICU patients with bacteraemia.
Methods: A retrospective cohort study was performed to identify risk factors for mortality in ICU patients with bacteraemia. Data from 76 patients with bacteraemia who admitted between January 2004 and December 2004 at ICU of Severance Hospital, Yonsei University College of Medicine in Seoul, Korea, a 1,300-bed tertiary-care teaching hospital, were analyzed to identify risk factors for in-hospital mortality. ICU patients with antimicrobial resistant pathogens such as MRSA, VRE, MDR P. aeruginosa, MDR A. baumannii, ESBL-producing E. coli, ESBL-producing Klebsiella sp., 3rd cephalosporin resistant Enterobacter sp were defined patients with antimicrobial resistant pathogens.
Results: The mean age of the 76 enrolled patients was 60 years, and mortality rate was 76.3% (58 of 76 patients). Univariate analysis revealed that the risk factors for mortality included; age, shock, elevated creatinine, duration from admission to bacteraemia and patients with antimicrobial resistant pathogens. Multivariate analysis revealed that the independent risk factors for mortality were age, elevated creatinine, duration from admission to bacteraemia and patients with antimicrobial resistant pathogens.
Conclusion: Isolation of antimicrobial resistant pathogens was independently associated with in-hospital mortality among critically ill patients with bacteraemia in ICU setting.
P958 Risk factors for inadequate antimicrobial treatment in patients with bloodstream infection
J. Vallés, E. Calbo, E. Anoro, D. Fontanals, M. Xercavins, E. Espejo, G. Serrate, M. Morera, N. Freixas, B. Font, F. Bella, F. Segura, X. Garau (Sabadell, Terrassa, ES)
Objective: Analyse the risk factors for ineffective antimicrobial therapy in patients with bloodstream infections (BI).
Methods: Prospective cohort study of adults with BI at three hospitals during one year. BI was defined as nosocomial-acquired (NA), community-acquired (CA), and healthcare-associated (HA).
Results: Of 1157 BI, 581 (50.2%) were CA, 295 (25.5%) were NA, and 281 (24.3%) were HA. Patients with NA received inadequate antimicrobial therapy in 28% of episodes, compared with 8% in CA (OR: 4.27; 95%CI, 2.8–6.3; p <0.001). Patients with HA received inadequate therapy in 16% of episodes compared with 8% in CA (OR: 2.07; 95% CI, 1.3–3.2; p = 0.001). The difference between NA and HA episodes (28% vs 16%) was also different (OR: 2.06; 95% CI, 1.3–3.1; p = 0.001). Mortality among patients with adequate treatment was 18% vs 34% with inadequate treatment (p < 0.001). Multivariate logistic regression using community-acquired status as reference, identified NA episodes (OR, 1.7; 95%CI, 1.1–2.7), and BI of unknown origin (OR, 2.3; 95% CI, 1.4–3.7) as independent predictors of inadequate therapy. C. albicans (OR, 13.9; 95%CI, 3.7–52.2), methicillin-resistant S. aureus (OR, 6.9; 95%CI, 2.4–19.6), P. aeruginosa (OR, 2.4; 95%CI, 1.3–4.3), and coagulase-negative staphylococci (OR, 1.9; 95%CI, 1.06–3.5) were associated with an increased likelihood of receiving inadequate therapy. Infection due to S. pneumoniae (OR, 0.14; 95% CI, 0.03–0.59) and E. coli (OR, 0.2; 95% CI, 0.1–0.3) were associated with a decreased risk of ineffective therapy.
Conclusions: Among patients treated for BI, nosocomial-acquired episodes, unknown origin of infection, and infection due to multiresistant pathogens were an independent predictor of inadequate antimicrobial therapy.
P959 Serum total cholesterol as a predictor for clinical outcome of bacteraemia
T. Kitazawa, Y. Ota, K. Tatusno, S. Okugawa, A. Fukushima, S. Yanagimoto, K. Tsukada, K. Koike (Tokyo, JP)
Objectives: Infection and inflammation induce the acute phase response, leading to multiple alterations in lipid metabolism. There have been few reports, however, about changes in blood cholesterol levels at the onset of bloodstream infection (BSI) and association between the changes and prognosis of the BSI. The aim of this study was to elucidate the factors which were related with the changes in blood cholesterol levels at the onset of BSI. In addition, it was also examined whether the changes or the levels of the serum cholesterol were associated with the prognosis of bacteraemia.
Methods: From April 2003 to March 2006, all the patients aged ≥20 years whose blood cultures were positive at the University of Tokyo Hospital were enrolled in this study. We selected the patients whose cholesterol levels were tested during both of the two periods; the first period, between 30 days and 10 days before the onset of BSI, and the second period, ≤2 days from the onset. A retrospective chart review was performed to collect demographics including medical history, invasive procedures and medications.
Results: Average total cholesterol levels before the onset of bacteraemia was 168.9±46.7 mg/dL, and decreased to 135.7±45.0 mg/dL at the onset of bacteraemia, with statistical significance (p < 0.001). Non-survivors had lower cholesterol levels than survivors, before the onset of BSI (130±45 vs 172±46, p = 0.022) and at the onset of BSI (103±40 vs 138±45, p = 0.044). The lower cholesterol group had higher 30-day mortality than the higher cholesterol group, but not significantly (p = 0.10). The lower cholesterol group had longer durations of treatment than the higher cholesterol group with statistically significant (P = 0.042).
Conclusion: Serum cholesterol level at the onset of BSI is an important predictor for the outcome of BSI.
P960 Prospective evaluation of risk factors for catheter-related bloodstream infections in two Swiss hospitals
A. Templeton, G. Rettenmund, B. Schöbi, K. Diethelm, M. Schlegel, F. Fleisch, S. Henz, G. Eich (St. Gallen, Zurich, CH)
Background and Objectives: Catheter related bloodstream infections (CRBSI) are a leading cause of nosocomial infections associated with significant mortality and costs. The objective of this study was to determine the rate of CRBSI in two large Swiss hospitals and to identify risk factors for this condition.
Methods: During one year all central venous catheters (CVC) inserted in patients admitted for visceral, orthopaedic, or urologic surgery to the cantonal hospital St. Gallen and all CVCs inserted in patients at the cantonal hospital Chur were included in the study. Catheters were followed for the duration of their insertion. Blood cultures and semiquantitative cultures from catheters were drawn in the presence of local or systemic signs of infection. Primary endpoint was CRBSI. CRBSI was defined as definite if (a) the same pathogen grew in at least one blood culture and from the distal segment of the catheter or (b) the same pathogen grew in at least one peripherally and centrally drawn blood culture and the differential time to positivity of central blood culture vs. peripheral blood culture was >120 minutes. CRBSI was defined as probable when at least one blood culture was positive with a recognized pathogen in the absence of an other site of infection. Data were analysed using univariate and multivariate time-to-event methods.
Results: During the study period 1396 CVCs in 1162 patients were prospectively studied. Incidence density of all CRBSIs (definite n = 29, probable n = 7) was 2.51 (95%CI; 1.81–3.49) per 1'000 catheter-days. The lowest rate of CRBSI was found in subclavian catheters, the adjusted hazard ratio (HR) for jugular catheters was 2.2 (95%CI: 1.1– 4.3; p = 0.03) and for femoral catheters 2.9 (95%CI: 0.6–14.4; p = 0.19). Each additional lumen increased the risk (HR = 4.4, 95%CI: 2.5–7.7, p< 0.001), whereas the blocking of additional lumens was protective (HR = 0.3, 95%CI: 0.1–0.7, p = 0.006). The most commonly isolated organism were coagulase negative staphylococci with a rate of 28%.
Conclusion: Number of lumens and site of access were independent risk factors for catheter-associated blood-stream infections. The use of catheters with multiple lumens should therefore be restricted as far as possible. If a catheter cannot be removed, the permanent closure of unneeded lumens may reduce the risk of CRBSI.
P961 Septicaemia secondary to Vibrio vulnificus
I. Villamil-Cajoto, L. Rodriguez-Otero, S. Cortizo, M.A. Garcia-Zabarte, A. Aguilera-Guirao, M. Treviño-Castellano (Santiago de Compostela, ES)
Introduction: Vibrio vulnificus is a halophilic species which has demonstrated its pathogenic power by causing septicaemia and bacteraemia in patients who had ingested contaminated seafood (especially raw oysters) and infections of soft tissues exposed to sea water. The mortality rate in the case of septicaemia can reach values higher than 50%. Hepatic diseases and inmunodepression are the main risk factors.
Clinical case: A 37 years old male arrives at the emergency service of the hospital because of a temperature of 38.4°C, local pain and a one week old inflammation in the right lower limb. Physical examination revealed an oedematous and erithematous infected wound in the external malleolar region of the right lower limb which was caused by a sharp vegetal object and which got considerably worse after swimming in the sea. The hemogramme showed 18.500 neutrophilic leukocytes (91%). VHS 72 mm. Coagulation tests revealed a protrombine time of 33%, INR of 2.6, fibrinogen level of 585 mg/dl and an activated partial thromboplastin time of 43 seconds. The patient was diagnosed with cellulitis and lymphangitis in the right lower limb. The molecular analysis of the HFE gen revealed a heterozygosis of the C282Y mutation. Six days after the admission to hospital the blood cultures became positive. The biochemical identification of the colony was realised by the API 20 NE system (bioMérieux, Marcy l'Etoile, France).
The result obtained showed the numeric code 7–6-3–0-0–4-4 which could be identified as Vibrio vulnificus with a T-value of 0.51. The isolated strain was resistant to ampicillin and amoxicillin-clavulanic and sensitive to third and fourth-generation cephalosporins, monobactams, carbapenems, tetracycline, cotrimoxazole, and fluorquinolones. On the 10th hospital day and after a favourable clinical progress the patient was discharged.
Conclusions: Vibrio vulnificus shows high affinity for iron, which can explain its connection to diseases with increased iron deposit, such as chronic liver problems caused by hemochromatosis. Our group describes the second case exposed in scientific literature of a patient whose biochemical markers for iron metabolism show normal values and who is carrier of heterozygosis of the C282Y mutation in the HFE gen of the hemochromatosis.
P962 Mannose-binding lectin promoter gene variants in Korean patient with bacteraemia
Y.S. Park, J.Y. Choi, S.Y. Shin, M.S. Kim, Y.H. Kim, Y.G. Song, J.M. Kim (Seoul, KR)
Mannose-binding lectin is a calcium-dependent, liver-derived serum protein involved in innate immune defence. Variant alleles causing low serum levels of functional mannose-binding lectin have been shown to be associated with an increased risk of infections. Also, there is the existence of additional polymorphisms at positions −550 (H/L variants) and −221 (Y/X variants) in promoter regions of the gene.
Blood samples were obtained from patients with bacteraemia, at Severance hospital between March, 2003 and September, 2004. For inclusion, patients were required to have at least one positive blood culture results. PCRs were performed using genomic DNA and DNA sequence analysis of the MBL promoter region was performed on PCR products (spanning position −801 to +77 of the MBL gene).
Each allele H, L, Y, X has 0.37, 0.63, 0.58, 0.42, of frequency, respectively and Haplotype HY, LY, HX, LX has 0.26, 0.32, 0.11, 0.32 of frequency. Correlation with SAPS II score is shown in Table 1 .
Table 1.
| total | H/Y | L/Y | L or H/X | |
|---|---|---|---|---|
| SAPS II score | 37.7±15.4 | 36.9±15.1 | 38.4±16.3 | 42.1±10.6 |
| Mortality predicted | 20.8% | 19.3% | 22.6% | 28.7% |
| Mortality observed | 27.1% (16) | 26.7% (5) | 21.1% (4) | 28.0% (7) |
The promoter haplotypes, HY, LY, and LX, have been known to associations with high, medium, and low levels of MBL serum concentrations, respectively. These gene and haplotypes frequencies in these bacteraemic patients were relatively similar to other normal Caucasoid. So, we need to examine Korean normal population and linkage disequilibrium between promoter and structural variants.
Fungal infections in the immunocompromised host
P963 Empirical antifungal therapy in febrile neutropenic patients – systematic review
E. Goldberg, A. Gafter-Gvili, M. Paul, E. Robenshtok, L. Vidal, L. Leibovici (Petah-Tikva, IL)
Objectives: The practice of administering empirical antifungal therapy to persistently febrile neutropenic patients has become a standard of care. This study aims to evaluate if empiric antifungal treatment reduces mortality and prevents invasive fungal infections (IFI).
Methods: Systematic review and meta-analysis including randomised controlled trials (RCTs) comparing empirical antifungal treatment with placebo or no intervention (control), or another regimen, in persistently neutropenic patients. Search was conducted until 2006. Outcomes assessed were: All-cause mortality, documented IFI, fungal-related mortality, composite failure, invasive mould infections, invasive yeast infections, resolution of fever during neutropenia, adverse events. Relative risks (RR) with 95% confidence intervals (CIs) were estimated and pooled.
Results: Our search yielded 25 trials, 5 of which compared polyenes or azoles to control, and 20 compared between different regimens of polyenes, azoles or glucan synthesis inhibitors. Compared to control, there was no difference in all-cause mortality (RR 0.93; 95%CI 0.55– 1.58, 5 trials, Fig. 1 ). The risk for developing documented IFI was lower (RR 0.30; 95% CI 0.11–0.84, 4 trials, 4 events in the treatment group versus 15 in the control). When azoles (fluconazole, ketoconazole, itraconazole, voriconazole) were compared to polyenes (amphotericin B in 7 trials, liposomal ampho B in 1 trial) there was a trend in favour of azoles for decreased all-cause mortality (RR 0.88; 95% CI 0.71–1.08, 8 trials) and for decreased documented IFI (RR 0.67; 95% CI 0.44– 1.02, 7 trials). Adverse events which required discontinuation were less frequent in the azole group (RR 0.52; 95%CI 0.39–0.69, 6 trials).
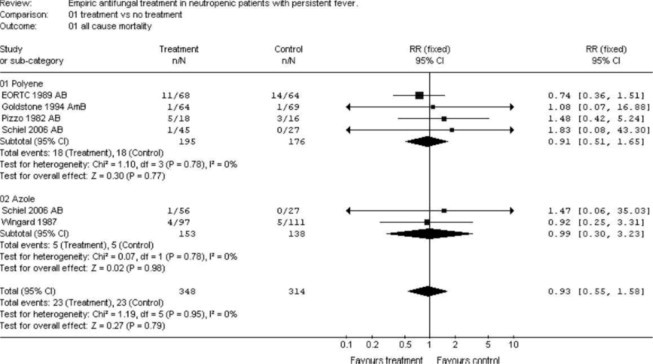
Conclusions: Our review demonstrates that empirical antifungal therapy does not reduce mortality. Although it reduces IFI, data are based on a small number of trials and events. Future trials should pursue a pre-emptive approach using improved diagnostic tools (such as galactomannan testing, high resolution CT), to identify the patients for whom antifungal treatment is warranted. The use of amphotericin B as empirical therapy seems unwarranted since it appears to be less effective than azoles, with no mortality benefit and an increased rate of side effects
P964 Antifungal prophylaxis for patients undergoing chemotherapy or haematopoeitic stem cell transplantation – meta-analysis
E. Robenshtok, A. Gafter-Gvili, M. Paul, E. Goldberg, L. Vidal, L. Leibovici (Petah-Tikva, IL)
Objectives: Fungal infections are a major cause of morbidity and mortality following intensive chemotherapy. This study aims to evaluate the prophylactic use of antifungal agents to prevent invasive fungal infections (IFI) and mortality in patients with malignancies or hematopoeitic stem cell transplantation (HSCT).
Methods: Systematic review and meta-analysis including randomised controlled trials (RCTs) comparing prophylactic antifungal treatment with placebo, no intervention, or oral polyenes in patients following chemotherapy or hematopoeitic stem cell transplantation (HSCT). Search was conducted until 2006. Primary outcomes were: all cause 30-day mortality, fungal related mortality, and invasive fungal infections (IFI). Relative risks (RR) with 95% confidence intervals (CIs) were estimated and pooled.
Results: Forty-nine RCTs were included in the analysis. Azoles (fluconazole, itraconazole, ketoconazole) or intravenous polyenes (Amphotericin B or liposomal Ampho B) were compared with placebo or no treatment in 29 trials, and with oral polyenes in 20 trials. Compared to control or oral polyene, there was a significant reduction in 30 day all-cause mortality (RR 0.80; 95% CI 0.70–0.91, fig. 1 ), fungal related mortality (RR 0.55; 95% CI 0.41–0.74), and documented IFIs (RR 0.49; 95% CI 0.40–0.60). Overall mortality was significantly reduced in HSCT recipients (RR 0.77; 95% CI 0.40–0.97, 9 trials, 1096 patients), and there was a strong trend toward reduced mortality in leukaemia patients (RR 0.84; 95% CI 0.70– 1.02, 26 trials, 3497 patients). There was a significant reduction in IFIs and fungal related mortality in all patient subgroups (HSCT, acute leukaemia, others).

Conclusions: Our review demonstrates that prophylactic antifungal therapy reduces overall mortality, fungal related mortality, and invasive fungal infections following chemotherapy or HSCT. This is the most updated systematic review of antifungal prophylaxis and the first to demonstrate a reduction in mortality. Prophylactic antifungals should be administered routinely to HSCT patients as recommended by guidelines, and should probably be considered for acute leukaemia patients.
P965 Prospective, multicentre study of caspofungin for prophylaxis in high-risk liver transplantation
J. Fortún, M. Montejo, P. Martín-Dávila, P. Muñoz, J.M. Cisneros, C. Aragón, M. Blanes, A. Ramos, J. Gavaldá, R. San Juan, P. Llinares on behalf of the Spanish Transplantation Infection Study Group (GESITRA-SEIMC)
Objective: There is significant morbidity and mortality related to invasive fungal infections (IFI) in patients undergoing liver transplantation (LT). Prevention remains elusive, especially for IFI caused by moulds. The aim of this study was to evaluate the efficacy and safety of caspofungin (CAS) as prophylaxis for IFI in the subset of LT recipients at high risk of developing IFI (HR-LT) (study sponsored by GESITRA, funded by MSD Spain).
Methods: A prospective, non-comparative, open label trial was conducted in HR-LT adult patients who received CAS prophylaxis. HR-LT included second LT due to primary dysfunction of a previous graft, renal failure and/or kidney replacement techniques, LT post-fulminant hepatitis, or the coincidence of ≥2 of the following: all-other-cause second LT, fungal colonisation, high transfusion requirements, choledocojejunostomy, biliary leak, or reintervention. Patients were expected to receive CAS at 50 mg qD (with 70 mg load) for 21 days. A succesful treatment outcome was defined as the absence of breakthrough IFI (proven or probable per EORTC/MSG criteria) during the first 100 days after the onset of caspofungin in the absence of premature discontinuation of prophylaxis because of toxicity or lack of efficacy.
Results: An interim analysis was performed on the first 41 patients enrolled in the study (enrollment plan: 70 patients). The median duration of CAS prophylaxis was 21 days (range, 5–54). The dose of CAS was lowered to 35 mg qD due to LT-related liver dysfunction in 16 (39.0%) patients. Four (9.8%) more patients discontinued CAS due to drug-related altered liver function tests after 8 to 19 days of therapy. CAS was otherwise well tolerated. Eight (19.5%) patients died of CAS-unrelated, LT-related complications other than IFI. Among survivors (up to 100-day follow-up), one (2.4%) patient experienced an invasive Mucor surgical wound infection 41 days post-CAS prophylaxis. No other patient met diagnostic criteria for possible, probable or proven IFI during CAS therapy or during the predefined 100-day follow-up period post-CAS discontinuation. Thus, the overall incidence of documented IFI was 1/41 (2.4%). Overall, CAS prophylaxis was successful in 36 patients (87.8%).
Conclusion: In this interim analysis, the outcome of CAS prophylaxis was successful in 87.8% (36/41) of the patients, with an overall incidence of documented IFI of 2.4% (1/41). These results suggest promise for the prophylactic use of CAS in HR-LT recipients.
P966 Prophylaxis for Pneumocystis pneumonia in non-HIV immunocompromised patients: systematic review and meta-analysis
H. Green, M. Paul, L. Vidal, L. Leibovici (Petah Tikva, IL)
Objectives: Pneumocystis pneumonia (PCP) is associated with significant mortality and morbidity among immunocompromised patients. Data on the benefit of PCP prophylaxis are available for HIV patients, but were not previously quantified for non-HIV immunocompromised patients. We performed a systematic review and meta-analysis to assess the effectiveness of PCP prophylaxis among non-HIV immunocompromised patients.
Methods: We searched for randomised controlled trials comparing prophylaxis using an antibiotic effective against Pneumocystis jirovecii (formerly P. carinii) versus placebo, no intervention or an antibiotic/s with no activity against P. jirovecii, among immunocompromised patients and assessing clinical PCP infections as an outcome. In addition, we included trials comparing different PCP prophylaxis regimens or administration schedules. The search included CENTRAL, PubMed, LILACS and relevant conference proceedings. No language, year or publication status restrictions were applied. Two reviewers independently searched, filtered the trials and extracted data and performed methodological quality assessment. Relative risks (RR) with 95% confidence intervals (CI) are reported. Meta-analysis was performed using the random effects model.
Results: 12 randomised trials performed between 1974–1997 were identified, including 1245 patients (50% children) with haematological cancer, bone marrow or solid organ transplantation. Compared to no PCP prophylaxis, there was a 91% reduction in the occurrence of PCP in patients receiving prophylaxis with cotrimoxazole, RR 0.09 (95%CI 0.02–0.32), numbers needed to treat 15 patients, with no heterogeneity (Figure ). No significant reduction was found in all cause mortality. PCP-related mortality was significantly reduced, RR 0.17 (95%CI 0.03–0.94). Adverse events requiring discontinuation occurred in 3.1% of adults and 0% of children and all were reversible. No differences between once daily and thrice weekly administration schedules were found.
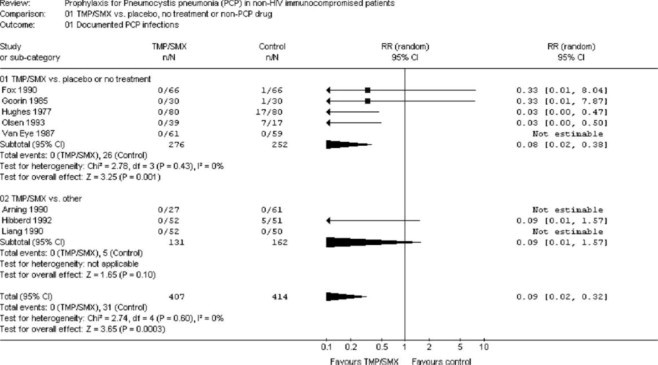
Conclusions: Balanced against severe adverse events, PCP prophylaxis is warranted when the prevalence of PCP in the treated population is above 3% for adults. The expected NNT to prevent one PCP with this baseline probability is 35 patients. Adverse events are less frequent in children, thus prophylaxis might be warranted with lower PCP prevalence rates.
P967 Association between systemic inflammation and Pneumocystis jirovecii colonisation in patients with chronic obstructive pulmonary disease
R. Morilla, L. Rivero, F. Muñoz-Lobato, M. Montes-Cano, V. Friaza, N. Respaldiza, F. Medrano, J. Varela, E. Calderón, C. de la Horra (Seville, ES)
Objectives: Chronic obstructive pulmonary disease (COPD) is characterised by significant chronic inflammation in the pulmonary compartment as well as in the circulation. Smoking is considered the major cause of the disease. However, only a small portion of all smokers develops the disease. In COPD, airway and systemic inflammatory markers increase over time and high levels of these markers are associated with a faster decline in lung function. Interest has focused on the potential role of infectious agents as cofactors in accelerating the progression of airway obstruction through increasing inflammatory response. It is known that Pneumocystis jirovecii “carriers” is present among COPD patients. In animal models, Pneumocystis causes an increase in proinflammatory cytokines. All these evidences raises the possibility that P. jirovecii colonisation might influence the natural history of this disease. This study was undertaken to identify possible inflammatory changes induced by P. jirovecii in COPD patients
Methods: The study included 51 COPD patients. The identification o f P. jirovecii was performed by Nested PCR of respiratory samples. Inflammatory responses were analyzed by ELISA commercials Kit (R&D systems) for IL-8, IL-6, and TNF-α in serum samples.
Results: Of the 51 individuals with COPD included in the study, 28 (54%) had P. jirovecii colonisation. No other infection were detected. No differences were detected due to age, gender, smoking habit, and functional respiratory parameters (FEV-1%) when comparing P. jirovecii carriers with non-carriers. However, patients with COPD colonised by P. jirovecii showed a higher level of proinflammatory cytokine than non colonised subjects (Table ).
Serum cytokine levels in chronic obstructive pulmonary disease patients with and without Pneumocystis jirovecii colonisation
| Cytokine level (pg/mL) | COPD without/with P. jirovecii colonisation |
P-value (Student's T) | |
|---|---|---|---|
| Without (N = 23) | With (N = 28) | ||
| IL-8 (mean±SD) | 13.89±13.87 | 21.26±9.25 | 0.028 |
| TNF-α (mean±SD) | 3.57±2.03 | 8.15±10.6 | 0.047 |
| IL-6 (mean±SD) | 5.34±5.45 | 16.95±25.06 | 0.038 |
Conclusions: Pneumocystis jirovecii colonisation is associated with higher proinflammatory cytokines levels. These findings suggest that P. jirovecii is an infectious agent that may play a role in the pathophysiology of COPD.
This work was funded by research grants SAF2003–06061 and FIS CP-04/217.
P968 Pulmonary surfactant-associated protein alterations in subjects colonised by Pneumocystis jirovecii with idiopathic interstitial pneumonia
V. Friaza, C. de la Horra, N. Respaldiza, M.A. Montes-Cano, F. Munoz-Lobato, R. Morilla, L. Rivero, J. Martin-Juan, J.M. Varela, E. Rodriguez-Becerra, F.J. Medrano, E.J. Calderon (Seville, ES)
Background: Idiopathic Interstitial Pneumonia (IIP) is a heterogeneous group of poorly understood diseases with uncertain etiology. All of them feature an initial inflammatory response that can lead to pulmonary fibrosis in some cases. This initial inflammatory response could be somehow trigger by an infectious agent. It is well known that pulmonary surfactant-associated proteins SP-A and SP-D play an important role in lung host defence. In the same manner, there are evidences that SP-A and SP-D regulate NF-kB and, by this pathway, are able to enhance or suppress inflammatory mediators production. It had been previously reported in vitro and in animal models, that Pneumocystis binds to surfactant proteins and could alter their expression and distribution.
Objectives: To identify possible alterations in Pulmonary Surfactant-associated Protein components in IIP Pneumocystis colonised patients.
Patients and Methods: Bronchoalveolar lavage fluid (BALF) from 40 IIP patients was collected and the identification of P. jirovecii was performed by mt LSU RNAr gene Nested-PCR. SP-A levels were determined by western-blotting following densitometry analysis using 1D Manager Software. SP-D was assayed by commercial available ELISA Kits (Human SP-D ELISA BioVendor). All data obtained were normalised with total protein concentration (quickstart Bradford dye reagent, Biorad) of each sample.
Results: P. jirovecii was identified in 14 out of 40 (35%) IIP patients. The results obtained for surfactant-associated proteins are showed in the following table :
| IIP colonised | IIP non-colonised | p | |
|---|---|---|---|
| SP-A (CN*/mg prot) | 11.77 | 15.25 | 0.295 |
| SP-D (ng/mg prot) | 406.24 | 904.86 | 0.026 |
aNormalised relative concentration.
Conclusions:
-
1
A high P. jirovecii colonisation rate in IIP patients has been found.
-
2
There is a statistical significant SP-D decrease in colonised IIP patients.
-
3
P. jirovecii could play a role in the physiopathology of this disease through interaction with pulmonary surfactant-associated proteins.
P969 Mucocutaneous manifestations of disseminated histoplasmosis in AIDS patients: special aspects in a Latin American population
V. Cunha, T. Cestari, M. Zampese, L. Goldani (Porto Alegre, BR)
Objectives: Mucocutaneous lesions in HIV-infected patients with disseminated histoplasmosis have a wide spectrum of clinical manifestations making the diagnosis very difficult. Studies have been restricted to case reports and series with small number of patients not specifically focusing on dermatological aspects of histoplasmosis. The authors describe the mucocutaneous lesions of disseminated histoplasmosis found on 36 HIV-infected patients with special attention to its morphology and clinical distribution. Diagnostic, clinical, and therapeutic implications were also analyzed in this study.
Methods: A retrospective and prospective study in 36 HIV-infected patients with mucocutaneous histoplasmosis in a tertiary-care hospital in Brazil.
Results: Eleven patients (30%) were taking antiretrovirals when diagnosed and their CD4+ cell counts ranged from 2 to 103 cells/mm3. The average number of lesions was 3 per patient, and each patient presented 1 to 7 different morphological types of lesions. Despite the variability of the lesions, papules (50%) and crusted papules (64%), and oral mucosa erosions or/and ulcers (58%) were the most frequent dermatolological lesions. A diffuse pattern of distribution of the skin lesions was found in 58% of the cases. There was a significant association between the CD4+ cell counts and the morphological variability of lesions per patient. Polymorphism of lesions was associated with higher counts of CD4+ cells.
Conclusion: Papules with or without crusts, and mucous erosions or ulcers in HIV-infected patients, along with general symptoms such as fever, malaise, anorexia, weight loss and fatigue, and CD4+ cell counts lower than 50 cells/mm3, are highly suspicious for histoplasmosis. In spite of being easily confused with other dermatological conditions, it is important to raise the possibility of histoplasmosis, especially in patients who live in endemic regions. Biopsy with microscopy and cultures of muco-cutaneous lesions are important tools for definitive diagnosis. Physicians caring for HIV-infected patients should be aware of the wide spectrum of dermatological lesions observed in disseminated histoplasmosis, besides the importance to detect and isolate the fungus in mucocutaneous tissues.
P970 Itraconazole does not affect the pharmacokinetics of micafungin (FK463)
N. Undre, P. Stevenson, A. Brooks (Munich, DE; Leeds, UK)
Objectives: To determine the pharmacokinetic interaction of intravenous micafungin and oral itraconazole following repeated administration to healthy male subjects.
Methods: This was an open-label non-randomised study in 24 healthy male subjects aged between 18 and 55 years. Single doses of 200 mg micafungin were administered as a 1-hour infusion on Days 1 to 5 and on Days 31 to 35. Oral doses of itraconazole (200 mg) were administered twice daily on Days 19 and 20 and once daily on Days 21 to 35. Subjects then received concurrent daily administration of micafungin and itraconazole for 5 days. Pharmacokinetics were evaluated on Day 5 (micafungin alone), Day 30 (itraconazole alone) and Day 35 (micafungin and itraconazole).
Results: Repeated daily administration of itraconazole had no effect on the extent of systemic exposure to micafungin as measured by AUC0–24 and Cmax, compared with that observed following administration of micafungin alone. The geometric mean ratio (90% CI) for micafungin plasma AUC0–24 and Cmax on the day of co-administration relative to administration alone were 0.958 (0.937, 0.980) and 0.991 (0.962, 1.02), respectively. Systemic exposure (AUC0–24) to itraconazole and hydroxyitraconazole, at steady state, showed a 22% and 20% increase, respectively, following co-administration of micafungin with itraconazole compared with itraconazole administered alone. The majority of adverse events reported during the study were mild in severity, and none were serious or severe.
Conclusion: There was no discernible influence of repeated daily administration of itraconazole on the pharmacokinetics of micafungin. The AUC0–24 of itraconazole and hydroxyitraconazole at steady state was approximately 20% higher following co-administration of micafungin with itraconazole compared with itraconazole administered alone. Repeated intravenous doses of micafungin were safe and well tolerated. No dose adjustments of either micafungin or itraconazole are required when the two compounds are co-administered.
P971 Amphotericin B does not affect the pharmacokinetics of micafungin (FK463)
N. Undre, P. Stevenson, D. Wilbraham (Munich, DE; London, UK)
Objectives: To investigate the pharmacokinetic interaction of intravenous (i.v.) micafungin (MICA) when given concomitantly with amphotericin B (AmB) following repeated administration to healthy male subjects.
Methods: This was an open-label non-randomised study. A total of 26 healthy male subjects aged between 18 and 55 years received a daily i.v. infusion of MICA (200 mg) over 1 hour for 5 days. After a 3-day washout, subjects received a daily 4-hour i.v. infusion of AmB (250 μg/kg/day) for 5 days. Subjects then received concurrent daily administration of MICA (200 mg) and AmB (250 μg/kg/day) for 5 days.
Results: Following repeated once-daily i.v. co-administration of MICA and AmB, there was no difference in the extent of systemic exposure to MICA, as measured by AUC0–24 and Cmax, compared to that observed following administration of MICA alone. The geometric mean ratio (90% CI) for MICA plasma AUC0–24 and Cmax after 5 days of coadministration relative to administration alone were 0.98 (0.93, 1.03) and 1.02 (0.99, 1.06), respectively. The systemic exposure to AmB increased over time by approximately 30%. This accumulation was expected based on its long half-life. These data suggest that repeated administration of MICA does not affect the pharmacokinetics of AmB.
Conclusion: There was no discernible pharmacokinetic interaction between MICA and AmB following repeated daily co-administration of both medications. Repeated i.v. doses of MICA were safe and well tolerated. Concurrent dosing with MICA and AmB was less well tolerated than either drug alone, probably owing to an accumulation of AmB towards steady state.
P972 A review of the safety of micafungin compared with liposomal amphotericin B in a large, phase III trial in patients with invasive candidiasis and candidaemia
A.J. Ullmann, J. De Waele, R. Betts, C. Rotstein, M. Nucci, O.A. Cornely (Mainz, DE; Ghent, BE; Rochester, US; Hamilton, CA; Rio de Janeiro, BR; Cologne, DE)
Objective: Liposomal amphotericin B (L-AmB) is associated with nephrotoxicity. The objective of this review is to present data on the safety profile of micafungin (MICA) compared with L-AmB in patients with invasive candidiasis and candidaemia.
Methods: In a phase III, randomised, double-blind study, patients with confirmed candidaemia or invasive candidiasis were randomised 1:1 to receive MICA (100 mg/day) or L-AmB (3 mg/kg/day). Safety was assessed in terms of observed adverse events (AEs) of any causality and AEs related to treatment. There were two predefined safety endpoints: estimated glomerular filtration rate (eGFR) and incidence of infusion-related reactions.
Results: In the adult population, the treatment groups (MICA: n = 264; L-AmB: n = 267) were well balanced at baseline with respect to age, gender and race. Neutropenia was present at baseline in 12.9% of MICA and 10.5% of L-AmB patients. At baseline, APACHE II scores of ≤20 were recorded in 72.5% of MICA patients and 76.0% of L-AmB patients. Treatment groups were well balanced with regard to underlying conditions, the most common being haematological disorders, solid organ tumours and diabetes mellitus. Mean treatment days for MICA and L-AmB were 14.1±8.0 and 13.9±8.5 days, respectively. AEs (any causality) were seen in 92.8% of MICA patients and 94.4% of L-AmB patients. Common treatment-related AEs are summarised in the table .
Common treatment-related AEs (>3 patients in either group)
| MICA (N = 264), n (%) | L-AmB (N = 267), n (%) | P-value | |
|---|---|---|---|
| Fever | 23 (8.7) | 36 (13.5) | NS |
| Chills | 3 (0.8) | 17 (6.4) | <0.05* |
| Back pain | 1 (0.4) | 12 (4.5) | <0.05* |
| Nausea | 12 (4.5) | 10 (3.7) | NS |
| LFT abnormal | 11 (4.2) | 5 (1.9) | NS |
| Vomiting | 9 (3.4) | 9 (3.4) | NS |
| Hypokalaemia | 18 (6.8) | 32 (12.0) | NS |
| Increased creatinine | 5 (1.9) | 17 (6.4) | <0.05* |
| Increased SGPT | 5 (1.9) | 1 (0.4) | NS |
| Increased LD | 4 (1.5) | 3 (1.1) | NS |
| Dyspnoea | 3 (1.1) | 6 (2.2) | NS |
| Rash | 4 (1.5) | 7 (2.6) | NS |
| Skin erythema | 1 (0.4) | 4 (1.5) | NS |
NS: not significant;
Fisher exact test.
Treatment-related renal AEs with respect to creatinine increase were seen significantly less often with MICA than with L-AmB (p < 0.05), and a lower incidence of chills and back pain was observed with MICA compared with L-AmB (p < 0.05). On the predefined safety endpoint of eGFR, the mean peak decrease compared with baseline was −1.3 ml/min/1.73m2 for MICA and −19.1 ml/min/1.73m2 for L-AmB, a difference which was statistically significant (p < 0.001). Furthermore, 88.0% of L-AmB patients experienced a decrease in eGFR during week 1. Significantly fewer MICA patients than L-AmB patients experienced treatment-related infusion-related reactions (MICA 17.0% versus L-AmB 28.8%; p ≤ 0.001).
Conclusion: This study showed overall safety advantages for MICA over L-AmB in the treatment of invasive candidiasis/candidaemia, with significant differences in eGFR and in acute infusion-related reactions.
P973 An analysis of the efficacy of micafungin in Candida infections caused by non-albicans Candida species in phase III trials
O.A. Cornely, J. De Waele, R. Betts, C. Rotstein, M. Nucci, L. Kovanda, C. Wu, A.J. Ullmann (Cologne, DE; Ghent, BE; Rochester, US; Ontario, CA; Rio de Janeiro, BR; Deerfield, US; Mainz, DE)
Objective: In recent years, the prevalence of non-albicans Candida species has increased. Some antifungal agents, such as fluconazole, are limited by intrinsic and acquired resistance in non-albicans Candida species. The objective of this study is to evaluate the efficacy of micafungin (MICA) against fungal infections caused by non-albicans Candida species in two comparative phase III trials.
Methods: Two phase III, randomised, multicentre, active-controlled trials conducted in patients with invasive candidiasis/candidaemia were examined to assess the efficacy of MICA in non-albicans Candida infections. Both studies were conducted in adult patients with confirmed Candida infection at baseline. One trial compared MICA at 100 mg/day (MICA100) with liposomal amphotericin B (L-AmB) at 3 mg/kg/day; the other trial compared MICA100 and MICA at 150 mg/day (MICA150) with caspofungin (CAS) at 50 mg/day (70 mg loading dose on Day 1). The primary efficacy endpoint was treatment success at the end of therapy, defined as both positive clinical and positive mycological response as assessed by the investigator.
Results: The primary efficacy population (per protocol set; PPS) of the two-arm trial included 392 patients (MICA100: n = 202; L-AmB: n = 190). In the three-arm trial, the primary efficacy population was the full analysis set (FAS), which included 593 patients (MICA100: n = 191; MICA150: n = 199; CAS: n = 188). The key efficacy results are shown in the table . In the two-arm trial, MICA showed equivalent treatment success rates to L-AmB in all Candida species isolated. A similar result was seen in the three-arm trial, with the two MICA doses being at least as effective as CAS for all species isolated. There were no marked differences in treatment success with MICA between infections caused by C. albicans and those caused by non-albicans Candida species.
| Treatment success, n (%) |
|||||
|---|---|---|---|---|---|
| Two-arm trial (PPS) |
Three-arm trial (FAS) |
||||
| MICA100 | L-AmB | MICA100 | MICA150 | CAS | |
| C. albicans | 76/86 (88.4) | 75/84 (89.3) | 71/93 (76.3) | 71/102 (69.6) | 62/84 (73.8) |
| Non-albicans Candida species | 113/126 (89.7) | 100/112 (89.3) | 84/111 (75.7) | 74/107 (69.2) | 85/122 (69.7) |
| C. glabrata | 19/23 (82.6) | 12/15 (80.0) | 24/28 (85.6) | 30/34 (88.2) | 22/33 (66.7) |
| C. tropicalis | 48/52 (92.3) | 41/43 (95.3) | 21/31 (67.7) | 20/33 (60.6) | 24/32 (75.0) |
| C. parapsilosis | 33/37 (89.2) | 26/30 (86.7) | 23/30 (76.7) | 15/21 (71.4) | 27/42 (64.3) |
| C. krusei | 5/6 (83.3) | 6/7 (85.7) | 6/8 (75.0) | 5/8 (62.5) | 3/4 (75.0) |
| C. guilliermondii | 4/4 (100) | 4/4 (100) | 3/3 (100) | 2/2 (100) | 1/1 (100) |
| C. lusitaniae | 1/1 (100) | 2/2 (100) | 3/3 (100) | 0/1 | 3/3 (100) |
| C. famata | 3/3 (100) | 1/1 (100) | − | − | − |
Only Candida species observed in ≥3 patients in either study are included in the table. Patients could be diagnosed with more than one organism at baseline.
Conclusion: MICA demonstrated broad-spectrum efficacy against Candida infections. MICA is a valuable treatment option in invasive candidiasis and candidaemia.
Clinical mycology I
P974 Antifungal chemoprophylaxis in the very low birth weight neonates: a prospective randomised controlled study
N. Klimko, A. Kolbin, N. Shabalov (Saint-Petersburg, RU)
Objectives: Invasive candidiasis is associated with high morbidity and mortality in preterm neonates. We have carried out analysis of clinical efficacy and safety of fluconazole as used for prophylaxis of invasive candidiasis in the very low birth weight neonates.
Methods: We conducted a prospective, analytical, randomised, non-parallel, controlled, clinical trial over 12-month period in 104 preterm neonates with birth weight <1500 g. Historical control group consist of 249 preterm neonates with birth weight <1500 g. The 104 neonates were randomly assigned during the first five days of life to receive either intravenous fluconazole at a dose of 3 mg/kg or at a dose of 6 mg/kg. On an average, one course of prophylaxis required 14 or 15 administrations of fluconazole in the course of 30 days.
Results: Analysis of the effect of the studied antifungal prophylaxis on the incidence of invasive candidiasis in very low birth weight neonates has not revealed any significant differences between the used doses ((3 mg/kg and 6 mg/kg) of fluconazole (2% vs 3.7%, p = 0.67). On the other hand, the prophylactic use of fluconazole in the specified doses was accompanied by significant decrease in the invasive candidiasis incidence as compared to control group (2.9% vs 10.8%, p = 0.004). The study has demonstrated that the prophylactic use of fluconazole at a dose of 3 or 6 mg/kg was accompanied by a significant decrease in mortality due to invasive candidiasis (0% vs 10%, p < 0.001). No pt had a serious AE or discontinued fluconazole due to toxicity. Evaluation of the isolated Candida species has shown that C. albicans dominated in both the prophylactic groups and the control group. In the course of the study, no effects of the specified doses of fluconazole prophylaxis on the selection of resistant strains of Candida were revealed.
Conclusion: Prophylactic use of fluconazole at a dose of 3 in the very low birth weight newborns is safe and decrease incidence of invasive candidiasis and attributable mortality.
P975 Liposomal amphotericin B followed by voriconazole for secondary antifungal prophylaxis in paediatric allogeneic haematopoietic stem cell recipients
K. Allinson, H. Kolve, H.G. Gumbinger, H.J. Vormoor, K. Ehlert, A.H. Groll (Münster, DE)
Objectives: Presumed or proven invasive pulmonary aspergillosis (IPA) is an important cause of infectious morbidity in patients with acute leukaemia. Although prior IPA is no longer a contraindication for subsequent allogeneic hematopoietic stem cell transplantation (HSCT), its successful management during granulocytopenia and immunosuppression remains challenging.
Methods: In the absence of an evidence-based approach, 11 adolescents (11 to 18 yrs) with acute leukaemia and a history of antecedent possible (4) or probable (7; modified EORTC/MSG criteria, Herbrecht et al. 02) IPA received liposomal amphotericin B (LAMB; 1 mg/kg QD) from the start of the conditioning regimen until engraftment and ability to take oral medication, followed by oral voriconazole (VCZ; 200 mg BID) until the end of the at-risk period. Nine patients had a good partial (>50% reduction in pulmonary infiltrates), and two a complete response to standard antifungal agents prior to admission for HSCT.
Results: Granulocyte engraftment occurred after a median of 16 days (r, 13–22). In addition to standard post transplant immunosuppression with methotrexate and cyclosporin A, 3 patients received short (<14), and 4 prolonged courses (r, 32–89 days) of methylprednisolon for acute grade I or II GVHD. The median duration of IV treatment with LAMB was 30 days (r, 19–36), followed by a median of 152 days (r, 19– 210) of oral VCZ. LAMB was discontinued early in 1 patient and VCZ was transiently or permanently discontinued due to adverse events/new contraindications in 2 and 2 patients, respectively. At +180 days post transplant, 8 patients were alive, 6 with complete, and 1 each with near complete and ongoing resolution of pulmonary infiltrates; all but one were in continuing haematological remission. Three patients had succumbed either to recurrent leukaemia (2) or refractory graft failure (1); while one of these patient had maintained a complete response, 2 died with secondary possible (1) or probable (1) IPA. Both patients had discontinued VCZ early and developed IPA in lung areas involved during the primary episode.
Conclusions: This paediatric series supports the notion that successful secondary antifungal prophylaxis for possible or probable IPA can be safely and effectively achieved in allogeneic HSCT. In the absence of chronic GVHD, reactivation or breakthrough infection appeared to be associated with recurrent leukaemia/graft failure and shorter duration of post-engraftment prophylaxis.
P976 Prospective, randomised, double-blind, placebo-controlled phase III trial on voriconazole as primary antifungal prophylaxis during induction chemotherapy for acute myelogenous leukaemia
J. Vehreschild, S. Mousset, M. Hummel, D. Arenz, G. Wassmer, U. Harnischmacher, A. Ullmann, D. Buchheidt, A. Böhme, O. Cornely (Cologne, Frankfurt, Mannheim, Mainz, DE)
Background: Invasive fungal infections remain a frequent cause of morbidity and mortality in long-term neutropenic patients. The availability of tolerable broad-spectrum antifungals like voriconazole reignited the discussion about the best timing of antifungal therapy. We therefore conducted a trial to analyze the efficacy and safety of voriconazole in the prevention of lung infiltrates during induction chemotherapy for acute myelogenous leukaemia (AML).
Methods: Prospective, randomised, double-blind, placebo-controlled phase III trial in AML patients undergoing first remission induction chemotherapy. Voriconazole 200 mg BID or placebo were administered until detection of a lung infiltrate or end of neutropenia. Primary objective was the incidence of lung infiltrates until day 21, i.e. the start of 2nd induction chemotherapy.
Results: 25 patients were randomised to voriconazole (N = 10) or placebo (N = 15). Incidence of lung infiltrates until day 21 was 0 (0%) in the voriconazole vs. 5 (33%) in the placebo treatment arm (chi2 test, p = 0.04). Incidences of chronic disseminated candidiasis at 4 week follow-up was 0 (0%) in the voriconazole vs. 4 (27%) in the placebo treatment group (chi2 test, p = 0.07).
Conclusion: In this randomised, double-blind, placebo-controlled trial voriconazole 200 mg BID PO reduced the incidence of lung infiltrates during AML induction chemotherapy. There was a trend towards a lower incidence of chronic disseminated candidiasis in the voriconazole arm.
P977 Safety and tolerability of voriconazole compared to amphotericin B followed by fluconazole in patients with candidaemia and baseline renal insufficiency
A.M.L. Oude Lashof, J.D. Sobel, M. Ruhnke, P.G. Pappas, C. Viscoli, H.T. Schlamm, J.H. Rex, B.J. Kullberg (Nijmegen, NL; Berlin, DE; Genoa, IT; Birmingham, New York, Houston, Detroit, US)
Background: Acutely ill patients with candidaemia frequently have other co-morbidities such as renal insufficiency. Voriconazole is approved for treatment of candidaemia in non-neutropenic patients but use of the intravenous formulation with sulfobutyl ether β-cyclodextrin (SBECD) is restricted in patients with renal insufficiency. We evaluated the use of i.v. voriconazole (formulated with SBECD) in candidaemic patients with moderate to severe renal insufficiency, and we compared treatment outcome and safety to patients who received a short course of amphotericin B followed by fluconazole (AmB/flu).
Methods: We retrospectively reviewed data from the previously published trial (Lancet 2005; 366: 1435–42) of voriconazole compared to a regimen of AmB followed by fluconazole for first-line treatment of candidaemia in 370 non-neutropenic patients. From this database we collected treatment outcome, survival, safety, and tolerability data from the patients with moderate to severe renal insufficiency at study entry, defined as a baseline creatinine clearance of 30–50 ml/min (moderate) or <30 ml/min (severe).
Results: 58 patients were included; 41 were randomised to intravenous voriconazole with moderate (32) or severe (9) renal impairment at baseline, and 17 patients were randomised to amphotericin B with moderate (13) or severe (4) renal impairment. Median duration of treatment was 14 days for voriconazole (of which 7 days iv) and 3 days for AmB, followed by 11 days of flu. Despite the short duration of exposure, worsening of renal function or newly emerged renal adverse events were reported in 53% of amphotericin B-treated patients compared to 39% of voriconazole-treated patients.
During treatment, median serum creatinine decreased in the voriconazole arm, whereas creatinine increased in the AmB/flu arm, before return to baseline at week 3. Outcome at end of therapy was successful in 63.4% on voriconazole and 64.7% on AmB/flu, and sustained success rate at 12 wks follow-up was 31.7% (voriconazole) vs. 23.5% (AmB/flu). All-cause mortality at 14 weeks was 49% in the voriconazole arm compared to 65% in the amphotericin B arm.
Conclusions: Intravenous voriconazole (with SBECD) was effective in patients with moderate to severe renal insufficiency and candidaemia, and tended to be associated with less acute renal toxicity and improved survival compared to a short course of conventional amphotericin B.
P978 Efficacy and safety of caspofungin in solid organ transplant recipients
J. Petrovic, A. Ngai, S. Bradshaw, A. Williams-Diaz, A. Taylor, C. Sable, N. Kartsonis (Rome, IT; West Point, US)
Objective: Efficacy & safety data for the echinocandins in solid organ transplant (SOT) recipients are limited. We reviewed the experience from all SOT patients (pts) receiving caspofungin (CAS) therapy (Rx) for an invasive fungal infection (IFI).
Methods: Data in SOT pts receiving CAS are available from 3 clinical trials, including 2 noncomparative clinical trials (a salvage invasive aspergillosis [IA] study [Protocol 019, P019] & a non-fungemic invasive candidiasis [IC] study [P045]) & 1 comparator-controlled study (CAS vs. amphotericin B [AmB] in IC [P014]). CAS was administered at doses ranging from 50 to 100 mg/day. Efficacy was assessed in all pts at end of CAS Rx (EOT). For IC pts, a favourable overall response required complete symptom/sign resolution & Candida eradication (or radiographic resolution). For IA pts, a favourable response required clinically meaningful improvement in attributable symptoms/signs & relevant radiographic findings. Adverse events (AE) & lab data were collected from all pts. Investigators identified the seriousness, causality, & action on study Rx for all clinical & lab AE noted during Rx & for ≥14 days postRx.
Results: 22 SOT pts, aged 34–67 years, were identified. All had proven IC (6 pts) or proven/probable IA (16 pts) & received ≥1 dose of CAS Rx. IC: Mean APACHE II score was 18 (range 6–27). All pts received CAS as primary Rx. Sites of IC included blood (4), peritoneal fluid (1), & liver abscess (1). CAS success at EOT was 83% (5/6), with responses seen across Candida spp. Success by SOT type was kidney 4/5 & liver 1/1. IA: All 16 pts had pulmonary IA (4 definite, 12 probable) & received CAS as salvage Rx. CAS success at EOT was 50% (8/16), with responses seen for both definite (3/4) & probable IA (5/12). Success by SOT type was: heart 2/2, heart/lung 0/2, kidney 3/3, liver 1/3, & lung 2/6. Outcome was not influenced by CAS dose. Safety: CAS, dosed for 2 to 162 (mean 36.8) days, was well tolerated. No pt had a serious drug-related (DR) adverse event (AE) or discontinued CAS due to toxicity. Incidence of DR clinical AE was 9% & DR lab AE 23%. The 30-day overall mortality was 32% (IC 0% & IA 41%).
Conclusion: Based on the limited data, CAS appears to be an effective & well tolerated option for the treatment of IC & IA in SOT recipients.
P979 Safety of high-dose caspofungin in 80 cancer patients and haematopoietic stem cell transplant recipients with invasive fungal infections
A. Safdar, G. Rodriguez, F. Al Akhrass, D. Kontoyiannis, H. Kantarjian, R. Champlin, I. Raad (Houston, US)
Background: We recently reported safety of high-dose caspofungin (HD-CAP) (100 mg daily) in 34 cancer patients (Safdar A, et al. ICAAC 2005). This study was performed to evaluate safety of HD-CAP in additional 80 patients at our cancer centre.
Methods: Retrospective review of adverse events (AE) was undertaken after obtaining IRB approval in patients who have received 3 or more days of HD-CAP between August 2004 and April 2006. All values are given in median ± standard deviations.
Results: Leukaemia was common underlying malignancy (63%) and age was 52±14 years. Over half of the patients (54%) had refractory or relapsed cancer. Among 37 (46%) haematopoietic stem cell transplant (HSCT) recipients, 29 (36%) had received allogeneic graft transplantation; 12 of 17 patients with GVHD were receiving treatment for chronic GVHD. APACHE II score was 12±5 (range, 2 to 25) and 34% had proven invasive fungal infections (IFI). Ninety-one percent received HD-CAP in combination with another antifungal agent including, voriconazole (34%) and AmBisome (36%). The duration of HD-CAP was 12±24 (range, 3 to 161) doses. In attached table , interval increase in values between start and end of HD-CAP therapy are shown. In 2 patients (3%) serum bilirubin level doubled after receiving 5 and 22 doses of HD-CAP therapy; levels return to normal after treatment was discontinued. One patient (1%) developed reversible paresthesia.
| Parameter | <4 week therapy | 4–23 weeks therapy | P value |
|---|---|---|---|
| Creatinine (mg/dL) | 0.5±0.7 | 0.2±0.3 | 0.8 |
| Total bilirubin (mg/dL) | 0.2±0.9 | 0.5±1.8 | 0.9 |
| Alanine aminotransferase (IU/L) | 4±10 | 38±29 | 0.6 |
| Alkaline phosphatase (IU/L) | 46±201 | 26±39 | 0.3 |
Conclusions: Extended therapy with HD-CAP was tolerated without serious hepatic or renal impairment. Reversible hyperbilirubinaemia may infrequently occur during HD-CAP therapy.
P980 Liposomal amphotericin B standard dose in combination with caspofungin versus liposomal amphotericin B high dose regimen for the treatment of invasive aspergillosis in immunocompromised patients: randomised pilot study (Combistrat Trial)
D. Caillot, A. Thiébaut, R. Herbrecht, A. Pigneux, S. de Botton, M. Attal, F. Bernard, J. Larché, S. Alfandari, F. Monchecourt, L. Mahi (France, Lyon, Strasbourg, Bordeaux, Lille, Montpellier, Nancy, Paris, FR)
Objectives: Antifungal Monotherapy for Invasive Aspergillosis (IA) treatment is still associated with substantial mortality. Preclinical studies data suggest that combination therapy with polyenes and echinocandins may have additive activity against Aspergillus species.
Methods: We assessed the efficacy of Liposomal Amphotericin B standard dose (3 mg/kg/d) in combination with caspofungin standard dose (COMBO) versus Liposomal Amphotericin B high dose (10 mg/kg/d) regimen (L-AMB HD). Modified EORTC-MSG criteria were used to include patients with proven or probable IA. Study drug was administered until investigator-defined end of treatment (EOT). The primary endpoint was favourable overall response (FOR) defined as partial or complete response assessed at EOT. Survival was followed up to 12 weeks. Centrally data review committee confirmed IA diagnosis.
Results: 30 patients with pulmonary IA diagnosis were assessed: 15 received COMBO and 15 L-AMB HD. Groups were well matched in terms of demographics and neutropenia. Median duration of study drug treatment was COMBO 18 days [10–35] and L-AMB HD 17 days [4–24]. At EOT, FOR was COMBO 67% vs. L-AMB HD 27% (p = 0.028). At week 12, FOR was 80% for COMBO and 67% for L-AMB HD (NS) and survival 100% COMBO vs. 86.7% L-AMB HD (NS). Infusion related reactions were observed in 3 L-AMB HD patients (20%) (Flush, cervical or thoracic pain, chills, nausea). Nephrotoxicity (serum creatinine >2x baseline) occurred in 4 L-AMB HD patients (23%) vs. 1 COMBO (7%). Hypokalaemia (K+ <3.0 mmol/L) developed in 3 L-AMB HD (20%) vs. 2 COMBO (13%).
Assessment of volume and number of lesions observed in chest CT-Scan showed an increase and worsening at day 7 then a decrease until EOT (see Table 1 ). These data support the initial diagnosis and allowed an optimal follow up of the pulmonary IA.
Table 1.
Characteristics of chest CT-scan evolution
| Screening | D7 | D14 | EOT | |
|---|---|---|---|---|
| Numbers of patients with reviewed scan | 29 | 25 | 13 | 25 |
| Median number of IA lesions (range) | 2.0 (1.0; 12.0) | 3.0 (1.0; 10.0) | 2.0 (1.0; 10.0) | 2.0 (0.0; 7.0) |
| Median volume of IA lesion (cm3) (range) | 20.8 (0.9; 216.8) | 48.5 (3.0; 471.8) | 33.0 (9.3; 361.2) | 25.5 (1.9; 216.6) |
| Median volume of lesions/screening (%) | 100 | 165.8 (27.1; 5110.9) | 143.7 (9.8; 1323.5) | 75.3 (4.5; 23113.1) |
Conclusion: In immunocompromised patients with underlying haematological malignancies, Liposomal Amphotericin B at 3 mg/kg/d in combination with caspofungin showed promising results in initial treatment of invasive aspergillosis. A large randomised controlled trial is required to confirm these data. In addition the systematic use of chest CT-Scan appeared to be a useful tool to follow the evolution of pulmonary IA.
P981 Efficacy of micafungin in patients with deep, invasive Candida infections
A.J. Ullmann, J. De Waele, R. Betts, C. Rotstein, M. Nucci, L. Kovanda, C. Wu, D. Buell, H. Diekmann, S. Koblinger, O.A. Cornely (Mainz, DE; Ghent, BE; Rochester, US; Hamilton, CA; Rio de Janeiro, BR; Deerfield, US; Munich, Cologne, DE)
Objectives: The objective of this study was a post hoc analysis of efficacy data from two double-blind, randomised, phase III trials with the echinocandin micafungin (MICA) in patients with invasive candidiasis (IC).
Methods: We reviewed data on patients with a baseline diagnosis of IC other than candidaemia enrolled in two double-blind, randomised, phase III MICA trials. A three-arm trial compared intravenous MICA 100 mg/day (MICA100) or 150 mg/day (MICA150) with intravenous caspofungin (CAS) (70 mg/day loading followed by 50 mg/day). A two-arm trial compared MICA100 with liposomal amphotericin B (L-AmB) 3 mg/kg/day. The studies shared similar endpoints: the primary efficacy endpoint was treatment success at the end of therapy, defined as both clinical and mycological response as assessed by the investigator. In this analysis, we reviewed data from the modified full analysis set (mFAS) for each of the trials, which for the three-arm trial included all randomised patients who received at least one dose of study drug and had documented Candida infection at baseline, confirmed by the data review panel. In this trial, the mFAS excluded patients with Candida-associated endocarditis, osteomyelitis or meningitis. For the two-arm trial, the mFAS included all patients who received at least one dose of study drug and had a confirmed diagnosis of IC or candidaemia.
Results: IC was observed in 85 of 578 patients (14.7%) in the three-arm trial and in 84 of 494 patients (17.0%) in the two-arm trial. Treatment success rates by infection site are shown in the table . MICA150 disseminated results were influenced by APACHE scores (2, 13, 14 for successes vs 12, 13, 16, 21, 23, 26, 29, 33 for failures). Otherwise, MICA was at least as effective as CAS in the three-arm trial in IC patients and not affected by the site of infection. In the two-arm trial, MICA was at least as effective as L-AmB in IC patients, and showed consistent efficacy independent of the infection site.
| Infection site | Treatment success (%) |
|||||
|---|---|---|---|---|---|---|
| Three-arm trial |
Two-arm trial |
Pooled data on MICA 100 | ||||
| MICA100 (n = 199) | MICA150 (n = 191) | CAS (n = 188) | MICA100 (n = 247) | L-AmB (n = 247 | ||
| Invasive candidiasis | 22/28 (78.6) | 16/30 (53.3) | 17/26 (65.4) | 28/40 (70.0) | 22/38 (57.9) | 50/68 (73.5) |
| Abscess | 5/5 (100) | 5/6 (83.3) | 6/9 (66.7) | 5/5 (100) | 4/6 (66.7) | 10/10 (100) |
| Bone | − | − | − | − | 1/1 (100) | − |
| Chorioretinitis | 0/2 | 1/1 (100) | − | − | − | 0/2 |
| Disseminated | 6/8 (75.0) | 3/11 (27.3) | 5/8 (62.5) | 7/10 (70.0) | 6/10 (60.0) | 13/18 (72.2) |
| Endocardium | − | − | −1/2 (50.0) | 3/5 (60.0) | 12 (50.0) | |
| Endophthalmitis | 4/4 (100) | 1/3 (33.3) | 1/1 (100) | − | − | 4/4 (100) |
| Kidney | − | − | − | − | 0/1 | − |
| Organ | −1/1 (100) | − | − | − | − | |
| Other | 3/3 (100) | 1/1 (100) | 3/3 (100) | 1/3 (33.3) | 0/1 | 4/6 (66.7) |
| Peritonitis | 4/6 (66.7) | 4/7 (57.1) | 2/5 (40.0) | 14/20 (70.0) | 8/14 (57.1) | 18/26 (69.2) |
Conclusion: In these trials, MICA was associated with efficacy comparable to both CAS and L-AmB in patients with IC and thus represents a valuable treatment option.
P982 Invasive cryptococcosis associated with treatment with adalimumab, a tumour necrosis factor alpha-antagonist agent
J.P. Horcajada, J.L. Peña, V.M. Martinez-Taboada, T. Pina, I. Belaustegui, M.E. Cano, J.D. García-Palomo, M.C. Fariñas (Santander, ES)
Objectives: Adalimumab is a human monoclonal anti-TNF-alpha antibody approved in 2002 for treatment of rheumatoid arthritis (RA). There is some evidence of an increased risk of serious infections in patients treated with anti-TNF antibody therapy. We report the first case of invasive cryptococosis in a patient under adalimumab therapy for RA.
Methods: Chart review and case report.
Results: A 69-year-old caucasian woman with RA was referred for severe acute inflammation of the second finger of the left hand. She was receiving adalimumab 40 mg sq every other week since a year ago. Examination revealed severe tenosynovitis of the digital flexor tendon with intense edema and compartimental signs. Early surgical decompression was performed. Intraoperative findings were extensive subcutaneous celullitis with infiltration of both vasculonervous bundles, and flexor tendon synovitis. Culture of extracted material identified Cryptococcus neoformans susceptible to amphotericin B, azoles and flucytosine, in four separated samples, including biopsy of subcutaneous tissue. Cerebrospinal fluid analysis was normal. Serum cryptococcal latex and HIV serology were negative. Nuclear Magnetic Resonance of the finger did not show arthritis nor osteomyelitis. Adalimumab therapy was discontinued. Intravenous liposomal amphotericin-B 300 mg qd and intravenous flucytosine 2.5 gr every 8 hours were administered for 7 days. Once systemic and central nervous system involvement were discarded, therapy was changed to intravenous fluconazole 400 mg bid, with a slow clinical response, but with reduction of inflammatory signs. However as the extent of residual soft tissue necrosis was very important reconstructive surgical solution was rejected and amputation of the second finger was performed on the third week after admission. Pathologic exam revealed chronic necrotising granulomatous inflammation with typical encapsulated fungal forms of Cryptococcus spp. inside multinucleated giant cells These forms were observed with hematoxillin-eosin and Mayer mucicarmine staining.
Conclusions: The first case of cryptococosis in a patient taking adalimumab is reported. Local manifestations were severe and response to antifungal drugs was slow, with loss of the affected finger. Active surveillance is required to detect severe opportunistic infections in these patients in order to start antimicrobial therapy earlier.
P983 Is the determination of galactomannan in non-haematological patients helpful for the diagnosis of invasive aspergillosis? The value of determination in serum of patients with clinical isolation of aspergillus
J. Guinea, J. Jensen, M. Rivera, R. Alonso, T. Peláez, P. Muñoz, M. Torres-Narbona, E. Bouza (Madrid, ES)
Objectives: Haematological patients have traditionally been the highest risk population for developing invasive aspergillosis (IA) and the detection of galactomannan (GM) in serum in these patients has been widely evaluated. However, the value of the test in non-haematological patients has received little attention. We studied the value of detecting GM in the serum of non-haematological patients with isolation of Aspergillus in clinical samples for the diagnosis of IA.
Methods: We followed up those patients admitted to non-haematological wards with one or more clinical isolations of Aspergillus in our institution from March 2003 to August 2006. The patients were classified according to EORTC standards (Ascioglu, CID 2002). A serum sample for GM detection was requested under non-surveillance conditions in those patients with a positive culture for Aspergillus. GM was detected by the Platelia Aspergillus test (Bio-Rad). A detection of GM ≥1 ng/mL was considered positive.
Results: During the study period, 75 different patients had an isolate of Aspergillus and a serum sample processed for GM detection. Admitted patients were located as follows: intensive care units 28 (37.3%), medical wards 38 (50.7%), and surgical wards 9 (12%). Of the 75 patients studied, 11 (14.7%) had proven or probable invasive aspergillosis. The underlying conditions of the patients were COPD 5 (50%), HIV infection 1 (10%), lymphoma 1 (10%), liver transplant 1 (10%), solid malignancies 1 (10%) and corticosteroid treatment 1 (10%). The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the detection of GM in serum for the diagnosis of IA were 50%, 98.5%, 83.3% and 92.7%, respectively (P < 0.001).
Conclusions: The detection of GM in the sera of non-haematological patients with a clinical isolate of Aspergillus is useful for the diagnosis of IA with PPV and NPV above 83% and 92%, respectively. Jesús Guinea Pharm D, PhD, is contracted by the Fondo de Investigación Sanitaria (FIS), contract number CM05/00171.
P984 111-indium labelled leukocyte renal scintigraphy in patients with candiduria: preliminary results of a prospective study
M. Gutiérrez-Cuadra, J.P. Horcajada, I. Martínez, C. Salas, D. García-Palomo, J.M. Carril, M.C. Farinas (Santander, ES)
Objectives: to study possible renal parenchyma involvement in patients with candiduria by means of 111-Indium labelled leukocyte scintigraphy.
Methods: prospective study of all candidurias detected between March 2006 and October 2006 in an university tertiary care hospital. Candiduria was defined as the presence of >10.000 CFU/mL of Candida sp. in urine culture. A second urine culture with Candida sp. and/or an urinalysis with abundant yeasts was needed to include the patients. Exclusion criteria: presence of bacteria in urine culture, ICU patients, advanced nephropathy, antifungal treatment, and pregnancy. 111-Indium labelled leukocyte scintigraphy was performed 24–48 h. after confirmation of candiduria. Positive scintigraphy was considered when renal area uptake was higher than the background. A renal CT scan was performed in patients with positive scintigraphy. Patients with positive scintigraphy were treated with oral fluconazole 200 mg qd for 14 days. A control scintigraphy was performed one month later.
Results: 192 candidurias in 153 patients were analyzed. 139 episodes were excluded and in the rest 53 candiduria was confirmed in 13 patients. Scintigraphy was performed in 8 patients. 5 were men and 3 women. Mean age was 68 years. Urinary tract symptoms were absent in all studied patients. 4 (50%) showed bilateral renal uptake before antifungal treatment. CT scan was performed in 2 of them and it was normal. Control scintigrapy after treatment was positive in the 3 patients in which it was performed. Positive and negative patients were comparable in terms of baseline conditions and the presence of indwelling urinary catheters. 3 (75%) of positive patients and 1 (25%) of the negative patients had low-grade fever (p = 0.2).
Conclusions: Half of the studied patients with candiduria showed renal uptake in 111-Indium labelled leukocyte scintigraphy. Uptake persisted one month later and after antifungal treatment. Although more data are needed, subclinical pyelonephritis could be very frequent in patients with candiduria. This could explain the high frequency of recurrencies in these patients and could have therapeutical implications.
P985 Pharmacokinetic interaction and dose adjustment for combined use of voriconazole and efavirenz
B. Damle, R. LaBadie, P. Crownover, P. Glue (New York City, US)
Objectives: Efavirenz (EFV) is an inducer of CYP450 isozymes while voriconazole (VORI) is a substrate and inhibitor of CYP450s. Coadministration of standard doses of VORI and EFV results in a substantial decrease in VORI levels while increasing EFV levels. Hence concomitant use of standard doses of VORI and EFV is contraindicated. Our aim was to determine dose adjustments for coadministration of VORI and EFV that result in exposures similar to respective monotherapy.
Methods: This study used an open-label, four-treatment, multiple-dose, fixed-sequence design in 16 healthy males. Steady-state PK parameters were assessed following two different test combinations (VORI 300 mg q12h + EFV 300 mg q24h and VORI 400 mg q12h + EFV 300 mg q24h). These parameters were compared to steady-state PK parameters for reference monotherapy with VORI 200 mg q12h or EFV 600 mg q24h, using an ANOVA model to determine point estimates and 90% CI. VORI and EFV levels were determined by validated LC/MS/MS and HPLC/UV assays, respectively. PK parameters were determined by non-compartmental analyses.
Results: PK parameters are listed in the table , presented as the mean ratio for each test combination relative to respective reference monotherapy. VORI 300 mg + EFV 300 mg resulted in a significant decrease in VORI AUC (–55%) and Cmax (–36%), making it an unsuitable dose combination. Dosing of VORI 400 mg + EFV 300 mg resulted in a minor decrease in VORI AUC (–7%) and increased Cmax (23%), while slightly increasing EFV AUC (17%) and not changing Cmax. VORI and EFV, administered alone or combined, were safe and well tolerated.
| Parameter | Geometric mean ratio (90% CI) |
|
|---|---|---|
| VORI 300 mg + EFV 300 mg | VORI 400 mg + EFV 300 mg | |
| VORI Cmax | 64% (51, 79) | 123% (99, 153) |
| VORI AUC | 45% (38, 55) | 93% (77, 113) |
| EFV Cmax | 86% (79, 93) | 96% (88, 104) |
| EFV AUC | 101% (92, 111) | 117% (106, 129) |
Conclusions: These data suggest that systemic exposures similar to those seen with monotherapy can be attained during coadministration with VORI and EFV by making adjustment to the dose regimen. To achieve exposure levels similar to monotherapy, VORI dose should be increased to 400 mg q12h and EFV dose should be decreased to 300 mg q24h when the two drugs are coadministered.
P986 Anidulafungin: pharmacokinetics and tissue distribution in rats
B. Damle, M. Stogniew, J. Dowell (New York City, US)
Objectives: Anidulafungin (ANID), a novel echinocandin antifungal agent, is used in the treatment of oesophageal candidiasis and candidaemia. For systemic fungal infection, drug levels in the infected tissues are crucial for pharmacodynamic activity. Little is known about the tissue distribution of ANID, so this study was conducted to assess the pharmacokinetics and tissue distribution of ANID in rats.
Methods: Male F344 rats were given a single IV bolus dose of 14C-ANID (5 mg/kg). Plasma and tissue (kidney, liver, lung, spleen, quadricep, skin) samples were collected at 0, 0.083, 0.5, 1, 2, 4, 8, 24, 48 and 72 h after dosing (n = 3 rats/time), and the levels of parent drug by HPLC/UV and total radioactivity were determined. Additionally, whole blood and cerebrospinal fluid samples were collected and analysed for total radioactivity. In another study, distribution of radioactivity was assessed in rats by whole body autoradiography for up to 168 h after a single dose.
Results: Pharmacokinetics of ANID (ie, parent drug) are given below (Table ).
| Tissue | AUC (0–∞) (ng×h/gm or mL) | Cmax (ng/gm or mL) | Tmax (h) | T1/2 (h) |
|---|---|---|---|---|
| Kidney | 658,926.9 | 16,087.9 | 0.5 | 32.3 |
| Liver | 767,221.0 | 15,854.5 | 0.083 | 33.9 |
| Lung | 638,209.7 | 31,105.8 | 0.083 | 24.4 |
| Quadriceps | 57,349.2 | 3,839.7 | 0.5 | 17.1 |
| Plasma | 61,643.8 | 5,253.6 | 0.083 | 18.5 |
| Skin | 124,106.6 | 4,021.0 | 0.5 | 28.2 |
| Spleen | 567,950.4 | 24,195.6 | 0.5 | 25.2 |
Parent drug distributed rapidly into the tissues (Tmax ≤0.5 h). Based on T1/2 values, the elimination of parent drug from most tissues was slower than from plasma. The AUC of parent drug in liver, lung, kidney, and spleen was 12.4-, 10.4-, 10.7-, and 9.2-fold greater than in plasma. The tissue/AUC ratio of parent drug in skin and quadriceps was 2.0 and 0.9, respectively. Levels of drug-derived radioactivity, which represents parent drug and degradation products, were approximately 2- to 3-fold higher than parent drug in plasma and tissues. Drug-derived radioactivity was eliminated from plasma (T1/2 = 37.4 h) and tissues (T1/2 = 37.5 to 69.2 h) at a slower rate compared to parent drug. Cerebrospinal fluid had minimal (0.4%) radioactivity relative to whole blood. In the autoradiography study, high levels of radioactivity were observed in the adrenal gland, bone marrow, caecal and intestinal walls, kidney, liver, lung, lymph nodes, salivary gland, spleen, stomach wall, and thyroid glands.
Conclusion: Following IV administration in rats, ANID distributes well into the tissues that are considered clinically relevant for systemic fungal infection.
P987 Steady-state bioavailability of oral isavuconazole in healthy volunteers
A. Schmitt-Hoffmann, B. Roos, M. Heep (Basle, CH)
Objectives: This was a double-blind, randomised, parallel-group, multiple-dose, placebo- and active-controlled study carried out with 80 male and female subjects at a single study centre. The primary objective of the study was to determine whether isavuconazole (BAL4815) affects cardiac repolarisation, assessed by measurement of the QT interval. Because pharmacokinetics of isavuconazole were also evaluated after oral and intravenous administration, a sub-analysis was carried out to estimate oral bioavailability.
Methods: Subjects received 400 mg oral moxifloxacin on day 1 as a positive control. Afterwards, the 39 male and female subjects in the verum group received isavuconazole at loading doses of 400, 300, 200 mg orally (days 4–6) followed by maintenance doses of 100 mg orally (days 7–10) and 100 mg intravenously (day 11), and loading doses of 300, 250, 200 mg orally (days 12–14) followed by maintenance doses of 150 mg orally (days 15–18) and 150 mg intravenously (day 19). Serial blood samples on days 10, 11, 18 and 19 and pre-dose (trough) samples on days 6, 7, 8, 9, 10, 14, 15, 16, 17 and 18 were obtained. Pharmacokinetic parameters on days 10, 11, 18 and 19 were derived by non-compartmental analysis using WinNonlin Version 4.0.1.
Results: The mean trough levels of isavuconazole indicated that, after the loading doses, steady-state was achieved for the two maintenance doses. Mean trough levels of 2024±552.7 ng/mL and 1954±590.0 ng/mL were observed on days 7 and 10, respectively during the 100 mg maintenance oral dose sequence. The corresponding values for the 150 mg maintenance sequence were 2886±843.3 ng/mL and 2815±737.6 ng/mL on days 15 and 18.
The mean AUC0–24h were 46956 (day 10) and 46976 ng.h/mL (day 11) after 100 mg orally and intravenously, respectively. The corresponding AUC-values on days 18 and 19 were 73016 and 71109 ng.h/mL after 150 mg orally and intravenous, respectively. The mean within-subject bioavailability of oral isavuconazole for the 100 mg dose was 100±8.63% and for the 150 mg dose was 102±13.2%.
Conclusion: Stable and dose proportional levels were achieved for oral once daily maintenance doses of 100 mg and 150 mg of isavuconazole. Comparison of the pharmacokinetic profiles following oral and intravenous dosing at steady state demonstrated complete bioavailability of isavuconazole oral formulation.
P988 Pharmakokinetics of voriconazole in an intensive care unit patient with liver cirrhosis
S. Weiler, R. Bellmann-Weiler, R. Bellmann (Innsbruck, AT)
Objectives: Voriconazole (VRC) is a second-generation triazole with a broad antimycotic spectrum including non-albicans Candida and Aspergillus species. Oral and parenteral formulations have similar pharmacokinetics, however interpatient variation in plasma concentrations is considerable. Plasma concentrations of >6 μg/mL were associated with occasional liver function abnormalities. Neurologic toxicity was found in 50% of the patients with peak levels >5.5 μg/mL.
Methods: We report a 45 year old male (body weight 100 kg) suffering from a fatty liver cirrhosis, who was admitted to the ICU because of unconsciousness. The patient had received an oral dose of 2 mg/kg bid of VRC for 30 days for pulmonary aspergillosis. In addition, he was treated with pantoprazole, risperidone, vancomycin, naloxon and furosemid. VRC plasma concentrations were determined starting 15 hours after the last intake, using a quantitative analysis via HPLC and UV detection. Pharmacokinetic parameters were calculated with Kinetica 2000 using a non-compartmental model.
Results: The plasma level determined at admission amounted 13.9 μg/mL. Therefore VRC therapy was discontinued. The patient's condition, particularly CNS symptoms, gradually improved while VRC levels slowly declined and he could be transferred to the ward after 2 days (VRC concentration ∼10 μg/mL). A half life of 53.1 h (in healthy volunteers: 4.71 h), an apparent volume of distribution at steady state of 0.132 L/kg (in healthy volunteers: 2.04 L/kg) and a VRC clearance as low as 1.39 mL/h/kg (in healthy volunteers: 253.94 mL/h/kg) were calculated. Even after 11 days VRC was detectable (0.66 μg/mL).
Conclusion: The elimination of VRC appears to be markedly prolonged in severe liver cirrhosis leading to potentially toxic plasma levels. Therapeutic drug monitoring of plasma concentrations could greatly improve therapeutic safety.
Resistant streptococci
P989 Identification and antimicrobial susceptibility of viridans streptococci isolated from blood cultures in Leipzig, Germany
C. Friedrichs, K. Fickweiler, P. Nitsche-Schmitz, S. Chhatwal, A. Rodloff (Leipzig, Braunschweig, DE)
Objectives: Viridans streptococci (VS) are commensal bacteria of the human oral cavity, the respiratory, the gastrointestinal and the genitourinary tract. On the other hand, they are responsible for several systemic diseases including bacteraemia. The aim of the present study was to identify clinically relevant VS isolated from blood cultures to the species level, determine their susceptibilities to different antimicrobial agents and analyse the macrolide resistance mechanisms.
Methods: A total of 51 strains of VS were recovered from blood cultures from March 2004 to July 2006. The isolates were identified with phenotypic and genotypic methods. Phenotypic classification of erythromycin resistance were determined by double-disk diffusion test. All strains were characterised by PCR to detect the presence of mef(A), erm(B) and erm(A) genes.
Results: 37 strains were identified as mitis group; 10 as anginosus group and 4 strains as salivarius group by PCR. The comparison of the species identification results of the phenotypic and genotypic identification systems showed a consistency of 94% on the group level. Macrolide resistance was found in nearly 40% of all strains with typical resistance mechanism and only one S. salivarius showed a high penicillin resistance.
Conclusions: The present study has shown a preponderance of mitis group species and indicate that the ID 32 STREP is an acceptable method for the identification VS on a group level while the species-specific differentiations were not reliable. The differentiation of the mitis group species still remained difficult. The high macrolide resistance indicate that macrolides can not be considered an optimal alternative to penicillin in the treatment of VS infections.
P990 Presence of plasmid pA15 correlates with prevalence of constitutive MLSB resistance in group A Streptococci isolates at a university hospital in southern Taiwan
Y-F. Liu, H-M. Fu, H-M. Wu, R.P. Janapatle, C-H. Wang, J-J. Wu (Tainan, TW)
Background: In streptococci resistance gene erm mediates high level macrolide, lincosamide and group B streptogramins (MLSB) resistance. In mid 1990s constitutive MLSB resistance phenotype was replaced by M phenotype in group A streptococci (GAS) clinical isolates in southern Taiwan. Our lab collection of GAS isolates from 1990 to 2006 consists of the 215 erythromycin resistance isolates, 56 were with MIC ≥ 256 μg/mL and 159 with MIC < 256 μg/mL.
Objective: To determine the mechanism and prevalence rate of erythromycin resistance mediated by plasmid and transferability of the plasmid.
Methods: Colony blot hybridisation, PCR, plasmid curing and transformation and PFGE techniques were used in our study.
Results: By using zeta gene of GAS plasmid pSM19035 as a probe and restriction enzyme digestion analysis we found that in the 56 isolates with high erythromycin resistance 53 isolates harbour a plasmid, pA15, of 19 kb, in one isolate a plasmid pA768, of 16 kb was detected and in the remaining two isolates plasmid was not detected. We observed that ermB was on pA15 and it confers constitutive MLSB resistance phenotype. Prevalence rate of the pA15 harbouring erythromycin resistance isolates was high in 1993 (60%), 1994 (36%) and 1996 (33%), but the plasmid could not be detected from 2004 to 2006. To link the high level resistance to the pA15, clinical isolate A15 was selected and pA15 was cured by novobiocin. In the plasmid cured isolate SW503, the erythromycin MIC decreased from 256 μg/mL to 0.032 μg/mL. By electroporation pA15 was introduced into plasmid cured erythromycin sensitive isolate and we found that high level erythromycin resistance was restored. Interestingly, pA15 was also transferred to GBS and GCS by electroporation, acquiring erythromycin resistance. In all the pA15 containing isolates emm1 type was present and pulse type J was predominant (48 of 54 isolates) which demonstrates that these isolates had evolved from a single clone.
Conclusion: the plasmid pA15 mediated cMLSB resistance in mid 1990s, but pA15 cannot be detected from 2004 in the clinical isolates, which correlates with the absence of cMLSB resistance in this region.
P991 Serotypes distribution and gene homology of penicillin-nonsusceptible Streptococcus pneumoniae isolated from children in Beijing, China, 2000–2004
K. Yao, X. Shen, S. Yu, Y. Yang (Beijing, CN)
Objective: The study was designed to investigate the situation of antibiotic resistance and serotypes distribution, as well as the genetic homology of penicillin-nonsusceptible Streptococcus pneumoniae (PNSP) isolated from children in Beijing, China from 2000 to 2004.
Method: Nasopharageal swab specimens were collected from children up to 5-year-old with upper respiratory infection. The antibiotic susceptibility was tested by E-test MIC method. The serotypes were tested by simplified chessboard system. Chromosomal macrorestriction patterns of the strains isolated from 2000 to 2002 were detected by PFGE.
Results: One hundred and twenty-nine PNSP strains were collected. Seven serotypes were identified, which covered 104 strains. The other 25 strains could not typed by the system. There were 99 strains (76.7%) covered by the common serotypes 19, 23, 6 and 14. From 2000 to 2004, the isolates of serotype 19 with higher penicillin MIC was increasing when the isolates of serogroup 23 with lower penicillin MIC was decreasing. The covering rate of the heptavalent conjugate vaccine in total strains was about 76.7%. Two clones, P.01 and P.02, defined by the PFGE detection, were founded in the serogroup 23. The strains included in the two types, which accounted for 70.8% (17/24) of the group, were isolated mainly in 2000 and 2001 and ≤0.19 μg/mL for penicillin MIC value. Meanwhile, about 62.5% of serogroup 19 strains were ≥0.5 μg/mL for penicillin MIC value, and 62.5% of the group were nonsusceptible to cefrotaxime, cefaclor and/or cefrotriaxone. Three clones, P.03, P.04 and P.05, were founded in the serogroup 19. The strains covered by the P.03, which accounted for 31.3% (5/16) of the group were isolated mainly in 2002 and most of them were resistant to penicillin.
Conclusions: The data on serotype prevalence could be a useful guide to application of the vaccine, which should be an effective way in Beijing to control the carriage, infection and spreading of PNSP. The higher penicillin resistant clones isolated in latest period are not developed from the lower resistant clones spreading in the early study stage.
P992 Streptococcus pneumoniae strains isolated from laboratory animals and pets
M. van der Linden, A. Al-Lahham, I. Seegmüller, W. Nicklas, P. Kopp, R. Reinert (Aachen, DE; Amman, JO; Heidelberg, Ludwigsburg, DE)
Objectives: Streptococcus pneumoniae is the major causative pathogen of many childhood community-acquired respiratory tract infections (RTIs), including community-acquired pneumonia, acute otitis media and acute maxillary sinusitis. In the present study S. pneumoniae strains isolated from mastomys, guinea pigs, mice, rats and a cat were characterised.
Methods: Species diagnosis was performed using optochin- and bile solubility testing. Serotyping was performed using the Neufeld Quellung reaction. MICs to antibiotics were determined with the microdilution method according to the CLSI recommendations. Multilocus Sequence Typing was performed according to standard methods.
Results: Between 1986 and 2006 S. pneumoniae was isolated from 33 laboratory animals during routine control checks in the animal facility in the German Cancer Research Center and 2 pets treated in a medical veterinary laboratory. Isolates were obtained from mastomys (n = 24), rats (n = 4), guinea pigs (n = 3), mice (n = 3) and a cat. S. pneumoniae were isolated from nose, lung, trachea, abdomen and eye. Only four animals showed disease symptoms. Two guinea pigs were C4-immune deficient and suffered from severe peritonitis. The third guinea pig and the cat were pets and had severe respiratory problems. Serotypes detected were: 14 (n = 29), 19F (n = 3), 33A (n = 2) and 7C (n = 1). One isolate was rough and two were non typable. The guinea pigs isolates had serotypes 19F. All isolates were sensitive to penicillin, telithromycin and levofloxacin. One isolate was clarithromycin resistant. Twenty-six isolates were ST 15 (serotype 14 and n.t.) a sequence type commonly found in human isolates. The cat isolate was ST 180 (serotype 3). Twelve isolates had new MLS-types, seven of which are close variants of known MLSTs. The three guinea pig isolates showed the same completely new combination of known alleles.
Conclusions: S. pneumoniae strains could be routinely isolated from non-human hosts, showing animals to be a reservoir for S. pneumoniae.
P993 High prevalence and persistence of the Streptococcus pneumoniae Taiwan19F-14 clone among children in Greece
A. Mavroidi, I. Paraskakis, A. Pangalis, E. Kirikou, A. Charisiadou, T. Athanasiou, P. Tassios, L. Tzouvelekis (London, UK; Athens, GR)
Objectives: Molecular characterisation of the multi-drug resistant serotype 19F Streptococcus pneumoniae strains, prevalent amongst Greek children during 2001–2006, i.e. before and after introduction of the heptavalent vaccine in 2004.
Methods: A representative subset of 143 of 241 (34%) serotype 19F pneumococci from invasive (IPD), non-invasive infections (NIPD) and healthy children during January 2001–April 2006 were studied with respect to antimicrobial susceptibility (penicillin, amoxicillin, cefotaxime, erythromycin, clindamycin, co-trimoxazole, tetracycline, chloramphenicol, rifampicin, ofloxacin), by disk diffusion and E-test, and subtyped by PFGE and MLST.
Results: Serotype 19F – representing 31.8% and 26.0% of all pneumococcal isolates from children during 2001–2004 and 2005–2006, respectively – was a leading cause of NIPD in both periods, and more important in IPD in the second (19% vs. 6.5%; chi2 = 6.51, p< 0.02). Apart from 18 fully susceptible isolates, the remaining 125 (87.4%) were resistant to three or more antimicrobial classes (MDR). PFGE yielded 18 types (a total of 55 subtypes); 93% of all isolates belonged to only eight types. The dominant type, G (110 isolates, of which 55 belonged to one of 30 subtypes), had been seen first in 1996, in a common UK-Greek erythromycin-resistant clone. All type G isolates were MDR (including resistant to erythromycin and non-susceptible to penicillin (PNS)), in contrast to only 15 (45%) of the remaining 33 isolates (chi2 = 68.64, p<0.001). Thirteen (12%) of the PNS isolates also showed decreased susceptibility or resistance to cefotaxime (MIC 2–8 mg/L). MLST followed by eBURST analysis identified 12 STs – of which five were novel – grouped in six clonal complexes (CC). PFGE type G isolates were either ST236, corresponding to the Taiwan19F-14 (CC271) international clone, or one of four new single locus variants (SLV). A novel SLV of ST177 was also found.
Conclusion: First observed in Greece in 1996, the Taiwan19F-14 (CC271) international clone – so far considered minor in Europe – has now become dominant amongst Greek paediatric S. pneumoniae, even after introduction of the heptavalent vaccine in 2004. Over 75% of these isolates belonged to CC271 and a single PFGE type, yet both phenotypic and genotypic variation suggest a locally actively evolving clone.
P994 Characterisation of fluoroquinolone non-susceptible clinical isolates of Streptococcus pneumoniae in Germany: 1992–2005
A. Al-Lahham, M. van der Linden, C. Heeg, I. Seegmueller, R. Reinert (Amman, JO; Aachen, DE)
Objectives: S. pneumoniae belongs to the main human infectious agents worldwide. In several nationwide studies on resistance development in pneumococcal disease fluoroquinolone resistant pneumococcal isolates were collected and charachterised.
Methods: Strains were serotyped using Neufeld's Quellungreaction and MICs of penicillin G (PEN), cefotaxime (CEF), amoxicillen (AMOX), erythromycin A (ERY), clindamycin (CLI), tetracycline (TET), ciprofloxacin (CIP), levofloxacin (LEV), sparfloxacin (SPA), grepafloxacin (GRE), clinafloxacin (CLX), moxifloxacin (MOX), gatifloxacin (GAT) and gemifloxacin (GEM) were determined using the microdilution method according to the CLSI guidelines. Fluoroquinolone resistant genotypes were checked using PCR followed by DNA-sequencing of gyrA, gyrB, parC and parE according to standard methods. MLST was performed according to standard methods.
Results: Between Jan. 1992 and Dec. 2005, 13.230 isolates from 315 laboratories were included in this study. A total of 42 fluoroquinolone resistant strains (MIC CIP >2 μg/mL) were identified (0.2–1.2%). The predominant serotypes were 6A (14.3%), 6B (11.9%) and 9V, 23F and 11A (7.1% each). MIC50 and MIC90 for 17 different antibiotics were as follows (μg/mL): Penicillin (0.016, 1), cefotaxime (0.016, 1), amoxicillin (0.016, 0.5), erythromycin (0.125, 32), clindamycin (0.125, 32), tetracycline (0.5, 32), chloramphenicole (<4, <4), telithromycin (0.016, 0.016), quinupristin-dalfopristin (1, 1), ciprofloxacin (16, >32), levofloxacin (4, 32), sparfloxacin (4, 32), grepafloxacin (1, 32), clinafloxacin (0.25, 0.5), moxifloxacin (0.25, 4), gatifloxacin (1, 4) and gemifloxacin (0.125, 2). Predominant mutations were S81F (11.9%) and S114G (11.9%) for gyr(A), S79F (33.3%) and S79Y (23.8%) for par(C) and I460V (47.8%) for par(E). One isolate possesses a combination of both gyr(A) (E85G) and gyr(B) (R571T) mutations. A fourfold reduction of the ciprofloxacin MIC in the presence of reserpine was only observed in 14 isolates. In multi locus sequence typing, 4 clones were observed for the first time in Germany ST1551, ST1983, ST2007, and ST2008.
Conclusions: The overall incidence of fluoroquinolone resistance in Germany remains low, nevertheless the spread of FQ resistance to multi-drug resistant clones with worldwide distribution is worrisome. A clonal relatedness of strains could not be affirmed using MLST.
P995 Characterisation of the genetic structure of the invasive macrolide-resistant pneumococci in Portugal
R. Dias, D. Félix, C. Pissarra, M. Caniça on behalf of the Antimicrobial Resistance Surveillance Program in Portugal (ARSIP)
Objectives: The Streptococcus pneumoniae resistant to macrolides had increased consistently in Portugal since 1997, mainly associated to the consumption of that drug. To a better understand about the increase of this resistance, a molecular study was undertaken to characterise the genetic structure of the macrolide resistant strains.
Methods: A total of 191 invasive S. pneumoniae strains resistant to macrolides were recovered from 1994 to 2004 in 24 hospitals and public health institutions across Portugal. Genetic characterisation was undertaken by PFGE (n = 186) and MLST (n = 73). Genetic Clustering was performed by PFGE; one strain from each cluster was randomly chosen to be further characterised by MLST (with selection of a greater number of isolates for larger PFGE clusters). Linage assignment was performed by e-BURST analysis.
Results: The combination of PFGE and MLST data, permitted to identify the genetic lineages of the majority of macrolide resistant isolates. We found 41 Sequence Types (ST) integrated in 16 clonal complexes (CC) and 4 singletons. In this study, 24 STs were also found elsewhere and 17 were described only in Portugal including 13 novel STs. Seven Pneumococcal Molecular Epidemiology Network (PMEN) clones were found and 10 STs were Single Locus Variant of PMEN clones. In the period 1994–1998, were found different CCs, with the following putative founders: ST156, ST15, ST90, ST63, ST81, ST315, and ST97. In the following years (1999–2004), in which was observed an increase of macrolide resistance, we identified the main previous CCs, and the emergence of new ones, with the following putative founders: ST230, ST177, ST717, ST193, ST176, ST90, ST180, ST191, ST88 and ST271. It was also observed the emergence of new single genetic lineages such as: ST2360, ST2357 and ST2359. However, in the period 2002–2004 the ST180 and ST191 disappeared and the number of isolates from the ST315 decreased significantly. Among the CCs, which emerged within the macrolide resistance, 5 were described as susceptible in the earlier years; 2 were described internationally in resistant strains and 3 were described in both susceptible and resistant strains.
Conclusion: Our results suggest that macrolide resistance among pneumococci increased in Portugal due to the expansion of clonal complexes (which appeared before the emergence of macrolide), and due to the acquisition of resistance by susceptible circulating clones and to the import of international resistant clones.
P996 Pneumococcal invasive isolates of non-vaccine serotypes in Italy, 1999–2003 (pre-vaccine era)
G. Gherardi, M. Monaco, R. Camilli, F. D'Ambrosio, P. D'Ancona, R. Manganelli, G. Dicuonzo, A. Pantosti (Rome, Padua, IT)
Objectives: The introduction of the 7-valent conjugate vaccine (PCV7) for children has caused a dramatic decrease of pneumococcal infections due to vaccine serotypes (VS) and a relative increase of infections due to vaccine-related (VRS) or non-vaccine serotypes (NVS). The aim of this study was to characterise invasive pneumococci belonging to serotypes non included in PCV7 before the implementation of this vaccine in Italy.
Methods: All pneumococcal invasive isolates recovered from children and adults in an Italian nation-wide surveillance in 1999–2003 were studied. All isolates were serotyped and examined for susceptibility to penicillin, cefotaxime, erythromycin, clindamycin, tetracycline and chloramphenicol following the CLSI standard procedures. Pulsed-Field Gel Electrophoresis and Multilocus Sequence Typing (MLST) were used to define clonal groups among penicillin non-susceptible (PNSSP) and/or multi-drug resistant isolates and selected susceptible isolates. The sequence types (ST) were related to those reported in the MLST website (www.mlst.net) and described in the Pneumococcal Molecular Epidemiology Network (PMEN) as international clones.
Results: Among 790 invasive isolates, mostly obtained from adult patients, 392 (49.6%) belonged to VS; the other group included isolates belonging to VRS (93 isolates), to NVS (297 isolates) or that were non-typable (8 isolates). Among these isolates, 19 (5.7%) were PNSSP, all in the intermediate range, and 63 (15.8%) were erythromycin-resistant. The most prevalent serotypes, were 6A, and 19A among VRS and 1, 3, 7F, 8, 10A, 11A, 12F, 15B/C, 20 and 22F among NVS. Among the PNSSP the most frequent serotypes were 19A and 35F (5 isolates each). Several PMEN clones were identified, such as Sweden15A-25/ST63, Greece21–30/ST193, Netherlands3–31/ST180, Netherlands8–33/ST53, Netherlands15B-37/ST199, Netherlands7F-39/ST191 and Sweden1–40/ST304. Some cases of capsular switching were detected. Some isolates belonged to clonal groups that had not been previously identified, such as serotype 15B/C isolates (ST1577) and serotype 6A isolates (ST675 and ST1833).
Conclusions: In the pre-vaccine era, serotypes not contained in PCV7 (VRS or NVS) show a lower rate of penicillin or erythromycin resistance than VS. Both international clones and newly-described clones were detected and capsular switching was observed. Future monitoring of serotypes and clones in the vaccine era is of paramount importance.
P997 Population structure of Spanish mef-PCR positive Streptococcus pneumoniae clinical isolates
E. Gómez G-de la Pedrosa, M. van der Linden, M.I. Morosini, J.C. Galán, F. Baquero, R.R. Reinert, R. Cantón (Madrid, ES; Aachen, DE)
Objectives: To determine the population structure of a collection of mef-PCR positive Streptococcus pneumoniae clinical isolates recovered in different Spanish hospitals. Prevalence of associated erythromycin resistance determinants and phenotypes were also screened.
Methods: A PCR assay was performed to identify erythromycin resistance genes (ermB or mef) in a collection of 712 clinical isolates recovered in Spain from 1999 through 2003. All mef-PCR positive S. pneumoniae strains (n = 46) were selected for further population structure analysis (serotyping with Neufeld's Quellung method, PFGE with SmaI digestion, and MLST with the standard 7 housekeeping loci scheme). Susceptibility testing was performed by the standard microdilution technique (CLSI) and resistance phenotypes by diffusion assay using commercial erythromycin, clindamycin, and rokitamycin disks. A multiplex-PCR was designed to distinguish between mef(A) and mef(E). msr(D) determinant (mel gene) was also detected by PCR.
Results: Resistance values among 46 selected mef positive isolates were: penicillin, 67.3% (intermediate + resistant), clindamycin, 52.2%, and tetracycline 56.5%. No telithromycin resistance was found (MIC range, 0.03–1 mg/L) but one isolate was resistant to levofloxacin (MIC, 8 mg/L). Interestingly, 4 mef positive isolates (8.7%) showed erythromycin MICs in the susceptible range (0.12 mg/L), but increased ≥32 mg/L endowing the M phenotype when plated on increased concentrations of erythromycin. The M and MLSB phenotypes in erythromycin resistant isolates were observed in 45.2% and 54.72% of isolates, respectively. The presence of both erm and mef determinants was found in 50% of isolates. All mef determinants belonged to mef(E) subclass and all isolates presented the msr(D) gene. Serotype distribution was as follows: 14, 21.7%; 19F, 19.5%; 19A, 15.2%; 6B, 6.5%; 9V 6.5%; and others, 30.6%. Most isolates belonging to the same serotype showed similar PFGE patterns. England∧14–9 and Taiwan∧19F-14 multiresistant clonal complexes were represented within mef positive isolates and Spain∧23F-1, Poland∧6B-20, Sweden∧15A-25 and Spain 6B among erm and mef positive isolates.
Conclusions: A high proportion of S. pneumoniae isolates harbouring the mef gene in our collection also presents the ermB determinant. Nearly all of them displayed the MLSB phenotype. A complex population structure was found in our mef positive S. pneumoniae collection.
P998 Antimicrobial resistance patterns and genotypes of Streptococcus agalactiae and Streptococcus dysgalactiae ssp. dysgalactiae from bovine mastitis in Portuguese dairy farms
M. Rato, D. Rolo, R. Bexiga, S.F. Nunes, L.M. Cavaco, C.L. Vilela, I. Santos-Sanches (Caparica, PT; Glasgow, Cambridge, UK; Lisbon, PT)
Objectives: To evaluate the profiles of antimicrobial (ATB) resistance and genotypes of the contagious bovine mastitis pathogen Streptococcus agalactiae (GBS) and of Streptococcus dysgalactiae ssp. dysgalactiae (GCS), which is considered to be either a contagious or an environmental bovine pathogen.
Methods: Among 459 milk samples collected during 2002/2003 from mastitis quarters of 377 bovines in 11 Portuguese herds, 13.9% of the bacteria were GBS and 3.7% were GCS. A total of 32 GBS and 17 GCS of these field isolates were studied. ATB resistance was evaluated by disk diffusion against macrolides-M (erythromycin-E), lincosamides-L (pirlimycin-PRL) and tetracycline-T. The ATB resistance genes tet(M), tet(O), tet(W), tet(L), tet(Q), tet(K), tet(S), mef(A), erm(A) and erm(B) were detected by PCR. The human pathogen S. pyogenes (GAS) virulence phage-encoded speA, speC and ssa genes were also tested by PCR in all GCS. Clonality among isolates was evaluated by pulsed field gel electrophoresis (PFGE). GBS representative clones, defined by PFGE, were further characterised by Multi Locus Sequence Typing for identification of Sequence Types (ST) and of Single and Double Locus Variants (SLV and DLV) of ST common in human isolates.
Results: Resistance to E was 21.9% in GBS and 23.5% in GCS, while resistance to PRL was 21.9% in GBS and 35.3% in GCS. Resistance to T was 65.6% in GBS and 100% in GCS. Three GCS resistant to E and T carry erm(B)/tet(O), six GBS carry erm(B)/tet(O)/tet(K) and one GCS carries erm(A)/tet(M). Most (79%) GCS and GBS resistant only to T carry tet(M), tet(O), tet(K) or tet(S). Out of the 20 GBS clones, six were herd-specific and none was disseminated in more than one herd. Out of the 15 GCS clones only one was herd-specific and one was disseminated in two herds. Among the 17 GCS, 29.4% are speC-positive and all are speA/ssa-negative. The six GBS representative strains were of ST-2 and ST-61. ST-2 is a SLV of ST-1 and ST-61 is a DLV of ST-17 and ST-18, all of human origin.
Conclusion: Results confirmed the contagious nature of GBS and suggested an environmental origin of GCS in the Portuguese herds. A putative novel phenotype of susceptibility to M and resistance to L was detected in GCS. To our knowledge, tet(S) and tet(K) genes, detected among this collection, were not reported yet in bovine GBS and GCS. The finding of speC in bovine GCS suggests that GAS prophages may also be critical contributors to animal GCS genetic diversity and virulence.
P999 Molecular characterisation of Streptococcus pneumoniae strains isolated from children with acute otitis media in Barcelona (1992–2005)
C. Ardanuy, A. Gene, L. Calatayud, E. Palacín, A. Fenoll, J. Linares (Barcelona, Esplugues de Llobregat, Majadahonda, ES)
Objectives: S. pneumoniae (Spn) is a frequent cause of acute otitis media (AOM) in children. The aims of this study were: (a) To analyse the distribution of Spn clones causing AOM in children in Barcelona (1992–2005). (b) To analyse the impact of the heptavalent pneumococcal conjugated vaccine (PCV7).
Methods: Three hundred and twenty pneumococci isolated from children (1992–2005) with AOM were studied. Serotyping was performed by Quellung reaction. The antibiotic susceptibility was performed by agar dilution and disk-diffusion methods to penicillin (PEN), erythromycin, clindamycin, tetracycline, chloramphenicol and cotrimoxazole. The isolates were typed by PFGE (SmaI and/or ApaI). Representative isolates of each dominant clone were studied by MLST.
Results: The most frequent serogroups were: 19 (31%), 6 (23%), 14 (14%), 23 (11%) and 3 (6%). Among 300 pneumococci studied by PFGE, 114 different band patterns were found. Four multiresistant clones, Spain6B-2 (14%), Spain9V-3 (12%), clone ST88 of serotype 19F (9%) and Spain23F-1 (9%) accounted for 44% of strains. Among 221 PEN-R Spn, 66 different PFGE patterns were found. Eight of them (Spain6B-2, Spain9V-3, ST88 clone of serotype 19F, Spain23F-1, Sweden15A-25, Spain14–5, Poland6B-20, and ST276 clone of serotype 19A) accounted for 73.7% of PEN-R strains. Among 79 PEN-S strains, 52 PFGE patterns were found, 39 (49%) of them had a single isolate. Multidrug-resistance was observed in 81% of PEN-R strains and 17% of PEN-S strains (p < 0.001). Capsular switching was observed in three clones: Spain9V-3 (25/36 were serotype 14 and 1/36 was serogroup 19); Spain23F-1 clone (3/26 were serogroup 19); Sweden15A-25 clone (10/13 were serogroup 19 and 1/13 was serotype 23F). Comparing 1996–2000 and 2001–2005 periods the following changes were observed: (1) A decrease in the rate of PEN-R isolates (76% vs 61%). (2) A decrease in the vaccine serotypes (6B, 14, 23F). (3) An increase in the vaccine-related serotype 19A. (4) A significant decrease of Spain6B-2 clone (17% vs 4%), Spain23F-1 clone (11.0 vs 1.3) and ST88 clone of serotype 19F (9% vs. 4%) observed. (5) The emergence of a new clone related to serotype 19A (ST276) (0% vs 5%).
Conclusion: The proportion of episodes of AOM caused by PEN-R Spn decreased after the introduction of the PCV7 in Barcelona, coinciding with a decrease of multiR clones of vaccine related serotypes. Capsular switching was frequently observed.
P1000 Molecular characterisation of penicillin non-susceptible invasive strains of Streptococcus pneumoniae prevalent in the Czech Republic
V. Jakubu, H. Zemlickova, P. Urbaskova, J. Kasik (Prague, CZ)
Objectives: To study the clonal diversity of penicillin non-susceptible invasive strains of S. pneumoniae isolated in the Czech Republic. By use of multilocus sequence typing (MLST) the present study allow to determine the epidemiological relationship between the various PNSP serotypes and establish the prevalence of international penicillin-resistant clones in the Czech Republic.
Methods: All strains (n = 1054) isolated from blood or cerebrospinal fluid between 1996 to 2004 were submitted to the National Institute of Public Health by 63 microbiology labs from 45 cities. Serotyping was performed by Quellung reaction and the minimal inhibitory concentrations (MICs) of penicillin, erythromycin, tetracycline and chloramphenicol were determined by CLSI broth microdilution method among all submitted strains. All viable PNSP isolates (n = 37) were analysed further by MLST.
Results: Overall 38 (3.5%) of strains had reduced susceptibility to penicillin but only five of these were highly resistant. The prevalence rate of invasive S. pneumoniae strains with reduced susceptibility to penicillin remains stable for the study period. The PNSP serotypes were as follows: 9V (26), 23F (4), 14 (3), 6B (2), 19A (1), 19F (1) and 22F (1). Except of decreased susceptibility to penicillin, some strains were also concomitantly resistant to other antibiotics tested. Twelve (31.6%) of PNSP strains were resistant to erythromycin, five (13.2% both) of strains were resistant to tetracycline and chloramphenicol. MLST typing has been performed on 37 of viable PNSP isolates. Isolates harbouring STs identical to that of two globally spread penicillin resistant clones of S. pneumoniae, Spain∧9V-3 and Spain∧23F-1, were detected among isolates of serotype 9V, 14, 22F and 23F, respectively. Sequence type (ST) 271, the single locus variant (SLV) of the Taiwan∧19F-14 clone (ST 236) was found in one isolate of 19F serotype. Two 6B isolates were not related to any major international drug-resistant clones already described.
Conclusion: Despite the low prevalence of penicillin non-susceptible S. pneumoniae in the Czech Republic, multidrug resistant clones, which attribute to high rates of resistance to β-lactams in many countries, were identified in our country. The Spain∧9V-3 clone (ST 156) to have a major contribution among our invasive PNSP. Apparently, the genetic background of this clone is not restricted to serotype 9V, but is also disseminated within serotype 14 and 22F.
P1001 Clonal relationships of Streptococcus pneumoniae isolates harbouring multiple antibiotic resistance from Belgium
F. P. O'Hara, J. Van Eldere, R. Mera, L. Miller, J. Poupard, H. Amrine-Madsen (Collegeville, US; Leuven, BE; Research Triangle Park, Philadelphia, US)
Objectives: Antibiotic resistance has been increasing in many European countries and strains that are non-susceptible to two or more classes of antibiotics (multiple resistance or MR) are particularly troubling. In order to better understand the evolution of resistance in S. pneumoniae, we have identified and characterised the clones present in isolates obtained from a surveillance dataset collected in Belgium.
Methods: A sample of 190 S. pneumoniae isolates from 1997–2004 was selected at random from the larger Belgian surveillance set. For each isolate, PCR was performed on seven housekeeping genes based on the universal Multi Locus Sequence Typing methods. Internal fragments were sequenced, compared to published alleles, and assigned allele numbers. Allele numbers were used to generate allelic profiles, which were compared to published allelic profiles to determine a sequence type (ST). The eBURST algorithm was used to assign STs to clonal complexes (CCs).
Results: The 190 isolates contain 95 unique STs which are divided into 17 CCs and 44 singletons. The 62 MR isolates represent 31 sequence types, but half of these are found within two major clonal complexes (CC156 and CC81). MR is not evenly distributed among CCs; within CCs, the rate of MR varies from 0% (CC53) to 100% (CC81). The clonal composition of the S. pneumoniae population changed over time. In 1997–1999, two CCs, CC156 (representing the PMEN clone Spain9V-3) and CC81 (representing the PMEN clone Spain23F-1), accounted for 25.8% and 13.4% of all isolates, respectively. By 2001–2004, the frequencies of these CCs had dropped to 7.5% and 3.2%. In 1997–1999, CC81 and CC156 accounted for 67.6% of all MR isolates. In 2001–2004, those CCs accounted for only 28.6% of MR isolates. The clonal structure of the different serotype groups varied. Of the 10 most common serotype groups, four were highly clonal, while six contained a variety of CCs representing genetically diverse STs. For example, serogroup 19 covered 7 CCs while serogroup 8 covered 1 CC. MR was much higher in the genetically diverse serotype groups (48.4%) than in those that were highly clonal (4.3%).
Conclusions: The PMEN clones Spain9V-3 and Spain 23F-1 decreased from 1997–2004 in Belgium despite the level of MR increasing over time. The frequency of CCs that harbour MR changes over time. Serotype groups that are genetically diverse such as 19 are more likely to harbour MR.
P1002 Streptococcus pneumoniae clonal relationships over time in isolates from Belgian provinces bordering countries with low and high levels of antibiotic resistance
H. Amrine-Madsen, J. Van Eldere, R. Mera, L. Miller, J. Poupard, F. P. O'Hara (Research Triangle Park, US; Leuven, BE; Collegeville, Philadelphia, US)
Objectives: Belgium is located between countries with low (Netherlands) or high (France) levels of antibiotic resistance. Border provinces from Belgium afford a unique opportunity to look at the clonal structure of isolates with non-susceptibility to two or more antibiotic classes (multiple resistance) across time and place. The clonal distribution of S. pneumoniae was analyzed in a sample of isolates collected from 1997–2004 in Limburg which borders the Netherlands and West-Vlaanderen which borders France.
Methods: A sample of 190 S. pneumoniae isolates collected in the provinces of Limburg and West-Vlaanderen was selected at random from a larger Belgian surveillance set. For each isolate, PCR was performed on seven housekeeping genes based on the universal Multi Locus Sequence Typing methods. Internal fragments were sequenced, compared to published alleles, and assigned allele numbers. Allele numbers were used to generate allelic profiles, which were compared to published allelic profiles to determine a sequence type (ST). The eBURST algorithm in conjunction with phylogenetic trees was used to assign STs to clonal groups.
Results: The 190 isolates contain 95 unique STs which are divided into 24 clonal groups. More than half of all isolates are contained within the 8 largest clonal groups. The vast majority of the clonal groups occur in both Limburg and West-Vlaanderen, however, the frequency of clones in each province is different. For example, clonal group 17 (representing the PMEN clone Spain 23F-1) is more likely to be present in West-Vlaanderen (94%) than in Limburg (6%) as is clonal group 1 (representing the PMEN clone Spain 9V-3). The distribution of multiple resistance (MR) varies across provinces and across clones with West-Vlaanderen harbouring 77.4% of the MR and Limburg only 22.6%. The variation in clonal group proportions explains much of the variation in MR levels between the provinces.
Conclusions: Although the same clonal complexes of S. pneumoniae are circulating in both Limburg and West-Vlaanderen, the frequency of clones varies significantly between the two provinces. A higher proportion of clonal groups harbouring MR such as Spain 23F-1 and Spain 9V-3 is seen in the Belgian province of West-Vlaanderen which borders France than in the Belgian province of Limburg which borders the Netherlands. There is evidence of a flow of clones and resistance from West-Vlaanderen to Limburg.
P1003 Macrolide resistance in Streptococcus pyogenes in the United Kingdom is encoded by a variety of resistance determinants associated with a small number of clones
G. Moore, K. McGregor (Slough, UK)
Objective: Identify and characterise the elements that harbour resistance genes in a collection of Streptococcus pyogenes from the United Kingdom.
Methods: The minimum inhibitory concentration of 17 antimicrobial agents was determined by agar dilution for 119 S. pyogenes isolates which had previously been characterised by MLST. PCR amplification of fragments of published resistance genes, their flanking genes, resistance element junction regions and chromosomal insertion sites was performed using specific primers.
Results: The majority (77%) of isolates were susceptible to all of the agents tested. Six isolates (5%) were resistant to erythromycin. Of these, two isolates (both ST150, tetracycline susceptible and with a M macrolide-resistance phenotype) harboured a mef(A) gene within a Tn1207.3 element that had been inserted into the chromosomal comEC gene. One isolate (ST36, and carrying tet(M)) demonstrated a cMLS phenotype that was encoded by an erm(B) gene. The remaining three isolates (one ST63-tet(O), two ST89-tet(M)) carried an erm(A) (subclass erm(TR)) gene which gave rise to the iMLS phenotype. Analysis of the genes flanking the erm(TR) gene revealed that the two ST89 isolates contain an element similar to the 48,853-bp erm(TR)-containing element present in the genome of US isolate MGAS10750 (ST39). However, while the ST63 isolate had the same DNA directly downstream of erm(TR), the upstream DNA differed. Furthermore, unlike the US isolate, the erm(TR) element in the UK strains did not insert into the chromosomal hsdM gene. Twenty isolates were resistant to tetracycline and erythromycin susceptible. A single ST63 isolate harboured tet(O), and 19 isolates (representing 14 STs) harboured tet(M). Four isolates (ST117-tet(M)) were resistant to rifampicin.
Conclusion: In this collection of S. pyogenes, macrolide resistance was associated with four distinct resistance elements, including a previously undescribed erm(TR)-containing element. The association of macrolide resistance with a relatively small number of clones, in the UK and other European countries, suggests that these clones may have a greater propensity for acquiring resistance elements.
Epidemiology of resistant non-fermenters
P1004 Comparison of Acinetobacter baumannii from human and animal isolates by pulsed-field gel electrophoresis in Scotland
A. Hamouda, L. Vali, D. Walker, A. Mateus, J. Dave, A. Gibb, S. Dancer (Edinburgh, Glasgow, UK)
Objectives: Several studies have shown that fluoroquinolone (FQ) use in food animals contributes to the selection of antimicrobial resistance in zoonotic bacteria and poses a risk to humans. However, the main problem in hospitals is not zoonotic bacteria but other species such as Acinetobacter baumannii, Pseudomonas aeruginosa, and Methicillin resistant Staphylococcus aureus. Preserving the efficacy of FQ in the treatment of A. baumannii infections in human is essential to reduce the current dependence on the carbapenems. The purpose of this study was to investigate a possible link between ciprofloxacin resistance in A. baumannii clinical and animal isolates.
Methods: A total of 264 samples collected from pig, cattle and sheep were screened for A. baumannii. Five clinical isolates were obtained from two Scottish hospitals. Acinetobacter selective medium was used for isolation of this species. Suspected isolates were phenotypically characterised by the API 20NE test, and identification to the species level was carried out by restriction analysis of the 16S-23S rRNA intergenic spacer sequences (RAI16rRNA). The minimum inhibitory concentration of antibiotics was determined by the agar dilution method following the BSAC Guidelines. Pulsed-field gel electrophoresis (PFGE) typing was performed using ApaI restriction endonuclease.
Results: Out of 264 animal samples processed, 18 were confirmed as A. baumannii by RAI16rRNA. Two isolates were resistant to ciprofloxacin and two were resistant to gentamicin. All isolates were resistant to ampicillin, trimethoprim, chloramphenicol and tetracycline but were sensitive to imipenem. Dendrogramme analysis showed that most (8 out 9 and 6 out 8) A. baumannii isolated from pig and cattle faecal samples respectively, fell into one major cluster. The clinical isolates grouped in two different clusters and were much more closely related to each other than to those of the animal isolate clusters.
Conclusion: This study has shown that PFGE revealed a certain degree of heterogeneity among ciprofloxacin-resistant-A. baumannii from both animal and human isolates, and that they were genetically different. It has also shown that there was no correlation between PFGE profile and ciprofloxacin resistance in both animal and clinical isolates.
P1005 Resident OXA-51-like carbapenemases in Acinetobacter baumannii: a useful marker for genotyping?
T. Giani, M.M. D'Andrea, R. Migliavacca, M. Spalla, E. Nucleo, G. Fina, F. Luzzaro, L. Pagani, G.M. Rossolini (Siena, Pavia, Varese, IT)
Objectives: Multi drug resistant (MDR) Acinetobacter baumannii is a problem of a great concern in nosocomial settings. Hospital outbreaks have been described worldwide and this organism has become endemic in some areas. Comparison of isolates obtained from different investigators is not straightforward, mostly because requires the interchange of isolates between laboratories and the standardisation of the genotyping techniques used. Recently resident blaOXA-51 type determinants have been proposed as a convenient marker for species identification, in alternative to previous molecular approaches. The aim of this study was to investigate the correlation between OXA-51-like carbapenemases and genotypes in a collection of A. baumannii isolates from Italian hospitals.
Methods: A. baumannii analyzed in this work included 151 clinical isolates, collected from 14 different Italian hospitals. Identification and susceptibility testing were carried out following standard procedures. Genotyping was carried out by RAPD, REP-PCR and PFGE analysis. OXA-51-like determinants were investigated by PCR, followed by direct amplicon sequencing.
Results: Genotyping analysis defined 5 major clusters of MDR A. baumannii, with consistent results using the various typing methods. Each cluster was characterised by a specific resident blaOXA-51-type carbapenemase, namely OXA-66, OXA-69, OXA-90, OXA-98 and OXA-100. The last three are new variants which differ from OXA-51 by 4, 7 and 6 amino acids respectively.
Conclusions: Consistent relationship between the blaOXA-51-like variants and genotyping results was found. Present results suggest that OXA-51 typing could be a useful marker for genotyping of A. baumannii clinical isolates.
This work was funded in part by the research grant “MIUR-PRIN 2005061894_004”.
P1006 Imported cases of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in Belgium: characterisation and unusual loss of resistance
P. Bogaerts, T. Naas, C. Bauraing, A. Deplano, D. Piérard, P. Nordmann, Y. Glupczynski (Yvoir, BE; Paris, FR; Brussels, BE)
Objectives: International travel contributes to the worldwide spread of multi-drug resistant (MDR) bacterial strains. Carbapenems are the drugs of choice for the treatment of nosocomial infections due to MDR Acinetobacter baumannii (Ab). However, their efficacy is actually compromised through the emergence of carbapenem-hydrolysing β-lactamases (CHBL) in that species. We report here, three independent cases of CHBL OXA-23-producing Ab strains documented after international travel and the unusual spontaneous loss of OXA-23 gene in the absence of any antibiotic treatment.
Methods: Over the last 24 months, three severely injured patients were transferred from Cairo (Egypt), Cape Town and Johannesburg (South Africa) to three unrelated hospitals in Belgium. Carbapenem-resistant clinical isolates of Ab were recovered upon patient arrival and for one patient 3 sequential isolates were obtained. The five strains were referred centrally for molecular investigation of their resistance mechanism, using DNA analysis, isoelectric focusing (IEF), conjugation assays and PFGE. BlaIMP, blaVIM, blaOXA-23-like, blaOXA-26-like, blaOXA-51-like, blaOXA-58 genes, blaTEM gene, blaAmpC and the presence of ISAba1 inserted upstream of different genes were sought by PCR.
Results: All 5 MDR-Ab isolates were positive blaOXA-51-like and blaAmpC genes and all but one were positive for blaOXA-23-like genes. ISAba1 was detected upstream of blaAmpC, blaOXA-23-like and blaOXA-51-like in 5, 4 and 1 isolates respectively. blaTEM was detected in two isolates. Sequencing of the amplicons revealed identical blaOXA-23 gene but different blaAmpC and blaOXA-51-like genes. Ab isolates from the different patients were unrelated as shown by PFGE. For one patient with a diagnosis of complicated urinary tract infection, the three consecutive isolates presented identical PFGE patterns, though only the two first isolates were found blaOXA-23-positive. The third isolate was susceptible to carbapenem by loss of the blaOXA-23 gene allowing the patient to be successfully treated by meropenem.
Conclusions: Our data further support the role of inter-country transfer in the spread of MDR-Ab and underline the importance of ISAba1 in the expression of β-lactam resistance genes. The spontaneous loss of carbapenem resistance along with blaOXA-23 gene in the absence of antibiotic treatment highlights the importance of judicious and well controlled usage of antibiotics for such MDR isolates.
P1007 Plasmid location of blaOXA-40 gene in Acinetobacter baumannii and Acinetobacter haemolyticus
S. Quinteira, F. Grosso, L. Peixe (Porto, PT)
Objectives: The aim of this work was to investigate the location and genetic background of the carbapenem-hydrolysing oxacillinase gene blaOXA-40 in Acinetobacter spp. isolated from a University Hospital with a high rate of imipenem resistance, mainly due to the endemic presence of the Iberian A. baumannii blaOXA-40 producing clone.
Methods: An isolate of A. baumannii and two A. haemolyticus producing OXA-40 recovered from a University Hospital were studied. Susceptibility to antibiotics was determined by standard agar diffusion method and Etest. Carbapenemase producing strains were detected by a bioassay using imipenem discs (10 μg) with enzymatic extracts. Clonality was established by PFGE. The presence and sequencing of blaOXA gene was determined using the blaOXA-24-like primers. Sequencing of flanking regions of blaOXA-40 was conducted in plasmidic extracts. Hybridisation assays were performed using blaOXA-40 probe after S1 nuclease digestion and plasmidic extraction. Furthermore, transfer of resistance by conjugation and transformation assays was also attempted.
Results: The A. baumannii and the two A. haemolyticus isolates presented resistance to imipenem, meropenem, ertapenem (MIC of ≥32 μg/mL), to amoxicillin and its association with clavulanic acid, ureidopenicillins and their associations (MIC of ≥256 μg/mL). Resistance to ceftazidime, cefepime and aztreonam (MIC of ≥256 μg/mL) was only observed in the A. baumannii isolate.
Hybridisation experiments with blaOXA-40 probe in S1 nuclease resulting bands and in plasmids extracts revealed a plasmid of about 30 Kb carrying blaOXA-40 in the A. baumannii and the two clonally related A. haemolyticus. These plasmids showed an identical sequence upstream of blaOXA-40.
Attempts to transfer resistance by conjugation of plasmid DNA into either an A. baumannii or an E. coli recipient were unsuccessful.
Conclusions: In this study we describe a plasmidic location for the blaOXA-40 gene in both an A. baumannii and A. haemolyticus isolates. Although the spreading of OXA-40, both in the Iberian Peninsula and France, has been correlated with the progressive dissemination of a single A. baumannii clone, the observation of this enzyme in different genomic species could be explained by a plasmid location.
The plasmid-mediated OXA-40 described in this study could contribute to a rapid spread of carbapenem-resistance in Acinetobacter spp.
P1008 Epidemiology and mechanism of resistance of an outbreak of multidrug-resistant Acinetobacter baumannii at in a Lebanese hospital
A. Di Popolo, A.U. Khan, Z. Daoud, M. Bagattini, C. Afif, M. Triassi, N.I. Hakimé, R. Zarrilli (Naples, IT; Aligarh, IN; Beirut, LB)
An epidemiological study to investigate an outbreak of multidrug-resistant Acinetobacter baumannii conducted at the Saint George University Hospital, Beirut, Lebanon. The first cases were observed in November 2004, when A. baumannii was isolated from 17 patients, 11 from medical-surgical intensive care unit (ICU), 6 from other wards. Since then, more than 30 cases were identified and the strain of Acinetobacter baumannii became a frequent isolate in our lab.
Epidemiological data
| Nr. | Date of admission | Date of discharge | Age | Gender | Transfer to ICU (duration) | Intubation (duration) | Recent surgery | Previous therapy | Clinical presentation of culprit A. baumannii |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 25/11/04 | 16/11/05 | 35 | M | 26/11/04 (27 days) | Yes (14 days) | Yes | No previous therapy | Effusion |
| 2 | 06/12/04 | 26/01/05 | 85 | M | 06/12/04 (14 days) | Yes (19 days) | No | No previous therapy | Pneumonia (RTI) |
| 3 | 18/12/04 | 27/12/04 | 77 | M | No | No | Yes | No previous therapy | Bacteraemia (BSI) |
| 4 | 14/12/04 | 26/12/04 | 64 | M | 14/12/04 (13 days) | Yes (13 days) | Yes | No previous therapy | Pneumonia (RTI) |
| 5 | 05/01/05 | Still in hospital | 44 | F | 05/01/05 (17 days) | Yes (15 days) | Yes | Antibiotic Prophylaxis | Pneumonia (RTI) |
| 6 | 02/01/05 | 27/01/05 | 17 | M | 03/01/05 (11 days) | Yes (8 days) | No | No previous therapy | Pneumonia (RTI) |
| 7 | 18/12/05 | Still in hospital | 30 | M | Not in ICU | No | Yes | Antibiotic treatment | Abscess |
| 8 | 27/11/04 | 08/02/05 | 66 | M | 27/01/05 (21 days) | Yes (3 days) | Yes | Antibiotic treatment | Pneumonia (RTI) |
| 9 | 16/01/05 | 06/02/05 | 70 | F | 19/01/05 (14 days) | Yes (9 days) | No | Corticosteroids | Pneumonia (RTI) |
| 10 | 15/02/05 | 23/02/05 | 86 | M | No | Yes (5 days) | Yes | Corticosteroids | Pneumonia (RTI) |
| 11 | 05/02/05 | 04/03/05 | 88 | F | 05/02/05 (10 days) | Yes (2 days) | Yes | Antibiotic treatment | Pneumonia (RTI) |
| 12 | 29/01/05 | Still in hospital | 79 | F | No | No | No | No previous therapy | UTI |
| 13 | 02/02/05 | 17/03/05 | 75 | F | Yes (16 days) | Yes (15 days) | No | Corticosteroids | Pneumonia (RTI) |
| 14 | 14/03/05 | Still in hospital | 57 | M | 14/03/05 (11 days) | Yes (9 days) | No | No previous therapy | Wound Infection (SSI) |
| 15 | 24/03/05 | 67 | M | No | No | Yes | Antibiotic treatment | Wound Infection (SSI) | |
| 16 | 24/09/04 | 26/01/04 | 64 | F | 24/09/04 (60 days) | Yes (55 days) | Yes | No previous therapy | Pneumonia (RTI) |
| 17 | 01/04/05 | Still in hospital | 71 | M | No | No | Yes | Antibiotic treatment | Pneumonia (RTI) |
Genotype analysis of all A. baumannii strains isolated during the outbreak identified one major PFGE type B, that differed in more than 6 bands from one additional strain isolated from ICU of the hospital six months before (PFGE type A). All A. baumannii strains of PFGE type B showed an identical multi-resistant antibiotype, being susceptible to colistin and trimetropim-sulphomethoxazole, of intermediate susceptibility to ampicillin-sulbactam and meropenem, while resistant to all other antimicrobial tested. In these isolates, inhibition of OXA enzymes by 200 mM of NaCl reduced imipenem MIC by up to 8-fold. Molecular analysis of antimicrobial resistance genes showed that all epidemic A. baumannii strains harboured in their genomic DNA a class 1 integron containing the aacA4, orfX, and blaOXA-20 gene cassettes, an ampC gene and a blaoxa-51-like allele. Moreover, a blaoxa-58 gene surrounded by regulatory insertion sequence elements ISAba1 and ISAba3 was identified in a 21 kb plasmid DNA from A. baumannii strains of PFGE type B, but not PFGE type A. No amplification products were obtained from genomic DNA of epidemic strains of PFGE type B for blaIMP-type, blaVIM-type or blaSIM-type metallo-β-lactamase genes or blaoxa-23 or blaoxa-24 carbapenem-hydrolysing oxacillinases. Also, both carbapenem-susceptible A. baumannii strains of PFGE type A and carbapenem-resistant strains of PFGE type B expressed 26-kDa outer membrane protein CarO. Conjugation experiments demonstrated that resistance to imipenem, along with the blaoxa-58 gene, was transferred from A. baumannii strains of PFGE type B to those of PFGE type A.
The selection and the spread between different wards of a single A. baumannii clone producing OXA-58 carbapenem-hydrolysing oxacillinase were responsible for the increase of A. baumannii infections that occurred in Saint George University Hospital of Beirut, Lebanon. The epidemiologic pattern of the resistant organism vary from hospital to hospital, and control measures or therapeutic approaches should be customised to each institution.
P1009 Carbapenemase producing clinical isolates among non-fermenting bacteria from a tertiary hospital
D. Perimeni, K. Trifynopoulou, P. Giakouppi, S. Vourli, A. Alevizos, O. Zarkotou, I. Tsimettas, D. Gianneli, V. Kalapothaki, A. Vatopoulos (Athens, Piraeus, GR)
Objectives: The aim of our study was to investigate the distribution of metallo-β-lactamase producing strains and their molecular characteristics in non fermenting bacteria isolated from clinical specimens from a tertiary General Hospital in Piraeus, Greece in an 18-month period.
Methods: 110 strains of Pseudomonas aeruginosa and 57 strains of Acinetobacter calcoaceticus baumannii have been obtained mainly from blood and wound cultures, between 2002 and 2004. Antibiotic resistance was identified by disc diffusion tests and by determination of the MICs. All carbapenem-resistant P. aeruginosa and A. baumannii isolates were tested for the possible presence of MBL by placing a home-made disc containing 750 g EDTA 20 mm away from one containing imipenem 10 μg, confirmed with E-test MBL combination strips when the expansion was marginal. The presence of the carbapenemase-encoding genes blaVIM-1 and blaVIM-2 was tested by PCR. Genomic DNA of the blaVIM producing P. aeruginosa isolates, was digested with SpeI enzyme and processed for typing by Pulsed-field gel electrophoresis.
Results: 49 (44.5%) of P. aeruginosa and 24 (42%) of A. calco. baumannii isolates were carbapenem resistant. Of those, after further testing, 19 P. aeruginosa isolates were found to contain a blaVIM gene, while no A. calco. baumannii strain carried a metallo-β-lactamase. 18 P. aeruginosa isolates carried a blaVIM-2 gene and PFGE analysis showed a single pattern with two subtypes, while one strain carried a blaVIM-1 gene. All blaVIM genes were located in class 1 integrons.
Conclusion: Almost 39% of the carbapenem resistant P. aeruginosa isolates were blaVIM positive, while carbapenemase production did not seem to contribute in the resistance of infections caused by carbapenem resistant A. baumannii isolates in our hospital. The clonality of blaVIM-2 P. aeruginosa isolates implies that strict measures should be implemented in order to control the further spread of resistance.
P1010 Increasing diversity of metallo-β-lactamases produced by Gram-negative bacilli at a Korean hospital in 2006
C.K. Kim, E.M. Ko, J.H. Yum, D. Yong, K. Lee, Y. Chong (Seoul, KR)
Objectives: Carbapenems have been the most successful β-lactam antibiotics in evading bacterial resistance. However, carbapenem-resistant Gram-negative bacilli (GNB), particularly Pseudomonas spp. and Acinetobacter spp., have been increasingly isolated at a Korean hospital, and some of them produced VIM-2, IMP-1 and SIM-1 type metallo-β-lactamases (MBLs). We determined trend of isolation of MBL producing GNB in 2006.
Methods: GNB were isolated from January to September, 2006, from clinical specimens at a tertiary-care hospital in Seoul, Korea. Imipenem susceptibility was tested by the CLSI disk diffusion test. Imipenem-nonsusceptible isolates were screened for MBL production by the imipenem-disk Hodge (cloverleaf) test, and a double disk synergy test using imipenem disks and EDTA (750 ug) plus sodium mercaptoacetic acid (SMA, 2 mg)-containing disks. Screening-positive isolates were further tested by PCR to detect blaIMP-1, blaSIM-1 and blaVIM-2 alleles.
Results: Alleles of MBL gene were detected in 13.4% (81 of 603) of imipenem-nonsusceptible GNB isolates. It was interesting that 86.4% (19 of 22) of imipenem-nonsusceptible P. putida isolate had MBL genes. The types detected were blaVIM-2 in 50, blaIMP-1 in 26, and blaSIM-1 in 5 isolates. Relatively more prevalent MBL types were blaVIM-2 in P. aeruginosa (71.9%) and in P. putida (100%), blaIMP-1 in Acinetobacter sp. (56%). blaSIM-1 was detected only in A. baumannii isolates. blaVIM-2 was detected first time in one Ralstonia pickettii isolate. Relative increase in blaIMP-1 was observed in P. aeruginosa, blaIMP-1 and blaSIM-1 in Acinetobacter spp. The MBL types in P. putida remained to be only blaVIM-2. MBL gene was not detected in any of the imipenem-nonsusceptible isolates of 3 Achromobacter xylosoxidans ssp. xylosoxydans, one each of Alcaligenes faecalis, Enterobacter cloacae, Proteus mirabilis, Providencia rettgeri, and 4 Klebsiella pneumoniae. MBL-producing GNB were mostly isolated from urine (42; 51.9%) and sputum (23; 28.4%), but 4 (4.9%) were from blood.
Conclusion: MBL genes were detected in 13.4% of 603 imipenem-nonsusceptible GNB isolates. Increasing diversity of MBL gene types was noted in Acinetobacter spp. from which all three types of MBL genes including blaSIM-1 were detected.
Spread of E. coli and Enterobacteriaceae
P1011 Carbapenem resistance in an epidemic CTX-M-15 β-lactamase-producing Escherichia coli strain in the United Kingdom
E. Karisik, M.J. Ellington, R. Pike, M.F. Palepou, S. Mushtaq, J.S. Cheesbrough, P. Wilkinson, D.M. Livermore, N. Woodford (London, Preston, UK)
Objectives: E. coli strain A is the most widespread lineage with CTX-M-15 β-lactamase in the UK, and is dominant among ESBL producers in some locales. Cephalosporin MICs are lower than for most other isolates with CTX-M-15 enzyme, owing to an IS26 element between blaCTX-M-15 and its normal promoter. In one locale, this strain has additionally acquired a CIT-type AmpC β-lactamase, conferring resistance to all β-lactams except carbapenems and temocillin. We report here a further variant from this centre, with ertapenem resistance.
Method: MICs were determined by BSAC methodology. XbaI-digested genomic DNA profiles were compared by PFGE. Culture sonicates were used for outer membrane protein (OMP) profiling by SDS-PAGE. Beta-lactamase-encoding genes, including blaCTX-M and blaCIT, were identified by PCR.
Results: An ertapenem-resistant E. coli (MIC 4 mg/L) was isolated from a catheter urine specimen from an 86-year-old female nursing-home resident. She had had a catheter in situ for over 4 years, but had no recent hospital admissions and minimal exposure to antibiotics, with just one course of oral cephalexin in the preceding 12 months. None of the other residents had received carbapenems in the home, though they may have done so during periodic hospitalisations. This isolate was resistant to most β-lactams including cephalosporin/clavulanate combinations, fluoroquinolones, and to aminoglycosides except gentamicin (MIC 1 mg/L). Susceptibility to imipenem (MIC 1 mg/L) and to meropenem (MIC 0.5 mg/L) were reduced compared with typical E. coli strain A isolates. blaCTX-M-15 linked to IS26 was identified by PCR, and PFGE confirmed this isolate as being strain A. blaCIT was also detected by PCR, correlating with the lack of cephalosporin/clavulanate synergy. SDS-PAGE showed loss of OmpC.
Conclusion: We report ertapenem resistance in a representative of the most successful CTX-M-15 β-lactamase-producing E. coli strain in the UK; this may be explained by the loss of OmpC, together with the production of AmpC and CTX-M-15 enzymes. The continuous adaptation of epidemic strain A remains alarming, as treatment options are further reduced.
P1012 First cases of infection with Escherichia coli containing plasmid-mediated fluoroquinolone resistance (qnrA and qnrS) in Scandinavia
L.M. Cavaco, D.S. Hansen, F.M. Aarestrup, N. Frimodt-Møller (Copenhagen, DK)
Objectives: To investigate plasmid mediated flouroquinolone resistance in human and animal E. coli strains in Denmark.
Methods: Escherichia coli strains from humans and swine in Denmark were selected based on nalidixic acid resistance (MIC ≥ 32 mg/L) or low level resistance to ciprofloxacin (MIC ≤0.125 mg/L). Screening for qnrA, qnrB and qnrS genes was performed by PCR in pooled samples, on strains with PCR positive strains were further tested for resistance genes and transfer studies performed. Chromosomal mutations were detected by PCR and sequencing of QRDR (quinolone resistance determining regions) of topoisomerase genes.
Clinical data for each case was collected from the patient records.
Results: By screening of 130 strains by PCR for qnr genes, two positive isolates were identified, one strain (H88) contained the qnrS gene, another the qnrA gene (H93). Both strains showed low level ciprofloxacin resistance (MICs = 0.5 mg/L), although susceptible to nalidixic acid (MICs = 4–8 mg/L).
First case was an 88 year old woman who previously treated for breast cancer, with recurrent pneumonia, that was admitted to the hospital on suspicion of pneumonia and cystitis. An urine culture showed growth of 105 CFU/mL of an ESBL-producing multi-resistant E. coli (H88). The patient was treated successfully with mecillinam orally and was returned to the nursing home.
Second case was a 72 year old man with septicaemia and colon cancer. From blood, an ESBL-producing multiresistant E. coli (H93) was cultured. The patient was treated with i.v. piperacillin+tazobactam and ciprofloxacin, on which he improved clinically and became afebrile. During the subsequent months he deteriorated and finally succumbed to his cancer disease.
Conclusion: These are to our best knowledge the first two cases of infections with plasmid mediated fluoroquinolone resistant strains reported from Denmark as well as from Scandinavia. In both cases the isolates were ESBL-producing multi-resistant strains. The clinical impact of this low level fluoroquinolone resistance is difficult to assess, as the one patient treated with ciprofloxacin also received piperacillin+tazobactam towards which the strain was found susceptible. Further studies on optimisation of treatment in these cases are needed, to avoid treatment failures and/or spread of resistance.
P1013 Detection and characterisation of ESBLs in German Escherichia coli, isolated from animal, foods, and human origin between 2001–2006
B. Guerra, M.D. Avsaroglu, E. Junker, A. Schroeter, L. Beutin, R. Helmuth (Berlin, DE; Ankara, TR)
Objectives: Detection and characterisation of ESBLs in E. coli isolates, from animal, foods, and human origin and isolated between 2001–2006. Characterisation of the host structures.
Methods: About 2000 E. coli isolates (2001–2006) mostly from animal and food origin have been analysed in the NRL-Salm (BfR) for their susceptibility to 17 antimicrobial agents (including the β-lactams ampicillin and ceftiofur, and amoxycillin/clavulanic acid) by broth microdilution. Isolates showing ceftiofur-R were further tested for a panel of 11 β-lactams by disc diffusion. According to the R-patterns found, the strains were screened for different ESBL genes. The complete R-repertoire of the strains was characterised by molecular methods (R-genes, class 1 and 2 integrons and mutations in the quinolone-R determining regions). Strains were typed by XbaI-PFGE and plasmid profile analysis. Location of ESBLs was determined by hybridisation.
Results: Only two isolates were completely resistant (MIC ≥ 8 μg/mL) to ceftiofur. Three isolates showed intermediate resistance values (MIC = 1–2 μg/mL) and their resistance phenotypes to other β-lactams indicated mechanisms different than ESBLs. The two resistant isolates showed resistance patterns which suggested the presence of CTX-M enzymes (cefotaxime 22 and 21 mm, ceftriazone 20 and 17 mm, cefuroxime-R, cefpodoxime-R, aztreonam 25 and 26 mm, respectively). After PCR and sequencing CTX-M15 was found in one human isolate (Ont:H4) which also showed resistance to kanamycin-neomycin, nalidixic acid, ciprofloxacin, streptomycin, spectinomycin, sulfadiazine, tetracycline, trimethoprim and trimethoprim/sulfamethoxazole. The other isolate (Ont:H-) was isolated from milk from a cow with mastitis, and carried CTX-M1. The strains showed different PFGE patterns and plasmid profiles. Both CTX-M15 and CTX-M1 were located on big plasmids (aprox. sizes 160–180 kb). These plasmids could not be transferred.
Conclusions: While in many countries the presence and spread of ESBLs is of concern, their prevalence still remains low among German E. coli isolates. However, presence of ESBLs in multiresistant strains (more than 7 different classes of antimicrobials) including fluoroquinolone resistance is worrying and should be prevented by prudent use of extended-spectrum β-lactams.
P1014 Characterisation of Escherichia coli isolates producing extended-spectrum β-lactamases from a hospital and its surrounding healthcare centres in Stockholm
H. Fang, F. Ataker, K. Dornbusch (Stockholm, SE)
Objectives: Nosocomial outbreaks of Escherichia coli producing extended-spectrum β-lactamases (ESBLs) were observed at South General Hospital and its surrounding healthcare centres in Stockholm in 2002. One of the endemic clones had already circulated in the area as early as 2001. In recent years, the number of new ESBL-cases has increased. The present study is aimed at identifying β-lactamase genes and investigating clonal diversity among the ESBL strains detected in the area during the period 2001 to 2006.
Methods: Consecutive, non-repeated ESBL-producing E. coli isolates from South General Hospital and its surrounding healthcare centres in Stockholm were included in the study. The presence of ESBL was first phenotypically determined by either the double-disk method or Etest. Then all the isolates were subjected to a multiplex PCR assay targeting SHV, TEM, CTX-M and OXA type β-lactamases, and analysed by pulsed-field gel electrophoresis (PFGE). At least one isolate of each genotype, as determined by the PFGE pattern combined with the PCR profile of β-lactamase genes, was further analysed by sequencing the specific β-lactamase genes.
Results: Eighty-one strains were collected during the period 2001 to Oct. 2006, while 53% (43/81) of them were isolated during the latest two years. CTX-M gene was present in 93% (75/81) of the strains, followed by TEM (64%) and OXA (59%). Among the strains tested, three types of CTX-M ESBLs were identified, belonging to CTX-M-1 group, CTX-M-2 group and CTX-M-9 group, respectively. It was common that one strain harboured two or three different β-lactamase genes. The nosocomial outbreaks in the area in 2002 were represented by two clones, clone 1 and clone 2. Both clones produced CTX-M-1 group ESBLs. The initial clone 1 isolates in 2001 harboured three β-lactamase genes (TEM, CTX-M and OXA). In 2002, one of the clone 1 ESBL strains was found to have lost the TEM β-lactamase gene. Moreover, isolates of clone 1 losing both TEM and OXA genes were then detected in 2004.
Conclusion: The frequency of ESBL-producing E. coli has increased in Stockholm. CTX-M ESBLs were the main type of β-lactamases produced by the strains. One endemic ESBL-E. coli clone was persistent in the area through the years.
P1015 Genetic diversity of SHV-12 producing isolates from the community and nosocomial settings (Madrid, Spain)
A. Valverde, R. Canton, A. Novais, F. Baquero, T. Coque (Madrid, ES)
Objectives: To study the epidemiological features and diversity of genetic elements surrounding blaSHV-12 in isolates harbouring this enzyme from hospitalised and non-hospitalised patients, and non clinical samples from healthy volunteers, environment and food.
Methods: Forty three producing SHV-12 isolates (28 Escherichia coli and 15 Klebsiella pneumoniae) obtained from 1999 to 2005 were studied. Clonal relatedness was established by XbaI-PFGE. Phylogenetic groups among E. coli isolates were identified by a multiplex PCR assay. Transfer of bla genes was searched by broth and filter mating. Plasmid characterisation was accomplished by Barton's method, RFLP and incompatibility group was identified by the method described by Carattoli et al. Presence of IS26 and its association with blaSHV-12 was elucidated by PCR. Presence of integrons and associated gene cassettes within the variable region was determined by PCR and further sequencing.
Results: SHV-12 isolates were recovered from hospitalised (32.5% medical, 11.7% surgery and 9.3% ICU) and community patients (28%), healthy volunteers (14%) and environmental samples (4.5%). Fifty four percent of isolates were resistant to ciprofloxacin and/or nalidixic ac., whereas 90% were resistant to sulphonamide. E. coli isolates belonged to phylogenetic groups D (46%), A (43%) and B1 (11%). High clonal diversity was observed in E. coli isolates (D = 0.99) while most K. pneumoniae isolates belonged to a single epidemic clone recovered from 2001 to 2003. Most isolates were able to transfer blaSHV-12 gene (83%), located in plasmids of 48, 97 and 145 kb. Analysis of incompatibility groups showed the predominance of IncI1 and IncN plasmids with a no clear trend of dominance in different settings. A high diversity of resistance gene cassettes within the integron variable region was found (dhfr1-aadA1, sat, aacA4-qacEdelta1, IS1-orf, dhfr16-aadA2). Presence of IS26 was detected in most of SHV-12-isolates.
Conclusions: Strains producing blaSHV-12 were found both in the nosocomial (mainly associated to an epidemic K. pneumoniae clone) and in the community (mainly associated to different E. coli clones) settings. Presence of integrons with different genes cassettes in conjugative plasmid might contribute to the dissemination and persistence of this enzyme.
P1016 Most Escherichia coli strains overproducing AmpC β-lactamase belong to phylogroup A
S. Corvec, A. Prodhomme, C. Giraudeau, A. Reynaud, N. Caroff (Nantes, FR)
Objectives: AmpC overproducers represent about 1% of the E. coli strains isolated from clinical specimens in our hospital. Overproduction of AmpC enzyme is mostly caused by mutations in the ampC promoter, particularly at position −42. The aim of this study was the classification of the strains presenting this specific resistance mechanisms in the different phylogenetic groups.
Methods: 54 E. coli strains, isolated over a 12-year period, and previously identified as AmpC overproducers by increased MICs for third-generation cephalosporins without ESBL production (negative double disk synergy test) and spectrophotometric data were phylogrouped by multiplex PCR. As a comparison, 82 E. coli clinical isolates, susceptible to all β-lactams were also tested by the same method. The ampC promoter sequence was determined for all these isolates. ERIC-2 PCR was used to compare the isolates.
Results: 45 (83%) of the 54 AmpC overproducers tested belonged to phylogroup A. Among the 43 isolates presenting a −42 mutation in the ampC promoter, 37 (86%) belonged to phylogroup A, 5 to phylogroup B1 and one to phylogroup D. Among the 6 isolates presenting mutations at position −32 (box-35) in the promoter, 5 were from phylogroup A. Three other isolates, which were also from phylogroup A, harboured a −11 mutation. These −32, −42 and −11 mutations, responsible for AmpC overproduction were usually associated with DNA polymorphisms at position −88, −82, −18, +1 and +58 in the ampC promoter.
In the control susceptible isolates, these polymorphisms were detected in 10 ampC promoters (8 phylogroup B1 and 2 phylogroup A). These polymorphisms were never associated with the main phylogroup B2, representing 70% of the isolates.
Conclusion: In this study, a well-known chromosomal mechanism of resistance, overproduction of AmpC cephalosporinase, was clearly linked to E. coli phylogroupA. The strains belonging to this phylogroup are usually considered as less virulent. Studies of virulence factors from these AmpC overproducing strains are in progress in our lab.
P1017 The first identification of an extended-spectrum β-lactamase-producing Escherichia coli from Norwegian livestock
M. Sunde, H. Tharaldsen, H. Solheim, M. Norström (Oslo, NO)
Objectives: Until now no ESBL-producing bacteria from Norwegian livestock have been detected. The situation regarding antimicrobial resistance in food producing animals in Norway is very good in an international perspective. The frequencies of resistance are low to moderate and the situation has been stable since the start of the monitoring programme (NORM-VET) in year 2000. The NORM-VET 2006 programme susceptibility tested 250 E. coli isolates from faecal samples from broilers collected within the Norwegian salmonella control programme, as well as 150 E. coli isolates from broiler meat (retail stores). One E. coli isolate from a faecal sample showed reduced susceptibility to cephalosporins (MIC ceftiofur 4 mg/L, MIC cefotaxime 1 mg/L), this isolate was subjected to further investigations.
Methods: Rosco synergi test and Etest® ESBL tests were carried out. Polymerase chain reaction (PCR) and sequencing was performed for detection of genes encoding ESBL, conjugation was carried out in broth with E. coli DH5á as recipient strain.
Results: The strain was positive in the ESBL tests. The β-lactamase produced was not resistant to clavulanic acid. Further investigations showed the presence of the blaTEM-20 gene. Resistance to cephalosporins was transferred when conjugation was carried out, indicating that blaTEM-20 was located on a mobile DNA element.
The finding of an ESBL positive E. coli from the faecal flora of a broiler on a Norwegian farm is surprising. Cephalosporins are not used in the Norwegian broiler production and the presence of selection pressure is therefore absent. Interview with the farmer confirmed that there had been no use of cephalosporins to any animals or humans at the farm.
The origin of the strain (or the plasmid encoding resistance to cephalosporins) is unknown. One explanation might be that the strain (or the plasmid) could be part of the normal flora of imported animals taken into Norway for breeding purposes.
Conclusion: ESBL-producing E. coli can be present in the normal flora of a healthy domestic animal not exposed to the antimicrobial agent in question. Furthermore, genes encoding ESBL can probably maintain within a bacterial population without the involvement of direct selection force.
P1018 Relationship between clonal variability, phylogenetic groups and multiresistance amoung extraintestinal isolates of Escherichia coli lacking extended-spectrum β-lactamases
B. Ruiz Del Castillo, L. Martinez-Martinez (Santander, ES)
Objectives: To evaluate the clonal relationship among multiresistant clinical isolates of E. coli not producing ESBL and to determine the phylogenetic group of these isolates.
Methods: E. coli(n = 100) obtained consecutively from clinical samples (1 per patient), resistant to at least three of the following agents: amoxicillin (AMX), amoxicillin-clavulanate (AMC), nalidixic acid (NAL), gentamicin (GEN) and cotrimoxazole (SXT) and not producing ESBL, have been studied. Preliminary identification and susceptibility testing was done with the WalkAway system (Dade Behring). MICs were confirmed by reference microdilution (CLSI guidelines). Phylogenetic groups were defined by a multiplex PCR (Clermont et al. Appl. Environm. Microbiol 2000: 4555), using 5 E. coli strains from the ECOR collection as controls. Clonal relationship was evaluated by REP-PCR using primers REP1(5′IIIGCGCCGICATCAGGC-3′) and REP-2 (5′ACGTCTTATCAGGCCTAC-3′). Haemolysis production was assessed in sheep blood agar.
Results: Organisms were obtained from urine (79) or from other samples (21). Sixty-one isolates were from samples of patients admitted to the hospital and 39 from outpatients. Percentages of resistant+intermediate organisms were: AMX (92%), AMC (41%), GEN (96%), NAL (98%), SXT (96%). Forty-five isolates were in phylogenetic group D, 26 in group A, 12 in group B1 and 17 in group B2. Forty REP-PCR patterns were observed, with 24 patterns including one single isolate. Patterns I, II, XXIII and XXIV included 18 (all group D; 9 from inpatients), 14 (all group B2; 9 from inpatients), 8 (all group D and from neonates) and 4 (all group A; 1 from inpatient) isolates, respectively. All organisms were non-haemolytic. The more frequent phenotypes of resistance were AMX/GEN/NAL/SXT(AGNS; n = 49)and AMX/AMC/GEN/NAL/SXT (ACGNS; n = 38). AGNS/ACGNS resistant isolates included 17/21, 16/8, 8/7, and 8/2 organisms in groups D, A, B2 and B1.
Conclusions: A great clonal variability among multiresistant ESBL-negative extraintestinal E. coli has been observed, but some clones are distributed in both hospitalised and non-hospitalised patients. While most isolates correspond to phylogenetic group D, almost one third of them are of groups A and B1, being isolates of group B2 the less frequent ones.
P1019 Epidemiology and molecular characterisation of clinical isolates of Escherichia coli producing extended-spectrum β-lactamases from Rome, Italy
A. Carattoli, A. Garcia, P. Varesi, D. Fortini, S. Gerardi, A. Penni, C. Mancini, A. Giordano (Rome, IT)
Objectives: Extended-spectrum β-lactamase (ESBL)-producing Escherichia coli are an emergent cause of infections in the Policlinico Umberto I, a large University hospital of Rome (1500 beds), making more difficult managing carrier patients. Our aim is to analyse the epidemiology and to characterise the E. coli ESBL producers in the population attending our hospital.
Methods: A perspective study was carried out from 1st April to 15th September 2006. A total number of 5674 various clinical samples were sent for bacterial isolation to the Microbiology Laboratory from 51 different wards, for suspected hospital acquired infections. 209 E. coli strains were isolated from 166 inpatients. Antimicrobial susceptibilities were carried out by the VITEK 2 system. Double-disk synergy test were used for the confirmation of ESBL producers. ESBL genes were characterised by PCR and sequencing. Clonal relationships among the isolates were determined by ERIC-PCR. Plasmid extraction was performed by alkaline lyses. Plasmids were typed by the PCR-based replicon typing method.
Results: A total of 51 non-repetitive (one for each patient) E. coli ESBL positive strains were obtained indicating a prevalence of 30% (51/166) of E. coli ESBL producers in our hospital. 29 isolates were genotyped by ERIC-PCR and analysed for the SHV-, TEM-, CTX-M-type ESBL genes. Nine different clonal lineages were evidenced and two slightly prevalent clones were identified. The most diffused ESBL was CTX-M-15, being detected in 27/29 E. coli strains of different genotypes. The two strains lacking the CTX-M-15 were SHV-12 producers. Three strains carried both the SHV-11 and CTX-M-15. Most of the strains were also positive for the blaTEM gene (22/29). The blaCTX-M-15 was detected on similar plasmids in all selected isolates and their transconjugants, belonging to the FII incompatibility group. Further plasmid characterisation is under way.
Conclusion: The eruptive worldwide dissemination of the CTX-M-15 in geographically distinct hospitals emphasizes the necessity of immediate intervention and epidemiological monitoring. E. coli with CTX-M-15 are a major problem in the our hospital causing outbreak as well as many sporadic infections. The impressive prevalence of this genetic trait in our hospital seems associated to the diffusion of epidemic plasmids and the current knowledge on the spread of these plasmids is still limited but it should be taken into account for the design of control measures.
P1020 Genotypic characterisation of Norwegian Escherichia coli clinical isolates with an AmpC-resistance profile
B. Haldorsen, B. Aasnaes, K. Dahl, A-M. Hanssen, G. Simonsen, T.R. Walsh, M.A. Toleman, A. Sundsfjord, E. Lundblad (Tromso, NO; Cardiff, UK)
Introduction: Resistance to extended-spectrum cephalosporins in E. coli can be meditated by mobile AmpC-type β-lactamases and over-expression of native AmpC enzymes normally repressed. Herein, we describe a full genetic characterisation of AmpC-mediated resistance and report novel insertion promoter sequences.
Methods: The Norwegian Reference Centre for Detection of Antimicrobial Resistance (K-res) received 414 clinical isolates of E. coli from different Norwegian laboratories with a reduced susceptibility to third-generation cephalosporins, over a 2-year period 2003–2005. Of these, 23 isolates showed an AmpC-mediated phenotype using substrate profiling. Strains were molecularly characterised and typed by IEF and PFGE. AmpC-type β-lactamase genes and adjacent sequences were analysed using PCR, sequence analysis, endonuclease plasmid analysis and hybridisation.
Results: The 23 isolates all possessed pI values of pI 9.0–9.2 indicative of AmpC expression. Multiplex blaampC-PCR and southern blot analysis of S1 nuclease digested total DNA separated by PFGE, suggests that 10 of the 23 E. coli isolates have the blaampC gene located on plasmids. Sequencing analysis showed that 9 of these had the blaCMY-2 gene, while one strain had the blaCMY-7 gene. All plasmid-mediated blaampC genes were found to be linked to an insertion sequence ISEcp1-like element by an ISEcp1-blaampC gene linkage PCR. The bla CMY-2 genes possessed blc, sugE and dsbC downstream suggesting that they belong to Type 1 and Type 2. The 13 non-plasmid mediated blaampC strains, revealed promoter and attenuator alterations compared to E. coli wild-type promoters. Two blaampC promoter regions were interrupted by the insertion sequence ISEc10 of the IS21-family. PFGE analysis suggests that the majority of the 23 E. coli isolates are unrelated.
Conclusion: These findings suggest there are two different populations of AmpC-type resistant E. coli clinical isolates in Norway – plasmid-mediated blaCMY genes and cognate AmpC promoter mutations. The genetic context of the blaCMY gene suggests they are identical to international strains.
P1021 A comparison of antimicrobial susceptibility in non-clinical and clinical isolates of Escherichia coli
M. Sjölund, J. Bonnedahl, S. Bengtsson, G. Kahlmeter (Växjö, Kalmar, SE)
Objectives: The frequent use of antimicrobial agents during the last decades has resulted in an increased selective pressure, driving resistance development in pathogenic, commensal as well as environmental microorganisms. To assess the effect of the host's environment and the role that human use of antimicrobials play in selecting for resistance in the normal microbiota, we have performed a unique comparison of antimicrobial susceptibility in isolates of E. coli originating from two widely different areas – one area with a standard use of antimicrobial agents, County of Kronoberg, Sweden, and one area totally devoid of antimicrobial agents; the Arctic.
Methods: In 2005, the Swedish Polar Research Secretariat organised a scientific expedition to the Arctic. During this expedition, cloacal swabs from Arctic birds were collected at three different geographical regions; Northern Siberia, Alaska and Greenland. In the present study, 97 E. coli isolates from birds, and 100 clinical isolates from blood cultures performed at Växjö hospital in 2004–2005, were included. The antimicrobial susceptibility to 17 antimicrobial agents was determined by disk diffusion. In addition, 25 copies of E. coli ATCC 25922 were included and served as quality-control in a blinded fashion. For each antimicrobial agent, zone inhibition histograms were established. The wild type distributions were defined by the median zone diameter and the NRI (Normalised Resistance Interpretation) value.
Results: The wild type distributions of avian and clinical isolates of E. coli showed complete agreement. Among the avian isolates, resistance (defined by the epidemiological cut-off) occurred most often to ampicillin, sulfamethoxazole, trimethoprim, chloramphenicol and tetracycline. Among the clinical isolates, resistance was most common to ampicillin, nalidixic acid, streptomycin, sulphamethoxazole and trimethoprim. The most pronounced difference between human and avian isolates was observed for fluoroquinolone resistance.
Conclusion: The wild type distributions of avian and clinical patient isolates of E. coli are identical. However, clinical isolates of E. coli were, as expected, significantly more often resistant although resistance as well as multi-drug resistance was observed among the avian isolates.
P1022 Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in a Portuguese university hospital
G.J. Da Silva, A.T. Simões, A. Tavares, L. Boaventura, C. Vital, G. Ribeiro (Coimbra, PT)
During the past decade CTX-M extended-spectrum β-lactamases (ESBL) emerged in many countries. In Portugal only sporadic CTX-M-producers isolates has been reported.
Objectives: The aim of the study was to investigate the prevalence of CTX-M producers among ESBL-producing isolates of Escherichia coli and Klebsiella pneumoniae as well as the molecular epidemiology and the mechanism of dissemination of these emerging resistance determinants.
Methods: Between November 2003 and February 2004 nonduplicate isolates of E. coli (n = 25) and K. pneumoniae (n = 17) collected in the University Hospitals of Coimbra were screened for CTX-M, TEM and SHV enzymes production. They were identified as ESBL producers by the automatic VITEK 2 AES. ESBL production was further confirmed by ESBL E-test strips and disk diffusion synergy test. CTX-M alleles were detected in whole DNA and plasmids by PCR with specific primers for phylogenetic groups 1, 2 and 9. Transfer of blaCTX determinants was assayed by conjugation using E. coli K102 (NalRRifR) as recipient cell. Genetic relatedness was assessed by RAPD.
Results: ESBL production was confirmed for ALL isolates by the synergy method using CAZ, CTX, AZT, CPM and AMC. These results were corroborated by E-test ESBL strips except for 3 K. pneumoniae isolates. Only CTX-M-1 cluster enzymes were detected: 23 (92%) E. coli and 2 (11.7%) K. pneumoniae isolates. TEM determinants were amplified in the CTX-M-negative E. coli isolates. None E. coli expressed SHV enzymes. The nature of these determinants was not further determined. Two major lineages were observed among E. coli isolates by RAPD. Clone A included the majority of the isolates (72%) while clone B comprise 7 isolates with minor band variations. By sequence analysis we found CTX-M-15 in clone A and CTX-M-1 in 2 isolates of clone B. CTX-M-1 was detected in one of the CTX-M-positive K. pneumoniae isolates. The other, a mutant found in the inhibition zone of cefotaxime disk, exhibited a CTX-M-15. The determinants were carried by plasmids >10 kb and it was possible to transfer blaCTX-M-1 determinant, but no transconjugants were obtained with transference of blaCTX-M-15.
Conclusions: A prevalence of the emergent CTX-M-15 was observed mainly in E. coli isolates from urinary tract infections. The lower genotypic diversity among E. coli CTX-M-15 producers and lower propensity of transfer by conjugation of blaCTX-M-15 suggest that dissemination of CTX-M-15 was more related to clonal expansion.
P1023 Clonality and presence of antibiotic resistance genes of E. coli isolated in recurrent urinary tract infections
K. Truusalu, E. Sepp, J. Shchepetova, S. Koljalg, I. Vainumäe, K. Stroo, K. Sepp, M. Mikelsaar (Tartu, EE)
E. coli is the main causative agent of recurrent urinary tract infections (rUTI). The antibacterial treatment can facilitate the transmission of resistance gene cassettes e.g. integrons and sul genes encoding resistance to sulfamethoxazole (SMX). However, it is not known if the persistence of clonal isolates can be predicted by antibiotic resistance pattern. We aimed to investigate the molecular identity of consecutive E. coli strains in rUTI patients and its association with geno- and/or phenotypically changed antibiotic resistance pattern.
Material and Methods: A total of 22 E. coli strains were isolated from urine of 8 children (aged 2 mo – 8 y) out of 64 with phenotypically changed antibiotic resistance pattern in 2–4 episodes of rUTI. The index episode was acute pyelonephritis but in recurrences cystitis was also found. During consecutive UTI episodes the changes in antibiotic resistance to trimetroprim-sulfamethoxazole (TMP-SMX) in 3, to ampicillin in 2, to cefuroxime in 1 and to a combination of both TMP-SMX and ampicillin in 2 patients were detected with disk-diffusion method. The MIC values to ampicillin, cefuroxime, cefotaxime, meropenem, gentamicin, ciprofloxacin and SMX were measured by E-test. Microbial DNA was extracted using QIAamp DNA Mini Kit. Genes of Int1, sul1, sul2 and sul3 were detected by PCR. Pulsed Field Gel Electrophoresis (PFGE) with Qiagen kit was applied to detect E. coli strain identity.
Results: In three patients index case and recurrences had similar PFGE patterns while in remaining 5 patients they varied. There was no relationship between clonality and phenotypically expressed changed antibiotic resistance and the presence of resistance genes. Eight strains isolated from six patients contained intI gene and the MIC values to cefuroxime were higher than in intI negative strains (p = 0.003). Phenotypical resistance to TMP-SMX was found in 6/22 strains, while sul genes were present in 17/22 strains (p = 0.001). The MIC to SMX was higher in sul positive than in sul negative strains (p = 0.001).
Conclusions: RUTI episodes are more frequently due to different individual clones than one persisting clone. Persistence of particular rUTI causing E. coli clones can not be detected by the stability of either phenotypic nor genotypic resistance pattern.
P1024 Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains isolated from drinking and recreational, salmaster waters
R. Cernat, C. Balotescu, D. Ivanescu, D. Nedelcu, V. Lazar, M. Bucur, D. Valeanu, R. Tudorache, M. Mitache, M. Dragoescu (Bucharest, RO)
Aim: To delineate the clonal diversity and transmission patterns, and to investigate the occurrence and distribution of various resistance genes in multiple antibiotic resistant E. coli strains isolated from various aquatic sources.
Methods: 100 environmental Escherichia coli isolated from drinking water (50) and marine water (50) in Constanta and Bucharest were previously characterised for their virulence, pathogenicity, antibiotic resistance and plasmid profile. 30 strains from this group showed multiple-antibiotic resistance and have been further investigated by PCR, multiplex PCR and sequencing, for the presence of TEM-, SHV- and CTX-M-like extended-spectrum β-lactamases, blaIMP1, blaOXA, sul1, sul2 and sul3 genes, dfr-A like genes, tet efflux resistance genes, class 1, class 2 and class 3 integrons with the corresponding gene cassettes, respectively. Clonal diversity was assessed through PFGE of XbaI-digested genomic DNA.
Results: E. coli isolates showed 16 unrelated PFGE patterns. All strains were ampicillin resistant, blaTEM1a and blaTEM1b genes being identified in 40% of the strains while group 1 and group 8 of blaCTX-M genes were present in 90% of the strains. Most of the retrieved sequences were identical or very similar to β-lactamase genes previously characterised from clinical isolates. None of blaSHV, blaOXA and blaIMP1 genes retrieved. 20% of E. coli strains were kanamycin resistant, and the aphA1 and aphA2 genes were detected. The tetA, tetB and tetD genes were found in all the strains. A relatively high prevalence has been also obtained for dfrA17 (70%), dfrA12 (50%), sul1 (50%) and sul2 (60%) genes. Class 1 integrons were the only found in resistant bacteria and harboured 6 different cassette arrays, most common being dfrA17-aadA5.
Conclusions: All aquatic multiresistant E. coli strains showed a wide viariety of antibiotic resistance genes. Therefore, the aquatic ecosystem may play a key role as acceptor and donor of transmissible antimicrobial resistance mechanisms. The inclusion of some resistance genes inside class 1 integrons constitutes an effective means to spread antibiotic resistance among bacteria from different ecosystems.
P1025 Isolation of quinolone-resistant CTX-M-producing Escherichia coli from raw chicken meat sold in retail outlets in the West Midlands, UK
V. Ensor, R. Warren, P. O'Neill, V. Butler, J. Taylor, K. Nye, M. Harvey, D. Livermore, N. Woodford, P. Hawkey (Birmingham, Shrewsbury, Colindale, London, UK)
Objectives: To identify genotypes of CTX-M β-lactamase-producing Escherichia coli isolated from raw chicken on sale in the UK West Midlands.
Methods: Fresh and frozen raw chicken fillets (originating from the UK and other countries) were purchased from 18 different retail outlets, including 13 major supermarkets in Shropshire and around Birmingham, UK. Samples were cultured onto CLED agar with 8 mg/L ciprofloxacin. A cefpodoxime disc (30 μg) was placed on the agar surface. Resistant colonies identified as E. coli using chromogenic agar were investigated for ESBL production using the combination disc test; those found positive isolates were screened for blaCTX-M by multiplex PCR. Reverse line hybridisation was used to identify blaCTX-M genotypes.
Results: See Table 1 .
Table 1.
CTX-M-producing E. coli isolated from chicken meat by country
| Origin | Total positive/tested | CTX-M enzyme present |
|||
|---|---|---|---|---|---|
| CTX-M-1 | CTX-M-2 | CTX-M-8 | CTX-M-14 | ||
| British | 1/62 | 1 | 0 | 0 | 0 |
| Irish | 0/3 | 0 | 0 | 0 | 0 |
| Brazil | 5/10 | 0 | 5 | 0 | 0 |
| Brazil/Poland/Francea | 3/4 | 0 | 3 | 0 | 0 |
| Poland | 0/4 | 0 | 0 | 0 | 0 |
| Netherlands | 2/2 | 0 | 2 | 0 | 0 |
| Spain | 0/1 | 0 | 0 | 0 | 0 |
| France | 0/1 | 0 | 0 | 0 | 0 |
| Denmark | 0/1 | 0 | 0 | 0 | 0 |
| German | 0/1 | 0 | 0 | 0 | 0 |
| Unknown | 6/40b | 0 | 1 | 1 | 4c |
| Total | 17/129 | 1 | 11 | 1 | 4 |
Precise country of origin not stated on packaging.
Country of origin not identified on packaging.
All chicken meat containing E. coli with CTX-M-14 was purchases from 2 major supermarkets and was processed at 2 UK cutting stations.
Conclusions: E. coli with CTX-M enzymes were infrequently isolated from chicken meat originating from EU countries, except that from the Netherlands. CTX-M-15, the predominant ESBL type in clinical isolates in the UK (and many other countries), was not found, suggesting contaminated chicken is an unlikely continuing source for current human gut colonisation with this type.
E. coli with CTX-M-2 were isolated from 2/2 and 5/10 chicken meat samples originating from the Netherlands and Brazil, respectively; CTX-M-2 is the most common CTX-M type from clinical E. coli in Brazil but is rare in clinical disease in the UK, although it has been isolated from faecal samples. CTX-M-14 is the second commonest UK isolate and common in other EU countries.
Analysis was complicated by the fact that countries of origin of 40 raw chicken samples was not identifiable on packaging, however, it was apparent that there may be significant differences in the CTX-M enzyme found in chicken originating from different countries. Our findings suggest that chicken meat may be a potential source of bowel colonisation with E. coli producing CTX-M enzymes, but do not identify a link to the main CTX-M type (CTX-M-15) from clinical infections in the UK.
P1026 Occurrence and diversity of genetic resistance determinants among Enterobacteriaceae clinical isolates in Aveiro, Portugal
S. Ferreira, M. Toleman, T. Walsh, S. Mendo (Aveiro, PT; Cardiff, UK)
Objectives: The aim of the present study was to assess the occurrence of antimicrobial resistance determinants and their genetic apparatus among clinical isolates from Aveiro, Portugal.
Methods: The isolates population was composed by Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa, Providencia stuartii, Stenotrophomonas maltophilia, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, Morganella morganii and Proteus mirabilis. PCR, sequencing and sequence analysis was used to assess β-lactamase encoding sequences, class 1 and class 2 integrases, integron variable regions and ISCR elements. The Biological Sequence Alignment Editor, BioEdit version 7.0.0 was used for DNA and amino acid sequence alignments. Sequences obtained were compared with others deposited in the EMBL Genebank.
Results: PCR specific for CTX-M encoding sequence showed the presence of this gene in 33.3% of the isolates bearing an ESBL. The intI1 gene was present in 20.7% of the Enterobacteriaceae isolates. The intI2 gene was present in 3 isolates possessing the same cassette array (dfrA1, sat1, aadA1). A total of 14 cassettes included in 6 different cassette arrays were identified – the most frequently found genes were aadA variants. The ISCR elements were present in 8.1% of the isolates and were present in 9 E. coli strains and 1 Enterobacter cloacae.
Conclusion: This is the first report of ISCR elements from Portugal and as they are intrinsically linked to antibiotic resistant genes, their high incidence, particularly among E. coli strains, is disconcerting. The high percentage of CTX-M genes confirms that these have now become the dominant ESBL genotype within this region.
P1027 Plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking a chromosomal ampC gene and E. coli: prevalence at a Swiss university hospital and distribution of the different molecular types in the northern part of Switzerland
H. Adler, L. Fenner, D. Hohler, R. Frei (Basle, CH)
Objectives: (1) to determine the prevalence of plasmid-mediated AmpC β-lactamases in isolates of Enterobacteriaceae lacking a chromosomal ampC gene and E. coli at the University Hospital Basel. (2) to determine which types of plasmid-mediated AmpC β-lactamases occur in the northern part of Switzerland.
Methods: Between January 27 and September 27, 2006, a total of 2,100 consecutive clinical isolates of E. coli and Enterobacteriaceae naturally lacking a chromosomal ampC gene were screened for resistance to cefoxitin with the disk diffusion test. Furthermore, clinical isolates suspected to harbour a plasmid-mediated AmpC β-lactamase were collected from laboratories in the northern part of Switzerland. Multiplex AmpC PCR (Perez-Perez and Hanson, 2002, J Clin Microbiol 40: 2153–62) was performed on all cefoxitin-resistant isolates. The ampC genes of the PCR-positive isolates were sequenced.
Results: (1) One hundred of the consecutive clinical isolates were cefoxitin-resistant. Plasmid mediated AmpC β-lactamases were found in three isolates of E. coli. Thus, the prevalence of plasmid-mediated AmpC β-lactamases was 0.14%. (2) Plasmid-mediated AmpC β-lactamases were found in 16 isolates from 5 laboratories in the northerm part of Switzerland. Thirteen of the isolates were E. coli, 2 were Klebsiella pneumoniae and one was Proteus mirabilis. CMY-2 and CMY-2 like AmpC β-lactamases were detected in 11 isolates of E. coli, 2 isolates of K. pneumoniae and one isolate of P. mirabilis. DHA-1 was detected in 2 isolates of E. coli.
Conclusion: (1) The prevalence of 0.14% of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking a chromosomal ampC-gene plus E. coli at the University Hospital Basel is very low. (2) CMY-2 and CMY-2 like AmpC β-lactamases are the predominant plasmid-mediated AmpC β-lactamases (87.5%) in the northern part of Switzerland. DHA-1 was the only other AmpC β-lactamase that was found (12.5%).
P1028 Countrywide spread of CTX-M-3 and CTX-M-15 extended-spectrum β-lactamases among Enterobacteriaceae in Bulgaria
R. Markovska, I. Schneider, E. Keuleyan, M. Sredkova, D. Ivanova, K. Rachkova, B. Markova, T. Kostyanev, E. Savov, I. Mitov, A. Bauernfeind (Sofia, BG; Munich, DE; Pleven, Stara Zagora, BG)
Objectives: (1) To characterise the distribution of CTX-M producing isolates among eight clinical centres from three towns in Bulgaria. (2) To investigate their epidemiology and mechanism of spread.
Methods: Antibiotic susceptibility was determined by disc diffusion method (CLSI standards 2005). The extended-spectrum β-lactamase (ESBL) production was confirmed by CLSI double disc ESBL confirmatory method. Conjugation was performed on solid medium. Isoelectric focusing, followed by bioassay, ESBL-group specific PCR and sequencing of ESBL genes of representative isolates were carried out. RAPD with ERIC-1A and ERIC-2 primers and plasmid fingerprinting with PstI restriction were performed with representative strains.
Results: During a survey on ESBLs in Bulgaria from 1996 to 2003, CTX-M-type ESBL-producing Klebsiella spp. (47), E. coli (113), S. marcescens (9), Enterobacter. (5), C. freundii (6) from eight centres in three different towns were detected. The most widespread enzyme was CTX-M-15 (in all centres), followed by CTX-M-3 (in six centres). CTX-M-15 was produced mainly from E. coli strains (83%) and associated with urinary tract infections (49%). CTX-M-3 was produced from K. pneumoniae – in 52%. The rate of CTX-M enzyme harbouring strains has increased rapidly after first detection in 2001 to 56% in 2003. Resistance of CTX-M-3 producing strains was: AUG – 39%, TOB – 89%, GEN – 100%, AMI – 87%, CIP – 0%, TET – 33%, SXT – 85% and CHL – 26%.
For CTX-M-15 producers the resistance was: AUG – 87%, TOB – 95%, GEN – 88%, CIP – 84%, TET – 94%, SXT – 52% and CHL – 51%.
Epidemiological analysis by RAPD revealed a broader diversity of RAPD-types among CTX-M-3 producing strains in comparison with CTX-M-15 producers. The PstI plasmid fingerprinting showed a broader variety of CTX-M-15 encoding plasmids than CTX-M-3 harbouring plasmids. One of CTX-M-3 carrying plasmid was dominant, we detected it in four different bacterial species, isolated from patients in three towns.
Conclusions: In eight clinical centres in three Bulgarian towns CTX-M-15 and CTX-M-3 were the only CTX-M-type β-lactamases found. Our data suggest that plasmid transfer is the main mechanism of CTX-M-3 spread, while for CTX-M-15 producers spread by clonal dissemination is more prevalent.
P1029 Explosive emergence of CTX-M-15 extended-spectrum β-lactamase in Enterobacteriaceae in Kuwait
L. Poirel, V. Rotimi, S. Bernabeu, W. Jamal, P. Nordmann (Le Kremlin Bicetre, FR; Kuwait City, KW)
Objectives: Emergence of expanded-spectrum β-lactamases (ESBL) of the CTX-M group in Enterobacteriaceae has been reported worldwide. Our goal was to identify ESBL genes from a collection of clinically-significant ESBL-producing enterobacterial isolates recovered from the Mubarak hospital in Kuwait City, Kuwait from 2002 to 2004.
Methods: PCR with primers specific for several ESBL genes (blaTEM, blaSHV, blaCTX-M, blaVEB, and blaPER) were used for screening followed by sequencing of PCR products. Primers specific for mobile elements known to be at the origin of blaCTX-M genes acquisition were also used. Strains (one per patient) were collected during a three-year period. ESBL production was suggested by results of liquid medium VITEK 2 analyzer and confirmed by synergy tests performed on solid agar plates. Plasmid profiles analysis was determined by the Kieser method followed by Southern hybridisation with blaCTX-M specific probes.
Results: 598 enterobacterial isolates were recovered during the studied period with 63 ESBL-producers (10.5%). Noteworthy, in 2002 and 2003 respectively, only 2 out of 57 (3.5%) and 6 out of 265 (2.3%) enterobacterial isolates were ESBL producers, whereas they were 55 out of 276 (20%) in 2004. ESBL producers were mostly Escherichia coli (n = 29), Klebsiella pneumoniae (n = 19). In addition, there were several Enterobacter cloacae (n = 4), Enterobacter aerogenes (n = 3), Proteus mirabilis (n = 6), Citrobacter freundii (n = 2) and Serratia marcescens (n = 1). They were from different hospital units, but mostly from Medicine, Surgery and Nephrology departments. The ESBL determinants were mostly of the CTX-M group (50 out of 63), with CTX-M-15 being the main ESBL (46 out of 50). Insertion sequence ISEcp1 was identified systematically upstream of the blaCTX-M-15 gene. The other CTX-M variants were CTX-M-2 and CTX-M-9. The other ESBL-type were mostly SHV-type enzymes, and one isolate was VEB-1 positive.
An heterogeneous pattern of blaCTX-M-borne plasmids was observed that indicated that the dissemination of those resistance determinants were not related to dissemination of a single plasmid.
Conclusion: This study constitutes the first survey on ESBL spread in Kuwait. Dissemination of CTX-M-15 producers was identified as observed worldwide with a threatening increasing detection rate.
P1030 Low prevalence of TEM-type ESBLs among nosocomial strains of Enterobacteriaceae in Russia
L.N. Ikryannikova, E. Shitikov, E.N. Ilina, M.VEdelstein, V.M. Govorun (Moscow, Smolensk, RU)
Objectives: Production of extended-spectrum β-lactamases (ESBLs) in Enterobacteriaceae is one of the most prevalent mechanisms of resistance to oxyimino-β-lactams. Among others enzymes, the numerous mutant variants of TEM-type penicillinases (e.g. TEM-1 and TEM-2) possess ESBL phenotype. In this study we evaluated a contribution of TEM-type ESBLs to the development of resistance to oxyimino-β-lactams in nosocomial strains of Enterobacteriaceae collected in Russian Federation.
Methods: A total of 718 ESBL-producing nosocomial isolates of 10 different genera of the family Enterobacteriaceae, mainly Klebsiella pneumoniae (47.4%) and Escherichia coli (40.5%), collected in the hospitals from Central (n = 235), North-West (n = 52), Volga (n = 39), South (n = 95), Ural (n = 62), Siberian (n = 175) and Far-Eastern (59) regions of Russian Federation as part of the national surveillance of antimicrobial resistance in ICU pathogens in 2002–2004 were investigated. The ESBL phenotype of these isolates was inferred based on the MIC of at least one of the oxyimino-cephalosporins (cefotaxime, ceftazidime and cefepime) ≥2 mg/L and positive results of the MIC synergy test (according to CLSI), or of the double-disk synergy with clavulanic acid. The complete coding sequences of TEM β-lactamase-coding genes were amplified by PCR and directly sequenced using the automated fluorescent dideoxy terminator sequencing.
Results: Three hundred and seventy three (51.9%) of the studied isolates were PCR-positive for blaTEM genes. Of these, 372 were found to possess TEM-penicillinase genes, including 368 – TEM-1 (98.7%), two – TEM-2, one – TEM-57 (Gly92Asp), and one – TEM-117 (Leu21Phe). Only one strain had a single Arg164His substitution consistent with the TEM-29 ESBL.
Conclusion: This is a first report of a distribution of TEM-type ESBLs in Russian nosocomial strains of Enterobacteriaceae expressing ESBL phenotype. Our results show that TEM-type ESBLs are not a common cause of resistance in Russian Federation.
P1031 Prevalence of faecal carriage of metallo-β-lactamase producing Enterobacteriaceae in a Spanish hospital
T. Curiao, M. Tato, A. Valverde, A. Novais, F. Baquero, T.M. Coque, R. Canton (Madrid, ES)
Background: Metallo-β-lactamase (MBL) producing Enterobacteriaceae have been recognized in different European countries, including Greece, France, and Spain. We recently described a polyclonal outbreak involving these isolates in our hospital in Madrid (Tato et al. 46th ICAAC, 2006; abstract C2–66). In this study we investigated the intestinal colonisation by MBL producing isolates of hospitalised and ambulatory patients in our geographic area.
Methods: A total of 600 faecal samples from 569 hospitalised (20%) and ambulatory (80%) patients who attended in our hospital (January-April 2006) were studied. Aliquots of saline faecal suspension were cultured in MacConkey supplemented with ceftazidime (4 mg/L) agar plates and in Luria-Bertani (LB) supplemented with imipenem (2 mg/L) broth. Isolates from positive cultures were screened for MBL production by a double disk synergy test (DDST) using imipenem (10 μg), ceftazidime (30 μg) and EDTA (1900 μg) disks. Bacterial identification and antimicrobial susceptibility were performed using the WIDER system (Fco. Soria Melguizo, Madrid, Spain). MLB characterisation (blaVIM and blaIMP PCR, sequencing, and IEF) and PFGE were performed in isolates with positive DDST.
Results: A positive faecal carriage with MLB producing isolates was observed in four patients (0.7%) hospitalised (3.5%) at surgical (n = 1) and medical (n = 1) wards and ICUs (n = 2). No colonised patients were detected in outpatients. Proposed method for detection of MBL producers was efficient. All MBL producers were recovered on both supplemented MacConkey agar plates and LB broth and identified as Enterobacter cloacae (n = 2) and Klebsiella pneumoniae (n = 2). All of them produced VIM-2 enzyme and belonged to epidemic clones detected in our hospital. Unlike E. cloacae, patients with K. pneumoniae colonisation were also infected by this clone.
Conclusions: MBL (VIM-2) producing Enterobacteriaceae have emerged in our geographic area in hospitalised patients. Prevalence of colonised patients (3.5%) in our institution represents a clinical concern for the maintenance of epidemic clones and source for infections. Intervention strategies of containment of MBL producing isolates should include faecal carriage studies.
P1032 Extended-spectrum β-lactamase producing Enterobacteriaceae in a Swiss university hospital: molecular characterisation and susceptibility to ertapenem
T. Bruderer, A.F. Widmer, M. Flury, S. Oezcan, M. Jutzi, R. Frei (Basle, CH)
Objectives: First, to determine the molecular types of extended-spectrum β-lactamases (ESBLs) in Enterobacteriaceae at the University Hospital Basel, Switzerland. Secondly, to assess the activity of ertapenem against ESBL-producing strains.
Methods: Between 1/1/2003 and 31/12/2005, all consecutive clinical Enterobacteriaceae isolates were screened by use of 4 ESBL markers (ceftazidime, ceftriaxone, cefpodoxime, and aztreonam) according to Clinical and Laboratory Standards Institute (CLSI) guidelines. As phenotypic confirmatory test, three different ESBL Etest strips (AB Biodisk, Sweden) were used, i.e., ceftazidime, cefotaxime, and cefepime, each with and without clavulanic acid. Repetitive patient isolates and K1 hyperproducing Klebsiella oxytoca were excluded. To determine the molecular types, PCR amplifying SHV, TEM, and CTX-M genes was performed and amplicons were sequenced. Minimal inhibitory concentrations (MICs) of ertapenem were determined by Etest and interpreted according to the standards of CLSI.
Results: Among 8,384 isolates screened, 57 (0.68%) phenotypically confirmed ESBLs were identified. The ESBL rate was 1.1% and 3.4% in Escherichia coli and Klebsiella pneumoniae, respectively. Among ESBL strains, 59.6% were E. coli, 31.6% K pneumoniae, and 8.8% other genera. The majority (49.1%) of ESBL strains was found in urine samples. PCR and DNA sequencing analysis demonstrated that 61.4% of the ESBL strains expressed a CTX-M type, 15.8% an SHV ESBL type, and 5.3% a TEM ESBL type. CTX-M-15 was the predominant (29.8%) β-lactamase followed by CTX-M-3 and SHV-12. There was no epidemiological and microbiological evidence for an outbreak situation. In 9 strains, no CTX-M, SHV or TEM ESBL could be detected. The MIC50 and MIC90 of ertapenem were 0.06 mg/l and 0.25 mg/l, respectively. Overall, 98.2% of the strains were susceptible to ertapenem (MIC ≤ 2 mg/l).
Conclusions: Despite an extensive screening strategy, the prevalence of ESBLs is low in our medical institution. The distribution of the ESBL types is similar to the distribution in institutions with high ESBL prevalence, being CTX-M the predominant ESBL class. The new compound ertapenem is highly active against the ESBL-producing Enterobacteriaceae tested.
P1033 Prevalence of the plasmid-mediated quinolone resistance determinants qnrA, qnrB and qnrS in Enterobacteriaceae isolates causing bacteraemia
J. Sanchez-Cespedes, S. Marti, M. Ruiz, S.M. Soto, M. Almela, F. Marco, J. Vila (Barcelona, ES)
Objectives: The main objective of this study was to investigate the prevalence of the qnrA, qnrB and qnrS genes in Enterobacteriaceae isolates causing bacteraemia.
Methods: Hundred and ninety-five Enterobacteriaceae isolates from blood samples were collected: 54 Klebsiella pneumoniae (27.7%), 28 Proteus mirabilis (14.4%), 25 Enterobacter cloacae (12.8%), 22 Serratia marcescens (11.3%), 21 Salmonella enteritidis (10.8%), 16 Klebsiella oxytoca (8.2%), 8 Salmonella typhimurium (4%), 6 Enterobacter aerogenes (3%), 5 Citrobacter freundii (2.6%), 4 Citrobacter koseri (2%), 3 Morganella morganii (1.5%), 2 Pantoea agglomerans (1%) and 1 Citrobacter werkmanii (0.5%). Screening of the qnrA, qnrB and qnrS genes was performed by multiplex PCR using a cocktail of specific primers. Bacterial strains positive for each qnr gene were used as positive controls, and were run in each batch of tested samples. Positive reactions were confirmed by direct sequencing of the PCR products. The sequences obtained were compared in the GenBank to determine the correspondent qnr variant. In order to characterise the genetic environment of these QNR determinants a PCR with an universal integron primer and a qnr multiplex primer was also performed.
Results: Five isolates (3%) carrying a qnr gene were found. Four of them corresponded to qnrB and the other one to qnrS; qnrA was not detected. The qnrS determinant corresponded to the variant qnrS2 and was found in K. pneumoniae 46408. Two variants of the qnrB gene were detected: qnrB2 in C. freundii 62778, C. freundii 21112 and 14.0 and qnrB6 in C. freundii 72857. The three isolates of C. freundii were not epidemiologically related by REP-PCR. Results of the PCR with the combination of integron and qnr primers showed that the qnrB2 determinant in the isolate C. werkmanii 14.0 was integrated in a qnr-containing complex sul1-type integron.
Conclusions: In our Hospital, the prevalence of the qnrB gene was higher than for the qnrA and qnrS determinants, differing to the situation in Spain where the qnrB determinant had not been detected until now. Also it is important to mention the high prevalence of the qnr gene in C. freundii (60% of the five studied isolates) and the presence of this gene in C. werkmanii given that this is the first time that the presence of a qnrB determinant has been reported in these species.
P1034 International dissemination of extended-spectrum β-lactamase TEM-24 among Enterobacteriaceae species is caused by spread of both epidemic IncA/C2 plasmid and strains
A. Novais, R. Cantón, E. Machado, L. Peixe, F. Baquero, T.M. Coque (Madrid, ES; Oporto, PT)
Objectives: TEM-24 is one of the TEM-type ESBL most widespread in Europe. The aim of this study was to characterise TEM-24-producing Enterobacteriaceae isolates recovered in different European countries and genetic elements participating on this ESBL dissemination.
Methods: Twenty five TEM-24-producing isolates recovered during a period of six years (1998–2004) from Spain (n = 11), Portugal (n = 11), and France (n = 3) were studied. Only one isolate/phenotype was included. They were Escherichia coli and Klebsiella pneumoniae (n = 6 each), and Enterobacter aerogenes (n = 13). Isolates were obtained from patients at medical wards (57.1%), ICUs (19.1%), surgical wards (9.5%), and 14.3% were outpatients. ESBL characterisation was performed by IEF, PCR and further sequencing. Relationship among isolates was established by PFGE and E. coli phylogenetic groups were searched as reported. Antibiotic susceptibility testing and conjugation assays were performed using standard methods. Integrons were determined as described. Plasmid characterisation included analysis of RFLP patterns and determination of incompatibility group by PCR, hybridisation and sequencing.
Results: E. coli isolates (n = 6) were assigned to six PFGE types whereas K. pneumoniae and E. aerogenes isolates were each associated with an epidemic clone. E. coli isolates belong to phylogroups D (75%) and A (25%). Most strains were resistant to tobramycin, amikacin and nalidixic acid. blaTEM-24 was located in a 170kb-IncA/C2 conjugative plasmid in all cases. Related plasmid RFLP patterns were observed among distinct Enterobacterial isolates, suggesting a common plasmid background involved in TEM-24 dissemination. This plasmid contain class 1 integron carrying aacA4 gene cassette.
Conclusions: Intercountry spread of blaTEM-24 is determined either by E. aerogenes or K. pneumoniae epidemic strains or by epidemic plasmids among Enterobacteriaceae species. Location on broad host range IncA/C2 plasmid might enables further transmission to other hosts.
P1035 ESBL-producing Enterobacteriaceae from non-human sources (poultry and swine) in Portugal
E. Machado, T. Coque, R. Cantón, F. Baquero, J. Silva, J.C. Sousa, L. Peixe (Porto, PT; Madrid, ES; Fafe, PT)
Objectives: To investigate the occurrence and diversity of ESBL-producing Enterobacteriaceae among healthy animals and raw poultry meat samples from Portugal.
Methods: We studied (i) 35 faeces samples from healthy swines (HS) in 1998 and 2004, (ii) 20 samples from uncooked poultry carcasses (PM) of two poultry retail markets (2003), and (iii) 20 faecal samples from healthy poultry (HP) in 2004 (n = 20). HS and HP were from distant geographic farms. Animals were not exposed to antibiotics in the 3 months preceding sample recovery. Samples were plated on MacConkey agar with ceftazidime (1 mg/L) or cefotaxime (1 mg/L). ESBL characterisation was accomplished by double disk diffusion, PCR, and sequencing. Bacterial identification was performed using API ID 32GN and antibiotic susceptibility tested by the disk diffusion method (CLSI). Clonal relatedness was established by RAPD and E. coli phylogenetic groups were identified by a multiplex PCR. Conjugation experiments were performed as described.
Results: ESBL were identified in PM (60%, 12/20), HS (5.7%, 2/35) and HP (10%, 2/20). ESBL-producing isolates were identified as C. freundii (2HS), E. coli (n = 12; 2HP, 10 PM) and K. pneumoniae (n = 4; 4PM), and ESBLs as TEM-52 (n = 12), SHV-2 (n = 3), SHV-12 (n = 2), and CTX-M-1 (n = 1). TEM-52 was found among E. coli. SHV-2 only detected in K. pneumoniae, SHV-12 only observed among C. freundii isolates and CTX-M-1 in one E. coli. Isolates showed decreased susceptibility patterns to other antimicrobial classes. Most bla genes were transferred by conjugation. RAPD analysis revealed that 2 TEM-52-producing E. coli from PM, 2 TEM-52-producing E. coli from HP, 2 SHV-2-producing K. pneumoniae and 2 SHV-12-producing C. freundii were clonally related (4 distinct RAPD-types). The remaining isolates showed unique RAPD patterns. These isolates and ESBL-producing clinical isolates spreading in Portuguese hospitals during 2003 and 2004 were clonally unrelated. E. coli phylogenetic group A was predominant (58.3%). Less represented phylogroups were D (33.3%) or B1 (8.3%).
Conclusions: This work constitutes the first report of some ESBLs from Enterobacteriaceae from livestock. The frequent presence of ESBL-producing Enterobacteriaceae in poultry meat in Portugal highlights the risk of transmission to humans via food chain. Horizontal gene transfer could be responsible for ESBL dissemination in our country.
P1036 Clinical urinary tract isolates of Enterobacteriaceae in the Arkhangelsk region, Russian Federation: antimicrobial resistance profiles and characterisation of ESBL-strains
V. Vorobieva, T. Bazhukova, N. Semenova, B.C. Haldorsen, A. Bettina, G. Simonsen, A. Sundsfjord (Tromso, NO; Arkhangelsk, RU)
Objectives: (i) Antimicrobial resistance in urinary tract isolates of Enterobacteriaceae in the Arkhangelsk, Russia. (ii) The molecular epidemiology of ESBL-strains.
Material and Methods: A total of 136 non-duplicate urinary isolates of Enterobacteriaceae were collected from September 2005 to September 2006; 112 (82%) and 24 (18%) strains from in- and out-patients were identified with standard microbiological methods. Antimicrobial susceptibility testing was performed by agar disc diffusion (AB, BIODISK, Solna, Sweden) to 15 different antibiotics. Clinical strains with reduced susceptibility to oxyimino-cephalosporins were analysed by oxyimino-cephalosporin Etests, blaTEM/SHV/CTX-M ESBL-genes, inhibitor-based AmpC-detection (boronic acid), ESBL phenotypic tests (ESBL Etest; combined disk method), susceptibility to non-betalactam antibiotics by VITEK 2 (bioMérieux, France). Interpretation criteria were according to EUCAST and NWGA. Strains with resistance and intermediate susceptibility were classified as non-susceptible.
Results: The rates of non-susceptibility to 110 (81%) Escherichia coli, 16 (12%) Klebsiella pneumoniae, 10 (7%) other Enterobacteriaceae were: ampicillin 99%, cefuroxim 98%, trimethoprim 32%, trimethoprim-sulfamethoxazole 32%, piperacillin-tazobactam 24%, nalidixic acid 23%, ciprofloxacin 21%, gentamicin 21%, aztreonam 20%, ceftazidime 19%, cefotaxime 18%, mecillinam 19%, nitrofurantoin 16%; meropenem 0%. Twenty-five (18%) clinical strains of Enterobacteriaceae: E. coli (n = 17), K. pneumoniae (n = 5), Enterobacter cloacae (n = 2), Serratia marcescens (n = 1) with reduced susceptibility to oxyimino-cephalosporins were detected among 22 (88%) and 3 (12%) in- and out-patients. Twenty-four were confirmed as ESBL-positive by blaTEM/SHV/CTX-M PCR; 24 blaCTX-M (E. coli n = 16; K. pneumoniae n = 5; E. cloacae n = 2; S. marcescens n = 1), blaSHV (E. coli n = 3; K. pneumoniae n = 5), blaTEM (E. coli n = 12; E. cloacae n = 2; S. marcescens n = 1). Sequence typing of amplicons is ongoing.
Conclusions: (i) High non-susceptibility rates to β-lactam and non-β-lactam antibiotics are found. (ii) ESBL-production was detected in 16/110 (15%), 5/16 (31%), 3/10 (30%) isolates of E. coli, K. pneumoniae, and other Enterobacteriaceae from 22 (92%) and 2 (8%) in- and out-patient. (iii) blaCTX-M was the most prevalent ESBL-type. (iv) Inhibitor-based detection of AmpC-production in E. coli and K. pneumoniae strains with reduced susceptibility to oxyimino-cephalosporins was negative.
Emerging infections
P1037 Evaluation of Platelia™ Dengue NS1 Antigen assay for early diagnosis of acute dengue infection
M. Tabouret, C. Salanon, P. Sarfati (Marnes la Coquette, FR)
Objectives: In tropical and subtropical region, endemic dengue disease is the most important arbovirosis in terms of morbidity and mortality. To implement appropriate treatment, there is a need for a direct, rapid and specific detection test during the acute phase of the infection. For that purpose, viral culture or nucleic acid detection (performed mainly in reference laboratories) require specialised environment but sometimes with delays to report results. Dengue specific antibodies detection is commonly used even if these antibodies appear several days after onset of symptoms. The objective of the study is to evaluate the performances of Platelia™ Dengue NS1 Ag (Bio-Rad Laboratories) for early diagnosis of acute dengue infection.
Methods: Platelia™ Dengue NS1 Ag is a one step sandwich ELISA assay for qualitative detection of Dengue virus NS1 antigen in human serum or plasma. Specificity and sensitivity were determined on 554 healthy blood donors and 297 dengue virus infected patients with accessible clinical information. 157 infected patients (52.9%) were selected based on positive PCR results and because the delay after onset fever was available. Serological status was determined using Dengue IgG indirect and Dengue IgM Capture Panbio assays.
Results: All 554 sera from blood donors were found negative for NS1 giving a specificity of 100%. Sensitivity of NS1 was 91.0% on the 157 PCR positive samples with no significant difference observed between the 4 dengue virus serotypes. NS1 sensitivity was significantly higher (98.5%) in patients with primary dengue infection (PCR+/IgG-) compared to 85.6% in patients with secondary (PCR+/IgG+) dengue infection (Chi2, p <0.05). During the first 5 days after clinical onset, detection rates ranged from 100.0 to 87.0% for NS1 Ag and from 0.0 to 30.8% for IgM serology. When considering all dengue infected patients, NS1 sensitivity was significantly higher for IgM negative (88.7%) than for IgM positive (66.2%) samples (Chi2, p<0.0001).
Conclusion: Platelia™ Dengue NS1 Ag assay is an easy-to-use ELISA for dengue antigen detection. The assay allows early and specific diagnosis of both primary and secondary dengue acute infections. This new test should be considered in diagnostic algorithms to improve patient care monitoring.
P1038 Seroprevalence of IgG antibodies against flaviviruses in German soldiers
S. Friedewald, A. Jäger, M. Faulde, S. Essbauer, M. Pfeffer, R. Wölfel, G. Dobler (Munich, Koblenz, DE)
Objectives: West Nil virus (WNV) is an emerging pathogen causing increasing numbers of epidemics and human cases in Europe, Africa, Asia and America. While WNV was detected in France, Slovak and Czech Republic, there is no evidence for circulation of this flavivirus in Germany thus far. Seroepidemiological studies for a particular flavivirus infection are hampered because of cross-reacting antibodies against other autochthonous or travel-related and vaccination-induced flavivirus antibodies, e.g. tick borne encephalitis virus (TBEV) Dengue virus (DENV), Japanese encephalitis virus (JEV). Hence two commercially available ELISA assays for the specific detection of antibodies against WNV were compared using well defined sera from German soldiers.
Methods: 715 sera from soldiers of different parts of Germany were tested for IgG antibodies (AB) against WNV using a commercial ELISA (Focus Diagnostics, USA). Reactive sera were retested in a second WNV AB ELISA, against DENV AB and TBEV AB (all Euroimmun, Germany). Positive sera were also tested for avidity (Euroimmun, Germany) to detect possible reduction of cross reactivity of AB. All tests were conducted according to the manufacturers' instructions.
Results: 142/715 sera (19.9%) reacted positive in the WNV AB ELISA (Focus Diagnostics). Re-testing by WNV AB ELISA (Euroimmun) showed positive results in 73 of these 142 sera. However 123/142 sera reacted positive in TBEV AB ELISA and 59/142 sera showed positive results in DENV AB ELISA. Results of only 6/715 sera (0.8%) were consistent with past WNV or DENV infection. These results are consistent with data of earlier studies and it can be assumed that these 6 soldiers acquired a WNV or DENV during travelling in endemic areas. The use of WNV AB avidity test did not reduce cross-reactivity.
Conclusions: Our data do not provide any evidence for autochthonous human WNV infection in Germany. However, huge differences in sensitivity and specificity of two commercial ELISA assays were found with the ELISA (Focus Diagnostics) showing the lowest specificity. The second ELISA (Euroimmun) showed higher specificity. The use of avidity testing had no effect on cross-reactivity at all. Only 6/715 sera showed evidence of past infection with WNV or DENV. In seroepidemiological studies or in serological diagnosis of WNV infections or vaccinations against other flaviviruses have to be excluded using tests with higher specificity, i.e. immunofluorescence and neutralisation test.
P1039 Antibody response in Crimean-Congo haemorrhagic fever, and association between antibodies and clinical outcome
Z. Özkurt, K. Özden, A. Kadanali, S. Erol, N. Yilmaz, M. Parlak (Erzurum, Ankara, TR)
Objectives: Crimean-Congo haemorrhagic fever is a serious viral disease. Fatality rate of the disease has been reported as 5–50%. The neutralising antibody response is weak and difficult to demonstrate in CCHF infection. In this study we aimed detection of antibody response in the disease and association between clinical outcome.
Methods: We evaluated 90 confirmed CCHF cases. Microbiological diagnosis and confirmation of the disease were made by demonstrating seroconversion, by IgM antibody, and/or polymerase chain reaction (PCR). IgM and IgG antibodies were detected by ELISA.
Results: Anti-CCHFV IgM antibody was detected in 4 of 10 (40%) fatal cases versus 54 of 90 (60%) survivors (p < 0.05). Anti-CCHFV IgG antibody was not detected in any fatal case (0/10) but it was found positive in 35 of 76 (46%) survivors (p<0.05).
Conclusion: These data show that antibody response is low in fatal cases than those of survivors. Neutralising activity may have an important role of limitation of the disease, and the value of immune sera in therapy should be investigated in further studies.
P1040 Clinical and laboratory features of Crimean-Congo haemorrhagic fever: predictors of fatality
M.A. Cevik, A. Erbay, H. Bodur, E. Gulderen, A. Bastug, A. Kubar, E. Akinci (Ankara, TR)
Objectives: To determine the predictors of fatality among patients with Crimean-Congo haemorrhagic fever (CCHF) based on epidemiological, clinical and laboratory findings.
Methods: Among the patients who were referred from surrounding hospitals to Ankara Numune Education and Research Hospital as a possible CCHF case during the spring and summer of 2003–2006, patients with IgM antibodies or PCR results positive for CCHF virus in blood and who had received only supportive treatment were included to the study.
Results: Sixty-nine patients were admitted from various cities of northeast of central Anatolia and the southern parts of the Black Sea region. Male patients accounted for 45 (65%) of the patients and the mean age was 50 years. Eleven (15.9%) cases were died with massive haemorrhage. The age, gender, days from the appearance of symptoms to admission and initial complaints were similar between fatal and survived cases (p > 0.05). Forty patients were IgM-positive, and 50 patients were PCR-positive. Among the clinical findings, ecchymosis (p = 0.007), hematoma (p = 0.023), haematemesis (p = 0.030), melaena (p<0.001) and somnolence (p<0.001) were more common among fatal cases. Almost all of the patients had leukopenia, thrombocytopenia, and elevated AST, ALT, LDH and CPK levels at admission. Mean thrombocyte level was 47000/mm3 in survived patients and 12000/mm3 in died patients (p = 0.003). Among the fatal cases the mean PT (18 s vs. 13 s; p<0.001) and mean aPTT were longer (69 s vs. 43 s; p = 0.001), the mean ALT (1688 vs. 293; p < 0.001) and the mean AST (3028 vs. 634; p < 0.001) were higher. All patients received intensive clinical supportive measures, including platelets, fresh frozen plasma, and packed erythrocyte infusions, when indicated. None of the patients received ribavirin. Fatal patients received significantly more fresh frozen plasma (p<0.001) and thrombocyte suspensions (p = 0.017) than survived patients. Cox proportional hazard model was used for fatality analysis, the starting time point was beginning of the complaints, and the ending time point was either death or discharge from the hospital. Thrombocytopenia of <20000/mm3 (hazard rate [HR], 9.8; 95% confidence interval [CI], 1.17–81.4; p = 0.035) and somnolence (HR, 5; 95% CI, 1.4–17.5; p = 0.012) were independently associated with mortality.
Conclusion: Thrombocytopenia of <20000/mm3 and somnolence were independent predictors of fatality among patients with CCHF.
P1041 Crimean-Congo haemorrhagic fever infection among children
G. Tanir, O. Ergonul, N. Tuygun, P. Golabi, V. Korten (Ankara, Istanbul, TR)
Objective: The epidemiological, clinical, and laboratory findings of the children diagnosed as Crimean-Congo haemorrhagic fever (CCHF) were described.
Methods: The children infected with CCHF virus in 2005 and 2006, and hospitalised in Ankara Dr. Sami Ulus Children's Hospital and Marmara University Hospital were included. All the patients had positive results of IgM and/or PCR results for CCHF virus in their blood.
Results: Thirteen cases were included, and all were from the northeastern Anatolia and the southern parts of Black sea region. The mean age was 8 (3–13), the rate of males was 62%. The majority (77%) of the cases had the history of tick bite. There was only one fatal case. All the patients had the history of fever. Rash (58%), myalgia (50%), and abdominal pain (40%) were common. The mean AST and ALT levels on the admission were 141 U/l and 85 U/l. The mean platelet count was on admission was 117000/mm3, and the lowest was 16100. The mean of the lowest white blood cell count was 3200/mm3. The mean of the highest lactate dehydrogenase was 925 IU/L. Ribavirin was given to three cases.
Conclusions: CCHF among children was not reported in detail before. Despite one fatal case, the clinical course of the infection among the children was milder and the duration of illness was shorter compared to adult cases.
P1042 Sero epidemiology of Crimean-Congo haemorrhagic fever in domestic animals in central area of Iran
M. Izadi, H. Salehi, K. Mostafavi, S. Chinikar, B. Ataei, F. Khorvash, M. Darvish, N. Jonaidi jafari (Tehran, Isfahan, IR)
Objectives: Crimean-Congo Haemorrhagic Fever is a viral zoonotic infection which several cases of that have been reported in Iran and during these recent years the nature of the pathogen has been revealed. The purpose of this study was determination of the seroprevalence of CCHF IgG among local and imported domestic animals of the Isfahan province during the year 2004.
Methods: This cross sectional study has been performed among 232 animals regarding to the presence of IgG antibody of Crimean-Congo Haemorrhagic Fever. The study was made with the special helps of Arbovirus laboratory Pasteur Institute of Iran on 2004.
Results: 88 (37.9%) of animals had seropositive results in which the most prevalent was within ovine (53.3%) and after that bovine (38.5%) and caprine (18.6%). With increasing age, the chance of seropositivity was increased and the most common range of age belonged to 4–5 years old animals. The most infected grassland was Borkhar region and after that Qom, Natanz, Fereydan, Falavarjan and Isfahan, respectively. No infected case was noticed among animals belonging to the Kordestan region grasslands.
Conclusion: The results of this study revealed the endemic spreading of CCHF in the animals in Isfahan province and it needs special attention to prevent the infection in the communities and occupational exposure.
P1043 Risk factors among patients with Crimean-Congo haemorrhagic fever
N. Tasdelen Fisgin, E. Tanyel, N. Tulek (Samsun, TR)
Objectives: Crimean-Congo haemorrhagic fever (CCHF) is a potentially fatal disease caused by Nairovirus in the Bunyaviridae family.
Methods: Between 2004 and 2006, patients with typical clinical findings and positive IgM for CCHFV in blood sample were enrolled in the study. Patients were divided 2 subgroup; group I (n: 19) alive patients, group II (n: 5) excites patients. Risk factors including demographic and laboratory data were investigated among 24 patients with CCHF.
Results: The median platelet count was significantly lower in excitus group (9400/mm3) when compared to alive group (43942/mm3). Aspartate transferase (AST) and alanin transferase (ALT) levels were significantly higher in excites group (AST: 2555 U/L, ALT: 1675 U/L) when compared to alive group (AST: 667U/L, ALT: 295U/L).
Conclusion: Low platelet count, increased AST and ALT levels could be poor prognostic factors in patients with CCHF
P1044 Serology and immunological study on the infectivity of host animals and ticks (Ixodidae, Argasidae) to CCHF virus in Ardabil Province, Iran
Z. Telmadarraiy, S. Chinikar, H. Vatandoost, K. Holakoui, F. Faghihi, Z. Zarei, M. Oshaghi (Tehran, IR)
Ardabil province is located in North western Iran. The main activities of people in this region is agriculture and cattle rearing. A comprehensive study was carried out in year 2004–2005 for determination of presence in virus in ticks and antibody against CCHF in their hosts. The villages selected randomly and ticks with their host sampled according to standard method. Results showed that the majority of ticks host were cow, sheep, goat, camel, poultry, and buffalo, At the same time all the collected ticks were searched for the presence of CCHF virus using RT-PCR method. Among 56 sera of cow and sheep and 3 goats, it is found that 48.7% of sheep, 21.4% of cow and 33.3% of goats was found IgG positive. Different species of soft and hard ticks including Rhipicephalus, Hyalomma, Argas and Ornithodoros was collected from various types of houses, stable and animal shelters.
Results of RT-PCR revealed that 33.3% of Hyalomma collected from buffalo was positive to virus, Rhipicephalus bursa and Hyalomma was found positive when they were collected from camel. The figures for Rhipicephalus bursa on goat was 40%. Argas reflexus which was collected on poultry was not infected to CCHF virus. The main species of infected ticks to CCHF was: Rhipicephalus bursa, Haylomma aegypticom, Hyalomma asiaticum, Hylaomma detritum, Hyalomma marginatum, Hyalomma shculzei and Ornithodoros lahorensis.
P1045 Clinico-epidemiologic feature and outcome analysis of Crimean-Congo haemorraghic fever in Iran (1999–2006)
M. Mardani, M. Goya, M. Zainali, M. Keshtkar Jahromi (Tehran, IR)
Background: Crimean Congo Haemorrhagic Fever (CCHF) is a tick borne viral disease reported from more than 30 countries in Africa, Asia, southeast Europe, and the Middle East. The disease has been reported in Iran since 1999. This study is performed to define the last data of CCHF cases in Iran.
Methods: Based on records of Ministry of Health of Iran, the epidemiological features and clinico-epidemiological manifestations of confirmed cases since 1999 upto 2006 in Iran have been studied.
Results: 287 out of 666 probable cases were confirmed to have CCHF by positive IgM and/or IgG for CCHF virus. The disease was more prevalent in middle aged men. The maximum incidence was in August and September. The clinical findings were severe headache, myalgia, nausa and fever. Epistaxiy, bleeding from the gums, nose and venopuncture sites, petechia, purpura, melena and hematemesis were common. Large ecchymotic areas developed on trunks, arms and legs. The most common laboratory findings were hematuria, proteinuria, prolonged partia thromboplastin time and AST greater than 100 IU/dl. Most of patients received oral Ribavirin in addition to supportive care including volume expanding intravenous fluids, packed cells, fresh frozen plasma and platelet, management of shock and renal failure and intensive care. 51 confirmed patients died (Mortality rate = 17.7%). The cause of death in the most of patients was intractable haemorrhage, multiorgan failure and shock.
Conclusion: It should be noted that case fatality rate has been increased despite the decrease in the incidence of disease recently and providing laboratory facilities in order to have rapid and accurate diagnosis for early treatment of the disease in endemic areas such as our country (Iran) is a priority.
P1046 Reappearance of CCHF and other tick-borne arboviruses in the Syrdarya region of the Republic of Uzbekistan
A. Kadyrov, D. Shermukhamedova, N. Komilov, S. Umurzakov, E. Bryanseva, M. Nazarbekova (Tashkent, UZ)
Objective: In the Syrdarya region of Uzbekistan, vast virgin lands were cultivated for agriculture during the last half of the 20th Century. As a result, natural-foci of arbovirus infections apparently disappeared. For example, there were not any cases of Crimean Congo Haemorrhagic Fever (CCHF) reported there in the last 35 years. However; since 2001, lethal cases of CCHF have been registered annually. It has become necessary to reevaluate the presence of, and determine the prevalence of human illness caused by such tick-borne arboviruses in the Syrdarya region.
Methods: Specimens were collected during the spring and summer of 2001–2004. The collected field material included: serum of patients with febrile illness of unknown etiology (n = 1238), serum of patients with suspicion of CCHF (n = 167 paired sera), serum from apparently healthy people (n = 3630), serum from contacts of suspected CCHF patients (n = 201), serum of cattle (n = 650), serum of other animals (n = 131), and Hyalomma spp. (H. spp.) ticks (n = 1130). Diagnostic assays for CCHF, Karshi, Tamdy, and Syrdarya Valley Fever viruses (SDVF) included: precipitation reaction in agar (RDPA), complement fixation (CFR), and indirect hemaglutination (IH).
Results: Among the patients with fevers of unknown etiology, CCHF was detected in 8, Tamdy in 9, Karshi in 24, SDVF in 5, and a mixed infection was detected in 5 patientsy Among healthy people we detected precipitating antibodies against CCHF virus in 11 samples. Antigens of CCHF, Karshi, Tamdy, and SDVF viruses were detected in H. anatolicum and H. detriticum. Infection of ticks with these viruses was relatively stable during the study period (19.7% (2001), 53.8% (2002), 21.7% (2003), 25% (2004). In cattle, precipitating antibodies against CCHF were detected in 8.6%, Karshi in 7.2%, Tamdy in 5.7%, and SDVF in 4.6% of samples.
Conclusions: Tick-borne arbovirus foci were detected in the Syrdarya region. H. anatolicum and H. detriticum are known to parasitise domestic animals and livestock, and are therefore likely contact humans and act as putative vectors for CCHF. Detection of antibodies against CCHF virus among healthy people – 0.35%, shows, that inapparent illness due to infection with CCHF is very low.
P1047 The situation of Crimean-Congo haemorrhagic fever in the last years in Iran
S. Chinikar, F. Ahmadnejad, A. Fayaz, N. Hosseini, N. Afzali, M. Gooya, M. Zeinali, B. Hooshmand, A. Lundkvist, M. Nilsson, A. Mirazimi, R. Flick, A. Grolla, H. Feldmann, M. Bouloy (Tehran, IR; Stockholm, SE; Galveston, US; Winnipeg, CA; Paris, FR)
Objectives: Crimean-Congo haemorrhagic fever (CCHF) is an acute zoonotic tick-borne viral disease caused by the CCHF virus. The virus belongs to the genus Nairovirus (family Bunyaviridae) and causes severe haemorrhagic symptoms in man with a mortality rate of 10–50%. Humans are usually infected with CCHF virus either through the bite of infected ticks or by direct contact with virus contaminated tissues or blood.
Methods: From June 2000 to August 2006, sera were collected from Iranian suspected patients for CCHF and have been sent to the Arboviruses Lab (National Center) of the Pasteur Institute of Iran. The sera have been analyzed with specific ELISA for detecting antibodies (IgM and IgG), and also with RT-PCR to investigate the genome of the virus.
Results: Between 854 suspected human cases, 335 were confirmed cases. Between the 335 cases, 298 were IgM positive and 37 cases only RT-PCR positive. Between the 298 IgM positive, 75 persons were also RT-PCR positive. The number of suspected, confirmed and dead cases according to the year respectively is as follows: 2000 (55, 20, 4), 2001 (167, 66, 11), 2002 (247, 111, 14), 2003 (144, 57, 12), 2004 (82, 26, 6), 2005 (84, 18, 7), up to August 2006 (75, 37, 3).
54.3% of the positive cases were in the age range 21–40 years. The Sistan-Baluchestan province, by having 64.8% of positive cases, was the most infected province and the Isfahan province (10.4%) and Fars province (5.7%) were the second and third infected province. The most exposed professions were: Farmer (19.4%), worker (18.2%), housewife (19%) and butcher (12.5%).
Conclusion: CCHF is the most important haemorrhagic fever in most parts of Iran and confirmed cases have been found in 21 provinces of Iran. The most infected province is Sistan-Baluchistan in the southeast of Iran near the border of Pakistan and Afghanistan where the disease is endemic.
As the most involved age range are people between 21–40 years old and the majority of confirmed cases professions in Iran are related to animals; such as farmer, slaughterer and butcher, so informing the high-risk groups about the transmission routes of the disease is very useful.
P1048 Pandemic flu preparedness – role of antibiotics
E. McIntosh, D. Wu, P. Paradiso, H. Tucker, B. Burlington, R. Brenner, J. Clarke, J. Cooke, P. Puz, M. Temple, D. Papiernik, R. Mallick, A. Kuznik, L. Barrett, A. Woodrow (Maidenhead, UK; Collegeville, US)
Objectives: This work analyses the potential role of antibiotics in pandemic flu preparedness using examples drawn from past pandemics, interpandemic flu and SARS, and makes recommendations to improve antibiotic availability in the event of a pandemic.
Methods: Commercial data for antibiotic sales during interpandemic flu and SARS was obtained. The following assumptions were made: 50% of the population would be affected by pandemic flu, 10% would be diagnosed with post-flu bacterial CAP, 20% of those will be hospitalised and 85% would require IV antibiotics. It was also assumed that 20% would be infected by MRSA and that 30% of the patients would ideally received ICU management.
Results: Sales of some IV antibiotics were found to follow the flu season by up to 3 weeks. The sales of 7 IV antibiotics were significantly related to the number of individuals testing positive for flu, an overall 4.1% incremental effect. About 40 to 60% of hospitalisations in the USA (350,000 to 1.2 million) for secondary CAP are projected to optimally warrant IV antibiotic therapy with coverage for MRSA. In a “moderate” pandemic there would be 209 thousand and in a “severe” pandemic 1.9 million flu-related deaths in the USA alone. In Hong Kong there was a very sharp rise in antibiotic use associated with SARS.
The expected successes of the following antibiotics in flu-related CAP are: ceftriaxone 39.3%, piperacillin/tazobactam 39.9%, azithromycin + ceftriaxone 63.8%, vancomycin + azithromycin + ceftriaxone 79.5%, levofloxacin 79.6%, ceftriaxone + levofloxacin 80.8%, azithromycin + ceftriaxone + levofloxacin 80.8%, piperacillin/tazobactam + levofloxacin 83.1%, tigecycline 85.1%, vancomycin + levofloxacin 93.2%.
Conclusions: In a flu pandemic, there will be a requirement for broad-spectrum hospital intravenous antibiotics to cover common pathogens, especially those resistant to conventional antibiotics. Further efforts should be made to incorporate antibiotic preparedness into pandemic flu preparedness plans.
P1050 Estimation of the incubation period for pandemic influenza during 1918–19
H. Nishiura (Tübingen, DE)
Background: The incubation period of influenza has not been precisely understood and is yet to be clarified.
Methods: This study reassessed an epidemiologic record of pandemic influenza observed on the ocean during voyages. The original data is based on observations of 92 voyages from 1918–19, departing from several ports in Australia with incubating individuals. Although infection event is not directly observable, the incidence (i.e., time of onset) is recorded according to the date after departure. Whereas Dr. Anderson Gray McKendrick (1876–1943) previously used a daily probability of onset to infer the incubation period, I show how to interpret the data and improve the estimation method assuming lognormal distribution for the incubation period.
Results: Maximum likelihood estimates of the mean incubation period and coefficient of variation were 1.34 days and 53.1%, respectively. The estimate was roughly consistent with the suggestion of Dr. McKendrick proposing 32.71 hours as the mean. An explicit distribution of the incubation period was obtained. Dr. McKendrick's method appeared to have assumed the complementary cumulative distribution function for the observed data.
Conclusions: Although it is often difficult to determine the incubation period without explicit information of the time of exposure, a hint to limit most probable time of exposure enables the estimation. Whereas the precision might be still limited, the length and variance of the incubation period would be directly relevant to public health interventions (i.e., determination of the time to quarantine exposed individuals).
P1051 New emerging human infection caused by Bartonella vinsonii ssp. arupensis in Russia
A. Kruglov, G. Barmina, V. Osadchaya, G. Frolova, A. Markov, G. Smirnov, V. Bashkirov, M. Kirillov, M. Kosoy (Moscow, RU; Ft. Collins, US)
Objective: First case of human illness caused Bartonella vinsonii ssp. arupensis was described in Wyoming, 1999. The cattle rancher suffered from bacteraemia and high fever. The second case was recorded in France in 2005 where a patient was hospitalised with endocarditis.
Methods: Surveyed 95 patients with infective endocarditis (IE) and a fever of unknown origin (FUO) in the clinics of Moscow Medical Academy during 2004–2006 years. The basic lab methods of research were: inoculation of blood samples to Vero E6 cell culture and incubation at 37°C for 10 days; culturing of samples on plates with hard medium in an atmosphere of 5% CO2; estimation of antibody titers by indirect immunofluorescence (IFA; PCR with genus-specific primers; RFLP and sequence of obtained amplicones.
Results: High frequency of the positive serological reactions to bartonellae antigenes was detected (10.3% – IE, 5.1% – FUO). Two cases of cultural positive B. vinsonii ssp. arupensis infections were detected. Both patients were young women. First had FUO. Second – IE. Blood samples from the patients were obtained on day 39 (1st case) and on day 73 (2nd case) after admission. Main clinical manifestations of the illness were: the acute onset of disease and the symptoms of expressed intoxication. The patients did not have typical clinical attributes of classical Bartonella infection; the inflammation traits in blood were slightly expressed, and low-grade bacteraemia symptoms were observed; the disease had positive dynamics of after the nonspecific antibacterial therapy. Typical colonies appeared on agar plates after 7 days of incubation at 37°C in 5% CO2 atmosphere. Small Gram-negative bacteria were seen after staining. Biochemical properties and antimicrobial susceptibility of the both strains were studied and found typical for Bartonella genus. Antibodies titers to B. vinsonii in serum of both patients were 1/64.
In both cases PCR from bacterial isolates with Bartonella-specific primers resulted in strong positive signals with 5 targets such as partial sequences of the gltA, ribC, ftsZ, 23S rRNA, and sucB genes. RFLP and sequence analysis demonstrated that both strains belong to B. vinsonii ssp. arupensis.
Conclusion: The high frequency of positive serological reactions to Bartonella amongst patients with IE was detected. This is a first report about the bartonelloses caused by B. vinsonii ssp. arupensis in humans in Russia.
P1052 Epidemiological, clinical and serological features of human leptospirosis in Bulgaria in 2005
E. Taseva, I. Christova, T. Gladnishka (Sofia, BG)
Objectives: Leptospirosis is a worldwide distributed zoonosis that affects humans and is now identified as one of the emerging infectious diseases. The present study aims to summarise incidence, epidemiology, clinical symptoms, circulating serovars, mode of transmission, and source of infection in laboratory confirmed human leptospirosis cases reported to the Bulgarian Ministry of Health for 2005 year.
Methods: All patients were diagnosed using the reference method for serological diagnosis, i.e. microscopic agglutination assay. Suspensions of live serovars from 9 different Leptospira serogroups were used as antigens.
Results: A total of 43 confirmed and reported leptospirosis cases for 2005 year, were analyzed. The average incidence of the disease was 0.55/100 000. The overall fatality rate was 11.63%, which is higher than in the previous study. The most cases were reported from north-eastern regions (Shumen – 27.91%) and north-western (Montana – 11.63% and Lovech – 9.30%) regions of the country. The disease affected mainly men. Cases occurred in all age groups, but were more common the men of working age (51–60 years) – 31.75%. The typical leptospiral seasonal course, with a peak in October, was observed. The infection was acquired through occupational (48.84%), recreational (30.24%) and accidental (20.93%) exposure. The most frequently reported symptoms were fever (38.46%), icterus (36.54%), myalgia, headache, hepatomegaly and splenomegaly. Serovars belonging to 6 different serogroups caused infection during the study period. Two serogroups, Icterohaemorrhagiae (48.70%) and Pomona (32.17%) accounted for most of leptospirosis cases. The third main cause of infection was serogroups Bataviae and Sejroe (6.96%).
Conclusions: The analysis of the data showed that measures for rodent control are still inefficient. Clinical data showed prevalence of the icteric clinical form of the disease and serological finding revealed prevalence of serogroup Icterohaemorrhagiae. Knowledge of epidemiological, clinical, and serological features of leptospirosis is an appropriate base to outline measures for successful prevention and early diagnosis of the disease.
P1053 Epidemiology of human leptospirosis in Malaysia
A. Fairuz, B. Abdul Rani, M. Ayu, I. Hishamshah (Kuala Lumpur, MY)
Leptospirosis is endemic in Malaysia. The epidemiological data for the past decade is very limited and the true disease burden in Malaysia is unknown. A hospital based study was carried out to estimate the most recent incidence and prevalence rates of the disease and also to descibe some epidemiological characteristics of the disease in Malaysia.
All sera of patients suspected to have leptospirosis admitted to the hospitals in Malaysia were tested for presence of antibody to leptospira. Isolation of leptospires and Polymerase chain reactions were also carried out to augment the diagnotic sensitivity. The clinical data of patients confirmed to have leptospirosis were obtained from their clinical case notes.
A total number of 5652 sera were received throughout 2003 till 2005 and 1060 were confirmed positive leptospirosis. The annual incidence rates for 2003, 2004 and 2005 were 0.9, 1.18 and 2.13 per 100000 population respectively. The case fatality rate 10.2%. The cases occurred throughout the year but there were more cases during the intermonsoon period that is between June to September and December to January. The male to female ratio was 4:1. The mean age of patients was 40.1±19. Ninety five percent of the cases were those in the productive ages between 20–60 years old. The disease was most common amongst the Malays, the largest ethnic group in Malaysia. The common clinical features were fever (98%), chills (64.2%), Jaundice (44%), Abdominal pain (42.9%), cough (56.5%) and hepatomegaly (40.5%). The most worrying trend is the increasing number of cases(50.8%) with severe pulmonary involvement which include pneumonia, pulmonary haemorrhage, ARDS, and pulmonary failure. Pulmonary failure was the major contributory factor causing death in 85% of the fatal cases.
In conclusion, leptospirosis still remain to be a significant cause of morbidity and mortality. There should be a concerted effort to assess the health as well as economic impact of leptospirosis in this country. A comprehensive database needs to be created in order to obtain more data on the current trend of leptospirosis in Malaysia. Research in this field should also be done via collaborative efforts between the human and veterinary health institutions to address major issues related to the disease and facilitate in designing of strategies for control and prevention of the disease in this country.
P1054 Tularaemia outbreak due to consumption of natural spring water in Karamürsel, Turkey
M. Meric, M. Sayan, A. Willke, S. Gedikoglu (Kocaeli, Bursa, TR)
Objective: To describe a recent tularaemia outbreak resulted from consumption of spring water in a village of Karamürsel/Turkey and to present the anti-epidemic measures implemented.
Methods: After diagnosis as oropharyngeal tularaemia of two patients come from the village, a field investigation was performed at the region in March 2005. Clinical samples from the patients and water samples from the natural spring water were obtained and microbiological investigation was performed. A questionnaire designed to determine the risk factors for the infection.
Results: Totally 17 patients were diagnosed as tularaemia with their clinics and serological results. The patients were treated with streptomycin, or ciprofloxacin, or doxycycline, or streptomycin+ doxycycline. All the patients recovered. Francisella tularensis PCR was found to be positive in water samples. After then spring water collected in a storage, chlorinated and then allowed to delivery. The outbreak was controled after hygienic measures.
Conclusion: Oropharyngeal tularaemia is dominant in our region and Turkey which could be associated with the consumption of the contaminated water. In this small outbreak, we showed the F. tularensis DNA by real-time PCR assay. After establishing hygienic regulations the outbreak was stopped. Surveillance and early alert system for tularaemia and field investigation teams are nessesary for detection and prevention of the outbreaks.
P1055 First case of nosocomial Clostridium difficile toxinotype III, PCR-ribotype 027-associated disease in Switzerland
A.F. Widmer, L. Fenner, M. Rupnik, A. Trampuz, R. Frei (Basle, CH; Maribor, SI)
Objectives: to describe the first case of Clostridium difficile toxinotype III, PCR-ribotype 027-associated disease in Switzerland, characterised by an 18 bp deletion in the tcdC gene and increased production of toxins A and B in vitro.
Methods: Since 2001, stool specimens are processed by testing for toxin A/B, and a routine culture for C. difficile is performed, followed by repeated toxin testing from the isolate. All isolates are saved and stored at −70°C, and patient data are stored in a standardised database.
A random sample of all isolates from 2003–2005 were screened for toxinotype III (Rupnik M J Clin Microbiol. 1998). and underwent ribotyping (Stubs et al. J Clin Microbiol. 1999). Regulator of toxin production (TcdC) was analyzed using the method by Spigaglia P. J. Clin. Microbiol 2002. A total of 388 cultures were positive for C. difficile during the study period. Of those, 102 isolates were further analyzed with the methods mentioned above.
Results: One 73 year old patient with advanced lymphoma was positive for Clostridium difficile toxinotype III, PCR-ribotype 027-associated disease. She had 2 relapses and suffered from prolonged disease over three months, only partly responding to metronidazole. The isolate was resistant to ciprofloxacin, levofloxacin and susceptible to moxifloxacin (MIC 0.75 mg/L). The strain had an MIC of clindamycin 3 mg/L, metronidazole 0.125 mg/L and vancomycin 0.75 mg/L. It was positive for binary toxins cdtA and cdtB, and showed a 18 bp deletion in tcdC gene. In addition, the strain exhibited a single nucleotide mutation at position 117, likely a more specific marker for highly-virulent epidemic C. difficile. No evidence for spread of this strain was observed during a one year follow-up in our institution.
Conclusions: This is the first documentation of the highly virulent strain of C. difficile in Switzerland. This strain demonstrates all characteristics of the epidemic strain observed in Canada, USA and few other European countries.
P1056 Emergence of a new epidemic Clostridium difficile strain (ribotype 017) resistant to newer fluoroquinolones in Poland
H. Pituch, D. Wultanska, P. Obuch-Woszczatynski, G. Nurzynska, W. van Leeuwen, F. Meisel-Mikolajczyk, M. Luczak, A. van Belkum (Warsaw, PL; Rotterdam, NL)
Outbreaks due to new hypervirulent C. difficile (ribotype 027, toxinotype III) strains that are highly resistant to fluoroquinolones were detected in The Netherlands between April 2005 and February 2006. One hospital in The Netherlands experienced an outbreak due to a toxin A negative C. difficile strains (ribotype 017 toxinotype VIII). Toxin A negative/toxin B positive C. difficile, PCR ribotype 017 strains lack a part of the toxin A gene and were first recognized as a cause of CDAD in Poland in 1995. Between 2001 and 2005 a growing epidemic of C. difficile-associated diarrhoea (CDAD) caused by toxin variant strains producing only toxin B (A–B+) was noted in our Polish university hospital. All C. difficile strains ribotype 017 isolated before 2001 were susceptible to the newer fluoroquinolones. The new Polish A–B+ isolates belonging to PCR ribotype 017 have a characteristic antimicrobial susceptibility pattern, since they are highly resistant to ciprofloxacin, moxifloxacin, gatifloxacin and show resistance to clindamycin and erythromycin (MLSB type resistance). Macrolide, lincosamide, and streprogramin B resistance is due to the presence of an ermB gene. All PCR ribotype 017 strains were resistant to gatifloxacin and moxifloxacin, but not the historical isolates (obtained before 2001). Exposure of patients to fluoroquinolones in Poland is recognized as a risk factor for CDAD caused by C. difficile strains ribotype 017.
This work was supported by Polish Ministry of Education and Science, Grant No. 2 P05D 074 27.
P1057 Transfusion-transmitted virus, injection drug users, hepatitis
A. Soudbakhsh, M. Hajabdolbaghi, M. Mohraz, M. Abdollahi Nami, P. Khairandish, H. Emadi Koochak (Tehran, IR)
Objective: Transfusion-Transmitted Virus (TTV) is a nonenveloped, single-stranded and circular DNA virus which is belongs to the genus Anellovirus of circuviridae family. It was discovered by Nishizawa in 1997. One of the most important ways of transmission is use of common syringes. An increase in the population of Injection drug users (IDU) can be dangerous to other persons who are not IDUs. The objective of this study was to determine the prevalence of TTV in the IDU population of Iran.
Materials and Methods: In this cross-sectional study which was performed at the Infectious ward of Imam Khomeini hospital, 60 IDU patients were studied. Blood samples were dispatched to the lab in citrated test tubes for Virus isolation, using the boiling method, and then PCB assay performed based on available primers. The patient's information gathered by interview and questionnaire methods. Statistical tests such as Pearson Chi-Square, Mann Whitey and T-test used to analyze these data.
Results: All our 60 patients were men and their age average was 35.3 years (SD±9.68). Twenty-six patients had positive TTV PCR and 92.3% of them had history of imprisonment. Of these 26 patients, 88.5% had positive HCV Ab, 65.4% had positive HIV Ab and 30.8% had positive HBS Ag. In the 60 patients studied, 80% had positive HCV Ab, 71.7% had positive HIV Ab, 43% had TTV PCR and 26.7% had positive HBS Ag.
Out of 26 patients who had TTV, 34.60% of them had no overt sickness and 11.5% of them displayed signs of hepatitis (fever, abdominal pain, nausea, vomiting, right upper quadrant tenderness and icter). Liver function test was abnormal in 34.6% The average years of injection in the 26 TTV patients was 9 years (SD±7.16) and the patient's age average was 36.35 years (SD±9.2).
Conclusion: One of the most important ways of TTV infection is needle sharing. By this rout chance of TTV infection is less than HIV and HCV but more than HBV. Due to the high prevalence of TTV infection in the IDU population in our country and the fact that this population is not limited and there is no comprehensive information about pathogenesis of this virus, which is faecal–oral, there is a need to make plans and adopt policies to decrease the danger of transmission of this virus to healthcare workers and their families as well as other near relatives.
P1058 Detection of toxigenic Vibrio cholerae O1 from a freshwater lake near Tbilisi, Georgia: a potential source for disease
M. Tediashvili, C.A. Whitehouse, A. Huq, E. Jaiani, T. Kokashvili, N. Janelidze, C. Grim, R.R. Colwell (Tbilisi, GE; Fort Detrick, Baltimore, US)
Objectives: Strategies for the prevention and control of cholera and other vibrio-related infections depend on regular monitoring of natural aquatic environments, as well as understanding the origin, pathogenicity, transmission dynamics, and other characteristics associated with the occurrence and spread of epidemic strains. As part of ongoing studies to examine the presence and diversity of pathogenic Vibrio spp. in aquatic environments in Georgia, we sampled three freshwater lakes, Lisi, Kumisi, and Tbilisi Sea near Tbilisi.
Methods: Lakes were monitored for various biological and physical-chemical characteristics including temperature, pH, and salinity. Samples were taken from 2 sites from each lake biweekly from May to October 2006. Because Vibrio spp. can survive in the environment in a viable but nonculturable state, direct detection methods, such as PCR and direct fluorescent antibody (DFA) assays were used.
Results: Of 375 bacterial isolates obtained, 191 were presumptively identified as Vibrio spp. by growth on selective media and typical biochemical profile. Complete characterisation of these isolates is ongoing. Total DNA was extracted from water samples and tested for the presence of V. cholerae and selected virulence factor genes. Samples from Lisi Lake were positive for V. cholerae-specific ISR rRNA (encoding the intergenic spacer region of the 16S-23S rRNA gene), ctxA, tcpA, zot, ompU, and toxR by PCR. Furthermore, DFA testing, using antibodies specific for either the O1 or O139 serotypes, were positive for V. cholerae O1. None of the samples tested were positive for V. cholerae O139.
Conclusion: These data provide initial evidence for an environmental reservoir for toxigenic V. cholerae O1 in a freshwater lake in Georgia. Since this lake is often used for recreational purposes, it should be considered a potential source for disease.
P1059 Detection of Dobrava hantavirus infections in rodents (Apodemus agrarius, A. flavicollis) in the Transdanubian region of Hungary
F. Jakab, G. Horváth, E. Ferenczi, G. Szucs (Pécs, Budapest, HU)
Background: Dobrava hantaviruses belong to the genus Hantavirus, family Bunyaviridae and carried by striped field (Apodemus agrarius) and yellow necked (Apodemus flavicollis) mice. Dobrava hantavirus causes severe haemorrhagic fever with renal syndrome (HFRS) in many European countries.
Objectives: The goal of this pilot study was to determine the prevalence of Dobrava hantavirus in A. flavicollis, A. agrarius, A. sylvaticus rodents in the Transdanubian region of Hungary.
Methods: The rodents were trapped in three different location of the Transdanubian region of Hungary (Gyékényes, Görcsöny and Kis-Balaton area) during the summer and autumn seasons of 2005. The rodents were dissected and lung tissues were used for hantavirus detection. The viral RNA was extracted from lung suspensions with TRIzol reagent according to the manufacturer's recommendation. Dobrava hantaviruses were detected by SYBR Green-based real-time PCR, using newly designed virus specific primers. Positive samples were selected for sequence and phylogenetic analysis.
Results: During the study period 22 Apodemus sp. (11 A. agrarius, 10 A. flavicollis and 1 A. sylvaticus) were tested for the presence of hantaviruses. Out of the 22 Apodemus rodents 3 A. agrarius were infected with Dobrava hantavirus. Based on the phylogenetic analysis at least two genetic lineages circulate and represent in the region, according to the different geographic locations.
Conclusion: In this study we reported the occurrence of Dobrava hantaviruses in the Transdanubian region of Hungary. Based on the clinical experience and our new data from the region we concluded that extended reservoir studies as well as serological investigations might be important in the future.
P1060 Burden of zoonotic diseases in Venezuela during 2004 and 2005
J. Benitez, A. Rodriguez-Morales (Maracay, Trujillo, VE)
Introduction: Emerging zoonotic diseases have assumed increasing importance in public and animal health, as the last few years have seen a steady stream of new diseases, each emerging from an unsuspected quarter and causing severe problems for animals and humans.
Objectives: For this reason surveillance, analyses, prevention and control of these diseases are of utmost importance. In this report we described the importance and burden of zoonotic diseases in Venezuela during 2004 and 2005.
Methods: Records reported from different divisions and states to the Ministry of Health compiled and summarised are analyzed for years 2004 and 2005. Zoonoses included are: rabies, equine encephalitis, leptospirosis, leishmaniasis, cysticercosis, teniasis, brucellosis, toxoplasmosis and yellow fever.
Results: During this period 3,859 cases of suspected zoonotic diseases were recorded, being confirmed 52.8%, 772/2031 in year 2004 (38%) and 1265/1828 in year 2005 (69%). Leishmaniasis was the first in incidence among this group, 458 cases (22.5%) (p < 0.05) [63 (3.1%) in humans], followed by rabies with 299 cases (14.7%) [5 (0.2%) in humans], 175 (8.6%) cases of cysticercosis in humans, 97 (4.8%) cases of leptospirosis in humans, 39 (1.9%) cases of brucellosis in humans, VEE 20 cases (1%), EEE 13 cases (0.6%), 18 cases of teniasis, 17 of yellow fever and 14 of toxoplasmosis.
Conclusion: The reasons for zoonoses emergence are multiple being the expansion of the human population, animals' mobilisation and close human contact, the most important. Current issues such as the increasing movement of a variety of animal species, ecological disruption, uncultivatable organisms, and terrorism, all imply that emerging zoonotic diseases will in all probability, not only continue to occur, but will increase in the rate of their emergence. The recurring nature of the crises dictates that closer integration of veterinary and medical communities is warranted, along with improved education of the general public and policy makers to reduce the impact of this important public health problem.
P1061 International Circumpolar Surveillance interlaboratory quality control programme, 1999–2004
T. Zulz, A. Reasonover, M. Lovgren, L. Jette, M.S. Kaltoft, M. Bruce, A.J. Parkinson (Anchorage, US; Edmonton, Sainte-Anne-de-Bellevue, CA; Copenhagen, DK)
Background: The International Circumpolar Surveillance (ICS) system conducts population-based surveillance of invasive pneumococcal disease in Finland, Greenland, N. Canada (N Can), N. Sweden, Norway and in the U.S. Arctic (AK). Reference laboratories in N. Can [National Centre for Streptococcus (NCS) and Laboratoire de santé publique du Québec (LSPQ)] and AK [Arctic Investigations Program (AIP)] began an interlaboratory quality control (QC) programme in 1999 for pneumococcal serotyping and antibiotic susceptibility testing. Staten Serum Institute (SSI) in Copenhagen, Denmark, joined the QC programme in 2004. The objectives of the QC programme were to provide participating laboratories with external proficiency testing that monitors routine protocols as they are applied to the characterisation of Streptococcus pneumoniae (Sp), to identify and correct potential problems and to continually improve services.
Methods: Each participating laboratory sent one panel per year. A panel contained 7 Sp isolates with a variety of serotypes and antibiotic resistance patterns. Pneumococcal type specific antisera from SSI was used at AIP, NCS and SSI to classify all 90 Sp serotypes; LSPQ maintained antisera to the most common serotypes and less common ones were referred to NCS. Minimum inhibitory concentrations were determined for each isolate for those antibiotics routinely tested in each lab. Results were considered to be in agreement if within one log(2) of each other. Each lab submitted a completed standard form to the distributing laboratory where a summary report was prepared that included discussion of any problems encountered.
Results: A total of 18 panels were tested. The overall serotype agreement was 91% (range 87.5% – 100%); there were only 2 (11%) out of the 18 panels with less than 100% agreement. Overall antibiotic susceptibility correlation was 96.3% (range 83%–99%) with 8 or 9 antibiotics tested per panel. Delays encountered during shipment of panels during 2001 prompted labs to consider using charcoal culturettes or to lyophilise the cultures for future shipments to maintain isolate viability.
Conclusion: The ICS QC programme is a successful international interlaboratory quality control testing programme. Correlation of serotyping and antibiotic susceptibility testing was high. The QC programme should be expanded to other participating regions in ICS to ensure interlaboratory comparability.
P1062 Brucellosis in Thailand: emerging and enigmatic
W. Kulwichit, B.K. De, O. Putcharoen, S. Nilgate, N. Hiransuthikul (Bangkok, TH; Atlanta, US)
Objectives: Brucellosis is a worldwide emerging disease. As per a recently-updated global map, our country belongs to the group of ‘nonendemic/no data', with fewer than 10 human cases. Our cases may be low in number, but certainly not in clinical attractiveness.
Methods: We gathered data of interesting cases of brucellosis in the past few years. CASE 1&2 are spouses who suffered from systemic brucellosis several months apart. Epidemiologic investigation pointed towards a probable sexual transmission. The isolates were examined at the US CDC by standard phenotypic assays, AMOS PCR, and multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) with 7 Brucella species as controls. CASE 3 developed vertebral osteomyelitis and psoas abscess following back injury, compatible with the principle of locus minoris resistentiae.
Results: CASE 1&2 – The 37-year-old husband had been exposed to goats and drinking unpasteurised goat milk. Blood cultures grew Brucella melitensis. Four months after his ‘recovery', his wife started a prolonged febrile illness. Blood cultures grew B. melitensis. Periodically visited by her husband, the wife lived in the national capital with no history of goat exposure or goat milk consumption. Isolates from the couple (BM1 & BM2) were identical phenotypically and identified as Brucella melitensis biovar 2. AMOS PCR revealed that both isolates co-migrated with a reference strain of B. melitensis. MLVA targeting 15 loci exhibited same-size amplicons for 14 of them, and allele sizes of 631 and 638 for each isolate for the other locus (Figure ).

CASE 3, a farmer presented with lumbosacral osteomyelitis and right psoas abscess by Brucella abortus. She had consumed raw beef for years without a previous problem. A few weeks prior to this illness, however, she had been kicked by a cow and fell to the ground, hurting the lower back area for about a week. This reminds us of locus minoris resistentiae, or a place of less resistance.
Conclusion: For case 1&2, both isolates were very similar and probably originated from a single source. With epidemiologic data, the organism could be transmitted from husband to wife, probably sexually. Case 3 is an example of food-borne zoonosis induced by physical injury. Brucellosis is not only emerging, it also demonstrates an ability to manifest and be transmitted in an extended spectrum. Clinicians need to maintain a suspicious mind to cope with the current world of dynamic infectious diseases.
P1063 Performance evaluation of a novel enzyme immunoassay for detection of human metapneumovirus
M. Sheridan (Dublin, IE)
Objectives: Human Metapneumovirus (hMPV) is a causative agent of acute respiratory tract infections and is emerging as a significant cause of hospitalisation for such infections in young children and the immunocompromised worldwide. Here we describe the performance evaluation of an antigen detection enzyme immunoassay (EIA) system based on a unique combination of monoclonal antibodies directed to major hMPV proteins.
Methods: The diagnostic performance characteristics (sensitivity and specificity) of the hMPV EIA were evaluated using panels of confirmed hMPV positive or negative respiratory samples that were recovered from patients by a range of typical collection methods. Analytical performance parameters were estimated using viral cultures of known TCID50. Analytical specificity was evaluated by testing panels of potentially interfering viruses, bacteria and medications.
Results: The EIA exhibited close correlation to PCR in terms of diagnostic sensitivity. A high level of specificity was demonstrated against a panel of potentially cross-reactive, non-hMPV, specimens. The EIA was found to be reactive with hMPV subgroups 1 and 2 of genotypes A and B from viral culture.
Conclusions: The hMPV antigen detection EIA is comparable to PCR in terms of key diagnostic performance characteristics is an easy-to-use, cost-effective alternative to PCR suitable for use in the clinical laboratory.
P1064 Fusobacterium necrophorum infections in Denmark from 1998 to 2001 – a prospective epidemiological and clinical survey
L.H. Kristensen, J. Prag (Viborg, DK)
Objectives: The aim of this study was to describe the epidemiology and clinical spectrum of F. necrophorum infections.
Methods: In a prospective countrywide 3-year-study in Denmark from 1998 to 2001 a total of 391 human isolates of F. necrophorum were registered and epidemiological and clinical data collected from the clinical records.
Results: Fifty-seven patients had Lemierre's syndrome, which gave an incidence of Lemierre's syndrome of at least 3.5 per mill. per year, without any increase during the study period or seasonal variation. Lemierre's syndrome was seen in 5 small children with otitis media, 38 youngsters with tonsillitis and 7 young adults with e.g. parapharyngeal abscess, meningitis or sinusitis and 7 elderly adults with presumably primary head focus. The patients with Lemierre's syndrome were previously healthy. They were obviously sick on admission with leukocytosis, band neutrophilia, elevated C-reactive protein, metastatic infections especially to the lungs and rapid progress, if they were not immediately treated with long-term antibiotics, often combined with surgery/drainage. Nine percent died.
Disseminated F. necrophorum infections caudally of the head were seen in 36 elderly patients, who often had predisposing diseases, e.g. abdominal or urogenital cancers. They had a mortality of 28%, predominantly due to age and predisposing diseases rather than the infection.
Localised F. necrophorum infections in the head were seen in 215 youngsters with peritonsillar abscess, 7 small children with otitis media, 9 small children with solitary abscess formation in cervical lymphadenitis, 26 youngsters with tonsillitis and 10 middle-aged with sinusitis or tooth problems. F. necrophorum was found in 20% of all peritonsillar cases cultured on routine anaerobic media. All localised cases were previously healthy. They were clearly less sick on admission than the patients with Lemierre's syndrome, and all recovered without sequelae.
Conclusion: The incidence of Lemierre's syndrome may be higher than previous thought. F. necrophorum seems to play a role in tonsillitis. Increased awareness of Lemierre's syndrome by the clinicians and the clinical microbiologists is mandatory, along with long-term antibiotic therapy, often combined with aggressive surgery and repeated drainage, to lower mortality and morbidity.
Table 1.
Prospective study of Fusobacterium necrophorum infections in Denmark, 1998—2001
| Male | Female | Total no. | Mortality |
||
|---|---|---|---|---|---|
| no. | (%) | ||||
| Lemierre's syndrome | 33 | 24 | 57 | 5 | (9%) |
| Other disseminated cases | 24 | 12 | 36 | 10 | (28%) |
| Local infections | |||||
| In the head and neck | 160 | 107 | 267 | 0 | |
| Caudally from the head | 13 | 9 | 22 | 0 | |
| Unknown primary focus | 5 | 1 | 6 | 1 | (17%) |
| No signs of infection | 1 | 2 | 3 | 0 | |
| Total | 236 | 155 | 391 | 16 | (4%) |
P1065 S. agalactiae invasive infections in male adults
M. Makarona, N. Tsagarakis, H. Moraitou, N. Kentrou, A. Gioga, C. Kapotis, A. Pefanis, S. Kanavaki (Athens, GR)
Objectives: To investigate S. agalactiae invasive infections in our hospital. In particular, S. agalactiae has been considered a major pathogen in neonates and pregnant women. However, there is accumulating concern about its significance in non-pregnant adults, because it has been recently recognized as an important and increasingly common cause of invasive diseases in the previous. Specifically, it can cause disease in adult patients with underlying medical conditions, including diabetes mellitus, malignancies and liver disease. Skin and soft tissue infections are common presentations of invasive S. agalactiae infections, while nosocomial infections and polymicrobial bacteraemia occur in a significant proportion of patients. The annual incidence of invasive disease has been estimated at 4.4 per 100,000 non-pregnant adults, whereas the incidence of infections due to S. agalactiae has increased among diabetic patients. Mortality rates in invasive diseases are particularly high (20–70%).
Materials: We retrospectively studied 9 cases of systemic infections due to S. agalactiae in male adults, during the last five-year period.
Results: From a total of nine cases of S. agalactiae infection, 6 (66.6%) concerned male patients that had all been hospitalised within this recent year, 2006, in various departments of our hospital. S. agalactiae was isolated and identified in various clinical specimens: blood, pus, cerebrospinal fluid, sputum, urine, pleural effusion and synovial fluid. Five out of nine strains (56%) were responsible for bacteraemia, from which 2/5 where responsible for bacteraemia only, 2/5 for bacteraemia as well as for diabetic foot, while 1/5 was simultaneously isolated in blood and cerebrospinal fluid. Among the others, 1/9 (11.1%) was isolated from pleural effusion, 1/9 (11.1%) from synovial fluid, 1/9 (11.1%) from urine and 1/9 (11.1%) from sputum. Eight out of nine patients (88.9%) were suffering from a serious underlying disease: 4 from diabetes mellitus, 2 from diabetes mellitus and malignancy, 1 had malignancy only, 1 suffered from chronic renal failure, while only 1 had a free previous medical history. One patient died, while all the rest had an improved clinical outcome.
Conclusion: S. agalactiae invasive infections seem to gain ground as a significant problem in male adults, especially in those with chronic diseases, such as diabetes mellitus. Efforts should be made to be identified and treated early for a successful clinical outcome.
P1066 Nineteen cases of Actinobaculum schaalii identified in Viborg County, Denmark
T.F. Jakobsen, H.L. Nielsen, K.M. Søby, J. Prag (Viborg, DK)
Objectives: Actinobaculum schaalii is a Gram-positive, non-spore-forming, facultative anaerobic, catalase-negative, rod-shaped bacillus, usually associated with urinary tract infection, and human infections are currently limited to a few reports. A. schaalii was identified in blood and/or urine from 18 patients in 18 months by the Department of Clinical Microbiology at Viborg Hospital, Denmark. Nine cases with clear pathological findings are described.
Methods: In our department urine cultures are routinely incubated in a CO2-enriched atmosphere and this practice has proved to facilitate the finding of A. schaalii, since it grows poorly or not at all in ambient air. Identification of A. schaalii is initially done by use of the API Coryne and Rapid ID32A test systems eventually followed by 16S rRNA sequencing to verify the identity of isolated bacteria for publication.
Results: Between February 2005 and July 2006, we identified A. schaalii in blood and urine from 18 patients (age range 60–92 years) in Viborg County, with a population of 230,000. Clinical data and predisposing conditions from nine patients are summarised in Table 1 .
Table 1.
In vitro activities of some antibiotics against multi-resistant bloodstream infections of Acinetobacter spp.
| Antibiotics | MIC |
% Resistance |
|||
|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | 1996–2000 (n = 60) | 2001–2005 (n = 104) | |
| Amikacin | 128 | 256 | 0.25–256 | 86.0 | 74.0 |
| Netilmicin | 4 | 32 | 0.125–64 | 46.7 | 25.0 |
| Imipenem | 32 | >32 | 0.064->32 | 65.0 | 63.5 |
| Piperacillin-tazobactam | 256 | >256 | 0.016->256 | 81.7 | 84.6 |
| Cefoperazone-sulbactam | 256 | 256 | 0.125->256 | 68.3 | 63.5 |
| Colistine | 2 | 1024 | 0.19–1024 | 63.3 | 25.0 |
| Tigecycline | 1 | 2 | 0.047–16 | 1.0 | |
| Ciprofloxacin | 32 | >32 | 0.064->32 | 66.7 | 83.7 |
| Tetracycline | 32 | 256 | 0.125->256 | 91.7 | 86.5 |
Although A. schaalii was found in urine from the remaining 9 patients as well, they had no clear clinical symptoms of urinary tract infection at the time of sampling. The asymptomatic patients suffered from recurrent UTIs and/or conditions, such as prostate cancer, urethrastenosis, cystocele and diabetes mellitus, that may predispose bacteuria.
Conclusion: Our findings confirm the importance of A. schaalii as a pathogen in UTIs as it can cause serious infections, such as pyleonephritis and urosepsis, but may also be present in cystitis as well as in asymptomatic bacteuria. The fact that A. schaalii was found in a patient in Viborg each month strongly suggests that A. schaalii infections are more common than previously recognized. Clinicians should be aware of A. schaalii in cases of unexplained pyuria, especially if the initial microscopic bacterial findings differ from the growth results under aerobic conditions.
P1067 Fusobacterium necrophorum severe sepsis associated with Lemierre's syndrome and pulmonary metastatic septic embolisation
G. Magnani, E. Gabbi, A. Catania, L. Ricci, C. Guidetti, L. Vecchia (Reggio Emilia, IT)
Objectives: Fusobacterium necrophorum is an unusual but potentially lethal pathogen, associated with Lemierre's syndrome, an oropharyngeal infection complicated by internal jugular vein (IJV) thrombophlebitis and metastatic septic embolisation. We report a case of this disease with a secondary pulmonary abscess, following F. necrophorum severe sepsis.
Case report: A 34-year-old female was admitted because of fever, abdominal pain and diarrhoea. She referred an acute tonsillitis 2 weeks before. Tonsils hypertrophy, but normal aspect of the neck was observed. Laboratory analysis demonstrated WBC 12.4×109/L (85% N), platelets 97×109/L, C-reactive protein 27.41 mg/dl and procalcitonin 9.06 ng/mL. Abdominal US showed splenomegaly and slight peritoneal fluid. Normal was chest x-ray. Empirical therapy with ceftriaxone 2 gr qd was initiated, but 24 hours later her clinical conditions worsened with hypotension (90/40 mmHg) and respiratory distress. Auscultation revealed bilateral chest crepitations and thoracic radiograph confirmed focal infiltrates and slight pleural fluid. Multiple blood cultures yielded β-lactamase negative F. necrophorum, susceptible in vitro to penicillin, metronidazole and clindamycin. So, metronidazole 500 mg i.v. qid was added. Six day after the admission, she developed painful left upper neck mass along anterior border of the sternocleidomastoid muscle. The ultrasound and the CT scan of the neck demonstrated a parapharingeal infiltrate with thrombosis of left IJV. The surgical approach was excluded. Anticoagulant treatment with warfarin was started, but ten day into therapy she worsened with dyspnoea and left pleuritic chest pain. The CT scan revealed an abscess of 3×3 cm in size in the left paramediastinal side with pleuric effusion. Surgical drainage revealed a purulent material, negative at the bacterial cultures. The patient completed a 4-week course of intravenous antibiotics; and was discharged 30 days after hospitalisation.
Results and Conclusions: We describe a case Lemmiere's syndrome, complicated by lung abscess and empyema, due to F. necrophorum. Note-worthy, the postanginal sepsis developed after two week from the initial episode of oropharingeal infection. Moreover, F. necrophorum was isolated from the blood several days before the initiation of the typical manifestation, underlining the importance of culture of anaerobic bacteria and the interdisciplinary collaboration
P1068 From insects to man: identification of virulence factors involved in the adaptation of the emerging pathogen Photorhabdus asymbiotica to human hosts
M. Forquin, J. Esque, A. Pimenta (Cergy Pontoise, FR)
Photorhabdus are entomopathogenic bacteria currently used for the biological control of crop pests. Recently, an increasing number of Photorhabdus strains have been isolated from human clinical specimens, associated with locally invasive soft tissue infections and disseminated bacteraemia.
Objectives: In view of their growing use in biological control, which increases the human potential rate of exposure to these pathogens, we undertook a comparative study between insect and human pathogenic strains of Photorhabdus, in an attempt to understand the genetic mechanisms involved in their recently acquired capacity to infect humans.
Methods: Three Photorhabdus strains, two human (P. asymbiotica SN and ATCC43949) and one insect pathogen (Photorhabdus sp Q617), were tested for their ability to bind to two major components of the human extracellular matrix (ECM), fibronectin (Fn) and vitronectin (Vn), to infect or invade human epithelial cells in culture, and to interact with the cellular cytoskeleton. Adherence, infection and invasion experiments were performed in 96-well microtiter plates coated either with Fn, Vn or with monolayers of human epithelial HaCaT cells. Bacterial invasion was quantified by the gentamicin survival assay. In inhibition assays, anti-Fn, anti-Vn antibodies, or genistein were added to epithelial cell monolayers. For immunofluorescence, HaCaT monolayers on glass coverslips were infected and labelled with phalloidine-TRITC and DAPI.
Results: Only P. asymbiotica human pathogenic isolates ATCC43949 and SN were able to adhere to human fibronectin and/or vitronectin, and to infect/invade HaCaT cells. This was inhibited in the presence of antibodies against human Fn or Vn. Only strain ATCC43949 induced cytotoxic effects after 6h co-culture with human cells. Cytoskeleton involvement in the P. asymbiotica infective process was indicated by blockage of infection and invasion by genistein, and by immunofluorescence analysis of infected cells.
Conclusion: The acquisition of virulence factors enabling P. asymbiotica to interact with human ECM and to human cells is a key factor in its evolution towards the colonisation of human hosts. ECM proteins are involved in the primary events during P. asymbiotica colonisation of human epithelial tissues, and the host cytoskeleton is solicited during the P. asymbiotica-epithelial cells interaction. Possible mechanisms used b y P. asymbiotica to colonise human tissues are discussed.
Vaccines
P1069 Clustered epitopes within the Gag-env HIV-1 fusion proteins DNA vaccine enhance immune responses in mice
F. Roodbari, F. Sabahi, M.N. Sarbolouki, F. Barkhordary, M.A. Mahdavi, R. Sarami Forooshani, T. Bamdad, A. Adeli, F. Mahboudi (Tehran, IR)
Objectives: The global HIV epidemic continues to expand and exceeding previous predictions. An effective vaccine represents the best hope to curtail the HIV epidemic. DNA vaccines induce conformational-dependent humoral and cellular response and mimic live vaccines without their pathogenic potential. The importance of CD8+ CTL responses in controlling HIV and SIV viraemia has led to a series of vaccine candidates that effectively induce these responses. It is now widely believed that an HIV vaccine strategy must stimulate both a strong humoral (antibody) as well as cell-mediated (CTL) immune response. Neutralising antibodies reduce viral burden by latching onto and destroying HIV circulating in body fluids. CTLs eliminate HIV-infected cells, thereby inhibiting viral replication. Both types of immune response are invoked by the presence of antigens, such as HIV envelope glycoproteins, in the body.
The P24 and gp41 play many of important roles in host-virus-interaction and pathogenesis. These proteins are considered as attractive vaccine candidate in which their immungenecity and immunomodulatory effects have been confirmed.
Methods: In this study, an expression vector (PCDNA3.1 hygro) containing P24-gp41 immunogenic sequences under the control of IE HCMV promoter was designed. The expression of the recombinant peptides was analyzed in an eukaryotic systems, COS-7 and Hela cells. Immunofluorescence and western blotting confirmed the presence of expressed proteins The cited P24-gp41 fusion gene that is able to express in a proper folding form. This fusion gene was studied as DNA vaccine in mice(BALB/c) for evaluation and generation of effective immune responses. For immunising, we used dendrosome, a novel family of vehicles for transfection and therapy. Some groups of mice were immunised with our construct and pCAGGS-IL-12 as co-injection. Humoral response and IFN-γ and IL-2 was detected by ELISA. Lymphoprolifration assay was detected by MTT.
Results: ELISA and MTT assays confirmed, the cited P24-gp41 fusion gene that is able to enhance immune responses in mice.
Conclusions: Our construct is a good candidate for DNA vaccine against HIV-1.OF course, it needs much more research for clearing some things that they are unclear.
P1070 Susceptibility to rubella in females born 1996–2003: a potential congenital rubella syndrome iceberg?
A. Vyse, R. Pebody, L. Hesketh (London, Preston, UK)
Objectives: Rubella immunisation programmes are designed to prevent maternal infections and thus congenital rubella infection. Since 1970 the UK has implemented both selective and universal rubella vaccination strategies. The WHO European Regional Office has established a target to eliminate rubella from Europe by 2010 and recommends susceptibility among women of childbearing age to be <5%. Whilst rubella infection has rarely occurred in the UK since 1996, decreasing MMR uptake in infants since the late 1990s and increased immigration from countries without routine rubella vaccination programmes suggest rubella susceptibility at the population level should be reviewed.
Methods: A total of 2883 serum samples collected in 2004 across the complete age range reflecting the general population were screened for rubella specific IgG by ELISA (Behring Enzygnost). Serological data were stratified by birth cohort and gender and described using a four component mixture model, exploiting differences in the distribution of quantitative results in samples from previously infected, previously vaccinated and previously unexposed individuals. Vaccine coverage data for MMR and laboratory confirmations of rubella obtained in 2004 were included.
Results: During 2004 only 14 cases of rubella were laboratory confirmed (ages <1->35 yrs) and vaccine coverage for MMR was estimated to be ∼80% by the second birthday for those born 2000–2003. Serological estimates of susceptibility to rubella in males and females born 2000– 2003 reflected vaccine coverage estimates. Whilst serological estimates of rubella susceptibility in females of childbearing age were very low (0–2%) in 2004, susceptibility in females born 1996–1999 who have had opportunity for a second dose of MMR was estimated at 9%. Susceptibility estimates were also high in males born 1956–1980 (range 7%-12%).
Conclusions: Most rubella susceptibility is found in those born post 1995 reflecting the current suboptimal uptake of MMR, and in males born 1956–1980 who have never had an opportunity for vaccination and not yet acquired the infection naturally. Susceptibility in women of childbearing age is currently estimated to be very low in the general population. It is important that those females born 2000–2003 receive a second dose of MMR and those females born 1996–1996 who are now too old to receive a routine second dose should be further monitored, to ensure low susceptibility levels are obtained in these cohorts before they reach childbearing age.
P1071 Measles sero-epidemiology in England in 2004
R. Pebody, A. Vyse, L. Hesketh (London, Preston, UK)
Objectives: The WHO European Regional Office has established a target of measles elimination by 2010. Since the late 1990s uptake of MMR in infants in the UK has declined, and recently the UK has experienced localised measles outbreaks, particularly during 2006. The aim of this study was to measure susceptibility to measles in the general population of England to assess progress towards measles elimination.
Methods: A total of 2883 serum samples collected in 2004 across the complete age range reflecting the general population were screened for measles specific IgG by ELISA (Behring Enzygnost). Serological data were stratified by birth cohort and described using a five component mixture model, exploiting differences in the distribution of quantitative results in samples from previously infected, previously vaccinated and previously unexposed individuals. Susceptibility was compared to WHO age-specific measles susceptibility targets of <15% in 1–4 year olds, <10% in those aged 5–9 years and <5% in those aged >10 years. Historical vaccine coverage data for MMR and laboratory confirmations of measles were also analysed.
Results: During 2004 vaccine coverage for MMR was estimated to be ∼80% by the second birthday in those born 2000–2003. Reported laboratory confirmed cases stand at 191 in 2004 (51% in those aged <5 years), 77 in 2005 (56% in those aged <5 years) and 438 during the first five months of 2006. The serological estimates of measles susceptibility in 2004 were 21% in those born 2000–2003, 5% in those born 1996–1999, 8% in those born 1991–1994, 3% in those born 1986– 1990 and <3% in those born before 1986. A similar study carried out using samples collected in 2000 estimated measles susceptibility in those born 1996–1999 to be 21%.
Conclusions: Compared to the WHO age-specific targets measles susceptibility in pre-school aged children is high but is within acceptable limits for older age groups, congruent with the low uptake of first dose MMR that has been observed. This highlights the potential for measles outbreaks in this age-group and is consistent with recent laboratory confirmations, indicating the need to improve first dose MMR vaccine coverage. In 2004 the estimate of measles susceptibility in those born 1996–1999 was considerably lower than the estimate made in 2000 for this cohort before they had opportunity for a second dose of MMR, reinforcing the value of including a second dose in the programme at school entry.
P1072 Interpreting the mumps epidemic in England in the early 21st century: a disease iceberg that is likely to continue?
A. Vyse, R. Pebody, L. Hesketh (London, Preston, UK)
Objectives: Currently in the United Kingdom, outbreaks of mumps are running at their highest level since the introduction of MMR vaccine in 1988. These have mainly affected young adults attending tertiary-level educational institutions, many of whom have had opportunity for at least one dose of MMR vaccine. This study draws on key surveillance data to investigate underlying reasons behind this epidemic.
Methods: A total of 2883 serum samples collected in 2004 across the complete age range reflecting the general population were screened for mumps specific IgG by ELISA (Behring Enzygnost). Serological data were stratified by birth cohort and described using a four component mixture model, exploiting differences in the distribution of quantitative results in samples from previously infected, previously vaccinated and previously unexposed individuals as the basis for analysis. Comparison was made with vaccine coverage data for MMR and laboratory confirmations of mumps obtained in 2004.
Results: During 2004, 8130 cases of mumps were laboratory confirmed with ∼80% in those born 1980–1989 and <1% in those born 2000–2003. Vaccine coverage for MMR was estimated to be ∼80% by the second birthday for those born 2000–2003. Serological estimates of those with no evidence of mumps specific IgG were highest in those born 2000– 2003 (15%) and low in all other birth cohorts/age groups (0%-3%). Strongest antibody responses occurred in older age groups (born pre-1980) most likely to have experienced natural mumps infection. Large proportions of those born 1981–2003 were estimated to have only low levels of mumps antibody. Those born 1986–2003 have had opportunity for at least one dose of MMR, with those born 1991–1999 opportunity for a second dose. There was no evidence that an opportunity for a second dose of MMR boosted mumps IgG levels.
Conclusions: The current mumps epidemic is occurring in a cohort where few have no evidence of mumps specific IgG but large proportions are estimated to have only low levels of antibody, suggesting a lack of protection. Behavioural mixing patterns, transmissibility of the virus and the relative proportions with low or no mumps antibody are likely to combine to determine where the burden of infection lies. Similar proportions observed with low IgG levels in those born 1991–2003 may therefore fuel future mumps outbreaks as they enter tertiary educational settings, suggesting mumps may be an ongoing problem in the UK.
P1073 Outcome of pregnancy in pregnant women vaccinated in the mass campaign against measles/rubella in Tehran, Iran
H. Emadi, M. Hajabdolbaghi, M. Rasoolinejad, S. Jafari, P. Khairandish, L. Taheri, A. Karami (Tehran, IR)
Introduction: Rubella is one of the most important diseases in pregnancy because of its ability to cause fetal malformations. Fetuses of women who acquire rubella in the first trimester of pregnancy may have congenital rubella syndrome (CRS) which includes sensorineural deafness, cataracts, microphtalmos, glaucoma, microcephalos, and other problems. Pregnancy is a contraindication for rubella vaccination because of concerns regarding the theoretical possibility of adverse effects on the developing fetus. To decrease the chance of acquiring this infection during pregnancy, mass campaign for measles/rubella vaccination was performed in December 2003 throughout Iran. During this time period, there were women who received this vaccine during pregnancy mistakenly as they were not aware of their pregnancy at the time of vaccination. In this study, we have followed these women who referred to the infectious disease clinic at Imam Khomeini Hospital to determine the outcome of pregnancy.
Materials and Methods: Women with history of rubella vaccination that were possibly pregnant and had referred to the infectious disease clinic at Imam Khomeini Hospital were included. Pregnancy test and anti-rubella IgG avidity were performed in all cases. According to the result of the avidity test, they were grouped into two groups:
-
1
High avidity (titer higher than 53% which is indicative of immunity)
-
2
Low avidity (titer lower than 53% which is indicative of susceptibility) Neonates born from susceptible women were tested for rubella-specific IgM in cord blood sera.
Results: Out of 325 women, 3 were excluded because they were not actually pregnant and the outcomes of conception were known only for 235 of the 322 pregnancies. Two-hundred-five (87.2%) had high avidity and 30 (12.8%) had a low avidity titer. The outcome of pregnancy in the two groups is depicted in the graph.
Discussion: In this study rubella vaccination did not increase the rate of still birth, congenital malformation and abortions in rubella susceptible vaccinated mothers as compared to immune mothers.
There was also no significant difference in still birth, congenital malformation and abortions between non-vaccinated and vaccinated pregnant women.

P1074 Evaluation of seroconversion to measles vaccine in vaccinated infants in Ibadan, Nigeria
O.M. Akinloye, O. Oyedele, O. Akinloye, F. Adu (Shagamu, Ibadan, Osogbo, NG)
Objectives: Measles account for more mortality than any other vaccine preventable diseases and responsible for over 10% deaths of children aged below 5 years worldwide. The use of measles vaccine over the last 30 years has however reduced global measles morbidity and mortality. This study was designed to investigate the efficacy of vaccination in Nigeria and to determine if the presence or absence of maternal antibody at the time of vaccination had any influence on the level of IgG produced after vaccination.
Methods: Using ELISA kits, the level of maternally derived IgG was determined in 24 infants at age of nine months who reported at the vaccination centre and 6 cord blood samples from the maternity ward of University College Hospital, Ibadan as control group. The children were bled prior to vaccination and six weeks after vaccination.
Results: Five of the infants had protective levels of maternally derived IgG while 19 had lost it at the time of sample collection while the control group shows high IgG levels. Six weeks after vaccination all the infants tested had post vaccination protective levels of measles IgG (400–21966 mlU/mL). However, we observed a drop in the IgG level of children with high maternal antibody level after vaccination, although the post-vaccination IgG levels were still higher than others. One of the infants had a dramatic rise in post vaccination IgG (from 367 to 21966 mlU/mL) this prompted the evaluation of pre and post vaccination measles IgM. Two of the children had detectable measles IgM before vaccination and 4 children after vaccination.
Conclusion: It is concluded from this study that there is a high vaccine efficacy with measles vaccination in Nigerian infants. The presence of measles IgM in some children reporting for vaccination shows possibility of recent exposure before vaccination. Sustenance of measles vaccine coverage may therefore result to eradication of measles from Nigeria population.
P1075 Mumps outbreak in a highly vaccinated school population: a question of vaccine failure?
D. Park, J. Kim, M. Nam, H. Kim, J. Sohn, K. Song, M. Kim (Ansan, Seoul, KR)
Objective: Despite MMR vaccination, mumps epidemics have increased. The aim of this study is to describe and identify possible reasons for mumps outbreak in a highly vaccinated high school population.
Methods: Survey and cohort study of 41 students in a class of high school, Gyeonggi province, Korea were carried out from Mar. to Apr., 2006. Mumps was clinically diagnosed as an illness with 2 or more days of parotid or any other salivary gland swelling. Vaccination records were achieved by questionnaire. Serologic screening study for mumps IgG and IgM antibody (Ab) and mumps IgG avidity test was done with serum samples collected from 2–5 weeks after onset of symptoms. Virus culture and PCR detection for the mumps virus from throat swab specimen, and sequence analysis were performed.
Results: Clinical mumps developed in 15 students (attack rate, 36.6%). Among them, 11 patients had been vaccinated single dose, 3 had second dose, and 1 had not. Twenty six students had not developed clinical illness, of which 23 had been vaccinated single dose, 2 had second dose, and 1 is unknown. All of 41 students had mumps IgG Ab. Two of 15 patients had IgM Ab. Virus was isolated in 3 patients. Sequence analysis of virus isolated revealed 99% similarity with Dg1062/Korea/98 strain. Among 15 patients, 11 vaccinated patients have been considered as secondary vaccine failure with high IgG titer and high avidity index (AI, ±32%), an unvaccinated patient as primary infection with high IgG titer and low AI, and 3 vaccinated patients as other infections with low IgG titer and low AI. Among 26 healthy students, 5 vaccinated patients were retrospectively diagnosed as inapparent infection with high IgG titer and high AI, one student as past infection with high IgG titer and high AI, and the remaining students showed low IgG titer and low AI.
Conclusions: The overall attack rate of this study is higher than rates of previous reports for a population with vaccine coverage. Secondary vaccine failure did play an important role in this mumps outbreak. Additionally, booster immunisation for mumps should be considered in immunised adolescent persons to prevent further outbreaks.
P1076 Immunological evaluation of Vi-CPS-BSA conjugate of Salmonella typhi
H. Ahmadi, B. Tabaraiee, S. Siadat, F. Poor Mirza Gholi, D. Norouzian Shamasbi, M. Hedayati (Tehran, IR)
Introduction: The role of Vi-capsular polysaccharide (Vi-CPS) in human immunity against infection caused by Salmonella typhi is well known. Purified Vi-CPS can be regarded as a reliable immunogen to control typhoid fever in man. Moreover BSA can be selected as a T-dependent macro-carrier protein in order to conjugate with Vi-CPS.
Methods: Vi-CPS was extracted from standard strain of Salmonella typhi Ty6s (CSBPI-B-191) according previously described, by Ahmadi et al, 1999. The Vi-CPS of Salmonella typhi Ty6s was conjugated to Bovine Serum Albumin (BSA-Sigma) according to Mattox et al, 1989. The conjugate was prepared with 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide (EDAC). The conjugate, Vi-CPS and BSA were injected intramuscularly into three group of three rabbit with boosters on days 15, 30 after the primary immunisation. The serum collected on days 0, 15, 30, 45 and 180 were tested by Passive Hemaglutination test for evaluation of antibody production responses in rabbit model.
Results: In comparison purified Vi-CPS with Vi-CPS-BSA conjugate, the titer of hemagglutination passive of anti Vi-CPS-BSA conjugate was 512 which shows 30 times increase in titer when 3 I/M injection of 50 μg of conjugate were given to animals.
Discussion: This finding reveals that I/M injection of covalently bound purified Vi-CPS to a T-dependent proteins elicited much higher levels of antibodies than Vi-CPS alone besides, this conjugate it induce a booster responses in human. But, unfortunately the level of conjugation in this process is low due to les number of protein binding sites on Vi-molecules.
Though the Vi-CPS conjugates elicits higher levels of antibodies, immunoprophylactic activities of the conjugate must be as curtained by clinical evaluation and more research should be conducted to increase the degree of conjugation of the two macromolecules.
P1077 Immunogenicity of tetanus component in monovalent and combined vaccines
W. Janaszek-Seydlitz, B. Bucholc, U. Czajka, A. Wiatrzyk (Warsaw, PL)
Objectives: Vaccination with combined vaccines is without any doubt beneficial, allowing for decreasing number of injections and costs connected with vaccination. However, there is a possibility that the interaction between components of combined vaccines may influence their immunogenicity and reactogenicity. The immunogenicity of tetanus component in monovalent tetanus toxoid adsorbed and combined vaccines DTP and DTP+IPV was evaluated on Balb/c mice.
Methods: Animals: Six weeks old Balb/c mice were immunised with tetanus toxoid adsorbed; DTP or DTP+IPV. Animals (10 mice in each group) were injected subcutaneously three times with dose 0.25 ml at 6 weeks intervals. Control was unvaccinated.
Enzyme-linked immunosorbent assay (ELISA): The antibody responses to tetanus toxoid in sera 14 days after the immunisation were determined using in house ELISA method. The level of antibodies was expressed in arbitrary units.
Production of IFN-γ and IL-5: The levels of IFN-γ and IL-5 were evaluated in supernatants of cell culture of splenocytes stimulated by non-adsorbed tetanus toxoid or PHA.
Results: Geometric mean titre for anti-tetanus antibodies evaluated 14 days after the immunisation in animals vaccinated with tetanus toxoid adsorbed was lower (2650.2 a.u./mL) than in animals vaccinated with DTP (3968.6 a.u./mL) or DTP+IPV (4070.7 a.u./mL). There was no statistical differences between groups of animals receiving only DTP or simultaneously DTP and IPV. The mean concentration of IFN-γ in supernatants from specific stimulated cell cultures derived from animals vaccinated with tetanus toxoid was 720.45 pg/mL, from animals vaccinated with DTP – 2579.7 pg/mL and from animals vaccinated with DTP+IPV 1812.05 pg/mL.
The mean concentrations of IL-5 are about 20-times lower than concentration of IFN-γ in cell cultures of splenocytes derived from all evaluated groups of animals.
Conclusions: Whole-cell pertussis component influences as adjuvant on tetanus component in combined vaccines. Simultaneously administrated IPV vaccine not influences on immunogenicity of tetanus component. Pertussis component significantly increases level of specific tetanus antibodies and production of IFN-γ in cell cultures derived from vaccinated animals.
P1078 Vaccination of healthcare workers against hepatitis B virus in a teaching hospital
C. Ataman Hatipoglu, M.A. Yetkin, F. Ergin, K. Ipekkan, F.S. Erdinc, C. Bulut, N. Tulek, G. Demir, Y. Yigit, A.P. Demiroz (Ankara, Samsun, TR)
Objectives: Hepatitis B virus infection which can be prevented by active immunisation is one of the common health problems among the healthcare workers (HCW). The aim of this study was to vaccinate our HCWs in order to protect them against hepatitis B virus.
Methods: A total of 1009 HCWs who were working in Ankara Teaching and Research Hospital during January 2002-June 2006 were included into the study. HBsAg and antiHBs results, demographic data (gender, age, occupation and department) and immune status against hepatitis B virus were recorded for each HCW. HCWs that were negative for HBsAg and antiHBs were included into the vaccination programme.
Results: Among 1009 HCWs, 448 were male (44.4%), 561 female (55.6%) and the mean age was 32.02±7.74 years. Of the HCWs, 209 were working as doctors (20.7%), 246 as nurses (24.4%), 322 as domestic workers (31.9%) and 232 (23.0%) as other works (laboratory technician, radiology technician, officer, etc). Thirty-one percent of HCWs were working in the surgical departments whereas 20.6% of them in internal medicine departments. The rest of them were working in laboratories, emergency department, outpatient clinics and various other parts of the hospital with a rate of 8.4%, 7.4%, 7.3% and 25.5%, respectively. HBsAg was negative in all HCWs. AntiHBs was positive in 439 (43.5%) and negative in 570 (56.5%). A total of 570 HCWs were included in the vaccination programme. One hundred and fifteen (20.2%) were vaccinated with three-dose regimen in 0, 1 and 6 months, 349 (61.2%) with four-dose regimen in 0, 1, 2 and 12 months and 106 (18.6%) with single booster dose with yeast derived hepatitis B vaccine (Euvax-B, LG Chemical, S. Corea). Four hundred and sixty-nine of 1009 HCWs had a history of vaccination before, 170 (36.3%) of them had negative value of antiHBs at the time of the study. These 170 HCWs were vaccinated with a single dose hepatitis B vaccine. One hundred and six of them became positive for anti HBs after this vaccination. A complete vaccination schedule was applied to the remaining 64 HCWs who were negative after a single dose. Three hundred and sixty HCWs (63.1%) were compatible with the vaccination programme whereas 210 (36.9%) of them were incompatible.
Conclusion: We think that systematic surveillance and vaccination against hepatitis B will reduce the risk of transmission of this infection to the HCWs.
P1079 Comparing the immunogenecity of hepatitis B vaccine in two different methods of intramuscular and intradermal injection in infants of Bandar Abbas city, Iran
P. Davoodian, M. Jamshidi, Z. Shamandi, K. Kargar, K. Mahouri (Bandar Abbas, IR)
Objectives: The efficacy and safety of recombinant DNA hepatitis B vaccine is well known. However, few data are available on comparasion of different injection methods of interadermal and intra muscular.
This study aimed to make a comparasion between immunogenicity of these two methods of injection in infants.
Methods: In a clinical trial 47 cases entered the study who were infants randomly selected and received Intra-dermal(ID) immunisation against Hepatitis B at the ages of 0, 1.5 and 9 months old. The blood samples were obtained from our cases at the age of one year. The control group was consisted of 100 blood samples which were obtained from 1 year old infants who had given blood sample for other purposes. These infants were immunised according to the national vaccination routines applying intra-mascular(IM) injection method, 3 times (0, 1.5 and 9 months old), against HBV. After collecting the blood samples from case and control groups all samples were tested with ELISA method to determine anti-HBs Ab level.
Results: 84% of cases in IM and 87/3% in ID method had protective immunity (p = 0.791). In IM method 41.7% of cases had complete immunity whereas only 19.5% of those with ID method had complete immunity against HBV (p = 0.024). There was no relation between gender and Ab level in any of the injection methods.
Conclusion: We found out that both IM and ID methods have the same immunogenecity feature but IM method resulted in more complete immunity in infants.
P1080 Enzyme-linked Immunospot testing to assess the cell-mediated immune response to anthrax vaccine precipitated booster vaccination
M. Hepburn, J. Eyles, G. Paterson, E. Dyson, A. Simpson (Porton Down, UK)
Objectives: Vaccination remains one of the crucial countermeasure strategies against the intentional use of Bacillus anthracis. Assessment of the cell-mediated immune response to the licensed UK vaccine, Anthrax Vaccine Precipitated (AVP), particularly in persons receiving a booster vaccination, is needed for determining the optimal utilisation of this vaccine. We are conducting a prospective, observational study using an Enzyme-Linked Immunospot (ELISPOT) assay to assess the cell-mediated immune response before and after booster vaccination.
Methods: Laboratory workers scheduled to receive their yearly AVP booster vaccination had blood samples obtained before vaccination, then at 1, 2, 4 and 8 weeks after vaccination. Peripheral blood mononuclear cells were extracted from whole blood samples, and BD™ ELISPOT kits (Becton Dickinson) for IFN-γ, IL-4 and IL-5 were used to assess cellular responses to antigen-specific stimulation (5×105 cells per well). Antigens included protective antigen (PA), lethal factor (LF), and edema factor (EF), with concanavalin-A as a positive control, and tetanus toxoid as an additional control. A positive response was defined as a stimulation index greater than three times the number of positive cells in the medium only (negative control) wells.
Results: Prior to booster vaccination, the following number of subjects had positive responses with IFN-γ: 2/5 for PA stimulation, 3/4 for LF, and 2/5 for EF. Two subjects had more than a three-fold increase in their IFN-γ spot counts with PA stimulation by day 56 after vaccination (compared to pre-vaccination spot counts), while three other subjects had no change. These subjects had similar trends in IFN-γ spot counts with LF and EF stimulation. Four subjects had low baseline responses to IL-5 and IL-4 across all of the anthrax antigens, with some increase in IL-5 noted after vaccination but minimal changes observed in IL-4 counts.
Conclusions: In this ongoing study, cell-mediated responses to anthrax antigens were measurable in approximately 1 2 of subjects one year after their last anthrax vaccination. Some subjects were observed to have a substantial increase in their cell-mediated response to these antigens, while other subjects had no response. Further testing is needed to assess the factors that influence these immune responses.
P1081 Preparation of the herpes simplex virus type 2 glycoprotein D subunit and DNA vaccines and evaluation of their efficacy in guinea pigs
F. Fotouhi, H. Soleimanjahi, M. Roostaee, A. Dalimi (Tehran, IR)
Objective: Herpes simplex virus type 2 (HSV-2) is highly prevalent world-wide and major cause of genital herpes in humans. The life-long nature of infection, the increasing prevalence of genital herpes, and the disease severity in neonates and immunocompromised individuals imply that vaccination is the best strategy for controlling the spread of infection and limiting HSV disease. Herpes simplex virus glycoprotein D (gD) is one of the most important viral immunogen which has an essential role in virus infectivity and can induce humoral and cellular immune responses. In this study, full length HSV-gD2 gene was used to develop DNA and subunit vaccines. Their immunogenicity and protectivity was evaluated in a genital guinea pig model.
Method: In the first step, an Iranian isolate of HSV-2 was propagated in HeLa cell line and its DNA was extracted and used as template in polymerase chain reactions (PCR), to amplify gD2 gene. The PCR product was confirmed by restriction enzyme analysis, cloned into a cloning vector and then sequenced. The gene of interest was subcloned into an eukaryotic expression vector (pcDNA3) to construct DNA vaccine which was transfected into mammalian cells and protein expression was confirmed using indirect immunofloerscent test. In the next step, the gD2 gene was subcloned into a donor plasmid to construct recombinant baculovirus and express the desired protein in insect cells as sununit vaccine which was confirmed with SDS-PAGE and western blot analysis. In the last stage, the prepared vaccines were used alone or in combination to immunise guinea pigs.
Results: Immunisation with the above materials elicited potent humoral responses as measured by neutralisation test and ELISA, protected guinea pigs from induced HSV-2 genital disease and decreased viral replication in genital tract as much as that achieved with live HSV-2. The results also showed that DNA priming-protein boosting induced a neutralising antibody titer higher than that obtained with DNA-DNA vaccination.
Conclusion: Although immunisation with recombinant DNAs encoding antigenic proteins can induce cellular and humoral responses by providing antigen expression in vivo, but higher immune response has been shown to occur when the recombinant proteins followed DNA inoculation. The findings confirmed that vaccine formulation containing gD2 protein and pcDNA-gD2 is more effective than DNA alone.
P1082 Concomitant administration of Zostavax® and influenza vaccine in adults ≥50 years old
B. Kerzner, A. Murray, E. Cheng, R. Ifle, P. Harvey, M. Tomlinson, K. Rarrick, J. Barben, J. Stek, M. Chung, J. Xu, I. Chan, K. Schlienger, F. Schödel, J. Silber (Baltimore, Greensboro, Brooklyn, US; Waterland-Oost, NL; Surrey, Ledbury, UK; North Wales, US)
Background: Zostavax® is licensed for prevention of herpes zoster (HZ) and its complications. This study evaluated Zostavax® administered concomitantly with influenza (flu) vaccine in adults ≥50 yrs old.
Methods: Randomised, double-blind, placebo-controlled study. Evaluated safety and immunogenicity profiles of Zostavax® given concomitantly or sequentially with flu vaccine. Primary safety endpoints: incidence of vaccine-related serious adverse experiences (AEs) within 28 days postvaccination (PV); and vaccination report card prompted systemic and injection-site AEs. Primary immunogenicity endpoints: geometric mean titer (GMT) and geometric mean fold rise (GMFR) of varicella-zoster virus (VZV) antibody (Ab) at 4 wks PV by glycoprotein enzyme-linked immunosorbent assay (gpELISA); and GMT of flu Ab for the 3 vaccine strains at 4 wks PV by hemagglutination inhibition assay. Secondary immunogenicity endpoint: flu seroconversion rates (titer ≥1:40 4 wks PV in baseline seronegative subjects).
Results: No serious AEs related to Zostavax® were observed during the study. VZV Ab GMTs 4 wks PV for the concomitant (N = 382) and nonconcomitant (N = 380) groups were 554 and 597, respectively. Estimated VZV GMT ratio was 0.9 (95% CI, 0.8, 1.0), indicating non-inferior (p < 0.001 that decrease is <1.5 fold) responses. Estimated VZV GMFR from baseline in the concomitant group was 2.1 (95% CI: 2.0, 2.3), indicating acceptable range of fold-rise. Estimated GMT ratios (concomitant/nonconcomitant) for flu strains AH1N1, AH3N2, and B were 0.9 (95%CI: 0.8 to 1.1), 1.1 (95%CI: 0.9 to 1.3), and 0.9 (95%CI: 0.8 to 1.1), respectively. Seroconversion rates for each of the 3 flu strains were comparable across the 2 groups, with >85% of subjects achieving ≥ 1:40 PV Ab titers, meeting EMEA criteria.
Conclusions: ZOSTAVAX® and flu vaccine administered concomitantly are generally well tolerated in adults ≥50 yrs old. VZV and flu Ab responses were similar whether Zostavax® and flu vaccine were given concomitantly or sequentially.
P1083 Investigation of IFN-γ and IL4 cytokine responses in vaccinated mice with DNA vaccine containing E7 gene of human papillomavirus type 16
Z. Meshkat, H. Soleimanjahi, Z. Hassan, M. Mahmoudi, H. Mirshahabi, M. Meshkat (Tehran, Mashhad, IR)
Objectives: Papillomaviruses are small, non-enveloped viruses, their sizes is 55 nm in diameter, and widely induce cervical cancer. Cervical cancer is the second most common malignancy in women, and an estimated 470,000 new cases are diagnosed annually. Human Papillomaviruses (HPVs), particularly HPV 16, are associated with most cervical cancers. For immunotherapy of HPV16 associated disease, the E7 gene is considered a prime candidate, as it is expressed in all HPV16-positive tumours.
Methods: Previously, we designed and constructed an expression vector containing E7 gene of HPV16 as a DNA vaccine. In this study, we used the vaccine and injected it to BALB/c mice for evaluation of cytokine responses. IFN-γ and IL4 production were measured in the mice by ELISA.
Results and Conclusion: Our results showed the vaccine can induce production of IFN-γ but not IL4. Therefore, the mice immune system showed CD8 than CD4 responses. It is very important in cancer therapy because CD8 responses can eliminate tumour cells.
P1084 Long-term public health effects of vaccination against cervical cancer in Germany: results from a Markov model
T. Schwarz, A. Schneider, R. Rogoza, N. Ferko, T. Hammerschmidt, U. Siebert (Würzburg, Berlin, DE; Burlington, CA; Munich, DE; Hall i.T., AT)
Objective: To assess the public health impact of vaccinating 10 year old girls with an HPV-vaccine on precancerous cervical lesions (CIN = cervical intraepithelial neoplasia) and invasive cervical cancer (ICC) in Germany.
Methods: A Markov model which replicates natural history of HPV infection to ICC is calibrated for Germany in order to reflect the current German epidemiologic situation and screening/treatment patterns using German cancer registry data, literature data and official statistics. Vaccination added to routine cervical cancer screening is compared with screening alone. Screening is maintained constant (∼47% of the German women aged 20 years and older screened each year). A cohort of 399,400 ten year old girls is completely vaccinated. The vaccine covers HPV-genotypes 16 and 18. Based on clinical evidence, it is assumed to provide 95% protection against HPV-16 and HPV-18 as well as cross protection against related genotypes HPV-45 (90%) and HPV-31 (50%) (Harper et al. 2006). Sensitivity analysis is performed.
Results: The screening alone strategy results in 9,088 detected CIN1 lesions (4,239 (46.6%) due to oncogenic HPV), 3,181 treated CIN1 lesions (1,484 (46.7%) due to oncogenic HPV), 40,023 detected CIN2/3 lesions (29,014 (72.5%) due to oncogenic HPV), 39,253 treated CIN2/3 lesions (28,464 (72.5%) due to oncogenic HPV), 4,425 ICC cases and 1,139 ICC-related death. Adding vaccination to screening reduces the number of treated CIN1 by 853 (–26.8% of any cases/–57.5% of cases due to oncogenic HPV), treated CIN2/3 by 16.808 (–42.8% of any cases/–59.1% of cases due to oncogenic HPV), ICC cases by 3,462 (–78.2%) and ICC deaths by 885 (–77.7%). 27.8%, 14.2%, and 5.1% of the overall effectiveness against CIN 1, CIN2/3, and ICC, respectively, can be attributed to cross protection.
The model is most sensitive regarding variations in age at vaccination, vaccination coverage and screening interval, but less sensitive against variations on vaccine efficacy, cross protection, or a theoretically possible waning of immunity.
Conclusions: Despite opportunistic cervical cancer screening, a high disease burden from precancerous lesions and invasive cervical cancer including related mortality remains in Germany. Adding HPV vaccination can significantly reduce this burden. Cross protection plays a remarkable role in the primary prevention of cervical cancer and precancerous lesions.
P1085 Administration of DNA vaccine containing E6 gene of HPV16 in order to evaluate cellular immunity
H. Soleimanjahi, Z. Hassan, Z. Poorpak, Z. Meshkat, H. Mirshahabi, M. Meshkat (Tehran, Mashhad, IR)
Objective: Cervical cancer is the second most common cause of cancer deaths in women worldwide. Human papillomaviruses (HPVs) are thought to be the primary causative agent in >90% of cervical cancers, with HPV 16 being the type most frequently found in these tumours. HPVs are thought to induce cervical carcinoma most likely through the expression of E6 and E7 genes presumably by inactivating the tumour suppressor protein, p53 and p Rb, respectively. E6 is a multifunctional protein that affects cell growth and proliferation. The E6-mediated activities include cell immortalisation, transformation, tumour formation, and apoptosis. E6 and E7 oncoproteins are the most logical target molecules for therapeutic vaccines. In the present study E6 was chosen as target gene in order to evaluate CMI responses.
Methods: The HPV16 E6 ORF was obtained by PCR amplification using designed specific primers and cloned into pcDNA3 expression vector. The constructed was used for immunisation of mice after confirmation. Animal receiving to injection of target expression vector (pcDNA3-HPV16 E6) and pcDNA3 vector as a negative control were evaluated for specific T helper immune responses.
Results: Mice receiving pcDNA3-HPV16 E6 demonstrated high level production of IFN-γ versus negative control group where as IL4 production was not statistically significant.
Conclusion: Based on the results, the injection of pcDNA3-HPV16 E6 examination plasmid could efficiently induce IFN-γ, an indicator of Th1 responses, with has an important role in cancer therapy. The ability of the vaccine to elicit specific T cell responses and modulate a relevant cytokine secretion pattern is the key to DNA vaccine efficacy.
P1086 Serum bactericidal antibody titres against serogroup C meningococci in Turkish children: serological basis of the need for conjugated meningococcal vaccine
I. Yildirim, M. Ceyhan, P. Balmer, N. Andrews, R. Borrow, A. Aydogan, C. Ecevit, M. Turgut, N. Kurt, G. Uysal (Ankara, TR; Manchester, London, UK)
Like many other developing countries; there is no accurate information about the distribution of Neisseria meningitidis serogroups and the potential need for a conjugated meningococcal C vaccination programme in Turkey. To evaluate the need for this vaccine in our country we collected serum samples from four health centres located in different geographic regions and stratified according to age. Sera were tested for serum bactericidal antibodies (SBA) to serogroup C meningococci using rabbit serum as the complement source. It was observed that 98 (29.8%) and 40 (12.2%) of 329 individuals within the study population achieved SBA titer of >8 and >128, respectively. Overall; at least 70.2% of the population are susceptible (SBA titer <8) to meningoccal serogroup C disease. The rate of susceptibility was highest in infants aged 7–12 months and young children (1–4 years). Although the true incidence of meningococcal serogroup C disease is not well-known in Turkey, conjugated meningococcal C vaccine should be in mind as candidate vaccine for routine immunisation schedule given the high rate of susceptibility of the Turkish population.
P1087 Comparison among opsonic activity and serum bactericidal activity against Meningococci in rabbit sera from vaccines after immunisation with the Neisseria meningitidis serogroup A capsular polysaccharide-serogroup B outer membrane vesicle conjugate vaccine
S. Siadat, H. Najafi Barzegar, Q. Behzadiyannejad, B. Tabaraiee, H. Ahmadi, D. Nourouzian Shamasbi, S. Najar Peerayeh, M. Nejati, M. Zangeneh, B. Adibi Motlagh (Tehran, IR)
Introduction: Production of effective vaccine formulations is dependent on the availability of assays for the measurement of protective immune responses. Antibody- and complement-mediated phagocytosis is the main defence mechanism against Neisseria meningitidis.
Methods: Therefore, a newly developed phagocytosis assay based on flow cytometry (flow assay) and the serum bactericidal assay (SBA) were using sera obtained from rabbit postvaccination with a bivalent conjugate of Neisseria meningitidis serogroup A capsular polysaccharide (CPSA) to serogroup B outer membrane vesicle containing PorA (OMV-PorA), an OMV-PorA of Neisseria meningitidis serogroup B and the CPSA (as control), was done in order to evaluation of the potential efficacy of (experimental) meningococcal vaccines. The conjugate and control were injected intramuscularly into groups of five rabbit with boosters on days 14, 28 and 42 after the primary immunisation. The following groups were used as control: 1: CPSA; 2: OMV-PorA; 3: normal saline. The serum on days 0, 14, 28, 42 and 56 were collected and stored at −20°C for next analysis.
Phagocytic function of and intracellular oxidative burst generation by rabbit PMN, against Neisseria meningitidis serogroup A and B, were measured with flow cytometer (Coulter Epics-XL-Profile USA), using dihydrorhodamine-123 as probes, respectively. In these experiments non-heat-inactivated standard strain Neisseria meningitidis serogroup A (CSBPI, G-243) and B (CSBPI, G-245) were used. SBA titers are given as reciprocal Log2 values of the dilution giving at least 50% killing of the inoculum measured as colony forming units.
Results: The results of SBA titers and quantitative flow cytometric analysis of rabbit PMN function in hyperimmun sera with the glycoprotein conjugate revealed a highly significant increase in opsonophagocytic responses and bactericidal antibody against serogroup A meningococci after 56 day in comparison with the CPSA and OMV-PorA control group (P < 0.05). Opsonophagocytic responses and bactericidal activity against serogroup B meningococci of the conjugate showed no significant difference in comparison with the OMV-PorA containing control (P > 0.05).
Conclusion: Both SBA and opsonic activity are crucial for the protection against meningococcal disease, Our results indicated that the CPSA–OMV-PorA conjugate could be as a candidate for bivalent vaccine toward serogroup A and B meningococci.
P1088 Evaluation of serum bactericidal activity specific for Neisseria meningitidis serogroup A and B: effect of immunisation with N. meningitidis serogroup A polysaccharide and serogroup B outer membrane vesicle conjugate as a bivalent vaccine candidate
B. Adibi Motlagh, S.D. Siadat, Q. Behzadiyannejad, B. Tabaraiee, H. Ahmadi, S. Najar Peerayeh, D. Nourouzian Shamasbi, M. Nejati, M.H. Hedayati, A. Moshiri (Tehran, IR)
Objective: Bacterial meningitis caused by different groups of Neisseria meningitidis is still one of the Serious health problems world wide. The serum bactericidal assay (SBA) to meningococci is the most important test in immunological evaluation of meningococcal infection after vaccination. The SBA has been adapted as the gold standard for immunity against different serogroups of Neisseria meningitides after immunisation.
Methods: Neisseria meningitidis serogroup A capsular polysaccharide (CPSA) was conjugated to serogroup B OMV-PorA in order to test the possibility of obtaining a bivalent serogroup A and B meningococcus immunogen. The conjugate was prepared with adipic acid hydrazid (ADH) and 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide (EDAC). The conjugate and control were injected intramuscularlly into groups of five rabbit with boosters on days 14, 28 and 42 after the primary immunisation. The following groups were used as control: (1) CPSA plus OMV-PorA; (2) CPSA; (3) OMV-PorA; and (4) normal saline. The serum collected on days 0, 14, 28, 42 and 56 were tested by complement mediated bactericidal assay according to the World Health Organization protocol.
Results: The results of SBA in the glycoprotein conjugate group revealed a significant increase in serum bactericidal titer against serogroup A meningococci after 56 day in comparison with the CPSA and OMV-PorA control group. Bactericidal titer against serogroup B meningococci of the conjugate showed no significant difference in comparison with the OMV-PorA containing control.
Conclusion: The results indicate that when polysaccharide A and OMV are in a covalent conjugate form, the complex is able to induce a high level of bactericidal antibody response. Therefore, this paper shows that the CPSA–OMV-PorA conjugate could be a candidate for bivalent Vaccine toward serogroup A and B meningococci.
P1089 Comparative studies of conjugated capsular polysaccharide of Neisseria meningitidis serogroup A with outer membrane vesicle of N. meningitidis serogroup B
S.D. Siadat, D. Nourouzian Shamasbi, B. Tabaraiee, H. Ahmadi, Q. Behzadiyannejad, S. Najar Peerayeh, B. Adibi Motlagh, M. Nejati, M.H. Hedayati (Tehran, IR)
Objective: Many strategies are being explored to manipulate the immune system of humans/animals to fight better against a pathogenic organisms. The incidence of endemic meningitis and the frequency of epidemic meningitis caused by group A Neisseria meningitidis (GAM) are increasing in Africa and Asia. Although GAMP vaccine confers immunity at all ages, the improved immunogenicity of a conjugate and its compatibility with the World Health Organization's extended programme on immunisation offers advantages over GAMP alone. Henceforth, disease caused by serogroup B strains remains an unsolved health problem in many part of the world and the lack of a serogroup B meningococcal vaccine is a serious public health limitation since these strains account for approximately one-third of meningococcal disease in North America and up to 80% in North Europe.
Method: In this study, polysaccharide from Neisseria meningitides serogroup A was purified according to the World Health Organization protocol. Then outer membrane vesicle (OMV) of serogroup B meningococci was also extracted by deoxycholate reagent using ultracentrifuge. Conjugates of group A meningococcal polysaccharide (GAMP) bound to OMV, with adipic acid dihydrazide (ADH) as a linker and 1-ethyl-3-(3 dimethylaminopropyl) carbodiimide (EDAC) as a coupling agent, were synthesized and characterised. The obtained polysaccharide was activated by cyanogens bromide (CNBr) and 1-cyano-4(dimethylamino)pridium tetrafloroborate (CDAP) separately. First ADH with EDAC was bound to GAMP activated with CNBr to form GAMPCNBr AH. Second ADH with EDAC was bound to GAMP activated with CDAP to form GAMPCDAP AH. Then these derivatives were conjugated to OMV by EDAC to form GAMPCNBr AH-OMV and GAMPCDAPAH-OMV.
Result: Thus, GAMPCDAPAH-OMV with immunogenicities improved over that of GAMP have been prepared and standardised. The yields of GAMPCDAPAH-OMV was 45 to 48%. GAMPCDAPAH-OMV was higher than that of the other conjugate (15–17%). The glycoconjugates were shown to induce hyperimmunity in rabbits and formed antibodies against the above mentioned conjugates were detected by immune diffusion technique.
Conclusion: The average yield of conjugation should be improved through a useful activating reagent. CDPA seemed to be a more useful activating reagent, because the treated GAMP had a higher molecular weight and content of O-acetyl than other activator(CNBr). Therefore, the development of a A/B bivalent anti-meningococcal vaccine could be a good candidate.
P1090 Development of novel recombinant group B streptococcal vaccine based on surface expressed polypeptides
A. Suvorov, L. Meringova, K. Grabovskaya, A. Simbirtcev, G. Leontieva, E. Vorobieva, I. Koroleva, A. Totolian (St. Petersburg, RU)
Objectives: Group B streptococcus (GBS) being a common cause of newborn mortality raises numerous problems to the healthcare system due to extremely high level of carriage and inefficiency of conventional antibiotic therapy. Recently several attempts to generate GBS vaccine had been made in different laboratories however at present there is no GBS vaccine on the market. Employing the strategy of cloning of the epitope containing regions of the surface expressed polypeptides several constructs had been made and the resultunt products were tested for immunogenicity and protection.
Goals of investigation: Analysis GBS surface proteins as vaccine component against infection.
Materials and Methods: GBS gene fragments encoding for proteins Bac, ScaAB, GS and ScpB had been cloned from the strains 090R (1a) and 219 (1bc) and expressed in E. coli. Affinity purified polypeptides were used for immunisation of mice and rabbits. Mice antibodies were used for protection and antibody dynamics studies. For opsonophagocytosis assay serum from mice or rabbits had been used. Four different types of adjuvants (Feund, Alum, Bestim and IfnG) had been used for stimulating immune response. Evaluation of the results of immunisation had been done by ELISA with the following computer analysis.
Results: It was determined that protein antigens depending on their kind and molecular weight differ dramatically regarding the immune response and protective force. Interestingly, in spite of the correlation between the antibody titer and the level of protection antibodies to some of the peptides were protective with relatively small concentrations of specific IgG. We could determine protective antibody titers for the most of the peptides. Results of in vitro opsonophagocytosis assay well correlated with data obtained after mice protection experiments. Alum and Freund adjuvants were providing most effective immune response stimulation. Results of the analysis allowed selecting most promising peptide antigens for the future vaccine development. Study had been supported by RFFI grants 06–04–08026 and 06–04–48949.
P1091 Trends in pneumococcal bacteraemia in adults after the beginning of a pneumococcal vaccination programme
G. Peralta, B. Ceballos, M. Sanchez, C. Fariñas, I. De Benito, M. Roiz, L. Ansorena (Torrelavega, ES)
Objectives: In Cantabria (Spain) a polysaccharide pneumococcal vaccination programme has been initiated in the year 2000, directed to people older than 65 years and/or with specific comorbidities. We analyze changes in incidence and characteristics of pneumococcal bacteraemia (PB) in our hospital after the beginning of the pneumococcal vaccination programme.
Methods: Retrospective review of all charts of patients with PB identified by using the microbiology database, attended in our hospital from January 1997 to December 2005. We compared the periods 1997– 2000 (prevaccination) and 2001–2005 (postvaccination). In statistical analyses, Mann-Whitney U test was used for the comparison of mean values and Chi-square test for the comparison of categorical data.
| Prevaccine (N = 65) | Postvaccine (N = 53) | p | |
|---|---|---|---|
| Age (years±SD) | 65±17.8 | 63.4±20.2 | 0.9 |
| Diabetes n (%) | 6 (9.2) | 5 (9.4) | 0.61 |
| Immunosuppression n (%) | 5 (7.7) | 1 (1.9) | 0.16 |
| Cirrhosis n (%) | 9 (9.2) | 2 (3.8) | 0.21 |
| Renal Chronic failure n (%) | 1 (1.5) | 0 | 0.56 |
| Charlson index | 0.74±0.94 | 0.94±1.14 | 0.49 |
| Pneumonia n (%) | 54 (83) | 41 (77) | 0.3 |
| Meningitis n (%) | 4 (6.2) | 6 (11.3) | 0.25 |
| Exitus n (%) | 6 (9.2) | 7 (13.2) | 0.35 |
| Penicillin susceptible n (%) | 50 (76.9) | 50 (94.3) | 0.007 |
| Incidence ×106 year | 9.22±3.48 | 6.12±2.04 | 0.14 |
| BP× 103/hospital admissions | 2.13±0.17 | 1.14±0.39 | 0.03 |
| BP× 103/blood cultures | 10.54±3.56 | 4.96±1.6 | 0.02 |
Results: In the period of study we detected 118 PB. The annual incidence 8.2 cases per 100000 population. No significant differences were detected among prevaccine and postvaccine periods in the age, the presence of any comorbidity, the origin of the bacteraemia or the outcome of patients with PB. A decrease in the proportion of PB caused by penicillin non-susceptible pneumococci in the postvaccination period was detected. Although no significant difference was detected in the global PB incidence among prevaccine and postvaccine period, both the number of annual PB respect to the hospital admissions and the number of annual PB respect blood cultures performed diminished in the postvaccine period.
Conclusions: Our data suggest that PB has decreased after the beginning of a pneumococcal vaccination programme, with an associated decline in penicillin non-susceptibility. No change in other characteristics of patients with PB has detected after the beginning of the programme.
P1092 Effect of the introduction of the pneumococcal conjugate vaccine on invasive disease produced by Streptococcus pneumoniae
E. Cercenado, O. Cuevas, A. Fenoll, M. Marin, D. Vicioso, E. Bouza (Madrid, Majadahonda, ES)
Objectives: The heptavalent pneumococcal conjugate vaccine (PCV7) reduces carriage of pneumococci and prevents the disease. The PCV7 was licensed in Spain in 2001. We evaluate the effect of PCV7 on invasive disease and the impact of non-PCV7 serotypes in the area cared for our institution.
Methods: We identified episodes of bacteraemia and meningitis due to pneumococci in patients admitted to our institution from 1999 to 2005. Serotype distribution and antibiotic resistance were compared.
Results: A total of 726 episodes of bacteraemia (9% children) and 46 cases of meningitis (22% children) were identified. Over the period of study, the most frequent serotypes were 14 (10.2%); 3 (9%); 19 (8.9%); 8 (8.5%); 1 (7.2%); 4 (6.6%); and 23F (3.5%). By comparing data from 1999–2001, with data from 2002–2005, the percentage of isolates non-susceptible to penicillin showed a slight decrease (from 31.47% to 30.2%) and isolates resistant to erythromycin increased (from 17% to 24.3%). The incidence of pneumococcal bacteraemia was unchanged (from 1.90 cases to 1.92 cases/1,000 admissions). Vaccine serotypes caused 38% of episodes of bacteraemia and meningitis in the first period, compared with 27.4% of episodes in the second period (p = 0.037). Infections due to vaccine serotypes 4, 9V, and 23F decreased significantly during the second period (p < 0.002) but not these caused by serotypes 6B, 14, 18C, and 19F. The percentage of episodes of invasive disease caused by vaccine-related serotypes (same serogroup but not the same serotype as PCV7) showed a tendency towards the increase from 10% of episodes in the first period to 14%of episodes in the second (p = 0.135). Episodes due to VRST 19A increased (p< 0.001). Infections caused by non-PCV7 serotypes 1 and 8 increased from 11% to 19% (p < 0.05). In 2005, all invasive infections in children were produced by non-vaccine serotypes, and the estimated coverage of the PCV7 in adults was 23.6%.
Conclusions: The overall incidence of invasive pneumococcal disease and the rate of antibiotic-resistant invasive infections has not decreased in our area after the introduction of the PCV7. There were significant decreases in the incidence of invasive infections caused by several vaccine serotypes, however, invasive infections due to vaccine-related serotype 19A, and to non-vaccine serotypes increased.
P1093 Immune response to serotype 19A induced by serotype 19F in pneumococcal protein conjugate vaccine
H.J. Lee, S.Y. Lim, K.-H. Kim (Seoul, KR)
Objectives: Pneumococcal 19F and 19A are important serotypes in invasive infection worldwide as well as in Korea. Pneumococcal vaccine containing 19F seemed to be protective against infection by 19A serotype. To elucidate immunogenicity to 19A serotyppe by 19F polysaccharide in pneumococcal conjugate vaccine, we studied antibody titer and opsonisation titer to 19F and 19A.
Methods: Twenty healthy adults were immunised with 7-valent pneumococcal conjugate vaccine which contained 19F polysaccharide antigen. Antibody titers and opsonisation titers to 19F and 19A serotypes were studied in preimmune and postimmue sera.
Results: Geometric mean titer of antibody to 19F serotype was increased significantly from 3.66 (95%CI 2.57, 5.13) μg/mL to 12.60 (95% CI 6.76, 23.44) μg/mL after immunisation. Geometric mean antibody titer to 19A serotype also increased from 4.43 (95% CI 3.02, 6.46) μg/mL to 8.43 (95%CI 4.90, 14.45) μg/m (P > 0.05). Antibody titer to 19F and 19A were not significantly different in preimmune sera. In postimmune sera, antibody titers to 19F were significantly different with those to 19A. Geometric mean opsonisation titer to 19F and 19A were increase from 30.74 (95% CI 13.18, 70.79) and 72.32 (95%CI 24.84, 181.97) to 1142.00 (95% CI 630.96, 2089.30) (P<0.05) and 948.41 (95% CI 512.86, 1737.80), respectively. Opsonisation titers were not significantly different between 19F and 19A after immunisation.
Conclusion: 19F polysaccharide in pneumococcal conjugate vaccine elicited antibody responses to 19A serotype, quantitatively and qualitatively in healthy adults. Further studies are needed in children and elderly people.
P1094 Prophylactic potential of high molecular weight antigenic fractions of a recent clinical isolate of L. donovani against experimental visceral leishmaniasis
P. Tripathi, S. Naik (New Delhi, Lucknow, IN)
Objectives: Identification of potential T cell stimulatory antigens of a recent clinical isolate of L. donovani against Visceral Leishmaniasis (VL) for prophylactic efficacy in hamster model of VL.
Methods: We have recently reported the presence of T cell immunostimulatory antigens in the high molecular weight (MW) fractions (F1-F4; MW range 134–64.2 Kd) of whole LD antigen [strain 2001; (F1-F11; MW range 139–24 Kd)], which stimulated variable amounts of IFN-γ, IL-12 and IL-10 in exposed immune individuals. The present study was undertaken to evaluate high molecular weight antigenic fractions of LD for potential protective efficacy. The high molecular weight region of the parasite was resolved into five antigenic fractions (Prep A-E; MW range >100–60 Kd) using continuous elution gel electrophoresis. Prior to in vivo protection studies in hamsters, these fractions were used to evaluate in vitro cellular responses (defined by their ability to induce in vitro T-cell proliferation and induction of cytokines: IFN-γ, IL-12 and IL-10) in eight leishmania exposed individuals. The protective efficacy of prep A+B, C, D and E in combination with BCG was evaluated in inbred hamsters using standard immunisation protocol.
Results: Proliferative responses were seen in all eight of 8 exposed individuals to prep D and E, 5 of 8 individuals to prep B and C and 3 of 8 to prep A. The median proliferative responses of eight exposed individuals to prep D (median SI: 5.2 [range 3.9–7.1]) and E (median SI: 5.6 [range 4.4–8.2]) were significantly higher than to fraction A (median SI: 0.2 [range 0.1–7.2]; P < 0.05) but not to prep B and C. However, prep A-E induced equivalent levels of IFN-γ, IL-10 and IL-12 cytokines. Fractions D and E also exhibited marked parasite inhibition in spleen (52.5% and 73.7%) and liver (65% and 80.2%) as compared to prep A+B (23% in spleen and 24% in liver) and prep C (38% in spleen and 24% in liver). Prep D and E vaccinated animals showed higher in vitro stimulatory responses (Mean SI: 6.6 and 8.8) and nitric oxide (NO) induction (mean NO levels: 6.4 and 10.7 mg/mL) against whole cell extract as compared to other groups. The protection also correlated with presence of suppressed leishmania specific IgG levels in prep D and E immunised hamsters.
Conclusions: These studies indicate the presence of immunostimulatory and protective molecules in 60–80 Kd region of LD, which may be further exploited for developing a subunit vaccine.
P1095 Dose sparing with intradermal injection of influenza vaccine
M. Metanat, M. Salehi, B. Sharifi-Mood, M. Safai (Tehran, IR)
Objective: The most effective measure available for the control of influenza is the annual administration of inactivated influenza vaccine. Inactivated influenza vaccine has been shown to be effective in the prevention of influenza A and relatively high efficacy(70% to 90%), is observed in healthy young people, who were vaccinated intramuscularly with dose of 15 μg of antigen. Intradermal admiminstration of antigens is expected to facilitate their exposure to antigens – presenting cells, such as maorophages and dendritc cells, which are present at higher level in skin than in muscle. Therefore, intradermal vaccination may induce similar serum antibody responses with a smaller quantity of antigen. Dose sparing with intradermal administration could be used to expand the supplies of influenza vaccine.
In order to determine the antibody response of an intradermal injection of trivalent inactivated influenza vaccine, with 40 percent of the usual dose and then to compare with the immunogenicity and safety of usual influenza vaccine, we conducted this study.
Materials and Methods: In this randomised control trial, we evaluated the antibody response of intradermal vaccine among 97 cases who were selected randomly from healthcare workers and then results compared with results in 94 cases of healthcare workers who were received intramuscular influenza vaccine with usual dose. One hundred ninety one cases with age range of 22–50 years, have been vaccinated by one experienced person then blood samples were evaluated for titers of hemagglutination – inhibition (HAI) antibody before and after injection of vaccine. Also, local and systemic adverse events assessed.
Results: Our study showed that there was no significant difference in seroconversion and seroprotection rates after vaccination, between two groups (p > 0.05). Local reactions (induration and redness) were significantly more common, among recipients of intradermal injections than among recipients of intramuscular injection, but such reactions were mild and transient.
Conclusions: We conclude that reduced dose of influenza vaccine given by the intradermal route in healthy adults was safe and immunogenic, similarly to intramuscular injection with usual dose and dose sparing with intradermal injection could be to increase the number of available doses of vaccine.
Patterns and determinants of antibiotic usage
P1096 Outpatient antibiotic utilisation in the city of Zagreb in 2005
D. Štimac, J. Culig, I. Vukušic, Z. Šostar, M. Bucalic (Zagreb, HR)
Objective: According to financial indicators, antibiotics ranked fourth, accounting for a high share of 9.44% in the overall drug utilisation in the City of Zagreb. The aim was to identify the causes of such a high utilisation and distribution within the group of antibiotics (J01).
Methods: Data on the number of packages and wholesale price were obtained from all pharmacies in the City of Zagreb for each individual drug registered in Croatia. Based on these data, the number of defined daily doses (DDD) and DDD per 1000 Zagreb inhabitants per day (DDD/1000/day) in 2005 were calculated for each individual drug at all levels of the Anatomic-Therapeutic-Chemical (ATC) drug classification system.
Results: In the City of Zagreb, total utilisation of antibiotics in 2005 was 34.62 DDD/1000/day, yielding a 16% increase from the preceding year. B-Lactam antimicrobials, penicillins (J01C) accounted for more than a half of total antibiotic utilisation with 17.99 DDD/1000/day Within J01C group, J01CR subgroup of penicillin combinations including β-lactamase inhibitors accounted for 53%. The entire consumption of the latter referred to a combination of amoxicillin and clavulanic acid, the most frequently prescribed antibiotic in the City of Zagreb with 9.61 DDD/1000/day. The subgroup of broad-spectrum penicillins (J01CA), led by amoxicillin with 4.33 DDD/1000/day, accounted for 25% of penicillin utilisation, followed by J01CE with 21%, J01D with 13.85% (predominated by second generation cephalosporins), J01F with 12.34% (predominated by azithromycin with 2.27 DDD/1000/day), J01A (systemic anti-infective agents) with 7.82% (predominated by doxycycline with 2.71 DDD/1000/day, J01M (quinolones) with 6.23% (predominated by norfloxacin with 1.6 DDD/1000/day), J01E with 4.76%, J01X with 2.61%, and J01G (aminoglycosides) with the lowest rate of 0.42%.
Conclusion: Broad-spectrum antibiotics predominated in the utilisation of antibiotics. Amoxicillin with clavulanic acid accounted for almost one third (28.26%) of antibiotic utilisation in the City of Zagreb, in spite of the guidelines preferring the use of narrow-spectrum antibiotics. The rather frequent empiric prescribing of broad-spectrum antibiotics entails the pending risk of the microorganism resistance associated with an array of unfavourable consequences for both the patients and healthcare system in general, ultimately resulting in treatment cost increase and therapeutic inefficiency.
P1097 Trends in antibiotic consumption over a 7-year period in a general hospital, Athens, Greece
C. Loupa, I. Tzannou, V. Tsolaki, K.J. Karnezi, A. Dimizas, M. Lelekis (Athens, GR)
Objective: The aim of the present study was to record the trends in antibiotic consumption in our 300-bed general hospital over a 7 year period.
Methods: Antibiotic consumption for the period 1999 to 2005 was studied retrospectively using data from the pharmacy computer. Antibiotic use was calculated in DDDs per 1000 patient days (ABC Calc 3.0). We used linear regression for statistical analysis (SPSS 11.5).
Results: Antibiotic consumption was 572, 647, 678, 675, 710, 785 and 892 DDDs/1000 patient days (years 1999, 2000, 2001, 2002, 2003, 2004, 2005 respectively) and the trend was significant (p = 0.001). Concerning consumption of major classes of antibiotics, only penicillins had a decrease (from 83.4 to 67.9). The consumption of other classes of antibiotics had the following trend over time: cephalosporins/aztreonam from 152.6 to 253.8 (p = NS), penicillins+inhibitors from 133.9 to 209.1 (p = 0.034), carbapenems from 3.8 to 15.2 (p = 0.029), aminoglycosides from 45.4 to 46.1 (p = NS), macrolides from 35.9 to 81.8 (p = NS), quinolones from 28.6 to 109.7 (p = 0.001) and glycopeptides from 1.3 to 18 (p = 0.014). It is worth noting that consumption of carbapenems, quinolones and glycopeptides had the above mentioned significant increase despite the fact that these antibiotics are included in the restricted list antibiotics since late ‘80s, together with 3rd-4th generation cephalosporins/monobactams. The heavily marketed, newly introduced in Greek market ertapenem and “respiratory quinolones” are at least partly responsible for the significant increase in carbapenem and quinolone consumption respectively. Furthermore, due to high prevalence of MRSA and MRSE strains in our hospital, glycopeptides are currently widely used for treatment of staphylococcal infections as well as for chemoprophylaxis in serious operations such as hip replacement.
Conclusions: During the study period, consumption increased significantly for almost all major classes of antibiotics, including ones under restriction. It is obvious that besides the existence of a restriction policy, additional measures are needed in order to control the increasing antibiotic consumption. The impact of this increase on resistance is currently under investigation.
P1098 Ambulatory care self-medication with antibiotics within countries in the south-eastern Mediterranean region: results from the ARMed project
E.A. Scicluna, M.A. Borg and ARMed Project Collaborators
Objective: Until recently, data on the degree of self medication with antibiotics in the southern and eastern Mediterranean was unavailable, contrary to the situation in the northern part of this region, where data has already been published. The ARMed project aimed to fill in this void and elucidate the situation in the countries under study.
Methods: Short structured interviews were held in out-patient clinics or primary health centres in Cyprus, Egypt, Jordan, Lebanon, Libya, Tunisia and Turkey between November 2004 and July 2005, using the country's own language. One person accompanying a patient was approached and, after approval, asked to correctly define an antibiotic. If the reply was in the affirmative, the full interview were then conducted,
Results: A total of 2109 subjects were approached and 1705 completed the full interview. Actual self-medication was reported by 326 respondents, which ranged from 0% in Cyprus to 37% (95% CI: 28.3% – 45.6%) in Lebanon. In the total group of antibiotic users, there was no association between self-medication and either gender or age, and a borderline association (p = 0.049) with education level. At the country specific level, self-medication was associated with age in Jordan (p = 0.016) and in Lebanon (p<0.001) only. Self-medication increased with age in both countries. There was no association with gender or education level in any of the participating countries.
Intended self-medication ranged from 1.3% (95% CI: 0% – 3%) in Cyprus to 70.7% (95% CI: 64% – 77%) in Jordan and for children ranged from 0% in Cyprus and 62% (95% CI: 55%, 69%) in Jordan. Upper respiratory tract infections were the most frequent reason for intended self-medication. Almost half of interviewees (48.4%) reported that they had antibiotics at home, reaching 60% (95% CI: 51% – 69%) in Lebanon. We found a statistically significant association between antibiotic hoarders and intended users of antibiotics for self-medication in all the participating countries, except Cyprus.
Conclusion: Ambulatory care consumption of antibiotics without prescription is significant in many southern and eastern Mediterranean countries and appears higher than that previously reported by other European counterparts in the region. Actual self-medication with antibiotics is strongly predicted by intended self-medication, which exists at much higher levels, implying that the public health risk is higher than indicated by actual self-medication rates.
P1099 Using directed graphs for examining transitions in antibiotic usage
S. Natsch, F. Baart, P. van der Linden, H. de Neeling on behalf of the SWAB's working group on Surveillance of Antimicrobial Use
Objective: to develop a method to analyse consecutive prescriptions for antibiotics. To describe how animated directed graphs can be used to gain insight into the subsequent use of antibiotics.
Method: The database of the Dutch Foundation of Pharmaceutical Statistics was used for this study, consisting of 10 million prescriptions from 2 million patients from 2000 to 2004. The prescriptions were sorted by patient and prescription date. These series were transformed into four kinds of transitions. First the transition from no antibiotic to an antibiotic, second from an antibiotic to no antibiotic, third from one antibiotic to the same antibiotic and fourth from an antibiotic to another antibiotic.
Results: A graph was created for each month in the 5 year period. The resulting 60 graphs were rendered into an animation. It can be viewed at www.swab.nl/wool/faster_transitions.svg. This animation shows that if a patient started using an antibiotic, it was most often amoxicillin. Doxycycline, amoxicillin and amoxicillin/clavulanic acid were often prescribed two or more times in a row. The most frequent transition was from amoxicillin to amoxicillin/clavulanic acid. Among the antibiotics used primarily for urinary tract infections, nitrofurantoin is most often used as first choice. Norfloxacin was the most prominent secondary drug subsequent to nitrofurantoin.
Conclusions: The method will allow more in-depth analysis of patterns of usage, choices for first and second-line treatments and adherence to guidelines and policies. A big advantage is the possibility for detection of changes over time as well as seasonal influences. In the future, linking these data to surveillance of antimicrobial resistance data will provide further insight into the relationship between antibiotic usage and development of resistance.
P1100 ESAC II hospital care subproject 2005–2007: improving quality indicators of hospital antibiotic prescribing within standardised data, longitudinal study
F. Ansari, H. Goossens, P. Davey on behalf of the ESAC IIHC Subproject Working Group
Objectives: The ESAC I project demonstrated the lack of unified hospital information on antibiotic use and standardised denominator of clinical activity across Europe. In 2004 DG SANCO supported the ESAC II Hospital Care subproject to improve the knowledge of antibiotic consumption, to consolidate the continuous collection of comprehensive data, to develop and validate health indicators based on antibiotic consumption data, and to use a set of core indicators to provide feedback to the participating countries. The ESAC II HC subproject consists of Longitudinal Survey and Point Prevalence Survey. By the end of the project we wish to produce standardised data from at least one hospital per country and to develop a strategy for regional and national roll outs. The main objective of longitudinal survey is to establish a platform for statistical analysis of time trends within and between the hospitals and countries and to compare results using bed days or admissions as denominators.
Methods: Monthly data were collected from each hospital for a 6-year period from 2000 for antibacterials dispensed to inpatient destinations in DDD as numerator, and bed days and admissions as denominators. Databases were assigned for ATC/DDD and analysed centrally in Dundee. Statistical analysis was conducted using regression modelling of time series.
Results: We have analysed longitudinal data from 14 hospitals so far. In 11 hospitals the direction of time trends was the same using bed-days or admissions (increasing in 8 which were significant for 3, decreasing in 3). In the 3 remaining hospitals the trends were different; DDD/100 bed days showed increase over time whereas DDD/100 admissions showed a decrease. In 3 hospitals there was significant increase over time in DDD/100 bed days whereas there was a non-significant increase in DDD/100 admissions. The proportion of parenteral antibacterials used in 15 hospitals in 2005 was 8–78% and the rank order of parenteral use was very different from total use.
Conclusion: The results show similar significant trends with the 2 denominators in 6 of the 14 hospitals. Using additional denominators of clinical activity especially in longitudinal studies could provide set of indicators of prescribing quality and resource use. Including prescription at discharge in some hospitals might explain differences in use of parenteral antibacterials from total use.
Parenteral use might be a more meaningful indicator of inpatient treatment than total use.
P1101 A comparative study on the antibiotic consumption of two Hungarian county hospitals
I. Almási, G. Ternák, F. Rozgonyi (Szekszárd, Pécs, Budapest, HU)
Objectives: The aim of this study was to analyse the changes in the antibiotic consumptions and indications of antibiotic prescription in two similar size hospitals in order to see the quality of antibiotic usage.
Methods: Case histories and the conditions of antibiotic treatment of patients such as considerations of indication, choice and name of antibiotics, treating dose and durations, microbiological examinations, etc were put in a computer database. Date of patients discharged from the hospitals in January 1995 and 2005, respectively, were analysed and compared in both hospitals. An antibiotic policy and infection control was implemented in both hospitals in 1995.
Results: Data of 2230 and 3031 patients from Hospital 1 (H1) and data of 1240 and 1809 patients from Hospital 2 (H2) in the year 1995 and 2005 were compared. Here we focus on the features of the antibiotic consumption. In the two periods, systemic antibiotic treatment was given to 24.7% and 18.4% of the patients in H1, while to 28.7% and 27.5% of the patients in H2. Surgical proplylaxis became the most frequent indication for antibiotic treatment: from 11.7% to 34.7% in H1, and from 6.7% to 35.6% in H2. Lower respiratory and urinary tract infections, respectively, were the next frequent indications. Rates of fever or unidentified indications for antibiotic treatment decreased remarkably. The usage of β-lactams increased in both H1 and H2: from 55.2% to 68.2% and from 51.0% to 58.2%. The usage of an aminopenicillin+β-lactamase inhibitor increased significantly: from 3.5% to 30.1% in H1 and from 5.5% to 36.5% in H2. The rates of both the prescription of multiple antibiotics under the same hospital stay and the combined antibiotic therapy decreased in both hospitals: from 33.8 to 24.0% in H1 and from 36.0% to 22.9% in H2 (concerning No. of patients), as well as from 16.0% to 11.2% in H1 and from 13.9% to 12.6% in H2 (concerning antibiotic courses).
Conclusion: The antibiotic policy and infection control guidelines were followed by the clinicians in both hospitals. These resulted in a better structure of the antibiotic consumption. These were mainly due to the more appropiate surgical antibiotic prophylaxis during the ten years.
P1102 Population approach to antibiotic use for children in Lithuania
A. Berzanskyte, R. Valinteliene (Vilnius, LT)
Survey in 18 European countries and Israel revealed Lithuania as the country with the highest rate of self-medication (22% of all respondents). It prevailed in almost the half of questioned residents, who used antibiotics during the last 12 months. However this figure reflected adults approach only towards their selves.
Objective: The aim was to find out if the attitudes of population about antibiotic use for children are different.
Methods: The study was performed as part of European survey – Self-Medication with Antibiotics and Resistance Levels in Europe (SAR project). 3000 questionnaires were sent by mail to one urban and one rural area. Response rate after reminders reached 25.4%.
Results: 3.9% of complete sample self-medicated their kids, what was much more seldom than in adults (22%). In general there were 181 of the responders, who mentioned about children up to 16 years old living together. During the last 12 months 59.6% of them pointed about used antibiotics for children. 86.9% of the latter used prescriptions, but nearly one third (27.1%) did not. Rural people tended to give antibiotics without consulting the doctor more often than urban people – relatively 31.3% and 22.5%, concerning gender – men more often than women – 36.5% and 14%. However only 1 person mentioned about intended self-medication for the children. The most frequent diseases for which children were self-medicated corresponded to the ones among adults – tonsillitis (28.2%), upper respiratory infections (23.6%), bronchitis (16.4%). 86.2% of self-medicators indicated pharmacy as the most often source to get antibiotics without prescription, 17.2% from leftovers, 3.4% from family members and friends.
Conclusion: All the determinants and tendencies in self-medication with antibiotics for children are mostly the same as in adults. However the general numbers, particularly intended self-medication, are lower. Although Lithuanian people consider antibiotics as “universal” drug, they are still cautious concerning children treatment with them. It proves need of education in this area.
P1103 Variability in the number and pattern of the most frequently used antibiotics to treat outpatients in 28 European countries
A. Muller, M. Ferech, S. Coenen, E. Hendrickx, D.L. Monnet, H. Goossens on behalf of the ESAC project
Objectives: Studies have shown country differences in outpatient use and class distribution of antibiotics. However, most of the use is constituted by a limited number of antibiotics, which may differ among countries. The objective of this study was to identify similarities or differences in the patterns of the most used antibiotics among European countries.
Methods: Data on outpatient antibiotic use were collected from 28 European countries in the ESAC network. For each country and each of the 210 antibiotics (ATC J01, WHO, 2005), we calculated the no. of Defined Daily Doses (DDD). Antibiotics representing 90% of total no. DDD of a country (DU90%) were labeled “routinely used”. A hierarchical cluster analysis was performed to study the similarity of the antibiotics that were routinely used in these countries.
Results: In 2004, the total no. of different antibiotics used in outpatients in Europe was an average of 57 (min: 33 in Iceland, max: 94 in Italy and Germany). The no. routinely used antibiotics was an average of 14 (min: 9 in Slovenia and Israel, max: 21 in France). Two antibiotics, i.e. amoxicillin (J01CA04) and doxycycline (J01AA02), were routinely used in all countries. Five other antibiotics, i.e. amoxicillin and enzyme inhibitor (J01CR02), ciprofloxacin (J01MA02), clarithromycin (J01FA09), azithromycin (J01FA10) and sulfamethoxazole+trimethoprim (J01EE01) were routinely used in more than 2/3 of the countries and 25 in only one country. The three mostly used antibiotics represented an average of 52% of the total no. DDD (min: 35% in Finland, max: 64% in Israel). Clustering of countries according to routinely used antibiotics is presented in the Figure .

