Summary
Insights into the host antiviral strategies as well as viral disease manifestations can be achieved through the elucidation of host‐ and virus‐mediated transcriptional responses. An oligo‐based microarray was employed to analyse mRNAs from rhabdomyosarcoma cells infected with the MS/7423/87 strain of enterovirus 71 (EV71) at 20 h post infection. Using Acuity software and LOWESS normalization, 152 genes were found to be downregulated while 39 were upregulated by greater than twofold. Altered transcripts include those encoding components of cytoskeleton, protein translation and modification; cellular transport proteins; protein degradation mediators; cell death mediators; mitochondrial‐related and metabolism proteins; cellular receptors and signal transducers. Changes in expression profiles of 15 representative genes were authenticated by real‐time reverse transcription polymerase chain reaction (RT‐PCR), which also compared the transcriptional responses of cells infected with EV71 strain 5865/Sin/000009 isolated from a fatal case during the Singapore outbreak in 2000. Western blot analyses of APOB, CLU, DCAMKL1 and ODC1 proteins correlated protein and transcript levels. Two‐dimensional proteomic maps highlighted differences in expression of cellular proteins (CCT5, CFL1, ENO1, HSPB1, PSMA2 and STMN1) following EV71 infection. Expression of several apoptosis‐associated genes was modified, coinciding with apoptosis attenuation observed in poliovirus infection. Interestingly, doublecortin and CaM kinase‐like 1 (DCAMKL1) involved in brain development, was highly expressed during infection. Thus, microarray, real‐time RT‐PCR and proteomic analyses can elucidate the global view of the numerous and complex cellular responses that contribute towards EV71 pathogenesis.
Introduction
Enterovirus 71 (EV71) was first identified in 1969 in California, when it was isolated from the feces of an infant suffering from encephalitis (Schmidt et al., 1974). Subsequently, EV71 was reported as the agent involved in severe neurological diseases such as meningitis, encephalitis, monoplegia and acute flaccid paralysis. The virus is also associated with non‐neurological diseases like hand, foot and mouth disease (HFMD), herpangina and pulmonary oedema. Among young children, EV71 is a notable cause of central nervous system (CNS) disease that usually results in rapid clinical deterioration and death, the molecular pathogenesis of which is still elusive (Kehle et al., 2003). In Singapore, the major EV71 outbreak in 2000 involved 6402 cases with four deaths (Singh et al., 2002a), and prompted the closure of pre‐school centres for about 2 weeks. In 2001, there were 5187 cases of HFMD with EV71 as the predominant agent, with 75% of afflicted children below the age of four.
Belonging to the Picornaviridae, which comprises a large complex family of small non‐enveloped, positive‐strand RNA viruses with a genome size of about 7–9 kb, EV71 is known to induce an apoptotic response via its viral proteins such as 2A (Kuo et al., 2002) and 3C (Li et al., 2002). This is also observed in the related and widely studied poliovirus (Barco et al., 2000; Goldstaub et al., 2000; Calandria et al., 2004), which belongs to the same genus as EV71. Apoptosis is a complex mechanism that involves a network of cross‐talk and multiple specifically controlled pathways. The process may be triggered by the interactions of the pro‐apoptotic stimuli with various sensors such as the receptor‐mediated pathway through caspase 8, mitochondrial‐related pathway through caspase 9 (Desagher and Martinou, 2000) and endoplasmic reticulum (ER) stress‐triggered pathway through caspase 12 (Nakagawa et al., 2000). Apoptosis in viral infection is the host response to lyse prematurely in order to curtail the reproductive cycle of the virus (Clem and Miller, 1993). In addition, the significance of apoptosis is to enable macrophages to phagocytose dead cells in order to prevent dysregulated inflammatory reactions, and to initiate specific immune responses in the infected host (Sun and Shi, 2001). However, the immune response can also result in apoptosis of uninfected cells, which causes enhanced immunosuppression or specific organ toxicity (Ahr et al., 2004).
Besides apoptosis, the other host response is the development of canonical cytopathic effect (CPE) following productive poliovirus infection, which often leads to inflammation. Thus, the interplay of the two cell death processes highlights both apoptotic and antiapoptotic effects of poliovirus (Agol et al., 2000; Belov et al., 2003; Romanova et al., 2005). It is postulated that in early infection during which the poliovirus RNA genome is translated, sufficient quantities of pro‐apoptotic proteins are synthesized to trigger an early apoptotic response. However, with the onset of viral replication, the apoptotic response is interrupted by the apoptosis‐suppressing effect of the virus, which dominates the entire period of productive infection. Only at the later stages of infection such as after the development of CPE, can some signs of apoptosis be obvious (Carthy et al., 1998). Although many RNA viruses do not encode antiapoptotic genes and the mechanism of this apoptosis‐suppressing effect is still unclear, enterovirus 2B protein which exhibits membrane permeabilizing activity (Agirre et al., 2002), may have a significant role. Campanella et al. (2004) also demonstrated that coxsackievirus 2B protein is able to suppress apoptotic host responses by manipulating intracellular calcium ion homeostasis.
In addition to the variability in apoptotic response, enteroviral infection instigates multiple cascades of host responses, especially mechanisms pertaining to its neuropathogenesis. These responses are also evident in infections with other viruses, e.g. dengue virus (Warke et al., 2003; Liew and Chow, 2004), herpes simplex virus (Kramer et al., 2003), JC virus (Radhakrishnan et al., 2003) and severe acute respiratory syndrome (SARS) coronavirus (Leong et al., 2005). Understanding the molecular basis of the host response to microbial infection particularly antiapoptotic responses, is essential for identifying targets to prevent disease and tissue damage resulting from the inflammatory response. In our study of EV71 infection of rhabdomyosarcoma (RD) cells, DNA microarray and two‐dimensional (2‐D) proteomic analyses were employed to probe into these molecular changes. The expression of the p53 tumour suppressor gene and its alternative splice variant in EV71‐infected cells was also investigated.
Results
Growth kinetics of EV71‐infected RD cells
In order to expand the scope of study, two batches of RD cells, each comprising four experimental models, were analysed at 8 and 20 h post infection (p.i.). The four models included uninfected control, mock‐infection control with UV‐inactivated EV71 reference strain MS/7423/87, EV71 strain MS/7423/87 infection, and EV71 strain 5865/Sin/000009 infection. The cell growth kinetics did not differ much at 8 h p.i. However, at 20 h p.i., a decrease in viable cell population was apparent in both EV71‐infected models (Fig. 1). Visible CPE was also observed in the infected cell culture, indicative of a full‐blown infection. It was speculated that in poliovirus‐infected cells, such CPE formation may be attributed to expression of the putative apoptosis‐preventing effect (Agol et al., 2000). Viable cell counts for the uninfected and mock‐infected controls were comparable without any potential ‘spillover’ effect from the inoculum itself, which may contain interleukins or interferons derived from the virus‐infected cell culture. There was also no visible effect in the cells subjected to the UV‐inactivated EV71. Thus, any evident changes that occurred in the infected cells were caused by the live virus itself.
Figure 1.
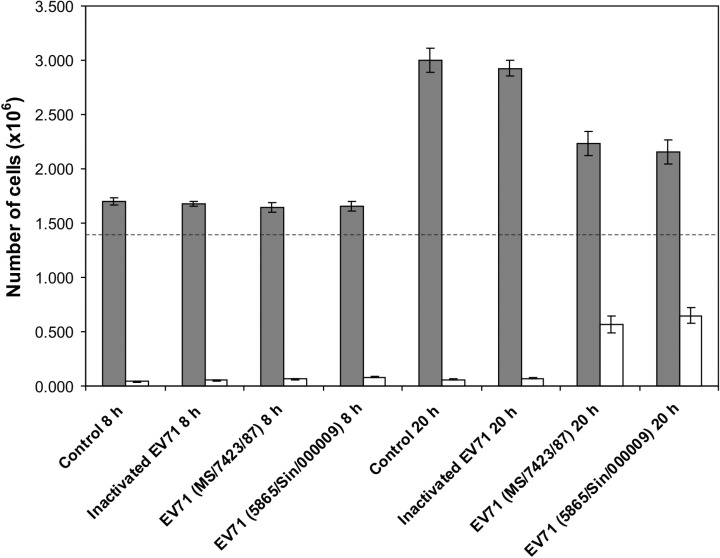
Kinetics of human rhabdomyosarcoma cell proliferation at 8 and 20 h following infection with two EV71 strains. Untreated control and inactivated EV71‐treated cells were included. Viable cell count (grey bars) was assessed by the trypan blue exclusion technique. Non‐viable cell counts are represented as unfilled bars. The dashed line depicts the starting number of cells at the point of virus inoculation.
Immunofluorescent analysis of EV71‐infected RD cells
Immunostaining of the cells with EV71‐specific monoclonal antibody at 8 h p.i. revealed only about 3–5% of the cells infected with either strain of EV71 (Fig. 2). Mature virions were reported to assemble in the cytoplasm at 12 h after infection (Rangel et al., 1998). At 20 h p.i., 50–60% of the cells were infected with either strain, especially those infected with strain 5865/Sin/000009 (Fig. 2). This observation reflected the virulence of the latter strain isolated from a fatal case of EV71 encephalitis during the outbreak in Singapore in 2000.
Figure 2.
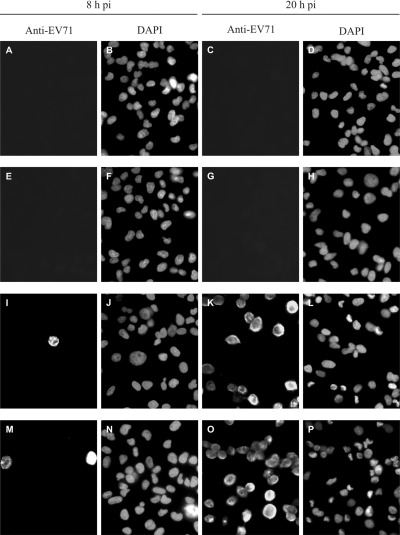
Immunofluorescence microscopy of uninfected and EV71‐infected RD cells at 8 and 20 h p.i. Infected cells were detected by anti‐EV71 monoclonal antibody, while the cell nuclei were displayed by DAPI staining. A–D. Uninfected RD cells. E–H. RD cells mock‐infected with inactivated EV71 strain MS/7423/87. I–L. RD cells infected with EV71 strain MS/7423/87. M–P. RD cells infected with EV71 strain 5865/Sin/000009.
