Summary
Because of their limited coding capacity, viruses are not able to encode all proteins that are required for their replication. Therefore, they depend on a wide variety of cellular functions and structures, such as the host cell nucleus. It has been shown that DNA, as well as RNA viruses, exploit the nucleus because it provides essential machinery for viral replication. On the other hand, the nucleus undergoes significant remodelling during viral usurpation or exploitation. Moreover, it is becoming increasingly clear that some subnuclear structures, such as promyelocytic leukaemia nuclear bodies, act as an antiviral defence mechanism, and several viruses antagonize this intracellular defence by modifying subnuclear structures. This article reviews the main alterations that take place in nucleus during viral infections.
Introduction
Viruses, even the largest ones, are minute compared with host cells and cannot encode all the proteins required for their replication because of limitations in their coding capacity. For this reason, viruses rely on the utilization of host cell proteins and cellular structures to facilitate their own replicative processes. Most DNA viruses have evolved the ability to replicate in the host nucleus (with the exception of poxviruses) and to exploit nuclear compartments. On the other hand, many RNA viruses are thought to replicate only in the cytoplasm. However, it has become increasingly apparent that many RNA as well as DNA viruses use nuclear compartments or components for their replication. The nucleus provides the machinery for viral DNA and RNA polymerization, processing and stabilization, and it also is stocked with essential factors for viral egress and maturation. At the same time, the nucleus is the most important organelle, which contains the cell's genetic material and several subnuclear structures that ensure normal cell function. Therefore, the usurpation of the nucleus by viruses constitutes an enormous stress for the cell that may lead to a profound disturbance of physiological functions.
Recent research based on cellular imaging techniques is beginning to increase our understanding of the mechanisms that viruses use to hijack and alter the host cell nucleus. In this article, we will review recent advances on how several viruses change the nucleus for their own needs.
Nuclear structure: nucleolus and promyelocytic leukaemia nuclear body
The heterogeneous nature of the nucleus became apparent after pioneering observations under well‐constructed light microscopes. At present, we know that the nucleus contains the chromatin territories that mainly consist of DNA‐protein complexes, as well as several distinct interchromosomal compartments (bodies) that support molecular activities, including replication, transcription and processing of nucleic acids (Fig. 1). The best‐characterized subnuclear structures are the nucleolus, the splicing speckles, the paraspeckles, as well as the Cajal, the Gem and the promyelocytic leukaemia nuclear bodies (PML NB, also known as ND10) whose precise functions, in spite of extensive characterization, remain unclear. However, much progress has been made in studies concerning the nucleolus and PML NBs, and the properties of these bodies are briefly summarized below.
Figure 1.
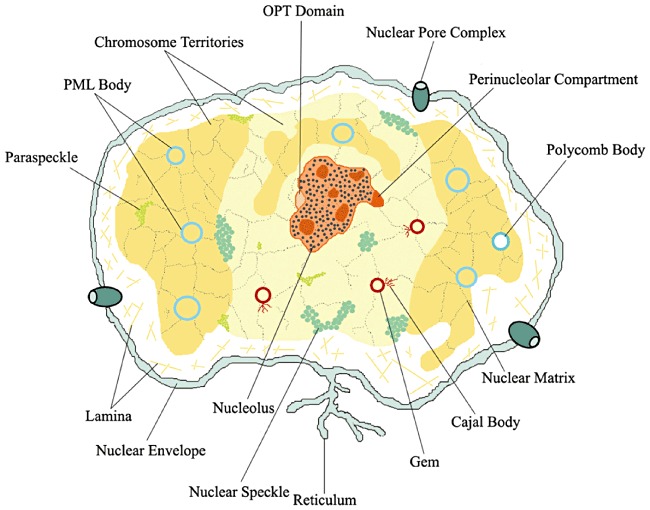
Schematic representation of the compartmentalized nucleus. The nucleus is bounded by the double‐membrane nuclear envelope. Nuclear pore complexes regulate nucleocytoplasmic exchange of macromolecules. Chromatin binds to the nuclear lamina that in turn is connected to the nuclear envelope. Nuclear matrix regulates spatial and functional organization of chromatin and nuclear bodies. The main nuclear bodies are represented: Nucleolus; the perinucleolar compartment; Cajal bodies, Gem, Polycomb body, Promyelocytic leukaemia (PML) bodies, Oct1/PTF/Transcription (OPT) domain, speckles and paraspeckles.
Nucleolus
Detailed analysis of the nucleolus by electron microscopy indicated that this subnuclear structure can be subdivided into three regions formed by the association of macromolecules that comprise fibrillar centres, surrounded by dense fibrillar components, which in turn are bordered by granular components (Scheer and Hock, 1999). For many years the classical view of these components focused ontheir crucial role for the biogenesis of ribosomal subunits, but proteomic analysis of human nucleoli showed the presence of about 4500 nucleolar proteins with different functions, thus revealing the multifunctional nature of the nucleolus (Ahmad et al., 2009). Nucleolin, fibrillarin and B23 (nucleophosmin) are three major proteins that represent approximately 20% of the total nucleolar protein content and that are highly conserved in structure and function within eukaryotes (Dumbar et al., 1989; Tollervey et al., 1993; Allain et al., 2000).
