Abstract
Severe acute respiratory syndrome (SARS) is a novel human illness caused by a previously unrecognized coronavirus (CoV) termed SARS‐CoV. There are conflicting reports on the animal reservoir of SARS‐CoV. Many of the groups that argue carnivores are the original reservoir of SARS‐CoV use a phylogeny to support their argument. However, the phylogenies in these studies often lack outgroup and rooting criteria necessary to determine the origins of SARS‐CoV. Recently, SARS‐CoV has been isolated from various species of Chiroptera from China (e.g., Rhinolophus sinicus) thus leading to reconsideration of the original reservoir of SARS‐CoV. We evaluated the hypothesis that SARS‐CoV isolated from Chiroptera are the original zoonotic source for SARS‐CoV by sampling SARS‐CoV and non‐SARS‐CoV from diverse hosts including Chiroptera, as well as carnivores, artiodactyls, rodents, birds and humans. Regardless of alignment parameters, optimality criteria, or isolate sampling, the resulting phylogenies clearly show that the SARS‐CoV was transmitted to small carnivores well after the epidemic of SARS in humans that began in late 2002. The SARS‐CoV isolates from small carnivores in Shenzhen markets form a terminal clade that emerged recently from within the radiation of human SARS‐CoV. There is evidence of subsequent exchange of SARS‐CoV between humans and carnivores. In addition SARS‐CoV was transmitted independently from humans to farmed pigs (Sus scrofa). The position of SARS‐CoV isolates from Chiroptera are basal to the SARS‐CoV clade isolated from humans and carnivores. Although sequence data indicate that Chiroptera are a good candidate for the original reservoir of SARS‐CoV, the structural biology of the spike protein of SARS‐CoV isolated from Chiroptera suggests that these viruses are not able to interact with the human variant of the receptor of SARS‐CoV, angiotensin‐converting enzyme 2 (ACE2). In SARS‐CoV we study, both visually and statistically, labile genomic fragments and, putative key mutations of the spike protein that may be associated with host shifts. We display host shifts and candidate mutations on trees projected in virtual globes depicting the spread of SARS‐CoV. These results suggest that more sampling of coronaviruses from diverse hosts, especially Chiroptera, carnivores and primates, will be required to understand the genomic and biochemical evolution of coronaviruses, including SARS‐CoV.
© The Willi Hennig Society 2008.
Severe acute respiratory syndrome (SARS) is a recently described human infectious disease caused by a previously unrecognized coronavirus, SARS‐CoV (Ksiazek et al., 2003). Between November 2002 and August 2003, there were 8422 cases and 916 deaths from SARS (WHO, 2003). These numbers are not on the scale of major epidemics such as seasonal forms of influenza infecting humans, but in an era of rapid globalization, the potential for a pandemic was significant. SARS‐CoV infection has not been reported among humans since the early days of 2004. However, there remain conflicting reports on the animal reservoir of SARS‐CoV. Guan et al. (2003) and Kan et al. (2005) implicate small carnivores whereas Li et al. (2005) and Lau et al. (2005) asserted that Chiroptera are the animal reservoir of SARS‐CoV. In a comprehensive review of CoV among Chiroptera, Tang et al. (2006) argued that the origin of SARS‐CoV remains unknown.
Among humans, serological surveys indicate that SARS‐CoV viruses were circulating in subepidemic levels in 2001 in residents of Hong Kong (data from mainland China is not available) (Zheng et al., 2004). Also, in describing the world's largest SARS epidemic in Beijing, Pang et al. (2003) point out that “It is possible that some SARS cases were not counted before mid‐April 2003 when the extent of the outbreak was fully recognized.”
In a search for the animal reservoir of SARS‐CoV outside of urban areas Kan et al. (2005) surveyed farmed Parguma larvata (Himalayan palm civet) in 25 farms spread over 12 provinces in South‐east China and found no evidence of SARS‐CoV infection. SARS‐CoV in carnivores was isolated to animals in the Xinyuan market, in the suburbs of Guangzhou, China. Vijaykrishna et al. (2007) make the argument that Chiroptera are a reservoir for a wide variety of coronaviruses (SARS and non‐SARS) that affect humans and animals. Before the SARS outbreak, coronaviruses were known primarily from animals of agricultural importance in which they cause respiratory and enteric infections (Siddell et al., 1983). The human strains CoV‐229E and CoV‐OC43, which are distantly related to SARS‐CoV, cause mild respiratory illnesses similar to the common cold (Mahony and Richardson, 2005). Recently Dominguez et al. (2007) have shown that Chiroptera (Myotis occultus and Eptesicus fuscus from the Rocky Mountains of Colorado, USA, carry group 1 coronaviruses. Our preliminary analyses show that these CoVs from Rocky Mountain Chiroptera are very closely related to group 1 CoV that infect humans (e.g., CoV‐229E and CoV‐OC43).
Genomic sequence data
The genome of a coronavirus is comprised of a single‐stranded, positive‐sensed RNA molecule 27–31 kilobases in length (Lai, 1990). Before the SARS‐CoV outbreak coronavirus diversity was poorly documented, especially at the genomic level. However, coronavirus research has been invigorated since the sequencing of the first SARS‐CoV isolate (Marra et al., 2003; Rota et al., 2003). For example, in the wake of SARS, two novel human coronaviruses were found [HKU1, GenBank (http://www.ncbi.nlm.nih.gov) accession AY597011 (Woo et al., 2005); and NL63, GenBank accession NC_005831 (van der Hoek et al., 2004)]. Also notable are the release of new genomic sequences for SARS‐CoV among carnivores, artiodactyls, humans and Chiroptera (Guan et al., 2003; Chinese SARS Molecular Epidemiology Consortium, 2004; Tu et al., 2004; Chen et al., 2005; Lau et al., 2005; Li et al., 2005; Tang et al., 2006).
Guan et al. (2003) sequenced several partial and complete genomes from SARS‐CoV isolated in 2003 from two small carnivore hosts Parguma larvata and Nyctereutes procyonoides (raccoon dog) that were for sale in live animal markets in Shenzhen, Guangdong Province, China. Complete and partial genomes of the coronaviruses isolated from P. larvata[SARS‐CoV SZ1, SZ16, SZ3; GenBank accessions AY304489, AY304488 and AY304486] and Nyctereutes procyonoides (SARS‐CoV SZ13; GenBank accession AY304487) became available publically in September 2003 but were updated in November 2003. A complete genome of a SARS‐CoV isolated from P. larvata host was released in January, 2005 (SARS‐CoV HC/SZ/61/03; GenBank accession AY515512). A complete genome of SARS‐CoV isolated from Melogale moschata, the Chinese ferret badger, was released in March, 2005 (SARS coronavirus CFB/SZ/94/03; GenBank accession AY545919).
Several, but not all of the genomes of the coronaviruses isolated from small carnivores contain a specific 29‐nucleotide region (CCTACTGGTTACCAACCTGAATGGAATAT, e.g., positions 27869–27897 in the of AY304488) in a protein with an unknown function. It was initially reported that this 29‐nucleotide region was absent from all human SARS‐CoV isolates sequenced with the notable exception of one isolate from Guangdong that contains the 29‐nucleotide region (GD01 GenBank accession AY278489) (Guan et al., 2003); however, several human isolates were later discovered to contain the region. Owing to the perceived potential of the 29‐nucleotide region as a clue to the animal origins and subsequent adaptation of SARS‐CoV to human hosts, this 29‐nucleotide region garnered media attention as early as May 2003 as a “29‐nucleotide deletion” in human SARS‐CoV that enabled animal to human transmission (Bradsher and Altman, 2003; Enserink, 2003).
SARS‐CoV isolates from Chiroptera contain a different 29‐nucleotide sequence (CCAATACATTACTATTCGGACTGGTTTAT, e.g., positions 27866–27894 in DQ648857, Bat coronavirus BtCoV/279/2005) in a protein with an unknown function. This fragment from isolates of SARS‐CoV derived from Chiroptera is in an orthologous genomic position to the 29‐nucleotide region described above for some SARS‐CoV isolated from small carnivores and humans. When the 29‐nucleotide regions from Chiroptera versus human and carnivore hosts are compared, 12 nucleotide positions are polymorphic (Lau et al., 2005). Under the current sampling of SARS‐CoV, this fragment is exclusive to SARS‐CoV isolated from Chiroptera.
The Chinese SARS Molecular Epidemiology Consortium (2004) published an analysis of molecular evolution of SARS‐CoV within humans during the 2002–03 epidemic. This study included the release of many new genomic sequences of SARS‐CoV from humans infected in the early stages of the outbreak in southern China 1 .
A human SARS‐CoV associated with a re‐emergent case of SARS in Guangzhou, Guangdong Province, China was isolated December 22, 2003. The sequence of this SARS‐CoV spike gene was released in February 2004 (SARS‐CoV GD03T0013; GenBank accession AY525636).
Song et al. (2005) released many full and partial genome sequences of SARS‐CoV isolated from human and palm civet cats collected in southern China into the public domain in 2005 2 .Kan et al. (2005) released many spike gene and three full genome sequences for SARS‐CoV isolated from human, raccoon dog and civet cat hosts into the public domain in July, 2006 3 .
Li et al. (2005) 4 published SARS‐CoV nucleoprotein and spike gene sequences (some recently updated as whole genomes) isolated from Chiroptera: Rhinolophus sinicus, Rhinolophus ferrumequinum, Rhinolophus macrotis and Rhinolophus pearsoni. Lau et al. (2005) 5 published three complete SARS‐CoV genomes isolated from the bat Rhinolophus pearsoni and a SARS‐CoV polymerase sequences from Rhinolophus sinicus. Poon et al. (2005) 6 published sequences of RNA‐dependent RNA polymerase (RdRp), polyprotein, and spike genes of a non‐SARS‐CoV isolated from the bat Miniopterous pusillus. Tang et al. (2006) 7 published a review of bat coronaviruses in August, 2006 and released three genomes and 70 gene fragments in July, 2006.
Receptor binding studies
Li et al. (2006) provide a review of the structural biology of the SARS‐CoV spike protein and the variation of the receptor for spike protein on host cells, angiotensin‐converting enzyme 2 (ACE2), among human and carnivore hosts. These authors point out via pairwise alignment that the spike protein of SARS‐CoV isolated from Chiroptera lack a stretch of amino acid residues and have mismatches among other residues that form the receptor‐binding motif for the human variant of ACE2.