Microarray data reveal a novel set of differentially expressed genes
Scanning of the microarray slides for Cy3 and Cy5 signals with a dual‐colour image scanner revealed that the majority of signals were of relatively high intensity. Stringency was exercised by flagging individual spots of poor quality (i.e. extreme unevenness in signals and streaks) that were omitted from Acuity software analysis. With the statistical program, we initially conducted a low‐intensity filtering based on the sum of medians of the foreground intensities of both channels that were less than 200. We selected this criterion as it allowed us to effectively disregard the low intensity spots while retaining 3736 spots of sufficient quality to be analysed. The filtered data were normalized by LOWESS analysis (Yang et al., 2002). Finally, we defined a Z‐score threshold to be greater than 1.96 based on the duplicated set, which permitted us to identify 191 genes that were differentially expressed at the 95% confidence level. Of these, 39 transcripts exhibited an increase while 152 demonstrated a decrease in expression level greater than twofold based on Acuity analysis (Table S1). Table 1 lists 58 representative transcripts that were significantly altered at 20 h p.i. according to their known functions.
Table 1.
Microarray data of 58 representative human genes exhibiting significantly altered transcription profiles in response to EV71 strain MS/7423/87 infection of RD cells at 20 h p.i.
| GenBank | Gene | Fold change |
|---|---|---|
| Transcriptional regulators | ||
| NM_004387 | Cardiac‐specific homeo box (NKX2‐5) | −5.035 |
| NM_002096 | General transcription factor IIF, polypeptide 1 (74 kDa subunit) (GTF2F1) | +2.454 |
| NM_002699 | POU domain, class 3, transcription factor 1 (POU3F1) | −5.988 |
| NM_006195 | Pre‐B‐cell leukaemia transcription factor 3 (PBX3) | −4.179 |
| Cell cycle | ||
| NM_001274 | CHK1 (checkpoint, S. pombe) homologue (CHEK1) | −4.252 |
| NM_001259 | Cyclin‐dependent kinase 6 (CDK6) | −3.048 |
| Cell adhesion | ||
| NM_005102 | Fasciculation and elongation protein zeta 2 (zygin II) (FEZ2) | −4.284 |
| NM_203487 | Protocadherin 9 (PCDH9) | −39.152 |
| NM_007008 | Reticulon 4 (RTN4) | −3.834 |
| Extracellular transport proteins | ||
| NM_006432 | Niemann‐Pick disease, type C2 (NPC2) | −2.310 |
| NM_002305 | Lectin, galactoside‐binding, soluble, 1 (galectin 1) (LGALS1) | −3.546 |
| Oncogenes and tumour suppressors | ||
| NM_006875 | pim‐2 oncogene (PIM2) | −2.601 |
| NM_000321 | Retinoblastoma 1 (including osteosarcoma) (RB1) | −3.858 |
| NM_004985 | v‐Ki‐ras2 Kirsten rat sarcoma 2 viral oncogene homologue (KRAS2) | −4.211 |
| Stress response proteins | ||
| Y00371 | Heat shock 70 kDa protein 8 (HSPA8) | −2.373 |
| NM_000454 | Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1, adult) (SOD1) | −2.478 |
| Membrane channels and transporters | ||
| NM_001689 | ATP synthase, H + transporting, mitochondrial F0 complex, subunit c (subunit 9) isoform 3 (ATP5G3) | −3.717 |
| NM_000593 | ATP‐binding cassette, subfamily B (MDR/TAP), member 2 (TAP1) | +3.816 |
| Trafficking and targeting proteins | ||
| NM_001283 | Adaptor‐related protein complex 1, sigma 1 subunit (AP1S1) | +3.571 |
| NM_004835 | Angiotensin receptor 1B (AGTR1) | −3.288 |
| NM_007033 | Similar to S. cerevisiae RER1 (RER1) | −3.633 |
| NM_003100 | Sorting nexin 2 (SNX2) | −4.464 |
| Cellular metabolism | ||
| NM_000672 | Alcohol dehydrogenase 6 (class V) (ADH6) | −8.140 |
| NM_004718 | Cytochrome c oxidase subunit VIIa polypeptide 2 like (COX7A2l) | −3.893 |
| NM_000769 | Cytochrome P450, subfamily IIC (mephenytoin 4‐hydroxylase), polypeptide 19 (CYP2C19) | −4.014 |
| NM_001931 | Dihydrolipoamide S‐acetyltransferase (E2 component of pyruvate dehydrogenase complex) (DLAT) | −3.918 |
| NM_002539 | Ornithine decarboxylase 1 (ODC1) | −2.526 |
| NM_000367 | Thiopurine S‐methyltransferase (TPMT) | −5.111 |
| Post‐translational modification | ||
| NM_006585 | Chaperonin containing TCP1, subunit 8 (theta) (CCT8) | −2.934 |
| NM_000801 | FK506‐binding protein 1 A (12 kDa) (FKBP1A) | −2.626 |
| NM_015929 | Lipoyltransferase (LIPT1) | +2.288 |
| Translational machinery | ||
| NM_001404 | Eukaryotic translation elongation factor 1 gamma (eEF1G) | −2.619 |
| NM_001967 | Eukaryotic translation initiation factor 4 A, isoform 2 (eIF4A2) | −2.565 |
| NM_000998 | Ribosomal protein L37a (RPL37A) | −3.224 |
| M77234 | Ribosomal protein S3A (RPS3A) | −3.519 |
| Apoptosis‐associated proteins | ||
| NM_004052 | BCL2/adenovirus E1B 19 kDa‐interacting protein 3 (BNIP3) | −2.603 |
| M74816 | Clusterin (complement lysis inhibitor, SP‐40,40, sulphated glycoprotein 2, testosterone‐repressed prostate message 2, apolipoprotein J) (CLU) | +2.083 |
| D42055 | Neural precursor cell expressed, developmentally downregulated 4 (NEDD4) | −4.371 |
| NM_002787 | Proteasome (prosome, macropain) subunit, alpha type, 2 (PSMA2) | −3.422 |
| RNA processing, turnover and transport | ||
| NM_004396 | DEAD/H box polypeptide 5 (RNA helicase, 68 kDa) (DDX5) | −6.570 |
| NM_001363 | Dyskeratosis congenita 1, dyskerin (DKC1) | −7.402 |
| DNA binding and chromatin proteins | ||
| NM_006026 | H1 histone family, member X (H1FX) | −6.325 |
| NM_004538 | Nucleosome assembly protein 1‐like 3 (NAP1L3) | −7.653 |
| Cellular receptors | ||
| NM_000739 | Cholinergic receptor, muscarinic 2 (CHRM2) | +2.509 |
| NM_000208 | Insulin receptor (INSR) | −15.573 |
| NM_004736 | Xenotropic and polytropic retrovirus receptor (XPR1) | −4.135 |
| Cell signalling, extracellular communication | ||
| NM_000882 | Interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) (IL12A) | −3.840 |
| NM_002415 | Macrophage migration inhibitory factor (glycosylation‐inhibiting factor) (MIF) | −2.815 |
| NM_003236 | Transforming growth factor, alpha (TGFA) | −4.287 |
| Intracellular transducers/effectors/modulators | ||
| NM_004734 | Doublecortin and CaM kinase‐like 1 (DCAMKL1) | +14.805 |
| NM_001946 | Dual specificity phosphatase 6 (DUSP6) | −4.022 |
| NM_003479 | Protein tyrosine phosphatase type IVA, member 2 (PTP4A2) | −4.475 |
| Cytoskeleton/motility | ||
| NM_001613 | Actin, alpha 2, smooth muscle, aorta (ACTA2) | +2.196 |
| NM_005159 | Actin, alpha, cardiac muscle (ACTC) | −2.339 |
| NM_000384 | Apolipoprotein B (including Ag(x) antigen) (APOB) | +5.184 |
| NM_005690 | Dynamin 1‐like (DNM1L) | −2.277 |
| NM_003283 | Troponin T1, skeletal, slow (TNNT1) | −3.514 |
| NM_003295 | Tumour protein, translationally controlled 1 (TPT1) | −3.511 |
Upregulated and downregulated transcripts are indicated as ‘+’ and ‘–’ values respectively.
Genes with altered transcriptional patterns belonged to a wide range of functional classes. For example, those involved in the host translational machinery, and mRNAs encoding RNA processing proteins were notably downregulated. This is also reflected in EV71‐infected human neural SF268 cells (Shih et al., 2004). In contrast to transcripts altered in EV71‐infected SF268 cells and SARS coronavirus‐infected Vero E6 cells (Leong et al., 2005), mRNAs of several stress response and apoptosis‐related genes were significantly downregulated, possibly to evade host immune responses and to facilitate virus multiplication. However, transcripts of other functional categories exhibited variable responses to EV71 infection.
Real‐time reverse transcription polymerase chain reaction (RT‐PCR) analysis confirms modified transcription of interesting cellular genes during EV71 infection
To verify the microarray data of cells infected with EV71 reference strain MS/7423/87, real‐time RT‐PCR was performed for 15 genes, which represented transcripts of the various functional clusters. Mock‐infected cells and those infected with EV71 strain 5865/Sin/000009 were also included. Only changes in relative expression levels greater than twofold and with P‐values less than 0.05 were considered significant. With the GAPDH gene serving as a normalization control, the changes in expression at 8 and 20 h p.i., and the corresponding P‐values are listed in Table 2. Our real‐time analysis of GAPDH gene expression also revealed that the difference in threshold cycle (CT) between infected and mock‐infected samples was negligible, thereby justifying the use of GAPDH for normalization (data not shown). Overall, the expression trends for all the genes tested by real‐time RT‐PCR concurred with the microarray data.
Table 2.