Conclusion: There were remarkable differences in the no. and pattern of antibiotics routinely used to treat outpatients in Europe. This inter-country variability may reflect cultural differences as well as national drug policies.
P1104 Antimicrobial consumption in Southern and Eastern Mediterranean hospitals: final results from the ARMed Project
P. Zarb, M.A. Borg, M. Ferech, H. Goossens (Gwardamangia, MT; Antwerp, BE)
Objectives: Data on antimicrobial consumption in southern and eastern Mediterranean countries has been sorely lacking. The Antibiotic Resistance Surveillance and Control in the Mediterranean Region (ARMed) project addressed this by studying antibiotic use in hospitals in the region.
Methods: Data was collected prospectively for 2 years: 2004–2005, using Anatomical Therapeutic Chemical (ATC) classification from 24 hospitals: Cyprus (5); Egypt (8); Jordan (1); Malta (1); Tunisia (3); Turkey (6). Defined Daily Doses was used as numerator, and 100-occupied-bed-days as denominator (DDD/100BD).
Results: Overall consumption exhibited a median of 84.3 DDD/100BD (IQR 43.0–310) with a range of 22.8 to 1900 (a bone marrow transplant centre) DDD/100BD. The ARMed median correlates well with the Antimicrobial Resistance Prevention and Control (ARPAC) median of 82 DDD/100BD for Southern European Hospitals.
Penicillins were the most commonly consumed antibiotics, (range 2.45–1465.3, median 28.8, IQR 12.6–120.9, DDD/100beddays): broad-spectrum penicillins (amoxicillin and ampicillin) were predominant in Cyprus, Egypt, Jordan and Tunisia as opposed to combinations with β-lactamase inhibitors in Malta and Turkey. Considerable variability was evident for cephalosporins (range 2.67–167.7, median 16.21, IQR 10.34–38.21, DDD/100BD). Hospitals in Cyprus utilised mainly 1st generation, Jordan and Malta 2nd generation and 3rd generation in Egypt, Tunisia and Turkey.
Remarkable variability in patterns of use was evident between hospitals in the same country, especially in Egypt, as well as between countries. The specialised hospitals in ARMed contributed to a predominance of high consumption outliers (>120DDD/100BD), (10/23), as opposed to ARPAC Southern European hospitals (2/28).
In 2005, antibiotic use increased in 14 hospitals (inter-year change from −41% to +127%). Contrarily, the proportion of parenteral drugs decreased in all countries (range: −10% to −32%).
Conclusion: The variations observed are likely to be the consequence of multifactorial causes: the range of antibiotics licensed; national/hospital formulary; type/size of hospital, as well as region specific drug supply mechanisms (donations, purchase outside hospital). However, in comparison with available European data, the region seems to utilise a higher quantity of broad-spectrum antimicrobials. This could be a risk factor for the high prevalence of resistance already documented by ARMed.
P1105 Shortage of antimicrobial agents in Europe: results of an international survey
S. Harbarth, P.M. Filius, S. Natsch, F. Mackenzie, I. Gyssens on behalf of the ESCMID Study Group on Antibiotic Policies (ESGAP)
Objectives: There is little information available regarding the shortage of antimicrobial agents in Europe. This survey attempted to determine to what extent shortages have already influenced prescribing in European hospitals.
Methods: The study was conducted as an international, 9-item survey sent out via email in October 2006 to 360 hospitals throughout Europe. The subjects were infectious disease physicians, clinical microbiologists or pharmacists. Data were entered in a spreadsheet, checked for accuracy, and transported to SPSS for descriptive analysis.
Results: Participants from 23 countries and 55 hospitals responded. 38/55 (69%) of the respondents had experienced a shortage of antimicrobial agents within the last 12 months. Shortages were not evenly distributed among countries. Only 4 countries did not report any shortage. The most frequently affected agents were cefepime (n = 15); mupirocin ointment (n = 15); meropenem (n = 13); piperacillin/tazobactam (n = 11) and piperacillin alone (n = 4); semi-synthetic penicillins (n = 7); aztreonam (n = 6); and 1st- or 2nd-generation cephalosporins (n = 5). Shortages of penicillin, ampicillin, colistin, doxycycline, rifampicin, vancomycin, nitrofurantoin, fosfomycin, amphotericin B, amikacin and tobramycin occurred less frequently. There was great variability in the duration of the shortages (range: 1 week – 18 months). Substitutes were either more expensive (e.g. carbapenems instead of cefepime), more broad-spectrum (e.g. cefuroxim, ceftriaxone or co-amoxiclav instead of oxacillin and penicillin), or less efficacious (e.g. neomycin and chlorhexidine instead of mupirocin ointment). 19 hospitals (35%) had trouble finding equivalent drugs for substitution. Only 10 hospitals reported that they had always been adequately informed by the producer or distributor about the shortage.
Conclusions: A substantial number of hospital pharmacies in Europe have experienced antimicrobial drug shortages within the last 12 months. Shortage of meropenem, mupirocin and cefepime were considered to have the largest impact on patient care. Shortage-triggered policy changes may negatively affect antimicrobial prescribing patterns and expenses. This survey provides important information that will guide further ESGAP initiatives to document and ultimately prevent shortages.
P1106 Hospital consumption of antibacterials for systemic use in Slovenia in 2005
M. Cizman on behalf of the Slovenian ESAC group
Objective: To collect data on Hospital consumption (HC) of antibiotics in all Slovenian hospitals and departments aggregated at national level in the year 2005.
Methods: Consumption data of systemic antibiotics in anatomical therapeutic chemical (ATC) (WHO version 2006) class J01 were collected, expressed in Defined Daily Doses (DDD). The consumption was expressed in DDD per 100 bed-days. Hospital pharmacist provided the data in the number of packages. The coverage of bed days were in all three years 100%.
Results: The results are shown in Table 1, Table 2 .
Table 1.
Hospital use (DDD/100 Bed-days) of antibacterials in Slovenian hospitals (n = 29) by type of hospital
| Departments/units | General hospitals mean (min–max) | UMC |
|---|---|---|
| Medical (n = 10) | 66.71 (45.29–85.40) | 80.40 |
| Surgical (n = 10) | 56.38 (39.28–75.84) | 80.85 |
| Paediatric (n = 11) | 41.88 (33.24–63.09) | 44.45 |
| Medical ICU (n = 6) | 143.46 (45.56–221.93) | 136.23 |
| Surgical ICU (n = 6) | 199.0 (132.97–261.82) | 238.33 |
| Medical and surgical ICU (n = 2) | 152.42 (146.76–178.07) | − |
| Gynaecological (n = 10) | 30.71 (16.81–46.85) | 22.08 |
Table 2.
Hospital use (DDD/100 bed-days) of antibacterials in UMC (teaching hospital) and departments of general hospitals in Slovenia in 2005
| Type of hospital | 2005 mean |
|---|---|
| UMC (teaching (n = 1) | 61.17 |
| General (n = 11) | 58.80 (42.66–77.47) |
| Orthopaedic (n = 1) | 32.61 |
| Private (n = 3) | 32.61 |
| Maternity (n = 2) | 27.07 (25.81–28.33) |
| Lung (n = 2) | 68.23 (58.50–77.96) |
| Psychiatric (n = 5) | 6.96 (6.23–7.15) |
| Nursery (n = 1) | 28.42 |
| Rehabilitation centres (n = 2) | 8.06 (6.42–9.75) |
| Oncology Institute (n = 1) | 51.51 |
Conclusion: total consumption of antibacterials in Slovenia is moderate. High variations of antibacterials use was found among similar types of some hospitals and departments. The data provide the rationale for an intervention.
P1107 Antibiotic use in German intensive care units
E. Meyer, F. Schwab, P. Gastmeier, H. Rueden, D. Jonas (Freiburg, Berlin, Hannover, DE)
Objective: To analyse antibiotic use in German ICUs participating in the project SARI (Surveillance of Antimicrobial use and antimicrobial Resistance in German Intensive Care Units).
Methods: Antibiotic use density (AD) was calculated in defined daily doses (DDD)/1000 patient days. DDDs according to the WHO in 2006 include 81 single antibiotics and 25 antibiotic groups.
Results: In 2005 mean AD was 1282 DDD/1000 pd in 43 ICUs. AD ranged from 559 to 2409. The top 3 antibiotic groups used were quinolones, the fixed combination of penicillins with lactamase inhibitor and 3rd generation cephalosporins (AD 165, 150 and 110, respectively); The top 3 single antibiotics were ciprofloxacin, cefuroxim and ampicillin-sulbactam (114, 89 and 74, respectively). The median of single antibiotics used was 27 (range 13–21). This did not differ by type or size (>1000 beds) of hospital or by type of ICU. Antibiotic use did not change significantly from 2000–2002 to 2002–2005 with the exception of a switch from the fixed combination penicillins with lactamase inhibitor to the free combination.
Conclusion: Antibiotic use in German ICUs is highly heterogeneous, but does not differ by type or size of hospital. Quinolones are the antibiotic group with the highest use.
P1108 Antimicrobial consumption and resistance in Staphylococcus aureus and Pseudomonas aeruginosa isolates from 47 French hospitals
A. Rogues, C. Dumartin, H. Boulestreau, A. Venier, J. Gachie, N. Marty, P. Parneix (Bordeaux, Toulouse, FR)
Objective: To investigate relationships between antimicrobial consumption and resistance in Staphylococcus aureus and Pseudomonas aeruginosa isolates from hospitals.
Methods: Hospital were asked to collect consecutive S. aureus and P. aeruginosa (Pa) isolates, consumption of antibiotics (J01A) and hospital characteristics (size, length of stay, number of beds: total and for each hospital areas) during one year. For methicillin-resistant S. aureus (MRSA) and for ceftazidime, ciprofloxacin and imipenem susceptibility of Pa, resistance (R) were expressed as percentage of nonsusceptible isolates and as incidence [nonsusceptible isolates per 1000 patient-days (PD)]. Antimicrobial consumption were expressed as number of defined daily doses per 1,000 PD). Association between continuous variables was tested in univariate analysis with the Spearman correlation test. Multiple linear regression was performed to control for hospital characteristics.
Results: Data were obtained from 47 hospitals with a total of 12188 S. aureus and 6370 P. aeruginosa tested. Resistance in percentage and incidence were, respectively, MRSA:42% and 0.8, ciprofloxacinR Pa:43% and 0.9, ceftazidimeR Pa:16% and 0.2, imipenemR Pa:14% and 0.2. In the multivariate analysis, significant association with antimicrobial consumption was less frequent with percentage than with incidence of resistance. The incidence of MRSA increased with use of ciprofloxacin and levofloxacin and with percentage of beds of intensive care in the hospital (R2a:0.30. Total antibiotic consumption minus glycopeptides explained 13% of the variance of MRSA incidence. For P. aeruginosa, incidence of ceftazidime resistance were higher in hospitals with higher consumption of ceftazidime, levofloxacin and gentamicin (R2a:0.37). The incidence of ciprofloxacin resistance increased with use of fluoroquinolones and percentage of beds of intensive care in the hospital (R2a:0.35). There was a positive and independent association between the incidence of imipenemR and consumption of carbapenem, cefotaxime, and macrolides (R2a:0.41).
Conclusion: A significant relationship occurred between fluoroquinolone use and resistance in S. aureus and Pa isolates. After adjusting for hospital characteristics, the associations between antimicrobial consumption and resistance found in our ecologic study were consistent with those observed on an individual patient level. Percentage of resistance was less interesting than incidence to study those relations.
In vitro antibacterial susceptibility
P1109 Antimicrobial and antioxidant actions of the clubmoss Lycopodium clavatum L
B. Özçelik, I. Orhan, S. Aslan, M. Kartal, T. Karaoglu, B. Sener, M. Choudhary (Etiler, Ankara, TR; Karachi, PK)
Purpose of the present study was to evaluate antioxidant, antibacterial, antifungal, and antiviral activities of the petroleum ether, chloroform, ethyl acetate and methanol extracts as well as the alkaloid fraction of Lycopodium clavatum L. (LC).
Antioxidant activity of the LC extracts was evaluated by 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical-scavenging method at 0.2 mg/mL using microplate-reader assay. Antiviral assessment of LC extracts was evaluated towards the DNA virus Herpes simplex and the RNA virus Parainfluenza using Madin-Darby Bovine Kidney and Vero cell lines. Antibacterial and antifungal activities of the extracts were tested against standard and isolated strains of the bacteria Escherichia coli, Pseudomonas aeruginosa, Proteus mirabilis, Acinetobacter baumannii, Klebsiella pneumoniae, Staphylococcus aureus, Bacillus subtilis, as well as the fungi C. albicans and C. parapsilosis. We evaluated the in vitro activity with standard antimicrobial powders as ketoconazole, fluconazole, amphotericin B, flucytosine, voriconazole (triazole derivative), and caspofungin (echinocandin).
All of the extracts possessed noteworthy activity against ATCC strain of S. aureus (4 μg/mL), while the LC extracts showed reasonable antifungal effect. On the other hand, we found that only the chloroform extract was active against HSV (16–8 μg/mL), while petroleum ether and alkaloid extracts inhibited potently PI-3 (16–4 μg/mL and 32–4 μg/mL, respectively). However, all of the extracts had insignificant antiradical effect on DPPH. In addition, we also analysed the content of the alkaloid fraction of the plant by capillary gas chromatography-mass spectrometry (GC-MS) and identified lycopodine as the major alkaloid.
P1110 In vitro study of antibiotic diffusion from bioceramic cylinders (hydroxyapatite and wollastonite)
M. Jorge, R. Seoane, F. Guitian, A. Perez-Estevez, M. Trevino-Castellano, L. Moldes-Suarez, C. Garcia-Riestra (Santiago de Compostela, ES)
Objectives: The use of antibiotics embedded in biomaterials is very common in orthopaedic surgery. The aim of the present study is to evaluate the delivery kinetics of teicoplanin from bioceramic cylinders vs two control antibiotics (ofloxacin and ceftriaxone).
Methods: Cylinders of hydroxyapatite (HA) and wollastonite (WA) 100 mg, 9 mm diameter and 3 mm thick, containing 3% each antibiotic were made at the Ceramic Institute of University of Santiago. Delivery was evaluated by bioassay using Mueller-Hinton plates inoculated with Bacillus subtilis ATCC 6633 spores. Inhibition diameters were measured daily. Once measured cylinders were transferred to new plates ever on the same side. When no inhibition could be detected cylinders were deposited on the other side. The residual activity was evaluated by extraction with PBS from cylinders no longer active. Delivered amounts were calculated by interpolation with standard curves made with paper disks containing known amounts of antibiotic.
Results: Results show a clear difference in delivery patterns from the two assayed bioceramics. Ceftriaxone delivery was quicker from hydroxyapatite opposite to observed with ofloxacin and teicoplanin. According to this fact ceftriaxone cylinders showed activity for 19 days from HA and 38 days from WA; on the contrary, teicoplanin and ofloxacin showed longer activity from HA, specially ofloxacin (50 days from HA vs. 26 days from WA).
Conclusions:
-
1
Hydroxyapatite delivers high levels of antibiotic for long periods and appears to be better material than wollastonite to carry antibiotics.
-
2
Wollastonite allows a quick delivery of both teicoplanin and ofloxacin. Nevertheless, ceftriaxone delivery was longer from this bioceramic.
-
3
The different delivery patterns detected suggest that bioceramic election could depend of antibiotic selected and desired delivery pattern.
P1111 Comparative in vitro susceptibility of bacterial isolates from Tbilisi maternity hospitals to antibiotics, specific bacteriophages and plant antibacterials
S. Tsertsvadze, M. Mshvildadze, A. Tskhvediani, N. Mitaishvili, T. Koberidze, M. Tediashvili, I. Davitaia (Tbilisi, GE)
Broad-spectrum antibiotics still remain the major treatments for neonatal bacterial infections. The same time occurrence of resistant bacterial strains increases every year. Application of bacteriophages and antibacterial substances of plant origin may be considered as effective alernative strategy.
The aim of the study was to isolate and study the bacterial microflora in neonatal departments and to perform screening of isolated strains on susceptibility to antibiotics, bacteriophages and tea antibacterials (TAB). Specimens were collected at neonatology departments of A and B maternity hospitals in Tbilisi during 4 months period in 2006. Women's vaginal microflora was studied before labour, while smears from the neonates oral cavity and nose were taken immediately after birth and within 24, 48, 72 and 96 hours. Processing of specimens was performed by standard bacteriological methods followed by identification using API-tests (bioMérieux, France). Sensitivity of the isolated strains to 12 antibiotics and 3 TAB preparaions was studied by disc diffusion and serial dilution methods. Lytic efficacy of specific phage clones as well as to mono- and complex bacteriophages (Pyophage, Intestyphage, Enterophage, SPS-phage, production of the Eliava IBMV) was estimated by spot test followed by titration using double layer method.
Specimens were taken from 32 mothers and newborns in parallel with collecting material from hospital environment. 128 bacterial isolates were characterised to reveal the prevalent microflora, the focus was made on isolation and identification of Staphylococcus spp., Streptococcus spp., Pseudomonas aeruginosa and Klebsiella spp. Resistance to 2–5 antibiotics was registered in >60% of isolated strains while susceptibility to different bacteriophages varied significantly within different bacterial groups and comprised in average 45%. It must be stressed that no correlation between phage- and antibiotic resistance was observed. High efficacy of experimental TAB preparations (Institute of Plant Biochemistry, Georgia) was shown as well, especially in case of Gram-positive bacteria.
P1112 Effect of polysorbate 80 on oritavancin binding to plastic surfaces – implications for susceptibility testing
F. F. Arhin, I. Sarmiento, T.J. Parr Jr., G. Moeck (Saint Laurent, CA)
Objectives: Oritavancin (ORI) is a lipoglycopeptide (LG) with activity against most Gram-positive bacteria including drug-resistant pathogens. Broth microdilution (BMD) assays with dalbavancin, another LG, require addition of 0.002% polysorbate 80 (P80). To investigate whether P80 affects ORI susceptibility test results, we performed BMD assays for ORI with and without P80 and quantitated 14C-ORI recovery from assay plates containing cation-adjusted Mueller Hinton Broth (CAMHB) ± P80.
Methods: For recovery assays, 14C-ORI and 14C-ciprofloxacin (14C-CIP) were dissolved in water ± P80, diluted in CAMHB or CAMHB plus 2% lysed horse blood (LHB) ± P80, and dispensed into 96-well polystyrene plates (Sensititre) without cells. Recovery of radiolabeled agents was assessed by scintillation counting of supernatant over time to yield residual counts relative to input label at time 0. BMD assays with Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212 and Streptococcus pneumoniae ATCC 49619 followed CLSI guidelines. P80, where present, was at 0.002%.
Results: In the absence of P80, 14C-ORI was rapidly lost from solution: at 1 mg/L, <10% of input ORI was recovered at 1 h. ORI loss was concentration dependent: proportionately greater losses were observed at lower ORI concentrations, suggesting saturable binding of ORI to surfaces. Inclusion of P80 or LHB promoted recovery of 80–100% of 14C-ORI at all concentrations tested up to 24 h. Quantitative recovery of input 14C-CIP +/– P80 was observed for all concentrations and time points tested. Concordantly, minimal inhibitory concentrations (MICs) of ORI for S. aureus and E. faecalis were 16- to 32-fold lower in the presence of P80 whereas P80 had no impact on MICs of vancomycin, teicoplanin or CIP. Assay plates with low-binding surfaces afforded 4- to 8-fold reductions in ORI MICs without P80, suggesting that ORI binding to plastic in the absence of P80 may underestimate in vitro potency. ORI MICs for S. pneumoniae tested in CLSI-recommended CAMHB + 2% LHB were identical ± P80.
Conclusions: ORI exhibits rapid, saturable binding to susceptibility test plates. P80 minimises binding of ORI to microtitre plate surfaces, thereby maintaining ORI available in solution for growth inhibition. 2% LHB in CAMHB promoted ORI recovery, helping to explain the lack of ORI MIC shifts for S. pneumoniae with P80. Current literature MIC data for ORI significantly underestimates ORI in vitro potency.
P1113 Antimicrobial susceptibility of anaerobic and facultative aerobic bacteria isolated from pus specimens of orofacial infections and β-lactamase production
G. Blandino, I. Milazzo, D. Fazio, S. Puglisi, A. Speciale (Catania, IT)
Objectives: Informations on susceptibility of the microorganisms involved in oro-facial infections can be useful for an effective antibiotic therapy. In the study we determine the antimicrobial susceptibility of 235 anaerobic and facultative aerobic bacteria isolated from pus specimens of orofacial infections during 2006 and the β-lactamase production.
Methods: The bacteria isolated from pus specimens were identificated using conventional methods and packages system. Minimum inhibition concentrations (MICs) were determined by the microdilution method in accordance with the guidelines ofthe NCCLS (1997) (anaerobic and facultative aerobic bacteria) and the CLSI (2005) (viridans streptococci).
Results: The table shows bacteria isolated, β-lactamase production, antimicrobials tested, and percentages of susceptibility.
| Species (# β-lactamase+a/# isolates) | Susceptibility (%)b |
|||||||
|---|---|---|---|---|---|---|---|---|
| P | A/C | CX | IP | E | DA | LEV | MTZ | |
| Viridans streptococci (0/18) | 100 | − | 100c | 100 | 83.3 | 88.8 | 100 | − |
| G. morbillorum (0/5) | 100 | − | 100 | 100 | 100 | 100 | 100 | 100 |
| Actinomyces sp. (0/35) | 97.1 | − | 100 | 100 | 85.7 | 94.2 | 100 | 40 |
| Peptostreptococcus sp. (0/60) | 93.3 | − | 93.3 | 100 | 53.3 | 93.3 | 100 | 100 |
| Eubacterium sp. (0/6) | 83.3 | − | 100 | 100 | 83.3 | 100 | 100 | 100 |
| Propionibacterium sp. (0/2) | 100 | − | 100 | 100 | 100 | 100 | 100 | 0 |
| P. gingivalis (2/32) | 93.7 | 100 | 100 | 100 | 90.6 | 93.7 | 93.7 | 100 |
| Prevotella sp. (8/30) | 73.3 | 100 | 100 | 100 | 90 | 100 | 93.3 | 100 |
| F. nucleatum (1/18) | 94.4 | 100 | 100 | 100 | 38.8 | 100 | 100 | 100 |
| C. rectus (0/7) | 100 | 100 | 100 | 100 | 85.7 | 100 | 100 | 100 |
| B. forsythus (3/9) | 66.6 | 100 | 100 | 100 | 88.8 | 100 | 100 | 100 |
| B. ureolyticus (1/7) | 85.7 | 100 | 100 | 100 | 42.8 | 85.7 | 100 | 100 |
| Veillonella sp. (1/6) | 83.3 | 100 | 100 | 100 | 83.3 | 100 | 100 | 100 |
Nitrocefin test (penicillin MIC ≥ 0.25 mg/L).
P: penicillin; A/C: amoxiclav, CX: cefoxitin; IP: imipenem, E: erythromycin; DA: clindamycin; LEV: levofloxacin, MET: metronidazole.
Cefotaxime.
Conclusion: Based on our findings amoxicillin-clavulanate and cefoxitin, and to less extent clindamycin and levofloxacin, remain good empirical choices against the polymicrobial aetiology of oro-facial infections, where anaerobes and viridans streptococci are predominant.
P1114 Increase of antimicrobial activity of β-lactam antibiotics in combination with sodium dioctylsulfosuccinate
S. Roveta, A. Marchese, E. Debbia (Genoa, IT)
Objectives: Previous studies (Simonetti et al., 2004) demonstrated that the dioctyl ester of sodium sulfosuccinate, Sodium dioctylsulfosuccinate (SDSS), at sub-inhibitory concentration, increases the activity of some antimicrobial agents. In the present study we attempted to examine the effect of SDSS in combination with various β-lactam antibiotics on the viabiliy of some bacterial species.
Methods: β-lactam resistant strains of Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Pseudomonas aeruginosa, Staphylococcus aureus and Enterococcus faecium (10 of each specie) were employed in this study. The antibiotics tested in combination with the surfactant were ampicillin (AMP), ceftazidime (CAZ), ceftriaxone (CRO), aztreonam (ATM) and imipenem (IPM). Preliminary screening to verify increasing of antibiotic activity by SDSS (ranging from 250 to 2,000 mg/L) was performed employing a disk-diffusion test. Time kill experiments (CLSI 2006) were performed on representative strains of E. coli, P. aeruginosa, S. aureus and E. faecium (1 strain each) employing selected antibiotics (AMP, CAZ, IPM) in association with 1,000 mg/L of surfactant.
Results: In preliminary screening SDSS extended the inhibition zones of the antibiotic disks in both Gram-negative and positive strains. The time-kill results confirmed those obtained with the disk-diffusion assay. The association of surfactant with a β-lactam antibiotic produced, in all strains tested, significative reductions of CFU/mL in comparison to each compound alone. SDSS in combination with AMP produced reductions greater than 104 CFU/mL in both E. coli and E. faecium. The surfactant associated with CAZ and with IPM was tested on S. aureus and P. aeruginosa respectively. Synergistic effect was noted against both pathogens. The best result was obtained against S. aureus with a reduction of 105 CFU/mL within 2 hours. No bactericidal activity was observed on the microorganisms tested employing SDSS alone.
Conclusion: SDSS is able to increase the antimicrobial activity of β-lactams on both Gram-negative and Gram-positive β-lactam resistant microorganisms. This phenomenon might be explained by interferences with transmembrane proteins, alterations of bacterial membrane or increase in cell permeability. These results suggests a new approach to the drug use in topical formulations with a significant improvement of antibiotic activity and a reduction of allergic and toxic reactions.
P1115 Evaluation of the efficacy of disinfectants used in hospital aseptic dispensing against Bacillus spores
M. Mehmi, J. Smith, P. Lambert (Birmingham, UK)
Objectives: To evaluate disinfectants currently used for transfer sanitisation in hospital aseptic dispensing processes. These disinfectants were assessed for efficacy against spores of Bacillus subtilis ATCC 6633 in suspension and carrier tests. Techniques of disinfectant application such as spraying, wiping or both were investigated.
Methods: Suspension testing: An aqueous spore suspension of B. subtilis ATCC 6633 was added to the disinfectant solution for a 2 minute contact time. The mixture was then neutralised, exposed to germinant solution and viable numbers determined by plating.
Hard-surface testing: Stainless steel carriers were aseptically inoculated with B. subtilis ATCC 6633 spores. After drying the carriers were sprayed, wiped, or sprayed and then wiped with disinfectant. After 2 minutes any surviving spores were recovered by agitation following neutralisation. Recovered spores were exposed to germinant solution and a dilution series was carried out.
Time-kill testing: As the suspension test, but samples were removed and enumerated over 24 hours. All dilutions were plated out onto tryptone soya agar and incubated overnight at 37°C. Any resulting colony forming units observed were counted. A control was used in each of the experiments.
Results: Of the 6 disinfectants tested a quaternary ammonium compound/chlorine dioxide formulation and 6% hydrogen peroxide were the more consistently effective biocides (approximate log 2 reduction after 24 hours). The disinfection method found to be the most effective in killing of the Bacillus spores was spraying followed by wiping, with the least effective being spraying alone. Overall, wiping as a method of disinfection was found to be very effective, with all disinfectants achieving similar reductions in spore levels. Disinfectants generally performed better in the carrier tests than the suspension tests. For carrier tests, spraying alone with a disinfectant was not as effective as wiping alone, or both spraying and wiping.
Conclusion: Disinfectant efficacy increased on an increase in contact time. However, longer contact times are not feasible and therefore a compromise may be required. The results obtained re-enforce research that indicates the insufficiency of current aseptic transfer disinfection protocols. Spores are very hard to kill with the use of chemical disinfectants currently available and therefore other methods of disinfection such as rapid gassing techniques need to be considered.
P1116 Antimicrobial activity of ozonised water and 0.9% NaCl
E. Bocian, D. Bialoszewski, B. Bukowska, M. Czajkowska, S. Tyski (Warsaw, PL)
Objectives: Antimicrobial activity of ozon is widely known and such property is sometimes utilised e.g. for water disinfection and for production of water intended for human consumption. However, in such application ozon concentration is rather low and bactericidal effect is not very high. Specially developed apparatus which allows ozon “in statu nascendi” production from oxygen and preparation of ozonised solutions, has been applied in this study. Ozonised water and 0.9% NaCl were prepared and such obtained solutions might be used as antiseptic and disinfectant agents, during surgical operations.
The aim of this study was to analyse bactericidal and yeasticidal activity of ozonised water and 0.9% NaCl, according to European standards EN 1040 and EN 1275.
Materials and Methods: Two European standards designated for estimation of antimicrobial activity of antiseptics and disinfectants were applied to analyse basic bactericidal activity – EN 1040 and basic yeasticidal activity EN 1275.
Freshly obtained ozonised water and phosphate saline were mixed with microbial cells suspensions (bacteria: 108 cfu/mL, yeasts: 107 cfu/mL). Test microbial strains: S. aureus ATCC 6538, P. aeruginosa ATCC 15442, E. coli ATCC 10538, Enterococcus hirae ATCC 10541 and C. albicans ATCC 10231 were used in order to analyse bactericidal and yeasticidal activity. Different contact times were applied and log reduction factors of viable bacteria count and viable yeast count, were calculated. Ozone concentration in tested liquids was also determined by titration. Chemically obtained data were compared with antimicrobial effect.
Results: It was shown, that 20 min of contact time caused at least a 105 log reduction in viable bacterial cells count of all tested strains and at least 104 log reduction in viable yeast cells count. Neutralisation method was effective in ozone antimicrobial effect liquidation. Test method was validated.
It means that prepared ozonised liquids complied the EN Standards: basic bactericidal and basic yeasticidal activities.
Conclusion: Applied apparatus produced high ozon concentration solutions and freshly prepared ozonised water and 0.9% NaCl, which could be used as effective antiseptic and disinfection preparations.
P1117 Qualitative and quantitative evaluation of micro-organisms present in excised human skin used for ex vivo assessment of topical antimicrobials
T.J. Karpanen, T. Worthington, B. Conway, P.A. Lambert (Birmingham, UK)
Introduction: Efficacy of topical antimicrobial agents is frequently assessed in vitro using suspension or carrier tests. Evaluation of the efficacy of antimicrobials applied topically should ideally incorporate in vivo studies, however these are often laborious and human volunteers are required. A method which overcomes the problems associated with in vivo studies is ex vivo assessment whereby excised skin is inoculated with microorganisms in laboratory conditions. However, the number and type of microorganisms already present in skin models used for ex vivo assays should be taken into consideration when evaluating antimicrobial efficacy data.
Objectives: Qualitative and quantitative assessment of the microbial load of excised human skin.
Methods: Full thickness human skin was obtained from ten patients undergoing breast reduction. Full thickness skin samples were cut aseptically to 2 cm × 2 cm squares in duplicate, weighed, diluted 1:10 with sterile phosphate buffered saline and ground in a stomacher. For quantitative assessment the suspensions were serially diluted and 1 mL aliquoted in duplicate into pour plates with molten 5% blood agar. For qualitative evaluation 100 microliters of suspensions were aliquoted in duplicate onto 5% blood agar. All the culture plates were incubated at 37°C in aerobic and anaerobic conditions for 48 hours. Microorganisms recovered were identified by VITEK compact 2 bacterial identification system (bioMérieux, Basingstoke, UK).
Results: All excised skin samples were contaminated with microorganisms [mean aerobic count 2,219 CFU/g (range 7–8,230), and mean anaerobic count 8,573 CFU/g (range 14–37,205)]. There was no correlation between weight of the skin sample and microbial load. The most common microorganisms recovered from the skin samples were Staphylococcus epidermidis and other coagulase negative staphylococci. Other organisms recovered include Kocuria spp., Bacillus spp., Acinetobacter spp., Klebsiella spp. and Rhizobium spp.
Conclusion: These results highlight the importance of taking into account the resident and contaminating microbial flora of excised human skin when undertaking ex vivo antimicrobial studies which use artificial microbial inoculation of the skin in laboratory.
P1118 Antimicrobial susceptibility of Nocardia spp. – experience in England and Wales
R.L.R. Hill, R. Pike, M. Warner (London, UK)
Objectives: Nocardia are responsible for a variety of cutaneous, lung and central nervous system infections. Although antibiotic therapy with co-trimoxazole is put forward as the treatment of choice, the reliability of such an agent is uncertain. We have therefore investigated the susceptibility of patient-isolates from a variety of lesions.
Methods: During 2004–06, all Nocardia spp. referred to the Antibiotic Resistance Evaluation Unit, a national reference service for England and Wales, were characterised by MIC profiling. Demographic and clinical data were also collated.
Results: Of 48 referred patient-isolates, 22 were unspeciated whilst there were 7 each of N. farcinica and N. cyriacigeorgica. There were three N. asteroides; other species amounted to 1 or 2 isolates each. MICs of co-trimoxazole were 0.25–0.03 for N. farcinica (sensitive), 0.03–32 for N. cyriacigeorgica (2/7 isolates resistant), 4–>256 for N. asteroides (3/3 resistant), 0.25–>32 for other speciated isolates (3/12 resistant) and 0.06–>32 for unspeciated Nocardia (18/29 resistant), Overall, 26/58 (44.8%) Nocardia spp. were resistant to co-trimoxazole. Of other antibiotics, 5/48 (10.4%) isolates were resistant to linezolid (MICs 0.25–64), 24/48 (50%) resistant to Amikacin, 8/48 (16.7%) resistant to imipenem.
The majority of patients were male (36, 75%); ages ranged from 28 to 87y. 27.1% presented with cerebral abscesses, followed by 18.9% with lung lesions. Other abscesses accounted for 12.6% whilst Nocardia were isolated from blood in 10.4%. Skin/wound infections accounted for 8.3% isolates, whilst eye lesions, multiple abscesses, synovial fluid and osteolysis accounted for 6.3, 4.1, 4.1%, respectively. Of 6 isolates from cerebral abscesses tested for susceptibility to moxifloxacin, MICs were 0.25–4 (66.6% sensitive). Only one isolate from cerebral pus was sensitive to doxycycline but only one was resistant to linezolid (MIC = 64); 2 were resistant to co-trimoxazole (MIC 4–>32).
Conclusion: Whilst about half of all isolates were resistant to cotrimoxazole, susceptibility amongst isolates from cerebral abscesses was more common. Overall, linezolid was the most reliably active agent although moxifloxacin may be useful for cerebral abscesses. The variability in antibiotic susceptibility highlights the need for continued surveillance of the susceptibility of these organisms to determine effective therapeutic strategies.
P1119 Antimicrobial effect of antibacterial essential oils and three common antiseptic products
J. Karbach, S. Königkamp, P. Warnke, E. Behrens, B. Al-Nawas (Mainz, Kiel, DE)
Objectives: Essential oils are used in cosmetics and healthcare products and have re-emerged as an effective antiseptic in patients with incurable head and neck cancer and associated malodorous necrotic ulcerations. The aim of the study was to evaluate the antimicrobial effect of a combination of the oils compared to a single application and standard antiseptics.
Methods: The efficacy of different oils was evaluated by agar diffusion test. Tea tree oil (TTO), Eucalyptus oil (EO), Lemon grass oil (LMO), and one Eucalyptus-based oil mixture (KM-PT 70, Klonemax®, Central Tilba, NSW, Australia) were tested and the results were compared with Paroex® (Chlorhexidindigluconat 0.2%, CHX), Octenisept® (octenidine dihydrochloride 0.1 g/100 mL) and Betaisodona® (Povidon-Iod 10 g/100 mL). 10 different cultured aerobic and facultative anaerobic bacteria and two yeasts were tested for the ability for proliferation (L. buchneri, L. brevis, S. gordonii, S. mutans, S. aureus, S. epidermidis, E. faecium, E. faecalis, M. micros, A. actinomycetemcomitans, C. albicans, C. glabrata).
Results: The results of the agar diffusion tests are shown in Table 1 . In the common antiseptic group Paroex® showed the highest antimicrobial effect. All tested oils also have antimicrobial effects against the biological indicators. The four oils showed a significantly higher antimicrobial effect compared to Betaisodona® (p = 0.001). Lemon grass oil (p = 0.005) and the mixture of oils (p = 0.039) had also a higher antimicrobial effect than Octenisept®, Lemon grass oil (p = 0.021) revealed a higher effect than Paroex®. Eucalyptus oil (p = 0.018) had a lower success than Paroex®. The four tested oils prevented bacterial growth similar to Paroex®. The results of Leman grass oil showed the best inhibitory effect of all tested products.
Table 1.
Diameter of the inhibition area of different pathogens [mm]
| Mean | Standard deviation | Minimum | Maximum | |
|---|---|---|---|---|
| TTO | 14 | 7 | 8 | 36 |
| EO | 12 | 4 | 9 | 22 |
| LGO | 22 | 9 | 11 | 39 |
| Mixture | 16 | 8 | 11 | 39 |
| 40% alcohol | 6 | 0 | 6 | 6 |
| Paroex® | 15 | 3 | 11 | 20 |
| Octenisept® | 12 | 2 | 10 | 16 |
| Betaisodona® | 7 | 2 | 6 | 12 |
Conclusion: The tested oils prevent bacterial growth in our in-vitro study and exhibit potency comparable to three well known standard products. In the agar diffusion test, the size of the microbial inhibition zone depends on the solubility and diffusion of the test substance and therefore, they may not express its full effective potential. The efficacy of the tested oily lipophyle and alcoholic hydrophyle antimicrobial agents and the results in antiseptic reaction should be evaluated in further studies.
P1120 Antimicrobial susceptibility testing for Mycobacterium kansasii in Taiwan
T.S. Wu, M.H. Lee, H.S. Leu, T.L. Wu, P.C. Chiang, C.H. Chiu (Taoyuan Hsien, TW)
Objectives: Although Mycobacterium kansasii had been found for decades, infections cause by this organism is newly emerging in Taiwan. We know some first-line antituberculous agents such as isoniazid, rifampin, and ethambutol are effective to treat M. kansasii infections, but we want to explore the efficacy of other antimycobacterial agents.
Methods: Bacterial strains: Thirty seven clinical isolates were collected from August, 1999 to January, 2003 at the Clinical Mycobacteriology Laboratory of Lin-Kou Chang Gung Medical Centre, Taoyuan, Taiwan. They are stored in skim milk containing 50% glycerol at −70°C until they are subcultivated.
PRA (PCR-restriction enzyme analysis): We differentiate M. kansasii from other mycobacterial species by using molecular biology methods described by Telenti et al.
Susceptibility tests: We use 96 wells plates for microdilution methods of susceptibility tests described by Wallace et al. We made bacterial 7H9 broth equal to 0.5 McFarland turbidity, then diluted to 1:1,000, and delivered 0.01 mL into each well containing 0.1 mL broth. Each antimicrobial agent was added by twofold dilution by sequence.
Breakpoints: Isoniazid (INH) >1 μg/mL; rifampin (RIF) >1 μg/mL; rifabutin (RFN) >2 μg/mL; ethambutol (EMB) >5 μg/mL; Clarithromycin (CLR) >16 μg/mL; ciprofloxacin (CIP) >2 μg/mL; moxifloxacin (MFX) >2 μg/mL; streptomycin (SM) >10 μg/mL; amikacin (AN) >32 μg/mL; sulfamethoxazole >32 μg/mL.
Results: The most active antibiotic is clarithromycin, the least is ethambutol. The intermediate sensitive antimicrobial agents are isoniazid, ciprofloxacin, and moxifoxacin; others are between 80% to 90% sensitive.
Conclusion: Ethambutol should not be contained in first-line antimycobacterial agents because most of M. kansasii isolates are resistant to ethambutol in Taiwan. M. kansasii isolates is not so sensitive to newer fluoroquinolones because they are 70% and 55% sensitive to ciprofloxacin or moxifloxacin respectively. The first line antimycobacterial regimens for M. kansasii infections should contain of clarithromycin, isoniazid, and rifampin at least. The alternative drugs for M. kansasii infections are rifabutin, aminoglycosides (streptomycin or amikacin), and sulfamethoxazole. The results of susceptibility tests are variable among nations; we must choose the correct drugs to improve the mortality and morbidity of patients according to local susceptibility tests data.
Susceptibility testing for M. kansasii isolates
| Antimicrobial agent | Sensitivity (%) | MIC (mg/L) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| INH | 64.86 | 1 | >32 | 0.25–>32 |
| RIF | 81.08 | 0.5 | 16 | 0.125–16 |
| RFN | 97.30 | 0.125 | 0.5 | 0.125–4 |
| EMBa | 12.12 | 8 | 16 | 4–>64 |
| CLR | 100 | 2 | 4 | 0.5–8 |
| CIP | 70.27 | 1 | 8 | 0. 5–>16 |
| MFXa | 54.55 | 2 | >8 | 0.06–>8 |
| SM | 94.59 | 4 | 8 | 0. 5–>64 |
| AN | 97.30 | 8 | 8 | 1–>64 |
| SXT | 81.08 | 4 | 128 | 1–>128 |
Only 33 isolates were done.
P1121 Patterns of susceptibility of Gram-negatives/positives isolated in the United Kingdom and Ireland
B. Johnson, S. Bouchillon, M. Hackel, R. Badal, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: The rapid emergence of multi-drug resistant pathogens has undermined the efficacy of many widely used broad spectrum antibacterials and prompted the development of newer antimicrobials. Tigecycline is a new glycylcycline shown to have broad spectrum activity against many hospital pathogens. The purpose of this study was to examine the activity of tigecycline and comparators to nosocomial pathogens isolated in the UK and Ireland between 2004–06.
Methods: A total of 1,131 nosocomial pathogens were identified at each site and confirmed at a reference laboratory. MICs were determined at each site utilising supplied broth microdilution panels and interpreted according to EUCAST guidelines.
Results: See the tables:
|
E. coli, K. oxytoca/pneumoniae (n = 294) |
Acinetobacter spp. (n = 86) |
P. aeruginosa (n = 119) |
||||
|---|---|---|---|---|---|---|
| %S | MIC90 | %S | MIC90 | %S | MIC90 | |
| Tigecycline | 95.2 | 1 | n/a | 1 | n/a | 16 |
| Amikacin | 99.7 | 4 | 88.4 | 16 | 93.3 | 8 |
| Cefepime | 86.1 | 4 | n/a | 32 | 83.2 | 32 |
| Imipenem | 100 | 0.5 | 91.9 | 2 | 89.1 | 8 |
| Levofloxacin | 79.3 | >8 | 76.7 | 8 | 65.5 | >8 |
| Minocycline | n/a | 8 | n/a | 4 | n/a | >16 |
| Pip/Tazo | n/a | 16 | n/a | >128 | n/a | 64 |
|
S. aureus (n = 136) |
Enterococcus spp. (n = 79) |
S. pneumoniae (n = 79) |
||||
|---|---|---|---|---|---|---|
| %S | MIC90 | %S | MIC90 | %S | MIC90 | |
| Tigecycline | 100 | 0.26 | 100 | 0.25 | n/a | 0.5 |
| Levofloxacin | 75.7 | 8 | n/a | >32 | 100 | 1 |
| Linezolid | 100 | 4 | 100 | 2 | 100 | 1 |
| Minocycline | n/a | 0.6 | n/a | >8 | n/a | 2 |
| Vancomycin | 100 | 1 | 92.4 | 2 | 100 | 0.5 |
n/a: breakpoints not yet available.
Conclusions: Tigecycline was as active as comparator agents against most Enterobacteriaceae spp., displayed the lowest MICs against Acinetobacter spp., and had minimal activity against P. aeruginosa. Against Gram-positives, tigecycline was as active as vancomycin and linezolid, and superior to levofloxacin or minocycline.
P1122 In vitro susceptibility of Actinobaculum spp. isolated from urinary tract infections against nine antibiotics
T. Schülin-Casonato, F. D'Ancona, H. Willemse, P. Sturm (Nijmegen, NL)
Introduction: Gram-positive rods have been described as cause of urinary tract infections (UTI) and urosepsis. Recently, with more sensitive detection and more advanced identification methods, Actinobaculum spp. have been identified to cause UTIs and urosepsis. The majority of UTIs is treated without microbiological diagnosis with antibiotics (AB) directed against Gram-negative rods.
Material and Methods: We tested the susceptibility of 21 Actinobaculum spp. isolated from urine for 9 antibiotics which are used as empirical therapy in UTIs or have known activity against Gram-positive organisms. MICs for 9 antibiotics (penicillin [PEN], ceftriaxone [CRO], moxifloxacin [MOX], ciprofloxacin [CIP], clindamycin [CLI], erythromycin A [ERY], vancomycin [VAN], tetracycline [TET], nitrofurantoin [NFT]) of 21 unique strains of Actinobaculum spp. (17 A. schaalii, 1 A. massiliae, 1 A. urinale and 2 A. species) isolated over a two-year period from urinary tract infections were determined by means of agar-dilution technique on lysed sheep blood agar with a final inoculum of 5×104 CFU per spot. Plates were incubated for 18 hours under anaerobic conditions.
Results: Distribution of MICs are presented in table 1 .
Table 1.
Number of strains for different MICs:
| Agent | MIC (mg/L) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.015 | 0.015 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | >32 | |
| PEN | − | 21 | − | − | − | − | − | − | − | − | − | − | − | − |
| CRO | − | − | 10 | 6 | 5 | − | − | − | − | − | − | − | − | − |
| CLI | − | − | − | − | 1 | 3 | 3 | 11 | 2 | − | − | − | − | 1 |
| ERY | − | 18 | 1 | 1 | − | − | − | − | − | 1 | − | − | − | − |
| TET | − | − | − | 3 | 4 | 11 | 2 | 1 | − | − | − | − | − | − |
| MOX | − | − | − | − | − | 1 | 6 | 13 | 1 | − | − | − | − | − |
| CIP | − | − | − | − | − | − | − | 1 | 4 | 14 | − | 1 | 1 | − |
| VAN | − | − | − | − | 5 | 16 | − | − | − | − | − | − | − | − |
| NFT | − | − | − | − | − | − | 1 | 1 | 13 | 5 | − | 1 | − | − |
Discussion: PEN and ERY were the agents with the lowest MICs. MICs for CRO, a third generation cephalosporin, which is often used as empirical therapy in urosepsis, were also in the range of available urine concentrations of this drug. Interestingly, one strain was resistant for CLI and ERY, which is suggestive for an MLSb resistance mechanism. MICs for CIP, frequently used in UTIs, were between 1 and 32 mg/L, which makes it less useful to treat UTIs due to Actinobaculum species. MICs for MOX were lower, but this drug does not reach high concentrations in urine.
Actinobaculum spp. can be a cause of UTIs and should be considered in patients who do not respond to therapy with antimicrobial agents directed solely against Gram-negative bacteria.
P1123 Antimicrobial susceptibility and phage types of Salmonella typhimurium human strains isolated during 2002–2005 in the Slovak Republic
L. Majtánová, V. Majtán (Bratislava, SK)
Objectives: Salmonella Typhimurium is a common cause of human salmonellosis in many countries. This is the second most common salmonella serovar isolated from human sources. The extensive use of antimicrobials in human and veterinary medicine has led to an increase in multidrug resistance strains (MDR). A MDR S. Typhimurium strain, with definitive phage type 104 (DT104), has emerged and spread over the world since 1984. The aim of this study was to analyse the antimicrobial resistance patterns of 330 S. Typhimurium human isolates and to investigate the distribution of their phage types.
Methods: Antimicrobial susceptibility tests were performed by disk-diffusion method using 10 different antibiotic disks according to the method of CLSI/NCCLS. Phage types were determined by the extended phage typing scheme of Anderson et al. (1977).
Results: Of total 330 strains analysed, MDR was identified in 192 (58.2%) isolates. They were the most frequently resistant to ampicillin, streptomycin, sulfisoxazole, tetracycline, chloramphenicol, sulfamethoxazole-trimethoprim and trimethoprim. None of the all isolates were resistant to ceftriaxone and only one isolate was resistant to ciprofloxacine. Three hundred and thirty strains encompassed 25 definitive phage types, 38 strains were atypical types (RDNC) and 76 ones were untypable. The predominant phage types were DT104 (22.4%), DT20a (22.1%) and DT120 (5.6%).
Conclusion: The strains studied expressed MDR and belonged to different phage types. The MDR was ascertained mainly in the predominant phage types DT104, DT20a, DT120 and also in untypable strains originating from sporadic cases of infection. Continual surrveillance is necessary to monitor the dissemination of these and similar MDR strains as well as their phage types in order to implement appropriate control measures.
P1124 In vitro antimicrobial activity of azithromycin against Mycoplasma genitalium and the bacteriological efficacy in male M. genitalium-positive non-gonococcal urethritis
M. Yasuda, N. Hagiwara, M. Ito, S. Ishihara, S. Maeda, S. Ito, H. Narita, T. Deguchi (Gifu, Minokamo, Toyota, Sendai, Nagoya, JP)
Objectives: Many studies have shown Mycoplasma genitalium to be a pathogen responsible for non-gonococcal urethritis (NGU). However, there are no published guidelines or recommendations for treating M. genitalium-positive NGU. So dosing regimens of antimicrobial agents active against Chlamydia trachomatis are usually used to treat M. genitalium-positive NGU. In the CDC and EAU guidelines, azithromycin or doxycycline treatment is recommended for management of patients with NGU. Thus we investigated the minimal inhibitory concentration (MIC)s of azithromycin and other antimicrobial agents for M. genitalium and whether azithromycin are clinically efficacious as antibacterial therapy against M. genitalium.
Methods: Seven strains of M. genitalium were obtained from the American Type Culture Collection. Susceptibility testing was performed by the broth microdilution method in 96-well microtiter plates with SP-4 medium. We then enrolled a total of 19 men with M. genitalium-positive NGU who were treated with azithromycin 1,000 mg orally as single dose. PCR-based assay was performed to evaluate the microbiological efficacy of eradication therapy in these patients.
Results: Azithromycin (MIC range: 0.004–0.008 mg/L) and clarithromycin (MIC range: 0.008–0.015 mg/L) were very active against M. genitalium. Doxycycline (MIC range: 0.06–0.125 mg/L) and minocycline (MIC range: 0.06–0.125 mg/L) had a moderate activities against M. genitalium-positive non-gonococcal urethritis. Gatifloxacin (MIC range: 0.125–0.25 mg/L) and sparfloxacin (MIC: 0.25 mg/L) were more active than levofloxacin (MIC: 1.0 mg/L). In 17 of 19 (89.5%) patients treated with azithromycin, M. genitalium was eradicated from the urethra.
Conclusions: Susceptibility testing suggests that azithromycin are suitable for treating M. genitalium-positive NGU. Clinical results suggest that azithromycin given orally at 1,000 mg as single dose is useful in treating M. genitalium-positive NGU.
P1125 Susceptibility pattern of Escherichia coli associated urinary tract infection: a comparison between spinal cord and nosocomial induced UTIs
F. Khorvash, K. Mostafavizadeh, S. Mobasherizadeh, A. Javadi, M. Behjati (Isfahan, IR)
Evolving resistant uropathogenes have been associated with the elevated risk of morbidity and mortality, both in nosocomial and spinal cord (SCI) associated urinary tract infections (UTI). Both of these complicated UTIs have concomitant risk factors for UTI confliction, as urinary catheterisation, probable poor hygiene, prolonged or intermittent antibiotic consumption and others. Regarding to this susceptibility, antibiotic resistance emerges, so more clinicoepidemiological studies are needed to facilitate empirical therapy and decrease time and costs toward diagnosis and treatment.
Method and Material: During 18 months period of this study (2005 and early 2006) 100 and 80 E. coli specimens from cases of diagnosed nosocomial UTI and spinal cord injury associated (SCI) UTI have been isolated by standard microbiological methods respectively. MIC of 10 antibiotics on isolated bacteria was determined by gradient concentration method (E-Test®; AB BIODISK Co. Sweden). Quality control was tested by E. coli ATCC 25922. Data was analysed by SPSS 13 and Whonet 5 software.
Results: Antibiotic resistant uropathogenes were more prevalent among nosocomial UTIs rather than SCI associated UTI (P <0.05). According to break point used for susceptibility meet CLSI M7-A6 (Clinical and Laboratory Standard Institute) criteria, resistance of isolated E. coli in nosocomial and SCI associated UTIs was 13.9% and 5.9% in amikacin (P<0.01), 65% and 40.5% in ceftazidim (P<0.05), 69.8% and 32.4% in ceftriaxone (P<0.005), 50% and 41.2% in ciprofloxacin (P<0.05), 56.9% and 13.6% in gentamicin (P < 0.005), 2.1% and 0% in imipenem (P< 0.005), 3.9% and 0% in meropenem (P< 0.005), 73.6% and 63.6% in trimethoprim/Sulfamethoxazole (P<0.05) respectively. Only resistance to nalidixic acid was mildly higher in SCI E. coli (79.1%, 73.6%). MIC50 of all antibiotics were higher in nosocomial E. coli (p < 0.05).
Conclusion: Despite more risk factors for antibiotic resistant E. coli in SCI population, the emerged antibiotic resistant E. coli was more diagnosed in nosocomial UTIs. Although this difference can be attributed to the hospital environment, more studies are needed to uncover the causes of these findings.
P1126 Emergence of Salmonella spp. isolates with reduced susceptibility to ciprofloxacin in Kuwait and the United Arab Emirates
V.O. Rotimi, W.Y. Jamal, T. Pal, A. Sonnevend, T.S. Dimitrov, M.J. Albert (Safat, KW; Al Ain, AE; Andolus, KW)
Objective: Kuwait and UAE are of sentinel significance in the context of global epidemiology of antimicrobial resistance because of a large expatriate population of workers from different parts of the world. Our main objective was to evaluate the antimicrobial susceptibility of clinical isolates of Salmonella spp. in these countries over a 2-year period and determine the extent of resistance problem.
Methods: A total of 410 Salmonella spp. isolated from symptomatic patients in various hospitals in Kuwait and UAE, were studied. The problem of drug resistance was investigated by determining the antimicrobial susceptibility of 410 Salmonella spp. from both countries by the E-test method.
Results: The MIC90s of amikacin, cefotaxime, ceftriaxone and ciprofloxacin were 1.9, 0.47, 0.47, and 0.14 μg/mL, respectively (Kuwait isolates), and 2.5, 0.22, 0.22, and 0.056 μg/mL, respectively (UAE). Resistance rates among Kuwait and UAE isolates to ampicillin were 25.1% and 15.5%, cefotaxime/ceftriaxone 1.6% and 1.6%, ciprofloxacin 1.2% and 0.8%, chloramphenicol 5.6% and 5.7%, and trimethoprim 25.5% and 7.4%, respectively. In Kuwait and UAE, 14.2% and 7.3% of the non-typhoidal Salmonella, respectively, and 44% of S. Typhi and 46.7% S. Paratyphi demonstrated reduced susceptibility (MIC; 0.125–0.5 μg/mL) to ciprofloxacin.
Conclusion: A large number of Salmonella spp. isolates, many of which were multi-drug resistant, were resistant to the first-line antibiotics. A high proportion of isolates also exhibited reduced quinolone susceptibility, a phenomenon that appears to be spreading rapidly throughout the world.
P1127 Enhancement of antimicrobial activities of cefteram or clavulanic acid/amoxicillin against cefixime-resistant Neisseria gonorrhoeae in the presence of clarithromycin or azithromycin
H. Kiyota, S. Onodera (Tokyo, JP)
We investigated the enhancement of antimicrobial activities of β-lactams against cefixime (CFIX)-resistant Neisseria gonorrhoeae in the presence of macrolides. Ten strains of CFIX-resistant N. gonorrhoeae, isolated from the patients with male urethritis between 2000 and 2003 at Jikei University Affiliated Hospital and its related clinics in the Tokyo metropolitan area, were tested. The fractional inhibitory concentrations of clavulanic acid/amoxicillin (CVA/AMPC), CFIX, or cefteram (CFTM) in the presence of clarithromycin (CAM) or azithromycin (AZM) against these strains were determined. Synergism, partial synergism or additivity between CVA/AMPC or CFTM and macrolides against 9 strains were recognized. Additivity or partial synergism between CFTM and macrolides against 9 or 10 strains were also recognized. On the other hand, antagonism between CFIX and macrolides was recognized. These results indicate that the combination of antimicrobial chemotherapy of CFTM or CVA/AMPC with macrolides is a possible alternative treatments against CFIX-resistant N. gonorrhoeae infections.
P1128 Microbial diversity and resistance of pathogens in prostatitis
M.S. Lapatschek, A. Hartinger (Munich, DE)
Objectives: Only very few types of antibiotics are efficient for the treatment of acute and chronic prostatitis. Moxifloxacin is a new fluoroquinolon with an extended antibacterial spectrum including Gram-positive bacteria and anaerobes. In this study we determined the activity of moxifloxacin against pathogens isolated from 100 patients with prostatitis in comparison with other antibiotics used for the treatment of this disease.
Methods: Prostatic fluid of patients suffering from clinically diagnosed prostatitis was cultured for potentially pathogenic microbes. In addition, a PCR for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae was performed. The conventionally cultured bacteria were tested for resistance by Etest against ampicillin, ampicillin/sulbactam, sulfamethoxazole/trimethoprim, doxycycline, levofloxacin and moxifloxacin.
Results: We received 147 specimens from 100 patients. We were able to cultivate 93 potential pathogens, distributed over 21 different species; 33% of these isolates were Gram-positive cocci, 45% Gram-negative rods and 2% anaerobic Gram-negative rods. Chlamydia and Gonococci were not found. The results of the sensitivity testing show an overall sensitivity of 59% for ampicillin, 83% for ampicillin/sulbactam, 79% for sulfamethoxazole/trimethoprim, 48% for doxycycline, 92% for levofloxacin and 91% for moxifloxacin.
Conclusions: Prostatitis is caused by a broad spectrum of different pathogens. Ampicillin without inhibitor, sulfamethoxazole/trimethoprim and doxycycline are not covering a sufficient spectrum of bacteria for calculated monotherapy. Ampicillin/sulbactam, levofloxacin and moxifloxacin have a comparable overall in vitro activity towards potentially pathogenic bacteria. Thus, moxifloxacin seems to be a feasible alternative for the treatment of prostatitis presuming a good tissue penetration into the prostate gland.
P1129 Antimicrobial susceptibility of gonococci isolated in Greece in 2005. Dramatic increase in quinolone resistance rate due to a single serovar
M. Stathi, H. Avgerinou, V. Miriagou, A. Flemetakis, M. Daniilidou, K. Kyriakis, E. Tzelepi (Athens, Thessaloniki, GR)
Objectives: Surveillance of antibiotic susceptibility trends of gonorrhoea in Greece.
Methods: Microbiological and epidemiological data for 168 gonococci received by the Greek National Reference Centre for Neisseria gonorrhoeae during the year 2005 were evaluated. The strains were isolated consecutively from gonorrhoea cases attended either the Andreas Sygros STD Hospital in Athens (90%) or other hospitals throughout Greece (10%). Susceptibilities of the isolates were determined in terms of Minimal Inhibitory Concentrations (MICs) of eight antimicrobial agents using Etest strips (AB Biodisk). Serological classification was performed with the Phadebact GC Serovar Test (Boule Diagnostics). Plasmid content analysis was performed for penicillinase producing strains (PPNG) and for strains exhibiting tetracycline MICs >8 mg/L (TRNG). Epidemiological data were obtained through a standard questionnaire, which included demographic data and information for sexual behaviour and source of infection.
Results: Only cefotaxime and spectinomycin were fully active against all of the 168 isolates. Whereas, high rates of resistance (R) and intermediate susceptibility (I) were observed for penicillin G (R 23.2%, I 70.8%), tetracycline (R 30.4%, I 68.4%), erythromycin (R 45.2%, I 50.6%), chloramphenicol (R 39.9%, I 56.5%), norfloxacin (R 33.3%) and ciprofloxacin (R 33.3%, I 0.6%). PPNG and/or TRNG isolates accounted for 5.4% of the total sample, including four strains that were simultaneously quinolone-resistant (QRNG). The isolation frequency of QRNG strains (33.3%) was found dramatically raised as compared to the rate of 11.3% which was recorded in the previous year. Serotyping results revealed a strong association (P <0.001) of quinolone resistance with a single serovar (Bropyst), present in 75% of the QRNG isolates. Interestingly, Bropyst/QRNG isolates exhibited cross-resistance to all antibiotics tested, except cefotaxime and spectinomycin. Information ensued by the questionnaires indicated that QRNG/Bropyst isolates derived from cases of infection acquired within Greece.
Conclusion: Resistance profiles in the Greek gonococcal sample of 2005 point out as drugs of choice for the treatment of gonorrhoea 3rd generation cephalosporins and spectinomycin. Typing results and epidemiological data suggest that triplication in QRNG frequency observed between 2004 and 2005 is probably due to a single, endemic strain.
P1130 The panhellenic study on the antimicrobial susceptibility of community-acquired uropathogens: preliminary data report
I. Katsarolis, G. Poulakou, S. Athanasia, F. V. Kontopidou, P. Panagopoulos, E. Karaiskos, D. Voutsinas, L. Zarkotou, P. Gavra, G. Koratzanis, M. Kanellopoulou, G. Adamis, E. Vagiakou, I. Matthaiopoulou, E. Vogiatzi, P. Perdikaki, G. Panou, E. Kremastinou, H. Giamarellou (Haidari, Piraeus, Marousi, Athens, Tripoli, Nafpaktos, Astros Kynourias, Karpenissi, GR)
Purpose: To present preliminary microbiological data from the first panhellenic survey investigating the prevalence and susceptibility of pathogens causing community-acquired acute uncomplicated urinary tract infections in 2005, with a view to guide empirical treatment.
Methods: Midstream urine samples were cultured from women <65 years, without known complicating factors, presenting with symptoms of acute cystitis at public and private community healthcare centres across Greece in 2005. Pathogens were identified and tested for their susceptibility to 13 antimicrobials by disk diffusion assay, using CLSI 2005 methodology and breakpoints. Patients with known complicating factors (urinary tract abnormalities, comorbidities, pregnancy, recent hospitalisation, antibiotic usage or UTI history in the past 2 weeks) were excluded.
Results: Preliminary microbiological data (March–July 2005) were derived from 418 consecutive positive cultures from women with acute uncomplicated cystitis, out of 1,532 patients tested in the same period. Escherichia coli accounted for 78.7% of isolates, Proteus mirabilis 8.9% and Klebsiella pneumoniae 2.4%. Staphylococcus saprophyticus was isolated in only two cases. Full susceptibility to all investigated antimicrobials was recorded among 39.2% of E. coli isolates, whereas resistance was most common to ampicillin (27.1%) and cephalothin (24.8%), followed by cotrimoxazole (21.1%), nitrofurantoin (19%) and amoxicillin/clavulanate (7.7%). Resistance to nalidixic acid and ciprofloxacin, fosfomycin and cefuroxime was 7.8%, 4%, 3.1% and 2.1%, respectively. Proteus mirabilis resistance rates per antimicrobial were: ampicillin (25%), cotrimoxazole (27.8%), nalidixic acid/ciprofloxacin (10.8/0.0%), amoxicillin/clavulanate (5.4%) and cefuroxime (2.7%). Gentamycin resistance rates for E. coli and P. mirabilis were similar (2.2/2.8% respectively). No ESBL-producing strains were detected.
Conclusions: From the up till now analysed preliminary data, it is evident that: (1) Empirical use of first-line antimicrobials in uncomplicated community-acquired UTIs, such as ampicillin, cotrimoxazole and nitrofurantoin, should be reevaluated, due to high rates of resistance. (2) Quinolones are still highly efficacious in this setting, but increasing resistance rates may advocate caution in their use in uncomplicated infections. (3) Low-level resistance to amoxicillin/clavulanate, cefuroxime and fosfomycin must be taken into account in the empirical treatment strategy.
P1131 Antimicrobial susceptibilities and prevalence of qnrA in UTI isolates from hospital and the community
P.G. Higgins, M. Lohr, F. Wisplinghoff, H. Seifert, H. Wisplinghoff (Cologne, DE)
Objectives: To determine the prevalence of the quinolone resistance gene qnrA and to compare species distribution and antimicrobial susceptibility of nosocomial and community acquired urinary tract infections.
Methods: Over a three-month period, isolates were prospectively collected from hospitals and private practices in the Cologne metropolitan area. 557 Gram-negative isolates were included in the study. Species identification and antimicrobial susceptibility testing were performed using the VITEK Two and the MicroScan-Walkaway system. Presence of qnrA was investigated by PCR.
Results: E. coli was most the commonly isolated organism, accounting for 59% of all isolates, followed by Klebsiella spp. and Pseudomonas aeruginosa (13% and 8% respectively). Species distribution was similar in nosocomial and community acquired UTI, except for Klebsiella spp. and Morganella morganii that were more commonly seen in nosocomial UTI, while E. coli was more frequently observed in community acquired UTI. Prevalence of qnrA was low; the gene was detected in 2 Enterobacter cloacae isolates, both from nosocomial UTIs. One of these strains showed intermediate resistance to ciprofloxacin while the other was fully susceptible. Nosocomial isolates were in general more resistant to the antimicrobial agents than the community isolates. For example, 90% of P. aeruginosa from the community were susceptible to ceftazidime and imipenem compared to 83% and 71% respectively of the hospital isolates. Also 93% of community E. coli were susceptible to cefuroxime compared to 78% of hospital isolates. Of note, 88% of nosocomial Proteus species were susceptible to ciprofloxacin compared to only 59% in community isolates.
Conclusions: As expected, nosocomial Gram-negative UTI pathogens are less susceptible to antimicrobials compared to those that are community acquired. However, the notable exception to this is ciprofloxacin activity against Proteus species. The qnrA gene is still rarely found in Germany and to date has not been found in community isolates.
P1132 Occurrence and susceptibility rates among urinary tract infection pathogens from Europe: a seven-year report from the SENTRY Antimicrobial Surveillance Program (1997–2000, 2003)
G. Moet, M. Stilwell, R. Jones (North Liberty, US)
Objectives: To report the occurrence and susceptibility (S) rates for pathogens causing urinary tract infection (UTI) isolated from medical centres in Europe, Turkey, and Israel. The S rates were compared by CLSI and EUCAST breakpoints. The SENTRY Antimicrobial Surveillance Program was utilised as the platform for collecting urine culture isolates during 5 years out of 10 year period that the programme has been in existence.
Methods: A total of 4,507 strains (50 consecutive, non-duplicate per site) were collected from 42 medical centres in 19 countries in Europe, Turkey and Israel (1997–2000, 2003). During the 5 years 31 locations participated in 3 or more years. All isolate identifications were confirmed and S testing performed in a central laboratory using reference broth microdilution methods (M7-A7) and interpretive criteria of CLSI and EUCAST (2006). ESBL phenotype rates were as determined by CLSI criteria.
Results: The 5 most frequent pathogens accounted for 83.3% of the total CA-UTI and the top 7 for 90.0%. These 7 pathogens displayed little change in occurrence rates over the 7 year period. Escherichia coli remained the dominant UTI pathogen at nearly 50% while enterococci showed a slight decrease from 11.7 to 10.1%. Variations in 1999 were greater due to only 9 participating sites (small sample size). Among commonly isolated Enterobacteriaceae, carbapenems were the most active agents ranging from 99.1% to 100.0% S; lower S was noted for ciprofloxacin (77–89%) and trimethoprim/sulfamethoxazole (T/S; 63–78%). Ceftazidime S rates for Klebsiella, E. coli, and Proteus mirabilis were 80.7, 97.2, 95.5% (CLSI) and lower at 76.9, 94.7, 92.5% (EUCAST), respectively. ESBL phenotype rates were 24.0, 5.3, and 7.2% for the same organism groups. Polymyxin B was active against Pseudomonas aeruginosa at 99% S, followed by carbapenems and piperacillin/tazobactam at 84% and amikacin at 83%. Vancomycin, teicoplanin, and linezolid remained active against enterococci (99% S).
Variation of rank order in SENTRY Program UTI pathogens by year for Europe
| Rank | Organism | All years | 1997 | 1998 | 1999 | 2000 | 2003 |
|---|---|---|---|---|---|---|---|
| 1 | E. coli | 2,175(48.3) | 505(51.0) | 548(48.0) | 68(39.5) | 361(46.1) | 693(48.8) |
| 2 | Enterococci | 510(11.3) | 116(11.7) | 127(11.1) | 23(13.4) | 100(12.8) | 144(10.1) |
| 3 | Klebsiella spp. | 445(9.9) | 83(8.4) | 120(10.5) | 25(11.6) | 69(8.8) | 153(10.8) |
| 4 | P. aeruginosa | 355(7.9) | 65(6.6) | 91(8.0) | 16(9.3) | 71(9.1) | 112(7.9) |
| 5 | P. mirabilis | 265(5.9) | 52(5.2) | 70(6.1) | 3(1.7) | 56(7.2) | 84(5.9) |
| 6 | Enterobacter spp. | 194(4.3) | 49(4.9) | 36(3.2) | 11(6.4) | 33(4.2) | 65(4.6) |
| 7 | Indole + Proteus spp. | 109(2.4) | 21(2.1) | 31(2.7) | 5(2.9) | 27(3.1) | 28(2.0) |
| Total | 4,507 | 991 | 1,142 | 172a | 783 | 1,419 |
Small sample.
Conclusions: This comprehensive report from the SENTRY Program covering 7 years of UTI sampling in Europe, reports minor variation in the most common pathogens between the sample intervals but with significant S differences noted among monitored countries. Emerging resistance is limiting the usefulness of commonly prescribed UTI agents including T/S and FQ, forcing reliance on more potent parenteral broad-spectrum agents and the additional problems inherent in their use.
P1133 Increasing ciprofloxacine resistance in the bacteria isolated from urine samples: 1999–2004
O. Alici, Z. Acikgoz, S. Gocer, S. Gamberzade, M. Karahocagil (Ankara, TR)
Purpose: The aim of the study was to determine the ciprofloxacin resistance of various bacterial species responsible for UTIs according to IDSA guidelines for UTIs between 1999 and 2004, retrospectively.
Materials and Methods: Overall 3,520 unduplicated urinary isolates were screened for their ciprofloxacin resistance between 1999 and 2004, retrospectively. Of the isolates 3,276 (93.1%) were from outpatients and 244 (6.9%) from the hospitalised ones. The screening was made regardless of age, gender and other demographic parameters. The isolates were identified by using conventional biochemical tests and the semi automated API systems (bioMérieux, France) when it was required. Antimicrobial susceptibility assays were performed using the disc diffusion method according to the recommendations of Clinical and Laboratory Standards Institute (CLSI) criteria. The data were analysed by Chi-square test in SPSS 10.0 programme and p values lower than 0.05 were considered significant.
Results: Of the screened isolates 3,216 (91.4%) were Gram negative bacilli and 304 (8.6%) were Gram positive cocci. Totally 29.9% (73/244) of in-patients isolates and 11.8% (281/2,376) of outpatient isolates had ciprofloxacin resistance. Out-patient and in-patient distributions of the resistant isolates according to main microorganism groups were as follows: For Gram positives 34.7% (n.8) and 65.3% (n.15); for Gram negatives 19.6% (n.65) and 80.4% (n.266), respectively. While there was no statistically significant difference between Gram positive and Gram negative microorganisms in terms of ciprofloxacin resistance (p > .05), the resistance rates of isolates from in-patients and out-patients differed significantly (p < 0.001). Ciprofloxacin resistance rates over the six years (1999–2004) were: 3.3% (17/509), 4.2 (33/551), 6.7 (40/593), 13% (102/786), 16.1% (124/768) and 15.3% (48/313) respectively. the differences between 1999 and 2001, 1999 and 2002, 2000 and 2001, 2000 and 2002 and 2001 and 2002 were significant (p<0.05).
Conclusions: We concluded that a prominent increase in ciprofloxacin resistance has emerged in bacteria isolated as UTI agents, especially in E. coli. This problem of resistance is a considerable issue for in-patients and has the potential to affect out-patients in long term periods.
Table 1.
Distribution of bacterial species and their ciprofloxacin resistance rates
| Species | No. of isolates | Resistance |
|
|---|---|---|---|
| n | (%) | ||
| E. coli | 2,444 | 305 | 12.5 |
| Klebsiella spp. | 480 | 21 | 4.4 |
| Proteus spp. | 190 | − | − |
| Enterococcus spp. | 137 | 18 | 13.1 |
| Gr. B streptococci | 85 | 1 | 1.1 |
| Staphylococcus spp.a | 82 | 4 | 4.9 |
| Enterobacter spp. | 51 | − | − |
| Pseudomonas spp. | 51 | 5 | 9.8 |
| Total | 3,520 | 354 | 10.1 |
Including Staphylococcus aureus and coagulase-negative staphylococci.
P1134 Antimicrobial resistance of Mycoplasma hominis and Ureaplasma urealyticum among women with vaginal discharge
A. Kurti, G. Mulliqi-Osmani, L. Raka, L. Berisha, A. Jaka, X. Jakupi, L. Begolli (Pristina, Kosovo, RS)
Objectives: The aim of study was to isolate Mycoplasma hominis (Mh) and Ureaplasma urealyticum (Uu) and determine the antimicrobial resistance in cervico-vaginal samples of women, who had complaints of vaginal discharge, using Mycoplasma IST 2 (bioMérieux).
Methods: There were 471 patients with vaginal discharge included in the study. Vaginal samples were taken from endocervikal region after exocervikal mucus had been swabbed clean. Mycoplasma IST 2 used for investigation of Mh and Uu provided information about the presence or absence of Mh and Uu and also their antimicrobial susceptibility to doxycycline, josamycin, ofloksacin, erythromycin, tetracycline, ciprofloxacin, azythromycin, clarythromycin and pristinamycin.
Results: Uu has been isolated in 235 (49.90%) patients, while Mh has been isolated in 42 (8.91%) of them. Uu and Mh have been both isolated in 36 (7.46%) of the patients.
The Uu has shown the lowest sensitivity in ciprofloxacin with 98 (41.70%) of isolates being resistant. The isolates of Mh and the mixt Mh/Uu isolates have shown low sensitivity on Erythromicin with 6 (100%) and 27 (75%) respectively being resistant.
Conclusion: A higher prevalence of Uu versus Mh is noted in the vaginal swabs as well as higher resistance in the chinolone. In addition, Mh and mixed isolates are more resistant in macrolides further explaining that the macrolide resistence is attributed to Mh.
P1135 Ten-year surveillance of antimicrobial susceptibility of community-acquired uropathogens in northern Israel
R. Wasseem, B. Chazan, M. Elias, R. Colodner, R. Raz (Afula, IL)
Background: Community acquired urinary tract infection (CA-UTI) is very common. Knowing local antibiotic sensitivity patterns is essential for empiric therapy.
Aim: Compare susceptibility patterns of community acquired uropathogens in northern Israel over a 10 year period.
Methods: All community urinary isolates processed by our laboratory during 1995, 1999, 2002 and 2005 were included. Distribution and susceptibility of Gram negative isolates were assessed. Antibiotic Consumption over 2000–2005 was evaluated.
Results: Over the study years approximately 10% of cultures grew Gram-negative isolates. E. coli remained the leading uropathogen (61%) without a significant change in the prevalence of other Gram-negative isolates. The susceptibility rates of E. coli for ampicillin (45%), cefuroxime (97%) and ceftriaxone (98%) remained stable. Susceptibility to amoxicillin-clavulanate (89% vs. 96%), TMP-SXZ (70.99% vs. 71.88%) and nitrofurantoin (93.9% vs. 97%) increased significantly. However, susceptibility of E. coli and other Gram–negative pathogens to ciprofloxacin (∼94% vs. ∼89.5%) decreased significantly. MIC90 of E. coli for amoxicillin-clavulanate, cephalothin, cefuroxime, ceftriaxone and ciprofloxacin remained stable. A significant decrease in the use nitrofurantoin and TMP-SMX and a significant increase in the use of ampicillin, cephalothin and ceftriaxone were seen by DDD/1,000/day.
Conclusions: Over a 10-year period susceptibility of E. coli and other Gram negative uropathogens responsible for CA-UTI in northern Israel has mostly remained stable or showed a small increase. However, there was a constant decrease in susceptibility of uropathogens to fluroquinolones.
We didn't find a marked correlation between bacterial susceptibility and antibiotic consumption.
P1136 Community coliforms in Cambridgeshire: resistance patterns in urinary isolates from 1997 to 2006
J.S. Cargill, H.A. Payne, D.A. Enoch, H. Ludlam, N.M. Brown (Cambridge, UK)
Objective: To demonstrate variations in resistance patterns for urinary isolates in the community over a 9 year period to the end of June 2006.
Methods: The laboratory results database was searched for urine samples obtained from general practice patients, and the susceptibility rates to ampicillin (AMP), co-amoxiclav (AMC), cephalexin (CLN), norfloxacin (NOR) and trimethoprim (TMP) against selected Enterobacteriaceae (grouped as Escherichia coli, Citrobacter–Enterobacter–Klebsiella–Serratia spp. [CEKS] and Proteus–Providencia–Morganella spp. [PPM]) were examined. Susceptibility was assessed by a breakpoint method prior to 1998 and by BSAC methods since. Resistance to cefpodoxime was introduced as a marker for extended-spectrum β-lactamase production in September 2005, and the resistance rates to this are also included.
Results: Of 58,823 positive isolates identified, 48,317 (82.1%) were E. coli, and so the resistance patterns are dominated by changes in this organism. Table 1 shows antibiotic susceptibilities at selected time points, showing that overall sensitivities to CLN have remained stable over the study period, while AMC (and AMP) sensitivity shows a cycling pattern. TMP showed constant susceptibilities until 2002, followed by a decline, while NOR has shown a gradual decrease in susceptibility throughout the time period, with an acceleration in resistance rates after 2004.
Overall susceptibility rates (% of isolates)
| Jul–Dec 1997 | Jan–Jun 1999 | Jul–Dec 2000 | Jan–Jun 2002 | Jul–Dec 2003 | Jan–Jun 2005 | Jan–Jun 2006 | |
|---|---|---|---|---|---|---|---|
| Ampicillin | 55.7 | 54.7 | 57.3 | 57.2 | 57.4 | 49.7 | 51.1 |
| Co-amoxiclav | 84.7 | 89.0 | 93.9 | 90.6 | 89.8 | 83.4 | 93.3 |
| Cefalexin | 94.4 | 93.9 | 92.5 | 95.2 | 96.5 | 95.9 | 94.8 |
| Norfloxacin | 98.5 | 98.2 | 96.8 | 96.4 | 93.6 | 90.4 | 87.3 |
| Trimethoprim | 78.8 | 79.3 | 80.0 | 82.2 | 78.4 | 76.0 | 74.7 |
For E. coli NOR sensitivity first dropped below 95% in early 2003, and below 90% in early 2005. For CEKS these levels were reached in early 2002 and late 2004 respectively, and for PPM late 2000 and early 2006 respectively. For E. coli TMP sensitivity dropped below 75% for the first time in early 2005, having been below 85% throughout the study period. For CEKS TMP sensitivity dropped below 85% for the first time in late 1998, recovering to 85–93% with a drop below 80% in early 2004, while for PPM TMP has shown less than 75% sensitivity rates throughout the study period.
Cefpodoxime resistance is present in 2.7–4.1% of E. coli, 7.6–12.1% of CEKS and 0.0–2.2% of PPM per quarter.
Conclusions: The resistance rates to antibiotics commonly prescribed in the community for urinary tract infection is increasing. This is potentially serious in cases of complicated or ascending infections. Local resistance rates should be taken into account when preparing empirical treatment guidelines, both for community and hospital use.
General microbiological tools: from swabs to species
P1137 Prevalence of syphilis and an examination of two syphilis rapid tests in the MCA, Cameroon
F. Oudenaarden, M. Siemerink (Rotterdam, NL)
Objectives: A two-fold cross-sectional field study was performed at Manyemen Presbyterian Medical Institutions, Cameroon. First, the prevalence of syphilis among people in the reproductive age (men: 15–60 years, women: 15–50 years) living in the Manyemen Catchment Area (MCA) was assessed. Second, the Abbott determine Syphilis TP® rapid test (Abbott test) was compared to the syphilis rapid test that is presently used in the MCA (local test).
Methods: The studied population was tested on syphilis with two different rapid tests: the Abbott test and the local test. People with at least one positive test were treated with benzathin penicillin. A questionnaire was used to obtain additional information about the studied population. Blood was collected on filter paper in order to perform the FTA-abs test in The Netherlands, which was used both for determination of the prevalence and as a confirmation test in order to differentiate between the two rapid tests. All positive blood samples according to the rapid tests performed in Cameroon, and a control group of randomly selected negative blood samples were (re)tested with the FTA-abs test.
Results: A total of 1,128 (539 men and 589 women) participated in this study. Of this population, 114 people (10.1%) had at least one positive rapid test. After cross-reference using FTA-abs a syphilis prevalence of only 2.9% was determined for the MCA.
The Abbott test has a greater positive predictive value (18% versus 16%) and a slightly greater specificity (93% versus 92%). The local rapid test on the other hand is more sensitive (52% versus 48%). Using the ASSURED method, the Abbott test showed advantages over the local test mainly in affordability, user-friendliness, and robustness.
Conclusions: This study shows that the syphilis prevalences world-wide might be much lower than is believed at this moment. The striking higher number of positives that was found using the rapid tests can be explained by the appearance of false positive tests due to cross-reactions that have occurred in these non-specific rapid tests.
The Abbott test can be considered superior to the currently used local test, although the local test is slightly more sensitive. However, the Abbott test has many advantages over the local test in the field. Hospitals and health centres in the MCA are advised to switch to the Abbott test.
P1138 Rash and fever in adults: the aetiology and clinical features
F. Tabak, A. Murtezaoglu, R. Ozaras, B. Mete, O. Tabak, A. Mert, R. Ozturk (Istanbul, TR)
The patient with fever and rash often poses an urgent diagnostic and therapeutic challenge for the clinician. The nonspecificity of many fever and rash syndromes mandates a systemic approach to diagnosis that includes a thorough history and physical exam as well as appropriate use of laboratory tests.
In this study, we aimed to determine the aetiology of fever and rash in 100 consecutive adult patients followed-up as in- or out-patients between May 2001 and April 2004 prospectively. Median age of the cases was 35 (range: 14–79); 45 were female and 55 male. The patients were divided into three groups according to the aetiology of fever and rash: infectious causes (50%), non-infectious causes (40%) and undiagnosed ones (10%). The most common type of rash was maculopapular and the most common five causes were measles, cutaneous drug reactions, varicella, adult-onset Still's disease and rickettsial disease. Viral diseases among infectious causes and cutaneous drug reactions among noninfectious causes were determined as the leading aetiologies among the aetiologies of fever and rash in adults.
Headache and conjunctivitis did relate strongly with infectious causes (p = 0.012, p = 0.011, respectively) and drug use and arthritis did so with non-infectious causes (p = 0.010, p = 0.037, respectively). The laboratory studies did not predict the aetiological categories. However, the increase in levels of acute phase reactants was greater in bacterial diseases when compared with those in viral diseases (ESR, p = 0.015; CRP, p = 0.006). The mortality rate was 5% and the causes of death were as follows: toxic epidermal necrolysis (2), adult-onset Still's disease (1), staphylococcal toxic shock syndrome (1) and graft-versus-host disease (1).
The main aetiologies of the patients admitted with rash and fever are measles, drug reactions, varicella, adult-onset Still's disease and rickettsial disease. However the diagnosis for every case includes use of epidemiology, detailed history, physical exam, and appropriate use of laboratory tests.
P1139 Presence of STa, STb and LT enterotoxin genes in Klebsiella strains isolated in different hospitals from gastrointestinal tracts
B. Maczyñska, U. Kasprzykowska, A. Zychowicz, A. Przondo-Mordarska, M. Bartoszewicz, D. Smutnicka (Wroclaw, PL)
Objectives: Klebsiella infections pose a significant problem, especially in paediatric wards where they can cause various nosocomial infections, such as serious diarrhoea in children and infants. Due to the fact that the microbes can also constitute normal flora of the alimentary canal, the key factor in considering them as an aetiological factor in diarrhoea is the determination of the presence of heat-stable and heat-labile enterotoxins. Therefore, the purpose of the paper was to investigate the genetic basis of the expression of enterotoxins (STa, STb and LT) among the selected strains isolated from diarrhoea cases in the hospitalised children. Because enterotoxin genes are encoded on the plasmids, which facilitates their transmission and spreading in nosocomial strains of Enterobacteriaceae, the paper also aimed to determine the frequency of occurrence of these genes in the Klebsiella species isolated in various clinical centres in Poland.
Methods: The research included 61 clinical Klebsiella strains isolated from faeces samples taken from children with diarrhoea symptoms hospitalised in various paediatric wards. Klebsiella strains not originating from diarrhoea cases but isolated from the blood and respiratory tract of hospitalised patients served as a control group. The presence of genes encoding ST and LT enterotoxins was detected using the PCR method with primers designed for sta, stb and lth genes described for E. coli.
Results: Research conducted indicates that in the plasmid DNA of the majority (85%) of Klebsiella strains isolated from diarrhoea cases, enterotoxin-encoding genes (sta, stb, lth) were detected. The 43% of the strains had genes for one enterotoxin, 39% genes for two endotoxins and 3% of strains showed the presence of three genes (sta, stb, lth). In the Klebsiella strains isolated from blood and the respiratory tract (negative controls), no sta, stb or lth genes were found.
Conclusions: The presence of enterotoxin-encoding genes have very important significance in diagnosing of aetiological agent of children diarrhoea. A direct correlation was shown between the frequency of individual gene detection and the clinical centre in which the given strain was isolated. Detection of enterotoxin-encoding genes in Klebsiella using the PCR method was regarded as useful in routine diagnosing of Klebsiella related diarrhoea cases.
P1140 Immunoreactivity studies on synthetic peptides deriving from variable domain IV of Chlamydia trachomatis major outer membrane protein
S. Klimashevskaya, T. Ulanova, A. Burkov, A. Obriadina (Nizhnij Novgorod, RU)
Objectives: The amino acid sequence of the major outer membrane proteins (MOMPs) from Chlamydia trachomatis serovars are predominantly conserved, but have four variable domains (VDs). The major neutralising and serotyping antigenic determinants are located in VD-I, VD-II and VD-IV.
The aim of this study was to identify the location of immunodominant regions and to study the relationship between the sequence heterogeneity and immunoreactivity by detecting antigenic reactivity of synthetic peptides derived from VD-I, VD-II and VD-IV of the C. trachomatis MOMP.
Methods: Unique 25-mer long (n = 281) overlapped by 4aa peptides derived from VD-I (aa65–127), VD-II (aa139–202) and VD-IV (aa286– 362) sequences corresponding to 9 different C. trachomatis serovars (A, C, D, E, F, H, L1, L2, L3) were designed and synthesized. The antigenic reactivity was detected by indirect enzyme immunoassay with known anti-C. trachomatis positive (N = 30) and negative (N = 30) sera.
Results: A strong antigenic region was identified within the MOMP VD IV at amino acids 302–345. The percent amino acid homology between different serovar sequences encompassing this region varied from 43.2% to 95.5%. All except one peptide (serovar C) contained the common motif TTTLNPTIA previously described as the minimal size of the antigenic epitope. However, the range of immunoreactivity with positive serum specimens varied from 50% to 93% for peptides with the common sequence and was 25% for synthetic peptide with substitution of two amino acids in this sequence. The most immunoreactive peptide (QPKSATAIFDTTTLNPTIAGAGDVK) detected 93% of the positive sera with the highest signal to cutoff ratio. This peptide was derived from the C. trachomatis serovar E.
Conclusion: The reactivity of the antigenic epitopes (s) may be differentially affected by neighbouring amino acids. Diagnostic test development requires careful selection of sequence variants, but the length and exact position of diagnostic targets also must be chosen carefully.
P1141 Rapid diagnosis of experimental meningitis by measuring bacterial heat flow in cerebrospinal fluid samples
A. Trampuz, A. Steinhuber, M. Wittwer, S.L. Leib (Basle, Berne, CH)
Background: Rapid diagnosis and therapy improves the outcome of bacterial meningitis. Growth of microorganisms generates heat flow that can be detected by calorimetry. Here we used isothemal calorimetry (detection limit ∼0.3 μW) to assess bacterial growth in cerebrospinal fluid (CSF) from rats with pneumococcal or meningococcal meningitis.
Methods: 11-days-old Sprague-Dawley rats were infected intracisternally with 10 μL saline containing 1 × 107 cfu/mL S. pneumoniae (n = 5) or 1×109 cfu/mL N. meningitidis (n = 5). Controls were injected with heat-inactivated N. meningitidis (n = 2) or sterile saline (n = 2). CSF was obtained at 18 h after infection and cultured quantitatively (5 μL). For calorimetry, 10 μL of CSF were added to 3 mL brain heart infusion for S. pneumoniae or tryptic soy broth for N. meningitidis. Heat flow was monitored at 37°C for 4 days using a 48-channel microcalorimeter (3102 TAM III, Thermometric, Järfälla, Sweden). Serial dilutions of known bacterial counts were used to determine the calorimetric detection limit. Positivity was defined as heat flow ≥10 μW; time to positivity (TTP) was calculated from the start of calorimetric measurement.
Results: The table shows mean values ± SD of CSF bacterial counts at 18 h after infection, calorimetric TTP and peak heat flow. The distinct heat flow curves were characteristic for the causative pathogen und independent from the initial concentration. The calorimetric detection limit per mL was 2 cfu S. pneumoniae (TTP = 13.9 h) and 4 cfu N. meningitidis (TTP = 19.5 h).
| Intracisternal injection | Bacterial count in CSF (cfu/mL) | Calorimetric TTP (h) | Peak heat flow (μW) |
|---|---|---|---|
| S. pneumoniae (n = 5) | 1.5×108 ±5.5×107 | 1.5±0.2 | 331±31 |
| N. meningitidis (n = 5) | 1.3×106 ±2.6×105 | 1.7±0.3 | 79±9 |
| heat-inactivated (n = 2) | 0 | Negative | <10 |
| Saline control (n = 2) | 0 | Negative | <10 |
Conclusion: Microcalorimetry is a rapid and sensitive method for the diagnosis of bacterial meningitis, allowing detection and potentially identification of pathogens from 10 μL CSF in <2 h.
P1142 Identification of Helicobacter pylori in gastric biopsy and resection specimens
T. Babic, H. Basic, B. Kocic, P. Stojanovic, B. Miljkovic Selimovic (Nis, RS)
Objectives: To compare the sensitivity of detecting H. pylori in gastric biopsy and resection specimens using classical morphologically oriented detection methods (haematoxylin and eosin (HE) stain, modified Giemsa stain) with immunohistochemistry using a commercially available anti-H. pylori antibody (Dako, Denmark).
Methods: Gastric antral biopsy specimens showing chronic gastritis (26 cases) together with tissue blocks from gastrectomy specimens for duodenal ulcer were histology reviewed. The paraffin sections were stained with traditional histological HE and modified Giemsa and immunoenzymatic by alkaline phosphatase antialkaline phosphatase (APAAP) method for the identification of H. pylori.
Results: The presence of chronic gastritis was confirmed in the 26 gastric biopsy specimens. A diagnosis of duodenal ulcer was confirmed in the mucosa from the gastrectomy specimens. The HE, modified Giemsa and immunoenzymatic treated sections were carefully examined for the presence of H. pylori. HE-stained H. pylori appeared as slightly basophilic, spiral-shaped organisms attached to the apical surface of the surface mucus cells. However, curved bacteria were only detected when found in great numbers. Using a modified Giemsa stain, the spiral-shaped bacteria of H. pylori stained blue, were attached to the brush border of the gastric foveolar epithelial cells and inside gastric pits. In some cases masked bacteria hidden within mucus were obvious only in immunostained preparations (red deposits). In addition, on sections stained with modified Giemsa coccoid forms, which were particularly seen in secions from resection specimens, caused some uncertainty. These coccoid H. pylori were obvious in immunostained preparations. Immunoenzymatic staining can be performed on cryostat and paraffin tissue sections, but reaction was more intense and diffuse in a cryostatic sections. H. pylori was identified in 34.6% sections stained with HE, but it could be identified with greater frequency in sections stained with modified Giemsa (73.0%). It could be detected at a still greater frequency in staining with APAAP (92.3%). Immunohistochemistry was positive in all cases in which H. pylori was detected by other methods.
Conclusion: Immunohistochemical identification of Helicobacter pylori by the APAAP procedure is a highly sensitive and easy to use method for detecting this organism.
P1143 Gastric juice urease test for Helicobacter pylori detection
P. Pirpiris, P. Sergouniotis, F. Sergouniotis, E. Papoylia (Amfissa, GR)
Objectives: The purpose of our study was to demonstrate the sensititivity, specificity and accuracy of gastric juice urease test and anti H. pylori IgG antibodies compared to the histological examination for HP.
Methods: We studied 55 patients age range 20–85 years, during the years 2005 and 2006. Forty-eight out of 55 patients underwent upper gastrointestinal endoscopy and had up to three gastric biopsy specimens taken for histological examination for HP. For each patient, 2–3 mL gastric juice was collected and 1 mL in the supernatant was tested for rapid urease reaction. We use per os thin “paediatric” Levin N°10 or N°11 or the gastroscopio to obtain gastric juice samples. Also blood samples were taken for the estimation of anti H. pylori IgG antibodies.
Results: The results are shown in Table 1 . Based on histology, the sensitivity of gastric juice urease test was found 97.4%, specificity 77.8%, positive predictive value 95% and negative predictive value 87.5% (48 patients) and for anti H. pylori IgG antibodies 65.0%, 83.3%, 92.8%, 41.6%, respectively (26 patients).
Table 1.
| Gastric juice CLO test | Biopsy | Anti H. pylori IgG | |
|---|---|---|---|
| Positive | 45 | 39 | 17 |
| Negative | 10 | 9 | 15 |
| Total | 55 | 48 | 32 |
Conclusions: It seems that the gastric juice urease test is an alternative non-invasive method by using Levin for the collection, to the invasive gastric biopsy specimens with little inconvenience to the patient, accurate, simple, cheap, rapid and reliable method for the detection of H. pylori. It should be used for detecting H. pylori infection in patients with coagulopathy, in children under the age of 5 years, for large epidemiologic studies, for the confirmation of H. pylori eradication and in several cases where 13-C-urea breath test contraindicated. In contrast the anti H. pylori IgG antibodies test has a low sensitivity and the results should always be interpreted in connection with other methods.
P1144 Antibody decay profiles during Yersinia enterocolitica O:3 infections: a follow-up study of 94 patients
E. Rasmussen, P. Schiellerup, T. Dalby, K. Krogfelt (Copenhagen, DK)
Objective: An enzyme-linked immunosorbent assay (ELISA) was employed to measure immunoglobulin G (IgG), IgM and IgA classes of human serum antibody in sera from adult Danish patients with a culture proven Yersinia enterocolitica O:3 infection at various intervals after debut of symptoms.
Methods: Yersinia enterocolitica O:3 LPS was used as coating antigen in the ELISA test. A total of 220 sera drawn between 19 and 1,038 days after onset of disease from 94 patients with verified Yersinia enteritis were examined. A control group of 100 sera from healthy Danish blood-donors were tested to determine the cut-off for negative results.
Results: With a 95th percentile of specificity, measurement of IgG, IgM, and IgA specific for Yersinia enterocolitica O:3 LPS within a month after infection showed a sensitivity of 50%, 100% and 83% respectively. 31 to 60 days after infection a sensitivity of 79% was found when combining all three immunoglobulin classes (IgG 40%, IgM 77% and IgA 56%) whereas a combined sensitivity of 61% was found 61 to 90 days after infection (IgG 29%, IgM 61% and IgA 25%). At 91 to 180 days after infection the combined sensitivity were 51% (IgG 38%, IgM 32% and IgA 30%). The antibody decay profiles for the three different immunoglobulin classes for the individual patients exhibited a large degree of variation.
Conclusion: At follow up of the patients, IgM and IgA levels were elevated up to 2 months from onset of infection. IgG levels were not elevated during infection, the highest sensitivity 50% was found within a month from infection. A rapid decrease in antibodies specific for Yersinia enterocolitica O:3 were shown. Elevated levels of antibodies in serum following Yersinia gastroenteritis were limited to the two first months after infection.
P1145 Development of hybridomas secreted Streptococcus suis 2 serotype-specific monoclone antibody
X.Z. Pan, J.C. Ge, C.J. Wang, X.F. Li, F. Zheng, J.Q. Tang (Nanjing, CN)
Objective: For the development of a diagnostic reagent and vaccine of Streptococcus suis (S. suis), monoclonal antibodies (MAbs) against S. suis were obtained by the fusion of SP2/0 murine myeloma cells and spleen cells from BALB/c mice immunised with a whole-bacterial-antigen of S. suis ZY/H33 (serotype 2).
Methods: Formalin-killed, 12 h cultures of ZY/H33 strain were used to immunise mice. After the booster inoculation, the spleen of the mouse was recovered and fused with myeloma cell line SP2/0. Hybridoma supernatants were screened for S. suis type 2 antibodies by the indirect ELISA. The isotypes of MAbs were determined. For Western blot, the sample of sonicated bacteria antigen was subjected to 12% SDS-PAGE, then transferred to nitrocellulose membrane. After blocked with 5% skim milk, the membrane was incubated with anti-S. suis type 2 (ZY/H33) ascite fluid in a 1:100 dilution in TBST for 1 h, followed by the incubation with HRP-conjugated anti-mouse antibody IgG in a 1:5,000 dilution in TBST for another 1 h. After washings in TBST and using DAB as substrate, the reacting bands were visualised.
Results: A total of 201 hybridomas were tested in ELISA, out of which 6 hybridomas, namely 1A8, 1B4, 2A9, 3C2, 4B4, and 4B9, showing a positive reaction were selected for further characterisation. The immunoglobulin classes of Mabs 1B4, 2A9, 4B4 and 1A8, 4B9, 3C2 were IgM and IgG3, respectively.
In the Western blot, MAb 2A9 reacted with a major band of an estimated molecular mass of 90 kDa. As show in Fig. 1 , MAb 2A9 reacted with a single 90 kDa protein of S. suis type 2 such as ZY/H33, Habb and S10, while S. suis type 3, S. faecalis, Pneumococcal Pneumonia without reaction. And in the following test, all S. suis type 2 including international and Chinese isolates were shown to react with MAb 2A9 using immunoblotting, while negative results were obtained in the other serotypes of S. suis and non-S. suis strains.

Conclusion: In conclusion, 2A9 is a serotype-specific monoclone antibody. It is a robust tool for S. suis type 2 test.
P1146 Fine-needle aspiration has a very low sensitivity in the diagnosis of tuberculous cervical lymphadenitis in Iran
M. Hasibi, J. Rezaei, Z. Madani Kermani, M. Asadollahi, M. Abouzari, A. Rashidi (Tehran, IR)
Objectives: To assess the issue of fine-needle aspiration (FNA) sensitivity for early tuberculous cervical lymphadenitis in Iran for the first time.
Methods: The results of FNA in 46 consecutive patients (12 Afghan, 34 Iranian) with cervical lymphadenopathy who had a definitive diagnosis of tuberculous cervical lymphadenitis were retrospectively evaluated. Definitive diagnosis was established on the presence of caseous necrosis with epitheloid-cell granulomas at histopathologic evaluation of the nodes after excisional biopsy. Smear (with Giemsa staining) and culture (with Ziehl–Neelsen's staining) for acid-fast bacilli on FNA specimens were available in 35 and 29 cases, respectively. All patients had a negative serology for HIV infection. A skin tuberculin test (PPD) had been performed on all patients.
Results: There were 13 males and 33 females ranging in age from 12 to 70 years with a mean age of 39.6 years. Lymphadenopathy ranged in duration from 15 days to 2 years with a mean of 55 days. PPD test, smear and culture were positive in 41 (89%), 1 (2.8%) and 1 (3.4%) cases, respectively. FNA results were nonspecific acute and/or chronic inflammation without necrosis or granuloma in 34 (73.9%), granuloma without necrosis in 3 (6.5%), malignancy in 5 (10.8%) and granuloma with caseous necrosis in 4 (8.7%) patients. Based on these results, if one defines a positive test as one revealing granuloma with caseous necrosis which is the characteristic finding of tuberculous lymphadenitis, then the sensitivity of FNA will be only 8.7%. Even if we regard the samples with granuloma but without necrosis as positive, the sensitivity will still be only slightly higher than 15%. Of particular note, more than 84% of samples did not show a granuloma, which made TB involvement seem less likely. Also, smear and culture could not increase the sensitivity significantly, each being positive in less than 4% of patients only.
Conclusion: The advantages of an early definite diagnosis even with the aid of an invasive procedure such as excisional biopsy should be carefully assessed against the disadvantages of high false negative rates obtained from a relatively non-invasive procedure like FNA.
P1147
Prevention of perinatal GBS infections: impact of ChromID® StreptoB, a new chromogenic medium on suscpetibility testing
W. Chaker, S. van den Wijngaert, A. Dediste (Brussels, BE)
Objective: Compare the results of antimicrobial susceptibility testing (AST) of Group B Streptococcus (GBS) by disk diffusion performed on the colonies directly picked from chromID® Strepto B agar (bioMérieux) with AST performed on colonies grown on Columbia agar with sheep blood supplemented with nalidixic acid (CNA).
Material and Methods: ASTs were performed on Mueller Hinton II agar enriched with 5% sheep blood (MHB) inoculated with a 0.5 McFarland suspension and incubated overnight at 37°C in 5% CO2. The Kirby-Bauer disk diffusion AST method was performed for penicillin, vancomycin, erythromycin, clindamycin, and levofloxacin. Zone diameters were measured, and the GBS were categorised as susceptible (S), intermediate (I) or resistant (R) according to CLSI guidelines (supplement M100-S14). Antimicrobial disks and plates were provided by Oxoid and CNA agar plates by bioMérieux.
Results: For the 18 strains preliminarily tested, we observed an overall concordance of 100% for penicillin, vancomycin, clindamycin and levofloxacin independently from the susceptibility or resistance of the bacteria to the antibiotic. The same diffusion-zone was observed with clindamycin by using both methods for the GBS strain that harboured the MLSb resistance. With erythromycin, only one minor discrepancy has been obtained (I vs. S) with respective diameters of 20 and 22 mm with colonies from chromID® Strepto B agar and CNA.
Conclusions: These first results are promising and will be confirmed by extending the sample size. This methodology with chromID® Strepto B agar directly associated with AST could avoid subculture on blood agar and permit an earlier result for the clinicians.
P1148 Impact on viral pathogen yield of the absence of cells in nasopharyngeal aspirate specimens
D. Piérard, D. Stevens, J. Mignolet, L. Moriau, K. Vergalle, S. Lauwers (Brussels, BE)
Objectives: To evaluate the impact of the absence of cellular material on the pathogen yield in nasopharyngeal aspirate specimens (NFAS) submitted for viral examination, as possible criterion for specimen rejection.
Methods: To check for the presence of a visible cell pellet, all NFAS submitted during winter 2005–06 were centrifugated after mixing with DMEM with 2% fetal calf serum and vortexing (and removal of excessive mucus if needed), before being resuspended and processed. Specimens showing a pellet were classified as C+, those without as C–. All specimens were examined by RSV Respi-Strip® antigen test (Coris, Gembloux, BE), by a multiplex in-house PCR protocol for respiratory viruses and by viral isolation on cell culture. Results were compared for C– and C+ specimens.
Results: Out of 634 samples, 537 were C+ and 97 C-. RSV antigen detection was positive in 33% C+ and 19% C– (p = 0.005). PCR was positive for RSV in 42% C+ and 30% C– (p = 0.029), for any virus in 59% C+ and 43% C– (p = 0.005). Culture was positive for RSV in 26% C+ and 16% C– (p = 0.021) and for any virus in 33% C+ and 19% C– (p = 0.005).
Conclusion: If we assume that the percentage of truly infected patients is similar in those with C– and C+ specimens, we conclude that the yield of viral pathogens is significantly diminished in C– samples, culture being mostly influenced, followed by antigen detection and PCR. However, since it is generally not possible to obtain a replacement sample, rejecting those samples could have a negative impact on patient care.
P1149 Evaluation of a new automated urine cell analyser (Sysmex UF-1000i) for bacteriological urinalysis
H. von Wulffen (Hamburg, DE)
Objectives: We wanted to assess to what extent the UF-1000i (SYSMEX) automated urine cell analyser can support bacteriological urinalysis, in particular to find answers to the following questions: can it replace Gram stain? Can we dispense with culture in case of negative results for bacteria, leukocytes and yeast-like cells? Is there a suitable cut-off for number of bacteria to select urine specimens for direct susceptibility testing?
Methods: Urine specimens with a request for urine culture were processed using the UF-1000i in parallel with Gram stain and culturing overnight of 0.001 mL undiluted urine each onto UTI, CNA, and Sabouraud agar using calibrated loops. Specimens were also tested for inhibitory activity using spore plates.
Results: To select out negative specimens UF-1000i results were considered positive if white blood cells exceeded 20/μl and/or bacterial particles were counted over 50/μl and/or yeast-like cells exceeded 25/μl. In comparison, cultures yielding organisms at 103 CFU/mL or higher were considered positive. On this basis out of 363 urine specimens evaluated 42% of the UF-1000i results were true positive, 30% true negative, 22% false positive, and 6% false negative. However, only 2/24 false negative results yielded 106 CFU/mL, all others had 104 CFU/mL or lower, most of them with mixed flora, thus reducing significantly false negative results to less than 0.6%. Only looking at the specimens without inhibitory activity (n = 232) and using a cut-off of 105 CFU/mL or higher for predicting a urinary tract infection, the UF-1000i yielded a sensitivity of 97.6%, a specificity of 86.7%, a positive predictive value of 80.0%, and a negative predictive value of 98.5% when using a cut-off of 100 bacterial particles/μl. This cut-off also proved useful for selecting those specimens suitable for direct susceptibility testing.
Conclusions: For our purposes the UF-1000i could replace Gram stain of urine specimens and thereby reduce time-consuming microscopy. The low rate of truly false negative results around 0.6% appears to be tolerable and would allow dispensing with culture in case of negative UF-1000i results and thus further reduce workload in the laboratory. Specimens from certain patients (e.g. haematology patients with cytopenia) or certain materials (e.g. suprapubic bladder aspirates) should be cultured in any case.
P1150 Community-acquired Legionnaire's disease in the urinary antigen era
G. Tolchinsky, L. Mateu, M. Pedro-Botet, N. Sopena, M. Garcia-Nuñez, M. Sabria, C. Rey-Joly (Barcelona, ES)
Objectives: At present urinary antigen detection is the hallmark of the diagnosis of Legionnaires' disease and may have influenced the demographic and the clinical data of this disease. The aim of this study was to describe demographic data, risk factors, clinical characteristics and outcome of patients with community-acquired pneumonia (CAP) by Legionella pneumophila serogroup 1 diagnosed using urinary antigen test.
Method: Patients were selected from a database of Legionella pneumonia prospectively collected from 1983 to 2006 in a 630-bed university hospital. Only patients with CAP diagnosed by Legionella urinary antigen detection were included.
Results: We studied 168 patients, most cases (97.1%) being sporadic. Males predominated (81%) with a mean age of 57.5 years (range 19–95 years). 85.1% had at least one risk factor, smoking habit (48.2%), alcohol intake (24.4%) or underlying diseases (56.5%) including chronic respiratory diseases (18%), diabetes (16.1%), cancer (10%), immunosuppressive therapy (9.5%) and HIV (9.5%). At presentation 94% had fever, 69.6% cough, 44.6% dyspnea and 42.9% expectoration. 48.8% presented neurological (35.1%) or gastrointestinal (26.8%) symptoms. Analytical data showed leucocytosis (46%), increased AST (42%), hyponatraemia (20.5%) and increased creatine phosphokinase (24.1%). 48.2% patients (81/130) had Fine score ≥ III. 92% were hospitalised, 17.3% being admitted to the ICU and 13% needing mechanical ventilation. 32.1% had received previous β-lactams and 34% began appropriate treatment within the first 3 days of symptom onset. Complications occurred in 84 patients (50%), with respiratory failure (46%), renal insufficiency (10.7%) and septic shock (8.4%) being of note. Radiological complications appeared in up to 17.5%, the most frequent being pleural effusion in 13.1% and bilateral progression in 16 (9.5%). 15 patients (8.9%) died due to the pneumonia and only one patient relapsed.
Conclusion: Nearly 15% of patients with Legionella CAP had no risk factor and almost 50% no underlying diseases. When faced with CAP, the “atypical pneumonia” syndrome should still suggest Legionella infection. Legionella CAP continues to be a severe disease based on hospitalisation, respiratory failure and ICU admission.
P1151 Utility and clinical impact on the reported aetiology of a urinary antigen-detection test for Streptococcus pneumoniae in community-acquired pneumonia at the emergency department
G. Ezpeleta, C. Ezpeleta, J.A. Alava, J. Unzaga, V. Cabezas, I. Gerediaga, B. Amezua, R. Blanco, R. Cisterna (Bilbao, ES)
Objectives: The diagnosis of severe pneumococcal infections is inadequate, relying heavily on culture of Streptococcus pneumoniae from blood or other normally sterile fluids, and is severely limited by prior administration of antibiotics. We evaluated prospectively the Binax NOW S. pneumoniae urinary antigen test, a rapid immunochromatographic (ICT) assay, for the diagnosis of bacteraemic pneumococcal infections in adult patients with community lower respiratory tract infection (CLRTI) at the emergency room.
Methods: One thousand, two hundred and thirty three adult patients with CLRTI who were admitted to the hospital were studied prospectively between January 2004 and September 2006. The ICT test was performed on fresh unconcentrated urine following the manufacturer instructions. The results of the ICT test were compared with blood culture ones, and sensibility, specificity and likehood positive and negative ratios were calculated.
Results: The ICT assay was positive in 298 (24.15%) of 1,233 patients enrolled into the study and in 78 (79.59%) of 98 patients with pneumococcal pneumonia confirmed by conventional methods. The test revealed a sensitivity of 75.9% (71.2–83.1) and a specificity of 94.0% using conventional microbiological criteria as the gold standard. The likehood positive ratio value was 12.65, and the negative one was 3.9. The diagnostic yield of pneumococcal pneumonia was increased using ICT combined with conventional methods. Antigen was still detectable in 53% of patients during treatment
Conclusion: The ICT to detect S. pneumoniae urinary antigen is therefore a rapid and useful method for diagnosing pneumococcal pneumonia. Detection of urinary antigen is a valuable, sensitive, and rapid test for the early diagnosis of bacteraemic pneumococcal infections in adult patients, even after antibiotic treatment has commenced
P1152 Sampling respiratory epithelial cells by nasal or nasopharyngeal flocked swabs
S. Castriciano, G. So, S. Buracond, M. Savarese, D. Triva, M. Smieja (Brescia, IT; Hamilton, CA)
Objective: Nasal swab (NS) collection is less invasive than nasopharyngeal swab (NPS) collection, and may facilitate wider surveillance and diagnosis of respiratory viral infections. We studied NS collection of two new swab designs to determine whether NS could sample respiratory epithelial cells as well as NPS.
Methods: A total of 52 volunteers were studied. 17 had one Copan pernasal Flocked Swab NPS and three different Copan Nasal Flocked Swabs (regular, round, and flat) administered and collected by a physician or trained research staff. The remaining 35 subjects had 3 NS performed without an NPS. For all individuals, the order of the three NS was randomised, while the NPS was done last for the subgroup of 17 volunteers. The swabs were placed in a 1-mL tube of Copan Universal Transport Medium, vortexed, and centrifuged, with the residual cell pellets resuspended in 1 mL of PBS. 25 μL of suspension were placed in wells on a glass slide, dried, fixed and counterstained with FITC labelled monoclonal antibody. Respiratory epithelial cells were quantitated using a fluorescent microscope at 400× magnification by an experienced microscopist blinded to swab type while enumerating. An average count from 4 fields was calculated when 10 or more epithelial cells were present per high-powered field (hpf); 10 fields were averaged if there was fewer than 10 cells/hpf.
Results: Among the 17 subjects with parallel NS and NPS specimens, mean(SD) cells yields were: 73.3(31.2), 83.1(35.5), and 92.5(35.1) cells/hpf for the regular, round, and flat NS, respectively; and 105.9(44.0) cells/hpf for NPS. The difference between the regular NS and NPS was statistically significant (P = 0.02), whereas the two new NS designs (round and flat) were not different from the NPS (P = 0.11 and 0.34, respectively). Among all 52 subjects, the mean(SD) cell yields were 65.2(31.3), 81.8(42.4), and 81.8(41.7) for regular, round, and flat swabs, respectively. The two new NS designs improved cell yield by 16.5 cells/hpf (95% CI: 8.0, 25.0, P < 0.001) compared with the regular NS. The 3 NS swabs were equally well tolerated and caused less discomfort than the NPS.
Conclusions: An improved flocked design for NS allows sampling equivalent to NPS with less discomfort. If validated in symptomatic patients, this will have important implications for surveillance swabbing or diagnosis in patients currently not benefiting from respiratory diagnostics.
P1153 Comparative study of Copan Venturi Transystem and Medical Wire and Equipment Transwab microbiological swabs, using the roll-plate method
K. Birrell (Hastings, NZ)
Objectives: To compare/contrast Copan with Medical Wire and Equipment (MWE) swabs to determine the differences/similarities in numbers of organisms retrieved from each swab type. Swabs which maintain viability of organisms are preferable over those which do not (avoiding overgrowth). Systems maintaining organisms for extended time, without overgrowth, are beneficial due to potentially prolonged transport times for example. The systems which best maintain organisms, representing the clinical picture will aid the laboratory in the correct determination of results.
Methods: The roll-plate method was used in these viability and overgrowth studies. This is qualitative/semi-quantitative method rather than quantitative (the elution method). Many clinical laboratories use the roll-plate method to directly inoculate media. Organisms tested: Streptococcus pyogenes ATCC 19615, Streptococcus pneumoniae ATCC 6305, Pseudomonas aeruginosa ATCC 27853, Haemophilus influenzae ATCC 10211, Neisseria gonorrhoeae ATCC 43069, Escherichia coli ATCC 25922 and Candida albicans ATCC 60193. This study was performed following the Clinical and Laboratory Standards Institute (CLSI) Quality Control of Microbiological Transport Systems; Approved Standard M40-A (Volume 23, Number 34).
Results: A significant variance in the number of colony forming units (CFU) was seen between the systems. In most cases twice as many CFU were released by the MWE Transwab than by the Copan Transystem. Two of the organisms tested showed a greater than a four-fold difference between the release of CFU from the Copan Transystem and the MWE Transwab.
Conclusions: The MWE Transwab performed better in this study. Both systems employed Amies Clear media. A possible reason for the results may involve the amount of media; the MWE Transwab released more transport media from the system onto the agar plates than the Copan Venturi Transystem swabs. The weave of the MWE swab tips tends to open after the swab had been in the transport media aiding in media transfer. In contrast there was very little change in the weave of the Copan Transystem swab tips. This is potentially a factor in the difference between the results obtained in this study.
P1154 Rapid pathogen identification by MALDI-TOF mass spectrometry/SARAMIS database in clinical microbiological routine diagnostics
J. Gielen, M. Erhard, W. Kallow, M. Krönke, O. Krut (Cologne, Potsdam, DE)
Automated identification systems are commonly used in medium-to-high-throughput clinical microbiology laboratories. However, such systems are relatively slow because they depend on a bacterial growth and metabolic activity. Bacterial identification by mass spectrometry provides a promising way to accelerate pathogen identification, since it can be performed in a few minutes. In this study we compared performance of mass fingerprinting with established methods (VITEK 2/API, BioMérieux) in the clinical microbiology laboratory settings. A part of a colony grown on solid culture medium was transferred on the sample plate of a MALDI-TOF apparatus, disrupted by addition of matrix followed by the generation of mass spectra. The acquired spectrum was used for a search in the SARAMIS database (Anagnostec), which holds reference spectra for common pathogens. Within three months we investigated 1,616 independent pathogen isolates representing more then 50 bacterial and fungal species. MALDI/SARAMIS correctly identified 94% and 96% of samples to species and genus level, respectively. The most exact identification was achieved for Staphylococci (99%) and Enterobacteriaceae (98%), followed by nonfermenter (92%) and Streptococci (91%). Remarkably, MALDI/SARAMIS method produced no misidentifications. The rate of samples identified as “unknown” was about 3%. In 22 cases where results of MALDI/SARAMIS identification were in discrepancy with VITEK the correctness of the mass spectrometry method was confirmed by sequencing. The typical time for the measurement and identification of a single isolate was about 3–5 minutes. In conclusion, the application of MALDI-TOF/SARAMIS represent a simple, rapid, robust and in the long run inexpensive method for the bacterial and fungal identification in the clinical microbiological routine laboratory.
P1155 Flow-cytometric assay to detect group B streptococci in vaginal swabs
L. Zabel, M. Anliker, S. Nuding, H. Müller (Goppingen, Stuttgart, DE)
Objectives: Group B streptococci (GBS) are the leading cause of neonatal sepsis in the USA and appear to be a growing problem world-wide. The CDC guidelines and the German AWMF guidelines recommend screening pregnant women for GBS. Collection of cultures between 35 and 37 weeks gestation is recommended to improve the sensitivity and specificity of detection of women who remain colonised at the time of delivery. Women at risk should receive antibiotic prophylaxis during delivery. The screening-cultures from vaginal swabs require at least two days for culture and identification. Unfortunately many women are hospitalised for delivery, when the woman is already in labour and a screening result is not available by the time.
Methods: Using flow-cytometry and a monoclonal mouse antibody (C55560M Dunn laboratories, Germany) directed against GBS an assay was established, which detected GBS from liquid cultures (Todd Hewitt broth) of vaginal swabs after a short incubation time of 90 min. Results were received within 2.5 hours. The assay was applied to 121 vaginal swabs of women selected independent of diagnosis or hospitalisation and compared to conventional culture methods.
Results: The detection limit was determined to be approximately 1.88×103 cfu/mL, using serial dilutions of the reference strain Streptococcus agalactiae ATCC 13813.
A total of 15 (12.4%) were identified as carriers of BGS by both, culture and flow-cytometry. Taking only the culture into account, 18 (14.9%) of the vaginal specimens were positive for GBS. Flow-cytometric analysis showed a more increased positivity rate with 44 (36.4%) GBS-positive specimens.
Conclusion: The flow-cytometric assay provided physicians with results within 2–3 hours and was more sensitive than conventional culture. It therefore may be used to detect GBS in sufficient time, even if the patient is already in labour.
P1156
Phenotypic characteristics of 357 consecutively collected human isolates of Fusobacterium necrophorum ssp. funduliforme from Denmark
A. Jensen, L.H. Kristensen, J. Prag (Viborg, DK)
Objectives: Fusobacterium necrophorum causes severe infections like Lemierre's syndrome. However, F. necrophorum is often not correctly identified or the identification process may be very slow. The aim of this study was to phenotypic characterise human isolates of F. necrophorum for the purpose of establishing a rapid procedure for identification of F. necrophorum. In addition, we examined whether there were any variation in phenotypic characters between strains causing Lemierre's syndrome and strains causing localised infections.
Methods: Three-hundred and fifty-seven clinical isolates of Fusobacterium necrophorum from human infections were collected from all 16 clinical microbiological departments in Denmark and identified by routine phenotypic characters through a three year period. In addition the first 40 isolates collected were subjected to an extended phenotypic characterisation. Fifty-three isolates were from confirmed Lemierre's syndrome cases, 249 from localised infections in the head and neck, including cases of tonsillitis and peritonsillar abscesses and 55 isolates were from other localised F. necrophorum infections.
Results: All human isolates, but one, which differed in cellular and colony morphology, were identical in all phenotypic characters used in routine identification (Table 1 ). The first 40 isolates that were subjected an extended phenotypic characterisation were identical. However the isolate, which differed in routine characters, agglutinated chicken and human erythrocytes (Table 1). All isolates, but the outlier, were identified as F. necrophorum ssp. funduliforme. The outlier was identified as F. necrophorum ssp. necrophorum.
Table 1.
Phenotypic characteristics of Fusobacterium necrophorum ssp. funduliforme and F. necrophorum ssp. necrophorum from 357 human isolates, four animal isolates and the type strains of ssp. necrophorum ATCC 252S6 and ssp. funduliforme ATCC 51357.
| Characteristic | Subspecies |
|
|---|---|---|
| funduliformea | necrophorumb | |
| Routine phenotypic characters | n = 358 | n = 5 |
| Colony margins | Entire | Irregular |
| Colony surface | Glistening | Dull |
| Colony elevation | Pulvinate | Convex |
| Colony consistency | Creamy | Waxy |
| Cellular morphology | Coccoid pleomorphic rods | Pleomorphic rods |
| Gram stain | ÷ | ÷ |
| Smell of butyric acid | + | + |
| Fluorescence in Wood's light | + | + |
| β-haemolysi s on horse blood | + | + |
| Metronidazole susceptible | + | + |
| Kanamycin susceptible | + | + |
| Penicillin susceptible | + | + |
| Additional phenotypic characters | n = 41 | n = 5 |
| Catalase | ÷ | ÷ |
| Indole | + | + |
| Lipase | + | + |
| β-galactosidase | ÷ | ÷ |
| DNase | + | + |
| Fermentation of mannose | ÷ | ÷ |
| Fermentation of fructose | ÷ | ÷ |
| Fermentation of glucose | + | + |
| Hydrolysis of esculin | ÷ | ÷ |
| Propionic acid from lactate (GLC) | + | + |
| Propionic acid from threonine (GLC) | + | + |
| Butyric acid from glucose (GLC) | + | + |
| Chicken erythrocyte agglutination | ÷ | + |
| Human erythrocyte agglutination | ÷ | + |
356 human isolates, 1 type strain, 1 animal isolate
1 human isolate, 1 type strain, 3 animal isolates.
Conclusion: F. necrophorum may be identified and differentiated from other Fusobacterium spp. in less than 72 hours reliable on basis of cellular and colony morphology, Gram stain, sensitivity to kanamycin and metronidazole, the smell of butyric acid, chartreuse colour fluorescence and beta-haemolysis on horse blood agar. Subspecies funduliforme can be differentiated from ssp. necrophorum on basis of cellular and colonial morphology and the agglutination of human and chicken erythrocytes. Human isolates of F. necrophorum infections in Denmark are phenotypic identical with no differences between isolates from Lemierre's syndrome and other localised infections.
P1157 Dirofilaria and Wolbachia antigens to detect naturally acquired dirofilariosis in usual and occasional hosts
S. Gabrielli, G. Cancrini (Rome, IT)
Zoonotic dirofilarioses are mosquito-borne diseases due to several species of animal filarial worms present in many regions of the world. Dirofilaria immitis and D. repens are usual parasites of dogs and other carnivores that develop heartworm disease and subcutaneous dirofilariosis, respectively. Humans, unsuitable hosts, are occasionally infected, and develop abortive infections or syndromes due to migrating larvae that cause pulmonary nodules (mainly if the worm is D. immitis), and ocular pathologies or subcutaneous lesions (mainly if the worm is D. repens). These lesions always give rise to the suspicion of a tumour, so requiring, as primary measure, differential diagnosis. Dirofilariosis is easily diagnosed in dogs by microscopy/molecular methods, or by serology, whereas in humans it is possible only by microscopy/molecular methods applied to the worm surgically removed, due to the lack of reliable serological tests.
Recent researches evidenced that most pathogenic filariae, dirofilariae included, harbour bacterial endosymbionts belonging to the genus Wolbachia as stable components of the body. Experimental infections suggested that the continuous release of bacterial antigens could stimulate the host immune response and even switch the response against Wolbachia rather than against the worm. Moreover, it has been hypothesised a possible role of these bacteria in the filariosis pathogenesis. Aim of the present study was to confirm in humans and dogs naturally infected by D. repens and D. immitis the hypothesis that also Wolbachia-derived proteins can be antigenic, to compare response patterns against Dirofilaria and Wolbachia antigens in suitable and unsuitable hosts, and to evaluate the possible use of an ELISA test to diagnose human dirofilariosis before surgery.
To evaluate the antibody response against filarial polypeptides and Wolbachia surface protein we tested the sera of 86 dogs and 88 humans positive for the two dirofilariae, and showing different clinical status. The response against filarial polypeptides was mainly observed in humans affected by D. repens (both ocular and abortive infections), and in dogs with chronic disease. As far the anti-Wolbachia response, it was evidenced mainly in dogs affected by heartworm occult disease and in humans with pulmonary nodules (coin lesions), so confirming, in natural infections, its relationship with severe pathology, and suggesting a possible diagnostic use of the test developed.
P1158 Helcococcus kunzii isolation from prosthetic joint infection
M. Balejová, M. Fialová, N. Piskunová, P. Trubac, J. Motlová (Ceské Budejovice, Prague, CZ)
Objective: Identification of a rare isolate – Helcococcus kunzii – from prosthetic joint infection is presented.
Methods: Perioperative specimens from prosthetic joint infection were processed according to standard laboratory practice. Tissue specimens were homogenised in thioglycolate broth, components of explanted prosthesis were sonicated in Ringer solution. Homogenised and sonicated fluids were centrifuged and the sediment used for Gram staining and culture. Culture media, both solid and liquid, were incubated at 35–37°C aerobically and anaerobically for 7 days, while daily examined. The identification of the microorganism was performed by conventional methods and API 20 Strep set (bioMérieux). DNA sequencing of the 16S rRNA gene was performed using ABI Prism 310 Genetic Analyser (Applied Biosystems, Foster City, CA, USA). Nucleotide sequences were analysed with MegAlign software (DNASTAR, Regent St. Madison, WI, USA) and aligned to EMBL/GenBank. Antimicrobial susceptibility testing was determined using disc diffusion method and E-test system (AB Biodisk).
Results: Direct smear examination from the tissue specimen showed only erythrocytes. After 72-hour of incubation, growth of pinpoint, translucent, non-haemolytic colonies appeared on 5% sheep blood agar. A subculture from the thioglycolate broth yielded the same bacterial growth. A Gram stain of the isolate revealed Gram-positive cocci of various sizes in clusters. The identification in API 20 Strep (bioMérieux) resulted in profile no. 4102411 – Aerococcus viridans (75.6%). DNA sequencing of the 16S rRNA gene of the isolate showed 99.8% similarity with H. kunzii. The strain was susceptible to ampicillin, erythromycin, clindamycin, vancomycin, rifampin and resistant to ciprofloxacin, gentamicin. The MICs of penicillin and clindamycin, determined by the E-test system were 0.500 and 0.032 mg/L, respectively.
Conclusions: H. kunzii is a rare isolate in clinical specimens. Identification of this bacterium on the basis of phenotypic tests is difficult. Absence in databases in the most of the commercial products makes impossible to identify this organism by those means. DNA sequencing of the 16S rRNA gene identified the aetiologic agent of prosthetic joint infection in our case.
P1159 In vitro evaluation of hand-held test kits, immunofluorescence microscopy, ELISA, and flow cytometric analysis for the rapid presumptive identification of Yersinia pestis
H. Tomaso, P. Thullier, E. Seibold, V. Guglielmo, A. Buckendahl, L. Rahalison, H. Neubauer, H. Scholz, W. Splettstoesser (Munich, DE; Grenoble, FR; Antananarivo, MG; Jena, DE)
Plague caused by Yersinia pestis is a zoonosis mainly circulating among rodents and their fleas, which can transmit the disease to humans. Due to the continuing threat of outbreaks in endemic regions and the potential use of Y. pestis as a biological agent rapid diagnostic tools are needed. The aim of this study was to evaluate the performance of immunological assays targeting the fraction 1 capsular antigen (F1 CA) for the rapid identification of Y. pestis.
An in house developed immunochromatographic test strip (ICT), the Plague BioThreat AlertTM test strip (BTA; Tetracore, USA), ABICAPTM columns (Senova, Germany), an ELISA (Seramun, Germany), immunofluorescence microscopy and flow cytometric analysis were compared. The specificity of the assays was determined using a representative collection of Y. pestis strains and clinically relevant bacteria. Detection limits were assessed using serial dilutions of F1 CA. Additionally, ease of handling, costs, and turn-around time were evaluated. All assays identified Y. pestis isolates correctly and detected no other bacteria. The in-house developed ICT strip proved to be specific, highly sensitive detecting 3.3 ng of F1 CA and was the most convenient and economic test. In contrast to immunofluorescence microscopy, ELISA can be used as a screening tool and for quantification of F1 CA. Flow cytometric analysis is sophisticated and too expensive for laboratories in most areas where plague is endemic. Commercially available products will become more attractive when certified for diagnostic purposes and available at lower costs.
P1160 Development of a rapid fluorescence assay in 96 well plate assay for multiple antibiotic resistant Salmonella typhimurium
N. Coldham, L. Randall, L. Piddock, M. Woodward (Surrey, UK)
Objectives: Multiple antibiotic resistant (MAR) mutants of Salmonella enterica and E. coli are characterised by MIC increases of 2–8 fold to unrelated antibiotics including tetracycline, chloramphenicol, some penicillins and quinolones. They have been associated with clinical disease in their own right and provide a platform for progression to high level antibiotic resistance. Previous studies have indicated that the AcrA/B-TolC efflux pump system and certain porins cooperate to reduce the uptake of antibiotics and are important effectors of MAR. Many substrates of AcrA/B-TolC are divalent cations and include fluoroquinolone antibiotics and ethidium bromide. In the present study, intracellular concentration of the fluorescent probe bis benzimide (H33342) was evaluated as a rapid single test for MAR Salmonella Typhimurium.
Methods: S. Typhimurium (SL1344), isogenic MAR mutants (n = 4) and isogenic mutants with defined deletions in acrB, tolC, ompC and ompF were cultured overnight in LB broth and diluted with phosphate buffered saline to an optical density of 0.1 (600 nm). The strains were incubated in 96 plate wells with bis benzimide (2.5 mM) and the fluorescence recorded for 30 minutes using a Fluostar plate reader at excitation and emission wavelengths of 350 and 460 nm respectively. Sensitivity of these strains to the efflux pump inhibitors Phe-Arg-b-naphthylamide (166 ug/mL) and carbonyl cyanide m-chlorophenylhydrazone (2 ug/mL) was also evaluated. The cyclohexane tolerance of all strains was also determined.
Results: Only the MAR mutants were tolerant to cyclohexane. Significantly (P < 0.0001) reduced uptake of bis benzimide was observed by the MAR mutants and strains with defined gene deletions in ompC and ompF compared to their parent (SL1344). The uptake of bis benzimide was unchanged in the acrB deletion mutant presumably due to increased compensatory expression of AcrE and AcrF efflux pump proteins. Increased uptake of bis benzimide was observed in the tolC gene deletion mutant and heat inactivated controls. Uptake by all strains was inhibited by the efflux pump inhibitors. Reduced sensitivity of the MAR mutants to CCCP further amplified the difference in bis benzimide uptake compared to the parent strain.
Conclusion: A rapid test in 96 well plate format has been developed and partially validated for the detection of MAR mutants. Differential sensitivity to efflux pump inhibitors enables further concurrent test amplification and phenotypic analysis.
P1161 Comparison of selective media for the detection and enumeration of lactobacilli and bifidobacteria from human faecal samples
V. Vankerckhoven, C. Lammens, O. Chonan, K. Oishi, H. Goossens (Wilrijk, Ghent, BE)
Objectives: Methods for the detection and enumeration of faecal bacteria should be properly validated. The aim of this study was to compare selective media for the enumeration of lactobacilli and bifidobacteria from human faecal samples.
Methods: Serial dilutions of 15 human faecal samples were plated, using the spiral enter apparatus Eddy Jet, on two commonly used selective media, LAMVAB and LBS for the detection and enumeration of lactobacilli, and transgalactosyloligosaccharide agar (TOS) and modified TPY agar (MTPY) for bifidobacteria. Colonies were counted based on colony morphology and Gram stain after a 72 h anaerobic incubation at 37°C. The number of lactobacilli and bifidobacteria per gram wet weight of faeces was estimated from the number of colonies.
Results: Total colony counts ranged from <2.0 to 5.8 log10 cfu/g faeces for LAMVAB and from <2.0 to 5.5 log10 cfu/g faeces for LBS. Mean bacterial counts for lactobacilli were 5.0 log10 cfu/g faeces on LAMVAB compared to 4.9 log10 cfu/g faeces on LBS (P > .05). Using LAMVAB, lactobacilli were found in 11 of 15 (73%) faecal samples compared to 9 of 15 (60%) using LBS (P > .05). For the bifidobacteria, total colony counts ranged from 7.1 to 9.6 log10 cfu/g faeces for TOS and from 7.0 to 9.4 log10 cfu/g faeces for MTPY. Mean bacterial counts for bifidobacteria were 8.3 log10 cfu/g faeces on both TOS and MTPY (P > .05). Bifidobacteria were found in all (100%) faecal samples using both media (P > .05).
Conclusions: It can be concluded that both lactobacilli and bifidobacteria could reliably be detected on each of the two selective media. For lactobacilli, LAMVAB and LBS showed comparable results and could be used to quantify lactobacilli in human faeces. The same holds for the media TOS and MTPY, which showed comparable results and could be used for the quantification of bifidobacteria from human faecal samples.
P1162 Retrospective analysis of Staphylococcus lugdunensis isolates
K. Chatzigeorgiou, N. Siafakas, A. Tarpatzi, S. Chaniotaki, S. Kouvardas, L. Zerva (Athens, GR)
Objectives: Staphylococcus lugdunensis is an emerging pathogen of a yet undefined clinical importance. The aim of the present study was the retrospective analysis of all S. lugdunensis isolates recovered in a Microbiology Laboratory and the assessment of their clinical significance.
Methods: Twenty-five Gram (+) cocci initially identified by Phoenix (Becton Dickinson) as S. lugdunensis during a 31 month period were obtained from 12 patients. Thus, 12 strains (1 strain per patient) were thawed and retested by Phoenix. API Staph system (bioMérieux), 16S rRNA gene sequencing, ornithine decarboxylase (ODC), PYR (Remel), slide (BD) and tube (Remel) coagulase tests were additionally performed. Susceptibility tests included disk diffusion (penicillin, oxacillin, cefoxitin, vancomycin, teicoplanin, tetracycline, gentamicin, erythromycin, clindamycin, ciprofloxacin, norfloxacin, rifampin and nitrofurantoin), E-test (penicillin and oxacillin), mecA gene (PCR) and PBP2a protein (slide latex, bioMérieux) detection and β-lactamase production (Oxoid). All relevant clinical information was retrieved from the medical records.
Results: By 16S rRNA sequencing, 10 of the 12 strains were identified as S. lugdunensis; the rest were not further considered. API Staph testing produced identical results with sequencing, while Phoenix misidentified 1 as S. capitis ssp. ureolyticus. All were ODC and PYR (+), tube coagulase (–) and half bound coagulase (+). All were mecA and PBP2a (–) and one produced β-lactamase. They all exhibited a multi-susceptible phenotype except for 1 gentamicin- and 1 quinolone-resistant strain. Clinical presentation included 1 case of endocarditis and 2 bacteraemias with excellent clinical outcome. The other isolates were obtained from pus (6 cases) or urine (1 case) specimens and their clinical significance was doubtful. Finally, analysis of laboratory records demonstrated that S. lugdunensis represented 1.3% of all coagulase negative isolates.
Conclusion: S. lugdunensis can be misidentified during routine laboratory practice and supplementary biochemical asays, especially ODC and PYR, are crucial for its recognition. It is rather a multi-susceptible species and may cause severe infections, albeit with good prognosis, or simply represent a contaminant.
P1163 Emergence of optochin resistance among Streptococcus pneumoniae in Portugal
S. Aguiar, M.J. Frias, L. Santos, J. Melo-Cristino, M. Ramirez (Lisbon, PT)
Objective: Characterise optochin-resistant strains of Streptococcus pneumoniae associated with pneumococcal infections in Portugal.
Methods: All strains submitted to our laboratory during 1999–2003 were optochin susceptible. Since then, 2 isolates (0.3%) in 2004 and 30 isolates (3.2%) in 2005, recovered from 10 geographically dispersed hospitals, were optochin-resistant. These 32 optochin-resistant S. pneumoniae strains consisted of mixed populations of optochin-susceptible and optochin-resistant isolates. For each isolate, both optochin-resistant and susceptible subpopulations were characterised using a combination of bile solubility, antimicrobial susceptibility, serotyping and macrorestriction profiling, using SmaI and pulsed field gel electrophoresis (PFGE). The BioNumerics software was used to make UPMGA (unweighed pair group method with arithmetic mean) dendrograms of PFGE patterns. The Dice similarity coefficient was used with optimisation and position tolerance settings of 1.0 and 1.5%, respectively.
Results: Subculture of individual colonies of each strain exposed a uniformly optochin-resistant subpopulation and a uniformly optochin-susceptible subpopulation. The optochin-resistant subpopulations were bile soluble and reacted with sera targeting pneumococcal capsular polysaccharides, confirming their identification as S. pneumoniae. The antimicrobial susceptibility profile and serotype was identical in both optochin-susceptible and resistant subpopulations suggesting that they corresponded to variants of the same clone. PFGE analysis of each subpopulations revealed indistinguishable PFGE profiles, further supporting the suggestion that these constitute two variants of the same strain. The 32 strains included in this study presented 19 different serotypes, all previously found among invasive and colonisation pneumococcal strains in Portugal. Furthermore, the diversity of PFGE profiles observed and the identification of internationally disseminated clones (ex: clone England14–9) excludes a clonal origin for these strains.
Conclusion: Accurate identification of S. pneumoniae isolates is essential for the correct diagnosis and adequate therapy of patients with pneumococcal infections. In view of the findings described here we recommend that at least the bile solubility test should be routinely performed in cases of suspected pneumococcal ethiology, even if the isolates are optochin-resistant, to exclude the possibility of S. pneumoniae.
P1164 Evaluation of 4 chromogenic media for detection and identification of clinically relevant yeasts
M. Ieven, Z. Wouters, E. Vercauteren, H. Goossens (Edegem, BE)
Background: Rapid detection and identification of yeasts is essential for an immediate and correct therapeutic management.
Objectives: Four commercially available chromogenic agars for the detection and identification of yeasts were evaluated on strains and directly on clinical specimens.
Methods: A total of 94 well characterised clinical isolates were included (31 C. albicans, 22 C. glabrata, 9 C. parapsilosis, 6 C. krusei, 8 C. tropicalis, 5 C. dubliniensis, 1 C. guilliermondii, 5 Cryptococcus neoformans, 3 Geotrichum spp. and 1 Saccharomyces cerevisiae) as well as 100 clinical specimens, identified positive for yeasts by conventional methods, including a wide range of sample types (wound swabs, sputa, urine blood cultures). All isolates and clinical specimens were cultured onto Candida ID2 (bioMérieux), Oxoid Chromogenic Candida Agar (OCCA, Oxoid) BD CHROMagar Candida (Becton Dickinson) and Cromogen Candida (Biomedics) and examined after both 24 h and 48 h incubation at 37°C or 30°C for the OCCA agar. Conventional non-chromogenic media, commercial identification systems and biochemical tests were used as gold standard to compare the results of the chromogenic media.
Results: The sensitivity for identification of isolates of C. albicans on chromogenic media varied between 13% and 100% after 24 h and increased to 97%-100% for all four media after 48 h. Both BD CHROMagar Candida and OCCA media correctly identified all C. albicans, C. krusei, C. tropicalis and C. dubliniensis after 48 h. Sensitivity for identification of C. glabrata, the second most prevalent species recovered from all clinical specimens, was 100% and 95.5% for OCCA and BD CHROMagar Candida respectively. Specificity for all 4 media was 100%: no single isolate was misidentified. The table presents sensitivity (%) for isolation and identification directly from clinical specimens.
Sensitivity for isolation and identification directly from clinical specimens
| Species (n = 114) | Sensitivity (%)a |
|||
|---|---|---|---|---|
| OCCA | BD CHROMagar | Cromogen Candida | Candida ID2 | |
| C. albicans | 91 | 82 | 84 | 96 |
| C. glabrata | 94 | 80 | NA | NA |
| C. krusei | 100 | 100 | 100 | 100 |
| C. tropicalis | 100 | 100 | 100 | NA |
| C. parapsilosis | 67 | NA | NA | NA |
NA, not applicable.
Conclusions: C. albicans was detected and identified with the highest sensitivity on the Candida ID2 but the OCCA chromogenic agar allowed to isolate and identify 98.2% of all species isolated from clinical samples with sensitivities varying between 91% and 100%.
P1165 Evaluation of various carbon sources on conidia production among clinical isolated moulds
P. Santanirand, S. Thanavanitnam, P. Chongtrakool, K. Angkananukool (Bangkok, TH)
Differentiation of conidia structure is one of the keys for fungal identification. The conventional slide culture technique has difficulty in certain culture conditions particularly in humidity levels which lead to uncertainty in sporulation. A new slide culture technique performed in 24-well tissue culture plate was introduced in order to overcome previous limitations. It showed several advantages over the conventional method, including simplicity and convenience in procedures, using less time and space, better humidity control and higher safety for workers. The latter advantage was due to double-coverglass technique. Apart from the adaptation of slide culture technique, the correlation of sporulation time and colony size or incubation period was studied. Fifty seven isolates of dermatophytes and forty eight black moulds from 16 genera were tested. In dermatophytes, approximately 90% of the isolates produced conidia when colony diameter reached 3 cm. which was equally to 7 days of incubation. For rapid and slow grower black moulds, the colony diameter of 1.4 cm. (or 5 days of incubation) and 6 cm. (or 6 days of incubation), respectively, were the optimal sporulation time. Lastly, the effect of various carbon sources on sporulation was studied. Among various flours, rice, brown rice and glutinous rice have showed the most promise in inducing conidia production. Therefore, these 3 flours were combined in a new media called triple flours agar (TFA). A comparison of TFA and potato dextrose agar and Sabouraud dextrose agar (SDA) in the dermatophyte group showed no differences but TFA was significantly better than SDA in the induction of sporulation in black moulds. Addition of fatty acids into TFA did not enhance induction capacity of sporulation in black moulds and dermatophytes. However, Nigrospora spp. responded to fatty acids addition by producing of significantly larger amount of conidia compared to TFA alone.
P1166 Diagnostics of fungal infections in the Nordic countries: we still need to improve!
M.C. Arendrup, E. Chryssanthou, P. Gaustad, P. Sandven, M. Koskela, V. Fernandez on behalf of the Nordic Reference Group for Methods in Medical Mycology (NRMM)
Objectives: A Nordic External Quality Assessment programme in medical mycology was established in 2005. In order to monitor not “best practice” but the level of routine diagnostics, the specimens were designed to resemble clinical samples and laboratories were asked to handle the samples like routine samples.
Methods: Five simulated clinical samples were distributed to 59 participating Nordic laboratories of clinical microbiology. The specimens contained the following microorganisms: (1) Candida glabrata and C. albicans in a ratio of 1:20; (2) Cryptococcus neoformans; (3) Aspergillus fumigatus, C. albicans and Enterobacter cloacae; (4) C. tropicalis, Klebsiella pneumoniae and Enterococcus faecium, (5) None.
Results: Sixty-six percent of the laboratories failed to detect the C. glabrata isolate in sample 1. Thirty-four percent of the laboratories reporting susceptibility results incorrectly reported the Cryptococcus neoformans isolate as fluconazole susceptible. Twenty-four percent of the laboratories failed to detect Aspergillus fumigatus in specimen 3 although the accompanying clinical information notified that it was a BAL sample from a neutropenic patient in an ICU.
Conclusion: This distribution of simulated clinical samples illustrates that the traditional quality assessment programmes may give a false sense of well performance, that mycological diagnosis is difficult, and that there is a need of further improvement and attention.
Viral serology
P1167 Evaluation of InfluA&B Respi-Strip for the rapid detection of influenza A and B viruses in respiratory samples
S. Degallaix, L. Denorme, O. Lefèvre, V. Labrune, T. Laurent, T. Leclipteux, P. Mertens (Gembloux, BE)
Influenza virus is the causative agent of flu, a highly contagious infection of the upper respiratory tract. Flu is characterised by the antigen variability, the seasonality and the impact on the general population.
There are two main (A and B) types of influenza viruses. H1N1, H2N2 and H3N2 InfA subtypes as well as InfB strains can cause outbreaks of variable intensity annually in humans. Although influenza A are the most prevalent, type B viruses can represent up to 50% of circulating virus during one season, and is often more abundant at the end of the season.
Flu is most often a mild viral infection, but influenza can cause severe complications such as bronchitis or pneumonia, particularly in children and elderly people. Antivirals are available, but these treatments are effective only if administered early requiring a rapid diagnostic test. There are many other viral infections that can mimic influenza making laboratory tests necessary to distinguish it from other acute respiratory infections. The widespread of H5N1 wildbird strains that can infect humans and the threat of a pandemic makes the rapid diagnosis still more valuable.
We have developed a new rapid dipstick test (InfluA&B Respi-Strip) based on monoclonal antibodies (MAb) directed agaisnt non-variable proteins of influenza A and B viruses. The final device has two sides, one for the detection of influenza A and the opposite side for the detection of influenza B. For each virus, MAbs are coated on the nitrocellulose, while other Mabs are conjugated to colloidal gold particles. A build-in control is used as migration control.
An evaluation on 113 frozen nasopharyngeal samples has been conducted to compare the InfluA&B Respi-Strip to a market leader rapid immunochromatographic test (Test N), with RT-PCR as the reference method. Both tests showed excellent specificities for both InfA and InfB detection (98.6–100%). InfluA&B Respi-Strip was slightly more sensitive than Test N, for both InfA and InfB detection. InfluA&B Respi-Strip will be further validated on fresh samples during the epidemic season.
In conclusion, the Coris' Influ-A&B Respi-Strip shows sensitivity and specificity comparable to one of the bests currently available rapid tests. The use of this test for diagnosis of influenza disease should enhance patient healthcare by enabling rapid and appropriate use of antiviral treatments and lowering inappropriate use of preventive antibiotic treatment still often prescribed by mistake.
P1168 Evaluation of an automated chemiluminescent immunoassay for the detection of anti-rubella virus IgM antibodies
B. Pustowoit, U.G. Liebert (Leipzig, DE)
Background and Objectives: Rubella, commonly known as German measles, occurs worldwide. In an acute rubella infection acquired either naturally or via vaccination, IgM antibodies appear in the patient serum at the onset of clinical illness and persist for a period of 4 to 5 weeks. However, low IgM levels may not necessarily indicate a recent rubella infection. A fully automated assay for the qualitative detection of anti-Rubella IgM in human serum developed on the Access® immunoassay systems (Beckman Coulter Inc.) was studied including clinical specificity and sensitivity as well as performance with potential cross reactivity panels. A study was performed with a panel named “long persistent Rubella IgM” referring to positive results obtained with other anti-Rubella IgM method but not considered as acute infection or recent vaccination.
Methods: The specificity was studied with a population of 96 samples considered negative in IgM (>360 days after exanthema or vaccination). Sensitivity was studied with 31 samples from a Rubella outbreak and 12 samples from recently vaccinated people. The evaluation of potential cross reactivity was based on 30 samples from individuals who had serological evidence of acute infections with Mumps (n = 9), EBV (n = 5), CMV (n = 3), Influenza A (n = 4) and other pathogenic microorganisms (n = 10). “Long persistent IgM” were studied with samples from persons with positive IgM with an other method but with high level of IgG avidity and/or presence of E2 band in native immunoblot (inconsistent with acute Rubella). Discrepant results were confirmed using additional EIA tests, avidity of anti-Rubella IgG as well as native immunoblot test for determination of IgG antibodies against Rubella virus proteins.
Results: In this study, Access®Rubella IgM exhibited a specificity of 98.09% and a sensitivity of 97.61%. A sensitivity of 100% was noted with seroconversion panels. Two samples (one positive in anti-E. coli and one positive in Influenza A IgA) presented a cross-reaction. Referring to the “long persistent” rubella IgM panel, 8 samples were negative with Access Rubella IgM in accordance with the sample status (no acute infection or recent vaccination).
Conclusion: The Access® anti-Rubella IgM assay combines clinical performance, with good sensitivity, specificity with the advantage of a rapid, automated, random access immunoassay system.
P1169 Evaluation of a fully automated Multiplexed Bead assay for assessment of Epstein-Barr virus immunologic status
M. Baccard, J.M. Seigneurin (Grenoble, FR)
Detection of clinically relevant circulating Epstein-Barr virus (EBV) specific and heterophile antibodies in serum are essential for the diagnosis of EBV infection and determination of the patient EBV serology status. Currently, specific serological assays using either indirect immunofluorescence or enzyme-linked immunosorbent (ELISA) are performed to evaluate the status of Epstein-Barr virus (EBV) infection in humans. Even if reliable, these methods are limited to testing an antibody response to a single viral antigen per reaction, thus necessitating a panel of assays to complete the evaluation.
The aim of this study was to evaluate the performance of the BioPlex 2200 EBV IgG and IgM Panels (Bio-Rad Laboratories, Hercules, CA), a new fully automated multiplex method that can simultaneously detect in a single tube VCA, EBNA-1 and EA-D IgG and VCA IgM and heterophile antibodies.
610 serum samples from 508 patients including seronegative and positive clinically documented samples (119 Infectious Mononucleosis [IM] and other stages of the infection) were tested both with the BioPlex 2200 panels and the assays routinely used in the lab.
Specificity on negative samples was 98.2% and ranged from 96 to 100% on clinical samples depending on the analyte. The clinical sensitivity on acute and IM samples was up to 95% for VCA IgM and 96.4% for heterophile. Combining those two analytes improved the clinical sensitivity on those samples. For past infection samples, VCA IgG clinical sensitivity was 97%. Sensitivity on clinical samples for EBNA-1 and EA-D IgG was respectively 98 and 87%. Concordance calculation demonstrated overall agreement of 90% for VCA IgM, 93% for heterophile antibody and 77% for VCA IgG (especially due to difference in the antibodies kinetic during primary infection). The correlation between samples classification based on BioPlex 2200 results and status determined with the lab routine assays was of 83% including atypical profiles and immunocompromised patients.
Conclusion: the BioPlex 2200 EBV IgG and IgM Panels showed very good interpretation concordance with the routine lab assays. Furthermore, the BioPlex 2200, the first and only fully-automated, random access multiplex EBV testing platform offers practical advantages that allow for rapid and simultaneous evaluation of the five most clinically relevant antibodies including heterophile for accurate assessment of EBV Immunologic Status.
P1170 Rapid, sensitive and specific lateral-flow immunochromatographic device to measure antibodies to HSV-2 in serum and whole blood
E.I. Laderman, E. Dumaual, E. Whitworth, M. Jones, J. Gonzalez, A. Hudak, J. Carney, J. Groen (Cypress, US; Cardiff, UK)
Objectives: Herpes simplex virus-2 is a common human pathogen that causes a variety of clinical manifestations. Adequate identification of anti-HSV-2 IgG is important for clinical management and seroepidemiological studies. HSV-2 serology tests currently used include Western Blot, Immunoblot and ELISA; however, all are laboratory based. In order to provide near-patient results to allow for faster counseling and treatment, a rapid assay that is accurate and simple to use is desirable. Here we describe the development of a lateral flow immunochromatographic assay (LFIA) using gold colloid nanoparticles as the detection reagent.
Methods: Samples: A panel of 359 patient sera consecutively submitted for HSV-1 and −2 type-specific serology tests was collected. In addition, sera positive for CMV (n = 11), EBV (n = 16), VZV (n = 14), rubella (n = 34) or HSV-1 (n = 196) but negative for HSV-2 IgG by ELISA were also selected for cross-reactivity testing. One hundred volunteer donors were recruited and heparinised-fingerstick blood, venous whole blood, and serum samples were collected to measure concordance of results across sample types. Samples positive or negative for HSV-2 were spiked with 10 mg/mL triglycerides, 60 mg/mL albumin, 0.2 mg/mL bilirubin or 220 mg/mL haemoglobin for interference testing. Reference ELISA: Samples were tested using the HerpeSelect-2 type-specific IgG ELISA per the package insert. An HSV-2 inhibition test was used to evaluate specimens with discordant ELISA versus LFIA results. LFIA: After addition of sample and buffer, the antibody-gold conjugate and sample migrate across the test strip. Antibodies (if present) and gold conjugate sequentially contact the test and control lines and cause specific coloured lines to form within 15 minutes.
Results: The sensitivity of the HSV-2 LFIA compared with ELISA was 100% (89/89), and the specificity was 97.3% (257/264). Cross-reactivity in HSV-1 positive samples was 2.6% (5/196), 2.9% (1/34) for Rubella and 6.2% (1/16) for EBV. No cross-reactivity in VZV or CMV positive samples was observed. No interference was observed from bilirubin, triglycerides, albumin or haemoglobin added to samples. Concordance of LFIA results between capillary whole blood, EDTA and heparin venous whole blood and serum was 99% (99/100).
Conclusion: The lateral flow immunochromatographic assay for HSV-2 IgG demonstrated excellent sensitivity, specificity, and concordance compared to type-specific ELISA assay.
P1171 Multiplex bead-based assays for Epstein-Barr virus, IgG and IgM serum antibody detection
H. Gregson, N. Liu, H. Truong, X. Su, E. Laderman, J. Groen (Cypress, US)
Objectives: Epstein-Barr Virus (EBV) is a common virus that can cause a wide variety of symptoms from asymptomatic to severe infectious mononucleosis. The symptoms of the disease can be similar to that of other viruses or haematologic cancers. Most important for EBV diagnosis is the distinction of acute infection, past infection, and no infection for differential diagnosis. EBV serology methods currently include IFA, ELISA and multiplex assays; however, IFA and ELISA are labour intensive and each has their limitations. A multiplexed assay with better performance is highly desirable. Here we describe the development of an IgG and an IgM EBV multiplexed assay using the xMAP technology (Luminex Inc., Austin, TX).
Methods: A panel of 426 patient sera consecutively submitted for EBV testing and 159 patient sera samples that were classified as EBV negative or EBV IgM positive were collected. EBV IgG assay: Recombinant EA-D and EBNA-1, and native affinity purified VCAgp-125, and goat-anti-human IgG (internal control) were covalently linked to the xMAP polystyrene bead sets. EBV IgM assay: Native affinity purified VCAgp125 and heterophile antigen were covalently linked to polystyrene bead sets. The bead mix was incubated with test serum followed by washing, and then incubated with anti-human IgG or IgM conjugated with phycoerythrin (PE). The fluorescence intensity of PE, representing the amount of IgG or IgM in the target sample, was measured by a Luminex 100 system. The same samples were tested using the EA IgG, EBNA IgG, VCA IgG and VCA IgM ELISA system and a rapid test for heterophile antibodies per the manufacturer's suggested protocol.
Results: Because reactivity to each antigen represents a specific stage in the EBV infection, each sample was classified by a FDA approved ELISA and rapid test for heterophile as acute, no infection, past infection, or indeterminate using the defined FDA algorithm. The sensitivity and specificity of each antigen in the multiplex based bead assay was determined by comparison to the relevant ELISA classification. See attached table for the results of sensitivity and specificity analysis.
| Antigen | Acutea |
Past Infectiona |
No infectiona |
|||
|---|---|---|---|---|---|---|
| % agreement | 95% CI | % agreement | 95%Cl | % agreement | 95%Cl | |
| VCA IgG | Sens. 100% (49/49) | 92.7–100% | Sens. 94.8% (307/324) | 91.8–96.7% | Spec. 94.8% (92/97) | 88.5–97.8% |
| EA IgG | Sens. 97.7% (42/43) | 87.9–99.6% | Spec. 96.3% (233/242) | 93.1–98.0% | Spec. 96.7% (88/91) | 90.8–98.9% |
| EBNA IgG | Spec. 100% (77/77) | 95.3–100% | Sens. 97.5% (316/324) | 95.2–98.7% | Spec. 100% (97/97) | 96.2–100% |
| VCA IgM | Sens. 93.5% (72/77) | 85.7–97.2% | Spec. 92.6% (300/324) | 89.2–95.0% | Spec. 95.9% (93/97) | 89.9–98.4% |
| Heterophile IgM | Sens. 98.0 (50/51) | 89.7–99.7% | Spec. 98.8% (320/324) | 96.9–99.5% | Spec. 96.9% (94/97) | 91.3–98.9% |
Abbreviations: Sens., sensitivity; Spec., specificity; 95% CI, 95% confidence interval.
Conclusion: The multiplex bead-based assay for EBV IgG and IgM demonstrated excellent sensitivity, specificity, and concordance compared to type-specific ELISA assays.
P1172 New enzyme immunoassay “DS-EIA-HIV-AG-SCREEN” for early diagnostics of HIV infection
I. Sharipova, E. Baranova, M. Levina, A. Burkov, A. Oriadina, T. Ulanova (Nizhniy Novgorod, RU)
Objective: The problem of early diagnostics of HIV infection still remains very important and can be solved by using highly sensitive tests for detection HIV-1 p24 antigen. Improvements in assay sensitivity have been achieved constantly since introduction of the first commercial test. The mostly sensitive current EIA for the detection of HIV p24 have an analytical sensitivity equal 3 pg/mL. Despite the high performance of screening assays, transfusion-associated HIV infection is still reported. The aim of this present study was to develop and evaluate EIA test for p24 detection with analytical sensitivity equal 0.5 pg/mL.
Methods: New EIA one-step diagnostic test based on biotin-streptavidin amplification. Sensitivity of new test was estimated by testing serial dilutions of the HIV 1 ANTIGEN STANDARD (Bio-Rad Laboratories, Hercules, CA, USA) and serum samples from HIV Antigen Sensitivity Panel No. 801 (Boston Biomedica Inc. (BBI), West Bridgewater, MA, USA). Seroconversion panels No. 62238 and 64578 (ZeptoMetrix Corp. (ZMC), Buffalo, NY, USA), No. 931, 939, 942, and 948 (BBI) have been additionally tested. To challenge the specificity of the new assay, serum samples of healthy blood donors (n = 1,990), pregnant women (n = 400), patients with other infections (n = 256) and patients with noninfectious diseases (n = 305) were investigated.
Results: Analytical sensitivity of the «DS-EIA-HIV-AG-SCREEN» assay was estimated equal 0.5 pg/mL. All positive samples from commercial panels demonstrated much higher signal to cutoff ratios than with alternative tests. The results obtained during the study of sera samples from seroconversion panels No. 64578 (ZMC, USA), No. 931, 939, 942, and 948 (BBI, USA) showed that the test “DS-EIA-HIV-AG-SCREEN” able to detect p24 antigen either simultaneously or two days later (ZMC, USA, panel No. 62238) than HIV RNA by PCR, but on an average 3 days earlier than lower sensitive tests. General specificity of the new test among different samples cohort was equal 99.95%.
Conclusion: Development of EIA tests for detection an HIV p24 antigen in concentration lower than 3–5 pg/mL is not technically insuperable problem. The obtained results showed high diagnostic efficiency of the “DS-EIA-HIV-AG-SCREEN” test which can be used for blood donor screening and examination of patients from risk groups of with the aim of early diagnostics of HIV infection.
P1173 Evaluation of a competitive quantitative RT-PCR-ELISA system for quantification of HIV-1 RNA in plasma: comparison with COBAS Amplicor HIV-1 monitor test
S. Amel jamehdar, F. Sabahi, M. Forouzandeh, M. Haji Abdolbaghi, A. Kazemnejad, F. Mahboudi, R. Edalat, M. Ravanshad (Tehran, IR)
Objectives: Quantification of the human immunodeficiency virus type 1 RNA levels (viral load) has become an indispensable tool for the assessment of patient prognosis and for the monitoring of antiretroviral therapy. The access to commercial assays for viral load monitoring in IRAN is still limited due to the high costs. The aim of this study was to design a competitive quantitative RT-PCR-ELISA system for sensitive amplification of HIV-1 pol gene and evaluation of the treatment of 80 HIV-1 seropositive patients. Results of this assay were compared with the COBAS Amplicor HIV-1 monitor test.
Methods: Whole blood samples from 80 HIV-1 seropositive and 40 seronegative blood donors were collected in vaccutainer tube. Plasma was separated and stored at −80°C. Total RNA was extracted according to the protocol supplied by the commercial viral RNA kit (Qiagen). A quantitative HIV-1 test is described based on competitive quantitative RT-PCR assay combine with hybridisation as a detection system. The internal RNA standard (IS) was designed by SOEing mechanism, specifically to be competitive during the amplification step. Sample viral load determination was carried out with one RT-PCR in the presence of 103 IS copies. The HIV-1 copy number was calculated by reference to an external standard curve performed on known and increasing amounts of the reference HIV-1 RNA co-amplified with a constant amount of the IS RNA.
Results: The results demonstrated that this technique detected 5 HIV-1 RNA copies per milliliter of plasma. The assay had a linear range from 103 to 106 HIV-1 copies. A positive linear correlation between the two tests (r2 = 0.88) was found. Overall, no significant differences were found in plasma viral load quantitation between both assays, therefore this assay is suitable for viral load quantitation of HIV-1 plasma samples.
Conclusions: Our data demonstrates that our competitive quantitative RT-PCR-ELISA system is a fast and reliable method for the detection and quantitation of HIV-1 RNA and this technique could be applied during the monitoring of HAART treatment.
P1174 Analysis of AtheNA Multi-Lyte HIV-1 Test for confirmation of HIV-1 infection
Y. Wang, A. Suantio, Y. Pogue (Atlanta, US)
Background: The Western Blot test for Human Immunodeficiency Virus Type 1 (HIV-1) is routinely required to detect antibodies against one or more HIV-1 antigens in the human serums that were tested positive by using the enzyme immunoassay (EIA) screening method for HIV-1/2. New HIV-1 Bead Blot (AtheNA Multi-Lyte HIV-1) test is a multiplexed, microparticle-based immunoassay for confirmation of antibodies against HIV-1 viral proteins in the Luminex system.
Methods: A total of 115 blood samples that were tested positive by Abbott HIV-1/2 EIA method were used for comparison of the HIV-1 confirmation by using the AtheNA HIV-1 method on Luminex system with the conventional manual HIV-1 Western Blot (WB) method.
Results: Of 56 non-duplicated samples tested positive by HIV-1 WB, all 56 (100%) were tested positive by the AtheNA HIV-1 method. Of 59 HIV-1 unconfirmed or negative samples, 46 (78%) resulted in the same interpretation by both methods: 41 samples were confirmed as negative, and 5 samples as indeterminant, respectively. Minor discrepant results were identified in 13 out of 59 samples (22%). Of 9 samples negative by the AtheNA method but were indeterminant by WB method, 7 were either positive or weak positive for p24. Of 4 samples tested negative by WB method but were indeterminant by AtheNA method, 3 were positive for p55.
Conclusions: Even with minor discrepancy noticed in the HIV-1 unconfirmed or negative samples, the new multiplexed AtheNA Multi-Lyte HIV-1 Bead Blot assay can provide the same confirmation as the HIV-1 Western Blot method in the HIV-1 positive serum samples.
P1175 Multi-centre evaluation of a new hepatitis B surface antigen (HBsAg) assay on the family of Access® immunoassay systems
A. Pivert, A. Ducancelle, T. Levayer, F. Descamps, M. Maniez-Montreuil, A. Artus, C. Heinen, D. Woodrum, F. Lunel-Fabiani (Angers, Montpellier, Lille, Marseilles, Marnes la coquette, FR; San Diego, US)
Background: Accurate detection of Hepatitis B Surface Antigen (HBsAg) is an important goal for the diagnosis of patients infected with the hepatitis B virus (HBV). A multi-centre study was conducted to characterise the clinical performance of the new HbsAg assay on the family of Access immunoassay systems from Beckman Coulter currently in development. The Access HBsAg and HBsAg Confirmatory assays are based on paramagnetic particle, solid phase technology and chemiluminescent signal detection.
Methods: Percent negative agreement (relative specificity) was calculated from 5,020 prospectively enrolled blood donors from two blood donation centres and 566 subjects from a university hospital for whom the HBsAg assay was requested by their physician as a part of their routine medical evaluation (due to suspected exposure or increased risk of HBV, signs and symptoms of infection, or prenatal screening of pregnant women). Percent positive agreement (relative sensitivity) was calculated from 290 subjects with a diagnosis of acute or chronic HBV infection who were prospectively enrolled or whose samples had been archived in a sample repository. Percent negative and positive agreements were calculated relative to the Abbott PRISM® and Abbott AxSYM® immunoassay systems.
Results: Negative percent agreement for the Access HBsAg assay in a blood donor population was 99.96% (5,018/5,020) [95% CI: 99.86– 100%]. Negative percent agreement for the Access HBsAg assay at the university hospital was 99.65% (564/566) [95% CI: 98.73– 99.96%]. Discrepant samples were not confirmed in the Access HBsAg Confirmatory assay and agreement between methods after confirmatory testing was 100%. Positive percent agreement for subjects with known HBV infection in the Access HBsAg assay was 100% (290/290) [95% CI: 98.69–100%]. All positive samples were confirmed in the Access HBsAg Confirmatory assay. Excellent separation of positive and negative populations was observed.
Conclusion: The Access HBsAg and the Access HBsAg Confirmatory assays provide excellent sensitivity and specificity for the detection of HBsAg in hospital and blood donor populations with the advantage of a rapid, automated, random-access immunoassay system.
®All trademarks are property of their respective owners.
P1176
Performance of an automated chemiluminescent assay for the detection of anti-hepatitis A virus IgM antibodies: Access® HAV IgM assay
Y. Buisson, C. Masson, F. Bouniort (Paris, FR; Nyon, CH)
Background: The diagnosis of acute hepatitis A is based on the detection of anti-HAV IgM antibodies. A fully automated assay for the qualitative detection of anti-HAV IgM in human serum and plasma developed on the family of Access immunoassay systems from Beckman Coulter was studied and included imprecision, clinical sensitivity and specificity, comparison with a reference method and negative samples distribution.
Methods: Intra-assay imprecision based on 30 replicates of 4 samples (1 negative and 3 positive). Inter-assay imprecision based on the same 4 samples tested in 5 replicates, twice a day, 5 different days. Sensitivity was studied with a population of 210 individual samples from patients with acute hepatitis A and with 157 samples from 26 cases of clinical follow-up or seroconversion. Results obtained with the Access HAV IgM assay were compared with a commercially available EIA technique; discrepant results were confirmed using a third EIA method. The specificity of the Access HAV IgM assay was evaluated on a normal population (n = 1,535) and a population with a potentially interfering pathologies (HBV Ab, HIV Ab, HCV Ab, CMV, EBV, HSV, VZV, Yellow fever, Mumps, Measles, Polio, Rubella, Toxoplasmosis or autoimmune disorders (n = 429). A comparison study with a commercialised automated HAV IgM assay was performed with 620 samples. Discrepant results were confirmed using a third EIA method. The negative samples repartition was studied with 706 blood donor samples.
Results: Imprecision was less than 10% CV for the positive samples. Sensitivity was 99.04% with individual samples and 100% with seroconversion or clinical follow-up cases. The Access HAV IgM assay demonstrated a specificity of 99.93% with a normal population and 99.53% with the population of potentially interfering pathologies. The comparison study exhibited an overall agreement of 98.40%, a relative specificity of 99.35% and a relative sensitivity of 97.40%. After further analysis with a third EIA method, 7 from the 10 discrepant results were correctly tested with the Access HAV IgM assay. The negative sample distribution showed that 93.90% of the results are lower than 0.3 S/CO.
Conclusion: The Access HAV IgM assay combines robust analytical and clinical performance with good precision, sensitivity, specificity and a good distribution of the negative samples far away from the cut-off value: with the advantage of a rapid, automated, random access immunoassay system.
P1177 Detection of hepatitis B surface antigen (HBsAg) subtypes, genotypes, mutants and evaluation of potential hook effect using the new Access® HBsAg and confirmatory assays
X. Duburcq, A. Clement, D. Duhamel, D. Leirens, F. Margotteau, F. Bouniort, S. Taskar, O. Flecheux (Paris, FR; Nyon, CH; Chaska, US)
Background and Objectives: Detection of genetic variability of the HBsAg protein represents significant challenges to the accurate detection of HBsAg. Genetic variants may escape immunoassay detection leading to erroneous results which could endanger patients' health and the blood supply. Another risk for misdetection in an immunological assay is a false negative result due to high concentration of HBsAg (hook effect). A study was conducted to characterise the detection of subtypes, genotypes and mutants; and to investigate the potential hook effect of the new HBsAg and HBsAg Confirmatory assays on the family of Access immunoassay systems from Beckman Coulter currently in development. The Access HBsAg and HBsAg Confirmatory assays are based on paramagnetic particle, solid phase technology and chemiluminescent signal detection.
Methods: The STFS 2004 Subtype Panel, containing 8 different HBsAg subtypes (adw2, adw4, adr, ayw1, ayw2, ayw3, ayw4, and ayr), was tested to investigate reactivity towards subtype recognition. Seven genotypes, A-G from the Teragenix HBV Genotype Panel (HBVGTP-002), were tested to investigate reactivity towards genotype recognition. A panel of 15 recombinant mutant proteins, each containing single or multipoint mutations located at 22 different amino acid positions in the S protein ectodomain, was tested to investigate reactivity towards genetic mutations. Five very high positive samples, containing 200 to 300 μg/mL of HBsAg and a spiked mixture of Ad + Ay HBsAg (approximately 5 mg/mL), were tested to investigate interference due to high concentration of antigen.
Results: All 8 subtypes in the STSF 2004 Subtype Panel, all 7 genotypes of the Teragenix HBV Genotype Panel, and all 15 recombinant HBsAg mutant proteins were detected without ambiguity with the Access HBsAg assay and confirmed positive with the Access HBsAg Confirmatory assay using multiple lots of assay kits. No false negative results, due to high amount of antigen (∼5 mg/mL in the highest case), were observed among the 6 samples tested and all samples were confirmed positive after dilution.
Conclusion: The Access HBsAg and Confirmatory assays provide excellent sensitivity for the detection of HBsAg mutants, subtypes and genotypes; and are not impacted by high concentration of antigen.
P1178 A comparative study of the new Access® HBs Ag assay and the HBs Ag confirmatory assay from Beckman Coulter
X. Duburcq, A. Pouzet, L. Marant, A. Clément, D. Duhamel, Y. Leirens, F. Margotteau, R. Falcou-Briatte, F. Bouniort, S. Taskar, O. Flecheux (Paris, FR; Nyon, CH; Chaska, US)
Background: Hepatitis B virus infection is a worldwide problem of clinical significance. The virus is transmitted via parenteral contact, through the exchange of blood and blood products, by sexual contact, and by perinatal spread from mother to child. In this study, we compared the relative sensitivity and specificity performances of the new Access HBs Ag assay currently in development and the Abbott AxSYM§ HBsAg V2.
Methods: Analytical sensitivity was compared by testing the BBI PHA 106 panel composed of 15 points with low HBs Ag titer within 0.05 and 0.6 IU/mL. Clinical sensitivity was investigated using commercialised panels for subtypes and genotypes or in-house panel for variants, 30 commercialised seroconversion panels (including 177 samples), and 100 positive samples from recent, acute or chronic phase infected patients. Negative sample repartition was compared with 577 negative samples collected in a hospital in France.
Results: All low titer panel members up to 0.05 IU/mL were recognized by the Access HBs Ag assay, whereas AxSYM HBs Ag failed to recognize 2 of them up to 0.1 IU/mL. All subtypes, genotypes and variants tested were recognized by both methods. The Access HBs Ag assay showed equivalent results to AxSYM HBs Ag for 18 panels among the 30 tested. The Access HBs Ag assay was more sensitive with 10 panels and less sensitive with 2 panels. Among the 100 positive samples, all were found positive, displaying 100% of agreement between the Access HBs Ag and AxSYM assays. The specific margin (SM), defined as the number of SD between the negative sample mean and the cut-off, was determined equal to 21 for the Access HBsAg assay and 5.5 for AxSYM HBs Ag.
Conclusion: The performance of the Access HBsAg and the Access HBsAg Confirmatory assays are at least equivalent to that of the AxSYM HBsAg V2. The Access HBsAg and the Access HBsAg Confirmatory assays can be used either with the Access Immunoassay System or with the high-throughput UniCel® DxI 800 Immunoassay System offering rapid and accurate results whatever the throughput requirement of the lab.
®All trademarks are property of their respective owners.
P1179 Specificity studies of the new Access® HBs Ag and HBs Ag confirmatory assays from Beckman Coulter
X. Duburcq, A. Pouzet, A. Clement, L. Marant, D. Duhamel, Y. Leirens, F. Margotteau, R. Falcou-Briatte, F. Bouniort, S. Taskar, O. Flecheux (Paris, FR; Nyon, CH; Chaska, US)
Background and Objectives: The challenge for a new HBs Ag assay is to detect genetic variability in the HBs Ag protein (mutation, subtypes, and genotypes) in order to avoid false negative results. Nevertheless, assay specificity remains essential, as false positives lead to erroneous results and trouble for the patient. A specificity study was conducted using blood bank samples, and the potentially interfering reactions were investigated using the new Access HBs Ag and HBs Ag Confirmatory assays currently in development. The Access HBsAg and HBsAg Confirmatory assays are based on paramagnetic particle, solid phase technology and chemiluminescent-signal detection.
Methods: Internal evaluation of specificity was performed with 4,343 samples from blood donors collected by blood banks in France. Potential cross-reactivity was evaluated by testing a panel of 293 samples from patients with 21 potentially cross-reactive infections and autoimmune disorders. Samples with high concentrations of human albumin, haemoglobin, bilirubin, triolein and biotin were also tested. To evaluate anticoagulant and SST impacts, matched sets of negative samples containing serum, EDTA K2, EDTA K3, Citrate, Lithium Heparin, ACD and SST were tested.
Results: The specificity for blood donor samples was determined at 99.98% [99.87–100%] (CI 95) and the specific margin, defined as the number of SD between the negative samples mean and the cut-off was equal to 22. Among all the samples tested for cross-reactivity, two (0.68%) gave positive results in the screening assay, but were not confirmed with the Access HBs Ag Confirmatory assay. Sample components and different anticoagulants tested had no impact on the assay specificity.
Conclusion: The Access HBsAg and the Access HBsAg Confirmatory assays provide excellent specificity without cross reactivity with potentially interfering components. Moreover, the assay showed an excellent specific margin that prevents specificity issue due to negative sample repartition close to the cut off.
P1180 Sensitivity studies of the new Access® HBs Ag and HBs Ag confirmatory assays from Beckman Coulter
X. Duburcq, A. Pouzet, L. Marant, A. Clement, D. Duhamel, Y. Leirens, F. Margotteau, R. Falcou-Briatte, F. Bouniort, S. Taskar, O. Flecheux (Paris, FR; Nyon, CH; Chaska, US)
Background and Objectives: Accurate detection of hepatitis B surface antigen (HBsAg) is important to aid in the diagnosis of individuals infected with hepatitis B virus. An automated assay, for qualitative detection of HBsAg in human serum and plasma on the family of Access® immunoassay systems from Beckman Coulter is currently in development. The new Access HBsAg and HBsAg Confirmatory assays are based on paramagnetic particle, solid phase technology and chemiluminescent-signal detection. The purpose of this study was to evaluate the analytical and the clinical sensitivity of the new assays.
Methods: The analytical sensitivity was determined using different commercialised panels (French SFTS 2004 panel, PEI Ad panel, PEI Ay panel, WHO 80/549 standard, WHO 00/588 standard). The clinical sensitivity was verified with the low titer panel BBI PHA 106 (containing 15 samples with concentration between 0.05 IU/mL and 0.6 IU/mL), 100 commercial HBsAg positive samples and 30 seroconversion panels.
Results: Analytical sensitivity was estimated to be ≤0.1 ng/mL (French SFTS panel), 0.020 PEI Units/mL (ad panel), 0.024 PEI Units/mL (ay panel), 0.092 IU/mL with WHO 80/549 HBsAg standard and 0.056 IU/mL with WHO 00/588 HBsAg standard. All low titer panel members up to 0.05 IU/mL were recognized by the Access HBsAg assay and confirmed by the Access HBs Ag confirmatory assay. Among the 100 isolated positive samples, all were found positive displaying 100% of sensitivity for the Access HBsAg assay and all were confirmed by the Access HBsAg Confirmatory assay. The Access HBsAg assay demonstrated equivalent results compared to the Abbott AxSYM§ HBsAg V2 assay for 18 panels among the 30 tested. For the remaining panels, the Access assay was more sensitive than 10 out of 12 cases.
Conclusion: The Access HBsAg and HBsAg Confirmatory assays provide excellent analytical and clinical sensitivity performance and can be used either with the Access Immunoassay System or with the high-throughput UniCel® DxI 800 Immunoassay System for rapid and accurate results in the management of HBV infection.
§All trademarks are property of their respective owners.
P1181 Comparison of real-time PCR with conventional bDNA based assay for quantification of HBV, HCV and HIV-1: possible clinical implications
A. Amendola, M. Solmone, A. Garbuglia, C. Angeletti, N. Lauria, M. Capobianchi (Rome, IT)
Objectives: We compared two commercial assays [Versant 3.0, System 340 (Bayer Diagnostics), based on branched DNA technology, and COBAS Ampliprep TaqMan, CAP-TCM (Roche Diagnostics), based on real-time PCR] in measuring HBV, HCV and HIV viraemia.
Methods: 150 consecutive serum samples from patients with chronic hepatitis B (divided in HBeAg-positive or Anti-HBeAb-positive), 92 clinical samples from patients eligible to HCV therapy and 46 clinical samples from HIV-seropositive individuals have been analysed. Moreover, commercially available standard panels of HBV, HCV and HIV-1 were used to compare the linearity of two assays.
Results: In the overlapping range, an elevated degree of correlation between the tests was measured for all viruses concentrations:
-
•
HBV: r = 0.9389, p<0.001;
-
•
all HCV genotypes: r = 0.7702, p < 0.001. HCV genotypes 1–4: r = 0.721, p < 0.001; r = 0.753, p<0.001; r = 0.0.873, p < 0.001; and r = 0.783, p < 0.001, respectively;
-
•
HIV-1: r = 0.9885, p<0.0001.
The analysis of mean values indicated that HBV DNA values obtained with bDNA-based assay were higher than those obtained with TaqMan. On the contrary, HCV and HIV-1 viral load values obtained in CAP/CTM were generally higher than those obtained in bDNA. Comparisons conducted according to Bland and Altman method showed that differences of measurement occurred at higher magnitude of viral load. Moreover, for all viruses analysed, a considerable proportion of clinical samples gave results discordant by more than 0.5 or even 1 log that seemed not genotype-dependent.
Conclusion: These results suggest that quantitative results obtained by different assays may not be interchangeable, even though expressed in the same (international) units. In general, CAP/CTM shows higher sensitivity and broad linear range, but at high magnitudo of viral load HBV-DNA was under estimated while HCV and HIV-1 were over estimated.
Therefore, it is recommendable that the same quantitative assay be used longitudinally in monitoring therapeutic response of individual patients. Longitudinal studies are necessary to evaluate the impact of the higher viral load results obtained in the real time PCR-based tests in clinical decision-making.
P1182 Differences of immunoreactivity of selected antigenic epitope of tick-borne encephalitis virus envelope protein gE modelled by different variants of recombinant fusion proteins
A. Obriadina, N. Saveleva, Y. Zagryadskaya, N. Oreshko, V. Pimenov, V. Puzyrev, A. Burkov, T. Ulanova (Nizhny Novgorod, RU)
Objectives: Tick-borne encephalitis virus (TBEV) is a human pathogenic member of the genus Flavivirus. TBE virus cause severe encephalitis with serious sequelae. Envelope protein (E) of the flaviviruses is a major target of neutralising antibodies and used as a diagnostic reagent for ELISA-format tests. The aim of present study was to evaluate the effect of different N-end tags on immunoreactivity of antigenic epitope(s) of gE protein modeled by recombinant fusion proteins.
Methods: Artificial gene encoded 296–414 aa region of gE TBEV have been synthesized from oligonucleotides by using PCR reaction. Recombinant antigen was expressed in E. coli cells as a fusion protein with four different N-end tags variants: 6-histidines (6His), glutathione-S-transferase (GST), GST and 6-histidines (GST+6His) and maltose binding protein (MBP). Recombinant fusion proteins were purified by affinity chromatography and tested in enzyme immunoassay with previously well defined 36 anti-TBEV positive and 56 anti-TBEV negative serum samples.
Results: Different levels of the protein expression, solubility and immunoreactivity was observed for all four recombinant constructions. GST-N-tagged protein was the most immunoreactive: it has detected IgG anti-TBEV in 97.2% positive serum samples and did not react with any anti-TBEV negative sera. Average signal to cut off (S/C) level with positive samples was equal toa 5.4. Fusion proteins with MBP and 6His also had high enough level of immunoreactivity. IgG anti-TBEV activity was detected in 86.1% and 72.2% of positive sera samples, and average S/C level with positive samples was somewhat higher – 6.3 and 6.1, respectively. But the specificities of these fusion variants were only 96.4% for MBP fusion antigen and 98.2% for 6His fusion protein. The GST+6His N-end tagged antigen demonstrated lower level of immunoreactivity. IgG anti-TBEV activity has been detected only in 5.6% of positive sera samples. Average S/C level with positive samples also was lowest – 2.3.
Conclusions: The achieved results suggest that selection of protein expression system, location and characteristics of fusion tag plays important role in successful modeling of antigenic epitopes by recombinant proteins. Fusion of recombinant antigen comprising TBEV gE regions at 296–414 aa position with glutathione-S-transferase at the N-end demonstrated significant diagnostic potential for the development of anti-TBEV ELISA diagnostic assays.
Respiratory tract infections and their management
P1183 Initial hospital management at emergency departments of community-acquired pneumonia in Spain
V. Alvarez-Rodriguez, D. Martinez, M.J. Gimenez, J. Barberan, L. Aguilar, M.J. Ruiz-Polaina, J. Prieto (Madrid, ES)
Objectives: To study the initial management of patients with community-acquired pneumonia (CAP) admitted to hospital through Emergency departments.
Methods: Clinical records of patients with ≥ 14 years of age and CAP diagnosis admitted to hospital through Emergency departments in a three-month period (January–March 2003) in 24 Spanish hospitals were retrospectively reviewed in order to obtain demographic data, clinical and analytical data necessary to classify them according to the Pneumonia Severity Index (PSI) category, comorbidity data, microbiological diagnostic tests performed, initial antibiotic treatment and outcome. Patients sent for treatment on an ambulatory basis were excluded from the study.
Results: 341 patients (67.0 ± 24.6 years; 65.3% males) were admitted to hospital trough the Emergency department: 24.0% were managed at Short Stay Medical Units (SSMU), 36.4% at Internal Medicine, 30.2% at Pneumology, 1.7% at Intensive Care Units (ICU), 2.1% at Infectious Diseases and 5.6% were derived to a different hospital. Patients in high-risk classes (IV-V) were 65.7%, 14.7% were class III and 19.6% were classes I-II. Blood culture was performed in 50.1% patients (41% patients class I-III, 53.8% patients class IV, and 56.2% patients class V), while sputum culture was performed in 35.8% patients (39.3% patients class I-III, 36.1% patients class IV, and 31.4% patients class V). Pneumococcal or Legionella urinary antigen detection was performed in 34% and 42.2%, respectively. Most frequent comorbidities were: chronic obstructive airway disease (37.2% patients), heart disease (24.6%), hypertension (17%), diabetes (10.8%), and malignancies (10%). Initial treatment was fluoroquinolone in 37.5% patients, 3rd generation cephalosporin + macrolide in 19.4%, amoxicillin/clavulanic acid in 17.9%, amoxicillin/clavulanic acid + macrolide in 7.0%, 3rd generation cephalosporin in 5.0%, macrolides in 4.7%, and others in 8.5%. Eleven patients died (3.2%), 100% of them within the high-risk class IV-V.
Conclusions: Microbiological diagnostic tests may be scarcely used. As severity increased, request of blood culture increased but of sputum culture decreased. Most frequent treatments applied were fluoroquinolones as monotherapy followed by 3rd generation cephalosporin + macrolide or amoxicillin/clavulanic acid. These empirical treatments may reflect resistance rates in Streptococcus pneumoniae in Spain, and/or coverage of both typical and atypical aetiologies.
P1184 Comparison of first with second-line antibiotics for acute bacterial exacerbations of chronic bronchitis: a meta-analysis of randomised controlled trials
G. Dimopoulos, I.I. Siempos, I.P. Korbila, K.G. Manta, M.E. Falagas (Athens, GR)
Background: Although acute bacterial exacerbations of chronic bronchitis (ABECB) are common, there has been no meta-analysis that focused on the optimum regimen.
Methods: To evaluate the comparative effectiveness and safety of the first- and second-line antimicrobial agents for the treatment of patients with acute bacterial exacerbation of chronic bronchitis (ABECB), in an era of increasing antimicrobial resistance among the responsible microbes for ABECB, we performed a meta-analysis of randomised controlled trials (RCTs) retrieved through searches of the PubMed and the Cochrane databases.
Results: Twelve RCTs, enrolling 2,261 patients, were included in the analysis. First-line antibiotics were associated with lower treatment success compared to second-line ones in the clinically evaluable patients [odds ratio (OR) = 0.51, 95% confidence intervals (CI): 0.34–0.75]. There was no difference between the compared regimens regarding mortality (OR = 0.64, 95% CI: 0.25–1.66) or treatment success in microbiologically evaluable patients (OR = 0.56, 95% CI: 0.22–1.43) or adverse effects in general (OR = 0.75, 95% CI: 0.39–1.45) or diarrhoea in particular (OR = 1.58, 95% CI: 0.74–3.35). Treatment success was lower in penicillin- than in macrolide-recipients (CE: OR = 0.36, 95% CI: 0.17– 0.75). On the contrary, in the subgroup analysis of RCTs that enrolled mainly hospitalised patients, less drug-related adverse effects were attributed to first- than to second-line antibiotics (OR = 0.34, 95% CI: 0.23–0.51). Second-line antibiotics are more effective, but less safe in the subset of hospitalised patients, than first-line ones when administered in patients with ABECB.
Conclusions: Compared to first-line antibiotics, second-line antibiotics are more effective but less safe in the subset of hospitalised patients, when administered in patients with ABECB. Although, the implications of our findings seem to contradict with the attempt to preserve new antibiotics and prevent induction of antimicrobial resistance, treatment directed toward resistant pathogens with inadequately effective antimicrobials and unnecessary administration of antibiotics in patients with mild ABECB are more important problems regarding this issue.
P1185 Community-acquired pneumonia: doctors do not follow guidelines
M.B.J. Beadsworth, P. Collini, P. Deegan, T. Neal, P. Burnham, J. Anson, N.J. Beeching, A.R.O. Miller (Liverpool, UK)
Objectives: Appropriate assessment of CAP allows accurate severity scoring and hence optimal management, leading to reduced morbidity and mortality. British Thoracic Society (BTS) guidelines provide an appropriate score. We assessed adherence to BTS guidelines in our medical assessment unit (MAU) in 2001/2. We re-assessed adherence in 2005/6, 3 years after introducing an educational programme.
Methods: A retrospective casenote study comparing diagnosis, management and outcome of CAP during admission to MAU during 3 months of winter in 2001/2 and 2005/6. (BTS, CURB-65, scoring was used for 2005/6)
Results: Over 3 months in 2001/2, of 165 patients coded as CAP, 65 were wrongly coded and 100 included in the study. In 2005/6 this had not significantly changed. 130 were coded as CAP, of these 43 were wrongly coded and 87 enrolled. In 2001/2, 48% did not receive a severity score, in 2005/6 this had significantly increased to 87%. (p < 0.0001) Parenteral antibiotics were used in 79% in 2001/2 and 77% in 2005/6. 3rd generation cephlosporins were used in 63% in 2001/2 and 54% in 2005/6.
In 2001, 15 different antibiotic regimes were prescribed, in 2005/6 this had increased to 19.
Conclusions: Coding remains poor. Adherence to CAP management guidelines was poor and has significantly worsened. Educational programmes, alone, do not improve adherence. Restriction of antibiotic prescribing should be considered.
P1186 Implementation of national guidelines for good antibiotic policy of respiratory tract infection in hospitalised children with community-acquired pneumonia
V. Maresova, Z. Blechova, M. Podojilová, B. Horová, K. Soltysova (Prague, CZ)
Objectives: The consumption of antibiotic (AB) increased in the Czech Republic in 1993 significantly and increased the resistance of pathogens too. Members of the Czech Medical Association prepared guidelines for treatment of community-acquired respiratory infections. The aim of our study was to evaluate the implementation of these guidelines in hospitalised children with community-acquired pneumonia (CAP).
Methods: 241 children (mean age 6.6 years) were hospitalised in Bulovka University Hospital (1997–2006) with diagnosis CAP. We investigated from this group two subgroups: first consists of children admitted before publication of the guidelines in 1997 and the second consists of children hospitalised in 2005–2006. We evaluated both the initial antibiotic therapy administered in accordance with the guidelines.
Results: The 1st group were evaluated 33 children (9 m and 24 f); in the 2nd second sub-group were 36 children (22 m and 14 f). All children were treated with antibiotics. In none patient was taken haemoculture, and in 6 patients was performed serologic examination: in 3 patients were positive antibodies against Mycoplasma pneumoniae, in 1case influenza virus and in 1child RSV antibodies. Initial therapy with penicillins antibiotics was used in 14 children, cephalosporins a macrolides in 5 children, 1 child was treated with co-trimoxazol, 8 children were treated with combination of antibiotics and in 4 cases antibiotic therapy changer. In the second group was haemoculture performed in 34 and in 3 patients was detected S. pneumoniae, in one case pneumococcal Ag in urine. S. pneumoniae was good sensitivity to penicillins and macrolides. Serologic examination was performed in 14 children. In 5 cases was detected M. pneumoniae, in 2 cases viral infections. Initial therapy with penicillin antibiotic was used in 18 children, with co-aminopenicillins in 4, macrolides in 6, cephalosporins in 3, in 2 cases doxycycline, 1 patient had co-trimoxazol. In 6 children was the AB therapy changer during hospitalisation, in other 6 cases was used combination of AB.
Conclusion: Initial therapy with penicillins used more frequently (22 vs. 14), prescription of macrolides didn't increase, and the use of cephalosporins decreased in 2nd group. Significantly higher was the number of aetiologically verified CAP. Quality of prescription ABs were better in the 2nd group than the 1st in 1997.
Acknowledgements: This study is supported by grant IGA Ministry of Health NR/8297–3.
P1187 Physician adherence to pneumonia guidelines regarding initial regimen choice
T.A. Peppas, P. Matsoukas, H. Vlastari, A. Sotiropoulos, I.D. Binikos, B.D. Kandyla, I. Tamvakos, S.I. Pappas (Piraeus, GR)
Objective: To assess physicians adherence on pneumonia guidelines (G) regarding the initial regimen chosen.
Methods: All pneumonia admissions were prospectively entered in database and all regimens chosen by the attending physicians were compared to the G available and taught to them by admission date, namely IDSA 2003, BTS 2004, Hellenic IDS 2005 and ERS 2005, as well as AJRCCM 2005; 388 for nosocomial and healthcare-associated pneumonia Outcome and hospital stay (HS) was compared for patients with regimen G concordant (GCR) and non-concordant (GNCR). SPSS programme, time: October 2004 to September 2006
Results: A total of 72 patients (M: 44, F: 28) mean age 72.1 yrs were included. Hospital or nursing home pneumonia consisted 14 of them and previous antimicrobial usage was recorded in 32 (44%) Concordance to any G was noted in 47 (65.3%) and the respective percentages were IDSA G: 65%, BTS G: 64% ERS G: 62% and Hellenic IDS: 46%. Main reason for low concordance to Hellenic G was choice of respiratory quinolone, without prior antimicrobial use. Hospital stay was higher for GNCR (10.2 v 8.4, NS) but mortality was higher (28% v 7% of GCR, p = 0.03, Yates corrected chi-square).
Conclusions: Physicians choice of regimen in pneumonia was concordant to existing G at 65% of cases. There was higher mortality among patients with non concordant regimen, and though confounding factors (mainly severity index) can easily bias a small sample size, this fact underlines guidelines utility and value, warranting a more strict adherence
P1188 The hospital admission decision for patients with community-acquired pneumonia; factors in low-risk PORT-score categories associated with hospitalisation: would low risk for death have applied in case of outpatients care?
S. Soto, E. García-Vázquez, J. Gómez, J.A. Herrero, V. Baños, J. Ruiz, T. Hernandez, M. Valdés (Murcia, ES)
Objectives: Pneumonia Patient Outcomes Research Team (PORT) has developed a prediction rule to identify patients with Community-acquired pneumonia (CAP) who are at risk for death and other adverse outcomes. The authors recommend outpatient management for Fine categories I and II and parenteral antimicrobial therapy at home or a short stay (<24 hours) in a hospital observation unit for group III. However, many of these patients are hospitalised.
Patients and Methods: Observational study of patients with CAP (class I, II and III according to PORT-score) admitted at a tertiary general hospital. Data collection from clinical records has been done according to a standard protocol study of CAP. We analysed epidemiological, clinical, radiological and laboratory data and outcome of low-risk hospitalised patients.
Results: ninety nine (46.9%) out of 211 patients with CAP were classified in Fine categories I, II and III (26, 33 and 40 respectively); 99 (100%) were hospitalised in general medical wards. Mean age was 50 years (range, 13 to 85 years); <65 years in 72%, 65–75 years in 19% and >75 years in 8%. A coexisting condition was present in 32% patients (congestive heart failure 15%; chronic lung disease 14%). More frequent physical-examination findings were systolic blood pressure <90 mmgHg (8%), pulse >125/min (7%) and respiratory rate >30/min (6%). More prevalent laboratory and radiologic findings were partial pressure of arterial oxygen <60 mmHg (26%), pleural effusion (9%) and glucose >250mg/dl (8%). Only 1 patient (1%) in group III died. Mean hospital stay was 6 days (range, 1–24 days).
Conclusions: In our series, CAP patients in PORT-Score categories I, II and IIII who were admitted to the hospital represent a very high rate (46.9%) of all CAP admissions. Our findings suggest that additional consideration is provided by physicians to the presence of coexisting conditions some of which are not considered in the PORT-score (chronic lung disease and diabetes) and certain findings in physical-examination (homodynamic instability and high respiratory rate) and laboratory/radiology studies (hyperglycaemia, hypoxaemia and pleural effusion). Mortality rate was very low (1%). Whether this low risk for death would have applied in case of outpatients care is a question that remains to be answered.
P1189 Simpler criteria to assess mortality in patients with community-acquired pneumonia
S. Soto, E. García-Vázquez, J. Gómez, J. Herrero, V. Baños, J. Ruiz, T. Hernandez, M. Valdés (Murcia, ES)
Objectives: Pneumonia Patient Outcomes Research Team (PORT) has developed a prediction rule to identify patients with Community-acquired pneumonia (CAP) who are at risk for death and other adverse outcomes. This score considers too many variables. Simpler criteria are needed to evaluate the risk of mortality in patients with CAP.
Patients and Methods: Observational study of patients with CAP admitted at a tertiary general hospital. Data collection from clinical records has been done according to a standard protocol study of CAP. We analysed epidemiological, clinical, radiological and laboratory data associated with mortality.
Results: two hundred and eleven (n = 211) patients with CAP were evaluated; severity distribution according to PORT score was 12.3% in group I, 15.6% in group II, 19% in group III, 35.5% in group IV and 17.5% in group V. Mean age was 63 years (range, 13 to 100 years); <65 years, 43.6%; 65–75 years, 25.6% and >75 years, 30.8%. One or two coexisting conditions were present in 54.5% patients and more than 2 in 7% (congestive heart failure 33.6%; chronic lung disease 29.9%). Mortality rate was 0% in groups I-II, 2.5% in group III, 5.3% in group IV and 27% in group V. All variables considered in PORT-score were included in a mortality predicting model; factors significantly associated with death were altered mental status, respiratory rate >30/min, arterial pH <7.35, glucose >250 mg/dl, and age >75 years; 99% of patients with CAP who did not present any of these abnormalities died.
Conclusions: In our series, simpler criteria to assess mortality in patients with Community-acquired pneumonia were identified. The non-existence of altered mental status, respiratory rate >30/min, arterial pH <7.35, glucose >250 mg/dl, and age >75 years predicted a non-fatal outcome in 99% of patients. Presence of these clinical or laboratory findings should be considered as mortality predictors and can be used as a severity adjustment measure and therefore may help physicians make more rational decisions about hospitalisation for patients with CAP.
P1190 Pre-admission use of statins and mortality within 90 days after hospitalisation with pneumonia: population-based cohort study of 29,900 adult patients
R.W. Thomsen, A. Riis, J.B. Kornum, S. Christensen, S.P. Johnsen, H.T. Sørensen (Aarhus, DK)
Objectives: Statins may improve outcomes of severe infections due to antithrombotic, anti-inflammatory, and immuno-modulatory effects. We examined preadmission use of statins as a predictor of 30-day and 90-day mortality among adult Danish patients hospitalised with pneumonia.
Methods: All patients recorded with a first-time discharge diagnosis of pneumonia between 1997 and 2004 were included in this population-based cohort study in two Danish counties (population, 1.15 million). Information on statin use, comorbidities, laboratory findings, occurrence of bacteraemia and death was obtained from medical and administrative databases. We computed mortality rate ratios in preadmission statin users vs. non-users, adjusted for potential confounders in a regression analysis.
Results: A total of 29,900 adult patients were hospitalised with pneumonia (median age 73 years); of these, 1,516 patients (5.1%) were preadmission statin users. Laboratory findings on admission were similar among users and non-users of statins. Among patients who were blood cultured (64% of statin-users and 60% of non-users), the adjusted RR for bacteraemia in statin-users vs. non-users was 0.99 (95% CI: 0.66–1.50). Overall mortality in statin-users vs. non-users was 10.8% vs. 15.7% after 30 days and 17.0% vs. 22.4% after 90 days, corresponding to adjusted 30- and 90-day mortality rate ratios of 0.65 (95% CI: 0.56–0.76) and 0.69 (95% CI: 0.61–0.79).
Conclusion: Preadmission use of statins was associated with a substantially decreased mortality following hospitalisation with pneumonia. Randomised trials are needed to determine if this association is causal or caused by uncontrolled confounding.
P1191 Association of proinflammatory cytokines with fibrinolytic enzymes and biochemical parameters in childhood infectious parapneumonic effusions
C.Y. Chiu, J.L. Huang, K.S. Wong, M.H. Tsai, T.Y. Lin (Keelung, Taoyuan, TW)
Objectives: To evaluate the relationship of proinflammatory cytokines (tumour necrosis factor-a (TNF-α), interleukin-1b (IL-1β) and IL-6) with intrapleural fibrinolytic enzymes [tissue-type plasminogen activator (tPA) and plasminogen activator inhibitor type 1 (PAI-1)] and common biochemical parameters (pH, glucose, LDH), and to compare various pleural variables for predicting the need of subsequent aggressive interventions.
Methods: A total of fifty-seven patients were enrolled in the present study. Chest ultrasound was performed and results were stratified into anechoic fluid (stage 1, n = 28), with floating fibrin strands (stage 2, n = 16) and with septated fibrin (stage 3, n = 13). The correlation of various pleural variables with the formation of complicated parapneumonic effusions (CPE) was performed and factors for predicting the need of intervention procedures were also analysed.
Results: Univariate analysis revealed that pH, glucose and LDH in biochemical parameters, proinflammatory cytokines (TNF-α, IL-1β, IL-6), and fibrinolytic enzymes (tPA, PAI-1) were significantly different between these three stages. Additionally, the trends of these variables were all significantly associated with the ordered stages of parapneumonic effusions (Ptrend <0.05). Proinflammatory cytokines of TNF-α, IL-1β and IL-6 were all significantly correlated with PAI-1 in non-CPE and CPE. However, only TNF-α and IL-1β were significantly correlated with tPA in non-CPE. The pleural variables of pH, IL-1β and PAI-1 were the most significant factors for predicting the need of intervention procedures (P<0.05).
Conclusion: The increased release of proinflammatory cytokines, such as TNF-α, IL-1β, and IL-6 in pleural fluid caused by bacteria may result in an imbalance of tPA and PAI-1, which may subsequently lead to fibrin deposition and the development of empyema. In addition to pleural fluid pH, IL-1β and PAI-1 in pleural fluid may be used as alternative markers for predicting the need of subsequent aggressive interventions.
P1192 Systemic TNF-α production and severity of disease in severe pneumococcal pneumonia
E. Calbo, M. Alsina, M. Rodríguez-Carballeira, M. Xercavins, E. Cuchi, J. Garau and The Spanish Pneumococcal Infection Study Network
Introduction: Tumour necrosis factor alpha (TNF-α) is one of the earliest mediators of the inflammatory response. It induces a second wave of pro and anti-inflammatory cytokines which are mediators of the inflammatory process. The aim of our study was to analyse the correlation between TNF-α and other cytokines plasma concentrations, clinical variables and severity of disease in patients with pneumococcal pneumonia (SPP) at hospital admission.
Material and Methods: Consecutive adults with pneumonia classes III-V of the Pneumonia Severity Index (PSI) developed by the Pneumonia Outcome Research Team and with a confirmed pneumococcal aetiology were included. At admission, demographic characteristics, smoking and alcohol habits, co morbidities (Charlson score), prognosis measured by PSI and APACHE II scores, immunosuppressive conditions, previous or current therapy with statins, use of non steroidal or steroidal anti-inflammatory drugs, and time between pneumonia onset and hospital admission were prospectively recorded.
Vital signs, haematological and biochemical parameters were also assessed. Circulating levels of CRP, serum amyloid A (SAA), C3a, C5a and cytokines TNF-α, IL-1β, IL-6, IL-8, IL-10, IL-1ra were measured. Comparisons were done among patients with TNF-α above and below the median values.
Results: 28 patients with SPP were included; 14 had TNF-α concentrations <20.8 pg/mL and the other half had TNF-α concentrations above this figure. Both groups were homogeneous in terms of of demographics, clinical characteristics, presence of comorbidities, previous use of statins, severity of disease at presentation, radiological involvement and time between pneumonia onset and admission. The group with higher concentrations of TNF-α presented more frequently bacteraemia (64.3% vs 21.4%; p = 0.027), higher axillary temperature (38.4[SD0.4] vs 37.7[SD0.8]; p = 0.046) and higher concentrations of IL-1 (7.8[SD8.5] vs 2.2[SD3]; p = 0.034), IL-6 (4,952[SD6,498] vs 871[SD1,453]; p = 0.037), and IL-1ra (11,029[SD7,669) vs 5,327[SD616]; p = 0.044).
Conclusions: In pneumococcal pneumonia, higher TNF-α levels were associated with the degree of fever and the frequency of bacteraemia; IL-1, IL-6 and IL-1ra increased in parallel with TNF-α levels. No correlation could be established between TNF-α and severity of disease.
P1193 Systemic inflammatory response and time from onset to hospital admission in severe pneumococcal pneumonia
E. Calbo, M. Alsina, M. Ródriguez-Carballeira, E. Cuchi, M. Xercavins, J. Garau and the Spanish Pneumococcal Infection Study Network
Introduction: The inflammatory response triggered by the presence of invading bacteria is a dynamic process changing over time, and many factors can influence the magnitude of this response. The aim of our study was to analyse the impact of time from onset of symptoms to hospital admission on the systemic plasmatic cytokine concentrations in patients with severe pneumococcal pneumonia (SPP).
Material and Methods: Consecutive adults with pneumonia classes III-V of the Pneumonia Severity Index (PSI) developed by the Pneumonia Outcome Research Team and with a confirmed pneumococcal aetiology were included. At admission, demographics, smoking and alcohol habits, comorbidities (Charlson score), immunosuppressive conditions, previous or current therapy with statins, use of non-steroidal or steroidal anti-inflammatory drugs, and prognosis measured by PSI and APACHE II scores were prospectively recorded. Special attention was paid to record time between pneumonia onset and hospital admission, collection of initial blood samples and the initiation of antimicrobial therapy. Vital signs, haematological and biochemical parameters were also assessed. Circulating levels of CRP, serum amyloid A (SAA), C3a, C5a and cytokines TNF-α, IL-1β, IL-6, IL-8, IL-10, and IL-1ra were measured at inclusion.
Results: 32 patients with SPP were included; 13 patients were seen and included within the first 48 h and the other 19 patients after 48 h from onset of symptoms. Both groups were homogeneous in terms of demographics, clinical characteristics, presence of comorbidities, bacteraemia, previous use of statins, severity of disease at presentation and radiological involvement. The group with a longer time of evolution at entry presented higher plasmatic levels of TNF-α (19.1[SD8.5] vs. 35.5[SD26] pg/mL; p = 0.035), fibrinogen (6[SD1.8] vs. 9[SD2]; p = 0.001); CRP (130[SD85] vs. 327[SD131]; p = 0.000), SAA (678[SD509] vs. 678[SD391]; p = 0.025) and lower albumin concentrations (40[SD5] vs. 35[SD4]; p = 0.043).
Conclusions: In pneumococcal pneumonia, the sustained release over time of pneumococcal antigens strongly correlates with a higher pro-inflammatory pattern and a higher expression of acute phase protein synthesis.
P1194 C-reactive protein in community-acquired pneumonia among adults in India: association with aetiology, severity, treatment and outcome
A. Manoharan, A. Kumar, M.K. Laliltha, D. Mathai (Vellore, IN)
Objectives: To study the utility of serum C-reactive protein (CRP) as a marker for aetiology, management and prognosis of CAP.
Methods: Adult patients (n = 104) with a diagnosis of acute community-acquired pneumonia (CAP) by clinical (<2 weeks history of cough with fever and any two respiratory signs), radiological (new infiltrate) and microbiological (sputum/blood or pleural fluid culture) methods at a tertiary care hospital in South India during the period January 2002–December 2004. Patients outcome research team (PORT) score was used to assess the severity of CAP on admission. Acute and convalescent sera were evaluated for IgM and IgG response to Chlamydophila pneumoniae (Thermolab system, Finland), Mycoplasma pneumoniae (IBL, Germany). Presence of IgM or four fold rise in titre in IgG antibody levels between acute and convalescent sera was considered to be positive for these atypical pathogens. Detection of Legionella pneumophila antigen in acute phase urine samples done by a immunochromatographic method (Binax, Scarborough, USA). CRP was measured by QuikRead CRP (Orion Diagnostica, Finland) method in acute phase serum samples available from 64 patients.
Results: In 64 patients in whom CRP levels were determined, mean age was 57.6 years (SD-15.9years), with 66% being males. According to receiver operator curve (ROC), a cut-off point of 40 mg/L appeared to be the best CRP value for definitive diagnosis of bacterial aetiology of CAP. Fifty patients had CRP ≥40 mg/L (Group A) while 14 had had CRP < 40 mg/L (Group B). Mean days of hospitalisation was similar in both these groups. Two patients in group A were blood culture positive for Streptococcus pneumoniae (mean CRP 49 mg/L) while two grew this pathogen in pleural fluid (CRP > 180 mg/L). In group A, 32% and 20% were positive for C. pneumoniae and M. pneumoniae while in group B it was 27% and 7%. None in both groups were positive for L. pneumophila. No significant difference in mean duration of antibiotic therapy and correlation to severity of pneumonia existed between both the groups to their CRP levels. Mortality in group A was 24% (n = 12; meanCRP-138 mg/L; p = 0.042), while none died in group B.
Conclusion: High CRP levels are associated with bacterial aetiology of CAP. Significant association exists between high CRP levels and mortality. CRP could be used as a surrogate marker for the management of patients with CAP
P1195 Low functional levels of mannose-binding lectin despite normal genotypes in Legionella pneumophila pneumonia
B.L. Herpers, E.P.F. IJzerman, B.A.W. de Jong, J.P. Bruin, S. Kuipers, K.D. Lettinga, E.J. van Hannen, G. T. Rijkers, H. van Velzen-Blad, B.M. de Jongh (Nieuwegein, Haarlem, Apeldoorn, Amsterdam, NL)
Objectives: Deficiency of mannose-binding lectin (MBL) has been associated with an increased risk of infection, compatible with its role as a pivotal protein in early complement activation. In infections with intracellular pathogens this association is more ambiguous, as some pathogens use MBL to enter their host cell. To study this relation with Legionella pneumophila, we determined MBL levels and genotypes in a well-described outbreak of legionellosis at a flower show in the Netherlands. Since this clonal outbreak has no pathogen variability, this patient cohort is very suitable to study genetic host factors.
Methods: In a retrospective patient-control study MBL levels were determined in a haemolytic assay in acute phase serum samples from 122 patients, 59 asymptomatic seroconversion controls and 447 blood bank donors. Serum levels were classified as deficient (<0.2 ug/mL) or normal (>0.2). If multiple serum samples were available from a subject, the highest measurement was used for classification.
Genotyping of MBL was performed with denaturing gradient gel electrophoresis (DGGE) of an amplicon harbouring the three polymorphic sites in exon 1 (“0” vs wildtype “A” allele) and a SNP-PCR determining the promotor X/Y polymorphism. Genotypes were classified as deficient (0/0, XA/0) or sufficient (YA/0, A/A). Whole blood DNA isolates were available from 77 patients, 53 seroconversion controls and 223 healthy adults. Two different nested PCR-DGGE protocols were performed on serum DNA isolates from 112 patients. Serum genotyping results were only considered in analysis when both protocols had corresponding results (72 patients). No discrepancies were found between serum and whole blood genotyping of exon 1 (38 patients). In this way 111 patients could be genotyped reliably.
Results: Compared to 30% of patients only 2% of seroconversion controls and 3% of blood bank donors had MBL levels less than 0.2 ug/mL (X2 p<0.01). Deficient genotypes were found in only 14% of patients compared to 7% and 17% of both control groups (X2 p > .05). Significantly more patients than seroconversion controls with sufficient genotypes had MBL levels below 0.2 ug/mL (20% vs 0%, X2 p< 0.01).
Conclusion: Patients had significantly lower MBL levels at the acute phase of legionellosis than both control groups. This difference was not found in genotypes. This discrepancy suggest either an exhaustion of MBL or an inability to produce MBL in legionellosis, even in patients with sufficient genotypes.
P1196 Mycoplasma pneumoniae infection – 63 case studies
A. Topan, M. Lupse, V. Zanc, D. Tatulescu, M. Flonta (Cluj-Napoca, RO)
Objective: To evaluate the clinical and laboratory findings in Mycoplasma pneumoniae infection.
Methods: A retrospective study was performed upon 63 cases hospitalised in the university hospital of infectious diseases from February 2005 to October 2006. The clinical diagnosis was confirmed by a positive enzyme-linked immunosorbent assay (ELISA) test for IgM antibodies.
Results: An almost equal gender distribution was found, 55.55% female (n = 35) and the median age was 25 within the range 1.3–63 years (18 children, 45 adults). The highest proportion of patients was found between ages 25 and 44 years (36.55%). At the onset of the disease the most frequent signs and symptoms were: fever in 53 cases (84.12%), cough in 38 cases (60.31%), headache 18 cases (28.57%), myalgias in 10 cases (15.87%), nonspecific arthralgias in 5 cases (7.93%). The pulmonary auscultation was normal in 28 cases (44.44%). Abnormal findings on chest X–ray examination were depicted in 42 cases (66.66%). Laboratory findings indicated the presence of leukocytosis in 23 cases (36.5%) and elevated erythrocyte sedimentation rate in 51 cases (82.25%). Extrapulmonary complications were found in 23 cases (36.5%): nervous system involvement was present in 6 cases (9.52%), skin rashes of different severity were found in 8 cases (12.69%) and cardiac involvement (pericarditis) was present in one single case (1.58%) and thrombocytopenia also in one case (1.58%).
Conclusions: Mycoplasma pneumonia infection was frequent in young adults. The onset with fever and cough, respiratory signs and laboratory findings are not specific for atypical pneumonia. The presence of extrapulmonary complications for more than one third of patients suggests the aetiology. The empiric antibiotic treatment with macrolides is strongly required for young adults with respiratory infections mainly when they have extrapulmonary manifestation. The aetiological diagnosis remains a problem in the absence of some quick and cost affordable methods of detection.
P1197 Microbial pathogens and antibiotic susceptibility in community-acquired lower respiratory tract infection in Korea
H-J. Kim, Y-J. Park, K-G. Park, E-J. Oh, M.W. Kang (Seoul, KR)
Introduction: Community acquired lower respiratory tract infection is recognized as a major cause of morbidity, mortality and antibiotic use in the community. We investigated major microbial pathogen and antibiotic susceptibility in community acquired lower respiratory tract infection in Korea.
Methods: This study was conducted using sputum specimens sent to community hospitals between December, 2002 and February, 2004. The sputum specimens were tested for adequacy by Gram staining. Microbial pathogens were identified by use of VITEK (bioMérieux, France), API ® strips (bioMérieux, France) and conventional biochemical tests.
Results: Of the 371 sputum specimens tested, 145 (39.1%) were inadequate, 226 (60.9%) were adequate. Of the 226 adequate specimens, 107 (47.3%) grew microbial pathogens; Streptococcus pneumoniae 18.7% (n = 20), Klebsiella pneumoniae 13.1% (n = 14), Pseudomonas aeruginosa 13.1% (n = 14), Haemophilus influenzae 12.1% (n = 13), Pseudomonas spp. 12.1% (n = 13), Staphylococcus aureus 12.1% (n = 13), Moraxella catarrhalis 9.4% (n = 10), Streptococcus spp. 5.6% (n = 6), and Acinetobacter spp. 3.7% (n = 4). Susceptibility of S. pneumoniae to penicillin, erythromycin, trimethoprim-sulfamethoxazole, cefotaxime and vancomycin was 60%, 30%, 25%, 85%, and 100%, respectively. K. pneumoniae showed high susceptibility (90–100%) to most antibiotics. Susceptibility of P. aeruginosa to ceftazidime, aminoglycoside, ciprofloxacin, imipenem, meropenem was as high as 92.9–100%. P. fluorescens/putida was 100% susceptible to cipirofloxacin, gentamicin, piperacillin, piperacillin/tazobactam and 84.6%, 92.3%, 92.3% susceptibility to imipenem, ceftazidime, sulbactam, respectively, but showed a low susceptibility of 23.1 and 69.2% to aztreonam and meropenem, respectively. S. aureus showed low susceptibility (15.4%) to penicillin, but was 84.6%, 92.3%, 92.3% susceptible to erythromycin, ciprofloxacin, oxacillin, respectively, and 100% susceptible to clindamycin, teicoplanin, and vancomycin.
Conclusion: In addition to S. pneumoniae, H. influenzae, and M. catarrhalis, the traditional common pathogens of community acquired lower respiratory tract infections, various pathogen such as K. pneumoniae, P. aeruginosa, S. aureus were detected. Antibiotic susceptibility of S. pneumoniae to erythromycin (30%), trimethoprim-sulfamethoxazole (25%) and S. aureus to penicillin (15.4%) was low. Except for these 3 pathogens, most pathogens showed high antibiotic susceptibility.
P1198 The aetiological agents in adult patients with community-acquired lower respiratory tract infections in Turkey
I. Koksal, T. Ozlu, O. Bayraktar Saral, K. Aydin, R. Caylan, F. Oztuna, Y. Bulbul, G. Yilmaz, N. Sucu and the Pneumonia Study Group, Turkey
Objectives: Lower respiratory-tract infections, community-acquired pneumonia (CAP) and acute exacerbation of chronic obstructive pulmonary disease (COPD), is recognized as a major cause of morbidity, mortality, and antibiotic use in the community. The microbiologic agents of CAP and COPD vary considerably not only worldwide, but regionally as well.
In this study, it is aimed to investigate typical and atypical aetiological agents of community-acquired lower respiratory-tract infections.
Methods: The study which was led by Karadeniz Technical University was conducted prospectively between November 2003 and March 2005. Eight universities hospitals from seven different geographical regions of Turkey participated in this study. Sputum, nasopharyngeal aspirate, blood and urinary samples were taken from the 218 patients diagnosed lower respiratory-tract infections clinically and radiologically. The samples were inspected by culturing, Gram's stains and immunofluorescence methods.
Results: Aetiological agents were identified in the 62.8% of the patients. While 24.3% of them were typical, 26.6% were atypical and 11.9% mix agents. Major pathogens identified in lower respiratory-tract infections were %23.4 S. pneumoniae, 21.9% M. pneumoniae, and 16% RSV. 37.6% of the cases were in the winter, 30.3% in the spring. 56.9% of the patients were under age 65. While 29.1% S. pneumoniae, 23.6% RSV and 14.5% M. pneumoniae were identified in the patients above age 65, 26.8% M. pneumoniae, 19.5% S. pneumoniae, 10.9% H. influenzae and 10.9% RSV identified in those under the age of 65. While the major pathogens in CAP were 24.4% M. pneumoniae, 20.9% S. pneumoniae and 10.4% RSV, those in COPD were 27.4% S. pneumoniae, 25.5% RSV, 17.6% M. pneumoniae and 17.6% H. influenzae.
Conclusion: When the lower respiratory-tract infection agents were inspected, S. pneumoniae, M. pneumoniae and RSV were found out in the first three positions. Most of the cases are seen in the winter. CAP and COPD agents are similar. S. pneumoniae is, by the ratio of 23.4%, the most frequent agent.
P1199 The microbiological isolates in patients with non-CF bronchiectasis in stable clinical situation and in disease exacerbation
P. Kecelj, E. Music, V. Tomic, M. Kosnik, R. Erzen (Golnik, SI)
Background: Isolation of potential pathogenic microorganisms (PPM) help in decision about first line antibiotic therapy in exacerbation of inflammation in bronhiectasis.
Methods and Patients: 68 patient (26 female, average age 68.2 years) with non CF bronchiectasis were included in two years retrospective study. Bronchectasis were diagnosed with CT scan. Patients were divided in group with (high CRP and WBC) and without exacerbation of the disease. Amount, quality and microbiological isolates of sputum were analysed in all patients. Antibiotic therapy of patients was recorded from medical documentation. Lung function was determinated by FEV1 and DLco.
Results: 35 patients (52%) were in group with no signs of inflammation. Average amount of sputum was 17 mL per day and purulence grade 1.0 of 3. Average FEV1 was 64% of normal values. Normal flora in sputum was found in 11% of patients. PPM were isolated in 48% of patients. Most frequent isolates were: P. aeruginosa in 11.4%, H. influenzae in 8.6%, Streptococcus spp. in 8.6%, MSSA in 8.6% of patients. Bacteria: A. baumannii, B. cepacia, A. xylosoxidans, M. catarrhalis, K. oxytoca were isolated in single patients. 33 patients (48%) were in group with exacerbation of disease. Average amount of sputum was 45 mL per day with purulence grade 2.5 of 3. Average FEV1 was 38% of normal. Normal flora in sputum was found in 24% of patients. Most frequent isolates were: P. aeruginosa in 30%, H. influenzae in 6%, Streptococcus spp. in 3%, MSSA in 15%, MRSA in 6% of patients. P. aeruginosa together with second bacteria was isolated in 10% of patients. In one patient H. influenzae and MSSA was isolated. Bacteria: A. baumannii, B. cepacia, A. xylosoxidans, M. catarrhalis, K. oxytoca, E. coli, E. cloacae were isolated in single patients.
Ciprofloxacin in combination with cephalosporins of 3th and 4th generation was therapy for all patients with isolated P. aeruginosa. In other patients AMC was used in 27%, ciprofloxacin in 20% of cases, others antibiotics were used seldom.
Conclusions: Potential pathogenic microorganisms were present in sputum in 48% of patients with clinically stable bronchiectasis, comparing to pathogenic microorganisms in 74% of patients with exacerbation of bronchiectasis. In exacerbation of the disease the empirical antibiotic therapy should be selected according to pervious antibiotic resistance pattern of PPM in patient, until the real pathogenic microorganisms are isolated.
P1200 An epidemiological analysis of Streptococcus pneumoniae infection in outpatient acute exacerbations of chronic bronchitis
P. Weber, A. Anzueto, J. Garau, D. Guillemot, J. Gaillat, S. Sethi (Vaires-sur-Marne, FR; San Antonio, US; Terrassa, ES; Paris, Annecy, FR; Buffalo, US)
Objectives: Streptococcus pneumoniae (SP) is a major bacterial cause of acute exacerbations of chronic bronchitis (AECB). This ad hoc analysis of a large cohort of AECB patients recruited to an ongoing clinical trial in a usual-care setting (>1,000 sites/8 countries) was conducted to determine the association between SP carriage and patient demographics and AECB symptoms.
Methods: Patients (n = 3,869) enrolled in the trial were ≥35 years of age with a history of chronic bronchitis and a clinical diagnosis of AECB. Bacteriologic testing of sputum samples obtained on Day 1 was conducted centrally. Patients were considered SP-positive (SP+) whenever SP was isolated, irrespective of the number of colony-forming units. Statistical comparisons of demographic data and AECB symptoms (mild/moderate/severe) were made between SP+ and SP-negative (SP-; including negative sputum cultures) patients using Wilcoxon, Fisher, and Cochran-Mantel-Haenszel tests.
Results: Overall, 13.88% (537/3,869) of evaluable patients were SP+. The median patient age was 61.0 years in both SP+ and SP– groups. More patients in the SP+ group were male (78.8% vs 69.5%; p< 0.0001). The distribution (%) of non-smokers/ex-smokers/current smokers was 9.7/35.8/54.6 in the SP+ group and 17.2/36.3/46.5 in the SP– group (p < 0.0001). The median number of packets/year smoked by current/ex-smokers was higher in the SP+ group than in the SP– group (36 vs 33; p = 0.005). Of the AECB-related symptoms assessed, the severity of dyspnoea, haemoptysis, cyanosis, and wheezing, rales, and rhonchi were not significantly different between the groups, while cough, sputum production, and sputum purulence were all more severe in the SP+ group (p = 0.012, p < 0.0001, and p<0.0001, respectively). Fever was observed in a greater proportion of patients in the SP+ group than in the SP– group (36.8% vs 29.2%; p = 0.0004).
Conclusions: A higher than expected proportion of usual-care AECB patients may be colonised or infected with SP. SP+ patients are more likely to be male, current or ex-smokers. SP-related AECB appears to be more symptomatic than other bacterial AECB, as SP+ patients present with more severe AECB symptoms, purulent sputum and fever than SP– patients.
P1201 Quantitative detection of Streptococcus pneumoniae from sputum samples for diagnosis of community-aquired pneumonia
N. Johansson, M. Kalin, J. Hedlund (Stockholm, SE)
Objectives: To assess sensitivity of real-time quantitative PCR (RQ-PCR) applied on induced sputum samples to identify Streptococcus pneumoniae (SP) aetiology in patients admitted to hospital with community-acquired pneumonia (CAP).
Methods: During a 12-month period, patients admitted with CAP were included prospectively. Induced sputum samples were obtained with assistance from a respiratory physiotherapist and analysed by culture and RQ-PCR.
Results: In total, 70/184 patients (38%) were diagnosed with SP as aetiologic agent. Cultures from blood, sputum, and nasopharyngeal secretions were positive in 27/179 (15%), 19/128 (15%), and 42/158 (27%) cases. Positive findings in urine antigen assays were obtained in 33/169 (20%), and in sputum analysed by RQ-PCR in 34/127 (27%) cases.
Of these 34 sputum samples positive by RQ-PCR, half (17) were negative by culture. Most of these patients (14/17) had been started on antibiotic therapy when the samples were collected.
Conclusion: SP pneumonia may be rapidly diagnosed by analysing induced sputum samples by RQ-PCR, especially in patients in whom antibiotic therapy has been initiated.
P1202 Severe invasive Streptococcus pneumoniae infection with cardiac involvement: report of two household cases
H. Hyvernat, P.M. Roger, T. Fosse (Nice, FR)
Objectives: We report two related cases of adults presenting severe community-acquired invasive infection with the same strain of Streptococcus pneumoniae and discuss the initial cardiologic presentation mimicking a myocarditis.
Method: Case report
Results: An 81-year-old man presented at the emergency unit with severe dyspnea. Ultrasound examination demonstrated systolic dysfunction with septal hypokinesis and 30% left ventricular ejection fraction. Laboratory values showed elevated C-reactive protein, myoglobin and BNP but no troponin elevation. Shortly after admission, the patient presented cardiac arrest with ineffective resuscitation. His 77-year-old wife was admitted on the following day because of a thoracic oppression and hypoxaemia. The ECG had low voltage in the limb leads and showed ST-segment elevation in the anterior septal apex. Laboratory data showed no troponin neither myogobin elevation but high CRP level. Ultrasound examination show major anterior and inferior left ventricular dysfunction but coronarography was normal. Subsequently, hyperthermia and unstable haemodynamic condition appeared. Blood culture allowed isolation of a S. pneumoniae serotype 9v strain susceptible to penicillin. The patient had a favourable course with amoxicillin treatment and mechanical ventilation. Cardiac function was gradually restored and the ECG reverted to normal. Pneumococcal antigens by agglutination test was found in the initial patient's serum which had been kept at 4°C in a dry tube and culture of the blood clot led to isolation of an identical S. pneumoniae (same serotype and identical biochemical phenotype).
Conclusion: These cases are unusual in the severity of initial presentation mimicking myocarditis and in the particular bacterial isolation method. Pneumococcal infections are rarely localised to cardiovascular system. Myocarditis seems to be exceptional. Cardiac dysfunction in the first case evokes myocardial depression of septic shock and the second case could evoke stress cardiomyopathy (takotsubo). But the two cases evoke both an alteration of myocardium directly by S. pneumoniae or indirectly in a toxinic way. The patients we present were not related genetically, suggesting that virulence is solely dependent on the bacterial strain. The lack of myocardium biopsy does not allow us to conclude. Nevertheless, this case report has the interest to describe potential differential diagnosis of myocarditis for invasive pneumococcal infection.
P1203 Pneumococcal empyema in Toronto, Canada, 1995–2006
T. Lee, K. Green, A. McGeer, D. Low for the Toronto Invasive Bacterial Diseases Network
Background: Several recent studies have documented increases in the rate of pneumococcal empyema (PEMP). We examined trends in the occurrence of PEMP in Toronto, Canada from 1995 to 2005.
Methods: Population based surveillance for invasive pneumococcal disease (IPD) in residents of Toronto and Peel region, Ontario, Canada (population 3.9M) has been on-going since 1995. PEMP includes cases with a positive pleural fluid culture, and bacteraemic cases with a clinical diagnosis of empyema. Data are collected from patients and their physicians. Antibiotic histories are available for patients from 2000 on.
Results: From 1995 to 2005, 4,496 episodes of IPD have been identified: 114 (2.5%) are PEMP. The incidence of empyema increased from 0.22/100,000/y in 1995–7 to 0.34/100,000/y in 2003–5, while the incidence of all IPD decreased from 13.5 to 7.9/100,000/y. Patients with PEMP were not significantly different from others with IPD in age (median 56y vs 50y, P = .14), gender (61% vs 55% male, P = .22), or proportion that were healthcare acquired (6.0% vs 4.2%, P = .34), but were somewhat more likely to have a chronic underlying illness (68% vs 59%, P = 0.05). Isolates from PEMP were more likely to be of serotype (ST) 1 (5/32 PEMP vs 96/3,902 other, P = 0.001) and ST12F (6/80 vs. 95/3,854, P = 0.02); they were also more likely to be resistant to levofloxacin (4/101 vs 29/3,933, P = 0.004) and erythromycin (17/101 vs 439/3,956, P = 0.08) but not penicillin (5/101 vs. 154/3,956, P = 0.60). Patients with PEMP were more likely to failing out-patient antibiotic therapy when admitted with IPD (11EMP of 127 cases failing antibiotics, vs 52EMP of 1,343 other cases, P = 0.01). Patients who were failing fluoroquinolone (FQ) outpatient therapy were more likely to have a diagnosis of empyema than other patients (4/26, 15% vs 65/2,275, 3%, P = 0.008). All 4 EMP patients with FQ resistant isolates were failing FQ: none of these patients died, compared to 14/58 (24%) other patients (P = .26). Similarly, 2/3 EMP patients with erythromycin resistant isolates were failing macrolides (ML); neither of these two patients died, compared to 14/62 (23%) other patients.
Conclusion: The incidence of PEMP is increasing. Some serotypes are more likely to cause PEMP than others. Patients with pneumococcal infection who are failing ML or FQ therapy because their isolate is resistant are more likely to present with empyema, but less likely to die than other patients, perhaps because these antibiotics control systemic but not local disease.
P1204 Community-acquired meningococcal pneumonia and empyema: case report
A. Kyratsa, I. Graphakos, Z. Salem, A. Zaphiropoulou, S. Tragaras, T. Tzanakaki, T. Kremastinou, E. Trikka-Graphakos (Athens, GR)
The primary clinical manifestations of meningococcal disease are meningitis and meningococcaemia. However, less frequent primary presentations may occur such as arthritis, pericarditis, pneumonia and empyema. We present a rare case of pneumonia and empyema due to Neisseria meningitidis, serogroup B.
A diabetic, debilitated 64-year-old man was admitted to the emergency department because of severe dyspnea and left pleuritic chest pain. Past medical history included congestive heart failure. On admission physical examination revealed jaundice, ascites, edema of the lower limbs and signs of left pleural effusion. Laboratory values included WBC 19.100/μL (90.6% PMNs), Hb 17.4 g/dL, Ht 52.9% and PLT 107,000/μL, glucose 296 mg/dL, T.Bil 7.4 mg/dL, SGOT 87IU/L SGPT 174IU/L, LDH 591IU/L, Na 126 mmol/L, K 5.2 mmol/L and CRP 85 mg/L. Chest X-ray revealed bilateral pleural effusion more prominent at the left side. Thoracocentesis was performed and pleural fluid cultures were drawn. The pleural fluid was turbid, ph 6.97, WBC 80.000/μL (93.1%PMNs), glucose 3 mg/dL, protein 2.4 g/dL and LDH 3.640IU/L. Gram stain revealed polymorphonuclear leukocytosis and Gram(–) diplococci. Latex examination test confirmed the presence of N. meningitidis antigen in pleural fluid. Lumbar puncture was performed and blood cultures were drawn simultaneously. Intravenous antibiotic treatment with ceftriaxone was initiated. On the third day of hospitalisation pleural fluid culture revealed N. meningitidis. Molecular typing (multiplex-PCR) and antibiotic succeptibility testing (E-test) was performed in the Reference Center. The isolate belonged to serogroup B, serotype 4:P1.14 and was sensitive to all tested antimicrobials. CSF and blood cultures were negative. Improvement of patient's clinical condition and laboratory findings permitted discharge after 20 days of hospitalisation.
N. meningitidis should be included in the differential diagnosis of pneumonia. An accurate and rapid aetiologic diagnosis due to the the severity of the condition, as well as the risk of spread to healthcare personell and family contacts, is mandatory.
Meningitis
P1205 Pneumococcal meningitis is a cause of death in patients with severe liver cirrhosis
P. Pagliano, U. Fusco, V. Attanasio, M. Rossi, M. Conte, F. Faella (Naples, IT)
Bacterial infections are frequently observed in patients with cirrhosis because of depression of neutrophil granulocyte functions and humoral and cell-mediated immunity. Poor data are available about pneumococcal meningitis in such patients. Aim of the study was to evaluate characteristics of pneumococcal meningitis in cirrhotic patients.
Methods: Fourteen patients with cirrhosis and pneumococcal meningitis (mean age 58.5±11 years; 9 males), referred to our tertiary care centre in the period 1997–2005, were evaluated as part of a prospective surveillance study of pneumococcal meningitis in adult. Pneumococcal meningitis was defined by CSF pleocytosis (>10 cells/μL) and positive CSF and/or blood culture. Diagnosis of cirrhosis had to be recorded by liver histology or unequivocal clinical, laboratory, ultrasonographic and endoscopic findings. Data about clinical presentation, laboratory investigations, treatment and neurological outcome were collected.
Results: All patients had viral cirrhosis. Other conditions predisposing to pneumococcal meningitis were present in 10 cases: 6 had diabetes mellitus, 5 recurrent otitis, 3 asplenia, 3 previous neurosurgery. Three patients had pneumonia. All patients were comatose at admission. Twelve patients reported fever before admission. Meningeal signs were present in 11 cases. Exam of the cerebrospinal fluid showed always pleocytosis, low CSF/serum glucose ratio and elevated protein. White blood cell count was 11,100/μL (range 3,900–30,000). Three of 5 patients presenting with CSF cell count below 500/μL died. Penicillin-susceptible strains of S. pneumoniae grew in 10 cases; in the remaining 4 cases penicillin-nonsusceptible strains were cultured. Patients received combined treatment with ceftriaxone associated to ampicillin (5 cases), vancomycin (5 cases), rifampin (2 cases) or linezolid (2 cases) as empirical treatment. Four patients died, 2 had neurological sequelae. Three deaths occurred in patients with Child–Pugh C cirrhosis. Child–Pugh score improved in surviving cases.
Conclusions: Pneumococcal meningitis should be suspected in cirrhotic patients with fever and changes in mental status, particularly when predisposing factors are present. Outcome and mortality are related to the severity of liver disease. The increasing incidence of penicillin-nonsusceptible strains must be considered in establishing the empirical treatment.
P1206 Repeated lumbar puncture in patients with pneumococcal meningitis: practical or anxiolytic relevance?
A. Alvarez, B. Mourvillier, H. Bout, C. Bruel, L. Ferreira, K. Lakhal, O. Pajot, L. Bouadma, B. Regnier, M. Wolff (Paris, FR)
Introduction: Repeated lumbar puncture (RLP) is recommanded in patients with pneumococcal meningitis who have not responded clinically after 48 h of appropiate antimicrobial therapy or in pneumococcal meningitis caused by penicillin- or cephalosporin-resistant strains (PNSP). Nevertheless the clinical impact of this practice is uncertain.
Patients and Methods: Retrospective analysis of charts of patients admitted in our 25-bed university medical ICU between January 2000 and December 2005, for pneumococcal meninigitis. All patients were appropiately treated according to international guidelines. The following data were collected at admission: clinical features, CSF characteristics: leukocyte count, protein and glucose concentrations, Gram stain and MIC of penicillin G. RLP data and changes in antibiotics dosage after results, were also noted.
Results: Among 51 included patients, 32 (63%) underwent RLP (RLP+) and were compared to 19 (37%) without RLP (RLP-). The mean delay between the first lumbar puncture and RLP was 4±2.5 days. RLP+ patients were older (59±14 vs 48±17 years), more severe at admission (SAPS II: 43±13 vs 29±4); had a more severely altered mental status (GCS 8±3 vs 12±3); and ICU length of stay was longer (23±15d vs 4±3 d). The number of PNSP strains was 7 and 2 respectively (p = 0.52). Characteristics of the first lumbar puncture were not different in the two groups, except for leucocyte count (1,766±2,190 vs 4,064±4,080, p<0.05). There was no difference in mortality (9% in RLP+ group vs 5% in RLP– group, p = 1) but a tendency toward a larger number of sequelae in the RLP+ group (34.4% vs 9.5%, p = 0.08). Gram stain was positive in 47% of RLP but cultures were always sterile. The comparison between admission and RLP is shown in the table .
| Day 1 | Repeated lumbar puncture | p | |
|---|---|---|---|
| Temperature (°C) | 39.2±1.2 | 37.7±1.1 | <0.05 |
| Leukocytes/mm3 | 1,773±2,155 | 3,713±6,334 | 0.11 |
| Protein (g/L) | 6.7±4.3 | 3.3±2.6 | <0.05 |
| Glucose level (mmol/L) | 1.1±2 | 2.7±2.5 | <0.05 |
Cefotaxime concentrations in CSF were measured in 14 patients and showed a level >5MIC in 4 and ≥10MIC in 9 patients, respectively. In one case cefotaxime concentration was <5MIC but the outcome was favourable. In none of the patients, the results of the RLP led to modifications of the antibiotic treatment.
Conclusion: When internationals treatment guidelines for pneumococcal meningitis are applied, based on pneumococcal resistance epidemiology, bacteriological failure seems improbable. Therefore, RLP may be unnecessary in this context. This hypothesis needs to be confirmed by a larger prospective study.
P1207 Treatment outcomes in adult purulent meningitis: a specialised centre experience in the Czech Republic
O. Dzupova, M. Helcl, J. Prihodova, J. Benes, H. Rohacova (Prague, CZ)
Objectives: Despite the availability of highly effective antimicrobials and intensive care in the last decades purulent meningitis continues to be a life-threatening disease burdened with an average mortality rate of 20–40%. The aim of the study is to report on outcomes of patients with purulent meningitis treated in a specialised centre.
Methods: A prospective study performed in years 1997–2004 which included all consecutive patients older than 16 years with diagnosis of purulent meningitis, both community-acquired and hospital-acquired, treated at the Infectious Diseases Department in Bulovka University Hospital, Prague. Patients with purulent meningitis are concentrated at the intensive care unit (ICU) of the Department from the subregion of c. 2-million population and account for ∼20% of all patients admitted at ICU. The outcomes were classified using the Glasgow Outcome Scale (GOS) score: 1, death; 2, vegetative state; 3, severe disability; 4, moderate disability; 5, mild or no disability.
Results: The study group consisted of 241 patients (134 men, 107 women), mean age 47.2 years (range 16–85 years). During the primary hospitalisation 44 patients died (18.3%). One hundred forty five patients (60.1%) were discharged to their homes; 52 patients (21.6%) were transferred to other healthcare facilities for rehabilitation or long-term hospital care.
Six months after the onset of disease the outcomes were reassessed. Twelve patients were lost for the follow-up for various reasons. One hundred and forty five patients (58.1%) were assessed with GOS 5; 27 patients (11.2%) with GOS 4; 10 patients (4.2%) with GOS 3; 2 patients (0.8%) with GOS 2; and 50 patients (20.7%) with GOS 1. Thus, in 62 patients (25.7%) the disease had got unfavourable outcome (GOS 1–3).
Conclusion: Relatively low rate of patients with unfavourable outcome in our study in comparison with data reported in the literature lead us to recommendation to centralise and treat patients with purulent meningitis in specialised centres with fully equipped intensive care unit and experienced personnel.
P1208 Community-acquired Listeria monocytogenes meningitis in adults
R. Amaya-Villar, E. Garcia Cabrera, E. Sulleiro, M.E. Jiménez-Mejías, D. Roriguez, P. Fernández-Viladrich, A. Coloma, P. Catalan, C. Rodrigo, D. Fontanals, F. Grill, M.L. Julia, J.A. Vazquez, J. Pachón, G. Prats (Seville, Barcelona, Madrid, Valencia, ES)
Methods: A descriptive, prospective, and multicentre study carried out in 9 hospitals of the Spanish Network for the Research in Infectious Diseases (REIPI) between 1/11/2003 and 31/07/2006. Patients: Three hundred and twenty-four adult patients with acute community-acquired bacterial meningitis were included. Forty-five episodes of Listeria monocytogenes meningitis were identified in 45 adult patients. All these cases were diagnosed on the basis of a compatible clinical and biological picture and a positive cerebrospinal fluid (CSF) or blood cultures.
Results: All 45 patients were either immunocompromised or aged over 60 years. The classic triad of fever, neck stiffness and altered mental status was present in 21 (46.7%) patients, however, almost all patients (95%) had at least 1 or more of these symptoms. CSF samples obtained through lumbar puncture showed a median WBC count of 550 per mm3 (interquartile range 0–70.000 per mm3); a glucose level of 39 (0.4–158)mg/dL; a ratio of CSF glucose to blood glucose of 0.27 (0.0–0.65); and a protein level of 181 (20–674) mg/dL. Gram stain of CSF samples was performed for 97.8% patients and it was positive in 12 (27.7%) of 44 cases. CSF and blood cultures were positive in 88.8% and 67.4%, respectively. The serotype more frequently found was the 4b in 16 (72.2%) of 22 cases. In 18 (40%) of 45 cases, the patients received empirical antimicrobial therapy. The initial antimicrobial therapy was ampicillin based for 35 (77.7%) of 45 patients, in 12 (34.3%) of them associated to aminoglycosides. Twenty-one patients (46.6%) received adjunctive therapy with dexamethasone; in 16 cases, the first dose was given previously or concomitantly to the first antibiotic dose. The median length of hospital stay was 20 (7–34) days. The mortality rate was 28.9% (13 of 45 patients) and 4 (8.9%) developed adverse clinical outcome (neurological and/or auditory sequels). Inadequate initial antimicrobial therapy was not related to outcome.
Conclusions: Listeria monocytogenes meningitis predominantly occurs in elderly or immunocompromised patients. Patients with Listeria monocytogenes meningitis often present with classic symptoms of bacterial meningitis. This clinical process also remains a serious disease that carries high morbidity and mortality rates.
P1209 A risk score for unfavourable outcome in adults with bacterial meningitis
M. Weisfelt, D. van de Beek, L. Spanjaard, J.B. Reitsma, J. de Gans (Amsterdam, NL)
Objectives: Clinical deterioration in bacterial meningitis can occur rapidly and is often difficult to predict. Identification of high-risk patients would be helpful in decision-making regarding the management of individual patients. The aim of the present study was to derive and validate a simple risk score to predict the risk for an unfavourable outcome in individual adults with community-acquired bacterial meningitis.
Methods: We derived a score for the risk of an unfavourable outcome by performing a multivariate logistic regression analysis of data from a prospective nationwide cohort study (Dutch Meningitis Cohort, DMC; n = 696). A key set of independent prognostic variables was selected from a starting set of 22 potential predictors by backward elimination procedure using bootstrap techniques to avoid the inclusion of spurious variables. A nomogram based on these key variables was constructed to facilitate the use in clinical practice. For validation, we used data from a randomised controlled trial of adjunctive dexamethasone therapy for adults with bacterial meningitis (European Dexamethasone Study, EDS; n = 301).
Results: Of the 696 episodes in the DMC, 237 (34%) had an unfavourable outcome, including 143 patients (21%) who died. The risk score was based on 6 routinely available variables: age, heart rate, score on the Glasgow Coma Scale, cranial nerve palsies, cerebrospinal fluid leukocyte count, and Gram-positive cocci in cerebrospinal fluid Gram's stain (figure 1 ). The concordance index for the final risk score in the EDS was 0.73 (95% confidence interval 0.65–0.80).
Fig. 1.

Conclusion: This bedside risk score can be used to reliably predict the individual risk for unfavourable outcome in adults with community-acquired bacterial meningitis.
P1210 Antibiotic resistance among Gram-negative bacilli causing meningitis in an adult neurosurgical centre in Manchester, UK: implications for effective antibiotic therapy
N. Doshi, T. Wilson, C. Subudhi (Salford, UK)
Objectives: Gram negative bacillary meningitis (GNBM) is an uncommon infection in adults and often occurs as a complication of complex neurosurgical procedure or following head injury. The treatment of Gram-negative bacillary meningitis remains a major therapeutic challenge with emergence of antibiotic resistance in many of the causative organisms and the restricted choice of antibiotics. We reviewed the aetiology and antimicrobial susceptibility pattern of GNBM in our centre over a 7 year period.
Methods: Hope Hospital, Salford is a 900 bed tertiary referral hospital, which houses the regional neurosurgical centre for Greater Manchester. The case notes and microbiological records of all patients with GNBM between 2000 and 2006 inclusive were reviewed retrospectively. Only patients with positive CSF culture and clinical features compatible with meningitis were included.
Results: During the study period, there were 74 patients who had GNBM from whom 92 different Gram-negative bacilli were isolated in CSF. The most common causative organisms were Enterobacter spp. (27.2%), E. coli (21.7%) and Klebsiella spp. (16.3%).
More than one third (38%) of Gram-negative bacillary isolates were resistant to third generation cephalosporins. 17% of isolates were resistant to gentamicin and 10% were resistant to ciprofloxacin. All the isolates were sensitive to meropenem and amikacin.
Conclusions: Third generation cephalosporins have been the mainstay to treat GNBM over the past 20 years because of high CSF penetrance. This has resulted in dramatic fall in meningitis related mortality. Emergence of strains resistant to third generation cephalosporins is a significant cause of concern. This retrospective study has highlighted that surveillance of local pathogens and resistance patterns are essential to guide empirical therapy. This study has prompted us to consider initiating empiric therapy for post neurosurgical GNBM with meropenem pending culture and susceptibility results.
P1211 Nosocomial meningitis
E. Garcia-Cabrera, M.E. Jiménez-Mejías, J. Serra-Vich, V. Pintado, F. Grill, M. Portillo, J. Colomina, G. Prats, J. Pachón-Diaz (Seville, Barcelona, Madrid, Valencia, ES)
Objectives: Study of epidemiology, demography and risk factors, clinical and cerebrospinal fluid (CSF) variables, aetiology and prognosis of a cohort of patients with nosocomial meningitis/ventriculitis (NM).
Methods: Multicentre and prospective study in six tertiary REIPI hospitals, from September 2004 to July 2006. Inclusion criteria: adults undergoing neurosurgical or spinal or facial skull surgical, or head trauma, with NM. Diagnosis NM: clinical features, CSF findings (pleocytosis, hypoglycorrachia or CSF/blood glucose ratio <40%, and increased protein level) and microbiological criteria (microbial presence in Gram smears and/or isolation from CSF). Monitoring: 30 days after treatment withdrawal.
Results: A total of 106 patients (57% males) were studied. Median age: 49.5±26.9 years (15–75). Risk factor: temporal shunt 83 patients (79%), neurosurgery 75 (71%), CSF fistula 26 (25%), permanent drainage of CSF 17 (16%), cranial trauma 10 (9%). Cause of admission: 33 (31%) brain tumour resection, 22 (21%) subarachnoid haemorrhage, 15 (14%) brain haematoma, 10 (9%) cranial trauma, 9 (8%) meningitis, 9 malfunction V-P shunt, and others 13. Clinical features: fever (mean 38.5±1°C) 92 (87%) patients, headache 50 (47%), decrease of Glasgow coma score 39 (37%), and meningismus 26 (24.4%), The classic triad was present in 13 (12.3%) cases. Microbiological studies: positive CSF cultures in 89 (84%) patients (71 monomicrobial and 18 polymicrobial infections), CSF Gram's stain 44 (42%); positive blood cultures 27 of 77 (35%) and no isolation in 16 (15%). Aetiological agents: Gram-negative bacilli 53 (49%) (Acinetobacter baumannii 25, Enterobacter spp. 8, Klebsiella pneumoniae 8, E. coli 6, Serratia marcescens 2, Citrobacter freundii 2, and others 3); Gram-positive cocci 49 (45%) (coagulase-negative Staphylococcus spp. 34, S. aureus 7, Enterococcus spp. 4; Streptococcus viridans 2, others 2); anaerobes 4 (Propionibacterium acnes 2, Shewanella spp., Provetella spp.); and others 2. Outcome: mortality 27 cases (25%) (only 6 related with NM); morbidity: 34 patients (32.1%). Relapse 2 cases.
Conclusions: Neurosurgery and temporal CSF shunt are the first causes of nosocomial meningitis. The sensitivity of classic triad is very low. The rate of positive CSF cultures is high. Gram-negative bacilli are the more frequent aetiological agents. Morbidity and mortality of nosocomial meningitis are very important; although the related mortality is lower than others series.
P1212 Hippocampal apoptosis in pneumococcal meningitis: contribution of decreased cellular NAD+ levels?
C. Bellac, R.S. Coimbra, S. Christen, S. Leib (Berne, CH)
Objectives: Apoptosis in the hippocampus (HC) is a characteristic feature of neuronal damage in pneumococcal meningitis (PM) and represents the histomorphological correlate for learning and memory deficits as a sequel of PM. Depletion of intracellular NAD+ and the resulting decrease in ATP is known to trigger cell death. Whether decreased NAD+ levels contribute to apoptosis in PM has not been demonstrated. NAD+ is either recycled from nicotinic acid or nicotinamide or is synthesized de novo through the kynurenine (KYN) pathway, which we have previously shown to be induced in PM. Here we measured NAD+ levels in the HC during acute PM and modulated the biosynthetic pathways of NAD+ to assess the role of NAD+ in the pathogenesis of hippocampal apoptosis in PM.
Methods: An established infant rat model of PM was used to determine hippocampal NAD+ levels at defined time points of acute PM. NAD+ was measured by fluorescence assay. Pharmacologic interventions were done with specific inhibitors of the KYN pathway (Ro-61–8048 for inhibition of kynurenine 3-hydroxylase, and oMBA for kynureninase) and nicotinamide. Apoptotic damage in the HC was evaluated by histomorphometry at 36 h after infection.
Results: Compared to uninfected littermates, PM led to a significant reduction in NAD+ levels at 24 h after infection (100% in mock-infected controls (n = 6) vs. 45 ± 18% in infected animals (n = 10), p<0.01). Simultaneous inhibition of the two major enzymes of the KYN pathway resulted in increased apoptotic damage in the HC of infected animals (apoptotic score of 1.5 ± 0.6 in infected double inhibited animals (n = 8) vs. score 0.5 ± 0.6 in infected vehicle treated animals (n = 31), p < 0.01). Inhibition of the KYN pathway had no effect on NAD+ levels in infected animals. Treatment with nicotinamide increased apoptotic damage, despite high NAD+ levels at 36 h p.i. (195 ± 29% in infected treated animals (n = 6) vs. 100% in mock-infected controls (n = 7), p < 0.01).
Conclusion: While pneumococcal meningitis causes a significant depletion in hippocampal NAD+, the exact consequence of this decrease is presently not clear. Pharmacologic inhibition of de novo synthesis of NAD+ increased hippocampal apoptosis without affecting NAD+ levels compared to untreated infected animals at 24 h and 36 h after infection. We conclude that the activation of the KYN pathway is insufficient to restore cellular NAD+ in the hippocampus during acute PM.
P1213 Nosocomial meningitis in the NICU population: perinatal risk factors, aetiology and outcome
J. Behrendt, M. Stojewska, M. Wasek-Buko, B. Krolak-Olejnik, U. Godula-Stuglik (Zabrze, PL)
Neonatal hospital-acquired meningitis (n.h.m.) is a major cause of morbidity with high risk of neurological consequences in infants treated in the NICU.
Aim: to assess the correlation between CSF and blood cultures in n.h.m. and the outcome according to gender, birth weight, aetiology, perinatal complications associated with high-risk pregnancy and delivery.
Methods: The study population consisted of 48 infants (36 boys, 12 girls) from 7 to 80 days of life, 38 premature and 10 full-term treated in NICU due to respiratory or cardiovascular disorders. Birth asphyxia in 21%, obstetric complications in 96%, VLBW in 54%, VLGA (<29 weeks) in 29% were noted. Urgent cesarean sections were carried out in 62%. N.h.m. was diagnosed after 7th day of life and confirmed by CSF pleocytosis over 30/mm3 with predominance of PMNs and increase of protein concentration >1.0 g/L.
Results: N.m. was confirmed by CSF culture in 24 (50%) neonates: Staph. epidermidis MR (14), Staph. warneri (1), Streptococcus faecalis (1), Klebsiella pn. (2), E. coli (2), Serratia marcescens (1), Pseudomonas aeruginosa (1), Candida albicans (1) and Candida sake (1) were isolated. Among 48 sick neonates 24 had positive blood culture. Only 8 (17%) neonates had a concomitant-positive (Staph. epidermidis MR 4, Streptococcus faecalis 1, Serratia marcescens 1, Klebsiella pn. 2) and 12 concomitant-negative blood cultures. In neonates with both positive blood and CSF cultures the isolated microorganisms were discordant in 5 (10%) of 48 cases. Three (6%) neonates died – all asphyxiated premature born in 25, 26 and 29 weeks of gestation, with birth weight 800–1,050 g, in the course of nosocomial sepsis due to Staph. epidermidis MR, Serratia marcescens, Streptococcus faecalis and with infectious perinatal risk factors (chorionamnionitis), admitted to the NICU because of RDS and congenital pneumonia. In 19 (40%) neonates with n.h.m. severe neurological consequence (hydrocephalus 10, PVL 5, hydrocephalus+PVL 4) was noted. In 26 (54%) infants the outcomes at age 6 month were satisfactory. The bad prognosis (death or brain injury) was not related to sex, birth asphyxia, birth weight and mode of delivery. The significant correlation between low GA of newborns with n.h.m. and neurological sequel was found.
Conclusions: (1) Nosocomial meningitis in neonates may occur in the absence of bacteraemia and positive CSF cultures. (2) Outcome of nosocomial meningitis depends on neonatal gestational age and mother's perinatal risk factors.
P1214 Succesful treatment of multidrug-resistant Acinetobacter baumannii meningitis with colistin and rifampicin
K.O. Memikoglu, F. Alver Alkaya, Z. Köken, H. Serdaroglu, T. Çakir, A. Azap (Ankara, TR)
Introduction: The treatment of multidrug-resistant A. baumannii infections is a serious therapeutic problem, especially in patients with meningitis because antibiotics have a limited ability to penetrate cerebrospinal fluid (CSF). Meningitis due to Acinetobacter spp. has an associated mortality rate of 20–27%. We described here a case of meningitis caused by multidrug-resistant Acinetobacter baumannii, which was suspectible to colistin and treated succesfully with intravenous and intrathecal use of colistin with rifampicin.
Case Report: A 38-year-old male was operated because of schwannoma. On day 9 external CSF shunt was installed. On day 11 the patient presented fever, headache, nause and vomitting and somnolance. Physical examination revealed a severly ill patient with lethargy and meningism. CSF showed pleocytosis (250 cells/μl; with polymorphonuclear neutrophil, dominance (100%), a protein level of 321 mg/dl, and a glucose level of 20 mg/dl (blood glucose level was 105 mg/dl). Meropenem (1 g q8h), vancomycin (1 g q12h) and rifampicin 10 mg/kg q12h po) were began. Multidrug-resistant A. baumannii was isolated from the CSF. Meropenem was stopped and the treatment changed to colistin intravenous (2 g q8h) and intrathecal (50.000U for the first 3 day and 50.000 U on alternative days). On day 2 of treatment the CSF was sterile. Treatment was maintained for 21 days and the patient was discharged.
Discussion: High use rates of broad-spectrum antibiotics in critically ill patients have been corraleted with the emergence of resistance in Acinetobacter strains. In a large serious of adults with acute bacterial meningitis Acinetobacter spp. were found to be responsible for approximately 10% of Gram negative bacillary and 4% of all nosocomial meningitis. Alternative therapies in multidrug-resistant A. baumannii infections such as colistin are being increasingly employed. Intrathecal colistin has been used with good results in case of multidrug-resistant A. baumannii meningitis. Cure seems to be more frequent among patients receiving combination systemic and intrathecal therapy.
Conclusion: Intravenous and intrathecal use of colistin with rifampicin may be a potentially effective therapy in cases of meningitis caused by A. baumannii resistant to carbapenems and other β-lactam agents.
P1215 Endemic serogroup W135 meningococcal meningitis possibly originated from 2000 and 2001 Hajj epidemics: a prospective study for the aetiology of childhood acute bacterial meningitis in Turkey
M. Ceyhan, I. Yildirim, P. Balmer, R. Borrow, G. Secmeer, B. Dikici, M. Turgut, N. Kurt, A. Aydogan, C. Ecevit, Y. Anlar, O. Gulumser, G. Tanir, N. Salman, N. Gurler, N. Aydin, M. Hacimustafaoglu, S. Celebi, Y. Coskun, E. Alhan, U. Celik, Y. Camcioglu (Ankara, TR; Manchester, UK)
Determination of bacterial meningitis aetiology is important for decisions about vaccine policies. Cerebrospinal fluid (CSF) samples taken from patients with a clinical and laboratory diagnosis of acute bacterial meningitis were obtained from 12 health centres in 7 different geographical regions that provide health service to 32% of the population of Turkey. Streptococcus pneumoniae, Haemophilus influenzae type b (Hib) and Neisseria meningitidis were detected by PCR. During a period of one year, 408 CSF samples were included and bacterial meningitis was detected in 243 samples. N. meningitidis was detected in 56.5%, S. pneumoniae in 22.5% and Hib in 20.5% of PCR positive samples. Among N. meningitidis positive CSF samples, 42.7%, 31.1%, 2.2% and 0.7% belonged to serogroups W135, B, Y, and A, respectively. Serogroup W135 was the predominant serogroup in Turkey, except for the south coast and northwest of the country. This study showed a shift to serogroup W135 whereas serogroups A and B previously predominated. Vaccines to prevent meningococcal disease should include serogroup W135 in Turkey and possibly in other countries where many muslim pilgrims are travelling annually to the Hajj.
P1216 Epidemiologic study of paediatric bacterial meningitis in Greece (1998–2006)
E. Lebessi, M. Papadimitriou, N. Paleologou, K. Kallergi, I. Niniou, K. Malliou, G. Antonaki, S. Ioannidou, M. Foustoukou (Athens, GR)
Objectives: To study the epidemiologic data of bacterial meningitis in children treated in “P. & A. Kyriakou” Children's Hospital, one of the two major paediatric hospitals in Greece, during a nine-year period (1998– 2006).
Methods: The laboratory archives were retrospectively reviewed. Positive CSF cultures of children hospitalised with diagnosis of meningitis were recorded. Children with history of neurosurgical procedures were excluded from the study. Cultures were performed in blood, chocolate, Levinthal agar and BHI broth and strains were identified by standard methods. Susceptibility to antimicrobial agents was tested according to the CLSI guidelines and MICs of penicillin and cefotaxime for Neisseria meningitidis and Streptococcus pneumoniae were determined by E-test.
Results: Overall, 104 strains were isolated from CSF cultures received from 104 patients (males, 63 and females, 41) aged from 3 days to 14 years (median age; 1.33 y). The most frequent pathogens were N. meningitidis (n = 61, 58.6%) and S. pneumoniae (n = 21, 20.2%), followed by Escherichia coli (n = 8, 7.7%), Streptococcus agalactiae (n = 5, 4.8%), Haemophilus influenzae (n = 4, 3.8%), Streptococcus pyogenes (n = 2, 1.9%), Enterobacter cloacae (n = 1, 0.9%), Bacillus cereus (n = 1, 0.9%) and Candida albicans (n = 1, 0.9%). Gram negative rods and S. agalactiae are the most frequent isolates in neonates. About two thirds of cases occurred in infants and children younger than 5 years. There were not significant changes in the annual distribution of cases due to N. meningitidis, although the widespread vaccination. Of the meningococcal strains, 78.7% were typed as serogroup B, 18.1% serogroup C, 1.6% A, and 1.6% W135. Two H. influenzae isolates were type a and two type b. About one quarter of N. meningitidis and 11% of S. pneumoniae isolates showed moderate resistance to penicillin (≥0.094–1 mg/L). High-level resistance to penicillin and other antibiotics was not detected. H. influenzae isolates were found to be susceptible to all antibiotics tested. Results from 2006 will be completed at the end of this year.
Conclusion: N. meningitidis is the most frequent pathogen in bacterial meningitis in infants and children; serogroup B is far more common than serogroup C. S. pneumoniae and H. influenzae strains remain susceptible to currently used antibiotics. However, N. meningitidis strains with reduced susceptibility to penicillin accounted for 25% of isolates, warranting continued monitoring of this problem.
P1217 Pneumococcal meningitis in Taiwanese children: emphasis on clinical outcomes and prognostic factors
M.H Tsai, S.H. Chen, C.Y. Hsu, D.C. Yan, M.H. Yen, C.H. Chiu, Y.C. Huang, T.Y. Lin (Keelung, Taoyuan, Taipei, TW)
Background: Pneumococcal meningitis still causes high morbidity or mortality in childhood despite the progress in medicine. Factors related to poor outcomes are needed to be investigated.
Methods: From January 1984 to December 2002, Children with pneumococcal meningitis admitted to Chang Gung Children's Hospital were included in this study. Pneumococcal meningitis was defined as isolation of S. pneumoniae from cerebrospinal fluid (CSF) or a blood culture positive for S. pneumoniae in combination with a CSF pleocytosis (CSF white blood cell count >10/mm3). Demography, clinical manifestations, laboratory findings, antimicrobial sensitivity patterns, managements and clinical outcomes were retrospectively reviewed. Features and outcome parameters were also compared between survivors and nonsurvivors.
Results: Forty-four children were eligible, with mortality in 20.5% of all and neurological sequelae in 37.1% of survivors. In the comparison of clinical profiles between survivors and nonsurvivors, only initial white blood cell count (/mm3) (mean: 7,666.7 vs 16,187.5, p = 0.022) and cerebral spinal fluid (CSF) leukocyte count (/mm3) (mean: 187.3 vs 1,408.0, p = 0.001) were found to be significantly lower in nonsurvivors. Features of clinical course associated with neurological sequelae in survivors were also analysed, and the results revealed that patients with longer duration of hospital stay (days) (mean: 45.3 vs 19.1, p < 0.001) or intensive care unit admission (days) (mean: 19.6 vs 3.0, p = 0.031) may have more neurological sequelae. Besides, initial medication including vancomycin or dexamethasone usage and severity of illness did not significantly affect the occurrence of neurological sequelae. In multivariate logistic regression analysis for outcome variables, only CSF leukocyte count ≤200/mm3 was a significantly risk factor for mortality.
Conclusions: In Taiwan, pneumococcal meningitis causes high morbidity and mortality. We investigated risk factors associated with mortality and only CSF leukocyte count ≤200/mm3 (p = 0.021) was found to be significant. Physicians should monitor such patients closely to decrease possible morbidity or even mortality.
Viral hepatitis: epidemiology and clinical manifestations
P1218 Hepatitis A infection in central Tuscany in children and youths
C. Gentile, I. Alberini, I. Manini, E. Montomoli, S. Rossi, T. Pozzi (Siena, IT)
In Italy, seroepidemiological studies have demonstrated a decrease in the circulation of the Hepatitis A virus (HAV). The notified cases in the SEIEVA system (Integrated Epidemiological System of Acute Viral Hepatitis) of HAV show a reduction in cases beginning in 1985 (10/100.000) up to 2004 (3.6/100.000) with a peak in 1997 connected to the epidemic that occurred in Puglia (19/100.000).
The decline in the circulation of HAV in Italy is linked to the improvement in hygienic conditions and life style, and above all to the implementation of vaccination, determining the decrease in the risk of acquiring infection during childhood.
In order to verify the trend in the diffusion of HAV infection in the geographical area of Central Italy, a seroepidemiological study on cohorts of subjects, children and youths, was carried out; the years 1992, 1998 and 2004 were considered, and for each of these years two samples from around 100 subjects aged, respectively, 0–5 and 15–20 years, were examined. All sera were refrigerated at −20°C. Each sample of sera was tested for the presence of total antibodies against HAV by the ELISA method (HAV antibody, DIESSE Diag. Senese SpA).
The obtained results show a higher prevalence of anti-HAV in youths compared to children in both 1992 (2.7% in 0–5 year-olds and 7.4% in 15–20 year-olds) and in 1998 (6.0% in 0–5 year-olds and 11.1% in 15–20 year-olds), while in 2004, infection was 16.4% in children and 5.4% in youths. The high antibody rate against HAV noted in children in 2004 could be correlated to a higher use of vaccine in this age group as a consequence of small epidemics in the infant community that occurred in Central Italy during that period. The statistical analysis of the results thus shows a significant tendency of increase in seroprotection over the years in children (χ2 for linear trend = 10.7; p = 0.0011). However, the overall decrease of prevalence in subjects protected from HAV in youths (χ2 for linear trend = 0.48; p = 0.49) up to the values shown in 2004 (5.4%), besides confirming the national trend of the past few years, does not allow us to hypothesise a trend toward an increase in the risk of acquiring infection in young adults.
This trend must be considered with caution because too much time probably passed between the intervals of blood collection compared to the period that is the object of our analysis.
P1219 Seroprevalence of hepatitis E virus antibody in 15–40 year-old Turkish women
T. Demirdal, N. Demirturk, D. Toprak, O. Aktepe, S. Orhan (Afyonkarahisar, TR)
Objectives: Hepatitits E virus (HEV) infection is the major cause of enterically transmitted Non-A Non-B hepatitis in many parts of the world. In this study, which was conducted in Central Anatolia in Turkey, we investigated the seroprevalence of HEV in women between 15 and 40 years of age.
Methods: The study was done in urban and rural areas of Afyonkarahisar province in Turkey. The study design was cross-sectional and people selected randomly in 15–40 years old Turkish women. Samples were tested for anti-HEV IgG by commercial enzyme-linked immunoassay (ELISA) tests.
Results: We tested 1,194 women whose ages were between 15 and 40 years. The overall seroprevalence of hepatitis E was 7.1% (85/1,194). We found 95.5% (81/85) of the low educated people HEV seropositive and the educational level was statistically significant (p <0.05). According to the salary with HEV seropositivity there was no significant correlation (p > .05). When we compared urban and rural, the distribution of HEV seropositivity was 48.2% and 51.8% respectively.
Conclusion: The prevalence rate of HEV seropositivity differ in various regions of Turkey. Protection from HEV infection, which transmitted faecal–oral route; education, sanitation precautions and environmental hygen must be actually considered.
P1220 Cardiomyopathy in patients with chronic hepatitis C virus
T. Demirdal, H. Saglam, N. Demirturk, Z. Asci (Afyonkarahisar, TR)
Objectives: A high prevalence of cardiomyopathy has been reported in patients with chronic hepatitis C virus (HCV). In this study, we investigated the relationship between cardiomyopathy and HCV infection.
Methods: Sixty subjects were included in the study. These were divided into two groups. In both groups anti-HCV and HCV RNA were detected and ALT levels were measured. Thirty consecutive anti-HCV and HCV RNA positive patients who were admitted to infectious diseases clinic and provided informed consent were evaluated as study group. The other thirty subjects were assumed control group. Healty volunteers who were anti-HCV and hCV RNA negative and whose ALT was normal were included in the control group. Clinical, electrocardiographic and transthorasic echocardiographic evaluations were performed to all subjects. Valve functions and sistolo-diastolic performance were evaluated. The subjects with diastolic disfunction were evaluated cardiomyopathic.
Results: The age of subjects in both groups ranged from 23 to 78 years. In each group, there were 15 man and 15 women. The groups displayed no significant differences for age and sex distribution. Anti-HCV was positive in all subjects in the study group. In this group, average serum alanine transferase (ALT) and HCV-RNA levels were 73.68±46.60 U/L and 1,289,243±2,042,026 copies/mL, respectively. Valve functions and systolic performance of the subjects in both groups were normal. Diastolic disfunction was observed in 18 patients in the study group and in four patients in the control group. The number of subjects with diastolic disfunction in the study group (n = 18) were significantly higher than the control group (n = 4) (p<0.05).
Conclusion: Diastolic disfunction is the finding of early stage cardiomyopathy. In this study, we observed diastolic disfunction in HCV infected patients. This result reveals that there may be a relationship between HCV infection and cardiomyopathy. However, larger scale studies to investigate this relationship are required for a conclusive result.
P1221 The anicteric form of viral hepatitis A
I. Mehmedi, R. Mehmedi, G. Temelkoski, G. Mehmeti, Z. Xhambazoski (Kercove, MK)
Objectives: The Anicteric Form of Viral Hepatitis A (HA) is an uncommon form of benign, where the main symptoms are the same as those associated with icteric HA, except that there is an absence of jaundice. The presentation of same clinical-epidemiological characteristics of anicteric form of HA during the epidemics 1998–2000.
Methodes: The material is taken from the Infectious unit at the G. H. of Kërçovë during the period of September 1st, 1999 to March 30th, 2000. There were 175 cases of hepatitis treated at the Infectious unit at this G. H. The diagnosis was determined on the basis of: anamnesis, epidemiological date, Physical examination, functional test of the liver (the serum aminotransferases AST, ALT) and serological testes anti HAV IgM done by the ELISA (ORGANONR) method.
Results: HA is a disease found mostly in children of a young age. In our study we found 175 cases with acute viral hepatitis (avh) at an average age of 8.86 years (from the age of 3 to 43). Based on the place of the residence of the patients, the patients from urban area dominate with 54.9%. Based on the gender of the patients, the masculine gender dominates with 52.6%. The morbidity for the last ten years is an average of 60 cases per 100,000 inhabitants (6). According to the aetiological moment the cases are as follows: anti HAV IgM+, 175 cases (100%); 53 (30.3%) were of anicteric form of HA, and 4 patients (2.02%) were subclinical forms of HA. The symptoms and the clinical examination in the prodromal stage of the anicteric form of HA are similar to the icteric form of HA, except that the symptoms of the anicteric form are shorter and less intense. Clinical–biochemical recuperation periods are shorter in the anicteric form of HA, but the differences were not significant according to statistics of the χ2 test (p > 0.1).
Conclusion: There were 53 patients (30.3%) with the anicteric form of HA. The clinical–biochemical symptoms of the anicteric form are the same as in the icteric form of HA, except that there is an absence of jaundice.
P1222 Hypercalcaemia associated with chronic viral hepatitis C
A. Yesilkaya, K.O. Memikoglu, A. Azap, Ö. Demir, I. Balik (Batman, Ankara, TR)
Introduction: Hypercalcaemia has been reported in the course of some cases of cirrhosis complicated by hepatocellular carcinoma or cholangiocarcinoma and in patients with hepatic tumours in the absence ofcirrhosis. However in the absence of liver tumour, this metabolic complication has been rarely reported in the course of chronic liver disease. We described here hypercalcaemia associated with chronic hepatitis C.
Case report: A 71 year old man was admitted in Febuary 2006 for exploration of back pain. He had had HCV chronic hepatitis for 15 years. Tests showed hypercalcaemia, bicytopenia and mild increase in aminotransferases. Serum calcium, ionised calcium were elevated and serum phosphorus, 24-hour urinary calcium excretion were normal. The level of serum immunoreactive parathyroid hormone (PTH) was normal. Nephrogenic cAMP 2.38 μmol/L, PTH-related peptide <0.2 pmol/L and α-fetoprotein were normal whereas plasma 1,25 (OH)2 vitamin D3 (25 pg/mL) was slightly high. There were none of the major causes of true hypercalcaemia such as any malignancy, hyperthyroidism, hyperparathyroidism and chronic bone disaese. Serum protein electrophoresis showed an increase in ammaglobulins with no monoclonol paraprotein lectrophoresis, total body 99-techntium radionuclide scan showed no increased uptake; chest X-ray and kidney ultrasonography were normal. There was no history of excessive aluminium derivates, milk or alkali intake, thiazide diuretic or vitamin A and D supplementation.
Discussion: Hypercalcaemia has been reported by Gerhardt et al. in eleven patients waiting for liver transplantation with chronic liver disease without hepatic tumour. Resorptive factors like interleukin-1, transforming growth factor, tumour necrotising factor, osteoclast activating factor, or prostaglandin could be responsible for the hypercalcaemia.
Conclusion: Hypercalcaemia could be a rare metabolic feature of chronic viral hepatitis C; the underlying mechanisms remain to be elucidated.
P1223 HCV-associated hereditary haemochromatosis and porphyria cutanea tarda in monozygotic twins
D. Telehin (Lviv, UA)
Objectives: The aim of this study is to investigate a link between inherited factors, phenotypic expression of hereditary haemochromatosis (HH), porphyria cutanea tarda (PCT) and course of chronic hepatitis C (CHC) and to evaluate the efficiency of antiviral therapy on HCV-associated PCT compared to the course of a similar disease without antiviral treatment.
Methods: We had a unique opportunity to compare a course of CHC and PCT in two monozygotic twins. Liver biopsy was performed for histologic evaluation and iron grading (with Perls Prussian stain). Iron status was determined from transferrin saturation, serum iron, serum ferritin and hepatic iron. The degree of porphyrin metabolism disturbance was evaluated according to the concentration of urine uro- and coproporphyrins. HCV-RNA and its genotype were detected by RT-PCR. HLA-type was determined by lymphocyte serologic identification method. We explored four generation of the twins' family.
Results: 2 patients, monozygotic male twins aged 42, while still being infants were simultaneously infected with HCV. The first symptoms of the disease appeared after 35 years: marked fatigue, increased skin fragility, bullons lesions, hypertrichosis and hyperpigmentation of the face, red colour of the urine. Chronic hepatitis C, (METAVIR score A2F3) with extrahepatic manifestations: PCT, primary haemochromatosis was established. Genealogic exploration of probands revealed clinical minifestation of HH and PCT only in the first generation suggestive of autosomal recessive inheritance. HLA-typing showed HLA-A3 phenotype of twins. Considering low concordance of monozygotic twins in relation to HH and PCT, it may be assumed that HCV-infection became the trigger of the latent forms of HH and PCT. One of twins received antiviral therapy (IFN-α-2b + Rbvn). A persistent remission of PCT and discontinued viraemia was achieved. The other one was only given pathogenetic and phlebotomy treatment. The symptoms of CHC and PCT in this case have undulating course.
Conclusions: The current study showed that HCV-infection probably acts as a main triggering factor of HH and PCT in genetically predisposed subjects. The management of PCT in HCV-infected patients should include antiviral therapy. An association is established between HH, PCT and HLA-A3.
P1224 The trend of screening tests from 2000 to 2006 and the relationship of these with age, gender, occupation and education of healthy blood donors in Turkey
A. Candevir, Y. Tasova, N.B. Kilic, B. Kurtaran, A.S. Inal, G. Seydaoglu (Adana, TR)
Introduction: Screening tests are mandatory for blood donations in Turkey. In this study we aimed to review the annual test results and look for a relationship between positive test results and age, gender, occupation and education at healthy blood donors in Turkey.
Material and Methods: Between January 2000 and December 2006 totally 141,928 donors' data was investigated. ELISA was used for HBsAg, antiHCV and anti HIV1/2, while VDRL was used for syphilis. Linear trend qui-square test was used for statistical analysis. A more thorough research was conducted for the year 2005 to investigate the relationship between screening test and demographic data like age, gender, occupation and education. Student t-test was used for continuous variables and qui-square for categorical variables. SPSS v11.5 was used for statistical analysis.
Results: The annually change of screening test results are shown on table 1 . There were no confirmed positive HIV results during these years. There was a statistically significant decrease at HBsAg positiveness and decrease at antiHCV and VDRL positiveness (p = 0.0001, p = 0.03, p = 0.002 respectively). In the year 2005, of the 23,368 donors of whom 95.7% was male, 64.5% was single and the mean age was 33±8. There was no relationship between seropositivity and gender (P values are 0.926, 0.224, 1.000 respectively for HBs Ag, anti HCV and RPR) or marital status (P values are 0.128, 0.051, 0.807 respectively for HBs Ag, anti HCV and RPR). There was a negative correlation between HBs Ag positiveness and educational status (p = 0.0001). There was also a relationship between HBs Ag seropositivity and occupation (p = 0.0001). Mean age was significantly higher at HBs Ag and RPR positive donors (p = 0.002 and p = 0.013 respectively).
HBsAg, anti-HCV and RPR positiveness according to year
| Year (N) | HBsAg |
Anti-HCV |
RPR |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (+) |
(−) n | (+) |
(−) n | (+) |
(−) n | |||||||
| n | (%) | (OR) | n | (%) | (OR) | n | (%) | (OR) | ||||
| 2000 (20,126) | 791 | (3.93) | (1.00) | 19,335 | 81 | (0.40) | (1.00) | 20,045 | 21 | (0.1) | (1.00) | 20,105 |
| 2001 (20,802) | 725 | (3.49) | (0.98) | 20,077 | 87 | (0.42) | (1.04) | 20,715 | 43 | (0.21) | (1.98) | 20,759 |
| 2002 (21,418) | 594 | (2.77) | (0.70) | 20,824 | 116 | (0.54) | (1.35) | 21,302 | 61 | (0.28) | (2.73) | 21,357 |
| 2003 (21,598) | 552 | (2.56) | (0.64) | 21,046 | 134 | (0.62) | (1.54) | 21,464 | 34 | (0.16) | (1.51) | 21,564 |
| 2004 (21,571) | 505 | (2.34) | (0.59) | 21,066 | 134 | (0.62) | (1.54) | 21,437 | 27 | (0.13) | (1.20) | 21,544 |
| 2005 (23,368) | 527 | (2.26) | (0.56) | 22,841 | 105 | (0.45) | (1.12) | 23,263 | 18 | (0.08) | (0.74) | 23,350 |
| 2006 (13,045) | 277 | (2.12) | (0.53) | 12,768 | 72 | (0.55) | (1.37) | 12,973 | 13 | (0.1) | (0.96) | 13,032 |
| Linear trend χ | 177.864 | 4.486 | 8.865 | |||||||||
| p value | 0.0001 | 0.03 | 0.002 | |||||||||
Conclusion: We found a negative trend for HBs Ag seroprevalence and a positive trend for anti HCV and RPR prevalence. We may say that we are having positive results of national vaccination and education programme. And also we may say that, as the educational status improves decreasing HBs Ag prevalence shows the benefit of education on development of conciseness for viral hepatitis and other blood borne infections. But it's clear that education at schools may not be enough and more efforts should be consumed for informing public.
P1225 The relationship between serum fibrosis markers and fibrosis in liver tissue in patients with chronic viral hepatitis
A. Candevir, H.S.Z. Aksu, B. Kurtaran, A.S. Inal (Adana, TR)
Introduction: In management of chronic hepatitis, measuring the degree of hepatic fibrosis and inflammation is crucial. Today the golden standard is showing the fibrosis and inflammation at the liver tissue via liver biopsy and pathological scoring systems. In this study, two of the noninvasive markers of fibrosis, hyaluronic acid (HA) and matrix metalloproteinase-2 (MMP-2), which can be used instead of liver biopsy, will be investigated.
Material and Methods: 42 patients of chronic hepatitis (mean age is 38±12, of which 69% male, n = 29) with activation criteria were included to our study. Routine biochemical assays and liver biopsies were performed. The relationship between serum HA, MMP-2 levels and liver fibrosis and histological activity was investigated.
Results: Serum HA levels were found to be increased by increasing liver fibrosis. But it was not statistically significant (p = 0.148). There was no significant relationship between MMP-2 levels and stage (p = 0.583). There was an increase, which is not significant, at serum HA and MMP-2 levels while histological activity was increasing from mild to severe (p = 0.117, p = 0.168 respectively). There was a positive correlation between HA and age (p = 0.002). HA levels were greater at cirrhotic patients than non cirrhotic ones (p = 0.075). The sensitivity and specificity of the HA assay at a cut off value of 50 ng/mL was 66.6% and 80.5% respectively. The likelihood ratio (LR) was 2.95 while positive predictive value (PPV) and negative predictive value (NPV) were 36.4% and 93.6%.
Relationship between serum fibrosis markers (hyaluronic acid [HA] and MMP2) and pathological assessment
| HA (ng/mL) | MMP2 (ng/mL) | ||
|---|---|---|---|
| Cirrhosis | |||
| Cirrhotic | N | 6 | 4 |
| Median (min–max) | 61 (15–160) | 100 (100–4,200) | |
| Mean±SD | 72.1±50.8 | 1,125±2,050 | |
| Non-cirrhotic | N | 36 | 35 |
| Median (min–max) | 25 (5–375) | 130 (100–18,000) | |
| Mean±SD | 57±88.7 | 2,699±4,688.7 | |
| Total | N | 42 | 39 |
| Median (min–max) | 26 (5–375) | 100 (100–18,000) | |
| P-valuea | Mean±SD | 59.1±84.0 0.075 | 2,537.0±4,498.4 0.407 |
| Histological activity | |||
| Minimal/absent hepatitis | N | 6 | 6 |
| Median (min–max) | 20 (10–45) | 150 (100–9,000) | |
| Mean±SD | 22.5±12.6 | 2,066.7±3,573.6 | |
| Moderate–severe hepatitis | N | 30 | 29 |
| Median (min–max) | 26 (5–375) | 130 (100–18,000) | |
| Mean±SD | 63.9±95.7 | 2,830.0±4930.6 | |
| Total | N | 36 | 35 |
| Median (min–max) | 25 (5–375) | 130 (100–18,000) | |
| P-valuea | Mean±SD | 57±88.7 0.406 | 2,699±4,688.7 0.871 |
Mann–Whitney U-test.
Conclusion: As a conclusion, serum HA and MMP-2 levels can not be used instead of liver biopsy and histological assessment which is composed of staging fibrosis and grading inflammation. For routine use of serum fibrosis markers, standard assays and new procedures are needed.
P1226 Oral manifestations of lichen planus in patients with hepatitis C virus and HIV/AIDS
E. Giovani, R. Souza, J. Cavasin, S. Egashira, J. Melo, P. Armonia, R. Andia-Merlin (São Paulo, BR)
Objective: The aim of this study was to investigate the prevalence of oral lichen planus (OLP) in patients with hepatitis C virus (HCV), and or in the patients with HIV/Aids.
Material and Method: All the patients were investigated on the presence of antibodies anti-HCV and anti-HIV. The parameters analysed included prevalence of the oral lichen planus, age, gender, race, and the counting of the lymphocytes T-CD4.
Results: A total of 165 patients have anti-HCV antibodies and or with co-infection for HIV/Aids, being 91 patients (55.2% male), and 74 patients (44.8% female). The medium age was 37 years for both sexes, and they were divided in 2 groups. Group I: 59 patients (35.8%) have anti-HCV antibodies and negative HIV antibodies, 32 were male (54.2%) and 27 (45.8%) were female, 5 patients (8.5%) exhibited OLP: 2 patients in the erosive pattern (1 male and 1 female), 1 in the mixed pattern (male) and 2 patients with the reticular pattern (1 male and 1 female). Group II: 106 patients (64.2%) have anti-HCV antibodies and positive HIV antibodies, 59 were male (55.7%), 47 were female (44.3%), 14 (13.2%) patients exhibited OLP: 5 patients with the erosive pattern (3 male and 2 female), 4 patients with the mixed form (2 male and 2 female), and 5 patients with the reticular pattern (2 male and 3 female), and all the patients make therapy only or combined of interferon and or ribavirina. As for the counting of the lymphocytes T-CD4, 33 patients (31.1%) with values below 200 cél/mm3 of blood, 55 patients (51.9%) they presented values among 200 to 499 cél/mm3 of blood, 18 patients (17%) with values above 500 cél/mm3.
Conclusion: The patients with counting of the lymphocytes low T-CD4 and that use ribavirina and the interferon exhibited more exuberant OLP in the several patterns. We still observed the presence of OLP in the erosive pattern in patients of the masculine sex.
P1227 Arthritis and chronic hepatitis C virus infection
J. Puerto, M.A. Escobar, A.A. Garcia-Egido, F.J. Fernandez, F.M. Gomez-Soto, J.A. Bernal, S.P. Romero, F. Gomez (El Puerto Santa Maria, ES)
Arthritis is one of the numerous manifestations of hepatitis C virus infection (HCV).
Objectives: To determine the prevalence of HCV in patients presenting with chronic peripheral inflammatory artritis (CPIA) and, the clinical forms of CPIAs associated with HCV in a 425 beds teaching hospital reference of the Bay of Cadiz (over 250.000 inhabitants).
Methods: Prospective case-control study in 1,972 patients presenting with CPIA as their predominant symptom (excluding cristal induced and degenerative arthritis), mean age (± SD) 47.6 ± 17.3 years, 64% of them women (n = 1,080). Control groups: 759 consecutive patients with non-inflammatory chronic arthritis (NICA), and 6,102 consecutive first time blood donors (BD) matched for age and sex. HCV status was determined (3rd generation ELISA confirmation by RT-PCR, and HCV genotype).
Results: The prevalence of HCV antibodies (5.6%) in patients with CPIA was higher than that in patients with NICA (2.6%) or BD (0.88%) (p < 0.01 in both cases). The prevalence of chronic HCV infection was higher in patients with CPIA (4.3%) than in BD (0.88%). The prevalence of chronic HCV infection by final diagnosis as compared to BD was: 3.1% for Rheumatoid arthritis (RA) (p < 0.01), 6.4% for seronegative RA (p < 0.005), 11.9% for intermitent mono-oligoarthritis (p < 0.001), 6.0% for Sjögren's syndrome (p < 0.005), and 9.8% for systemic lupus erythematosus (p<0.001). The prevalence of chronic HCV infection in psoriasic arthritis, seronegative spondyloarthropathies, and other final diagnosis was not different than that in BD. CPIA associated to HCV infection was non erosive/non deforming in 76% of the patients. Type II cryoglobulins and, decreased levels of C4/C3 were detected in 37% and 48%/22% of the patients with CPIA associated to HCV infection, respectively.
Conclusions: An increased prevalence of HCV infection exists in patients presenting with chronic peripheral inflammatory arthritis. HCV antibodies (3rd generation ELISA) overestimates the prevalence of HCV infection in those patients. A search for HCV infection should be performed in the diagnostic process of both inflammatory arthritis and collagen vascular diseases.
P1228 Seroprevalence of blood-borne infections among blood donors in Boushehr, Iran
Hossein Esmaeili, Z. Hamidiya, S. Mirlatifi, A. Mankhian, G. Hajiyani, Hassan Esmaeili, M. Azizzadeh (Tehran, Boushehr, IR)
Background and Objectives: In recent years there have been special interest in the donor selection strategies in blood banks in order to provide safer blood supply. Important steps are exclusion of paid and replacement donation and increase in the number of regular donors. In this survey the seroprevalence of blood-borne infections among regular, sporadic (lapsed) and first-time blood donors were compared (from March 2005 to February 2005).
Materials and Methods: In a retrospective study the prevalence of transfusion transmitted infections was compared among regular, sporadic (lapsed) and first-time blood donors of Boushehr city. The total of 19,627 blood donors donated blood during March 2005 to February 2005 in Boushehr; out of this number, 7,282 were regular donors (37.1%), 728 sporadic donors (3.7%), and 11,617 first-time donors (59.18%). Data were collected from the computerised data source of Boushehr Blood Transfusion Center and were then compared by the Chi-square statistical test.
Results: Out of 7,282 regular donors, 6 (0.082%) were HBsAg positive, 6 (0.08%) were HCV-Ab reactive, and all were HIV-Ab negative. Whereas, out of 728 sporadic donors one (0.13%) was HBsAg positive, and all were HCV-Ab and HIV-Ab negative. Out of 11,617 first-time blood donors, 65 (0.55%) were HBsAg positive, 41 (0.35%) HCV-Ab reactive. and 3 (0.02%) were HIV-Ab reactive. The prevalence of blood-borne infections (hepatitis B, hepatitis C and HIV) was less in regular than sporadic and first-time blood donors. P value of HBsAg factor was p = 0.0000 for regular and first-time donors, also P value of HCV-Ab factor was p = 0.0004 for regular and first-time donors that these difference were significant.
Conclusions: Regular blood donation is one of the important steps in blood safety; hence, retention of regular donors, and awareness-raising and recruitment of sporadic and first-time donors can increase the rate of regular donation leading in turn to higher blood safety.
Community-acquired bacterial infections
P1229 New concerns with human brucellosis in France in the beginning of the 3rd millenium
A. Mailles, B. Garin-Bastuji, M. Maurin, V. Vaillant (Saint Maurice, Maisons-Alfort, Grenoble, FR)
Context: Human brucellosis is mandatorily notifiable in France. Due to a veterinary policy based on stamping out and vaccination carried out since the 1970s, France is considered “officially brucellosis free” for cattle since 2005 and no cases have been identified in sheep nor goats since 2003. In this context, we studied human brucellosis diagnosed in France to assess remaining risk factors and make specific recommendations.
Methods: Our descriptive study included all human cases notified in mainland France from 1st June 2002 to 31st May 2004. A case was defined as any patient with clinical signs consistent with brucellosis. A confirmed case had a bacterial isolation from any biological sample, or a fourfold increase in anti-Brucella antibodies in 2 samples taken at 2-week interval or a seroconversion. A probable case had a single elevated titre in anti-Brucella antibodies and no alternative diagnosis. For all patients, data were collected using a standardised questionnaire.
Results: During the 2-year period of the study, 105 patients were notified, including 72 cases and 26 false cases who did not meet the case definitions' criteria. The annual incidence of human brucellosis was 0.05 cases per 100,000 inhabitants. Of all cases, 32% had localised infections, mainly arthritis. The disease was diagnosed by bacterial isolation in 49 cases (65%). Eighty percent of cases were imported, having been infected while travelling in an enzootic country or by eating raw milk products imported from an enzootic country. The countries where cases were most frequently contaminated were Portugal (n = 14), Algeria (n = 7) and Turkey (n = 6). Cases had more frequent contacts with animals in enzootic countries than false cases (p = 0.05) and had eaten more frequently raw milk products from enzootic countries than false cases (p = 0.004). One case had a laboratory acquired brucellosis.
Conclusions: Our study confirmed the efficiency of the veterinary policy against animal brucellosis. The incidence of human brucellosis has dramatically decreased and most cases are now imported. Considering the very low prevalence of brucellosis, the direct diagnosis must be preferred to serology to avoid false cases. Recommendations are made to increase the specificity of the surveillance, such as modifying the case definition and consider only bacterial isolation and the increase of serological titre as reliable tests. Specific recommendations are made to prevent the increase of laboratory acquired cases.
P1230 Risk factors for human brucellosis in Iran: a case-control study
M. Soofian, A. Ramezani, M. Banifazl, M. Mohraz, A.A. Velayati, A. Aghakhani, A. Eslamifar (Tehran, IR)
Objective: Brucellosis is a health problem in the central province of Iran (Arak city). The major aim of the present study was to evaluate the risk factors for acquiring brucellosis in Arak.
Methods: A matched case–control study was conducted in the central part of Iran. A total of 300 subjects (150 cases and 150 controls) were enrolled in our investigation. Brucellosis cases were defined on the basis of epidemiologic, clinical, and laboratory criteria using the Standard Tube Agglutination (STA) test and 2-mercaptoethanol agglutination. Subjects were interviewed using a standard questionnaire acquiring demographic and risk factor information. Data were analysed calculating the odds ratio and the confidence intervals for the studied variables. A conditional logistic regression model was used to explore the association between disease and the studied variables.
Results: The age of patients varied between 2 and 86 years (mean: 33.37±21.3 years); 55.3% were males. There were no significant differences in age, gender, marriage situation, residence area (rural/urban), education level, knowledge about prevention routes of brucellosis and occupation between case and control subjects. There was no statistically significant correlation between acquisition of brucellosis, infertility and abortion in sheep and cattle kept at home. Significant risk factors for infection related to existence of (an)other case(s) of brucellosis at home (OR = 7.55; CI: 3.91–14.61; P<0.0001) and consumption of unpasteurised dairy products (OR = 3.7; 95% CI: 1.64–8.3; P<0.014). Knowledge of the mode of brucellosis transmission by fresh cheese appeared to be protective against disease transmission (OR = 0.44; CI: 0.23–0.85; P = 0.01).
We observed significant difference about keeping infected cattle, number of cattle in the house, cattle vaccination, positive family history of brucellosis, number of infected members in household, rate of relapse in family between case and control subjects (P < 0.05).
Conclusion: It is concluded that pasteurisation of dairy products and education regarding fresh cheese must be pursued for eradication of human brucellosis from rural areas. The greatest risk factor for acquiring brucellosis is existence of infected family members.
P1231 Antagonistic activity of Lactobacillus acidophilus toward clinical strains of Helicobacter pylori
E. Andrzejewska, A. Szkaradkiewicz (Poznan, PL)
Objectives: Clinical observations document antagonistic activity of probiotic bacteria of the Lactobacillus genus in the course of Helicobacter pylori infection. Therefore, in present study the in vitro effects was evaluated of L. acidophilus strains which produced or did not produce hydrogen peroxide on clinical isolates of H. pylori.
Methods: The material for studies involved 20 clinical isolates of H. pylori obtained from mucosa of patients with endoscopically confirmed duodenal ulcers. The biopsies were plated on Columbia agar (bioMérieux) with 7% shepp blood agar and incubated in microaerophilic conditions at 37°C for 4 days. The isolated H. pylori strains were identified on the basis of Gram staining and by their production of urease, catalase and oxidase.
Strains of Lactobacillus acidophilus originated from vaginal smears or saliva samples of healthy women. Species identification was conducted using API 50 CHL test (bioMérieux). For detection of hydrogen peroxide production by the isolated Lactobacillus acidophilus TMB-Plus agar was applied. In studies on antagonism the so called columnar technique was employed measuring H. pylori growth inhibition zone around an agar column containing 48 h culture of the analysed L. acidophilus strain.
Results: The range of obtained zones of H. pylori growth inhibition in presence of hydrogen peroxide producing strains of L. acidophilus ranged from 4.9 to 14.2 mm (mean: 9.6±3.6 mm). The range of zones of H. pylori growth inhibition in presence of L. acidophilus strains which did not produce hydrogen peroxide ranged from 5.7 to 11 mm (mean: 8.0±2.0 mm). Analysis of the obtained results disclosed no significant differences between H. pylori growth inhibition induced by L. acidophilus strains which produced or did not produce hydrogen peroxide (p > 0.05)
Conclusions: Strains of L. acidophilus induce variable inhibition of growth in clinical isolates of H. pylori and the effect is not related only to hydrogen peroxide production by the former strains.
P1232 Oral lactobacilli in dental caries
A.K. Szkaradkiewicz, J. Stopa (Poznan, PL)
Objectives: Caries is a unique multifactorial infectious disease, inducing local destruction of dental tissues. Bacteria of the Lactobacillus genus have been well documented to be involved in progress of the carietic process. Present study aimed at evaluation involvement of Lactobacillus spp. producing or not producing hydrogen peroxide in active caries.
Methods: Material for the studies involved samples of the full saliva obtained from 120 patients (20–49 years of age), in whom clinic, according to World Health Organization criteria, permitted to diagnose active caries in 45 patients (group 1) or diagnose patients as caries free (75 patients; group 2). The samples of saliva were plated on Rogosa agar and the cultured isolates obtained in anaerobic conditions were identified using API 50 CHL (bioMérieux). In parallel, for detection of hydrogen peroxide production by the isolated Lactobacillus spp. TMB-Plus agar was applied.
Results: In group 1 with active caries saliva of all patients was found to contain Lactobacillus spp. unable to produce hydrogen peroxide. On the other hand, in 42 (56%) patients in the caries free group (group 2) presence of hydrogen peroxide-producing Lactobacillus spp. was disclosed while no such Lactobacillus spp. were detected in the remaining 33 (44%) patients of group 2.
Conclusions: Development of caries seems to be linked to presence of oral lactobacilli which produce no hydrogen peroxide. Detection of hydrogen peroxides, produced by Lactobacillus spp. in saliva, may be of diagnostic importance in evaluation of the risk of intense caries.
P1233 Methicillin-resistant Staphylococcus aureus infection in a community hospital and the impact of a nursing home opening in its influence area
J.A. Díaz-Peromingo, S. Cortizo-Vidal, F. Pardo-Sánchez, J. Sánchez-Leira, M.F. García-Suárez, S. Molinos-Castro, P. Pesqueira-Fontán, M. Iglesias-Gallego, J. Saborido-Froján, E. Padín Paz, J. Naveiro-Soneira (Riveira, Santiago, ES)
Objective: To study the incidence of Methicillin resistant Staphylococcus aureus (MRSA) infection in a new opened hospital and the impact of a nearby nursing home opening.
Methods: Retrospective analysis of patients infected with MRSA from November 1998 to October 2006. Our hospital covers a population of about 66,000 people and lacks of Intensive Care Unit (ICU). Age, sex, concomitant diseases and mortality were analysed. A nursing home was opened in the hospital influence area (April 2002). We analysed the impact of this in terms of hospital admissions due to MRSA infections. Nasal culture of the nursing home residents was performed to know the prevalence of MRSA carriers.
Results: 64 patients were admitted to the department of Internal Medicine suffering MRSA infections. Blood culture was positive in 10 cases; skin and subcutaneous tissue culture in 7 cases and sputum culture in 47 cases. Middle age was 73 (range 43–91) years old; 45 patients were men and 19 women. During 1998 and 1999 there were no SAMR infections reported. In 2000 (3), 2001 (5), 2002 (3), 2003 (8), 2004 (13), 2005 (20), and 2006 (12) cases. The annual incidence of MRSA infection ranged from 4.54/100,000 (years 2000 and 2002) to 30.30/100,000 habitants (year 2005). The most common associated diseases were chronic obstructive pulmonary disease (COPD, 40 patients), hypertension (22 patients), anaemia (16 patients), hyperlipidaemia (15 patients) and smoking, heart failure and atrial fibrillation (12 patients each). Thirteen patients (20.31%) died. When examined nasal culture, a total of 89 cultures were performed (98% of the total number of residents). From these, 9 were positive for MRSA (carrier prevalence of 10.11%).
Conclusions:
-
1
There is an increasing rate of MRSA infections in our hospital, especially respiratory infections.
-
2
The most frequent underlying clinical conditions in our patients are COPD, hypertension, anaemia and hyperlipidaemia. MRSA infection leads to a considerable death rate (20%).
-
3
The opening of a nursing home in our hospital influence area, probably explains the increasing rate in the following years.
-
4
The relatively old age of our patients all together with the high rate of MRSA carriers and the probable previous admissions to other hospitals (with ICU) of the nursing home residents, points to the need of eradicating this carriers in order to reduce the rate of hospital infections for MRSA related conditions in our area.
P1234 Use of an AFLP technique for the discrimination of source (human or animal) of E. coli
A. Vantarakis, M. Chondrou, M. Efstratiou, M. Papapetropoulou (Alexandroupolis, Patras, Athens, GR)
Objectives: The development and evaluation of a molecular method Amplified Fragment Length Polymorphism technique to fingerprint and differentiate the source of E. coli was the original scope of this study.
Methods: 50 samples of human and 50 samples of animal E. coli were used for the development of the method. The method for the AFLP was adapted by Gibson et al. (1998) with minor modifications. DNA extraction of the E. coli isolates was performed by Prot K extraction method. DNA was digested by Hind III in the buffer provided with the enzyme with 5mM spermidine trihydrochloride. A 5-mL aliquot of the reaction was used to a ligation reaction containing 0.2 μg. The ligated DNA was precipitated with ammonium acetate and ethanol. Then PCR was performed in a total volume of 50 μL as Gibson et al. (1995) described. Amplified fragments were electrophoresed in 1.5% agarose gel. Bands were statistically evaluated to select the specific bands representative of the source.
Results: Four primers with sequences complementary to the adapter sequence were tested. The ability of each primer was assessed on the basis of the results for ease differentiation of specific bands from animal and human E. coli strains isolated from human faeces (hospital) and animal faeces. The human faeces derived from humans (adults and babies) hospitalised for gastroenteritis; the animal faeces came from poultry, bovine and goats. Two primers were judged to produce the most satisfactory results for ease differentiation of origin (human or animal) of E. coli.
Conclusion: AFLP as described can be applied to E. coli without the need for expensive equipment or reagents and is relatively rapid, technically simple, reproducible and specific. It can be a useful a method for the discrimination of source of E. coli. It can be proved a very powerful tool for the evaluation of the origin of unknown E. coli and help to the sanitary survey of an area as well as to the risk assessment.
Aknowledgements: We thank the European Social Fund (ESF), Operational Program for Educational and Vocational Training II (EPEAEK II), and particularly the Program PYTHAGORAS II, for funding the above work.
P1235 Efficacy of long-term co-trimoxazole in Q fever during pregnancy
X. Carcopino, D. Raoult, L. Boubli, A. Stein (Marseille, FR)
Background: Q fever is a worldwide zoonosis caused by the obligate intracellular bacterium Coxiella burnetii. During pregnancy it may result in obstetrical complications such as spontaneous abortion, low birth weight, oligoamnios, fetal death and premature delivery.
Objectives: To evaluate the efficacy of long-term co-trimoxazole treatment in cases of Q fever during pregnancy.
Material and Methods: We evaluated the prognosis of 53 pregnant women who developed Q fever with and without long-term cotrimoxazole treatment defined by the oral taken of trimethoprim and sulfamethoxazole during at least 5 weeks.
Results: In the 37 pregnant women who did not received long-term cotrimoxazole treatment, we observed a 81.1% obstetrical complications rate: 5 (13.5%) spontaneous abortion, 10 (27%) low birth weight, 4 (10.8%) oligoamnios, 10 (27%) intra uterine fetal death and 10 (27%) premature deliveries. Outcome of the pregnancy was found to depend on the trimester in which patients had been infected (p = 0.032) and on the placental infection by Coxiella burnetii (p = 0.013). Long-term co-trimoxazole treatment protected against chronic Q fever (p = 0.001), placental infection (p = 0.038), and obstetrical complication (p = 0.009) especially fetal death (0/16) (p = 0.018).
Conclusions: Our results show that Q fever during pregnancy causes severe obstetrical complications including fetal death. Because of its ability to protect against obstetrical complications and chronic Q fever, long-term treatment by co-trimoxazole should be used to treat women who develop Q fever during pregnancy, specifically when infected during the first trimester.
P1236 Listeriosis in pregnant women
I. Dumitru, S. Rugina, E. Gorun (Constanta, RO)
Background: Listeriosis is relatively rare and occurs primarily in newborn, elderly patients, and immunocompromised persons. Incidence is 7.4 cases per million population. Pregnant women account for 27% of all cases. Hormonal changes during pregnancy have an effect on the mother's immune system that lead to an increased susceptibility to listeriosis in the mother. According to the CDC, pregnant women are about 20 times more likely than other healthy adults to get listeriosis. Listeriosis can be transmitted to the fetus through the placenta even if the mother is not showing signs of illness. This can lead to premature delivery, miscarriage, stillbirth, or serious health problems for her newborn.
Objective: To established the incidence of listeriosis in pregnant women in Constanta and the consequence concerning to fetus and newborn.
Material and Method: Retrospective study on 280 women with abortive disease serologically investigated for Listeria monocytogenes (serotype 1a) in the last five years (2001–2006).
Results: The most affected age group proved to be that in the range of 20–30 years: 206 (73.9%) cases. 112 (40%) female had the diagnostic titer (≥ 1/320); among these, 64 (22.85%) had miscarriages in the IVth-VIIIth month and 38 (13.57%) gave birth to dead foetuses. During pregnancy, only 10 female (3.57%) was diagnosticated with listeriosis and received treatment with Ampicillin. The rest of female (168 cases) was diagnosticated later to abortion and the titer was low. All Ampicillin treated female gave birth healthy newborn.
Conclusions: Listeriosis is a serios diseases during pregnancy. In our study only 10 female patients was diagnosticated with listeriosis during pregnancy, received early treatment and gave birth healthy newborn.
P1237 Cutaneous anthrax in a teaching hospital, Turkey: a review of 39 cases
A. Engin, N. Elaldi, I. Dokmetas, M. Bakici, M. Bakir (Sivas, TR)
Objectives: To evaluate the epidemiological, clinical and laboratory findings and treatment protocols of patients with cutaneous anthrax retrospectively.
Methods: Patients diagnosed with cutaneous anthrax and followed up in the Department of Clinical Bacteriology and Infectious Diseases in the Cumhuriyet University Hospital, an 800-bed teaching hospital in Sivas, central Turkey, between January 1983 and December 2005 were included in the study. The diagnosis of cutaneous anthrax was based upon clinical findings and/or microbiological procedures, including Gram stain and culture of materials obtained from lesions. Patients' charts were reviewed and age, sex, occupation, exposure to a sick animal or to animal products, symptoms, location and type of lesion, clinical and laboratory findings and choice of treatment were recorded for each patient.
Results: In this period 39 patients with cutaneous anthrax were diagnosed and followed up in our clinic. Mean age of the patients was 44 (15 to 74) years, 12 (31%) of whom were females and 27 (69%) of were males. Twenty-five cases (64%) had a history of recent animal slaughtering activity. The clinical presentations were malignant oedema in 10 (26%) of 39 patients and malignant pustule in 29 (74%). The lesions were mostly located on the hand and forearm. The diagnosis was confirmed by bacterial isolation in 17 (44%) cases, by direct examination from the lesion material in 8 (21%) cases. The diagnosis was established in 14 (36%) cases by the lesion presentation. Thirty-six patients were treated with penicillin G. Because of the fact that three patients had a history of penicillin allergy, chloramphenicol, ciprofloxacin and doxycycline was used respectively. A patient with malignant oedema died on the first admission day due to asphyxiation caused by tracheal compression from the extensive oedema.
Conclusions: Although the incidence of anthrax is decreasing worldwide, it is still encountered in Turkey. It manifests sporadically in the eastern, south-eastern and central parts of Turkey where the disease is endemic. Cutaneous anthrax should be considered in any patient with a painless ulcer with vesicles, oedema and a history of exposure to animals or animal products. We think that penicillin is the antibiotic of choice for the treatment of anthrax in endemic regions.
P1238 New perspectives on the bacteriology and antimicrobial susceptibility of dog bite wounds
B. Meyers, J. Schoeman, A. Goddard, E. Seakemela, J. Picard (Pretoria, ZA)
Objectives: In spite of dog bite wounds being a common reason for dogs requiring veterinary care, there is surprisingly little data on the bacteriology of bite wounds. Thus, a prospective study was performed on dogs presenting at the Onderstepoort Veterinary Academic Hospital, University of Pretoria, and a nearby animal shelter with various grades of bite wound.
Methods: Fifty dogs with bite wounds inflicted within the previous 72 hours were selected. This represented 104 wounds. Wounds were clinically graded according to severity and evaluated cytologically. Swabs were collected from all wounds for bacterial culture. Wounds were classified as infected or non-infected. Infection was diagnosed if 2 of the following 3 criteria were met: macroscopic purulence, phagocytosed bacteria present or if the wounded dog had pyrexia. Non-infected wounds were either sterile (established by culture) or contaminated (culture positive but bacteria not phagocytosed on cytology). All wounds were cultured aerobically and anaerobically and all aerobic cultures were evaluated for antibiotic susceptibility.
Table 1.
Percentage antibiotic susceptibility of the most common bacteria
| Susceptibility (%)a |
|||||
|---|---|---|---|---|---|
| Pasteurella multocida | Pasteurellaceae | Staphylococcus intermedius | Pyogenicb streptococci | Escherichia coli | |
| n = 30 | n = 13 | n = 23 | n = 27 | n = 10 | |
| Amoxycillin-clavulanate | 87 | 100 | 91 | 78 | 80 |
| Cloxacillin | 64 | 83 | 90 | 70 | 22 |
| Penicillin G | 93 | 92 | 65 | 81 | 10 |
| Cephalothin | 93 | 92 | 100 | 86 | 20 |
| Ceftiofur | 93 | 44 | 65 | 81 | 44 |
| Enrofloxacin | 93 | 85 | 91 | 7 | 50 |
| Orbifloxacin | 64 | 90 | 74 | 48 | 50 |
| Doxycycline | 93 | 85 | 57 | 67 | 10 |
| −Sulphamethazole + trimethoprim | 90 | 100 | 74 | 89 | 60 |
| Gentamicin | 43 | 92 | 91 | 19 | 60 |
| Amikacin | 65 | 90 | 100 | 7 | 89 |
| Kanamycin | 83 | 92 | 95 | 11 | 50 |
| Lincomycin | 17 | 31 | 40 | 0 | 10 |
| Lincospectin | 33 | 50 | 33 | n/a | 0 |
| Tylosin | 76 | 92 | 95 | 77 | 10 |
Shaded areas indicate susceptibility of 50% or less.
Pyogenic streptococci included: S. canis, S. pyogenes, S. agalactiae, Group-C streptococci.
Results: Of the 104 wounds, 21 were judged to be infected and 83 non-infected. Seventeen (16%) of all wounds were sterile were also classified as non-infected. This was statistically significant (P = 0.02). Of the 84% that were culture positive, 16% grew aerobes, 1% anaerobes and 67% a mixture of aerobes and anaerobes. A total of 211 isolates were cultured representing a mean of 2.1 isolates per wound. Of the aerobes cultured, 22%, 20% and 17% belonged to Pasteurella, Streptococcus and Staphylococcus species, respectively. Within these groups, Pasteurella multocida (65%) and Staphylococcus intermedius (70%) were predominant. Pasteurella canis and pyogenic streptococci were common in infected wounds, whereas Bacillus spp., Actinomyces spp. and the oral streptococci were usually found in contaminated wounds. Three anaerobic genera were cultured, namely, Prevotella, Clostridium and Peptostreptococcus, and were usually associated with wounds with dead space. This is also the first recorded case of Capnocytophaga canimorsus in an infected dog bite wound. Significantly, clinical examination and cytological assessment were capable of establishing whether antibiotics were required or not. Although no single antibiotic was considered to be effective against all the bacteria, in vitro, potentiated sulphonamides, ampicillin and amoxycillin plus clavulanic acid gave the best results.
P1239 Patients with chronic fatigue syndrome have higher numbers of anaerobic bacteria in the intestine compared to healthy subjects
B. Evengård, C.E. Nord, Å. Sullivan (Stockholm, SE)
Objectives: To compare the intestinal microflora in patients with chronic fatigue syndrome with the microflora in healthy subjects.
Methods: Ten patients (6 females and 4 men, mean age 38.7, range 30–53 years), fulfilling the criteria according to the US Centers for Disease Control and Prevention (1994) for chronic fatigue syndrome, were included in the study. The patients had high fatigue severity scores and high disability scores. Ten healthy subjects, matched for gender and age, were included as a control group (6 females and 4 men, mean age 37.6, range 29–48 years). None of the patients or the healthy subjects had taken any antimicrobial agents within the preceding 3 months. Two stool samples (2 to 5 days apart) were collected from each subject. The stool specimens were suspended, diluted and inoculated on non-selective and selective media. The aerobic agar plates were incubated for 24 hours and the anaerobic plates for 48 h at 37°C. After incubation different colony types were counted, isolated in pure cultures and identified to genus level. Streptocoocci and aerobic Gram-negative rods were identified to species level. Median values from the two samples were used in the comparisons.
Results: The aerobic genera alpha-haemolytic streptococci, Gram-positive bacilli, enterobacteria and Candida as well as anaerobic lactobacilli, bifidobacteria, clostridia, veillonella and bacteroides were found in higher numbers in patients with chronic fatigue syndrome than in healthy subjects. However, only for bifidobacteria the difference was statistically significant (P < 0.05). The difference between the two groups in total numbers of anaerobic microorganisms was also statistically significant (P < 0.001).
Conclusion: In this study, higher numbers of anaerobic intestinal microorganisms were identified in patients suffering from chronic fatigue syndrome than in healthy subjects. Disturbances in the intestinal ecology have been implicated as a part of the pathogenesis in chronic fatigue syndrome. Further work is needed to increase our understanding of the role of the intestinal microflora in this disorder.
P1240 Detection and characterisation of enterotoxigenic Clostridium perfringens type A isolates in Japanese retail meats
Y. Miki, I. Kaneko-Hirano, K. Miyamoto, K. Fujiuchi, S. Akimoto (Wakayama, JP)
Objectives: Clostridium perfringens is an important pathogen causing food-borne gastrointestinal disease. In Japan, C. perfringens is ranked as the third or forth greatest cause of the disease and sickened approximately 4,000 people, while the outbreak annually occurred 20 to 40 cases. Previous surveys found that the prevalence of enteropathogenic C. perfringens in Japanese retail food was approximately 0–4%, which was similar to that in USA. It is thought that food poisoning isolates carry cpe on their chromosome, while isolates from other gastrointestinal diseases, such as antibiotic-associated diarrhoea, carry cpe on transferable plasmid. In Japan, recently two food-borne outbreaks by plasmid-cpe isolates were reported, while several C. perfringens food poisoning isolates carry chromosomal cpe. In this survey, we investigated the prevalence of enterotoxigenic C. perfringens in Japanese retail meat samples, and then cpe-positive isolates were tested with multiplex PCR, which could differentiate chromosomal cpe isolate versus plasmid cpe isolate.
Methods: We collected two hundred meat samples from grocery stores and meat shops in Wakayama City. Approximately 100 g of food samples were anaerobically incubated at 45°C overnight with TGC II medium in stomacher bag, and then cultured samples were tested for the presence of cpe-positive C. perfringens followed by further incubation on SFP agar plate for bacterial isolation. DNA were prepared from overnight culture with InstaGene matrix (BioRad) and then used for PCR reaction for cpe detection.
Results: In PCR reaction, we detected cpe from 8 samples (4%), from three of which, we isolated cpe-positive C. perfringens. Interestingly, all three cpe-positive isolates carry cpe on the plasmid and it is combined with downstream IS1470-like structure in PCR cpe-genotyping assay.
Conclusion: From these results, C. perfringens carrying cpe on the plasmid might be a possible pathogen of C. perfringens food poisoning in Japan.
P1241 Isolation and characterisation of Salmonella from Turkish avian food samples
D. Avsaroglu, M. Jaber, M. Akcelik, F. Bozoglu, A. Schroeter, B. Guerra, R. Helmuth (Ankara, TR; Berlin, DE)
Objective: To assess the prevalence of Salmonella (S.) in avian food samples from Turkey and to characterise the strains according to their serotypes, phage types and antimicrobial resistance (R) patterns.
Methods: 76 chicken meat samples were collected from different markets in Ankara, Turkey (2005–2006). Strains were isolated according to ISO 6579. Biochemical tests were performed with the API20E system. Serological analyses, and phage typing of S. Enteritidis and S. Typhimurium, were carried out. All isolates were tested for their susceptibility to 16 antimicrobial agents by the disk diffusion method. All resistant isolates were further tested by broth microdilution and for an extra panel of 11 β-lactams, as well.
Results: 45% of the chicken meat samples analysed (34 out of 76) were positive for Salmonella. From these samples, 71 isolates were obtained. After phenotypic characterisation, they were considered as 42 epidemiologically unrelated strains. The 42 strains belonged to 12 different serotypes. The most prevalent was S. Enteritidis (10 strains, 24%) followed by S. Infantis and S. Virchow (7, 17% each); S. Group C1 with antigenic formula [6,7: k: -] (5); S. Kentucky, S. ssp. I rough form (3); S. Thompson (2); S. Agona, S. Corvallis, S. Nchanga, S. Senftenberg, and S. Typhimurium (1). Among S. Enteritidis, phage type PT21 was predominant (6 of 10), followed by PT6, PT1 and PT3 (2, 1, and 1 strain). Forty per cent (17 strains) of the strains were resistant to some antimicrobial agent. From these, 13 strains were multiresistant and four monoresistant. The most frequent resistance was to nalidixic acid (36% of all strains, 88% of the resistant). All these strains showed also decreased susceptibility to ciprofloxacin. Multidrug resistance to more than 6 antimicrobials was observed in S. Infantis (6 strains showed KAN-NEO-NAL-STR-SPE-SUL-TET-TMP-SXT resistance), and S. Virchow (1 strain AMP-CEF-NAL-STR-SUL-TMP-SXT). This Virchow strain showed resistance to several extended-spectrum β-lactams. All S. Enteritidis were susceptible, except one PT21 NAL-resistant strain.
Conclusion: Almost half of the food samples analysed were contaminated with Salmonella, especially serotypes S. Enteritidis, S. Infantis and S. Virchow. Antibiotic resistance is also a problem in the contaminated food from avian origin (40% resistance). The quinolone resistance in all serotypes and multi-resistance in S. Infantis and S. Virchow need further attention.
P1242 Is a cholera outbreak a natural disaster? Overview on a cholera outbreak in Iran in 2005
N. Jonaidi jafari, M. Radfar, H. Ghofrani, M. Izadi (Tehran, IR)
Objectives: There has been a sharp increase in the number of cholera cases reported to WHO during 2005. A total of 131,943 cases, including 2,272 deaths, have been notified from 52 countries. Overall, this represents a 30% increase compared with the number of cases reported in 2004. During the summer of 2005, an outbreak of choler a struck Iran, infecting 1,118 individuals and killing 11 patients. The epidemic started from the Southern regions and rapidly disseminated across the country.
Methods: In this descriptive, epidemiological research, the related data about all individuals Diagnosed with cholera during the epidemic of summer 2005 has been gathered from the Disease Management Center (DMC). The number of patients in each province, their demographic data, and the death toll were included.
A stool sample from all patients with the clinical diagnosis of cholera was sent to the provincial medical laboratory for confirmation, serotyping, and antibiogram. All confirmed samples were then sent to the central reference laboratory in Tehran for further confirmation. If both these tests were positive for V. cholera, the patient was considered a confirmed case.
Results: It is difficult to pinpoint the first case of the recent outbreak. In 25 June 2005, a 26 month-old Iranian child was admitted in the Southern city of Bushehr with a possible diagnosis of cholera. Ten days later, two Pakistanis were reported to have the same symptoms in the Southeastern city of Chabahar, near the Iran-Pakistan border. Serological studies showed that the former was caused by O1 biotype El Tor serotype Ogawa, while in the two latter cases, the Inab a serotype was isolated. Most cases were reported in Tehran (216 cases), Hamadan (187 cases) and Qom (152 cases) provinces – all located in central Iran. Almost all Iranian provinces were struck by the epidemic. The total number of confirmed cases at the end of the epidemic was 1,118, of which 11 had died.
Conclusion: There is clear trend that cholera is re-emerging in parallel with the ever-increasing proportion of vulnerable populations who live in unsanitary situations. Globally cholera remains a threat for social development. Almost all of the developing countries in the out world are facing either a cholera outbreak or the threat of an epidemic.
P1243 Contamination of toothbrushes with Helicobacter pylori, a possible source of infection transmission
A. Helaly (Alexandria, EG)
Introduction: Helicobacter pylori (HP) infection is a common aetiology in some gastric disorders such as peptic ulcers. The route of infection transmission is not clear. The natural reservoir for HP is not known. HP in dental plaque and saliva had been detected with nested PCR.
Objectives: The aim of this work was to examine the presence of the organism in the toothbrushes of patients with positive infection and compare that with controls without HP infection, in an attempt to search for a possible source of reservoir.
Methods: Forty volunteer patients (10 female, 30 male, mean age 41.2 years) with a positive Helicobacter pylori (HP) infection were included in the study, and ten negative HP volunteers of matched age and sex. All patients had positive test for HP during routine endoscopic examination, those with positive test and those with negative test (biopsy from the antrum) were included in the study. The toothbrush (at least used for one weeks by the patients for 3 times per day) was rinsed in 50 mL of 0.9% NaCl for about 30 minutes, then centrifugation was done, the sediment was examined for HP by CLO test. The result was analysed using standard statistical analysis and student t table.
Result: positive test is considered when any sign of the presence of the infection is present. Positive test was in 28 patients with positive HP infection, no one was positive in the negative volunteers (P > 0.005).
Conclusion: toothbrushes may be a source of HP infection, so that the use of a new one is essential during HP treatment to avoid re-infection. Also the change of toothbrushes is a good hygienic habit as regards HP infection. There may be a possible role for anti HP infection solution to handle the toothbrushes with it.
P1244 Point prevalence study of the spectrum of infectious diseases and antimicrobial drugs use in homogeneous slum population of Mukuru in Nairobi
K. Kandrava, V. Svabova, M. Bartkovjak, K. Kutna, A. Kolenova, M. Krumpolcova, M. Pivarnik, A. Svitkova, Z. Njambi, S. Kamoche, M. Kiwou, V. Krcmery, P. Kisac (Bratislava, Trnava, SK)
Introduction: Aproximately 20–40% of the population of big African cities lives in slums with limited acces to drinking water, sewage removal, electricity and healthcare. Our tropical health centre in Mukuru slum in Nairobi (when aproximately 5 million live in slums) serves about 50,000 people. Since 1999, more than 386,600 patients visits were recorded, and about 105,000 children and 55,000 adults have been registered within last 8 years. Because of the medical service being free of charge, extremely high patient flow (100–120 daily) for physicians, 2 nurses and 2 lab technicians is noted, and it is difficult to perform other than routine outpatient medicine.
Methods: We performed prospective point prevalence study met a simple protocol and analysed spectrum of infections (ID) aetiology and antimicrobial therapy in all cases coming for one month (June 2006).
Results: Majority of cases (312) were RTI, which represented 33% of all visits, followed by diarrhoeal infections (197 cases) and sexually transmitted disease (86), skin and soft tissue infection (68), AIDS (40) and malaria (26). The majority of isolates were Staphylococcus aureus (of which only 3 MRSA), C. albicans and NAC (19; of which only 2 Fluconazol resistant) and S. pneumoniae (8; 2 penicillin resistant).
Including penicillin resistance in pneumococci (25%), resistance profiles were higher (10% of FLU-R among non-albicans Candida and 13% of MRSA) in comparison to data published previously. Despite of that, after treatment with simplex and “old” antimicrobials for RTI, most cases responded clinically. Second commonest infection was gastroenteritis, when Giardia lamblia followed by Entamoeba histolytica, followed by Ascaris lumbricoides, Ankylostoma duodenale and Schistosoma mansonii. Only few cases of GIT infections were of bacterial origin (S. typhi). AIDS appeared in about 11% of visit and malaria in nontravelers about 5%, which is interesting since our clinic is at an altitude of 1,700 m a.s.l. However so called Highlands malaria has been sporadically refuld, also in Nairobi and even in Eldored (2,200 m a.s.l.).
Conclusion: Because of free medical service, extremely high patient flow (100–120 daily) for physicians, 2 nurses and 2 lab technicians makes it difficult to perform other than routine outpatient medicine.
P1245 Increased frequency of isolation of Vibrio cholerae O1 serotype Inaba replacing prevailing serotype Ogawa in Karachi, Pakistan
K. Jabeen, A. Zafar, R. Hasan (Karachi, PK)
Background and Aims: V. cholerae O1 strains can undergo serotype conversion related to a mutation in the wbeT region. Predominant cholera serotype in Pakistan is Ogawa and serotype Inaba is rare. However there is significant increase in isolation of serotype Inaba in our laboratory outnumbering serotype Ogawa and we are now reporting this observation. Moreover we further analysed previous cholera data from years 1993–2005 at our institution to assess the trend of occurrence and resistance pattern of Vibrio cholerae strains.
Material and Methods: This study was conducted at the clinical laboratory of a tertiary care centre located in Karachi, Pakistan which receives specimens from all over country. Cholera data was retrieved from a computerised database at our institution. During the study period (1993–Sept 2005) all stool samples yielding growth of V. cholerae were included. V. cholerae isolates were isolated and identified using standard methodology. Antimicrobial sensitivities were performed by Kirby Bauer technique. Data was analysed using SPSS version 10.
Results: In year 2005 up to September, 245/3,292 (7.4%) specimens yielded growth of V. cholerae. Out of which 243 were Vibrio cholerae serotype Inaba (VCI) which have outnumbered serotype Ogawa (VCO). This recent VCI strain is 100% resistant to cotrimoxazole, 3% resistant to chloramphenicol and no resistance were observed against ampicillin, tetracycline and ofloxacin. This sensitivity pattern is almost similar to that of previous predominant serotype Ogawa.
Conclusion: We are for the first time reporting increased isolation of VCI from Pakistan and suggest that these VCI strains are wbeT mutants from the previously predominant VCO and presumably have arisen as result of selection due to immune response against VCO in Pakistani population. The effective transmission of this strain in the community suggests decrease immune response of the population against this mutant VCI strain.
P1246 EpiScanGIS: a geographical information system for laboratory surveillance of meningococcal disease in Germany
M. Reinhardt, J. Elias, J. Albert, M. Frosch, U. Vogel, D. Harmsen (Würzburg, Münster, DE)
Objectives: The early detection of meningococcal disease outbreaks is important in order to minimise morbidity and mortality through timely enforcement of prevention and control measures. The availability of temporal, spatial and molecular typing data of cases allows the implementation of an online geographic information system (GIS) including objective cluster detection routines. This service should enable members of the public health community to make more informed decisions based on the latest surveillance data.
Methods: Specimens or strains from patients with IMD are typed at the NRZM using a well-proven highly discriminatory approach including serogrouping and DNA sequence typing of the outer membrane proteins PorA and FetA (Elias et al., EID 2006). Case data are stored in a protected, local, custom designed database (MS Access). On a weekly basis, an anonymised fraction of the data is transferred to a PostgreSQL database server (Linux OS). The automated visualisation of the data is handled by the PostGIS and UMN MapServer software components. OpenLaszlo generates Flash-code to deliver these maps in an interactive graphical user interface for a geographical information system (GIS) web server. Additionally, data are scanned weekly for finetype-specific spatiotemporal clusters in a prospective fashion employing the freely available software SaTScan by Michael Kulldorff (www.satscan.org). Moreover, the system generates interactive slide shows within freely selectable time frames.
Results: An online GIS has been set-up, which currently includes data of 2,300 cases of invasive meningococcal disease. To date, the system has 55 registered users, mainly from the public health sector. 25–30 visitors of the site are counted daily. The system optimises the flow of information between public health services, laboratories, healthcare workers, and the public. This proves to be increasingly useful for the evaluation of regional and national epidemiologic changes of invasive meningococcal disease.
Conclusion: The innovative web service EpiScanGIS provides the health community and the general public with easily accessible enriched maps displaying current surveillance data from the NRZM. Furthermore, it implements a national early outbreak detection system. Its inclusion of several non-commercial software packages following a modular “model-view-controller” design makes it easily expandable and adaptable to the surveillance of other infectious agents.
P1247 Clonal spread of Staphylococcus aureus among soldiers in Latvian contingent in Iraq
M. Pinke, L. Cupane, A. Balode, E. Pujate, U. Dumpis, K. Aksenoka, E. Miklasevics (Riga, LV)
Aim: Staphylococcus aureus is an important human pathogen. Although S. aureus is considered to be an opportunistic pathogen, it is possible that certain clones are more prone to cause invasive disease than others due to the presence of virulence factors that increase chance of gaining access to normally sterile sites. The aim of the present work was to study S. aureus carriers rates, resistance levels and dissemination in Latvian army.
Methods: S. aureus strains were isolated from 337 military persons dislocated either in Latvia (Alûksne, 126) or being on Peacekeeping Missions in Iraq (125) and Kosovo (86). S. aureus strains isolated from nasal swabs were tested antimicrobial susceptibility (PEN, FOX, GEN, CIP, CHL, CLI, ERY, RIF, STX, TET, VAN) according to CLSI standard. The presence of the mecA and toxin (PVL, HLG, TSST) genes was determined by PCR. Strain typing was performed by repetitive extragenic palindromic sequence PCR (rep-PCR).
Results: While the rates of asymptomatic S. aureus carriers in Alûksne and Kosovo contingents were similar (30.9% and 34.8%, respectively) the percentage of carriers in Iraq unit was almost doubled (57.6%). From 141 S. aureus isolates obtained none was methicillin-resistant. 104 isolates were resistant to penicillin, and 5 to erythromycin. As MSSA was found more often among members of Iraq contingent these isolates were subjected to more detailed investigation. Molecular analysis revealed 46 strains harbouring no toxin genes, 11 were hlg positive, 7 were hlg-v positive and two strains produced PVL. Rep-PCR revealed three major groups of strains with identical chromosomal background. This strongly points toward clonal spread which may explain high carrier rate in this group.
Conclusions: Antimicrobial resistance levels of analysed strains were low. Our study revealed clonal spread of S. aureus among members of closed community. Personal hygiene is of great importance to reduce dissemination of S. aureus.
P1248 Outbreaks of shigellosis in England and Wales, 2004: use of phenotypic and molecular typing for strain differentiation
T. Cheasty, J. Threlfall (London, UK)
Objectives: In the summer of 2004 a number of apparently unrelated outbreaks of Shigella sonnei infection were observed amongst patients throughout England and Wales. Infections were identified throughout the country but were concentrated in the London area, and the northwest and north-east of England. Outbreaks were in both religious communities and in homosexual men. The outbreak among homosexual men was centred in the London area but outbreaks in the religious community occurred in London, the north-west and the north-east. For meaningful epidemiological investigations in real-time discrimination within the serovar was essential. With this in mind a hierarchical approach based on phenotypic subdivision by phage typing and antibiogram, supplemented by molecular typing using plasmid profile and macrorestriction fingerprinting by pulsed-field gel electrophoresis (PFGE) has been adopted.
Methods: Isolates of Sh. sonnei from infections in England and Wales in 2004 have been phage typed using the scheme of Hammerstrom, Kallings and Sjoberg. Isolates have been tested for resistance to a range of antimicrobials, plasmids identified and sized following extraction of plasmid DNA according to Kado and Liu, and PFGE performed by standard protocols according to Pulse-Net USA.
Results: In all outbreaks the only resistance pattern (R-type) identified was that of ASSuSpTTm (A, ampicillin; S, streptomycin; Su, sulfonamides, Sp, spectinomycin; T, tetracyclines; Tm, trimethoprim). The outbreak among homosexual men was characterised by a new Sh. sonnei phage type (PT), designated PT Q. The isolates from this outbreak were further characterised by a distinctive plasmid-and pulsed-field profile. Isolates from outbreaks among members of a religious community were characterised by three other phage types distinct from PT Q, and by different plasmid- and PFGE profiles, indicating the presence of temporally-associated but independent incidents.
Conclusions: These results indicate that outbreaks of Sh. sonnei can be subdivided by a combination of phenotypic and molecular typing. Results can be obtained rapidly, and can be used in real-time for epidemiological investigations.
P1249 Virulence, pathogenicity, antibiotic resistance and plasmid profile of Escherichia coli strains isolated from drinking and recreational waters
C. Balotescu, E. Panus, M. Bucur, R. Cernat, D. Nedelcu, C. Bleotu, D. Valeanu, V. Lazar (Bucharest, Constanta, RO)
The aim of this study was to investigate the antibioresistance profile and the virulence and pathogenicity hallmarks of Escherichia coli acquatic strains.
Material and Methods: 116 environmental Escherichia coli were isolated from drinking water and marine, salmaster water in Constanta, Romania. They were identified both by biochemical and serological tests. Both disc diffusion susceptibility test and microplate dilution technique were used to investigate the antibiotic resistance profiles. The rapid test to nitrocephine and isoelectrofocusing techniques were used for the confirmation of the presence and type of β-lactamases. The analysis of plasmid DNA was performed using Wizard extraction kit. The virulence tested features were: adherence and invasion of HeLa cells, adherence on inert substrata quantified by slime test, production of extracellular enzymes and exotoxins (haemolysins and other pore-forming toxins, amylase, mucinase, gelatinase, caseinase, aesculin hydrolysis).
Results and Discussion: The tested strains, irrespective to their source of isolation exhibited resistance to ampicillin (28% vs 23%), ticarcillin (7.7% vs 19.4%), tetracyclines (33.3% vs 37.6%) and sulphametoxazole (2.5% vs 18.2%) and were susceptible to all other tested antibiotics. 21 strains exhibiting resistance to β-lactam antibiotics (7 isolated from drinking and 14 from marine waters) proved to be positive for the presence of β-lactamases when tested by nitrocephine rapid test. The β-lactamases of the periplasmic extract exhibited an isoelectric point ranging from 7.5 to 9.6. 10% of the drinking water strains exhibited 1 to 3 plasmids, while 5% of marine strains only 1 plasmid. As concerning the virulence hallmarks, 90% of the strains isolated from drinking water exhibited high capacity of adherence to the cellular substrate (adherence indexes of 85–100% with localised, aggregative and diffuse patterns) demonstrating the potential of these strains to colonise the animal and human tissues and to initiate an infectious process as compared to the marine strains showing low adherence potential. All strains showed colonisation ability of the inert substrate as demonstrated by the high positivity rate of slime test. All tested strains produced lipase, which could act as pore-forming toxin in case of tissue colonisation. Our results are pointing out the importance of detecting specific virulence factors before incriminating water as a source of human diseases.
P1250 Influence of Lactobacilli probiotic strains on apoptosis of colon cancer cells lines
A.K. Gonet-Surówka, M. Strus, P.B. Heczko (Cracow, PL)
Objectives: Cancerogenesis is often associated with disregulation of the apoptosis process. Many external stimuli like cytokines, viruses or bacteria can regulate this process, during which activation of several different proteins takes place within the cell. The central component of apoptosis is a cascade of proteolytic enzymes called caspases. The aim of this study was to investigate how different species of Lactobacilli influence this process on human colon cancer cell lines.
Methods: We studied the effect of live or heat-killed Lactobacilli strains on apoptosis by the pan-caspases activation. We used ESP's green fluorescent-labeled inhibitor, FAM-VAD-FMK, to detect active caspase in living cells. To distinguish apoptosis from necrosis we stained the cells with propidium iodide. As a positive control we used staurosporine, a well-known caspase activator. The apoptosis profiles were determined by FACS analysis.
Results: Our findings indicate that ability to induce apoptosis or necrosis in cancer cell lines vary between different species of Lactobacilli. Many of them have positive effect on cell survival even in serum-starved conditions. On the other hand cell incubation with other Lactobacilli species strongly activate pan-caspases.
Conclusion: This result suggests that some species of Lactobacilli, many used as probiotics, may have positive effect on cancer cell apoptosis. This process is strongly strain specific and depends on the species of Lactobacilli used.
P1251 Prevalence of thermotolerant Campylobacter species in broilers, eggs, chicken abattoir and human samples in a Hungarian county
G. Kardos, J. Mészáros, Z. Galántai, I. Turcsányi, A. Bistyák, Á. Juhász, I. Damjanova, J. Pászti, I. Kiss (Debrecen, Budapest, HU)
Objectives: We monitored the prevalence of thermotolerant campylobacters in chickens and chicken-derived food by following a broiler premise to the abattoir as well as a laying stock and the eggs produced. We also examined the incidence of human campylobacteriosis in Hajdú-Bihar county at the same period.
Methods: Study period was April to October 2006. Twelve pools of five droppings were taken at the age of one, three and immediately prior to slaughter from five consecutive stocks of a broiler premise. A laying stock was sampled bimonthly. From the latter fifty and six pools of five commercial and reject eggs, respectively, were examined weekly. Environmental and abattoir samples were collected following the official guidelines. Human data originated from routine faecal samples, only individuals older than one year were considered. Animal, egg, environmental and abattoir samples were enriched in Preston broth and inoculated onto CAT or CCDA agar. Human samples were inoculated onto solid medium directly. Identification of isolates was performed using genus- and species-specific PCRs.
Results: Environmental as well as chicken feed samples were always negative. Pooled faecal samples collected from three- and six-week-old broilers were almost uniformly culture as well as PCR positive, excepting the April rotation. Those collected at the age of one week were negative. All isolates, excepting two Campylobacter coli, were C. jejuni, though PCR revealed four cases of mixed carriage. All sample sets from layer hens were also positive, but not all individual pools; C. jejuni, C. coli and mixed carriage were found in approximately equal proportion. Out of 300 reject eggs 17 C. jejuni and 8 C. coli, while out of 570 pools of five commercial eggs 10 C. jejuni and 2 C. coli were isolated. From 70 meat samples (including 30 collected from retail shops) 35 C. jejuni and 23 C. coli isolates were obtained. Twenty C. coli isolates originated from processing of one stock. Two faecal samples taken from abattoir staff yielded 2 C. jejuni isolates. Human outbreaks were not detected, the incidence was 2–5%, the monthly proportion of C. coli was between 13% and 36%.
Conclusions: Both eggs and broilers were contaminated by campylobacters, leading to high contamination of meat, which also passed to the shops. Human incidence could be lowered considerably by decreasing Campylobacter carriage of broilers and laying hens.
The work was supported by the GVOP-3.1.1.-2004–05–0472/03 grant.
P1252 Risk factors for nasal carriage of Staphylococcus intermedius in dog owners and their pets
D. Ko, M. Boost (Kowloon, HK)
Objectives: Recently, attention has been drawn to the role of dogs as a reservoir of resistant staphylococci. S. intermedius is the most frequently isolated staphylococcal species from the nares of dogs. We investigated carriage of S. intermedius in dog owners and their pets, and investigated risk factors for carriage.
Methods: A cross-sectional study of owners and their dogs was performed using a convenience sample of 800 pairs recruited at six veterinary practices. Subjects provided information on demographics of owner and dog, amount of contact between dog and owner, and recent antibiotic treatment of the dog. S. intermedius was isolated from nasal swabs collected from both the owner and dog by culture on blood and mannitol salt agar and enrichment in 5% salt meat broth and further subculture. Colonies were confirmed as S. intermedius by catalase and coagulase tests, VP, polymyxin susceptibility and trehalose fermentation. Association between colonisation and a range of demographic and contact factors was determined.
Results: S. intermedius was isolated from 64 of 815 dogs (7.9%) and 8 of 740 owners (1.1%). Of the 8 colonised owners, 4 had colonised dogs. One owner and two dogs were colonised by both S. aureus and S. intermedius. Colonisation of the owner was not associated with multiple dog ownership or with age or sex of the owner, or the number of persons in the household. Dog colonisation was not associated with the presence of multiple dogs or the presence of other animals. Sex of the dog did not influence carriage, but carriage was significantly higher in small dogs and dogs less than one year old, as well as in dogs with chronic health problems. Carriage was significantly higher in dogs that spent more time outside. Owners of puppies were more likely to be colonised than those of older dogs, but occupation was not associated with colonisation of either dogs or owners. S. intermedius was more frequently isolated from owners permitting the dog to lie on the bed, but was not associated with licking the face or carrying.
Conclusion: S. intermedius carriage was higher in younger animals and may spread to their owners more frequently than from older dogs. Unlike S. aureus, which more frequently colonises bitches, there was no difference in sex distribution of carriage. Healthcare workers or their dogs did not have increased risk of colonisation. Co-colonisation with S. aureus and S. intermedius highlights the possibility of transfer of resistance determinants.
P1253 School education in Europe: what are children being taught about antibiotic use and hygiene?
D.M. Lecky, P. Kostkova, C.A.M. McNulty for the e-Bug Working Group
Objectives: e-Bug is a European-wide European Commission (DG SANCO) funded antibiotic and hygiene teaching resource. It aims to reinforce an awareness of the benefits of antibiotics, prudent use and how inappropriate use can have an adverse effect on an individual's good bugs and antibiotics resistance in the community. Prior to production, it is essential to
-
a
examine the educational structure across each partner country
-
b
assess what children are being taught about micro-organisms, good and bad bugs, hand and respiratory hygiene and antibiotic use across Europe
Methods: A questionnaire was devised for distribution to each European partner (Belgium, Czech Republic, Denmark, England, France, Greece, Italy, Poland, Portugal and Spain), exploring the educational structure across Europe and examining educational resources/campaigns currently available.
Results: From the data collected it is evident that the majority of European schools have structured hand hygiene practices in place from a young age. The curricula in all countries cover the topic of human health and hygiene however limited information is provided on antibiotics and their prudent use. Resources comparable to the Bug Investigators Pack in England, which reinforce and build on teaching and implement National Advice to the Public (NAP) campaigns in the classroom, are limited. The ‘Microbes en question’ mobile health education campaign in France is an example of a successful children's education campaign and an innovative programme. Evaluation of the impact of school education on attitude and behaviour change is also limited throughout many European countries.
Conclusion: Not enough is currently being done across Europe to educate children on the importance of appropriate antibiotic use and antibiotic resistance. The data from this research will be used to develop e-Bug: a pan-European EU funded antibiotic and hygiene teaching resource.
P1254 Differences between early and late prosthetic valve endocarditis in a series of 169 cases
A. Plata, J.M. Reguera, A. Alarcon, J. Galvez, J. Ruiz, J. de la Torre, J.M. Lomas, C. Hidalgo-Tenorio, J.L. Haro, J.D. Colmenero for the Andalusian Group for the Study of Cardiovascular Infections
Objectives: To determine the clinical, epidemiological, diagnostic, and therapeutic differences between early (EP) and late (LP) prosthetic valve endocarditis in a series of 169 cases of prosthetic valve endocarditis.
Method: Descriptive study of 169 cases of prosthetic valve endocarditis from a series of 696 left-sided infectious endocarditis from six second- or third-level Andalusian hospitals from 1985 to 2005. Early prosthetic endocarditis was considered up to 12 months after surgery.
Results: No major differences in age, gender or valve affected were found. Clinical presentation was more acute in EP (15.7±18 days) than in LP (41±107 days) and there were more congestive heart failure (NYHA III/IV: 22.7% vs 14.8%). Other clinical signs like fever, splenomegaly and constitutional syndrome were more frequent in LP than EP.
Clinical complications (CNS affectation, systemic embolisms, ocular or skin involvement) were more frequently registered in LP, but the complications related to poorer prognosis (renal failure, septic shock or distress) were more frequent in EP.
Microbiology: EP: S. coagulase negative (50%), S. aureus (13%). LP: S. coagulase negative (22.8%), S. viridans (20.8%). Vancomicin was needed in 63% of EP and 27.7% of LP. Transthoracic echocardiography reaveled diagnosis more frequently in EP than in LP (66.6% vs 41.2%) and so did transoesophageal echocardiography (91% vs 84.7%). 63.2% of the EP and “only” 41.6% of the LE needed surgery. The most frequent indication for surgery for both was right ventricular failure. The complications found in ultrasound or surgery (abscess, fistula, …) were more frequent in EP (66%) than in LP (27.7%). Related mortality was 55.9% in EP versus 31.7% in LP.
Conclusions: Attending the different physiopathology between EP and LP we have found the following differences:
-
1
LP is more insidiosus and needs more days of simptoms for its diagnosis.
-
2
Transoesophageal echocardiography is better than transthoracic echocardiography for the diagnosis in EP and LP.
-
3
S. coagulase negative is still the most frequent microrganism in EP and in LP the aetiology is similar to native endocarditis.
-
4
EP patients develop more complications (annular abscess, fistula, …) leaving them in a worse basal situation (NYHA) and a greater need for surgery together with an increased mortality.
P1255 Risk factors for polymicrobial blood stream infections of biliary origin
M. Salvadó, V. Arauzo, E. Calbo, P. Vazquez, N. Freixes, M. Riera, M. Xercavins, C. Nicolás, J. Garau (Terrassa, ES)
Introduction: Biliary tract infection is an important source of bacteraemia. Frequently, polymicrobial blood stream infections (BSI) are also of biliary origin. The aim of our study was to compare monomicrobial (M) with polymicrobial (P) BSIs of biliary origin in terms of epidemiology, aetiology, severity and outcome.
Material and Methods: From Jan 2000 to June 2006, all adult cases of BSI from a biliary source were identified through records of the Clinical Microbiology Laboratory in a 450-bed acute care teaching hospital. Medical charts were retrospectively reviewed. Variables included demographics, aetiology, comorbidities, severity of disease, and mortality.
Results: During the study period, a total of 2,260 BSI were recorded; of these, 106 (4.7%) were polymicrobial, and one third (31 episodes) of them were of biliary origin. These 31 episodes represent a 14.8% of all BSI of biliary origin seen during this period. Men (66% vs 45%; p = 0.028), older age (76 y vs 70 y; p = 0.019), and the presence of stones (41% vs 19.2%; p = 0.046) were more frequent in the M group. Comorbidities (Charlson score mean 3.3 vs 2.3; p = 0.039), CPR levels (198 vs 115; p = 0.008), the presence of biliary neoplasia (14.8% vs 4.2%; p = 0.005) or biliary prostheses (25% vs 6.3%; p = 0.006) were more frequent in the polymicrobial bacteraemia group. E. coli was the most frequently isolated microorganism in both groups. Previous surgery or endoscopic retrograde cholangiopancreatography were identified in 18.8% in the M group vs. 26.9% in P (NS), and in 35% in M vs. 22.2% in P (NS), respectively. Severity, defined by the presence of shock, ICU admission or vasoactive drugs requirement, was similar in both groups. Mortality was higher in the polymicrobial bacteraemia group [10% in M vs 19% in P; p = 0.217)].
Conclusions: Biliary tract infection is an important cause of polymicrobial bacteraemia, a third of all cases of polymicrobial bacteraemias seen during a six year period were of biliary origin. It is associated with higher co morbidity as compared with the cases of monomicrobial bacteraemia from the same source. Finally, biliary neoplasm and biliary prosthesis appear to be risk factors for polymicrobial bacteraemia among patients with BSI of biliary origin
P1256 Outbreaks of invasive meningococcal disease caused by Neisseria meningitidis group C in two regions of southern Poland
A. Pisula, E. Janczewska-Kazek, A. Szczerba-Sachs, U. Posmyk, K. Kosciow (Chorzow, Katowice, Opole, PL)
Neisseria meningitidis (Nm) is still an important cause of life-threatening infections. Infections with group B strains are the most common in Poland, but infections with group C strain are rare.
Between June and November 2006 in two neighbouring regions, Silesia and Opole, 13 cases of invasive meningococcal disease (IMD) caused by Nm group C were noted. First cases were observed in Silesia region in 3 teenagers (all 16 years old, 1 male, 2 females) and 25-year old woman who meet together in the same discotheque. The IMD form in one woman and the man from this group was meningitis, but in 2 women it was sepsis with disseminated intravascular coagulation (DIC). The epidemiological investigation has shown the connection between sharing of glasses or intranasal inhalations of illegal drugs and IMD development in this group. Prevalence of Neisseria meningitidis group C carriage in nasopharyngeal cavity among habitual guests of the discotheque was 24%. One discotheque worker was also a Nm carrier.
The next cases were noted among 3 children (5, 10 and 2 years old) from families living in the same house in poor household condition. Nobody from their environment was Nm carrier.
In Opole region 4 cases of meningitis (3 men and 1 woman) and 2 cases of sepsis with disseminated intravascular coagulation (1 man, 1 woman) were noted in patients aged 16–57. One case of sepsis and one case of meningitis caused by C ST11 strain resulted in patients' death. The epidemiological investigation has shown no connections between these patients and their environments. In 2 persons from these patients families carriage of Nm group C was confirmed.
86 persons from environment received the chemoprophylaxis (ciprofloxacin, rifampicin). There were no new infections till half of November 2006.
Conclusions: (1) An important risk factor of invasive meningococcal disease is intranasal inhalation of illegal drugs using the same straw. (2) Vaccination against Neisseria meningitidis group C should be widely conducted in children and teenagers.
P1257 Assessment of risk factors in continuous ambulatory peritoneal dialysis-related peritonitis attacks
G.R. Yilmaz, R. Ozturk, C. Bulut, H. Parpucu, H. Irmak, S. Kinikli, M. Duranay, A.P. Demiroz (Ankara, TR)
Objectives: Peritonitis is a common clinical problem that occurs in patients with end stage renal disease and treated by peritoneal dialysis. The aim of this study was to evaluate the potential risk factors for continuous ambulatory peritoneal dialysis (CAPD) related peritonitis
Methods: The study included a study group and a control group. CAPD related peritonitis patients treated in Infectious Diseases and Clinical Microbiology Department between February 2006 and July 2006 formed the study group. And the control group included CADP patients without peritonitis. Demographic characteristics of patients and control group were recorded. Complete blood count, serum protein, albumin, procalcitonine (PCT) and C-reactive protein (CRP) levels were tested in both groups. Cloudiness of the peritoneal dialysis fluid with or without abdominal pain was considered as a suggestive finding of peritonitis and was confirmed by cell count and culture. Patients with more than 100 white blood cells/mL in their dialysate fluid were accepted as having peritonitis. SPPS 13.0 for Windows was used for statistical analysis.
Results: There were 50 patients in the study group and 50 patients in the control group. The mean age of the patients was 48.4 years (range 18–83 years) and 44.8 years (range 19–79 years) in the study and control groups, respectively. The median peritoneal dialysate fluid white blood cell count was 1,275/mm3 (170–17,900) in study group. There were 56 episodes of peritonitis in 50 patients.
Age, gender, education, living in rural area and profession have not been found as risk factors for peritonitis attacks. However, ESR, CRP, PCT, anaemia, hypoalbuminaemia, and hypoproteinaemia were found as risk factors for attacks (p < 0.05). Serum CRP level was found high in 88% of study and 26% of control group (p<0.05). Serum PCT level was found high in 42% of patient and 16% of control group (p < 0.05). Serum CRP level greater than 5 mg/mL and serum PCT level greater than 2 ng/mL were found to have 100% positive predictive value for the diagnosis of CAPD related peritonitis.
Conclusion: In several previous studies, high serum CRP and PCT levels were reported in CAPD patients without peritonitis. In follow up period of CAPD patients with anaemia, hypoalbuminaemia, and/or hypoproteinaemia, high serum CRP and/or PCT level may imply peritonitis.
P1258 Some aspects of treatment of pneumonia complicated with the development of septic shock
D. Piskun, V. Semenov, A. Kutuzova (Vitebsk, BY)
The aim of the work was the development of recommendations on optimisation of treatment of pneumonia complicated with septic shock.
Methods: We used statistical and biochemical methods for the determination of characteristics of nitrosative (nitrites/nitrates) and oxidative (diene conjugates) stress in blood taken from 32 patients on 1, 3 and 5–6 days from the moment of hospitalisation. The control group consisted of 32 healthy donors.
Results: On assessment of kinetics of nitrites/nitrates level reflecting nitrosative stress it was found that on the 1st day of admission its concentration in the blood serum was 41.10 μmol/L, much higher than in the control group – 21.8 μmol/L, on the 3rd and 5–6 days – 39.9 and 38.93 μmol/L. The level of nitrites/nitrates on the 1st day from the moment of admission in the serum of patients receiving cephalosporine of 3rd generation (C3) and macrolide (M) was 40.1 μmol/L, on the 3rd and 5–6 days – 39.4 and 38.45 μmol/L. At the same time, the concentration of nitrites/nitrates in serum of patients receiving C3 and fluoroquinolone (F) was higher than in the group of patients receiving C3 and M: it was 42.09 μmol/L on the 1st day of the disease, 44.89 μmol/L on the 3rd day, and 41.98 μmol/L on the 5–6 days.
The level of diene conjugates in the experiment group on the 1st day of blood collection was 124.52 nM/g lip., and on the 3rd and 5–6 days – 127.5 and 139.42 nM/g lip. In the donor group the level of diene conjugates was 69.18 nM/g lip. Level of diene conjugates in the group of patients receiving C3 and M on the 1st day was 123.52 nM/g lip., and on the 3rd and by the 6th day of the disease was 129.4 and 132.8 nM/g lip. Meantime, in the group of patients receiving C3 and F the level of diene conjugates was higher: on the 1st day of hospitalisation it was 131.7 nM/g lip., by the 3rd and 6th days – 173.8 and 172.9 nM/g lip.
Conclusions: Due to the development of nitrosative stress in patients with septic shock the prescribing of iNO-synthase inhibitors (such as vitamin B12 and trental) is reasonable, especially for the first several days of the disease. Taking into account the trend to increase of the characteristics of oxidative stress by the 6th day after shock occurrence, it is necessary to prescribe antioxidants with the duration of the treatment course not less than 6 days to this group of patients. We recommend the combination of C3 and M for treatment of pneumonia complicated by the development of septic shock.
P1259 Host susceptibility to accute community-acquired bacterial meningitis: multicentre study of genetic markers
E. García-Cabrera, R. Amaya-Villar, S. Barroso, E. Sulleiro, D. Rodriguez, P. Fernández-Viladrich, A. Coloma, P. Catalan, C. Rodrigo, D. Fontanals, F. Grill, M.L. Juliá, J.A. Vázquez, J. Pachon, G. Prats (Seville, Barcelona, Madrid, Valencia, ES)
Objective: The aim of this study was to investigate the influence of host genetic factors in the susceptibility and clinical outcome in patients with acute community-acquired bacterial meningitis (ACABM).
Methods: An observational, prospective, and multicentre study carried out in 9 hospitals of the Spanish Network for the Research in Infectious Diseases, between the 1/11/2003 and 30/09/05. A total of 102 Spanish Caucasian patients (paediatrics and adults) with ACABM and 135 healthy controls (blood donors) from the same ethnical origin were included in the study. The following polymorphisms were genotyped using PCR-ARMS or PCR-RFLP: promoter and allelic variants of mannose-binding lectin gene and G1166C polymorphism of complement component C7 gene. At admission to the hospital, we studied the presence of the following predicting variables of unfavourable evolution (Ann Inter Med 1998; 129: 862–869): hypotension, altered mental status, and/or seizures. Unfavourable outcome was considered in the presence of neurological and/or auditory sequels to the hospital discharge and hospital mortality.
Results: Altered mental status was more frequent in patients homozygous for the C allele of C7 than for the other genotypes (76.7% vs 49%, respectively; p = 0.014). Patients homozygous for the G allele developed more neurological sequels or auditive deficits at the hospital discharge than the rest of genotypes (41% vs 16.4%; p = 0.0287). No association was found between C7 polymorphism and intrahospitalary mortality (p = 0.841).
Conclusions: Our results show that the presence of genotype CC for polymorphism G1166C of C7 is associated significantly with alteration of the level of consciousness at admission to the hospital and developing of neurological or auditive deficit in patients with acute community-acquired bacterial meningitis, although we have found no association with increased mortality.
P1260 Existence of Bartonella henselae reservoir in Poland
T. Chmielewski, R. Marczak, E. Sochon, E. Podsiadly, S. Tylewska-Wierzbanowska (Warsaw, PL)
Infections due to Bartonella species are recently considered emerging diseases. Bartonella organisms are well adapted to facilitate intracellular persistence in a wide variety of animal species, including humans.
The aim of the study was to determine the existence of Bartonella henselae reservoir and vectors of infection in the close surroundings of human beings. This is the first report about the existence of B. henselae in natural reservoirs and vectors in Poland.
The studied groups included mammals (54 dogs, 137 cats) and arthropods collected from both cats and dogs (32 fleas, 107 ticks).
Blood samples were collected from each animal and cultured on chocolate agar plates (Choc V, Oxoid) and in mouse fibroblasts L-929 cell line culture. The levels of Bartonella henselae IgM and IgG antibodies were determined by indirect immunofluorescence assay (Focus diagnostics, USA). Dogs and cats antibodies bound to the antigen were developed with anti-cat and anti-dog FITC IgG Conjugates (Sigma, Germany).
Isolated Bartonella spp. strains grown on chocolate agar and in cell line were detected and identified by PCR methods. Identification of all PCR products was confirmed by DNA sequencing. Bartonella spp. strains were isolated from blood cultures of 14 cats (10.2%). Isolates were identified as: B. henselae 18 isolates, B. clarridgeiae 1 isolate. No positive Bartonella spp. cultures were obtained from any of the 54 samples of dog blood.
Fifty-nine (45.0%) of 131 cats had specific B. henselae antibodies. Twenty seven of tested dogs (50%) had B. henselae antibodies. The majority (96.2%) of seroreactive dogs had low titers of the specific antibodies (1:32, 1:64).
Bartonella DNA was not found in 32 fleas collected from cats and from dogs. None of the ticks collected from cats were infected with Bartonella spp., whereas B. henselae DNA was detected in five ticks collected from five dogs.
These data demonstrate that in the area of central Poland B. henselae is distributed in environmental sources to which humans might be exposed. This concerns pets and their arthropod parasites.
Epidemiology of fungal infections
P1261 Epidemiology trends of candidaemia in an adult intensive care unit in a teaching hospital
J. Garbino, P. Rohner, V. Alencar Nobre, N. Vernaz, C. Garzoni, J-A. Romand (Geneva, CH)
Objectives: A large proportion of nosocomial infections are acquired in intensive care units (ICU). Candida spp. are the leading non-bacterial nosocomial pathogens, and candidaemia carries a high risk of mortality in critically ill patients. Candida spp. are responsible for the majority of nosocomial fungal infections.
The study objectives were to describe the local epidemiology of candidaemia in the ICU of a tertiary care hospital, and to analyse the impact of the use of the newer antifungal compounds.
Methods: Hospital medical records of adult patients with candidaemia were reviewed from January 1989 to December 2005. Demographic information, overall mortality, and predisposing factors for the development of fungal infections were retrieved. We tested the susceptibility of Candida strains after the introduction of the newer antifungal drugs in the institution.
Results: A total of 443 episodes of candidaemia were reviewed. 150 (34%) developed in critical care: 96 (64%) in surgical ICU and 54 (36%) in medical ICU. The median age of patients with candidaemia was 56.5 years (range 17–92); 92 (61%) were males. 87 patients (58%) died within 30 days. Candida albicans was responsible for 73% (n = 109) of infections acquired in ICUs. Non-albicans Candida species (27%) were equally distributed throughout the study period. C. glabrata was identified in 14 episodes, followed by C. parapsilosis (n = 8), C. tropicalis (n = 6), C. krusei (n = 2), C. guilliermondii (n = 1), C. lusitaniae (n = 1), C. pseudotropicalis (n = 1), and unspecified Candida species (n = 9). No change in the distribution of C. albicans versus non-albicans Candida species identified over years was noted. We observed a trend toward a decrease in candidaemia from 1989–1994 (n = 77) to 1995–2004 (n = 62). Most strains of C. albicans (94%) remained highly sensitive to fluconazole and all the newer antifungals tested.
Conclusion: Fungal infection remains a severe disease associated with high crude mortality. We observed a trend toward a decrease in candidaemia during the study period out of last year. C. albicans was the most commonly isolated species, and no shift was observed towards non-albicans Candida species.
P1262 Ongoing semi-national surveillance of fungaemia in Denmark: species distribution and antifungal susceptibility
M. Arendrup, K. Fuursted, H. Schønheyder, A. Holm, J. Knudsen, I. Jensen, B. Bruun, J. Christensen, H. Johansen (Copenhagen, Aarhus, Aalborg, Odense, Hillerød, DK)
Objective: A seminational surveillance programme of fungaemia was initiated in 2003 and now includes 2/3 of the Danish population.
Methods: Participants are 8 departments of clinical microbiology serving the greater Copenhagen area and major parts of Zealand, Funen and Jutland (population 3,440,000). From each episode of fungaemia, defined as a unique blood culture isolate within a 21-day period, a specimen is sent to the National Reference Laboratory of Mycology for verification of species identification and susceptibility testing. Completeness was ensured by comparing lists of referred isolates with local laboratory records. Species identification was done by use of micro and macro morphology and ID32C (bioMérieux, France). Susceptibility testing for amphotericin B, caspofungin, fluconazole, itraconazole, and voriconazole (since 2004) was performed according to the CLSI 27-A2 document in 2003–4 and according to the EUCAST discussion document 7.1 in 2004–6.
Results: A total of 1,068 episodes of fungaemia were recorded during the 3-year study period corresponding to a rate of 11/100,000 population. Candida species accounted for 98% of the fungal pathogens. Although C. albicans was the predominant species (64%) the proportion was lower than previously described (85% in 1994–5). The relative proportion varied considerably among participating departments, i.e. from 55% at Hvidovre University Hospital, Copenhagen and in the county of Frederiksborg to 74% in North Jutland County. C. glabrata was the second most frequently isolated fungus (20%) but again with considerable variation in frequency (8–32%). C. krusei was rarely isolated (3%) and not encountered at 3 of the 8 sites. MIC distributions for amphotericin B and caspofungin were in close agreement with patterns predicted by species diagnosis. However, decreased susceptibility to voriconazole defined as MIC >1 μg/mL was detected in 1/54 (1.9%) C. glabrata isolate in 2004–5 and in 8/73 (11.0%) in 2005–6 (P = 0.08); Overall, 25% of the isolates showed decreased susceptibility to fluconazole and or itraconazole.
Conclusion: The incidence rate of fungaemia in Denmark is 3 times higher than reported in other Nordic countries. Decreased susceptibility to fluconazol and/or itraconazole is frequent and a novel trend towards more C. glabrata isolates with elevated MICs to voriconazole was observed. This should be kept in mind when prescribing antifungal agents before species identification is known.
P1263 Candida lusitaniae fungaemia in cancer patients: risk factors, amphotericin B-mutational frequency and killing and outcome
B. Atkinson, R. Lewis, D. Kontoyiannis (Houston, US)
Introduction: Candida lusitaniae, a Candida species frequently resistant to amphotericin-B (AMB), is a rare cause of candidaemia, seen typically in immunocompromised patients (pts). The clinical significance of this in vitro resistance phenotype as well as the risk factors for and clinical presentation of C. lusitaniae fungaemia in comparison with those of C. albicans have not been completely characterised.
Methods: We reviewed 13 consecutive cases of C. lusitaniae fungaemia and compared them with 41 consecutive cases of C. albicans fungaemia (1990–2004). We also determined mutational frequency of an AMB-susceptible C. lusitaniae isolate as well as time-kill rates to AMB (1 μg/mL, 2 μg/mL) in comparison to C. albicans and C. glabrata reference strains (controls).
Results: In univariate analysis, pts having C. lusitaniae fungaemia were more likely to have leukaemia as an underlying disease (54% vs 15%, P = 0.007), higher APACHE II score (median 15.5vs 12, 95% CI; 0.68–8.05), profound neutropenia (38% vs 15%, P = 0.007), sustained neutropenia (38%vs 7%, P = 0.001), concurrent infections (85%vs 54%, P = 0.05), received growth factors (70% vs 22%, P = 0.005) and systemic high dose steroids (38% vs 5%, P = 0.006). These pts had more frequent exposure to antifungal prophylaxis (38%vs 11%, P = 0.048), higher treatment failure (38% vs 19%, P = 0.028) and need for ICU transfer (54%vs 22%, P = 0.04). The two groups did not differ in frequency of catheter-associated fungaemia. In multivariate analysis, pts having C. lusitaniae fungaemia were more likely to have underlying neutropenia (P = 0.001), stem cell transplantation (P = 0.014) and to have received prior antifungals (P = 0.04). Mutational frequency to AMB for the C. lusitaniae isolate was 8×10–5 and <1×10−9 for both C. albicans and C. glabrata isolates. In addition, kill rates of AMB were low for C. lusitaniae compared to the other isolates.
Conclusion: (1) Host determinants associated with greater susceptibility to C. lusitaniae are underlying haematologic malignancy and prolonged neutropenia, (2) pts having C. lusitaniae fungaemia appeared to be more ill than those having C. albicans fungaemia, (3) C. lusitaniae isolates, even originally susceptible to AMB, might be less amenable to AMB therapy.
P1264 Changing pattern of candidaemia 2001–2005 and consumption of antifungal agents at a university hospital, Vienna
E. Presterl, B. Willinger, R. Gattringer, F. Daxboeck, H. Lagler, W. Graninger (Vienna, AT)
Background: Invasive fungal infections contribute substantially to the morbidity and mortality of immunocompromised patients. Although C. albicans has been the predominant species over years isolated from blood cultures, a shift to so-called non-albicans Candida species has taken place varying from hospital to hospital.
Methods: We performed a retrospective survey on candidaemia over a 5-year period at the University Hospital of Vienna, a 2,200-bed centre with large organ transplantation and haemato-oncology units. The numbers of admissions range between 94,000 and 96,000 patients/year without significant change over the years.
Results: The isolation of Candida spp. from blood increased significantly from n = 25 in 2001, to n = 38 in 2002, n = 60 in 2004 and n = 55 in 2005 (p<0.05). Although C. albicans remained the most frequently isolated pathogen, emergence of non-albicans Candida species, primarily C. glabrata and C. parapsilosis, was observed. C. albicans (17/22 isolates; 77%) was still the leading pathogen at the haemato-oncology unit, followed by C. tropicalis (2/22; 9%), C. lusitaniae, C. sake and C. glabrata (1/22; 5%). At the surgical wards, C. albicans was still the leading pathogen, but the percentage of the non-albicans Candida species (33%) was significantly higher than at any other specialty. The consumption of antifungal agents increased continuously from 2001 to 2005 (p < 0.05). Fluconazole was abundantly used followed by amphotericin B (2005: 24701.5 DDD and 24701.5, respectively). The overall increase in the consumption of antifungals was due to increased use of voriconazole and caspofungin while the consumption of fluconazole and amphotericin B was declining.
Conclusion: Although the number of patients remained unchanged, the net increase of candidaemia in the University Hospital of Vienna, and the consequently increased consumption of antifungal agents warrant the identification of new risk factors.
P1265 Candida blood stream infection in paediatric haematology oncology patients at a Saudi Arabian hospital and research centre
I. Bin-Hussain, A. Al Aidaroos, D. Sharani, I. Barron, M. Jamal, A. Belgaumi, H. El Solh (Riyadh, SA)
Background: Candida blood stream infections are the second most frequent isolates from blood cultures in hospital with large populations of immunocompromised patients.
Objectives: To study the trend of candida blood stream infection at KFSH&RC in the haematology oncology patients and to evaluate the risk factors, the response to antifungal therapy and the outcome of candidaemia in this patient population.
Method: Retrospective chart review utilising the microbiology & Infection Control Data-Bases from Jan 1996 till Dec 2004.
Results: Total of 229 patients with positive blood for Candida spp. Seventy paediatric patients with haematology oncology disorders (30.5%), 57% are males and the majority in the age group 2–5 years (38.6%). Forty percent of candidaemia occur in patients with diagnosis of ALL which constitute the majority of our patients.
Forty eight percent during induction phase of chemotherapy, the majority (68.6%) were neutropenic. Other risk factors like TPN recorded in 25.7% of the patients and 38.6% of them on greater than equal 3 antibiotics. 85% had central venous lines, which were removed in 80%. Dissemination of fungal infection occur in 38.6% of the patients and the mortality because of candidaemia alone is 2.9% and due to candidaemia and other causes is 8.6%.
Conclusion: Candida is not uncommon blood stream isolates in our patient population and Candida albicans is the common Candida species isolated, followed by C. tropicalis in paediatric patients with haematology oncology disorders. Previous hospitalisation is a risk factor for candidamia. Presence of CVL and prolonged use of broad spectrum antibiotics are major risk factor for candidaemia.
P1266 The Scedosporium/Pseudallescheria Database and Culture Collection
R. Horre, V. Robert, G.S. de Hoog, W. Becker, F. Symoens, D. Garcia-Hermoso, E. Goettlich for the ECMM Working Group on Pseudallescheria and Scedosporium Infections
Pseudallescheria and Scedosporium are among the very few fungi that are truly emerging as opportunistic agents of disease. An analysis of 528 published cases of P. boydii infections in humans revealed marked changes in its clinical spectrum. Initially the fungus was observed mainly as a cause of mycetoma after traumatic inoculation. Since the seventies of the previous century, systemic disease entities have become preponderant. Two disease entities are particularly notable: the species regularly colonises lungs of patients with cystic fibrosis, and a unique syndrome is delayed cerebral infection after near-drowning.
The ECMM/ISHAM Network on Pseudallescheria/Scedosporium Infections (PSI) unites partners from 20 countries and includes clinicians, microbiologists and fundamental scientists. With a multidisciplinary approach we aim to understand virulence and routes of infection, and develop isolation, detection and treatment protocols.
An infrastructure has been implemented to collect and preserve large numbers of clinical and environmental strains. These are made freely available for members of the Working Group by IHEM (Brussels) and CBS (Utrecht). In addition, a web-accessible database is being built up (www.Scedosporium-ECMM.com), with a number of interesting features such as on-line identification tools, an interactive phenetic tree option, a picture gallery, and full literature data. Ample options for regular updating will be provided. Strain-specific clinical information combined with susceptibility and microbiological data will enable retrospective monitoring of disease processes for precise understanding of virulence and therapeutic success. The database will be demonstrated.
P1267 Meningitis caused by Candida spp. in a tertiary-care hospital: a 14-year review
E. Calabuig, J. Pemán, M. Salavert, L. Amselem, M. Bosch, A. Viudes, M. Gobernado (Valencia, ES)
Introduction: Meningitis caused by Candida species is a serious condition that may result in significant morbidity and morbility if not recognized and treated effectively. Although meningitis due to Candida spp. remains rare, the frequency of this life-threatening infection has increased in the last years, particularly in patients undergoing neurosurgical procedures (6–17%).
Methods: We have reviewed retrospectively demographic, clinical and microbiological data of patients with isolation of Candida spp. from cerebrospinal fluid (CSF) at the La Fe University Hospital, a tertiary-care hospital in Valencia, Spain, during the period of 1993–2006.
Results: A total of 18 patients, (10 children and 8 adults, with ages ranging from 15 days to 78 years) had positive CSF cultures for Candida spp. accompanied by symptoms consistent with infection. Five patients were female. All cases of meningitis had a nosocomial origin. The species isolated (n = 20) were: C. albicans (n = 12), C. parapsilosis (n = 4), C. tropicalis (n = 3) and C. glabrata (n = 1). Two patients had two different episodes. All patients presented one or more underlying conditions predisposing to meningitis (9 hidrocephalus, 12 recent neurosurgical procedures and 12 CSF shunt). Shunt removal was performed in 10 episodes, but all the patients were treated with different antifungal regimens, including fluconazole, flucytosine, deoxicolate or liposomal amphotericin B. Ten patients (55.6%) survived the infection, six patients (33.3%) died from infection (attributable mortality), whereas two patients (11.1%) died from unrelated causes.
Conclusions: Candida meningitis represents a significant nosocomial infection in at-risk patients and is associated with high mortality rates. It is more common in children (55.5%) than in adults. C. albicans is the predominant causative agent (60%), although other Candida spp. are also associated with these infections. Adequate management of Candida meningitis includes instauration of adequate antifungal therapy and removal of contaminated shunts.
This study has been partially financed by a Grant (2/2005/0140) from the Fundación Investigación del HU La Fe and a Grant from the ISCIII (CM04/00248).
P1268 Invasive mould infections in paediatric cancer and transplant patients
A. Al-Rezqi, M. Hawkes, J. Doyle, S. Richardson, U. Allen (Toronto, CA)
Objectives: As the number of iatrogenically immunocompromised patients rises with improved survival after cancer diagnosis, increasing rates of haematopoietic stem cell transplantation (HSCT) and solid organ transplantation (SOT), the incidence of mould infections has also increased. However, mould infections in the paediatric population have not been well studied. The purpose of this study is to document the incidence, clinical presentation, risk factors, diagnosis, treatment and outcomes of invasive mould infections in paediatric patients with cancer, HSCT and SOT.
Methods: A retrospective chart review was performed of all patients with a microbiologically confirmed (culture-proven) diagnosis of invasive mould infection over an eight-year period (1997–2004) at the Hospital for Sick Children, Toronto, Canada.
Mortality in the first year after diagnosis (cancer) or transplantation (HSCT or SOT).
Results: Twenty-eight patients developed proven invasive mould infections over the eight-year study period (10 cancer, 12 HSCT, 6 SOT). Mould infection occurred in 2.2% and 2.0% after a median time of 60 days and 71 days, among HSCT and SOT recipients, respectively. Aspergillus spp. infection was diagnosed most commonly (23 patients); other moulds included zygomycetes (2), Alternaria spp. (2) and Fusarium oxysporum (1). The most common site of infection was the lung (21 patients), followed by sinus (4) and soft tissue (3). Recipients of allogeneic unrelated HSCT had 7.7-fold higher risk of infection compared to allogeneic related and autologous HSCT patients (RR = 7.7; 95% CI: 2.1–27.9; p < 0.001). Lung transplant patients had a 21.2-fold higher risk compared to other SOT patients (RR = 21.2; 95% CI: 4.7–95.2; p = 0.001). The mortality after one year was 60% among cancer patients, 58% among HSCT patients and 16% among SOT patients (Figure ). For treatment, amphotericin B (conventional or liposomal formulation) was used most commonly (93% of patients); caspofungin (29%) and voriconazole (18%) were also used. Combination antifungal therapy was used in 25% of patients. The total number of patient-days of antifungal therapy was 2,409 patient-days. Fifteen patients with mould infection were admitted to the intensive care unit, for a total duration of 325 patient-days.