The main function of nucleolin is to facilitate the first cleavage step of rRNA. Nucleolin also functions as a chaperon for correct folding in pre‐rRNA processing. Fibrillarin is involved in post‐transcriptional processes, such as pre‐rRNA processing, pre‐rRNA methylation and ribosome assembly. B23 is widely distributed among different species. It has two isoforms (B23.1 and B23.2) that have multiple functions. They have been implicated in ribosome assembly (Dumbar et al., 1989), nucleocytoplasmic shuttling (Li et al., 1996) and regulation of transcription of rDNA (Okuwaki et al., 2001).
PML NB
As for several other subnuclear structures, PML NBs were originally observed through electron microscopy in the early 1960s, which revealed the presence of spheric nuclear bodies (0.2–1.0 µm in diameter) in most cell types and many tissues (de The′et al., 1960). The frequency of PML NBs depends on cell type and condition, varying from 3 to 30 per cell (Ascoli and Maul, 1991). Several studies have shown that PML NBs are dynamic, macromolecular structures that represent accumulations of multiple cellular proteins, whose only common known feature is their ability to be SUMOylated (Bernardi and Pandolfi, 2007). Probably the most important components of PML NBs are Sp100, hDaxx and PML. These proteins have several isoforms and motifs, such as SUMOylation sites (PML, hDaxx and Sp100), a RING finger motif (PML), SUMO‐interacting motifs (PML, hDaxx and Sp100) and a SAND domain (Sp100) that are involved in PML formation and functionality (Fig. 2).
Figure 2.
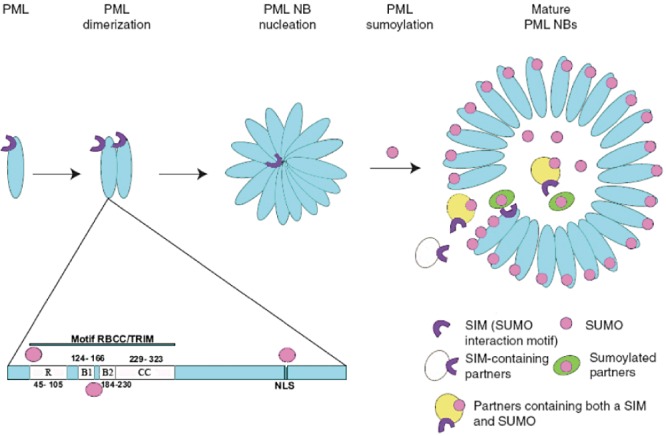
Schematic representation of PML and biogenesis of PML NB. Promyelocytic leukaemia (PML) protein is the key organizer of the PML nuclear bodies (NB). It belongs to the RING–B‐box–coiled‐coil/tripartite motif (RBCC/TRIM) protein family and is modified by small ubiquitin‐like modifier (SUMO) (pink circles) at 65, 160 and 490 amino acid positions. PML proteins dimerize through RBCC domains and then multimerize. The SUMOylated PML proteins form a spherical body (0.2–1.0 µm in diameter) with inner core. Other SUMOylated or SUMO interaction motif (SIM)‐containing proteins, such as speckled protein 100 (Sp100) and death associated protein (hDaxx), are recruited by SUMOylated PML into inner core (Bernardi and Pandolfi, 2007).
The previous decade of extensive studies implicated PML NBs in stress response (Maul et al., 1995), gene regulation (Zhong et al., 2000), oncogenesis (Salomoni and Pandolfi, 2002), cell senescence (Bischof et al., 2002), DNA damage repair (Dellaire and Bazett‐Jones, 2004), apoptosis (Takahashi et al., 2004) as well as in antiviral defence mechanisms (Everett and Chelbi‐Alix, 2007).
Hijacking the nucleolus
The first description of an interaction between the nucleolus and viruses was the observation of the nucleolar changes caused by influenza virus in chick embryo fibroblasts (Compans and Dimmock, 1969). Many other studies showed that viral interaction with the nucleolus is a widely distributed phenomenon among different viral families. Whereas the interaction with the nucleolus is a logical event for DNA viruses that replicate in the nucleus, the interaction between the nucleolus and RNA viruses whose site of replication is far from the nucleus is currently not well understood. On the other hand, the recruitment of nucleolar proteins with defined functions in RNA metabolism can be a logical step for RNA viruses that have a limited coding capacity. However, recent studies revealed a profound reorganization of nucleolar components in virus‐infected cells that contributes to the functional remodelling of the entire nucleus.
