Summary
Rubella virus (RUB) assembles its replication complexes (RCs) in modified organelles of endo‐lysosomal origin, known as cytopathic vacuoles (CPVs). These peculiar structures are key elements of RUB factories, where rough endoplasmic reticulum, mitochondria, and Golgi are recruited. Bicistronic RUB replicons expressing an antibiotic resistance gene either in the presence or the absence of the RUB capsid (C) gene were used to study the structure of RCs in transfected cells. Confocal microscopy showed that the RUB replicase components P90 and P150 localized to CPVs, as did double‐stranded RNA (dsRNA), a marker for RNA synthesis. Electron microscopy (EM) showed that replicons generated CPVs containing small vesicles and large vacuoles, similar to CPVs from RUB‐infected cells and that the replicase proteins were sufficient for organelle recruitment. Some of these CPVs contained straight membranes. When cross‐sectioned, these rigid membranes appeared to be sheets of closely packed proteins. Immuno‐EM revealed that these sheets, apparently in contact with the cytosol, contained both P150 and P90, as well as dsRNA, and thus could be two‐dimensional arrays of functional viral replicases. Labelling of dsRNA after streptolysin‐O permeabilization showed that replication of viral genome takes place on the cytoplasmic side of CPVs. When present, C accumulated around CPVs. Mitochondrial protein P32 was detected within modified CPVs, the first demonstration of involvement of this protein, which interacts with C, with RCs.
Introduction
Enveloped RNA viruses replicate and assemble in association with cellular membranes, often in a complex structure, a ‘viral factory’, to which the participating cell and viral factors are recruited. Mitochondria, cytoskeletal elements and cellular endomembranes are frequent participants in the factory architecture. Knowledge about the signals involved in organelle recruitment and modification, as well as mechanisms operating in spatial connection between viral replication and morphogenesis, is very limited. One of the earliest events in the construction of the factory is the formation of the replication complexes (RCs). In this sense, the replication of viral genomes in association with intracellular membranes is a common feature of many viruses, particularly of positive‐stranded RNA viruses (Salonen et al., 2005). A variety of membranes are used by different viruses, including membranes of secretory pathway organelles (picorna‐, arteri‐, bromo‐ and flaviviruses) (Schlegel et al., 1996; van der Meer et al., 1998; Mackenzie et al., 1999; Suhy et al., 2000; Dubuisson et al., 2002; Westaway et al., 2002), mitochondria (nodaviruses) (Miller et al., 2001), peroxisomes and chloroplasts (tombusviruses) (Rochon, 1999), endosomes and lysosomes (togaviruses) (Froshauer et al., 1988; Lee et al., 1994; Magliano et al., 1998; Kujala et al., 2001) and autophagosomes (arteriviruses, coronaviruses, poliovirus and rhinovirus) (reviewed by Wileman, 2006). It appears that targeting to the specific organelle is mediated by viral replicase components (Salonen et al., 2003; 2005).
A very unique feature of togaviruses is that their RCs assemble in modified organelles of endo‐lysosomal origin that have been termed ‘cytopathic vacuoles’ or CPVs (Froshauer et al., 1988; Lee et al., 1994; Magliano et al., 1998). CPVs contain internalized, membrane‐bound spherules of 50–70 nm which are thought to be the sites of viral RNA synthesis. In some electron micrographs, these spherules appear to be connected with the organelle membrane by slender threads or ‘necks’. It is probably through these necks that newly synthesized RNA is transported to the cytoplasm. While the two togavirus genera, alphaviruses (e.g. Semliki Forest virus and Sindbis virus) and rubiviruses (rubella virus, RUB), generate CPVs with a similar morphology and have rough endoplasmic reticulum (RER) cisternae and mitochondria around, Golgi stacks are additionally recruited to RUB CPVs (Lee et al., 1992; Magliano et al., 1998; Risco et al., 2003) forming a ‘viral factory’ where the virus replicates, assembles and matures (Lee and Bowden, 2000; Novoa et al., 2005a).
The study of intracellular RUB infection has been hampered by the slow rate of virus replication and lack of synchronous infection in culture cells. The RUB genome is a single‐stranded RNA of plus‐polarity of ∼10 kb in length that contain two open reading frames (ORF), a 5′ proximal ORF (the NS‐ORF) that encodes the two non‐structural replicase components, P150 and P90, and is translated from the genome RNA and a 3′ proximal ORF (the SP‐ORF) that encodes the virion structural proteins, capsid (C) and envelope glycoproteins E2 and E1 and is translated from a subgenomic RNA synthesized in infected cells (Frey, 1994). Replicons which substitute reporter genes for the SP‐ORF have been developed and used to study replication in the absence of the virion proteins (Tzeng et al., 2001). Recently, genomic and replicon constructs have been generated which express an additional gene under control of an internal ribosome entry site (IRES) (Pugachev et al., 2000). When the additional gene is an antibiotic resistance gene, populations of stably transfected cells that uniformly harbour the replicon can be selected (Chen and Icenogle, 2004) enhancing the ability to study intracellular replication events.
In this study we have initiated the structural examination of RCs and CPVs in stable replicon‐transfected cells by confocal and electron microscopy (EM). Although it is not required for RNA synthesis, the virus C protein has recently been shown to both enhance and modulate virus RNA synthesis (Tzeng and Frey, 2005; Tzeng et al., 2006) and rescue a variety of deleterious viral mutants, an effect manifested at the level of RNA synthesis (Tzeng and Frey, 2003; Chen and Icenogle, 2004; Tzeng et al., 2006). Furthermore, the C protein binds to a mitochondrial protein, P32, and has been reported to alter the cellular distribution of mitochondria (Beatch et al., 2005). Therefore, it was of interest to study the effect of expression of the C protein on RCs in replicon‐transfected cells. For EM, specimens were prepared by freeze substitution, allowing for superior structural preservation, prior to embedding and sectioning. This is the first study of togaviruses RCs and CPVs in which freeze substitution has been employed for sample preparation and differences in CPV morphology were noted, included discovery of a novel CPV architecture in replicon‐transfected cells that is also present in CPVs assembled in virus infected cells.
Results
Formation of RCs in RUB replicon‐transfected cells
As shown in Fig. 1A, the replicons used in this study expressed two heterologous genes, GFP or a C‐GFP fusion under control of the subgenomic promoter followed by the neomycin resistance gene under control of a picornaviral IRES (translation can be from either the genomic or subgenomic RNA). The former construct was termed RUBrep/GFP/neo and the latter was termed RUBrep/C‐GFP/neo. Confocal microscopy of Vero and BHK‐21 cells transfected transiently (2–4 days post transfection with no drug selection) revealed that P150 (Fig. 1B) and P90 (not shown), the two replicase components, localize in organelles labelled with antibodies against the lysosomal marker LAMP‐1, GFP localizes in both the nucleus and the cytoplasm (Fig. 1B and C) while the C‐GFP fusion localizes in the cytoplasm (Fig. 1D and E), the site of accumulation of C protein in RUB‐infected cells. Double‐stranded RNA (dsRNA), an intermediate of RNA replication, also localized with lysosomes (Fig. 1C and D), but interestingly was also sometimes on the cell periphery as well, either in association with lysosomes or without apparent association with lysosomes (Fig. 1E). Many cells exhibited both patterns of dsRNA distribution, depending on the plane of the micrograph within the cell (Fig. 1F and G). Similar findings were made with stably transfected cell cultures obtained following drug selection (not shown). Replicons thus assemble RCs in association with lysosomes, similarly to virus‐infected cells.
Figure 1.