Conclusions: Mould infection is an unusual but frequently fatal complication among iatrogenically immunocompromised children. Aspergillus spp. predominate, and patients at increased risk include allogeneic unrelated HSCT recipients and lung transplantation recipients.
P1269 Species distribution in bloodstream isolates of Candida species in Bulgaria and their susceptibility to antifungal agents
L. Boyanova, T. Kantardjiev, A. Kouzmanov, Z. Ivanova, T. Velinov, M. Petrov (Sofia, BG)
Objectives: The aim of this study is to evaluate species distribution and antifungal susceptibility of Candida bloodstream isolates in Bulgaria. This is a part of surveillance programme, called BulSTAR, which researches the aetiological structure of microorganisms and their susceptibility to antimicrobial agents from 1997 to 2005.
Methods: In the National Reference Laborarory of Mycology, we collected and investigated 277 Candida blood isolates. Species identification was confirmed with VITEK YBC system or with API 20 C AUX, followed by antifungal susceptibility testing of isolates, performed with E-test and microdilution kit Micronaut-AM (Merlin). The results were confirmed with the referent broth microdilution method NCCLS (currently CLSI) M27-A2.
Results: The data from BulSTAR showed that, for the period of 1997 to 2005 from all bloodstream isolated microorganisms, Candida strains are: 2.6% (1997), 1.9% (1998), 2.7% (1999), 3.3% (2000), 2.8% (2001), 2.1% (2002), 2.8% (2003), 2.9% (2004), 5.1% (2005).
In the current study overall species distribution is: 53% Candida albicans, 21% C. parapsilosis, 10% C. glabrata, 3.2% C. krusei, 3.6% C. tropicalis and other non-albicans species. We report one interesting case of infective prosthetic endocarditis, caused by C. rugosa, isolated only from the mitral valve.
Minimal inhibitory concentrations (MICs) of fluconazole, read at 24–48 h are: 0.75–2 mg/L for C. albicans, 12–32 mg/L for C. glabrata, 1.5–3 mg/L for C. parapsilosis, 48–64 mg/L for C. krusei. MIC of voriconazole is: 0.023–0.032 mg/L for C. albicans, 3–6 mg/L for C. glabrata, 1–2 mg/L for C. parapsilosis, 2–3 mg/L for C. krusei.
Conclusion: These data suggest that, in Bulgaria the species distribution of Candida bloodstream infections and their antifungal susceptibility rate are similar to those reported in Europe and North America. The highest percentage of non-albicans species identified are C. parapsilosis and C. glabrata.
P1270 Candida albicans versus non-albicans bloodstream infection in critically ill patients: differences in risk factors and outcome
G. Dimopoulos, F. Ntziora, G. Rachiotis, A. Armaganidis, M.E. Falagas (Athens, GR)
Objective: To identify differences of the risk factors and outcome in critically ill patients with Candida albicans and non-albicans candidaemia.
Methods: A prospective observational study was performed in a medical-surgical ICU at a tertiary academic hospital in Athens, Greece. Non-immunosuppressed, non-neutropenic patients with candidaemia diagnosed after ICU admission were included in the study.
Results: During the study period (01/2001–12/2005) 56 candidaemia episodes among 1,037 ICU admissions were included (5.4%). Among these patients 36/56 (64.3%) had candidaemia due to C. albicans and 20/56 (35.7%) due to non-albicans species [8/56 (14.3%) C. glabrata, 6/56 (10.7%) C. tropicalis, 3/56 (5.4%) C. parapsilosis, 1/56 (1.8%) C. lusitaniae, 1/56 (1.8%) C. krusei and 1/56 (1.8%) C. dubliniensis]. Administration of glucocorticosteroids, central venous catheter (CVC) placement and preexisted candiduria were shown to be independently associated with candidaemia due to C. non-albicans species (OR 45.1, 95% CI 3.0–669.9; OR 26.2, 95%CI 2.1–334.8; and OR 16.5, 95% CI 1.6–173.9, respectively). Mortality was higher in patients with non-albicans spp. than C. albicans blood stream infection [18/20 (90%) vs 19/36 (52.8%), p = 0.005]. The multivariable logistic regression analysis revealed that candidaemia due to non-albicans species was independently associated with death (OR 6.7, 95%CI 1.2–37.7).
Conclusions: In the subset of critically ill non-immunosupressed patients, candidaemia caused by non-albicans species occurred more likely in those with medical devices or receiving steroids. Candidaemia blood stream infection due to non-albicans species was also associated with higher mortality.
P1271 An outbreak of fungaemia in a neonatal intensive care unit in Kuwait caused by Candida haemulonii resistant to amphotericin B, itraconazole and fluconazole
Z. Khan, N. Al-Sweih, S. Ahmad, N. Al-Kazemi, S. Khan, L. Joseph, R. Chandy (Kuwait, KW)
Objective: Candida haemulonii is an uncommon species of Candida that is occasionally isolated from foot lesions, and is rarely reported as a cause of invasive disease. Here, we describe an outbreak of C. haemulonii fungaemia in a neonatal intensive care unit (ICU) of a maternity hospital in Kuwait.
Methods: The diagnosis of C. haemulonii fungaemia involving four neonates was made by isolating the yeast from blood cultures. Repeat isolates were obtained from two neonates. Since the phenotypic characteristics of the isolates were not helpful, molecular methods were used for species-specific identification. The amplification of species-specific internally transcribed spacer (ITS)-1 and ITS-2 regions of rDNA and/or D1/D2 regions of 26S rRNA was performed by using panfungal primers and the amplicons were sequenced. The antifungal susceptibility of the isolates was determined by E-test using RPMI 1640 medium supplemented with 2% glucose.
Results: Of the 4 neonates (2 males and 2 females) diagnosed with C. haemulonii fungaemia, 3 were born pre-term weighing 710–1,035 g, and one full-term but with congenital abnormality. All were exposed to risk factors for Candida infections that included use of broad-spectrum antibiotics, administration of total parenteral nutrition and placement of central venous catheters. All the 7 isolates did not assimilate cellobiose, melibiose, melezitose and salicin but assimilated trehalose, esculin, and glycerol. The ITS and/or D1/D2 region sequences of all the 7 isolates matched nearly completely (only 2 and 1 nucleotide differences in the ITS and D1/D2 regions, respectively) with the corresponding sequences from the reference C. haemulonii strain CBS 5149, thus confirming their identification as C. haemulonii. They were uniformly resistant to amphotericin B, fluconazole and itraconazole, but susceptible to 5-flucytosine (0.008–0.064 μg/mL), voriconazole (0.047–0.38 μg/mL) and caspofungin (0.023–0.5 μg/mL). Despite antifungal therapy, 2 of the neonates died.
Conclusion: To the best of our knowledge, this is the first report describing an outbreak of C. haemulonii fungaemia in neonates. The report underscores the emergence of new or rare Candida species, resistant to polyenes and azoles, as a cause of invasive disease in neonates or immunocompromised patients.
Supported by KURA grant MI04/05, Department of Microbiology, Faculty of Medicine Kuwait University and Ministry of Health, Kuwait
P1272 In vitro activity of newer antifungal agents against Candida albicans biofilms
A. Katragkou, M. Simitsopoulou, E. Diza-Mataftsi, C. Tsantali, E. Roilides (Thessaloniki, GR)
Objectives: Candida albicans biofilms have been shown to be resistant to a variety of antifungal agents. We aimed to determine the in vitro activities of the newer antifungal agents voriconazole (VRC), posaconazole (PSC), caspofungin (CAS) and anidulafungin (AND) against C albicans biofilms and compare them to those against planktonic C albicans cells.
Methods: C albicans GDH2346 and M61, two documented biofilm-producing clinical isolates, were used. Biofilms were grown by incubation on silicone elastomer disks in 12-well (macro-method) or 96-well (micro-method) plates at 37°C under constant shaking for 48 h. Mature biofilms were then incubated with no drug (controls) and with two-fold dilutions of VRC (0.125–256 mg/L), PSC (0.06–64 mg/L), CAS (0.015–16 mg/L) or AND (0.0078–16 mg/L) for 24 h. Percent biofilm damage was then assessed colorimetrically by XTT assay as reduction in the metabolic activity of biofilms. Biofilm MIC was defined as the least antifungal concentration causing ≥50% reduction in the metabolic activity of biofilm compared to controls. Planktonic MIC was determined according to CLSI M27-A2 microdilution method. Statistical analysis of 5 experiments for each strain included ANOVA and post hoc analysis (P <0.05).
Results: Biofilm MIC of VRC was >256, of PSC >64 mg/L for both strains tested, of CAS 0.031 for GDH2346 and 0.0625 mg/L for M61, and of AND 0.125 for GDH2346 and 0.5 mg/L for M61. By comparison, planktonic MICs of VRC were 4 for GDH2346 and 0.015 mg/L for M61, of PSC ?0.0018 mg/L for both strains, of CAS 0.031 for GDH2346 and 0.0625 mg/L for M61 and of AND 0.003 mg/L for both strains. The maximum VRC-, PSC-, CAS- and AND-induced % biofilm damage for GDH2346 was 30±2.3% (at 256 mg/L), 42±9.5% (at 16 mg/L), 70±3.7% (at 0.0625 mg/L) and 60±3.8% (at 0.5 mg/L), respectively. For M61 strain the corresponding values were 24.6±3% (at 256 mg/L), 47±4.3% (at 32 mg/L), 62.3±11% (at 2 mg/L) and 52±3.4% (at 0.5 mg/L). Complete sterility of C. albicans biofilms on silicone substrate was not achieved even at the highest concentrations of antifungal agents used.
Conclusions: C. albicans biofilms are >1,000-fold less susceptible to azole-class antifungal agents than their planktonic counterparts. On the contrary, echinocandin-class antifungal agents appear to have enhanced activity against C albicans biofilms. These findings may assist better understanding antifungal drug activity against biofilms and have therapeutic implications.
P1273 Activity of posaconazole against clinical isolates of Candida albicans with decreased sensitivity to fluconazole from autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy patients
R. Rautemaa-Richardson, M. Richardson, M. Pfaller, P. Koukila-Kähkölä, J. Perheentupa, H. Saxen (Helsinki, FI; Iowa City, US)
Objectives: Most patients with APECED (autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy) suffer from chronic oral candidosis from early childhood. Thus most patients receive repeated treatment and maintenance courses of azole antifungals, principally ketoconazole and fluconazole, throughout their lives. This has resulted in both mycological and clinical resistance. Our aim was to determine the susceptibility of patient isolates from Finnish patients with APECED to the new triazole antifungal posaconazole, recently approved for the treatment of oropharyngeal candidosis, including infections refractory to fluconazole. In previous studies posaconazole has been shown to be active against most fluconazole-resistant C. albicans isolates.
Methods: The antifungal susceptibility profiles and antifungal usage of all 56 APECED patients followed in our centre were reviewed for the period 1994–2004. C. albicans isolates of 11 patients reported to have decreased fluconazole sensitivity (n = 27, MIC range: 8–32 mg/L) were tested for their sensitivity to posaconazole using a broth dilution technique as detailed in the CLSI document M23-A2. C. albicans strains of 11 patients APECED patients reported to be sensitive to azoles (n = 16, MIC range: 0.12–2 mg/L) were tested in a similar manner.
Results: C. albicans isolates previously shown to be of decreased susceptibility to fluconazole were uniformly sensitive to posaconazole (MIC range: 0.03–1 mg/L). Isolates previously scored as fluconazole sensitive were equally sensitive to posaconazole (MIC range: 0.12–1 mg/L). The upper limit of posaconazole sensitivity has been tentatively been set at ≤1 mg/L.
Conclusions: Decrease in the susceptibility of the colonising C. albicans strains to antifungals was noted in the mid-1990s. Currently, symptomatic patients are prescribed topical polyenes. In some patients eradication treatment of the fluconazole resistant strains with iv caspofungin or liposomal amphotericin B has been successful in combination with topical polyenes and professional oral hygiene procedures. The present data, which highlights the fungistatic and fungicidal activity of posaconazole against strains with decreased fluconazole sensitivity, suggests that oral posaconazole would be effective for treatment of candidosis caused by strains of C. albicans with decreased sensitivity to fluconazole in APECED patients. Furthermore, posaconazole could be used as a first-line drug for eradication treatment.
P1274 Decreased sensitivity of Candida albicans for azole antifungals a complication of long-term prophylaxis in APECED patients
R. Rautemaa-Richardson, M. Richardson, M. Pfaller, P. Koukila-Kähkölä, J. Perheentupa, H. Saxen (Helsinki, FI; Iowa, US)
Objectives: Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) is an autosomal recessive disease exceptionally common in Finland. Most patients have chronic oral candidosis since early childhood and the infection has been suggested to be carcinogenic. Thus, most patients receive repeated treatment and prophylactic courses of antifungals throughout their lives. Since the late 1980s topical and systemic azole antifungals, mainly ketoconazole and fluconazole have been used. In Finland 92 patients have been diagnosed with APECED and 66 of them are alive. Our aim was to study the effect of long term azole treatment on candidal colonisation of APECED patients and changes in antifungal susceptibilities.
Methods: We gathered and evaluated the routine fungal culture reports, antifungal susceptibility profiles and antifungal usage from 1994 to 2004 based on patient records of all 56 APECED patients followed in our institution. Candida albicans strains of eleven patients reported to have decreased azole antifungal sensitivity (n = 27) were re-analysed for their species and sensitivity to fluconazole. Candida albicans strains of another eleven APECED patients reported to be sensitive to azole antifungals (n = 16) were also re-analysed.
Results: A total of 162 fungal cultures had been performed of the 56 APECED patients during 1994 to 2004. Of these 42 (75%) had been reported to be positive for Candida and 35 (63%) for C albicans. Eleven patients had been found to be colonised with a C albicans strain with a decreased sensitivity to fluconazole. Seven patients had been reported to harbour a non-C. albicans strain but only two of them with a decreased fluconazole sensitivity. Re-analysing the stored strains with decreased sensitivity to fluconazole showed that colonisation had evolved already by the mid 1990s and seems to be persistent. The cessation of the use of azoles and favouring topical polyenes since the year 2000 has not induced colonisation with sensitive strains in the eleven patients. However, new patients have not been reported to have become colonised with Candida strains with decreased sensitivity to fluconazole.
Conclusions: Due to the life long need for antifungal treatment and the risk of selection of azole resistant Candida strains the use of topical polyenes should be preferred and the use of repeated, low dose courses all azoles should be avoided.
Clinical mycology II
P1275 Longitudinal trends in candidaemia in England and Wales
G. Gardiner, T. Lamagni (London, UK)
Objectives: In light of reports from many countries documenting increases in the incidence of candidaemia, data from England and Wales were analysed to characterise epidemiological patterns and trends in these infections.
Methods: Laboratories across England and Wales participate in population-based surveillance of infectious diseases. These data are collated, managed and stored by the Health Protection Agency. Data were extracted retrospectively on episodes of candidaemia voluntarily reported between 1980 and 2005 and analysed for epidemiological patterns and trends. Rates were calculated using mid-year resident population estimates based on a national census carried out in 2001.
Results: Between 1990 and 2005, the rate of candidaemia across England and Wales increased almost five fold from 0.6 to 3.1 per 100,000 population. In 2005, candidaemia reports accounted for 81% of all invasive fungal infections. The proportion of bloodstream infections (bacterial and fungal) due to Candida species increased steadily between 1980 and 2005, from 0.74% to 1.79%. Candidaemia rates were higher in males than females overall (3.4 vs 2.9/100,000; RR = 1.19; 95%CI: 1.08–1.31). Cases were concentrated in infants (15.4 and 8.7/100,000 in <1 year old males and females respectively) and the elderly (12.1 and 7.1/100,000 in ≥75 year old males and females respectively). Candida albicans accounted for 55% of candidaemia reports in 2005 with the other common species including C. glabrata (18%) and C. parapsilosis (10%). The distribution of Candida species changed substantially between 1986 and 1998 where the proportion of candidaemia reports due to C. albicans fell from 75% to 54%, with increases observed amongst C. glabrata, C. parapsilosis and C. tropicalis species.
Conclusion: Analysis of trends in candidaemia in England and Wales has shown marked increases in the rates of disease, more pronounced than generally seen for bacteraemic infections over the past two decades. Surveillance has shown changes in the epidemiology of these infections, in particular a decrease in the proportion of reports due to C albicans between 1986 and 1998. This may be of clinical importance with many non-albicans species being inherently less susceptible to antifungal agents.
P1276 Ten years of Candida glabrata and C. krusei fungaemia: epidemiology and susceptibility patterns in a tertiary-care hospital
M. Bosch, E. Calabuig, J. Pemán, A. Valentín, A. Viudes, E. Cantón, M. Gobernado (Valencia, ES)
Objective: To study the incidence, clinical features and susceptibility patterns of candidaemia caused by Candida glabrata (CG) and Candida krusei (CK), two species with diminished fluconazole susceptibility, in a tertiary care hospital during the last ten years.
Methods: Retrospective study of all candidaemia episodes by CG and CK diagnosed from January 1995 to December 2004 at La Fe University Hospital. Mycology, laboratory and clinical records were reviewed. The susceptibility study was carried out using the M27-A2 document.
Results: In the period studied, 567 episodes of candidaemia were found, 23 of them were due to CG and 26 to CK. The 93.9 and 69.4% of patients were adults and male, respectively. The most important underlying conditions were haematological malignancy (49%), solid organ transplantation (6.1%) and pancreatitis (6.1%). The major predisposing factors were, respectively, previous antibiotic therapy (100%), intravascular line (89.5%; 31.6 and 57.6%), neutropenia (55.3%; 5.3 and 50%) and previous chemotherapy (55.26%; 2.63 and 52.63%), antifungal prophylaxis (44.74%; 2.63 and 42.11%) and parenteral nutrition (28.9%; 18.4 and 10.5%). 28.6% episodes of candidaemia occurred in ICU patients, 65.31% in medical department patients (46.9% haematological) and 4.1% in surgical patients. After diagnosis, 84.2% of patients received antifungal treatment. Crude mortality rate of candidaemia due to CG and CK were 50% and 63.64%, respectively. Risk factors associated with death most frequently observed were previous chemotherapy (59.1%), acute myelogenic leukaemia (45.4%), pneumonia in HIV (13.6%) and coronary by-pass (13.6%).
Fluconazole MICs ranges (mg/L) for CK/CG, respectively, were 16–32/0.5–32; itraconazole, 0.06–2/0.06–4; voriconazole, 0.03–1/0.06–4; flucytosine, 4–32/0.06–2 and amphotericin B, 0.06–2/0.03–1; and the respective MIC90 (mg/L) for the same drugs were 32/32, 1/1, 1/1, 32/0.5 and 1/1.
Conclusions: CG plus CK are the third cause of candidaemia in our hospital. Most of candidaemia episodes occur in adults. Major risk factors for CG or CK candidaemia include broad-spectrum antibiotics, intravascular line, neutropenia and chemotherapy. The crude mortality in patients is high (57.9%). All isolates were susceptible to voriconazole and amphotericin B.
This study has been partially financed by a Grant (2/2005/0140) from the Fundación Investigación del HU La Fe and a Grant from the ISCIII (CM04/00248).
P1277 Caspofungin for the treatment of invasive mycoses in critically ill patients (ProCAS study)
C. Leon for the ProCAS Study Group – Grupo de Trabajo en Enfermedades Infecciosas (GTEI), SEMICYUC
Introduction: Caspofungin is an echinocandin with proven efficacy against invasive candidiasis (IC) and aspergillosis (IA). The ProCAS study is an ongoing multicentre, prospective, noncomparative observational study co-sponsored by GTEI-SEMICYUC and PETHEMA and funded by MSD Spain, aimed at estimating the effectiveness and safety of caspofungin in adults with IC or IA under everyday conditons.
Methods: The interim analysis herein presented focused on the first 64 critically ill patients enrolled. Caspofungin was generally dosed as recommended in the package insert. Favourable outcomes included complete and partial responses on the last day of caspofungin therapy. Safety was assessed up to 14 days post-caspofungin.
Results: Thirty-three [52%] medical, 26 [41%] surgical, and 5 [8%] trauma critically ill patients were analysed. The median age was 58 years (range, 21–83). Nineteen (30%) patients had candidaemia, 16 (25%) other types of IC, 14 (22%) suspected IC (11 had a Candida Score [Crit Care Med. 2006 Mar;34(3):730–7] of >2.5), 14 (22%) suspected or proven IA, and one (2%) mucormycosis. Candida albicans was the single most frequently isolated species causing documented IC (54%), followed by C. glabrata (16%), C. krusei (11%), C. tropicalis (8%), and C. parapsilosis and Candida spp. (5% each). The median duration of caspofungin therapy was 15 days (range, 2–337). Caspofungin was given as first line therapy to 40 (63%) patients. The remaining patients were either refractory (21 [33%]) and/or intolerant (2 [6%]) of other drugs. Eleven (17%) patients received it in combination with other antifungals. Favourable response rates were 83% (29/35) for proven IC and 50% (4/8) for documented (proven or probable per EORTC criteria) mould infections. Overall, 4 (6%) patients had an adverse reaction to caspofungin (three non-serious, one serious). No significant changes in serum liver function tests and creatinine occurred during caspofungin therapy. No relapse was detected among patients with documented IC or IA who responded favourably to caspofungin (median follow-up, 61 days; range, 1–102). Overall and infection-related mortalities were 44% (28/64) and 31% (20/64).
Conclusion: The results of this interim analysis of the ProCAS Study suggest that, under everyday conditions, caspofungin is an effective and safe alternative for the treatment of invasive mycoses in critically ill patients.
P1278 Prevalence and epidemiology of onychomychosis in diabetic patients in Duzce, Turkey
A. Gulcan, I. Sahin, M. Yildirim, S. Oksuz, E. Gulcan, D. Kaya (Duzce, Kutahya, TR)
Objectives: In comparison with the general population, patient with diabetes mellitus are predisposed to mycotic infections. The aim of this study was to determine frequency of onychomycosis in diabetic patients, to identify causative agents, and to evaluate diabetic and epidemiologic risk factors.
Materials and Methods: Three hundred and thirty one diabetic patients were included in the study. Of 331 diabetic patients, there were 217 (65.6%) women and 114 (34.4%) men with an age range of 18 to 88 years (mean: 55.6 ±11.8). Type 1 diabetes was present in 10 (3%) patients (6 men, 4 women) and type 2 diabetes was in 321 (97%) patients (108 men, 211 women). The demogaphic and clinical data of the diabetic patients were interrogated by face to face interview. Samples from clinically abnormal nails were collected by vigorously scraping the distal portion of the nail. All specimens collected were analysed by direct microscopy and culture. Onychomycosis frequences relation with demographic, epidemiologic and diabetic features in diabetic patients were evaluated Logistic Regression analysis.
Results: Forty-two (12.6%) patients were diagnosed as onychomycosis mycologically. Of the isolated fungi, 24 (57.1%) were yeasts (19 Candida spp. and 5 Rhodotorula spp.), 18 (42.9%) dermatophytes (13 T. rubrum, 4 T. mentagrophytes, and 1 T. tonsurans). The significant correlation was found between the frequency of onychomycosis and obesity (B = 1.446, p = 0.002), the time of nail growth (B = 1.124, p = 0.006), wearing nylon socks (B = 1.151, p = 0.009), not drying extremities after ritual ablution (B = −1.430, p = 0.011), family history (B = 1.044, p = 0.012), and duration of diabetes (B = 1.002, p = 0.038). The data are shown in Table 1 .
Table 1.
Relation between onychomycosis and the risk factors
| Risk factor | N | Onychomycosis |
P | B | Exp(B) |
||
|---|---|---|---|---|---|---|---|
| n | % | Exp(B) | 95% CI | ||||
| Duration of diabetes (years) | |||||||
| 0–10 | 244 | 29 | 11.9 | 0.038 | 1.002 | 2.724 | 1.056–7.026 |
| >10 | 87 | 13 | 14.9 | ||||
| Not drying after ablution | |||||||
| yes | 87 | 5 | 5.7 | 0.011 | −1.430 | 0.239 | 0.079–0.720 |
| no | 187 | 35 | 18.7 | ||||
| Wearing nylon socks | |||||||
| yes | 157 | 32 | 20.4 | 0.009 | 1.151 | 3.161 | 1.338–7.470 |
| no | 174 | 10 | 5.7 | ||||
| BMI (kg/m2) | |||||||
| <30 | 133 | 9 | 6.8 | 0.002 | 1.446 | 4.244 | 1.694–10.631 |
| ≥30 | 139 | 33 | 23.7 | ||||
| Family History | |||||||
| Yes | 104 | 23 | 22.1 | 0.012 | 1.044 | 2.840 | 1.260–6.402 |
| No | 227 | 19 | 8.4 | ||||
| Time of nail growth (week) | |||||||
| <2 mm | 242 | 19 | 7.9 | 0.006 | 1.124 | 3.076 | 1.391–6.800 |
| >2 mm | 89 | 23 | 25.8 | ||||
Conclusions: These results indicated that education of the diabetic patients about the importance of the simple preventions, foot and nail care should be essential components of diabetes management.
P1279 Candida dubliniensis in a general hospital over an 8-year period
M. Guembe, T. Peláez, C. Matesanz, M. Torres-Narbona, J. Guinea, L. Alcalá, E. Bouza, P. Muñoz (Madrid, ES)
Objectives: C. dubliniensis is a recognized fungal pathogen causing mucosal disease in AIDS patients. We investigated the incidence and antifungal susceptibility of C. dubliniensis isolated from clinical samples during an 8-year period in our hospital (1999–2006).
Methods: The antifungal susceptibility patterns for amphotericin B (AB), fluconazole (FZ), itraconazole (IZ), ketoconazole (KZ), voriconazole (VZ), posaconazole (PZ), flucytosine (FC), and caspofungin (CS) were determined by the broth microdilution method (Sensititre YeastOne YO7) and E test. The isolates were identified as C. dubliniensis by their inability to grow at 45°C and by rapID 32 C (bioMérieux).
Results: A total of 76 isolates of C. dubliniensis (49 patients) were recorded. The distribution of isolates and patients (isolates/patients) during the study period was as follows: 1999 (1/1), 2000 (0/0), 2001 (4/4), 2002 (11/5), 2003 (6/6), 2004 (30/23), 2005 (6/2) and 2006 (18/8). Clinical isolates were obtained from mucocutaneous sites (17/22%), respiratory tract samples (51/67%) and normally-sterile sites (8/11%). Overall, 35 of our 49 patients were HIV-infected. The MIC ranges (mg/L) of antifungal agents tested against C. dubliniensis were as follows: AB (0.008–0.06), FZ (0.06–1), IZ (0.008–0.125), KZ (0.008–0.03), VZ (0.008–0.016), PZ (0.002–0.25), FC (0.003–0.5), and CS (0.016–0.5). The overall geometric mean and MIC90 (mg/L) were as follows: AB (0.034/0.06), FZ (0.117/0.25), IZ (0.020/0.06), KZ (0.010/0.016), VZ (0.008/0.008), PZ (0.018/0.125), FC (0.043/0.006), and CS (0.096/0.25.
Conclusions: C. dubliniensis is increasingly isolated in our microbiology laboratory and remains susceptible “in vitro” to the most commonly used antifungal agents.
P1280 A study on Candida blood stream infection at university hospitals, Leicester, UK
S.C. Hardman, N. Perera (Leicester, UK)
Objectives: To follow up all new episodes of Candida blood stream infections (CBSI) with a view to inform the antifungal treatment guidelines in the Trust.
Methods: All patients with a new episode of Candida blood stream infection between January 2005 and June 2006 were identified. Isolates were identified by AUXACOLOURTM2, Bio-Rad France and were tested for antifungal susceptibility to six antifungal drugs by YeastOne™ Sensititre, Trek Diagnostic Systems Ltd, UK. All relevant clinical information was collated by clinical and case note review.
Results: A total of 60 patients with 61 episodes of CBSI were identified. Ages ranged from 28 days to 90 years, and 36 patients were males. 59 isolates were identified to species level, of which 28 were C. albicans. The rest were non-albicans: 15 C. glabrata, 9 C. parapsilosis and 2 C. krusei isolates. All C. albicans and 19 non-albicans isolates were susceptible to fluconazole (MIC ≤ 8). The latter included 8 C. glabrata and 8 C. parapsilosis. Resistance to amphotericin (MIC ≥ 2) was seen in non-albicans isolates only and included one each of C. krusei and C. inconspicua. 90% of patients with CBSI were adults, of whom 27 were from the intensive care units (ICU), 13 from haematology and oncology and 5 from the renal ward. 10% were paediatric. These included 3 from paediatric haematology and oncology, and one each from the paediatric ICU, the neonatal unit and general paediatrics. 10/27 isolates from the adult ICU were non-albicans, of which two isolates were resistant to fluconazole, both from haematology patients. 11/13 isolates from adult haematology and oncology were non-albicans, with 45% resistant to fluconazole. There were several host factors associated with CBSI. These included colonisation with Candida, central venous catheter in situ, neutropenia and antifungal prophylaxis. From the data available at the time of writing 25 patients were alive at 30 days follow up. Data on associated factors including the timeliness of line removal, antifungal prophylaxis, treatment including duration and mortality will be presented.
Conclusion: The drug of choice for empirical antifungal therapy for non haematology oncology patients should be fluconazole pending sensitivities. In view of the higher number of non-albicans isolates with a significant percentage of resistance to fluconazole in adult haematology and oncology the current practice of fluconazole for empirical therapy needs to be revisited.
P1281 Analysis of clinical characteristics in 53 patients with Crypotococcus neoformans meningitis
X.J. Lu (Chengdu, CN)
Objective: To analyse the clinical characteristics of Crypotococcus neoformans meningitis and provide evidence for clinical diagnosis and therapy.
Method: The clinical manifestations, data of laboratory examination, features of CT and MRI, antifungal therapy and prognosis of Crypotococcus meningitis were analysed retrospectively in 53 patients.
Results: All patients were confirmed by cerebral spinal fluid smear staining 31 cases were positive at first puncture, 6 cases were positive just at the 6th puncture time, the others were positive at 2nd or 3rd time and culture. The main clinical manifestations were intracranial hypertension and signs of meningeal irritation in 53 patients. Thirty out of 53 (56%) patients had underlying diseases and 30 had the history of contact with pigeons or poultries. The intracranial pressure increased in 51 (96%) cases, 49 (54%) cases had intracranial pressure over 200 mmH2o. For treatment, 30 patients accepted intravenus amphotericin B combining with fluconazole. When patients discharged from hospital, 4 out of 53 cases were cured (the therapy duration were 24 days, 55 days, 76 days and 245 days respectively), 23 were improved, 5 died and 18 giving up any further treatment due to poor economic condition.
Conclusion: The clinical manifestations of Crypotococcus meningitis are mainly progressive intracranial hypertension. When progressive intracranial hypertension and meningeal irritation syndrome occur, cerebral spinal fluid should be examined repeatedly by Indian ink staining and culture so as to prevent misdiagnosis and mistreatment. It is indicated that amphotericin B combining with fluconazole are effective and well tolerated in treatment of Crypotococcus meningitis.
P1282 Risk factors and outcome of critical ill patients with candidaemia
M. Pratikaki, E. Platsouka, C. Routsi, C. Sotiropoulou, A. Priovolos, E. Angelopoulos, O. Paniara, C. Roussos (Athens, GR)
Objective: To determine the incidence and risk factors of Candida bloodstream infection in non-neutropenic critically ill patients and to define whether nosocomial candidaemia is associated with increased mortality in intensive care unit (ICU) patients.
Methods: This is a retrospective cohort study, conducted in the 30-bed multidisciplinary ICU of Evangelismos Hospital in Athens, during a 18-month period (August 2004–January 2006). All patients with ICU stay more than 48 hours were included. Patients with microbiologically documented candidaemia were matched to control patients (1:4). Matching was based on an equivalent APACHE II score (±2 points), diagnostic category, and length of ICU stay (±2 days).
Results: During the study period, 33 of 855 consecutively admitted patients developed one or more episodes of candidaemia, giving an incidence of 3.86 per 100 patients. Candida albicans was identified in 11 patients (33.3%) and Candida non-albicans species in 22 patients (66.7%). Population characteristics, included age, gender, underlying disease, hospitalisation before ICU admission, length of ICU stay and in-hospital mortality rates of patients with candidaemia and their controls were compared. We used APACHE II score on ICU admission to compute the total estimated risk of death for all patients; this was 23% (12.15–50.5), median value. Mortality rates for cases and controls were 60.6% and 22.0% respectively, giving an attributable mortality of 38.6%. The best independent prognostic factor for the development of candidaemia was the presence of ARDS (OR, 3.995; 95% CI 1.332– 11.983, p = 0.013). By multivariate analysis, independent predictors of mortality for cases and their controls were: admission APACHE II score (OR, 1.168; 95% CI 1.105–1.236, p < 0.001), and the presence of candidaemia (OR, 9.374; 95%CI 3.479–25.259, p<0.001). Also multivariate analysis, showed that independent predictors of mortality for candidemic patients, were: SOFA score (OR, 1.568; 95% CI 1.000– 2.457, p = 0.05), and hypoalbuminaemia (OR, 0.049; 95% CI 0.005– 0.515, p = 0.012), both on candidaemia day.
Conclusion: The incidence of bloodstream infections caused by yeasts was 3.86% in our ICU. There was a predominance of Candida species other than Candida albicans, and candidaemia was associated with an excess mortality rate.
P1283 Candidaemia in an intensive care unit
L. Rodriguez-Otero, I. Villamil-Cajoto, S. Cortizo, L. Moldes, M.A. Garcia-Zabarte, A. Aguilera-Guirao, C. Garcia-Riestra, B. Regueiro (Santiago de Compostela, ES)
Objectives: The incidence of candidaemia – a common and potentially fatal nosocomial infection – has risen dramatically, and this increase has been accompanied by a shift in the infecting pathogen away from Candida albicans to treatment-resistant non-albicans species (NAS). In this study, we attempted to identify the risk factors for candidaemia caused by C. albicans and NAS.
Methods: We reviewed the clinical data on 80 inpatients with candidaemia at an Intensive Care Unit of our tertiary University Hospital in Spain, over a 5-year period. We defined mortality as occurring between days 3 to 30 after candidaemia.
Results: Candida albicans, C. parapsilosis, C. glabrata, C. tropicalis and C. krusei caused 46%, 38%, 5%, 3% and 3% of the candidaemia episodes, respectively. The overall mortality was 51% and it was highest in patients suffering from candidaemia of the albicans species (73%) compared NAS (40%). The risk factors to candidaemia caused by C. albicans and NAS respectively included: keeping the catheter in place more than 5 days 78%, 72%; use of total parenteral nutrition 32%, 51%; postoperative state of gastrointestinal tract surgery 35%, 44%; administration of broad-spectrum or combination antibiotics 5 days or more 78%, 77%; under corticosteroid therapy 47%, 36% and antifungal prophylaxis 19%, 28%. The medium length of hospital stay were 61 and 127 days.
Conclusions: Prophylactic azole antifungals, and the use of parenteral nutrition, may play an important role not only in the management of candidaemia but also in the proliferation of hard-to-treat candida species.
P1284 Efficacy and safety of voriconazole in real-life setting in Belgium: results from the Voribel-1 trial
F. Jacobs, D. Selleslag, M. Aoun, A. Sonnet, A. Gadisseur, P. Damas, P. Laterre, O. Van Der Meeren (Brussels, Brugge, Yvoir, Antwerp, Liège, BE)
Objective: Voriconazole (VRC) is standard of care for invasive aspergillosis (IA) and a key treatment option for candidaemia and invasive candidiasis (CC). VRC has been granted reimbursement by the Belgian Health System in 2003 under the condition to confirm its safety, efficacy and cost-efficacy through an observational study in a local real-life setting. The clinical results are presented here.
Methods: A national multicentre prospective non-interventional study was initiated after approval by ethics committees. All patients (p) receiving VRC and presenting with proven/probable IA or CC were allowed to enter a 12-week follow-up. A sample size of at least 120p was needed to test the hypothesis that based on average cost, the real-life setting is not inferior to the pharmacoeconomic model assumed in the reimbursement dossier.
Results: 141p started treatment with VRC at 14 sites between 3/2004 and 9/2005. Most p were adults with proven/probable IA (116); 9 adults had CC, 5p were children (all with CC) and 11p had a primary diagnosis outside the scope of the study. Most p (90%) were immunocompromised and suffered from haematological conditions (leukaemia 43%, stem-cell transplantation 21%) or had undergone solid organ transplantation (9%). In adult p with IA, a complete or partial clinical response was observed in 50% of p after median treatment duration of 50 days; the mortality rates by day 14, 42 and 84 were 14%, 27% and 42% respectively. Most patients (64%) were treated with VRC only and didn't switch, combine or interrupt therapy; 41% of p received the oral formulation only and 29% received both the IV and oral forms sequentially. Serious adverse events regarded as possibly drug-related were reported in 3p (2%); 23p (16%) discontinued VRC for safety issues. No new or unexpected adverse event was reported.
Discussion: In p with proven/probable IA, the response rate of 50% in this real-life setting confirms the results of the landmark randomised trial by Herbrecht et al (53%); this result is achieved despite a shorter median treatment duration (50 vs 77 days). Not surprisingly, the death rate at week 12 in our non-selected p population is slightly higher than in the randomised series (42% vs 29%).
Conclusions: In this large real-life cohort, VRC demonstrates very similar safety and efficacy for patients with IA than in published randomised trial. The shorter treatment duration and the broad use of oral therapy are remarkable from an economic perspective.
P1285 Empirical treatment of Candidaemia in intensive care units: fluconazole or broad-spectrum antifungal agents?
J. Guinea, T. Peláez, M. Torres-Narbona, P. Muñoz, L. Alcalá, E. Bouza (Madrid, ES)
Objectives: The fear of candidaemia by a fluconazole-resistant species of Candida is causing many ICUs to switch empiric therapy from fluconazole to broad-spectrum antifungal agents, such as amphotericin B or caspofungin. Our hypothesis was that the risk of having an episode of candidaemia or invasive candidiasis by an “in vitro” fluconazole-resistant strain was low and did not justify the empiric selection of broad-spectrum antifungal drugs.
Methods: We determined the species of Candida involved in episodes of invasive candidiasis and/or candidaemia in patients admitted to the three adult ICUs of our institution and evaluated the fluconazole antifungal susceptibility of each isolate. We estimated the proportion of episodes produced by fluconazole-resistant species of Candida from 1988 to 2006.
Results: 271 patients admitted to ICUs had proven invasive candidiasis or candidaemia and 520 strains were isolated from normally sterile sites from them. Candida albicans (60.9%), C. tropicalis (5.2%), C. parapsilosis (15%), and a miscellany of other species (3.7%) represented 84.8% of the isolates and were uniformly susceptible to fluconazole. Of the remaining strains, C. glabrata (13.3%) and C. krusei (1.9%) were responsible for invasive candidiasis in 38 patients, but only in nine were the isolates fluconazole-resistant in vitro (MIC ≥ 64 μg/mL). Overall, only 3.3% of the episodes of candidaemia or invasive candidiasis in our institution had an isolate resistant to fluconazole in vitro (the 6 C krusei isolates and 3 of 32 C glabrata isolates).
Conclusion: The generalised use of broad-spectrum antifungal agents for the empirical treatment of invasive candidiasis in ICUs may not be universally justified.
Jesús Guinea Pharm D, PhD, is contracted by the Fondo de Investigación Sanitaria (FIS), contract number CM05/00171.
P1286 Association between oral candidiasis and immunity level in a population of drug addicts in a centre for prevention and treatment of drug addictions in Rome, Italy
L. Rocchi, D. D'Alessandro, M. Fabiani, J.F. Osborn, C. Leonardi, G. Tarsitani, R. Grillot, N. Vescia (Rome, IT; Grenoble, FR)
Objectives: To evaluate the prevalence of Candida infections in the oral cavity of a population of drug addicts, in order: (1) to test the susceptibility of Candida isolates to antifungal compounds; (2) to evaluate the relationship between history of drug abuse, Candida infection and CD4 levels.
Methods: Fifty-seven drug addicts were enrolled in the prospective study: oral samples were collected from each one and cultured for yeast detection on selective medium. The Candida isolates were identified by API 20C AUX® (bioMérieux, France) and C dubliniensis by BICROMO-DUBLI® (Fumouze Diagnostic, France). Using E-test (AB Biodisk, Sweeden), the strains' susceptibility to fluconazole was determined. Regression analysis and significance test were used to compare means and distributions.
Results: Heroin (55 subjects, 96.5%) and/or cocaine (31 subjects, 54.4%) were the drugs most used, and 51 subjects (89.5%) were addict to more than one drug. The duration of the drug addiction varied from 1 to 26 years (mean 13.4). Nine subjects (15.8%) were HIV-infected. The prevalence of oro-pharyngeal candidiasis was 31.6% (18 subjects): 11 cases >250 CFU and 7 cases 50–250 CFU. C. albicans was the most common species (n = 16, 88.8%) and infection due to C. dubliniensis was identified in 4 subjects (22.2%). 87% of C. albicans and 86% of C. dubliniensis isolates were susceptible to fluconazole (MIC ≤ 8 microg/mL). The CD4 count was less than 500/mL in 8 subjects (14.03%) and for 3 of them, HIV positive, it was less than 200/mL. The difference between the mean values of PMNs was statistically significant (mean difference 1,479.3; P = 0.04) in subjects with >150 CFU of Candida in oral cavity. There was no evidence of an association between CD4 count and years of use of heroin. However the percentage level of CD4 reduced on average by 0.033 for each additional year of heroin use (P = 0.03). The difference between the mean years of use of heroin in subjects infected or not by C. dubliniensis (difference: 6.35 years) was statistically significant (P < 0.05).
Conclusion: This work shows an association between oral candidiasis (>150 CFU) and reduction in the level of PMNs. The percentage of CD4 is lower in patients who abuse heroin together with other drugs, and the reduction is proportional to the number of years of drug use. The finding of C. dubliniensis in the oral cavity is associated with the duration of heroin abuse.
P1287 Transmission of Pneumocystis jirovecii from the grandparent immunocompetent carriers to his susceptible granddaughter
L. Rivero, M. Montes-Cano, C. de la Horra, N. Respaldiza, V. Friaza, F. Munoz-Lobato, R. Morilla, F. Medrano, J. Varela, E. Calderon (Seville, ES)
Objectives: Pneumocystis jirovecii causes pneumonia in immunocompromised host but was first identified as an interstitial pneumonia involving premature and malnourished children. Asymptomatic forms of Pneumocystis infection have been described in immunocompetent persons like patients with chronic respiratory diseases who could play a role for the transmission to susceptible persons. Clusters of Pneumocystis pneumonia (PCP) cases among immunosuppressed patients suggesting person to person transmission have been described. This study was undertaken to determine if PCP could have been caused by familiar transmission.
Methods: A six months old female was hospitalised for acute bilateral pneumonia associated with chronic dry cough. Diagnostic test revealed severe malnutrition and PCP. Oral wash samples were collected from: mother, father, brother, grandmother and grandfather who referred a history of rheumatoid arthritis and chronic bronchitis respectively. None of them had symptoms at the moment of the study. DNA was extracted using a commercial kit. The mt LSU rRNA gene was identified by nested-PCR; polymorphism at nucleotide position 85 and 248 were stablishing by direct sequencing.
Fig. 1.
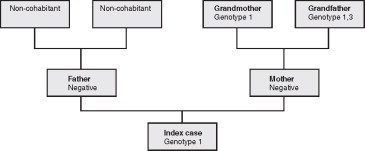
Probable transmission of P. jirovecii infection from grandparents to infant.
Results: The mt LSU rRNA gene of P. jirovecii was identified in three cases: the infant, the grandmother and the grandfather. The child and grandparents showed the same cluster of P. jirovecii, genotype 1 (85C/248C) was identified in the three cases, and genotype 3 (85T/248C) was also detected in the grandfather.
Conclusion: Our findings support a probable transmission of Pneumocystis infection from the grandparent asymptomatic carrier to his susceptible granddaughter.
P1288 Trichosporon asahii in hospitalised patients
M. Kanellopoulou, A. Velegraki, D. Adamou, M. Martsoukou, A. Milioni, N. Skarmoutsou, E. Papafrangas (Athens, GR)
Trichosporon asahii is implicated in a variety of clinical manifestations ranging from simple superficial cutaneus to severe invasive disease.
Objectives: To assess the incidence of T. asahii in clinical specimens from patients with different underlying conditions and to retrospectively compare the molecular subtypes of isolates in a period of three years (2002–2005) in a tertiary Greek hospital.
Patients and Methods: Isolates originated from 12 males and 5 females, (mean age 69 years) with cerebral completed stroke (n = 5), diabetes mellitus (n = 4), ureterostomy-nephrostomy (n = 3), renal dysfunction (n = 2), COPD (n = 2), rheumatoid arthritis (n = 1). Indwelling catheter was present in all patients. T. asahii identification was performed by API ID32C, bioMérieux, France). Complementary biochemical and physiological tests confirmed characterisation of the strains. PCR fingerprinting by a minisatellite specific primer M13 [5′-GAGGGTGGCGGTTC-3′] was used for strain delineation.
Results: During the study period, 17 T. asahii strains were isolated from an equal number of patients. Fifteen strains were recovered from urine specimens, and two from respiratory secretions. The two patients with T. asahii isolated from respiratory secretions were intubated. Eleven strains were isolated from the same clinic at different time intervals. Minisatellite fingerprinting grouped the in time and space related isolates in a single cluster.
Conclusion: (1) Although T. asahii is classed as an emerging opportunistic agent, in this study was exclusively associated to colonisation of the urinary tract. (2) Risk factors for colonisation included indwelling/endotracheal catheter, diabetes mellitus, and old age. (3) The low T. asahii prevalence among these high-risk groups of patients over a period of three years is in accordance with the prevalence recorded internationally. (4) Pending examination of a larger sample size, M13 minisatellite fingerprinting can be recommended for rapid prospective identification of hospital outbreaks.
P1289 Candida kefyr: a new emerging pathogen?
S. Agudo, J. Navarro, C. De Las Cuevas, M. Moreno, B. Buendia, D. Domingo, T. Alarcon, M. Lopez Brea (Madrid, ES)
Introduction: Candida species are present as normal microflora in humans, alterations in host defences can lead to the development of the disease. Candida infections include a wide range of infections, they are common in immunodepressed patients. Infections with non-albicans Candida species are becoming more frequent. Candida kefyr is an uncommon fungal pathogen. It is found in dairy products and in humans as normal flora of the respiratory tract, but it is rare in the skin. This organism has been isolated from multiple milk products, for example bovine milk contains at least 16 different species of yeasts including C. kefyr. In this study we report 6 cases of C. kefyr isolation.
Methods: All the specimens received at the Department of Microbiology (Hospital Universitario de la Princesa, Madrid) from January 1st 2004 to November 8th 2006 were cultured according to standard microbiological procedures. Yeast identification was performed using ChromAgar®, morphology in Corn Meal Agar and Auxacolor®. Clinical files of C. kefyr positive patients were checked.
Results: 6 cases of C. kefyr isolation were found, 5 of which were obtained in the last year. It was only a 0.1% incidence rate of all Candida species isolated during these three years. Case 1 was a 82 year-old woman who had multiorganic failure in the context of respiratory failure after pulmonary thromboembolism. Case 2 was a 91 year-old woman who suffered a stroke with important neurological consequences. Case 3 was a 83 year-old who had several repetitions of urinary tract infections. In the first and the second cases, C. kefyr was isolated from urine cultures in May 2006. In the third case, it was found in urine culture in January 2005. Case 4 was a 73 year-old man with multiorganic failure caused by hepatic encephalopathy. This case of C. kefyr was detected in bloodstream in September 2006. Case 5 was a 77 year-old man with acute renal failure and respiratory failure; C. kefyr was found in bronchoaspirate in April 2006. Case 6 was a 87 year-old woman with gastric cancer; C. kefyr was isolated from peritoneal liquid in October 2006.
Conclusion: C. kefyr is an uncommon human pathogen. The three patients reported here were elderly and had severe physical deterioration; however the clinical significance of these isolates remains unclear. C. kefyr infections reflect the increase in Candida species as responsible of disseminated infections in immunodepressed patients.
P1290 Fungal keratitis in healthcare area of Santiago de Compostela (Galicia, NW Spain)
F. Pardo, P. Romero, M. Pérez del Molino, L. Martínez, S. Cortizo, B. De Domingo, T. Rodríguez-Ares (Santiago de Compostela, ES)
Objective: The aim of this study is to analyse risk factors, incidence, visual signs and symptoms, treatment in fungal keratitis diagnosed in the Health Care Area of Santiago de Compostela between November 1998 and May 2006.
Methods: Retrospective study of the 22 fungal keratitis diagnosed in the Health Care Area of Santiago de Compostela between November 1998 and May 2006. Medical records were reviewed for all patients. Demographic data, clinical signs and symptoms, microbiologic isolations and treatment were analysed in SPSS programme.
Results: 22 patients were included in this study, 10 males (45.5%) and 12 females (54.5%). The mean age were 60.32 years (28–87). Corneal infiltrate: 31.8% (7/22) were 1–2 mm and 68.2% (15/22) >2 mm. Peripheric lesions were observed in 22.7% and 77.3% in central corneal zone. Two patients showed satellite lesions. All patients (100%) had Thyndall in anterior chamber and 81.8% (18/22) had hypopyon and keratic precipitate at the same time. Principal local risk factors were: topical corticoid treatment (27.3%), previous ocular surgery (22.8%), ocular trauma (18.2%), ocular herpes infection (18.2%), contact-lents user (9.1%), previous ocular disease (9.1%).
Fungal isolation: 57.1% (13/22) were Candida species, and the most frequent isolates were Candida albicans (53%, 7/13), followed by C. parapsilosis (38.4%, 5/13) and C. glabrata (7.6%, 1/13). Filamentous fungi were isolated in 42.9% (9/22), Fusarium spp. (55.6%, 5/9), Aspergillus spp. (22.2%, 2/9), Dactilaria spp. (11.1%, 1/9), Alternaria spp. (11.1%, 1/9). Topical and systemic antifungal treatment was performed in all cases. Amphotericin B was used in 77% of cases, and in 3 cases inside the anterior chamber. Voriconazol was used in two cases with no improvement with conventional treatment and showed clinic amelioration.
Conclusion: (1) Annual incidence in this study was 2.7 cases/year, higher than European average. We observed the major number of cases in autumn, probably due to high relative humidity and temperate temperatures. (2) The most frequent isolate was Candida albicans. (3) The more frequent local risk factors were: local corticoid therapy, previous ocular surgery and ocular trauma. (4) Pharmacological and surgery treatment together were necessary in 25% of the patients to control the infection. (5) 76% had no posterior relapse due to an appropiate control of the infection.
P1291 Aspergillus keratomycosis at an eye hospital in south India: a retrospective study
L. Kredics, P. Manikandan, S. Kocsubé, I. Dóczi, V. Narendran, J. Varga, Z. Antal, C. Vágvölgyi, R. Revathi, E. Nagy (Szeged, HU; Coimbatore, IN)
Objectives: The objectives of this study were to analyse the epidemiological features of Aspergillus keratitis in South India, to perform morphology-based and molecular identification of Aspergillus strains isolated from keratomycosis patients, to determine the antifungal susceptibilities of the isolates and to study their genetic diversity.
Methods: 26 Aspergillus strains isolated from keratomycosis in the Aravind Eye Hospital between August 2005 and February 2006 were involved in the study. Epidemiological data were recorded for the patients. Morphological examination was performed by microscopy, culture characteristics were studied on malt extract agar. Molecular identification was carried out by sequence analysis of the ITS region. Antifungal susceptibilities were determined by the E-test method.
Results: The 26 patients included 17 males and 9 females. Rural, semiurban and urban populations were represented among the patients with 23, 50 and 27%, respectively. Farmer was the most frequent occupation among the male patients, while most of the female patients were housewives. Corneal trauma was reported as the potential predisposing condition of the infection for 57.7% of the patients, the traumatising agents were dust or iron particles, insects or plant seeds. Among the further possible predisposing conditions, systemic diseases like diabetes mellitus and hypertension proved to be frequent. The antifungal drugs applied for the therapy of patients included natamycin, itraconazole, ketoconazole, econazole, clotrimazole and amphotericin B. Therapeutic keratoplasty was performed in the case of 9 patients. Most of the isolates proved to belong to the species A. flavus, however, other species of the genus including A. terreus, A. fumigatus and A. tamarii were also represented in the sample. The initial identification of the isolates based on conidial- and colony morphology could be confirmed by ITS sequence analysis. All of the examined strains proved to be resistant to fluconazole with MIC values above 256 mg/L. MIC-ranges of the other examined antifungal drugs were in the following ranges (mg/L): 0.064–4 for amphotericin B, 0.25–1 for ketoconazole, 0.064–32 for itraconazole and 0.064–1 for voriconazole.
Conclusions: Our data provide important information on the current incidence of Aspergillus species in corneal ulcers in South-India confirmed by molecular diagnosis, as well as useful ideas for the selection of the appropriate antifungal agents.
P1292 Fatal case of antritis caused by mucormycosis in patient with diabetes mellitus
S. Tsoutsos, Z. Alexiou, A. Kallitsis, P. Konstantopoulos, T. Loukas, G. Liappi, G. Panoutsopoulos (Elefsina, GR)
Objectives: Mucormycoses are potentially fatal infections unless they receive immediate diagnosis, followed by surgical and pharmaceutical treatment.
Methods: Description of the case. M, 75 y.o., was admitted with symptoms of diabetic ketoacidosis and fever. He developed within 24 hours a strong headache, confusion, amaurosis of vision on both sides and aglutition.
Results: Physical examination findings: Edema and proptosis of orbits, opthalmoplegia on both sides. ENT examination found an extensive necrosis of the quadrangular septal cartilage, extending to the perpendicular plate of the ethmoid, enlargement of nasal chambers due to necrosis of the inferior and middle nasal conchae on both sides. An extended necrosis of the hard palate and inhibition of the pharyngeal rephlex were found from the orphopharynx. The CT imaging protocol: visceral cranium: Enlargement of nasal chambers to a large extent, both the anterior and posterior ethmoidal air cells, the sphenoid sinuses and the left antrum maxillare were affected, cerebral leukoencephalopathy lesions, microinfarcts of periventricular white matter, cerebellum atrophy. Thorax findings: ascending thoracic aorta aneurysm. Laboratory examination: WBC 18,900, PMN 89%, MO 7.5%, HCT 39.9%, ESR 124 mm, CRP 108 mg/dl, lgG 1,130 mg/dl, lgA 903 mg/dl, lgM 219 mg/dl, C3 167, C4 32.6. CSF examination: Protein <20 mg/dl, Glu 118 mg/dl, LDH 75, cells 35. The nasal mucosa tissue biopsy revealed a hyphomyces of the genus Mucor. Histological evaluation show necrotic changes of the superficial layers of the nasal mucosa and necrosis of many vessels caused by mucor. Alcian blue-PAS stain accentuated the wide-angle branching hyphae characteristic of mucor. Diabetic ketoacidosis and respiratory infection were treated with ceftriaxon and amphotericin B. Surgical treatment of the disease was contraindicated. The patient died 23 days after admission.
Conclusion: Prognosis of mucormycosis is not favourable despite early diagnosis and aggressive combined surgical and medical therapy.
P1293 Comparison of plasma trough concentrations of voriconazole in patients with or without comedication of ranitidine or pantoprazole
W.J. Heinz, C. Kloeser, A. Helle, C. Guhl, S. Scheuermann, H. Einsele, H. Klinker (Würzburg, DE)
Objectives: Voriconazole is a broadly used antifungal agent which has demonstrated superior efficacy compared with amphotericin B for long term therapy of invasive aspergillosis. It is often used especially in haematological patients. The azole is metabolised by the cytochrome P450 (CYP) isoenzymes 2C9, 2C19 and 3A4. In addition it shows inhibiting and inducing activity. Due to its good bioavailability voriconazole is often administered orally. On the other side severely ill patients often receive proton pump-inhibitors (PPI) or H2-receptor antagonists (H2RA). These agents may reduce absorption of azoles by increasing gastric pH, or inhibit hepatic metabolisation by CYP. In previous trials with healthy volunteers no effect with ranitidine could be detected. For omeprazole an increase of the AUC could be shown, but PPIs differ in their affinity to CYP isoenzymes. We therefore analysed plasma through concentrations (PC) of voriconazole from patients with or without pantoprazole or ranitidine.
Methods: Plasma samples from haematological patients with oral voriconazole treatment have been collected and voriconazole concentrations determined. The results were analysed in correlation to the comedication of H2RA and PPI.
Results: We analysed 134 samples from 44 patients, 20 samples without comedication (group 1), 10 samples from patients receiving ranitidine (group 2), and 104 from patients also receiving pantoprazole (group 3). Plasma trough levels showed a broad range from undetectable to a maximum of 12,708 ng/mL. 37 PC have been below 200 ng/mL, 9 (45%) of group 1, 2 (10%) of group 2 and 26 (25%) of group 3 and only 4 above 10,000 (all group 3). Average plasma concentrations have been 1,212 ng/mL (1), 923 (2) and 1840 (3).
Conclusion: In this retrospective analysis PC of voriconazole have shown to be lower in patients receiving ranitidine and higher in patients with comedication of pantoprazole. The average PC for voriconazole is twice as high in patients taking pantoprazole than in those with ranitidine. The increase of voriconazole PC stands in parallel to the results previously shown for omeprazole. Although few values are rather high (above 10,000 ng/mL) no dosing recommendation can be made up to now.
P1294 Determination of caspofungin plasma concentrations in surgical intensive care unit patients
H. Nguyen, T. Hoppe-Tichy, H. Geiss, M. Weigand (Heidelberg, DE)
Objectives: The aim of the present study was to investigate plasma caspofungin concentrations in surgical intensive care unit [SICU] patients.
Methods: According to their demographic data (Table ) caspofungin concentrations of ten adult subjects were compared in a pairwise manner. All patients received an initial caspofungin loading dose of 70 mg followed by 50 mg/day. Caspofungin trough concentrations [C24h] were determined daily for the duration of therapy using High-Performance-Liquid-Chromatography.
| Pair | Age | BW(kg)/BS(m2) | Mean C24 h (μg/mL) [range] | p-value |
|---|---|---|---|---|
| 1 (F) | 47 vs. 46 | 47/1.45 vs. 48/1.46 | 1.6 [0.91–2.45] vs. 3.48 [1.54–5.01] | 0.028 |
| 2 (F) | 41 vs. 40 | 72/1.89 vs. 75/1.82 | 1.04 [0.7–1.74] vs. 2.22 [2.08–2.36] | 0.068 |
| 3 (M) | 72 vs. 65 | 75/1.93 vs. 75/1.92 | 1.24 [0.85–1.62] vs. 2.43 [1.97–2.77] | 0.028 |
| 4 (M) | 60 vs. 69 | 80/1.98 vs. 80/1.65 | 0.93 [0.55–1.47] vs. 2.0 [1.68–5.1] | 0.043 |
| 5 (M) | 54 vs. 56 | 77/19.5 vs. 70/1.76 | 1.34 [0.74–1.97] vs. 3.0 [1.63–3.63] | 0.005 |
aF, female; M, male.
bBW, body weight; BS, body surface.
Results: Plasma caspofungin C24h differed significantly between paired subjects (Table). Importantly, on more than one day, caspofungin C24h for five patients were found to have fallen below the target concentration of 1 μg/mL (see data ranges on Table).
Conclusion: Our data indicate the importance of C24h monitoring in SICU patients. Further investigations are needed to evaluate the parameters which influence plasma caspofungin concentrations in this patient group.
Typing MRSA
P1295 Clonal characterisation of methicillin-resistant Staphylococcus aureus isolates at an Innsbruck university hospital using five different molecular typing methods
K. Grif (Innsbruck, AT)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most prevalent nosocomial pathogens throughout the world, capable of causing a wide range of hospital infections. In recent years, several genotyping methods have been developed for the characterisation of MRSA strains. Of these, one of most suitable for epidemiological investigation is PFGE. However, a combination of different typing methods is often needed to obtain reliable analysis of strains that are suspected to be epidemiologically related.
The aim of our study was to characterise the MRSA strains at the University Hospital of Innsbruck and the nearby rehabilitation facilities and to compare these with the simultaneously circulating MRSA clones using 5 different molecular typing methods.
Material and Methods: A group of 109 MRSA isolates, collected from patients at the University Hospital of Innsbruck during 2003–2005 was tested for their antimicrobial susceptibility profiles. Clonality was determined using a combination of PFGE, automated ribotyping, spa-typing and SCC-mec and agr determination.
Results: Thirteen ribotypes and 18 PFGE types were found. Two main ribotypes, as well as two PFGE types dominated among MRSA isolates collected. The majority of isolates gained spa-type t001 (67.9%) and arg type 2 (85%).
Conclusions: Clonal distribution was found of MRSA isolates collected at the University Hospital of Innsbruck.
P1296 Molecular characterisation of methicillin-resistant Staphylococcus aureus in South Africa
W. F. Oosthuysen, A.G. Dusé, E. Marais and the South African MRSA Surveillance Group
Objective: The clonal prevalence, relatedness and distribution of clinical MRSA isolates have not yet been determined in the Republic of South Africa (RSA). This study is the first ever undertaken to elucidate the molecular epidemiology of MRSA in South Africa.
Methods: Clinical MRSA isolates were collected from private and state laboratories from each of the 9 provinces throughout South Africa. Standard microbiological techniques were used to confirm both the identity and their resistance to methicillin. The SCCmec type of each isolate was investigated using multiplex PCR. All isolates were screened by PCR to detect the genes for the toxin Panton-Valentine leukocidin (PVL). spa typing was performed according to previously described techniques.
Results: Of the 342 isolates confirmed to be S. aureus, 314 were methicillin-resistant. The SCCmec types of 302 MRSA isolates were determined. 36.7% were type IV (Paediatric clone/CA-MRSA); 25.2% were type II (NY/Japan clone) or a variant thereof; 23.2% were type III (Hungarian clone) or a variant thereof, including type IIIA (Brazilian clone); 13.2% were SCCmec type I (Archaic clone), including type IA (Iberian clone); and 1.7% were non-typeable according to this method.
PVL genes were detected in 2/314 (0.6%) of the MRSA strains.
Of 218 MRSA isolates that were subjected to spa typing, 27 different spa types were identified that could be grouped in 5 spa clonal complexes (spa-CC) (table 1 ). 53% were clustered in spa-CC 012; 35% were clustered in spa-CC 064; 8% were clustered in spa-CC 045; 2% were clustered in spa-CC 032 and 1% were clustered in spa-CC 015. Only 1 singleton was found. Nine spa types identified were novel.
Table 1.
spa types identified in South Africa
| spa type | Number of isolates (n = 128) | Percentage of total |
|---|---|---|
| spa-CC 012; Brazilian/Hungarian clonality | ||
| 1012 | 54 | 23 |
| 1037 | 50 | 23 |
| 1018 | 4 | 2 |
| 1021 | 3 | 1 |
| 1046 | 2 | 1 |
| 1399 | 2 | 1 |
| 1840 | 1 | 0.45 |
| t1504 | 1 | 0.45 |
| spa-CC 064; Archaic/Iberian clonality | ||
| 1064 | 44 | 20 |
| t1257 | 20 | 9 |
| t951 | 3 | 1 |
| t1443 | 3 | 1 |
| t104 | 1 | 0.45 |
| t1256 | 1 | 0.45 |
| t1555 | 1 | 0.45 |
| t1774 | 1 | 0.45 |
| t1779 | 1 | 0.45 |
| spa-CC 045; NY/Japan clonality | ||
| t04514 | 6 | |
| t0013 | 1 | |
| t1154 | 1 | 0.45 |
| spa-CC 032 | ||
| t032 | 2 | 1 |
| t891 | 1 | 0.45 |
| t1468 | 1 | 045 |
| spa-CC 015 | ||
| t015 | 1 | 0.45 |
| t050 | 1 | 0.45 |
| t456 | 1 | 0.45 |
| Singleton | ||
| t174 | 1 | 0.45 |
Conclusion: These results indicate that the population structure of MRSA is composed of a variety of epidemic clones. To our knowledge is this the first report to document epidemic clones circulating in South Africa. Furthermore, to the best of our knowledge, this is the first report of MRSA isolates shown to carry genes for the toxin PVL in South Africa.
P1297 Double locus sequence typing, a new fast and unambiguous typing method to study the epidemiology of MRSA at a regional level
D.S. Blanc, G. Kuhn, A. Wenger, J. Bille, P. Francioli, G. Zanetti (Lausanne, CH)
We developed a fast and highly discriminatory typing method for MRSA, based on partial sequences of 2 polymorphous genes (clfB and spa) (JCM 2006, in press).
Objectives: To use this method to investigate the epidemiology of MRSA in Western Switzerland, and to compare the results with those obtained with PFGE.
Material and Methods: From Jan. 05 to June 06, one isolate per antibiogram profile and per patient was prospectively stored in 96-well plates (N = 1,126). DNA extraction, PCR and sequencing was also performed in 96-well plates. Each allele at each sequence was attributed a number. The composition of the two alleles defined a DLSType labelled with 2 numbers. Considering single locus variant (SLV), e-BURST was used to analyse the relationship between DLSTypes. For comparison with previous MRSA surveillance performed with PFGE, a selection of 153 isolates representative of the major PFGE strains recovered previously was analysed. In addition, the first 150 isolates of 2005 were typed by PFGE and DLST. MLST was performed on isolates of predominant DLSTypes.
Results: DLST results showed the presence of the four predominant genotypes already identified with PFGE: DLSTypes 1–1 (20 pts, ST45), 2–2 (560 pts, ST105), 3–3 (130 pts, ST247), and 4–4 (83 pts, ST228). A good congruence was found between PFGE and DLST: 95%, 98%, 85% and 100% of isolates belonging to PFGE clones A, B, C and D, respectively, were clustered within the respective predominant DLSTypes and their SLVs. However, different PFGE types could be observed within the same DLSType, and the reverse was also true. This suggests that the evolution of both markers was different. The discriminatory index, calculated from 150 unselected isolates of 2005 typed by both methods, were 0.82 (41 types) for PFGE and 0.76 (37 types) for DLST.
Snapshot analysis with e-BURST revealed that DLSTypes 2–2, 3–3, and 4–4 were linked by SLVs 3–2, 2–36 and 4–36, suggesting either a common ancestor, recombination of alleles, or homoplasy. Analysis of allele sequences and frequency of types in the population, together with analysis of ST of predominant DLSTypes, lead us to hypothesise a recombination in one case (DLSType 3-2), and a clonal evolution from DLSType 2-2 to DLSType 4-4 in the second case.
Conclusion: DLST gave results comparable with those of PFGE, and appeared suitable for the epidemiological surveillance of MRSA at a regional level. Moreover, it allowed to hypothesise the origin of some predominant genotypes.
P1298 Increase of Simpson's diversity index parallels the control of a major regional MRSA outbreak in the Netherlands
P. Gruteke, M.E.O.C. Heck, A.J. de Neeling, W.J.B. Wannet, H.A. Verbrugh (Amsterdam, Bilthoven, Rotterdam, NL)
Objectives: Health authorities need appropriate incidence figures to monitor MRSA control policies. UK national health authorities for instance set targets for local MRSA bacteraemia rates, although rate reductions may be difficult to interpret (Spiegelhalter 2005). Typing of strains is vital in directing MRSA control efforts since it can help discriminate outbreaks from spurious associations. Because the genetic diversity among isolates is lower in epidemic versus endemic MRSA situations scoring the Diversity Index (DI) of consecutive collections of MRSA may provide an additional monitoring tool.
Methods: All 5,300 MRSA isolates were typed by PFGE at the RIVM. MRSA strain PFGE typing (PT) patterns differing 1 band from preceding patterns were ascribed a new PT. The 4 years period from April 1st 2002 was divided into 9 time slots (slots 1 and 9: 3 months, slots 2 through 8: 6 months). During the first part of this period the province of Zuid Holland experienced a large outbreak of healthcare associated MRSA. One isolate per patient per referring laboratory was included. Isolates were labeled according to the province of origin. Simpson's DI's were calculated per time slot for Zuid Holland and compared with data from the other 11 provinces of the Netherlands.
Results: MRSA isolates in the Netherlands have high DI scores, reflecting the small sizes of most outbreaks. In contrast, the DI's of the isolates referred from South Holland in the first time slots were significantly below those of the rest of the Netherlands. As the MRSA epidemic became controlled in South Holland, its DIs increased and became similar to the rest of the Netherlands.
Conclusions: Simpson's DI clearly reflected the existence and the control of a large MRSA outbreak in a province of the Netherlands. In situations where typing results are available and sampling strategies are comparable, trends in DI scores allow monitoring another dimension of MRSA epidemiology and control.
P1299 The genetic organisation of staphylococcal cassette chromosome mec type IVA from invasive methicillin-resistant Staphylococcus aureus isolates in Korea
C. Park, H-S. Chun, D-G. Lee, S.H. Park, S-M. Choi, J-H. Choi, J-H. Yoo, W-S. Shin (Seoul, KR)
Objective: It was reported that type II of SCCmec was the most prevalent in Korea. Our previous study on bacteraemic MRSA showed type IVA of SCCmec was the second most prevalent type following type II and type IVA isolates were more commonly isolated among community acquired MRSA (CA-MRSA) than type II isolates. So we investigated the genetic organisation and variation of SCCmec IVA by sequencing and their other epidemiologic characteristics from invasive CA-MRSA isolates.
Material and Method: From invasive CA-MRSA isolates (n = 84), SCCmec PCR typing was performed by multiplex- and single-PCR method described by Oliveira and Okuma et al. The genetic organisation and variation of SCCmec IVA was analysed by long PCR based sequencing. The prediction of assembled sequencing results was performed by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and the homology of each open reading frames (orfs) was analysed by BLAST search (BLASTN 2.2.13). Each variations of class mecA complex were compared. Other molecular characteristics and antibiotics susceptibility were also analysed.
Results: Type IVA isolates accounted for approximate 50% by multiplex PCR typing and were further classified into three major groups by single PCR typing. One group was composed of ccr2-typical class B mec complex, the other ccr2-class A mec complex, and the third group ccr2-atypical class B mec complex. The sequencing and assembly results of the third group showed that its class B mec was composed of IS1272-tnp-ÄmecR1–mecA IS431 (∼1kb insertion compared to other class B). The total length was ∼27 kb, and pUB110 (ble, and aadD) was incorporated into the right region of SCCmec. There were two kinds of STs (72 and 1) in IVA isolates. And orfs of left region had the high homology of orfs in the left region of IVc. Antibiotics susceptibility test showed that IVA isolates of the third group were not multi-drug resistant (mainly resistant to oxacillin, penicillin, erythromycin, tobramycin) but isolates of first group with typical class B were resistant to more antibiotics (oxacillin, penicillin, erythromycin, tobramycin, tetracycline, gentamicin).
Conclusion: Although its SCCmec type was classified to type IVA in multiplex PCR, type IVA was subsequently classified into several groups. In each groups, there were some variations in the genetic organisation of SCCmec. Future studies should determine the epidemiologic and molecular factors that might contribute to the high occurrence.
P1300 Characterisation of SCCmec type of methicillin-resistant Staphylococcus aureus strains from hospitalised patients admitted in Belgian hospitals
O. Denis, A. Afsar, R. De Mendonça, A. Deplano, C. Nonhoff, M.J. Struelens (Brussels, BE)
Background: The analysis of SCCmec structure is a key typing tool for classification of clones of methicillin-resistant Staphylococcus aureus. Several strategies based on multiplex PCR have been developed for this purpose. We compared the results of two PCR protocols for SCCmec typing with MRSA strains collected during a national survey in Belgian hospitals in 2005.
Methods: 321 MRSA isolates collected from 116 hospitals were identified by multiplex PCR for mecA, nuc and 16S rDNA genes. SCCmec type was determined by methods described by Oliveira DC (PCR A) (AAC 2002;46:2155) and Zhang K (PCR B) (JCM 2005;43:5026). Discrepant results between two methods were resolved by multiplex PCR for ccr and mec complex determination.
Results: The proportion of non typeable SCCmec was 4% by PCR A and 26% by PCR B (p < 0.001). Only, 210 (64%) isolates had concordant results by both methods and were SCCmec type I (n = 7), type II (n = 44) and type IV (n = 159). 26 isolates were incorrectly typed as SCCmec type I by PCR B but were classified as type IV by PCR A as confirmed by ccr and mec complex analysis. Of 75 isolates which were not typeable by PCR B, 73 were determined of type IV by PCR A, and confirmed by ccr and mec complex determination. Two isolates which were not typeable by PCR A were determined as type IV and type V by PCR B, ccr and mec complex analysis. Ten isolates were untypeable by both methods.
Conclusion: The method of Oliveira et al. showed the best performance for SCCmec typing. However, the presence of untypeable SCCmec variants warrants the combination of methods for SCCmec characterisation.
P1301 Comparative molecular analysis of veterinary, dairy, and clinical Staphylococcus aureus isolates by spa typing and amplification of the mecA and the PVL genes
A. Stöger, M. Gonano, A. Pietzka, F. Allerberger, M. Wagner, W. Ruppitsch (Vienna, AT)
Objectives: The aim of this study was to compare Staphylococcus aureus isolates of veterinary, dairy and human origin to evaluate a possible animal-to-food, human-to-animal, and/or human-to-food transmission or vice versa.
Methods: 1,058 isolates were collected by screening cows suspected of having mastitis, from vat milk and fresh cheese, from wounds of ambulant patients and from hospitalised patients. Sequencing of the variable X-region of the protein A gene (spa typing), detection of the methicillin resistance gene (mecA), and detection of the Panton-Valentine leukocidin gene (PVL) were used for molecular comparison of these isolates.
Results: All tested veterinary isolates (n = 60), dairy isolates (n = 64), and 11 isolates from the ambulant patients, and 57 isolates from hospitalised patients were negative for mecA (MSSA). The remaining 860 clinical isolates were positive for mecA (MRSA). The PVL gene as a marker for CA-MRSA was detected in a single veterinary isolate of spa type t042, which is closely related to CA-MRSA t044. Typing yielded 152 spa types. Thirty-four spa types were identified for the 60 veterinary isolates, 35 for the 64 dairy isolates, 28 for the 59 human MSSA isolates, and 134 for the 860 clinical MRSA isolates. 24 new spa types were found among the veterinary and dairy isolates that were not described in the spa database (RIDOM). In contrast to the clinical MRSA isolates, where the majority belonged to spa complexes SC I and SC II the majority of veterinary, dairy and also the MSSA isolates belonged mainly to spa complexes SC VI and SC VII. 11 spa types of the human population including t190 and t008 were also found in the veterinary or in the dairy isolates. Spa type t529 was the only one that was detectable in all populations. Approximately 50% of veterinary and dairy isolates belonged to nine common spa types which were different from that found in the human isolates.
Conclusion: Typical and very frequent human spa types i.e. t190 or t008 rarely occurred in veterinary and dairy isolates. However, the occurrence of typical human isolates on cow udders and in milk and fresh cheese might reflect a possible transmission from human to animals and food. No MRSA was detected among the veterinary and dairy isolates. The high number of identical spa types in veterinary and dairy isolates indicated a transmission of Staphylococci from animal reservoirs into the dairy chain.
P1302 Spa sequence-based typing of MRSA starting directly from multiplex PCR for grouping of SCCmec elements
C. Cuny, W. Witte (Wernigerode, DE)
Objective: The discriminatory power of sequence based typing of MRSA can be enhanced by grouping of SCCmec elements. Here we report about grouping of SCCmec elements by PCR and amplification of the relevant sequence of spa for subsequent sequencing starting immediately from the same multiplex PCR.
Methods: Multiplex PCR with primers for ccrA1, ccrA2, ccrA3, ccrC, and kdp for grouping of SCCmec elements, and for amplification of the X region of spa.
The multiplex PCR was designed in such a way that the size of amplimers indicating the SCCmec group would be either below or above the size of the spa-amplimers (200 bp for the smallest with only 2 repeat units and 512 for the largest with 15 repeat units).
Results and Conclusion: We found a complete congruence of results of spa-sequence typing and of grouping of SCCmec elements by separate PCR experiments and by multiplex PCR for 75 isolates representing major lineages of epidemic nosocomial MRSA and of community MRSA. These isolates are epidemiologically unrelated and originate from various kinds of infections in different geographical areas of Germany. The deduction of SCCmec groups is especially useful in cases where isolates exhibiting the same MLST-types and even spa-types (the same core genome) contain different SCCmec-elements (e.g. STS with SCCmec I, II, III, V described so far).
P1303 clfB typing in Staphylococcus aureus: online database for display, clustering analysis and storage of clfB DNA sequences
A.R. Gomes, J. Carriço, S. Vinga, A.L. Oliveira, M. Zavolan, H. de Lencastre (Oeiras, Lisbon, PT; Basle, CH; New York, US)
Objectives: Typing approaches based on DNA sequencing have provided robust and unambiguous data for Staphylococcus aureus: multilocus sequence typing (MLST) is useful for global epidemiology and evolutionary studies and spa typing has been shown to be efficient not only for evolutionary studies but also for outbreak investigations. Recently, typing of S. aureus based on sequencing of the repeat region of the clumping factor B gene (clfB) was shown to be more discriminatory than spa typing and capable of distinguishing among seemingly related strains. However, the nomenclature for this new typing methodology yielding long sequences of repeated units has not been defined yet. Here, we propose a free-access online interface for the identification of repeated units in DNA sequences, display of the degree of variation in the nucleotide sequence of the repeats, and for clustering analysis and storage of clfB DNA sequences.
Methods: An encoding was devised for the representation of sequences in which each of the four nucleotides was represented by a three-dimensional floating-point vector and a sequence composed of n-nucleotide repeated units was represented by a set of 3n-dimensional vectors. In order to visualise the sequences, the 3n-dimensional vectors were projected onto a 3-dimensional space by single value decomposition and the three coordinates were used to colour-code each occurrence in terms of hue, brightness and contrast as well as with a numerical profile. Sequencing of the clfB repeated region was performed in ninety-six S. aureus isolates representing some of the major clones recovered from infection or carriage in several countries over a period of more than forty years. DNA sequences were subsequently grouped using hierarchical clustering algorithms based on the numerical values obtained, allowing for various differences from the consensus for the repeating units. All this information is stored in a free-access database for comparison with future submissions.
Results: A clfB typing database containing clfB sequences from the major S. aureus clones is available online. clfB DNA sequences may be submitted for both a numerical and a graphical colour representation, clustering analysis and storage in the database.
Conclusion: The online clfB typing database provides a free-access interface for representation, analysis and storage of clfB DNA sequences, and constitutes a highly discriminatory typing tool for the characterisation of S. aureus isolates.
P1304 Array-based typing of community- and healthcare associated MRSA and occurrence of epidemic strains in eastern Saxony
S. Monecke, L. Jatzwauk, S. Weber, R. Ehricht (Dresden, Hoyerswerda, Jena, DE)
Objectives: Objective of the study was to test a diagnostic microarray under routine conditions and to obtain insights into the epidemiology of Community- and Healthcare Associated MRSA (CA- and HA-MRSA) in eastern Saxony.
Methods: Multiplex linear DNA-amplification and subsequent microarray hybridisation were performed which allowed to detect 144 genes or distinct allelic variants simultaneously. All relevant exotoxins, antimicrobial resistance determinants, the allelic variants of agr, as well as staphylococcal exotoxin-like genes were included. Hybridisation profiles were used for assignment to epidemic strains. The study was performed in eastern Saxony including an university hospital, a long-term rehabilitation facility and three county hospitals. For the study of HA-MRSA, random samples from routine diagnostics as well as isolates from outbreak investigations were tested. Additionally, all cases were included in which PVL-positive CA-MRSA were suspected to be involved.
Results: Most prevalent HA-MRSA strains are South German EMRSA, New York/Japan Clone (“Rhine-Hessen EMRSA”), Berlin EMRSA, and EMRSA-15 (“Barnim Epidemic Strain”). Other strains are by comparison rather rare. Isolated cases of infections with PVL-negative ST1 and ST8 MRSA were detected. ST8 Hannover ERMSA which used to be very common in the 1990s virtually disappeared, and EMRSA 16 was completely absent.
CA-MRSA were not found prior to 2004. Still now, cases are rather sporadic. The most prevalent strain is the PVL-positive ST80-MRSA IV (four detected cases in 2004, one outbreak including four cases in 2005, two cases in 2006). First infections with PVL-positive ST8-MRSA IV (USA300, one case in 2005 and one in 2006) and ST30-MRSA IV (WSPP strain, one case in 2006) were also diagnosed. PVL-positive ST1-MRSA IV (USA400) were not detected yet.
Conclusions: The DNA-microarray proved to be a powerful tool for epidemiological monitoring and identification of epidemic strains. In contrast to other typing methods it facilitated the assessment of virulence and antibiotic susceptibility of a given isolate within a single experiment. In the region studied, the majority of HA-MRSA infections was caused by a few epidemic strains. Emerging strains (New York/Japan Clone as well as SCCmecIV strains EMRSA-15 and Berlin EMRSA) play a major role. PVL-positive CA-MRSA are still very rare, and the most important strain is ST80-MRSA IV.
P1305 Double locus sequence typing a new high through put typing method for MRSA. Results from a 1.5-year surveillance in a university hospital
I. Nahimana-Tessemo, G. Kuhn, J. Bille, A. Wenger, P. Francioli, G. Zanetti, D.S. Blanc (Lausanne, CH)
We developed a high through put and highly discriminatory typing method for MRSA, based on partial sequences (500bp) of 2 highly polymorph genes (clfB and spa) (JCM 2006, in press). This method was used to investigate the molecular epidemiology of MRSA in a University tertiary care hospital of 800 beds during a 1.5-year period of time (Jan 2005 to June 2006).
Isolates, one per antibiogram profile per patient and per year, were prospectively stored in 96-well plates. Once a plate was filled up, DNA extraction, PCR and sequencing of the two genes were performed. Each allele at each sequence was attributed a number. Thus, the composition of the two alleles defined a DLSType.
From January 2005 to June 2006. 288 isolates from 264 patients were typed by the new method. Results showed that 67 different DLSTypes were observed. One predominant DLSType (2–2, 125 pts) and 2 less predominant DLSTypes (4–4 and 3–3, in 33 and 19 pts, respectively) accounted for 67% of the patients. These 3 major DLSTypes corresponded to clones B, D and C previously identified by PFGE surveillance. Among the 23 patients for whom 2 isolates were typed, one harboured single locus variants (SLV): the first isolate was 2–65 and the second was 66–2; and another patient harboured two different DLSTypes (134–59 and 3–3). Transmission was suspected from an index case to roommates at 20/374 occasions. Among these, 11 harboured the index case DLSType. Two clusters of cases were identified (hospitalised in the same unit during overlapping period of time) accounting for 10 possible transmissions, from which only 7 could be confirmed by DLST These results are concordant with previous results in our institution using PFGE as molecular markers. The value of DLST was found to be similar to PFGE for epidemiological investigation at a local level, with the major advantage that the method give unambiguous data.
P1306 Direct repeat unit typing to differentiate methicillin-resistant Staphylococcus aureus isolates in Ireland indistinguishable by pulsed field gel electrophoresis
A. Rossney, R. Goering (Dublin, IE; Omaha, US)
Objectives: In Ireland, the prevalence of a single MRSA strain exhibiting antibiogram-resistogram (AR) type AR06, multilocus sequence type (ST) ST22 and staphylococcal cassette chromosome (SCC) mec type IV has increased from 22% in 1999 to 80% in 2003. In any one year, 50% of these isolates yield a single chromosomal pattern by pulsed field gel electrophoresis (PFGE). The present study investigated whether sequence analysis of the direct repeat unit (dru) in the hyper-variable region of the SSCmec cassette could aid in differentiating isolates of this strain.
Methods: Twenty-three MRSA isolates from six suspected outbreaks (Clusters A-E) in four different human hospitals and two veterinary practices in geographically distinct areas of Ireland were investigated. For dru analysis, 5′-GTTAGCATATTACCTCTCCTTGC-3′ and 5′-GCCGATTGTGCTTGATGAG-3′ forward and reverse PCR primers, respectively, were employed with an initial denaturation step at 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min. The resulting DNA sequences were aligned and interrelationships analysed using BioNumerics v. 4.61 (Applied Maths, Sint-Martens-Latem, Belgium).
Results: All isolates exhibited AR type AR06 (or a closely related AR pattern) and PFGE types (PFT) that were either indistinguishable or not differing by >6 bands. By dru typing, isolates from two geographically distinct veterinary practices (Cluster A and B; all PFT 01018) were differentiated. In Cluster C, dru analysis grouped all four fusidic acid resistant isolates together, separated them from fusidic acid susceptible isolates and further differentiated one isolate in the cluster that originated from a different source. In Cluster D, dru typing differentiated the isolate with the least similar PFT from the remaining isolates. In Cluster E, two PFT 01039 isolates were separated while, in Cluster F, four isolates exhibiting three PFTs were found to be indistinguishable.
Conclusions: The mec-associated dru region has the potential for extensive variability both in sequence and in number of 40-bp tandem repeats yet specific sequence types appear to be very stable over time. On the basis of this preliminary study, analysis of dru sequences appears very promising as a means of tracking difficult to differentiate epidemic MRSA isolates in Ireland such as AR06, warranting further investigation with this and potentially other difficult groups of isolates.
MRSA: infection control and nosocomial infections
P1307 Does a preliminary surveillance study enhance formulation of a comrehensive MRSA containment action plan? Study from a large district hospital in northwest England
S. Ambalkar, R. Palmer, A. Guleri (Blackpool, UK)
Objectives: Hospital MRSA control policy is based on national guidelines. Certain specialities are associated with higher than desirable MRSA bacteraemia rates. This requires targeted action plans to be designed for such areas. This study was carried out to evaluate if conducting a preliminary surveillance study would help in optimal planning and design of action plans for MRSA hotspots in the hospital.
Method: 53 MRSA bacteraemia episodes over 1-year (April 2005 through March 2006) were analysed.
Results: 95% of cases were over 50-years of age. Majority (60%) of cases were from medical wards, 25% from surgical, and 12% from intensive care. Majority (75%) were males. Clear information from 37 cases revealed 81% were hospital acquired and remaining from community. Out of 30 cases, 27% were previously known MRSA carriers. 53% of patients died either with or due to MRSA bacteraemia, of whom 38% died sometime within 14-days of bacteraemia. 20% of deaths were before anti MRSA treatment was commenced.
Conclusions: MRSA bacteraemia was more common in >50-years age group, males, hospital acquired and in medical wards. Medical ward has a large number of patient throughput and patients are frequently shifted between wards.
Salient points considered while formulating action plan were: Need for risk stratification, screening of high-risk patients, role of empirical anti MRSA agents in high-risk group, role of rapid diagnostic tests, decolonisation regimes for known/identified MRSA carriers, strict isolation policy, strict intravenous catheter policy, appointing local consultant/matron as infection prevention lead with support from hospital infection control team.
It emerged that conducting a preliminary surveillance study definitely helped identify local deficits and risk factors that contribute to the higher than acceptable MRSA rates in that area.
P1308 Improving the evidence base for the effectiveness of Staphylococcal healthcare-associated infection interventions
B. Cookson, S.P. Stone, B.S. Cooper, C.C. Kibbler, J.A. Robert, G.F. Medley, G. Duckworth, R. Lai, S. Ebrahim, P.G. Davey, E.M. Brown, P.J. Wiffen (London, Warwick, Dundee, Bristol, Oxford, UK)
Introduction: The last decade has seen a move towards evidence based medicine. The publication of “Consolidated Standards of Reporting Trials” (CONSORT) statement (1996) did much to improve matters regarding the transparency needed to improve the quality of reporting, so that information critical to synthesis of research was not missing. However, it did not provide items or descriptors easily translatable into the wide variety of interventions, settings, designs and statistical issues relating to infectious diseases.
Methods: Our systematic review of MRSA Isolation presented at a previous ISSS revealed many flaws in study design and presentation. A working party of the British Society of Antimicrobial Chemotherapy had used our approach in the field of antimicrobial prescribing and we decided to join forces and form “ORION” (Outbreak Reports and Intervention studies Of Nosocomial infection). This is presented here.
Results: It comprises a list of items or descriptors easily translatable into the wide variety of interventions, settings, designs and statistical issues relating to infectious diseases (including for example Staphylococcal) intervention studies. They are aimed at researchers, editors, reviewers, and grant assessment panels. It is intended that they facilitate well-designed interventional studies to help choose which methods are effective in reducing antimicrobial resistance and healthcare associated infection. This list has been available on the www: http://www.bsac.org.uk/_db/_documents/Orion_5_(3).doc) and been sent to several editors of journals for comment. ORION consists of a 22 item checklist, a summary table is recommended for description of the population, clinical setting, and the precise nature and timing of all interventions and outcomes and a graphical summary of the main results is recommended when outcomes are not independent. For intervention studies we strongly recommend a flow chart to track participants through each stage of the study.
Conclusion: The ORION Collaboration has produced proposals to improve the evidence base for infection control and antimicrobial stewardship interventions. They have been widely consulted and will be reviewed again with the scientific community in ∼3y time.
P1309 Reduction in the incidence of staphylococcal infections in hospital environment after the starting of the guideline of perioperative antibiotic prophylaxis. Confirmation of the decrease in 2006
G. Gattuso, D. Tomasoni, L. Palvarini, D. Berra, C. Chiarelli, R. Stradoni, A. Scalzini (Mantua, IT)
Objectives: Comparing the general incidence of staphylococcal and MRSA infections in the “C. Poma” Hospital versus the incidence of staphylococcal and MRSA in the surgical wards after the introduction of guidelines for perioperative antibiotic prophylaxis (PAP).
Methods: The hospital is a 530-bed tertiary care teaching hospital in Mantova with 30,000 patients admission per year. Two periods have been compared: 2nd Six Months (S) of 2002–1st S of 2003 (period A) and 2nd S of 2003–1st S of 2004 (period B), respectively before and after the introduction of the guideline of PAP. Two years later the we re-evaluated the data, considering the 1st semester 2006, to verify the maintenance of the trend.
Results: In the period A 10,407 microbiological examinations have been executed; 637/10,407 cases (6.1%) were positive for S. aureus and 119/637 (18.6%) were MRSA. In the period B 8,421 microbiological exams have been performed: 315/8,421 (3.7%) S. aureus and 81/315 (25.7%) MRSA.
In the period A a significant increase of MRSA was found in the surgical wards comparing to the other wards: 58.6% vs 22.9% (p = 0.001), whereas in the period B was checked a relevant decrease of MRSA in the surgical wards compared to the general data (25.0% vs 25.7%; p = ns). In the second period the data of MRSA in the surgical wards were found significantly reduced (58.6% vs 25.0%; p = 0.001).
Moreover in the period B the global antibiotic consumption was reduced with an increment of the cephalosporins of 1st generation (+11.5% of cephazolin) used correctly in surgical prophylaxis. In the first semester 2006 we noticed that the value regarding MRSA keeps unchanged, as in medical wards and ICU MRSA were 37.7% of the whole and in the surgical ones were 21.7%. The hospital general data in 2006 show an average value of MRSA equal to 35% of all the S. aureus.
Conclusions: 12 months after the introduction of PAP a significant reduction of incidence of MRSA infections was found in the surgical wards; moreover such result can be ascribed to a greater adhesion to norms of “good clinical practice”. The evaluation after two years (1st semester 2006) confirms the decreasing trend, pointing out an MRSA incidence of 21.7% of the whole S. aureus isolates, decreasing in comparison with 25% of the 2004.
P1310 Healthcare workers – Source, vector or victim of MRSA?
W.C. Albrich, S. Harbarth (Johannesburg, ZA; Geneva, CH)
Objectives: There is ongoing controversy about the role of healthcare workers (HCWs) in transmission of methicillin-resistant Staphylococcus aureus (MRSA). We performed a systematic literature search of electronic databases from 1980 through March 2006 to determine the likelihood of MRSA colonisation in HCWs, their role in MRSA transmission and their own subsequent risk of infection.
Methods: We scanned the English, French and German literature and excluded articles that did not present quantitative data. Two reviewers independently assessed the relevance of the included studies.
Results: In 121 studies, the prevalence of MRSA carriage among 33,318 screened HCWs was 4.6% (range: 0–65%). Average MRSA carriage in HCWs working in settings with endemic MRSA was 8.1%, compared to 3.9% in outbreak settings. Risk factors of MRSA carriage included chronic skin diseases, poor hygienic practices, close contact with infected or colonised patients and having worked abroad. Transient or intermittent colonisation predominated, but persistently colonised HCWs were found to be responsible for several nosocomial MRSA clusters. Almost 80 reports supported a causal role of HCWs in transmission of MRSA to patients. Success rates of eradication therapy were high (461/521 HCWs; 88.5%), and independent from duration of therapy. Subclinical infections and colonisation of extranasal sites were associated with persistent carriage after eradication therapy. Eradication of the MRSA carrier status and/or removal of personnel from patient care were successful in terminating MRSA transmission in most cases. About 4.9% of colonised HCWs developed infections, primarily of the skin and soft tissues. In settings with a high prevalence of community-associated MRSA, HCWs carriage of MRSA was 3.2% (92/2,832) with a high proportion of clinical infections (4/63; 6.3%).
Conclusion: A non-negligible number of HCWs become colonised with MRSA and 5% of those develop clinical disease. HCWs most frequently act as vectors and not as main sources of MRSA transmission. HCWs as persistent reservoir of MRSA are rare but nonetheless important. Aggressive screening and eradication policies seem justified in outbreak investigations or when MRSA has not reached highly endemic levels yet.
P1311 Controlling an outbreak of Staphylococcus aureus in a vascular and cardiac surgery ward
K. Kristóf, K. Antmann, Z. Szalay, N. Pesti, K. Nagy, F. Rozgonyi (Budapest, HU)
Objectives: Since January 2001 a fluctual increase in S. aureus infections has been recognized in a vascular and cardiac surgery ward of the Semmelweis University. The outbreak has been characterised and the S. aureus infections were monitored both phenotypically and genotypically.
Methods: From January 2001 to September 2006 a total of 511 S. aureus isolates were cultured. Identification of S. aureus strains was carried out on the basis of catalase positivity, gold pigment, beta-haemolysis and clumping factor. The presence of mecA and nucA genes was detected by PCR. Both the disc diffusion and the microdilution methods were used according to NCCLS/CLSI recommendations to determine the antibacterial sensitivity to vancomycin, teicoplanin, gentamicin, tobramycin, amikacin, netilmicin, ciprofloxacin, trimethoprim/sulphamethoxasole, doxycycline and linesolid. The clonal relationship of the isolates was determined by pulsed field gel-electrophoresis using SmaI enzyme digestion. Multilocus sequence typing of the representative MRSA isolates is in process.
Results: 511 cases of S. aureus infection were identified during the investigation period. The incidence of MRSA was the highest in the year 2005 (27.6% of all S. aureus isolates), while the earlier frequency was 7.14%, 5.66%, 7.5% and 6.24% from 2001 to 2004. Twelve antibiotic resistance patterns were found among the MRSA isolates. The commonest was the multiple resistance to all aminoglycosides, erythromycin, clindamycin and ciprofloxacin. This phenotype showed a mean incidence of 59% of all MRSA isolates in 2005–2006. Its evolution ranging from 0% in 2003 to the alarming proportion of 63.9% in 2005. The spread of this epidemic strain was stopped after introducing strict hygienic measures with a close cooperation between microbiologists and hygienists. In 2006, the MRSA incidence decreased to 10.7%. PFGE analysis revealed MRSA strains form four types with different frequency. PFGE type C was the most common.
Conclusions: Although the outbreak was sufficiently controlled in the point of view of hospital hygiene, however multiple resistant MRSA strains of similar clonality could select causing a disadvantage in the entire microbiological eradication of them.
Supported by OTKA No.: T 46186
P1312 The MRSA point-prevalence: an important tool in the infection control programme
I. Chaberny, A. Bindseil, F. Mattner, P. Gastmeier (Hannover, DE)
Background: Due to the enormous increase in the number of MRSA-patients, an extended admission screening on ICUs and surgical wards was implemented at the Hannover Medical university hospital since July 2004 (18.4 nares cultures per 1,000 patient days).
Objectives: To determine the prevalence of MRSA and Panton-Valentine leukocidin (PVL) among inpatients, to identify other patients at risk for MRSA colonisation and to evaluate the compliance with the established admission screening.
Methods: Inpatients were screened by cultures from nose, throat and broken skin in a point-prevalence survey in the year 2005. S. aureus isolates were tested for oxacillin susceptibility and for PVL. MRSA were tested for antimicrobial susceptibility and were analysed by staphylococcal protein A (spa) typing.
Results: Of 510 inpatients, 145 (28%) were S. aureus carriers. 27 (19% of all S. aureus isolates) were MRSA, i.e. 5.3% of all inpatients were MRSA carriers (95% CI: 3.49; 7.70) which was the MRSA prevalence. With 67% was the spa type t032 or so called “Barnimer” strain predominant followed by t001 (15%), t004 (4%), t008 (4%), t022 (4%) and t067 (4%). The PVL gene was present in one (0.2%) methicillin-susceptible strain. Compared to our MRSA database the MRSA status of 37% of the patients (pts) was unknown (ten out of 27 pts), e.g. in 2.0% (ten out of 510 pts) of all inpatients the MRSA colonisation was identified for the first time. In three out of these ten cases the compliance with the established admission screening had failed. Although the admission screening had previously led to negative results concerning three other patients, a nosocomial acquisition was identified. Four other newly MRSA patients had been detected on wards without a routinely admission screening (three cases on neurological and one case on an internal medicine ward).
Conclusion: The results of our study pointed to a 5.3% prevalence of MRSA, which proved higher than our previous observations had suggested and it pointed to a 0.2% prevalence of PVL-positive S. aureus. Despite an extended admission screening practice 37% of all MRSA-positive inpatients had been missed. The neurological patients were identified as a further risk group for admission screening and were included in the established admission screening practice. In addition, a weekly surveillance cultures practice seems to be useful in order not to miss further MRSA cases.
P1313 Topical treatment with mupirocine/chlorexydine and long-term risk of methicillin-resistant Staphylococcus aureus infection
A. Pan, L. Soavi, P. Mondello, P. Catenazzi, S. Lorenzotti, L. Signorini, S. Testa, G. Carnevale, G. Carosi (Brescia, Cremona, IT)
Objective: To evaluate the long term effect of topical treatment with mupirocine and chlorexidine on the risk of colonisation/infection in patients with a previous microbiological isolation of MRSA.
Methods: This prospective study was carried out at the hospital of Cremona, an 850-bed community hospital, where a MRSA control programme including topical treatment was ongoing. All hospitalised patients identified as being MRSA positive and for whom data regarding topical treatment with 2% nasal mupirocine ointment and 4% chlorexydine baths/showers and shampoo for 5 days were available, were included in the study.
Results: from January 1, 1997 through July 30, 2006, we identified 1,000 MRSA positive patients, for a total of 1,101 admissions. Data regarding topical treatment were available for 768 patients (77%) for a total of 853 admissions: 479 (62%) were treated with mupirocine on their first admission while 289 (38%) where not. Sixty-three treated and 78 untreated patients died during the first admission. MRSA was identified during a second admission in 41/416 treated patients (9.9%) and in 35/211 untreated subjects (16.6%) (P = 0.02). Six patients, 4 previously treated and 2 not treated, had more than one admission. We observed 17 infections in treated patients (4%) as compared with 7 in the untreated group (3.3%) (P = 0.8). The 17 infections in treated patients were as follows: 7 blood-stream infections (BSI), 3 respiratory infections (RTI), 6 skin and soft tissue infections (SSTI), and 1 other infection. Untreated patients had 7 infections: 1 RTI, 4 SSTI, 2 other infections. No significant difference was observed for any type of infection between treated and untreated subjects (P > 0.1 in all cases).
Conclusions: Patients treated with topical mupirocine and chlorexydine have a reduced risk of being identified as MRSA positive during after previous infection or colonisation (P = 0.02). Despite this, no reduction in the overall risk of infection in the follow-up was identified among treated vs untreated subjects.
P1314 Susceptibility patterns to oral antimicrobials of PVL+ MRSA from Austria
C. Jebelean, K. Pfeil, C. Luger, S. Stumvoll, M. Stammler, B. Sammer, H. Mittermayer (Linz, AT)
Objectives: The prevalence of PVL+ MRSA is rising in many European countries.
The aim of our study was to investigate the prevalence, the genetics and the susceptibility to oral antimicrobials of PVL + MRSA strains from Austria.
Materials and Methods: We screened 904 strains out of our collection of 1,742 strains of MRSA going 10 years back for PVL gene using a PCR method. For all strains we determined the mecA and femA gene using a multiplex PCR. For subtyping we performed coagulase gene PCR with restriction digestion of PCR products with AluI, and we further analysed the PFGE patterns. We then determined the susceptibility to: erythromycin (E), clindamycin (LC), rifampin (RI), tetracycline (T), minocycline (MH), trimethoprim/sulfamethoxazole (TMP/SXT), fusidic acid (FU) and linezolid (L) using the agar diffusion method as recommended by CLSI.
Results: We found 78 PVL+ MRSA strains out of the 904 strains tested. The percentage of PVL+ strains varied along the years 2001 to 2006 as follows: 1.5%, 1.9%, 7.1%, 5.2%, 3.6%, 5.9%, respectively. The strains were isolated mainly from abscesses, wounds and noses mainly in Upper Austria, Salzburg and Vienna. The patients were adults: 51%, older persons: 31% and children 13%. Among the PVL+ strains we found the following sequence types (ST): ST8 (33%), ST80 (18%), ST30 (17%), ST152 (15%), ST5 (10%), ST22 (3%), ST777 (3%), non-typable (1%). The usual encountered MRSA ST in Austria are: ST228, ST8, ST22 and ST5. Overall the L, RI, TMP/SXT, MH, LC, FU, E, and T susceptibility was 100%, 100%, 99%, 99%, 99%, 79%, 59%, 54%, respectively. The resistance to E was higher in Vienna and Salzburg, to FU and T in Salzburg and Upper Austria.
Conclusions: This is the first report on PVL+ MRSA from Austria to date. The PVL+ MRSA strains encountered in Austria were isolated mainly from abscesses in adults. The most encountered ST were: ST8, ST80, ST30, ST152, and ST5. L, RI, TMP/SXT, MH and LC showed a good in vitro activity on these strains.
P1315 The emergence of methicillin-resistant Staphylococcus aureus as a major cause of surgical site infection in England
J. Wilson, S. Elgohari, M. Lillie (London, UK)
Objectives: to explore the role of MRSA as a cause of surgical site infection (SSI).
Methods: The surgical site infection surveillance service (SSISS) in England collects data on a complete set of patients undergoing specified surgical procedures. Hospitals participate in the surveillance for minimum 3-month periods in one or more of the 13 categories of procedures. Trained staff in each hospital use a standard methodology to identify those patients that develop a surgical site infection (SSI) meeting a specific set of criteria during the patients in-patient stay. Between 1998 and 2005 data on 7,366 SSI from 254,607 surgical procedures by were collected by 247 hospitals participating in the surveillance. Data on the causative micro-organisms were available for 5,957 (81%) of SSI reported.
Results: S. aureus was the most common pathogen causing SSI, accounting for 52% of infections where a causative organism was reported. Of these 66% were caused by methicillin-resistant S. aureus (MRSA), although this proportion varied by type of surgery from 20% in abdominal hysterectomy to 82% in bowel surgery. One third of the SSI caused by MRSA involved the deep tissues and MRSA was more likely to cause deep SSI than methicillin sensitive S. aureus (p = 0.001). The risk of an SSI being caused by MRSA increased with patient age; 18% of SSI in patients under 45 compared to 46% of SSI in patients of 85 years or over (p < 0.001). MRSA has accounted for more than 60% of S. aureus SSI since the inception of SSISS in 1997. The proportion that were MRSA fluctuated from year to year with no evidence of an increasing or decreasing trend. This may reflect between year variations in hospitals participating in the surveillance.
Conclusions: This analysis has provides evidence for the role of MRSA as a pathogen of surgical wounds from a large national dataset. The finding that MRSA accounts for almost two-thirds of S. aureus SSI across a range of surgical procedures in England has important implications. This proportion of MRSA is significantly higher than that reported for bacteraemia. Serious MRSA infections can be difficult to treat, more so in the presence of prostheses, increasing the risks of chronic sequelae. The prevalence of MRSA as a cause of SSI identified by this study emphasizes the need to identify and treat colonised patients prior to surgery where possible, and review antimicrobial prophylaxis for patients at high risk of developing post-operative wound infection.
P1316 Incidence and significance of methicillin-resistant Staphylococcus aureus in ventilator-associated pneumonia: a prospective study
M. Torres, C. Radice, P. Muñoz, E. Bouza (Madrid, ES)
Objectives: MRSA is a growing cause for concern in many hospitals. Ventilator-associated pneumonia (VAP) is one of the most severe infections in ICUs at present. The incidence and clinical significance of MRSA-VAP are not well known. We aimed to assess the incidence of MRSA-VAP and to define its clinical relevance in our institution.
Method: Prospective cohort study in the 3 ICUs of a general reference hospital. Lower respiratory tract (LRT) samples of all patients with suspicion of VAP were cultured from Dec 2003 to Dec 2005. Standard diagnostic criteria were followed.
Results: Over a two-year period, 1,220 consecutive patients were cultured (2,779 LRT samples) and VAP was diagnosed in 250 patients. S. aureus was the most frequent causal agent of the VAP episodes (104: 41.6%). Of these, 66 VAP were caused by MRSA (26.4%) and 38 by MSSA (15.2%).
MRSA-VAP (66 episodes) was compared with MSSA-VAP (38 episodes). MRSA-VAP patients were older (63 vs. 51 years, p< 0.01), had a higher Charslon comorbidity score (4 vs. 2.7, p = 0.01), more severe underlying diseases (48.5% fatal vs. 21.1%, p = 0.01), more frequent multiorganic failure or septic shock (23.7% vs. 5.3% p = 0.01), a longer hospital admission before VAP diagnosis (13 vs. 6 days, p = 0.01), a higher Apache II score on admission to the ICU (13.1 vs. 10.3 p = 0.01), and had consumed more antibiotics before the VAP episode (1.9 vs. 0.7 p < 0.01). MRSA-VAP had a higher mortality rate than MSSA-VAP (74% vs. 31%; p<0.01). However, MRSA was not found to be an independent risk factor for mortality in our study.
Conclusions: MRSA causes 26.4% of VAP episodes and 64% of VAP by S. aureus. MRSA is a marker of severity VAP but resistance is not in itself independently associated with a worse outcome.
P1317 Where is MRSA in an intensive care unit and does it matter?
L. White, S. Dancer, C. Robertson (Glasgow, UK)
Objective: This project set out to evaluate the hygiene status of a six-bedded surgical intensive care unit (SICU) environment alongside the occurrence of hospital-acquired infection over a ten-week period.
Methods: Ten sites within SICU were sampled twice weekly using dipslides applied to a range of hand-touch and other environmental surfaces. The results were analysed using recently proposed standards for surface level cleanliness, which specify enumeration of aerobic colony counts from hand-touch sites as well as the identification of specific indicator organisms.
Results: The audit exposed hygiene fails in over a quarter of the 200 samples obtained. Most occurred at near-patient hand-touch sites, particularly curtains, beds and cardiac monitor buttons, and suggested insufficient cleaning of these areas. Sites three or more metres away from the patients, such as computer keyboard, telephone and staff chair, also demonstrated hygiene failures but at less than half the number. In contrast, non-hand-touch sites (sink and floor) rarely failed the hygiene standards. Fourteen staphylococcal isolates associated with SICU-acquired infection were collected from patients during the study period. Pulsed-field gel electrophoresis revealed five patient strains to be indistinguishable from eight environmental strains, with timescales supporting dynamic staphylococcal transmission in both directions between patients and the environment in this SICU.
Conclusion: These findings suggest that hand-touch sites closest to patients may act as a reservoir for potential pathogens.
P1318 Enhanced surveillance of MRSA bacteraemia in English hospitals: comparison of risk factors in patients with community onset bacteraemia with cases whose onset was in hospital
A. Pearson, K. Wagner, M. Murray, M. Painter, J. Giltrow, M. Fleming (London, UK)
Objective: The English Department of Health introduced mandatory reporting of methicillin resistant (MRSA) and sensitive (MSSA) Staphylococcus aureus bacteraemia in April 2001. In 2004 a performance target for a 50% reduction in the numbers of MRSA bacteraemia was set for April 2008. Enhanced mandatory MRSA bacteraemia surveillance was introduced in October 2005. This paper compares the patient characteristics of admission diagnosed cases of MRSA bacteraemia with hospitalised MRSA bacteraemia cases in acute NHS hospitals in England.
Methods: The analysis presented was based on information sent to a web enabled database as part of a national mandatory surveillance scheme between October 2005 and September 2006. Standardised screen formats were used to record the mandatory enhanced surveillance data. From May 2006 an optional data collection screen became available on the system which enabled entry of additional data on source, risk factors and prior healthcare contacts for each MRSA bacteraemia case.
Results: During October 2005 to September 2006 approximately 7,000 reports of MRSA bacteraemia were received from 172 English Trusts. Early enhanced surveillance data collected on a voluntary basis suggest that the most common source of MRSA bacteraemia in patients with community onset bacteraemia was skin/soft tissue infection (23%), followed by UTI (17%). In contrast the most common source of MRSA bacteraemia in patients who had been admitted for 2 or more days before their bacteraemia was detected was CVC associated (23%), followed by PVC associated (20%). The most common risk factors for both groups were peripheral IV devices, followed by central IV devices.
Conclusion: Data from the MRSA bacteraemia enhanced surveillance scheme provide an important means of identifying risk factors and sources of bacteraemia in English patients. Skin/soft tissue infections (the most common source of bacteraemia in the admission diagnosed group) may result from the use of peripheral IV devices, the most common risk factor in this group. It is hypothesised that a large proportion of those patients presenting to the acute trust hospital with an MRSA bacteraemia developed this condition as a result of a prior healthcare contact. Identification of key risk factors and sources of bacteraemia will allow effective targeting of infection control interventions.
P1319 Clonal analysis of methicillin-resistant Staphylococcus epidermidis strains isolated from bloodstream infections in Greece
A. Foka, S. Leveidiotou, S. Vamvakopoulou, E.D. Anastassiou, I. Spiliopoulou (Patras, Ioannina, GR)
Objectives: Methicillin resistant-coagulase negative staphylococci constitute a major cause of hospital infections. Staphylococcus epidermidis is the predominant species associated with clinically relevant infections. The purpose of the present study was to investigate any clonal relationship among methicillin-resistant S. epidermidis (MRSE) isolated from different patients hospitalised in two University Hospitals located in Western and North-western part of Greece.
Methods: A total of 73 MRSE isolated from bloodstream infections of inpatients were tested for their antibiotic resistance against representative antibiotics by the disk diffusion method: cefoxitin (Cef), tobramycin (Tm), gentamicin (Gm), linezolid (Lin), erythromycin (Em), clindamycin (Cm), ciprofloxacin (Cip), fusidic acid (FA), sulfamethoxazole/trimethoprim (SXT), vancomycin (Va). The MIC of oxacillin was determined by the agar dilution method with Mueller-Hinton agar containing 2% NaCl. PBP2a was detected by a Latex agglutination test in all S. epidermidis. The presence of mecA gene was investigated by PCR with specific primers. Slime production was detected by the qualitative method. Clonal types were identified by PFGE of chromosomal DNA SmaI digests.
Results: All 73 MRSE were mecA-positive while 57 (78%) produced slime. The PFGE analysis revealed five PFGE types, common between the two hospitals. Twenty three out of 73 (31.5%) isolates had the same PFGE type, named a. Thirteen MRSE (18%) belonged to PFGE type b, six (8%) to type c, five (7%) to type d and three (4%) to type e. The rest 23 (31.5%) were classified into 23 different PFGE types. All MRSE of PFGE type a exhibited the same resistance phenotype to Cef, Tm, FA. PFGE type b isolates showed a common resistance phenotype to Cef, Tm, Cip, Gm, SXT. The isolates of the PFGE type c were resistant to Cef, Em, Cip, Cm, Tm, FA. MRSE of PFGE type d were multi-resistant to Cef, Em, Cip, Gm, Cm, Tm, FA, while strains of PFGE type e were resistant to Cef, Em, Cm, FA.
Conclusions: Among hospitalised patients with bloodstream infections in two University Hospitals, located in adjacent regions of the country, common MRSE clonal types exhibiting multi-resistant phenotypes are distributed.
P1320 Mortality following invasive MRSA infection in England, 2004–5
N.A.C. Potz, D. Powell, T. Lamagni, R. Pebody, C. Rooney, C. Griffiths, D. Bridger, P. Goldblatt, J. Dobbs, G. Duckworth (London, UK)
Objectives: To evaluate the public health burden associated with MRSA infection, information is needed on the incidence of infection and patient outcome. Given that national surveillance of MRSA bacteraemia in England does not capture patient mortality, a method was developed to obtain this information from an external source. Preliminary analyses of mortality following invasive MRSA infection were undertaken using this augmented dataset.
Methods: A probabilistic record linkage method was developed to link death registrations from the Office for National Statistics and laboratory confirmed invasive MRSA records from the Health Protection Agency (HPA) as unique personal identifiers are not uniformly available in the surveillance data. Surveillance reports from laboratories in England of MRSA isolated from blood and/or CSF from 1st January 2004 to 31st December 2005 were linked to deaths occurring between 1st January 2004 and 31st March 2006. Individuals with multiple episodes of invasive MRSA infection were identified and analysed separately. Descriptive and multivariable analyses of factors associated with mortality following invasive MRSA infection were undertaken.
Results: Preliminary results identified that 97% of the 10,449 invasive MRSA infection records from 1st January 2004 to 31st December 2005 had sufficient identifying variables available to permit linkage. Of these, 59% were identified as having died up to 31st March 2006 (maximum 27 months follow-up). The 7-day case fatality ratio following a single episode of invasive MRSA infection was 0.20 rising to 0.38 within 30 days. With adjustment for age, there was no significant difference in 30 day mortality for males compared to females (OR = 1.03: 95% CI 0.94–1.12). Age was a strong predictor of 30 day mortality: compared to a baseline age group of 0–50 years, the likelihood of mortality was four-fold higher in 71–80 year olds (OR = 3.80: 95%CI 3.21–4.49), and six-fold in 81–90 year olds (OR = 6.19: 95% CI 5.22–7.33).
Conclusion: Mortality following invasive MRSA infection was found to be very high, almost 40% of individuals dying within 30 days of diagnosis, with the elderly being substantially more likely to die. However, it is known that these patients are likely to have other underlying conditions and it is recognized that both the risk of contracting an invasive MRSA infection and the risk of dying are related to a patients' underlying disease and treatment.
P1321 Regional variation in Panton-Valentine leukocidin positivity smong S. aureus isolates in complicated skin and skin structure infections
R. Strauss, K. Amsler, M. Jacobs, K. Bush, G. Noel (Raritan, US)
Objectives: Panton-Valentine leukocidin (PVL)-positive methicillin-resistant S. aureus (MRSA) has been recognized as a virulent pathogen that can cause severe skin and respiratory tract infections. PVL-positive methicillin-susceptible S. aureus (MSSA) has also been increasingly described in severe skin infections. The prevalence of PVL-encoding genes among S. aureus isolates were analysed from a multicentre trial comparing ceftobiprole, in investigational broad-spectrum cephalosporin with anti-MRSA activity, to vancomycin in the treatment of patients with complicated skin and skin structure infections (cSSSI).
Methods: S. aureus isolates were analysed for PVL by multiplex PCR (n = 415). In vitro susceptibility (CLSI methodology) and a mecA probe were used to identify MRSA.
Results: The trial involved 92 sites from 15 countries on 4 continents. Of the 784 patients enrolled, S. aureus was isolated in 494 baseline samples obtained from pus, leading edge of cellulitis, or debrided tissue. Overall, 47% (195/415) of S. aureus isolates were PVL positive. There were large regional differences in the prevalence of MRSA and MSSA among S. aureus isolates. In Europe, 40/239 (16.7%) S. aureus were MRSA, compared to the USA, where 129/185 (69.7%) of S. aureus were MRSA (P<0.001). PVL results were available for 447/497 S. aureus isolates (89.9%). PVL-positive MRSA isolates were most commonly seen in the USA, with significantly lower rates of PVL-positive MRSA in Europe (P<0.001) (Table ). In contrast, PVL-positive MSSA most commonly occurred in Asia/Africa and Europe. Clinical cure rates of cSSSI due to PVL-positive S. aureus were comparable between ceftobiprole- and vancomycin-treated subjects (95.0% vs 95.0%). Cure rates of cSSSI due to PVL-positive MRSA were 93.9% for ceftobiprole-treated subjects and 86.2% for vancomycin-treated subjects. Frequency of PVL positivity among S. aureus isolates is shown in the table.
| Population | PVL (+) |
|
|---|---|---|
| % MRSA | % MSSA | |
| Europe | 5.3 (2/38) | 37.4 (70/187) |
| USA | 86.8 (92/106) | 26.7 (12/45) |
| Asia/Africa | 0.0 (0/3) | 70.8 (17/24) |
| Central/South America | 25.0 (1/4) | 12.5 (1/8) |
Conclusion: Prevalence of PVL-positive MRSA and PVL-positive MSSA differed widely among geographic regions. In this phase 3 cSSSI trial, PVL-positive S. aureus isolates from US subjects were substantially more likely to be MRSA than MSSA. This may be reflective of the high prevalence of USA300 isolates as an important cause of cSSSI in the USA. Outside the USA, the PVL toxin gene was more likely to be found in MSSA than in MRSA isolates.
P1322 Environmental contamination in the rooms of MRSA-colonised patients
H. von Baum, F. Raychevska, A. Möricke (Ulm, DE)
Objectives: The presence of MRSA in the rooms of MRSA colonised patients and its relationship to the contamination of healthcare workers' (HCW) gloves and clothing was examined.
Methods: 10 standardised environmental samples from patients' rooms as well as samples from the gloved hands and clothing of HCW were examined. Samples were incubated for 48 h at 37°. Presumptive positive colonies were confirmed as S. aureus. Resistance to methicillin was determined using the Slidex® MRSA Detection (bioMérieux). All environmental MRSA isolates as well as the corresponding patients' isolates were genotyped using pulsed-field-gel-electrophoresis (PFGE).
Results: MRSA was present in the rooms of 18 of 25 MRSA patients (72%). Contamination of HCWs' gloved hands and/or clothing was identified in 15 of 25 nurses (60%); 3 of 6 physicians (50%), 3 of 3 physiotherapists (100%) and 3 of 15 members of the housekeeping staff (20%). In the majority of patients more than one MRSA strain was present.
Environmental MRSA strains indistinguishable from the strains colonising the patients were detected in only half of the cases.
Conclusion: Healthcare workers must be aware that contamination of their gloved hands and clothing whilst caring for a MRSA colonised patient is highly probable. In the environment of MRSA colonised patients not only the patients' own strains but additional genotypically distinct MRSA strains were detected.
P1323 Who's carrying MRSA? Prevalence of MRSA amongst staff at a district general hospital and associated risk factors
J. Clark, J. Archibald, A. Kearns, S. Barnass, M. Kyi (Isleworth, London, UK)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) infections are important nosocomial infections in the UK. In efforts to reduce infection rates, interest has focussed on patient colonisation and transmission. The aim of this study was to determine the rate of and risk factors for staff carriage of MRSA in a district general hospital setting.
Methods: Staff who enrolled gave written signed consent. They were required to complete an anonymous questionnaire and a nasal swab was taken for MRSA culture (also anonymous, but linked to the questionnaire). MRSA was identified using standard laboratory methods, and tested for antimicrobial susceptibility using the British Society for Antimicrobial Chemotherapy method. Phage typing and pulsed field gel electrophoresis (PFGE) was conducted at the Centre for Infections, Colindale, London. Statistical analysis was done using Fisher's Exact Test. Approval for the study was granted by the Local Research Ethics Committee.
Results: Of 120 members of staff (21 medical staff, 54 nursing staff, 11 healthcare assistants, 15 student nurses, 19 non-clinical staff) who took part in the study, 10 (8.33%) were found to be carrying MRSA. All the isolates were from nursing staff of varying grades. Previous MRSA carriage was associated with current MRSA colonisation (P value = 0.02), with other symptoms of upper respiratory tract infection (sore throat, runny nose, cough, “cold”, sneezes, fever and hay fever) showing no statistically significant association. A history of sinusitis or facial surgery, or having been a hospital in-patient in the previous year, were not associated with MRSA colonisation. Typing of the isolates showed most (8/10) were UK EMRSA-15 and the other 2 were EMRSA-16. On PFGE, four isolates were found to be a common pulsotype, EMRSA-15 variant B3, whilst the other 6 were unique to the hospital.
Conclusion: MRSA is an important cause of nosocomial infection and its control is a priority for infection control teams. This study has shown that even when small numbers of staff are screened for nasal carriage of MRSA, carriers are found. Therefore, good infection control practice by staff remains a top priority.
P1324 Control of an outbreak of an unusual ciprofloxacin-susceptible variant of methicillin-resistant Staphylococcus aureus EMRSA-15 on a neonatal unit
J. Otter, J. Klein, T. Watts, A. Kearns, G. French (London, UK)
Objectives: To report an outbreak and subsequent control of an unusually antimicrobial-susceptible strain of healthcare-associated MRSA on a NNU, which was initially mistaken for community-associated MRSA (CA-MRSA) and was not detected by standard local screening methods. We also investigated whether environmental contamination may have contributed to the outbreak.
Methods: All babies are screened for MRSA on admission to our NNU using ciprofloxacin-containing broth media. Isolates were characterised by antimicrobial susceptibility (by disc diffusion), pulsed-field gel electrophoresis (PFGE), phage type and Panton-Valentine leukocidin (PVL) status. Forty environmental sites were sampled, plating onto Baird-Parker agar directly and after broth enrichment.
Results: The index case was a premature baby who developed MRSA bacteraemia with associated tibial osteomyelitis and multiple subcutaneous abscesses. Screening with non-selective media identified three further colonised babies. All four patient isolates were UK EMRSA-15 by PFGE, PVL-negative and susceptible to all non-β-lactam antimicrobials tested, including ciprofloxacin. This pattern is rare in our hospital. Seven (17.5%) of the 40 environmental sites yielded MRSA, including three that matched the phage type and unusual antibiogram of the outbreak strain. The outbreak strain was cultured from the door of the milk expressing room and, in the same room, a different MRSA strain was isolated from a composite sample from a breast-pump and associated chair arm.
Outbreak interventions included cohort isolation of affected babies, thorough cleaning of the unit with a chlorine-containing compound, introduction of a new cleaning regime for the milk expressing room and increased emphasis on hand hygiene amongst staff and mothers. No further cases were reported.
Conclusions: The outbreak strain was an unusual ciprofloxacin-susceptible variant of EMRSA-15 and was initially mistaken for CA-MRSA. MRSA was identified on hand-touch sites in the NNU including communal milk expression areas, and mothers may have contributed to transmission. Non-outbreak MRSA strains were also found in the environment and may have survived from previous incidents. The outbreak was controlled by multiple interventions aimed at improving hand hygiene and reducing environmental contamination. Ciprofloxacin-containing MRSA screening media are sub-optimal in paediatrics, where ciprofloxacin susceptible strains are increasing in prevalence.
P1325 Methicillin-resistant Staphylococcus aureus carriage in patients discharged from a teaching hospital in endemic area
V. Tomic, V. Ursic (Golnik, SI)
Introduction: After introducing an aggressive infection control programme of transmission of methicillin-resistant Staphylococcus aureus (MRSA) to our tertiary care, teaching hospital the proportion of MRSA cases acquired in our institution decreased from 50% to 6.1%. To better evaluate efficiency of infection control measures (screening on admission, use of alcohol hand rub, barrier precautions) we decided to determine the MRSA carriage rate at discharge which, to our knowledge, was not yet performed in a highly endemic setting.
Methods: We screened all patients admitted to our hospital in the periods April 1–30, 2004 and April 1–30, 2006. Swabs were obtained from the anterior vestibule of the nose within 24 hours of admission. We performed nasal swabs in all these patients on the day of their discharge from hospital regardless of the month of discharge.
Results: In April 2004 673 patients and in April 2006 643 patients were admitted to our hospital. Both screening swabs (on admission and discharge) were available for evaluation in 521/673 patients (77.4%) in 2004 and in 403/643 (62.7%) in 2006. On admission 8 (1.5%) and 4 (0.6%) patients were already colonised with MRSA in April 2004 and 2006, respectively. Two of 8 patients in 2004 were successfully decolonised. In 2004 we detected MRSA at discharge in 3 (0.5%) patients not harbouring MRSA on admission. In April 2006 no such patients were detected. Patients who acquired MRSA in our institution during stay in April 2004 were hospitalised on the same ward and the dates of their stay on the ward partially overlapped. After screening the staff on the ward a medical nurse with history of non-compliance to hand disinfection protocol was found colonised with MRSA in the nose and external ear and was successfully decolonised afterwards. PFGE pattern of MRSA strain detected in the nurse was undistinguishable from the PFGE patterns of MRSA strains detected in 3 patients. Through screening of all medical staff on other wards we detected no additional MRSA carriers.
Conclusions: When screening of patients for MRSA carriage, use of an alcohol hand rub when necessary and barrier precautions are strictly implemented even in an endemic setting transmission of MRSA can be prevented. Undetected colonised healthcare workers pose a great risk, even more so if hand disinfection is not flawless.
P1326 MRSA spa-typing reveals a newly imported hospital endemic strain
F. Huenger, S. Klik, H. Haefner, V. Krizanovic, S. Koch, S.W. Lemmen (Aachen, DE)
Background: One aim of hospital infection control is to minimise emergence and spread of Methicillin-resistant Staphylococcus aureus (MRSA). Typing of isolates is used to support infection control measures by revealing transmission and outbreaks. The analysis of the staphylococcal protein A (spa) gene repeat sequence is a typing method to discriminate MRSA strains.
Objective: Evaluation of spa-typing as a tool for hospital MRSA surveillance.
Methods: Non-replicate MRSA isolates detected by screening or clinical cultures between January 2004 and September 2006 from patients admitted to the University Hospital Aachen (1,500 beds) were assayed by spa-gene repeat sequence analysis and spa-types were assigned using the Ridom StaphType™ software. Typing results were analysed and used for standard infection control measures.
Results: Among 573 analysed MRSA isolates 46 different spa-types were identified. The majority of 324 (57%) isolates belonged to spa-type t003. The second most frequent strain was t838 with 58 (10%) isolates followed by 25 t038; 19 t037; 18 t032; 17 t045; 13 t004; 8 t001; 8 t002; 7 t264; 7 t030; 5 t018; 4 t014; 4 t161; 4 t111; 3 t035. In 3 epidemiological not related patients the Panton-Valentine leucocidin (PVL) positive Community-Acquired (CA) MRSA strain t044 was detected. Strains t109, t216, t457 and t740 were isolated twice. The remaining isolates belonged to 25 different spa-types.
During an outbreak in a surgical ICU in June/July 2005 the previously not detected strain t838 was isolated from 8 patients. The first isolate was detected in blood cultures of a patient transferred from the United Arabic Emirates with multiple traumatic injuries. Screening of staff and environment was negative. Although the outbreak was controlled by immediate standard isolation and prevention measures, this rare strain t838 was isolated regularly during the following 14 months.
Conclusions: Since the majority of 56% of hospital MRSA isolates belonged to the regional endemic strain t003, spa-typing showed limitation in differentiation between transmission/outbreak and accidental accumulation. Nonetheless this method turned out to be a useful tool in hospital infection control, as 3 CA-MRSA strains could be identified by spa-typing and transmission within the hospital was excluded. In addition the typing revealed that the most likely imported strain t838 has established as a second hospital endemic strain and underlines the risk of international MRSA spread.
P1327 Spread of a vancomycin-intermediate, methicillin-resistant Staphylococcus caprae in a neonatal intensive care unit
M. John, D. Kenny, D. Diagre, Z. Hussain (London, CA)
Objectives: We report spread and control of a strain of a methicillin-resistant and vancomycin intermediate Staphylococcus caprae strain in a 38 bed neonatal intensive care unit (NICU), following isolation from a blood culture of one of a triplet.
Methods: Specimens from the nose and respiratory tract were collected from all neonates and cultured. Coagulase negative staphylococci (CoNS) were screened for vancomycin resistance using BHI plates with 6mg/L vancomycin. Susceptibility to vancomycin was confirmed by E-test (AB Biodisk) according to the manufacturer's instructions. Susceptibility to other antimicrobials was determined using the VITEK system and the presence of the mecA gene was ascertained by an in-house PCR. CoNS that grew on screen plates were identified using long set sugars (Kloos method) and were typed using restriction enzyme digestion (Sma1) and Pulsed Field Gel Electrophoresis (PFGE). Infected and colonised neonates were cohorted in the same room and placed on contact precautions. Staff and families were instructed regarding good hand hygiene.
Results: CoNS from five babies grew on screen plates and were identified as most closely resembling Staphylococcus caprae. Three were blood isolates, one was from an endotracheal aspirate and one was a colonising organism. All displayed vancomycin intermediate resistance and carried the mecA gene. The isolates had an MIC of 8 mg/L of vancomycin by E-test. The profile of PFGE of all five isolates was identical. All five babies with this strain, including a sibling of the index case were linked epidemiologically and had been in the same room at the same time.
Conclusion: CoNS resistant to β-lactam antimicrobials have emerged as important causes of bloodstream infection in NICUs. Vancomycin is commonly chosen in the treatment of these infections. The outbreak in our NICU demonstrates the importance of screening isolates of CoNS for decreased vancomycin susceptibility, which automated methods and disc diffusion methods may fail to detect. Institution of strict hand disinfection and contact precautions was successful in preventing further spread of this organism within our NICU
P1328 Assessing Staphylococcus aureus nasal colonisation in a cohort of healthy volunteers
M.E. Wood, D.J. Morse, T.M. Nordby, L.K. Olson, P.A. Mach, C.J. Jacobson (St. Paul, US)
Objectives: The objective of this analysis was to understand S. aureus nasal carriage levels and patterns (persistent vs. intermittent) among healthy volunteers.
Methods: A screening study was conducted to obtain nasal samples from healthy volunteers for use in the development and assessment of a new rapid in vitro diagnostic device for S. aureus. Right and left naris of subjects were sampled separately at two time periods 30 days apart. Nasal specimens were analysed for S. aureus by standard quantitative culture methods using a selective and differential medium. Confirmation testing was conducted using the Tube Coagulase Test. Culture data from all studies was entered into a database for analysis of persistence vs. intermittent carriage and log count variability.
Results: A total of 151 subjects were enrolled with a total 147 evaluable subjects (having samples at both time points). 28% of evaluable subjects were persistent carriers (S. aureus detected on both sampling days), 16% were intermittent (S. aureus detected on one sampling day) and 56% were non-carriers. Mean log count for a persistent carrier was 3.67 logs while the mean log count for an intermittent carrier was 1.67 logs. Mean log count difference between the initial and 30 day screening was approximately 0.96 logs.
Not all subjects were found to be colonised in both nares. 27% of persistent carriers were colonised in only one naris. 70% of intermittent carriers were colonised in only one naris. When counts were observed in both nares, the average difference between nares was approximately 1.19 logs.
29 persistent carriers were sampled repeatedly over a 3 year period. 83% of the time S. aureus was detected at subsequent samplings. 45% of the persistent carriers had S. aureus detected at every sampling.
Conclusions: Staphylococcus aureus accounts for approximately 20% of surgical-site infections. Different nasal colonisation patterns (persistent, intermittent, non-carrier) may play a key role in defining the epidemiology and pathogenesis of infection. Understanding the relationship between quantitative nasal cultures and nasal carriage patterns in healthy volunteers will allow for the development of relevant clinical diagnostics to determine patients at risk. New rapid diagnostic tests for S. aureus will enable clinicians to quickly identify those patients who may be at elevated risk of developing healthcare acquired infections due to their nasal S. aureus carriage status.
P1329 A quantitative analysis of clincal nasal swab samples testing positive for methicillin-resistant Staphylococcus aureus
L. Mermel, J. Lonks, S. Gordon, T. Perl, S. Fadem, M. Acharya, D. Dacus, S. Eells, G. Maxey, J. Cartony, D.J. Morse, M.E. Wood, P.A. Mach, C.J. Jacobson (Providence, Cleveland, Baltimore, Houston, Hudson, Delray Beach, Atlanta, St. Paul, US)
Objective: Approximately 25–30% of the population is asymptomatically colonised with S. aureus, which is an important pathogen that accounts for approximately 20% of surgical-site infections. Over the past couple of decades, antibiotic treatment of infections due to S. aureus has become more difficult because of increasing methicillin resistance. The objective of this study was to quantify methicillin-resistant Staphylococcus aureus (MRSA) on clinical nasal swab samples, which were obtained from patients known to be positive for MRSA, or suspected to be at increased risk for MRSA carriage.
Methods: This is a prospective, multi-centre US clinical study. Swab samples were collected from in-patient and out-patient populations, using a sterilised rayon swab. Samples were obtained from both nares of patients at 11 sites. Samples were analysed at a central microbiology laboratory by a standard quantitative method using a selective medium and enrichment broth. Primary isolate slants were generated from MRSA samples for strain analysis via pulse field gel electrophoresis.
Results: A total of 771 subjects were enrolled, of which 219 (31%) were positive for MRSA. Fourteen of the 219, (6%), yielded MRSA growth only by broth enrichment. Quantification of MRSA samples resulted in a mean of 2.9 logs, range 0.4–6.8 logs. MRSA strain analysis of 115 isolates identified 7 unique MRSA strains, with 72% percent identified as USA100. Sites with active surveillance yielded 81% of cultures with MRSA, due to the fact that potential carriers were flagged upon hospital admittance. For sites without active surveillance yielded 15% of cultures yielded MRSA, from potential carriers in general high risk populations.
Conclusion: Based on swab samples from both nares, there was an average of 103 logs of MRSA colony forming units (cfu's) found in patients from multiple regions of the USA. The USA100 strain predominated in patients from multiple sites. To increase confidence in the diagnoses of MRSA, it may be beneficial to include a broth enrichment step. The yield of MRSA cultures was greater in sites using active surveillance, compared to sites obtaining samples from a general high risk population.
Community-acquired genito-urinary tract infections
P1330 Prevalence of Chlamydia trachomatis and Neisseria gonorrhoeae among female university students in Novosibirsk, Russian Federation
A. Khryanin, O. Reshetnikov, A. Anpilogova (Novosibirsk, RU)
Objective: Epidemiology of sexually transmitted infections (STIs) is largely based on surveillance data. The aim of the present study was to examine the prevalence of Chlamydia trachomatis and Neisseria gonorrhoeae infections and risk factors for these infections among young women in Novosibirsk, Western Siberia.
Methods: Female university students were invited to undergo free gynecological examination. One hundred students (mean age 20.6 years, range 16–24) voluntarily participated in the study. After a confidential interview, a gynecologic examination was performed with collection of endocervical specimens. A nucleic acid amplification method (APTIMA Combo 2 assay, Gen-Probe Inc, USA) was used for N. gonorrhoeae and C. trachomatis testing.
Results: The prevalence of C. trachomatis infection was 12.0%, and N. gonorrhoeae prevalence was 2.0%. Both N. gonorrhoeae-positive females were also infected with C. trachomatis. Infected women reported younger age at first sexual intercourse (16.1 years vs 17.5 years among non-infected, p = 0.01) and a higher number of lifetime sexual partners (6.1 vs 2.8, p = 0.01). Other variables (age, new sexual partner, condom use, history of STDs etc.) did not differ between infected and non-infected subjects. Among reported symptoms, vaginal discharge (p = 0.04) and intensive menstrual bleeding (p = 0.04) were more frequent in infected women, whereas dysmenorrhea, genital pruritus, burning, pain, and dyspareunia were not.
Conclusion: The prevalence rates of C. trachomatis and N. gonorrhoeae infections in young females in Novosibirsk, Russia are higher than those previously published, possibly due to higher sensitivity of the method used in our study. As in other populations, age at first intercourse and the number of lifetime sexual partners had a significant effect on the risk of bacterial STIs.
P1331 Can we use erythromycin or clindamycin in penicillin-allergic patients with vaginal and anorectal carriage of Streptococcus agalactiae in late pregnancy?
F. Khorvash, A. Javadi, M. Izadi, S. Mobasherizadeh, K. Mostafavizadeh, M. Ahmadi (Isfahan, Tehran, IR)
Objective: The aim of this study was to establish the incidence and frequency of Group B Streptococcus (GBS) colonisation in pregnant women and to determine antibiotic susceptibility and policies for prevention of neonatal GBS infection in Isfahan, Iran.
Methods: A total of 170 pregnant women at 35–37 weeks of gestation were screened by placement of a swab in the lower third of the vagina with subsequent passage across the perineum and anus. GBS was identified via colonial morphology, beta-haemolytic, and biochemical reactions. The antimicrobial susceptibility test was performed by the agar dilution method.
Results: The prevalence of vaginal and rectal GBS colonisation was 18.2%. Colonisation rates were significantly increased among multi parity (p = 0.04), and antecedent abortion (p = 0.01). There were no significant differences between age, socioeconomic condition and GBS colonisation. All strains were susceptible to penicillin, ampicillin, vancomycin, cephazolin and imipenem. Thirteen (41.9%) isolates were resistant to erythromycin (MIC ≥ 1 g/L) and eight (25.8%) were resistant to clindamycin (MIC ≥ 1 g/L). Two strains (6.4%) showed intermediate susceptibility to clindamycin (MIC = 0.5 g/L). 87% of strains were resistant to gentamicin.
Conclusion: The colonisation rate of GBS among pregnant women in the present study is high, and on the basis of documented benefits of antenatal screening in western countries we recommend routine screening, especially for our at risk patients. According to high resistant to erythromycin and clindamycin in our population we recommended that for allergic penicillin patients use of these antibiotics should be done after identifying of sensitivity in laboratory.
P1332 Chlamydial infections of non-pregnant women in a gypsy population
R. Zafirovska, G. Bosevska, S. Panev, S. Kuzmanovska (Skopje, MK)
Objective: To assess the incidence of chlamydial infections among non-pregnant women with genital infections of gypsy population, as a part of the Program of Ministry of Health of R. Macedonia, for HIV/AIDS prevention supported by Grant of Global Fund to fight AIDS, Tuberculosis and Malaria.
Material and Methods: During period of 1 April to 1 November 2006, from 471 gynecological examined non-pregnant women with STDs problems, age of 15 to 64 years, 325 cervical samples were tested for Chlamydia trachomatis infections with ELFA VIDAS bioMérieux method.
Results: See the Table .
| Age | Gynecological examined for STDs | Cervical samples |
Chlamydia test |
||
|---|---|---|---|---|---|
| (+), ≥80 | (+/−), >60 <80 | (−), ≤60 | |||
| <19 | 30(64%) | 20(6.1%) | 0 | 0 | 20 (100%) |
| 20–25 | 87 (18.5%) | 67 (20.6%) | 4 (6%) | 0 | 63 (94%) |
| 26–40 | 218 (46.3%) | 146 (44.9%) | 6 (4.1%) | 1 | 139 (96%) |
| >40 | 136 (28.8%) | 92 (28.3%) | 1 (1%) | 2 | 89 (96.7%) |
| Total | 471 | 325 | 11 (34%) | 3 | 311 (95.6%) |
Conclusion: Low incidence of detected genital chlamydial infections, probably is a result of low prevalence (25%) of adolescent group of women (up to 25 years of age) as a result of gypsy population tradition for early marriages, average 4 births and termination of reproduction around 25 years of age.
P1333 Coinfection of genital mycoplasmas among men with gonococcal urethritis and their roles in post-gonococcal urethritis
T. Deguchi, M. Yasuda, S. Maeda, M. Tamaki, S. Ito, M. Nakano, S. Yokoi (Gifu City, Toyota City, JP)
Objectives: F We determined the prevalence of coinfection with genital mycoplasmas, including Mycoplasma genitalium, Mycoplasma hominis, Ureaplasma parvum and Ureaplasma urealyticum, among men with gonococcal urethritis (GU) and analysed roles of the genital mycoplasmas in post-gonococcal urethritis (PGU).
Methods: We examined first-voided urine samples from 390 men with gonococcal urethritis (GU) for the presence of C. trachomatis, M. genitalium, M. hominis, U. parvum and U. urealyticum, by polymerase chain reaction-based assays. The patients were treated with cefixime, ceftriaxne, or spectinomycin. PGU was judged to occur if, in spite of the eradication of Neisseria gonorrhoeae, the urethral smear showed significant numbers of polymorphonuclear leucocytes from 7 days to 14 days after treatment.
Results: C. trachomatis and/or the genital mycoplasmas were coinfected in 132 (33.8) of 390 men with culture-confirmed N. gonorrhoeae. In 85 (21.8%), C. trachomatis was detected with or without the genital mycoplasmas. One or two species of the genital mycoplasmas were detected in 15 men coinfected with C. trachomatis (3.8%) and in 47 men without chlamydia cofinfection (12.1%). Of these 47 men, M. genitalium and U. urealyticum were detected in 12 and 23, respectively. In 291 men, the eradication of N. gonorrhoeae was confirmed by culture after treatment. Of these 291 men, 103 men (35.4%) were judged to have PGU. PGU occurred in 51 (77.3%) of 66 men with chlamydia-positive GU and in 14 (46.7%) of 30 men with chlamydia-negative but mycoplasma- and/or ureaplasma-positive GU, whereas it was observed in 38 (19.5%) of 195 men with GU negative for all of C. trachomatis and the genital mycoplasmas. C. trachomatis, M. genitalium and U. urealyticum were significantly associated with PGU, but M. hominis or U. parvum was not.
Conclusion: The prevalence of coinfection with C. trachomatis among men with GU was 21.8%, whereas that with the genital mycoplasmas was 15.9%. M. genitalium and U. urealyticum were associated with PGU as well as C. trachomatis. In clinical settings, the detection of genital mycoplasmas is frequently difficult so that men with NGU should be treated presumptively with antimicrobial agents active against C. trachomatis and genital mycoplasmas.
P1334 Epidemiology of urinary pathogens in adults during 2005–2006
I. Apostolidou, M. Zoumberi, K. Papaefstathiou, E. Koloka, G. Kouppari (Athens, GR)
Objectives: To find out the frequency and antimicrobial susceptibility of urinary tract pathogens during 2005–2006.
Methods: 10,323 urine samples were examined from 9,399 patients with median age 75 years. The identification of the isolated bacteria was performed by standard methods and the API systems (bioMérieux). The antimicrobial susceptibility testing was carried out by disk diffusion method and interpreted according to NCCLS and the MIC with E-test when was needed.
Results: Positive urine cultures were found in 2,183 patients (23.2%). The microbial strains most frequently isolated from the positive urine cultures were: Escherichia coli (52.6%), Enterococcus spp. (11.1%), Pseudomonas aeruginosa (8.8%), Candida spp. (8.6%), Proteus mirabilis (6.3%), Klebsiella pneumoniae (4.6%), others (7.9%). Of Escherichia coli isolates 54.6% were resistant to ampicillin, 19.3% to amoxicillin+clavulanic acid, 25% to co-trimoxazole, 13.1% to cefaclor, 4.4% to cefotaxime, 4.8% to gentamicin, 8.9% to norfloxacin, 0% to imipenem and 13 (0.9%) produced ESBL. Of Enterococcus spp. isolates 9.7% were resistant to vancomycin and teicoplanin (VanA). In 43 patients the same strain was isolated and from blood culture (2%).
Conclusions: The most common pathogen among isolates from positive urine cultures was E. coli followed by Enterococcus spp. and Pseudomonas aeruginosa. Of E. coli isolates 0.9% produced ESBL. In 2% of the patients with urine infection there was bacteraemia and the same strain was isolated in both urine and blood cultures. Of Enterococcus spp. isolates 29 (9.7%) were resistant to vancomycin and teicoplanin (VanA).
P1335 Antimicrobial resistance of Escherichia coli urinary isolates from primary care patients in Greece
M.E. Falagas, M. Polemis, V.G. Alexiou, I. Sarantou, J. Kremastinou, A.C. Vatopoulos (Athens, Kyparissia, GR)
Objective: Most of antimicrobial susceptibility surveillance studies focus on isolates from hospitalised patients. We performed a retrospective analysis of microbiological data of the antimicrobial susceptibility of Escherichia coli urinary isolates from primary care patients in Greece.
Methods: The in vitro susceptibility to ampicillin, amoxicillin/clavulanate, cefaclor, cefprozil, trimethoprim-sulfamethoxazole (cotrimoxazole), amikacin, and norfloxacin of 2,460 E. coli isolates (01/2005–06/2005) from the urine specimens of patients tested at the laboratories of 3 Greek primary care diagnostic centres were analysed. Only the first isolates per patient (2,074 females and 386 males) were included in the analysis.
Results: The proportion of E. coli urinary isolates that were resistant to cotrimoxazole was 20.8% and 26.4% for females and males, respectively. There were noteworthy differences between age groups; 37.8% isolates from females <15 years old were resistant to cotrimoxazole compared to 21.9% and 16.7% for >45 years old and 15–45 years old females, respectively (P < 0.001). The proportion of isolates resistant to ampicillin was very high (from 32.5% to 43.3% and 31% to 63% for urinary isolates from females and males, respectively, in the different age groups examined) while it was relatively low for amikacin (up to 4.8%); 17.8% and 5.5% of the isolates from males and females, respectively, were resistant to norfloxacin (20.7% for males >45 years).
Conclusions: These findings offer help to clinicians in deciding the appropriate empirical treatment for primary care patients with urinary tract infection and emphasize the increasing problem of antimicrobial resistance in the primary care setting in Greece.
P1336 A meta-analysis of risk factors for mortality in patients with emphysematous pyelonephritis
M.E. Falagas, V.G. Alexiou, K.P. Giannopoulou, I.I. Siempos (Athens, GR)
Objectives: Emphysematous pyelonephritis (EPN), an infection most commonly seen in patients with diabetes mellitus, is associated with considerable case fatality. We sought to identify the factors associated with mortality in patients with EPN and estimate the magnitude of the associations.
Methods: Pub Med was searched to identify studies reporting on risk factors of mortality in patients with EPN. A meta-analysis of the eligible studies was performed.
Results: Seven study cohorts, representing 175 patients with EPN, were included in the meta-analysis. The overall mortality was 25%, ranging from 11% to 42%. Conservative treatment alone [odds ratio (OR) = 2.85, 95% confidence interval (CI): 1.19–6.81), bilateral EPN (OR = 5.36, 95% CI: 1.41–20.33), type I EPN (OR = 2.53, 95% CI: 1.13–5.65), and thrombocytopenia (OR = 22.68, 95% CI: 4.4–116.32) were associated with increased mortality. Systolic blood pressure <90 mmHg, serum creatinine >2.5 mg/dl, and disturbance of consciousness were also found to be associated with increased mortality, based however on limited data. On the other hand, there was no association between mortality and diabetes mellitus (OR = 0.32, 95% CI: 0.05–1.99) in patients with EPN.
Conclusion: Based on the available evidence, conservative treatment, type I EPN, bilateral EPN and thrombocytopenia are significant risk factors for mortality in patients with EPN. These data may be taken under consideration when managing patient with devastating infection.
P1337 Diabetes increases relapses and recurrences of acute urinary tract infections in adult women but does not evoke a difference in the pattern of antibiotic prescriptions in primary care
K.J. Gorter, D. van Oostveen, E. Hak, N.P.A. Zuithoff, A.I.M. Hoepelman, G.E.H.M. Rutten (Utrecht, NL)
Objective: Women with diabetes (DM) have an increased risk for urinary tract infections (UTI). Aim was to assess diabetes care associated risks of recurrent episodes of UTI in adult women with diabetes (DM) in primary care and the pattern of prescribed antibiotics.
Design: Retrospective study. Setting: All women of 30 years or over registered in two primary healthcare centres (n = 22,000 patients) during a period of 43 months (1998–2001).
Patients: We compared incidence rates of single and recurrent urinary tract infections (UTI) in women with diabetes (n = 340) and women without diabetes (n = 6,618).
Main outcome measures: Independent risk of diabetes characteristics, indicators of care and complications for relapsed and recurrent UTI; odds ratio with 95% confidence interval [OR; 95% CI]. The diabetes–no-diabetes associated pattern of antibiotic prescriptions.
Results: Of the women with diabetes and those without diabetes relapses and (recurrences) were reported in 7.1 (16.9%) and 2.0 (4.2%) respectively. After adjustments in a multivariate logistic regression analysis for age, socio-economic status and history of vaginitis, the risk of a recurrent episode was 2.0 times higher in women with diabetes than in controls (95% CI: 1.4–2.9). Risks were higher in women who used oral hypoglycaemic medication (odds ratio [OR]: 2.1; 95% CI: 1.2–3.5), insulin (3.0; 1.7–5.1), who had >5 years diabetes (2.9; 1.9–4.4) and who had retinopathy (4.1;1.9–9.1). A similar pattern of antibiotics was prescribed in women with and without diabetes, both for one episode of UTI and for relapses and recurrences.
Conclusions: Women with diabetes of longer duration, on treatment and with retinopathy have an elevated risk for a recurrent episode of acute symptomatic UTI's, but this does not evoke difference in the pattern of antibiotic prescriptions. Research focussing on preventive and antibiotic management of UTI is needed in women with DM.
Epidemiology of multi-drug resistant Gram-negative organisms
P1338 Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a university hospital, Split, Croatia
I. Goic-Barisic, B. Bedenic, M. Tonkic, S. Katic, A. Novak, S. Kalenic, V. Punda-Polic (Split, Zagreb, HR)
Objectives: Acinetobacter baumannii is an opportunistic pathogen that is frequently involved in outbreaks of infection, occurring mostly in intensive care units. Carbapenem resistance is now being reported increasingly in A. baumannii isolates in association with the production of carbapenem-hydrolysing class D β-lactamases or oxacillinases that have now emerged worldwide. The aim of the present study was to analyse and compare genotypes of clinical isolates of carbapenem resistant Acinetobacter baumannii collected from three different Intensive Care Units in University Hospital Split, Croatia.
Methods: During 2004, twenty-two non-repetitive A. baumannii isolates with an unusual resistance profile were obtained from patients hospitalised at three different Intensive Care Units (two adults ICU and one children ICU) inside University Hospital Split. All collected isolates of A. baumannii displayed intermediate (MIC > 8 mg/L) or resistant (MIC> 16 mg/L) profile to imipenem and/or meropenem. Minimum inhibitory concentrations were also determined for ceftazidime, cefepime, ceftriaxone, amikacin, gentamicin, ciprofloxacin and piperacillin-tazobactam by broth microdilution according to CLSI (formerly NCCLS) recommendation. All isolates were multidrug-resistant exhibiting high resistance to tested antimicrobials. The isolates of A. baumannii were genetically characterised using pulsed-field gel electrophoresis (PFGE). Strain typing was performed by macrorestriction analysis of chromosomal DNA by use of PFGE (Apa I enzyme, in a CHEF DR III drive module).
Results: We report the clonal dissemination of pulsotype A between two different adult intensive care units in University Hospital Split, belonging to the same pulsed-field gel electrophoresis (PFGE) profile, probably by hospital staff during medical procedures. The strain characterised as pulsotype B was the only strain isolated from children intensive care unit without expanding inside the hospital.
Conclusion: The infection control team of the hospital implemented restriction of carbapenem usage and strict antiseptic techniques, which included the rigorous use of alcohol-clorhexidine solutions before and between patient and equipment contact and before leaving the units. Consequently, incidence and spread of multidrug-resistant A. baumannii nosocomial infections suggest the necessity of a surveillance programme and enforcing adequate control measures in different hospital settings.
P1339 Molecular characterisation of carbapenem-resistant Acinetobacter baumannii isolated in two Turkish medical centres in 2006: report from the SENTRY Antimicrobial Surveillance Program
L. Deshpande, H. Sader, D. Gur, V. Korten, G. Soyletir, R. Jones (North Liberty, US; Ankara, Istanbul, TR)
Objectives: To evaluate the mechanisms of resistance (R) to carbapenems (CARB) and the epidemiologic typing of CARB-R Acinetobacter spp. (ASP) isolated in Turkey through the SENTRY Program in 2006. We also evaluate the antimicrobial susceptibility (S) of ASP strains collected in Turkey over the last 7 years (2000–2006).
Methods: A total of 304 ASP strains were submitted by two Turkish medical centres (MCs; Ankara and Istanbul) from 2000 to 2006, 57 in 2006. The isolates were mainly from bloodstream (64.1%) and respiratory tract (26.0%) infections and were S tested by reference broth microdilution methods to >30 antimicrobials according to CLSI guidelines (2006). Strains R to imipenem and meropenem (MIC > 16 mg/L) collected in 2006 were screened for production of carbapenemases (IMP, VIM, SPM and OXA groups) by PCR and epidemiologically typed by PFGE.
Results: The most active antimicrobials overall were: polymyxin B (99.2% S), imipenem (51.3% S), meropenem (51.0% S) and tobramycin (42.4% S). S to imipenem decreased from 80.4% in 2000 to only 29.8% in 2006. Forty of 57 (70.2%) ASP strains collected in 2006 were CARB-R with high rates of cross-R to all other antimicrobials tested, except polymyxin B (100.0% S) and tigecycline (100.0% inhibited at ≤2 mg/L), and 23 of those were randomly selected for molecular characterisation. All 23 strains were PCR-positive for blaOXA-51, indicating A. baumannii. Five strains were from one site and these strains showed identical or similar PFGE patterns and were PCR-positive for blaOXA-58. The remaining 18 CARB-R strains also showed PFGE pattern identical/similar to each other and distinct from that of MC 068. All 18 strains had PCR positive results for blaOXA-23.
Conclusions: Clonal dissemination of R strains caused a significant increase in the prevalence of CARB-R ASP in the Turkish medical centres evaluated by the SENTRY Program. CARB-R was found to be largely driven by OXA-type carbapenemases, OXA-58-type being prevalent in Ankara and OXA-23-like in Istanbul. Polymyxin B and tigecycline represent the only antimicrobials with reasonable in vitro activity against ASP in the Turkish MCs evaluated.
P1340 Comparative results of the in vitro efficacy of colistin and carbapenems against multidrug-resistant strains of Acinetobacter baumannii isolated from clinical specimens
V. Koulourida, A. Papa, M. Moskophidis (Thessaloniki, GR)
The infectious diseases caused by Acinetobacter baumannii are severe and need instant and proper therapy. Since 1970, the constantly increasing resistance of A. baumannii strains to antibiotics, sets up a great problem, as the “weapons” for the treatment of these infections have been decreasing dramatically.
The aim of the present study was to determine the in vitro sensitivity of imipenem and meropenem, as well as colistin.
Material and Methods: Material consisted of 109 non replicated MDR strains A. baumannii, isolated from patients from various clinics of Papageorgiou General Hospital in Thessaloniki during the period 2001–2006: 50 from the ICU, 35 from the surgery wards and 24 from the pathological wards. Strains were isolated from various clinical specimens: 22 from blood cultures, 23 from respiratory samples, 14 from intravenous catheters, 10 from urines, 26 from surgery wounds, 14 from other materials. For the culture of the specimens MacConkey agar, Mueller Hinton agar and Blood agar were used. Identification and determination of susceptibility to 20 antibiotics were performed by the automatic system WIDER using the MIC/ID Gram-negative panels. The MIC in colistin, imipenem and meropenem, was determined, by the dilution of antibiotics in agar method in according the NCCLS standards. PCR for the integrase genes int1, int2 and int3 was performed in al the strains.
Results: Only 15/109 (14%) strains of A. baumannii were sensitive to imipenem, and 2/109 (2%) were sensitive to meropenem. The resistance in the other antibiotics ranged from 87 to 100%. All the strains were found in vitro sensitive to colistin. Concerning integrons, 89% of the strains were carrying the int1 gene, while int2 and int3 were not detected.
Conclusions: Most of the A. baumannii strains tested were resistant to carbapenems, while all of them were sensitive to colistin, suggesting that clinicians have to take these results into consideration for the antibacterial treatment.
P1341 Emergence of carbepenem-resistant Acinetobacter baumannii in a Bulgarian university hospital
R. Dobrewski, T.J.K. van der Reijden, E. van Strijen, E. Keuleyan, H. Hitkova, E. Savov, M. Lesseva, P.J. van den Broek, L. Dijkshoorn (Sofia, Pleven, BG; Leiden, NL)
Objectives: From 2000 to 2002 a predominant multidrug resistant (MDR) but carbapenem susceptible Acinetobacter baumannii strain was identified in the Military Medical Academy (MMA), a teaching hospital in Sofia, Bulgaria. Since, carbapenem resistant A. baumannii has emerged in this hospital and in other Bulgarian hospitals. The purpose of our study was to investigate whether this phenomenon was due to the spread of a single strain.
Methods: A total of 72 MDR A. baumannii isolates from 72 patients was investigated including 58 isolates from the MMA and 14 from three other hospitals (A–C). Sixty-six isolates were carbapenem resistant; six were carbapenem susceptible. The 58 MMA isolates comprised 53 carbapenem resistant and five carbepenem susceptible isolates from epidemic episodes from 2004 to 2006. The 14 isolates from the hospitals A–C were from 2005 to 2006; all but one were carbapenem resistant. Presumptive identification and antibiotic susceptibility determination was done with the VITEK 2 system (bioMérieux). AFLP analysis was used to identify the organisms at species, clone and strain level using clustering levels of 50%, 80% and 90% respectively.
Results: All isolates were identified to A. baumannii as they clustered with A. baumannii reference strains at 50% or above. At the strain delineation level of 90%, the isolates were allocated to seven clusters and a single strain, coded 1–8. Two major clusters, no. 1 and no. 7 included 47 and 11 isolates respectively. Five other clusters contained 2–4 isolates each. Eleven isolates from hospital A–C grouped together in the major cluster 1 with isolates from the MMA; three other isolates from hospital A–C were in cluster 3, 4 and 7, respectively. Thus, there was no clear correlation between AFLP clustering and the hospitals of origin. The four isolates of cluster 4 belonged to EU clone II; three were carbapenem resistant, one was susceptible. The four carbapenem resistant isolates that grouped together in cluster 5 belonged to EU clone I. None of the currently found strains corresponded to the strains found in the MMA from 2000–2002.
Conclusion: Eight different carbapenem resistant A. baumannii strains were distinguished by AFLP fingerprint analysis. Most isolates belonged to two predominant clusters and were considered to represent two major strains. In particular one strain (corresponding to AFLP cluster 1) was widely prevalent in the MMA and was also in the three other hospitals.
P1342 Molecular differentiation of multidrug-resistant Acinetobacter baumannii clinical strains in a Greek hospital
A. Papa, V. Koulourida, M. Moskophidis, A. Antoniadis (Thessaloniki, GR)
Objectives: Acinetobacter baumannii is an important cause of nosocomial infections especially among critical ill patients. The number of multidrug-resistant (MDR) strains of A. baumannii is increasing, resulting in great problems for choosing the proper treatment. Aim of the present study was the molecular typing of MDR A. baumannii strains isolated during the last year in a tertiary Greek hospital.
Material and Methods: Thirty-five non-replicated multiresistant A. baumannii strains isolated from patients from various wards of Papageorgiou General Hospital were tested: 14 patients were hospitalised in the ICU, 11 in the internal medicine wards and 10 in the surgery wards. Susceptibility to 20 antibiotics was tested by microdilution method. RAPD and PFGE using ApaI were used for molecular typing of the strains. In addition, PCR was applied for the detection of integrase genes int1, int2 and int3.
Results: The 35 strains were divided into three clonal groups using RAPD and PFGE. The two methods gave similar results. The first group consisted of 18 strains, the second group of 12 strains, while the third one of 5 strains. Strains of the first two groups were dispersed in all wards, while the majority (80%) of the strains of the third group was isolated from the internal medicine ward. All strains presented similar multi-resistant patterns, with the exception of the third group which was susceptible to aminoglycosides. Class 1 integrons were missing from the strains of the third group, while it was present in all other strains. Class 2 or 3 integrons were not detected.
Conclusions: Three different clonal groups of A. baumannii strains are present in the hospital during the last year. The main difference among them is the resistance to aminoglycosides combined with the presence of intI gene in strains of two of the three groups. Results of this study gave an insight into the molecular diversity of multiresistant A. baumannii strains isolated in Papageorgiou hospital during the last year.
P1343 Molecular epidemiology of pandrug-resistant Acinetobacter baumannii infection at a university hospital in Taiwan
L.H. Su, T.L. Wu, J.H. Chia, A.J. Kuo, C.F. Sun (Taoyuan, TW)
Objectives: Acinetobacter baumannii has constituted as an important clinical pathogen causing an increasing number of respiratory infections at the Chang Gung Memorial Hospital, Taiwan. Moreover, approximately 50% of the isolates were multidrug-resistant A. baumannii (MDRAB) that were multiply resistant to all antibiotics commonly used to treat infections caused by Gram-negative bacteria except imipenem. The incidence of pandrug-resistant A. baumannii (PDRAB), which is resistant to all antibiotics including imipenem, had been kept low at 2.7% before 2002, but a sudden increase to 16%-20% was noticed during 2003–2006.
Methods: A total of 597 isolates of A. baumannii collected from 287 patients during 1998–2006 were retrospectively studied. Antimicrobial susceptibilities of A. baumannii isolates to amikacin, aztreonam, ceftazidime, ciprofloxacin, cefepime, gentamicin, imipenem, and piperacillin were analysed by a standard disk diffusion method and interpreted according to the criteria suggested by the Clinical and Laboratory Standard Institutes. Genotypes of the isolates were determined by pulsed-field gel electrophoresis (PFGE) and infrequent-restriction-site PCR (IRS-PCR).
Results: A total of 303 (50.8%) PDRAB isolates and 135 (22.6%) MDRAB isolates were identified from the 597 isolates studied. Genotyping analysis by PFGE and IRS-PCR produced compatible results and 82 genotypes, including 17 from the PDRAB isolates and 13 from the MDRAB isolates, were identified. The majority (78.4%) of the isolates belonged to a predominant genotype 7, the prevalence of which was significantly higher among the PDRAB (93.4%) and MDRAB (88.1%) isolates compared to isolates of other antibiograms (41.5%; P<0.00000005). Among 50 of the 56 patients from whom multiple isolates of different antibiograms were available for comparison, genotypes of their isolates remained the same when MDRAB became PDRAB (in 38 patients) or vice versa (in 22 patients) during their hospital stay.
Conclusions: The recent upsurge of PDRAB isolates at this hospital was closely associated with a predominant clone, genotype 7, which had been prevalent among MDRAB isolates prior to the PDRAB era. During the hospital stay, MDRAB from the same patients may become PDRAB, some of which may subsequently turn back to MDRAB again. Infection control strategies to contain PDRAB infections may not be effective if patients with imipenem-susceptible MDRAB infections were not included in the isolation policies.
P1344 Multidrug-resistant Acinetobacter baumannii susceptible only to colistin outbreak in a cardiac surgical intensive care unit
A. Mastoraki, E. Douka, I. Kriaras, G. Stravopodis, G. Saroglou, H. Manoli, S. Geroulanos (Athens, GR)
Objectives: The aim of this study was to determine the incidence and mortality of Multi-Drug Resistant Acinetobacter baumannii (MDR-AB) in patients undergoing cardiac surgery, to elucidate the effectiveness of treating them with colistin and to identify if the additional measures to the recommended procedures were able to control the dissemination of MDR-AB isolates in our institution.
Methods: A prospective study was conducted among 1,451 patients who were submitted to cardiovascular surgery from 1 September 2005 to 31 August 2006. All case histories of infected patients were objected to meticulous analysis. We reviewed the prophylactic measures of the SICU and implemented a two scale multiple programme. Scale 1 included classical infection control measures, i.e. strict contact and droplet isolation, surveillance of throat, nasal and anal flora for MDR pathogens on all patients transferred from other hospitals, separate nursing staff for each infected or colonised case and strict antibiotic policy, while Scale 2 referred to the geographic isolation of all positive MDR-AB cases in distinct hospital units with exclusive medical and nursing personnel, use of separate supplies and facilities and environmental intense surveillance.
Results: Among 121/1,451 infected patients 15 were colonised by strains of MDR-AB susceptible only to colistin. All patients were mechanically ventilated; 13 presented respiratory tract infection, 1 patient suffered deep surgical site infection and 1 patient catheter related infection. Transmission of the pathogen into the hospital occurred via 2 patients transferred from 2 other institutions. They were all treated with colistin. Cure or clinical improvement was observed only in 4 patients (27%). Scale 1 measures were implemented for the whole 12-month period while Scale 2 for 2 separate periods of 3 weeks. Environmental specimens (n > 350) sampled on several occasions proved negative.
Conclusions: The increasing prevalence of MDR-AB in SICU patients creates demand on strict screening and contact precautions. Following this infection control strategy we were able to achieve intermittent eradication of the pathogen during a 12-month period with continuous function of the SICU. Despite the significant “in vitro” activity of colistin against MDR-AB the results were discouraging. Due to the significant mortality of MDR-AB infected patients additional measurements are highly recommended.
P1345 Multidrug-resistant Acinetobacter baumannii prosthetic joint infection treated with colistin
A. Rodríguez Guardado, V. Asensi, M. Lantero, M. Castillo, J. Carton (Oviedo, ES)
Background: Description of the characteristics of the prosthetic joint infection by A. baumannii treated with colistin emphasizing the factors influencing their outcome
Methods: All the episodes of prosthetic joint infection by multidrug-resistant A. baumannii diagnosed between 2004–2006 treated with colistin were retrospectively reviewed. The Acinetobacter isolate was defined as MDR if it was resistant to ≥ 3classes of antimicrobial agents as tested by commercial system. The dose of parenteral colistin was 160 mg/8 h.
Results: Eight cases were reviewed. All the patients had underwent surgery. Five patients were carriers of a knee prosthesis and the rest hip prosthesis. The mean time between the surgery and the onset of the infection was 26.5 days. Five patients had deep wound cultures and the rest had bone tissue cultures. Three patients had mixed infections [methicillin-resistant Staphylococcus aureus (two cases), E.faecium (one case)] coccus aureus (and case each). All the patients had received previous antibiotic treatment which were inadequate in all cases Four patients received IV colistin (160 mg/8 hours) monotherapy. Three received a combined IV therapy with colistin and vancomycin. One patient development a infection by a A. baumannii resistant to colistin and he was treated with a combination of IV colistin 160 mg/8 hours, IV rifampicin (600 mg/day) and IV imipenem (1 g/8 hours) with good evolution. In 3 cases the treatment was associated with removal of the prosthesis and in the rest a surgical drainage was performed. The mean of treatment was 59.7 days. The follow-up period was 9 months (range 6–18 months). Only one patient died a consequence of the infection and the rest cured. The renal function was normal in all the cases.
Conclusions: The use of intravenous colistin is safe and effective treatment for prosthetic joint infection due to multiresistant A. baumannii.
P1346 Seventy Enterobacter cloacae bloodstream infections in central Taiwan during 2001 and 2003: evaluation of the roles of extended-spectrum β-lactamases and class 1 integron
C. Chen, Y-J. Chang, C-E. Liu, C-C. Huang (Changhua, Taichung, TW)
Objectives: Enterobacter cloacae (E. cloacae) bloodstream infection is an important cause of morbidity and mortality, with an increasing incidence at Changhua Christian Hospital (CCH) of Central Taiwan since 1995. The development and spread of the extended-spectrum β-lactamases (ESBLs) is one of the most important issues in infection control. Jiang et al. disclosed specific clonally related E. cloacae isolates possessed a novel type β-lactamase, which was located in an integron. So, we designed this study to investigate the clinical epidemiology of E. cloacae and to elucidate the roles of the ESBLs and integron.
Methods: We analysed the clinical and microbiological data from E. cloacae bloodstream infection during January 1st 2001 to December 31st 2003 to delineate the epidemiology as an aid in control. Antimicrobial susceptibility testing for confirmation of ESBL was performed on the VITEK 2 System (bioMérieux, Hazlewood, MO). Koeleman's previously described method was used to amplify the target IntI1 genes. We also performed the pulse field gel electrophoresis (PFGE).
Results: Seventy E. cloacae strains isolated from 70 patients (56 nosocomial infections) between January 1st 2001 and December 31st 2003 were subjected to standard microbiological testing and to methods of class 1 integron detection and PFGE analysis. Case records were reviewed and statistical analysis was carried out using the chi-square test and Fisher's exact test. The significant difference was found between ESBL-positive and ESBL-negative isolates with regard to risk factors, including the diseases severity (p = 0.03), category of nosocomial infection (p = 0.04), prior use of antibiotics (p = 0.023), and prior use of a ventilator (p = 0.037). Also, there was a significant difference in mortality among two groups (p = 0.004; OR = 4.750; 95% CI: 1.573–14.344). The predominant clone A in ESBL-positive strains was associated with a higher mortality rate but not with the presence of the integron.
Conclusion: The study disclosed four major risk factors for ESBL-positive E. cloacae bloodstream infection and the predominant clone A in ESBL-positive strains for higher mortality. It is necessary to review antibiotic prescription practices, and we needed to consider ESBL-producing strains in empirical treatment of nosocomial bloodstream infection.
P1347 Prevalence of multiple-resistant Escherichia coli strains in southern and eastern Mediterranean countries
M.A. Borg, E.A. Scicluna, M. de Kraker, N. van de Sande-Bruinsma, E.W. Tiemersma, J. Monen, H. Grundmann and ARMed Project Collaborators
Objective: Resistance to Escherichia coli is fast becoming a major challenge in many countries. The Antibiotic Resistance Surveillance and Control in the Mediterranean Region (ARMed) project [www.slh.gov.mt/armed] provided a first time opportunity for a longitudinal multi-year study of trends of antimicrobial resistance amongst this species within countries of the southern and eastern Mediterranean.
Methods: ARMed collected routine susceptibility test results of more than 4,000 invasive isolates of E. coli from blood cultures tested by laboratories in Algeria, Cyprus, Egypt, Jordan, Lebanon, Malta, Morocco, Tunisia and Turkey. Identical protocols were used as those of the European Antimicrobial Resistance Surveillance System (EARSS), including testing of aminopenicillin, third generation cephalosporins (3GC), fluoroquinolones (Fq) and aminoglycosides.
Results: The proportion of aminopenicillin resistant E. coli in 2005 varied between 49% (Malta) and 83% (Lebanon), with a significant increase observed in Turkey: 68% to 75% between 2003 and 2005. The highest proportion of aminoglycoside resistance was observed in Egypt (57% in 2005), whereas the lowest proportion was reported by Malta (7%). Between 2003 and 2005 a significant increase was seen in Morocco from 12% to 33%, and a significant decrease was reported by Malta from 18% to 7%.
Resistance to 3GC varied widely from 1% in Malta to 70% in Egyptian centres in 2005. Between 2003 and 2005, Morocco and Turkey showed a significant increase in resistance from 2% to 28% and 26% to 31% respectively. Levels of Fq resistance in excess of 40% were reported by Egypt and Turkey where a significant increase was observed.
In excess of 15% of isolates from Turkey, Morocco, Jordan and Egypt were multi-resistant to all 4 classes of antimicrobials. The Egyptian centres, in particular, showed consistent increase in multiresistant E. coli during the 3 years of the project to reach a level in excess of 38% in the final year.
Conclusion: Several southern and eastern countries in the Mediterranean show evidence of significant resistance within E. coli, especially to 3GC and Fq, which are considerably higher than that reported by EARSS in the European counterpart countries of the same region. Of particular importance appear to be strains which are multiresistant to several major antimicrobial groups and which can potentially spread outside of the region due to the heavy human traffic in this area of the world.
P1348 Prevalence of an epidemic ESBL-producing Escherichia coli strain in LTCFs in Belfast
A. Loughrey, P. Rooney, M. O'Leary, M. McCalmont, M. Warner, E. Karisik, P. Donaghy, B. Smyth, N. Woodford, D. Livermore (Belfast, London, UK)
Objective: ESBL-producing E. coli (ESBLEC) were isolated rarely in the Belfast area prior to 2003, but have since increased, particularly from residents of long-term care facilities (LTCFs). We investigated the epidemiology of ESBLEC among LTCF residents.
Method: LTCFs in the Belfast area were invited to participate. Residents consented to supply medical data and a single sample of faeces for ESBLEC culture. To qualify for inclusion the LTCFs had to provide complete data plus a faecal sample from at least 10 residents between July 05 and September 06. Samples were screened on CLED agar containing 1 mg/L ciprofloxacin with a cefpodoxime disc. Suspected ESBLEC colonies were confirmed as E. coli on TBX agar and ESBL production was confirmed by double disc synergy testing. Antibiotic susceptibilities were determined and interpreted using BSAC guidelines. Isolates were grouped broadly by antibiogram. PCR was used to screen selected isolates for an IS26-blaCTX-M link, which is characteristic of CTX-M-15 ESBL-producing UK epidemic strain A.
Results: Overall, 120/307 (39%) samples from 13 homes yielded at least one ESBLEC; range, 0% (2 LTCFs) to 75% (18 of 24 positive samples from 1 LTCF). Sixty (50%) of 120 ESBLEC-positive residents had no hospital admissions since January 04. The majority of ESBLEC had phenotypes consistent with production of a CTX-M enzyme. Isolates assigned presumptively to strain A by PCR accounted for 59/120 (49%) ESBLEC. Although distinct from strain A, most of the other 61 isolates also produced a group 1 CTX-M ESBL; these isolates had varying antibiograms, suggesting multiple strains. In the eastern district of Belfast, 50/175 samples were ESBLEC-positive, and 38 (76%) of these were strain A; in the other districts 70/132 samples were positive, but only 22 (31%) were strain A. The proportion of strain A isolates varied widely in different LTCFs, ranging from 0/11 ESBLEC in one centre to 9/9 in another.
Conclusions: Epidemic strain A was the predominant ESBLEC strain among LCTF residents in Belfast. ESBLEC were found in many residents with no history of recent hospital admission. LTCF residents requiring hospital admission tended to be referred to their nearest hospital. Differences in the distributions of strain A and other ESBLEC in LTCFs from particular districts are consistent with acquisition in hospital and introduction into the LTCF on discharge. Our study highlights the importance of spread within LTCFs in the epidemiology of ESBLEC.
P1349 Risk factors for ciprofloxacin resistance among ESBL-producing Escherichia coli isolated from non-hospitalised patients in Spain
J. Alcalá, J. Rodríguez-Baño, J. Cisneros, C. Llanos, R. Cantón, F. Grill, G. Navarro, M. Cuenca, C. Peña, M. Esteve, A. Oliver, B. Mirelis, M. Almela, T. Tórtola, A. Pascual for the Red Española de Investigación en Patología Infecciosa (REIPI)
Objectives: ESBL production is frequently associated with quinolone (QUIN) resistance. We studied the risk factors associated with ciprofloxacin (CIP) resistance among ESBLEC isolated from non-hospitalised patients in Spain.
Methods: A double case-control study performed in 11 centres in Spain. Cases: non-hospitalised patients with isolation of ESBLEC in a clinical sample. Controls: 2 per case, non-hospitalised patients without ESBLEC. Variables analysed: demographics, comorbidities, healthcare relation and previous antimicrobials. Susceptibility was assessed by microdilution (NCCLS). ESBL were characterised by isoelectric focusing, PCR and sequencing.
Results: We included 121 cases. In 78 cases (68%) ESBLEC strains were resistant to CIP (MIC50 = 4 mg/mL, MIC90 = 64 mg/mL). Resistance to CIP was higher among strains also resistant to co-amoxiclav (AMC) (89% vs 60%, p = 0.001), co-trimoxazol (SXT) (80% vs 50%, p = 0.001), gentamicin (GNT) (85% vs 65%, p = 0.06) and tobramycin (TOB) (92% vs 65%, p = 0.04). Resistance to CIP was more frequent among patients with renal insufficiency (100% vs 65%, p = 0.012). No significant differences were found with respect to previous antimicrobial use, including QUIN, or with the type of ESBL produced. When compared to controls, the following risk factors (OR, 95% CI) for CIP-resistant ESBLEC were found: female gender (2.32, 1.30–4.16), renal insufficiency (3.12, 1.30–7.49), diabetes mellitus (5.52, 2.80–10.87), recurrent UTI (7.88, 4.43–14.02), healthcare relation (2.11, 1.24–3.58), urinary catheterisation (5.11, 2.12–12.29), previous use of QUIN (5.79, 3.15–10.64) and previous use of AMC (7.97, 3.66–17.36). Risk factors for CIP-susceptible ESBLEC were: recurrent UTI (4.15, 1.09–9.08), previous use of QUIN (4.73, 2.09–10.71) and previous use of AMC (5.44, 1.95–15.22).
Conclusions: Resistance to CIP among ESBLEC from non-hospitalised patients is associated with resistance to other antimicrobial agents, such as AMC, SXT, GNT and TOB. There is association with renal insufficiency but no association with any other variable, including previous use of QUIN. Previous use of antimicrobial agents and recurrent UTI are risk factors for both CIP-resistant and CIP-susceptible ESBLEC; and female sex, diabetes mellitus, renal insufficiency, healthcare relation and urinary catheterisation are risk factors just for CIP-resistant ESBLEC. More studies on the epidemiology of this organism are needed to identify factors related to quinolone resistance.
P1350 Study of an outbreak of imipenem-resistant Klebsiella pneumoniae infections in the intensive care unit of a Greek hospital
G. Totos, K. Pappas, V. Mpekos, S. Vourli, M. Polemis, A. Vatopoulos (Athens, GR)
Objectives: Infections due to imipenem resistant K. pneumoniae, due to the production of metaloenzymes, particularly in the ICU environment, represent an emerging major Public Health problem in Greece. In this report we describe an outbreak of infections due to imipenem resistant K. pneumoniae that occurred in Athens Naval Hospital from May 2006 to June 2006 and involved six patients, three of whom eventually died.
Methods: K. pneumoniae strains were isolated from all patients; three strains were isolated from bronchial secretions, one from blood, one from a central venous catheter and one from urine. Hand cultures were performed on all ICU personnel and samples from various environmental sources were also collected. All strains were identified and tested for susceptibility using the VITEK® 2 system (bioMérieux, France). Metallo-β-lactamase (MBL) and extended-spectrum β-lactamase (ESBL) production were screened by the imipenem+EDTA disk synergy test and the double disk synergy test, respectively. The presence of genes of the blaVIM family was detected by PCR. PFGE (Xba I) was used for typing.
Results: All isolates exhibited the same antibiotic resistance profile and were found resistant to all major classes of antibiotics and sensitive only to colistin. The imipenem+EDTA disk synergy test was positive in all six isolates and all isolates were found to harbour the blaVIM gene. All six isolates were also ESBL positive. PFGE revealed that five out of the six isolates were indistinguishable, whereas the sixth seemed genetically different. None of the staff members harboured imipenem-resistant K. pneumoniae and all samples from environmental sources were tested negative.
Upon recognition that the outbreak is possibly due to the spread of a single clone of K. pneumoniae in the ICU, strict infection control measures were implemented which resulted in no further, so far, isolation of imipenem-resistant K. pneumoniae.
Conclusion: The emergence of MBL producing K. pneumoniae infections is a new threat that can lead, particularly in the ICU, to serious infections and increased mortality. Early recognition of the problem, collaboration of Clinicians and Laboratory personnel and timely application of infection control measures are imperative for addressing this problem.
P1351 Comparison of phenotypic methods for identification of AmpC β-lactamase-producing Escherichia coli and Klebsiella spp. clinical isolates
A. Poulou, F. Markou, V. Kolliou, A. Karakolios, T. Kallinikidis, P. Rozi (Serres, Athens, GR)
Objectives: Early and accurate detection of the resistance mediated by class C β-lactamases in Escherichia coli and Klebsiella spp. is of utmost importance for the proper antibiotic therapy but also for providing infection control information. We compared three phenotypic methods for the detection of AmpC-producing E. coli and Klebsiella spp. strains in a region with increasing incidence of plasmid-mediated class C enzymes.
Methods: A total of 2,023 E. coli and 235 Klebsiella spp. nonduplicate isolates were consecutively recovered from clinical infections. Cefoxitin insusceptibility (MIC >8 μg/mL) was used as a screen for plasmid-mediated AmpC enzymes. The three dimensional test, the AmpC disk test with EDTA disks and the boronic acid disk test were used for the detection of AmpC enzymes. Additionally, the DDST was used for detection of ESBLs according to CLSI guidelines.
Results: Forty-seven E. coli and 25 Klebsiella spp. isolates with reduced susceptibility to cefoxitin were further tested with phenotypic confirmatory methods. The three dimensional test revealed AmpC enzymes in 21 (29.1%), the AmpC disk test in 30 (41.6%) and the boronic acid disk test in 36 (50%) isolates. In addition, the latter test in accordance with the DDST confirmed coexistence of ESBL in 26 of 36 (72.2%) AmpC-producing isolates. Interestingly, 13 of the 36 (36.1%) AmpC-producing isolates were recovered from community-acquired infections. Antimicrobial resistance rates among the AmpC producers were: 55.5% to amikacin, 63.8% to ciprofloxacin and 72.2% to co-trimoxazole. Resistance rates among the AmpC non-producers were: 9.2% to amikacin, 12.9% to ciprofloxacin and 17.8% to co-trimoxazole.
Conclusions: Detection of the resistance mediated by class C enzymes is a challenging issue, particularly when plasmid-mediated AmpC β-lactamases have been disseminated in the community, as it was detected herein. In our study, the three dimensional test was the most time consuming, the AmpC disk test was the easiest to be interpreted while the boronic acid disk test was the most sensitive assay and allowed the simultaneous detection of ESBLs, which in our population was associated with multiple antibiotic resistances.
P1352 Influx of metallo-β-lactamase- and AmpC-producing Klebsiella pneumoniae clinical isolates in a regional hospital
A. Poulou, F. Markou, S. Pournaras, S. Sideri, A. Tsakris, P. Rozi (Serres, Larissa, Athens, GR)
Objectives: Acquired metallo-β-lactamases (MBLs) have been recently emerged in Klebsiella pneumoniae clinical isolates in our region and elsewhere. Such strains are mainly detected among ICU patients, have reduced susceptibility to carbapenems and constitute the cause of nosocomial infections with high rates of mortality. We report the emergence of K. pneumoniae clinical isolates that produce both MBLs and AmpC β-lactamases in a Greek hospital.
Methods: During a two month period (August–September 2006), four K. pneumoniae isolates were recovered from blood and/or bronchial samples of separate patients hospitalised in the ICU (two cases) and in medical wards (two cases). The latter two patients have been previously hospitalised in the ICU and recently transferred to the medical wards. Identification of the isolates and antimicrobial susceptibility testing was performed by Microscan system (Dade Behring). MBLs were phenotypically detected with the E-test MBL (AB Biodisc) and the Double Disc Synergy Test using disks of IPM and EDTA, while AmpC were detected with the three dimensional test and AmpC disk test. All isolates were screened for blaVIM and blaCMY genes using PCR assays. Random amplified polymorphic DNA (RAPD) was used for typing.
Results: All four isolates exhibited resistance to imipenem, meropenem, broad spectrum cephalosporins, cephamycins, fluoroquinolones, aztreonam and amikacin but susceptibility to gentamicin, colistin and tigecycline. In all cases, the phenotypic methods showed production of MBL and AmpC β-lactamases, while PCR confirmed the presence of blaVIM and blaCMY genes. RAPD analysis proved the dissemination of a single clonal strain. One of the patients died from septicaemia despite the therapeutic treatment with gentamicin and colistin.
Conclusions: The genetic similarity of the isolates in conjunction with the history of previous ICU hospitalisation suggest that a single MBL- and AmpC-producing K. pneumoniae strain has been disseminated within the ICU and subsequently in other hospital wards. Continuous surveillance and accurate recognition of resistance phenotypes are needed for the early detection of new patterns of antimicrobial resistance and new combinations of antimicrobial resistance genes.
P1353 Emergence of metallo-β-lactamase-producing Proteus mirabilis in the community setting
A. Poulou, F. Markou, V. Kolliou, H. Iliopoulou, A. Tsakris, P. Rozi (Serres, Athens, GR)
Objectives: Metallo-β-lactamases (MBLs) constitute a growing class of β-lactamases that readily hydrolyse most β-lactams including carbapenems. Acquired MBLs have emerged among nosocomial Pseudomonas aeruginosa isolates and more recently among other Gram-negative pathogens such as Acinetobacter baumannii and several enterobacterial species. However, their dissemination in the community setting has not been described. In the present study we report the emergence of MBL-producing Proteus mirabilis recovered from outpatients in a rural area.
Methods: A total of 202 P. mirabilis isolates were consecutively recovered from patients with community-onset clinical infections (87% from urinary tract infections) in a rural Greek region. Identification and susceptibility testing was performed with the Microscan system (Dade Behring). Among isolates with reduced susceptibility to imipenem (MIC> 4 mg/L), phenotypic detection of MBL production was performed using the E-test MBL (AB Biodisk). The genes encoding the MBLs were characterised by PCR and sequencing analysis.
Results: Nine (4.5%) P. mirabilis isolates exhibited reduced susceptibility to imipenem (MICs ranged from 32 to >256 mg/L). All but one of the isolates, were susceptible to aminoglycosides and aztreonam but resistant to the other β-lactams, quinolones and trimethoprim; the remaining isolate was pandrug-resistant. Production of MBL was detected in all 9 isolates with E-test MBL. Furthermore, genetic analysis identified a blaVIM-1 gene cassette in a class 1 integron in all cases. The majority of the isolates (8/9) were recovered from urine specimens of outpatients who had a history of hospitalisation in the previous three months. They had treated during their hospitalisation with aminoglycosides, ticarcillin/clavulanic and fluoroquinolones. Aztreonam was used for the treatment of MBL-positive infections with successful results.
Conclusions: We detected a high prevalence of infections associated with MBL-producing P. mirabilis in the community setting of our region. Previous hospitalisation and exposure to antimicrobials were predictive of MBL-producing P. mirabilis community-onset infection. Continuous surveillance is essential for the early recognition and for the preventation of dissemination of such strains by indicating the proper treatment.
P1354 Prevalence of extended-spectrum β-lactamase-producing Enterobacteriacae isolates at two Belgian hospitals: a descriptive study
P. Bogaerts, G. Claeys, C. Bauraing, A. Piette, A. De Vleeschauwer, G. Verschraegen, Y. Glupczynski (Yvoir, Ghent, BE)
Objectives: To study the prevalence and distribution of extended-spectrum β-lactamase-producing Enterobacteriacae (ESBLE) isolates found at two university hospitals geographically located in two different regions of Belgium.
Material and Methods: Centres A and B are respectively a 1,050-bed hospital located in North Belgium (Province of East Flanders) and a 300-beds unit located in the Southern part (Province of Namur). During a 4 (centre A) and 20 month (centre B) period, respectively 1,589 and 2,695 consecutive non-duplicated Enterobacteriaceae isolates were screened retrospectively for ESBL production. Antimicrobial resistance phenotypes were confirmed by double disk synergy test. Characterisation of ESBLs was performed by IEF, multiplex PCR assays targeting blaTEM, blaSHV and blaCTX-M genes and sequencing when needed. In centre A, ESBLE were typed to detect the occurrence of clonal outbreaks during the analysed period.
Results: Ninety-four (6%) and 74 (3%) ESBLE were recovered from centre A and centre B respectively.
In centre A, the presence of ESBL was equally distributed among Klebsiella pneumoniae (Kp) (34%), Escherichia coli (Ec) (31%) and Enterobacter aerogenes (Ea) (30%). In centre B, almost half of the ESBLE were Ec (47%), followed by Ea (32%) with only very few Kp isolates (5%). In both centres, ESBLs were only rarely found among Enterobacter cloacae, K. oxytoca and C. freundii (1 to 7% of the total ESBLE population). TEM, SHV, and CTX-M type ESBLs were detected in both centres albeit at a variable frequency depending on the ESBLE species. In Ea, TEM-24 accounted for 75% of more of all ESBLs in both centres. For Kp, SHV were found in more than 50% and CTX-M in about 25% in centre A, whereas only TEM-derived ESBLs were detected in centre B. Concerning Ec, no significant differences were found between the two centres, with CTX-M types accounting for more than 50% of the ESBLs.
In centre A, PFGE typing revealed the occurrence of limited clonal outbreaks of ESBLE in Ea, Kp and also in Ec.
Conclusions: This study confirms the variable prevalence and distribution of ESBLE in different hospitals within the same country and highlights the importance of the knowledge of the epidemiology of ESBL-mediated resistance in order to optimise therapy and limit the spread of such organisms inside and outside the hospital. In addition, the global emergence of CTX-M enzymes is confirmed and the reason of this spread should be urgently elucidated.
P1355 Epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae in Belgium: preliminary results of a national multicentre survey in 2006
Y. Glupczynski, C. Berhin, H. Rodriguez-Villalobos, M. Struelens, B. Jans on behalf of the Belgian Infection Control Society
Objectives: As part of a national two-yearly surveillance, we evaluated the species distribution of ESBL-producing Enterobacteriaceae (ESBLE) isolated in Belgian hospitals and the in vitro activity of 11 antimicrobials against these isolates.
Materials and Methods: Consecutive, unduplicated clinical isolates of ESBLE collected in the participating centres (maximum of 5 strains per centre) between 01/2006 and 06/2006 were sent to the reference laboratory. All strains were confirmed as ESBL producers by double combination disk test (DDT) and/or by ESBL E-tests (cefotaxime and ceftazidime + clavulanic acid). The presence of ESBL among AmpC hyperproducers was assessed by DDT in the presence of cloxacillin 500 μg disks or on cloxacillin (250 μg/mL) agar. E-test MICs were determined and results were interpreted according to CLSI and EUCAST clinical MIC breakpoints.
Results: 86 hospitals (regional distribution: 42 from Flanders, 29 from Wallonia and 15 from Brussels) sent 401 potential ESBLE isolates. Of these, 330 (82%) were confirmed as ESBL producers. The ESBLE strains originated from patients (mean age 72 years; range 1–97 years) hospitalised in medical wards (39%), ICU (21%), geriatric or surgical units (13% each); they were mainly isolated from urinary (50%) and respiratory tracts (25%), wound swabs (19%) and blood (3%). Enterobacter aerogenes (Ea), Escherichia coli (Ec) and Klebsiella pneumoniae were the most frequent ESBLE and represented respectively 44%, 39% and 9% of all isolates. 72% of all ESBLE were considered as nosocomially acquired; however almost 50% of Ec ESBL producers were deemed to be community-acquired.
97% of all ESBLE were resistant to ceftazidime, 86% to cefotaxime, and 78% to ciprofloxacin. Carbapenems (meropenem, imipenem) were the most active agents (98–99% susceptibility) followed by temocillin (93% susceptibility), gentamicin and amikacin (85% susceptibility each) and tigecycline (82% susceptibility). Higher resistance rates and co-resistance to ceftazidime and ciprofloxacin were observed among Ea while almost two-third of the Ec ESBL producers had a resistance phenotype compatible with CTX-M ESBLs. Most Ec strains originated from urines of elderly patients and were co-resistant to ciprofloxacin.
Conclusions: These in vitro data provide new insights in the epidemiology of resistance among Belgian ESBLE isolates and highlight the rising importance of community-acquired ESBLs, especially among Ec strains.
P1356 Epidemiology of ESBL-positive Enterobacteriacae in Copenhagen, Denmark
H. Fjeldsøe-Nielsen, A. Friis-Møller, D. Schrøder Hansen, N. Frimodt-Møller, K. Schønning (Hvidovre, Hillerød, Copenhagen, DK)
Objectives: The aim of this study was to describe the epidemiology of ESBL-producing Enterobacteriacae and the initial molecular typing of prevailing β-lactamases in Copenhagen, Denmark.
Methods: 79 consecutive incident ESBL positive isolates were detected between January 1st and June 30th 2006. A 10 μg cefpodoxime disc and other 2nd- and 3rd-generation cephalosporins were used to screen for β-lactamase production. 32/79 (41%) isolates have subsequently been further characterised by PCR.
Results: 33/79 (42%) infections were community acquired (diagnosed in specimens from the primary care sector or emergency departments) the remaining 46/79 (58%) being nosocomial infections. There was as expected a gender difference with 58% women and 42% men. Age ranged from 1–99 years with a median 70 years. Infections were dominated by the urinary tract 65/79 (82%). 32 incident isolates were characterised by PCR during this period of which 25 were found CTX-M type positive. CTX-M was the single β-lactamase detected in 16 of these, the remaining 9 isolates harbouring CTX-M and other β-lactamases as well such as TEM and SHV. 2/32 isolates remained uncharacterised.
Conclusion: Many ESBL positive Enterobacteriacae infections are acquired in the community. The urinary tract alone accounts for 82% of all incident isolates. Older people and women seem to be more prevalent. 25/32 (78%) isolates were PCR positive for CTX-M type β-lactamase. CTX-M type β-lactamase seems to be the most prevalent among ESBL-producing Enterobacteriacae in Copenhagen, Denmark.
P1357 Incidence density of highly resistant Gram-negative bacteria in a Dutch hospital
I. Willemsen, M. van der Wiel, J. Kluytmans (Breda, NL)
Objective: The objective of this study was to determine the incidence density of highly resistant Gram-negative bacteria (HRGNB) in a Dutch hospital.
Method: Between 1 Jan 2005 and 30 Jun 2006 all HRGNB (Enterobacteriaceae, Acinetobacter spp. and Pseudomonas spp.) from hospitalised patients in the Amphia hospital were included. The definition for highly resistant micro-organisms according to the national guideline (www.wip.nl) was used. From all patients one isolate of each species was selected. When multiple isolates from one species were found in one patient the selection was made based on the susceptibility pattern. All isolated with major (S<>R) differences in susceptibility for amoxicilline-clavulanic acid, cefalosporines, quinolones, meropenem, aminoglycosides or sulfa-trimethoprim were included. HRGNB isolated more than 72 h after admission or less than 72 h after admission but within 30 days after discharge were defined as nosocomial. Patients who were on the ICU or who had been on the ICU in a period of 30 days before culture were defined as ICU-related. The number of bed days was collected from the hospital administration.
Results: In a period of 1.5 years, 121 patients with 139 HRGNB were identified, including: E. coli (95), Klebsiella spp. (11), Proteus spp. (9), Enterobacter spp. (7), Citrobacter spp. (5), Pseudomonas aeruginosa (5), Acinetobacter spp. (3), Morganella spp. (2), Salmonella spp. (1) and Serratia spp. (1). 106 HRGNB were nosocomial, of which 59 were ICU related and 47 were non-ICU related. The ICU accounts for 15,427 bed days and the rest of the hospital for 383,070 in 1.5 years. The incidence density for nosocomial HRGNB on the ICU was 38.2 and that in the rest of the hospital 1.2 per 10.000 bed days (RR: 31.1; 95% CI: 21.3–45.5).
Conclusion: Estimating the incidence density of HRMO according to well-defined criteria can be used as a benchmarking method. Considering the enormous difference between the ICU and the rest of the hospital most attention to control the development and spread of HRMO should be given to the ICU.
P1358 Prevalence of Mex-mediated resistance in Pseudomonas aeruginosa isolates from patients with ventilator-associated pneumonia in 4 Belgian hospitals
N. Mesaros, Y. Glupczynski, D. Pierard, A. Dediste, Y. Van Laethem, F. Jacobs, M. Struelens, D. De Vos, J. Pirnay, F. Van Bambeke, P. Tulkens (Brussels, BE)
Objectives: Efflux mediated resistance is difficult to detect in Pseudomonas aeruginosa (Pa) by routine susceptibility testing. Yet, it may confer cross-resistance to unrelated classes of drugs and contribute to selection of other resistance mechanisms. Our aim was to determine the prevalence of Mex efflux pumps in Pa isolates obtained from patients with ventilator-associated pneumonia (VAP).
Methods: Pa isolates were collected as pairs from each patient (first isolate before initiation of antibiotic treatment [pre]; second isolate, after 5 to 10 days of treatment [post]). In three hospitals (A-C), isolates were collected consecutively from all eligible patients; in the 4th hospital (D), isolates were selected on the basis of compatible resistance phenotypes. mexA and mexX transscription levels were quantified by real time PCR; mexC and mexE transcription was detected by semi-quantitative PCR (their basal expression being undetectable in wild-type strains). Isolates typing was performed by fAFLP.
Results: The table shows the number of isolates in which overexpression of mex genes was detected (pre and post) by hospital. DNA-based typing globally confirmed the clonality of the sucessive isolates in each patient, and excluded the occurrence of epidemic strains in the non-selected isolates.
| Hospitala (no. of pairs) | No. of strains showing gene overexpression |
|||||||
|---|---|---|---|---|---|---|---|---|
|
mexA |
mexC |
mexE |
mexX |
|||||
| pre | post | pre | post | pre | post | pre | post | |
| Random sampling | ||||||||
| A (n = 9) | 0 | 1 | 1 | 0 | 0 | 0 | 4 | 7 |
| B (n = 7) | 0 | 1 | 5 | 6 | 2 | 3 | 1 | 3 |
| C (n = 6) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (n = 22) | 0 | 2 | 6 | 6 | 2 | 3 | 5 | 10 |
| Strains collected on the basis of compatible resistance phenotype | ||||||||
| D (n = 17) | 2 | 8 | 4 | 5 | 0 | 4 | 7 | 9 |
A, AZ-Vrije Universiteit Brussels, B, CHU Saint-Pierre; C, Hôpital Erasme; D, Clinique universitaire UCL Mont-Godinne.
Conclusions: A variable prevalence of Mex efflux pumps is found before treatment in clinical isolates, and increases in several cases following antibiotic exposure, particularly for MexX. In samples collected based on antibiograms, prevalence was very high, confirming the value of the interpretive algorithms used to detect mechanisms of efflux resistance. These data highlight the need of detecting efflux-mediated resistance in Pa clinical isolates originating from hospitalised ICU patients.
P1359 Clonal types and serotypes of multidrug-resistant Pseudomonas aeruginosa isolates spread in a university hospital in Greece
E. Drougka, T. Panagea, V. Chini, A. Foka, M. Christofidou, I. Spiliopoulou (Patras, GR)
Objectives: Despite the introduction of a wide variety of antimicrobial agents, Pseudomonas aeruginosa remains a common cause of nosocomial infections. Typing techniques are essential to investigate clonal relationship among isolates in the hospital setting, to recognize outbreaks and to elucidate the source of infection. We have studied by phenotypic and genotypic methods the spread of multi-resistant P. aeruginosa isolates recovered during one-year period from different patients at a University Hospital.
Methods: A total of 126 P. aeruginosa isolates recovered from different patients admitted at the University Hospital of Patras, Greece during 2004 were studied. Antibiotic susceptibility testing was performed by the Etest according to CLSI guidelines and the production of metallo-β-lactamases (MBL) was tested by the use of the strip containing imipenem and imipenem/EDTA. Serotyping was performed by 17 monovalent antisera against the O antigen according to the International Antigenic Typing Scheme. Clones were defined by PFGE of chromosomal DNA SpeI digests.
Results: Fifty-six isolates (45%) derived from the Dept. of Internal Medicine (IM), 33 (26%) from the Intensive Care Unit (ICU), 24 (24%) from the Dept. of Surgery (GS), 13 (10%) from the Dept. of Outpatients (OUTP). Overall, 53.2% of the isolates were resistant to tobramycin, 52.4% to ciprofloxacin, 52.4% to meropenem, 49.2% to imipenem, 36.3% to piperacillin, 33.9% to aztreonam, 28.2% to piperacillin/tazobactam, while 61 isolates (48%) were MBL-positive. By serotyping 58 (46%) isolates were classified to O:11, 19 (15%) to O:12, 12 (10%) to O:3 and 11 (9%) to O:1. PFGE revealed the presence of 19 clones. Clone A predominated including 10 strains with 7 strains of serotype O:11; clone B included 9 strains with 6 of O:11; clone C included 7 strains with 4 of O:12; clone D included 6 strains with 5 of O:11, while the rest strains were classified into different clones. Strains of serotype O:11 were mainly isolated from patients hospitalised at the IM and the ICU. Clone A was mainly spread in IM and ICU, clone B in GS, clone C in IM and clone D in GS.
Conclusions: Serotype O:11 predominated but it was not classified in one PFGE type, neither MBL-positive strains belonged to a common clone. Even though multi-resistant P. aeruginosa strains are spread in our hospital, we have identified four main clones and a variety of clonal types related to different departments.
P1360 Evaluation of metallo-β-lactamase producing Klebsiella pneumoniae epidemiology in a Greek hospital (2004–2006)
H. Moraitou, N. Kentrou, I. Galani, N. Tsagarakis, M. Makarona, A. Gioga, S. Kanavaki (Athens, GR)
Objectives: Nosocomial infections due to the dissemination of multi-resistant metallo-β-lactamase producing K. pneumoniae within the hospital setting are of growing concern worldwide. The aim of this study was to retrospectively analyse all MBL-positive K. pneumoniae strains isolated in our laboratory, from 2004 (first isolation) to 2006.
Methods: A total of 52 carbapenem resistant K. pneumoniae strains were isolated and collected since March 2004 to September 2006. 39/52 (75.0%) derived from ICU patients and the rest 13/52 (25.0%) from patients in different wards of our Hospital. 18/52 (34.6%) strains were isolated in 2004, 23/52 (44.2%) in 2005 and 11/52 (21.2%) in 2006. All isolates were tested for imipenem resistance by E-test, and for MBL production by imipenem/EDTA disk synergy test and PCR. Molecular typing was performed by REP-PCR.
Results: All isolates showed reduced susceptibility (MIC >0.25) or resistance to imipenem (MIC> 8) as well as resistance to all cephalosporins. In addition, all strains were positive for MBL production by EDTA synergy test and PCR experiments detected blaVIM gene. Molecular typing by REP-PCR revealed 14 K. pneumoniae clones (A-N), with predominant the clone A (17/52, 32.7%) throughout the period studied. It is interesting that clusters of the same or related clones were noted in ICU, being responsible for infections appeared in March 2004 (7 strains, clone A), March 2005 (6 strains, clone B), and May 2005 (6 strains, clone J). In addition, the Oncology Unit revealed one small cluster of infections in July 2006 (2 strains, clone F).
Conclusions: All carbapenem-resistant K. pneumoniae isolated in our laboratory were blaVIM positive. We observed three clusters of clonal K. pneumoniae infections as well as six single unrelated clones throughout the study period. The emergence of MBL-producing K. pneumoniae is of great concern and measures should be taken in order to avoid dissemination of highly resistant strains within hospital setting.
P1361 Metallo-β-lactamase producing P. aeruginosa in a tertiary hospital, 2004–2006
H. Moraitou, N. Tsagarakis, I. Galani, M. Makarona, N. Kentrou, A. Gioga, S. Kanavaki (Athens, GR)
Objectives: MBL production has been increasingly reported as a mechanism conferring carbapenem resistance to P. aeruginosa clinical isolates. The aim of this study was to retrospectively detect the MBL-producing multi-drug resistant P. aeruginosa strains isolated and collected in our Hospital from January 2004 to September 2006 and determinate the type of MBL they carry.
Methods: A total of 128 MDR P. aeruginosa clinical isolates were retrospectively investigated for MBL production. The strains were isolated either from ICU (54/128, 42.2%), or non-ICU (74/128, 57.8%) patients, from different kinds of clinical samples, from January 2004 to September 2006. The resistance level to imipenem was determined by MIC, using E- test (AB Biodisk, Sweden). The isolates were tested for MBL production by imipenem/EDTA disk synergy test as well as the MBL E-test (AB Biodisk, Sweden). PCR and PCR-RFLP analysis were further performed to analyse the blaVIM gene.
Results: All isolates exhibited high level resistance to imipenem (MIC > 16). Among all MDR P. aeruginosa strains 23/128 (18.0%) were found blaVIM positive, by PCR. Imipenem/EDTA combined synergy test identified 22/23 (95.7%) positive isolates, while only 5/23 (21.7%) found positive by MBL E-test. PCR-RFLP analysis revealed the presence of a blaVIM-2-like gene in all positive isolates. 9/23 (39.1%) MBL (+) strains were isolated from ICU patients (6 bronchial secretions, 2 blood cultures, 1 intravenous catheter) and 14/23 (60.9%) from non-ICU patients (7 sputum, 4 bronchial secretions, 3 blood cultures, 1 pus).
Conclusion: MBL-producing isolates are prevalent among MDR P. aeruginosa clinical isolates in our Hospital. Phenotypic tests may not be entirely accurate, but are indicative enough for the presence of MBL production. We found that imipenem/EDTA combined synergy test is more sensitive in indicating MBL (+) isolates. The high incidence of MBL-producing strains among non-ICU patients is of great concern, and measures should be taken to avoid dissemination of these MDR strains in the community.
P1362 The first isolation of PER-1-producing Pseudomonas aeruginosa and Acinetobacter baumanii strains in Hungary
D. Szabó, L. Rókusz, Z. Juhász, J. Szentandrássy, K. Katona, K. Nagy (Budapest, HU)
Objectives: A 45-year-old Hungarian tourist injured in terror-attack was transferred with serious burn injury from Egypt to the Central Military Hospital, Budapest, Hungary. Our aim was to characterise the β-lactamases produced by the Gram-negative bacteria isolated from the patient.
Methods: The identification and the antimicrobial susceptibility profile of the Gram-negative bacteria were evaluated using a VITEK 2 system. The isolates were screened for extended-spectrum β-lactamase (ESBL) and metallo-β-lactamase (MBL) production. The minimal inhibitory concantrations (MICs) were determined using E-test according to Clinical and Laboratory Standards Institute (CLSI) recommendations. Isoelectric focusing was performed using crude β-lactamase extracts in polyacrylamide gels containing ampholines with a pH range of 3.5 to 9.5. Pulsed-field gel electrophoresis (PFGE) was done with SpeI enzyme to compare the isolates. DNA amplification by PCR was carried out using primers specific to the blaTEM gene, the blaSHV gene, the blaPER gene and the blaVIM gene. Both strands of the amplification products were sequenced by the standard Sanger dideoxynucleotide method.
Results: The ESBL phenotype was detected in Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae and Enterobacter cloacae strains. The PER-1-producing (pI:5.3) P. aeruginosa and A. baumannii strains were isolated from the bloodculture and the wound of the pateint on the admission day suggesting that these strains were transported from Egypt. The PER-1 producing A. baumanii also produced TEM-1 enzyme. The PER-1 producing P. aeruginosa and A. baumannii were resistant to the ceftazidime, cefotaxime, cefepime, aztreonam, but were sensitive to imipenem, meropenem. On the first day TEM-1 and SHV-type ESBL-producing K. pneumoniae strain was also isolated.
MBL phenotype was also detected in a P. aeruginosa strain. This VIM-2 producing P. aeruginosa showed resistance to almost all β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, but it was sensitive to colistin. The PER-1 producing P. aeruginosa and the VIM-2 producing P. aeruginosa strains were unrelated as determined by PFGE.
Conclusion: This is the first report of the PER-1 and VIM-2-producing P. aeruginosa and PER-1 producing A. baumannii strains from Hungary. This work illustrates the inter-country and the inter-continent spread of PER-1-producing P. aeruginosa and A. baumannii isolates.
Supported by OTKA F048410.
P1363 Carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii isolates in an Iranian 1,000-bed hospital
M. Rahbar, P. Islami, S. Molanaee, M. Deldari (Tehran, IR)
Objective: Carbapenems, including imipenem and meropenem, are β-lactam antibiotics which widely are used recently in our country. The aim of this study was to determine resistance of P. aeruginosa, A. baumannii and Klebsiella pneumoniae to carbapenems in our hospital.
Methods: From April 2006 to October 2006 one hundred nosocomial isolates including 29 strains of P. aeruginosa, 27 Acinetobacter baumannii and 44 isolates of Klebsiella pneumoniae were collected from specimens obtained from hospitalised patients. The MIC f of meropenem for all isolates was determined by E-test (AB/Bio disk Sweden). Susceptibility to other antibiotics including Pipracillin/Tazobactam, Cefepime Ciprofloxacin, Amikacin, Imipenem and Ceftazidim were performed by disk diffusion method as recommended by Clinical Laboratory Standards institute (CLSI).
Results: MIC for meropenem by E-test ranged from 0.25 μg/mL to 32 μg/mL. Of 29 isolates of P. aeruginosa, 7 (24%) were resistant to meropenem (MIC > 32 μg/mL). These isolates also were resistant to imipenem. In Acinetobacter baumannii, of 27 isolates 8 (27%) were resistant to meropenem. All isolates of K. pneumoniae were susceptible to meropenem and imipenem. Only one strain had intermediate resistance (MIC = 6 μg/mL). Resistance to other tested antibiotics was very high and more than 80% of organisms were resistant to Piperacillin/Tazobactam, Cefepime, Ciprofloxacin, Amikacin, and Ceftazidim. All isolates of Klebsiella pneumoniae were resistant to Ceftazidim.
Conclusion: These studies showed that nearly one fourth of non-fermenter tested organisms in our study were resistant to carbapenems. However resistance of K. pneumoniae to carbapenem is not a serious problem already in our hospital.
P1364 The first metallo-β-lactamase producing clinical isolate of Pseudomonas aeruginosa in Norway
Ø. Samuelsen, M.A. Toleman, N.O. Hermansen, B. Aasnæs, B. Haldorsen, A. Sundsfjord, T.R. Walsh (Tromsø, NO; Cardiff, UK; Oslo, NO)
Objectives: MBL are of great clinical significance as they are able to hydrolyse virtually all β-lactams limiting therapeutic options. Here we describe the first clinical isolate of MBL-producing Pseudomonas aeruginosa in Norway and its subsequent genetic characterisation.
Methods: A clinical isolate of P. aeruginosa (K34–7) was recovered from a patient recently admitted from an African country with a high-level resistance to carbapenems and was examined for MBL production. Susceptibility testing was performed using disc diffusion, VITEK and Etest. The presence of MBL was confirmed by PCR (custom oligonucleotides for all major blaMBL genes) and by spectrophotometric analysis of crude cell extracts (with and without EDTA – 25 mM for 1 hr) for the ability to hydrolyse imipenem. Characterisation of the MBL gene and its genetic support was evaluated by its association with integrons, transposons and insertion elements (ISCR) using PCR and sequencing.
Results: Susceptibility testing showed that P. aeruginosa K34–7 was multi-drug resistant, highly resistant to imipenem and meropenem (MIC > 32 mg/L) and positive on the MBL Etest (ratio MIC imipenem/imipenem + EDTA ≥8). MBL production was also verified by hydrolysis/inhibition assays on crude cell extracts ± EDTA. Sequencing analysis of the positive blaVIM amplicon revealed it was 100% identical to blaVIM-2. PCR and sequencing analysis revealed that K34–7 contained 2 very different integrons. The class 1 integron contains 4 antibiotic resistant genes including blaOXA31 and genes encoding novel multi-drug resistant pumps and aminoglycoside modifying enzymes. The second integron contained TniCR at the 3′ end instead of qac/sul and the gene cassette sequence: aacA7-blaVIM-2-aacA5. Although standard transposons was not detected adjacent to the integrons, we detected a new ISCR element, designated ISCR10, and the first to be found in P. aeruginosa.
Conclusion: This is the first report of an MBL-producing P. aeruginosa identified in Norway. Genetic analysis revealed the “new” TniCR integron associated with the blaVIM-2 MBL gene and the first ISCR10 found in P. aeruginosa.
P1365 Dissemination of a VIM-positive Pseudomonas aeruginosa clone in a university hospital, Ankara, Turkey
N. Gurkan Aydin, G. Metan, B. Altun, P. Zarakolu (Ankara, Kayseri, TR)
Objectives: The aim of this study was to report the dissemination of a VIM-positive Pseudomonas aeruginosa clone in Hacettepe University Hospital in Ankara, Turkey.
Methods: P. aeruginosa strains (n = 26) isolated from various clinical specimens (4 blood, 7 urine, 12 pus, 1 cerebrospinal fluid, 1 bronchoalveolar lavage, 1 sputum) of patients with nosocomial infections in Hacettepe University Adult Hospital during January and July 2005 were included in the study. The isolates were identified by Phoenix System (Becton Dickinson, USA) and the antimicrobial susceptibility testing was performed by Etest (AB Biodisk, Sweden) method according to CLSI criteria. All the isolates were screened for carbapenem hydrolysing metallo-β-lactamase activity by imipenem-EDTA disk method phenotypically and genotypic detection was performed by PCR with bla-IMP and bla-VIM primers. Clonal diversity was examined by PFGE of SpeI-digested genomic DNA.
Results: The antimicrobial resistance of the isolates to ceftazidime was 27% where as 31% to piperacillin-tazobactam, 35% to imipenem, 38% to meropenem, 46% to tobramycin, 58% to ciprofloxacin. All the isolates were carrying the blaVIM gene and the blaIMP gene was not detected in any of them. However, only 8 (31%) were phenotypically positive for metallo-β-lactamase activity. There were 3 clones with one dominant clone (n = 22).
Conclusion: P. aeruginosa is a multidrug resistant pathogen of nosocomial infections. Although metallo-β-lactamase activity as a resistance mechanism has been reported increasingly worldwide, there are few reports from Turkey. Our finding of dissemination of this clone is pointing out the emergence of this resistance pattern in our country.
P1366 Multiresistant Pseudomonas aeruginosa serogroup O:12 outbreak in an intensive care unit
R. Rodríguez, M.M. Gallardo, M.V. García, A. Vindel, E. Granados, F. Ropero, I. Viciana, A. Pinedo (Malaga, Madrid, ES)
Objective: Research on an multirresistant P. aeruginosa outbreak among patients hospitalised in the Intensive Care Unit.
Methods: A retrospective study was carried out where the clinical reports of those patients with a presence of P. aeruginosa of the same resistant phenotype, were revised. Antimicrobial suceptibilities were carried out by the MicroScan automised system and by disk difussion test and E-test. The results interpretation were according NCCLS guidelines. The epidemiological characterisation by serotyping was analysed using an agglutination technique with the antisueros of the IATS. Clonal relationship of strains were carried out with Pulse Field Gel Elctrophoresis (PFGE) of the restriction fragments obtained with XbaI and PCR for blaVIM for the detection of metallo-β-lactamases.
Results: Between September 2002 and February 2003, 45 P. aeruginosa serotype O:12 were isolated with identical resistant phenotype only sensitive to amikacin. These 45 strains were obtained from 13 patients out of which 10 (76.9%) were men and 3 (23.07%) were woman with an average age of 60 years old. Isolates were produced in two intensive care units from different hospital areas: General Intensive Care Unit (ICU) and Anaesthesia Intensive Care Unit (recovery). 9 patients were admitted in ICU, 3 in recovery and one in both wards. Average hospitalisation days were 49.23 days. 76.9% of the patients were smoker and the base pathology most frequently found were cardiopathy, diabetes mellitus and hypertension (46.2%). Mostly of patients were no fatal underlying condition (76.9%). Samples where P. aeruginosa was isolated more frequently were blood in 37.7% of the cases followed by bronchial aspirations (26.6%). Index case was a patient hospitalised in ICU for 49 days. The last 4 cases were recovery patients, one of them had been admitted previously in ICU. Bronchial aspiration had been done in all of them as well as mechanical ventilation and SNG. Mortality was of 46.15% and the mortality attributable was of 23.1%. Restriction patterns obtained through PFGE were identical in all strains confirming isolation clonal relationships. PCR blaVIM for carbapenemas detection was negative in all cases.
Conclusions: Antibiotic pressure to which these patients are submitted plus the control measures relaxation of healthcare workers, probably are the cause of the outbreak appearance and dissemination. Molecular characterisation is an useful tool for epidemiologic surveillance programmes.
P1367 Epidemiological and clinical investigation of Gram-negative anaerobic infections in Greece
A. Katsandri, A. Avlamis, N.J. Legakis, G.L. Petrikkos, A. Pantazatou, J. Papaparaskevas for The Hellenic Study Group for Gram-Negative Anaerobic Bacteria*
Objective: Epidemiological and clinical investigation of Gram(–) anaerobic infections.
Materials and Methods: A prospective multicentre study of 206 Gram(–) anaerobic infections was conducted during the period 2003– 2006. Data collected included gender, age, ward type, duration of hospitalisation, underlying disease, clinical manifestation, outcome, prior antimicrobial therapy, infection treatment, and microbiological data (clinical specimen, species identification, other pathogens from the same specimen). Analysis was performed with the STATA 6.0 programme.
Results: Bacteroides spp. isolates were more frequent in intra-abdominal infections (p<0.001). Infections due to B. fragilis compared to other species of the B. fragilis group were more frequent in surgical ward patients (p = 0.008), and those who received treatment with second generation cephalosporins (p = 0.017). In bacteraemic cases, Bacteroides non-fragilis spp. were more frequent than B. fragilis group spp. (p = 0.02). B. fragilis group spp. were isolated more frequently than Bacteroides non-fragilis spp. in polymicrobial infections (p = 0.05), and younger patients (p = 0.049, for median ages of 47.4 and 61.3 years). Prevotella spp. strains were more frequent in pulmonary infections compared to bacteraemias, soft tissue, or intrabdominal infections (p < 0.001), from outpatients than inpatients (p < 0.05), and from patients with shorter hospitalisation duration (p = 0.033). Fusobacterium spp. were isolated more frequently from patients with lung abscess (p = 0.038), and from those that did not receive any form of antimicrobial therapy (p = 0.024). Mortality was higher among bacteraemic patients (p < 0.001), those that were hospitalised in internal medicine dpts. or ICU (p = 0.001), or those that were treated with aminoglycosides (p = 0.008). Among polymicrobial infections an association was detected between Staphylococcus and Prevotella spp. (p = 0.006), and between Enterobacteriaeae and Bacteroides spp. (p = 0.007).
Conclusions: Epidemiological and clinical differences were detected among cases of infections due to different Gram(–) anaerobic species, differences that provide useful information for optimisation of the respective empirical treatment guidelines.
Acknowledgements: The study was funded by European Social Fund and National Resources (grant EPEAEK II–Pythagoras II).
*Drs M. Foustoukou, M. Kanellopoulou, C. Koutsia-Karouzou, H. Malamou-Ladas, A. Pangalis, E. Papafrangas, M. Toutouza, E. Trika-Grafakos, and A. Vogiatzi.
P1368 Correlation between colonisation and infection with antibiotic-resistant Gram-negative bacilli in the neonatal intensive care unit
A. Dedeic Ljubovic, D. Bekic, M. Hukic (Sarajevo, BA)
Objectives: Infection due to Gram-negative bacilli which are often resistant to multiple antibiotics usually occurs in neonates already colonised in the pharynx or intestine.
We studied bacterial colonisation in the intestinal and nasopharyngeal flora and development of severe infection (septicaemia and meningitis) in colonised neonates admitted to a neonatal intensive care unit.
Methods: The study included 255 neonates admitted to a neonatal intensive care unit at Clinical Center University of Sarajevo in a six-month period. Cultures of nose, throat and stool were obtained on admission and once weekly if the lenght of stay was more than seven days. A total of 923 nose and throat cultures and 450 stool cultures were performed. Antibiotic sensitivity pattern and ESBL production were determined with disc-diffusion methods and E test (AB Biodisk, Solna Sweden) according to the NCCLS.
Results: 85% (217/255) of patients became colonised in throat/intestine with antibiotic-resistant Gram-negative bacilli. 37.3% (81/217) of neonates became colonised early, in the first week of hospitalisation. Klebsiella pneumoniae, Pseudomonas spp., Acinetobacter baumannii, Serratia marcescens were isolated more often. 92% (47/51) of colonised neonates had intestinal colonisation, and 45.7% (76/166) nasopharyngeal colonisation with Klebsiella pneumoniae. 42.3% (52/123) of isolates were with same antibiotic sensitivity pattern and biotype (1 and 4). All Klebsiella pneumoniae isolates from stool, and 69.7% (53/76) from throat and nose were ESBL producers. Pseudomonas and Acinetobacter were also isolated from stool, throat and nose in 21% and 12% of patients, respectively. 47% (22/46) of Pseudomonas and 20% (5/25) of Acinetobacter isolates were imipenem resistant. Serratia marcescens was isolated from nose and throat in 13.3% (22/166) of patients, but not in the stool specimens. Severe infection (septicaemia and meningitis) occurred in 23% (50/217) of colonised neonates with the same strain of antibiotic resistant Gram-negative bacilli. A mean of 11.6 days was between colonisation and infection.
Conclusion: The high percentage of patients in our study developed infection after colonisation. Multi drug resistant hospital strain of Gram-negative bacilli are the most frequently isolated in colonised and infected patients. Screening cultures are necessary to detect colonised neonates when nosocomial Gram-negative bacilli become epidemic in the neonatal intensive care unit.
Pharmacokinetics and pharmacodynamics
P1369 Modification of cephalexin serum concentration by grapefruit juice co-administration in rats
M. Cuciureanu, C. Tuchilus, A. Poiata, M. Nechifor, R. Cuciureanu (Iasi, RO)
Acute ingestion of usual quantities of grapefruit juice or whole fresh fruit segments can alter oral drug pharmacokinetics by different mechanisms (including the inhibition of enteric cytochrome P450 3A4 isoenzymes). As a result, metabolism of co-administred drugs is reduced and oral bioavailability is increased.
Objective: To assess the interactions between grapefruit juice and cephalexin, we compared the serum concentration of cephalexin given alone and in association with grapefruit juice after oral administration in rats.
Methods: Mature male Wistar rats weighing 180–200 g were randomised into two groups of 10 animals each. The animals were housed at room temperature (22±1°C) and had free acces to food and water except during the course of the experiment. Group I received cephalexin (30 mg/kg, p.o.) and to group II was administred cephalexin (30 mg/kg, p.o.) and grapefruit juice (5 mL/kg, p.o.). Blood sample were collected at 0, 1, 2, 3, 8 and 24 hours after oral administration. Microbiological assay was performed by the agar plate diffusion technique using Sarcina lutea ATCC 9341 as test organism. Based on the results of the above assay and on known concentrations of antibiotic solutions we realised the serum concentration/time curve and we determined the rats' serum cephalexin levels at various times after administration.
Results: The mean cephalexin serum concentrations for group I were 14; 6.7; 2; 0,5; 0 μg/mL and for group II were 19.5; 8.2; 2.5; 0.7; 0 μg/mL at respectively 1, 2, 3, 8 and 24 hours after the administration.
Conclusion: The serum concentrations of cephalexin of group which received grapefruit juice are higher compared to group which received only cephalexin, at all determination times.
P1370 Pharmacokinetics of penicillin G in infants with a gestational age of less than 32 weeks
A.E. Muller, J. DeJongh, Y. Bult, W.H. Goessens, J.W. Mouton, M. Danhof, J.N. van den Anker (The Hague, Leiden, Assen, Rotterdam, Nijmegen, NL; Washington, US)
Objectives: Infectious complications in the immediate postnatal period are common and especially premature infants are vulnerable. Penicillin G is often given in such cases, but the pharmacokinetics in premature infants may differ substantially and is largely unknown, in particular in infants with a gestational age of less than 32 weeks. We investigated the pharmacokinetics of penicillin G in this age group.
Methods: Infants with a gestational age of less than 32 weeks on day 3 of life were eligible for the study. They received 50.000 EH/kg penicillin G every 12 h intravenously. From each patient 6–9 blood samples were taken. Antibiotic concentrations were determined by a validated HPLC method. Pharmacokinetic parameters were estimated by population pharmacokinetic modeling using NONMEM. Various models were tested. To discriminate between models the minimum value of objective function (MVOF) was used.
Results: Twenty intants were included. A two-compartment pharmacokinetic model best described the time course of penicillin G. Clearance (CL), volume of the central compartment (Vc) and intercompartimental clearance were estimated at respectively 0.103±0.01 L/h, 0.359±0.06 L and 0.774±0.277 L/h (mean±SE). The residual error was found to be both proportional and additive to the blood concentrations. Interindividual variability was found for CL and Vc. The volume of distribution at steady state was 0.54L and terminal half-life was 3.9 h. CL increased significantly with body weight. The relationship between gestational age and CL was examined, but found to be inferior to the relationship between body weight and CL.
Conclusions: The pharmacokinetics of penicillin in infants with a gestational age of less than 32 weeks differs significantly from that in adults and children with a prolonged mean terminal half-life of approximately 4 hours. These results warrant a dosing schedule of two times daily dosing in this age group. CL significantly increased with body weight, rather than with gestational age.
P1371 The influence of uterine contractions during labour on the pharmacokinetics of intravenously administered amoxicillin
A.E. Muller, P.J. Dörr, J.W. Mouton, J. DeJongh, P.M. Oostvogel, E.A.P. Steegers, R.A. Voskuyl, M. Danhof (The Hague, Nijmegen, Leiden, Rotterdam, NL)
Objectives: Amoxicillin is widely used in pregnant women, both in those who are in labour and those who are not. An effect of labour on the pharmacokinetics of ampicillin has been shown previously, but similar data are not available for amoxicillin. Changes in the pharmacokinetics due to uterine contractions may pose these patients at risk for inappropriate dosing. We investigated the effects of uterine contractions on the pharmacokinetics of amoxicillin.
Methods: Healthy women at 30–42 weeks of gestation were eligible for the study. They received amoxicillin following the local guidelines. From each patient 5–28 serial blood samples were taken and demographic data recorded. Serum antibiotic concentrations were determined by a validated HPLC method. Pharmacokinetic parameters and the influence of uterine contractions on these parameters were estimated by population pharmacokinetic modelling using NONMEM. Various models were tested. To discriminate between models the minimum value of the objective function (MVOF) was used.
Results: Thirty women were included, 20 without uterine contractions and 10 who were in labour. A three-compartment pharmacokinetic model best described the time course of amoxicillin. The estimated values (mean ±SE) for the entire population for clearance, volume of distribution of the peripheral compartments (V2 and V3) and the intercompartmental clearances (Q1 and Q2) were 21.6±0.99 L/h, 6.63±0.53, 8.65±0.99L, 35.8±5.15 and 5.95±0.65 L/h, respectively. The terminal half-life was 1.16±0.26 h. There was no effect of uterine contractions on the parameters, except on the volume of distribution of the central compartment (Vc). Uterine contractions slightly increased Vc from 6.93±1.73L to 8.64±2.21L (p<0.05; mean ±SD). The residual error was found to be proportional to the blood concentrations. Interindividual variability was mainly due to differences in clearance and Vc. The main demographic characteristics did not influence the individual posthoc estimates for clearance.
Conclusion: A 3-compartment model best described the data. The consequential slow elimination is beneficial for the efficacy of amoxicillin, especially when it is used to prevent Streptococcus agalactiae infections in the neonate, because of the relatively low MICs. Both the absence of effects of uterine contractions and the lack of influence of patient characteristics indicate that dose-adjustments in this critical situation are not necessary.
P1372 Optimal piperacillin/tazobactam dosing against Pseudomonas aeruginosa: prolonged or continuous infusion?
A. Kim, C. Sutherland, J. Kuti, D. Nicolau (Hartford, US)
Objectives: As a result of increasing resistance among Pseudomonas aeruginosa (PSA), alternative dosing strategies to optimise piperacillin/tazobactam (TZP) pharmacodynamics, such as prolonged (PI) and continuous infusion (CI), have been advocated; however, comparative data to aid in deciding which to use are sparse. Monte Carlo simulation was utilised to compare the bactericidal cumulative fraction of response (CFR) between PI and CI regimens against a local population of PSA.
Methods: TZP pharmacokinetics were extrapolated from a published population pharmacokinetic study in surgical and neutropenic patients. A 2-compartment model was used to calculate steady-state concentration time profiles for the piperacillin component of the following TZP dosages: 4.5 g q6h (30 minute infusion), 3.375 g q8h (4 h PI), 4.5 g q8h (4 h PI), 4.5 g q6h (3 h PI), 10.125 g CI, 13.5 g CI, 18 g CI, 20.25 g CI, and 22.5 g CI. A 5,000 Monte Carlo simulation with covariance matrix was conducted. Pharmacodynamic targets were 50% free time above the MIC (T>MIC) for the 30 minute infusion and PI regimens, and 100% free T>MIC for CI regimens. MICs for 416 consecutive nonduplicate PSA collected from our institution over a 6 month period during 2006 were tested via E-test.
Results: 92.3% of PSA were TZP susceptible; the MIC50 and MIC90 were 8 and 32 mg/mL, respectively. CFR was 79.6 (4.5 g q6h, 30 minute infusion), 83.1 (3.375 g q8h, 4 h PI), 86.9 (4.5 g q8h, 4 h PI), 89.6 (4.5 g q6h, 3 h PI), 82.0 (10.125 g CI), 86.3 (13.5 g CI), 89.2 (18 g CI), 90.0 (20.25 g CI), and 90.6 (22.5 g CI).
Conclusion: Both PI and CI dosing strategies improve the pharmacodynamics of TZP over that of standard infusion regimens; moreover, PI and CI regimens that contain the same daily piperacillin dose will have similar likelihood of bactericidal exposure. Thus, the choice of administration strategy depends on the convenience of once daily dosing versus line access availability. Finally, dosing regimens that contained greater than 18 g TZP daily did not appear to provide any meaningful additional exposure for this PSA population.
P1373 Clinical significance of pharmacokinetics of ertapenem in an outpatient setting
S. Garazzino, F.G. De Rosa, A. D'Avolio, M. Michelazzo, G. Di Perri (Turin, IT)
Objectives: Ertapenem is a novel parenteral carbapenem active against most pathogens associated with community-acquired and mixed infections. Given its once daily administration, it is an appealing option in an outpatient setting. Aim of our study was to evaluate pharmacokinetic parameters of ertapenem in a cohort of outpatients with a variety of infections and to compare data with those obtained in healthy volunteers.
Methods: Outpatients treated with intravenous ertapenem at standard dosage were analysed. Plasma samples for pharmacokinetic analysis were obtained after multiple dosing at steady state conditions immediately before and at the end of the infusion, at 2 h and 24 h. Plasma concentrations of ertapenem were measured by a validated HPLC-UV method and Cmax, Cmin and AUC0–24 were determined by non compartmental analysis (Win Nonlin Professional 4.1).
Results: Twenty-five patients were studied; 14 of them had a complicated skin/skin structure infection, 8 an osteo-articular infection (chronic osteomyelitis, diabetic foot or septic arthritis), 1 a mastitis and 2 had a genito-urinary tract infection. Median age was 55.5 years (range 27–85). All patients received intravenous ertapenem 1 g once daily over a 30-min infusion. Serum creatinine levels were within normal range in all patients but one (1.3 mg/dl). Mean Cmax and Cmin were respectively 97.6 mg/L and 1.8 mg/L (range 19.4–438 and 0.1–11.6 respectively); mean AUC was 536.3. Cmin values were associated with age (p<0.05); no statistical association was found between ertapenem concentrations and either body weight, BMI, sex or creatinine levels.
Conclusion: Mean plasma concentrations of ertapenem exceeded the MIC of susceptible bacteria for the entire dosing interval, satisfying the pharmacokinetic requirements of a once daily dosing. Data were similar to those reported in healthy volunteers: no dose adjustment is required by body weight and creatinine levels. Higher high trough concentrations observed in patients with advanced age are probably accounted for by a reduced renal excretion of ertapenem.
P1374 Comparison of oral dosing regimes for pivampicillin by Monte Carlo PK/PD simulation
K.S. Jensen, N. Frimodt-Møller (Copenhagen, DK)
Objectives: Compartmental models employed in a Monte Carlo simulation scheme are frequently used to study PK/PD aspects of antibiotics given intravenously. In contrast, very few modeling studies of that kind seem to have been reported for drugs when taken as oral doses. We therefore wanted to test the ability of a compartmental model to predict concentration-time profiles in serum after PO uptake of pivampicillin when population PK parameters are estimated with the NPAG computer programme. Furthermore, we wanted to use Monte Carlo simulation to find optimal dosage regimes for pivampicillin with respect to Streptococcus pneumoniae and S. pyogenes, since aminopenicillins are commonly used drugs for treatment of upper-respiratory tract infections.
Methods: PK-data for ampicillin concentrations after dosing of pivampicillin 500 mg (equivalent to 360 mg ampicillin) were kindly provided by Leo-Pharma Denmark. 7 (n = 12) or 8 (n = 5) measurements of serum concentration within 8 hours from intake were recorded in 17 individuals (mean age: 38 years, mean weight: 76 kg). The NPAG programme was used to fit a linear compartment model (seven state variables, six PK parameters) to the data whereby mean values as well as covariance matrix for the PK parameters were obtained. Monte Carlo simulations (Nsubjects = 100,000) were then made with the compartment model assuming lognormal as well as nonparametric (empirical) frequency distributions for the PK parameters. Results were obtained on the probability of target attainment with respect to T>MIC 50% for a range of MIC values covering Streptococcus pneumoniae and S. pyogenes (distribution of MIC's from the EUCAST website).
Results: After the parameter estimation process, the compartment model was able to reproduce the measured serum concentrations with good precision. For MIC targets of 0.063 mg/L (streptococci), T>MIC 50% (free fraction eq. to 100%) could be reached for 99% of the simulated population on a 250 mg q8h pivampicillin dose (breakpoint: 0.125 mg/L), while the same population coverage (99%) required 1,000 mg doses if using a q12h schedule (breakpoint: 0.125 mg/L).
Conclusion: Monte Carlo simulation of oral dosage of antibiotics based on estimation of population PK parameter values can be applied for pivampicillin. The simulations indicate that for streptococcal infections the total daily dose can be reduced to less than 40% if distributing the intake on three instead of two occations per day.
P1375 Pharmacodynamic comparison of linezolid, teicoplanin, and vancomycin against clinical isolates of Staphylococcus aureus and coagulase negative staphylococci collected from hospitals in Brazil
C. Kiffer, J. Kuti, C. Mendes, S. Sinto, P. Koga, D. Nicolau (Sao Paulo, BR; Hartford, US)
Objectives: The association of MIC distribution and pharmacokinetic (PK) data derived from microbiological and human studies through the use of pharmacodynamic (PD) models offers a more developed tool for prediction of infection outcome. The probability of obtaining critical PD targets was determined and compared by the use of Monte Carlo simulation for Linezolid (L), Teicoplanin (T), and Vancomycin (V) against non-duplicate clinical isolates of Staphylococcus spp. collected from 3 hospitals in São Paulo, Brazil between 2003 and 2005.
Methods: MICs were determined for 201 Staphylococcus aureus (SA) and coagulase negative staphylococci (CNS) isolates by Etest method for L, T, and V. Interpretations were done according to established CLSI criteria. Methicillin resistance (MR) was confirmed by oxacillin and cefoxitin disc diffusion. PD exposures, measured as steady-state total drug AUC/MIC, were modeled via a 5,000 patient Monte Carlo simulation for the following: L 600 mg q12h, T 400 mg q24h (T6), T 800 mg q24h (T8), V 1 g q12h, and V 1 g q8h using PK from patients. PD targets included AUC/MIC >82.9 for L and >345 for T and V. The bactericidal cumulative fraction of response (CFR) was calculated for each regimen against each specific population: methicillin-susceptible (MS) and MR, for both SA and CNS.
Results: There were 40/119 MRSA and 74/82 MR-CNS isolates. The SA isolates were fully susceptible to T (MIC90 = 2 μg/mL) and L (MIC90 = 1 μg/mL). There was one SA isolate with intermediate susceptibility to V (MIC = 3 μg/mL) (MIC90 = 1.5 μg/mL). The CNS isolates were fully susceptible to V (MIC90 = 3 μg/mL) and L (MIC90 = 1 μg/mL), and 13.4% resistant to T (MIC90 = 32 μg/mL). CFR against all SA were 96.0%, 30.1%, 71.6%, 48.0%, and 65.1% for L, T6, T12, V q12h, and V q8h, respectively. CFR against all CNS were 97.8%, 13.4%, 34.6%, 10.9%, and 31.3% for L, T6, T12, V q12h, and V q8h, respectively. The CFR was reduced for all compounds among the MR versus the MS strains, except for L against the MR-CNS (94.9% MS-CNS and 98.1% MR-CNS).
Conclusion: Although MR was not expected to affect MIC, these isolates did present with higher MICs than the MS isolates, thereby contributing to lower CFR for MR strains. These data suggest that L has a greater probability of attaining its requisite PD target than V and T against these staphylococci. Although higher doses of V and T increased the CFR for both SA and CNS, the bactericidal targets achieved still appeared insufficient.
P1376 Ceftazidime vs cefepime: comparison of two drugs as empirical therapeutic drugs for febrile neutropenia by Monte Carlo simulation method
Y. Sumitani, Y. Kobayashi (Tokyo, JP)
Objectives: To evaluate of ceftazidime and cefepime as the empirical therapeutic drugs for Febrile neutropenia based on pharmacokinetics/pharmacodynamics theory.
Methods: The probabilities of attaining targets (TA%) of time above the MIC (%T>MIC) were calculated by Monte Carlo Simulation method with Crystal Ball 2000, and compared TA% of the targets for bactericidal effects in available maximum dosage of ceftazidime (1,000 mg every 6 h) and cefepime (2,000 mg every 12 h) in Japan. The bactericidal target of Craig was adapted: cefems (70%T>MIC).
The distribution of MICs of antibiotics against 12 strains of P. aeruginosa isolated from blood of patients admitted to Keio University Hospital between 2002 and 2004 was used for pharmacodynamic data. Pharmacokinetic data were obtained from healthy volunteers in each of interview form of ceftazidime and cefepime.
Results: The results of calculation of TA% of 70%T>MIC was 80.26% with ceftazidime in maximum dosage (1,000 mg every 6 h) and TA% of 70%T>MIC was 56.86% with cefepime maximum dosage (2,000 mg every 12 h) against P. aeruginosa.
Conclusion: Cefepime is the only authorised agent as therapeutic drug for Febrile neutropenia in Japan. But ceftazidime is also one of the first choice agents for FN in the guideline of HCR (The Infectious Diseases Society of America). In our study, TA% of 70%T>MIC with CAZ was much higher than that of CFPM and adaptation expansion as therapeutic drug for FN in Japan is expected.
P1378 In vivo PK/PD of linezolid against extra- and intracellular Staphylococcus aureus
A. Sandberg, K.S. Jensen, R.L. Skov, N. Frimodt-Møller (Copenhagen, DK)
Objectives: Intracellular persistence of S. aureus has been acknowledged for years but regarding antibiotic treatment, S. aureus infections are still considered as mainly extracellular. Using antibiotic regimens resulting in poor intracellular activity might therefore be a cause of treatment failure. Only few have studied this issue in vivo.
The purpose of this study was to characterise the PK/PD of both the intra- and extracellular anti-staphylococcal activity of linezolid (LZ) using a newly developed in vivo model.
Methods: PK-studies: plasma concentrations over time, in female NMRI mice, were measured by bio-assay after s.c. application of different doses, to test linearity between doses and the PK-parameters. The protein binding in plasma from mice was determined by ultrafiltration. PD-studies: infection was induced by i.p. inoculation of 2.5×107 CFU S. aureus E19977 (MIC-LZ = 0.5 mg/L) in 5% mucin in mice. Two hours later mice received the first of 1–6 doses of 13 different dosing regimens designed to give various fAUC/MIC24 (342–30 hr) and fT>MIC 24 (24–82%). 24 hrs after infection, mice were killed, a peritoneal wash in saline was performed, and the fluid retracted. After quantitative culture of the total bacterial count (TOT) the cell fraction was isolated by centrifugation and the extracellular bacterial count (EX) determined in the supernatant. The remaining extracellular bacteria in the pellet were killed by resuspension in HBSS containing 100 μg/mL of lysostaphin. The cells were lysed in water and the intracellular count (IN) quantified. Data were analysed by the Hill equation.
Results: Linearity between doses and PK-parameters was found. The protein binding was estimated to around 30%. There was no correlation between effect and the fT>MIC, while significant correlations (R2) were found between bacterial killing and fAUC/MIC: 0.73 (TOT), 0.75 (EX) and 0.62 (IN). The Emax (log CFU/mL) for fAUC/MIC ranged from 1.12 (TOT), to 1.9 (EX) and 0.85 (IN), respectively. EC50 (AUC/MIC [hr]) for fAUC/MIC ranged from 147.6 (TOT) to 91.0 (EX) and 144.9 (IN), respectively. The maximum effect in all compartments was reached at around an AUC/MIC ratio of 160.
Conclusion: The majority of the effect of LZ was seen in the extracellular compartment. As expected, best correlations for bacterial killing were found for fAUC/MIC in all three compartments. fT>MIC shows no significant correlation with anti-staphylococcal killing, neither in total, nor in extra- or intracellular counts.
P1379 Pharmacokinetics of azithromycin in serum and sinus fluid after administration of extended release and immediate release formulations in patients with acute bacterial sinusitis
A. Ehnhage, M. Rautiainen, A. Fang, S. Sanchez (Nacka, SE; Tampere, FI; New York, US)
Objectives: High drug levels at the site of infection are desirable for optimal activity. The pharmacokinetic (PK) advantages of azithromycin (AZ) extended release (AZ-ER) versus azithromycin immediate release (AZ-IR) have been shown previously in human lung and animal models (Danesi et al., ATS, 2006; Girard et al., JAC 2005). Hence, the aim of this study was to determine if a single dose of AZ-ER (2 g) achieved higher AZ levels in the sinus fluid compared to AZ-IR (500 mg daily for 3 days). The invasive nature of repeated puncture makes sinus fluid sampling difficult. In this study, an indwelling sinus catheter was used (Ambrose et al., CID, 2004) for repeated sampling to determine serial levels of AZ for the first time.
Methods: This was a randomised, open-label, multi-centre study. Subjects received either a single 2-g dose of AZ-ER (n = 5) or AZ-IR 500 mg once daily for 3 days (n = 4). Serum samples and sinus aspirates were collected for up to 120 h after the first dose. AZ levels were analysed by a validated LC/MS/MS method. AZ levels in sinus fluid were corrected by urea dilution method. A non-compartmental method was used for PK analyses.
Results: Within the first 24 h after dosing, the mean serum AUC24 and Cmax values were 4-fold higher for AZ-ER than for AZ-IR. Similarly, the mean AUC24 and Cmax in sinus fluid were 3-fold higher for AZ-ER than for AZ-IR. After the first 24 h, the average AZ levels in the sinus fluid were higher with AZ-ER than with repeated AZ-IR dosing. The AUC24 values in the sinus fluid were approximately 4–5-fold higher than serum with both formulations. The mean(SD) PK parameters are shown in the table .
| Matrix | Parametera | AZ-ER (n = 3–5) | AZ-IR (n = 3–4) | ER/IR ratio |
|---|---|---|---|---|
| Serum | AUC24 | 7.35 (2.93) | 1.77 (0.57) | 4.2 |
| Cmax | 1.15 (0.38) | 0.30 (0.11) | 3.8 | |
| Sinus fluid | AUC24 | 27.1 (15.5) | 8.78 (4.57) | 3.1 |
| Cmax | 7.34 (10.4) | 2.58 (1.71) | 2.8 | |
| AZ conc. | ||||
| 48 h | 5.34 (9.49) | 2.00 (1.91) | 2.7 | |
| 72 h | 5.57 (11.1) | 1.69 (0.85) | 3.3 | |
| 96 h | 3.22 (3.09) | 2.11 (1.09) | 1.5 | |
| 120 h | 3.39 (4.09) | 1.68 (1.89) | 2.0 |
Units: AZ conc, ug /mL; AUC, ug·h/mL.
Conclusions: AZ demonstrated good penetration into the sinus fluid regardless of the formulation. Despite the small sample size, this study suggests that administration of AZ-ER as a single dose results in increased exposure in the first 24 h and concentrations were maintained in the sinus fluid for up to 120 h.
P1380 Intraperitoneal penetration of imipenem in patients undergoing abdominal surgery
K. Ikawa, N. Morikawa, K. Sakamoto, K. Ikeda, H. Ohge, Y. Takesue, T. Sueda (Hiroshima, JP)
Objectives: Imipenem is used for surgical intra-abdominal infections, however, the penetration into the abdominal cavity remains unclear. This study was conducted to examine the intraperitoneal pharmacokinetics of imipenem to rationalise the dosing regimen.
Methods: Ten patients with inflammatory bowel diseases who underwent abdominal surgery were enrolled. Imipenem-cilastatin sodium 500 mg was administered by intravenous infusion over 30 minutes before the surgery. Plasma and peritoneal fluid samples were collected at the end of the infusion and every hour for 6 hours. Pharmacokinetic data were analysed by the multi-compartment model method, and used for Monte Carlo simulation with MIC data in postoperative intra-abdominal infections.
Results: Imipenem in peritoneal fluid reached a peak concentration of 16.7 ± 4.3 (mean ± SD) (mg/L) at 0.72 ± 0.07 (h), whereas the maximal concentration in plasma was 40.5 ± 13.8 (mg/L) at 0.50 (h), and both concentrations were similar between 1 and 6 hours after the end of infusion. The volume of distribution and total clearance of imipenem were 8.58 ± 3.31 (L) and 9.50 ± 2.38 (L/h), respectively. The penetration ratio from plasma to peritoneal fluid in the area under the drug concentration-time curve was 0.64 ± 0.11, ranging from 0.49 to 0.92. The probabilities of achieving bacteriostatic/bactericidal pharmacodynamic target (20/40% of the time above MIC for 24 hours) were 99/96% in peritoneal fluid and 99/97% in plasma using a regimen of 500 mg every 8 hours against an Escherichia coli population, whereas a regimen of 1,000 mg every 8 hours was needed to produce a values of 90/76% in peritoneal fluid and 93/83% in plasma against a Pseudomonas aeruginosa population.
Conclusion: This study characterised the intraperitoneal pharmacokinetics of intravenously administered imipenem, and provided a pharmacokinetic-pharmacodynamic rationale for the dosing regimen for prophylactic or empiric therapy of surgical intra-abdominal infections.
P1381 Pharmacokinetics of teicoplanin in Chinese patients undergoing CAPD
H. Qi, X. Ma, P. Zhang, L. Gao, H. Cui, F. Hou (Beijing, CN)
Objective: To define a dose regimen of teicoplanin for Chinese patients undergoing CAPD (continue ambulatory peritoneal dialysis) so that they achieve trough drug serum levels above 10 mg/L.
Methods: Eight CAPD patients with acute respiratory system infection (seven with upper respiratory tract infection) of Gram-positive cocci were included in this study. At least seven dose of 400 mg teicoplanin were administered intravenously. After three 400 mg teicoplanin q12h loading doses, patients were given a repeated dose at 24 hour interval, then q72h for at least three times. Concentrations of teicoplanin in samples collected from peripheral vein and excluded dialysis fluid after the fourth dosages and 48, 72, 96 hours after the last dosage were determined by a microbiological assay.
Results: Mean Cmax was 63.57±16. 90 mg/L, mean AUC0–72 at steady state (AUCss) was 921.51±147.22 mgh/L, mean half life (t1/2) was 76.89±21.06 h, mean total clearance rates (CLt) was7.69±0.61 mL/h/kg clearance rates of peritoneal dialysis (CLPD) was 1.66±0.62/h/kg Mean volumes distribution at steady state (Vss) was 40.16±9.10 1, mean serum concentration at steady state (Cav) was 12.69±1.82 mg/L, Trough serum levels above 10 mg/L of most patients were found at 48 hr after the administration (10.03±2.41 mg/L), and mean serum concentration at 72 hr after the administration was 9.05±3.81 mg/L. Concentration of 96 h after last dose was 7.70±0.87 mg/L. The serum drug concentration of one patient whose total creatinine clearance rate was 38l/week (3.77 mL/min) showed that the concentration at 72 hours after administration was higher (17.94 mg/L) than the concentration at 48 hours (12.75 mg/L). The Clinical efficacy rate and bacterial eradication rates was 100%. Two patients experienced anorexia one might be related to primary diseases. There was no adverse effect observed on the anuric patient who had been given teicoplanin for a period of 22 days because of complicating G+ coccus panaritium and pneumonia.
Conclusion: It is proposed that teicoplanin is safety and efficacy on CAPD patient when administered according to the dose recommended by Sanofi-Aventis for renal failure patients with serious infection.
P1382 Concentrations of moxifloxacin (400 mg) in prostatic secretions and ejaculate after a single oral dose in healthy volunteers
F. Wagenlehner, F. Kees, W. Weidner, K. Naber (Giessen, Regensburg, Straubing, DE)
Objectives: The spectrum of bacterial prostatitis comprises Gram-negative, Gram-positive and atypical pathogens. Prostatic concentrations of most antibiotics are far below serum levels. Because of its broad spectrum of activity, moxifloxacin might be a suitable antibiotic for the treatment of bacterial prostatitis. In this study serum concentrations and the penetration of moxifloxacin into prostatic secretions and ejaculate of healthy volunteers after a single oral dose of 400 mg were investigated.
Methods: In a prospective phase I study 12 healthy male volunteers received a single oral dose of 400 mg moxifloxacin and at the same time 3.24 g iohexol intravenuously to determine urinary contamination. Serum concentrations were determined at 0, 0.5, 1, 2, 3 and 4 h, prostatic secretions and ejaculate were determined 3 to 4.5 h after administration of drugs. Urinary concentrations were determined in the collecting urine 0 to 4.5 h. Concentrations of moxifloxacin and iohexol in serum, secretions and urine were determined by HPLC.
Results: Serum and urine concentrations of moxifloxacin were evaluated in 11, prostatic secretions in 8 and ejaculate in 10 volunteers. Interassay and intraassay precisions varied between 3.0 to 5.4% and 5.5 to 7.8%, respectively. The pharmacokinetic parameters of moxifloxacin are shown in table 1 . The possible urinary contamination of prostatic secretions and ejaculate (determined with iohexol) was less than the precision of the analytical method. Concentrations were therefore not corrected.
Table 1.
| Pharmacokinetic parameter | N | Mean | SD | Median | Min | Max |
|---|---|---|---|---|---|---|
| Serum | ||||||
| Cmax (mg/L) | 11 | 2.84 | 0.52 | 2.74 | 2.08 | 3.55 |
| Tmax (h) | 11 | 1.59 | 0.86 | 1.00 | 0.50 | 3.00 |
| AUC0–4 (mg/L*h) | 11 | 9.04 | 1.56 | 9.02 | 6.52 | 11.55 |
| Prostatic secretion | ||||||
| Concentration prostatic secretion (mg/L) | 8 | 3.79 | 1.24 | 3.99 | 2.00 | 5.57 |
| Ratio prostatic secretion/seruma | 8 | 1.61 | 0.51 | 1.57 | 0.97 | 2.32 |
| Ejaculate | ||||||
| Concentration ejaculate (mg/L) | 10 | 2.46 | 0.35 | 2.49 | 1.94 | 2.97 |
| Ratio ejaculate/seruma | 10 | 1.02 | 0.18 | 1.01 | 0.77 | 1.31 |
Corresponding serum concentrations were linearly extrapolated from serum concentrations at 3 and 4 h.
Conclusion: After a single oral dose of 400 mg moxifloxacin the concentrations in prostatic secretion were 60% higher than in serum and in ejaculate comparable to serum, 3 to 4 h after administration. Therefore, moxifloxacin might be a good alternative for the treatment of chronic bacterial prostatitis.
P1383 Penetration of moxifloxacin and lomefloxacin into the prostatic tissue and fluid
S. Yakovlev, R. Komarov, G. Izotova, V. Yakovlev (Moscow, RU)
Objectives: The successful treatment of chronic bacterial prostatitis depends to a large extent on the effective antimicrobial concentrations in the prostate against the causative microorganisms. Fluoroquinolones are recommended as first-line treatment of bacterial prostatitis due to its good penetration into the prostate. In the present study we examined the prostatic tissue and fluid penetration of moxifloxacin (MOX) in comparison with lomefloxacin (LOM) in patients with chronic bacterial prostatitis.
Methods: The concentrations of fluoroquinolones in blood and prostatic fluid were determined in 28 patients with chronic bacterial prostatitis after a first oral 400-mg dose. Blood samples were taken prior to and 1, 2, 3, 6, 12 and 24 h after drug administration. Prostatic fluid was obtained after prostatic massage prior to and 3, 6 and 24 h after drug administration. Thirty patients with benign prostatic hypertrophy undergoing transurethral resection of the prostate received a single oral 400-mg dose of MOX or LOM 3 or 24 h before surgery for prophylaxis. Concentrations of MOX and LOM were measured using a microbiological plate assay. Minimum inhibitory concentrations (MIC) of MOX and LOM for causative pathogens were determined by broth microdilution method.
Results: Maximum serum concentrations of MOX and LOM were 3.4 and 1.3 μg/mL. Three, 6, 12 and 24 hours after administration, the mean serum concentrations of MOX were 3.4, 3.3, 2.2 and 1.1 μg/mL and those of LOM were 1.1, 1.0, 0.7 and 0 μg/mL. Mean prostatic fluid concentrations of MOX at 3, 6 and 24 hours after administration were 3.3, 4.7 and 3.3 μg/mL and those of LOM were 4.5, 2.3 and 0.4 μg/mL. Mean prostatic tissue concentrations of MOX and LOM at 3 hours after administration were 0.3 and 0.5 μg/g and at 24 hours – 0.3 and 0.3 μg/g. MIC90 of MOX and LOM for most common pathogens were: E. coli 0.25 and 0.25 mg/L, Klebsiella spp. 4 and 16 mg/L, Enterobacter spp. <0.125 and 0.25 mg/L, Staphylococcus spp. 0.25 and 1 mg/L, E. faecalis <0.125 and 0.5 mg/L, Streptococcus spp. 0.5 and 2 mg/L.
Conclusion: The concentrations of MOX were higher in both serum and prostatic fluid than those of LOM. Prostatic tissue levels of both quinolones were similar. The concentration of MOX in the prostatic fluid and tissue in contrast with LOM is high enough to eradicate the majority of pathogens causing chronic bacterial prostatitis.
P1384 PK-PD parameters ratio of antibiotics released from antibiotic-loaded spacers in drainage fluids
E. Bertazzoni Minelli, A. Benini, B. Magnan, P. Bartolozzi (Verona, IT)
Objectives: Infection remains a difficult problem in total hip or knee replacement. Polymethylmethacrylate (PMMA) cements impregnated with aminoglycosides and/or vancomycin are currently utilised as local antibiotic carriers in orthopaedic prosthetic infections. Few data are available on antibiotics release in vivo at the moment of implant as well as their antimicrobial inhibitory activity.
Methods: The local and systemic release of gentamicin (G) from temporary spacers (hip and knee) loaded with 2.5% gentamicin (Spacer-G®) was studied in 13 patients undergoing two-stage revision surgery. We currently combine the use of spacers with systemic therapy for the treatment of prosthetic infections. Vancomycin (V) was parenterally (1 g, b.i.d.) or locally administered (cement, 2.5%). The concentrations of G and V were determined in drainage fluid (DF) and serum collected in the first 24 hours after implantation of spacers. Microbiological method and FPIA were utilised. Their inhibitory activity was evaluated against multiresistant clinical isolates (S. aureus, S. epidermidis, S. hominis, S. haemolyticus, E. coli, P. aeruginosa) by bactericidal titer and continuous turbidimetric recording of bacterial growth (Bioscreen, Labsystem).
Results: The release of G from spacers in the infection site seems prompt and effective, attaining high local concentrations. Serum levels of G were very low. Parenteral V showed good penetration in the infection site. Higher levels in drainage fluids were attained by local administration. G and V were present in the implant site at inhibitory concentrations. Drainage bactericidal levels presented great intersubject variability. Highest concentrations of DFs (from 80% to 20%) inhibited bacterial growth of all strains tested for 24 h, 10% for 12 h. G and V in combination showed an additive effect against multiresistant strains when tested at the same concentration of bactericidal titer; lower levels of G and V inhibited bacterial strains for 10–14 h. DFs maintained their inhibitory activity also after filtration and proteins precipitation.
Conclusions: G and V are released from cement at inhibitory concentrations in the infection site for prolonged periods. V presents a good and prompt penetration in the infection site after i.v. administration. The combination of G and V exerts an effective effect against multiresistant bacteria. PK/PD parameters ratio seems good and can be considered a predictive value for therapeutic efficacy.
P1385 Antibiotic diffusion in pseudoarthrosic infected bone: glycopeptides, fluoroquinolones and carbapenems
S. Garazzino, A. Aprato, D. Aloj, F.G. De Rosa, A. D'Avolio, A. Massé, G. Di Perri (Turin, IT)
Objectives: Antibiotic concentration in infected bone is a major determinant of clinical response. As glycopeptides, fluoroquinolones and carbapenems are widely used for the treatment of bone infections, aim of our study was to assess their diffusion in infected human bone.
Methods: Patients with a septic pseudoarthrosis undergoing debridement of infected tissue and treated with either a glycopeptide, a fluoroquinolone or a carbapenem iv for >1 week, were studied. Plasma and bone specimens were collected intraoperatively for PK and microbiologic assays at a mean of 4.7 h after antibiotic administration. Antibiotic concentrations were measured by HPLC-UV method.
Results: Twenty patients (pts) were studied. Five received vancomycin: bone cultures grew E. Faecalis, MRSA and MRSE; bone concentrations (mean) were 2.4 mg/L in cortical and 7.1 mg/L in cancellous bone, with a bone/plasma extraction of 12% and 36%, respectively.
Five pts were treated with teicoplanin for MRSA infection: bone concentrations were 3.1 mg/L and 7.7 mg/L respectively for cortical and cancellous bone (16% and 39% of plasma levels).
Five pts received a fluoroquinolone for E. coli or Enterobacter spp. infection. Ciprofloxacin concentrations were 1.8 mg/L in cortical, and 30.2 mg/L in cancellous and newly formed bone (respective bone/plasma ratios: 1.06, and 8.4). Levofloxacin concentrations were 0.3 and 2.69 mg/L in cortical and cancellous bone, with diffusion rates of 12% and 108% respectively.
Five pts received a carbapenem. Imipenem diffusion rates were respectively 7.5% and 58.3% for cortical and cancellous bone (bone concentrations: 0.09 mg/L and 0.7 mg/L). Meropenem bone levels were 1.2 mg/L and 5.2 mg/L in cortical and cancellous bone, with respective diffusion rates of 3.6% and 15%.
Conclusion: Both vancomycin and teicoplanin provided concentrations exceeding the MIC of infecting agents and displayed a satisfactory bone diffusion. Ciprofloxacin provided tissutal levels higher than levofloxacin, and both showed a good bone diffusion; ciprofloxacin concentrations in cancellous bone and in bony callus were far higher than in plasma, suggesting an accumulation into highly vascularised bone. Imipenem had higher diffusion rates than meropenem, but bone levels were under the MIC of susceptible agents. Glicopeptides and fluoroquinolones appear an excellent option for bone infections, while carbapenems should be considered a second choice treatment.
Measuring and restricting antibiotic usage
P1386 Successful reduction of the use of intravenous ciprofloxacin using a simple protocol
I. Willemsen, C. van Buitenen, M. Winters, P. van Hattum, J. Kluytmans (Breda, NL)
Objectives: The objective of this study was to reduce the hospital use of intravenous (iv) ciprofloxacin (CIPR) with 50%.
Methods: This project was performed in a 1,400 bed teaching hospital including all medical specialities. The intensive care unit was excluded from the intervention. There were three criteria to qualify patients for a switch to oral medication. First patients should be able to take oral medication. Second, no switch was allowed when patients were haemodynamically unstable (systolic bloodpressure <100 mmHg or pulse >100 beats/min) and third, when patients were given parenteral nutrition. A pharmacy assistant assessed all iv recipes for a switch to oral medication. The medical staff of the hospital approved with the switch protocol. The attending physician was informed when a switch was possible. In complicated cases and when the pharmacy assistant considered the switch questionable, the microbiologist was consulted. The intervention started on January first 2006. The use was registered per month in Prescribed Daily Doses (PDD) CIPR iv therapy.
Results: The use in 2006 between January and October was 623 PDD of CIPR iv. In 2005 in the same period the use was 1,281 PDD. The reduction was achieved after one month and stable from that moment on.