Nucleolar proteins
The multifunctional nature of many nucleolar proteins may explain the usurpation and incorporation of these proteins into replication and translation complexes formed by RNA as well as DNA viruses. The best example for this is the interaction between picornaviruses and nucleolin followed by blocking of nuclear import pathways (Gustin and Sarnow, 2002). These dramatic changes in the nuclear pore complex result in an accumulation of nucleolin in the cytoplasm of infected cells. After accumulation, nucleolin interacts with the 5′ non‐coding region of the poliovirus genome known as internal ribosome entry site (IRES). IRESes are RNA structural elements that function in the binding of ribosomes to the mRNA and initiate translation of mRNA in a cap‐independent manner (Hellen and Sarnow, 2001), and therefore nucleolin interaction with the IRES element stimulates IRES‐dependent translation (Izumi et al., 2001). Nucleolin can also interact with the poliovirus 3′ non‐coding region, which is implicated in the regulation of negative‐strand RNA synthesis (Waggoner and Sarnow, 1998). Moreover, interaction of picornaviruses with the nucleolar proteins not only facilitates viral replication but also leads to the shutdown of host cell transcription. Aminev and co‐authors showed that encephalomyocarditis virus proteins 2A and 3BCD localize in the nucleoli and inhibit cellular mRNA transcription but not rRNA transcription (Aminev et al., 2003). Another study done by Aminev showed that human rhinovirus 3C protease precursors also localize in the nucleoli of infected cells and inhibit cellular RNA transcription through proteolytic mechanisms (Amineva et al., 2004). Virus‐induced shutdown of the host‐cell transcription may inhibit intracellular defence mechanisms and may free the translational machinery for viral replication.
Other RNA viruses that replicate and assemble in the cytoplasm also induce nucleolar response in the host cell as viruses alter nucleolar protein content. Quantitative proteomics was used to take an overview of the protein changes in epithelial cells infected with human respiratory syncytial virus. In addition to the alterations in mitochondrial and nuclear pore complex proteins content, Hiscox and co‐authors described depletion of nucleolin and B23 in human respiratory syncytial virus‐infected cells (Munday et al., 2010). Similarly, high throughput proteomics was used to analyse protein content of Vero cells infected with the coronavirus infectious bronchitis virus (IBV) and protein content of 293‐T cells infected with influenza A virus. Proteomic analyses revealed that global nucleolar proteome change did not occur in influenza A virus and IBV infected cells, and only a subset of nucleolar proteins changed in abundance (2010a, 2010b).
Some DNA viruses, including herpesviruses, papillomaviruses and adenoviruses can also interact with the nucleolus. For example, several nucleolar proteins are relocalized during herpes simplex virus 1 (HSV‐1) infection. Nucleolin and B23 are dispersed throughout the nucleus in a manner dependent on expression of the viral protein UL24 (Lymberopoulos and Pearson, 2007; Calle et al., 2008; Lymberopoulos et al., 2011). Unlike nucleolin and B23, fibrillarin is redistributed in spots throughout the nucleus during infection in a manner independent of UL24 (Lymberopoulos and Pearson, 2007).
Interaction between adenoviruses and the nucleolus is another well‐studied example. It has been shown that adenoviruses inhibit the synthesis, processing and exit of the 18S and 28S species of rRNA (Castiglia and Flint, 1983) and disrupt the nucleolus at late time of infection (Walton et al., 1989). More recent studies done by Matthews and colleagues showed that the adenovirus core protein V is delivered to the nucleus and is associated with the nucleolus, leading to the mass relocation of nucleolin to the cytoplasm (Matthews and Russell, 1998; Matthews, 2001). Examination of the adenoviral genome revealed potential binding sites for nucleolin. Thus, nucleolin may interfere with adenoviral replication via its ability to bind and repress transcription (Yang et al., 1994). Therefore, adenovirus‐induced displacement of nucleolin appears to constitute an important step towards the antagonization of host obstacles. It has also been shown that B23.1, B23.2 and UBF (upstream binding factor) interact with viral proteins in replication centres and stimulate adenovirus DNA replication independently (Okuwaki et al., 2001; Lawrence et al., 2006; Hindley et al., 2007). And finally, proteomic analyses identified quantitative changes in 351 proteins of the nucleolus during adenovirus infection, finally corroborating that the nucleolus is extensively targeted and modified by adenoviruses (Lam et al., 2010).
Localization of RNA virus proteins to the nucleolus: is there a benefit?
It has been shown that many capsid and RNA‐binding proteins, including influenza virus nucleoprotein (Dimmock, 1969), Newcastle disease virus matrix protein (Peeples et al., 1992), dengue virus core protein (Wang et al., 2002) or coronavirus nucleocapsid protein (Dove et al., 2006) localize to the nucleolus, and for many years these localizations seemed only phenomenological without any known consequence for cells or viruses. It was speculated that viral (particularly RNA viral) proteins might accumulate at the nucleolus as a result of diffusion through the nuclear pore complex and association with the nuclear compartments that have high RNA content, and therefore within the nucleolus because it is transcriptionally active. Although the characterization of viral life cycles determined the accumulation of several specific viral proteins within the nucleolus, until recently there was huge gap in our knowledge about the consequences.