Real‐time RT‐PCR analyses of 15 selected genes expressed in RD cells at 8 and 20 h following infection with EV71 strains MS/7423/87 (MS) and 5865/Sin/000009 (SIN).
| Gene | Fold change | ||||||
|---|---|---|---|---|---|---|---|
| Microarrayanalysis | RT‐PCR 8 h p.i. | RT‐PCR 20 h p.i. | |||||
| Mock | EV71 (MS) | EV71 (SIN) | Mock | EV71 (MS) | EV71 (SIN) | ||
| CDK6 | −3.05 | −1.02 | +1.37 | +1.80 | 1.00 | −2.94 (0.013) | −3.43 (0.031) |
| eEF1G | −2.62 | +1.08 | +1.50 (0.034) | +1.80 (0.023) | −1.08 | −2.85 (0.039) | −3.56 (0.025) |
| IL12A | −3.84 | +1.23 | −1.01 | +1.28 | −1.14 | −2.60 (0.009) | −3.15 (0.018) |
| NPC2 | −2.31 | −1.08 | +1.24 | +3.19 (0.013) | +1.04 | −2.58 (0.004) | −3.07 (0.002) |
| ODC1 | −2.53 | +1.08 | +1.11 | +1.25 | +1.10 | −2.99 (0.007) | −2.35 (0.017) |
| PIM2 | −2.60 | +1.10 | +1.11 | +1.30 | +1.20 | −2.69 (0.027) | −3.17 (0.008) |
| PSMA2 | −3.42 | +1.04 | +1.33 | +1.27 | +1.17 | −4.61 (0.001) | −4.71 (0.001) |
| TPT1 | −3.51 | −1.05 | +1.09 | +1.78 (0.036) | +1.27 | −3.16 (0.008) | −3.07 (0.006) |
| ACTA2 | +2.20 | 1.00 | +4.07 (0.020) | +5.05 (0.006) | +1.21 | +7.75 (0.001) | +8.25 (0.011) |
| AP1S1 | +3.57 | −1.23 | −1.12 | −1.16 | +1.03 | +3.59 (0.030) | +3.31 (0.022) |
| APOB | +5.18 | +1.02 | −1.02 | −1.03 | −1.02 | +8.97 (0.005) | +6.57 (0.007) |
| CHRM2 | +2.51 | +1.04 | +3.11 (0.028) | +3.05 (0.004) | −1.23 | +2.27 (0.033) | +2.14 (0.037) |
| CLU | +2.08 | −1.25 | −1.45 | −1.32 | −1.11 | +3.59 (0.015) | +3.31 (0.030) |
| DCAMKL1 | +14.81 | +1.12 | +1.20 | +1.35 | +1.09 | +35.88 (0.013) | +36.25 (0.033) |
| GTF2F1 | +2.45 | +1.03 | −1.08 | +1.41 | +1.04 | +2.73 (0.004) | +2.80 (0.004) |
Samples were normalized with GAPDH as the control housekeeping gene, and with uninfected RD cells at each respective time‐point as the reference sample (value of 1). The P‐values denoting significant differences (P < 0.05) are shown within parentheses.
Real‐time RT‐PCR targeting a 269 bp fragment within the EV71 VP1 gene provided an indication of the intensity of virus replication and viral load. At 8 h p.i., we observed a 32.67‐fold and 41.07‐fold increase in viral transcripts for EV71 strain MS/7423/87 and strain 5865/Sin/000009 respectively. However, 20 h after infection, the transcript levels rose to 1552‐fold and 2646‐fold, respectively, reflecting the profuse EV71 replication process within RD cells.
Western blot analyses correlate alterations in protein expression with transcriptional changes
In order to correlate our findings at the transcriptional level with protein expression, four representative genes evaluated by real‐time RT‐PCR were selected for Western analysis (Fig. 3). We selected DCAMKL1 as it was highly transcribed in EV71‐infected RD cells. Western blot analysis revealed strong expression of DCAMKL1 in infected cells at 20 h p.i., which was also reflected in the transcript level. Similarly, APOB and CLU proteins were also upregulated in EV71 infection. However, ODC1 protein level decreased in infected cells at this time point. Interestingly, the VP1 structural protein of EV71 was shown to interact with ODC1 by coimmunoprecipitation and coimmunolocalization experiments (W. Yeo and V. Chow, unpubl. data). Although only four gene products were assessed, it was possible to correlate the transcriptional modification with protein expression. The GAPDH housekeeping protein was included as a control to ensure equal loading of protein samples.
Figure 3.
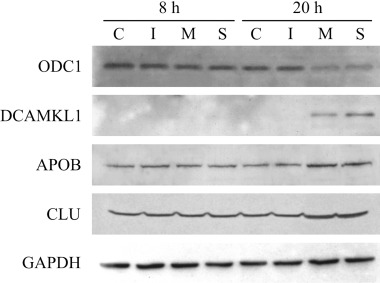
Western blot analysis of ODC1, DCAMKL1, APOB and CLU expression at 8 and 20 h p.i. Total protein extracts (20 µg each) of uninfected control (C), mock‐infected (I), EV71 MS/7423/87 strain‐infected (M), and EV71 5865/Sin/000009 strain‐infected (S) RD cells were tested against the relevant specific antibodies. The GAPDH housekeeping protein was included to ensure equal loading of protein samples.
The p53 splice variant is expressed in EV71‐infected cells
The p53 tumour suppressor gene is known to play a role in viral infection (Grayson et al., 2001; Garden et al., 2004; Takemoto et al., 2004). Moreover, it is post‐translationally modified during infection (Kao et al., 2004), thus prompting us to further investigate p53 expression. From the uninfected, mock‐infected and EV71‐infected RD cells, we amplified a 353 bp p53 gene fragment spanning exons 7–10, sequencing of which corresponded to nucleotides 928–1280 of the wild‐type human p53 gene (GenBank Accession number NM_000546). Intriguingly, at 20 h p.i., an additional larger 486 bp fragment was also amplified especially in the infected samples (Fig. 4), sequencing of which revealed that it spanned nucleotides 928–1244 and 1245–1280 of the p53 gene. Between these two segments was found an insertion of 133 nucleotides from intron 9 as previously reported in the MOLT‐4 human acute lymphoblastic leukaemia cell line (Chow et al., 1993), which can generate a truncated p53 product lacking the C‐terminal amino acids encoded by exons 10 and 11. This p53 alternative transcript was more evident in EV71‐infected cells, and may represent a stress‐related response (Lainez et al., 2004).
Figure 4.
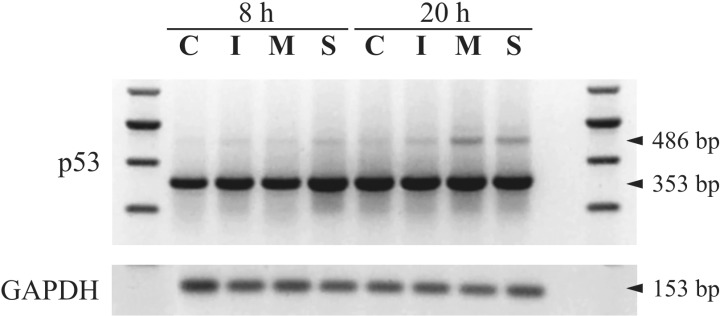
RT‐PCR amplification of the p53 gene at 8 and 20 h p.i. A 353 bp fragment representing the wild‐type p53 transcript was amplified in uninfected control (C), mock‐infected (I), EV71 MS/7423/87 strain‐infected (M), and EV71 5865/Sin/000009 strain‐infected (S) RD cells. An additional 486 bp fragment corresponding to the p53 splice variant appeared more distinctly in cells infected with the two EV71 strains after 20 h. GAPDH (153 bp fragment) served to ensure equal amounts of starting template in the samples.
Two‐dimensional proteomic analysis of EV71‐infected RD cells
In order to expand the scope of studying EV71 infectomics, limited global cellular protein expression in response to infection with EV71 strain MS/7423/87 was also investigated by 2‐D gel electrophoresis. Using Delta2D analysis, the gels were normalized by their total intensity of all spots because we did not have an internal control for normalization. However, one‐dimensional separation and Western blot analysis of GAPDH in the same samples displayed equal intensities (data not shown). Each spot was expressed as relative volume with respect to all detected spots (% vol), and as absolute volume of the ID spot. The fold change was calculated as the percentage vol of infected divided by percentage vol of uninfected samples. Six distinct differentially expressed protein spots with greater than twofold changes in expression were identified (Fig. 5). Through MALDI‐TOF/TOF analysis of each sample, a score was generated based on −10*log(P), where P was the probability that the observed match was a random event. Only protein scores greater than 76 were considered as significant (P < 0.05). The analyses also correlated with the estimated molecular masses and isoelectric points of the corresponding spots, thereby further strengthening the validity of the results. Three of the six spots were downregulated during EV71 infection, i.e. chaperonin containing TCP1, subunit 5 (epsilon) (CCT5); stathmin 1 (STMN1); and proteasome (prosome, macropain) subunit, alpha type, 2 (PSMA2). Interestingly, PSMA2 was also identified in the microarray as a downregulated transcript albeit at a different magnitude of fold change, thus lending further support to the microarray data. Moreover, PSMA2 was also identified as a downregulated transcript in our previous study (Leong et al., 2002). The three upregulated proteins included heat shock 27 kDa protein 1 (HSPB1); cofilin 1 (non‐muscle) (CFL1); and enolase 1 (ENO1).
Figure 5.
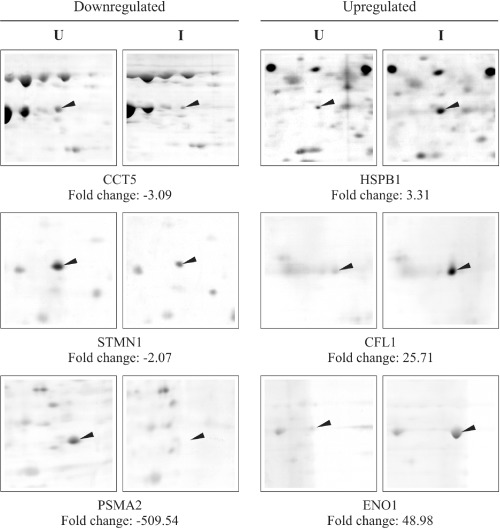
Identification of six differentially expressed proteins by two‐dimensional proteomic analysis of uninfected (U) and EV71 MS/7423/87 strain‐infected (I) RD cell samples at 8 and 20 h p.i. The arrowheads indicate the protein spots identified as differentially regulated at greater than twofold changes (P < 0.05) using Delta2D analysis.