Conclusion: Ten months after the start of the project the aim of the project, reduction of the CIPR iv medication with 50% has been achieved. This will result in a net saving of approximately €50.000. This is conservative estimate of the true savings, as it does not include the costs associated with giving IV treatment, e.g. the infusion system.
P1387 Is treatment with sulfamethizole before admission associated with poor outcome in bacteraemia patients? A Danish population-based cohort study
M. Søgaard, M. Nørgaard, H. Sørensen, H. Schønheyder (Aalborg, Århus, DK)
Objectives: In contrast to most other countries, sulphamethizole is still used in Denmark for uncomplicated urinary tract infections. Concern has been raised that sulphamethizole may be associated with a poor outcome if the clinical course is complicated by bacteraemia. We compared the impact of preadmission treatment with either sulphamethizole or a commonly used alternative, pivmecillinam, on the 30-day mortality following community-acquired bacteraemia (CAB).
Methods: We conducted a 10-y population-based cohort study in North Jutland County (1995–2004) based on all patients with a first-time episode of CAB. We obtained data from the County Bacteraemia Registry, Prescription Database, and Hospital Discharge Registry. All patients were followed for 30 days after hospitalisation through the Danish Civil Registration System. We compared cumulative mortality and mortality rates in patients who did and did not redeem at least one prescription for sulphamethizole or pivmecillinam within 30-days before admission and controlled for gender, age, comorbidity, and type of bacteraemia in a Cox regression analysis.
Results: During the study period we identified 2,858 eligible patients, 108 (3.8%) and 86 (3%) had redeemed at least one prescription for sulphamethizole or pivmecillinam, and 11 had redeemed prescriptions for both. Overall cum. 30-day mortality in patients exposed to sulphamethizole alone or in combination with other antibiotics was 13.4% and 12.2%, respectively, compared with 16.2% among patients not exposed to antibiotics [adjusted mortality rate ratios, MRRs, 0.9 (95%CI: 0.5–1.7), and 1.0 (95%CI: 0.4–1.5)]. The corresponding figures for patients exposed to pivmecillinam alone or pivmecillinam and other antibiotics was 15.4% and 8.5%, respectively [adj. MRRs 1.1 (95%CI: 0.4–3.0), and 0.6 (95%CI: 0.2–1.4)].
Conclusion: In this study treatment with either sulphamethizole or pivmecillinam prior to admission did not influence 30-day mortality.
P1388 Aminoglycosides, mortality and increase of serum-creatinine in patients with bacteraemia given appropriate empirical therapy. A Danish hospital-based cohort study
H. Schønheyder, M. Freundlich, L. Pedersen, H. West, R. Thomsen (Aalborg, DK)
Objectives: Aminoglycoside (AG)-β-lactam combination therapy has been discouraged after the appearance of metaanalyses which did not find significant advantages compared to β-lactam monotherapy. Whether this conclusion is valid to all patient groups is questionable and therefore we examined the association between AG therapy, mortality and increased serum-creatinine in patients with bacteraemia given appropriate empirical antibiotic therapy.
Methods: Cohort study based on prospective registration of bacteraemias 1996–2002 at a Danish hospital. AG+β-lactam was the recommended empirical therapy for severe sepsis. We identified 1,257 patients, 969 of whom received gentamicin or tobramycin (AG cohort), while 288 patients who were not exposed to AGs formed the non-AG cohort. We performed Cox regression analysis to compare mortality rates adjusted for potential confounders including comorbidity; the association between AG therapy and an increase of serum-creatinine >45 mmol/L was analysed by logistic regression.
Results: The cumulative 30-day mortality in the AG cohort was 17.3% vs. 18.1% in the non-AG cohort (adjusted mortality rate ratio (MRR) 1.02; 95% CI 0.74–1.39). For patients alive after 30 days the cumulative mortality 31–180 days after the bacteraemia was 20.3% in the AG cohort vs. 12.3% in the non-AG cohort (adj. 31–180 day MRR 1.72; 95% CI 1.15–2.55). AG therapy was associated with lower 30-day mortality in patients with an abdominal focus (adj. 30-day MRR 0.52; 95%CI 0.24–1.10) or a urinary tract focus (adj. 30-day MRR 0.48; 95%CI 0.22–1.08), but with a worse prognosis in patients with a respiratory tract focus (adj. 30-day MRR 2.06; 95%CI 0.93–4.53). The higher mortality associated with AG therapy beyond day 30 was observed for all major foci except for the urinary tract where the MRR was close to one. Serum-creatinine concentration was available at baseline in 983 (78.2%) patients and repeated measurements within 14 days in 422. An increase of serum-creatinine >45 mmol/L was observed less often in AG treated patients (5.9% vs. 15.0%, adjusted OR 0.54; 95%CI 0.26–1.15).
Conclusion: Among patients with bacteraemia given appropriate empirical therapy, AG therapy was neither associated with increased 30-day mortality nor risk of rising serum-creatinine. AG therapy was associated with a survival benefit during the first weeks of infection in patients with a urinary tract or an abdominal focus. The long-term outcome warrants further consideration.
P1389 Mortality risk factors in patients admitted to an emergency unit with severe infection treated with imipenem
N.R. Berenguer Torrijo, S. Grau Cerrato, J. Mateu-de Antonio, S. Luque Pardos, O. Ferrández, D. Conde-Estévez, E. Salas (Barcelona, ES)
Objectives: To assess the mortality risk factors in patients with severe infections treated empirically with imipenem (IMP) when they were admitted to an emergency unit (EU).
Methods: Prospective observational study carried out in 104 patients consecutively admitted in a EU from September 2003 to March 2006 in a 450-bed university hospital. Patients included where those who presented severe infections at admission and started an empirically antimicrobial therapy with IMP. For the analysis we selected those patients in which a positive microbiological culture was further obtained (67 patients). Simplified Acute Physiology Score II (SAPS-II) was used to stratify the severity of patients at admission.
In statistical analysis, Fischer exact Test was employed for dichotomic variables and “U”Mann-Whitney test for continuous variables. A regression binary logistic was used to evaluate the risk factors of mortality.
Results: Isolated microorganisms were susceptible to imipenem in 63 (94%) patients. Statistical differences were not observed when the infection source was compared (p = 0.466).
| Surviving patients | Deceased patients | p | |
|---|---|---|---|
| No. of patients | 59 | 8 | |
| SAPS-II at admission | 38.3 (95% CI: | 47.4 (95% CI: | 0.038 |
| 35.05–41.58) | 39.15–55.60) | ||
| Presence of septic shock | 5 (8.5%) | 3 (37.5%) | 0.048 |
| Multiresistant microorganisms isolated | 20 (33.9%) | 3 (37.5%) | 1.00 |
| Type of multiresistant microorganisms | 0.652 | ||
| EBSLa-producing | 9 (45%) | 1 (33.3%) | |
| Pseudomonas aeruginosab | 2 (10%) | 0 | |
| Other multiresistant Gram(−)b | 7 (35%) | 1 (33.3%) | |
| Gram(+) microorganismsc | 2 (10%) | 1 (33.3%) | |
| Polymicrobial infections | 12 (20.3%) | 3 (37.5%) | |
| Evaluation of mortality | Odds ratio | 95%CI | p |
| SAPS-II at admission | 1.060 | 0.997–1.127 | 0.061 |
| Presence of septic shock | 6.480 | 1.184–35.452 | 0.031 |
| Multiresistant microorganisms | 1.170 | 0.253–5.401 | 0.841 |
EBSL, extended-spectrum β-lactamase.
Resistant to more than 3 antibiotic families.
Presence of methicillin or vancomycin resistance.
Conclusions: Died patients presented a higher SAPS-II at admission and a higher frequency of septic shock. The presence of septic shock was the only factor associated with mortality when logistic regression was applied.
Neither the isolation of Gram-positive or Gram-negative microorganisms nor the presence of multirresistant microorganisms was related with mortality.
Imipenem showed an appropriate microbiological coverage against most of the isolated microorganisms, including Gram-negative bacilli and Gram-positive cocci, even in the presence of multirresistant strains.
P1390 Impact of antibiotic referral and management system on antibiotic expenditure in a hospital
E. Fleet, G. Gopal Rao (London, UK)
Background: Excessive expenditure on antibiotics is frequently a cause for concern in many hospitals. Prudent and cost-effective prescribing of antibiotics is necessary to minimise this expenditure.
Objective: To describe the impact of Antibiotic Referral and Management System (ARMS) on antibiotic expenditure in a general hospital.
Methods: ARMS was implemented in 2005/6 to promote prudent and cost effective prescribing of antibiotics in in-patients in our hospital. ARMS consists of a series of practical measures used to aid in such prescribing.
The key measures included in ARMS were:
-
•
Restriction of empirical use of antibiotics not included in the hospital formulary.
-
•
Screening of all antibiotic prescriptions to identify those that did not comply with hospital guidelines.
-
•
Referral of non-compliant prescriptions to ‘antibiotic pharmacist'.
-
•
Review of non-compliant prescriptions by antibiotic pharmacist and medical microbiologist (The Antibiotic Management Team [AMT]).
-
•
Discussion with prescribing doctor and recommendation to change to appropriate antibiotics in line with patients' latest laboratory results.
-
•
Monitoring by Ward pharmacists to ensure that changes are implemented and intravenous antibiotics are switched to oral as per guidelines.
Financial impact was determined by comparing the antibiotic expenditure before and after ARMS.
Results and Conclusion: There was €211,000 (16%) reduction in expenditure compared to previous financial year (2004/5). Considerable reductions were seen in the use of IV antibiotics, cephalosporins, IV ciprofloxacin, teicoplanin and linezolid.
Conclusion: Implementation of ARMS leads to substantial reduction in expenditure on antibiotics. Review of clinical outcomes is necessary to reassure clinicians.
P1391 The HARMONY antibiotic policy and process tools and SWAB collaboration
B. Cookson, J. Prins, J. Kullberg, I. Gyssens (London, UK; Rotterdam, NL)
Objectives: To assess the applicability of the HARMONY tools for international education and helping to produce a national prescribing policy.
Methods: The HARMONY tools were developed by BC interacting with 19 hospitals in the UK. It was an iterative process and much was learnt about differing opinions on prescribing practices and policy writing. For example, some were of an enormous size and unlikely to be used, authors were opposed to changing them and often did not seek comments from users. In a second HARMONY phase the tools were further progressed in the EU with 14 experts in antibiotic stewardship in ESCMID ESGAP and 20 others in UK, Malta and German workshops. We describe here their further progression at international educational meetings and in the National Netherlands Antibiotic Policy Group (SWAB). 76 professionals representing 44 hospital committees were sent the tool. Analysis was performed before the meeting and results were presented at the SWAB meeting, which coincided with the ARPAC Consensus meeting.
Results: Parts 1 and III of the tool were found to be particularly helpful in multi-disciplinary discussions at two International Federation of Infection Control meetings, national consensus meetings in Malta, Oman and Dubai. In multi-disciplinary meetings there are issues relating to stop dates for surgical prophylaxis and treatment, rotating of antibiotics and switching to less expensive antibiotics, these being common audit targets.
The whole tool also informed the design of the ARPAC and ARMed EU projects.
64% of SWAB participants responded: this is very high considering the large size of the tool and its orientation for local rather than national policy design. The tool was found to be very useful in progressing a consensus: for example 52% of prescribing effectiveness statements, and 70 to 85% of other sections were agreed with. There were interesting discussions including the use of costing data in a national policy. The spreadsheet approach and Likert qualitative scales were particularly successful in facilitating the rapid production of useful results that enabled further discussions at the meeting.
Conclusion: The HARMONY tools have now matured into a stage where they should be of use in many other countries including the EU and further discussions are underway to explore this possibility.
P1392 Effects on quality prescription of a prospective interventional study based on a restrictive antibiotic policy
F. Jover, J.M. Cuadrado, V. Sánchez, V. Ortiz de la Tabla, M. Gonzalez, R. Martinez, E. López-Calleja, C. Martin (Alicante, ES)
Objective: To assess the impact of a programme to evaluate the quality of an antibiotic restriction policy at an University hospital.
Methods: A multidisciplinary antimicrobial management team (AMT) composed by Infectious Diseases, Microbiology and Pharmacy members daily evaluated restricted antibiotic prescriptions. We designed a prospective study with two periods: observational (October 1st 2003– March 31th 2004) and interventional (April 1st 2004–February 28th 2006). A specific prescription formulary aiming to evaluate the usage of broad spectrum antibiotics was introduced. Revision criteria were according to ID Comittee guidelines and Sanford Guide to Antimicrobial Therapy. Clinical, microbiological and pharmaceutical data were collected from each patient. Antibiotic therapy appropiateness, restricted therapy elegibility and economic outcomes were evaluated. During interventional period, antibiotic recommendation prescriptions were formulated. Statistical analysis with non-parametric tests were performed. A p value of <0.05 was accepted as significant.
Results: 1,212 patients were evaluated (21% observational/79% interventional period). Overall, restricted agents prescribed were imipenem (26.1%), ceftazidime (23.5%), piperacilin-tazobactam (15.3%), amikacin (12.3%), vancomicin (10.6%), cefepime (5%). Most frequent diagnostic groups were sepsis (36.3%), respiratory (16.2%), prophylaxis (14.6%), gastrointestinal (14.4%), soft-tissue and bone infections (13.7%) and UTI (4.9%). Any microbiological sample was taken in 78.8% of cases. Comparing both periods, adequate prescription of antibiotherapy increased from 90.7% to 97.3%. Restricted antibiotic prescription significantly improved from 59.4% to 88.5%, while concordance to AMT prescription increased from 54.8% to 81.1%. We found a significant reduction in DDD/1,000 patients-day in two periods (52.36 vs 46.21: p <0.05). Other several statistically significant diferences in DDD/1,000 patients-day were detected: decrease in ceftazime (15.3 vs 8.9) and teicoplanin (3.11 vs 0.5). Economic outcomes were significantly reduced. In the interventional period, 329 written recommendations were formulated (27.1%), while 73% of them were accepted.
Conclusions: The development of prophylactic and therapeutics local guidelines reached by consensus, and the evaluation and interventional policy by an AMT group have significantly improved the quality and economical costs of restricted antibiotic prescription in our hospital.
P1393 The evaluation of a fluoroquinolone restriction policy at a university teaching hospital
A. Hilts, D. Fish, M. Barron (Denver, US)
Objectives: Gram-negative bacterial (GNB) resistance has been associated with overuse of fluoroquinolones (FQ). At the University of Colorado Hospital, increasing resistance in Pseudomonas aeruginosa has occurred concurrent with increased use of FQs. In 1/2005, FQs were placed on 100% restriction in an effort to curb resistance, with approval by an Infectious Diseases (ID) physician or Pharm.D. required prior to use. The objective of this study was to evaluate the impact of this restriction policy on FQ use [defined daily doses (DDD)/1,000 admissions] and appropriateness, patient outcomes, and GNB resistance.
Methods: Hospital records were used to evaluate FQ use, cost, and GNB resistance. Randomly selected charts of 139 hospitalised patients receiving FQs (65 pre- and 74 post-restriction) were reviewed for indications and appropriateness of FQ use and patient characteristics. Appropriateness was determined by a consensus panel of 2 ID PharmDs and 1 ID MD based on indications, pathogen susceptibilities, dose, route of administration, and availability of other suitable drugs. FQs were also deemed appropriate if allergies or renal dysfunction made the use of other agents undesirable. Statistical significance was defined as P<0.05.
Results: Compared to 2004 (pre-restriction), patients receiving FQs in 2005 (post-restriction) were more often transferred from outside facilities (P = 0.04), had previously received antimicrobials (P = 0.001), were treated for hospital-acquired pneumonia (P = 0.01), were infected with P. aeruginosa or MRSA (P = 0.03), and had history of penicillin allergy (P = 0.01). FQ use in 2005 was reduced 66% compared to 2004 (64 DDD/1,000 vs. 188 DDD/1,000). Appropriateness of empiric FQ use was 92% post-restriction vs. 68% pre-restriction (P = 0.0008). FQ acquisition cost was reduced 60%, a savings of US$151,000. Other outcomes (length of FQ therapy, hospital length of stay, mortality) were not significantly different. Resistance of P. aeruginosa to FQ decreased from 49% to 39% (P = 0.02) while susceptibilities of other drug classes remained stable.
Conclusion: Implementation of a rigorous restriction policy significantly increased appropriateness of FQ use while decreasing overall use, cost, and P. aeruginosa resistance. Other patient outcomes were unchanged despite apparently higher-risk patients receiving FQ in the post-restriction period. Increased FQ susceptibility of P. aeruginosa was achieved with no adverse effects on susceptibility to other drug classes.
P1394 Therapeutic approach to upper respiratory tract infections among primary healthcare physicians
W.L. Lukas, L.P. Panasuik, I.S. Szymczyk, J.S. Skorupka (Zabrze, Lublin, PL)
Objective: Appraisal of factors underming the decision about antibiotic use.
Methods: 152 Primary Health Care (PHC) physicians (70% of women), mean age 42y, mean PHC work's span 12y participated in the questionnaire-based study, namely: 38 physicians without any specialty degree, 67 family physicians, 5 general practitioners, 17 internists, 18 paediatricians, 7 other specialits. Subjects were asked about their work-span in the PHC, area of practising (village, town, city), awareness of current guidelines on antibiotic usage, previous attending of any training courses concerning antibiotic-therapy. Two case report questions with options for further treatment were presented.
Results: As for the factors enhancing antibiotic use of highest consequence were: clinical picture, previous experience, awareness of guidelines, while of least were: patients' claims, pharmaceutical companies' suggestions, consultation with specialists. Acute upper respiratory tract infections (URI) was found to be an indication for antibiotic-therapy in 46% of repondents (only 26% considered viral aetiology) while its purulent stage did in 92%. Gender, work span in PHC, previous attending of training courses did not affected the therapeutic decision significantly. Physicians under 40 administered antibiotics at the purulent stage rarely, were more adherent to guidelines and attentive to patients' demands. Rural doctors prescribed antibiotics for URIs significantly more frequently than those from urban areas. Comparing to colleaques without any specialty degree family physicians showed less regard to recommendations and consultations and aimed at optimal duration of therapy.
Conclusions: Despite the introduction of relevant recommendations (2003) and common participation in training courses, vast majority of PHC physicians proved not to adhere to current guidelines.
P1395 Controlling costs in the intensive care unit: role of daily microbiology rounds
A. Kothari, V. Sagar, B. Panigrahi, N. Selot, A. Bhan (New Delhi, IN)
Objective: Spiralling costs of procedures and drugs have led to an increasing emphasis on controlling costs in hospitals, with a special focus towards ICUs. Historically, antimicrobial control policies have been used to control costs, with a side benefit of reduced selective pressure and thus less antimicrobial resistance.
We did a retrospective analysis looking at the impact of daily clinical microbiology bedside rounds in our cardiothoracic surgical ICU on antibiotic consumption patterns of expensive, broad spectrum agents.
Methods: Data regarding usage of antimicrobial agents before (Jan–Feb 2005) and after (Mar–Dec 2005) the introduction of clinical microbiology rounds was analysed. Defined daily doses (DDD) per 1,000 patient bed days was calculated for carbapenems, antipseudomonal penicillins, glycopeptides, and 3rd generation cephalosporins using the National Nosocomial Infection Surveillance definitions. Drug acquisition costs of these antibiotics were used to calculate antibiotic cost (in USD) per patient day in the ICU. The statistical significance of the difference between the 2 groups was calculated using the unpaired t-test.
Results: The DDD per 1,000 patient bed days in the period before the introduction of clinical microbiology rounds was 146.4 for carbapenems, 308.1 for antipseudomonal penicillins, 127.8 for glycopeptides and 235.2 for 3rd generation cephalosporins. After the introduction of clinical microbiology rounds, the DDD per 1,000 patient bed days was 35.6 (p = 0.0153) for carbapenems, 150.9 (p = 0.0137) for antipseudomonal penicillins, 140.4 (p = 0.78) for glycopeptides and 119.5 (p = 0.3) for 3rd generation cephalosporins. The antibiotic cost per patient day reduced from 48.5 USD before introduction of clinical microbiology rounds to 20.6 USD in the subsequent period (p = 0.0023). Sepsis associated morbidity and mortality did not change during the same period.

Antibiotic cost in USD per patient day in Cardiothoracic ICU.
Conclusions: There was a significant decrease in the costs associated with antibiotic usage after the introduction of clinical microbiology rounds in the ICU. Our experience, which is a novel experiment in India, showed that daily clinical microbiology bedside rounds helped in optimising antibiotic therapy of patients, reducing the cost of therapy with no adverse impact on morbidity and mortality from sepsis in the same period.
P1396 Impact of an infectious disease service on the quality of the management of infections and the usage of antimicrobials
S. Lemmen, S. Koch, F. Huenger, H. Haefner (Aachen, DE)
Objective: The aim of the study was to evaluate the impact of an infectious-disease consult service on the quality of medical care especially concerning the usage of antimicrobials in mainly severe infections.
Method: From January till June 2006 1,000 consults were documented by 4 infectious-disease specialists at the University Hospital Aachen, Germany with an elaborated standardised questionaire. Since 1997 the infectious disease service is implemented and written guidelines for the usage of antibiotics exist.
Results: 92% of the consults were performed at 7 different ICUs and 8% at normal wards, respectively. In 78% the consult was performed during the routine weekly infectious disease ward round together with the attending physicians; 15% were telephone consults; and in 7% the ID specialist was called. The locations of the infections were as follows: 310 respiratory tract, 201 sepsis, 101 S-ST, 83 endocarditis, 60 abdominal, 35 CNS, 33 each urogenital and bone and joint. In 201 cases there was no obvious infection diagnosed and in 117 patients there was definitely no infection present.
In 430 consults additional microbiological work up and in 170 further diagnostic procedures (e.g. ultrasound, echo, endoscopy or radiology) were recommended, respectively. In 49 patients antimicrobials were started due to the advice of the ID specialist, however in 50 cases the physician could be convinced not to start any antimicrobials.
In 837 patients antimicrobials were already administered at the time of the consult; in 336 cases the therapy was considered adequate. However in 501 cases interventions were performed: 134× stopping of the therapy, 129× deescalation to monotherapy, 60× decrease and 71× increase of the antimicrobial spectrum, 60× decrease and 21× increase of the duration, and 10× increase and 16× decrease of the dosage.
It could be calculated that a total of 3,500 antibiotic days were not given in comparison to if no intervention had happened. According to the data of our pharmacy the average cost for one antibiotic day is 45€, thus a total of 157,500€ could be saved.
Conclusion: This study documents that an infectious-disease service had an impact on the quality of medical care by suggesting further diagnostic procedures and reducing the usage of antimicrobials. Since antibiotics are a major driver for bacterial resistance, their limited administration is necessary. The saving outweighs by far the expenses for the personnel and the recommended diagnostic.
P1397 The economical impact of new anti-infective drugs on the drug budget of a surgical intensive care unit
T. Hoppe-Tichy, M.A. Weigand (Heidelberg, DE)
Background: New antiinfective drugs are usually expensive. In many cases the new antiinfectives will replace older and cheaper drugs. We studied antiinfective drug consumption in a surgical ICU with 14 beds to show the impact of new antiinfectives on the overall drug costs and to show the changes in consumption of defined daily doses (DDD; according to the WHO ATC/DDD methodology), costs and costs per DDD in the years 2000 to 2005 regarding different ATC classes, respectively.
Methods: Antiinfective drug consumption data were taken from the pharmacy files. New antiinfective drugs were defined as Tigecycline (first appearance in hospital: 2005), Ertapenem (2005), Synercid (2000), Linezolid (2001), Voriconazole (2003) and Caspofungin (2002). Changes in consumptions were calculated on the basis of data for 2000.
Results: There is an increase in overall drug costs in the ICU of 140% (years 2000 to 2005). The costs of antiinfective therapy increased by 570%. More than 50% of the additional costs were due to antifungal drug therapy. In contrast numbers of DDD only increased slightly to 44%. The rate in costs for new antiinfectives within all antiinfectives rose from 16 to 45%, the rate in DDD only from 0 to 16%. The cost of DDD of new antibiotics is 4 times higher, of new antifungals 20 times higher than the old drugs in those groups. The use of new antiinfectives is highly connected to the length of stay in the ICU (Regression coefficient: 0.977).
Conclusion: New antiinfective drugs are responsible for disproportinately higher costs in the ICU setting due to the high costs of DDD. Primarily, the pricing for new antibiotics is based upon the cost of DDD for Linezolid and for the new antifungals to Caspofungin. Within the group of antiinfectives the new antifungal agents are the primary cost drivers. Therefore new antiinfective drugs should be controlled by methods such as patient based prescribing, counselling by multidisciplinary antibiotic management teams, restricted prescribing or written recommendations. Length of stay is associated with the use of new antiinfectives in our surgery ICU.
P1398 The impact of empiric antimicrobial treatment and the clinical microbiological guidance in sepsis
U.S. Jensen, D.L. Monnet, N. Frimodt-Møller, J.D. Knudsen (Copenhagen, DK)
Objectives: The recommendations for antibiotic therapy are based on local findings of microorganisms and the resistance patterns found. The recommendations for empiric sepsis treatments are of especial importance for the outcome and of patients. The recommended empiric treatment for community-acquired sepsis with undocumented focus is cefuroxime, usually 1.5 g tid, together with gentamicin 3–5 mg/kg od (for max three days), and metronidazole 500 mg tid if an abdominal focus is suspected.
Evaluation of the coverage of the recommended empiric treatment and the impact of microbiological guidance is sought by continuous electronic registration.
Materials and Methods: The findings in blood cultures at a centralised clinical microbiology laboratory serving 2,500 beds in 5 somatic hospitals in Copenhagen were electronically recorded together with information regarding the signs and symptoms and the treatment guidance given by the clinical microbiological consultants, in a new electronic recording system from January 2006. Only cases of sepsis with findings of significant microorganisms were included, and cases with coagulase negative staphylococci, Corynebacteria, and other possible less important findings were excluded. Only one episode per patient was included.
Results: In the six month period, Jan–Jun 2006, the findings of a total of 582 sepsis patients were considered significant, and the electronic records were evaluated for 362 (70%) of these patients. The overall 30 days mortality was 18%, varying from 62% for fungaemia, and 8% for pneumococcus sepsis, respectively. Escherichia coli was the most frequent finding, in 39% of all cases, followed by Staphylococcus aureus in 10% and Pneumococcus in 9%. Among the E. coli, 98% were fully susceptible to cefur, 5% of the Pneumococci were intermediate susceptible to penicillin, and all S. aureus were MSSAs. Cefur alone covered 77% of the blood culture findings. The guidance by the consulting microbiologist modified the treatments if necessary, in almost all cases within two days after the blood cultures were drawn. The group of patients (N = 277) with bacteria in blood cultures susceptible for cefuroxime had a significantly better outcome with 30 days mortality of 16% compared to patients (N = 64) with bacteria not susceptible to cefur (Fisher's exact test, p = 0.03).
Conclusions: The coverage of the empiric antimicrobial treatment was sufficient and clinical microbiological guidance had a major impact on the outcome of bacteraemia.
P1399 Cost-effectiveness of ertapenem vs. piperacillin/tazobactam in the treatment of diabetic foot infections in the United Kingdom accounting for development of antibiotic resistance
J. Jansen, R. Kumar, Y. Carmeli (Houten, NL; Whitehouse Station, US; Tel Aviv, IL)
Objective: Increased antibiotic use is associated with a greater likelihood of reduced effectiveness due to resistance development in the future. The objective of this study was to evaluate the cost-effectiveness of ertapenem versus piperacillin/tazobactam in the treatment of diabetic foot infections in the UK, accounting for development of antibiotic resistance over time.
Methods: A decision tree model was developed to estimate the cost-effectiveness of ertapenem vs piperacillin/tazobactam at different time points following introduction of treatment. Development of antibiotic resistance was incorporated in the analysis using a previously published compartment (SIS) model (Laxminarayan and Brown et al., 2000). The development of resistance was a function of the eradication rate of pathogens and prescription of the antibiotic. The microbiological eradication rate and clinical success rates were assumed to be related and were obtained from the SIDESTEP study (Lipsky et al., 2005). These data included the impact of resistant pathogens (either acquired or intrinsic resistance) such as Enterobacteriaceae, MRSA, enterococci and P. aeruginosa. Model outcomes over time included quality-adjusted life years (QALYs), direct medical costs and cost per QALY saved. Clinical efficacy of second-line treatment, direct medical costs, and utilities were derived from other existing studies. Probabilistic sensitivity analyses were undertaken to estimate the uncertainty of model outcomes.
Results: The model suggested savings of £406 (95% uncertainty interval £–381; £1,468) per patient when diabetic foot infections are treated with ertapenem instead of piperacillin/tazobactam. Probabilistic sensitivity analysis found a 93% probability of the incremental cost per QALY saved being within £20,000. After 3 years, it is expected that the antibiotic resistance profile with piperacillin/tazobactam has increased with a greater rate than with ertapenem. As a result, the cost-savings with ertapenem are expected to increase to £2,694 (£1,856–3,616), and ertapenem will additionally result in greater clinical success rates and QALY savings (0.90; 0.34–1.58).
Conclusion: Ertapenem appears to be an economically dominant therapy over piperacillin/tazobactam for the treatment of patients with diabetic foot infections in the UK.
P1400 Pharmaco-economic impact accounting for future resistance of ertapenam vs. piperacillin/tazobactam in the treatment of community-acquired complicated intra-abdominal infections in the Netherlands
J. Jansen, R. Kumar, Y. Carmeli (Houten, NL; Whitehouse Station, US; Tel Aviv, IL)
Objective: Ertapenam and piperacillin/tazobactam are commonly used in the treatment of complicated intra-abdominal infections. However, increased antibiotic use is associated with a greater likelihood of resistance development. The objective of this study was to evaluate the cost-effectiveness of ertapenem versus piperacillin/tazobactam accounting for development of antibiotic resistance using treatment costs from the Dutch setting.
Methods: A decision tree model was developed to estimate the cost-effectiveness of ertapenem vs piperacillin/tazobactam. Development of resistance was incorporated in the analysis using a previously published compartment model (Laxminarayan and Brown et al., 2000). Resistance was a function of the eradication rate of pathogens and prescription of the antibiotic. Model outcomes over time included quality-adjusted life years (QALYs), direct medical costs and cost per QALY saved. Microbiological eradication rate, clinical success rates of first and second line treatment, direct medical costs, and utilities were derived from the literature. These included pathogens intrinsically resistant to ertapenam such as enterococci and P. aeruginosa, and those with intrinsic or acquired resistance to piperacillin/tazobactam. Probabilistic sensitivity analyses were undertaken to estimate the uncertainty of the model outcomes.
Results: The model suggested overall savings of €355 (95% uncertainty interval €–501; €1,189) per patient when abdominal infections are treated with ertapenem instead of piperacillin/tazobactam. These results were derived based on including first line treatment (ertapenam or piperacillin/tazobactam) and switches in therapy for resistant pathogens. Probabilistic sensitivity analysis found a 94% probability of the incremental cost per QALY saved being within the generally accepted threshold for cost-effectiveness (€20,000). After 5 years, it is expected that the antibiotic resistance with piperacillin/tazobactam has increased with a greater rate than with ertapenem. As a result, the cost-savings with ertapenem are expected to increase to €430 (€–286; €1129), and ertapenem will result in greater clinical success rates and QALY savings (0.12; 0.04–0.24).
Conclusion: Given the underlying assumptions and data, ertapenem is an economically dominant therapy over piperacillin/tazobactam for the treatment of community acquired complicated abdominal infections in the Netherlands.
P1401 Pharmaeconomics of aspergillosis after liver transplantation: a descriptive evaluation
M. Rosenhagen, R. Feldhues, H.K. Geiss, T. Hoppe-Tichy (Heidelberg, DE)
Objectives: Invasive aspergillosis (IA) causes significant morbidity and mortality. New antifungal agents have been introduced which improve the management of invasive fungal infections. But therapy of IA is still associated with poor outcome and extremely high costs.
We evaluated the costs associated with cases of IA in patients undergoing liver transplantation in a university hospital.
Methods: According to a retrospective investigation of all liver transplant patients (Dec 01 to Dec 04) in the transplantation unit of the University of Heidelberg, Germany, patients developing IA were pair wise matched with controls on the basis of SAPS II and age.
IA was classified in accordance with EORTC/MSG criteria. The economic impact of antiinfective treatment was assessed taking into account the prices of defined daily doses (DDD). Prices were taken from the hospital formulary.
Results: A total of 195 liver transplantations were performed on 170 patients. 14 patients developed IA (8.2%). One patient was excluded from economic analysis due to poor documentation. The mortality rate for patients with IA (n = 13) and matched controls (n = 46) were 92.8% and 19.2%, respectively (p = 0.001). There was no significant difference between the age and SAPS II score of cases and controls.
The mean costs of antifungal plus antibacterial treatment in patient with IA was 19.006 € vs 1.136 € in the control group, with an average increase of 59% per patient.
In two patients IA was proven. The costs of these patients were 49.692 € and 10.759 €, respectively (controls: 454 €/2.324 €). The criteria for probable IA were fulfilled by six patients, recieving medication for €8,486 on average (controls: €1,844). The percentage of antifungal drugs in patients with proven and probable IA mostly exceeded 70%.
Conclusions: IA is associated with increased use of antibiotic and antifungal drugs which invoke high costs. Considering this data the primary focus should be the prevention of IA in high risk patients.
Techniques in molecular bacteriology
P1402 Comparison of two commercial multiplex and two commercial single PCR Kits for detection of atypical pathogens causing pneumonia
V. Ursic, J. Stokic, V. Tomic (Golnik, SI)
Objectives: Atypical bacterial pathogens Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila cause lower respiratory tract infections (LRTI) and the rate of atypical LRTI is probably underestimated due to detection difficulties. In our study the presence of these pathogens was investigated in patients with clinical symptoms of atypical pneumonia, by single PCR kits (Onar® Ls and Venor® Mp kits, Minerva Biolabs, Germany, GeneKam Biotechnology AG, Germany) and multiplex PCR kits (Pnemoplex, Prodesse Inc., USA and Chlamylege, Argene Inc., France).
Methods: We collected 250 sputum samples and throat swab specimens and used three single and two multiplex PCR kits available commercially for detection. QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany) was used to extract DNA and was performed as recommended by the manufacturer. PCR was performed with conventional cycler (Biometra T3000, Biometra, Germany), PCR products were detected by standard agarose gel electrophoresis (single PCR kits) or by hybridisation in microtiter plate (multiplex PCR kits). Every specimen was analysed in parallel with two different kits. For analysis and evaluation, each testing method was considered a separate experiment.
Results: 135 sputum samples (54%) and 115 throat swab specimens (46%) were tested prospectively for the presence of the DNA of C. pneumoniae, M. pneumoniae and Legionella spp. The overall agreement between the four tests was 100%. Negative samples showed no inhibition. No discrepancies were found. Since Pneumoplex assay allows detection of the DNA of Bordetella pertussis, screening for the presence of the DNA of that pathogen was performed, as well. 32 samples (12.8%) were positive for at least one pathogen and 218 (87.2%) samples were negative. Among positive, 17 (53.1%) were sputum sample, 15 (46.9%) were throat swabs. 20 (62.5%) samples showed presence of M. pneumoniae, 6 (18.8%) showed presence of Legionella spp., 6 (18.8%) were positive for B. pertussis. No specimen showed presence of C. pneumoniae. Serological data wasn't avaible for all patients.
Conclusion: Multiplex PCR offers a rapid, sensitive, and easy method to simultaneously detect several atypical bacterial pathogens, while with single PCR we detect only one causative microbial agent. Furthermore, in our hands Pneumoplex multiplex assay proved to be faster and less labour intensive than Clamylege multiplex assay.
P1403 Extended evalution of the NucliSens easyMAG sytem (BioMérieux) for automated extraction of microbiological DNA/RNA
K. Beuselinck, E. Verschueren, J. Van Eldere (Leuven, BE)
Objective: Nucleic acid (NA) extraction is the most critical and labour-intensive step in NA based diagnostics. Performance of NA based diagnostics is primarily dependent on NA extraction yield, purity and the amount of sample that can be extracted. Recently, BioMérieux introduced the Nuclisens easyMAG, an automated extraction system with magnetic micro-particle processing allowing extraction of up to 24 samples in 1 hour.
Methods: Performance of the easyMAG as a front-end extraction system for our ‘in-house’ Taqman PCR assays (HSV-1, HSV-2, VZV, CMV, EBV, enterovirus, polyomavirus, Toxoplasma gondii, Chlamydophila pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila and Pneumocystis jirovecii) was analysed and compared to manual Qiagen extraction for the following sample matrices: cerebrospinal fluid, amniotic fluid, phosphate buffer (swabs), plasma and bronchoalveolair lavage samples.
Results: Dilution series of the evaluated micro-organisms were spiked in the appropriate sample matrices and divided into two aliquots. One aliquot was manually extracted with QIAamp Viral RNA kit (enterovirus) or QIAamp DNA Mini kit (all other organisms) simultaneously with automated extraction of the other aliquot with the generic protocol on the easyMAG. Of 38 dilutions, 10 aliquots were made to test reproducibility of both extraction methods on 5 different days. Out of 424 duplicate extractions, 289 were positive and 113 negative for both extractions (94.8% concordance). 22 discrepant results all had one weak positive result close to the detection limit of the PCR with 4 manual extractions leading to a false positive result. The correlation coefficient between automated and manual extraction was 0.98; the mean difference in cycle treshold (Ct) was + 0.08 Ct for the easyMAG. The mean variability with 5 repeats for the 38 dilutions was 0.50 Ct (SD: 0.38 Ct) for easyMAG and 0.57 Ct (SD: 0.55 Ct) for manual extraction. No cross-contamination was observed with easyMAG when strongly positive polyomavirus samples (15.4 Ct) and polyomavirus negative samples were tested. Turn-around time on easyMAG for 24 samples was one hour and hands-on time was 15 minutes compared to 95 minutes for manual extraction. The reduced hands-on time partially compensates the higher easyMAG reagents list price.
Conclusion: Automated easyMAG extraction offers NA yield similar to manual Qiagen extraction with reduced cross-contamination risk and reduced hands-on time.
P1404 A semi-automated procedure for monitoring of cytomegalovirus
V. Törmänen, J. Prieto, N. Finnström, B. Jansson (Bromma, SE)
Objectives: Monitoring of cytomegalovirus (CMV) in transplanted and other immunocompromised patients can efficiently be performed using molecular diagnostics based on real-time PCR, such as the CE-labelled affigene® CMV trender assay (Sangtec, Bromma, Sweden). In high throughput laboratories it is not always possible or convenient to use manual sample preparation systems for the analysis of large amount of samples. Today, there are a number of automated sample preparation systems on the market. An interesting instrument for automated sample preparation is the easyMAG from Biomérieux (Lyon, France) which is CE-labelled for in-vitro diagnostic use. The aim of this study is to evaluate the easyMAG instrument in combination with affigene® CMV trender.
Methods: The parameters limit of detection (LOD) and quantitative range were determined using the AD169 strain from the well recognized VQC (Acrometrix, The Netherlands). The strain was diluted to different concentrations in human plasma.
For LOD five different levels of CMV in the range from 5 c/mL to 100 c/mL were prepared and amplified in each of 24 replicates. The probit statistical analysis was used for determination of LOD. For quantitative range six different levels of CMV, in the range from 100 c/mL to 2×107 c/mL were prepared and amplified in twelve replicates. The imprecision was determined by repeating four replicates at two different CMV levels on three consecutive runs.
For each preparation 500 microliter of plasma sample was used and the elution volume was 60 microliter.
Results: As determined using probit analysis the LOD of the semi-automated system was 57 c/mL (confidence interval 36–134 c/mL). The quantitative range of the system is from at least 100 c/mL to 2×107 c/mL with a total imprecision of less than 0.2 log standard deviation.
Conclusions: Automation of sample preparation, using the easyMAG instrument, in combination with the standardised affigene® CMV trender PCR assay is an attractive option for quick CMV monitoring in laboratories requiring high throughput.
P1405 Unique pre-analytic tool for microbial diagnosis
S. Sachse, K.H. Schmidt, M. Lehmann, H.P. Deigner, S. Russwurm, E. Straube (Jena, DE)
Objectives: Speed is the essence when evaluating a patient with symptoms consistent with sepsis. Because of the high morbidity and mortality associated with sepsis, patients receive multiple, broad-spectrum antibiotics before receiving finalised blood culture results. Rapid assay to detect bacteria directly from Blood would facilitate timely diagnosis and appropriate care.
That is why culture independent pathogen detection methods become increasingly interesting. However, suitable methods for DNA isolation is direfully important precondition for further nucleic acid-based detection (NA) methods. DNA samples can show reduced sensitivity due to very low amounts of bacterial DNA compared to the human DNA background as well as inhibitors like hemin or immunoglobulin. Therefore, an improvement of NA methods can be achieved by an innovative sample preparation procedure, which reduce the human DNA background significantly and enrich the bacterial DNA simultaneously as well as inhibitors are removed.
Methods: A recombinant protein with specific binding affinity for prokaryotic DNA is the keystone of our innovative and easy-to-use sample preparation method. To validate the properties of this protein, we performed classical gel retardation, radioactive experiment, classical as well as real time PCR experiments using artificial DNA mixtures containing pro- and eukaryotic DNA as well as spiked blood samples.
Results: Our experiments showed that prokaryotic DNA is specifically retained by the described protein. The protein can be immobilised to matrices and transferred to spin columns for enrichment experiments. The DNA preparation procedure using such columns enables a change of the eukaryotic–prokaryotic DNA ratio in favour for the bacterial DNA. Furthermore, by means of radioactive and real time PCR experiments, it could be shown that the application of this procedure decreases the background of human DNA significantly, leading to an increased sensitivity of the PCR methods for bacterial detection in blood samples.
Conclusion: The proposed unique pre-analytic method improves NA methods for the detection of small amounts of bacterial DNA in complex samples (e.g. human whole blood), by a significant reduction of the eukaryotic DNA background. This might be a milestone allowing “same day diagnosis” of sepsis and SIRS as well.
P1406 Direct detection of bacterial DNA in clinical samples; evaluating a 4½ year period of sample collection and analysis
K.R. van Slochteren, E. van Zanten, G.J. Wisselink, R.F. de Boer, T. Schuurman, A.M.D. Kooistra-Smid (Groningen, NL)
Objective: Four and a half years ago the department of Research and Development (R&D) of the Laboratory for Infectious Diseases (LvI) started to analyse direct clinical specimens on request for the presence of bacterial DNA.
This study shows an evaluation of 4½ years of detecting bacterial DNA in direct clinical specimens.
Methods: During the last 4½ years 480 specimens (that contain no bacteria in healthy individuals) were send to the LvI with suspicion of bacterial involvement, where culturing bacteria was not possible or unlikely to yield results, e.g. because antibiotic treatment had already commenced.
Putative bacterial DNA was isolated from these samples by means of mechanical disruption of cells followed by a Boom extraction. If the specimen consisted of tissue it was first digested in order to form a cell homogenate. A PCR was performed on these samples to detect the presence of bacterial DNA coding for 16S ribosomal RNA. Positive PCR products were subsequently sequenced to determine which organism was present in the clinical specimen. (ref. T. Schuurman et al., JCM 42;2, 2004, p734)
Results: Of these 480 specimens 101 were found to contain a single bacterial species. In 351 specimens no bacterial DNA was found. In the remaining 28 specimens a single bacterial micro-organism could not be identified either because of the presence of more than 1 strain in which case sequencing yields an uninterpretable signal, or the PCR remained negative due to inhibiting factors in the specimen.
Specimens had a wide variety of origins but 67% of requests pertained to only 4 biological locations: joints 25%, blood 19%, central nervous system 16% and lung 7%. From these organs came 62% of the positive results with a single bacterial strain. None of the specimen types from the different organs submitted for bacterial DNA detection produced only negative results.
Many different species were found: the identified strains belonged to 30 different known genera. Streptococci were found by far the most: Of the positive specimens 33% contained streptococci, whereas the next most frequently found genus was Bacteroides with only 7% of all positive results.
Conclusion: Even though the number of positives isolated from various biological sites differs, every organ could yield a positive result. Therefore in cases where bacterial infection of normally sterile biological sites is suspected, determining whether bacterial DNA is present provides useful information.
P1407 Cultivation versus PCR analysis of joint fluid samples in prosthetic joint infection
J. Gallo, M. Kolar, D. Koukalova, P. Sauer, Y. Loveckova, J. Zapletalova (Olomouc, CZ)
Objectives: Successful treatment of prosthetic joint infection (PJI) requires correct diagnosis, preferably established presurgically. At this time, aspiration of joint fluid is easily performed and for this reason widely practised. The goal of this study was to evaluate the role of cultivation and PCR in joint fluid analysis.
Methods: Joint fluid samples (JFS) were obtained as a part of a prospective study of 115 failed total joint arthroplasties between January 2003 and June 2005. Of these 49 were considered to be PJI according to strictly fulfilled inclusion criteria. The PCR assay aimed at 16S rRNA was performed in 35 PJI and 66 controls while cultivation of JFS was done in 46 PJI and 48 controls. Standard techniques were used for microbial identifications of pathogen.
Results: PCR was positive in 71% of PJI cases (25/35) and in 2 controls (3%). Cultivation of JFS was positive in 44% of investigated PJI (20/46) while the false positive rate was 6% (3/48). A significantly higher sensitivity, accuracy and negative predictive value was calculated for PCR analysis of JFS compared to cultivation of JFS (p = 0.014, p = 0.0014, and p = 0.0016). In 42 cases (85.7%), infectious pathogens were identified by cultivation of intraoperative samples. Coagulase-negative staphylococci and Staphylococcus aureus strains were proved to be aetiologic agents in 62% of all strains. The concordance between PCR and intraoperative cultivation methods with regard to identification of causative pathogens could be evaluated in 23 patients. In 19 cases (82.6%) the same bacteria was reported by the methods under study at the species-level whereas in 4 patients the results from PCR and cultivation assays were different.
Conclusion: In this study we found a very low sensitivity of joint fluid cultivation in contrast to PCR analysis based on detection of 16S rDNA in the same material. Furthermore, PCR was more accurate and had much higher negative predictive value and likelihood ratio for positive result. For this reason, the PCR protocol must be regarded as progress. In addition, more than 80% of PJI pathogens were correctly identified by means of PCR detection.
The study was supported by the Internal Grant Agency of the Czech Ministry of Health projects No. NR/9065–3.
P1408 Demonstration of the application of the tmRNA transcript of the bacterial ssrA gene as a molecular diagnostic target using a combination of NASBA and BiaCore technologies
B. Glynn, K. Lacey, P. Palta, L. Kaplinski, M. Remm, T. Barry, T. Smith, M. Maher (Galway, IE; Tartu, EE)
Objectives: The tmRNA transcript of the bacterial ssrA gene exhibits several properties that make it suitable as a molecular target in Nucleic Acid Diagnostics (NAD). Sequence homology at the 5′ and 3′ ends of the gene facilitate universal PCR amplification for characterisation of tmRNA from different species, while sequence heterogeneity in the internal portions of the gene enable the design of oligonucleotide probes to identify bacteria at genus and species level. We have previously demonstrated that tmRNA is present at approximately 1,000 copies/cfu in a range of pathogens. The natural amplification of this target has the potential to improve the detection capability of an NAD test.
The objective of this study was to demonstrate the utility of tmRNA as a molecular diagnostic target using a combination of BiaCore biosensor and Nucleic Acid Sequence Based Amplification (NASBA) technologies. Streptococcus pneumoniae was chosen as a model organism for these evaluations.
S. pneumoniae is a leading cause of morbidity and mortality worldwide. Infants and young children are most at risk, presenting with meningitis, pneumonia, bacteraemia, and other infections.
Methods: The ssrA gene was sequenced in geographically distinct S. pneumoniae strains and in related species of the streptococcus genus and other bacterial species that may be present in the same clinical environment as S. pneumoniae. Species-specific probes were designed by sequence alignment and by the SLICsel bioinformatics software package. Initial evaluation of probe specificity was performed on microarrays using fluorescently labelled in vitro transcribed cRNAs. Selected probes were immobilised onto Biacore sensor chips and further evaluated using a series of unlabelled tmRNA containing targets including; cRNA, total RNA, and NASBA products.
Results: Using selected S. pneumoniae specific probes, 1012 cRNA copies of the tmRNA target were detected on the BiaCore. It was not possible to detect tmRNA from total S. pneumoniae RNA using this system. Inclusion of a NASBA step enabled BiaCore detection of a single S. pneumoniae cell. Specificity was demonstrated using NASBA products from a panel of clinically relevant bacteria.
Conclusion: We have demonstrated that the tmRNA transcript of the ssrA gene in S. pneumoniae is suitable for use as a molecular diagnostic target. Application of a combination of NASBA and BiaCore biosensor technologies enabled specific detection of a single S. pneumoniae cell.
P1409 PCR of the 16S rRNA gene and DNA sequencing in establishing the aetiology of bacterial infections in children
A. Gleesen, C. Grarup (Copenhagen, DK)
Objectives: Bacterial infections are common in children and an exact microbiological diagnosis is crucial for specific antimicrobial treatment, especially in serious cases. Standard culture of clinical specimens may not always result in determination of the aetiological organism. In these cases PCR of bacterial DNA has shown to be an excellent supplement to culture. As the literature on the use of molecular diagnostics of bacterial infections in individuals of young age is sparse, we found it relevant to review results of samples from this population, obtained in our laboratory.
Methods: The samples were examined using the QIAamp DNA Mini Kit and the Quantitect SYBR Green kit. Samples were tested in real-time PCR. Both DNA strands of the amplicons were sequenced using BSF-8 and BSR-534 as sequencing primers and the BigDye v.3.1 sequencing kit. Data were compared using the BLAST search engine to deposited sequences in the NCBI database.
Results: During a three year period between 2003 and 2006, 62 samples from 58 patients less than 16 years old were analysed. Bacterial DNA was detected in 20 of the samples, displayed in Table 1 . These included nine different pathogens of which Streptococcus pyogenes (GAS) and Streptococcus pneumoniae were the most frequent. The other pathogens identified were Streptococcus agalactiae, Kingella kingae, Neisseria meningitidis, Streptococcus intermedius, and Francisella tularensis.
Table 1.
Bacterial identification of the 20 samples positive by PCR
| ID | Age | Specimen | Clinical tentative diagnosis | Bacterial identifcation |
|---|---|---|---|---|
| 1 | 11 years | Pus | Osteomyelitis | GAS |
| 2 | 13 years | Joint fluid | Coxitis | GAS |
| 3 | 19 months | Pus from a lymph node | Not specified | GAS |
| 4 | 2 years | Joint fluid | Coxitis | GAS |
| 5 | 20 days | Joint fluid | Coxitis | S. agalactiae |
| 6 | 8 years | Biopsy, brain | Not specified | S. intermedius |
| 7 | 2 years | Spinal fluid | Not specified | S. pneumoniae |
| 8 | 15 years | Spinal fluid | Meningitis | S. pneumoniae |
| 9 | 37 days | Inoculation from eye | Not specified | S. pneumoniae |
| 10 | 9 months | Joint fluid | Septic arthritis | K. kingae |
| 11 | 3 years | Spinal fluid | Not available | N. meningitidis |
| 12 | 9 years | Biopsy, lymph node | Tularaemia | F. tularensis |
| 13 | 4 years | Pus | Abscess | F. necrophorum |
| 14 | 8 years | Joint fluid | Septic arthritis | P. acnes |
| 15 | 10 years | Spinal fluid | Not available | P. acnes |
| 16a | 7 months | Joint fluid | Septic arthritis | P. acnes |
| 16b | 7 months | Joint fluid | Septic coxarthritis | Polymicrobial |
| 17a | 10 years | Pus, Femur | Ewings sarcoma, ostitis | Polymicrobial |
| 17b | 10 years | Biopsy, Femur | Ewings sarcoma, ostitis | Polymicrobial |
| 18 | 12 months | Spinal fluid | Meningitis | Polymicrobial |
DNA from Propionibacterium acnes was detected in three samples. In four samples a positive PCR reaction was found, but a particular identification could not be determined by DNA sequencing as examination of the electropherogram suggested the presence of DNA from more than one bacterial species.
Conclusion: In the present material PCR of the 16S rRNA gene could detect bacterial DNA in 20 out of 62 clinical specimens from paediatric patients. A definite microbial diagnosis of a recognized pathogen was obtained in 13 of these. The bacterial identifications were in agreement with characteristic clinical presentations, age of the patients, and sites from which the specimens were obtained: meningococci and pneumococci in cerebrospinal fluid with meningitis, S. intermedius in a brain abscess, S. agalactiae in a 20 days old baby, and K. kingae in joint fluid.
We conclude that PCR amplification with subsequent DNA sequencing is a useful diagnostic supplementary tool on various specimens in establishing the cause of bacterial infections in paediatric patients.
P1410 Comparison of real-time PCR and blood culture for the diagnosis of bloodstream infections in onco-haematologic patients: microbiological and clinical assessment
S. Varani, M. Stanzani, L. Nardi, M. Paolucci, N. Vianelli, M. Baccarani, M. Landini, P. Dal Monte, F. Cavrini, B. Giannini, F. Melchionda, A. Pession, V. Sambri (Bologna, IT)
Objectives: Sepsis is a syndrome that results from a dysregulated host response to infection which can rapidly lead to organ dysfunction and death. The aim of this study was the microbiological and clinical assessment of a new real-time PCR test (SeptiFast: SF, Roche Diagnostics) for the diagnosis of blood stream infections (BSI) in a population of neutropenic patients. The results of SF were compared with blood cultures (BC) obtained on the same specimens and the clinical significance of the two methods was assessed.
Methods: This single-centre study was conducted from the beginning of May 2006 until the end of July 2006. Eighty-six samples were obtained simultaneously for culture and SF from peripheral blood of 51 oncohaematologic patients with clinical suspicion of bacterial or fungal BSI. All patients included in the study exhibited leukopenia (WBC< 200/mL). The SF was performed following manufacturer's instructions. BC was performed by using an automated system (BacTAlert, bioMérieux). Standard laboratory procedures were employed for identification and antimicrobial susceptibility testing of the bacterial isolates from BC.
Table 1.
Comparative identification of bacteria from blood culture and SeptiFast (febrile episodes/blood specimens)
| PCR | Bacteria | Blood culture |
|||||
|---|---|---|---|---|---|---|---|
| Negative | Positive |
||||||
| CoNS | S. aureus | S. pyogenes | Acinetobacter spp. | Total positive | |||
| Negative | 41/57 | 6/6a | 0 | 1/1b | 1/1c | 8/8 | |
| Positive | CoNS | 1/1 | 9/11 | 0 | 0 | 0 | 9/11 |
| S. aureus | 1/3 | 0 | 3/3 | 0 | 0 | 3/3 | |
| E. coli | 1/1 | 0 | 0 | 0 | 0 | 0 | |
| S. maltophilia | 1/1 | 0 | 0 | 0 | 0 | 0 | |
| A. baumannii | 1/1 | 0 | 0 | 0 | 0 | 0 | |
| Total positive | 5/7 | 9/11 | 3/3 | 0 | 0 | 12/14 | |
*in 3 out of 6 cases a contamination by CoNS was indicated by SeptiFast; **SeptiFast indicated the presence of contamination by CoNS; ***SeptiFast indicated the presence of contamination by streptococci spp.
Results: Fourteen and 57 samples were identified, respectively, as positive and negative by both the methods. 8 specimens were found positive by BC, but negative by SF and 7 samples were detected as negative by BC, but positive when tested by SF. The agreement between the two tests was 16.3% and 66.3% for the positive and the negative specimens, respectively. The overall agreement between the two methods was 82.6%. In 3 out of 6 cases that resulted positive for coagulase-negative staphylococci (CoNS) at BC, SF showed the presence of contamination by CoNS. A clinical surveillance of these cases suggested that the detection of CoNS by culture was a false positive result probably due to contamination in the collection procedure. A comparative identification of bacteria from BC and SF was performed and we found that SF identified Gram-negative bacteria more efficiently than BC. The average time to identification of different bacteria was 85.1±7.9 hours (mean±standard error mean) for BC versus 27.7±4.3 for SF (p<0.000001).
Conclusion: The capability to identify “contaminating” low bacterial load, together with the fast availability of clinical relevant results suggest that SF should be performed together with standard BC in neutropenic febrile patients to improve the sensitivity and specificity of the laboratory diagnosis of BSI.
P1411 PCR with universal primers targeting the small ribosomal RNA (16S rRNA) gene of bacteria as a diagnostic tool in 15 hospitalised patients with infectious diseases
E. Longhi, A. Foschi, G. Bestetti, V. Acquaviva, A. Radice, C. Parravicini, S. Antinori, M. Corbellino (Milan, IT)
Objectives: To evaluate the utility of 16S rRNA gene PCR followed by sequencing of the amplified products for the in vivo detection and identification of bacteria in clinical specimens and to compare it with conventional in vitro isolation.
Patients and Methods: Presence of bacterial DNA was sought for by 16S rRNA gene PCR with broad-range primers in 18 clinical specimens collected from 15 selected patients hospitalised at Luigi Sacco Hospital (Milano, Italy). The patients had miscellaneous clinical syndromes at presentation: sepsis, sepsis and meningitis, lymphadenitis, localised abscess, osteomyelitis, pleural effusion and latero-cervical mass. The biological specimens analysed were: EDTA-anticoagulated whole blood (n = 5), cerebro-spinal fluid (n = 1), pleural fluid (n = 2), lymph-node aspirate (n = 1), lymph-node biopsy (n = 1), skeletal muscle (n = 2), bone (n = 1) and purulent exudate obtained from abscess (n = 5). In 13 of the 15 patients, coeval samples were tested both by PCR and by traditional in vitro isolation.
After DNA extraction, PCR was carried out using primers 515F-806R that amplify a 328 base pair region of the bacterial 16S rRNA gene. PCR products were directly sequenced and analysed using the BLAST programme.
Results: 16S rRNA gene PCR allowed the identification of the bacterial pathogen in all cases while in vitro isolation was diagnostic in 8 out of 13 (61%) cases. Bacterial culture and molecular identification were concordant in 7/13 (54%) patients while PCR was the only method that yielded a diagnosis in 5/13 patients (40%). 8/15 (53%) individuals were receiving antimicrobial treatment when PCR and culture were performed: 16S rRNA gene PCR allowed pathogen identification in all of them, while in vitro isolation was positive in only 3/8 cases (37%).
Conclusions: Broad-Range 16S rRNA PCR allowed rapid identification not only of common bacterial pathogens such as Neisseria meningitidis, Streptococcus pneumoniae, Staphylococcus aureus, Streptococcus pyogenes and other Streptococci but also of fastidious bacteria such as Bartonella henselae and Mycobacterium genavense.
PCR proved to be superior to bacterial culture in two clinical situations: infections caused by bacteria with unusual growth requirements and specimens collected during antimicrobial treatment of the patient.
P1412 Use of denaturing gradient gel electrophoresis for polymicrobial sample analysis
B. Zaloudikova, M. Slany, T. Freiberger (Brno, CZ)
Objectives: In clinical microbiology most polymerase chain reactions (PCRs) for the detection of bacteria are designed specifically to one organism. This provides high sensitivity and specificity but detects only the microorganism we are looking for. To screen for a wide spectrum of bacterial pathogens, broad-range PCR assays based on the detection of the 16S rRNA gene were designed. The identity of the bacterium is revealed by nucleotide sequencing of the PCR product and comparison of the obtained sequence with the GenBank database. In contrast to standard culturing techniques, the methods of broad-range PCR and sequencing have a potential to rapidly detect also fastidious or unculturable bacteria. However, applying this approach to clinical samples containing more than one bacterial species may result in multisequence signal, which disables exact identification of particular bacteria. Denaturing gradient gel electrophoresis (DGGE) might overcome this problem. This method is based on separation of fragments of the same length with different base-pair sequences in polyacrylamide gel containing a linearly increasing gradient of DNA denaturant. DGGE results in a pattern of bands each corresponding to specific bacterial species. The aim of our study was to optimise the method of universal molecular detection of pathogens in polymicrobial materials.
Methods: Samples of DNA extracted from different bacteria were mixed and used as a template for broad-range PCR. The PCR product was subsequently analysed using DGGE. Separated bands were cut from polyacrylamide gel, reamplified and sequenced. Basic Local Alignment Search Tool (BLAST) programme has been used to compare the sequences with the GenBank database. The approach was tested on synovial fluid sample that showed a multiple signal after routine broad-range PCR and sequencing analysis.
Results: All sequences obtained from arteficial template corresponded to GenBank sequences of bacteria used for DNA extraction. DGGE analysis of the clinical material resulted in two bands. Their sequences showed 99% similarity with Enterobacter cloacae (Accession number DQ202394) and 98% similarity with Staphylococcus aureus (Accession number DQ997837), respectively.
Conclusion: Broad-range PCR combined with DGGE and sequencing seems to be a promissing tool for molecular detection of pathogens in polymicrobial clinical samples.
P1413 Analysis of mixed microbial species in clinically relevant biofilms using denaturing HPLC
F. Jury, A. Bueid, W. Ollier, A. Fox, M. Upton (Manchester, UK)
Objective: Many infections have a multi-species aetiology with causative agents existing in bacterial biofilms. Differentiation of these organisms usually is possible using culture based approaches. However, in an age when molecular detection and typing methods are being heralded as the future of diagnostic microbiology, they are not optimal for analysis of mixed species populations. We have previously shown that denaturing HPLC (DHPLC) has utility for rapid identification and typing of Staphylococcus aureus strains. Here we investigate the application of DHPLC to analysis of mixed bacterial species from culture and clinical swabs.
Methods: DNA recovered from cultured cells and wound or screening swabs was used as template in PCR using universal primers. The primers target the 16S rRNA genes and the forward primer carried a 40bp GC clamp to allow optimal separation during DHPLC. Conditions for DHPLC were based on those shown to be useful for separation of amplicons from environmental samples. DHPLC separation was carried out using fragments amplified from swabs known to carry single and mixed species.
Results: Amplification of 16S rDNA from strains and swabs was successful following some minor modifications. Separation of multiple peaks representing individual rDNA species was possible.
Conclusions: DHPLC has potential to be a powerful tool for separation of mixed amplification products for species identification and for analysing population dynamics in bacterial biofilms. The method could have a significant impact on direct detection of clinically relevant organisms and rapid identification for targeted patient management and antimicrobial therapy selection.
P1414 Use of FTA paper and PCR for detection of bacterial DNA in clinical swabs
A. Bueid, A. Barhameen, J. Jones, M. Upton (Manchester, UK)
Objective: Detection of bacterial pathogens in clinical samples is being greatly enhanced by the use of real-time PCR based approaches. The rate limiting step in such approaches is often extraction of DNA. The aim of the current study was to investigate possible means of reducing sample processing time and labour.
Methods: FTA paper was originally developed for storage of human DNA samples and has been widely used in forensic science. The paper cards have recently been shown to stabilise bacterial and viral DNA. Following a simple wash procedure, DNA can be stored at room temperature for prolonged periods.
We have assessed the utility of FTA paper cards for extraction of bacterial DNA from clinical swabs carrying between one and four bacterial species. Swabs were applied to the surface of the cards and washed, dried and stored prior to use in PCR. Data from PCR reactions were compared with those obtained by conventional culture based approaches.
Results: DNA was suitable for amplification by PCR using universal and specific primers and the approach killed all organisms that were present. Good correlation was seen between results from the molecular and culture based approaches. This approach has significant implications for making safe samples that contain highly pathogenic organisms.
Conclusions: It is possible that FTA paper cards could be introduced into routine molecular diagnostic assays to greatly reduce sample preparation time and make high risk samples safe for handling by lab personnel.
P1415 Improvements in the molecular detection of pathogens through participation in international external quality assessment schemes
W. MacKay, C. Scott, C. Steel, H.G.M. Niesters, P. Wallace, A.M. van Loon (Glasgow, UK)
Objectives: Quality Control for Molecular Diagnostics (QCMD) is an independent not-for-profit organisation whose primary aim is to establish and develop External Quality Assessment (EQA) Programmes for the evaluation of nucleic acid amplification techniques (NATs) in the diagnostic laboratory. QCMD collects and analyses a wealth of information on the performance of NATs and is uniquely placed to comment on trends in their performance worldwide.
Methods: Data for all of the EQA programmes offered by QCMD (including those for MRSA, Mycobacterium tuberculosis, Neisseria gonorrhoeae, Chlamydia trachomatis and Legionella pneumophila) were analysed to determine trends in the number of participants contributing to the EQA programmes, changes in the types of NATs being used and performance over time.
Results: The number of EQA programmes offered increased from eight in 2001 to 32 in 2006. Panels were distributed to over 2,500 participants in more than 50 countries worldwide in 2006. Commercial assays, when available from major diagnostic companies, were the favoured method of choice. However a large number of laboratories developed and used in-house methods. A move towards the use of real time assays was also evident whilst the use of non-PCR methods was uncommon.
Conclusion: Considerable differences in performance were noted between laboratories, particularly when in-house methods were used. False-positive results are still a problem. Nevertheless, the general trend was towards an improvement in performance.
QCMD is now the primary provider of EQA programmes for the evaluation of NATs in molecular diagnostics throughout Europe and the rest of the World. We continue to maintain and develop a comprehensive portfolio of programmes covering a wide range of pathogens.
P1416 Statistical modelling of the performance of nucleic acid amplification technologies in clinical diagnostic applied to quality control for molecular diagnostics hepatitis B virus programmes
L.M. Garcia Fernandez, P. Wallace, H. Staines (Glasgow, Dundee, UK)
Objectives: Pathogen load estimation provided by Nucleic Acid Technologies (NATs) used to diagnose and manage patients with infectious disease gives more information than positive/negative results available from earlier techniques. Generalised Linear Models (GLM) are currently used to analyse NAT users' performance. However, these ignore the censored quantities lying outside the detection limits of the assays. We introduce an approach that identifies significant factors associated with lab performance including censored values. The model is tested on data from 4 years of QCMD HBV programmes.
Methods: We propose a GLM allowing a censored mechanism and Bayesian parameter inferences using Markov Chain Monte Carlo (MCMC) methods. The model assumes that the log10 copies/mL pathogen load estimates are normally distributed.
Results: Potential explanatory variables in the model include NAT technology used, year of programme and sample genotype. Lab performance was assessed by the difference between the lab's and the target estimate of the pathogen load
The proportion of censored data was higher for samples with lower viral load.
Users of Commercial PCR technologies were compared with other technology groups and results depended on viral load. HBV genotype was a significant factor for some sample categories whilst programme year was significant for almost all sample categories.
Conclusions: The model deals with multiple parameters and censored values in a simpler way than traditional statistical techniques. Information from the censored values (outside the assay limits of detection) is incorporated in the model and further modelling of the censored values can be made.
P1417 Modelling performances of quality control for molecular diagnostics participants in Enterovirus programmes over time
L. Garcia Fernandez, P. Wallace, H. Staines, A. van Loon (Glasgow, Dundee, UK)
Objectives: To analyse data from Enterovirus (EV) Quality Control (QC) programmes over time to provide a better feedback to participants and improve the design of future QC programmes. These should help improve the performance of molecular diagnostic technologies users.
Methods: Homogeneity tests and Generalised Linear Models (GLM) are used to model the positive/negative responses provided in the QC programmes. Homogeneity tests are performed on the ratio of correct results over time for the different category of samples. GLM (logistic regression) is used to find significant factors on the estimated probabilities of correct/incorrect results over time and sample categories.
Results: Data from the 1999 to 2005 QCMD EV programmes were analysed. The EV proficiency panel compositions varied by year although they contained series of similar samples (sample category): negative, non-EV and EV samples with different serotype and viral load.
Labs were categorised on whether they had been in previous EV programmes, if they had returned a correct result in that previous programme and whether they were accredited. The technology used was one amongst other factors included as potential explanatory variables.
The difference in the proportions of false positives and false negatives results over time varied depending on sample and lab category. The proportion of false positives for non-EV samples varied on the virus included in the sample. Laboratories that had a correct result in the previously panel are significantly less likely to obtain a false positive that those that are new to the programme. However, no significant differences were found when analysing negative samples. No significance difference was found between the performances of accredited and non-accredited participants. Performances from different technology users varied over time and sample category. In 2004/05 commercial assay users were less likely to detect low dilution samples than in 2002/03. The proportion of correct result over time decreased, as the dilution series are lower.
Conclusions: Performance of participants to the EV QC programme depended on the virus. These results suggest that participating in an EV QC programme helps improve the performance of laboratories. However no difference was found between the performance of accredited and non-accredited labs.
P1418 Evaluation of fluorescence in situ hybridisation for the identification of clinically relevant Candida species
S. Poppert, A. Lakner, M. Bartel, A. Essig (Ulm, DE)
Objectives: Rapid identification of Candida albicans is highly warranted in order to adapt antifungal therapy. Aim of the study was to establish and evaluate a Fluorescence in situ Hybridisation (FIAH) assay for this purpose.
Methods: We evaluated FISH using a previously published and a newly developed C. albicans probe for the identification of C. albicans in comparison to API 20 C, the chromogenic Candida ID-Agar and determination of filamentous colony morphology. 17 reference strains, 182clinical isolates and 41 blood cultures showing yeasts in the Gram stain were included.
Results: All tests produced highly concordant results. The Candida ID-Agar however was not able to discriminate C. albicans and C. dubliniensis. Reliable identification of C. albicans by API and by determination of filamentous colony morphology took 48 hours. FISH allowed identification of C. albicans within 3 hours with a specificity and sensitivity of 100% from subculture as well as directly from blood cultures.
Conclusion: As a robust, simple and over all extraordinarily fast method, FISH is an appropriate tool for the identification of C. albicans isolates, especially from blood cultures.
P1419 Tri-colour PNA FISH detection of five Candida species directly from positive blood culture bottles
F. Wu, S. Whittier, P. Della-Latta (New York, US)
Objectives: Candidaemia is associated with significant morbidity and mortality. C. albicans has historically been the most frequent cause of candidaemia. However, the frequency of candidaemia due to non-albicans species such as C. tropicalis, C. glabrata, C. parapsilosis and C. krusei has risen dramatically. Rapid detection of the most common Candida species can optimise selection of antifungal therapy.
Methods: A peptide nucleic acid probe fluorescence in situ hybridisation (PNA FISH) assay was developed and evaluated by using multicolour labeled fluorescent PNA probes targeting specific 26S rRNA sequences of C. albicans (CA), C. glabrata (CG), C. krusei (CK), C. parapsilosis (CP) and C. tropicalis (CT). The probe reagent was applied to smears made directly from positive blood culture (BC) bottles. After a 90 min. incubation at 55° C, unbound probe was removed by washing at 55°C for 30 min. Smears were then examined by fluorescence microscopy. CA and CP were identified as bright green, CG and CK as red and CT as gold fluorescent cells, respectively. Results were compared to identification by conventional methods. PNA FISH was also challenged with lab strains of Candida, phylogenetically related species and other organisms frequently recovered from BC.
Results: A total of 50 blood cultures were tested, including 40 positive for yeast and 10 positive for bacteria. The assay accurately identified 14 CA, 13 CG, 2 CK, 2 CP and 6 CT. Positive and negative predictive values were 100% for all yeast isolates. One BC was negative for CA by PNA FISH but reported as culture positive for C. albicans. Sequencing analysis identified the isolate as Candida dubliniensis. All challenge strains were PNA FISH negative except for one blood culture positive for Acinetobacter haemolyticus which initially tested positive for CG/CK but was negative upon repeat testing. Further investigation is in progress.
Conclusion: The current evaluation of the Yeast Traffic Light PNA FISH assay demonstrates that this new test can provide rapid and reliable detection results for five clinically relevant Candida species, leading to improvements in antifungal therapy.
P1420 Expression of IgA-specific endopeptidases from Neisseria meningitidis in E. coli
V. Khamiankou, T. Kazeeva, A. Shevelev (Moscow, RU)
Objective: Immunoglobulin A1-specific proteinase (Igase) is a pathogenicity factor of meningococcus (Neisseria meningitidis). These enzymes belong to trypsin-like clan of serine proteinases. They exhibit a high substrate selectivity being able to discriminate between IgA1 and IgA2 and, on the other hand, to distinguish human IgA1 and IgA1 from non-primate species. Taking in account conservancy of hinge region subjected to cleavage in human and other vertebrates, these means that Igases exhibit two-level substrate-recognition control which can be denoted as a primary and non-primary specificity. Structural basis of such a high specificity remains obscure due to unavailability of native recombinant IgA1 good for crystallographic and enzymatic studies.
Methods: We carried out expression in E. coli of two separate fragments of Igase corresponding to theoretically predicted domains, refolded and purified the proteins.
Results: N-terminal domain harboured the clearly recognized triad of residues homologous to the active centre of kallikrein, a serine protease involved to haemostasis. After two-step chromatographical purification and refolding, this protein exhibited a protease activity towards human myeloma IgA1. C-terminal portion of IgAse corresponds to a putative substrate-binding domain. This protein was expressed as a peptide with N-terminal 6His-tag in form of inclusion bodies, purified by to-step anion-exchange chromatography and refolded. As expected, the refolded protein exhibited a high binding capacity towards IgA1 from both normal and myeloma patient plasma. Besides, the binding domain tightly bound human IgA2 as confirmed in both ELISA-type and affinity chromatography tests. Likely to the native IgAse, its separated catalytic domain attacked a single peptide bound in the native IgA1, as visualised by immunoblotting with anti-IgA heavy chain antibodies under non-reducing conditions. However, similar experiment accompanied with immunoblotting under non-reducing conditions revealed slight shifting of the processing site of the isolated catalytic domain versus ones of the native enzyme.
Conclusion: We first succeeded to predict occurrence of separate domains in IgA-protease, to refold and purify them as separate proteins and to demonstrate their activity.
P1421 Expression of human serum albumin–L7/L12 (Brucella abortus ribosomal protein) fusion protein in Saccharomyces cerevisiae
I. Pakzad, A. Rezaee, M. Rasaee, A. Hosseini, A. Kasemnejad, B. Tabaraiee (Ilam, IR)
Brucella abortus is a facultative interacellular Gram-negative bacterial pathogen that causes abortion in pregnant cattle and undulant fever in humans. The immunogenic B. abortus ribosomal protein L7/L12 is a promising candidate antigen for the development of subunit vaccines against brucellosis. It has already been expressed in several bacteria and has been used as DNA vaccine. In order to construct yeast expressing vector for the tHSA-L7/L12 fusion protein, the l7/L12 ribosomal gene was amplified by PCR. The expression plasmid pYtHSA-L7/L12 was constructed by inserting the L7/L12 gene into the pYHSA5 shuttle vector (Containing inulinase signal sequence, HSA gene and Gal10 promoter). The recombinant vector was transformed into S. cerevisiae and was then induced by galactose. The secreted recombinant fusion protein was detected in supernatant by SDS-PAGE and confirmed by western blot analysis using anti-HSA and Anti-L7/L12 antibodies. Fusion protein was purified by affinity chromatography and its amount was approximately 500 μg/L. These results indicate that both fragments have antigenic property and they do not have structural (or antigenic) overlapping.
Molecular diagnosis of streptococci
P1422 Development and validation of a real-time PCR assay for the detection of group B Streptococcus in pregnant women
M. Wernecke, C. Mullen, M. Maher, T. Barry, T. Smith (Galway, IE)
Objectives: Group B Streptococcus (GBS) (Streptococcus agalactiae) is one of the leading causes of neonatal sepsis, meningitis and death in the developed world. Approximately 10–40% of pregnant women carry GBS in their vagina or rectum. Transmission to the infant occurs vertically during labour. Of the colonised infants, 1–3% develop disease, usually within the first 24 h after birth.
The objective of this study was to develop a rapid diagnostic test for GBS that can be performed in a wide variety of labour and delivery settings using a proprietary NUI Galway technology. Nucleic acid diagnostic tests for GBS have the potential to overcome the limitations associated with culture-based detection enabling timely assessment of GBS colonisation status in pregnant women at the time of delivery. As a result, intrapartum antibiotic prophylaxis can be administered more effectively leading to lower infant morbidity and mortality rates.
Methods: The real-time PCR test, performed on the Roche LightCycler® instrument, uses a proprietary National University of Ireland, Galway target, RiboSEQ based on the bacterial ssrA gene and includes a GBS-specific hybridisation probe. An internal amplification control (IAC) has been incorporated in the test. Sensitivity of detection was established using serially diluted GBS genomic DNA in the real-time PCR test. Specificity studies were performed using a panel of 48 related streptococci and other species found in the genital tract environment. The performance of the test is currently being evaluated in a clinical study and compared to the CDC-approved microbiological method for the detection of GBS. For this study, sample preparation is performed using commercially available nucleic acid extraction kits followed by PCR amplification and detection on the LightCycler®.
Results: The detection limit for the GBS real-time PCR test was determined to be 1–10 cell equivalents. The test was 100% specific for GBS. Clinical evaluation of the PCR test is ongoing and a progress report on test performance will be presented.
Conclusion: We have developed a rapid, sensitive and specific real-time PCR test based on an NUI Galway proprietary target, RiboSEQ for the detection of GBS in pregnant women.
P1423 The importance of signal recognition particle for viability of Streptococcus pneumoniae
E. Pinto, P. Andrew, L. Faleiro (Faro, PT; Leicester, UK)
S. pneumoniae is an important Gram-positive bacteria responsible for high levels of human morbidity and mortality, at all ages, around the world, causing pneumonia, meningitis, otitis media and septicaemia.
The dramatic increase in worldwide incidence of antibiotic resistant pneumococcal strains required a better understanding of bacterial physiology to facilitate the development of new antibacterial drugs. The best targets are those essential for cell function. In other Gram-positive and Gram-negative bacteria, deletion of components of the signal recognition particle (SRP) has been shown to cause cell death.
The aim of this research is to confirm that the SRP is essential for pneumococcus viability and its exploitation as a new target for therapy. To achieve this genes ffh and ftsY were amplified by PCR and cloned using the vector pGEM-T Easy. For S. pneumoniae mutagenesis these genes were subcloned into the integrational vector pRKO2 under the control of a tetracycline inducible promoter. S. pneumoniae transformants of strains D39 and TIGR 4 were analysed in order to confirm the plasmid integration and viability dependence of tetracycline. Forward the mariner mutagenesis in vitro was used for transformation of S. pneumoniae D39 to confirm the previous results. A mutant of S. pneumoniae D39Äffh was obtained and no phenotypical differences with the wild type strain were found. The mutation on gene ftsY seems to have a crucial role in S. pneumoniae D39 viability.
This work is supported by the project POCTI/58702/2004.
P1424 Cloning and purification of a sortase-like protein in Streptococcus suis type 2
C.J. Wang, Y.Q. Dong, J.Q. Tang (Nanjing, CN)
Objective: Streptococcus suis 2 is an important zoonotic pathogen, lead to two serious outbreaks with hallmark of human STSS in Jiangsu and Sichuan province in China in 1998 and 2005, respectively. The molecular mechanism of STSS is poorly understood. Sortase, an enzyme cleaves the LPXTG sequence in surface proteins and catalyses the transfer of the processed protein to target site. There is little doubt that some surface proteins of bacteria might play important roles during infection. As an enzyme which functions during some cell wall surface proteins covalently anchored to the cell wall, sortase might help some virulent factors to function during infection. To define the role of sortase in the pathogenesis of S. suis infections, cloning and expressing the sortase gene of S. suis type 2 ZYH33 that isolated from Streptococcal Toxic Shock Syndrome outbreak patients in China.
Methods: The srtA gene of S. suis was cloned and sequenced. The recombinant vector pGEX-4T-2-srtA was constructed and the expression of recombinant protein was analysed by SDS-PAGE and Western blotting.
Results: For the first time, we obtained the recombinant vector pGEX-4T-2-srtA and a recombinant sortase protein of S. suis2 ZYH33 in China.
Conclusion: From the results of sequencing and homology searches, we found srtA shows strong conservation and exhibits a high level of homology with the sortases of most European and domestic isolates. Polyclonal antiserum from rabbit immunised with S. suis2ZY05719 was used against the Sortase protein antigen, a brown band of approximately 55 kDa was visualised on the nitrocellulose membranes by Western blot, demonstrating that the expressed Sortase protein possessed good immunogenicity (Fig. 1 ). On account of a transmembrane domain existing within the Sortase protein, the protein produced by prokaryotic expression mainly presented in the form of inclusion bodies. A recombinant Sortase protein removing of the transmembrane domain was presented by Docter Youjun Feng of Chinese Academy of Sciences, that the recombinant protein was highly expressed as the form of solvend. a protein with an apparent molecular mass of 22 KDa by Western blot that was reacted strongly with polyclonal antiserum from swine and rabbit immunised with ZY05719, suggesting that the recombinant Sortase protein had specific antigenicity. Further work will be needed to really appreciate the role of sortase in the infections generated by S. suis.

P1425 Distribution of emm genotypes among S. pyogenes isolates from Serbia
V. Mijac, N. Opavski, L. Ranin, C. Heeg, R. Reinert (Belgrade, RS; Aachen, DE)
Objective: S. pyogenes is a major human pathogen that causes suppurative and nonsuppurative infections, including pharyngitis, necrotising fasciitis, and streptococcal toxic shock syndrome. The emm gene which encodes the major virulence factor, the M protein, is used as the basis for the characterisation of S. pyogenes. The aim of this study was to investigate the distribution of emm genotypes in Serbia.
Methods: Seventy seven isolates of S. pyogenes collected from various regions throughout Serbia were included in the study. Fifty-five isolates originated from patients with pharyngitis, while remaining 22 were isolated from patients with skin and soft tissue infections. The presence of emm genes was determined by PCR using “all M” primers following a previously published protocol by Podbielski and co-workers.
Results: A total of 17 different emm types were identified among the tested isolates. The most frequent ones were emm 6 (25%), emm 12 (16%), emm 1 (13%) and emm 50 (9%). We observed an association between emm types and isolation site: among pharyngeal strains the most common types were emm 6 (33%), followed by emm 12 (22%) and emm 1 (18%), whereas in skin and soft tissue group the most frequent isolates were emm 50 (27%), emm 58 (27%) and emm 78 (14%).
Conclusion: To the best of our knowledge, this is the first report on the emm type distribution in Serbia. Our results confirm the association between particular types and isolation site. The high incidence of emm 6 in patients with pharyngitis is a remarkable finding of the present investigation and not in concordance with reports from other parts of the world. Moreover, the present investigation underscores the need for a future active population-based surveillance programme in Serbia.
P1426 Microbiological aspects of a Streptococcus pyogenes outbreak causing invasive disease in children attending a day care centre in Cantabria, Spain
J. Agüero, M. Ortega, M. Cano, A. González de Aledo, J. Calvo, L. Viloria, P. Mellado, T. Pelayo, L. Martínez-Martínez (Santander, Cantabria, ES)
Objectives: To study microbiological aspects of S. pyogenes isolates cultured from children (5 to 40 month old) attending a day care centre in Cantabria (Northern Spain), and to compare these isolates with those from contacts of the affected children and with epidemiologically unrelated isolates from the same area.
Methods: Pharyngotonsillar swabs from 40 children, 4 day care staff and 258 contacts were cultured according to standard methods. Samples from multiple organs were cultured from a child who died from streptococcal toxic shock syndrome (STSS). Blood cultures were obtained from 2 other children with STSS. Nine S. pyogenes isolated from pharyngotonsillar samples unrelated with the outbreak were included for comparison. Clonal relationship of the isolates was assessed by PFGE. Protein M typing was performed by sequencing of the region of the emm gene coding for the N-terminal zone of the protein. Susceptibility testing was performed with Etest strips on Mueller-Hinton blood agar plates. A multiplex PCR was used for detecting the presence of genes: speA, speB, speC, speF, speG, speH, speJ, ssa and smeZ.
Results: S. pyogenes was isolated from pharyngotonsillar samples of 11 children and 14 contacts, and from invasive samples of 2 out of 3 children with STSS. All isolates from children at the day care centre were of emm type 4, presented the same PFGE pattern and contained speB, speC, speF, ssa and smeZ. Isolates from contacts corresponded to emm types 1, 2, 12 or 22 and to 5 PFGE patterns unrelated to that of the children isolates. Isolates from epidemiologically unrelated patients were of emm types 3, 11, 12, 18 or 22 and presented 5 PFGE unrelated to that of the outbreak strain. All isolates were susceptible to penicillin, clindamycin, levofloxacin and tetracycline and, except 1 isolate from an unrelated patient, to erythromycin.
Conclusions: A strain of S. pyogenes of emm type 4 caused an outbreak in children attending a day care centre. The organism was also cultured from two children who developed STSS, one of which died. This strain was unrelated to other S. pyogenes isolated from contacts of the children or from epidemiologically unrelated patients from the same area
P1427 Molecular identification of Streptococcus bovis group isolates causing bacteraemia by sodA gene (superoxide dismutase gene) PCR and sequencing
M. Marin, M.J. Goyanes, E. Cercenado, M. Rodríguez-Créixems, P. Muñoz, E. Bouza (Madrid, ES)
Objective: To describe the distribution of species in our S. bovis-group isolates causing bacteraemia and to evaluate the utility of sodA PCR associated with sequencing to identify them.
Methods: 36 S. bovis-group strains (36 patients) isolated from blood cultures or heart-valve tissue between 2000 and 2006 were studied. Phenotypic identification was by conventional microbiological methods and API 20 Strep. Molecular identification was performed by sodA gene PCR (superoxide dismutase gene) followed by sequencing. The sequences obtained were compared with those included in Genebank. Only alignments with similarities higher than 99% were considered. Sequences of our S. bovis-group isolates were compared with type strains deposited in Genebank and well characterised phenotypically using Clustal X 1.8 software.
Results: The distribution of species after sodA PCR and sequencing was S. gallolyticus (19), S. pasteurianus (13), S. lutetiensis (3) and S. infantarius (1). A review of the clinical records of patients revealed 11 cases of infective endocarditis (all but one caused by S. gallolyticus) and 10 cases of S. bovis bacteraemia associated with abdominal malignancies (5 S. gallolyticus and 5 S. pasteurianus). For 6 patients with infective endocarditis, S. gallolyticus and S. infantarius were detected by sodA PCR directly in heart-valve tissue, but traditional culture was positive in only two patients.
Conclusions: sodA PCR followed by sequencing is a useful and rapid molecular tool for the identification of S. bovis-group clinical isolates. This method could provide more accurate results than the conventional methods used routinely in our laboratory. Sequence analysis of the sodA gene has enabled us to recognize recently described species among our S. bovis-group isolates that would have gone unnoticed using non-sequencing-based methods. S. gallolyticus was the most frequently identified species.
P1428 Preliminary evaluation of a PCR protocol to directly detect Streptococcus equi ssp. zooepidemicus in equine specimens
S. Preziuso, L. Bastianini, F. Laus, V. Cuteri (Matelica, Perugia, IT)
Objectives: Streptococcus equi ssp. zooepidemicus (S. zooepidemicus) are beta-haemolytic streptococci of the Lancefield group C. These bacteria are responsible for several diseases of animals and are though to be infrequent in humans, although cases of cervical lymphadenitis, pneumonia, meningitis, endocarditis, nephritis, septicaemia, cellulitis, and deaths due to S. zooepidemicus are not rarely described in humans. Animal to human transmission by respiratory or milk route are strongly suggested, but few studies in veterinary sciences are aimed at S. zooepidemicus detection, although reproductive disorders and strangle-like diseases due to S. zooepidemicus infection in horses are increasing in importance.
The objective of this work was to evaluate a PCR protocol to detect S. zooepidemicus in clinical equine specimens.
Methods: Twenty uterine swabs, 60 nasopharyngeal swabs and 4 milk samples collected from symptomatic horses were tested blindly by bacteriology and PCR.
After incubation of swabs in enrichment broth for 6 hours, two aliquots of the broth were used respectively for culture on equine blood agar (Oxoid-Milan) or for DNA extraction and PCR. Latex-agglutination test (Oxoid-Milan) was used to identify the Lancefield group of the beta-haemolytic streptococci grown on agar, and API Strep was used for the biochemical identification of the group C colonies. Furthermore, a loop of beta-haemolytic streptococci of group C was processed for DNA extraction and PCR. Primers for sodA and for seeI genes were used as previously reported by Alber et al (2004). PCR conditions were optimised using reference strains.
Results: S. zooepidemicus was cultured from 5/20 (25.0%) uterine swabs, 4/60 (6.7%) nasopharyngeal swabs and 1/4 (25.0%) milk samples. The same results were obtained by PCR on broth samples, with the difference that 7/60 (11.7%) nasopharyngeal swabs were positive. PCR on colonies confirmed API Strep (bioMérieux-Milan) identification in all cases except 1 case where a wrong API lecture was done.
Conclusions: PCR represents a rapid, sensitive and specific method to directly detect S. zooepidemicus in respiratory and genital samples of symptomatic horses. This protocol could represent a basis for the development of multiplex PCR method to differentiate the respiratory and genital pathogenic streptococci.
Further investigation will be aimed at verifying if this PCR protocol can be used also on human samples for diagnosis or epidemiological studies.
P1429 Serotyping of Streptococcus pneumoniae by PCR – expansion of the method to identify the Hungarian serotypes
O. Dobay, K. Nagy, S. Amyes, F. Rozgonyi (Budapest, HU; Edinburgh, UK)
Objectives: Streptococcus pneumoniae has 90 different serotypes, and as certain serotypes are linked with antibiotic resistance, virulence or different types of diseases, it is essential for the epidemiological studies to determine the serotype of the isolates, in addition to genotyping. The conventional method is to use antisera, but it is time consuming, expensive and difficult to evaluate. Some workers have started determining the serotype by PCR and we have expanded this methodology to characterise the strains normally found in Hungary.
Methods: Conventional serotyping was performed with MAST antisera on glass slides. For the PCR, the grouping primers and primers for several individual serotypes described by Brito et al. (1, 3, 4, 6, 14, 18C, 19F, 19A, 23F) were used, but we designed a set of new primers to other serotypes that are also relatively common in Hungary (9V, 6A, 6B, 7F, 11A, 15A, 15B). The PCR was done in two stages. The first multiplex PCR reaction divided the strains into 6 different groups, based on the gel pattern. Then a second multiplex PCR reaction, which included the primers for the individual serotypes of each group (3 or 4), was performed.
Results: Isolates of known serotypes were tested first with the individual primers in single PCR reactions. Then the grouping reactions were tried with these strains. The multiplex PCR worked best with Tth polymerase, but in Taq buffer. We had to alter the annealing temperature and other parameters as well. After improving the method, we designed the new primers. These had to be suited for the existing reaction, i.e. had to work under the same cycling parameters that had originally been used; however these primers also had to be unique for the new serotype within the group and provide PCR products of different sizes for each serotype. When invasive isolates of unknown serotypes were tested by this method and the results compared with the conventional method, the same serotypes were identified.
Conclusions: The new 7-valent conjugate vaccine was introduced in Hungary very recently, therefore the examination of invasive strains is of great importance. We have extended the method of Brito et al. to identify Hungarian isolates, by designing primers for new serotypes. Serotyping of large numbers of isolates by the conventional method requires expensive resources, while the capability to identify serotypes by PCR is much simpler, is as sensitive and is more cost effective than conventional serotyping.
P1430 Quantification of Streptococcus pneumoniae DNA in blood samples from patients with invasive pneumococcal infection
R.P.H. Peters, R.F. de Boer, T. Schuurman, M.A. van Agtmael, M. Kooistra-Smid, C.M.J.E. Vandenbroucke-Grauls, M.C. Persoons, P.H.M. Savelkoul (Amsterdam, NL)
Objectives: Streptococcus pneumoniae is the most prevalent cause of community-acquired pneumonia (CAP) and meningitis (CAM) in adults in the Netherlands. Invasive infection is associated with increased mortality, and rapid diagnosis is essential. Blood cultures, the diagnostic gold standard, have a relatively low yield and may take two days to become positive. We evaluated the clinical use of real-time PCR for direct detection and quantification of bacterial DNA in blood samples, without prior cultivation.
Methods: Whole blood samples were obtained simultaneously with blood cultures from patients suspected of CAP or CAM. Bacterial DNA was isolated blood samples with the QIAamp DNA Mini Kit (Qiagen, Germany). PCR amplifications were done on a TaqMan 7000 System (Applied Biosystems, USA) with a PCR assay that targeted the autolysin A gene of Streptococcus pneumoniae. A quantitation curve was included to calculate the bacterial DNA load (BDL). PCR results were compared to blood culture outcome and clinical data.
Results: In total, 130 blood samples from 63 patients were tested with PCR. The sensitivity of PCR for detection of Streptococcus pneumoniae in blood samples was 80% and the specificity 94%, Table 1 . PCR amplifications were positive in 11/14 patients (79%) with bacteraemia and negative in 44/48 patients (92%) without bacteraemia. With PCR, 4 additional cases were identified compared to blood culture, all of whom had a microbiologically proven source of Streptococcus pneumoniae infection (including 1 case of meningitis). Blood samples from patients with a positive PCR signal and negative blood culture result had been obtained more frequently under antimicrobial treatment than blood samples from patients with positive blood cultures (p = 0.04). When PCR results were combined with blood culture outcome, the detection rate of invasive Streptococcus pneumoniae infection increased from 22% to 29%. The median BDL was 24 cfu equivalents/mL (range 3.9–683). No association was found between BDL and clinical characteristics.
Table 1.
Comparison of PCR and blood culture for detection of Streptococcus pneumoniae in blood samples.
| PCR result | Blood culture result |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 16 | 6 | 22 |
| Negative | 4 | 101 | 105 |
| Total | 20 | 107 | 127 |
Conclusion: Quantification of bacterial DNA in blood samples adds to the diagnosis of invasive pneumococcal disease, also in patients receiving antibiotics, but sensitivity of the PCR assay should be improved to ensure clinical applicability. The clinical value of BDL in invasive pneumococcal infection seems promising but needs to be further evaluated.
P1431 Quantification of bacterial DNA during bacteraemia: a possible marker of severity of infection
R.P.H. Peters, M.A. van Agtmael, C.M.J.E. Vandenbroucke-Grauls, P.H.M. Savelkoul (Amsterdam, NL)
Objectives: We evaluated the clinical value of determination of Staphylococcus aureus and Enterococcus faecalis bacterial DNA load (BDL) during bacteraemia in critically ill patients. BDL was related to clinical and laboratory variables, and to microbiological culture results.
Methods: Blood samples for PCR were obtained whenever blood was drawn for culture from patients admitted to the intensive care unit of our hospital. After extraction of DNA from 200 mL blood with the QIAamp DNA Mini Kit, real-time PCR amplification was performed on a TaqMan 7000 System with specific primers and probe targeting the 16S rRNA gene of S. aureus and E. faecalis. A quantitation curve was included to determine the BDL.
Results: In total, 175 blood samples for S. aureus PCR and 180 for E. faecalis PCR, both from 80 patients, were included. The S. aureus PCR was positive in 31 blood samples from 18 patients; the E. faecalis PCR in 18 blood samples from 14 patients. Blood cultures were negative in 7/18 (39%) patients with a positive S. aureus PCR and 4/14 (29%) with a positive E. faecalis PCR result. Antimicrobial therapy had been administered to 10/11 patients with a positive PCR and negative blood culture result prior to blood sampling, which is significantly more frequent than patients with growth-positive blood cultures (p < 0.01). The median (range) S. aureus BDL was 440 (3–3,370) cfu equivalents/mL; median E. faecalis BDL was 130 (12–6,582). The E. faecalis BDL was significantly lower in patients with negative blood cultures compared to a positive culture result (p = 0.03), while a tendency was observed for S. aureus BDL in this regard (p = 0.07). Furthermore, patients with only a positive PCR result for S. aureus were less likely to have a microbiologically proven source of infection (p < 0.01) than patients with a positive blood culture.
We observed a significant correlation between BDL and C-reactive protein concentration, figure 1 .
Fig. 1.

Correlation between bacterial DNA load and C-reactive protein.
Individual correlations of S. aureus BDL (r = 0.60) and E. faecalis BDL (r = 0.75) with CRP were also significant (p<0.05). No other associations were observed between BDL and clinical or laboratory variables.
Conclusion: Determination of BDL can be used to quantify bacteraemia in ICU patients, also when blood samples are obtained under antimicrobial treatment or when there is no clear focus of infection. The positive correlation that we observed between BDL and CRP suggests that BDL may be a promising marker of severity of infection.
General molecular and viral diagnostics
P1432 Comparative analysis of three different regions of hepatitis C virus for genotyping Romanian strains
A. Oprisoreanu, C. Szmal, V. Thiers, G. Oprisan (Bucharest, RO; Paris, FR)
Objective: to compare the three different genomic regions (5'UTR, core and NS5B) for hepatitis C virus (HCV) identification and characterisation to analyse the strains in relation to those circulating worldwide in order to have a larger view of the epidemiology of HCV infection in Romania
Methods: the RNA from sera was extracted using QIAmp viral RNA extraction kit (Qiagen) and SV Total RNA Isolation System (Promega). The RT-PCR was done in 20ul with random primers and AMV (4U/ul) revers transcriptase enzymes. The obtained cDNAs were amplified in three genomic regions (5'UTR, core and NS5B) by nested-PCR. RFLP was realised in 5'UTR region with three restriction enzymes: Mva I, Mvn I and Sau3A I. The sequencing was performed on an automated sequencer ABI Prism 3100 Avant (Applied Biosystems). The analysis of sequences was performed using BLAST, BioEdit 7.0, Mega3 softwares.
Results: We analysed 153 sera from patients and blood donors in three genomic regions by all three methods: PCR, RFLP, sequencing and philogenetic analysis. The molecular analysis in the 5'UTR by RFLP gave very good information about the discrimination between 1b (the predominant subtype) and other genotypes and subtypes. The sequencing was done in all three regions (5'UTR, core and NS5B). In all three sequenced region the results indicated the same genotype the most reliable being NS5B and core region. Some samples identified as subtype 1a by RFLP were identified later by sequencing as genotype 1b in all regions. In Romania, so far we have identified genotype 1b 96%, 1a 2.6% and 4a 1.4%. In relations to other 1b genotype circulating in Europe, our strains do not form a specific cluster. Strains of genotype 1a are becoming more and more predominant in our country due to drug abuse, piercing, tattooing, etc. The presence of genotype 4a might indicate a possible root of circulation from Egypt.
Conclusions: In conclusion RFLP in 5′UTR region is the first test that allows a rapid and relatively cheap method for identification of the genotype but it has a low sensibility towards some subtypes. The distribution of genotype 1 in Romanian population reveal that genotype 1b is predominant in all ages while genotype 1a is found especially in younger populations, which might be an evidence of a shift between these two genotypes. Interestingly for Romania is the presence of genotype 4a, which is an endemic subtype for Egypt.
P1433 In vitro replication of hepatitis D virus using a new construct containing a dimer of HDV genome
F. Behzadian, F. Sabahi, M. Karimi, M. Sadeghi Zadeh, N. Maghsodi (Tehran, IR)
Objectives: There is no cell line susceptible to Hepatitis D Virus (HDV) infection. Therefore several genetic based models have been introduced being able to initiate HDV RNA replication which bypass needs of HDV particle to attachment, entrance and uncoating stages. Because of circular nature of HDV genome and its resemblance to plants viroids, the cloned HDV cDNA generally employed were constructed in tandem repeats of dimer or trimer genomes. Two ribozyme sequences on the HDV cDNA are necessary to generate a unit-length HDV RNA after transcription.
Methods: in this study a replicative dimeric construction of HDV cDNA was made from a single copy of full-length genome through a four-step cloning scheme (Figure 1 ). In the resulting recombinant plasmid named pcDNA3.1D2, two head-to-tail tandem repeats of genomic HDV cDNA transcript under CMV promoter control. pcDNA3.1D2 was transfected into the Huh7 and Cos7 cell lines, as cells with liver and non-liver origin respectively. The cells then were subjected for analysis of replication markers. Initiation of HDV replication was assessed by two markers, formation of antigenomic RNA strands and expression of HDAg, the only HDV encoded protein. RT-nested PCR was performed on DNA-removed total RNA samples from transfected cells and untransfected ones (as negative control) to show presence of HDV antigenomic RNA. Immunofluorescence and Western blotting also were done to detect expressed HDAg in the mentioned cell lines.

Results: Seven days after transfection, both replication markers in Cos7 and Huh7 were positive.
Conclusion: Since pcDNA3.1D2 is made from genomic HDV cDNA, antigenomic RNA would be raised exclusively after HDV genome replication. The results indicate that pcDNA3.1D2 could serve as a template for DNA-directed RNA synthesis and the transcribed RNA molecules were processed via ribozyme regions to form circular genomic RNAs which in turn proceeded HDV replication events. pcDNA3.1D2 can be used conveniently in transfection experiments to explore HDV biology further. Since this model is made of single unique HDV genome, there is possibility of using different isolates from different genotypes or pre-mutated HDV genomes to construct this kind of model and studying the capacity of their corresponded dimeric vectors for replication efficiency.
P1434 Incidence of double positive amplifications from immunocompetent patients screened for active human herpesvirus replication
A. Manna, A. Di Girolamo, D. Racciatti, L. Manzoli, G. D'Amico, E. Pizzigallo, A. Balbinot, F. Febbo, D. D'Antonio, G. Parruti (Pescara, Chieti, IT)
Objectives: In 2003 we introduced into clinical practice the use of molecular tools for diagnosing active Human Herpes Virus replication on various clinical specimens from immunocompetent patients. Since 2005, parallel quantitative amplifications were carried out for each virus by Real Time PCR. We report on the incidence of double-positive amplifications and their possible clinical significance.
Methods: DNA extraction from plasma, urine, saliva and vesicular eluates was performed using the QIAamp DNA Mini Kit (QIAgen GmbH, Hilden, Germany) according to manufacturer's instructions. A qualitative PCR amplification was carried out for VZV (Nested–PCR; Amplimedical Bioline), whereas for the remaining Herpes Viruses screened (HSV1, HSV2, EBV, CMV, HHV6 and HHV8) quantitative amplifications were carried out, using TaqMan probes and primers in an ABI PRISM 7000 Sequence Detection Systems (Real Time System – Applied Biosystems).
Results: Since January, 2005 through October, 2006, 820 samples from immunocompetent patients were collected. Thirty-one double positive samples (3.7%) were detected from 22 patients, 8 of whom with persistent fever, 8 with recurrent stomatitis, the remaining 6 with other herpetic recurrences. All double positive reactions were repeated from the same source, with 100% confirmatory results; for 9 patients parallel or serial samples could be analysed, showing >80% concordance. Double positive amplifications showed a non-random pattern. The 3 patients with atypical zoster lesions yielded VZV associated with either HSV1 or HSV2; patients with stomatitis yielded HSV1 or HSV2 in association with either EBV or HHV6; in patients with persistent fever, HSV1 and CMV were more frequently isolated from plasma, co-isolated in 2 cases and associated with either EBV or HHV6 in the other 6 patients. HHV8 was never amplified. Molecular diagnosis had a clinical impact in patients with atypical zoster-like lesions and in most cases of stomatitis.
Conclusion: In immunocompetent patients, double positive amplifications of Herpes Viruses seem to be rather frequent from selected clinical specimens. Our results suggest that they reflect true and complex patterns of viral re-activation, associated with frequently encountered clinical conditions. Molecular analysis of these patterns by Real Time PCR amplifications may be useful to guide clinical management in selected cases.
P1435 Monitoring of Epstein-Barr virus DNA in whole-blood samples of paediatric liver transplant recipients
M.P. Romero, M.P. González, E. Frauca, P. Jara, M.P. Peña, A. Gutierrez (Madrid, ES)
Introduction: Quantitative analysis of the Epstein–Barr virus (EBV) genome has been recently reported to be helpful for early identification of EBV viraemia which could reduce the risk of post-transplantation lymphoproliferative disorder (PTLD). The aim of this study was to determine circulating EBV DNA after transplantation in paediatric patients in relation to primary EBV infection and developing of PTLD.
Methods: We retrospectively analysed EBV-DNAemia in paediatric liver transplant patients by a quantitative real-time PCR assay (affigene® EBV trender, Sangtec, Sweden).
The EBV genome load in whole blood was measured serially in a total of 9 recipients of liver transplantation. The mean number of samples analysed per patient was 6.4 (range 4–8).
All patients were treated prophylactically with ganciclovir IV during the first month after transplantation and continued with oral Aciclovir or Valganciclovir during three months. Only in the patients with positive EBV-DNAemia were continued with treatment oral. The immunussuppressive protocol including tacrolimus and steroids was used during the first year post-transplantation.
Results: Before transplantation 3 patients were EBV seropositive and 6 were EBV seronegative. Five of the six patients who were EBV seronegative before transplantation developed positive EBV DNA samples and the levels were high (mean level 2.38×105 copies/mL, range 6.88×103–4.67×105 copies/mL). In these patients the primary infection developed between 12–16 weeks after transplantation.
In patients who were seropositives before the transplantation the EBV DNA levels were low (mean 1.48×103 copies/mL, range 7.14×102–1.90×103 copies/mL).
Patients with symptomatic EBV infections had higher EBV DNA levels than those with asymptomatic infection (median 3.70×105 vs 1.74×103 copies/mL). The frequent symptoms were fever, elevated aminotransferases and neutropenia.
One of the patients developed PTLD grade 1. In this patient the EBV DNA levels, retrospectively investigated were the highest (mean 1.48×106 copies/mL, range 5.24×105–2.00×106 copies/mL). In this patient when the levels of immunosuppressives agents were gradually lowered the hypertrophic amygdala disappeared.
Conclusions: Serial measurement of EBV viral load by quantitative PCR is a useful tool for monitoring the course of EBV infection and to help detect the patients who are at the highest risk for PLTD. Frequent monitoring can by used to make modification in the levels of immunosuppressives and/or antiviral agents.
P1436 Cytomegalovirus reactivation in inflammatory bowel disease
F. Hanses, B. Salzberger, J. Schölmerich, C. Franzen (Regensburg, DE)
Objectives: Clinical significant cytomegalovirus infection or (re)-activation is rare in immunocompetent patients, however, much more frequent in immunocompromised patients. CMV reactivation occurring in patients with inflammatory bowel disease (IBD) has been insufficiently characterised so far concerning frequency and clinical significance. Patients with IBD (ulcerative colitis, Crohn's disease) are supposed to be less immunodeficient than patients after solid organ or bone marrow transplantation or patients with HIV. However, previous studies suggest a more severe course of the disease in CMV positive patients, eventually pointing to a local immune imbalance.
Methods: After establishing highly sensitive PCR methods to detect both CMV DNA and mRNA of immediate-early, early and late CMV genes (using RT-PCR after DNA digestion), we investigated tissue from IBD patients and controls. So far, 12 IBD patients (7 of whom reveived immunosuppressive therapy) were studied.
Results: IBD patients with disease activity under moderate to strong immunosuppressive therapy (n = 7, on local or systemic steroids, methotrexate, azathioprin, TNF-α-inhibitor) showed a higher degree of CMV activity than IBD patients with routine control biopsies or patients with disease activity under no immunosuppressive therapy or mesalazine alone (n = 5). Both DNA positivity in tissue biopsies and the degree of CMV specific mRNA transcripts were higher in the patient group under immunosuppressive therapy (83% vs. 60% for CMV DNA and 43% vs. 20% for CMV mRNA.
Conclusion: CMV (re-)activation seems to be associated with IBD disease activity and immunosuppressive therapy. It is not clear yet whether this is due to immunosuppression or a refractory disease course. Although the total number of patients studied so far is small, these findings warrant the investigation of the role of CMV associated complications in a larger patient cohort.
P1437 Real time PCR for monitoring herpes simplex virus load in tracheal aspirates of intensive care unit patients
N. De Vos, A. Vankeerberghen, A. Boel, I. Demeyer, L. Van Hoovels, H. De Beenhouwer (Aalst, BE)
Objectives: Detection of Herpes Simplex Virus (HSV) in respiratory secretions of ventilated patients by culture is described but the significance of these findings remains unclear. Publications with fast and reliable methods for quantitative detection of HSV in lower respiratory tract samples are lacking. We started an observational study in which HSV viral load was monitored in tracheal aspirates of ventilated ICU patients by a quantitative real time PCR (Q-PCR).
Methods: Successive tracheal aspirate samples of ventilated adults (n = 65) staying at the ICU were collected. Sampling was started on day 3 of intubation. Nucleic acid extraction was performed on the NucliSens-EasyMAG® (bioMérieux). DNA-extract was used for Q-PCR with primers and probe targeting the DNA polymerase gene of HSV-1 as well as HSV-2 (ABI 7000 Sequence Detection System).
Results: The Q-PCR gave very stable and reproducible results with minimal intra- and inter-run variability. The assay could detect 102 copies/mL of HSV and no interferences were observed with bacteria, fungi or other viruses.
Of the included patients 32% were mechanically ventilated for at least 12 days. In 65% of these patients HSV was detected at any point. In these long term intubated patients often a sharp increase was observed in consecutive samples coinciding with clinical deterioration and bacterial Ventilator Associated Pneumonia (VAP). Q-PCR could demonstrate in some of these patients extremely high viral loads (>108 copies/mL tracheal aspirate). Administration of acyclovir treatment resulted in gradual decrease of the viral load.
Conclusion: Q-PCR can be used to monitor the HSV viral load quantitatively in tracheal aspirates of intubated patients. An increase in HSV viral load is often observed in ICU patients who are intubated for a long time. Under acyclovir administration a decrease of HSV viral load can be observed. Further studies need to be done to investigate whether antiviral treatment can influence the incidence of VAP or the final outcome in ICU long term intubated patients.
P1438 Seroprevalence and genotyping of Varicella zoster virus strains in the Russian Federation
O. Vankova, T. Ulanova, V. Loparev (Nizhny Novgorod, RU; Atlanta, US)
Background: Varicella zoster virus (VZV) is distributed worldwide and is associated with diseases both in children and in adults. In Russia, data on VZV seroprevalence in adults and children in different parts of the country is limited, and the genetic variability of circulating VZV strains is unknown.
Objectives: We aimed to study the seroprevalence of VZV in population in various age groups, and characterise VZV strains from known chickenpox and varicella zoster cases. Further, all known genotyping methods were compared.
Materials and Methods: The IgG antibodies to VZV were measured from 2,000 serum samples by using commercial EIA test for VZV. Specimens for VZV genotyping were obtained from skin lesions of 61 paediatric and adult patients. PCR amplified fragments of virus DNA were examined with three different genotyping methods. Genotyping was being carried out by sequencing analysis on ABI 3100 Avant Genetic Analyzer instrument (ABI, Foster City, CA, USA) and by Real-Time PCR with Light Cycler instrument (Roche, Indianapolis, IN, USA).
Results: Seroprevalence to VZV varied from 45% in children aged 2 months (maternal antibodies) or 30% in children of 1 year old rising to more than 90% towards the age of 10 years and remaining then about at the same level to older age. All VZV DNA samples were Pst+Bgl– (in ORF38 and 54, accordingly) and did not have Sma I site in VZV ORF 62. These data showed that Russian VZV strains are not related to Japanese (J) VZV vaccine-like variants. All analysed VZV strains belonged to European (E1 and E2) genotype identical to Europenian reference Dumas and HJO strains or to C and B genotype according to Breuer classification system. Similar VZV strains were found in countries which border Russian Federation in Europe and Asia.
Conclusion: These data gave a reference to immunisations strategies and can be useful to predict possible consequences after carrying out mass vaccination against VZV, with vaccine created on the basis the Japanese genotype strain.
P1439 Mutagenesis analysis of the NS2B determinants of a tick-borne flavivirus NS2B-NS3 protease activation
B. Pastorino (Marseille, FR)
The genus Flavivirus, family Flaviviridae, contains more than 70 viruses, many of which are arthropod-borne pathogens of humans and animals. Being critical for virus replication, the viral protease NS2B-NS3 is an interesting target for inhibitors with possible use as antiviral drugs especially by inhibiting the complex formation. Alkhurma virus (ALKV) is a tick-borne class-4 flavivirus responsible for several human cases of haemorrhagic fever in Saudi Arabia and a protease assay, using a p-nitroanilide substrate (BAPNA) had been previously developed in our laboratory (Bessaud et al., 2005).
In the present work, in an attempt to identify conserved residues and motifs essential for the NS2B-NS3 protease activity, we first compared ALKV NS2B sequence with the corresponding sequences of other flaviviruses. The deltaNS2B sequence alignment analysis revealed that only a few residues (L50, W60, G68, G81 and E89) are conserved and the differences observed between viruses studied were regularly distributed along the deltaNS2B region affecting all the motifs previously described for NS3 protease activation. Based on this sequence comparison, alanine substitutions were introduced in the NS2B sequence at residues W60, G68, L73, Q77, G81 by site-directed mutagenesis. In addition, substitution mutants V88D and V88K were generated as well as a deletion one called ÄV88. Two chimeric proteases were also constructed, with the NS2B cofactor from Langat virus and Dengue virus type 3 replacing the ALKV sequence. The kinetic parameters of the deltaNS2B-NS3pro mutant proteases was measured and compared with the activity of the wild-type ALKV enzyme. Finally, four mutants and the two chimeric proteins exhibited reduction of protease activity against BAPNA showing that tight complementarity of the proteins sequences is necessary for NS2B-dependant activation of NS3.
In conclusion and for the first time, the enzyme-cofactor interactions of a tick-borne flavivirus were analysed and two critical NS2B residues for ALKV NS3 activation V88 and Q77 were identified. This study also highlighted the need to enlarge and accumulate the experimental identification of NS2B important residues with protease complex belonging to different flavivirus cluster
P1440 Human papillomavirus genotyping by DHPLC and sequencing
E. Dimitriadis, N. Pandis, E. Kada, F. Lazaraki, E. Kouri-Bairaktari, A. Kourtis, M. Karakoulaki, S. Kanakakis (Athens, GR)
Objectives: Since HPV is the central causal factor in cervical cancer, understanding the epidemiology and geographical area distribution of the most prevalent HPV genotypes constitutes an important step towards development of prevention strategies. This study was aimed to a detailed HPV genotyping with a combination of molecular techniques, in women attended the Cervical Pathology Department of “Helena Venizelou” Hospital.
Methods: HPV Hybrid Capture applied to all samples. All the samples were also genotyped using PCR with a set of consensus primes (L1C1/L1C2M, Gp5+/Gp6+, PU-1M/PU-2R), combined with High-Performance Liquid Chromatography and direct sequencing.
Results: Up to now, 154 samples have been tested. 22.7% was found positive for High Risk genotypes with the Hybrid Capture. 39.6% was positive by the PCR methods. 15 different genotypes were identified, 14 of these being High-risk genotypes. HPV56 was the most frequent genotype (22.9%) followed by HPV33 (14.7%), HPV31 (9.8%), HPV39 (9.8%) and HPV58 (8.1%). In 14.7% of the samples double infection was found.
Conclusion: HPV genotyping by extensive molecular techniques allows a more precise estimation of the most frequent HPV genotypes in a given population. To the best of our knowledge this is the first study in a Greek population with these techniques.
P1441 Oncogenic human papillomavirus type distribution by L1 and E6/E7 sequencing
S. Perez, I. Lopez, M.J. Lamas, M.J. Alvarez, F.J. Vasallo, F. Ulloa, J. Torres (Vigo, ES)
Objectives: A subgroup of human papillomavirus (HPV) termed “high-risk” is associated with more than 90% of cervical cancers. The aim of this study was to know the high-risk HPV distribution in women and its relationship with cytology in our environment.
Methods: 197 samples of endocervical scrapes (January to September, 2006) from 193 patients with recent cytological/histological data were selected. 3 PCRs per sample were carried out for the amplification of L1 and E6/E7 regions and the human β-globine gene, using consensus primers (MY09/MY11, HPCF/HPCR, HBBF/HBBR respectively). The PCR products of L1 and E6/E7 regions were analysed by sequencing using the same primers (Big Dye Terminator sequencing kit. Applied Biosystems. GenBank database).
Results: 2 samples (1%) were excluded because they were inhibited. HPV was detected in 59/195 samples (30.3%): 45/195 (23.1%) high-risk HPV, 2/195 (1%) low-risk HPV, 6/195 (3.1%) undetermined risk, 2/195 (1%) non-genotypable and 4/195 (2.1%) co-infections. The distribution of high risk genotypes according to cytological data was: normal cytology 17/140 (12.1%), atypical squamous cells of undetermined significance (ASC-US) 7/21 (33.3%), low grade squamous intraepithelial lesion (LSIL) 11/22 (50%), high grade squamous intraepithelial lesion (HSIL) 8/10 (80%), carcinoma 2/2 (100%). 20 of these 45 high-risk HPV (44.4%) were detected by only one of the two especific PCRs used. 50 patients were infected by only one HPV type: HPV 16: 17/50 (34%), HPV 31: 5/50 (10%), HPV 52: 3/50 (6%), HPV 56: 3/50 (6%), HPV 33: 3/50 (6%), HPV 66: 3/50 (6%), HPV 58: 2/50 (4%), HPV 82: 2/50 (4%), HPV 35, 39, 45, 53, 68: 1 of each type (2% each). It is important that certain genotypes undetected by commercial methods (HPV 53, 66, 82) were found in 12% of the patients. HPV 16 was the most frequent genotype detected in the patients with high-grade lesions (7/10 with HPV 16), however we have not found one predominant genotype in the patients with low-grade lesions (10/32 with HPV 16).
Conclusions: (1) PCR and DNA sequencing of part of L1 and E6/E7regions of HPV have a good sensitivity and diagnostic utility for the screening of uterine cervix cancer. (2) The amplification of both E6/E7 and L1 genes was necessary for the accurate detection and genotyping of HPVs. (3) HPV 16 was the most frequent genotype found in our population, followed by HPV 31. (4) HPV 16 represents most of the high-risk HPVs associated with HSIL and carcinoma.
P1442 Amplification and sequencing of a 190bp segment containing nucleotide 6240 from prototype human papillomavirus16 L1 gene and 10 clinical isolates from the UK
H. Al Zoubi, P. Vallely, A. Turner, M. Guiver (Manchester, UK)
Previous studies have shown that all HPV16 clinical isolates, for which sequence data is available, have a cytosine to guanosine (C to G) nucleotide change at position 6240 relative to the prototype HPV16 sequence. The coordinate L1 amino acid 202 resulting from this change is aspartic acid instead of histidine (D202H). This change has been suggested to be important to escape the immune response and was also found to be important for the efficient assembly of HPV16L1 recombinant proteins into virus-like particles which are currently being used as a vaccine against HPV infections. An HPV16 genome cloned into pAT153 plasmid was obtained by our laboratory in 1986. However, no information on the source of the cloned isolate was available. In an attempt to identify the cloned HPV16 isolate for further L1 proteins expression study, we amplified and sequenced a 190bp segment of L1 gene which contained the coding region for the nucleotide of interest (6240). In this isolate nucleotide 6240 was found to be C and the correspondent amino acid at codon 202 in which the C nucleotide is found was therefore histidine. Based on the previously published data and on the NCBI BLAST alignment, we identified the cloned HPV16 to be the prototype HPV16 isolate. To find an appropriate HPV16 isolate for protein expression and to confirm the presence of the D202H observation in clinical isolates, 10 clinical isolates collected from UK patients with different degrees of cytological abnormality were also amplified and sequenced using the same PCR and sequencing conditions. All clinical isolates were found to contain nucleotide G at position 6240 coding for aspartic acid at position 202 in the L1 gene.
The established PCR and sequencing conditions could be used to confirm the presence of D202H in HPV16 isolates before proceeding to protein expression of HPV16L1. It could also be used to sequence larger numbers of clinical isolates from patients with different degrees of cytological abnormality, which will further help in understanding the HPV16 associated pathology.
P1443 Isolation of a canine-like human rotavirus strain G3P[3] from a child with acute gastroenteritis hospitalised in Palermo, Italy
S. De Grazia, G.M. Giammanco, S. Ramirez, A. Cascio, V. Martella, S. Arista (Palermo, Messina, Bari, IT)
Objectives: Group A rotavirus are the most common aetiologic agents of acute gastroenteritis in children and infants worldwide. They are ubiquitous viruses, also infecting the young of many animal species. There are a number of reports of atypical human rotavirus strains, that show some genetic and antigenic features of rotavirus isolated from various animal species. Such features suggest that the occurrence of rotavirus genetic reassortment and interspecies transmission may occur in nature. In the present study, we report the molecular analysis of VP7, VP4, VP6 and NSP4 genes of an unusual human rotavirus strains G3P[3], detected in a young child with acute gastroenteritis in Palermo, South of Italy, during 1997.
Methods: The rotavirus strain PA260 was isolated from a 2 year-old child admitted with severe diarrhoea at the “G. Di Cristina” Children's Hospital of Palermo, Italy during an epidemiological surveillance study. As it showed a G3 type, VP7 related, a P, VP4 related, not determinable type, and a subgroup I specificity, it was considered atypical and it was submitted to VP7, VP4, VP6 and NSP4 characterisation by sequence analysis.
Results: The viral strain was characterised as G3P[3] rotavirus, showing a 99% and 98% identity in the VP7 and VP4 proteins, respectively, to the canine rotavirus RV52/96 strain isolated in Bari in 1996. Also the analysis of VP6 and NSP4 proteins revealed an high amino acid identity, 95% and 98.8% respectively, of PA260/97 to the canine RV52/96 strain. No human G3 strain, available in GenBank, appeared as closely related to PA260 strain as RV52/96 strain.
Conclusions: Since the VP7, VP4, VP6 and NSP4 sequences of the PA260/97 strain are located very close to canine RV52/96 strain sequences, it is likely that the infection of the Italian child was the result of canine-to-human transmission. The PA260 strain is the third rotavirus strain with G3P[3] type described throughout the world and the first in Italy.
P1444 Evaluation of a real-time RT-PCR assay for the diagnosis of measles virus infections during a measles outbreak in Greece
S. Kokotas, M. Giannaki, E. Horefti, D. Sgouras, M. Logothetis, P. Panagiotopoulos, T. Georgakopoulou, A. Kansouzidou, M. Theodoridou, A. Pangalis, A. Mentis (Athens, Thessaloniki, GR)
Objectives: The aim of the study was to evaluate a real-time RT-PCR (RRT-PCR) for the detection of measles virus (MeV) RNA in clinical specimens collected during a measles outbreak in Greece.
Materials and Methods: Serum samples and at least one of the following samples, namely, heparinised blood, pharyngeal exudate and urine, were collected from suspected measles cases. All cases came from the measles outbreak in Greece, (October 2005–August 2006). The presence of MeV-specific IgM in patient sera was analysed by a commercial indirect ELISA (Enzygnost, Dade Behring). Equivocal results were re-tested with an IF method (BIOS). Viral RNA was extracted from peripheral blood mononuclear cells (PBMC), pharyngeal exudates and urine by Qiagen Viral RNA mini kit. A quantitative RRT-PCR (Schalk et al, 2004) was utilised for the detection of MeV-RNA in all three types of samples, based upon amplification of a 413 bp amplicon in the carboxyterminal region of measles nucleoprotein gene, detected by a FRET hybridisation probe pair on the Light-Cycler.
Results: In total, 94 specimens from measles suspected cases were included in the study. In 87 patients, at least one method (serology and/or RRT-PCR) was positive for MeV infection. Of those cases, 78 were positive and 7 negative by both methods, while 9 cases were positive solely by RRT-PCR. However, in six of the 9 cases positive by RRT-PCR and negative by serology, all specimens had been collected during the first two days after onset of the rash, which might lead to false-negative results by IgM serology. After exclusion of these cases, sensitivity, specificity, positive and negative predictive values for the RRT-PCR were calculated at 100% (95% CI 95.4–100.0), 70% (95% CI 34.8–93.3), 96% (95%CI 89.5–99.2) and 100% (95% CI 59.0–100.0) respectively. Pharyngeal exudates yielded a higher proportion of MeV RNA detection, followed by PBMC and urine samples.
Conclusion: A higher detection rate was observed for MeV RNA in pharyngeal exudates and PBMC than for specific IgM in serum. RRT-PCR is a useful method for MeV infection confirmation and should supplement serology, especially when the specimens for MeV infections are collected within 2–3 days after rash onset and a second visit of the patient is not practical.
P1445 Molecular diagnostics of noroviruses detected in Apulia, Italy
M. Santantonio, C. Rizzo, M.F. Coscia, D. Rizzi, G. Favia, M. Barile, R. Bavarese, D. De Vito (Bari, IT)
Objectives: Human caliciviruses have been considered one of the most common causes for nonbacterial acute gastroenteritis outbreaks and sporadic cases in adults and children worldwide.
A research network set up to study food-borne viruses covering ten European countries found that norovirus was responsible for over 85% of all non-bacterial outbreaks. Their role as the second cause of sporadic cases of viral gastroenteritis has also been reported by several studies carried out in other European countries.
Italy has no specific surveillance system for viral gastroenteritis and laboratory diagnosis is only carried out in a few cases. Therefore the impact of NV infection is currently unknown and little information is available about circulating strains.
The aim of this study was to evaluate the relative contribution of noroviruses to sporadic cases in adults and children of acute nonbacterial gastroenteritis in Apulia and to investigate the genotype of the strains involved.
Methods: A panel of 273 stool specimen collected over a two years period of time (2004–2005), were derived from cases of gastroenteritis and included in this study.
Information, collected by means of a standardised questionnaire, included: age, sex, hospital and division, diagnosis.
All faecal samples have been tested for NLVS by RT-PCR.
The primer used in this study were JV12 and JV13 located in the ORF1 region, and amplify region of 327 bases, of the RNA-dependent RNA polymerase gene of NLVs.
Results: Among the 273 stool specimens studied, 43 cases (15.7%) were associated to noroviruses infection.
The seasonal distribution of the episodes showed that acute gastroenteritis caused by norovirus were distributed in winter months.
The age of the individuals was one of the factors analysed: norovirus were more common among children under 5 years and in those 15 to 19 years old.
The frequent diagnosis associated were gastroenteritis (38%) and diarrhoea (23%), and the virus was more frequently detected in paediatric division.
Sequence analysis using the database Food-borne Viruses Europe revealed strains belonging to the Lordsdale genotype (GGII.4) and Rotterdam genotype (GGII.3).
Conclusion: The high detection rate of norovirus as the cause of gastroenteritis in adults and children reported in this study supports their inclusion in routine screening to diagnose sporadic cases of acute gastroenteritis.
P1446 Detection of polyomaviruses and herperviruses in human adrenal tumours
L. Barzon, M. Trevisan, G. Masi, A. Porzionato, M. Iacobone, G. Palu' (Padua, IT)
Objectives: The presence of polyomaviruses and herpesviruses in adrenal tumours and their role in adrenal tumorigenesis has never been investigated, even though the adrenal gland seems to be a preferential site of infection by these viruses and adrenal steroid hormones have been shown to activate their replication. Aim of this study was to investigate whether there is an association between viral infection and adrenal tumours.
Methods: A total of 128 human adrenal tissue samples, including 20 normal adrenal tissues, 37 nonfunctioning adrenocortical adenomas, 16 cortisol-producing adenomas, 12 aldosterone-producing adenomas, 3 androgen-producing adenomas, 16 adrenocortical carcinomas (14 functioning and 2 nonfunctioning), and 24 adrenal medullary tumours (including 21 benign pheochromocytomas, 1 malignant pheochromocytoma, 2 ganglioneuromas) was investigated for the presence of viral DNA sequences of the herpesviruses EBV, HCMV, HSV1, HSV2, VZV, HHV6, HHV7, and HHV8 and the polyomaviruses JCV, BKV, and SV40 by real-time quantitative PCR assays on DNA estracted from frozen or formalin-fixed paraffin-embedded samples. Expression of viral genes was investigated by immunohistochemistry.
Results: Herpesviruses and polyomaviruses sequences and gene expression, which were detected in a high proportion of both normal and neoplastic adrenal samples (overall, viruses were found in 15% normal adrenals, 25% benign adrenal tumours, and 31% malignant tumours). The polyomaviruses SV40 and BKV were more frequently found in adrenocortical carcinomas and in the malignant pheochromocytoma, whereas herpesviruses, especially EBV and HCMV, were more frequently detected in functioning benign adrenocortical tumours, often as coinfection. Moreover, tumours from patients with severe hypercortisolism frequently showed herpesvirus co-infections at high viral copy number. In positive samples, immunohistochemical analysis demonstrated EBV and CMV lytic gene expression, but no expression of latent genes, suggesting virus reactivation. As for polyomavirus-positive tumours, expression of T antigen was confirmed by immunostaining.
Conclusions: Our study suggests that the adrenal gland could be a reservoir of infection for these viruses and that hormone overproduction by the adrenal gland could represent a trigger for virus reactivation. On the other hand, these viruses could also contribute to adrenal cell proliferation and tumorigenesis.
P1447 Prospective evaluation of a commercial enzyme immunoassay for the rapid antigenic detection of influenza A and B virus from paediatric respiratory samples in five consecutive influenza epidemics seasons
J. Reina, E. Ruiz de Gopegui, A. Mena, M. Macia (Palma de Mallorca, ES)
Objectives: We report a prospective study of the efficacy of a commercial enzymoimmunoassay (EIA) compared with the shell-vial culture method (MDCK cell line) in the rapid antigen detection of Influenza A (IA) and Influenza B (IB) viruses in the respiratory samples of paediatric patients obtained in five consecutive flu seasons (2000–2005).
Material and Methods: Each of the respiratory samples (nasopharyngeal aspirates) was subjected to antigen detection against IA and IB viruses using the rapid differential EIA membrane test (Directigen FluA+B, Becton & Dickinson Co., Sparks Maryland, USA) following the manufacturer's recommendations. At the same time each sample was inoculated in two shell vials of the MDCK cell line, incubated at 36°C for 3 days and stained with specific monoclonal antibodies against each virus (Monofluokit Influenza A and B, Sanofi Diagnostics, France).
Results: In this period we studied 1,077 samples (30.8% were considered as positive) and we isolated 331 influenza viruses (234 IA, 70.7% and 97 IB, 29.3%) (p < 0.05). The percentage of influenza viruses isolated in the different epidemic periods varies significantly. The overall data of EIA method compared with the shell-vial culture show for the overall antigenic assay (IA + IB) a sensitivity of 65.8%, specificity 100%, positive predictive value 100% and negative predictive value 86.8%. There were no stastically significant differences between the overall sensitivity values (62.4% vs 74.2%) nor between the negative predictive values (89.4% vs 96.7%) for the IA and IB viruses. Statistically significant differences were observed only in the sensitivity values for the IA and IB viruses in the 2000–2001 (18 IA and 4 IB isolates) and 2001–2002 (50 IA and 20 IB isolates) seasons. We detected higher overall sensitivity overall values for the detection of IB viruses than for IA viruses. In the 2003–2004 season we only isolated IA viruses (29.7% of positivity).
Conclusions: In spite of the low overall sensitivity of the commercial EIA method studied, it must be regarded as an acceptable (high specificity), simple and rapid method for use in the antigenic detection of IA and IB viruses in paediatric respiratory samples.
P1448 Flocked swabs and UTM-RT are pre-analytical tools suitable for rapid antigen kits, direct immunofluorescence, culture and PCR diagnostics assay for viral infections
S. Castriciano, A. Petrich, M. Smieja, M. Chernesky (Brescia, IT; Hamilton, CA)
Objective: Detection of antigens, nucleic acids, and isolation of microbes depend on pre-analytical devices used for specimen's collection. Diagnostic sensitivity varies with the number of cells and free organisms released in the transport system. It was reported that Flocked Swabs (FS) and UTMRT (Copan, BS, Italy) enhances analytical sensitivity of antigen detection, culture and nucleic acid amplification assays. To compare the Copan FS and UTMRT to the Remel Dacron swabs (DS) and M4RT for virus culture, epithelial cells recovery for direct immunofluorescence assay (DFA), antigens and nucleic acids stability for rapid kits and amplification assays from nasopharyngeal swabs (NPS) for the diagnose of respiratory viruses.
Methods: 5,002 consecutive NPS, collected with FS and UTMRT, submitted to the Virology Laboratory from November 1, 2004 to April 30, 2006, were compared to 4,288 NPS collected with DS and M4RT from November 1, 2002 to April 30, 2004. Of the NPS collected with FS and UTMRT, 261 were analysed for Flu A/B and 375 for hMPV by PCR, 291 were tested with 4 rapid antigen kits for RSV, flu A and B. All NPS were tested by DFA, cells pellets were spotted on glass slides, fixed and stained with the para 1, 2 and 3, flu A and B, RSV, adenovirus and hMPV FITC antibodies. NPS were inoculated into R-Mix shell vial cultures. After 48 h, cells were fixed, and stained with the Pool and hMPV reagents. Pool positives were typed. For PCR, nucleic acids were extracted with the miniMAG system. 5 ul purified nucleic acid was tested with the RealArt® kit for Influenza A/B or with an hMPV specific RT-PCR.
Results: 1,318/4,288 were positive in the NPS collected with DS and M4RT: DFA/culture had 538 flu A, 635 RSV, 10 para 1–3, 35 adenovirus; 2,099/5,002 were positive in the NPS collected with FS and UTMRT: DFA/culture had 663 flu A, 277 flu B, 879 RSV, 171 para 1–3, 109 adenovirus; 101 positive out of 868 tested for hMPV. Antigen tests had 121 RSV, 33 flu A, 37 flu B and 100 negatives. The PCR detected 102/261 Flu, 66/375 hMPV. NPS collected with FS and UTMRT detected 2,009/5,002 (42%) positive compared to 1,318/2,288 (31%) positive detected in NPS collected with DS and M4RT.
Conclusions: NPS collected with flocked swabs in UTMRT detected a higher number of positives than NPS collected with DS and M4RT. The Copan flocked swabs and UTMRT collection and transport system is a universal system compatible with rapid antigen kit, DFA, culture and PCR and supports the detection/growth of hMPV.
P1449 Assessment of the Immulite systems CMV IgM assays in comparison to the Vidas assay in a clinical evaluation
F. Vlaspolder, P. Singer (Alkmaar, NL)
Objective: Cytomegalovirus (CMV) belongs to the herpesvirus family with other well-known viruses such as Herpes Simplex Virus (HSV), Varicella Zoster Virus (VZV) and Epstein-Barr Virus (EBV). CMV is found throughout the world and although the infection can be asymptomatic in the general population, it can cause severe infections in immunocompromised individuals and in newborns of infected pregnant women. The objective of this study was to evaluate the clinical performance of the DPC IMMULITE® systems' CMV IgM assay to that of the bioMérieux VIDAS® for detection of IgM antibodies to CMV.
Methods: A total of 523 samples were analysed on the two DPC IMMULITE platforms and the bioMérieux VIDAS system. Included in the sample population were 158 pregnancy samples, 37 samples from transplant cases, 68 samples defined as primary CMV infections, 153 known negatives, 15 BBI panel, 44 cross-reactives (RFand toxoplasma IgM) and 48 samples defined as EBV infections.
Results: The overall agreement to the VIDAS assay was 92.3% for IMMULITE and 92.8% for IMMULITE 2000. In assessing the performance of the IMMULITE assays within the specific populations tested, the Mikrogen® CMV recombinant immunoblot, clinical findings if available, and EBV serology were used to further analyse the true IgM reactivity to CMV. Upon resolution of discordant results with these additional methods, the IMMULITE assays and the VIDAS assay showed acceptable agreement within the specified patient populations.
Based on the blot results, some samples originally identified with the VIDAS assay as CMV reactive cases, were reclassified as EBV infections. Of the 48 samples identified as reactive to EBV and non-reactive to CMV, the IMMULITE (2000/One) assays resulted correctly in up to 37/39 of the 48 cases, while the VIDAS assay resulted correctly in up to 23 of the 48 cases
Conclusion: Overall analysis of the IMMULITE and VIDAS results indicates that the IMMULITE CMV IgM assays are comparable in performance to the VIDAS assay in all sample populations tested. Additionally, the IMMULITE assays have a lower level of cross reactivity interference to EBV, suggesting a higher level of accuracy in reporting a true CMV IgM result.
P1450 Evaluation of real-time PCR based assays for the detection and quantitation of CMV DNA
M. Rudolph, B. Willert, T. Grewing, J. Fuhrmann (Hamburg, Berlin, DE)
Introduction: Infections with human cytomegalovirus (CMV) are usually asymptomatic or related with harmless symptoms. But severe problems occur if embryos, transplant patients, patients under immunosuppressive therapy or AIDS patients get infected. CMV infection in this risk group can even be fatal. The aim of this work was the evaluation of real-time PCR assays for the fast, sensitive and specific detection of CMV DNA and the reliable quantitation of the viral load. These assays are important and useful for testing organ donors, monitoring of CMV patients under antiviral therapy, and for the protection of unborn children.
Materials and Methods: The artus® PCR Kits were designed and optimised for different real-time PCR instruments: LightCycler®, the Rotor Gene™ 3000 and the ABI PRISM®7000–7900 Instruments. The analytical sensitivity was determined by probit analysis. Using negative human EDTA-plasma was spiked with pre-quantified CMV. Serial dilutions were prepared, purified using the QIAamp® DSP Virus Kit and analysed with the respective artus CMV PCR Kit. The analytical specificity was tested with isolates of related viruses and other human pathogens. The diagnostic specificity and sensitivity was determined by analysing 177 samples in comparison with the COBAS AMPLICOR™ CMV MONITOR System.
Results: The determined analytical sensitivity was 65, 57 and 64 copies/mL for the LightCycler Instrument, the Rotor-Gene 3000, and the ABI PRISM assay, respectively. Cross reactivity was excluded testing isolates of other pathogens. The diagnostic sensitivity and specificity of the artus CMV PCR Kits was determined by comparison with the COBAS AMPLICOR CMV MONITOR System. Clinical sensitivity of 100% was determined.
Conclusions: The results demonstrate that the artus CMV PCR Kits, fulfil all requirements for a fast, sensitive, and specific detection of CMV DNA and the reliable quantitation of the CMV viral load. Therefore, the artus CMV PCR Kits can be perfectly used for the successful management of CMV disease, especially in immunocompromised patients and pregnant women.
Trademarks: QIAGEN®, artus®, QIAamp®, (QIAGEN Group), ABI PRISM® (Applera Corporation), CORBAS AMPLICORTM, LightCycler® Instruments (Roche Group), Rotor-GeneTM (Corbett Research)
P1451 Pitfalls in molecular detection of human respiratory syncitial virus
A. Vankeerberghen, A. Boel, H. De Beenhouwer (Aalst, BE)
Objective: Molecular detection techniques take, due their speed, specificity and sensitivity, a major place in the detection of viruses on clinical samples. We started 2 years ago with the detection of human respiratory syncitial virus (hRSV) by real time PCR (RT PCR) on aspirates. The PCR technique proved to be more sensitive than the antigen test. In the autumn of 2006 hRSV antigen positive samples that were negative by RT PCR were found. Further experiments were done to resolve the problem.
Methods: Antigen testing was performed with the hRSV Respistrip® (Coris Bioconcept). RNA was extracted (EasyMAG, bioMérieux) from the aspirates, converted in cDNA (Applied Biosystems) and analysed by RT PCR on an ABI 7000 (Applied Biosystems) with primers and probe located in the Nucleoprotein (N) gene of hRSV-A and hRSV-B. Primers and probe were designed on the basis of BLAST analysis of all sequences available in Genbank (October 2004). PCR products of antigen positive, RT PCR negative samples were analysed by agarosegel electrophoresis and sequenced with the CEQ Dye terminator cycle sequencing Quick start kit on the CEQ 8000 capillary sequencer (Beckman Coulter).
Results: Agarosegel electrophoresis of the RT PCR products of hRSV antigen positive, RT PCR negative samples showed the presence of a PCR product indicating that the RT amplification was normal. Sequence analysis of the PCR product revealed the presence of 3 extra mismatches in the binding site of the probe which apparently results in insufficient binding of the probe. BLAST-analysis showed that the “new” hRSV variant sequence was not present in Genbank (November 2006). The RT PCR products of 5 hRSV antigen positive, RT PCR negative samples were sequenced and they all showed the same nucleotide sequence. RT PCR with a new probe complementary to the binding site of the “new” hRSV variant gave a strong positive amplification signal. All together, these results indicate that a new hRSV variant arose with mutations in the binding site of the probe in the N-gene. This gene is thought to be one of the more conserved genes of hRSV.
Conclusion: PCR is a powerful technique that detects targets with a high specificity and sensitivity. Exactly this specificity can also be a drawback. All living creatures are prone to evolution and the mutagenesis rate of viruses is high. Therefore, care must be taken with the use of PCR as golden standard. The use of a two-target PCR might decrease the chance of missing mutated viruses.
P1452 Microarray analysis of liver cells containing a full-length hepatitis C virus replicon
A. Ciccaglione, C. Marcantonio, R. Bruni, E. Tritarelli, A. Costantino, P. Tataseo, M. Rapicetta (Rome, Sulmona, IT)
Objectives: Infection with hepatitis C virus (HCV) represents the major cause of liver disease, affecting more than 170 million individuals worldwide. Several data indicate that HCV replication can influence the progression and the severity of liver diseases by a modulation of cellular gene expression. To evaluate HCV-induced modification in the global expression profile, we compare gene expression of the human hepatoma cell line 21–5 carrying the full-length HCV replicon with the parental cell line Huh7 and the cured replicon cells in which the HCV replicon was removed by IFN-á treatment.
Methods: Gene expression was examined using Applied Biosystems Human Genome Survey Arrays containing 31,700 60-mer oligonucleotides probes representing a set of 27,868 individual human genes and more than 1,000 control probes. The study was designed to analyse gene expression for three biological replicates for each cell line and two technical replicates for each biological replicate, for a total of 18 microarrays. The fold change of HCV replicon vs. parental or cured cell lines was analysed by filtering the dataset of normalised signals (using p value <0.01 and <0.05 and a signal-to-noise ratio >3) and performing ANOVA statistical analysis. PANTHER Classification system (Celera Genomics) was used to interpret gene expression data obtained comparing HCV replicon vs parental or cured cells. Alteration of specific genes was confirmed by Real-time PCR.
Results: Data obtained from this study indicated that HCV replication results in a specific and significant alteration of genes involved in cellular process such as endoplasmic reticulum (ER) and oxidative stress, apoptosis, lipid metabolism, immune defence and cell cycle.
Conclusion: The data from the microarray analysis are relevant for the definition of HCV pathogenesis and possibly related molecular markers.
P1453 Enterovirus meningitis in south west Greece: clinical manifestations and CSF laboratory findings
F. Frantzidou, A. Agelatou, K. Dumaidi, J. Kaleyias, A. Papa, A. Antoniadis, A. Spiliopoulou (Thessaloniki, Patras, GR)
Introduction: Enteroviruses are common human viruses associated with various clinical syndromes, from minor febrile illness to severe, potentially fatal conditions such as aseptic meningitis, myocarditis and neonatal enterovirus sepsis. The method of molecular identification not only provides rapid diagnosis of enterovirus infection, but also information about the genetic character of the viruses.
Objective: The aim of the study was the assessment of the enteroviral (EV) RNA detection by reverse transcription polymerase chain reaction (RT-PCR) in the diagnosis of enteroviral meningitis.
Methods: All consecutive cases of aseptic meningitis that were admitted at the Children Hospital of Patras during the study period (December 2004–June 2006) were including in the study. Clinical data were analysed. Rapid detection of EV RNA was directly carried out in the cerebrospinal fluid CSF using RT-PCR targeting to VP1–2A region and nucleotide sequence analysis.
Results: Thirty cases of aseptic meningitis were including during the study period. RT-PCR for EV RNA was positive in 12/30 (40%) of cases. The ratio male: female was 2:1. The main clinical manifestations were: fever (100%), headache (42%) nuchal rigidity (40%), skin rash (30%) and vomiting (30%). The CSF leukocyte count was elevated in all the cases and revealed a lymphocytic-monocytic predominance. The identified virus were found to be HEV-B group: 1 strain echovirus 11 (8.3%), 1 echovirus 14 (8.3%), 2 strains echovirus 9 (16.7%), 3 strains echovirus 6 (25%) and 5 new strains (41.7%) of the HEV-B species. Five cases (41.7%) were taken place at the end of October-November 2005 (HEV-B) and three cases (25%) with echovirus 6 in January.
Conclusions: The predominant strains during the study period were the new strain of the species HEV-B and echovirus 6. The new strain of the species HEV-B had prominent fall seasonality; in addition, there was a perennial distribution of EV meningitis cases. The molecular techniques (RT-PCR/VP1–2A) seem to be accurate and rapid methods for the detection and identification of enterovirus.
Molecular detection of rare pathogens
P1454
First molecular detection of Rickettsia sibirica ssp. sibirica in skin biopsies from patients with north Asian tick typhus
S. Shpynov, P.-E. Fournier, M. Granitov, I. Arsen'eva, N. Rudakov, A. Matushchenko, I. Tarasevich, D. Raoult (Omsk, Barnaul, Moscow, RU; Marseille, FR)
Objectives: The tick-borne North Asian tick typhus (NATT) caused by Rickettsia sibirica ssp. sibirica (R. sibirica), has been described in Russia in 1935. Currently, active foci of this communicable disease are spread in 18 administrative territories of Siberia and Far East, in the Asiatic part of Russia. Approximately one of half registered cases occur in the Altay region. From 1936 to 2005, more than 64,000 NATT cases have been registered in Russia. However, the diagnosis is mostly based on clinical and serological data, and no direct confirmation is available.
Methods: In Spring 2005, 12 inoculation eschar biopsies were taken from patients with clinically typical NATT in the city of Barnaul, Altay. Rickettsiae were detected using partial amplification of the 5'-end of the ompA gene with the 190–70 and 190–701 primers, and amplification of the gltA gene using the CS1d-CS535r and CS409d-RP1258n primer pairs. PCR reactions were carried out in a PTC-200 thermal cycler (MJ Research, Inc, Watertown, MA). Positive PCR products were sequenced using the d-Rhodamine Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Warrington, UK) and an ABI 3100 PRISM automated sequencer (Applied Biosystems). Obtained sequences were compared to the GenBank database.
Results: Ten of the 12 skin biopsies were positive by PCR amplifying both genes. Inferred sequences were identical and were 100% similar to those of R. sibirica ssp. sibirica (gltA, GenBank accession number U59734; ompA, U43807).
All 10 proven NATT cases were characterised by a mild clinical picture different from that of Far-Eastern rickettsiosis caused by Rickettsia heilongjiangensis in Russian Far East.
Conclusions: We demonstrate for the first time by direct identification of the aetiological agent in skin biopsies the aetiological role of R. sibirica ssp. sibirica in NATT in the Altay territory of Russia. Other tick-borne members of the order Rickettsiales, such as R. heilongjiangensis, R. helvetica, R. slovaca, R. aeschlimannii, and Anaplasma phagocytophilum are distributed in natural NATT foci in the Altay territory as well as in territories free from NATT. Our results highlight the need to pursue molecular identification of tick-borne human infections in different regions of Russia.
P1455 Learning from mistakes: Taq-polymerase contaminated with β-lactamase sequences provides false findings of Streptococcus pneumoniae containing TEM
R. Koncan, A. Valverde, M. Morosini, R. Cantón, G. Cornaglia, F. Baquero, R. del Campo (Verona, IT; Madrid, ES)
Background: In 2004 an Asiatic group published in a local journal an article entitled “Study on the molecular epidemiology of β-lactamase TEM gene in isolated Streptococcus pneumoniae“ (PMID: 15769331). In this article the authors referred about S. pneumoniae isolates carrying TEM genes, never described before. Furthermore, the authors described new TEM sequences (TEM-129) which were included in the GenBank data base under the numbers AY452662 and AY392531. The aim of this work was to explore the reasons for such abnormal findings.
Methods: We used total DNA extracted from a S. pneumoniae collection of 24 isolates with MIC values for penicillin of 2–4 μg/mL. The strains were serotyped, PFGE-typed, and the PBP2x, 2b and 1a were characterised. Total DNA from TEM-4 harbouring Klebsiella pneumoniae F40 and S. pneumoniae R6 strains were used as positive and negative controls. To evaluate the presence of TEM genes, PCR experiments with specific primers were performed with two different Taq-polymerase enzymes: FastStart (12032945001 from Roche) and Taq-Core (Qbiogene, MpBIO). PCR products were further sequenced. Hydrolysis assays using nitrocefin as subtract were also performed in crude extracts of S. pneumoniae isolates.
Results: Using the FastStart polymerase, a clear positive amplification corresponded to the bla-TEM size were observed in all strains tested, including the negative control. With this enzyme, amplifications occurred also in absence of pneumococcal DNA. Four different amplicons were sequenced, which corresponded to the ESBL TEM-116 previously described. When the other Taq-polymerase was used, positive amplifications were not detected in any of the isolates, except the positive control. Nitrocefin test was consistently negative in all isolates.
Conclusion: Caution should be required to accept data about the presence of TEM-β-lactamases based only on PCR-based results in the absence of controls that might reveal TEM sequences contamination of amplification reagents.
P1456 Periodontal pathogenic bacteria in oral microflora of halitosis patients in Latvia
D. Rostoka, J. Kroica, A. Reinis, V. Kuznecova (Riga, LV)
Objectives: Halitosis, commonly referred to as bad breath, may be due to physiological and/or pathological causes of oral, gastroinstenal, nasopharingeal or other origin. Oral bacteria hydrolyse proteins and further degrade the amino acids, which leads to halitosis. Oral anaerobes, such as Porphyromonas, Prevotella, Treponema spp., produce volatile sulphur compounds (VSC) from amino acids.
Methods: The recent investigation began in 1997 and included 260 untreated halitosis patients (43.1 year, 167 females, 93 males). Due to the fact that a tactful approach to a patient is very important detailed patient's questionaire was prepared. This questionnaire was approved by the Committee of Ethics of the Ministry of Welfare of Latvia and covered questions about the use of antibiotics and other medicine, especially which affect the quality and quantity of saliva, as well as questions concerning smoking, the use of alcohol, etc. The oral odour or bad breath was confirmed by the measurements made by the portable sulphide monitor or halimeter (Interscan Corporation, Model RH-17E). The halimeter quantifies breath measurements in parts-per-billion (ppb) of VSC. Periodontal pocket and dorsal part of tongue microflora was analysed by quantitative PCR (micro-IDent®, Hain Lifescience) for amounts of periodontal pathogenic bacteria: A. actinomycetemcomitans, P. gingivalis, T. forsyth, T. denticola, and P. intermedia. The patients were examined for individual genetic periodontal risk. Polymorphisms of the IL-1A – 889 and IL-1B + 3953 gene cluster was checked (GenoType®PST, Hain Lifescience). ANOVA, t-test, and chi-square were used to detect statistically significant differences.
Results: Halimeter measurements of 260 patients showed increased VSC (210–340 ppb compared with control group 40–70 ppb).
GenoType®PST results showed that individual genetic periodontal risk of chronic periodontal diseases was positive in 66.9% out of 260.
Conclusions: Halitosis is mainly an oral problem (94.7%) in Latvia. Halitosis is a disease which can be diagnosed by a proper examination. Halitosis can be diagnosed and controlled using different methods. Since protein degradation is present in oral cavity and increased amount of VSC, there are changes in oral microflora with big prevalence of oral anaerobes. That can explain often incidence of periodontal diseases in Latvia. Periodontal disease diagnosis can be based on presence of periodontal pathogens.
P1457 Streptobacillus moniliformis endocarditis diagnosed by 16S rRNA gene PCR and direct sequencing applied to a resected heart valve
D.H. Forster, A. Becker, P. Stahlhut, M. Elgas, E. Kniehl (Karlsruhe, DE)
Objective: S. moniliformis is a facultatively anaerobic, fastidious Gram-negative rod that may develop into long filaments containing granules, bulbs and bands; coccal forms may also be observed. It occurs naturally in the nasopharynx of rats and other rodents. Human infections result either from bites of rodents (rat bite fever) or from consumption of contaminated food or water. The most common manifestations are fever, arthralgias and rash. Endocarditis is a rare complication of S. moniliformis infection, underlying cardiac valve abnormalities have been reported in most cases. Though S. moniliformis is a fastidious organism, diagnosis is usually made by culture. We describe a case of S. moniliformis endocarditis leading to valve replacement. Diagnosis was made by 16S rRNA gene PCR and direct sequencing applied to the resected heart valve and a typical Gram stain finding of the resected valve.
Case report: A 74-year-old female patient was admitted for mitral valve replacement. She presented with a history of fever since 3 weeks and reduced general condition due to mitral valve insufficiency grade IV with endocarditis. The cause of endocarditis was unknown at the time of admission.
Gram stain of the resected heart valve revealed pleomorphic, filamentous Gram-negative rods, culture remained sterile. Partial 16 S rRNA gene PCR was performed on the resected valve, the received amplicon was sequenced. Comparison of the sequence (GenBank accession number: DQ914526) with the GenBank database showed a 100% sequence homology with two published S. moniliformis sequences. Together with the typical Gram stain finding, this can be considered as unambiguous identification of S. moniliformis as causative agent of endocarditis. Empirical broad antibiotic therapy was then changed to penicillin G 4×5 Million U for 4 weeks. The postoperative course was uneventful. Rodent contact or a rodent bite was denied by the patient, the modus of aquiring the agent is therefore unclear.
Conclusion: Identifying the causative agent of endocarditis cases by 16S rRNA gene PCR and direct sequencing applied on resected heart valves is in the meantime an established method in culture-negative endocarditis. The sequencing result in this unusal case is supported by the Gram stain finding.
To the best of our knowledge, this is the first report of S. moniliformis endocarditis in Germany and the first worldwide diagnosed by the means of molecular biology.
P1458 Molecular and serologic evidence of Anaplasma phagocytophilum and Borrelia burgdorferi sensu lato in dogs in the Czech Republic
K. Kybicova, Z. Kurzova, L. Uherkova, P. Schanilec, S. Spejchalova, Z. Svobodova, D. Hulinska (Prague, Brno, CZ)
Objectives: Dogs have been considered important domestic host of Anaplasma phagocytophilum and Borrelia burgdorferi sensu lato. The aim of this study was to assess the prevalence of these microorganisms among dogs from the Czech Republic.
Methods: A total of 192 samples of dog blood and 65 ticks (43 female, 12 male) from 35 dogs collected in Moravia and North Bohemia over the year 2006 were investigated. Blood samples were obtained from (i) health dogs naturally exposed to ticks (38.5%), (ii) imunosuppressed dogs (30.7%), (iii) dogs with oncological disease (5.7%) or (iv) dogs with neurological disorder or arthritis (25%). DNA was isolated from blood plasma and buffy coat or from whole ticks. All DNA samples were analysed by PCR with the LD primer set (Borrelia) and the Ehr 521–790 primer set (Anaplasma) targeting 16S rDNA. The samples PCR positive for Borrelia were retested by real-time PCR with primer sets also targeting 16S rDNA (VS, BG, BB) and with a recA primer set to confirm the positivity and to identify the Borrelia species. The samples PCR positive for Anaplasma were retested with G3-G4 primers. Serological study was carried out using ELISA detection of specific IgG and IgM antibodies against Borrelia afzelii. Antibodies against A. phagocytophilum were examined by IFA test with the fluorescein isothiocyanate-conjugated goat anti-dog IgG diluted at 1:320.
Results: One dog (0.5%) and nine ticks (13.8%) were positive for B. burgdorferi sensu lato while four dogs (2.1%) and four ticks (6.2%) were positive for A. phagocytophilum. ELISA demonstrated that 9.7% and 5.6% of samples contained anti-Borrelia IgG and IgM antibodies, respectively. If borderline values were considered, the positivity would be 11.1% and 13.2% for the IgG and IgM antibodies, respectively. The A. phagocytophilum-specific IgG were found in 11.4% of dogs.
Conclusions: The findings of the present study partly confirm results of other European studies. The difference between the PCR results for ticks and dogs may come from the fact that most ticks are from healthy hunting dogs.
P1459 Application of hybridisation probe-based real-time PCR assay to monitor the Brucella DNA load in patients with brucellosis throughout different stages of the disease
G. Vrioni, E. Priavali, G. Pappas, C. Gartzonika, S. Levidiotou (Ioannina, GR)
Different methods have been described for diagnosis of human brucellosis. Among them, peripheral blood real-time PCR assay (RTPCR) seems to be a very promising technique not only for the initial diagnosis of the disease, but also for the post-treatment follow-up by the accurate quantification of Brucella DNA. A quantitative RTPCR assay using the LightCycler instrument (Roche Diagnostics, Mannheim, Germany) was employed for the monitoring of bacterial DNA load in patients with brucellosis throughout different stages of the disease. A total of 130 peripheral blood specimens were examined from 39 patients with acute brucellosis as determined by blood culture, serologic tests, and the patients' clinical picture. A minimum of 3 specimens (one at diagnosis, one at the end of treatment and at least one during the post-therapy follow-up period) were obtained per patient. RTPCR assay was based on direct amplification of a 207-bp DNA sequence of a gene that codes for the synthesis of an immunogenetic 31-kDa protein specific for Brucella genus (BCSP31). The amplification product was detected by using as fluorescence technique hybridisation probes labelled with LightCycler Red 640 (detected in channel F2). Primers and probes were designed by TIB MOLBIOL (Berlin, Germany) and fluorescence curves were analysed with LightCycler software v. 3.5. Following amplification, melting curve analysis was performed to verify the specificity of PCR products. A standard curve, comprising 10-fold dilutions of Brucella BCSP31 DNA from 101 to 107 target equivalents, allowed quantification of unknown samples.
RTPCR assay was 100% sensitive and specific for B. melitensis at the time of diagnosis. Despite the decrease of bacterial DNA load at the completion of treatment, the RTPCR results for 87% of patients remained positive. Six months after therapy, 77% of patients presented with residual bacterial DNA without experiencing relapse. From 21 patients who were monitored for >1 year after therapy, 13 continued to have positive RTPCR results, albeit asymptomatic.
In conclusion, B. melitensis DNA may persist on finalising and after therapy, despite of the appropriate treatment and apparent recovery from infection. The significance of these findings remains to be elucidated in reference with their pathogenic and therapeutic implications.
P1460 Proof of specific DNA using nested-PCR in patiens with different clinical form of Lyme borreliosis
D. Picha, L. Moravcova, D. Holeckova, K. Sedivy, E. Zdarsky, V. Maresova (Prague, CZ)
Objectives: This study was aimed to the direct proof of borrelial DNA with polymerase chain reaction (PCR) in patients with lyme borreliosis (LB) and to estimate the specifity of individual primers in the diagnostic of its clinical forms.
Methods: Into the prospective study 71 patients with LB were enrolled (35 patients with neuroborreliosis, 14 with joint involvement and 22 with skin form). The criteria for inclusion were recent clinical symptoms of LB and the presence of specific antibodies in the blood, CSF, joint fluid (confirmed by Western blotting) or proof of DNA in body fluids. Before the treatment the presence of borrelial DNA was tested in plasma, CSF, joint fluid and urine, after the treatment in plasma, urine and joint fluid. A system with five newly derived primers has been used, with three primers specific for chromozomal genes coding 16SrDNA, flagellin and p66 and two for plasmid genes which are coding the outer membrane's protein OspA, OspC. The presence of borrelial DNA was tested by nested-PCR in parallel with all the primers.
Results: From the total number of 71 examined patients the borrelial DNA was proved in 42 patients (59.2%) before the treatment; 23 positive patients (65.7%) were found in group of neuroborreliosis, 10 positives (45.5%) showed signs of skin involvement and 9 patients (64.3%) were positive in arthritis. After the treatment the borrelial DNA was proved in half of the patients with neuroborreliosis (13×) and in the skin form patients (4×). In patients with the joint form the proof was the same as before the treatment. From the 105 positive amplifications the most frequently positive primer was 16SrDNA 54× (51.4%). Less sensitive were primers OspA (20×;19%), OspC (14×;13.3%) and flagellin (14×;13.3%). In 14 patients (31.1%) with skin and joint form of LB only borrelial DNA without specific antibodies was proved.
Conclusion: This study has shown that in the period before treatment the sensitivity of PCR was 59.2% using 5 primers and including all the clinical forms of borreliosis. The most positive result has been found by using the primer 16SrDNA (51.4%). The same reactivity was being shown in plasmid sequences coding genes OspA and OspC and chromosomal sequence for flagellin. Examination of the borrelial DNA in urine displayed the same sensitivity as in CSF.
The study was supported by grants of Ministry of Health and Education (IGA-8293, MSM 0021620812)
P1461 Performance of a new molecular assay for detection and differentiation of Bordetella spp. DNA
C. Koidl, M. Bozic, H.H. Kessler, E. Marth (Graz, AT)
Objective: To investigate the performance of the new molecular artusTM Bordetella LC PCR Kit (QIAGEN Hamburg GmbH, Germany) assay that provides detection and differentiation of Bordetella spp. DNA in a routine diagnostic laboratory.
Methods: The assay was performed according to the manufacturer's package insert. A total of 85 nasopharyngeal swabs obtained from patients with a clinical presentation compatible to B. pertussis infection were tested with the new assay in an International Standards Organization (ISO9001, 2000)-certified routine clinical laboratory.
Results: Sixteen of 85 samples were found positive for Bordetella DNA. Differentiation by melting curve analysis yielded B. pertussis in 7 samples, B. parapertussis in 1 sample, and B. bronchiseptica in another one. In the remaining 7 samples, Bordetella DNA was detected by analysis of fluorescence curves of IS481 and IS1001 amplification products only. The heterologous internal control was consistently detected. In 7 specimens inhibition of the assay was found.
Conclusion: In conclusion, the new molecular assay includes all features required for molecular detection and differentiation of Bordetella spp. DNA. It proved to be suitable for the routine diagnostic laboratory allowing a rapid and safe diagnosis of pertussis.
P1462 Sensitivity of Legionella pneumophila DNA detection in serum samples in relation to disease severity
B.M.W. Diederen, J.P. Bruin, J.W. den Boer, M.F. Peeters, E.P. F. Yzerman (Tilburg, Haarlem, NL)
Objectives: Legionella pneumonia can be difficult to diagnose. Existing laboratory tests all have shortcomings, especially the ability to diagnose all Legionnella spp. in a specimen that is readily obtainable. The aim of this study was to assess the sensitivity of PCR as a rapid diagnostic method and to investigate a possible relation between test sensitivity and the severity of disease.
Methods: In this study 68 outbreak-related patients (Bovenkarspel, The Netherlands) with a confirmed Legionnaires' disease (LD) according to the European Working Group for Legionella Infections (EWGLI) criteria were investigated. To investigate the relation between test sensitivity and severity of disease, the patients were divided into two clinical categories for pneumonia. Patients were classified as category 1 (mild pneumonia and moderately severe) and category 2 (severe pneumonia). LD was defined as severe when two or more of the following conditions were present: (1) respiratory rate >30/min, (2) chest radiograph showing bilateral involvement or involvement of multiple lobes, (3) shock, (4) PaO2 < 60 mmHg or arterial oxygen saturation <92%. A real-time assay targeted at specific regions within the 5S rRNA gene was used.
Results: Samples included 136 serum samples from 68 patients with proven LD. In one sample inhibition of PCR occurred; this patient was excluded from the analysis. Among the patients with LD 39% (26/67) tested positive in PCR in the first available serum sample, and this number increased to 54% (36/67) if all serum samples were included in the calculations. The detection rate for all non-inhibited samples was 35% (47/135). For 58 patients data on disease severity was available. In patients with severe pneumonia, 49% (19/39) tested positive in the first available serum sample, increasing to 67% (26/39) if all serum samples were included in the calculations. In patients with mild and moderately severe pneumonia, 37% (7/19) tested positive in the first available serum sample, increasing to 53% (10/19) if all serum samples were included.
Conclusion: The sensitivity of PCR on serum samples found in our study was relatively low compared to previous studies. Although we observed a higher sensitivity in patients with more severe disease (49% vs 37%, p = 0.4), these differences did not reach statistical significance.
P1463 A real-time PCR protocol to detect Neisseria meningitidis in formalin-fixed paraffin-embedded tissues
A. Fernandez-Rodriguez, B. Alcala, G. Vallejo, J. Gómez, R. Alvarez-Lafuente (Madrid, ES)
Objectives: Invasive meningococcal infection is a well-established cause of sudden death. However, symptoms are often vague and postmortem findings may be either indistinguishable from any other infection or overlooked. For these reasons, the differential diagnosis in sudden deaths should include Neisseria meningitidis detection. Since cultures may not be available in fulminant fatal cases, the diagnosis of meningococcus has to rely on PCR assays. CSF and serum are not always taken as postmortem samples, and sometimes the only specimens collected at autopsy are tissues. The aim of this study was to evaluate the usefulness of a real-time PCR protocol to detect meningococcus in formalin-fixed paraffin-embedded tissues from patients with meningococcal disease.
Methods: Our protocol included: (i) detection of meningococcal DNA, targeting the ctrA gene, (ii) genogrouping to identify serogroups B and C (siaD gene) in a multiplex format, and (iii) a rapid confirmation of PCR products with a microchip CE device. We analysed 15 cases of sudden death due to meningococcal disease where formalin-fixed paraffin-embedded tissues were available. In all those cases meningococcus had been detected by real-time PCR in fresh samples. Formalin-fixed paraffin-embedded tissues from 5 cases with other infections and from 5 healthy individuals were considered as negative controls. Optimisation of real-time PCR conditions was performed, as well as validation studies with other microorganisms.
Results: In all positive cases Meningococcus was detected in at least one tissue sample. Meningococcus could be detected in skin, spleen, heart, liver, lung, kidney, bone marrow, brain and adrenal. Positive results were obtained in ten-year old paraffin-embedded tissues. No false-positive results were detected in negative controls.
Conclusion: Meningococcus can be detected in formalin-fixed paraffin-embedded tissues from fulminant fatal cases when no other samples are available. However, a thorough protocol is needed to deal with DNA degradation and PCR inhibition.
Bone, joint, skin and soft tissue infections
P1464 Trends in the diagnosis of osteomyelitis at the National Pathology Reference Center of Venezuela, 1996–2006
S. Dickson-Gonzalez, J. Mota-Gamboa, A. Rodriguez-Morales (Caracas, Trujillo, VE)
Objectives: To describe and analyse the patterns and trends in the diagnosis of osteomyelitis at the Instituto Anatomopatológico José A. O'Daly (the National Pathology Reference Center of Venezuela), Caracas, Venezuela, 1996–2006.
Methods: A total of 613 patients from our pathological database were included in this report, since January 1996 to May 2006 (10 years). The samples processing procedure was through paraffin blocks, with tissular sections (3 um), stained with haematoxylin-eosyn, Ziehl-Nielsen, Gram and Grocott, then the microscopic evaluation is made. Epidemiological, clinical and pathological features are also described.
Results: During the study period there was a clear increase in the diagnosis of osteomyelitis, from 25 cases in 1996 to 92 cases in 2005 (r2 = 0.85, p < 0.05), with a mean of 59±21 cases per year. According to the trends model (y = 6.497x + 23.267), we could expect for year 2006 a record of 95 cases, during the first 5 months of this year we have diagnosed 23 cases. The sex distribution of these cases is predominantly male (68.1%) over female cases (31.9%) (p < 0.05). The histopathological features will furtherly discussed, as well the aetiological agents.
Conclusion: As we have observed, the incidence of osteomyelitis is increasing; although the sex and age distribution not. Different reasons could be attributable to this increase, the number of orthopaedic surgeries, vehicle accidents and new type of injuries which are more often seen.
P1465 Long-term clinical and radiological (MRI) outcome of abscess-associated spontaneous pyogenic vertebral osteomyelitis under conservative management
G. Euba, J.A. Narvaez, J.M. Nolla, O. Murillo, J. Narvaez, C. Gomez, J. Ariza (Barcelona, ES)
Objectives: Spontaneous Pyogenic Vertebral Osteomyelitis (SPVO) is classically considered a medical management disease. Surgical approach is widely accepted for spinal cord compression or vertebral instability, but controversial for abscess-associated SPVO. Magnetic Resonance Imaging (MRI) is the test of choice at diagnosis, most patients (pts) showing Soft Tissue Involvement (STI); it seems also helpful for follow-up, but MRI and clinical outcome interrelation is often unclear. The aim of this study is to evaluate clinical and MRI outcome of SPVO with STI under conservative treatment.
Methods: Prospective study (PS, period 2000–05) and retrospective review (RR, period 1995–99) including pts with SPVO in a 1,000 bed tertiary hospital, treatment according to medical protocol and follow-up evaluation considering clinical, biological and MRI findings at diagnosis, Early Response (ER, end of planned antibiotic therapy) and Late Response (LR, ≥ 6 months from end of therapy). Inclusion criteria: MRI at diagnosis showing STI; follow-up MRI; initial non-surgical approach. MRI images were reviewed by an expert radiologist; STI was classified as inflammatory reaction (IR) or abscess, and abscesses were measured.
Results: Twenty-seven pts (19 PS, 8 RR) were included, 20 men (74%), mean age 65 ± 14. All had pain, 17 (63%) fever and 6 (22%) mild neurological impairment. Main aetiology was Staphylococcus spp. (11, 41%); no microorganism was identified in 4 (22%). Bacteraemia was documented in 21 (81%) and epidural/paraspinal abscess in 18 (67%). Antibiotics were given for median of 9 weeks, orally for 6 weeks. ER: only 3 pts were considered as therapeutic failure (1 underwent laminectomy for paraparesis development; 2 had therapy extended for persistent clinical findings) with good final outcome; clinical and biological improvement was seen in the remainder; MRI showed persistent STI, which diminished in all but 2 pts, whereas bone and disc findings hardly improved or worsened. LR: median follow-up was 29 months, no relapse or related death occurred; all pts were considered clinical and biologically cured, with mild sequels in 8 (pain/dysesthesia); MRI still showed bone/disc abnormalities, but residual IR was infrequent (6 pts).
Conclusion: Most pts with abscess-associated SPVO cure with a medical approach and 8–10 week antibiotic therapy, STI reduction being found in MRI at ER point. Bone/disc MRI worsening alone during follow-up should not suggest therapeutic failure.
P1466 Comamonas testosteroni spondylodiscitis
G. Carolo, M. Ganau, E. De Micheli, M. Gerosa, M. Solbiati (Verona, IT)
Introduction: We describe the first case in literature of spondylodiscitis caused by Comamonas testosteroni. It is a Gram-negative rod, present in soil, water and animal; rarely involved in human pathology, to our knowledge until now were described 23 cases in the world medical literature.
Case: A 63 year-old male, following lumbar discectomy for a L2-L3 disc herniation (prophylaxis with Cefazoline 1 g 1 hour before surgery), presented with a persistent five-week untreatable lumbar pain, accentuated by walking. On admission, he was apiretic, neurological examination was normal. Plain Rx rays of lumbar spine showed reduction of L2 body; MRI of the spine confirmed crush of the L2 body with vertical line of fracture from the superior to the inferior edge plate of the vertebra accompanied by an uptake of gadolinium in the epidural and paraverterbal areas of the L2-L3 interbody space. Full blood, and urinary tests, showed a normocromic normocitic anaemia, associated to sideropenia, elevated ESR (72 s) and CPR (1.82 mg/dl). Tumour markers were negative, as well as Total Body CT. Blood culture was positive for Comamonas testosteroni, therefore he started antibiotic therapy with teicoplanine (600 mg e.v./day) and ciprofloxacin (400 mg 2times/day) (therapy supported by the evidence at antibiogram of sensibility to aforementioned drugs). He performed a CT guided biopsy of L3: anatomopathologic, immunohystochemical and cytological examination of the biopsy were all negative; from cultures grew Comamonas testosteroni. Teicoplanin was stopped and the therapy prosecuted with ciprofloxacin and cotrimoxazole After 3 months of sustained therapy a control MRI of the lumbar spine showed initial resolution of the L2-L3 discitis and Xrays confirmed the stability of the L2 body. The patient kept on observing rest, and drug protocol until the 9th month, serial blood test confirmed CPR and ESR within normal ranges, MRI confirmed the completely resolution of the inflammation and X-rays showed good healing of the L2 vertebral body fracture.
Conclusion: Discitis is a rare complication of disc operation (0.2–0.8%); direct contamination during surgical time is far more frequent than haematogenous contamination. In our patient the pathogenic role of C. testosteroni was suggested by two facts: first the organism was isolated from both the peripheral blood and CVC cultures; second the evolution was favourable when the patient was treated with adapted antimicrobial agents.
P1467 Mixed infection of the lower limb caused by rare bacterial and fungal pathogens in a patient with multiple traumatic injuries
G. Corti, N. Mondanelli, M. Losco, L. Bartolini, A. Fontanelli, F. Paradisi (Florence, IT)
Objectives: Enterobacter amnigenus and Leclercia adecarboxylata are Gram-negative aerobic bacilli of the family Enterobacteriaceae that have been isolated from water and, rarely, from various clinical specimens. Absidia is a filamentous fungus of the class Zygomycetes that is ubiquitous in nature and can cause infection, primarily in compromised hosts. Here we describe a case of mixed infection of the left lower limb caused by E. amnigenus and L. adecarboxylata and subsequent superinfection by Absidia in a patient with severe traumatic injury.
Methods: Multiple surgical samples of the wound were collected for microbiological cultures. The identification of bacteria was carried out by standard techniques (isolation on blood agar plates and subsequent biochemical identification). The susceptibility testing was performed by an automated method (VITEK 2, BioMeriéux, France). The identification of the fungus was carried out after direct microscopic examination and inoculation onto Sabouraud dextrose agar for culture.
Case report: A 37-year-old male with multiple traumatic injuries (open fractures of the left distal femur and tibia with bone loss, multiple fractures of the left foot with soft-tissue involvement) following motorcycle accident was admitted to our emergency department after first aid on the road. He was treated with irrigation and debridément, open reduction and internal fixation of the tibia and foot fractures, and external fixation of the femur fracture. Intravenous antibiotics (tobramycin, teicoplanin, metronidazole) were empirically initiated on the basis of a hospital protocol. After 14 days, a below-knee amputation was performed as there was no possibility for limb salvage. Following isolation of two isolates of E. amnigenus and L. adecarboxylata, piperacillin/tazobactam was substituted for tobramycin. After seven days, a novel amputation on a more proximal level was needed for control of stump infection. Following isolation of a strain of the genus Absidia, a 15-day course of liposomal amphotericin B was started, and control of stump infection was finally achieved. At five months from final surgery, no signs of infection are present.
Conclusion: E. amnigenus and L. adecarboxylata are multi-susceptible environmental Enterobacteriaceae and Absidia an ubiquitous zygomycete, and all of them are rare agents of infection. Nevertheless, they can cause severe life-threatening infections that require complex and sometimes devastating treatments.
P1468 Predictive risk factors for amputation in diabetic fetid foot infections
J. Barberan, B. Sanchez, M.A. Menendez, J.R. Toral, A. Fe, I. Fraile, R. Alguacil, D. Martinez, J. Prieto (Madrid, ES)
Objective: Lower extremity amputation (LEA) is one of the most disabling complications of diabetes. We report a serie of diabetic foot infections treated with antibiotic therapy and surgery, to examine the association of usual tests in its management with incidence of amputation.
Methods: All enrolled patients had a previously history of diabetes and infection defined by clinical symptoms and signs. Infections were categorised by size and depth. Usual test in management were determined at baseline in all patients.
Results: Seventy eight patients (68.8 ± 2.3 years) were included (55 had vascular insufficiency, 7 neuropathy, and 15 both). Amputation was performed in 26 (33.3%) cases, and it was associated significantly more frequently with previous ulcers (p = 0.001), vascular insufficiency (P = 0.011), size depth of tissue involeved (P = 0.001), Wagner's classification (P = 0.00001), purulent secretions (P = 0.048), and fetid smell (p = 0.033). In a multivariant analysis doppler arterial pressure, Wagner's classification, and ESR (erythrocyte sedimentation rate) were found the best variables to indicate the risk of LEA.
Conclusions: With this logistic model and once known three usual tests, easy to obtain (ESR, doppler arterial and Wagner's classification), we could predict correctly, in about 90% of cases, the survival of lower limb afected. These results suggest that single usual tests the low cost can be very useful to find out the risk of amputation in the diabetic fetid foot.
P1469 Liver abscesses: changes in aetiology associated with inmigration
A. Salinas, J. Troya, M. De Górgolas, I. Gadea, M. Fernández-Guerrero (Madrid, ES)
Background: Liver abscesses are relatively common intrabdominal infections which aetiology, diagnosis and treatment are well-known. However, changes on the microbiology and prognosis over the years have not been assessed and specifically, the role of immigration of people from South America on microbial aetiology has not been assessed.
Methods: Retrospective study of 60 patients seen by the authors from 1985–2005. For comparison, patients were classified into 2 groups: those studied from 1985–1995 and those seen from 1996–2005. Demographics, clinical findings, microbiology, treatment and prognosis were compared.
Results: Patients were 40 men and 20 women (male/female ratio 2:1) with a mean age of 53.7 years; 85% were spaniards and 15% from other countries, mainly from Central and South America. The most frequent symptons were fever (80%) followed by abdominal pain (71.6%) in both periods. Diagnosis was made by sonography in 83.3% of cases. A single abscess was found in 70%. The right lobe of the liver was the most frequent involved site (55%). The mean diameter of abscess was 7.83 cm. A bacterial aetiology was found in 61.6% of cases and the microorganisms most frequently found were Escherichia coli (9 cases) and Streptococcus anginosus (9 casos). Amoebic abscess were diagnosed in 5 cases. Diagnosis was reached by serology in most of these cases (91.6%). Empirically patients were treated with a combination of β-lactam and metronidazol. A surgical or percutaneous drainage was performed in 76.6% of cases. Complete cure was attained in 93.3% of cases. However, 4 patients died in spite of therapy (2 sepsis, 1 gall bladder adenocarcinoma and 1 perforated diverculitis with sepsis). The clinical presentation was almost identical in the two periods. The time from presentation to diagnosis was 3.5 weeks (minimum 3 days; maximum 8 weeks). Eight out of 9 cases seen in foreigners occurred from 1996–2005 and all amoebic hepatic abscesses occurred in this period. The most common drainage procedure in the first period was open laparotomy (50%), whereas percutanous ultrasound-guided drainage was most frecuent in second period (94%).
Conclusions: Advances in knowledge of liver abscess has led a more rapid diagnosis of infection and drainage by means of percutaneous techniques with less morbidity and mortality. Immigration may introduce changes in the microbial aetiology of liver abscess in Europe.
P1470 Changing clinical features of severe odontogenic infections
L. Seppänen, A. Lauhio, C. Lindqvist, R. Rautemaa-Richardson (Helsinki, FI)
Objectives: The number of patients requiring hospital care due to odontogenic infection has increased in the Helsinki and Uusimaa Hospital District, Finland. Our aim was to evaluate the clinical features of the patients hospitalised due to an odontogenic infection.
Methods: In this retrospective study two patient cohorts (years 1994–1996 and 2004) admitted to the Helsinki University Central Hospital due to an odontogenic infection were analysed. This patient series covers all cases of severe odontogenic infections in the region of Helsinki University Central Hospital (1.4 million inhabitants in year 2004). 99 (1994–1996) and 102 (2004) consecutive patients were included. Age, gender, fever at admission, WBC counts, CRP levels, focus, fascial spaces involved, need for re-operation and length of hospital stay were reviewed.
Results: The incidence of these severe infections increased from 5.3 to 7.2 per 100,000 inhabitants since 1994 to 2004. The two patient cohorts (1994–96 and 2004) did not differ in age (mean 40 and 43 years, respectively), gender (60.6% and 63.7% males, respectively), WBC count on admission (mean 12.4 and 12.5×103/μL, respectively), focus (mandibular molar 76.8% and 81.4%, respectively) or fascial spaces (submandibular space involvement 46.5% and 49.0%, respectively). However, the proportion of patients with WBC greater than 11 × 103/μL (49.5% and 53.9%, respectively) and CRP levels greater than 90 mg/L (36.3% and 54.9%, respectively), CRP level on admission (mean 89.2 and 112.0 mg/L, respectively) and maximal CRP levels (mean 93.4 and 153.7 mg/L, respectively), fever on admission (37.4% and 43.1%, respectively), need for re-operation (8% and 16.7%, respectively) and intensive care (18.2% and 31.4%, respectively) were markedly higher in 2004. In addition, the proportion of patients with mental disorders, cardiovascular diseases and diabetes had also increased.
Conclusion: This study shows that in the Hospital District of Helsinki and Uusimaa odontogenic infections require hospital care more often although the admission criteria have not changed. The increase in the need for intensive care implicates that the infections have become potentially more severe. The incidence of underlying systemic diseases has also increased.
P1471 Pyomyositis. A current review
F.J. Fernandez-Fernandez, S. Pérez-Fernández, M. Rubianes-González, L. González-Vázquez, R. Puerta-Louro, J. De la Fuente-Aguado (Vigo, ES)
Objectives: To describe the epidemiological, clinical, microbiological and therapeutical features of pyomyositis.
Methods: Retrospective descriptive study of the medical records of all patients diagnosed of pyomyositis in POVISA and CHUVI between 1996 and 2005. Patients with gas gangrene and necrotising fasciitis were excluded, as well as psoas abscesses due to its different characteristics.
Results: Forty-one patients were included, 27 males and 14 women, with a mean age of 50 years. An underlying disease was present in 63% of the patients: diabetes mellitus in 12 patients, human immunodeficiency virus (HIV) infection and injection drug use in 6, chronic kidney disease in 5, alcoholism in 4 and cancer in 3. The most common affected sites were thigh (49%) and gluteal regions (22%). Primary pyomyositis was diagnosed in 36 patients (88%). Evidence of endocardial involvement was not demonstrated in any patient in whom echocardiography was performed. The most frequent clinical manifestations at diagnosis were muscle pain (90%), swelling (66%) and fever (66%). Clinical symptoms were present between 2 and 60 days before diagnosis. In laboratory studies leukocytosis was found in 58%, anaemia in 46% and elevated erythrocyte sedimentation rate in 96% of patients in which they were analysed.
Diagnosis was performed by ultrasound (with one false negative result), computed tomography or magnetic resonance imaging. Aetiologic agent was known in 31 patients (76%). Staphylococcus aureus was found in 23 patients, streptococci in 6 and Salmonella in 3. Four patients have a polymicrobial infection. Culture of the abscess was positive in 78%, and blood cultures in 38%. Drainage of the muscle abscess was performed in 31 patients, open surgical in 14 patients, percutaneous in 13, and both types in 4 patients, all of them combined with antimicrobial therapy. Mortality rate was 7%. Advanced age was the only poor prognostic factor.
Conclusion: Pyomyositis usually has a primary origin and it is caused by S. aureus. HIV infection and injection drug use are important predisposing factors. In some patients there are neither leukocytosis nor fever, so diagnosis can delay. Patients with pyomyositis and bacteraemia seem not to require echocardiography to rule out infective endocarditis. We report a good outcome only with antibiotics in 10 patients, though more studies should be performed in this direction because of the small number of patients. Advanced age was related with higher mortality.
Endocarditis
P1472 Enterococcal endocarditis: a comparison of patients with native and prosthetic valve endocarditis and analysis of risk factors of mortality
M. Fernández-Guerrero, A. Goyenechea, A. Salinas, R. Fernández Roblas, J. Fraile, M. De Górgolas (Madrid, ES)
Background: Enterococci are the third leading cause of infectious endocarditis. Mortality of enterococcal endocarditis although relatively low, has not changed in the last decades. We investigated the risk factors of mortality in a single institution and whether infections occurring on prosthetic valves are associated with a poorer prognosis.
Methods: Retrospective review of 44 consecutive episodes of enterococcal endocarditis with strict case definitions according to the modified Duke's criteria from a prospective observational cohort of cases of infectious endocarditis. The main outcome measure was in-hospital mortality. Stepwise logistic regression analysis was applied to identify risk factors for mortality.
Results: 27 patients (61.3%) had NVE and 17 (38.6%) PVE. Predisposing heart conditions were observed in 77.7% of patients with NVE. A portal of entry was suspected or determined in 38.2% and the genitourinary tract was the most common source of the infection (29.7%). Comorbidities were present in 52.2% of cases. Twelve episodes (25.5%) were acquired during hospitalisation (7 NVE and 5 PVE). Only 3 isolates of Enterococcus faecalis were highly-resistant to aminoglycosides. Eighteen patients (40.9%) needed valve replacement due to cardiac failure or relapse. The comparison between cases of NVE and PVE did not show differences regarding symptoms, complications, or the need of surgical treatment (12/44% vs 6/35%; ns). The mean time to diagnosis was shorter in patients with PVE than NVE (30±18 days vs 9±6 days) and murmurs were more commonly found in patients with NVE (25/96% vs 10/59%; p 0.02). Eight out of 47 (17%) episodes were fatal. The mortality rate of patients with PVE and NVE were not significantly different (2/11.7% vs 6/22% respectively; ns). Age over seventy (p 0.05), heart failure (OR 1.61;CI:1.15–2.25; p 0.001) and nosocomial acquisition (OR 8.05; 95% CI:1.50–43.2; p 0.01) were associated with mortality. In the multivariate analysis, only nosocomial acquisition increased the risk of mortality.
Conclusions: Enterococcal endocarditis occurring on prosthetic valves are not associated with an increased need of valve replacement nor mortality. In this series, nosocomial acquisition of endocarditis was the most important factor determining outcome.
P1473 Current trends of infective endocarditis caused by Staphylococcus aureus and coagulase negative staphylococci in Greece: results from the Hellenic Endocarditis Study Group, 2000–2004
E. Giannitsioti, A. Antoniadou, S. Tsiodras, I. Skiadas, K. Kanavos, H. Triantafyllidi, H. Giamarellou on behalf of the Hellenic Endocarditis Study Group
Objectives: Infective endocarditis (IE) of staphylococcal aetiology emerges worldwide. Current trends of Staphylococcus aureus IE and coagulase negative staphylococci (CNS) IE were investigated through a large epidemiological survey in Greece.
Patients and Methods: Cases of IE fulfilling the Duke criteria were prospectively recorded in 20 general hospitals at the metropolitan area of Athens from 2000–2004. Both S. aureus and CNS cases of IE were assessed regarding epidemiological issues, and outcome. Chi-square test was used for statistical analysis of categorical variables.
Results: From 195 eligible IE cases, 34 (17.4%) and 19 (9.7%) were caused by S. aureus and CNS respectively. S. aureus IE more often than other causative agents of IE affected patients younger that 40 years old (11/34, p < 0.001), intravenous drug users-IVDU (9/34, p < 0.001) and the tricuspid valve (8/34 p < 0.001. Cardiac predisposition for S. aureus IE was not frequent (5/34, p = 0.005). CNS (11/19, p < 0.001) but not S. aureus (5/34, p = 0.3) IE cases were nosocomially acquired. CNS more often affected aortic valve and patients with prosthetic valve IE (p < 0.001). Mortality at day 28 was higher for CNS (10/19, p = 0.009) than for S. aureus (11/34, p = 0.2). Surgical rates for CNS were lower than for other IE causes (1/16 vs 52/118, p = 0.04). On the contrary, there was no difference in rates of surgery for S. aureus IE (10/24, p = 1.0).
Conclusion: CNS IE was mostly hospital acquired, affecting prosthetic valves and presenting higher mortality rates. S. aureus IE affected native valves in younger patients and IVDU. These trends generate the need for a continuous investigation of risk factors of acquisition of nosocomial and community acquired staphylococcal IE in Greece in order to better apply prevention and treatment.
P1474 Risk factors for infective endocarditis and outcome of patients with Staphylococcus aureus bacteraemia
E. Hill, S. Vanderschueren, J. Verhaegen, P. Herijgers, P. Claus, M-C. Herregods, W. Peetermans (Leuven, BE)
Objectives: Staphylococcus aureus bacteraemia (SAB) carries a risk for the development of infective endocarditis (IE). This study investigated risk factors for S. aureus IE (SAIE) and 6-month mortality in patients with SAB.
Methods: A nested case-control study including 66 patients with S. aureus IE (39 community-acquired (CA) and 27 nosocomial) and 132 ad random selected patients with SAB (78 CA and 54 nosocomial) from June 2000 to December 2005.
Results: The median age of patients with SAB and SAIE was 66 and 68 years respectively. Bivariable analysis showed that unknown origin of SAB, cutaneous origin, a foreign body, a valvular prosthesis, a pacemaker, persistent fever, persistent bacteraemia and prior IE were significantly associated with SAIE. In multivariable analysis, unknown origin of SAB (OR, 4.8; 95% CI, 1.7–13.1; P = 0.003), a valvular prosthesis (OR, 8.8; 95% CI, 2.8–28.6; P < 0.001), persistent fever (OR, 3.4; 95% CI, 1.1–10; P = 0.03) and persistent bacteraemia (OR, 7.2; 95% CI, 2.4–21.7; P < 0.001) remained independently associated with SAIE. Six-month mortality was 8% in patients with SAB versus 35% in SAIE (P < 0.001). In bivariable analysis, MRSA and persistent bacteraemia were associated with 6-month mortality in patients with SAB. In multivariable analysis, persistent bacteraemia (OR, 4; 95% CI 1.6–10.1; P = 0.004) remained independently associated with mortality.
Conclusion: Persistent bacteraemia was independently associated with SAIE and 6-month mortality in patients with SAB. Unknown origin of SAB, a valvular prosthesis and persistent fever were associated with SAIE. SAIE has a significantly higher mortality than SAB. The optimal management of SAB and SAIE deserves further study.
P1475 PET-CT scan in patients with infective endocarditis for early detection of embolisation and metastatic infection
E. Hill, E. D'Hondt, J.H.G. Crevits, S. Vanderschueren, M-C. Herregods, P. Herijgers, S. Dymarkowski, L. Mortelmans, W.E. Peetermans (Leuven, BE)
Objectives: In infective endocarditis (IE), the risk for embolisation and metastatic infection is the highest during the first 2 weeks after initiation of therapy. Before surgical intervention, it is important to exclude metastatic foci to reduce the risk of relapse bacteraemia in the presence of a prosthetic valve.
Methods: From March to August 2006, 13 patients (14 episodes) with definite IE according to modified Duke criteria were included. PET-CT scan was performed within 2 weeks after diagnosis of IE. This study investigated whether PET-CT should be recommended in all patients with IE to detect early peripheral embolisation and metastatic infection.
Results: The mean age was 62 years and 54% were males. The causative microorganisms were Staphylococcus aureus in 3 episodes, streptococci in 5, enterococci in 4 and others in 2. Nine episodes involved native valves and 5 prosthetic. In 3/14 episodes (21%) PET-CT was positive for peripheral embolisation without clinical suspicion. In 5/14 episodes (36%), PET-CT was positive for metastatic infection, whereof in 1 episode without clinical suspicion. In 7/14 episodes with clinical suspicion of metastatic infection, it was not confirmed by PET-CT in 3 episodes. In 1 patient with clinical suspicion of metastatic infection and a positive PET-CT (spondylodiscitis), it was not confirmed on MRI. Overall, PET-CT was positive in 4/14 episodes (29%) without previous clinical suspicion.
Conclusion: Overall, 50% of IE episodes had a positive PET-CT result for peripheral embolisation and/or metastatic infection. Early detection of peripheral embolisation and/or metastatic infection by PET-CT in episodes without clinical suspicion occurred in nearly one third.
P1476 Streptococcus agalactiae endocarditis: improved prognosis of a severe infection
A. Salinas, A. Goyenechea, M. De Górgolas, R. Fernández Roblas, M. Fernández-Guerrero (Madrid, ES)
Background: Streptococcus agalactiae endocarditis is not commonly found but its mortality has ranged from 34 to 56%, similar to that observed in most severe forms of endocarditis. However, a better knowledge of the disease and a more aggressive use of cardiac surgery may have improved prognosis.
Methods: Review of cases of Streptococcus agalactiae endocarditis seen by the authors between 1990 and 2005. The diagnosis of endocarditis was done following the modified Duke' criteria. Identification of S. agalactiae and susceptibility tests were performed by conventional methods.
Results: 10 cases were reviewed: 3 men and 7 women; mean age 61.3 years. Streptococcus agalactiae endocarditis accounted for 1.7% of all the cases of endocarditis seen during the study period. Fifty per cent of patients had underlying valvulopathy: 2 previous endocarditis, 1 rheumatic mitral disease, 1 mitral mixoid, 1 mitral calcified. 80% had a chronic disease: liver cirrhosis 3, cancer of cervix 2, breast cancer 1, diabetes mellitus 1, parenteral drug abuse 1. The origin of the infection was the skin (4), cervix (2) and peritoneum (1). The presentation was acute in all the cases (average duration: 3.3 days) with fever and heart murmur. Skin lesions and brain embolism were detected in 30%. 4 patients had heart failure. TTE showed vegetations in 50% of cases. TEE showed vegetations in all the cases that was performed. The size of vegetations ranged from 5 to 16 mm in diameter (average 11.5 mm). The heart valves involved were: mitral valve 60%, aortic 40% and mitralaortic 10%. All the isolates were penicillin-susceptible (MIC < 0.1 mg/L) and patients were treated with β-lactam (penicillin or 3rd-generation cephalosporins) and gentamicin was associated in 60% of cases. Heart surgery was indicated in all patients with heart failure. Mortality was 20%.
Conclusions: Endocarditis due to Streptococcus agalactiae is an uncommon, opportunistic infection in patients with severe chronic diseases. In comparison with historical series, mortality has decreased. A rapid diagnosis and a judicious use of cardiac surgery are of paramount importance to improved prognosis
P1477 Nosocomial endocarditis due to Gram-negative bacteria
A. Demitrovicova, L. Findova, V. Krcmery, V. Hricak, M. Karvaj (Bratislava, Trnava, Nove Zamky, SK)
Objectives: Common cause of infective endocarditis (IE) are Gram-positive bacteria: staphylococci, viridans streptococci and enterococci, which represent 50 to 70% of all bacterial isolates from blood cultures in patients with IE. Culture negative IE represents 25 to 40% and Gram-negative bacteria 5 to 10%, 90% of them are of nosocomial origin. Objective of the study was to investigate IE due to Gram-negative bacteria.
Methods: We assessed the proportion, aetiology, risk factors and mortality of IE due to Gram-negative bacteria (GNB) within our database of IE in 23 years in Slovakia. Forty-two (42) cases of IE due to GNB appeared within 606 cases (6.9%). Most frequently isolated organisms in 42 cases were Pseudomonas aeruginosa (8), Haemophilus influenzae (7), Acinetobacter baumannii (6), Klebsiella pneumoniae (6), S. enteritis (5), E. agglomerans (4), E. coli (4), Citrobacter freundii (1), Serratia marcescens (1), Pseudomonas fluorescens (1). All of the cases appeared in hospital.
Results: Comparison of 42 cases (one was polymicrobial) to all 606 cases from the national database in univariate analysis showed that diabetes mellitus type 1 (6.2% vs. 11.4%, p < 0.045), previous cardiosurgery (19.4% vs. 9.9%, p < 0.05), prior endoscopy (31% vs. 8.1%, p < 0.04) and vitium cordis (11.9% vs. 3.3%, p < 0.04) were significantly more frequent, and vice versa, prosthetic valve insertion (2.4% vs. 14.9%, p < 0.01) less frequently observed in IE due to GNB than among all the cases. Mortality rate was similar in both groups (11.5% vs. 15%). Proportion of IE due to GNB (6.9%) within 23 years in Slovakia was similar to proportion of GNIE (5 to 10%) from other national studies in Europe.
Conclusion: Patients with diabetes mellitus type 1, after cardiac surgery and/or endoscopy of gastrointestinal or genitourinal tract are at higher risk of GNIE. Therefore, initial therapy with anti-Gram-negative antibiotic until aetiology/susceptibility is not determined is advisable.
P1478 Case report: Granulicatella elegans causing native valve endocarditis
S. Nayman Alpat, I. Ozgunes, S. Nemli, N. Erben, E. Doyuk Kartal, G. Usluer (Eskisehir, TR)
Objectives: Infective endocarditis (IE) is a life threatening disease of endocardium of the heart. It is reported that viridans streptococci as the most common isolates among patients with IE. Abiotrophia and Granulicatella species form part of the normal flora of the oral cavity. They are infrequently isolated in patients with IE.
Case report: We report a case of native valve endocarditis attributed to Granulicatella elegans. A 60 year old man was admitted to our hospital with malaise and history of fever. He experienced fever 6 days prior to admission. Fever was maximal in the afternoons without chills and sweating. He had no history of underlying heart disease. He had a history of cholecystectomy due to cholelithiasis 3 weeks ago. He was admitted to another hospital with fever (40°C) and unconsciousness 4 days prior to admission. Transthoracic echocardiography was performed and an intracardiac motile mass was noted on the posterior part of mitral valve. On physical examination he had a temperature of 37.8°C, a 3/6 systolic murmur was heard at all cardiac focuses on chest. No organomegaly and rashes were detected during examination. Two sets of blood cultures were taken with BACTEC blood culture bottles and a urine culture was taken because of pyuria. Both of blood cultures yielded Gram-positive coccoids. They were identified as Granulicatella elegans by VITEC II identification system. Urine culture was negative. Physical examination and laboratory data showed 1 major and 3 minor criteria according to DUKE criteria for IE. He was successfully treated with ceftriaxone and gentamycin.
Conclusion: Twelve cases of IE caused by G. elegans have been reported in the literature up to October, 2006. Senn et al. reported that bacteraemia caused by immunosuppressed patients compared to other NVS species. Therefore our patient was immuncompetant and had no underlying problem that can suppress immune system. It was our limitation not to perform antimicrobial susceptibility tests for our clinical G. elegans isolates. We believe that clinical and laboratory departments should work closely and communicate continuously especially when microorganisms such as G. elegans that can also be a part of normal flora are isolated in specimens. Laboratory department should warn clinicians, get clinical information about patient and consider it can also be pathogen not a contaminant.
P1479 Propionibacterium acnes endocarditis
A.L. Gray, M.R. Sohail, L.M. Baddour, A. Virk (Rochester, US; Al Ain, AE)
Objectives: Propionibacterium acnes is a rare cause of bacterial endocarditis. When identified in blood cultures, it is usually considered a skin contaminant. The purpose of this study is to describe the clinical presentation, management and outcome of the patients with P. acnes endocarditis based on our institutional experience and review of the published medical literature.
Methods: We retrospectively reviewed all cases of P. acnes endocarditis identified at Mayo Clinic Rochester and searched the English language medical literature for all previously published reports.
Results: We identified 34 cases in the medical literature and 5 cases from our institutional database (clinical details were available in only 27 of these cases). Mean age of the patients was 53 years (range15–78) and 89% were males. Most of the cases (90%) presented with non-specific symptoms including fever, malaise and weakness while 3 patients had severe valvular insufficiency and 2 presented with embolic strokes. Blood cultures were positive in 81% (22/27) of the cases. All 5 cases with negative blood cultures were confirmed by positive valve tissue cultures. In 85% of the cases (29/34), the infection was in associated with presence of a foreign body (25 patients with prosthetic valves (PV), 1 left ventricular Teflon patch, 1 Carpentier mitral valve ring, and 2 involved pacemaker leads). Prosthetic valve infection was complicated by abscess formation in 50% of the cases and 17% demonstrated valve dehiscence on echocardiogram. Infection of the Carpentier mitral ring resulted in detachment of the ring in one case. Only 4 cases had native valve endocarditis with P. acnes. Majority of the patients (85%) required surgery; either valve replacement or removal of the pacemaker apparatus. Eleven percent (3/27) cases died secondary to the infection. In two of these, death was described as being secondary to splenic rupture and valve dehiscence. Penicillin resistant isolated was recovered in only one case. Median duration of post-operative antibiotics was 6 weeks (mean 7.1 weeks). Patients were typically treated with a penicillin derivative alone or in combination with gentamicin.
Conclusions: P. acnes endocarditis should be considered when this organism is isolated from multiple blood cultures especially in the presence of a foreign cardiovascular device. Morbidity is high, and aggressive medical and surgical interventions are required to achieve cure.
P1480 Persistent bacteraemic infective endocarditis
L. Findova, A. Demitrovicova, V. Krcmery, P. Kisac, P. Marks (Trnava, Bratislava, SK; London, UK)
Objectives: Persistent bacteraemic infective endocarditis (PBIE) is defined as persistence of positive blood cultures for 3 or more days despite antibiotic therapy. The reason for persistence of bacteraemia is deficient or inappropriate selection of antimicrobials, resistant organism, foreign body or prosthetic endocarditis. Objective of the study was to analyse persistent IE.
Methods: Within 606 cases of IE in last 23 years in Slovakia, 85 (14%) fulfilled the definition and had positive blood cultures for 3 or more days (3 to 7 positive blood cultures). We compared these 85 cases of PBIE to 606 cases of all IE from the database of the national survey.
Results: Several risk factors were observed more frequently among persistent IE: elderly age (60% vs. 33%, p < 0.045), diabetes mellitus (26.9% vs. 11.4%, p < 0.03), prior cardiosurgery (20% vs. 9.9%), prior surgery (19.4% vs. 42.7%), p for both is 0.04, right side IE (17.6% vs 11.4%, p < 0.01) and prosthetic valve (32.9% vs 2.4%, p < 0.001). Also aetiology was different in persistent IE in comparison to all IE. Viridans streptococci (37.5% vs. 15.2%, p < 0.01), coagulase-negative staphylococci (51.8% vs. 21.9%, p < 0.02) and enterococci (23.5% vs. 7.6%, p < 0.01) were observed more frequently among persistent IE and vice versa, embolisation (7.1% vs. 35.5%, p < 0.01), rheumatic fever (9% vs 22.3%, p < 0.05) were less frequent. However, mortality and type of intervention (surgery, antimicrobials only) were similar in both groups. This surprising finding is difficult to explain because persistent bacteraemia in IE should be logically related with increased death rate.
Conclusion: Diabetics or elderly patients after prior surgery as well as those with prosthetic or right side endocarditis may suffer from PBIE with multiple positive blood cultures. However, risk of embolisation as well as mortality rate were surprisingly lower.
P1481 10-year epidemiology of operated endocarditis patients at a tertiary university hospital in Germany – a preliminary analysis
F. Zauner, T. Glück, B. Salzberger, B. Ehrenstein, D. Birnbaum, H.J. Linde, F.X. Audebert (Regensburg, DE)
Objectives: To describe retrospectively the epidemiology and clinical presentation among patients receiving heart valve surgery for endocarditis at the Regensburg University Medical Center and to compare these data with other national and international cohort studies. To correlate pathogen types with anatomic location and histopathologic damage patterns of affected heart valves.
Methods: 211 intraoperatively confirmed endocarditis episodes of 205 patients were reviewed from September 1994 to February 2005. Data was obtained from surgical records, microbiology results, histopathology reports, in-house charts, and medical charts of admitting hospitals.
Results: Altogether 252 valves were replaced or reconstructed. The median age at the time of operation was 61 years with female patients accounting for 25% of all patients. Heart valves were affected as follows: aortic valve (49.3%), mitral valve (28%), aortic and mitral valve (17.5%), and valves of the right heart (5.2%). Mechanical valves were inserted in 58%, biological valves in 19% of the episodes. Valve reconstruction without replacement was performed in 13% and any kind of combined multiple valve operation was done in 9% of the operations. Postoperative diagnoses were classified into florid natural valve endocarditis (NVE; 58%), florid prosthetic valve endocarditis (PVE; 12%), status after NVE (26%), and status after PVE (4%). As predisposing factor, diabetes mellitus was elicitable in 24% of all patients. Causative organisms could be delineated in 70% of the episodes. Streptococcus viridans spp. accounted for 30.8%, Staphylococcus aureus for 28.7%, Enterococcus spp. for 14.7%, Staphylococcus epidermidis for 9.8%, infections with any two pathogens at a time for 5.6%, Streptococcus spp. for 3.5% and other species for the remaining 7% of episodes with delineation of a causative organism. Among the episodes with confirmed Staphylococcus aureus, the aortic or mitral valves were approximately equally affected. In-hospital mortality after cardiac surgery was 10.4% from the day of surgery and the following 14 days. After 30 days, the mortality rate slightly increased by 3.3% and accounted for 13.7%.
Conclusions: The distribution of affected valves and the 30-days mortality rate in our cohort is consistent with results from current international studies. The spectrum of causative organsims represents spectra observed in similar studies, although Staph. aureus was delineated at a lower rate.
P1482 Risk factors for systemic emboli in infective endocarditis
N. Esmailpour, M. Rasoolinejad, A. Ghoochani (Tehran, IR)
Objectives: As one of the complications of infective endocarditis, embolisation has a great impact on prognosis. This study was undertaken to analyse the risk factors for systemic emboli in infective endocarditis.
Methods: A retrospective study was conducted during 2001–2004 in two teaching hospitals in Iran, included patients with infective endocarditis as defined by Duke Criteria. Demographic, clinical, echocardiographic and microbiological data were entered in data base, and the relationship between emboli and these variables was reviewed.
Results: We studied 76 cases (80.5% male, 19.7% female) with infective endocarditis according to Duke Criteria. Mean age of patients was 37.42 years. In 27.6% mitral valve was induced. The most common organism was Staph. aureus (14.5%). Systemic embolus was seen in 32.9% of patients. Most emboli (15.8%) affected the central nervous system. Mortality in patients with emboli was higher than cases without emboli (36% vs. 12.8% respectively, P = 0.03). The risk of emboli was 80% when the vegetation measured larger than 10 mm and only 15% when vegetation size was smaller than 10 mm (P < 0.05). There was no relationship between age, gender, kind of induced valve, site of vegetation, kind of pathogen and duration of symptoms and embolus formation.
Conclusion: Mortality was higher in infective endocarditis patients with emboli. According to this study, vegetation size (larger than 10 mm) was the only factor that was associated with an increased risk of embolic episodes.
P1483 Permanent pacemaker and implantable cardioverter-defibrillator-related infective endocarditis
M. Sohail, D. Uslan, A. Khan, P. Friedman, D. Hayes, W. Wilson, J. Steckelberg, S. Stoner, L. Baddour (Al Ain, AE; Rochester, Atlanta, US)
Objectives: Endocarditis is an uncommon but serious complication of permanent pacemaker (PPM) and implantable cardioverter-defibrillator (ICD) implantation. Purpose of this study is to describe the management and outcome of patients with PPM/ICD-related endocarditis.
Methods: Retrospective review of all cases of PPM/ICD infection presenting to Mayo Clinic Rochester between 1991 and 2003. Device-related endocarditis was defined using modified Duke's criteria.
Results: We identified 189 PPM/ICD infections during study period. Forty-four (23%) of these met the case-definition for device-related endocarditis (33 PPM, 11 ICD). Mean age of patients was 68 years. (77% were males). Mean duration from device implantation to infection was 739 (median 419) days. Coagulase-negative staphylococci (18, 41%) and Staphylococcus aureus (18, 41%) were the most commonly isolated organisms. Blood cultures were positive in 77% of cases. Twenty-six (59%) patients had lead vegetations, 6 (14%) had valvular vegetations and 12 (27%) had both lead and valvular vegetations (tricuspid valve 11, pulmonic valve 1, mitral valve 3, aortic valve 5). The mean diameter of vegetations was 16 mm (median 11 mm). Forty-three (98%) patients were treated with a combined approach of hardware removal and parenteral antibiotics. Mean duration of systemic antibiotics after device explantation was 45 days (median, 28 days). Electrode leads were percutaneously removed in 34 (77%) cases using manual traction (7), locking stylet (13) or laser sheath (16); 7 (16%) cases underwent surgical lead extraction. Fifteen patients had lead diameter ≥10 mm and percutaneous lead extraction was complicated by pulmonary embolism in 5 cases as demonstrated per radiographic procedures. However, none of these patients were clinically symptomatic. Mean time from removal of an infected device to placement of new system was 14.7 days (median, 9.5 days). A replacement device was not necessary in 17 (39%) cases. Follow-up data were available for 33 (75%) cases (mean duration, 75 weeks); 27 (82%) of them were cured with device removal and antibiotics. Six (14%) patients died during index hospitalisation (4 sepsis, 1 nosocomial pneumonia, 1 tricuspid valve rupture).
Conclusions: Device-related endocarditis is associated with severe morbidity and mortality. Electrode leads can be removed percutaneouly even in cases with large sized (≥10 mm) lead vegetations without increasing the risk of symptomatic pulmonary embolism.
P1484 Diagnosis difficulties of infective endocarditis in children, adolescents and young adults without cardiac predisposing factors results of a case series
A. Radulescu, A. Slavcovici, M. Flonta, D. Tatulescu, V. Zanc (Cluj-Napoca, RO)
Objectives: To evaluate infective endocarditis (IE) in previously healthy children, adolescents and young adults within a retrospective case study in a tertiary infectious diseases hospital.
Methods: Between 1988 and 2006, 212 cases of infective endocarditis were documented based on the Duke criteria (range of age 7–83 years). Medical charts were reviewed for demographic and clinical data (age, clinical status at admission, signs and symptoms of endocardial infection, predisposing factors, echocardiographic documentation, predisposing factors, treatment and outcome).
Results: There were 166 episodes of native valve IE (78%), the aetiology dominated by streptococci, staphylococci and Gram-negative bacteria being established in 122 cases (57%). Sixteen cases of IE occurred in children and young adults without known predisposing factor (median: 25.5 years, range 7 to 35 years, gender ratio1:1). Persistent fever, fatigue, malaise and heart murmurs were present at admission in all cases, vascular and immunological phenomena in 4 (splinter haemorrhages, Osler's node). A 7 year old boy presented a meningitis clinical picture with petechial lesions. Dental procedures, poor dental condition and mild upper respiratory infections were described in all cases. All patients underwent transthoracic two-dimensional and Doppler transthoracic echocardiography, 10 patients underwent additional transoesophageal echocardiography. The initial echocardiogram suggested IE in 14 cases. Main findings were vegetations in 14, perivalvular abscesse in 2 and valvular leaks in 3 cases. Despite repeated blood cultures the aetiology was established in only 7 cases (44%): oral streptococci (4), Streptococcus bovis (1), S. agalactiae (1) and polymicrobial endocarditis (1), mainly due to empirical antibiotic treatment. Mitral and aortic valve involvement was prominent – 13 cases. Antimicrobial therapy was completed by cardiac surgery in 5 cases. All patients went well, one relapse was documented. Applying the modified Duke criteria to our patient series 11 were considered definite and 5 possible. Major echographic findings became particularly helpful in the cases without bacteriologic evidence.
Conclusion: Diagnosis of IE is challenging especially in the absence of predisposing cardiac factors. Delayed diagnosis, empirical antibiotic treatments significantly limit the chance of positive blood cultures. Modified Duke classification is to be used in children and young adults.
Hepatitis B virus
P1485 Performance evaluation of VIDIA HBs Ag, a new automated immunoassay test for the qualitative detection of HBs antigen in serum and plasma human samples
B. Seignères, N. Ripoll, P. Gradon, P. Desmottes (Marcy l'Etoile, FR)
Objective: The VIDIA system is a new automated, primary tube immunoassay instrument designed to reinforce traceability, simplifying the daily workload for routine testing. The qualitative detection of HbsAg is used as a first-line diagnostic test for hepatitis B infection or for the follow-up of HBV chronic carriers.
Then, we performed an evaluation of VIDIA HbsAg (bioMérieux) compared to VIDAS HbsAg Ultra (bioMérieux) and AxSYM HBsAg (V2) (Abbott).
Methods: The VIDIA HbsAg assay principle combines a two-step enzyme immunoassay sandwich method with a final chemiluminescence detection. HBsAg present in the sample binds with monoclonal anti-HBs antibodies coated on the magnetic particles and with a mixture of monoclonal anti-HBs conjugates used in the revelation phase. The antibodies have been selected for their binding ability towards wild type and variant HBs antigen.
The comparative study was performed by testing 180 positive samples from Chronic HBV carriers and 213 negative samples including blood donors and pregnant women. The serological status of 3 samples was established according to the results obtained with 3 CE-Marked immunoassays. In case of discrepant samples, the positive samples were retested and confirmed if repeatedly positive.
Results: In the comparative study, specificity and sensitivity of VIDIA HBsAg was assessed according to the clinical status of the samples and compared to VIDAS HBsAg Ultra and AxSYM HbsAg (V2). VIDIA HBsAg sensitivity was tested on 30 seroconversion panels. According to NCCLS recommendations, VIDIA HBsAg precision was evaluated inferior at 15%.
| 180 Positive samples | 213 Negative samples | ||
|---|---|---|---|
| VIDIA | Positive results | 179 | 3 |
| Negative results | 1 | 210 | |
| MIDAS | Positive results | 179 | 0 |
| Negative results | 1 | 213 | |
| AxSYM | Positive results | 178 | 25 |
| Negative results | 2 | 189 | |
| MONOLISA | Positive results | 179 | 1 |
| Negative results | 1 | 210 | |
| Doubtful results | − | 2 |
Conclusions:
-
–
VIDIA HBsAg display a better specificity than Abbott AxSYM HbsAg (V2) and performed similarly to VIDAS and MONOLISA.
-
–
The evaluation of VIDIA HBsAg sensitivity tested on seroconversion panels and the comparative data results obtained on positive samples demonstrated that VIDIA HbsAg is as sensitive as other already CE-Marked test and present good precision performances.
P1486 Associated factors and viral DNA in isolated hepatitis B core antibody group among healthcare workers
S. Chiarakul, S. Vuttiopas, K. Eunumjitkul, J. Kaewkungwal, Y. Poovorawan (Bangkok, TH)
Objectives: To determine the factors associated with healthcare workers (HCWs) who had isolated hepatitis B core antibody (anti-HBc), and to determine HBV-DNA result of isolated anti-HBc group.
Methods: HCWs at Prasat Neurological Institute (PNI), Thailand who had serology HBV profiling (HBsAg, anti-HBs and anti-HBc) result of isolated anti-HBc were confirmed by repeated Microparticle Enzyme Immunoassay (MEIA) methods. Associated factors were evaluated by using interview and self-administered questionnaires covering historical, personal and occupational risk factors. Alanine aminotransferase (ALT), aspartate aminotransferase (AST) and HBV DNA by nested PCR were performed in isolated antiHBc group.
Results: Among 548 HCWs, 39 were in isolated anti-HBc group consisting of 20 had anti-HBc positive and 19 had anti-HBc positive with low anti-HBs. Twelve (30.8%) were male and 27 (69.2%) were female, age ranges between 27 and 60 year (mean 47.1). Twenty-three were in the high occupational risk group, 4 were in moderate risk, 12 in low risk group. Comparing between isolated anti-HBc group and the other groups, there were statistically significant differences in sex (male: 15.2% vs. female: 5.7%, p = 0.002), marital status (single: 3.8% vs. married/divorce: 9.1%, p = 0.021), education (undergraduate: 10.4% vs. postgraduate: 3.9%, p = 0.038), history of HB vaccine (vaccinated: 2.5% vs. non-vaccinated: 9.1%, p = 0.041), history of viral hepatitis (25.9% vs. 6.1%, p = 0.000), alcohol drinking (drinker: 15.8% vs. non-drinker: 4.7%, p = 0.000).
In isolated anti-HBc group, mean AST was 30.1 u/L (range from 16 to 84 u/L) and mean ALT was 27.4 u/L (range from 4 to 114 u/L). Six (15.4%) had elevated transaminase levels. Five had elevated both AST and ALT. 1 with heavy alcohol drinking had elevated only ALT.
Four cases of 39 (10.3%) had HBV DNA detected; 2 were male, 2 were female; 1 in high occupational risk group with no history of hepatitis; 1 had hepatitis C cirrhosis and post liver transplantation; 1 had past history of acute hepatitis; and 1 was a heavy alcohol drinker.
Conclusion: Significant factors associated with isolated anti-HBc were found among male, non-vaccinated, history of viral hepatitis, low level of education, and alcohol drinking. The study results indicated that 15.4% had elevated liver transaminase; and occult HBV infection (HBV DNA) were detected in 10.3% of isolated anti-HBc group.
P1487 Studying the association between X gene mutations with hepatocellular carcinoma and liver cirrhosis in HBsAg-positive patients referring to medical centres in Tehran
S. Hekmat, M. Ahmadi Pour, S. Amini, F. Roohvand, S. Alavian, M. Joulaie, S. Andalibi Mahmoudabadi (Tehran, IR)
Objectives: The viral transactivator HBVX (HBx) gene plays a critical role in the molecular pathology of HBV-related HCC. This study investigated the possible HBx mutations in Iranian patients with HCC or liver cirrhosis, especially identification of a point mutation at codon 31 from xORF.
Methods: Sera from 30, 10 and 12 patients with acute hepatitis B, HBV-related HCC and liver cirrhosis, respectively, were analysed by nested-PCR and RFLP methods to detect (HBx-A31) mutation. In addition we also cloned the PCR products from second round of nested PCR in TA cloning vector from 3 HCC, 2 cirrhosis and 2 acute hepatitis subjects followed by sequencing and aligned them with the downloaded sequences from GeneBank. We use neighbour-joining method to make phylogenetic trees.
Results: Two common HBx variants in wild-type HBV, serine-31 (TCG) and praline-31 (CCG), were digested by AvaI in the presence of (T) or (C) at the first nucleotide position of codon 31, the same PCR products were not digested by MwoI in lack of Alanine amino acid (GCG) (a common point mutation at the East Asia). We found two point mutations (nt.1762, nt.1764) and one point mutation into HBV genome from serum of all patients with cirrhosis and HCC. These mutations at both codons 130 (AAG to ATG, lysine to methionine) and 131 (GTC to ATC, valine to isoleucine) were observed in all available HCC cases and located at Basal Core Promoter (BCP) which regulate transcription and production of core and precore RNA. All cases had D genotype and D1 subgenotype (The common genotype in Iran).
Conclusions: Core promoter region have been associated with more-severe liver damage, inflammation and fibrosis. But the implications of these findings in the pathogenesis of HBV are discussed.
P1488 Prevalence of hepatitis B virus genotypes in patients with chronic hepatitis B virus infection in Madrid
A. Fernandez-Olmos, M. Mercadillo, V. Moreira, M. Mateos (Madrid, ES)
There is evidence that HBV genotypes play a role in several aspects of HBV infection, such as disease profile or response to treatment. The eight genotypes of HBV (A-H) have a distinct geographic distribution.
Objectives: To study the prevalence of hepatitis B virus genotypes among hepatitis B chronic patients and their relationship with age and the presence of HBeAg and anti-HBe in serum.
Methods: HBV genotypes were determined in serum from 76 patients, by direct sequencing of a 137-nucleotide fragment from the S region of the HBV genomal (Trugene HBV Genotyping kit, Bayer Health Care, New York, USA).
Results: Prevalence of HBV genotypes in the 76 cases was as follows: 41 genotype D (53.9%), 23 genotype A (30.3%), 8 genotype F (10.5%) and 4 genotype E (5.3%). After stratification of patients by age, we found that genotype D was significantly more prevalent among patients over 45 years (71.4% vs. 37.5%, P = 0.021) Comparison of the distribution of the most common genotypes, A and D, showed that genotype D was present more often in HBeAg(–) (ratio A:D = 0.5) than in HBeAg(+) (A:D = 1.6), although the difference was not significant (P = 0.142). Distribution of genotypes F and E was similar among the two groups.
Conclusion: Genotype D is the most common HBV genotype found in our patients in Madrid, in keeping with other studies in Western Europe. Interestingly, genotype F was found in a high number of Spanish patients and this finding points to the introduction of strains from South America to the Spanish population and posibly to Europe due to the high immigration rate. In contrast, no genotype E was found among autochthonous patients. All four genotype E infected patients were immigrants from Africa. Our study did not include Asian patients, therefore, genotype B or C were not found as expected. We did not find genotype G. In agreement with other studies, we conclude that genotype D is the most prevalent, the presence of genotypes E and F may be due to demographic movements and genotype G is uncommon in Spain. As stated by other authors genotype D was more prevalent than A in patients aged over 45 years.
P1489 Primary resistance and treatment associated resistance to adefovir in four chronic hepatitis B patients
A. Fernandez-Olmos, M. Tato, V. Moreira, M. Mateos (Madrid, ES)
Background and Objectives: Adefovir dipivoxil is used for the treatment of patients with chronic hepatitis B infection who are infected with lamivudine-resistant hepatitis B virus. Mutations A181V, N236T, L217R, I233V, V214A and Q215S confer resistance to adefovir. We studied four patients in whom adefovir associated mutations were detected (N236T, V214A and Q215S). Two of these mutations were considered as primary resistance since patients had never received adefovir.
Methods: Serum samples of patients were studied by sequencing a 137-nucleotide fragment of the retrotranscriptase region of the hepatitis B virus (Trugene HBV Genotyping kit, Bayer Health Care, New York, USA). Clinical data are shown in Table 1 .
| Case | AST (U/I) | ALT (U/I) | Genotype | Adefovir resistance mutation | Viral load (UI/mL) | Treatment |
|---|---|---|---|---|---|---|
| 1 | 129 | 76 | D | N236T | 30,000,000 | Lamivudine, adefovir |
| 2 | 47 | 29 | D | N236T | 5,484 | Lamivudine, adefovir |
| 3 | 32 | 66 | F | N236T | 3,284 | Lamivudine |
| 4 | 43 | 53 | D | V214A, Q215S | 39,152 | No |
Discussion: In cases 1 and 2 adefovir was ineffective since no decrease in the viral load was observed after 16 and 17 months of treatment, respectively. Resistance to adefovir is considered unusual but it must be considered in patients who do not respond to adefovir. In cases 3 and 4, mutations N236T, V214A and Q215S were found when studying the genotype of the HBV in the patients. These patients had not received adefovir for the chronic hepatitis B and, therefore, these variants should be considered as primarily resistant to adefovir. In conclusion, the detection of resistance mutations is necessary to monitor and anticipate possible treatment failure.
P1490 Lamivudin treatment in patients with acute severe hepatitis B
L. Roznovsky, I. Orsagova, A. Kloudova, J. Mrazek, I. Lochman (Ostrava, CZ)
Objectives: Severe acute hepatitis B in immunocompetent patients may progress to fulminant hepatitis and death. Lamivudin has been approved for the treatment of chronic hepatitis B but experience with lamivudin treatment for acute severe hepatitis B is still limited.
Methods: In the period of 1999–2006 years, 15 immunocompetent patients (10 women, 5 men, age 18–83 years) with severe acute hepatitis B were treated with lamivudin. Fourteen patients received lamivudin at a dose 100 mg per day, one patient 150 mg daily. Prior to treatment, all patients were positive for hepatitis B surface antigen (HBsAg) and IgM antibodies to hepatitis B core antigen; 12 patients were positive for hepatitis B e antigen (HBeAg); all patients had evidence of severe hepatocyte lysis; all patients had total bilirubin 230 micromole per litre or higher (above 14 mg per decilitre). Nine patients had rapid increase of total bilirubin and contemporary decrease of alanine aminotransferase level, which escalate risk of development of fulminant hepatitis B.
Results: Fourteen patients responded to treatment. Therapy induces a prompt clinical and biochemical response and was well tolerated in all cases. HBsAg disappearance was criterion for termination of lamivudin treatment. HBsAg was undetectable in 11 patients, and protective anti-HBs antibodies developed in 4 of them. Lamivudin was administered in these 11 patients for 4–13 months. Three patients still receive lamivudin, but the therapy is shorter than 6 months. The 12 patients who were HBeAg positive before treatment seroconverted within 2 months, and anti-HBe antibodies developed in 11 of them.
One patient with severe encephalopathy developed fulminant hepatitis B and underwent urgent liver transplantation 7 days after initiation of lamivudin treatment. Seven years after transplantation, the patient is without recurrence of hepatitis B and the treatment with lamivudin and hepatitis B immunoglobulin still continues.
Conclusion: Lamivudin induces a prompt clinical, biochemical and serological response in immunocompetent patients with severe acute hepatitis B. Timing of lamivudin administration is crucial, early lamivudin treatment of severe acute hepatitis B may prevent the progression to fulminant hepatitis.
P1491 Interferon plus lamivudine in treatment of chronic hepatitis B in patients unresponsive to lamivudine monotherapy
M. Panahi, A. Heydari (Mashad, IR)
Introduction: Interferon α-2b has anti-viral and immunomedulatory effects that can induce virologic and biochemical remission in chronic hepatitis B (CHB) infection.
Methods: Lamivudine 100 mg per day and interferon α-2b 10 mega ut three times per week were given for 26 weeks to 22 adult patients with liver biopsy-proved chronic hepatitis B who had been treated with lamivudine for at least 52 weeks. The virologic, immunophoresis, liver function tests and haematologic assays were evaluated.
Aim: The aim of this study was to evaluate the efficacy of high dosages of interferon α-2b combined with standard dosages of lamivudine in precore mutant or lamivudine resistant chronic hepatitis B.
Results: We selected patients to three groups:
Group I: HBeAg(–), HBeAb(+), HBV DNA PCR(+) CHB (6 patients).
Group II: HBeAg(+), HBeAb(–), HBV DNA PCR(+) CHB (11 patients).
Group III: HBeAg(–), HBeAb(–), HBV DNA PCR(+) CHB (5 patients).
Clearance of HBV DNA occurred in three patients in Group II and in one patient in each of Groups I and III. Aminotrasferase normalisation occurred in nine patients in Group II and four patients in Groups I and III.
Conclusions: A six-month course administration of 10 mega ut of interferon α-2b 3 times per week in combination with lamivudine in patients with chronic hepatitis B infection made a sustained depression of CHB in HBeAg(+) patients. Significant studies with more patients are needed to confirm this study.
P1492 Acute hepatitis B virus infection by genotype F despite successful vaccination in an immune-competent German patient
S. Amini-Bavil-Olyaee, A. Heim, T. Luedde, M. Manns, C. Trautwein, F. Tacke (Aachen, DE)
Background and Aims: Hepatitis B Virus (HBV) infection is a leading cause of chronic hepatitis and liver cirrhosis worldwide, and efficient protection can usually be achieved by vaccination that is based on recombinant HBsAg protein from HBV genotype A and D. We report the case of a fully immune-competent German patient that acquired a symptomatic acute HBV infection during adulthood despite a complete and formally successful vaccination, which had resulted in anti-HBs titers considered protective.
Case presentation and Methods: A 32 year old, fully immune-competent German male was referred with classical symptoms of acute hepatitis, including jaundice, fatigue and generalised skin rash. Initial aminotransferase activities were AST 525 U/L (normal <50) and ALT 567 U/L (normal <30), HBsAg and HBeAg were positive, and the copy number of HBV DNA was 26,700 copies/mL by real-time PCR (Roche). The HBV infection was most likely acquired in Spain during a business trip as a mining engineer, and resolved without specific therapy within 6 months. The patient had completed a full three-doses-course of vaccination about 10 months prior to infection. HBV DNA was extracted, amplified, sequenced and subjected for sequence and phylogenetic analyses.
Results: At the time of acute HBV infection, ‘protective’ anti-HBs antibody titers of 50 IU/L (AxSYM AUSAB, Abbott Diagnostics) or 82 IU/L (Enzygnost, Dade Behring) were determined using two independent test systems. Phylogenetic analysis revealed that the isolated HBV strain classified into genotype F, sub-genotype F1 and cluster Ib with bootstrap values of 99, 98 and 85%, respectively. Interestingly, this genotype is extremely rare in Europe. The HBsAg sequence showed no apparent aberrations in the immunodominant ‘a’ determinant domain of the envelope gene. However, sequence comparisons revealed that all reported genotype F isolates display marked differences from the other genotypes in this domain which serves as an epitope for humoral immune responses.
Conclusions: Current vaccines may not confer full protection against rare genotypes, even if no specific mutations in the ‘a’ determinant epitope are present. It should be considered for future HBV vaccines to comprise immunodominant proteins from all HBV genotypes to cover genetic variability flaw among different HBV genotypes.
P1493 Hepatitis B virus genotypes and the response to antiviral treatment in children with chronic hepatitis B
K. Dzierzanowska-Fangrat, M. Woynarowski, J. Socha, D. Dzierzanowska (Warsaw, PL)
Objectives: Hepatitis B virus infection remains a serious health problem worldwide. One of the viral factors believed to be associated with the clinical outcome of the infection is the genetic variability of the virus. The aim of this study was to analyse the correlation between HBV genotype and the response to antiviral therapy in children with chronic hepatitis B.
Patients and Methods: A total of 92 children and adolescents who completed IFN-a therapy due to chronic HBV infection 1–13 years (mean 6 years) earlier were included in the study. A blood sample was collected from each child for biochemical (ALT) testing and serological markers of HBV infection, including HBV DNA levels (Cobas Amplicor HBV Monitor). HBV genotyping was performed in archive serum samples by a nested-multiplex-PCR.
Results: One to thirteen years after IFN-a therapy 77/92 (84%) patients were positive for HBsAg, but only 5 (5%) had detectable HBeAg. ALT levels ranged from 15 to 208 U/L (mean 45.7 U/L). HBV DNA testing was performed in 83 children, and 66 (80%) were found positive. HBV DNA levels ranged from <300 (10 patients) to >200,000 copies/mL (5 patients). HBeAg was detected only in children with high viraemia (>200,000 copies/mL). HBV genotypes were available in 49 patients, 41 (84%) of whom were infected with the genotype A, whereas the remaining 8 had the genotype D. There was no association between the viral genotype and either serological or biochemical parameters in the long post-interferon therapy follow-up.
Conclusions: The response to IFN treatment in children with chronic hepatitis B is not significantly influenced by HBV genotypes A or D, and the rate of serum HBV DNA clearance a few years after treatment remains low.
P1494 Viral clearance, lamivudine resistant mutants appearance and HBV reactivation in patients with chronic hepatitis B in end-stage renal disease treated with lamivudine
D. Telehin, I. Tychka, M. Lulchuk (Lviv, Kiev, UA)
Objectives: The fact of HBV-infection in patients with end-stage renal disease (ESRD) is common and ranges 20–30% in some eastern European countries. Schemes and effectiveness of treatment of chronic hepatitis B (CHB) in this cohort were not established. The aim of our study was to evaluate course of CHB at the patients with ESRD treated with Lamivudine in the haemodialysis department.
Methods: 55 patients with HBV-related chronic liver disease (CLD) and ESRD were studied: 28 males, 15 females. Mean age 41 years. Median ALT was 98 IU/L, all of them HBV-DNA+. 72% patients were with wild-type, 28% with mutant HBV (HBeAg-negative). High virulence HBV was characterised by high level virus circulation (21.5%). 37 patients have taken Lamivudine in creatinine clearance depending doses (10–50 mg/day) during 24 weeks. Serum HBV-DNA was detected using a PCR by “Amplisens HBV” (>1,000 cp/mL). Lamivudine resistance was presumed by virological and biochemical worsening during treatment. HCV-, HDV-superinfection was excluded.
Results: At the end of treatment with Lamivudine 17 patients (46%) achieved HBV-DNA clearance. Decreasing of ALT was notified in 52%. HBeAg seroconversion took place in 19% of HBeAg-seropositive patients. Sustained virological response (SVR) within 12 months after treatment was revealed in 13 patients (35%). In 4 patients (24% of responders) HBV reactivation was noted. Kidney transplantation was made in 12 patients with SVR. In this group 8 patients (66%) develop HBV reactivation within the first year after surgery. In 18 patients without Lamivudine therapy late CHB sequences were present: mild and average CLD – 28%, severe CLD and cirrhosis – 50%, fatal outcome in 22% of patients was due to chronic liver failure. In patients nonresponders and/or in those with HBV reactivation the late sequences rate was accordingly 55%, 45%, 0%.
Conclusions: Peculiarities of clinical course and treatment in immunocompromised patients with ESRD strictly depend on the fact of HBV virulence. In this cohort SVR (6 month duration Lamivudine therapy course) is 35%. In 66% kidney recipients reactivation of CHB is experienced even if surgery was after HBV disappearance. Severe course and mortality rate in Lamivudine treated patients, including those without confirmed virologic response were more rear then in non treated patients.
P1495 One-year follow-up of adefovir dipivoxil treatment in chronic hepatitis B patients
B. Kurtaran, H. Aksu, Y. Tasova, N. Saltoglu, A. Inal, A. Candevir, G. Seydaoglu (Adana, TR)
Objective: The aim of this study is to evaluate the results of adefovir dipivoxil treatment in hepatitis B e antigen-positive and negative chronic hepatitis B patients. We conducted this study to investigate the efficacy of 48 week treatment with this drug at a dose 10 mg once daily.
Methods: This study was conducted in Çukurova University Hospital. Patients were enrolled between March 2005 and July 2006. Patients were manner to receive 10 mg of oral adefovir dipivoxil daily for 48 weeks. Virologic and biochemical assessments (serum HBV DNA and alanine aminotransferase [ALT] levels) were conducted every four weeks.
Results: The mean age of 36 patients was 34.8±10.3 years and 26 (72.2%) were male. Of the total, 32 (84%) had interferon and/or lamivudin treatment before administration. Of patients, 17 were HBeAg-negative and 19 were HBeAg-positive. Both the median of level of HBV DNA and median of ALT levels was significantly decreasing both in hepatitis B e antigen-positive and negative group during 48 weeks. The median change of HBV DNA was higher in HBeAg(+) group than HBeAg(–) group at every four weeks, however no significant differences between two groups at the end of 48 weeks. At the end of the 48 weeks, 28 of 36 (77.7%) patients had improvement in serum HBV DNA levels. Reductions in serum HBV DNA levels were 1.7, 2.3 and 2.7 log at 12th, 24th and 48th week respectively. Hepatitis B e antigen seroconversion rate in HBeAg(+) group was 15.8% (3/19). Treatment has been found to be safe and effective and well tolerated, with a low incidence of adverse events.
Conclusion: In patients with HBeAg-negative and positive chronic hepatitis B, 48 weeks of adefovir dipivoxil treatment resulted in significant virologic, and biochemical improvement.
P1496 Determination of lamivudine and adefovir resistance in acute hepatitis B virus infections
C. Eroglu, H. Leblebicioglu (Samsun, TR)
Introduction: The most widely used antivirals for treatment of hepatitis B virus (HBV) infection is lamivudine and adefovir. Reduced virologic response to therapy with nucleos(t)ide analogues can occur due to the emergence of mutation. Mutations conferring resistance to lamivudine and adefovir occur particularly in the reverse transcriptase gene. Importance of antiviral resistance to nucleos(t)ide analogues in the naive HBV infected patients is not known.
Methods: Sequencing, restriction fragment length polymorphism, line probe assay, and real-time PCR can be used in the detection of drug resistance. In this study, the reverse transcriptase region of polymerase gene has been sequenced to detect antiviral resistance. 158 serum samples of patients with acute HBV infection have been investigated to detect antiviral resistance to lamivudine and adefovir. Mutations at the codons rtM204, rtL180, and rtV173 for lamivudine resistance, and mutations at the codons rtN236 and rtA181 for adefovir resistance have been examined.
Results: Sequences have been obtained in 118 samples. Resistance for lamivudine in codon V173L has been revealed only in one case. No resistance to lamivudine at codons 180 and 204 or to adefovir at codons 181 and 236 has been detected.
Conclusion: This study demonstrated the presence of mostly wild type strains in patients with acute HBV infection. It has been concluded that lamivudine or adefovir resistance is not an important problem in acute naïve HBV infected patients.
P1497 Cytokine levels in patients with chronic hepatitis B infection
F. Yildiz, H. Irmak, G. Tuncer Ertem, M.A. Yetkin, U. Onde, C. Bulut, S. Kinikli, A.P. Demiroz (Ankara, TR)
Objectives: Hepatitis B virus (HBV) infection may progress to liver failure and cancer, for this reason it is a serious health problem. The mechanisms of chronicity are poorly understood. The balances in cytokine production profiles may play a crucial role in determining the resolution or persistence of infection. In this study, we aimed to determine various cytokine levels in patients with chronic HBV infection and to reveal the relationship between levels of cytokines and different clinical phases.
Methods: HBsAg positive patients with a duration of positivity longer than six months who applied to outpatient clinic of Infectious Diseases and Clinical Microbiology Department of Ankara Training and Research Hospital between October 2005 and June 2006 were included into the study. There were 57 cases in patient group and 24 healthy individuals in control group. Serum of interleukin (IL)-4, IL-6, IL-10 and interferon (IFN)-γ levels were detected by ELISA and compared between patients and controls. Additionally, correlation between serum alanine aminotransferase (ALT) levels and cytokine levels were also examined. SPSS version 11.5 for Windows was used for the analyses.
Results: In patient group, there were 25 chronic hepatitis and 32 inactive carrier patients. The mean of serum IL-6 levels in patients with chronic HBV infection (8.496 pg/mL) was higher than those of the control group (6.850 pg/mL) and the difference was statistically significant (p = 0.022). Serum IL-10 levels were 0.784 pg/mL and 0.052 pg/mL in patient and control group, respectively. There was a statistically significant difference between these groups (p = 0.019). Although, serum IL-4 and IFN-γ levels were higher in patients than controls, the difference was insignificant (p > 0.05). There was not any correlation between the levels of IL-4, IL-6, IL-10, IFN-γ with ALT in patients.
Conclusion: As a conclusion, increased IL-10 levels may be associated with persistence of hepatitis B infection. Testing the level of serum IL-6 may be helpful to determine the liver inflammation in patients with chronic hepatitis B infection.
P1498 The value of interleukin-6 as a predictor of tissue injury due to hepatitis B virus replication
M. Cojocaru, I. Cojocaru, I. Matei, S. Iacob, G. Ene, M. Delagramatic (Bucharest, RO)
Objectives: Interleukin 6 (IL-6) is a critical mediator for various host responses. IL-6 also plays the principle roles in the induction of the hepatic acute phase responses. The purpose of the present study is to clarify (1) the relationship between the magnitude of tissue injury and the elevation of serum IL-6, and (2) the change in serum IL-6 and acute phase reactants.
Methods: Serum samples were obtained from 33 patients with chronic HBsAg positive hepatitis diagnosed by ultrasonography, liver biopsy and histopathology. Their ages ranged from 29 to 58 years with a mean of 46±5 years. Another 20 healthy, age and sex matched adults with no evidence of liver disease were also included as a control group. For all cases and controls, full history taking, thorough clinical examination and the following routine laboratory tests were done: Complete blood count; liver function tests including measurement of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase, γ-glutamyltransferase (GGT), total proteins, albumin and bilirubin serum. Hepatitis B surface antigen (HbsAg) and HCV antibody were detected in the sera of patients and control subjects by an enzyme linked immunosorbent assay (ELISA).
Results: Serum IL-6 was measured with an ELISA using a monoclonal antibody against IL-6. The normal range of serum IL-6 was less than 4 pg/mL, CRP less than 0.5 mg/dL. Serum IL-6 was elevated in 28 patients (1,400.8±383.4 pg/mL), and serum CRP was elevated in 30 patients (68±8 mg/dL). The elevation of serum IL-6 was significantly correlated with the elevation of serum CRP (r = 0.84).
Conclusion: Serum IL-6 is an informative and sensitive marker to show the extent of tissue injury due to hepatitis B virus (HBV) replication.
Borreliosis
P1499 Comparison between Enzygnost® Lyme link VlsE/IgG and LIAISON® Borrelia IgG for the laboratory diagnosis of Lyme disease
A. Marangoni, A. Moroni, V. Mondardini, S. Accardo, R. Cevenini (Bologna, Belluno, IT)
Objectives: We evaluated diagnostic performances of Enzygnost® Lyme link VlsE/IgG (DADE Behring, Marburg, Germany), in comparison with those obtained by LIAISON® Borrelia IgG (DiaSorin, Saluggia, Italy).
Methods: In this study a total of 183 human serum specimens were retrospectively studied. Sera were serially collected from 72 Lyme disease patients suffering from EM following a tick bite.
A skin punch biopsy was obtained from each patient when entering the study and cultivated in BSKII medium; the tubes were examined weekly by dark-field microscopy over a period of 45 days.
PCR was performed by using five different sets of primers: FL6-FL7 (amplifying a fragment of the flagellin gene, conserved in all the Borrelia burgdorferi s.l. strains); LD (amplifying a 16S rRNA genomic fragment common to the three genospecies); BB, BG, BA (each one amplifying a species-specific 16S rRNA genomic fragment).
Enzygnost® Lyme link VlsE/IgG is a quantitative immunoenzymatic method based on a mix of native Borrelia antigens from B. afzelii strain PKo and recombinant VlsE obtained from all the three genospecies pathogenic to humans (B. burgdorferi s.s., B. garinii, B. afzelii).
LIAISON® Borrelia Screen is a one step sandwich CLIA. Recombinant specific VlsE antigen obtained from the PBi strain of B. garinii is used for coating magnetic particles.
Results and Conclusion: 66/72 cultures obtained from the biopsies were identified as B. afzelii by PCR; the remaining 6 cultures presented borreliae belonging to more than one genospecies.
Enzygnost® Lyme link VlsE/IgG was more sensitive than LIAISON® Borrelia IgG. In particular, considering the 66 sera obtained at first presentation that were available for this study, Enzygnost® Lyme link VlsE/IgG scored 27 sera as reactive, 10 as border line and the remaining 29 as negative. LIAISON® Borrelia IgG identified 21 sera as positive, 5 as border line and 40 as negative. No seroconversion was observed by both methods during the follow-up of all the patients found initially negative. Moreover, all the patients initially IgG positive showed a decrease in the serological titre within 3–6 months.
Results obtained by Enzygnost® Lyme link VIsE/IgG and LIAISON® Borrelia IgG, respectively, when the 66 sera obtained at the first presentation were tested.
| Enzygnost® Lyme link VlsE/IgG | LIAISON® Borrelia IgG | |
|---|---|---|
| Positive | 27 (40.9%) | 21 (31.8%) |
| Border line | 10 (15.2%) | 5 (7.6%) |
| Negative | 29 (43.9%) | 40 (60.6%) |
| Total | 66 (100%) | 66 (100%) |
P1500 Neuroborreliosis and tick-borne encephalitis – same vector, but different clinical course
K. Soltysova, D. Smiskova, D. Picha, N. Sojkova, M. Podojilova, K. Roubalova, K. Sedivy, V. Maresova (Prague, CZ)
Objectives: Aetiology of aseptic meningitis in childhood remains often unclear. Czech Republic is endemic area for Lyme borreliosis and tick born encephalitis. Both of them belong to the most common neuroinfections in children and have the same vector – a tick.
Aim of our retrospective study was to characterise dominant neurological symptoms in children with neuroborreliosis (NB) in comparison with tick born encephalitis (TBE).
Methods: 38 children (mean age 10 years) hospitalised in Bulovka University Hospital (2002–2006) with final diagnosis neuroborreliosis and corresponding number of age-matched children suffering from TBE were enrolled to the study. The diagnosis was established on the presence of intrathecal antiborrelial IgG synthesis and/or PCR positivity in cerebrospinal fluid (CSF). TBE diagnosis was determined on the presence of anti-TBE IgM in blood and signs of aseptic inflammation in CSF. Patients were screened for history of tick bite, ECM, clinical course, results of laboratory testing including CSF examination, EEG, neurological complication and sequelae. Patients were monitored at admission and after 1, 3 and 6 months. Statistical analysis was performed using a paired t-test.
Results: All NB children except one have signs of aseptic inflammation in CSF. NB patients were more likely to have mononeuritis (NVII in most cases), afebrile or subfebrile course of the disease, shorter duration of headache and absence of meningeal irritation. When compared to the children with NB and TBE, there was no significant difference in white blood cell count, C reactive protein level, red blood cells sedimentation rate, number of lymphocytes and neutrophils in CSF and total protein level in CSF. Length of hospitalisation was longer in the NB group due to antimicrobial therapy. No permanent sequelae were observed in both groups.
Conclusion: NB had mild afebrile course in children and facial palsy was the most frequent clinical syndrome. No permanent sequelae where observed when treated with correct antimicrobial therapy.
Supported by grant No. 1166/2005-IV-GAUK of the Charles University Grant Agency
P1501 The level of morbidity of Lyme borreliosis after antibiotic preventive treatment
A. Yagodina (Perm, RU)
Objectives: Intensive research into the problem of ITBB has started just after the discovery of the causative agent since 1980s. These diseases hold the leading place by their level of morbidity and dissemination compared with the other zoonotic diseases with the particular locality of distribution. The purpose or my research work is to compare the efficiency of doxycycline and azythromycin for antibiotic preventive treatment of Lyme disease.
Materials and Methods: In Perm region where Lyme disease is hyperendemic we conducted a trial of treatment with antibiotics in subjects who had removed attached ticks from their bodies within the previous 72 hours. Direct microscopic analysis of live preventions of the material obtained from the gut of a tick removed from the patient's body after tick bite helps to reveal the borrelia contamination of the vector and is the indication for medical prevention. During the period of 2001 to 2006 year 1,782 patients received antibiotic preventive treatment of ITBB, 1,421 patients bitten by borrelia-infected ticks received 100 mg of oral doxycycline twice daily for 5 days, 381 patients received 500 mg azythromycin once daily for 3 days.
Results: The data obtained after antibiotic preventive course have shown that 19 out of 1,782 patients developed ITBB. The disease developed at the site of the tick bite in a significantly smaller proportion of the subjects in the group that received preventive treatment (1%) than of those in the placebo group (12%). In the group receiving preventive doxycycline 13 patients fell ill, in the group receiving preventive azythromycin 6 patients fell ill. The clinical characteristics of the cases of the disease are as follows: I. Localised stage, manifested form, associated with erythema migrans – 7 patients; II. Disseminated stage, erythema free form – 12 patients. The patients with unfavourable premorbid background developed the disease. The disease developed in a same proportion of the subjects in the doxycycline group (0.9±0.2%) and those in azytromycin (1.5±0.6%) group.
Conclusion: The trial revealed the efficiency of antibiotic preventive treatment. Both antibiotics are efficient in preventive treatment. In the group receiving preventive doxycycline treatment the level of adverse reactions is higher.
P1502 Epidemiological aspects of Borrelia spielmanii from Germany with special respect to its genetic heterogeneity
V. Fingerle, U. Schulte-Spechtel, E. Ruzic-Sabljic, F. Strle, S. Leonhard, H. Hoffmann, K. Weber, K. Pfister, B. Wilske (Munich, DE; Ljubljana, SI)
The Borrelia burgdorferi sensu lato complex comprises at least 11 different species. Three of them – B. burgdorferi sensu stricto (s.s.), B. afzelii, and B. garinii – are known to cause human disease. In 1999 Wang et al. isolated a new B. burgdorferi genospecies – designated A14S – from a patient with erythema migrans which was recently confirmed as a new species and named B. spielmanii. Here we give data on the prevalence of this new species among European human isolates and in the tick vector collected from several Bavarian regions and provide data on its genetic heterogeneity.
A total of 2,155 ticks (136 larvae, 612 nymphs, 1,407 adults) from 8 Bavarian regions and 242 European patient isolates, primarily from Germany, were investigated by ospA-PCR followed by RFLP and/or sequencing of the amplicon. Out of 507 B. burgdorferi infected ticks 28 (6%) harboured B. spielmanii. Notably this genospecies was exclusively present in adult ticks. The by far highest B. spielmanii prevalence (18% of infected ticks) was present in the English Garden, a recreational area in the centre of Munich. In the 242 European patient isolates 4 (2%) B. spielmanii could be identified. Interestingly all isolates were cultured from Erythema migrans and all from patients from the Munich area.
To gain deeper insight into the genetic diversity of B. spielmanii, patient isolates (4 from Germany and 2 additional from Slovenia) and tick materials (n = 28) were subjected to further genetic analysis. Sequence identities were 99–100% for rrs, the inner part of fla and ospA, respectively, 96–100% for rrf-rrl, 66%-100% for dbpA, and 84%-100% for ospC. Four separate ospC and two dbpA clusters were distinguishable. Compared to the few sequences which have been to our disposal this heterogeneity is noteworthy and reveals that the new species B. spielmanii did not evolve just recently.
The results show that B. spielmanii is present possibly in a focal manner in Southern Germany and Slovenia and is able to cause human disease. Further studies on the prevalence of this species in ticks and clinical material are necessary to clarify its role.
P1503 Protein profile determination of Borrelia afzelii and Borrelia garinii isolated from skin and cerebrospinal fluid
U. Glinsek, T. Udovic, E. Ruzic-Sabljic, F. Strle (Ljubljana, SI)
Lyme borreliosis is endemic in Slovenia. B. afzelii is most often isolated species, associated with erythema migrans. Other clinical disorders like nervous system invasion and musculoskeletal manifestations are usually associated with B. garinii. The aim of our study was to compare protein expression of B. afzelii and B. garinii isolated from skin of patients with erythema migrans and from cerebrospinal fluid of patients with neuroborreliosis. In the study we analysed 187 samples of borrelia isolates, 74 were B. afzelii and 113 B. garinii. Protein profile was performed by sodium dodecyl sulfat polyacrilamid electrophoresis (SDS-PAGE). We found differences between protein profiles of B. afzelii and B. garinii isolates and between skin and cerebrospinal fluid isolates. The protein profiles showed differences in the number and the amount of analysed proteins. OspB was expressed by 70 (95%) B. afzelii strains and by 32 (28%) B. garinii strains. OspC was expressed by 36 (49%) B. afzelii strains and by 93 (82%) B. garinii strains. There were differences in protein expression regarding to the origin of isolates. OspB was expressed by 68 (69%) isolates from skin and by 34 (39%) isolates from cerebrospinal fluid. OspC was expressed by 58 (59%) isolates from skin and by 71 (81%) ioslates from cerebrospinal fluid. We found heterogeneity in expression of borrelial proteins, heterogeneity was found between species as well as within particular species. B. afzelii from skin and from cerebrospinal fluid had 11 different protein profiles. B. garinii from skin and cerebrospinal fluid had 19 different profiles. Differences in expression of Osp proteins can be important for bacterial dissemination and colonisation of the host.
P1504 Evaluation of PlateliaTM Lyme IgM and PlateliaTM Lyme IgG assays for Lyme borreliosis diagnosis
B. Jaulhac, S. De Martino, M. Tabouret (Strasbourg, Marnes la Coquette, FR)
Objectives: Lyme borreliosis (LB) is a non contagious infection caused by Borrelia burgdorferi sensu lato and is transmitted by ticks of the Ixodes genus. It is the most prevalent tick-borne zoonosis and is considered as an important emerging infection in Europe, North America and Far Eastern countries. Due to limitations of direct detection, antibodies screening remains the mainly used method to support a clinical diagnosis of LB. Objective of the study is to evaluate the performances of Platelia™ Lyme IgM and Platelia™ Lyme IgG (BioRad Laboratories) for an accurate diagnosis of Lyme disease.
Methods: Platelia™ Lyme IgM and IgG assays are two step assays using respectively immunocapture and indirect ELISA formats. Obtained results were compared to Enzygnost® Borreliosis IgM and IgG (Dade Behring).
Specificity was calculated on 200 healthy blood donors from eastern region of France and on 206 patient samples with potential cross-reactive conditions (syphilis, leptospirosis, rheumatoid factor, CMV or EBV infections, etc…). Sensitivity was determined on 70 confirmed LB sera from early and late stages of the disease.
Results: By comparison with reference ELISA methods, specificities on blood donors were 99.0% and 99.5% for respectively IgM and IgG assays. On potential interfering samples, specificities were 94.6% for IgM and 97.5% for IgG (100% and 97.8% for syphilis IgM and IgG positive samples). Sensitivity results are presented in the table below for assays considered individually or in combination.
| LB stage | IgM | IgG | IgM+IgG |
|---|---|---|---|
| Erythema migrans (n = 17) | 58.8% | 66.7% | 82.4% |
| Neuroborreliosis (n = 33) | 40.0% | 100.0% | 100.0% |
| Lyme arthritis/Acrodermatitis (n = 20) | 5.0% | 100.0% | 100.0% |
Conclusion: Performances of Platelia™ Lyme assays are adequate for diagnosis of infected LB patients and fulfill recommended criteria for screening. Using same protocols, both IgM and IgG assays can be run in parallel.
P1505 Detection of antibodies to Anaplasma phagocytophilum in Lyme disease patients
J. Chmielewska-Badora, J. Zwolinski, E. Cisak, A. Wójcik-Fatla (Lublin, PL)
The aim of this study was to evaluate the possibility of coinfection with Anaplasma phagocytophilum in Lyme disease patients.
Material and Methods: 180 persons with diagnosed Lyme disease were examined for serum antibodies to Borrelia burgdorferi with ELISA kits (Borrelia IgM Rekombinant and Borrelia IgG Rekombinant, Biomedica Medizinprodukte, Austria) and to Anaplasma phagocytophilum with IFA kit (Anaplasma phagocytophilum IFA IgG, Focus Diagnostics, USA). Chi-square test was used for statistical analysis.
Results: In 159 persons (88.3%) antibodies to Borrelia burgdorferi were found and in 38 persons (21.1%) antibodies to A. phagocytophilum were detected. 34 (18.9%) persons possesed antibodies to both infectious agents. A chi-square analysis was used to test the association between prevalence of antibodies to Borrelia and antibodies to Anaplasma and it was found as statistically insignificant.
Conclusion: Results of these findings indicate the possibility of coinfection with Anaplasma phagocytophilum in Lyme disease patients.
P1506 Prevalence of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti in Ixodes ricinus ticks collected from the Lublin region (eastern Poland)
E. Cisak, J. Chmielewska-Badora, A. Wójcik-Fatla, J. Zwolinski (Lublin, PL)
The objective of the study was to evaluate the risk of acquaring infection of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti for residents and visitors of the Lublin region
Material and Methods: 1,813 ticks were examined for the presence of B. burgdorferi sensu lato by PCR and for particular genospecies by nested PCR, in 1,378 ticks the prevalence of Babesia microti DNA was determined by PCR and nested PCR and 1,079 ticks were checked for Anaplasma phagocytophilum by PCR. All ticks were collected from forest areas of six districts of the Lublin region.
Results: The total infection rate of Borrelia burgdorferi s. lato, Babesia microti and Anaplasma phagocytophilum amounted: 5.4%, 5.4% and 8.4% respectively.
The total occurrence of individual Borrelia burgdorferi genospecies in infected ticks with regard to the summarised single and mixed infection amounted 62.8% for Borrelia burgdorferi sensu stricto, 39.8% for Borrelia afzelii and 17.7% for Borrelia garinii.
Conclusion: The results confirm that Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti are present in forest areas of the Lublin region and indicate a potential risk for the residents of surrounding territories as well as for visitors to acquire infections caused by these pathogens.
The research were supported financially by Ministry of Science and Higher Education (grant no. 2PO5D 054 27).
P1507 Serologic and molecular-genetic data for infection with Borrelia burgdorferi sensu lato, Anaplasma phagocytophilum and Francisella tularensis in wild rodents
T. Gladnishka, I. Christova, E. Taseva (Sofia, BG)
Objectives: The aim of this study was to generalise serologic and molecular-genetic data for infectious of wild rodents with aetiological agents of Lyme disease (B. burgdorferi sensu lato), Human granulocytic anaplasmosis (A. phagocytophilum) and tularaemia (F. tularensis).
Methods: A total of 70 rodents from whole country from the species Rattus rattus, Mus musculus and Apodemus agrarius were studied by PCR and serological methods – ELISA and agglutination.
Results: Our PCR results demonstrated that the most common pathogen was B. burgdorferi sensu lato – a total of 18 positive samples (18/70; 25.71%). Infection with F. tularensis was on the second place only from rodents in Meshtitza – a total of 9 positive samples (9/42; 21%) of spleens of the rodents. The infection with A. phagocytophilum was the most rare – a total of 5 positive samples (5/70; 7.14%). Twenty-three positive samples (23/70; 32.86%) of B. burgdorferi sensu lato and 15 positive samples (15/70; 21.43%) of A. phagocytophilum were detected by ELISA. Nine positive samples (9/42; 21%) of F. tularensis were detected by agglutination in Meshtitza.
Conclusions: Molecular-genetic methods demonstrated DNA of the infectious agents before antibody-responses of the macroorganism. Combining of PCR and serological methods of diagnosis was adequate tool for quick and effective surveillance on circulation of different infectious agents among populations of the rodents. Data for infections of wild rodents with B. burgdorferi sensu lato, A. phagocytophilum and F. tularensis showed prevalence with different pathogens among main reservoirs of tick borne infections and warning possible contraction for human population in different regions in the country.
P1508 Usefulness of C6 peptide antibody assay in early localised Lyme borreliosis
R.W. Vreede (Delft, NL)
Objectives: In early localised Lyme borreliosis (erythema migrans, EM), ELISA's detecting antibodies against flagellin of Borrelia burgdorferi, followed by immunoblot, usually have a sensitivity of about 50%. Recently, ELISA's have been developed detecting antibodies against C6 peptide, a conserved and immunodominant region of the VlsE antigen of B. burgdorferi. Initial reports showed a specificity of this assay equal to that of immunblot and a sensitivity in disseminated Lyme borreliosis of more than 90%. In a recent Dutch study, the sensitivity in 40 patients with EM was 22.5% (AP van Dam et al Abstract. Bussiness Meeting Dutch Society of Medical Microbiology, April 2004).
Methods: We evaluated the performance of the C6 peptide (Immunetics Inc, Boston, USA) six months after introduction in our laboartory, which is connected to a large, regional hospital in the Western part of the Netherlands. In a preliminary study using 39 sera of patients with disseminated Lyme-borreliosis, the correlation between C6 assay and flagellin ELISA (Dade Behring, Marburg, Germany) combined with immunoblot (Mikrogen GMBH, Neuried, Germany) was 90%. All aforementioned assays were performed in 90 selected sera.
Results: Thirteen of the 15 patients showing a seroconversion in the flagellin ELISA were also positive in the C6 assay. In 39 patients with EM, the C6 assay showed a sensivity of 69%. Sensitivity in 20 patients with disseminated Lyme-borreliosis was 100%. Specificity of the C6 assay was 95%. No crossreactive antibodies were detected in 20 sera with positive rheumafactor, IgM antibodies to Epstein-Barr virus or cytomegalovirus, or specific antibodies against Treponema pallidum.
Conclusion: The C6 assay showed a high specificity, an excellent sensivity in late Lyme-borreliosis and a sensitivity of nearly 70% in EM, making immunoblot unneccasary and the C6 assay suitable for use in diagnostic laboratories to detect both disseminated and localised stages of Lyme disease. Performance of the updated version of the C6 peptide ELISA and validation of the assay for cerebrospinal fluid are under evaluation.
Travel medicine
P1509 Impact of pretravel medical consultation on behaviour and health status of travellers
A. Wagenknecht, D. Drescher, A. Knaust, M. Eikenberg, T. Eikmann, C. Herr (Giessen, DE)
Background: The Center for Travel Medicine and Yellow Fever Vaccination, Institute for Hygiene and Environmental Medicine, University of Giessen (Germany) offers special consultation and immunisation for travellers. This centre advises approximately 400 travellers every year. A trained physician provides standardised advice about their particular destination, general as well as local health-risks, and administers any necessary vaccination. Some travellers use further information sources to find out more about their destination (for example the internet, books and locals), therefore status of information varies interindividually.
Methods: We study the quality of pretravel information and measures in relation to behaviour during travel as well as post travel health status and medical consultations.
Two trained medical students recruit travellers during pretravel consultation with an informed consent and a first data assessment using a specially developed form on general travel and personal information, the content of medical consultation/measures in the Center for Travel Medicine and Yellow Fever Vaccination and from other information sources. Travellers are followed-up six to eight weeks after their journey to capture post-travel health status. The questionnaire applied at this time point has some similarities to the pretravel assessment form and contains additional items on during travel health status, physician consultations, medication and risk behaviours as well as preventive measures. Post travel medical history is also assessed. The questionnaire comprises forty items offering the option of completition via telephone, print-out or internet.
Results: About seventy-five percent of the travellers agreed to participate in the study. The posttravel follow-up questionnaire has been completed by twenty-five travellers so far. The travellers were 17 to 64 years old, eight men and 17 women. Travel destinations were in and outside Europe (e.g. Africa, Uganda). The mean duration of travel was 29 days. Results of this first assessment will be presented.
P1510 Traveller diarrhoea. Aetiological study in a tropical medicine unit
R. De Julián, J. Ruiz, M. Lago, S. Puente, M. Baquero, J. Gascón, J. G. ónzalez-Lahoz (Madrid, Barcelona, ES)
Introduction: The number of international travels is increasing continuously. 12 million Spaniards travelled abroad last year; 900,000 of them had tropical countries as final destination. Over 10% of travellers went to a doctor at their arrival. Diarrhoea was the most frequent cause (over 40%).
Material and Method: 43 patients incoming to Tropical Medicine Unit were under study, in a four months period, showing diarrhoea after a travel to the tropic as main symptom. 24 (55.8%) female and 19 (44.2%) male. Final travel destination by continent was 25 (58.4%) to Africa, 10 (23.26%) to Asia and 8 (18.6%) to Latin America. 3 faeces samples from different times were collected from all of them, and microbiological study (bacteriologic and parasitic) was done. When Escherichia coli was diagnosed by coproculture we tried to identify the strain diarrhogenic character using PCR, looking for ETEC, EAEC and EPEC-EHEC strains.
Results: 100% exhibit diarrhoea, with a duration of: <7 days, 9 cases (20.9%, average 4.8 days). Aetiology: E. coli (8): 1 ETEC + Entamoeba histolytica, and 1 EAEC. Parasites (1): Giardia intestinalis.
7–30 days, 19 cases (44.2%, average 18 days). Aetiology: E. coli (14): 2 ETEC (1+ Blastocystis hominis); 4 EAEC (1 + Cyclospora cayetanensis, 1 + B. hominis) and 1 EPEC. Parasites (2): Cryptosporidium parvum, Endolimax nana + B. hominis. Others: Shigella flexneri (1), Dengue (1), Rickettsiosis (1).
>30 days, 15 cases (34.9%, average 54 days). Aetiology: E. coli (11): 1 ETEC + E. histolytica and 1 EAEC. Parasites (4): G. intestinalis (1 + B. hominis).
In 3 cases of the last group diarrhoea lasted more than 300 days.
From the beginning of the travel, diarrhoea started in <7 days in 10 cases (23.3%), between 7–15 days in 21 cases (48.8%) and >15 days in 12 cases (27.9%). 20 cases (46.5%) developed invasive diarrhoea. Fever in 16 cases (37.2%) and abdominal pain in 31 cases (72.1%) appeared as associated symptom.
Conclusions: Escherichia coli was the most frequent isolated germen in a important number of parasite-associated cases (multifactorial aetiology?). E. coli diarrhogenic strain was not able to be confirmed by PCR in some cases. In this study we cannot conclude that chronic diarrhoeas are of parasitical aetiology. Microbiological study of faeces of travellers with diarrhoea arriving from tropical countries must be done to rule out potentially treatable bacterial and parasitical aetiology.
P1511 A retrospective study of 230 consecutive patients hospitalised after transcontinental travel
H. Leroy, C. Arvieux, J. Biziragusenyuka, J. Chapplain, C. Guiguen, C. Michelet, P. Tattevin (Rennes, FR)
Objectives: With the current craze for transcontinental travels, infectious diseases consultants practicing in Europe or Northern America hospitals need to be well aware of the distribution of diseases in hospitalised returning travelers and migrants. Our study adds to the previous which tried to characterise causes of consultation and/or hospital admission after traveling. Its purpose is to better define preventive measures in patients who consult before traveling and to guide diagnostic investigations in patients who return ill after traveling abroad.
Methods: We reviewed 230 consecutive patients who were hospitalised in our infectious diseases unit between January 2000 and March 2006 after traveling to another continent. We conducted a retrospective observational study. Cases were identified by our computerised database system. Data were extracted from medical charts, including demographic variables, travel conditions, diagnostic workout, treatment and outcome.
Results: Male to female ratio was 1.58; median age was 33 years and median time from travel to hospitalisation was 13 days. Among the travelers, 52% were tourists, 28% were long term expatriates, 8% worked for humanitarian organisations, 10% visited their relatives or friends, and 26% couldn't be classified. The main destination of travel was sub-Saharan Africa (70.9%), much more frequent than Southern Asia (8.2%) or Northern Africa (8.2%). Malaria was the most common diagnosis (49.1%), followed by gastroenteritis (13.0%), viral hepatitis (6.1%), and bacterial pneumonia (4.3%). A significant proportion of patients were diagnosed with tuberculosis (5.2%) or HIV infection (4.3%) but the link between recent travel and these pathologies could not be ascertained. Rare diseases with potential nosocomial transmission were also observed, including severe acute respiratory syndrome (SARS) and Crimean-Congo haemorrhagic fever.
Conclusion: Even if the study was not limited to febrile illnesses, malaria remains the first cause of hospitalisation after traveling to another continent. Owing to the diversity of tropical pathologies and to the increase of travel to developing countries, this study underscores the need to maintain tropical expertise for physicians in charge of patients who return ill from travel. A substantial proportion of these diseases may be prevented by validated protective measures.
P1512 Wave of undocumented migrants in the Canary Islands: new challenges for European microbiologists
I. Montesinos, M. Miguel, S. Campos, Y. Pedroso, A. Torres, A. Sierra (La Laguna, ES)
Introduction: From June to October 2006, 25,000 undocumented immigrants from West Africa have reached the Canary Islands by boat from the coast of Senegal and Mauritania, i.e. 3 times the number during the first 5 months of 2006 and around five times the total for the whole of last year. Most have arrived in Tenerife. The aim of this work was to evaluate the impact of this immigrants' wave for the laboratory of microbiology of the University Hospital of Canary Islands (HUC).
Methods: The samples received and the results obtained in the section of parasitology were assessed differentiating two periods: January–May and June–October, 2006. Parasite examination was performed in faecal specimens, urine and broncho-alveolar lavage (BAL) after concentration by microscopic exam with iodine, trichrome and Kinyoun staining. Graham test to pinworm screening was also performed. Blood samples were analysed by thin and thick blood films stained with Giemsa stain.
Results: During the first period of study (January–May) approximately 8,000 undocumented immigrants arrived to Canary Islands. During this period the parasitology section of the HUC received a total of 953 samples for detection of parasites: 783 stool samples, 160 Graham tests and 10 blood samples. Only 3 samples were positives for Giardia lamblia (1) and Entamoeba coli (2) and no cases of malaria were detected. From June to October the number of immigrants rose 25,000 individuals. The total number of samples received for detection of parasites ascended to 1,527: 1,239 stool samples, 221 Graham tests, 7 urines, 2 BAL and 58 blood samples. Significant increase in the number of isolates and diversity of pathogens were observed comparing to the first period: Ancylostoma duodenale (2), Ascaris lumbricoides (7), Giardia lamblia (6), Entamoeba coli (29), Endolimax nana (8), Iodamoeba bütschlii (2), Enterobius vermicularis (5), Schistosoma haematobium (2) and Plasmodium falciparum (12).
Conclusion: Both the number and the variety of samples and techniques realised as well as the parasites observed have increased in the last five months coinciding with a new wave of undocumented migrants arrived this summer to the Canary Islands. This should make us think about the need to guarantee the necessary human and material resources as well as training and update courses in parasitology.
P1513 KAP evaluation about travel medicine in international travellers and medical students in Chile
L. Guerrero-Lillo, J. Medrano-Diaz, C. Pérez, R. Chacón, J. Silva-Urra, A. Rodriguez-Morales (Antofagasta, CL; Trujillo, VE)
Objectives: To determinate knowledge, attitude and practices (KAP) about Travel Medicine on International Travelers and Medical Students.
Methods: A KAP instrument was designed, constituted by 24 questions (questionnaire), answered by 100 international travelers (at the “Arturo Merino Benítez” International Airport of Santiago), and 100 Antofagasta's Medical Students (randomly selected), exploring knowledge about Travel Medicine, traveler diseases, morbidity and prevention of them among interviewed people. Analyses were made with SPSS 10 and Epi Info 6 (confidence 95%).
Results: Mean age was 44.9±12.3 y-old (travelers) and 22.6±3.0 y-old (medical students) (p < 0.001). In travelers 83% were universitary professionals. Medical students, 49% were at 3° year. From the total, 78.5% stated as unknowners of Travel Medicine (90% travelers, 67% medical students; p < 0.001); 92% perceived health risks traveling (94% and 90% respectively). Malaria was the disease recognized as more risky (10%; 13% medical students and 7% travelers). From the total, 5% stated get sick during international trips (6% and 4%, respectively). Travelers stated in 56% being non-informed about sanitary conditions of their destinations and 3% have some vaccination (2% YF, 1% HBV). Travelers were 93% chilean going in 60% to tropical countries.
Conclusions: These results indicated the importance of Travel Medicine in those countries where the knowledge of tropical diseases (malaria, dengue, yellow fever, etc), is lower given their inexistence, and where the traveling pattern indicates a great proportion of people visiting risk zones for the acquisition of these pathologies, and without the prevention due to lack of knowledge and immunisation against them.
P1514 Recurrent pyoderma caused by Staphylococcus aureus with Panton-Valentine leukocidin production in a young, male traveller who infected his non-travelling partner and in a young, female traveller returned from a tropical country
R. Schleucher, M. Gaessler, B. Schulte, M. Marschal, J. Knobloch (Tübingen, DE)
Objectives: Pyoderma associated with visits to tropical countries is well known. In most of the cases pyoderma is caused by Staphylococcus aureus. After a sufficient antibiotic treatment the skin infections resolve without any sequelae. But several patients suffer from recurrent pyoderma. The reason is not elucidated so far. Panton-Valentine leukocidin is a cytotoxin produced by Staphylococcus aureus. It is associated with severe necrotic lesions involving skin and mucosa. Inter alia Panton-Valentine leukocidin was isolated in several outbreaks of community-acquired methicilin-resistant Staphylococcus aureus caused infections which were associated with a high contagiosity.
Methods: We describe two cases of travellers returned from tropical countries with recurrent pyoderma. In the first case within nine month after returning from Fiji a young, male, healthy patient relapsed seven times on different parts of the body. Seven month after the first skin infection the girl friend of this patient developed a pyoderma as well. In the second case a young, female, healthy patient suffered eight times from a recurrent conjunctivitis accompanied by pyoderma within one year after returning from Ghana.
Results: In all cases Staphylococcus aureus producing Panton-Valentine leukocidin was isolated. In the skin lesions of the first patient and his girl friend Staphylococcus aureus of the same strain was detected. In the second case the same strain and Panton-Valentine leukocidin production were found in Staphylococcus aureus of the nasal mucosa and conjunctival swab.
Conclusion: For the first time recurrent pyoderma in young, healthy travellers caused by Staphylococcus aureus with Panton-Valentine leukocidin production is described. The relapses and the infection of the partner demonstrate the high contagiosity and virulence. Additional nasal carriage represents a source of reinfection. Further studies are necessary to elucidate the part of Panton-Valentine leukocidin positive Staphylococcus aureus in the increasing problem of recurrent pyoderma in healthy travellers. The results could be highly important for the public health worldwide.
Infections in the tropics
P1515 Relapses versus reactions in multibacillary leprosy: proposal of new relapse criteria
K. Linder, M. Zia, W. V. Kern, R.K.M. Pfau, D. Wagner (Freiburg, DE; Karachi, PK)
Objective: The WHO criteria for the diagnosis of relapses in leprosy rely on a marked increase in the bacterial index at any single site and evidence of clinical deterioration. Confirmation of the diagnosis would require growth of Mycobacterium leprae in the mouse footpad system (MFP) which rarely is available in the field. Relapses and isolated late reactions have different times of onset after the end of treatment, but are difficult to distinguish by clinical examination. A simple, practical and sensitive scoring system that circumvents these problems is needed for a reliable diagnosis of relapses in leprosy. In 2006 the WHO suggested to include time after stopping treatment as distinguishing feature for the diagnosis of paucibacillary leprosy. We hypothesised that criteria to diagnose multi-bacillary (MB) relapses should also include a time criterion.
Methods: We collected data on all relapses diagnosed between 1998 and 2004 at the Marie-Adelaide-Centre in Karachi, Pakistan, including case histories, clinical manifestations, follow-up, bacterial indices, treatment and contacts. We developed a simple scoring system for the diagnosis of MB-relapses and validated it on a data-set of MFP-confirmed relapses (Shetty et al. 2005; Lepr Rev 76, 241). Its sensitivity was further evaluated in the Karachi relapse cohort. The p value was calculated with McNemar's test with continuity correction.
Results: The new scoring system that combines time factor, risk factors and clinical presentation at relapse (see table ), had a higher sensitivity in MFP-confirmed relapses than the WHO-criteria (95% versus 65%, p < 0.01). The sensitivity of the scoring system was also significantly higher than the WHO criteria in the 57 cases of MB-relapses diagnosed in Karachi (75% versus 54%, p < 0.05).
Proposed diagnostic scoring system for MB-relapses in leprosy
| Factor | Scorea |
|---|---|
| I. Time factor | |
| Time after release from treatment [months] | |
| ≤12 | 0 |
| 12–24 | 1 |
| 25–60 | 2 |
| >60 | 3 |
| II. Risk factor | |
| If the initial bacterial index [BI] is ≥3+ | 1 |
| Ill. Clinical presentation at relapse | |
| If the BI in a single lesion is ≥2+ higher than expectedb | 1 |
| If the average BI is ≥2+ higher than the expected BI | 1 |
| If no signs of a reactionc are present | 1 |
Relapses are diagnosed with a score of ≥3; the maximal score = 7: the maximal score for the time factor = 3, the maximal score for the additional factors = 4, these scores are added to give the final score.
Expected BI = calculated BI with an assumed fall of 1 log-unit/year; if the initial BI was negative, a positive BI at relapse is sufficient to score.
Clinical signs of inflammation of the nerve or the skin or erythemata nodosa leprosa.
Conclusion: This new simple scoring system for diagnosing MB-relapses in leprosy should be further validated in a prospective study to confirm its superior sensitivity and to evaluate the specificity of these criteria by using MFP-confirmation for patients presenting with signs of activity after treatment.
P1516 Spectrum of gastrointestinal infections among 420 cases in population without regular access to drinking water in Kenya
Z. Kniezova, V. Švábová, M. Bartkovjak, M. Kiwou, J. Johnson, Z. Njambi, V. Krcmery, M. Krumpolcova, S. Kamoche (Bratislava, SK)
Objectives: The purpose of the study was to describe aetiology, risk factors, therapy and outcome of gastrointestinal infections (GI) in slum population in Mukuru without adequate access to drinking water.
Patients and Methods: Community clinic in Mukuru slum serves the poorest population. The number of patients visiting the clinic is approximately 30,000 pts per year and 2,500 per month. Within June 420 cases of diarrhoeal diseases have been documented. Microscopic evaluation of stool for ova and cysts was performed in every patient with diarrhoea or anaemia.
Results: Majority of cases (120) were due to Giardia (29%), 40 (10%) due to Entamoeba histolytica. Serology for Salmonella typhimurium was positive in 42 patients (10%). Ascaris lumbricoides (20 cases, 5%) was the most common worm pathogen, followed by Ankylostoma duodenale (9 cases, 2%).
Metronidazol for giardia and amoeba diarrhoea (243 pts) and mebendazol (45 pts) were the most commonly used antiparasitic agents and TMP/SMX in 65 cases and chloramphenicol in 40 cases resulted to 100% efficacy in treated cases.
Conclusion: Only 23 of 420 cases were HIV positive (5%), however only 120 of 420 pts tested (14% HIV prevalence among tested). Surprisingly, despite of high HIV prevalence, no cryptosporidium cysts were found.
P1517 An unusual presentation of acute brucellosis with thrombocytopenia and maculopapular rash
O. Alici, B. Kasapoglu, R. Alkan, E. Sarifakioglu, R. Akgedik, R. Bozalan, A. Kosar, H. Sahin (Ankara, TR)
Purpose: We report a case of brucellosis together with severe thrombocytopenia and skin rash that does not related to thrombocytopenia.
Case report: A 69-year-old woman, who was a housewife, was seen for complaints of extensive pruritic skin lesions over the trunk and extremities, knee pain and generalised myalgia over the previous one month. These symptoms were accompanied by sweating, fatigue, anorexia, bleeding of gum tissues and nosebleed. There was no preceding episode of any kind of infection or any medication use. She used to eating cheese made from raw milk.

Physical examination revealed that her temperature was 36.7°C. She had modarate splenomegaly (2 cm palpable below the costal margin). The characteristic feature of skin rash was as maculopapular exanthema and covered the trunk, arms and legs. The intensity of the rash was more over trunk than extremities and the lesions were pruritic.
Laboratory analysis yielded a platelet count of 15,000 g/L. Erythrocyte sedimentation rate of 8 mm/h, C-reactive protein level of 9.21 mg/L (0 to 8), alanine aminotransferase level of 56.34 IU/L (1 to 31), and aspartate aminotransferase level of 33 IU/L (1 to 32). Abdominal ultrasonography showed only splenomegaly. Bone marrow biopsy was normocellular. Because of the patient's sweating, generalised myalgia and being from a rural community Brucella agglutination test was performed and revealed 1/2,560 positivity. Blood cultures were sterile. Doxycycline (200 mg/day) and rifampin (600 mg/day) were started. According to dermatology consultation, skin rash was thought as brucellar dermatitis. The patient rejected skin biopsy.
The rash began to disappear within two days and thrombocytopenia began to resolve within three days of therapy commencement. The patient was discharged from the hospital 7 days later in good health on rifampicin and doxycycline therapy. At the 6-week follow up, the patient had came to control and had made a full clinical recovery; skin rash recovered completely and platelet count increased to normal levels.
Conclusions: It is known that cutaneous lesions and thrombocytopenia are not specific to brucellosis. Although this event could be considered rare, brucellosis should be kept in mind in the investigation of aetiology of thrombocytopenia and maculopapular rash with absence of fever in Brucella-endemic areas.
P1518 Serum copper and zinc levels in patients with brucellosis
M.F. Geyik, C. Ustun, I. Tegin, S. Hosoglu, C. Ayaz (Diyarbakir, TR)
Objective: The aim of this study is to compare the serum Copper (Cu) and Zinc (Zn) levels of patients with brucellosis and healthy individuals.
Material and Methods: In this study, 43 patients with brucellosis, and 32 healthy individuals in the control group were included. From totally 75 individuals, 5 mL of venous blood was taken after fasting 10 hours at night. Serum samples were decomposed by centrifugeting at 5,200 turn for 5 minutes. Serum samples were diluted with deionised water. Cu and Zn levels were measured in all serum samples by using Unicam 929 Atomic Absorption Spectrophotometer. All data entry and analysis were performed using SPSS 10.0 for Windows Version programme.
Results: The average age of 43 patients with brucellosis was 39 years; 21 were males (49%), 22 were females (51%). The average age of 32 healthy individuals was 32 years; 16 were males (50%), 16 were females (50%). Serum Cu and Zn levels of patients with brucellosis were found to be 89 μg/dL and 38.6 μg/dL, respectively. Serum Cu and Zn levels of healthy individuals were found to be 57.2 μg/dL and 55.9 μg/dL respectively. A statistically significant (p = 0.0001) increase at serum Cu levels and decrease in Zn levels of patient with brucellosis were detected, when compared with the control group.
Conclusion: The current evidence suggests that, Cu and Zn levels may be helpful as indicator in the course of brucellosis.
P1519 Brucella as a rare cause of cirrhosis in a southern Italy endemic area
T. Ascione, M. Iannece, P. Rosario, M. Onofrio, S. Quaranta, R. Pempinello (Naples, IT)
Background: Brucella infection may be manifest by nonspecific hepatic inflammation or by granulomatous hepatitis. Cirrhosis may be a rare complication of an untreated Brucellosis. Liver and spleen involvement is commonly described during Brucella infection. We reported a case of cirrhosis whose only cause was Brucella infection.
Case report: A 60-year-old woman was referred to our hospital because of gastrointestinal bleeding caused by use of FANS for a 10 months history of unknown origin fever. At admission the patient presented elevated liver enzymes (ALT, GGT, ALP) and elevated bilirubin levels. Low level of albumin (2.5 g/dl) and prothrombin were present. PLT count was 97,000/mmc. HBsAg, HBcAb, anti-HCV, anti-HIV, HBV-DNA and HCV-RNA were negative. Immunologic markers of AIH (ANA, AMA, ASMA, p-ANCA, anti-LKM1) were negative too. Haemochromatosis was excluded of the basis of genotyping testing. No history of alcohol abuse was reported. Abdominal ultrasonography showed enlarged spleen (longitudinal diameters of spleen was 157 mm) and ascites. An endoscopic exam showed gastric ulceration related to FANS use and F2 varices of the distal tract of the oesophagus. Serologic tests for Brucellosis was positive (1/640) and Brucella melitensis grew from blood cultures. The patient was treated with doxycycline and ciprofloxacin for six weeks. Fever disappeared after 2 weeks of therapy. Liver enzymes and bilirubin level improved returning within the normal range after 8 weeks. Three months after the end of therapy an ultrasonographic exam showed disappearance of ascites, reduced spleen longitudinal diameter (132 mm), low platelets (150,000/mmc) and an improved level of albumin (3.7 g/dl).
Conclusions: Although liver involvement is frequent in patients with brucellosis because of both the direct brucella infection effect and the host immune response, progression to cirrhosis is rarely reported. In this case diagnosis of Brucellosis was obtained only after a severe impairment of liver function and portal hypertension appeared. We suggest that brucellosis has to be considered both in cases with criptogenetic cirrhosis or when a progressive worsening of liver functions occurs in patients with an otherwise stable liver disease, if a history of exposure in endemic areas is reported.
P1520 The evaluation of 23 anthrax patients: dilemmas in diagnosis and our experiences from 2004 to 2006
H. Ozcan, U. Kayabas, Y. Bayindir, M. Bayraktar (Malatya, TR)
Objectives: Anthrax is a potentially fatal, zoonotic disease. Naturally occurring anthrax in humans is acquired from contact with infected animals or contaminated animal products. The diagnosis of cutaneous anthrax (CA) may be very difficult especially in rarely seen regions or in atypical presentations. The aim of this study is to evaluate the clinical features of 23 CA cases between May 2004 to September 2006.
Methods: Twenty-three patients with CA were included in this study. The diagnosis of the CA was based upon clinical findings and/or microbiological procedures.
Results: Of the 22 cases followed up with CA and one patient with CA resulted to anthrax sepsis, 13 (56.5%) were male and 10 (43.5%) were female. The mean age of the patients was 36.04±14.67 years. All of the patients have been living in the rural area of Malatya province in Turkey. Twenty-two patients had a history of slaughtering, skin peeling, meat and bone processing or helping in one of these activities. One patient did not have any history of contact with animals or animal products. He was a construction worker in the village where an attack of CA was seen a year ago. He had only contacted with soil. Clinical presentation of CA was typical in 20 patients. Two patients were misdiagnosed as insect bite and one patient as angioedema.
The vesicular fluid cultures were positive for Bacillus anthracis in seven (30.43%) cases and Gram stain revealed Gram-positive rods in eight (34.78%) cases. The diagnosis of CA in remained 15 (65.21%) cases was made by clinical presentation and history of contact with the sick animal. Nine of these cases have history of contact with same sick cow. And also, B. anthracis was detected in the spleen of this cow.
Conclusion: CA is a very contagious and important infectious disease in worldwide. Early and accurate diagnosis can dramatically affect the prognosis of the disease. So, CA should be kept in mind especially in endemic regions.
P1521 Enteric fever due to Brucella melitensis: a case report
A. Erbay, H. Bodur, E. Akinci, A. Bastug, M.A. Cevik (Ankara, TR)
Objectives: Although pathogenetically qualifying as an enteric fever, the gastrointestinal manifestations of brucellosis in humans are relatively uncommon. Alimentary tract complaints such as anorexia, nausea, vomiting, abdominal pain, diarrhoea or constipation are elicited in patients with brucellosis but systemic symptoms generally are more common than symptoms localised to the gastrointestinal tract. A patient with enteric fever caused by Brucella melitensis is reported.
Case: A previously healthy 16 year old male was admitted with fever, vomiting, diarrhoea and skin rash. His initial complaint was started with fever six days before admission. He was living at a village and had a history of consumption of unpasteurised dairy products. Physical examination revealed an acutely ill boy with a temperature of 39.9°C. Blood pressure was 110/60 mmHg and pulse was 96/min. He had abdominal tenderness and hepatosplenomegaly. Maculopapuler rashes with a diameter of 1–2 mm were present on the trunk and arms. After admission, vomiting, abdominal pain and melena were observed. Pertinent laboratory findings were as follows: white blood cell (WBC) count 3,000/mm3 haemoglobin 12.6 g/dl, platelet count 44,000/mm3, erythrocyte sedimentation rate 9 mm/h, ALT 230 U/L, AST 169 U/L, CRP 77.4 mg/dl. The direct microscopic examination of faecal smear showed prevalent leukocytes. A faecal occult blood test was positive. Initial and follow up Widal tests were negative. Serological tests for acute viral hepatitis were negative. A brucella serum agglutination test was positive, with a titre of 1:1,280. Stool culture revealed no evidence of bacterial pathogens. Brucella melitensis was isolated from the cultures of blood. An upper gastrointestinal endoscopic examination revealed bulbitis. Combined therapy with rifampicin (600 mg/d p.o.) and doxycycline (100 mg p.o., b.i.d.) was started. On the 3rd day of therapy the patient became afebrile. In the following days the patient improved clinically. The faecal occult blood test became negative on the 7th day. A repeated CRP was 18.9 mg/dl. On the 12th day of the therapy laboratory test were as follows; WBC count 5,100/mm3, haemoglobin 12.8 g/dl, platelet count 275,000/mm3, alanine aminotransferase 19 U/L and aspartate aminotransferase 41 U/L. Antibiotic therapy was stopped on day 42.
Conclusion: Brucellosis is an infection with multiple presentations, and in an endemic region a thorough history of exposure and clinical suspicion are required.
P1522 Preferential sites of eschar for scrub typhus
D.M. Kim, S.H. Lee (Gwang-Joo, Nam-won, KR)
Background: Eschar has been shown to be an important finding for the diagnosis of scrub typhus patients, and other mite-born rickettsiosis patients. However, any reports on systemic studies, including the difference of the gender regarding the preferential site of eschar as well as a schematic diagram showing the preferential site of eschar, have been rarely reported.
Materials and Methods: IFA test using serum for scrub typhus was performed on the adult patients who visited Chosun University hospital and its branches for diagnosing their acute febrile diseases that developed within the previous 4 weeks. The presence or absence of eschar was thoroughly examined.
Results: Eschars were preferentially formed on the front of body. The sites in the male where eschars were primarily detected was the area within 30 cm below the umbilicus (19 patients, 34.5%). The lower extremities and the front chest above the umbilicus were 23.6% (13 patients) and 20.0% (11 patients) respectively. However, on the females and different from the males, the most prevalent area was the front chest above the umbilicus for 40.4% (46 patients) of all the detected eschars.
Conclusions: For both the males and the females, the front and the rear 30 cm below the umbilicus should be examined thoroughly, and it is thought to be important to carefully examine the lower extremities of the males, and the front chest as well as the back area of the females. Our study is the first report that shows the difference between the males and the females in regard to the area of developing eschars on scrub typhus patients.
P1523 Diagnosis of scrub typhus by immunohistochemical staining of Orientia tsutsugamushi in cutaneous lesions
D.M. Kim, S.H. Lee, S.C. Lim (Gwang-Joo, KR)
Introduction: The aim of this study is to assess the clinical usefulness of an immunohistochemical staining on paraffin-embedded skin biopsy specimens for the diagnosis of scrub typhus with a comparison of indirect immunofluorescent antibody (IFA) assay.
Method: A prospective study of patients with possible scrub typhus was conducted from September, 2005 to August, 2006. The patients participating in the study were thoroughly examined for the presence or absence of eschar like lesions and maculopapular lesions. Skin biopsy specimens for immunohistochemical staining (IHC) were obtained using a 3 mm punch upon the consent of the patient.
Result: 125 potential scrub typhus patients were studied prospectively. Skin biopsy specimens were obtained from 63 patients. To minimise the effects caused by antibiotics on the IHC, 46 patients were assessed prior to the administration of antibiotics (except for the 4 patients who had received prior antibiotic therapy before admission and 13 patients who were underwent skin biopsy after administration of antibiotics at our hospital). Compared with the result of IFA which is the definite diagnostic method for scrub typhus, the result of IHC of maculopapular skin lesions demonstrated a sensitivity of 65% (95% confidence interval 0.38–0.85) and a specificity of 100% (95% confidence interval 0.6–1). The result of IHC of eschars demonstrated a sensitivity of 100% (95% confidence interval 0.82–1) and a specificity of 100% (95% confidence interval 0.40–1). IHC of eschar from one of the 13 patients who was administered antibiotics at our hospital showed negative results on the 13th day of the initial antibiotic administration. IHC done within 3 to 4 days following the initial antibiotic treatment in the remaining 12 patients showed almost the same results as that of the patients who did not receive prior antibiotics.
Oriential antigens, discrete coccobacilli were primarily located within the cytoplasm of the infected endothelial cells, and they were also identified within macrophages around the blood vessels on IHC staining. Intracytoplasmic positive staining of the lining epithelia of the sweat ducts and glands in the mid- and deep-dermis were also found.
Conclusion: The immunohistochemical staining of skin biopsy specimens, particularly from eschars is sensitive and specific, which can be a reliable test for confirmation of the diagnosis of scrub typhus.
P1524 Preparation of recombinant antigen from Orientia tsutsugamushi Ptan strain and development of rapid diagnostic reagent of scrub typhus
J. Tang (Nanjing, CN)
Objectives: Preparation of recombinant antigen from O. tsutsugamushi Ptan strain and then a mixture of the r56 of Gilliam and Ptan strain was used as the diagnostic antigen to develop a rapid colloidal-gold immunochromatographic assay (CIA) for diagnosis of scrub typhus.
Methods: A 1,352 bp fragment encoding a truncated 56kD outer membrane protein of Ptan strain, was amplified by PCR and then was cloned into the pET28a vector for expression. The recombinant protein containing six his tag was purified by Ni2+ chromatography column, analysed by SDS-PAGE and identified by western-blot. A rapid colloidal-gold immunochromatographic assay (CIA) was developed to detect the serum anti-O. tsutsugamushi total antibodies, IgG and IgM by employing the mixture of the truncated 56kD recombinant antigen from Gilliam and Ptan strain as the diagnostic antigen.
Results: Resulted by SDS-PAGE the expression of a protein with approximately 52 kD MW was detected when the recombinant plasmid was transformed into the host E. coli BL21 and no similar protein was observed when the empty plasmid of pET28a was transformed into the same host.
Resulted by western-blot the recombinant protein can be recognized by patient's positive serum while the negative serum cannot have the same blot band, showing a good antigenicity. The performance of CIA was evaluated with a panel of 112 sera from clinical confirmed cases of scrub typhus. The CIA's detection sensitivities against anti-O. tsutsugamushi total antibodies, IgM and IgG were 98.2%, 81.2% and 94.6% respectively while IFA's sensitivity against IgG was 85.7%. The specificity of CIA against anti-O. tsutsugamushi total antibodies, IgM and IgG were 98.1%, 100% and 98.9% respectively while the specificity of IFA against IgG was 98.9%.