At present, it is apparent that nucleolar localization of viral proteins may disrupt the nucleolus and may lead to serious consequences for the cell (Matthews and Olson, 2006). For example, the HIV‐1 Rev protein localizes to the dense fibrillar component and is able to alter the nucleolar architecture, which in turn may lead to a dysregulation of the cell cycle and of cytokinesis (Miyazaki et al., 1996; Cannavo et al., 2001). Similarly, IBV proteins interact with nucleolar proteins and cause damage to the nucleolus and failure of cytokinesis (Wurm et al., 2001; Dove et al., 2006).
On the other hand, the localization of viral proteins to the nucleolus seems a prerequisite for replication of some viruses. For example, it has been shown that the nucleolar localization of HIV‐1 regulatory proteins, Rev and Tat, serves an important role in virus life cycle. The function of Rev is to bind HIV RNA and assist the transportation of spliced and unspliced RNA to the cytoplasm, and therefore it is likely that the nucleolus plays a critical role in HIV‐1 RNA export (Michienzi et al., 2000). The Tat is a key regulator of HIV‐1 replication, and studies strongly suggest that the nucleolus is implicated in the regulation of Tat activity and consequently in HIV‐1 replication (Wei et al., 1998; Michienzi et al., 2002).
Several other studies also showed that viral proteins may accumulate at the nucleolus and this phenomenon is important for viral life cycle (Tijms et al., 2002; Kim et al., 2007; Pei et al., 2008). However, the precise role of the nucleolus in viral pathogenesis remains poorly understood and further investigations will be necessary to shed light on the all aspects of interaction between nucleolus and viral proteins.
Overcoming the PML NBs
As mentioned above, PML NBs are implicated in antiviral response. Early studies indicated that many PML NB proteins, such as PML and Sp100, are IFN (interferon) inducible (Guldner et al., 1992; Stadler et al., 1995). IFN treatment of cells greatly increases the number, intensity and size of PML NBs and thus, this contributes to an enhanced antiviral response (Grotzinger et al., 1996; Everett and Chelbi‐Alix, 2007). However, many viruses have evolved a variety of mechanisms to avoid the antiviral function of PML NBs. Below we discuss how DNA as well as RNA viruses overcome the PML NB barrier, leading to striking changes of these nuclear bodies.
DNA viruses
It was shown that the parental genomes of herpes simplex virus type 1 (HSV‐1), human cytomegalovirus (HCMV) and adenovirus are closely associated with PML NBs (Ishov and Maul, 1996). The consequences of association were unclear for many years, but recent studies revealed the importance of this event for intrinsic defence against DNA viruses (Everett, 2006). In turn, DNA viruses, especially herpesviruses, counteract this intracellular defence by modifying PML NBs.
Herpes simplex virus 1 was the first virus to be shown to disrupt PML NBs early during infection. The observation that HSV‐1 affects PML NBs, whereas an ICP0‐deficient mutant does not, led to the identification of the importance of ICP0 for this effect (Maul et al., 1993; Everett and Maul, 1994). The ICP0 protein contains a RING finger domain with ubiquitin E3 ligase activity, which induces the degradation of PML, leading to the release of other proteins of PML NBs (Parkinson and Everett, 2000). Proteins with similar, ND10‐modifying functions have been identified for other herpesviruses, including human cytomegalovirus (pp71 and IE1) and Epstein‐Barr virus (EBNA LP). The cytomegalovirus structural protein pp71 accumulates at PML NBs and is able to induce the proteasomal degradation of hDaxx (Saffert and Kalejta, 2006). Thus, pp71 can antagonize the cellular repression of immediate early gene expression. Consequently, the IE72 protein is synthesized, which then localizes to PML NBs and dissociates them through an abrogation of PML SUMOylation (Kang et al., 2006). In case of Epstein‐Barr virus, EBNA LP dislocates Sp100 from PML NB, leading to an alteration of this nuclear body (Ling et al., 2005).
Infection with human adenovirus type 5 (Ad5) results in a repartition of PML NBs into elongated tracks during the early hours of infection (Doucas et al., 1996). By screening a variety of Ad5 mutants, viral E4 ORF3 early protein was identified as key protein that induces the rearrangement of PML NBs into track‐like structures. Until recently the significance of E4 ORF3‐induced PML NBs track formation was obscure. Recent studies by Ullman and colleagues showed that the replication of an E4 ORF3 deletion virus is restricted in cells that are pretreated with IFN‐α or IFN‐γ, thus revealing the role of E4 ORF3 in viral counteraction against IFN‐induced antiviral response (Ullman et al., 2007). The exact mechanism by which E4 ORF3 helps Ad5 to overcome PML NBs is still unclear. Instead, it was reported that another Ad5 protein, E1B‐55K, may induce the degradation of hDaxx by a proteasome‐dependent pathway, thus antagonizing the antiviral activity of PML NBs (Schreiner et al., 2010). This finding demonstrates that E4 ORF3 is most probably not the only adenoviral protein involved in viral countermeasures against ND10, and further studies may reveal additional proteins like E1B‐55K.