DCAMKL1 and ODC1 were not detected in this proteomic analysis, possibly due to loss during the fractionation step or poor resolution of spots at basic pH of greater than 8 (especially for DCAMKL1 with its predicted isoelectric point of 8.84).
Discussion
In order to better understand the pathophysiology of EV71 infection and host cellular responses particularly the apoptotic responses, we employed DNA microarray and 2‐D proteomic analyses to probe into the cellular gene expression changes during the infection process. The RD cell line served as the host cells for infection as it is widely used for isolation and detection of enteroviruses with characteristic CPE as observed in our study (Kok et al., 1998). RD cells are continuous, stable and can be cultivated in‐house without decreasing sensitivity even as the passage number increases. Notably, RD cells are derived from muscle tissue, and may offer some insights into the neuromuscular diseases caused by EV71, thus further warranting their use. Signs of late apoptotic responses detectable after the phase of active virus growth and the development of CPE (Romanova et al., 2005) in RD cells allowed us to probe into the apoptosis‐evading mechanism during productive infection. Pelletier et al. (1998) stated that the multiplicity of infection (moi) also plays an important role in the outcome of infection. A lower moi can leave the infected cell sufficient time to produce certain factors necessary for its survival. Thus, this prompted us to use an moi of 0.1 to investigate such a phenomenon. From the immunofluorescence staining of cells infected at an moi of 0.1, we observed huge aggregates of virions inside the infected cells as early as 8 h p.i. even though there was no visible CPE, which may imply a possible apoptosis‐evading mechanism to allow virus multiplication.
We were also interested to observe whether the Singapore EV71 isolate showed any temporal differences in inducing cellular gene expression or in eliciting host phenotypic changes compared with EV71 strain MS/7423/87. However, real‐time RT‐PCR did not reveal significant differences except for the upregulation of NPC at 8 h p.i. for cells infected with the Singapore isolate from a fatal case. NPC is involved in the transport of cholesterol through the late endosomal/lysosomal system (Garver and Heidenreich, 2002). An increase in NPC may be related to dysfunctional cholesterol homeostasis. However, the downregulation of NPC at 20 h p.i. for both strains may eventually lead to cognitive impairment and focal frontal atrophy (Klunemann et al., 2002). Nevertheless, the significance of such alterations of NPC expression in viral pathogenesis awaits further functional investigation.
From the microarray analysis, 191 differentially regulated genes were classified according to their functional roles. The most noteworthy genes downregulated by the infection were those involved in the host translational machinery, including 40S ribosomal proteins, 60S ribosomal proteins, eukaryotic translation elongation factors and eIF4A2, which were also observed in our previous study (Leong et al., 2002). Such host translational shutoff is frequently observed, as evidenced by infections with influenza virus type A (Garfinkel and Katze, 1992), herpes simplex virus (Everly et al., 2002), and poliovirus (Kuyumcu‐Martinez et al., 2004). The poliovirus protease 2A‐mediated cleavage of eIF4G, which is part of the host cap‐dependent translation initiation complex, causes a rapid shutoff of cap‐dependent translation (Krausslich et al., 1987), thereby shifting the host cell machinery from cellular to viral processes. The downregulation of eIF4A2 which is also part of the host translation initiation complex (Sudo et al., 1995), may contribute to host translational shutoff.
Transcripts involving protein folding such the chaperonin T complex family were also downregulated. CCT8 forms part of the T complex polypeptide 1 (TCP1) that appears to be involved in the folding of actin and tubulin in an ATP‐dependent fashion (Kubota et al., 1995). Poliovirus infection is usually accompanied by rearrangement of the intermediate filament framework (Weed et al., 1985). Thus, its downregulation in EV71 infection may play a role in the disruption of cytoskeletal structure to aid viral replication, characterized by the large accumulation of membranous vesicles to which viral replication complexes are attached. Another member, CCT4, was also downregulated in our previous study (Leong et al., 2002).
Several other genes pertaining to the cytoskeleton and motility were also differentially expressed, which may adversely affect structural integrity or even lead to growth arrest. In EV71‐infected cells, the downregulation of TNNT1, which maps to the myotonic dystrophy region of human chromosome 19 (Novelli et al., 1992), may be implicated in the chronic inflammatory muscle disease or fibromyalgia of infected patients (Douche‐Aourik et al., 2003). Expression of the TPT1 microtubule‐stabilizing protein was also downregulated, which may promote an increase in microtubule dynamics (Yarm, 2002) observed in enterovirus infection. The guanine nucleotide dissociation inhibitor activity of TPT1 may also prevent eEF1A activation into eEF1A‐GTP before it is recruited by other components of the translational machinery to form a complex with a de novo‐aminoacylated tRNA (Cans et al., 2003). Thus, decreasing TPT1 expression in infected cells may be a means to decrease the efficiency of host protein synthesis, to stifle cell proliferation, which allows the host's resources to be channelled to virus replication. In addition, ODC1 enzyme which is involved in the cell cycle and possesses growth‐promoting activity (Moshier et al., 1990; Nemoto et al., 2002) exhibited reduced expression in infected cells that may serve to limit cell proliferation in support of virus replication.
The expression of another tubule network‐associating protein, DNM1L, was also decreased in EV71‐infected cells. DNM1L is involved in peroxisomal fission and in the maintenance of peroxisomal morphology in mammalian cells (Koch et al., 2003). Its disruption will lead to an increase in hydrogen peroxide which can cause oxidative damage, and a decrease in cholesterol synthesis that may affect the integrity of cellular membranes. Furthermore, the synthesis of lipids for myelin production may also be compromised, and be potentially linked with the neurodegenerative disorders observed in certain EV71‐infected patients. On the other hand, the decrease in DNM1L will also limit its activity in the scission of the outer mitochondrial membrane, thus preventing caspase activation and subsequent apoptosis (Breckenridge et al., 2003). This notion is further supported by the downregulation of FKBP1A that modulates the activity of type 1 TGF‐β receptor (Wang et al., 1994) as well as ryanodine receptor. Possible consequences may be diminished apoptosis, disruption in muscle contractility, and cardiac defects (Van Acker et al., 2004), characteristic of enteroviral infection.
Alpha actin mRNAs were also differentially transcribed during EV71 infection. ACTC is highly expressed in heart muscle, and is a major constituent of the contractile apparatus. Defects in this gene are associated with idiopathic dilated cardiomyopathy and familial hypertrophic cardiomyopathy (Mogensen et al., 1999). Thus, reduced ACTC expression level may be associated with myocarditis experienced by some EV71‐infected patients. ACTA2 is a smooth muscle α‐actin whose expression is transformation‐sensitive to growth signals in normal cells. Actively proliferating fibroblasts and smooth muscle cells have low levels of ACTA2, but the inhibition of cell proliferation by density arrest or treatment with antimitotic agents, induces the smooth muscle α‐actin promoter (Kumar et al., 1995). Thus, ACTA2 upregulation may be a result of growth arrest induced by EV71 infection, and was observed as early as 8 h p.i. by real‐time RT‐PCR.
The upregulation of LIPT1, which transfers the lipoyl moiety to apoproteins, was also associated with increased expression of two apolipoproteins APOB and CLU as revealed by microarray and Western blot analyses. APOB is a major component of low‐density lipoproteins (LDLs) whose upregulation may promote the myelination of the CNS. However, LDL is considered as an undesirable cholesterol, and APOB results in platelet activation (Relou et al., 2002). An increase in both processes may enhance the risk of atherosclerosis and eventually myocardial infarction (Qureshi et al., 2002). Indeed, enteroviruses have been detected in some patients with atherosclerosis (Kwon et al., 2004). In contrast, CLU is a controversial protein whose accumulation may culminate in antiapoptotic (Miyake et al., 2004) as well as pro‐apoptotic (Trougakos and Gonos, 2004) events. However, Chen et al. (2004) showed that the action of CLU is dependent on its two different isoforms, one nuclear, and the other secreted and cytoplasmic. Notably, overexpression of CLU is also observed in neurodegenerative diseases (Duguid et al., 1989), which is relevant to the CNS disease model of enterovirus infection. Given the hypothesis that nuclear CLU function is pro‐apoptotic, it would be interesting to investigate its isoforms as well as the localization of the upregulated CLU in EV71 infection.
DCAMKL1, a protein involved in neuronal development, was very highly expressed during EV71 infection. It is a probable member of the Ser/Thr protein kinase family that may be involved in a calcium signalling pathway that controls neuronal migration in the developing brain (Burgess and Reiner, 2002), and may also participate in functions of the mature nervous system. Although its significance in EV71 infection is yet to be elucidated, it may represent a scrambled effort by the host to salvage the hostile infection. In order to understand its significance, one could knock‐down DCAMKL1 expression by the small interfering RNA approach and observe the effect on the disease process induced by EV71.
Other cell‐signalling proteins involved in immune responses that were downregulated during EV71 infection include IL12A and MIF. The altered expression of IL12A in EV71 infection may serve to downmodulate the recruitment of T‐cells and natural killer cells as demonstrated in HIV infection in which it facilitates virus replication (Ayyavoo et al., 1997). The same effect may apply to MIF which is proinflammatory and involved in the recruitment of inflammatory cells (Onodera et al., 2004). Notably, poliovirus 3A protein has also been shown to prevent the cell surface expression of MHC class I molecules (Deitz et al., 2000), suggestive of its role in regulating these genes and inhibiting the cellular immune mechanisms. In general, many cytokine‐related genes were not transcriptionally regulated in the infected cells, possibly as part of the immunoevasive strategy of EV71.
Apoptosis‐associated proteins were regulated to various degrees, notably to support the apoptosis‐evasive effects of EV71. BNIP3 is an apoptosis‐inducing dimeric mitochondrial protein (Chen et al., 1997), whose decreased expression may serve to delay the onset of apoptosis to favour virus multiplication.