Left: CIA strip for detecting anti-O. tsutsugamushi total antibodies in serum: (a) positive and (b) negative result. Right: CIA strip for diagnosis of anti-O. tsutsugamushi IgG and IgM in serum: (a) IgG+/IgM+, (b) IgG+/IgM–, (c) IgG–/IgM+, (d) IgG–/IgM–.
Conclusion: CIA was a good assay and could substitute the conventional IFA method in the diagnosis of scrub typhus.
P1525 Brucella multi-organ involvement in a patient with negative serologic tests: case report
F. Khorvash, A. Hassanzadeh Keshteli, M. Behjati, M. Salehi, B. Ataei (Isfahan, IR)
Background: Brucellosis is a multi-system disease that may present with a broad spectrum of clinical manifestations. Whilst hepatic involvement in brucellosis is not rare, it may rarely, involve the kidney or present with cardiac manifestations. Central nervous involvement in brucellosis some times can cause demyelinating syndromes. Here we present a case of brucella hepatitis, myocarditis, acute disseminated encephalomyelitis, and renal failure.
Case presentation: A 26 year old man presented with fever, ataxia, and dysarthria. He was a shepherd and gave a history of low grade fever, chilly sensation, cold sweating, loss of appetite, arthralgia and 10 Kg weight loss during previous 3 months. He had a body temperature of 39°C at the time of admission. On laboratory tests he had elevated levels of liver enzymes, blood urea nitrogen, Creatinine, Creatine phosphokinase (MB), and moderate proteinuria. He had abnormal echocardiography and brain MRI too. Coombs test and enzyme-linked immunosorbent assay for IgG, IgM were negative. Standard tube agglutination test (STAT) and 2-mercaptoethanol (2-ME) titers were 1:80 and 1:40 respectively. Finally he was diagnosed with brucellosis by positive blood culture and polymerase chain reaction for Brucella melitensis.
Conclusion: Brucella infection should be ruled out in patients with multi-organ involvement, especially in ones from endemic regions. We can come to this conclusion that we may not be in the safe side from brucellosis only with low STAT and 2-ME titers and other diagnostic methods like blood culture and PCR should be administered to rule out brucellosis.
P1526 Two immunological test to diagnostic Carrion's disease (bartonellosis) in patients of endemic areas in Peru
S. Palma, M. Villar, M. Gonzales, C. Prause, P. Ventosilla (Lima, PE)
Bartonella bacilliformis is the aetiological agent of human bartonellosis, which has the acute phase (with fever, severe haemolytic anaemia) and chronic phase (with bacillary angiomatosis-like lesions). The methods of diagnosis are smears (27% of sensibility) and culture (take from 7 to 45 days).
The main goal of this study was detected inmunoglobulins anti-Bartonella bacilliformis using enzyme linked inmunoabsorbent assay (ELISA) and Western Blot.
Methods: Bartonella bacilliformis strain ATCC 35685 was cultured on Columbia Agar supplemented with 10% sheep desfibrinated blood. The cells were harvested, sonicated and diluted in solution buffer, this was the antigen. For ELISA (to detection of Ig G) we had 34 sera of acute, chronic patients and control negative. And for Western Blot, we used pool sera of 9 acute and chronic patients, and 8 negative control sera.
Results: In ELISA the sensitivity was 82% and specificity was 70% in acute phase patients, and in chronic phase patients was 91% and 76% respectively.
For Western Blot, we found 26 protein bands (2.5–132 kDa), and 3 antigenic bands (12–47 kDa) for acute phase and 2 antigenic bands (11, 12 kDa) for chronic phase.
Conclusions: ELISA could be used as screening test because the sensitivity is higher than current methods employed for the diagnosis of bartonellosis. In Western blot founded differences between acute and chronic phase as preliminary results.
Emerging viral infections
P1527 A new type of TT virus in eastern Taiwanese indigenes
H.-F. Liu, K.-L. Hsiao, H.-H. Lee, C.-L. Lin, Y.-J. Lee, L.-Y. Wang (Taipei-Tamshui, Keelung, Hualien, TW)
Objectives: Torque Teno virus (TTV), the first circular negative-stranded human DNA virus, has a very high prevalence among general population. Early studies of prevalence were mostly based on the N22 region which has now been shown to strongly underestimate the prevalence of TTV infection. Although TTV infection has been reported in Taiwanese populations, they were all based on the N22 region. In the present study, we investigated the prevalence of TTV in eastern Taiwanese indigenes by using newly designed PCR primers deduced from a highly conserved untranslated region (UTR) and performed phylogenetic analysis using almost entire open reading frame (ORF).
Methods: Serum or plasma samples from 140 eastern Taiwanese indigenes were included in this study. PCR were performed on the highly conserved UTR region (about 150 nucleotides) to detect the prevalence and a long one amplifying the entire ORF (around 3.2Kb) for phylogenetic analysis. Phylogeny reconstruction was performed using the Phylip software package, with the neighbour-jointing, the maximum parsimony, and the maximum likelihood methods. The robustness of the neighbour-jointing and maximum parsimony trees was statistically evaluated by bootstrap analysis with 1,000 bootstrap samples.
Results: A very high prevalence (95%) of TTV infection was found in the eastern Taiwanese indigenes based on the highly conserved UTR region. Direct sequencing of these PCR products found a severe mixed infection of many different TTV strains. By cloning, at least 13 isolates belonging to 3 different TTV groups could be identified from one individual. Phylogenetic analysis showed a consistent tree topology by all 3 methods suggesting that TTV could be classified into 6 monophyletic groups including a new one consisted of 11 isolates from an eastern Taiwanese indigene.
Conclusion: TTV are highly prevalent in eastern Taiwanese indigenes and a new TTV group was identified from this population. Severe mixed infection of many different TTV strains in one individual was observed. Since TTV can cause persistent infection and various different strains can be found within one individual, this suggested that TTV might have a quasispecies-like nature which was only observed in some RNA viruses before.
P1528 Comparison of human metapneumovirus strains from Alto Adige with strains from surrounding regions in Italy and Austria
E. Pagani, C. Petters, D. Secolo, P. Rossi, M. Casini, I. Cavattoni, E. Morello, L. Pescolderungg, G. Campanini, E. Percivalle, H.P. Huemer, C. Larcher (Bolzano, Pavia, IT)
Human Metapneumovirus (HMPV), in addition to paediatric patients, has been increasingly described in elderly people and has been found to cause more severe infections in immunosuppressed patients. In our laboratory in Bolzano serving a regional hospital routine HMPV diagnosis is based on direct immunofluorescence assay (DFA) using monoclonal antibodies. Species diagnosis is confirmed by reverse transcriptase-PCR. During 2005 we performed a preliminary epidemiological survey to compare the local strains to those of surrounding regions by doing also sequencing analysis of the N-Genes as well as the L-genes of the obtained isolates. By alignment with established Dutch reference strains we identified three different genotypes circulating in our region with predominantly type B strains being detected in this season. This is in contrast to published data from nearby Italian regions from the 2003/2004 winter season where predominantly A2 strains have been characterised. Also our study in the Austrian Tyrol region from the same season detected A-type strains only. Whether this is a further indication for similar strains cycling in a wider geographic area with the respective types replacing each other on a seasonal basis remains to be clarified although absolute sequence identity due to the high variability of RNA viruses was rarely detected. Similar HMPV types circulated in both the paediatric and adult patients and, of interest, the two isolates originating from bone marrow transplant recipients (BMT) were of different genotypes. Both findings rather exclude hospital transmission in our BMT patients as has been suspected in lung transplant patients (Larcher et al., J Heart & Lung Transplant, 2005).
P1529 The distribution of human papillomavirus infection in the male urogenital tract in Russian urological patients
V. Smelov, A. Gorelov, J. Pleijster, A. Savicheva, S. Morre (Saint-Petersburg, RU; Amsterdam, NL)
Objectives: The prevalence of symptomless genital human papillomavirus (HPV) infection among men is not well established. This is partly due to a poor understanding of which sites should be tested. Up to now there is no published study regarding the HPV distribution in Russian male population. HR-HPV types are accepted to be involved in the aetiology of cervical cancer and this has lead to new vaccine approaches. The influence of HPV infection on the risk of cancers among men, i.e. prostate cancer, is still under investigations. The goal of this study was to investigate the HPV infection presence in the urethral and prostate samples in the Russian urological patients.
Methods: The urethral and prostate samples of 210 men (mean age 31.7, range 16–60 years), who visited 2 urology clinics in St-Petersburg, were sequentially collected from May through to October 2006 to be tested against sexually transmitted infections and HPV. No specific selection was made to the obtained this study group.
The presence of high- and low-risk HPV (HR- and LR-HPV, respectively) DNA in the distal urethra and expressed prostate secretion (EPS) was investigated in Amsterdam by the GP5+/6+ PCR assay followed by subsequent HR- vs LR-HPV typing using EIA (enzyme immunoassay). The oligoprobe cocktails for 24 HR-HPVs (i.e., 16, 18, 31, 33, 35) and 21 LR-HPVs (i.e., 6, 11, 32, 40, 42) were used for the group-specific detection.
Results: The results are presented in the Table . The HPV distribution in the urethra and prostate was detected as 16.2% and 13.3% for HR- and 6.2% and 5.7% for LR-HPV infection, respectively. Concordance between LR- and/or HR-HPV distribution in both anatomical sites was observed in only 20–25%. We observed “grey zone” values in 5.7%; these samples will be re-tested.
Table 1.
HPV distribution and HR/LR concordance in the distal urethra and expressed prostate secretion (EPS) samples in the Russian male urological patients
| HPV distribution, n (%) |
||
|---|---|---|
| HR | LR | |
| Urethra | 34 (16.2%) | 13 (6.2%) |
| EPS | 28 (13.3%) | 12 (5.7%) |
| HR/LR concordance |
||
|---|---|---|
| Urethra/EPS | HPV+/210 | |
| HR/LR | 4 | |
| HR/HR | 11/49 (22.5%) | 23.3% |
| LR/HR | 2 | |
| LR/LR | 5/21 (23.8%) | 10.0% |
| HR+LR/HR+LR | 2 | |
Conclusions: The presence of HR- and LR-HPV types in the distal urethra and EPS of men was investigated and compared in this study for the first time.
The total HR- and LR-HPV prevalence was detected in this group as 23.3% and 10.0%, respectively; with a low concordance rate between the two anatomical sites studied.
At the moment we are in progress to: (1) determine HPV types distribution; (2) extend the study group; and to investigate the HPV distribution in men with: (3) co-infection, i.e. Chlamydia trachomatis, and (4) chronic urological disease, i.e. chronic prostatitis.
P1530 CMV reactivation and antiviral immune response in patients with septic shock
L. von Müller, M. Principi, A. Klemm, N. Durmus, M. Schneider, M. Weiß, H. Suger-Wiedeck, T. Mertens (Homburg/Saar, Ulm, DE)
Cytomegalovirus (CMV) is discussed as a pathogen of emerging evidence in critically ill patients without immunosuppressive therapy. Sixty-five patients patients with septic shock were recorded and prospectively monitored for active CMV infections using quantitative pp65-antigenaemia and viral isolation. Forty-three CMV seropositive patients with a prolonged ICU stay (<7 days) were included. Within two weeks fourteen patients (32.6%) developed an active CMV infection with low pp65-antigenaemia (median 3 positive per 500,000 WBCs). CMV reactivations occurred despite CMV specific Th1-cell functions. The active CMV infections turned negative without antiviral therapy after twenty days, on average. Following active CMV infection the frequency of CMV and of superantigen (SEB) reactive Th1-cells significantly increased which means that patients with septic shock were capable to mount an antiviral immune response and to repress active CMV infection. In patients without active CMV infection the frequencies of reactive Th1-cells remained low. In parallel active herpes simplex virus (HSV) infections were detected in sixteen patients using bronchial aspirates. The HSV and CMV reactivations were associated and occurred at the same time. ICU treatment and the need of mechanical ventilation was significantly prolonged in the group with active CMV infection and we suggest that viral reactivation could increase morbidity of patients with septic shock. Early antiviral therapy aimed at preventing viral associated morbidity of CMV seropositive patients with septic shock should be evaluated in future clinical as a new treatment option.
P1531 Persistence of the poliovirus genome in the cerebrospinal fluid of patients affected by post-polio syndrome
A. Baj, S. Monaco, G. Zanusso, F. Molteni, A. Toniolo (Varese, Verona, Costamasnaga, IT)
Objectives: Polioviruses (PV; three types) are small, non-enveloped, positive-strand RNA viruses belonging to the Enterovirus genus of Picornaviridae. Twenty to thirty years after being hit by paralytic poliomyelitis, over 50% of patients develop the so called “post-polio syndrome” (PPS). PPS is characterised by slowly progressing muscular weakness, chronic pain, fatigue, and other symptoms. The cause of PPS is likely due to distal degeneration of enlarged post-poliomyelitis motor units. Virus persistence in the central nervous system (CNS) has long been investigated with controversial results.
Methods: PPS patients aged 50–65 years have been investigated. The PV genome has been searched for in cerebrospinal fluid (CSF) samples using RT-PCR with primers directed to different genomic regions. The utilised amplification methods were capable of detecting <10 genome equivalents per reaction tube. Direct sequencing of purified amplicons allowed identifying the persisting PV serotype. CSF samples from patients with non-infectious pathologies were used as controls.
Results: All investigated patients (11/11) were positive for the presence of PV genomic fragments. The 5′ untranslated sequence represented the most sensitive target for molecular detection. Sequencing of VP1 and 2A tracts revealed that the PV type-1 genome was present in most patients. Infectious virus was isolated in cell culture from a single patient undergoing orthopaedic surgery. Complete sequencing of the isolate is expected to shed light on molecular mechanisms of PV persistence. No PV amplicons were detected in CSF samples from control patients.
Conclusion: By gene amplification and genome sequencing, we have shown that PV genomes are able to persist for long periods of time (i.e., >30 years) in the CNS of PPS patients. Though the contribution of viral persistence to PPS pathogenesis is still undefined, our findings may contribute to introducing new molecular tools for PPS diagnosis. The finding of persistent PVs also indicates the need of exploring innovative therapeutic methods for PPS patients. PPS, in fact, represents the most prevalent motor neuron disease today.
Support from Cariplo Foundation, Varese and Milano (Italy) is gratefully acknowledged.
P1532 Serological evidence of hantavirus and arbovirus infections among acute febrile patients in Uzbekistan
A. Soliman, K. Earkart, E. Mohareb, E. Musabaev, S. Vafakulov, N. Yarmuhamedova, M. Monteville (Cairo, EG; Tashkent, Samarqand, RU)
Objectives: Hantavirus and arbovirus infections are known to occur in Eastern Europe and Central Asia, particularly in Kazakhstan and Turkmenistan. These viral agents have not, however, been reported in neighbouring Uzbekistan.
Methods: We are currently conducting hospital based surveillance for undifferentiated fever in Tashkent and Samarqand, Uzbekistan. Following study enrollment, acute sera was collected from 817 patients presenting with acute febrile illness between October 2004 and May 2006. Convalescent sera were collected upon discharge or 2–3 weeks after the first specimen was drawn. Sera were screened at 1:100 dilutions for IgM antibodies to Hantavirus, West Nile (WN), Sindbis (SIN), Sandfly Naples (SFN) and Sandfly Sicilian (SFS) viruses using Focus® IgM-EIA kits (Cypress, California) for Hantavirus and an IgM capture-ELISA developed at the U.S. Naval Medical Research Unit-3, Cairo, Egypt. Samples testing positive by ELISA for arbovirus were confirmed by a plaque reduction neutralisation test (PRNT).
Results: A total of 127 (15.5%) patients were determined IgM positive to Hantavirus with rates of 6.1% (11/181) and 18.2% (116/636) in Tashkent and Samarqand, respectively. Six patients (0.7%) had anti-SFS IgM antibodies (titers ranged 1:400 to 1:6,400), all confirmed by PRNT. All specimens had no IgM titers against the other viruses tested. Clinical and epidemiological data of patients will be described.
Conclusions: Hantavirus infection may constitute a significant proportion of undiagnosed acute febrile illness particularly in Samarqand (rural area) in Uzbekistan. This is the first report of Hantavirus and SFS virus infections in Uzbekistan.
P1533 Crimean-Congo haemorrhagic fever and haemorrhagic fever with renal syndrome in Kazakhstan
B. Ospanov, R. Hewson, J. Hay (Almaty, KZ; Porton Down, UK; Buffalo, US)
Objectives: We are undertaking a survey of the incidence of two endemic viral diseases in Kazakhstan. Both diseases are caused by bunyaviruses: CCHF is spread by ticks, while haemorrhagic fever with renal syndrome (HFRS), caused by hantaviruses, is contracted through contact with infected rodent host excreta. Our plan is to integrate studies of human disease with viral incidence in ticks and reservoir animals.
Methods: To achieve this, we are examining the strain characteristics of viruses (CCHFV and hantaviruses) that we identify through PCR, and are assessing incidence of antibody positivity by ELISA against viral antigens.
We have developed ELISA assays for six strains of both viruses using cloned, purified nucleocapsid protein, and PCR assays using detailed sequence analysis of strains that are likely to be circulating in this part of central Asia.
Results: We have collected field samples as follows: 350 human serum samples, 2,500 organ samples from over 1,000 rodents and 15,000 ticks (13,800 H. asiaticum and 1,200 other Ixoides spp.). In preliminary screening experiments, approximately 40% of ticks tested were positive for CCHF viral antigen by ELISA. Five percent of rodents were positive for hantavirus (Puumala) antigen by ELISA.
Conclusion: We anticipate that these numbers are substantial underestimates of the true incidence of viral infection in both ticks and rodents, and our current PCR and ELISA (for antibodies) studies are designed to be both more sensitive and specific than tests used in the past.
Paediatric bacterial infections
P1534 Comparison of sulbactam/ampicillin and tazobactam/piperacillin for the treatment of lower respiratory tract infection in paediatric patients
N. Ishiwada, T. Hoshino, Y. Kohno (Chiba, JP)
Objectives: Paediatric lower respiratory tract infections (LRTI) are common causes of morbidity and mortality worldwide. Haemophilus influenzae (Hi), Streptococcus pneumoniae (Pn) and Moraxella catarrhalis (Mc), the major bacterial aetiologic agents are responsible for cases of paediatric LRTI. The prevalence of penicillin resistant Pn and ampicillin (ABPC) resistant Hi has remarkably increased in recent years in Japan. Furthermore, almost all Mc isolated from patients with LRTI are β-lactamase-producing strains. Therefore, it becomes more difficult to treat with antibiotic therapy for LRTI in children. The purpose of this study is to evaluate the efficacy and safety of two types of penicillin with an addition of a β-lactamase inhibitor (tazobactam/piperacillin – TAZ/PIPC – and sulbactam/ampicillin – SBT/ABPC) for the initial treatment of hospitalised infants and children with LRTI.
Methods: A total 108 paediatric inpatients with LRTI were enrolled in the study. Of 108 patients, 64 treated with TAZ/PIPC and 44 treated with SBT/ABPC. We retrospectively analysed clinical efficacy, adverse events and antibiotic susceptibility of the causative agents isolated from sputum samples of the patients.
Results: For the patients who received TAZ/PIPC, the overall favourable clinical response rate was 89.1% (57/64). On the other hand, for the patients who received SBT/ABPC, the overall favourable clinical response rate was 86.4% (38/44). Adverse events were reported by two TAZ/PIPC treated patients and one SBT/ABPC treated patient, but no severe incidence was observed. In the TAZ/PIPC group, the three most frequently isolated bacteria from sputum samples were Hi (34 samples), Pn (14 samples), and Mc (8 samples). Of the 35 Hi strains cultured from 34 samples, 24 strains (66.7%) were ABPC resistant strains including 6 β-lactamase producing strains. In the SBT/ABPC group, the three most frequently isolated bacteria from sputum samples were Hi (16 samples), Mc (13 samples), Pn (10 samples). Of the 16 Hi strains, 5 strains (31.3%) were ABPC resistant strains including 1 β-lactamase producing strain.
Conclusions: Both SBT/ABPC and TAZ/PIPC produced a satisfactory therapeutic outcome and they are good candidate for initial treatment for LRTI in children. Especially, TAZ/PIPC was effective for the treatment of LRTI due to β-lactamase non-producing ABPC resistant Hi. Therefore, TAZ/PIPC could be a useful agent for treatment of LRTI due to antimicrobial resistant Hi in children.
P1535 Risk factors for Stenotrophomonas maltophilia isolation from patients with cystic fibrosis
H. Alexandrou-Athanasoulis, S. Doudounakis, I. Loucou, A. Sergounioti, M. Georgara, A. Pangalis (Athens, GR)
The number of patients with cystic fibrosis (CF) whose airway secretions yielded Stenotrophomonas maltophilia (Sm) has been increasing during the last years.
The aim of our study is to determine risk factors for the initial recovery of Sm from respiratory secretions of patients with Cystic Fibrosis.
Material and Method: We reviewed retrospectively the clinical and laboratory data of 87 patients with CF who have been colonised with Sm between 01/2000 and 11/2006 and 87 matched controls of similar age (±1 yr) who never had a positive culture for Sm. Variables included age, gender, mean values of FEV as well as the use of oral, intravenous or inhaled antibiotics in the three months prior to the first isolation of S. maltophilia from airway secretions.
Results: (1) Eighty-seven out of the 428 patients who have attended our departments were found to be colonised with Sm. The incidence rate of Sm acquisition has increased from 3.6% in 2000 to 4.5% in 2006. (2) The mean age at the initial recovery of Sm was 6.9±4.6 years. (3) Patients positive for Sm were not found to have significantly worse spirometric values than Sm-negative matched controls. (4) Consumption of oral antibiotics (amoxicillin + clavulate/ciprofloxacin/macrolides) as well as the number of days on intravenous antibiotics were significantly higher in Sm-positive patients. (5) Treatment with inhaled antibiotics was not found to be a significant risk factor for Sm acquisition in our patients.
Conclusions: Patients with CF with Sm in their airway secretions do not have an overall worse clinical status at the time of initial Sm acquisition than Sm-negative patients. Although oral antibiotics or days of intravenous antibiotic therapy seems to be a significant risk factor for initial Sm acquisition the inhaled antibiotic therapy was not.
P1536 High occurrence of staphylococcal toxins among S. aureus isolated from children with cystic fibrosis
H. Alexandrou-Athanasoulis, E. Petinaki, S. Nikolaou, S. Doudounakis, A. Sergounioti, A. Maniatis, A. Pangalis (Athens, GR)
Objectives: S. aureus exerts its pathogenicity by production of staphylococcal toxins such as toxic-shock syndrome toxin (TSST-1), Panton-Valentine leucocidin, enteroto-xins etc. In this study, the occurrence of staphylococcal toxins in S. aureus stains isolated from children with cystic fibrosis was evaluated.
Material and Methods: A total of 65 S. aureus were collected between 1/1/2005 and 30/6/2006 from sputum of 65 children with cystic fibrosis (CF), aged 6.9y SD±3.7. Cultures were performed using conventional methods, while the identification of isolates was done by Gram-stain, catalase and Dnase-activity. Antimicrobial susceptibility to various antimicrobial agents was tested by the automated VITEK2 system and the Kirby-Bauer method according to CLSI recommendations. Methicillin resistance was confirmed by the PB2a detection (Slidex bioMérieux). The detection of genes coding the toxins TSST-1, PVL, and enterotoxins (seu, sei, seg, sen, seo) was assessed by PCR. A total of 100 S. aureus, collected from different specimens of patients free cystic fibrosis were also examined.
Results: All 65 isolates were MRSA. The great majority of isolates expressed a sensitive phenotype (only resistance to oxacillin); only twelve isolates were resistant to more than three different antimicrobial agents. All isolates were found to carry at least one gene of the enterotoxin gene cluster (egc), including the genes sem, sei, seg, sen, seo; 42.5% were found to carry entire the egc operon with the co-existence of tsst-1 gene. However, none isolate was found to carry the PVL-gene. Among S. aureus from patients free cystic fibrosis, 13.7% were found to carry entire the egc operon with the co-existence of tsst-1 gene, while the presence of PVL-gene was detected in 20% of isolates.
Conclusions: These results clearly indicate that S. aureus from cystic fibrosis are potentially more virulent than other S. aureus isolates.
P1537 Antimicrobial susceptibility of the pathogens of bacteraemia in a tertiary neonatal intensive care unit in Bogota, Colombia, 2001–2006
G. Contreras, R. Prieto, A. Leal (Bogota, CO)
Objectives: Determine the most common pathogens isolated from blood and characterised their antimicrobial susceptibility pattern
Methods: A descriptive retrospective study was performed. The data of the Neonatal Intensive Care Unit was collected from the Microbiology laboratory. Bacterial identification and the antimicrobial susceptibility were determined by the MicroScan system. The data was transferred to WHONET 5.4 with the aid of BacLink software and were interpreted according to the criteria of the CLSI. Duplicated isolates were identified and deleted. Quality assurance was performed by the National Institute of Health.
Results: Of 1,210 blood cultures examined in this investigation, 206 (17%) were positive for bacterial growth. The frequency of isolation of Gram-positive bacteria was 72% (149 of 206) of all isolated pathogens, 26% (621 of 206) were for Gram-negative and 2% for fungi (2 of 206). The 45% of all Gram-positive organisms recovered from our blood cultures coagulase negative staphylococci were the group of organisms most frequently isolated, the 19% of all Gram-negative organism Enterobacter cloacae were the most common organism isolated and Candida albicans was the fungi most common isolated. Table 1 shows the antimicrobial susceptibility rates for Gram-positive and Gram-negative bacterial species recovered from blood cultures.
Table 1.
| S. aureus | CoNS | E. faecalis | K. pneumoniae | E. coli | K. oxytoca | E. cloacae | S. marcescens | |
|---|---|---|---|---|---|---|---|---|
| Antibiotic | %S | %S | %S | %S | %S | %S | %S | %S |
| Ampicillin | N/A | N/A | 100 | 0 | 50 | 100 | 40 | 0 |
| Aztreonam | N/A | N/A | N/A | 100 | 100 | 100 | 70 | 100 |
| Cefepime | N/A | N/A | N/A | 100 | 100 | 100 | 100 | 100 |
| Cefotaxime | N/A | N/A | N/A | 100 | 100 | 100 | 60 | 100 |
| Ceftazidime | N/A | N/A | N/A | 100 | 66.7 | 100 | 80 | 100 |
| Ceftriaxone | N/A | N/A | N/A | 100 | 100 | 100 | 70 | 100 |
| Ciprofloxacin | 68.2 | 80.2 | 100 | 100 | 100 | 100 | 100 | 100 |
| Clindamycin | 54.5 | 52.5 | N/A | N/A | N/A | N/A | N/A | N/A |
| Erythromycin | 50 | 32.7 | N/A | N/A | N/A | N/A | N/A | N/A |
| Gentamicin | 50 | 31.7 | 0 | 77.8 | 75 | 100 | 90 | 33.3 |
| Gentamicin-high | N/A | N/A | 88.9 | N/A | N/A | N/A | N/A | N/A |
| Imipenem | N/A | N/A | N/A | 100 | 100 | 100 | 100 | 100 |
| Meropenem | N/A | N/A | N/A | 100 | 100 | 100 | 100 | 100 |
| Oxacillin | 54.5 | 19.8 | N/A | N/A | N/A | N/A | N/A | N/A |
| Piperacillin/tazobactam | N/A | N/A | N/A | 71.4 | 100 | 100 | 80 | 100 |
| Rifampin | 95.5 | 94.1 | N/A | N/A | N/A | N/A | N/A | N/A |
| Trimethoprim/sulfamethoxazole | 86.4 | 76.2 | N/A | N/A | 8.7 | N/A | N/A | 100 |
| Vancomycin | 100 | 100 | 100 | N/A | N/A | N/A | N/A | N/A |
N/A, not applicable.
Conclusions: This study show the greatest importance of CoNS, which have higher percentage of resistance to oxacillin and the progressive increase in the resistance in Staphylococcus aureus in our Unit. The development of this type of studies in developing countries will facilitate to guide the empirical antimicrobial treatment in our Institution and stables control strategies to antimicrobial use.
P1538 The severity of Streptococcus pyogenes infections in children is significantly associated with plasma levels of inflammatory cytokines
S. Wang (Tainan City, TW)
Objectives: Cytokines are intimately involved with the innate and adaptive immune response to bacterial infections and sepsis. This study was designed to determine the expression of pro-inflammatory and regulatory cytokines in children according to the severity of infections caused by Streptococcus pyogenes (group A streptococcus, GAS).
Methods: The study population consisted of 81 children. This included 20 with noninvasive (pharyngitis 7; scarlet fever 13), 16 with invasive GAS infections (toxic shock syndrome 11; necrotising fasciitis 4; pneumonia 1), 24 with GAS pharyngeal colonisation and 21 healthy controls with negative pharyngeal cultures. Plasma levels of interleukin (IL)-1b, IL-2, IL-6, IL-8, IL-10, IL-12, IL-18, tumour necrosis factor (TNF)-α, and interferon (IFN)-γ were measured by ELISA and flow cytometry in a particle-based immunoassay.
Results: Patients with invasive GAS diseases had significantly higher IL-1-b, IL-6, IL-8, IL-10, IL-18, and IFN-γ than those with noninvasive diseases, colonisation, and healthy controls. There was no difference in IL-2, IL-12 and TNF-α levels among the groups. Elevated levels of white blood cell counts, C-reactive protein and C3 were detected only patients with invasive diseases.
Conclusions: Children with invasive GAS infections exhibited significant up-regulation of plasma levels of IL-1-b, IL-6, IL-8, IL-10, IL-18, IFN-γ during the acute phase of their illness compared to those with noninvasive infections, pharyngeal colonisation and controls. An exuberant cytokine response was associated with the severity of illness.
P1539 Laboratory investigation of acute otitis media in children
K. Papavasileiou, H. Papavasileiou, A. Makri, I. Varzakakos, E. Nika, A. Voyatzi (Athens, GR)
Introduction: Acute Otitis Media (AOM) is a common disease of childhood, and quite often the children attend the outpatient clinic for this reason. Risk factors forthe development of AOM are: low socioeconomic status, gender and the use of various hygienic items.
Purpose: To evaluate the incidence of bacterial pathogens responsible for AOM in children and to monitor the change of their antimicrobial susceptibility through the five years of testing (2001–2005).
Material and Methods: A total of 960 middle-ear fluid samples were collected from infants and children with AOM symptomatology, who either were hospitalised or attended the ENT outpatient Clinic. The examined material study was divided in two different time periods: (A) 2001–2002: 260 samples and (B) 2003–2005: 700 samples. Identification and susceptibility tests were based on classical laboratory methods, according to NCCLS instructions. In all S. pneumoniae strains the MICs were defined with automated system (VITEK2, bioMérieux).
Results: Out of 960 cultures, 725 (75.5%) were positive to one of more bacteria. The most prevalent bacteria were: S. aureus (25%), Ps. aeruginosa (16%), H. influenzae (14%), S. pneumoniae (10%) and Streptococcus pyogenes (GAS) (9%). A seasonal distribution has been detected in the incidence of AOM. In winter-spring time, especially S. pneumoniae, GAS and H. influenzae were most predominant.
Comparing the A and B periods, the resistance of S. pneumoniae to penicillin increased from 36% to 40%, to erythromycin from 26% to 28% whereas to cotrimoxazole it decreased from 52% to 40%. The determination of MICs showed that there has been a steady increase in the high-level resistance to penicillin of S. pneumoniae (MIC ≥ 2 mg/L) and frequently the strains were multiple resistant. Among S. aureus a significantly lower prevalence of resistance to oxacillin (36% versus 15%) and to erythromycin (25% versus 21%) was observed. The incidence of erythromycin resistance reduced among H. influenzae (25% versus 3%) and GAS (16% versus 14%) during the five-year period.
Conclusions: 1. S. aureus was the most predominant causative organism of AOM in children, followed by H. influenzae and S. pneumoniae. 2. Significant increased resistance of S. pneumoniae strains was not noted in the last years. 3. The variation on the pattern of antimicrobial resistance of H. influenzae and GAS is probably due to the more prudent use of antibiotics.
P1540 Evaluation of Haemophilus influenzae isolates from children with respiratory tract infections
A. Makri, K. Papavasileiou, E. Panagiotaki, H. Papavasileiou, I. Varzakakos, A. Voyatzi (Athens, GR)
Purpose: The aim of this study was to characterise isolates of Haemophilus influenzae (H.i) with respect to serotype and the in vitro susceptibility to first-line antibiotics used for lower respiratory tract infections.
Material and Methods: A total of 518 samples were collected during a three-year period (2004–2006) from the lower respiratory tract of children (327 sputum and 191 bronchoalveolar lavage). Isolates of H.i were confirmed by using conventional criteria and serotyping was performed by a slide agglutination procedure with all types antiserum (Denka seiken). Susceptibility testing was carried out by the disk diffusion method and the E test (AB Biodisk), while the interpretation was performed according to CLSI breakpoints. H.i ATCC 49247 and H.i ATCC 49766 were used as quality control strains. Beta-lactamase production was assessed by the nitrocefin stick assay.
Results: A total of 212 (40.9%) isolates of H.i were characterised in this study. Biotypes II and III were the most prevalent types (31.2% and 25.8% respectively) followed by biotypes I (20.5%), IV (12.9%), V (4.3%), VIII (3.3%), VI (1%) and VII (1%). The majority of these isolates (59.4%) were not typeable, whereas serogroups b, a, and d represented 18.9%, 7.9% and 6%, respectively. The serotypes c, e and f were rarely found (1%). All isolates were susceptible to amoxicillin-clavulanate, cefotaxime, meropenem and ciprofloxacin. Ampicillin resistance was observed in 36 strains (17%) and all isolates were identified as β-lactamase producers. Resistance to trimethoprim-sulfamethoxazole and clarithromycin was 26% and 13.7% respectively. The rank order of activity based on the MIC50 and MIC90 of the macrolide agents was: azithromycin (0.50–1.5 mg/L) > erythromycin (1.5–4 mg/L) > roxithromycin (2–4 mg/L) > clarithromycin (2–6 mg/L).
Conclusions: (1) Azithromycin was consistently four-fold more active than the others macrolides. (2) No BLNAR (β-lactamase-negative ampicillin resistance) isolates were found in our study. (3) The vast majority of our isolates were non-typeable after the widespread of conjugate vaccines. (4) Continued surveillance is necessary to monitor trends with the H.i disease.
P1541 Characteristics of Staphylococcus aureus isolated from the respiratory tract of cystic fibrosis patients
K. Semczuk, K. Dzierzanowska-Fangrat, D. Borowiec, D. Dzierzanowska (Warsaw, PL)
Objectives: Staphylococcus aureus is one of the most important causes of respiratory tract infections in cystic fibrosis (CF) patients. Formation of small colony variants (SCVs) is usually associated with recurrent infections refractory to antimicrobial treatment. The aims of this study were: (1) to determine the frequency of spontaneous and inducible SCVs formation in CF patients, and (2) to analyse the relation between antibiotic therapy used and SCVs development.
Material and Methods: During 2003–2005 a total of 107 S. aureus strains were isolated from 56 deep throat swabs and 61 sputum samples of 20 CF patients with respiratory tract infections. S. aureus was identified by positive catalase, coagulase, DNAase and latex agglutination tests (Staphytect, Oxoid). Bacteria growing exclusively as small, non-haemolytic, non-pigmented colonies were considered SCVs. The ability to form SCVs was evaluated by an in vitro passage in the medium containing subinhibitory concentration of gentamicin (1 mg/L). Susceptibility to antimicrobial agents was determined by the Etest method. Forty S. aureus isolates obtained from the upper respiratory tract of patients without CF were used as control strains.
Results: Eight S. aureus isolates growing as SCVs were cultured from 61 sputum samples. They came from 5 CF patients chronically colonised with S. aureus. All SCVs isolates were resistant to aminoglycosides (MIC range 8–32 mg/L), 5 isolates were resistant to macrolides (MIC range 2–256 mg/L, 4 had MLSB phenotype), 2 were resistant to tetracycline (MIC range 8–12 mg/L) and 1 was resistant to ciprofloxacin (MIC-8 mg/L). Among the remaining 99 isolates, 69 (64%) formed SCVs in the subinhibitory gentamicin concentration, as compared to 20% of control strains.
Resistance rates of all S. aureus isolates cultured from CF patients to methicillin, erythromycin, tetracycline, clindamycin, gentamicin, ciprofloxacin and co-trimoxazole were 11, 38, 35, 21, 8, 4 and 3%, respectively. All isolates were susceptible to vancomycin. Macrolides, β-lactams and inhaled aminoglycosides were the most frequently used antimicrobials in CF patients studied.
Conclusions: Chronic S. aureus colonisation in CF patients may be associated with the bacterial ability to form SCVs. Inhaled gentamicin may stimulate SCVs formation since these variants were cultured predominantly from CF patients treated with this form of the drug.
P1542 Arcanobacterium haemolyticum as a cause of pharyngotonsillitis in Santander (Spain): incidence, clinical features and antimicrobial susceptibility
C. Garcia, A.B. Campo-Esquisabel, F. Unda, C. Ruiz de Alegría, J. Ag üero (Santander, ES)
Objectives: Arcanobacterium haemolyticum (AH) is considered to be a cause of infections clinically similar to those caused by beta-haemolytic streptococci. There have been few published studies on the prevalence of AH in patients with pharyngotonsillitis. To our knowledge, no data regarding its frequency in Spain have been published. We reviewed the 46 cases of positive AH isolations from throat samples which occurred at our institution from Jan 2005 to Oct 2006.
Methods: Throat samples (n = 10.346) from patients with pharyngotonsillitis attending primary healthcare centres and emergency services in our area (215.000 inhabitants) were studied. Samples were cultured on sheep blood agar plates for 48 h at 35°C in 5% CO2. Identification of AH was based on Gram stain, catalase test, α-mannosidase, reverse CAMP and API-Coryne (bioMérieux, France). Definitive identification was accomplished by sequencing of 16S rDNA. Susceptibility testing was performed by microdilution with cation-adjusted Mueller-Hinton broth supplemented with 3% laked horse blood (CLSI guidelines).
Results: AH was recovered from 46 patients, representing 1.54% of all samples yielding clinically relevant pathogens (n = 2.973). The median age was 13 years (range 4 to 32). Tonsillar exudate was detected in 78.2% of patients, rash in 86.9%, fever in 76% and cervical lymphadenopathy in 78.2%. Recurrent episodes of pharyngitis and/or exantema occurred in 91.3%. One patient, who presented a peritonsillar abscess required hospitalisation. Beta-haemolytic streptococci were concomitantly isolated from 6/46 (13%) samples positive for AH. All AH strains were susceptible to: penicillin (MIC90 0.06 μg/mL), cefuroxim (MIC90 0.25 μg/mL), erythromycin (MIC90 0.01 μg/mL), clindamycin (MIC90 0.03 μg/mL), vancomycin (MIC90 0.5 μg/mL) and levofloxacin (MIC90 1 μg/mL). Fourteen strains were tetracycline resistant (range of MICS from 8 to >16 μg/mL).
Conclusions: Pharyngotonsillitis by AH tended to occur more frequently in adolescents, being the incidence similar to those reported from other European countries. The symptoms of infection by this organism closely mimiced those of acute streptococcal pharyngitis, including a scarlatiniform rash in most patients. A history of recurrent pharyngitis was also present in many cases.
P1543 Anaerobic microbiology of peritoneal cavity specimens in childhood
M. Vlachou, D. Garantziotou, J. Kaleyias, L. Galanopoulou, V. Rouli, T. Apostolaki, O. Koureli, A. Spiliopoulou, A. Gatopoulou (Patras, GR)
Introduction: Even though anaerobic organisms are part of the normal flora, they are associated with several infections with significant morbidity and mortality.
Objectives: The aim of the study was the assessment of incidence of the anaerobic organisms isolated from peritoneal cavity specimens in children.
Material and Methods: Setting: Children Hospital of Patras “Karamandanio”. Study period: June 2003–June 2006 (3 years). Material: Smears from the peritoneal cavity and the lumen of the appendix vermiformis. All the positive anaerobic culture were included in the further analysis of the study. The anaerobic bottles were incubated with the automatic system BacT-Alert (bioMérieux). The culture of specimens was performed using the appropriate material (Schaedler) for Gram(+) and Gram(–), incubated under anaerobic conditions for 48–72 hours. The identification of the organisms was performed with API-20A (bioMérieux) and BBL Crystal Anaerobe (BD).
Results: One hundred forty-two positive anaerobic cultures during the three years study period were identified and included in the further analysis. The bacteria isolated were: Bacteroides spp. 46/142 (32.39%), Peptostreptococcus 45/142 (31.69%), Clostridium spp. 21/142 (14.78%), Bifidobacterium 15/142 (10.6%), Actinomyces israeli 9/142 (6.33%), Propionibacterium 6/142 (4.22%).
Conclusions: Among the isolated anaerobic microorganisms the most common Gram-negative was Bacteroides spp. (32.39%), while the most common Gram-positive was Peptostreptococcus (31.69%). Even though the anaerobic microorganisms constitute a part of the normal flora of the human, they may cause severe and sometimes lethal infections. The appropriate conditions of sampling and transportation of the specimens are mandatory for the accurate evaluation of each patient.
P1544 Survey of Clostridium difficile infection and faecal lactoferrin in Polish paediatric patients with inflammatory bowel disease
D. Wultanska, A. Banaszkiewicz, A. Radzikowski, P. Obuch-Woszczatynski, J. Brazier, A. van Belkum, F. Meisel-Mikolajczyk, H. Pituch, M. Luczak (Warsaw, PL; Cardiff, UK; Rotterdam, NL)
A total of 64 faecal samples submitted from 28 paediatric outpatients with inflammatory bowel disease (IBD) such as Crohn disease (CD) and ulcerative colitis (UC) between January 2005 and February 2006 were screened simultanously for detection of C. difficile toxins TcdA/TcdB, culture of C. difficile strains and faecal lactoferrin (FL). Occurrence of C. difficile TcdA/TcdB and FL was determined as 70% and 44% of all cases, respectively. The proportion of positive specimens as well as for toxins and faecal lactoferrin was not different in CD and UC. Isolates of C. difficile strains (16 in total) obtained over a 1 year period were characterised by toxigenicity profile, PCR-ribotyping and susceptibility to 7 antimicrobial agents. PCR-ribotyping showed six different ribotypes among 16 C. difficile strains: three toxigenic (014, 018, 046) and four non-toxigenic (010, 035). The most predominant ribotype in paediatric patients with IBD was ribotype 014 (A+B+) accounting for 50% (8/16). No resistance to metronidazole and vancomycin was found. Investigation of C. difficile infection should be taken into account in group of paediatric patients with IBD and reccurences of diarrhoea.
This work was supported by Polish Ministry of Education and Science, Grant No. 2P05D 074 27.
P1545 Prevalence and antimicrobial resistance of bacterial isolates in neonatal blood-stream infections
E. Ikonomopoulou, M. Vlachou, P. Michail, S. Paratiras, G. Stavropoulou, C. Verra, A. Regli (Patras, GR)
Objectives: The aim of this study was to determine the prevalence and the antibiotic resistance of bacterial isolates from blood-stream infections in the neonatal care unit of our hospital.
Methods: During a three year period (2003–2005) a total of 1,152 blood cultures were obtained from neonates that were suspect of infection. Blood cultures were performed using the Bactec 9120 (Becton Dickinson) and Bact–alert (bioMérieux) during 2003 and 2004–2005, respectively. The identification and the antimicrobial resistance of bacterial isolates were carried out by the VITEK system (bioMérieux) or the mini API system (bioMérieux).
Results: A total of 90 (7.8%) neonates had positive blood culture. The most common pathogens were: Coag(–) staphylococci 38, Escherichia coli 24, Klebsiella pneumoniae 10. Other Gram-negative bacteria were: Serratia marcescens 3, Proteus mirabilis 2, Enterobacter cloacae 1, Citrobacter freundii 1 and Stenotrophomonas maltophilia 1. Other Gram-positive bacteria were: Staphylococcus aureus 3, Enterococcus faecalis 2, Streptococcus viridans 2, Streptococcus group C 1, and Candida krusei 1. E. coli strains were 50% resistant to ampicillin, 29% to piperacillin, 33% to ticarcillin, 17% to cefalothin, 8% to nalidixic acid and ciprofloxacin, 12.5% to gentamicin and trimethoprim/sulfamethoxazole and 0% to amikacin, netilmicin, tobramycin, amoxycillin/clavulanic acid, cefotaxime, ceftriaxone ceftazidime, imipenem, meropenem and aztreonam. K. pneumoniae strains were 100% resistant to ampicillin and ticarcillin, 50% to cefalothin and 0% to piperacillin, amoxycillin/clavulanic acid, cefotaxime, ceftriaxone ceftazidime, imipenem, meropenem, aztreonam, nalidixic acid, ciprofloxacin, gentamicin, amikacin, netilmicin, tobramycin and trimethoprim/sulfamethoxazole. E. coli and K. pneumoniae strains were ESBL negative. Although Coag(–) staphylococci sometimes considered to be a contaminant were 17% resistant to oxacillin, 29% to erythromycin, 5% to gentamycin, 16% to fusidic acid and trimethoprim/sulfamethoxazole.
Conclusions: Considering the most common pathogens and their antibiotic resistance especially to ampicillin perhaps we should revise the present choise of empirical treatment of ampicillin and one aminoglycoside until a pathogen has been isolated.
P1546 Serum chemokin RANTES as a immunological indicator of early-onset neonatal sepsis
M. Wasek-Buko, J. Behrendt, M. Stojewska, B. Krolak-Olejnik, J. Karpe, U. Godula-Stuglik (Zabrze, PL)
Objectives: Intensive studies on improving the early diagnosis of neonatal sepsis are ongoing, but only a few of them have focused on the role of serum chemokine RANTES (s.ch.R). Aim: to establish the s.ch.R. concentrations in septic neonates, to find the relationship between their gestational age, gender, birth asphyxia, mode of delivery, selected haematological and biochemical parameters and ch.R. in sick neonates.
Methods: The study comprised 41 septic neonates (28 boys, 13 girls; 18 full-term and 23 preterm). Low birth weight (LBW) was noted in 13 neonates, extremely LBW in 3, and birth asphyxia in 11. Cesarean section in 53%, chorioamnionitis in 10% and high-risk pregnancy in 78% of cases were stated. The most common isolate from blood was Staph. epidermidis MR (51%). Pseudomonas aeruginosa was cultured in 9 cases, Klebsiella pn. in 4, E. coli in 3, Staph. aureus in 2, and Serratia marcescens in 2. Septic neonates presented: pneumonia (65%), severe gastrointestinal disorders (60%), jaundice with hepatosplenomegaly (41%), shock (49%), purulent meningitis (17%), metabolic acidosis and thrombocytopenia in 21%, elevated serum CRP in 63%. The control group consisted of 40 newborns (25 full-term, 15 preterm, 21 boys, 19 girls) healthy, born vaginally, without perinatal risk factors. The ch.R. concentration in peripheral vein blood was measured between the 2nd and 4th day of life by ELISA method using the Quantikine set (R&D Systems, Minneapolis, USA).
Results: The mean s.ch.R. concentration in full-term septic neonates was 73.9±25.9 (31.5–17.9) ug/mL and differed significantly (p < 0.001) from the mean value in healthy neonates (28.25±14.0, 12.6–68.4). Septic prematures had significantly (p<0.001) higher mean s.ch.R. concentration (59.2±28.2, 12.2–118.2) than healthy preterm newborns (25.6±8.2, 14.3–41.2). There were no differences between the mean s.ch.R. value in neonates with Gram(+) sepsis (64.5±22.0, 12.2–96.3) and neonates with Gram(–) sepsis (67.5±33.9, 22.9–118.2), both in full-term and preterm neonates. No statistically significant difference was noted in mean s.ch.R. value regarding gender, birth weight, mode of delivery, or Apgar score in septic neonates. A positive correlation was found between s.ch.R. concentration and band cells count in peripheral vein blood (r = 0.249) and serum CRP value (r = 0.301).
Conclusion: Serum chemokine RANTES concentrations increase in early-onset sepsis in neonates, independently of their gestational age, gender, Apgar score or mode of delivery, and may be useful in diagnosis of this congenital bacterial disease.
P1547 Antimicrobial susceptibility of bacteria causing early onset sepsis
E. Zisovska, L. Lazarevska, J. Zivkovik (Skopje, MK)
Introduction: Neonatal sepsis has high morbidity and mortality, so rapid and accurate detection and treatment with initial antibiotic therapy, before the microbiological evaluation finished, is necessary and is based on empirical data in regard to sensitivity of prevalent bacterial strains. AIM of our investigation was to present the spectrum of prevalent bacteria and their susceptibility to the antibiotics used by the standard protocols, based on evidence.
Methods: Our prospective study included all neonates with suspected sepsis born in our Department of neonatology in a two year's period (2004 and 2005). Microbiological identification and susceptibility were performed with routine methods.
Results: A total of 8,356 neonates were evaluated during this study period and 167 patients fulfilled the eligible criteria. Three most frequent bacterial strains were Staphylococcus epidermidis, Escherichia coli and Group B streptococci. Less frequent were Staphylococcus aureus, Klebsiella pneumoniae and Pseudomonas aeruginosa. In 21/167 (12.57%) patients initial antibiotic treatment was inappropriate, and the most resistant strains were S. aureus (8) followed by K. pneumoniae (7), E. coli (4) and P. aeruginosa (2).
Discussion: the initial use of appropriate antibiotics, before the detection of bacteria, is very useful, it reduces the time of therapy, length of stay and costs of treatment.
Conclusion: It is recommended for each neonatal department to determine their own spectrum of bacteria and to adjust the initial antibiotic treatment to the findings which are typical for the unit. But, also repeated surveillance is necessary in order to follow the longitudinal changes of the bacterial spectra due to the resistance acquired.
P1548 Rapid molecular detection of bacterial species and resistance determination in paediatric blood cultures
D. Menichella, P. Bernaschi, B. Lucignano, C. Manfredini, C. Russo (Rome, IT)
Bacteraemia is one of the most serious and potentially life-threatening infection disease in children, specially in case of septic shock. Signs or symptoms of bacteraemia are highly variable in childhood and diagnosis is routinely supported by blood culture.
In our study, we evaluated a new molecular genetic assay (GenoType BC Gram-positive® and GenoType BC Gram-negative® – Hain Lifescience) as rapid tool for identification of most of bacteria involved in bloodstream infections in children suspected to have bacteraemia, hospitalised at “Bambino Gesu‘” Children Hospital (Rome, Italy) and to improve the work-flow TAT in a microbiology laboratory.
At instrumental positive time (BACTEC 9240 Becton Dikinson) all the bottles (PEDS PLUS/F) were evaluated with microscopy Gram stain and subcultured in agar standard media. Phenotypic identification was obtained on colonies grown after 24 hours of incubation by using the VITEK® 2 (bioMérieux) automated system. For molecular identification 20 μL of blood positive culture were spotted on GENO-CARD® and after a drying step, a punch of 1 mm in diameter was transferred into the PCR mix followed by thermal-cycling amplification. Hybridisation protocol was performed as recommended using a shaking incubator and results were obtained by comparison of the hybridisation pattern present on the test strip with the ones of the provided evaluation sheet.
148 positive blood culture bottles representing 106 Gram-positive cocci, 28 Gram-negative bacilli and 14 mixed infections were included in this study. At start time we performed all of the proceduring steps as manufacturing instructions and the GenoType BC system correctly amplified and identified 41/49 Gram-positive cocci (sensitivity 83%); 8 Staphylococci were not amplified. 17/17 Gram-negative bacilli were correctly amplified and identified (sensitivity 100%).
In the effort to optimised the GenoType assay we improved by in house steps the direct DNA extraction. Finally all of 53 Gram-positive cocci, 11 Gram-negative bacilli were correctly amplified, 4 K. kristinae were correctly not amplified (sensitivity and specificity 100%). About 14 mixed cultures were correctly identified and for 1 of them the GenoType BC assay recognized a double infection (P. aeruginosa and S. maltophilia) demonstrated only after this result by a new subculturing. GenoType BC assay is very helpful in the rapid diagnosis of paediatric blood stream infections in effort to prevent severe sepsis and mortality.
P1549 Genetic factors in the development of unfavourable effects of intrauterine infections on the course of neonatal periods in neonates
N. Gorovenko, Z. Rossokha, S. Podolskaya (Kiev, UA)
Objectives: Intrauterine infection in neonates is an important factor of perinatal risk in the development of complications during neonatal period. The latent forms of these infections are not always when effecting neonates. In a great number of studies the genes deletion polymorphism of glutathione-S-transferases was associated with higher sensitivity to the influence of many factors. The aim of this research was to study the prevalence of intrauterine infections and GSTT1 genes allelic polymorphism in neonates.
Methods: The prevalence of Toxoplasma gondii, Chlamydia trachomatis, Mycoplasma hominis, Herpesvirus I/II types, Cytomegalovirus in the blood of neonates with perinatal pathologies and neonates from the comparative group (clinical healthy neonates) was tested by polymerase chain reaction (PCR) method. The allelic polymorphism of GSTT1 genes was investigated by means of multiplex PCR in neonates. Differences in these groups were assessed by the Pearson chi-square analyses.
Results: There were detected latent forms of intrauterine infections in 74 of 117 neonates with perinatal pathologies (63.24%) as compared to the comparative group of the neonates where the frequency of latent forms accounted for 9 of 70 neonates (12.85%). In our investigation we have observed an increased frequency of GSTT1 null genotypes among infected neonates (31.32%; 26 of 83 individuals were homozygous for GSTT1 deletion genotype) as compared to uninfected neonates (16.98%; 18 of 104 individuals were homozygous for GSTT1 deletion genotype). The frequency of deletion polymorphism of GSTT1 genes was significantly increased among infected neonates of both groups (4.59, P < 0.05), i.e. GSTT1 null genotype was associated with the development of intrauterine infections. The GSTT1 frequency of deletion polymorphisms was significantly (6.66, P < 0.01) increased among infected neonates with perinatal pathologies (33.78%; 25 of 74 individuals were homozygous for GSTT1 deletion genotype) as compared to uninfected clinical healthy neonates (13.11%; 8 of 61 individuals were homozygous for GSTT1 deletion genotype).
Conclusion: Thus, the deletion polymorphism of GSTT1 genes in neonates stipulates the predisposition to the intrauterine infections causes its effect with further development of perinatal pathologies.
Paediatric viral infections
P1550 Features of norovirus and rotavirus infections in small children
I. Narkeviciute, I. Tamusauskaite, G. Bernatoniene (Vilnius, LT)
Objectives: No comparison studies of clinical presentation of norovirus and rotavirus infection in small children were ever made. We investigated norovirus infection features in children under three year's age and compared the results with rotavirus infection in the children of the same age.
Methods: The random selection and retrospective analysis of 70 norovirus and 70 rotavirus-infected children's case notes were done. All children were treated in Vilnius University Children Hospital in 2005. The norovirus antigen was assayed using ELISA, rotavirus – using immunochromatography diagnostic assay.
Results: In small children norovirus infection manifested as vomiting (94.3%), diarrhoea (81.4%) and pyrexia (65.7%). It manifested as either gastroenteritis with pyrexia (47%) or gastroenteritis alone (30% of all cases) while 18.6% cases were without diarrhoea. Granulocytosis was found in 72.7% of blood results. Pyrexia was present in 97.1% case of rotavirus infection and 80.9% were >38°C. However, in norovirus infection accordingly 65.7% and 47.8% (p < 0.0001). Diarrhoea (≥4 times/d) more frequently appeared in children with rotavirus infection than with norovirus (p < 0.0001). Repeated vomiting (≥4 times/d) has been more common for children with norovirus infection. As opposed to norovirus infection, rotavirus infection usually presented just as one syndrome of gastroenteritis with pyrexia (p < 0.0001).
Conclusion: Norovirus infection can present either as gastroenteritis or gastroenteritis with pyrexia. However, diarrhoea was absent in almost 1/5 of all patients. Norovirus and rotavirus infections had statistically significant differences in a degree of pyrexia, intensity of diarrhoea and vomiting as well as syndromes frequency.
P1551 Clinical profile of dengue infection in children versus adults
N. Othman (Selangor, MY)
Objectives: To compare the clinical profile between children and adults with serologically confirmed dengue infection.
Methods: A cross-sectional retrospective study comprising 581 adults and 372 children admitted with dengue infection was carried out from July to December 2004. Children was defined as <13 years old and adults as ≥13 years. Diagnosis of dengue infection was established by detection of specific dengue IgM ELISA.
Results: Out of 953 cases, there were 528 (55.4%) males and 425 (44.6%) females. The mean ages were 26.5 and 7.00 years for adults and children respectively. Children were found to be more prone to have dengue shock syndrome (DSS). Fever, respiratory symptoms (cough and coryza) and abdominal pain were commonly present in children. On the other hand, adults were more likely to complain of retro-orbital pain, myalgia, athralgia and anorexia. Reported bleeding manifestations were significantly higher in adults. In terms of signs, hepatomegaly and lymphadenopathy were commoner in children, while flushing and abdominal tenderness were seen more in adults. The most remarkable finding on laboratory investigation was adults had higher alanine transferase levels as compared to children, implicating a higher tendency for liver impairment. Both groups showed thrombocytopenia on admission (71.4×109/L in adults versus 99.05×109/L in children); thrombocytopenia was found to be statistically lower in adults. No mortality outcome was noted.
Conclusion: Children tend to be inflicted with a more severe form of dengue infection. Both age groups present different clinical features. In addition, complication such as liver impairment is more seen in adults
P1552 Myocarditis heart failure associated with Adenovirus infection
O. Valdes, B. Acosta, C. Savon, A. Piñón, A. Goyenechea, L. Palerm, G. Gonzalez, V. Kouri, A. Martínez, L. Sarmiento, D. Rosario, M. Guzman (Havana City, CU)
Objective: Rapid detection of aetiological agent causing myocarditis using molecular tools.
Methods: During an atypical outbreak of acute febrile syndrome of viral aetiology, 8 children ranged between 5 months aged to 12 years old have died in cardiogenic shock due to myocarditis, on July and August, 2005, in Havana city, Cuba. Nested polymerase chain reaction (nPCR) was used to detect the genomic sequences of Enterovirus, Adenovirus, Cytomegalovirus (CMV), Herpes simplex virus (HSV), Epstein Barr virus (EBV), Varicella zoster virus (VZV), Human Herpes virus 6 (HHV6), Flavivirus, Alphavirus, human Respiratory syncytial virus (hRSV) and Influenza virus (A, B and C) in tissue samples (heart, lung, brain and tonsil), cerebrospinal fluid and faeces specimens.
Results: We found evidence for the Adenovirus genome in a significant proportion of patients, 6 (75%) of them. Non others viruses were detected. The predominant clinical picture was characterised by acute cardiac decompensation and progress to death. In these cases the organs most common detected were lung (83%) and myocardium (67%). In three cases adenovirus was detected in both organs. Electrocardiography changes suggestive of myocarditis were detected in all patients. Histopathology findings characterised by an infiltrate of inflammatory cells and myocardial necrosis, established the anatomapathologic diagnosis of myocarditis post-mortem.
Conclusion: After exclusion of other possible causes of death, our results indicate that viral myocardial affection is the cause of death in Adenovirus positive cases. It should be noted that this is the first report about the molecular diagnosis of Adenovirus in myocarditis cases in Cuba.
P1553 Rotaviral and noroviral infections among children inpatients of a clinic of infectious diseases in Pilsen, Czech Republic
P. Pazdiora, M. Švecová, J. Táborská (Plzeo‘, Plzen, CZ)
Objectives: Rotaviruses are the most frequent aetiological agent among patients with diarrhoeal diseases. Data about community-acquired noroviral infections is sporadic, there is the first data from the Czech Republic.
Methods: The importance of rotaviruses was analysed among childrenœ inpatients during years 1986–2005, the importance of noroviruses during years 2003–2006.
Results: In years 1986–2005 were examined 6,416 younger children with gastrointestinal diseases. Rotaviruses were observed among 26.4% patients, their occurrence changed in different years between 14–41%. The incidence of rotavirus acute gastroenteritis among different age groups was as follows: children under 7 months 16.3%, 7–12 months 28.7%, 13–24 months 32.2%, 25–36 months 28.0%, older 24.4%. The highest incidence rate was during March (44.8%), the lowest during September (17.3%). Among 1,385 patients without gastrointestinal symptoms rotaviruses were detected 57x (4.1%). Nosocomial rotaviral infection was laboratory detected in 290 of 5,714 infants repeatedly examined during the hospitalisation (i.e. 5.1%). These infections protracted hospitalisation on average by 4 days. In years 2003–2006 were examined 248 children with gastrointestinal diseases. Noroviruses were detected EIA test among 6.8% patients. The highest incidence of norovirus acute gastroenteritis was among children 7–12 months 18.5%.
Conclusion: The results confirm frequent occurrence of rotaviral and noroviral infections among hospitalised children.
Grant support: MSM 0021620819 VZ 6096
P1554 Increased prevalence of genotype G2P(4) among children with rotavirus-associated gastroenteritis in Honduras
A. Ferrera, D. Quan, F. Espinoza (Tegucigalpa, HN; Leon, NI)
Background: Rotavirus is a major cause of gastroenteritis among young children worldwide and is associated with significant morbidity and mortality, particularly in developing countries. From January to March 2006, a large outbreak of gastroenteritis occurred in Honduras that was associated with hospitalisation and death among children.
Objective: To characterise by molecular methods the rotavirus A genotypes G and P involved in the outbreak of 2006.
Methods: Faecal samples positive for rotavirus by enzyme-linked immunosorbent assay were collected from 45 children under 5 years of age from two geographically distinct regions of Honduras where the epidemic had occurred. G- and P-types were determined in a two-step procedure using reverse transcription followed by a standard multiplex PCR.
Results: The infection G2P(4) (43.3%) appeared to be the most frequent cause of the rotavirus outbreak, occurring mainly in children less than one year of age. Other genotypes encountered were G4P(8) (16.7%), G9P(8) (13.3%) and G2P(10) (3.3%). Co-infection with more than one G genotype occurred in 5 children, and co-infection with more than one P genotype occurred in 2 children.
Conclusion: We found for the first time genotypes not reported before in the population, suggesting a great diversity of rotavirus strains circling in the country. Further molecular surveillance could provide a sound basis for improving the response to epidemic gastroenteritis and could provide data needed for the introduction of vaccination programmes in the country.
P1555 Human herpes virus 6 infection in children in Northern Greece
P. Sidira, S. Mitka, D. Karabaxoglou, K. Politi, A. Kansouzidou, A. Papa (Thessaloniki, GR)
Objective: Human Herpes virus 6 (HHV-6) is a human pathogen of emerging clinical significance. Two genetically distinct variants of the virus exist, HHV-6A and HHV-6B. HHV-6 has been identified as a causative agent of exanthem subitum. HHV-6 infection is common in the first 3 years of life and the disease has a variety of clinical presentations. The aim of this study was the estimation of the incidence of the HHV-6 infection in children in northern Greece and the evaluation of the contribution of PCR in the early and specific diagnosis of the disease.
Materials and Methods: A total of 27 patients (children aged under 3 years) with signs and symptoms suggestive of exanthem subitum were studied. A total of 39 serum specimens from the patients were examined for the detection of specific IgM and IgG antibodies against HHV-6 by ELISA and 27 blood specimens were tested for the HHV-6 DNA by polymerase chain reaction (PCR). In the PCR, a fragment of U67 gene (homology 96.5% between HHV-6A and HHV-6B) was used as DNA target (fragment 223 bp). HHV-6A U1102 strain was used as a positive-control.
Results: In total, HHV-6 primary infection was diagnosed in 10 patients out of 27 patients examined, who presented seroconvertion and/or had HHV-6 DNA detectable in the acute blood specimen. Past HHV-6 infection was diagnosed in 6 patients who had IgG antibodies against HHV-6 in the acute serum, while PCR was negative in the acute blood specimen. Absence of HHV-6 infection was diagnosed in 11 patients who presented negative results in HHV-6 PCR and IgG serology.
Conclusions: HHV-6 primary infection is a significant causative agent of illness in children aged under 3 years presenting with fever and exanthem. PCR assay contributes significantly in the early diagnosis of HHV-6 primary infection, as serology alone proved to have many limitations.
P1556 Varicella-related hospitalisation in children: a seven-year retrospective study in the pre-vaccine era, in Ankara, Turkey
N. Belet, A. Tapisiz, E. Çiftçi, E. Ince, Ü. Dogru (Ankara, TR)
Objectives: Varicella is predominantly a childhood disease in non-vaccinated populations. Although varicella infection is more severe in immunocompromised people, most cases of severe morbidity and mortality are seen in healthy individuals. The purpose of the study was to evaluate the indications of hospital admissions and complications of varicella infection in a tertiary care hospital, in Ankara, Turkey.
Methods: Hospital records of children hospitalised for varicella between January 2000 and October 2006 were reviewed. Demographics, immune status, clinical features, microbiological findings, informations on varicella complications and the outcomes of patients were analysed.
Results: A total of 30 children were hospitalised during the study period. The mean age was 53.1±42.1 months. The mean hospital stay was 7.0±6.5 days. There was a male predominance with 20 (66%) males and 10 (34%) females. The indications of hospitalisation were immunocompromised status in 46%, varicella-related complications in 40%, malnutrition in 4%, and isolation requirement during hospitalisation for another reason in 10%. At admission, 63% of patients had fever, 80% profused vesicular rash, 20% skin lesions less than 10, and 16% mucosal involvement. There were 29 complications in 14 patients and the majority of patients had more than one complications. Pneumonia and secondary skin or soft tissue infections were the most common complications in 6 (20%), followed by hepatitis in 5 (17%), thrombocytopenia and dehydration in 3 (10%), arthritis and sepsis in 2 (6%), and DIC and feeding difficulty in 1 (3%). All patients who developed complications were healthy except for two. Four patients (13%) had positive bacterial isolates, including 2 Staphylococcus aureus, one group A beta-haemolytic Streptococcus, and one Haemophilus influenzae type b. Sixteen patients (53%, 14 immunocompromised and two immunocompetent children) were treated with acyclovir and the duration of therapy was 7.2±2.2 days. No fatality was reported during the study period. Only one patient who had pneumonia, hepatitis and DIC was admitted to the intensive care unit.
Conclusion: Varicella complications are believed to be rare in immunologically healthy children. However, our results indicate that varicella complications occurred mainly in immunocompetent children. Thus, a universal childhood varicella immunisation may reduce the rate of varicella-related complications and admissions in our country.
P1557 Human metapneumovirus in paediatric viral respiratory infections
J. Alexandre, M. Alves, G. Rocha, A. Magalhães-Sant Ana, A. Meliço-Silvestre, C. Nogueira (Coimbra, PT)
Objectives: Respiratory infection in infants and young children is a significant public health problem worldwide. Infection with respiratory syncytial virus (RSV) is the most common known cause of lower respiratory tract infection in young children. Human metapneumovirus (hMPV), a member of Pneumovirinae subfamily of the Paramyxoviridae family, first discovered in 2001, has recently been described as a casual agent of acute respiratory disease. The virus is widely distributed and has been detected in Japan, China, Australia, USA and several European countries, and is responsible for about 2 to 25% of cases of acute respiratory tract infections in infants. This relatively high incidence and the fact that hMPV associated disease may be severe, emphasized the need for a reliable, sensitive and rapid diagnostic test for detection of hMPV
Methods: From November 2005 to October 2006, 157 respiratory samples were collected from paediatrics patients with symptoms of respiratory tract disease. For the hMPV study total RNA was extracted by MagNA Pure Compact (Roche) using the MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche), according to the manufacter's instructions. cDNA was synthesized with random hexamer primer from the Transcriptor cDNA synthesis Kit (Roche) according to the manufacter's instructions. The detection of hMPV was performed by a real time PCR assay using primers targeting the nucleoprotein gene (N) and a fluorogenic endonuclease oligoprobe (Taqman) for detection. Samples were also tested for screening and identification of Adenovirus (ADV), Parainfluenza (P) 1, 2 and 3, and RSV by an indirect immunofluorescence assay (Respiratory Virus Panel, Biotrin) according to the manufacter's instructions.
Results: hMPV was detected in 20 (13%), RSV in 87 (55%), ADV in 6 (4%), P3 in 3 (2%), respiratory samples. Co infections were also identified: 3 hMPV +/RSV + and 1 RSV +/ADV+.
Conclusions: RSV is still the leading cause of respiratory tract illness in infants and children and our findings agree with detections rates (50–90%) published in literature. Nevertheless, hMPV was found in a significant number of cases (13%) as reported by other authors.
P1558 Human bocavirus infections in children in Hong Kong
S.K.P. Lau, P.C.Y. Woo, C.C.Y. Yip, R.A. Lee, L.Y. So, T.L. Que, K.H. Chan, K.Y. Yuen (Hong Kong, HK)
Objectives: Human bocavirus (HBoV) is a recently discovered human parvovirus associated with respiratory tract infections in children. We carried out a one-year prospective study on the clinical spectrum of disease and epidemiology of HBoV infections in children in Hong Kong.
Methods: A total of 1,200 nasopharyngeal aspirates, negative for respiratory viruses by immunofluorescence tests, from patients ≤ 18 years old with acute respiratory tract infections were subjected to PCR for HBoV Clinical records of patients positive for HBoV were analysed to determine the clinical presentations and outcome.
Results: HBoV was detected in 96 (8%) patients. Of the 96 patients, 11 were ≤ 9 years old. The male to female ratio was 1.5: 1. HBoV infections were associated with both upper and lower respiratory tract disease, including pneumonia, acute bronchiolitis and asthmatic exacerbation. All patients survived. As in other reports, HBoV infections in children peaked in winter, although cases also occurred in other seasons.
Conclusions: HBoV circulates in children in Hong Kong and is detected in nasophayngeal aspirates of those with acute respiratory tract infections. Further studies should be performed o assess the role of HBoV in respiratory tract infections in children.
Public health aspects of viral infections
P1559 Biological risk stemming from the long-term burial of NDV vaccinal strains
S. Bianchi, A. Amendola, M. Canuti, A. Zappa, E. Tanzi, R. Koncan, G. Cornaglia (Milan, Verona, IT)
Objective: Large areas are often used as uncontrolled pits for pharmaceutical waste. No worldwide-accepted criteria define the extent and persistence of the consequent biological risk, related, for instance, to vaccines and their by-products. The objectives of the present study were:
-
1
To study the residual viability of Newcastle Disease Virus (NDV) vaccine strains stored in sealed vials and recovered after burial for 30 years.
-
2
To determine the persistence of the NDV genome in embryonated chicken eggs, i.e. the substrate used for producing the vaccine.
Methods: Vaccines against Newcastle Disease (avian pseudoplague) were analysed both in lyophilised form and as specimens of eggs recovered from the same area. To evaluate the competence and integrity of the viral genome, gene amplification was performed by nested RT PCR specific for a 275 bp fragment of NDV gene F. The PCR-positive specimens were submitted to viability tests through preparation of permissive cell cultures (Vero, African green monkey kidney) and detection of the cytopathic effect. To confirm the specificity of the cytopathic effect, an haemadsorption test was performed. As a positive control, we used a vaccine currently being used against the avian pseudoplague (Izovac, Brescia, Italy) containing live NDV strains (titre, >106 EID50).
Results and Conclusions: Nested RT PCR permitted detection of the NDV genoma in the lyophilised vaccine vials, but not in the egg specimens. This suggests that the NDV viral RNA can persist for several years even under uncontrolled environmental conditions if preserved in sealed vials, but is not able to persist in substrates undergoing chemical and bacteriological modifications, such as the eggs. The recovered viruses were endowed with the specific NDV cytopathic effect, thus showing that they retained their replicative ability. As a consequence, an NDV infection risk is actually involved in the manipulation of pharmaceutical waste. The lack of studies aimed at evaluating the environmental infection risk from the uncontrolled burial of industrial bio-material, such as vaccines and their by-products, constitutes a serious gap in our knowledge.
P1560 Reappraisal of the Berg method to assess viral contamination of the soil
S. Bianchi, A. Amendola, M. Canuti, A. Zappa, E. Tanzi, R. Koncan, G. Cornaglia (Milan, Verona, IT)
Objectives: Some viruses are able to contaminate the soil and persist in it. Nevertheless, at present, no widely accepted standards exist for the evaluation of viral contamination in this environment. The recent scientific literature is not particularly informative, since most studies on this subject date back to roughly 30 years ago and focus on soil contamination by sewage and on controlling the circulation and transmission of oro-faecal agents. The Berg method was developed in 1980 for ‘recovering viruses from sludges', and the following years did not witness any evolution in that technique or its application to other types of viral contamination. Thus, we have re-examined it in order to extend its applicability to soil.
Methods: Known amounts of enteric viruses (namely Coxsackie B6 virus and Echovirus 12) were used to infect the dry soil. The viral particles were subsequently eluted with 10% beef extract, by means of centrifuging and serial passages through 3-μm, 0.45-μm and 0.22-μm filters, respectively. All tests were performed in the absence of sterility and under standardised environmental conditions, namely aerobiosis, 23°C and pH 7. Following every step of the elution procedure, the presence of the virus was tested by means of gene amplification assays specific for the enteroviruses used. The viability of the viruses in the eluates was assessed on the basis of their cytopathic effect on permissive cells (human rhabdomyosarcoma cell line RD).
Results and Conclusions: Optimal parameters were identified as regards quantity of soil and beef extract (100 g and 100 mL, respectively). Titration of the filtrates obtained from each single step made it possible to assess the efficiency of viral recovery, which exceeded by 50% that envisaged in the original Berg method (reported recovery values, 35–40%). The application of this revised technique to the soil may permit effective monitoring of different viruses with alleged high environmental resistance.
P1561 Study of immunity level to measles and mumps in women of reproductive age in Northern Greece
A. Fylaktou, M. Goutaki, S. Kalamitsou, A. Haidopoulou, G. Atmatzidis, E. Papadopoulou (Thessaloniki, GR)
Objectives: The aim of our study was to examine the immunity status to measles and mumps in women of reproductive age.
Methods: Our specimen consists of 198 women between 16–45 years old who were examined in our hospital during the years 2004–2006. Our specimen was divided in two groups according to vaccination status. IgG and IgM antibodies titers for measles and mumps was determined in blood serum by Elisa Virotech (genzyme diagnostics) semiquantitive method.
Results: 91.9% of women are immune to measles whereas for mumps the percentage presents significant decrease to 75.76%. In the overwhelming majority the immunity is due to infection. This is justified because the MMR vaccine was introduced to the National Immunization Program at 1989 while previously the immunisation rate either with MMR (after the mid seventies) or with the monovalent vaccines for measles and mumps was less than 30%.
| IgG+ |
IgG− |
|||
|---|---|---|---|---|
| n | (%) | n | (%) | |
| Measles | ||||
| Group A (after vaccination) | 29 | (14.65) | 0 | (0) |
| Group B (no vaccination) | 163 | (77.27) | 16 | (8.08) |
| Total | 182 | (91.92) | 16 | (8.08) |
| MUMPS | ||||
| Group A (after vaccination) | 29 | (14.65) | 0 | (0) |
| Group B (no vaccination) | 121 | (61.11) | 48 | (24.24) |
| Total | 150 | (75.76) | 48 | (24.24) |
Conclusion: A significant percentage of women of reproductive age (24.24% of specimen) is susceptible to mumps. Although these women are protected by the community's immunity it would be wise to be alert since mumps during the first trimester of pregnancy is associated with an increased rate of spontaneous abortion. Also we note that infants of these mothers will be at high risk for serious infection from mumps and measles virus with an increased frequency for complications, having obtained no immunity by transported maternal IgG.
P1562 Role of exposure to HPV infection and genetic susceptibility for cervical neoplasia in Italy
A. Agodi, M. Barchitta, N. La Rosa, R. Cipresso, M. Guarnaccia, M. Cultrera, M. Caruso, M.G. Castiglione, G. Ettore, S. Travali (Catania, IT)
Objectives: Although cervical infection sustained by high-risk human papillomaviruses (HPV) is considered a necessary event in the development of cervical cancer, it is clear that it is not sufficient. Thus, other factors are likely involved in influencing the persistence and progression of an HPV infection towards malignant disease. One such influence may be genetic susceptibility, although this issue continues to be a focus of debate. A cross-sectional study was conducted in order to: i) estimate the prevalence of cervical HPV infection and ii) investigate the role of the p53 codon 72, GSTM1 and GSTT1 polymorphisms in cervical neoplasia.
Methods: During the period 2004–2006, all women attending the Service for Colposcopy of an Italian Hospital were consecutively enrolled. Cases were women with high-grade cervical intraepithelial lesion (H-SIL). Controls were women without a history of cervical abnormalities or with low-grade cervical intraepithelial lesion (L-SIL). The presence of various HPV types was examined using the HPV Screen Kit (Nuclear Laser Medicine). Polymorphisms of p53 gene were evaluated with PCR with primers encompassing the polymorphic site at codon 72. The GSTM1 and GSTT1 genetic polymorphisms were evaluated using multiplex PCR techniques. In all subjects, gene polymorphisms were determined and results were analysed according to HPV infection status and histological feature, in comparison to the control group.
Results: A total of 232 women were enrolled. The overall HPV prevalence was 71.1% patients. High-risk types were found in 89.7% of infected women and multiple infections in 47.9% of infected women. HPV 16 was by far the most common type (51.5% of HPV positive women), followed by HPV 6 (32.1% of HPV positive women), and HPV 56 (29.7% of HPV positive women); type 18 was found in only 2.4% of HPV positive women. No significant differences in the p53 and GST polymorphisms distribution were found in relation to the infection with HPV. On the contrary, our study showed a significant association between the arginine homozygous genotype and H-SIL, corresponding to an odds ratio of 5.6 (95% CI: 1.515–20.880).
Conclusion: Our study showed a higher HPV prevalence compared with other European populations. Besides, it revealed a possible susceptibility role of the p53 codon 72 polymorphism at an early carcinogenetic stage in cervical cancer. However further research is needed with close attention to study design and methodological features.
P1563 An investigation of a cluster of parapoxvirus cases in Missouri, February–May 2006
E. Lederman, M. Tao, H. Pue, M. Reynolds, S. Smith, Y. Li, H. Zhao, L. Sitler, A. Mahmutovic, G. Emerson, C. Hutson, D. Bensyl, R. Regnery, B. Zhu, I. Damon (Atlanta, Hyattsville, Jefferson City, Troy, US)
Background: Human infections with “barnyard parapoxvirus” (i.e. orf virus and pseudocowpox virus) are rarely reported to health departments. From February to May 2006, four cases were reported in Missouri, two of which were evaluated for anthrax infection.
Methods: Survey of the four cases, family members, county veterinarians and a convenience sample of local farmers were conducted. Samples were collected from implicated animals and animal environments (barnyards) for laboratory analysis.
Results: Two additional cases of human orf infection were identified at a community farm. Five of the six cases sought medical care; none of the primary care providers included parapoxvirus infection in their initial diagnosis. The presence of parapoxvirus DNA was confirmed by real time polymerase chain reaction (DNA polymerase gene-ORF25) in 100% of symptomatic animals (n = 9) and 21% of asymptomatic animals (n = 14). Phylogenetic analysis was performed on virus strains, utilising the virion core protein gene (ORF-27) and the extracellular envelope protein genes (ORFs 109 and 110). Twenty-two percent of local farmers surveyed (n = 58) reported having a parapoxvirus infection at least once in their lifetime. Of these 39% sought medical care for their lesion and 67% of them received antibiotic therapy. Of local veterinarians surveyed (n = 14), 43% were consulted for human cases of orf during their practice and 29% reported that they had been consulted for orf virus infections in sheep/goats at least once a year. Fewer veterinarians were consulted for pseudocowpox virus infections in dairy cattle (0% in the last year; 21% ever).
Conclusions: Parapoxvirus infections are common in Missouri livestock handlers and their animals. The use of nonporous gloves while handling any animals in an infected herd/flock is prudent as parapoxvirus DNA was detected in the oral cavities of asymptomatic animals. Human infections may have a clinical appearance similar to cutaneous anthrax, however the mild illness associated with parapoxvirus infected animals provides a key point of differentiation.
P1564 Mortality and hospitalisations related to influenza and respiratory syncytial virus in the Netherlands
A. Jansen, E. Sanders, A. van Loon, A. Hoes, E. Hak (Utrecht, NL)
Objective: To estimate influenza virus- and respiratory syncytial virus (RSV)-related excess mortality and hospitalisation in the Dutch population, with special attention to influenza-virus-related excess among 0–1-year-olds and non-high-risk 50–64-year olds.
Methods: Retrospective study over the years 1997–2003. Influenza-virus- and RSV-active periods were defined according to virus surveillance data. Nationwide all-cause mortality and hospitalisation figures were collected. The average excess mortality and hospitalisation were determined during periods of influenza-virus and RSV predominance (i.e. weeks with 5% or more of the season's total number of reported influenza-virus- or RSV-positive patients) over peri-seasonal and summer baseline periods.
Results: No evident excess mortality was found among persons younger than 50 years during influenza-virus active periods. In all 50–64-year-olds together, influenza-virus-active periods were associated with excess mortality (on average yearly around 1.3–4.7 per 100,000 persons over respectively peri-seasonal and summer baseline period). Among elderly excess mortality was on average around 102–152 per 100,000 persons yearly. Influenza-related excess hospitalisation was highest in 0–1-year-olds, namely around 79–271 per 100,000 children yearly. For non-high-risk 50–64-year olds and elderly these figures were 18–38 and 111–208 per 100,000 persons yearly. The highest RSV-related excess hospitalisation was demonstrated in 0–1-year-olds (on average around 523–699 per 100,000 yearly). Among elderly this excess was on average around 50–140 per 100,000 persons yearly.
Conclusion: Influenza-virus-active periods were associated with excess mortality in 50–64-year-olds but not in 0–1-year olds. Also a clear influenza-virus-related excess hospitalisation was demonstrated among both 0–1- and non-high-risk 50–64-year-olds. Influenza-virus-related mortality among elderly appeared however far highest. RSV-active periods were associated with excess hospitalisation particularly in the youngest and to a lesser extent also in the oldest.
P1565 Seroprevalence of HIV, hepatitis B and hepatitis C among intravenous drug users, men who have sex with men and commercial sex workers in Kosova
G. Mulliqi, A. Kurti, L. Raka, L. Begolli, L. Berisha, A. Jaka, H. Fejza, S. Mehmeti (Pristina, RS)
Objectives: Intravenous drug users (IDU), men who have sex with men (MSM) and female commercial sex workers (CSW) are known to be at high risk of acquiring blood-borne infections. The objective of this study was to determine the seroprevalence of HIV, hepatitis B (HBV) and hepatitis C (HCV) infections among these risk groups.
Methods: A prospective study was performed among total of 422 IDU, MSM and CSW at three cities of Kosova (Prishtina [capital], Prizren and Ferizaj). The study was carried out during the year 2006. The subjects consisted of randomly selected 199 intravenous drug users (IVDU), 154 commercial sex workers (CSW) and 69 men who have sex with men (MSM). The blood samples were tested for HBsAg, anti-HCV antibodies and anti-HIV antibodies using ELISA commercial kits (Nubenco Diagnostics, USA).
Results: Of the 199 IDU, 161 (80.1%) were male and 38 (19.9%) female. The mean age was 23.2 years. The mean length of self-reported injecting drug use was 3.4 years. Among 199 IDU, in 36 (18.1%) of them were detected anti-HCV antibodies, whereas HBsAg as a marker of HBV was found in 29 of them (14.6%). All tested persons among this risk group were HIV negative. One fourth of them shared needles. Prevalence rates of hepatitis B among MSM and CSW were 8.7% and 18.1%, respectively. Five CSW were positive for anti-HCV antibodies (3.2%). Of all tested samples 48% were positive for Chlamydia infection.
Conclusion: Prevalence rates of HIV and hepatitis in Kosova remain low at present, but risk behaviours are present already and renewed efforts are necessary to intervene effectively in these groups.
P1566 HIV infection and the immigration phenomenon. Consequences on inpatient hospitalisation in a large Italian hospital, 2000–2006
R. Manfredi, L. Calza (Bologna, IT)
Introduction: Immigration is a recent phenomenon in Italy, mainly caused by the sudden and unexpected arrival of wawes of foreign citizens, refugees, and individuals escaping from war. This phenomenon is of great concern due to its serious socio-economic and healthcare impact.
Patients and Methods: A prospective survey of all charts of patients (p) hospitalised or followed on day-hospital (DH) basis at our Infectious Disease ward until mid-2006, allowed us to assess the frequency of admission of immigrants from extra-Western Europe (eWE), and to analyse multiple variables related to epidemiologic-clinical features.
Results: The rate of p immigrated from eWE showed a significant increase among our inpatients, and at a lesser extent and later for DH admissions: 7.7% and 3.1% during the year 2000, 10.1% and 4.6% in 2001, 13.2% and 6.2% in 2002, 17.9% and 7.9% in 2003, 21.3% and 8.9% in 2004, 17.7% and 10.8% in 2005, up to 17.9% and 11.3% in the year 2006 (p<0.0001 for inpatients; p < 0.001 for DH p). Over 60% of p came from Africa, followed by Eastern Europe, Asia, and Central-Southern America. When comparing the admission features of WE citizens with those of p coming from abroad, no differences were found as to duration-intensity of assistance, with HIV disease prevailing among regular admissions (35.1%), and DH access (33.8%), followed by acute-chronic hepatitis, pulmonary or other-site tuberculosis, central nervous system and respiratory tract infection, and sexually-transmitted diseases. HIV-infected immigrants were frequently (60.1% of cases) AIDS presenters, and less than 5% of them were already on anti-HIV therapy upon admission. While the frequency of HIV-associated admissions did not show differences in the considered 7-year period, p from eWE had an increasing frequency of tuberculosis, skin-soft tissue infection, exanthems, gastroenteric-parasitic diseases, and malaria (p < 0.05 to <0.0001).
Conclusions: A continued monitoring of this phenomenon is strongly warranted, to improve a sustainable social-cultural network, to plan health resource allocation, and to define adequate and well-targeted prevention and public health measures.
P1567 Improving handling of blood exposure incidents: a 3-year prospective study
P.T.L. van Wijk, M. Pelk-Jongen, C. Wijkmans, A. Voss, P.M. Schneeberger ('s-Hertogenbosch, Nijmegen, NL)
Objectives: Handling and counselling of blood exposure incidents can be complicated, time costing and cause a lot of anxiety and stress for the injured. Incidents occur at all hours, both inside and outside the hospital. Depending on the kind of injury and the infectivity of the source, they pose a risk for transmission of hepatitis B (HBV), hepatitis C (HCV) en HIV.
Methods: To assess quality improvement, all blood exposure incidents registered throughout 2003–2005 by a 24 hour accessible expert telephone centre in a Dutch region (500.000 inhabitants) were analysed. The expert-centre was established for blood exposure incidents in and outside the hospital. Infection control practitioners make a risk-assessment and arrange all logistic requirements, treatment and follow-up according to standardised procedures. Weekly feed back was given to counsellors to achieve adherence to the protocol.
We analysed occurrence of injuries, HBV vaccination level of the injured, identification of sources, the time in which a HIV test could be accomplished when risk of HIV transmission was involved and the adherence to the protocol by the expert centre.
Results: In average 465 reports were registered per year, 50% took place outside the hospital and 33% of the reports were not during office hours. Sources for blood analysis could be identified in 85% of the cases. HBV vaccination level in healthcare workers outside the hospital rose from 34% to 70% during the three years. This caused reduction of administering immunoglobulines and unnecessary laboratory testing. HIV tests could progressively be performed more rapidly during the registration period due to earlier reporting and improvement of logistics. No PEP had to be administered unnecessary. In assessing the quality of the handling by the expert centre, in 2003, in 37% of the reports flaws in the protocol were identified, in 2005 this was 8%.
Conclusion: Handling of blood exposure incidents should logistically also be optimised outside office hours and outside hospitals. Higher level of HBV vaccination makes handling far more simple. Faster reporting of incidents extends the necessary time for laboratory tests and administering medication. Quicker HIV testing prevents unnecessary administering of medication. Less flaws in applying the protocol, prevents unnecessary interventions and is cost-effective. Through a 24/7 centralised counselling facility for handling of blood exposures we achieved significant improvements.
P1568 Enrollment of women in HIV clinical trials in Rwanda
L. Ackerson, F. Abalikumwe, M. Goble, R. Horsburgh (Kigali, RW)
Objective: To evaluate the proportion of women enrolled in clinical trials of investigational HIV therapies and assess factors that may influence enrollment of women.
Methods: Protocols were identified from a search of the AIDS Clinical Trials Information Service (ACTIS), a database that provides detailed information, including entry and exclusion criteria, on federally and privately sponsored clinical trials for HIV/AIDS therapies initiated between 2000 and 2004. Demographic and enrollment information was requested by letter from trial sponsors of all protocols.
Results: A total of 221 privately sponsored protocols were identified from the ACTIS database. Sponsors of 156 protocols provided demographic data stratified by gender. Enrollment of women in this sample ranged from 0% to 64% (mean 11.6%). Twenty-four trials enrolled no women. All were in phase 1 or 2 of development. All 24 protocols permitted enrollment of non-pregnant women. Sixteen of 24 studies required some form of birth control, including barrier methods, abstinence, and/or oral contraceptives. Eight of 24 protocols did not specify any reproductive-related inclusion/exclusion criteria. None of the 24 protocols had laboratory-based exclusion criteria that appeared to exclude women.
Conclusion: Factors other than gender-based eligibility criteria appear to be responsible for the failure of women to enroll in this sample of HIV clinical trials. To further explore this, The Project San Francisco (psf), Project UBUZIMA (with microbicide) and We-ACT, all located in Kigali, are developing an HIV investigational new drugs database to assess the recruitment and to promote the retention of women in current and future HIV clinical trials.
P1569 Strategies for containment of biological risk in reclaiming a large urban area used as a waste pit by a serum and vaccine factory
A. Amendola, S. Bianchi, M. Canuti, R. Koncan, A. Zanetti, G. Tridente, G. Cornaglia (Milan, Verona, IT)
Objectives: The 12,000 m2 waste pit of the former Istituto Sieroterapico Milanese, in the urban centre of Milan, Italy, has recently been reclaimed after nearly a decade of complete abandonment. Since the early 1900s, different bio-pharmacological products – vaccines, prophylactic sera, blood flasks, waste of various animal origins – had been buried at a depth not exceeding 1.5 m. Some material, such as animal waste, had decomposed in direct contact with the soil for a variable number of years, and therefore dispersed pathogens are likely to have interacted with the soil's own microbicidal properties. Other materials, such as human or veterinary vaccines, were recovered as sealed vials, and there was presumably a risk that their contents (either liquid or lyophilised) might have been totally or partially preserved. The aim of the whole exercise was to carry out a thorough reclamation whilst minimising the potential biological risks.
Methods: A preliminary electro-magnetometric investigation was performed on the whole area, identifying those sub-areas with uneven soil homogeneity attributable to the alleged presence of heterologous materials. A pilot intervention zone, the so-called ‘first experimental module’, was used to set up and test all the procedures. All the operations were conducted under a special tensostructure (base, 30×30 m), with air exchangers and extractors endowed with both ‘absolute’ (i.e., high-specification fluid filters used to remove small solid particles) and activated carbon filters. Removal of the soil was done using excavators over the whole area.
Results and Conclusions: Given the obvious impossibility of treating the whole area either by autoclaving or incineration, vibratory sifting of the possibly contaminated soil was necessary, permitting gross separation of the foreign materials. The sifted soil was subsequently put back in the original areas and submitted to 120-days' quarantine. The bio-pharmacological products isolated were stored in special containers and incinerated, after careful sampling for future studies of their residual biological activities related to both bacteria (Brucella abortus, Salmonella spp., Streptococcus spp., Bacillus anthracis) and viruses (Newcastle Disease Virus, Rhabdovirus, Canine Distemper Virus, Pestivirus, Poliovirus). Despite the lack of similar experience worldwide, the whole procedure was carried out under optimal conditions and with adequate levels of both environmental and human safety.
Epidemiology of MRSA
P1570 Isolation of methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leukocidin genes in a tertiary hospital in Greece
G. Ganderis, E. Vagiakou, M. Orfanidou, D. Petropoulou, S. Vourli, A. Vatopoulos, H. Malamou-Lada (Athens, GR)
Objectives: To isolate and investigate strains of methicillin-resistant Staphylococcus aureus for the presence of the lukS-PV, lukF-PV determinant of Panton-Valentine leukocidin from specimens obtained from adult patients with soft tissue abscesses and purulent skin infections.
Methods: Between 4 January and 17 October 2006, a total of 135 S. aureus strains were revealed from soft tissue abscesses and purulent skin infections.
S. aureus was identified by conventional methods. Antimicrobial susceptibility was tested by Kirby–Bauer method and the MICs by automated system VITEK 2. Polymerase chain reaction (PCR) was performed to detect the genes for PVL from the MRSA strains (lukS-PV, lukF-PV).
Results: Fifty-nine MRSA out of 135 S. aureus strains (59/135, 44%) were isolated from specimens and 12 MRSA strains were PVL-positive (12/59, 20%) shared a type IV SCCmec cassette (lukS-PV, lukF-PV genes). The sources of MRSA-PVL-positive strains were: soft tissue abscesses (8), furunculosis (2), cellulites (1), pyomyocitis (1). Eight MRSA-PVL positive strains were isolated from immunocompetent patients, two from patients with diabetes mellitus (all were community-acquired) and two from patients with persistent purulent infection in the site of previous orthopaedic surgery and history of hospitalisation during the last six months (hospital-acquired). The mean age of 12 patients (10 male/2 female) was 41 years (range 22–58 years). All MRSA-PVL positive strains were susceptible to macrolides, fluoroquinolones, tobramycin and glycopeptides, while they were resistant to oxacillin, kanamycin, tetracycline, and intermediate-susceptible to fusidic acid. The MICs of 12 strains to resistant antibiotics were: oxacillin ≥16 mg/L, tetracycline ≥16 mg/L, and fusidic acid between 8 mg/L and 16 mg/L.
Conclusions: (1) Twelve MRSA-PVL positive strains (carrying lukF, lukS genes, SCC mec type IV) were isolated from 12 patients during the last 10 months from soft tissue abscesses, furunculosis, cellulites, pyomyocitis. (2) The MRSA-PVL-positive strains had the same unique resistance pattern to antibiotics (resistance to kanamycin, oxacillin, tetracycline, and intermediate susceptibility to fusidic acid). (3) MRSA-PVL-positive strains is now an emerging problem in Greece.
P1571 Characterisation of methicillin resistant Staphylococcus aureus strains isolated in a tertiary care hospital in the United Arab Emirates
A. Sonnevend, T. Pal, E. Ungvari, J. Paszti, P. Jumaa, K. al-Dhaheri (Pecs, Budapest, HU; Al Ain, AE)
Objectives: The incidence of methicillin resistant Staphylococcus aureus (MRSA) is generally perceived to be low in the United Arab Emirates (UAE) compared to that of most western countries where its high prevalence is due, to a large extent, to the spread of epidemic MRSA (E-MRSA) clones. Our main aim was to type the local MRSA isolates and to compare them to the worldwide known E-MRSA strains. We investigated 50 MRSA strains isolated from individual patients between July 2002 and July 2003 at a tertiary care referral hospital, Tawam Hospital, Al Ain, UAE.
Methods: The presence of the mecA gene, and those coding for enterotoxin A-E, toxic shock syndrome toxin and exfoliativ toxin A and B were investigated by PCR. Furthermore, the antibiotic sensitivity and phage type of the isolates and the macro-restriction profile subsequent to SmaI digestion of the genom were also determined.
Results: All isolates harboured the mecA gene, 54% of them possessed at least one of the toxin genes. None of the strains exhibited reduced susceptibility to vancomycin, while resistance to other antibiotics varied. Surprisingly, as pheno-, and genotyping revealed, 26% of the isolates (13 strains) were identical or nearly identical to E-MRSA15, and 12% (6 strains) showed more than 80% similarity by PFGE to the USA100 clone, i.e. the most widespread MRSA clones in the United Kingdom and the United States, respectively.
Conclusion: The lack of detailed epidemiological data prevented us to determine the origin of these MRSA strains. However, patients receiving medical care outside of the UAE may serve as potential sources, although the extent of local spread remains to be investigated. The possibility of carrying E-MRSA strains either by patients or by healthcare workers between continents and countries calls for a well co-coordinated effort of authorities to limit the further spread of this pathogen.
P1572 Emergence of EMRSA-15 in a university hospital, Canary Islands
I. Montesinos, T. Delgado, B. Castro, M. Cuervo, M. Lecuona, M. Ramos, A. Sierra (La Laguna, ES)
Objectives: Since 2000, the rate of MRSA infections has increased at the University Hospital of Canary Islands (HUC) coinciding with the emergence and spread ofEMRSA-16 clone and replacement of the Iberian one. In 2003, a new digestion profile by PFGE was observed. The aim of this study is the characterisation of this new PFGE type emerged in the HUC.
Methods: Between 2003 to 2004 29 MRSA isolates showed this new PFGE profile and were obtained from clinical samples from hospitalised patients and out-patients at the HUC. Susceptibility testing and bacterial identification were performed using VITEK2 System (bioMérieux, France) and susceptibility testing for vancomycin, teicoplanin, linezolid and quinupristin-dalfopristin was assessed with E-test (AB BIODISK, Sweden). Molecular typing was performed by PFGE, MLST, SCCmec typing and protein A gen amplification (spa).
Results: All MRSA isolates studied were susceptible to gentamicin, tetracycline, rifampim, trimethoprim-sulfamethoxazole and tobramycin. They were all resistant to ciprofloxacin and 45% of them were susceptible to clindamycin and erythromycin. All of them were also susceptible to vancomycin, teicoplanin, linezolid and quinupistrin-dalfopristin by E-test. The SmaI digestion patterns obtained by PFGE were different to those obtained previously and included 4 PFGE subtypes. The sequence type of this clone was 22 by MLST and it revealed cassette IV by SCCmec typing, this corresponding with the pandemic clone EMRSA-15. The number of repeats obtained by spa typing was also different to those obtained up to date. Thirteen isolates had 17 repeats, five had 16 repeats and one of them 18 repeats. The first isolation of EMRSA-15 appeared in the HUC in April, 2003 isolated from a patient hospitalised in internal medicine with a respiratory colonisation. In 2003 and 2004 the 10% and the 13% of the MRSA isolated in the HUC were EMRSA-15 respectively.
Conclusions: Starting 2000, EMRSA-16 emerged at our hospital and soon became the predominant clone. In April 2003, a new MRSA clone, EMRSA-15, appeared in this changing panorama of MRSA clones at HUC. The EMRSA-15 and EMRSA-16 are the two predominant clones circulating in the UK.
P1573 Characterisation of an MRSA clone in Denmark similar to USA300 in PFGE pattern but with different properties: a potential pitfall when identifying USA300 isolates
A. Larsen, R. Goering, M. Stegger, K. Boye, M. Sørum, H. Westh, R. Skov (Copenhagen, DK; Omaha, US)
Objectives: In recent years, Denmark has experienced a dramatic increase in the incidence of MRSA, from 100 to 864 cases in 2002 and 2005, respectively. A comparison of PFGE patterns of the Danish MRSA isolates with the USA100–1100 reference isolates identified a large number of MRSA isolates with a PFGE profile similar to USA300. However, further analysis revealed that a significant number of these isolates were actually of a previously undescribed strain variant being indistinguishable from the classical USA300 by PFGE. The aim of this study was to characterise this USA300 variant with respect to epidemiology and a variety of important genetic markers.
Methods: All Danish MRSA isolates from 1999–2005 were collected at Statens Serum Institut. Discharge summaries for each patient/carrier were screened for epidemiological information regarding acquisition (hospital or community) and types of infection. All MRSA isolates were characterised by PFGE, SCCmec type, spa type, analysis of the mec-associated direct repeat unit (dru), presence or absence of the arginine catabolic mobile element (ACME), and genes encoding Panton-Valentine leukocidin (PVL).
Results: A total of 90 MRSA isolates was found to have PFGE patterns identical to USA300. This group, although indistinguishable by PFGE, was found to contain two distinctly different subtypes. Forty-two isolates were classical USA300, spa type t008, PVL positive, and SCCmec type IVa. However, 48 isolates were spa type t024 and lacked the PVL genes although still SCCmec type IVa. Further examination revealed that the t008 and t024 subtypes differed substantially, with dru type 9g and ACME present in all examined t008 isolates while the t024 isolates were dru type 10a with ACME absent from several isolates. In addition, a majority of the t008 (typical USA300) isolates were community acquired, while the t024 isolates were primarily associated with hospitals and residential homes.
Conclusion: An MRSA clone that could not be differentiated from USA300 by PFGE was detected in high numbers as a cause of infections in Denmark. However, this clone was found to differ substantially from typical USA300 in epidemiology. Proper identification of these isolates requires the use of tests beyond PFGE, such as spa typing with analysis of pvl genes, dru and ACME as additional markers.
P1574 Molecular epidemiology of methicillin-resistant Staphylococcus aureus from Swiss healthcare institutions in Northeastern Switzerland, 1992–2006
A. Imhof, G. Senn, C. Ruef (Zurich, CH)
Objectives: The emergence of methicillin-resistant Staphylococcus aureus (MRSA) has generated considerable concern among medical and public health professionals. We conducted this study to investigate the antimicrobial susceptibility and the molecular epidemiology of MRSA strains from Swiss healthcare institutions of Northeastern Switzerland between 1992 and 2006.
Methods: MRSA isolates (n = 1982) were susceptibility tested according to CLSI guidelines. PCR for Panton-Valentine leukocidin (PVL) genes were done from MRSA isolates from outpatient clinics and from isolates belonged to the 9 predominant PFGE types. Analysis of epidemiological and molecular typing data (PFGE) of MRSA strains were used to examine epidemiologic trends for MRSA.
Results: The proportion of MRSA strains resistant to gentamicin was 30%. 48% of the MRSA isolates were resistant to erythromycin and 46% to clindamycin. 17% revealed macrolide-lincosamide-streptogramin B resistance (MLS). More than 75% of strains were susceptible to tetracycline, rifampicin and cotrimoxazole; 62% of isolates were resistant to ciprofloxacin. Annual trends of the susceptibility pattern showed a significant decrease of resistance of ciprofloxacin (100% (1992) to 60% (2006), p < 0.001), tetracyline (98% (1992) to 16% (2006), p < 0.001), and rifampicin (83% (1992) to 8% (2006), p < 0.001). A significant increase of MLS was found (12% (1993) to 25% (2006), p = 0.029). All isolates were susceptible to linezolid and glycopeptides. Fifty-one percent of the isolates belonged to 9 PFGE types, of which three were predominant: USZ19 (20%), USZ20 (12%) and USZ21 (5%). PVL gene was found in 3% of the tested MRSA isolates. Interestingly, no PVL was found in a MRSA clone, which is epidemic in drug addicted people in the north-eastern Switzerland. However, PVL gene was found in a MRSA clone which was isolated during an outbreak in a dermatological clinic.
Conclusions: Specific clones of MRSA are circulating in hospitals and communities in north-eastern Switzerland. The susceptibility pattern of the isolates changed significantly during the observation period: Fewer isolates with resistance to non-β-lactam antibiotics was found. However, MLS resistance increased significantly during the last years.
P1575 Molecular characterisation of hospital-acquired methicillin-resistant Staphylococcus aureus in a Turkish university hospital
H. Akoglu, P. Zarakolu, B. Altun, G. Hascelik, S. Unal (Ankara, TR)
Objective: The aim of this study is to evaluate the molecular characterisation of hospital-acquired (HA) methicillin-resistant Staphylococcus aureus (MRSA) isolates by analysing the presence of PVL genes and SCCmec typing.
Methods: A total of 110 MRSA isolates from clinical samples (35 blood, 37 pus, 23 deep tracheal aspiration, 5 catheter, and 10 other samples) of inpatients with nosocomial infections at Hacettepe University Adult Hospital between January 2004 and December 2005 were included in the study. Only one isolate from each patient was included. The identification of the isolates was made by Sceptor automated system (Becton Dickinson, USA). The isolates were retested for methicillin resistance by oxacillin (1 μg, Oxoid, UK) disk diffusion test according to CLSI and for the presence of mecA gene by polymerase chain reaction (PCR). The presence of PVL genes and SCCmec types were detected also by PCR analysis. Isolates were stored at −80°C until studied. S. aureus ATCC 29213, S. aureus 27R and S. aureus 8328 were included as control strains.
Results: All 110 HA-MRSA isolates were positive for mecA gene. Of the isolates, 68 (56%) were harbouring SCCmec type III, 38 (30%) SCCmec variant IIIB, and 3 (2.5%) SCCmec type IV. One isolate positive for mecA gene, could not be typed for SCCmec gene. PVL was positive in 14 (12.7%) of 110 HA-MRSA isolates. Ten of PVL positive strains carried SCCmec III; whereas 4 of them carried SCCmec type variant IIIB. None of the strains harbouring SCCmec type IV were positive for PVL genes.
Conclusion: Majority of the HA-MRSA isolates in our hospital are carrying SCCmec III, or variant IIIB. Although PVL is a common virulence factor of community acquired-MRSA, HA-MRSA isolates in our centre have a considerable rate of PVL positivity pointing out the fact of changing epidemiology of MRSA isolates.
P1576 Molecular epidemiology of hospital-acquired methicillin-resistant Staphylococcus aureus in Russia
M.V. Afanas'ev, S.V. Karakashev, E.N. Ilina, S.A. Grudinina, A-S. Salem, S.V. Sidorenko, V.M. Govorun (Moscow, RU)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) is a common cause of nosocomial infection. The purpose of this study was to evaluate clonality of hospital-acquired MRSA (HA-MRSA) circulating in Russian Federation and to compare different multiplex PCR techniques with SNP-based approach for MRSA typing.
Methods: Epidemiologically unrelated MRSA isolates (n = 62) from Moscow hospitals have been selected for typing. Genomic DNA from clinical isolates was purified using “DNA express” kit (Lytech Ltd, Russia). Staphylococcus chromosomal cassette mec (SCCmec) typing and detection of Panton-Valentine leukocidin (PVL) gene were performed by PCR using previously described methods. Seven loci from five housekeeping genes (arcC162, arcC210, aroE132, gmk123, tpi241, tpi243 and yqiL333) were used for SNP-typing. Detection of particular nucleotides in selected loci was carried out in thermocyclic primer extension reaction, followed by mass-spectrometric detection of the products. Standard MLST-typing procedure was preformed as reference method.
Results: Results of SCCmec, SNP-typing and MLST-typing are presented in the Table . The majority of MRSA isolates (93.6%) belong to world-wide disseminated clonal complex (CC) 8, Three isolates (4.8%) belong to CC 1. All ST 239 isolates were found to carry SCCmec type III, and ST 8 isolates carried SCCmec type IV. No PVL genes were detected in SCCmec type IV isolates included in this study.
| No. of isolates (%) | Traditional typing | Olivera's M-PCR typing | Zhang's M-PCR typing | SNP-typing | MLST |
|---|---|---|---|---|---|
| 51 (82.3) | Type IV | Type IV | mecA gene | T-A-T-G-T-A-C | ST 8 |
| 3 (4.8) | Type IV | Type IV | mecA gene | C-A-T-G-C-A-C | ST 1 |
| 1 (1.6) | mecA gene | mecA gene | Type III | C-A-T-G-T-A-T | n/d |
| 1 (1.6) | Type III | Type IIIa | Type III | T-A-A-G-T-A-C | ST 239 |
| 6 (9.7) | Type III+V | Type IIIa | Type III | T-A-A-G-T-A-C | ST 239 |
ST: sequence type; n/d: not determined
Conclusion: Among Russian MRSA CC8 isolates carrying SCCmec IV type are predominant. SNP-typing is powerful tool for studies of molecular epidemiology of MRSA.
P1577 SCCmec type and epidemiological characteristics of MRSA isolates in the Netherlands
I. Van den Broek, X. Huijsdens, E. Tiemersma, I. van Loo, J. Kluytmans (Bilthoven, Tilburg, Amsterdam, NL)
Objectives: To investigate the development of SCCmec types of Methicillin Resistant Staphylococcus aureus (MRSA) in the Netherlands in relation to epidemiological characteristics.
Methods: 76 MRSA isolates were randomly selected from the national MRSA database at the National Institute for Public Health and the Environment, in the period January 2003 to September 2005. We compared the epidemiological characteristics of the SCCmec types I, II and III ('Hospital Acquired') with types IV and V ('Community Acquired'). Patient information was provided by the referring hospitals.
Results: Of the 76 isolates, 43 could be typed by SCCmec and 17 of these had the ‘HA'-type (4 I, 7 II and 6 III) whereas 26 had the ‘CA'-type (21 IV and 5 V). The development of these two groups over the years is shown in the figure . Of the HA-type isolates 82% were attributable to healthcare exposure while this was the case for only 35% of CA-type isolates; the first were more likely to originate from patients who had been admitted to a foreign hospital than the latter (65% versus 12%, p < 0.01). HA-isolates were found by intentional MRSA screening more frequently than CA-isolates (88% vs 42%, p < 0.01). CA-type isolates were more often associated with clinical signs of MRSA infection than HA-type isolates (46% vs 18%, p = 0.1), most of which were skin infections (92% vs 100%, ns). The living circumstances (degree of urbanisation) and occupation of the patient had no clear link to SCCmec type, though CA-type isolates more frequently originated from jobless persons than HA-type isolates (39% vs 18%).

MRSA isolates from the Netherlands, 2003–2006.
Conclusion: The increase of MRSA in the Netherlands seems to be attributable to the CA-type isolates. The HA-type isolates are mainly derived from patients who have been hospitalised abroad and found by intentional screening. The majority of CA-types cannot be related to the healthcare setting and are coincidental findings indicating that the patients were not considered to be at increased risk. Further studies are warranted to identify the patients at risk for CA-MRSA.
P1578 Comparative genome hybridisation of epidemic and sporadic methicillin-resistant Staphylococcus aureus strains did not reveal a specific genetic factor
G. Kuhn, T. Koessler, P. Francois, A. Huyghe, J. Schrenzel, P. Francioli, D.S. Blanc (Lausanne, Geneva, CH)
Introduction: Staphylococcus aureus has proven an outstanding capacity to adapt to hospital environmental conditions. This behaviour included the evolution of Methicillin resistant S. aureus (MRSA) clones which have spread among hospital settings worldwide with fatal consequences for health institutions.
Objectives: Identification of the specific presence or absence of genetic factors associated with epidemic behaviour of Methicillin resistant Staphylococcus aureus.
Methods: We have collected 12 genetically diverse strains which have at local or at international level caused epidemic outbreaks, and 8 sporadic isolates which have never been transmitted between patients despite colonisation during several weeks. Genetic analysis was performed using comparative genome hybridisation with an S. aureus multi-strain oligo microarray, which was constructed on the basis of published genome sequences of strains COL, MW2, Mu50, N315. The datasets obtained for both collections were subtracted.
Results and Discussion: No genetic factor was associated by its specific presence or absence with epidemic behaviour. This was also true, when stringency for data collection was relaxed, i.e. when values of estimated probability of presence (EPP, an estimate for the risk of false positive signals) were increased from 1% to 20%. Several hypotheses can be derived to explain this finding. First, specific factors might have been missed during analysis, because no epidemic strains have been considered for construction of the microarray. Second, epidemic behaviour might be associated with specific alleles or a specific transcriptional pattern and not with accessory genetic elements. Third, epidemic behaviour might have evolved independently and with different genetic determinants in different phylogenetic lineages.
P1579 Low prevalence of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in two West German hospitals
K. Kösters, I. Greiffendorf, E. Schemken-Birk, E. Becker-Boost, C. Becker (Krefeld, DE)
Objectives: To determine the prevalence of reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated in two German teaching hospitals between January 2002 and October 2006.
Methods: Vancomycin and teicoplanin minimal inhibitory concentrations (MIC) of clinical MRSA strains were determined with the Etest macromethod on brain heart infusion agar in addition to routine susceptibility testing with VITEK2.
Results: 671 MRSA strains collected between January 2002 and October 2006 were analysed. MICs for vancomycin and teicoplanin ranged from 1.5 to 6 μg/mL and 0.5 to 8 μg/mL, respectively. According to Etest interpretation guidelines no glycopeptide resistent or intermediate strains were found. Mean vancomycin MIC was consistently higher than teicoplanin MIC. An increase in mean glycopeptide MICs over the study period could not be demonstrated.
Conclusion: Reduced glycopeptide susceptibility could not be detected with the Etest macromethod in MRSA strains isolated from patients in two West German hospitals over a four and a half year period. Additional quantitative determination of glycopeptide resistance such as the Etest macromethod should be reserved for cases where unusual susceptibility is suspected or VITEK2 detects high glycopeptide MICs (≥ 4 μg/mL).
P1580 Characterisation of hospital- and community-acquired methicillin-resistant Staphylococcus aureus in Hong Kong
G. Siu, M. Boost, M. O'Donoghue (Kowloon, HK)
Objectives: In the past, methicillin-resistant Staphylococcus aureus (MRSA) was mainly a pathogen associated with hospital acquired infections (HA-MRSA), but in recent years community-acquired strains (CA-MRSA) causing infections with high morbidity and mortality have emerged. In Hong Kong, although almost 50% of hospital isolates of S. aureus are HA-MRSA community isolation of HA-MRSA remains low. However, in the last year, there have been several reports of infections with CA-MRSA. This study aimed to characterise both hospital and community isolates of MRSA.
Methods: The presence of the mecA resistance gene was confirmed in 139 hospital isolates (clinical isolates) and 13 community isolates (nasal carriage) of MRSA. The mecA gene was further characterised by multiplex PCR. Those found to harbour the SCCmec IV element were investigated for the presence of genes coding for Panton-Valentine leucocidin and for agr to determine their prototypic or archaic type.
Results: 95% of HA-MRSA strains were found to be multi-drug resistant, displaying resistance to an average of 6 non-β-lactam drugs. 91 strains (65%) harboured SCCmec variant IIIA and 22 (16%) SCCmec III. One strain was PVL positive. CA-MRSA remains uncommon in hospital isolates with only 3 (2%) of isolates harbouring the SCCmec IV element. These belonged to the less virulent archaic type only one being a PVL producer. In contrast, over 50% of community isolates were CA-MRSA, 3 strains with the SCCmec IV element and 4 with new SCCmec patterns. Fusidic acid and erythromycin resistance was present in >80% of community isolates. Characterisation revealed the presence of both archaic strains and a highly resistant PVL-producing prototypic strain.
Conclusions: CA-MRSA remains rare in hospital infections in Hong Kong, with most HA-MRSA strains harbouring type III subtypes of the SCCmec element. One healthy community dwelling person was found to be colonised by a highly virulent agr type 3 positive strain, which has not previously been reported from nasal carriage. As colonisation typically precedes infection, the potential of increased incidence of severe CA-MRSA-associated infections is likely.
P1581 Genetic analysis of high- and low-level Mupirocin-resistant MRSA isolated in Kuwaiti hospitals
E. Udo, N. Al-Sweih, B. Mathew, C. Noronha (Kuwait, KW)
Background: Mupirocin resistance in Staphylococci is expressed either as high-level (MIC >512 mg/L) or low-level (MIC 8–256 mg/L) resistance. The prevalence of high-level mupirocin resistance has been increasing in MRSA isolated in Kuwait in recent years. This necessitated monitoring MRSA isolated in Kuwaiti hospitals at regular intervals for changes in their mupirocin resistance patterns.
Objective: To monitor the prevalence of mupirocin resistance in MRSA isolated in Kuwait hospitals and determine its genetic location.
Methods: MRSA isolated from six hospitals between January 2000 and December 2005, were tested for susceptibility to antibacterial agents by disk diffusion. Mupirocin MICs was determined with Etest strips. The presence of mupA genes that encodes high-level mupirocin resistance (mupH) was investigated by PCR. Pulsed-field gel electrophoresis (PFGE) was used to study their genetic relatedness. Curing and transfer experiments were used to determine the plasmid or chromosomal location of mupirocin resistance.
Results: A total of 4,982 MRSA isolates were investigated. The prevalence of mupH declined from 22% in 2000 to 1.8% in 2005. In contrast, the prevalence of low-level resistance increased from 3.6% in 2000 to 90.8% in 2004 before decreasing to 65.7% in 2005. Transfer experiments revealed that mupH was located on a 38-kb conjugative plasmid while the low-level resistance was chromosomal. The mupA gene was detected only in the mupH isolates. PFGE analysis revealed that the low-level mupirocin-resistant isolates belonged to six different clones. Two of these clones, constituting 87.6% of the isolates, were detected in all six hospitals and were related to isolates that expressed high-level resistance.
Conclusions: This study revealed a dramatic shift in the prevalence of high- and low-level mupirocin resistance among Kuwait MRSA isolates. The relatedness of some low-level mupirocin-resistant isolates to those expressing high-level resistance suggests that these isolates have arisen by the loss of the 38-kb mupA plasmid. The study also highlights the value of regular surveillance in detecting changes in resistance and clonal patterns of MRSA isolates in healthcare environments
P1582 Molecular characterisation of community-associated and healthcare-associated methicillin-resistant Staphylococcus aureus in Korea
E.S. Kim, H.B. Kim, J.S. Song, K.U. Park, S. Shin, H.J. Choi, D.M. Kim, M.S. Lee, W.B. Park, N.J. Kim, M.D. Oh, E.C. Kim, K.W. Choe (Goyang, Seoul, Gwangju, KR)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA), a leading cause of nosocomial infections worldwide, has become a big problem in the community as well. Despite the high prevalence of MRSA in tertiary hospitals, there were only a few reports of MRSA infections in the community in Korea. This study was performed to determine the molecular characteristics of community-associated MRSA (CA-MRSA) in Korea.
Methods: Seventy-two CA-MRSA strains and 72 healthcare-associated MRSA strains (HA-MRSA) were collected from 7 hospitals between January and June 2005. To ascertain the presence of the risk factors for MRSA acquisition, the medical records and the database of the Health Insurance Review Agency were reviewed. Antibiotic susceptibility tests were done by the disk diffusion method. Staphylococcal cassette chromosome mec (SCCmec) typing and multilocus sequence typing (MLST) were performed as previously described, and the genetic relationship was analysed by the eBURST programme. The Panton-Valentine leukocidin (PVL) gene was screened by polymerase chain reaction.
Results: Nineteen and 24 CA-MRSA strains were classified as pathogens and colonisers, respectively. Twenty-nine CA-MRSA isolates from patients with chronic otitis media (COM) were categorised as a separate group due to the uncertainty of their clinical significance. Multi-drug resistance was observed in 9 (47%), 9 (38%), and 28 (97%) isolates of the 3 above-mentioned groups, respectively. Sequence type 72 and SCCmec type IVA (ST72-MRSA-IVA) were most prevalent among CA-MRSA. However, most of the COM isolates belonged to the ST5-MRSA-II or the ST239-MRSA-IIIA strains, which are common among HA-MRSA in Korea. While only 1 of 25 ST72-MRSA strains showed multidrug resistance, all of the ST5-MRSA and the ST239-MRSA strains were multi-drug resistant. None of the CA-MRSA and the HA-MRSA strains had the PVL gene.
Conclusion: This study shows that there are some CA-MRSA clones in Korea which are different from HA-MRSA clones in Korea and CA-MRSA clones in other countries.
P1583 Molecular patterns of community-acquired methicillin-resistant Staphylococcus aureus strains in Malaysian carriers
V. Neela, N. Mariana, S. Zamberi (Selangor, MY)
Objective: The ultimate aim of the study is to perform molecular characterisation of methicillin-resistant Staphylococcus aureus (CA-MRSA) strains to understand the drug resistance and virulent properties. Knowledge on the molecular properties is important for the correct treatment and efficient management of CA-MRSA strains. Specific objectives are to determine the molecular patterns of local strains focusing on the SCCmec (Staphylococcal cassette chromosome mec element) types, presence of Panton-Valentine leukocidin (PVL) gene and the strain types (ST).
Methods: Twenty-two S. aureus isolates isolated from 100 university students screened during November 2005 to February 2006 were identified as MRSA. Phenotypic confirmation was carried out by antibiotic susceptibility test against various antibiotics (vancomycin, gentamicin, erythromycin, and mupirocin). All isolates were confirmed genotypically as MRSA using mecA primers. Each isolate was further investigated for CA-MRSA by SCCmec typing using published primers. Presence of PVL genes in all CA-MRSA isolates was also determined. Multi locus sequence typing (MLST) analysis of each CA-MRSA isolates was carried out to identify the strain type (ST).
Results: Of the twenty-two MRSA isolates, three isolates were identified as CA-MRSA. The molecular pattern of the three CA-MRSA isolates analysed by SCCmec typing revealed two SCCmec types. Two isolates were positive for SCCmec type V and one isolate was positive for SCCmec type IVa. The two SCCmec type V isolates were sensitive to non β-lactams while the SCCmec type IVa positive isolate was resistant to non β-lactam antibiotics and also carried the PVL gene. Positive signal for PVL gene was not observed in the SCCmec type V isolates. MLST analysis of the three CA-MRSA strains showed the strain type for SCCmec type 1Va isolate as ST-80, while SCCmec type V isolates belonged to ST-34.
Conclusion: The molecular patterns of the local CA-MRSA strains studied give valuable information on the SCCmec type, virulent gene carried and the clonal type circulating in local communities. The molecular information of the local strains could be very useful in prescribing correct antibiotics enhancing appropriate treatment and successful management of the CA-MRSA strains in Malaysia.
P1584 Community-acquired methicillin-resistant Staphylococcus aureus in Stockholm with increased genetic diversity
H. Fang, G. Hedin, G. Li, C. Nord (Stockholm, SE)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) has become a worldwide problem. Infections caused by MRSA have now also been found in the community. The first community-acquired MRSA (CA-MRSA) in the southern Stockholm region was documented in 2000. The present study is aimed to investigate the molecular epidemiology of CA-MRSA in the southern Stockholm region during the period 2000 to 2005.
Methods: One hundred and fourteen consecutive non-repeated CA-MRSA isolates from the southern Stockholm region between 2000 and 2005 were included in the study. All strains were analysed by pulsed-field gel electrophoresis (PFGE) and staphylococcal chromosomal cassette mec (SCCmec) typing. At least one isolate of each PFGE pattern combined with varied SCCmec types was selected for multilocus sequence typing (MLST). The presence of the Panton-Valentine leukocidin (PVL) genes was detected by PCR.
Results: During the 6-year period, 114 CA-MRSA isolates were collected, and the majority (66%) of them were isolated in the recent 2 years (32 strains and 44 strains for 2004 and 2005, respectively). Thirty-six different PFGE patterns and 16 sequence types (STs) were identified among the strains. Among these genotypes, ST80-MRSA-IV carrying the PVL genes was the most prominent MRSA clone (34%) during all 6 years. However, increased genetic diversity of the CA-MRSA strains in the region was observed during the last years. Only eight sequence types were identified during 2000–2003 when MRSA was emerging in the community, while 13 STs were observed during 2005. Furthermore, the MRSA clones previously regarded as hospital-associated have also been identified in the community, i.e., STs 22 and 45. Regarding SCCmec typing, types IV and V were detected in 83% (95/114) and 15% (17/114) of the strains, respectively. One SCCmec pattern found in two isolates with ST 150 could not be classified as a known type. The same PFGE banding profile was presented by both ST 150 isolates with SCCmec V and the new SCCmec pattern. The PVL genes were found in 53% (60/114) of the strains.
Conclusion: The frequency of CA-MRSA has been increasing in Stockholm during the last years. ST 80 predominated while a polyclonal tendency was observed. SCCmec IV and V were present with different genetic backgrounds. The study also indicated that cross-talking between the hospital and community strains of MRSA have started in the region.
P1585 First cases of infections caused by community-associated methicillin-resistant Staphylococcus aureus in Bulgaria
D. Nashev, L. Bizeva, K. Toshkova, I. Alexiev, D. Beshkov (Sofia, BG)
Objectives: We report first cases of skin infections caused by Panton-Valentine leukocidin (PVL) positive cMRSA strains in Bulgaria.
Methods: During 2005 and 2006 five cases of infection caused by MRSA with unusual susceptibility patterns and community onset were documented in our institution. The patients were young people with no history of previous MRSA isolation, no history of hospitalisation or invasive manipulations in the past year and without permanent medical devices. The skin infections were represented by furunculosis in 4 patients and cutaneous abscesses in one patient. MRSA were isolated from skin lesions as well as from anterior nares of 4 patients with furunculosis. Antibiotic susceptibility testing was performed according to CLSI. Methicillin-resistance was confirmed by latex agglutination test for PBP 2a and PCR detection of mecA gene. The presence of PVL genes (lukS-PV-lukF-PV) were assessed by PCR. The isolates were further analysed by spa-sequence typing, spa types were designated using Ridom Staphtype software®.
Results: All isolates in the present study were found positive for PVL genes. MRSA isolates from 4 patients belonged to spa type t008 and from one patient to spa type t044. In all 4 cases caused by t008 strain, nasal carriage of the strain with the same spa type was detected. The isolates belonging to t008 showed resistance to erythromycin, kanamycin, tetracycline and ciprofloxacin and were susceptible to fusidic acid. The isolate belonging to t044 were resistant to kanamycin, tetracycline as well as to fusidic acid. The existing sequence databases for spa types allowed specific spa types to be associated with particular MLST sequence type (ST). The isolates belonging to spa type t044 represent the major European cMRSA clone ST 80. MRSA isolates with spa type t008 are frequently associated with ST8. However, cMRSA isolates belonging to ST8 cannot be discriminated from hospital MRSA isolates of this lineage by spa sequence typing alone; they also exhibit t008. All patients were succefully treated with clindamycin or trimethoprim-sulfamethoxazole in combination with rifampin. Nasal carriage was supressed with intranasal mupirocin, but 3 months later in one patient recolonisation with the same t008, PVL positive strain occurred.
Conclusion: The appearance of the first cMRSA strains should raise a great concern about diagnosis and treatment of these infections in Bulgaria.
P1586 Community-associated MRSA clone encoding toxic shock syndrome toxin 1 (TSST-1) in England
M.J. Ellington, M. Ganner, C. East, M.A. Ganner, M. Warner, A.M. Kearns (London, UK)
Objectives: TSST-1 is a superantigenic toxin associated with toxic shock syndrome and staphylococcal scarlet fever amongst other diseases. Traditionally, TSST-1 has been associated with ST30, agr type 3 methicillin-susceptible S. aureus but, recently, new community-associated MRSA (CA-MRSA) clones have been reported as TSST-1-positive in Europe (ST30, agr3 & ST5 agr2). We aimed to detect and characterise TSST-1-positive MRSA referred to the England and Wales Staphylococcal Reference Unit (SRU).
Methods: All putative CA-MRSA isolates referred to SRU in 2005 were tested by PCR for the presence of toxin genes including for PVL and TSST-1. TSST-1 (encoded by tst) positive isolates were characterised further by spa typing and SmaI PFGE. Further PCRs were used to detect: mecA and the SCCmec cassette type, ccr type and agr allotype. MICs of a range of antibiotics were determined. Patient demographic and clinical data were retained for analysis.
Results: Ten tst-positive MRSA isolates were similar to a new clone described in France and Switzerland; they were agr type 2, harboured SCCmecIVa, were related by pulsotype and spa typing; 8/10 were spa type t002, the remaining two were closely related to spa type t002 (t1737). All 10 isolates were β-lactam resistant and 9/10 were fusidic acid resistant. Unlike tst-negative, PVL-positive spa type t002 CA-MRSA isolates, these 10 isolates were susceptible to tetracycline in addition they were susceptible to ciprofloxacin, gentamicin, clindamycin, trimethoprim, vancomycin, linezolid and mupirocin. Five of the 10 patients were community based, 5 were females, 4 were male and 1 patient was of undisclosed sex. The age range was <1 to 76 y (mode ≤1y, median 33 y). Four isolates were from nasal swabs, the remainder were taken from: bacteraemia, wound infection, urinary tract infection, red umbilicus, line related infection and a skin and soft tissue infection. Three of the 10 cases were sporadic, a further three were from a healthcare associated outbreak, the remaining four were related geographically.
Conclusion: TSST-1-positive MRSA similar to the ST5 clone found in Europe, are present, albeit in small numbers, in England. Currently, these isolates show low levels of antibiotic resistance. Worryingly, like PVL positive MRSA, the emergence of these TSST-1 positive MRSA strains portends higher MRSA rates among the young with attendant impact on infection control and empiric therapeutic strategies.
P1587 The molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus at a London teaching hospital
J. Otter, G. French (London, UK)
Objectives: Community-associated MRSA (CA-MRSA) are common in some parts of the world such as the USA, but comparatively few have been reported in the UK. We retrospectively analysed a collection of MRSA from our London Teaching Hospital to determine whether CA-MRSA have emerged in our patient population.
Methods: We used ciprofloxacin-susceptibility as a phenotypic marker of CA-MRSA because nearly all healthcare-associated MRSA in our hospital are ciprofloxacin-resistant (CIP-R) and most CA-MRSA reported in the UK have been ciprofloxacin-susceptible (CIP-S). We therefore analysed surviving CIP-S MRSA isolates for antimicrobial susceptibility, possession of mecA, staphylococcal cassette chromosome mec (SCCmec) type, spa type and presence of Panton-Valentine leukocidin (PVL) genes. Multilocus sequence typing (MLST) was performed on representative spa types.
Results: CIP-S MRSA were isolated from 414 unique patients during 2000–2005. Only 122 (29%) were available for testing and therefore the results should be interpreted with caution. However, compared with CIP-R isolates, CIP-S MRSA were associated with significantly younger patients (average age 39.8y, (95%CI 35.3–44.3) vs. 62.6y (95%CI 62.1–63.1), unpaired t-test p < 0.0001) (figure ), were isolated more frequently from wound infections and were more frequently susceptible to non-β-lactam antimicrobials. These features are consistent with CA-MRSA. CIP-S MRSA were isolated from adult in-patients (40% of cases), outpatients (23%), patients from A&E (16%) and paediatric in-patients (14%).

Age distribution of 122 ciprofloxacin-susceptible MRSA isolates from 2000–2005 compared with all 27,422 MRSA isolates at Guy's and St. Thomas' NHS Foundation Trust from 2001–2004.
The majority of the isolates were SCCmec type IV, with IVa accounting for 54% of the total. The most frequent spa type was t127 (which was PVL-negative, ST1 and matches the Australian WA-MRSA-1 community-associated clone), accounting for 39% of isolates. 7% of the isolates matched the European CA-MRSA clone (PVL-positive ST80-t044 isolates). 16% of all isolates were PVL-positive, 80% of which were from wounds. PVL-positive isolates also included CA-MRSA-containing genotypes such as ST8-t008, ST1-t131 and ST30-t012.
Conclusions: Although many of these CIP-S isolates were from in-patients, a combination of epidemiological, microbiological and molecular data defined most of them as CA-MRSA, including common clones previously reported in the UK and Europe. Thus, CA-MRSA have emerged in our patient population over the last six years. Prospective studies are needed to define the true size of the problem in London.
P1588 Community-acquired methicillin-resistant Staphylococcus aureus in Austria: prevalence and clinical features
A.J. Grisold, G. Feierl, E. Leitner, A. Badura, L. Masoud, U. Wagner, I. Wendelin, E. Marth (Graz, AT)
Objective: Infections due to community acquired Staphylococcus aureus (CA-MRSA) are a recent worldwide phenomenon. Aim of the present study was to determine the current prevalence of CA-MRSA in Austria.
Methods: A total of 2,433 non-duplicated S. aureus strains, isolated in 2005 at the Institute of Hygiene, Medical University Graz, Austria, were investigated. The isolates were characterised by routine laboratory standard methods and determination of the resistance pattern according the CLSI (formerly NCCLS) guidelines. Additionally polymerase chain reaction tests for S. aureus-specific genes, the mecA gene and the Panton-Valentine leukocidin (PVL) gene were performed. Patients with MRSA were analysed for their clinical features, with a main focus on the PVL-gene positive MRSA.
Results: 83 (3.4%) out of the 2,433 S. aureus isolates were MRSA, approved by the resistance to oxacillin, evidence of S. aureus specific genes and detection of the mecA-gene. 29 (34.9%) of the MRSA originated from non-hospitalised patients, including patients from long-term care facilities, from practitioners or from ambulances. 17 (20.5%) out of the 83 MRSA-strains were defined as CA-MRSA, affirmed by a positive reaction for the PVL gene and the clinical appearance. Clinical features of the CA-MRSA patients were superficial to deep soft tissue abscess formation with single through multiple abscesses. After surgical intervention and/or antibiotic therapy all patients recovered without sequelae. Pneumonia could not be observed in any case.
In addition to the resistance to oxacillin CA-MRSA strains exhibited resistance to tetracycline, ciprofloxacin and gentamycin with 35.3%, 29.4% and 17.6%, respectively. Resistance to fusidic acid could be detected in 6 CA-MRSA strains (35.3%).
Conclusions: The results of this study indicate that MRSA even in Austria more and more spread into the community, with 34.9% of the detected MRSA originated from non-hospitalised patients in the year 2005. The occurrence of PVL-positive CA-MRSA is more frequent than suspected, with 20.5% of the investigated MRSA isolates containing the PVL gene. In consideration of these facts the worldwide trend of emergence of CA-MRSA could be confirmed in Austria, which supports once more the necessity of systematic surveillance.
P1589 Report of a hospital outbreak of CA-MRSA
I.M. Gould, F.M. MacKenzie, D. Morrison, R. Browning, G. Edwards (Aberdeen, Glasgow, UK)
Objectives: True community-acquired methicillin resistant Staphylococcus aureus (CA MRSA) with the type IV Staphylococcal Chromosomal Cassette mec (SCCmec) is rarely reported as being acquired in hospital. We report a hospital outbreak of 8 cases of skin and soft tissue infections due to such a strain.
Methods: From 23/09/03 to 11/03/04 a cluster of skin and soft tissue infections due to MRSA involved neonatal and maternity patients in a Grampian community hospital 30 miles north of Aberdeen. After recognition of the outbreak, nursing and medical staff were reminded of the ways in which staphylococci spread and contact precautions were reinforced. The availability of alcohol hand gel was increased and the importance of hand washing stressed. All patients had been in the labour, delivery and maternity units of a small community hospital during a 7-month period. Routine clinical cultures on blood agar identified all cases. Contact tracing, environmental cultures and follow up cultures were plated on ORSAB agar. PFGE and PCR for toxin genes were carried out at the Scottish MRSA reference laboratory.
Results: 8 patients presented with minor skin infections, mostly blistering. None of the patients was seriously ill and their condition only necessitated the use of systemic antibiotics in one case. None required surgical draining. None of the patients had relevant past hospital exposure, predisposing medical conditions for MRSA carriage or had received prior antibiotic therapy except for one infected mother. Typing by Pulsed Field Gel Electrophoresis showed the isolates to be a single strain closely related to the USA800 lineage (Paediatric Clone) and additional typing confirmed it as ST5-MRSA-IV. Genes for exfoliative toxin A (ETA) and enterotoxin D were detected in all the isolates although none carried the Panton-Valentine Leukocidin gene. The uniforms of 7 staff members were cultured and found to be MRSA negative – as were the hands of staff, all contact screens and all 45 environmental and equipment swabs.
Conclusions: CA-MRSA strain ST5-MRSA-IV with the ETA gene has been present in the region since 1999 at the latest but has not been seen elsewhere in Scotland. We describe the spread of this strain within a group of healthy newborns and one of their mothers. It was probably a self-limiting outbreak.
P1590 Prevalence of methicillin-resistant Staphylococcus aureus in veterinarians: an international view
M. Wulf, M. Sorum, A. van Nes, R. Skov, W. Melchers, C. Klaassen, A. Voss (Nijmegen, NL; Copenhagen, DK; Utrecht, NL)
Objective: In the Netherlands, veterinarians in contact with pigs and pig farmers were found to have a higher risk of methicillin resistant S. aureus (MRSA) carriage than the general population. Identical strains were found in pig farmers, pig veterinarians and pigs. The objective of this study was to investigate if contact with pigs is a risk factor for MRSA carriage in an international setting.
Design: 272 attendants of an international conference on pig health were screened for MRSA carriage by nasal and throat swabs. A questionnaire was included about the type of animal contacts and possible exposure to known MRSA risk factors and the protective measures taken while entering stables.
Results: Thirty-four participants from nine countries carried MRSA (12.5%). Thirty-one of these strains were non-typable by PFGE using SmaI. With the exception of five isolates with spa-type 899, the non-typable strains belonged to closely related spa-types (t011, t034, t108, t571, t567) which correspond to MLST 398. All of the above mentioned spa-types have also been found either in Dutch pigs, pig farmers and/or veterinarians. Protective measures such as masks, gowns and gloves did not protect against acquiring MRSA.
Conclusions: Transmission of MRSA from pigs to care-takers appears to be an international problem, creating a new reservoir for community acquired MRSA in humans in Europe, and possibly worldwide. The rise of a new zoonotic source of MRSA can have a severe impact on the epidemiology of community acquired MRSA and may have consequences for the control of MRSA, especially in the current low prevalence countries using search and destroy policies.
P1591 Increase of pig- and calf-related MRSA in a Dutch hospital
M. van Rijen, P. van Keulen, J. Kluytmans (Breda, NL)
Objectives: Methicilline-resistant Staphylococcus aureus (MRSA) has become an increasingly important pathogen in hospitals and recently also in the community. In Dutch hospitals the ‘Search and Destroy’ policy is applied successfully. Recently a new risk category was added to this control policy. This category includes patients who have professionally direct contact with living pigs or fatting calves. Patients who live at a cattle farm are included too. The objective of this study was to determine the sources of the MRSA's found in the Amphia hospital over the years 2002 until November 2006.
Methods: Data of the Infection Control Department was used to retrieve the number of newly introduced MRSA's and the sources of these MRSA's.
Results: During 2002 until November 2006 61 newly introduced MRSA's were found. Thirty-six percent was related to a foreign hospital, 10% to another Dutch hospital, in 31% the source was unknown and in 23% of the cases the MRSA was related to pig/cattle breeding. This last category was seen for the first time in 2005. In total, 28 patients with direct contact with pigs/fatting calves were screened. 14 of them were found to be MRSA positive (50%). Eight pig farmers, one pig farmer's wife, one veterinary surgeon, one artificial inseminator, one calf farmer and one calf farmer's daughter were colonised with MRSA and 1 calf farmer suffered from an MRSA-related osteomyelitis.

Conclusion: The incidence of MRSA over the year 2006 increased more than 100% compared to previous years. This is caused by MRSA related to pig and calf farmers. Patients in this risk category have an extremely high carriage rate of MRSA (14/28 positive).
P1592 Low prevalence of non-typable methicillin-resistant Staphylococcus aureus in meat products in the Netherlands
B.M.W. Diederen, P. Savelkoul, J.H.C. Woudenberg, R. Roosendaal, C. Verhulst, P.H.J. Verkeulen, J.A.J.W. Kluytmans (Tilburg, Amsterdam, Breda, NL)
Objective: A new clone of methicillin resistant Staphylococcus aureus (MRSA) emerged in the Netherlands that was related to pigfarming. A survey in pigs showed that nearly 40% carried this new clone. This study was undertaken to determine the prevalence and genetic relationship of S. aureus and MRSA in meat products.
Methods: Samples were collected between February and May 2006. A total of 79 raw pork and cow meat products were collected from 31 different shops (butcheries n = 5, supermarkets n = 26) in the South of The Netherlands. The samples were cultured using three procedures. Identification of the strains as S. aureus and methicillin resistance were determined by Martineau PCR assay for species identification and PCR for the presence of the mecA gene. Susceptibility to cefoxitin and doxycycline was determined using disk diffusion according to CLSI standards. All isolated S. aureus strains were genotyped by amplified fragment gel electrophoresis (AFLP).
Results: Direct plating yielded no MRSA positive isolates. Single-enrichment microbial culture identified 8 (10%) S. aureus isolates, of which 1 isolate was an MRSA. Combining the results of single-and double-enrichment broth culture systems identified a total of 35 (44%) S. aureus isolates of which 2 isolates (5%) were found to be methicillin resistant. A total of 19 shops (61%) were found to be positive for S. aureus in at least one meat sample. The AFLP results indicated that there were 8 genetic lineages, covering 80% [28/35] of the isolated strains, and a smaller number of unique sporadic isolates (20% [7/35] of isolated strains). In 5 out of 6 shops (83%) in which more than one S. aureus isolate was found, there was evidence for a clonal relationship within the shop. One MRSA isolate was untypable by PFGE using Sma1 digestion and identical to a pigfarming related isolate found in a patient. The other was identical to the USA 300 clone.
Conclusion: This is the first report of MRSA prevalence that is available for meat products in the Netherlands. 2.5% of the meat contained MRSA. Furthermore, S. aureus is found regularly in low amounts in meat as it is sold to consumers. The high rate of clonal relatedness of different strains within a shop indicates cross contamination of the meat at some point during processing. Considering the low amounts of contamination these findings suggest that under normal conditions meat consumption is very unlikely to be a hazard to consumers for the acquisition of MRSA.
P1593 Investigation of the genetic basis of erythromycin resistance in Staphylococcus intermedius and MRSA isolates of household pets
M. Rich, L. Roberts, A.M. Snelling, J.I. Ross (Wetherby, Bradford, UK)
Objectives: There is increasing concern about the potential transfer of antibiotic-resistant bacterial strains between the human and veterinary world. The aim of this study was to determine the prevalence of the main genes responsible for erythromycin (ERY) resistance in coagulase-positive staphylococci from household pets, and compare in-vitro antibiotic susceptibility profiles.
Methods: Fifty strains of S. intermedius and 68 strains of MRSA isolated from clinical infections of UK household pets were studied. Strains were isolated at a veterinary diagnostic laboratory between 2005–6. All isolates demonstrated resistance to ERY (≥8 mg/L). Resistance profiles were determined for a range of antibiotics. Isolates were tested for the presence of erm(A), erm(B), erm(C) and msr(A) genes by PCR.
Results: At least one ERY resistance gene was detected in all isolates. The predominant resistance gene in S. intermedius was erm(B) (72%), whereas only a single MRSA strain carried erm(B) (1.5%). In MRSA isolates erm(C) predominated (97%), whereas just 18% of S. intermedius carried erm(C). The distribution of erm(A) was similar in both groups (10–12%). There was a higher prevalence of msr(A) amongst the S. intermedius (14%) than MRSA strains (1.5%). Only one S. intermedius isolate contained msr(A) as the only ERY resistance gene. Multiple ERY resistance genes were detected in 14% of S. intermedius and 12% of MRSA strains. Incidence of resistance in the S. intermedius group was higher for clindamycin, tetracycline and neomycin compared to the S. aureus group (P < 0.001). Resistance to the fluoroquinolones, marbofloxacin and ciprofloxacin was 100% in the MRSA group versus just 4% in S. intermedius. Multiple resistance (resistance to ≥3 drug classes in addition to the pre-selected ERY-resistance) was detected in 19% of MRSA strains and 32% of S. intermedius strains.
Conclusion: The ERY resistance determinant most commonly associated with animals strains, erm(B), is rare in MRSA isolates from household pets. There is evidence of erm(A) and erm(C), genes usually associated with ERY-resistance in human strains, amongst S. intermedius isolates. Surprisingly, twice as many S. intermedius strains were multiply resistant as MRSA strains. The possibility of exchange of resistance determinants between these two groups, and the potential for MRSA strains to acquire resistance determinants from S. intermedius in pets exits.
P1594 Evaluation of MLVA and spa-typing as a rapid method for studying relationships of human and bovine Staphylococcus aureus
R. Ikawaty, J. Verhoef, A.C. Fluit (Utrecht, NL)
Objectives: At present, there is a need for a simple, efficient and high-resolution molecular typing method for S. aureus to rapidly detect outbreaks and to study evolutionary relationships. Therefore, we developed and evaluated a Multiple Locus Variable-number tandem repeat Analysis (MLVA) system combined with spa-typing on human and bovine S. aureus (HSA and BSA respectively).
Methods: Six loci (SIRU01, 05, 07, 13, 15 and 21) of 103 and 88 S. aureus isolated from humans and bovine mastitis were amplified. The number of repeat units of each locus was calculated from each amplicon. Multi Locus Sequence Typing (MLST) was used as a reference method. All HSA isolates have a known sequence type (ST) and they were well distributed within the population of S. aureus. MLST of BSA isolates was performed as part of this study. Spa-type was determined by PCR and sequencing.
Results: Almost all isolates yielded a product for the MLVA PCR. Some isolates yielded no locus-specific products. These loci were assigned an X. Variations in repeat units were observed in all loci. Large variation in the number of repeat units suggests a greater discriminatory power as shown for SIRU05, 13 and 21 of HSA isolates, and for SIRU21 of BSA isolates. It seems that HSA and BSA can be differentiated by the presence or absence of SIRU05 and 07. The absence of locus-specific PCR products and the presence of repeat numbers for each locus are shown in the table . HSA isolates revealed 46 spa-types and 3 new spa-types, whereas BSA isolates showed 5 spa-types and 1 new spa-type. When we compared the data obtained from MLVA combined with spa-type and the data from MLVA, variations within sequence types (ST) were observed indicating the MLVA combined with spa-type has better discriminatory power.
| LOCUS | HAS (n = 103) |
BSA (n = 88) |
||
|---|---|---|---|---|
| % of PCR neg. | No. of repeats | % of PCR neg. | No. of repeats | |
| SIRU01 | 6.8 | 0–5 | 0 | 1–6 |
| SIRU05 | 23.3 | 1–22 | 100 | |
| SIRU07 | 1.9 | 1–3 | 92 | 2–3 |
| SIRU13 | 1.9 | 0–26 | 42 | 1–5 |
| SIRU15 | 0.9 | 0–5 | 0 | 0–3 |
| SIRU21 | 0 | 1–18 | 0 | 2–12 |
Conclusions: The majority of S. aureus isolates can be typed by MLVA and spa-typing. The discriminatory power to detect outbreaks for HSA is good, but questionable for BSA.
P1595 Antimicrobial resistance in Staphylococcus intermedius from dogs in Norway
M. Norström, H. Tharaldsen, H. Solheim, T. Mørk, B. Bergsjø, M. Sunde (Oslo, NO)
Objectives: Staphylococcus intermedius is the most common bacterial pathogen involved in skin and ear infections in dogs. It is questioned whether antimicrobial resistance genes in S. intermedius from dogs might be transferred to other staphylococci, including those causing infections in humans, and thereby lead to further problems with resistance in such bacteria. The Norwegian monitoring programme for antimicrobial resistance in bacteria from feed, food and animals, NORM-VET, investigated S. intermedius causing skin and ear infections in dogs in the years 2000, 2002 and 2004. The results suggest an increased frequency of antimicrobial resistance among S. intermedius from dogs in Norway (NORM-VET 2004).
The aim of the present study was to examine if the high occurrence of resistance in S. intermedius from dogs in Norway is due to a limited number of resistant clones circulating among dogs in Norway, and to examine the genetic background for resistance.
Methods: A total of 59 S. intermedius isolates from 95 samples obtained in NORM-VET 2004 were subjected for further analyses. These isolates were sampled from dogs with no antimicrobial treatment during the last six months prior sampling. The isolates were sampled from dogs spread geographically over the whole country. In total, 18.6% of the isolates were susceptible to all antimicrobial agents included, 28.8% were resistant to one (mainly penicillin), 23.7% to two (mainly penicillin and oxytetracycline/fusidic acid), 18.6% to three (penicillin, oxytetracycline and fusidic acid) and 10.2% to four or more antimicrobial agents. The isolates were subjected to polymerase chain reaction (PCR) for detection of resistance genes and to pulsed field gel electrophoresis (PFGE).
Results: A multitude of S. intermedius clones are involved in skin and ear infectious in dogs. PCR showed that resistance to tetracycline was mediated by tet(M) in all strains expressing resistance to tetracycline, and that all penicillin resistant strains, except one, harboured the blaZ β-lactamase gene. Strains resistant to fusidic acid were screened for the fusB gene, all strains were negative.
Conclusion: The many different clones found in this study indicate a high diversity of S. intermedius from skin and ear-infections in Norwegian dogs. The occurrence of antimicrobial resistance is widespread among all clones.
P1596 Occurrence and enterotoxic characterisation of methicillin-resistant Staphylococcus aureus from raw milk in southern Italy
A. Dambrosio, G. La Salandra, M. Corrente, N.C. Quaglia, V. Lorusso, G.L. Germinario, G. Mula, G. Normanno, G.V. Celano (Valenzano, Foggia, Barletta, IT)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) strains are a global health concern. Infection could be associated with the ingestion of contaminated foods. In addition the ingestion of food contaminated by enterotoxins (SEs) synthesized by S. aureus is responsible of one of the most common foodborne intoxication. Since S. aureus is often involved in subclinical mastitis of ruminants, milk may results contaminated. In fact, the dairy products are frequently related to cases of Staphylococcal Food Poisoning (SFP), expecially in areas characterised by a high level of consumption of dairy products. Consequently an active microbiological surveillance of milk and dairy products is desirable in order to control the risk of MRSA foodborne infections and SFP and to allow the improvement of the public health standards. In this work are reported the results of a survey conducted on the occurrence and enterotoxic characterisation of MRSA in bulk milk from Apulia region (Southern Italy).
Methods: In the present study are reported the results obtained from the investigation of 560 bulk milk samples produced in Apulia region in order to detect MRSA strains. The S. aureus isolates (one per positive sample) were characterised by detecting the mecA gene by polymerase chain reaction, and the production of type A to D staphylococcal enterotoxins (SEs), using the reverse passive latex agglutination method.
Results: Of the 560 samples 90 (16%) were contaminated with S. aureus and among the positive samples one showed the presence of a MRSA (mecA positive) strain. The MRSA strain resulted able to produce staphylococcal enterotoxin C plus enterotoxin D (SEC plus SED).
Conclusions: Despite its low prevalence in the samples analysed, the presence of MRSA constitutes a serious health hazard, especially for immunocompromised individuals. Such a finding calls for better hygiene in milk production along the food-chain, especially at the primary production level. The presence of antibiotic resistance in potentially harmful bacteria in foods of animal origin warrants further investigations into the role of antibiotics when they are used for therapeutic purposes or as growth promoters in food-producing animals.
P1597 Prevalence of methicillin-resistant Staphylococcus aureus carriage in Italian long-term care facilities
P. Brugnaro, J. Mantero, R. Cazzaro, U. Fedeli, P. Mantoan, G. Pellizzer, A. Tomasini, M. Zoppelletto, P. Spolaore (Castelfranco Veneto, Vicenza, IT)
Objective: Data on the prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) carriers in long term care facilities are lacking in Italy, being 11% the prevalence reported in the only available investigation conducted in a single facility. Aim of the study is to determine prevalence and risk factors for MRSA carriage in nursing home residents in Vicenza (North-Eastern Italy).
Methods: Nasal swabs were obtained in late June-early July 2006 from residents of the two largest long term care facilities of the city. Laboratory screening for MRSA was performed by means of the MRSA Select Agar (Bio-Rad) and full antibiotic susceptibility was assessed in MRSA isolates. Demographic and clinical data, dependency, cognitive function, length of stay, current and previous antibiotic treatment, presence of medical devices, previous hospital admission, presence of infection according to Association for Professionals in Infection Control criteria were assessed in each subject on the same day of sample collection. The factors significantly associated to MRSA carriage at univariate analysis were introduced in a multiple logistic regression model, and the corresponding odds ratio (OR) with 95% Confidence Intervals (CI) were estimated.
Results: Out of 570 residents nasal swabs were obtained in 551 subjects (96.7%). Among the latter, 73% were females; the mean age was 83 years (31% of residents being aged 90 or more). 118 residents (21%) had at least one hospital admission in the previous year. 63% of subjects received systemic antibiotic treatments in the previous 12 months: 37% were treated with fluoroquinolones, 26% with cephalosporins. Overall 43 MRSA carriers were detected (7.8%; CI: 5.7–10.4%). All MRSA isolates showed fluoroquinolones resistance. At logistic regression the risk of MRSA carriage was increased in patients with cancer (OR = 6.1; CI: 2.5–15.0), with previous hospitalisation (OR = 2.0; CI: 1.0–4.0), and raised with the number of previous antibiotic treatments, reaching an OR = 3.9 (CI: 1.6–9.1) in those with 3 or more treatments.
Conclusion: To date the present study is the largest Italian survey of MRSA carriage in elderly people outside the hospital setting. The prevalence resulted higher than that reported from nursing homes in other European countries like Germany. Both comorbidities (cancer) and pattern of care (previous hospitalisation and antibiotic treatment) were associated with MRSA carriage.
P1598 Tracking the epidemiology of methicillin-resistant Staphylococcus aureus strains producing Panton-Valentine leukocidin in Greece
V. Chini, A. Foka, E. Petinaki, H. Meugnier, F. Kolonitsiou, D. Garantziotou, M. Bes, J. Etienne, I. Spiliopoulou (Patras, Larissa, GR; Lyon, FR)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) is recently associated with an increasing number of community-acquired infections (CA-MRSA), including superficial, deep-seated infections and pneumonia. CA-MRSA harbours the Staphylococcal Cassette Chromosome IV or V (SCCmec) and the genes encoding the Panton-Valentine leukocidin (PVL). We have investigated the distribution and clonal evolution of CA-MRSA during 2005–2006 in Greece.
Methods: S. aureus isolates from three hospitals in Western and Central Greece were identified by conventional tests, followed by the determination of MIC of oxacillin by the agar dilution method. PBP2a production was investigated by a Latex agglutination test (bioMérieux). SCCmec types, agr groups and the presence of lukS and lukF genes (encoding PVL) were defined by PCRs. Clonal types were determined by MLST. CA-MRSA were isolated from patients without any predisposing risk factors and identified on the basis of their genotypes.
Results: A total of 1,541 S. aureus isolates were collected from different patients from January 2005 until June 2006. Among them 697 isolates (45%) were MRSA (495 from the Department of Outpatients, OUTP) and 490 (70%) PVL-positive (443 from the OUTP). An increasing rate of the total S. aureus infections was observed with a parallel increase of PVL-positive CA-MRSA. The great majority of CA-MRSA was associated with skin and soft tissue infections, but five cases of acute osteomyelitis and one pneumonia were diagnosed. The majority of CA-MRSA (432) belonged to the major European clone, which is ST80, agr 3, SCCmec IV, while the rest 11 belonged to a new emerging clone, spread also in Europe, ST377, agr 1 and SCCmec type V. Most of the PVL-positive MRSA were associated with community-acquired infections, but 47 PVL-positive MRSA of ST80 were isolated from hospital-acquired infections, mainly at the departments of Orthopaedics and Surgery.
Conclusions: There is an unusual epidemiology of PVL-positive CA-MRSA in Greece where, we are encountering an increasing rate of S. aureus infections, due to CA-MRSA producing PVL and causing mainly superficial but also deep-seated infections. The European CA-MRSA clone seems to be spread rapidly, but a new clone is also showing up.
P1599 A CA-MRSA strain with decreased vancomycin susceptibility as a cause of serious invasive infection in an immunocompetent adolescent
M. Tronci, G. Parisi, A. Pantosti, M. Monaco, P. Valentini (Rome, IT)
Obiectives: CA-MRSA are primarily associated with skin and soft tissue infection although they can also cause more serious infections such as necrotising pneumonia and septicaemia. This report describes a severe invasive infection in a immunocompetent adolescent with isolation of an CA-MRSA strain with decreased susceptibility to vancomycin.
Methods: Blood and CSF cultures were obtained from a 16-year old Italian boy admitted to the Paediatric Emergency room with a temperature of 40.5°C, stiff neck, headache and vomiting. The patient was previously healthy with no recent hospitalisation story; he played football and had a purulent skin lesion in the back. An empiric therapy with a 3rd generation cefalosporin was started. The identification and susceptibility tests were performed by PHOENIX instrument. Susceptibilities were confirmed by E-test. The presence of the Panton-Valentine Leucocidin (PVL) toxin genes lukS-PV-lukF-PV was determined by PCR. The structural type of the Staphylococcal chromosomal cassette mec (SCCmec) was performed by multiplex PCR. Multi-locus sequence typing (MLST) was performed to characterise the clonal group.
Results: Blood and CSF cultures grew an MRSA. In vitro susceptibility tests showed that both the isolates from blood and from CSF were resistant to oxacillin, and were sensitive to gentamycin, kanamycin, erythromycin, clindamycin, rifampicin, tetracycline, ciprofloxacin, chloramphenicol and linezolid. MIC to vancomycin was 4 μg/mL. The isolates contained SCCmec type IV and were positive for PVL. By MLST the isolates were found to belong to the European clone. The patient was transferred to another hospital where vancomycin was started. On the third day, his condition deteriorated and he was admitted to the ICU for respiratory distress and hypoxaemia. The chest X-ray revealed infiltrates and the CT scan showed severe necrotising pneumonia. The antimicrobial regimen was switched to linezolid, rifampicin and teicoplanin. His conditions improved and he was discharged after four weeks.
Conclusions: To our knowledge this is the first case of an invasive infection due to a CA-MRSA strain with decreased susceptibility to vancomycin. We emphasize the importance of an early laboratory diagnosis for an appropriate and successful therapy. An epidemiological surveillance system is needed to characterise and monitor CA-MRSA infections and to understand how MRSA are transmitted in the community.
P1600 Methicillin-resistant Staphylococcus aureus from the lower respiratory tract: a cause for concern?
J. Wadula, S. Vally, T. Adams, M. Khoosal (Johannesburg, ZA)
Background: Methicillin-resistant Staphylococcus aureus has emerged, as a significant cause of pneumonia and infection caused by this organism has been associated with high mortality.
Recent reports allude to the fact that Staphylococcus aureus strains carrying the Panton-Valentine Leukocidin gene are highly associated with lethal necrotising pneumonia in healthy individuals. Community-associated methicillin-resistant Staphylococcus aureus strains have been implicated specifically to harbour the Panton-Valentine Leukocidin gene.
Objectives: To substantiate Methicillin-resistant Staphylococcus aureus (MRSA) strains in relation to whether, they are community-associated or hospital-acquired as defined by molecular definition of specifically looking for the panton-valentine leukocidin gene.
To document the reported antimicrobial sensitivity patterns of MRSA strains.
Methods: Retrospective laboratory data collection of all MRSA cultured from good quality sputum and tracheal aspirates during a nine-month period commencing July 2005 through to March 2006. Majority of these MRSA cultures were from patients in intensive care unit.
Sixty-seven cultures of MRSA from July 2005 to March 2006 were evaluated to confirm the identification, establish the presence of mecA and PVL genes (Hain Lifescience, Germany).
Results: Sixty-four (97%) of the 66 strains harboured the MecA gene while none of the 66 carried the PVL gene.
These strains exhibited multiple antibiotic resistance in keeping with hospital-acquired MRSA strains.
Conclusion: There is a predominance of hospital-acquired MRSA strain in our hospital, which highlights the need to constantly adhere to infection control measures.
“Classical” MRSA detection
P1601 Evaluation the cefoxitin 30 μG disk diffusion method for detection of methicillin-resistance in selected Staphylococcus aureus isolates
M. Lara, N. Batista, F. Laich, S. Méndez-Álvarez (Santa Cruz de Tenerife, ES)
Objective: To evaluate the discriminative capacity of the CLSI disk diffusion method with a cefoxitin 30 μg disk on S. aureus isolates with unusual phenotypic characteristics of antimicrobial resistance.
Material and Methods: We studied 53 clinical S. aureus isolates from N.S. Candelaria Hospital, Tenerife, Spain. The antimicrobial susceptibility of all isolates was routinely studied by the VITEK 2 System (bioMe‘rieux). The methicillin resistance was also studied by CLSI oxacillin method, and confirmed by a previously described multiplex PCR method which permitts S. aureus identification and simultaneous detection of methicillin resistance.
MecA positive isolates presenting a diffuse growth within the zone of inhibition when exposed to oxacillin, were considered heteroresistant; mecA negative, oxacillin intermediate or resistant isolates were considered borderline.
All the isolates were tested with a cefoxitin 30 μg disk, according to the CLSI guidelines (susceptibility: ≥20 mm; resistance: ≤19 mm). Control strains for all assays included MRSA strain ATCC 43300, MSSA ATCC 25923, and MRSA B/3/02 SEIMC. Secondly, we also considered the criterium of Skov et al (J Clin Microbiol, Oct 2006) who proposes a zone diameter for cefoxitin susceptibility (≥22 mm).
Results: The isolates formed four groups. The first one comprised 20 multi-resistant but oxacillin susceptible and mecA negative isolates; the second group
was a series of 16 borderline isolates; the third group comprised a selection of 11 heteroresistant isolates, and the fourth group was a series of six mecA positive isolates with atypical resistance profiles (penicillin and oxacillin, or ciprofloxacin and erythromycin resistance).
The 36 mecA negative isolates included in groups 1 and 2 showed susceptibility to cefoxitin disk. Fifteen mecA positive isolates from groups 3 and 4 showed inhibition zones ≤ 19 mm; two isolates from these groups (one of each), showed 20 and 21 mm respectively. So, sensitivity was 96.3% (51 of 53 isolates). After applying the Skov proposal, the susceptibility was 100%.
Conclusions: The 30 μg cefoxitin disk diffusion method is proposed as an efficient method for the detection of methicillin resistance and permitts a clear determination for S. aureus isolates with atypical antimicrobial characteristics.
P1602 Comparison of chromogenic agar in the detection of MRSA
A. Curry, M. Walsh, M. Matheson, A. O'Reilly, S. Knowles (Dublin, IE)
To compare two different chromogenic agars for the isolation of MRSA in order to find the most appropriate agar for use in our laboratory. Nasal, umbilical and groin screening swabs from each neonate in the NICU were pooled together. Each screen was put in Brain Heart Infusion enrichment broth, which was incubated for 24 hours aerobically at 37°C and subsequently subcultured onto both MRSA ID agar and MRSA Select agar. Both agars were incubated for 48 hours aerobically at 37°C and checked for the presence of green (MRSA ID) or pink (MRSA Select) colonies at 24 and 48 hours. All green or pink colonies had confirmatory tests for MRSA.
In total 300 screens were tested on both agars. The same 10 screens were identified MRSA positive on both agars after 24 hours. Incubation for 48 hours did not yield any additional MRSA isolates. Therefore both agars were comparable in sensitivity and specificity for MRSA identification. Other factors influenced our decision to choose a chromogenic agar for our laboratory. MRSA ID agar grew as large green colonies on a clear agar and was considered easy to read. MRSA Select grew as small pink colonies on an opaque agar, which were easy to read in large numbers. However they could easily be missed when a scanty growth was present under our laboratory lighting conditions. Also on both agars, Enterococci grew as a green or pink fine growth after 24 hours and it distorted white colonies that grew coagulase negative Staphylococci (CoNS) giving them a green or pink hue. On MRSA ID agar it was easy to distinguish between true MRSA and CoNS that appeared green due to the presence of Enterococci, however on MRSA Select, as CoNS were also small colonies, the pink from the Enterococci distorted the CoNS, giving them the appearance of MRSA and resulted in them having to be confirmed as CoNS. Cefoxitin resistant Gram-negative bacilli also grew on both agars after 48 hours incubation and in some cases a wet prep was required to exclude them as possible MRSA. One strain of methicillin sensitive Staphylococcus aureus also grew on both agars after 48 hours incubation.
In conclusion, both agars were comparable in sensitivity and specificity for identifying MRSA. MRSA ID agar was chosen as the preferred agar in our laboratory due to the ease in distinguishing true MRSA from CoNS affected by the presence of Enterococci and also the ease of reading the agar under our laboratory conditions.
P1603 Performance of MRSA ID chromogenic medium for detection of methicillin-resistant Staphylococcus aureus directly from blood cultures and clinical specimens
S. Colakoglu, H. Uncu, S. Senger, T. Turunc, Y. Demiroglu, H. Arslan (Ankara, TR)
Objectives: Rapid identification of methicillin-resistant Staphylococcus aureus (MRSA) in clinical samples are an important part of the infection control measures taken to control the spread of MRSA. The purpose of this study is to evaluate the performance of MRSA ID chromogenic medium compared with nonspecific media routinely used for the detection of MRSA directly from blood cultures and clinical specimens.
Methods: We analysed 774 blood cultures from 471 patients and 94 clinical samples (wounds or abscesses) from 71 patients. Each positive blood culture bottle (Bactec Plus/F;BD) and clinical sample were directly inoculated to primary culture media and MRSA ID (bioMérieux, France). All cultures were incubated in aerobic condition at 370C for 24–48 h. Suspect colonies were identified as MRSA based on positive reaction on the tube coagulase test with rabbit plasma, the detection of DNase and growth on Mueller-Hinton (MH) oxacillin agar (6 μg of oxacillin/mL, according to CLSI). Inoculated MRSA ID plates were interperated in accordance with the manufacturer' s instructions. Growth of colonies showing distinctive green coloration was considered to be positive. No growth or colonies with colours other than green were considered negative. Discordant results were confirmed by mecA gene PCR.
Results: The results obtained with MRSA ID are summarised in Table 1, Table 2 . Three methicillin-susceptible S. aureus (MSSA) isolates gave false-positive results on MRSA ID and these strains gave negative result with the mecA PCR.
Table 1.
Results for MRSA ID medium after 24 h and 48 h of incubation
| Organism | No. strains with a positive test result/total no. of strains (%) after: |
|
|---|---|---|
| 24 h | 48 h | |
| MSSA | 2/34 (5.9) | 3/34 (8.8) |
| MRSA | 54/56 (96.4) | 56/56 (100) |
Table 2.
Strains producing green colonies which have a different appearance from MRSA colonies
| Organism | No. of strains detected on medium |
|---|---|
| Aerococcus viridans | 1 |
| Acinetobacter baumannii | 1 |
| Bacillus sp | 1 |
| Brucella sp | 2 |
| Candida sp | 4 |
| Enterococcus sp | 2 |
| Enterobacter gergovias | 1 |
| Stenotrophomonas maltophilia | 4 |
Conclusions: (1) MRSA ID is highly effective for the isolation and presumptive identification of MRSA directly from wound samples and blood cultures. (2) The use of MRSA ID with primary culture media should decrease the time (18–24 h) to reporting positive results compared with conventional methods.
P1604 Evaluation of a new chromogenic medium SaSelect for identification of Staphylococcus aureus
C. Daurel, R. Leclercq (Caen, FR)
Objectives: The objectives were to compare the sensitivities and specificities of SaSelect, a new chromogenic medium, and Mannitol salt agar medium (MSA, Chapman) associated with a slide coagulase test, for rapid and direct identification of Staphylococcus aureus from cultures of various clinical samples.
Methods: 322 clinical specimens of various natures were studied: suppurations (n = 126), upper and lower respiratory tract secretions (n = 54), faecal samples (n = 31), blood cultures with mixed Gram strains (n = 30), nasal samples (n = 41), urines (n = 20), catheters (n = 8) and others (n = 12). All samples were isolated on SaSelect (incubation 24 h) and MSA (incubation time: 24 and 48 h). The identification of pink colonies on SaSelect and colonies with a yellow halo on MSA was confirmed by a slide coagulase test.
Results: Most clinical samples were polymicrobial. 152 samples contained S. aureus with SaSelect (24 h of incubation) and 144 with MSA (48 h of incubation). 153 S. aureus were isolated on either medium. Specificity and sensitivity were calculated, considering that a sample was positive if any of the two tested media showed the presence of S. aureus. When only the pink colour of the colonies at 24 h of incubation on SaSelect was taken as a criterion of identification for S. aureus, the specificity (100%) was similar to that obtained on MSA combined with coagulase test at 24 and 48 h of incubation (99.4%). The sensitivity values were different. At 24 h, the sensitivity was 90.8% for MSA and 98% for SaSelect. At 48 h incubation time, the sensitivity determined on MSA increased to 94.7%.
Conclusion: The sensitivity and specificity of the SaSelect medium were close to 100% in our study. This medium can be used in routine for the identification of S. aureus on the basis of the pink colour of the colonies without requiring a complementary coagulase test for confirmation. SaSelect makes it possible to reduce the cost of the test and the working time.
P1605 Comparison of three chromogenic media for rapid detection of methicillin-resistant Staphylococcus aureus from screening swabs in hospitalised patients
C. Nonhoff, O. Denis, A. Brenner, P. Buidin, C. Thiroux, M. Struelens (Brussels, BE)
Objectives: To evaluate the performance of MRSA-ID (bioMérieux), MRSA Chromogenic Agar (MRSA-Screen, Oxoid) and MRSA-Select (Bio-Rad) media for detection of methicillin-resistant Staphylococcus aureus (MRSA) in muco-cutaneous swab samples from patients admitted to a 860-bed teaching hospital.
Methods: Hospitalised patients (n = 639) were screened for MRSA carriage by sampling swabs from nares (n = 726), throat (n = 116), perineum (n = 109 and skin (n = 51). Swabs were inoculated into nutrient broth (SB) supplemented with 7.5% NaCl, MRSA-ID, MRSA-Screen and MRSA-Select agar at 35°C. SB broths were sub-cultured after 24 h onto the three chromogenic media. S. aureus isolates were identified by coagulase test. Susceptibility to oxacillin was determined by cefoxitin disk method according to CLSI. Identification and oxacillin resistance were confirmed by PCR for 16S rRNA, nuc and mecA genes.
Results: MRSA strains were isolated from 68 (6.8%) specimens from 45 patients: nares (n = 28), throat (n = 16), perineum (n = 13), skin (n = 11). Performance results of the three chromogenic media are shown in Table 1 . Seven (10.8%) MRSA isolates grew only on primary agar plates while 10 (15.4%) were only isolated after the enrichment procedure. The specificity of MRSA-Screen decreased after 36 h incubation and with the enrichment step because of growth of pigmented methicillin-susceptible S. aureus (MSSA) (23 and 24 isolates, respectively).
| Medium | Sensitivity (%) |
Specificity (%) |
||||
|---|---|---|---|---|---|---|
| 16–18 h | 36 h | Enrichment | 16–18 h | 36 h | Enrichment | |
| MRSA-ID | 45.6 | 75.0 | 85.3 | 100 | 99.9** | 99.8 |
| MRSA-Screen | 44.1 | 80.9* | 85.3 | 99.4 | 97.5** | 97.4 |
| MRSA-Select | 45.6 | 72.1* | 85.3 | 100 | 100** | 100 |
p = 0.045
p < 0.001
Conclusion: The three chromogenic media demonstrated equivalent performance after 16–18 h. The use of an enrichment broth was necessary to optimise sensitivity but was associated with decreased specificity for MRSA-ID and MRSA-Screen. In this study, we observed equivalent sensitivity after 16–18 h and marginal difference in sensitivity after 36 h between the three media but significant lower specificity for MRSA-Screen after 36 h and enrichment (p < 0.001).
P1606 In vitro evaluation of MRSA screening methods
S. Böcher, R. Smyth, G. Kahlmeter, R. Leo Skov (Copenhagen S, DK; Växjö, SE)
Objectives: Chromogenic agars for detection of MRSA carriers are now widely available. Enrichment broths have been shown to increase sensitivity. Studies of enrichment broths in combination with these new selective agars, using diverse MRSA strains, are needed. This study has been designed to meet that need.
Materials and Methods: Isolates: 99 well characterised MRSA strains, representing 13 clonal complexes, including many highly heterogenic isolates, 50 methicillin sensitive Staphylococcus aureus (MSSA) and 50 methicillin resistant coagulase negative staphylococci (MRCNS) were tested. Selective agars: MRSA SSI (Statens Serum Institut), MRSA ID® (bioMérieux), MRSA Select® (Biorad), Mannitol salt agar with 4 mg/L cefoxitin (MSA) and 5% blood agar (BA). Enrichment broths: TSB with 2.5% salt, 20 mg/L aztreonam and 3.5 mg/L cefoxitin (TSB), phenol red mannitol broth with 75 mg/L aztreonam and 5 mg/L ceftizoxime (MB).
Method: 20 CFU of each strain was inoculated directly onto each of the agar media and into the broths. After 18 and 48 h incubation at 35–36°C, colonies on the solid media were counted. After 18–24 h incubation of the broths, 20 μL was subcultured onto each of the selective agars and a blood agar control.
Results: For direct inoculation and 18–24 h incubation the sensitivities and specificities were: MRSA SSI: 97%/100%, MRSA ID: 96%/95%, MRSA Select: 95%/99%, MSA 75%/71%. (88%/71% after 48 hours). For chromogenic agars 48 h incubation only served to lower selectivity, especially for MRCNS. Using turbidity or colour change as a growth indicator, the sensitivities and specificities of the broths were: TSB: 97%/74%, MB: 47%/100% and after subculture to BA: TSB: 97%/57%, MB: 74%/58%
After enrichment in broths the sensitivity of MRSA ID and MRSA Select rose by 1–2%. The sensitivity of MSA increased from 75% to 97% after enrichment in TSB.
All strains grew on blood agar.
Discussion
and Conclusions: In this in vitro evaluation, all 3 chromogenic MRSA agars supported growth of this diverse collection of MRSA using low inocula and had high specificity. Enrichment using TSB gave no overall advantage. MB enrichment displayed low sensitivity suggesting that the antibiotic content is too high. Incubation of the chromogenic agars beyond 24 hours had no effect on sensitivity. For the MSA plate, comparable sensitivity was obtained only after preenrichment.
P1607 Evaluation of a direct cefoxitin disc diffusion test for the presumptive identification of MRSA from clinical specimens
J. Fuller, B. Lui, L. Rosmus, J. Johnson, R. Rennie (Edmonton, CA)
Objective: Despite intensive patient screening programmes, the prevention of methicillin-resistant Staphylococcus aureus (MRSA) nosocomial transmission continues to challenge many Canadian hospitals. This study evaluated the predictive capacity of a non-standardised direct cefoxitin disk test to presumptively identify MRSA isolates from routine clinical specimens.
Methods: For a six-month period routine clinical isolates collected from any patient site that resembled S. aureus morphologically at 24 hr of incubation were subcultured onto sheep blood agar, as per laboratory protocol. A 30 mg cefoxitin disk (Oxoid) was placed in the first quadrant of the subculture plate, which was incubated at 35 degrees C for 18 to 24 hours. The inhibitory zone size of each isolate was measured and categorised as susceptible (S) or resistant (R) according to current Clinical Laboratories Standards Institute (CLSI) breakpoint values (S ≥ 20 mm, R ≤ 19 mm). Comparatively, a standard cefoxitin Kirby-Bauer disk diffusion (CLSI) was performed for each S. aureus isolate. Confirmatory MRSA identification was performed using a PBP2A assay (Denka Seiken).
Results: 518 S. aureus isolates were collected from 372 patients that were evenly distributed across common specimen sites, including blood, respiratory, skin, and wounds. The direct and CLSI disk methods showed 100% breakpoint agreement (S, 79%) and cefoxitin R isolates were confirmed MRSA by PBP2A. However, the direct method provided presumptive MRSA results 24 hours sooner. Interestingly, only one isolate (0.2%) that was S by the CLSI method had an inhibitory zone between 20 and 23 mm. In contrast, using the direct cefoxitin method 72 isolates (14%) had a reduced S zone between 20 and 23 mm. Similar results were observed when the data was restricted to one isolate per patient.
Conclusion: In this study, a non-standardised cefoxitin disk test reliably predicted cefoxitin susceptibility for clinical S. aureus isolates 24 hours sooner than that provided by CLSI disk diffusion. If adopted, this would allow faster laboratory reporting and infection control interventions for presumptive MRSA-positive patients. As a precautionary measure, infection control precautions could also be considered for patients with an isolate exhibiting a reduced S inhibitory zone size (20 to 23 mm) until confirmatory testing was complete.
Tuberculosis and other mycobacterial infections: clinical epidemiology
P1608 Tuberculosis case finding and treatment in the central prison of Qazvin province
S. Salehi Shahidi, M. Assefzadeh, R.Gh. Barghi (Qazvin, IR)
Objectives: This study is aimed at determining tuberculosis prevalence in the central prison of Qazvin province.
Methods: It was a cross-sectional study started from April 2004 to July 2005 which use active case finding based on clinical signs and symptoms (persistent cough more than 2 weeks, weight loss, fever, night sweats, decreased appetite) and a minimum of 3 morning sputum samples of TB suspects for microscopic exam.
Results: A total number of 768 prisoners including 95% males and 5% females were examined. 75% of them were in 21–40 age-group. 5.3% of total examined were tuberculosis suspects. 7 smear-positive TB cases were found who were all males. 6 cases were in 21–40 age group. All of the cases were addicted to cigarette and drug. In general the tuberculosis prevalence was 911 per 100,000. Directly observed short course treatment was started for all 7 patients. 4 cases were cured, 1 case treatment completed and 2 cases were failed who one of them was cured after prescription of Category2 regimen, the other one interrupted his treatment after having been released from the prison.
Conclusion: In our study, the tuberculosis prevalence in evaluated prisoners was 911 per 100,000. It was 227 times more than smear positive tuberculosis prevalence and 113 times more than total tuberculosis prevalence of the province in the same year. In a separate study in Central Qazvin prison in 1999, it was found out that the tuberculosis prevalence was 136 per 100,000. This increasing trend of tuberculosis in Central Qazvin Prison from 136 to 911 per 100,000 in about 5 years is possibly because of an increased accuracy of active case finding and increased predisposing factors for tuberculosis infection/reactivation in the prison. Since there are no limitation and physical barriers for tuberculosis as restricting factors this increasing trend can be an alarm for spreading the disease in the society. Therefore there should be continuous training programmes for both staff and prisoners.
We had two cases of treatment failure (28.5%) which were higher than the same result in non-prisoner patients (2.9%) may be due to noncompliance of the prisoners to the treatment, multiple lung cavities which were found in chest X-rays of both failure of treatment cases, smoking and drug addiction. Therefore arrangements should be made in such a way that all inmates must refrain from smoking and drug abuse and their treatment should be performed under more supervision and care.
P1609 Extrapulmonary tuberculosis in central Tunisia: a review of 114 cases
N. Kaabia, F. Bellezreg, M. Khalifa, E. ben Jazia, A. Braham, F. Bahri, A. Letaief (Sousse, TN)
Tuberculosis continues to be a serious public health problem in Tunisia. The aim of this study was to identify epidemiological and clinical characteristics of extrapulmonary tuberculosis (EPT) in a tertiary care setting.
Methods: A retrospective case series was carried out of all cases diagnosed and treated as EPT during January 2000 to September 2006 at Farhat Hached Hospital, Sousse, Tunisia. Demographic, clinical, laboratory data and outcome were abstracted from medical records.
Results: 114 cases of EPT were included. The average age of patients was 47.5 years, 61 (53.5%) were men and 59% were rural dwellers. Isolated EPT was identified in 86 (75.5%) patients, 28 cases had both pulmonary and EPT. 41 patients (36%) had co-morbidities, most commonly diabetes mellitus in 9.5% of patients. Diagnosis delay was 16.5 days. The most frequent form was lymphadenitis in 30 cases (26.5%), followed by digestive (23), osteoarticular (17), genitourinary (13), pleural (11) and meningeal (11) forms. More than one location was noted in 24 patients (21%). EPT was diagnosed by histopathologic methods in 64 (56%) cases and by microbiologic methods in 33 (29%). Tuberculin skin test was significantly positive in 35% of patients. The average hospital stay was 24.5 days. No case had a relapse or recurrence of disease in the period of follow up, 4 patients with meningeal tuberculosis died. All patients were treated by antituberculosis drug, 21 (18.5%) presented adverse effects, most commonly hepatitis in 16 cases.
Conclusion: Although this study did not show the real frequency of EPT in our region, it noted the high frequency of lymph nodes localisation and the principal diagnostic method was the biopsy.
P1610 Clinical analysis of tuberculous meningitis cases hospitalised from 1993 to 2006
M. Bociaga-Jasik, A. Garlicki, A. Kalinowska-Nowak (Cracow, PL)
Objectives: The aim of the study was the retrospective analysis of TB meningitis cases.
Material: On the basis of medical records 25 cases of TB meningitis hospitalised in the Department of Infectious Diseases CMUJ from 1993 to 2006 were analysed (1.98% of all meningitis cases hospitalised at this time).
Results: 18 (72%) men and 7 (28%) women, age range 21–76 years (mean age 39) with TB meningitis were hospitalised. 5 (20%) were infected with HIV. Other risk factors included: alcoholism 6 (24%), diabetes mellitus 2 (8%). 6 (24%) had previous TB lung infection, 1 had involvement of perihilar lymph nodes. There was 1 case (4%) of milliary TB and 3 (12%) of UT tuberculosis. Mean time since the onset of symptoms to final diagnosis was 13.3 days (min 6, max 26). Most common complains were: headache 24 (96%), fever 23 (92%), nausea 22 (88%), upper RTI 20 (80%), haematuria 3 (12%). On PE: meningeal signs 23 (92%), changes in consciousness 2 (88%), hemiparesis 4 (16%), Horner sign 3 (12%), cranial nerve palsy 3 (12%), spastic paresis of lower extremities 2 (8%) were detected.
Changes in CSF differ significantly among patients. Number of cells in CSF range from 40 to 1,117 cells/mm3, with lymphocyte predominance. Also protein, glucose and chlorine concentration differ significantly and range from 1.0 to 27.7 g/L, 0.69 to 2.2 mmol/L and 94 to 119.2 mmol/L. The confirmation from CSF (PCR, BACTEC) was achieved in 13 (52%) cases. 8 (32%) patients had positive cultures from respiratory tract, 3 (12%) from urinary tract and 1 (4%) from blood. In the rest of cases the diagnosis was established on the basis of clinical picture, changes in CSF and response for therapy with antituberculous drugs. All patients were treated with 4 drugs and steroids. The mean hospitalisation time was 107 (min 17, max 188). 3 (12%) patients died. 11 (44%) developed hydrocephalus, 5 of whom required shunts. In 2 (8%) cases brain ischaemia was detected. 2 (8%) developed epilepsy, 1 (4%) tuberculomas of the brain, and 1 (4%) mental retardation. 9 (36%) patients recovered completely.
Conclusions: (1) Tuberculous meningitis in Poland is not forgotten disease, and should be taken into consideration in diagnostic work up also of immunocompetent individuals. (2) The diagnosis is usually delayed because of uncharacteristic clinical presentation and different pattern of changes in CSF. (3) In spite of therapy tuberculous meningitis is associated with high risk of complications.
P1611 Clinical and paraclinical aspects of tuberculous meningitis in a hospital of infectious diseases, Iasi, Romania
C. Luca, C. Dorobat, D. Mihalache, C. Petrovici, E. Nastase, R. Nemescu, O. Caliman-Sturdza, V. Luca (Iasi, RO)
Objectives: The aim of this study is presentation of tuberculous meningitis in the department of Iasi Infectious Diseases Hospital, where patients from all North-East Romania are attended to.
Materials and Method: We retrospectively studied the cases of tuberculous meningitis during 2005–2006 regarding clinical and laboratory methods of diagnosis. We used the classic laboratory methods together with new ones, the quantification of gamma-interferon in blood, in presence of M. tuberculosis specific antigens.
Results: A total of 58 patients (41 male [71%] and 17 female [29%]) were included. The mean age was 31 years (age range: 10 months–80 years) with 12 patients (21%) over 50 years old and 15 under 14 years old (26%). 15 (26%) patients had a history of tuberculosis. The clinical symptoms and signs on the admittance were compatible with tuberculous meningitis: consciousness alteration in 18 cases (31%), focal neurological signs in 11 cases (19%), paraplegia and/or hemiplegia in 7 cases (12%) and extra-neurological tuberculosis was associated in 32 cases (55%).
The patients fulfilled some of following diagnosis criteria: CSF (cerebrospinal fluid) pleocytosis (with <100 elements/mL for 15 cases, 101–300 elements/mL for 25 cases and >300 elements/mL for 17 cases); high CSF protein level (45 cases, 78%); low CSF glucose level (32, 55%); favourable response to antituberculous therapy and steroids (51 cases, 88%); and a mortality of 12% (7 cases).
Microbiological diagnosis was done through culture in CSF with positive results in 18 cases: 6 cases (10%) were with negative CSF cultures and 12 (21%) had positive M. tuberculosis CSF cultures and positive Quantiferon TB Gold results. Also, in 3 cases (5%) we had multidrug resistant strains of M. tuberculosis.
We used quadruple association of antituberculous drugs for treatment, with favourable response in 48 cases (83%) and with a mortality of 12% (7 cases). Sequels were present in 18 cases (31%), with: hydrocephalus in 10 cases (56%), arachnoiditis in 3 cases (16%) and tuberculomas in 5 cases (28%).
Conclusion: Tuberculous meningitis is still a very serious disease in Romania, with high morbidity and mortality, especially among children and elders. Early diagnosis should be considered to improve prognosis. This study indicates that a new and rapid diagnosis method is highly needed and improves the prognosis of tuberculous meningitis, shorting the time needed for an early and accurate treatment initiation.
P1612 Impact of pneumonia in pregnancy on maternal and neonatal outcomes in Durban, South Africa
M. Khan, J. Moodley, M. Adhikari (Durban, ZA)
Introduction: Pneumonia in pregnancy occurs in <1% of antenatal attendees; an incidence rate not dissimilar to that reported in non-pregnant adults. However the pregnant state and risk factors associated with the development of pneumonia adversely influence the outcome of pregnancy. It is the third leading cause of indirect maternal mortality during pregnancy, labour and the puerperium in the USA and in South Africa, it was the leading cause of maternal deaths due to non-pregnancy related infections.
The contribution of pneumonia and HIV-1 co infection in pregnancy has been poorly documented in South Africa. The aim of our study was to determine the clinical and demographic profile of women admitted to the maternity unit of a tertiary centre in Kwa Zulu-Natal, South Africa and to evaluate the association between pneumonia, HIV-1 infection and maternal and perinatal outcomes.
Methods: The study was performed between January and December 2000 in the antenatal clinic and labour ward of a tertiary centre. The investigation of pneumonia by the attending obstetrician was prompted by symptoms and clinical signs. All women were offered pre and post test counseling for HIV-1.
A control arm of HIV infected and uninfected women without pneumonia were randomly selected from the antenatal population.
Results: Twenty nine pregnant women were diagnosed with pneumonia. Mycobacterium tuberculosis (MTB) was the only causative organism isolated from sputum samples. Of the 14 MTB samples, 10 were smear positive and 8 were culture positive, with one case of drug resistant MTB. EPTB accounted for 9 of the 14 MTB cases. As a result, 3 categories developed: women with probable or confirmed PTB, women with suspected PTB and women with suspected bacterial pneumonia. There were 7 maternal deaths which occurred in women with pneumonia. There were 18 live born neonates, one intra uterine death and 6 stillbirths. In the control arm of 112 women, a greater number of obstetric co morbidities occurred. There were no maternal or neonatal deaths.
Conclusion: Pneumonia in pregnancy has a low incidence rate yet carries significant maternal and neonatal morbidity and mortality. Risk factors for the development of pneumonia were identified in our study and almost two thirds of the mothers had a chronic cough. In resourced constrained settings with high HIV and TB prevalence, it might be cost effective to do screening chest radiographs in pregnancy.
P1613 Tuberculosis in the area of Barcelona: the increasing influence of immigration
R. Güerri, R. Perelló, F. Sánchez, J.L. Gimeno-Bayon, G. Vallecillo, J.L. López, J.A. Kaylá (Barcelona, ES)
Introduction: The epidemiology of tuberculosis (TB) in urban populations is on dynamic change. The density of TB in a certain country or area is determined by (1) the TB prevalence background over the previous 40 years; (2) the socio-economic situation of the new diagnosed patients; (3) the migratory movements, and (4) the programmes for TB control.
Aim: To describe the new epidemiological profile of tuberculosis in Barcelona (Spain).
Methods: A retrospective-prospective study of the TB cases declared in the 2002–2005 period has been performed in the University-affiliated Hospital del Mar. The variables included are epidemiological as well as microbiological.
Results: Within 399 cases registered, 47.8%t were foreign-born, 19.5% of them coming from south-eastern Asia (India, Pakistan, Afghanistan) and 12.6% from South America. HIV co-infection was present in 16.8% of all patients, and in 35% of those coming from South America. The most frequent clinical presentations were toxic syndrome (22.7%), prolonged fever (17.9%) and respiratory symptoms (14.6%); 6.5% of patients presented haemoptisis. Those presentations were different between autochthonous and foreign-born, being the pulmonary form identifiable in the 59.1% of cases. From those, 27.6% presented cavities, and 52.6% presented abnormal radiography without cavities. The culture for mycobacteria was positive in 71.7% of cases, and negative in 28.3% (M. tuberculosis was identified in 62.7%). The susceptibility tests showed 5% strains resistant to rifampin among patients coming from South America, and 4.8% strains resistant to pyrazinamide in those coming from south-eastern Asia.
Conclusion: TB is now an emerging epidemic in Barcelona, and it is strongly related to increasing immigration. It is necessary to improve the diagnostic tools that could help to rapidly identify new TB cases among foreign-born presenting fever and toxic syndrome, as well as to prescribe accurate treatments in patients with high suspect of resistance to first-line drugs.
P1614 Tuberculosis among immigrants: our experience
A. Antypa, M. Pouyiouka, E. Spanos, K. Petrochilou, V. Karabassi, K. Paraskeva, N. Alexandropoulos, M. Toutouza (Athens, GR)
Tuberculosis is an airborne pathogen, propelled through tiny droplets via sneezing, talking or coughing. The disease is extremely widespread: one person in three worldwide has the infection and two million die annually from TB. Mass immigration from developing countries entails the risk of an increased rate of tuberculosis in our country.
Aim: The aim of this study was to assess the prevalence of tuberculosis among legal immigrants who asked for lawful permanent residency Green Cards.
Methods: The study population consisted of 1,460 (638 males and 822 females) immigrants aged 15–65 years old, who came to our hospital for examination, the period from 12.10.2005 to 02.05.2006. Their countries of origin were mainly Albania, Bulgaria, Romania, ‘the former Soviet Union', countries of Africa and South-Eastern Asia.
Tuberculin skin testing was performed on all study subjects upon enrollment, according to the routine procedures. A tuberculin skin test reaction size >10 mm was considered positive for all study participants. They were all examined with a posterior-anterior chest radiograph to detect chest abnormalities. A short history of TB exposure, infection, or disease was asked though in many cases it was difficult to communicate with them.
Results: 96 persons found tuberculin skin test positive with negative history of B.C.G. and 69 (4.9% of all the examined) of them had positive findings for pulmonary active Tb, from their chest radiograph. We sent the positives to the hospital of Chest diseases to be cured.
Conclusions: Screening for TB on arrival should be strengthened, and preventive therapy for those with recent TB infection should be considered. Healthcare professionals need to be aware that immigrants from countries with a relatively high prevalence of TB are a risk for countries with low prevalence of TB.
P1615 No evidence for hypoadrenalism as a cause of early mortality in a high HIV/TB prevalence population starting anti-tuberculosis treatment in Malawi
M.B.J. Beadsworth, J.J. van Oosterhout, M. Diver, A. Shenkin, H.C. Mwandumba, S. Khoo, T. O'Dempsey, S.B. Squire, E.E. Zijlstra (Liverpool, UK; Blantyre, MW)
Mortaliy is high in the first 2 months after commencing anti-tuberculosis treatment (ATT), especially in areas with high HIV prevalence, such as Malawi, where case fatality rates are 23–26%. (40% of these are in the first month.) Various causes have been proposed, including delayed treatment, other co-infections, advanced HIV disease and severe malnutrition. Another proposed aetiology is adrenal crisis precipitated by rifampicin. The commonest cause of hypoadrenalism worldwide is still described as TB and in addition rifampicin induces cytochrome p450, which metabolises endogenous glucocorticoid. We tested the hypotheses; that corticosteroid metabolism is deranged after starting ATT and that this disturbance is linked to early mortality.
Methods: Consenting consecutive smear positive adults admitted to Queen Elizabeth Central Hospital, Malawi were enrolled. Adrenal function was assessed using a short Synacthen (synthetic ACTH) test. Adequate adrenal function was defined as a rise above 550 nmol/L at 30 minutes. Testing was carried out at baseline and two weeks after commencing rifampicin (as part of ATT). Follow-up was for 3 months. All patients were offered HIV testing, as per national guidelines. Treatment for TB followed national guidelines.
Results: 51 smear-positive patients were enrolled of whom 29 (57%) were female, median age 32 years (range 18–62). 41 patients consented to HIV testing, of whom 88% were seropositive. The median time from onset of symptoms to starting ATT was 48 days (range 7–365 days). Case fatality in the first month was 16%. Hypoadrenalism was found in 2 patients on admission and none on day 14. Both of these were discharged and one was followed up at 3 months, the other was well, but declined further follow up.
Conclusions: Hypoadrenalism is not common and is not a cause of early mortality after commencement of ATT. Further studies are urgently required to identify other possible aetiologies.
P1616 XDR tuberculosis in Lisbon health region
L. Brum, R. Macedo, P. Cristovão, L. Reis, E. Fernandes, E. Pereira, A. Garcia (Lisbon, PT)
Introduction: Portugal is still the European country with the highest incidence of notified tuberculosis cases (31/100,000 in 2005 compared to 12.8/100,00 habitants in Central and Eastern E.U.). Despite MDR-TB strains isolated in hospital and public health laboratories being currently notified to Portuguese Health authorities, the true magnitude of this problem remains unknown. In fact, even though only 17 MDR-TB cases were notified in 2003, the National Institute of Health Dr. Ricardo Jorge in Lisbon, usually receives around 80 MDR-TB strains per year from the Health Region of Lisboa e Vale do Tejo (which represent 6–8% of the total of isolated strains in this region).
Recently it has been described MDR-TB strains that have mutated into an even stronger forms designated as extremely drug resistant tuberculosis (XDR-TB). This form of TB is defined as resistance to both isoniazid and rifampicin (definition of MDR-TB) in addition to resistance to any fluoroquinolone and to at least one of the three injectable second-line anti-TB drugs used in treatment (capreomycin, kanamicin and amikacin).
Material and Methods: In this study we evaluated the prevalence of XDR-TB strains among strains isolated in Lisbon laboratories and hospitals, in the years 2002 to 2006.
Susceptibility testing to first line drugs (Isoniazid, rifampicin, ethambutol, pyrazinamid, streptomycin) and second line drugs (ethionamide, capreomycine, ofloxacyn, kanamycin, amikacin and PAS), was performed in Bactec 460 Tb in the critical concentrations indicated in the Bactec manual or according to Pfyffer et al.
Results: Among the MDR-TB strains isolated in our laboratory we found that about half of them are also XDR-TB (table 1 ).
Table 1.
Number of XDR/MDR patients per year
| Year | No. of XDR | No. of MDR | %(XDR/MDR) |
|---|---|---|---|
| 2002 | 41 | 86 | 48 |
| 2003 | 50 | 87 | 57 |
| 2004 | 49 | 86 | 57 |
| 2005 | 49 | 94 | 52 |
| 2006a | 18 | 46 | 39 |
Data until September 2006.
Conclusion: These numbers are extremely high comparing to other European countries such as France, Belgium or UK that have 3–11% of XDR-TB cases. This problem is particularly worse for the HIV population as the rate death for HIV/AIDS patients with XDR-TB is nearly 100%.
The emergence of XDR-TB calls for a vigorous international response to fully implement proper TB control, to limit their public health impact and to ensure that countries at risk from the disease are prepared for it.
P1617 Impact of latent and overt tuberculosis on HIV-infected patients in southern Iran
M. Davarpanah, A.A. Sohrabpour, R. Neirami, M. Darvishi (Shiraz, IR)
Objectives: Tuberculosis (TB) is an important and potentially fatal opportunistic infection in the HIV-infected population. It is especially prevalent in the developing countries, including Iran. This study was aimed at determining the impact of latent TB infection and overt tuberculosis among a group of HIV infected clients attending a university affiliated clinic caring for voluntarily participants with high risk behaviours.
Methods: 572 HIV-infected persons including 60 females (11.4%) were registered. They all inhabit in the city of Shiraz, with around one million population located in the Southern Province of Fars, Iran. All had baseline and annual standard 5-tuberculin unit PPD (Purified Protein Derivative) skin tests performed. The patients were followed for a mean period of 29.2 ± 5.1 months.
Results: 114 patients (19.9%) had positive PPD tests (defined as >5 mm induration) at baseline. 10 more patients (1.7%) with negative baseline PPD developed positive skin test in follow-up annual tests. During the follow-up period, 13 patients (2.3%) suffered clinical tuberculosis including 9 patients with smear-positive pulmonary TB, 2 with biopsy-proven TB lymphadenitis, and 2 with TB pericarditis as evidenced by pericardial fluid analysis and complete response to anti-tuberculosis therapy. 6 out of 13 patients with overt tuberculosis died during the follow up. Death was directly TB-associated in four, all of whom had smear positive pulmonary tuberculosis.
Conclusion: There is a high rate of tuberculin reactivity in HIV-infected patients in southern Iran. The importance of PPD testing and providing isoniazid preventive therapy deserves more attention in the HIV-infected population.
P1618 Treatment outcome of tuberculosis patients diagnosed with human immunodeficiency virus infection in Iran
P. Tabarsi, P. Baghaei, M. Amiri, H. Emami, D. Mansouri, M. Masjedi (Tehran, IR)
Background: Concomitant treatment of tuberculosis (TB) and human immunodeficiency virus (HIV) faces many problems due to pill burden, drug/drug interactions and toxicity. As there are yet no reports made on the treatment outcome of TB-HIV patients in Iran, this study aims to evaluate the outcome of treatment at the Masih Daneshvari Hospital, the National Referral Center for Tuberculosis and Lung Diseases in Tehran.
Methods: TB patients who were admitted to the hospital between the years 2002–2004 and tested positive for HIV were included in the study. 61 variables were chosen for comparison. All analysis was done using SPSS and significance was deduced via chi-square testing.
Results: 56 patients were included in the study. The mean CD4 count was 193 + 181 while the mean count at which death occurred was 128 + 41. Fifty percent of the cases were cured while mortality occurred in 15 cases (26.8%). The rate of mortality in patients who developed AIDS defining illnesses (ADI) was higher (35.1%). The most common adverse drug effect observed was hepatitis (14.3%). A Kaplan Meier of survival showed 50% of deaths occurring in the first four months of TB treatment. Comparison of patients who were receiving highly active antiretroviral therapy (HAART) and mortality did not yield significant results.
Conclusion: Even though the sample size of this study was limited, it can be suggested that mortality of TB/HIV patients is high. Starting HAART early for TB-HIV patients whose CD4 count is less than 130 can reduce the rate of mortality.
P1619 Unusual association of tuberculous meningitis and cerebral toxoplasmosis in a HIV-negative patient. Case report
A. Slavcovici, C. Marcu, M. Lupse, C. Itu, D. Carstina (Cluj-Napoca, RO)
Objective: To underline the concordance/discordance of aetiologic diagnosis with neuroimagistic assesments.
Case report: A 7-year-old child, with prolonged neonatal jaundice, non-investigated and without any subsequent results, was admitted in our clinic for high fever, headaches, vomiting, second degree coma, nuchal rigidity and left hemiparesis. The cerebral magnetic resonance imaging (MRI) was negative. The examination of the cerebrospinal fluid (CSF) revealed discreet lymphocytic pleocytosis, a moderately increased protein concentration and a very low glucose level (12 mg%), suggesting a tuberculous meningitis. The usual CSF cultures, Gram stain and Ziehl– Neelsen stain examinations were negative, as well as the HIV test. After doing CSF culture using MB/BacT system, a specific tuberculostatic therapy was initiated (isoniasid, rifampicin, ethanbutol, pyrazinamide), associated with cerebral depletion (manitol), dexamethasone, and vitamins B1 and C. 30 days later, the M. tuberculosis strain was isolated and identified with susceptibility to classical antituberculosis drugs. The clinical status worsened with the persistence of the fever, coma and the neurologic signs, and with the occurrence of seizures. After 14 days of treatment, the neuroimaging assesment (cranial CT) described important cerebral edema and multiple round hypodensity areas in the frontal and parietal lobes, and in the caudatus and lentiform nucleus; these findings needed other serological examinations. Toxoplasmosis Ig M antibodies were detected in high level (ELISA method). We reevaluated the diagnosis as a proven tuberculous meningitis and a probable cerebral toxoplasmosis, with unknown time of infection, which might be either a recent one, or a reactivation of a congenital one. The treatment was continued with antituberculosis drugs in association with trimethoprim/sulfamethoxazole, pyrimethamine and folinic acid. The adjunctive therapy consisted of cerebrolisine, and vitamins B1 and C. After 4 days of antituberculosis and antitoxoplasmosis treatment, the outcome was rapidly favourable, without fever and with substantial remission of the neurological signs.
Conclusions: The risk factor for cerebral toxoplasmosis was the immunodepression induced by M. tuberculosis. The association of mycobacterial infection with cerebral toxoplasmosis in a HIV negative patient is rare. The importance of cranial CT in an unfavourable evolution of the central nervous system infections is relevant.
P1620 Primary tuberculosis of the breast mimicking breast cancer
B. Nasr, B. Borzouei, A. Nasserabadi, M. Ahmadi (Hamedan, IR)
Background: Tuberculosis of the breast is seen quite rarely. It is mainly classified as primary and secondary forms. Primary form is rarer. The gold standard for diagnosis of the disease is detection of the aetiologic agent, Mycobacterium tuberculosis, by using Ziehl–Neelsen staining or culture. However, smear positivity for acid-fast bacilli by Ziehl–Neelsen staining is low, and in most cases, tuberculosis of the breast can only be accurately diagnosed by histological identification of the typical necrotising granulomatous lesion.
Case report: A 49-year-old woman with complaints of breast lump and swelling of her right breast, which had appeared 40 days earlier, was admitted in our hospital. Her medical history was unremarkable. On physical examination, a hard mass (5×5 cm), which caused nipple retraction in the upper outer quadrant of the right breast, was observed. In laboratory findings she had WBC = 9,000 (neu = 38%, lym = 60%, mono = 2%), Hb = 12.7 g/dL and HCT = 38. Ultrasonography examination revealed hypoechoic mass with significant ductal dilatation, without collection and lymphadenopathy, which suggested mastitis. Mammography, which was performed in another clinical setting, suggested a malignant tumour (70×50 mm) with irregular border. Tc-99m sestamibi scan revealed abnormal large focal sestamibi uptake with T/N ratio = 1.45 which suggested malignant lesion.
The mass was excised by using surgery. After surgery, pathologic examination reported necrotising granulomatous lesion, which confirmed breast tuberculosis. The patient had no abnormal findings on computer tomography (CT) of chest but the result of a tuberculin skin test was positive. After confirmation of the diagnosis, the patient received antituberculosis therapy (rifampin, isoniazid, ethambutol, and pyrazinamide). She was currently being followed-up and the disease was showed no signs of recurrence one year after treatment.
Discussion: Breast tuberculosis is classified as primary and secondary forms. CT is useful in differentiation of primary and secondary breast tuberculosis. Pathologic confirmation is required for diagnosis. Although tuberculosis of the breast is rare, it should be considered as a possible diagnosis, particularly in countries where tuberculosis is endemic. The prognostic of breast tuberculosis is favourable.
P1621 Early effective treatment of cutaneous infection with Mycobacterium chelonae complex after rapid molecular identification
J. Hopman, A. Langewouters, T. Kroese, P.D.J. Sturm (Nijmegen, NL)
Objectives: Slow-growing as well as rapid-growing Mycobacteria can cause cutaneous infection. The latter specifically in immunocompromised patients. The clinical diagnosis may be delayed due to recurrent skin lesions. Rapid diagnosis and identification aids in the initiation of appropriate therapy.
Methods and Results: A 50 y.o. male patient had received a renal transplant 10 years ago for which he used azathioprine and prednison. He presented to a dermatologist with a 2 month history of recurrent skin lesions on his right lower leg and pain of his right foot. A diagnosis of gout was suspected and allopurinol was administered. After a few days, brownish nodules appeared on the right leg which eventually ulcerated with purulent discharge. Subsequently, the lesions healed slowly, leaving behind violaceous maculae with crusts. Flucloxacillin and clindamycin respectively were started empirically with improvement of the skin lesions. New purulent nodules developed after cessation of therapy. Now, a clinical diagnosis of pyoderma gangrenosum was considered which was confirmed histologically. However, prednison improved symptoms only temporarily. Histological examination of new skin biopsies showed acid-fast bacilli. Meanwhile the foot was swollen with increased redness and tenderness and an abscess was suspected. Acid fast staining of surgically drained pus showed numerous PMNs and AFBs. A molecular identification of M. chelonae complex was available within 48 hours and treatment could be initiated with clarithromycin. Amikacin was not considered an option with respect to his renal function and tigecycline was added as second agent.
Conclusion: Spontaneously resolving lesions have been described previously in cutaneous mycobacterial infection and unfamiliarity with this clinical presentation may result in a delayed diagnosis. Because a rapid identification was established with molecular methods, effective antimicrobial therapy could be started early. Newer drugs, such as tigecycline and linezolid, with in-vitro activity against rapid-growing mycobacteria may be useful in future treatment of these infections.
P1622 Mycobacterium mucogenicum pulmonary infection in an immunocompromised patient
Z. Gitti, I. Neonakis, E. Stylianoudaki, E. Papadopoulou, S. Maraki, D. Spandidos (Heraklion, Crete, GR)
Background: Infections caused by non-tuberculous mycobacteria are continuously increasing. Mycobacterium mucogenicum is a recently characterised, rapidly-growing mycobacteria, rarely seen in human infections. Here we report the clinical and laboratory findings of a M. mucogenicum pulmonary infection in an immunocompromised patient.
Case presentation: The patient was a 70-year-old male with non-Hodgkin's lymphoma. He was admitted with a low-grade fever of 37.7° C and dyspnea of two months duration. Clinical examination revealed pulmonary involvement. A chest X-ray showed pulmonary infiltrates, multiple nodular lesions, and perihilar adenopathy. Mantoux skin test was negative. His past medical history was significant for chronic obstructive pulmonary disease and arterial hypertension. Ziehl–Neelsen acid fast staining of the gastric fluid was positive. Liquid and solid cultures of the sample yielded mycobacteria. The identification of the isolate as M. mucogenicum was performed by Genotype AS diagnostic reverse hybridisation strip test assay (Hain Lifescience, Germany). Antimicrobial susceptibility was determined by the E-test. The isolate was sensitive to cefoxitin, imipenem, clarithromycin, azithromycin, doxycycline, linezolid, cotrimoxazole, ciprofloxacin, sparfloxacin, moxifloxacin, tobramycin. Combined therapy with clarithromycin and ciprofloxacin for 4 months was proved effective.
Conclusion: Non-tuberculous mycobacteria can cause considerable pulmonary infections in immunocompromised patients. M. mucogenicum is most frequently found in the environment but is also incriminated for human infections. Early diagnosis is important for the determination of the appropriate treatment.
P1623 Granulomatous hepatitis due to intravesical therapy by BCG for superficial bladder cancer
A. Hristea, V. Arama, R. Moroti, D. Nae, E. Angelescu, R. Sandu, C. Tiliscan, V. Molagic, D. Munteanu (Bucharest, RO)
Between January 1996 and January 2006, 680 patients with superficial urinary bladder cancer were admitted to the Urology Department of Clinical Institute Fundeni, Bucharest. After endoscopic resection they received BCG immunotherapy according to international guidelines. The following side effects have been noted: cystitis 92.3%, early macroscopic haematuria (24–48 h after instillation) 53.8%, low-grade fever (24–36 h after instillation) 46.1%, fever and chills for more than 7 days 7.6%, BCG-related visceral granulomas or BCG sepsis 1.8%.
We report a case of disseminated BCG infection causing granulomatous hepatitis in a 58-year-old man. His medical history included a noninvasive transitional-cell carcinoma treated by surgery followed by intravesicular instillation of BCG. The first 6 weekly and the following 5 monthly BCG treatments were well tolerated. Two days after his 12th instillation he developed 39°C fever and weakness followed by jaundice. On admission he had 38.5°C fever, a pulse rate of 104/min, jaundice and hepatosplenomegaly. Liver enzymes were elevated (ALT 2N, AP 2.4N), bilirubin level of 7.6 mg/dl (conjugated 7.3), moderate inflammatory syndrome, proteinuria above 3 g/L, albumin level of 2.2 g/L. Abdominal CT scan showed hepatosplenomegaly without changing of bile ducts. He was started on empiric broad-spectrum antibiotic. We noted the persistence of his symptoms and an increasing cholestasis. A hepatic biopsy was performed and antibiotic therapy was changed to corticosteroids. The patient's condition improved over the next 7 days. The result of the hepatic biopsy showed non-caseating granulomas, suggesting mycobacterial spread, although the acid-fast bacillus stains on tissue specimens were negative. In situ PCR confirmed the mycobacterial dissemination, despite the negative culture of blood and urine for M. bovis. Antituberculous drugs were added to his treatment and prednisone was discontinued within 3 weeks. Clinical and laboratory improvement were marked and sustained so the patient could be discharged home for a six-month course with an isoniazid–rifampin combination.
Conclusion: The development of granulomatous lesion in distant organs is an uncommon complication of immunotherapy with BCG vaccine. Clinical and laboratory findings in our patient suggest the haematogenous spread of mycobacteria to liver, as well as a hypersensitivity response.
P1624 Tenosynovitis caused by Mycobacterium haemophilum in a patient with rheumatoid arthritis
P.C.A.M. Buijtels, P.A. vd Lubbe, E.J. Kuijper, L.E. Bruijnesteijn van Coppenraet, P.L.C. Petit (Rotterdam, Schiedam, Leiden, NL)
Introduction: M. haemophilum can present with an array of symptoms ranging from focal involvement to widespread disease. M. haemophilum is a fastidious organism. Due to its specific growth requirements in vitro, infections were not recognized until 1978.
Objective: Description of M. haemophilum infection diagnosed by real-time PCR on clinical material.
Methods and Results: We describe a 71-year-old female patient with rheumatoid arthritis who presented with chronic dorsal tenosynovitis of the finger one month after intra-articular injection of steroids. Two months after the manifestation of the tenosynovitis, a synovectomy was performed. Histological examination revealed a granulomatous necrotising inflammation by acid-fast bacteria. The culture for mycobacteria remained negative, but application of a Mycobacterium genus-specific real-time polymerase chain reaction (PCR) in combination with a M. haemophilum-specific PCR resulted in the recognition of M. haemophilum as the causative agent.
The patient recovered completely after synovectomy and a 6 months treatment by clarithromycin, ciprofloxacin and rifampicin.
Conclusion: M. haemophilum infections may be relatively under-recognized and underreported. Only three patients with rheumatoid arthritis and M. haemophilum infection have been reported in the literature. The development of tenosynovitis after intra-articular injection in the patient we described, is very suggestive for inoculation of M. haemophilum via contaminated fluid. The real-time PCR has shown to be a significant assay for detecting and identifying M. haemophilum in clinical samples and may provide insight in the prevalence of this nontuberculous mycobacterial infection and its clinical relevance. Because autoimmune diseases like rheumatoid arthritis are increasingly treated with immunosuppressive drugs clinicians should be aware of this infection as it may become clinically more important in the future.
P1625 Using ELISPOT to expose false positive skin test conversion in tuberculosis contacts
P. Hill, D. Jeffries, R. Brookes, A. Fox, D. Jackson-Sillah, M. Lugos, S. Donkor, B. de Jong, T. Corrah, R. Adegbola, K. McAdam (Banjul, GM)
Objectives: Repeat tuberculin skin tests may be false positive due to boosting of waned immunity to past mycobacterial exposure. We evaluated whether an the booster ELISPOT test could be used to distinguish new Mycobacterium tuberculosis infection from phenomenon.
Methods: We conducted tuberculin and ELISPOT tests in 1,665 TB contacts: 799 were tuberculin test negative and were offered a repeat test after 3 months. Those test positive with ≥6 mm increased induration had an ELISPOT, chest X-ray and sputum analysis if appropriate. We compared converters with non-converters and assessed the probability of each of 4 combinations of ELISPOT results.
Results: 704 (72%) contacts had a repeat tuberculin test; 176 (25%) had test conversion, which increased with exposure to a case (p = 0.002), increasing age (p = 0.0006) and BCG scar (p = 0.06). 114 tuberculin test converters had ELISPOT results: 16 (14%) were recruitment positive/follow up positive, 9 (8%) positive/negative, 34 (30%) negative/positive, and 55 (48%) were negative/negative. Approximately 38% of skin test converters were estimated to have boosting. There was a significant non-linear effect of age for ELISPOT results in skin test converters (p = 0.038). Three converters were diagnosed with TB, 2 had ELISPOT results: both were positive, including one at recruitment.
Conclusions: We estimate that approximately 38% of tuberculin skin test conversion in Gambian TB case contacts is due to boosting. This varies with age. Use of ELISPOT only in tuberculin converters may lead to delayed diagnosis of M. tuberculosis infection and disease. Further studies are required to determine the relevance of ELISPOT reversion.
Methods for antimicrobial susceptibility testing
P1626 Detection of resistance to linezolid in Enterococcus spp. by fluorescence in situ hybridisation using locked nucleic acid probes
G. Werner, A. Essig, M. Bartel, N. Wellinghausen, I. Klare, W. Witte, S. Poppert (Wernigerode, Ulm, DE)
Objectives: Resistance to linezolid, caused by mutation of one or multiple alleles of the 23S rRNA gene, is emerging in Enterococcus spp. especially in vancomycin-resistant Enterococcus faecium. Aim of the study was to establish and evaluate a fluorescence in situ hybridisation (FISH) assay for the detection of the resistance mutation.
Methods: FISH probes containing a locked nucleic acid at the mutation locus were labelled with FITC respectively Cy3 and used in conjunction (LNA wild type: CCCAGCTCGCGTGC; LNA resistant: CCAGCTAGCGTGC).
In a first step a panel of 30 linezolid-sensitive (15) and -resistant (15) Enterococcus strains was tested two times in a blinded manner. The minimal inhibitory concentration (MIC) of enterococci to linezolid was determined by microbroth dilution using Iso-Sensitest broth as nutrient medium. In resistant isolates the number of mutated alleles was determined using LabChip technology and BioAnalyzer 2100.
In the second step 83 clinical isolates from blood cultures and VRE screening agar plates were investigated by FISH in parallel to broth dilution using the Merlin Micronaut System.
Results: FISH consistently correctly recognized all 30 well characterised isolates as susceptible respectively resistant. Concerning the clinical isolates seventy-nine isolates were rated susceptible and one isolate as resistant by both methods. Three E. faecium isolates that were measured repeatedly susceptible by broth dilution, gave a positive result with the linezolid-resistance as well as with the wild type probe indicating presence of mutated and non mutated alleles. All three isolates showed a single mutated 23S rDNA allele when pooled 23S rDNA PCR fragments were digested with NheI and resolved on a LabChip 1000.
Conclusion: FISH using LNA probes is a rapid, cheap and easy performable method that showed 100% sensitivity and specificity for determination of presence of resistance genes to linezolid in E. faecium.
P1627 Comparison between MicroScan system and agar dilution method for quinupristin/dalfopristin susceptibility of Enterococcus faecium
Y-R. Kim, Y.J. Kim, S.I. Kim, Y.J. Park, M.W. Kang (Seoul, KR)
Objectives: Quinupristin/dalfopristin (Q/D) is one of the effective antimicrobials in treating vancomycin resistant E. faecium, but Q/D resistant E. faecium was observed frequently using MicroScan as a tool for routine antibiotic susceptibility test. We compared the results of Q/D susceptibility test by the Positive Combo Panel Type11 of the MicroScan Walk Away 96 (Dade Behringer) with those obtained by reference agar dilution method.
Methods: We collected consecutively 65 E. faecium isolates nonsusceptible to Q/D (42 resistant, 23 intermediate), and selected randomly 32 E. faecium isolates susceptible to Q/D by MicroScan system from September 2003 to August 2004. All isolates were identified in Kangnam St. Mary's Hospital Clinical Microbiology Laboratory using MicroScan system. For Q/D non-susceptible isolates, MIC of Q/D, vancomycin and teicoplanin for were determined by agar dilution method according to the CLSI guidelines, and for Q/D-susceptible (by MicroScan), MIC of Q/D was measured by agar dilution method.
Results: The agreement rates are shown in Table 1 . The MIC of Q/D resistant isolates by agar dilution method were all 4 ug/mL except one, that is low-grade resistant. Of the Q/D-resistant isolates, most were susceptible to vancomycin and teicoplanin (78% and 82%) respectively.
Table 1.
Comparison of Q/D susceptibility test by Microscan and agar dilution method
| No of isolates according to susceptibility category by agar dilution method | Susceptibility category by Microscan (No. of isolates) |
||
|---|---|---|---|
| Susceptible (32) | Intermediate (23) | Resistant (42) | |
| Resistant | 0/32 (0%) | 14/23 (60.9%) | 36/42 (85.7%) |
| Intermediate | 0/32 (0%) | 6/23 (26.1%) | 1/42 (100%) |
| Susceptible | 32/32 (100%) | 3/23 (13.0%) | 5/42 (11.9%) |
Conclusion: Of the Q/D resistant E. faecium, the major error rate was 11.9%, which is over the acceptable range (≤3%). If the Q/D susceptibility test to Q/D results ‘resistance’ by MicroScan, it is needed to confirm the results using reference method. There may be another factor except Q/D use in hospitals or virginiamycin use in animals for high rates of glycopeptide-susceptible and Q/D resistant E. faecium.
P1628 Fluoroquinolone resistance in nalidixic acid-resistant Escherichia coli
S. Bengtsson, M. Sundqvist, O. Samuelsen, N. Krrh, A. Sundsfjord, G. Kahlmeter (Växjö, SE; Tromsoe, NO)
Objectives: The Swedish Reference Group on Antibiotics (SRGA) and the Norwegian Working Group on Antibiotics (NWGA), recommend the use of a nalidixic acid (NAL) 30 μg disc for the detection of fluoroquinolone (FQ) resistance in Enterobacteriacae. Recently the European Committee on Antimicrobial Susceptibility Testing (EUCAST) harmonised European fluoroquinolone breakpoints. We wanted to determine to what extent NAL overestimates clinical resistance to FQ in Escherichia coli as defined by EUCAST and Clinical Laboratory Standards Institute (CLSI) and compare these breakpoints with EUCAST epidemiological cut-off values.
Methods: 149 consecutive NAL-resistant E. coli from urine samples collected in Kronoberg county, Sweden, June 2004-January 2005 were investigated. The antibiotics included were ciprofloxacin (CIP), levofloxacin (LVX), moxifloxacin (MXF), norfloxacin (NOR) and ofloxacin (OFX). The minimum inhibitory concentrations (MIC) were determined by Etest (AB Biodisk, Solna), qnr genes and aac(6')-Ib with PCR and all strains were typed with PhP-typing. EUCAST and CLSI breakpoints (2006) and epidemiological cut-off values were used to define resistance.
Results: All NAL-resistant isolates showed reduced susceptibility (higher MIC than epidemiological cut-off values) to the studied FQs except for LVX and MXF where 37% and 66% of the isolates showed MIC values inside respective wild type distribution. NAL-resistant strains would be reported susceptible for CIP in 52% of the isolates, LVX 55%, MXF 52%, NOR 24% and OFX 27% using EUCAST clinical breakpoints. With CLSI clinical breakpoints, CIP would be reported susceptible in 54% of the isolates, LVX 55%, MXF (no CLSI approved breakpoints), NOR 54% and OFX 52%.
Conclusion: The EUCAST and CLSI breakpoints showed good agreement for CIP and LVX. However, using CLSI breakpoints for NOR and OFX would result in reporting 30% and 25%, respectively, as susceptible while EUCAST would have classified them as nonsusceptible. In situations where there are few treatment alternatives, the FQ-susceptibility should be further investigated in NAL-resistant isolates. FQ-resistance is increasing rapidly and the risk that during therapy these strains may develop more pronounced FQ resistance must be weighed against the therapeutic benefits. The fact that so many of the NAL-resistant E. coli isolates appeared devoid of all signs of any resistance mechanisms to LVX and MXF is probably due to the higher activity against topoisomerase IV of these agents.
P1629 Comparison of antimicrobial susceptibility tests for the detection of KPC-positive strains of Klebsiella pneumoniae
S. Whittier, F. Wu, P. Della-Latta (New York, US)
Objective: The emergence of multi-drug resistant organisms is a tremendous challenge for New York City hospitals. As the armamentarium of antibiotics for Gram-negative bacteria [GNB] remains stable, resistance is on the rise, leaving clinicians limited choices for treatment regimens. Accurate and rapid detection of these multi- or pan-resistant GNB by the microbiology laboratory is critical both for therapy and infection control purposes. We examined the performance of 3 antimicrobial susceptibility tests [AST] and one screening method for the detection of carbapenem resistance due to KPC production by Klebsiella pneumoniae [KP].
Methods: Forty clinical isolates of KP were identified as KPC positive by PCR, which was designed to detect two mutation points in the purported blaKPC coding region. These isolates were tested for imipenem [IM] and meropenem [MP] susceptibilities by the following methods: Microscan Walkaway (Dade Behring, West Sacramento, CA), VITEK2 (bioMeriuex, Durham, NC) and E test (AB Biodisk, Solna, Sweden). 2006 CLSI guidelines were utilised for interpretation of MIC data. The ertapenem disk (BD, Sparks, MD) was also used as a screen for carbapenem resistance. Ten KPC negative isolates of KP were also included as negative controls.
Results: Of the 40 KP isolates, 36 were identified by PCR as KPC type 2 and 4 were KPC type 3. Ten carbapenem susceptible isolates were KPC negative by PCR. When IM and MP results were examined, the abilities of Microscan, VITEK and E test to correctly determined resistance were 90%, 85% and 20%, respectively. The ertapenem disk screen correctly identified 100% of KPC positive strains. All systems accurately determined MIC values for the 10 KPC negative isolates.
Conclusions: We evaluated 3 AST systems for their ability to accurately detect carbapenem resistance. This study demonstrated that the Microscan and E test performed favourably when compared to PCR, however VITEK 2 did not. We also found that the ertapenem disk test is a reliable predictor of CB resistance.
P1630 Moraxella catarrhalis susceptibility testing; development of a CLSI disc diffusion method
J.M. Bell, L.J. Walters, J.D. Turnidge (North Adelaide, AU)
Objectives: At present there is only CLSI susceptibility testing standards for the species Moraxella catarrhalis (MCAT) for a broth microdilution (BMD) method. MCAT is isolated frequently from respiratory specimens, and resistance to a variety of agents has emerged or is emerging. It is implicated in a range of clinical syndromes, especially acute otitis media, acute bacterial sinusitis and acute exacerbations of chronic bronchitis. It has established that broth microdilution susceptibility testing works reliably. The aim of this study was to develop a disk diffusion method, and interpretative zone diameter correlates for the tentative MIC breakpoints, including molecular data on resistance mechanisms.
Methods: Three hundred well characterised MCAT isolates were tested against penicillin, ampicillin, amoxicillin/clavulanic acid (2:1 ratio), ampicillin/sulbactam (2:1 ratio), meropenem, cephalexin, cefaclor, tetracycline, trimethoprim/sulfamethoxazole, erythromycin, clarithromycin, azithromycin, telithromycin, cefuroxime, rifampin, chloramphenicol, moxifloxacin, ceftriaxone, ciprofloxacin, gentamicin; using CLSI susceptibility standards for broth microdilution and disc diffusion. Custom made dry-form BMD panels (Trek Diagnostic Systems) were used. The disk diffusion method used Mueller-Hinton agar without blood supplementation, CLSI standard disk strengths and incubation in 5% CO2 for 20–24 h.
All isolates were confirmed as MCAT and β-lactamase enzymes typed by PCR. Isolates with elevated tetracycline MICs were tested for the presence of tet(B) or tet(M) efflux genes.
Results: A significant number of isolates did not grow on Mueller-Hinton Agar in air. Disc diffusion tests were therefore incubated in 5% CO2. On-scale MIC values were obtained for all antimicrobials tested, except for meropenem where the majority of MICs were ≤0.004 mg/L. Using the current CLSI disc strengths many of the antimicrobials tested produced large zone diameters. Eighteen isolates were β-lactamase negative. Of the β-lactamse positive isolates, 18 typed as BRO-2; all had lower ampicillin/penicillin MICs than those strains typed as BRO-1. All isolates with tetracycline MIC > 1 mg/L harboured tet(B). Twelve isolates had elevated (≥0.25 mg/L) ciprofloxacin MICs. No erythromycin resistant strains were found.
Conclusion: Although incubation in 5% CO2 was necessary, and large zones were obtained, zone diameter correlates could be estimated based on the current CLSI breakpoints.
P1631 Moxifloxacin MIC results for Neisseria gonorrhoeae (ATCC 49226) for an eight lab study by CLSI agar dilution methodology
L. Koeth, J. DiFranco (Westlake, US)
Objectives: This study was performed in order to establish a MIC quality control range for N. gonorrhoeae (ATCC 49226) according to CLSI M23-A2 for agar dilution methodology.
Methods: Moxifloxacin was tested at concentrations of 0.002–0.25 μg/mL using 3 different manufacturers of GC agar base media (Remel, Accumedia and PML) and 1% defined growth supplement (Remel). Ciprofloxacin was also tested as the control drug at concentrations of 0.0025 to 0.03 ug/mL using 1 lot of media. 10 replicates of the QC strain were tested for each of the media lots by eight U.S. laboratories on each of two days.
Results: Of the total 434 moxifloxacin MICs evaluated, 100% were within a four well range of 0.004 to 0.03 ug/mL and 97% were within a three well range of 0.008 to 0.03 μg/mL. All ciprofloxacin MICs were within the CLSI established range of 0.001 to 0.008 μg/mL (90.6% of MICs at 0.004 ug/mL). The modal moxifloxacin MIC for 7 of the 8 labs and 2 of the 3 media lots was 0.016 μg/mL. The moxifloxacin MICs for 1 lab and PML media were slightly lower (mode of 0.008 μg/mL.). There was also a higher incidence of poor or no growth with the PML media. The average inoculum concentration was 5×104 CFU/spot. MIC distributions for all labs are shown in the table .
| Antimicrobial agent | MIC (μg/mL) number of occurrences: |
Total n | |||||
|---|---|---|---|---|---|---|---|
| 0.001 | 0.002 | 0.004 | 0.008 | 0.016 | 0.030 | ||
| Moxiflioxacin | 13 | 58 | 334 | 29 | 434 | ||
| Ciprofloxacin | 1 | 12 | 145 | 2 | 160 | ||
Conclusions: When performing agar dilution MIC testing according to CLSI methodology, a quality control range for N. gonorrhoeae (ATCC 49226) versus moxifloxacin of 0.004 to 0.03 μg/mL is recommended.
P1632 Comparison of BD Phoenix Automated System and Etest to broth microdilution method for antimicrobial susceptibility testing of multiple resistant Pseudomonas aeruginosa and Acinetobacter spp
P. Zarakolu, O. Ozcakir, G. Hascelik, S. Unal (Ankara, TR)
Objectives: To compare the BD Phoenix Automated System (Becton Dickinson Diagnostic Systems, Sparks, MD, USA) and Etest (AB Biodisk, Solna, Sweden) to broth microdilution method for antimicrobial susceptibility testing of multiresistant Pseudomonas aeruginosa and Acinetobacter spp. for some of the commonly used antibiotics in our hospital.
Methods: Pseudomonas aeruginosa (n = 61) and Acinetobacter spp. (n = 61) strains isolated from different clinical samples of patients with hospital infections in Hacettepe University Adult Hospital in January 2004-July 2005 were included in the study. The isolates were identified by BD Phoenix Automated System and stored at −80°C until study time. Antimicrobial susceptibility testing by Phoenix System and Etest method were performed according to the manufacturers' instructions and interpretive categories (resistant, intermediate, susceptible) were determined. Broth microdilution method, according to CLSI guidelines was used as the reference method. The antimicrobials tested in the study were piperacillin, cefepim, ceftazidim, meropenem, ciprofloxacin and tobramycin. For evaluation, essential and categorical agreements were determined.
Results: The isolates were multiple (≥ 3) resistant to the antimicrobials tested. The overall category agreement was 63.7%; with the very major, major and minor error rates 2.8%, 19.1%, 14.5%, respectively when Phoenix System were compared to microdilution method. However, when Etest antimicrobial susceptibility results were compared to the reference method, the overall essential agreement was 58.9%; the category agreement was 69.0% with the very major, major and minor error rates 3.3%, 15.3%, 12.3% respectively.
Conclusion: Clinicians should be aware of the conflicting antimicrobial susceptibility results by different testing methods while facing difficulties with the infections due to multiresistant nonfermentative bacteria.
P1633 Accuracy of β-lactam susceptibility testing results from VITEK and VITEK2 systems when testing Pseudomonas aeruginosa
V. LaBombardi, V. Rekasius, P. Schreckenberger, R. Jones (New York, Maywood, North Liberty, US)
Objectives: To critically assess VITEK and VITEK2 accuracy for determining β-lactam susceptibility (S) when testing P. aeruginosa (PSA). Recent publications have questioned the interpretative and quantitative agreement of various automated systems especially for piperacillin/tazobactam (P/T), cefepime (FEP), and aztreonam (AZM). A collaborative multi-site study tested local (LOC) clinical strains and selected challenge (CH) strains.
Methods: This study was performed in 3 sites (Emory University/CDC, Atlanta, GA; St. Vincent's Hospital [STV], New York, NY; Loyola University Medical Center [LUMC], Maywood, IL) with each site testing strains by one or more automated methods e.g. VITEK by CDC and LUMC; and VITEK 2 by CDC and STV. Each processed 30 PSA (15 LOC and 15 CH) that included an equal distribution of isolates S and resistant (R) to β-lactams. The tested agents were AZM, FEP, ceftazidime (CAZ), imipenem (IPM), piperacillin (PIP) and P/T. Utilised cards/software were: for VITEK (GNS-122/WSVTK-R10.01) and for VITEK 2 (AST-6N09/WSVT2-R04.02). Reference methods were reference frozen-form panels, Etest and disk diffusion methods using standardised CLSI procedural details or manufacturer instructions. Quality Control (QC) was assured via concurrent testing, and all presented data were associated with acceptable QC results.
Conclusions: PSA were tested by VITEK and VITEK 2 and results were observed as unacceptable for all 6 agents. VME was common with IPM, PIP and P/T (15.0–21.7%), and mE with significant skewing toward false R (11.0–21.0%) was noted for AZM, FEP and CAZ. These systematic errors adversely affect local antibiograms (empiric choices), individual patient care, and require corrective action by the manufacturer.
Results: Analyses compared VITEK and VITEK 2 results to those produced by the reference broth microdilution test, and consensus categorical results from all reference methods. A significant bias toward S or R was defined as a shift of ≥10% for S rate when using the commercial product. Unacceptable levels of intermethod error were found using both applied analyses. Elevated minor error (mE; limit 10%) was noted for both systems with AZM (18.3–33.3%), FEP (13.3–36.7%), CAZ (16.7–23.3%) and IPM (13.3–26.7%). Serious, very major error (VME; false-S) several-fold greater than acceptable limit (1.5%) was detected for P/T with lesser degrees of ME and VME for PIP. Error rates by each system were distributed among LOC and CH strains and test sites.
Errors produced when testing 30 P. aeruginosa against 6 β-lactam agents by VITEK and VITEK 2 automated systems.
| System/Antimicrobial agent (no. tested) | Percentage of errora |
|||||
|---|---|---|---|---|---|---|
| Compared to BMD result |
Compared to consensus result |
|||||
| VME | VM | ME | VME | VM | ME | |
| VITEK | ||||||
| Aztreonam (60) | 0.0 | 3.3 | 18.3 | 0.0 | 5.0 | 31.7 |
| Cefepime (60) | 1.7 | 0.0 | 36.7 | 1.7 | 0.0 | 36.7 |
| Ceftazidime (60) | 1.7 | 0.0 | 20.0 | 1.7 | 3.3 | 16.7 |
| Imipenem (60) | 8.3 | 0.0 | 13.3 | 6.7 | 0.0 | 10.0 |
| Piperacillin (60) | 0.0 | 8.3 | NA | 0.0 | 6.7 | NA |
| Piperacillin/tazobactam (60) | 15.0 | 5.0 | NA | 15.0 | 5.0 | NA |
| VITEK2 | ||||||
| Aztreonam (60) | 1.7 | 0.0 | 28.3 | 0.0 | 0.0 | 33.3 |
| Cefepime (60) | 0.0 | 0.0 | 13.3 | 1.7 | 0.0 | 16.7 |
| Ceftazidime (60) | 3.3 | 0.0 | 23.3 | 1.7 | 0.0 | 21.7 |
| Imipenem (60) | 6.7 | 0.0 | 25.0 | 5.0 | 0.0 | 26.7 |
| Piperacillin (60) | 5.0 | 0.0 | NA | 6.7 | 0.0 | NA |
| Piperacillin/tazobactam (60) | 21.7 | 1.7 | NA | 20.0 | 0.0 | NA |
Unacceptable levels of error are underlined. NA = not applicable.
P1634 Critical assessment of P. aeruginosa susceptibility testing results for six β-lactams using the BD Phoenix system
S. Juretschko, T. Beavers-May, C. Essmyer, R. Jones (Little Rock, Kansas City, North Liberty, US)
Objectives: To study the accuracy of commercial automated systems when testing P. aeruginosa (PSA) due to escalating and recent reports of interpretive error. Documented high error rates were recorded for β-lactams tested by MicroScan WalkAway, VITEK and VITEK2 when compared to CLSI methods ranging from false-resistant (ME; major) to false-susceptible (VME; very major) results. Although in depth studies comparing automated systems have been reported, the BD Phoenix System has limited intermethod data for PSA, with susceptibility (S) test results often interpreted by non-CLSI criteria; thus, a 2-laboratory study was designed to validate Phoenix PSA results.
Methods: 60 tests were produced by local processing of 15 recent clinical strains (RCS) from each site (Arkansas Children's Hospital and St. Luke's Regional Laboratories) and 15 challenge strains (CS) representing equal numbers of PSA that were S and resistant (R) to the tested β-lactams (aztreonam[AZM], cefepime[FEP], ceftazidime[CAZ], imipenem[IPM], piperacillin[PIP] and piperacillin/tazobactam[P/T]). Each strain was tested by Phoenix (panel NMIC-112; software V5.15A/04.11B) and compared to CLSI broth microdilution (BMD) results or to the consensus result from BMD, disk diffusion test (DD) and Etest. Errors were defined as VME, ME and minor (mE; intermediate by one method) guided by M23-A2.
Results: The table compares Phoenix categorical results and the consensus results from 3 validated methods. Unacceptable rates of mE (16.7–36.7%) occurred with AZM, FEP and CAZ, regardless of reference result utilised for analysis. Results were consistent between sites, RCS or CS subsets, and with prior publications that showed compromised categorical agreement. For AZM, the mE level was associated with systematic bias toward false-R. In contrast, IPM, PIP and P/T had generally acceptable error rates or were only marginally elevated.
Types of intermethod errors when testing 30 P. aeruginosa isolates by the automated Phoenix system in two laboratoriesa.
| Antimicrobial agent (no. tested) | % error types compared to consensusb |
||
|---|---|---|---|
| Very Major | Major | Minor | |
| Aztreonam (60) | 0.0 | 1.7 | 36.7 |
| Cefepime (60) | 0.0 | 1.7 | 18.3 |
| Ceftazidime (60) | 1.7 | 0.0 | 16.7 |
| Imipenem (60) | 0.0 | 0.0 | 1.7 |
| Piperacillin (30) | 0.0 | 3.3 | NA |
| Piperacillin/tazobactam (60) | 1.7 | 5.0 | NA |
Results from two laboratories (Arkansas Children's Hospital and St. Luke's Regional Laboratories).
Consensus of broth microdilution, disk diffusion and Etest categorical results. Unacceptable levels of error are underlined and mE was associated with a systematic trend toward false-R. NA = not applicable.
Conclusions: This rigorous challenge of the Phoenix to assess the PSA susceptibility versus key β-lactams displayed error rates indicating a modest need for re-evaluation (AZM). Automated systems have been documented to produce inaccurate results with trending toward false S as well as R. Clinical laboratories should be aware of these interpretive problems for PSA testing and seek alternative, validated methods such as simple agar diffusion tests (DD and Etest). Among the automated systems evaluated to date, the Phoenix appears to possess the fewest β-lactam/PSA testing discords.
P1635 An evaluation of MicroScan WalkAway results for broad-spectrum β-lactams when testing Pseudomonas aeruginosa
S. Lerner, H. Salimnia, L. Steed, J. Bosso, R. Jones (Detriot, Charleston, North Liberty, US)
Objectives: To compare contemporary clinical (CL) and challenge (CH) strains of Pseudomonas aeruginosa (PSA) susceptibility (S) test results from an automated system (MicroScan WalkAway; 2 laboratories) against 6 broad-spectrum β-lactams-to-results of reference broth microdilution (BMD) and consensus results from 3 validated methods (BMD, Etest, disk diffusion[DD]). Previous reports have documented high minor (mE; 10–32%) and very major (VME; false-S; 19%) errors among anti-PSA β-lactams.
Methods: Each centre tested CH (15) strains of PSA that included equally represented S and resistant (R) MICs across a wide range for tested agents (aztreonam[AZM], cefepime[FEP], ceftazidime[CAZ], imipenem[IPM], piperacillin[PIP], piperacillin/tazobactam[P/T]). 15 CL strains were also tested by MicroScan WalkAway (panel no. NEGMIC30 or NC32 and software no. LabPro 2.01) and 3 reference methods (BMD, Etest, DD). Categorical results from MicroScan WalkAway were compared to CLSI BMD results (see Table ) as well as the consensus of all tests. Error limits for acceptability were those listed in M23-A2 guideline.
Error rates for the MicroScan WalkAway system when testing 30 P. aeruginosa isolatesa
| Antimicrobial agent (no. tested) | Percentage of errorb |
|||||
|---|---|---|---|---|---|---|
| Compared to BMD result |
Compared to consensus result |
|||||
| VME | ME | mE | VME | ME | mE | |
| Aztreonam (60) | 0.0 | 3.3 | 21.7 | 0.0 | 3.3 | 23.3 |
| Cefepime (60) | 0.0 | 3.3 | 48.3 | 0.0 | 3.3 | 45.0 |
| Ceftazidime (60) | 1.7 | 6.7 | 23.3 | 0.0 | 6.7 | 20.0 |
| Imipenem (60) | 0.0 | 1.7 | 11.7 | 1.7 | 1.7 | 10.0 |
| Piperacillin (60) | 10.0 | 3.3 | NA | 15.0 | 3.3 | NA |
| Piperacillin/tazobactam (60) | 5.0 | 1.7 | NA | 10.0 | 0.0 | NA |
NA, not applicable.
Results from University of South Carolina (Charleston, SC) and Wayne State University, Detroit Medical Center (Detroit, MI).
BMD: reference broth microdilution MIC results. Consensus result was determined from the BMD, disk diffusion and Etest values. Unacceptable levels of error are underlined.
Results: The table shows the comparisons of categorical agreement and the listed error rates occurred equally between participant sites and organism populations (CH, CL). Markedly elevated rates of mE were observed for AZM (21.7–23.3%), FEP (45.0–48.3%) and CAZ (20.0–23.3%), regardless of reference result selected for comparative analysis. More serious errors by MicroScan WalkAway of false-R (major-error; ME) were also noted for the same agents, but false-S (VME) was detected (5.0–15.0%) for PIP and P/T. For drugs showing unacceptable mE or serious interpretive errors (ME), a systematic bias toward false-R MicroScan WalkAway results was found for AZM (10.0%), FEP (48.3%) and CAZ (16.7%). The skewing toward R was most extreme for FEP (3-fold greater than CAZ). PIP and P/T results trended toward false-S (VME) at a level of 10.0–11.7%.
Conclusions: These results corroborate findings of others that some automated susceptibility testing systems perform poorly when testing β-lactam agents against PSA. Error rates documented in our medical centres for MicroScan WalkAway, using diverse PSA collections clearly indicates false-S and -R trends that seriously compromise patient care as well as misdirecting empiric treatment choices and formulary decisions. We recommend that our colleagues employ alternative methods such as agar diffusion tests (DD, Etest) to more accurately assess antimicrobial S among PSA.
P1636 Pilot evaluation of Innovotech bioFILM PA susceptibility test for clinical utility in the treatment of cystic fibrosis patients infected with Pseudomonas aeruginosa
R.P. Rennie, L. Turnbull, C. Brosnikoff, N.E. Brown, D.C. Lien, B. Rawal, M. Olson (Edmonton, Alberta, Calgary, Alberta, CA)
Objective: To compare the Innovotech bioFILM PA susceptibility kit designed for planktonic and sessile susceptibility testing with standard broth micro-dilution susceptibility testing for application in the treatment of cystic fibrosis (CF) patients infected with Pseudomonas aeruginosa (Pa).
Methods: 25 strains of Pa isolated from CF patients were tested by CLSI broth microdilution for susceptibility. The same strains were then tested with the Innovotech bioFILM PA kit. Isolates were grown first in trypticase soy broth (TSB) and inoculated into wells of the Innovotech bioFILM PA kit. The sessile inoculating peg lid was placed on top of the tray containing inoculum and TSB and rocked gently on a rocker overnight at 35°C. The sessile inoculating 96 peg lid was then placed into a 96 well tray containing anti-pseudomonal antimicrobials in cation-adjusted Mueller Hinton broth (CAMHB) at CLSI breakpoint concentrations. This tray also contained combinations of agents commonly used for Pa therapy. After overnight incubation, the planktonic susceptibility was recorded, and the 96 peg lid was then placed on a second recovery tray containing CAMHB. After overnight incubation, the biofilm susceptibilities were determined.
Results: Standard CLSI broth microdilution MICs and Innovotech planktonic breakpoints (S, I, R) were within the same range. There were no major or very major errors. For sessile organisms, the Innovotech MICs were often greater than for the planktonic forms. Tobramycin combined with colistin, ciprofloxacin or meropenem were active against many of the biofilm cultures. These combinations may be useful for treatment of biofilm infections in these CF patients. When tested alone, standard planktonic MICs often indicated resistance to these agents. The breakpoints of the susceptible combinations were within the ranges clinically achievable with standard therapeutic doses of these agents.
Conclusions: These first attempts to utilise a susceptibility test system (Innovotech bioFILM PA kit) that better mimics the biofilms of Pa in the respiratory tract of CF patients provide early support for the use of this device to assist with appropriate antimicrobial therapy in CF treatment. In a small number of CF patients our results have suggested that combination biofilm MICS may have clinical utility. Further studies are underway to confirm these observations. (This device has been approved for use by Health Canada; FDA clearance is pending.)
P1637 Evaluation of the in vitro activity of levofloxacin and moxifloxacin tested against Stenotrophomonas maltophilia: can moxifloxacin activity be predicted by levofloxacin MIC results?
A. Gales, R. Jones, M. Janechek, H. Sader (Sao Paulo, BR; North Liberty, US)
Objective: To evaluate the in vitro activity of levofloxacin (LEV) and moxifloxacin (MXF) tested against S. maltophilia and the correlation between MIC values for these fluoroquinolones (FQ) compounds in order to assess if LEV susceptible (S) strains could be categorised as MXF-S.
Methods: A total of 763 unique S. maltophilia strains collected worldwide through the SENTRY Antimicrobial Surveillance Program (2002–2005) were tested for S against LEV, MXF and selected antimicrobials by broth microdilution methods according to CLSI guidelines. MIC results were interpreted according to CLSI and EUCAST breakpoints. CLSI has LEV breakpoints of (S/resistant [R] in mg/L) ≤2/≥8 for S. maltophilia and other Gram-negative bacilli (GNB) and no MXF breakpoints except for Gram-positive pathogens; while EUCAST has GNB breakpoints for LEV (≤1/≥4 for Enterobacteriaceae [ENT], Acinetobacter and P. aeruginosa) and MXF (≤0.5/≥2 for ENT only).
Results: LEV showed good in vitro activity against S. maltophilia (MIC50, 1 mg/L and MIC90, 2 mg/L) with 85.8% S at the CLSI breakpoint, but only 70.3% if EUCAST breakpoints were applied. Scattergram with the correlation between the MXF and LEV MIC results is shown in the comparison Figure .

Scattergram showing correlation between levofloxacin and moxifloxacin MIC results for S. maltophilia. Solid lines indicate CLSI breakpoints, dashed lines indicates EUCAST breakpoints.
By applying the CLSI LEV breakpoints (≤2/≥8 mg/L) and MXF breakpoints at ≤1/≥4 mg/L, the overall categorical agreement was 95.5% with only 0.1% very major (VM; false-S), no major (MA; false-R) and 4.3% minor (MI) errors. Using EUCAST ENT breakpoints (LEV includes P. aeruginosa and Acinetobacter spp.), the overall agreement was 93.3% with no VM or MA error and 7.7% MI error. S rates for ciprofloxacin were 29.5 and 10.0% when GBN S breakpoints of CLSI and EUCAST were applied, respectively.
Conclusions: The spectrum of LEV against S. maltophilia decreases significantly if EUCAST breakpoints are used in preference to CLSI breakpoints. There was an excellent correlation (r = 0.93) between LEV and MXF MIC results and categorical results for LEV may be used to predict categorical results for MXF if breakpoints were one doubling dilution lower than that of LEV. However, clinical studies may be necessary to establish the role of these FQs in the treatment of S. maltophilia infections, but achieving a critical number of case studies would be difficult.
P1638 Comparison of methods for penicillinase detection in S. aureus
M. Kaase, S. Lenga, F. Szabados, T. Sakinc, A. Anders, S. Friedrich, S.G. Gatermann (Bochum, DE)
Background: Penicillin resistance in S. aureus is mediated by four types of Penicillinases encoded by the blaZ gene. Because penicillin is considered superior to oxacillin in those rare isolates with true penicillin susceptibility, correct determination of penicillinase is important. Laboratories using the VITEK2 for susceptibility testing of S. aureus have to use an additional test for β-lactamase detection for those isolates with an MIC of ≤ 0.12 mg/L.
Methods: We investigated several methods for penicillinase detection, like size and character of penicillin inhibition zones, nitrocefin testing, cloverleaf assay, starch iodine plates and an in-house blaZ-PCR for isolates with an MIC ≤ 0.12 mg/L in the VITEK 2.
An isolate was considered penicillinase positive if either the blaZ-PCR, the nitrocefin test read after 15 min, the cloverleaf assay or the penicillin zone edge appearance suggested the presence of a penicillinase.
Results: Of 274 isolates 50 (18.2%) were penicillinase positive. Sensitivities were 80% for penicillin zone edge appearance, 62% for an penicillin inhibition zone diameter ≤28 mm, 52% for the nitrocefin test read after 15 min, 74% for the cloverleaf assay, 58% for starch iodine plates and 88% for the blaZ-PCR. Isolates with a MIC of 0.12 mg/L and 0.06 mg/L in the VITEK 2, respectively were penicillinase positive in 24.2% and 6.2%.
Conclusion: Cloverleaf assays, penicillin zone edge determination and blaZ-PCR are superior to starch iodine plate and nitrocefin testing for the detection of penicillinase in S. aureus.
P1639 Assessment of cefoxitin MICs with the BD Phoenix® system to detect methicillin-resistant Staphylococcus aureus isolates
M.I. Morosini, M. García-Castillo, E. Loza, F. Baquero, R. Cantón (Madrid, ES)
Cefoxitin (FOX) disks have demonstrated to enhance MRSA detection when compared with oxacillin (OXA) disks but information with FOX MICs is scarce. Automated systems are often used in clinical laboratories but, in some cases, they are not proficient enough to recognize heterogeneous MRSA (especially low level resistant populations).
Objectives: BD Phoenix was evaluated for the detection of methicillin resistance in clinical S. aureus with both OXA and FOX MICs. Results were compared with those of reference microdilution and disk diffusion (CLSI), and Etest methods. In addition, BD Phoenix was challenged to detect OXA resistance (R) in artificially mixed cultures containing different proportions of OXA-susceptible (S) and OXA-R populations.
Methods: A total of 159 S. aureus isolates prospectively recovered from blood cultures along 1998–2005 were studied. S. aureus ATCC (29213, 43300, 33591, and 25923) strains were used as quality controls. PCRs for mecA detection were performed in isolates with OXA MIC range of ≥1–≤16 mg/L representing isolates close to the OXA susceptibility breakpoints. Detection of PBP2a with the MRSA latex agglutination test (Slidex®, bioMérieux) was carried out. Mixed broth cultures with different proportions [75:25, 90:10 and 99:1] of OXA-S (29213) and OXA-R (43300, heterogeneous) or (33591, homogeneous) strains, respectively, were used to challenge the BD Phoenix.
Results: A total 27% of MRSA isolates was detected. When considering OXA results, 100% of categorical agreement was observed between all methods. All OXA-R strains were mecA positive. However, a false methicillin–S strain (but mecA and PBP2a positive) was detected with FOX data obtained only with BD Phoenix and disk diffusion. Overall mean time to methicillin results availability was of 6 hours. Interestingly, BD Phoenix was able to detect the presence of methicillin-R bacteria even in the 99:1 mixture (lowest proportion of OXA-R population) through OXA and FOX MICs.
Conclusion: BD Phoenix compared favourably to reference tests. Although all methods failed to detect a MRSA strain, only BD Phoenix and disk diffusion FOX results phenotypically revealed such resistance. BD Phoenix, when challenged with mixed OXA-S/OXA-R populations, was able to detect methicillin-R irrespective of bacterial proportions. BD Phoenix system enhances routine detection of MRSA, particularly those with low level expression of resistance.
P1640 Evaluation of cefoxitin disks to detect methicillin-resistant coagulase negative staphylococci
J.W. Mouton, M.T. Bohne, C.H. Klaassen, A.M. Horrevorts, H.A. de Valk, A. Voss (Nijmegen, NL)
Objectives: For S. aureus, the use of cefoxitin disks is the recommended method to screen for methicillin resistance (MetR). A similar method has been suggested for coagulase negative staphylococci (CNS) but it is not clear which method performs best. Specificity is compromised since CNS consists of multiple species and every species has its own characteristics, including the discriminatory effect on zone diameters. We investigated 4 different methods (2 disks and 2 media) to determine the cut-off zone diameters to achieve a high specificity and sensitivity to distinguish metR from metS CNS. These values were also determined at the species level.
Methods: 250 CNS were isolated from blood cultures or other relevant sites of infection and stored at −70°C. Strains were revived by plating on blood agar. The next day, colonies were suspended to obtain an adequate inoculum to result in semiconfluent growth (Iso Sensitest agar) or confluent growth (Mueller Hinton agar). A 10 and 30 μg cefoxitine disk (Oxoid) was placed on each 9 cm agar plate and incubated for 24 h at 35°C. Zone diameters (mm) were read in two directions to one decimal. Identification was performed by AFLP and partial 16S rRNA gene sequencing. The mecA gene was identified by a LightCycler method.
Results: 167 strains were metR and 83 metS. Of the four methods, the 30 μg disk on MH performed best, but there was considerable overlap in zone size between metR and metS strains. Setting the sensitivity at 100% resulted in a specificity of 67%. At the species level, the median zone diameter for metS S. epidermidis (N = 128) and S. hominis (N = 38) was 30.7 mm and 26.6 mm respectively. A 100% sensitivity as well as specifity at the species level was obtained at cut offs of 27.5 mm and 17 mm, respectively.
Conclusion: The method best discriminating between metR and metS strains was MH with confluent growth and a 30 μg disk but with a relatively poor specificity of 67%. Speciation increased both sensitivity and specificity to 100% for S. epidermidis and S. hominis.
P1641 Validation of Etest for susceptibility testing of moulds with posaconazole
A. Yusof, A. Engelhardt, P. Ho, Å. Karlsson, A. Wanger (Solna, SE; Texas, US)
Background: Posaconazole is a new broad spectrum triazole shown to be highly active against many species of filamentous fungi including Aspergillus, Fusarium, Penicillium and Trichophyton. The performance of Etest Posaconazole was compared to the CLSI broth microdilution method (BMD) using a study design to fulfil the CE marking requirements according to the European Directive for in vitro diagnostic devices.
Method: Etest Posaconazole was compared to BMD in a two site study involving clinical, stock and challenge organisms. A total of 217 strains were tested: A. fumigatus (52), A. flavus (40), A. nidulans (6), A. terreus (16), A. niger (18), A. versicolor (9), A. ustus (5), A. sydowii (3), Aspergillus spp. (10), F. oxytoca (5), F. solani (4), Fusarium spp. (13), Rhizopus spp. (11), P. boydii (7), S. apiospermum (12), Acremonium spp. (3), Alternaria spp. (2) and Cladosporium spp. (1). 25 identical strains were used to examine inter-laboratory reproducibility, and 4 ATCC reference strains to generate quality control specifications. Inoculum density checks to verify CFU/mL were done using the quality control and reproducibility organisms and 10% of clinical isolates. Etest was used according to the manufacturer's instructions and BMD according to M38A, Vol 22, No16.
Results: Essential agreement found between Etest and BMD are shown in the table .
| Species | N | MIC range | EA ± 2 dilution |
|---|---|---|---|
| Aspergillus spp. | 10 | 0.002–0.125 | 100 |
| A. niger | 18 | 0.008–0.38 | 100 |
| A. terreus | 16 | 0.008–0.19 | 100 |
| A. flavus | 40 | 0.032–0.25 | 100 |
| A. fumigatus | 52 | 0.023–1.5 | 98 |
| A. nidulans | 6 | 0.047–0.125 | 100 |
| A. versicolor | 9 | 0.023–32 | 100 |
| A. sydonii | 3 | 0.064–0.125 | 100 |
| A. ustus | 5 | 0.25–32 | 100 |
| F. oxytoca | 5 | 0.125–2 | 100 |
| F. solani | 4 | 0.047–32 | 100 |
| Fusarium spp. | 13 | 0.5–32 | 100 |
| S. apiospermum | 12 | 0.094–2 | 100 |
| Rhizopus spp. | 11 | 0.047–32 | 91 |
| P. boydii | 7 | 0.125–2 | 100 |
| Aeremonium spp. | 3 | 0.023–2 | 100 |
| Cladosporium spp. | 1 | 0.094 | 100 |
| Alternaria spp. | 2 | 0.047–0.094 | 100 |
Conclusion: Etest Posaconazole was found to be reproducible for testing of filamentous fungi and MIC results were substantially equivalent to the CLSI reference method. In the absence of interpretive breakpoints for new agents such as posaconazole, Etest provides a practical alternative for full range MIC testing.
P1642 Evaluation of the BACT/ALERT 3D system for testing susceptibility of Mycobacterium tuberculosis to first-line antituberculosis agents: comparison with BacTec MGIT 960 and Lowenstein-Jensen proportion method
C. Guney, A. Suel, T. Karagoz, Y. Yorulmaz (Istanbul, TR)
Objectives: The performance of the BACTALERT 3D (BTA) system for the testing of Mycobacterium tuberculosis susceptibility to streptomycin (STR), isoniazid (INH), rifampin (RIF) and ethambutol (ETH) was evaluated with 101 clinical isolates and compared with BACTEC MGIT 960 (M960) system and Lowenstein-Jensen (L-J) proportion method (currently used) from clinical specimens at Sureyyapasa Chest Disease Hospital.
Methods: A series of different samples recovered from patients in hospital were cultivated in three systems at the same time according to NaOH %3+NALC decontamination protocol. BTA and M960 susceptibility testing was performed as described by instructions of manufacturers. M. tuberculosis H37Rv (ATCC 27294) standard strain was used for quality control of the all tests.
Results: In all, there were 404 test combinations with a total of 19 discrepancies (4.7%) between M960 and BTA results. Total agreements were 95.3%. The discrepancies resolved in favour of the M960 system in 11 cases and in favour of the BTA system in 8 cases according to L-J proportion method. The BTA system produced 3 very major errors (VME) and 8 major errors (ME), while the M960 showed 2 VME and 6 ME. No statistically significant differences were found. The mean times of detection for BTA and M960 were 16.3 days and 12.2 days, respectively. Two smear negative sputums detected by BTA in 26 and 36 days were not detected by M960. Turnaround times from detection to reporting SIRE results for BTA and M960 were 12.8 and 11.4 days, respectively. No statistically significant differences were found. The contamination rate for BTA, MGIT and L-J were 2%, 4%, 3% respectively.
Conclusion: Some discordant results between the standard test and the test method could be attributed to the presence of borderline-resistant strains, mainly in relation to LJ tests where the final results depend on an accurate count of colonies. These results suggest that BACTALERT 3D system is a valid alternative to the BACTEC MGIT 960 for M. tuberculosis susceptibility testing.
P1643 Comparison of quality of drug susceptibility testing of Mycobacterium tuberculosis by absolute concentration method and BACTEC 460
A. Baranov, S. Presnova, N. Nizovtseva, I. Trekin, L. Gvozdovskaja, L. Yendaurova, A. Mariandyshev, G. Bjune, E. Heldal, T. Mannsaker (Oslo, NO; Murmansk, Archangel, Syktyvkar, Petrozavodsk, RU)
The study has been carried out in 4 administrative territories of the North-western federal region of Russia. Namely, in Archangel oblast, Murmansk oblast, Republic of Karelia and Republic of Komi.
Objectives: The aim of our study was to find rates of agreement of DST between absolute concentration method, routinely performed at the Russian laboratories, and the reference method – BACTEC 460, performed at the National Reference Laboratory for Mycobacteria of the Norwegian Institute of Public Health (NIPH).
Laboratory at the Archangel Regional Anti-tuberculosis dispensary has been involved in DST quality assurance programmes on different levels for many years. The main component of these programmes was the collaboration with the reference laboratory at NIPH. Data on this collaboration from previous years is not presented in this manuscript.
Methods: In total, 88 strains of M. tuberculosis were analysed and results for these strains were compared for both methods. In addition to the computing of the overall percent agreement for each drug, kappa test has been used. The following arbitrary divisions for interpreting the results of kappa test have been used: under 0.20 is negligible; from 0.21 to 0.40 is minimal; from 0.41 to 0.60 is fair; from 0.61 to 0.80 is good; and over 0.81 is excellent.
Results: The highest rates of agreement were observed for isoniazid and rifampicin, 95.5% (CI; 88.8–98.8) and 97.7% (CI; 92.0–99.7), respectively. They also had the highest k-values 0.90 (CI; 0.80–1.0) and 0.96 (CI; 0.89–1.0), which correspond to excellent agreement beyond chance between investigating methods. Streptomycin and kanamycin had agreements in 93.2% (CI; 85.8–97.5) and 90.9% (CI; 82.9–96.0) in all tested strains, respectively, with k-values 0.85 (CI; 0.73–0.97) and 0.78 (CI; 0.64–0.92). These k-values mean from good to excellent agreements. The lowest rate of agreement was found for ethambutol, in 85.2% (CI; 76.1–91.9) of cases. This is consistent with relatively low k-value for this drug 0.70 (CI; 0.56–0.85), which means from fair to good agreement between investigating methods.
Conclusion: These findings suggest that, currently, the results of absolute concentration method for isoniazid and rifampicin can serve as a reliable diagnostic tool. This is very important for diagnosis and treatment of MDR tuberculosis.
P1644 Sensitivity to colistin: evaluation of available interpretive criteria of disc diffusion test
I. Galani, F. Kontopidou, P. Rekatzina, J. Deliolanis, M. Souli, A. Antoniadou, H. Giamarellou (Athens, GR)
Objectives: Increased antibiotic resistance in Gram-negative bacteria isolated in ICUs of tertiary-care hospitals, has augmented the use of colistin (CL) in recent years, since it is the only available active agent against these isolates. However CLSI and EUCAST does not currently provide interpretive criteria for disc susceptibility as there is very little data on the accuracy of disc testing methods for CL. In this study, we evaluate the accuracy of two available interpretive criteria of disc susceptibility testing for CL, compared to E-test.
Methods: A total of 409 clinical isolates of Acinetobacter baumannii [AB] (178), K. pneumoniae [KP] (185) and Escherichia coli [EC] (46) were included in the study. All strains were isolated from hospitalised patients in 11 different tertiary hospitals. Isolates were tested by E-test for susceptibility to CL, and results were compared with those obtained by disc susceptibility testing method (DD). Two breakpoints were evaluated, Gales et al. (JCM, 2001; 39: 183–90) (≤11 and ≥14 mm) and BSAC 2006 (≤14 and ≥15 mm) and compared to MIC breakpoints established by BSAC (>4 and ≤4 mg/L) for Enterobacteriaceae and Acinetobacter spp.
Results: CL displayed good activity against AB (MIC90 0.5 mg/L), and EC (MIC90 0.625 mg/L) but was less active against KP (MIC90 32 mg/L). Forty-six (24.9%) KP, six (3.4%) AB and two (4.3%) EC strains were resistant (R) to CL according to the E-test method. Totally, 16.4% and 5.5% of CL-resistant isolates were falsely reported as susceptible (S) when evaluated by the two available criteria, respectively. Applying the BSAC breakpoints decreased the very major error (R by E-test but S by DD) to 1.5% from 2.9% compared to Gales et al criteria. On the contrary major discrepancies (S by E-test but R by DD) were found in 45% of strains tested with BSAC recommendations while 11.5% of minor errors (R by E-test and intermediate by DD) were found during application of Gales et al criteria.
Conclusion: Use of the BSAC criteria decreased the very major errors but significantly increased the major errors when compared with the breakpoints proposed by Gales et al. Disc susceptibility testing methods are unreliable at detecting CL resistance. E-test or dilution methods should be the method of choice for susceptibility testing of CL as zone diameter interpretive criteria exhibiting good correlation with MICs seems difficult to establish.
P1645 Comparative evaluation of the VITEK2, disk diffusion, E-test, and agar dilution susceptibility testing methods for colistin in clinical isolates (including heteroresistant E. cloacae and A. baumannii strains)
J. Lo-Ten-Foe, A. de Smet, B. Diederen, J. Kluytmans, P. van Keulen (Tilburg, Utrecht, Breda, NL)
Objectives: The increasing use of colistin necessitates the availability of rapid and reliable methods for colistin susceptibility testing. We compared four methods of colistin susceptibility testing.
Methods: Disk diffusion (Rosco), agar dilution, E-test (Biodisk) and VITEK2 (bioMérieux) colistin susceptibility testing methods were used in 102 clinical isolates collected from patient materials during a Selective Digestive Decontamination or Selective Oral Decontamination trial in an intensive care unit. The agar-based tests were performed on both Mueller-Hinton (MH) agar and Isosensitest (IS) agar. All isolates were tested and compared to the agar dilution reference method. BSAC breakpoints for colistin were used for media containing IS agar. For MH agar containing media CLSI breakpoints were used for interpretation.
Results: The error rates of the different colistin testing methods compared to the reference agar dilution method are summarised in table 1 .
Table 1.
Error rates compared to the reference agar dilution methoda.
| VM | M | MI | |
|---|---|---|---|
| Disk diffusion MH | 18% | 4% | 25% |
| E-test MH | 4% | 0% | 0% |
| E-test Isosensitest | 0% | 3% | 7% |
| VITEK 2 | 8% | 0% | 0% |
VM, very major error; M, major error; MI, minor error.
High error rates and low levels of reproducibility were observed in the disk diffusion test. The colistin E-test and the VITEK 2 showed >90% agreement with the agar dilution reference method. We found no significant differences in the performance of either of these test media. Heteroresistance for colistin was observed in 6 E. cloacae isolates and in 1 A. baumannii isolate. Heteroresistant isolates could be detected in the agar dilution, E-test, or disk diffusion tests. The VITEK 2 displayed a low sensitivity in the detection of heteroresistant subpopulations of E. cloacae. Heteroresistance was easier detected using Isosensitest agar and seemed to be induced upon exposure to colistin rather than being caused by stable mutations.
Conclusion: Disk diffusion method is an unreliable method to measure the susceptibility to colistin. The VITEK 2 colistin susceptibility test is a reliable tool to determine the susceptibility to colistin in isolates that do not exhibit heteroresistance. The E-test is also a reliable method to measure colistin susceptibility and detect heteroresistant isolates. Heteroresistance was observed in several E. cloacae and in A. baumannii isolates. It is not clear whether these colistin resistant subpopulations are truly clinically significant or are representing in vitro artifacts.
P1646 Detection of low-level resistance to carbapenems using the VITEK2 and the advanced expert system
Z. Afkou, K. Kontopoulou, D. Sofianou, M. Tsivitanidou (Thessaloniki, GR)
Objectives: Metallo-β-lactamases (MBLs) are plasmid mediated β-lactamases conferring resistance to all β-lactams except aztreonam. These acquired MBLs recently have been spread among Enterobacteriaceae. Many of enterobacteria, possessing MBL genes exhibit low-level resistance to carbapenems or appear susceptible by the routine techniques and pass unnoticed in most cases. Our objective was the evaluation of VITEK 2 automated system to detect low-level resistance to carbapenems.
Methods: A total of 33 Enterobacteriaceae isolates (2 Escherichia coli, 9 Enterobacter cloacae, 22 Klebsiella pneumoniae) recovered from clinical specimens at Hippocration General Hospital were studied. The identification and susceptibility testing were performed by using the VITEK2 automated system (bioMérieux, France).
All isolates resistant to ceftazidime, cefoxitin and cefotaxime were characterised as resistant or intermediate to imipenem (IMP) by the automated system. The strains were tested by disk-diffusion method according to CLSI breakpoint criteria. IMP-EDTA double disk synergy test was carried out to screen the production of MBL. Molecular investigation was used to confirm the presence of plasmid-mediated MBL.
Results: By the VITEK2 automated system 18 isolates (54.5%) were characterised as intermediate to imipenem (MIC = 8 μg/mL) while the remaining 15 (45.5%) were resistant (MIC > 16). By the disk diffusion method 1 isolate (3%) was intermediate (diameter 14–15 mm), 9 strains (27.2%) were resistant (diameter ≤13 mm) and 23 strains (69.8%) were susceptible (diameter >16 mm). Double disk synergy test using an imipenem disk and an EDTA disk was positive in all strains indicating the MBL production. Additionally, all isolates were positive in PCR assays with blavim specific oligonucleotides.
Conclusions: This study demonstrated the capacity of VITEK 2 to detect low-level resistance to carbapenems. This automated system combined with EDTA synergy test is able to detect MBL producers in clinical laboratories. The early detection of MBLs permits the selection of appropriate treatment and the implementation of strict infection control measures in order to prevent their spread in the hospital.
P1647 Evaluation of Etests for determining tigecycline MICs by BSAC methodology
R. Hope, S. Mushtaq, D. Livermore (London, UK)
Introduction: Etests are calibrated against MICs determined using Clinical and Laboratory Standards Institute (CLSI) methods, with Mueller Hinton media. Most UK laboratories however use British Society for Antimicrobial Chemotherapy (BSAC) susceptibility testing methods, with IsoSensitest agar. We tested the agreement between tigecycline MICs determined by Etest and BSAC agar dilution methods.
Method: Isolates (n = 558) were selected from the BSAC 2004 bacteraemia surveillance collection and other collections held at the reference laboratory. They comprised: 63 Acinetobacter spp., 44 alpha-haemolytic streptococci, 45 beta-haemolytic streptococci, 35 coagulase-negative staphylococci, 55 Enterobacter, 40 enterococci, 35 E. coli, 46 Haemophilus, 54 Klebsiella, 55 Proteeae, 21 Serratia, 35 S. pneumoniae, and 30 S. aureus, selected to cover the broadest range of tigecycline MICs and to span the EUCAST/BSAC S/I/R breakpoints. Tigecycline MICs were determined in parallel using Etests, with confluent growth on IsoSensitest agar, and by the BSAC agar dilution method.
Results: MICs by agar dilution and Etest were within experimental error (i.e. one doubling dilution) in 527/558 (94%) of cases and within two dilutions in all cases. MICs by Etest nevertheless tended to be slightly lower than by agar dilution (lower in 33% of cases vs. higher in 10.5%), with this effect most evident for S. pneumoniae and other alpha-haemolytic streptococci, all of which were very susceptible, (MIC ≤ 0.5 mg/L). alpha-haemolytic streptococci accounted for 22/27 cases where MICs by Etest were 2 tubes lower than by agar dilution, despite using a hand lens to seek microcolonies and the final termination of growth. Among Gram-negative isolates (n = 113) with agar dilution MICs in the range 1–4 mg/L (i.e. spanning the “top” end of susceptible, through intermediate, to the “bottom” end of resistant with respect to EUCAST/BSAC breakpoints), catergorisation agreement between agar dilution and Etest MICs was 86%, with 14% minor errors and no major or very major errors.
Conclusion: Tigecycline Etests used on IsoSensitest agar gave good agreement with MICs determined using BSAC methodology. Categorisation agreement was good for isolates with borderline susceptibility or resistance – a group where Etests are likely to be used to verify disc based results. MICs for highly-susceptible alpha-haemolytic streptococci tended to be underestimated by Etest, but this seems unlikely to be consequential.
P1648 A new mass-spectrometric approach to antibacterial susceptibility testing
V. Vereshchagin, E. Ilina, M. Atroshkina, M. Serebryakova, T. Maier, M. Kostrzewa, V. Govorun (Moscow, RU; Leipzig, DE)
Objectives: Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) is an established technique for profiling bacterial proteins directly from cell extracts, providing a reproducible spectrum within minutes. This technique is applied for different microorganisms identification and subtyping. We report potentialities of this technique for the antibiotic susceptibility testing remaining one of the principal assays of clinical microbiology.
Methods: All experiments were performed using laboratory strain of Escherichia coli DH5α (E. coli). After cultivation during 14 h at 37°C using SOB agar, bacterial cells were transferred into liquid media and cultured during 3 h. Then 1 mL of bacterial culture (106–107 cells/mL) was exposed to different concentrations (10–100 μg/mL) of the antibiotics ampicillin (AMP), ceftriaxone (CEF) or tetracycline (TET) during 1, 2 or 3 hours. A sample without antibiotics was used as control. Protein extraction was performed as follows: (1) samples were centrifuged, pellet was solved with pure water and then ethanol was added; (2) after centrifugation pellet was added with 70% formic acid and pure acetonitrile, solution was mixed well and centrifuged again. Supernatant was co-crystallised with matrix (α-cyano-4-hydroxycinnamic acid) on a steel target plate. Using a microflex MALDI-TOF mass-spectrometer (Bruker Daltonics, Germany) reproducible spectra for different cultivating conditions were collected.
Results: After antibiotic treatment both small and significant changes in E. coli profile were observed. They depend on the time of exposure, antibiotic concentration and differ for different antibiotics. E. coli spectra after AMP treatment show a significant new peak (m/z 3822). Similar results were obtained for TET: spectra show another new peak (m/z 6970). The more exposure time increased, the worse spectra (AMP, TET, CEF) were obtained. As for CEF, dramatic suppression of spectra for 2- and 3-hour exposures was observed.
Conclusion: The suggested approach has great advantages such as requiring only a small amount of biological material, easy sample preparation, highly accurate measurement with low cost per analysis. Mass spectra have specific alterations under antibiotics exposure. Very likely these changes reflect some molecular events accompanying cell death. Thus direct mass-spectrometric analysis of whole bacterium cell has great potential to be used as rapid technique for antibiotic susceptibility testing.
P1649 Determination of antibiotic breakpoints using a novel electrochemical respiratory activity assay
T. Mann, O. Hammarstrom, D. Burca, L. O'Hagan, D. Sparkes, S. Mikkelsen (Kitchener, Waterloo, CA)
Objective: We have developed a rapid respiration-based electrochemical method for determining antibiotic breakpoint susceptibility of microorganisms, including Gram-positive, Gram-negative, aerobic and anaerobic bacteria. Respiratory activity measurements made on a suspended bacterial isolate in the absence/presence of an antibiotic provide results in 35 minutes.
Methods: Measurements of respiratory activity have been made on 25 microorganisms to date using a biamperometric method and an electrochemical mediator cocktail containing ferricyanide and dichlorophenolindophenol. A bacterial isolate was initially incubated for 20 minute with various biochemicals and the antibiotics. Ferricyanide was then added to the suspension where it was reduced to ferrocyanide by the microorganism's respiratory activity, during a 10 minute reaction time. The ferrocyanide was quantitated by a 2 minute biamperometric measurement made with two platinum electrodes and a 100 mV applied potential. In parallel with these measurements, standardised disk-diffusion tests were undertaken to determine whether the organism was to be classified as resistant, intermediate or susceptible. These standard microbiological results were then compared to electrochemical results obtained with the new method, and were used to build up a correlation database. Predictive algorithms were created to determine susceptibility results, based on the database values.
Results: Classification of the bacteria as susceptible, resistant or intermediate was possible based on the new respiratory activity measurements. Preliminary results (n = 2,000) using simple discriminate models predicted either susceptibility or resistance with greater than 80% categorical agreement between the biamperometric respiratory activity method and the disk-diffusion method for a range of 25 bacteria to 30 antibiotics from several classes having different modes of action. The scientific basis for the new respiratory measurements and the results obtained with the new respiratory activity–disk diffusion correlation database will be presented along with the statistical methods for breakpoint susceptibility determination.
Conclusions: Antibiotic breakpoint testing based on respiratory activity, measured electrochemically following exposure of bacteria to an antibiotic, allowed classification with greater than 80% categorical agreement. Further development is underway to examine statistical methods that will improve classification accuracy.
P1650 Use of clustering techniques for the prediction of microbiologic breakpoints, in well established bacteria-antibiotic pairs and in the Acinetobacter spp.–tigecycline combination
F. Fernandez, D. Curcio, M. Galas (Capital Federal, AR)
Objectives: Evaluate the potential of data-mining clustering techniques in the detection of strains with different behaviour with respect to its susceptibility measured by size of inhibition zone (SIZ), and in the prediction of microbiologic breakpoints. Secondarily, we explore its application to set a proposed breakpoint for Acinetobacter spp.–Tigecycline (ASP-TIG).
Methods: We used 5 datasets, 4 containing the values of SIZ of bacteria-antibiotic pairs with well-defined breakpoints, Escherichia coli-ciprofloxacin (ECO-CIP; n = 19,857).
Staphylococcus aureus-oxacillin (SAU-OXA; n = 7,006), enterococciampicillin (ENT-AMP; n = 2417), Klebsiella pneumoniae-ceftazidime (KPN-CAZ; n = 3,064) and 1 with no established breakpoint, ASP-TIG (n = 336).
K-means algorithm was used to divide subpopulations and a normal probability distribution was fitted to each one. The 99% probability inferior limit of the subpopulation with greater values was selected to predict susceptibility breakpoint. First, we had evaluated that established breakpoints was near that limit in the distribution of susceptible bacteria-antibiotic pairs.
Results: We could obtain 2 clusters clearly defined in the 5 subsets. Values of mean, standard deviation (SD), and predicted breakpoints for susceptible subpopulations sharply corresponded to the values of subpopulations defined by well-established breakpoints, except for KPN-CAZ (Table 1 ). For ASP-TIG we show predicted values since actual ones are not available.
| Subpopulation | Pair | Value type | Mean | SD | Breakpoint |
|---|---|---|---|---|---|
| Susceptible | SAU-OXA | Predicted | 20.6132 | 3.2112 | 12.34 |
| Actual | 20.5380 | 3.2834 | ≥13 | ||
| ECO-CIP | Predicted | 32.6523 | 4.0805 | 22.14 | |
| Actual | 32.6520 | 4.0806 | ≥21 | ||
| ENT-AMP | Predicted | 26.8536 | 3.7885 | 17.09 | |
| Actual | 26.8540 | 3.7894 | ≥17 | ||
| KPN-CAZ | Predicted | 28.6576 | 2.4451 | 22.35 | |
| Actual | 26.3330 | 4.3723 | ≥18 | ||
| ASP-TIG | Predicted | 21.5549 | 1.8532 | 16.78 | |
| Actual | NAa | NA | NA | ||
| Resistant | SAU-OXA | Predicted | 6.3353 | 1.2834 | 9.64 |
| Actual | 6.1299 | 0.6584 | ≤10 | ||
| ECO-CIP | Predicted | 7.6686 | 3.5242 | 16.75 | |
| Actual | 6.9649 | 2.1790 | ≤15 | ||
| ENT-AMP | Predicted | 6.2493 | 1.3373 | 9.69 | |
| Actual | 6.2493 | 1.3391 | ≤16 | ||
| KPN-CAZ | Predicted | 16.2532 | 7.6563 | 35.97 | |
| Actual | 8.4537 | 2.8475 | ≤14 | ||
| ASP-TIG | Predicted | 17.8589 | 1.3420 | 21.32 | |
| Actual | NA | NA | NA |
NA, not available.
Conclusion: By this method we could discriminate susceptible from resistant subpopulations when the histogram of SIZ shows a normal shape by visual inspection. When the shape is not normal, predicted values were far from actual ones, as in the case of KPN-CAZ. In the case of ASP-TIG pair, the histogram has normal shape, and the predicted value was between 16–17.
In vitro susceptibility of Gram-negatives/ESBLs/anaerobes
P1651 Determination of PK/PD parameters of moxifloxacin against Bacteroides thetaiotaomicron in vitro at static and at varying drug concentrations
A. Mayer-Bartschmidt, J. Schuhmacher, S. Dörlemann, M. Haas (Wuppertal, DE)
Objectives: Moxifloxacin (MXF) is the only marketed fluoroquinolone approved by the FDA as monotherapy for complicated intra-abdominal infections. This study evaluated the antibacterial effect of MXF against B. thetaiotaomicron, an obligate anaerobic bacterium implicated in intraabdominal abscesses. The PD parameters determined from in vitro data were used to simulate the clinical therapeutic effect.
Methods: Time-kill curves with B. thetaiotaomicron ATCC 29741 were performed with a range of static concentrations between 0.25 and 20 μg/mL of MXF under anaerobic conditions. Using a sigmoidal Emax model, the PD parameters (maximal kill rate, EC50 value, Hill slope) as well as derived parameters e.g. static concentration were calculated. In addition, PD parameters were determined at varying drug concentrations by using an in vitro one-compartment PK model. Starting concentrations ranged between 0.25 and 4 μg/mL, t 1/2 was set to 13 h. Simulation of number of colony-forming units (CFU) vs time-course in man following MXF 400 mg IV was performed using the PD parameters as determined in vitro and the known PK parameters in man.
Results: MXF showed an MIC of 2 μg/mL against B. thetaiotaomicron ATCC 29741 representing the susceptible breakpoint recommended by CSLI. PD parameters for MXF derived from kill curves at static drug concentrations or from a one-compartment model at varying drug concentrations are summarised in the table . Simulation of the CFU vs time course in humans after MXF 400 mg IV predicts clinical success against B. thetaiotaomicron ATCC 29741: an approximate 1,000-fold reduction in CFU is expected after 13 h.
| PD parameter | One-compartment |
Kill curves |
||
|---|---|---|---|---|
| Estimate | CV (%) | Estimate | CV (%) | |
| k0 (h−1) | 0.781 | 4.54 | 0.84 | 9.74 |
| zmax (CFU/mL) | 2.66×109 | 37.1 | 2.4×109 | 10.8 |
| kmax (h−1) | 1.81 | 3.96 | 2.48 | 7.28 |
| EC50 (μg/mL) | 0.561 | 3.52 | 1.10 | 20.5 |
| S | 2.74 | 9.98 | 1.19 | 20.5 |
| Static concentration (μg/mL) | 0.507 | n.c. | 0.626 | n.c. |
Conclusions: MXF is highly active against B. thetaiotaomicron ATCC 29741. PK/PD simulations indicate likely clinical efficacy in man.
The MXF susceptible CSLI breakpoint of 2 μg/mL for anaerobes is supported by this study.
Research funding: Bayer HealthCare Pharmaceuticals
P1652 Susceptibility of extended-spectrum β-lactamase-producing Escherichia coli strains causing nosocomially- and community-acquired bacteraemia
E. Picon, J. Rodriguez-Bano, P. Gijon, J.R. Hernandez, M. Ruiz, C. Pena, B. Almirante, M. Almela, R. Canton, A. Guerrero, M. Gimenez, A. Oliver, J. Horcajada, G. Navarro, A. Coloma, A. Pascual on behalf of the Red Española de Investigacion en Patologia Infecciosa (REIPI)
Background: Community and nosocomially acquired bacteraemias caused by extended-spectrum β-lactamase-producing Escherichia coli (ESBL-Ec) are increasing in Spain. Therapeutic options for these infections are scarce.
Methods: Prospective cohort study including all episodes of bacteraemia due to ESBL-Ec from October/04 to January/06 in 13 hospitals from 6 areas in Spain. E. coli isolates were identified with API20E and ESBL production was confirmed according to CLSI guidelines. Susceptibility to 18 antimicrobial agents including ceftazidime, cefotaxime, cefepime, piperacillin-tazobactam, amoxicillin-clavulanate, ticarcillin-clavulanate, imipenem, meropenem, ertapenem, ciprofloxacin, nalidixic acid, amikacin, gentamicin, tobramycin, trimethoprim-sulfamethoxazole, fosfomycin, and tigecycline was determined by broth microdilution following CLSI recommendations. For tigecycline EUCAST guidelines were followed.
Results: We included 190 episodes of bacteraemia; 49.5% were nosocomially-acquired and 50.5% were community-acquired. No differences were found in the MIC and clinical category distribution between community and nosocomial isolates. All the strains were susceptible to the three carbapenems studied (imipenem, meropenem and ertapenem) and tigecycline. The MIC50 and MIC90 of tygecycline were 0.06 mg/L. Among β-lactam/β-lactamase inhibitor combinations piperacillin-tazobactam was the most active agent with 92% susceptible strains, followed by amoxicillin-clavulanate and ticarcillin-clavulanate (61% and 18% respectively). Aminoglycosides activity was as follows: amikacin 98%, tobramycin 82% and gentamicin 80% of susceptible strains, respectively. Ciprofloxacin and trimethoprim-sulfamethoxazole were the less active agents with 33% and 39% of susceptible strains, respectively.
Conclusions: ESBL-Ec is a relevant cause of community and nosocomially acquired bacteraemia in Spain. The most active agents against ESBL-Ec were carbapenems and tigecycline with 100% of susceptible strains. No difference in susceptibility to antimicrobial agents were found between nosocomial or community acquired isolates.
P1653 Determination of the minimum inhibitory and mutant prevention concentration of tigecycline against clinical isolates of E. coli and Klebsiella species
J. Blondeau, S. Borsos, C. Hesje (Saskatoon, CA)
Objectives: Tigecycline is the first of the glycylcycline class of antimicrobials and has reported in vitro activity against E. coli and Klebsiella spp. The mutant prevention concentration (MPC) defines the antimicrobial drug concentration threshold that block the growth of resistant bacterial subpopulations that may be present in high density bacterial populations such as those causing infection. We determined MIC and MPC values for tigecycline against clinical isolates of E. coli and Klebsiella spp.
Methods: Minimum inhibitory concentration (MIC) was in accordance with the recommended Clinical and Laboratory Standards Institute procedure by microbroth dilution using 105 cfu/mL tested against doubling drug dilutions in appropriate media. For MPC testing 1010 CFUs were added to drug containing agar plates and incubated in ambient temperature and atmosphere. The lowest drug concentration preventing growth was recorded as the MIC or MPC depending on method.
Results: For 24 E. coli clinical isolates, MIC50 (mg/L), MIC90 (mg/L) and MIC (mg/L) range values were 0.031, 0.063, 0.031–0.125 with 96% of strains having MICS ≤0.063; MPC50 (mg/L), MPC90 (mg/L) and MPC (mg/L) range values were 0.5, 1, 0.05–1 with 96% of strains having MPCs ≤ 1. For 24 Klebsiella spp. clinical isolates, MIC50, MIC90 and MIC range values were 0.125, 0.5, ≤ 0.016–1 with 96% of strains having MICs ≤ 0.5; MPC50, MPC90 and MPC range values were 8, 8, 2–8.
Conclusion: Tigecycline was active in vitro against E. coli and Klebsiella spp. isolates having MIC90 values of 0.063 and 0.5 mg/L respectively. MPC90 values were 8-fold lower for E. coli (1 mg/L) than for Klebsiella spp. (8 mg/L), however, therapeutic drug concentrations exceed both of these values. Tigecycline appears to have a low propensity to select for resistance for the organisms studied.
P1654 Determination of the minimum inhibitory and mutant prevention concentrations of moxifloxacin against clinical isolates of E. coli and Klebsiella spp
J. Blondeau, S. Borsos, C. Hesje (Saskatoon, CA)
Objective: The fluoroquinolone (FQ) moxifloxacin has in vitro potency against Gram-negative bacilli – particularly the Enterobacteriaceae. The mutant prevention concentration (MPC) defines the antimicrobial drug concentration threshold that would require an organism to simultaneously acquire two resistance mutations for growth in the presence of a FQ. We measured the minimum inhibitory concentration (MIC) and MPC values of moxifloxacin against clinical isolates of E. coli and Klebsiella spp.
Method: MIC was determined in accordance with the recommended CLSI procedure by microbroth dilution using 105 CFU/mL tested against doubling drug dilutions in appropriate media. For MPC testing 1010 CFU were added to drug-containing agar plates and incubated at ambient temperature and atmosphere. The lowest drug concentration preventing growth was recorded as the MIC or MPC depending on the method used.
Results: Data are shown in the Table . For E. coli, 91% of strains had MIC values ≤0.031 and 86% of strains had MPC values ≤0.5. For Klebsiella spp., 96% of strains had MIC values ≤0.25 mg/L and 92% of strains had MPC values ≤ 1 mg/L.
| MIC (mg/L) |
MPC (mg/L) |
|||||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MPC50 | MPC90 | Range | |
| E. coli (n = 23) | 0.016 | 0.031 | ≤0.008–2 | 0.12 | 1 | 0.12–2 |
| Klebsiella spp. (n = 24) | 0.031 | 0.12 | 0.016–2 | 1 | 1 | 0.25–2 |
Conclusion: Moxifloxacin is highly active in vitro against E. coli and Klebsiella spp. with MIC90 values of 0.031 and 0.12 mg/L respectively. By MPC testing, MPC90 values were 1 mg/L – a value that is below achievable serum drug concentration over the duration of the dosing interval. Moxifloxacin shows low propensity to select for resistance in these organisms based on MPC measurements.
P1655 Time trends in odds ratios of antimicrobial susceptibility in nosocomial and non-nosocomial Gram-negative infections in Israel
A. Zalounina, M. Paul, S. Andreassen, L. Leibovici (Aalborg, DK; Petah Tikva, IL)
Objectives: To study the changes in difference between antimicrobial susceptibilities in hospital environment and community over time.
Methods: Clinically significant, patient and episode unique, bloodstream isolates (n = 5,790) were collected prospectively from adults between 1988–2004 at Rabin Medical Center, Israel. Bacteraemia was defined as nosocomial if it occurred 48 h or more following hospital admission. We analysed in-vitro susceptibilities of Escherichia coli, Klebsiella spp., Pseudomonas spp. and Acinetobacter spp. For each pathogen and antibiotic we calculated the Odds Ratios (OR) with 95% confidence intervals (CI) for susceptibility comparing non-nosocomial vs. nosocomial isolates. ORs were plotted in blocks of 3 years. Heterogeneity of the ORs across time was assessed by the Breslow-Day test.

Results: Nosocomial infections comprised 47% of all cases. The results for the most prevalent pathogen E. coli (44%) and 5 antibiotics are shown in the chart below. The highest OR for non-nosocomial E. coli being more susceptible than nosocomial was obtained for ceftazidime during 1997–1999 (9.9, CI: 3.4–29.0). We observed 2 main patterns of ORs changes over time. In the first, ORs increased over time followed by a decrease (observed for E. coli and all antibiotics except quinolones; Acinetobacter and amikacin; Klebsiella spp. and ceftriaxone or aminoglycosides). In the second pattern, ORs decreased with time (observed for Pseudomonas and piperacillin; Acinetobacter and imipenem). ORs for quinolones fluctuated over time. The test on heterogeneity disclosed significant changes in ORs over time for E. coli with ceftazidime and ceftriaxone, Pseudomonas spp. and ciprofloxacin (p < 0.05).
Conclusion: For the majority of the pathogen/antibiotic combinations the OR starts relatively low, indicating low resistance both in community and in the hospital, then rises due to the appearance of resistance in the hospital and finally descends again, indicating diffusion of resistance from the hospital to the community.
P1656 Susceptibility of Acinetobacter spp. to antimicrobial agents with respect to their susceptibility to carbapenems
M. Sredkova, S. Mihaylova, V. Edreva, S. Pachkova, V. Grigorova, H. Hitkova, V. Popova, K. Dragoev (Pleven, BG)
Objective: To compare susceptibility of carbapenem-resistant and carbapenem-susceptible isolates of genus Acinetobacter to nine antimicrobial agents.
Methods: Four hundred and eighty-five clinical isolates of genus Acinetobacter were tested. The in vitro activities of ampicillin/sulbactam, azlocillin, piperacillin, ceftazidime, cefepime, imipenem, meropenem, gentamicin, tobramycin, amikacin and ciprofloxacin were determined by the disk diffusion method according to guidelines of the CLSI. In order to be included in the study, Acinetobacter strains had to meet the following criteria: to be isolated during the period 2002–2006, to be the first isolate from a patient and to be either susceptible or resistant to both imipenem and meropenem. Student's t test was used to compare groups.
Results: Carbapenem-resistant Acinetobacter spp. were more resistant to all antibiotics tested in comparison to carbapenem-susceptible isolates (p < 0.05). In general, A. baumannii showed superior resistance than A. non-baumannii. The difference was significant between carbapenem-resistant A. baumannii and carbapenem-resistant A. non-baumannii in case of susceptibility to ceftazidime, cefepime, gentamicin, tobramycin and ciprofloxacin (p < 0.05). Carbapenem-susceptible strains of A. non-baumannii possessed greater susceptibilities to azlocillin, piperacillin, ceftazidime, cefepime, gentamicin, tobramycin, amikacin and ciprofloxacin than did strains of carbapenem-susceptible A. baumannii (p < 0.05). Ampicillin/sulbactam was the only agent towards which carbapenem-susceptible isolates of A. baumannii were resistant in lower percentage than carbapenem-susceptible isolates of A. non-baumannii (p < 0.05).
Conclusion: Carbapenem-resistant Acinetobacter spp. tend to exhibit multidrug resistance. Beta-lactams, aminoglycosides and fluoroquinolones demonstrate different activity to carbapenem-susceptible Acinetobacter spp., with respect to their species affiliation.
P1657 Antimicrobial susceptibility of Peptostreptococcus anaerobius and Peptostreptococcus stomatis
E. Könönen, P. Niemi, A. Kanervo-Nordström (Helsinki, Kotka, FI)
Objective: Peptostreptococcus anaerobius ‘sensu lato’ currently includes two closely related species, P. anaerobius and P. stomatis. In the present study, we tested their in vitro antimicrobial susceptibilities to eight antimicrobials potentially used to treat anaerobic infections.
Methods: Sixty-five Gram-positive anaerobic coccal isolates sensitive to SPS (inhibition zone around the disk >12 mm) were available from various human sources. Thirty isolates were confirmed as P. anaerobius and 32 isolates re-identified as P. stomatis based on phenotypic differential characteristics described by Downes and Wade (Int J Syst Evol Syst Microbiol 2006;56:751–4), whereas the identification of three isolates remained uncertain. The susceptibilities were tested by Etest (AB Biodisk) and drugs included amoxicillin (AC), amoxicillin/clavulanic acid (XL), cefoxitin (FX), ertapenem (ETP), azithromycin (AZ), clindamycin (CM), metronidazole (MZ), and moxifloxacin (MX). Beta-lactamase production was tested using nitrocefin (AB Biodisk).
Results: In general, the drugs exhibited good activities against most isolates. However, P. anaerobius had constantly higher MIC50 and MIC90 values (mg/L) than P. STOMATIS: 0.125 and 0.38 vs. <0.016 and 0.064 to AC, 0.125 and 0.5 vs. <0.016 and 0.094 to XL, 0.38 and 0.75 vs. 0.064 and 0.38 to FX, 0.25 and 0.38 vs. 0.047 and 0.19 to ETP, 0.5 and 1.0 vs. 0.125 and 0.75 to AZ, 0.19 and 0.75 vs. <0.016 and 0.016 to CM, 0.094 and 0.19 vs. <0.016 and 0.047 to MZ, and 0.125 and 0.125 vs. 0.094 and 0.125 to MX, respectively. None of the isolates produced β-lactamases. Three (10%) P. anaerobius isolates were resistant to several drugs.
Conclusion: The possibility of antimicrobial resistance should be taken into account when treating infections with the involvement of P. anaerobius ‘sensu stricto'.
P1658 Occurrence and mechanisms of resistance to β-lactam antibiotics in multiresistant Acinetobacter spp. clinical isolates
E. Sodomova, D. Michalkova-Papajova, B. Gottwaldova, D. Rovna (Bratislava, SK)
Objectives: The aim of the study was to determine the occurrence and mechanisms of resistance to β-lactam antibiotics in multiresistant clinical isolates of the genus Acinetobacter.
Methods: Clinical isolates included in the study (n = 111) were obtained from University Hospital Ruzinov, Bratislava (Slovakia) and identified by NEFERMtest24 (Pliva-Lachema, Czech Republic). Resistance to antimicrobial agents was determined by standard disk diffusion method according to NCCLS. The production of ESBL was detected by combination disk method with disk combinations: ceftazidime (30 μg) and ceftazidime-clavulanic acid (30/10 μg), cefotaxime (30 μg) and cefotaxime-clavulanic acid (30/10 μg), cefepime (30 μg) and cefepime-clavulanic acid (30/10 μg). The presence of the genes coding for TEM- and SHV-type β-lactamases was determined by PCR.
Results: In the set of 111 Acinetobacter spp. isolates 100.0% were resistant to mezlocillin, 81.1% to ticarcillin, 99.1% to piperacillin, 99.1% to carbenicillin, 35.1% to ampicillin-sulbactam, 55.9% to piperacillin-tazobactam, 64.0% to ticarcillin-clavulanic acid, 81.1% to ceftazidime, 47.7% to cefepime, 100.0% to cefoperazone, 91.9% to cefotaxime, 91.0% to ceftriaxone, 94.6% to ceftizoxime, 91.9% to moxalactam, 0.0% to imipenem, 20.7% to meropenem, 73.9% to aztreonam, 96.4% to gentamicin, 55.0% to amikacin, 15.3% to tobramycin, 20.7% to netilmicin, 96.4% to tetracycline, 72.1% to doxycycline, 74.8% to minocycline, 98.2% to ciprofloxacin, 91.9% to levofloxacin, 99.1% to lomefloxacin, 99.1% to norfloxacin, 80.2% to ofloxacin, 92.8% to gatifloxacin, 99.1% to chloramphenicol, 77.5% to trimethoprim-sulfamethoxazole and 90.1% to sulfonamides. In the presence of cloxacillin (inhibitor of class C cephalosporinases, 200 μg/mL) at least 5 mm increase in a zone diameter in 67.6% of the isolates for ceftazidime, 82.9% for cefotaxime and 17.1% for cefepime was observed. Only one isolate (0.9%) was detected as ESBL-producer. The presence of the genes coding for TEM-type β-lactamases was determined in all clinical isolates tested, but the presence of the genes coding for SHV-type β-lactamases was determined only in two clinical isolates (1.8%).
Conclusion: Most of the Acinetobacter spp. clinical isolates (91.0%) were resistant to 20 or more from 33 antimicrobial agents tested. Only imipenem was effective to all isolates. The production of β-lactamases contributed to frequent occurrence of resistance to β-lactam antibiotics.
P1659 Resistance to a wide spectrum of antimicrobial agents in ESBL-producing Klebsiella pneumoniae clinical isolates
S. Resetarova, D. Michalkova-Papajova, E. Sodomova, B. Gottwaldova, D. Rovna (Bratislava, SK)
Objectives: The purpose of the presented study was to determine the occurrence of in vitro resistance to 37 antimicrobial agents in the set of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae clinical isolates obtained from University Hospital Ruzinov, Bratislava (Slovak Republic) during the years 2004–2006.
Methods: Clinical isolates included in the study (n = 47) were identified by ENTEROtest24 (Pliva-Lachema, Czech Republic). The production of extended-spectrum β-lactamases was verified by combination disk method (NCCLS 2003) with following disk combinations: ceftazidime (30 μg) and ceftazidime-clavulanic acid (30/10 μg), cefotaxime (30 μg) and cefotaxime-clavulanic acid (30/10 μg). Resistance to selected antimicrobial agents was determined by standard disk diffusion method according to the NCCLS recommendations.
Results: All of the 47 ESBL-producing Klebsiella pneumoniae clinical isolates were in vitro resistant to ampicillin, mezlocillin, piperacillin, carbenicillin and minocycline, 93.6% of the isolates were in vitro resistant to amoxicillin-clavulanic acid, 85.1% to ampicillin-sulbactam, 2.1% to piperacillin-tazobactam, 93.6% to ticarcillin-clavulanic acid, 97.9% to cefazolin, 38.3% to cefepime, 89.4% to cefoperazone, 6.4% to cefoxitin, 83.0% to cefotaxime, 10.6% to ceftizoxime, 85.1% to ceftriaxone, 55.3% to ceftazidime, 97.9% to cefpodoxime, 0.0% to imipenem, 2.1% to meropenem, 83.0% to aztreonam, 78.7% to gentamicin, 0.0% to amikacin, 14.9% to netilmicin, 97.9% to tobramycin, 89.4% to tetracycline, 31.9% to doxycycline, 85.1% to ciprofloxacin, 68.1% to levofloxacin, 23.4% to gatifloxacin, 95.7% to lomefloxacin, 87.2% to ofloxacin, 51.1% to trimethoprim-sulfamethoxazole, 40.4% to sulfonamides, 51.1% to trimethoprim, 31.9% to chloramphenicol and 80.9% to nitrofurantoin.
Conclusion: The production of extended-spectrum β-lactamases in Klebsiella pneumoniae is often associated with resistance to other antibiotics. In the presented study, 87.2% of the isolates were resistant to 20 or more from 37 antimicrobial agents tested. Most of the isolates (93.6%) were simultaneously resistant to some of the β-lactams, aminoglycosides and fluoroquinolones. Imipenem and amikacin were the only antibiotics effective to all ESBL-producing Klebsiella pneumoniae.
P1660 The activity of mecillinam vs Enterobacteriaceae resistant to 3rd-generation cephalosporins in Bristol, UK
G. Weston, K. Bowker, A. Noel, A. MacGowan, R. Howe (Bristol, Cardiff, UK)
Objectives: Resistance in coliforms to 3rd generation cephalosporins (3GC) is an increasing problem both in hospitals and the community. Oral options for the treatment of these organisms is often limited due to resistance to multiple antimicrobial classes. Mecillinam, an amidinopenicillin that is available in Europe as the oral pro-drug pivmecillinam, is stable to many β-lactamases. We aim to establish the activity of mecillinam against Enterobacteriaceae resistant to 3GC.
Methods: Coliforms isolated from urine samples routinely submitted to North Bristol NHS Trust were screened for resistance to 3GC by disc testing with cefpodoxime on Isosensitest agar. Resistant isolates were identified using the BBL Crystal ID system. Further susceptibility testing was performed by agar dilution MIC using an inoculum of 104 cfu/spot on Mueller-Hinton agar. Phenotypic characterisation of 3GC resistance was determined from MICs for cefotaxime (CTX) and Ceftazidime (CAZ) with and without clavulanate at a fixed concentration of 2 mg/L.
Results: 127 isolates were identified by screening of which 123 were confirmed as resistant to either CTX or CAZ by BSAC criteria. The majority of 3GC-resistant strains were E. coli 74/123 (60.2%), followed by Enterobacter spp. 16.2%, Klebsiella spp. 12.2%, and others 11.4%. Overall 98.4% of 3GC-resistant coliforms tested sensitive to mecillinam (BSAC breakpoint S≤64 mg/L). Of 30 strains that were multiply-resistant to 3GC, trimethoprim, nitrofurantoin, gentamicin, and ciprofloxacin, 96.7% tested sensitive to mecillinam. As shown in the figure , all 67 ESBL-producing E. coli tested sensitive to mecillinam with a 4-fold or greater decrease in MIC in the presence of clavulanate in most cases.