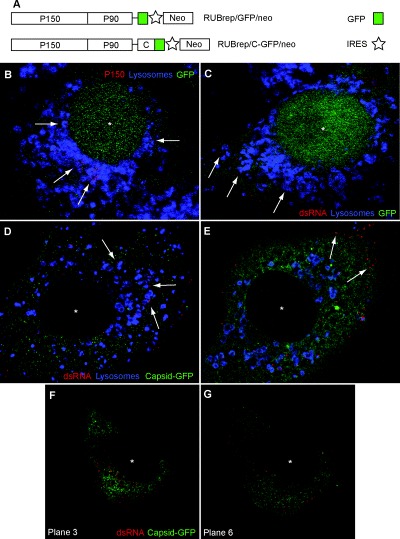
Immunofluorescence and confocal microscopy localization of RUB replication components in replicon‐transfected cells (Vero cells, 2 or 4 days post transfection). Single optical sections are shown. An asterisk marks the centre of the cell nucleus. A. Schematic diagrams of the two replicons used in this study, RUBrep/GFP/neo and RUBrep/C‐GFP/neo. ORFs are represented by boxes and untranslated regions by lines. B. Localization of P150 non‐structural protein (red) and lysosomes (blue) in cells transfected with RUBrep/GFP/neo. Lysosomes were labelled with anti‐LAMP‐1 antibodies. Arrows indicate P150 signal within lysosomes. With this construct, GFP localizes in both the cell nucleus (*) and the cytoplasm. C. Localization of dsRNA (antibody labelled in red) and lysosomes (blue) in cells transfected with RUBrep/GFP/neo. Arrows indicate dsRNA signal within lysosomes. D and E. Localization of dsRNA (red) and lysosomes (blue) in cells transfected with RUBrep/C‐GFP/neo. In D, arrows indicate dsRNA signal within lysosomes while in E, arrows indicate localization of dsRNA signal on cell periphery. With this construct, the C‐GFP fusion accumulates in the cytoplasm. F and G. Two confocal single sections of the same cell transfected with RUBrep/C‐GFP/neo and stained for dsRNA (red) showing dsRNA concentrating in the perinuclear region in the central section (F) or on the cell periphery in the lower section (G).
Effect of replicons on organelle distribution
Confocal microscopy was used to study the global distribution of Golgi, mitochondria, lysosomes, RER in control and replicon‐transfected cells. No differences were observed between cells transfected with RUBrep/GFP/neo and RUBrep/C‐GFP/neo and thus images from cells transfected with only the latter replicon are shown (Fig. 2). Lysosomes are shown in Fig. 2A–C, Golgi and mitochondria are shown in Fig. 2D–F, and RER in Fig. 2G–J; in panels C, F and J, the GFP signal has been subtracted so that organelle distribution can be compared. No significant alterations in the global distribution of lysosomes, Golgi or mitochondria were observed between control and transfected cells. However, the RER signal was weaker in transfected cells, suggesting a fragmentation and spreading of this compartment. EM was used to study regional organelle distribution. As shown in Fig. 2K, in RUBrep/C‐GFP/neo‐transfected cells, electron‐dense, lysosome‐like organelles, putative CPVs (as shown below, these contain both RUB replicase proteins and dsRNA) were surrounded by mitochondria, RER elements and Golgi stacks. Similar results were obtained in RUBrep/GFP/neo‐transfected cells. No differences in global or regional organelle recruitment were observed between transiently transfected or drug‐selected cells.
Figure 2.
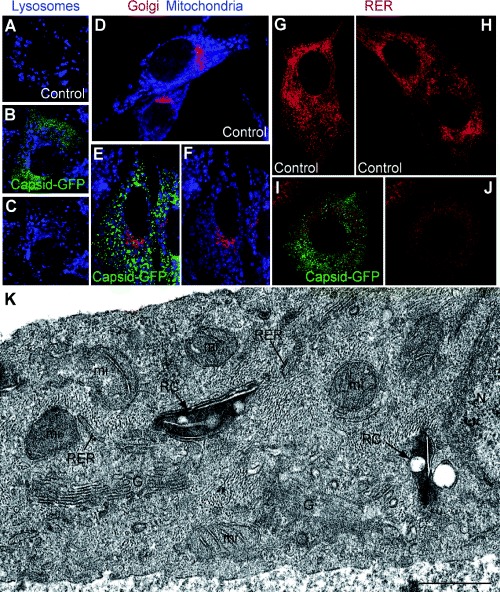
Organelle distribution and ultrastructure in replicon‐transfected cells (control non‐transfected BHK‐21 cells, BHK‐21 cells stably transfected with RUBrep/C‐GFP/neo, and Vero cells transiently transfected with this replicon at 2 days post transfection). Single optical sections are shown. Vero cells are shown in A–C while the rest of fields are of BHK‐21 cells. The cells were stained for lysosomes (blue) (A–C), Golgi (red) and mitochondria (visualized with Mitotracker blue) (D–F) and RER (red) (G–J). Panels B/C, E/F and I/J show the same field; in C, F and J, the green signal has been removed to allow better resolution of the organelle staining pattern. (K) EM of perinuclear areas of cells transfected with RUBrep/C‐GFP/neo. Dense, modified lysosome‐like organelles or CPVs (‘RC’ for replication complex) locally recruit mitochondria (mi) in the proximity of the cell nucleus (N) and are surrounded by RER, and Golgi stacks (G). Bar: 1 μm.
Colocalization of C protein with replicase components
In cells transfected with RUBrep/C‐GFP/neo the C‐GFP fusion protein colocalized with both P150 and P90 replicase components (Fig. 3A and B, drug selected cells are shown in these images). Interestingly, in cells with a moderate level of C‐GFP expression, the C‐GFP signal outlined lysosomes visualized with Lysotracker (this reagent was used because we found that anti‐LAMP antibodies labelled Vero lysosomes, but not lysosomes in BHK‐21 cells). C‐GFP exhibited a more localized pattern around lysosomes in stably transfected BHK‐21 cells and apparently the Lysotracker probe had difficulty penetrating some lysosomes that were surrounded by C‐GFP (Fig. 3C). Colocalized C‐GFP and replicase occasionally exhibited a filamentous pattern reminiscent of cytoskeletal architecture (Fig. 3D). While no colocalization of microtubules or intermediate filaments with replicase components was detected (not shown), actin filaments did colocalize with replicase components in both RUBrep/GFP/neo‐ (Fig. 3E) and RUBrep/C‐GFP/neo‐ (not shown) transfected cells and with C‐GFP in cells transfected with the latter replicon (Fig. 3F).
Figure 3.
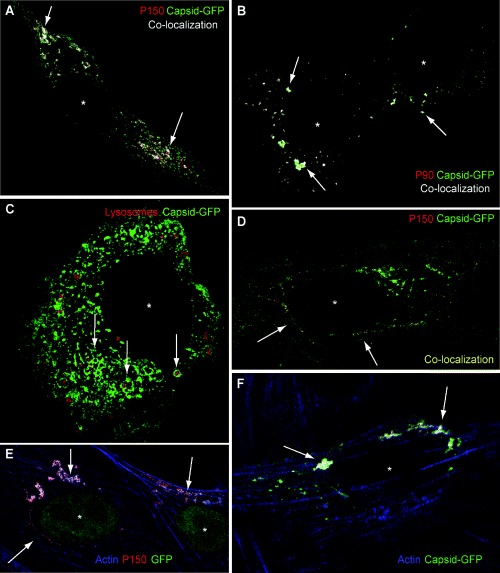
Cellular distribution of replicase components, C, lysosomes and actin in replicon‐transfected cells (replicon‐transfected BHK‐21 cells maintained for several weeks after drug selection). Single optical sections are shown. Cells were transfected with RUBrep/C‐GFP/neo in A–D and F and with RUBrep/GFP/neo in E. An asterisk marks the centre of the cell nucleus in the micrographs. A, B and D. Antibody labelling of P150 (A and D) or P90 (B), both in red, with C‐GFP (green), with colocalization shown in white (A and C) or yellow (D) and indicated by arrows. C. Arrows indicate C‐GFP (green) surrounding lysosomes (red, visualized with Lysotracker). Filamentous patterns of colocalization are denoted in D. E and F. Colocalization (indicated by arrows) of actin (labelled with phalloidin‐blue) and P150 (E, antibody labelled in red) or C‐GFP (F, green).