RNA viruses
Examination of the functional role of PML in vivo demonstrated that fibroblasts derived from PML −/− mice are much more sensitive to some RNA viruses, such as rhabdoviruses vesicular stomatitis and rabies virus, and the arenavirus lymphocytic choriomeningitis virus. This and several other findings implicate PML in an intrinsic antiviral response of the cell that targets not only DNA but also RNA viruses (Djavani et al., 2001; Bonilla et al., 2002; Blondel et al., 2010). Like DNA viruses, RNA viruses have evolved different ways to impair the cellular antiviral response. For example, the lymphocytic choriomeningitis virus RING finger protein Z with an unknown function targets and redistributes PML to the cytoplasm (Borden et al., 1998). Rabies virus phosphoprotein P and four other amino‐terminally truncated products (P2, P3, P4 and P5) are involved in virus‐induced reorganization of PML NBs (Blondel et al., 2002). Encephalomyocarditis virus 3C protease co‐localizes with PML within nuclear bodies and induces the degradation of PML in a proteasome‐ and SUMO‐dependent manner (El McHichi et al., 2010). Within the family of retroviruses human T‐cell leukaemia virus type 1 encodes the Tax oncoprotein that has been demonstrated to alter the localization of a PML NB‐associated protein known as Int‐6 (Desbois et al., 1996). Although PML NBs are thought to be implicated in retroviral pathogenesis, their role for retroviruses is still not fully understood.
Other nuclear alterations
Above, we discussed two subnuclear structures that are mostly by viruses. However, during viral infections several other subnuclear structures, including the transport machinery (Fontoura et al., 2005), Cajal bodies (Kim et al., 2007), the nuclear lamina (Camozzi et al., 2008), as well as chromatin (You, 2010), are also targeted. Moreover, viruses can induce profound changes of the overall nuclear architecture. For example, African swine fever virus, a double‐stranded DNA virus that mainly replicates in the monocyte‐macrophage cell lineage and causes a lethal haemorraghic disease in domestic pigs, is prone to split the host cell nucleus. Recently, we described the presence of an additional nucleus in cells that were infected with African swine fever virus. The cytophotometry revealed that the DNA content in both nuclei did not exceed the diploid standard, which indicates that the additional nucleus was a consequence of fragmentation of cell nucleus (Karalova et al., 2011). However, the mechanism of fragmentation remains obscure and warrants further investigation.
Concluding remarks
Until recently, with some exceptions, research into the interactions between viruses and the nucleus has been descriptive. The functional implications of these interactions are now being realized and progress is being made to clarify how and why viruses modify subnuclear structures. At the same time, many questions concerning virus–nucleus interactions remain open. For example, it is still unclear why RNA virus proteins localize to the nucleolus or whether the PML NBs play a role for establishing herpesviral latency. Thus, further studies are likely to provide new and exciting insights into viral and cellular biology as well as to uncover new avenues of therapeutic interests.
Acknowledgements
The authors would like to thank Drs Robin Weiss and Yana Galoyan for critical review and Armenak Keshishyan for help with the illustrations. The work of H.Z. was supported by ANSEF grant. T.S. was supported by the DFG (SFB796).