MIC of mecillinam +/− clavulanate for ESBL-producing E. coli.
Conclusions: Mecillinam maintains good activity against Enterobacteriaceae resistant to 3GC. This includes prevalent strains of ESBL-producing E. coli. Oral formulations of mecillinam should be considered as possible agents for the treatment of uncomplicated urinary tract infections caused by such organisms.
P1661 Beta-lactamase production and antibiotic susceptibility testing of anaerobic Gram-negative bacilli isolated from oral and maxillofacial infections
G. Bancescu, A. Bancescu, S. Dumitriu, E. Preoteasa, M. Pana, M. Andrei (Bucharest, RO)
Objectives: The aim of the present study was to identify at species level and to test in vitro the susceptibility to antibiotics of the anaerobic Gram-negative bacilli strains isolated from 60 pus samples collected by needle aspiration from Rumanian patients with different oral and maxillofacial infections.
Methods: Microscopy of Gram-stained direct smears and cultures on selective and non-selective media incubated both aerobically and anaerobically, were performed in each pus specimen. The strictly anaerobic bacteria were identified to genus and species level using the conventional methods of diagnosis and the Rapid ID 32 A system (bioMérieux, Marcy-l'Etoile, France). The anaerobic Gram-negative rods isolates were tested for β-lactamase production by nitrocefin disk. The susceptibility of these strains was tested against: metronidazole, penicillin G, ampicillin, amoxicillin/clavulanic acid and clindamycin by disk diffusion method and E-test (Ab Biodisk, Solna, Sweden).
Results: The Gram-negative bacilli isolates represented 73% of the 64 strictly anaerobic bacteria strains and belonged to Fusobacterium nucleatum and different species of Prevotella and Bacteroides. The Prevotella isolates were the most frequently identified anaerobic Gram-negative bacilli (31 strains), with P. melaninogenica predominating. Correlating the direct microscopy with the culture results, Prevotella was found to be the only bacteria involved in the aetiology of two cases of submandibular space abscess. All anaerobic Gram-negative bacilli isolates were susceptible to metronidazole, clindamycin and amoxicillin/clavulanic acid. The β-lactamase production was detected among the Prevotella isolates (13%), while the other Gram-negative bacilli strains were susceptible to all tested β-lactam antibiotics.
Conclusion: The most frequently isolated anaerobic species was P. melaninogenica. As antibiotics are often prescribed empirically in the treatment of acute oral and maxillofacial infections – which are usually mixed infections, involving anaerobic bacteria that may be β-lactamase producers – it is recommended to choose an association of a penicillin with a β-lactamase inhibitor. Clindamycin might be a good alternative, especially in patients allergic to penicillin. Monitoring of local susceptibility patterns of anaerobic bacteria is necessary for antibiotic selection in the initial therapy.
P1662 Emerging colimycin resistance in Gram-negative strains isolated from intensive care unit patients
F. V. Kontopidou, A. Antoniadou, G. Poulakou, E.D. Papadomichelakis, I. Galani, A. Armaganidis, H. Giamarellou (Athens, GR)
Background: Infections due to multidrug resistant (MDR) Gram(–) microorganisms in the ICUs has prompted the use of Colimycin (COL) an antibiotic forgotten for decades. The mechanism of COL resistance (COLR) has not been well defined. Until now resistance to COL is uncommon. While emergence of COL R in vivo, has not yet been reported.
Patients and Methods: The study was performed in a new 6-bed general ICU from November 2003 to October 2006. Empirical antimicrobial treatment was guided by weekly active surveillance of patients floras (faecal and respiratory). Resistance of the isolated pathogens prompted COL use as part of empirical antimicrobial regimens or in microbiologically documented infections. COLR was defined as <14 mm diameter by the disk diffusion method and was confirmed as an MIC > 4 mg/mL by the agar dilution and E test method (according to BSAC).
Results: Among 146 patients with more than 10 days hospitalisation, 72% was colonised by A. baumannii, 61% by P. aeruginosa, 50% by K. pneumoniae, 17% by Enterobacter spp., 28% by E. coli, 27% by S. maltophilia. The resistant strains to COL were 3.8%, 4.5%, 32.4%, 4%, 7.3%, 35% respectively. Using REP-PCR six distinct clones were identified among K. pneumoniae Col resistant strains. Most of K. pneumoniae strains were pan-drug resistant, produsing ESBL and MBL susceptible only to tetracyclines or and gentamycin. The majority represented colonisation; only 7 cases of infection were recorded (six with K. pneumoniae and one with Enterobacter sp), all of them with fatal outcome. 83 patients had exposure to colistin, 29 of them harbouring a colistin resistant Gram(–) strain. All patients treated prolonged (>20 d) with Col developed normal floras colonisation with K. pneumoniae, P. aeruginosa, S. maltophilia R to Col plus of intrinsically resistant to Col enterobacteriaceae (ie. Pr. mirabilis, M. morgani, Serratia spp., and Providencia spp.). Colonisation with more than one Col resistant pathogen correlated with length of stay, severity of illness and a significant exposure to colistin.
Conclusions: Selective pressure due to extensive COL use seems to lead to the emergence of COL resistance among Gram(–) strains, jeopardising treatment choices in the ICU, increasing morbidity and mortality. Therefore it is urgent intensivists to avoid unecessar or/and prolonged therapy with colistin.
P1663 Baseline profile of RO4908463 (CS-023) against recent isolates of target Gram-negative pathogens exhibiting β-lactam-resistant phenotypes from Europe, 2003–2006
N. Brown, D. Draghi, M. Jones, D. Sahm, B. Dannemann (Herndon, US; Breda, NL; Nutley, US)
Objectives: RO4908463 (CS-023), a carbapenem with a unique guanidine-pyrrolidine side chain, has broad-spectrum activity against both Gram-positive and Gram-negative pathogens. As such, this study was done to establish an in vitro activity baseline of RO4908463 against Europe (EU) isolates of P. aeruginosa (PA), Acinetobacter spp. (AS), Enterobacteriaceae (EN), and H. influenzae (HI), including those exhibiting resistance to currently available β-lactams.
Methods: RO4908463 and comparator agents were tested by broth microdilution (CLSI; M7-A7, 2006) against 735 Gram-negative isolates selected to include organisms resistant to β-lactams. Isolates were collected (2003–2006) from hospitals in 18 European countries and included ceftazidime-resistant EN and PA, imipenem-resistant PA, and β-lactamase positive HI.
Results: For ceftazidime-susceptible (n = 83) and ceftazidime non-susceptible (NS; n = 23) PA isolates, the RO4908463 MIC range/MIC90 (mg/L) was 0.06–16/2 and 0.25–>32/32, respectively. Among imipenem-S AS (n = 45) and imipenem-NS AS (n = 8) the MIC ranges were 0.12–16 and 4–>32 mg/L, respectively. The MIC90 for all EN species combined (n = 523) was 0.12 mg/L. For ceftazidime NS EN (n = 90) the MIC range was ≤0.015 to 1 mg/L. The MIC range for HI (n = 53) was ≤0.008 to 0.5 mg/L with an MIC90 of 0.12 mg/L for both β-lactamase-positive and -negative strains combined.
Conclusion: RO4908463 demonstrated good in vitro activity against a wide spectrum of Gram-negative pathogens, including Enterobacteriaceae with resistance to ceftazidime. These baseline data will provide an important reference for detecting any changes in the activity of RO4908463 among target Gram-negative pathogens from European countries as the clinical development proceeds.
P1664 Baseline surveillance profile of doripenem against key Gram-negative pathogens encountered in Europe
M.K. Aranza, D.C. Draghi, M.E. Jones, C. Thornsberry, D.F. Sahm (Herndon, US; Breda, NL)
Background: DOR, is an investigational parenteral broad-spectrum 1-β-methyl carbapenem that is refractory to hydrolysis by most Gram-negative β-lactamases. It is under clinical development for complicated urinary tract infections, complicated intra-abdominal infections, and hospital acquired pneumonia (including ventilator-associated pneumonia). As a key component of the development programme, this surveillance initiative was done to establish baseline data on DOR activity against recent Gram-negative pathogens relative to other key β-lactam agents.
Methods: During 2005–2006, Citrobacter spp. (CP; n = 387), E. cloacae (EA; n = 406), E. coli (EC;n = 1213), K. pneumoniae (KP;n = 854), P. mirabilis (PM;n = 443), S. marcescens (SM; n = 291), P. aeruginosa (PA;n = 621), and Acinetobacter spp. (AC;n = 278) collected from 31 institutions in 12 European countries were centrally tested by broth microdilution (CLSI; M7-A6). Testing included DOR, imipenem (IMP), ceftazidime (CAZ), cefepime (FEP), piperacillin-tazobactam (PTZ) and other relevant agents.
Results: See the table .
| Orga | MICb (mg/L) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DOR |
IMP |
CAZ |
FEP |
PTZ |
||||||
| Range | M90 | Range | M90 | Range | M90 | Range | M90 | Range | M90 | |
| CP | ≤0.015–8 | 0.06 | 0.06–32 | 2 | ≤0.03–>32 | >32 | ≤0.015–>32 | 1 | ≤0.25–>128 | 64 |
| EA | ≤0.015–1 | 0.12 | ≤0.015–4 | 2 | ≤0.03–>32 | >32 | ≤0.015–>32 | 8 | ≤0.25–>128 | >128 |
| EC | ≤0.015–0.5 | 0.03 | ≤0.015–4 | 0.5 | ≤0.03–>32 | 2 | ≤0.015–>32 | 1 | ≤0.25–>128 | 64 |
| KP | ≤0.015–8 | 0.06 | 0.06–16 | 1 | ≤0.03–>32 | >32 | ≤0.015–32 | >32 | 0.5–128 | >128 |
| PM | ≤0.015–2 | 0.25 | 0.06–32 | 4 | ≤0.03–>32 | 0.12 | ≤0.015–>32 | 0.12 | ≤0.25–128 | 2 |
| SM | ≤0.015–1 | 0.25 | 0.25–4 | 4 | ≤0.03–>32 | 2 | ≤0.015–>32 | 1 | 1–>128 | 64 |
| PA | ≤0.015–>32 | 4 | 0.25–>32 | 32 | 0.06–>32 | >32 | ≤0.015–>32 | 16 | 0.5–>128 | >128 |
| AC | ≤0.015–>32 | 32 | ≤0.015–>32 | >32 | 0.06–>32 | >32 | 0.06–>32 | >32 | ≤0.25–>128 | >128 |
Org, Organism
M90: MIC90.
Conclusions: Based on MIC90s, for every organism group surveyed, including PA, DOR's in vitro activity was comparable or more active than a carbapenem (IMP), the cephalosporins (CAZ, FEP), and a β-lactam (PTZ). These findings establish an important baseline for continued tracking of DOR activity as clinical development continues.
New antimicrobials II
P1665 Mode of action and resistance of the new fluoroquinolone BOL-303224-A
E. Cambau, J.F. Lepage, S. Matrat, X.S. Pan, R. Roth dit Bettoni, C. Corbel, T. Daniel, N. Darchy, A. Aubry, C. Lascols, L.M. Fisher (Créteil, Montpellier, Paris, FR; London, UK)
Objectives: To investigate the mode of action and mechanisms of resistance to the new quinolone BOL-303224-A.
Methods: The new quinolone BOL-303224-A was investigated for (1) its inhibition of DNA gyrases and topoisomerases IV from Streptococcus pneumoniae, taken as the targeted bacterial species, and Escherichia coli, taken as the reference species, and induction of DNA cleavage by these type II topoisomerases, (2) mutations in the type II topoisomerase genes occurring in resistant mutants of S. pneumoniae, S. aureus and E. coli selected in vitro, and (3) activity on gyrase and topoisomerase IV defined mutants. These experiments were conducted in comparison to ciprofloxacin.
Results: At low concentrations, BOL-303224-A inhibited supercoiling activity of DNA gyrases of S. pneumoniae and E. coli (IC50s of 1 mg/L for both species) and decatenation by topoisomerase IV (IC50 of 0.4 and 10 mg/L, respectively), and induced formation of cleavable complexes at concentrations of 2.5 and 1 μM for DNA gyrase and topoisomerase IV of S. pneumoniae and 0.1 and 1.4 μM for those of E. coli, respectively. IC50s of BOL-303224-A were 5- to 16-fold lower than those of ciprofloxacin against pneumococcal topoisomerases. The main mechanism of resistance to SS734 was mutation in the DNA gyrase genes, mostly gyrA mutation at position 81, 83 and 87, but also gyrB mutation at positions 426 and 466 (numbering system used in E. coli). From the results on in vitro selection of resistant mutants, DNA gyrase was the primary target in E. coli, S. aureus and S. pneumoniae, and topoisomerase IV was the secondary target. However, a dual targeting is likely since the selection of resistant mutants of S. aureus could not be obtained in a single step and inhibitory concentrations were in the same range for DNA gyrase and topoisomerase IV of S. pneumoniae. MICs of BOL-303224-A were similarly affected by parC or gyrA mutations in the defined mutants of these two species, and they remained below 1 μg/mL in double gyrA-parC mutants of S. aureus and S. pneumoniae whereas ciprofloxacin MICs were up to >32 mg/L.
Conclusion: BOL-303224-A is a good inhibitor of bacterial type II topoisomerases in vitro, and is one of the rare quinolones to be as active against DNA gyrase as against topoisomerase IV of S. pneumoniae.
P1666 Daptomycin therapy for Gram-positive infections in cancer patients
H. Hogan, I. Raad, K. Rolston (Texas, US)
Objectives: To assess the clinical outcomes of daptomycin in cancer patients.
Methods: Daptomycin is a novel cyclic lipopeptide antibacterial agent approved in the USA for treatment of Staphylococcus aureus bacteraemia and skin and skin structure infections caused by susceptible Gram-positive organisms. Clinical data in cancer patients are limited. Daptomycin usage for documented and/or suspected Gram-positive infections in cancer patients was retrospectively reviewed at a comprehensive cancer centre.
Results: Forty-eight patients, 26 males and 22 females were treated from 1/04 to 5/06. Nineteen (40%) patients received 6 mg/kg/day, 27 patients received 4 mg/kg/day, and 1 patient received 6 mg/kg/Q48h. Twenty-five (52%) had haematological malignancies and 23 (48%) had solid tumours (17 leukaemia, 8 lymphoma, 7 breast cancer were the most common). The average age was 50 years (range 12–74). Seventeen (35%) were neutropaenic. The infections treated were cellulitis (23; 48%), abscess (8; 17%), bacteraemia (11; 23%), UTI (2; 4%), cellulitis + bacteraemia (1; 2%), abscess + bacteraemia (2; 4%) and UTI + bacteraemia (1; 2%). The organisms isolated were S. aureus (18, including 14 MRSA), Enterococcus spp. (10, including 7 VRE), coagulase-negative staphylococci (7) and alpha-haemolytic streptococcus (1). Twenty-three patients (48%) had failed vancomycin (14 cellulitis, 9 bacteraemia), 4 failed linezolid (2 cellulitis, 1 VRE bacteraemia, 1 E. faecalis bacteraemia) and 1 discontinued linezolid to prevent toxicity. All isolates from clinical failures were susceptible to prior antimicrobial regimens. The overall response to daptomycin (42 of 48 patients) was 87.5%. Thirteen of the seventeen neutropaenic patients responded to daptomycin therapy. The mean duration of therapy was 13.5 days. No CPK elevations were observed. One patient developed a rash on day 13 of treatment. Six patients failed to respond to daptomycin (2 leukaemics with cellulitis/abscess that responded to alternative agents; 2 Sweet's Syndrome; 2 died due to pancytopenia/pneumonia/refractory leukaemia).
Conclusions: Daptomycin appears to be a promising agent for the treatment of Gram-positive infections in neutropaenic and non-neutropaenic cancer patients and needs further clinical evaluation in this setting.
P1667 In vitro activity of daptomycin and tigecycline against Staphylococcus epidermidis blood isolates from bone marrow transplant patients
E. Presterl, F. Thalhammer, S. Reichmann, W. Rabitsch, A.M. Hirschl, W. Graninger (Vienna, AT)
Background: Daptomycin (DAP), a new lipopeptide antibiotic, and tigecycline (TGC) have activity against Gram-positive bacteria including methicillin-resistant staphylococci and vancomycin-resistant enterococci. Staphylococcus epidermidis isolated from bone marrow transplant (BMT) recipients were multiresistant due to exposure to multiple antibiotics and protection by biofilm formation on implanted catheters. Moreover, they are significant pathogens in this particularly vulnerable patient population. We present a study on the in-vitro activity of DAP and TGC against S. epidermidis isolates from blood of 74 bone marrow transplant patients.
Methods: 107 non-duplicate blood stream isolates of S. epidermidis were collected in 2000–2006. All patients had the clinical signs and symptoms of sepsis. All isolates were susceptibility (S) tested using CLSI methods against DAP, TGC, vancomycin (VAN) and fosfomycin (FOS).
Results: The MIC50 and MIC90, were 0.125 μg/mL and 0.125 μg/mL for DAP, 0.25 and 0.25 μg/mL for TGC, 1 μg/mL and 1 μg/mL for VAN, and 8 μg/mL and >256 μg/mL for FOS, respectively. The cumulative numbers inhibited at MIC are given in the table . At established CLSI breakpoints for VAN (4 μg/mL) and for TGC (1 μg/mL), 100% and 98%, respectively, of the S. epidermidis blood stream isolates were susceptible. For FOS, 41% of the isolates exhibited a MIC ≥64 μg/mL, considered to be resistant. For DAP, the MICs of all isolates) were ≤1 μg/mL.
Inhibition of S. epidermidis isolates (n = 106)
| Cumulative numbers inhibited at MIC (μg/mL) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.01 | 0.03 | 0.06 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 64 | 128 | ≥256 | |
| Vancomycin | 5 | 9 | 72 | 105 | |||||||||
| Fosfomycin | 1 | 2 | 8 | 20 | 41 | 55 | 56 | 57 | 60 | 63 | |||
| Tigecycline | 1 | 7 | 25 | 48 | 84 | 101 | 105 | ||||||
| Daptomycin | 7 | 10 | 16 | 65 | 103 | 105 | |||||||
Conclusions: Despite considerable antibiotic pressure at a BMT unit S. epidermidis blood isolates are still susceptible to VAN. Caution and resistance testing is warranted with TGC. DAP will be useful for the treatment of S. epidermidis infection in case of hypersensitivity or emergence of resistance against VAN.
P1668 Evaluation of NAD+-dependent DNA ligase of mycobacteria as a potential target for antibiotics
E. Rychta, M. Korycka-Machala, A. Brzostek, H. Sayer, A. Rumijowska-Galewicz, R. Bowater, J. Dziadek (Lodz, PL; Norwich, UK)
Objectives: The aim of this study was verification of the essentiality of NAD+-dependent DNA ligase (LigA) in mycobacteria and its evaluation as a putative target for new antituberculosis drugs.
Methods: A homologous recombination system was used to assess essentiality of the mycobacterial gene for NAD+-dependent DNA ligase (ligA). Conditional mutants carrying a single ligA gene controlled with chemically inducible promoters were constructed. The protein level was measured with western blot using antibodies raised against LigA of Mycobacterium tuberculosis.
Results: Since NAD+-dependent DNA ligase was postulated as a useful target for new antibiotics, we evaluated if ligA is essential for the viability of mycobacteria. Using a homologous recombination system we found that wild type ligA gene cannot be deleted from the chromosome of Mycobacterium smegmatis. We were able to delete the native ligA in M. smegmatis by integrating in to the attB site of chromosomal DNA an extra copy of M. smegmatis or M. tuberculosis ligA, with expression controlled by chemically inducible promoters. The resultant conditional mutants allow studies of the effect of LigA depletion on growth of mycobacteria. Interestingly, the LigA protein from M. smegmatis can be substituted with the NAD+-dependent DNA ligase of Escherichia coli or the ATP-dependent ligase of bacteriophage T4. The investigation of LigA conditional mutants revealed that mycobacterial cells are not affected by large changes to the levels of LigA. The strong overproduction or depletion of LigA did not affect the growth or survival of mycobacteria.
Conclusion: NAD+-dependent DNA ligase is essential for mycobacterial viability, but only low levels of protein are required. Thus, further investigations are required to determine if NAD+-dependent DNA ligase could be useful as an antibiotic target in mycobacteria.
P1669 In vitro activity against Staphylococcus aureus, including MRSA, of soluble copper silicate, a new topical antimicrobial
K.C. Carson, J.G. Bartlett, T-J. Tan, T.V. Riley (Perth, AU)
Objectives: People who are colonised on the skin or in the nares with Staphylococcus aureus have an increased risk of subsequent infection. Topical mupirocin has been used for decolonisation therapy, but with the emergence of both low- and high-level resistance, and increased nasal carriage of community acquired methicillin resistant S. aureus (MRSA), impetus has been given to the search for alternatives. Mankind has used copper as an antimicrobial for thousands of years, and a novel solubilised form of copper silicate (CS) has been developed by Conve plc for use as a topical antimicrobial.
Methods: The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of CS were determined for 200 isolates (100 MRSA and 100 methicillin susceptible S. aureus (MSSA) in Mueller Hinton Broth with 2% NaCl following Clinical and Laboratory Standards Institute methodology. Time-kill studies, post antibiotic effect (PAE) and resistance were also investigated.
Results: All 200 S. aureus isolates had a MIC for CS of 175 mg Cu/L. The MBC range was 175–700 mg Cu/L, with an MBC for 90% of isolates of 350 mg Cu/L. In time-kill assays of MRSA strain 101, CS at a concentration of 700 mg Cu/L gave a 3-log reduction at approx. 30 min. For S. aureus ATCC® 29213, 350 mg Cu/L and 700 mg Cu/L a 3-log reduction was predicted to be achieved in approx. 30 min and ≤1 min, respectively. A PAE was seen in two MRSA isolates after exposure to CS at 350 mg Cu/L (0.99 h and 2.53 h), while with S. aureus ATCC® 29213 a PAE was seen at 175 mg Cu/L (1.65 h) and 350 mg Cu/L (2.10 h). The S. aureus mutation frequency for resistance to CS was <1 per 109. Rifampicin at a concentration of 0.5 mg/L gave frequencies of 5–17×10−8.
Conclusion: CS had similar in vitro antimicrobial activity against all S. aureus isolates. In time kill and PAE assays, MRSA appeared slightly less susceptible than MSSA; however these differences may be due to minor variations in susceptibility between the isolates tested. In S. aureus, there was no evidence of individual cells being resistant, or becoming tolerant, to CS. Clinical trials are needed to determine if CS is an efficacious therapy for decolonisation of and dermatological infections with S. aureus.
P1670 PZ-601 is active against both Gram-positive and Gram-negative bacterial pathogens causative for complicated skin and skin structure infections, and community-acquired pneumonia
J. Pace, T. Stevens, B. Ruth, A. Chua, K. Kanazawa, C. Young, L. Xerri (Malvern, Schaumberg, US; Osaka, JP)
Background: PZ-601 (SMP-601) is an investigational parenteral carbapenem with potent activity against both Gram-positive and Gram-negative pathogens. The projected role for this antibiotic is empirical monotherapy of infections where drug-resistant staphylococci and enterococci or Gram-negative pathogens may be encountered. Indications might include complicated skin and skin structure infections (cSSSI), and community-acquired pneumonia (CAP).
Methods: Numerous Staphylococcus aureus, coagulase-negative staphylococci, Streptococcus pyogenes, Streptococcus agalactiae, Enterococcus faecalis, Escherichia coli, and Klebsiella pneumoniae, as well as strains of Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae were obtained from various U.S. and European hospitals. These bacteria were subjected to susceptibility testing with PZ-601 and comparator antibiotics using broth microdilution minimal inhibitory concentration (MIC) method according to CLSI guidelines.
Results: PZ-601 was highly active against all bacterial species tested. This exploratory carbapenem was most active against Staphylococcus spp., Streptococcus spp., H. influenzae, and M. catarrhalis. Modal MICs of 1 mg/L were observed against the least susceptible species E. faecalis and K. pneumoniae. When bacteria were grouped by susceptibility to antibiotic classes, PZ-601 was substantially more active than imipenem against the methicillin-resistant S. aureus (MRSA) for example with modal MIC values of 2 and >32 mg/L, respectively.
Table 1.
Modal MIC (mg/L) values and range for PZ-601 and comparator antibiotics against bacterial Gram-positive species cumulative for cSSSI and CAP
| Species (no.) | PZ-601 | Vancomycin | Linezolid | Daptomycin | Imipenem |
|---|---|---|---|---|---|
| S. aureus (208) | 0.03 | 1 | 2 | 0.5 | ≤0.06 |
| ≤0.004–4 | ≤0.5–1 | 1–4 | 0.15–1 | ≤0.06–>32 | |
| S. epidermidis (201) | 0.015 | 2 | 1 | 0.5 | ≤0.06 |
| ≤0.004–3 | ≤0.5–2 | 0.25–4 | 0.25–1 | ≤0.06–>32 | |
| S. pyogenes (105) | ≤0.004 | 0.5 | 1 | ≤0.03 | ≤0.06 |
| ≤0.004–0.015 | ≤0.03–0.5 | 0.5–2 | ≤0.03–0.5 | ≤0.015–0.03 | |
| S. agalactiae (98) | 0.015 | 0.5 | 1 | 0.12 | ≤0.015 |
| ≤0.004–0.5 | 0.25–0.5 | ≤0.03–2 | ≤0.03–1 | ≤0.015–1 | |
| E. faecalis (101) | 1 | 1 | 2 | 2 | 1 |
| 0.25–2 | ≤0.5–>32 | 1–4 | 0.5–4 | 0.25–8 | |
| S. pneumoniae (106) | 0.008 | 0.25 | 1 | 0.06 | ≤0.015 |
| ≤0.004–1 | ≤0.03–0.5 | 0.25–2 | ≤0.03–0.25 | ≤0.015–1 |
Table 2.
Modal MIC (mg/L) values and range for PZ-601 and comparator antibiotics against bacterial Gram-negative species cumulative for cSSSI and CAP
| Species (no.) | PZ-601 | Ceftriaxone | Amp/Sulb | Levofloxacin | Imipenem |
|---|---|---|---|---|---|
| H. influenzae (107) | 0.03 | ≤0.03 | ≤0.5 | ND | 0.5 |
| 0.008–0.12 | ≤0.03–0.6 | ≤0.5–8 | 0.08–8 | ||
| M. catharralis (98) | ≤0.004 | ≤0.03 | 4 | ND | 0.06 |
| ≤0.004–0.06 | ≤0.03–4 | ≤0.12–16 | ≤0.008–0.12 | ||
| E. coli (105) | 0.25 | ≤0.25 | >32 | >8 | ≤0.12 |
| 0.06–32 | ≤0.25–>64 | 2–>32 | ≤0.5–>8 | ≤0.12–0.5 | |
| K. pneumoniae (103) | 1 | >64 | >32 | ≤0.5 | ≤0.12 |
| ≤0.015–>64 | ≤0.25–>64 | ≤1–>32 | ≤0.5–>8 | ≤0.12–64 |
Conclusion: PZ-601 is broadly active in-vitro against antibiotic-susceptible and -resistant Gram-positive and Gram-negative pathogens causative for cSSSI and CAP.
P1671 PZ-601 antibiotic and bactericidal activity against multidrug-resistant Gram-positive bacteria
J. Pace, L. Xerri (Malvern, US)
Background: PZ-601 (SMP-601) is an investigational parenteral carbapenem with potent activity against both Gram-positive and Gram-negative pathogens. Its enhanced affinity for PBP2a has extended its spectrum to include multidrug-resistant Gram-positive bacterial pathogens. The projected role for this antibiotic is empirical monotherapy of infections where drug-resistant staphylococci and enterococci may be encountered.
Methods: Numerous Staphylococcus aureus strains including methicillin-resistant (MRSA) and community acquired (CAMRSA), and vancomycin-intermediate susceptible and resistant (VISA and VRSA) strains, coagulase-negative staphylococci, both vancomycin-susceptible and -resistant enterococci including ampicillin-susceptible and -resistant strains, and select linezolid-, daptomycin-, and tigecycline-resistant isolates were obtained from NARSA or various U.S. and European hospitals. These bacteria were subjected to susceptibility testing with PZ-601 and comparator antibiotics using broth microdilution minimal inhibitory concentration (MIC, mg/L) method according to CLSI guidelines. In some instances minimal bactericidal concentrations (MBCs) and bactericidal killing by time-kill assay were also evaluated.
MIC (mg/L) values and range for PZ-601 and comparator antibiotics against select clinical isolates and reference strains of Gram-positive species
| Species | Phenotype | MIC (mg/L) |
|||||
|---|---|---|---|---|---|---|---|
| PZ-601 | Vancomycin | Linezolid | Daptomycin | Tigecycline | Imipenem | ||
| Staphylococcus aureus | |||||||
| ATCC 29213 | MSSA | ≤0.015 | 0.5 | 2 | 1 | 0.5 | 0.09 |
| COL | MRSA | 1 | 1 | 4 | 2 | 1 | 32 |
| USA300 | CA-MRSA | 0.5 | 0.5 | 8 | 1 | 1 | 4 |
| USA400 | CA-MRSA | 0.25 | 1 | 4 | 2 | 0.5 | 0.5 |
| GC7647 | TIG-R | 2 | 2 | 4 | 4 | 16 | >32 |
| HIP5836 | VISA | 1 | 8 | 4 | 4 | 1 | 32 |
| Mu-50 | VISA | 2 | 8 | 4 | 4 | 1 | 32 |
| VRSA1 | VRSA | 1 | >256 | 4 | 1 | 2 | >32 |
| Enterococcus faecium | |||||||
| J4026 | DAP-R | 4 | 128 | 8 | 32 | ND | >32 |
| I225 | LIN-R | 4 | 128 | 32 | 1 | ND | >32 |
Results: MICs of PZ-601 were superior to other carbapenems tested against most MRSA strains and enterococci. PZ-601 modal MIC was 2 and MIC range was ≤0.004–4 for MRSA, and 1 and 0.25–2 for Enterococcus faecalis, respectively. PZ-601 was active against bacteria intermediate-susceptible or resistant to vancomycin, linezolid, daptomycin, or tigecycline.
Conclusion: PZ-601 is broadly active in-vitro against drug-resistant Gram-positive pathogens including MRSA and CAMRSA, VISA, VRSA, VRE, and those isolates resistant to linezolid, daptomycin, or tigecycline. PZ-601 is bactericidal and reduces bacterial counts to a greater degree than vancomycin at early time-points.
P1672 PZ-601 antibacterial activity against well-characterised and fresh Enterobacteriaceae isolates
M. Mezzatesta, M. Trovato, F. Gona, M. Santagati, F. Campanile, S. Stefani (Catania, IT)
Objectives: The in vitro activity of PZ-601, formerly SMP-601, an investigational broad-spectrum carbapenem antibiotic, with potent activity against multi-drug resistant Gram-positive and Gram-negative bacteria, was tested in comparison with other drugs against a group of well-characterised and fresh Enterobacteriaceae isolates.
Methods: The strains included in the study – Escherichia coli, Klebsiella spp. and Enterobacter spp. – were the first 70 Enterobacteriaceae consecutive non-repetitive strains collected from urine and respiratory tract specimens of inpatients and outpatients, isolated from the Policlinico of Catania (I) in a 4-month period, and 23 strains of Enterobacteriaceae previously molecularly characterised for their ESBL production (Luzzaro et al. JCM 2006; 44: 1659). All bacterial isolates were identified at the species level using standard procedures. Antibiotic susceptibility was determined by the standard broth microdilution method (MICs) following CLSI guidelines. The antibiotics used were: PZ-601, imipenem (IMI), ceftriaxone (CRO), cefepime (CPE), piperacillin/tazobactam (PIP/TAZ), levofloxacin (LEVO) and tobramycin (TOBRA). PZ-601 was freshly prepared and maintained in ice.
Results: PZ-601 was highly active against Enterobacteriaceae and ESBL-producing strains with MIC90 of 2 mg/L, comparable with that of imipenem, and superior to that of the other drugs tested. Among ESBL isolates, PZ-601 showed a variable activity against the strains possessing different enzymes, with a MIC90 value of 0.5 mg/L in SHV-containing strains, while it showed MIC90 values of 2 in strains containing CTX-M-type enzyme.
Table 1.
MIC90 (mg/L) of PZ-601 and comparators against Enterobacteriaceae
| Strains | MIC90 (mg/L) |
||||||
|---|---|---|---|---|---|---|---|
| PZ-601 | CRO | CPE | PIP/TAZ | LEVO | TOBRA | IMI | |
| Enterobacteriaceae (n = 70) | 2 | 128 | 4 | 512 | 4 | 32 | 2 |
| ESBLs producers (n = 23) | 2 | >128 | 128 | >512 | 16 | 64 | 2 |
| SHV strains (n = 7) | 0.5 | 64 | 4 | 256 | 16 | 64 | 1 |
| CTX-M (n = 16) | 2 | >128 | 128 | >512 | 16 | 64 | 1 |
Conclusion: In this small sample, PZ-601 was highly active against Enterobacteriaceae and ESBL-producing isolates, demonstrating that this carbapenem is a potentially outstanding alternative drug in the empiric treatment of infections caused by these multiresistant microrganisms resistant to cephalosporins, protected penicillins, quinolones, and aminoglycosides.
P1673 Safety and pharmacokinetics of single-dose intravenous administration of PZ-601: a novel investigational carbapenem
C. Young, A. Rusca, A. Di Stefano, M. Radicioni, D. Binelli, L. Xerri (Malvern, US; Arzo, CH)
Objective: PZ-601 (SMP-601) is a novel carbapenem with an antimicrobial spectrum that includes multidrug-resistant Gram positive (methicillin-resistant Staphylococcus aureus, Enterococcus faecium) and extended-spectrum β-lactamase producing Gram negative organisms. We studied the pharmacokinetics and safety of single ascending doses in otherwise healthy male volunteers.
Methods: Fifty-six subjects (42 PZ-601, 14 placebo) were randomised in seven cohorts (6 PZ-601, 2 placebo) to ascending doses of PZ-601: 30, 100, 300, 600, 1,000, 1,500, 2,000 mg or placebo by infusion over 30 minutes. Assessments included adverse events, laboratory, ECGs and PZ-601 plasma levels and urinary excretion.
Results: Adverse events were infrequently noted in 5/42 PZ-601 and in 1/14 placebo recipients. The most common events were headache (3 PZ-601, 1 placebo) and dyspepsia (1 PZ-601). There were no serious adverse events. There were no laboratory or ECG findings of clinical concern. PZ-601 exposure was linear for the administered doses with a half-life of 1.4 hours at a dose of 1,000 mg. A summary of key PZ-601 pharmacokinetic parameters is presented.
| Parameter | PZ-601 dose (mg) |
||||||
|---|---|---|---|---|---|---|---|
| 30 | 100 | 300 | 600 | 1,000 | 1,500 | 2,000 | |
| Cmax (mg/L) | 2.1 | 7.8 | 20 | 51 | 72 | 122 | 143 |
| AUC0–∞ (mg·h/L) | 3 | 12 | 26 | 68 | 97 | 165 | 220 |
| Volume of distribution (L) | 15.5 | 13.6 | 16.5 | 14.2 | 16.5 | 14.1 | 14.7 |
| Clearance (mL/min) | 175 | 137 | 195 | 152 | 250 | 154 | 154 |
| T1/2 (h) | 1.1 | 1.2 | 1.2 | 1.3 | 1.4 | 1.4 | 1.5 |
Conclusion: PZ-601 in single doses up to 2 g was generally well-tolerated in normal healthy male subjects. These results suggest the potential for twice daily dosing and support progression to repeat dosing.
P1674 Broad spectrum anti-Candida activities of a new indolyl-triazole, NL114
F. Pagniez, N. Le Bouvier, Y. Na, M. Le Borgne, M. Pfalzer, R. Hartmann, P. Le Pape (Nantes, FR; Saarbrücken, DE)
Objectives: During the last decades, the frequency of fungal infections has increased. Azole drug activities against systemic fungal infections have not to be demonstrated. However, azole resistance of Candida spp., such as C. krusei and C. glabrata, has been reported. In this work we present the potent anti-Candida activity of NL114, an indolyl-triazole, against C. albicans, C. krusei and C. glabrata.
Azole antifungals are known to interfere with fungal but also mammalian cytochrome P450 (CYP). In view of future development, inhibition of these CYPs was investigated as well as the cellular toxicity of NL114.
Methods: 39 strains of Candida belonging to the species C. parapsilosis, C. krusei, C. albicans, C. glabrata and C. lusitaniae were used. Antifungal drugs were NL114 [2-(2,4-dichlorophenyl)-3-(1H-indol-1-yl)-1-(1,2,4–1H-triazol-1-yl)-propan-2-ol], ketoconazole, fluconazole and voriconazole. Antifungal activity was evaluated with a fluorometric microdilution method. MICs are the concentrations that inhibit 80% of fungal growth and MIC90s are the concentrations that inhibit growth of 90% of strains. Cytotoxicity of compounds was studied with human fibroblast (MRC5) with a fluorometric microdilution method. In vitro human cytochrome interactions were tested on aromatase (CYP19) and 17 α-hydroxylase/17,20-lyase (CYP17).
Results: Antifungal activity of NL114 is higher than fluconazole and voriconazole on C. albicans (MIC90 = 1.5, 20 and 2.8 μg/mL, respectively), C. glabrata (MIC90=0.26, 31 and 2.1 μg/mL, respectively) and C. krusei (MIC90 = 0.23, 20 and 0.23 μg/mL, respectively). Fluconazole-resistant or less-susceptible Candida strains were susceptible to NL114. Results of the cellular toxicity assay showed that NL114 is more toxic than fluconazole and voriconazole. Nevertheless geometric mean of MICs against Candida is 190 times lower than the toxic concentration on MRC5 cells.
Human cytochrome interaction study showed that NL114 at a concentration of 36 μM did inhibit CYP19 as well as ketoconazole. NL114 did not interact with CYP17 at a concentration of 2.5 μM.
Conclusion: In conclusion, these promising results point out that NL114 is a new lead antifungal candidate. Cellular toxicity and interaction with human P-450 cytochrome studies let predict a good safety.
P1675 QTc measurements during a placebo- and actively controlled multiple dose study of two different dosing regimens of isavuconazole
M. Heep, B. Roos, M. Sochor, A. Schmitt-Hoffmann, S. Van Merle, D. Kappers, P. Voiriot (Basle, CH; Zuidlaren, NL; Nancy, FR)
Background: Isavuconazole (BAL8557/BAL4815, ISA) is a new water-soluble triazole which has broad in-vitro activity against yeasts, moulds (including zygomycetes) and dermatophytes. The oral formulation is almost completely bioavailable and the i.v. formulation can be administered to renally impaired patients because it does not contain cyclodextrins. ISA is currently in phase III clinical development. This study was designed to investigate the cardiac safety of ISA at two dose levels.
Methods: A total of 80 healthy subjects received a single dose of 400 mg moxifloxacin as a positive control for prolongation of QTc, and subsequently were randomised to groups A and B:
Group A: Staggered oral loading doses of 400, 300 and 200 mg ISA were administered on Days 4, 5 and 6, followed by 100 mg qd ISA on Days 7 to 10, and one IV infusion of 100 mg ISA on Day 11. Three staggered oral loading doses of BAL8557 of 300, 250 and 200 mg ISA equivalents were administered on Days 12 to 14, followed by once daily oral maintenance doses of 150 mg ISA on Days 15–18, and one i.v. infusion of 150 mg ISA as a 1-hour infusion on Day 19.
Group B: 40 subjects receiving placebos.
Time-matched, baseline-subtracted QTc measure was used for analysis.
Results: Moxifloxacin effect was statistically significant and increased QTcF in both groups (6 ms in Group A, 7 ms in Group B) compared to baseline, as expected. During the remainder of the study period a reduction of QTcF from baseline was observed both after ISA and in the placebo control arm. Reduction of QTc from baseline was generally more pronounced in the ISA group. There was no apparent difference in QTc results between the oral or i.v. treatment phase, nor between the 100 mg steady state or 150 mg steady-state dose level. Dosing was well tolerated.
Conclusion: In contrast to the moxifloxacin positive control, no QTc increase was observed for two dosing regimen of isavuconazole. The decrease of QTcI in comparison with placebo was slight (between 5 and 10 msec) and does not preclude of the safety of the drug.
P1676 Evaluation of isavuconazole (BAL8557/BAL4815) Etest compared to broth microdilution antifungal susceptibility testing against quality control strains and fluconazole-susceptible clinical Candida isolates
M. Heep, P. Grover, N.P. Brown, D. Sahm, M.E. Jones (Basle, CH; Herndon, US)
Background: Isavuconazole (ISA, BAL8557/BAL4815) is a new broad-spectrum triazole. In preparation of clinical trials in primary and salvage treatment of invasive fungal infections, options allowing susceptibility testing were evaluated. Agar-based methods such as Etest may be preferred by some clinical laboratories for their simplicity and ease of use. Broth microdilution (BMD) is a reference method for antifungal susceptiblity testing (AFST) against which the Etest method was validated.
Methods: Etest was performed according to manufacturer's instructions (AB Biodisk) and BMD according to CLSI M27-A2 guideline. Organisms tested were C. parapsilosis ATCC 22019, C. krusei ATCC 6258 and 4 clinical isolates of C. parapsilosis, C. glabrata, C. tropicalis, C. krusei. On three separate days a total of 14 individual inoculum suspensions for each test organism was prepared and used concomitantly for Etest and broth microdilution; voriconazole and fluconazole were included as reference. Both Etest and BMD results were read at 24 and 48 hours incubation. Concordance between methods was defined as within 1 doubling dilution of the BMD MIC.
Results: For ISA 100% concordance was detected between BMD and Etest at 24 h for C. parapsilosis ATCC 22019 (BMD MIC range 0.015 ug/mL; Etest MIC range 0.012–0.016 mg/L) and C. krusei ATCC6258 (BMD MIC range 0.06–0.12 mg/L; Etest MIC range 0.0125 mg/L); and at 48 h for C. parapsilosis ATCC22019 (BMD MIC range 0.03 mg/L; Etest MIC range 0.016–0.023 mg/L) and C. krusei ATCC6258 (BMD MIC range 0.25 mg/L; Etest MIC range 0.0125–0.19 mg/L).
Similarly for all clinical isolates 100% of results were concordant at both 24 h and 48 h with the exception of one 24 h inoculum suspension (BMD 0.06 mg/L versus Etest 0.25 mg/L), one 48 h inoculum suspension (BMD 2 mg/L versus Etest 0.75 mg/L) of C. glabrata 1547507; and two inoculum suspensions (both BMD 1 mg/L versus 0.38 mg/L Etest) of C. krusei 1547509.
Conclusions: For the ATCC quality control strains and susceptible clinical isolates ISA Etest results correlated well with the CLSI M27-A2 BMD method. This agar-based method represents a simple and reliable method for determining ISA susceptibility of Candida spp. Additional testing will establish concordance between methods for ISA-resistant isolates.
P1677 Natural coumarins from the plants Seseli devenyense and Peucedanum luxurians as in vitro antibacterial agents
J. Widelski, K. Craikou, K. Glowniak, E. Chinou, I. Chinou (Lublin, PL; Athens, GR)
Objectives: Coumarins constitute a major category of secondary plant products that are widely distributed in the plant Kingdom. Very interesting subclasses of this category are the linear or angular pyrano and furanocoumarins that have been found to possess antiproliferative, antiviral and antibacterial activities. The antimicrobial activity of a series of 24 natural coumarins isolated from the plants Seseli devenyense and Peucedanum luxurians from Poland, is evaluated against six pathogenic bacteria.
Methods: The antimicrobial activities of the tested compounds were determined, using the diffusion and dilution techniques and measuring their MICs against Gram-positive bacteria (S. aureus, S. epidermidis), Gram-negative bacteria (E. coli, E. cloacae, K. pneumoniae, P. aeruginosa) all of them strains of ATCC. Standard antibiotics were used in order to control the sensitivity of the test organisms.
Results: Through the antimicrobial screening, the 24 tested compounds showed a broad diversity regarding growth inhibitory activity. Twelve compounds among the tested ones (cis-khellactone, laserpitine, 3'-angeloyl-khellactone, praeroside II, devenyol, denyosides A, B and C, peucedanin, xanthotoxin, stenocarpin and isoimperatorin) appeared to be significantly active against all six tested bacteria (zones of inhibition 16–20 mm, MIC values 0.05–3.5 mg/mL), six others showed moderate activity while a variety of six medium chain (C6-C12) aliphatic esters of (+)-lomatin and (+)-cis-khellactone coumarins appeared completely inactive.
Conclusions: The results of the assays suggest that the activity of the tested coumarins can be attributed, to the existence of linear furan ring or to oxygenated substituents to simple coumarins, while the derivatives of the angular pyranocoumarins showed increased activities in comparison with all the rest coumarins possessing another pattern of substitution.
P1678 New antiprotozoal drugs refered as antimicrotubular haloacetamidobenzoates
P. Le Pape, A. Fellous, A. Valentin, S. Andrade, J. Kalil, M. Veauvy, M. Le Borgne (Nantes, Toulouse, FR; Salvador de Bahia, Sao Paulo, BR)
Objectives: Protozoal diseases constitute a major world public health problem. Malaria conveyed in the yearly deaths of 2,000,000 children and there would be 300 millions of new cases per year. Concerning leishmaniasis, it is estimated that approximately up to 20–30 million people in 88 countries are infected. Chagas' disease is an anthropozoonosis due to Trypanosoma cruzi. Chagas' disease is widespread in Latin America where it strikes 20 millions of people. The growing problem of drug resistance has greatly complicated the treatment for these parasitic diseases. There is still a real need for active new compounds that would provide therapeutic benefits with less side effects and a defined cell target.
Methods: A new family of patented antimicrotubule drugs named haloacetamidobenzoates (MF and GR derivatives) was designed and evaluated against three protozoa, Plasmodium, Leishmania and Trypanosoma. In vitro and in vivo models were developed. Mechanism of action was studied by scanning electron microscopy, immunocytochemistry and microtubule assembly assay.
Results: MF29 and GR37 were found to be cytotoxic in vitro on chloroquine resistant Plasmodium falciparum strains, Leishmania spp. and Trypanosoma with IC50 values about 0.1 μM. These compounds were shown to be very poor inhibitors of in vitro mammal brain microtubule assembly indicating parasite specificity. Parasite MAP2-like proteins may increase strongly the sensitivity of parasite tubulins to the drug. Concerning Plasmodium, the in vitro data have been confirmed in an in vivo murine model using Plasmodium berghei showing a high reduction in mortality percentage and a moderate activity on parasitaemia. In the visceral and cutaneous murine leishmaniasis models, the reference drug antimoniate meglumine (Glucantime) showed a less marked reduction in parasite organ load that the MF 29 compound. Moreover a significant reduction of mortality was observed when infected mice by Trypanosoma cruzi are treated by MF 29 compound.
Conclusion: These data demonstrated the high level of antiprotozoal activity of MF derivatives compared to other antimicrotubule agents or other antileshmanial drugs. An interesting point of our work is related to the fact that we have established the mechanism of action of haloacetamidobenzoates with the precise identification of the cellular target.
P1679 In vitro activity of SS734, a novel fluoroquinolone, against pathogens associated with bacterial conjunctivitis
L.S. Brunner, S.E. Norton, J.M. Blondeau (Rochester, US; Saskatoon, CA)
Objectives: SS734 is a novel fluoroquinolone (FQ) compound being investigated for the treatment of bacterial conjunctivitis (BC). We tested the in vitro activity of SS734 against Gram-positive and negative bacterial pathogens – including those resistant to various antimicrobial agents – associated with BC.
Methods: Minimum Inhibitory Concentrations (MICs) were determined on 627 clinical isolates following the recommended procedure of the Clinical and Laboratory Standards Institute. MICs were performed by broth microdilution: Briefly, approximately 105 CFU/mL were exposed to doubling dilutions of drug and following incubation in the appropriate atmosphere and temperature for 18–24 hours, the lowest concentration preventing growth was the MIC. Neisseria gonorrhoeae was tested by agar dilution.
Results: The following MIC50 (μg/mL), MIC90 (μg/mL) and MIC range (μg/mL) values respectively were recorded: N. gonorrhoeae (n = 107) 0.008, 0.015, 0.004–0.03; MICs to 2 quinolone resistant strains were 2 (μg/mL); H. influenzae (n = 105) 0.03, 0.06, 0.015–0.25; H. influenzae β-lactamase (BL) positive, ampicillin resistant strains (n = 25) 0.03, 0.12, 0.015–0.25; E. cloacae extended-spectrum β-lactamase (ESBL) negative (n = 92) 0.25, 8, 0.06–>8 and MICs tended to be higher against ESBL positive isolates; S. aureus methicillin sensitive (MS) and ciprofloxacin sensitive (CS) (n = 28) 0.03, 0.06, 0.015–0.25; S. aureus methicillin resistant (MR) and ciprofloxacin resistant (n = 24) 1, 8, 0.5–8; S. aureus MR CS (n = 25) 0.03, 0.06, 0.015–1; S. epidermidis MS (n = 38) 0.03, 0.5, 0.03–1; S. epidermidis MR (n = 64) 0.5, 8, 0.015–>8; S. aureus vancomycin intermediate (n = 23) 1, 2, 0.03–2; S. pneumoniae (n = 78), 0.12, 0.12, 0.06–2, MIC values were not different for penicillin susceptible or resistant strains and 1, 2, 0.5–8 against levofloxacin resistant strains (n = 25).
Conclusion: SS734, a novel FQ being investigated for BC, is active in vitro against Gram-negative and positive organisms including those resistant to other antimicrobial agents. MICs for SS734 were well below the expected drug concentrations delivered to the eye during therapy even for ciprofloxacin or levofloxacin resistant organisms.
P1680 Bactericidal activity of SS734, a novel fluoroquinolone, against pathogens associated with bacterial conjunctivitis
L.S. Brunner, PA. Walsh, S.E. Norton, J.M. Blondeau (Rochester, US; Saskatoon, CA)
Objectives: SS734 is a novel fluoroquinolone (FQ) currently under development for the treatment of bacterial conjunctivitis. We performed time kill studies on SS734, norfloxacin (NRFX), and ciprofloxacin (CPFX) against a variety of ocular isolates from clinical sources.
Methods: Two isolates of each organism were chosen for time kill analysis based on their range of NRFX and CPFX activity. SS734, NRFX, and CPFX at concentrations of 1 (1×), 2 (2×), and 4 (4×) times the MIC90 values were inoculated with approximately 105 cfu/mL challenge organisms. At each sampling time point, the test samples were diluted in physiologic saline and spread plated onto appropriate agar. After aerobic incubation, the colonies were counted and the viable cells per milliliter were calculated.
Results: SS734 has rapid bactericidal activity against Streptococcus pneumoniae (Sp), Staphylococcus aureus (Sa), coagulase-negative Staphylococcus (cnS), and Haemophilus influenzae (Hi). The percent reduction in viable cells (and log10 reduction) following exposure of 2–4× the MIC90 for SS734, NRFX and CPFX, respectively, was as follows: Sp at 60 minutes (min) 57–77 (0.4–0.7), 43–68 (0.3–0.5), 46–56 (0.3–0.4); Sa at 30 min 74–97 (0.6–1.8), −7–+7 (0–0.1), 38–43 (0.3–0.4); cnS at 30 min 88–90 (0.9–1), 5–10 (0.1–0.1), 27–23 (0.2–0.1); and Hi at 45 min 63–88 (0.5–1), 35–59 (0.2–0.4), 68–74 (0.6–0.6). MIC90 values for SS734 were 4–128 fold lower than CPFX and NRFX for Gram-positive organisms and equivalent for Haemophilus influenzae.
Conclusion: SS734 was rapidly bactericidal against all strains tested and showed a more rapid killing rate than NRFX and CPFX. Based on the time kill studies against these pathogens associated with bacterial conjunctivitis, SS734 looks to be a suitable fluoroquinolone compound for further clinical investigation.
P1681 In vitro activity of three echinocandins (caspofungin, micafungin and anidulafungin) against Aspergillus spp
A.I. Martos, C. Martin de la Escalera, E. Lopez-Oviedo, A.I. Aller, A. Romero, C. Serrano, A. Valverde, E. Martín-Mazuelos (Seville, ES)
Objective: Echinocandins are a novel class of antifungal agents with an excellent activity against Aspergillus spp. The in vitro activity of caspofungin (CF), micafungin (MF) and anidulafungin (AF) were tested against clinical isolates of Aspergillus spp.
Material and Methods: Forty-one clinical isolates of Aspergillus spp. were used in the study: 28 isolates of A. fumigatus, 10 isolates of A. flavus and 3 A. terreus. The susceptibility testing to these three echinocandins were performed by the broth microdilution (MD) procedure according to the CLSI M-38A document. The ranges of drugs dilution were 0.03–8 mg/L. Inoculum suspensions were prepared recovering the conidia from a 7-day culture growth on Potato Dextrose Agar at 35°C (final inoculum: 0.5–5×104 CFU/mL). MECs (Minimal Effective Concentrations) were determined and the readings were performed at 48 h (Kurtz et al. 1994, AAC 38: 1480–1489). Candida parapsilosis 22019, C. krusei 6258, A. fumigatus 204305 and A. flavus 204304 were included as Quality Control strains.
Results: MECs50, MECs90 and ranges of MECs (mg/L) of CF, MF and AF are shown in Table 1 . The MECs were ≤ 0.25 mg/mL for all strains tested. MF and AF shown similar MECs (≤ 0.03 mg/L) while CF MECs range were from 0.03 to 0.25 mg/L. The Quality Control strains were within the published range.
Table 1.
| Speciesa | MEC (mg/L) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Caspofungin |
Micafungin |
Anidulafungin |
|||||||
| Range | MEC50 | MEC90 | Range | MEC50 | MEC90 | Range | MEC50 | MEC90 | |
| A. fumigatus (28) | ≤0.03–0.25 | 0.06 | 0.125 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 |
| A. flavus (10) | ≤0.03–0.25 | 0.06 | 0.06 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 |
| A. terreus (3) | 0.125 | ≤0.03 | ≤0.03 | ||||||
| Total (41) | ≤0.03–0.25 | 0.06 | 0.25 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 |
Number of isolates given in parentheses.
Conclusion: These three echinocandins shown an excellent in vitro activity against all clinical isolates of Aspergillus spp. tested.
P1682 In vitro activity of tigecycline against multiresistant Acinetobacter baumannii clinical isolates
A. Stylianakis, A. Koutsoukou, S. Tsiplakou, S. Sardiniari, A. Vourtsi, A. Kolokouri (Athens, GR)
Objectives: The purpose of our study was to evaluate the in vitro activity of tigecycline against multiresistant Acinetobacter baumannii clinical isolates.
Methods: We examined 276 non duplicated multiresistant Acinetobacter baumannii isolates recovered from blood cultures, bronchoalveolar excretions and pus and urine samples derived from ICU-patients of our hospital during a one year-time period. The identification of the isolates and the susceptibility testing were performed by the automated VITEK 2 system (bioMérieux, France). The susceptibility testing for tigecycline was performed by Kirby-Bauer disk diffusion method and done by direct colony suspension according to CLSI guidelines. Paper disks containing tigecycline at 15 mg per disk were used (Becton-Dickinson, USA). The determination of tigecycline-MIC values was performed by E-test strips according to the manufacturer's guidelines (AB-Biodisk, Sweden). Isolates with an inhibition zone diameter of ≥19 mm and with an MIC level ≤ 1 mg/L were considered as susceptible to tigecycline (MIC-susceptibility limits determined by EUCAST).
Results: All the Acinetobacter baumannii clinical isolates were resistant to aminoglycosides, β-lactams, carbapenems, monobactames, furanes, cinolones except colistin. The tigecycline was found absolute active to all the examined multiresistant isolates. The inhibition diameter zones were found of ≥ 20 mm and the MIC levels of ≤ 0.5 mg/L
Conclusion: Tigecycline is absolute active to all the multiresistant Acinetobacter baumannii isolates and it is consisted a very important option for treatment of serious infections caused by these isolates. Clinical laboratories should be aware of testing tigecycline susceptibility in Acinetobacter baumannii isolates; so that any emergence of tigecycline-resistance may be detected as soon as possible.
P1683 Analysis of reports of Clostridium difficile infection in phase 2 and 3 studies of tigecycline
M. Wilcox, J. Freeman (Leeds, UK)
Objectives: Antibiotics vary in their propensity to induce C. difficile infection (CDI). Anaerobic and/or broad spectrum activity may be associated with increased risk of CDI, although these associations are not clear. We reviewed the reports of possible CDI in tigecycline preregistration studies to determine whether this extended broad spectrum activity antibiotic has increased risk of inducing CDI.
Methods: Clinical trial database searches were performed on all completed Phase 2 (n = 4) and 3 (n = 9) studies; 7 of the 9 phase 3 studies were originally blinded. To identify subjects who may have had CDI associated with tigecycline or comparator therapy, searches were performed for adverse event terms, indications for receipt of concomitant antibiotics, and the results of local hospital faecal sample investigations. A diagnosis of CDI required symptoms (diarrhoea, abdominal pain/distension, colitis) to be present for at least 48 h; 2 cases (1 tigecycline and 1 comparator), who also received no antibiotic treatment for CDI, were excluded on this basis. Cases who had symptoms of (n = 1) or were being treated for (n = 5) CDI at the time that study antibiotic was started were also excluded.
Results: Records of 2,402 tigecycline and 1,892 comparator (825 imipenem, 589 vancomycin ± aztreonam, 422 levofloxacin, 52 ciprofloxacin and 4 linezolid) recipients were searched for evidence of CDI. After the above exclusions there were 9 possible cases of CDI. Of these, 5 received tigecycline, 3 comparator and 1 tigecycline (5 days) and then oral comparator (levofloxacin) for 5 days. The latter case developed symptoms of CDI 4 days after stopping levofloxacin, and therefore causality with either antibiotic could not be established. The tigecycline associated CDI cases received 3–6 (median 3) other antibiotics before onset of symptoms. Of the comparator associated CDI cases, 2 (imipenem) each received 3 other antibiotics before onset of symptoms; the remaining case received only the comparator antibiotic (levofloxacin). All of the cases were mild-moderate in severity.
Conclusion: CDI was uncommon following tigecycline administration, was associated with prior exposure to other antibiotics, and was of a similar frequency to that seen with comparator antibiotics (∼0.2% of study recipients). The extended broad spectrum activity of tigecycline does not appear to result in increased risk of CDI.
P1684 Tigecycline in vitro activity against often difficult to treat European pathogens
D. Hoban, B. Johnson, R. Badal, S. Bouchillon, M. Hackel, J. Johnson, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Enhanced activity agents such as tigecycline (TIG) may offer antibacterial coverage of common pathogens including multi-drug resistant Gram-negatives/positives. The T.E.S.T. programme monitored the in vitro activity of TIG and comparators against current European pathogens.
Methods: 62 hospital sites in 19 European countries collected over 11,494 clinical isolates from community/hospital infection sites. MICs were determined to TIG and comparators using broth microdilution panels, interpreted according to EUCAST guidelines.
Results: Selected European pathogens tested against tigecycline are shown in the table .
| Organism (#) | Tigecycline MIC |
% inhibited at MIC |
%S* | ||||
|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | ≤0.5 | 1 | 2 | 4 | ||
| A. baumannii (669) | 0.25 | 1 | 80.3 | 96.3 | 99.0 | 100 | na |
| E. faecalis (679) | 0.12 | 0.25 | 100 | 100 | |||
| EC, KO, KPa (3,056) | 0.25 | 1 | 89.8 | 95.5 | 97.8 | 99.7 | 95.5 |
| ESBLb (276) | 0.5 | 2 | 75.4 | 86.2 | 93.1 | 97.8 | 86.2 |
| Enterobacter spp. (1,455) | 0.5 | 2 | 74.9 | 88.5 | 94.6 | 99.2 | 88.5 |
| H. influenzae (814) | 0.12 | 0.25 | 96.6 | 98.6 | 100 | na | |
| P. aeruginosa (1,236) | 8 | >16 | 1.5 | 3.2 | 7.8 | 25.4 | na |
| S. agalactiae (520) | 0.03 | 0.12 | 100 | 100 | |||
| S. aureus (MR) (1,340) | 0.12 | 0.25 | 100 | 100 | |||
| S. pneumoniae (752) | 0.03 | 0.5 | 100 | na | |||
c EUCAST approved breakpoints (2006) where available; na = not yet available.
EC: E. coli, KO: K. oxytoca, KP: K. pneumoniae.
ESBL-producing EC, KO and KP.
Conclusions: European isolates of both Gram-positive and -negative community/hospital pathogens (excluding Pseudomonas) demonstrated very good TIG MIC90s and % susceptible. For most organisms/phenotypes, TIG MIC90s were ≤1 μg/mL and % susceptible >95%. TIG activity was reduced against European ESBL-producing Enterobacteriaceae isolates (MIC90 = 2; %S = 86.2). TIG promises expanded broad spectrum activity against multiply resistant European pathogens.
P1685 In vitro activity of tigecycline and 10 common therapeutic agents against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus species Global Data, 2004–2006
R. Badal, J. Johnson, D. Hoban, B. Johnson, S. Bouchillon, M. Hackel, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline (TIG), a member of a new class of antimicrobials (glycylcyclines), has been shown to have potent activity against community and hospital acquired staphylococcal and enterococcal pathogens. The T.E.S.T programme determined the in vitro activity against methicillin-resistant S. aureus and vancomycin-resistant Enterococcus spp. of TIG and 10 antimicrobials commonly prescribed for serious Gram-positive infections: amoxicillin-clavulanic acid (AUG), piperacillin-tazobactam (PT), levofloxacin (LVX), ceftriaxone (CAX), linezolid (LZD), minocycline (MIN), vancomycin (VAN), ampicillin (AMP), penicillin (P), and imipenem (IMP). Study strains were collected from 272 laboratories in 34 countries globally throughout 2004–2006.
Methods: A total of 9,806 clinical isolates (3,925 enterococci, 5,881 S. aureus) were identified to the species level at each participating site and confirmed by a central laboratory. Minimum Inhibitory Concentrations (MICs) were determined by the local laboratory using broth microdilution panels. Antimicrobial resistance was interpreted according to CLSI breakpoints with TIG susceptible breakpoints defined as ≤0.5 μg/mL for S. aureus and ≤0.25 μg/mL for enterococci.
Results: 14.4% (565/3,925) of enterococci were resistant to vancomycin (VRE), and 45.4% (2,669/5,881) of S. aureus were resistant to oxacillin (MRSA). Among the VRE, % resistance rates to other study drugs were LVX 99.6, P 99.1, AMP 97.9, VAN 96.6, MIN 11.9, and LZD 0.0. Resistance rates for MRSA were P 100, AMP 100, AUG 99.8, LVX 100, PT 99.3, CAX 98.9, IMP 100, MIN 0.7, LZD 0.0, and VAN 0.0. TIG inhibited 100% of the enterococci and S. aureus resistant to other drugs. Modal TIG MICs for VRE/nonVRE were 0.03/0.06, and 0.12/0.12 for MRSA/MSSA).
Conclusions: TIG retained potent activity against drug-resistant S. aureus and enterococcal isolates, inhibiting 100% of all strains tested at their defined breakpoints of 0.5 and 0.25 μg/mL, respectively. TIG should prove to be a useful drug for therapy of infections with these resistant Gram-positive pathogens.
P1686 Resistance to tigecycline and other non β-lactam antimicrobials of MRSA strains collected in Austria in 2005/2006
C. Jebelean, K. Pfeil, C. Luger, S. Stumvoll, M. Stammler, B. Sammer, H. Mittermayer (Linz, AT)
Objectives: The prevalence of MRSA is rising in many European countries.
The aim of our study was to investigate the epidemiology and the prevalence of resistance to tigecycline (TG) and other non β-lactam antimicrobials of Austrian MRSA strains.
Materials and Methods: We determined the resistance to: erythromycin (E), clindamycin (LC), gentamicin (G), fosfomycin (FOS), fusidic acid (FU), tetracycline (T), vancomycin (V), teicoplanin (TE) using the agar dilution method as recommended by CLSI, the resistance to linezolid (L) and TG by E-Test method and the resistance to mupirocin (MUP) by disk diffusion method of 128 MRSA strains collected allover Austria during 2005–2006.
Additionally, we screened for heterogeneous VISA using a BHI agar plate containing 4 mg/L TE and the E-Test Macromethod with V and TE. For all the strains we determined the mecA and femA gene using a multiplex PCR. For subtyping we performed coagulase gene PCR with restriction digestion of PCR products with AluI, and we further analysed the PFGE patterns.
Results: Overall the E, LC, G, FOS, T, FU, L, V, TE, TG and MUP resistance was 71%, 54%, 59%, 27%, 11%, 7%, 0%, 0%, 0%, 0%, 12%, respectively, with higher prevalence in Carinthia and Lower Austria and lower prevalence in Upper Austria and Salzburg. 3 MRSA strain were sent for hVISA confirmation by PAP-analysis. Among the MRSA strains we found following sequence types (ST): ST228 (30%), ST8 (29%), ST22 (15%), ST5 (12%), and other types under 3% each.
Conclusions: In comparison with resistance data up to 2003 from Upper Austria we observed an increase in G and T resistance, a decrease in FOS resistance and a stable resistance rate to LC and FU. All strains were susceptible to L and TG. The ST distribution was more diverse in Vienna and Salzburg and with strong dominant types in Carynthia and Lower Austria were the resistance rate was also higher.
P1687 Marked tigecycline and colistin synergistic bactericidal effect against a nosocomial epidemic carbapenemase (VIM-1) producing Klebsiella pneumoniae clone
M.I. Morosini, M. García-Castillo, M. Tato, V. Pintado, F. Baquero, J. Cobo, R. Cantón (Madrid, ES)
Tigecycline, a new available glycylcycline derivative, is one of the few alternatives for the treatment of Gram negative bacilli harbouring extended-spectrum β-lactamases and its role against carbapenemase-producing strains is also advocated. Colistin comprises another option in these cases.
Objectives: The activity of tigecycline and colistin, alone or in combination, was assessed in time-kill studies against a multiresitant VIM-1-producing Klebsiella pneumoniae epidemic clone recovered in Madrid (Spain) in 2005.
Methods: Tigecycline and colistin sulphate MICs were determined by the standard broth macrodilution method (CLSI). Duplicate killing curves (Lorian, 1991) with each compound alone or in combination at 1xMIC and 2xMIC and at the serum peak concentration achievable after standard dosing regimens (1 mg/L for tigecycline and 4 mg/L for colistin) were performed.
Results: MICs were 0.5 mg/L for tigecycline and 1 mg/L for colistin. Tigecycline was bacteriostatic at all tested concentrations whereas colistin exhibited concentration-dependent bactericidal activity. Tigecycline and colistin combinations were markedly synergistic at 3 hours (≥106 drop in CFU per mL). Moreover, consistent regrowth at 6 hours observed with single-colistin exposure with all tested concentrations was delayed with tigecycline until 12 hours, even with the lowest tigecycline concentration.
Conclusion: Tigecycline and colistin act synergistically rendering complete killing of a VIM-1 producing K. pneumoniae epidemic clone at 3 hours. Tigecycline abolishes consistent regrowth observed with single-colistin exposure. In vitro synergistic activity of both compounds gives support to the possible treatment of infections caused by carbapenemase-producing isolates. Addition of tigecycline may allow reducing colistin doses lowering secondary toxicity. Optimal regimens remain to be determined.
P1688 In vitro activity of tigecycline and other antimicrobial agents against extended-spectrum β-lactamase E. coli producers
C. Rodriguez-Avial, O. López, I. Rodriguez-Avial, E. Hernandez, J. Picazo (Madrid, ES)
Objective: Tigecycline is likely to have a role in the treatment of infections due to multirresistant pathogens. ESBL-producing E. coli are being recognized as an increasing problem. When they are isolated from patients with urinary tract infections (UTI), they leave few therapeutical options. We sought to determine the incidence and susceptibility patterns of ESBL-E. coli isolated in urine samples in our laboratory in 2005.
Methods: A total of 5.053 E. coli were isolated from urine samples. The MICs of the β-lactamic agents: cefotaxime, ceftazidime, aztreonam and cefepime, with and without clavulanic acid, amoxycillin-clavulanate, cefoxitin, imipenen (I) and ertapenem (E), and the non β-lactamic: ciprofloxacin (CIP), tetracycline (T), tigecycline (TY), gentamycin (G) and nitrofurantoin (N) were determined by agar dilution method (CLSI guidelines), fosfomycin (F) and co-trimoxazol (SXT) were tested by Etest method. The origin (hospital or community) was also determined.
Results: Of the 5.053 E. coli isolates, 216 (4, 2%) were ESBL producers (CLSI Confirmatory test). ESBL-E. coli (n = 192, one isolate per patient) showed also elevated resistance percentages to non-β-lactamic agents: CIP 83%, T 69%, SXT 58% and G 15%. Resistance was low for N 3% (MIC90: 64 mg/L), and F 3, 7% (MIC90: 16 mg/L). No resistance was found to the β-lactamic agents I (MIC90: 0.25 mg/L) and E (MIC90: 0.12 mg/L) and to TY (MIC90: 2 mg/L). We observed a greater proportion (67%) of ESBL-E. coli isolates from non-hospitalised patients.
Conclusions: F and N showed good activity against ESBL-E. coli, and are options for the treatment of uncomplicated UTI in the greater proportion of ESBL-E. coli isolated in non-hospitalised patients. I and E showed very high activity, making carbapenems a good option for the treatment of complicated UTI. TY was very active against our ESBL-E. coli producers. Tigecycline is likely to have a role in the treatment of infections due to multirresistant pathogens including ESBL-E. coli producers.
P1689 Antimicrobial activity of tigecycline and daptomycin against Gram-positive cocci: a multicentre study in Greek hospitals
F. Kolonitsiou, A. Pratti, I. Spiliopoulou, M. Karanika, S. Alexiou-Daniel, D. Bakola, M. Vergi, M. Oeconomou, C. Koutsia-Karouzou, H. Malamou-Lada, L. Zerva, E. Petinaki (Patras, Larissa, Thessaloniki, Lamia, Athens, GR)
Objectives: During the last decades, the emergence of Gram-positive strains resistant to different antimicrobial agents has become a serious medical problem. New compounds that are effective against infections caused by multi-resistant Gram-positive cocci are urgently needed. In the present study we tested the activity of tigecycline and daptomycin against a large number of recent Gram-positive cocci, collected in seven Greek hospitals located in different regions
Methods: From January 2005 to August 2006 a total of 3,160 Gram-positive cocci (870 Staphylococcus aureus, 700 coagulase-negative staphylococci, 400 Enterococcus faecalis, 320 Enterococcus faecium, 200 Streptococcus pneumoniae, 200 Streptococcus agalactiae, 300 Streptococcus pyogenes and 170 viridans streptococci) were isolated in seven different Greek hospitals, from clinically significant specimens (blood, pus, pleural fluid, etc). MICs to tigecycline and daptomycin were determined by the E-test. Susceptibility for tigecycline and daptomycin was defined as approved by US-FDA.
Results: Table 1 summarises the MICs modal, MIC50 and MIC90 of the isolates tested. According to the US-FDA criteria, six MSSA were not susceptible to tigecycline, having MICs 0.75 mg/L; on the other hand among E. faecalis, twelve isolates had MICs greater than the breakpoints proposed (four with MICs 0.5 mg/L, five with MICs 0.75 mg/L and three with MICs 1 mg/L). Among E. faecium, 30 isolates had MICs greater than 0.25 mg/L, while among coagulase-negative staphylococci 35 isolates had MIC greater than 0.5 mg/L. Based on the FDA breakpoints for daptomycin, all S. pyogenes, S. agalactiae, and E. faecalis were susceptible to daptomycin. However, 52 MRSA and 59 MSSA had MICs greater than 1 mg/L.
Table 1.
MICs of isolates tested against tigecycline and daptomycin
| Tigecycline |
Daptomycin |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolates | No | MICmodal | Range | MIC50 | MIC90 | MICmodal | Range | MIC50 | MIC90 |
| MRSA | 348 | 0.125 | 0.016–0.5 | 0.069 | 0.109 | 0.909 | 0.125–3 | 0.438 | 0.791 |
| MSSA | 522 | 0.133 | 0.064–0.38 | 0.077 | 0.112 | 0.8 | 0.19–3 | 0.234 | 0.624 |
| E. faecalis | 400 | 0.311 | 0.047–1 | 0.16 | 0.255 | 1.49 | 0.19–3 | 1.005 | 1.363 |
| E. faecium | 320 | 0.26 | 0.064–1.5 | 0.118 | 0.204 | 2.17 | 0.5–4 | 1.36 | 1.905 |
| CoNS-ME | 120 | 0.187 | 0.064–0.75 | 0.104 | 0.141 | 0.317 | 0.023–1.5 | 0.123 | 0.241 |
| CoNS-MR | 580 | 0.215 | 0.094–1.5 | 0.076 | 0.151 | 0.57 | 0.125–5 | 0.244 | 0.436 |
| S. pneumoniae | 200 | 0.185 | 0.094–0.5 | 0.125 | 0.155 | 0.78 | 0.125–1 | 0.5 | 0.62 |
| S. agalactiae | 200 | 0.137 | 0.047–0.5 | 0.073 | 0.103 | 0.15 | 0.064–0.75 | 0.089 | 0.118 |
| S. pyogenes | 300 | 0.157 | 0.047–0.5 | 0.082 | 0.132 | 0.148 | 0.125–1 | 0.125 | 0.35 |
| S. viridans | 170 | 0.123 | 0.015–0.38 | 0.114 | 0.120 | 0.86 | 0.06–1 | 0.5 | 0.7 |
Conclusions: This is the first multi-centre study in Greece, confirming the good in vitro activity of tigecycline and daptomycin against a range of Gram-positive pathogens.
P1690 In vitro activity of tigecycline against multidrug resistant A. baumannii isolates
G. Yilmaz Bozkurt, K.O. Memikoglu, H. Kutlu, A. Azap, F. Çokça (Ankara, TR)
Objectives: Acinetobacter baumannii is one of the important pathogens of the hospital infections. The evaluation of the in vitro activity of tigecycline in comparison to imipenem, meropenem, tazobactam-piperacillin and cefoperazone-sulbactam against Acinetobacter baumannii strains.
Methods: A total of 100 Acinetobacter baumannii clinical strains isolated during the period of September 2005-September 2006 were tested. The isolates originated from a wide variety of clinical specimens including tracheal aspirate (30), sputum (6), blood (17), central venous catheter (4), abscess-wound swabs (30), abdominal drain fluid (4), cerebrospinal fluid (1), pleural fluid (1), bile (1), urine (5). Sixty percent of these isolates was from clinics of surgery where as 40% was from internal medicine clinics. Their Minimum Inhibitory Concentration (MICs) were determined by using E-Test according to CLSI (Clinical Laboratories Standarts Institute) guidelines.
Results: Overall MIC90 to tigecycline, imipenem, meropenem, tazobactam-piperacillin and cefoperazone-sulbactam were 6 mg/L, >32 mg/L, >32 mg/L, >256 mg/L, >256 mg/L, respectively, whereas MIC50 were 2 mg/L, >32 mg/L, >32 mg/L, >256 mg/L, 32 mg/L, respectively.
Conclusion: In general, tigecycline showed better in vitro activity than imipenem, meropenem, tazobactam-piperacillin and cefoperazone-sulbactam against Acinetobacter baumanii strains. The results show that tigecycline should be considered as a possible alternative for the treatment of Acinetobacter baumannii infections.
P1691 Activity of tigecycline against ESBL-producing enterobacteria spreading in Italy
F. Luzzaro, C. Mugnaioli, S. Gualandris, L. Pallecchi, G. Brigante, B. Pini, G.M. Rossolini, A. Toniolo (Varese, Siena, IT)
Background: Tigecycline is a new glycylcycline active against a variety of Gram-positive and Gram-negative resistant pathogens. We evaluated tigecycline activity against a collection of clinical isolates of multidrug-resistant (MDR) enterobacteria producing different types of extended-spectrum β-lactamases (ESBL) from a recent Italian nationwide survey.
Methods: ESBL-producing enterobacteria were tested for susceptibility to tigecycline and other agents by broth microdilution (Microscan panels, Dade-Behring). The collection included isolates of Escherichia coli (n = 138), Klebsiella pneumoniae (n = 64), Klebsiella oxytoca (n = 14), Enterobacter aerogenes (n = 25), Enterobacter cloacae (n = 12), Serratia marcescens (n = 12), and Citrobacter spp. (n = 15), producing TEM, SHV and/or CTX-M type ESBLs. Detection of tetracycline resistance genes [tet(A), tet(B), tet(C) and tet(D)] was performed by PCR.
Results: Overall, 275/280 ESBL-positive isolates (98.2%) were susceptible to tigecycline (MIC ≤2 mg/L). Susceptibility rates were 100% among E. coli, including members of the CTX-M-15-producing E. coli clones causing major outbreaks in some Italian hospitals. Susceptibility rates were 97.4% among Klebsiella spp. and 95.3% among other Enterobacteriaceae. Overall, MIC50/90 of tigecycline were 0.25/1.0 mg/L. E. coli showed MIC50/90 values (0.25/0.5 mg/L) lower than those of other species (0.5/2.0 mg/L). The presence of tet(A), (B), and/or (D) determinants did not affect susceptibility to tigecycline (MIC50/90 0.25/1.0 mg/L for both tet-negative and tet-positive isolates).
Conclusion: Tigecycline showed potent in vitro activity against the vast majority of ESBL-producing enterobacteria circulating in Italy. It might thus represent a valid therapeutic option for treating selected infections caused by these MDR pathogens.
Support from Wyeth Lederle Italia SpA is gratefully acknowledged.
Infection control and nosocomial infections
P1692 Environmental decontamination of an intensive care unit to control outbreaks of multidrug-resistant Gram-negative rods using hydrogen peroxide vapour
M. Schouten, J. Otter, A. van Zanten, G. Houmes-Zielman, M. Nohlmans-Paulssen (Ede, NL; London, UK)
Objectives: 25 patients on our 12-bed intensive care unit (ICU) acquired multidrug-resistant Gram-negative rods (MDR-GNR) over an 8-month period, comprising Enterobacter cloacae (7) and Acinetobacter baumannii (18). Transmission continued despite the implementation of standard infection control measures including an emphasis on hand and environmental hygiene. We investigated the microbiological and clinical impact of hydrogen peroxide vapour (HPV) decontamination of the entire ICU.
Methods: All patients were temporarily relocated from the ICU and the entire unit was decontaminated using HPV. 100 cm2 sterile cotton gauzes were moistened in sterile water and used to sample 41×1 m2 areas after cleaning (which included 1,000 ppm sodium hypochlorite for surfaces and 70% alcohol for equipment) but before HPV. Nine matched adjacent areas were sampled after HPV decontamination. Gauzes were subcultured on blood and MacConkey agar after overnight broth enrichment. Thirty commercially-available Geobacillus stearothermophilus biological indicators (BIs) with an inoculum of >1.0×106 were located around the periphery of the unit, collected at the end of the HPV cycle and cultured according to the manufacturer's instructions.
Results: HPV decontamination of the unit took approximately 12 hours and was completed without incident or damage to the materials and equipment in the ICU. 9 (22%) of 41 areas cultured after cleaning but before HPV yielded MDR-GNR, including patient strains of E. cloacae and A. baumannii. These areas were sampled after HPV and yielded no MDR-GNR; however Bacillus sp. was cultured from 5 sites and Staphylococcus aureus (sensitive to methicillin) from 1 site. All BIs were killed by the process. No acquisition of MDR-GNR occurred on the unit for approximately two months after HPV decontamination. However, transmission of MDR-GNR has since been identified on the unit.
Conclusions: HPV proved to be more efficacious than conventional cleaning methods for reducing the bio-burden on our ICU. Although several sites remained contaminated following HPV, none were contaminated with MDR-GNR and we sampled large areas, including surfaces which may have been occluded from HPV. HPV appeared to break the cycle of transmission on the unit, but MDR-GNR re-occurred a few months later. The source of reintroduction is currently unknown.
P1693 Comparative incidence of fungal and bacterial infections in haematological patients occurring before, during and after the installation of HEPA filters in individual patient rooms
R. Araujo, A. Carneiro, J. Guimaraes, A. Rodrigues (Porto, PT)
Objective: Extensive intervention was undertaken in the Haemato-Oncology Unit of the Department of Clinical Haematology, Hospital S. Joao, in June 2005: 8 rooms with high efficiency particulate air (HEPA) filters and two 3-bed rooms were built, in order to prevent infections in high risk neutropenic patients. The objective of this study was to evaluate the incidence of fungal and bacterial infections in haematological patients admitted before, during and after the installation of the new wards.
Methods: All patients admitted in the department from April 2004 to December 2005 were enrolled. Data regarding haematological disease, length of stay, reason of admission, hospital acquired-infections, therapeutic options and, whenever, origin of infection were registered. Air surveillance was simultaneously performed using the Andersen one stage sieve impactor.
Results: A total of 287 admissions were registered in the department, corresponding to 171 patients, during the surveillance period. The incidence of fungal and bacterial infections was higher in the group of patients with acute leukaemia and aplastic anaemia, infections being associated to prolonged neutropenia. Mould infections, mostly due to Aspergillus spp., were reduced after the renovation works and installation of HEPA filters, as well as Staphylococcus infections. The period after renovation works also revealed a reduction of 16.7% of central venous catheter related infections and of 47.9% of microbial agents isolated from bronchial secretions of patients with acute leukaemia and aplastic anaemia. Interestingly, in the last period Candida infections were more common in patients admitted in wards with no air filtration system versus rooms with HEPA filters. The new rooms with HEPA filters showed large improvements on air quality, especially after the first week.
Conclusions: Patients with hospital stays longer than 3–4 weeks and with prolonged neutropenia are more susceptible to develop fungal infections, usually associated to the isolation of other microbial agents and the administration of wide spectrum antibiotics. The improvement of indoor air quality in hospital environment after the installation of HEPA filters in individual rooms effectively reduced invasive infections in haematology patients, mainly caused by Aspergillus spp. and Staphylococcus spp.
P1694 Risk of occupational infections due to occupational exposures in healthcare workers
S.A. Nemli, I. Ozgunes, S.N. Alpat, E.D. Kartal, N. Erben, G. Usluer (Eskisehir, TR)
Healthcare workers (HCWs) are exposed to the risk of occupational infections due to accidental exposures such as needlestick injuries (NSIs), sharps injuries (SIs) and contaminated blood or body fluids (CBBFs).
The objectives of this study were to determine the rates of bloodborne exposures and related risk factors experienced by HCWs at a University Hospital during 2005–2006.
One hundred and thirty HCWs filled out a questionnaire devised to determine the numbers of occupational exposures they had experienced and reported, condition of hepatitis B virus (HBV) immunisation, behaviours on performing procedures that can cause occupational exposures. 26.2% of them were male, 73.8% were female. 50% of participants were physicians and rests of them were nurses. 60% of participants had been working in internal medicine departments, 40% in surgical departments. 95.4% of participants determined at least one or more occupational exposure during their whole career. 79.2% of participants determined at least one or more occupational exposure during the preceding 1 year. 31.5% of them experienced SIs, 71.5% NSIs, 33.8% mucosal exposure of CBBFs. 90.4% of participants working in surgical departments experienced any occupational exposure during the preceding year and this was statistically significantly more compared to internal departments. Therefore 17.5% of participants exposed to accidental exposure during last one year reported these exposures, 12.3% of NSIs, 4.6% of CBBFs.
10% of participants had a history of previous HBV infection. 83.8% of participants had a history of vaccination and 70.7% were vaccinated three doses of HBV vaccine. 73.8% of participants were tested for HBV and 92.7% of them had protective anti HBV level 10 IU/mL or above. 59.6% of vaccinated participants had a history of vaccination after they started working.
Despite 83.1% of HCWs agreeing that using gloves decreases the risk of transmission of bloodborne pathogens, only 16.2% of HCWs reported they always wore gloves prior to any IV applications. 79.2% of HCWs reported that they feel themselves at risk about transmission of bloodborne pathogens. The ratio was significantly higher for nurses (58/65; 89.2%) compared to physicians (45/65; 69.2%).
These findings show that HCWs are at high risk for occupational injuries. Providing education about universal precautions and bloodborne pathogen exposure is crucial for the safety of HCWs. But it is difficult to change behaviours.
P1695 Hepatitis C virus kinetics during the acute phase of infection after occupational exposure in healthcare workers
M. Pape, K. Mandraveli, A. Politou, I. Isticoglou, S. Alexiou-Daniel (Thessaloniki, GR)
Objective: The aim of our study was to estimate the host immune response in the initial phase of infection after occupational exposure in healthcare workers (HCWs) and to evaluate HCV-RNA, anti-HCV and ALT levels as markers of acute infection and reliable predictors of infectivity.
Methods: Over a 4-year period (1998–2002), 134 occupational exposures of HCWs to possible infectious for HCV material, were reported in AHEPA University Hospital. Blood specimens from HCWs and source patients were drawn for the identification of their immune status to HBV, HCV and HIV. The HCWs were also followed up at 3, 6, 9, 16 weeks with repeated tests for anti-HCV, HCV-RNA and ALT. Anti-HCV immunoreactivity was confirmed by a line immunoassay. HCV-RNA was detected by using the Cobas Amplicor HCV v2.0 assay. HCV genotyping was performed in all HCV-RNA positive HCWs.
Results: Forty-six source patients were found to be anti-HCV positive. None of these were positive for HIV or HbsAg. In the remaining 88 of the injuries the source of the contact was not known. Among the 46 anti-HCV(+) patients, twelve (26%) had detectable HCV-RNA levels, with 1b genotype being the predominant one. One HCW demonstrated elevated ALT on 6th, 9th and 16th, two on 6th and one on 9th week, respectively. ALT levels were normalised during the remaining follow-up period. One HCW seroconverted for anti-HCV antibodies on 16th week and 6 months after exposure. Anti-HCV antibodies were also detected in two others HCWs on 6th and 9th and on 9th week, respectively. Two HCWs had detectable HCV-RNA on 6th, 9th, 16th and on 6th and 9th week, respectively. HCV RNA was detected in four HCWs only on the 6th week after exposure. All HCWs 6 and 12 months after exposure were HCV-RNA negative. Three HCWs were assigned to genotypes 1b, 2 and 3 respectively. Genotype could not be determined in the remaining HCWs. One HCW with detectable HCV-RNA received antiviral therapy (combination of ribavirin and interferon) and sustained virological response was defined by a negative HCV-RNA measurement until the end of the follow up.
Conclusion: The risk of acquiring HCV through occupational exposure is very low. It is difficult to establishe diagnosis of acute HCV infection with both serological and virological tests. HCV RNA is temporarily detectable during the follow up examination. Treatment decisions cannot be based only on HCV RNA measurements since a self-limiting infection is a possible outcome.
P1696 Prevalence and diversity of bacteria in root-filled teeth associated with periradicular lesions
A. Al-Ahmad, A-L. Liebenow, A. Wittmer, K. Pelz, E. Hellwig, J-F. Schirrmeister (Freiburg, DE)
Objectives: Persistence of microorganisms or reinfection are the main reasons for failure of root canal treatment. The knowledge of bacterial diversity in root-filled teeth can help to find the adequate treatment plan to eradicate microorganisms associated with periradicular lesions. The aim of this study was to isolate and identify microorganisms of twenty different root-filled teeth associated with periradicular lesions.
Methods: All teeth studied had been previously root-filled and showed radiographic evidence of periradicular disease. After removal of the root-filling material, samples of 20 cases undergoing retreatment were collected using paper points which were immediately transferred to reduced transport fluid (RTF). A quality control was performed to exclude contamination of the samples with saliva or dental plaque. Serial dilutions were plated on Columbia Blood Agar (CBA) and on Yeast-Cystein Blood Agar (HCB) to isolate aerobic and anaerobic microorganisms, respectively. Bile Esculin Agar was used to cultivate enterococci. The bacteria were identified by morphological and biochemical analysis. Additionally, identification by 16S rRNA-gene sequencing was applied.
Results: The samples of eight patients did not contain bacteria. In samples of 12 patients microorganisms could be isolated. The colony forming units/mL (CFU/mL) were in the range of 103–107 on CBA and of 103–109 on HCB, respectively. A mixed culture was found in the samples of 10 patients. Species of Enterococcus, Streptococcus, Peptostreptococcus, Campylobacter, Veillonella, Fusobacterium, Dialister, Bulleidia, Bacteroides, Eubacterium, Actinomyces, Propionibacterium, Serratia, Klebsiella, Porphyromonas, Mycoplasma, Gemella, Corynebacterium, Capnocytophaga, Vagococcus, Megasphaera, Atopobium, and Flexistipes were detected. In samples of two patients E. faecalis was the only isolated species. In two samples similar bacterial species were also found in the quality control which indicates a contamination of the revision samples by saliva or dental plaque.
Conclusions: Microorganisms could not be isolated from all teeth. The majority of positive samples revealed a mixed culture of 2 to 9 species. E. faecalis was the only detected agent in two patients underlining its suggested important role in endodontic infections. A quality control was essential to avoid false positive results which could be easily caused the contamination with saliva or dental plaque.
P1697 Reduced rates of vancomycin-resistant enterococci colonisation after implementation of infection control measures
L. Galani, V. Sakka, S. Tsiodras, M. Pantelaki, I. Galani, M. Souli, A. Antoniadou, S. Athanasia, F. V. Kontopidou, H. Nikolaou, H. Fytrou, N. Siafakas, L. Zerva, H. Giamarellou (Athens, GR)
Objective: The aim of the study was to evaluate the effect of infection control measures that were applied during one year period after an outbreak of ancomycin resistant enterococci (VRE) colonisation in our hospital.
Methods: A hospital wide prevalence study was performed during the 20 April–30 May 2005 period, recording faecal carriage and clinical VRE isolation. Presence of vancomycin resistance genes and species identification was assessed by multiplex PCR and clonality of isolates by PFGE. A case-control design, using two randomly selected VRE(–) controls for each positive case, was performed to identify cases with VRE colonisation and to evaluate risk factors for such colonisation. Control measures including patient cohorting, education efforts about hand hygiene and control of vancomycin use were instituted. An active surveillance problem was established for high risk patients identified through the case control study. A new survey was conducted in October 2006 in order to evaluate all control efforts.
Results: During the initial outbreak of colonisation 460 samples were evaluated from 367 patients. Total mean VRE carriage was initially 19.7%. All isolates were identified as E. faecium, with vanA genotype. Multivariate analysis identified immunodeficiency (OR 3.7, 95% CI: 1.4–9.3), any invasive device (OR 5.5, 95% CI: 2.2–13.9), and duration of antimicrobial treatment prior to VRE isolation (OR 1.2, 95% CI: 1.1–1.3) as the most important predictors for VRE positivity. During the 2nd screening 132 patients were screened and 11 were found to be positive for VRE colonisation (8.3%, p = 0.01). Of these, 4 (3%) were hospitalised in the orthopaedics department, 4 (3%) in the haematology unit and the other 3 in internal medicine wards. VRE colonisation was detected only in patients at high risk who had been already put in cohort for other multidrug resistance pathogens.
Conclusions: The implementation of infection control measures and an active surveillance screening programme targeting high risk patients resulted in a significant decrease of VRE isolation rates. High rates of VRE colonisation can be controlled by timely and intensively applied infection control and antibiotic use measures, preventing the emergence of clinical infections.
P1698 Intravenous cannulation in patients: room for improvement?
A.T. George, P. Turner, D. Defriend (Torquay, UK)
More than 90% of all patients admitted to hospital will have a peripheral cannula inserted. Complications associated with this procedure include extravasation, thrombosis and infection. The latter represents one of the most common preventable causes of MRSA bacteraemia.
To investigate both compliance with national guidelines on cannula insertion and care, as well as complications associated with use, 130 randomly selected medical/surgical patients with a peripheral venous cannula in situ were monitored daily for 8 days or until removal of the cannula. A non-interventional study was undertaken by gathering information on all aspects of care including site, size, use and indications, level of documentation, status of dressings, infection/thrombosis risk using the VIPS (Visual Infusion Phlebitis Score) as well as cannula changes.
Analysis demonstrated poor compliance with choice of non-dominant hand, cannula size, continuing indications for use. 95% of cannulae had no documentation regarding insertion, removal or changes. Compliance of cannulae maintenance was poor, with most of the cannula dressings becoming contaminated with blood and fluid or the cannulae being inappropriately secured with non-sterile adhesive tape and bandage dressings. A non-compliance with best practice for cannulae removal was seen even when the VIPS score was 2 or more, with a significant group being left in with a score of 3 – indicating active thrombophlebitis. Frank pus was seen on 2 dressings – in spite of which it was still being used. 22% of the inserted cannulae were never used. Most cannulae were left in situ for 48 or more hours after its use had stopped with a significant minority being left in for more than 120 hours.
This study shows that though protocols and national guidelines exist for use, maintenance and removal of peripheral cannulae, they may be overlooked and tend not to be followed. As a result, the potential for serious consequences such as nosocomial MRSA bacteraemia remain. The lack of documentation is also concerning, given that there is a legal requirement to do so. An improvement in peripheral cannula use, care and documentation is essential to ensure best clinical practice and reduce complications in vulnerable patients.
P1699 Inexpensive interventions can significantly reduce the number of unnecessary intravenous devices present in medical inpatients
D. Partridge, S. McBride, D. Scott, S. Briggs (Sheffield, UK; Auckland, NZ)
Objectives: Local and bloodstream infections are common complications of intravascular devices with significant associated morbidity and mortality. Many such devices are either inserted unnecessarily or their removal is delayed. Previous attempts to reduce the number of unnecessary devices have commonly centred around the education of medical and nursing professionals but rapid staff turnover and a multitude of other priorities impact negatively upon their effectiveness. We assessed the impact of the following inexpensive interventions on the prevalence of unnecessary intravenous devices amongst medical inpatients:
-
1
A short notice handed to patients with their breakfast menus emphasizing the potential need for intravenous cannulae but also mentioning the associated risks and urging the recipient to ask their physician whether the device could be removed.
-
2
A sticker inserted in the medical notes requiring the medical team to indicate whether a device inserted en route to hospital or in the Emergency Department should or should not be removed.
Methods: Strict criteria were defined to determine whether individual intravenous devices were necessary. Inpatients occupying a total of 100 beds on 4 General Medical wards were assessed on a daily basis for two weeks to identify the number of intravascular devices present and to determine how many of these devices were unnecessary. After a 5 week period, the above interventions were instigated and the assessment process was repeated for a further two weeks.
Results: Data was gathered on 1,144 patient days in the pre-intervention period. 592 of these were associated with a vascular access day, 233 of which were deemed unnecessary. Data was gathered on 1,153 patient days during the intervention. 436 of these were associated with a vascular access day, 144 of which were deemed unnecessary. The relative reduction in unnecessary vascular access device days was 39%, and an absolute reduction of 8% (p < 0.01), with an NNT of 12.
Conclusion: Inexpensive interventions targeted at patients as well as medical staff can significantly reduce the number of unnecessary intravenous devices present in general medical inpatients. We would suggest that these interventions would also, therefore, be likely to reduce the morbidity and mortality associated with intravascular device associated infections.
P1700 Use of intravenous catheters in a general hospital: results of a day-prevalence survey
C. Loupa, K. Vamvakoula, H. Demetriadi, E. Katsiki-Divari, K. Romanos, G. Petrou, M. Lelekis (Athens, GR)
Objectives: Intravenous (IV) catheters are considered a potential cause of problems such as thrombosis, fever and/or septicaemia. The aim of the present study was the evaluation of the conditions under which they are used, as well as of the appropriateness of their use in a general hospital of Athens, Greece.
Methods: On a specific day all hospitalised patients were examined for use of IV catheters. For every patient with such a catheter, an investigation was performed, concerning reason for use, duration of catheterisation, appropriateness of use and possible complications.
Results: Peripheral IV catheters were found in 116/203 hospitalised patients (age 70.9+15.4 years, 52% men). Eight patients had 2 catheters. There were no central venous catheters. The majority (69%) of the 116 catheterised patients were hospitalised in medical wards. The catheterised/hospitalised patients ratio was 63% vs. 57% for medical and surgical wards, respectively. Total days of catheter presence were 1–25 [5.4+5.1 in all catheterised patients, 6.0+5.6 and 3.9+3.2 for medical and surgical wards respectively, (x+SD, p = 0.01)]. Current catheter was in place already for 1–5 days (1.7+0.8 in total, 1.8+0.7 and 1.5+0.9 for medical and surgical wards, NS). Eighty-six per cent of catheterised patients had no restriction for food by mouth and 27% were not receiving IV medications or IV fluids on that day. In 27 IV catheters (23%) the date of insertion was not recorded, and in 11 cases (9%) local inflammation was observed. No significant difference regarding proportion of patients with inflammation signs was recorded, but insertion date was more frequently recorded in medical wards (p < 0.001).
Conclusions: (1) Peripheral IV catheter was present in more than half of hospitalised patients and (2) in a significant proportion of cases it was there “just in case …”. (3) IV catheters were used in a larger proportion of patients and remained for more days in medical wards compared to surgical ones. (4) Inflammation signs were noted in approximately 10% of IV catheters. (5) Education of hospital staff on proper use of IV catheters is probably needed.
P1701 Implementation of chlorhexidine gluconate for central venous catheter site care at a Bangkok hospital, Thailand
B. Balamongkhon, V. Thamlikitkul (Bangkok, TH)
Background: Central venous catheter-related blood stream infections (CRBSIs) are an important cause of patient morbidity, mortality, and increased healthcare costs. A meta-analysis and cost-effectiveness analysis of randomised controlled trials comparing chlorhexidine gluconate with povidone-iodine solutions for venous catheter site care found that the use of chlorhexidine gluconate significantly reduced the risk for CRBSIs and it was cost-effective. The objective of the study was to implement chlorhexidine gluconate for central venous catheter (CVC) site care in intensive care units (ICUs) at Siriraj Hospital.
Methods: The study was conducted in three ICUs at Siriraj Hospital from January to July 2006. One hundred and twenty adult patients who needed CVC insertions and received 2% chlorhexidine gluconate in 70% alcohol as the antiseptic solution for CVC site care were followed up daily for CVC related infections until the CVC had been removed for 48 hours and for any adverse effects of 2% chlorhexidine gluconate in 70% alcohol.
Results: The incidence of CRBSIs in the indwelling CVC patients who received 2% chlorhexidine gluconate in 70% alcohol was less than those who received 10% povidone-iodine during the same period, 3.2 vs 6.4 episodes per 1,000 CVC days (p = 0.06). The incidence of CRBSIs in the indwelling CVC patients who received 2% chlorhexidine gluconate in 70% alcohol was reduced by 36% to 43%. No adverse effects related to using 2% chlorhexidine gluconate in 70% alcohol were observed.
Conclusion: The locally produced 2% chlorhexidine gluconate in 70% alcohol was safe, effective and efficient for CVC site care in ICUs at Siriraj Hospital.
P1702 Prospective, randomised trial of three antiseptic solutions for prevention of central venous or arterial catheter colonisation in intensive care unit patients
J. Vallés, I. Fernandez, D. Alcaraz, E. Chacon, A. Cazorla, M. Canals, D. Mariscal, D. Fontanals, A. Moron (Sabadell, ES)
Objectives: Povidone-iodine is the most common catheter-site disinfectant, but accumulating evidence has indicated that chlorhexidine gluconate may be a more effective agent. On the other hand, 2% aquous clorhexidine gluconate is not available in our country.
The aim of this study was to compare the effectiveness in preventing central venous and arterial catheter colonisation of three protocols of skin antisepsis using 10% aqueous povidone-iodine (PI), 2% aqueous chlorhexidine gluconate (AC), and 0.5% alcoholic chlorhexidine gluconate (ALC) solutions.
Methods: Prospective and randomised trial in a medical-surgical intensive care unit (ICU) in a teaching hospital. All patients admitted to the ICU and requiring the insertion of a central venous and/or arterial catheter from January 1, 2005 to June 30, 2006 were included in the study. Patients were randomised assigned to each group according to the antiseptic solution used for insertion the catheter. Catheter distal tips were quantitatively cultured when catheters were no longer necessary and if there was a suspicion of catheter-related infection. Rates of catheter colonisation and catheter-related bacteraemia were compared in the three groups.
Results: A total of 631 catheters were included in the study (194 in PI group, 211 in AC group, and 226 in ALC group). The incidence of catheter colonisation was significantly lower in the ALC and AC groups than in PI group (14.2% vs 24.7% [RR 0.5, 95%CI 0.3 to 0.8, p < 0.01] and 16.1% vs 24.7% [RR 0.6, 95%CI 0.4 to 0.9, p:0.03], respectively). The incidence-rate per 1,000 catheter-days were also significantly lower in the ALC and AC groups than PI group (19.8 vs 31.8; p:0.02 and 21.8 vs 31.8; p:0.02, respectively). There were not difference among ALC and AC groups. Catheter-related bacteraemia were similar in the three groups. Both types of chlorhexidine solutions were superior to povidone-iodine solution in preventing catheter colonisations due to Gram-positive bacteria (p:0.03), whereas there were not difference in preventing Gram-negative bacteria colonisation.
Conclusions: We conclude that use of both ALC and AC solutions for disinfection of intravascular catheter sites reduces the incidence of catheter colonisation compared with 10% aqueous PI solution in an adult ICU. This effect seems to be related to the more efficacious prevention of colonisation with Gram-positive bacteria.
P1703 Mortality associated with bloodstream infection
R.A. Leth, J.K. Møller (Aarhus, DK)
Objectives: To analyse the seven-, thirty-days, and one year crude mortality rate of patients having had a blood-stream infection (BSI) with major groups of micro-organisms. Furthermore, to construct a survival curve for patients having had BSI with selected micro-organisms.
Material and Methods: All patients with a positive blood-culture at Skejby Hospital during the period 2001–2005 were included in the study. An episode of BSI was defined as a positive blood-culture with a significant pathogen and concomitant antibiotic treatment of the patient. A new BSI episode was defined as a new culture of the same microorganism after seven days or later or as growth of a new microbial species two days or more after previous positive blood-culture. Crude mortality was analysed in relation to the date of the first positive blood-culture. Seven, thirty-days and one year mortality rates were calculated. Blood-cultures representing positive repeat cultures or contaminants were excluded. Patient charts were checked for prescription of antibiotics and indications that the patient was regarded as having a true BSI. The date of death of patients was compiled from the patient administrative system. For the survival curves the Kaplan-Meier method was chosen.
Results: A total of 1,711 episodes of BSI were found. Thirteen patients were lost to follow-up. The overall crude mortality rates were: seven-days 5.4%, thirty-days 11.1%, and one year 24.4%. For E. coli (319 episodes) and S. aureus (300 episodes), the rates were: seven-days 7.8% and 5.7%, thirty-days 11.9% and 14.7%, and one year 26.6% and 30.7%, respectively. For fungi (81 episodes) the mortality rates were: seven-days 14.8%, thirty-days 27.2%, and one year 42.0%.
The crude mortality rates in relation to BSI episodes from various medical specialities A: cardiology and nephrology; B: urology and thoracic surgery; C: intensive care were: seven-days A: 4.7%, B: 7.3%, and C: 14.7%; thirty-days A: 11.3%, B: 15.2%, and C: 27.5%; and one year A: 32.7%, B: 33.4%, and C: 42.2%.
Conclusions: BSI is still associated with substantial short and long-term crude mortality rates. BSI caused by fungi seems to be associated with a higher crude mortality compared to E. coli and S. aureus. The mortality rates seem partly to reflect the population of the patients (medical specialities). The high mortality rates of patients in the intensive care unit coincide with a high rate of fungal BSI.
P1704 First outbreak of carbapenem-resistant Klebsiella pneumoniae in an Israeli university hospital
K. Hussein, H. Sprecher, T. Mashiach, G. Rabino, O. Eluk, I. Kassis, E. Braun, I. Oren, R.A. Fenkelstein (Haifa, IL)
Objective: To describe a preliminary data related to the emergence and spread of a Klebsiella pneumoniae (CRKP) strain in a large tertiary care university hospital in Haifa, Israel
Methods: Case-control study.
Results: Starting in January 2006, the prevalence of carbapenem-resistance (CR) among isolates of K. pneumoniae (KP) increased significantly in our hospital. Among 867 patients infected or colonised with KP during the first 10 months of this year, 48 (5.5%) were CR (cases). All isolates were susceptible to colistin and gentamicin but resistant to all other tested antibiotics. Fifteen isolates that underwent pulse-field gel electrophoresis were genetically related and different from two CRKP isolates previously identified at our institution. The outbreak was first identified in April 2006. Four possible index cases included 3 patients hospitalised in 2 geographically close orthopaedic wards and one transferred to our institution after renal transplant abroad. This patient had on admission a surgical site infection due to CRKP. By October 2006, the outbreak has spread to 16 wards. CRKP was isolated during or after admission to the General ICU in 32 of 48 cases (66.6%). Several characteristics (longer length of stay (LOS) before isolation of Klebsiella, admission to the General ICU, mechanical ventilation, central venous catheterisation, imipenem and vancomycin treatment) were found to be significantly (p < 0.05) more common among cases than among 48 infected or colonised carbapenem-susceptible KP control patients. In multivariable analysis, admission to the General ICU (OR, 3.5; [95% CI, 1.3–9.5]; p = 0.01), LOS of >7 days (OR, 3.2; [95% CI, 1.0–11.0]; p = 0.05), and previous treatment with vancomycin (OR, 5.1; [95% CI, 1.5–17.9]; p = 0.01) remained significantly associated with acquisition of CRKP.
Conclusions: CRKP has emerged as an important pathogen in our institution. Despite implementation of several important infection control measures, the outbreak has not been contained. Because these isolates are resistant to almost all commonly used antibiotics, control of their spread is crucial. In this study, prolonged LOS, admissions to ICU and previous use of vancomycin were significantly associated with the acquisition of CRKP at our hospital.
P1705 A pseudo-outbreak with Mycobacterium intracellulare
M. Van De Vyvere, J. Vanpellicom, M. Verschueren, K. Camps, L. Winnepenninckx, P. Bomans (Antwerp, BE)
Problem: In the autumn of 2005 we noticed a sharp increase in the isolation of Mycobacterium avium complex (MAC).
Objectives: Finding the cause of this increase to obtain a solution to stop the outbreak or pseudo-outbreak.
Methods:
-
•
investigation of the culture data
-
•
mycobacterial culture of the bronchoscopes (secondary on the results of the data investigation)
-
•
mycobacterial culture of the rinsing water of our two washers (decontamination procedure with peracetic acid)
Results: Because of the slow growth of the strain and the isolation of it in samples of immunocompromised patients (which is not exceptional) it took us a (too) long time before we were aware of the problem. Meanwhile a lot of other samples were taken. 32 MACs were isolated from September 2005 until February 2006, with a peak in november. All strains (minus 1) were isolated from samples of patients of the same hospital (delivering 35% of the samples for mycobacterial culture). All samples were acquired by bronchoscopy. All MACs were later identified as Mycobacterium intracellulare (MI). Only samples taken from the bronchoscopes after decontamination in the most recently bought washer showed growth of MAC(MI). In the same period a warning of possible contamination of the rinsing water of this type of washer was given by the manufacturer. It was recommended to extend the length of the procedure until the washer was ‘upgraded’ by the company. The automatic decontamination procedure was changed by a manual procedure. Samples of the rinsing water of the new washer showed growth of MAC(MI). After the upgrading of the washer all cultures taken from scopes and rinsing water were negative. No growth of MAC(MI) in patient samples was seen from the time the cause of the problem was suspected and measures were taken.
Conclusion: Rinsing water of automatic washers can be contaminated with mycobacteria. This contamination can be the cause of a pseudo-outbreak. It is not excluded that it could be the cause of real infection by installing mycobacteria directly in the lungs during bronchoscopy, especially in immunocompromised patients. An automatic warning system should be implemented to give an early warning of an increase in isolation of specific strains. If surveillance cultures of scopes and washers are done: culture of mycobacteria in the rinsing water is most important.
P1706 A case of Ralstonia paucula septicaemia in an intensive care patient following blood transfusion via a level-1 fast flow fluid warmer, successfully treated with meropenem
B.M. Clark, R. Townsend, P. Norman (Sheffield, UK)
Case: An 80-year-old woman on warfarin therapy underwent emergency surgery for a suspected leaking aortic aneurysm. Instead a tear in the lower pole of the right kidney was found and repaired. She had 2 cardiac arrests during the operation and was resuscitated with fluid infused via a Level-1 Fast Flow Fluid Warmer, including 7 units of blood. Following the operation it was noted that the water bath of the fluid warmer, which warms the infused fluid, had been contaminated with blood. IV (Intravenous) cefuroxime and metronidazole were commenced.
She was transferred to the ICU (Intensive Care Unit) for invasive ventilation and inotropic support. On the second day she became pyrexial and blood cultures were taken, from which a Gram-negative bacteria was grown. Antibiotics were changed to IV pipericillin-tazobactam. On the third day she developed septic shock. The bacteria had now been identified as a likely environmental organism, but given the possible contamination of the patient's blood transfusion this was thought to be relevant and IV meropenem was commenced. She improved clinically and received a total of 6 days of meropenem. She was eventually discharged home after a lengthy hospital stay, including 41 days on the ICU.
Subsequently the organism was identified as Ralstonia paucula using API and confirmed by the reference laboratory (Colindale, UK). Unfortunately the fluid warmer used in the operation was sterilised immediately after the operation. However, the water bath from a second fluid warmer was sampled. This was in the same operating theatre as the other fluid warmer, and had been filled from the same water source and maintained in the same way. From this we also cultured R. paucula.
Discussion: We propose that contamination of transfused blood possibly occurred during the operation via the Level-1 Fast Flow Fluid Warmer. The water bath of the fluid warmer used in the operation was blood stained, and we believe conversely that fluid from the non-sterile water bath, containing R paucula, was infused into the patient. We will illustrate how this may have occurred and will also describe the organism R. paucula in greater detail.
This case has wide reaching implications as fast flow fluid warmers are used in hospitals across the UK and Europe. Whilst they have contributed to the saving of countless lives during medical and surgical emergencies we believe that this case illustrates an important infection control issue regarding the use of this equipment.
P1707 Report of multicentre outbreak of Buffalopox virus infection in burn units, Karachi, Pakistan
A. Zafar, R. Swanepoel, R. Hewson, M. Nizam, A. Ahmed, A. Husain, A. Grobbelaar, K. Bewley, V. Mioulet, B. Dowsett, L. Easterbrook, R. Hasan (Karachi, PK; Sandringham, ZA; Salisbury, UK)
Objectives: Sporadic outbreaks of Buffalopox (BPV) and its transmission among farmers have been reported from the subcontinent. We describe the first multi-centre nosocomial outbreak of BPV infection amongst burns patients in a major city of Pakistan.
Method: Notification of outbreak: Atypical lesions involving burns wound were first noted in a patient at a burns unit in Karachi. Similar lesions rapidly affected other patients within this unit. During infection control investigation, patients with similar infections were being reported from other burns centres in the city. Investigation: Clinical specimens from the lesions were submitted for routine culture and for histopathology. Impression smears, material from the lesions and biopsy tissue were used for electron microscopy, DNA sequencing and in-vitro culture.
The epidemiological finding suggested a common origin of infection. To investigate this, practices at the affected burns units were observed and materials in common use for cleaning and dressing wounds at all the various centres were collected for testing.
Results: Clinical samples submitted for bacteriological and mycological culture proved negative. On histopathological examination no Molluscum bodies were found. Electron microscopy revealed the presence of orthopox virus particles. Examination of ultrathin sections of infected Vero cells demonstrated classic orthopox virus maturation with intracytoplasmic ‘virus factories'. Portions of the gene were sequenced and analysed. The resulting phylogenetic tree grouped the sample within the Buffalopox sequences. All surveillance was reported negative. Nevertheless, clinical samples submitted from several centres revealed the existence of the same pox virus.
To control nosocomial transmission in the units, a number of measures were taken including staff education, reinforcement of infection control practices and strict isolation measures which successfully controlled the disease spread within the primary unit.
Conclusion: We report first nosocomial outbreak of BPV infection among patients admitted to different burns units located significant distances apart. Initial investigation could not reveal the source but it is very likely that nosocomial transmission of the viral infection was due to frequent transfer of exposed patients between the different units. This hypothesis is supported by the fact that strict infection control measures in one unit succeeded in controlling transmission.
P1708 Colonisation by resistant Staphylococcus epidermidis strains in patients qualified for routine vascular reconstructions
E. Mazur, J. Niedzwiadek, A. Niwinska, P. Terlecki, J. Wronski, M. Koziol-Montewka (Lublin, PL)
Objectives: The aim of our study was to examine S. epidermidis colonisation of the anterior nares in patients qualified for routine vascular reconstructions on the day of admission to the hospital and after a week of staying there and to evaluate antimicrobial susceptibility of isolated strains, including detection of resistance mechanisms.
Methods: The study included 35 patients with PAOD (peripheral occlusive arterial disease) qualified for routine vascular reconstruction. Nasal swabs were obtained on the day of admission and on 8th day of hospital stay. Classification to the species was done on the ground of the biochemical properties of isolated strains. Susceptibility to antibiotics was assessed by disc-diffusion method.
Results: S. epidermidis was present in all but one admission culture, in 8th day cultures this species was isolated from 33 patients. In every performed culture 1 to 3 different strains of S. epidermidis were isolated. In 6 patients no changes in S. epidermidis strains were noticed comparing first and second culture, in 7 ones a total change of isolated strains occurred during hospital stay. As a total 73 strains of S. epidermidis were isolated, 26 strains were present in both cultures, 23 strains were acquired in the hospital, 24 strains were lost during hospitalisation. 17 resistant strains were brought into the hospital, among them 10 MR (methicillin-resistant), 10 MDR (multidrug-resistant) and 6 MLSB (some resistance mechanisms were present in the same strain simultaneously). During hospital stay the examined patients were colonised with 12 resistant strains of S. epidermidis, among them 11 MR, 8 MDR, 4 MLSB. 28.8% of strains were resistant to methicillin (MRSE), all of them were fully susceptible to vancomycin, teicoplanin, and mupirocin. 13.7% of isolated S. epidermidis strains possessed MLSB resistance mechanism. 24.7% of S. epidermidis strains were multiresistant, 72.2% of them were simultaneously methicillin-resistant.
Conclusions: Due to high rate of resistant strains among S. epidermidis isolated from patients prepared for routine vascular reconstructions it seems useful to evaluate nasal colonisation in every subject before and after such intervention to:
-
1
have the opportunity to verify perioperative antibiotic prophylaxis in selected cases and to prevent possible complications;
-
2
limit the expansion of resistant strains of S. epidermidis in the hospital.
P1709 The activity of five commonly used topical antiseptic agents on epidemic and sporadic strains of Acinetobacter baumannii
E. Demertzi, G. Avraamides, B. Azadian, R.L.R. Hill, J. Turton, T.L. Pitt (London, UK)
Objectives: Acinetobacter baumannii is increasingly being recognized as a cause of outbreaks of opportunist infections in intensive care patients in hospitals. These organisms contaminate the inaminate environment and may survive for prolonged periods on surfaces. Contact with these surfaces by healthcare personnel may lead to contamination of their hands and facilitate the transmission of the organisms to others. A number of topical antibacterial agents are used for hand hygiene or as surgical disinfectants and so we questioned whether selected agents were equally effective against strains of A. baumannii prevalent in British and European hospitals.
Materials and Methods: A panel of 30 strains was assembled comprising 11 characterised epidemic strains (SE clone, Oxa-23 clones 1 and 2, European clones, etc.) 12 representatives of more minor outbreak strains, and 7 sporadic isolates. The following products (active agent) were tested:
-
1
Purell gel (62% ethyl alcohol),
-
2
Desderman gel (78.2% ethanol + 0.1% phenylphenol),
-
3
Sterzac bath liquid (2% Triclosan, isopropyl alcohol etc.),
-
4
Hydrex (0.5% chlorhexidine gluconate w/v in 70% v/v DEB),
-
5
Betadine (10% povidone-iodine).
Agents were tested for activity using the prEN 12054 European standard method. Standardised bacterial test suspensions (1 mL) were mixed with products (9 mL) and survival at 30 s, 1 min and 5 min was determined by viable counts on agar.
Results: The minimum requirement for bactericidal activity for compliance with prEN 12054 is a 10(5)-fold reduction in viable count within 1 min. Each product tested fulfilled prEN 12054.
We conclude that A. baumannii are killed by commonly used topical antiseptics and hand hygiene should be effective in limiting their spread in hospitals.
P1710 Infection risk for filamentous fungi in water in a paediatric haematology/oncology ward
H. Kennedy, C. Williams (Glasgow, UK)
Objectives: Filamentous fungi from the air, particularly Aspergillus spp., cause severe opportunistic infection in immunocompromised patients. However, there is now some data to suggest that hospital water may also be a potential source of these organisms. The present study examined tap water in a paediatric haematology/oncology unit for the presence of fungi.
Methods: Tap water (hot and cold) samples from three rooms in the haematology/oncology ward were collected and passed through sterile 0.45um filters (Millipore). Filters were then transferred to Sabouraud dextrose agar plates supplemented with chloramphenicol and incubated for one week. Fungal isolates were identified by macroscopic and microscopic characteristics. This procedure was repeated one month later. Any fungal isolates, which could not be identified in-house, were sent to the HPA Mycology Reference Laboratory, Bristol, UK for identification.
Results: Filamentous fungi were cultured from eleven of the twelve water samples. Exophiala was isolated from 83% of the water samples, Fusarium (not F. solani or F. oxysporum) from 25%, Verticillium from 17%, Phoma from 25%, Penicillium from 8% and a phialidic mould which could not be identified precisely but which resembled Fusarium was isolated from 92%. The distribution of fungi from the two sampling periods was similar. There was no evidence of Aspergillus spp.
Conclusion: This study demonstrated that various filamentous fungi were present in the water system but that the major opportunistic pathogen Aspergillus was absent. Exopiaila, Fusarium and Verticullium spp. are widely distributed in soil and plants and may be found in aqueous environments. The absence of Aspergillus spp. is desirable and is probably influenced by the water storage and distribution system (following chlorination), which prevents exposure to large volumes of air from which Aspergillus spores could potentially cause contamination. Further studies on filamentous fungi in hospital water systems are required to investigate the organisms' ability to persist, their role in biofilm formation and their clinical significance.
P1711 Reduction of healthcare-associated infection rates in a Swiss university hospital following hand hygiene promotion
I. Uçkay, D. Pittet, C. Ginet, H. Sax (Geneva, CH)
Background: Hand hygiene is considered the single most effective measure to reduce healthcare-associated infection.
Objective: To measure the prevalence of healthcare-associated infection at the University of Geneva Hospitals before and during participation in a national hand hygiene promotion campaign (www.swisshandhygiene.ch).
Methods: Two hospital-wide period-prevalence studies including all healthcare-associated infections according to CDC definitions were conducted in May 2004 and May 2006. The assessment took place two years before and four month after the launch of a multimodal promotional hand hygiene campaign. An experienced infection control team collected data from all hospitalised patients (except for the psychiatric service) according to a standard, unchanged protocol. Other major infection control strategies were ongoing unchanged in parallel since 2003.
Results: In 2004, 1,494 patients were included in the study; in 2006, there were 1,560 patients. Overall compliance with hand hygiene recommendations improved from 57% to 64% (p < 0.001). Patient's case mix between the periods was equivalent based on McCabe score (p = 0.2). The prevalence of nosocomial infections decreased by 20% hospital-wide, from 12.8% to 10.2% (OR, 0.77; p = 0.03; 95% CI, 0.61–0.97). It decreased from 11.8% to 9.6% in the acute care sectors, and from 13.9% to 11.0% in the long-term sectors. The three most prevalent infections in 2006 were urinary tract (26%), respiratory tract (26%) and surgical site infections (19%). By assuming an average attributable cost of €2,200 per infection, saving equalled at least €1.8 million in 2006.
Conclusion: A multimodal hand hygiene promotion campaign was associated with a successful reduction of healthcare-acquired infections, in both acute and long-term care sectors. Long-term follow-up is required to assess the continuous impact of the strategy.
P1712 Layperson's technique of hand washing: is it effective in reducing bacterial carriage?
Z. Sekawi, F. Jamal (UPM Serdang, MY)
Introduction: In spite of major advances in the diagnosis, treatment and prevention of infectious diseases, cross infections in hospitals continue to be a major problem worldwide. Frequent hand washing among healthcare personnel using a proper prescribed technique has been shown to reduce the spread of causative agents of these infections. More often, this technique comprising several systematic steps is considered a nuisance to some personnel and therefore, compliance to this technique may not be desirable and will pose problems such as increase in hospital acquired infections. To encourage compliance, a simple routine household technique of hand washing may be considered as an alternative albeit less superior. Is a layperson's technique of hand washing effective in reducing bacterial carriage?
Objectives: To determine bacterial flora of hands of staff and students in Faculty of Medicine and Health Sciences, Universiti Putra Malaysia and to determine the effectiveness of layperson's hand washing on bacterial carriage.
Method: A cross-sectional study with non-probability convenience sampling was carried out at the faculty premises. Two hundred and twenty eight participants were asked to inoculate their right hand fingertips on the blood agar plates before and after washing their hands with antimicrobial soap using their own usual routine technique. Instruction on proper hand washing was not given. The agar plates were then incubated at 37°C for 18–24 hours and inspected by standard methods.
Results: All participants but four (98.2%) had Staphylococcus aureus on their hands while 80 (35.1%) participants had Bacillus spp. before washing. Other organisms such as Streptococcus spp., Corynebacterium spp., unidentified Gram-negative bacteria and fungus were found on four participants. One hundred and sixty-three (71.5%) of 228 participants had reduction in bacterial hand flora after hand washing. Socio-demographic factors such as gender, age, ethnic groups, occupation and income were analysed and found to be statistically insignificant.
Conclusion: It is shown that simple routine household technique of hand washing can reduce the carriage of bacteria on the hands considerably. This implies that healthcare worker's ignorance on the proper hand washing technique but using their routine layman's technique of hand washing is better than not washing hands at all during their course of their duty.
P1713 Evaluation of a new ultra-rapid hand drier in relation to hand hygiene
A.M. Snelling, T. Saville, D.G. Stevens, C.B. Beggs (Bradford, Malmesbury, UK)
Objectives: Handwashing is a key element of infection control in the clinical and domestic settings. Whilst much research focuses on antibacterial wash products, or increasing compliance, little attention has been paid to the drying step. However, washing does not always remove 100% of contaminants, and can increase levels of commensal bacteria on the skin surface. The new AirbladeTM drier uses two high pressure air ‘knives' to strip water from still hands. Drying takes ∼10 seconds. Conventional warm air driers rely on evaporation from the skin and often take ≥30 s with rigorous hand rubbing to achieve a satisfactory effect. The study aimed to compare the two types of drier in terms of residual bacterial load on hands.
Methods: The Airblade (Dyson Ltd) was tested against 2 conventional warm air driers widely used in the UK. 14 (7M, 7F) volunteers took part. In study (A) hands were contaminated by handling uncooked chicken. Washing was performed using the Euro-standard handwash (EN1499:1997) + non-medicated liquid soap. After using a drier, the fingers of each hand were pressed onto a strip (10×4 cm) of sterile aluminium foil. Bacteria transferred to foil were eluted into MRD + soap inactivator, and enumerated on TSA plates. Volunteers repeated the protocol with each machine, in random order. Study (B): clean hands were washed in the same way but without soap. Drying was performed with the machines +/– hand rubbing. Agar contact plates were used to enumerate bacteria on palms, fingers, fingertips before and after drying.
Results: In study (A) bacterial transfers of 0–107 cfu/hand were observed. For a set drying time of 10 s the Airblade led to significantly less bacterial transfer than with the other driers (p < 0.05). When the latter were used for longer (30–35 s) the trend was for the Airblade to still perform better, but results were not as significant (p > .05). In study (B) rubbing hands whilst using the driers counteracted the reduction in overall bacterial numbers at all anatomical sites. When hands were held still, there was no statistical difference between the driers.
Conclusions: If hands are not dried properly after washing, transfer of commensals or remaining contaminants to other surfaces is more likely to occur. The Airblade was superior to the conventional driers for reducing transfer. Its short drying time should encourage greater compliance with hand drying and hence help reduce the spread of infectious agents by the hand-borne route.
P1714 Compliance of healthcare workers with hand hygiene rules in the emergency room of two tertiary hospitals in the area of Athens
S. Athanasia, E. Giannitsioti, A. Kolias, O. Koulouri, H. Fitrou, H. Giamarellou (Athens, GR)
Background: The aim of this study is to compare the influence of hand hygiene policies on Healthcare workers (HCWs) in the emergency rooms between two tertiary general hospital at the area of Athens. Hand hygiene policies were launched in both Institutions.
Patients and Methods: To evaluate the impact of hand hygiene policies in the emergency rooms of two tertiary hospitals named A (400 beds) and B (330 beds), behaviour of physicians and nurses regarding the use of gloves and the use of alcohol-based hand-rubs was assessed by two independent observers via a standardised registry form. The study was conducted during a two month period. Alcohol based hand-rubs have been installed on the tables in every room. Results were analysed by chi-square and fisher exact tests for categorical variables.
Results: Physicians of Hospital A were more prompt to use gloves than physicians of Hospital B [26/53 (49%) vs 55/182 (30.2%), p = 0.01]. No difference in the use of gloves was observed between nurses of Hospital A (27/47, 57.4%) and nurses of Hospital B (72/109, 66%). Nearly all HCWs were changing gloves between each patient. Among physicians who did not wear gloves, the rates of those that did not use antiseptic were similar in both hospitals [Hospital A, 13/27 (48.2%), Hospital B, 87/127 (68.5%), pNS]. Nurses were equally not using antiseptic before and after contact with the patients in both Hospitals. However, in Hospital A, nurses applied more frequently antiseptic after contact with the patient than nurses in hospital B [Hospital A, n =11/20 5 (55%), Hospital B, 2/109 (1.8%), p < 0.001].
Conclusions: Despite the same policies regarding hand hygiene which were applied in both hospitals A and B, compliance of HCWs with the appropriate use of antiseptic on hands was extremely low in the emergency rooms. Special issues for the emergency room and continuous surveillance and education are required in order to change compliance of HCWs with hand hygiene and enhance the infection control.
P1715 Questionnaire results of a survey on health workers' knowledge and attitudes to nosocomial infections
A. Dogru, P. Kaymaz, S. Baris, N. Kilicarslan, T. Zengin Elbir (Istanbul, TR)
Objectives: Our aim is to search the knowledge capacity and behaviour types of healthcare workers about nosocomial infections and indicate the wrongs and defects in applications.
Materials and Methods: We applied our questionnaire of 24 questions on 200 health workers of whom 70 (35%) were doctors, 107 (53.5%) were nurses and 23 (11.5%) were other healthcare workers (pharmacists, biologists, technicians, laboratory asistants).
Results: Most of the participants (98%) defined AIDS as the most important blood-transmitted illness, and 66.3% of them said “i'll be more careful about the hygiene rules” while giving care to the patients with blood-transmitted infectious diseases. 55.1% of nurses express that they “always” adhere to the rules aimed at avoiding nosocomial infections, and 55.74% of doctors responded to this question as “frequently”. It was thought that (77.04%) intensive care units were the most risky units about nosocomial infections, and handwashing with 55.73% was thougth to be the most important precaution in avoiding the nosocomial infections. According to participants, with 82.67% pneumonia was the most important nosocomial infection and most important agent was Pseudomonas genus bacterias with 86.95%. 35.71% of doctors and 40.75% of nurses defined that they “always” wash hands before and after touching a patient. 62.12% of participants indicated that “they are not able to find material everytime when it is needed” as a reason for not using barriers while contacting with patients. 62.31% of doctors and 77.58% of nurses expressed that they had contact with patients' blood and/or extracts during their professional life. With 62.5% “tapping the injectors plastic cover” became the most important process as injury/contact reason.
Conclusions: Participants have enough knowledge about nosocomial infections and precautions. We determined that those who had education about the subject gave correct answers in higher proportions. Thus, we decided to increase the portion of education and seminars in addition to the routine duties of infection control committee, aiming to avoid the defects that we determined during our applications. Additionally, using notice boards we will hang in every service, we are planning to acquaint the healthcare workers with different subjects every month. So, in one year duration, we are planning to lower the nosocomial infection rates of our hospital.
P1716 The impact of an intervention on nurses' HIV/AIDS knowledge and compliance with universal precaution procedures in emergency department in a university hospital, Shiraz, Iran
N. Pasyar, S. Gholamzadeh (Shiraz, IR)
Introduction: Emergency department nurses are potentially exposed to blood in the course of their work and therefore are at risk of infection with bloodborne pathogens, including HIV.
Methods: A quasi-experimental survey was carried out amongst 120 nurses. The intervention consisted of 1-day training workshop, that consisted of lecture, and focus group discussion. Each of these nurses was asked to answer pre- and postsession knowledge questions during three periods of time (before training, immediately after training and three months later). Compliance with universal precaution (UP) was measured through 11 items and data were gathered from observation. Paired t-tests were used to compare differences between the pre- and postsession knowledge scores and compliance with UP.
Results: The comparison in three periods revealed that the knowledge of personnel significantly increased immediately and three months after the intervention compared with before (P < 0.0001). The nurses' knowledge scores increased from 68.9% before training to 100% immediately after and 95% 3 months after the training programme (p < 0.0001). There was a statistically significant difference in the knowledge of HIV and the implementation of universal precaution (P < 0.0001). Observed compliance with universal precaution procedures before and after training workshop ranged from 71.7% to 98% for glove use, 75.5% to 99% for handwashing after glove removal, 53.8% to 83% for wearing mask, 78.3% to 87.7% for not making use of needle cutter. The results also indicated that some nurses (37.7%) still recapped needles.
Conclusion: The training session significantly improved the nurses' knowledge and implementation of universal precautions among nurses in emergency department.
Automation in bacteriology
P1717 Comparison of the Plus Aerobic/F and Plus Anaerobic/F media using the BacTec 9240 system with an in-house agar-broth biphasic selective fungal culture media (modified from the BACTEC Myco/F Lytic media) for the diagnosis of fungaemia
C.Y. Low, A.L. Tan, B.H. Tan (Singapore, SG)
Objectives: To compare the Plus Aerobic/F and Plus Anaerobic/F media using the BACTEC 9240 system with our in-house agar-broth biphasic selective fungal culture media (modified from the BACTEC Myco/F Lytic media) for the diagnosis of fungaemia.
Methods: This is a retrospective study of patients admitted to the Singapore General Hospital over a 27-month period, from April 2004 to June 2006, who had at least one positive blood culture for fungi using the Plus Aerobic/F, Plus Anaerobic/F or the agar-broth biphasic fungal culture media. Comparative analyses were made exclusively on blood culture sets, a set being made up of one pair of BACTEC Aerobic and Anaerobic vials and one agar-broth biphasic fungal culture vial, sampled from the patient at the same time, from the same site, and processed simultaneously in the laboratory. We compared the following: Positivity rate by bottles – either BACTEC bottle in the set detecting fungaemia was taken as positive and compared with the selective fungal culture media. Positivity rate by patient – cases of fungaemia by the patient were compared. Time to positivity based on preliminary laboratory reporting – the earliest day reported by the laboratory for the detection of yeast or mould was compared. Time to positivity based on final laboratory reporting – the day of the final report by the laboratory for the detection of fungaemia was compared.
Results: There were 141 sets of blood cultures from 86 patients available for comparison. There were no statistical differences in the positivity rates between the BACTEC media and the selective fungal culture media, either by bottle (75.9% versus 69.5%, p = 0.285) or by patient (69.8% versus 75.6%, p = 0.494). There was poor agreement (kappa, measure of agreement = −0.369) between the two culture media for the diagnosis of fungaemia. The mean time to positivity on the BACTEC media was significantly shorter than on the selective fungal culture media, both in preliminary (3.3±1.9 days versus 8.0±6.2 days, p < 0.0005) and in final laboratory reporting (5.6±1.9 days versus 9.7±6.1 days, p < 0.0005). The fungaemia-related mortality in the same admission was 54.7% (47/86).
Conclusion: The Plus Aerobic/F and Plus Anaerobic/F BACTEC media have similar sensitivities to our in-house agar-broth biphasic fungal culture media in detecting fungaemia but does so in a significantly shorter time. The poor agreement between the two culture media suggests that they complement each other in the diagnosis of fungaemia.
P1718 Evaluation of the VITEK® 2 system for testing of Gram-negative bacilli directly from BacT/Alert Fan blood cultures
M. Ullberg, J. Bergström (Uppsala, SE)
Objectives: The usefulness of the VITEK 2 system for rapid diagnosis of Gram-negative bacilli in blood cultures was evaluated. The aim was to deliver species identification and susceptibility results to the wards one day earlier than would be feasible using conventional methods.
Methods: Blood cultures were processed by a BacT/ALERT 3D system using FA and FN bottles. Bacteria from cultures showing Gram-negative bacilli on the smears were concentrated by centrifugation in two steps (150g and 2200g for 10 min respectively) and then analysed in the VITEK2 system. The new colorimetric GN cards were used for identification and AST-N029 cards for antibiotic susceptibility testing. Results were compared with standard procedures using an in-house fermentation system for identification and disc diffusion for susceptibility testing.
Results: A total of 67 Gram-negative cultures were investigated. 96% of the collected strains were correctly identified. Complete susceptibility results were obtained within the time limit of 8 hours for 76% of the strains. Susceptibility results in general correlated well with disc diffusion data. Nineteen of 67 (28%) patients were found to receive an inappropriate antibiotic therapy considering Gram smear and VITEK 2 results. Twelve of those patients would have remained on improper therapy for one more day if standard therapeutic regimen had been applied. These cases included 5 ESBL-producing Klebsiella pneumoniae, 2 multiresistant Escherichia coli, one multiresistant Citrobacter braakii, one Stenotrophomonas maltophilia, one Pseudomonas aeruginosa and 2 Salmonella sp.
Conclusions: The VITEK 2 system proved to be a valuable tool for direct identification and susceptibility testing of Gram-negative bacilli recovered from BacT/ALERT FAN blood cultures. By using the VITEK 2 system, 12 of 67 patients could receive an appropriate antibiotic treatment one day earlier than if standard in-house diagnostic procedures had been implemented.
P1719 Validation of VITEK2 GN Cards and VITEK2 Version 4.02 Software for identification and antimicrobial susceptibility testing of nonfermenting Gram-negative bacilli from cystic fibrosis patients
I.M. Otto-Karg, S. Jandl, T. Müller, B. Stirzel, U. Vogel, M. Frosch, H. Hebestreit, M. Abele-Horn (Würzburg, DE)
Objectives: Accurate identification (ID) and antimicrobial susceptibility (AST) testing of bacterial isolates from cystic fibrosis (CF) patients is known to pose problems. Commercially automated test systems are available, but have not been recommended so far. Aim of this study was to evaluate the VITEK 2 GN card for ID and the new VITEK 2 version 4.02 software for AST of nonfermenting Gram-negative bacilli from CF-patients compared to reference methods.
Methods: A total of 168 strains for ID and 117 strains for AST were investigated. Clinical isolates from CF-patients were identified with the VITEK 2 GN card and with the API NE system. If ID results differed, molecular methods were used for definitive identification. Broth microdilution method served as reference method for AST against nine antibiotic agents including cefepime, ceftazidime, piperacillin, imipenem, meropenem, gentamicn, tobramycin, ciprofloxacin, and cotrimoxazole. Interpretation of the AST results was performed according to the DIN 58940 standard.
Results: Of the 168 strains tested 143 yielded concordant results. The discordant 25 strains underwent 16S rRNA gene sequencing. The VITEK2 GN card identified 12 of 25 strains correctly to the species level (155/168, 92%) and nine strains correctly to the genus level (164/168; 98%); 4 (2%) strains were misidentified. In contrast, the API NE correctly identified 4 of those strains to species level (147/168; 88%), 9 strains to genus level (154/168; 93%) and misidentified 12 (7%) strains. Regarding AST, overall categorical agreement of VITEK 2 compared to the reference method was 97% for all antibiotics. Minor errors were found in 8%.
Conclusion: The new VITEK2 GN card appears to be a reliable method for identification of nonfermenting Gram-negative bacilli from CF patients. Moreover, the new VITEK2 version 4.02 software showed good results when compared to reference method for AST and appears to be applicable for routine clinical use in AST of nonfermenting Gram-negative bacilli of CF patients.
P1720 Evaluation of the new VITEK2 NH identification card
G. Valenza, C. Ruoff, U. Vogel, M. Frosch, M. Abele-Horn (Würzburg, DE)
Objectives: The purpose of this study was to evaluate the ability of the new VITEK2 NH card (bioMérieux, Nürtingen, Germany) to identify fastidious Gram-negative bacteria in comparison to reference methods.
Methods: A total of 149 strains belonging to the species listed in the data base of the VITEK 2 software were investigated, including Neisseria spp., Haemophilus spp., Campylobacter spp., Capnocytophaga spp., Cardiobacterium hominis, Eikenella corrodens, Gardnerella vaginalis, Kingella denitrificans, and Moraxella catarrhalis. Thirty-one strains were originated from the strain collection of the German National Reference Centre for Meningococci (NRZM), from the American Type Culture Collection (ATCC), and from the German Collection of Microorganisms and Cell Culture (DSM). The remaining 118 strains were own clinical isolates, which previously had been identified by API NH system or other conventional methods. The new VITEK2 NH cards were used according to the manufacturer's recommendations. The cards were filled with organism suspensions made in 0.45% aqueous NaCl to turbidity equivalent to a McFarland #3 standard. Identification results were generated by a computer-assisted algorithm. In the case of conflicting results, 16S rRNA gene sequencing (16 S) method was used for genetic identification.
Results: The new VITEK 2 NH card was able to identify 142 (95%) strains to the genus level and 138 (92.7%) strains to the species level. Six (4%) strains were misidentified and five (3.3%) strains could not be identified. 19 of 21 pathogenic Neisseria isolates were correctly identified.
Conclusion: The new VITEK2 NH card appears to be a useful method for the identification of fastidious Gram-negative bacteria and seems to be an improvement over the conventional diagnostic methods of some fastidious microorganisms. More work is needed to finally evaluate the performance of VITEK 2 with regard to pathogenic Neisseria.
P1721 Species identification with the BD Phoenix® System at low and high inoculum in a routine laboratory
F. Geppert, N. Kempter (Vienna, AT)
Objectives: Most automated systems require inoculation with McFarland 0.5 (1.5×108 CFU/mL). In the routine laboratory sometimes only few colonies resulting in lower applicable inocula are available. Becton & Dickinson has thus developed a new software allowing a lower density of McFarland 0.25 (7.5×107 CFU/mL). We compared the results obtained with these two inoculation modes under routine conditions.
Methods: 181 Gram-positive and Gram-negative isolates including fastidious and difficult to identify strains from clinical material were analysed. Reference diagnoses were made by conventional methods using the API- and ID 32 system, tube coagulase (RAPIDEC®) and molecular typing (GenoType MRSA®, Hain Lifescience). Using the Phoenix®, all strains were tested in parallel at the standard inoculum of McFarland 0.5 (P 0.5) and 0.25 (P 0.25), respectively, after suspension in 4.5 ml of identification broth. The P 0.25 panels were marked for automatic identification by the system and the results were evaluated in separate data bases.
Results: 80% of the ID results were available within 4 hours. Accurate identification (ID) at species level was achieved in 94.5% at P 0.5 and in 93.4% at P 0.25, respectively. Details of discordant results (n = 18) are presented in the table .
| Reference ID | Phoenix 0.5 | Phoenix 0.25 |
|---|---|---|
| Enterobacter amnigenus | Enterobacter cloacae | no ID |
| Klebsiella ozeanae | Klebsiella ozeanae | no ID |
| Kluyvera ascorbata | Kluyvera ascorbata | no ID |
| Salmonella abortus equi | Salmonella typhi | Salmonella paratyphi A |
| Shigella boydii | Shigella sonnei | Shigella boydii |
| Yersinia enterocolitica | Yersinia frederiksenii | Yersinia frederiksenii |
| Bordetella bronchiseptica | no ID | Bordetella bronchiseptica |
| Bordetella bronchiseptica | Moraxella spp. | Bordetella bronchiseptica |
| Pasteurella pneumotropica | Pasteurella multocida | Pasteurella multocida |
| Pasteurella multocida | Pasteurella multocida | no ID |
| Delftia acidovorans | Delftia acidovorans | no ID |
| Pseudomonas stutzeri | Pseudomonas spp. | Pseudomonas stutzeri |
| Staphylococcus aureus SCV | Staphylococcus aureus | no ID |
| Staphylococcus aureus SCV | Staphylococcus warneri | Staphylococcus aureus |
| Staphylococcus intermedius | Staphylococcus aureus | Staphylococcus intermedius |
| Staphylococcus lentus | Staphylococcus lentus | Macrococcus caseolyticus |
| Streptococcus equi | Streptococcus equi | no ID |
| Streptococcus pneumoniae | Streptococcus pneumoniae | no ID |
Conclusion: The new software accurately identified strains at a lower inoculum, interestingly in some cases even better than with McFarland 0.5. Since susceptibility testing can be done simultaneously and there is no need for subculturing in case of low biomass, a report is completed 18–24 hours earlier.
P1722 Accuracy of the VITEK2 system to identify coagulase-negative staphylococci
A. Sáez, B. Ruiz del Castillo, J. Agüero, L. Martínez-Martínez (Santander, ES)
Objective: We have determined the reliability of the identification of coagulase-negative staphylococci (CoNS) with the VITEK 2 system at species level, considering as a reference the results of molecular identification.
Methods: One hundred and sixty-six clinical isolates of CoNS (Oct. 2003-May 2005) and 7 ATCC type strains were evaluated. Bacteria were identified with the VITEK2 system using ID GP cards. Absence of coagulase was determined with a latex assay (Pastorex® staph-plus, BioRad). Reference identification was established by 16S rDNA sequence analysis; when identification with VITEK2 and 16S rDNA disagreed, definitive identification was defined after sequencing of the sodA and tuf genes, as previously described (Drancourt et al. JCM 2000; 38: 3623–30 and Heikens el al. JCM 2005; 43: 2286–90). For species assignation, the sequences of 16S rDNA, sodA and tuf were compared with those in GenBank. Homology values above 97% were considered reliable.
Results: All 7 ATCC strains were correctly identified by 16S rDNA sequencing. Among the 166 clinical isolates, the molecular method identified the following species (number): S. hominis (39), S. haemolyticus (35), S. saprophyticus (30), S. epidermidis (25), S. lugdunensis (12), S. schleiferi (7), S. capitis (7), S. simulans (4), S. pasteuri (2), S. warneri (2), S. intermedius (2) and S. equorum (1). VITEK2 correctly identified 6 out of the 7 ATCC strains and 151/166 (91%) clinical isolates, including 38/39 S. hominis, 11/12 S. lugdunensis, 5/7 S. capitis, 3/4 S. simulans. Most misidentifications occurred with S. epidermidis (4/25: 3 as S. hominis and 1 as S. intermedius/S. chromogenes), S. saprophyticus (3/30: 1 as S. hominis, 1 as S. warneri and 1 as S. cohnii) and S. haemolyticus (3/32: 1 as S. hominis, 1 as S. warneri and 1 as S. warneri/S. haemolyticus). All isolates of S. schleiferi, S. intermedius, S. pasteuri, S. warneri and S. equorum were correctly identified by the VITEK2 system.
Conclusions: The VITEK 2 system allows reliable identification of CoNS at species level, but additional improvement would be necessary for identifying S. epidermidis, S. saprophyticus and S. haemolyticus.
P1723 Evaluation of the 3M Rapid Detect Staph aureus: an in vitro diagnostic device for direct detection of Staphylococcus aureus nasal colonisation
L.G.M. Bode, Y. Hendriks, M.C. Vos, H.A. Verbrugh, J.A.J. WKluytmans, D.J. Morse, M.E. Wood (Rotterdam, Breda, NL; St. Paul, US)
Objectives: Staphylococcus aureus is the leading cause of surgical site infection, and roughly 25–30% of the population are colonised. In general, colonised patients have a 2–14 fold increased risk of infection, but those patients that carry at higher levels are at highest risk. The patient's own nasal flora account for 80% of S. aureus infections. Rapidly identifying colonised patients provides the ability to proactively manage S. aureus carriers for the prevention of infection. The objective of this pilot study was to investigate the clinical performance of a new rapid diagnostic test system (for proposed introduction in 2007 under the trade designation 3M Rapid Detect Staph aureus) to identify high level carriage of S. aureus directly from nasal swabs.
Methods: This was a multi-centre, prospective, comparative pilot study. There were a total of four sites, two in Europe and in the United States that screened and sampled healthy subjects. One nasal sample was collected from both nares using a swab. The sample was analysed by the 3M Rapid Detect diagnostic test system, which provides results in approximately 20 minutes and a standard quantitative culture method using a selective medium. Confirmation testing was conducted using the Tube Coagulase Test.
Results: A total of 1044 subjects were enrolled across four sites with a total of 999 evaluable subjects (having both culture and detector results). 33.6% of all evaluable subjects were S. aureus carriers, while 12.4% carried ≥5000 cfu (high level carriers). The mean level of carriage for those who were positive was 3.23 Logs (Range 0.63–6.50 Logs). The sensitivity of the test at ≥5000 cfu was 87% (95%CI: 80–92%). The overall sensitivity of the test system was 51% (95%CI: 46–57%). The overall specificity was 92% (95%CI: 90–94%). At ≥5000 cfu the negative predictive value was 98% (95% CI: 97–99%) while the positive predictive value for all levels of carriage was 76% (95% CI: 70–82%). Based on a positive test result 77% of all patients are not treated preemptively.
Conclusions: The proposed 3M Rapid Detect diagnostic test system proved to be a sensitive tool to identify subjects carrying high numbers of S. aureus in the nares. Especially, the high negative predictive value provides a screening tool to rapidly and accurately identify patients who are at low risk of developing a S. aureus infection after surgery, thus allowing for more targeted use of antibiotics and other infection control measures.
P1724 Performance evaluation of the Vidia™ toxoplasmosis IgG and IgM assays
A. Calderaro, G. Piccolo, S. Peruzzi, G. Dettori, C. Chezzi (Parma, IT)
Objective: The aim of the present study was to compare the clinical performance of the new VIDIA® Toxoplasmosis assays (bioMérieux, France) to VIDAS (bioMérieux, France), Axsym (Abbott, USA) and Liaison (DiaSorin, Italy) assays for both IgG and IgM, using clinical specimens.
Methods: A total of 405 frozen and fresh serum samples from 243 pregnant women, 6 pregnant women-HIV infected, 39 healthy adults, 31 infants, 71 immuno-compromised patients and 15 blood donor was used for this retrospective and prospective study.
The samples for which at least one equivocal result have been found whatever the method have been excluded for the analysis, as well as the non resolved samples.
Sensitivity and specificity of the VIDIA TOXO IgM and IgG were determined, on all sera, in comparison with the VIDAS (routinely used in our laboratory), Axsym and Liaison systems.
Results: For clinical samples, sensitivity was found to be 100% for all the four systems for the detection of Toxoplasmosis IgG; regarding Toxoplasmosis IgM, sensitivity was established at 100% for both VIDIA and VIDAS systems, with Axsym we obtained 82.35 and 94.12% with Liaison.
The specificity was 99.25% for VIDIA, 98.49% for Axsym and 100% for both VIDAS and Liaison for the detection of toxoplasmosis IgG; regarding TOXO IgM. Regarding the specificity we obtained 100% for VIDIA, VIDAS and LIAISON and 99.73% for Axsym.
Conclusion: According to the results obtained during this study, the VIDIA TOXO IgG and VIDIA TOXO IgM assays show an excellent sensitivity and a good specificity.
P1725 Comparison of two automated systems (WalkAway and VITEK 2) for identification of Gram-negative bacteria
K. Becker, B. Grünastel, G. Peters, C. von Eiff (Münster, DE)
Objectives: The major advantages of automated systems widely used for species identification in the clinical laboratory are their speed, high throughput, and expedient interfaces with laboratory and hospital information systems. Since each system has its inherent strengths as well as recognized restrictions, the performance of two automated systems was studied with special respect to the time necessary for processing the isolates.
Methods: A total of 375 clinical isolates comprising 277 enterobacterial, 94 non-fermenter and other Gram-negative isolates (e.g. Pasteurellaceae) were used to compare the identification accuracies of the Dade Behring MicroScan WalkAway 96 and the Biomérieux VITEK 2 automated systems. Isolates showing differences in identification were subjected to 16S rRNA gene sequencing.
Results: Overall, 81.6% (n = 306) of the identification results were identical in both systems on the species level, reaching an accordance of 93.9% if the genus level was considered. Differences on the family level (n = 10) occurred for Alcaligenaceae, Enterobacteriaceae, Flavobacteriaceae, Moraxellaceae, and Pseudomonadaceae, whereas differences on the genus and species level were mainly observed for enterobacterial genera, such as Citrobacter, Enterobacter, Klebsiella, and Serratia. In contrast to the fixed processing time of the MicroScan WalkAway system (2.5 hrs), the time of the VITEK 2 system spanned from 2.5 to 9.75 hrs necessary for identification. However, the MicroScan WalkAway system required in some cases repeats to get a final identification result.
Conclusion: Both automated systems allow a timely identification of Gram-negative isolates. Moreover, they constitute an accurate and reliable method for identification of Gram-negative rods. Nevertheless, the use of automated systems does not supersede the need for determination of basic criteria for bacterial identification in order to discover major errors in identification.
P1726 Direct identification and susceptibility testing of staphylococci from positive BacTec blood cultures with WalkAway system
E. Garduño, Ó. Herráez, I. Márquez-Laffón, R. Sánchez-Silos, J. Blanco-Palenciano (Badajoz, ES)
Objectives: To evaluate the combination of the automated BACTEC 9240 blood culture system (Becton Dickinson) and the WalkAway/96 instrument (Dade Behring) for the direct identification and antimicrobial susceptibility testing of staphylococci from positive blood cultures without subculture.
Methods: Positive blood cultures collected between Nov 2005 and March 2006 from patients at our institution were examined by Gram stain. Specimens containing Gram-positive cocci in clusters were included in the study. The direct method was done using a suspension made by two differential centrifugations of positive blood culture broth for inoculation of the MicroScan Pos Combo Type 2SA panels. Standard identification and susceptibility were done using an inoculum made from an overnight culture on solid media.
Results: A total of 88 positive blood cultures were evaluated. 81 isolates showed concordant identification between the direct and standard methods, and the discrepancies were 3 S. epidermidis, 2 S. hominis and one isolate of S. simulans identified by the standard method that were respectively identified as S. hominis, S. epidermidis and S. epidermidis by the direct method.
We found a rate of 92.04% correct identification of staphylococci. There were 87.4% categorical agreements; the major error (false resistance) rate was 2.1%, and the very major error (false susceptibility) rate was 3.7%. The drugs with the most very major errors were trimethoprim-sulfamethoxazole and ampicillin, and only vancomycin and cefazolin showed total agreement with the reference method. Overall, 97.72% blood cultures found to be positive for oxacillin-resistant were correctly detected by direct method, while all oxacillin-resistant S. aureus isolates were correctly detected.
Conclusion: Our results showed that the direct method provided acceptable bacterial identification, but cannot replace the standard method for susceptibility testing due to unacceptable major and very major errors rates. Oxacillin resistance is reliably detected by direct method and could provide earlier information to clinicians.
P1727 Analysis of the comparative work-flow and accuracy of the VITEK2 Compact and the combination mini-API/Agar diffusion SIRSCAN method
V. Doat, M. Roubille (Bourgoin-Jallieu, FR)
Objectives: The aim of this study was to analyse the impact of introducing the VITEK®2 Compact, (bioMérieux, France), an automated identification (ID) and susceptibility testing (AST) system into our laboratory that is currently using a semi-automated method (identification with mini-API® and AST by agar diffusion with results expertised by SIRSCAN® (Dos version 1999, I2A).
Methods: This study was performed in the laboratory of a 390 bed hospital from January to April 2006. A total of 300 strains isolated from various clinical specimens were tested in parallel with the VITEK®2 Compact and our routine technique. The distribution of the strains, representative of our routine workload, is as follows: 190 Gram-negative bacilli (15% non-fermenters) and 110 Gram-positive cocci (66% staphylococci). No fastidious organisms were included in the study The workflow and productivity of each system were evaluated by measuring the manipulation time for one operator to test 5 strains repeated 3 times on each system. The total cost to perform one test, including materials, repeats and waste disposal, were calculated. The patient benefit of rapid results was estimated by a retrospective analysis of 10 patient files in collaboration with an Infectious Disease physician.
Results: The identification agreement obtained between the systems was 98% for the Enterobacteriaceae, 97% for the non-fermenters, and 95% for Staphylococcus and Streptococcus spp. The AST category agreement was 93% with both systems.
The benefits of introducing VITEK®2 Compact are as follows:
-
–
Rapid results and rapid on-line result validation
-
–
One third less manipulation time
-
–
Reduced cost of reagents and consumables
-
–
Reduction of waste disposal
-
–
Reduced risk of biohazard exposure
Rapid result availability to the clinician is of interest especially in cases of S. aureus and for P. aeruginosa in severe infections. Time saved is dependent on laboratory organisation and direct communication with the ward.
Conclusion: Both systems gave good results for the majority of strains encountered in our medium size hospital. The reduced manipulation times, rapid time to results, as well as the easy to use platform of the VITEK2 Compact provide benefits for both the laboratory and the patient.
Automation of molecular viral diagnostics
P1728 Multiplexed molecular testing for the detection of human respiratory viruses using the TM Bioscience ID-Tag RVP assay
R. Manji, M. Lotlikar, C. Ginocchio (Lake Success, US)
Introduction: The effective, reliable, and timely identification of respiratory viral infections is of tremendous importance for diagnosis, infection control, and for epidemiology and surveillance studies. This study evaluated Tm Bioscience's (Toronto, CA) respiratory virus test, ID-TagTM RVP for the detection of the majority of strains and subtypes of respiratory viruses, including respiratory syncytial virus A and B, influenza A (H1, H3), influenza B, parainfluenza 1, 2, 3, 4, adenovirus, coronavirus (229E, OC43, NL63, HKU1), human metapneumovirus, and enterovirus/rhinovirus (E/R).
Methods: In total, 253 respiratory samples (nasopharyngeal [NP] washes, NP aspirates, NP swabs in VTM and bronchial alveolar lavages) collected during the 2005–2006 viral respiratory season were tested. Samples (0.2 ml) were extracted using the NucliSENS easyMAG instrument (bioMérieux, Durham, NC). RVP results were compared to direct immunofluorescence (DFA) and/or viral culture (VC). Discordant samples were analysed by additional nucleic acid amplification assays and sequencing.
Results: Detection rates (% of total tested) for the 3 comparator methods (DFA, VC, RVP) for each virus are as follows (ND=not done): Influ A: 13.4, 17.8, 21.0; Influ B: 2.8, 3.2, 4.8: RSV: 18.2, 15.0, 17.9; Para-1: 2.0, 2.8, 2.8; Para-2: 1.2, 2.8, 2.8; Para-3: 0.8, 0.8, 0.8; Para-4: ND, ND, 2.4; Adeno: 2.8, 7.5, 6.0 (RVP increases to 7.2 with 3 low level equivocal results): Corona: ND, ND, 1.6; hMPV: ND, ND, 1.2; E/R: ND, ND, 9.5. The overall detection rate for each test for all viruses was: DFA = 29%, VC = 49.9%, RVP = 60.4%. Mixed infections were found in 11 samples (4.35%) including 3 E/R+RSV; 2 E/R+Para-1; 1 E/R+ Para-2; 2 E/R+Adeno; 1 RSV+NL63, 1 RSV+Adeno; 1 Adeno+NL63.
Conclusions: ID-TagTM RVP assay can efficiently detect the major respiratory viruses, including mixed infections. This assay will provide insight on other respiratory pathogens (ex. coronavirus and metapneumovirus) that are not routinely cultured in most laboratories, the prevalence of mixed viral infections, and their role in disease severity and outcome.
P1729 Clinical evaluation of the NucliSENS analyte specific reagents for the detection of respiratory syncytial virus and human metapneumovirus in paediatric respiratory specimens
R. Manji, F. Zhang, M. Lotlikar, C. Ginocchio (Lake Success, US)
Background: Respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) are significant causes of lower respiratory tract infection in infants and young children with bronchiolitis and pneumonia. The rapid diagnosis of RSV and hMPV infections is important for patient management and infection control measures. This study evaluated the NucliSENS RSV and hMPV analyte specific reagents (ASRs) (bioMérieux, USA) for the detection of RSV and hMPV in paediatric respiratory samples. The prevalence of RSV and hMPV was determined by age group and compared to other common viral respiratory pathogens.
Methods: Samples included: frozen respiratory specimens (nasopharyngeal aspirates, washes or swabs in VTM) from 653 children (<2 yr: n = 433; 2–5 yr: n = 220) evaluated for respiratory disease. Specimens and target specific RNA inhibition controls (IC) for RSV and hMPV were co-extracted using the NucliSENS miniMAG and/or easyMAG instruments (bioMérieux). NASBA amplification and molecular beacon detection of either RSV or hMPV and their specific IC were performed using a NucliSENS EasyQ analyser (bioMérieux). Results for RSV were compared to direct immunofluorescence (DFA) and/or viral culture (VC) using R-Mix cells (DHI, USA). hMPV and RSV discordant results were arbitrated using Pro-hMPV and Pro-flu 1 ASRs (Prodesse, USA). Pathogen prevalence was determined by patient age and location (outpatient [OP], emergency department discharge (EDD), paediatric unit [PU], intensive care unit [PICU]).
Results: For the comparison methods (DFA, VC, RSV ASRs) sensitivity was 85.5%, 45.4%, 96.4%; specificity was 98.6%, 100%, 98.6%, PPV was 92.9%, 100%, 93.9%, NPV was 96.2%, 91.4%, 99.0%.
NucliSENS hMPV positives were confirmed with Pro-hMPV assay.
Prevalence rates (%) by age were as follows:
-
–
Age <2 yr: Influ A =4.84; Influ B =2.77; Parainflu-1 =1.15; Parainflu-2 =0; Parainflu-3 =1.15; Adenovirus =2.77; Enterovirus =0; RSV =27.48; hMPV =5.77.
-
–
<2–5 yr:Influ A =8.18; Influ B =2.73; Parainflu-1 =0.91; Parainflu-2 =0; Parainflu-3 =0; Adenovirus =6.36; Enterovirus =0.45; RSV =12.73; hMPV =4.55.
Distribution (%) of positive samples by patient location for RSV: OP =43.9, EDD =35.4, PU =9.2, PICU =11.5; for hMPV: OP =2.9, EDD =37.1, PU= 40, PICU =20.
Conclusions: The NucliSENS ASRs provided rapid (<3.5 hr) and sensitive methods for the detection of RSV and hMPV in respiratory samples. RSV remains the most isolated virus among those tested in this study. RSV and hMPV infections required hospitalisation in 21% and 60% of the patients in which virus was detected.
P1730 Efficiency of RNA isolation from respiratory samples using the NucliSENS miniMAG and easyMAG extraction procedures
R. Manji, F. Zhang, C. Ginocchio (Lake Success, US)
Objectives: The semi-automated NucliSENS miniMAG (MM) and the automated NucliSENS easyMAG (EM) (bioMérieux, Durham, NC) are platforms for the extraction of total nucleic acids (NAs) from clinical samples. The method used by both is based on the established Boom chemistry but utilises magnetic silica particles. The purpose of this study was to evaluate both systems for the removal of inhibitors and the efficiency of isolation of respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) RNA from paediatric respiratory samples.
Methods: NAs were extracted from 656 samples (nasopharyngeal [NP] washes, NP aspirates, NP swabs in VTM) using either MM (n = 390) or EM (n = 181) or both systems (n = 85). Samples (0.2 ml) were pretreated with DNAse, transferred to a NucliSENS lysis buffer tube, and target specific Inhibition Controls for RSV and hMPV were added to assess the efficiency of extraction, amplification and the presence of inhibitors. Each NA extract was tested for RSV and hMPV using a laboratory validated protocol and NucliSENS analyte specific reagents (bioMérieux). Sensitivity of each method was compared using aliquots of serially diluted in vitro transcribed RSV RNA.
Results: For RSV testing, initial inhibition rates for samples extracted with EM was 0% (0/266) and for MM 2.11% (10/475). Inhibition was resolved for all samples after repeat MM extraction from a new aliquot. For hMPV testing, initial inhibition rates for samples extracted with EM was 1.88% (5/266) and for MM 2.95% (14/475). Inhibition was resolved for all EM samples and for 10/14 MM samples after repeat extraction from a new sample aliquot for a final inhibition rate of 0% for EM and 0.84% for MM. RSV assay sensitivity using MM and EM extraction was 66.7% and 87.5%, respectively, for detecting 10 RSV RNA copies/isolation. RSV and hMPV were detected in 22.5% and 5.36% of the samples, respectively.
Conclusions: MM and EM provided highly purified NAs and efficiently removed inhibitory substances from difficult respiratory samples. A slightly better performance seen with EM may relate to the fuller extent of the automation and less operator error as compared to the semi-automated MM. Other benefits of the EM were better throughput per run (EM:24 samples vs MM:12 samples) and less hands on time (EM:15 min vs MM:45 min). In addition, better assay sensitivity was achieved with EM extraction. Both systems provide a generic extraction method for a variety of analytes and sample types.
P1731 HBVDNA extraction from serum using the NucliSENS easyMAG platform
P. van Deursen, G. Onland, A. van den Brule, A. Verhoeven, M. Jacobs (Boxtel, Veldhoven, NL)
Background: The NucliSENS easyMAG platform (bioMérieux) is designed for nucleic acid extraction from a broad range of different sample specimens. However, for some specific applications additional pre-extraction procedures and/or a specific extraction protocols might be needed. Recently a new extraction protocol (Specific A) was introduced for whole blood specimens.
Objectives: The aim of the study was to measure the recovery efficiency of HBV DNA from serum using five different extraction protocols. Four of these protocols were performed in combination with the NucliSENS easyMAG platform and one other manual method was included as reference.
Methods: A panel of 12 samples, including 4 standards (obtained from VQC, Amsterdam), 7 HBVDNA positive serum samples and 1 negative control, were tested with five different extraction protocols. Identical input (200 ul) and output volumes (110 ul) were used for the different methods. Briefly, for the NucliSENS easyMAG the following procedures were tested; (1) no pre-incubation and Generic extraction protocol, (2) pre-incubation with proteinase K and Generic extraction protocol, (3) no pre-incubation and Specific A extraction protocol, and (4) pre-incubation with proteinase K and Specific A extraction protocol. The High Pure PCR Template Preparation kit (Roche diagnostics) was used as reference method. To all samples control virus (PhHV) was added to measure overall extraction performance. Extracted samples were analysed by real time PCR.
Results: PhHV DNA was detected in all samples. Mean Ct values were 29.6, 29.8, 29.5, 29.3, and 31.4 for protocols 1, 2, 3, 4, and 5, respectively. HBV DNA was not detected in the negative control samples. For the remaining samples HBV DNA was detected in 100% (protocols 1, 2, 4, and 5) and 91% (protocol 3). For the samples scored positive with all five methods the mean Ct values were 32.8, 31.0, 32.9, 30.5, and 31.3, respectively.
Conclusion: Best results, for both HBV DNA and control PhHV DNA detection, were obtained in combination with protocol 4 that uses proteinase K pre-incubation, followed by extraction with the NucliSENS easyMAG platform using the Specific A protocol. The proteinase K pre-incubation resulted in on average −2.1 lower Ct values for HBV DNA detection, whereas for this application the Specific A protocol contributed minimal (on average −0.2 Ct) to the overall improvement measured.
P1732 Evaluation of an automated sample preparation system (MDx) in combination with affigene CMV trender
I. Abbate, N. Finnstrom, A. Garbuglia, M. Solmone, S. Selvaggini, E. Bennici, S. Neri, C. Brega, M. Capobianchi (Rome, IT; Bromma, SE; Milan, IT)
Background: Monitoring of Cytomegalovirus (CMV) viral load is efficiently performed by using molecular diagnostics based on PCR, such as the CE-labelled affigene CMV trender assay (Sangtec, Bromma, Sweden). Today, a number of automated nucleic acid sample preparation systems are available for a high throughput laboratory. One of them is the BioRobot MDx Workstation (MDx) from Qiagen (Hilden, Germany). This study aims to evaluate the MDx in combination with the PCR system affigene CMV trender. Moreover, on a small number of clinical samples a comparative analysis was conducted between affigene CMV trender assay and an house nested PCR method.
Materials and Methods: For the evaluation, the 2005 QCMD panel for CMV was used. To mimic the laboratory routine for analysing whole blood patient sample, freeze-dried samples from the QCMD panel were diluted into whole blood, or in water and diluted in the lysis buffer of the extraction kit. Each of the panel member was extracted twice on the MDx and thereafter each extraction was amplified in duplicates using affigene CMV trender.
Fiftyfour clinical whole blood samples were also included in the study. Extraction was performed as above and the amplification was done in parallel with affigene CMV trender assay and with nested PCR.
Results: The average results from each of the QCMD samples showed a less than 0.25 log difference compared to the average reported value in the QCMD report based on 30 different datasets. One sample varied more than 0.25 log compared to the reported values. However, this sample was in the area of the limit of detection for the assay. The negative sample was reported negative in all four replicates. The comparison between affigene CMV trender assay and nested PCR showed a good concordance. Only 3 out of 54 patients showed discordant results, being 2 of them CMV DNA positive only in nested PCR and 1 positive only when using CMV trender. Moreover, the affigene CMV trender assay was able to quantify the CMV DNA present in the positive samples with a mean value of 43750 ± 26265 CMV DNA copies/mL.
Conclusions: The BioRobot MDx Workstation works well in combination with the affigene CMV trender PCR assay as verified on both QCMD panel and clinical samples. This opens up the possibilities for quicker and accurate CMV monitoring for high-throughout laboratories.
P1733 Comparison of three different diagnostic tests for HCV-RNA viral load: Cobas Amplicor Monitor HCV 2.0, Versant β-DNA HCV 3.0, and Cobas TaqMan HCV
P. Ravanini, A. Nicosia, M. Crobu, E. Grossini, V. Quaglia, M. Cagliano, C. Cusaro, F. Milano, F. Caviglia (Novara, Vercelli, IT)
Objectives: The correct quantification of HCV-RNA viral load is an important parameter for the assessment and monitoring of antiviral therapy.
In this work, the possibility to obtain comparable results by different standardised tests for HCV-RNA viral load has been verified. To perform this aim we verified if there is an acceptable difference (<0.5 Log) among three different tests and if there are differences of quantification due to viral genotypes.
Methods: In the last years we performed two similar studies of comparison. The first one compared Bayer Versant HCV 3.0 and Roche Cobas Monitor HCV 2.0. 44 HCV-RNA positive samples of different genotype were collected and analysed by both methods between October 2001 and May 2002. A new study was performed between November 2005 and May 2006 on further 70 samples, comparing Bayer Versant HCV 3.0 and Roche Cobas TaqMan HCV.
Results: In the first study the median ratio between the results obtained by the two tests showed a value of 1.22 with a light overestimate of Cobas Monitor in comparison to Versant HCV. In the second study the median ratio was clearly higher: Cobas TaqMan showed an overestimate of 7.39 folds in comparison to Versant HCV. The cases with not acceptable differences (>0.5 Log) were 5/44 (11%) in the first study, and 60/70 (86%) in the second one.
The first study showed a good correspondence for genotype 1 (ratio 0.99), a light overestimate for genotypes 2 and 3 (ratio 1.86 and 1.46) and an underestimate for genotype 4 (ratio 0.79).
In the second study the only genotype with acceptable differences (<0.5 Log) was genotype 4 (ratio 2.0). The other genotypes showed poorer acceptability because of greater differences (genotype 1 ratio 7.87; genotype 2 ratio 6.30; genotype 3 ratio 13.11).
Conclusions: According to these data there is an important difference between the viral loads by Versant HCV and Cobas TaqMan. In the 86% of the cases these differences are not acceptable. The scattering of the results appears wider in the second study than in the first one. The comparison of different tests underwent a worsening.
Moreover there are great differences of quantification according to genotype. The only genotype with acceptable differences, in the second comparison, was genotype 4.
In our opinion the standardisation of molecular methods for HCV-RNA viral load testing is not yet concluded, and have to be resumed for new Real Time methods which seem to be not perfectly comparable with the other standardised methods.
P1734 Performance evaluation of the QIAGEN EZ1 DSP virus kit with Abbott RealTime HIV-1, HCV and HBV assays
G. Schneider, K. Kuper, K. Abravaya, C. Mullen, M. Schmidt, M. Sprenger-Haussels (Des Plaines, US; Hilden, DE)
Objectives: Automated systems must meet the demands of routine diagnostics laboratories with regard to performance characteristics and compatibility with downstream assays. The EZ1 DSP Virus system is a generic sample preparation system utilising QIAGEN's second generation magnetic-bead technology on the QIAGEN BioRobot EZ1 DSP workstation for isolation and purification of viral nucleic acids from human plasma or serum. The improved EZ1 DSP Virus protocol is taking advantage of optimised lysis conditions as well as increased binding and washing efficiencies. The performance of the QIAGEN EZ1 DSP Virus Kit was evaluated in combination with the Abbott RealTime HIV-1, HCV and HBV assays.
Methods: RNA viruses (HIV-1 and HCV) and a DNA virus (HBV) were spiked into negative human plasma or serum and processed in batches of six. An internal control was added to each sample to control for recovery and inhibition. Nucleic acids were extracted from 0.4 ml of plasma or serum using the EZ1 DSP Virus Kit and were eluted in a volume of 90 microliters. The resulting nucleic acids were analysed by the appropriate RealTime assay on the Abbott m2000rt thermalcycler/reader according to the package inserts. Run controls were included on every thermalcycler run.
Results: RealTime assay results were linear over the ranges tested. The observed inter-assay SDs were less than or equal to 0.13 log except at the lowest concentration tested in each assay. The EZ1 DSP Virus kit enabled detection of low levels of virus. Good correlation between the BioRobot EZ1 and the Abbott m2000sp was observed. EZ1 DSP Virus kit reagent lot-to-lot variability was evaluated. No detectable false positives among negative samples were detected in six carryover challenge runs. No inhibition was observed in the presence of potentially interfering substances.
Conclusions: The QIAGEN EZ1 DSP Virus system enables reproducible purification of viral nucleic acids from human plasma or serum and allows for reliable, sensitive and specific detection and quantification of RNA and DNA viruses by the Abbott RealTime HIV-1, HCV and HBV assays.
*Under development for in-vitro-diagnostic use in Europe. The EZ1 DSP Virus Kit as described in this poster will not be available in the USA.
** Abbott RealTime HIV-1 and HCV are CE marked, not available in the USA. Abbott RealTime HBV is in development. Abbott Molecular makes no Abbott RealTime assay performance claims using the EZ1 DSP Virus Kit.
P1735 The EZ1 DSP Virus Kit for use in molecular viral diagnostics: performance verification of kit and instrument
M. Schmidt, C. Kupfer, M. Hess, A. Burmeister, M. Sprenger-Haußels (Hilden, Hamburg, DE)
Objectives: The generic EZ1 DSP Virus Kit* utilises magnetic-bead technology on the QIAGEN BioRobot EZ1 workstation for isolation and purification of viral nucleic acids from human plasma and serum samples. In this study, the performance of the EZ1 DSP Virus Kit was evaluated in combination with several diagnostic downstream assays. Validation parameters determined for DNA and RNA viruses are limit of detection, linear ranges, lack of cross-contamination, robustness and stability of eluates.
Methods: Known quantities of viruses were spiked into negatively tested human plasma or serum pools. Diagnostic downstream assays used during the performance verification included in-house HCV RT-PCR assay, artus CMV PCR Kits§ (LC, RG, and TM), artus HBV PCR Kits§ (LC, RG and TM), artus Parvo B19 RG PCR Kit§, COBAS Amplicor Monitor HIV-1§, COBAS TaqMan HBV§, HCV§, and HIV-1§ (Roche).
Results: We present some of the results of the performance evaluation of the EZ1 DSP Virus Kit. The 95% probit value for HIV-1 was determined to be 114.5 IU/mL using the COBAS Amplicor Monitor HIV-1 Test (v1.5) and the preliminary 95% probit value for HBV 6.4 IU/mL using the COBAS TaqMan 48 HBV Test. Plasma samples generated from different blood collection tubes performed within the acceptance ranges using an in-house HCV RT-PCR assay and the artus HBV RG PCR Kit. Cross-contamination was not detected using plasma samples containing 1.00E+08 IU/mL parvovirus B19 processed in a checkerboard pattern in 9 runs on the BioRobot EZ1 DSP workstation.
Conclusion: The results of the performance verification of the EZ1 DSP Virus Kit confirm the generic application of the EZ1 DSP Virus Kit for fully automated sample preparation in combination with different diagnostic downstream assays.
The EZ1 DSP Virus Kit and all of the artus PCR Kits as described in this poster will not be available in the USA.
*Under development for in-vitro-diagnostic use in Europe. §CE-marked for in-vitro-diagnostic use in Europe.
Pathogenesis of intracellular bacterial infections
P1736 A proteomics approach to understanding the pathogenesis mechanisms of the obligate intracellular pathogen Coxiella burnetii
G. Samoilis, A. Psaroulaki, K. Vougas, A. Gikas, Y. Tselentis, G. Tsiotis (Heraklion, Athens, GR)
Objectives: Coxiella burnetii is an obligate intracellular pathogen with nearly worldwide distribution. It is the pathogenic agent of Q-fever in man [1], and it has been classified as a Class-B possible agent for bioterrorism [2]. Coxiella burnetii enters target cells by phagocytosis and forms within the infected cell phagosomes that fuse with lysosomal compartments to form the phagolysosome. Within the harsh environment of the phagolysosome Coxiella burnetii manages to survive and replicate. In this work we have attempted to construct a 2-dimensional map of the proteome of intracellular Coxiella burnetii Phase II strain Nine Mile. Identifying proteins being expressed by the intracellular form of the bacterium can provide us with vital information concerning its pathogenesis.
Methods: Intracellular Coxiella burnetii Phase II strain Nine Mile was cultivated in Vero cell lines, isolated and separated from host cell components in a renographin density gradient [4]. Total protein extracts were analysed by conventional 2-dimensional PAGE. Trypsinised protein spots were analysed in a MALDI-TOF MS instrument. Peptide analysis and protein identification were performed as previously described [5]. Peptide matching and protein searches were performed automatically using the Mascot Software.
Results: We have detected 600 protein spots in 2-DE gels, and MS allowed us the identification of 168 Coxiella burnetii proteins. Identified species seem to be involved in a wide range of bacterial processes, whereas 12% of them were hypothetical.
Conclusion: The present study allowed the identification of 168 different proteins and, as far as we know, represents the first proteome analysis of Coxiella burnetii Phase II strain Nine Mile. Genome analysis has predicted that Coxiella burnetii possesses components of the Secretion IV System [3], and in our work we have confirmed the expression of DotB. This pathogen expresses an extensive network of chaperones, which also seems to be coupled to the secretory pathway. We have also found several other proteins that may be essential virulence or survival factors for Coxiella burnetii. This work opens the way to characterise the proteome of Coxiella burnetii, and to compare protein profiles of different isolates.
P1737 Identification of Chlamydophila pneumoniae heat shock protein 60 on atherosclerotic carotid plaque by RT-PCR
R. Cultrera, V. Crapanzano Minichello, S. Seraceni, M. Coen, F. Mascoli, C. Contini (Ferrara, IT)
Objectives: Some difficult microorganisms, including Chlamydophila pneumoniae or Mycoplasma spp., are associated with the atheroscletotic tissue damage. The aim of this study was to evaluate the employment of culture together with PCR and RT-PCR to identify C. pneumoniae in atheromatous carotid plaques (ACP) in order to define their possible role in the pathogenesis of the arterial tissue damage.
Materials and Methods: ACPs were obtained by endoarterectomies from 10 patients with severe stenosis of the internal carotid artery. Each specimen was divided in three parts: a proximal tract to heart, healthy, without stenosis, a medial tract, corresponding to the atheromatous plaque, and a distal tract above the plaque. Each sample obtained in aseptic conditions was immediately homogenised by freezing to −80°C. Aliquots were employed to perform cultures for C. pneumoniae on Hep-2 cell line in DMEM. DNA and total RNA were obtained from aliquots of each tissue sample and from Hep-2 cultures. C. pneumoniae 16S, momp and hsp60 genes were investigated.
Results: The PCR and RT-PCR for momp and 16S genes of C. pneumoniae resulted negative in all samples. PCR and RT-PCR for the hsp60 gene of C. pneumoniae resulted positive in the proximal portion of one ACP with haemorrhagic evolution of a patient with a retinal tromboembolic outcome. The molecular analyses on C. pneumoniae growing in the culture are in progress.
Conclusions: The DNA and RNA amplification of different portions from ACP seems to be useful to evidence the effective localisation of C. pneumoniae in the atheromatous arterial tissue. The evidence of the gene expression of C. pneumoniae hsp60 in a patient with acute haemorrhagic evolution of the carotid plaque may suggest that C. pneumoniae might partecipate in the atherogenesis and to induce atherosclerosis complications by inflammatory pathways (activation of cytokines, endothelial factors and matrix-degrading metalloproteinases).
P1738 The role of Chlamydophila pneumoniae in pathogenesis of adenoiditis in children
I. Choroszy-Król, M. Frej-Madrzak (Wroclaw, PL)
Objective: The aim of the study was to determine a correlation between the presence of Chlamydophila pneumoniae EB antygen and the appearance of the ompA gene in pharyngeal tonsil tissue.
Materials and Methods: 162 different samples such as: tonsil swab (n = 57), sera (n = 57), tonsil tissue (n = 48) were analysed. The samples were taken from 57 patients aged from 3 to 15 years hospitalised in Paediatric Otorhinopharyngeal Department of The Lower Silesian Specialistic Hospital – Center of Medical Emergency in Wroclaw. The control group consisted of 31 chldren aged from 3 to 15 years with no symptoms from respiratory tract. Pharyngeal tonsil swabs were analysed by direct immunofluorescent method by Cp IFT reagent manufactured by Cellabs Pty Ltd. and by fluorescent microscope made by Olympus. The level of specific anti-Cp IgG antibodies was determined by ELISA assay using diagnostic test made by Vircell SL, Cp IgG ELISA Granada, Spain. The opmA gene fragment in tonsil tissue was detected by nested PCR using PCR diagnostic made by DNA – Gdańsk II s.c.
Results: Infection caused by Chlamydophila pneumoniae was detected in tonsil swabs by direct immunofluorescent method in 13 of 57 (22.8%) examinde children whereas in control group the presence of Cp was not observed. The increased level of anti-Chlamydophila IgG antibodies was indicated by ELISA in 10/57 (17.5%) examined subjects while in control group this parameter was not performed for reasons of economy. OmpA gene of Chlamydophila pneumoniae was detected by PCR in 28 of 48 (58.3%) patients; in control group this parameter was not evaluated for reasons already mentioned. The analysis of the frequency of Cp infection in children of different age showed that the highest rate of infection was present in group of age ranging from 6 to 10 years: 26.9% of cases with detection of Cp antigen and 72.2% of cases with detection of Cp DNA respectively.
Conclusions: In children with adenoiditis the presence of Cp antigens in tonsil swabs did not correlate with the appearance of IgG antibodies in sera. The association between the C. pneumoniae antigens observed in examined patients with the presence of ompA gene in tonsil tissue of those patients was also excluded. A common appearance of C. pneumoniae in adenoids could suggest that tonsil tissue may act as natural reservois for many pathogens of respiratory tract. The significance of detection of Cp in children who underwent adenoidectomy remains still difficult to define.
P1739 Looking for evidence of Chlamydia pneumoniae presence in atherosclerotic and non-atherosclerotic segments of arteries obtained from autopsies at a forensic unit in a Kuala Lumpur hospital, by both rtPCR to detect omp A VD4 base sequences and immunohistoch
P.S. Puvaneswaren, M.I.A. Mustafa, N.B. Abdullah, M. Inuzukam, Q. Haque, A.H. Mansor (Kuantan, MY; Fukui, JP; Kuala Lumpur, MY)
Background and Objective: A plethora of conflicting reports regarding the link between Chlamydia pneumoniae (CP) and atherosclerosis exist, however few are available from Malaysia and Southeast Asia. This study was performed to look for evidence of CP presence in atherosclerotic as well as non-atherosclerotic arteries of the same human cadaver.
Methods: Both a molecular method using real-time PCR to look for the presence of VD4 sequences, a highly conserved domain of CP ompA gene, as well as immunohistochemical method using CP species-specific antibody ab13941 purchased from DakoCytomation were conducted. Amplification of human beta actin gene, a housekeeping gene, acted as an endogenous control. Fifty-two atherosclerotic arterial segments and thirty-three non-atherosclerotic segments were obtained from 33 fresh cadavers at the Forensic Unit, Hospital Kuala Lumpur for the purpose of the study.
Results: Both the immunohistochemistry assay as well as the real-time PCR assay failed to detect any C. pneumoniae from all the tested tissue samples.
Conclusion: Thus, this study could not demonstrate any molecular or immunohistochemical evidence for the presence of CP in any of the arterial tissues tested whether atherosclerotic or non-atherosclerotic.
P1740 Beta-2-microglobulin-deficient mice: increased susceptibility to Mycobacterium avium infection
S. Gomes Pereira, P. Rodrigues, R. Appelberg, S. Gomes (Porto, PT)
Iron availability is critical for mycobacterial survival and multiplication, with host iron overload favouring bacterial growth and increasing susceptibility. Previous studies have shown that beta-2-microglobulin-deficient mice (b2m KO) are highly susceptible to M. tuberculosis infection, which was in part due to the absence of CD8+ T cells but also to the iron overload typical of b2m KO hepatocytes. In M. avium infection, CD8+ T cells are not fundamental for the control of bacterial growth, whereas an excess of iron is beneficial for its proliferation. Hfeknock-out animals, on the other hand, represent a model of systemic iron overload, in the absence of any known immune deficiency.
In this study, we evaluated the susceptibility to M. avium infection of both b2mKO and HfeKO mice.
Age-matched animals from C57Bl/6 (wild-type), b2m KO and Hfe KO mouse strains were infected with M. avium strain 2447 SmT by the intravenous route. At different times post-infection, blood was collected for the quantification of serum iron and transferrin saturation. Spleen and liver were recovered for mycobacteria CFU counts, iron content determination and histological analysis (Ziehl-Nielssen, Perl's staining). Two months after infection, b2m KO mice began to show higher hepatic mycobacteria counts when compared to wild-type animals. This difference was even more pronounced 4 months post-infection. In the liver, non-hemic iron values were about four-times higher in b2m KO animals than in wild-type controls, Serum iron and transferrin saturation were also higher in b2m-deficient mice. Spleen iron content did not show significant differences between these two strains. The increased susceptibility of b2mKO mice to M. avium infection does not result exclusively from the iron overload status of b2m KO mice. Hfe KO mice, which show similar iron levels and tissue distribution, also exhibited higher mycobacterial counts than wild-type animals, but did not show the same degree of susceptibility as b2m KO mice. In order to understand if CD8 T cells deficiency is contributing to the increased susceptibility in b2m KO animals, experiments with CD8 T cells depletion in Hfe KO mice are being carried out.
P1741 Association of apolipoprotein E polymorphism with tuberculosis
T.N. Farivar, P. Johari (Zahedan, IR)
Apolipoprotein E (ApoE) is a 34-kDa glycoprotein involved in lipoprotein transport through interaction with the low-density lipoprotein and related receptors and exists as three isoforms, designated E2, E3, and E4. Recently, it has become clear that ApoE binding to its receptors plays a role both in development and in control of the immune system. Also, it has been showed that ApoE modulates the rate of uptake of apoptotic cells by macrophages. Studies in several populations have indicated that genetic variation at the apolipoprotein E structural locus influences amplitude to infectious diseases. The possible role of apolipoprotein E polymorphism in the development of Tuberculosis has not been investigated sufficiently. In this study, we aimed to determine the significance of association between Tuberculosis and apolipoprotein E genotypes.
The apolipoprotein E genotypes were assayed in 82 tuberculosis patients by polymerase chain reaction followed by enzymatic digestion with Hha I and compared with previously reported Apo E genotype frequencies. Our results showed that the incidence of E2 and E4 allelic frequencies were significantly higher in patients (0.34 and 0.12) than in controls (0.03 and 0.06 respectively). Also, the incidence of E3 allelic frequency was lower in patients (0.54) than in controls (0.91). Also, our study revealed an association of apolipoprotein E locus with Tuberculosis. However, large population-based studies are needed to understand the exact role played by the locus in causing the condition.
P1742 Rapid systemic propagation of Legionella longbeachae lung infection in A/J mice
I. Gobin, M. Sarec, K. Selenic, M. Doric, M. Susa (Rijeka, HR)
Objectives: Legionnaire's disease was considered to be exclusively a lung disease and limited informations were available concerning the progression of the disease. It was shown that mice intratracheally inoculated with 105 CFU L. longbeachae serogroup 1 developed an acute disease and died within six days. Pathohistological changes in the lungs of infected animals were typical for multifocal bronchopneumonia. Therefore, we were interested to explore whether a systematisation of the primary process in the lungs may occur. In this study we analysed the intensity of bacterial multiplication and pathohistological changes in liver, spleen and kidney.
Methods: Pathogen-free female 6- to 10-weeks-old A/J mice were infected by intratracheal inoculation with L. longbeachae serogroup 1 using a dose of 105 CFU. We determined the CFU of bacteria in the lung, liver, spleen and kidney of A/J mice 2, 24, 48 and 72 hours post infection. We also followed the patohistological changes in these organs 72 hours post infection (HE, PAS, Mallory and Gomory stain).
Results: Increasing multiplication of L. longbeachae in the lung was associated with increased colony counts in analysed organs reaching the concentration between 103 CFU (kidney) and 105 CFU (liver) at 72 hours post infection. We never detected an excessive proliferation of bacteria in these organs, which would be independent of the bacterial proliferation that has been seen in the lungs. Pathohistology of liver, spleen and kidney analysed 72 hours post infection showed mainly mild inflammatory cellular infiltrates and degenerative changes. In the liver, beyond the degeneration of the hepatocytes, the most prominent observation was focal infiltrations within portal triads by mononuclear and polymorphonuclear leukocytes. The architecture of the white and red pulp of the spleen showed all signs of severe destructive splenitis. In kidney the glomerules were reduced in size and the juxtaglomerular apparatus showed decreased cellularity. Additionally, a significant degeneration of particular proximal tubule and collagen increase in the intertubular spaces was observed.
Conclusion: We confirmed that L. longbeachae serogroup 1 rapidly disseminate in the liver, spleen and kidney causing severe systemic disease in mice. At the moment we do not know if the observed pathohistological changes are caused by local bacterial multiplication and their virulent factor(s) or by uncontrolled host immune response.
Sepsis and sepsis markers
P1743 Predicting sepsis mortality in the emergency department: a comparison among mortality in emergency department sepsis (MEDS) score, procalcitonin, and C-reactive protein
C-C. Lee, S-Y. Chen, P-R. Hsueh (Taipei, TW)
Objectives: To compare the prognostic value of a validated clinical prediction score, Mortality in Emergency Department Sepsis (MEDS) score, and two labratory markers, procalcitonin (PCT), and C-reactive protein (CRP), in ED sepsis patients.
Methods: The study is a prospective observational cohort design, carried out at the emergency department (ED) of an urban, university-based medical centre. Consecutive adult patients admitted to the ED fulfilling the ACCP/SCCM Consensus Conference definition of sepsis were prospectively enrolled. PCT and CRP were evaluated for each enrolled patient on ED admission. Clinical characteristics and laboratory results were recorded using a standardised form. Each patient was followed for at least 30 days. The main outcome was 30-day all cause mortality. The PCT level and MEDS scores were correlated with clinical severity of sepsis. Areas under the receiver operating characteristic curve (ROC curve) were calculated for each prognostic marker.
Results: A total of 525 patients were included in the analysis. The overall 30-day mortalty was 10.5% (55/525). All three markers, PCT, CRP, and MEDS score, were higly correlated with the severity classification of sepsis (sepsis, severe sepsis, septic shock). The area under the ROC curve in the prediction of mortality was 0.82 for MEDS, 0.73 for PCT, and 0.64 for CRP. Controlling for MEDS score variables, elevated serum level of PCT (≥ 0.5 ng/mL) was independently associated with 30-day mortality (odd ratio 2.82; 95%CI, 1.26–6.33). Incorporation of PCT as an item of MEDS score increases the area under curve from 0.82 to 0.88.

ROC curves of three predictive markers for mortality in patients with sepsis.
Conclusions: MEDS score was the best predictor of mortality in patients with sepsis in the ED setiing. PCT provided a better estimate of clinical severity and prognosis than CRP. Incorporation of PCT into the MEDS scoring system will enhance its capability to predict mortality in patients with sepsis.
P1744 Procalcitonin as a parameter for the early recognition of severe bacterial infections
A. Georgouli, E. Chinou, D. Manavoglou, P. Ntavlouros, E. Skouteli, P. Golemati (Athens, GR)
Objectives: The aim of our study was to confirm the diagnostic value of procalcitonin (PCT) levels in the discrimination between severe bacterial infections and other conditions such as localised infections, superficial bacterial colonisation or viral infections.
Methods: We examined 166 patients divided into four groups: control group (n = 85, patients with no sign of infection); patients with confirmed sepsis (n = 12); patients with localised infections (n = 32); patients with bacterial colonisation (n = 26); and patients with confirmed viral infection (n = 11). Plasma PCT levels were determined by an immunochromatographic test for the semi-quantitative detection of PCT (normal range of PCT values <0.5 ng/mL).
Results: The following results were obtained:
-
–
Control group (n = 85): PCT levels <0.5 ng/mL.
-
–
Sepsis (n = 12): PCT levels ≥10.0 ng/mL.
-
–
Localised infection (n = 32): PCT levels 0.5–2.0 ng/mL.
-
–
Bacterial colonisation (n = 26): PCT levels 0.0–2.0 ng/mL.
-
–
Viral infection (n = 11): PCT levels 0.0–0.5 ng/mL.
Conclusions: Our study confirmed that PCT is a sensitive parameter that is higly elevated only in severe bacterial infections as opposed to viral infections, localised infections and superficial bacterial colonisation.
P1745 Are procalcitonin and C-reactive protein accurate prognostic markers in sepsis?
N. Komitopoulos, E. Giakoymaki, V. Tsiavos, D. Charitos, A. Komitopoulou, A. Kanavou, E. Varsamis (Athens, GR)
The initial presentation of sepsis is often non specific and its severity is sometimes underestimated. Procalcitonin is the prohormone of calcitonin produced by several cell types and many organs in response to proinflamatory stimuli, in particular by bacterial products. High values of Procalcitonin (PCT) and C-Reactive Protein (CRP) are considered useful and reliable markers for the diagnosis of sepsis, but their association with sepsis severity is still debated.
Aim: To evaluate whether PCT and CRP serum levels could be correlated with APACHE II score in septic elderly patients.
Subjects and Methods: 48 patients, 23 females and 25 males, with sepsis of various origin, aged 74 ± 15 years, were included in this study. PCT levels (semi-quantitative immunochromatographic test, DIACHEL), high sensitivity CRP levels (DADE BEHRING) and APACHE II score were measured within 6 hours after hospital admission.
Results: PCT values were found >0.5 ng/dl in 78% of the patients. The mean value of APACHE II score in patients with PCT >2 ng/dl (cut off of severe sepsis) was compared with that of patients with PCT < 2ng/dl and no statistically significant difference was found (20.7 ± 8.2 vs 17.9 ± 8.3, respectively, student t-test, p = 0.3). Although the CRP values in septic patients were high (150 ± 107 mg/L), no correlation between CRP and APACHE score was observed (Regression analysis, p = 0.19). No significant differences were also found in the PCT and CRP levels between patients who survived and succumbed.
In conclusion, the high levels of PCT and CRP could contribute to the diagnosis of sepsis in elderly patients but might lack prognostic accuracy.
P1746 Serum procalcitonin and organ failure in critically ill patients with sepsis
M. Kompoti, C. Kostopoulos, M. Daganou, P. Bakakos, A. Gonis, A. Rassidakis, J. Nanas (Athens, GR)
Sequential Organ Failure Assessment (SOFA) score is applied by internists for daily description of organ dysfunction in critically ill patients.
Objective: Our study investigates any association between SOFA score and serum procalcitonin during the first 48 h of clinically detectable sepsis in critically ill patients.
Patients and Methods: Forty-four consecutive patients with sepsis admitted in the intensive care unit (ICU) of the first hospital were included in our study. At 0 h (baseline), 24 h and 48 h serum procalcitonin was measured and SOFA score was calculated in all patients. Pearson's correlation coefficients were used to quantify the association between serum procalcitonin and the corresponding SOFA scores. Statistical significance level was set at p < 0.05
Results: Age (mean±SD) was 66.7±11.2 years and APACHE score at admission in the ICU was 15.2±5.8. SOFA score was 5.5±2.8 at baseline, 5.5±2.4 at 24 h and 5.5±2.6 at 48 h. Serum procalcitonin [median (interquartile range)] was 0.53 (0.23–0.85) ng/mL at baseline, 0.48 (0.22–0.76) ng/mL at 24 h and 0.44 (0.25–0.61) ng/mL at 48 h. SOFA score at all three time-points was linearly associated with the natural logarithm of serum procalcitonin (Pearson's correlation coefficients: r(0 h)=0.564 (p < 0.001), r(24 h) = 0.486 (p < 0.001) and r(48 h)=0.611 (p < 0.001).
Conclusions: In critically ill patients with sepsis, serum procalcitonin serial measurements reflect the degree of organ failure and are linearly associated with the corresponding SOFA scores.
P1747 The impact of insulin therapy on inflammatory markers in critically ill patients with sepsis
M. Kompoti, C. Kostopoulos, M. Daganou, P. Bakakos, A. Rassidakis, J. Nanas (Athens, GR)
Objective: To assess the impact of insulin therapy on serum concentration of C-reactive protein (CRP) and fibrinogen (FBG) in intensive care unit (ICU) patients.
Patients and Methods: We studied 44 consecutive patients who were admitted with sepsis in the ICU and who had a known history of type 2 diabetes or presented with stress hyperglycaemia at admission. Blood glucose was controlled with continuous intravenous insulin infusion. Serum levels of FBG and CRP were measured at 0 h, 24 h and 48 h. Regression models were fitted describing the association between changes of CRP and FBG serum concentrations from baseline to 24 h and 48 h and the total insulin dose administered during the corresponding time periods. Data analysis was performed with repeated measures test and linear regression. Statistical significance level was set at p < 0.05.
Results: CRP (mean±SD) at 0 h, 24 h and 48 h was 13.9±8.5, 13.8±8.1 and 13.8±8.8 mg/L, respectively and FBG was 561±190, 544±181 and 567±196 mg/dl, respectively. Insulin dose [median (interquartile range)] administered over the first 24 h and over the first 48 h was 40 (13–80) and 98 (28–179) IU, respectively.
Serum concentration changes of CRP and FBG were linearly associated with the administered insulin dose after adjustment for age, BMI and mean blood glucose (CRP: adjusted R-square = 0.199, p = 0.025 for 0–24 h and adjusted R-square = 0.124, p = 0.022 for 0–48 h; FBG: model for 0–24 h not significant and for 0–48 h adjusted R-square = 0.111, p = 0.023).
Conclusions: Insulin administered with continuous intravenous infusion in critically ill patients during the first 48 h of sepsis produces a significant effect on CRP and FBG serum concentrations.
P1748 Protein Z, other natural anticoagulan and procalcitonin levels in adult patients with sepsis
N. Tasdelen Fisgin, T. Fisgin, F. Ulger, S. Esen, E. Tanyel, M. Zivalioglu, H. Leblebicioglu (Samsun, TR)
Objectives: Severe sepsis is one of the major causes of mortality in intensive care unit patients. Protein Z (PZ) is a vitamin K-dependent glycoprotein synthesized in the liver that serves as a cofactor for the inhibition of factor Xa (FXa) in the presence of Ca+2. In some reports low PZ levels were reported to be a risk factor for thrombosis including coronary artery disease, ischemic stroke, but, there is no data about PZ levels in adult with sepsis. Elevated procalcitonin levels have been reported in acute bacterial infections and sepsis. It is known that a long term sustained elevation of PCT associated with poor prognosis. In this study we evaluated levels of protein Z, other natural anticoagulants (PC, PS, antithrombin III) and procalcitonin levels in patients with sepsis.
Methods: From 2004 through 2005, eligible patients were included in this study. Twenty patients were diagnosed as having sepsis. Patients with sepsis (group 1, n:20) and healthy control (group 2, n:16). Patients were evaluated for age, clinical findings, laboratory data, treatment, and prognosis. Two groups were described in this study.
Results: Protein C levels were significantly decreased than normal range in sepsis group (48.9 ± 14.4%, n:50 – 150%). Protein Z levels were significantly decreased in sepsis group (76.9 ng/mL) when compared to control patients (98.1 ng/mL) (p = 0.02). Procalcitonin levels were significantly increased in sepsis group (98.1 ng/mL) when compared to control patients (0.23 ng/mL) (p = 0.000).
Conclusions: We found that PZ and PC levels were significantly decreased in patients with sepsis, but we did not observe correlation between mortality and decreased levels of PZ, PC and increased procalcitonin levels. In additionally, we found that there was no positive or negative correlation between PZ and, PC, procalcitonin levels
P1749 Detection of the Factor V Leiden mutation in septic patients
E. Gorodnova, S. Grachev, V. Beloborodov, V. Malov (Moscow, RU)
Objective: To investigate clinical and pathogenic value of identification of activated protein C (APC) resistance associated with Factor V Leiden mutation in adult patients with bacterial sepsis. Study design is “Case–Control” = 2(23):1(10), “Control” was the group of comparison, CI=95, power= 80% (EPI INFO 6, CDC.gov). Outcome: severe bacterial sepsis; bias: sampling; confounders: icteric, lipemic and haemolyze plasma samples; deficiencies of coagulations factors VII, VIII, IX, XII; limitations: enrollment.
Methods: Case: 23 patients with meningococcal sepsis, serotypes of N. meningitidis- – A 78%, B 18%, C 4% (mortality 22%, average age 44, m/f 13/10). Control:10 patients with sepsis caused by other bacterias, like Kl. pneumoniae 34%, Str. pneumoniae 22%, E. coli 22% and Staph. aureus (mortality 50%, average age 53, m/f 7/3). Inclusion criteria: adults from 18 to 75 years old with bacterial sepsis; intensive Care Unit patients; human blood samples were obtained after informed consent, genders – both. Exclusion criteria: heparin therapy, viral infections; cancer; pregnancy and lactation. 100% Cases and part of Control patients displayed skin and mucous haemorrhagic rash, and they were bleeding of various sites. All patients were characterised by APACHE II score as severe (M = 21.5; SD = 7.3). Exposure: APCr – Quik test bioMérieux, France.
Results: The Leiden mutation is revealed in 30.5% (n = 7) of Cases and in 40% (n = 4) of Controls (OR = 0.7). Irrespective of sepsis aetiology, in case of sepsis lethal outcome, the given genetic defect came to light in 60%. All of these cases are revealed at patients of 40 years or senior. Its in 100% of cases has been registered in Control group only at persons of a male.
Conclusion: Detection of Factor V Leiden mutation in adult patients with severe sepsis is an additional and potent criterion of estimation of DIC severity (one important component of MOF). Presence of this genetic defect is an adverse background for development of severe DIC. Our study data provide the expediency and utility of Leiden mutation detection to choose the proper medical approach for DIC correction in patients with sepsis, because whether patients with the mutation would benefit from more intense or prolonged anticoagulation is unknown. It includes the replaceable therapy by an APC drug; and administration of different anticoagulants for septic patients with Leiden mutation is not proved because of pathogenetical, clinical and cost-effectiveness reasons.
P1750 Community-acquired Gram-positive sepsis
M. Lupse, M. Flonta, A. Slavcovici, A. Bica, V. Zanc, D. Tatulescu (Cluj-Napoca, RO)
Community acquired sepsis is an important cause of morbidity and mortality. Clinical characteristics of Gram-positive sepsis could suggest the aetiology, being useful for empirical antimicrobial treatment.
Material and Method: We performed a retrospective study based on case records over a 4-year period (2001–2005). We selected adult patients having at admittance in ICU Teaching Hospital of Infectious Diseases the diagnosis criteria of community-acquired sepsis, according to ACCP/SCCM expert panel recommendation and Gram-positive aetiology. We made a descriptive statistics regarding: SIRS criteria, multiple organ dysfunction syndrome (MODS), aetiology and survival using Excel statistical analysis.
Results: From 209 adult patients non-HIV with community acquired sepsis we selected 53 cases (25%) with confirmed Gram-positive aetiology, median age 51.6 years, 26 males (49%). The aetiology was: 66% S. aureus (34% of these were methicillin-resistant MRSA), 14% Enterococcus spp., 9% Streptococcus pneumoniae, 9% Streptococcus viridans, 2% others. SIRS criteria were: fever in 74% patients, tachypneea in 62%, tachycardia in 62% and leukocytosis/leukopenia in 42% patients. The percent of patients with Gram-positive sepsis was: 40% for 2 SIRS criteria, 40% for 3 SIRS criteria and 20% for 4 SIRS criteria. The organ dysfunction: 70% respiratory dysfunction (most of them acute lung injury), septic shock 22%, renal dysfunction 20%, metabolic dysfunction 18%, neurological dysfunction 18%, liver dysfunction 12% and coagulopathy 8%. 47% of patient had MODS: 21% with 2-organ dysfunction, 17% with 3-organ dysfunction and 9% with more than 3 organ dysfunction. Fatality rate was 28%.
Conclusions: S aureus is the most important aetiology of community acquired Gram-positive sepsis with an important percent of methicillin resistance. Clinical course of community acquired Gram positive sepsis is characterised by: 2 or 3 SIRS criteria, frequent with respiratory involvement, few severe criteria and a good prognostic.
Immunology and host defences
P1751 DARC and MBL2 polymorphisms in Kenya
J.K. Nganga, J.N. Waitumbi (Nairobi, KE)
Objectives: The clinical outcome of malaria infection depends on multiple factors. Certain polymorphisms within specific parasite recognition receptors (PRRs) confer a selective advantage during malaria infection. Mannose binding rectin (MBL2) which activates complementis an important PRR. The Duffy antigen receptor for chemokines (DARC) on the other hand acts as a receptor for P. vivax and mops up cytokines during a malarial infection. This study therefore sort to determine the degree and frequency of single nucleotide polymorphisms within DARC and MBL2 genes in subjects with severe malarial anaemia versus age matched controls, determine SNP allele distribution within the promoter and coding domains within each of the selected genes and finally determine the effect of selection pressure on the distribution of the alleles.
Methods: In this Case-control study, 30 children typed severe malaria anaemia were recruited as well as 30 of each age and sex-matched asymptomatic P. falciparum infected and healthy children. Venous blood was collected for DNA extraction and malaria parasites counted on Giemsa-stained thick blood films. P. falciparum infection was aslso ascertained by specific PCR assays. A 376 base pair fragment in the neighbourhood of MBL2 Arg52Cys, MBL2 Gly57glu and MBL2 Gly54Asp mutations was amplified and cycle sequenced. Data was analysed against the human genome data base for this and other mutations. Frequencies and proportions were compared by χ2 test for trend. The study was approved by Kenya Medical Research Institute Ethics Committees and had written consent from the participants' parents and or guardians.
Results: Initial analysis indicate that all the individuals genotyped were fixed for the FY0 allele as expected of a population with an African descent. This is in agreement with the earlier described effects of intense selection pressure exerted by malaria in the region of western Kenya leading to fixation of the null B allele. Though the three MBL2 mutations were common they did not seem to segregate differently in the symptomatic P. falciparum infected and healthy children except for one novel mutation as represented in Figure 1 .
Fig. 1.

Conclusion: Carrier trait analysis involving a combination of SNPs will help to determine the role of different SNPs in disease susceptibility and severity as a result of their synergistic action. the specific SNP identified on MBL2 will be further analysed for it role in malaria pathogenesis.
P1752 Mannan-binding lectin in neonates
A. Szala, S. MacDonald, I. Domzalska-Popadiuk, A. Atkinson, M. Cedzynski, A. Swierzko, J. Szemraj, M. Borkowska, J. Szczapa, M. Matsushita, M. Turner, D. Kilpatrick (Lodz, PL; Edinburgh, UK; Gdansk, PL; Poznan, PL; Kanagawa, JP)
Objectives: The mannan-binding lectin (MBL) protects the host from infection by lysis of microorganisms involving complement activation via the lectin pathway (LP). MBL may enhance phagocytosis by direct opsonisation. MBL deficiency may lead to the increased susceptibility to infection. It has been suggested to contribute to unexplained miscarriages and preterm births. We report some preliminary findings from ongoing prospective study of MBL levels, LP activities and mbl2 genotypes in Polish neonates.
Methods: MBL level in cord serum samples was determined in ELISA using specific monoclonal antibody (HYB 131–01, AntibodyShop). LP activity was determined as the ability to C4d deposition on mannan-coated plates. Genotyping was performed with the help of INNO-LIPA MBL2 kits (Innogenetics NV).
Results: The median MBL level in babies with suspected perinatal infections was 965 ng/mL (n = 203), and did not differ from that determined in newborns with no symptoms of infection (1093 ng/mL, n = 890). However, LP activity was significantly lower in the first group (181 mU/mL vs 267 mU/mL, p = 0.0017). The incidence of mbl2 gene defective alleles' (O/O) homozygosity (5.9%) was found to be almost twice that of the cohort in general (3.4%). The median MBL level in low birthweight (≤ 2500 g) babies (n = 86) did not differ from that of neonates with birthweights over 2500 g (n = 1006) (1048 vs 1076 ng/mL), however, LP activity was lower in low birthweight babies (161 vs 258 mU/mL, p = 0.006). Neither MBL nor LP differed between preterm and term babies, however the frequency of O/O genotypes among premature neonates was almost twice that of the cohort in general (6.2% and 3.2%, respectively).
Conclusion: The low LP activity may increase the risk of infection in perinatal period.
This study is partially supported by Ministry of Science and Higher Education (Poland), grant 2 P05E 011 28. Authors are very grateful to Innogenetics NV for providing INNO-LIPA MBL2 tests.
P1753 Mortality and time for detection of blood growth in patients with Staphylococcus aureus bacteraemia; implication of mannose-binding lectin
A. Smithson, A. Muñoz, B. Suarez, R. Perello, J.A. Martinez, J. Mensa, F. Lozano (Barcelona, ES)
Background: Structural and promoter MBL2 gene polymorphisms, responsible for low mannose-binding lectin (MBL) levels, are associated with a higher incidence and severity of infections. The aim of the study is to asses the possible association between mortality and time for detection of blood growth in patients with S. aureus bacteraemia and the presence of low expression MBL2 genotypes (O/O and LXA/O).
Methods: Blood samples from 49 Caucasoid patients with S. aureus bacteraemia (mean age 63 ± 16.7 years) admitted in our hospital were prospectively collected. For further comparison samples from 104 blood donors were also included in the study. Forty-two bacteraemic patients (85.7%) had associated comorbidity: 1 (2%) was a liver transplant recipient, 7 (14.2%) had cirrhosis, 17 (34.7%) had past history of heart failure or chronic lung disease, 1 (2%) had AIDS, 13 (26.5%) had underlying neoplasic diseases, 8 (16.3%) had chronic kidney failure, 13 (26.5%) had diabetes mellitus and 6 (12.24%) were receiving immunossupressive or corticosteroid therapy. Six single nucleotide polymorphisms (–550 G/C, −221 C/G, +4 C/T, codon 52 CGT/TGT, codon 54 GGC/GAC and codon 57 GGA/GAA) in the MBL2 gene were genotyped using a sequence-based typing technique.
Results: No significant differences were observed in the frequencies for low expression MBL2 genotypes between healthy controls (15.4%) and patients with S. aureus bacteraemia (10.2%) (P = 0.38; Chi square test). Nine patients (19.1%) died due to the infection, 1 of whom had a low expression MBL2 genotype (LXA/O). In the univariate analysis, chronic kidney failure was the only variable associated to death (P = 0.004; Fisher test). When time for detection of growth in blood culture vials was analysed, the existence of underlying neoplasic disease was the only variable associated to a lower average time to hemoculture positivity (P = 0.022; U Mann-Whitney), while a tendency towards a lower time was observed in patients with low expression MBL2 genotypes (P = 0.078; U Mann-Whitney).
Conclusions: Low expression MBL2 genotypes have no significant impact in survival of patients with S. aureus bacteraemia and have no relationship to the time to positivity of blood growth.
P1754 Pathogen-related association of mannose-binding lectin deficiency with community-acquired pneumonia
B.L. Herpers, H. Endeman, B.A.W. de Jong, E.J. van Hannen, G.T. Rijkers, H. van Velzen-Blad, D.H. Biesma, J.C. Grutters, G.P. Voorn, B.M. de Jongh (Nieuwegein, NL)
Objectives: Mannose-binding lectin (MBL) is an activating protein of the lectin complement pathway. As a pattern recognition receptor of innate immunity, it binds to a variety of microorganisms including respiratory pathogens like pneumococci and influenza A virus. The association of MBL deficiency and pneumococcal disease has been studied with varying outcome. Less is known about the association with other pathogens in pneumonia. We studied the role of MBL deficiency as a risk factor for community acquired pneumonia (CAP) in relation to the causative agent and clinical course of disease.
Methods: In a prospective case-control study 193 immunocompetent patients with confirmed CAP were included. Severity of disease at presentation and clinical endpoints (intubation, death) were scored. Blood cultures, respiratory specimen cultures for bacteria and viruses, urinary tests for pneumococcal and Legionella antigen, serology and PCR for atypical pathogens were performed. MBL genotypes were determined by denaturing gradient gel electrophoresis of an amplicon harbouring the three polymorphic sites in exon 1 (“0” versus wildtype “A” allele) and a SNP-PCR determining the promotor X/Y polymorphism. Genotypes were classified deficient (0/0, XA/0) or sufficient (YA/0, A/A). 223 healthy adults served as a control population.
Results: S. pneumoniae was found in 60 patients, viruses in 16 and Legionella and Mycoplasma both in 9. Significantly more patients with viral pneumonia were MBL deficient compared to controls (40% vs 17%). In contrast, none of the atypical pneumonia patients showed MBL deficiency. Patients with pneumococcal pneumonia did not differ from controls in MBL deficiency (18% vs 17%). However, MBL deficiency was associated with more severe disease at presentation in this subgroup. More MBL deficiency was found in patients with concomitant viral and pneumococcal infections than with pneumococcal infections alone. No effect of MBL was seen on mortality of CAP, independent of the pathogen.
Conclusion: MBL deficiency is associated with susceptibility to community acquired viral pneumonia. No association was found with susceptibility to pneumococcal pneumonia, although MBL deficient patients presented with more severe disease. The absence of MBL deficiency in 18 patients with atypical pneumonia is intriguing, as some other intracellular pathogens use MBL to enter their host cell. A larger study is required to confirm this relationship.
P1755 Coding and non-coding polymorphisms in the lectin pathway activator L-ficolin gene in 188 Dutch blood bank donors
B.L. Herpers, M.M. Immink, B.A.W. de Jong, H. van Velzen-Blad, B.M. de Jongh, E.J. van Hannen (Nieuwegein, NL)
Objective: Human L-ficolin (FCN) is a serum lectin characterised by a collagen-like and a fibrinogen-like domain that can activate the lectin pathway of complement. Structural and functional similarities to mannose-binding lectin (MBL) suggest a role for L-ficolin in innate immunity similar to MBL. Structural polymorphisms in the MBL2 gene, leading to functional deficiency of MBL, have been associated with disease. We screened the coding regions of the FCN2 gene for similar polymorphisms and determined their frequencies in a Dutch population.
Methods: We developed 10 denaturing gradient gel electrophoresis (DGGE) assays to screen a total of 188 Dutch Caucasians for polymorphisms in all exons of FCN2 and their flanking regions. Sequence variations detected with DGGE were confirmed with sequencing.
Results: Total gene screening in this large cohort revealed 10 single nucleotide polymorphisms (SNPs) in the FCN2 gene (table ; (a) location and allele names, (b) nucleotide substitutiuon, (c) frequency and allele count). Non-coding SNPs were found in the 5'-UTR and 3'-UTR, in introns 2, 3 and 6 and in exons 3 and 8. Two highly conserved coding SNPs were found in exon 8, leading to amino acid substitutions within the fibrinogen-like domain
Location, amino acid substitution and frequency of SNPs in FCN2
| Locationa | SNPb | Substitution | Frequencyc |
|---|---|---|---|
| 5'-UTR (1B) | c.–4A>G | − | 0.27 (101) |
| Intron 2 (3B) | c.213–7A>G | − | 0.33 (123) |
| Exon 3 (3B) | c.222T>C | Synonymous | 0.33 (123) |
| Intron 3 (3C) | c.269+10G>A | − | 0.26 (91) |
| Exon 5 (5B) | c.337C>T | p.His113Tyr | 0.01 (2) |
| Intron 6 (6B) | c.560+44G>A | − | 0.14 (52) |
| Exon 8 (8.1B) | c.707C>T | p.Thr236Met | 0. 277 (103) |
| Exon 8 (8.1C) | c.772G>T | p.Ala258Ser | 0.14 (51) |
| 3'-UTR (8.2B) | c.987T>G | − | 0.45 (169) |
| 3'-UTR (8.2C) | c.987_988insG | − | 0.01 (2) |
Conclusion: A total of ten polymorphisms were identified. Non-coding SNPs were found in the 5'-UTR and the 3'-UTR, in the introns 2, 3 and 6 and in exons 3 and 8. Although such polymorphisms have been described to influence translation or splicing events in other genes, the relevance of these SNPs in this particular gene has to be further investigated. Three coding SNPs were found in exon 5 and exon 8 encoding the fibrinogen-like domain of L-ficolin. Fibrinogen-like domains are highly conserved throughout several proteins in many species. These domains consists of approximately 220–250 residues with 26 invariant, mostly hydrophobic residues and at least 46 highly conserved residues. The two SNPs found in exon 8 respectively result in the substitution of threonine with the hydrophobic methionine and alanine with serine in the near proximity of several conserved residues.
As the fibrinogen-like domain of L-ficolin is responsible for pattern recognition, it is of interest to investigate whether the genetic variation in this domain alters the affinity or specificity of carbohydrate binding. Such possible influence could affect the recognition of invading microorganisms and thereby influence one of the first lines of defence in innate immunity.
P1756 Low mannose-binding lectin complement activation function is associated with predisposition to Legionnaires' disease
D. Eisen, J. Stubbs, D. Spilsbury, J. Carnie, J. Leydon, B. Howden (Parkville, Brisbane, Melbourne, Clayton, AU)
Objectives: Innate immune system deficiency may predispose to severe infections such as Legionnaires' disease. Mannose-binding lectin (MBL) is a pattern recognition molecule of the innate immune system that mediates killing of microorganisms through complement activation and opsonophagocytosis. There is a high frequency of MBL deficiency states. We have investigated whether MBL deficiency affects susceptibility to Legionella pneumophila through study of the Melbourne Aquarium Legionnaires' disease outbreak.
Methods: Serum samples from patients and controls that were exposed but shown to be uninfected from the Melbourne Aquarium Legionnaires' disease outbreak were tested for MBL function (C4 deposition) and level (mannan-binding). The absence of stored cellular material precluded determination of MBL2 genotype.
Results: MBL function was lower in Legionnaires' disease cases than in age and sex matched uninfected, exposed controls (0.09 U/ul vs. 0.13 U/ul, p < 0.02). The frequency of MBL deficiency with C4 deposition <0.2 U/ul was significantly higher in Legionnaires' disease cases (OR 2.60 [95% CI, 1.28–5.36]). This also applied to Legionnaires' disease cases requiring hospital care. Amongst all the Legionnaires' disease cases, the hospitalised patients had a higher frequency of MBL deficiency (OR 4.10 [95%CI 1.08 – 17.11]) than those who were well enough to not require hospitalisation. There was no difference in age between these two groups. There was no difference in MBL mannan-binding levels between Legionnaires' disease patients and controls (0.81 ug/mL vs. 0.86 ug/mL) and no significant interval change in MBL function or level after a mean of 46 days.
Conclusions: In contrast to the role MBL may play in protecting against other intracellular pathogens like TB, MBL complement activation functional deficiency appears to predispose to Legionnaires' disease. There was no acute phase change in MBL found in these patients with severe infection. MBL-mediated opsonophagocytosis may be shown to increase intracellular killing, potentially through efficient delivery of L. pneumophila to lysosomes. Additionally, lectin pathway complement activation may directly kill L. pneumophila. More study of these pathways is required.
P1757 Role of Toll-like receptor 2 in immunomodulation by lactobacilli
V. Vankerckhoven, S. Van Voorden, H. Goossens, E. Wiertz (Wilrijk, BE; Leiden, NL)
Objectives: Different lactobacilli have been shown to differentially influence immune responses. The aim of this study was to evaluate Toll-like receptor 2 (TLR2) signalling of clinical and probiotic Lactobacillus strains.
Methods: A total of 37 Lactobacillus strains (17 L. paracasei, 19 L. rhamnosus, and 1 L. murines) of different origin (18 probiotic, 2 faecal, 15 clinical, and 2 unknown) were tested for TLR2 signalling. Strains were grown in MRS broth to OD620=1 and killed by exposure to UV light. TLR2 signalling was measured as relative IL-8 promotor activation in transfected human embryonic kidney (HEK) 293 cells. IL-8 concentrations were measured using an enzyme-linked immunosorbent assay. Heat-killed Listeria monocytogenes (HKLM) and PAM3 were used as positive controls. All assays were performed at least in duplicate. Statistical analysis was performed using an unpaired T-test. P < 0.05 was considered statistically significant.
Results: Lactobacilli were shown to signal through TLR2. However, the production of IL-8 was shown to be variable for the different Lactobacillus isolates. IL-8 production ranged from 374 pg/mL to >22,000 pg/mL. More specifically IL-8 production ranged from 733 pg/mL to 21372 pg/mL for the probiotic isolates and a similar range was seen for the clinical isolates (from 1021 pg/mL to 18946 pg/mL). No significant differences in IL-8 production were seen between clinical and probiotic isolates. Also when analysing L. paracasei and L. rhamnosus isolates separately, no significant difference was seen in IL-8 production between clinical and probiotic isolates. On the contrary, L. rhamnosus isolates induced a significantly higher IL-8 production compared to L. paracasei isolates.
Conclusions: The results indicate that different lactobacilli may induce different levels of IL-8 production through TLR2. Lactobacilli have been found to modulate the immune response in various ways. This variation may be related to differential signalling through TLRs, including TLR2.
P1758 IL-8 and anti-Chlamydia trachomatis specific mucosal IGA as the best marker of Chlamydia trachomatis chronic prostatitis
S. Mazzoli, T. Cai, R. Bartoletti (Florence, IT)
Introduction: Chronic infections of the upper genital tract in males are impacting pathologies leading to severe sequelae up to infertility. Chronic prostatitis/chronic pelvic pain syndromes (Cat III NIH) remains the most frequently detected especially in young fertile men. Chlamydia trachomatis (CT) DNA was associated with “chronic abacterial prostatitis” starting from 1996. Cytokines content has been analysed in prostatitis patients.
Aim of our study was to correlate IL-8 production to CT infection in a population of 78 CP/CPPS patients positive for C.t. DNA and in a negative control group.
Materials and Methods: Selected patients (N. 78) were positive for CT genomic DNA and some for plasmidic DNA and/or secretory specific IgA in total ejaculate, but negative for common bacteria, yeasts, mycoplasmas and viruses. Control population (N. 20) was a non prostatitis healthy population. Natural human produced IL-8 was detected in total ejaculate by Quantikine IL-8 Immunoassay, (R&D Systems, MN, USA). CT plasmidic DNA was detected by Cobas Amplycor CT/NG test, (Roche Molecular systems, NJ, USA); chromosomal DNA by an in-house omp-1 Real-Time PCR. Mucosal IgA were detected by IPAzyme Chlamydia IgG/IgA by Savyon Diagnostics, Ashdod, Israel. Westen Blot anti-CT IgA was performed by CT Westen Blot kit by AID Autoimmun Diagnostika GmbH, Straberg, Germany.
Results: 98 subjects were enrolled, 78 with prostatitis and 20 healthy negative controls. 35 subjects were pDNA positive (35.7%), 54 were sIgA positive (55.1%)
IL-8 was detected in 59 out of the studied subjects (60.2%). IL-8 showed an efficient separation between patients with CT prostatitis and those without prostatitis (sensitivity 75% and specificity 100%; PPV 100% and NPV 51%) (p < 0.001). Patients with negative pDNA showed a more presence of IgA than those with pDNA positive (p < 0.001). The same correlation for IL-8 was reported (p < 0.001).
Conclusion: Mucosal immune system activation may represent a critical point in the determinism of adverse consequences associated with chronic Chlamydial infection. This activation was proved in these CT prostatitis patients by high production of specific anti-CT IgA and iperproduction of IL-8 which positively correlated with the IgA response. The continuous inflammatory reaction leads to persistent tissue damages, fibrosis and pelvic pains in these patients. In conclusion IL-8 and sIgA to CT antigens are the best immunological markers of chronic CT prostatitis.
P1759 Host defence control in children with chronic sinusitis: recolonisation of nasal microflora
I. Kuznecovs, I. Joksta, K. Jegina (Riga, LV)
Objective: Staphylococcus aureus (SA) enterotoxins are associated with host defences mechanism disturbances by elevated specific IgE production in nasal polyps (NP). It is the main cause of disbacteriosis in upper airways. Certain alpha-streptococci have been shown to have growth inhibitory activity against common pathogens in the upper respiratory tract in vitro (K. Roos, 2000). The role of normal flora of upper respiratory tract and its interference with SA in host defences in children with chronic sinusitis (CS) and NP was the aim of present study.
Methods: Nasopharyngeal swabs and nasal tissue samples were taken from 120 patients with NP and CS. Antibiotic resistance of the strains was determined with the disc diffusion method. Recolonisation with selected alpha-streptococcal strains was used in 58 patients with nasal polyposis. Method of Nonlinear diagnostic was used for demonstration
Results: The rates of nasal carriage of SA were found to be 15% in control group and 89% in patients with NP and 58% with CS. A lower number of alpha-streptococci have been found in patients with NP and CS, compared to control. Colonisation rates of alpha-streptococci strains were 90% in control and 10% in patients with nasal polyposis. MRSA was found in 2% in control and in 51% in patients with NP and 74% with CS. It was demonstrated that nasal carriage of MRSA in patients recolonised with alpha-streptococci was lower (8%) than in nontreated patients with NP and CS.
Conclusions: The results, obtained in this investigation can be used in chronic staphylococcal sinusitis treatment in children by recolonisation of nasal microflora with selected alpha-streptococcal strains. It was shown that patients recolonised with alpha streptococci with interfering activity against Staphylococcus aureus, get less recurrences of NP and CS symptoms than nonrecolonised persons. Recolonisation of microflora is a new effective method for host defence control in children with chronic sinusitis.
P1760 Immune evasion mechanisms of Aspergillus sp. in cerebral aspergillosis
G. Rambach, I. Mohsenipour, M. Dierich, C. Speth (Innsbruck, AT)
Objectives: Cerebral aspergillosis is the most dangerous complication of human Aspergillus infections with a death rate of more than 90%. The central nervous system (CNS) is separated from the periphery by the blood-brain-barrier, leaving the local innate immunity with its main components complement, cytokines/chemokines and resident immune cells (microglia, astrocytes) the only defence system. We examined putative evasion strategies of Aspergillus against these weapons: reduction of antigen presentation, inhibition of phagocytosis and degradation of complement proteins.
Methods: Immune cells were incubated with fungal secretory factors. Antigen presentation was analysed by quantification of subsequent T-cell activation. Phagocytosis was measured microscopically. Complement degradation was investigated by incubation of purified complement proteins with cerebrospinal fluid (CSF) wherein Aspergillus was grown.
Results: The fungal factor patulin significantly reduced the capacity of cells to efficiently present antigens and thus to stimulate immune cells. Furthermore, the presence of different fungal toxins resulted in diminished phagocytosis by brain-resident astrocytes. The growth of Aspergillus sp in CSF resulted in secretion of proteolytic factors which degraded various complement factors. The extent of the proteolysis was dependent of the time period of fungal growth and the used Aspergillus species. A. fumigatus, the predominant cause of cerebral aspergillosis, showed a rather quick and strong degradation, whereas the proteolysis by A. terreus was weaker and rather slow.
Conclusion: These data indicate a broad spectrum of immune evasion mechanisms executed by Aspergillus species. These processes may significantly contribute to the pathogenesis of cerebral aspergillosis. Diminished antigen presentation, attenuation of phagocytosis and probably of further immunological activities of resident defence cells as well as the degradation of complement proteins act as striking weapons against the local immunity of the CNS. On the one hand, these facts in addition to other known escape mechanisms explain the high fatality of this fungal disease, otherwise this plurality shows new optional therapeutical targets to support the established antifungal treatments. Neuroprotective substances or antagonists might neutralise or decrease the harmful effects of mycotoxins; specific protease inhibitors could strengthen the attack of complement against the fungal pathogens.
P1761 Extraction, purification and detoxification of Brucella abortus lipopolysaccharide and biological activity evaluation
I. Pakzad, A. Rezaee, M. Rasaee, A. Hosseini, A. Kasemnejad, T. Bahman, R. Saied (Ilam, IR)
Lipopolysaccharide (LPS) of Brucella abortus is important in brucellosis diagnosis and as one of the components for developing a subunit vaccine against brucellosis. Biological evaluation of B. abortus LPS and it's protection have been considered as the goal of this research. Brucella abortus LPS was extracted by n-butanol, then it was purified by ultracentrifugation and it was detoxified by alkaline treatment. Toxicity of LPS and detoxified LPS (D-LPS) by LAL (Limulus Amoebocyte Lysate) method, and pyrogenicity in Rabbit was compared. Immunological evaluation in animal model was carried out.
Purified LPS from B. abortus by butanol extraction was shown to have <2% (wt/wt) contamination by protein and <1% (wt/wt) contamination by nucleic acids. Pyrogenicity test of B. abortus LPS (10 μg/mL) and E. coli LPS (0.5 μg/mL) was positive, but for D-LPS (10, 50 μg/mL) it was negative. In LAL test, 10 ng/mL of D-LPS was negative, but 0.04 ng/mL of B. abortus LPS was positive and endotoxin unit of B. abortus LPS was less than E. coli LPS. Antibody titer of LPS group was higherthan that of D-LPS group. The difference of protection among LPS group, DLPS group comparing with negative control was significant (p≤0.05), In addition, the difference of protection ratio between LPS and D-LPS groups was not significant.
Results show that D-LPS toxicity is severely decreased; we can use several as many as B. abortus LPS for stimulating immune system. Besides, ability of B. abortus LPS is likely much less than the LPS from E. coli to evoke endotoxic shock, and it can be used directly as immunogen. In addition, protection of LPS and D-LPS probably is due to humoral important role in secondary infection of brucellosis.

P1762 Effect of moxifloxacin and human beta-defensin 2 on ICAM-1 and cytokines expression in human lung epithelial cell line
M. Tufano, I. Paoletti, B. Perfetto, M. Iovene, L. Tudisco, G. Donnarumma (Naples, IT)
Interaction of pathogenic bacteria with airway epithelium is usually an essential step in the infectious process. Epithelia in the human airways, from the nasal aperture to the alveoli, are covered in a protective film of fluid containing a number of antimicrobial proteins.
Antimicrobial peptides have been identified as key elements in the innate host defence against infection. Defensins are single chain strongly cationic peptides of molecular weight 3000–4500 and are one of the most extensively studied classes of antimicrobial peptides. In the airways, the antimicrobial peptide human beta-defensin 1 (HBD1) is constitutively expressed by epithelia at low levels while the related peptide human beta-defensin 2 (HBD2) is predominantly induced at sites of inflammation. A number of antibiotics have been found to have significant immunomodulatory properties both in vitro and in vivo in animal models. Moxifloxacin (MXF) is a fluoroquinolone with activities against both Gram-positive and Gram-negative bacteria. It has been suggested that MXF has inhibitory and stimulatory effects on the immune system. Our data demonstrate that HBD2 stimulation produces a remarkable increase of ICAM-1 protein expression, but does not induce proinflammatory cytokines expression. In contrast, the costimulation with MXF/HBD2 induces a different production of proinflammatory cytokines (IL-8, IL-1 and IL-6). Moreover, the adhesion of PMN to airway epithelium is increased over either baseline when stimulated with HBD2 or with MXF/HBD2. These data indicate that the association of MXF/HBD2 can play a crucial role during the infectious processes determining a balanced release of proinflammatory cytokines and adhesion molecules, possessing chemotactic activity for neutrophils.
Experimental models of infectious disease
P1763 Bacillus smithii TBMI12 spores as a potential competitive exclusion agent against Salmonella enteritidis
I. Suitso, E. Jõgi, E. Talpsep, P. Naaber, A. Nurk (Tartu, EE)
Objectives: Our work-group has isolated thermopilic sporogenous bacteria Bacillus smithii TBMI12 (B. TBMI12) from human gut and supposed it to be a potential probiotic. An objective of the current research was to investigate the following hypothesis:
-
1
B. TBMI12 spores are able to colonise intestinal tract of mice.
-
2
Mice colonised previously with B. TBMI12 spores will not become infected with pathogen Salmonella enteritidis wt (S. enteritidis).
Methods: In total 25 mice were divided into three groups. The group A was intragastrically inoculated with one dosage of S. enteritidis cells 106 CFU. The group B was treated daily for three days with B. TBMI12 spores 108 CFU and after that inoculated with one dosage of S. enteritidis cells 106 CFU. The group C was inoculated intragastrically with B. TBMI12 spores 108 CFU on the 1st and the 8th day. Samples of faeces were taken maximally over 72 hours and plated on selective media to count B. TBMI12 or S. enteritidis colonies. Whole A and B group were executed on the 18th day (group C: 30th day) and the samples from liver and spleen were plated on selective media too.
Results: All mice from the group A were infected with S. enteritidis. The first colony was detected from faeces on the very next day after inoculation.
Meantime the group B mice were successfully colonised by B. TBMI12 but the count of B. TBMI12 found in faeces decreased during two weeks about ten times. S. enteritidis was detected in faeces first time on the 13th day and 40% of mice were infected.
First dosage of B. TBMI12 spores colonised the group C mice only for a couple of days. However, stabile population of B. TBMI12 in intestinal tract of mice was caused by the second dosage. B. TBMI12 did not cause any harm to mice.
Conclusion: The current research showed that it is possible to colonise intestinal tract of mice with several dosages of B. TBMI12 spores. Unfortunately the number of bacteria fell about ten times in two weeks. 60% of the mice colonised with B. TBMI12 spores were not infected with S. enteritidis. Therefore we suggest Bacillus smithii TBMI12 spores as a potential competitive exclusion agent against Salmonella enteritidis.
Figure 1.

Infected mice. Infection of the group A (triangle) and the group B (quadrate) mice with pathogen S. enteritidis was detected by plate count of faeces, liver or spleen onto selective media XLD.
P1764 In vitro efficacy of moxifloxacin, levofloxacin, piperacillin/tazobactam and metronidazole in a murine model of complicated intra-abdominal infection
R. Endermann, K. Ehlert (Wuppertal, DE)
Objectives: Complicated intra-abdominal Infections cIAIs) are usually polymicrobial, with aerobic and anaerobic bacteria. Moxifloxacin (MXF), a broad-spectrum antibiotic given once daily orally or intravenously, has recently been approved in the US and other countries for the treatment of cIAIs. Here we studied the efficacy of MXF, levofloxacin (LVX), piperacillin/tazobactam (TZP) and metronidazole in the murine cecal ligation and puncture model (CLP). CLP represents a peritonitis model with clinical features comparable to human peritonitis.
Methods: Mice were anaesthetised and the peritoneum opened with a small cut. The cecum was taken out of the peritoneum without harming the surrounding intestine. A ligation was set at the proximal end of the cecum and thereafter the ligated part punctured using an injection needle (21G). Then, the ligated part of the intestine was put back in the peritoneum and the wound closed. Mice were treated at 4, 18 and 24 h after operation (10 animals/group). Survival of treated and non-treated mice was determined daily.
Results: After ligation and puncture, 70% of the untreated control animals died by day 3, in the stage of early sepsis. A further 20% of the control group died on day 8–9. No improvement in survival was achieved with 3×10 or 3×25 mg/kg metronidazole. In contrast, treatment with MXF, LVX or TZP at dosages of 3×10 mg/kg IV resulted in a survival rate of 70–80% on day 3 and 50–60% on day 14.
Conclusions:
-
–
MXF is effective in this model of polymicrobial peritonitis.
-
–
The efficacy of MXF is comparable to broad spectrum antibiotics like LFX and TZP.
-
–
Metronidazole showed no therapeutic efficacy.
-
–
The failure of this strictly anti-anaerobic agent and the successful outcomes with levofloxacin might be because the early mortality in this model is due to Gram-negative aerobic infection.
Research funding: Bayer HealthCare Pharmaceuticals
P1765 Clarithromycin enhances the effect of piperacillin/tazobactam in an experimental model of empyema by multidrug-resistant Pseudomonas aeruginosa
T. Tsaganos, T. Adamis, I. Txepi, I. Tsovolou, G. Zografos, H. Giamarellou, E. Giamarellos-Bourboulis (Athens, GR)
Objective: Clarithromycin (CL) is considered to present immunomodulatory properties for chronic inflammatory disorders of the lung. Its effect was tested in combination with piperacillin/tazobactam (PT) in experimental pleuritis by multidrug-resistant P. aeruginosa (MDRPA).
Methods: 0.3 ml of tuberntine were injected in the right pleura of 32 rabbits followed by the inoculation of 7log10 cfu/mL of one isolate of MDRPA on the next day. Isolate was resistant to PT, ceftazidime, imipenem and ciprofloxacin. Rabbits were then equally divided into four groups: A, controls; B, intravenous 80 mg/kg CL followed by a second dose of 30 mg/kg two hours after; C, intravenous 400 mg/kg of PT; D: both agents. Therapy was administered only the next day of bacterial challenge. Pleura was punctured each second day; fluid was quantitatively cultured. On the seventh day, rabbits were sacrificed and the segments of the lower lobe of the right lung were quantitatively cultured.
Results: Mean values of bacteria are given in the Table . Mean log10 of bacteria in lung after sacrifice of groups A, B, C and D was 4.63, 5.53, 4.23 and 2.77.
| Time (days) | log10 cfu/mL |
|||
|---|---|---|---|---|
| A | B | C | D | |
| 1 | 5.12 | 6.15 | 6.18 | 4.17 |
| 3 | 6.68 | 6.00 | 4.64 | 6.59 |
| 5 | 3.98 | 4.80 | 5.00 | 2.22 |
| 7 | 4.10 | 4.06 | 3.95 | 1.49 |
Conclusions: Co-administration of CL and PT decreased considerably bacterial load in lung and pleural fluid. There co-administration is proposed for the management of nosocomial pneumonia and empyema by MDRPA.
P1766 Echinococcus cyst of liver and lung in animal models treated by radio-frequency thermal ablation. An ex vivo pilot experimental study
V. Lamonaca, L. Maruzzelli, M. Minervini, N. Virga, R. Di Stefano, P. Tagliareni, A. Luca, G. Sardina, G. Vizzini, U. Palazzo, B. Gridelli (Palermo, IT)
Background: Echinococcus cyst (EC) usually involves liver and lung. Treatment is surgery or PAIR (Percutaneous Aspiration, ethanol Injection and Reaspiration); infective complications and biliary fistulae, mainly due to proligera detachment, easily occur. Radio-frequency thermal ablation (RITA) is currently used for treatment of neoplasms, primarily HCCs.
Aim: evaluate RITA for treatment of EC of liver and lung, in animal models (explanted organs).
Materials and Methods: infected livers and lungs from slaughtered animals were studied. Cysts were photographed, classified, measured and had ultrasounds (US). Some of the cysts had RITA: 150 watt, 80°C, 7 minutes; just 1 liver and 2 lung cysts: 70°C, 7 minutes. During RITA, temperature was monitored in and outside. A second needle, placed inside the cyst, stabilised pressure. Both treated and untreated cysts were sectioned and examined by histology. Viability definition; alive cyst: clear fluid plus well preserved scolici and/or preserved germinal layer at histology; killed cyst: necrotic scolici and/or necrotic germinal layer at histology.
Results: 28 cysts were studied, 16 hepatic and 12 lung. US showed proligera adhese in 100% of hepatic and 75% of lung; focally detached in 25% of lung cysts. 17 cysts, 9 hepatic (average volume 40.5 ml) and 8 lung (average volume 137 ml) had RITA. The average of highest temperatures outside the cyst was 41.2°C (22.3–68.3) for liver and 49.3°C (38.8–55.0) for lung. The average extension of parenchymal necrosis outside the cyst was 0.64 cm (0–2) for liver, 1.57 cm (0.8–2.5) for lung. After RITA pathology showed: rate of killing 100% in hepatic (9/9) and 100% in lung cysts (8/8); proligera adhese in 67% (6/9) of hepatic and 75% (6/8) of lung cysts; focally detached in 33% (3/9) of hepatic and 25% of lung. 2 cysts, adjacent to treated ones, were killed despite no direct treatment; 1 more cyst was partially alive but the procedure was non-optimal (cyst broken). 11 cysts, 7 hepatic and 4 lung (average volume 30 ml), had no treatment. Pathology showed all of them alive and with proligera adhese.
Conclusion: Our study showed RITA is very effective in killing EC of explanted liver and lung. It causes limited damage to normal tissue, biliary duct damage, and keeps proligera attached. In vivo, RITA could be therapeutic with a lower incidence of post-procedure complications and probably also effective on external daughter/adjacent cysts. In vivo studies are required to confirm and validate such a new therapeutic approach.
P1767 Probiotic influence of Bacillus smithii TBMI 12 against Clostridium difficile infection in mice and hamster model
E. Jõgi, I. Suitso, E. Talpsep, P. Naaber, K. Lõivukene, A. Nurk (Tartu, EE)
The objective of our study is a investigation about the possibilities to use sporogenic lactic acid producing bacilli as preventive agency to avoid Clostridium difficile associated diseases in murine and hamster model. Possible probiotic bacteria Bacillus smithii TBMI 12 (sporogenic, lactic acid producing) was isolated from human gut and so we try this train probiotic ability against Clostridium difficile associated diseases.
Clostridium difficile (CD) is a common nosocomical pathogen that is the causative agent of pseudomembraneous colitis and a major cause of antibiotic-associated diarrhoea. Usual therapy in these cases is metronidazole or vancomycine oral treatment. But in this way there's also a chance of recurrences of therapy, estimably up to 50% of cases In vivo were used for Clostridium difficile infection two models: cefoxitine treated mice model and ampicilline treated hamster model.
In mice model experimental animals were treated during 5 days with cefoxitine, and then inoculated intragastrically with C. difficile strain VPI 10463 (107 cells). Next day 4 groups were treated; B. smithii TBMI 12 (108 spores); B. smithii TBMI 12 (108 spores) + metronidazole; metronidazole and control group.
In hamster model experimental animals were treated with single dose ampicilline, inoculated with B. smithii TBMI 12 (108 spores) and after 4 hours were group infected with C. difficile strain VPI 10463 (105 cells). After C. difficile infection hamsters received again 2 doses spores next 2 days. Control group was infected without probiotic treatment (without B. smithii TMBI 12 spores).
Results in CD mice model does not indicate good effectivity of B. smithii TBMI 12 spores. Significant positive influense was only in between comparison of metronidazol or metronidazol+ TBMI 12 groups. This indicates that mouse model is complicated in such investigation.
Bacillus smithii TBMI 12 influence in antibiotic-compromised hamsterCD infection model was more promising. As hamsters are extremly sensitive to CD toxins, the in vivo experiments are more trustable than experiments with mice. B. smithii TBMI 12 was maintained in hamsters during a month, and all hamsters colonised with spores (3 days 108), survived the infection with 104 CD cells. Hamsters in control group who received only 104 CD cells died with 48 hours.
Conclusion: Bacillus smithii TBMI 12 spores could be demonstrate probiotic effect against Clostridium difficile infection especially in hamster model.
P1768 Pertactin evaluation as immunogen in murine model
E. Rosales, A. Fernandez-Ramirez, G. Barcenas-Morales, J. Montaraz-Crespo (Cuautitlan Izc., Edo de Mexico, MX)
Pertactin (PRN) is an antigen involved in protective immunity against whooping cough, caused by Bordetella pertussis. The presence of antibodies against pertactin correlates with protection in humans. B. pertussis is closely related with B. bronchiseptica, aetiological agent of respiratory tract infections in animal species. The aim of the present work was to obtain and purify pertactin from B. bronchiseptica and evaluate it as an immunogen in a murine model.
Previously, infection murine model has been employed using three different mouse strains (NIH, Hsd:ICR and BALB/c) and four B. bronchiseptica strains by aerosol via. In the present work, three methods were tested in order to obtain PRN from two different B. bronchiseptica strains: I. By sonicate and acetone precipitation; II. By acid extraction method (pH = 3) and, III. By ultracentrifugal series and ethanol precipitation. The product of each method was corroborated by SDS-PAGE and western blot (BB07 and BB05 monoclonal antibodies) assays. Groups of mice were immunised with PRN, PRN+FCA or B. bronchiseptica bacterin (day zero) and immunisations were repeated at day 15. All mice were challenged with B. bronchiseptica at 21st day; cfu/lung were determined at days 0, 3, 5, 7, 10, 15 and 21 after challenge. Additionally, the two groups of immunised mice with each pertactin preparation was tested in a cross-challenge experiment.
Hsd:ICR and BALB/c mouse strains were susceptible to infection by LBF (isolated from pig) and ESP1 (from human) B. bronchiseptica strains, respectively. PRN was found to be protective for both mouse strains, and this protection was even better than that conferred by whole cell bacterin. Finally, cross protection effect was not observed when the animals were immunised with the pertactin preparations.
In conclusion, the animal test described in this work could be considered as a good option for future studies of comparison the protection grade using different commercial pertactin vaccines.
P1769 Comparative efficacies of telavancin, vancomycin and linezolid in a rabbit model of methicillin-resistant Staphylococcus aureus osteomyelitis
L-Y. Yin, J. Calhoun, T. Thomas, E. Wirtz (Columbia, US)
Objectives: Staphylococcus aureus is the most common pathogen isolated in osteomyelitis. However, therapeutic options are limited for osteomyelitis infections caused by methicillin-resistant S. aureus (MRSA) strains. Telavancin is a novel, rapidly bactericidal lipoglycopeptide with a multifunctional mechanism of action against Gram-positive bacteria, including MRSA. The aim of the current study was to evaluate the efficacies of telavancin, vancomycin and linezolid in a rabbit model of MRSA osteomyelitis.
Methods: Localised osteomyelitis was induced in New Zealand White rabbits by percutaneous injection of 106 colony-forming units of MRSA clinical isolate 168–1 through the lateral aspect of the left tibial metaphysis into the intramedullary cavity. The infection was allowed to progress for 2 weeks, at which time rabbits with radiographically confirmed, localised proximal tibial osteomyelitis were randomised into four treatment groups (n = 15 per group): untreated controls, telavancin 30 mg/kg subcutaneously every 12 hours, vancomycin 30 mg/kg subcutaneously every 12 hours, and linezolid 60 mg/kg orally every 8 hours. Antibiotic treatment was continued for 4 weeks, followed by 2 weeks without treatment. The rabbits were then euthanised, and the left and right tibias were recovered from each animal. Bone matrix and bone marrow from each tibia were cultured on blood agar plates, and bacterial counts per gram of tissue were determined.
Results: For the MRSA strain used in the study, the MIC was 0.25 mg/mL for telavancin, 0.5 mg/mL for vancomycin and 0.5 mg/mL for linezolid. The left tibias of rabbits from each treatment group yielded positive cultures for MRSA as follows: untreated controls, 9 of 15 (60%); telavancin-treated, 3 of 15 (20%); vancomycin-treated, 3 of 15 (20%), and linezolid-treated, 4 of 14 (29%).
Conclusion: Rabbits with tibial MRSA osteomyelitic lesions treated with either telavancin or vancomycin showed 80% infection clearance, and those treated with linezolid showed 71% clearance. Untreated controls demonstrated only 40% clearance. These results indicate that telavancin is efficacious in a rabbit model of MRSA osteomyelitis, with efficacy comparable to that of vancomycin and to linezolid.
P1770 Lack of robust neuroprotection by erythropoietin in acute experimental pneumococcal meningitis
D. Grandgirard, O. Steiner, R. Gäumann, S. Leib (Berne, CH)
Objectives: Erythropoietin (EPO) is neuroprotective in models of brain injury e.g. ischaemia, hypoxia, experimental autoimmune encephalitis, and subarachnoid haemorrhage. Bacterial meningitis causes brain injury including cortical necrosis and hippocampal apoptosis. Here, the effect of erythropoietin was evaluated in an infant rat model of acute pneumococcal meningitis (PM).
Methods: PM was induced in 11 day old Wistar rats by intracisternal injection of Streptococcus pneumoniae (serotype 3). Eighteen hours after infection, antibiotic therapy (ceftriaxone, 100 mg/kg s.c. bid) was initiated. Animals were randomised for treatment with EPO or an identical volume (150 microL) of saline. Different treatment regimens were tested i.e. pre-treatment studies (administration of EPO at the time of infection) and post-treatment studies (administration of EPO simultaneous to antibiotic therapy) and different doses were evaluated i.e. 5, 50 and 500 Units/kg/d EPO i.p). Survival was monitored and hippocampal apoptosis and cortical necrosis were assessed by histomorphometry at 36 h after infection.
Results: Survival, evaluated by Kaplan-Meier curves, was not significantly influenced by administration of EPO, for all regimens tested. While pre-treatment regimens showed a non-significant trend towards a decrease in hippocampal apoptosis (Table 1 ), post-treatment regimens had no effect on brain injury.
Table 1.
Hippocampal apoptosis score (mean±SD), in animals 36 h after infection. Animals were randomised for different dosage regimens of EPO or saline.
| Dosage of EPO in pre-treatment | Hippocampal apoptosis score (n) |
t-test P-value | |
|---|---|---|---|
| Saline | EPO | ||
| 5 U | 1.0±0.8 (12) | 0.8±0.6 (13) | 0.46 |
| 50 U | 0.8±0.5 (21) | 0.5±0.5 (14) | 0.096 |
| 500 U | 0.6±0.4 (27) | 0.5±0.4 (25) | 0.21 |
Conclusions: In contrast to other models of brain injury, EPO had no robust neuroprotective effect in this infant rat model of PM. The multifactorial disease mechanisms that include inflammatory, ischemic, excitotoxic and pathogen-derived mechanisms may explain the lack of neuroprotection. A beneficial effect of EPO in the regenerative phase of PM is however not excluded.
P1771 The effects of probiotics on allergy
R. Can, B. Aridogan, M. Demirci (Samsun, Isparta, TR)
In this study, we aimed to investigate the effects of the probiotic mixture which we prepared from Lactobacillus and Enterococcus strains isolated from healthy human gut flora and the standard Lactobacillus GG (ATCC 53103) probiotic strain on immune responses in allergy in an experimental animal model.
We prepared a mixture from a total of 13 probiotic strains composed of 10 Lactobacilli (8 Lactobacillus plantarum, 2 Lactobacillus rhamnosus) and 3 Enterococcus faecalis which are isolated from the gut flora of healthy people. We administered the probiotic mixture, standard strain LGG and skim milk suspension orally to Balb-c mice in the study and control groups for 28 days. The mice in the study groups were immunised by intraperitoneal injection of Ovalbumin on days 14 and 21 and the mice in the control groups were injected PBS only.
The OVA specific IgE levels were assayed by ELISA method in serum in each group. The OVA specific IgE levels of the study groups which were administered probiotic and standard strain were found lower than the skim milk fed groups. The difference was statistically significant (p < 0.001).
The OVA specific IgE levels in the group which was given the probiotic mixture were found lower than the group which was fed by a standard probiotic diet but the difference was insignificant. There was statistically no difference between the OVA specific IgE levels of the control groups which were not immunised (p > 0.05). The OVA specific IgE levels of the study groups were found significantly higher than the control groups (p < 0.001).
In conclusion, probiotics may put forward to treatment of allergic diseases as a new alternative but specific strains of probiotics with immunoregulatory effects need to be clearly identified and their mechanisms of action will have to be well characterised.
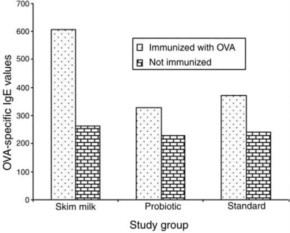
OVA-specific IgE levels in study and control group.
P1772 Pathogenicity of viridans streptococci in three murine in vivo models
G. Genzel, J. Beer, T. Rüdiger, R. Schaumann, P.D. Nitsche-Schmitz, G.S. Chhatwal, A.C. Rodloff (Leipzig, Braunschweig, DE)
Objectives: To establish a murine model of infection, which allows to study the pathogenic potential of various viridans streptococci.
Methods: Three different models of infection were first established using Staphylococcus aureus (ATCC 25923) as reference: (i) intravenous (i.v.) application into the tail vein; (ii) intramuscular (i.m.) application into the thigh; (iii) application into a preformed pouch. The same models were then used to assay the pathogenic potential of Streptococcus oralis, S. anginosus, and S. salivarius that had been isolated from relevant clinical samples. The infection dose was 108 cfu. Seven days post infection surviving mice were killed and bacterial loads in liver, spleen, kidney, blood, abscesses in the muscles and in granulomas were determined. If not indicated otherwise, 20 mice per infection model and bacterial species were infected.
Results: In the i.v.-model, S. aureus was isolated in different organs of 11 mice: liver (n = 9), kidney (n = 4), spleen (n = 6), blood (n = 2) while S. oralis was detected in organs of 4 animals: spleen (n = 3); blood (n = 1). In the i.m.-model S. aureus was isolated out of 18 muscles, infections of other organs occurred in 9 animals. In this model, S. oralis was isolated from the thighs of 15 animals and S. anginosus in 15 and S. salvarius in 19 cases, respectively. CFU found in the abscesses ranged from 106 to 108 for S. aureus while only 102 to 106 viridans streptococci were recovered. In the granuloma pouch-model (n = 18) S. aureus was detected in 16 granulomas. Six mice had additional organ infections. By comparison S. oralis was isolated from the granulomas of 17 animals.
Conclusion: The i.m.-model and the granuloma pouch-model showed that viridans streptococci have the potential for abscess formation in immuno-competent mice. However, all 3 models showed that the virulence of the tested strains was drastically reduced as compared to S. aureus. The i.m.-inoculation seems to be a suitable model for studying abscess formation of viridans streptococci and is easy and timesaving to perform.
P1773 The role of interleukin-1 during Pseudomonas aeruginosa bacteraemia in compromised host
T. Horino, T. Matsumoto, M. Uramatsu, M. Tanabe, K. Tateda, S. Miyazaki, Y. Aramaki, Y. Iwakura, K. Yamaguchi (Tokyo, JP)
Objectives: The roles of Interleukin (IL)-1 in infectious diseases are fluctuated by the experimental conditions, especially in the difference of pathogens and immunological disorders. Therefore in this study, we evaluated the role of IL-1 in bacteraemia due to Pseudomonas aeruginosa, by comparing IL-1-deficient mice and WT (WT) mice, with or without cyclophosphamide pretreatment.
Methods: P. aeruginosa bacteraemia was induced by intravenous injection of P. aeruginosa strain D4 via retro-orbital plexus. To evaluate the role of IL-1 under various immune conditions, we used compromised host following cyclophosphamide treatment, neutropenic mice by anti-Gr-1 antibody and macrophage-depleted mice by liposomes containing dichloromethylene diphosphonate. Furthermore, to investigate bacterial clearance under macrophage-depleted condition, mice were sacrificed after bacterial inoculation, and cardiac blood, liver and spleen samples were obtained aseptically. In addition, we compared Tumour necrosis factor alpha, IL-6 and interferon-gamma productions between IL-1-deficient mice and WT mice in early and late phase sepsis.
Results: Survival rates after P. aeruginosa bacteraemia did not show significant difference between IL-1-deficient mice and WT mice. On the other hand, cyclophosphamide pretreatment in both groups of mice cause significantly higher mortality in IL-1-deficient mice compared to WT mice (P < 0.01). Bacterial counts in the blood, liver, spleen, kidney and lungs were significantly higher in IL-1-deficient mice than in WT mice following cyclophosphamide treatment. These significant differences were represented in macrophage-depleted mice, but not in neutropenic mice. Under macrophage-depleted condition, bacterial counts in blood, liver, and spleen were higher in IL-1-deficient mice than in WT mice, and Tumour necrosis factor alpha in the liver in early phase sepsis were significantly lower in IL-1-deficient mice than in WT mice.
Conclusion: Our results indicated that the role of IL-1 during bacteraemia due to P. aeruginosa was enhanced in immunocompromised conditions, especially in the devastation of macrophage functions.
P1774 Vitamin B6 attenuates cellular energy depletion and hippocampal apoptosis in experimental pneumococcal meningitis
M. Wittwer, C. Bellac, R. Coimbra, S. Leib (Berne, CH)
Objectives: In pneumococcal meningitis (PM) the cellular energy status may contribute to neuronal apoptosis in the hippocampal dentate gyrus. In experimental PM we evaluated the effect of vitamin B6 on cellular energy status in the hippocampus by assessment of NAD+ levels and on apoptosis in the hippocampal dentate gyrus.
Methods: Eleven day old Wistar rats were infected intracisternally with 10 microL of saline containing 1.00E+06 cfu/mL Streptococcus pneumoniae (SP; serogroup 3). Animals were randomised for treatment with vitamin B6 (600 mg/kg s.c. 0 and 18 h after infection, n = 20) or an equal volume (360 microL) of saline (n = 17). Brains were analysed for the extent of hippocampal apoptosis by histomorphometry at 36 h after infection. For assessment of cellular energy status NAD+ levels were measured at 0, 12, 18, 24 and 36 h after infection in hippocampal tissue from animals with PM treated with vitamin B6 or with saline and in uninfected age matched littermates (n = 3 for each experimental group and time).
Results: Compared to saline, vitamin B6 significantly (p <0.02) increased cellular NAD+ in hippocampal tissue at 24 h after infection from (mean ± SD) 1756.4 ± 350.0 to 2484.4 ± 146.5 (figure 1A ) and significantly (p < 0.03; figure 1B) reduced hippocampal apoptosis from [median score (range)] 0.44 (0–1.67) to 0.12 (0–0.79).
Fig. 1.

Conclusion: In experimental PM vitamin B6 prevents the decrease in cellular energy in the hippocampus during the acute disease phase, when hippocampal apoptosis develops. This improved energy status is associated with a significant attenuation of neuronal apoptosis in the hippocampal dentate gyrus at 36 h after infection. Thus, vitamin B6, a well tolerated and readily available compound may represent a promising candidate for the adjunctive therapy of PM.
P1775 Efficacy of telavancin in a murine model of pneumonia induced by methicillin-susceptible Staphylococcus aureus
S. Hegde, N. Reyes, R. Skinner, S. Difuntorum (South San Francisco, US)
Objectives: Telavancin, a novel, rapidly bactericidal lipoglycopeptide with a multifunctional mechanism of action against Gram-positive bacteria (including methicillin-resistant Staphylococcus aureus [MRSA]), exhibited efficacy in a murine model of pneumonia caused by MRSA in a previous study. The objective of the current investigation was to evaluate the efficacy of telavancin in a murine model of pneumonia induced by methicillin-susceptible S. aureus (MSSA) in comparison with three other antimicrobial agents.
Methods: Female Bagg inbred albino c-strain (BALB/c) mice, weighing 18–20 g, were rendered neutropenic with two intraperitoneal doses of cyclophosphamide 250 mg/kg administered 4 and 2 days prior to infection. Mice were infected with an intranasal inoculation (50 microL) of 107 colony-forming units of MSSA strain ATCC 29213. Infected mice were then allocated to one of five treatment arms: telavancin 40 mg/kg subcutaneously (SC) every 12 hours, nafcillin 40 mg/kg SC every 4 hours, linezolid 80 mg/kg intravenously every 12 hours, vancomycin 110 mg/kg SC every 12 hours, or no drug (control group). Treatment was initiated at either 4 or 8 hours post-inoculation. Drug doses were calculated to simulate human exposures (area under the curve or time >MIC) at therapeutic doses. Lungs were harvested and homogenised 24 and 48 hours after inoculation to determine the bacterial titre using trypsin-soy agar plates containing aztreonam 1.0 μg/mL (to select for MSSA).
Results: Telavancin demonstrated potent bactericidal activity against MSSA strain ATCC 29213 in vitro in terms of MIC, and in vivo in terms of reduction in lung bacterial titre, compared with nafcillin, linezolid and vancomycin (Table ).
Table.
MICs and lung bacterial titresa
| Treatment | MIC (μg/mL) | Change in lung bacterial titre (log CFU/g) (n = 6) |
|
|---|---|---|---|
| 4 hb | 8 hb | ||
| Telavancin | 0.5 | −4.3†‡ | −3.2†§ |
| Nafcillin | 0.25 | −1.4† | −1.8† |
| Linezolid | 2 | −0.4 | 0.3 |
| Vancomycin | 1 | −2.9† | −2.2† |
MIC, minimum inhibitory concentration; CFU, colony-forming units.
Time (post-inoculation) of treatment initiation.
P < 0.05 vs pre-treatmen titre
P < 0.05 vs nafcillin, vancomycin and linezolid
P < 0.05 vs nafcillin and vancomycin.
Conclusion: The data from the present study demonstrate the efficacy of telavancin in reducing lung bacterial burden in an MSSA model of pneumonia in neutropenic mice. The reduction in lung bacterial titre with telavancin was significantly greater than with nafcillin, vancomycin and linezolid.
P1776 Efficacy of linezolid alone and in combination with rifampin in staphylococcal experimental foreign-body infection
O. Murillo, G. Euba, A. Domenech, A. Garcia, F. Tubau, C. Cabellos, F. Gudiol, J. Ariza (Barcelona, ES)
Objectives: To test the efficacy of linezolid alone (L) or combined with rifampin (R) against methicillin-susceptible Staphylococcus aureus (Sa) in comparison with cloxacillin (O) +R and R alone in vitro and in vivo in a rat tissue cage model of foreign-body infection.
Methods: In vitro studies: MICs (mg/L) of Sa (ATCC 29213) were L 4, O 0.5 and R 0.015. MBCs and 24 h kill-curves were performed in both the log-phase (EXP) and stationary phase (STP). Drugs were tested for a range of concentrations according to their MICs and achievable levels in human serum. Animal studies: Two Teflon tissue cages with 2 PMMA cover-slips each were subcutaneously implanted in rats. Three weeks after surgery, tissue-cage fluid (TCF) was inoculated with Sa and three weeks later, therapy was started and administered for 7 or 10 days. Therapeutic groups were (mg·kg/h): L (35/12), R (25/12), O (200/12) + R, L+R and controls (C). All groups were compared at days 7 and 10 (except for the O+R group, which was only evaluated at day 7) after the initiation of therapy. Quantitative cultures of TCF before and at the end of therapy were performed and differences between bacterial counts used as criteria of efficacy. Screening of R- or L-resistant strains was performed with culture of samples in agar plates containing R or L at 4 μg/mL.
Results: MBCs (mg/L) in the EXP and in the STP of Sa were respectively: 1, >256 (O); 64, >256 (L) and >8, >8 (R). Time-kill studies in the STP showed indifference and no bactericidal activity for any combination of L+R and O+R. Bacterial killing in TCF (means of decrease in log cfu/mL) at day 7 was (n): L 0.63 (26), R 1.2 (26), L+R 1.01 (28), O+R 1.29 (20) and C 0.33 (20). All groups showed better result than C (p < 0.05, ANOVA) and O+R and R were also better than L (p < 0.05). After 10 days of therapy the killing in TCF was L 1.22 (15), L+R 1.44 (16), R 0.1 (13) and C 0.4 (18); L and L+R were better than R and C (p < 0.05). Neither R- nor L-resistant strains were detected in TCF for combined therapies, whereas R-resistant Sa using R alone was observed in 90% and 100% of cases at days 7 and 10, respectively.
Conclusions: Combinations L+R and O+R were likely efficacious and as active as R alone, but both prevented the emergence of R-resistant Sa. L+R increased the efficacy with time in contrast with the decreased activity of R alone due to the growth of resistant strains. L alone was less effective although development of L-resistant Sa was not observed.
P1777 Investigation on efficacy of garlic extract (Allium sativa) against burn wound infection by Pseudomonas aeruginosa in burned animal model
M. Erzanlou, R. Arab, R. Alaei (Ardabil, IR)
Objective: Despite advances in medical cares, infections are the major causes of death in patients with sever burn. Gram negative rod, P. aeruginosa, is among the leading causes of burn wound infection. The situation for patients with P. aeruginosa infections is particularly problematic, because this organism rapidly evolves to multi-drug resistant forms and spreads through hospital wards and burn units. So it could not be eradicated readily with common antibiotic therapies.
Methods: Burn was induced on the back of anaesthetised animals by heated brass bar, after the hair removal. Bacterial infection was established by topical applying of highly pathogenic clinical isolate of P. aeruginosa. Potential of garlic extract on reduction of mortality was evaluated by topical application of 10 percent (V/V) garlic extract on burned and infected animals, and was compared with two control groups: burned and infected animals either treated with topical silver sulfadiazine (SSD) (1%) or left untreated. The same groups were subjected to evaluate bacterial counts in organs (blood, liver, spleen and skin).
Results: Our results indicated that topical administration of garlic extract (10%) extended the survival of mice for 3–6 days, compared with survival of the untreated group. Both garlic extract and SSD treatments reduced the microbial loads in vital organs (blood, liver, spleen), compared to that of untreated control group (P< 0.05).
Conclusion: The preliminary results presented in this study suggest that garlic extract may offer a promising and novel means for the treatment o f P. aeruginosa burn wound infection and open new insight into further investigation on this area.
P1778 Different frequency of N-myc2 activation in liver tumours from male and female woodchucks chronically infected by WHV
R. Bruni, A. Paganelli, I. Conti, U. Villano, R. Giuseppetti, M. Rapicetta (Rome, IT)
Objectives: WHV/woodchuck is a model of HBV infection. In liver tumours induced by chronic WHV infection, the N-myc2 gene is frequently activated. In most cases, transcriptional activation is due to WHV insertion either nearby the gene, or in two downstream loci, b3n and win. N-myc2 is located into the X chromosome. Mutation of several X-linked genes involved in cancer is known to show different outcome according to the gender. No previous studies investigated the distribution of N-myc2 positive (i.e. showing transcriptional activation and/or WHV integration in known target loci of the N-myc2 chromosomal domain) liver tumours in relation with woodchuck gender. Aim of the present work was to investigate the frequency of N-myc2 positive liver tumours from male and female woodchucks.
Methods: Tumours were classified as N-myc2 positive or negative by studying (A) rearrangement and WHV insertion in N-myc2, b3n and win loci by Southern blotting and (B) N-myc2 transcription by Northern blotting.
Results: A survey of 46 not clonally related liver tumours naturally developed in chronically WHV infected woodchucks was analysed. 16 tumours were from 8 male and 30 from 14 female animals. Southern and Northern blotting data showed that N-myc2 positive tumours were 7/16 (44%) from male animals vs. 27/30 (90%) from females (p = 0.002 with Fisher's exact test).
Conclusions: Though statistically significant, the biological reasons for the observed gender related difference remain to be investigated. As N-myc2 is an X-linked gene, two gene copies per diploid genome are present in female cells vs. one copy in male cells: thus, as the simplest explanation, female cells would provide a two-fold more abundant target for WHV integration. However, in female cells one X chromosome is functionally inactivated, heterochromatic and, thus, expected to be a poor substrate for host enzymatic machinery responsible for illegitimate WHV integration. So, the actual X chromosome target for WHV integration would not differ between male and female cells. Because of the multistep nature of tumour development, it is possible that other female specific factors might select for N-myc2 positive cells, leading to the observed higher frequency of tumours with N-myc2 activation in females.
P1779 Rapid insurgence of a viral-resistant mutant in WHV chronically infected woodchucks treated with lamivudine and a PRE-S/S CHO-derived hepatitis B virus vaccine
E. D'Ugo, L. Kondili, A. Canitano, S. Catone, R. Giuseppetti, B. Gallinella, G. Palmieri, S. Orobello, C. Argentini, M. Rapicetta (Rome, IT)
Objectives: The woodchuck hepatitis virus (WHV) and its host, the Eastern woodchuck (Marmota monax) are predictive models for evaluating HBV antiviral therapy. To determine whether the addition of a pre-S/S human vaccine, previously shown to protect woodchucks from productive WHV infection, could increase the antiviral activity of a high-dosage of lamivudine in the woodchuck model.
Methods: The 4 woodchucks were administered a daily dose of 100 mg/kg body weight of lamivudine for 40 weeks and were vaccinated with four doses (50 μg each) of CHO–HBsAg vaccine at 14, 18, 22, and 34 weeks after starting lamivudine. WHV-DNA were determined using a TaqMan Real-Time PCR (sensitivity of 10 genome equivalents/mL); the positive samples were considered for the sequence analysis. The A, B, C and D domains of the WHV polymerase region were amplified by specific primers. Serial liver biopsies were performed and numerical score according to the Ishak system was used.
Results: WHV DNA titres decreased up to two logarithms in 3 woodchucks. Since week 4 the sequence analysis in 3 of the 4 animals showed a heterogeneous viral population, with the predominant strain containing a single nucleotide change at position 1696 (from A to T) in the sequence coding of FLLA motif (domain B). In 2 of the last 3 woodchucks, the increase in WHV DNA titres was observed, though not immediately after the FLLT mutation appearance. No mutations in the other domains were observed. Vaccination did not further suppress WHV DNA, despite anti-HBs production in three animals. In one animal WHV DNA levels increased immediately after the first dose of pre-S/S vaccine and the grading score increased from 2 to 10.
Conclusions: The emergence of the lamivudine-resistant mutation in the B domain was observed within 4 weeks of starting lamivudine, which is much earlier than previously reported. Vaccination could trigger the immune system and liver inflammation in the context of high WHV DNA levels resistant to lamivudine. Under the conditions used, a transient effect of combination lamivudine and anti HBs vaccine therapy was observed.
P1780 Antimicrobial biodegradable composition based on high-molecular weight polyvinylpyrrolidone for the prophylaxis of experimental osteomyelitis
G. Afinogenov, R. Tikhilov, A. Mironenko, T. Bogdanova, A. Afinogenova, I. Kozlov, M. Krasnova, L. Anisimova (St. Petersburg, RU)
Objectives: The problem of the post-surgical osteomyelitis prophylaxis after metal-osteosuture is still actual now. The local application of collagen with gentamycin used to prolong the antibiotic activity in tissues of wound area. Modern technologies allow to obtain high-molecular weight PVP with different biodegradation terms to use it as a matrix for various medicines.
The aim was to evaluate the efficacy of antimicrobial composition of amikacin with activator and polyvinylpyrrolidine as a biodegradable prolonging agent for the post-surgery infections prophylaxis.
Methods: The rabbit shin-bone was bared under the aseptical intravenous anaesthesia and two bone defects were formed with 0.1 N NaOH to obtain surface necrosteosis. The microbial culture of S. aureus (0.5 ml of 2×08 CFU/mL) was introduced into the bone defects twice before and after the K-wires were placed. 5 ml of amikacin with activator and PVP were placed directly on the bone and wound area before made a suture. The total condition, body temperature and wound view were valued. The animals were sacrificed on the 7th, 14th and 21st days after the surgery. The clinical evidences of irritation, the pus and tissue necrosis presence were valued as well as bacteriological and histological researches were made with muscle and marrow biopsies and implanted K-wires.
Results: An acute osteomyelitis with expressed tissue microbial contamination was obtained in the control group. The animals with prophylaxis introduction of antibacterial composition didn't display clinical evidences of osteomyelitis. The tissues microbial contamination proved to be the same during the experiment in the control group. The experimental animals showed a reliable decreased microbial contamination of tissues involved in infectious process. The whole eradication of S. aureus was obtained by the 21st day in muscle and marrow biopsies. Testing the K-wires the microbial growth was displayed in control group and only by the 7th day – in experimental group. The bactericidal composition showed a residual antimicrobial activity in animal tissues during the experiment.
Conclusions: The data obtained show the efficacy of the amikacin combination with activator based on high-molecular weight polyvinylpyrrolidine and an opportunity to it's application for the post-surgery infections prophylaxis in traumatology, orthopaedics and other surgery areas.
P1781 Efficacy of oritavancin in a mouse model of Streptococcus pneumoniae pneumonia
D. Lehoux, G. McKay, I. Fadhil, K. Laquerre, M. Malouin, V. Ostiguy, G. Moeck, T. Parr Jr. (St-Laurent, CA)
Objective: Streptococcus pneumoniae is one of the leading causes of community-acquired bacterial infections such as pneumonia. Oritavancin (ORI) is a semi-synthetic glycopeptide with bactericidal activity against Gram-positive cocci. We investigated the activity of ORI in vitro in the presence of surfactant and in vivo in a mouse model of S. pneumoniae pneumonia.
Methods: 1) Broth microdilution assays with ORI, daptomycin (DAP) and ceftriaxone (CTX) used Staphylococcus aureus ATCC 29213 following CLSI guidelines either without modification or including 0.5 to 5% commercially-available surfactant in parallel. 2) In vivo efficacy studies were performed in an immunocompetent mouse pneumonia model. CD-1 mice (n = 10/group) were challenged intranasally with 106 colony-forming units (CFU) of penicillin-sensitive S. pneumoniae (PSSP) ATCC 6303. In a study to assess efficacy resulting from administration of two doses of ORI and comparators, agents were administered at 1 and 4 h post-infection as follows: ORI, 32 mg/kg, intravenously; DAP, 100 mg/kg subcutaneously; CTX, 50 mg/kg, subcutaneously. In a dose-ranging study, ORI was administered at 1 h post-infection in single intravenous doses ranging from 0.25 to 32 mg/kg. For both the single- and two-dose studies, lungs were harvested 24 h post-infection, homogenised, and plated for bacterial counts. A maximal effect (Emax) model was used to examine the relationship between dose and efficacy.
Results: (1) The addition of 5% surfactant resulted in a 2- to 4-fold increase in ORI MIC compared to a ≥64-fold increase for DAP MIC and no shift for CTX MIC. (2) ORI (2 × 32 mg/kg) was more active in the mouse pneumonia model than CTX (2 × 50 mg/kg) and DAP (2 × 100 mg/kg): no detectable CFU were recovered from the lungs of 100% of mice treated with ORI while 40% and 0% of mice treated with CTX and DAP, respectively, had sterile lungs. Single-dose dose-response studies of ORI yielded an Emax of 6.49±0.18 mg/kg and an ED50 (dose resulting in 50% of the maximal killing) of 2.76±0.3 mg/kg.
Conclusions: (1) Surfactant affected only minimally the antibacterial activity of ORI in vitro. (2) ORI is highly active in the mouse pneumonia model and it may have potential utility in the treatment of pneumonia caused by PSSP.
P1782 Pneumococcal meningitis: is disease severity related to capsular or clonal types?
S. Ribes, F. Taberner, C. Cabellos, J. Gerber, F. Tubau, C. Ardanuy, J. Linares, R. Nau, F. Gudiol (Hospitalet de Llobregat, ES; Gottingen, DE)
Objectives: Capsular serotype of Streptococcus pneumoniae (Sp) strains is an important virulence factor. A single pneumococcal serotype includes a number of genetically divergent clones. Using the rabbit model of meningitis, we tested whether five different pneumococcal clones belonging to serotypes 23F and 3 caused different disease severity.
Methods: Meningitis was induced by intracisternal inoculation of 106 CFU/mL of five invasive strains of Sp in five groups of rabbits (n ≥10/group). We used three strains of serotype 23F with different degree of penicillin (PEN) susceptibility and sequence type (ST): strain A (ST311, PEN-S), strain B (ST81, PEN-R) and strain C (ST37, PEN-I) and two PEN-S strains of serotype 3: strain D (ST260) and strain E (ST180). Eighteen hours after infection, serial CSF samples were collected over 24 h to determine bacterial counts and inflammatory data levels: lactate, protein and lipoteichoic/teichoic acids (LTAs). Bacteraemia at 0 h and mortality at 24 h were also assessed. Fisher's exact test was used in bacteraemia/mortality studies. ANOVA/T-Student test or Kruskal Wallis/Mann-Whitney tests were performed dependent on whether data were normally distributed or not.
Results: Bacteraemia at 0 h was found in 10/10, 11/13, 8/12, 5/13 and 3/11 of rabbits infected with strains A, B, C, D and E, respectively. Bacterial concentrations at 0 and 6 h were lower in serotype 3 strains than in serotype 23F isolates (p < 0.05). Bacterial counts at 24 h were above 4.5 log CFU/mL in all strains. Lactate at 0, 6 and 24 h, and protein levels at 6 and 24 h were higher in serotype 23F than in serotype 3 strains. Mortality was 50, 30.8, 58.3, 15.4 and 0% for strains A, B, C, D and E, respectively. Among strains belonging to serotype 23F, there were neither marked differences in bacterial counts nor in inflammatory activity. On the contrary, there were statistical differences in CSF bacterial concentrations and in lactate, protein and LTAs levels among strains belonging to serotype 3.
Conclusions: In the rabbit meningitis model, serotype 23F strains (independently of their PEN susceptibility patterns), produced higher bacteraemia and mortality rates, and higher CSF levels of inflammatory parameters than serotype 3 strains. On the other hand, differences noted between two clones of serotype 3 indicated that additional genetic loci other than capsular serotype could be involved in pneumococcal disease severity.
P1783 Levofloxacin-rifampin combination is highly bactericidal in experimental methicillin-susceptible Staphylococcus aureus prosthetic joint infection
C. Muller-Serieys, A. Saleh Mghir, L. Massias, A. Andremont, B. Fantin (Paris, FR)
Objectives: Staphylococcus aureus is the most common pathogen responsible for bacterial joint infections. Levofloxacin (LEV) and rifampin (RIF) are highly active in vitro against susceptible strains of MSSA; nevertheless no data are available on the in vivo relevance of this combination for treating MSSA prosthetic joint infection.
Methods: A partial knee replacement was performed in rabbits with a silicone implant fitting into the intramedullary canal of the tibia, and 107 CFU of a MSSA strain (MICs and MPCs: 0.125 and 1 mg/L for LEV and ≤ 0.016 and 256 mg/L for RIF, respectively) were injected into the knees. Treatment was started 7 days after inoculation and lasted for 7 days. Infected animals were randomly assigned to: either no Rx (controls), or LEV (25 mg/kg IV b.i.d.), or RIF (10 mg/kg IM b.i.d.) or LEV+RIF. Surviving bacteria were counted in bone 2 days after the end of Rx, and resistant mutants were detected on antibiotic-containing agar.
Results: LEV AUC was comparable to that achieved in human after a 750 mg daily dose. The mean log10 CFU/g of bone were significantly reduced with LEV (2.92 ± 1.33, 5/12 sterile animals) and RIF (3.20 ± 2.12, 5/11 sterile animals) versus controls (6.36 ± 1.33, 0/10 sterile animals) (p < 0.05). RIF+LEV combination (mean log10 CFU/g of bone: 1.99 ± 0.52, 6/12 sterile animals) was significantly more effective than RIF alone (p < 0.05) and prevented the emergence of RIF-resistant mutants as observed in 4/6 positive bones in RIF treated animals and 2/6 controls. No LEV-resistant mutants were detected in any group of animals.
Conclusion: In this MSSA joint prosthesis infection, LEV combined with RIF was highly bactericidal and prevented the selection of mutant resistant to RIF. LEV+RIF should be of interest for treating MSSA prothetic joint infection in human.
P1784 Standarisation of a rabbit model of methicillin-susceptible, methicillin-resistant and glycopeptide intermediate Staphylococcus aureus meningitis
F. Taberner, S. Ribes, E. Force, C. Cabellos, M.A. Dominguez, J. Ariza, F. Gudiol (Hospitalet, ES)
Staphylococcus aureus meningitis is a rare but severe disease with high mortality rates. It is usually nosocomial-acquired and associated with neurosurgical procedures.
Objective: To study the ability of three strains of S. aureus with different susceptibility to β-lactams and glycopeptides to induce meningitis and inflammatory activity in CSF and to standardise the rabbit meningitis model to study the efficacy of different antibiotics against them.
Methods: Three different staphylococcal strains were used: Strain A, methicillin-susceptible S. aureus (MSSA) with MICs (in mg/L) to cloxacillin (CLOXA) 0.25 and to vancomycin (VAN) 1; strain B, methicillin-resistant S. aureus (MRSA): CLOXA 512, VAN 2; and strain C, glycopeptide-intermediate S. aureus (GISA): CLOXA 1024, VAN 4. Meningitis was induced in New Zealand white rabbits (n ≥ 6 per group) by intracisternal inoculation of an inoculum of 107 CFU/mL for MSSA strain and of 108 for MRSA and GISA strains. Bacterial growth curve was assessed in CSF of rabbits after 10 h and 20 h of inoculation. Additionally, CSF samples were used to quantify CSF leukocyte counts, lactate and protein concentrations. Statistical analysis was performed using T-student for bacterial counts and for inflammatory activity.
Results: All tested animals developed meningitis after 10 hours of inoculation. Bacterial titers (median ± SD) with all tested strains at different time points are shown in the table . Some inflammatory activity is shown in the table. All blood cultures were negative. Without treatment, all animals survived at least 24 h after the inoculation point.
| 10 h after inoculation |
20 h after inoculation |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 2 h | 6 h | 24 h | 0 h | 2 h | 6 h | 24 h | |
| Log cfu/mL | ||||||||
| Strain A | 4.1±0.8 | 4±0.8 | 3±0.81 | 3±1.9 | 4.7±1.4 | 4.2±1.6 | 3.8±2.9 | 4.8±2.0 |
| Strain B | 5.2±0.4* | 4.8±0.7* | 4.2±0.4* | 3.7±1.1* | 3.8±0.5 | 3.5±0.8 | 2.8±0.7 | 2.2±1 |
| Strain C | 4.8±0.4* | 4.7±0.4* | 4.5±0.6* | 2.9±0.7* | 2.6±1.3 | 2.2±1.7 | 2±1.6 | 1.4±1.5 |
| [Lactate] mmol/L | ||||||||
| Strain A | 10.1±3* | 10.7±3* | 10.3±2.5* | 5.6±2.7 | 6±0.3 | 4.2±0.2 | 6±3.2 | 3.5 |
| Strain B | 12.5±1.4* | 11.1±1.7* | 9.3±1.1* | 7.2±2.7* | 6.2±2.4 | 5.1±1.5 | 4.7±1.6 | 4.4±1.3 |
| Strain C | 11.6±1.6* | 9.7±1.5* | 7.5±1* | 5.9±1.2* | 5.7±0.9 | 4.9±1.6 | 4.1±1.6 | 3.5±1.1 |
Statistical difference 10 h vs 20 h.
Conclusion: All three Staphylococcus aureus strains were able to induce meningitis and inflammatory activity without statistically significant differences at 10 h. Log CFU/mL and inflammatory parameters were higher (with statistical significance) and more homogeneous at 10 h time point than at 20 h. This time point has been choosed to begin therapeutic schedules.
P1785 Efficacy of voriconazole versus anidulafungin alone and in combination in a guinea pig model of invasive pulmonary aspergillosis
W.R. Kirkpatrick, L.K. Najvar, R. Bocanegra, A.C. Vallor, M.C. Olivo, D. Molina, J.R. Graybill, T.F. Patterson (San Antonio, US)
Objectives: Combination therapy for invasive aspergillosis with an echinocandin and a triazole antifungal is attractive due to distinct mechanisms of action. Possible antagonism of some drug combinations and the potential paradoxical “eagle” effect of higher doses of echinocandins have been suggested; thus, combination therapy remains controversial. The antifungal activities of voriconazole (VRC) and anidulafungin (ANID) alone or in combination were evaluated in a guinea pig model of invasive pulmonary aspergillosis (IPA).
Methods: Guinea pigs were immunosuppressed with cortisone acetate and made temporarily neutropenic with cyclophosphamide 2 days prior to, and 3 days following aerosolised challenge with 12 ml 109 CFU/mL Aspergillus fumigatus AF293 in an acrylic chamber. Therapy with oral VRC at 2.5 or 10 mg/kg/bid, ip ANID at 1, 6 or 12 mg/kg/d, or combinations of VRC 2.5+ANID 1 or VRC10+ANID6 or 12 was begun 24 hr after challenge and continued for 8 d.
Results: Both doses of VRC and the low dose combination of VRC2.5+ANID1 showed significantly increased survival vs controls. Decreased mortality, expressed as mean day of death, was also shown in a dose dependent manner with all 3 doses of ANID with longer survival seen with higher doses of ANID. Semi-quantitative assessment of lung tissue burden by colony forming units (CFU) revealed that therapy with VRC10 and both VRC10/ANID combinations yielded significant reductions in counts as compared to controls. Similarly, therapy with VRC10 or VRC10+ANID12 yielded significant reductions in counts as compared to either ANID6 or 12. Assessment of lung tissue burden by Q-PCR (Conidial equivalents “CE”) revealed that VRC10 and both VRC10/ANID combinations yielded significant reductions in counts as compared to controls. Similarly, Q-PCR showed that therapy with VRC10 yielded significant reductions in counts as compared to ANID12 as did VRC10+ANID12 as compared to either ANID 1 or 12.
Conclusion: Therapy with VRC yielded greater survival than did ANID, and VRC10 or the combination of VRC10+ANID12 were superior in reducing tissue burden as assessed by either CFU or CE. In this model of IPA, VRC10 alone was superior to any dose of ANID tested alone. ANID combined with low dose VCR reduced lung CE. Combination therapy with the higher VRC doses demonstrated similar efficacy to VRC alone although no evidence of a paradoxical effect in tissue burden was seen.
P1786 Dispersin B therapy of Staphylococcus aureus experimental port-related bloodstream infection
A. Serrera, J.L. del Pozo, A. Martinez, M. Alonso, R. Gonzalez, J. Leiva, M. Vergara, I. Lasa (Pamplona, ES)
Objectives: S. aureus is a common cause of port-related bloodstream infections (PRBI). Treatment of these infections typically consists of surgical removal of the port and prolonged therapy with adequate antimicrobials. Dispersin B (Dsp-B) is an enzyme produced by Actinobacillus actinomycetemcomitans that cleaves beta 1–6 N-Acetylglucosamine polymers in S. aureus biofilm. We analysed the activity of Dsp-B (administered as a lock solution inside the port with teicoplanin) in a sheep model of PRBI.
Methods: Experimental PRBI was established in 24 female sheeps. Three days after initialing the infection, animals were randomised into three equal groups and antimicrobial therapy was initiated as follows: (i) no treatment, (ii) iv teicoplanin (loading dose of 6 mg/k/12 h × 3 doses and then 4 mg/k/24 h) plus teicoplanin locks (10 mg/mL) and (iii) iv teicoplanin plus Dsp-B-teicoplanin locks (40 μg/mL and 10 mg/mL respectively). This treatment was administered every day for ten days. Blood cultures were extracted (venipuncture and port) every day. Three days after completion of therapy, sheeps were sacrificed and ports were aseptically removed and cultured (swabbing of the internal port lumen, septum sonication and catheter tip sonication).
Results: All the animals in no treatment group died due to septic complications and all blood and port cultures were positive for S. aureus. In group ii, 75% of the animals got sterile blood cultures during treatment (mean: 7, range: 3–10 days), but all the port cultures were positive after removal. In group iii 100% of the animals got sterile blood cultures during treatment (mean: 3, range: 3–8 days), and catheter cultures were completely negative in 50% of the animals.
Conclusion: Our results indicate that a combination lock of Dsp-B with teicoplanin besides systemic antibiotherapy is active in a sheep model of PRBI. Application of this therapy in the human setting could eliminate the need for port removal in related infections.
This work was supported by a grant from Proy. Inst. Carlos III ref. 03/1102
P1787 Morphological substantiation of antibacterial therapy in acute cystitis
T. Perepanova, J. Kudrjavtcev, V. Kirpatovski, P. Hazan (Moscow, RU)
Objectives: To study the impact of antibacterial medications on the mucus of bladder in the cases of acute bacterial cystitis in animal testing.
Methods: The experiments were made on 15 white outbreed female rats with a mass of 280–320g. Acute bacterial cystitis was modeled in all 15 rats. The rats were divided into 5 groups, 3 rats to a group. The first group was a control one – without antibiotics; in the second group the rats were given furazydin (0.5–1 mg to a rat; 1.5 mg/kg) with food 3 times a day during 7 days; the third group took fosfomycin (15 mg to a rat; 45 mg/kg) in one dose; in the forth group – norfloxacin (2 mg to a rat; 6 mg/kg) 2 times a day during 3 days; the fifth group was given norfloxacin in the same dose during 5 days. By means of microscope studies of the bladder preparations morphometrics of a number of parameters was carried out which allowed to evaluate quantatively the expressed inflammatory reaction. All rats were done away with by putting to sleep on the 7th day from the beginning of the experiment.
Results: 3 rats from the 1st group revealed visual evidence of cystitis. The 2nd-5th groups didn't show any visual evidence of cystitis. Epithelium alteration showed minimal evidence in groups 4th and 5th. Development of mesenhematic reaction revealed itself by the evidence of inflammation of hystiocytic cells. The biggest concentration of these cells showed in the first group. The contents of these cells dropped sharply in the 4th and the 5th groups. There was practically no difference in the concentration of lymphocytes and macrophages in different groups. The biggest concentration of polymorphonuclear leucocytes was revealed in the 4th and the5th groups.
Conclusion: The necessity of antibacterial therapy of acute bacterial cystitis proved to be crucial. Norfloxacin showed better bacterial efficiency compared to fosfomycin and furazydin. Alteration of urothelium in treatment with norfloxacin decreases to a more extent than that with fosfomycin and furazydin. The application of norfloxacin shortens the reparation phase while long-term antibacterial therapy course shows moderate suppression of mesenhematic reaction. Three-day treatment course with norfloxacin has certain advantages compared to five-day treatment course by this medication to reparation phase.
P1788 Experimental studies with T. pallidum Nichols strain – Efficacy of treatment in late infection
A.C. Magalhães Sant'Ana, G. Rocha, J. Poiares da Silva (Coimbra, Porto, PT)
Objective: More than one hundred years since the identification by Schaudinn and Hoffman of Treponema pallidum, the inability to grow it has limited the knowledge of syphilis physiopathology. In the last decades, there has been a marked increase in the number of cases of syphilis and there are still doubts about the total elimination of T. pallidum in the late syphilis. These factors justify the need to continue the studies on T. pallidum infection. To study the efficacy of penicillin and ampicillin treatment in infected
Methods: Rabbits not infected with Treponema paraluiscuniculli were intratesticular inoculated with T. pallidum Nichols strain. They were treated with intramuscullar penicillin and/or ampicillin and monitored by qualitative and quantitative FTAabs. Dark-Field and Transmission Electron Microscopy (ME) was done in testis, popliteus adenitis and ocular aqueous humour.
Results: We detected: (i) In early treatment, FTAabs reverts to negative and in testis the T. pallidum shows lytic aspects with total cure of orchitis. (ii) If the treatment was initiated after 3 months, FTAabs persist positive. However with the maintenance of treatment with ampicillin the title of Ig G decreases to 1/5. (iii) In rabbits that survive until 3 years FTA abs persists positive, but in autopsy we can see in popliteus adenitis but not in testis or in aqueous humor some cells, that in ME shows the morphology of T. pallidum.
Conclusion: We don't understand the complete meaning of persistence of T. pallidum in late infection because it was not possible to obtain infection with this products in other rabbits. In patients with HIV infection we find an unusual incidence of neurosyphilis, despite adequate therapy. It is of utmost importance to proceed with these studies with new biothecnology.
P1789 Moxifloxacin versus ampicillin+gentamicin in the therapy of experimental Listeria monocytogenes meningitis
O. Sipahi, T. Turhan, H. Pullukcu, S. Calik, M. Tasbakan, H. Sipahi, B. Arda, T. Yamazhan, S. Ulusoy (Izmir, TR)
Objective: Listeria monocytogenes (LM) is an important pathogen causing meningitis in immunocompromised and and immunocompetent host, The main therapeutic choice is ampicillin+gentamicin which is shown to be synergic in experimental meningitis model in rabbits, Moxifloxacin is effective against LM in vitro and in machrophages, There is no human or animal study comparing ampicillin+gentamicin versus moxifloxacin in LM meningitis, In this study it was aimed to compare the antibacterial activity of moxifloxacin and ampicillin+gentamicin in the treatment of LM meningitis in rabbit meningitis model.
Methods: Meningitis was induced by direct inoculation of a clinical strain isolated from an immunocompromised patient (107cfu/mL), into cisterna magna of New Zealand rabbits, After 16 h of incubation time, rabbits were separated into three groups moxifloxacin (M), ampicillin+gentamicin (A) and control, (C), Group M received 20 mg/kg moxifloxacin (Bayer, Germany) at the end of the incubation time and 5 h later by IV route, Ampicllin (Mustafa Nevzat, Turkey, 30 mg/kg/h) and gentamicin (IE Ulagay, Turkey, 2.5 mg/kg/h) were also given by IV route with continuous infusion for 8 h in 36 cc 0.9% NaCl and group C did not receive any treatment, CSF samples (0.1–0.25 mL) were obtained 16 h and 24 h after induction of meningitis, At the end of the study period (24 h after the induction of meningitis), animals were humanely killed by intravenous infusion of high dose nembutal, The bacterial count in CSF was measured by standard serial dilutions of 10 μL CSF in 0.9% NaCl and incorporation into sheep blood agar (Oxoid, Basingstoke, UK) pour plates, Data were evaluated by SPSS 11.0 package programme using Mann-Whitney U-test, Kruskal-Wallis test and Fisher's χ2 test, A P-value less than 0.05 was considered significant.
Results: At the end of 16 h of incubation time, CSF bacterial counts were similar in all groups (p > 0.05), When three groups were compared (table 1 ), bacterial counts in both treatment groups 24 h after the induction of meningitis were significantly lower (p< 0.05) than control group, When two treatment groups were compared bacterial counts were similar (Table 1).
Table 1.
Bacterial counts in CSF of rabbits during study period.
| Group | No. of rabbits | Log10 cfu.mL |
|
|---|---|---|---|
| 16 h after induction of meningitis | End of treatment | ||
| Group C | 9 | 5.309±0.461 | 6.346±0.623 |
| Group M | 9 | 5.375±0.356 | 3.830±0.518 |
| Group A | 7 | 5.512±0.712 | 4.193±0.794 |
Conclusion: These data suggest that antibacterial activity of moxifloxacin is similar with ampicillin+gentamicin in the treatment of experimental LM meningitis of rabbits
P1790 Dose-dependent effects of octreotide on oxidant-antioxidant imbalance during experimental sepsis in rat
A. Seydanoglu, M. Gül, M. Ayan, I. Erayman, H. Toy, S. Girisgin (Konya, TR)
Objective: Sepsis remains one of the leading causes of death in intensive care units, despite recent acquired knowledge on pathophysiology and treatment. Oxidant-antioxidant balance involves different pathways and yet, there is no ideal drug able to affect all of them. The aim of this study was to investigate whether Octreotide (OCT), a synthetic somatostatin analogue, has antioxidative effects in experimental sepsis.
Methods: Forty Sprague-Dawley rats were randomly divided into four groups. Group I was the control group. Group II received no treatment. Group III and group IV received subcutanously (sc) OCT 50 μg/kg and 100 μg/kg after Cecal Ligation and Perforation (CLP) respectively. White blood cell (WBC) count, erythrocyte glutathione (GSH), leucocyte myeloperoxidase (MPO) and plasma malondialdehyde (MDA) were assessed in all groups.
Results: In group sepsis, while MDA increased in sepsis periods, GSH decreased when compared with group Sham (p< 0.05). Increase in MDA levels and decrease in GSH levels after CLP-induced sepsis was significantly prevented by OCT 100 μg/kg sc administration (p <0.05).
Conclusion: Octreotide seems to have a dose-dependent antioxidative effect in CLP-induced sepsis in rats. As a drug with the wide magrin of safety and less adverse reaction profile merits consideration as a choice of treatment in sepsis and septic shock.
P1791 Comparison of the effects of erdosteine and n-acetylcysteine on the levels of glutathione, myeloperoxidase, plasma and tissue malondialdehyde in experimental sepsis model
M. Ayan, M. Gül, A. Seydanoglu, I. Erayman, S. Erdem, B. Cander (Konya, TR)
Objective: The aim of this study was to determine the effects of N-Acetylcysteine (NAC) and Erdosteine as antioxidant agents on the free radicals and their plasma levels.
Methods: Forty Sprague-Dawley rats were randomly divided into four groups. Group I was the control group. Group II received no treatment. Group III and group IV received orally NAC (20 mg/kg/24 h) and Erdosteine (20 mg/kg/24 h) after Cecal Ligation and Perforation (CLP) respectively. White blood cell (WBC) count, erythrocyte glutathione (GSH), leucocyte myeloperoxidase (MPO) and plasma malondialdehyde (MDA) were assessed in all groups.
Results: In groups III; WBC, erythrocyte GSH, and plasma MDA levels were measured higher than control group (p < 0.05). However, in groups IV; WBC, erythrocyte GSH, and plasma MDA levels were measured higher than control group, but not statistically significant (p > 0.05). The lung tissue MDA levels decreased in group III and IV according to control group (p < 0.05).
Conclusion: By the administration of NAC and Erdosteine as antioxidant agents al lower doses many meaningful positive effects were detected on the levels of WBC, erythrocyte GSH, and plasma MDA. These findings suggest that NAC and Erdosteine could be a possible therapeutic agent for sepsis and its mortality. However, further studies are needed to elucidate the effects of these drugs at higher doses.
P1792 The effect of Ipsat P1A, a novel drug for prevention of antibiotic-induced intestinal pathogenic colonisation, on the gut antibiotic levels after intravenous administration of β-lactam antibiotic/β-lactamase inhibitors in the dog
K. Raatesalmi, T. Korkolainen (Helsinki, FI)
Background: Novel therapies for the prevention of antibiotic (AB)-induced intestinal pathogenic colonisation and gastrointestinal side-effects such as diarrhoea, colitis and abdominal discomfort are needed. In preclinical studies and clinical trials, Ipsat P1A, a recombinant class A β-lactamase, was shown to prevent ampicillin-induced intestinal pathogenic colonisation and diarrhoea after intravenous (i.v.) administration of AB.
Objective: The aim of the study was to test the effect of Ipsat P1A treatment to prevent the increase in intestinal AB levels after i.v. β-lactam/β-lactamase inhibitor administration.
Methods: Experiments were carried out on 6 beagle dogs (appr.17 kg), which were fed with food pellets 20 minutes before AB administration. During the first treatment the dogs received two doses of AB/β-lactamase inhibitor i.v. six hours apart. In the second treatment the dogs received a dose of Ipsat P1A (0.1 mg/kg, per os) and after 10 minutes an i.v. dose of AB/β-lactamase inhibitor. After six hours the treatment was repeated. The chyme samples were collected at different time points for 9 h by inserting a silicon hose through the jejunal fistula (170 cm from the pylorus). Jejunal antibiotic concentrations were determined by an HPLC method.
Results: Administration of ampicillin-sulbactam (40+20 mg/kg), amoxicillin-clavulanic acid (25+5 mg/kg) and piperacillin-tazobactam (100+ 12.5 mg/kg) resulted in AUC0–360 values of 38,100, 5,700 and 441,000 μg min/g, respectively, for the intestinal concentration/time curves of the ABs. When the dogs were treated with Ipsat P1A the AUC0–360 of ampicillin-sulbactam, amoxicillin-clavulanic acid and piperacillin-tazobactam were statistically significantly reduced by 89, 77 and 36%, respectively.
Conclusion: Ipsat P1A degraded effectively the residues of ABs in the intestine after i.v. AB administration.
P1793 Emergence of fluoroquinolone resistance in the murine model of tuberculosis
A. Aubry, A. Chauffour, C. Truffot-Pernot, V. Jarlier, N. Veziris (Paris, FR)
Objective: Fluoroquinolone (FQ) are key antibiotics in the treatment of multidrug-resistant tuberculosis, and are under investigation for first line treatment. However, FQ resistance is emerging in M. tuberculosis. We aim to determine whether selection of FQ-resistant mutants occurs in the murine model among mice treated with clinically used FQ.
Methods: 80 swiss mice were inoculated in the tail vein with 107 CFU of M. tuberculosis virulent strain H37Rv. When the CFU count reached 7.9 log10 in the lungs, mice were randomly allocated to 4 groups: a negative control group in which mice were infected but untreated (10 mice), and 3 tests groups in which 10 mice were treated with ofloxacin (OFX 200 mg/kg), levofloxacin (LVX 200 mg/kg) or moxifloxacin (MXF 100 mg/kg) for 6 months. All mice surviving after 6 months of treatment were sacrificed. Lungs were plated on medium containing or not OFX 2 mg/L. Resistant strains selected in vivo were submitted to sequencing of the QRDRs of gyrA and gyrB genes.
Results: Survival rates after 1 and 6 months of treatment were 1/10 and 0/10 for OFX, 9/10 and 3/10 for LVX, and 9/10 and 7/10 for MXF. Bacilli recovered from mice treated with OFX were still susceptible to FQ, suggesting that OFX failed to prevent multiplication of the bacilli, consistently with low in vivo efficacy.
Among, the 13 strains resistant to FQ isolated from the lungs of mice treated with LVF (5/6) and MXF (8/9), 11 harboured already described mutations in GyrA (A83V, S84P or D87G, N, Y), 1 harboured an unusual mutation in GyrB (D426N) and one strain harboured no mutation in GyrA or GyrB. Among mutated strains, the majority (83%) were constituted by a mixture of different clones, 2 different single mutants in 4 cases and mutant plus wild-type clones in 6 cases.
Conclusion: In the murine model of tuberculosis, under FQ therapy, we observed the emergence of heterogeneous populations of bacilli with different resistance mutations, as well as mixtures of drug-susceptible and drug-resistant genotypes. These data, that are in concordance with recent observation in human tuberculosis, are informative for the understanding of the selection of FQ-resistant mutants in tuberculosis and can be used as a model to assess how to prevent selection of resistant mutants by second line antituberculous agents. These data will also be useful for designing molecular diagnosis of resistance to FQ in case of mixture of resistant and sensitive strains.
P1794 The protective effect of doxycyclin against Mycobacterium avium infection in mice
S. Ikegame, M. Fujita, E. Harada, H. Ouchi, I. Inoshima, Y. Nakanishi (Fukuoka, JP)
Objective: Mycobacterium avium causes chronic and progressive respiratory infection. Therapeutic regimen including clarithromycin, rifanpin and ethambutol has been commonly employed, but the effect of such anti-bacterial therapy is not satisfactory and new approach for this disease is needed. Doxycyclin (DOXY) is an antibiotic known to have immunomodulating effects as well as anti-bacterial activity. In this study, we investigated the effect of DOXY administration on M. avium infection in mice. It is well known that TNF-alpha plays central roles in immunity against mycobacteriosis. We investigated the effect of DOXY in TNF-alpha 1 receptor knock out mice as well.
Methods: Clinically isolated strains of M. avium were used. Either wild type of C57Bl/6 mice (wt) or TNF receptor 1 knock out (TNF-R1 KO) mice on same background was administered of M. avium (1×107 cfu/body) intratracheally. 2mg/kg/day of DOXY was administered orally to mice after M. avium administration. Mice were sacrificed on day 21, 60 after M. avium administration. The lung homogenates were inoculated on Middlebrook 7H10 agar plates for counting the number of colonies. Tissue sections of the lungs were stained by haematoxylin and eosin or Ziehl-Neelsen (Z-N) methods.
Results: Administration of DOXY attenuated lung inflammation caused by M. avium. Moreover, DOXY administration improved the survival rate of M. avium infected TNF-R1 KO mice. However, DOXY administration did not affect the number of M. avium in the lungs.
Conclusion: This study suggests that DOXY administration may have protective effects against M. avium infection in mice. The effects of DOXY are probably due to its biological effect apart from its antimicrobial function.
Pathogenesis of Gram-positive bacterial infections
P1795 The prevalence of Staphylococcus aureus biofilm formers in Scotland
A. Perez, S. Lang, C. Gemmell, G. Ramage (Glasgow, UK)
Background: Staphylococcus aureus has the ability to attach and colonise implanted biomaterials such as catheters and indwelling medical devices. These biofilm structures are notoriously difficult to treat with antibiotics, and this characteristic combined with an increased prevalence of methicillin resistant S. aureus clones has thwarted our ability to successfully treat these infections.
Objectives: The aim of this study was to investigate the ability of both methicillin sensitive and resistant S. aureus (MSSA and MRSA) strains to form biofilms in an in vitro model.
Methods: One thousand characterised clones of MSSA and MRSA from the Scottish healthcare environment were obtained from the Scottish MRSA Reference Laboratory. Each isolate was grown overnight in brain heart infusion (BHI) broth in a 96-well microtitre plate, which also contained an appropriate control (8 replicates). Subsequently, a Nunc Immuno Maxisorp 96 peg plate was immersed in the cultures to inoculate the individual pegs. This was transferred to a microtitre plate containing BHI broth, which was incubated for 48 h under shear-force conditions. Pegs containing biofilms were removed, washed and air dried prior to staining in 0.5% crystal violet (CV). Excess CV was removed and each peg decolourised in 100% ethanol which was measured at 570 nm in a microtitre plate reader. Biofilms were compared to the positive control (100%): ≥ 75% = +++; 25 – 75% = ++; ≤ 25% = +. Representative strains from each section were also examined in a clinically relevant catheter model using this quantitative methodology.
Results: Of the 1000 isolates screened approximately 82% were MRSA isolates. Of these 20% displayed an optimal biofilm phenotype (+++), 50% displayed a good biofilm (++), 30% displayed a less than average biofilm (+). On closer inspection of the MRSA phenotypes in relation to their genotypes the following was observed: +++ (67% = EMRSA15, 20% = EMRSA16); ++ (65% = EMRSA15, 30% = EMRSA16); + (52% = EMRSA15, 41% = EMRSA16). In addition, each strain tested in a catheter model displayed biofilm growth similar to that of the peg plate model.
Conclusions: This study has shown that EMRSA15 clones from Scotland appear to have a greater propensity than other MRSA clones to form tenacious biofilm structures. This may indicate that these strains have a novel virulence factor that confers a selective advantage for adhesion and biofilm formation. This is the subject of ongoing investigations.
P1796 A comparative study on the frequency of genes encoding cytotoxins in Austrian, Hungarian and Macedonian methicillin-resistant and sensitive Staphylococcus aureus strains
E. Kocsis, N. Pesti, K. Kristóf, K. Nagy, Z. Cekovska, H. Lagler, K. Stich, W. Graninger, F. Rozgonyi (Budapest, HU; Skopje, MK; Vienna, AT)
Objectives: The aim of this study was to detect the presence of genes encoding cytotoxins in Staphylococcus aureus strains isolated from invasive infections, and compare according to country and methicillin resistance.
Methods: The phenotypical identification of the strains was done by classical microbiological methods. The genetical confirmation of the strains was done by detecting genes for thermostabile endonuclease (nucA) and 23S rRNA. According to the presence of the gene encoding methicillin resistance (mecA) 48 MRSA and 128 MSSA from Austria (AT), 110 MRSA and 94 MSSA from Hungary (HU), 73 MRSA and 29 MSSA strains from Macedonia (MK) were examined. The genes responsible for cytotoxins were detected by polymerase chain reaction. The pulsed-field gel electrophoresis of the strains was also done, evaluation of results is in progress.
Results: The Panton-Valentine leukocidin genes (lukS-PV, lukF-PV) were only found in 2.3% of AT MSSA, in 4.3% of HU MSSA and in 1.4% of MK MRSA strains. The genes of the alpha- and delta-haemolysins (hla, hld) were present in all HU and MK strains. AT MRSA and MSSA strains harboured the hla gene in 94% and 86%, respectively. The hld gene was carried in 96% and 98% of AT MRSA and of MSSA strains. The hlb gene coding beta-haemolysin was found in 33% and 52% of HU MRSA and MSSA, in 68% and 76% of MK MRSA and MSSA strains, and in 58% and 40% in AT MRSA and MSSA strains, respectively. The gamma-haemolysin gene (hlg) carriage was 96% and 93% in MK MRSA and MSSA, 88% and 68% in HU MRSA and MSSA, 69% and 74% in AT MRSA and MSSA strains. The haemolysin gamma variant gene (hlgv) was found in 100% in the MK strains, 100% and 84% in HU MRSA and MSSA, 100% and 56% in AT MRSA and MSSA strains. Comparing all the MRSA with MSSA strains independently from country origin the presence of hlg and hlgv genes was significant for MRSA strains (p< 0.05). The presence of 5-gene combination pattern composed of hla, hlb, hlg, hlgv, hld genes was significant in both MSSA and MRSA strains (p < 0.05).
Conclusions: Significant differences in harbouring cytotoxin genes were found between S. aureus strains of different countries and between MRSA and MSSA strains. The low frequency of beta-haemolysin gene in MRSA strains can explain the high organ persistence experienced in our former study and the delayed or incomplete development of haemolytic zone around the MRSA colonies.
Supported by ÖAAD and TéT grant No.: A-19/02 and OTKA T046186
P1797 Prevalence of different virulence factors and in vitro adherence of Enterococcus spp. to urinary catheters
J. Dlugaszewska (Poznañ, PL)
Objectives: There is little information about the virulence properties of enterococci. Bacterial adherence to plastic biomaterials is the initial step necessary for the colonisation of the catheter surface. Hemolysins, gelatinase and some phenotypic markers have been proposed as possible virulence factors of Enterococcus strains. This study aimed to determine the incidence of hemolysin, gelatinase, lipase among enterococi isolated from patient with UTI and their adherence to urinary catheters.
Methods: For detection of hemolysin activity brain heart infusion agar supplemented with 5% horse blood was used. Gelatinase activity was detected using BHI broth containing 3% gelatin. For detection of lipase activity enterococci were inoculated on Spirit Blue agar supplemented with Lipase reagent.
Adherence to latex siliconised Foley and PCV Nalaton catheters were analysed. For the adherence assays, it was used the method by Joyanes et al., in which the number of adherent bacteria is estimated by agar plate method.
Results: 78 enterococcal strains: E. faecalis (43) and E. faecium (35), from UTI were investigated. Sixty-six (84%) isolates were hemolysin producing (70% of E. faecalis; 79% of E. faecium), twelve (15%) were gelatinase producing (E. faecalis). The production of lipase was observed in 13 (17%) isolates (7% of E. faecalis, 14% of E. faecium).
Adherence of enterococci to biomaterials occurred very rapidly. It was observed after 1 h already but the highest number of bacteria was isolated after 24 h. The adherence of E. faecalis to both biomaterials was significantly greater (p < 0.05) than that of E. faecium. Moreover, after 24 h of incubation the adherence of both E. faecalis and E. faecium to PCV was significantly greater (respectively: mean 8.4×106 cfu/mL and 6.2×105) than that to siliconised latex (respectively: mean 5.7×105 cfu/mL and 1.2×105).
Conclusion: Further studies of the prevalence of hemolysin, gelatinase, lipase production and adherence to biomaterials in enterococcal isolates from infectious and noninfectious sites are necessary to elucidate the role of these factors in human infections.
P1798 Association between opaque variants and serotypes in invasive isolates of Streptococcus pneumoniae
I. Serrano, J. Melo-Cristino, M. Ramirez (Lisbon, PT)
Objectives: Determine the opacity phenotype of Streptococcus pneumoniae responsible for invasive human infections and probe the relationship between serotype and opacity phenotype.
Methods: The phenotype of 304 invasive isolates from Portugal (1999–2002) analysed previously and representing the 10 most prevalent serotypes was determined. Three possible phenotypes were considered, opaque (O), transparent (T) and intermediate (I). For the scoring of the dominant phenotype, 104 cfu were plated on a transparent medium and incubated at 37°C in a candle extinction jar for 16 h. The colony morphology was determined using a stereomicroscope and oblique transmitted illumination.
Results: The visual inspection of the plates allowed unambiguous identification of the phenotype of the strain, which was considered to be the phenotype exhibited by >70% of the colonies. 18 strains could not be classified according to this criterium. The majority of the isolates classified (52%, n = 158) were O variants and only 26% (n = 79) were T variants while the remaining 16% (n = 49) were I phenotype. We found a striking difference in prevalence of each phenotype among serotypes, therefore serotype specific empirical odds ratio (OR) and 95% confidence intervals (CI) were calculated by reference to all other serotypes. In this analysis strains classified as I variants were excluded. Serotypes 1, 4 and 23F presented a higher association with the opaque phenotype than expected by chance (OR > 1), whereas serotypes 3 and 14 were more associated with the T phenotype than expected (OR < 1). The serotypes 9V, 8, 19A and 7F were not statistically associated with the O phenotype and the calculation of OR was not applicable to serotype 12B. We found no relationship between the O phenotype and the different genetic backgrounds of the isolates as determined by PFGE and MLST.
Conclusion: The O phenotype dominated on a 2:1 ratio over T phenotype among our collection of invasive isolates representing 10 serotypes, suggesting a role for phase variation in pneumococcal human infections. This observation highlights the heterogeneity of opacity phenotypes in invasive isolates and lends further support to the proposal that other factors, in addition to the site of isolation, determine the opacity phenotype of a given isolate. The association between serotype and colonial opacity could help explain epidemiological differences observed among pneumococcal serotypes such as a higher invasive disease potential.
P1799 Antibodies against LTA isolated from E. faecalis 12030 recognize LTA from heterologous enterococcal strains but mediate opsonophagocytic killing only to CPS-A and CPS-B strains
I. Toma, C. Theilacker, I. Sava, A. Kropec, F. Hammer, J. Huebner (Freiburg, DE)
Objectives: Enterococci are among the most common causes of infections in hospitalised patients. Alternative treatment strategies are needed since these bacteria commonly exhibit resistances to multiple antibiotics. We have shown previously that LTA is the target of opsonic antibodies against Enterococcus faecalis strain 12030. Here we present new data extending our knowledge of the basis of serodiversity of E. faecalis and E. faecium.
Methods: LTA from E. faecalis and E. faecium strains was prepared as described previously. Rabbit antiserum raised against LTA from strain 12030 was tested by ELISA using LTA isolated from the four prototype strains of the serotyping system by Hufnagel et al. as coating antigens. An opsonophagocytic assay (OPA) was used to asses killing mediated by the anti-LTA serum. Inhibition of opsonophagocytic killing with purified LTA was employed to confirm that LTA was the target of opsonic antibodies. Protection mediated by rabbit anti-LTA serum was evaluated in a mouse bacteraemia model.
Results: Rabbit anti-LTA serum promoted killing of E. faecalis strain 12030 in the OPA (>90% killing at a serum concentration of 1:200). Mice immunised with rabbit anti-LTA serum and challenged i.v. with E. faecalis strain 12030 had significantly lower bacterial counts (99% reduction). Anti-LTA serum was broadly cross-reactive with LTA isolated from heterologous E. faecalis serotypes by ELISA. The antiserum also reacted with heterologous strains of E. faecalis and E. faecium in a whole cell ELISA. In the OPA, anti-LTA serum killed not only strains belonging to the same serogroup as strain 12030 (CPS-A), but also strains of the CPS-B serotype and two vancomycin-resistant E. faecium strains. Killing activity of anti-LTA serum against these strains was fully inhibited by homologous LTA as well as LTA derived from a CPS-B strain. Strains of the CPS-C and CPS-D serotype were not opsonised by anti-LTA. The opsonophagocytic killing mediated by antisera raised against CPS-C and CPS-D prototype strains were not inhibited by LTA from strain 12030 or homolgous LTA despite the presence of anti-LTA antibodies in these antisera.
Conclusions: Antibodies against LTA bind ubiquitiously to enterococcal LTA, but mediate opsonophagocytic killing only to E. faecalis CPS-A and CPS-B strains as well as to some E. faecium strains. Despite binding to LTA of CPS-C and CPS-D strains these strains are not killed, maybe due to the presence of an antiphagocytic capsule.
P1800 Do enterotoxins and toxic shock syndrome toxin-1 of Staphylococcus aureus contribute in the pathogenesis of septic arthritis?
M. Chitsaz, M. Barton, M. Bazargan, N. Badami, R. Sedaghat (Tehran, IR; Adelaide, AU)
Background: Septic arthritis is one of the most destructive joint diseases in humans. Staphylococcus aureus is the most common cause of infectious arthritis and its rate is about 60–80% in many reports. To elucidate the probable involvement of enterotoxins and toxic shock syndrome toxin-1 (TSST-1) in the pathogenesis of S. aureus septic arthritis, production of these superantigens (SupAg) were determined in 42 clinical isolates of S. aureus obtained from patients with septic arthritis and the arthritogenicity of SupAg positive and SupAg negative isolates were studied.
Methods: Reversed passive latex agglutination, and isoelectricfocusing were used for detection of enterotoxins A, B, C, and D, and TSST-1 in culture supernatants. Arthritogenicity was evaluated by using an established rat model of septic S. aureus arthritis. Experimental groups of Sprague Dawely rats (n = 5) for each type of toxin producing strains (enterotoxins A, B, C, and D, and TSST-1), and non-SupAg producing strains were evaluated (totally 6 group). A suspension of bacterial strains (107 live cells/rat) were injected through tail (intravenously) and rats were evaluated clinically and histopathologically for development and severity of arthritis.
Results: From 42 S. aureus isolates, 35 (83.3%) were positive for SupAg production. Among SupAg positive strains, 24 strains (57.1%) produced only one type of toxin and 11 strains (26.2%) produced a combination of two types of superantigen toxins and we did not detect any of these super-antigens in 6 strains. Almost all rats who received SupAg positive strains injection, developed arthritis (93.75%). In contrast, only 43.5% of rats with SupAg negative strains displayed arthritis. Also there was significant difference in the severity of arthritis developed by SupAg positive (Arthritic Index = 7.2) with SupAg negative strains (Arthritic Index = 2.65).
Conclusion: The results indicate that super-antigen production is possibly an important virulence factor in the development and severity of S. aureus septic arthritis.
P1801 Significance of Streptococcus bovis biotype II/2 bacteraemia in patients with chronic liver diseases
O. Putcharoen, G. Suwanpimolkul, M. Pongkumpai, S. Nilgate, W. Kulwichit, T. Tantawichien, M. Hanvanich, C. Suankratay (Bangkok, TH)
Background: The association between bacteraemia caused by Streptococcus bovis and colonic malignancy is well documented. However, most studies have not focused on the relationship between S. bovis biotypes and colonic malignancy and hepatobiliary diseases. We conducted a retrospective study of patients with S. bovis bacteraemia in King Chulalongkorn Memorial Hospital, Bangkok, Thailand from 2002 to 2005. S. bovis biotypes were determined with the API Rapid Strep system (Analytab Products, Plainview, NY). Demographic data, clinical manifestations, and mortality were obtained for statistical analysis.
Results: During the study period, a total 106 S. bovis strains were isolated. Of 88 isolates causing bacteraemia, 8 (8.9%) were biotype I, 1 (1.1%) were biotype II/1, and 79 (90%) were biotype II/2. The sex and age distributions of patients infected with different S. bovis biotypes were not different. Data of 52 patients with clinically significant bacteraemia were reviewed. Primary bacteraemia was the most common clinical feature (67%), followed by biliary tract infection (13.5%), spontaneous bacterial peritonitis (7.7%), infective endocarditis (4%), meningitis (4%), and skin and soft tissue infection (4%). Of 52 patients, the prevalence of S. bovis from each biotype was 90%, 7.6%, and 2.4% for biotype II/2, 1, and II/1, respectively. Of 48 patients infected with S. bovis biotype II, 28 (53%) had underlying hepatobiliary diseases, and 12 (25%) had hepatocellular carcinoma. In addition, no patients infected with biotype II in our study had colonic malignancy. All isolates were susceptible to penicillin and the third-generation cephalosporins. Overall in-hospital mortality rate from S. bovis bacteraemia was 37%. The mortality rate was 50% and 24% in cirrhotic and non-cirrhotic patients, respectively (p < 0.05).