Ultrastructure of replicon CPVs
Stably transfected cultures after drug selection were used for transmission EM after both conventional and freeze‐substitution processing and for immunolabelling. Two views of CPVs in RUBrep/C‐GFP/neo stably transfected cells are shown in Fig. 4A and B which likely represent a cross‐section and a side view respectively. The CPV in Fig. 4A is electron‐dense and contains a number of hollow vacuoles while the CPVs in Fig. 4B are characterized by a large vacuole and a long straight element. CPVs in RUBrep/GFP/neo stably transfected cells had a similar morphology (not shown). As a control for the replicon‐transfected cells, uninfected and RUB‐infected BHK‐21 cells (24 and 48 h post infection) were similarly analysed (Fig. 4C and D). Lysosomes in uninfected cells (inset in Fig. 4D) were electron‐dense with some small vesicles occasionally distinguishable (but at a much lower number than in CPVs in replicon‐transfected cells) and usually devoid of vacuoles and straight elements and without local recruitment of mitochondria and RER (not shown). However, RUB‐infected cells (Fig. 4C and D) contained electron‐dense CPVs with internal vacuoles and straight elements, although these were usually smaller and in lower abundance than in replicon stably transfected cells. Immunogold labelling with antibodies against P150 and P90 replicase components (Fig. 5) demonstrated the presence of both proteins in CPVs, which were also labelled by anti‐lysosome antibodies (not shown). Thus, these CPVs contain RUB replicon RCs. Both P150 and P90 were associated with both vacuoles and straight elements in the CPVs. Side views of CPVs suggest that straight elements and their replicase proteins are in contact with the cytoplasm (Fig. 5B and C). To study if RNA replication takes place on the cytosolic or on the lumenal side of the CPVs, transfected cells were treated with streptolysin‐O (SLO) to open pores in the plasma membrane without altering intracellular membranes before immunolabelling with anti‐dsRNA antibodies (Fig. 5E–G). Saponin, which permeabilizes all endomembranes, was used in some of the samples as a control. Labelling of the cytosolic domain of TGN38 and the mitochondrial matrix protein Hsp‐60 (not shown) served as controls for the permeabilization procedures and confirmed the topological interpretation of dsRNA localization. After saponin treatment an intense signal associated to dsRNA is observed as shown in Fig. 5D. After SLO treatment and immunolabelling before fixation, similar dsRNA signals were obtained but only in permeabilized cells, where free cytosolic GFP has been released (Fig. 5F and G). These results indicate that RNA replication is taking place on the cytoplasmic side of CPVs.
Figure 4.
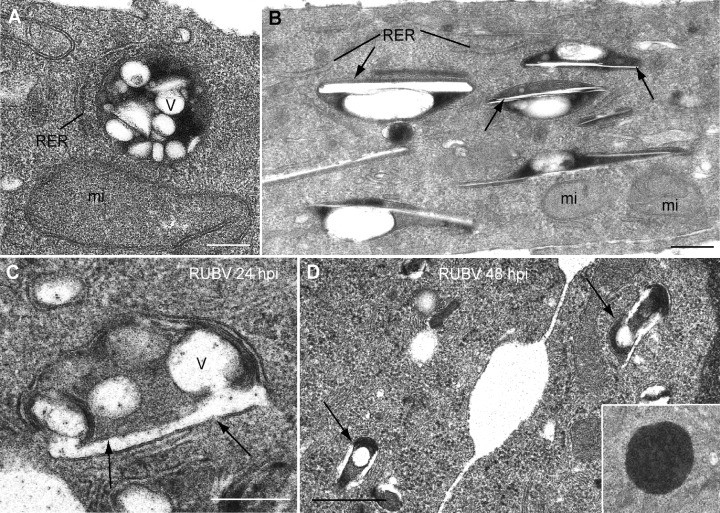
Ultrastructural analysis of CPVs in replicon‐transfected and RUB‐infected cells by conventional processing for thin‐section and EM (BHK‐21 cells transfected with RUBrep/C‐GFP/neo after drug selection). A and B. Conventional EM shows two views of electron‐dense CPVs in perinuclear areas of replicon‐transfected cells. Internal vesicles (V) and straight elements (arrows) are denoted as are locally recruited organelles, RER and mitochondria (mi). The CPVs in B appear to be side views of the CPVs while in A, a frontal view of the CPV is shown. C and D. CPVs in RUB‐infected BHK cells 24 (C) and 48 (D) hours post infection. C is a higher magnification while D is a lower magnification. CPVs in infected cells contain both vesicles and straight elements (arrows). Inset in D shows a characteristic dense lysosome in a control cell. V, vesicles; N, nucleus. Bars: 200 nm in A–C; 0.5 μm in D.
Figure 5.
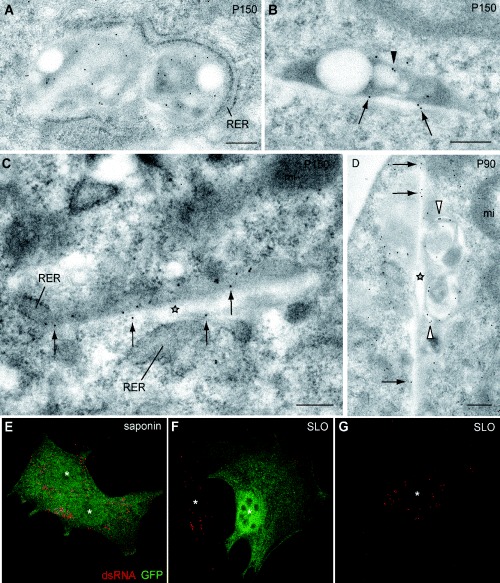
Association of RUB replicase components with CPVs by immuno‐EM in BHK cells transfected with RUBrep/C‐GFP/neo after drug selection (A–D) and confocal microscopy detection of dsRNA after saponin or SLO permeabilization in BHK cells transfected with RUBrep/GFP/neo after drug selection (E–G). Immunogold labelling with anti‐P150 (A–C) and anti‐P90 (D) antibodies demonstrate the presence of these replicase components in CPVs at the EM level. Labelling of vacuoles within the CPVs is denoted by arrowheads, the straight elements in side views by stars, and labelling of straight elements by arrows. (E–F) Confocal microscopy localization of dsRNA (red) after total permeabilization with saponin (E) or plasma membrane permeabilization with SLO (F and G). Z‐series projections of single optical sections are shown. The cell on the right side in F, which was not permeabilized with SLO treatment, maintained the cytosolic GFP and did not show dsRNA labelling. Under these conditions only the molecules exposed to the cytoplasmic side are detected. White asterisks mark cell nuclei. Bars: 200 nm.
