Abstract
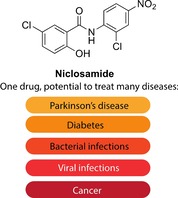
Niclosamide is an anthelmintic drug that has been used for over 50 years mainly to treat tapeworm infections. However, with the increase in drug repurposing initiatives, niclosamide has emerged as a true hit in many screens against various diseases. Indeed, from being an anthelmintic drug, it has now shown potential in treating Parkinson's disease, diabetes, viral and microbial infections, as well as various cancers. Such diverse pharmacological activities are a result of niclosamide's ability to uncouple mitochondrial phosphorylation and modulate a selection of signaling pathways, such as Wnt/β‐catenin, mTOR and JAK/STAT3, which are implicated in many diseases. In this highlight, we discuss the plethora of diseases that niclosamide has shown promise in treating.
Keywords: cancer, disease, niclosamide, repurposing, screening
Old dog, new tricks! Many recent drug repurposing studies have identified the anthelmintic drug niclosamide as a potential treatment for numerous diseases. Although in many cases the mechanism of action of niclosamide is not fully understood, its in vitro and in vivo therapeutic effects have resurrected this old drug and generated some excitement about its potential repurposing for various diseases that are currently difficult to treat.
Introduction
Following its discovery in the early 1950s, niclosamide (Figure 1) was initially used as a molluscicide to kill snails,1 and around a decade later, it was found to be effective against human tapeworm infections.2 Its use in humans to treat tapeworm infections started in 1982 following approval by the US Food and Drug Administration (FDA) and has since been on the World Health Organization's essential list of medicines. Structurally, niclosamide belongs to a large group of lipophilic, weakly acidic molecules called salicylanilides (Figure 1).3
Figure 1.
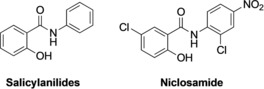
Chemical structures of salicylanilides and niclosamide.
Despite its wide use as an anthelmintic drug, the mechanism of action of niclosamide is yet to be completely understood. Early studies have linked niclosamide's activity to the uncoupling of oxidative phosphorylation.4 However, over the last decade or so, niclosamide has been shown to act on other targets such as Wnt/β‐catenin, mTOR and JAK/STAT3 signaling pathways (reviewed by Chen et al.5), which inevitably linked niclosamide's therapeutic potential to many diseases that involve these important signaling cascades. The increasing interest in niclosamide over the last decade is reflected in the significant increase in the number of publications regarding the study of this drug (Figure 2). Herein, we briefly discuss the numerous diseases niclosamide has shown promise in treating.
Figure 2.
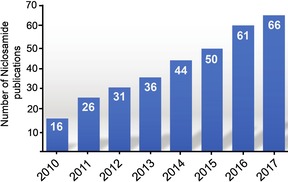
The number of niclosamide papers published since 2010. The data were obtained from PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) using the search term “niclosamide” and fixing the dates from January 1st to December 31st of each indicated year.
Pharmacological Activities of Niclosamide
1. Parkinson's disease (PD)
As mutations that impair the catalytic activity of the mitochondrial serine/threonine protein kinase PINK1 (PTEN‐induced kinase 1) were found to cause early onset PD6 through neuronal apoptosis, it became apparent that small molecules that activate PINK1 would have neuroprotective effects and thus have the potential to treat PD.7 Equipped with the fact that PINK1 is activated by mitochondrial depolarization, Barini et al.8 investigated whether niclosamide,9 which is known to cause mitochondrial depolarization, could activate PINK1. The results showed that niclosamide is a potent activator of PINK1 in cells. The study showed that niclosamide is not a direct activator of PINK1, as no activation was observed when recombinant PINK1 was incubated with PINK1 in vitro. However, it was shown that niclosamide caused mitochondrial depolarization, a phenomenon that was found to be reversible following removal of niclosamide from the cell media.8 Impressively, niclosamide was also shown to activate PINK1 in cultured neurons. Together, this study highlights the PINK1‐mediated neuroprotective promise niclosamide may hold. Further in vivo studies in PD models are needed to establish the ability of niclosamide to treat this disease.