There is also empirical evidence concerning the relative affinity of various spike proteins to ACE2 from various hosts. The SARS‐CoV spike proteins tested include: an early epidemic, 2002–03, human isolate (SARS‐CoV, TOR 2), a human isolate tied to sporadic infections in 2003–04 (SARS‐CoV, GD03T0013), and a carnivore isolate (P. larvata, SZ3) from 2003 to 2003 (Li et al., 2005). 2006, 2005) describe and “expected” result for SZ3 and an “unexpected” result for GD03T0013 that both of these spike proteins bound P. larvata ACE2 better than they bound human ACE2. Spike protein from TOR 2 bound ACE2 from P. larvata and human equally well. The unexpected nature of their results is tied to the perception that the SARS‐CoV virus was adapting from carnivore to humans as suggested by prevailing phylogenetic studies of the time (e.g., Guan et al., 2003; Chinese SARS Molecular Epidemiology Consortium, 2004; Kan et al., 2005; Song et al., 2005).
Methods
Demarcation of sequence characters
We compared nucleotide sequences for whole and partially sequenced genomes that were in the public domain as of January 1, 2005. This data set included 83 viruses from a wide host and geographic range (Table 1). First, we compared these genomes with ClustalW under default settings (i.e., gap opening penalty 15 gap extension penalty 6.66, DNA transition weight 0.5) (Thompson et al., 1994) and developed a set of fragment boundaries that accommodated both sequence similarity and unequal sequencing coverage. We then split the genomes along these boundaries and remove all gaps inserted by ClustalW, thus forming 62 sequence fragment characters for POY3 (Wheeler et al., 2006).
Table 1.
GenBank accession numbers and descriptions of genomes and partial genomes of virus exemplars considered in the 83 isolate data set
| GenBank accession no. | Name of virus |
|---|---|
| AF124986 | Canine coronavirus |
| AF124987 | Feline infectious peritonitis virus |
| AF124988 | Porcine hemagglutinating encephalomyelitis virus |
| AF124989 | Human coronavirus OC43 |
| AF124990 | Rat sialodacryoadenitis coronavirus |
| AF124991 | Turkey coronavirus |
| AF201929 | Murine hepatitis strain 2 |
| AF207902 | Murine hepatitis virus ML11 |
| AF208066 | Murine hepatitis virus Penn 971 |
| AF208067 | Murine hepatitis virus ML10 |
| AF220295 | Bovine coronavirus Quebec |
| AF304460 | Human coronavirus 229E |
| AF391542 | Bovine coronavirus LUN |
| AJ271965 | Transmissible gastroenteritis virus |
| AY278487 | SARS coronavirus BJ02 |
| AY278488 | SARS coronavirus BJ01 |
| AY278489 | SARS coronavirus GD01 |
| AY278490 | SARS coronavirus BJ03 |
| AY278491 | SARS coronavirus HKU39849 |
| AY278554 | SARS coronavirus CUHK W1 |
| AY278741 | SARS coronavirus Urbani |
| AY279354 | SARS coronavirus BJ04 |
| AY282752 | SARS coronavirus CUHK Su10 |
| AY283794 | SARS coronavirus SIN 2500 |
| AY283795 | SARS coronavirus SIN 2677 |
| AY283796 | SARS coronavirus SIN 2679 |
| AY283797 | SARS coronavirus SIN 2748 |
| AY283798 | SARS coronavirus SIN 2774 |
| AY291315 | SARS coronavirus Frankfurt1 |
| AY291451 | SARS coronavirus TW1 |
| AY297028 | SARS coronavirus ZJ01 |
| AY304486 | SARS coronavirus SZ3 civet cat |
| AY304487 | SARS coronavirus SZ13 civet cat |
| AY304488 | SARS coronavirus SZ16 civet cat |
| AY304489 | SARS coronavirus SZ1 raccoon dog |
| AY304490 | SARS coronavirus GZ43 |
| AY304491 | SARS coronavirus GZ60 |
| AY304492 | SARS coronavirus HKU 36871 |
| AY304493 | SARS coronavirus HKU 65806 |
| AY304494 | SARS coronavirus HKU 66078 |
| AY304495 | SARS coronavirus GZ50 |
| AY313906 | SARS coronavirus GD69 |
| AY321118 | SARS coronavirus TWC |
| AY323977 | SARS coronavirus HSR1 |
| AY345986 | SARS coronavirus CUHK AG01 |
| AY345987 | SARS coronavirus CUHK AG02 |
| AY390556 | SARS coronavirus GZ02 |
| AY394978 | SARS coronavirus GZ B |
| AY394979 | SARS coronavirus GZ C |
| AY394980 | SARS coronavirus GZ D |
| AY394981 | SARS coronavirus HGZ8L1 A |
| AY394982 | SARS coronavirus HGZ8L1 B |
| AY394983 | SARS coronavirus HSZ2 A |
| AY394984 | SARS coronavirus HSZ A |
| AY394985 | SARS coronavirus HSZ Bb |
| AY394986 | SARS coronavirus HSZ Cb |
| AY394987 | SARS coronavirus HZS2 Fb |
| AY394989 | SARS coronavirus HZS2 D |
| AY394990 | SARS coronavirus HZS2 E |
| AY394991 | SARS coronavirus HZS2 Fc |
| AY394992 | SARS coronavirus HZS2 C |
| AY394993 | SARS coronavirus HGZ8L2 |
| AY394994 | SARS coronavirus HSZ Bc |
| AY394995 | SARS coronavirus HSZ Cc |
| AY394996 | SARS coronavirus ZS B |
| AY394997 | SARS coronavirus ZS A |
| AY394999 | SARS coronavirus LC2 |
| AY395000 | SARS coronavirus LC3 |
| AY395001 | SARS coronavirus LC4 |
| AY395002 | SARS coronavirus LC5 |
| AY395003 | SARS coronavirus ZS C |
| AY395004 | SARS coronavirus HZS2 Bb |
| AY515512 | SARS coronavirus HC SZ 61 03 civet cat |
| AY525636 | SARS coronavirus GD03T0013 |
| AY567487 | Human Coronavirus NL63 |
| AY654624 | SARS coronavirus TJF pig |
| BCU00735 | Bovine coronavirus Mebus |
| NC_001451 | Avian infectious bronchitis virus |
| NC_001846 | Murine hepatitis virus MHVA59 |
| NC_003045 | Bovine coronavirus |
| NC_003436 | Porcine epidemic diarrhea virus |
| NC_004718 | SARS coronavirus TOR2 |
| NC_005147 | Human coronavirus OC43 NL |
We use the same ClustalW settings to produce an updated aligned data set of whole and partially sequenced genomes that were in the public domain as of July 21, 2006. The updated data set includes 157 viruses many of which were isolated from Chiroptera and small carnivore hosts (Table 2). We then split the genomes along 66 boundaries and removed all gaps inserted by ClustalW, thus forming an updated set of 67 sequence fragment characters for POY3.
Table 2.
GenBank accession numbers and descriptions of genomes and partial genomes of virus exemplars considered in the 157 isolate data set
| GenBank accession no. | Name of virus |
|---|---|
| AF124986 | Canine coronavirus |
| AF124987 | Feline infectious peritonitis |
| AF124988 | Porcine hemagglutinating encep |
| AF124989 | Human coronavirus strain OC43 |
| AF124990 | Rat sialodacryoadenitis CoV |
| AF124991 | Turkey coronavirus |
| AF201929 | Murine hepatitis 2 |
| AF207902 | Murine hepatitis ML 11 |
| AF208066 | Murine hepatitis Penn 97 1 |
| AF208067 | Murine hepatitis ML 10 |
| AF220295 | Bovine coronavirus Quebec |
| AF304460 | Human coronavirus 229E |
| AF391542 | Bovine CoV LUN |
| AJ271965 | Transmissible gastroenteritis |
| AP006557 | SARS coronavirus TWH |
| AP006558 | SARS coronavirus TWJ |
| AP006559 | SARS coronavirus TWK |
| AP006560 | SARS coronavirus TWS |
| AP006561 | SARS coronavirus TWY |
| AY278487 | SARS coronavirus BJ02 |
| AY278488 | SARS coronavirus BJ01 |
| AY278489 | SARS coronavirus GD01 |
| AY278490 | SARS coronavirus BJ03 |
| AY278491 | SARS coronavirus HKU 39849 |
| AY278554 | SARS coronavirus CUHK W1 |
| AY278741 | SARS coronavirus Urbani |
| AY279354 | SARS coronavirus BJ04 |
| AY282752 | SARS coronavirus CUHK Su10 |
| AY283794 | SARS coronavirus Sin2500 |
| AY283795 | SARS coronavirus Sin2677 |
| AY283796 | SARS coronavirus Sin2679 |
| AY283797 | SARS coronavirus Sin2748 |
| AY283798 | SARS coronavirus Sin2774 |
| AY291315 | SARS coronavirus Frankfurt 1 |
| AY291451 | SARS coronavirus TW1 |
| AY297028 | SARS coronavirus ZJ01 |
| AY304486 | SARS coronavirus SZ3 |
| AY304487 | SARS coronavirus SZ13 |
| AY304488 | SARS coronavirus SZ16 |
| AY304489 | SARS coronavirus SZ1 |
| AY304490 | SARS coronavirus GZ43 |
| AY304491 | SARS coronavirus GZ60 |
| AY304492 | SARS coronavirus HKU 36871 |
| AY304493 | SARS coronavirus HKU 65806 |
| AY304494 | SARS coronavirus HKU 66078 |
| AY304495 | SARS coronavirus GZ50 |
| AY310120 | SARS coronavirus FRA |
| AY313906 | SARS coronavirus GD69 |
| AY321118 | SARS coronavirus TWC |
| AY323977 | SARS coronavirus HSR |
| AY338174 | SARS coronavirus Taiwan TC1 |
| AY338175 | SARS coronavirus Taiwan TC2 |
| AY345986 | SARS coronavirus CUHK AG01 |
| AY345987 | SARS coronavirus CUHK AG02 |
| AY345988 | SARS coronavirus CUHK AG03 |
| AY348314 | SARS coronavirus Taiwan TC3 |
| AY350750 | SARS coronavirus PUMC01 |
| AY357075 | SARS coronavirus PUMC02 |
| AY357076 | SARS coronavirus PUMC03 |
| AY390556 | SARS coronavirus GZ02 |
| AY394850 | SARS coronavirus WHU |
| AY394977 | SARS coronavirus GZ A |
| AY394978 | SARS coronavirus GZ B |
| AY394979 | SARS coronavirus GZ C |
| AY394980 | SARS coronavirus GZ D |
| AY394981 | SARS coronavirus HGZ8L1 A |
| AY394982 | SARS coronavirus HGZ8L1 B |
| AY394983 | SARS coronavirus HSZ2 A |
| AY394984 | SARS coronavirus HSZ A |
| AY394985 | SARS coronavirus HSZ Bb |
| AY394986 | SARS coronavirus HSZ Cb |
| AY394987 | SARS coronavirus HZS2 Fb |
| AY394988 | SARS coronavirus JMD |
| AY394989 | SARS coronavirus HZS2 D |
| AY394990 | SARS coronavirus HZS2 E |
| AY394991 | SARS coronavirus HZS2 Fc |
| AY394992 | SARS coronavirus HZS2 C |
| AY394993 | SARS coronavirus HGZ8L2 |
| AY394994 | SARS coronavirus HSZ Bc |
| AY394995 | SARS coronavirus HSZ Cc |
| AY394996 | SARS coronavirus ZS B |
| AY394997 | SARS coronavirus ZS A |
| AY394998 | SARS coronavirus LC1 |