At the EM level dsRNA is also localized in discreet spots of the rigid membranes (Fig. 6A). Although some C‐GFP was detected inside the CPVs in RUBrep/C‐GFP/neo stably transfected cells (Fig. 6B) most of it accumulated in neighbouring cytoplasmic areas (Fig. 6C). Some C‐GFP was also present on the periphery of mitochondria (arrowhead in Fig. 6C). Views at higher magnification revealed moderate labelling with C‐GFP of the internal straight elements inside CPVs (Fig. 6D). Antibodies specific for the mitochondrial protein P32 concentrated in two locations: a moderate labelling of the interior of mitochondria (Fig. 6E, main field) and weaker labelling of internal vesicles and straight elements of CPVs from RUBrep/C‐GFP/neo (Fig. 6E, main field and inset) and RUB/GFP/neo stably transfected cells (not shown), consistent with small foci of colocalization of P32 and dsRNA observed by confocal microscopy (not shown).
Figure 6.
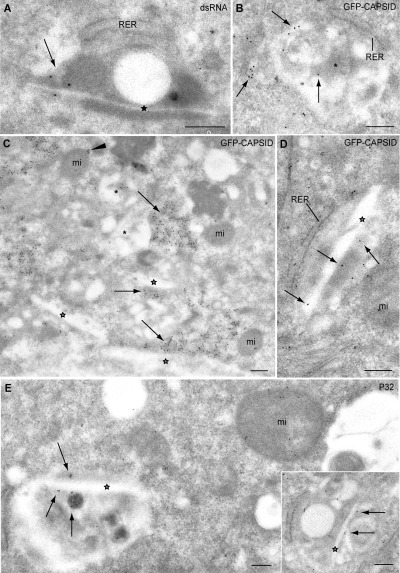
Association of dsRNA, C and mitochondrial protein P32 with CPVs in replicon‐transfected cells by immuno‐EM (BHK cells transfected with RUBrep/C‐GFP/neo after drug selection). A. Immunogold labelling with anti‐dsRNA antibodies. dsRNA signal associated with a straight element (star) is indicated with an arrow. B–D. Immunogold labelling with anti‐GFP antibodies to detect C‐GFP. Labelling around vacuoles (asterisks) and straight elements (stars) within CPVs is indicated by arrows. In C, exterior labelling of a mitochondrion (mi) is indicated by an arrowhead. E. Immunogold labelling with anti‐P32 antibodies. Labelling of the interior of a CPV is shown with arrows in both the main field and inset (straight elements are indicated by a star). Note the labelling of a neighbouring mitochondrion (mi). Bars: 200 nm.
When replicon stably transfected cells were processed by freeze substitution, the interior of CPVs was found to have a complex organization, with vacuoles, straight elements and vesicles of variable sizes, some of which appeared to be filled or to contain a central dense core (Fig. 7A–C). The vesicles were located both along the interior and on the periphery of the CPV. Higher magnification views of the internal straight elements revealed that they are bordered by intensely stained membranes (Fig. 7D) which sometimes formed closely apposed stacks (Fig. 7E). When the section plane was parallel to one of these membranes, structures compatible with close‐packing in two dimensions (2D) were observed (Fig. 7F and G), strongly suggestive of the existence of 2D arrays of proteins in these membranes.
Figure 7.
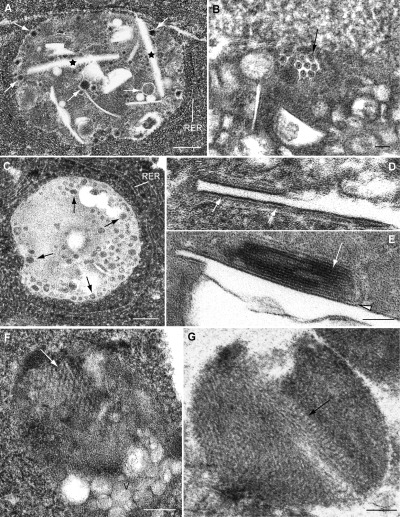
Ultrastructural analysis of modified lysosomes in replicon‐transfected cells prepared by freeze substitution (BHK cells transfected with RUBrep/C‐GFP/neo after drug selection). A. Large CPV with numerous filled vesicles (arrows) and straight elements (stars). B. Higher magnification of a CPV with small, filled vesicles that appear to contain an internal core (arrow). C. Small vesicular elements are abundant inside the dense content of some CPVs, both on the periphery (arrows) and internal areas. D and E. High magnification of straight elements showing dense, rigid membranes (arrows in D, arrowhead in E). White arrow in E points to a stack of rigid membranes. F and G. Sections along the straight sheets (rigid membranes) show features compatible with a close packing of particles (arrows). V: vesicles. Bars: 200 nm in A; 50 nm in B, D and E; 100 nm in C, F and G.
Vacuoles and straight elements were characteristic of CPVs in stably transfected cells; however, they were also occasionally observed in lysosomes in control cells. The presence of these elements in lysosomes from control cells, and in CPVs from transiently and stably transfected cells were quantified and reported in Table 1. Vacuoles and straight elements were present in an about three‐ to fivefold higher percentage of CPVs in replicon‐transfected cells in comparison with lysosomes in control cells. RUBrep/GFP/neo transiently transfected cells had a lower percentage of CPVs with vacuoles and straight elements than did RUBrep/C‐GFP/neo transiently transfected cells, although the percentage of CPVs in cells stably transfected with these two replicons was similar.
Table 1.
Percentage of lysosomes in control and transfected cells containing vacuoles and straight elements.
| Cells a | Vacuoles a | Straight elements a |
|---|---|---|
| Control BHK | 13% | 15% |
| 2 days post transfection | ||
| BHK RUBrep/C‐GFP/neo | 51% | 38% |
| BHK RUBrep/GFP/neo | 77% | 61% |
| 1 month post transfection | ||
| BHK RUBrep/C‐GFP/neo | 63% | 83% |
| BHK RUBrep/GFP/neo | 72% | 89% |
Lysosomes in control BHK cells or BHK cells transiently transfected (2 days post transfection) or stably transfected (1 month post transfection, including drug selection) were screened for the presence of large vacuoles and straight elements and the percentage containing each of these elements was determined. Fifty‐five to 65 lysosomes were included in the calculation of each percentage.
Discussion
Viral factories are one of the most interesting examples of viruses manipulating cell organization. It is thought that these complex structures are built by viruses to concentrate key components and increase the efficiency of replication and assembly of new viral particles (Novoa et al., 2005b; Wileman, 2006). Modified organelles that are specifically labelled for lysosomal markers are the central elements of RUB factories, and RER cisternae, mitochondria and Golgi stacks are among the recruited cellular factors. The architecture of the RCs inside the modified lysosome or CPV and the mechanism of transfer of replicated genomes to the assembly sites in Golgi membranes remain to be elucidated. Towards this end, in this study we investigated RCs in cells transfected with RUB replicons. With replicons, the contribution of the structural proteins can be dissected from the non‐structural replicase proteins. Additionally, as the replicons used expressed a GFP reporter gene and an antibiotic resistance gene, transfected cells could be readily identified and drug selection could be used to select a population of cells that uniformly harboured the replicon. We found CPVs in replicon‐transfected cells that were modified lysosomes similar in structure to those found in RUB‐infected cells, although they were present in greater abundance and appeared to have a more developed internal architecture of vacuoles and membrane‐bound straight elements. These elements were also present in a small percentage of lysosomes in control cells and thus the generation of these structures in CPVs may be a normal process stimulated by virus/replicon replication. Interestingly, viral replicase proteins reach the same intracellular location during infection and transfection. The use of replicons is then a valuable strategy to study these early interactions that trigger the formation of the RUB factory and the beginning of viral genome replication in cell membranes.