2. Type 2 diabetes miletus (T2DM)
Niclosamide ethanolamine, the salt form of niclosamide, which exhibits improved aqueous solubility, was shown to improve the symptoms of T2DM in mice.10 Indeed, niclosamide treatment of mice fed on a high‐fat diet led to improved metabolism, increased lipid oxidation, and high energy expenditure. Additionally, in db/db mice wherein diabetes is induced as a result of mutations causing deficiency in leptin receptor activity, niclosamide ethanolamine was shown to be effective in improving glycemic control, treating hyperglycemia, and slowing the progression of the disease. In a subsequent study, niclosamide piperazine, which is another salt form of niclosamide, was also found to have similar efficacy in treating T2DM.11 It has been suggested that niclosamide's antidiabetes therapeutic potential is due to the inhibition of the glucagon signaling PKA pathway.12
3. Bacterial infections
Niclosamide has also shown antimicrobial activities against various bacterial infections such as methicillin‐resistant Staphylococcus aureus (MRSA),13, 14 tuberculosis (TB),15 and anthrax.16 In a drug repurposing study using a library of FDA‐approved drugs, niclosamide was found to specifically inhibit growth of Gram‐positive bacteria and to display strong in vivo and in vitro activities against MRSA (MIC=0.125 μg mL−1).13 Such findings showed that niclosamide is as effective as vancomycin, the current drug of choice for treating MRSA.13 Subsequently, niclosamide was investigated for its potential as an antimicrobial surface coating against device‐associated and hospital‐acquired infections.14 Highly versatile niclosamide‐based antimicrobial coatings were developed and were shown to clear existing infections and prevent biofilm formation at very low concentrations.14
Niclosamide was also shown to have significant antituberculosis (TB) activity (MIC=0.5–1 μg mL−1).15 Interestingly, niclosamide exerted pharmacological activity against stationary‐phase tubercle bacilli and multidrug‐resistant tuberculosis. However, potential toxicity to mammalian cells was noted when niclosamide was used at MIC, and hence topical use of niclosamide against surface‐located tuberculosis may hold better therapeutic promise.
Furthermore, niclosamide was identified to be able to significantly protect cells from anthrax lethal toxin as well as from lethal factor Pseudomonas exotoxin fusion protein and diphtheria toxin at low micromolar concentrations.16 This activity was linked to obstructing the process of anthrax toxin internalization and directly linked to endosome acidification.16
4. Viral infections
Niclosamide has been suggested for potential use as a broad‐spectrum antiviral to target host pathways used by viruses for infection, unlike the current antiviral strategies, which are directed against a specific viral target.17 This broad antiviral activity of niclosamide has been linked to its ability to neutralize endosomal pH and thereby disrupt the pH‐dependent membrane fusion required for viral entry.
To date, several studies have indicated the potential use of niclosamide to treat coronaviruses (SARS‐CoV and chikungunya virus),18, 19 Zika,20 and Ebola viruses.21 Niclosamide was discovered to be able to inhibit SARS‐CoV replication at low micromolar concentrations. Viral antigen synthesis was totally abolished at a niclosamide concentration as low as 1.56 μm.18 The exact mode of action is not clear, but niclosamide was found not to interfere with the virion's attachment and entry into cells, as it was still active even when added three hours after viral infection of cells. Some in silico studies suggested the potential binding of niclosamide to the SARS‐CoV main proteinase.22 However, only modest activity of niclosamide against this proteinase was reported (IC50=40 μm).23 Moreover, niclosamide exhibited antiviral activity against chikungunya virus (CHIKV) using a CHIKV 26S‐mediated insect cell fusion screening assay and was able to inhibit virus entry and cell‐to‐cell transmission of CHIKV infection.19 In light of the recent outbreak of Zika and Ebola infections, focus was on the drug repurposing screen of FDA‐approved drugs for rapid identification of potential therapeutic agents to meet the urgent medical need to treat these infections. Using such an approach, niclosamide was identified as an inhibitor of the Zika virus.20 Further validation assays confirmed the ability of niclosamide to significantly decrease Zika RNA levels and suppress the production of infectious Zika particles at sub‐micromolar concentrations.21 Subsequently, niclosamide emerged as a positive hit from another drug repurposing screen aimed at identifying clinically used agents that may have activity against the Ebola virus (EC50=1.5 μm).21