In addition to DNA microarray analysis, 2‐D proteomic analyses allowed us to identify proteins that were differentially regulated. ENO1 is involved in the induction of cell death in fibroblasts and in the tumorigenicity of human breast carcinoma cells (Ray, 1995; Ray et al., 1995). Its cell growth regulation is further evidenced by the identification of functional domains involved in transcriptional repression (Ghosh et al., 1999). Therefore, ENO1 upregulation may enhance growth arrest in EV71‐infected cells culminating in cell death and CPE. In contrast, introduction of recombinant HSPB1 into neutrophils delays apoptosis (Sheth et al., 2001). Disruption of the interaction between HSPB1 and Akt impairs Akt activation, leading to an enhanced rate of constitutive neutrophil apoptosis (Rane et al., 2003). Hence, the upregulation of HSPB1 during EV71 infection may delay neutrophil apoptosis, and result in enhanced host defence. However, inappropriate delay of neutrophil apoptosis is associated with generalized inflammation and multi‐organ failure of the systemic inflammatory response syndrome (Keel et al., 1997).
PSMA2 is part of a multicatalytic proteinase complex (proteasome), and also of the immunoproteasome whose essential function is the processing of class I MHC peptides (Coux et al., 1996). Its transcriptional downregulation coincided with the 2‐D analysis as well as with a previous study (Leong et al., 2002), and may be suggestive of viral immunoevasion. Another downregulated protein was CCT5, which shares similar functions with CCT8 (Kubota et al., 1994; 1995). STMN1 is involved in the regulation of the microtubule filament system by destabilizing and preventing assembly of microtubules (Gavet et al., 1998). Thus, its decreased expression is expected to induce growth arrest in EV71‐infected cells. Moreover, a low level of STMN1 in adult brains is also associated with Down's syndrome and Alzheimer's disease (Cheon et al., 2001), which may be linked to the CNS complications of EV71 infection.
However, another cytoskeleton‐related protein, CFL1 was upregulated. CFL1 is a member of the CFL/actin depolymerizing factor (ADF) family, which regulates actin dynamics by increasing the rate of actin depolymerization and facilitating actin filament turnover (Chen et al., 2000). In neutrophils and neutrophil‐like cells, the dephosphorylation of CFL1 and its translocation to F‐actin‐rich regions, correlate with the production of superoxide and hydrogen peroxide (Nagaishi et al., 1999). This is congruent with the downregulation of DNM1L, which leads to an increase in hydrogen peroxide level. In cultured hippocampal and cortical neurons, mitochondrial dysfunction leads to the rapid dephosphorylation of ADF and CFL1, which coassemble with actin to form rods that span the diameter of the neurite and disrupt the normal cytoskeleton (Minamide et al., 2000). Such persistent rods cause degeneration of the distal neurite without killing the neuron. These findings suggest a common pathway that can lead to loss of synapses, which mirrors the neurodegenerating disorder seen in some EV71‐infected patients. Dephosphorylated CFL1 binds to actin and translocates into mitochondria, and actin cytoskeletal changes may enhance mitochondrial translocation of pro‐apoptogenic proteins resulting in mitochondrial dysfunction, release of cytochrome c and apoptosis (Chua et al., 2003). Thus, enhanced CFL1 level in EV71 infection and its dephosphorylation may lead to destruction of neuronal processes and the initiation of apoptosis in the late phase of infection, similar to poliovirus infection (Agol et al., 2000). It will be interesting to investigate the effect of silencing CFL1 on neuronal plasticity during EV71 infection, and whether the neuropathologic fate can be attenuated.
In EV71‐infected RD cells, the alternate p53 transcript was expressed at 20 h p.i. Its presence in normal, non‐malignant Vero E6 cells infected with SARS coronavirus (Leong et al., 2005), implies that this splice variant is not an exclusive by‐product of cancerous cells. Various alternatively spliced p53 isoforms are expressed at relatively low levels and tend to be restricted to particular cell types and/or physiologic conditions (Courtois et al., 2004). Compared with wild‐type p53, the C‐terminally altered p53 protein inhibits both p53‐dependent apoptosis and transactivation (Almog et al., 2000); binds more efficiently to DNA in a sequence‐specific manner; and is more efficient in concentration‐dependent transcriptional repression of the promoter of the p21 cyclin‐dependent kinase inhibitor gene. The presence of this p53 isoform with a truncated C‐terminus is therefore expected to modify the activity of the wild‐type p53, and exert a role in the pathogenesis of EV71 infection. Although the expression of this alternative transcript was lower than the wild‐type p53, this isoform does not possess ubiquitination sites as in the wild type. Thus, it may be more stable, allowing it to exert its function longer in infected cells.
Enteroviruses are known to shut off the host translational machinery, which may limit the expression of genes, and may cast doubt on studies that hinge on transcriptional levels. However, poliovirus regulates host cell transcription by translocation of the viral 3C protein into the nucleus in which it exerts its effect on the PolI and II transcription system (Weidman et al., 2003). Furthermore, poliovirus also regulates host cell transcription in neighbouring uninfected cells, possibly through accumulation of poliovirus proteins in host cell nuclei (Bossart et al., 1984). Thus, viruses may preprogram uninfected cells prior to virus infection in order to promote favourable metabolic conditions to ensure cell survival during the early phase of infection and to allow rapid multiplication of progeny virus before CPE occurs. Koyama et al. (2000) also demonstrated that poliovirus can replicate considerably in apoptotic cells.
From our analysis of EV71‐infected RD cells, it is apparent that there is a general trend to inhibit cell proliferation coupled with mechanisms which delay the apoptotic process to permit virus replication. However, Shih et al. (2004) observed upregulation of apoptotic genes in EV71‐infected human SF268 cells, which may be attributed to different conditions such as moi, infection time, and strain of EV71 used. For example, the infectivity of and CPE induction by different EV71 strains varies in different cells. Our study also alluded to the possible roles of cytoskeleton‐related genes during EV71 infection of RD cells, compared with EV71‐infected SF268 cells (Shih et al., 2004). The present study offers clues to a global view of the dynamic balance between EV71 and the host immune system which attempts to control the infection, resulting in modulation of apoptosis by the virus. This complex network is host‐specific as well as virus‐specific. The consequences of such modulation can contribute to the outcome of infection and to disease pathogenesis. For example, CPE (but not apoptosis) is the contributory factor for an inflammatory reaction, and the occurrence of inflammation will determine the clinical manifestation of the disease. Thus, molecular insights into the pathophysiologic mechanisms can provide a better understanding of such relationships, and lead to novel and viable strategies for intervention of the process of EV71 infection (Huang et al., 2002).
Experimental procedures
Cell culture and virus infection
Rhabdomyosarcoma cells (ATCC number CCL‐136) were cultured in minimum essential medium supplemented with 5% fetal bovine serum, 1.05 g l−1 sodium bicarbonate, 20 mM HEPES, 0.1 mM non‐essential amino acids at 37°C with 5% CO2 in a humidified incubator. When cells reached ∼90% confluency, one batch of uninfected RD cells in 75 cm2 culture flask served as the control, while another three batches were infected with UV‐inactivated EV71 strain MS/7423/87, live EV71 strains MS/7423/87 and 5865/Sin/000009 (Singh et al., 2002b) at an moi of 0.1, with 4 ml of virus inoculum diluted with maintenance medium. After adsorption for 2 h, the inoculum was removed, and 15 ml of maintenance medium was added. Following incubation at 37°C for 8 and 20 h, both uninfected and infected RD cells were harvested for extraction of total RNA and proteins.
Immunofluorescence staining
Rhabdomyosarcoma cells were seeded at a density of 1 × 105 cells onto pretreated coverslips in a 6‐well plate. After overnight incubation at 37°C, the cells were inoculated at an moi of 0.1 with the two live EV71 strains and inactivated EV71 for 2 h. After adsorption, the inoculum was removed, replaced with 3 ml of maintenance medium, incubated for 8 and 20 h, and fixed with 3.7% paraformaldehyde for 30 min at room temperature. The cells were then permeabilized with 0.2% Triton X‐100 for 5 min, and incubated with mouse anti‐EV71 IgG2b monoclonal antibody (Chemicon, Temecula, CA, USA) for 30 min at 37°C. After washing excess antibody, the cells were incubated for 30 min with anti‐mouse IgG conjugated with FITC (Chemicon) with Evans blue counterstain. Stained coverslips were mounted with mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA), and visualized under fluorescence microscopy.
Total RNA preparation
Total cellular RNA was extracted from uninfected and EV71‐infected RD cells using the SV Total RNA isolation system (Promega, Madison, WI, USA). The RNA was suspended in nuclease‐free water, and the concentration quantified by UV spectrophotometry at 260 nm. The integrity of the extracted RNA was confirmed by electrophoresis in a 1% denaturing agarose gel.
Microarray hybridization and analysis
Total RNA (25 µg) extracted from each batch of uninfected and EV71‐infected RD cells at 20 h p.i. was reverse‐transcribed, and labelled with Cy3‐ and Cy5‐dCTP, respectively, using a CyScribe first‐strand cDNA labelling kit (Amersham, Little Chalfont, UK) with anchored oligo(dT) and random nonamers. Labelled cDNAs were purified using a CyScribe GFX purification kit (Amersham) and resuspended in 60 µl of elution buffer. The amounts of Cy3‐ and Cy5‐labelled cDNAs were measured at absorbances of 550 nm and 650 nm, respectively, and calculated based on the formula: (absorbance/extinction coefficient) × volume of cDNA × dilution factor × 106. The extinction coefficients for Cy3 and Cy5 were 150 000 and 250 000 l mol−1 cm−1 respectively. The experiments were performed using the Altas™ glass human 7.6K microarray set (BD Clontech, Mountain View, CA, USA) which comprises a total of 7600 human genes spotted onto two slides each consisting of 3800 genes. This oligo‐based array consists of ∼80 bp fragments representing a wide range of known genes. The Cy3‐ and Cy5‐labelled cDNA samples (100 pmol each) in 2.1 ml of BD GlassHyb hybridization solution (BD Clontech) prewarmed at 50°C, were hybridized to each microarray slide in an upright position at 50°C for 17 h. The slides were washed according to the manufacturer's recommendations. Each slide was scanned with the GenePix 4000B image scanner, and the data were collected with the GenePix Pro 4.1 software (Axon Instruments, Union City, CA, USA). The experiment was repeated from two independent samples with reversal of Cy3 and Cy5 dye labelling. The raw data were analysed using Acuity software version 3.1 (Axon Instruments).
cDNA synthesis and real‐time SYBR Green RT‐PCR detection
Total RNA (4 µg) was reverse‐transcribed in a 40 µl reaction mix containing 1 × first‐strand buffer, 10 mM dithiothreitol, 600 ng random hexamers, 0.5 mM deoxyribonucleoside triphosphates, 10 U RNase inhibitor and 200 U SuperScript reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Synthesized cDNA was aliquoted and stored at −80°C.