The ultrastructure of CPVs reminds some of the EM images of cellular autophagosomes found in the bibliography (Teckman et al., 2004). Some viruses replicate in autophagosomes (Jackson et al., 2005; Wileman, 2006), that are double‐membrane vacuoles where cytosol and organelles are sequestered and delivered to lysosomes (Klionsky, 2005). However, when treating cells transfected with RUB replicons with 3‐methyl‐adenine, a specific inhibitor of early stages of the autophagic process (Seglen and Gordon, 1982) no decrease in dsRNA signal was observed (not shown). Viruses that use the autophagic machinery induce an increase in the number of autophagic vacuoles and consequently in the signal provided by monodansyl‐cadaverine (MDC), a fluorophore specific for autophagic vacuoles under certain fixation conditions (Jackson et al., 2005). However, in cells transfected with RUB replicons there was no increase in the number of autophagosomes as observed after incubation with MDC (not shown) while Lysotracker signal colocalized with the CPVs (Fig. 3C). These data would suggest that CPVs are not related to autophagosomes but with modified lysosomes as reported (Magliano et al., 1998). However, a detailed study should be done at very short times post transfection to study if autophagy is induced by the virus at the very beginning of the life cycle.
One of the main aspects of this study is trying to understand the complex structure of the CPVs, in particular where the functional elements are located and working. In order to be able to transfer the replicated genomes to the assembly sites, RCs must have an exit to the cytoplasm. The observation that the rigid membranes containing the replicase proteins and dsRNA are apparently in contact with the cytosol (4, 5, 6) but mainly the detection of dsRNA on the cytosolic side of the CPVs after SLO permeabilizaton (Fig. 5) confirm that viral genome RNA replicated in CPVs must have a direct access to the cytoplasm and consequently to the assembly sites in Golgi membranes that surround the CPVs. C molecules that accumulate around CPVs probably participate in the transfer of viral genomes.
The P150 and P90 RUB replicase components and dsRNA localized within CPVs and the RER, mitochondria and Golgi stacks were locally recruited to the CPVs. Similar observations have been made in RUB‐infected cells (reviewed in Lee and Bowden, 2000). As lysosome modification to generate CPVs and organelle recruitment was observed with replicons lacking the C protein, the non‐structural replicase proteins by themselves can carry out these functions. Although local organelle recruitment was observed, global organelle recruitment in replicon‐transfected cells was not, independently of whether the replicon expressed the C protein or not. Beatch et al. (2005) recently reported a global recruitment of mitochondria to perinuclear regions in cells expressing the RUB C protein. However, as such a global recruitment is not observed in RUB replicon‐transfected cells, this observation appears to be specific for expression of the C protein alone. Although expression of the C protein by replicons was not necessary for generation of CPVs, in cells transfected with replicons expressing the C protein, colocalization of the C protein with P150 and P90 was detected and the C protein accumulated on the exterior of CPVs. Small amounts of C were also detected inside CPVs. Confocal microscopy suggested that C covered the exterior of some modified lysosomes. The replicon expressed C protein is actually a fusion protein with GFP and we cannot rule out a different behaviour of C‐GFP when compared with C alone. However, C‐GFP fusions are functional in modulation of replicon RNA synthesis and mutant rescue (Tzeng and Frey, 2003; 2005) and Lee et al. (1999) detected C protein around CPVs by immuno‐EM of RUB‐infected cells (Lee et al., 1999). With regard to C‐mediated enhancement and modulation of replicon RNA synthesis and mutant rescue, it was recently found that P150 and C can be co‐immunoprecipitated (Tzeng et al., 2006) and our finding of colocalization of C, P150 and P90 further confirms an interaction, which may play a role in these C‐mediated phenomena. Colocalization of C with P150 was previously reported in RUB‐infected cells (Kujala et al., 1999).
In replicon‐transfected cells, we also observed colocalization of C, P150 and P90 with actin microfilaments. Kujala et al. (1999) also observed the colocalization of P150 with actin and a considerable disruption of the actin cytoskeleton in RUB‐infected cells. It is known that viruses frequently use actin filaments for intracellular transport while some viruses disrupt the actin cytoskeleton to facilitate infection (Cudmore et al., 1997; Dohner and Sodeik, 2005). Actin could also be a structural component of herpesviruses (Grünewald et al., 2003) and the participation of actin in organizing the sites of mRNA accumulation and synthesis of viral structural proteins has been reported for HIV (Kimura et al., 1996). The potential role of actin in RUB replication remains to be established. We also detected small amounts of the mitochondrial P32 protein in modified lysosomes and, conversely, C located on the exterior surface of mitochondria. The association of C and P32 was reported by two groups (Beatch and Hobman, 2000; Mohan et al., 2002) and Beatch and Hobman (2000) demonstrated that C and P32 interacted on the periphery of mitochondria in transfected cells expressing C protein. The function of this interaction in RUB replication has not been elucidated although Beatch et al. (2005) recently reported that mutations in the C protein that disrupt the interaction adversely affect virus infectivity. Our results confirm the localization of C to the exterior of mitochondria and are the first demonstration of P32 associated to RUB CPVs. We cannot discard that the presence of P32 inside CPVs could be due to autophagy. However, as P32 is able to leave the mitochondria (Matthews and Russell, 1998; Brokstad et al., 2001) its presence inside CPVs could have a different meaning. It has been proposed that RUB C could interact with P32 shortly before it is translocated into mitochondria or alternatively as it leaves it (Beatch et al., 2005). On the other hand, our results indicate that C protein would not be necessary for P32 association with RUB RCs because P32 is detected in CPVs when cells are transfected with RUBrep/GFP/neo. These data suggest that P32 could be a structural component of the RUB RC and it is tempting to speculate that P32 could participate in directing C protein to the RC for packaging of the newly synthesized RNA before virus assembly. Further studies will be necessary to clarify these key aspects of RUB replication.
Interestingly, confocal microscopy showed that dsRNA, a marker for RNA replication, was not only present in perinuclear lysosomes, but also at the cell periphery, either associated with lysosomes or in the apparent absence of lysosomes. It has been previously reported that both alphaviruses and RUB RCs are secreted by infected cells at extended times post infection (Froshauer et al., 1988; Lee and Bowden, 2000). It is known that an increase in the intracellular concentration of Ca2+ above 1 μM induces secretion of conventional lysosomes in many cell types (Rodríguez et al., 1997; Martínez et al., 2000). This mechanism could be upregulated in infected/transfected cells. dsRNA detected on the cell surface could be actually contained in CPVs from which specific markers had disappeared on the way to the plasma membrane. This may be a mechanism for cells ridding themselves of virus‐specific dsRNA.
Ultrastructural analysis of CPVs in replicon‐transfected cells revealed an electron‐dense interior with variety of internal vesicles and vacuoles, the former of which were filled and the latter of which appeared empty, as well as straight elements bounded by rigid membranes. Although size, shape and organelle association are similar, CPVs in RUB‐infected cells described in previous studies (Lee et al., 1994; Magliano et al., 1998; Lee and Bowden, 2000) generally appeared empty. However, the electron density of the CPVs in this study was consistent with the electron density of lysosomes in control cells and the difference is likely due to fixation techniques employed because we used conventional processing with very short dehydration steps or freeze substitution, both of which have been shown to yield superior structural preservation (Risco and Carrascosa, 1999). The filled vesicles we observe likely correspond to the spherules in the CPVs in RUB‐infected cells described in previous studies while the large vacuoles and straight elements were absent in CPVs described in these previous studies. In addition to differences in processing, it is also possible that we could have visualized stages in the biogenesis of the CPVs that were missed in these earlier studies.