| AY394999 | SARS coronavirus LC2 |
| AY395000 | SARS coronavirus LC3 |
| AY395001 | SARS coronavirus LC4 |
| AY395002 | SARS coronavirus LC5 |
| AY395003 | SARS coronavirus ZS C |
| AY395004 | SARS coronavirus HZS2 Bb |
| AY427439 | SARS coronavirus AS |
| AY461660 | SARS coronavirus SoD |
| AY463059 | SARS coronavirus Shanghai QXC1 |
| AY485277 | SARS coronavirus Sino1 11 |
| AY485278 | SARS coronavirus Sino3 11 |
| AY502923 | SARS coronavirus TW10 |
| AY502924 | SARS coronavirus TW11 |
| AY502925 | SARS coronavirus TW2 |
| AY502926 | SARS coronavirus TW3 |
| AY502927 | SARS coronavirus TW4 |
| AY502928 | SARS coronavirus TW5 |
| AY502929 | SARS coronavirus TW6 |
| AY502930 | SARS coronavirus TW7 |
| AY502931 | SARS coronavirus TW8 |
| AY502932 | SARS coronavirus TW9 |
| AY508724 | SARS coronavirus NS 1 |
| AY515512 | SARS coronavirus HC SZ 61 03 |
| AY525636 | SARS coronavirus GD03T0013 |
| AY545914 | SARS coronavirus HC SZ 79 03 |
| AY545915 | SARS coronavirus HC SZ DM1 03 |
| AY545916 | SARS coronavirus HC SZ 266 03 |
| AY545917 | SARS coronavirus HC GZ 81 03 |
| AY545918 | SARS coronavirus HC GZ 32 03 |
| AY545919 | SARS coronavirus CFB SZ 94 03 |
| AY559082 | SARS coronavirus Sin852 |
| AY559084 | SARS coronavirus Sin3765V |
| AY559085 | SARS coronavirus Sin848 |
| AY559086 | SARS coronavirus Sin849 |
| AY559093 | SARS coronavirus Sin845 |
| AY559095 | SARS coronavirus Sin847 |
| AY559096 | SARS coronavirus Sin850 |
| AY567487 | Human Coronavirus NL63 |
| AY568539 | SARS coronavirus GZ0401 |
| AY572034 | SARS coronavirus civet007 |
| AY572035 | SARS coronavirus civet010 |
| AY572038 | SARS coronavirus civet020 |
| AY613947 | SARS coronavirus GZ0402 |
| AY613948 | SARS coronavirus PC4‐13 |
| AY613949 | SARS coronavirus PC4‐136 |
| AY613950 | SARS coronavirus PC4‐227 |
| AY613951 | SARS coronavirus PC4‐127 |
| AY613952 | SARS coronavirus PC4‐205 |
| AY613953 | SARS coronavirus GZ0403 |
| AY627044 | SARS coronavirus PC4‐115 |
| AY627045 | SARS coronavirus PC4‐137 |
| AY627046 | SARS coronavirus PC4‐145 |
| AY627047 | SARS coronavirus PC4‐199 |
| AY627048 | SARS coronavirus PC4‐241 |
| AY654624 | SARS coronavirus TJF |
| AY686863 | SARS coronavirus A022 |
| AY686864 | SARS coronavirus B039 |
| AY864197 | Bat coronavirus strain 61 |
| BCU00735 | Bovine coronavirus Mebus |
| DQ022305 | Bat SARS coronavirus HKU3 1 |
| DQ071613 | Bat SARS coronavirus Rp1 |
| DQ071614 | Bat SARS coronavirus Rp2 |
| DQ071615 | Bat SARS coronavirus Rp3 |
| DQ084199 | Bat SARS coronavirus HKU3 2 |
| DQ084200 | Bat SARS coronavirus HKU3 3 |
| DQ412042 | Bat SARS coronavirus Rf1 |
| DQ412043 | Bat SARS coronavirus Rm1 |
| DQ648857 | Bat coronavirus BtCoV 279 2005 |
| NC_001451 | Avian infectious bronchitis |
| NC_001846 | Murine hepatitis virus |
| NC_003045 | Bovine coronavirus |
| NC_003436 | Porcine epidemic diarrhea virus |
| NC_004718 | SARS coronavirus Toronto 2 |
| NC_005147 | Human coronavirus OC43 |
We produced a data set of 113 whole genomes of SARS‐CoV from human, Chiroptera, swine and carnivore hosts (Table 3) that were available to the public as of July 21, 2006. We used a single outgroup, human coronavirus NL63 (GenBank accession no. AY567487). The sequences in this data set were similar enough to align without splitting them into sequence fragment characters. Together these 114 complete genome sequences were aligned using default settings in ClustalW. This alignment was analyzed with standard tree search methods.
Table 3.
GenBank accession numbers and descriptions of whole genomes of virus exemplars considered in the 114 isolate data set
| AP006557 | SARS coronavirus TWH |
| AP006558 | SARS coronavirus TWJ |
| AP006559 | SARS coronavirus TWK |
| AP006560 | SARS coronavirus TWS |
| AP006561 | SARS coronavirus TWY |
| AY278487 | SARS coronavirus BJ02 |
| AY278488 | SARS coronavirus BJ01 |
| AY278489 | SARS coronavirus GD01 |
| AY278490 | SARS coronavirus BJ03 |
| AY278491 | SARS coronavirus HKU 39849 |
| AY278554 | SARS coronavirus CUHK W1 |
| AY278741 | SARS coronavirus Urbani |
| AY279354 | SARS coronavirus BJ04 |
| AY282752 | SARS coronavirus CUHK Su10 |
| AY283794 | SARS coronavirus Sin2500 |
| AY283795 | SARS coronavirus Sin2677 |
| AY283796 | SARS coronavirus Sin2679 |
| AY283797 | SARS coronavirus Sin2748 |
| AY283798 | SARS coronavirus Sin2774 |
| AY291315 | SARS coronavirus Frankfurt 1 |
| AY291451 | SARS coronavirus TW1 |
| AY297028 | SARS coronavirus ZJ01 |
| AY304486 | SARS coronavirus SZ3 |
| AY304488 | SARS coronavirus SZ16 |
| AY304495 | SARS coronavirus GZ50 |
| AY310120 | SARS coronavirus FRA |
| AY313906 | SARS coronavirus GD69 |
| AY321118 | SARS coronavirus TWC |
| AY323977 | SARS coronavirus HSR |
| AY338174 | SARS coronavirus Taiwan TC1 |
| AY338175 | SARS coronavirus Taiwan TC2 |
| AY345986 | SARS coronavirus CUHK AG01 |
| AY345987 | SARS coronavirus CUHK AG02 |
| AY345988 | SARS coronavirus CUHK AG03 |
| AY348314 | SARS coronavirus Taiwan TC3 |
| AY350750 | SARS coronavirus PUMC01 |
| AY357075 | SARS coronavirus PUMC02 |
| AY357076 | SARS coronavirus PUMC03 |
| AY390556 | SARS coronavirus GZ02 |
| AY394850 | SARS coronavirus WHU |
| AY394978 | SARS coronavirus GZ B |
| AY394979 | SARS coronavirus GZ C |
| AY394981 | SARS coronavirus HGZ8L1 A |
| AY394982 | SARS coronavirus HGZ8L1 B |
| AY394983 | SARS coronavirus HSZ2 A |
| AY394985 | SARS coronavirus HSZ Bb |
| AY394986 | SARS coronavirus HSZ Cb |
| AY394987 | SARS coronavirus HZS2 Fb |
| AY394988 | SARS coronavirus JMD |
| AY394989 | SARS coronavirus HZS2 D |
| AY394990 | SARS coronavirus HZS2 E |
| AY394991 | SARS coronavirus HZS2 Fc |
| AY394992 | SARS coronavirus HZS2 C |
| AY394993 | SARS coronavirus HGZ8L2 |
| AY394994 | SARS coronavirus HSZ Bc |
| AY394995 | SARS coronavirus HSZ Cc |
| AY394996 | SARS coronavirus ZS B |
| AY394997 | SARS coronavirus ZS A |
| AY394998 | SARS coronavirus LC1 |
| AY394999 | SARS coronavirus LC2 |
| AY395000 | SARS coronavirus LC3 |
| AY395001 | SARS coronavirus LC4 |
| AY395002 | SARS coronavirus LC5 |
| AY395003 | SARS coronavirus ZS C |
| AY395004 | SARS coronavirus HZS2 Bb |
| AY427439 | SARS coronavirus AS |
| AY461660 | SARS coronavirus SoD |
| AY463059 | SARS coronavirus ShanghaiQXC1 |
| AY485277 | SARS coronavirus Sino1 11 |
| AY485278 | SARS coronavirus Sino3 11 |
| AY502923 | SARS coronavirus TW10 |
| AY502924 | SARS coronavirus TW11 |
| AY502925 | SARS coronavirus TW2 |
| AY502926 | SARS coronavirus TW3 |
| AY502927 | SARS coronavirus TW4 |
| AY502928 | SARS coronavirus TW5 |
| AY502929 | SARS coronavirus TW6 |
| AY502930 | SARS coronavirus TW7 |
| AY502931 | SARS coronavirus TW8 |
| AY502932 | SARS coronavirus TW9 |
| AY508724 | SARS coronavirus NS 1 |
| AY515512 | SARS coronavirus HC SZ 61 03 |
| AY545914 | SARS coronavirus HC SZ 79 03 |
| AY545915 | SARS coronavirus HC SZ DM1 03 |
| AY545916 | SARS coronavirus HC SZ 266 03 |
| AY545917 | SARS coronavirus HC GZ 81 03 |
| AY545918 | SARS coronavirus HC GZ 32 03 |
| AY545919 | SARS coronavirus CFB SZ 94 03 |
| AY559082 | SARS coronavirus Sin852 |
| AY559084 | SARS coronavirus Sin3765V |
| AY559085 | SARS coronavirus Sin848 |
| AY559086 | SARS coronavirus Sin849 |
| AY559093 | SARS coronavirus Sin845 |
| AY559095 | SARS coronavirus Sin847 |
| AY559096 | SARS coronavirus Sin850 |
| AY567487 | Human Coronavirus NL63 |
| AY568539 | SARS coronavirus GZ0401 |
| AY572034 | SARS coronavirus civet007 |
| AY572035 | SARS coronavirus civet010 |
| AY572038 | SARS coronavirus civet020 |
| AY613947 | SARS coronavirus GZ0402 |
| AY613948 | SARS coronavirus PC4 13 |
| AY613949 | SARS coronavirus PC4136 |
| AY613950 | SARS coronavirus PC4227 |
| AY654624 | SARS coronavirus TJF |
| AY686863 | SARS coronavirus A022 |
| AY686864 | SARS coronavirus B039 |
| DQ022305 | Bat SARS coronavirus HKU3 1 |
| DQ071615 | Bat SARS coronavirus Rp3 |
| DQ084199 | Bat SARS coronavirus HKU3 2 |
| DQ084200 | Bat SARS coronavirus HKU3 3 |
| DQ412043 | Bat SARS coronavirus Rm1 |
| DQ648857 | Bat coronavirus BtCoV 279 2005 |
| NC_004718 | SARS coronavirus Toronto 2 |
Sensitivity analysis plus tree fusion under direct optimization
Direct optimization (Wheeler, 1996) works by creating parsimonious hypothetical ancestral sequences at internal nodes of a cladogram. The key difference between direct optimization and multiple alignment is that in direct optimization evolutionary differences in sequence length are accommodated, not by the use of gap characters, but rather by allowing insertion–deletion events between ancestral and descendant sequences. In direct optimization, evolutionary base substitution and insertion–deletion events are treated with the same edit costs that are used in standard studies using static alignment followed by search for a set of optimal tree(s). However, in direct optimization, alignment is dynamic in that a novel set of putative sequence homologies is considered each time a novel topology is considered. The best set(s) of homologies is discovered by searching for the topology(ies) that minimizes the global cost of substitution and indel events.