5. Cancer
The disease that has received by far the most attention in the repurposing of niclosamide is cancer. To date, niclosamide has been shown to exhibit anticancer activity against colon,24 breast,25 prostate,26 glioblastoma,27 osteosarcoma,28 ovarian,29 leukemia,30 adrenocortical carcinoma,31 lung,32 and oral33 cancers.5
These observed anticancer activities have been linked to niclosamide's ability to damage tumor cell mitochondria, induce apoptosis, inhibit tumor cell proliferation, and inhibit various aberrant tumor signaling pathways including Wnt/β‐catenin, mTORC1, STAT3, nuclear factor‐κB (NF‐κB) and Notch pathway (reviewed in detail by Moskaleva et al.34). In addition to this broad‐spectrum anticancer activity, niclosamide was also shown to inhibit cancer stem cells,35 which are considered an attractive and promising anticancer therapeutic target, as accumulating evidence has established their crucial role in cancer initiation, progression, establishment, recurrence, and drug resistance.36 Beyond its use as a single agent, niclosamide has been studied in combination with conventional chemotherapeutic agents. For instance, the combination of niclosamide with cisplatin led to the inhibition of cisplatin‐resistant triple‐negative breast cancer37 and exhibited synergistic effects in cisplatin‐resistant lung cancer cells.38 Enhanced sensitivity of chronic myeloid leukemia cells to dasatinib39 and that of cervical cancer cells to paclitaxel40 was also achieved when these drugs were used in combination with niclosamide. Synergistic effects with erlotinib in colon cancer,41 head and neck cancer,42 and in erlotinib‐resistant non‐small lung cancer cells32 have also been described. While the combination of niclosamide with bicalutamide led to overcoming enzalutamide‐ and bicalutamide‐resistant prostate cancer,43 the combination with abiraterone was found to be effective in treating advanced castration‐resistant prostate cancer.44 Furthermore, the use of niclosamide in conjunction with radiotherapy resulted in significant radiosensitizing effects in triple‐negative breast cancer cells45 and overcame radio‐resistance in lung cancer.46
Considerations for the Use of Niclosamide as a Therapeutic
Along with the interest in repurposing niclosamide for these various diseases, it must be noted that niclosamide's ability to influence many signaling pathways may work to its disadvantage, as it would lead to diverse side effects. Also, the favorable safety profile of niclosamide in treating humans with tapeworm infection could be due to the fact that the site of action in this case is the gut and hence the drug is not absorbed to modulate various signaling cascades and cause side effects. With the newly discovered diseases for which niclosamide may have a therapeutic effect, systemic delivery maybe required and hence the safety profile of niclosamide in this case is still largely unknown. Current and future in vivo studies on niclosamide will provide a clearer picture of its efficacy and toxicity, which will determine the success of these repurposing initiatives.
Conclusions
Niclosamide has shown great promise in treating many diseases through the uncoupling of oxidative phosphorylation and the manipulation of various cell‐signaling cascades. With this some safety concerns may arise following its systemic exposure. Insight into these safety issues will be attained in the near future from the various ongoing clinical investigations of niclosamide.
Statement: As this is a brief highlight of niclosamide's pharmacological activities, it was impossible to include all of the studies published on niclosamide in this area. The studies discussed in this work are just a representative sample of the myriad of niclosamide's therapeutic activities.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank Khadeejah Mehellou for her design of the Table of Contents graphic.