The differential expression of 15 genes selected after the microarray analysis was validated by real‐time quantitative RT‐PCR using SYBR Green‐based detection with a LightCycler system (Roche, Basel, Switzerland) (Table 3). Each 10 µl reaction included 1 µl of cDNA, 0.5 µM of each primer, 2.5–3 mM MgCl2, and 1 µl of 10 × Hot Start reaction mix (Roche). The reactions were subjected to an initial denaturation of 95°C for 10 min, followed by 40 cycles each of 95°C for 10 s, 58–62°C for 5 s and 72°C for 10 s, before subjecting to melting curve analysis. Amplified products were also analysed for specificity by agarose gel electrophoresis, and further verified by automated cycle sequencing. The conserved GAPDH gene served as the housekeeping gene transcript to normalize the samples as its expression level was relatively constant. To ensure consistency in CT values, duplicate reactions were performed (i.e. from each of the two cDNA preparations of each RNA sample), and the mean CT values were used for calculating the relative expression levels. The CT values were analysed as described previously (Leong et al., 2002; , 2005), and the normalized CT values of each gene were subjected to Student's t‐test with two‐tailed distribution to determine the significance at the 95% confidence level.
Table 3.
Sequences of primers for real‐time RT‐PCR amplification of selected genes.
| Gene | Primer sequence (5′→3′) | Target size(bp) | |
|---|---|---|---|
| Sense | Anti‐sense | ||
| ACTA2 | ATTGACATCAGGAAGGACCT | TGTGTGCTAGAGACAGAGAG | 305 |
| AP1S1 | TGCCAGCCTCTACTTCTGCT | CGATGGCTTTCAGCACACTC | 221 |
| APOB | GCTTCAGCCATTGACATGAG | CTTTGTGTTCAAGAGCTGCA | 277 |
| CDK6 | AAGAATGTTGGCAGGTGACT | TTCAAGAGGGCTTTCTGTGT | 258 |
| CHRM2 | AACCCAGGATGAAAACACAG | GTGATGATGAAAGCCAACAG | 294 |
| CLU | ATTCTGACCAAGGTGCGACA | TGGACAGGAAATGCCACAGT | 265 |
| DCAMKL1 | GCCTGAGTTTCTTCTGGAAC | TACTGTTCAGCCACCAGATG | 209 |
| eEF1G | TCAGACCTTCATGAGCTGCA | TACTCTCGAACCAGCGTCTG | 249 |
| EV71VP1 | TTCAGTAGGGCAGGCTTGGT | GTCTGCCAAGCGAGTGATTC | 269 |
| GAPDH a | AGGTGAAGGTCGGAGTCAAC | CATGGGTGGAATCATATTGG | 153 |
| GTF2F1 | GACCACTAAGGACCTGCTGA | AGTCAGGGAGCAAGCAGGAT | 303 |
| IL12A | CCAGGTGGAGTTCAAGAC | ACAACGGTTTGGAGGGAC | 285 |
| NPC2 | AGCTCTGCTGCTTCAACAAC | AGGTGTAGAAAGAGGCCACA | 223 |
| ODC1 | ATACTCTATGACCACGCACA | GCCCTGACATCACATAGTAG | 250 |
| PIM2 | GGAGATTCTGGAAGCTGAG | TGTCCATCTATCCCTGTGA | 286 |
| PSMA2 | AAGGGCAAATGACAGAGGAT | TGGAAGGTGGGTTTAACAGT | 276 |
| TPT1 | ACCGAAAGCACAGTAATCAC | CACGGTAGTCCAATAGAGCA | 274 |
. Housekeeping gene serving as the normalization control.
Classical RT‐PCR amplification of p53 splice variant
Each cDNA sample was subjected to classical RT‐PCR amplification of the p53 splice variant using primers P53U and P53D spanning exons 7–10, and the products electrophoresed in an agarose gel (Chow et al., 1993). The target cDNA fragment size of the alternative p53 transcript was expected to be 133 bp larger than the wild‐type p53 transcript. Amplified fragments were cloned into pGEM T‐Easy vector (Promega), and directly sequenced.
Western blot analysis
Uninfected RD cells and EV71‐infected cells at the two time points were lysed using cold lysis buffer (containing 50 mM Tris pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X‐100 and protease inhibitor cocktail) with sonication. Cell lysates (20 µg each) were separated by SDS‐PAGE alongside BENCHMARK protein ladder markers (Invitrogen). The separated proteins were transferred to polyvinylidine difluoride membranes, reacted with 1:500 dilutions of the corresponding primary specific antibodies in TBST with 0.1% gelatin. This was followed by incubation with secondary anti‐rabbit or anti‐goat IgG conjugated with horseradish peroxidase at dilutions. The blots were visualized by the ECL Western blotting detection system (Amersham), and exposed to autoradiography film for several seconds until the band intensities were satisfactory. The films were then scanned with ImageScanner (Amersham).
Protein analysis via 2‐D gel electrophoresis
The protein samples were fractionated using the 2‐D fractionation kit (Amersham) to yield a total of five fractions, which reduced the number of protein species present in each fraction. This strategy increased the mass of each protein that could be loaded onto the gel in order to bring the low abundance proteins into the dynamic range of detection, and also produced less crowded individual protein maps thus simplifying analysis and interpretation. The fractions were dialysed overnight against 30 mM Tris pH 8.5 and 7 M urea in mini‐dialysis tubes (Amersham). The dialysed samples were concentrated using the 2‐D clean up kit (Amersham). The proteins were then dissolved in rehydration buffer comprising 2% CHAPS, 2 M thiourea, 8 M urea and 0.5% IPG buffer. An equal amount of each protein fraction (∼250 µg) was loaded onto each 24‐cm Immobiline DryStrip with pH range 4–7 or 6–9 (Amersham). The strips were later focused on the Ettan IPGphor isoelectric focusing system (Amersham) according to the manufacturer's recommendation. Prior to second dimension separation, each IPG strip was equilibrated with 10 ml of SDS equilibration buffer containing 50 mM Tris pH 8.8, 2% SDS, 30% glycerol, 6 M urea and 0.002% bromophenol blue. The SDS‐PAGE separation process was performed at 18°C, and then stained with the silver stain plus kit (Bio‐Rad, Hercules, CA, USA). The gel images were captured as 400 dpi TIFF format using ImageScanner (Amersham), and analysed using Delta2D v3.2 (DECODON, Greifswald, Germany). The differentially expressed protein spots were subjected to MALDI‐TOF‐TOF mass spectrometry analysis with automated database (Mascot) search for protein identification based on the peptide mass fingerprinting and partial sequence data.
Acknowledgements
We thank M. A. Pallansch (Centers for Disease Control, Atlanta, GA, USA) and K. P. Chan (Singapore General Hospital) for kindly providing the EV71 strains. W. F. Leong was supported by a research scholarship from the National University of Singapore. This project was funded by a grant from the Biomedical Research Council, Singapore.