References
- Ahmad, Y. , Boisvert, F.M. , Gregor, P. , Cobley, A. , and Lamond, A.I. (2009) NOPdb: Nucleolar Proteome Database‐2008 update. Nucleic Acids Res 37: 181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain, F.H.‐T. , Bouvet, P. , Dieckmann, T. , and Feigon, J. (2000) Molecular basis of sequencespecific recognition of pre‐ribosomal RNA by nucleolin. EMBO J 19: 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminev, A.G. , Amineva, S.P. , and Palmenberg, A.C. (2003) Encephalomyocarditis virus (EMCV) proteins 2A and 3BCD localize to nuclei and inhibit cellular mRNA transcription but not rRNA transcription. Virus Res 95: 59–73. [DOI] [PubMed] [Google Scholar]
- Amineva, S.P. , Aminev, A.G. , Palmenberg, A.C. , and Gern, J.E. (2004) Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J Gen Virol 85: 2969–2979. [DOI] [PubMed] [Google Scholar]
- Ascoli, C.A. , and Maul, G.G. (1991) Identification of a novel nuclear domain. J Cell Biol 112: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, R. , and Pandolfi, P.P. (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016. [DOI] [PubMed] [Google Scholar]
- Bischof, O. , Kirsh, O. , Pearson, M. , Itahana, K. , Pelicci, P.G. , and Dejean, A. (2002) Deconstructing PML‐induced premature senescence. EMBO J 21: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, D. , Regad, T. , Poisson, N. , Pavie, B. , Harper, F. , Pandolfi, P.P. , et al (2002) Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21: 7957–7970. [DOI] [PubMed] [Google Scholar]
- Blondel, D. , Kheddache, S. , Lahaye, X. , Dianoux, L. , and Chelbi‐Alix, M.K. (2010) Resistance to rabies virus infection conferred by the PMLIV isoform. J Virol 84: 10719–10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla, W.V. , Pinschewer, D.D. , Klenerman, P. , Rousson, V. , Gaboli, M. , Pandolfi, P.P. , et al (2002) Effects of promyelocytic leukemia protein on virus‐host balance. J Virol 76: 3810–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K. , Campbell‐Dwyer, E.J. , Carlile, G. , Djavani, M. , and Salvato, M. (1998) Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol 72: 3819–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle, A. , Ugrinova, I. , Epstein, A.L. , Bouvet, P. , Diaz, J.J. , and Greco, A. (2008) Nucleolin is required for an efficient herpes simplex virus type 1 infection. J Virol 82: 4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camozzi, D. , Pignatelli, S. , Valvo, C. , Lattanzi, G. , Capanni, C. , Dal Monte, P. , et al (2008) Remodeling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J Gen Virol 89: 731–740. [DOI] [PubMed] [Google Scholar]
- Cannavo, G. , Paiardini, M. , Galati, D. , Cervasi, B. , Montroni, M. , De Vico, G. , et al (2001) Abnormal intracellular kinetics of cellcycle‐dependent proteins in lymphocytes from patients infected with human immunodeficiency virus: a novel biologic link between immune activation, accelerated T‐cell turnover, and high levels of apoptosis. Blood 97: 1756–1764. [DOI] [PubMed] [Google Scholar]
- Castiglia, C.L. , and Flint, S.J. (1983) Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol Cell Biol 3: 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans, R.W. , and Dimmock, N.J. (1969) An electron microscopy study of single‐cycle infection of chick embryo fibroblasts by influenza virus. Virology 39: 499–515. [DOI] [PubMed] [Google Scholar]
- Dellaire, G. , and Bazett‐Jones, D.P. (2004) PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26: 963–977. [DOI] [PubMed] [Google Scholar]
- Desbois, C. , Rousset, R. , Bantignies, F. , and Jalinot, P. (1996) Exclusion of Int‐6 from PML nuclear bodies by binding to the HTLV‐I Tax oncoprotein. Science 273: 951–953. [DOI] [PubMed] [Google Scholar]
- Dimmock, N.J. (1969) New virus‐specific antigens in cells infected with influenza virus. Virology 39: 224–234. [DOI] [PubMed] [Google Scholar]
- Djavani, M. , Rodas, J. , Lukashevich, I.S. , Horejsh, D. , Pandolfi, P.P. , Borden, K.L. , et al (2001) Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J Virol 75: 6204–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucas, V. , Ishov, A.M. , Romo, A. , Juguilon, H. , Weitzman, M.D. , Evans, R.M. , et al (1996) Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev 10: 196–207. [DOI] [PubMed] [Google Scholar]
- Dove, B.K. , You, J.H. , Reed, M.L. , Emmett, S.R. , Brooks, G. , and Hiscox, J.A. (2006) Changes in nucleolar architecture and protein profile during coronavirus infection. Cell Microbiol 8: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbar, T.S. , Gentry, G.A. , and Olson, M.O. (1989) Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry 28: 9495–9501. [DOI] [PubMed] [Google Scholar]