Moreover, we varied alignment parameter sets across five sets of edit costs ranging from unitary costs for nucleotide insertion–deletions, transversions and transitions to costs with upweighted insertion–deletions and transversions (4, 5) (Wheeler, 1995). This process of parallel direct optimization across many edit costs not only allows for analysis of whether the results are sensitive to parameter choice, but when also coupled with a genetical algorithm can shorten the computation time necessary to find satisfactory results (treated below).
Table 4.
Phylogenetic position of carnivore and swine relative to human SARS‐CoV isolates in trees calculated under various edit costs under direct optimization for the 83 isolate data set
| Indel cost | TV cost | TS cost | Tree length | Position of SARS CoV isolated from carnivores and swine in tree |
|---|---|---|---|---|
| 1 | 1 | 1 | 44737 | Terminal, nested within SARS CoV isolated from humans |
| 2 | 2 | 1 | 71583 | Terminal, nested within SARS CoV isolated from humans |
| 2 | 1 | 1 | 51209 | Terminal, nested within SARS CoV isolated from humans |
| 4 | 2 | 1 | 82802 | Terminal, nested within SARS CoV isolated from humans |
| 8 | 2 | 1 | 96851 | Terminal, nested within SARS CoV isolated from humans |
Table 5.
Phylogenetic position of carnivore and swine relative to human SARS‐CoV isolates in trees calculated under various edit costs under direct optimization for the 157 isolate data set
| Indel cost | TV cost | TS cost | Tree length | Position of SARS CoV isolated from carnivores and swine in tree | Position of SARS CoV isolated from Chiroptera in tree |
|---|---|---|---|---|---|
| 1 | 1 | 1 | 60614 | Terminal, nested within SARS‐ CoV isolated from humans | Basal to SARS‐CoV isolated from humans, carnivores and swine |
| 2 | 2 | 1 | 98057 | Terminal, nested within SARS‐ CoV isolated from humans | Basal to SARS‐CoV isolated from humans, carnivores and swine |
| 2 | 1 | 1 | 74521 | Terminal, nested within SARS‐ CoV isolated from humans | Basal to SARS‐CoV isolated from humans, carnivores, and swine |
| 4 | 2 | 1 | 123885 | Terminal, nested within SARS‐ CoV isolated from humans | Basal to SARS‐CoV isolated from humans, carnivores, and swine |
| 8 | 2 | 1 | 154549 | Terminal, nested within SARS‐ CoV isolated from humans | Most basal to SARS‐CoV isolated from humans, carnivores, and swine. Two isolates from Chiroptera are terminal |
Initial tree build strategies under direct optimization
We analyzed the 83 (1, 4; Table 1) and 157 (2, 5; Table 2) isolate data sets with direct optimization into phylogenetic trees as implemented in POY3 on a 16 processor cluster of Linux PC based workstations running in parallel over a gigabit Ethernet switch. We used both parallel build and multibuild strategies (Janies and Wheeler, 2001). (POY3 parallel build commands: ‐parallel ‐replicates 9 ‐fitchtrees ‐quick ‐staticapprox ‐notbr ‐maxtrees 10). (POY3 multibuild commands: parallel ‐multibuild ‐buildsperreplicate 16 ‐approxbuild ‐nodiscrepancies ‐norandomizeoutgroup ‐sprmaxtrees 2 ‐tbrmaxtrees 2 ‐fitchtrees ‐holdmaxtrees 2 ‐quick ‐staticapprox ‐replicates 2 ‐buildmaxtrees 2).
Figure 1.

Phylogenetic tree produced by direct optimization of 83 coronavirus isolates based on whole and partial genomes (sampling in Table 1). Branches with black traces indicate presence of the 29‐nucleotide region, CCTACTGGTTACCAACCTGAATGGAATAT (e.g., positions 27869–27897 in AY278489) in an uncharacterized protein of variants of the SARS‐CoV that infect small carnivores and humans. White traces indicate the absence of this region. In this analysis, the evolution of insertions and deletions of this region is labile and complex.
Figure 4.

Phylogenetic tree produced by direct optimization of 83 coronavirus isolates based on whole and partial genomes (sampling in Table 1). The evolution of hosts is optimized on the genome‐based tree as shown by the colors traced on the branches. Note that the SARS‐CoV isolates from carnivores (purple trace: civet cat Parguma larvata, raccoon dog Nyctereutes procyonoides, and ferret badger Melogale moschata) and artiodactyls (light blue trace: pig, Sus scrofa) are nested within a large clade of SARS‐CoV isolates from humans (yellow trace: Homo sapiens), which are basal among SARS‐CoV. The search method for the genomic data was direct optimization. Parsimony optimization was used for the host data. The edit costs were indels 1, transversions 1, transitions 1.
Figure 2.

Phylogenetic tree produced by direct optimization of whole and partial coronavirus genomes produced of 157 isolates (sampling in Table 2). Branches with black traces indicate presence of the 29‐nucleotide region, CCTACTGGTTACCAACCTGAATGGAATAT (e.g., positions 27869–27897in AY278489) in an uncharacterized protein of variants of the SARS‐CoV that infect small carnivores and humans. Branches with green traces indicate the presence of the 29‐nucleotide region CCAATACATTACTATTCGGACTGGTTTAT (e.g., positions 27866–27894 in DQ648857) in an uncharacterized protein of all SARS‐CoV isolated from Chiroptera. White traces indicate the absence of either region. In this analysis, the evolution of insertions and deletions of these regions is labile and complex.
Figure 5.
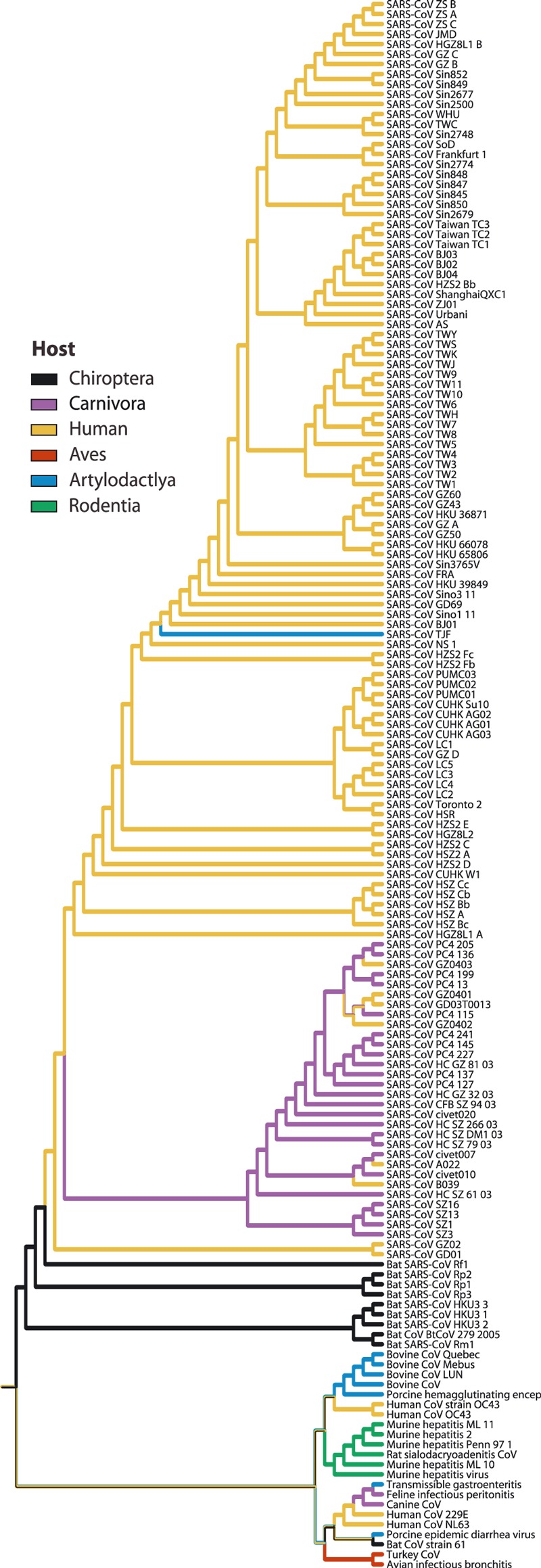
Phylogenetic tree produced by direct optimization of whole and partial coronavirus genomes produced of 157 isolates (sampling in Table 2). Note that the SARS‐CoV isolates from Chiroptera (black trace: Rhinolophus sinicus, Rhinolophus ferrumequinum, Rhinolophus macrotis and Rhinolophus pearsoni) are basal among the entire SARS‐CoV clade. SARS‐CoV isolates from small carnivores (purple trace) and artiodactyls (light blue trace) are nested within a clade of SARS‐CoV isolates from humans (yellow trace), although there were several exchanges between humans and carnivores. The search method for the genomic data was direct optimization. Parsimony optimization was used for the host data. The edit costs were indels 1, transversions 1, transitions 1.
Genetical algorithms under direct optimization
Next, we used POY3 to perform tree fusion, a search heuristic first presented in a phylogenetic context by Goloboff (1999) to address the problem of composite optima. With a set of various near suboptimal trees such as produced during direct optimization analysis, often some taxa are in an optimal configuration in some of the trees but no one tree is optimal for all taxa. We applied the following POY3 commands to a concatenated file named “ALL.TREES” containing trees collected under various edit costs (POY3 commands: ‐parallel ‐fitchtrees ‐treefuse ‐fusemingroup 5‐fusemaxtrees 10‐fuselimit 100‐slop 5‐checkslop 10‐maxtrees 10‐topofile ALL.TREES ‐molecularmatrix $ALIGNMENTPARAMETERS).