H. Kadri, O. A. Lambourne, Y. Mehellou, ChemMedChem 2018, 13, 1088.
References
- 1. Andrews P., Thyssen J., Lorke D., Pharmacol. Ther. 1982, 19, 245–295. [DOI] [PubMed] [Google Scholar]
- 2. Pearson R. D., Hewlett E. L., Ann. Intern. Med. 1985, 102, 550–551. [DOI] [PubMed] [Google Scholar]
- 3. Terada H., Environ. Health Perspect. 1990, 87, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Weinbach E. C., Garbus J., Nature 1969, 221, 1016–1018; [DOI] [PubMed] [Google Scholar]
- 4b. Frayha G. J., Smyth J. D., Gobert J. G., Savel J., Gen. Pharmacol. 1997, 28, 273–299. [DOI] [PubMed] [Google Scholar]
- 5. Chen W., R. A. Mook, Jr. , Premont R. T., Wang J., Cell. Signalling 2018, 41, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., Gonzalez-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W., Science 2004, 304, 1158–1160. [DOI] [PubMed] [Google Scholar]
- 7. Hertz N. T., Berthet A., Sos M. L., Thorn K. S., Burlingame A. L., Nakamura K., Shokat K. M., Cell 2013, 154, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barini E., Miccoli A., Tinarelli F., Mulholand K., Kadri H., Khanim F., Stojanovski L., Read K. D., Burness K., Blow J. J., Mehellou Y., Muqit M., ChemBioChem 2018, 19, 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khanim F. L., Merrick B. A., Giles H. V., Jankute M., Jackson J. B., Giles L. J., Birtwistle J., Bunce C. M., Drayson M. T., Blood Cancer J. 2011, 1, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tao H., Zhang Y., Zeng X., Shulman G. I., Jin S., Nat. Med. 2014, 20, 1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo J., Tao H., Alasadi A., Huang Q., Jin S., Eat. Weight Disord. 2017, 10.1007/s40519-017-0424-7. [DOI] [PubMed] [Google Scholar]
- 12. Chowdhury M. K., Turner N., Bentley N. L., Das A., Wu L. E., Richani D., Bustamante S., Gilchrist R. B., Morris M. J., Shepherd P. R., Smith G. C., Sci. Rep. 2017, 7, 40159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rajamuthiah R., Fuchs B. B., Conery A. L., Kim W., Jayamani E., Kwon B., Ausubel F. M., Mylonakis E., PLoS One 2015, 10, e0124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gwisai T., Hollingsworth N. R., Cowles S., Tharmalingam N., Mylonakis E., Fuchs B. B., Shukla A., Biomed. Mater. 2017, 12, 045010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Z., Zhang Y., Tuber. Lung Dis. 1999, 79, 319–320. [DOI] [PubMed] [Google Scholar]
- 16. Zhu P. J., Hobson J. P., Southall N., Qiu C., Thomas C. J., Lu J., Inglese J., Zheng W., Leppla S. H., Bugge T. H., Austin C. P., Liu S., Bioorg. Med. Chem. 2009, 17, 5139–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U. F., PLoS Pathog. 2012, 8, e1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C. J., Jan J. T., Chen C. M., Hsieh H. P., Hwang D. R., Liu H. W., Liu C. Y., Huang H. W., Chen S. C., Hong C. F., Lin R. K., Chao Y. S., Hsu J. T., Antimicrob. Agents Chemother. 2004, 48, 2693–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y. M., Lu J. W., Lin C. C., Chin Y. F., Wu T. W., Lin L. I., Lai Z. Z., Kuo S. C., Huo Y. J., Antiviral Res. 2016, 135, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu M., Lee E. M., Wen Z., Cheng Y., Huang W. K., Qian X., Tcw J., Kouznetsova J., Ogden S. C., Hammack C., Jacob F., Nguyen H. N., Itkin M., Hanna C., Shinn P., Allen C., Michael S. G., Simeonov A., Huang W., Christian K. M., Goate A., Brennand K. J., Huang R., Xia M., Ming G. L., Zheng W., Song H., Tang H., Nat. Med. 2016, 22, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Madrid P. B., Panchal R. G., Warren T. K., Shurtleff A. C., Endsley A. N., Green C. E., Kolokoltsov A., Davey R., Manger I. D., Gilfillan L., Bavari S., Tanga M. J., ACS Infect. Dis. 2015, 1, 317–326. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X. W., Yap Y. L., Bioorg. Med. Chem. 2004, 12, 2517–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wen C. C., Kuo Y. H., Jan J. T., Liang P. H., Wang S. Y., Liu H. G., Lee C. K., Chang S. T., Kuo C. J., Lee S. S., Hou C. C., Hsiao P. W., Chien S. C., Shyur L. F., Yang N. S., J. Med. Chem. 2007, 50, 4087–4095. [DOI] [PubMed] [Google Scholar]