Underlying diseases of 52 patients with Streptococcus bovis bacteraemia by biotype. HCC, hepatocellular carcinoma; DM, diabetes mellitus; CRF, chronic renal failure.
Discussion: This study demonstrated that S. bovis biotype II, especially biotype II/2, is the most common isolate in our institute, compared with the previous studies conducted in western countries. All patients with S. bovis biotype II bacteraemia had underlying diseases, and hepatobiliary diseases are the commonest underlying condition.
P1802 Detection of Panton-Valentine leucocidin gene in some clinical isolates of methicillin-resistant Staphylococcus aureus: relation to virulence
N. Rizk, S. El-Fiky, E. El-Gamal, E. Badawy (Alexandria, Al Mansoura, EG)
Objectives: To assess the prevalence of Panton-Valentine leucocidin (PVL) encoding genes lukF and lukS among methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates, and to investigate its association with various types of staphylococcal diseases.
Methods: Forty seven MRSA isolates were recovered from unique patient clinical specimens: skin and soft tissue infections (SSTI), n = 18; broncho-alveolar lavage (BAL) from clinically diagnosed cases of pneumonia, n = 20; surgical site infections (SSI), n = 7; and central venous line (CVL) catheter tip, n = 2. Specimens were collected during the period between June 2005 and May 2006. All isolates were tested for the presence of mecA and PVL genes (lukS/F) by single target polymerase chain reaction (PCR).
Results: All isolates were mecA gene positive. The PVL genes were detected in 8 (17.02%) of the 47 MRSA isolates included in the study. The prevalence of PVL genes varies with the type of specimen from which MRSA isolates were recovered; 27.78% (5/18) among isolates associated with SSTI, 15% (3/20) among isolates responsible for community-acquired pneumonia; one of them was recovered from a case of acute necrotising pneumonia. No PVL genes were detected from MRSA isolates recovered from cases of hospital acquired infections including SSI or CVL catheter tips.
Conclusion: PVL genes are prevalent among community-acquired MRSA strains with no prevalence among hospital acquired strains. The presence of PVL genes is related to disease severity especially acute necrotising pneumonia. Close surveillance of these strains is essential to monitor their spread, antimicrobial resistance profile, and association with disease severity.
P1803 Capsular and surface polysaccharide serotypes of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains currently circulating in Germany
C. von Eiff, K.L. Taylor, A. Mellmann, A.I. Fattom, A.W. Friedrich, G. Peters, K. Becker (Münster, DE; Rockville, US)
Objectives: Due to its ability to cause serious and fatal infections, Staphylococcus aureus remains one of the most feared microorganisms. The severe consequences of staphylococcal infection heighten the importance of prevention. Currently, a capsular polysaccharide (CP) bivalent conjugate vaccine with type 5 and type 8 polysaccharides chemically conjugated is being developed as a new tool for infection prevention of S. aureus infections. In addition, a S. aureus 336-conjugate vaccine is aimed to prevent those infections which are caused by serotype 336 and were previously referred to as nontypeable.
Methods: To analyse the potential applicability of a putative S. aureus polysaccharide conjugate vaccine, 714 German methicillin-susceptible (MSSA) and methicillin-resistant (MRSA) S. aureus strains collected in various multi-centre studies were tested for their capsular and surface polysaccharide serotype by slide agglutination with specific antibodies (anti-T5-DT, anti-T8-DT, anti-336-rEPA). The strain serotypes were confirmed by immunodiffusion using lysostaphin-digested cell lysates.
Results: Studying MRSA strains representing 86 unique spa types and thus covering >90% of European MRSA spa types registered, 39 (45.3%) were type 5 and 36 (41.9%) were type 8. Eleven isolates (12.8%) were serotype 336. Among 400 MRSA isolates collected from ten different laboratories covering university hospitals, general hospitals and clinics, serotype 336 was the second most common serotype (16.5%) throughout Germany. In a multi-centre study mostly comprising MSSA, type 8 positive strains were more prevalent among isolates recovered from anterior nares of patients who did not subsequently develop S. aureus bacteraemia compared to those who became bacteraemic and S. aureus could be isolated from blood.
Conclusion: Capsular and surface polysaccharide serotyping showed that the majority of German MSSA and MRSA isolates are comprised of serotypes 5 and 8, although a higher than expected percentage of type 336 strains were observed. The addition of the newly described type 336 to a capsular polysaccharide-protein conjugate vaccine could extend the coverage substantially and would include virtually all MSSA and MRSA strains currently circulating in Germany.
P1804 The ACME-associated arcA gene occurs in diverse strain types of MRSA in England and Wales
M.J. Ellington, L. Yearwood, M. Ganner, C. East, M.A. Ganner, M. Warner, A.M. Kearns (London, UK)
Objectives: The ST8-SCCmecIVa (USA300) MRSA clone is highly successful and prevalent, most notably, in the USA. This clone can harbour the Arginine Catabolism Metabolic Element (ACME) suggested as contributing to pathogenicity. To date, the ACME associated arcA gene has not been reported outside the ST8 genetic background of S. aureus. We sought to determine the distribution of ACME-arcA amongst common community associated (CA)- and healthcare associated (HA)-MRSA strains in England and Wales.
Methods: Previously characterised CA-MRSA isolates, and representatives of HA-MRSA clones occurring in the UK (total n = 215), were PCR screened for ACME-arcA. MICs of a range of antibiotics were determined; SmaI PFGE, spa sequence based typing and toxin gene profiling were also performed. Patient demographic and clinical data were collated.
Results: The ACME-arcA gene was detected in 23 isolates. Seventeen isolates were spa type t008 and the same pulsotype as known ST8 isolates. These 17 isolates were resistant to β-lactams with variable ciprofloxacin and erythromycin resistance. Four of the remaining six ACME-arcA positive isolates were closely related to each other by PFGE (≤ 2 band changes) and spa typing (t359 and t267 – differing by only one repeat). These four isolates were resistant to β-lactams, and had variable resistance to erythromycin, ciprofloxacin, clindamycin, gentamicin, tetracycline and fusidic acid. One of the remaining two isolates was ST1 by pulsotype, agreeing with the t127 spa type, the other was an EMRSA-15 variant; spa type t020 and ST22 by pulsotype. The ST1 isolate was resistant to β-lactams only, whilst the ST22 isolate was resistant to β-lactams, ciprofloxacin and erythromycin. The 17 ACME-arcA, ST8 isolates were PVL-positive and mostly associated with skin and soft tissue infections. In addition PVL-positive ST1 isolate and the four PVL-negative spa t359 isolates were associated with skin and soft tissue infections.
Conclusions: Whilst the ACME-arcA gene is most prevalent amongst ST8-like (equivalent to USA300) isolates in the UK, to our knowledge this is the first time it has been identified in other genetic backgrounds. The emergence of strains with acquired elements associated with the ‘fit’ USA300 strain is a significant public-health concern.
Resistance surveillance – general
P1805 Susceptibility analysis of gastroenteritis and beta-haemolytic streptococcal pathogens collected during a decade of SENTRY Antimicrobial Surveillance Program Monitoring in Europe (1997–2006)
D. Biedenbach, T. Fritsche, H. Sader, R. Jones (North Liberty, US)
Objectives: To determine the antimicrobial susceptibility (S) of beta-haemolytic streptococci (BHS) and gastroenteritis (GI) pathogens in Europe (EUR) over 10 years. Infections caused by BHS can be relatively mild or severe and invasive due to suppurative sequelae. Although infections caused by GI pathogens are mostly self-limiting (diarrhoea), invasive disease such as bacteraemia due to Salmonella spp. (SAL) or Aeromonas. (ASP) can be more common among the very young, elderly and immunocompromised persons.
Methods: During 1997–2006, the following pathogens (no.) were collected from SENTRY Program (EUR) objectives and tested for S: ASP (154), Y. enterocolitica (YET; 52), Shigella spp. (SHIG; 283), SAL (1,526) and BHS (2,553). CLSI broth microdilution test methods and interpretive criteria were utilised with appropriate concurrent quality assurance practices.
Results: ASP/YET isolates were resistant (R) to ampicillin (AMP) but S to penicillin with a β-lactamase inhibitor (85/>99%), cefuroxime (CXM; 80/95%) and ceftriaxone (CRO), cefepime (FEP), and gentamicin (GENT) at >95%. Tetracycline (TET) and trim/sulfa (T/S) R in ASP was 15%, and nalidixic acid (NA)-R was 28% associated with a high ciprofloxacin (CIP) MIC90 to 1 mg/L compared to the wildtype population (≤0.03 mg/L). The AMP-S of SHIG in 2001 (63%) was only 23% in 2003 influenced by Russian epdemic isolates (>95% AMP-R). R to NA was 0% in 2001 but 4.3% in 2003; and TET-R increased >7% during the sampled interval. S among SAL and BHS were compared during the first and the second five year samples. There was a decline in S among SAL during the last five years with AMP-R (24.7%), NA-R (17.4%) and TET-R (20.3%). A significant increase in ESBL phenotypes (2002–2006) followed a clonal S. typhimurium (CTX-M-5) outbreak in Russia. We noted a 5–7% decline in S to erythromycin (ERY) among Group A and B BHS during the second sampled period. ERY-R rates for BHS varied between nations with highest rate in Italy (37%). TET-S was stable between years, but varied among serogroups (A and C/70–80%, G/50% and B/20%). All BHS were S to linezolid, vancomycin, quin/dalfo and were very S to levofloxacin and penicillin.
Antimicrobial activity of 9 drugs versus Salmonella spp. (1,526)
| Salmonella spp./Antimicrobial | Year (no.) |
|||
|---|---|---|---|---|
| 1997–2001 (664) |
2002–2006 (862) |
|||
| %Sa | %Ra | %Sa | %Ra | |
| AMP | 77.4 | 21.8 | 74.6 | 24.7 |
| CXM | 69.6 | 0.2 | 53.1 | 2.7 |
| CRO | 100.0 | 0.0 (0.2)b | 98.1 | 1.9 (2.7)b |
| FEP | 100.0 | 0.0 | 98.4 | 0.3 |
| NA | 85.4 | 14.6 | 82.6 | 17.4 |
| CIP | 99.9 | 0.1 (13.7)c | 99.7 | 0.1 (16.1)c |
| TET | 76.8 | 22.0 | 78.7 | 20.3 |
| T/S | 93.8 | 6.2 | 93.6 | 6.4 |
| GENT | 98.3 | 1.4 | 97.1 | 1.3 |
Criteria as published by the CLSI (M100-S16).
% of strains with ESBL screen-positive (MIC ≥ 2 mg/L for CRO).
% strains with elevated CIP MIC (≥0.12 mg/L).
Conclusions: GI pathogens and BHS contribute a significant amount of morbidity in EUR and worldwide. Antimicrobial surveillance of these species in EUR shows a trend toward declining S to several drug classes and continued monitoring is necessary to track the R profiles of these important bacterial species.
P1806 Prevalence and antimicrobial susceptibility profiles of skin and skin structure infection pathogens in Europe: report from the SENTRY Antimicrobial Surveillance Program (1997–2005)
J. Ross, R. Jones, H. Sader, M. Stilwell, T. Fritsche (North Liberty, US)
Objectives: To present a 9-year summary of bacterial pathogens (prevalence and S trends; SENTRY Program) recovered from EUR patients experiencing skin and skin structure infection (SSSI). Rising resistance (R) rates are being observed globally in both Gram-positive and -negative SSSI pathogens, challenging accepted approaches to clinical management, especially empiric antimicrobial therapy guidelines.
Methods: Non-duplicate, clinically-significant SSSI isolates (6,828) were collected from >25 medical centres in Europe participating in the SENTRY Program from 1997–2005 (exception, 2001). Identifications were confirmed by the central monitoring laboratory and all isolates were S tested using CLSI methods and interpretive criteria (M100-S16) against antimicrobial agents commonly utilised as empiric or directed therapy.
Results: The ten ranking EUR SSSI pathogens for all years were: S. aureus (SA; 38.8%), P. aeruginosa (PSA; 11.8%), E. coli (EC; 10.4%), Enterococcus spp. (ENT; 5.8%), coagulase-negative staphylococcus (5.0%), Enterobacter (ESP; 4.9%), Klebsiella spp. (KSP; 4.3%), beta-haemolytic streptococcus (BHS; 3.8%), P. mirabilis (3.1%), and Acinetobacter spp. (ASP; 2.6%). Analysis of intervals 1997–2000 and 2002–2005 showed that BHS moved from 10th to 4th in prevalence and erythromycin (ERY)-R increased from 12.3 to 22.5%. While prevalence of SA varied widely between countries (highest in the UK [86.1%] and lowest in Turkey [40.5%]) overall EUR methicillin-R (MRSA) rates remained unchanged between intervals (22.8 and 24.3%, respectively) and ERY-R decreased slightly (31.2 to 27.2%). Vancomycin-R rates in ENT increased slightly between intervals (2.7 and 3.9%, respectively). R increases were most notable for imipenem (IPM) with PSA and ASP, and also with levofloxacin (LEV) for KSP and EC. ESBL-phenotypes were detected in 12.7% of EC and 20.0% of KSP in 2002–2005; both were increased significantly from the previous monitored interval.
| Organism (no.) | R pattern | % Inhibited at CLSI breakpoints |
|
|---|---|---|---|
| 1997–2000 | 2002–2005 | ||
| SA (2,652) | MRSA | 22.8 | 23.0 |
| PSA (807) | IPM-R | 9.5 | 12.4 |
| Ceftazidime (CAZ)-R | 17.8 | 16.8 | |
| LEV-R | 28.2 | 26.2 | |
| Amikacin (AMK)-R | 10.4 | 5.5 | |
| EC (711) | CAZ-R | 2.2 (8.3)b | 5.7 (12.7)b |
| LEV-R | 6.5 | 12.3 | |
| ESP (338) | IPM-NSa | 0.6 | 1.7 |
| CAZ-R | 22.4 | 17.5 | |
| LEV-R | 7.5 | 6.2 | |
| KSP (283) | CAZ-R | 14.9 (20.3)b | 11.9 (20.0)b |
| LEV-R | 3.4 | 9.6 | |
| AMK-R | 1.4 | 1.5 | |
| ASP (179) | IPM-R | 7.9 | 11.5 |
| CAZ-R | 46.5 | 41.0 | |
| LEV-R | 28.7 | 30.8 | |
| AMK-R | 55.4 | 28.2 | |
NS, non-susceptible.
Number in parentheses reflects the ESBL-phenotype rate (MIC values ≥2 mg/L).
Conclusions: With key exceptions (ERY-R in BHS; IPM-R in PSA and ASP; LEV-R in Enterobacteriaceae), R profiles for leading SSSI pathogens did not change significantly during the first decade of SENTRY Program data collection. The continued spread of virulent MRSA into the community setting and increases being detected in ESBL and carbapenem-R rates are, however, cause for concern and require continued monitoring to guide contemporary antimicrobial therapies and searches for new compounds.
P1807 Resistance rates among selected Gram-positive and -negative isolates from European medical centres: a decade of SENTRY Program Surveillance (1997–2006)
D. Biedenbach, T. Fritsche, H. Sader, R. Jones (North Liberty, US)
Objectives: To study the prevalence and resistance (R) trends among selected Gram-positive and -negative pathogens. The increase in R to antimicrobials can occur soon after introduction into clinical practice, evolve slowly, or be sporadic without a clear trend. Mupirocin (MUP)-R occurred soon after it was released onto the market (associated with increased use). A high level of quinupristin/dalfopristin (Q-D)-R in some countries has been said to be associated with the use of related agents in animal husbandry. Trimethoprim/sulfamethoxazole (T/S) is the drug of choice for the treatment of S. maltophilia (SM) and T/S-R SM are perceived as rare in occurrence. Carbapenemases and high-level class C chromosomal cephalosporinase production combined with altered outer membrane permeability are primarily responsible for carbapenem [CARB]-R among enteric bacilli which occur at varying frequency.
Methods: Countries in the European region (14–16) contributed isolates for reference broth microdilution testing using CLSI (2006) methods and interpretive criteria (2007). MUP was tested over a seven-year period (2000–2006) against S. aureus (SA) and coagulase-negative staphylococci (CoNS), and Q-D (1997–2006) against SA, CoNS and E. faecium (EFM). T/S was analysed against 736 SM and CARB (imipenem [IPM]) against >30,000 enteric bacilli during 1997–2006.
Results: The highest % of Q-D non-susceptible (S) SA/CoNS were recovered in France and Austria (10/3%) and Greece (2/4%) compared to other countries (≤1%). All monitored countries detected (Q-D)-non-S EFM at a rate of 10–>70%. Clonal dissemination influenced the rate of non-S Q-D SA, CoNS and EFM during some monitored years. High MUP-R rates among MRSA were noted in Belgium, Ireland, Sweden and the UK (18.2–25.6%) and CoNS in Belgium, Ireland and Turkey (≥40%). T/S-R SM (4.5% overall) were detected in nearly all countries, but at very low prevalence and CARB-R among enterics was also rare in Europe with IPM-R at <0.1%.
Occurrences of resistance to Q-D and MUP among staphylococci and E. faecium (1997–2006)
| Org./Antim.-R (no.) | Year (%non-S)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | |
| SA | ||||||||||
| OX-S/Q-D (11.967) | 0.3 (1.0)b | 0.1 | 0.0 | 0.3 | 0.2 | <0.1 | <0.1 | 0.3 | 0.2 | 0.2 |
| OX-R/Q-D (4,813) | 4.3 | 4.9 | 5.9 | 4.3 | 3.7 | 2.2 | 0.8 | 1.1 | 1.3 | 1.3 |
| OX-S/MUP (9,399) | − | − | − | 0.6 | 0.5 | 1.0 | 0.9 | 1.0 | 0.6 | 0.7 |
| OX-R/MUP (3,903) | − | − | − | 14.6 | 11.1 | 12.1 | 10.6 | 7.7 | 7.2 | 6.2 |
| CoNS | ||||||||||
| OX-S/Q-D (1,822) | 1.0c | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| OX-R/Q-D (5,517) | 1.3 | 1.0 | 0.5 | 3.1 | 0.6 | 1.1 | 0.7 | 0.7 | 0.7 | 2.1 |
| OX-S/MUP (1,269) | − | − | − | 5.1 | 10.5 | 5.4 | 4.2 | 3.5 | 5.2 | 2.0 |
| OX-R/MUP (3,994) | − | − | − | 16.7 | 21.8 | 25.4 | 17.7 | 18.8 | 18.3 | 21.4 |
| EFM/Q-D (1,361) | 20.3 | 26.1 (50.0)d | 22.7 | 18.7 | 25.5 | 26.3 | 23.6 | 27.4 | 29.5 | 30.2 |
Non-susceptibility % based upon the CLSI interpretive criteria for Q-D (M100-S16) and MUP at MIC values ≥16 mg/L (high- and low-level rates combined).
Clonal outbreak in France increased the prevalence to 1%.
Three isolates from different countries with intermediate MIC values (2 mg/L).
A large number of clonally disseminated strains in Austria, Germany, Italy, Portugal and Spain produced a higher percentage of non-susceptible isolates.
Conclusions: MRSA and CoNS were more resistant to Q-D and MUP compared to methicillin-S strains with R rates remaining stable or declining over the tested SENTRY Program time period. In contrast, (Q-D)-non-S among EFM has steadily increased in Europe and is presently over 30%. Continued monitoring of regional pathogens is essential to determine the longitudinal efficacy of older and more recently introduced antimicrobial agents.
P1808 Update on potency and spectrum of activity of meropenem and selected broad-spectrum agents: testing results from the USA MYSTIC Program (2006)
P. Rhomberg, J. Kirby, T. Fritsche, H. Sader, R. Jones (North Liberty, US)
Objectives: To monitor the potency and spectrum of meropenem (MEM) and 10 other broad-spectrum agents against pathogens collected from hospitalised patients within USA medical centres actively using carbapenems (CARBs) for the treatment of serious infections. The Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Program is a longitudinal resistance (R) surveillance network with >100 sites worldwide. In the USA (2006), 15 sites continued participation by submitting 200 consecutive, non-duplicate clinical isolates from defined pathogen groups.
Methods: 2,841 isolates (95% compliance) including 1,260 Enterobacteriaceae (ENT), 606 P. aeruginosa (PSA), 456 oxacillin-susceptible S. aureus (MSSA), 300 streptococci, 149 E. faecalis, and 70 Gram-positive anaerobes were tested using CLSI reference broth microdilution or agar dilution susceptibility (S) methods and associated interpretative criteria. Extended-spectrum β-lactamase (ESBL) phenotype strains were confirmed using Etest methods, negative strains were then screened for acquired AmpC genes (8 classes) by PCR. Serine carbapenemase (C-ase) production was screened by disk approximation and characterised by PCR. All fluoroquinolone (FQ)-R ENT isolates were tested for a qnr using PCR.
Results: The CARB class had the lowest R rates against ENT (3.3–4.0%) and the FQs had the highest R rates (21.7–22.5%). MEM was the most potent agent tested with an 8-fold lower MIC90 compared to imipenem against ENT strains and was 2-fold more potent against PSA strains (MIC90, 8 vs. 16 mg/L). Among Klebsiella spp. (KSP), 9.5% (59) possessed KPC-type serine C-ase from sites in New York (2), New Jersey, and Ohio at rates of 49.2, 22.2, 32.5, and 2.4%, respectively. Confirmed ESBL rates among E. coli (EC) and KSP were only 4.8 and 5.0%. Unconfirmed ESBL R was due to FOX (5) and CMY (8) in 57% of strains. The qnr gene was detected in only 2.5% of 283 FQR ENT strains. Among PSA, cefepime, MEM, and tobramycin had lowest R rates (5.6–7.9%). All comparator agents were >98% S against MSSA except the FQs (only 88.6–90.4%).
Ten monitored USA MYSTIC Program pathogens in 2006
| Organisma (no. tested) | Meropenern MIC (mg/L) |
%Sb | %Rb | ||
|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | |||
| EC (641) | ≤0.015 | 0.03 | ≤0.015–0.06 | 100.0 | 0.0 |
| KSP (619) | 0.03 | 0.5 | ≤0.015–>32 | 91.0 | 8.2 |
| PSA (606) | 0.5 | 8 | ≤0.015–>32 | 86.5 | 6.4 |
| MSSA (456) | 0.12 | 0.12 | 0.03–0.25 | 100.0 | 0.0 |
| EF (149) | 4 | 8 | 0.5–16 | − | − |
| SPN (138) | ≤0.015 | 1 | ≤0.015–1 | 78.3 | 10.1 |
| BST (113) | 0.03 | 0.06 | ≤0.015–0.25 | 100.0 | − |
| VGS (49) | 0.03 | 0.25 | ≤0.015–4 | 95.9 | − |
| CSP (37) | 0.06 | 2 | ≤0.015–4 | 100.0 | 0.0 |
| PEP (33) | 0.06 | 0.25 | ≤0.015–0.5 | 100.0 | 0.0 |
E F, E. faecalis; SPN, S. pneumoniae; BST, beta-haemolytic streptococci; VGS, viridians group streptococci; CSP, Clostridium spp.; PEP, Peptostreptococcus spp.
CLS1 M100-S16 or M11-A6 breakpoints applied.
Conclusions: MYSTIC Program MIC results demonstrate continued CARB potency and spectrum of activity against ENT, PSA and MSSA, however the escalating presence of serine C-ase among KSP is a serious concern. Continued surveillance in the USA MYSTIC sites appears warranted to monitor the activity of the CARBs and other broad-spectrum agents used as empiric or directed therapy against key pathogens for the treatment of serious infections.
P1809 Ongoing activity of meropenem against European isolates: report on the MYSTIC 2005 results
P. Turner (Macclesfield, UK)
Objectives: MYSTIC (Meropenem Yearly Suceptibility Test Information Collection) is a longitudinal global surveillance Study that has examined annually the in vitro activity of meropenem and other broad spectrum agents against clinically significant, non-copy isolates since 1997. The data presented here covers the year 2005 and deals with isolates from Europe.
Methods: A total of 7710 isolates from centres in Belgium, Bulgaria, Croatia, Czech Republic, Finland, Germany, Greece, Malta, Poland, Russia, Spain, Sweden, Turkey and the UK were tested by CLSI reference methodology (either agar dilution or broth microdilution) and associated interpretative criteria. Extended-spectrum β-lactamase (ESBL) were confirmed by synergy with clavulanic acid and AmpC by lack of synergy with this compound.
Results: See the table .
| Organism (n) | Percent suceptibility |
||||||
|---|---|---|---|---|---|---|---|
| MEM | IPM | P+T | CAZ | CPM | CIP | GM | |
| Staphylococci (1326) | 99 | 99 | 96 | 58 | 97 | 86 | 93 |
| Enterobacteriaceae (3718) | 99 | 98 | 82 | 82 | 87 | 82 | 87 |
| P. aeruginosa (1098) | 75 | 67 | 78 | 72 | 61 | 66 | 62 |
| A. baumannii (408) | 60 | 58 | 23 | 22 | 20 | 27 | 41 |
aMEM, meropenem; IPM, imipenem; P+T: piperacillin + tazobactam; CAZ, ceftazidime; CPM, cefepime; CIP, ciprofloxacin; GM, gentamicin.
Conclusions: Meropenem, as in 1997, remains the antibiotic with the broadest spectrum of activity followed by imipenem. Many of the other comparators are experiencing considerable drops in their in vitro effectiveness indicating the need for continued surveillance.
P1810 Bulgarian Surveillance Tracking Antimicrobial Resistance – BulSTAR: bloodstream infections in 2005
M. Petrov, T. Velinov, T. Kantardjiev, A. Bachvarova, N. Hadjieva (Sofia, BG)
Bulgarian Surveillance Tracking Antimicrobial Resistance – BulSTAR monitors the isolation and antimicrobial susceptibility of all clinically significant microorganisms from blood cultures, cerebrospinal fluid, upper and lower respiratory tract, urine and wound samples. One hundred and thirty six (136) microbiological laboratories – 25 public, 70 hospital and 41 private laboratories from all 28 counties of the Republic of Bulgaria participated in BulSTAR in 2005.
Objectives: The aims of this study are to analyse the aetiological structure of bloodstream infections and the susceptibility to antimicrobial agents of the most frequently isolated bacteria from blood among the participants.
Methods: All the participating laboratories used the CLSI methodology.
Results: Data on bloodstream infections in 2005 are based on 68 083 samples, 2 980 from which were positive. Most frequently isolated bacteria from blood are: S. aureus – 19.3% followed by E. coli – 15.4%, Coagulase-negative staphylococci – 9.9%, A. baumannii – 7.2%, P. aeruginosa – 6.3%, K. pneumoniae – 6.2%, Candida spp. – 5.1% etc. In 2005 the prevalence of methicillin-resistant Staphylococcus aureus was 18.8% and the resistance of the species to erythromycin, clindamicin, gentamicin and ciprofloxacin was higher than 20%. Among the Gram-negative bacteria: the prevalence of ESBL-producing E. coli was 14.1% and among K. pneumoniae was 28.8%. Ceftazidime-resistant P. aeruginosa reached 32.9% and the resistance to ciprofloxacin among the species was similar – 31.5%. Resistance to imipenem among A. baumannii was 16%.
Conclusion: The aetiological structure of bloodstream infections is similar to those in most European countries and also with that in the previous years in Bulgaria. Remarkable is the stable presence of P. aeruginosa and A. baumannii among the leading pathogens causing sepsis in the country. There is a decrease in the proportion of S. aureus and E. faecalis and an increase in the proportion of Candida spp. compared with 2004. Main therapeutical problems are ESBL-positive K. pneumoniae and multiresistant P. aeruginosa and A. baumannii.
P1811 In vitro activity of tigecycline against Gram-positive and Gram-negative bacteria cultured from blood samples, wound swabs and intra-abdominal specimens: results of the German T.E.S.T. Surveillance Program 2005
M. Kresken, J. Brauers, H. Geiss, E. Halle, E. Leitner, G. Peters, H. Seifert (Rheinbach, Heidelberg, Berlin, Münster, Cologne, DE)
Objectives: Tigecycline (TGC), the first glycylcycline antibacterial agent, is indicated for the treatment of complicated intra-abdominal and complicated skin and skin structure infections. G.-T.E.S.T. is a surveillance programme comprising 15 German laboratories which monitors the susceptibility of bacterial pathogens to TGC. The objective of this study was to evaluate the in vitro activity of TGC against bacterial isolates cultured from blood samples, wound swabs and intraabdominal specimens.
Methods: A total of 1,464 isolates including Staphylococcus aureus (Sa, n = 204), S. epidermidis (Se, n = 162), Enterococcus faecalis (Es, n = 107), E. faecium (Em, n = 112), Escherichia coli (Ec, n = 170), Enterobacter (En, n = 126), Klebsiella spp. (Kl, n = 125) were studied. Susceptibility testing with TGC, doxycycline (DOX), cefotaxime (CTX), ciprofloxacin (CIP), oxacillin (OXA), vancomycin (VAN) and other drugs was performed by broth microdilution according to German DIN guidelines in a central laboratory. MICs of TGC were interpreted by EUCAST and FDA criteria.
Results: The majority of isolates was recovered from wound swabs (52.3%), followed by blood samples (29.4%) and intra-abdominal specimens (18.2%). Among Sa and Se, 54% and 77% of the isolates, respectively, were resistant to OXA. The rate of VAN resistance in Em was 7%. TGC was highly active against Gram-positive species. All strains were inhibited at 0.5 mg/L, except two OXA-resistant staphylococci (MICs 1 mg/L). In case of Ec, all strains were inhibited by TGC at the EUCAST breakpoint (bp) of ≤1 mg/L. In contrast, 41% of strains were resistant to DOX, 19% were resistant to CIP, and 6% were putative ESBL producers (MIC of CTX >1 mg/L). Among the Kl isolates, 87% were susceptible to TGC, 2% were intermediate and 10% were resistant, when EUCAST bp were applied. Similar results were observed for En isolates, of which 87% were susceptible, 4% intermediate, and 9% resistant to TGC. However, only two strains one each of Kl and En were resistant at the FDA bp of resistance (≥4 mg/L), with MICs of 8 mg/L. Kl strains intermediate or resistant to TGC were all resistant to DOX, as were 12 of 16 En isolates.
Conclusion: TGC demonstrated excellent in vitro activity against organisms recovered from blood, wounds and the intra-abdominal tract, including multiple resistant organisms. Therefore, TGC seems to be a suitable drug for the treatment of complicated intra-abdominal as well as complicated skin and skin structure infections.
P1812 In vitro activity of tigecycline against pathogens from Turkey – T.E.S.T. Program 2006
V. Korten, G. Söyletir, H. Leblebicioglu, G. Usluer, D. Dündar, D. Ögünç on behalf of the TEST Study Group – Turkey
Objective: The Tigecycline Evaluation Surveillance Trial (TEST) is a global surveillance programme to determine in vitro activity of Tigecycline (TIG), a new glycylcycline, compared to various antibiotics for Gram-positive/negative species. As part of global TEST programme a multicentre surveillance study was performed to evaluate the activity of TIG in Turkey.
Methods: A total of 994 non-duplicate clinically significant isolates collected in 2006 were analysed in this survey. The isolates were identified to the species level at 10 participating centres and confirmed by the central laboratory. MICs were determined by each centre using supplied E-test and interpreted according to CLSI guidelines for comparators (vancomycin, ampicillin, amoxicillin/clavulanate, moxifloxacin, linezolid, erythromycin, ceftazidime, piperacillin/tazobactam, amikacin, imipenem, levofloxacin). Breakpoints defined by FDA were used for tigecycline where applicable.
Results: Selected pathogens tested against tigecycline are shown in the table . No resistance was observed against TIG and linezolid in Gram-positive isolates. Tigecycline presented MIC50/MIC90 of 0.125/0.5 mg/L against MRSA isolates and MIC90 of ≤ 0.25 mg/L against Enterococcus spp. including isolates with high-level gentamicin and vancomycin resistance. Tigecycline, at an MIC of 2 mg/L, inhibited 100% and 97% of E. coli and K. pneumoniae respectively, including ESBL producers. Of 153 acinetobacter isolates, 88 and 57 were non-susceptible to imipenem and all antibiotics tested, respectively. TIG, with an MIC90 of 2 mg/L, was especially active against Acinetobacter spp. including imipenem- and pan-resistant isolates.
| Organism | n | Tigecycline (mg/L) |
|||
|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | Susceptibility (%) | ||
| S. aureus (MR) | 99 | 0.125 | 0.5 | 0.03–0.5 | 100 |
| S. aureus (MS) | 58 | 0.125 | 0.38 | 0.03–0.38 | 100 |
| Enterococcus spp. | 86 | 0.064 | 0.19 | 0.01–0.19 | 100 |
| S. pneumoniae | 57 | 0.023 | 0.094 | 0.01–0.19 | 100 |
| E. coli | 164 | 0.19 | 0.5 | 0.047–1 | 100 |
| Klebsiella spp. | 155 | 0.5 | 1 | 0.047–12 | 97 |
| Enterobacter spp. | 127 | 0.5 | 1 | 0.032–6 | 94 |
| Citrobacter spp. | 25 | 0.19 | 0.5 | 0.047–2 | 96 |
| Morganella morganii | 29 | 1 | 1.5 | 0.38–3 | 97 |
| Serratia spp. | 41 | 1 | 2 | 0.032–4 | 93 |
| Acinetobacter spp. | 153 | 1 | 2 | 0.064–8 | na |
MR, methicillin-resistant; MS, methicillin-susceptible; na, not applicable.
Conclusions: The study validates the potent inhibitory activity of TIG against hospital pathogens in Turkey including strains resistant to antimicrobials currently used to treat nosocomial infections. The data suggest that tigecycline may be a therapeutic option for Acinetobacter spp. including multidrug resistant strains that pose a particular problem in Turkish hospitals.
P1813 A pharmacodynamic analysis of resistance trends in pathogens from patients with infection in intensive care units in the USA between 1993 and 2004
K. Eagye, D. Nicolau, K. Heilmann, J. Quinn, G. Doern, G. Gallagher, M. Abramson (Hartford, Iowa City, Chicago, Upper Gwynedd, US)
Objectives: Increasing resistance (R) to available antimicrobial agents is an area of concern, particularly with respect to nosocomial pathogens. While higher minimum inhibitory concentrations (MICs) can inhibit antimicrobial effectiveness, dose adjustments can often mitigate this effect. The purpose of this study was to ascertain whether reported R in intensive care units (ICUs) to several commonly used agents has increased enough to significantly impact their ability to achieve a bactericidal effect.
Methods: We evaluated 74,235 Gram negative bacilli obtained in the Merck ICU Surveillance Program in the United States of America (USA) between 1993 and 2004. MICs to cefepime (FEP), ceftriaxone (CRO), imipenem (IMP) and piperacillin-tazobactam (TZP) were grouped into four 3-year periods. Patient-derived pharmacokinetics with Monte Carlo simulation were used to predict microbiological success in each period, as measured by cumulative fraction of response (CFR). Trends in CFR over the four 3-year periods of this survey were assessed using the Cochran-Armitage trend test. The primary analysis included all organisms aggregated; P. aeruginosa (PSA) and Acinetobacter spp. (ACNB) were also evaluated individually.
Results: In the primary analysis, IMP 1g q8 h showed %CFRs from 87 to 90 across all four study periods, a trend toward slightly improved susceptibility (p < 0.0001). %CFR for FEP 2g q12 h declined from 87 to 85, showing increasing R over time (p <0.01). The %CFR for TZP 4.5g q6 h declined from 80 to 78, showing increasing R (p < 0.05). CRO had <52% CFR for all regimens in all periods, with no significant trend. Against PSA, significant declines in CFR were seen for (%CFR range, p-value): IMP 1g q8 h (82–79, <0.0001), FEP 1g q12 h (70–67, <0.01), FEP 2g q12 h (84–82, <0.05), TZP 3.375g q6 h (76–73, <0.01), TZP 4.5 q8 h (71–68, <0.01), and TZP 4.5 q6 h (80–77, <0.01). Against ACNB, all regimens of IMP, FEP and TZP showed significant declines in CFR over time (p < 0.0001).
Conclusions: These data suggest that increasing R in ICU pathogens is indeed present in the USA, and that drug effectiveness as measured by ability to attain pharmacodynamic targets has declined. Declines against PSA are significant, though FEP 2g q8 h remains potent and more aggressive dosing of IMP and TZP can preserve some viability for those compounds. The most relevant declines were seen against ACNB; only IMP remains a viable option against this pathogen.
P1814 Susceptibility to tigecycline and to an extended panel of antimicrobials of contemporary Gram-positive and Gram-negative Portuguese isolates
C. Silva-Costa, M. Ramirez, J. Melo-Cristino (Lisbon, PT)
Objectives: Antibacterial resistance is a frequent finding in common bacterial isolates from Portugal. Availability of new agents offer clinicians novel options for therapy. Tigecycline has a broad spectrum of activity, including strains resistant to other drugs. Strains collected in Portugal in 2005 were evaluated for susceptibility to tigecycline and several other antimicrobials.
Methods: A total of 1575 clinically significant isolates were collected and identified at 15 hospitals in Portugal. MICs were determined at the coordinating laboratory using E-tests, and interpreted according to CLSI guidelines. The antimicrobials tested against Gram positive bacteria were: tigecycline, vancomycin, teicoplanin, linezolid, quinupristin/dalfopristin, levofloxacin, gentamicin, streptomycin, rifampicin, erythromycin, benzylpenicillin, ampicillin, cefotaxime. The antimicrobials tested against Gram negative bacteria were: tigecycline, imipenem, trimethoprim/sulphamethoxazole, ciprofloxacin, levofloxacin, gatifloxacin, gentamicin, amikacin, ampicillin, cefotaxime, ceftazidime, cefepime, piperacillin/tazobactam, amoxicillin/clavulanic acid, ampicillin/sulbactam and sulbactam.
Results: Among Gram-positive isolates (n = 825), all Staphylococcus spp. isolates (n = 375), including methicillin-resistant strains, had tigecycline MIC90 ≤0.5 mg/L. All Enterococcus spp. isolates (n = 150), including vancomycin-resistant strains, had tigecycline MIC90 ≤ 0.25 mg/L. Streptococcus pneumoniae (n = 150), Streptococcus pyogenes (n = 75) and Streptococcus agalactiae (n = 75) had tigecycline MIC90 ≤ 0.25 mg/L.
Among Gram-negative isolates (n = 750), ESBL-producing or quinolone resistant Escherichia coli (n = 225), Klebsiella spp. (n = 225) and Enterobacter (n = 75) isolates had tigecycline MIC90 ≤ 2 mg/L. Haemophilus inlfuenzae (n = 75) had tigecycline MIC90 ≤ 0.5 mg/L, Stenotrophomonas maltophilia (n = 75) had tigecycline MIC90 ≤ 1.5 mg/L and Acinetobacter baumannii (n = 75) had tigecycline MIC90 ≤ 4 mg/L.
Conclusion: Most of the pathogens analysed in this study were resistant to many broad-spectrum antibiotics. Tigecycline presented consistently low MIC90 values and broad spectrum of activity, including otherwise resistant strains. These characteristics should make tigecycline a useful option for difficult-to-treat infections.
P1815 Micronet: an Italian automatised laboratory based surveillance and early warning system for infectious diseases
F. D'Ancona, C. Rizzo, V. Alfonsi, M.L. Ciofi degli Atti on behalf of the Micronet Group
Objectives: In Italy several diseases specific surveillance systems have been in use to support mandatory notification of infectious diseases. These systems are mainly based on reporting by clinicians and they require additional resources also from clinical microbiologists. In 2005, the Istituto Superiore di Sanità, supported by Ministry of Health, began to create Micronet, the first Italian automatised laboratory based surveillance system.
Methods: Micronet is designed to be a sentinel surveillance system that collect all laboratory test results (positive and negative) from a convenience sample of peripheral microbiological laboratories. The approach is based on the clinical requests. All data are collected from the informative system (LIS) of each laboratory. Before the transmission to the central server, all data are converted automatically in Micronet data format using standardised tables. The data transmission is designed to be on daily basis.
Results: A group of microbiologists and epidemiologists produced 11 standardised tables, regularly updated. The Micronet team released the specifics for the exchange of the data in XLM format. Seven laboratories, as pilot test, implemented the tables and the specifics at local level, sending 3 months of data, corresponding, removing duplicates, to more than 50,000 records. All the data are stored into the Micronet central database and a web site was set up to provide feedback in terms of analysis on aggregated data. For specific requirements, data could be exported on statistical packages for analysis.
Conclusion: The potential users of Micronet are Regional Authorities (integrating existing clinical and laboratories surveillance system) National authorities (trend analysis, alert and support of the infectious diseases notification system) and participant laboratories (comparing local data with regional/national average). Micronet could also represents an important a national network providing instruments for rapid detection of outbreaks and assessment of microbiological trends. Micronet should be fully operative from January 2007 when it is also planned to recruit other laboratories in order to improve the representativeness of the system. The project is now facing some difficulties and criticalities such as the representativeness, comparability of data and methods for duplicates clearing, management of the standardised tables at local level, but the results obtained in the pilot phase show its potentialities.
P1816 Matching criteria in case-control studies in the field of antimicrobial resistance
M.E. Falagas, E.G. Mourtzoukou, K. Giannopoulou, V.G. Alexiou, P.I. Rafailidis (Athens, GR)
Objectives: Although the effect of confounding factors on the studied outcome may be taken under consideration in stratified or multivariable analysis of the data, another approach is to adjust for the effect of such factors in the design of the study. We evaluated the available evidence from case-control studies in the field of antimicrobial resistance to identify the degree that matching was performed and the criteria used to do so.
Methods: We performed a systematic search of the PubMed database (articles archived by 08/2006) to identify relevant studies. Studies that used the individual matching technique were further analysed.
Results: 115 case-control studies with a focus on antimicrobial resistance were identified; 28 regarding A. baumannii, 25 regarding P. aeruginosa, and 62 for other bacteria. Individual matching was performed in 32 (27.8%) out of the 115 studies. Age was the most frequently used matching criterion in 22 of 32 (69%) evaluated matched case-control studies, while gender was used in 11 (34%), presence of underlying illness in 8 (25%), site of infection in 5 (16%), and area of residence in 4 studies (12.5%). Other criteria were used in less than 10% of the studies.
Conclusion: The available evidence from the analysed data from case-control studies in the field of antimicrobial resistance shows that individual matching is employed only in a proportion of such studies and only for a few characteristics, when it was used. The practical difficulties in finding appropriately matched control patients may contribute to this finding. Although there is a discussion regarding the risks of adjustment, we believe that methodological advances and more utilisation of the individual matching technique in case-control studies will help in answering important specific research questions in the field of antimicrobial resistance.
Resistance surveillance of staphylococci
P1817 Changes in antibacterial resistance patterns in Staphylococcus aureus strains which are related to microbiologically confirmed nosocomial bacteraemia in a tertiary care educational hospital in Turkey
O. Sipahi, H. Pullukcu, S. Aydemir, M. Tasbakan, A. Tunger, B. Arda, T. Yamazhan, S. Ulusoy (Izmir, TR)
Objective: In this study it was aimed to evaluate the resistance patterns of microbiologically confirmed nosocomial bacteraemia (MCNB) related S. aureus strains between 2001–2005 retrospectively.
Method: Any patient in whom S. aureus was isolated in at least one set of blood cultures (Sent to the bacteriology laboratory 72 h after hospital admission) was considered to have MCNB. Data of antibacterial resistance and hospital admission dates were extracted from hospital patient record database. Double or more isolates during each episode were counted as one episode. Resistance patterns in the 2001–2002 and 2004–2005 periods were compared by Chi-square test. 2003 data was excluded to see the probable effect governmental antibiotic restriction policy, which was started in March 2003. Blood cultures were performed on Bact/Alert (bioMérieux, Durham, NC). Bacterial identifications were performed by automated API (bioMérieux, Durham, NC). Oxoid antibiotic dics (England) were used to test antibacterial susceptibility by disc diffusion method following the recommendations of CLSI. Results were interpreted as described by CLSI.
Results: Oxacillin resistance in 2001 was 73.8% and 55.2% in 2005. When 2001–2002 and 2004–2005 periods were compared, resistance to oxacillin, levofloxacin, gentamicin, erythromycin and clindamycin decreased significantly (p < 0.05) (Table 1 ). There was no glycopeptide resistance.
Table 1.
Resistance patterns of S. aureus strains and comparison of resistance patterns in 2001–2002 and 2004–2005 periods
| 2001 | 2002 | 2003 | 2004 | 2005 | 2001–2002 | 2004–2005 | P | |
|---|---|---|---|---|---|---|---|---|
| Methicillin | 73.8% (186/252) | 70.5% (124/177) | 76.7% (89/116) | 63.2% (119/188) | 55.2% (115/208) | 72.2% (310/429) | 59.0% (234/396) | <0.001 |
| Levofloxacin | 76.1% (118/155) | 76.2% (93/122) | 64.4% (67/104) | 57.5% (99/172) | 51.4% (106/206) | 76.1% (211/277) | 54.2% (205/378) | <0.001 |
| Gentamicin | 61.4% (153/249) | 61.0% (108/177) | 69.5% (80/115) | 54.2% (102/188) | 49.0% (100/208) | 61.2% (261/426) | 51.0% (202/396) | <0.001 |
| Erythromycin | 66.9% (168/251) | 61.0% (108/177) | 62.9% (73/116) | 48.4% (91/188) | 33.6% (70/208) | 64.4% (276/428) | 40.6% (161/396) | <0.001 |
| Clindamycin | 43.4% (109/251) | 36.7% (65/177) | 50.8% (59/116) | 37.9% (71/187) | 28.8% (60/208) | 59.3% (254/428) | 33.0% (131/396) | <0.001 |
| Penicillin | 92.8% (233/251) | 93.2% (165/177) | 96.5% (112/116) | 93.0% (175/188) | 92.3% (192/208) | 92.9% (398/428) | 92.6% (367/396) | NS |
Conclusion: Decrease in the resistance rates after the 2003 budget application suggests that the application is useful not only in decreasing the cost of antibiotics but the antimicrobial resistance in several bacteria.
P1818 Emergence of linezolid-resistant methicillin-resistant Staphylococcus aureus in Japan
H. Hanaki, C. Yanagisawa, Y. Hososaka, K. Ishibashi, S. Yamamoto, Y. Imamura, S. Taniwaki, T. Nakae, K. Sunakawa (Tokyo, Hukuoka, JP)
Objectives: The linezolid (LZD)-resistant methicillin-resistant Stapylococcus aureus (MRSA) has been reported from USA (8 cases), UK (1), and Brazil (1) yet not from Japan. We isolated the LZD-resistant MRSA from a case of septicaemia patient in June 2006 only four months after LZD had been introduced to clinical use in Japan. Five strains of LZD-resistant MRSA were isolated from the same patient and they were characterised by means of bacteriology and DNA technology.
Methods: Minimum inhibitory concentration (MIC) of antimicrobial agent was determined according to the guideline of Clinical and Laboratory Standard Institutes. Occurrence of the genes encoding staphylococcal cassette chromosome mec (SCC mec), hemolysin (hla, hlb), exofoliative toxin (eta, etb) enterotoxin (sea, seb), toxic shock syndrome toxin-1 (tss, TSST-1), panton-valentain leukocidin (pvl, PVL) and β-lactamase (blaZ) were examined by polymerase chain reaction (PCR). Identity of the strains was verified by pulsed-field gel electrophoresis (PFGE) of the SmaI treated DNA.
Results: MIC of oxacillin, imipenem, pazufloxacin, vancomycin, teicoplanin, arbekacin, minocycline, rifampin, sulfametoxazole/torimetoprim was >128, 64, 1, 1, 4, 16, ≤0.25, and 2 μg/mL, respectively, and that was identical in all five strains. MIC of LZD was highest in the latest isolates of MRSA, 32 μg/mL, though that in all the LZD-unexposed MRSA was 2 μg/mL. SCCmec in all five strains isolated from the patient was type II, which is common in the MRSA in Japan. Chromosomal DNA encoding enterotoxin, heamolysine, and β-lactamase was detected in all the strains, but that was undetectable encoding TSST-1, exofoliative toxin and panton-valentine leukocidin. PFGE of SmaI-treated DNA in all the LZD-resistant MRSAs showed identical DNA profile suggesting the identical origin for all. Only the difference from the DNA profile of the LZD-susceptible strains was that 120 bp band was shifted to 160 bp in the resistant strain.
Conclusion: LZD was licensed in clinical use in Japan since April 2006 as an anti-MRSA drug, and the LZD-resistant MRSA emerged only four months later. LZD was intermittently administered intravenously or orally for 48 days to the patient with MRSA septicaemia, and the LZD-resistant MRSA was isolated from blood on the day the patient passed away. We reported a very first case of LZD-resistant MRSA in Japan and called for precaution for proper use of this powerful antimicrobial agent.
P1819 Heterogeneous vancomycin intermediate resistance within methicillin-resistant Staphylococcus aureus clinical isolates in Alexandria province, Egypt
N. Rizk, S.A. Zaki (Alexandria, EG)
Objectives: To explore the prevalence of heterogeneous vancomycin intermediate resistant Staphylococcus aureus (hVISA) among methicillin resistant S. aureus (MRSA) strains isolated from hospitalised patients in Alexandria University Hospital over a 2 years period, and to investigate the mechanism of vancomycin resistance.
Methods: Sixty two MRSA isolates were screened by using brain heart infusion agar supplemented with 4 ug/mL vancomycin (BHI-V4) and macro Etest. Minimum inhibitory concentrations (MICs) of vancomycin was determined by broth microdilution and standard Etest. Population analysis profile (PAP) was performed for detecting the frequency of heterogeneous resistance for isolates grown on BHI-V4. Vancomycin intermediate resistant subpopulations of hVISA were viewed with scanning electron microscopy, and tested for the presence of vanA gene by PCR.
Results: Twenty one (33.87%) MRSA isolates grew on BHI-V4, 7 (11.29%) isolates were suspected of having reduced susceptibility to vancomycin by the macro Etest. PAP confirmed 6 (9.68%) isolates as hVISA since they produced subpopulations with MIC of vancomycin of more than 4ug/mL at frequency of 1 in 106 cfu/mL or higher. Vancomycin MIC values for all isolates were ≤ 4ug/mL. Electron microscopy of vancomycin intermediate resistant subpopulations of hVISA showed enhanced cell wall thickness with evidence of increased extra-cellular material and irregular shape compared to vancomycin susceptible cells. All hVISA isolates were vanA gene negative by PCR.
Conclusion: This study is an early warning that MRSA strains with full resistance to vancomycin might emerge in Egypt in the future. Due to the increased use of vancomycin for treatment of MRSA infections, screening for hVISA in MRSA strains should be considered as a necessary part of infection control practice emphasizing the importance of a laboratory capability of its identification.
P1820 High prevalence of inducible clindamycin resistance among community-associated methicillin resistant Staphylococcus aureus in Europe
D. Farrell, J. Shackloth, L. Cossins, A. Quintana, P. Hogan (London, UK; New York, US)
Background: Community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) is known to cause serious skin and soft structure infections (SSTI) and is increasingly reported in Europe. Although CA-MRSA are often susceptible to many antibiotic classes, clindamycin (CLIN) is often the preferred antibiotic because of previously demonstrated clinical efficacy. CLIN resistance is underestimated because testing for inducible clindamycin resistance (ICR) is not performed routinely in many clinical laboratories. The aim of this study was to assess the prevalence of ICR in CA-MRSA from Europe.
Methods: A total of 750 clinically significant S. aureus from the UK (n = 163), Germany (n = 187), Italy (n = 139), Spain (n = 133), and France (n = 128) were selected and were collected either in 2003–2005 (n = 522) or 1994 (n = 228). Isolates were classified as CA-MRSA, healthcare associated MRSA (HA-MRSA), or MSSA based on antibiotic susceptibility patterns. MICs were determined and interpreted according to CLSI methodology. ICR was determined by the double disc diffusion method.
Results: ICR was prevalent in both 1994 and 2003–2005. All 750 isolates were 100% susceptible to vancomycin, teicoplanin and linezolid.
Erythromycin resistant isolates
| N | Ery-R |
% CLIN-S | % ICR | ||
|---|---|---|---|---|---|
| n | % | ||||
| CA-MRSA | 227 | 108/227 | 48 | 95.4 | 38 |
| HA-MRSA | 360 | 360/360 | 100 | 7.8 | 6.7 |
| MSSA | 163 | 22/163 | 13.5 | 63.6 | 63.6 |
Conclusions: ICR prevalence was very high in macrolide resistant CA-MRSA (and MSSA) in 5 European countries suggesting that empirical treatment with CLIN for serious SSTI may not be appropriate. Because recent research provides evidence that, as with CLIN, linezolid also suppresses toxin production in S. aureus whilst vancomycin and teicoplanin do not, linezolid may be a more suitable empiric therapeutic option to consider for serious SSTI in Europe until ICR is excluded. This data also suggests that testing for ICR should be routinely performed in Europe.
P1821 Staphylococcus aureus bacteraemia in the south-east of Ireland 2002–2006
K. Burns, S. Fitzgerald, D. O'Brien, E. O'Neill, D. Keady, A. Moloney, M. Hickey (Waterford, IE)
Background and Objectives: There are 1236 acute hospital beds in the southeast of Ireland. During the period from 1998 to 2001, our laboratory surveillance recorded an annual increase in the number of cases of S. aureus bacteraemia. A decision was made to establish a database and prospectively record clinical information and outcome on all cases of S. aureus bacteraemia commencing in July 2002 for a four-year period.
Methods: Each patient with a blood culture positive for S. aureus was included in a database. We recorded demographic and clinical information, source of bacteraemia, laboratory findings, clinical outcome, whether or not there was evidence of relapse, reinfection and whether complications arose following each infection. Only patients with genuine S. aureus bacteraemia were included in the database. Centre for disease control definitions of infection were utilised in this study. Bacteraemia was defined as either community or hospital-acquired based on recognized definitions.
Results: There were 317 cases of S. aureus bacteraemia over the forty-eight months of the study. 182 cases were due to methicillin-sensitive S. aureus (MSSA). Age range was 3 days to 97 years with a mean age of 67 years. 59% of patients were male. We recorded 261 cases of hospital-acquired infection and 56 cases of community-acquired infection. There were no cases of bacteraemia due to community-acquired methicillin-resistant S. aureus (MRSA) in our study. The underlying source of each episode of bacteraemia was recorded as follows:
-
–
33% secondary to infection of central venous catheter
-
–
15% due to infection of peripheral vascular catheter
-
–
9% due to skin or soft tissue infection
-
–
4.5% due to septic arthritis
-
–
4% due to pneumonia
-
–
3% due to surgical site infection
-
–
3% due to urinary tract infection
In 20% of cases, the underlying source of infection was not uncovered despite investigation or because a patient was unfit for invasive investigation.
62% of patients recovered fully without complications. 25% of patients died. For 22% of total deaths, S. aureus was the underlying cause of death. In 10% of cases, complications arose secondary to the infection. In 3% of cases, the outcome was unknown.
Conclusion: In our patient population, 47% of all cases of S. aureus bacteraemia were due to infection of either a central or peripheral venous catheter. We have targeted measures to prevent catheter-related bloodstream infection. We hope to reduce the incidence of S. aureus bacteraemia in our region in the future.
P1822 Resistance pattern of community-acquired S. aureus infections in children
T. Panagea, G. Ikonomopoulos, A. Makri, A. Vatopoulos, P. Koukou, A. Voyatzi (Athens, GR)
Introduction: Community-acquired (CA) methicillin-resistant S. aureus (MRSA) infections in healthy children and young adults are a recent worldwide phenomenon. Most CA-MRSA strains are producing Panton Valentine Leukocidin (PVL), a highly potent toxin, associated with skin lesions, soft tissue infections and necrotising pneumoniae.
Objectives: The purpose was to determine the incidence, the antibiotic resistance pattern and the prevalence of PVL producing S. aureus isolates in children suffering from skin or soft tissue infections in the community.
Material and Methods: A total of 285 strains S. aureus were isolated from 960 samples obtained from children aged 0–14 years old, during the last 2 years (October 2004–October 2006). All children suffered from skin or soft tissue infections and attended the outpatient department for medical advice. Samples were obtained from skin lesions (520), abscesses (290), wounds (120), and umbilical swabs (30).
Identification of S. aureus was achieved by conventional methods. Antibiotic susceptibility testing was performed according to the CLSI guidelines. Twenty randomly selected CA-MRSA strains were further examined by PCR technique for the presence of lukF-PV and lukS-PV genes.
Results: S. aureus was responsible for 29.7% of community acquired skin and soft tissue infections in children. Antibiotics resistance pattern was as follow: penicillin 91%, methicillin 22.9%, fusidic acid 19%, tetracycline 23.6%, clindamycin 2.3%, gentamicin 0.9%, erythromycin 13.1%, rifampicin 0.9%, trimethoprim-sulfamethoxazole 0.9%, chloramphenicol 1.2% and ciprofloxacin 0.9%. None of the strains was resistant to vancomycin, teicoplanin and amikacin. MRSA accounted for 22.9% of all CA-S. aureus infections. Out of 65 MRSA strains, 48 (73.8%) were also resistant to fusidic acid and tetracycline. The genes lukF-PV and lukS-PV were detected in 18 out of 20 (90%) CA-MRSA strains.
Conclusion: In our paediatric population, 22.9% CA-MRSA infections were observed. Ninety percent of examined CA-MRSA isolates were found to be PVL producers. A significant number of MRSA isolates (73.8%) were resistant to fusidic acid and tetracycline. This unusual antibiotic resistance pattern has been associated with the presence of PVL producing ST80 CA-MRSA clone, that has previously been described in Greece.
P1823 The impact of multiple-resistant Staphylococcus haemolyticus in intensive care units – acquired infection
K. Kristóf, S. Kardos, V. Cser, É. Volter, K. Nagy, F. Rozgonyi (Budapest, HU)
Objectives: The aims of the study was to investigate the susceptibility to antistaphylococcal agents and the presence of genes encoding adhesins of S. haemolyticus strains isolated from invasive infections between January 2001 and December of 2005.
Methods: Identification of S. haemolyticus strains was performed by the API Staph 32 (bioMérieux). Disc diffusion test, minimum inhibitory concentration (MIC) of antibiotics and analysis of heteroresistance to glycopeptides was carried out according to the NCCLS/CLSI guidelines. Oxacillin resistance was confirmed showing the presence of mecA by PCR technique. The icaA, icaC, icaD, icaR, fbnA, fbnB genes responsible for adhesion were detected by PCR.
Results: S. haemolyticus is the second (after S. epidermidis) most frequently isolated coagulase negative Staphylococcus from clinically verified bacteraemias in patients treated at the intensive care units of our University. During the investigation period 356 strains were isolated from blood cultures amounting to 5.8% of all positive cases. Most of the strains were methicillin resistant (94%). The MIC90 was 512 mg/L. Each of them had the mecA gene. 98%, 96%, 57%, and 41% of the strains were resistant to gentamicin, tobramycin, netilmicin and amikacin, MIC90 for them were 256 mg/L, 256 mg/L, 128 mg/L and 64 mg/L respectively. Only 12% of the strains were sensitive to fluoroquinolones, ciprofloxacin, levofloxacin and moxifloxacin. All strains were sensitive to vancomycin. The rate of heteroresistant strain to teicoplanin was 79% (MIC90 = 16 mg/L). Concerning genes for adhesins icaC was present in 8.6% and icaD in 1.2% of the strains. None of the strains contained genes for fibronectin binding proteins.
Conclusions: The proportion of multiresistant S. haemolyticus strains in blood stream infections of patients treated at the ICU was relatively high. The quantitative antibiotic resistance of these strains was extraordinary high. Vancomycin is the only choice of the treatment. Intercellular polysaccharide adhesins and fibronectin binding protein genes seem not to be responsible for the attachment of these strains to indwelling devices. Supported by OTKA No.: T 46186
P1824 Present situation of antimicrobial resistance of Staphylococcus in Spain: Sixth Nationwide Prevalence Study and the in vitro activity of new antimicrobial agents
O. Cuevas, E. Cercenado, M. Goyanes, M. Marin, A. Vindel, E. Bouza (Madrid, ES)
Objective: Data regarding the evolution of Staphylococcus resistance in a whole country have a definite influence in the design of empirical treatment regimens. For the last 20 years we have been performing prevalence studies in order to ascertain the situation of the antimicrobial resistance of Staphylococcus in our country. In this study we present the results of the sixth point prevalence study performed in 2006.
Methods: In a selected day of October 2006, we collected all staphylococci isolated in 145 Spanish hospitals. All microorganisms were sent to a reference laboratory where identification and antimicrobial susceptibility testing was performed against 15 antimicrobial agents using and automated microdilution method (MicroScan). Additional E-test susceptibility testing was performed for the new antimicrobials daptomycin and tigecycline. We present the data of 75 hospitals, and 260/223 S. aureus/coagulase negative staphylococci (CoNS) isolates analysed to date and compare the results with these of the fifth study performed in 2002.
Results: The percentages of resistance of S. aureus/CoNS against selected antimicrobials (PEN = penicilin, OXA = oxacillin, ERY = erythromycin, CLI = clindamycin, GEN = gentamicin, TOB = tobramycin, CIP = ciprofloxacin, RIF = rifampin, T/S = trimethoprim/sulfamethoxazole, VAN = vancomycin), are summarised in the table . Resistance to linezolid, daptomycin, and vancomycin was found among CoNS (one isolate each). All strains were susceptible to tigecycline. The most frequent phenotypes of resistance among methicillin-resistant S. aureus were: ERY+CIP+TOB (75%); ERY+CIP+TOB+CLI (11%); ERY+CIP+TOB+CLI+GEN (14%). Fenotype M was present in 40% of isolates.
Percentage of resistance of S. aureus/CoNS to selected antimicrobials
| Year | Resistance (%),S. aureus/CoNS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PEN | OXA | ERY | CLI | GEN | TOB | CIP | RIF | T/S | VAN | |
| 2002 | 88/79 | 31/63 | 33/64 | 20/35 | 18/35 | ND/ND | 37/50 | 2/7 | 2/22 | 0/0 |
| 2006 | 88/90 | 28/70 | 32/65 | 18/44 | 10/43 | 26/50 | 37/53 | 1/6 | 0/30 | 0/0.5 |
Conclusions: The resistance of S. aureus to methicillin in Spain seems to be stabilised (28% in 2006 versus 31% in 2002), and strains are more susceptible to other antimicrobials, mainly to gentamicin and clindamycin. However, resistance of CoNS to all antimicrobials is increasing and emerging resistance to the new antimicrobial agents has been detected
P1825 Antimicrobial resistance of S. aureus strains isolated in 2005 from nosocomial infections in Romania
C.C. Dragomirescu, I. Codita, M. Oprea (Bucharest, RO)
Objectives: obtain information on antimicrobial resistance patterns of nosocomial S. aureus (S.a.) strains; provide feed-back to participant laboratories; build a national data base for further action.
Methods: Nosocomial infections case definitions recommended by the National Centre for Prevention and Control of Communicable Diseases; Identification of S.a. by tube coagulase test and APIStaph; disc difusion AST cf. CLSI (formerly NCCLS) standard M100-S15; confirmation of methicillin resistance by cefoxitin disc difusion and oxacillin MIC; vancomycin susceptibility by testing for MIC; statistical analysis only when studied cathegory numbered >30 items.
Results:
-
–
6 from 17 sentinel hospitals in the National Nosocomial Infections Sentinel Surveillance System sent 1 to 34 S. aureus strains; the total number of S.a. strains was 84.
-
–
78% of S.a. strains were MRSA.
-
–
Distribution by specimen type of MRSA strains: 18 blood, 27 wound, 7 catheter, 5 umbilical cord and 8 other sites isolates.
-
–
Distribution of all strains by oxacillin MIC's was trimodal, with peaks at: 4 mg/L (heteroresistant strains), 64 mg/L and 512 mg/L
-
–
86% of MRSA strains harboured a KTG aminoglycoside phenotype
-
–
MRSA strains were co-resistant to other antimicrobials: 90.76% to erythromycin; 52.30% to ciprofloxacin, 50.76% to rifampicin, 9.23% to trimethoprim-sulfamethoxazole and 6.15% to chloramphenicol.
-
–
It was not possible to make separate analysis by hospital, but for 3 hospitals. Proportion of MRSA, MLSBi strains, distribution of oxacillin MIC's varied between these hospitals, as did KTG phenotype, ciprofloxacin and rifampicin resistance proportions.
Conclusions:
-
1
Functioning of the sentinel system in the first year yielded to heterogenous participation of hospitals expressed in heterogeneous composition of S.a. strains sample received at the national laboratory.
-
2
Only 3 from 17 hospitals sent a sufficient number of S.a. to be analysed for the local level.
-
3
Differences were noticed between the 3 hospitals as to the MRSA proportions, distribution of oxacillin MIC's, MLSBi strains, KTG phenotype, ciprofloxacin and rifampicin resistance proportions. These differences suggest different ways to implement antimicrobials use policies.
-
4
Taking precaution on the limited number of strains, these very first data could play a cathalist role to continue the surveillance at the local, county and national level in order to formulate and adopt antimicrobial resistance use policies.
P1826 Investigation of erythromycin-resistant staphylococci from the skin of Egyptian acne patients
T.S.M. El-Mahdy, M.M. Abd-Elaziz, M.H. El-Sayed, J.I. Ross, S. Abd-Allah, H. Radwan, A.M. Snelling (Bradford, UK; Cairo, Ismailia, EG)
Objectives: Long courses of antibiotics remain the mainstay of acne treatment. Whilst Propionibacterium acnes is being targeted, selective pressure is also exerted on other members of the commensal skin flora, including coagulase negative staphylococci (CNS) such as Staphylococcus epidermidis. These bacteria may then act as reservoirs of resistance genes for more pathogenic strains or species. Erythromycin (ERY) is widely used in Egypt to treat acne, and available over the counter, but it is not known how this is affecting the commensal flora. This study sought to determine the prevalence of ERY resistant CNS, and the genes responsible, from acne patients attending clinics in Cairo.
Methods: Fifty-two acne patients, 13 dermatology staff and 36 controls were studied. Facial skin swabs were taken, and inoculated onto Mueller Hinton agar containing 4 mg/L ERY (breakpoint concentration). Plates were incubated at 35°C for 48 hrs. ERY resistant staphylococci were identified by standard tests. MIC profiles for other drugs commonly used in dermatology were determined using CLSI criteria. Resistance determinants were identified by PCR using primer pairs specific for erm(A), erm(B), erm(C), and msr(A).
Results: The prevalence of ERY resistant CNS amongst patients, clinic staff, and controls was 80.8%, 69.2%, and 66.7%, respectively. These differences were not significant (p > 0.05) and patients on current or very recent antibiotic treatment were no more likely than those using other or no medication to carry ERY-R strains. A total of 42 ERY-R strains were isolated from 42 different patients. 54.5% of these had high level resistance (MIC > 1024 mg/L). The most widely-distributed erythromycin resistance determinants were erm(C) alone (42.9% strains), msr(A) (40.5%) or both together (16.7%). Erm(A) and erm(B) were not detected. Amongst the 42 strains, cross-resistance to tetracycline (64.3%), clindamycin (9.5%), or fusidic acid (45.2%) was detected.
Conclusions: Although high (80.8%) carriage of ERY resistant CNS amongst Egyptian acne patients is comparable to that reported from Western countries, and was not significantly different from carriage amongst the non-patients. Prevalence of msr(A) (>half strains tested) was much higher than expected, and the driver for this remains to be elucidated. Carriage of CNS with multiple resistances to drugs used to treat dermatological conditions was common, and is of concern for treatment of staphylococcal skin infections in Egypt.
P1827 Prevalence of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in Belgian hospitals: results of a multicentre survey
Y. Glupczynski, C. Behrin, D. Allemeersch, M. Andre, H. De Beenhouwer, R. Cartuyvels, J. Frans, D. Govaerts, R. Joseph, I. Phillipart, I. Surmont, A-M. Van den Abeele, J. Verhaegen, C. Nonhoff, O. Denis, M. Struelens (Yvoir, Brussels, BE)
Objectives: MRSA isolates with decreased susceptibility to glycopeptides (GISA/h-GISA) have been reported worldwide. Several studies have shown that such strains could be associated with a poor clinical outcome. The aim of the present study was to determine the prevalence of GISA/h-GISA strains among Belgian MRSA isolates collected from hospitalised patients.
Methods: Consecutive non duplicate MRSA clinical isolates prospectively collected between 01/2005 and 07/2005 in 13 hospitals were screened locally for reduced susceptibility to glycopeptides by the modified Macromethod Etest according to Walsh et al. (JCM 2001) and by teicoplanin (5 mg/L) agar screen (TAS). Etest MICs of both vancomycin (VA) and teicoplanin (TP) ≥ 8 mg/L or of TP alone >12 mg/L and growth on TAS were indicative of potential GISA/h-GISA or of hetero-teicoplanin intermediate S. aureus (h-TISA). All suspected strains were sent to the reference laboratory for confirmation by Etest MIC determination on MH agar as well as by VA and TP population analysis profiles (PAP). Confirmed GISA/h-GISA isolates were also analysed by PFGE in order to delineate their clonality.
Results: Of 573 MRSA strains tested (mean number of isolates by centre: 44; range: 30–54), 31 (5.4%) met the phenotype screen criteria of GISA/h-GISA. Putative GISA strains were found in 10 of the 13 participating labs and their frequency by centre ranged between 0 and 13%. 24 isolates (4.2%) were confirmed as h-TISA and 2 (0.3%) as GISA or h-GISA by MIC testing (VA MIC of 4 and 8 mg/L) and by PAPs. Origins of the isolates were: lower respiratory tract (11), wounds (7), blood (4) and urine (2). Fourteen of the 24 h-TISA strains were resistant to gentamicin by disk diffusion. By molecular analysis, all h-TISA isolates clustered in two epidemic PFGE groups (A and D) which were previously reported in h-GISA or in h-TISA isolates in Belgium. The two GISA strains were clearly unrelated to each other and to the h-TISA strains. Dissemination of h-TISA isolates belonging to the same clone resulted in small local outbreaks in six of the centres.
Conclusion: Overall GISA/h-GISA isolates were found in 4.5% of the Belgian MRSA isolates from hospitalised patients. Most of the isolates had a h-TISA phenotype. Although uncommon, the occurrence of GISA/h-GISA among MRSA strains is higher than reported previously in other surveys in Belgium and could possibly be attributed to the spread of certain epidemic clones within and across hospitals.
Resistance surveillance in Gram-positives
P1828 Update on daptomycin activity and spectrum when tested against Gram-positive strains collected in European medical centres (2006)
H. Sader, P. Rhomberg, T. Fritsche, R. Jones (North Liberty, US)
Objective: As part of the Daptomycin Surveillance Program, we evaluated the in vitro activity of daptomycin against recent clinical isolates collected in Europe (2006). Daptomycin is a novel cyclic lipopeptide recently approved by European Medicines Agency (EMEA) for the treatment of complicated skin and soft structure infections (cSSSI).
Methods: A total of 2,907 consecutive strains were collected in 24 medical centres located in nine European countries, Turkey and Israel. The organisms were isolated mainly from bloodstream infections (52%) and cSSSI (18%). The following pathogens were evaluated: S. aureus (SA; 31% oxacillin [OXA]-resistant [R]); coagulase-negative staphylococci (CoNS; 75% OXA-R), E. faecalis (EF; 1% vancomycin [VAN]-R), E. faecium (EFM; 14% VAN-R), beta-haemolytic Streptococcus spp. (BHS; 235), and viridans group Streptococcus spp. (VGS; 119). The strains were susceptibility (S) tested by broth microdilution methods in cation-adjusted Mueller-Hinton broth, additionally supplemented to 50 mg/L of calcium for daptomycin tests. Numerous comparators were also tested.
Results: Daptomycin activity is summarised in the table . Daptomycin was highly active against SA and CoNS (MIC90, 0.5 mg/L) and its activity was not adversely affected by resistance to OXA or VAN. All staphylococcal strains were inhibited at the daptomycin S breakpoint of ≤1 mg/L. Daptomycin and linezolid were the only compounds active against all enterococci. Daptomycin was highly active against BHS (MIC90, 0.25 mg/L) as were most comparison agents tested. Daptomycin was also very active against VGS (MIC90, 0.5 mg/L).
| Organism (no. tested) | Cumulative % inhibited at daptomycin M IC (mg/L) of: |
%Sa | ||||
|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | ||
| SA OXA-S (1,014) | 88.4 | 99.5 | 100.0 | − | − | 100.0 |
| OXA-P (465) | 75.7 | 99.4 | 100.0 | − | − | 100.0 |
| CoNS OXA-S (132) | 72.0 | 96.2 | 100.0 | − | − | 100.0 |
| OXA-R (388) | 68.3 | 98.7 | 100.0 | − | − | 100.0 |
| Enterococcus spp. | ||||||
| VAN-S (526) | 4.0 | 44.6 | 87.1 | 99.4 | 100.0 | 100.0 |
| VAN-R (28) | 7.1 | 14.3 | 71.4 | 100.0 | − | 100.0 |
| BHS (235) | 100.0 | − | − | − | − | 100.0 |
| VGS (119) | 75.8 | 97.5 | 100.0 | − | − | 100.0 |
US-FDA/CLSI interpretive criteria.
Conclusions: Daptomycin showed significant potency and broad-spectrum activity against recent clinical isolates of Gram-positive organisms isolated in European medical centres, including MDR subsets. All organisms tested were S to daptomycin and R to other compounds did not adversely influence the daptomycin potency against staphylococci, enterococci or streptococci.
P1829 Activity of daptomycin and selected antimicrobial agents tested against Gram-positive organisms isolated from European patients with complicated skin and skin structure infections
H. Sader, T. Fritsche, P. Strabala, R. Jones (North Liberty, US)
Objective: To evaluated the activity of daptomycin tested against Gram-positive organisms isolated from patients with complicated skin and skin structure infections (cSSSI) hospitalised in European hospitals in the last 4 years (2003–2006). Daptomycin has been used for the treatment of cSSSI in the United States since October 2003 and was recently approved for clinical use in European countries by the European Medicines Agency (EMEA).
Methods: A total of 2,725 bacterial strains causing cSSSI were collected from 24 medical centres located in 12 European countries, Turkey and Israel. The strains were susceptibility (S) tested against daptomycin and numerous comparator agents by reference broth microdilution methods performed according to CLSI documents (M7-A7; M100-S16). Test medium was supplemented with calcium (50 mg/L) for testing daptomycin only. All quality control results were within published CLSI ranges.
Results: Daptomycin and selected comparator activities (vancomycin [VAN], quinupristin/dalfopristin [Q/D], linezolid [LZD] and levofloxacin [LEVO]) are summarised in the table . Rates of oxacillin-resistant S. aureus (MRSA) varied widely from 0.7% in Sweden to more than 40% in Greece (41.0%) and Ireland (47.8%). Vancomycin-resistant Enterococcus (VRE) was observed in Germany, Greece, Ireland, Italy, Turkey and the United Kingdom, and prevalence remains low in most European countries relative to the USA. Daptomycin was active against all strains at the S breakpoint and its activity remained stable over the 4 years evaluated. Daptomycin and LZD showed the broadest spectrum of activity (100.0% S) among the antimicrobials tested, but daptomycin was generally more potent (lower MIC50 and MIC90) than LZD. Non-S to Q/D has increased significantly among E. faecium (42.2%; 27.5% at 2 mg/L) and also emerged among staphylococci (0.4–0.5%). LEVO showed limited activity against staphylococci and enterococci. Resistance (R) to other antimicrobial classes did not adversely affect daptomycin activity against staphylococci, enterococci or streptococci. Daptomycin was very active against ORSA (MIC50, 0.25 mg/L) and VAN-R E. faecium (MIC50, 2 mg/L).
| Organism (no.) | MIC90 (mg/L)/% S |
||||
|---|---|---|---|---|---|
| Daptomycin | VAN | Q/D | LZD | LEVO | |
| S. aureus (1,878) | 0.5/100.0 | 1/100 | 0.5/99.5 | 2/100.0 | >4/75.5 |
| Coag.-negative staphylococci (235) | 0.5/100.0 | 2/100 | 0.5/99.6 | 1/100.0 | >4/56.2 |
| E. faecalis (198) | 1/100.0 | 2/98.0 | >2/0.0 | 2/100.0 | >4/61.1 |
| E. faecium (51) | 4/100.0 | 1/90.2 | >2/58.8 | 2/100.0 | >4/27.5 |
| Beta-haemolytic streptococci (306) | 0.25/100.0 | 0.5/100.0 | 0.5/100.0 | 1/100.0 | 1/100.0 |
| Viridans group streptococci (38) | 0.5/100.0 | 1/100.0 | 1/100.0 | 2/100.0 | 1/97.4 |
Conclusions: Daptomycin was highly active (100.0% S) against the most clinically important Gram-positive pathogens causing cSSSI in European medical centres, including multi-drug resistant organisms. Continued wide geographic monitoring would be preferred to detect emerging daptomycin-R strains, especially among strains with compromised VAN activity.
P1830 Susceptibility of Gram-positive pathogens to daptomycin and other antimicrobial agents: first results from the DAPTOGERM Surveillance Study
M. Kresken, J. Brauers (Rheinbach, DE)
Objectives: Drug resistance in Gram-positive bacteria has become an increasing problem over the past 20 years. New antibacterial agents such as the lipopeptide daptomycin (DPC), which was recently approved in the EU, have been developed to address this problem. Shortly after the approval, a surveillance study was started by a network of clinical microbiology laboratories to monitor the susceptibility of Gram-positive pathogens to DPC in Germany. This report comprises the results of isolates of five frequently encountered Gram-positive species collected in 49 laboratories during the time period from March to June 2006.
Methods: A total of 1,520 isolates including oxacillin-susceptible Staphylococcus aureus (MSSA, n = 360), oxacillin-resistant S. aureus (MRSA, n = 286), Staphylococcus epidermidis (Se, n = 262), Enterococcus faecalis (Es, n = 209), Enterococcus faecium (Em, n = 208), and Streptococcus pyogenes (Sp, n = 195) were tested against DPC, linezolid (LZD), vancomycin (VAN) and other drugs. First isolates obtained from hospitalised patients with skin and soft tissue infections, respiratory tract infections, foreign body/catheter infections, or sepsis were included. MICs were determined in a central laboratory using the broth microdilution procedure according to the standard of the German DIN (Deutsches Institut für Normung) 58940 guidelines. MICs of DPC were interpreted by the EUCAST criteria.
Results: The majority of isolates was recovered from wound swabs (57.7%), blood samples (21.9%) and respiratory tract specimens (8.2%). MIC50/90 values of DPC for MSSA, MRSA, and Se each were 0.5/1 mg/L. All strains were inhibited by DPC at the EUCAST breakpoint of ≤1 mg/L. LZD and VAN were also active against all staphylococci. However, based on MIC50/90 values, DPC was up to 4 fold more active than LZD or VAN. Of the Em isolates, 23 (10.9%) were resistant to VAN. In contrast, none of the Ef isolates exhibited resistance to VAN. DPC inhibited all strains at 4 mg/L. MIC50/90 values of DPC for either enterococcal species were 2/4 mg/L. One Em strain was resistant to LZD (MIC 32 mg/L). DPC was more active than LZD or VAN against Sp. MIC50/90 values were 0.125/0.25 for DPC compared to 0.25/0.5 and 1/1 for VAN and LZD, respectively. The highest MICs of DPC for Sp isolates were 0.5 mg/L.
Conclusion: DPC demonstrated excellent in vitro activity against frequently encountered Gram-positive species including multi-resistant isolates such as MRSA and VRE. Resistent strains were not detected.
P1831 Comparative in vitro activity of tigecycline against staphylococci and enterococci from patients on ICU- and non-ICU wards: results of the German T.E.S.T. Surveillance Program 2005
M. Kresken, J. Brauers, H. Geiss, E. Halle, E. Leitner, G. Peters, H. Seifert on behalf of the German T.E.S.T. Surveillance Program
Objectives: Tigecycline (TGC), the first glycylcycline antibacterial agent, has been shown to be highly effective against a wide range of bacteria including methicillin (oxacillin)-resistant staphylococci and vancomycin-resistant enterococci. G.-T.E.S.T. is a surveillance programme comprising 15 German laboratories which monitors the susceptibility of bacterial pathogens to TGC. The objective of this study was to evaluate the in vitro activity of TGC against both ICU- and non-ICU isolates of four clinically important Gram-positive pathogens, namely Enterococcus faecalis (Es), Enterococcus faecium (Em), Staphylococcus aureus (Sa) and Staphylococcus epidermidis (Se).
Methods: A total of 271 ICU isolates (52 Ef, 67 Em, 77 Sa, 75 Se) and 452 non-ICU isolates (88 Ef, 69 Em, 208 Sa, 87 Se) were tested against TGC, doxycycline (DOX), oxacillin (OXA), moxifloxacin (MXF), gentamicin (GEN), linezolid (LZD), vancomycin (VAN) and other drugs. MICs were determined by broth microdilution according to German DIN guidelines in a central laboratory. The MICs of TGC and LZD were interpreted by EUCAST criteria. DIN breakpoints were applied to the other drugs.
Results: The rates of OXA resistance in ICU/non-ICU isolates of Sa and Se were 61/57% and 81/74%, respectively. The susceptibility rates of ICU/non-ICU isolates to MXF and GEN were 47/45% and 81/87% for Sa compared to 57/60% and 32/39% for Se. In contrast, all staphylococci were susceptible to VAN and LZD. TGC exhibited excellent in vitro activity against all staphylococci including OXA-resistant isolates. It was equally active against ICU- and non-ICU isolates. Only one OXA-resistant Sa strain isolated from an ICU patient was resistant to TGC (MIC 1 mg/L). Compared to DOX, MIC90s of TGC were 16- and 2fold lower for ICU- and non-ICU isolates of Sa, respectively, and 8fold lower for both ICU- and non-ICU isolates of Se. TCG also exhibited excellent activity against enterococci. All strains were inhibited by TGC at 0.25 mg/L. In contrast, 7% and 14% of the ICU/non-ICU Em isolates were resistant to VAN. High-level resistance to GEN in ICU/non-ICU isolates of Ef and Em ranged from 30 to 54%.
Conclusion: TGC demonstrated excellent in vitro activity against staphylococci and enterococci isolated from both ICU- and non-ICU patients. TGC seems to be a useful drug for the treatment of infections caused by multiple resistant enterococci and staphylococci, even in patients on intensive care units.
P1832 Gram-positive bacteraemia due to Enterococcus spp. in Russian intensive care units: results of a 2-year surveillance
D. Galkin, A. Veselov, O. Kretchikova, E. Riabkova, A. Dekhnich (Smolensk, RU)
Objectives: Bacteraemia caused by Gram-positive microorganisms is increasing in prevalence and antimicrobial resistance surveillance is necessary to optimise antibiotic therapy of the corresponding condition both at the local and national levels. ICUs are characterised by accumulation of patients with severe infections and compromising underlying condition. Hence, it makes adequate empiric therapy crucial for favourable clinical outcomes.
Methods: A multicentre study was conducted in the period of 2002–2004 in ICUs of 17 Russian cities to collect Gram-positive blood isolates and evaluate resistance of Enterococcus spp. to commonly used antimicrobials. Strains were tested using agar dilution method in accordance with CLSI 2006 (M100-S16) performance standards.
Results: A total of 205 Gram-positive microorganisms were isolated, of which Enterococcus spp. accounted for 29.7% (n = 61). Enterococcus faecalis and Enterococcus faecium constituted for 44.3% (27) of strains each. Enterococcus species were isolated primarily from the patients with sepsis – 54.1% (33), pneumonia – 13.1% (8), and bacterial endocarditis – 8.2% (5). Susceptibility of the strains was evaluated to ampicillin, chloramphenicol, gentamicin, ciprofloxacin, levofloxacin, moxifloxacin, tetracycline, streptomycin, vancomycin, teicoplanin, linezolid. All the strains (100%) were susceptible to glycopeptides and linezolid. Resistance rates (I+R) of E. faecalis to ampicillin was 11% with MIC50/MIC90 of 1/16. Gentamicin, tetracycline, ciprofloxacin and chloramphenicol were not active against these strains with resistance rates of 100%, 74.1%, 77.7%, and 55.6%, respectively. Levofloxacin and moxifloxacin were slightly more active with resistance rates of 25.9% and 18.5%. All (100%) E. faecium strains were resistant to ampicillin, ciprofloxacin, gentamicin and streptomycin. Resistance to levofloxacin, moxifloxacin, chloramphenicol, and tetracycline was 92.5%, 88.8%, 62.9%, and 37.1%, respectively.
Conclusion: Glycopeptides and linezolid are recommended to use for empiric therapy of bacteraemia caused by Enterococcus species in ICU patients. Ampicillin is another option for the therapy of E. faecalis bacteraemia.
P1833 A cost-effective study of VRE surveillance in a Greek university hospital
N. Charalambakis, M. Pantelaki, K. Chatzigeorgiou, M. Kapi, N. Siafakas, L. Galani, L. Zerva (Athens, GR)
Objectives: Enterococci with acquired resistance to the glycopeptides (VRE) have emerged in European hospitals necessitating prompt identification. Enterococcal speciation and susceptibility determination by automated systems (including Phoenix by Becton Dickinson) remains problematic. While performing a VRE surveillance study, a cost-effective approach of rapid and accurate pathogen identification was developed.
Methods: A rectal swab was collected from all consenting hospitalised patients (n = 132) and was inoculated into vancomycin (6 μg/mL) containing Enterococcosel Agar and Broth (BD). Phoenix was used for speciation and susceptibility testing supplemented by PYR, motility and pigment production. When indicated, vanco and teico E-tests were performed (Biodisk). A PCR assay confirmed speciation and glycopeptide resistance of VRE strains. Reagent cost and number of performed tests were recorded; test results were analysed in order to develop the most cost-effective approach for performing a VRE surveillance study.
Results: A total of 99 isolates were obtained but only 57 were Enterococci (44.4% colonisation rate). They were speciated as E. faecalis (n = 4), E. faecium (n = 11) and E. cass.//gallinarum (n = 42) by Phoenix. All E. faecium strains demonstrated high level glycopeptide resistance and were confirmed as E. faecium vanA by E-test and PCR. VRE carriage rate corresponded to 8.3%. The majority of strains (81%, including all VRE strains) were isolated from both agar and broth cultures. Among E. cass./gallinarum strains 78% were motile and 21% produced pigment. All E. faecalis isolates were vanco-S and teico-S by Phoenix, however, one dilution lower vanco MICs were obtained by the E-test. Non-motile E. cass./gallinarum strains demonstrated vanco-S, I or R and teico-S MICs by Phoenix, while vanco E-testing showed lower MICs. Three E. cass./gallinarum isolates revealing high level glycopeptide resistance by Phoenix were vanco-I and teico-S MICs by the E-test. Total reagent cost for “extended testing” corresponded to 2,760 Euros, but the analysis of obtained results allowed the development of a “minimally required testing” approach reducing costs by 29%.
Conclusion: These results show high rates of enterococcal and non-enterococcal strain isolation on selective media, an overestimation of vanco MICs by Phoenix, while solid media inoculation suffices for VRE strain isolation. Significantly, cost reduction for performing a VRE surveillance study is possible.
P1834 Detection of linezolid-resistant, vancomycin-resistant Enterococcus strain in Iran
R. Hosseini Doust, S. Yasliany Fard, A. Mohabati Mobarez (Tehran, IR) Enterococci, and especially the vancomycin-resistant enterococci (VRE), are recognized as one of the most important causes of nosocomial infections in seriously ill and immunocompromised patients. Linezolid has been used in clinical practice for a relatively short period of time; there are already several reports of linezolid-resistant enterocci. The sensitivity of Enterococci, isolated from clinical samples, to vancomycin, linozolide and teicoplanin were determined by disk diffusion. Then the existence of VanA and vanB genes were assessed by PCR.
We isolated 195 Enterococci of human clinical specimens. Forty (20.5%) isolates were resistant to vancomycin. Fifteen percent of VRE isolates were resistant to linezolide. Eighteen percent of VRE strains possessed the VanA and 10% of them possessed the VanB gene, which confers high level resistance to vancomycin. When the PCR reactions were carried out specific and predicted size amplicons of vanA (734 bp) and VanB 420bp were observed. Of six linezolid resistant isolates, 2 (33.3%), 1 (16.6%) and 1 (16.6%) were E. casseliflavus, E. gallinarum and E. faecium respectively.
These results are important for developing control strategies point of view. The spreading of glycopeptide and lineozolid resistance strains of Entrococci is becoming more serious in the healthcare institutions.
Molecular typing in clinical bacteriology
P1835 Genotypic characterisation of Mycobacterium tuberculosis isolates from Bogotá, Colombia based on IS6110-based restriction fragment length polymorphism
J. Hernandez, F. de la Hoz, M. Murcia (Bogota, CO)
Objective: The aim of this study was to provide the first insight of the molecular epidemiology of Tuberculosis in Bogotá – Colombia.
Methods: A total of 137 M. tuberculosis complex strain obtained were systematically cultured from pathological samples from 1995 to 2006. The data for each patient included sex, age, HIV serological status, and the following microbiological data: data of sampling, smear microscopy results and strain identification. All isolates were tested for susceptibility to antimicrobial agents according to the proportional method. Extraction of DNA from mycobacterial strains and DNA fingerprinting with standard IS6110-based RFLP method were performed on all isolates. RFLP fingerprint images were analysed by using Gelcompar II software (Applied Maths) version 4.6. Clusters were defined as isolates showing identical restriction fragment length polymorphism patterns.
Results: RFLP analysis was performed on 129 isolates from Bogotá. The numbers of IS6110 copies per isolate varied from 5 to 25. A total of 96 (74%) different IS6110 fingerprints patterns were identified and 35 isolates (26%) were clustered into 17 groups consisting of two to four isolates. Direct epidemiological links could no be established from most of the cases in clusters. Age, sex, drug resistance and positive sputum smear were not significant associated with molecular clustering (p > 0.05). In contrast HIV-1 was associated with molecular clustering (p < 0.05).
Conclusion: The results of our study show a high genetic variation of M. tuberculosis in Bogotá Colombia, suggesting that the transmission results from reactivation of infection adquired in the distant past. In consequence additional molecular epidemiologic studies are necessaries to improve the knowledge of transmission dynamics of tuberculosis in Colombia
P1836 Characterisation of multidrug-resistant Mycobacterium tuberculosis isolates from Johannesburg, South Africa, by spoligotyping
C. Mlambo, R. Warren, T. Victor, A. Duse, E. Streicher, E. Marais (Johannesburg, Tygerberg, ZA)
Objective: The development of drug resistance and the emergence of multidrug-resistant tuberculosis (MDR-TB) is a threat to TB control globally. The estimated proportion of MDR-TB cases in South Africa ranges from a median of 1.6% (range 1–2.6%) among new cases to a median of 6.6% (range 4–13.9%) among re-treatment cases, depending on the Province. There is however, limited information on Mycobacterium tuberculosis genotypes currently circulating in the country. The objective of the study was to determine the population structure of MDR-TB strains in the greater Johannesburg area, using molecular techniques.
Methods: Spacer oligonucleotide typing (spoligotyping) was used to analyse 270 consecutive MDR-TB isolates collected from June 2004 to February 2006 at the National Health Laboratory Service TB division, which receives the bulk of samples in the region. The spoligotyping results were compared to the World Spoligotyping Database of the Institut Pasteur de Guadelope.
Results: A total of 247 (92%) different spoligotypes were found in the spoligotyping database, whereas the remaining 23 (8%) were orphan types. A major proportion of the strains (23%) belonged to the LAM family. The proportions of the strains that belonged to the remaining families are as follows: 20% to T, 14% to Beijing, 9% to EAI and 8% to the S/F28 family. The remaining strains belonged either to the CAS family, X family or the U family. Strains from the Beijing family are spread throughout the world and are often associated with drug resistance.
The drugs recommended for treatment of TB in South Africa are isoniazid, rifampicin, ethambutol and streptomycin. Our study showed that the majority of strains (57%) from patients who have MDR-TB were resistant to all the first-line drugs.
Conclusion: This is the first study to detail the molecular types of MDR-TB strains in Johannesburg. It highlights the diversity of the strains that are circulating in the area and the high proportion of MDR-TB strains that are resistant to all the first line anti-TB drugs. These findings could have a significant impact on TB disease control programmes in the region, especially as these strains are the precursors to extensively resistant strains (XDR TB).
P1837 Three years of universal molecular epidemiology of tuberculosis in Almeria (Spain), a setting with a high proportion of TB in immigrants
M. Martínez, D. García de Viedma, M. Sanchez, M. Rogado, M. Cabezas, W. SánchezYebra, M. Herranz, R. Fernandez, J. Martínez, M. Lucerna, P. Barroso, I. Cabeza-Barrera, F. Díez, M. Rodriguez, M. Escámez, P. Marín, A. Lazo, J. Gamir, J. Vazquez, C. Gutierrez on behalf of the INDAL-TB group
Objective: To evaluate the TB transmission patterns in a population with a high proportion of immigrant cases.
Background: Almeria has a high intensive agriculture production, responsible of a calling effect for immigrants.
Methods: 469 TB cases were communicated in the period 2003–2005, among them 283 (60.3%) microbiologically confirmed and 233 (49.7%) in foreigners [Africa-Magreb 94 (40.3%) cases, Africa Sub-Saharan 56 (24.0%), East-Europe 38 (16.3%), Latin-America 33 (14.2%)]. Microbiology: Samples were inoculated on to Löwenstein and BacT/ALERT®MP (bioMérieux) media. The isolates were identified by means of a genomic probe (Gen Probe, USA). Genotyping: M. tuberculosis (MTB) isolates were fingerprinted by IS6110 RFLP, Spoligotyping, and MIRU-VNTR with the set of 15 loci. MTB isolates were considered to be clustered if their MTB genotypes shared identical RFLPtypes (for patterns with more than 6 bands) or also identical spoligotypes and MIRUtypes (for cases with less than 6 RFLP bands). Cases with suspicious of being false positive due to laboratory cross-transmission were excluded. Epidemiology: data were obtained from the Register of the “Sistema de Vigilancia Epidemiológica de Andalucía” and from standardised interviews of the clustered cases.
Results: At least one MTB isolate from each of 256 (90.5%) patients were genotyped. Seventy-eight cases (30.6%) were grouped into 26 clusters (range 2–6 members) and the remaining cases (69.4%) were infected with isolates not shared by other members in the population. Thirty-eight (27.0%) of 141 foreign-born patients and 40 (34.8%) of 115 Spaniard cases were clustered (p 0.176). Nine clusters (34.6%) included only autochthonous cases; eight (30.8%) only foreign patients and the remaining nine clusters included both autochthonous and foreign cases (34.6%). The standard contact tracing identified epidemiological links in only 2 of the 8 clusters constituted by four or more members. However, the standardised interview of clustered cases could reveal links for 7 clusters.
Conclusions: Our data suggest the occurrence of TB transmission among the foreign-born population and between the autochthonous and foreign cases in Almeria. Molecular epidemiology is required in addition to the traditional contact tracing to reveal novel TB transmission routes in this area with a high socio-epidemiological complexity.
Finance: Fondo Inv. Sanitaria (PI030986-PI030654), J. Andalucía (248–03, 151–05) and F. Progreso y Salud (14033)
P1838 Molecular analysis of Clostridium difficile by PCR-ribotyping
N. Sadeghifard, B. Kazemi, M. Salari (Ilam, Tehran, IR)
Objectives: This study was aimed to determine the prevalence and molecular typing by PCR amplification of rRNA intergenic spacer regions (PCR ribotyping) of C. difficile in patients and the hospital environment in three teaching hospitals in Tehran, Iran.
Methods: During the study period, the stool samples of 1822 hospitalised patients with nosocomial diarrhoea and 100 environmental samples were cultured and tested by stool cytotoxin assay, toxigenic culture and enzyme immunoassay. Biochemical identification of the isolates was performed by conventional tests then Chromosomal DNA was extracted from colonies of C. difficile and PCR amplification of the ISR was carried out using two universal primers complementary to conserved regions in the 16S and 23S rRNA genes.
Results: Toxigenic C. difficile was isolated from 124 of 1822 (6.8%) patients analysed and recovered from 17 of 100 (17%) environmental sites cultured. Among patients with Clostridium difficile –associated diarrhoea, 69 patients were male (7.1%) and 55 patients were female (6.5). There was no significant relationships between C. difficile associated diarrhoea and age of patients. According to PCR-ribotyping results, 27 different PCR-ribotyping patterns were observed among clinical isolates, 4 different PCR-ribotyping patterns among environmental toxigenic C. difficile isolates and 1 PCR-ribotyping pattern was common among clinical and environmental isolates. Ribotypes 1–5 were isolated only from hospitalised patients and some of environmental isolates at Emmam hospital; ribotypes 6–9 were detected only at Children Medical Center and ribotypes 10–12 were restricted at Shariati hospital. Ribotypes 13–17 were four distinct clones that were circulating in all three hospitals. The following ribotypes were common among two hospitals; ribotypes 18–21 were common among Emmam and Shariati hospitals, 22–25 among Emmam hospital and Children Medical Center and ribotypes 26–28 among Shariati hospital and Children Medical Center. Ribotypes 29–32 were observed among environmental isolates.
The predominant ribotypes from the clinical isolates were types 13 and 14, which accounted for 29.9% of all isolates in three hospitals.
Conclusion: Our results suggest that CDAD is an emerging nosocomial problem in Iran and it will be necessary to evaluate the epidemiology and measures to control nosocomial spread. These findings show that the isolates associated with CDAD in Iran have different PCR-ribotyping patterns.
P1839 Characterisation of virulence factors in Vibrio cholerae isolates from the recent cholera outbreak in Senegal (Sub-Saharan Africa)
M.H. Diallo, A.I. Sow, C.S. Boye, A. Aidara Kane, A. Gassama Sow (Dakar, SN)
Cholera is an epidemic diarrhoeal disease generally caused by toxigenic O1 or O139 Vibrio cholerae. Senegal has in last two years experienced major cholera epidemic with a number of cases totalling more than 23, 325 with approximately 303 fatal outcomes. The most important virulence factor produced by V. cholerae is cholera toxin (CT) encoded by ctx gene. The genes encoding zonula occludens toxin (Zot) and accessory enterotoxin (Ace) are located upstream to the ctx gene on a dynamic region of V. cholerae chromosome termed virulence cassette. Toxin co-regulated pilus (Tcp) is a colonisation factor. The aim of our study is to characterise virulence genes among 50 of O1 clinical V. cholerae isolates from the last V. cholerae outbreak occurring in 2004–2005 in Senegal. Virulence genes were characterised by polymerase chain reaction (PCR) using specifics primers. Ctx, zot, ace genes were present respectively in 98%, 88%, and 92%. Most of V. cholerae isolates have an intact virulence cassette (86%) (ctx, zot, ace genes). This virulence cassette is absent in one isolate; it was deleted (zot, ace genes) in two isolates. Both ZOT and ACE play synergetic role with CT in the epidemic V. cholerae toxicity. Strains did not harbour Tcp gene.
P1840 Double digest selective label: a fast and discriminatory molecular typing technique for clinical isolates of Pseudomonas aeruginosa
V. Terletskiy, G. Kuhn, P. Francioli, D. Blanc (Lausanne, CH)
Objectives: To develop a new molecular typing technique for P. aeruginosa, Double Digest Selective Label (DDSL), which could be used as an alternative to currently widely accepted pulsed-field gel electrophoresis (PFGE).
Methods: In the DDSL approach, large DNA fragments (produced by SpeI restriction) which were normally analysed by PFGE were trimmed by a second frequently cutting restriction enzyme (StuI) to give smaller fragments. Fragments containing SpeI enzyme overhangs were selectively tagged with biotinylated dCTP (single-tube digestion/labelling reaction) and then separated on a conventional agarose gel. Transfer of resolved double-stranded DNA fragments to a nylon support was performed immediately after electrophoresis in a vacuum blotter. Subsequent detection of DNA fragments was mediated with alkaline phosphatase by colour reaction.
Results: 40 isolates collected during a period of 8 years at the intensive care units of the hospital were typed by both DDSL and PFGE. Turn around time was only 12 hours in contrast to 4 days required by PFGE typing. DDSL gave rise to bands which were generally sharper than those observed by PFGE typing. Both methods discriminated isolates into 34 different types, resulting in discriminatory power values of 0.990 and 0.989 for DDSL and PFGE, respectively. Two major clusters (8 and 6 related isolates in each) were recognized by both methods, segregation of subclusters within these groups was however different. Minor clusters containing two closely related isolates observed with PFGE were also identified by DDSL.
Conclusion: DDSL typing of P. aeruginosa in a hospital setting was found to be as discriminatory as PFGE while time required for analysis was significantly reduced. The technique does not require expensive specialised equipment and is easy to set up in a microbiological laboratory.
P1841 On-site molecular typing of Pseudomonas aeruginosa as an adjunct to infection control in a paediatric cystic fibrosis centre
C. Lucas, M. McMillan, D. Brown, C. Williams (Glasgow, UK)
Introduction: Cystic Fibrosis (CF) is a genetically inherited disease affecting around 1 in 3,000 children and young adults in the UK. It is characterised by a progressive decline in pulmonary function, exacerbated by the acquisition of specific micro-organisms, in particular Pseudomonas aeruginosa. To date, no cure is available and the life span of sufferers is directly influenced by the effectiveness of local infection control practices. However, at the present time, detection of cross-infection episodes is entirely dependent on the genotyping service provided off-site.
Methods: Isolates of Ps. aeruginosa grown from CF patients were collected throughout 2005. An in-house molecular typing method using the Random Amplification of Polymorphic DNA (RAPD) technique was developed and genetic profiles of the isolates were analysed compared using BioNumerics v.4.0 computer software.
Results: DNA extracts from 90 Ps. aeruginosa strains grown from 19 CF patients have been analysed using RAPD. Strains determined to be unique to a single patients by the Reference Laboratory clustered together, demonstrating >80% homology using RAPD. Transmissible strains known to be genotypically related, which were cultured from different patients, demonstrated a similar degree of homology. The advantage for infection control purposes is the rapid availability of results with on-site molecular screening.
Conclusions: Initial data indicate that RAPD allows recognition of unique Ps. aeruginosa strains and allows discrimination between diverse strains, to a level which is adequate for routine screening purposes. Introduction of on-site RAPD typing will clarify the relatedness between local strains, hasten detection of cross infection episodes and facilitate timely and effective infection control action.
P1842 Epidemiology of Candida krusei and Candida inconspicua infections in haematology-oncology ward
M. Sikora, E. Swoboda-Kopec, E. Stelmach, B. Interewicz, A. Domaszewska, A. Majewska, W. Olszewski, M. Luczak (Warsaw, PL)
The need for adequate molecular typing is especially relevant in cases of pathogens that either increase in clinical prevalance or gain specific features increasing their disease-causing capacity. Both factors became appearent for fungy upon the rise in number of infections in immuno-incomponent.
Objectives: Analysis of epidemiology of the clinical isolates of C. krusei and C. inconspicua cultured in 2004 from patients hospitalised in haematology-oncology ward of the Central Clinical Hospital of the Medical University in Warsaw and molecular characterisation of selected strains by RAPD (Random amplification of polymorphic DNA) technique.
Methods: The fungal strains were cultured from stool specimens, sputum, urine, blood, bile as well as swabs of the throat, mouth, wounds and perioneum of hospitalised patients. The isolation and identification of cultured fungi was done according to standard mycological procedures and commercially available tests (bioMérieux, Sanofi Diagnostics Pasteur). Sixty nine selected strains of C. krusei and seventeen strains of C. inconspicua were further characterised by RAPD technique. PCR products were analysed by PAGE electrophoresis, silver stained and evaluated using Syngene software.
Results: The strains were most often isolated from stool samples 60.8%, sputum 15.7%, and throat swabs 9.8%. Molecular analysis of C. krusei strains revealed three groups (14, 12 and 7 isolates) with 98% of relatedness between strains, and 80% for 13 strains. 6 and 2 isolates of C. inconspicua were similar in 98% and 4 in 80%.
Conclusions: We report a high frequency of C. krusei isolations from patients hospitalised in the haematology–oncology ward. Molecular analysis of selected strains points to nonsocomial spread of this pathogen.
P1843 Distribution of Ure C, cagA and vacA genes in Helicobacter pylori isolated from patients with gastroduodenal disease in Tehran, Iran
F. Jafari, M. Aslani, L. Shokrzadeh, K. Baghai, H. Dabiri, H. Zojaji, V. Razavilar, P. Kharaziha, M. Haghazali, M. Molaei, M.R. Zali (Tehran, IR)
Objective: The aim of this study was to investigate the prevalence of H. pylori infection and the frequency of UreC, cagA and vacA genes in H. pylori infection in Iran.
Methods: A total of 280 gastric biopsies from patients who were qualified for endoscopies of the upper gastrointestinal tract from February 2006 to September 2006 were included in this study. These samples were cultured for H. pylori by conventional methods. The genomic DNA was extracted from biopsy samples by standard method. UreC (glmM) gene was amplified by polymerase chains reaction (PCR), then genotyping was done based on cagA and vacA (s1/s2/m1a/m1b/m2) genes.
Results: Out of 280 biopsy samples, 179 (63.9%) patients with gastroduodenal disease were infected with H. pylori. The number of positive culture in patients with gastric cancer and duodenal ulcer were 8 (4.5%) and 27 (15.1%) respectively. The cagA gene was amplified in 133 (74.3%) of the isolated strains. The frequency of vacA genotypes for s1 (signal region) and m2 (middle region) genotypes were 63.5% and 61.2%, respectively. In 67 (38%) of strains s1m2 genotype was found.
Conclusion: The study demonstrated that vacA s1and m2 genotypes dominant in Iranian strains were also predominant strains in North America (60–70%), but not in Middle-East (39%), suggesting the relation between Iranian strains and European strains. The VacA s1 genotype is the most frequent genotype and there was a correlation between infections with H. pylori strains carried cagA gene. Moreover vacA s1/m2 genotypes found the frequent genotype responsible for the development of gastroduodenal diseases.
P1844 Detection and genotyping of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected in Lithuania
D. Ambrasiene, A. Paulauskas, J. Turcinaviciene (Kaunas, Vilnius, LT)
Lyme borreliosis is the most common tick-borne infectious disease in humans and domestic animals. The Ixodes ricinus ticks are well known as major vector of the causative agents of Lyme borreliosis and others infections diseases. In Europe, Lyme borreliosis is associated with three genospecies of Borrelia burgdorferi sensu lato: B. burgdorferi sensu stricto, B. garinii and B. afzelii. These species can cause distinct clinical manifestations of Lyme disease. B. burgdorferi s.s. can cause arthritis, B. garinii serious neurological manifestations and B. afzelii a distinctive skin condition known as acrodermatitis chronica atrophicans. I. ricinus is a widely distributed tick in Lithuania and may transmit pathogens to mammalian hosts, including human beings.
The aim of this study: to genotype B. burgdorferi genospecies in I. ricinus ticks and to determinate dominant Borrelia genospecies by molecular genetics methods.
Methods: More than 2000 I. ricinus ticks were collected by the flagging method in Lithuania. Examination of ticks (n = 1679) for the presence of B. burgdorferi s.l. DNA was performed by PCR with fla gene specific primers. For Borrelia genotyping (B. burgdorferi s.s., B. garinii and B. afzelii) were used multiplex PCRs with genospecies-specific primers.
Results: The overall prevalence of B. burgdorferi s.l. was 13.3% (223/1679). 190 samples positive for B. burgdorferi s.l. were used for genotyping. B. afzelii was found in 141 ticks (74%), B. garinii in 19 (10%), B. burgdorferi s.s. in 14 (7%). Double infections were observed in 1% of the infected ticks, other 14 samples (7%) of the Borrelia infections were non typed.
Conclusions: The molecular tools allow identifying different Borrelia genospecies which can be maintained in nature and are important for pathogenesis, diagnostic and preventative implications. It was detected B. afzelii was the dominant genospecies in Lithuanian ticks. Prevalence of genospecies is similar to neighbour country Latvia (B. afzelii 64.9%), but different from western Germany (B. afzelii 39.9%) and Slovakia and Southern Poland, where B. garinii is predominant genospecies (45.5%). It is necessary to have a closer look at the local vertebrate biocenosis in epidemiological studies of Lyme borreliosis, because reservoir host population is to be held responsible for special distribution of certain genospecies.
This study was supported by ENLINO (Environmental Studies in Lithuania and Norway) and the Lithuanian State Science and Studies Foundation.
P1845 Borrelia burgdorferi species isolated from different natural sources in Slovenia
A. Zore, E. Ruzic-Sabljic, F. Strle, T. Trilar, T. Avsic-Zupanc (Ljubljana, SI)
Objectives: Species of Borrelia burgdorferi sensu lato complex are differently disposed in nature. The aim of this study was to determinate Borrelia burgdorferi species in various natural sources: humans with Lyme borreliosis, vectors – Ixodes ricinus ticks found in woods or on birds, different reservoir animals: small mammals and birds.
Methods: We tested 103 Borrelia human strains isolated from skin, 148 free living ticks from the meadows or forests and 40 ticks removed from birds, 12 strains isolated from birds (skin or blood) and 141 urinary bladders from small mammals. For detection of Borrelia species we used direct methods: PCR or cultivation and determination with PFGE or RFLP of PCR products.
Results: In our human isolates we identified frequently B. afzelii (84%), rarely B. garinii (14%) and only exceptionally B. burgdorferi sensu stricto (1%) or B. spielmanii (less than 1%). From ticks collected from meadows or forests we determined with PCR most frequently B. garinii (56%), often together with B. afzelii (41%), rarely with B. garini and B. burgdorferi sensu stricto (32%). Engorged ticks from birds were infected most frequently with B. garinii, exceptionally with B. afzelii in only one species of birds (Prunella modularis). In birds we found also B. valaisiana in blackbird (Turdus merula) and in ticks on the blackbirds (53%). The most frequently infected birds species were blackbirds (50%). In this species we detected also mixed infections with B. garinii and B. valaisiana. B. valaisiana is probably not pathogenic species for human. We never isolated B. valaisiana from human specimens. In urinary bladders of small mammals we detected B. burgdorferi sensu lato in 60% of animals with PCR, frequently B. afzelii (39%), in almost the same frequency B. burgdorferi sensu stricto (33%) and only exceptionally in mixed infections B. garinii (8%). In isolates of B. burgdorferi from urinary bladder in small mammals we determinate with PFGE always only B. afzelii.
Conclusion: Our study demonstrated that different Borrelia burgdorferi sensu lato species are adapted to specific reservoir species. PFGE or RFLP of PCR products are good tools for determination of different species of Borrelia burgdorferi sensu lato.
P1846 A real-time PCR assay for typing emerging pneumococcal serotypes
D. Tarrago, A. Fenoll, I. Obando, D. Sanchez-Tatay, L. Arroyo, J. Casal (Majadahonda, Madrid, ES)
The need for rapid, reliable, and highly sensitive methods for the diagnosis and serotyping of invasive pneumococcal disease (IPD) is becoming more urgent due to the emergence of non-vaccine serotypes after the introduction of 7-valent conjugate vaccine. Conventional methods of serotyping as the gold standard Quellung Reaction are hampered by the need of clinical isolates. PCR based assays could be used in typing Streptococcus pneumoniae directly from clinical samples and may be helpful in the epidemiological surveillance of invasive pneumococcal disease.
Objectives: Development of PCR assays based on Streptococcus pneumoniae capsular genes for typing pneumococci directly from clinical samples. Assessment of the assay as diagnostic and pronostic marker of IPD.
Methods: DNA was extracted from blood, pleural fluid and cerebrospinal fluid using Qiamp DNA kit (Qiagen). Pneumococcus DNA was detected using a previously described PCR based on pneumolysin gene. Aligment of cpsb, cpsf and, cpsk genes of the capsular operon of pneumococci from most of the 90 serotypes was used to design primers and probes with Primer Express software v3.0 (Applied Biosystems). TaqMan probe-based chemistry was used for Real-Time PCR. Amplifications were performed on an Applied Biosystems 7300 Fast Real-Time PCR System and absolute quantification plate documents were created and analysed by Sequence Detection Software v1.3.1
Results: A real-time PCR assay using TaqMan-MGB probes based on the amplification of the capsular genes fragments directly from blood, pleural fluid and cerebrospinal fluid has been developed for detecting and serotyping Streptococcus pneumoniae in IPD caused by 1, 5, 14, 19A and 19F serotypes.
The assay is able to detect 10 DNA copies/PCR tube and it has been tested with clinical isolates and clinical samples including blood and pleural fluid with reliable results for 1, 3, 5, 19A and 19F serotypes.
In a pilot study to investigate the usefulness of the assay, 55 negative-culture pleural fluids from empyema cases in children <14 who were admitted to hospitals in southern Spain were analysed by real-time PCR. Pneumococcus DNA was detected and typed by PCR in 89% (49/55) and 58% (32/55) of samples, respectively.
Conclusions: Molecular epidemiological analyses are imperative for the surveillance of IPD. The results indicate that PCR based assays are needed to know the actual contribution of the different serotypes to IPD.
P1847 Detection of phage-associated virulence/resistance genes in induced prophages of Streptococcus pyogenes clinical isolates
S. D'Ercole, D. Petrelli, S. Rombini, M. Prenna, S. Ripa, L. Vitali (Camerino, IT)
Objectives: The genome of each sequenced Streptococcus pyogenes strain shows to contain many prophages encoding proven or putative extracellular virulence factors and antibiotic resistance determinants. In this work we determined the presence of prophage-associated virulence/resistance genes (F-vir) and the induction profile of prophages harbouring virulence/resistance genes in fifty-nine S. pyogenes pharyngeal isolates.
Methods: PCR was used to assess the presence of those genes (i.e. speA, speC, speH, speI, speK, speL, speM, ssa, spd1, spd3, spd4, sdn, sda, sla, mefA and TetO) known to be prophage-associated in S. pyogenes.
Each strain was treated with mitomycin C to induce release of functional phages, and the corresponding unrestricted total DNA was then analysed by pulsed field gel electrophoresis (PFGE). S. pyogenes SF370 and strain-6 (mefA/TetO) were used as control strains. Single or multiple bands, corresponding to phage DNA, were obtained only in mitomycin-treated cells. They were excised and analysed by PCR to detect the specific F-vir.
Results: Five percent of the strains did not contain any of the known F-vir, while 76% had at least two. The distribution of F-vir was greatly variable and the overall mean number of F-vir per isolate was 3.8 (± 2.3). The release of phage DNA was achieved in 17 strains. Among these, PCR analysis detected a single F-vir in the phage DNA released by 35.3% of the strains, and three F-vir in 17.6%. One strain released phage DNA containing two F-vir, whereas another strain released phage DNA positive to five F-vir.
The F-vir most frequently associated with released phage DNAs were sdn, spd4, speC, spd1 and spd3.
Conclusion: The pharynx is colonised by S. pyogenes harbouring a variable number and assortment of F-vir. A limited number of virulence genes are hosted by functional prophages and, therefore, have the potentiality to be horizontally transferred. Many strains possess different important F-vir that, at least in the conditions used, cannot be detected in released phage DNAs. These results suggest that the population of F-vir-lacking functional prophages is vast and that the polylysogeny would be the product of the accumulation of temperate phages that eventually become defective.
P1848 Molecular epidemiology of Streptococcus pneumoniae associated with paediatric invasive disease in the Czech Republic
H. Zemlickova, V. Jakubu, P. Urbaskova, M. Musilek, J. Motlova (Prague, CZ)
Objectives: To characterise invasive isolates of S. pneumoniae associated with paediatric invasive disease in Czech children under 6 years of age by multilocus sequence typing (MLST) and to study the clonality of the causative serotypes collected prior to the introduction of the pneumococcal conjugate vaccine into the Czech Republic.
Methods: The strains (n = 124) isolated from blood or cerebrospinal fluid of children under 6 years of age between 1996 and 2003 were referred to the National Institute of Public Health by 39 microbiology labs from 28 cities. Serotyping was performed by Quellung reaction. The minimal inhibitory concentrations (MIC) of penicillin, erythromycin, tetracycline and chloramphenicol were determined by the CLSI broth microdilution method. MLST typing was carried out for all available isolates (n = 80) of the major predominant serotypes 6B, 14, and 23F.
Results: Overall, 5 (4.0%) strains exhibited reduced susceptibility to penicillin but only one of these was highly resistant. The resistance rates to erythromycin, tetracycline and chloramphenicol were 5.6%, 8.1% and 4.8%, respectively. The most frequent serotypes were 6B (19), 14 (15), 23F (12), 19F (11), 9V (9), 18C (9) and 1 (8). MLST typing was performed on 80 (64.5%) viable isolates of the above named serotypes. Nine of 32 revealed sequence types (STs) were newly identified. Isolates of serotype 6B were quite diverse: 17 strains yielded 8 clonal lineages (10 STs). In contrast, isolates of serotypes 1, 9V and 14 were highly homogenous. Serotypes 1 and 9V were classified into a single clone each. Thirteen of 15 isolates of serotype 14 had the same sequence type (ST124).
Conclusion: The vaccine-included serotypes 6B, 14, 23F, 19F, 9V and 18C are most frequently associated with paediatric invasive disease in the Czech Republic. Clonality varies between serotypes. The continued presence of previously described invasive clones of S. pneumoniae was demonstrated for serotypes 1, 9V and 14.
P1849 Molecular typing of clinical isolates of Escherichia coli by a new PCR MP method
B. Krawczyk, A. Sledzinska, A. Samet, M. Bronk, J. Komarnicka, J. Leibner, J. Kur (Gdansk, PL)
Background: Epidemiological typing of bacteria is a routine procedure used in the investigation of infectious outbreaks and requires accurate molecular typing methods with high discriminatory power. We had performed evaluation of a new PCR melting profile (PCR MP) technique for E. coli strain differentiation. Results were compared with PFGE method.
Methods: Epidemiologic investigation was shown for E. coli isolates from patients of the Hematological Unit of Clinical Hospital in Gdansk. A total of 90 isolates from 36 patients were included. The E. coli were recovered from blood (66), stool (15), urine (8) of patients with suspected bloodstream infection. To study the genetic similarities between E. coli isolates, a new PCR MP method and REA-PFGE were used.
Results: PCR MP analysis distinguished 44 types (H1-H44) among all isolates studied. One genotype was markedly predominant (H2), as this was represented by 10 isolates from 4 patients. This genotype probably represents E. coli strain from hospital infection. The molecular typing by REA-PFGE found also 44 unique profiles. Clustering of REA-PFGE fingerprinting data matched exactly PCR MP data. The PCR MP was next investigated for the similarities of the E. coli isolated from different sites of examined patients. PCR MP fingerprinting analysis exhibited usefulness for bacterial spread monitoring. As expected, the related epidemiologically isolates show a high degree of similarity. Genotype similarity was documented in 10 of 36 patients with E. coli bacteraemia for whom paired blood and faecal or urine isolates were available for genomic typing, and probably was the major source of these bacteraemias.
Conclusion: We found that PCR MP technique is a rapid method that offers good discriminatory power, excellent reproducibility and may be applied for epidemiological studies. We suggested that there is at least a similar power of discrimination between the present gold-standard REA-PFGE and a PCR MP method. Data presented here demonstrate the complexity of the epidemiological situation concerning E. coli that may occur in a hospital.
P1850 The analysis Y. pestis strains from Kazakhstan and USA by PFGE method
T. Meka-Mechenko, R. Tashmetov, B. Atshabar, L. Nekrassova (Almaty, KZ)
Objectives: Y. pestis strains from plague foci of Republic of Kazakhstan differ on phenotypic properties. The differences in plasmid profile and structure of genes are marked also. The comparative analysis Y. pestis strains isolated from the natural plague foci of Kazakhstan and USA by PFGE method are of interest.
Methods: Are investigated by PFGE method Y. pestis 48 strains isolated in 1938–2002 from the natural plague foci of Kazakhstan and 12 strains from USA. Strains are isolated from fleas, ticks, wild rodents, domestic animals and from the plague patients.
Results: Y. pestis strains isolated from autonomous plague of Central Asian desert natural focus, Volga-Ural steppe autonomous focus, Volgo-Ural sandy natural focus have identity at the genetic level from 82.6% up to 100%, biotype of these strains is mediaevalis. Only 1 strain from Ustyurt autonomous focus (strain's biotype is antigua) is generically close (84.5%) to strain from Tian-Shanian natural plague focus (Sarydzhas autonomous focus). Two plague strains by genetic structure are closer to strains isolated in USA (83%) in comparison with others strains isolated from plague foci of Kazakhstan (77.2%).
In strains isolated from Sarydzhas autonomous focus (Tian-Shanian natural focus) percent of similarity rather low – 77.2%. One strain with positive fermentation of glycerine and negative denitrification 95.7% of similarity of genetic picture with strain isolated from Moinkum autonomous focus (Central Asian desert natural plague focus). Others investigated strains are typical and percent of genetic identity makes 84.5%. The similarity of genetic structure strains isolated from USA varies from 88.8% up to 100%. Rather low percent of identity plague strains isolated in plague foci of Kazakhstan (77.2% up to 100%), is caused landscape-epizootological feature of sites and characteristic properties of circulating populations of the activator of plague.
Conclusion: The identity plague strains on genetic level depend on geographical region and from strain's biotype. In strains concerning to one biotype and isolated from one natural plague focus percent of identity at molecular level is high. The received results of computer analysis and dendrogram of phylogenetic connections between investigated strains allow to determine their origin that increases efficiency of epidemiological of supervision of plague.
P1851 Molecular diversity of Bacillus anthracis in Kazakhstan
A. Aikimbayev, L. Lukhnova, S. Zakaryan, G. Temiraliyeva, Y. Pazylov, T. Meka-Mechenko, W. Easterday, M. Van Ert, P. Keim, T. Hadfield, S. Francesconi, J. Blackburn, M. Hugh-Jones (Almaty, KZ; Flagstaff, Palm Bay, Washington D.C., Baton Rouge, US)
Objectives: In the central Asian republic of Kazakhstan, anthrax is endemic and represents a public health concern. In this study we used high resolution genotyping to examine Bacillus anthracis strain dynamics among historical outbreaks in Kazakhstan and to understand Kazakh B. anthracis genetic diversity on a regional and global scale.
Methods: Isolates were cultured from clinical and environmental samples from anthrax outbreaks in Kazakhstan spanning a 55 year period. Archival Bacillus anthracis cultures (N = 92) were grown on Hottinger blood agar and a QIAamp DNA mini kit (Qiagen) was used to extract DNA from colonies using the manufacturer's protocol. Eight VNTR (MLVA-8) markers were amplified using primers and conditions according to Keim et al. (2000). Electrophoresis of PCR products was performed on an ABI 310 genetic analyser and data was analysed using Genemapper software (ABI, Foster City, CA). UPGMA cluster analysis of VNTR data from the 92 Kazakh isolates and the ‘diverse 89’ genotypes presented by Keim et al. was used to establish genetic relationships (Mega 3.1).
Results: UPGMA cluster analysis of isolates with complete MLVA-8 data (N = 88) identified 12 MLVA subtypes and comparison with the ‘diverse 89’ genotypes revealed that the majority of isolates (n = 74) belonged to the A1a genetic cluster, 6 isolates belonged to the A3b cluster, and 2 isolates belonged to the A4 cluster. Over half of the A1a isolates belong to previously described genotypes (38/74), including G3 (N = 15), G6 (N = 2) and G13 (N = 21). Two Kazakh MLVA types (N = 6) appear to represent new sub lineages in the ‘A’ branch.
Conclusion: This study provides the first description of the genetic diversity of B. anthracis in this Central Asian region. The data presented here are useful for retrospective epidemiological analyses and are valuable baseline data for future epidemiological investigations. The genetic data describes novel lineages and genotypes and furthers our understanding of the global diversity and evolutionary history of B. anthracis.
This research was made possible by financial support from the U.S. Defense Threat Reduction Agency (DTRA) under the project KZ-1 and administered by U.S. Civilian Research and Development Foundation.
P1852 Direct comparison of pulsed-field gel electrophoresis and multilocus sequence typing
O. Dobay, F. Rozgonyi, F. Walsh, M. Diggle, K. Nagy, S. Amyes (Budapest, HU; Edinburgh, Glasgow, UK)
Objectives: Pulsed-field Gel Electrophoresis (PFGE) and Multilocus Sequence Typing (MLST) are thought to have similar discriminatory power. However, although the results from MLST may be more transportable which may be useful in global epidemiological studies, PFGE is more sensitive to smaller genomic arrangements and potentially more discriminatory. In this study, we compare the two methods directly with Streptococcus pneumoniae.
Methods: PFGE was performed on 30 routine Scottish clinical isolates of serotype 14 with identical MLST pattern (sequence type, ST). MLST was performed on 26 Hungarian isolates of very similar PFGE patterns, 12 of serotype 6A and 14 of 23F. Serotyping was performed with the MAST antisera.
Results: PFGE examination of the 30 isolates with identical ST revealed small differences in the banding pattern indicating very little diversity in this group. On the other hand, MLST of the 26 strains with similar PFGE pattern resulted in two main STs (i.e. differences in 6 loci out of 7). One ST comprised the 12 isolates of serotype 6A with identical PFGE type. The other ST contained 11 isolates of serotype 23F, but these had 1–3 PFGE band differences. The remaining 3 strains (23F) belonged to two further STs, but these had also only 1–3 band differences from the others.
Conclusions: Strains identified as a single ST, did show some diversity when tested by the more sensitive discriminator of PFGE. Also in the other way around, strains with 1–3 PFGE band differences, resulted mostly in identical ST. Our results suggest that for more diverse bacteria such as pneumococci, MLST categorises them into sharply distinct groups, as it is based on sequences, while PFGE allows a bit more genetic variation, being based on fragment differences. Therefore, PFGE seems to be better at identifying smaller changes that occur at a certain geographical region within a shorter time period, while MLST is probably more useful for international comparisons. Interestingly clustering by both methods showed little diversity within individual serotypes. However, although the two techniques are very different in principle, they are equally important in epidemiology.
P1853 Chlamydia trachomatis genotypes and infection rates in Lisbon, 1991–2005
S. Viegas, J. Gomes, C. Florindo, A. Nunes, M. Ferreira, A. Paulino, M. Borrego (Lisbon, PT)
Objective: To establish the infection rate of Chlamydia trachomatis (CT) and CT genotype distribution in Lisbon.
Methods: Urethral, cervical or urine samples were collected (1991– 2005) from 10550 (3810 men, 6740 women) attendees of the major Portuguese sexually transmitted diseases (STD) clinic.
9783 (1578 men, 8205 women) non-STD attendees from general practice, family planning or urology clinics of the Lisbon area were also evaluated.
The laboratory diagnose of CT was performed in Instituto Nacional de Saúde Dr. Ricardo Jorge (INSA), Lisbon, through direct immunofluorescence or culture in McCoy cells (1991–1994), and PCR-Amplicor (Roche) (1994–2005). Isolates were genotyped through a modified protocol of the nested-PCR first described by Lan et al, 1994.
Results: During the study period, CT positivity rates [9.1% (965/10550)] were 2.1-fold higher among STD attendees, than in non-STD attendees [4.3% (421/9783)]. Overall, this bacterial STI were found more often in men (9%) than in women (5%).
The mean positivity of CT infection didn't exhibit a major evolution since 1991 in both STD (7.8% in 1993 to 7.7% in 2004) and non-STD populations (7.4% in 1991 to 7.1% in 2004).
Genotype E was the most frequent among STD and non-STD attendees (41.8% and 42.9% respectively) followed by F (13.1% and 19.7% respectively). Genotype D/Da was the 3rd most frequent in STD attendees (12.3%) and G (12.3%) among the non-STD's. The 4th most frequent was G for STD attendees (11%) and D/Da for the non-STD's (11.3%). Genotypes H, I, and J/Ja were found at lower percentages and genotypes A, B/Ba, C, K and L2 were rarely found. Mixed and untypeable infections were found more often among STD's (3.1%) than in non-STD's (1%).
Conclusions: Surprisingly, the number of diagnosed chlamydial infections didn't exhibit a major evolution since 1991, despite a slight decline more obvious among STD-attendees.
The distribution of CT genotypes is similar to the described in the literature and to previous results of our laboratory (Borrego et al, 1997); however, in the 1991–7 period there was an unusual prevalence of genotype H among STD clinics female attendees that is no longer observed. We speculate that the higher occurrence of mixed infections among STD attendees could relate to sexual risk behaviours. Genotype E remains the most frequent either because is the best adapted to the host or because is the most disseminated; this issue is currently under evaluation in our laboratory.
P1854 Rethinking the applicability of Tenover criteria: a model algorithm and a new dendrogram approach for the direct and wider application
I.C. Acuner, C. Eroglu (Samsun, TR)
Objectives: Modified Tenover criteria (MTC) propose the difference that is greater than two genetic events (i.e. >eight band difference) as the criterion for unrelated isolates. However, in the investigation of outbreaks, PFGE patterns of the isolates are mostly evaluated on the basis of various similarity indexes to construct similarity matrices, and then to generate dendrograms through various methods. One of the most used dendrogram generation methods, UPGMA, together with the assured statistical significance by bootstrapping, is claimed to produce results in concordance with MTC. Although their biological basis is more relevant than the other methods, Tenover criteria didn't find much ground for direct application. In this study, direct applicability of MTC by the use of an algorithm to detect the related isolates within the limit of one genetic event difference has been investigated.
Methods: Based on the variables such as genetic variation mechanism with or without affecting the restriction site existence or emergence, relative event position and length, genetic event number, band difference, position, and intensity, in a set of hypothetical PFGE genotypes and in the Escherichia coli strains with known whole genome sequences, random and calculated PFGE patterns are generated, respectively. Consequently, related genotypes up to three genetic event differences have been investigated by the application of an algorithm that compares each PFGE genotype, to the “index” PFGE genotype and a new dendrogram approach has been adapted.
Results: One genetic event differences that may occur with different mechanisms as described by MTC were succesfully detected by the developed algorithm and shown to be distinguishable from each other. MTC display a special pairwise symmetry in the genetic event types. MTC cause triangle inequality and hidden histories like the other above mentioned methods. The selection of the index isolate of an epidemic seems critical. MTC can not exclude the probability of the occurrence of PFGE genotypes that may have greater than two genetic event differences with lower than nine band differences. Direct application of MTC in two and more genetic events may become so complex that it does not allow clear-cut interpretation.
Conclusion: It has been shown that MTC can be applicable for automated direct use in the analysis of large sets of epidemic isolates by PFGE through an algorithm within the limit of one genetic event difference.
P1855 Heterogeneity of Chlamydia trachomatis L2 strains involved in the current outbreak of Lymphogranuloma venereum
T. Meyer, H.J. Stellbrink, S. Fenske, G. Stary, A. Geusau, A. von Krosigk, A. Plettenberg (Hamburg, DE; Vienna, AT; Munich, DE)
Objectives: Since 2003 an outbreak of lymphogranuloma venereum (LGV) among men having sex with men (MSM) was recognized, which has been reported first in Rotterdam, and then was also detected in other European countries and in North America. The disease mainly represented with anal/rectal symptoms, less frequently with genital/inguinal manifestations, and rarely with oral/pharyngeal lesions. The aim of the study was to analyse whether one or multiple Chlamydia trachomatis strains are involved in the current LGV outbreak.
Methods: C. trachomatis infections were diagnosed by detection of bacterial DNA using PCR (TaqMan, Roche) and SDA (ProbeTec, Becton-Dickinson). C. trachomatis genotypes (serotypes) were identified by sequence analysis of the VS4 region of outer membrane protein (Omp) 1. To characterise different strains of LGV genotypes the VS1 and VS2 region of Omp1 were also analysed.
Results: Between July 2003 and October 2006 we have identified 126 patients with LGV, confirmed by detection of genotype L2. Most of these patients resided in Hamburg (n = 65), followed by Berlin (n = 29), Munich (n = 19), and Vienna (n = 10). The majority of the patients were infected with HIV (n = 69), only two patients were HIV negative. Of the remaining 55 patients no information about HIV-status was available. Most of the patients had anal or rectal disease manifestations (98/126, 77.8%). Inguinal lesions (genital ulcer or inguinal lymph node swellings) occurred in 11/126 (8.7%) of the patients. In one patient LGV manifested as oral ulcerative lesion. For 16 patients we did not receive informations about disease manifestations. In 10 patients with anorectal symptoms the same L2 sequence was identified, which was 100% identical to L2b, previously described in Amsterdam. The L2b sequence was also identified in one patient with inguinal disease manifestation, but in another 4 patients with genital or inguinal lesions the L2 sequence was different from L2b.
Conclusion: Sequence analysis of Omp1 regions indicates that more than one L2 strain is involved in the current LGV outbreak. Our preliminary data may further indicate preferential association of individual L2 strains with different disease manifestations.
P1856 INCF-replicon typing: a study on a widely spread group of plasmids carrying both virulence and resistance genetic determinants
L. Villa, A. Garcia, A. Bertini, A. Carattoli (Rome, IT)
Objectives: Plasmids belonging to the incompatibility (Inc) group F are widespread in the family of Enterobacteriaceae, providing advantageous traits to their host, encoding antibiotic resistance and virulence determinants. Recently, they have been found associated to clinically relevant β-lactamases such as CTX-M-15, CTX-M-9, SHV-5, SHV-12, DHA-1. The PCR-based replicon typing method (Carattoli et al. 2005) allows a rapid identification of the FII, FIA, FIB and FIC replicons, however the high level of heterogeneity of the IncF plasmids requires a further characterisation to better identify and trace their spread.
Methods: Alignment analyses in the region of the FII replicons were performed on thirty IncF plasmid DNA sequences, including E. coli, Salmonella, Shigella and Yersinia virulence plasmids, obtained from reference and clinical strains. Primer pairs were devised on the highly conserved region of the regulatory antisense RNA, CopA and on the replicase repA gene to set up a specific IncF-replicon typing. Plasmids were typed by these PCRs and DNA sequence analysis of the amplicons.
Results: Three different groups of IncF plasmids were analysed, classified and compared by the IncF-replicon typing: (1) CTX-M-15 carrying plasmids isolated in UK and Italy; (2) plasmids from a collection of human clinical E. coli strains from Spain related to those previously described in avian pathogenic E. coli; (3) Salmonella, Shigella, Yersinia, E. coli O157:H7 virulence plasmids. Interesting correlations were obtained among emerging plasmids circulating in human clinical isolates with respect to well known reference plasmids or to plasmids isolated from E. coli from animals.
Conclusion: Resistance plasmids have been described to carry virulence factors (bacteriocins, siderophores, cytotoxins, or adhesion factors) and virulence plasmids have been described to carry resistance genes. For plasmids carrying virulence and resistance linked determinants, an infective population may be selected for antimicrobial resistance, and antimicrobial resistance pressure will select the virulence traits. The acquisition of antimicrobial resistance genes on virulence plasmids, could represent a novel tool in bacterial evolution, implementing adaptive strategies to explore and colonise novel hosts and environments. A rapid method to detect these kind of plasmids could be of great relevance to monitor their spread and diffusion in relevant clinical isolates.
P1857 Molecular epidemiology of extended-spectrum β-lactamases-producing organisms of the family Enterobacteriaceae in Sofia, Bulgaria
D. Ivanova, R. Markovska, D. Iordanov, I. Mitov (Sofia, BG)
Objectives: High prevalence of nosocomial ESBL-producing Enterobacteriaceae in Bulgarian hospitals has been shown previously, but epidemiological typing of such strains has never been conducted. The aims of this study were to investigate ESBL-producing isolates of the family Enterobacteriaceae collected in Queen Joanna hospital, Sofia for phenotypical and genotypic characteristics.
Methods: 160 ESBL-producing clinical isolates: K. pneumoniae (n = 80), E. coli (n = 68) and S. marcescens (n = 12) were selected and were characterised by Sceptor MIC/ID system. The ESBL production was confirmed. The detection and molecular characterisation of the ESBL-encoding genes of the SHV and CTX-M family were performed by PCR and PCR-RFLP. RAPD analysis has been used to type a diversity of microorganisms.
Results: Antimicrobial profiles were demonstrated high degree of diversity of resistance phenotype. All K. pneumoniae strains were examined by PCR for the presence of blashv genes and a positive amplification was observed for 60 tested isolates. The data from PCR-RLFP analysis with NheI were showed point mutation at position 238 in all cases. PCR amplification with CTX-M-type specific primers was positive from 68 E. coli, 30 K. pneumoniae and 12 S. marcescens. As was demonstrated PCR-RLFP analysis with PstI and PvuII, all CTX-M β-lactamases found in tested strains was belonged to the CTX-M-1 cluster. 22 distinct RAPD pattern were obtained for K. pneumoniae, 18 for E. coli and 2 for S. marcescens, respectively. Isolates from the same wards mostly belonged to 1 or 2 major clones, but we also observed a number strains with unique patterns. Cases of clonal relatedness between strains from distinct wards were found.
Discussion: In this study we presented the results of typing ESBL-producing Enterobacteriaceae strains by combination of phenotypic and genomic markers. Antimicrobial profiles have revealed the high degree of diversity of resistance phenotype of ESBL-producing strains. SHV ESBLs have been found predominantly in K. pneumoniae, while CTX-M in E. coli and S. marcescens. All CTX-M-producung strains were belonged to the CTX-M-1 cluster. The results of genotypic typing (RAPD) were indicated that several clones of ESBL-producing Enterobacteriaceae were associated with either sporadic infection or cross-infections or outbreaks. The results indicate that ESBL-producing bacteria are present a significant problem and that clonal spread within hospital may be occurred.
P1858 Molecular typing and pathogenic potential of Listeria monocytogenes isolates from food and clinical origin
N. Papageorgiou, E. Scoulica, C. Panoulis, S. Maraki, A. Christidou, Y. Tselentis (Heraklion, GR)
Objectives: The purpose of this study was to correlate the molecular characters and pathogenic phenotype of Listeria monocytogenes isolates, of both food and clinical origin which are temporarily and geographically related. Typing included Pulse Field Gel Electrophoresis (PFGE), ribotyping and genetic polymorphism of the prfA, plcA, actA and hly genes. The haemolytic activity of the strains was used for the assessment of pathogenicity.
Methods: 92 Listeria monocytogenes strains isolated from food and food processing environment from Massive Feeding Systems in the island of Crete in Greece and 5 clinical isolates cultivated from 5 patients hospitalised in the University Hospital of Heraklion Crete. For the PFGE, the AscI and ApaI restriction enzymes were used. Automated ribotyping was performed with EcoRI restriction enzyme and the region that includes the prfA, plcA, hly, and actA virulence genes was completely sequenced.
The haemolytic activity of the strains was investigated by measuring spectrophotometrically the lysis of human red blood cells.
Results: A total of 11 different Ribotypes and 23 Pulsotypes were found among 93 different Listeria monocytogenes isolates analysed.
ActA gene was highly polymorphic. Phylogenetic analysis of ActA protein sequences showed that Listeria monocytogenes isolates arranged into 2 main clusters that could be further divided into 12 subclusters.
According to their haemolytic activity strains grouped into a high, intermediate and low cytotoxicity.
Strains with zero haemolysis had accumulated mutations in the PrfA protein.
Conclusion: Listeria monocytogenes strains belong to different clonal groups. Both food and clinical isolates showed differences in haemolytic activity in vitro. As actA gene was highly polymorphic the DNA sequencing of this gene can be used as a useful tool for epidemiological studies. Mutations in the PrfA protein seem to correlate with variability in haemolytic potential of the strains.
P1859 Variable number of tandem repeats of S. typhimurium in Ireland
G. Doran, N. DeLappe, J. O' Connor, D. Morris, M. Cormican, B. Lindstedt (Galway, IE; Oslo, NO)
Objectives: The discriminatory power of variable number of tandem repeats (VNTR) was compared to that Pulsed Field Gel Electrophoresis (PFGE) for the analysis of isolates of DT104 and related phage types. The current “gold standard” method for molecular typing of Salmonella spp. is PFGE. This has poor discriminatory power within certain phage types e.g. DT104 and DT104b. As these phage types account for most S. Typhimurium isolates a more discriminatory method is desirable.
Methods: Isolates of DT104 (161) DT104b (107) and U302 (2) were analysed by PFGE according to the Pulse-Net standardised procedure and by VNTR according to Lindstedt et al. Isolates were collected from 2000 to 2006 and were from human (207), poultry (27), bovine (19), swine (13) and other sources (4). Fifty isolates were from 15 known small outbreaks. Profiles were analysed by Bionumerics. VNTR profiles were forwarded to the Norwegian Institute of Public Health, for comparison with known VNTR profiles.
Results: PFGE of the 270 isolates yielded 7 patterns, with 95% belonging to a single pattern. VNTR yielded 88 distinct profiles. All isolates from within a particular outbreak were of the same VNTR profiles. The number of isolates in each profile ranged from single isolates to 34 isolates. No VNTR profile was confined to a particular animal source. Some VNTR profiles encompassed isolates which were isolated during a short period of time and therefore may represent previously undetected outbreaks. VNTR profiles were compared to those in the Norwegian database. Thirty eight Irish profiles were new to the Norwegian bank. Irish profiles were indistinguishable to human isolates from Norway (10), Cyprus (2), Spain (5), Romania (1), Poland (1) and a canine isolate from Norway.
Conclusion: VNTR was more discriminatory, less time consuming and less labour intensive than PFGE. It clustered together known outbreak strains and highlighted other clusters which were also probable outbreaks. The profiles were comparable to profiles from other European countries and many were unique to Ireland.
P1860 Use of multilocus enzyme electrophoresis to examine genetic relationships among isolates of Klebsiella pneumoniae
M. Rahmati, G. Etemadi, S. Asadi, A. Abdi-Ali, M. Feizabadi (Tehran, IR)
Objectives: Multilocus enzyme electrophoresis was carried out in order to epidemiological studies and to evaluate the genetic relationships of Klebsiella pneumoniae in Tehran. The characterisation of electrophoretic variants of metabolic enzymes reflects allelic variations at the corresponding structural gene locus, and defines as electrophoretic types (ET) which correspond to genotypes. The aim of this study was to determine the antibiotic susceptibility pattern, detection of ESBLs and analyse the genetic relationships between the strains by Multilocus Enzyme Electrophoresis (MEE). No information to date is available on the population genetic analysis of K. pneumoniae in Iran
Methods: 100 isolates of K. pneumoniae were collected from different clinical samples in two hospitals in Tehran. In addition to antibiotic susceptibility testing, Extended spectrum β-lactamase enzymes were detected by both Double Disk Synergy Test (DDST) and Phenotype Confirmatory Test (PCT). Population genetic of isolates was determined by Multilocus Enzyme Electrophoresis.
Results: Of 100 isolates, 56% and 20% were from urinary and respiratory tracts infections, respectively. The other 24% were from other sources such as blood, wound, CSF, ear, eye, vagina, bone and stool infections. The highest resistance of isolates was to ampicillin (97%) and aztreonam (77%), respectively. All strains were susceptible to imipenem and meropenem. 46% of isolates were ESBLs positive and the resistance of these strains to ampicillin and cephalexin was 97.83% and 80.43% respectively. Analysis of the strains with 17 metabolic enzymes produced 51 electrophoretic types (ETs) There was more than one strain in 13 ETs, whereas other strains classified in separate ETs. Genetic diversity among strains varied from leucin-tyrisin peptidase (0.733) to Glucose 6-phosphate dehydrogenase (0.387). Mean genetic diversity among these strains were determined 0.515.
Conclusion: This present work shows high rate of infections with ESBL strains in Tehran. MEE revealed the close relationship among isolates of K. pneumoniae from patients at Tehran hospitals. However polymorphic loci such as leucin-tyrosin peptidas with a high genetic diversity (0.733) were observed, MEE differentiated the strains with ESBL phenotype. MEE as a chromosomal marker is a reliable technique for typing and discriminating of bacterial population.
P1861 Genetic profiles of intestinal Escherichia coli isolates from Romanian subjects
C. Usein, A. Palade, N. Popovici, L. Grigore, D. Tatu-Chitoiu, S. Ciontea, M. Damian, M. Nica (Bucharest, RO)
Objectives: The intestinal microbiota is routinely regarded as a natural reservoir of strains that might influence morbidity in humans. Escherichia coli is a complex species comprising several clones that could either harmlessly colonise or aggress the host, causing intestinal and extraintestinal illness. The bulk of E. coli population is divided into four main phylogenetic groups, namely A, B1, B2 and D, and a link between phylogeny and virulence was observed.
In an attempt to gain insight into the composition of autochthonal E. coli human population, we investigated some genotypic characteristics of strains isolated from the intestinal reservoir of 27 healthy subjects (135 isolates), and 49 subjects with intestinal symptoms (141 isolates).
Methods: The phylogenetic grouping, and the presence of determinants specifically associated with E. coli intestinal pathovars were assessed by PCR-based methods. Pulsed-field gel electrophoresis of XbaI macrorestriction DNA fragments was used for evaluating the clonal relatedness among circulating strains.
Results: Only 4 of the subjects with diarrhoea carried pathogenic E. coli strains identified as EPEC and VTEC. These strains belonged to B1 and A phylogroups. Strains of phylogenetic group A significantly predominated in both healthy subjects and individuals with acute diarrhoea (66% vs. 63%). In contrast, strains belonging to B1 phylogenetic group were rare (7%), and were restricted to faecal isolates from diarrhoea stools. A comparable number of group B2 (14% vs. 20%) and D (18% vs. 11%) strains was identified in both intestinal collections. The PFGE patterns analysis revealed the genomic diversity present among the intestinal isolates of E. coli.
The PCR negative results obtained when targeting virulence determinants associated with intestinal pathogenicity of E. coli could exclude this cause of the symptoms, and the great majority of the tested E. coli faecal isolates in this study could be considered as representing the normal colonic flora. At the same time, if ExPEC strains are considered to belong mainly to group B2, and to a lesser extent to group D, one sixth of the studied isolates could possess the capability of causing infections when exitting the intestinal niche.
Conclusion: The specific phylogenetic distribution of E. coli isolates from Romanian subjects supports the importance of understanding the structuring of E. coli population as a result of adaptation to various hosts and geographic environments.
P1862 Evaluation by molecular tools of Brucella persistence in soil
R. Koncan, R. del Campo, S. Bianchi, A. Amendola, A. Mazzariol, R. Cantón, F. Baquero, G. Cornaglia (Verona, IT; Madrid, ES; Milan, IT)
Background: Persistence in soil of pathogens such as Brucella has never been explored with the newly available molecular tools. The aim of this work was to investigate the biological risk in the waste pit of a vaccine-producing factory, in which a number of vials originally containing viable Brucella were being buried over 30 years and subsequently disrupted and spilt in the soil during the reclamation procedures.
Methods: The contents of the unbroken vials found in the waste pit were inoculated in Brain-Heart broth, and incubated for 7 days. Positive cultures were sub-cultivated onto Columbia agar plates, and a morphological analysis of the resulting colonies was performed. For bacterial identification, DNA was extracted from the different colonies and amplified by PCR with general primers based on conservative region of the 16S ribosomal. Positive amplicons were sequenced and compared by Clustal analysis with the Genbank data. Specific primers for Brucella spp. were also used with the DNA extracted from the vials' content.
Total soil DNA was extracted by the FastDNA® SPIN Kit for soil (Qbiogene, Carlsabd, CA, USA). Qualitative bacteriological analysis of the soil was performed by DGGE experiments, using clinical isolates of Brucella as the positive controls. Dot-blot assays were also performed with both the total DNA from soil and the bacteria grown on Brain-Heart.
Results: A total of sixty-nine unbroken vials were studied, six of them labelled as containing Brucella. Positive cultures were obtained in twenty-nine vials (42%), and they yielded in all cases contaminant environmental bacteria, namely Paenibacillus barengoltzii, Paenibacillus dendritiformis Bacillus thuringensis, Bacillus licheniformis, Bacillus benzoeorarus, and Ochrobactrum spp. Positive amplification with the specific primers for Brucella was not successful in any case. This result was also confirmed by the dot blot experiments. In the DGGE experiments with the DNA extracted from soil, no band corresponding to the Brucella size was detected.
Conclusion: Modern molecular tools might help to carry out a thorough reclamation of putative contaminated areas whilst minimising the potential biological risks. In particular, they allow assessment of possible soil contamination by highly infectious bacterial pathogens such as Brucella, avoiding risk exposure related to the classical culture techniques.
Molecular fungal diagnosis
P1863 Specific detection of Candida albicans using real-time PCR on the LightCycler
L. O'Connor, M. Ruttledge, M. Maher (Galway, IE)
Objectives: Candida albicans is the fourth most common cause of bloodstream sepsis in humans. It is estimated that the annual incidence of severe sepsis in Europe is in the range of 44000–95000 cases. The ability to diagnose and identify sepsis arising from C. albicans blood stream infections in the clinical setting is greatly enhanced by the use of nucleic acid diagnostics (NAD). It is important to rapidly identify C. albicans as the cause of sepsis in order to rule out bacterial infection and quickly facilitate the administration of appropriate therapy. Traditional diagnosis of C. albicans infections can take from 3 to 5 days and requires specialised and experienced staff. Broad spectrum antibiotics are often administrated before these results are available, and along with their ineffectiveness against C. albicans, they also increase the risk of microbial antibiotic resistance characteristics. To aid in rapid detection of C. albicans a nucleic acid diagnostic test using real-time PCR on the LightCycler™ has been developed.
Methods: A specific labelled primer-probe combination was designed to amplify and detect a segment of the HWP1 gene. The specificity of the test was verified using DNA extracted from geographically distinct C. albicans reference strains and clinical isolates. In addition, DNA from other Candida and non-Candida fungal species was also tested. The specificity of the test was further cross-checked against DNA from a panel of bacterial species and human DNA. The detection limit of the test was established using as template both DNA extracted from serially diluted overnight culture of C. albicans and DNA from blood spiked with known numbers of C. albicans cells. DNA for all experiments was extracted on the MagNA Pure system using DNA isolation kit III (Roche).
Results: The test was 100% specific for C. albicans with no cross reaction with any other non-albcians Candida species, other fungal species, bacterial or human DNA. The test was also found to be highly sensitive, reliably detecting between 1 and 10 copies of the target gene from both culture and spiked blood samples.
Conclusion: A rapid, sensitive nucleic acid test for detection of the clinical pathogen C. albicans has been developed for use on the LightCycler™. The key advantage of the test is its specificity for C. albicans since over 50% of fungal infections are caused by the organism.
P1864 A real-time PCR assay for detection of six common species of Candida from blood, validated in adult critically ill patients
R. McMullan, L. Metwally, P. Coyle, S. Hedderwick, B. McCloskey, H. O'Neill, H. Webb, R. Hay (Belfast, UK)
Objectives: One of the major obstacles to improving patient outcomes in invasive Candida infection is lack of a sensitive, specific and rapid test to confirm the diagnosis and provide an early guide to the species implicated. Such a tool, from which prescribers may infer the likely antifungal susceptibility profile within a few hours of collecting a specimen of blood, has the potential to influence therapy at an early stage and may improve patient outcomes. We have developed such a diagnostic test and have evaluated its reliability in adult critically ill patients.
Methods: Three Taqman-based real-time PCR assays were developed for identifying up to six Candida species. Target sequences in the rRNA gene complex were amplified, using a consensus nested PCR protocol, to identify Candida albicans, C. parapsilosis, C. tropicalis, C. dubliniensis, C. glabrata and C. krusei. These take approximately six hours to complete.
The assays were validated in a single adult intensive care unit (ICU) over a 16-month period. All patients remaining in the ICU for at least 48 hours were eligible for inclusion and consent was sought from all. Whole blood and clotted blood specimens were obtained from participants twice weekly and on each occasion, participants were categorised either as ‘unlikely’ fungal infection, ‘probable’ or ‘proven', based on published consensus definitions endorsed by an EORTC group for use in clinical trials.
Results: During the study period, 203 patients were evaluated and were categorised as follows: unlikely = 178, probable = 6, proven = 19. All patients in the unlikely and probable categories tested PCR negative. Fifteen of the 19 patients with proven Candida infection tested PCR positive with four testing negative, although one of these had C. famata infection. Using the definitions of ‘proven’ and ‘unlikely’ infection as the reference standards (excluding the more ambiguous ‘probable’ group) the assays had sensitivity of 79% and specificity of 100%. In this population, when no stringent selection criteria for applying this diagnostic test were applied, the NPV was 98%, with a PPV of 100%.
Conclusions: These assays provide a reliable diagnostic modality for rapidly detecting invasive Candida infection in this population as an adjuct to standard care. Furthermore, speciation, which allows prediction of azole susceptibility is available on the same day, helping prescribers to optimise antifungal therapy at an early stage.
P1865 Detection and identification of Aspergillus spp. and Candida spp. by real-time PCR in clinical samples
M.J. Torres, A. Aller, M. Ramírez, C. Castro, M. Ruiz, J.M. Cisneros, I. Espigado, J. Aznar, E. Martín-Mazuelos, J.C. Palomares (Seville, ES)
Objective: To develop and evaluate prospectively a real-time PCR assay for detection and identification of Aspergillus spp. and Candida spp. in patients with suspected IFI.
Methods: Referenced cultures of Aspergillus species and Candida species were included to determine the analytical sensitivity and specificity of the assay.
The amplification method used the 18S rRNA genes and the identification of the genus was made by two specific pair of probes. The analytical sensitivity of the process was evaluated with suspensions of A. fumigatus and Candida albicans in five concentrations (101 to 105 cfu. per ml), and serially diluted DNA of the referred species.
Total genomic DNA extracted from 948 blood samples obtained from 127 patients suspected of having IFI were used.
Results: No cross-reactivity was obtained with any of the collection of pathogenic and non-pathogenic bacteria and fungi used in the study.
Species identification was determined by analysing the Tm of the melting curves obtained with the specific probes, ranging from 67.34°C to 70.7°C for Aspergillus spp. and from 51.3°C to 64.5°C for Candida spp. Analytical sensitivity for Aspergillus fumigatus was 60 fg using DNA and 15 conidia using conidial suspensions; and 100 fg and 3 cells for Candida albicans.
There were 2 cases of proven IA, 3 cases of probable IA and 14 cases of possible IA. One patient was classified as having probable Candidiasis, and 6 cases possible Candidiasis. Only patients with serial positive results were considered to be PCR positive.
The 2 IA proven cases and 2 of 3 IA probable cases were PCR positive for Aspergillus and all the cases with probable/possible Candidiasis had positive PCR results for Candida albicans.
Conclusions: The Light Cycler technique is rapid, accurate, and reproducible and combines rapid in vitro amplification with real-time species determination and quantification of the fungal load. The linear range of the assay was from 6 to 6×107 fg of Aspergillus DNA and from 10 to 107 fg of Candida DNA. The assay used provides high sensitivity and specificity for fungal detection but more cases are needed to elucidate the true potential of the technique.
P1866 Comparison of PCR-Reverse Line Blot analysis and traditional culture of dermatophytes in clinical samples
G. Wisselink, E. van Zanten, K. van Slochteren, M. Kooistra-Smid, A. Bergmans, R. Wintermans (Groningen, Roosendaal, NL)
Objectives: Traditionally, laboratory detection and identification of dermatophytes consists of culture on selective media and potassium hydroxide (KOH) tests. This process yields positive results within approximately 2–6 weeks, and negative results are generated after 6 weeks of incubation. Using PCR followed by Reverse Line Blot (PCR-RLB) analysis it becomes possible to obtain positive and negative results within 2–3 days. In this study we compared traditional culture with PCR-RLB analysis.
Methods: One hundred clinical samples (92 nail, 8 skin) were analysed retrospectively by PCR-RLB after traditional culture. Samples were processed using QIAamp® DNA mini kit (Qiagen, Germany) with a separate pre-lysis step. PCR targeted the ITS region between the genes coding for 18S and 5.8S rRNA. PCR products were analysed using RLB [Bergmans et al., in preparation]. The membrane harboured 13 different probes to identify and discriminate between 9 different dermatophyte species within 3 genera, namely; T. rubrum, T. mentagrophytes, T. interdigitale, T. tonsurans, T. violaceum, T. verrucosum, M. canis (complex), M. audouinii and E. floccosum.
Results: Culture, KOH and PCR-RLB analysis yielded 18/100, 39/99 and 58/100 positive results respectively. Six samples showed inhibition in the PCR-RLB; one of these samples was KOH positive and all were culture negative. All 18 culture positive samples scored positive in the PCR-RLB. Seventeen of these 18 samples were identified as T. rubrum both by PCR-RLB and by culture; one sample was identified as T. mentagrophytes by culture, whereas PCR-RLB identified this sample as T. interdigitale. Forty samples scored positive in PCR-RLB but negative in culture. In these samples PCR-RLB identified 27 T. rubrum, 8 T. interdigitale, 2 Trichophyton spp. and 3 unidentified fungi. In twenty samples PCR-RLB scored positive where both culture and KOH scored negative. Sensitivity for the PCR-RLB compared to culture is 100% (18/18), compared to the KOH the sensitivity of PCR-RLB is 92% (35/38)
Conclusion: These data show PCR-RLB to be a fast and very sensitive method to detect and identify dermatophytes compared to traditional culture methods. Molecular assays are known to be more sensitive than culture or microscopic techniques. Therefore it is very likely that the PCR-RLB positive, culture negative samples are in fact positive.
P1867 Detection of fungi in hospital water supplies using molecular beacons
P. Bowyer, L. Hoare, D. Denning (Manchester, UK)
Objective: Immunocompromised patients are susceptible to fungal disease and it is thus extremely important to both prevent exposure to fungi via hospital water supplies and monitor possible sources of exposure in units dealing with such individuals. Sensitive, rapid detection and identification of fungi in these environments is therefore important. Black materials were demonstrated in water in a single leukaemia ward and an investigation was launched to identify the source of the contamination.
Methods: Samples were prepared by filtration of 1l of water through a 0.4 micron filter followed by direct extraction from the filter using a MoBio Ultraclean Soil DNA kit and the presence of fungus was determined using RT-PCR with a fungal specific molecular beacon. The beacon was constructed so as to recognize all fungi and provide a basic measure of the level of fungal contamination. Fungal biomass was quantified using a standard curve based on cloned Aspergillus target DNA and a further curve based on serial dilutions of genomic DNA.
Results: 26 water samples were processed. Results show presence of high levels of fungal DNA in water from 3 particular tap and shower sources on particular days. The fungus was identified as Ochroconis by standard mycological methods. No infections were documented. Subsequently the mains water was provided from a separate uncontaminated source.
Conclusion: These results show the usefulness of beacon based molecular diagnostics for the rapid detection of contaminating fungi. This provides a rapid and sensitive method to identify fungal contaminants.
P1868 Identification of dermatophytes by using in-house databases of sequences of the ribosomal DNA internal transcribed spacers
H.C. Li, J.P. Bouchara, M.M. Hsu, R. Barton, T.C. Chang (Tainan, TW; Angers, FR; Leeds, UK)
Objectives: The aetiologic agents of the dermatophytoses are classified in three anamorphic (asexual) genera, Epidermophyton, Microsporum, and Trichophyton. The traditional identification of dermatophytes is sometimes delayed or problematic because of atypical microscopic structures or colony appearances. This study evaluated the feasibility of sequence analysis of the ribosomal DNA internal transcribed spacer 1 (ITS1) and 2 (ITS2) for identification of dermatophytes.
Methods: The ITS regions of 71 reference strains (19 species) were amplified by PCR and sequenced. The determined sequences and reference sequences available in GenBank were used to construct the in-house ITS1 and ITS2 databases. The efficacies of the databases were then validated by testing 131 clinical isolates of dermatophytes that were identified to species level by morphological characteristics. Strains producing discrepant identifications between the conventional methods and ITS sequence analysis were further analysed by sequencing of the D1-D2 domain of the large-subunit rRNA gene for species clarification.
Results: By sequence comparison with the in-house ITS1 and ITS2 databases, the correct identification rates of clinical isolates by ITS1 and ITS2 sequence analysis were 98.7% and 100%, respectively. In addition, signature sequences were found in the ITS1 regions of Trichophyton rubrum and T. violaceum, and in the ITS2 regions of Microsporum audouinii and M. ferrugineum. These signature sequences are useful for species delineation in case of the result of sequence comparison is ambiguous.
Conclusion: Identification of medically relevant dermatophytes by ITS sequencing, especially the ITS2 region, is reliable and can be used as an accurate alternative to the conventional identification methods. The present method could be finished within approximately 24 h starting from isolated colonies.
P1869 Evaluation of a new commercial real-time PCR for the detection of Aspergillus spp. in serum and respiratory samples
M.P. Hayette, C. Meex, R. Boreux, P. Huynen, P. Melin, P. De Mol (Liege, BE)
Objectives: Diagnosis of invasive aspergillosis is still disappointing and often delayed because of the lack of sensitivity of diagnostic tools. DNA detection based-methods have been developed, but differ widely and comparisons are difficult to assess. The objective of the study is to compare a new commercial real-time PCR kit, affigene® Aspergillus tracer assay, with an in house nested PCR targeting 18S rRNA Aspergillus sp. gene.
Methods: Twelve patients at risk for invasive aspergillosis were included in the study. They were classified to have possible (5 cases), probable (1 case) or proven (6 cases) invasive aspergillosis following E.O.R.T.C. criteria. Fifteen serum and respiratory paired samples were collected. The DNA extraction was performed by using the QIAmp DNA mini kit® (Qiagen, Germany). All samples were tested by both PCR assays and respiratory samples were cultured.
Results: Respiratory samples. A. fumigatus, A. niger and A. flavus were isolated from 10/15 samples; both PCR methods were positive for these samples except one that was positive for affigene® and equivocal for the nested PCR. The real-time PCR assay reported cycle thresholds ranging from 25 to 38. Three of the five culture-negative samples were negative by both PCR methods; one of three was negative in affigene® assay and equivocal by nested PCR; the last sample was positive in affigene® assay and negative by nested PCR. Serum. Thirteen of fifteen blood samples were negative by both PCR methods. One sample was equivocal by nested PCR and was inhibited in affigene® assay despite a culture-positive paired respiratory sample. The last case was inhibited by the real-time PCR assay and negative by nested PCR. Nor the nested PCR, nor affigene® assay could detect any Aspergillus DNA in serum. In total, there was 93% of agreement between the two PCR assays.
Conclusion: Both methods are in good agreement and can detect at least three different species of Aspergillus. However, the sensitivity of both assays does not permit the detection of Aspergillus DNA in serum. affigene® assay can easy replace the “in house” assay: it allows a fast and standardised detection of Aspergillus sp. DNA in respiratory samples without inconvenient due to the handling of PCR products.
Community-acquired bacterial respiratory tract infections
P1870 Prediction of the tuberculosis reinfection rate from the local incidence
P.R. Hsueh, J.Y. Wang, L.N. Lee, H.C. Lai, H.L. Hsu, Y.S. Liaw, P.C. Yang (Taipei, TW)
Background: Reinfection is a major contributor to the development of tuberculosis (TB). It seems that the higher the local incidence, the higher the frequency of reinfection. Based on a systematic review of the literature, we established a regression model to predict the reinfection rate from the local incidence and verified it using our local data.
Methods: We first reviewed the literature for population-based studies. The relationship between the reinfection rate and local incidence was examined to generate a regression model. We then used our local data, obtained by genotyping the clinical isolates from recurrent patients from 1999 through 2004, to verify the algorithm developed.
Results: Of the 24 studies reporting the contributions of reinfection in recurrent TB, five were population-based. Statistical analysis showed that the reinfection rate was significantly correlated with the local incidence (Reinfection Rate = −32.636+44.061 Log Incidence) (p = 0.010, R2 = 0.906). Accordingly, the reinfection rate in Taiwan (incidence: 62.4 per 100,000 people) was estimated to be 46.5%. In our hospital from 1999 through 2004, 25 (51.0%) of the 49 recurrent patients were reinfection.
Conclusions: The regression model we have developed could possibly predict the TB reinfection rate from the local incidence. This reinfection prediction algorithm is important and probably helpful in planning and policy making for TB control programmes. In endemic areas, such as Taiwan, reinfection might be responsible for >50% of TB cases, and aggressive surveillance to detect asymptomatic carriers (case finding) could be a more important strategy than direct observed therapy for controlling the disease.
P1871 Fluctuation in Legionella pneumophila counts and persistence of DNA subtypes in 15 cooling towers over a one-year period
S. Ragull, M. Garcia-Nuñez, N. Sopena, M. Pedro-Botet, M. Esteve, R. Montenegro, M. Sabria (Badalona, Barcelona, ES)
Introduction: The presence of Legionella in cooling towers has been well documented and implicated in most community outbreaks. However, Legionella counts seem to vary over time. The genotypic variability and the persistence of Legionella DNA subtypes in cooling towers can help to identify the source of the outbreak.
Objectives: The aims of this study were to describe the fluctuation in Legionella counts in cooling towers over a one-year period, the variability of the chromosomal DNA subtypes and their persistence.
Methods: Fifteen cooling towers, located within a 3-km radius were selected. All the cooling towers underwent the same maintenance regimen. The towers were sampled fortnightly over one year. Water samples were concentrated by filtration and seeded on agar Legionella GVPC. Four to 10 colonies were selected from each positive culture and analysed by Pulsed Field Gel Electrophoresis.
Results: Legionella pneumophila was isolated in 13 out of 15 cooling towers. Five cooling towers were positive only once (range: 66–306,666 CFU/L) and one cooling tower was positive twice (500 CFU/L, 3,333 CFU/L). In the remaining 7 towers, concentrations of Legionella fluctuated during the study period ranging from negative cultures up to 2×106 CFU/L in some samples.
Sixteen different DNA subtypes were obtained among the 13 positive cooling towers. Seven cooling towers had a single DNA subtype. Three showed two different DNA subtypes and another 3 showed three different DNA subtypes. The same DNA subtype persisted in the 8 cooling towers which were positive in more than one sample.
Conclusions: Concentrations of Legionella pneumophila in cooling towers fluctuate considerably over time. The temporal variation of Legionella counts in cooling towers limits the usefulness of infrequent sampling to know the Legionella status. In small geographical areas the same indistinguishable DNA subtypes may be shared among different cooling towers and persist over time.
P1872 Study of the possible casual effect of Chlamydophila pneumoniae on COPD exacerbation
S. Shokouhi, B. Hajikhani, M. Godazgar, M. Samanabadi, M. Sattari, H. Jamaati, S. Sazgar (Tehran, IR)
Objectives: Chlamydophila pneumoniae is one of the common causative agents of respiratory system infections. The present study aims to find out the role of Chlamydophila pneumoniae infection in infectious exacerbation of chronic obstructive pulmonary disease (COPD).
Methods: 65 nasopharyngeal swab specimens of chronic obstructive pulmonary disease (COPD) patients were studied using fluorescent antibody staining with Chlamydia specific conjugated antibody and with fluorescent microscopes. The patient's data was analysing by using SPSS Software (version 13).
Results: A total number of 65 COPD patients (as defined by the American thoracic society), 53 (81.5%) male and 12 (18.5%) female were included in the study. 46 (70.7%) subjects had COPD in exacerbation period, 19 (29.3%) were stable COPD patients. We found 4 positive cases of Chlamydophila infection (6.15%), 3 of which (2 men and 1 woman) belonged to the exacerbation group and 1 to the stable COPD patients.
Conclusion: Data analysis revealed that there wasn't any significant correlation between Chlamydophila infection and COPD exacerbation (P = 0.848).
P1873 Complaints of cough in patient older than 50 in general practice: an exacerbation of COPD
L. Broekhuizen (Utrecht, NL)
Objectives: to determine whether COPD (Chronic Obstructive Pulmonary Disease) is present in patients, older than 50, who present themselves with complaints of cough at the general practitioner and are not yet known with COPD.
This objective is one of the objectives of the FRESCO-project. The main objective of this project is to determine the cost-effectiveness of the most optimal diagnostic strategy to diagnose COPD in patients older than 50 with complaints of cough in general practice.
Methods: This is a an observational, prospective diagnostic study.
The study population consists of four hundred patients who are older than 50 years of age and present themselves with a new episode of cough at the general practitioner and are not known by their general practitioner with COPD. Inclusion of these patients started in January 2006 by 48 general practitioners in the eastern part of the Netherlands. Information on signs and symptoms as well as results of additional tests, namely lung function tests and repeated blood tests (C-reactive protein and Brain Natriuretic Peptide) are gathered by the general practitioners, both during the episode of acute cough as six weeks later, when the patient is in a stable condition concerning cough and other respiratory symptoms. In addition, the effect of a diagnostic treatment with prednisolone in stable condition is assessed. Finally, at day 90, all patients visit an out-patient clinic where state-of-the-art lung function measurements are done. After this, for all patients the final diagnosis, namely COPD or no COPD and/or other diagnoses like for example pneumonia or lung carcinoma, is determined by an expert panel, which consists of a pulmonologist and a general practitioner. This diagnosis is based on the results of all diagnostic tests.
Results: At this moment, 100 patients have been included in the study. The first results of these patients show that 40 percent of the included patients with cough have a chronic lung disease, namely mainly COPD and in fewer cases asthma, that was unknown before.
Other relevant diagnoses were heart failure in one patient, pneumonia in two patients and one patient had disseminated lung cancer.
Conclusion: in a large part of the patients that are not known with the diagnosis COPD that present themselves with cough in general practice, these complaints are in fact an exacerbation of COPD.
P1874 Clinical characteristics of patients with Haemophilus influenzae meningitis in Denmark, 1994–2005
T.I. Pedersen, M. Howitz, C. Østergaard (Herlev, Copenhagen, DK)
Objectives: When a new vaccine is introduced in vaccination programmes the epidemiology of the disease covered by the vaccine is expected to change (e.g. disease in other age groups, infection with other non-vaccine types). Our aim is to present clinical and paraclinical information on Haemophilus influenzae (Hi) meningitis cases in Denmark (pop. ∼5 mio.) after the introduction of the Hi type b vaccine in the Danish childhood vaccination programme in 1993.
Methods: Information on cases with Hi meningitis has been collected consecutively since 1980 in the Danish national notification system. Additional clinical and laboratory findings, including vaccination status, were collected retrospectively from medical records from the period 1994–2005.
Results: In the period 1994–2005 52 cases with Hi meningitis were notified. 24 cases (46%) were less than 5 years of age; median age 14 months (range 0–45), and 28 cases (54%) were 5 years or older; median age 54 years (range 5–88). No cases were in the age group 8 to 24 years. Significantly more cases under the age of 5 years were infected with Hi type b (54%, 13/24) as compared to cases 5 years or older (18%, 5/28, Fisher's exact test P <0.01). In the age group under 5 years of age 39% (5/13) of cases with Hi type b meningitis were known to have received one or more doses of Hi type b vaccine and 3 of these cases (60%, 3/5) could be defined as vaccine failures. The remaining 8 children had either not been vaccinated or had unknown status. Among children under 5 years of age, a predisposing condition was found in 29% (7/24) of cases; 6 cases with otogenic focus and 1 case with CSF leakage. Hi type b accounted for 71% (5/7) of these cases. In cases 5 years or older a predisposing condition was found in 46% (13/28); 9 with an otogenic focus, 1 with sinusitis and 3 with CSF leakage.
3 patients (6%) died due to meningitis; 1 premature infant and 2 adults at 84 and 86 years of age. None of these cases were due to Hi type b.
Hearing impairment among survivors was reported for 23% (6/26) of cases 5 years or older; 60% (3/5) after a type b and 14% (3/21) after a Hi type non b meningitis.
Conclusion: Meningitis caused by Hi type b is rare in Denmark after implementation of the vaccine in the childhood vaccination programme. Those cases with Hi meningitis we observed had a clinical presentation and disease outcome as expected, with regards to case fatality rate, frequently an otogenic focus and a significant risk of hearing loss.
P1875 Evolution of H. influenzae resistance in a chest diseases hospital, for the period 2001–2006
N. Tsagarakis, A. Gioga, M. Makarona, H. Moraitou, N. Kentrou, M. Panagi, A. Pefanis, S. Damianidou, S. Kanavaki (Athens, GR)
Objectives: The aim of this study was to investigate the resistance patterns of H. influenzae, from strains isolated in ‘Sotiria’ Chest Diseases Hospital of Athens, within the period September 2001 to September 2006.
Material and Methods: 700 H. influenzae strains were isolated from respiratory samples (sputum and bronchial secretions). The culture and antibiotic susceptibility tests (Kirby-Bauer) were performed according to NCCLS 2004 guidelines, including the following antibiotics: ampicillin, amoxicillin/clavulanic acid, cefalothin, cefuroxime, erythromycin, co-trimoxazole, tetracycline and ciprofloxacin. The β-lactamase production was tested with nitrocefine disks (OXOID, UK).
Results: Among all isolates: 152/700 strains were β-lactamase positive (21.7%), while a gradual increase of the percentage of β-lactamase positive strains was noted from 2001 (10.9%) to 2006 (27.93%). 167/700 strains were ampicillin-resistant (23.8%). The percentage of the ampicillin-resistant strains was gradually increased from 10.9% in 2001–02 to 29.73% in 2005–06. Within the total of ampicillin-resistant strains, 152/167 (91%) were β-lactamase positive (BLPAR), whereas 15/167 (9%) were β-lactamase negative (BLNAR). A consistent increase of the percentage of BLNAR strains is noted from 0% in 2001–02, to 9% in 2005–06. A low rate of ciprofloxacin resistance was noted for the study period (0.9%). The percentage of the strains resistant to erythromycin was almost doubled from 49.5% in 2001–02 to 90.01% in 2005–06
Conclusions: It is interesting that the prevalence of β-lactamase positive H. influenzae isolates has been significantly increased (3×) and BLNAR strains have also emerged throughout the decade. In addition, erythromycin resistance is of great concern, since it has been double raised recently.
The evolution of H. influenzae resistant patterns should be taken under consideration, when empiric antibiotic treatment is offered for upper respiratory system infections.
P1876 Epidemiology of Haemophylus influenzae and meningococcal invasive diseases in a Spanish hospital (1987–2006)
R. Blanco, M. Unzaga, C. Ezpeleta, J. Alava, I. Guerediaga, B. Amezua, V. Cabezas, R. Cisterna (Bilbao, ES)
Objective: To describe the epidemiology of serogroup C meningococcal and Haemophilus influenzae invasive diseases and evaluate the efficacy of vaccination against them in Basurto Hospital.
Material and Methods: In the Basque Country scheduled vaccination with meningococcal conjugate vaccine (MCV4) began in 2000, previous sporadic campaigns of vaccination were performed since 1997. In 1996 the vaccine against Haemophilus influenzae was implemented too.
We evaluated the number of cases of CSF and bloodstream infections in the pre-vaccination period compared with those in post-vaccination period.
Results: The number of cases of serogroup B meningococcal invasive disease was 62 and 77 in the prevaccination period and post-vaccination period, respectively. The number of cases of serogroup C meningococcal invasive disease (SCMID) in the pre-vaccination period were 59 and 29 in the post-vaccination period; only 5 of them were detected in CSF. Among persons of 18 years and younger, the number of cases of SCMID reduced from 49 cases in the pre-vaccination period to 9 cases during the post-vaccination period. Three patients out of them were vaccinated, one of them was not and in the remaining 5 patients no data were available. The number of SCMID cases among persons older than 18 years increased from 3 to 16 cases in the post-vaccination period.
The number of cases of Haemophilus Influenzae invasive infections (HIID) in the prevaccination period was 65 and 35 in post-vaccination period, only 2 of them were detected in CSF. Among persons of 18 years and younger the number of HIID cases reduced from 25 cases in the pre-vaccination period to 6 cases during the post-vaccination period. One patient out of them was previously vaccinated, no data of vaccination were available in the others. The number of HIID among persons older than 18 years was reduced from 39 to 24 cases in the post-vaccination period.
Conclusion: Since the introduction of vaccination, the incidence of meningitis and bloodstream infections by Haemophilus Influenzae and serogroup C meningococci in the Basque Country is declining, which is partly caused by the natural fluctuation in the incidence of serogroup B meningococci. Nevertheless, the vaccination against serogroup C meningococcal disease as well as Haemophilus were very effectives and contributed significantly to the decline in the incidence of meningitis.
P1877 Control programme of an outbreak of pneumococcal pneumonia among residents of a rest home in southern Spain
J. Delgado de la Cuesta, A. Luna, M. Chavez, R. de la Rosa, J. Fernandez Rivera, S. Gomez, S. Exposito, B. Romero (Seville, ES)
Objective: To assess the effectiveness of a prophylactic programme, including vaccination against Streptoccus pneumoniae and antimicrobial therapy, to control an pneumococcal pneumonia outbreak (PPO) among elder residents in a Spanish retirement home.
Methods: A prospective study to determine epidemiological characteristics of bacterial pneumonia is being developed in our secondary-care hospital. From January 24–February 24, 2006, seven (21.4%) out of twenty-nine patients admitted to our hospital with diagnosis of pneumococcal pneumonia were residents of the same retirement home. A positive urinary Streptococcus pneumoniae antigen test (BINAX-NOW®) was demonstrated in all of them. In order to control a possible PPO, on February 24, 2006 all residents of this rest-home underwent an epidemiological questionnaire and an exhaustive physical examination. All healthy 37 residents were treated with a prophylactic antimicrobial therapy with Amoxicillin (500 mg tid 14 days) and vaccinated with 23-valent pneumococcal capsular polysaccharide vaccine (Pneumovax®). Oro-nasal samples from all 24 retirement home workers were obtained for bacterial culture. All residents and workers were followed for three months after this intervention.
Results: Median age [range] of residents was 83 [61–94] years. Fourteen (40.4%) were males. None of patients admitted to hospital for pneumococcal pneumonia died. All of them were discharged to complete therapy as out-patients. None of the healthy residents had been ever vaccinated against Streptococcus pneumoniae. All of them accepted to be vaccinated and received prophylactic antimicrobial therapy (Intervention rate 100%). No statistical differences were found with respect to age (Student t-test; p = 0.95) or gender (Mann-Whitney U test; p = 0.43) between hospitalised patients and healthy residents. None of patients diagnosed with pneumococcal pneumonia were roommates or were attended by the same care-worker. All oronasal cultures were negative for Streptococcus pneumoniae. There were no further cases of pneumonia after these control measures during the follow-up period.
Conclusions: Prophylactic intervention programme performed, including antimicrobial chemotherapy and vaccination against Streptococcus pneumoniae, allowed to control an PPO in a retirement home for elder people. The present study enforced the vaccination againts Streptococcus pneumoniae among this epidemiological group in our environment strikingly poorly used instead of its high effectiveness.
P1878 Validation and optimisation of a prediction rule for hospitalisation or death from lower respiratory tract infections in elderly patients in primary care
J. Bont, E. Hak, A. Hoes, T. Verheij on behalf of ESPRIT
Objective: Acute lower respiratory tract infections (LRTIs) in elderly patients often follow a more complicated course. Prediction rules may support physicians in their treatment options. So far, most prediction rules have been derived and validated in a hospital setting, while risk stratification is also of great importance to general practitioners. Earlier we retrospectively derived and externally validated a prediction rule to estimate the probability of 30-day hospitalisation or death in elderly patients with LRTI in primary care. The objective of the current study is to prospectively validate and optimise this prediction rule.
Methods: 1,158 patients aged 65 years or older visiting the general practitioner with LRTI were included. The main outcome measure was 30-day hospitalisation or death. First the prediction rule was validated. Next the rule was optimised with data on signs and symptoms by multiple logistic regression analysis. A new scoring system was derived with the use of the regression coefficients of the original prediction rule and new data. The area under the receiver operating curve (AUC) was calculated for the validated and optimised prediction model.
Results: The original prediction rule performed almost as good in the prospective cohort as in the retrospective cohort, with an AUC of 0.73 and 0.75 respectively. The optimised prediction rule performed better with an AUC of 0.81. A low score (≤11 points) corresponded with a 96% probability of absence and a high score (≥19 points) with a 28% probability of presence of the outcome.
Conclusions: Validation shows that the original prediction rule differentiates well between high- and low-risk patients with LRTI in primary care. It may support general practitioners in their decision treating elderly patients at home or referring them to the hospital. Although the optimised prediction rule has better discriminative power, it should be validated in another cohort.
Hepatitis C virus
P1879 Aspartate aminotransferase to platelet ratio index for the evaluation of fibrosis in chronic viral hepatitis
M.A. Yetkin, C. Bulut, M. Çaydere, F.S. Erdinc, G. Ertem, S. Kinikli, N. Tülek, H. Üstün, A.P. Demiroz (Ankara, TR)
The aspartate aminotransferase to platelet ratio index (APRI), developed for the diagnosis of significant liver fibrosis, is calculated by the simple parameteres used for the routine follow-up of the chronic hepatitis patients. The aim of this study is to evaluate the value of APRI for predicting significant fibrosis in chronic hepatitis B and chronic hepatitis C patients.
Chronic hepatitis B and chronic hepatitis C patients who were admitted to our clinic and performed liver biopsy between 1999 to 2005 were inclueded into the study. APRI values of 0.50 or less and greater than 1.50 were evaluated for predicting significant fibrosis. Fibrosis was considered to be insignificant in cases with scores 0 to 1 and significant in cases with scores 2 and 3.
Ninty-seven male and 58 female patients were inclueded into the study. Chronic hepatitis B was present in 114 patients and chronic hepatitis C was present in 41 patients. Avarage Knodell and fibrosis score of the patients were 8.5±3.6 and 1.3±1.0, respectively. Significant fibrosis was detected in 50 of 155 patients and 34 of them were infected with chronic hepatitis B. Chronic hepatitis C were detected in rest of them. Avarage age, serum gama-glutamyl-transpeptidase, Knodell scores and trombocytopenia were detected statistically high in patients with significant fibrosis (p < 0.05). Significant fibrosis was observed in 8/43 patients with APRI ≤ 0.5, 29/87 (%33) patients with 0.50 < APRI ≤ 1.5 and 13/23 (%57) patients with APRI > 1.50 (p <0.05). The combination of APRI ≤ 0.50 and APRI > 1.50 classified correctly %31 of patients with and without significant fibrosis.
In conclusion APRI can not replace liver biopsy for the detection of significant fibrosis in chronic hepatitis patients, but it might be helpful in cases in which liver biopsy could not be performed.
P1880 Immunisation of mice with HCV core protein formulated in montanide-ISA720 and CpG primes CD+8 CTLs and elicits Th1-Th2 responses
M. Aghasadeghi, M. Sadat, A. Khabiri, F. Rouhvand (Tehran, IR)
Objectives: Hepatitis C (HCV) core protein (HCVcp) is a prime candidate for a HCV vaccine. HCVcp is the most conserved gene in HCV genotypes with several well characterised B cell, T cell and CTL determinants and target cells expressing HCVcp can be identified and destroyed by Core-specific CTLs. Immunisation with recombinant proteins however, requires formulation in proper adjuvants. Herein we describe results of evaluating adjuvant effect of M720 (Montanide ISA 720), CPG ODNs and F127 (Pluronic acid) in different formulations on induction of HCVcp-specific Th1/Th2 responses and CD+8 CTLs to HCVcp in BALB/C mice.
Methods: The HCVcp (amino acids 2–122) was expressed in a T7/arabinose induction system in E. coli and purified in native condition on Ni-NTA agarose. Mice were immunised with HCVcp as antigen (Ag) in following formulations; Ag, Ag-Freund (IFA), Ag-CpG, Ag-M720, Ag-F127, Ag-CpG-F127, Ag-CpG-M720. Total IgG, IgG-isotypes and cytokines were quantified by ELISA. Re-stimulated spleenocytes of immunised mice by CD8-specific-HCVcp peptides were analysed for CTLs by LDH release cytotoxic assay.
Results: expressed HCVcp was identified as a 21 kd band by SDS-PAGE and western blotting using anti-penta His and HCV-core mAb and sera of HCV infected patients. All Immunised mice developed anti HCV-core antibodies of IgG class but the mean value for M720 and IM720-CpG groups was significantly higher than for other groups. Cytokine measurement indicated that although CpG and F127-CpG groups showed the highest IFN-g:IL-4 ratio, the levels of each cytokine separately was highest for M720 and M720-CpG adjuvated groups. The CLTs of mice immunised with HCVcp formulated in M720-CpG, M720 and CpG shpwed the highest percent of specific lysis at the effector: target ratio of 20:1, respectively. In contrast, in other HCVcp formulated groups (F127, F127-CpG, IFA, free HCVcp and control adjuvats) no detectable HCVcp-specific CTL responses could be mounted.
Conclusion: HCVcp purified in native from is capable to elicit different level of Th1 and Th2 immune responses when administered in a combination with selected adjuvants. HCVcp administered with M720 or M720-CpG as adjuvants seem to elicit strong and balanced immune responses and HCVcp-specific CD+8 CTL.
P1881 High expression of HCV core protein: a comparative study on effect of 6xHis-tag location (N- versus C-terminal) and purification method for various properties
M. Sadat, M. Aghasadeghi, G. Bahramali, F. Rouhvand (Tehran, IR)
Objectives: Hepatitis C virus (HCV) core protein (HCVcp) as a multifunctional and conserved protein among various HCV genotypes is important for diagnosis, vaccine formulation experiments and studies on HCVcp-mediated pathogenesis. For such studies high amounts of pure HCVcp might be prepared by heterologous expression. Insertion of Histag for purification of recombinant proteins has found great application in different laboratories; however, this procedure may have unwanted effects on conformation and antigenic properties. In this study, characterisation of a new E. coli-derived HCVcp as N- or C-terminally His-tagged protein (N- or C-HCVcp), purified in both native and denatured condition is provided.
Methods: The hydrophilic section of HCVcp gene (aa 2–122) was inserted into pIVEX 2.3 and 2.4a which provided a 6xHis-tag at C- or N-terminal of the protein, respectively. E. coli BL21-AI harbouring constructs were induced by addition of L-Arabinose. HCVcp was purified in both native and denatured condition by NI-NTA agarose and characterised by SDS-PAGE, Immunoblotting and SELDI-TOF mass spectrometry. Antigenic and immunogenic properties of HCVcp were evaluated with HCV-infected human and immunised mice sera by ELISA respectively. Ability of particulate formation of proteins was examined by immuno-gold electron microscopy.
Results: The yields of protein expression were 25 and 16 mg/L in denatured versus 7 and 4 mg/L in native purification for N- versus C-HCVcp, respectively. N-terminal fragmented products of 9 and 11 Kd, which were not due to proteolytic activity but apparently result of ribosomal release were identified. However, these fragmented products were not purified with C-HCVcp. Diagnostic properties of natively purified proteins were predominant and still better for C-HCVcp, However N-HCVcp reacted with C-HCVcp-Immunised mice sera in lower titers. Only natively purified proteins were capable of particulate formation and assembling to generate VLPs.
Conclusion: Purification in denatured/refolding condition may not result to proper conformation of HCVcp, thereby native purification may be undertaken for any kind of applications. C-HCVcp which can be purified as a homogenous product is predominant for diagnostic and pathogenic studies while N-HCVcp that is purified as both fragmented and complete products may be used for generation of antibodies because of better presentation of linear epitopes which are mostly located on the N-terminal of HCVcp.
P1882 Hepatitis C virus genotyping: correlation between real-time PCR and probe hybridisation assays
A. Rossi, A. Bassani, A. Berrone, A. Baj, R. Pulvirenti, A. Toniolo (Varese, Rome, IT)
Background: In the US, HCV is responsible of 3.1–4.8 million people chronically infected and of 8–10 thousand deaths per year. Genetic heterogeneity of virus may account for differences in clinical outcome and response to treatment. Factors influencing treatment outcome are HCV genotype, baseline viral load, liver fibrosis and inflammation. Patients infected with HCV genotype 1 tend to have reduced response rates in comparison to patients infected with genotypes 2 or 3.
Study design: A conventional HCV genotype method (line probe hybridisation, LiPA assay) was compared with a real time PCR genotyping assay (Abbott-Celera) targeting the 5'UTR and NS5B genomic regions. In the latter method, HCV genotype is obtained by comparison of cycle threshold values obtained in three PCR reactions each containing different primer/probe combinations. Probes are labeled with FAM, VIC, or NED. In reaction 1, the HCV genome is detected by FAM, genotype 1a by VIC, 1b by NED. In reaction 2, genotype 2a by FAM, 2b by VIC, 3 by NED. In reaction 3, genotype 4 by FAM, 5 by VIC, 6 by NED. Genotypes other than those mentioned above, are detected in reaction 1 (FAM) and give an indeterminate result. Sera of chronically-infected Italian patients were investigated.
Results: 88 samples were genotyped by real-time PCR and conventional LiPA. Results are summarised in Figure 1 . Of 88 samples that had been genotyped by LiPA, 58 belonged to genotype 1, 11 to genotype 2, 8 to genotype 3, 9 to genotype 4, 1 to genotype 5, 1 contained the 1 & 4 genotypes. Real-time PCR and LiPA gave concordant results in 86/88 samples (97.7%). The real-time PCR method correctly identified (at the subtype level) 56/58 samples (1a and 1b genotypes). LiPA identified the above samples as genotype 1. The correct subtype (1a or 1b) was attributed in only 39/58 cases. One case with mixed infection (1 & 4) was identified by both methods. One case attributed to genotype 1 by LiPA was given as indeterminate by real time PCR. One case was identified as genotype 4 by real-time PCR and as genotype 1 by LiPA. By sequencing the 5'UTR region it was shown to contain both genotypes 1 and 4.
Figure 1.

HCV genotyping: agreement between results of LiPA and real-time PCR assays.
Conclusions: Results of HCV genotyping by real-time PCR were in consistent agreement with LiPA results (97.7% of cases). The Abbott-Celera genotyping assay appeared to allow better discrimination of subtypes 1a and 1b (p < 0.05). The assay was fast and easy to perform and allowed to detect mixed infections.
P1883 Evaluation of a new combined hepatitis C antigen/antibody assay for routine HCV testing of patient samples
P. Vermeersch, B. Van Meensel, M. Van Ranst, K. Lagrou (Leuven, BE)
Objective: To evaluate a new combined hepatitis C antigen/antibody assay (HCV Ultra, Bio-Rad) in patient samples that were borderline positive or positive with AxSYM anti-HCV EIA (version 3, Abbott) in a routine hospital setting. This test was shown to have a smaller window phase for the detection of acute HCV infection compared to anti-HCV assays that only detect antibodies.
Methods: The performance of HCV Ultra was determined in 257 sera that were borderline positive (S/CO = 0.8–1.0) or positive (S/CO > 1.0) on AxSYM. The group of positive sera consisted of 82 of the 2408 in-house sera tested over a 2.5 month period and 175 sera referred for confirmatory testing. We also tested 18 sera that were negative on AxSYM. All sera were tested with Monolisa Plus Anti-HCV EIA (version 2, Bio-Rad). Sera that were AxSYM S/CO > 1.0 and Monolisa S/CO > 3 were considered positive. Otherwise immunoblot analysis was performed with INNO-LIA HCV Score (Innogenetics). When INNO-LIA did not allow a conclusion (indeterminate), the sample was tested with PCR for the presence of HCV RNA when enough serum was available.
Results: All 118 sera that were positive with both AxSYM and Monolisa were positive with HCV Ultra. The results of the 111 other sera that were not positive with both AxSYM and Monolisa are shown in table 1 . A significant number of sera were undetermined with INNO-LIA (13.5%). These 15 sera were excluded for the calculation of the performance of HCV Ultra. In 13 of these samples, HCV Ultra gave a correct result. The sensitivity and specificity of HCV Ultra on AxSYM borderline and positive sera was 99% and 95%, respectively. The 18 sera that were negative on AxSYM were also negative on Monolisa and Ultra. All sera that were HCV Ultra S/CO ≥2.5 were from HCV-positive patients. The positive predictive value for in-house samples was 0% for AxSYM borderline positive sera and 54% for AxSYM positive sera. The only serum that was negative with HCV Ultra and positive with INNO-LIA was from a patient with normal liver enzymes and was negative with PCR. The 4 sera that were positive with HCV Ultra and negative with INNO-LIA could be from seroconverters as was confirmed by PCR in the only patient from which serum was available.
Table 1.
Results in samples that were positive with AxSYM and Monolisa Plus
| HCV Ultra\INNO-LIA | Negative | Positive | Undetermined |
|---|---|---|---|
| Negative | 78 | 1 | 12 |
| Positive | 4 | 13 | 3 |
Conclusion: The performance of HCV Ultra in sera that were AxSYM borderline or positive was excellent. Confirmation testing with INNO-LIA has little or no added value in sera tested with HCV Ultra.
P1884 Early prediction of response during high-dose interferon induction therapy in difficult-to-treat chronic hepatitis C patients
H.C. Gelderblom, H.L. Zaaijer, C.J. Weegink, M.G.W. Dijkgraaf, P.L.M. Jansen, M.G.H.M. Beld, H.W. Reesink (Amsterdam, NL)
Background and Aims: The aims of our study were to determine (i) if early viral kinetics could predict treatment outcome in “difficult-to-treat” hepatitis C patients during high dose interferon induction treatment, (ii) if fast-responders (≥ 3 log drop in HCV RNA at week 4) could stop treatment at week 24.
Methods: We treated 100 hepatitis C patients (46 previous non-responders/relapsers (any genotype), 54 treatment-naïve genotype 1 and 4) with triple antiviral therapy: Amantadine hydrochloride and ribavirin, combined with 6 weeks interferon alfa2b induction (week 1–2: 18 MU/day, week 3–4: 9 MU/day, week 5–6: 6 MU/day), thereafter combined with weekly peginterferon alfa-2b, for 24 or 48 weeks. Fast-responders (≥3 log drop in HCV RNA at week 4) were randomised to 24 or 48 weeks. Patients with <3 log drop in HCV RNA at week 4 (slow-responders) were treated for 48 weeks. Patients with HCV RNA detectable by PCR at week 24 stopped treatment.
Results: 36 patients achieved SVR: 19 fast-responders after 24 weeks of treatment, 9 fast-responders and 8 slow-responders after 48 weeks of treatment. 64 patients became non-SVR (27 non-response, 9 breakthrough, 15 relapse, 13 dropout). Predictive values of early viral kinetics were different for treatment naïve patients and previous non-responders/relapsers. In treatment-naïve patients, PPV for SVR was 100% if HCV RNA was <5 IU/mL at week 1 or 2; PPV for non-SVR was 100% if HCV RNA was ≥ 615 IU/mL at week 12, or ≥ 5 IU/mL at week 16. In previous non-responders/relapsers PPV for non-SVR was 100% if HCV RNA was ≥ 615 IU/mL at week 4, or ≥ 5 IU/mL at week 8. Relapse rates among fast-responders treated for 24 or 48 weeks were 27% and 20%, respectively (P = ns). SVR in fast-responders treated for 24 or 48 weeks was independent of baseline HCV RNA ≥ or <800,000 IU/mL.
Conclusion: With high dose interferon induction therapy: (i) early viral kinetics can predict SVR and NR in treatment naïve patients and previous non-responders/relapsers, (ii) SVR is independent of baseline HCV RNA.
P1885 Similar early viral kinetics of hepatitis C virus genotypes 1 and 4
H.C. Gelderblom, H.L. Zaaijer, C.J. Weegink, P.L.M. Jansen, H.W. Reesink, M.G.H.M. Beld (Amsterdam, NL)
Background: Patients infected with hepatitis C virus genotype 2 (HCV-2) or HCV-3 respond better to interferon alfa (IFN-alfa) treatment than HCV-1 or HCV-4 patients. The mean initial decline in HCV RNA during IFN-alfa therapy is faster for HCV-2 and HCV-3 compared to HCV-1 patients. Little is known about early viral kinetics in patients with HCV-4.
Aims: The aim of our study was to determine genotype specific differences in early viral kinetics in HCV-1 and HCV-4 patients during a modified treatment regimen with a high initial dose of interferon (induction).
Methods: We treated naïve patients with HCV-1 (n = 42) or HCV-4 (n = 12) with triple antiviral therapy consisting of amantadine hydrochloride and ribavirin, combined with 6 weeks of IFN-alfa2b induction (week 1–2: 18 MU/day, week 3–4: 9 MU/day, week 5–6: 6 MU/day), thereafter combined with weekly Peg-IFN-alfa2b, for 24 or 48 weeks. HCV RNA was assessed at baseline, day 1, 2, week 1, 2, 4, 6, 8, and then every 4 weeks until end of treatment by: quantitative bDNA (LLD 615 IU/mL), qualitative PCR (LLD 50 IU/mL), and TMA (Transcription-Mediated Amplification, LLD 5 IU/mL). Viral dynamics were estimated using the bi-phasic model for HCV during treatment with IFN-alfa.
Results: Baseline HCV RNA levels, and the 1st and 2nd phase decline in HCV RNA, were similar in HCV-1 and HCV-4 patients (Figure ). Mean time to reach a TMA negative status in patients with subsequent SVR was shorter in HCV-4 (4.3 ± 2.3 weeks) compared to HCV-1 (6.4 ± 4.5 weeks), this difference was not significant. SVR was achieved by 43% of HCV-1 and 50% of HCV-4 patients.
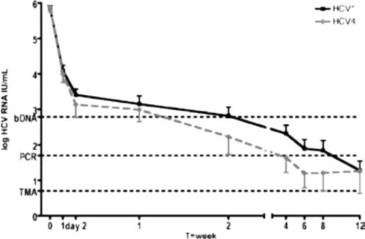
Conclusion: Early viral kinetics are similar in HCV-1 and HCV-4 patients, these results confirm that HCV-4 patients should be treated as HCV-1.
P1886 Meta-analysis of blood scores for liver fibrosis in chronic hepatitis C
F. Lunel-Fabiani, P. Calès, P. Halfon, Y. Bacq, V. Leroy, M.C. Rousselet, M. Bourlière, A. de Muret, Y. Gallois, N. Sturm, G. Penaranda, M.C. Bréchot, C. Trocme (Tours, Grenoble, Angers, FR)
Blood scores of liver fibrosis are alternative tools to liver biopsy or imaging. The aims of this meta-analysis with individual data were to evaluate the diagnostic accuracy, the centre effect (reproducibility) of scores and to compare them.
Methods: The populations from 4 independent centres (dosages, liver interpretation) included 300, 217, 159, and 149 patients with chronic hepatitis C, i.e. 825 patients. Blood scores included Fibrotest (FT), FibroMeter (FM), Hepascore (HS) and APRI.
Results: The global characteristics were the following: 44±12 yr, males: 59.5%, Metavir stages: F0: 4.8%, F1: 46.7%, F2: 25.0%, F3: 12.5%, F4: 11.0%. The 4 populations were significantly different for: age, sex, Metavir score and prevalence of clinically significant fibrosis (≥F2), severe fibrosis (≥F3), and cirrhosis (F4). AUROC are listed in the following table
| ≥F2 | ≥F3 | F4 | |
|---|---|---|---|
| FibroMeter | 0.831±0.014 | 0.887±0.014 | 0.923±0.013 |
| Fibrotest | 0.803±0.016 | 0.853±0.016 | 0.892±0.015 |
| APRI | 0.784±0.017 | 0.836±0.017 | 0.874±0.019 |
| Hepascore | 0.775±0.017 | 0.834±0.017 | 0.886±0.019 |
FM AUROC for ≥F2 was superior to that of FT (p = 0.049), APRI (p = 0.001) and HS (p < 10−3). AUROC were different according to centre, e.g. for FM from 0.773±0.042 to 0.883±0.026. The score profile significantly varied as shown by the comparison of disagreement rate between blood score and liver biopsy (misclassified patients) as a function of Metavir stage: this rate was significantly superior for FM vs FT in F1 (22.9 vs 14.7%, p < 10−3) but significantly inferior for FM vs FT in F2 (40.8 vs 62.8%, p < 10−3), F3 (13.3 vs 27.6%, p = 0.003) and F4 (1.3 vs 9.0%, p = 0.07). This disagreement rate blood vs liver significantly varied according to centre, e.g. for ≥F2 and FM from 18.3 to 28.6% (ANOVA, p = 0.02). By logistic regression, the centre had an independent role for this disagreement. Likewise, the disagreement rate between blood scores significantly varied according to centre, e.g. for ≥F2, FT vs FM from 16.9 to 26.3% (ANOVA, p = 0.05). By contrast, Metavir stage, but not centre, had an independent role for this disagreement.
Conclusion: This meta-analysis with individual data validates the published data of accuracy for blood scores of liver fibrosis (except for HS) and shows significant differences between blood scores for global accuracy and even more as a function of Metavir stage which explains a population effect.
P1887 Three-year study of hepatitis A, B and C infection prevalence in a major Greek hospital
V. Karabassi, M. Poujioyka, K. Petrochilou, C. Achilleos, V. Dimitriou, M. Toutouza (Athens, GR)
Objectives: The purpose of this study was to perform a serological analysis of Hepatitis A, B and C virus (HAV, HBV and HCV) infection prevalence in the patients of a Greek major hospital within a three-year period.
Methods: During the three last years 1/7/2003–30/6/2006, sera from 15,790 patients (13,090 in-patients and 2,700 out-patients), were tested for the detection of HAV, HBV and HCV markers. There were 4 groups of patients: Group A, 5,065 patients (3,745 in-patients, 1,320 out-patients) who were controlled for HBV surface antigen (HBsAg); Group B, 10,725 patients (9,345 in-patients and 1,380 outpatients) who were controlled for the HBV markers HBsAg, HBeAg, antiHBe, antiHBcore, antiHBs; Group C, 11,160 patients (9,600 in-patients, 1,560 out-patients) who were controlled at the same time for HCV-antibodies (HCVAb); and Group D, 1,610 patients (1,230 in-patients, 380 out-patients) who were controlled for HAV-antibodies (HAVAb, total and IgM). All tests were performed during 2003–2004 by MEIA methodology (AXSYM – ABBOTT) and during 2005–2006 by immunoluminometric assay method (ARCHITECT – ABBOTT).
Results: In Group A, of the 3,745 in-patients and 1,320 out-patients, 75 (2%) and 41 (3.1%). respectively, were found to be positive for HBsAg. In Group B, of the 9,345 in-patients and 1,380 out-patients there were, respectively, 4364 (46.7%) and 610 (44.2%) seronegative for HBV markers, 607 (6.5%) and 173 (12,5%) positive for HBsAg, 1,476 (15.8%), 87 (6.3%) positive for antiHBs due to HBV vaccination, 2,897 (31%) and 510 (37%) negative for HBsAg and positive for the rest HBV markers. In Group C, of the 9,600 in-patients and 1,560 out-patients, HCVAb-positivity was found in 221 (2.3%) and 94 (6%), respectively. 2% of them were at the same time positive for HBsAg. In Group D, of the 1,230 in-patients and 380 out-patients HAVAb total positivity was found in 652 (53%) and 194 (51%) and HAVAb IgM positivity was found in 25 (2%) and 5 (1.3%) respectively.
Conclusions: This study showed that there is a significant prevalence of HAV, HBV and HCV infection in the patients of our hospital (53%, 37.5%, 2.3% for in-patients; 51%, 49.5%, 6% for out-patients, respectively). The effort for the prevention must be continual and the control measures with the systematic Hepatitis A and Hepatitis B vaccination should be strictly enforced.
P1888 Spread of hepatitis B and C viruses among healthy people in Bulgaria
K. Mekouchinov, E. Marinova, A. Kalchev (Sofia, BG)
Background: The viral Hepatitis B and C are infections of great social importance because of their worldwide spread, severe complications and big economic loss. Bulgaria belongs to the Middle Endemic World Zone of Hepatitis B Virus (HBV) carriers, with a rate of 3.5%, and to the Zone with a low rate (1.11%) of Hepatitis C Virus (HCV). Some HBV and most HCV infections are subclinical or asymptomatic and remain undiscovered, causing chronic diseases with irreversible complications.
Objectives: To determine HBV and HCV carriers among healthy people and to udertake an appropriate treatment.
Methods: During the period of 1 year (2005) we investigated 3,387 healthy people, divided in 2 groups: 1,641 in Military Medical Academy, Sofia and 1,746 in Military Hospital, Plovdiv. The detection and confirmation of specific antigens and antibodies we performed with ELISA and Blot tests, respectively.
Results: We found
-
–
HBV carriers 3.05% and 1.78%, respectively in Sofia and Plovdiv, mean 2.39%.
-
–
HCV carriers 0.91% and 0.05%, respectively in Sofia and Plovdiv, mean 0.47%.
Conclusions: Compared with the results obtained in 2004, the 2.39% of HBV carriers is higher than 2004 (1.11%), but is still lower than whole Bulgarian population of 3.5%. The 0.47% of HCV arriers is the same as in 2004, lower than whole Bulgarian population 1.11%.
P1889 Pharmacokinetic profiles in chronic hepatitis C patients treated with high-dose pegylated interferon
A. van der Eijk, J. Vrolijk, R. de Knegt, S. Schalm, A. Osterhaus, B. Haagmans (Rotterdam, NL)
Objectives: Therapeutic drug monitoring has been implicated in many antiviral therapies, but not those involving interferon-alpha (IFN-alpha). We determined early pharmacokinetic and viral kinetic profiles in difficult-to-treat chronic HCV patients treated with high-dose pegylated (PEG)-IFN-alpha-2a.
Methods: Nine patients were treated with high dose PEG-IFN-alpha-2a (360 μg/week) during the first four weeks. We measured PEG-IFN-alpha levels using a quantitative sandwich IFN-alpha enzyme-linked immunosorbent assay and quantitated HCV RNA levels (detection limit 103 copies/mL) in plasma for each individual patient at 0, 1, 2, 4, 7, 14, 21 and 28 days after start of therapy.
Results: We observed large variation in pharmacokinetic profiles between individual patients. PEG-IFN-alpha-2a levels were virtually undetectable in one virological nonresponder (patient 2). In this case IFN-alpha specific antibodies were detected at low levels before start of treatment and increased after the first week of therapy. Using a sensitive HCV replicon-based IFN bioassay we confirmed that antiviral activity of PEG-IFN-alpha-2a was abrogated by serum from this patient.

Conclusion: Our study showed two important aspects. First, individual PEG-IFN-alpha PK profiles can be used to identify non-responders to antiviral therapy. Secondly, the presentation of the first patient with antibodies that neutralise PEG-IFN-alpha in vivo. Future studies need to address the extent of autoantibody induction by PEG-IFNs. Our preliminary results from different HBV and HCV PEG-IFN-alpha trials indicate that 10–20% of the patients developed antibodies against the different forms of PEG-IFN-alpha.
P1890 Serum adiponectin in chronic hepatitis C and B
D. Siagris, A. Lekkou, G. Vafiadis, M. Michalaki, I. Starakis, M. Makri, V. Margaritis, A. Tsamandas, C. Labropoulou-Karatza (Patras, GR)
Objectives: Adiponectin possesses anti-inflammatory, insulin sensitising, and anti-atherosclerotic properties. The aim of this study was to assess the levels of adiponectin in patients with chronic viral hepatitis C and B and correlate them with parameters exploring insulin resistance and indices of chronic liver disease.
Methods: Seventy-two patients with chronic HCV infection and 73 patients with chronic HBV infection, matched for age and sex were studied. All individuals were examined for serum concentrations of adiponectin, insulin, C-peptide and insulin resistance index (HOMA-IR). Viral parameters and liver histology were also evaluated.
Results: Serum adiponectin levels were found significantly higher in HCV compared to HBV-infected patients. Correlation analysis in the whole group, demonstrated that serum adiponectin was positively correlated with aspartate aminotransferase (AST), alkaline phosphatase, globulins, high-density lipoprotein cholesterol (HDL-C) and staging score, while it was negatively correlated with body mass index (BMI), insulin, C-peptide HOMA-IR. Logistic regression analysis identified type of infection (HCV versus HBV), alcohol consumption more than 25 g daily, serum total globulin and low C-peptide as significant predictive variables associated with high adiponectin levels.
Conclusion: Higher levels of serum adiponectin in HCV compared to HBV patients could have a role in the slower disease progression of chronic HCV infection. In addition, alcohol intake more than 25g daily seems to be a significant predictor for hyperadiponectinaemia in patients with chronic viral hepatitis C or B. Finally in this study, a clear positive association between adiponectin and hepatic necroinflammation or staging score was not found.
P1891 Effect of intrahepatic hepatitis C virus internal ribosomal entry site activity on the outcome of infection
M. Motazakker, E. McCruden, P. Preikschat, J. Elliott, C. Smith, K. Oien, E. Spence, R. Elliott, P. Mills (Uromieh, IR; Glasgow, UK)
Introduction: The course of chronic hepatitis C virus (HCV) infection is variable and unexplained. Hepatic steatosis has been linked to the progression of fibrosis in HCV and is known to be more common in genotype (gt) 3 infected patients. This study explores variation in intrahepatic HCV internal ribosomal entry site (IRES) nucleotide sequences and outcome of infection.
Method: The translation efficiencies of eight gt 1a and eight gt 3a 5™UTR sequences derived from 16 liver biopsies were measured in cell culture using a dual luciferase reporter system. The values obtained were then compared to the liver biopsy scores for steatosis and for inflammation and fibrosis (Ishak system) score and serum viral load.
Results: The steatosis scores correlated positively with the presence of gt 3a (Mann-Whitney test, p = 0.008), increasing viral load (correlation coefficient (r) = 0.49, p = 0.02) and fibrosis stage (r = 0.47, p = 0.03). The IRES activity did not correlate with severity of steatosis (r = −0.3, p = 0.1), inflammation grade (r = 0.09, p = 0.6), stage of fibrosis (r = 0.1, p = 0.5) or with serum HCV RNA levels (r = −0.11, p = 0.6).
Conclusion: The previously reported association of steatosis with genotype 3, viral load and stage of fibrosis is confirmed. No correlation was seen between IRES activity and these parameters. The efficiency with which the IRES directs translation of the polyprotein appears not to be the limiting factor governing the rate of replication of HCV and pathological features seen in the liver
P1892 Management of pegylated interferon alpha and ribavirin induced neutropenia and thrombocytopenia in patients with chronic hepatitis C
T. Htwe, D. Blessman, M. Spain, A. Adamski, A. Mushtaq, J. Koirala, N. Khardori (Springfield, US)
Objective: Pegylated IFN-alpha and ribavirin is the standard treatment for chronic Hepatitis C and bone marrow suppression is a major side effect of that combination treatment. Treating with GCSF (Granulocyte Colony Stimulating Factor) for Hepatitis C treatment induced neutropenia is an option while maintaining the effective dose of pegylated IFN-alpha and ribavirin to reach sustained virologic response. We use GCSF prospectively in those patients when the absolute neutrophil count becomes less than 1000/μL and evaluate its efficacy for neutropenia and its stabilising effect on platelet count.
Method: We did a retrospective chart review of our Hepatitis C patient population. We identified a total of 222 patients from our hepatitis C database and 60 patients with thrombocytopenia (platelet count less than 140,000/uL) at any point of treatment were identified. Of these 60 patients, 30 received GCSF. In GCSF group, only 27 became eligible for evaluation. One patient had incomplete medical records and two patients were started on GCSF prior to starting treatment. In the Non-GCSF group, 23 patients were evaluated. Two patients never received Hepatitis C treatment; one patient stopped treatment due to intolerance and four patients did not have pre treatment platelet value. Both groups were divided into sub groups according to prior to GCSF treatment platelet values (less than 100,000/uL, between 100,000/uL and 139,000/uL, between 140,000/uL and 200,000/uL, and greater than 200,000/uL). The monthly platelet values in GCSF group were plotted on a line graph and trends of platelet count variations were studied. The platelet values in Non-GCSF group were followed prior to and during Hepatitis C treatment and are also plotted on a line graph.
Results: See the graph.

Average platelet values in GCSF group.
Conclusion: As shown in the graph, GCSF treatment stabilises the platelet count in patients on pegylated interferon alpha and ribavirin treatment for chronic Hepatitis C. The comparison with control will be discussed also.
P1893 Changing of hepatitis C virus genotypes patterns in HIV-positive and -negative patients in Northern Spain
M. Basaras, M.J. Fernández, M. Sota, P. Liendo, R. Cisterna (Bilbao, ES)
Hepatitis C virus (HCV) genotypes show specific geographic distribution and association with particular risk factors. Nevertheless, differences in genotype distribution can be observed in different areas of the same region as well as different areas within a country.
The aim of the present study was to analyse differential distribution of HCV genotypes in Northern Spain along a six-year period in HIV-negative and -positive patients.
Patients and Methods: a retrospective analysis was carried out in 2301 serum samples from HCV RNA-positive patients during a six-year period (from 2000 to 2005). HCV genotyping was determined by reverse hybridisation of PCR amplicons (Versant HCV Genotype Assay, LIPA). Human immunodeficiency virus antigen was detected by INNOTEST and hepatitis B virus HBsAg and anti-HBs were detected by ADVIA Centaur (Bayer).
Results: More than a half of the study group (1248 serum samples, 54.23%) were HIV-negative patients. Of them, the predominant genotype was 1 (71.39%), followed by 3 (16.98%) and 4 (9.13%), being the most prevalent subtype 1b (50.64%). On the other hand, 1053 (45.76%) of serum samples were HCV-HIV co-infected. In that group, the most frequent HCV genotype was 1 (48.71%), followed by 3 (29.25%) and 4 (19.56%); however, subtype 3a was the most prevalent (29.15%). Co-infection with several genotypes was infrequent (1.61%). There was a decline in subtype 1a over time and conversely an increase in subtype 4c/d. In all the study group, subtype 1b was detected preferentially in patients with history of blood transfusion or no known exposure to risk factors, whereas subtypes 1a, 3a and genotype 4 are related with injection drug users. Year after year, subtype 4c/d was detected in younger patients than subtype 3a. On the other hand, genotype 2 was almost imperceptible (1.76% in HIV-negative and 0.85% in HIV-positive). Moreover, HBsAg was detected in 0.7% of serum samples and anti-HBs in 1.34%.
Conclusions: HCV-HIV co-infection was very frequent in the study group due to these viruses share similar transmission routes. Although genotype 1 is the most frequent HCV variant, subtype 1a has declined over time and subtype 4c/d has increased. This last subtype and subtype 3a were related with injection drug use. Subtype 1b was introduced before than subtype 1a or 3a and is related with history of blood transfusion or no known exposure to risk factors. Subtype 1a and 3a, and more recently 4 c/d, are related with HIV co-infection.
P1894 Therapeutical aspects and predictors of outcome for HIV/HCV co-infected patients treated with pegylated interferon plus ribavirin. Survey in an Italian university hospital
E. Righi, M. Bassetti, A. Di Biagio, C. Dentone, R. Rosso, A. Beltrame, G. Mazzarello, S. Ratto, C. Viscoli (Genoa, IT)
Introduction: One third of HIV-infected individuals worldwide suffers from chronic hepatitis C virus (HCV) infection. HCV therapy is a priority in coinfected patients because they have faster liver disease progression. We assess the impact of interferon and ribavirin combination therapy in a cohort of coinfected patients analysing specifically prognostic factors for outcome.
Methods: Patients enrolled in the study were all HIV/HCV patients who received HCV treatment between 2002 and 2006. Clinical data included: demographical data, HAART sitauation, HCV genotype and viral load (considering high rates >800,000 UI/mL). Primary outcome were early virological response (EVR) at week 12 and sustained virological response (SVR) 6 months after stopping treatment.
Results: Forty-four patients were treated for chronic hepatitis C during the four years period. Mean age was 41 years (SD±6.7) and 74% were males. Risk factors for co-infection were mainly related to intravenous drug abusers 32/43 (74%). Regarding immuno-virological situation, 51% had T-CD4+ count >500/mm3 at baseline, 79% showed no more than four HAART changes and only 19% had a CD4 nadir <200 cells/mm3. Genotype 3a represented 51%, while 40% were infected by genotype 1 and 9% by genotype 4. Only 16/43 (37%) were on HAART at baseline and half of the patients showed high HCV-RNA levels. Liver steatosis was present in 14/43 (33%) patients. Mean T-CD4+ count reduction was 175 cells/mm3 (33%), compared a T-CD4+ percentage reduction of 11%. High rates of HCV treatment discontinuation were present (63%), due to voluntary interruptions (52%), virological failure (26%), adverse events as cytopenia (11%) and others. Total EVR was 51%. The SVR was 30% in total, 38% and 24% for genotypes 3a and 1, respectively. Positive and negative predicting values of EVR were 60% and 90%. SVR was significantly lower in high HCV-RNA viral load group (χ2 = 6, P < 0.0025), CD4 nadir <350 cells/mm3 (χ2 = 3.26, P < 0.01) and <500 cells/mm3 (χ2 = 3.94, P < 0.005) and in patients with genotype 1 and high viral load (χ2 = 4.8, P < 0.005).
Conclusions: Factors as HCV viral load rates and genotype 1 have been confirmed to threaten the response to therapy. Patients with no EVR did not have any probability of SVR. There is a significant response rate if patients have a history of nadir >350 T-CD4+/mm3. High dropouts rates reported due to adverse event and voluntary discontinuations complicate HIV/HCV therapeutic management.
P1895 Low prevalence of HCV co-infection in HIV/HBV co-infected teenagers from Romania
L. Manolescu, C. Sultana, P. Marinescu, S. Ruta (Bucharest, Giurgiu, RO)
Objectives: In Romania there was the biggest paediatric HIV infection acquired by parenteral route. The prevalence of HBV markers at these children was of 78.3% at HIV infected versus 21.8% in HIV negative controls. The teenagers acquired HIV and HBV infection horizontally. This study was performed to examine the impact of HCV co infection in HIV/HBV co infected teenagers from Romania.
Methods: Two groups of patients with HIV infection, from different Romanian regions (161 from Constanta, 198 from Giurgiu), average age 13.8±1.3, were retrospectively analysed 2 years for clinical and virological parameters. Clinical liver function parameters (AST, ALT, total bilirubin, albumin), CD4 count, and virological parameters (HCV, HBV and HIV viral load by Roche Amplicor), HBsAg, HBeAg, HBsAb, HBcAb were performed twice a year.
Results: Prevalence of HCV antibodies was low and similar in both groups 2.04% respectively 2.5%. All HCV/HIV co infected patients were also HBV infected and treated with potent antiretroviral therapy (HAART), but never experienced specific antiviral anti HCV treatment. We noticed in both groups similar correlations. HBV/HCV/HIV co infection with active HBV replication revealed through the presence of HBeAg and DNA-HBV high titer (40.000.000 copies DNA-HBV/mL) in both groups: 33.3% respectively 40%, was associated with worsened HIV-related parameters (CD4 count under 100 cell/microL, average HIV titer above 750.000 copies RNA HIV/mL), and increased transaminase (average values 156 mg/dl) but undetectable HCV titers, compared to patients without active HBV replication that presented detectable HCV titers (average of 11550 copies RNA-HCV/mL) and undetectable HIV and HBV titers and transaminase. Patients co infected with HIV/HCV and without HBeAg appeared to have a good response to HAART in terms of CD4 count changes, with a CD4 count average of 532 cells/microL.
Conclusions: A surprisingly low prevalence of HCV co infection was found in HIV/HBV co infected patients despite the commune route of transmition. HIV disease outcomes following HAART do not appear to be adversely affected by HCV co infection and reveal good CD4 count responses. Active chronic HBV co infection in HCV/HIV co infected patients is associated with worsened liver disease and worsened HIV condition.
P1896 Relationship between virological evolution and acute exacerbation of chronic hepatitis C (A-E-CHC) and during the natural history of infection
E. Sagnelli, C. Argentini, D. Genovese, M. Pisaturo, C. Sagnelli, S. Taffon, C. Alteri, N. Coppola, M. Rapicetta (Naples, Rome, IT)
Objectives: To evaluate the relationship between symptomatic a-e-CHC and virological evolution during the natural course of the disease.
Methods: 49 consecutive patients with acute hepatitis C (AHC) (55% IVDA) and 57 with a-e-CHC (43.8% IVDA) were observed. AHC was diagnosed on the basis of seroconversion to anti-HCV and of the increase in ALT serum levels of at least 5 times the normal value. The diagnosis of a-e-CHC was based on the presence of anti-HCV and HCV-RNA in the blood at least 6 months before the onset of symptoms and during exacerbation and of increased ALT serum values of at leas 5 times the baseline at exacerbation. Several plasma samples were obtained and stored frozen at −80°C. Three patient followed-up for 40–68 months showed both AHC and a-e-CHC 3–24 months later. In these patients we evaluated clinical characteristics, viral variability and the variation of known predicted epitopes in the consensus sequence in the Core, E1/E2, and NS5b regions, during the different phases of infection.
Result: The first patient (genotype 3) presented a high rate of genetic and epitopic variability which could be related to the maintenance of a chronic HCV infection state. The second patient (genotype 1b) presented an apparent resolution of the infection with absence of detectable viraemia after a symptomatic acute phase; the phylogenetic and epitopic analysis of the virus isolate during a-e-CHC presented little variation with the isolate of AHC, indicating a phase of occult presence of the virus in the period between AHC and a-e-CHC. The third patient presented two different HCV genotypes, 2c and 2a, associated with the initial phase of AHC and with a-e-CHC, respectively.
Conclusion: The follow up analysis revealed that, despite similar clinical presentation, the viral variability and the differences in the ability to develop or select epitopic variants may drive HCV infection toward different outcomes.
P1897 Spontaneous virus clearance in perinatally HCV-infected children: is it a way to escape the risk of chronicisation?
A. Maccabruni, E. Minola, B. Mariani, G. Bossi (Pavia, Bergamo, IT)
Introduction: Vertical transmission of HCV is proved to be the most frequent route of infection in children, although only 5–7% of infected mothers are thought to transmit the virus to their offspring. Perinatal infections occur at a time when the immune system is still immature, so their chronicisation rate may be very high (>80%); although chronic HCV hepatitis is generally a mild disease in paediatric patients a slow and insidious progression of liver disease may occur over the course of years and cases of severe activity and cirrhosis may be observed in adolescents and young adults.
The clearance of viraemia, reported in about 16% of vertically infected children, could represent a spontaneous way out of the risk of chronicisation, but whether or not they have forever eliminated the virus is still controversial.
Methods: We conducted a retrospective study on paediatric HCV infected subjects followed from 1995 in our Department and in the Infectious Diseases Department of Ospedali Riuniti – Bergamo to evaluate how many cases of spontaneous clearance of viraemia occurred and how the natural history of infection has changed from then on.
Results: In our cohort, including 48 perinatally infected children, virus clearance occurred in 9 of them (17.8%), aged from 10 months to 6 years. In only one of them HCV-RNA was intermittently detected during a follow-up period of 36 months, while in three cases the clearance of viraemia was persistent and associated with loss of specific antibodies and normal ALT values, without any clinical manifestations (follow-up 38–60 months).
Conclusion: Although in our cases we weren't able to individuate any maternal or viral factors predictive of virus clearance in children we can conclude that perinatally infected children are likely to have successfully resolved their status of infection when spontaneous clearance of viraemia with antibody loss and ALT normalisation are persistent for at least 48 months.
As virus clearance can be observed also >5 years after the diagnosis a prolonged and close monitoring of vertically HCV-infected infants to evaluate the course of infection is mandatory; the concept of chronicity in this population should moreover be reconsidered.
Aspects in HIV/AIDS
P1898 HIV-related morbidity in the HAART era
M.A. Escobar, A.A. Garcia-Egido, S.P. Romero, J.A. Bernal, J.L. Puerto, J. Gonzalez-Outon, C. Asencio, F. Gomez (El Puerto Santa Maria, ES)
AIDS related hospital admissions and morbidity have decreased after the introduction of HAART.
Objective: To assess the changes in hospital admissions and morbidity with the improvement of HARRT.
Methods: We studied the hospital admissions and morbidity over three different periods pre-HAART (January 1992 to December 1994), early-HAART (January 1995 to December 1998) and post-HAART (January 1999 to October 2004), in HIV infected patients followed at our centre. Results are expressed for periods pre-HAART, early HAART, and post-HAART respectively. Mean (SD) CD4/mL were: 16±5, 37±11 and 118±33 (p < 0.0001). Undetectable plasma viral load (%) were: 0, 16 and 43 (p < 0.0001). Patients with a prior diagnosis of AIDS were: 91%, 68% and 36% (p < 0.001). The rate (%) of women/heterosexuals was: 6/2, 11/5 and 21/11 (p < 0.01) and, that of IVDA (%) was: 96, 88 and 75 (p = 0.03). Hospital admissions due to AIDS-defining illnesses decreased (p < 0.001) with a significant increase in the rate of respiratory tract infections (p < 0.005), digestive tract (p < 0.01) and liver diseases (p <0.001). The proportion of AIDS-defining illnesses decreased after HAART (p < 0.01), whereas the rate of liver diseases increased (p < 0.001).
Conclusions: The HAART era has been associated with a progressive decrease in hospital admissions due to AIDS-defining conditions, and a steady enhancement of the spectrum of admissions by non-AIDS-defining conditions has increased.
P1899 Assessment of liver cirrhosis by elastography in HCV/HIV co-infected patients
C. Castellares, L. Martín-Carbonero, P. Barreiro, B. Ramos, J. Sheldon, P. Labarga, S. Novoa-Rodríguez, J. González-Lahoz, V. Soriano (Madrid, ES)
Introduction: Chronic hepatitis C is now one of the leading causes of morbidity and mortality among HIV-infected individuals. The evaluation of liver injury in HIV/HCV co-infected patients should follow the same principles as monoinfected ones.
Methods: Retrospective analysis of all HCV/HIV co-infected patients in our institution assessed by FibroScan since Oct 2004 to June 06. Patients with F4 METAVIR fibrosis score estimates (>12.5 kPa) by elastography (FibroScan) were compared to patients without liver cirrhosis.
Results: A total of 772 HIV/HCV co-infected patients were examined, 20% of whom had F4. The mean liver stiffness in them was 25.7 kPa. Overall, 42% of cirrhotics admitted current and/or past alcohol abuse. Their distribution according to Child-Pugh score was: A 68%, B 28% and C 4%. Plasma HIV RNA was undetectable in 67% of cirrhotics and 62% of the rest [p = NS], and CD4 counts <200 cells/mL were present in 27% of cirrhotics vs. 12% of the rest [p <0.01]. Overall, 25% of patients who had received HCV treatment showed liver fibrosis improvement. During the study period, mortality was 16.5% in cirrhotics.
Conclusions: liver cirrhosis is quite prevalent in HCV/HIV-coinfected patients. Transient elastography may help to recognize subclinical cirrhosis and design more appropriate therapeutic and prophylactic strategies for this population.
| Cirrhotics | Non-cirrhotics | P | |
|---|---|---|---|
| No. | 158 | 614 | |
| Male gender (%) | 79 | 69 | 0.02 |
| Mean age (years) | 43±5 | 42±5 | NS |
| Mean HCV load (IU/mL) | 4.71±2.28 | 5.26±2.05 | NS |
| HCV genotype (%) | |||
| 1 | 64 | 56 | NS |
| 2 | 2 | 2 | NS |
| 3 | 21 | 26 | NS |
| 4 | 13 | 16 | NS |
| Any IFN-based therapy (%) | 69 | 47 | <0.01 |
| HCV clearance after | 6 | 30 | <0.01 |
| IFN-based therapy (%) | |||
| Currently on therapy | 7 | NA | NA |
| Chronic hepatitis B (%) | 7 | 1 | <0.01 |
P1900 HIV infection in an intensive care unit
S. Silva, A. Ferreira, C. Piñeiro, S. Xerinda, L. Santos, A. Sarmento (Porto, PT)
Objectives: Evaluation of demography, immunologic state, severity (SAPS II), causes of acute respiratory failure (ARF) and mortality rate in HIV infected patients (pts) admitted in an Infectious Diseases Intensive Care Unit (ICU).
Methods: Retrospective study of HIV infected pts admitted between January 1991 and July 2006. Mortality rates were compared using the Fisher test.
Results: 210 pts were admitted, 153 (73%) males. The mean age was 36.5±12.7 years (3 months–78 years). The routes of transmission of HIV were intravenous drug use in 129 (61.4%), sexual in 68 (32.4%), mother to child in 7 (3.3%), transfusional in 3 (1.4%) and unknown in 3 (1.4%). The mean CD4 count in admission was 116.8±162.4 cells/μL. SAPS II was 45.62±16.56 (median 43). 132 (63%) pts were admitted by ARF. The mortality rate in ICU was 48.5% and inside the hospital 51.5%.
Causes of ARF are listed in the table . PCP and cancer diagnosis were associated with a superior death risk (p = 0.003 and p = 0.002 respectively).
| Disease | Total | Mechanical ventilation | Death |
|---|---|---|---|
| Pneumocystis jiroveci pneumonia (PCP) | 43 (32.6%) | 36 (81.4%) | 29 (67.4%) |
| Tuberculosis | 28 (21.2%) | 19 (67.9%) | 10 (35.7%) |
| Viral and bacterial pneumonia | 17 (12.9%) | 16 (94.%) | 6 (35.3%) |
| Pneumonia by unidentified agent | 30 (22.7%) | 24 (80.0%) | 11 (36.7%) |
| Cancer | 8 (6.1%) | 6 (62.6%) | 8 (100%) |
| Others | 6 (4.5%) | 1 (16.7%) | 1 (16.7%) |
Conclusion: ARF is the main cause of admission in ICU. PCP is the most common cause of ARF.
P1901 Factors associated with the level of HIV-1 proviral DNA in HIV-infected persons
J.Y. Choi, Y.G. Song, Y.K. Kim, M.S. Kim, Y.S. Park, S.Y. Shin, Y.H. Kim, J.M. Kim (Seoul, KR)
Background: HAART does not eliminate the persistently replicating, latent HIV-1 reservoir in patients, despite the viral suppression associated with the therapy. The ability to generate anti-HIV-1 cytotoxic T lymphocyte response is considered to be a critical component of the host immune response to HIV-1.
Patients and Methods: To evaluate the factors associated with the level of HIV-1 proviral DNA and the relationship between the level of HIV-1 proviral DNA and CTL responses in chronically HIV-infected Koreans, we conducted cross-sectional study of eighty-two chronically HIV-1-infected Koreans who had been admitted to Severance Hospital, Yonsei University College of Medicine. In order to determine the proviral HIV DNA copies per 106 peripheral blood mononuclear cells, real-time PCR was carried out. An ELISPOT assay, using HIV-1 peptides, was done in order to evaluate the HIV-1-specific CTL response. Multiple linear regression analysis was used for analysing factors associated with the HIV-1 proviral DNA level.
Results: There was a statistically significant inverse correlation between the proviral DNA level and the CD4+ T cell counts in the peripheral blood of subjects (P = 0.01, r = −0.307). Among the patients receiving HAART, the proviral DNA level was lower in patients in whom viral load had been suppressed to levels below 25 copies/mL, than in patients in whom viral load had not been suppressed (P = 0.01). The proviral DNA level did not correlate with the CTL response (P = 0.36). However, using multivariate analysis, factors associated with the proviral DNA level were plasma HIV RNA, HAART, and CTL responses (P < 0.05).
Conclusions: The HIV-1 proviral DNA level was associated with plasma HIV RNA, HAART, and CTL responses. Intensification of HAART and cellular immune-based therapy could affect the proviral reservoir.
P1902 Relationship between the C3435T and G2677A/T single nucleotide polymorphisms of the P-glycoprotein in the mdr-1 gene and efavirenz plasma concentration in patients with HIV infection
G. Peralta, M.B. Sanchez, E. Valdizan, S. Echevarria, J. Nicolas, R. Segura, V. Badía, J.A. Armijo (Torrelavega, Santander, ES)
Objectives: P-glycoprotein is a drug transporter recognized to be important to drug disposition and response. This protein is of particular clinical relevance due to a broad substrate specificity and expression in multiple tissues. The C3435T AND G2677A/T single nucleotide polymorphisms (SNPs) have been associated with altered transporter function or expression (higher function/expression of P-glycoprotein for CC or GG genotypes, and less for TT genotype). Its role in antiretroviral drug pharmacokinetics is still to be clarified.
Methods: For the study we selected 40 patients in treatment with Efavirenz (600 mg qd) for at least six weeks. We analysed the relationship between the C3435T and G2677A/T SNPs genotypes and efavirenz plasma concentrations twelve hours after oral administration. We excluded those patients in treatment with other drugs with potential interaction with Efavirenz. The genotypes were determined with the use of polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP) assays and with allelic discrimination by PCR assays. Efavirenz plasma levels were measured by high pressure liquid chromatography. To compare means an ANOVA and t-test were used.
Results: For the 40 patients studied, the genotype distribution was: CC: 25%, CT: 52.5%, TT: 20%, for the C3435T SNP; and GG: 35%, GT: 55%, TT: 10% for the G2677A/T SNP. We did not find a statistically significant difference between the C3435T SNP genotypes and efavirenz plasma concentrations (CC: 1.73±0.80mg/L; CT: 3.11±2.42 mg/L; TT: 2.63±1.24 mg/L). But when we compared patients with genotype CC with patients with genotype CT and TT all together, the difference tended to significance (P = 0.083). We did find statistically significant difference between the G2677A/T SNP genotypes and efavirenz plasma concentrations (GG: 1.62±0.76 mg/L; GT: 3.20±2.28 mg/L; TT: 3.13±1.60 mg/L; P = 0.047).
Conclusions: Although the sample is small, in our patients the efavirenz concentrations seems to be in relation with the G2677A/T SNP, and probably with C3435T SNP too. These data have to be confirmed in a bigger group of patients.
P1903 Performance evaluation of VIDIA HIV DUO, a new automated immunoassay test for the qualitative HIV antigen/antibodies detection in serum and plasma human samples
H. Briand, F. Vayssié, O. Breton-Laval, J.M. Dugua, X. Lacoux, N. Battail-Poirot, P. Gradon, J. Piche (Marcy l'Etoile, FR)
Objective: The VIDIA system is a new automated, primary tube immunoanalyser designed to reinforce traceability and simplify the daily workload for routine testing. The qualitative 4th generation combined antigen/antibodies detection is the most appropriate solution for routine screening of HIV infection. During this study, we evaluated the performance of the VIDIA HIV DUO (bioMérieux, France) test in terms of sensitivity and specificity.
Methods: The VIDIA HIV DUO assay principle combines a two-step enzyme immunoassay sandwich method with chemiluminescence detection.
The kit uses:
-
–
HIV antigen, HIV synthetic peptides and HIV p24 monoclonal antibodies coated on microparticles,
-
–
biotin-labelled conjugates: HIV synthetic peptides and HIV p24 monoclonal antibodies,
-
–
and alkaline phosphatase conjugates: HIV antigen and streptavidin.
The raw materials selected enable us to detect HIV1, HIV1 group O, and HIV2 for IgG, IgM as well as Antigen. Specificity was studied using 800 blood donor samples collected from French blood banks. Sensitivity was established using:
-
•
255 confirmed HIV-positive samples from our collection (200 HIV1 group M, 30 HIV1 group O, 16 HIV 2 and 9 HIV 1 non B subtypes)
-
•
20 commercial seroconversion panels from BBI (Boston, USA) and Zeptometrix (Buffalo, USA)
-
•
HIV antigen sensitivity has been established according to the French SFTS (French Blood Transfusion Society) Panel
The performance characteristics of this new product were established in our R&D laboratory during the development of the test.
Results: The specificity determined using 800 blood donors was 99.88% [99.28%; 99.98%] for VIDIA HIV DUO. Sensitivity was 100% [98.46%; 100%] for the VIDIA HIV DUO assay on the 255 confirmed positive samples. As regards the seroconversion panels, the results with VIDIA HIV DUO were comparable to those provided by the panel manufacturer and other collected results. HIV antigen sensitivity of VIDIA HIV DUO was determined against the SFTS panel and found to be 16.5 pg/mL of HIV antigen.
Conclusion: This new HIV 4th generation assay shows excellent performance in terms of sensitivity as well as specificity. The kit's early detection was conclusively demonstrated on the seroconversion panels which is an added value for patient diagnosis. The combination of automation and high performance makes this product an excellent test for routine screening. These results must be confirmed on the final commercialised product.
P1904 Trends in HIV incidence in Siberia, Russian Federation (1987–2005)
A. Khryanin, O. Zhukovskaya, O. Reshetnikov (Novosibirsk, RU)
Objective: Currently the spread of HIV infection is acquiring an epidemic pattern in Russia. Thus, HIV incidence rates increased 24-fold in 2001, and the cumulative number of HIV cases reported in the country increased tenfold compared to 1997–1998. The aim of the present study was to analyse the epidemiology of HIV infection in Western (WS) and Eastern (ES) Siberia over the period from 1987 through 2005.
Methods: Siberia consists of 14 administrative regions with total population of 32,115,697 citizens, which is about one fourth of total Russian population. The data from district STD clinics were obtained and registered in the departments of social statistics of the regional committees of statistics.
Results: HIV incidence rate in Siberia accounts for 17.0% of all HIV infected persons in Russian Federation. Additionally, the HIV incidence rate is higher than average in Russia by 13.1 percent. Trends in the incidence of HIV infection are shown in the Table (number of reported cases in Russia). In 2002, HIV incidence rates in Irkutsk and Novosibirsk were 102.1 and 8.9 (per 100 000 inhabitants), respectively. More than 50% of all HIV cases were reported from intravenous drug users aged 19–29 years in 2004 (in comparison 80% in 2000). Males comprise 70% of newly diagnosed HIV cases. Sexual transmission was reported in 4.7% in 1999 and 54.3% in 2004. The sexual/intravenous routes ratio was 1/1.7 in 2004 (in comparison with 1/21 in 1999).
| 1987–1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
|---|---|---|---|---|---|---|---|---|---|
| Russia | 7,024 | 10,952 | 26,414 | 79,881 | 173,000 | 226,003 | 264,996 | 303,274 | 331,559 |
| Siberia | 226 | 493 | 5,456 | 12,881 | 24,396 | 31,287 | 35,693 | 39,968 | 44,406 |
| Novosibirsk (WS) | 16 | 29 | 48 | 167 | 361 | 600 | 768 | 918 | 1,080 |
| Irkutsk (ES) | 22 | 47 | 3,285 | 7,997 | 11,563 | 14,060 | 15,650 | 17,382 | 19,429 |
Conclusion: At present, HIV epidemic unevenly spreads in the Siberian region. The reservoirs of infection are already formed in some administrative territories such as Irkutsk (ES). As for Novosibirsk (WS), the process of formation is under way. Local intervention programmes are urgently needed to prevent the forthcoming HIV epidemic in Siberia.
P1905 HIV prevalence and risk behaviours among international truck drivers in Azerbaijan
B.A. Botros, Q. Aliyev, M. Saad, M. Monteville, A. Michael, Z. Nasibov, H. Mustafaev, P. Scott, J. Sanchez, J. Carr, K. Earhart (Cairo, EG; Baku, AZ; Rockville, Frederick, Baltimore, US)
Objectives: Assess HIV prevalence and high-risk behaviours among international truck drivers (TDs) traveling between Azerbaijan and Russia, Europe, China, Central/West Asia and Middle East.
Methods: Clinics were established near two major truck terminals in Baku. TDs who volunteered to participate signed consent in his language and received pre- and post-test counseling. Each participant completed a questionnaire that included demographics and information about his personal sexual behaviour, use of condoms and international travel history. A blood sample was obtained by finger stick and tested using 2 different rapid HIV tests: 1) Determine (Abbott Laboratories, Abbott Park, IL USA) and 2) Oraquick (Orasure Technologies Inc., Bethlehem PA USA). Test result was provided within 30 minutes. Enrollment was performed between July 2004 and October 2005. Data was analysed using SPSS version 11.1 statistical package (SPSS Inc, Chicago, IL, USA).
Results: A total of 3,763 TDs from 21 countries were enrolled. All were males between the age of 24 and 67 (median, 45 years). Majority came from Turkey (n = 1369), Russia (n = 1145), Ukraine (n = 662), Georgia (n = 234) and Azerbaijan (n = 184). 58 TD (1.54%) (95% CI: 1.17–1.99) tested HIV positive. Highest prevalence was among Russians (2.88%), then Ukrainians (1.66%), Azerbaijani (1.09%), Georgians (0.85%) and Turkish (0.73%). 1.9% of participants were injecting drug users (IDUs) and 60% of these tested HIV positive compared to 0.4% of non IDU (P < 0.001). Of those using condoms with FSW, 0.7% were HIV positive, whereas of those not using condoms 3.6% were positive (p < 0.001). Men having sex with men (MSM) had higher (p < 0.001) HIV prevalence (42.9%). TD that had STD had higher (P <0.001) HIV prevalence (4.4%) than those not having STD (1.2%). Circumcised TD had lower (p = 0.001) HIV prevalence (0.8%) than non-circumcised (2.2%).
Conclusions: TDs may engage in risky behaviours and are a bridge population for transmission of HIV to the community. This study demonstrates that HIV is present among international TDs in Azerbaijan. As is true for other countries of former Soviet Union, HIV infection was highly associated with IDU. Other associated factors for HIV included not using condoms, MSM and STDs. TDs transiting Azerbaijan should be considered a high risk group and should be a target for surveillance and prevention programmes. Prevention programmes in this region should target knowledge and risk behaviours of TDs.
P1906 No re-imbursement for statins, fibrates, and omega-3 derivatives for HIV-infected patients with HAART-related dyslipidaemia in Italy
R. Manfredi, L. Calza (Bologna, IT)
Introduction: The significant HAART-prompted advances achieved in the management of HIV disease may be frustrated by the modified reimbursement modalities of all lipid-lowering drugs (LLD) available in Italy. The remarkably increased life expectancy attained thanks to HAART, is borne by significant risks to develop an uncontrolled hypercholesterolaemia and/or hypertriglyceridaemia, often concomitant with insulin resistance and visceral adiposity, which strongly predispose to cardiovascular events and stroke.
Methods and Results: The present prescribing rules of LLD based on a computer-generated score, were matched with the present situation of around 1000 HIV-infected patients (p) treated with HAART, to assess the frequency and type of dyslipidaemia, and the estimated rate of need of LLD prescriptions. The rate of hypertrigyceridaemia and hypercholesterolaemia exceeded 28% and 19% of p respectively, while around 22% of p had a mixed dyslipidaemia. Over 200 p were currently treated with statins and/or fibrates, with the eventual adjunct of omega-3 fatty polyunsaturated acids (PUFA). When applying the risk score proposed for the general population, <10% of these p reached the threshold of a >20% risk of major vascular events in the next decade (due to the proportionally lower mean age, the absence of familial dyslipidaemia, diabetes, elevated systolic pressure, and anti-hypertension therapy, vs. the general population), while only very few p needed a secondary prophylaxis, due to a prior, major cardio-cerebro-vascular accident. As a result, more than 90% of HIV-infected p presently treated with LLD due to present antiretroviral therapy recommendations have lost all rights to LLD re-imbursement in Italy, and are at serious risk to give up LLD due to not sustainable linked costs.
Discussion: The recent dispositions of the Italian Health Care System ignore the situation of HIV-infected p, who are exposed to a frequent, severe, drug-induced dyslipidaemia, and an elevated major vascular risk despite their lower mean age, and the lack of multiple generic risk factors. At mid-term, the majority of HAART-induced benefits might be blunted by the sudden lack of LLD re-imbursement, which is estimated to regard most of treated HIV-infected p. A comparison with LLD reimbursement facilities in other countries is also warranted, to draw some epidemiologic and pharmacoeconomic issues suggesting a re-extension of reimbursement facilities of these life-saving drugs to HIV-infected p.
P1907 Immune markers of HIV disease progression are not modified by long-term statin administration
R. Manfredi, L. Calza (Bologna, IT)
Introduction: Statins were recently hypothesized to act unfavourably on the immune system through an altered cytokine pattern and a Th1/Th2 imbalance, as shown in non-HIV-infected patients (p) [JAIDS 2005;39:503], while a small experience claimed a p < 0.05 loss of absolute CD4+ lymphocyte count of p treated with a protease inhibitor (PI)-based HAART, in a 6–18-month period [HIV Clin Trials 2003;4:164].
Patients and Methods: From over 1,000 HIV-infected p, we identified all p on HAART since at least 12 months, with compliance levels of at least 90%, and with altered fasting cholesterol (>200 mg/dL) and/or triglycerides (>250 mg/dL), of at least six-month duration.
Results: When comparing the quarterly immunological trend of the 88 p prospectively taking statins (either pravastatin, fluvastatin, or rosuvastatin) for a predominant hypercholesterolaemia, versus the 103 p receiving fibrates (either bezafibrate, or fenofibrate, or gemfibrozil) for a prevailing hypertriglyceridaemia, versus the 76 p who underwent a diet/exercise programme only, no significant difference in mean-median CD4+ lymphocyte count occurred among the three p groups during a mean follow-up which now reachs 15.3±6.7 (range 6–26) months. All p remained evaluable when still on an HAART regimen ensuring virologic suppression, and an unchanged hypolipidemic therapy.
Discussion: Examining the broad-spectrum pleiotropic activities attributed to statins, also the virologic course of HIV infection received limited evidences from a study claiming a direct anti-HIV activity of statins by down-regulating the small signaling Rho protein [J Exp Med 2004;200:541], while neither virologic failure nor viral blips occurred among 78 HAART-stable p who started a statin [JAIDS 2005;39:637]. Also the anti-triglyceride fibrates may have extensive pleiotropic effects [Curr Vasc Pharmacol 2005;3:87]. In fact, both fibrates and other molecules interacting with the peroxisome proliferator-activated receptors (PPAR) (e.g. rosiglitazone and pioglitazone, whose use is clained by experimental studies on HIV-infected p suffering from the lipodystropy syndrome), may affect major T-lymphocyte cytokine mediators [Circ Res 2002;90:703; J Immunol 2004;172:5790]. Based on our preliminary, simple experience, and waiting for enlarged in vivo studies conducted on affordable p samples, we remain prudent and wait for further studies of both basic and clinical sciences in this field.
P1908 The assessment of knowledge about AIDS and its prevention on Isfahan high school students
I. Karimi, B. Ataei (Isfahan, IR)
Introduction: Since there isn't any effective vaccination against AIDS, the best way for prevention is to increase the knowledge in society; especially adolescents and students in high schools, as the most common location for accumulation of young men and women, has a special importance. The target of this study was to evaluate knowledge about AIDS in high school students of Isfahan, in 2003.
Methods: 1,200 validated questionnaires consisting of 20 questions divided between students of six regions of Isfahan education organisation. In each region 100 male and 100 female students from different fields and grades answered the questions. Then the data were obtained and the result analysed by SPSS 11 software with different statistical tests including spearman χ2 and Student's t.
Results: The knowledge score increased with increasing educational level of parents, especially mother (P < 0.001). There is a relationship between knowledge score and field of education and grade of students. The knowledge level in female students was significantly higher than in males. It was shown to increase with increasing age (P < 0.001) and mean scores of students (P = 0.006).
Among the resources of obtaining information, the most was radio and TV (83.6%) then newspapers and magazines (63.3%); the most uncommon sources were educational programmes in their schools (24.2%) and textbooks (24.3%).
Discussion: Overall the highest level of knowledge was among females, older, high grade, students of experimental sciences field, rich regions and educated parents.
HIV: antiviral treatment
P1909 Incidence and risk factors of nevirapine-associated skin rashes and hepatitis among naïve HIV-infected patients with CD4 cell counts <250 cells/μL
W. Manosuthi, A. Chaovavanich, Y. Inthong, W. Prasithsirikul, S. Chottanapund, C. Sittibusaya, V. Moolasart, P. Termvises, S. Sungkanuparph (Nonthaburi, Bangkok, TH)
Objectives: To determine incidences and risk factors of nevirapine (NVP)-associated skin rashes and clinical hepatitis that lead to NVP discontinuation among antiretroviral naïve HIV-infected patients who had baseline CD4 cell counts <250 cells/μL
Methods: A retrospective cohort study was conducted among naïve HIV-infected patients with baseline CD4 cell counts <250 cells/μL who were initiated NVP-based antiretroviral therapy (ART) between January 2001 and January 2004. All patients were followed until 180 days after initiation of ART. Incidence of skin rashes and clinical hepatitis after 1, 2, 3 and 6 months of ART were studied. Cox's proportional hazard model was used to analyse the possible risk factors.
Results: There were 785 patients with a mean age of 35.2±7.4 years and 44% female. Median (IQR) CD4 cell count was 26 (8–76) cells/μL and median (IQR) baseline plasma HIV RNA was 268,000 (101,250–548,000) copies/mL. Incidence of NVP-associated skin rash grade II-IV at 1, 2, 3 and 6 months after ART were 5.7%, 7.4%, 7.7% and 7.7%, respectively. Incidence of clinical hepatitis at 1, 2, 3 and 6 months after ART were 0.5%, 0.8%, 0.9% and 1.2%, respectively. Cox's proportional hazard was used after adjusting for gender, baseline CD4 cells, log plasma HIV RNA and serum alkaline phosphatase; the result showed that every increment of baseline CD4 50 cells/μL was associated with higher incidence of NVP-associated skin rashes that lead to NVP discontinuation (HR = 1.431, 95%CI: 1.006–2.036, P = 0.046). The number of CD4 cell counts was not associated with NVP-associated clinical hepatitis.
Conclusions: HIV-infected patients with baseline CD4 <250 cells/μL had incidences of NVP-associated skin rashes grade II-IV and hepatitis that lead to NVP discontinuation approximately 8% and 1.2%, respectively. Almost all of events occurred within the first 3 months after ART. Even in HIV-infected patients with baseline CD4 < 250 cells/μL, the higher number of baseline CD4 cells is associated with a higher risk for skin rashes
P1910 Potential role of TDM in dosing protease inhibtors in HIV-HCV co-infected patients with or without cirrhosis
F. Gatti, S. Pagni, P. Nasta, C. Boldrin, A. Matti, A. Loregian, L. Biasi, M. Puoti, S.G. Parisi, K. Prestini, G. Palù, G. Carosi (Brescia, Padua, IT)
Objectives: to evaluate the influence of liver cirrhosis on pharmacokinetics (PK) of the main protease inhibitors (PIs) in HIV/HCV co-infected patients (pts) treated with atazanavir/ritonavir (ATV/r), fosamprenavir/r (FAPV/r) or lopinavir/r (LPV/r).
Methods: 39 HIV/HCV co-infected pts receiving ATV/r (14), LPV/r (13) and FAPV±r (12) were included.
According to liver stiffness (LS) value obtained by fibroscan® at the moment of PK determination or histological diagnosis patients were classified in 2 groups:
-
•
NC, no cirrhosis (24) if LS < 12 kPa or Knodell fibrosis score (Kfs) 1–3;
-
•
C, cirrhosis (15) if LS ≥ 12 kPa or Kfs 4.
PIs plasma levels (PL) were determined by High Performance Liquid Chromatography. Samples for determination of Ctrough (Ct) PL were collected before the morning or the daily dose at the steady state. Results are expressed as median (interquartiles); parametric and non parametric tests have been used for comparison of continuous variables between groups when appropriate (p < 0.05 was considered as significant).
Results: 35 pts had HIV-RNA < 50 and 4 <2500 c/mL; 29/39 pts were on TDF+3tc/FTC. According to CHILD-PUGH score 10/15 C pts were classified as A5, 2 as A6 and 3 as B7.
LS in pts taking ATV/r, LPV/r and FAPV/r was respectively 6 (5–8), 6 (4–9) and 6.3 (5.9–6.9) kPa in NC and 17 (12–22), 34 (21–50) and 53.2 (48–75) kPa in C pts.
LS of C pts on FAPV was significantly higher than LS of C pts on ATV (p = 0.0004); C pts on FAPV were taking 700 mg BID according to DHHS guidelines.
Median Ct levels were 540 (170–990) in NC and 340 (100–460) ng/mL in C pts (p = 0.3) for ATV; 3020 (1020–4910) in NC and 3250 (1490–10100) ng/mL in C pts (p = 0.6) for LPV; 1350 (1020–1740) in NC and 210 (180–420) ng/mL in C pts (p = 0.01) for FAPV Moreover PIs Ct was above minimum target concentration for wild-type virus (as suggested by DHHS guidelines) in all pts taking ATV/r, LPV/r and FAPV/r; this concentration was reached only in 2/5 C pts taking unboosted FAPV, both with a LS < 50 kPa.
Conclusions: ATV and LPV Ct doesn't seem to be affected by liver cirrhosis, while Ct of FAPV was found significantly lower in C with respect to NC pts. Of note C pts on FAPV had a median LS significantly higher than others C pts and this may cause a reduction and/or diversion of the liver blood flow related to initial portal hypertension: dosing FAPV in C pts with a very high LS (>50 kPa) may therefore require TDM.
P1911 Immmune recovery in treatment-naïve patients under HAART according to baseline CD4 count: the Chilean AIDS Cohort (ChiAC) experience
J. Aranciabia, D. Gallardo, C. Beltrán, O. Morales, M. Wolff for the Chilean AIDS Group
Background: Immune recovery is one of the achievable goals of modern antiretroviral therapy (HAART) and is measured through CD4 counts which increase after successful viral suppression. The magnitude of recovery also seems to depend on baseline (BL) CD4 count, the higher the later, the better the results. Present guidelines recommend treatment before severe immunosuppresion is present but many patients begin HAART with CD4 levels much lower than the recommended due to advanced disease at diagnosis.
Objective: To evaluate the rate of immune recovery in treatment naïve pts (TxN) initiating HAART at various levels of clinical and immunologic disease cared for in the Chilean Public Health System and followed by the ChiAC.
Methods: ChiAC has 4,500 pts under follow up, 52% TxN who initiated HAART between 10/2001 and 01/2004. Results from 2,429 TxN with information updated to 08/2006, with an average follow up period of 3.5 years were available. Variables studied were BL CD4 and CDC clinical staging, (A, B and C) HAART regimens and results. CD4 response was compared according to BL CD4 (Group [G] 1: 0–100, G2: 101–200 and G3: 201–300/mm3) both in absolute number (median) and slope and CDC BL clinical stage; CD4 results were measured every 6 months; Patients on HAART with BL CD4 > 300/mm3 were excluded; for statistical analysis SPSS 14 was used.
Results: At the beginning of HAART the number of patients in G1 was 1106; in G2, 712 and in G3, 304. Median baseline CD4 counts for each group were: 36 (SD ±29.0), 151 (SD ±28.4) and 232 (SD ±27) cells/mm3, respectively for groups 1, 2, and 3. Median CD4 rise were 137, 125, 188, 242 and 260 cells/mm3, at 0.5, 1, 2, 3 and 4 years of treatment respectively for G1; 69, 95, 193 and 239 cells/mm3 for G2, and 72, 74, 148, 228, 280 cells/mm3 for G3, respectively for these periods. The higher the BL CD4 the higher the CD4 rise over time but the slopes of the CD4 rise curves were comparable between groups. There was no difference in CD4 rise according to BL clinical status.
Conclusions: CD4 rise after successful HAART in treatment naïve pts was obtained at different levels of BL CD4 count, without significant difference according to BL clinical staging or viral load. The magnitude of the rise but not its rate is dependent on the BL CD4.
P1912 Efficacy and safety of tenofovir, abacavir and efavirenz in treatment-naïve patients: 48-week results (The ABATE Trial)
A. Leon, J. Pich, E. Ferrer, J. Murillas, I. García, F. Segura, F. Vidal, F. Gutierrez, D. Podzamczer, J.M. Miro (Barcelona, Palma de Mallorca, Hospitalet, Sabadell, Tarragona, Elche, ES)
Background: There are few data on the activity of tenofovir (TDF) plus abacavir (ABC) as a nucleotide/nucleoside backbone regimen in combination with efavirenz (EFV). The aim of this study was to evaluate the efficacy and safety of this combination.
Methods: This is a prospective and multicentre cohort study performed in nine Spanish HIV Units. Patients came from a randomised, multicentre, open-label, induction-maintenance clinical trial (the ABATE trial) in naïve HIV-1-infected patients with >100 CD4 cells/mm3 designed in 2002 and started in May 2003. Induction therapy was performed with TDF (300 mg, QD) plus ABC (300 mg, BID) plus EFV (600 mg, QD). Randomisation to maintaining or stopping EFV was planned at 6 months in patients with undetectable plasma RNA HIV-1 viral load (PVL) (<100 copies/mL). However, the DSMB recommended not starting the maintenance phase in September 2003 due to the high rate of early virologic non-response in treatment-naïve patients with the combination of TDF plus ABC plus lamivudine (ESS300009 trial). Endpoints of the study were proportion of patients with PVL of <50 copies/mL, CD4 cell increase and drug-related adverse events (AE) leading to treatment discontinuation at 48 weeks.
Results: The 52 patients included in the study were followed for at least 48 weeks. Mean (IQR) age was 35 (32–43) years, 73% were males. Only 15% of patients were former drug users. At baseline, median (IQR) CD4 and PVL were 287 (238–391) cells/mm3 and 4.9 (4.3–5.3) log10/mL, respectively. None of the patients died or had C events. Three patients were lost to follow-up and 13 patients (26%; 95% CI, 14–39%) discontinued therapy due to AE. Two patients had virological failure. K65R mutation emerged in one patient. The proportion of patients (95% CI) with PVL below 50 copies/mL at week 48 by intention-to-treat (ITT) or per protocol (OT) analysis was 65% (51–78%) and 94% (81–99%), respectively. Median CD4 cell increase (ITT/OT) at 12 months was +201/+232 cells/mm3. AE were due to hypersensitivity reaction (HSR) to ABC in 5 cases, to either CNS disturbances or skin rash associated with EFV in 4 cases, to GI alterations in 2 cases and to skin rash (ABC or EFV) in another 2 cases. 85% of AE appeared within the first month of therapy.
Conclusions: This combination had a high rate of early AE but was highly effective in naïve patients who were able to tolerate it. Genetic screening (HLA-B*5701) to prevent ABC HSR could select in the future those patients who can benefit from this treatment.
P1913 Genotypic drug resistance mutations among HIV-1-infected patients failing an initial non-nucleoside reverse transcriptase inhibitor-based antiretroviral regimen in a resource-limited setting
S. Sungkanuparph, S. Kiertiburanakul, K. Malathum, N. Saekang, W. Pairoj, W. Chantratita (Bangkok, TH)
Objectives: To determine genotypic drug resistance patterns and optimal initial antiretroviral regimens in a resource-limited setting after failing an initial non-nucleoside reverse transcriptase inhibitor (NNRTI)-based antiretroviral therapy (ART).
Methods: A retrospective study was conducted among HIV-infected patients who failed an initial ART of NNRTI-based regimen and had genotype resistance testing between January 2002 and December 2005 in a medical school hospital.
Results: There were 63 patients with a mean (SD) age of 38.3 (7.4) years and 76% were male. Of these, 60% received nevirapine and 40% received efavirenz. For nucleoside reverse transcriptase inhibitor (NRTI) backbone, 79% received lamivudine (3TC) + stavudine (d4T) or zidovudine (AZT) and 21% received didanosine (ddI) + AZT or d4T Median (IQR) duration of ART was 21 (12–32) months. Median (IQR) CD4 cell count and HIV RNA at the time of virological failure was 193 (107–301) cells/mm3 and 4.0 (3.7–4.4) log copies/mL, respectively. The prevalence of patients with ≥ 1 major mutation conferring drug resistance to NRTIs and NNRTIs were 89% and 95%, respectively. For NNRTI resistance, K103N was found more often in patients receiving efavirenz (80% vs. 32%, p < 0.001) whereas Y181C was more common in patients receiving nevirapine (40% vs 4%, p = 0.002). For NRTI-resistance mutations, M184V, TAMs, K65R and Q151M were observed in 73%, 38%, 5% and 8%, respectively. Patients receiving 3TC+AZT or d4T had a higher frequency of M184V than patients receiving ddI + AZT or d4T (90% vs 8%, p < 0.001). Q151M, a multinucleoside resistance mutation, was found more often in patients receiving ddI+AZT or d4T than that in patients receiving ddI+AZT or d4T (23% vs 4%, p = 0.050). The frequencies of TAMs and K65R were not different between the two groups of NRTI backbone. By logistic regression, receiving ddI+AZT or ddI was the only factor associated with occurrence of Q151M (OR 13.4, p = 0.039). NNRTI drug, CD4 cell count, HIV RNA at failure and duration of ART were not associated with Q151M. No predicting factors for the occurrence of TAMs and K65R were found from the analyses.
Conclusion: In a resource-limited setting where the options for the second regimen are limited, 3TC with AZT or d4T should be a preferred NRTI backbone for an initial ART regimen. Genotype resistance testing is still needed to optimise the second ART regimen. The accessibility of this tool needs to be scaled-up along with accessibility of ART.
P1914 The coexistence of secondary PR mutations M36I, K20I and L63H predominates in CRF06-cpx and its next generation recombinant viruses circulating in Estonian treatment-naïve patients
R. Avi, K. Huik, T. Karki, M. Sadam, P. Paap, J. Smidt, K. Ainsalu, T. Krispin, I. Lutsar (Tartu, EE)
Objectives: The naturally occurring HIV secondary drug resistance mutations (DRM) in widely spread subtypes (A, B and C) have been adequately described and a high variability between different subtypes has been demonstrated. Such data on circulating recombinant forms (CRF), however, are often missing. We aimed to describe the profile of DRMs in Estonian treatment naive HIV positive subjects carrying predominantly CRF06-cpx viruses or their next generation recombinants.
Methods: A total of 76 treatment-naive subjects (median ages 26 y; 59 male) infected with HIV-1 between 2000 and 2005 were analysed. Of these 62 (82%) were or had been IV drug users and 14 acquired infection via heterosexual route. A direct sequencing of plasma viral RNA was performed in protease (PR), reverse transcriptase (RT), p7 and V3 regions. DRMs were detected by submitting PR and RT sequences to Stanford University HIV Drug Resistance Database; p7 and V3 sequences were subtyped using HIV-Blast application. A footprint analysis of sequenced regions was applied to map recombination patterns between CRF06-cpx and subtype A viruses.
Results: The majority of subjects (96%) carried rare recombinant viruses (CRF06-cpx by 53/76; various recombinant forms of former and A subtype [A06] by 20/76) and only 3/76 had Eastern European subtype A. On footprint analysis of A06 viruses almost all recombination breakpoints were situated in p7 region. A secondary PR resistance mutation M36I was present in all strains. The mutation L63H and K20I were seen in >90% of CRF06-cpx and in approximately 75% of A06 strains. In addition mutations L33V, I93L and L10I were found in 5%, 5%, 7% of CRF06-cpx viruses, respectively. Of all recombinant viruses more than 80% carried 3 or more PR mutations.
Conclusions: The vast majority of CRF06-cpx and its next generation recombinants with Eastern-European subtype A in Estonia carry number of secondary protease mutations of which the combination of M36I, K20I, L63H was most commonly seen.
P1915 Immunological reconstitution in severely immunosuppressed antiretroviral-naïve patients (<100 CD4+ T cells/mm3) using a non-nucleoside reverse transcriptase inhibitor-based or boosted protease inhibitor-based ART regimen: 3-year results
J.M. Miro, J. Pich, P. Domingo, D. Podzamczer, J.R. Arribas, E. Ribera, J. Arrizabalaga, M. Lonca, E. De Lazzari, M. Plana and the ADVANZ Study Group
Background: There are few randomised clinical trials involving severely immunosuppressed patients. The aim of this study was to analyse whether there are differences in the timing of immune reconstitution in patients with <100 CD4+ cells/mm3 on non-nucleoside reverse transcriptase inhibitor (NNRTI)- vs. boosted protease inhibitor (PI)-based antiretroviral therapies.
Methods: Multicentre, randomised, prospective, open-label clinical trial in naïve HIV-1-infected patients with <100 CD4 cells/mm3. Enrollment period: Nov. 2001 – Jan. 2003. Treatment regimens were two NRTIs plus an NNRTI (efavirenz [EFV] 600 mg QD, with nevirapine [NVP] as the second option for EFV-intolerant patients) or a boosted PI (indinavir/ritonavir [IDV/r] 800/200 mg BID, which, once approved in Spain, could be changed lopinavir/ritonavir [LPV/r] or atazanavir/ritonavir [ATZ/r] as the second option for IND/r-intolerant patients). Analysis was by intention-to-treat (ITT) and per protocol (OT), allowing changes of NNRTI or boosted PI within the same class of antiretrovirals.
Results: Sixty-six patients were randomised: 34 received an NNRTI-based regimen and 32 received a boosted PI-based regimen. 50% had had C events. Median (range) CD4+ and PVL at baseline were 40 (1–99) cells/mm3 and 5.5 (4.0–>6) log10/mL, respectively. All patients completed 3 years of follow-up. Five patients died (NNRTI, 4; PI, 1) and 12 developed a C event (NNRTI, 6; boosted PI, 6), most of them within the first six months. Seventeen patients changed EFV (NVP, 3; LPV/r 1) or IDV/r (LPV/r 9, ATZ/r 3, EFV 3, NVP 1) because of adverse events and eight patients stopped HAART or were lost for follow-up (NNRTI, 1; PI, 7). There were 12 virological failures (NNRTI, 5; PI, 7). PVL < 200 copies/mL for NNRTI/PI arms at 1, 2 and 3 years was 71%/65%, 68%/49% and 65%/29% (p = 0.05 at the last time point) by ITT (M = F) and 73%/77%, 73%/69% and 73%/49% (p = NS at all time points) by OT analysis, respectively. Median CD4+ cell increase after 1, 2 and 3 years by OT analysis was +186/+136, +226/+162 and +303/+220 cells/mm3 (p = NS for all time points) for NNRTI/PI groups, respectively. At 1 and 2 years, immune activation (CD8+CD38+) was significantly lower in the NNRTI group (p = 0.004). There were no differences in T-cell subsets, proliferative responses to mitogens and recall antigens between both study arms.
Conclusions: At 3 years, the virological response and the magnitude of the immune reconstitution induced by an NNRTI-based regimen was at least as potent as that induced by a boosted PI-based regimen.
P1916 The evolution of the avidity of HIV-1-specific antibodies is prevented by early treatment started during primary HIV-1 infection
M. Selleri, M. Zaniratti, N. Orchi, A. Corpolongo, P. Zaccaro, G. Ippolito, M. Capobianchi, E. Girardi (Rome, IT)
Objectives: Avidity of anti-HIV antibodies progressively increases after primary HIV-1 infection (PHI). It has been reported that an Avidity Index (AI) <0.80 identifies recent (<6 months) infections, whereas anti-HIV antibodies with an AI ≥ 0.90 indicates long-standing infections. We evaluated if the administration of highly active antiretroviral therapy (HAART) during PHI may affect AI evolution.
Methods: The AI and Western blot (WB) evolution patterns were retrospectively analysed on serial serum/plasma samples from 13 individuals with symptomatic PHI, defined as detectable viral load and negative or indeterminate HIV-1 antibody tests. Five patients have never been treated, eight individuals had initiated HAART at the time of diagnosis (range 0–46 days after presentation), of whom, 4 discontinued HAART after a variable time lapse from PHI (≥ 1 year).
Results: At diagnosis, the range of HIV viraemia was 0.003–38 ×106 copies/mL. In untreated patients viraemia reached the set-point in 4–6 months, while in all patients receiving HAART complete suppression of viraemia (<50 RNA copies/mL) was achieved early, lasting for the entire observation period (12 months). At presentation, the median AI was 0.42 (range 0.33–0.43) in untreated patients, and 0.44 (range 0.40–0.72) in subsequently treated patients. At 3, 6 and 12 months the median AI was 0.75, 0.89, 0.97 in untreated patients, and 0.42, 0.49, 0.54 in treated patients. In the 4 patients who stopped HAART, AI increased after interruption, reaching the value of 0.80 in 6 months. WB pattern transiently/partially reversed during HAART in 2 patients.
Conclusions: During PHI, early HAART, accompanied by rapid virologic suppression, prevents the gradual increase of AI, while resumption of viral replication is paralleled by a prompt increase of AI. This altered pattern of maturation of the antibody response in patients achieving rapid control of viral replication may suggest a complex mechanism of immune response to HIV not yet fully investigated.
P1917 Antiretrovirals adverse reactions from a prospective HIV/AIDS cohort study in Bogotá, Colombia
J.F. Vesga, C. Alvarez, J.R. Tamara, J.A. Cortes, P. Guerrero, F. Gil (Bogota, CO)
Background: Adverse Drug Reactions (ADR), lead not just to a high percentage of therapy abandonment, but also to a poor adherence to treatment instructions resulting in failure of therapy. The purpose of this study was to show the relation between the use of drugs and ADRs, their impact on treatment and the severity of reaction as defined by the DAIDS scale.
Methods: Making use of a database designed in Access (ver 11.5 2003, Microsoft, USA), information from ADR to Antiretrovirals from 384 HIV positive patients between June 2005 and June 2006 was analysed. Patients were seen routinely every 4 weeks, and data was collected from such consults. Patient was asked about the presence of ADRs, lot and drug manufacturer, the active commencement of the antiretroviral, the effect of the reaction on the overall treatment scheme, and laboratories related to the negative reaction. For abnormal laboratory results a DAIDS grade was established. Data was then analysed using programme Stata (Ver 9.0).
Results: In the consequent year (4608 month/patient), 672 ADRs were reported to have occurred with the following frequency: metabolic reactions 28.1%, haematologic 28%, gastrointestinal 26.5%, neurologic and psychological 13.5%, dermatological 3.28%, hepatic 2.39%, renal and urological 0.9%.
Regarding intensity of ADRs, 85.2% were mild, 14.4% moderate and 1.7% severe. The most frequent ADRs were macrocytosis, nausea, hypertrygliceridaemia, combined dyslipidaemia and diarrhoea. Among all ADRs 89.5% were classified as DAIDS grade I, 8.8% grade II, 1% grade III and 0.4% grade IV. The 89.9% of ADRs mean no change in the scheme of treatment, 6.3% required a change of the drug, 1.3% quit the treatment, and 2.4% of treatments were transitorily suspended.
Conclusions: ADRs were frequent in our study, however the most of it were reported as mild reactions and didnt necessitate change or suspension of the treatment. As expected, metabolic reactions like dyslipidaemia where most frequently found among protease inhibitors and NNRTs than in NRTs.
P1918 Pharmacokinetic parameters of 400/100 mg indinavir/ritonavir in HIV-infected Thai patients
S. Jaruratanasirikul (Songkhla, TH)
Introduction: The dosage recommendations of 800/100 mg indinavir/ritonavir in the Thai population may lead to more severe side effects. This may be due to higher drug levels.
Objective: The aim of this study was to assess the steady state pharmacokinetics of reduced doses of indinavir boosted with ritonavir in HIV infected Thai patients.
Methods: Our study was conducted in ten HIV infected patients. All patients received 400/100 mg indinavir/ritonavir combined with lamivudine and stavudine or zidovudine every 12 h. After 4 weeks of starting antiretroviral therapy, indinavir pharmacokinetics studies were carried out and HIV-1 RNA viral load was determined at 24 weeks.
Results: The pharmacokinetic parameters are shown as below and all patients had a HIV-1 viral load less than 400 copies/mL after 24 weeks follow up.
| Parameter | Mean (SD) | Units |
|---|---|---|
| Cmax | 7.51 (3.57) | mg/L |
| Cmin | 0.86 (0.59) | mg/L |
| AUC0–12 | 38.14 (15.17) | mg·h/L |
| T1/2 | 2.94 (0.90) | h |
| Tmax | 2.10 (1.31) | h |
Conclusion: The dosage of 400/100 mg indinavir/ritonavir can maintain adequate indinavir plasma concentrations and suppression of viral replication was observed in all ten patients at 24 weeks follow up.
P1919 Factors correlated with adherence to HAART: a multicentre study from an HIV-infected Greek population
N. Pitsounis, S. Peristeraki, N. Tsogas, N. Mangafas, M. Chini, A. Lioni, A. Skoutelis, P. Nikolaides, P. Gargalianos, G. Panos, T. Kordosis, V. Paparizos, A. Gikas, M. Moschou, G. Chrysos, G. Koratzanis, M. Lazanas (Athens, GR)
Objectives: Poor adherence to HAART can result in treatment failure and the development of resistant strains of HIV. We examined the association between adherence to HAART and various factors in an HIV infected population.
Methods: A structured questionnaire was distributed to the patients regularly attending 11 Infectious Diseases Units in Greece (∼2200 patients). Adherence to HAART was measured by self report. A single question measuring frequency of dose omission within the previous 4 months was administered. Adherence was defined as <10% of doses missed. Statistical analysis involved non parametric correlations (Spearman's-Rho and Pearson's χ2). Statistical significance was set at 0.05.
Results: 482 patients completed the questionnaire, of whom 409 (84.9%) were male and 73 (15.1%) were female. Mean age was 41 years (range 20–80 years). Age, gender, adjustment to their new health status, satisfaction with the number of pills taken and psychological support received were not associated with adherence.
Factors correlated with adherence were length of known HIV infection (rho = −0.16, p = 0.002), length of antiretroviral treatment (rho = −0.13, p = 0.001), side effects of HAART (rho = −0.11, p = 0.0035), belief about radical cure for HIV infection (rho = 0.11, p = 0.0034), and frequency of condom use (rho = 0.14, p = 0.010).
Conclusion: Adherence to HAART correlates behaviorally positively with the use of condoms (indicating risk aversion), as well as psychologically positively with optimism about radical cure for HIV. Simultaneously adherence to HAART is negatively correlated with disease duration, measured by length of HIV seropositivity and length of treatment, as well as with occurrence of side effects from HAART.
P1920 Increase in prevalence of primary mutations in naive HIV patients in Spain
M. Cianchetta, A. Paravissini, S. Raso, A. Machuca, A. Goyenechea, M. Gorgolas, R. García, M. Fernandez-Guerrero (Madrid, ES)
Objective: Recent studies demonstrate the increase in the prevalence of genotypic mutations of primary resistance to antiretroviral drugs among drug naive HIV patients. The objective of the present work is to evaluate the changes in the prevalence of primary and secondary mutations in protease (P) and reverse transcriptase (RT) in drug naïve HIV patients in one year in our country
Methods: In 85 HIV infected patients, with no previous antiretroviral treatment, viral RNA was extracted by Ampliprep system (Roche) and genotypic testing was performed using Trugene (Bayer).
Results: Mutations of primary resistance to anti-retrovirals were found in 9.4% of patients (5/53) till the end of 2005, with 4 patients with mutations in RT and 1 in IP. However, this number increases till 14% (12/85) at the end of 2006, with 5 patients with mutations in IP, 4 patients with mutations in RT and 4 in both. In patients with no evidence of primary mutations, secondary mutations in P were found in 28 out of 73 patients (38%), being the most common the changes in position L63. Secondary mutations in RT were found in 15 out of 73 (20%) patients, being changes in positions A98 and V179 the most frequents. Silent mutation and polymorphisms were found in all patients.
Conclusions: Primary mutations that confer drug resistance in drug naïve HIV infected patients increased in our country from 9.4% to 14% demonstrating a increment in the transmission of mutated virus
Clinical bacterial infections
P1921 Exogenous ventilator associated pneumonia incidence in a medical-surgical intensive care unit: a four-year surveillance
M. Lecuona, L. Lorente, M. Miguel, B. Castro, S. Campos, Y. Pedroso, A. Sierra (La Laguna, ES)
Objective: To evaluate the ventilator associated pneumonia (VAP) incidence in an ICU, their aetiology and the exogenous VAP incidence.
Method: A 51 month duration prospective study, between July 1, 2000 to September 30, 2004 in a 24-bed medical-surgical Intensive Care Unit (ICU) of a 650-bed tertiary hospital. Patients requiring mechanical ventilation during more than 24 hours were included. The following variables were taken from each patient: sex, age, APACHE II, length of stay in ICU, days of mechanical ventilation, mortality and diagnosis group. Diagnosis of VAP was established according CDC guidelines. Throat swab and tracheal aspirate were taken at the moment of admission and twice a week until discharge for bacterial flora study and/or respiratory infection and colonisation. Respiratory infections and colonisation events were classified regarding the throat flora, in endogenous and exogenous.
Results: A total of 1434 patients were included: 65.1% male, mean age 58.1±16.9, mean length of stay 19.14±21.49 days, APACHE II 15.28±6.82, mortality 23.8%. VAP incidence (I) was 18.96% (272/1434) and VAP incidence by 1000 days of MV (DI) was 15.69‰ (279/18926). Exogenous pneumonia was 7.22% of total VAP (20/277), with an I of 1.39% of total patients and DI of 1.05‰. When we evaluated the temporal evolution of exogenous VAP, we could appreciate a decrease in I (from 3% to 1.4%) and in DI (from 2.4‰ to 1.05‰). More frequently aetiological agents were non-fermenting Gram-negative bacilli (31.3%), Staphylococcus aureus (23.6%) (48% of them were MRSA) and Enterobacteriaceae (21.1%). There were no differences in the exogenous VAP aetiology.
Conclusions: In our ICU exogenous VAP represents a little part of the total amount of VAP. The decrease in the incidence could be due to the effects that continuous surveillance makes in the staff when they manipulate the patient airway.
P1922 Can routine microbiology data be used as a proxy marker for infection?
M. Heginbothom (Cardiff, UK)
Objectives: Currently MRSA bacteraemia data is used as a proxy marker for healthcare associated infections in England and Wales. However this association has not been validated. The aims of the study were to assess if routinely collected laboratory data could be readily retrieved from laboratory systems, and to validate whether such data could provide a proxy marker for infection.
Methods: The study was performed in six District General Hospitals in Wales over a 12 month period in 2004. All microbiology data was transferred into a relational database management system (DataStore) and a programme was developed to mark the infection class of isolates:
-
•
Invasive infection – blood culture, Cerebrospinal fluid, sterile fluid or tissue.
-
•
Non-Invasive infection – pus, sputum, urine, non-sterile fluid, medical device, mouth swab, ulcer swab, wound swab, genital swab, eye swab, or other (miscellaneous specimens).
-
•
Colonisation – ear swab, nose swab, screening swab or throat swab.
Clinical evaluations were performed by infection control nurses using CDC definitions for nosocomial infections (Garner et al. 1988) to assign a clinical infection class to each MRSA isolate. DataStore information was then compared with the ICN-validated data to determine the degree to which the data sets matched.
Results: Clinical evaluations were achieved for 3,924 MRSA isolates (43 were excluded from the analysis as they did not match the study criteria; data for 3,881 isolates were analysed). For those isolates classed by DataStore as ‘Invasive infection', clinical evaluation agreed with 86.7% of the classifications. For those classed as ‘Non-invasive infection’ the agreement was 55.6% and for those classed as ‘Colonisation’ the agreement was 85.7%.
Conclusions: This study shows that isolation of MRSA from specific specimen types can provide a reasonably accurate indication invasive infection or colonisation. Thus microbiological data from selected specimen types could be used as a proxy marker for infection. However the lack of agreement between clinical evaluation and specimen types classed as indicating non-invasive infection suggests that caution should be exercised in the selection of proxy infection markers.
P1923 Characterisation and management of healthcare onset Clostridium difficile associated diarrhoea in a hyperendemic region in Germany
H-P. Weil, F. Mattner, R. van den Berg, P. Gastmeier, E. Kuijper, U. Fischer-Brügge (Nordhorn, Hannover, DE; Leiden, NL)
Objectives: To investigate whether intensifying infection control measures or an antibiotic use programme is effective to reduce C. difficile incidence rates in three German hospitals (geriatrics [A], diabetology [B] and internal medicine [C]).
Methods: A case of CDAD was defined as a patient with diarrhoea and a positive assay for C. difficile toxin A/B. Medication use was subtracted from an electronic pharmacy database. The strains were analysed by PCR-ribotyping and pulsed-field gel electrophoresis (PFGE) and the antibiotic susceptibility was determined by E-test.
Results: In 2005, 120, 57 and 123 patients developed CDAD in hospital A, B, or C, respectively. Isolated strains (n = 54) belonged to the two PCR ribotypes 001 (A and C) and/or 046 (A and B), whereas typing by PFGE (n = 38) revealed 9 different strains. Three strains accounted for at least 6, 10, and 9 transmissions in A, B, or C, respectively. The strains were resistant towards erythromycin (89%), clindamycin (96%) and all quinolones (95%). In April 2006 A and B installed infection control measures and reduced the fluoroquinolone use, B also reduced the use of clindamycin. A reduced the incidence rate by 70%, whereas B could not lower the incidence rate. C switched from cefotaxime to ampicillin/sulbactam without special infection control measures, thereby reducing the incidence rate by 76%.
| Hospital | Incidence ratesa |
Transmission rateb |
Control measures |
Quinolonesc |
Clindamycinc |
Cephalosporinsc |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005 | p.i.d | PFGE | (%) | 2005 | p.i. | 2005 | p.i. | 2005 | p.i. | 2005 | p.i. | |
| A | 58 | 17 | 6/12 | (50) | no | yes | 106 | 46 | 54 | 36 | 131 | 142 |
| B | 48 | 51 | 10/13 | (77) | no | yes | 176 | 66 | 134 | 70 | 119 | 166 |
| C | 38 | 9 | 9/13 | (69) | no | no | 78 | 56 | 29 | 27 | 184 | 54 |
(CDAD/10,000PD)
(DDD/1000PD)
Estimated
After intervention.
Conclusions: Multiresistant C. difficile PCR ribotypes 001 and 046 are hyperendemic in northwest Germany. PFGE typing, rather than PCR-ribotyping, allows the study of nosocomial transmissions. Infection control measures should be installed and the use of fluoroquinolones, clindamycin and cephalosporins should be restricted.
P1924 Study on Clostridium difficile-associated diarrhoea in a hospital in Japan
Y. Ito, T. Matsushita, Y. Takahashi, T. Nakamura, K. Hayashi, E. Morita, H. Kato (Gifu, Tokyo, JP)
Objectives: In Europe and North America, several outbreaks due to NAP1/027 Clostridium difficile have been reported. In our hospital the number of patients with C. difficile-associated diarrhoea (CDAD) increased after January 2005. Our aim of this study is to know nosocomial transmission of specific strains including NAP1/027 among these patients.
Methods: Subjects were 15 patients who suffered from CDAD between January 2005 and March 2006 in Gifu Red Cross hospital, general hospital of 310 beds. The occurrence of CDAD did not concentrate on a single ward, and extended to 5 wards. Five patients had recurrent episodes of CDAD. CDAD was defined as diarrhoea associated with positive for toxigenic culture and/or toxin A assay. The toxin-producing type of isolates was determined by PCR and PCR ribotyping of isolates was performed.
Results: Twenty-two strains of C. difficile were isolated from 22 episodes. Twenty strains of C. difficile were toxin-A positive, toxin-B positive and 2 strains were toxin-A negative, toxin-B positive. PCR detecting the binary toxin gene was positive in 3 isolates from 2 patients. Twenty-two isolates were typed into 9 different PCR ribotypes. Type smz, which has been recognized as an epidemic type causing outbreaks in multiple hospitals in Japan, was recovered from 7 episodes of 4 patients and dominant in our hospital. Other three types were isolated from 2 or 3 patients each. Of 5 patients with recurrent CDAD, 4 patients relapsed with the original strain and 1 patient acquired a new strain. PCR ribotype gc8 (Brazier's PCR ribotype 027) was isolated from one patient with pseudomembranous colitis, who had recurrent diarrhoea caused by the same type. This type was recovered from no other patients.
Conclusions: Not a wide spread of a single C. difficile strain but a small scale transmission of several C. difficile strains was found to be related to the occurrence of CDAD. Although nosocomial spread of NAP1/027 strain was not recognized in our hospital, the isolation of the strain in Japan indicates the potential for wide spread of the strain in Japanese hospitals.
P1925 Teicoplanin versus cefuroxime as prophylaxis in prosthetic joint implant surgery
M. Galiè, E. Pistella, F. Napoleoni, G. Condarelli, C. Santini (Rome, IT)
Objectives: Infections after joint replacement require prolonged antibiotic therapy and often graft removal. Staphylococci cause up to 70–90% of these infections. Antistaphylococcal prophylaxis decrease the infection rate and I° or II° generation cephalosporins are routinary employed. Glycopeptides (teicoplanin and vancomycin) represent a valid alternative for the centres where methicillin-resistant staphylococci are prevalent. Due to the risk of reduced susceptibility to glycopeptides and to the lower activity against methicillin-susceptible strains the use of glycopeptides for prophylaxis is recommended only as alternative and for short periods. Aim of the study was to compare the efficacy of cefuroxime and teicoplanin in a hospital with high (60–80%) incidence of methicillin-resistant staphylococci.
Methods: All the patients undergoing elective total joint replacement over a 12-month period were randomly assigned to receive one of the following prophylactic regimen: teicoplanin (a single dose of 400 mg i.v. preoperatively) or cefuroxime (2g i.v. preoperatively and 1g i.v. every 8 h postoperatively for 24 hours). Patients where daily controlled for wound infection and antibiotics side effects. Gram stains and cultures of wound secretions were routinary performed.
Results: Ninety consecutive patients were enrolled; 48 received cefuroxime and 42 teicoplanin. The 2 groups were comparable with the exception of a higher rate of patients >75 years, with femoral fractures and with diabetes mellitus in the cefuroxime group. Three patients (7.1%) in the teicoplanin group and 1 patient (2.0%) in cefuroxime group developed a surgical wound infection. Methicillin-sensitive S. aureus (2) and Corynebacterium spp. (1) were isolated from wound infections in teicoplanin group and methicillin-resistant S. epidermidis (1) was isolated from wound infection in cefuroxime group.
Conclusion: Our study confirms the prevalent staphylococcal aetiology and the efficacy of cefuroxime. Two infections caused by methicillin sensitive S. aureus occurred in the teicoplanin group. The present study confirms that by virtue of their excellent activity against methicillin-sensitive Staphylococci, I° and II° generation cephalosporins should be preferred for routinary perioperative prophylaxis in orthopaedic surgery. Teicoplanin may represent a reasonable option when the risk of methicillin-resistant staphylococcal infection is high.
P1926 Surgical wound infections after median sternotomy: clinical and microbiologic preliminary results of a large multicentre study in Italy
C. Santini on behalf of the GIS-INCARD (Gruppo Italiano per lo Studio delle Infezioni in Cardiochirurgia)
Objectives: Sternal wound infections (SWIs) after median sternotomy are associated with high mortality and morbidity. Prevention, early diagnosis and adeguate treatment are essential, but consensus abiut the preoperative management and the best surgical treatment strategy is still lacking. The aim of the study was to evaluate the incidence of SWI, the occurrence of the known risk factors, the clinical and microbiologic features and the current therapeutic approaches in a large series of patients.
Methods: Nineteen divisions of Cardiac Surgery spread all over Italy participated in a prospective, observational study. Pre, intra and postoperative risk factors of all the patients undergoing median sternotomy in the first week of every month from October 2005 to July 2006 were recorded. Further data were recorded for all the patients who developed SWI.
Results: Overall, 1,746 patients were evaluated. Characteristics of the patients are reported in Table 1 . All patients received antibiotic prophylaxis for a mean duration of 46.5 hours. Overall 76.1% received cefazolin, cefuroxime or cefamandole, 24.7% had glycopeptides (8.1% as single dose combined to other regimens) and 4.8% received ampicillin-sulbactam or amoxicillin-clavulanate, 1.1% carbapenems and 1.2% other antibiotics. Forty-six patients developed SWIs (2.7%); 78.5% were due to Staphylococci. Deep SWIs were 26 (19 sternal osteomyelitis and 7 mediastinitis) and required reoperation (73%), mediastinal irrigation (30.7%), closed mediastinal drainage (19.2%), vacuum continuous aspiration (30.7%) and plastic surgery (23%). Forty-four patients recovered (95.6%) and 1 died for mediastinitis.
Table 1.
| Mean age (years) | 65.9 |
| Mean preoperative hospitalisation (days) | 5.5 |
| Mean ASA score | 2.95 |
| Coronary artery bypass graft/valve replacement (%) | 58.4/34.9 |
| Elective procedures (%) | 88 |
| Cardiopulmonary bypass (%) | 90.9 |
| Length of surgery >3 hours (%) | 86.3 |
| Preoperative hyperglycaemia (%) | 22.7 |
| Cigarette smoking (%) | 21.3 |
| Patients with cultures for staphylococcal nasal colonisation (%) | 52.1 |
| Staphylococcal nasal carrier (% of cultured patients) | 8.6 |
| Shaving of operative site (%) | 97.8 |
| Shaving of operative site at day of surgery (%) | 40.6 |
| Mechanical ventilation >24 hours (%) | 19.5 |
| Central venous catheter >72 hours (%) | 32.7 |
| MediastinaI drainage >72 hours (%) | 19.4 |
Conclusions: The rate of SWI was acceptably low. However, efforts for shorter hospitalisations before surgery, better control of preoperative hyperglycaemia and for preoperative shaving closer in time to surgery should be considered. The low rate of staphylococcal nasal carriers suggests further evaluations for the routinary preoperative nasal cultures. Although perioperative prophylaxis was mostly adequate for choice of regimen and duration, only 76% of our patients received I° or II° generation cephalosporins as recommended, the remaining receiving vancomycin. Deep involvement was frequent and required re-operation in 75% of cases. Mediastinal irrigation or drainage, vacuum aspiration and plastic surgery were frequently required. One out of 7 patients with mediastinitis died, confirming the severe prognosis of this event.
P1927 Matching criteria in case-control studies in postoperative infections
M.E. Falagas, E.G. Mourtzoukou, F. Ntziora, G. Peppas, P.I. Rafailidis (Athens, GR)
Background: Matching is commonly used in case-control studies to control for the effect of major confounding factors. We sought to evaluate the available evidence from case-control studies regarding postoperative infections to identify how frequently matching was performed and with what specific variables.
Methods: We searched for relevant case-control studies archived by PubMed until August 2006. Only studies regarding postoperative infections were included in our review. We further evaluated studies that used individual matching between cases and controls.
Results: We identified and evaluated 42 relevant studies. Age was used as a matching criterion in 27 of these 42 (64.3%) case-control studies. The specific type of surgical procedure was the second most commonly used criterion, in 17 of 42 studies (40.5%). Gender was used in 14 of 42 studies (33.3%) as a matching criterion between case and control patients. The period at risk for development of surgical site and/or other postoperative infections, that is the time from surgery to the diagnosis of infection was used in 9 of 42 studies (21.4%), as was the date of operation, and the primary diagnosis that led the case and control patients to surgery. Same surgeon or surgical team was used in 7 studies (16.7%), while matching according to the National Nosocomial Infection Surveillance system risk score was performed in 5 studies (11.9%).
Discussion: The findings of our analysis suggest that various characteristics have been used for matching in case-control studies of postoperative infections. A more consistent use of matching with the specific type of surgical procedure may help in increasing the internal validity of a case-control study in this field of clinical research.
P1928 Risk factor analysis for surgical site infections following vascular surgery
F. Mattner, E. Ott, I. Chaberny, P. Gastmeier, O. Teebken (Hannover, DE)
Objective: To determine surgical site infection (SSI) rates in abdominal and peripheral vascular surgery and to perform a risk factor analysis
Methods: Between Jan 2002 and Dec 2005 all patients (pts) receiving vascular surgery (thoraco-abdominal, abdominal, retroperitoneal, iliaco-femoral, femoral, femoro-distal replacements, patches, stents or bypasses) were included in our cohort study. SSIs were determined according to CDC definitions. Postdischarge data were included when available. Risk factor analysis was performed including indications for surgery, emerging procedures, intervention procedures, demographic data, and material of vascular grafts as variables.
Results: In 750 pts, 73 thoraco-abdominal, 276 abdominal, 24 retroperitoneal, 172 femoral and 53 femoro-distal surgical procedures, 99 stent-graftings and 112 embolectomies were performed (combined intervention are included). 42 (5.6%) pts developed SSIs in average 11.5 days (range 5 to 33 days) after surgery. 28 (3.7%) superficial, 10 (1.3%) deep SSIs and 4 (0.5%) organ space infections occurred. S. aureus [13 (31%), out of which 8 (19%) were MRSA], CoNS [17 (40%)], enterococci [14 (33%)], and P. aeruginosa [7 (17%)] were the most prevalent pathogens [other pathogens (21 (26%)]. Vascular surgery in pts with femoral interventions (relative risk (RR) 2.3, 95%CI 1.6;3.4, p < 0.001), operative revisions (RR 1.2, 95% CI 1.1;1.4, p < 0.001), peripheral vascular disease class Fontaine III (OR 3.5, 95% CI 1.5;8.2, p = 0.002) and the use of autologous vessels (RR 2.0, 95%CI 1.3;3.3, p = 0.008) increased the infection risk significantly. Elective aneurysm repair (OR 0.4; 95% CI 0.21;0.77, p = 0.008), and the use of stents (RR n.d., p = 0.01) was associated with a decreased infection risk.
Conclusion: In vascular surgery postoperative surgical site infections occur frequently (incidence 5.6%). The choice of surgical access, graft material and the grade of peripheral vascular disease at the time of surgery were found crucial risk factors for the development of surgical site infection in vascular surgery. Future studies should address modifications of surgical access, graft materials, and whether the decision for an intervention in earlier stages of peripheral vascular disease would prevent surgical site infections.
P1929 Joint implant infections in a general hospital
F. Moreno, J. Aguilar, M. Noureddine, J. De la Torre, J.L. Prada, A. Del Arco, L. Merida, J.D. Ruiz-Mesa (Marbella, ES)
Objectives: to analyse features and sources of joint implant (JI) infections and the medical and surgical treatment in our hospital.
Method: Retrospective study of patients admitted suffrering from JI infections from 1997 to 2005. Hospital data base and discharge reports were used.
Results: 89 patients were admitted. 15 had 2 admissions and 8 three admisions with a total of 121 cases of JI infection. The mean-age was 73 ± 12.39 years. 64% were female. 22% were diabetic, 25% heart patients and 11% COAF patients. In 57% artrosis was the cause of JI indication, and 32% by fracture. 60% were hips (96% by fracture), and 38% from knee (63% by artrosis). 85% were the first JI: 81% complete/16% parcial. In 45% first symptom started in first month after surgery, 34% between 1 month and 2 years and 20% more than 2 years after surgery. Only 20% had risk factors for infection, 10% surgical wound infection and 10% haemorrhage and haematoma. The more recurrent symptoms were local inflamation or wound drainage in 50%, joint pain in 31% and fever in 14%, a fistula in 8% and in 2% joint effusion. Bacteria were isolated in 57%, in 36% from fistula or injure 9% from joint fluid and the others from surgical samples or blood stream. Only a bacteria was isolated in 82%. In 12% were 2 bacterias and in 6% more than 2. In 57% Staphylococcus was isolated (37% S. aureus, and 20% S. coagulase negative), in 19% Bacillus Gram-negatives (P. aeruginosa and E. coli), and 14% Streptococcus (beta-haemolytic streptococcus, viridans and enterococcus), and 6% were with more than 2. About therapy, surgical cleaned and antibiotical treatments were performed in 66%. JI was removed in 15% with reinstatement of a new JI. JI removed without a new implant and antibiotics in 9%. In the others, parcial JI were removed, cronic antibiotical treatment was indicated or amputation was made. The mean length of stay was 24 days (TD 19.33). Only 2 died. The common antibiotical combination was aminoglicosides plus cloxacilin or cefazoline, vancomycin or ciprofloxacin (44%), and cocktail with ciprofloxacin plus cloxacilin, rifampicin or vancomycin (16%). One case was treated only with one antibiotic. Ciprofloxacin was the wide used (12%). After discharged therapy was prolonged between 3 and 16 weeks with a mean of 7 weeks.
Conclusions: More than 50% were late JI infections but in less than 50% JI was removed. In the causes, S. aureus is the main bacteria followed by Staph coagulase negatives and Gram-negatives.
P1930 A nosocomial outbreak due to Serratia marcescens in a West-Tallinn hospital
I. Zolotuhhina, R. Voiko, P. Albri, V. Adamson, K. Allik (Tallinn, Tartu, EE)
Objectives: A nosocomial outbreak due to S. marcescens in neonatal unit of West-Tallinn Central Hospital involving 7 neonate patients was investigated.
Materials and Methods: During 2005 and 2006 total of 1796 specimens (369 blood cultures and 1427 swabs taken from nasopharynx, eyes, ears and umbilical wounds) from patients with clinical signs of infection were examined. First isolation of S. marcescens from blood of twins, one of whom developed meningitis (S. marcescens was also isolated from the cerebrospinal fluid) and another sepsis was registered in November 2005. 6 following isolates were obtained in the period from April to May 2006 from 5 patients – 2 from blood cultures and 4 from nasopharyngeal swabs. All these patients manifestated clinical symptoms of true infection (purulent rhinitis, sepsis). A nosocomial infection was suspected. The epidemiological investigation included the review of medical records and of medical and nursing practices in the department. 40 environmental samples from surfaces and medical equipment were examined. All cultures were identified initially by BBL Crystal and confirmed by other biochemical tests. Susceptibility of all strains against 6 antibiotics was determined by the CLSI agar dilution method. Pulse-field gel electrophoresis (PFGE) was used for genotyping of 6 strains.
Results: During epidemiological investigations a single S. marcescens strain was isolated from the inner surface of the bottle with 0.05% xylomethazoline drops used for the patients with nasal congestion. The bottle had been in use during several months. All investigated strains of S. marcescens had similar antibiotic susceptibility profiles. 6 cultures recovered from clinical material and from the bottle showed indistinguishable PFGE patterns. This led to conclusion that the bottle with nasal drops contaminated with S. marcescens was the likely reservoir from which the spread of infection occurred. The bottle was eliminated and that put a rapid end to the outbreak. No new recovery of S. marcescens from patients and unit environment has been registered since May 2006.
Conclusions: The outbreak was related to the bottle with nasal drops contaminated with S. marcescens. From November 2005 till May 2006 7 neonates in the unit developed clinical manifestations of true S. marcescens infection. The outbreak stopped after the elimination of the bottle. The importance of following good medical practice and standard hygienic precautions should be stressed out.
P1931 Microbiological epidemiology of infections related to pace-makers and indwelling cardiac defibrillator
C. Tascini, M. Bongiorni, G. Gemignani, E. Soldati, A. Leonildi, G. Arena, S. Capolupo, F. Menichetti (Pisa, IT)
Background: The cardiology Unit at Cisanello Hospital in Pisa is the Italian reference centre for non invasive, transvenous removal of infected pace-makers (PM) or indwelling cardiac defibrillator (ICD). The aim of the study is to review microbiological findings of PM and ICD infections observed at this Unit in five years period (2000–2005).
Materials and Methods: Retrospective observational study on consecutive patients. All patients with clinically documented PM or ICD infections were enrolled in the study. To define the aetiology of the infections. Catheter leads and/or the infected material from the pocket upon removal were cultured in aerobic and anaerobic solid media.
Results: In the study period PM and ICD leads and generators were removed transvenously from 602 patients. Microbiological documentation was obtained from 451 patients (75%) and 560 different microorganisms were isolated; therefore we had negative culture in 151 patients. In 99 patients infections were polymicrobial. Staphylococci were the most frequently isolated pathogens (474/560–84%). The coagulase-negative staphylococci (CNS) resulted the first pathogen (380) followed by S. aureus (94), methicillin-resistant strains, among staphylococci, were 123/474 (26%). Gram negative rods and fungi are relatively rare. Overall susceptibility were also studied; the classes of antibiotic with good activity were newer quinolones (moxifloxacin and levofloxacin followed by aminoglycosides and glycopeptides).
| Year |
Total | ||||
|---|---|---|---|---|---|
| 2000–2001 | 2002–2003 | 2004 | 2005 | ||
| Patients | 121 | 228 | 136 | 127 | 602 |
| Microrganisms isolated | 132 | 180 | 128 | 120 | 560 |
| CNS | 93 (70%) | 117 (65%) | 89 (69%) | 81 (67%) | 380 (68%) |
| S. aureus | 19 (14%) | 35 (19%) | 19 (15%) | 21 (18%) | 94 (17%) |
| Total MR staphylococci | 33 (29%) | 36 (30%) | 29 (22%) | 25 (25%) | 123 (26%) |
| Gram-negative rods | 16 | 14 | 3 | 2 | 35 (6%) |
| Yeasts/moulds | 1 | 4 | 2 | 5 | 12 (2%) |
| Others | 4 | 10 | 15 | 11 | 40 (8%) |
Conclusions: In our experience PM and ICD infections were mainly caused by Staphylococci, especially CNS, other microorganisms (Gram-negative rods and fungi) were also documented in few cases. The knowledge of the epidemiology of these infections and the susceptibility to antibiotics might be useful for clinicians to start an adequate empiric antibiotic therapy.
P1932 REA-RAISIN: microbiology of national surveillance network of ICU-acquired infections, France, 2005
R. Gauzit, A. Savey, A. Lepape, B. Tressieres, N. Garreau, T. Lavigne, F. L'Heriteau, P. Parneix, J. Fabry, B. Coignard on behalf of the National Nosocomial Infection Alert, Investigation & Surveillance Network (RAISIN), France
Background: The objective of nosocomial infections (NI) surveillance in intensive care units (ICU) is to assess and compare rates, microbiology and resistance over time and amongst ICUs, in order to provide an evidence-based approach for improving infection control practices.
Methods: In France in 2004, surveillance methods were standardised through a national surveillance project (raisin). The national REA-RAISIN network conducts 6 months a year a patient-based NI surveillance in ICU. Surveillance (on a voluntary basis and using a standardised methodology) focuses on device related-infections: ventilator-associated pneumonia (PNE), urinary tract infections (UTI), central venous catheter colonisation (COL) with or without catheter-associated infections and bacteraemia (BAC). A maximum of 2 bacteria are recorded for each infection with, for the most relevant of them, resistance markers.
Results: From January to June 2005, 151 ICUs included 20,632 patients. A total of 5 159 events were documented concerning 2 569 patients (12.4%). The distribution of micro-organisms per site under surveillance is illustrated in the table . The resistance markers were: MRSA 47.5%, Enterococcus ampicillin-R 18.5%, Enterococcus vancomycin-R 1.8%, Enterobacteriaceae cefotaxime-R 17.2%, A. baumannii ceftazidime-R 78.6%, P. aeruginosa ceftazidime-R 22.6%, and S. maltophilia ceftazidime-R 48.5%.
| PNE (n = 2,314) | COL (n = 840) | BAC (n = 785) | UTI (n = 1,428) | |
|---|---|---|---|---|
| Enterobacteriaceae | 29.1% | 24.3% | 26.7% | 45.6% |
| Other GNB | 3.3% | 0.2% | 1.9% | 0.2% |
| P. aeruginosa | 19.7% | 14.4% | 8.5% | 15.9% |
| A. baumannii | 2.2% | 2.4% | 1.0% | 1% |
| Haemophilus sp. | 5.4% | − | − | − |
| S. aureus | 22.8% | 13.5% | 18.6% | 1.8% |
| CNS | 2.4% | 35.7% | 21.2% | 2.1% |
| Enterococcus sp. | 0.7% | 3.5% | 8% | 12% |
| Other GPC | 6% | 0.5% | 3.9% | 1.7% |
| Candida sp. | 5.4% | 3.7% | 5.2% | 18.1% |
| Anaerobes | 0.2% | − | 3.7% | − |
| Others | 2.7% | 1.5% | 0.9% | 1.4% |
Conclusion: This report on microbiology and resistance markers of NI surveillance in ICU from a large sample of French hospitals will serve as a national reference and will allow describing, evaluating and monitoring resistance level in ICUs. Feedback to ICUs provides them with relevant information to monitor and target antimicrobial use and infection control policies.
P1933 Clinical outcomes of Clostridium difficile ribotype 027 infection versus other ribotypes at a UK hospital
R. Whiting, J.C. O'Driscoll (Aylesbury, UK)
Objectives: Stoke Mandeville Hospital came to prominence due to a large outbreak of Clostridium difficile disease over a two-year period. It was the first site in the UK where ribotype 027 was identified, and the initial clinical impression was that infections due to this ribotype were more severe and resulted in more relapses and deaths than had been experienced previously.
The objective of this study was to study patient records retrospectively to determine whether or not infection with ribotype 027 resulted in a poorer outcome than that with other ribotypes grouped together.
Methods: A survey form was designed. Notes of patients whose C. difficile strains had been typed between February 04 and October 05 were reviwed. The results were analysed statistically.
Results: The notes of 77 patients were studied. Forty-five of these (58%) had ribotype 027 infection, infection was due to ribotype 001 in 13 cases, ribotype 106 in 12 cases, ribotype 026 in 2 cases, and the following ribotypes were found in 1 patient each; 005, 017, 050, 078 and 081. The median age of patients in both the 027 and non-027 groups was 79 years.
There was no statistical difference in any of the following between ribotype 027 strains and non-027 strains: duration of diarrhoea (first episode), number of relapses, and crude mortality. In addition, there was no statistical difference in any of the following parameters between the 2 groups (as measured on admission to hospital, at onset of symptoms, and 1 week after onset of symptoms): white cell count, haemoglobin, sodium, potassium, creatinine, C-reactive protein, or albumin.
Conclusion: This study suggests that, despite initial perceptions to the contrary, C. difficile ribotype 027 infection is no more severe than that due to other ribotypes.
P1934 Ointments as a source of methicillin-resistant Staphylococcus aureus spread
D. Orth (Innsbruck, AT)
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) has for several years been increasingly spreading at inpatient and outpatient healthcare facilities and constitutes a great epidemiological problem. Colonised and infected patients represent the most important reservoir and MRSA is transmitted by direct person-to-person contact, usually on the hands of healthcare workers. Long term carrier and recurrent infections/colonisations occur despite optimal preventive hygienic measurements. The source of MRSA spread is unknown in such cases. The aim of this study was to investigate the contamination of ointments as a source of MRSA spread in the University Hospital of Innsbruck.
Methods: During a six month period, ointments of MRSA-positive patients were screened for MRSA contamination. MRSA strains collected from patients and ointments were tested for in vitro susceptibility and the automated ribotyping was used for molecular typing. Ointments primary for skin care and other medical indications were applied for one patient only and used during hospitalisation.
Results: A total of 101 ointments from19 MRSA-positive patients were examined. Seven ointments specimens (6.9%) (Bepanthen®, Candio-Hermal® soft paste, Spoendlin'sche® nasal ointment and even Bactroban®) from 3 patients were found positive for MRSA. Four different ribotypes (RT 2, 6, 10, 12) were occurred in patients and 2 in ointments (RT2, 6). MRSA from the ointments were identical to MRSA collected from patients as shown by ribotypes and antimicrobial susceptibility profile in all cases.
Conclusions: Ointments, in use for patients care were contaminated with MRSA and could therefore contribute to MRSA spread in the patient-setting.
P1935 Retrospective analysis of multidrug-resistant Acinetobacter baumannii isolates recovered during a 3-year period in a Portuguese hospital
G.J. Da Silva, T. Ferreira, A. Duarte (Coimbra, Lisbon, PT)
In Portugal, an A. baumannii multi-resistant strain belonging to European clone II is disseminated since 1998 and is endemic in some hospitals.
Objective: In recent years, the Hospital dos Capuchos, a hospital located in Lisbon, has increasingly observed the isolation of resistant A. baumannii. The main goal was to study the epidemiology of A. baumannii infection during 2004–2006 and to determine whether the European clone II strain was, at present, endemic.
Methods: During the period of December 2003 until September 2006, 49 non-duplicate A. baumannii isolates were collected. Antibiotic susceptibility was determined by VITEK 2 Advanced Expert System. Clinical epidemiologic study of A. baumannii isolates was performed by combination of antibiotyping and randomly amplified polymorphic DNA analysis (RAPD).
Results and Conclusions: Isolates were more frequently recovered from older patients (55 to 70 years old, 26.5%; 71 and more, 55%). Fifty seven per cent of the isolates were collected from intensive care units samples, 32.7% from Medicine wards, 8.2% from Surgery wards and 2% from Haematology. They were isolated mainly from bronchial secretions (40.8%), urine (24.5%) and blood (22.4%). Ten different antibiotypes were found, according to antibiotic susceptibility profiles. The majority of isolates collected since January 2004 showed multidrug-resistance. Only one isolate from January 2004 showed a susceptibility profile, with a unique DNA fingerprint. During 2004, it was observed an increasingly higher genetic diversity (five genotypes), replaced at the end of the year, and subsequent years, by two major clusters. Both were multidrug resistant, including resistance to carbapenems and quinolones. Cluster A isolates were consistently susceptible to gentamycin and amikacin, showing intermediate susceptibility to ceftazidime, while isolates from cluster B were resistant to all antibiotics except to tobramycin and amikacin (variable susceptibility). Phenotypic and genotypic profiles of cluster B isolates were compatible with those of European clone II strain. At our knowledge, cluster A is found for the first time in Portugal in this hospital. Although, considering antibiogram and DNA fingerprinting, it cannot be considered as a clone. It is possible that these isolates may have evolved independently from a common ancestor, forming a recent lineage. The study shows that several multidrug-resistant clusters of EU clone II can coexist endemically for several years.
P1936 Assessment of the risk factors for mortality in hospital-acquired Acinetobacter baumannii infections in a teaching hospital
C. Bulut, M.A. Yetkin, F.S. Erdinc, C. Ataman Hatipoglu, U. Onde, E.A. Karakoc, N. Tülek (Ankara, Samsun, TR)
Objectives: Acinetobacters are increasingly involved as aetiological agents of hospital acquired infections in seriously ill patients, in recent years. The aim of this study was to evaluate the hospital acquired Acinetobacter baumannii infections and to assess the risk factors associated with mortality.
Methods: Prospective laboratory based active surveillance has been performed in our hospital between January 2000 and December 2004. Hospital acquired infections were diagnosed according to the CDC criterias. Conventional methods were used for the identification of Acinetobacter isolates. Disc diffusion method was used for antimicrobial susceptibility test of the microorganisms.
Results: Hospital acquired Acinetobacter baumannii infections were detected in 95 male and 71 woman patients. The mean age of the patients were 58.0±17.8 years. Acinetobacter baumannii was isolated mainly from the ICU patients (32.5%), from the patients on the orthopaedy wards (16.3%) and on the neurology ward (11.4%) consecutively. Predisposing factors detected in the patients infected with Acinetobacter baumannii were urinary catheter insertion (81.3%), mechanical ventilation (25.3%) and central venous catheter (21.1%) consecutively. The mean duration of time between hospitalisation and occurrence of infection were 17.2±14.8 days. Urinary tract infection, surgical site infection, primary bloodstream infection and hospital acquired meningitis were the most common infections and existed in 30.1%, 29.5%, 17.5% and 7.3% of patients respectively. Carbapenems and netilmicin were the most effective antimicrobials agents against Acinetobacter isolates and an evident decrease in amicasin resistance was detected overall the five year period.
Crude mortality rate was 36.5%. Older age (≥60 years), mechanical ventilation, presence of central venous catheter, tracheostomy, being an ICU patient, presence of coma and having a carbapenem resistance were found to be associated with mortality due to A baumannii infection by univariate analysis. Of these factors older age (≥ 60 years), mechanical ventilation, presence of coma and being an ICU patient were independently associated with mortality on multivariate analysis (p <0.05).
Conclusion: These data suggest that, clinical efforts aimed at the improvements in the medical management of ICU patients with severe A. baumannii infection especially those who need mechanical ventilation, are required for further improvement in patient outcome.
P1937 Multidrug-resistant Acinetobacter baumannii in a surgical intensive care unit
J.M. Sahuquillo Arce, E. Colombo Gainza, A. Gil Brusola, P. Ramirez Galleymore, M. Bosch Alepuz, J. Cordoba, M. Gobernado (Valencia, ES)
Objective: To study the changes in the population of Acinetobacter baumannii in a period of two years (2003–2005) in an endemic situation of a surgical ICU.
Materials and Methods: We contrasted the antibiotic susceptibility (Rosco Disk diffusion and bioMérieux VITEK 2) and the genetic polymorphism (Amersham Biosciences Random Amplified Polymorphic DNA [RAPD]) data of two environmental studies conducted in February 2003 and January 2005 in a surgical ICU to determine the colonisation pressure of multidrug-resistant A. baumannii. The antibiotic consumption of this department and the percentage of patients who suffered a bacteraemia by this microorganism were analysed as well.
Results: Nineteen strains of A. baumannii were isolated in the first study, this represented 20% of all the samples included. They were classified into three different clones by the RAPD: clone 1 (5 isolates) was susceptible to sulbactam/ampicillin (SAMP), colistin (CS), sulfamethoxazole trimethoprim (SXT), and two of the isolates were also susceptible to amikacin (AMK); clone 2 (13 isolates) was susceptible to SAMP, CS, AMK, and five isolates were intermediate to cefepime (CEFP); clone 3 (1 isolate) was only susceptible to CS.
Thirty seven strains of A. baumannii were isolated in the second study, this meant 31% of all the samples included. They were classified into the three original clones by the RAPD: clone 1 (18 isolates) was susceptible to SAMP, CS, SXT and two of these were also susceptible to AMK; clone 2 (3 isolates) was susceptible to CL, AMK, intermediate to CEFP and SAMP; clone 3 (16 isolate) was only susceptible to CS.
The consumption of SXT during that period of time did not vary, while that of CEFP decreased to half its previous value. On the other hand, the use of SAMP and AMK reached three times its initial level. Bacteraemia by A. baumannii increased constantly from 2.8% to 4%.
Conclusions: A population of multidrug-resistant A. baumannii has settled at our surgical ICU. Three different clones comprise all the isolates found in both studies with a two-year delay, which points to an endemic colonisation. The antibiotic pressure over this population due to the increased use of AMK and SAMP favoured the expansion of clones 1 and 3, whereas clone 2 almost disappeared. An extensive use of antibiotics against this microorganism did not improve the situation, moreover, the number of bacteraemia cases augmented and the treatment options dimished.
P1938 Use of nested PCR for evaluation of Helicobacter pylori transmission by endoscopies
H. Mikkelsen, L.P. Andersen (Copenhagen, DK)
Objective: Helicobacter pylori colonise the gastric mucosa of almost half the world population and have been recognized as the main cause of gastritis. Additionally, it is considered to play a major role in development of duodenal ulcers, gastric ulcers and gastric lymphoma as well as gastric cancer. Infection is usually acquired in childhood and it is speculated that infections occurs primarily from person to person via faecal-oral routes. However, the precise route of transmission remains undocumented and so far no reservoir for the bacteria has been identified outside the human stomach. Endoscopy is a potential source of nosocomical infection and several studies have described transmission of infectious agents via contaminated endoscopes. Different approaches have been applied in order to prevent such iatrogenic infections, and have resulted in the formulation of strict guidelines for cleaning and disinfections of endoscopes after use. At Copenhagen University Hospital, Rigshospitalet endoscopes are cleaned in endoscopic washing machines after use. The quality of cleaning and disinfections is monitored by routine inspections. During the last 5 years the measured germ number have been very low and H. pylori has not been identified in these samples.
Methods: To assess the efficacy of cleaning of endoscopes water samples were collected from both the air channel and biopsy channel of 20 endoscopes. In the latter case a brush was passed through the channel followed by cleaning of the brush in sterile water. The water samples were processed and a Helicobacter pylori-specific PCR targeting the 16S rDNA gene was conducted to determine the presence or absence of the bacteria in the water samples.
Results: Samples from all endoscopes were found to be negative with respect to the presence of H. pylori.
Conclusion: Based on the absence of H. pylori it is concluded that the present cleaning practise is sufficient to prevent iatrogenic infections in relation to endoscopes.
P1939 Catheter-associated urinary tract infections: distribution of uropathogens and patterns of antimicrobial resistance in an Italian hospital (2000–2006)
M. Bonadio, T. Tartaglia, S. Fondelli, G. Andreotti (Pisa, IT)
Objectives: To evaluate some risk factors which could affect the isolation rates of various uropathogens and their in vitro antibiotic resistance in patients with catheter-associated urinary tract infection (CAUTI) at an Italian teaching hospital.
Methods: A prospective study was conducted in an Urinary Tract Infections Clinic at Pisa General Hospital. Two hundred twenty-nine patients (72 males and 157 females) with documented CAUTI (≥104 CFU in a catheter sample of urine) were enrolled from January 2000 to October 2006. One hundred eighty-four (81.3%) of all patients were over the age of 70. The distribution of uropathogens and their in vitro susceptibility to antimicrobials were evaluated.
Results: The causative microorganism of CAUTIs were the following: Escherichia coli (28.4%), Enterococcus spp. (18.3%), Pseudomonas spp. (12.6%), KES group spp. (10.0%), fungi (10.0%). E. coli was more frequently isolated in women than in men (31.8% vs 20.8% respectively; p = 0.1). An increased trend of fungi (mostly Candida spp.) isolation rate throughout the whole study period was observed. E. coli patterns of antibiotic resistance were the following ones: cotrimoxazole (35.0%), ampicillin (51.6%), ciprofloxacin (18.6%), nitrofurantoin (12.7%). Enterococci were all susceptible to vancomycin and teicoplanin. Pseudomonas spp. were frequently resistant to ciprofloxacin (40.7%); the most active antibiotics against Pseudomonas were imipenem, amikacin, piperacillin-tazobactam and aztreonam. E. coli showed a significant increase of resistance (p = 0.03) to ciprofloxacin during the study period. E. coli and Enterococcus spp. isolated in patients who received antimicrobials within the 2 months prior to their enrollment were often in vitro resistant to ciprofloxacin and gentamycin. Nitrofurantoin remained the most active drug against E. coli and Enterococcus in both groups of patients whether or not they received antimicrobial drugs.
Conclusions: The type of uropathogens associated with CAUTI have changed over the last 7 yrs at our hospital. The gender of patients seems to influence the isolation rate of E. coli. An increased trend of isolation rate of fungi has been observed during the study period. A previous antimicrobial treatment in patients with CAUTI increases the risk of isolation of uropathogens resistant to the antimicrobials.
P1940 Epidemiology of MRSA colonisation in a university hospital and a cardiosurgical hospital in Hessen, Germany
A. Knaust, K. Madlener, N. Tschernych, J. Barekzai, T. Eikmann, C. Herr (Giessen, Bad Nauheim, DE)
Objectives: Infections caused by methicillin resistant Staphylococcus aureus (MRSA) are a major health problem in German hospitals, causing increased duration of hospital stay and incidence of mortality. In 2004 the Paul-Ehrlich Institute published a rate of 22.6 percent methicillin resistant isolates amongst all S. aureus isolates sent in by reference laboratories. Since rates of methicillin resistance have constantly increased during the last decades strategies to prevent a further spread of MRSA are urgently needed. Screening of incoming patients became more and more evident to be an effective method to decrease rates of colonisation and infection with MRSA in hospital patients. In previous studies various designs were used for MRSA screening and each strategy needs careful evaluation for each hospital or healthcare facility.
Methods: In the current study based in a university hospital and a cardiosurgical hospital, different strategies were used to screen incoming patients for colonisation or infection with MRSA: (i) patients who belong to risk groups (history or MRSA, wounds, haemodialysis, submission from healthcare facilities); (ii) patients who were admitted to selected departments or wards (intensive care units); (iii) all patients were screened upon admission. Microbiological diagnostics were performed either by bacterial cultures or by a real time PCR based method.
Results and Conclusion: The total prevalence of MRSA colonisation and/or infection was about 3 percent during the initial four month period comprising more than 1000 screening samples. Most of the classical risk factors showed only weak correlation to an enhanced risk of MRSA colonisation. Screening of risk groups will miss many asymptomatic MRSA carriers as could be shown when complete screening of all admissions was performed. A fast microbiological verification is of great advantage for patient management on the wards to reduce preventive isolation measures, i.e. single rooms. Each hospital needs to evaluate its individual patient population to establish an effective and save screening strategy to prevent MRSA transfer and infection.
P1941 A practical approach to post-discharge surveillance of surgical site infections
A. Knaust, A. Moussa, T. Eikmann, C. Herr (Giessen, DE)
Objectives: Surgical site infections account for about 30 percent of all nosokomial infections, resulting in prolonged hospital stay, increased rate of morbidity and mortality and additional costs for hospitals and public health systems. Systematic surveillance of infections has been shown to serve as an effective tool to improve hygiene management and consecutively to reduce infection rates and costs. As, in times of increasing cost pressure, duration of hospital stay after many surgical interventions is minimised to a few days, surveillance during hospital stay is suspected to miss a major part of infections.
Methods: In the current study surgical site infections in five surgical departments were monitored during hospital stay by a computer assisted on-ward surveillance system and after discharge from the hospital by a questionnaire and personal communication by telephone.
Results and Conclusion: The vast majority of surgical site infections were found by post-discharge surveillance. Infection rates were up to 8-fold higher than during on-ward surveillance. About 90 percent of infections became evident within 10 days after surgical intervention. The evaluation of the symptoms pain and tumour by the questionnaire were shown to be a fairly good screening tool. The combination of these two questions led to a positive likelihood ratio of 4.75 and negative likelihood ratio of 0.29 for the detection of a surgical site infection, taken the telephone interview as gold standard. These results demonstrate a simple and time saving approach for the surveillance of surgical site infections which overcomes the bias caused by shortened hospital stays after surgical interventions.
P1942 A low nurse-to-patient ratio is an independent risk factor for late-onset VAP
I. Uçkay, S. Hugonnet, H. Sax, D. Pittet (Geneva, CH)
Background: The economic and clinical burden of ventilator-associated pneumonia (VAP) is uncontested. The role of nurses' understaffing is poorly understood.
Objective: To assess whether low nurse-to-patient ratio increases the risk of VAP and whether this effect is similar for early-onset and late-onset VA P.
Methods: Prospective, observational, single-centre cohort study conducted in the medical intensive care unit (ICU) of the University of Geneva Hospitals. All patients at risk of ICU-acquired infection admitted from January 1999 to December 2002 were followed from admission to discharge. Collected variables included patient characteristics, admission diagnosis, APACHE II score, comorbidities, exposure to invasive devices, daily number of patients and nurses on duty, nurse training level and all-site nosocomial infections. VAP was diagnosed according to standard definitions. VAP was defined as early-onset when occurring 1 to 5 days after intubation, and late-onset when occurring from day 6. Respiratory infections other than VAP were excluded.
Results: Of 2470 patients who were followed during their ICU stay, 262 VAP episodes were diagnosed in 209/936 patients (22.3%) receiving mechanical ventilation. Late-onset VAP accounted for 61% of all episodes. Median duration of mechanical ventilation was 3 days (interquartile range, 3–11). VAP rate was 37.6 episodes per 1000 days at risk (95% CI, 33.2–42.4). ICU mortality among patients with and without VAP was 33.5% and 31.2%, respectively (p = 0.54), whereas hospital mortality was 45.0% and 39.5%, (p = 0.154). Neither the nurse training level nor the APACHE II score had any effect on the hazard of VA P.
The median daily nurse-to-patient ratio over the study period was 1.9 (interquartile range, 1.8–2.2). Results were similar in multivariate analysis for all VAP (adjusted HR 0.66, p = 0.11) and early-onset VAP (adjusted HR, 0.78, 95% CI, 0.42–1.45). For late-onset VAP, a higher nurse-to-patient ratio was independently associated with a reduced risk in multivariate Cox regression analysis (adjusted HR, 0.42, 95% CI, 0.18– 0.99).
Conclusions: A low nurse-to-patient ratio is an independent risk factor for late-onset VAP.
P1943 Intensity of care is the major determinant of coagulase-negative staphylococcal bacteraemia in NICU: implications for risk adjusted surveillance of hospital-acquired infections
P.H. Phillips, M. Cortina Borja, M. Millar, L. Thalib, S. Kempley, R. Gilbert (London, UK; Kuwait City, KW)
Coagulase-negative staphylococcus (CNS) is the commonest cause of bacteraemia in UK Neonatal Intensive Care Units (NICU), accounting for two-thirds of positive blood cultures[1]. CNS bacteraemia is related to the use of invasive devices and can be reduced by up to 50% by improved hygienic practices [2]. We propose that the rate of CNS bacteraemia could be used as a marker for the quality of care related to hospital acquired infection in NICUs. To be interpretable across different units, surveillance must be risk adjusted to take into account the vulnerability of babies and the invasive procedures used. We examined how risk factors including intensity of care predicted CNS bacteraemia in a London NICU.
Methods: The study population included all NICU inpatients in the hospital over 9 years. Admission data were linked with the laboratory database for all blood cultures. Levels of care, used for resource allocation throughout the UK, were defined as Special Care, High Dependency or Intensive Care, and were used as surrogate markers for invasive procedures. Dates of admission to each level were used to determine the total days at risk at each level of care. Analyses determined the rates of CNS infection stratified by various risk factors and intensity of care. Multivariate Poisson regression was then used to estimate adjusted incidence rate ratios for CNS bacteraemia.
Results: 2492 infants were admitted to NICU. 393 infections were recorded, of which 164 (42%) were recurrent. The overall rate of CNS bacteraemia was 5.79 per 1000 baby days, with crude rates of 2.95, 9.63 and 8.92 in Special Care, High Dependency and Intensive Care respectively. Intensity of care was strongly associated with CNS infection, with those in High Dependency being most at risk: the incidence rate ratios adjusted for gestation at birth were 3.48 (95% CI 2.43–4.96) and 2.24 (95%CI 1.54–3.29) for High Dependency and Intensive Care compared with Special Care. Babies born between 32 and 37 weeks were significantly less likely to develop bacteraemia than term infants, but the adjusted rate ratios for lower gestations were not significant.
Conclusion: Level of care was the main determinant of the incidence of CNS bacteraemia. These predictors warrant evaluation in other datasets with a view to developing a marker for risk adjusted monitoring of hospital acquired bacteraemia.
P1944 Reducing hospital infection rates in burn unit thanks to compliance with infection control measures
Z. Ozkurt, M.N. Akcay, S. Erol, A. Aras (Erzurum, TR)
Objective: Because infections are easily develop on burn wound, hospital infection rates usually high in burn units. The aim of the study is to show affect of compliance with infection control measures on burn wound infections.
Methods: The study was conducted a 14-bed burn unit hospitalised paediatric and adult case in a 1200-bed university hospital. Previously infection rates and antibiotic resistance rates were high. Sometimes educations had been made related infection control measures at unit. Imipenem resistant P. aeruginosa strains were increased and 5 pan-resistant strains observed. Environmental surveillance cultures were performed. Shutting the unity would be planned if pan-resistant strains can not be eradicated Education related infection control precaution was repeated and strictly adherence was advised the unity staff.
Results: Compliance with infection control measures was increased. Adherence of hand hygiene, asepsis and antisepsis improved. Environmental culture results show that hydrotherapy tank is hydrotherapy tank is an important source of P. aeruginosa infections. Because of previous experience we had learned that completely disinfection of the tank was very hard, and sometimes it may be impossible. For this reason hydrotherapy tank use was abandoned. Environmental cleaning and disinfections were made more frequently. Isolation and contact precautions were applied colonised and infected patients. Different rooms were used for surgical operation of colonised or infected patients and others. Empiric antibiotic use was not received at admission, antibiotics use was limited only by infectious diseases specialist at short periods.
At this period burn wound infection rates sharply decreased. Infection rates were reduced: 28.3% in 2003, 16.8% in 2004, 9.8% in 2005 and 7.4% in 2006. Pan resistant P. aeruginosa strains were not seen. Antibiotic susceptibility in the Gram-negative microorganisms was improved to some degree.
Conclusion: Adherence of universal infection control practice reduced the burn wound infections. Antibiotic use should be based on evidence, and duration of the antimicrobial drugs should be short. Hydrotherapy tank should use only if necessity is available.
P1945 European surveillance of surgical site infections (HELICS-SSI), 2004–2005
J. Wilson, C. Suetens, I. Ramboer, J. Fabry for the HELICS-SSI working group
Introduction: Surgical site infections (SSI) account for up to 26% of healthcare associated infection (HCAI) and cause considerable morbidity. There is a perception that the risk of acquiring HCAI varies between countries because of different standards in infection control practice. The Hospitals in Europe Link for Infection Control through Surveillance (HELICS) has formed a ‘network of networks' enabling data from hospitals contributing to national surveillance networks to also be submitted to the HELICS database. This database provides an opportunity to evaluate inter-country differences in rates and explore the problems associated with making such comparisons.
Methods: HELICS SSI surveillance is targeted at 7 defined groups of procedures. A standard protocol is used by partner countries to collect a defined set of demographic and operation data on all patients undergoing an eligible procedure (http://ipse.univ-lyon1.fr). Additional data is provided on those that subsequently develop an SSI that meets the case-definition.
Results: Data on a total of 255 999 operations were received from 15 countries from 2004 to 2005 (641 hospitals in 2004, 776 hospitals in 2005). The coverage of national networks varies and hence a considerable proportion of the HELICS dataset comprises data from countries with well-established networks (41% from Germany, 24% England and 15% France). All countries submitted data on hip prosthesis Rates of SSI are conventionally expressed as a cumulative incidence. However, there are important differences between countries in the use and intensity of post-discharge surveillance and length of post-operative inpatient stay. In hip prosthesis the length of postoperative stay varies from 7 to 14 days and therefore incidence density of SSI/1000 post-operative inpatient days is a more valid measure for making inter-country comparisons (see Figure 1 ). The incidence density can only be calculated when the discharge date is known. Despite the standard HELICS protocol, there is evidence of differences in interpretation of case definitions and sensitivity of case finding that need to be taken into account when making comparisons.
Fig. 1.