Standard tree search for aligned data
For the 114 isolate multiple alignment we ran a new technology search in TNT (Goloboff et al., 2003b) under equally weighted parsimony and stabilized the consensus 10 times (Fig. 6). We also ran these data under maximum likelihood under the GTR + GAMMA and CAT models of nucleotide substitution for 1000 randomly generated maximum parsimony trees in RAXML (Stamatakis, 2006) on a computing cluster.
Figure 6.

Note that the SARS‐CoV isolates from Chiroptera (black trace) are basal to the entire SARS‐CoV clade. The SARS‐CoV isolates from carnivores (purple trace) and artiodactyls (light blue trace) are nested within a large clade of SARS‐CoV isolates from humans (yellow trace), although there were exchanges of SARS‐CoV between humans and carnivores. The tree search and character optimization were conducted under equally weighted parsimony.
Character optimization on flat trees
We optimized the position of the animal SARS‐CoV isolates in the best tree(s) produced by tree fusion in each parameter set with the program MESQUITE (Maddison and Maddison, 2004) using the option: trace character history: parsimony ancestral states. All best trees from the parameter study were used for study of the relative topological position of isolates in various hosts (4, 5).
For flat tree presentation of the optimization of: various 29‐nucleotide fragments, key amino acid mutations, and host character states we used MESQUITE with trees for the 83 (1, 4) and 157 isolate datasets (2, 5, and supplemental data at http://supramap.osu.edu/cov) produced by direct optimization under unitary edit costs (indels = 1, transversions = 1, transitions = 1).
For flat tree and geographic visualization studies (treated next) we used a binary version (using the TNT command randtree*) of the 114 isolate strict consensus tree produced by ClustalW alignment and parsimony search (6, 3).
Figure 3.

Binary representation of strict consensus tree produced by multiple alignment followed by tree search under parsimony of 114 whole coronavirus genomes. Branches with black traces indicate presence of the 29‐nucleotide region, CCTACTGGTTACCAACCTGAATGGAATAT (e.g., positions 27869–27897 in AY278489) in an uncharacterized protein of variants of the SARS‐CoV that infect small carnivores and humans. Branches with green traces indicate the presence of the 29‐nucleotide region CCAATACATTACTATTCGGACTGGTTTAT (e.g., positions 27866–27894 in DQ648857) in an uncharacterized protein of all SARS‐CoV isolated from Chiroptera. White traces indicate the absence of either region. In this analysis the evolution of insertions and deletions of these regions is simple.
Projection of a tree, key mutations and metadata into a virtual globe
We used the methods described in Janies et al. (2007) to project a binary representation of the tree found for 114 isolates in TNT into a virtual globe (http://supramap.osu.edu/cov/janiesetal2008covsars.kmz). One subtle difference was that in this case we used an apomorphy list derived from PAUP* (version 4.0b10; Swofford, 2002) using the command describe trees:output list of apomorphies. We drew data on host and date of isolation from Lau et al. (2005; GenBank, or the International Committee on Taxonomy of Viruses database (http://www.ncbi.nlm.nih.gov/ICTVdb).
Spike protein mutations
Not all nucleotide records for coronaviruses in GenBank had translations to proteins. To get amino acid data of interest we translated nucleotide records into proteins in the Genetic Data Environment (http://www-bimas.cit.nih.gov/gde_sw.html) and checked these translations against reference amino acid sequences from GenBank. Amino acid sequences were aligned with ClustalW. Amino acid positions 479 and 487 of the spike protein were optimized on a tree using apomorphy commands of PAUP for tree projections. Optimizations of these amino acid positions were also conducted in MESQUITE for flat tree visualization (supplemental data at http://supramap.osu.edu/cov).
Genotype–phenotype correlation studies
We used the options: trace and chart of MACCLADE (Maddison and Maddison, 2000) to perform the concentrated changes test (Maddison, 1990) with the presence of the region CCTACTGGTTACCAACCTGAATGGAATAT as the independent character and the infection of carnivores as the dependent character. Any ambiguities in the optimization were resolved using the DELTRAN option. The CCT test was performed using simulation sample size of 100 000 iterations.
Sensitivity analysis of outgroup choice
Rooting an evolutionary tree is a critical step to polarize the temporal sequence of genomic and phenotypic changes and clarify the relationships of the organisms. Unlike Snijder et al. (2003) who used an equine torovirus outgroup (as the taxonomy suggests might be suitable http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/index.htm), we could not verify the suitability of an outgroup from outside the coronaviruses. Our investigation using BLAST (Altschul et al., 1997) [default values as implemented in GenBank http://www.ncbi.nlm.nih.gov (i.e., expect = 10)] indicated to us that no arterivirus or torovirus genome in GenBank bears significant nucleotide similarity with any coronavirus. As outgroups, we used genomes and partial genomes from non‐SARS coronaviruses (1, 2). We choose many candidate outgroup taxa to maximize host and antigenic diversity. Clades formed by antigenic group 1, group 2, and group 3 coronaviruses have significant branch lengths between each other and the SARS‐CoV clade. Finding the ingroup root when the available outgroups are markedly divergent can be challenging. The divergence can be a result of rapid mutation rates, recombination events, inadequate sampling, multiple evolutionary origins, or a combination of these phenomena. Thus we performed several experimental searches in which a random outgroup selected from non‐SARS taxa was used. The results of these searches were assessed to see whether our phylogenetic and host evolution results were affected by outgroup choice. To perform these randomization experiments, we output an implied alignment (Wheeler, 2003) resulting from each parameter set and best tree. (POY3 commands: ‐phastwincladfile $IMPLIEDALIGNMENT.phast ‐topodiagnoseonly ‐topofile $ALIGNMENTPARAMETERS.TREE). Next, for each implied alignment we used 1000 replicate new technology tree searches (TNT command: XMULT 2) (Goloboff et al., 2003b). In each search replicate, we randomly deleted a subset of the outgroup taxa and assessed: (1) whether the most basal taxon in the SARS ingroup was stable, and (2) whether the most basal taxon of the SARS ingroup was ever an isolate from an animal host (scripts available from the authors).
Resampling
We performed jackknife GC resampling in TNT (Goloboff et al., 2003a,b) on the ClustalW alignment of the 114 isolate data set and the implied alignment from unitary costs for the 83 and 157 isolate data sets as specified by the following commands: resample jak rep1000 [xm = lev5 rep5] from 0.
We performed 1000 bootstrap resampling replicates in RAXML (Stamatakis, 2006) with the following commands: ‐f d ‐m GTRCAT ‐# 1000 ‐b 12345 ‐n MultipleBootstrap.
Results
Direct optimization searches
Best tree lengths for the direct optimization searches under various parameters are reported for the 83 isolate data set in Table 4 and for the 157 isolate data set in Table 5. The resampling values are reported as supplemental data at http://supramap.osu.edu/cov/.
Multiple alignment to standard tree search
For the 114 isolate data set, a best score of 22 363 steps under equally weighted parsimony was hit 107 times and 87 trees were retained. A strict consensus of 59 nodes was stabilized 10 times (Fig. 6). The best RAXML tree for this alignment was found under GTRGAMMA at –ln likelihood of 111006.264984. RAXML trees with host character optimization and resampling values are available in supplemental data at http://supramap.osu.edu/cov/.
Evolution of host shifts among coronaviruses
In the 83 isolate data set in all parameter sets considered, we found the SARS‐CoV isolates from P. larvata, N. procyonoides (Carnivora) and Sus scrofa (Artiodactyla) to occur in terminal positions of the trees, nested well within a large clade of SARS‐CoV isolated from humans (Fig. 4, Table 4). Thus, based on genomic evidence, SARS‐CoV occurred in P. larvata, N. procyonoides and S. scrofa after SARS‐CoV occurred in humans (4, 3). The shift of SARS‐CoV from human hosts to S. scrofa host is independent of the shift from human host to small carnivore hosts (N. procyonoides and S. scrofa).
In the 83 isolate tree recovered under unitary costs, the polarity of host shift is ambiguous between the SARS‐CoV isolate from N. procyonoides (HC/SZ/61/03) and the SARS‐CoV isolate GD03T0013 from humans. GD03T0013 is closely related to SARS‐CoV isolated from civets served in a restaurant in Guangzhou, China in late 2003 and early 2004. No epidemiological data link the GD03T0013 human case to exposure to laboratory isolates of SARS‐CoV (Wang et al., 2005).
In the 157 isolate data set, under all parameters we found the SARS‐CoV isolates from P. larvata, N. procyonoides and S. scrofa were terminal, nested well within a large clade of SARS‐CoV isolated from humans (Fig. 5, Table 5). In the analysis of these data under most parameter sets the SARS‐CoV isolated from Chiroptera were basal to SARS‐CoV isolated from humans, carnivores and swine. A solitary minor exception to this pattern occurred under an extremely biased edit cost model of indels 8, transversions 2, transitions 1 (Table 5). In this analysis, two of four isolates of SARS‐CoV from Chiroptera occur in terminal rather than basal positions.
In the 157 isolate tree recovered under unitary costs, the human SARS‐CoV isolate GD03T0013 is closely related to civet as well as human isolates SARS‐CoV. This is consistent with the result that there were bidirectional exchanges of SARS‐CoV between humans and carnivores.
The 114 isolate trees that result from analyses using multiple alignment and standard tree searches under parsimony and maximum likelihood show a pattern of host shifts similar to those described for the direct optimization searches. SARS‐CoV isolated from Chiroptera are basal to SARS‐CoV under alignment plus parsimony search or alignment plus maximum likelihood search. In all results from the 114 isolate data set SARS‐CoV isolated from carnivores are terminal and nested within a large clade of SARS‐CoV isolated from humans and there is evidence of bidirectional exchange of SARS‐CoV between humans and carnivores (Fig. 6 and supplemental data at http://supramap.osu.edu/cov).
Evolution of a labile region of the SARS‐CoV genome
In all three isolate sampling regimes the first insertion of the 29‐nucleotide region, CCTACTGGTTACCAACCTGAATGGAATAT, occurs phylogenetically basal to the clade exhibiting the earliest hosts shift among humans and carnivores. However, the result of whether this region covaries with host shifts is dependent on isolate sampling regime.
Locus insertion and deletion among SARS‐CoV from various hosts in the 83 isolate data set
We present the phylogeny for 83 isolates found under unitary costs with tracing depicting the complex pattern of presence and absence of the 29‐nucleotide region CCTACTGGTTACCAACCTGAATGGAATAT (Fig. 1). The pattern of insertion and deletion of the 29‐nucleotide region region includes four to eight insertions and zero to four deletions. However, two host shifts from human to carnivore occur in concert with insertions of the 29‐nucleotide region (Fig. 4). Using Maddison's (1990) concentrated changes test, we find statistically significant correlation between this 29‐nucleotide region and host shifts (CCT = 0.0123).