References
- Agirre, A. , Barco, A. , Carrasco, L. , Nieva, J.L. (2002) Viroporin‐mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J Biol Chem 277: 40434–40441. [DOI] [PubMed] [Google Scholar]
- Agol, V.I. , Belov, G.A. , Bienz, K. , Egger, D. , Kolesnikova, M.S. , Romanova, L.I. , et al. (2000) Competing death programs in poliovirus‐infected cells: commitment switch in the middle of the infectious cycle. J Virol 74: 5534–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahr, B. , Robert‐Hebmann, V. , Devaux, C. , Biard‐Piechaczyk, M. (2004) Apoptosis of uninfected cells induced by HIV envelope glycoproteins. Retrovirology 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almog, N. , Goldfinger, N. , Rotter, V. (2000) p53‐dependent apoptosis is regulated by a C‐terminally alternatively spliced form of murine p53. Oncogene 19: 3395–3403. [DOI] [PubMed] [Google Scholar]
- Ayyavoo, V. , Mahboubi, A. , Mahalingam, S. , Ramalingam, R. , Kudchodkar, S. , Williams, W.V. , et al. (1997) HIV‐1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor kappa B. Nat Med 3: 1117–1123. [DOI] [PubMed] [Google Scholar]
- Barco, A. , Feduchi, E. , Carrasco, L. (2000) Poliovirus protease 3C (pro) kills cells by apoptosis. Virology 266: 352–360. [DOI] [PubMed] [Google Scholar]
- Belov, G.A. , Romanova, L. , Tolskaya, E.A. , Kolesnikova, M.S. , Lazebnik, Y.A. , Agol, V.I. (2003) The major apoptotic pathway activated and suppressed by poliovirus. J Virol 77: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart, W. , Egger, D. , Rasser, Y. , Bienz, K. (1984) Accumulation of poliovirus proteins in uninfected isolated HEp‐2 cell nuclei in vitro . Intervirology 21: 150–158. [DOI] [PubMed] [Google Scholar]
- Breckenridge, D.G. , Stojanovic, M. , Marcellus, R.C. , Shore, G.C. (2003) Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol 160: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, H.A. , Reiner, O. (2002) Alternative splice variants of doublecortin‐like kinase are differentially expressed and have different kinase activities. J Biol Chem 277: 17696–17705. [DOI] [PubMed] [Google Scholar]
- Calandria, C. , Irurzun, A. , Barco, A. , Carrasco, L. (2004) Individual expression of poliovirus 2Apro and 3Cpro induces activation of caspase‐3 and PARP cleavage in HeLa cells. Virus Res 104: 39–49. [DOI] [PubMed] [Google Scholar]
- Campanella, M. , De Jong, A.S. , Lanke, K.W. , Melchers, W.J. , Willems, P.H. , Pinton, P. , et al. (2004) The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J Biol Chem 279: 18440–18450. [DOI] [PubMed] [Google Scholar]
- Cans, C. , Passer, B.J. , Shalak, V. , Nancy‐Portebois, V. , Crible, V. , Amzallag, N. , et al. (2003) Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc Natl Acad Sci USA 100: 13892–13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthy, C.M. , Granville, D.J. , Watson, K.A. , Anderson, D.R. , Wilson, J.E. , Yang, D. , et al. (1998) Caspase activation and specific cleavage of substrates after coxsackievirus B3‐induced cytopathic effect in HeLa cells. J Virol 72: 7669–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Ray, R. , Dubik, D. , Shi, L. , Cizeau, J. , Bleackley, R.C. , et al. (1997) The E1B 19K/Bcl‐2‐binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J Exp Med 186: 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Bernstein, B.W. , Bamburg, J.R. (2000) Regulating actin‐filament dynamics in vivo . Trends Biochem Sci 25: 19–23. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Turner, J. , McCarthy, S. , Scaltriti, M. , Bettuzzi, S. , Yeatman, T.J. (2004) Clusterin‐mediated apoptosis is regulated by adenomatous polyposis coli and is p21 dependent but p53 independent. Cancer Res 64: 7412–7419. [DOI] [PubMed] [Google Scholar]
- Cheon, M.S. , Fountoulakis, M. , Cairns, N.J. , Dierssen, M. , Herkner, K. , Lubec, G. (2001) Decreased protein levels of stathmin in adult brains with Down syndrome and Alzheimer's disease. J Neural Transm Suppl 61: 281–288. [DOI] [PubMed] [Google Scholar]
- Chow, V.T. , Quek, H.H. , Tock, E.P. (1993) Alternative splicing of the p53 tumor suppressor gene in the Molt‐4 T‐lymphoblastic leukemia cell line. Cancer Lett 73: 141–148. [DOI] [PubMed] [Google Scholar]
- Chua, B.T. , Volbracht, C. , Tan, K.O. , Li, R. , Yu, V.C. , Li, P. (2003) Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol 5: 1083–1089. [DOI] [PubMed] [Google Scholar]
- Clem, R.J. , Miller, L.K. (1993) Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J Virol 67: 3730–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, S. , De Fromentel, C.C. , Hainaut, P. (2004) p53 protein variants: structural and functional similarities with p63 and p73 isoforms. Oncogene 23: 631–638. [DOI] [PubMed] [Google Scholar]
- Coux, O. , Tanaka, K. , Goldberg, A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65: 801–847. [DOI] [PubMed] [Google Scholar]
- Deitz, S.B. , Dodd, D.A. , Cooper, S. , Parham, P. , Kirkegaard, K. (2000) MHC I‐dependent antigen presentation is inhibited by poliovirus protein 3A. Proc Natl Acad Sci USA 97: 13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher, S. , Martinou, J.C. (2000) Mitochondria as the central control point of apoptosis. Trends Cell Biol 10: 369–377. [DOI] [PubMed] [Google Scholar]
- Douche‐Aourik, F. , Berlier, W. , Feasson, L. , Bourlet, T. , Harrath, R. , Omar, S. , et al. (2003) Detection of enterovirus in human skeletal muscle from patients with chronic inflammatory muscle disease or fibromyalgia and healthy subjects. J Med Virol 71: 540–547. [DOI] [PubMed] [Google Scholar]
- Duguid, J.R. , Bohmont, C.W. , Liu, N.G. , Tourtellotte, W.W. (1989) Changes in brain gene expression shared by scrapie and Alzheimer disease. Proc Natl Acad Sci USA 86: 7260–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everly, D.N. , Feng, P. , Mian, I.S. , Read, G.S. (2002) mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J Virol 76: 8560–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden, G.A. , Guo, W. , Jayadev, S. , Tun, C. , Balcaitis, S. , Choi, J. , et al. (2004) HIV associated neurodegeneration requires p53 in neurons and microglia. FASEB J 18: 1141–1143. [DOI] [PubMed] [Google Scholar]
- Garfinkel, M.S. , Katze, M.G. (1992) Translational control by influenza virus. Selective and cap‐dependent translation of viral mRNAs in infected cells. J Biol Chem 267: 9383–9390. [PubMed] [Google Scholar]
- Garver, W.S. , Heidenreich, R.A. (2002) The Niemann‐Pick C proteins and trafficking of cholesterol through the late endosomal/lysosomal system. Curr Mol Med 2: 485–505. [DOI] [PubMed] [Google Scholar]
- Gavet, O. , Ozon, S. , Manceau, V. , Lawler, S. , Curmi, P. , Sobel, A. (1998) The stathmin phosphoprotein family: intracellular localization and effects on the microtubule network. J Cell Sci 111: 3333–3346. [DOI] [PubMed] [Google Scholar]
- Ghosh, A.K. , Steele, R. , Ray, R.B. (1999) Functional domains of c‐myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol Cell Biol 19: 2880–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstaub, D. , Gradi, A. , Bercovitch, Z. , Grosmann, Z. , Nophar, Y. , Luria, S. , et al. (2000) Poliovirus 2A protease induces apoptotic cell death. Mol Cell Biol 20: 1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson, J.M. , Lanier, J.G. , Altman, J.D. , Ahmed, R. (2001) The role of p53 in regulating antiviral T cell responses. J Immunol 167: 1333–1337. [DOI] [PubMed] [Google Scholar]
- Huang, S.H. , Triche, T. , Jong, A.Y. (2002) Infectomics: genomics and proteomics of microbial infections. Funct Integr Genomics 1: 331–344. [DOI] [PubMed] [Google Scholar]
- Kao, C.F. , Chen, S.Y. , Chen, J.Y. , Wu‐Lee, Y.H. (2004) Modulation of p53 transcription regulatory activity and post‐translational modification by hepatitis C virus core protein. Oncogene 23: 2472–2483. [DOI] [PubMed] [Google Scholar]
- Keel, M. , Ungethum, U. , Steckholzer, U. , Niederer, E. , Hartung, T. , Trentz, O. , Ertel, W. (1997) Interleukin‐10 counterregulates proinflammatory cytokine‐induced inhibition of neutrophil apoptosis during severe sepsis. Blood 90: 3356–3363. [PubMed] [Google Scholar]
- Kehle, J. , Roth, B. , Metzger, C. , Pfitzner, A. , Enders, G. (2003) Molecular characterization of an Enterovirus 71 causing neurological disease in Germany. J Neurovirol 9: 126–128. [DOI] [PubMed] [Google Scholar]
- Klunemann, H.H. , Elleder, M. , Kaminski, W.E. , Snow, K. , Peyser, J.M. , O'Brien, J.F. , et al. (2002) Frontal lobe atrophy due to a mutation in the cholesterol binding protein HE1/NPC2. Ann Neurol 52: 743–749. [DOI] [PubMed] [Google Scholar]
- Koch, A. , Thiemann, M. , Grabenbauer, M. , Yoon, Y. , McNiven, M.A. , Schrader, M. (2003) Dynamin‐like protein 1 is involved in peroxisomal fission. J Biol Chem 278: 8597–8605. [DOI] [PubMed] [Google Scholar]
- Kok, T.W. , Pryor, T. , Payne, L. (1998) Comparison of rhabdomyosarcoma, buffalo green monkey kidney epithelial, A549 (human lung epithelial) cells and human embryonic lung fibroblasts for isolation of enteroviruses from clinical samples. J Clin Virol 11: 61–65. [DOI] [PubMed] [Google Scholar]
- Koyama, A.H. , Fukumori, T. , Fujita, M. , Irie, H. , Adachi, A. (2000) Physiological significance of apoptosis in animal virus infection. Microbes Infect 2: 1111–1117. [DOI] [PubMed] [Google Scholar]
- Kramer, M.F. , Cook, W.J. , Roth, F.P. , Zhu, J. , Holman, H. , Knipe, D.M. , Coen, D.M. (2003) Latent herpes simplex virus infection of sensory neurons alters neuronal gene expression. J Virol 77: 9533–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich, H.G. , Nicklin, M.J. , Toyoda, H. , Etchison, D. , Wimmer, E. (1987) Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol 61: 2711–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota, H. , Hynes, G. , Carne, A. , Ashworth, A. , Willison, K. (1994) Identification of six Tcp‐1‐related genes encoding divergent subunits of the TCP‐1‐containing chaperonin. Curr Biol 4: 89–99. [DOI] [PubMed] [Google Scholar]
- Kubota, H. , Hynes, G. , Willison, K. (1995) The eighth Cct gene, Cctq, encoding the theta subunit of the cytosolic chaperonin containing TCP‐1. Gene 154: 231–236. [DOI] [PubMed] [Google Scholar]