Incidence density of surgical site infections/1000 post-op inpatient days in HPRO (+95% CI) by country, HELICS-SSI, cumulative data for 2004–2005.
Conclusions: The HELICS network represents a unique opportunity to measure the occurrence of clinically defined HCAI across European countries using standard definitions of infection, methods of data collection and analysis. The data shows inter-country differences in rates of SSI but also provides evidence of variation in methodology that needs to be considered when comparing rates.
P1946 Effectiveness of the antibiotic policy by bed-side consultation
A.K. Alzubaidy, W. Bronsveld, A. Steenhoek, F. Vlaspolder, J.H. Sloos (Alkmaar, NL)
Objectives: Restrictive use of antibiotics is important in the prevention of resistance against these medicaments in hospitals. This study describes the effect of bed-side consultations on the maintenance of antibiotic guidelines. In the MCA a medical doctor under supervision of the consultant microbiologist is comparing the antibiotic use with the hospital guide lines of each individual admitted patient on the wards internal medicine, geriatrics, and cardiology. After physical examination a written advice is given to the clinician. In case of discrepancy between this advice and the initially started antibiotics, the consultant microbiologists advices the clinician additionally.
Results: From March-July 2005, and the same period in 2006 a total number of 469 consultations were performed (318 internal medicine ward, 97 geriatrics, 48 cardiology, and 6 of other wards). For 360 (76.8%) cases guidelines for antibiotic treatment were available, whereas for 28 (6.0%) cases no indication for antibiotics existed. 389 (82.9%) of the given advices were directly followed and 24 (5.1%) additional consultation were necessary for success. In 25 (5.3%) cases the clinician has deviated from the guidelines rightly. In 216 (46.1%) cases continuing the initially chosen antibiotics was the option chosen, whereas in 160 (34.1%) antibiotic has to be changed, and in 82 (17.5%) to stop the antibiotic treatment.
Conclusion: Bed-side consultations results in uniformity and reduction of the use of antibiotics on clinical wards.
P1947 Isolation of Microbacterium species in a case of dialysis-associated peritonitis
I. Wybo, D. Piérard, M. Ichiche, K. Vandoorslaer, O. Soetens, P. Rosseel, S. Lauwers (Brussels, BE)
Objectives: To present a case of dialysis-associated peritonitis due to Microbacterium species with a poor response to intraperitoneal vancomycin. This genus has occasionally been reported as human pathogen.
Methods: Case report. A 48-year-old woman with chronic renal failure treated by continuous ambulatory peritoneal dialysis since June 2002 was hospitalised in April 2004 with peritonitis. The patient improved with intraperitoneal aztreonam 2 g and vancomycin 1g in 2L dialysate and was discharged after four days with amoxicillin/clavulanic acid 875 mg twice a day orally. She was readmitted with recurrence of peritonitis on day 8. Cultures of peritoneal dialysate became positive with coryneform Gram-positive rods. In spite of a new course of intraperitoneal vancomycin, the cellular count of peritoneal effluent remained elevated and the implanted catheter was removed on day 15. Peritoneal dialysis was restarted without further problems 2 months later. Microbiological examination: Specimens were cultured after concentration of the effluent by Millipore filtration according to standard protocols. The following identification techniques were used: conventional phenotypic tests, whole-cell fatty acids (CFA) analysis with the Microbial identification System (MIS-Microbial ID, Newark, Del) and DNA sequencing of the 16 S rRNA gene. Susceptibility testing was performed by disk diffusion (Rosco) and by EtestR (Oxoid).
Results: A yellow pigmented, catalase positive, oxidase negative, coryneform Gram positive rod was repeatedly recovered from concentrated peritoneal dialysate effluent. The CFA profile showed predominance of anteiso C15:0 (35%) and anteiso C17:0 (40.1%). This pattern fitted with Microbacterium species as did presence of gelatinase activity and lack of xylose fermentation. 16 S rRNA gene sequence analysis (1393 bp) showed 99.8% similarity with M. oxydans DSM 20578T and 99.6% with M. liquefaciens DSM 20638T. According to disk diffusion the isolate was susceptible to ampicillin, ciprofloxacin and vancomycin. However, the vancomycin MIC determined by E-test was 8 mg/L, suggesting a diminished susceptibility.
Conclusions: The failure of vancomycin treatment in this case of CAPD-peritonitis caused by Microbacterium species was probably due to decreased susceptibility of this organism and presence of the foreign body. Cure was obtained after removal of the implanted catheter.
P1948 Infectious complications of monoclonal antibodies used in cancer therapy
P.I. Rafailidis, O.K. Kakisi, K.Z. Vardakas, M.E. Falagas (Athens, GR)
Objectives: The introduction of monoclonal antibodies in the treatment of cancer has led to improvement in patients'survival. However, little attention has been paid to the infectious complications associated with their use.
Methods: We performed a systematic review of the literature to identify randomised controlled trials (RCTs) that included in their outcomes comparison of the infectious complications of a monoclonal antibody plus a chemotherapy regimen versus the chemotherapy alone.
Results: 20 RCTs with relevant data were retrieved. They regarded the use of monoclonal antibodies in patients with haematological malignancies (10 RCTs) and solid tumours (10 RCTs). Six RCTs compared rituximab in conjunction with CHOP versus CHOP alone for the treatment of B-cell non-Hodgkin lymphoma (NHL). No significant additive effect to the frequency of development of infection was caused by the addition of rituximab to chemotherapy (data from 5 RCTs). However, in HIV seropositive patients rituximab seems to augment the possibility of a serious infection event or an opportunistic infection and was associated with a 12% higher infection-related mortality (data from 1 RCT). Six RCTs compared trastuzumab in addition to chemotherapy versus chemotherapy alone in patients with solid tumours (4 in breast cancer). The addition of trastuzumab to the various chemotherapy regimens caused a slight increase in the frequency of infectious complications when compared to chemotherapy regimens alone.
Conclusion: Monoclonal antibodies added to chemotherapy have comparable infectious complications with the chemotherapy alone regimen when administered for the treatment of patients with NHL, with the exception of HIV seropositive patients.
P1949 The utility of vitamin K3 for local injection therapy against advanced pancreas cancer
S. Osada, S. Komori, S. Matsui, Y. Tokuyama, F. Sakashita (Gifu, JP)
Background: The molecular mechanism of Vitamin K3 (VK3)-induced cellular growth inhibitory effect was characterised to evaluate its efficacy by local injection for application against unresectable pancreatic cancer.
Methods: Cell viability was determined by test method with MTT. Expressions of cellular proteins were evaluated by Western blott analysis.
Results: The IC50 of VK3 for pancreas cancer cells was calculated to be 42.1 ± 3.5 microM. By Western blot analysis, VK3 was shown to induce phosphorylation of extracellular signal-regulated kinase (ERK) and c-Jun NH2-terminal kinase (JNK) for 30 minutes. Expression of apoptosis by VK3, as shown by caspase-3 activation, and poly ADP-ribose polymerase cleavage is found. Treatment with the thiol-antioxidant (>0.2 mM) completely abrogated VK3-induced ERK but not JNK phosphorylation or inhibition of proliferation. A caspase-3 inhibitor had no inhibitory effect on the proliferative activity of VK3. As a comparison with agent to mediate cellular molecular phosphorylation, oxidative stress (H2O2) at concentrations >5.0 μM was found to inhibit cell proliferation at 24 hours. H2O2 also induced phosphorylation of JNK or ERK, and activation of Caspase-3. And H2O2-induced these activations were completely abrogated by non-thiol-antioxidant, but not thiol-antioxidant. Because H2O2 mediated growth inhibitory effect was quite hasty, high toxicity was anxious for in vivo study. By contrast, VK3 was found to induce extensive tumour tissue necrosis. By Western blot, ERK phosphorylation, but not JNK, was clearly detected only in response to VK3 injection into tumour tissue.
Conclusion: The local injection with VK3 may be useful in the curative treatment of unresectable tumours, and the detection of ERK phosphorylation in tissue is important to predict the effect.
New antifungals and antifungal resistance
P1950 In vivo efficacy of the triazole BAL8557 against disseminated Candida albicans in mice assessed by survival and tissue burden in temporarily and persistently neutropenic mice treated with 1–7 doses of drug over a dose range of 20–80% of Emax
P.A. Warn, A. Parmar, A. Sharp, M. Heep, J. Spickermann, D.W. Denning (Manchester, UK; Basle, CH)
Objectives: Isavuconazole (BAL8557/BAL4815, ISA) is a water-soluble broad-spectrum antifungal triazole. We determined the efficacy of ISA in temporarily (TN) and persistently neutropenic (PN) mice infected IV with C. albicans. Mice were given ISA covering 20–80% of the Emax, 3 to 50 mg/kg, determined by a dose response curve after a single dose of ISA.
Methods: Male CD1 mice and a clinical strain of C. albicans FA6862 were used. Mice were compromised using either 1 dose of 200 mg/kg cyclophosphamide (TN) or multiple doses (PN). Mice were infected IV with an LD50 (8×103 cfu/mouse) orLD90 inoculum (2×104 cfu/mouse) in the TN and PN models of C. albicans then were treated 5 hours later with solvent, 3, 7, 25 or 50 mg/kg ISA 1× daily (20, 40, 60 & 80% Emax). 3 mice had kidney burden assessed 24 hours post 1, 3, 5 and 7 days treatment. Groups of 6 mice were treated 1× daily for 1, 3, 5 & 7 days with ISA or solvent & survival assessed for 11 days post infection.
Results: (A) TN model: tissue burdens after 1 dose of 25 or 50 mg/kg were significantly lower (p < 0.05) than controls & 3 mg/kg at all time points up to 11 days post infection. Mortality was significantly lower with 25 & 50 mg/kg than solvent treatment even after 1 dose (p < 0.05). (B) PN model: tissue burden was significantly lower than solvent controls after a single dose 25 & 50 mg/kg (p < 0.05). Mortality in solvent treated controls was 100% reducing to 0% after 25 & 50 mg/kg. Survival in all ISA treatments (including a single dose of 3 mg/kg) was significantly superior to solvent controls.
Conclusions: ISA demonstrated impressive efficacy against disseminated C. albicans in both temporarily and persistently neutropenic models measured by both tissue burden and survival. Highly significant improvements in outcome were achieved by single doses of ISA which were still evident 10 days later indicating high efficacy and prolonged protection.
P1951 Dose response in neutropenic mice with disseminated infection of multiple strains of Aspergillus fumigatus with widely varying MIC and MFC values treated with isavuconazole
PA. Warn, A. Parmar, A. Sharp, D. W. Denning (Manchester, UK)
Objectives: Isavuconazole (BAL4815, ISA) a broad spectrum triazole antifungal is administered as its water soluble prodrug BAL8557. Dose response curves to treatment with ISA were determined in murine models against strains of Aspergillus fumigatus with low and high MIC (<0.5 and ≥mg/L respectively) and low and high MFC (<1 and >4 mg/L respectively). Strains were selected that had previously demonstrated either treatment success or failure when treated with itraconazole.
Methods: In vitro susceptibility tests were performed according to the CLSI guidelines. A. fumigatus strains isolated from clinical samples were selected with ISA MIC/MFC values classed as either Low/Low, Low/High or High/High (including AF91 which has previously been shown as resistant in vitro and in vivo to itraconazole). Male CD1 mice 21–24g were immunosuppressed with a single dose of 200 mg/kg cyclophosphamide IV then 3 days later infected IV with a sub-lethal inoculum of Aspergillus (4 strains of fumigatus). 5 hours after infection mice were treated orally once daily with either solvent or ISA (2.5–250 mg/kg) for 4 days (ISA 1.33–132 mg/kg active compound). 100 hours post infection mice were killed and the kidneys removed for quantitative culture.
Results: All strains generated moderately severe non-lethal infections with moderate/heavy tissue burdens in the kidneys of infected animals (Cmax 3.4–4.5 log10 cfu/g). Treatment resulted in Emax of 1.2–1.9 log10 cfu/g. Increasing efficacy occurred over the entire dose range. In all cases the steepest part of the dose response curve was in the range of 5.3–40 mg/kg ISA (regardless of the MIC or MFC of the isolate). The magnitude of the drug effect was up to 1.2 log10 cfu/g and similar regardless of the ISA MIC (AF10 MIC = 0.25 mg/L and AF91 MIC = 2 mg/L both had an Emax 1.2 log10 cfu/g).
Conclusion: ISA was effective in reducing the tissue burden of all isolates of A. fumigatus including a strain with an MIC of 4 mg/L that demonstrates in vivo resistance to itraconazole. Increased efficacy was noted over a wide therapeutic range (up to 250 mg/kg). In this model once a day therapy with ISA was effective despite a half life in mice <2 hours.
P1952 Identification of hsp90 in Cryptococcus neoformans and examination of the effect of Mycograb® on hsp90 gene expression in cells exposed to various stress factors
R. Al-Akeel, A. Curry, E. Hoyes, L. Nooney, R. Matthews, J. Burnie (Manchester, UK)
Cryptococcus neoformans is the causative agent of cryptococcosis, a chronic and life-threatening infection common in AIDS patients. Mycograb® is a recombinant antibody derived against the LKVIRKNIV epitope of candidal heat shock protein 90. Hsp90 is an important molecular chaperone in all eukaryote cells. Its interaction with groups of proteins involved in cellular regulatory processes helps to maintain their active conformation. Little is known about the function of Hsp90 in C. neoformans in protection against cryptococcosis. A monoclonal antibody specific for Hsp90 was used to detect Hsp90 in C. neoformans cells grown in sub-inhibitory concentrations of amphotericin B and to identify the effects of amphotericin B on Hsp90 expression. We have examined the expression of the hsp90 gene in C. neoformans and the ability of Mycograb® to inhibit its expression under various stress factors, including heat shock and treatment with various antifungal agents. Cells were exposed to sub-inhibitory concentrations of anti-fungal agents alone and plus three different concentrations of Mycograb®. The expression of Hsp90 in presence of these antifungal agents and the effect of Mycograb® on its expression was examined. Total RNA was isolated from stressed cells and analysed by RT-PCR. Immuno-electron microscopy showed that Hsp90 was mainly located on the outer membrane surface of the cells. The electrophoretic outer membrane protein patterns indicated 14 major protein bands within a molecular weight range varying from 19 to 188 kDa. All isolates displayed similar protein patterns irrespective of the test conditions although band intensity was clearer when cells were grown in the presence of amphotericin B than those were not. Western blotting with anti-Hsp90 monoclonal antibody revealed that the 90kDa was present in the outer membrane fraction of disrupted cells. Results of RT-PCR showed that the treatment of C. neoformans with sub-inhibitory concentrations of amphotericin B and fluconazole induced Hsp90 mRNA expression, while treatment with caspofungin did not. Hsp90 gene expression was inhibited by different concentrations of Mycograb®. The presence of Hsp90 in the capsule of C. neoformans supports the idea of using this protein as a target for new antifungal or antibody therapy. Inhibition of Hsp90 mRNA gives the potential to investigate the use of Mycograb® in the treatment of cryptococcosis.
P1953 In vitro activity of fluconazole and voriconazole against invasive Candida parapsilosis, Candida tropicalis and Candida glabrata isolates in patients with candidaemia from 2001–2005 in Hong Kong
M. Hui, M-L. Chin, L-K. Chan, K-C. Chu, C-Y. Chan (Shatin, HK)
Objectives: Candidaemia is one of the commonest nosocomial blood stream infection and is associated with high mortality. Non-albicans Candida organisms had been repeatedly reported to emerge in high risk patients. In particular, Candida parapsilosis, Candida tropicalis and Candida glabrata are among the most frequently encountered invasive isolates. These organisms often raised a concern on reduced antifungal susceptibility. Therefore, it is important to monitor for the trend of antifungal resistance. In this study, we aimed to investigate the in vitro activities of fluconazole and voriconazole against invasive Candida parapsilosis, Candida tropicalis and Candida glabrata isolates in patients with candidaemia from 2001 – 2005 in Hong Kong.
Methods: Non-duplicate Candida parapsilosis, Candida tropicalis and Candida glabrata isolates collected from candidaemic patients from 2001 – 2005 were tested against fluconazole and voriconazole in accordance to the NCCLS(CLSI) standard M44-A by disk diffusion method. Fluconazole and voriconazole susceptibility results were interpreted according to NCCLS(CLSI) criteria into susceptible, susceptible dose dependent (SDD) and resistant categories. Tests were done in duplicates.
Results: A total of 84 isolates were collected over the 5-year period. 27 were Candida parapsilosis, 32 were Candida tropicalis and 25 were Candida glabrata. All Candida parapsilosis were susceptible to fluconazole and voriconazole. Only 10 isolates (40%) of the Candida glabrata were susceptible to fluconazole, 12 (48%) showed decreased susceptibility (SDD) and 3 (12%) were resistant to fluconazole. Whereas 24 (96%) Candida glabrata were susceptible to voriconazole (p < 0.05). For Candida tropicalis, 24 (75%) isolates were susceptible to fluconazole, and 21 (65%) were susceptible to voriconazole (p > 0.05).
Conclusion: Candida parapsilosis remains very susceptible to both fluconazole and voriconazole. Voriconazole has significantly more activity than fluconazole against Candida glabrata. Both drugs showed similar activities against Candida tropicalis. Although some cross-resistance may be expected for the two azole agents, their activities were different with various species. Antifungal susceptibility testing is warranted to guide the choice of antifungal agents and to monitor the trend of resistance.
P1954 Trends in species distributions and susceptibilities of fungal pathogens recovered from European medical centres: a report from the SENTRY Antimicrobial Surveillance Program (1999–2006)
T. Fritsche, M. Stilwell, H. Sader, R. Jones (North Liberty, US)
Objectives: To understand changes occurring with the prevalence of fungal pathogens in European patients and characterise key susceptibility (S) profiles of the predominant species utilising results from the fungal surveillance component of the SENTRY Program (1999 to 2006). Development of standardised testing methodologies now allows the tracking of resistance (R) emergence to numerous old and new antifungal agents.
Methods: A total of 965 Candida spp. (CSP; 94.8%), 33 Aspergillus spp. (ASP; 3.2%), 8 C. neoformans (0.8%) and 12 other isolates (1.2%) from infected sterile-site sources (primarily bloodstream) were submitted from >20 European (EUR) medical centres to the central monitor (JMI Laboratories, IA) for identification and S testing. S for 5-fluorocytosine (FC), fluconazole (FLU), itraconazole (ITR) and voriconazole (VOR) were determined as part of the SENTRY Program for the intervals 1999–2001 and 2003 using NCCLS/CLSI reference methods and interpretive criteria (M38-A and M27-A2).
Results: Species rank order of CSP occurrence was: C. albicans (CA; 49.6%), C. parapsilosis (CP; 10.1%), C. glabrata (CG; 7.9%), C. tropicalis (CT; 6.9%), C. krusei (CK; 2.1%) and uncharacterised CSP (3.0%). Prevalence of CA declined slightly over the years 1999 to 2006 from 51 to 48%, whereas CP and CG both increased during the monitored interval (9.3 to 11.8% and 2.8 to 11.8%, respectively); CT and CK also increased slightly during the study (5.4 to 8.3% and 1.0 to 2.9%, respectively). Among ASP, A. fumigatus was predominant (63.6%). Comparison of MIC90 values for CA, CP and CT, respectively, demonstrated few differences between the intervals of 1999–2001 (13th ECCMID, abstract P713) and 2003 (44th ICAAC, abstract M1797) for FC (0.25–0.5, 0.12–0.5 and >64 mg/L), FLU (0.25–0.5, 1–2 and 2–4 mg/L), and ITR (0.06–0.12, 0.25–0.5 and 0.5 mg/L). Increases in MIC90 values (≥ four-fold) of VOR were seen between these intervals with CP (0.015 to 0.06 mg/L), CG (0.25 to 2 mg/L) and CT (0.06 to 0.25 mg/L). CG also displayed the highest MICs against FLU (MIC90 values, 16–128 mg/L) and ITR (2 mg/L).
Conclusions: CSP accounted for 94.8% of fungal isolates recovered from patients in EUR medical centres from 1999–2006. While most were CA, increases were detected among other species, particularly CG, that display reduced S to azoles. The potential for emergence of species with R to recently marketed antifungal agents (echinocandins) warrants continued monitoring.
P1955 Killing kinetics of caspofungin and anidulafungin against Candida krusei
E. Canton, J. Pemán, A. Viudes, A. Espinel-Ingroff, A. Valentín, M. Bosch, M. Gobernado (Valencia, ES; Richmond, US)
Introduction: C. krusei is isolated with low frequency from blood culture with the frequency of candidaemia caused by this species ranging from 2 to 3%. Treatment of these infections presents problems, especially in the immunocompromised patient. A fungicidal agent should be the optimal choice to treat these patients.
Objective: To determine the killing kinetics of caspofungin and anidulafungin against C. krusei.
Materials and Methods: Three strains of C. krusei were tested. MICs were determined following M27-A2 method. Caspofungin and anidulafungin MIC2 and MIC0 (minimum concentration that produces a growth reduction ≥50% and 100%, respectively) were determined at 24 h.
Time-killing studies were performed in RMPI 1640 medium (5 mL) by using an inoculum size of 1 to 7×105 CFU/mL. Concentrations tested ranged from 0.03 to 32 mg/L. Aliquots were removed to determine CFUs at 0, 2, 4, 6, 12, 24 and 48 h. Time-killing data were fitted to an exponential equation (Nt = N0 × e−kt) to determine the time (hours) to achieve 50, 90, 99 and 99.9% (T50, T90, T99 and T99.9, respectively) of growth reductions from the starting inoculum at each concentration tested.
Results: Caspofungin MICs0 were 0.5 to 2 mg/L and those of anidulafungin were 0.25 to 0.5 mg/L for the three strains tested. The fungicidal activity of both echinocandins against the three C. krusei isolates was strain dependent. With 2 mg/L, the T50 and T99.9 ranges for anidulafungin were 1.4 to 4.3 h and 14.1 to 42.9 h, respectively and for caspofungin 0.06 to 2.48 h and 24.73 to 48 h, respectively.
Conclusions: Anidulafungin and caspofungin display strain and concentration dependent fungicidal activity against C. krusei, but more than 24 h were required to reach a three log decrease in CFU with both agents. The in vitro vs. in vivo correlation of these results is yet to be determined in clinical trials.
P1956 In vitro activity of amphotericin B and anidulafungin against Candida albicans biofilms
A. Valentín, E. Canton, J. Pemán, M. Bosch, E. Eraso, M. Gobernado (Valencia, Bilbao, ES)
Objectives: A great number of Candida spp. infections are related to the formation of biofilms on inert or biological surfaces. One of the main consequences of Candida spp. biofilms is the increased resistance to antifungal agents. The purpose of this study was to determine the activity of amphotericin B (AMB) and anidulafungin (AND) against C. albicans free-living cells and biofilms.
Methods: Twelve C. albicans isolates from catheter were studied. MICs of planktonic cells were determined following the M27-A2 document. MIC0 corresponded to the lowest drug concentration that support no visible growth (a clear well) and MIC2 was the lowest drug concentration that showed a reduction of growth ≥ 50% compared with the growth control. Biofilms were formed following the method described by Ramage et al.* AMB and AND at concentrations ranging from 16–0.03 mg/L were added to microplates containing C. albicans biofilms of 24 and 48 h of maturation. Sessile MICs (SMICs) 50 and 80 corresponded to the lowest drug concentration that support an OD ≤ 50% and an OD ≤ 20%, respectively compared with the growth control.
Results: The 12 isolates were able to form biofilms. Geometric mean SMICs 50 and SMICs 80 of AMB against biofilms of 24/48 h of maturation were 0.5/0.7 mg/L and ≥16/≥ 16 mg/L, respectively. Geometric mean SMICs 50 and SMICs 80 of AND against biofilms of 24/48 h of maturation were 0.4/3.5 mg/L and ≥16/≥ 16 mg/L, respectively.
Conclusions: C. albicans biofilms showed more resistance to AMB and AND than their planktonic counterparts. Complete killing of biofilms was never achieved, even at the highest concentrations of the drugs tested. In general, similar results were obtained for 24 and 48 h of biofilm maturation.
AMB and AND exhibited activity against biofilms as indicated by SMICs 50, but SMICs 80 fell into the resistant range ≥16 in all cases except for one isolate for which the AMB SMICs 80 was 1/1 mg/L.
P1957 Growth inhibition of biofilm-associated Candida albicans by voriconazole in long-term continuous flow cultures
H. Bernhardt, M. Knoke, G. Schwesinger, J. Bufler, V. Czaika, K. Ludwig, J. Bernhardt (Greifswald, Karlsruhe, Bad Saarow, Rostock, DE)
Objectives: We investigated the antifungal effect of voriconazole (VORI) on biofilm and MIC values of C. albicans (C.a.) in a miniature flow cell system.
Methods: Several C.a. strains (SC5314 and 3 blood culture isolates) were grown in miniature flow cells (160 μL; Stovall Life Science Inc.) incubated at 37°C. Peptone-yeast extract (0.5%/0.2%) + 50 mM glucose ± VORI (16 mg/L) was supplied at very low flow rates (1.3 and 2.5 mL/h). Biofilm and biomass production, glucose metabolism, pH, morphological features and planktonic cell counts of C.a. were investigated. MIC testing was performed according to DIN 58940–84 at inocula of 0.5–2.5 × 104 cfu/mL. Etest® on RPMI or Mueller-Hinton methylene blue agar and NCCLS/CLSI methods were used for resuspended biofilms. These were removed from the flow cells and immediately transferred into phosphate-buffered saline. Statistical analysis (Wilcoxon rank test) was based on AUCs. Morphology and viability of fungal cells were assessed by FUN®1 staining.
Results: Each C.a. strain was incubated for up to 10 d in 12 flow cells with 3 channels. In all experiments, metabolic activities in the established biofilms (1 or 2 d old) ceased within 24 h after addition of VORI. We found a strong decrease of biomass production, elevated concentrations of glucose (p = 0.043) and an increase in pH. None of the C.a. recovered from resuspended biofilms subject to 1 passage on Sabouraud glucose agar (SDA) showed changes in their MICs vs. baseline (0.032 μg/mL). C.a. from resuspended biofilms remained susceptible 24 h and 48 h after input of VORI. After longer incubation, growth was so poor that reliable results could not be obtained. C.a. from the residual biofilms could be recultured by several SDA passages and remained sensitive to VORI as well.
Conclusion: The heterogeneity of C.a. biofilms in different test models results in divergent experimental results. Continuous-flow models should be preferred to stationary cultures for the evaluation of antifungal sensitivity of fungi in biofilms. This setting more closely mimicks the conditions encountered on catheter surfaces in vivo.
P1958 Parallel- and cross-resistance of the azoles fluconazole, itraconazole, kezoconazole, voriconazole, and of fluconazole to clinical yeast isolates
A.F. Schmalreck, K. Becker, W. Fegeler (Munich, Münster, DE)
Objectives: From a collaborative study of the antifungal study group of the German Speaking Society of Mycology (DMykG), a subset (n = 3703) of the database of clinical yeast isolates was evaluated for their susceptibility/resistance to antifungal agents (AM) fluconazole (FLC), itraconazole (ITR), ketoconazole (KET), voriconazole (VOR), and flucytosine (FCY) by analysis of susceptible-intermediate-resistant (S-I-R) strains and susceptibility pattern analysis (SPA) to determine the amount and type of parallel- (between azoles) and cross-resistance (between azoles and FCY).
Methods: Susceptibility testing was performed by the test centres according to a common test protocol, the same lot of culture media (YST medium, with 2% glucose and 0.5 mg methylene blue/L; Sifin GmbH, Berlin), by microdilution according to DIN. MIC assessment was performed by categorisation of MICs into susceptible (S), intermediate (I), or resistant (R) by applying the following S/R breakpoints: FLC, ≤4 mg/L and ≥32 mg/L; ITR, ≤0.125 mg/L and ≥1 mg/L; KET and VOR, ≤1 mg/L and ≥4 mg/L; FCY, ≤4 mg/L and ≥32 mg/L. SPA analysis was performed in order to compare specific susceptibility/resistance-patterns (SP) for each individual strain based on a pair-to-pair analysis of S-I-R results.
Results: Of the 3703 clinical yeast isolates, 30.4% demonstrated resistance to the individual antifungal agents (R Sigma-AM identical number or resistances by S-I-R), which resulted in 737 patterns with 1 to 5 AM in sequence (RSPA), of which 1% were parallel resistant to all 4 azoles (RPARA), and 0.4% cross-resistant to the azoles and flucytosine (Table 1 ).
Table 1.
Number (N) and percentage (%) of clinical isolates tested, together with the frequency of occurrence (n = number; % = percentage) of AM-resistances within the isolates (RΣ-AM), their SP, each containing 1 to 5 resistant AM (RSPA), and the amount of parallel- and cross resistance (RPARA; RCROSS)
| Organisms | Frequency |
RΣ-AM |
RSPA |
RPARA |
RCROSS |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | % | n | % | n | % | n | % | n | % | |
| Total isolates | 3703 | 100.0 | 1126 | 30.4 | 737 | 19.9 | 34 | 1.0 | 15 | 0.4 |
| Candida albicans | 1661 | 44.6 | 157 | 9.5 | 90 | 5.5 | 12 | 0.7 | 0 | 0.0 |
| non-C. albicans | 1505 | 40.6 | 702 | 46.6 | 466 | 31.0 | 8 | 0.5 | 4 | 0.3 |
| C. glabrata | 554 | 15.0 | 184 | 33.2 | 135 | 24.4 | 4 | 0.7 | 4 | 0.7 |
| C. guilliermondii | 97 | 2.6 | 99 | 102.1 | 54 | 55.7 | 0 | 0.0 | 0 | 0.0 |
| C. krusei | 254 | 6.9 | 314 | 126.6 | 178 | 70.0 | 4 | 1.6 | 4 | 1.6 |
| C. lusitaniae | 92 | 2.5 | 22 | 23.9 | 22 | 23.9 | 0 | 0.0 | 0 | 0.0 |
| C. parapsilosis | 196 | 5.3 | 2 | 1.0 | 2 | 1.0 | 0 | 0.0 | 0 | 0.0 |
| C. tropicalis | 312 | 8.4 | 81 | 26.0 | 75 | 24.0 | 1 | 0.3 | 0 | 0.0 |
| other Candida spp. | 118 | 3.2 | 8 | 6.8 | 8 | 6.8 | 0 | 0.0 | 0 | 0.0 |
| not-Candida spp. | 140 | 3.8 | 72 | 51.4 | 54 | 38.6 | 3 | 2.1 | 2 | 1.4 |
| Cryptococcus spp. | 287 | 7.8 | 185 | 64.5 | 117 | 39.4 | 9 | 3.1 | 9 | 3.1 |
| Cr. neoformans | 236 | 6.4 | 82 | 34.8 | 74 | 31.3 | 0 | 0.0 | 0 | 0.0 |
To the individual AM, 10.2% of the yeast collective were resistant to FLC, 7.3% to ITR, 2.8% to KET, 1% to VOR, and 9.1% to ITR. The amount of parallel- and cross-resistance for the individual AM was calculated by considering R Sigma-AM, and was in the following order – parallel: VOR(21.3%), KET (11.7%), FCY (6.4), ITR(5.7), and FLC (4.4%); cross: VOR (9.4%), KET (5.2%), FCY (2.8), ITR (2.5%), and FLC (2.0%).
Conclusion: As demonstrated, total parallel- (1.0%) and cross-resistance (0.4%) is very low, and found in a significant amount only in C. glabrata, C. krusei, and Cryptococcus albidus. The higher FLC-resistance rates are due to the “lower” DIN breakpoints. Overall, the isolates were in vitro most susceptible to voriconazole (99.0%).
P1959 Antifungal susceptibility of Candida clinical isolates in Canada: a ten-year review
J. Fuller, C. Sand, R. Rennie (Edmonton, CA)
Objective: The Canadian National Centre for Mycology (NCM) provides susceptibility testing of yeast and filamentous fungi for Canadian laboratories submitting specimens for routine mycology or isolates for confirmatory testing. The purpose of this study was to determine the antifungal susceptibility rates of clinical Candida isolates submitted to the NCM over the past ten years.
Methods: Susceptibility testing of Candida isolates was performed according to the Clinical Laboratories Standards Institute (CLSI) M27A-2 document for broth microdilution (BMD) testing of yeast. BMD panels containing RPMI broth with MOPS buffer, and doubling dilutions of antifungal agents, were inoculated with 10e3 CFU/mL of the test isolate and incubated at 35 degrees C for 48 hours. Antifungal agents included amphotericin B, 5-fluorocytosine, fluconazole, intraconazole, voriconazole, and caspofungin; the latter two agents were tested against 2004 and 2005 isolates only. The 90% minimum inhibitory concentration (MIC90) averaged from total annual isolates was recorded to monitor temporal trends.
Results: From 1995 to 2005 the most common species submitted to the NCM annually included C. albicans, C. glabrata, C. parapsolosis, and C. tropicalis with 801, 421, 188, and 96 total isolates, respectively, and primarily consisted of invasive isolates. Overall, MIC90 values remained unchanged and showed little evidence of reduced susceptibility to the antifungal agents tested; amphotericin B ≤ 4 mg/L, 5-fluorocytosine ≤ 8 mg/L, intraconazole ≤16 mg/L, voriconazole ≤ 2 mg/L, and caspofungin ≤ 2 mg/L. However, for fluconazole the C. albicans MIC90 decreased from 16 mg/L to 4 mg/L while C. glabrata MIC90 increased from 8 mg/L to 64 mg/L.
Conclusion: Despite reported increases of invasive candidiasis, this passive surveillance of Candida susceptibility rates indicates that recommended antifungal agents remain quite active against the most common infecting species. However, C. glabrata resistance to the azoles is increasing in Canada. As new antifungal agents become available, continued and increased surveillance is necessary to monitor the emergence of resistance.
P1960 Fluconazole resistance in C. parapsilosis, no evidence for ERG11 mutation
M.A. Alkhalaf, K.J. Dodgson, E. Sarvikivi, A.R. Dodgson (Manchester, UK; Helsinki, FI)
Objectives: C. parapsilosis is a well recognized cause of systemic infection in premature neonates, fortunately the isolates of this species demonstrate almost universal sensitivity to fluconazole. However, a 12 year clonal outbreak with a C. parapsilosis strain that developed fluconazole resistance has recently been described While many mechanisms of fluconazole resistance have been described in C. albicans, resistance mechanisms in C. parapsilosis have not been documented. We sequenced the ERG11 gene in a number of isolates from the outbreak to determine whether mutation in the target of fluconazole played a role in the development of resistance.
Methods: As only a fragment of the C. parapsilosis ERG11 gene is available in GenBank, we searched for fragments in the Genomic Sequence Survey database that displayed homology to C. albicans ERG11. These fragments were assembled to produce a template from which primers were designed. This putative CpERG11 gene was amplified and sequenced in seven isolates, representing fluconazole MIC's ranging from 0.5 to 64 ug/mL. Sequences obtained were compared to known ERG11 sequences and to each other to determine if any mutations were likely to contribute to resistance.
Results: Sequence was obtained in all seven isolates. Up to 1317 bp of the coding region of the putative CpERG11 was sequenced, comparison of the translated protein showed 73% identity and 86% similarity with C. albicans ERG11 (Accession no. XP716761) suggesting that the sequences obtained were indeed the C. parapsilosis ERG11 gene. However, no differences were detected in any of the sequences obtained.
Conclusions: Fluconazole resistance in other Candida species has been shown to be mediated by a number of mechanisms. Whilst upregulation of efflux pumps appears to be the most common mechanism of resistance in most species, mutations in the ERG11 gene that encodes the target of fluconazole has also been described. It appears that in the set of isolates used in this study, fluconazole resistance is not mediated by ERG11 mutations. A number of other possibilities such as ERG11 expression and efflux pump expression remain to be investigated.
P1961 Effects of vasoactive amines and albumin upon yeast susceptibility to antifungals
S. Costa-de-Oliveira, A. Silva Dias, C. Pina-Vaz, A. Gonçalves Rodrigues (Porto, PT)
Critically-ill patients are often administered a variety of therapeutical drugs namely vasoactive amines and albumin. Following a prospective study about fungaemia conducted at our University hospital, several risk factors for invasive fungal disease and unfavourable outcome were detected. A significant correlation was found between fungaemia and those prescriptions. Previous studies showed the promotion of germination of A. fumigatus and the increase of antifungal resistance following incubation with albumin (Rodrigues et al, 2005).
Objective: To evaluate the effect of albumine and vasoactive amines like adrenaline, noradrenaline, dopamine and dobutamine upon the susceptibility of Candida to several antifungals.
Methods: 15 clinical strains (3 C. glabrata, 4 C. parapsilosis, 5 C. albicans and 3 C. tropicalis) plus 3 control strains belonging to American Type Culture Collection (ATCC) were studied. Susceptibility tests, according by the microdilution protocol M27-A2 from CLSI, were performed to amphotericin B, fluconazole, voriconazole and posaconazole; minimal inhibitory concentrations (MIC) were determined. MIC values were re-determined in the presence of 2% and 4% of albumin (Octapharma, Viena, Austria), 10, 25 and 100 μg/mL of adrenaline (Braun), 2.5, 5 and 100 μg/mL of noradrenaline (Braun) and 10, 50 and 200 μg/mL of both dobutamine (Sigma, Germany) and dopamine (Sigma).
Results: All strains showed initial low MIC values to the antifungals tested. In the presence of 2% and 4% of albumin, MIC of all antifungal drugs increased from 2 to 10 dilutions. The MIC values of azoles increased from 2 to 4 dilutions in the presence of adrenaline and noroadrenaline. These effects were more consistent observed with the non-albicans strains tested.
Conclusion: Vasoactive and inotropic amines have the potential to promote antifungal resistance when administered to patients simultaneously receiving antifungal therapy, especially in non-albicans species. Such fact may help to explain the increasing incidence of invasive fungal disease caused by such species among critical care patients.
P1962 Comparison of Etest and microdilution method for anti-fungal susceptibility testing of C. neoformans to four azoles
A.I. Aller, R. Claro, E. López-Oviedo, A. Romero, E. Martín-Mazuelos (Seville, ES)
Objective: The purpose of this study is to compare the results obtained by the reference broth microdilution method (MDM) with those obtained with Etest system (ET) (Ab Biodisk, Solna, Sweden) for antifungal susceptibility testing of C. neoformans to fluconazole (Pfizer Central Research, UK), itraconazole (Janssen Pharmaceutica, UCA), voriconazole (Pfizer Central Research, UK) and posaconazole (Shering-Plough Research Institute, USA).
Methods: We have studied retrospectively 80 clinical isolates of C. neoformans. The susceptibility to four azoles were performed by the MDM according to the CLSI guidelines (M27-A2 document) modified by Ghannoum et al (J Clin Microbiol 1992; 30:2881–86). The final concentrations were of 0.12–64 mg/L for fluconazole and 0.015–8 mg/L for the rest of antifungal agents. The final inoculum size was 104 CFU/mL. The MICs were defined as the lowest concentration in which a prominent decrease in turbidity was observed.
The ET was performed according to manufacturer's instructions. The inoculum suspensions of C. neoformans isolates matched the turbidity of n°1 McFarland standard. The incubation time was 48–72 h. The MIC was read where the border of the elliptical inhibition zone intersected the scale on the antifungal strip. C. krusei ATCC 6258, C. parapsilosis ATCC 22019, C. neoformans 90112 and C. neoformans 90113 were included as control strains. Essential agreement (EA) was defined as MIC results by ET and MDM in exact agreement or within two dilutions.
Results: The MICs obtained by the two methods after different incubation times varied by no more than one two-fold dilutions. The MIC ranges, MIC50 and MIC90 obtained at 72 h are summarised in table 1 . The EA was 75% for fluconazole, 63.4% for posaconazole, 30% for voriconazole and 6.3% for itraconazole.
Table 1.
| Antifungal agent | Method | MIC (mg/L) |
||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Fluconazole | MDM | 0.5–32 | 4 | 8 |
| ET | 0.032–≥256 | 4 | 32 | |
| Itraconazole | MDM | 0.06–1 | 0.5 | 1 |
| ET | <0.002–0.25 | 0.064 | 0.25 | |
| Posaconazole | MDM | 0.03–4 | 0.25 | 0.5 |
| ET | <0.02–1 | 0.023 | 0.125 | |
| Voriconazole | MDM | 0.25–1 | 0.5 | 0.5 |
| ET | 0.008–1.5 | 0.094 | 0.19 | |
Conclusions:
-
1
Fluconazole MICs by ET showed good correlation with MDM (75%)
-
2
Itraconazole showed the lowest agreement (6.3%)
-
3
It would be necessary to carry out further studies including interlaboratory agreement and correlation of MICs by different methods with “in vivo” response.
P1963 Effects of MG3290, a selective histone deacetylase inhibitor, on azole resistance in Candida glabrata
B. Yeung, N. Georgopapadakou (Montreal, CA)
Objectives: To examine whether MG3290, a highly active potentiator of azole antifungals against Aspergillus and Candida species, potentiates azoles against azole-resistant C. glabrata mutants and lowers resistance frequency when combined with azoles.
Methods: Synergy of MG3290 with azoles was determined by twofold dilutions of each compound alone and in combination in 96-well plates (checkerboard format). Ergosterol synthesis was measured by the absorbance of lipid extracts at 281 nm; azole transport by the transport of Rhodamine123 (excitation max, 485 nm; emission max, 535 nm).
Results: C. glabrata mutants resistant to itraconazole and fluconazole were detected at 3×10−7 and 5×10−6 frequencies respectively on agar plates containing 4×MIC of the relevant azole. Mutants resistant to itraconazole/MG3290 were detected at 1×10−7 frequency on plates containing 4×MIC of itraconazole/MG3290 (none at 2×MIC), while mutants resistant to fluconazole/MG3290, at 2×10−7 frequency at 4×MIC of fluconazole/MG3290. Three of seven itraconazole- and 13 of 14 fluconazole-resistant mutants isolated and examined had altered transport and were cross resistant. Basal ergosterol levels in the azole-resistant mutants were unchanged from the parent. MG3290, at ≤4 μg/mL, lowered itraconazole MICs against itraconazole-resistant mutants and fluconazole MICs against fluconazole-resistant mutants.
Conclusion: MG3290 potentiates azoles currently used in the clinic against resistant mutants and lowers resistance frequency when combined with azoles. It is therefore an attractive candidate for further development in combination with azoles.
P1964 Enhancement of yeast susceptibility to antifungals
J. Subik, J. Cernicka, Z. Kozovska, M. Valachovic, I. Hapala, G. Hajos (Bratislava, Ivanka pri Dunaji, SK; Budapest, HU)
Objectives: Decreased susceptibilities to azole antifungals in Candida species were resulting from combination of several molecular mechanisms involving the enhanced expression of drug transporter genes (MDR1, CDR1/2) and over-expression or mutations in the ERG11 gene encoding the 14-a-lanosterol demethylase. To overcome multidrug resistance in yeast pathogens, antifungals with novel cellular targets as well as multidrug resistance reversal agents rendering drug resistant strains sensitive to commercially used antifungals are being developed. In this report one polyfused heterocyclic compound was found to enhance the activity of several antifungals in different yeast species.
Methods: A library of polyfused heterocyclic compounds was tested for multidrug resistance modulating activity in a screening system using the agar diffusion method. Drug susceptibilities were assayed by microbroth dilution method in 96-well plates according to the proposed NCCLS M27-A standard guidelines. RP-HPLC analysis of sterols was conducted on Eclipse XDB-C8 column using an Agilent 1100 series instrument. Accumulation of rhodamine 6G (Sigma) was measured by flow cytometry in a FACS Calibur fluorescence-activated cell cytometer (Becton Dickinson).
Results: We identified one compound able to enhance the susceptibility to cycloheximide and other drugs in the drug resistant Saccharomyces cerevisiae mutant strain. In the presence of this organic compound an increased antifungal activity of fluconazole was demonstrated in yeast mutant strains deleted in genes encoding the major multidrug resistance transcription factors Yap1p, Pdr1p and Pdr3p, as well as the drug efflux pumps Pdr5p and Snq2p in S. cerevisiae or their counterparts in C. albicans and C. glabrata, named Cdr1p and Mdr1p, respectively. Importantly, the compound increased the sensitivity to fluconazole also in multidrug resistant cells over-expressing the efflux pumps. Yeast cells grown in the presence of sub-inhibitory concentrations of the compound exhibited an altered sterol composition and a slightly enhanced accumulation of rhodamine 6G, which suggests that plasma membrane plays a role in yeast sensitisation to drugs.
Conclusion: The activity of a new compound that can overcome multidrug resistance in yeast may prove useful in combined treatment of infections caused by drug resistant fungal pathogens.
P1965 Antifungal drug susceptibility testing of Candida sp. by two methods: CLSI(M27-A2) broth microdilution and the image analyser method Wider-I
M. Rodriguez-Dominguez, G. Ayala, A. Fernandez-Olmos, M. Mercadillo, M. Alvarez, A. Sanchez-Sousa, A. Espinel-Ingroff (Madrid, ES; Richmond, US)
Introduction: Accurate as well as faster methods than CLSI methodology are needed for in vitro susceptibility testing of yeasts. WIDER I (Francisco Soria Melguizo S.A., Madrid, Spain) is a less labour-intensive and much simpler to perform assay than the CLSI broth microdilution M27-A2 method. Wider I is a computer-assisted image processing system, optimised for microdilution susceptibility testing of yeasts.
Objectives: To evaluate the suitability of the WIDER I system for testing the in vitro susceptibility of Candida spp. to four antifungal agents. We compared results by this automated method to those by the CLSI reference method for yeasts (M27-A2 document).
Methods: We tested 101 yeasts isolates from transplant recipients (C. albicans 71, C. glabrata 13, C. parapsilosis 8, C. lusitaniae 4, C. krusei 2, C. famata 1, C. guilliermondii 1, C. tropicalis 1) against amphotericin B, voriconazole, itraconazole and fluconazole with both CLSI (M27-A2) and WIDER I methods.
Results: Agreement (within three dilutions) between methods for voriconazole and fluconazole was 90% and 87%, respectively 74% for itraconazole. For amphotericin B, the agreement between both methods was excellent at 24 h (97%). By breakpoint category, the agreement was 91% for fluconazole (VME = 1% [very major errors, susceptible by new method and resistant by reference method], ME = 3% [major errors, resistant by new method and susceptible by reference method] and M = 5% [minor errors, shifting between categories]. The agreement by category was 93% for voriconazole (ME = 5%, M = 2%) and lower (87.1%) for itraconazole (VME = 3%, ME = 1% and M = 8.9%).
Conclusion: Although MIC results varied between the methods, we found a high level of agreement by breakpoint categories. Further evaluations are needed to assess the suitability of this method.
P1966 FKS mutation in echinocandin-resistant C. glabrata
K.J. Dodgson, B. Kaur, A.Y. Sharafadein, S.A. Messer, M.A. Pfaller, A.R. Dodgson (Manchester, UK; Iowa City, US)
Objectives: Taking a range of isogenic isolates from 43 year old AML patient, one of which developed resistance whilst on Caspofungin therapy, we aimed to elucidate the mechanism of resistance in this isolate. The isolate was shown to be resistant to all the available echinocandins (Caspofungin, Micafungin and Anidulafungin) and the resistance was demonstrated to be stable. Given other descriptions of mutations in the FKS genes we aimed to clarify whether similar mutations were responsible for the development of resistance in this clinical isolate of C. glabrata
Methods: To assess whether the echinocandin resistance was due to a mutation in a glucan synthase (FKS) gene, the portions of the FKS gene previously shown harbour mutations known to mediate resistance in C. glabrata and other species were sequenced from the each of the three FKS homologues in the C. glabrata genome (CAGL0G01034g, CAGL0K04037g and CAGL0M13827g). Sequences were obtained from and compared between the resistant isolate (145.088) and the sensitive isolates.
Results: No differences were found between the sequences obtained from the resistant or sensitive isolates in the sequenced portions of the CAGL0G01034g or CAGL0M13827g ORFs. However the sequence from the CAGL0K04037g locus revealed a single base pair mutation from T to C at position 1987, which resulted in a change of amino acid from serine to proline at position 663.
Conclusions: We describe the development of echinocandin resistance in a series of isogenic isolates of C. glabrata obtained from a patient treated with caspofungin. We detected a single base pair mutation in one of the glucan synthase genes sequenced from the resistant isolates, this mutation resulted in a change of amino acid for serine to praline. A number of point mutations in glucan synthase genes have been associated with echinocandin resistance in a number of Candida species. In C. glabrata, the only mutation described to date is F659V, however a S645P mutation has been described previously in a number of C. albicans isolates. The S663P mutation described in this study corresponds to that at position 645 in C. albicans and we believe this to the first description of this mutation in C. glabrata.
P1967 Mycograb increases Candida sensitivity to fluconazole in vitro
L. Nooney, R. Al-Akeel, S. Awad, I. AlShami, R. Matthews, J. Burnie (Manchester, UK)
Introduction: Hsp90 has been implicated in the development of anti-fungal drug resistance and has been shown to play an important role in the development and continuation of resistance. The role of hsp90 in resistance to fluconazole (FLC) is exerted through calcineurin. The blocking of either hsp90 or calcineurin results in enhanced drug activity or reversed drug resistance. We hypothesize that Mycograb, an inhibitor of hsp90, may act in a similar way to geldanamycin and delay or block the development of high level resistance to fluconazole in Candida.
Methods: Six isolates of Candida were exposed to FLC at a range of 0.03125 to 512ug/mL in the presence of 4, 8 and 16ug/mL Mycograb in a broth macrodilution assay. Colonies from each of the isolates that grew at the concentration just prior to culture sterilisation were also examined in the same assay.
Results: Five of the six isolates displayed increased sensitivity to FLC with the addition of 4, 8 or 16ug/mL Mycograb, the MIC-2 was reduced by one or more dilution. A fluconazole sensitive strain of Candida albicans was the most sensitive with a FLC MIC-2 of 2ug/mL before the addition of Mycograb and an MIC-2 of 0.5ug/mL in the presence of 8 and 16μg/mL Mycograb. Comparisons of CFU/mL between cultures with FLC only and those that contained Mycograb demonstrated additivity (>1 log reduction in CFU/mL) between fluconazole and Mycograb in four isolates and synergy, ≥2 log reduction in CFU/mL, in a single isolate. Repetition of the assay using resistant colonies also showed that Mycograb increased sensitivity to FLC in four isolates. A fluconazole resistant strain of C. albicans was the most sensitive isolate to the activity of Mycograb with an MIC-2 of 16μg/mL and an MIC-2 of 0.03125ug/mL in the absence and presence of Mycograb respectively. Additivity was observed in two isolates and synergism in a single isolate. No antagonism was demonstrated.
Conclusion: Candida species exposed to fluconazole (FLC) in combination with Mycograb were found to have increased sensitivity to fluconazole in vitro. The MIC-2 to FLC in fluconazole sensitive and in fluconazole resistant sub-populations of Candida species was found to be reduced
P1968 Determination of the antifungal susceptibility of zygomycetes by the ETest and Sensititre YeastOne: a comparison with the CLSI M-38 A procedure
M. Torres Narbona, J. Guinea, J. Martínez-Alarcón, T. Peláez, M. Guembe, E. Bouza (Madrid, ES)
Objectives: Zygomycetes are emerging as a main cause of invasive fungal infections in immunocompromised patients, but the in vitro antifungal susceptibility profiles have not been widely investigated. The few studies that have evaluated the antifungal susceptibility of clinical isolates of zygomycetes are mainly based on CLSI M38-A, a time-consuming procedure. We evaluated the correlation of two rapid methods (E-test and Sensititre YeastOne) with CLSI M38-A for zygomycetes susceptibility testing.
Methods: We studied 45 clinical strains of zygomycetes collected during an 18-year period in our institution. We evaluated the activity of amphotericin B (AMB), itraconazole (ITC), voriconazole (VC), caspofungin (CAS) and posaconazole (POS) by the E-Test procedure (AMB, ITC, VC, POS and CAS), by Sensititre YeastOne (AMB, ITC, VC and CAS), and by the CLSI M38-A procedure (AMB, ITC, POS, VC and CAS). The plates and trays were incubated at 35°C. We compared CLSI M-38 A results (incubation at 24 hours) with the alternative methods (incubations at 16, 24, 36, 48 and 72 hours). The E-Test minimal inhibitory concentration (MIC) was read as the drug concentration at which the border of the elliptical inhibition zone intercepted the scale on the strip. The microdilution MIC was defined as the lowest concentration producing complete inhibition of growth (MIC-0) for all the antifungal drugs studied. Both methods were considered to agree when the results of the MICs by Sensititre or the E-test were within 2 dilutions of the MICs obtained by CLSI M38-A.
Results: The highest agreement between AMB, ITC and POS E-test and the reference MICs (70.5%; 70.5%; 88.6%) was at 48, 16 and 16 hrs of incubation, respectively. The highest agreement between AMB and ITC Sensititre YeastOne and reference MICs (40.9%; 79.5%) was at 36 and 24 hrs of incubation, respectively. Although the E-test and Sensititre YeastOne presented a good correlation with CLSI M-38A for VC and CAS, both agents presented very poor activity.
Conclusions: The E-test and Sensititre YeastOne are rapid alternative methods to determine the susceptibility profiles of zygomycetes, but the correlation (within 2 dilutions) with CLSI M-38A was moderate (below 80%), with the exception of POS. The MICs of POS obtained by the E-test showed a good correlation at 16 hours of incubation.
P1969 Susceptibility of Candida spp. clinical isolates to antifungal drugs
S. Tyski, M. Staniszewska, B. Rozbicka, A. Rajnisz, E. Bocian, E. Wasinska, W. Kurzatkowski (Warsaw, PL)
Objectives: The purpose of this investigation was to determine the susceptibility of clinical isolates of different Candida species to antifungal therapeutics.
Methods: Altogether 106 clinical isolates of Candida spp. obtained from different clinical materials were under investigation: 53 C. albicans, 23 C. glabrata, 10 C. tropicalis, 10 C. parapsilosis, 5 C. krusei and single strains: C. kefyr, C. globosa, C. dubliniensis, C. lipolytica and C. inconspicua.
Strains susceptibility to: amphotericin B (A), 5-fluorocytosine (5), itraconazole (I) and fluconazole (F) was determined by ATB Fungus system, while susceptibility to ketoconazole (K), voriconazole (V), caspofungin (C) and posaconazole (P) was assayed with the usage of E-tests, according to manufacturer's recommendation.
Results: The strains were divided into 3 groups: C. albicans (53), C. glabrata (23), and other Candida isolates (30).
-
•
All C. albicans isolates were susceptible to (A) and (5), while 5 and 3 of them were resistant to (I) and (F), respectively.
(K) MIC (μg/mL) range (<0.002–>32); MIC50 = 0.012, MIC90 >32.
(V) MIC (μg/mL) range (<0.002 –>32) MIC50 = 0.016, MIC90 >32.
(C) MIC (μg/mL) range (0.006–0.25) MIC50 = 0.125, MIC90 = 0.19.
(P) MIC (μg/mL) range (0.003–>32) MIC50 = 0.047, MIC90 > 32.
-
•
All C. glabrata isolates were susceptible to (A), 2 were intermediate and 21 susceptible to (5), while 21 and 19 C. glabrata were not susceptible to (I) and (F), respectively.
(K) MIC (μg/mL) range (0.016–8) MIC50 = 0.75, MIC90 = 3.
(V) MIC (μg/mL) range (0.019–12) MIC50 = 0.38, MIC90 = 1.
(C) MIC (μg/mL) range (0.094–0.25) MIC50 = 0.19, MIC90 = 0.25.
(P) MIC (μg/mL) range (0.19 –>32) MIC50 = 12, MIC90 >32.
-
•
Among other Candida isolates, all but one were susceptible to (A), two were resistant to (5), while 16 and 21 were susceptible to (I) and (F), respectively.
(K) MIC (μg/mL) range (0.004–1.5) MIC50 = 0.016, MIC90 = 0.38.
(V) MIC (μg/mL) range (0.006–2) MIC50 = 0.023, MIC90 = 0.125.
(C) MIC (μg/mL) range (0.064–1) MIC50 = 0.25, MIC90 = 1.
(P) MIC (μg/mL) range (0.08–4) MIC50 = 0.047, MIC90 = 1.
Conclusion: Candida spp. isolates showed to be very susceptible to amphotericin B and 5-fluorocytosine. C. glabrata isolates were more frequently, then other strains, resistant to itraconazole and fluconazole. Among analysed strains, number of isolates, showed high resistance (>32 ug/mL) to ketoconazole, voriconazole and posaconazole.
P1970 In vitro susceptibility of clinical isolates of Cryptococcus gattii
A. Gomez-Lopez, L. Bernal-Martinez, A. Alastruey-Izquierdo, J. Rodriguez-Tudela, M. Cuenca-Estrella (Majadahonda, ES)
Background: A recent outbreak of C. gattii infection of Vancouver Island (Canada) emphasize the role of this specie as causal agent of infections in temperate climate. In addition, Cryptococcus gattii has shown less in vitro susceptibility than C. neoformans to amphotericin B and flucytosine although contradictory results have been obtained in other studies. We have analysed the antifungal susceptibility profile of clinical isolates of C. neoformans var. gatti and compared with C. neoformans var. neoformans.
Methods: A collection of 20 isolates of C. neoformans var. gatti was included in the study. Nineteen of the strains were obtained from colleagues. In addition, the in vitro susceptibility of 323 strain of C. neoformans var. neoformans received in our institution between 1995 and 2006 was evaluated.
The isolates were identified by routine physiological tests. The susceptibility testing followed strictly the recommendations proposed by the Antifungal Susceptibility testing Subcommittee of the European Committee on Antibiotic Susceptibility Testing for fermentative yeast. The antifungal agents used in the study were as follow: amphotericin B, flucytosine, fluconazole, itraconazole, voriconazole, ravuconazole, posaconazole and caspofungin.
Results: The table displays susceptibility results.
| Antifungal agent |
C. neoformans var. gattii (n = 20) |
C. neoformans var. neoformans (n = 333) |
||||
|---|---|---|---|---|---|---|
| GM | MIC90 | MIC50 | GM | MIC90 | MIC50 | |
| Amphotericin B | 0.09 | 0.125 | 0.125 | 0.24 | 1 | 0.25 |
| Flucytosine | 1.07 | 2 | 1 | 4.38 | 16 | 4 |
| Fluconazole | 14.93 | 16 | 16 | 7.44 | 16 | 8 |
| Itraconazole | 0.45 | 1 | 0.5 | 0.25 | 1 | 0.25 |
| Voriconazole | 0.47 | 1 | 0.5 | 0.12 | 0.5 | 0.125 |
| Ravuconazole | 0.53 | 2 | 0.5 | 0.15 | 1 | 0.125 |
| Posaconazole | 0.26 | 0.5 | 0.25 | 0.16 | 0.5 | 0.25 |
| Caspofungin | 16 | 16 | 16 | 18.02 | 32 | 16 |
Conclusions: (i) C. gattii was less susceptible in vitro to amphotericin B and fluocytosine than C. neoformans (p <0.01). (ii) C. gattii showed significantly higher MICs to azole agents (p <0.01). (iii) Both species seems intrinsically resistant to caspofungin (GM ≥ 16 mg/mL).
P1971 Induction of resistance and cross-resistance of fluconazole, voriconazole, caspofungin and cationic antimicrobials against Candida species
S. Tobudic, C. Kratzer, W. Graninger, A. Georgopoulos (Vienna, AT)
Objectives: The purpose of this study was to compare the in vitro selection of resistance and cross-resistance of fluconazole, voriconazole, caspofungin and the cationic antimicrobials chlorhexidine digluconate and Akacid plusR against Candida species.
Methods: For the in vitro selection of resistance Candida albicans ATCC 10231, C. albicans ATCC 90029, Candida glabrata ATCC 90030, Candida krusei ATCC 6258 and Candida tropicalis ATCC 750 were subcultured in liquid RPMI medium containing selected concentrations of active substances during 30 passages. Minimal inhibitory concentrations (MICs) of antimicrobials were performed after each passage using broth microdilution method according to the CLSI guidelines (M27/A2).
Results: Subcultivation of C. tropicalis in RPMI containing 32 mg/L fluconazole resulted in a 256–fold increase in the MICs of fluconazole and voriconazole after less than 10 passages. Following exposure to 4 mg/L voriconazole, comparable MIC increases of voriconazole and lower increases of fluconazole were found (128 and 16 times of the MIC) in C. tropicalis. Preexposure to subinhibitory concentrations of caspofungin induced a 64-fold MIC-increase of caspofungin in C. tropicalis following 10 passages, but no cross-resistance to azoles or cationic antimicrobials was observed. Subcultivation of both strains of C. albicans in 2 mg/L voriconazole induced a high MIC increase (>256×) after 10 passages, whereas no cross-resistance to other antimicrobials could be induced. MICs of active substances against C. glabrata and C. krusei remained constant during 30 passages. Development of resistance or cross-resistance to Akacid plusR or chlorhexidine was not observed in any of the tested isolates.
Conclusion: Repeated exposure to subinhibitory concentrations of cationic antimicrobials had no effects on selection of resistance or cross-resistance in Candida spp., whereas resistance and cross-resistance to azoles and resistance to caspofungin could be easily induced in C. tropicalis. In vitro resistance of voriconazole was also developed in both strains of C. albicans.
P1972 Broth microdilution, Etest and disk diffusion methods for Trichosporon asahii susceptibility testing against established and new antifungal agents
M. Kanellopoulou, D. Adamou, M. Martsoukou, N. Skarmoutsou, E. Alexopoulos, A. Milioni, G. Arsenis, A. Velegraki, E. Papafrangas (Athens, Patras, GR)
The genus Trichosporon is a basidiomycetous yeast phylogenetically close to Cryptococcus. T. asahii is an emerging cause of disseminated trichosporonosis. Isolation of T. asahii from patient indwelling devices and biological secretions, without related dissemination, is hitherto of ambiguous clinical significance.
Objectives: (a) To monitor over 3 years (2002–2005) T. asahii susceptibility to antifungals isolated from urinary tract and bronchial secretions of patients without trichosporonosis; (b) to compare broth microdilution (BMD), Etest and disk diffusion (DD) methods for determining susceptibility to established antifungals and to posaconazole.
Methods: Seventeen urine specimen isolates from patients with indwelling catheter (n = 15) and from respiratory secretions with endotracheal catheter (n = 2) were tested against posaconazole (POS; Schering Plough Research Institute, New Jersey, USA), amphotericin (AB; Sigma, St. Louis, MO, USA), flucytosine (FC; Sigma), itraconazole (IT; Janssen, Beerse, Belgium) fluconazole and voriconazole (FLU and VOR; Pfizer, Sandwich, Kent, UK). Strains were tested by BMD (CLSI, M-27A2) and Etest (AB Biodisk, Solna, Sweden). FLU and POS were also tested by DD (CLSI, M-44A, 2004) using commercially prepared disks (OXOID, Basingstoke, UK). Incubations were performed at 35°C and read at 24, 48 and 72 hours. The QC isolates used were: Candida krusei ATCC6258 and C. parapsilosis ATCC22019.
Results: MIC ranges (μg/mL) were AB: 0.5–16, FC: 2–64, IT: 0.25–2, FLU: 2–32, VO: 0.215–1 and POS: 0.125–0.5. No intrazonal growth was observed in the DD method. Inhibition zone diameters ranged from 5 to 35 mm (FLU) and 10 to 20 mm (POS). Interclass correlation coefficients (ICCs) and 95% confidence intervals for comparing the methods were calculated using log2 transformed data. ICC for BMD versus Etest ranged from 0.98 to 0.99 (P < 10−4) for all drugs. Pearson's correlation coefficient used as a measure of linear associations for FLU and POS BMD versus Etest, versus disk diffusion was 0.944 (P < 0.01).
Conclusion: High AB, FC, IT and FLU MICs warrant clinical alertness for breakthrough Trichosporon infections when used for prophylaxis. Pending clinical studies, the low VOR and POS MICs suggest that they may be a better alternative for prophylaxis in patients at risk. Disk diffusion is an option for rapidly testing T. asahii isolates, provided it is further evaluated. Standardised susceptibility testing can aid the selection of the most relevant antifungal therapy for the management of trichosporonosis.
P1973 Comparison of RPMI and AM3 medium for in vitro susceptibility testing of caspofungin against Candida spp. by EUCAST broth microdilution method
E. Dannaoui, O. Lortholary, D. Raoux, D. Hoinard, F. Dromer (Paris, FR)
Objectives: To compare two media, RPMI and AM3, for antifungal susceptibility testing of caspofungin against Candida spp.
Methods: A total of 863 Candida spp. isolates belonging to 25 different species were evaluated. All isolates were tested for caspofungin susceptibility in RPMI and AM3. All isolates were identified according to standard procedures. Susceptibility testing was performed according to the EUCAST broth microdilution reference method. Both media were supplemented with 2% glucose with a final inoculum size of 1×105 CFU/mL. Microplates were read spectrophotometrically after 24 h of incubation at 35°C. MICs were defined as the lowest concentration resulting in 50% or more reduction of growth compared to the drug-free growth control. Quality control strains were included in each set of MIC determination.
Results: Overall, lower caspofungin MICs were obtained in AM3 (CMI50 = 0.03 μg/mL) compared to RPMI (CMI50 = 0.5 μg/mL) and this was true for all tested species. There was a broader range (i.e. number of log2 dilutions between the highest and the lowest MIC) of MICs in AM3 than in RPMI for C. albicans (11, and 6 dilutions for AM3 and RPMI, respectively), C. glabrata (11, and 6), C. parapsilosis (6, and 4), and C. tropicalis (7, and 4). Moreover, more isolates exhibited MIC >MIC90 when AM3 was used compared to RPMI for C. albicans, C. glabrata, and C. parapsilosis, suggesting that AM3 is more suitable than RPMI for detection of resistance to caspofungin.
Conclusions: For caspofungin susceptibility testing with the EUCAST reference method, the use of AM3 instead of RPMI yielded lower MICs as well as a broader range of MICs. Our results suggest that AM3 could allowed better detection of caspofungin resistant isolates. Further studies are needed to confirm these preliminary results.
Table 1.
Detection of Candida spp. isolates with high caspofungin MICs in RPMI and AM3 medium
| Species (n) | Caspofungin |
|||
|---|---|---|---|---|
| MIC90 (μg/mL) |
No. of isolates with MIC>MIC90 |
|||
| RPMI | AM3 | RPMI | AM3 | |
| C. albicans (404) | 0.5 | 0.03 | 14 | 23 |
| C. glabrata (157) | 1 | 0.06 | 1 | 11 |
| C. parapsilosis (109) | 2 | 0.5 | 5 | 6 |
| C. tropicalis (62) | 1 | 0.06 | 3 | 2 |
| C. krusei (21) | 2 | 0.125 | 1 | 1 |
P1974 In vitro activity of caspofungin and micafungin against 1038 yeasts isolates from France by EUCAST reference method
E. Dannaoui, O. Lortholary, D. Raoux, D. Hoinard, F. Dromer and the YEASTS Group (Paris, FR)
Objectives: To assess the in vitro antifungal susceptibility to caspofungin and micafungin of yeast clinical isolates received at the National Center for Mycoses and Antifungals (CNRMA) from 2003 to 2006.
Methods: All isolates were identified according to standard procedures. Susceptibility testing was performed according to the EUCAST broth microdilution reference method. RPMI with 2% glucose was used as test medium with a final inoculum size of 1×105 CFU/mL. Microplates were read spectrophotometrically after 24 h of incubation at 35°C. MICs were defined as the lowest concentration resulting in 50% or more reduction of growth compared to the drug-free growth control. Quality control strains were included in each set of MIC determination.
Results: 1038 yeast isolates belonging to 31 different species were included. Most of the isolates were recovered from blood. Five species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and Cryptococcus neoformans) accounted for 86% of all isolates. Overall, there was a positive correlation between MICs of both drugs. C. albicans, C. glabrata, and C. tropicalis, were the most susceptible species with Gmean MIC of <1 and <0.1 μg/mL for caspofungin and micafungin, respectively. Among other Candida spp. C. parapsilosis, C. guilliermondii, and C. fermentati showed the highest MICs. No activity was found against C. neoformans with Gmean MIC of >9 μg/mL for both echinocandins. Overall, micafungin showed lower MICs than caspofungin.
Table 1.
MIC values for species with at least 10 tested isolates
| Species | MIC (μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Caspofungin |
Micafungin |
|||||||||
| Na | Range | MIC50 | MIC90 | GMICb | Na | Range | MIC50 | MIC90 | GMICb | |
| C. albicans | 404 | 0.125–4 | 0.5 | 0.5 | 0.38 | 161 | 0.015–1 | 0.03 | 0.03 | 0.027 |
| C. glabrata | 157 | 0.25–8 | 0.5 | 1 | 0.51 | 60 | 0.015–8 | 0.03 | 0.03 | 0.033 |
| C. parapsilosis | 109 | 0.5–4 | 2 | 2 | 1.57 | 49 | 0.25–2 | 1 | 2 | 1.29 |
| C. tropicalis | 62 | 0.25–2 | 0.5 | 1 | 0.60 | 23 | 0.03–0.125 | 0.06 | 0.06 | 0.052 |
| C. guilliermondii | 27 | 0.06–2 | 2 | 2 | 1.32 | 13 | 0.25–1 | 1 | 1 | 0.77 |
| C. krusei | 21 | 0.5–8 | 1 | 2 | 1.10 | 13 | 0.125–4 | 0.25 | 0.25 | 0.26 |
| Cr. neoformans | 158 | 1–16 | 8 | 16 | 9.59 | 64 | 2–16 | 16 | 16 | 9.62 |
Number of isolates tested.
Geometric mean MIC.
Conclusions: Caspofungin and micafungin exhibited good in vitro activity by EUCAST method in RPMI medium against most clinical isolates of Candida spp. Although micafungin showed lower MIC values, there was a positive correlation between MICs of both echinocandins.
P1975 Detection of Fusarium oxysporum DNA and (1–3)-β-D-glucan in serum specimens of experimentally infected mice
Z.U. Khan, S. Ahmad, A. Theyyathel (Kuwait, KW)
Objective: Invasive Fusarium infection is an emerging opportunistic mycosis associated with high mortality. Its diagnosis is difficult due to non-specific signs and symptoms and similarity in tissue morphology with Aspergillus species. The aim of this study was to develop a sensitive and specific diagnostic procedure based on amplification of Fusarium oxysporum DNA by nested (n) PCR and to compare the results with (1-3)-β-D-Glucan detection in serum samples obtained from experimentally infected mice.
Methods: Sixty mice immunosuppressed with intraperitoneal injections of cyclophosphamide were infected intravenously with a dose of 1×107 F. oxysporum conidia/mouse. Six mice were sacrificed every day post-infection. Blood specimens were collected by cardiac puncture. One hundred μL of blood was used for culture and the rest was processed for separation of serum. The lung homogenate was cultured and was also used for direct microscopic examination. The genomic DNA from the reference strains of F. oxysporum and 9 other fungi including Fusarium solani, Aspergillus fumigatus, A. flavus and A. terreus was isolated and used as template for nPCR specificity. The species-specific primers were derived from the internally transcribed spacer (ITS)-1 and ITS-2 regions of rDNA and their specificity was confirmed by BLAST searches. The DNA from serum specimens was extracted using standard procedures. The PCR amplicons were detected by agarose gel electrophoresis. The (1-3)-β-D-Glucan was detected by Fungitell (Cape Cod Inc. E. Falmouth, MA, USA).
Results: The nPCR was specific for F. oxysporum and detected nearly 440 fg of Fusarium DNA which is roughly equivalent to 10 genome copies. Of the 60 serum samples tested, 55 (92%) were positive for (1-3)-beta-D-Glucan and 47 (78%) were positive for F. oxysporum DNA. The lung homogenates of all the infected animals yielded F. oxysporum in culture. The fungus was also discernible in KOH-calcofluor mount of 40 (67%) of the animals.
Conclusions: A sensitive and species-specific nPCR assay has been developed for the detection of F. oxysporum DNA in serum samples of experimentally-infected mice. During a follow-up of 14 days post-infection, 92% and 78% of serum samples were positive for (1-3)-β-D-Glucan and F. oxysporum DNA, respectively. The data suggest that detection of F. oxysporum DNA by nPCR combined with (1-3)-β-D-Glucan may help in early diagnosis of invasive fusariosis. Supported by KURA Grant MI 04/02.
P1976 Comparative evaluation of Aspergillus terreus DNA and galactomannan in serum and bronchoalveolar lavage of intravenously infected mice
S. Ahmad, A. Theyyathel, Z. Khan (Kuwait, KW)
Objective: Invasive aspergillosis is a growing problem in immunocompromised individuals. Although Aspergillus fumigatus is the principal aetiologic agent of the disease, infections caused by Aspergillus terreus have shown an increased occurrence in recent years. Additionally, A. terreus is more resistant to amphotericin B and is probably also associated with higher mortality in humans. Therefore, an early diagnosis is imperative for reduced mortality and better prognosis. The aim of this study was to develop sensitive and specific diagnosis of invasive aspergillosis by detecting A. terreus-specific DNA by nested PCR (nPCR) in serum and bronchoalveolar lavage (BAL) specimens of intravenously infected mice and to compare the results with galactomannan (GM) levels.
Methods: Sixty BALB/c mice, immunosuppressed with four intraperitoneal injections of cyclophosphamide (200 mg/kg) on day −1, 0, +1, +3 were infected intravenously with 1×106 conidia of A. terreus. The mice were sacrificed on day 1, 3, 5, 7 and 9 post-infection in groups of twelve each and their BAL, blood, and lungs were cultured. The A. terreus-specific DNA and GM in serum and BAL were detected by nPCR and Platelia Aspergillus (BioRad, Marnes-la-Coquette, France), respectively.
Results: The nPCR developed with primers derived from internally transcribed spacer (ITS)-1 and ITS-2 regions of rDNA was specific for A. terreus as genomic DNA from other Aspergillus species or other fungi was not amplified. The lung homogenates of all the infected mice yielded A. terreus in culture, while the blood and BAL specimens were uniformly negative. The % positivity of serum samples for GM and nPCR were 78% and 73%, respectively. In contrast, among BAL samples, 71% were positive for GM and 81% for A. terreus-specific DNA. The combined detection of A. terreus DNA and/or GM enhanced the positivity to 95% in serum and 98% in BAL.
Conclusions: The data suggest that GM and A. terreus DNA are easily detected in both, serum and BAL, during the entire course of infection and their combined detection could be useful in the early diagnosis of invasive aspergillosis.
Supported by Kuwait University Research Grant MI04/02
P1977 In vitro activity of isavuconazole (BAL4815/8557) compared with six other antifungal agents against 180 Cryptococcus neoformans meningitis isolates from the Netherlands
I. Curfs-Breuker, M. Illnait-Zaragozi, J. Mouton, B. Janssen, F. Hagen, L. Spanjaard, T. Boekhout, J. Meis (Nijmegen, NL; Havana, CU; Utrecht, Amsterdam, NL)
Background: Data regarding the antifungal susceptibility of Cryptococcus neoformans (CNE) isolates from The Netherlands are limited. We determined and compared the in vitro activity of the new azole antifungal drug isavuconazole (BAL4815/8557) with six other antifungals against CNE isolates from patients with menigitis.
Methods: 180 CNE strains isolated from cerebrospinal fluid and blood were used. MICs were determined for amphotericin B (AMB), flucytocine (5FC), fluconazole (FLU), itraconazole (ITC), voriconazole (VOR), posaconazole (POS) and isavuconazole (ISA). Microdilution testing was done in accordance with CLSI M27-A2 guidelines in RPMI 1640 MOPS broth with L-glutamine without bicarbonate. Plates were incubated at 35°C for 72 hours. The MIC was determined visually and spectrophotometrically as the lowest concentration of drug showing absence of growth or ≥ 50% reduction of growth compared with that of the growth control for AMB, and 5FC and azoles, respectively.
Results: No significant differences between visual and spectrophotometric reading was observed. Over all strains, MIC90s of AmB, 5FC, FLU, IT, VOR, POS, CAS and ISA were 0.5, 8, 8, 0.25, 0.25, 0.125, >64, and 0.125 mg/L respectively.
Conclusions: Emergence of drug resistance to AmB, 5FC, FLC, and IT against CNE, as reported from other regions, were not confirmed in this study from the Netherlands. Overall, the activity of ISA was at least equal to FLU and equal to IT, VOR and POS. IT, VOR, POS, and ISA are the most active antifungal agents in vitro against CNE.
P1978 In vitro activity of isavuconazole (BAL4815) compared with seven other antifungal agents against 309 prospectively collected clinical Candida isolates from the Netherlands
I. Curfs-Breuker, J. Mouton, Y. Debets-Ossenkopp, H. Endtz, P. Verweij, J. Meis (Nijmegen, Amsterdam, Rotterdam, NL)
Background: Isavuconazole (ISA;BAL 4815/8557) is an antifungal drug of a new generation of triazoles including posaconazole, voriconazole and ravuconazole. We compared the in vitro activity of ISA against Candida species isolated from invasive infections with seven other antifungal drugs.
Methods: 309 strains were prospectively isolated during a 6-month period (07-2005 to 01-2006). MICs were determined for amphotericin B (AmB), flucytocine (5FC), fluconazole (FLU), itraconazole (ITC), voriconazole (VOR), posaconazole (POS), caspofungin (CAS) and ISA. Microdilution testing was done in accordance with CLSI M27-A2 in RPMI 1640 MOPS broth with L-glutamine without bicarbonate and incubated at 35°C for 24 hours. The MIC was determined visually and spectrophotometrically as the lowest concentration of drug showing absence of growth or ≥50% reduction of growth compared with that of the growth control for AMB, and 5FC and azoles, respectively.
Results: Species distribution was C. albicans (61%), C. glabrata (21%), C. tropicalis (5%) and C. parapsilosis (4%). Only 1% of C. albicans was resistant to FLU compared to 10% of the non-albicans Candida species isolates.
Among the FLU susceptible (MIC ≤ 8) non-albicans Candida species isolates (n = 103) the MIC90 and MIC range for ISA, CAS, FLU, ITC, POS, VOR, 5FC and AmB was 0.016, <0.002–0.125; 0.5, 0.016–2; 4, 0.125–4; 0. 25, <0.016–0.5; 0.125, <0.016–0.25; 0.125, <0.016–0.25; 0.063, <0.063–>64; 0.5, 0.063–1 mg/L, respectively.
Among the FLU less susceptible (MIC ≥ 16) non-albicans isolates (n = 10) the MIC90 and MIC range for ISA, CAS, FLU, ITC, POS, VOR, 5FC and AmB was 0.125, 0.004–0.125; 0.5, 0.016–0.5; 64, 16–64; 0.5, 0.031–0.5; 0.5, 0.031–0.5; 1, 0.125–1;4, <0.063–8;1, 0.125–1 mg/L, respectively.
Among the FLU susceptible C. albicans isolates (n = 193) the MIC90 and MIC range for ISA, CAS, FLU, ITC, POS, VOR, 5FC and AmB was <0.002, <0.002–0.63; 1, 0.016–2; 0.25, <0.063–8; 0.063, <0.016–0.5; <0.016, <0.016–0.5; <0.016, <0.016–0.125; 0.063, <0.063–2; 1, 0.125–2 mg/L, respectively.
Conclusion: Isavuconazole has a high in vitro activity against invasive Candida isolates with lower MICs than the seven comparator drugs. Isavuconazole is a potential attractive new agent to treat invasive Candida infections.
P1979 Risk factors for voriconazole resistance in non/albicans candida in HIV-positive children
A. Shahum, V. Krcmery, A. Liskova, P. Kisac, V. Hricak, M. Karvaj, A. Augustinova, E. Kalavsky, J. Benca, S. Seckova, K. Holeckova, Z. Havlikova, V. Sladeckova, D. Horvathova, H. Havrilova (Phnom Penh, KH; Bratislava, Trnava, Nove Zamky, SK)
Introduction: Resistance to voriconazole (VOR) has been observed in C. glabrata and mucorales. However in Candida spp. is still rare and represents less than 1% of all Candida spp.
Methods: We have analysed VOR and fluconazole (FLU) resistant candida in HIV-positive children in Cambodia. 8 cases occurred in 2006 after 3 years of HAART among 52 HIV-positive children at antiretroviral therapy (2 C. krusei, 2 C. glabrata, 2 C. famata, 1 C. lusitaniae, 1 C. kefyr).
Results: Non-albicans Candida (NAC) resistant to VOR appeared with no different distribution among children with different degree of immunosuppression. There was unproved significant association with HAART, antituberculotic drug, or pretreatment with antibiotics or antifungals. We did not record any resistance to VOR among C. tropicalis and C. parapsilosis, but resistance to both VOR and FLU were found among C. famata, C. glabrata, C. krusei and C. lusitaniae. VOR-resistant NAC were more likely resistant also to itraconazole and ketokonazole in contrast to FLU-susceptible NAC (P < 0.02 and P < 0.002 respectively). More frequent concomitant resistance to itraconazole, ketokonazole, 5-FC and voriconazole was also seen among FLU-resistant NAC (P <0.0175, P <0.004, P <0.011 and P <0.02).
Conclusion: VOR-R was related neither to low CD4 count nor to length of HAART or TMP/SMX prophylaxis and also not to antituberculotic and antibiotic therapy. We did not prove any risk factor for VOR resistance among NAC yeasts in HIV-positive children. Resistant NCA to VOR and/or FLU were more likely also resistant to other antifungals.
P1980 Identification of a mating-type region in the opportunistic fungus Pneumocystis carinii
B. Sanyal, P. Vohra, C. Thomas (Rochester, US)
Pneumocystis carinii (PC) is an intractable opportunistic fungus which causes severe pneumonia in patients with AIDS or other immunosuppressive conditions. PC alternates between distinct life cycle forms comprised of trophic forms and multinucleated cysts. The mechanism of cellular differentiation from the trophic form to the cyst is unknown. PC is phylogenetically related to the non-pathogenic ascomycetes S. cerevisiae and S. pombe. The mating-type locus of ascomycetes control key events in their life cycles. In S. pombe, full meiotic competence of haplotypes is conferred by the mating-type region which consists of mat1, mat2 and mat3 loci. The mat3-M loci contains 2 open-reading frames (ORFs) encoding mat-Mc and mat-Mi proteins. We identified and cloned this analogous region in PC using PCR techniques. This 4956-bp region contains 2 ORFs: PCmatMc is a 699-bp ORF encoding a predicted 232 amino acid protein and PCmatM1 is a 1080-bp ORF encoding a predicted 359 amino acid protein. Both ORFs are on opposite DNA strands similar to the configuration in S. pombe. PCmatMc contains an HMG-box domain postulated to bind DNA sequences necessary for life cycle regulation. We have expressed this gene as a recombinant protein and are determining the PC DNA sequences to which PCmatMc binds. Our investigations may provide new insights into the pathogenesis of PC and potential new therapeutics for treating PC pneumonia through identification of mechanisms involved in PC life cycle regulation.
Supported by NIH grant 2R01 AI-48409 to CFT.
P1981 Antifungal activity of miconazole against recent clinical isolates of Candida spp., as determined by minimum inhibitory concentration
M.A. Ghannoum (Cleveland, US)
Objectives: Miconazole (MICON) has long been used for the topical treatment of Candida vaginitis and other fungal infections of the skin. As such, existing susceptibility data for MICON is quite old and was generated before standard methodology was established. Currently there is an increased incidence of oropharyngeal candidiasis among immunocompromised patients, especially in the HIV positive population, for which a new delivery system of MICON has been developed. With Miconazole Lauriad® tablets currently in Phase III clinical trials, it is important to establish a susceptibility profile for MICON against recent clinical Candida isolates using standard methodology. The objective of this study was to test MICON against a large number of Candida isolates to determine whether there has been any development of resistance to this antifungal in the patient population. The antifungal activity of MICON, as determined by minimum inhibitory concentration (MIC) was compared to amphotericin B (AM), caspofungin (CAS), clotrimazole (CLOT), fluconazole (FLU), itraconazole (ITRA), nystatin (NYS), and voriconazole (VOR).
Methods: MICs were determined according to CLSI M27A2. Inhibition endpoints were read as a 50% reduction in growth compared to the growth control after 24 hours incubation (AM was read at 100% inhibition).
Results: MICON demonstrated potent inhibitory activity against all of the Candida strains tested. The overall MIC range for MICON was 0.004–1.0 μg/mL, while the MIC50 and MIC90, defined as the minimum concentration that inhibited 50% and 90% of isolates tested, were 0.06 and 0.5 μg/mL, respectively. With the exception of FLU, all of the comparator drugs demonstrated activity against the panel of test isolates. The MICON MIC90 for all species was comparable to that of AM, CAS, CLOT, ITRA, NYS, and VOR (within three dilutions), and was four dilutions lower than FLU. Importantly, MICON demonstrated MICs three to eight fold lower than FLU against FLU-resistant strains.
Conclusion: Our data showed that MICON demonstrated potent inhibitory activity against all of the Candida isolates tested, including those with known FLU resistance. This indicates that recent clinical isolates remain susceptible to this antifungal and do not reflect increased resistance. This has important implications to the new application of MICON in the treatment of oropharyngeal candidiasis.
Tuberculosis and other mycobacterial infections: diagnostic microbiology
P1982 Evaluation of partial 16S rRNA gene sequencing for identification of clinical isolates of non-tuberculous mycobacteria in a routine mycobacterial laboratory
R. Insa, M. Marín, M.J. Ruiz Serrano, M. del Rosal, M.J. Goyanes, D. García de Viedma, E. Bouza (Madrid, ES)
Objectives: Conventional identification of non-tuberculous mycobacteria (NTM) is based on biochemical characteristics, PCR-RFLP (PRA) or a combination of both. Both techniques are time consuming and occasionally produce discordant identification. The aim of this study is to evaluate the utility of the amplification and sequencing of the 5′ end 16S rRNA gene (16S-PCR) in the identification of clinical isolates of NTM in comparison with both conventional techniques.
Methods: NTM were characterised by: (i) biochemical and phenotypical characteristics, (ii) PRA and (iii) 16S-PCR. The 16S sequences were compared with those included in Genebank and Ridom. Only alignments with similarities higher than 99% were considered. Results obtained by 16S-PCR were compared with our gold standard (biochemical test plus PRA results)
Results: Overall we evaluated 52 isolates from different patients. Biochemical tests and PRA matched exactly in 42 isolates: M. fortuitum (6), M. abscessus (1), M. chelonae (2), M. xenopi (5), M. gordonae (6), M. avium–intracellulare (5), M. celatum (3), M. kansasii (5), M. peregrinum (1), M. lentiflavum (3), M. simiae (2), M. smegmatis (2) and M. thermorresistibile (1). Results obtained by 16S-PCR agree with that gold standard in 90.5% (38/42). In 10 isolates there was disagreement between biochemical test and PRA. Among these cases, 16S-PCR supported the result obtained by biochemical test in 5/6 and PRA in 1/6. In 4 isolates each of the three methods provided a different identification. 16S-PCR was able to recognize two new species: M. wolinskyi and M. mageritense.
Conclusions: 16S-PCR could be useful for a preliminary and rapid identification of NTM but needs to be complemented by one of the conventional methods, preferably biochemical tests. The analysis of other conserved genes could contribute to a more accurate identification.
P1983 Molecular detection of Mycobacterium tuberculosis by tHDA-ELISA DIG detection system
P. Gill, H. Abdul-Tehrani, A. Ghaemi, T. Hashempour, M. V-p Amiri, M. Ghalami, N. Eshraghi, M. Noori-Daloii (Tehran, IR)
Objectives: The recent resurgence in tuberculosis (TB) cases poses a serious public health problem. Effective TB management require the simple detection and rapid identification of the aetiologic agent. The nucleic acid amplification methods have proven to be very useful tools in the rapid diagnosis of Mycobacterium tuberculosis. Several PCR-based colorimetric methods such as PCR-ELISA have been previously described for molecular diagnosis of tuberculosis. However this study, we designed and developed a novel non-PCR-based colorimetric assay for specific and sensitive detection of rrs gene of Mycobacterium tuberculosis, is named enzyme-linked immunosorbent assay of thermophilic helicase-dependent DNA amplification (tHDA-ELISA).
Methods: In this procedure, like PCR-ELISA, the tHDA reaction selectively amplified a target sequence defined by two primers and simultaneously incorporated digoxigenin-deoxyuridine triphosphate (DIG-dUTP) in the resulting amplified DNAs. However, unlike PCR-ELISA, the tHDA uses an additional enzyme called a thermophilic helicase to separate DNA rather than heat. Thus the entire tHDA reaction can be performed at isothermal temperature which is optimised for synthesis and it needs no an expensive and power-hungry thermocycler. Then the DIG-labeled amplicons were detected using species-specific probe (biotin-modified), and colorimetric ELISA method.
Results: Our results were shown an equal sensitivity and specificity between this method and ELISA colorimetric assay of PCR products.
Conclusions: THDA-ELISA DIG detection system can be used more cost-effectively than PCR-ELISA for molecular detection of Mycobacterium tuberculosis in developing countries. The other formats of this technology are under study in real-time format and mobile format.
P1984 Utility of the Gen-Probe amplified MTD test for the tuberculosis diagnosis in formalin-fixed, paraffin-embedded histologic specimens
M. Ros, D. Mariscal, P. Ferreros, N. Combalia, R. Orellana, J.A. Vázquez, D. Fontanals, M. Torra, M. Rey (Sabadell, ES)
Objective: To value the relationship among the mycobacteria culture, clinical and histopathologic findings compatible with tuberculosis (TBC) and the results of the Gen-Probe amplified MTD test (MTDT) in formalin-fixed, paraffin-embedded histologic specimens.
Methodology: The affected anatomic sites of 24 biopsies were: lymph nodes (9); skin (3); synovial (2); colon (2); lung (1); pleural (1); peritoneal (1), parotid gland (1); bone marrow (1); spleen (1); breast (1); vertebral (1). All had presence of granuloma with multinucleated giant cells and/or caseous necrosis. To detect AFB, all smears were stained by Ziehl–Neelsen (ZN). Only 9 (37.5%) of the specimens were cultured for mycobacteria. A MTDT was considered positive when the result was greater than 500.000 RLU. The clinical information of the 24 patients was reviewed and those that have received antituberculous treatment or/and grew mycobacteria in the culture were considered cases of TBC.
Results: 23 of the biopsies had chronic granulomatous inflammation and 1 caseous necrosis. Only the specimen with necrosis presented ZN positive. In 7 specimens (29.1%) MTDT was positive, clinical information was suggestive of TBC and in 3 mycobacteria grew. One patient had MTDT negative, clinical information suggestive of TBC and a positive culture. In two cases, MTDT was negative, there was no culture but they were treated as TBC. Sensibility, specificity, PPV and NPV are 70%, 100%, 100% and 82.3% respectively.
Conclusions: Considering that there are cases in which the corresponding tissues are not sent for TBC culture, the MTDT can be a useful technique for the diagnosis of TBC in formalin-fixed specimens when clinical or histopathologic suspicion exists.
P1985 Direct identification of mycobacteria from BacTec MGIT 960 system tubes: comparative evaluation of INNO-LiPA Mycobacteria v2 and DNA AccuProbe assays
E. Mokaddas, S. Ahmad (Dasma, Kuwait, KW)
Objective: Species-specific identification of mycobacteria is of clinical relevance since treatment varies according to the species causing infection. This study evaluated the performance of two commercial probe assays; INNO-LiPA MYCOBACTERIA v2 (LiPA) and AccuProbe for species-specific identification of mycobacterial isolates in Kuwait directly from MGIT 960 system tubes.
Methods: Seventy-three respiratory and extrapulmonary specimens flagged positive for growth by MGIT 960 system and smear-positive for mycobacteria were evaluated for identification. The AccuProbe assay was performed and interpreted according to the manufacturer's instructions. The DNA extraction, PCR amplification and hybridisation steps were performed by following the instructions supplied with the LiPA kit. The results were validated by PCR amplification and DNA sequencing of 16S-23S internal transcribed spacer (ITS) region.
Results: Each of the 73 tubes grew one mycobacterial culture. Both AccuProbe and LiPA identified 47 isolates as Mycobacterium tuberculosis complex (MTC) members and ITS region sequences of 8 randomly selected isolates were concordant. AccuProbe assay identified 26 isolates as non-tuberculous mycobacteria (NTM) with species-specific identification of 9 isolates. The ITS region sequences were concordant with the NTM status for 17 isolates and species-specific identification of 7 of 9 isolates while two M. intracellulare strains were actually M. chimaera and M. avium complex sequevar MAC-C isolates. The LiPA identified 21 isolates as NTM with species (or complex)-specific identification of 19 isolates while no result was obtained for 5 isolates (no amplicons with LiPA primers). The ITS region sequencing confirmed the species (or complex)-specific identification by LiPA of all the 19 isolates while 2 isolates detected only as NTM by LiPA were identified as M. immunogenum and M. lentiflavum. The DNA sequencing also identified 4 isolates yielding no result in LiPA as M. kansasii and one isolate as M. chimaera.
Conclusions: All the MTC isolates are correctly identified by both, the LiPA and AccuProbe assays. Although, AccuProbe assay identified only some (9 of 26) while LiPA identified most (19 of 26) NTM isolates to the species or complex level, the 4 M. kansasii isolates correctly identified by AccuProbe assay were not even detected by LiPA indicating variations in sequences targeted for LiPA amplification in M. kansasii strains in Kuwait.
Supported by KURA grant MI02/04.
P1986 Evaluation of real-time PCR assays, based in molecular beacon technology, for detection and quantification of Mycobacterium tuberculosis DNA
D.P. Houhoula, A. Siatelis, N.J. Legakis, A. Tsakris, J. Papaparaskevas (Athens, GR)
Objective: The evaluation of a conventional and two real-time PCR assays, in comparison to the standard culture for tuberculosis (Tb), and the patients' clinical data.
Materials and Methods: A total of 60 clinical samples submitted for Tb diagnosis, were tested using three PCR assays targeting IS6110. The samples were 30 (7 pulmonary and 23 extra-pulmonary) from 22 patients, for which Tb diagnosis was based either on positive culture, or clinical presentation, and 30 from 30 patients with other non-Tb diagnoses. The assays were a previously reported (Kox et al, JCM 1994) conventional PCR (C-PCR), a real-time PCR (RT1) using previously reported nested-PCR inner primers (Caws et al, JCM 2000) with a newly designed molecular beacon, and a novel real-time PCR (RT2). The Beacon Designer 5.0 software was used for all newly designed beacons (RT1, RT2) and primers (RT2). DNA quantification was performed using a standard curve of five positive concentrations (3.4 pg/μL, and 340, 34, 3.4, and 0.34 fg/μL) of DNA extracted from M. tuberculosis H37Rv strain. All samples were tested neat and diluted 1/10 for PCR inhibition. Real-time PCR runs were acceptable when the negative controls had undetectable cycle threshold (CT) values, and the first two positive DNA concentrations had CT values of 27 to 30 cycles.
Results: Out of 30 samples from confirmed Tb cases, 28, 19 and 27 were positive using C-PCR, RT1, and RT2, respectively, corresponding to diagnostic sensitivities of 93%, 63% and 90%, respectively. Based on the DNA quantification performed, analytical sensitivities were 10 and 1 fg of DNA, for RT1 and RT2, respectively. When diluted specimens were examined, 5 inhibition cases were detected (1, 1 and 3 for C-PCR, RT1, and RT2, respectively). Combined performance of C-PCR+RT2 increased overall sensitivity to 100%. All 30 samples from non-Tb cases were negative with all assays (specificity 100%). RT2 quantification indicated that DNA extraction from tissue biopsies resulted in the lowest DNA yield compared to pleural/sputum samples (DNA load range 0.7–659 fg, and 4.4–576 pg, respectively). However, a single pus specimen with DNA load of 2.7ng was identified.
Conclusions: RT2 and C-PCR were the most sensitive assays tested, and their combination resulted in 100% diagnostic sensitivity. Specificity was 100%. Real Time PCR, based on molecular beacon technology, seems to be a rapid and sensitive procedure for Tb diagnosis and DNA quantification in clinical samples.
P1987 Rapid diagnosis of tuberculous pleuritis by real-time PCR
K. Baba, S. Pathak, L. Sviland, A.M. Dyrhol-Riise, N. Langeland, A.A. Hoosen, B. Asjø, T. Mustafa (Bergen, NO; Pretoria, ZA)
Tuberculosis is a public health problem. The hallmark of tuberculosis control is prompt diagnosis and treatment. This is even more important in suspected tuberculosis pleuritis (TP) for which accurate diagnosis is very crucial since delay in chemotherapeutic intervention is associated with poor prognosis. Diagnosis of TP requires microscopy and culture both of which has limitation, microscopy is not sensitive and culture takes time. The nested PCR (N-PCR) is used to detect M. tuberculosis complex organisms and genus specific real time PCR is used for detection of a broader spectrum of mycobacteria since atypical mycobacteria is common in HIV-TB coinfected patients. The real-time PCR (RT-PCR) has the advantage of not being prone to contamination and quicker because it does not require post amplification detection.
The aim of this study was to assess the diagnostic potential of nested PCR of the IS6110 gene to detect M. tuberculosis complex organisms and of RT-PCR for genus specific hsp65 detection in the diagnosis of T P.
Thirty-six paraffin-embedded pleural tissue samples were taken from the archives of the department of Pathology Dr. George Mukhari Hospital/Medunsa Campus University of Limpopo Pretoria South Africa. Twenty-five biopsies were from patients with clinical TP, and 11 biopsies from cases with other diseases were used as controls.
Nested PCR was performed using the Mycobacterium tuberculosis complex specific IS6110 primer and real-time PCR was performed using the genus specific hsp65 primer (ABI 7500 RT-PCR system).
Sixteen samples were positive by N-PCR and 15/16 was positive by RT-PCR. Out of the 11 non-TB cases 2 were positive by both N-PCR and RT-PCR while additional 2 cases were positive by only RT-PCR. Using clinical diagnosis as gold standard the sensitivity of N-PCR and RT-PCR were 64% and 76% while specificity was 82% and 64%, respectively. The sensitivity of RT-PCR using N-PCR as gold standard was 94%.
This study showed that RT-PCR is equally sensitive to N-PCR in diagnosing TP. Its lower specificity might be due to presence of atypical mycobacteria. Therefore RT-PCR is highly sensitive, not prone to contamination, and its quicker turn around time will lead to rapid diagnosis of TP.
P1988 Diagnosis of mycobacterium infection by TB-rapid-antigen-test in patients from Belarus
S. Zaker, L. Titov, A. Bahrmand, E. Sagalchyk, L. Surkova (Tehran, IR; Minsk, BY)
Background: Tuberculosis (TB) remains an important problem in the world, but TB diagnosis is often difficult. The routinely used procedures for TB diagnosis in clinical laboratory either show poor sensitivity (microscopy) or take several weeks (culture) before results are obtained. The aim of the study was to estimate sensitivity and specificity of developed for indication MTB in sputum kit “Rapid-Test”.
Method: In this study we tested 288 sputum samples from patients living in different regions of Belarus with aim to detect Mycobacterium tuberculosis by developed Rapid-Test. Sputum samples from patients were concentrated and washed in the specific cassettes containing nitrocellulose membrane with absorbing pads above it. MBT were detected by addition anti-TB monospecific rabbit serum and gold-protein A solution. Positive samples were characterised by displaying a red-to brown colour in central area of cassettes in the contrast to the blue-purple colour in negative cases. This new rapid test was compared with the results of acid-fast staining microscopy, specific culture for TB bacilli and specific PCR amplification of TB DNA segments.
Results: In this study 107 samples from healthy persons were used as control and 177 sputums from patients with tuberculosis confirmed by culture method. We found that developed Rapid-Test was more effective for M. tuberculosis detection compared with specimen microscopy. Efficiency of Rapid-test in compare with “gold standard” of culture investigation was 88.5% for Belarussian samples. Compared with culture for Belarussian samples Rapid-Test was false positive in 22 cases, false negative in 19 cases, sensitivity was 81%, specificity 93%, positive predictive value 66.3% and negative predictive value 89.9%. Meanwhile Ziehl–Neelsen microscopy did not give false positive results, false negative results were registered in 23 cases, efficiency of microscopy was estimated as 88%, sensitivity 68.5%, specificity 94.8%, PPV 82% and NPV 89.7%.
Conclusion: Therefore, we observed 25% more sensitivity and 20% specificity of rapid test compared with microscopy for Mycobacterium tuberculosis detection.
P1989 Evaluation of a rapid automated assay for the isolation of Mycobacterium tuberculosis DNA directly from clinical samples, using magnetic particles
J. Lysén, H.K. Høidal Berthelsen, M. Espelund, S. Jeansson, A.T. Mengshoel, U.H. Refseth (Oslo, NO)
Objectives: The objective of this study was to evaluate the performance of a fully automated DNA preparation system, the BUGS'n BEADS™ MYCOBACTERIUM (Genpoint, Norway), to isolate Mycobacterium tuberculosis complex DNA from clinical respiratory samples.
Method: The BUGS'n BEADS™ principle was used to develop an automated customised application. Briefly, the automatic isolation begins with addition of specially coated magnetic particles to Nalc-NaOH-treated clinical samples. Following bacterial capture, the sample matrix is removed. A rapid lysis at RT releases DNA, which is then adsorbed on to the same magnetic particles. Beads are washed and the purified DNA is robotically transferred for NAAT analysis. Samples were analysed both by using an in-house developed real-time PCR targeting the M. tuberculosis complex, and the Strand Displacement Amplification (SDA) part of the ProbeTec™ ET Mycobacterium tuberculosis Complex (DTB) Direct Detection (Becton Dickinson).
Results: Up to 48 samples can be simultaneously analysed using the automated BUGS'n BEADS™ system, with as little as 20 minutes of hands-on for each run, and requiring a total time to results of four hours. For smear positive samples, the results for the automated BUGS'n BEADS™ system in combination with real-time PCR were comparable to culture. The combination of SDA with the automated BUGS'n BEADS™ was comparable to the in-house Real-Time PCR. For the smear negative samples, we have observed so far that lowering the predefined BD cut-off from 3400 to 1000, enabled to improve sensitivity without worsening specificity. No inhibition was recorded for the SDA.
Conclusions: We here present a fully automated system for preparation of M. tuberculosis complex DNA from clinical respiratory samples, compatible with in-house developed and commercial downstream detection systems. Used with respiratory samples, the system will give clinically relevant results within one working day, requiring minimal hands-on.
The combination with SDA demonstrated successful removal of inhibition, while opening for further improvement of sensitivity. To further challenge the system, and evaluate lowering cut off, non-respiratory samples will be included in a more extensive study.
P1990 Genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis isolates
M. Unzaga, R. Blanco, M. Fernandez, A. Morla, L. Garcia, C. Ezpeleta, J. Alava, R. Cisterna (Bilbao, ES)
Objectives: The rapid determination of drug resistance in clinical isolates of Mycobacterium tuberculosis is essential for the initiation of effective chemotherapy and preventing further spread of drug-resistant isolates. Drug susceptibility testing by conventional methods takes several weeks although some new liquid medium-based systems have been commercially introduced with a shortened incubation time. A commercially DNA strip assay (Genotype MTBDR assay (Hain Lifescience Nehren, Germany) based on a multiplex PCR in combination with reverse hybridisation to identify either wild type or most common mutations in M. tuberculosis katG and rpoB was evaluated.
Material and Methods: Genotype MTBDR assay was evaluated by investigating 53 strains isolated in Basurto Hospital. The results were compared to data obtained by BACTEC MGIT 960 Becton Dickinson Microbiology Systems, Sparks, MD). Both assays were performed as recommended by the manufacturer.
Results: According to the results of BACTEC MGIT 960, 49 were fully susceptible, 2 were resistant to high level isoniazid (0.4μg/mL) (INH), 2 WERE resistant to low level INH (0.1μg/mL), and two was Multidrug-resistant (MDR).
All the isolates that were phenotypically resistant to high level INH had a S315T1 mutation in katG codon 315. No mutation was found in the 2 strains which showed a low level of resistance. None of phenotypically INH-susceptible isolates had the codon 315 mutation.
No mutations were detected in the phenotypically RIF-susceptible.
One of MDR isolate had an rpoB S531L mutation but did not hybridised with the Genotype MTBDR test probe (S531L), although the test with the WT 5 probe was negative. The other one had an rpoB S531L mutation that hybridised with the mutation-specific probe (MUT3). In both of MDR the mutation in katG of MDR was the same than the mentioned in the other strains.
Conclusions: Genotype MTBDR assay allows rapid and specific detection to INH and RMP although it cannot totally replace traditional cultured-based methods basically due there is not molecular test established targets all possible genes o mechanism and thus a variable proportion of resistant strains will not be detected.
P1991 Comparison of Gen-Probe A-MTD test and GenoType Mycobacteria Direct 3.0 test in detecting Mycobacterium tuberculosis complex and four nontuberculous mycobacteria in clinical samples
E. Fakiri, E. Papaefstathiou, K. Paulou, A. Charalampopoulou, D. Bassoulis, A. Kosmopoulou, E. Papafrangas (Athens, GR)
Aim: The comparison of two RNA-amplification bassed assays, Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test (A-MTD), Gen-Probe, Inc. San Diego, CA and a newly developed commercial kit GenoType Mycobacteria Direct 3.0 (GTMD), Hain Lifescience, Germany, for the detection of five mycobacterial species (M. tuberculosis complex, M. avium, M. intracellulare/M. scrophulaceum/M. interjectum, M. kansasii and M. malmoense) directly from respiratory and extrapulmonary clinical specimens.
Material and Methods: We tested a total of 120 clinical samples by both assays: 97 from lungs (sputa, BALs, bronchial washings) and 23 from extra-pulmonary sites (CSFs, pleural fluids and pericardial effusions, ascetic fluids, urines and lymph node aspirates)
Results: Positive M.tb complex: A-MTD 42, GTMD 40. False negative M.tb complex: A-MTD 0, GTMD 2. False positive M.tb complex: A-MTD 2, GTMD 0. Additionally GTMD assay correctly identified 6/120 samples as M. intracellulare and 2/120 as M. avium.
Conclusions: 1. The GTMD test detects in one step five mycobacterial species including M.tb complex. 2. The ribosomal RNA isolation and NASBA amplification in GenoType test monitors the presence of amplification inhibitors, since the A-MTD does not. 3. The A-MTD test is quicker (3.5 h) than GTMD test (5–6 h). 4. The A-MTD test seems to be more sensitive but less specific than GTMD test.
P1992 Molecular methodologies in identifing mycobacteria isolates and detecting Mycobacteria spp. directly in clinical samples
E. Fakiri, E. Papaefstathiou, D. Bassoulis, A. Charalampopoulou, K. Paulou, S. Mamouha, E. Papafrangas (Athens, GR)
Aim: The evaluation of commercially available molecular methodologies (Hain Lifesciences, Germany) in (1) The identification of mycobacteria from cultures in solid (LJ) and liquid (MGIT) media and (2) the detection of mycobacteria spp. directly in clinical samples.
Materials and Methods: (1) We tested 520 mycobacteria strains by PCR and reverse hybridisation in nitrocellulose strips. We used the MTBC kit and in case of failure consecutively the CM and AS kits.
(2) Additionally, 100 clinical samples (73 sputa, 2 BALs, 10 bronchial secretions, 3 CSFs, 4 pleural, 1 pericardial & 1 ascetic fluids, 4 urines and 2 lymphnode biopsies) from suspected TB patients, were tested directly for the five most frequent mycobacteria spp. by GenoType Mycobacteria Direct Test (VER 3.0).
Results: (1) 365 Mtb strains were identified as M. tuberculosis–M. africanum–M. canetti (363), M. bovis BCG (1) and M. africanum É (1). Of the 155 MOTTs the CM kit identified 102, of the remaining strains the AS kit identified 31, while the last 22 failed to be identified by all the three kits.
(2) Of the 100 clinical samples tested with the GenoType Mycobacteria Direct Test (VER 3.0) kit, 44 tested positive for mycobacteria (41 M. tuberculosis complex, 2 M. intracellulare, 1 M. avium), while the rest tested negative.
Conclusions: (1) The commercially available kits we used proved to be fast, reliable and relatively cheap in identifying mycobacteria species from solid and liquid media. (2) The new kit GenoType Mycobacteria Direct Test (VER 3.0) is a highly sensitive, specific and quick method for the detection of mycobacteria in clinical samples.
P1993 Extrapulmonary mycobacteria isolations
S. Karabela, S. Nikolaou, A. Raftopoulou, S. Anagnostou, I. Kouseris, N. Makrigiannis, M. Koletou, S. Kanavaki (Athens, GR)
Aim: To study the extrapulmonary mycobacteria isolations, in an eight years period (1998–2005).
Material: 21177 extrapulmonary clinical specimens.
Methods: Microscopy by Ziehl Neelsen stain. Culture by the classical method on solid Löwenstein-Jensen (LJ) medium, as well as, by the automated system Bactec MGIT 960 (Becton Dickinson). Identification by molecular hybridisation, using the commercial kits: InnoLipa V2 (Innogenetics), Accuprobe (Gen Probe, bioMérieux) and Genotype Mycobacterium CM and AS (Hain Life, Science).
Sensitivity testing by the classical method of proportion on LJ solid medium, as well as, by the automated system, Bactec MGIT 960 (Becton Dickinson) and the molecular hybridisation technique, Geno Type MTBDR, (Hain Life Science)
Results:
-
–
The number of extrapulmonary specimens tested increased through out the study period, from 2197 (13%) in 1998 to 3322 (18.2%) in 2005.
-
–
685/21177 (3.2%) clinical specimens grew a mycobacterium.
-
–
622/685 (90.8%) of mycobacteria were M. tuberculosis (MTB), while
-
–
63/685 (9.2%) were Non TB (NTB) mycobacteria.
-
a
Concerning MTB isolations, most common clinical sources were pleural fluid 243/622 (39%), urine 91/622 (14.6%), lymph nodes 83/622 (13.3%) and pus 81/622 (13%). Drug resistance of MTB isolates showed that 35/622 (5.6%) were resistant to isoniazide (INH), 15/622 (2.4%) to rifampicin (RIF) and 12/622 (1.9%) were MDR (INH+RIF)
-
b
Concerning NTB isolations, 52/63 (82.5%) were M. avium, 9/63 (14.3%) were M. chelonae, 1/63 (1.6%) was M. peregrinum and 1/63 (1.6%) was M. fortuitum. Most common clinical sites for the NTB isolates were for M. avium, lymph nodes 21/63 (33.3%), blood 20/63 (31.7%) and pleural fluid 8/63 (12.7%), while for M. chelonae blood 9/9 (100%), for M. peregrinum pleural fluid and for M. fortuitum a lymph node.
Conclusions: The vast majority of extrapulmonary specimens grew an MTB. Most common clinical sources for extrapulmonary MTB isolations were pleural fluid and urine, while for NTB extrapulmonary isolations, lymph nodes and blood. Concerning drug resistance and multidrug resistance of MTB, they are significantly lower in extrapulmonary compared to pulmonary specimens, according previous studies of our laboratory.
P1994 Extensively drug-resistant tuberculosis (XDR TB) in Athens
S. Karabela, S. Nikolaou, I. Kouseris, N. Makrigiannis, S. Kanavaki (Athens, GR)
Objectives: The Global XDR TB Task Force (Geneva, October 8–9, 2006) agreed upon a revised case definition of XDR TB, as those cases of TB patients, whose responsible M. tuberculosis isolates are resistant to isoniazide and rifampicin, plus any of the quinolones and at least one of three injectable second-line drugs (amikacin, kanamycin or capreomycin). Since transmission of XDR TB strains is an emerging threat for the world, countries should strengthen TB control measures. For this reason, we evaluated the presence of the extensively drug resistant M. tuberculosis (MTB) strains in TB patients in Athens (January 2004–August 2006).
Methods: 1,415 new TB patients resulted in an equal number of MTB isolations. Microscopy, culture by the classical method on solid Löwenstein-Jensen (LJ) medium, and by the automated system Bactec MGIT 960 (Becton Dickinson), identification by hybridisation methods and commercial kits: InnoLipa V2 (Innogenetics), Accuprobe (Gen Probe, bioMérieux) and Genotype Mycobacterium CM and AS (Hain Life, Science) and sensitivity testing by the method of proportion on LJ solid medium, by Bactec MGIT 960 (Becton Dickinson) and by hybridisation technique, Geno Type MTBDR (Hain Life Science).
Results: 40/1,415 (2.8%) were multi-drug resistant (MDR) and 9/1,415 (0.6%) were XDR, so that percentage of MDR which were XDR was 22.5% (9/40 MTB isolates). Regarding XDR TB patients' nationality, 3/9 patients (33.3%) were from NE European countries and 6/9 (66.7%) were native Greeks.
Conclusions: The isolation of XDR MTB strains in TB patients in our country, should alert public health authorities. Greece, being a common destination for a great number of immigrants coming from NE Europe, SE Asia and Africa and a portal to Europe, should immediately respond to this danger to prevent further spread of XDR TB isolates.
P1995 Incidence of cmaA2 gene in Beijing and non-Beijing strains of Mycobacterium tuberculosis strains isolated from clinical specimens
N. Amir Mozaffari (Tehran, IR)
Objectives: The purpose of this investigation was to determine the prevalence of cmaA2 gene in M. tuberculosis strains of both Beijing and non-Beijing types isolated and typed by spoligotyping. Related risk factors among patients with different nationalities residing in Iran were also determined.
Methods: The study population involved a total of 742 patients that referred to the NRITLD, the referral tuberculosis centre in Iran; during March 21, 2003, till March 21, 2005. The isolated M. tuberculosis strains have been characterised by performing susceptibility tests against four first-line antituberculosis drugs and were then subjected to spoligotype characterisation. PCR was used for detection of cmaA2 gene and its nucleotide sequence was also determined.
Results: Spoligotyping of M. tuberculosis strains resulted in 150 different patterns. One hundred thirty five (90%) of these spoligotype isolates were unique and reported for the first time. The remaining 15 (10%) spoligotype patterns were previously reported from other geographical regions of the world. East African Indian (EAI) family was most prevalent than the other genotypes. Interestingly, 6.1% of the strains belonged to the Beijing family. Multidrug-resistance M. tuberculosis isolates were obtained from 13.6% of the patients during this study. The cmaA2 gene was detected in M. tuberculosis clinical isolates, but not in saprophytic strains such as M. kansasi. The other risk factors such as sex and age were also contributing factors to the disease state.
Conclusion: The results showed that multidrug-resistant bacteria were more prevalent in Iranian tuberculosis patients. In addition, the spread of M. tuberculosis strains belonging to the Beijing family among Iranian patients has to be considered seriously. This study confirmed the widespread existance of cmaA2 gene in almost all clinical isolates. It is also important to undertake studies to identify which factors are the most significant to consider in tuberculosis control programme.
P1996 Molecular analysis of drug-resistant Mycobacterium tuberculosis isolates collected from patients with pulmonary tuberculosis in central Poland
T. Jagielski, E. Augustynowicz-Kopec, M. Kozinska, A. Zabost, M. Klatt, Z. Zwolska (Warsaw, PL)
Objective: Although the incidence of tuberculosis (TB) in Poland is steadily decreasing, the prevalence of primary drug-resistant TB, including multidrug resistance (MDR), has been on the rise over the last few years. Therefore, continued surveillance and monitoring of TB epidemiology are required. Currently, molecular typing methods have become a useful tool for tracking and control of transmission of drug-resistant TB. The aim of this study was the molecular characterisation of drug-resistant Mycobacterium tuberculosis clinical isolates.
Materials and Methods: A total of 48 clinical isolates of M. tuberculosis representing 48 non-related, adult patients with resistant pulmonary tuberculosis in central Poland in 2004 were analysed by spoligotyping and newly devised PCR-based method designated IS6110-Mtb1/Mtb2 PCR. Species identification and drug susceptibility testing had been performed beforehand.
Results: Among strains tested, a total of 26 distinct spoligotypes were identified. Unique spoligotype patterns were observed in 19 (39.6%) isolates and the remaining 29 (60.4%) isolates were grouped within 7 clusters, made up of 2–8 isolates. When compared with an international spoligotyping database SpolDB4, 13 (27.1%) of the 19 unique profiles shared already described spoligotypes, whereas the rest 6 (12.5%) did not match any existing spoligotype and were defined as orphans. Of the 7 recognized clusters, one had not been described in SpolDB4. This cluster, comprising 4 isolates, seem to be specific for the study setting. Interestingly, two members of the Beijing family were identified.
Of the 7 spoligotyping clusters, 3 were not confirmed and 2 were partially confirmed by IS6110-Mtb1/Mtb2 PCR. Only two spoligotyping clusters, comprising 2 and 4 isolates, respectively, were identical both with spoligotyping and IS6110-Mtb1/Mtb2 analysis.
Table 1.
Comparison of discriminating power among two genotyping methods used in this study
| Technique | No. of clustered isolates (%) | No. of clusters | Size of cluster |
|---|---|---|---|
| Spoligotyping | 29 (60.4) | 7 | 2–8 |
| IS6110-Mtb1/2 PCR | 12(25) | 4 | 2–4 |
Conclusions: Molecular analysis by using two genotyping methods, undertaken in 48 drug-resistant M. tuberculosis isolates, showed a variety of different genetic profiles. Spoligotyping overestimated by about 2.5 times the number of clustered isolates, compared to IS6110-Mtb1/Mtb2 PCR. A combination of spoligotyping and IS6110-Mtb1/Mtb2 PCR showed 12 (25%) strains to be clustered, suggesting possible recent transmission.
P1997 An IS6110-targeting fluorescent amplified fragment length polymorphism DNA fingerprinting alternative to IS6110 RFLP: a pilot study
N. Thorne, J. Evans, E.G. Smith, P. Hawkey, S. Gharbia, C. Arnold (London, Birmingham, UK)
Objectives: A rapid, simple, highly-discriminatory DNA fingerprinting methodology that produces data that can be easily interpreted, compared and transported is the ultimate goal for M. tuberculosis infection control professionals and epidemiologists. The current ‘gold standard', IS6110 RFLP, is far from meeting all these requirements; however its discriminatory power has not been matched by a rapid, alternative method that also analyses the IS6110 sequence.
Methods: A panel of 78 blinded isolates from TB cases that were either epidemiologically linked or distinct were selected for analysis by IS6110 RFLP and FAFLP. A pilot study for a novel Fluorescent Amplified Fragment Length Polymorphism (FAFLP) approach to M. tuberculosis DNA fingerprinting targeting the variable IS6110 marker was tested for specificity and reproducibility and compared with IS6110 RFLP typing results on the panel of isolates. IS6110 RFLP was carried out using the internationally standardised published method.
Results: IS6110 RFLP defined the 78 isolates into nine clusters containing 2–32 isolates, with a total of 63 clustered isolates and 15 unique isolates. There were seven low-copy number isolates included in which four clustered into two, each containing a pair of isolates. Comparison of in silico analysis with strain H37Rv found FAFLP detected fragments to be highly specific in vitro. The reproducibility of FAFLP fragment profiles for five repeated digestions with strain H37R was 100%. FAFLP and RFLP clustering results were found to be highly congruent. FAFLP clustering was highly similar to RFLP discrimination of the 78 isolates as unique or clustered strains. FAFLP allocated 97% of RFLP-clustered isolates to the same eight clusters as RFLP. FAFLP failed to cluster two single-copy strains that had been clustered by RFLP, however clustering based on a single IS6110 copy is not reliable and secondary fingerprinting is necessary.
Conclusion: The discriminatory power exhibited by FALP in this initial collection of isolates indicates that it would be a rapid and specific screening alternative to IS6110 RFLP. The accuracy of fragment sizing by FAFLP has allowed common fragments to be defined that are likely hot-spots for IS6110 transposition. FAFLP will enable rapid screening of strains for epidemiological investigations and initiate further research into the frequency, conservation and consequence of IS6110 hotspots and specific transposition sites.
P1998 Determination of the invasiveness of non-pigmented rapidly growing mycobacteria by means of two different in vitro assays
J. Esteban, N. Zamora, I. Gadea, R. Fernández-Roblas, A. Fernandez-Martínez (Madrid, ES)
Objectives: To determine the invasive capacity of non-pigmented rapidly growing mycobacteria (NPRGM) in clinical isolates and in collection strains of NPRGM using the fibroblast-microcolony assay and the HEp-2 monolayer invasiveness assay. To correlate the invasive capacity with the species, colony phenotype and clinical significance of the strains.
Methods: Clinical isolates were identified according recommended biochemical tests and PCR-RFLP analysis of the hps65 gene. Clinical significance of the strains was evaluated by analysis of the clinical charts according to commonly accepted criteria. Colony phenotype was evaluated by culture on Middlebrook 7H10 agar plates during 5 days. The fibroblast-microcolonies assay, previously described by Byrd and Lyons, was used with minimal modifications. The relationship between diameters of the individual microcolonies in fibroblast were used by determinate the invasive capacity. The invasion assay in HEp-2 monolayers was carried out according to the method described by Bermudez et al. with an inoculum of 106 CFU. Invasive bacteria in HEp-2 monolayers were quantified performing 1:10 serial dilutions of the sonicate and streaking each dilution in Tryptic Soy Agar with 5% sheep blood plates. These plates were incubated at 30°C during 7 days before counting the colonies.
Results: 92 strains of NPRGM were tested, including 19 collection isolates of 15 species and 73 clinical isolates of 5 species (M. abscessus, M. chelonae, M. fortuitum, M. peregrinum and M. mucogenicum). 35 colonies gave rough colonies, and 57 were smooth colonies. 12 strains showed elongated colonies in the fibroblast microcolonies assay, while all the other strains showed rounded colonies. All but one strain showed different degrees of invasiveness of the Hep-2 monolayers, with a range of 8–22,500 UFC recovered. No statistically significant differences were found in relation with colony phenotype, clinical soignificance and the presence of elongated colonies or higher counts in the Hep-2 invasiveness assay.
Conclusions: Almost all strains of NPRGM were able to invade Hep-2 cell monolayers, but only 13% of the strains were able to invade fibroblasts according to the results of the microcolonies assay. No relationship were found between these data and colony phenotype or clinical significance.
Acknowledgments: This study was supported by a grant from the Fondo de Investigaciones Sanitarias (FIS PI030146). N. Zamora was funded by a grant from the Fundación Conchita Rábago de Jimenez Díaz (Madrid, Spain).
P1999 pncA and rpoB mutations in Mycobacterium tuberculosis, a 3-year experiment in Lyon, France
S. Mignard, G. Carret, M. Chomarat, I. Fredenucci, M. de Montclos, J. Flandrois (Pierre Benite, FR)
The emergence of drug resistant Mycobacterium tuberculosis is a serious public health problem. Pyrazinamide and rifampicin are first line drugs and their efficacy can be evaluated by DNA sequencing. We realised rpoB and pncA sequencing for all MTBC isolated from 2003 to 2006 in order to check the different mutations existing in Lyon, France. The in vitro susceptibility testing was done in the same time. 330 isolates of MTBC were studied during this 3-year period. On the rpoB gene, 9 mutations have been reported, 6 have ever been described: H526L (n = 1), S531L (n = 3), L511P (n = 1) and the deletion of nucleotides 514–516. 3 mutations were unknown N518D (n = 1), S469L (n = 2) but for these strains isolates were susceptible to rifampicin by in vitro testing. On the pncA gene 22 mutations have been reported. Among the 17 ever known in the literature, 10 correspond to the well known H57D mutation of M. bovis strains and 7 were T87M mutations. This last mutation is described in the literature associated with a deletion conferring resistance to pyrazinamide. All our T87M mutated strains were susceptible to pyrazinamide in vitro (none presented the deletion). Five mutations were unknown: A146V (n = 1), R2W (n = 1), H43Y (n = 1), L19P (n = 1) and V21A (n = 1). Only the A146V strain appeared resistant to pyrazinamide. The conclusion of this study is the emergence of the T87M mutation on pncA gene among strains that remain susceptible to pyrazinamide in vitro. No relationships were observed between patients by spoligotypes and MIRU types' comparison.
P2000 The muralitic activity of Rpf proteins and their ability to resuscitate dormant mycobacterial cells
G Demina, G. Mukamolova, A. Kaprelyants (Moscow, RU)
Objectives: Micrococcus luteus secretes a small protein Rpf, which plays an important role in the resuscitation of dormant cells of several species of Gram-positive bacteria, e.g. M. luteus and pathogenic bacteria Mycobacterium tuberculosis. Latent tuberculosis infection is believed to connect with the dormant state of M. tuberculosis in the human host. Rpf-like proteins contain specific domain which adopts a lysozime-like fold however enzymatic activity of Rpf proteins is poorly understood.
The purpose of this study is to elucidate an enzymatic activity of Rpf proteins and its relation to biological activity.
Methods: The muramidase activity was measured fluorimetrically, 4-methylumbelliferyl-beta-D-N,N′N″-triacetylchitotrioside was used as a substrate. The efficacy of resuscitated “non-culturable” M. smegmatis cells in liquid medium was estimated by MPN assay.
Results: Recombinant M. luteus Rpf is able to hydrolise the artifical lysozyme substrate which is structurally similar to peptidoglycan of bacterial cell wall. The invariant catalytic glutamate residue found in lysozyme and lytic transglycosylases is also conserved in the Rpf proteins. Replacement of glutamate residue by amino acids with less structural similarity resulted in a decrease in enzymatic activity of Rpf and its ability to resuscitate dormant Mycobacterium smegmatis cells. Rpf possible cleaves glycosidic bonds with inversion of configuration, but in contrast to lytic transglycosylase, Rpf doesn't form 1,6-anhydromuro derivatives (Keep et al., 2006). The effect of the some metal ions on the enzymatic activity was studied; the results show that Mg2+ and Ca2+ slightly activate the enzyme, while Zn2+ inhibits enzymatic activity.
Conclusions: Rpf proteins are evidently peptidoglycan hydrolase enzymes. Rpf's muralitic activity correlates with its ability to stimulate resuscitation of dormant bacteria, consistent with the view that biological activity of Rpf molecule is connected directly or indirectly with its ability to cleave glycosidic bonds in bacterial peptidoglycan.
P2001 Evaluation of the BacT/Alert PZA test for susceptibility testing of Mycobacterium tuberculosis to pyrazinamide in comparison with BacTec 460TB
M. Garrigó, C. Moreno, L. Aragon, P. Coll (Barcelona, ES)
The importance of rapid availability of Mycobacterium tuberculosis susceptibility testing is universally acknowledged. Although BACTEC 460TB susceptibility testing is considered the gold standard, its major disadvantages are that it is semiautomatic and uses radioactive material, with the need for disposal of radioactive waste.
Objectives: The purpose of this investigation was to evaluate the performance of the BacT/ALERT PZA (Bio-Mèrieux) kit for testing M. tuberculosis susceptibility to Pyrazinamide (PZA) drug compared to the radiometric BACTEC 460TB PZA test (Becton-Dickinson)
Methods: A total of 50 M. tuberculosis isolates from human samples were included in this study. Twenty, from a culture collection of the Mycobacteria Study Group of Barcelona, were resistant to PZA having a pncA mutation. The selected clinical strains were tested in parallel with BacT/ALERT PZA kit and BACTEC 460TB PZA test. Two reference control strains, ATCC 27294 (PZA sensitive) and ATCC 35745 (PZA resistant) were included in order to validate the conditions in which the clinical strains were analysed. Control and clinical strains were prepared and tested according to the respective test procedure and the reading and interpretation of each test were performed in accordance with the manufacturer's recommendations. Strains with discrepant results were repeated in the two methods.
Results: No technical problems were detected. Five strains needed to be repeated: two because of initial discrepant results, two because of lack of growth in BACTEC 460 and one because of no respect of the test validation criteria of the BacT/ALERT PZA kit. Final overall agreement between the two systems was 100%. The total turnaround time was 13.87 days for BacT/ALERT PZA kit versus 12.39 days for BACTEC 460TB PZA test.
Conclusion: Final overall agreement between the two systems was 100%. That is, there were no differences between the two systems in the ability to detect the sensitivity to PZA or the expression of the different resistance molecular mechanisms included. There was no significant difference in the total turnaround time of the two systems. The main advantages of BacT/ALERT PZA kit are that it is fully automated with a nonradiometric detection and that it is less labour-intensive than the BACTEC 460TB system.
P2002 Drug resistance of Mycobacterium tuberculosis in Santiago de Compostela, Spain (2002–2005)
M. Pérez del Molino, V. Tuñez, F. Pardo, S. Cortizo, P. Romero (Santiago de Compostela, ES)
Objective: The aim of the study was investigate the rate of drugs resistant tuberculosis (DR-TB) in Santiago de Compostela (Galicia, north-west of Spain, 459.180 inhabitants). In our sanitary area, the total number of cases per 100.000 population dropping from 51.5 to 28.3 between 2002 to 2005. The rate of TB patients with human immunodeficiency virus (HIV) coinfection remain 1.7% (2002) and 1.5% (2005).
Methods: The study followed the methodology recommended by the WHO/IUATLD, and included isolates processed between January 2002 and December 2005. Susceptibility testing was performed using the Bactec 460 method (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD, USA) with Middlebrook 7H12 medium (Bactec 12B) and the following final drug concentrations: 0.1 μg/mL for INH, 2μg/mL for RMP, 6 μg/mL for streptomycin (SM), and 7.5μg/mL for ethambutol (EMB). Culture and detection procedures were performed in accordance with the manufacturer's instructions. All susceptibility testing was performed at the Reference Mycobacteria Laboratory in Galicia, was subjected to quality control checks by the corresponding supranational reference laboratory.
Results: The sample included 417 strains of M. tuberculosis (379 never treated cases and 38 previously treated cases). Isolates from 345 of the 379 never treated cases (91.03%) were susceptible to all of the drugs tested. This percentage was 86.84% for previously treated. Fifteen strains from never treated patients and four from previously treated patients were resistant to INH (3.96% and 10.53%, respectively). There were one case of MDR-TB among never treated patients (0.26%) and two among previously treated patients (5.26%). The resistance to SM from never treated patients remain in 4.75%.
Conclusion: The rate of primary resistance (8.97%) are also similar to the mean published by the WHO in its latest report 10.2% [range0– 57.1%]). The rate of primary MDR-TB (0.26) was slightly lower than the WHO mean of 1.1% [range 0–14.2%]. Although Santiago de Compostela has a high incidence of TB, the resistance levels detected by the study do not currently pose a serious problem for the region.
We believe in the importance of keeping the quality of the TB prevention and control programmes.
Tuberculosis: diagnostic immunology
P2003 Recommendations for the use of interferon-gamma assays for the diagnosis of tuberculous infection
H. Hoffmann, T. Bodmer, P. Kirschner for the Working Group on Mycobacteria (Arbeitskreis Mykobakterien – AKM) of Germany, Austria and Switzerland
Two years ago, CE certified interferon-gamma release assay (IGRA) has been launched on the German marked. Ever since, a multitude of studies have tested the IGRA. Guidelines have been elaborated by national expert committees for England, the USA and Switzerland. However, standards of diagnostics and management of tuberculosis may change from country to country.
This statement was drafted by members of the “Arbeitskreis Mykobakterien” (AKM), the expert committee for TB diagnostics of Germany, Austria and Switzerland on the basis of review of relevant literature and experience with CE certified IGRA available so far. The statement provides practice-relevant guidelines for indication, pre-analytics and interpretation of IGRA test results in different clinical conditions. The IGRA are integrated in existing guidelines of the DZK for TB management.
P2004 Serological rapid test for discrimination between latent tuberculous infection and active tuberculosis
H. Hoffmann, A. Bosse, A. Neher, M. Singh (Gauting, Braunschweig, DE)
Since 100 years, the tuberculin skin test (TST) is the diagnostic basis of tuberculous infection. Recently, the interferon-gamma release assays (IGRA) appeared as new tests with higher specificity than the TST. However, neither the TST nor the IGRA are able to discriminate between latent tuberculous infection (LTBI) and active tuberculosis (TB).
Very recently, the serologic rapid test TB-ST (Lionex GmbH, Braunschweig, Germany) has been launched functioning similarly to pregnancy tests. The seven antigens combined in the test are supposed to confine specifically active TB versus LTBI.
In the present study, we analysed the validity of the test. 260 patients with active TB, LTBI or different pneumological disorders were enroled. The TB-ST was able to detect active TB with a specificity of 98%. Overall sensitivity was 53%, rising to 80% with smear detection rate of acid fast bacilli.
TB-ST is easy to perform (hands-on time less than 2 minutes), lasts only about 20 minutes, and a positive test result is a strong hint for active TB. The test principle is presented and options for expedient use of TB-ST are put up for discussion.
P2005 Role of an interferon-gamma based assay (T-SPOT®.TB) for the diagnosis of tuberculosis in routine clinical practice
J. Santos, M. Cordeiro, T. Hanscheid, E. Valadas, F. Antunes (Lisbon, PT)
Objectives: Diagnosis of tuberculosis (TB) can be difficult and is often fraught with long delays, which makes the search for new diagnostic methods very important. This issue is even more relevant in HIV infected patients. We evaluated the role of a new commercial ELISPOT for the diagnosis of TB in routine clinical practice.
Methods: T-SPOT.TB was performed in consecutive patients admitted to the Department of Infectious Diseases at a large teaching Hospital in Lisbon, in 2006. Twenty healthcare workers (HCW), from the Departments of Infectious Diseases and Microbiology, agree to participate as control group. PBMCs, obtained using a Ficoll gradient, were incubated overnight in a cell culture containing ESAT-6 and CFP-10. The assay detects individual effector T cells through their secretion of interferon-gama which results in a spot. A sample was considered positive if a well showed at least 6 spots more than the negative control well.
Results: Patients (n = 79) were male, and all were BCG vaccinated. Mean age was 40 years and 55 (70%) were HIV infected. Fourteen patients (22.8%) had TB, latter confirmed by culture, and most were HIV infected (n = 12, 85.7%). Of those, ELISPOT was positive in 11 patients (78.6%), negative in two (14.3%), and indeterminate in one (7.1%). From the remaining 55 patients admitted for other infectious cause, 13 (23.6%) also had a positive ELISPOT. All 20 HCW had a negative result.
Conclusion: ELISPOT was positive in almost 80% of all TB cases, even in a population with a very high rate of HIV infection and severe imunodepression. As T-SPOT.TB uses ESAT-6 and CFP-10, Mycobacterium tuberculosis complex specific antigens, these results were expected in TB cases. Over the last few years, several studies have showed that this test might have an important role on the diagnosis of latent infection (LTBI). As Portugal has the highest incidence of TB in Western Europe (∼40/100.000), one would expect to find a high prevalence of latent infection. ELISPOT was positive in ∼1/3 of all patients with other diagnosis, results that support the role of this test in the diagnosis of LTBI. Therefore, T-SPOT.TB is highly likely to become a useful and sensitive assay for the diagnosis of TB in routine clinical practice, as well as to identify individual with LTBI. The capacity of this test to identify patients that might have a worse response to treatment, as well as those individuals that can progress from infection to disease, is still to be proven.
P2006 The use of QuantiFERON-TB Gold In-Tube to screen for tuberculosis infection in employees of a Swiss university hospital
A. Stebler, K. Franz, T. Bodmer (Berne, CH)
Objectives: Hospital employees and healthcare workers in particular have a professional risk for contracting Mycobacterium tuberculosis (Mtb) infection. Traditionally, screening for latent Mtb infection (LTBI) is done with the tuberculin skin test (TST). However, previous BCG vaccination has been shown to interfere with TST results. In contrast, novel interferon-gamma release assays (IGRA), such as the QuantiFERON-TB Gold In-Tube (QFT-GIT; Cellestis Inc., Carnegie, Australia) assay, remain unaffected by BCG vaccination. The aim of this study was to assess the prevalence of LTBI among employees of a Swiss University Hospital.
Methods: Between June 1, 2005 and May 31, 2006 all employees who entered service at the University Hospital and employees who had professional exposure to infectious forms of tuberculosis (TB) were screened for LTBI by QFT-GIT as recommended by the manufacturer. In addition, information on country of origin, BCG vaccination status, profession and potential risk factors for the progression of LTBI to overt TB disease were gathered. GraphPad Prism 4.0 was used for all data evaluations.
Results: Overall, 1261 employees were enrolled. BCG vaccination status and country of origin were available in 608; of these, 494 (81.3%) were Swiss. Thus, the overall BCG vaccination rate among Swiss was 90.3%. From 1261 employees enrolled, 768 (60.9%) were newly recruited personnel: 620 were born in countries with a TB incidence rate <10/100,0000 (group A) and 87 in countries with a TB incidence rate ≥10/100,000 (group B). In 61 cases this information was missing. Overall, 7.7% of newly recruited personnel had a positive QFT-GIT test result; the respective positivity rates for group A and group B were 5.9% (37/620) and 16.1% (14/87). This difference was significant (odds ratio = 3.01; 95%CI: 1.55–5.84; p = 0.0028).
Conclusions: We found a very high BCG vaccination rate among Swiss hospital employees. This finding limits the usefulness of the TST for the diagnosis of LTBI. The overall positivity rate of QFT-GIT in our study population was higher than expected. However, employees born in a country with a high TB incidence had a significantly higher positivity rate than employees born in a country with a low TB incidence rate. This indicates that QFT-GIT is a useful tool to screen for LTBI in this population with a very high background of BCG vaccination. The study of risk factors for a positive QFT-GIT result other than the country of origin is ongoing.
P2007 Comparison of T-SPOT.TB and QuantiFERON-TB-Gold In Tube testing with regular tuberculin skin testing in a cohort of haemodialysis patients
U. von Both, J. Trachsler, R. Zbinden, A. Zellweger, P. Wrighton-Smith, C. Ruef (Zurich, Lausanne, CH; Abingdon, UK)
Objectives: Tuberculin skin testing (TST) is the current standard for screening of persons exposed to tuberculosis. In recent years, a higher specificity and sensitivity of interferon-based blood testing methods was reported. Twenty-six hemodialysis patients had been exposed to a case of open pulmonary TB in our tertiary care hospital. The aim of this study was to evaluate the use of T-SPOT.TB and QuantiFERON-TB-Gold In Tube (QFT-G) testing in comparison with regular TST for detection of TB-infection among this cohort.
Methods: The cohort consisted of 26 adult patients (10 male, 16 female, median age 57 years, range 25 to 82 years). Eighteen patients were of Swiss origin, while eight were residents of southern European or developing countries. Five patients (19.2%) received immunosuppressive drugs. A total of 11 patients suffered from malignancies or other additional causes of immunosuppression. Reliable information on former BCG vaccination could only be collected for 16 individuals. All patients were screened by TST, T-SPOT.TB and QFT-G. One patient could not be tested in the T-SPOT.TB assay due to logistic reasons. A positive TST was defined as an induration of ≥5 mm in diameter.
Results: 19 patients (73%) were anergic to TST, while 7 individuals showed positive TST with three of them revealing skin induration of >10 mm in diameter. A positive QFT-G was obtained for 8 patients, while 5 individuals showed a positive T-SPOT.TB result. TST positivity was neither associated with positive results in QFT-G nor T-SPOT.TB testing (P > 0.05), as 5 of 8 positive QFT-G results and 2 of 5 T-SPOT.TB results were found in patients with negative TST. There was no correlation between age >60, gender, TST positivity and either of the two new tests (P > 0.05). Quantitative analysis of the QFT-G showed a stronger reaction in patients with a positive TST. After analysis of all tests, 9 patients were considered to have subclinical TB infection and underwent radiological investigation, 6 receiving INH prophylaxis. No case of active TB was detected.
Conclusions: In the absence of a gold standard to diagnose subclinical tuberculosis infection, the interpretation of our results is difficult, especially in light of immunesuppression. The comparison of the three tests gave discordant results in some patients. Combining TST with either one of the two interferon-based tests may enhance the sensitivity in detection of subclinical TB infection in patients with chronic underlying diseases.
P2008 Evaluation of three media for the cryopreservation of peripheral blood mononuclear cells for ELISPOT
C. Maier, S. Bigler, T. Bodmer (Berne, CH)
Background: ELISPOT is used in microbiology for the analysis of the cell-mediated immune response. Typically, this technique requires sample processing within hours in order to preserve the vitality of cells. This may limit its application in clinical settings. Therefore, cryopreservation of vital lymphocytes prior to testing would facilitate a wider use in clinical laboratories. The aim of this study was to evaluate an optimised protocol for cryopreservation of vital lymphocytes.
Methods: Blood was obtained from three healthy donors, and peripheral blood mononuclear cells (PBMC) were isolated using Vacutainer Cell Preparation Tubes (Becton Dickinson, Franklin Lakes, US). PBMC were split into four aliquots, one for direct processing and three for cryopreservation (liquid nitrogen, −160°C, 4 weeks) in medium A (FCS, 90%; DMSO, 10%), medium B (AIM-V, 50%; FCS, 35%; DMSO, 15%), and medium C (AIM-V, 90%; DMSO, 10%), respectively. Cell counts of the aliquots were assessed prior to and after four weeks of cryopreservation using a Cell Dyn 1200 System (Abbott, Laboratories, US); cell viability and background were tested using mitogen controls and negative controls of T-Spot.TB (Oxford Immunotec Ltd., Oxford, UK). The results were compared and the ideal medium formulation was identified. For the purpose of further validation, PBMC of three individuals with previous positive T-Spot.TB results were processed as described above, and tested including the TB-specific antigens of T-Spot TB after eight weeks of cryopreservation. Spot-forming cells were counted by the AID Elispot Reader System (AID GmbH, Strassberg, Germany).
Results: With respect of survival rate, cell viability, and technical requirements medium A was superior: survival rate was 61.7±12.7% and mitogen response as a surrogate for cell viability remained above the level of validity of T-SPOT.TB (≥20spots/well). Background signal was not influenced by FCS, as there was no increase in the extent of background signal (p = 0.19). All specimens, that had previously tested positive with the T-SPOT.TB, remained positive after cryopreservation (Panel A, p = 0.91; Panel B, p = 0.30).
Conclusions: We have identified a reliable protocol for the cryopreservation of PBMC to be tested in ELISPOT-assays. Successful cryopreservation of PBMC's facilitates a wider application of this technique in clinical laboratories, and will allow cell banking for prospective studies.
P2009 Diagnostic value of neopterin in patients with active pulmonary tuberculosis and relation with CRP
D. Tarhan, S. Aksaray, A. Toyran, D. Köksal, E. Guvener (Ankara, TR)
Objectives: Macrophages and T cells are responsible for the main immune response to tuberculosis by secreting many cytokines and other substances. Neopterin is a marker of cell-mediated immunity, and it has been demonstrated that in tuberculosis neopterin levels could be determinated in various body fluids. The aim of this study was to investigate diagnostic values of serum neopterin evaluation in tuberculosis infection.
Methods: Sixty-one male patients (ages 17–66 years) with active pulmonary tuberculosis, confirmed by clinical, radiological and bacteriologic examination findings who hadn't antituberculosis medication were included to this study. Sixty-two healthy male subjects (ages 18–67 years) were selected as a control group. Neopterin was measured by competitive enzyme-linked immunosorbent assay (IBL, Hamburg) in seras. CRP values were measured by nephelometric method (Dade Behring, Germany). Results were expressed as mean ±SD and ranges.
Results: Serum neopterin concentrations were 18.28±23.12 (0–83) nmol/L in patients with tuberculosis infection, 4.53±8.30 (0–37) nmol/L in the control group. CRP levels were 62.95±51.7 (4–198) mg/L in patients with tuberculosis infection and 3.13±21.7 (3–156) mg/L in control group. There was significant difference between patient and control groups on CRP and neopterin values (p <0.001).
Conclusion: These findings indicate that measurements of serum neopterin levels are useful in diagnosis of active tuberculosis.
P2010 Evaluation of cellular immune response against purified antigen 85 in patients with tuberculosis
I. Nikokar, M. Makvandi, K. Huygen (Langaroud, Ahwaz, IR; Brussels, BE)
Objective: Tuberculosis (TB) remains an important health problem throughout the world. Despite its significance in public health, mechanisms of protective immunity against Mycobacyerium Tuberculosis in humans have not yet been understood. In the this study we evaluated cell mediated immune response against purified Ag 85, PPDand Phytohemagglutinin (PHA) in patients with tuberculosis and healthy tuberculin positive and negative individuals.
Methods: Thirty patients with tuberculosis and 60 healthy tuberculin skin test positive and negative volunteers were participated in this study. Cell mediated immunity was assessed by measuring [3H]-thymidine uptake and detection of IFN-gamma in the culture supernatant using commercial ELISA test.
Results: In the present study, we showed that IFN-gamma production and cell proliferation response to Ag 85 were significantly higher in tuberculin positive than tuberculin negative individuals (P < 0.01). Among tuberculous patients, IFN-gamma production and cell proliferative responses to Ag 85 was significantly lower in contrast to healthy tuberculin positive individuals (P < 0.01). In addition, IFN-gamma response in patients with cavitary tuberculosis was lower than patients without cavitation (P < 0.05).
Conclusion: Based on the higher cell mediated immune responses to Ag 85 in healthy tuberculin positive volunteers compared to patients (especially with advanced disease), purified Ag 85 can be used as a sensitive marker for analysis of cell immune responses in tuberculosis
P2011 Expression of phagocyte Fcg receptors during anti-tuberculous treatment
M.A. Escobar, A.A. Garcia-Egido, F.M. Gomez-Soto, F.J. Fernandez, J.L. Puerto, P. Ruiz, C. Rivera, F. Gomez (El Puerto Santa Maria, ES)
Receptors for IgG (FcgRs) on phagocytic cells are important in host defence against infection.
Objectives: We have studied the expression of FcgRs by peripheral blood monocytes (M), M cultured for 72 hours (M/Mø), and granulocytes (G) in patients with active Tuberculosis (TB), during anti-TB therapy (anti-TB-Rx) and, after completition of anti-TB-Rx.
Methods: The expression of the three type of FcgRs, FcgRI, FcgRII and FcgRIII, on M, M/Mø and G were analysed by flow cytometry in 103 HIV-negative patients with TB (77 men and 26 women), at diagnosis, and monthly thereafter until completition of anti-TB-Rx. FcgRs expression was assessed on resting M, M/Mø and G, and on these cells after stimulation by culture with IFNg.
Results: The expression of FcgRI and FcgRIII by M, M/Mø and G was enhanced in patients with active TB by: 42±4% and 22±2% for M, respectively (p < 0.001), 56±6% and 41±4% for M/Mø, respectively (p < 0.001) and, 119±9% and 37±3% for G, respectively (p < 0.001). The expression of FcgRIIA by M, M/Mø and G was decreased by −31±1% (p = 0.02), −46±3% (p <0.001), and −23±1% (p = 0.002), respectively. These alterations of FcgRs expression normalised from the 8th week until the end of effective anti-TB-Rx. The expression of FcgRI, FcgRIIA and FcgRIII by M, M/Mø or G from patients with active TB was significantly increased by culture in the presence of IFNg (p < 0.001), returning to normal after 8 wks of anti-TB-Rx. Setting a cut-off value = 25% of the mean fluorescence intensity over controls for FcgRs surface expression and, assuming a prevalence range of active TB between 25 and 80% among patients undergoing confirmatory tests, results in a range of sensitivity, specificity, positive and, negative predictive values of: 57–96%, 48–97%, 34–74%, and 59–98%, respectively for M-FcgRIIA, 48–73%, 51–96%, 38–74% and 68–97%, respectively for M-FcgRIII, 31–58%, 64–93%, 37–79% and 69–95%, respectively for G-FcgRI and, 52–71%, 88–98%, 49–77% and 81–93%, respectively for G-FcgRIIB.
Conclusions: Mø and G from patients with active TB exhibit an altered expression of FcgRs that disappear after effective anti-TB-Rx. Thus, Mø and G FcgRs expression may help in predicting the response to anti-TB therapy.
P2012 Expression of phagocyte Fcg receptors in HIV infected patients with tuberculosis
M.A. Escobar, A.A. Garcia-Egido, J.A. Bernal, F.J. Fernandez, M. Rosety, P. Ruiz, C. Rivera, F. Gomez (El Puerto Santa Maria, ES)
Phagocyte receptors for IgG (FcgRs) are important in host defence against infection.
Objectives: We have studied the expression of FcgRs by peripheral blood monocytes (M), monocytes cultured for 72 hours (M/Mø), and granulocytes (G) in HIV infected patients with active Tuberculosis (TB), during anti-tuberculous therapy (anti-TB-Rx) and, after completition of anti-TB-Rx.
Methods: The expression of FcgRI, FcgRII and FcgRIII, on M, M/Mø and G (resting and cultured with IFNg) were analysed by flow cytometry in 68 HIV infected patients with active TB (45 men and 6 women), at diagnosis of TB, and monthly thereafter until completition of anti-TB-Rx.
Results: The expression of FcgRI and FcgRIII by M, M/Mø and G was enhanced in HIV infected patients with active TB by: 34±4% and 19±3% for M, respectively (p < 0.001), 43±6% and 29±3% for M/Mø, respectively (p < 0.001) and, 54±6% and 21±3% for G, respectively (p < 0.001). The expression of FcgRIIB by M, M/Mø and G was decreased by −24±3% (p = 0.02), −33±4% and −19±2% (p < 0.005), respectively. These alterations of FcgRs expression normalised from week 12th until the end of effective anti-TB-Rx. The expression FcgRI, FcgRIIA and FcgRIII by M, M/Mø and G from HIV patients with TB was significantly enhanced by culture with IFNg (p < 0.005). Setting a cut-off value = 25% of the mean fluorescence intensity over controls for FcgRs surface expression and, assuming a prevalence range of active TB between 25 and 80% among HIV patients undergoing confirmatory tests, results in a range of sensitivity, specificity, positive and, negative predictive values of: 61–86%, 58–96%, 64–89%, and 57–90%, respectively for M-FcgRIIA, 56–84%, 63–91%, 58–79% and 68–97%, respectively for M-FcgRIII, 61–88%, 54–87%, 44–87% and 69–91%, respectively for G-FcgRI and, 42–75%, 88–98%, 49–77% and 86–98%, respectively for G-FcgRIIB.
Conclusions: Our results suggest that, the alterations of Mø and G Fcg receptors expression in HIV infected patients with active TB can be used to predict the response to anti-tuberculous therapy.
Quinolones: clinical trials
P2013 A decade of levofloxacin: summary of the cumulative clinical trial safety database
J. Peterson, A. Fisher, M. Khashab, J. Kahn (Raritan, US)
Background: The comprehensive safety profile of a drug is not established until enough subjects are treated to identify rare adverse events. Levofloxacin was approved by the US Food and Drug Administration on December 20, 1996. Worldwide patient exposure to levofloxacin is estimated to be >300 million prescriptions. Of 9,083 patients who were evaluable for safety in comparator-controlled trials, 5200 were treated with levofloxacin. The safety experience with levofloxacin includes four dose regimens in oral and IV form and numerous disease states.
Methods: Adverse events (AEs) that were identified during clinical studies were coded using a WHOART dictionary. The incidence of TEAE from comparator-controlled studies is summarised by body system and/or preferred term for the overall populations, certain special populations (e.g., age > or < 65 years) or body systems or events of special interest (musculoskeletal, cardiac, CNS etc).
Results: In all active-controlled studies, treatment emergent adverse events occurred in 42.2% of levofloxacin-treated subjects and in 43.5% of comparator-treated subjects. At least one drug-related AE occurred in 6.6% of levofloxacin-treated patients and in 7.6% of patients who received control drugs. usculoskeletal system: Tendinitis or tendon disorder was reported for 5 (0.1%) and 2 (<0.1%) of levofloxacin-treated subjects and 0 comparator-treated subjects. Central and Peripheral Nervous System: Convulsions were reported in 0.1% of levofloxacin treated subjects and in 0.1% of comparator-treated subjects. Cardiac Arrhythmia: Arrhythmia occurred in 0.1% of levofloxacin-treated subjects and in 0.1% of comparator-treated subjects. Elderly Subjects: In subjects >65 years, the incidence of TEAE's was 45.4% with levofloxacin and 45.7% in comparator-treated subjects. As compared with 41.4% of levofloxacin-treated and in 42.7% of comparator-treated subjects <65 years of age.
Conclusion: After a decade of exposure in patients with a variety of infections, levofloxacin's safety profile is well established. In comparator-controlled studies, the incidence of adverse events, including ones that have been associated with fluoroquinolones, is similar to those seen with comparator agents, regardless of the levofloxacin dose or dosing regimen used.
P2014 Pooled analysis of retrospective and prospective data on levofloxacin vs. other antibiotics in the treatment of hospitalised patients with pneumonia
H. Lode (Berlin, DE)
Objectives: The main objective of this study was to compare the efficacy of levofloxacin with other antibiotic options in hospitalised patients suffering pneumonia.
Methods: This pooled analysis includes a retrospective data set based on records of hospitalised patients with hospital-acquired pneumonia (HAP) or community-acquired pneumonia (CAP) and a prospective data set of a non-interventional observational study on levofloxacin in the treatment of hospitalised CAP patients.
Results: 609 patients were included (261 CAP,112 HAP, 236 NOS [pneumonia not otherwise specified]). Levofloxacin was part of the initial antibiotic therapy in 27.8% of the patients (LEV-group) being compared to patients with an initial antibiotic therapy without levofloxacin (OTHER-group), who most frequently received a cephalosporin or an extended-spectrum penicillin. The results indicate a better clinical success in the LEV-group: In the total group (84%/66%) and in the subgroup CAP (93%/79%) and HAP (81%/58%) higher rates of patients with cure/improvement were found as well as lower mortality rates for LEV-patients. Analyses stratified on type of pneumonia and risk factors before treatment (PSI risk-classes) showed that the higher success rates of the LEV-patients were not only the result of an unbalanced risk distribution between treatment groups (adjusted treatment effect p = 0.0121). To eliminate the effect of treatment changes, the clinical success rates were evaluated additionally for patients without change of therapy: Even in this subgroup LEV-patients had better cure/improvements rates (adjusted treatment effect p = 0.0018) and lower mortality rates than OTHER-patients. A shorter hospitalisation in LEV- compared to OTHER-patients (means: CAP 14.7/17.8 d;HAP 22.7/31.6d; total 17.9/24.8 d) may be a hint for a pharmacooeconomic impact of an efficacious initial antibiotic treatment including a fluoroquinolone-like levofloxacin. Tolerability data were documented only in the prospective data set, showing a low incidence of adverse drug reactions in only 1 of 135 LEV-patients (0.74%).
Conclusions: The results of this pooled analysis confirm that levofloxacin compared to other therapeutic options is a successfull initial antibiotic treatment of hospitalised patients with pneumonia.
P2015 Evaluation of clinical dosage of gatifloxacin for respiratory tract infections in elderly patients
H. Yoneyama, Y. Niki, T. Matsushima (Kasaoka, Kurashiki, JP)
Objectives: We studied the efficacy and safety of gatifloxacin in elderly patients with respiratory tract infections, in addition of the pharmacokinetic analysis.
Methods: Thirty four patients, 65–93 years-old, with mild to moderate acute bronchitis (10 cases), pneumonia (16 cases) and acute exacerbation of chronic respiratory disease (8 cases) were administered 100 mg once or twice daily for 3 to 14 days. Gatifloxacin was dosed as below, on based of the body weight and age of patients.
Serum level of gatifloxacin (1 to 3 samples in each patient) was determined and the AUC and Cmax was calculated by using the Bayesian method with population pharmacokinetic parameters.
Results: Total efficacy rate was 84.8% (28/33) at day3 and 87.9% (29/33) at the end of treatment. For each dose, it was 75% (6/8) at 100 mg once daily and 92.0% (23/25) at 100 mg b.i.d. For each infection, it was 90.0% (9/10), 86.7% (13/15) and 87.5% (7/8) for acute bronchitis, pneumonia and acute exacerbation of chronic respiratory diseases, respectively. Fifteen pathogens such as S. pneumoniae, B. catarrhalis and H. influenzae were isolated from 13 patients. All strains except for MRSA and E. faecalis were eradicated after administration of gatifloxacin. Side effect and clinical abnormality was not observed.
The pharmacokinetic parameters were as follows: The AUCs of patients dosed 100 mg once daily and b.i.d. ranged 11.2 to 37.5 and 12.7 to 111 mg*h/L, respectively. AUC/MICs for 2 S. pneumoniae isolates were 39 and 67, and for all of H. influenzae and M. catarrhalis were >250. Cmax level of gatifloxacin ranged 0.72 to 3.36 mg/L, except for one patient with Cmax of 6.35 mg/L.
| Body weight (kg) | Age (yrs) |
||
|---|---|---|---|
| 65–74 | 75–84 | ≥85 | |
| ≤40 | 100 mg×1 (1) | 100 mg×1 (2) | 100 mg×1 (1) |
| 41–60 | 100 mg×2 (6) | 100 mg×2 (13) | 100 mg×1 (4) |
| ≥61 | 100 mg×2 (4) | 100 mg×2 (3) | 100 mg×2 (0) |
(): No. of cases.
Conclusion: It was suggetsted that gatifloxacin of 100 mg once or twice daily could achieve an adequate AUC and high clinical efficacy and tolerance in elderly patients with respiratory tract infections.
P2016 Comparative study of garenoxacin vs levofloxacin on bacterial pneumonia
H. Kobayashi, K. Fujimaki (Tokyo, JP)
Objectives: Garenoxacin, a des-F(6)-quinolone, has strong activity against respiratory pathogens including multi-drugs resistant S. pneumoniae (MDRSP). The objective of the study was to evaluate the clinical response of 400 mg QD (400 mg× 1) of garenoxacin (GRN) for 10 days comparing to 300 mg TD (100 mg ×3) levofloxacin (LVFX) for 10 days.
Methods: The double-blind, controlled study was conducted, and a total 253 subjects were randomised 1:1 to either study drug. Clinical efficacy was assessed upon completion of the treatment and 7 days post-treatment. Safety was assessed up to 7 days post-treatment.
Results: The efficacy rate upon completion of treatment was 99.1% (111/112) for GRN and 94.3% (82/87) for LVFX (95% CI, −0.3 to 10), indicating non-inferiority of garenoxacin. The efficacy rate at 7 days post-treatment was 94.9% (94/99) for GRN and 92.8% (77/83) for LVFX. The bacterial eradication rate was 100% (53/53) for GRN and 87.8% (36/41) for LVFX. Garenoxacin was particularly effective against S. pneumoinae including MDRSP where the eradication rate was 100% (22/22) for GRN versus 80.0% (16/20) for LVFX. After treatment with LVFX, 4 strains of S. pneumoniae (2 PRSP, 1 PISP and 1 PSSP) recurred. The incidence of drug-related adverse events (AEs), including laboratory abnormalities, was 35.6% (48/135) for GRN and 27.1% (32/118) for LVFX. The main AEs were diarrhoea (3.0% vs. 3.4%), headache (1.5% vs. 2.5%), ALT increase (15.9% vs. 14.8%) and AST increase (13.6% vs. 11.3%). The incidence of AEs was similar in the both study arms.
Conclusions: A 10-day course of oral GRN 400 mg QD showed a high efficacy rate and high eradication rate in the treatment of bacterial pneumonia.
P2017 Study of oral Garenoxacin in Japanese patients with secondary infection of chronic respiratory diseases and penetration into sputum
H. Kobayashi, A. Watanabe, K. Fujimaki (Tokyo, Sendai, JP)
Objectives: Garenoxacin (GRN), a des-F(6)-quinolone, has strong activity against respiratory tract infection pathogens including multi-drug resistant S. pneumoniae (MDRSP). The objective of this study was to evaluate the clinical response of GRN against secondary infection of chronic respiratory disease namely acute exacerbation of chronic bronchitis (AECB). Penetration into sputum was also examined.
Methods: An open-label study was conducted to evaluate clinical response and microbiological response to treatment with oral GRN 400 mg QD for 10 days in adults with secondary infection of chronic respiratory disease in Japan. The study followed another GRN AECB study conducted in the US and Europe. The clinical efficacy of GRN was assessed upon completion of treatment and 7 days post-treatment. Safety was assessed up to 7 days post-treatment. The drug concentration of GRN in sputum after administration of GRN 400 mg was also measured.
Results: 136 subjects were enrolled in this study. The efficacy rate of GRN 7 days post-treatment in subjects with secondary infection of chronic bronchitis was 91.3%. A similar efficacy rate (94.3%) was observed in the AECB study conducted in the US and Europe. The bacterial eradication rate of GRN against Gram-positive pathogens was 90.9%. Bacterial eradication rates against PRSP, PISP and BLNAR were 100%. Drug related adverse events (AEs) were observed in 14.0% of the subjects; no serious AEs were found among them. The main AEs were gastrointestinal disorders, and all events were mild to moderate. The main laboratory abnormalities were ALT increase and AST increase. Similar AEs were observed in the US/European study. The drug concentration of GRN in sputum 3 hours after administration of GRN 400 mg was 3.50 ± 1.17 micro g/g (fluid to plasma ratio: 0.536 ± 0.273). This was regarded as efficient penetration well over the susceptibility level of major pathogens causing respiratory tract infection.
Conclusions: A 10-day course of oral GRN 400 mg QD showed good efficacy in the treatment of secondary infection of chronic respiratory disease. Good penetration into sputum was also confirmed.
P2018 Study of oral garenoxacin against respiratory tract infection and otrhinolaryngological infection caused by multidrug-resistant S. pneumoniae
S. Kohno, H. Kobayashi, S. Baba, K. Fujimaki (Nagasaki, Tokyo, Nagoya, JP)
Objectives: Respiratory tract infections caused by multi-drug resistant S. pneumoniae (MDRSP) are increasing in frequency worldwide. The objective of this analysis was to evaluate the clinical response of garenoxacin (GRN) treatment against S. pneumoniae infection (including MDRSP) to examine GRN susceptibility against S. pneumoniae isolates, and to analyse the correlation between resistance and gene mutation in isolates of S. pneumoniae.
Methods: Eight studies conducted in Japan assessed the clinical and biological efficacy of GRN in subjects with respiratory tract and otorhinolaryngological infection caused by S. pneumoniae. Treatment consisted of oral GRN 400 mg QD for 7 to 10 days. The minimum inhibitory concentration (MIC) of GRN against 106 isolates of S. pneumoniae (27 PRSP, 28 PISP and 51 PSSP, including 3 quinolone-resistant, 47 cephalosporin-resistant, 84 macrolide-resistant, 85 tetracycline-resistant, 10 trimethoprim-resistant and 83 multi-drug resistant) was measured using the CLSI method. GRN was compared with levofloxacin (LVFX), gatifloxacin (GFLX), moxifloxacin (MFLX), cefroxime (CXM), erythromycin (EM), azithromycin (AZM), telithromycin (TEL), tetracycline (TC) and trimethoprim/sulfametozalole (TMP/SMX). In addition, the mutation of the penicillin-binding protein genes and macrolide-resistant genes of S. pneumoniae isolates was investigated using electrophoresis analysis.
Results: The clinical efficacy rate of GRN against S. pneumoniae in 106 subjects was 96.2%; the bacterial eradication rate was 100%. MIC90 values of the 106 isolates were GRN 0.125, LVFX 2, GFLX 0.5, MFLX 0.25, CXM 8, EM >128, AZM >128, TEL 0.25, TC 64 and TMP/SMX 2 μg/mL. In the PRSP/PISP, pbp1a + pbp2x + pbp2b mutations for PBP and ermB presence for the macrolide-resistant gene were observed most frequently. There appeared to be a correlation between drug-resistance and gene mutation.
Conclusions: GRN showed high efficacy and eradication rates in the treatment of respiratory tract infection caused by S. pneumoniae including MDRSP.
P2019 Study of oral garenoxacin in Japanese patients with otorhinolaryngological infections and penetration into otorhinolaryngological tissues
S. Baba, K. Fujimaki (Nagoya, Tokyo, JP)
Objectives: Garenoxacin (GRN), a des-F(6)-quinolone, has strong activity against otorhinolaryngological infection pathogens. The objective of this study was to evaluate the clinical response of garenoxacin (GRN) in the treatment of otorhinolaryngological infections. Additionally, penetration into otorhinolaryngological tissues was examined.
Methods: Clinical response was evaluated in 2 studies: GRN 400 mg QD 10-day treatment against sinusitis, laryngo-laryngitis and tosillitis, and GRN 400 mg QD 7-day treatment against otitis media. Clinical efficacy was assessed upon completion of the treatment and safety was assessed up to 7 days post-treatment after the last dose. Additionally, after the administration of GRN 400 mg, drug concentration in sinus mucosa, middle ear mucosa, tonsil tissue and plasma was measured in subjects who required surgery.
Results: In the study involving sinusitis, laryngo-laryngitis and tosillitis, 71 subjects were enrolled. Efficacy rates were 92.0% in sinusitis, 85.0% in laryngo-laryngitis and 95.2% in tosillitis. In the study for otitis media, 50 subjects including chronic otitis media were enrolled. The efficacy rate was 87.2%. Bacterial eradication rates were 100% in the sinusitis, laryngo-laryngitis and tosillitis study, and 93.3% in the otitis media study. All PRSP, PISP and BLNAR pathogens were eradicated in both studies. Drug-related adverse events (AEs) were observed 19.7% of the time in the sinusitis, laryngo-laryngitis and tosillitis study, and 14.0% in the otitis media study. The main AEs were gastrointestinal disorders, and all events were mild to moderate. The drug concentration (tissue to plasma ratio) after administration of GRN 400 mg was 6.01 micro g/mL (1.03) in sinus mucosa, 5.89 micro g/mL (1.04) in middle ear mucosa and 9.44 micro g/mL (1.61) in tonsil tissue. This result was regarded as efficient penetration well over the susceptibility level against major pathogens causing otorhinolaryngological infection.
Conclusions: A 7 to 10-day course of oral GRN 400 mg QD showed good efficacy in the treatment of otorhinolaryngological infections. Good penetration into otorhinolaryngological tissues was confirmed.
P2020 Assessment of electrocardiograms and garenoxacin plasma concentrations from subjects with acute bacterial infections
G. Krishna, H. Waskin (New Jersey, US)
Background: Treatment with certain fluoroquinolones may increase the risk of QT interval (QT) prolongation. Garenoxacin is a novel des-F(6)-quinolone with broad antimicrobial activity against clinically important pathogens including multidrug-resistant S. pneumoniae. The relationship between maximal plasma garenoxacin concentration (Cmax) and QT measurements has been studied in Phase I studies in healthy (predominantly young male) subjects but not in subjects treated with IV garenoxacin for an acute bacterial infection. This analysis assessed for possible correlation between peak plasma garenoxacin values and QT abnormalities in subject populations that included women, elderly, and those with underlying electrolyte disturbances.
Methods: Following informed consent, electrocardiographs (ECG) and plasma samples were collected from subjects enrolled in 5 randomised, comparative, multicentre, double-blind, Phase 3 studies of IV garenoxacin (400 mg or 600 mg once daily). Serial ECG data and blood samples were collected before the first dose of study medication on day 1 (baseline), within 3 h after the end of the first infusion, and once on study day 3, 4, or 5. For this analysis, only data from the garenoxacin cohorts (unblinded after data lock) are presented. Blood samples (5 mL) were collected immediately following ECG measurements. For this retrospective analysis, the potential association between changes in ECG parameters from baseline to post-dose, measured as maximal QTcF interval at anticipated Cmax, and plasma garenoxacin concentration was examined using linear regression analysis.
Results: Collectively, 800 subjects (52%/48% M/F, 24% ≥65 yr old) received garenoxacin treatment. No association was apparent between maximum QTcF and garenoxacin plasma concentrations ranging from 0 to >20 μg/mL. These results were consistent with data from a previous analysis of QTc versus garenoxacin concentration in Phase I studies. There were no trends in interval abnormalities or change from baseline of ≥60 ms in the various garenoxacin concentration strata, including the highest concentration stratum (>20 μg/mL). Similarly, no statistically significant temporal change in QTc from baseline was observed after the first dose of garenoxacin or after 3 to 5 days of treatment.
Conclusion: Garenoxacin does not demonstrate clinically significant concentration- or time-dependent effects on QT intervals in subjects receiving IV treatment for acute bacterial infections.
P2021 Clinical and bacteriological efficacy of sequential intravenous to oral moxifloxacin in hospitalised patients with community-acquired pneumonia and pneumococcal bacteraemia
S. Choudhri, P. Arvis, D. Haverstock (West Haven, US; Puteaux, FR)
Objectives: Streptococcus pneumoniae is the leading bacterial pathogen in patients with community-acquired pneumonia (CAP) and the presence of pneumococcal bacteraemia in this setting is associated with significant morbidity and mortality. Pooled data from 6 CAP trials were used to compare the clinical and microbiological efficacy of sequential intravenous (IV)/oral (PO) moxifloxacin (MXF) with comparator (COMP) therapy in patients with pneumococcal bacteraemia.
Methods: Data were pooled from all 6 of the sequential trials carried out on 400 mg (IV/PO) MXF in the treatment of CAP in hospitalised patients. COMP treatment consisted of ceftriaxone + erythromycin, amoxicillin/clavulanate ± clarithromycin, trovafloxacin, levofloxacin, ceftriaxone ± azithromycin ± metronidazole or ceftriaxone + levofloxacin. All patients had blood cultures performed prior to initiation of study drug therapy. Severe CAP was defined using the modified ATS criteria.
Clinical and bacteriological success rates of MXF and COMP therapy in patients with pneumococcal bacteraemia
| MXF n/N (%) | COMP n/N (%) | |
|---|---|---|
| Overall clinical success | 40/48 (83.3%) | 49/64 (76.6%) |
| Clinical success in patients with severe CAP | 13/19 (68.4%) | 18/31 (58.1%) |
| Overall bacteriological success | 42/48 (87.5%) | 50/64 (78.1%) |
Results: The 6 trials randomised 3015 patients (1494 MXF, 1521 COMP) of which 2288 were valid per protocol (PP) (1141 MXF, 1147 COMP). Of the valid PP patients, 342 MXF and 361 COMP-treated subjects were also microbiologically valid. Of these 178 (52.0%) MXF and 192 (53.2%) COMP-treated subjects had CAP due to S. pneumoniae with 48 (14.0%) MXF and 64 (17.7%) COMP patients having pneumococcal bacteraemia. Clinical and bacteriological success rates of MXF and COMP therapy in patients with pneumococcal bacteraemia.
Conclusions: Sequential therapy with IV/PO MXF resulted in higher clinical and bacteriological success rates than comparator therapy in patients with CAP associated with pneumocococcal bacteraemia, including patients with severe CAP.
Research funding: Bayer HealthCare Pharmaceuticals.
P2022 Clinical and bacteriological efficacy of sequential intravenous to oral moxifloxacin in hospitalised patients with community-acquired pneumonia due to Enterobacteriaceae
S. Choudhri, P. Arvis, D. Haverstock (Wuppertal, DE; Puteaux, FR; West Haven, US)
Objectives: Although S. pneumoniae is the most common bacterial cause of community-acquired pneumonia (CAP), Enterobacteriaceae are an important cause of CAP in patients with underlying comorbid illnesses such as chronic obstructive pulmonary disease. We used pooled data from 6 CAP trials to compare the clinical and microbiological efficacy of sequential intravenous (IV)/oral (PO) moxifloxacin (MXF) with comparator (COMP) therapy in patients with CAP due to Enterobacteriaceae.
Methods: Data were pooled from 6 sequential (IV/PO) trials of 400 mg IV/PO MXF in the treatment of mild, moderate and severe CAP in hospitalised patients. COMP treatment consisted of ceftriaxone + erythromycin, amoxicillin/clavulanate ± clarithromycin, trovafloxacin, levofloxacin, ceftriaxone ± azithromycin ± metronidazole or ceftriaxone + levofloxacin.
Results: The 6 trials randomised 3015 patients (1494 MXF, 1521 COMP) of which 2288 were valid per protocol (PP) (1141 MXF, 1147 COMP). Of the valid PP patients, 342 (30.0%) MXF and 361 (31.5%) COMP-treated subjects were also microbiologically valid. Of these 27 (7.9%) MXF and 15 (4.2%) COMP-treated subjects had Enterobacteriaceae isolated from sputum or blood cultures. The following individual organisms were isolated (MXF, COMP): E. coli (3, 6), Klebsiella spp. (19, 8), Proteus mirabilis (1, 0), Proteus vulgaris (1, 0), Serratia marcescens (0, 1) and Enterobacter cloacae (3, 0). The overall clinical and bacteriological success rates in these patients were 22/27 (81.5%) and 21/27 (77.8%) for MXF and 10/15 (67.0%) and 10/15 (67.0%) for COMP-treated patients.
Conclusions: Sequential therapy with IV/PO moxifloxacin was highly effective in the treatment of CAP due to Enterobacteriaceae.
Research funding: Bayer Health Care Pharmaceuticals.
P2023 Clinical and bacteriological efficacy of sequential intravenous to oral moxifloxacin in hospitalised patients with severe complicated skin and skin structure infections due to Enterobacteriaceae
J.R. Bogner, S.H. Choudhri, P. Arvis, P. Pertel, J-M. Breilmann (Munich, DE; West Haven, US; Puteaux, FR; Wuppertal, DE)
Objectives: Enterobacteriaceae species are an important cause of cSSSI in patients with underlying comorbid illnesses such as diabetes mellitus, and require empiric therapy with broad spectrum antimicrobials. Pooled data from 2 cSSSI trials were used to compare the clinical and microbiological efficacy of sequential intravenous (IV)/oral (PO) moxifloxacin (MXF) with comparator (COMP) therapy in patients with cSSSI due to Enterobacteriaceae.
Methods: Data were pooled from 2 trials of sequential MXF (400 mg q.d. IV/PO) in the treatment of cSSSI in hospitalised patients. COMP treatment consisted of IV piperacillin/tazobactam followed by PO amoxicillin/clavulanate and sequential IV/PO amoxicillin/clavulanate.
Results: The 2 trials enrolled 1421 patients (712 MXF, 709 COMP) of which 999 were valid per protocol (495 MXF, 504 COMP). Of these patients, 286 (57.8%) MXF and 291 (57.7%) COMP-treated subjects were also microbiologically valid. Of these, 87 (30.4%) MXF and 77 (26.5%) COMP-treated subjects had Enterobacteriaceae isolated from their skin infection sites. The following organisms were isolated (MXF, COMP): E. coli (38, 33), Klebsiella spp. (14, 10), Proteus spp. (14, 16), Morganella spp. (1, 3), Providencia spp. (2, 2), Serratia marcescens (4, 2) and Enterobacter (14, 11). The overall clinical and bacteriological success rates at test-of-cure among per protocol valid patients who had a positive skin culture for Enterobacteriaceae were 65/87 (74.7%) and 64/87 (73.6%) for MXF and 56/77 (72.7%) and 56/77 (72.7%) for COMP-treated patients.
Conclusions: Enterobacteriaceae were cultured frequently from hospitalised patients with cSSSI. Monotherapy therapy with IV/PO MXF had similar clinical and bacteriological efficacy as the standard treatment regimens in the management of cSSSI due to Enterobacteriaceae.
Research funding: Bayer HealthCare Pharmaceuticals.
In vitro susceptibility: cephalosporins
P2024 Profile of ceftobiprole activity against S. aureus, S. pneumoniae, Enterobacteriaceae, and P. aeruginosa: results of the 2005–2006 Surveillance Program
M.K. Aranza, D.C. Draghi, M.E. Jones, C. Thornsberry, D.F. Sahm (Herndon, US; Breda, NL)
Background: BPR is an investigational broad spectrum cephalosporin with activity against both Gram-positive organisms (including MRSA) and Gram-negative (including PA). BPR is under clinical development for hospital-acquired pneumonia and complicated skin and skin structure infections. Surveillance of BPR activity against current Gram-positive organisms, especially those resistant to β-lactams, and Gram-negative organisms is an important component of the development programme and was done to establish a database for longitudinal tracking of BPR activity throughout the development programme and beyond.
Methods: During 2005 – 2006, S. aureus (SA; including both methicillin-susceptible SA [MSSA] and methicillin-resistant SA [MRSA]), S. pneumoniae (penicillin-susceptible [PEN-S] and penicillin non-susceptible [PEN-NS]), Citrobacter spp. (CP), E. cloacae (EA), E. coli (EC), K. pneumoniae (KP), P. mirabilis (PM), S. marcescens (SM), and P. aeruginosa (PA) were collected from 31 institutions in 12 European countries and were centrally tested by broth microdilution (CLSI; M7-A6).
Results: See the table .
| Organism | Total n | BPR (mg/L) |
|||
|---|---|---|---|---|---|
| MIC Range | MIC Mode | MIC50 | MIC90 | ||
| SA all | 1203 | ≤0.12–>16 | 1 | 0.5 | 2 |
| MSSA | 404 | ≤0.12–8 | 0.25 | 0.25 | 0.5 |
| MRSA | 799 | 0.25–>16 | 1 | 1 | 2 |
| SP all | 526 | ≤0.002–1 | 0.015 | 0.015 | 0.25 |
| SP PEN-S | 406 | ≤0.002–0.06 | 0.015 | 0.008 | 0.015 |
| SP PEN-NS | 120 | 0.008–1 | 0.5 | 0.25 | 0.5 |
| CP | 387 | ≤0.015–>32 | 0.06 | 0.06 | 2 |
| EA | 406 | 0.03–>32 | 0.06 | 0.12 | >32 |
| EC | 1213 | ≤0.015–>32 | 0.03 | 0.03 | 0.25 |
| KID | 854 | ≤0.015–>32 | 0.03 | 0.06 | >32 |
| PM | 443 | ≤0.015–>32 | 0.03 | 0.03 | 0.12 |
| SM | 291 | 0.03–>32 | 0.06 | 0.12 | 2 |
| PA | 621 | 0.03–>32 | 2 | 4 | 16 |
Conclusions: BPR exhibited potent activity against Gram-positive organisms, including MRSA, and had activity very similar to currently available cephalosporins against Gram-negative organisms. Because all of these target pathogens can develop resistance to variety of antimicrobial agents, this baseline surveillance data will be important for continued tracking of BPR activity throughout clinical development and beyond.
P2025 Activity of ceftobiprole against US clinical S. pneumoniae isolates that are ceftriaxone non-susceptible
T. Davies, K. Bush, P. Appelbaum (Raritan, Hershey, US)
Objectives: Ceftobiprole (BPR) is an investigational cephalosporin with broad-spectrum activity against many Gram-negative rods and Gram-positive cocci, including methicillin-resistant Staphylococcus aureus and drug-resistant Streptococcus pneumoniae. In this study the antipneumococcal activity of BPR was further evaluated by testing recent clinical isolates that were non-susceptible to ceftriaxone.
Methods: BPR, ceftriaxone (CRO), cefotaxime (CTX), cefuroxime (CXM), and penicillin (PEN) MICs from 95 CRO-non-susceptible (CLSI non-meningitis breakpoints) S. pneumoniae isolates (94 clinical, 1 lab mutant) were determined using CLSI methodology. PCR and DNA sequencing of the penicillin-binding protein genes pbp1a, pbp2b, and pbp2x were carried out on all isolates with BPR MICs of 2–4 mg/L and some with BPR MICs <2 mg/L.
Results: BPR had MIC90s 2- to 8-fold lower than the other agents against CRO-intermediate isolates and 8- to 32-fold lower against CRO-resistant isolates. The highest BPR MIC was 4 mg/L (in two strains) compared to 16 to >64 mg/L for the other agents. Isolates with BPR MICs of 2 mg/L had similar mutations in the penicillin binding motifs of pbp1a, pbp2b, and pbp2x as isolates with BPR MICs of 1 mg/L. One isolate with a BPR MIC of 4 mg/L had a T338 to G change in the 337STMK motif of PBP2x while isolates with BPR MICs of 1 to 2 mg/L had a T338 to A and M339 to F change. A CRO-resistant lab mutant (CRO MIC 4 mg/L) had no changes in pbp1a, pbp2b, and pbp2x compared to the parent strain (CRO MIC 0.5 mg/L).
| Drug | MIC (gm/L) |
|||||
|---|---|---|---|---|---|---|
| CRO-Intermediate (n = 62) |
CRO-Resistant (n = 33) |
|||||
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| BPR | 0.5 | 1 | 0.12–1 | 1 | 2 | 0.5–4 |
| CTX | 2 | 2 | 1–4 | 8 | 16 | 2–32 |
| CXM | 8 | 8 | 2–16 | 32 | 64 | 4–>64 |
| CRO | 2 | 2 | 2 | 4 | 16 | 4–16 |
| PEN | 4 | 4 | 2–16 | 8 | 16 | 4–32 |
Conclusion: BPR exhibited excellent activity against most CRO-non-susceptible isolates with an MIC90 of 1 mg/L that was 8- to 32-fold lower than CRO, CTX, CXM, and PEN. Strains with elevated BPR MICs (2 to 4 mg/L) contained multiple mutations in pbp1a, pbp2b, and pbp2x.
P2026 Activity of ceftobiprole against Pseudomonas aeruginosa clinical isolates from Europe collected in 2005–2006
T. Davies, D. Sahm, R. Flamm, K. Bush (Raritan, Herndon, US)
Objectives: Ceftobiprole, an investigational parenteral cephalosporin exhibiting a broad spectrum of activity against many Gram-positive (including methicillin-resistant Staphylococcus aureus) and Gram-negative organisms, is currently under clinical development for pneumonia and complicated skin and skin structure infections. P. aeruginosa is one of the most common Gram-negative pathogens causing these infections. The activity of ceftobiprole and comparators was tested against P. aeruginosa isolates from Europe.
Methods: A total of 621 P. aeruginosa isolates were collected from 31 cites in 13 European countries during 2005–2006. Ceftobiprole, ceftazidime, cefepime, imipenem, piperacillin-tazobactam, gentamicin, levofloxacin, and tigecycline MICs were determined by broth microdilution according to CLSI methods.
Results: The ceftobiprole MIC50 (4 mg/L) was similar to the other cephalosporins, imipenem, and gentamicin; and was four-fold lower than piperacillin-tazobactam and tigecycline. Levofloxacin had the lowest MIC50 (1 mg/L), while ceftobiprole and cefepime MIC90s (16 mg/L) were the lowest of all the drugs tested.
| Drug | MIC (mg/L) |
||
|---|---|---|---|
| MIC50 | MIC90 | Range | |
| Ceftobiprole | 4 | 16 | 0.03–>32 |
| Cefepime | 4 | 16 | ≤0.015–>32 |
| Ceftazidime | 2 | >32 | 0.06–>32 |
| Piperacillin–tazobactam | 16 | >128 | 0.5–>128 |
| Imipenem | 4 | 32 | 0.25–32 |
| Gentamicin | 4 | >16 | ≤0.12–>16 |
| Levofloxacin | 1 | >8 | 0.008–>8 |
| Tigecycline | 16 | >16 | ≤0.015–>16 |
Conclusion: Ceftobiprole, in addition to its activity against methicillin-resistant staphylococci (MRS), had anti-pseudomonal activity in vitro, similar to other advanced generation cephalosporins and imipenem. Compared to piperacillin-tazobactam, or tigecycline (another anti-MRS agent with activity against Gram-negative pathogens), ceftobiprole had greater in vitro potency against P. aeruginosa as determined by the lower MIC50 and MIC90 values.
P2027 Ceftobiprole activity and resistance patterns in staphylococcal isolates from a recent complicated skin and skin structure infection study
K. Amsler, M. Jacobs, S. Bajaksouzian, A. Windau, M. Heep, K. Bush (Raritan, Cleveland, US; Basle, CH)
Objectives: Ceftobiprole (BPR) is a broad-spectrum investigational cephalosporin with demonstrated activity against methicillin-susceptible and methicillin-resistant staphylococci, as well as against most Enterobacteriaceae. In this study, the geographic variation of BPR activity and antimicrobial agent resistance patterns are reported from the staphylococcal baseline pathogens from a recent Phase 3 clinical trial to treat complicated skin and skin structure infections.
Methods: The baseline pathogens from 784 patients enrolled in a multicentre clinical trial involving complicated skin infections were examined for their susceptibility to BPR and selected antimicrobial agents. MICs were determined using CLSI methodology.
Results: Of the 461 baseline Staphylococcus aureus isolates available for testing, 178 isolates were from the USA and 283 isolates were from 16 non-USA countries, including 10 European countries. All staphylococcal isolates had BPR MICs ≤ 4 mg/L. Resistance patterns varied between the USA and non-USA isolates. A higher percentage of staphylococcal USA isolates were resistant to oxacillin (OXA), ceftazidime (CAZ), penicillin (PEN), erythromycin (ERY), and levofloxacin (LVX). The most common resistance patterns observed in the S. aureus USA isolates are shown in the table , including at least three different drug classes.
| Resistance profile | Source of S. aureus isolates | % Isolates (n) |
|---|---|---|
| OXA, CAZ, PEN, ERY | USA | 36.5 (65/178) |
| Non-USA | 1.1 (3/283) | |
| OXA, CAZ, PEN, ERY, LVX | USA | 20.8 (37/178) |
| Non-USA | 2.1 (6/283) |
Conclusion: Against staphylococcal isolates with methicillin resistance and multiple resistance phenotypes, ceftobiprole MICs were ≤4 mg/L. Multidrug-resistant strains were found more frequently in USA isolates compared to other countries.
P2028 Activity of cefditoren versus seven other antimicrobials against amoxicillin non-susceptible Streptococcus pneumoniae
E. Perez-Trallero, J. Marimon, M. Erzibengoa, M. Gimenez, P. Coronel, L. Aguilar (San Sebastian, Madrid, ES)
Objective: Amoxicillin has the highest intrinsic activity against S. pneumoniae but amoxicillin non-susceptibility is not rarely found among penicillin-resistant isolates. Antibiotic susceptibility testing against these isolates can be considered the workhorse to test new and old compounds.
Methods: Susceptibility to penicillin (PEN), amoxicillin/clavulanic acid (2:1) (AMC), cefuroxime (CXM), cefpodoxime (CPD), cefotaxime (CTX), cefditoren (CDN), erythromycin (ERY), and levofloxacin (LVX) was determined by microdilution with Mueller-Hinton broth supplemented with 3% lysed horse blood according to CLSI guidelines, against 244 isolates exhibiting non-susceptibility to amoxicillin (MIC ≥4μg/mL) and higher amoxicillin vs. penicillin MIC. CLSI susceptibility breakpoints (μg/mL) were considered.
Results: MIC distribution of the antimicrobials tested: no. of isolates inhibited (cumulative percentage) at concentration (μg/mL) are shown in the Table .
| MICa (μg/mL) |
||||||||
|---|---|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥16 | %S | |
| PEN | 51 (20.9) | 162 (87.3) | 31 (100) | 0 | ||||
| AMC | 124 (50.8) | 117 (98.8) | 3 (100) | 0 | ||||
| CXM | 64 (26.2) | 138 (82.8) | 42 (100) | 0 | ||||
| CPD | 1 (0.4) | 29 (12.3) | 141 (70.1) | 40 (86.5) | 29 (98.4) | 4 (100) | 0 | |
| CTX | 36 (14.8) | 146 (74.6) | 52 (95.9) | 9 (99.6) | 1 (100) | 74.6 | ||
| CDN | 58 (23.8) | 126 (75.4) | 51 (96.3) | 7 (99.2) | 2(100) | − | ||
| ERY | 67 (27.5) | 1 (27.9) | 3 (29.1) | 173 (100) | 27.5 | |||
| LVX | 7 (2.9) | 99 (43.4) | 102 (85.2) | 5 (87.3) | 1 (87.7) | 19 (95.5) | 11 (100) | 87.3 |
MIC90 and MIC50 values are in bold.
Conclusions: On a MIC90 basis, CDN (1μg/mL) exhibited the highest intrinsic activity followed by CTX (2μg/mL), AMC, CPD and LVX (8μg/mL), with a clear inadequacy of CXM and ERY (≥16μg/mL). CDN activity (twice that of CTX) may be useful against amoxicillin non-susceptible strains where only CTX and LVX offer significant activity (70–90% susceptibility rates).
P2029 In vitro efficacy of cefditoren against recent isolates of Streptococcus pneumoniae in Greece
G Poulakou, I. Katsarolis, L. Lianou, N. Labri, D. Hatzaki, H. Giamarellou (Haidari, GR)
Purpose: To evaluate the in vitro efficacy of cefditoren, an oral 3rd generation cephalosporin in licensure phase in Greece, against recently isolated Streptococcus pneumoniae strains.
Methods: A total of 261 S pneumoniae strains isolated during 2004–2006 from clinical specimens of adults and children as well as from nasopharyngeal swabs of preschool healthy children were tested against penicillin (PEN) and erythromycin (ERY) by E-test, according to CLSI 2005 methodology and breakpoints for non-susceptibility (NS) and resistance (R). Susceptibility to cefditoren (CFD) was tested by agar dilution. Strains with MIC >2 mg/L were considered resistant, according to recent literature.
Results: Against the total tested population of pneumococci, CFD MIC50/90 were 0.03/1 mg/L (range 0.008–4 mg/L). Only 1/262 tested strains (0.4%) exhibited R to CFD (MIC = 4 mg/L). For PEN R strains (N = 47) CFD MIC50/90 were 0.5/1 mg/L (range: 0.008–4 mg/L). Among 124 PEN NS strains CFD MIC50/90 were 0.5/1 mg/L (range 0.008–4 mg/L), while respective values for 137 PEN S strains were 0.015/0.034 mg/L (range 0.008–0.5 mg/L) (p < 0.001, t-test). Among 142 ERY R isolates CFD MIC50/90 were 0.06/1 mg/L (range 0.008–4 mg/L), whereas among 119 ERY S respective values were 0.015/0.5 mg/L (range 0.008–2 mg/L) (p < 0.001, t-test). CFD MICs for paediatric isolates did not differ statistically compared to adult isolates, and the same was also observed for invasive and non-invasive isolates.
Conclusions: CFD exerted an excellent in vitro antibacterial activity against recently isolated strains of S. pneumoniae in Greece, retaining low MICs against all strains displaying resistance to PEN and ERY, although statistically significant differences in MIC distribution were observed between PEN NS and S, as well as ERY R and ERY S isolates. Cefditoren could prove a valuable alternative to existing drugs for community acquired pneumococcal infections treated in the outpatient setting.
In vitro susceptibility: tigecycline
P2030 A prospective study of tigecycline susceptibility of bacteria isolated from complicated intra-abdominal as well as skin and soft tissue infections in a large UK hospital
L. Oliver, E. Watson, J. Hovenden, I. Budai, C. Marodi (Middlesbrough, UK; Budapest, HU)
Objectives: Tigecycline (TIG) is a glycylcycline antibiotic with expanded activity licensed in the UK for the treatment of complicated intraabdominal (cIAI) and skin/soft tissue infections (cSSTI) in 2006. No data have been available about its activity against regional isolates. TIG had not been used in our hospital prior to this study. We aimed to collect information about the TIG sensitivity of isolates from cIAI/cSSTI in our hospital.
Methods: Isolates from cIAI (infections that spread beyond the hollow viscus of origin into the peritoneal space associated with abscess formation/peritonitis) and cSSTI (infections involving deeper soft tissue/requiring surgical intervention) from both in- and outpatients, processed in line with the UK laboratory and local guidelines, were collected over 1 month. In polymicrobic infections, each isolate was tested. No duplicate isolates were included. Pseudomonads and anaerobic bacteria were not included in this study.
TIG sensitivity was tested using the standard E-test method on Isosensitest or Columbia blood agar (BSAC). MICs were read after an incubation of approximately 20 hours at 35–37°C on air or at 5% CO2 atmosphere (BSAC).
Results: Altogether 97 isolates were tested (21 from cIAI's, 76 from cSSTI's). See Table for results. Two ESBL-producing E. coli were isolated with TIG MICs of 0.19 and 0.38 mg/L respectively. We found two AmpC positive Enterobacteriaceae, an E. cloaceae and an S. marcescens, with MICs of 3.0 and 4.0, respectively.
Table 1.
Tigecycline sensitivity of clinical isolates recovered from intra-abdominal and skin/soft tissue
| Isolates | n | Breakpoints (mg/L) |
MIC (mg/L) |
Sensitive |
||||
|---|---|---|---|---|---|---|---|---|
| S ≥ | R< | Range | MIC50 | MIC90 | No. | (%) | ||
| Streptococcus beta haemolytic | 10 | 0.25 | 0.5 | 0.047–0.190 | 0.064 | 0.094 | 10 | 100 |
| Enterococcus faecalis | 1 | 0.25 | 0.5 | 0.125 | NA | NA | 1 | NA |
| MSSA | 37 | 0.5 | 0.5 | 0.047–0.47 | 0.125 | 0.19 | 37 | 100 |
| MRSA | 11 | 0.5 | 0.5 | 0.125–0.250 | 0.19 | 0.25 | 11 | 100 |
| Total Staphylococcus aureus | 48 | 0.5 | 0.5 | 0.047–0.47 | 0.125 | 0.19 | 48 | 100 |
| Escherichia coli | 13 | 1 | 2 | 0.125–1.5 | 0.25 | 0.5 | 12 | 92.3 |
| Enterobacter spp. | 5 | 1 | 2 | 0.5–3 | 0.75 | 0.75 | 4 | 80 |
| Klebsiella spp. | 7 | 1 | 2 | 0.380–3.0 | 0.75 | 0.75 | 6 | 85.7 |
| Proteus mirabilis | 2 | 1 | 2 | 1.5–3.0 | NA | NA | 0 | NA |
| Serratia marcescens | 4 | 1 | 2 | 2.0–4.0 | NA | NA | 0 | NA |
| Citrobacter koseri | 3 | 1 | 2 | 0.380–0.50 | NA | NA | 3 | NA |
| Total Enterobacteriaceae | 34 | 1 | 2 | 0.125–4.0 | 0.5 | 3 | 25 | 73.5 |
| Sphingomonas paucimobiIis | 1 | 0.25 | 0.5 | 0.32 | NA | NA | 1 | NA |
| Acinetobacter baumannii | 3 | NA | NA | 0.19–0.5 | NA | NA | NA | NA |
| Total isolates | 97 | NA | NA | 0.032–4.0 | 0.19 | 2 | NA | NA |
S, sensitive; R, resistant; n, number; NA, not applicable.
Conclusions: Our Gram-positive isolates were universally sensitive to TIG with generally low MIC50 and MIC90, although MRSA strains had slightly higher MICs than the MSSAs. Among Gram-negative isolates sensitivity was not universal; with only 73.5% of Enterobacteriaceae being susceptible. Despite the short study period, it was shown that Gram-negatives are frequently isolated in both cIAIs and cSSTIs.
We can conclude that despite its excellent activity against regional Gram-positive isolates TIG cannot be used universally in cIAI and cSSTI because of the significant number of Gram negatives tested as resistant. The excellent activity against MRSA makes TIG a good candidate for outpatient intravenous antibiotic treatment for MRSA infected chronic ulcers and pressure sores. At present the use of TIG is restricted in our hospital and can only be prescribed on the advice of a Consultant Microbiologist or Infectious Disease Physician.
P2031 In vitro activity of tigecycline against multidrug-resistant Acinetobacter baumannii and enhanced susceptibility to amikacin selected by exposure to tigecycline
E. Smith Moland, L. Hachmeister, S. Sayed, D. Craft, S. Hong, S. Kim, K. Thomson (Omaha, Washington DC, US)
Objectives: A study was designed to investigate the in vitro activity of tigecycline (TGC) and comparison agents against 170 isolates of multidrug-resistant Acinetobacter baumannii (MRAB). Three isolates were investigated for drug interactions between TGC and either polymyxin B (PB) or amikacin (AN) by time-kill methodology.
Methods: Susceptibility was determined in fresh Mueller-Hinton broth by CLSI microdilution methodology using TREK custom panels containing doubling dilutions of TGC, minocycline (MIN), imipenem (IPM), ampicillin/sulbactam (SAM), PB, ceftazidime (CAZ), levofloxacin (LEV), AN, piperacillin/tazobactam (TZP), and gentamicin (GM). Some isolates produced IMP-1, VIM-1, and OXA-23 β-lactamases. The bactericidal activities of TGC, AN, and PB, both alone and in combinations of TGC with either AN or PB, were evaluated by time-kill methodology. Three isolates with varying levels of susceptibility to TGC were investigated using broth cultures inoculated with >5×105 CFU/mL of each isolate in log phase growth. The concentrations investigated were chosen to avoid significant killing by any drug on its own.
Results: Based on MIC90 values the most potent agent (MIC90 in mg/L) was PB (2), followed by MIN and TGC (8), IPM and LEV (16). In time-kill studies, both combinations exhibited enhanced activity against the isolate that was most susceptible to TGC (MIC 0.06 mg/L). TGC and PB were synergistic and bactericidal against the isolate that was less susceptible to TGC (4 mg/L) and highly resistant to AN (>128 mg/L). Against the most TGC-resistant isolate (64 mg/L), TGC and PB exhibited enhanced activity, while the combination of TGC and AN was synergistic and bactericidal. It was noteworthy that TGC exposure caused the development of increased susceptibility of this isolate to AN.
Conclusion: TGC alone is a potential treatment option for infections caused by some, but not all, MRAB. In combination with an appropriate agent, it may be effective against most isolates. The capability of TGC exposure to cause increased susceptibility to potential co-drugs should be further investigated.
P2032 Determination of the minimum inhibitory concentration and mutant prevention concentration of tigeycline against clinical isolates of methicillin-susceptible and methicillin-resistant Staphylococcus aureus
J. Blondeau, C. Hesje, S. Borsos (Saskatoon, CA)
Objective: Tigecycline, a member of a new class of antimicrobials (glycocyclines) has been shown to have potent in vitro activity against clinical isolates of Staphylococcus aureus (SA) by MIC measurements. We were interested in determining if the low MIC values would translate into low mutant prevention concentration (MPC) values.
Methods: Minimum inhibitory concentration was in accordance with the recommended Clinical and Laboratory Standards Institute procedure by microbroth dilution using 105 cfu/mL tested against doubling drug dilutions in appropriate media. For MPC testing 1010 CFUs were added to drug containing agar plates and incubated in ambient temperature and atmosphere. The lowest drug concentration preventing growth was recorded as the MIC or MPC depending on method.
Results: For 68 clinical MSSA strains, MIC50, MIC90 and MIC range values were 0.063 mg/L, 0.125 mg/L and 0.125 mg/L. By MPC testing, the MPC50, MPC90 and MPCrange was 1 mg/L, 2 mg/L and 1–4 mg/L (1 strain). A total of 64% of strains had MPCs of 1 mg/L and 99% of strains had MPCs of 1–2 mg/L. For 63 MRSA strains MIC50, MIC90, and MIC range values were 0.125, 0.5 and 0.063–1 mg/L; MPC50, MPC90 and MPC range values were 1, 4 and 0.5–8 mg/L with 76% of strains having MPCs ≤2 mg/L.
Conclusion: By MIC testing, tigeycline was highly active in vitro against MSSA and MRSA strains with MICs ≤0.125 mg/L and ≤1 mg/L respectively. Tigecycline MPC values were ≤2 mg/L for 97% and 76% of MSSA and MRSA strains respectively. Tigecycline appears to be a promising agent for treating Staphylococcus aureus infections and appears to have a reduced likelihood for selecting for resistance based on MPC measurements.
P2033 Antimicrobial activity of tigecycline and other broad-spectrum agents tested against bacterial isolates collected in European hospitals in 2006
H. Sader, T. Fritsche, M. Stilwell, R. Jones (North Liberty, US)
Objectives: To assess the activity of tigecycline against recent bacterial isolates from Europe. Tigecycline is a novel glycylcycline antimicrobial recently approved by the European Medicines Agency for the treatment of complicated skin and skin structure infections (cSSSI) and intraabdominal infections.
Methods: Bacterial isolates (non-duplicates) were consecutively collected in 2006 from documented infections in patients hospitalised in 24 medical centres located in Europe (9 countries), Turkey and Israel. The isolates were obtained mainly from patients with bacteraemia (BSI; 61.9%), pneumonia (16.1%) and cSSSI (11.1%). Frequency of occurrence of pathogens was determined and their antibiograms assessed using reference broth microdilution methods according to the CLSI M7-A7 (2006). Tigecycline-susceptible (S) breakpoints (US-FDA/EUCAST) were defined as ≤2/≤1 mg/L for Gram-negative bacilli; ≤0.5/≤0.5 mg/L for staphylococci, and ≤0.25/≤0.25 mg/L for streptococci and enterococci.
Results: A total of 5,032 strains were evaluated and the frequency of pathogen occurrence and susceptibility rates to tigecycline are summarised in the table . Tigecycline was highly active against the top 8 pathogens, except for P. aeruginosa (PSA). Among the 4 most common indicated pathogens (3,430 strains; 68.0% of the total), tigecycline was active against >99% at the US-FDA/EUCAST S breakpoints. The main resistance phenotypes detected were methicillin-resistant (R) S. aureus (31.4%) and CoNS (74.6%), ciprofloxacin-R E. coli (20.6%), extended-spectrum β-lactamase (ESBL)-screen-positive Klebsiella spp. (22.8%) and E. coli (6.3%), imipenem-R PSA (IRPSA; 24.2%) and vancomycin-R enterococci (5.0%). Tigecycline showed excellent activity against these R pathogens, except IRPSA.
| Organism (no. tested) | Cumulative % inhibited at tigecycline MIC (mg/L): |
%Susceptiblea | ||||||
|---|---|---|---|---|---|---|---|---|
| ≤0.12 | 0.25 | 0.5 | 1 | 2 | 4 | >4 | ||
| S. aureus (1,479) | 80.0 | 99.5 | 99.5 | − | − | − | − | 99.5/99.6 |
| E. coli (854) | 77.0 | 98.0 | 99.9 | 100.0 | − | − | − | 100.0/100.0 |
| Enterococcus spp. (577) | 83.4 | 100.0 | − | − | − | − | − | 100.0/100.0 |
| Coagulase-negative staphylococci (520) | 64.0 | 97.3 | 99.8 | 100.0 | − | − | − | 99.8/99.8 |
| P. aeruginosa (302) | 0.3 | 1.0 | 1.7 | 3.0 | 9.3 | 49.3 | 100.0 | – b/– b |
| Klebsiella spp. (263) | 14.4 | 73.0 | 92.4 | 98.5 | 100.0 | − | − | 100.0/90.5 |
| Beta-haemolytic streptococci (236) | 100.0 | − | − | − | − | − | − | 100.0/100.0 |
| Enterobacter spp. (167) | 4.2 | 58.7 | 84.4 | 95.2 | 98.8 | 100.0 | − | 98.8/95.2 |
US-FDA/EUCAST criteria.
No breakpoints have been established by the US-FDA or EUCAST.
Conclusions: Tigecycline exhibited a wide-spectrum of activity and potency versus contemporary BSI isolates collected in Europe. R to tetracycline or other antimicrobial classes did not adversely influence tigecycline activity. Treatment options for serious infections in the nosocomial environments should benefit from the availability of the new antimicrobial class agent, tigecycline.
P2034 In vitro antibacterial activity of tigecycline in comparison with doxycycline, ciprofloxacin and rifampin against Brucella species
H. Turan, H. Arslan, H. Uncu, O. Azap, K. Serefhanoglu (Konya, Ankara, Adana, TR)
Objectives: Human brucellosis affecting any organ system is a prevalent public health problem with worldwide distribution. The high rate of relapse after treatment with conventional drugs underlines the need for new anti-Brucella antibiotics. We therefore measured the in vitro activity of a new antimicrobial agent, tigecycline and compared it with the activity of established antimicrobials. No data exists regarding this issue.
Methods: A total of 82 strains of Brucella species were tested. All of these were obtained from blood and bone marrow cultures. The in vitro antimicrobial activities of tigecycline, doxycycline, ciprofloxacin, and rifampin were determined by E-test method.
Results: Doxycycline demonstrated the lowest MIC50 and MIC90 values, followed by tigecycline, ciprofloxacin, and rifampin. The MIC50 and MIC90 values of antibiotics for the strains of B. melitensis are shown in the Table . While all isolates were susceptible to ciprofloxacin and doxycycline, intermediate resistance to rifampin was detected in 19 (23%) isolates.
In vitro activities of tigecycline, doxycycline, ciprofloxacin and rifampin against 82 strains of B. melitensis
| Antibiotic | Range (μg/mL) | MIC50 | MIC90 |
|---|---|---|---|
| Tigecycline | <0.016–0.125 | 0.064 | 0.125 |
| Doxycycline | <0.016–0.125 | 0.016 | 0.047 |
| Ciprofloxacin | 0.047–0.25 | 0.094 | 0.19 |
| Rifampin | 0.25–2 | 0.75 | 1.50 |
Conclusion: In conclusion, in the present in vitro study, the MIC values of tigecycline, a novel tetracycline analogue, were assessed for the first time against strains of Brucella, and the MIC values of tigecycline were found to be lower than those of rifampin and ciprofloxacin, but were greater than those of doxycycline, a major component of currently available regimens in Brucella infections. Considering the fact that Brucellae are intracellular pathogens, in vivo studies are needed to fully evaluate therapeutic potential of tigecycline in brucellosis.
P2035 In vitro activity of tigecycline and linezolid against nosocomial MRSA strains
O. Kaya, F. Akcam, E. Temel (Isparta, TR)
Objective: Methicillin-resistant Staphylococcus aureus (MRSA) has become one of the most widespread causes of nosocomial infections worldwide. Moreover, it has been recovered from community-acquired infections. Therapeutic options for these infections are limited.
Research for new antimicrobial agent is perpetuated for treatment of this infections. Tigecycline is the first drug in the glycylcycline class of antibiotics. Linezolid is a synthetic antimicrobial agent of the an oxazolidinone class. The aim of this study is to evaluate MIC50 and MIC90 values of tigecycline and linezolid against MRSA strains.
Methods: A total of 60 MRSA strains isolated from various clinical specimens and at different times were included in the study. All the isolates were stored at −80°C when they had been isolated. These isolates were identified as S. aureus by phenotypic characteristics, Gram staining, catalase and coagulase tests. Resistance to methicillin was confirmed using the Mueller-Hinton agar diffusion procedure with 1μg oxacillin disks, as recommended by the Clinical and Laboratory Standards Institute (CLSI). Minimal inhibitory concentrations (MIC) were determined by the E-test method, according to the CLSI criteria.
Results: All isolates were sensitive to linezolid and tygecycline. The MIC levels of both antibiotics are listed in the table .
| Antibiotic | Range (μg/mL) | MIC50 (pg/mL) | MIC90 (μg/mL) |
|---|---|---|---|
| Tigecycline | 0.032–1.0 | 0.125 | 0.38 |
| Linezolid | 0.094–4 | 0.50 | 1 |
Conclusion: In conclusion, given the limited therapeutic options for serious MRSA infections, tigecycline and linezolid may offer additional options as they demonstrated good activity against clinical strains of MRSA infections.
P2036 In vitro antibacterial activity of tigecycline against hospital isolates: comparison of two methods and proposal for zone diameter breakpoints in France: a multicentre study
C.J. Soussy, S. Bremont, J.D. Cavallo, M. Chomarat, H.B. Drugeon, V. Jarlier, M.D. Kitzis, C. Lascols, R. Leclercq, C. Muller-Serieys, J. Rolain, M. Roussel-Delvallez, A. Dubanchet (Creteil, Paris, Saint Mande, Lyon, Nantes, Caen, Marseille, Lille, FR)
Objectives: The aim of the study was to compare the MICs of tigecycline (TIG) by agar dilution (A) using the Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) method and by broth microdilution (B) using the Clinical Laboratory Standards Institute (CLSI) method and to propose zone diameter breakpoints for clinical categorisation in France.
Methods: MICs of TIG were determined by A and B against 1478 strains collected in 10 university hospitals in France in 2006. Antibiograms were performed with 15 μg disks, according to the CA-SFM method. An internal quality control was carried out with genetically defined strains.
Results: According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) MIC breakpoints (mg/L): Enterobacteriaceae ≤1/>2, Staphylococcus ≤0.5/>0.5; Enterococcus and Streptococcus A, B, C, G. ≤0.25/>0.5 and zone diameter breakpoints were (mm): Enterobacteriaceae ≥22/<20, Staphylococci: ≥22/<22, Enterococci: ≥22/<18, Streptococci: ≥21/<18. However in Enterobacteriaeceae, it was difficult to discriminate isolates with zone diameter <22 mm.
| Bacterial species (no. of strains) | MIC50 |
(mg/L) |
MIC90 |
(mg/L) |
|---|---|---|---|---|
| A | B | A | B | |
| Enterobacteriaceae except | 0.5 | 0.5 | 2 | 2 |
| Proteus spp. (400) | ||||
| Proteus spp. (100) | 2 | 1 | 8 | 4 |
| Pseudomonas aeruginosa (100) | 16 | 8 | 32 | 32 |
| Acinetobacter baumannii (100) | 0.5 | 0.25 | 2 | 2 |
| Stenotrophomonas maltophilia (50) | 0.5 | 0.25 | 2 | 2 |
| MSSA (100) | 0.12 | 0.06 | 0.5 | 0.5 |
| MRSA (150) | 0.12 | 0.06 | 0.5 | 0.5 |
| GISA (30) | 0.25 | 0.12 | 0.5 | 0.5 |
| MSCNS (50) | 0.12 | 0.06 | 0.5 | 0.5 |
| MRCNS (50) | 0.25 | 0.12 | 0.5 | 0.5 |
| Enterococcus faecalis (100) | 0.12 | 0.06 | 0.5 | 0.5 |
| Enterococcus faecium (50) | 0.12 | 0.06 | 0.25 | 0.25 |
| Streptococci A, C and G (100) | 0.06 | 0.03 | 0.25 | 0.25 |
| PSSP (50) | 0.03 | 0.03 | 0.12 | 0.12 |
| PRSP (45) | 0.03 | 0.03 | 0.12 | 0.12 |
Conclusion: MICs of TIG obtained by both methods (A and B) appeared identical or very similar. The susceptibility zone diameter breakpoint recommended in France by using the 15 μg TIG disks could be ≥22 mm for all species. However, for Enterobacteriaceae, except Proteus, Morganella and Providencia showing a poor susceptibility to TIG, MICs should be determined on isolates with zone diameter <22 mm.
P2037 In vitro antibacterial activity of tigecycline against anaerobic hospital isolates; comparison of two methods in France: a multicentre study
L. Dubreuil, L. Calvet, V. Jarlier, J. Cavallo, M. Chomarat, H. Drugeon, M. Kitzis, C. Soussy, C. Lascols, M. Roussel-Delvallez, C. Muller-Serieys, R. Leclercq, J. Rolain, A. Dubanchet (Lille, Paris, Saint-Mandé, Lyon, Nantes, Créteil, Caen, Marseilles, FR)
Objectives: The aim of the study was to compare the MICs of tigecycline (TIG) by agar dilution (A) using the Clinical Laboratory Standards Institute (CLSI) M11 A6 method and a disk-diffusion test using the Comité de l'Antibiogramme de la Sociéte Française de Microbiologie (CA-SFM) method and to propose zone diameter breakpoints for clinical categorisation in France.
Methods: MICs of TIG were determined by the reference agar method M11A6 against 230 strains collected in 10 university hospitals in France in 2006. Antibiograms were performed with 15 μg disks, according to the CA-SFM method on Brucella blood agar. An internal quality control was carried out with B. fragilis ATCC25285, B. thetaiotaomicron ATCC 29741 and Eggerthella lenta ATCC 43055.
Results: Results for TIG are shown in the table . By disk-diffusion test the range for the inhibition zones were 28–41 mm for GPAC, 21–37 mm for clostridia (with exception of C. perfringens inhibition zone 14–35 mm), and 11–40 mm for the B. fragilis group. Considering the 4 mg/L CLSI breakpoint, the susceptibility zone diameter breakpoint recommended in France by using the 15 μg TIG disks could be ? 21 mm for all species except C. perfringens. MIC should be determined on isolates with zone size diameter <20 mm.
| Bacterial species | MIC (mg/L) |
||
|---|---|---|---|
| Range | MIC50 | MIC90 | |
| Bacteroides fragilis (84) | 0.06–16 | 0.5 | 2 |
| Bacteroides thetaiotaomicron (14) | 0.06–16 | 2 | 16 |
| Other Bacteroides (19) | 0.06–2 | 0.25 | 1 |
| All Bacteroides of the fragilis group (117) | 0.06–16 | 0.5 | 2 |
| Prevotella spp. (4) | 0.06–0.125 | ND | ND |
| C. perfringens (48) | 0.06–2 | 0.5 | 1 |
| C. difficile (18) | 0.06–0.25 | 0.125 | 0.25 |
| Other clostridia (6) | 0.06–0.5 | 0.5 | 0.5 |
| Finegoldia magna (13) | 0.06–0.5 | 0.125 | 0.25 |
| Micromonas micros (10) | 0.06–0.5 | 0.125 | 0.25 |
| Other GPAC: Gram-positive cocci (14) | 0.06–0.5 | 0.125 | 0.25 |
| All Gram-positive anaerobes (109) | 0.06–2 | 0.125 | 1 |
| All anaerobes (230) | 0.06–16 | 0.25 | 2 |
Conclusion: Categorisation by disk-diffusion test remains difficult for anaerobes. Among the B. fragilis group, resistance to metronidazole or imipenem could not be detected, but decreased susceptibility to metronidazole (4.3%) was observed. TIG 4 mg/L inhibited 111/117 strains of the B. fragilis group meanwhile TIG 2 mg/L was able to inhibit all Gram-positive anaerobes. Thus, at a concentration of 4 mg/L, TIG inhibited 97.3% of the investigated strains.
P2038 In vitro activity of tigecycline against linezolid-resistant Staphylococcus cohnii ssp. urealiticum clinical isolates
A. Stylianakis, A. Koutsoukou, S. Tsiplakou, S. Sardiniari, A. Vourtsi, I. Merianos (Athens, GR)
Objectives: The purpose of our study was to evaluate the in vitro activity of tigecycline against linezolid-resistant Staphylococcus cohnii ssp. urealiticum clinical isolates.
Methods: We examined 18 non duplicated linezolid-resistant Staphylococcus cohnii ssp. urealiticum clinical isolates. These isolates were recovered from blood cultures of ICU patients of our hospital during a 12 months time period. The identification and the susceptibility testing of the isolates were performed by the automated VITEK 2 system (boiMérieux, France). The susceptibility testing for tigecycline was performed by Kirby-Bauer disk diffusion method and done by direct colony suspension according to CLSI guidelines. Paper disks containing tigecycline at 15 mg per disk were used (Becton-Dickinson, USA). The determination of tigecycline and linezolid MIC values was performed by using E-test strips according to the manufacturer's guidelines (AB-Biodisk, Sweden). Isolates with an inhibition zone diameter of ≥ 19 mm and with an MIC level ≤ 0.5 mg/L were considered as susceptible to tigecycline. (MIC-susceptibility limits determined by EUCAST). Isolates with an MIC level ≥ 8 mg/L were considered as resistant to linezolid.
Results: All the Staphylococcus cohnii ssp. urealiticum clinical isolates were resistant to linezolid with an MIC level ≥24 mg/L. The tigecycline was found absolute active to all the examined linezolid-resistant Staphylococcus cohnii ssp. urealiticum clinical isolates. The inhibition diameter zones for tigecycline were found of ≥22 mm and the tigecycline-MIC levels of ≤0.125 mg/L.
Conclusion: Tigecycline is absolute active to all linezolid-resistant Staphylococcus cohnii ssp. urealiticum clinical isolates. This new glycycline is consisted an alternative option for treatment of infections caused by these isolates. Clinical laboratories should be routinely testing tigecycline susceptibility in all clinically significant Staphylococci; so that any changes to the current status may be detected as soon as possible.
P2039 Evaluation of tigecycline in vitro activity against panresistant Klebsiella pneumoniae clinical isolates
A. Stylianakis, A. Koutsoukou, S. Tsiplakou, V. Garavella, M. Taxtatzis, I. Merianos (Athens, GR)
Objectives: The purpose of our study was to evaluate the in vitro activity of tigecycline against panresistant Klebsiella pneumoniae clinical isolates.
Methods: We examined 68 non duplicated panresistant Klebsiella pneumoniae isolates recovered from blood cultures, bronchoalveolar excretions and pus and urine samples derived from ICU-patients of our hospital during a one year-time period. The identification of the isolates and the susceptibility testing were performed by the automated VITEK 2 system (boiMérieux, France). The susceptibility testing for tigecycline was performed by Kirby-Bauer disk diffusion method and done by direct colony suspension according to CLSI guidelines. Paper disks containing tigecycline at 15 mg per disk were used (Becton-Dickinson, USA). The determination of tigecycline-MIC values was performed by E-test strips according to the manufacturer's guidelines (AB-Biodisk, Sweden). Isolates with an inhibition zone diameter of ≥ 19 mm and with an MIC level ≤ 1 mg/L were considered as susceptible to tigecycline (MIC-susceptibility limits determined by EUCAST).
Results: All the Klebsiella pneumoniae clinical isolates were resistant to aminoglycosides, β-lactams, carbapenems, monobactames, furanes, cinolones and colistin. The tigecycline was found absolute active to all the examined isolates. The inhibition diameter zones were found of ≥ 21 mm and the MIC levels of ≤0.5 mg/L
Conclusion: Tigecycline is absolute active to panresistant Klebsiella pneumoniae isolates and holds great promise as a therapeutic agent to combat or limit desperate infections caused by these panresistant isolates.
P2040 Activity of tigecycline versus carbapenem-resistant strains of Acinetobacter spp
P. Khanna, D. Krahe, D. Bean, D. Wareham (London, UK)
Objectives: Acinetobacter sp have emerged worldwide as important noscomial pathogens responsible for ventilator associated pneumonia, bacteraemia and sepsis in the critically ill. In the UK, infection and colonisation with carbapenem resistant strains have been reported since 2001 and have been responsible for 65 episodes of bacteraemia in our institution from April 2002 – September 2006. The glycylcyline antibiotic tigecycline has been proposed as a useful therapeutic agent for the treatment of multidrug Acinetobacter infection. In order to determine the potential usefulness of this agent in our population we carried out susceptibility testing using Etest versus 32 recent multidrug resistant isolates.
Methods: Multidrug resistant Acinetobacter were isolated from clinical specimens and identified to genus level by API 20NE. Antimicrobial susceptibility testing to ampicillin, cefuroxime, ceftriaxone, trimethoprim, amikacin, ceftazidime, ciprofloxacin, gentamicin, imipenem, piperacillin/tazobactam, aztreonam, colistin, sulbactam and minocycline was performed by the BSAC disc diffusion method and MIC's to imipenem, meropenem and tigecyline by Etest. Carbapenem resistant isolates were assayed for the OXA-51, OXA-23, OXA-58 and OXA-24 genes to identify A. baumannii isolates and strains belonging to the OXA-23 clonal group.
Results: All strains were sensitive to colistin. Sensitivity to ciprofloxacin, amikacin, sulbactam and minocycline varied. Imipenem MIC's ranged from 0.19–>32 mg/L and meropenem MIC's from 0.25–>32 mg/L. Tigecycline MIC's ranged from <0.016–2 mg/L with an MIC50 = 0.5 mg/L and an MIC90 = 1.0 mg/L. All but one isolate were PCR positive for the OXA-51 gene and negative for both OXA-58 and OXA-24. The presence of OXA-23 varied.
Conclusion: Tigecycline demonstrates good activity against the prevalent strains of multidrug resistant strains of Acinetobacter isolated in our institution. The majority of our isolates are A. baumannii exhibiting marked carbapenem associated with OXA carbapenemases. Although our practice has been to use colistin for the treatment of serious infections with these organisms, tigecyline is an attractive alternative option. Its broad spectrum of activity, in comparison to colistin, combined with its activity versus A. baumannii, in comparison to carbapenems, make it especially suitable for empirical treatment of nosocomial infections in critical care.
P2041 In vitro antimicrobial activity of tigecycline against multidrug-resistant Acinetobacter baumannii strains
O. Kurt Azap, F. Timurkaynak, S. Karaman, H. Arslan (Ankara, TR)
Introduction: Acinetobacter baumannii strains are important multidrug-resistant nosocomial pathogens especially in the intensive care unit. Tigecycline, member of glycylcylines, exhibits antibacterial activity against multidrug-resistant Gram-negative strains.
The aim of this study is to determine the antibacterial activity of tigecycline against the multidrug-resistant A. baumannii strains isolated from the blood cultures of the patients hospitalised in the intensive care unit.
Materials and Methods: Multidrug-resistant A. baumannii strains isolated from the patients hospitalised in the intensive care unit between the period January 2005-April 2006 were included in the study. Bacteria were identified to the species level by conventional methods and automated identification kits. Each isolate was obtained from one patient. Antimicrobial activities of ceftazidime, cefepime, piperacillin-tazobactam, amikacin, ciprofloxacin and imipenem were studied according to the CLSI criteria. Minimal inhibitory concentrations of tigecycline against 39 A. baumannii strains of which were resistant to at least four of the drugs above were studied by E test strips. ≤ 2 mg/L was chosen as the susceptibility breakpoint according to the other studies.
Results: MIC50 and MIC90 values were found to be 1.5 mg/L and 2 mg/L respectively, within a range of 0.75–4 mg/L. The susceptibility rate of the strains were determined to be 94.7%.
Conclusion: MIC50 and MIC90 values of tigecycline against multidrug-resistant A. baumannii strains were found to be high as in the other studies. However, tigecycline seems to be an alternative for the infections caused by resistant pathogens because of the high susceptibility rate (94.2%). Further studies including higher numbers of strains are needed to determine the value of tigecycline in the treatment of the infections caused by multidrug-resistant A. baumannii strains.
P2042 In vitro activity of tigecycline against the multidrug-resistant bacteria isolated from burn patients
Ö. Kurt Azap, F. Timurkaynak, S. Karaman, H. Arslan (Ankara, TR)
Introduction: Infections in burn patients are usually caused by multidrug-resistant microorganisms. Tigecycline, a derrivative of glycylcyclines, is an effective antibiotic against the resistant strains. The aim of this study is to determine the in vitro activity of tigecycline against the multi-drug resistant bacteria isolated from burn patients.
Materials and Methods: Forty-five bacteria isolated from 118 patients hospitalised in the burn unit during 2003–3005 were included in the study. Gram-negative bacteria that were resistant to at least two classes of antibiotics among the β-lactam, quinolone, aminoglycoside classes, methicillin-resistant staphylococci, ampicillin-resistant enterococci were studied. Minimal inhibitory concentration values of tigecycline against these bacteria were tested by E test strips. Susceptibility breakpoints were determined according to the previous studies; ≤0.25 mg/L for enterococci, ≤0.5 mg/L for staphylococci and ≤2 mg/L for Acinetobacter baumannii.
Results: Forty-seven percent of the materials were obtained from wound, 31% from tissue specimens, 27% from blood and 3% from catheters. MIC50 and MIC90 values were shown in the Table .
MIC values of tigecycline against the bacteria isolated from burn patients
| Bacteria (n) | MIC (mg/L) |
||
|---|---|---|---|
| Range | MIC50 | MIC90 | |
| Enterococcus spp. | 0.032–1 | 0.125 | 0.25 |
| Methicillin-resistant S. aureus | 0.19–2 | 0.25 | 1.5 |
| Acinetobacter baumannii | 0.50–2 | 1 | 2 |
Conclusion: Susceptibility rates were found to be 94.2%, 85.5%, 100% for MRSA, enterococci, A. baumannii respectively. These results are in accordance with the other studies. In conclusion, tigecycline seems to be a good alternative for the difficult-to-treat infections in burn patients.
P2043 Susceptibility to tigecycline among multidrug-resistant Acinetobacter baumannii clinical isolates
A. Capone, S. D'Arezzo, P. Visca, N. Petrosillo on behalf of GRAB: Gruppo Romano Acinetobacter baumannii (Italy)
Introduction: Acinetobacter baumannii is serious clinical challenge in Intensive Care Units (ICUs). Treatment options for infections caused by Multi Drug Resistant (MDR) strains are currently limited to colistin, or to a glycylcycline compound, namely tigecycline (TIG). TIG is a promising antibacterial agent, active in treatment of complicated intra-abdominal, skin and skin-structure infections and towards a wide range of pathogens, including A. baumannii. Few studies on limited numbers of A. baumanni strains on the in vitro activity of TIG have so far been conducted in Italy.
Objective: Aim of the present study was to assess the in vitro activity of tigecycline against 80 MDR clinical isolates of A. baumannii collected from ICU patients in Rome hospitals in the course of a large urban nosocomial epidemic.
Materials and Methods: 80 A. baumannii clinical strains were isolated from hospitalised patients in six ICUs in Rome from January 2004 to June 2005 and analysed at the Molecular Microbiology Unit of the National Institute for Infectious Diseases “L. Spallanzani”, Rome, for sensitivity assays. Organisms were identified to species level by conventional methods. The minimal inhibitory concentration (MIC) for TIG was determined by broth microdilution reference method recommended by the NCCLS. The antimicrobial range tested was 0.031–0.128 mg/L. MIC breakpoints used for this agent were ≤2, 4, ≥8 mg/L to designate susceptible, intermediate, and resistant strains, respectively.
Results: We identified 58 susceptible isolates (72.5%) and 22 unsusceptible isolates (27.5%) of which 3/80 (3.7%) with full resistance and 19/80 (23.7%) with intermediate susceptibility. Information on previous antibiotic treatment before isolating A baumannii was available on 42 patients. 16/36 (44.4%) and 2/6 (33.3%) were unsusceptible strains in patients with and without previous antibiotics, respectively (OR: 1.6, 95% CI: 0.25–9.87). Notably, all MDR A. baumannii strains were susceptible to colistin but one (1.3%) that showed intermediate susceptibility.
Conclusion: Study shows that about one quarter of MDR A. baumannii clinical strains analysed, were unsusceptible to TIG. Even though we did not provide evidence of a statistically significant association with the previous antibiotic regimen, due to the limited size of our series, low TIG susceptibility can partly be explained by induction of efflux-based multridrug resistance pathways due to previous antibiotic exposure.
P2044 Comparative in vitro activity of tigecycline according to carbapenem resistance of different sequential clones of Acinetobacter baumannii in bacteraemic critically ill patients
J. Camarena, R. Gonzalez, R. Zaragoza, J. Navarro, S. Sancho, J. Nogueira (Valencia, ES)
Objectives: Multiresistant A. baumannii (MRAb) remains during the last 10 years like the most important problem in our ICU. In order to know the activity of antimicrobial agents (ATB), the aim of this study was to compare, according to carbapenem resistance, in vitro activity of tigecycline and 11 other ATB against epidemiologically characterised MRAb isolates from bacteraemic critically ill patients.
Methods: MRAb isolates (196) were collected from 100 bacteraemic ICU patients (samples: 100 bloodstream, 52 respiratory, 22 catheter, 11 wound and 11 other samples) during the last 10 years. MICs of tigecycline and 11 other ATB (carbapenems, aminoglycosides, ampi-sulbactam, piperacillin-tazobactam, ceftazidime, colistin, polimixin, doxycycline and minocycline) were determined by E-test® (AB-Biodisk, Solna, Sweden), using CLSI breakpoints, except for tigecycline. All tested strains were previously characterised by genotipically methods (REP-PCR) in order to cluster related strains.
Results: Because all the isolates/case had the same feno-genotipically pattern, the results are showed like the 100 cases patient. We reported 10 different clones with a lot of fenotipically antimicrobial patterns. The 80% (158 strains) of epidemiologically defined MRAb isolates from clinically significant ICU bacteraemias were carbapenem (imipenem and meropenem) resistant (CR-MRAb). The in vitro activity of tigecycline according to this carbapenem resistance did not show any differences between both groups, CR-MRAb or carbapenem susceptible (CS-MRAb) strains: range 0.25–4/8 mg/L and MIC50–90 2–2 mg/L. Only 8 cases in CR-MRAb (10%) and 2 in CS-MRAb (10%) had MIC for tigecycline ≥4 mg/L. The MIC50–90 in both groups (CR-MRAb/CS-MRAb) of the 11 other ATB tested were different for minocycline (4–32/2–2 mg/L), ampi-sulbactam (16–64/2–4 mg/L), tobramycin (16-≥256/2-≥256 mg/L), and amikacin (≥256-≥256/ 8-≥256 mg/L). These differences related with the characterizated clonal outbreaks were not found in tigecycline, with the same MICs in the different clones.
Conclusion: Tigecycline appears to be a promising agent that remain active against MRAb and there are not any differences in the activity according to carbapenem resistance or clonallity of the strains. For A. baumannii the MIC values imply that bacteraemia infection in critically ill patients might be treated with tigecycline despite the breakpoint of this is not clearly allocated.
P2045 Evaluation of expected clinical success of tigecycline and other commonly used antimicrobials for empiric treatment of complicated skin and skin structure infections in Germany, Spain and the United States
R. Mallick, T. Fritsche, R. Jones, A. Kuznik, M. Stilwell, H. Sader (Collegeville, North Liberty, US)
Objective: To model the expected clinical success of tigecycline and commonly used antimicrobials for empiric treatment of complicated skin and skin structure infections (cSSSI) based on evaluation of antimicrobial susceptibility and frequency of pathogen occurrence from a global surveillance programme. Tigecycline is a novel semisynthetic glycylcycline recently approved for parenteral treatment of cSSSI and intraabdominal infections in the United States (US) and European countries.
Methods: Consecutive, nonduplicate bacterial isolates collected between 2000 and 2005 from patients with documented cSSSI in 48 medical centres located in Germany (7), Spain (3), and the US (38) were used to evaluate the frequency of pathogen occurrence and susceptibility rates of tigecycline and select parenteral antimicrobials. All isolates were tested using CLSI broth microdilution methods and interpretive criteria. Tigecycline breakpoints approved by the USA-FDA were used. By applying pathogen-specific susceptibility rates to the frequency of occurrence of pathogens in each country, we calculated the overall expected coverage for each antimicrobial agent or combination.
Results: The top 3 pathogens identified in Germany and Spain were (frequency [%] by country): S. aureus (35.1 and 33.3%, respectively), E. coli (11.3 and 12.8%), and P. aeruginosa (11.0 and 12.6%). In the US, the top 3 pathogens identified were S. aureus (48.1%), P. aeruginosa (9.4%) and enterococci (8.8%). Other frequently isolated pathogens included b-haemolytic streptococci, Enterobacter and Klebsiella spp., with some inter-country variation. The rates of oxacillin-resistance (MRSA) varied from 3.6% in Germany to 17.3% in Spain and 43.5% in the US. Tigecycline was highly active (>90% S) against the most common pathogens, except P. aeruginosa and P. mirabilis. The overall expected coverage of cSSSI for the antimicrobials evaluated is summarised in the table .
| Antimicrobial | Overall expected coverage (% susceptible of all pathogens) |
||
|---|---|---|---|
| Germany | Spain | USA | |
| Tigecycline | 87.5 | 85.1 | 90.6 |
| Vancomycin | 66.9 | 59.5 | 73.4 |
| Linezolid | 66.9 | 59.5 | 74.4 |
| Levofloxacin | 88.3 | 84.6 | 75.7 |
| Piperacillin/tazobactam | 96.0 | 90.0 | 75.6 |
| Imipenem | 95.8 | 92.1 | 74.1 |
| Cefazolin | 73.2 | 66.2 | 60.1 |
| Ceftriaxone | 80.2 | 74.3 | 64.0 |
| Vancomycin & Piperacillin/tazobactam | 98.2 | 95.8 | 95.7 |
| Vancomycin & Levofloxacin | 95.5 | 93.1 | 93.8 |
Conclusion: Vancomycin in combination with piperacillin/tazobactam had the highest overall expected empiric coverage of cSSSI in the countries evaluated. Among monotherapies, tigecycline had the highest expected coverage rate in the US, where the prevalence of MRSA was relatively high. Piperacillin/tazobactam and imipenem had the highest expected coverage in Germany and Spain, where the prevalence of MRSA was relatively low. Our results suggest that tigecycline might be a viable option for empiric treatment of cSSSI in these countries, especially in settings with high MRSA rates.
P2046 Analysis of tigecycline activity against Acinetobacter spp. and Enterobacteriaceae based on clinical specimen source
D. Draghi, M. Dowzicky, M. Jones, N. Brown, C. Thornsberry, D. Sahm (Herndon, Collegeville, US; Breda, NL)
Objective: TIG, the first in class glycylglycine was approved for clinical in 2005 in the US and 2006 in Europe for treatment of skin and intraabdominal infections. Because variations in the in vitro activity of an antimicrobial can occur against organisms (Org) isolated from different clinical specimen sources, the activity of TIG was analysed according to specimen source (SPEC) for recent isolates of Acinetobacter spp. (AC) and Enterobacteriaceae (EN).
Methods: AC and EN isolates were collected from 52 hospital sites distributed among all nine US Bureau of Census regions and 15 hospital sites distributed across 4 countries in Europe (EU; Germany, Italy, Spain, and France). All AC and EN were isolated from clinical specimens in 2005–2006 and centrally tested against TIG and comparators by broth microdilution (CLSI guidelines M7-A7). The SPECs studied were blood [BD], respiratory [RP], skin and soft tissue [SST], and urine [UN]). US FDA breakpoints (BPs; ≤2 mg/L, susceptible [S]; 4 mg/L, intermediate [I]; ≥8 mg/L, resistant [R]) were applied to results from EN isolates that originated from the US and EUCAST BPs (≤1 mg/L, S; 2 mg/L, I; ≥4 mg/L, R) were applied to results from EN isolates that originated from EU. Currently, BPs do not exist for AC.
Results: See the table .
| Org | SPEC | USA |
EU |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | MIC (mg/L) |
%S | %I | %R | N | MIC (mg/L) |
%S | %I | %R | ||||||
| Range | Mode | MIC90 | Range | Mode | MIC90 | ||||||||||
| AC | SST | 83 | 0.034 | 0.25 | 1 | − | − | − | NA | NA | NA | NA | − | − | − |
| RP | 64 | 0.03–2 | 0.5 | 2 | − | − | − | 15 | 0.06–2 | 2 | 2 | − | − | − | |
| UN | 35 | 0.03–2 | 2 | 2 | − | − | − | 16 | 0.06–2 | 0.5 | 1 | − | − | − | |
| BL | 25 | 0.03–1 | 0.12 | 1 | − | − | − | 15 | 0.06–1 | 0.25 | 1 | − | − | − | |
| EN | SST | 215 | 0.06–4 | 0.5 | 1 | 99.1 | 0.9 | 0 | NA | NA | NA | NA | NA | NA | NA |
| RP | 209 | 0.06–4 | 0.25 | 1 | 99.0 | 1.0 | 0 | 68 | 0.06–2 | 0.25 | 1 | 97.1 | 2.9 | 0 | |
| UN | 758 | 0.064 | 0.25 | 0.5 | 99.9 | 0.1 | 0 | 187 | 0.03–2 | 0.25 | 0.5 | 98.4 | 1.6 | 0 | |
| BL | 126 | 0.03–2 | 0.25 | 1 | 100 | 0 | 0 | 71 | 0.06–2 | 0.25 | 0.5 | 98.6 | 1.4 | 0 | |
Conclusions: Based on susceptibility patterns susceptibility patterns and MIC90s, TIG demonstrated a potent activity against EN and AC, regardless of either the geographic (US or EU) or clinical source of the isolates. While TIG currently exhibits potent activity against these important Gram-negative bacilli, the ability of these organisms to develop resistance to a variety of antimicrobials warrants continued surveillance as TIG use to treat infections increases.
P2047 Tigecycline Evaluation Surveillance Trial (T.E.S.T.) – United States in vitro antibacterial activity against selected species of Enterococcus spp
S. Bouchillon, M. Hackel, B. Johnson, J. Johnson, D. Hoban, R. Badal, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline, a member of a new class of antimicrobials (glycylcyclines), has been shown to have potent expanded broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. programme determined the in vitro activity of tigecycline compared to vancomycin, linezolid, ampicillin, imipenem, ceftriaxone, levofloxacin, minocycline, penicillin and piperacillin/tazobactam against members of Enterococcus spp. collected from hospitals in the USA.
Methods: A total of 2,486 clinical isolates were identified to the species level at each participating site and confirmed by the central laboratory. Isolates were collected throughout 2004. Minimum Inhibitory Concentration (MICs) were determined by the local laboratory using broth microdilution panels and interpreted according to NCCLS guidelines.
Results: Of 1,709 E. faecalis evaluated, vancomycin resistance was noted in 80 (4.7%) isolates. These isolates were all susceptible to linezolid, penicillin and tigecycline. Tigecycline presented the lowest MIC50/MIC90 (0.06/0.12 μg/mL) among all antimicrobial agents evaluated. As a typical profile of E. faecalis, fluoroquinolone (levofloxacin) and tetracycline (minocycline) had limited activities against this species. Among 713 E. faecium, 477 (66.9%) were resistant to vancomycin, of which 17 isolates were non-susceptible to linezolid. Tigecycline also presented the lowest MIC50/MIC90 of 0.03/0.12 μg/mL.
Conclusion: Tigecycline's in vitro activity was comparable to or greater than most commonly prescribed antimicrobials. The presented data suggest that tigecycline may be an effective and reliable therapeutic option against Enterococcus spp. including vancomycin-resistant strains.
P2048 Comparison of in vitro activity of tigecycline against pathogens from intensive care patients in Europe – T.E.S.T. Program 2006
S. Bouchillon, B. Johnson, R. Badal, M. Hackel, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline, a member of a new class of antimicrobials (glycylcyclines), has been shown to have potent broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. Program surveyed the in vitro activity of tigecycline from ICU and non-ICU pathogens during 2004 to 2006.
Methods: A total of 4,744 clinical isolates from 62 testing sites in 19 European countries. Minimum Inhibitory Concentration (MICs) were determined by each site using common broth microdilution panels and interpreted according to EUCAST guidelines.
Results: See the tables.
| Drug | Enterobacteriaceae (n = 1,206) |
Acinetobacter spp. (n = 221) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC50 | MIC90 | |
| Tigecycline | 0.5 | 2 | 0.25 | 1 |
| Amikacin | 2 | 8 | 4 | >64 |
| Cefepime | ≤0.5 | 8 | 8 | 32 |
| lmipenem | 0.5 | 1 | 0.5 | >16 |
| Levofloxacin | 0.06 | 8 | 2 | >8 |
| Minocycline | 2 | 8 | ≤0.5 | 4 |
| PipTazo | 2 | 128 | 16 | >128 |
| Drug |
S. aureus (n = 206) |
Enterococcus spp. (n = 209) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | MIC50 | MIC90 | |
| Tigecycline | 0.12 | 0.25 | 0.12 | 0.12 |
| Levofloxacin | 0.12 | 16 | 2 | >32 |
| Linezolid | 2 | 4 | 2 | 2 |
| Minocycline | ≤0.25 | 1 | 8 | >8 |
| Vancomycin | 1 | 1 | 1 | 2 |
Conclusion: Tigecycline's in vitro activity was comparable to or greater than most commonly prescribed broad spectrum antimicrobials for all ICU bacterial study strains encountered. Tigecycline's inhibitory against Enterobacteriaceae, and Acinetobacter spp. was comparable to imipenem. Against Gram-positive organisms, Tigecycline's activity was comparable to linezolid and vancomycin.
P2049 In vitro activity of tigecycline against pathogens isolated from cerebrospinal fluid – T.E.S.T. Program 2006
R. Badal, S. Bouchillon, M. Hackel, J. Johnson, D. Hoban, B. Johnson, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline (TIG), a new glycylcycline, has been shown to have potent broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. Program determined the in vitro activity of TIG and 10 comparators against respective Gram-positive/negative species.
Methods: 295 cerebrospinal fluid pathogens from 272 sites and 34 countries were analysed in this survey. The isolates were identified to the species level at the participating sites and confirmed by the central laboratory. MICs were determined by each site using supplied broth microdilution panels and interpreted according to CLSI guidelines.
Results: TIG activity against pathogens isolated from cerebrospinal fluid are shown in the table .
| Organisms (n = 296) | %Sus | Tigecycline MIC (μg/mL) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| Acinetobacter spp. (n = 36) | n/a | 0.25 | 1 | 0.03–4 |
| P. aeruginosa (n = 17) | n/a | 16 | >6 | 4–>16 |
| Enterobacter spp. (n = 29) | 100 | 0.5 | 1 | 0.25–2 |
| Enterococcus spp. (n = 22) | 100 | 0.12 | 0.12 | 0.03–0.25 |
| E. coli (n = 27) | 100 | 0.12 | 0.25 | 0.06–2 |
| Klebsiella spp. (n = 17) | 100 | 0.5 | 1 | 0.12–1 |
| ESBLs (n = 6) | 100 | 0.25 | 0.5 | 0.12–0.5 |
| H. influenzae (n = 8) | n/a | 0.12 | 0.25 | 0.06–0.25 |
| S. aureus (n = 30) | 100 | 0.12 | 0.25 | 0.06–0.25 |
| MRSA (n = 7) | 100 | 0.12 | 0.25 | 0.06–0.25 |
| S. pneumoniae (n = 79) | n/a | 0.03 | 0.5 | ≤0.008–0.5 |
| S. marcescens (n = 16) | 100 | 1 | 2 | 0.5–2 |
| S. agalactiae (n = 15) | 100 | 0.03 | 0.06 | 0.03–0.25 |
n/a, breakpoints not available.
Conclusions: Tigecycline showed excellent inhibitory activity against all pathogens invading the cerebrospinal fluid in this study with the exception of P. aeruginosa. Tigecycline demonstrated MIC90 values of ≤0.5μg/mL against Gram-positive pathogens (including resistant phenotypes) and MIC90 values of ≤1μg/mL against the Enterobacteriaceae and Acinetobacter spp. validate the potent inhibitory activity of TIG against these invasive pathogens.
P2050 Antimicrobial susceptibility of 2,537 bacteraemia causative pathogens: Tigecycline Evaluation Surveillance Trial (TEST) in Europe, 2004–2006
R. Badal, S. Bouchillon, B. Johnson, M. Hackel, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Bacterial resistance patterns vary over both time and geography. One of the goals of surveillance studies is to identify those patterns to help guide current therapy. The Tigecycline Evaluation Surveillance Trial (TEST) is an ongoing global study that can serve to help recognize current trends in resistance on many levels. This report evaluates differences in susceptibility of bacterial pathogens isolated from the blood stream, collected in Europe from 2004 to 2006.
Methods: 2,537 bacteraemic pathogens were collected and identified from 2004–2006 at 62 hospitals in 19 countries in Europe. MICs for each strain were determined per EUCAST guidelines at each facility using broth microdilution.
Results: Tigecycline MICs are recorded in the table .
| Organisms (n = 296) | %Susa | Tigecycline MIC (μg/mL) |
||
|---|---|---|---|---|
| MIC50 | MIC90 | Range | ||
| Acinetobacter spp. (n = 161) | IE | 0.25 | 1 | 0.03–4 |
| P. aeruginosa (n = 208) | 8 | 16 | 0.5–>1 | 6 |
| Enterobacter spp. (n = 324) | 91.0 | 0.6 | 1 | 0.06–8 |
| Enterococcus spp. (n = 227) | 100 | 0.06 | 0.12 | 0.015–0.25 |
| VREs (n = 20) | 100 | 0.06 | 0.12 | 0.015–0.12 |
| E. coli (n = 531) | 99.8 | 0.12 | 0.25 | 0.03–2 |
| Klebsiella spp. (n = 413) | 91.0 | 0.6 | 1 | 0.12–8 |
| ESBLs (n = 71) | 85.9 | 0.6 | 2 | 0.06–8 |
| Serratia spp. (n = 118) | 91.5 | 0.5 | 1 | 0.12–8 |
| H. influenzae (n = 19) | IE | 0.12 | 0.25 | 0.03–0.5 |
| S. aureus (n = 282) | 100 | 0.12 | 0.25 | 0.03–0.5 |
| MRSA (n = 75) | 100 | 0.12 | 0.25 | 0.03–0.5 |
| S. agalactiae (n = 72) | 100 | 0.03 | 0.12 | 0.015–0.25 |
| S. pneumoniae (n = 182) | IE | 0.03 | 0.5 | ≤0.008–0.5 |
IE: Insufficient Evidence that this species is a good target for therapy with this drug.
–– indicates this species is a poor target for therapy with this drug.
Conclusions: Tigecycline showed excellent inhibitory activity against all causative bacteraemic pathogens with the exception of P. aeruginosa. Tigecycline demonstrated MIC90 values of ≤0.5 μg/mL against Gram-positive pathogens (including resistant phenotypes) and MIC90 values of ≤2 μg/mL against the Enterobacteriaceae and Acinetobacter spp. validate the potent inhibitory activity of TIG against these invasive pathogens.
P2051 Antibacterial activity of tigecycline against H. influenzae and S. pneumoniae (2004–06) Global Population – T.E.S.T. Program, 2006
S. Bouchillon, M. Hackel, J. Johnson, R. Badal, D. Hoban, B. Johnson, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline (TIG), a new glycylcycline, has been shown to have potent broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. Program determined the in vitro activity of TIG and comparators against respective Gram-positive/negative species. Isolates were collected from 272 hospital sites in 34 countries from 2004 to 2006.
Methods: A total of 4,724 clinically significant respiratory isolates collected worldwide were analysed in this survey. The isolates were identified to the species level at the participating sites and confirmed by the central laboratory. MICs were determined by each site using supplied broth microdilution panels and interpreted according to CLSI guidelines.
Results: Activities of tigecycline and comparator antimicrobials are shown in the table . Overall, 22.6% of H. influenzae were β-lactamase producers and 36.5% of S. pneumoniae presented some degree to non-susceptibility to penicillin. Tigecycline demonstrated potent inhibitory activity with MIC90 of ≤0.5 μg/mL and ≤0.25 μg/mL against β-lactamase positive H. influenzae and penicillin non-susceptible S. pneumoniae, respectively.
| Drug |
H. influenzae (n = 3,044) |
S. pneumoniae (n = 3,264) |
||||
|---|---|---|---|---|---|---|
| %Sus | MIC50 | MIC90 | %Sus | MIC50 | MIC90 | |
| Tigecycline | n/a | 0.12 | 0.5 | n/a | 0.03 | 0.25 |
| AmoxClav | 99.7 | 0.5 | 1 | 95.7 | ≤0.03 | 1 |
| Ceftriaxone | 99.9 | <0.06 | <0.06 | 98.2 | <0.03 | 0.5 |
| Levofloxacin | 100 | 0.015 | 0.03 | 99.8 | 0.5 | 1 |
| Imipenem | 100 | 0.5 | 1 | 74.6 | ≤0.12 | 0.5 |
| Linezolid | − | − | − | 100 | ≤0.5 | 1 |
| Penicillin | − | − | − | 63.5 | ≤0.06 | 1 |
an/a: Breakpoints not yet available.
Conclusions: Tigecycline showed excellent inhibitory activity against H. influenzae and S. pneumoniae regardless of the presence of β-lactamase or penicillin-resistance mechanisms. The results of this study suggest that Tigecycline may be a reliable therapeutic option for the treatment of respiratory infections due to these species.
P2052 Variable resistance patterns among tigecycline and 10 comparators against multidrug-resistant Acinetobacter from the T.E.S.T. Program
B. Johnson, S. Bouchillon, M. Hackel, R. Badal, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline (TIG), a member of a new class of antimicrobials (glycylcyclines), has been shown to have potent expanded broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. Program determined the in vitro activity of TIG compared to piperacillin-tazobactam (PT), levofloxacin (LVX), ceftriaxone (CAX), cefepime (CPE), amikacin (AK), minocycline (MIN), ceftazidime (CAZ), and imipenem (IMP) against multi-drug resistant Acinetobacter strains collected from 272 investigational sites in 34 countries throughout 2004–2006.
Methods: A total of 3,334 clinical Acinetobacter were identified to the species level at each participating site and confirmed by the central laboratory. Minimum Inhibitory Concentrations (MICs) were determined by the local laboratory using broth microdilution panels. Antimicrobial resistance was interpreted according to CLSI breakpoints where applicable.
Results: Resistance rates to the comparator drugs against all Acinetobacter spp. were CAX 33.5%, CAZ 42.8%. LVX 38.3%, CPE 33.5%, PT 25.5%, AK 14.3%, IMP 12.1%, MIN 2.5%. Strains were grouped by presence of resistance to 0, 1, 2, 3, 4, or ≥5 drug classes. TIG inhibited ≥96% of all 930 multi-drug resistant strains (resistant to 3 or more drug classes) at 2 μg/mL with only 3/930 (0.3%) strains at 8 μg/mL. TIG MIC50/90 for Groups Resistant to 0–5 drug classes were 0.12/0.5, 0.5/1, 0.5/2, 1/2, 1/2 and 1/2 μg/mL, respectively.
Conclusions: It has been seen in some species that existing multidrug resistant efflux pumps may also pump TIG. In spite of this, TIG remained effective although resistance to one drug classes increased the TIG MIC90 by 2-fold, and resistance to ≥2 classes increased it by 4-fold, TIG remained active against the great majority (96%) of the multi-drug resistant Acinetobacter strains at MIC values ≤2 μg/mL. TIG's in vitro activity against multi-drug resistant Acinetobacter should prove useful in therapy of infections caused by such therapeutically challenging strains.
P2053 Evaluation of in vitro activity of tigecycline and ten comparators against methicillin-resistant Staphylococcus aureus from 34 countries: TEST Program 2004–2006
R. Badal, S. Bouchillon, M. Hackel, J. Johnson, D. Hoban, B. Johnson, M. Dowzicky (Schaumburg, Collegeville, US)
Background: Tigecycline (TIG), a member of a new class of antimicrobials (glycylcyclines), has been shown to have potent expanded broad spectrum activity against most commonly encountered species responsible for community and hospital acquired infections. The T.E.S.T. Program determined the in vitro activity of TIG compared to amoxicillin-clavulanic acid, piperacillin-tazobactam, levofloxacin, ceftriaxone, linezolid (LZD), minocycline (MIN), and vancomycin (VAN), ampicillin, penicillin and imipenem against methicillin-resistant S. aureus (MRSA) isolates collected from 272 sites in 34 countries throughout 2004–2006.
Methods: A total of 2,669 clinical isolates of MRSA were identified to the species level at each participating site and confirmed by the central laboratory. Minimum Inhibitory Concentration (MICs) were determined by the local laboratory using supplied broth microdilution panels and interpreted according to CLSI guidelines, except for tigecycline, which used the susceptible breakpoint <0.5 μg/mL for S. aureus (including MRSA) as defined in the FDA approved package insert.
Results: The %S for the study drugs with MRSA activity – TIG, VAN, LZD, and MIN – was 100, 100, 100, and 97.6 respectively. There were few significant differences among geographic regions, except for lower activity of MIN in Asia (%S = 80.4) and the Middle East (%S = 13.3) compared to Europe (97.3%) and North America (99.1%). IMP showed a broad range of MIC50 results, with North America and Europe at the low end (0.5 and 2 μg/mL, respectively), and all others ≥16μg/mL. TIG inhibited 100% of strains in all regions. MIC50/90 (μg/mL) for TIG, VAN, LZD, and MIN were 0.12/0.25, 1/1, 2/2, and ≤0.25/2, respectively.
Conclusions: Global susceptibility patterns of MRSA remain fairly consistent. TIG was as potent as VAN and LZD, inhibiting 2,669/2,669 (100%) of the MRSA isolates at their respective breakpoints. TIG's excellent expanded broad spectrum of activity against MRSA should make it a very useful drug in treatment of difficult staphylococcal infections.
P2054 Impact of different FDA and EU breakpoints on bacterial susceptibility patterns Analysis of data from the T.E.S.T Program
R. Badal, S. Bouchillon, B. Johnson, M. Hackel, J. Johnson, D. Hoban, M. Dowzicky (Schaumburg, Collegeville, US)
Background: The MIC testing methodology recommended by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee for Antimicrobial Susceptibility Testing (EUCAST) is nearly identical, but interpretive MIC breakpoints established by the US FDA, based on CLSI methods, often differ from those set by EUCAST, sometimes leading to significantly different interpretations of the same MIC. It is important to understand the impact of these differences when interpreting susceptibility data reported in the literature. This study evaluated the impact of discrepant S/I/R breakpoints on susceptibility data from the global Tigecycline Evaluation Surveillance Trial (T.E.S.T).
Methods: A total of 48,232 pathogens from 34 countries were identified at each site and confirmed at a reference laboratory. MICs were determined at each site utilising supplied broth microdilution panels and interpreted according to FDA and EUCAST guidelines.
Results: There were discrepancies for Gram-positive and -negative organisms with several drugs, but the most significant (FDA %S ≥ 90% S, EUCAST <90%) were seen only with the Gram-negative species summarised in the table .
| Organism | %S (FDA/EUCAST) |
|||
|---|---|---|---|---|
| Tigecycline | Cefepime | Ceftriaxone | Meropenem | |
| E. aerogenes | − | 95.7/87.4 | − | − |
| E. cloacae | 93.6/88.1 | 93.5/76.2 | − | − |
| E. coli | − | − | 92.3/89.7 | − |
| K. pneumoniae | 95.0/88.6 | 91.3/88.5 | − | − |
| K. oxytoca | − | − | 93.1/84.1 | −– |
| S. marcescens | 96.8/86.0 | − | 91.6/81.2 | − |
| A. baumannii | − | − | − | 95.3/84.9 |
Conclusions: Differences between FDA vs. EUCAST MIC interpretive breakpoints can lead to significantly different assessments of an antimicrobial's potency vs. various bacterial species. Although the quantitative differences in %S were usually relatively small, there were several drug/bug combinations in this analysis for which use of EUCAST breakpoints caused the %S to fall below 90%. Since ≥90% susceptibility is often viewed as the minimum for a drug to be considered useful vs. a given species of bacteria, it is essential when evaluating reports of a drug's activity to be aware of which interpretive breakpoints were used in the analysis, and to bear in mind that there can be significant differences when using EUCAST instead of FDA breakpoints.
P2055 The killing of E. coli by tigecycline using the minimum inhibitory and mutant prevention drug concentration
J. Blondeau, S. Borsos (Saskatoon, CA)
Objective: Tigecycline (TIG) is a new antibacterial agent with low minimum inhibitory concentration (MIC) and low mutant prevention concentration (MPC) against clinical isolates of E. coli. We were interested in determining the rate and extent of killing by Tig using MIC and MPC drug concentrations against high density inocula of E. coli.
Methods: For MIC testing, 105 cfu/mL of E. coli were exposed to doubling drug concentration of Tig in Mueller-Hinton (MH) broth and following incubation under ambient conditions, the lowest concentration preventing growth was the MIC. For MPC testing, ≥109 CFUs were exposed to doubling dilutions of TIG on drug containing MH agar plates and following incubation for 24–48 hours, the lowest drug concentration preventing growth was the MPC. For kill experiments, 106 cfu/mL and 107 cfu/mL were exposed to the measured MIC and MPC drug concentrations and the reduction in viable cells (log10 and % kill) were recorded at 30 min, 1, 2, 3, 4, 6, 12 and 24 hours. All experiments were conducted in triplicate.
Results: For 2 clinical E. coli isolates, MIC values were 0.063 mg/L and MPC values were 1 mg/L. When 106 or 107 cfu/mL were exposed to the MIC drug concentrate, positive growth was seen for the first 6 hours of drug exposure; at 106 cfu/mL, a 1.06 log10 reduction (13% kill) was seen by 12 hours and 1.64 log10 reduction (97.7% kill by 24 hours). Exposure of 106 cfu/mL to the MPC drug concentration yielded a 0.16 to 0.42 log10 reduction (31–62% kill) by 30 min to 4 hours and this increased to a 1.16–1.86 log10 reduction (93–98% kill by 12–24 hours). Exposure of 107 cfu/mL to the MPC drug concentration yielded similar results: 0.13–0.31 log10 reduction (26–51% kill) by 1–4 hours and 0.71–1.14 log10 reduction (81–99% kill) by 12 hours.
Conclusion: TIG MIC and MPC values were low against E. coli – 0.031 and 1 mg/L respectively. Killing using MIC drug concentrations against higher density inocula was slow and incomplete. Killing was faster and more complete following exposure to MPC drug concentrations – 51–62% by 4 hours and 81–99% by 12 hours. The low MIC and MPC values suggest that TIG is less likely to select for resistant E. coli and dosing to achieve MPC results in the efficient killing of high density bacterial burdens.
P2056 Determination of the minimum inhibitory concentration and mutant prevention concentration (MPC) of tigecycline against clinical isolates of Streptococcus pneumoniae; impact of media on MPC results
C. Hesje, S. Borsos, J. Blondeau (Saskatoon, CA)
Objective: Tigecycline is the first of a new class of compound – glycylcyclines – with reported potent in vitro activity against penicillin-susceptible and multi-drug resistant strains of Streptococcus pneumoniae (SP). The mutant prevention concentration (MPC) defines the antimicrobial drug concentration threshold that blocks the growth of resistant bacterial sub-populations that may be present in high density bacterial populations such as those present in infection. We measured the MIC and MPC values for tigecycline against clinical isolates of SP and compared MPC results on blood agar versus solidifying Todd-Hewett broth (THB).
Methods: For MIC testing, the recommended Clinical and Laboratory Standards Institute procedure was followed utilising 105 cfu/mL tested against doubling drug dilutions in THB with incubation at 35–37 degrees Celsius in 5% CO2 for 18–24 hours. For MPC testing, ≥109 CFUs were added to drug containing agar plates: 1) tryptic soy agar containing 5% sheep red blood cells (BA), 2) THB solidified with 1.5% agar. Inoculated plates were incubated as described for 24–48 hours and screened for growth. The lowest drug concentration preventing growth was the MIC or MPC depending on method.
Results: For 140 SP isolates (121/86.4% penicillin susceptible; 19/13.6% penicillin non-susceptible), MIC50, MIC90 and MIC range values were 0.016 mg/L, 0.016 mg/L and ≤0.008–0.31 mg/L respectively and these MIC values were not influenced by SP susceptibility or resistance to penicillin nor for isolates recovered from blood or respiratory tract specimens. MPC50, MPC90 and MPC values were 8 mg/L, 16 mg/L and 1–16 mg/L when tested on BA. MPC testing on solidified THB yielded MPC50, MPC90 and MPC range values of 0.063 mg/L, 0.12 mg/L and 0.063–0.25 mg/L.
Conclusion: Tigecycline was highly active in vitro against SP with MICs ≤0.031 mg/L and not different for strains of SP resistant to penicillin. MPC values ranged from 1–16 mg/L on BA but we were substantially lower on solidified THB – 0.063–0.25 mg/L. This data suggests that SP testing of tigecycline on BA yields falsely elevated values. Solidified THB appears to be a more suitable media for SP MPC testing against tigecycline. Tigecycline appears to have a low propensity to select for resistance.
Pharmacodynamics and untoward effects of antibacterials
P2057 Susceptibility of various β-lactams to the inoculum effect
V.H. Tam, K.R. Ledesma, K.T. Chang, T.Y. Wang, J.P. Quinn (Houston, Chicago, US)
Objective: Escherichia coli (EC) is part of the human gastrointestinal flora and a common pathogen implicated in intra-abdominal infections such as perforated appendicitis and peritonitis. A heavy bacterial burden is anticipated in this type of infection and the clinical utility of the β-lactams may be limited by the inoculum effect. We compared the bactericidal activity of various β-lactam sub-classes against a standard and heavy inoculum of EC.
Methods: A wild-type (EC ATCC 25922) and a clinical ESBL-producing (TEM-26) strain were used. Time-kill studies were performed using approximately 105 and 108 cfu/mL at baseline. A clinically achievable concentration range of piperacillin/tazobactam (PIPT), ceftriaxone (CRO) and ertapenem (ERT) were used, and the drug concentrations were normalised to multiples of MIC. Serial samples were obtained in duplicate over 24 hours; viable bacterial burden was determined by quantitative culture to examine the impact of a higher inoculum on the bactericidal activity of various β-lactams.
Results: MIC of the wild type strain to PIPT, CRO and ERT were 2/4, 0.125 and 0.008 mg/L; for TEM-26 the respective MIC values were 16/4, 64 and 0.06 mg/L. All 3 β-lactams demonstrated significant killing with the standard inoculum (except for CRO against TEM-26). However, with the higher inoculum, the activity of PIPT was drastically reduced in both strains. CRO remained reasonably bactericidal against the wild type strain only (at ≥16×MIC). ERT was the least affected by the inoculum effect in both strains; bactericidal activity was retained with concentrations ≥4–8 × MIC.
Conclusion: Our results suggest that different β-lactam sub-classes have a distinct killing profile against a dense EC population. ERT appeared to be the least susceptible to the inoculum effect, which might be more efficacious than other non-carbapenem β-lactams in the treatment of intra-abdominal infections. Comparative in-vivo/clinical investigations are warranted to validate our findings.
P2058 Extracellular and intracellular activities of quinupristin-dalfopristin (Synercid) against Staphylococcus aureus, with different resistant phenotypes (MSSA, MRSA, VISA)
P. Baudoux, Y. Glupczynski, P. Tulkens, F. Van Bambeke (Brussels, BE)
Objectives: S. aureus survives and thrives in mild acidic pH environments, such as found intracellularly in phagolysosomes. Synercid, a semi-synthetic streptogramin antibiotic composed of quinupristin and dalfopristin (30:70 w/w ratio), displays a synergistic antibacterial activity against S. aureus and other Gram-positive bacteria in vitro. Synercid also displays activity against S. aureus phagocytised by murine J774 macrophages (JAC, 1992; 30 Suppl A:107–15), but this has been examined after short term incubation (2 h) and at a single fixed concentration (10×MIC) only. Our objective was to assess the activity of Synercid in a newly developed model of S. aureus-infected human THP1 macrophages that allows longer exposure periods and a detailed analysis of dose-responses for extracellular and intracellular activities (AAC 2006; 50:841–851), and using strains of S. aureus with resistance phenotypes of clinical significance.
Methods: We used an erythromycin-susceptible MSSA strain (ATCC 25923) and two erythromycin-resistant (MLSB) MRSA (ATCC 33591) and VISA (NRS 126) strains. MICs were determined by arithmetic dilution in MH Broth adjusted to pH 7.4 and 5.4. Change in CFU, compared to controls, were examined for bacteria incubated in MH broth (extracelllar activity) or phagocytised by THP-1 macrophages (intracellular activity) after 24 h exposure to concentrations from 0.01 to 100× the MIC. Key microbiological and pharmacological parameters (static concentration [Cs]; concentration yielding 50% of the maximal effect [EC50]; and maximal effect for drug concentration at infinity [Emax]) were determined by non-linear regression (Hill equation; slope factor = 1).
Results: Results are shown in the Table .
| Strain | MIC at pH |
Extracellular activitya |
Intracellular activitya |
|||||
|---|---|---|---|---|---|---|---|---|
| 7.4 | 5.4 | EC50 | Cs | Emax | EC50 | Cs | Emax | |
| MSSA | 0.45 | 0.40 | 0.58 | 0.68 | −2.96 | 3.95 | 0.43 | −3.45 |
| MRSA (MLSB) | 0.40 | 0.40 | 0.40 | 0.49 | −2.94 | 0.71 | 0.16 | −1.40 |
| VISA (MLSB) | 0.30 | 0.30 | 0.31 | 0.43 | −2.36 | 0.22 | 0.27 | −1.19 |
EC50 and Cs in mg/L; Emax is change in CFU (log10) from the original inoculum (∼106 cfu per mL [extracell.] or per mg cell protein [intracell.]).
Conclusion: Synercid (a) shows no increase in MIC at acid pH; (b) shows comparable activities against extracellular erythromycin-susceptible MSSA and erythromycin-resistant MRSA and erythromycin-resistant VISA; (c) shows also activity against their intracellular forms (but a bactericidal effect [>3 log] is only observed with the erythromycin-susceptible MSSA).
P2059 Effect of human albumin physiological concentrations on the in vitro bactericidal activity of daptomycin vs. vancomycin Cmax concentrations against Gram-positive isolates exhibiting the main resistance phenotypes
F. Cafini, L. Aguilar, N. Gonzalez, M. Giménez, M. Torrico, L. Alou, D. Sevillano, P. Vallejo, J. Prieto (Madrid, ES)
Objectives: To study the effect of the presence of physiological concentrations of human albumin on the Time needed to obtain bactericidal activity (≥3 log10 – 99.9% – initial inocula reduction) by concentrations similar to the Cmax obtained in serum after i.v. 4 mg/kg of daptomycin and 1g vancomycin, against quinolone-susceptible (QS) and -resistant (QR) S. pneumoniae (SP), methicillin-susceptible (MS), -resistant (MR), and heterogeneous vancomycin-intermediate (hVI) S. aureus (SA), and vancomycin-susceptible (VS) and -resistant (VR) E. faecium (E).
Methods: Killing curves were performed with final inocula of approx. 107 cfu/mL, and a final concentration of daptomycin or vancomycin of 56 μg/mL and 25.5 μg/mL, respectively in different media: a) Mueller-Hinton broth with 5% lysed horse blood for S. pneumoniae or without blood supplement for S. aureus and E. faecium (Cmax-MH), and b) MH broth with 4g/dl human albumin (Cmax-HAlb). In parallel, killing curves with daptomycin or vancomycin concentrations (4.7 and 16.1 μg/mL, respectively) corresponding to free-drug Cmax were performed in Mueller-Hinton broth.
Results: MICs of daptomycin and vancomycin and Time (h) to obtain bactericidal activity are shown in the Table .
| Strains: | Time to bacterial activitya (h) |
|||||||
|---|---|---|---|---|---|---|---|---|
| QSSP | QRSP | QRSP | MSSA | MRSA | hVI | VSE | VRE | |
| DAP MIC (μg/mL) | 0.06 | 0.06 | 0.06 | 0.25 | 0.5 | 1 | 2 | 4 |
| Cmax-MH | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| Cmax-HAIb | 2 | 3 | 3 | 3 | 2 | 4 | 8 | 24 |
| 8.3% Cmax | 2 | 2 | 3 | 3 | 2 | 4 | − | − |
| VAN MIC (μg/mL) | 0.5 | 0.5 | 0.25 | 1 | 1 | 4 | 1 | 64 |
| Cmax-MH | 10 | 10 | 24 | 24 | 24 | − | − | − |
| Cmax-HAIb | 10 | 12 | 24 | 24 | 24 | − | − | − |
| 8.3% Cmax | 10 | 24 | 24 | 24 | 24 | − | − | − |
– no bactericidal activity achieved.
Conclusions: Daptomycin Cmax, despite its high protein binding, exhibited rapid bactericidal activity (time ≤ 2 h) against all strains tested regardless resistance phenotype, that is delayed in the presence of human albumin. In contrast, vancomycin Cmax exhibited slow bactericidal activity (obtained generally at 24 h) against SP, MSSA and MRSA, but never against hVI, VSE and VRE. Theoretical extrapolation of active drug from total drug by using the protein binding rate seems a non accurate method to study antibacterial activity considering the implications of protein binding.
P2060 Effects of cefditoren, cefuroxime, cefaclor and cefixime on the competitive growth of Streptococcus pneumoniae, as a model approach to selection of populations
F. Cafini, M. Torrico, N. Gonzalez, O. Echevarria, D. Sevillano, M.J. Gimenez, L. Alou, L. Aguilar, P. Coronel, M. Gimeno, J. Prieto (Madrid, ES)
Objectives: To explore how oral cephalosporins influence the evolution of S. pneumoniae populations sharing the same ecological niche.
Methods: A computerised pharmacodynamic model (J Antimicrob Chemother 2006;58:794–801) simulating physiological concentrations obtained after 400 mg b.i.d. cefditoren (CDN), 500 mg b.i.d. cefuroxime (CXM), 500 mg t.i.d. cefaclor (CEC), and 400 mg o.d. cefixime (CFM) was used to investigate its effect on a mixed culture of four S. pneumoniae serotypes (Ser) as an approach to ecology of population dynamics. MICs (μg/mL) of CDN, CXM, CEC and CFM were: 0.015, 0.03, 0.25, and 0.5 for Ser4 and Ser19A; 0.5, 4, >8 and >8 for Ser19F; and 1,4, >8 and >8 for Ser23F, respectively. Initial mixed inocula (time 0) of 5×106 to 107 cfu/mL, including similar percentages of each Ser were used.
Results: Mean colony counts in antibiotic-free plates (whole pneumococcal population) increased (from 0 to 12 h) from log10 7.0 to 6.3 in drug-free simulations (control), from log10 6.6 to 3.6 in CDN simulations, from log10 6.6 to 8.0 in CXM simulations, from log10 7.0 to 7.2 in CEC simulations, and from log10 6.7 to 7.4 in CFM simulations. At 12 h of control drug-free experiments, the dominant population was Ser4 (70.0%). The small final population after CDN simulations included 48% Ser23F and 20% Ser19F. With all other compounds, the final population was higher than initial inocula, and corresponded to 90% Ser19F after CXM simulations, 61% Ser19F and 35% Ser23F after CEC simulations, and 96% Ser19F after CFM simulations.
Conclusion: Strain distribution in antibiotic-free environment depends on bacterial fitness in mono- and multi-strain niches. The selective pressures of antimicrobial regimens eradicate some populations and unmask minor populations, thus redistributing the whole population. CDN was the only cephalosporin showing bactericidal activity (≥3 log10 reduction of initial inocula) at 12 h of the whole initial population, with a small remaining population mainly composed by Ser23F. All other oral cephalosporins selected populations higher than initial inocula mainly composed by Ser19F.
P2061 Bactericidal activity of simulated serum concentrations of bid regimens of 400 mg cefditoren and 2000/125 mg amoxicillin/clavunanic acid against ampicillin susceptible and resistant H. influenzae: an in vitro pharmacodynamic model
N. Gonzalez, O. Echeverria, M. Torrico, D. Sevillano, M. Gimenez, L. Alou, L. Aguilar, P. Coronel, J. Prieto (Madrid, ES)
Objective: To explore the serum bactericidal activity over 48 h of simulated serum concentrations obtained after bid regimens of 400 mg cefditoren (CDN) and 2000/125 mg amoxicillin/clavulanic acid (AMC) against ampicillin (AMP) susceptible and resistant H. influenzae.
Methods: Three strains were used: a) AMP-susceptible β-lactamase negative strain (AMP-S) with AMC and CDN MICs of 2 and 0.015μg/mL; b) β-lactamase negative AMP-resistant strain (BLNAR) with AMC and CDN MICs of 8 and 0.12μg/mL; and c) β-lactamase positive AMC-resistant strain (BLPACR) with AMC and CDN MICs of 8 and 0.06μg/mL. The changing free and total antibiotic concentrations were simulated over 48 h (4 doses) in a computerised model based on a two-compartment model described previously (J Antimicrob Chemother 2004;54:1148–51). Initial inocula was approx. 108 cfu/mL. Antibiotic concentrations and colony counts were determined (in triplicate) at 0, 1, 2, 3, 4, 6, 8, 10, 12, 24, 25, 26, 27, 28, 30, 32, 34, 36 and 48 h. Limit of detection was 50 cfu/mL.
Results: Mean differences between log10 cfu/mL initial inocula and log10 cfu/mL at 24 h and 48 h sampling times are shown in the Table .
| Strain | 24 h |
48 h |
||||||
|---|---|---|---|---|---|---|---|---|
| CDN |
AMC |
CDN |
AMC |
|||||
| Free | Total | Free | Total | Free | Total | Free | Total | |
| AMP S | >6.3* | >6.3** | 4.1 | 4.3 | >6.3* | >6.3** | 4.3 | 4.9 |
| BLNAR | 3.7 | 5.0** | 3.0 | 3.1 | 3.7 | 5.5** | 3.2 | 3.5 |
| BLPACR | 4.5* | >6.3** | 0.1 | 0.8 | 4.6* | >6.3** | 0.8 | 1.1 |
P < 0.001 free CDN vs. free AMC
P < 0.001 total CDN vs. total AMC.
Conclusions: 400 mg CDN total drug serum concentrations profile exhibited at 24 and 48 h significantly higher initial inocula reduction than 2000/125 mg AMC against the three strains tested regardless the resistance phenotype. CDN free-drug concentrations exhibited also higher initial inocula reduction than AMC against the three strains, with statistically significant reductions against the BLPACR phenotype and even against the AMP-S phenotype.
P2062 Bacterial strain-independent anti-staphylococcal effects of telavancin in an in vitro dynamic model that simulates multiple-dose pharmacokinetics in humans
S. Zinner, E. Strukova, M. Smirnova, I. Lubenko, S. Vostrov, A. Firsov (Cambridge, US; Moscow, RU)
Objective: Telavancin is a lipoglycopeptide antibiotic with potent, rapidly bactericidal activity against Gram-positive bacteria including methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. To test the hypothesis of its bacterial strain-independent pharmacodynamics, telavancin was studied with two strains of S. aureus.
Methods: A methicillin-resistant strain of S. aureus ATCC 43300 (MIC 0.25 mg/L) and a glycopeptide-intermediate strain of S. aureus (GISA) Mu-50 (MIC 0.5 mg/L) at a starting inoculum of 8 log CFU/mL were exposed to telavancin administered daily for five consecutive days. Mono-exponential concentration decays of telavancin with a half-life of 8 h were simulated over a wide range of ratios of the 24-h area under the curve (AUC) to MIC: from 30–50 to 1200 h. Depending on the simulated AUC/MICs, specimens were sampled from the central compartment of the dynamic model for 6–8 days. The cumulative antimicrobial effect was determined as an area between the level corresponding to the starting inoculum and the time-kill curve (ABBC) from time zero to 144 h.
Results: Killing kinetics of S. aureus ATCC 43300 and GISA Mu-50 exposed to telavancin were similar: initial reduction of the starting inoculum was followed by bacterial regrowth. An increase in the AUC/MIC ratio was accompanied by greater killing and delay in bacterial regrowth. However, the rate of initial killing did not correlate with the simulated AUC/MIC ratio. The ABBC versus log AUC/MIC plots were linear and virtually superimposed for both organisms. A linear regression fits the combined data with r2 of 0.92.
Conclusion: These findings suggest a bacterial strain-independent pattern of telavancin pharmacodynamics with staphylococci. The established AUC/MIC-response relationship allows clinically relevant prediction of telavancin effects on a hypothetical strain of S. aureus with MICs values equal to previously reported MIC50 or MIC90 values.
P2063 Dose ranging and fractionation of moxifloxacin against Stenotrophomonas maltophilia using an in vitro pharmacodynamic model
D. Sevillano, L. Alou, F. Cafini, M. Lopez, N. Gonzalez, O. Echeverría, M. Torrico, J. Prieto, M. Gómez-Lus (Madrid, ES)
Objectives: To explore the antibacterial activity of higher total daily doses of moxifloxacin (MXF) against S. maltophilia with or without expression of SmeABC or SmeDEF efflux pump systems.
Methods: Eight PFGE characterised isolates of S. maltophilia (MIC to MXF 0.03–8 mg/L) without QRDR mutations in gyrA and parC were exposed to MXF 200 mg o.d., MXF 400 mg o.d., MXF 400 mg b.i.d., and MXF 800 mg o.d. (AUC0–24/MIC ratio of 4.4–1186.3 h) using a two-compartment in vitro pharmacodynamic model with peripheral units containing a starting inoculum of 1–2 ×109 CFU of each strain. A sigmoid dose-response model was used to estimate the required AUC0–24/MIC to achieve a 50% (ED50) and 80% (ED80) of maximum antibacterial effect at 24 hours. Concentrations of MXF were determined by bioassay and were closed to target values.
Results: Only the strain with MIC to MXF of 0.03 (no SmeABC and SmeDEF expression) was eradicated at MXF 200 mg. The three strains with MIC to MXF of 0.12–0.5 mg/L (SmeDEF expression) was related to regrowth after MXF 400 mg administration but eradicated after MOX 400 mg b.i.d. or 800 mg o.d. These doses were more active (2.7–4.3 log cfu/mL reduction) than MXF 400 mg o.d. (0.6–1.2 log cfu/mL reduction) (p < 0.05) against strains with MIC to MXF of 1 mg/L (SmeDEF expression) or MIC to MXF of 2 mg/L (simultaneous SmeABC and SmeDEF expression) at 24 h. The activity of the MXF 400 b.i.d. regimen was higher than MXF 800 o.d. against strain with MIC to MXF >2 mg/L (1.7–2.4 vs. 0.7–0.8 log cfu/mL reduction). Sigmoid relationship between AUC/MIC and measurement of the antibacterial effect was more adequately described using AUBKC (r2 = 0.95). Values associated with ED50/ED80 at 24 h were 24/87 h.
Conclusions: Higher total daily doses of moxifloxacin (400 mg b.i.d. and 800 mg o.d.) than the usual moxifloxacin clinical dose (400 mg o.d.) improve significantly the antimicrobial activity against strains of S. maltophilia with MIC to MXF ≤ 2 mg/L independently of the efflux pump expression.
P2064 Evaluation of daptomycin activity against Staphylococcus aureus following vancomycin exposure in an in vitro pharmacodynamic model with simulated endocardial vegetations
W. Rose, M. Rybak, S. Leonard, G. Sakoulas, G. Kaatz, M. Zervos, A. Sheth, C. Carpenter (Detroit, Valhalla, Royal Oak, US)
Objective: Staphylococcus aureus (SA) remains clinically problematic due to multidrug resistance. Vancomycin (V) has been the primary therapy though its efficacy has recently come into question. Daptomycin (D) is bactericidal against SA and often used following V failure. The objective of this study was to use an in vitro pharmacodynamic model with simulated endocardial vegetations (IVPD-SEV) model to evaluate if V exposure affects D activity.
Methods: 5 clinical isolates were evaluated. 4 isolates (3 MRSA, 1 MSSA) were reported to demonstrate D non-susceptibility following V exposure and 1 MRSA that was not exposed to V but became D non-susceptible after suboptimal dosing of D. All pre-V isolates were susceptible to D with MICs of 0.125–0.5 mg/L. Two simulations were evaluated: (1) 4 d of V 1 g q 12 h (Cmax 30 mg/L, Cmin 10 mg/L) followed by 4 d of D 6 mg/kg q 24 h, and (2) 8 d of D6 q 24 h. Time to 99% kill (T99) was calculated for each regimen. Changes in MIC over 8 d were evaluated using D 1.5, 2 and 3×MIC screening plates. D Etest MICs were performed for all model samples including any growth on screening plates to confirm changes in MIC. If changes in MIC were demonstrated, model experiments of no V treatment × 4 d (growth control) were followed by D6 q 24 h × 4 d to confirm a possible relationship to V. Additional regimens of V×4 d followed by D 10 mg/kg q 24 h × 4 d and D10× 8 d were also analysed.
Results: Pre-exposure MICs for V/D were 2/0.25 for MRSA isolates and 1/0.125 for the MSSA. The MSSA demonstrated D heteroresistance at baseline; the MRSA isolates did not. No change in MIC was detected for any MRSA isolate treated with D following V for any regimen tested. T99 was 1.39–11.36 h for D6 regimens without V exposure and 3.43–9.41 h following V. MIC elevations to D > 1 mg/L for the MSSA isolate were noted after 96 h of D6 following V but not when D6 was used alone during the 8 d simulation. Increases in MIC remained stable to serial passage for 5 d. D maintained bactericidal activity against the MSSA isolate even during the post V period with T99 achieved at 9.46–9.91 h. No resistance was detected when D6 followed 4 d of growth control, D10 post V, or D10 alone × 8 d.
Conclusion: Exposure to V prior to D therapy resulted in no change in MIC for all MRSA strains. The MSSA isolate demonstrated MIC changes post V; however, effective D kill was maintained. The preexisting heteroresistance to D with the MSSA isolate suggests that the exposure to V may not have induced this phenomenon. Further investigation is warranted to confirm these results.
P2065 Evaluation of various fluoroquinolones against S. pneumoniae with 1st step mutations of parC
J. Chin, M. Rybak, G. Kaatz (Detroit, US)
Objectives: To compare the resistance (R) potential, AUC/MIC breakpoints, and activity of moxifloxacin (MOX), gemifloxacin (GEM), levofloxacin (LEV) and gatifloxacin (GAT) against 1st Step Mutations of parC (SP1), which have been shown in previous studies to be predisposed to 2nd-step gyrA mutations (MUT) upon re-exposure to older fluoroquinolones (FQ) leading to high-level R.
Methods: A clinical strain of SP1 (KD2138) was evaluated in an in vitro PK/PD model over 96 hours with a starting inoculum of 107 CFU/mL. MOX, GEM, LEV, and GAT were dosed to simulate from 50–800, 60–640, 500–1500, and 100–800 mg QD with their respective fAUC/MIC exposures. Samples were drawn from models to determine emergence/absence of further R. In the event of R, dose and fAUC/MIC exposure were increased until R was prevented or a dose was reached that was not clinically feasible. MICs for resistant organisms (RO) were determined by Etest or microdilution according to CLSI. ROs with elevated MICs post FQ-exposure underwent PCR amplification of the gyrA quinolone resistance-determining region for DNA sequence analysis.
Results: Pre-exposure MICs for LEV, MOX, GEM, & GAT were 2, 0.5, 0.125, & 0.5 μg/mL. R occurred with MICs up to 128 μg/mL from LEV 500–1500 mg QD (fAUC/MIC of 21–62 μg/mL/h). ROs emerged as early as 24 h and beyond with a delay in R in the higher fAUC/MIC (at 96 h with 62 μg/mL/h). This was also consistent with GAT 100–400 mg QD (fAUC/MIC of 12–41 μg/mL/h) with MICs up to 32 μg/mL. R also occurred with GEM 150–320 mg QD (fAUC/MIC 18–32 μg/mL/h), resulting in MIC increases at 1–2 μg/mL. However, R was prevented at 2X the therapeutic dose with GEM 640 mg QD (fAUC/MIC of 64 μg/mL/h) & GAT 800 mg QD (fAUC/MIC of 82 μg/mL/h). In addition, ROs were only derived from MOX 200 mg QD (fAUC/MIC of 24 μg/mL/h) with MICs up to 32 μg/mL, but not from the therapeutic dose of MOX 400 mg QD (fAUC/MIC of 48 μg/mL/h). All MIC increases ≤ 4-fold were associated with efflux. MIC elevations ≥32 μg/mL were associated with the following MUTs in gyrA: S81L, S81F, and E85A.
Conclusions: Additional R resulting in gyrA MUTs in SP1 occurred with LEV and GAT at therapeutic fAUC/MIC but not with MOX. Changes in MICs to GEM were observed, but this was attributed to efflux, not to gyrA mutations.
P2066 Influence of newer fluoroquinolones on phagocytosis and killing of Candida albicans by human polymorphonuclear neutrophils
C. Mörler, T. Grüger, K. Brandenburg, S. Nidermajer, N. Schnitzler, R. Horrè, J. Zündorf (Bonn, DE)
Objectives: The function of the Polymorphonuclear Neutrophils (PMN) is of high relevance for the prognosis of antibiotic treated patients. Especially phagocytosis and intracellular killing of pathogenic microorganisms have large impact on the outcome of infections. Many antiinfectives have been shown to influence the function of these cells. The aim of this project was to achieve more detailed information about the influence on phagocytosis and intracellular killing of pathogenic microorganisms such as Candida albicans by newer fluoroquinolones.
Methods: We analysed the effect of several fluoroquinolones on phagocytosis and killing of C. albicans by human PMN. C. albicans was chosen as pathogen for its naturally high resistance to fluoroquinolones thereby excluding direct influence of the substances tested on the vitality of the microorganism.
Whole blood samples from healthy volunteers were incubated with the fluoroquinolones in concentrations ranging from 0.5 μg/mL up to 1500 μg/mL and compared to a drug-free control. Fluorescent-labeled C. albicans cells were then added in a C. albicans/PMN ratio of at least 2.5:1. Phagocytosis was stopped after incubation of 5, 15, 30 and 60 minutes and the samples were measured by flow cytometry.
Intracellular Killing of C. albicans was analysed after phagocytosis (4 h) by plate counting the washed probes.
Results: Tested fluoroquinolones in clinical relevant concentrations (0.5, 5.0, and 100 μg/mL) did not have significant influence on phagocytosis and killing of C. albicans by human PMN. In contrast higher concentrations (1500 μg/mL) of some newer fluoroquinolones do have negative impact on phagocytosis without influencing viability of PMNs.
Conclusion: Newer fluoroquinolones in clinical relevant concentrations do not affect phagocytosis and killing of Candida albicans. Thus, antiinfective therapy with newer fluoroquinolones seems to have no impact on the main human defence mechanism against the opportunistic pathogenic fungus C. albicans.
P2067 Ciprofloxacin and doxorubicin are substrates of different multidrug resistance-related proteins efflux transporters in J774 macrophages
F. Clesse, M. Mingeot-Leclercq, P. Tulkens, F. Van Bambeke (Brussels, BE)
Objectives: Ciprofloxacin (CIP) and doxorubicin (DOX) are both substrates of the Multidrug Resistance-related Proteins efflux transporters (MRP), a subfamily of the ATP-Binding Cassette transporters (ABC) in eukaryotic cells. This contributes to a reduction of their cellular accumulation, and, therefore, of their intracellular activity (Int J Cancer. 2001;93:107–13; J Antimicrob Chemother. 2003;51:1167–73). Our objective was to determine whether CIP and DOX could share the same MRP transporter in macrophages since this could be the basis of potential, unanticipated drug interactions in cancer patients receiving both drugs either simultaneously (by direct competition for transport) or in succession (trough drug-induced overexpression of the transporter).
Methods: We used J774 macrophages, a cell line that spontaneously expel CIP (Antimicrob Agents Chemother. 2004;48:2673–82; wild-type cells) and a cell line derived thereof by continuous exposure to CIP (CIP-resistant cells) and which overexpresses the corresponding MRP transporter (Antimicrob Agents Chemother. 2006;50:1689–95). Cells were incubated with CIP or DOX alone, or in competition with each other, or in the presence of probenecid (PB), a non-specific inhibitor of MRP transporters. Cell-associated CIP and DOX were measured by fluorimetry and their accumulation recorded as cellular to extracellular concentration ratios (Cc/Ce).
Results: In wild-type cells, (i) PB increased both CIP and DOX Cc/Ce but not to the same extent; (ii) excess of CIP but not of DOX caused an increase in CIP Cc/Ce; (iii) excess of DOX had no effect on CIP and only a marginal effect on DOX Cc/Ce. In CIP-resistant cells, accumulation of CIP was markedly reduced compared to wild type cells (and only partially restored by PB), but not that of DOX (with PB exerting a similar effect to that observed in wild-type cells).
| Condition | Cc/Ce (% control in wild-type cells) |
|||
|---|---|---|---|---|
| wild-type cells |
CIP-resistant cells |
|||
| CIP | DOX | CIP | DOX | |
| Control (drug conc. 50 μM) | 100±1 | 100±8 | 10±2* | 98±10ns |
| + PB (5 mM) | 335±24* | 178±41* | 225±41* | 195±16ns |
| + DOX (total conc. 75 μMa) | 101±2ns | 135±16* | ||
| + ClP (total conc. 500 μMa) | 494±43* | 105±6ns | ||
Statistical analysis: (i) wild-type cells: differences with control values; (ii) CIP-resistant cells: differences with the corresponding values in wild-type cells;
nsnon-significant.
Highest testable conc. (limit of solubility).
p < 0.05
Conclusions: CIP and DOX are substrates of distinct MRP efflux transporters in J774 cells. Drug interactions related to competition for transport or overexpression of CIP transporter are, therefore, unlikely.
P2068 Adverse reactions to imipenem therapy in adult patients with sepsis
D. Ciubotariu, C. Ghiciuc, V. Luca, M. Nechifor (Iasi, RO)
Objective: To assess the incidence of side and adverse effects of imipenem treatment (alone or in associations with other antibacterial drugs) in adult patients with sepsis.
Method: The study was performed on 83 cases (48 male and 35 female, mean age 39.06 years) diagnosed with sepsis and hospitalised in the Infectious Diseases Hospital Iasi, Romania between 1999 and 2006, treated with imipenem-cilastatin (Tienam®). Only patients receiving therapy for at least 7 days were included. Mean therapy duration was 10.78 days and mean doses of imipenem were 0.5g tid. 52 patients (62.65% of total cases) received imipenem as single antibacterial therapy and 31 cases (37.34% of total cases) received imipenem associated with other antibacterial drugs, most frequently used being fluoroquinolones – 14 cases and aminoglycosides – 9 cases. Sepsis aetiological agent was identified in 67.46% of cases (56 patients), most frequently encountered bacteria being Staphylococcus aureus (16 patients, 19.28% cases), Pseudomonas aeruginosa (5 patients, 6.02% cases), Staphylococcus epidermidis and Klebsiella penumoniae (4 patients, 4.81% cases). All identified bacteria were sensitive to imipenem. Adverse reactions were evaluated both by clinical and paraclinical evaluation (main haematological and bio-chemical parameters).
Results: Adverse reactions occurred in 23 patients (27.71% of cases), most patients experiencing more than 1 adverse effect. Most frequently encountered reactions were vomiting (15 patients, 18.07% of cases), abdominal pain and diarrhoea (13 patients, 15.66% of cases), transient AST elevations (11 cases, 13.25% of cases), eosinophilia (6 patients, 7.22% of cases), coetaneous erythema (4 patients, 4.81% of cases) and convulsions (2 patients, 2.4% of cases). In patient treated with imipenem alone, the incidence of digestive effects (nausea and diarrhoea) was lower compared to the group treated with imipenem + fluoroquinolones: 16.07% vs. 35.71%, p < 0.01). In 55 patients (66.26% of cases) a transient black colouring of the teeth occurred. None of patients died due to adverse effects of imipenem.
Conclusions: In patients receiving only imipenem, the incidence of adverse effects was 18.07%, almost the same as that reported by Sanz et al., 2005 and other authors (22%). Association of imipenem with fluoroquinolones significantly increases the risk of adverse reactions. Septic state does not seem to significantly modify the incidence of imipenem adverse reactions.
P2069 Relationship of plasma concentration and changes in the QTc interval in hospitalised patients receiving intravenous moxifloxacin for the treatment of community-acquired pneumonia
M. le Berre, P. Arvis, H. Stass, S. Choudhri (Puteaux, FR; Wuppertal, DE; West Haven, US)
Background: It is well documented that moxifloxacin can prolong the QTc interval in some patients, although not to a clinically relevant degree. In Phase I studies, the magnitude of QTc prolongation showed a linear correlation with plasma moxifloxacin concentration, although with high variability. This analysis was designed to assess the relationship between plasma concentrations and changes in QTc in hospitalised patients with severe community-acquired pneumonia (CAP) requiring intravenous (IV) therapy.
Methods: All patients included in this analysis had CAP with a pneumonia severity index (PSI) Class of III to V and received IV moxifloxacin 400 mg (infused over 60 minutes) once daily. In all patients, 3 standard 12-lead ECG measurements were obtained within 6 hours prior to first infusion, within 30 minutes after the first infusion (day 1) and within 30 minutes after the third infusion (day 3). Plasma moxifloxacin concentrations were determined concomitantly using a HPLC validated method.
Results: 217 moxifloxacin-treated patients were valid for the analysis of QTcB changes between baseline and day 1 and 163 patients for the analysis between baseline and day 3. Mean QTcB (±SD) changes and the concentration effect relationship for the day 1 and day 3 groups are shown in the Table .
| Group | Mean ΔQTcB (ms, ±SD)a | Concentration effect relationship |
||
|---|---|---|---|---|
| Slope [s] | r2 | P | ||
| Day 1 | 10.9±20.8 | −0.0001 | 0.0001 | 0.88 |
| Day 3 | 8.5±27.9 | +0.0014 | 0.0134 | 0.15 |
QT interval corrected for heart rate using Bazett's formula.
Conclusions: The QTcB changes observed were similar to those observed in previous CAP studies. There was no significant correlation between changes in QTcB and plasma moxifloxacin concentrations.
P2070 DNA characteristics of male generative cells during treatment of urogenital chlamydiasis with tetracyclines, macrolides and fluoquinolones
A.G. Zakharenka (Vitebsk, BY)
Objectives: To evaluate the influence of tetracyclines, macrolides and fluoquinolones on chromatingeterogenic test (CT) results in young male patients with urogenital chlamidiasis.
Methods: 99 male patients (20–25 years old) with verified non-complicated urogenital chlamidiasis without initial changes in sperm characteristics were randomly treated with doxycycline (0.1g twice a day 7 days), or azytromycine (1.0 g once), or levofloxacine (0.5 f once a day 7 days). CT was performed before treatment, immediately after the treatment course ending and after 3 and 6 months.
Results: Before treatment the mean CT value was 22.3±4.6% (normal level is up to 30%). Immediately after treatment, this parameter was 56.9±3.2% in doxycycline group, 29.2±2.1% in azytromycine group and 29.1±1.5% in levofloxacine group. In 3 months, the mean CT values in these groups were 39.6±4.2%, 26.8±3.2% and 26.3±4.2%, respectively. In 6 months after the ending of a treatment course the mean CT values were 35.7±3.1% (after doxycycline), 21.3±1.5% (after azytromycine) and 23.4±3.2% (after levofloxacine).
Conclusion: The data obtained permit to conclude that azytromycine and levofloxacine have demonstrated the low level of toxicity to male generative cells and can be the drugs of choice for the treatment of urogenital chlamidiasis in young male patients. Doxycycline was characterised by the high level of male generative cells damage and therefore this fact should be considered during determination of the treatment strategy of this disease in this special population.
P2071 Characteristics of DNA condition from male generative cells under doxycycline and macrolides
U.A. Baliasau (Vitebsk, BY)
Objectives: The study of chromatingeterogenic test (CT) results in sperm of subjects taking doxycycline (D) and some macrolides [erythromycin (E), josamycin (J), and azythromycin (A)] in moderate therapeutic doses.
Methods: 40 healthy volunteers (20–23 years) were studied. Daily dose of D was 0.2; E was administered in dose 0.25 four times per day 10 days; J – 0.5 before meals twice daily 10 days; A – 0.25 before meals once daily 5 days. CT for evaluation of DNA condition in human spermatozoids was performed before treatment (twice), on the 5th and 10th days of treatment, as well as 1 and 2 months after treatment course completion.
Results: CT data analysis revealed that the mean amount of defective spermatozoids before treatment was 18.5+3.5%. By the 5th day of D treatment the index of de-natured DNA was 63.4+6.5% (p < 0.001), by the 10th day – 80.4+7.2% (p < 0.001). 1 month and 2 months after the D treatment course the amount of generative cells with denatured DNA was 54.5+4.6% and 42.5+3.8%, respectively (p < 0.001 in both case). Under E treatment the amount of defective spermatozoids changed as 38.2+4.2 (5th day), 42.4+2.7 (10th day), 35.6+2.5 (after 1 month), and 30.5+4.2% (after 2 months) (p < 0.05 in any case). Under A using the CT results at the same control points were 39.5+4.7, 46.5+3.2, 40.2+3.5, 25.6+3.2% (p < 0.05 in any case); and under J treatment – 18.2+2.5, 20.3+2.5, 17.2+3.2, 15.2+4.2%, respectively (p > 0.05 in any case).
Conclusions: The data obtained permit to conclude that D demonstrates a high level of toxicity to male generative cells. This effect persists during 2 month after the course of D treatment.
New antimicrobials III
P2072 Bactericidal effect of Ltx peptides against Staphylococcus aureus in vitro and in murine skin infection model
R.L. Fischer, N. Frimodt-Møller, W. Stensen, J.S. Svendsen (Copenhagen S, DK; Tromsø, NO)
Objective: Staphylococcus aureus is a major human pathogen with increasing resistance towards important antimicrobial agents. The Ltx micropeptides are a novel group of membrane-lysing antimicrobial compounds, and the aim was to investigate the effect against S. aureus in vitro and in vivo.
Methods: In vitro time-kill curves for Ltx-5 were tested against S. aureus FDA486 at 0.5×, 2× and 8×MIC including vancomycin and dicloxacillin at 8×MIC. The MIC of Ltx-5 was 16 mg/L. A suspension of fresh overnight colonies was grown in broth one hour before test solutions were added. CFU counts were determined at 0, 10, 30 minutes, 1, 2 and 5 hours after addition of the compounds.
Topical treatment with 2% Ltx-7, Ltx-8 or Ltx-10 formulations was investigated in the skin infection model including Fucidin®, Bactroban® and placebo formulation. BALB/c mice were anaesthetized before the fur was removed in a 2–3 cm2 area on the back of the mice. Next razor and tape stripping was used to give a final skin lesion. A suspension of S. aureus FDA486 was applied on the lesion, and treatment was initiated the next day. Mice were treated by topical application over the affected area with about 45g of formulation/mouse in groups of 9 mice. Each mouse was treated for one day at 9 a.m., 12 noon and 3 p.m., followed by sampling of skin biopsy at 6 p.m. The biopsies were homogenised in saline with a piston and the CFU was determined in the sample.
Results: Ltx-5 showed dose-dependent killing in the time-kill curve, and a 5 log reduction in CFU/mL was observed at 8×MIC after 2 hours. The killing of Ltx-5 at 2×MIC was comparable to 8×MIC for vancomycin and dicloxacillin. A bactericidal effect of the Ltx peptides was also observed in the skin infection model. The mean log reduction in CFU/mouse for the Ltx-treated groups compared to placebo was in the range of 3.1–4.8 after one day treatment. The corresponding reduction for Fucidin® was 1.2–1.3 log CFU/mouse, and for Bactroban® 0.7–0.9 log CFU/mouse.
Conclusion: A bactericidal effect of the Ltx peptides was observed in the time-kill curves at 2×MIC and 8×MIC. In addition, topical treatment with the Ltx petides showed excellent killing of S. aureus after one day treatment.
P2073 The effect of efflux pump inhibitor on MIC values of nalidixic acit, flouroquinolones and macrolide of multidrug-resistant Escherichia coli
Y. Tezer, I.T. Filik, O. Memikoglu, H. Kurt (Ankara, TR)
Objective: Efflux pumps are found in all living cells and drop toxic substances and antibiotics from inside to out. As a result, pump system is responsible for multidrug resistance. So we aimed to investigte the effect of this system on some antibiotics minimal inhibitor concentration (MIC) values.
Methods: In this study, we investigated phenyl-arginin-beta-napthylamide (PA-beta-N) effect as an efflux pump inhibitor on multidrug resistant Escherichia coli. For this study during 6 months periods, 100 clinical isolates which were multiple drug resistant pattern that had been determined via disc diffusion methods collected. With agar dilution method, MIC values of four antibiotics which were nalidixic acit, ciprofloxacin, levofloxacin, and erytromycin were determined.
After that, having efflux pump had been searched by organic solvent tolerance test. Then we examined nalidixic acit, flouroquinolones and macrolide MIC values via broth dilution method with PA-beta-N and without it.
After broth dilution method, different concentrations of PA-beta-N were studied in tolerant isolates. Results were analysed with Mann-Whitney U tests.
Results: Fifty five percent of isolates were tolerant to organic solvents. It means that, efflux pump was overexpressed in these isolates. For levofloxacin and erytromycin MIC values were decreased with PA-beta-N. On the other hand there were no change on ciprofloxacin and nalidixic acit MIC values. For ciprofloxacin and nalididixic acit there were no significant statistical value on MIC values. But on the other hand for levofloxacin and erytromycin there were significant statistical change on MIC values.
In addition PA-beta-N has homogenous effect in different concentrations and no concentration dependent effect was determined.
Conclusions: Effect of efflux pump on multidrug resistance mechanisms was rarely reported from our country and all over the world. So we thought that studies that made by specific inhibitors of efflux pump, like using β-lactam/β-lactamases might be innovator in clinical applications.
P2074 In vitro and in vivo antibacterial activity of novel nitrile-containing fluoroquinolones
M.D. Huband, P.J. Pagano, R.W. Murray, J.W. Gage, G. Gibson, S.T. Murphy, K.R. Marotti (Ann Arbor, US)
Objectives: The continuing emergence of resistance in Gram-positive bacterial species including multi-drug resistant Streptococcus pneumoniae, vancomycin-R Enterococci (VRE), ciprofloxacin-R MRSA (CRMRSA), and Vancomycin-intermediate Staphylococcus aureus (VISA) has created the need for new antibacterial compounds. Two new nitrile-containing fluoroquinolones (#1966 and #9402) were developed to treat infections caused by susceptible and multi-drug resistant Gram-positive and fastidious Gram-negative bacterial strains. This study investigated the in vitro antimicrobial activity of 1966, 9402, moxifloxacin, and conventional antibacterials against 1204 geographically diverse recent bacterial clinical isolates. Compound efficacy was also evaluated by in vivo testing (PD50s).
Methods: Microbroth dilution MICs followed CLSI guidelines. In vivo testing was performed in CD1 female mice using protocols approved by the Pfizer Animal Use Committee in compliance with NIH guidelines for proper care and use of laboratory animals.
Results: See the table . Both of the nitrile fluoroquinolones were highly active against Gram-positive (MIC90s 0.5–4 μg/mL) and fastidious Gram-negative strains (MIC90s 0.03–0.06 ug/mL). This potent activity carried over to efficacy in Streptococcus pneumoniae animal infection models. Acute systemic PD50s (oral dosing) were 3.2 and 1.1 mg/kg, respectively, and pneumonia model PD50s (oral dosing) were 5.93 and 5.48 mg/kg.
| Organism (# strains) | MIC90 (μg/mL) |
||
|---|---|---|---|
| #1966 | #9402 | Moxifloxacin | |
| S. aureus CRMRSA (34) | 1 | 1 | 8 |
| S. aureus VISAa (4) | 2 | 0.5 | 4 |
| S. epidermidis ORSE (23) | 2 | 1 | 32 |
| S. pneumoniae FQ-R (26) | 0.5 | 0.5 | 4 |
| E. faecalis (13) | 0.5 | 0.5 | 8 |
| E. faecalis Van A (14) | 1 | 0.5 | 16 |
| E. faecalis Van B (22) | 1 | 0.5 | 16 |
| E. faecium Van A (45) | 4 | 4 | 32 |
| Moraxella catarrhalis (30) | 0.06 | 0.06 | 0.06 |
| Haemophilus influenzae (67) | 0.03 | 0.06 | 0.03 |
MIC50 (μg/mL).
Conclusion: This study confirms both the in vitro antibacterial potency and in vivo efficacy of 1966 and 9402 against clinically relevant Gram-positive and fastidious Gram-negative organisms.
P2075 Screening for medicinal plants with broad spectrum of antibacterial activity
S.P. Voravuthikunchai, S. Limsuwan, S. Subhadhirasakul (Hatyai, Songkla, TH)
Objectives: Interest in the study of medicinal plants as a source of pharmacologically active compounds has increased worldwide. The objective of this study was to screen for effective plants from 31 plant species commonly used in Thai traditional medicine for bacterial infections.
Methods: Preliminary screening was performed using agar disc diffusion method. Minimal inhibitory concentration and Minimal bactericidal concentration were subsequently carried out on effective plants.
Results: Agar disc diffusion method showed that most plants were more active against Gram-positive than Gram-negative bacteria. Streptococcus pyogenes was the most sensitive organism inhibited by nearly all of the extracts (97.6%), followed by Staphylococcus aureus (61.0%), and Bacillus cereus (63.4%). We report two plants with very broad spectrum of activity. The ethanolic extracts from Quercus infectoria demonstrated significant activity against all important pathogens including Acinetobacter baumannii, Bacillus cereus, Enterococcus faecalis, Escherichia coli, Klebsiella pneumoniae, Listeria monocytogenes, Pseudomonas aeruginosa, Salmonella spp., Shigella flexneri, Staphylococcus aureus, Streptococcus mutans, and Streptococcus pyogenes. It inhibited the growth of all pathogens with the MIC values of 62.5 to 1000 ug/mL and the MBC values of 125 to >1000 μg/mL. The ethanolic extracts from Piper betle showed antibacterial activity against almost all species, except Enterococcus faecalis. It showed antibacterial activity with the same MIC and MBC values (125–500 μg/mL). The leaf extract of Rhodomyrtus tomentosa showed extremely good antibacterial activity on most Gram-positive bacteria with the MICs and MBCs ranging from 3.9 to 15.6 and 7.8 to 125 μg/mL.
Conclusion: Further studies on these plant species may result in discovery of novel natural medicine against pathogenic bacteria.
Acknowledgement: This work was supported by the Thailand Research Fund: Royal Golden Jubilee (PHD/0029/2548), Fiscal year 2005–2010, the Thailand Research Fund: Basic Research (BRG4880021), Fiscal year 2005–2008, and Natural Products Research Unit, Faculty of Science, Prince of Songkla University, Thailand.
P2076 Linezolid versus a glycopeptide or β-lactam for treatment of Gram-positive bacterial infections: a meta-analysis of randomised controlled trials
M.E. Falagas, I.I. Siempos, K.Z. Vardakas (Athens, GR)
Objectives: During the last decade, several new antibiotics have been released to the market for the treatment of patients with infections due to Gram-positive cocci resistant to traditionally used antibiotics, including glycopeptides. Among these antibiotics, linezolid has been reported to have excellent pharmacokinetics and effectiveness. We performed a meta-analysis of randomised controlled trials (RCTs) to further clarify the therapeutic role of linezolid.
Methods: Our data sources for relevant RCTS were PubMed, Current Contents, and Cochrane Central. A total of 12 RCTs comparing linezolid with vancomycin (6 RCTs), teicoplanin (2 RCTs), and β-lactams (amoxicillin/clavulanic acid, cephadroxil, and ceftriaxone in 2, 1, and 1 RCT, respectively) that studied 6,093 patients were included in our analysis.
Results: Two reviewers independently extracted data from published RCTs. Overall linezolid, as empirical treatment, was more effective regarding treatment success than the comparator antibiotics (OR = 1.41, 95%CI 1.11–1.81). Mortality was similar between the compared antibiotics (OR = 0.97, 95% CI 0.79–1.19). Linezolid was more effective in the subset of patients with skin and soft tissue infections (OR = 1.67, 95%CI 1.31–2.12) and bacteraemia (OR = 2.18, 95% CI 1.10–4.29). However, there was no difference in treatment success for patients with pneumonia (OR = 1.03, 95%CI 0.75–1.42). Although treatment with linezolid was not associated with more adverse effects in general (OR = 1.40, 95% CI 0.95–2.05), more episodes of thrombocytopenia were recorded in patients receiving this antibiotic (OR = 9.25, 95%CI 3.52–25.76).
Conclusion: Linezolid was more effective than a glycopeptide or a β-lactam for the empirical treatment of patients with skin and soft tissue infections and bacteraemia due to Gram-positive cocci. However, the lack of any benefit in the treatment of patients with pneumonia, the same all-cause mortality, and the higher probability of thrombocytopenia are major limitations of the antibiotic that should be taken under consideration and limit the use of linezolid to specific patient populations or infections that are difficult to treat with other antibiotics.
P2077 Bactericidal activity of ertapenem against A. baumannii
T. Schneiders, S. Amyes (Edinburgh, UK)
Ertapenem is a new carbapenem targetted for clinical use against fermenters such as E. coli and Klebsiella spp. but with reduced or weak activity against non-fermenting Gram-negative bacteria such as Acinetobacter baumannii and Pseudomonas aeruginosa. Both Acinetobacter spp. and Pseudomonas spp. are established pathogens in the hospital setting and are resistant to a range of antibiotics which include the carbapenems. Since the carbapenems (e.g. meropenem) represent the last line of antibiotics against both Acinetobacter and Pseudomonas spp., it is imperative that this antibiotic class is preserved. Due to its reduced activity the introduction and use of ertapenem, suggests a potential for resistance development in these bacteria. The combination of increasingly carbapenem-resistant bacteria and a less efficacious antibiotic such as ertapenem against a rapidly evolving bacterial population could result in the emergence of completely untreatable bacteria. The efficacy of ertapenem against three characterised strains of A. baumannii was determined using time-kill studies at 4 times the MIC. The three strains of A. baumannii were ATCC 19606, a carbapenem sensitive clinical A. baumannii isolate, SD16, and another carbapenem sensitive A. baumannii clinical isolate, E13, which harbours a mutS mutation. As a comparator antibiotic, meropenem, was used. Against ATCC 19606 and at 4 times the MIC, ertapenem reduced the viable count by 2 log10 reduction in 3 hours and extended the killing to greater than 4 log10 after 24 hours. Against the sensitive clinical isolate SD16, ertapenem exerted a 3 log10 reduction in 3 hours and after 24 hours reduced the viable count to a 4 log10 reduction. Of note, the levels of reduction in viable cell counts by ertapenem were comparable to meropenem. Interestingly against the mutS mutant, E13, the viable counts obtained with either meropenem or ertapenem, showed little or no reduction over the 3 hour period which was in contrast to that observed with ATCC19606 and SD16. However a reduction to 4 log10 was observed with both ertapenem and meropenem after 24hrs. The results obtained here clearly demonstrate that ertapenem inhibits non-fermenters such as A. baumannii with similar efficacy as meropenem. These findings are significant as it is the first to describe the effect ertapenem has on A. baumannii and serves to re-examine the role of ertapenem either as a potential therapeutic option or a selector of resistance.
P2078 Novel acyclic substituted (pyridin-3-yl)phenyl oxazolidinones: antibacterial agents with reduced activity against monoamine oxidase A and improved solubility
F. Reck, F. Zhou, G. Zainoun, D. Carcanague, C. Eyermann, M. Gravestock (Waltham, US)
Objectives: Oxazolidinones bearing a (pyridin-3-yl)phenyl moiety (e.g. 1) generally show increased antibacterial activity compared to linezolid, but suffer from potent monoamine oxidase A (MAO-A) inhibition and low solubility. New analogs with acyclic substituents on the pyridyl moiety were prepared and their antibacterial activity, MAO-A inhibition, and solubility were determined.
Methods: Compounds were synthesized by Suzuki Coupling of pyridyl-3-bromides with oxazolidinone phenylboronic acids. Antibacterial activity, MAO-A inhibition, and solubility were evaluated in vitro. A homology model of MAO-A was built and used to rationalise the SAR.
Results: Antibacterial potency for many analogs was excellent, with MICs for linezolid resistant Streptococcus pneumoniae for compound 2 and 3 at 1 and 0.5 ug/mL respectively. Activity against Staphylococcus aureus was also very good, with MICs of 0.25 ug/mL for 2 and 3. In addition, compound 2 displayed good activity against Haemophilus influenzae, with an MIC of 1 ug/mL. In general, bulkier substituents yielded significantly reduced MAO-A inhibition (Ki = 40 uM for 2; Ki = 19 uM for 3) relative to the unsubstituted parent 1 (Ki < 0.3 uM). Solubility was enhanced with incorporation of polar groups.
Conclusion: Acyclic 6-substituted pyridin-3-yl phenyl oxazolidinones with sufficiently bulky substituent such as 2 and 3 are potent antibacterial compounds against Gram-positive organisms, including linezolid resistant Streptococcus pneumoniae. The properties of these compounds include reduced MAO-A activity and increased solubility.

P2079 Synthesis and evaluation of novel (pyridin-3-yl) phenyl oxazolidinones with improved physicochemical properties
D. Carcanague, M. Gravestock, S. Hauck, F. Reck, G. Zainoun (Waltham, US)
Objectives: Isoxazolino oxazolidinone 1 has potent microbiological activity against Gram-positive pathogens, however its in vivo efficacy is limited by low bioavailability due to high crystallinity and poor aqueous solubility. Ester prodrugs of 1 with improved DMPK properties have been described previously. Our further studies of this compound series focused on the synthesis of novel derivatives that would retain microbiological potency but exhibit improved solubility. In order to achieve this goal, the hydroxyl group of 1 was replaced with various amines or ethers.
Methods: Compounds were synthesized from 1 using standard functional group manipulations. The antibacterial activity and solubility of these compounds were measured.
Results: Good antibacterial activity, including against linezolid-resistant strains, was retained in a number of new compounds with enhanced solubility, such as the N-methyl amine 16. MICs for linezolid resistant Streptococcus pneumoniae were in the range 0.5–1 mg/L for most compounds. Excellent activity against Staphylococcus aureus and linezolid-susceptible Streptococcus pneumoniae was observed, with MICs for compound 16 of 0.5 mg/L and <0.06 mg/L respectively.

Conclusion: Novel ether or amine isoxazolino oxazolidinone analogues of 1 show potent antibacterial activity against Gram-positive pathogens and significantly improved solubility.
P2080 Microbiological activity and efficacy of novel (pyridin-3-yl) phenyl oxazolidinones
R. Illingworth, K. Coleman, J. Jones (Waltham, US)
Objectives: We describe the in vitro antibacterial activity and in vivo efficacy of a novel series of oxazolidinones with potent activity against linezolid-susceptible and –resistant Gram-positive bacteria.
Methods: MICs were determined using CLSI standard broth microdilution methods on a total of 280 recent clinical isolates with a multitude of resistance markers. In vivo efficacy against S. pneumoniae was determined in a mouse lung infection model and against S. aureus in a neutropenic mouse thigh infection model. To determine the pharmacodynamic index of the series a dose fractionation study was carried out with compound II in the S. pneumoniae lung model where total daily doses up to 160 mg/kg p.o. were fractionated over 24 hours.
Results: Analogues in the series were consistently more potent than linezolid against all Gram-positive species tested and maintained good activity against linezolid-resistant isolates. Improved activity was also observed with most derivatives against strictly anaerobic Gram-negative bacteria such as B. fragilis, but activity was weak or absent against other Gram-negative bacteria, such as H. influenzae and E. coli. Compounds I and II yielded a >2 log reduction in cfu vs. untreated control at 20 mg/kg/day p.o. in the mouse S. pneumoniae lung infection model. Compound II yielded a 2-log reduction at 40 mg/kg in the mouse S. aureus thigh infection model. The PD index associated with efficacy was AUC/MIC, consistent with data published for linezolid.

Conclusions: The (pyridin-3-yl) phenyl oxazolidinones reported here were ≥ 4-fold more potent than linezolid against all Gram-positive organisms and ≥ 32-fold more potent against strains resistant to linezolid. Activity against most Gram-negative species was poor, but against B. fragilis, most analogues were ≥ 4-fold more potent than linezolid. Analogs in this series exhibited good in vivo efficacy against S. pneumoniae and S. aureus in mouse models of infection.
P2081 Evaluation of antibacterial activity and biofilm formation in Klebsiella pneumoniae in contact with essential oil and alcoholic extract of cumin seed (Cuminum cyminum)
S. Derakhshan, M. Sattari, M. Bigdeli (Tehran, IR)
Introduction: Klebsiella pneumoniae has special importance in nosocomial infection. Because of the acquisition of multiresistant plasmids, these organisms are resistant to a number of antibiotics, including extended-spectrum cephalosporins and aminoglycosides.
Methods: In this study essential oil of cumin seeds (Cuminum cyminum) obtained in a cleavenger system by hydrodistillation method. Antibacterial activity of essential oil and alcoholic extract of cumin against Klebsiella pneumoniae ATCC13883 and clinical Klebsiella pneumoniae (ceftazidime-resistant strain) were evaluated on the minimum inhibitory concentration by the broth-dilution method. Synergistic or antagonistic effect with antibiotic disks were tested in agar media involving sub-MIC concentration of oil and alcoholic extract. Biofilm assay were performed by colorimetric method as described by O'Toole in 1998.
Results: The MIC was 1/800 fold of essential oil for K. pneumoniae ATCC 13883 and 1/200 for clinical K. pneumoniae. Also MIC was 1/32 fold of alcoholic extract for two strain in sub-MIC concentration, synergistic effect was seen between ciprofloxacin and oil (significant increasing of inhibition zone diameter) and antagonistic effect was seen between trimethoprim and sulfamethoxazol (reversion of colonies around the antibiotic disk) for the clinical K. pneumoniae. The decrease or prevention of biofilm formation occurred when oil concentration increased. Alcoholic extract was low effect in antibiotical activity and biofilm formation. The oil was analysed by GC and GC/MS. Main constituent in oil of Cuminum cyminum is cuminaldehyde (4 isopropylbenzaldehyde).
Discussion: The results of this study suggested that the essential oil and alcoholic extract of cumin seed could be used in medicinal industries (desinfectant or antiseptic), but in-vivo tests are required to ascertain fully their medicinal properties and potential toxicity.
P2082 Galanin-message-associated peptide suppresses hyphal growth of Candida albicans
I. Rauch, L. Lundström, M. Hell, B. Kofler (Salzburg, AT; Stockholm, SE)
Objectives: Several epithelial surfaces are continuously exposed to potential pathogens but rarely become infected, because specific secretions protect them. Antimicrobial peptides (AMPs) participate in this innate immune response by providing a rapid first line defence against infection. Neuropeptides are expressed by neuronal and non-neuronal tissues in various organs including the skin, which constitutes the first barrier against external stress. These neuropeptides exert potent growth- and immunomodulatory effects and few have also been shown to possess antimicrobial activity. Recently, we have gained evidence that galanin precursor peptide mRNA (ppGAL) is one of the few neuropeptide mRNAs expressed in keratinocytes of the human skin. The galanin gene encodes a precursor peptide that is processed into the 29 amino acid peptide galanin and the 59 amino acid galanin-message-associated peptide (GMAP). Our objectives where to test whether one of these two peptides possesses antimicrobial activity.
Methods: A microbial cell viability assay and microscopical evaluation were used to examine influences on microbial growth. The effect of co-culture with C. albicans on ppGAL expression in human cultured keratinocytes was assessed by quantitative real time PCR.
Results: We were able to demonstrate that GMAP possesses growth-inhibiting activity against C. albicans, but not Gram-positive and Gram-negative bacteria. GMAP inhibits the yeast-to-hyphal transition, which is one of the most important virulence factors of C. albicans. After testing shorter variants of GMAP, we identified GMAP 16–41 as the essential part of the peptide for this antifungal activity. Co-culture of keratinocytes with C. albicans resulted in a 2-fold upregulation of ppGAL expression.
Conclusion: These studies establish GMAP as a new component of the innate immune system, which has implications for prophylactic and therapeutic strategies of Candida infections.
Supported by grants of the Jubiläumsfonds (Nr. 11996) and the Paracelsus Private Medical University (Nr.05/01/003).
P2083 Emergence of linezolid resistance of S. epidermidis in a critical care unit
L. Martínez, M. Treviño, S. Cortizo, M. Peñalver, M. García-Zabarte, F. Pardo, B. Regueiro (Santiago de Compostela, ES)
Objectives: Linezolid is the first member of the oxazolidinone class of antimicrobials, approved for clinical use in 2000, particularly appropriate for treating multiresistant staphylococci. In the present work, we report four cases of linezolid-resistant coagulase-negative staphylococci (CoNS).
Methods: CoNS strains were isolated from blood and/or wound cultures from patients belonging to the same clinical unit (postsurgical care unit) from July to October, 2006. Identification and susceptibility testing of the strains were performed by the VITEK 2 System (bioMérieux®, France) and E-test (ABBiodisk®, Sweden).
Results: All strains showed slower growth than linezolid-sensitive CoNS strains, were identified as Staphylococcus epidermidis and were sensitive to glycopeptides, quinupristin-dalfopristin, fosfomycin, tetracycline, erythromycin, clindamycin and moxifloxacin, and resistant to oxacillin, rifampicin, fusidic acid, trimethoprim-sulfamethoxazole, ciprofloxacin, gentamicin and tobramycin. CMI range for linezolid was 8–16 mg/L. Only one of these patients had been previously treated with linezolid.
Conclusions: Linezolid resistance in CoNS has emerged at our institution. We think that, probably, person-to-person spread of linezolid-resistant CoNS has led to establishment of skin colonisation with the strain, however, genotypical characterisation methods must be applied to comfirm this approach. In any case, in vitro susceptibility testing of linezolid is advisable for accurate therapy of CoNS in medical units where linezolid is habitually used.
P2084 Antimicrobial activity of apidaecin peptides
P. Czihal, A. Nimptsch, U. Möllmann, P. Zipfel, R. Hoffmann (Leipzig, Jena, DE)
In recent years there was a dramatical increase in bacteria strains resistance to one or even several antibiotics. Thus, the development of antimicrobial compounds with novel modes of action is a major focus of current pharmaceutical research. A very interesting and promising approach relies on antibacterial peptides, because bacteria do not develop any resistance to these antimicrobial peptide families. One of the most promising among these families are the short, proline-rich antibacterial peptides originally isolated from insects, such as apidaecin, drosocin, formaecin, and pyrrhocoricin. These peptides represent a viable treatment option for the major pathogens in urinary tract infections, that is, E. coli and K. pneumoniae, causing 90–95% of all urinary tract infections.
Based on the native sequences of apidaecin 1a and apidaecin 1b, we have identified all residues important for the antibacterial activity with an alanine scan. In a second round, the identified positions were selectively modified with other alpha-amino acids and unnatural amino acid derivatives to increase the antibacterial activity. Furthermore, the protease resistance was increased to elongate the peptide activity in blood. All peptides were synthesized on solid-phase using the Fmoc/tBustrategy and purified by reversed phase HPLC. The activities of the peptides were determined by an agar diffusion assay and the minimal inhibition concentration (MIC) was determined on a microtiter plate for pathogenic E. coli strains (e.g. E. coli 1103) and Klebsiella pneumoniae ssp. pneumoniae 11678. The first results on the antimicrobial activities and protease resistances of the new apidaecin analogs will be presented.
P2085 A novel antimicrobial peptide with anti-MRSA activity
R. Aunpad, K. Na-Bangchang (Pathumthani, TH)
Objectives: To search and characterise a new antimicrobial peptide against MRSA.
Methods: Forty-three different water and soil samples were collected from three different areas from Thailand. The single colony obtained from each sample was screened for its antagonistic activity against the indicator strain, methicillin resistant Staphylococcus aureus (MRSA) by co-culture method. The agar-well diffusion method was used to assess the antimicrobial activity spectrum of 20 selected bacterial strains against 20 Gram-positive and Gram-negative test microorganisms. The agent containing antimicrobial peptide (ACAP) prepared from the culture supernatant of one strain, WAPB4, was tested for its antimicrobial spectrum, enzyme sensitivity, heat and pH stability and mode of action. The antimicrobial peptide was purified from ACAP by a three-step procedure: solvent extraction, solid phase extraction and followed by reversed phase chromatography. The molecular mass was determined by mass spectrometry.
Results: A total of 20 water and soil isolated bacteria can inhibit the growth of MRSA. It was also found that three Gram-positive isolated bacteria strain WARY9–1, WARY6–6 and WASM9–25 showed a broad range of inhibition. Two Gram-negative bacteria strain WARY9–10 and WARY7–4 are active against both Gram-positive and Gram-negative bacteria. One strain, WAPB4, identified as Bacillus pumilus showed remarkable antibacterial activity against MRSA and VRE. The antimicrobial peptide produced by WAPB4 was heat stable up to 121°C, 15 min and active within the pH range of 3–9. Its activity disappeared when treated with pronase E, chymotrypsin and trypsin demonstrating its proteinaceous nature. ACAP showed bacteriostatic effect to MRSA in vitro. The antimicrobial peptide could be purified by a three-step procedure and the molecular mass as determined by mass spectrometry was 1994.62 Dalton.
Conclusion: Bacillus pumilus strain WAPB4 produced a new antimicrobial peptide with broad spectrum antibacterial activity including MRSA and VRE. The compound is thermal and pH stable and has potential for use as an alternative antibacterial agent for the treatment of infection with MRSA and VRE.
P2086 Clinical efficacy of retapamulin, 1% ointment, in subjects with antibiotic-resistant Staphylococcus aureus: pooled Phase III results from Europe, Costa Rica, India, Mexico, Peru, Russia and South Africa
A. Oranje, O. Chosidow, N. Scangarella, M. Twynholm, R. Shawar, T. Erb (Rotterdam, NL; Paris, FR; Collegeville, US; Greenford, UK)
Background: Antimicrobial resistance to the oral and topical agents used in treating skin infections is a growing problem globally, with specific regional differences in the prevalence and strains of resistant bacteria. For instance, 33–50% of S. aureus isolates from European subjects with skin infections were recently found to be resistant to fusidic acid [1, 2]. The problem of resistance demands new antibiotic choices. Retapamulin is the first antibiotic of the novel pleuromutilin class developed for the treatment of uncomplicated skin and soft tissue infections. It has a unique mode of action, no known target-specific cross resistance and a low propensity for resistance development in vitro. It has excellent in vitro activity against skin pathogens, including those resistant to existing antibiotics. Retapamulin, 1% ointment, has demonstrated good clinical efficacy in the treatment of adults and children with impetigo, secondarily infected dermatitis (SID) and secondarily infected traumatic lesions (SITL).
Objectives: To determine the clinical efficacy of retapamulin against antibiotic-resistant S. aureus recovered from European and international subjects in Phase III studies.
Methods: Clinical efficacy data for retapamulin were pooled from five worldwide studies: 469 (vs placebo) and 224 (vs topical fusidic acid) in subjects with impetigo; 032 in those with SID (vs oral cephalexin) and 030A and 030B in SITL (vs oral cephalexin). Only subjects from Europe (Austria, France, Germany, Greece, Italy, Poland, Spain, The Netherlands and UK) and Costa Rica, India, Mexico, Peru, Russia and South Africa were included in the analysis. In each study, subjects received retapamulin, 1% ointment, twice daily for 5 days, or placebo/comparator (2:1 randomisation). A total of 1527 subjects (ITT population) were considered.
Results: Pooled data from European and international subjects for the three indications show excellent clinical efficacy in subjects with fusidic acid-resistant (FusR), mupirocin-resistant (MupR) or methicillin-resistant (MR) S. aureus (SA) isolates.
Clinical efficacy of retapamulin by baseline pathogen (ITT population)
| Clinical success % (n/N)a |
|||
|---|---|---|---|
| MupRSAb | IFusRSAb | MRSAb | |
| Europe | 100 (2/2) | 92.3 (24/26) | 80.0 (4/5) |
| International | 92.9 (13/14) | 100 (6/6) | 92.0 (23/25) |
| Overall | 93.8 (15/16) | 93.8 (30/32) | 90.0 (27/30) |
n/N: number of clinical success/number of subjects with pathogen at baseline.
Resistance defined as MIC ≥ 8 g/L for mupirocin and ≥4 g/L for fusidic acid. Oxacillin disk test used to define methicillin resistance.
Conclusions: Retapamulin achieved high rates of clinical success against FusRSA, MupRSA or MRSA infections. Retapamulin, 1% ointment, is therefore a highly effective new treatment option for uncomplicated skin infections, including those involving pathogens resistant to current antibacterials.
P2087 Spectrum and activity of doripenem, an investigational carbapenem, tested against bacterial pathogens recovered from patients hospitalised with pneumonia (Europe; 2004–2006)
T. Fritsche, H. Sader, P. Strabala, R. Jones (North Liberty, US)
Objectives: To summarise the activity of doripenem (DOR), an investigational broad-spectrum parenteral carbapenem (CARB) in late stage clinical development, against leading bacterial pathogens recovered from European patients with HAP. Emerging resistance (R) among hospital-acquired pneumonia (HAP) pathogens, especially Gram-negative bacilli, compromises patient management and contributes to excess morbidity and mortality.
Methods: A total of 2,127 consecutive, non-duplicate isolates determined as the cause of pneumonia in hospitalised patients were submitted from 24 medical centres in Europe (2004–2006). Included HAP pathogens are summarised in the Table . Susceptibility (S) testing of DOR and comparator agents was performed using current CLSI methods and interpretive criteria, including those for characterisation of ESBL-phenotypes.
| Organism (no. tested) | DOR MIC (mg/L) |
|||
|---|---|---|---|---|
| 50% | 90% | Range | % ≤2/≤4 | |
| P. aeruginosa (PSA; 488) | 0.5 | 8 | ≤0.06–>8 | 72/85 |
| S. aureus (SA; Oxacillin [OXA]-S; 266) | ≤0.06 | ≤0.06 | ≤0.06–0.5 | 100/100 |
| E. coli (EC; 241) | ≤0.06 | ≤0.06 | ≤0.06–0.25 | 100/100 |
| Klebsiella spp. (KSP; 229) | ≤0.06 | ≤0.06 | ≤0.06–>8 | >99/100 |
| Enterobacter spp. (ESP; 146) | ≤0.06 | 0.12 | ≤0.06–1 | 100/100 |
| Acinetobacter spp. (ASP; 134) | 8 | >8 | ≤0.06–>8 | 38/46 |
| Serratia spp. (SER; 75) | 0.12 | 0.25 | ≤0.06–0.5 | 100/100 |
Results: Results for DOR are in the Table. Ranking of the top-eight occurring HAP isolates and key R characteristics were (see Table): PSA (21.5% meropenem-non-susceptible [MER-NS]) > SA (441 isolates; 39.7% OXA-R) > EC (11.2% ESBL) > KSP (31.4% ESBL) > ESP (31.5% ceftazidime-R) > ASP (66% MER-NS) > SER > S. maltophilia (SM; 68 isolates; not shown). OXA-R SA and SM are inherently R to CARB and are not discussed further. Overall, DOR inhibited 90.8% of the 7 top-ranked pathogens within its spectrum of activity at ≤ 4 mg/L (equivalent to breakpoints of peer CARB) compared with 86.3% for imipenem (IPM) and 88.6% for MER. Elevated DOR, MER and IPM MIC values were detected among PSA (MIC50/90 values, 0.5/8; 1/>8; and 1/>8 mg/L, respectively) and ASP (MIC50/90 values, 8/>8; 4/>8; and 8/>8 mg/L). Strains of metallo-β-lactamase producing PSA (IMP, VIM series) were detected in Italy, Greece and Turkey.
Conclusion: The described prevalences of R phenotypes among contemporary European HAP pathogens increasingly complicates therapeutic management decisions. DOR is a promising CARB that may represent an important choice among broad-spectrum agents for this indication (HAP) especially given its enhanced activity against ESBLs and PSA.
P2088 Variations and trends in the activity of doripenem and other broad-spectrum agents against leading bacterial pathogens: results from a European Surveillance Program (2003–2006)
T. Fritsche, H. Sader, P. Strabala, R. Jones (North Liberty, US)
Objectives: To summarise the results of a European-focused surveillance programme comparing activity of doripenem (DOR) and other agents tested against leading contemporary pathogens. DOR is a broad-spectrum investigational parenteral carbapenem (CARB) in late stage clinical development that displays enhanced activity against P. aeruginosa (PSA) compared to other marketed CARB. As we continue to evaluate DOR, regional data assessing resistance (R) patterns of targeted pathogens are needed.
Methods: Non-duplicate bacterial isolates (27,689; 54.5% bloodstream; 21.6% respiratory tract; 8.3% skin and skin structure; others, 15.6%) were collected from 24 medical centres in Europe during 2003–2006. Identifications were confirmed and all isolates were susceptibility (S) tested using CLSI broth microdilution methods and interpretive criteria against DOR, meropenem (MEM), imipenem (IPM) and comparators.
Results: At MIC values of 0.25 mg/L for SPN, 0.5 mg/L for BHS and HI, and 4 mg/L for all others (equivalent to peer agents), DOR inhibited 95.7% of the tabulated pathogens recovered from all sources. DOR was broadly active against staphylococci, streptococci, and HI, and at least 2-fold more potent against PSA than either MEM or IPM (MIC90/% ≤4 mg/L: 8/85, >8/81 and >8/76, respectively). Only polymyxin B (>99% S) and amikacin (89%) provided greater PSA coverage than DOR. While ESBL-phenotype rates varied considerably between 2003 and 2006 (EC, 4.9–8.3%; KSP, 17.8–26.9%), DOR inhibited 100% of strains confirmed as ESBL-producers at ≤2 mg/L.
| Organism (no. tested) | MIC (mg/L) |
Cum. % inhibited at MIC (mg/L) |
||||
|---|---|---|---|---|---|---|
| 50% | 90% | ≤1 | 2 | 4 | 8 | |
| S. aureus (OXA-S; 4,441) | ≤0.06 | ≤ | 0.06 | >99 | 100 | |
| CoNSa (OXA-S; 588) | ≤0.06 | ≤0.06 | 100 | |||
| BHSa (836) | ≤0.06 | ≤0.06 | 100 | |||
| S. pneumoniae (SPN; 2,146) | ≤0.06 | 0.5 | >99 | 100 | ||
| H. influenzae (HI; 1,594) | 0.06 | 0.25 | >99 | 100 | ||
| E. coli (EC; 4,979) | ≤0.06 | ≤0.06 | 100 | |||
| Klebsiella spp. (KSP; 1,621) | ≤0.06 | ≤0.06 | 98 | 99 | >99 | >99 |
| Enterobacter spp. (1,010) | ≤0.06 | 0.12 | 98 | 99 | >99 | >99 |
| P. aeruginosa (PSA; 1,934) | 0.5 | 8 | 69 | 77 | 85 | 92 |
| Acinetobacter spp. (568) | 2 | >8 | 48 | 57 | 66 | 81 |
CoNS, coagulase-negative staphylococci; BHS, beta-haemolytic streptococci.
Conclusions: Emerging global R has created a critical need for accelerated development and introduction of new antimicrobials. DOR inhibited 100% of ESBL-producing strains and was more potent than other CARB against PSA. DOR is a promising CARB displaying a broad-spectrum against most common hospital pathogens, especially Enterobacteriaceae and non-fermentative bacilli.
P2089 Extending structure-activity-relationship studies of bioactive natural products by novel iminonitroso cycloaddition chemistry
M. Miller, U. Moellmann, F. Li, B. Yang, T. Zoellner, P. Gebhardt, H. Dahse, P. Miller (Notre Dame, US; Jena, DE)
Objectives: Healthcare problems caused by multidrug-resistant microbes are increasing constantly. However, the pipeline for novel drug candidates is substantially unfilled. Natural products and their derivatives remain a significant source of molecular diversity for drug discovery. However, the development of highly efficient and versatile natural product derivatisation methods remains a challenge. Classical derivatisation methods are often limited to standard modification of nucleophilic or electrophilic functional groups and selectivity is problematic.
Here we report the use of stabilised iminonitroso Diels-Alder reactions for derivatisation of complex diene-containing natural products. This totally atom economical cycloaddition reaction produces conformationally restricted analogs and introduces new functionality to rarely modified centres, diene or polyene components, of bioactive natural products in a highly regio- and stereoselective and chemically efficient fashion.
Methods: Six natural products (turimycin H3, ergosterol, reductiomycin, isoforocidin, colchicine and thebaine) were selected for our initial studies using nitrosopyridine Diels-Alder reactions. The configuration of cycloadducts was assigned based on 1D and 2D high resolution NMR studies, 1H NMR analysis, NOE correlations from ROESY experiments and X-ray crystallographic analyses. Bioactivity of the cycloadducts was studied against bacteria, yeast and fungi. Different cell lines were used for antiproliferative and cytotoxicity assays.
Results: The reactions with nitroso dienophiles proceeded in 10–20 min to afford the corresponding oxazine cycloadducts as single isomers in ∼90% yield and with excellent regio- and stereoselectivity for all of the substrates examined.
Incorporation of the oxazine heterocycles altered the biological activity profile of the corresponding parent natural products. While the cycloaddition products of turimycin retained reduced, but did not eliminate, antibacterial activity, antiproliferative activity was introduced. Biological activity studies of related cycloadducts of all of the other natural products mentioned will be presented as well.
Conclusion: The methodology presented herein provides a highly efficient and atom economical tool for derivatisation and functionalisation of complex diene-containing bioactive natural products at rarely modified centres. It provides a new basis towards structural diversity for drug discovery.

P2090 In vitro activity of API-1252, a novel anti-staphylococcal agent, against 100 genotypically unique Staphylococcus aureus strains
F. Perdreau-Remington, G. Zhanel, D. Vaughan, N. Kaplan (San Francisco, US; Winnipeg, Toronto, CA)
Objectives: API-1252 is a novel anti-staphylococcal agent that targets the key component of bacterial fatty acid synthesis, the enoyl-ACP reductase (FabI). API-1252 is being developed as a new class of oral and intravenous agent with a novel mechanism of action. We selected 100 unique Staphylococcus aureus (SA) strains based on a combination of genotypes and antimicrobial resistance phenotypes and compared the activity of API-1252 to other antimicrobials commonly used in the treatment of SA infections
Methods: MRSA and MSSA were selected according to their sequence type (ST), presence or absence of mecA and Panton Valentine Leucocidine (PVL) genes, staphylococcal chromosomal cassette element (SCCmec), pulsed field gel electrophoresis (PFGE) profiles and antimicrobial phenotypes. MICs were determined using current CLSI guidelines.
Results: The 79 MRSA strains belonged to 12 ST's (SCCmecIV n = 62; SCCmecII n = 17; PVL+ n = 35) and 47 unique PFGE profiles including USA100–1100. The 21 MSSA strains represented nine ST's and 20 unique PFGE profiles. All strains represented a combination of 100 unique genotypic/phenotypic profiles. Seventy nine percent of the strains originated from non sterile sites. The percent of SCCmecIV, SCCmec II and MSSA strains susceptible to comparators were 92%, 29% and 100% (meropenem); 65%, 12% and, 81% (ciprofloxacin) and 97%, 76% and 100% (gentamicin). All strains were susceptible to vancomycin. The ranges of API-1252 MIC's (mg/L) were 0.004–0.125 for MRSA and 0.004–0.015 for MSSA. For MRSA, the MIC90 for API-1252 was 0.015 mg/L for all SCCmecII strains as well as all SCCmecIV USA300 strains (n = 26) while all other SCCmec IV (n = 36) recorded an MIC90 of 0.03 mg/L. All MSSA strains independent of ST or phenotype had an MIC90 of 0.015 mg/L.
Conclusions: API-1252 demonstrated highly potent in-vitro activity against the diverse genetic backgrounds included in this study. The equipotent activity against representatives of all common community and hospital acquired MRSA strains greatly encourage the further development of API-1252 for challenging staphylococcal infections.
P2091 In vitro–in vivo clearance correlation of EDP-420, clarithromycin and telithromycin
L.J. Jiang, D. Wachtel, T. Phan, A. Sonderfan, Y.S. Or (Watertown, US)
Objectives: A significant number of drug candidates entering clinical development are discontinued due to unacceptable pharmacokinetic (PK) properties and, thus, there is an urgent need to establish and validate the in vitro and interspecies scaling methods which can accurately predict human PK before clinical trials. Herein, the in vitro-in vivo clearance (CL) correlations of two marketed antibiotics, clarithromycin and telithromycin, are evaluated. Additionally, the human clearance of EDP-420, a first-in-class bicyclolide (bridged bicyclic macrolide) currently in clinical development, was predicted before clinical trials.
Methods: Clarithromycin, telithromycin, or EDP-420 (1 microM) were incubated separately in triplicate with human microsomes (0.5 mg protein/mL) in the presence or absence of NADPH at pH 7.4 and 37 degree. Reactions were stopped at designated times, and supernatant concentrations of clarithromycin, telithromycin and EDP-420 were determined by LC/MS/MS. Human hepatic CL was calculated from the intrinsic CL of human liver microsomes using the well-stirred model.
Results: Following 90-minute incubations with human liver microsomes, 98% of telithromycin and 84% of clarithromycin were metabolised while 52% of the initial amount of EDP-420 remained. The in vitro human liver microsomal half-life of telithromycin, clarithromycin and EDP-420 were calculated to be 18.1, 44.2 and 89.8 minutes, respectively. The calculated human CL values of clarithromycin and telithromycin based on in vitro human liver microsomal results were 30.3 and 55.6 L/h, respectively, which were very close to the observed human CL of 31.1 L/h for clarithromycin and 57.7 L/h for telithromycin. The predicted human CL of EDP-420 is 12.9 L/h, which is 4-fold and 2-fold lower than the corresponding values for telithromycin and clarithromycin.
Conclusion: An excellent in vitro-in vivo clearance correlation was demonstrated for both clarithromycin and telithromycin, indicating that in vitro microsomal incubations would be a useful tool for predicting human clearance for this drug class. EDP-420 is predicted to have low clearance in humans.
P2092 Preclinical pharmacokinetics and interspecies scaling of EDP-420, a first-in-class bicyclolide
L.J. Jiang, G. Wang, L. Phan, A. Sonderfan, M. Paris, Y.S. Or (Watertown, US)
Objectives: EDP-420 (EP-013420) is a first-in-class bicyclolide (bridged bicyclic macrolide) currently in clinical development for the treatment of respiratory tract infections (RTI). It exhibits potent activity against RTI pathogens, including multi-drug resistant streptococci and the atypical organisms. EDP-420 also demonstrates in vivo efficacy against S. pneumoniae, S. pyogenes, and S. aureus in mouse protection tests, and against H. influenzae in a rat lung infection model. Herein, preclinical pharmacokinetics (PK) and interspecies scaling of EDP-420 are reported.
Methods: The single-dose intravenous (i.v.) and oral PK of EDP-420 was examined in mice (15 mg/kg), rats (10 mg/kg), dogs (5 mg/kg) and monkeys (5 mg/kg). Plasma EDP-420 concentrations were determined by LC/MS/MS. Allometric scaling was used to predict human PK parameters from multiple species animal data by linear least-squares regression analysis.
Results: Following a single oral dose, the average bioavailability was low (38.6%) for rats but relatively high for monkeys (50.8%), dogs (63.8%) and mice (88.6%). The area under the plasma concentration-time curve in mice, rats, dogs, and monkeys was 16.7, 4.1, 8.9, and 6.8 mg-hr/mL after a single oral dose of 15, 10, 5, and 5 mg/kg, respectively. Plasma clearance was substantially slower in dogs (0.26 L/h/kg) and monkeys (0.36 L/h/kg) than in mice and rats (0.8 to 0.95 L/h/kg). Volume of distribution (Vd) exceeded 2.5 L/kg in all species. Dogs had substantially longer half-life (t1/2, 15.0 and 10.8 hours for i.v. and oral, respectively) than mice, rats and monkeys (t1/2 ranged from 2.7 to 5.2 hours). The predicted human clearance and Vd are 13 L/h and 300 L, respectively, based on multiple species animal data using allometric scaling (R2 > 0.97). A long t1/2 of ∼16 hours is predicted in humans.
Conclusions: EDP-420 is well absorbed and has optimal pre-clinical PK properties across multiple species. The excellent correlation of allometric plots of PK parameters in four preclinical species suggests that EDP-420 PK behaviour in humans can be well predicted.
P2093 Tissue distribution of [14C]-EDP-420 in the rat by quantitative whole-body autoradiography
L.J. Jiang, M. Takeuchi, R. Lewsley, A. Sonderfan, T. Baba, Y.S. Or (Watertown, US; Osaka, JP; Harrogate, UK)
Objectives: EDP-420 (EP-013420) is a first-in-class bicyclolide (bridged bicyclic macrolide) currently in clinical development for the treatment of respiratory tract infections (RTI). This study shows the tissue distribution of radioactivity in the male and female albino rat and the male pigmented rat following a single oral gavage administration of 10 mg/kg of [14C]-EDP-420 using QWBA.
Methods: Rats were euthanised by cold shock following deep anaesthesia under isoflurane at specific times after dosing. The carcasses were frozen in a mixture of hexane and solid carbon dioxide and were subjected to quantitative whole-body autoradiography (QWBA) procedures. Radioactivity concentrations in tissues were quantified from the whole body autoradiograms using a validated image analysis system.
Results: Radioactivity was widely distributed at 2 hours after administration to male albino rats with Tmax = 8–10 hours in most tissues. Cmax in most tissues was greater than plasma Cmax of 0.871 μg equiv/g. Highest concentrations of radioactivity in tissues were generally associated with the Harderian gland (unique to rodents), lung, pituitary and spleen, with Cmax = 20–60 μg equiv/g, and AUC0–∞ = 671–1940 μg equiv-h/g. Plasma AUC0–∞ was 14.5 μg equiv-h/g. The ratios of Cmax and AUC0–∞ in lung over the corresponding values in plasma were 32.3 and 46.3, respectively. The majority of tissues had terminal half-lives of radioactivity in the range of 10 to 20 hours. Plasma had the shortest terminal half-life of 6.15 hours. There was little evidence for any gender difference in the distribution of radioactivity. Absorbed radioactivity was eliminated by both the biliary and renal routes. Quantifiable radioactivity was present in melanin-containing tissues (uveal tract and pigmented skin) of pigmented animals and declined at late sampling times.
Conclusion: EDP-420 has demonstrated good tissue distribution (especially lung penetration) in rats, suggesting that EDP-420 may be an effective compound for the treatment of RTI.
Relationship between antibiotic use and resistance
P2094 Association between antibiotic use and incidence of methicillin-resistant Staphylococcus aureus in general intensive care unit
E. Pujate, A. Balode, P. Oss, U. Dumpis (Riga, LV)
Objectives: The use of 3rd generation cephalosporins and fluoroquinolones has been previously associated with increased risk for acquisition of MRSA. Active surveillance of all MRSA cases and infection control programme was started in March, 2003 when the first cases were identified in our hospital. Most of the outbreaks were associated with the ICU. Aim of this study was to examine possible correlation between aggregated antibiotic consumption data and MRSA incidence in the general ICU.
Method: Retrospective study on antibiotic usage and MRSA incidence was performed in ICU. Data on MRSA cases were taken from MRSA surveillance database. Number of new MRSA cases in ICU was calculated monthly. Antibiotic consumption was measured in DDD per 100 bed days and patient on antibiotic days (PAD) recorded every day for each antibiotic by the nurse. Statistical analysis was performed with SPSS 13 software package.
Results: Three hundred sixty five new MRSA cases were registered in the hospital by September, 2006. 34.3% (CI: 29.6; 39.3) of cases were detected in general ICU. 88.8% (CI: 82.1; 93.2) of them received antibiotics before the cultures were taken of whom 36.0% (CI: 24.5; 40.6) received ciprofloxacin, 31.5% (CI: 23.6; 40.07) ceftriaxone, and 8.1% (CI: 4.3; 14.7) ceftazidime. In contrast, for the study time period the proportion in total antibiotic consumption in DDD/bed days for ciprofloxacin was 11.1% (CI: 6.8; 1.8), ceftriaxone 11.7% (CI: 7.3; 18.2), and ceftazidime 0.83% (CI: 0.2; 4.2). Ciprofloxacin was the only fluoroquinolone used.
Average monthly antibiotic consumption in ICU was 134.37 (CI: 122.2; 146.6) DDD per 100 bed days or 230.1 (CI: 221.54; 252.3) PAD. There was no correlation or association between monthly variations in total consumption and MRSA incidence. Time trend analysis showed association of number of new MRSA cases and PAD for ciprofloxacin. We observed the peak of ciprofloxacin consumption measured in PAD before increase in MRSA cases with the time delay of 2–3 months. There was no such association with consumption of third generation cephalosporins and other antibiotics except vancomycin that was used for treatment.
Conclusions: Use of ciprofloxacin and 3rd generation cephalosporins was identified as a risk factor for acquisition of MRSA infection and did not reflect the general pattern of antibiotic use in our ICU. Measurement of antibiotic consumption by using PAD could have advantage for evaluation of the impact of antibiotic resistance selection pressure.
P2095 Carbapenem consumption and resistance of P. aeruginosa and K. pneumoniae to carbapenems: results from a 7-year study in a general hospital
M. Lelekis, C. Loupa, A. Karaitianou, H. Papadaki, I. Tzannou, E. Katsiki-Divari, K. Papaefstathiou, G. Kouppari (Athens, GR)
Objective: The aim of the present study was to record on one hand carbapenem consumption and on the other hand carbapenem resistance o f P. aeruginosa and K. pneumoniae strains isolated in our 300-bed general hospital over a 7 year period.
Methods: Antibiotic consumption for the period 1999 to 2005 was studied retrospectively using data from the pharmacy computer. Antibiotic use was calculated in DDDs per 1000 patient days (ABC Calc 3.0). On the other hand data concerning imipenem resistance of P. aeruginosa and K. pneumoniae strains isolated in our hospital during the same time period were taken from the microbiology department.
Results: Antibiotic consumption was 572, 647, 678, 675, 710, 785 and 892 DDDs/1000 patient days (years 1999, 2000, 2001, 2002, 2003, 2004, 2005 respectively). Carbapenem consumption was 3.8, 6.7, 6.2, 3.9, 7.1, 9.8, 15.2 DDDs/1000 patient days for the above mentioned years (figure ). During the same time period P. aeruginosa resistance to imipenem was 14.5%, 7.8%, 14%, 17.5%, 15.4%, 15.5%, 15.5% and that of K. pneumonia to imipenem 0, 2.1%, 0, 0, 2%, 2.3%, 7.3% (figure).

Conclusions: During the study period, total consumption increased significantly in our hospital and this was the case for carbapenem consumption as well. At the same time while P. aeruginosa resistance to carbapenems seems to have stabilised around 15%, it is worrisome that K. pneumoniae resistance increased significantly in the last years. Follow up studies will reveal if this will be the next serious resistance problem in our hospital. At the same time resistance mechanisms of carbapenem resistant Klebsiella are being investigated.
P2096 Are recommendations for antibiotic use related to antibiotic consumption in hospital? Results from a French surveillance network
K. Miliani, F. L'Heriteau, I. Arnaud, A. Carbonne, P. Astagneau (Paris, FR)
Objectives: In France, antibiotics (AB) consumption is dramatically high in parallel to high rate of multiresistant bacteria. For the last few years, a well-defined policy has been implemented at the national level to control and monitor antibiotic consumption. Since 2002, surveillance networks with voluntary hospitals were set up to evaluate AB policy and consumption. The aim of the study was to evaluate whether recommendations for good practices of AB use were related to reduction of AB consumption.
Methods: Based on data from the Northern France surveillance system, recommendations for AB use were collected annually on a standardised questionnaire including 21 items. AB consumption was expressed in defined daily dose (DDD) per 1000 patient-days. A multivariate logistic regression analysis was performed with AB consumption categorised according to low (≤ percentile 75th) and high consumption (>p75) as the dependent variable. Whole AB as well as fluoroquinolones and amoxicillin/clavulanic acid (amoxiclav) consumption were analysed separately.
Results: Among the 83 hospitals participating in the study, 75% had a whole AB consumption ≤ 669.5 DDDs/1000 patient-days. The less frequent practices were: educational AB programmes (17%), request for an antibiotic specialist for second-line antibiotics (26%), systematic reassessment of AB treatment after 72 hours (27%), and computerised prescription and dispensation (35%). In the multivariate analysis, nominative delivery form (whole AB and amoxiclav) and computerised prescription (fluoroquinolones only) were associated with lower AB consumption.
Conclusions: Control measure of AB prescription appears to be the key for lowering consumption. Sustained efforts should focus on AB with high potential for emerging bacterial resistance.
P2097 Development of resistance toward macrolides in oropharyngeal streptococci after administration of azithromycin SR or clarithromycin XL in comparison to a control group
G. Westphal, M. Schmidt-Ioanas, J. Wagner, H. Lode (Brandenburg, Berlin, DE)
Objectives: Macrolide antibiotics are thought to enhance the development of bacterial resistance. We assessed the resistance development of oroharyngeal streptococci after a single dose of azithromycin SR (sustained release fomrula, 2.0 g), a five day treatment of clarithromycin XL (extended release formula, 1.0 g/day) and a control group (no treatment).
Methods: 50 healthy subjects were randomised into three groups. One group was given a single 2g dose of azithromycin SR (20 volunteers): the other group received 1g/day of clarithromycin XL for 5 days (20 volunteers). The control group received no treatment and consisted of 10 healthy volunteers. In a surveillance period of 13 weeks post administration, oropharyngeal streptococci viridans were tested for their MIC (minimal inhibitory concentration) of macrolides at 0, 7, 21, 35, 49, 63, and 91 days post administration by eTest®. The bacterial flora was obtained by oropharyngeal swabs. For each subject and each visit, five distinct streptococci viridans strains were selected and tested. Compliance was confirmed by supervision of administration and measurement of urinal concentration.
Results: We tested 700 strains in the azithromycin SR group (20 subjects), 665 strains in the clarithromycin XL group (19 subjects) and 280 strains in the control group (8 subjects). The transient development of resistance was similar in both treated groups. The geometrical mean erythromycin MICs (in mg/L) in the azithromycin SR group were 0.42 at day 0, 1.82 at day 63 (maximum value) and 0.60 at day 91; in the clarithromycin XL group 1.40 at day 0, 5.66 at day 63 (maximum value) and 1.22 at day 91. The control group did not develop any significant changes in the resistance index. In comparison to baseline, the analysed streptococci showed no significant differences in the level of resistance for erythromycin at day 91 within the three groups (p = 0.09). Azithromycin SR and Clarithromycin XL showed no severe side effects.
Conclusion: Azithromycin SR administered in a single dose of 2 g has a similar transient development of resistance as clarithromycin XL. There is no significant development of resistance to erythromycin after 91 days. In comparison to a control group the changes of the resistance index in a period of 91 days are not significant either.
P2098 Tobramycin resistance in Pseudomonas aeruginosa isolates from children with cystic fibrosis on nebulised tobramycin – temporal relationship with antibiotic usage
M. Doyle, P. McNally, G. Leen, J. Dier, P. Greally, P. Murphy (Dublin, IE)
Objectives: High level resistance (HLR) isolates of P. aeruginosa with a Minimum inhibitory concentration (MIC) >1024 for Tobramycin have been cultured from patients on Tobramycin Inhaled Solution (TSI). In the patients with isolates with an MIC > 1024 μg/mL, there is a tendency towards a more rapid fall off in both weight z scores and FEV1 [1].
The aim of this study is to explore the temporal relationship between use of (TSI) and fluctuations in the (MIC) in P. aeruginosa isolated from sputum of children with cystic fibrosis receiving inhaled Tobramycin.
Methods: Sputum was collected prospectively from all children with a persistent growth of P. aeruginosa receiving (TSI). Isolates were collected from Oct 2005 to Oct 2006. Patients were requested to send sputum for culture before and after a course of TSI therapy. All isolates were tested to evaluate the MIC. The timing and duration of TSI usage was recorded prospectively. The data was examined to establish if a temporal relationship exists between use of TSI and the changes in Tobramycin MIC of the P. aeruginosa isolates cultured.
Results: Thirty patients were treated with TSI during the study period. Two-hundred and twenty isolates were tested. The Tobramycin MIC in P. aeruginosa isolates from each patient fluctuated during the study period. No relationship could be demonstrated between the Tobramycin MIC from the P. aeruginosa isolated and the timing of TSI. Ten of the patients had a high level resistant (HLR) strain of P. aeruginosa isolated with a Tobramycin MIC of >1024 μg/mL. Seven patients remained colonised with a HLR isolate. One patient discontinued therapy with TSI during the study period. This patient continued to culture P. aeruginosa with a Tobramycin MIC >1024 one year after discontinuing TSI.
Conclusions: Isolates of P aeruginosa cultured from patients on TSI have a fluctuating Tobramycin MIC. The fluctuations in MIC do not appear to be related to the timing of therapy with TSI. Once a patient is colonised with an isolate with high level resistance to Tobramycin, most will remain colonised (7/10). The isolation of HLR P aeruginosa didn't cease once TSI therapy was stopped.
P2099 Strong association between ciprofloxacin use and ciprofloxacin resistance within a hospital
I. Willemsen, A. Heijneman, D. Bogaers-Hofman, J. Kluytmans (Breda, NL)
Objectives: In the last years the use of ciprofloxacin in our hospital has increased significantly. The objective of this study was to determine the increase of resistance over time and to determine if the density of antibiotic use correlates with the resistance in E. coli within individual hospital units.
Method: The density of the use of ciprofloxacin (cip), amoxicillin + clavulanic acid (amcl), 1st- and 2nd-generation cephalosporins (cef) and trimethoprim/Sulfamethoxazole (sxt) was measured in 6 consecutive one-day prevalence-surveys between 2001 and 2004. The susceptibility patterns from E. coli were obtained from the Laboratory Information System between 2003 and 2006. The percentage of resistance to cip, amcl, cef and sxt in E. coli isolates was calculated per unit and over time.
Results: A total of 4105 patients were included in the prevalence-surveys. 23% (938) of the patients were on antibiotics, of whom 12.8% (120) were treated with cip, 39.1% (367) with amcl, 13.2% (124) with cef and 5.7% (53) with sxt. 4790 E. coli susceptibility patterns were obtained from hospitalised patients. There was a significant increase of resistance over time for all antibiotics, except for SXT, and a significant correlation between the prevalence of use and the percentage of resistance for cip (R = 0.795, p = 0.006), amcl (R = 0.860, p = 0.001) and cef (R = 0.828, p = 0.003). For cip the urology unit rose above the other units with a use of 9.9% and a resistance rate of 19.8%. Also cip resistance showed the strongest increase over time.
Conclusion: This study shows that cip resistance is associated with the density of its use even on the micro-level of a hospital unit, and its use has the strongest association with resistance of all frequently used groups of antimicrobial-agents in the hospital.
Quality of antibiotic prescribing
P2100 Appropriateness of antibiotic therapy on weekends vs weekdays
J. Bishara, D. Hershkovitz, S. Pitlik (Petah-Tiqwa, IL)
Background: Several recent reports have raised concerns about the adequacy of medical care provided by hospitals on weekends. Antibiotic resistance is an emerging and universal problem and one of its major contributors is the inappropriate prescription of antibiotics.
Objective: To compare the appropriateness of antibiotic treatment prescribed in an emergency department (ED) of a tertiary medical centre on weekdays and weekends.
Methods: During a one month period medical charts of 1029 ED visits, who were discharged from ED were reviewed. Data of patients that were discharged with antibiotics was blindly evaluated by two infectious diseases specialists, and an “appropriateness score” was given to the antibiotic prescription.
Results: Antibiotics were prescribed at discharge for182 (17.7%) patients. The appropriateness score was significantly better at the beginning of the week and declined progressively toward the weekend (p = 0.025). Appropriateness scores were higher for the surgical and urological wings (p = 0.011), and for diagnoses of pneumonia and urinary tract infection (p = 0.005). Time of the day, patients age and sex did not have a significant effect on the appropriateness score.
Conclusions: During the week, there is a progressive decay in the appropriateness of antibiotic prescriptions in the ED. More studies are needed to clarify measures improving appropriate antibiotic therapy in weekends.
P2101 Do antibiotic ward rounds improve antibiotic prescribing?
J. Hinton, M.S. Kyi, S. Barnass (Isleworth, Middlesex, UK)
Objectives:
-
–
To review all prescription charts for patients receiving restricted antibiotics to ensure they are in line with the Trust Antibiotic Guidelines or have been approved by one of the medical microbiologists – To ensure antibiotics are stopped appropriately.
-
–
To optimise timing and choice of switch from intravenous to oral antibiotics.
-
–
To provide ongoing education of staff during the review.
-
–
To ensure ward staff are aware that the prescriptions are reviewed.
-
–
To inform guidelines on review of antibiotics for pharmacists new to the Trust.
Methods: Ward antibiotic charts were reviewed weekly (on different days of the week and at different times) between May 2005 and October 2006 by the Senior Antibiotic Pharmacist and one of the Consultant Microbiologists. All patients' charts for whom restricted antibiotics had been prescribed were reviewed, and patients were often reviewed on several occasions. Changes were made or antibiotics stopped, as appropriate, after consultation with the doctors looking after the patient and explanation of the rationale. If the medical staff could not be contacted, a note was left in the medical notes or on the prescription chart to request review.
Results: A total of 403 patients' charts were reviewed (mean 22 per month, range 5–50). Of these, 212 (52.6%) had been approved by the medical microbiologist or were in line with the Trust Antibiotic Guidelines. 224/403 (55.5%) antibiotics were continued and 180/403 (44.5%) were changed or stopped. The mean number changed or stopped per month was 10 (45.5%), with a range of 1–24 (11–69%).
The percentage of restricted antibiotics approved by the medical microbiologists or in line with the Trust Antibiotic Guidelines, and the percentage of antibiotics changed or stopped, did not change over the course of the study or with the experience of the cohort of junior doctors, the majority of whom change posts at the beginning of August each year. Junior ward staff know who we are and we deal with medical and pharmacy enquiries on our round. Unused restricted antibiotics are taken back to pharmacy, decreasing the likelihood of their use. Other prescribing problems which are found are addressed.
Conclusion: We have not seen an improvement in prescribing of restricted antibiotics, but we expect a continuing need to review charts and educate staff. We plan to improve the prescription charts with a box for the indication and duration of treatment. Guidelines to assist new pharmacists have been compiled.
P2102 Pre-admission penicillin in patients with meningitis or meningococcal disease
J.M. Darville, A.M. Lovering, A.P. MacGowan (Bristol, UK)
Objectives: UK government guidelines recommend that patients with suspected meningitis or meningococcal disease receive intra-muscular penicillin before admission to hospital. This study assesses adherence to these guidelines by determining the frequency of such antibiotic use in the area served by North Bristol Trust hospitals in a 6 year period.
Methods: Laboratory computer records were searched for patients from whom Neisseria meningitidis was isolated from any sample, and for those from whom Streptococcus pneumoniae or Haemophilus influenzae were isolated from CSF samples. Hospital computer records were searched for patients for whom the final diagnosis was infectious meningitis. Personal, demographic, clinical and therapeutic data were extracted from patients' hospital notes. To estimate the extent of preadmission illness, 10 presenting symptoms consistent with meningitis or meningococcal disease were each allocated one point. The data were transferred to a data-base for analysis.
Results: Mean severity scores are in brackets. 57 patients (2.9) did not see a doctor (medical practitioner) pre-admission. 7 of these had meningococcal septicaemia, 46 had meningitis (15 meningococcal) and 2 had meningococcal infection. 149 patients (3.36) did see a doctor. 40 (3.75) of these had antibiotic and 109 (3.21) did not. Of the 40, 4 had septicaemia (all meningococcal) and 35 had meningitis (11 meningococcal). Of the 109, 13 had septicaemia (all meningococcal) and 90 had meningitis (29 meningococcal).
72 patients (3.15) had meningococcal disease. 48 (3.15) of these saw a doctor, 24 (3.17) did not. Of the 48, 15 (3.67) had antibiotic, 33 (2.91) did not.
46 (3.44) patients with rash saw a doctor.18 (3.72) of these had antibiotic, 28 (3.25) did not. Of 32 (3.28) patients with meningococcal disease and rash seeing a doctor, 13 (3.69) had antibiotic, 19 (3.0) did not.
175 patients (3.3) had meningitis. 125 (3.47) of these saw a doctor, and of these 35 (3.77) had antibiotic, 90 (3.36) did not.
Conclusions: Patients may be more likely to receive pre-admission antibiotic the more symptoms they present with, when a rash is present or when Neisseria meningitidis is subsequently isolated. However, the majority of patients with meningitis or meningococcal disease seeing a doctor before admission did not receive antibiotic. Our study suggests that the guidelines have not been adequately observed and that it may necessary to revise them.
P2103 Value of early serum assay for dosage adjustment of vancomycin in continuous infusion
A. Forgeot, A. Carricajo, J. Morel, C. Venet, P. Mahul, S. Guyomarc'h, N. Fonsale, C. Auboyer, F. Zeni, G. Aubert (Saint-Etienne, FR)
Objective: It is difficult to obtain rapid bactericidal effects with vancomycin (V) due to the increased MIC for staphylococcal strains (in particular hetero-glycopeptide intermediate Staphylococcus aureus). The aim of this study was to obtain a serum concentration for V (SCV) of between 25 and 30 mg/L and a serum concentration/MIC ratio of >8 within first 24 hours in patients receiving V in a continuous infusion.
Methods: In 2005 and 2006, all patients requiring treatment with V in the two study ICUs were given a loading dose of V of 30 mg/kg in a 1-hour infusion followed by V at a dosage of 30 mg/kg/24 h. Creatinine clearance (CLc) was required to be >50 mL/min. Dosage was adjusted daily using a predetermined algorithm according to results of SCV: SCV < 25 mg/L, dosage increased by 4 to 8 mg/kg/24 h; SCV > 30 mg/L, dosage reduced by 4 to 8 mg/kg/24 h. V concentrations were measured by FPIA kit (Abbott, USA) and the MIC for V was determined using the E-Test method (AB Biodisk, Sweden).
Results: 21 patients were included in the study, 11 of whom presented documented infection (coagulase-negative Staphylococcus sp and Staphylococcus aureus with MIC between 0.5 and 3 mg/L). Mean CLc was 105 mL/min (range 50 to 178), mean body weight was 83 kg (range 60 to 115), and SAPS II was 42.4 (range 22 to 64). A serum concentration of V ≥ 26.5 mg/L within 6 hours of the start of treatment (best cut-off) was predictive of a concentration of V in excess of 25 mg/L at 24 h. The positive predictive value was 100% for the 14 patients with CLc < 120 mL/min and 90% for all 21 patients.
Conclusion: The target concentration for V of between 25 and 30 mg/L was achieved within 24 hours using our protocol in patients with CLc < 120 mL/min. Concentrations of V below the target level were seen in patients with CLc > 120 mL/min. Assay of V 6 hours after the start of treatment allowed early dosage adjustment. The serum concentration target must be reassessed on achievement of the MIC for V with regard to the offending staphylococcal strain.
P2104 Surgical antibiotic prophylaxis in a Turkish university hospital
S. Serin Senger, T. Togan, O. Kurt Azap, F. Timurkaynak, H. Arslan (Ankara, TR)
Objective: Since surgical site infections are important causes of morbidity, mortality, and healthcare costs, we aimed to assess the appropriateness of use of surgical antimicrobial prophylaxis at our institution.
Methods: A retrospective evaluation of the use of antimicrobial prophylaxis in patients undergoing major surgery at Başkent University Hospital from July to September 2006 was carried out. The surgical operations included were liver and kidney transplantations, cardiovascular surgery, urological operations, neurosurgical operations, head and neck surgeries, orthopaedic surgery, breast surgery, general abdominal colorectal surgery, and abdominal and vaginal hysterectomy. Patients having infectious diseases prior to the surgery were excluded. The guidelines to assess choice of antimicrobial agent and duration of prophylaxis, were ASHP Therapeutic Guidelines on Antimicrobial Prophylaxis in Surgery (1999), IDSA (Antimicrobial Prophylaxis for Surgery: An Advisory Statement from the National Surgical Infection Prevention Project) (2004), and Sanford Guide (2005).
Results: A total of 148 patients with a mean age of 41.9 years (ranged 1–89 with SD = 24.5) were included in the study. General Surgery was the leading department where 50 (33.8%) of the operations were performed. Cefazolin was the most commonly used antibiotic (constituting 48.0% of all the antimicrobial agents) during the study period. In 77 (52.0%) of the cases the prophylaxis decisions were appropriate, with proper choices of antibiotics in 51 cases and omissions in 26, in accordance with the guidelines. Unnecessary prolongation of prophylaxis in the patients of appropriately chosen antibiotics was found to be 41.0%.
Conclusion: The results showed a significantly high level of inappropriate use of antimicrobial prophylaxis in our institution. To improve the proper use of prophylactic antimicrobials, the infection control committee planned to prepare a local guideline and to organise educational sessions for the surgeons.
Reference(s)
- 1.George SL, Xiang J. Virology. 2003;316:191–201. doi: 10.1016/s0042-6822(03)00585-3. [DOI] [PubMed] [Google Scholar]
- 2.Naito H, Abe K. J Virol Methods. 2001;91:3–9. doi: 10.1016/s0166-0934(00)00207-x. [DOI] [PubMed] [Google Scholar]
- 1.Marton A. Epidemiology of resistant pneumococci in Hungary. Microb Drug Resist. 1995;1:127–130. doi: 10.1089/mdr.1995.1.127. [DOI] [PubMed] [Google Scholar]
- 2.Dobay O. Antimicrobial susceptibility and serotypes of Streptococcus pneumoniae isolates from Hungary. J Antimicrob Chemother. 2003;51:887–893. doi: 10.1093/jac/dkg171. [DOI] [PubMed] [Google Scholar]
- 1.Talbot Clinical Infectious Diseases. 2006;42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 1.Huletsky A, Giroux R., Rossbach V., Gagnon M., Vaillancourt M., Bernier M., Gagnon F., Truchon K., Bastien M., Picard F.J., van Belkum A., Ouellette M., Roy P.H., Bergeron M.G. New Real-Time PCR Assay for Rapid Detection of Methicillin-Resistant Staphylococcus aureus Directly from Specimens Containing a Mixture of Staphylococci. J. Clin. Microbiol. 2004;42:1875–1884. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekmann S.E. Effects of rapid detection of bloodstream infections on length of hospitalisation and hospital charges. J Clin Microbiol. 2003;41:3119–3125. doi: 10.1128/JCM.41.7.3119-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt G. Influence of antimicrobial treatment on mortality in septicaemia. J Chemother. 1999;11:179–186. doi: 10.1179/joc.1999.11.3.179. [DOI] [PubMed] [Google Scholar]
- Hautala T. Blood culture Gram stain and clinical categorization based empirical antimicrobial therapy of bloodstream infection. Int J Antimicrob Agents. 2005;25(4):329–333. doi: 10.1016/j.ijantimicag.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 1.Baron E.J., Weinstein M.P., Dunne W.M., Yagupsky P., Welch D.F., Wilson D.M. Cumitech Blood Cultures IV. ASM Press; 2005. [Google Scholar]
- 1.Maurin M., Raoult D. Clin Micro Re. 1999;12:518. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurin M. Inf and Immu. 1992;60:5013. doi: 10.1128/iai.60.12.5013-5016.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seshadri R. PNAS USA. 2003;100:5455. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spyridaki I. J Anti Chem. 2002;49:379. doi: 10.1093/jac/49.2.379. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L. Amino Acids. 2003;25:49. doi: 10.1007/s00726-003-0356-6. [DOI] [PubMed] [Google Scholar]
- *Gordon Ramage, Kacy Vande Walle, Brian L. Wickles, and José L. López-Ribot. Standardised method for in vitro antifungal susceptibility testing of Candida albicans. [DOI] [PMC free article] [PubMed]
- Rodrigues Med Mycol. 2005;43:711–717. doi: 10.1080/13693780500129814. [DOI] [PubMed] [Google Scholar]
- 1.Shah Br J Dermatol. 2003;148:1018–1020. doi: 10.1046/j.1365-2133.2003.05291.x. [DOI] [PubMed] [Google Scholar]
- 2.Afset Scand J Infect Dis. 2003;35:84–89. doi: 10.1080/0036554021000026980. [DOI] [PubMed] [Google Scholar]
- 1.Longitudinal development of tobramycin resistance in Pseudomonas aeruginosa isolates from children with cystic fibrosis on nebulised tobramycin – effects on clinical outcomes. Doyle M, McNally P, Leen G, Dier J, Greally P, Murphy P. ECCMID, 2006.