Locus insertion and deletion among SARS‐CoV in the 157 isolate data set
We optimized the presence of 29 nucleotide sequence regions CCTACTGGTTACCAACCTGAATGGAATAT and CCAATACATTACTATTCGGACTGGTTTAT over the tree calculated for 157 isolates under unitary costs (Fig. 2). The region CCAATACATTACTATTCGGACTGGTTTAT occurs in all wholly sequenced genomes of SARS‐CoV isolated from Chiroptera and is well correlated with this host. In contrast, the region CCTACTGGTTACCAACCTGAATGGAATAT is inserted seven to eight times and deleted four to five times. In terms of host use in this tree, there are five shifts from carnivore to human hosts and two changes from human to carnivore hosts (Fig. 5). Among all these changes in the presence of the 29‐nucleotide region, CCTACTGGTTACCAACCTGAATGGAATAT, and changes in host use, there is only one branch where these two changes occur concurrently. This results in a CCT value of 0.108. Thus the CCTACTGGTTACCAACCTGAATGGAATAT region shows insignificant correlation with the host shift in the 157 isolate data set.
Locus insertion and deletion among SARS in the 114 isolate data set
We optimized the presence and absence of the 29‐nucleotide regions CCTACTGGTTACCAACCTGAATGGAATAT and CCAATACATTACTATTCGGACTGGTTTAT, on a binary representation of strict consensus tree resulting from parsimony search of the 114 isolate data set (Fig. 3). There are no branches where a host shift (Fig. 6) is coincident with an insertion or deletion of this fragment. This result indicates, that like the 157 isolate data set, the insertion of this 29‐nucleotide region is not significantly correlated with a host shift. Moreover, just as in the 157 isolate dataset, the region, CCAATACATTACTATTCGGACTGGTTTAT, occurs in all wholly sequenced genomes of SARS‐CoV isolated from Chiroptera and is well correlated with this host.
Mutations in the spike protein
Li et al. (2005) interpret the distribution of states and polarity of change of position 479 of the SARS‐CoV spike protein as follows. Viruses infecting carnivores contain a basic residue, arginine (R) or lysine (K). Next mutation to a small uncharged residue asparagine (N) allowed infection of humans.
However, in the 157 isolate tree we see a different distribution of genotypes and polarities of change. SARS‐CoV isolated from carnivores exhibit three genotypes at position 479: asparagine (N) arginine (R) or lysine (K). SARS‐CoV infecting humans have two genotypes at position 479: asparagine (N) and arginine (R). SARS‐CoV infecting Chiroptera contain exclusively serine (S) at position 479. SARS‐CoV isolated from the artiodactyl contain asparagine (N). Considering the tree in the 157 isolate data set, we observe the following mutations at in the spike protein: N479K, N479R, S479N, R479N (supplemental data at http://supramap.osu.edu/cov).
Li et al. (2005) also describe diversity and polarity of change for position 487 of the spike protein of SARS‐CoV. They describe SARS‐CoV isolated in 2002–03 to contain threonine (T) and SARS‐CoV isolated from humans and carnivores in 2003–04 to contain serine (S) at position 487.
We observe essentially the same diversity of genotypes at position 487 with some additions. SARS‐CoV infecting Chiroptera contain primarily valine (V) at position 487 with the exception of one isolate that contains an isoluceine (I). SARS‐CoV isolated from the artiodactyl exhibits a threonine (T). However, we observe different polarities of change than those inferred by Li et al. (2005). We observe the muations: V487I, V487T, T487S based on the tree from the 157 isolate data set (supplemental data at http://supramap.osu.edu/cov).
We found a statistically signifcant covariation of mutation T487S in the spike protein with carnivore hosts (Fig. 5 and supplemental data at http://supermap.osu.edu/cov). The CCT is 0.019 with DELTRAN optimization and 0.018 with ACCTRAN optimization.
We find no correlation of the mutations N479K and N479R in the spike protein with change from human to carnivore hosts (Fig. 5 and supplemental data at http://supramap.osu.edu/cov) as there are no branches that share these mutations and a shift in host.
Outgroup choice
As presented in 1, 4, 2, 5, 6, 3 and supplemental figures at http://supermap.osu.edu/cov, we rooted our phylogenies on non‐SARS coronaviruses. Due to the long internal branches (e.g., ranging from 1680 to 3332 steps in the 83 isolate data set) between any antigenic groups and SARS we decided to use this rooting only for visualization.
The rooting we can present in a figure does not fully represent the extent of our analyses. Our tests as to whether our results were sensitive to outgroup choice showed that our results were not affected by outgroup choice. SARS‐CoV isolates from human hosts were consistently basal to any SARS‐CoV isolate from a carnivore host irrespective of outgroup choice.
Discussion
Based on the SARS‐CoV data released as of July 2006, the polarity of host shifts from human to carnivore hosts and humans to artiodactyl host is clear. Simply put, the SARS‐CoV sequence data from animal hosts that has been released as of July 2006 are the results of two zoonotic events that occurred after the 2002–03 outbreak of SARS in humans: one major shift from human to carnivore hosts (with subsequent reversals that were not significant to human outbreaks) and one shift to an artiodactyl. SARS‐CoV isolated from Chiroptera are consistently basal to clades containing SARS‐CoV from human, carnivore and artiodactyl hosts.
Outgroup choice and presentation
Many of the reports that argue for carnivores as the original reservoir of SARS‐CoV use a phylogeny to support their arguments (Guan et al., 2003; Chinese SARS Molecular Epidemiology Consortium, 2004; Kan et al., 2005; Song et al., 2005; Zhang, C et al., 2006). However, the phylogenies in these studies lack outgroup and rooting criteria necessary to derive such evidence for the origins of SARS‐CoV. Outgroups chosen from outside of SARS‐CoV are necessary to test the monophyly of the SARS‐CoV ingroup (Barriel and Tassy, 1998). Moreover in optimal trees, non‐SARS‐CoV outgroups will join the region of the SARS‐CoV subtree that is closest to the ancestor of SARS and provide a point suitable for rooting and subsequent character analysis (Grandcolas et al., 2004).
In the case of Guan et al. [2003, see their figs 2 and S2) and the Chinese SARS Molecular Epidemiology Consortium (2004); see their fig. S7 of their supplemental materials] these researchers simply force the root position on their drawings such that they represent SARS‐CoV isolates from animal hosts as ancestral. In other drawings, no outgroup is designated (Chinese SARS Molecular Epidemiology Consortium, 2004, fig. 2) or a human SARS‐CoV outgroup is used and the animal SARS‐CoV isolates are omitted from the tree (Chinese SARS Molecular Epidemiology Consortium, 2004, fig. S6). In the case of Song et al. (2005a) human SARS‐CoV is designated as the outgroup. Regression methods are used to construct a rooted tree in which the date of the most recent ancestor is reconstructed as December 2002 (Song et al., 2005). Song et al. (2005) conclude that a source of disease common to humans and civets must be in the environment and further surveys of the CoV in the Guangdong region are warranted. In the case of Zhang, C et al., 2006, fig. 1; and pers. comm.) an outgroup was used for tree construction but not for tests of selection.
Many researchers agree that SARS represents a previously unrecognized fourth lineage of coronaviruses (Marra et al., 2003; Rest and Mindell, 2003; Rota et al., 2003). Thus, the non‐SARS coronaviruses can serve as outgroups to SARS‐CoV. This can be revisited if and when data on viruses closely related to SARS‐CoV become available. Alternatively, other researchers used a torovirus and/or okavirus outgroup(s) to place SARS‐CoV as sister to group 2 coronaviruses (Snijder et al., 2003; Lió and Goldman, 2004). However, based on the data in GenBank, toroviruses and okaviruses bear little sequence similarity to any coronavirus. The danger in use of such distant outgroups is well documented (Wheeler, 1990; Graham et al., 2002). In essence, distant outgroups act as if they are random sequences resulting in spurious attraction to the longest branch available among the ingroup. Indeed the branch lengths between the major clades of coronaviruses in the 83 and 157 isolate datasets of this paper are long. This problem is addressed in the 114 isolate data set. The best approach going forward is to extend sampling of diverse coronavirus genomes to search for outgroups of SARS‐CoV in humans, especially from Chiroptera, carnivores and non‐human primates.
Taxonomic sampling affects analyses
The lack of a good outgroup to SARS‐CoV is tied to (1) poor sampling of non‐SARS coronavirus genomes before the 2002–03 SARS outbreak, and (2) the preoccupation with animals in Chinese markets, farms and restaurants after the outbreak without regard to highly diverse species traded as bush meat in South‐east Asia (Bell et al., 2004). Before the SARS epidemic, the small number of animal coronaviruses that had been sequenced were selected primarily from animals of agricultural importance or model organisms. This lack of sampling of coronaviruses from wild animals is changing as viral surveys of Chiroptera, camelids and bovids are published and in preparation (Chu et al., 2006; Dominguez et al., 2007; Jina et al., 2007; Zhang, X et al., 2007).
Insertion of the 29‐nucleotide regions
Presence of the region CCTACTGGTTACCAACCTGAATGGAATAT is correlated with host switching beween human and carnivore hosts in the 83 isolate data set but is insignificantly correlated with switches from human to carnivore hosts in the larger (114 and 157 isolate) data sets. The concentrated changes test (CCT; Madison, 1990) whether a change in one character (e.g., insertion or deletion of the 29‐nucleotide region) and a change in another character (e.g., host phenotype) co‐occur on the same branches of a tree more often than expected by chance. In the case of the 83 isolate data set we observe a significant correlation between the presence of this 29‐nucleotide region and carnivore hosts. In the case of the 157 isolate data set we observe an insignificant correlation. In the case of the 114 isolate data set we do not observe changes that strictly co‐occur. However, we do observe that host shifts in the 114 and 157 isolate data set that host shifts occur in the region of the tree in which changes in the 29‐nucleotide region occurred more basally. Thus, the presence of the 29‐bp region may predispose or be part of a suite of genomic changes associated with host shifts. In light of these results, it is of interest to implement a relaxed concentrated changes test. This test could examine the branches in the vicinity of the change of interest for a correlated change in a second character.
Mutations of the spike gene
Our phylogenetic results shed fresh light on the polarity of mutations and diversity of genotypes in the spike protein of SARS‐CoV. Our results differ from the result of Zhang, C et al. (2006) who using CODEML (Yang, 1997) and HYPY (Kosakovsky Pond and Frost, 2005) for a tree‐based spike nucleotide sequence analysis show that the codon for amino acid position 479 was under positive selection and the codon for amino acid position 487 was not. The trees used to derive these results reflect the same bias seen in other studies—that transmission of SARS‐CoV was from carnivore to human hosts.