The straight elements frequently observed inside CPVs in replicon‐transfected cells were particularly striking. Their rigidity suggested a high protein content and immunolabelling demonstrated that both replicase components were particularly abundant in these membranes while dsRNA and C, while detected, were present in lower amounts. Sectioning along the plane of the membrane revealed an apparent two‐dimensional proteinaceous sheet. The potential formation of 2D arrays of viral polymerases has been previously proposed in studies with poliovirus using in vitro transcription assays (Hobson et al., 2001; Lyle et al., 2002) but to our knowledge this has not been previously observed in infected cells. The participation of such arrays in virus or replicon RNA synthesis remains to be established, although the presence of dsRNA indicates that they are active. The availability of drug‐selected, uniformly transfected cell lines will allow us to further explore the structure of RUB RCs in CPVs at the molecular level by 3D reconstruction based on serial sectioning and electron tomography.
Experimental procedures
Generation of replicons
Recombinant DNA manipulations were performed essentially as described (Sambrook et al., 1989) with minor modifications. E. coli JM109 and DH5α were used as the bacterial hosts. Restriction enzymes and T4 DNA ligase were obtained from New England BioLabs (Beverly, MA) or Roche Molecular Biochemicals (Indianapolis, IN) and used as recommended by the manufacturer. To generate the RUBrep/GFP/Neo and RUBrep/C‐GFP/Neo constructs, a SpeI site was first introduced between the IRES and SP‐ORF in siRobo402 (Pugachev et al., 2000). This intermediate construct, termed siRobo402‐SpeI, was generated by a three‐round asymmetric PCR followed by a three‐fragment ligation (Tzeng and Frey, 2002). In the first round of PCR amplification, mutagenic oligo 937 (5′‐GCTTGCCACAACCACTAGTATGGCTTCTACTAC‐3′; SpeI site underlined) was used to prime an asymmetric amplification on a PstI‐linearized siRobo402 template. The second round was an asymmetric amplification using the first round PCR product as template and oligo 941 (5′‐GCATTCTAGAGTCGCGCTGTCGCG‐3′; XbaI site underlined; the sequence is complementary to the nts 6630−6643 of the RUB genome). In the third round, the second round PCR product and oligo IR‐5 (5′‐CACAATGCATAATTCCGCCCCTCTCCCTC‐3′; NsiI site underlined) was used to prime PCR amplification on PstI‐linearized siRobo402 template. The NsiI‐NotI‐digested PCR amplification product was ligated with the NotI‐EcoRI fragment from Robo502 (Tzeng and Frey, 2002) and NsiI‐EcoRI fragment from siRobo402. The neomycin resistance gene was PCR‐amplified from BamHI‐restricted pCI‐neo template (Promega) with oligos 938 (5′‐GCATACTAGTATGATTGAACAAGATGGATT‐3′; SpeI underlined) and 943 (5′‐TATAAGGCCTTCAGAAGAACTCGTCAAGAA‐3′; StuI underlined). The amplified gene was restricted with SpeI and StuI and cloned into siRobo402‐SpeI, creating siRobo402‐Neo. To generate RUBrep/GFP/Neo and RUBrep/C‐GFP/Neo, an NsiI‐EcoRI fragment was removed in siRobo402‐Neo and was used to replace the corresponding fragments in RUBrep/GFP and RUBrep/C‐GFP (Tzeng and Frey, 2002; 2003) respectively.
In vitro transcription and transfection
In vitro transcription was carried out as previously described and Vero and BHK‐21 cells were transfected with the resulting in vitro transcripts using Lipofectamine 2000 as previously described (Tzeng and Frey, 2002). By monitoring GFP expression by direct examination of the transfected monolayer with a Zeiss Axioplan fluorescence microscope with epifluorescence capability, efficiency of transfection was determined to be roughly 30%. To select a population of cells uniformly harbouring replicons, at 4 days post transfection cultures were incubated with medium containing G‐418 (1.2 mg ml−1). After selection, the cultures were maintained in medium containing G‐418 at the same concentration.
Antibodies and fluorescent probes
Rabbit antisera against non‐structural proteins P150 and P90 (GU3 for P150, GU8 and GU10 for P90) were previously described (Forng and Frey, 1995). Rabbit anti‐GFP (green fluorescence protein) was provided by Dr David Shima (Imperial Cancer Research Foundation, London, UK). Anti‐LAMP‐1 antibodies (Carlsson et al., 1988) were kindly provided by Dr M. Fukuda (The Burnham Institute, La Jolla, USA). Anti‐giantin antiserum was a kind gift of Dr M. Renz (Institute of Immunology and Molecular Genetics, Karlsruhe, Germany). K2 monoclonal antibody (mAb) specific for dsRNA was kindly provided by Dr N. Lukács (Corvinus University, Budapest, Hungary). Anti‐dsRNA J2 mAb was purchased from Biocenter (Szeged, Hungary). Polyclonal antibody specific for P32 mitochondrial protein was a kind gift of Dr W.C. Russell (University of St Andrews, Scotland). mAb specific for PDI (protein disulfide isomerase) was from Sigma. Fluorescent secondary antibodies, Alexa 594, phalloidin 660 (blue), Lysotracker (red and blue) and Mitotracker blue were purchased from Molecular Probes/Invitrogen. Fluorescent secondary antibodies conjugated with Cy5 were from Jackson ImmunoResearch Laboratories.
Immunofluorescence and confocal microscopy
Cell monolayers grown on coverslips were washed with phosphate‐buffered saline (PBS) and fixed 30 min at 4°C with a mixture of 4% paraformaldehyde (PFA) in PBS. After washing with PBS, monolayers were permeabilized 10 min at room temperature (RT) with 0.25% saponin before treating 30 min with PBS containing 2% bovine serum albumin (BSA) and 0.25% saponin. Monolayers were incubated 45 min at RT with primary antibodies diluted in PBS containing 0.1% BSA and 0.25% saponin. When antibodies against the P150 and P90 proteins were used, the monolayers were pre‐incubated for 10 min with PBS containing 20% goat serum and 0.25% saponin and the primary antibodies diluted in PBS‐containing 2% goat serum were added and incubated overnight at 4°C. After washing with PBS samples were incubated 45 min with fluorescent secondary antibodies diluted in PBS/2% goat serum/0.25% saponin (for anti‐P150 and anti‐P90 primary antibodies) or in PBS/0.1% BSA/0.25% saponin (for the rest of primary antibodies). Coverslips were mounted on glass slides with ProLong gold anti‐fade reagent (Molecular Probes/Invitrogen). Mitotracker and Lysotracker were added to live cell cultures at a concentration of 200 nM in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. After 45 min at 37°C, the medium was removed and the cells were either washed and mounted on glass slides or fixed and processed for immunolabelling as described above. Specimens were observed using a Bio‐Rad Radiance 2100 confocal laser microscope. Colocalization studies were performed with the program LaserPix (Bio‐Rad). White and yellow indicate colocalization.