- Kumar, C.C. , Kim, J.H. , Bushel, P. , Armstrong, L. , Catino, J.J. (1995) Activation of smooth muscle alpha‐actin promoter in ras‐transformed cells by treatments with antimitotic agents: correlation with stimulation of SRF: SRE mediated gene transcription. J Biochem 118: 1285–1292. [DOI] [PubMed] [Google Scholar]
- Kuo, R.L. , Kung, S.H. , Hsu, Y.Y. , Liu, W.T. (2002) Infection with enterovirus 71 or expression of its 2A protease induces apoptotic cell death. J Gen Virol 83: 1367–1376. [DOI] [PubMed] [Google Scholar]
- Kuyumcu‐Martinez, N.M. , Van Eden, M.E. , Younan, P. , Lloyd, R.E. (2004) Cleavage of poly (A)‐binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, T.W. , Kim, D.K. , Ye, J.S. , Lee, W.J. , Moon, M.S. , Joo, C.H. , et al. (2004) Detection of enterovirus, cytomegalovirus, and Chlamydia pneumoniae in atheromas. J Microbiol 42: 299–304. [PubMed] [Google Scholar]
- Lainez, B. , Fernandez‐Real, J.M. , Romero, X. , Esplugues, E. , Canete, J.D. , Ricart, W. , Engel, P. (2004) Identification and characterization of a novel spliced variant that encodes human soluble tumor necrosis factor receptor 2. Int Immunol 16: 169–177. [DOI] [PubMed] [Google Scholar]
- Leong, P.W. , Liew, K. , Lim, W. , Chow, V.T. (2002) Differential display RT‐PCR analysis of enterovirus‐71‐infected rhabdomyosarcoma cells reveals mRNA expression responses of multiple human genes with known and novel functions. Virology 295: 147–159. [DOI] [PubMed] [Google Scholar]
- Leong, W.F. , Tan, H.C. , Ooi, E.E. , Koh, D.R. , Chow, V.T. (2005) Microarray and real‐time RT‐PCR analyses of differential human gene expression patterns induced by severe acute respiratory syndrome (SARS) coronavirus infection of Vero cells. Microbes Infect 7: 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.L. , Hsu, T.A. , Chen, T.C. , Chang, S.C. , Lee, J.C. , Chen, C.C. , et al. (2002) The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 293: 386–395. [DOI] [PubMed] [Google Scholar]
- Liew, K.J. , Chow, V.T. (2004) Differential display RT‐PCR analysis of ECV304 endothelial‐like cells infected with dengue virus type 2 reveals messenger RNA expression profiles of multiple human genes involved in known and novel roles. J Med Virol 72: 597–609. [DOI] [PubMed] [Google Scholar]
- Minamide, L.S. , Striegl, A.M. , Boyle, J.A. , Meberg, P.J. , Bamburg, J.R. (2000) Neurodegenerative stimuli induce persistent ADF/cofilin‐actin rods that disrupt distal neurite function. Nat Cell Biol 2: 628–636. [DOI] [PubMed] [Google Scholar]
- Miyake, H. , Hara, I. , Gleave, M.E. , Eto, H. (2004) Protection of androgen‐dependent human prostate cancer cells from oxidative stress‐induced DNA damage by overexpression of clusterin and its modulation by androgen. Prostate 61: 318–323. [DOI] [PubMed] [Google Scholar]
- Mogensen, J. , Klausen, I.C. , Pedersen, A.K. , Egeblad, H. , Bross, P. , Kruse, T.A. , et al. (1999) Alpha‐cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest 103: R39–R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshier, J.A. , Gilbert, J.D. , Skunca, M. , Dosescu, J. , Almodovar, K.M. , Luk, G.D. (1990) Isolation and expression of a human ornithine decarboxylase gene. J Biol Chem 265: 4884–4892. [PubMed] [Google Scholar]
- Nagaishi, K. , Adachi, R. , Kawanishi, T. , Yamaguchi, T. , Kasahara, T. , Hayakawa, T. , Suzuki, K. (1999) Participation of cofilin in opsonized zymosan‐triggered activation of neutrophil‐like HL‐60 cells through rapid dephosphorylation and translocation to plasma membranes. J Biochem 125: 891–898. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Zhu, H. , Morishima, N. , Li, E. , Xu, J. , Yankner, B.A. , Yuan, J. (2000) Caspase‐12 mediates endoplasmic‐reticulum‐specific apoptosis and cytotoxicity by amyloid‐beta. Nature 403: 98–103. [DOI] [PubMed] [Google Scholar]
- Nemoto, T. , Hori, H. , Yoshimoto, M. , Seyama, Y. , Kubota, S. (2002) Overexpression of ornithine decarboxylase enhances endothelial proliferation by suppressing endostatin expression. Blood 99: 1478–1481. [DOI] [PubMed] [Google Scholar]
- Novelli, G. , Gennarelli, M. , Zelano, G. , Sangiuolo, F. , Lo Cicero, S. , Samson, F. , Dallapiccola, B. (1992) Polymerase chain reaction in the detection of mRNA transcripts from the slow skeletal troponin T (TNNT1) gene in myotonic dystrophy and normal muscle. Cell Biochem Funct 10: 251–256. [DOI] [PubMed] [Google Scholar]
- Onodera, S. , Nishihira, J. , Koyama, Y. , Majima, T. , Aoki, Y. , Ichiyama, H. , et al. (2004) Macrophage migration inhibitory factor up‐regulates the expression of interleukin‐8 messenger RNA in synovial fibroblasts of rheumatoid arthritis patients: common transcriptional regulatory mechanism between interleukin‐8 and interleukin‐1beta. Arthritis Rheum 50: 1437–1447. [DOI] [PubMed] [Google Scholar]
- Pelletier, I. , Duncan, G. , Pavio, N. , Colbere‐Garapin, F. (1998) Molecular mechanisms of poliovirus persistence: key role of capsid determinants during the establishment phase. Cell Mol Life Sci 54: 1385–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi, A.I. , Giles, W.H. , Croft, J.B. , Guterman, L.R. , Hopkins, L.N. (2002) Apolipoproteins A‐1 and B and the likelihood of non‐fatal stroke and myocardial infarction – data from The Third National Health and Nutrition Examination Survey. Med Sci Monit 8: CR311–CR316. [PubMed] [Google Scholar]
- Radhakrishnan, S. , Otte, J. , Enam, S. , Del Valle, L. , Khalili, K. , Gordon, J. (2003) JC virus‐induced changes in cellular gene expression in primary human astrocytes. J Virol 77: 10638–10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane, M.J. , Pan, Y. , Singh, S. , Powell, D.W. , Wu, R. , Cummins, T. , et al. (2003) Heat shock protein 27 controls apoptosis by regulating Akt activation. J Biol Chem 278: 27828–27835. [DOI] [PubMed] [Google Scholar]
- Rangel, S.R. , Grief, C. , Da Silva, E.E. , De Filippis, A.M. , Taffarel, M. (1998) Ultrastructural and immunocytochemical study on the infection of enterovirus 71 (EV 71) in rhabdomyosarcoma (RD) cells. J Submicrosc Cytol Pathol 30: 71–75. [PubMed] [Google Scholar]
- Ray, R.B. (1995) Induction of cell death in murine fibroblasts by a c‐myc promoter binding protein. Cell Growth Differ 6: 1089–1096. [PubMed] [Google Scholar]
- Ray, R.B. , Steele, R. , Seftor, E. , Hendrix, M. (1995) Human breast carcinoma cells transfected with the gene encoding a c‐myc promoter‐binding protein (MBP‐1) inhibits tumors in nude mice. Cancer Res 55: 3747–3751. [PubMed] [Google Scholar]
- Relou, A.M. , Gorter, G. , Van Rijn, H.J. , Akkerman, J.W. (2002) Platelet activation by the apoB/E receptor‐binding domain of LDL. Thromb Haemost 87: 880–887. [PubMed] [Google Scholar]
- Romanova, L.I. , Belov, G.A. , Lidsky, P.V. , Tolskaya, E.A. , Kolesnikova, M.S. , Evstafieva, A.G. , et al. (2005) Variability in apoptotic response to poliovirus infection. Virology 331: 292–306. [DOI] [PubMed] [Google Scholar]
- Schmidt, N.J. , Lennette, E.H. , Ho, H.H. (1974) An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis 129: 304–309. [DOI] [PubMed] [Google Scholar]
- Sheth, K. , De, A. , Nolan, B. , Friel, J. , Duffy, A. , Ricciardi, R. , et al. (2001) Heat shock protein 27 inhibits apoptosis in human neutrophils. J Surg Res 99: 129–133. [DOI] [PubMed] [Google Scholar]
- Shih, S.R. , Stollar, V. , Lin, J.Y. , Chang, S.C. , Chen, G.W. , Li, M.L. (2004) Identification of genes involved in the host response to enterovirus 71 infection. J Neurovirol 10: 293–304. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Chow, V.T. , Phoon, M.C. , Chan, K.P. , Poh, C.L. (2002a) Direct detection of enterovirus 71 (EV71) in clinical specimens from a hand, foot, and mouth disease outbreak in Singapore by reverse transcription‐PCR with universal enterovirus and EV71‐specific primers. J Clin Microbiol 40: 2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Poh, C.L. , Chow, V.T. (2002b) Complete sequence analyses of enterovirus 71 strains from fatal and non‐fatal cases of the hand, foot and mouth disease outbreak in Singapore (2000). Microbiol Immunol 46: 801–808. [DOI] [PubMed] [Google Scholar]
- Sudo, K. , Takahashi, E. , Nakamura, Y. (1995) Isolation and mapping of the human EIF4A2 gene homologous to the murine protein synthesis initiation factor 4A‐II gene Eif4a2. Cytogenet Cell Genet 71: 385–388. [DOI] [PubMed] [Google Scholar]
- Sun, E.W. , Shi, Y.F. (2001) Apoptosis: the quiet death silences the immune system. Pharmacol Ther 92: 135–145. [DOI] [PubMed] [Google Scholar]
- Takemoto, M. , Mori, Y. , Ueda, K. , Kondo, K. , Yamanishi, K. (2004) Productive human herpesvirus 6 infection causes aberrant accumulation of p53 and prevents apoptosis. J Gen Virol 85: 869–879. [DOI] [PubMed] [Google Scholar]
- Trougakos, I.P. , Gonos, E.S. (2004) Functional analysis of clusterin/apolipoprotein J in cellular death induced by severe genotoxic stress. Ann N Y Acad Sci 1019: 206–210. [DOI] [PubMed] [Google Scholar]
- Van Acker, K. , Bultynck, G. , Rossi, D. , Sorrentino, V. , Boens, N. , Missiaen, L. , et al. (2004) The 12 kDa FK506‐binding protein, FKBP12, modulates the Ca (2+)‐flux properties of the type‐3 ryanodine receptor. J Cell Sci 117: 1129–1137. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Donahoe, P.K. , Zervos, A.S. (1994) Specific interaction of type I receptors of the TGF‐beta family with the immunophilin FKBP‐12. Science 265: 674–676. [DOI] [PubMed] [Google Scholar]
- Warke, R.V. , Xhaja, K. , Martin, K.J. , Fournier, M.F. , Shaw, S.K. , Brizuela, N. , et al. (2003) Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol 77: 11822–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed, H.G. , Krochmalnic, G. , Penman, S. (1985) Poliovirus metabolism and the cytoskeletal framework: detergent extraction and resinless section electron microscopy. J Virol 56: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman, M.K. , Sharma, R. , Raychaudhuri, S. , Kundu, P. , Tsai, W. , Dasgupta, A. (2003) The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res 95: 75–85. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H. , Dudoit, S. , Luu, P. , Lin, D.M. , Peng, V. , Ngai, J. , Speed, T.P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarm, F.R. (2002) Plk phosphorylation regulates the microtubule‐stabilizing protein TCTP. Mol Cell Biol 22: 6209–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]