Geographic visualization
The pattern of geographic spread of SARS‐CoV is similar to that of avian influenza (H5N1; Janies et al. 2007) in that both viral lineages that have caused recent outbreaks have their origins in Southern China. However, H5N1 and SARS‐CoV contrast in the rapidity in which they moved across the planet. The recent outbreak lineage of H5N1 has spread from Asia to Europe, the Middle East, and Africa during the period of 1996–2005 and has not yet arrived in North America. In contrast, SARS‐CoV spread not only from Asia to Europe but also North America in a matter of months (November 2002–March 2003). These differences are perhaps associated with the fact that SARS‐CoV infected carnivores in urban markets and a cosmopolitan human population with access to world travel. In contrast, H5N1 is currently infecting primarily avian populations and humans that live in rural settings and come into close contact with birds via subsistence farming and food processing.
Further directions
In order to better understand the molecular epidemiology of SARS‐CoV we must develop research programs that include comprehensive sampling and phylogenetic analyses of many whole viral genomes, including outgroups that are closely related to SARS‐CoV. As a result of the previously unrecognized zoonotic threat they pose, several groups have embarked on large‐scale sequencing projects on coronavirus genomes isolated from diverse animal hosts, especially Chiroptera, carnivores and primates. These efforts will help us pinpoint the zoonotic origins of SARS‐CoV, develop an understanding of the zoonotic potential of coronaviruses as well as the genomic changes that underlie host shifts among coronaviruses.
Acknowledgments
Research facilities and funding was provided by the Department of Biomedical Informatics of the Ohio State University College of Medicine. D.J. acknowledges the National Aeronautics and Space Administration (grant NAG 2‐1399). In addition, this material is based upon work supported by, or in part by, the US Army Research Laboratory and the US Army Research Office under contract/grant number W911NF‐05‐1‐0271. D.P. acknowledges support from the National Science Foundation via a grant to the Mathematical Biosciences Institute of the Ohio State University. B.A. was supported by Ohio State University's Research on Research Program. Computational equipment was provided by The Hewlett Packard Corporation (Advanced Technology Platforms Itanium2 Grant, 89910.1) and The Ohio Supercomputer Center (Resource Grant PAS0119). Thanks to Aaron Nile for proofreading.
Supporting information
spike.aa.pos479.pdf. Phylogenetic tree of 157 coronavirus isolates based on whole genomes (sampling in Table 2). This is the same tree as Figs 2 and 5 in the body of the paper except that in this instance the amino acid states at position 479 in the spike locus are traced.
spike.aa.pos487.pdf. Phylogenetic tree of 157 coronavirus isolates based on whole genomes (sampling in Table 2). This is the same tree as Figs 2 and 5 in the body of the paper except that in this instance the amino acid states at position 487 in the spike locus are traced.
cov114.host.raxmltree929.names.pdf. RAXML search under GTRGAMMA for 114 isolates. Character optimization was conducted under equally weighted parsimony
cov114.host.raxmltree929boot.nex. Tree with bootstrap values for RAXML search. To be viewed with MESQUITE.
r1000.cov114.jackknife.log. Jackknife values for 114 isolate data set under equally weighted parsimony. To be viewed with a text editor.
r1000.cov83.jackknife.log. Jackknife values for 83 isolate data set under equally weighted parsimony. To be viewed with a text editor
r1000.cov157.jackknife.log. Jackknife values for 157 isolate data set under equally weighted parsimony. To be viewed with a text editor
janiesetal2007covsars.kmz. Keyhole Markup file depicting the spread of 114 isolates of SARS‐CoV over geography. To be opened with Google Earth. See also readmesarskml.pdf.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Footnotes
GenBank accession numbers for SARS‐CoV sequences released in January 2004: AY394978AY394979AY394980AY394981AY394982AY394983AY394984AY394985AY394986AY394987AY394989AY394990AY394991AY394992AY394993AY394994AY394995AY394996AY394997AY394999AY395000AY395001AY395002AY395003AY395004.
GenBank accession numbers for SARS‐CoV sequences released in 2005: AY313906AY338174AY338175AY348314AY394850AY461660AY485277AY485278AY525636AY568539AY613947AY613948AY613949AY613950AY613951AY613952AY613953AY627044AY627045AY627046AY627047AY627048
AY687354 AY687357 AY687358AY687361 AY687365 AY687370 AY686863 AY572034 AY687372 AY687362 AY686864 AY687364 AY687367 AY572038 AY304486 AY687363 AY687355 AY687369 AY687366 AY687371 AY525636 AY687359 note erratum published to correct accession numbers and SNPs (Kan et al. (2005)
GenBank accession numbers for SARS‐CoV sequences released as nucleocapsid sequences in January 2006 and then as whole genomes in June 2006: DQ071611, DQ071612. Whole genomes released in January 2006: DQ071615. Nucleocapsid sequences released in January 2006: DQ071613, DQ071614. Spike sequences released in November 2005 revised in July 2006: DQ159956, DQ159957.
GenBank accession numbers for whole genomes released in September 2005 and later updated in October 2005: DQ022305, DQ084199, DQ084200.
GenBank accession numbers for RNA‐dependent RNA polymerase, polyprotein gene and spike gene: AY864196, AY864197, AY864198.
GenBank accessions for genomes DQ648794, DQ648856, DQ648857, various genes DQ648786DQ648786DQ648787DQ648788DQ648789DQ648790DQ648791DQ648792DQ648793DQ648795DQ648796DQ648797DQ648799DQ648800DQ648801DQ648802DQ648803DQ648804DQ648805DQ648806DQ648807DQ648808DQ648809DQ648810DQ648811DQ648812DQ648813DQ648814DQ648815DQ648816DQ648817DQ648818DQ648819DQ648820DQ648821DQ648822DQ648823DQ648824DQ648825DQ648826DQ648827DQ648828DQ648829DQ648830DQ648831DQ648832DQ648833DQ648834DQ648835DQ648836DQ648837DQ648838DQ648839DQ648840DQ648841DQ648842DQ648843DQ648844DQ648845DQ648846DQ648847DQ648848DQ648849DQ648850DQ648851DQ648852DQ648853DQ648854DQ648855DQ648858.
References
- Altschul, S., Madden, T., Schaffer, R., Zhang, J., Zhang, Z., Miller, W., Lipman, D., 1997. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriel, V., Tassy, P., 1998. Rooting with multiple outgroups: consensus versus parsimony. Cladistics 14, 193–200. [DOI] [PubMed] [Google Scholar]
- Bell, D., Robertson, S., Hunter, P., 2004. Animal origins of SARS coronavirus: possible links with the international trade in small carnivores. Philos. Trans. R. Soc. Lond. B. 359, 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradsher, K., Altman, L., 2003. Strain of SARS is found in 3 animal species in Asia. May 24. New York Times.
- Chen, W., Yan, M., Yang, L., Ding, B., He, B., Wang, Y., Liu, X., Liu, C., Zhu, H., You, B., Huang, S., Zhang, J., Mu, F., Xiang, Z., Feng, X., Wen, J., Fang, J., Yu, J., Yang, H., Wang, J., 2005. SARS‐associated coronavirus transmitted from human to pig Emerg. Infect. Dis. 11(3) Available from: http://www.cdc.gov/ncidod/EID/vol11no03/04-0824.htm/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium , 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303, 1666–1669. [DOI] [PubMed] [Google Scholar]
- Chu, D., Poon, L., Chan, K., Chen, H., Guan, Y., Yuen, K., Peiris, J., 2006. Coronaviruses in bent‐winged bats (Miniopterus spp.). J. Gen. Virol. 87, 2461–2466. [DOI] [PubMed] [Google Scholar]
- Dominguez, S., O'Shea, T., Oko, L., Holmes, K., 2007. Detection of Group 1 Coronaviruses in Bats in North America Emerg. Infect. Dis. 13(9) Available from: http://www.cdc.gov/EID/content/13/9/1295.htm/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink, M., 2003. Clues to the animal origins of SARS. Science 3001, 1351. [DOI] [PubMed] [Google Scholar]
- Goloboff, P., 1999. Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics 15, 415–428. [DOI] [PubMed] [Google Scholar]
- Goloboff, P., Farris, J., Kallersjo, M., Oxelmann, B., Ramirez, M., Szumik, C., 2003a. Improvements to resampling measures of group support. Cladistics 19, 324–332. [Google Scholar]
- Goloboff, P., Farris, S., Nixon, K., 2003b. TNT. http://www.zmuc.dk/public/phylogeny/TNT/
- Graham, S., Olmstead, R., Barrett, S., 2002. Rooting phylogenetic trees with distant outgroups: a case study from the commelinoid monocots. Mol. Biol. Evol. 19, 1769–1781. [DOI] [PubMed] [Google Scholar]
- Grandcolas, P., Guilbert, E., Robillard, T., D'Haese, C., Murienne, J., Legendre, F., 2004. Mapping characters on a tree with or without the outgroups. Cladistics 20, 579–582. [DOI] [PubMed] [Google Scholar]
- Guan, Y., Zheng, B.J., He, Y.Q., Liu, X.L., Zhuang, Z.X., Cheung, C.L., Luo, S.W., Li, P.H., Zhang, L.J., Guan, Y.J., Butt, K.M., Wong, K.L., Chan, K.W., Lim, W., Shortridge, K.F., Yuen, K.Y., Peiris, J.S., Poon, L.L., 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278. [DOI] [PubMed] [Google Scholar]
- Van Der Hoek, L., Pyrc, K., Jebbink, M.F., Vermeulen‐Oost, W., Berkhout, R.J.M., Wolthers, K.C., Dillen, P.M.E.W.‐V., Kaandorp, J., Spaargaren, J., Berkhout, B., 2004. Identification of a new human coronavirus. Nat. Med. 10, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janies, D., Wheeler, W., 2001. Efficiency of parallel direct optimization. Cladistics 17, S71–S82. [DOI] [PubMed] [Google Scholar]
- Janies, D., Hill, A., Guralnick, R., Habib, F., Waltari, E., Wheeler, W.C., 2007. Genomic analysis and geographic visualization of the spread of avian influenza (H5N1). Syst. Biol. 56, 321–329. [DOI] [PubMed] [Google Scholar]
- Jina, L., Cebra, C., Baker, A., Mattsona, D., Cohen, S., Alvarado, S., Rohrmann, G., 2007. Analysis of the genome sequence of an alpaca coronavirus. Virology 365, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, B., Wang, M., Jing, H., Xu, X., Jiang, X., Yan, M., Liang, W., Zheng, H., Wan, K., Liu, Q., Cui, B., Xu, X., Zhang, E., Wang, H., Ye, J., Li, G., Li, M., Cui, Z., Qi, X., Du Chen, K.L., Gao, K., Zhao, Y., Zou, X., Feng, Y., Gao, Y., Hai, R., Yu D., Guan, Y., Xu, J., 2005. Molecular evolution analysis and geographic investigation of Severe Acute Respiratory Syndrome coronavirus‐like virus in palm civets at an animal market and on farms. J. Virol. 79, 11892–11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond, S., Frost, D., 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. [DOI] [PubMed]