- 24. Suliman M. A., Zhang Z., Na H., Ribeiro A. L., Zhang Y., Niang B., Hamid A. S., Zhang H., Xu L., Zuo Y., Int. J. Mol. Med. 2016, 38, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Londono-Joshi A. I., Arend R. C., Aristizabal L., Lu W., Samant R. S., Metge B. J., Hidalgo B., Grizzle W. E., Conner M., Forero-Torres A., Lobuglio A. F., Li Y., Buchsbaum D. J., Mol. Cancer Ther. 2014, 13, 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu C., Lou W., Armstrong C., Zhu Y., Evans C. P., Gao A. C., Prostate 2015, 75, 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wieland A., Trageser D., Gogolok S., Reinartz R., Hofer H., Keller M., Leinhaas A., Schelle R., Normann S., Klaas L., Waha A., Koch P., Fimmers R., Pietsch T., Yachnis A. T., Pincus D. W., Steindler D. A., Brustle O., Simon M., Glas M., Scheffler B., Clin. Cancer Res. 2013, 19, 4124–4136. [DOI] [PubMed] [Google Scholar]
- 28. Liao Z., Nan G., Yan Z., Zeng L., Deng Y., Ye J., Zhang Z., Qiao M., Li R., Denduluri S., Wang J., Wei Q., Geng N., Zhao L., Lu S., Wang X., Zhou G., Luu H. H., Haydon R. C., He T. C., Wang Z., Curr. Cancer Drug Targets 2015, 15, 726–738. [DOI] [PubMed] [Google Scholar]
- 29. King M. L., Lindberg M. E., Stodden G. R., Okuda H., Ebers S. D., Johnson A., Montag A., Lengyel E., J. A. MacLean, II , Hayashi K., Oncogene 2015, 34, 3452–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang A. M., Ku H. H., Liang Y. C., Chen Y. C., Hwu Y. M., Yeh T. S., J. Cell. Biochem. 2009, 106, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satoh K., Zhang L., Zhang Y., Chelluri R., Boufraqech M., Nilubol N., Patel D., Shen M., Kebebew E., Clin. Cancer Res. 2016, 22, 3458–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li R., Hu Z., Sun S. Y., Chen Z. G., Owonikoko T. K., Sica G. L., Ramalingam S. S., Curran W. J., Khuri F. R., Deng X., Mol. Cancer Ther. 2013, 12, 2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X., Yang Z., Han Z., Wen Y., Ma Z., Wang Y., Oncol. Rep. 2018, 39, 827–833. [DOI] [PubMed] [Google Scholar]
- 34. Moskaleva E. Y., Perevozchikova V. G., Zhirnik A. S., Severin S. E., Biomed. Khim. 2015, 61, 680–693. [DOI] [PubMed] [Google Scholar]
- 35. Pan J. X., Ding K., Wang C. Y., Chin. J. Cancer 2012, 31, 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen K., Huang Y. H., Chen J. L., Acta Pharmacol. Sin. 2013, 34, 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu J., Chen X., Ward T., Pegram M., Shen K., Tumour Biol. 2016, 37, 9825–9835. [DOI] [PubMed] [Google Scholar]
- 38. Zuo Y., Yang D., Yu Y., Xiang M., Li H., Yang J., Li J., Jiang D., Zhou H., Xu Z., Yu Z., Mol. Med. Rep. 2018, 17, 3497–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Z., Li Y., Lv C., Wang L., Song H., Biochem. Biophys. Res. Commun. 2016, 478, 893–899. [DOI] [PubMed] [Google Scholar]
- 40. Chen L., Wang L., Shen H., Lin H., Li D., Biochem. Biophys. Res. Commun. 2017, 484, 416–421. [DOI] [PubMed] [Google Scholar]
- 41. Shi L., Zheng H., Hu W., Zhou B., Dai X., Zhang Y., Liu Z., Wu X., Zhao C., Liang G., Oncol. Targets Ther. 2017, 10, 1767–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li R., You S., Hu Z., Chen Z. G., Sica G. L., Khuri F. R., Curran W. J., Shin D. M., Deng X., PLoS One 2013, 8, e74670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu C., Armstrong C. M., Lou W., Lombard A. P., Cucchiara V., Gu X., Yang J. C., Nadiminty N., Pan C. X., Evans C. P., Gao A. C., Mol. Cancer Ther. 2017, 16, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu C., Armstrong C., Zhu Y., Lou W., Gao A. C., Oncotarget 2016, 7, 32210–32220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yin L., Gao Y., Zhang X., Wang J., Ding D., Zhang Y., Zhang J., Chen H., Oncotarget 2016, 7, 42126–42138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. You S., Li R., Park D., Xie M., Sica G. L., Cao Y., Xiao Z. Q., Deng X., Mol. Cancer Ther. 2014, 13, 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]