- El McHichi, B. , Regad, T. , Maroui, M.A. , Rodriguez, M.S. , Aminev, A. , Gerbaud, S. , et al (2010) SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J Virol 84: 11634–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott, E. , Rodgers, M.A. , Macdonald, A. , McCrory, S. , Ajuh, P. , and Hiscox, J.A. (2010a) Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol Cell Proteomics 9: 1920–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott, E. , Wise, H. , Loucaides, E.M. , Matthews, D.A. , Digard, P. , and Hiscox, J.A. (2010b) Quantitative proteomics using SILAC coupled to LC‐MS/MS reveals changes in the nucleolar proteome in influenza A virus‐infected cells. J Proteome Res 9: 5335–5345. [DOI] [PubMed] [Google Scholar]
- Everett, R.D. (2006) Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol 8: 365–374. [DOI] [PubMed] [Google Scholar]
- Everett, R.D. , and Chelbi‐Alix, M.K. (2007) PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89: 819–830. [DOI] [PubMed] [Google Scholar]
- Everett, R.D. , and Maul, G.G. (1994) HSV‐1 IE protein Vmw110 causes redistribution of PML. EMBO J 13: 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M. , Faria, P.A. , and Nussenzveig, D.R. (2005) Viral interactions with the nuclear transport machinery: discovering and disrupting pathways. IUBMB Life 57: 65–72. [DOI] [PubMed] [Google Scholar]
- Grotzinger, T. , Sternsdorf, T. , Jensen, K. , and Will, H. (1996) Interferon‐modulated expression of genes encoding the nuclear‐dot‐associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur J Biochem 238: 554–560. [DOI] [PubMed] [Google Scholar]
- Guldner, H.H. , Szostecki, C. , Grotzinger, T. , and Will, H. (1992) IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol 149: 4067–4073. [PubMed] [Google Scholar]
- Gustin, K.E. , and Sarnow, P. (2002) Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76: 8787–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U.T. , and Sarnow, P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hindley, C.E. , Davidson, A.D. , and Matthews, D.A. (2007) The relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J Gen Virol 88: 3244–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov, A.M. , and Maul, G.G. (1996) The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol 134: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, R.E. , Valdez, B. , Banerjee, R. , Srivastava, M. , and Dasgupta, A. (2001) Nucleolin stimulates viral internal ribosome entry site‐mediated translation. Virus Res 76: 17–29. [DOI] [PubMed] [Google Scholar]
- Kang, H. , Kim, E.T. , Lee, H.R. , Park, J.J. , Go, Y.Y. , Choi, C.Y. , et al (2006) Inhibition of SUMO‐independent PML oligomerization by the human cytomegalovirus IE1 protein. J Gen Virol 87: 2181–2190. [DOI] [PubMed] [Google Scholar]
- Karalova, E.M. , Sargsyan, K.V. , Hampikian, G.K. , Voskanyan, H.E. , Abroyan, L.O. , Avetisyan, A.S. , et al (2011) Phenotypic and cytologic studies of lymphoid cells and monocytes in primary culture of porcine bone marrow during infection of African swine fever virus. In Vitro Cell Dev Biol Anim 47: 200–204. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Ryabov, E.V. , Kalinina, N.O. , Rakitina, D.V. , Gillespie, T. , MacFarlane, S. , et al (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, Y.W. , Evans, V.C. , Heesom, K.J. , Lamond, A.I. , and Matthews, D.A. (2010) Proteomics analysis of the nucleolus in adenovirus‐infected cells. Mol Cell Proteomics 9: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, F.J. , McStay, B. , and Matthews, D.A. (2006) Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J Cell Sci 119: 2621–2631. [DOI] [PubMed] [Google Scholar]
- Li, Y.P. , Busch, R.K. , Valdez, B.C. , and Busch, H. (1996) C23 interacts with B23, a putative nucleolar‐localization‐signal‐binding protein. Eur J Biochem 237: 153–158. [DOI] [PubMed] [Google Scholar]
- Ling, P.D. , Peng, R.S. , Nakajima, A. , Yu, J.H. , Tan, J. , Moses, S.M. , et al (2005) Mediation of Epstein‐Barr virus EBNA‐LP transcriptional coactivation by Sp100. EMBO J 24: 3565–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymberopoulos, M.H. , and Pearson, A. (2007) Involvement of UL24 in herpes‐simplex‐virus‐1‐induced dispersal of nucleolin. Virology 363: 397–409. [DOI] [PubMed] [Google Scholar]
- Lymberopoulos, M.H. , Bourget, A. , Abdeljelil, N.B. , and Pearson, A. (2011) Involvement of the UL24 protein in herpes simplex virus 1‐induced dispersal of B23 and in nuclear egress. Virology 412: 341–348. [DOI] [PubMed] [Google Scholar]
- Matthews, D.A. (2001) Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J Virol 75: 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.A. , and Olson, M.O. (2006) What's new in the nucleolus? EMBO Rep 7: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.A. , and Russell, W.C. (1998) Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J Gen Virol 79: 1671–1675. [DOI] [PubMed] [Google Scholar]
- Maul, G.G. , Guldner, H.H. , and Spivack, J.G. (1993) Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol 74: 2679–2690. [DOI] [PubMed] [Google Scholar]
- Maul, G.G. , Yu, E. , Ishov, A.M. , and Epstein, A.L. (1995) Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem 59: 498–513. [DOI] [PubMed] [Google Scholar]