Permeabilization with SLO and immunofluorescence assays
Cells were processed following previously described procedures (Sodeik et al., 1993). SLO was purchased from Dr Sucharit Bhakdi's laboratory (Institute of Medical Microbiology and Hygiene, Hochhaus am Augustusplatz, Mainz, Germany). The optimal SLO concentration was determined by monolayers of BHK‐21 cells grown on coverslips (1 × 105 cells per 16 mm coverslip) with different concentrations of SLO (20, 10, 5, 3, 2.5, 2 and 1 μg ml−1) and determining the percentage and integrity of permeabilized cells with trypan blue exclusion and light microscopy. Cells were washed three times with ice‐cold SLO buffer (25 mM Hepes pH 7,0 with 250 mM sucrose, 5 mM magnesium acetate, 50 mM potassium acetate) containing 2 mM DTT, incubated 15 min on ice with 2 μg ml−1 SLO diluted in the same buffer, washed again with ice‐cold SLO‐buffer containing 2 mM DTT, incubated 15 min at 37°C to allow pore formation, and then washed three times with SLO buffer to remove the released cytosolic content.
For immunofluorescence, SLO‐treated cells were incubated 15 min at 37°C with SLO buffer containing 20% calf serum, and then incubated with anti‐dsRNA antibodies diluted 1/100 in SLO buffer supplemented with 2% calf serum at 37°C 45 min. After washing three times with PBS, cells were fixed 15 min with 3% PFA in PBS at RT. Cells were washed three times with PBS, and incubated again 15 min with SLO buffer containing 20% calf serum at 37°C, before incubation with the fluorescent secondary antibodies diluted in SLO buffer supplemented with 2% calf serum at 37°C, 45 min. After washing three times with PBS, cells were mounted and studied by confocal microscopy.
For a complete permeabilization cells were washed three times with PBS and fixed 15 min with 3% PFA at RT. After washing three times with PBS, cells were processed as above but including 0.25% saponin in all the steps. The rest of the procedure was as indicated.
Immunolabelling with antibodies against the cytosolic domain of TGN38 (kindly provided by Dr George Banting from the University of Bristol, UK) and Hsp‐60, a mitochondrial matrix protein (purchased from Stressgen Biotechnologies, Victoria, Canada) were used as controls for the two permeabilization procedures and the detection of epitopes exposed to the cytosol or oriented to the lumen of endomembranes.
Transmission EM: ultrastructural studies
Monolayers of control BHK‐21 cells, cells transfected transiently (2–4 days post transfection with no drug selection), stably transfected BHK‐21 cells (drug‐selected) and BHK‐21 cells infected 24 or 48 h with the F‐Therien strain of RUB at a multiplicity of infection (moi) of 1 PFU per cell were chemically fixed with a mixture of 2% glutaraldehyde and 2% tannic acid in Hepes buffer (pH 7.4). All samples were processed by embedding in the epoxy‐resin EML‐812 (Taab Laboratories, Adermaston, Berkshire, UK) as previously described (Novoa et al., 2005b). Ultrathin sections were collected on Formvar‐coated EM grids and stained with uranyl acetate and lead citrate. For freeze substitution, cells were fast‐frozen in liquid ethane and treated with a mixture of acetone and osmium tetroxide in a automated freeze substitution unit (AFS; Leica‐Reichert‐Jung, Vienna, Austria) (Risco et al., 2002; 2003). For immunolabelling of proteins and RNA, the substitution medium was methanol containing 0.1% uranyl acetate. Samples were embedded in the epoxy‐resin EML‐812 (for ultrastructural analysis) or Lowicryl K4M (for immunolabelling) (Salanueva et al., 1999). Ultrathin sections of approximately 30–40 nm were collected on EM grids and either stained with uranyl acetate and lead citrate or processed for immunogold labelling. Samples were studied in a JEOL 1200‐EX II electron microscope.
Immuno‐EM
Immunogold labelling of proteins was performed by incubating sections with primary antibodies and secondary antibodies, these conjugated with 10 nm colloidal gold (BioCell International, Cardiff, UK) (Risco et al., 2002). For detecting dsRNA, sections were treated 15 min at 37°C with proteinase K (1 μg ml−1 in Tris‐EDTA) before immunolabelling (Salanueva et al., 2003).
Acknowledgements
We express our gratitude to Sylvia Gutierrez Erlandsson for expert assistance with confocal microscopy. This work has been supported by Grants BMC2003‐01630 from the MEC of Spain and GR/SAL/0671/2004 from the Comunidad de Madrid (to C.R.), and AI21389 from NIH (to T.K.F.).
References
- Beatch, M.D. , and Hobman, T.C. (2000) Rubella virus capsid associates with host cell protein P32 and localizes to mitochondria. J Virol 74: 5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatch, M.D. , Everitt, J.C. , Law, L.‐J. , and Hobman, T.C. (2005) Interactions between rubella virus capsid and host protein P32 are important for virus replication. J Virol 79: 10807–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokstad, K.A. , Kalland, K.‐H. , Russell, W.C. , and Matthews, D.A. (2001) Mitochondrial protein P32 can accumulate in the nucleus. Biochem Biophys Res Commun 281: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Carlsson, S.R. , Roth, J. , Piller, F. , and Fukuda, M. (1988) Isolation and characterization of human lysosomal membrane glycoproteins, h‐lamp‐1 and h‐lamp‐2. J Biol Chem 263: 18911–18919. [PubMed] [Google Scholar]
- Chen, M.‐H. , and Icenogle, J.P. (2004) Rubella virus capsid protein modulates viral genome replication and virus infectivity. J Virol 78: 4314–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore, S. , Reckmann, I. , and Way, M. (1997) Viral manipulations of the actin cytoskeleton. Trends Microbiol 5: 142–148. [DOI] [PubMed] [Google Scholar]
- Dohner, K. , and Sodeik, B. (2005) The role of the cytoskeleton during viral infection. Curr Top Microbiol Immunol 285: 67–108. [DOI] [PubMed] [Google Scholar]
- Dubuisson, J. , Penin, F. , and Moradpour, D. (2002) Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol 12: 517–523. [DOI] [PubMed] [Google Scholar]
- Forng, R.‐Y. , and Frey, T.K. (1995) Identification of the rubella virus non‐structural proteins. Virology 206: 843–853. [DOI] [PubMed] [Google Scholar]
- Frey, T.K. (1994) Molecular biology of rubella virus. Adv Virus Res 44: 69–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froshauer, S. , Kartenbeck, J. , and Helenius, A. (1988) Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol 107: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald, K. , Desai, P. , Winkler, D.C. , Heymann, J.B. , Belnap, D.M. , Baumeister, W. , and Steven, A.C. (2003) Three‐dimensional structure of herpes simplex virus from cryo‐electron tomography. Science 302: 1396–1398. [DOI] [PubMed] [Google Scholar]
- Hobson, S.D. , Rosenblum, E.S. , Richards, O.C. , Richmond, K. , Kirkegaard, K. , and Schultz, S.C. (2001) Oligomeric structures of poliovirus polymerase are important for function. EMBO J 20: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, W.T. , Giddings, T.H. , Jr, Taylor, M.P. , Mulinyawe, S. , Rabinovitch, M. , Kopito, R.R. , and Kirkegaard, K. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, T. , Hashimoto, I. , Nishikawa, M. , and Fujisawa, J.I. (1996) A role for Rev in the association of HIV‐1 gag mRNA with cytoskeletal beta‐actin and viral protein expression. Biochimie 78: 1075–1080. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J. (2005) The molecular machinery of authophagy: unanswered questions. J Cell Sci 118: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala, P. , Ahola, T. , Ehsani, N. , Auvinen, P. , Vihinen, H. , and Kääriäinen, L. (1999) Intracellular distribution of rubella virus ninstructural protein p150. J Virol 73: 7805–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala, P. , Ikaheimonen, A. , Ehsani, N. , Vihinen, H. , Auvinen, P. , and Kääriäinen, L. (2001) Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75: 3873–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.‐Y. , and Bowden, D.S. (2000) Rubella virus replication and links to teratogenicity. Clin Microbiol Rev 13: 571–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.‐Y. , Marshall, J.A. , and Bowden, D.S. (1992) Replication complexes associated with the morphogenesis of rubella virus. Arch Virol 122: 95–106. [DOI] [PubMed] [Google Scholar]