- Ksiazek, T., Erdman, D., Goldsmith, C., Zaki, S., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J., Lim, W., Rollin, P., Dowell, S., Ling, A., Humphrey, C., Shieh, W., Guarner, J., Paddock, C., Rota, P., Fields, B., DeRisi, J., Yang, J., Cox, N., Hughes, J., LeDuc, J., Bellini, W., Anderson, L. and the SARS Working Group , 2003. A novel coronavirus associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 348, 1953–1966. [DOI] [PubMed] [Google Scholar]
- Lai, M., 1990. Coronavirus: organization, replication and expression of genome. Annu. Rev. Microb. 44, 303–333. [DOI] [PubMed] [Google Scholar]
- Lau, S., Woo, P., Li, K., Huang, Y., Tsoi, H., Wong, B.H.L., Wong, S.S.Y., Leung, S.Y., Chan, K.H., Yuen, K.Y., 2005. Severe acute respiratory syndrome coronavirus‐like virus in Chinese horseshoe bats. Proc. Natl Acad. Sci. USA 102, 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J.H., Wang, H., Crameri, G., Hu, Z., Zhang, H., Zhang, J., McEachern, J., Field, H., Daszak, P., Eaton, B.T., Zhang, S., Wang, L.F., 2005. Bats are natural reservoirs of SARS‐like coronaviruses. Science 310, 676–679. [DOI] [PubMed] [Google Scholar]
- Li, W., Wong, S., Li, F., Kuhn, J., Huang, I., Choe, H., Farzan, M., 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insights from ACE2–S–protein interactions. J. Virol. 80, 4211–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, C., Sui, J., Kuhn, J., Moore, M., Luo, S., Wong, S., Huang, I., Xu, K., Vasilieva, N., Murakami, A., He, Y., Marasco, W., Guan, Y., Choe, H., Farzan, M., 2005. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J. 24, 1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lió, P., Goldman, N., 2004. Phylogenomics and bioinformatics of SARS‐CoV. Trends Microbiol. 12, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W., 1990. A method for testing the correlated evolution of two binary characters: are gains or losses concentrated on certain branches of a phylogenetic tree? Evolution. 44, 539–557. [DOI] [PubMed] [Google Scholar]
- Maddison, W., Maddison, D., 2000. MACCLADE, Version 4.06. http://www.macclade.org/
- Maddison, D., Maddison, W., 2004. MESQUITE, Version 1.01. http://www.mesquiteproject.org/
- Mahony, J.B., Richardson, S., 2005. Molecular diagnosis of severe acute respiratory syndrome: the state of the art. J. Mol. Diagn. 7, 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M.A., Jones, S.J.M., Astell, C.R., Holt, R.A., Brooks‐Wilson, A., Butterfield, Y.S.N., Khattra, J., Asano, J.K., Barber, S.A., Chan, S.Y., 2003. The Genome sequence of the SARS‐associated coronavirus. Science 300, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Pang, X., Zhu, Z., Xu, F., Guo, J., Gong, X., Liu, D., Liu, Z., Chin, D.P., Feikin, D.R., 2003. Evaluation of control measures implemented in the severe acute respiratory syndrome outbreak in Beijing. JAMA 290, 3215–3221. [DOI] [PubMed] [Google Scholar]
- Poon, L.L.M., Chu, D.K.W., Chan, K.H., Wong, O.K., Ellis, T.M., Leung, Y.H.C., Lau, S.K.P., Woo, P.C.Y., Suen, K.Y., Yuen, K.Y., Guan, Y., Peiris, J.S.M., 2005. Identification of a novel coronavirus in bats. J. Virol. 79, 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest, J., Mindell, D., 2003. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host‐shifting. Infect. Genet. Evol. 3, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, P.A., Oberste, M.S., Monroe, S.S., Nix, W.A., Campagnoli, R., Icenogle, J.P., Peñaranda, S., Bankamp, B., Maher, K., Chen, M., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J.L., Chen, Q., Wang, D., Erdman, D.D., Peret, T.C.T., Burns, C., Ksiazek, T.G., Rollin, P.E., Sanchez, A., Liffick, S., Holloway, B., Limor, J., McCaustland, K., Olsen‐Rasmussen, M., Fouchier, R., Günther, S., Osterhaus, A.D.M.E., Drosten, C., Pallansch, M.A., Anderson, L.J., Bellini, W.J., 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399. [DOI] [PubMed] [Google Scholar]
- Siddell, S.G., Anderson, R., Cavanagh, D., Fujiwara, K., Klenk, H.D., Macnaughton, M.R., Pensaert, M., Stohlman, S.A., Sturman, L., Van Der Zeijst, B.A., 1983. Coronaviridae. Intervirology 20, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E.J., Bredenbeek, P.J., Dobbe, J.C., Thiel, V., Ziebuhr, J., Poon, L.L.M., Guan, Y., Rozanov, M., Spaan, W.J.M., Gorbalenya, A.E., 2003. Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, H., Tu, C., Zhang, G., Wang, S., Zheng, K., Lei, L., Chen, Q., Gao, Y., Zhou, H., Xiang, H., Zheng, H., Wang, S.C., Cheng, F., Pan, C., Xuan, H., Chen, S., Luo, H., Zhou, D., Liu, Y., He, J., Qin, P., Li, L., Ren, Y., Liang, W., Yu Y., Anderson, L., Wang, M., Xu, R., Wu, X., Zheng, H., Chen, J., Liang, G., Gao, Y., Liao, M., Fang, L., Jiang, L., Li, H., Di Chen, F.B., He, L., Lin, J., Tong, S., Du Kong, X.L., Hao, P., Tang, H., Bernini, A., Yu, X., Spiga, O., Guo, Z., Pan, H., He, W., Manuguerra, J., Fontanet, A., Danchin, A., Niccolai, N., Li, Y., Wu, C., Zhao, G., 2005. Cross‐host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl Acad. Sci. USA 102, 2430–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A., 2006. RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Tang, X.C., Zhang, J.X., Zhang, S.Y., Wang, P., Fan, X.H., Li, L.F., Li, G., Dong, B.Q., Liu, W., Cheung, C.L., Xu, K.M., Song, W.J., Vijaykrishna, D., Poon, L.L.M., Peiris, J.S.M., Smith, G.J.D., Chen, H., Guan, Y., 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80, 7481–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J., Higgins, D., Gibson, T., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C., Crameri, G., Kong, X., Chen, J., Sun, Y., Yu, M., Xiang, H., Xia, X., Liu, S., Ren, T., Yu, Y., Eaton, B.T., Xuan, H., Wang, L., 2004. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 10, 2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna, D., Smith, G.J.D., Zhang, J.X., Peiris, J.S.M., Chen, H., Guan, Y., 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 81, 4012–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Zheng, Z., Shang, L., Li, L., Cong, L., Feng, M., Luo, Y., Cheng, S., Zhang, Y., Ru, M., 2005. Molecular evolution and multilocus sequence typing of 145 strains of SARS‐CoV. FEBS Lett. 579, 4928–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, W.C., 1990. Nucleic acid sequence phylogeny and random outgroups. Cladistics 6, 363–367. [DOI] [PubMed] [Google Scholar]
- Wheeler, W.C., 1995. Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Syst. Biol. 44, 321–331. [Google Scholar]
- Wheeler, W.C., 1996. Optimization alignment: the end of multiple sequence alignment in phylogenetics? Cladistics 12, 1–9. [Google Scholar]
- Wheeler, W.C., 2003. Implied alignment: a synapomorphy‐based multiple‐sequence alignment method and its use in cladogram search. Cladistics 3, 261–268. [PubMed] [Google Scholar]
- Wheeler, W.C., Aagesen, L., Arango, C.P., Faivovich, J., Grant, T., D'Haese, C., Janies, D., Smith, W.L., Varon, A., Giribet, G., 2006. Dynamic Homology and Phylogenetic Systematics: A Unified Approach Using Poy3. American Museum of Natural History, New York. [Google Scholar]
- WHO , 2003. Summary table of SARS cases by country, 1 November 2002–7 August 2003. http://www.who.int/csr/sars/country/country2003_08_15.pdf/
- Woo, P.C.Y., Lau, S.K.P., Chu, C., Chan, K., Tsoi, H., Huang, Y., Wong, B.H.L., Poon, R.W.S., Cai, J.J., Luk, W., Poon, L.L.M., Wong, S.S.Y., Guan, Y., Peiris, J.S.M., Yuen, K., 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Hasoksuz, M., Spiro, D., Halpin, R., Wang, S., Vlasova, A., Janies, D., Jones, L., Ghedin, E., Saif, L.J., 2007. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 363, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Wei, J., Shao‐Heng, H., 2006. Adaptive evolution of the spike gene of SARS coronavirus: changes in positively selected sites in different epidemic groups. BMC Microbiol. 2006, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B.J., Guan, Y., Wong, K.H., Zhou, J., Wong, K.L., Young, B.W.Y., Lu, L.W., Lee, S.S., 2004. SARS‐related virus predating SARS outbreak, Hong Kong Emerg. Infect. Dis. 10(2) Available from: http://www.cdc.gov/ncidod/EID/vol10no2/03-0533.htm/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
spike.aa.pos479.pdf. Phylogenetic tree of 157 coronavirus isolates based on whole genomes (sampling in Table 2). This is the same tree as Figs 2 and 5 in the body of the paper except that in this instance the amino acid states at position 479 in the spike locus are traced.
spike.aa.pos487.pdf. Phylogenetic tree of 157 coronavirus isolates based on whole genomes (sampling in Table 2). This is the same tree as Figs 2 and 5 in the body of the paper except that in this instance the amino acid states at position 487 in the spike locus are traced.
cov114.host.raxmltree929.names.pdf. RAXML search under GTRGAMMA for 114 isolates. Character optimization was conducted under equally weighted parsimony
cov114.host.raxmltree929boot.nex. Tree with bootstrap values for RAXML search. To be viewed with MESQUITE.
r1000.cov114.jackknife.log. Jackknife values for 114 isolate data set under equally weighted parsimony. To be viewed with a text editor.
r1000.cov83.jackknife.log. Jackknife values for 83 isolate data set under equally weighted parsimony. To be viewed with a text editor
r1000.cov157.jackknife.log. Jackknife values for 157 isolate data set under equally weighted parsimony. To be viewed with a text editor
janiesetal2007covsars.kmz. Keyhole Markup file depicting the spread of 114 isolates of SARS‐CoV over geography. To be opened with Google Earth. See also readmesarskml.pdf.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