- Michienzi, A. , Cagnon, L. , Bahner, I. , and Rossi, J.J. (2000) Ribozyme‐mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV‐1 RNA. Proc Natl Acad Sci USA 97: 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi, A. , Li, S. , Zaia, J.A. , and Rossi, J.J. (2002) A nucleolar TAR decoy inhibitor of HIV‐1 replication. Proc Natl Acad Sci USA 99: 14047–14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, Y. , Nosaka, T. , and Hatanaka, M. (1996) The posttranscriptional regulator Rev of HIV: implications for its interaction with the nucleolar protein B23. Biochimie 78: 1081–1086. [DOI] [PubMed] [Google Scholar]
- Munday, D.C. , Emmott, E. , Surtees, R. , Lardeau, C.H. , Wu, W. , Duprex, W.P. , et al (2010) Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol Cell Proteomics 9: 2438–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki, M. , Iwamatsu, A. , Tsujimoto, M. , and Nagata, K. (2001) Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J Mol Biol 311: 41–55. [DOI] [PubMed] [Google Scholar]
- Parkinson, J. , and Everett, R.D. (2000) Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol 74: 10006–10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples, M.E. , Wang, C. , Gupta, K.C. , and Coleman, N. (1992) Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J Virol 66: 3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y. , Hodgins, D.C. , Lee, C. , Calvert, J.G. , Welch, S.K. , Jolie, R. , et al (2008) Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res 135: 107–114. [DOI] [PubMed] [Google Scholar]
- Saffert, R.T. , and Kalejta, R.F. (2006) Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate‐early gene expression. J Virol 80: 3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni, P. , and Pandolfi, P.P. (2002) The role of PML in tumor suppression. Cell 108: 165–170. [DOI] [PubMed] [Google Scholar]
- Scheer, U. , and Hock, R. (1999) Structure and function of the nucleolus. Curr Opin Cell Biol 11: 385–390. [DOI] [PubMed] [Google Scholar]
- Schreiner, S. , Wimmer, P. , Sirma, H. , Everett, R.D. , Blanchette, P. , Groitl, P. , et al (2010) Proteasome‐dependent degradation of Daxx by the viral E1B‐55K protein in human adenovirus‐infected cells. J Virol 84: 7029–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, M. , Chelbi‐Alix, M.K. , Koken, M.H. , Venturini, L. , Lee, C. , Saib, A. , et al (1995) Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11: 2565–2573. [PubMed] [Google Scholar]
- Takahashi, Y. , Lallemand‐Breitenbach, V. , Zhu, J. , and de The, H. (2004) PML nuclear bodies and apoptosis. Oncogene 23: 2819–2824. [DOI] [PubMed] [Google Scholar]
- de The′, G. , Rivie're, M. , and Bernhard, W. (1960) Examen au microscope e'lectronique de la tumeur VX2 du lapin domestique de'rive'e du papillome de Shope. Bull Cancer 47: 570–584. [Google Scholar]
- Tijms, M.A. , van der Meer, Y. , and Snijder, E.J. (2002) Nuclear localization of non‐structural protein 1 and nucleocapsid protein of equine arteritis virus. J Gen Virol 83: 795–800. [DOI] [PubMed] [Google Scholar]
- Tollervey, D. , Lehtonen, H. , Jansen, R. , Kern, H. , and Hurt, E.C. (1993) Temperature‐sensitive mutations demonstrate roles for yeast fibrillarin in pre‐rRNA processing, pre‐rRNA methylation, and ribosome assembly. Cell 72: 443–457. [DOI] [PubMed] [Google Scholar]
- Ullman, A.J. , Reich, N.C. , and Hearing, P. (2007) Adenovirus E4 ORF3 protein inhibits the interferon‐mediated antiviral response. J Virol 81: 4744–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner, S. , and Sarnow, P. (1998) Viral ribonucleoprotein complex formation and nucleolar–cytoplasmic relocalization of nucleolin in poliovirus‐infected cells. J Viro 72: 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, T.H. , Moen, P.T. , Fox, E. , and Bodnar, J.W. (1989) Interactions of minute virus of mice and adenovirus with host nucleoli. J Virol 63: 3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.H. , Syu, W.J. , Huang, K.J. , Lei, H.Y. , Yao, C.W. , King, C.C. , and Hu, S.T. (2002) Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J Gen Virol 83: 3093–3102. [DOI] [PubMed] [Google Scholar]
- Wei, P. , Garber, M.E. , Fang, S.M. , Fischer, W.H. , and Jones, K.A. (1998) A novel CDK9‐associated C‐type cyclin interacts directly with HIV‐1 Tat and mediates its high‐affinity, loop‐specific binding to TAR RNA. Cell 92: 451–462. [DOI] [PubMed] [Google Scholar]
- Wurm, T. , Chen, H. , Britton, P. , Brooks, G. , and Hiscox, J.A. (2001) Localisation to the nucleolus is a common feature of coronavirus nucleoproteins and the protein may disrupt host cell division. J Virol 75: 9345–9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T.H. , Tsai, W.H. , Lee, Y.M. , Lei, H.Y. , Lai, M.Y. , Chen, D.S. , et al (1994) Purification and characterization of nucleolin and its identification as a transcription repressor. Mol Cell Biol 14: 6068–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, J. (2010) Papillomavirus interaction with cellular chromatin. Biochim Biophys Acta 1799: 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, S. , Salomoni, P. , and Pandolfi, P.P. (2000) The transcriptional role of PML and the nuclear body. Nat Cell Biol 2: 85–90. [DOI] [PubMed] [Google Scholar]