- Lee, J.‐Y. , Marshall, J.A. , and Bowden, D.S. (1994) Characterization of Rubella virus replication complexes using antibodies to double‐stranded RNA. Virology 200: 307–312. [DOI] [PubMed] [Google Scholar]
- Lee, J.‐Y. , Marshall, J.A. , and Bowden, D.S. (1999) Localization of Rubella virus core particles in Vero cells. Virology 265: 110–119. [DOI] [PubMed] [Google Scholar]
- Lyle, J.M. , Bullitt, E. , Bienz, K. , and Kirkegaard, K. (2002) Visualization and functional analysis of RNA‐dependent RNA polymerase lattices. Science 296: 2218–2222. [DOI] [PubMed] [Google Scholar]
- Mackenzie, J.M. , Jones, M.K. , and Westaway, E.G. (1999) Markers for the trans‐Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in Flavivirus‐infected cells. J Virol 73: 9555–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliano, D. , Marshall, J.A. , Bowden, D.S. , Vardaxis, S. , Meanger, J. , and Lee, J.Y. (1998) Rubella virus replication complexes are virus modified lysosomes. Virology 240: 57–63. [DOI] [PubMed] [Google Scholar]
- Martínez, I. , Chakrabarti, S. , Hellevik, T. , Morehead, J. , Fowler, K. , and Andrews, N.W. (2000) Synaptotagmin VII regulates Ca2+‐dependent exocytosis of lysosomes in fibroblasts. J Cell Biol 148: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.A. , and Russell, W.C. (1998) Adenovirus core protein V interacts with P32, a protein which is associated with both the mitochondria and the nucleus. J Gen Virol 79: 1677–1685. [DOI] [PubMed] [Google Scholar]
- Van Der Meer, Y. , Van Tol, H.G. , Krijnse‐Locker, J. , and Snijder, E.J. (1998) ORF1a‐encoded replicase subunits are involved in membrane association of the arterivirus replication complex. J Virol 72: 6689–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.J. , Schwartz, M.D. , and Ahlquist, P. (2001)Flock house virus RNA replicates on outer mitochondria membranes in Drosophila cells. J Virol 75: 11664–11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, K.V. , Ghebrehiwet, B. , and Atreya, C.D. (2002) The N‐terminal conserved domain of rubella virus capsid interacts with the C‐terminal region of cellular P32 and overexpression of P32 enhances the viral infectivity. Virus Res 85: 151–161. [DOI] [PubMed] [Google Scholar]
- Novoa, R.R. , Calderita, G. , Arranz, R. , Fontana, J. , Granzow, H. , and Risco, C. (2005a) Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 97: 147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa, R.R. , Calderita, G. , Cabezas, P. , Elliott, R.M. , and Risco, C. (2005b) Key Golgi factors for structural and functional maturation of Bunyamwera virus. J Virol 79: 10852–10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugachev, K.V. , Tzeng, W.‐P. , and Frey, T.K. (2000) Development of a Rubella virus vaccine expression vector: use of a picornavirus internal ribosome entry site increases stability of expression. J Virol 74: 10811–10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco, C. , and Carrascosa, J.L. (1999) Visualization of viral assembly in the infected cell. Histol Histopathol 14: 905–926. [DOI] [PubMed] [Google Scholar]
- Risco, C. , Rodríguez, J.R. , López‐Iglesias, C. , Carrascosa, J.L. , Esteban, M. , and Rodríguez, D. (2002) Endoplasmic reticulum‐Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J Virol 76: 1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco, C. , Carrascosa, J.L. , and Frey, T.K. (2003) Structural maturation of rubella virus in the Golgi complex. Virology 312: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon, D.M. (1999) Tombusviruses In Encyclopedia of Virology. Granoff A., and Webster R.G. (eds). San Diego, CA: Academic Press, pp. 1789–1798. [Google Scholar]
- Rodríguez, A. , Webster, P. , Ortego, J. , and Andrews, N.W. (1997) Lysosomes behave as Ca2+‐regulated exocytic vesicles in fibroblasts and epithelial cells. J Cell Biol 137: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva, I.J. , Carrascosa, J.L. , and Risco, C. (1999)Structural maturation of the transmissible gastroenteritis coronavirus. J Virol 73: 7952–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva, I.J. , Novoa, R.R. , Cabezas, P. , López‐Iglesias, C. , Carrascosa, J.L. , Elliott, R.M. , and Risco, C. (2003) Polymorphism and structural maturation of Bunyamwera virus in Golgi and post‐Golgi compartments. J Virol 77: 1368–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen, A. , Vasiljeva, L. , Merits, A. , Manden, J. , Jokitalo, E. , and Kääriäinen, L. (2003) Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J Virol 77: 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen, A. , Ahola, T. , and Kääriäinen, L. (2005) Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol 285: 139–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schlegel, A. , Giddings, J.T.H. , Ladinsky, M.S. , and Kirkegaard, K. (1996) Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol 70: 6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen, P.O. , and Gordon, P.B. (1982) 3‐Methyladenine: specific inhibitor of the autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 79: 1889–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik, B. , Doms, R.W. , Ericsson, M. , Hiller, G. , Machamer, C.E. , Van't Hof, W. , et al. (1993) Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol 121: 521–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy, D.A. , Giddings, J.T.H. , and Kirkegaard, K. (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy‐like origin for virus‐induced vesicles. J Virol 74: 8953–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teckman, J.H. , An, J.‐K. , Blomenkamp, K. , Schmidt, B. , and Perlmutter, D. (2004) Mitochondrial autophagy and injury in the liver in α1‐antitrypsin deficiency. Am J Physiol Gastrointest Liver Physiol 286: G851–G862. [DOI] [PubMed] [Google Scholar]
- Tzeng, W.‐P. , and Frey, T.K. (2002) Mapping the rubella virus subgenomic promoter. J Virol 76: 3189–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, W.‐P. , and Frey, T.K. (2003) Complementation of a deletion in the Rubella virus p150 nonstructural protein by the viral capsid protein. J Virol 77: 9502–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, W.‐P. , and Frey, T.K. (2005) Rubella virus capsid protein modulation of viral genomic and subgenomic RNA synthesis. Virology 337: 327–334. [DOI] [PubMed] [Google Scholar]
- Tzeng, W.‐P. , Chen, M.‐H. , Derdeyn, C.A. , and Frey, T.K. (2001) Rubella virus DI RNAs and replicons: requirement for non‐structural proteins acting in Cis for amplification by helper virus. Virology 289: 63–73. [DOI] [PubMed] [Google Scholar]
- Tzeng, W.‐P. , Matthews, J.D. , and Frey, T.K. (2006) Analysis of Rubella virus capsid protein‐mediated enhancement of replicon replication and mutant rescue. J Virol 80: 3966–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway, E.G. , Mackenzie, J.M. , and Khromykh, A.A. (2002) Replication and gene function in Kunjin virus. Curr Top Microbiol Immunol 267: 323–351. [DOI] [PubMed] [Google Scholar]
- Wileman, T. (2006) Aggresomes and autophagy generate sites for virus replication. Science 312: 875–878. [DOI] [PubMed] [Google Scholar]


