Abstract
Introduction
Viral infection is a significant cause of chronic obstructive pulmonary disease (COPD) and acute exacerbation of COPD. Retinoic acid inducible gene I (RIG‐I)‐like receptors (RLRs), including RIG‐I and melanoma differentiation associated gene 5 (MDA‐5), are important pattern recognition receptors for viral elimination.
Objective
The study aims to investigate the role of RIG‐I and MDA‐5 in COPD pathogenesis.
Methods
We examined the expression of RIG‐I and MDA‐5 by immunohistochemistry, real‐time PCR and Western blots in COPD patients and control subjects.
Results
Our results showed that MDA‐5 expression was upregulated in lung tissues and peripheral blood mononuclear cells of COPD patients and there was a negative correlation between MDA‐5 mRNA levels and forced expiratory volume in 1 s %pred. COPD patients had higher interleukin (IL)‐1 and IL‐8 mRNA expression levels, and these inflammatory cytokines positively correlate with MDA‐5 levels. However, there was no difference in the expression of RIG‐I between COPD patients and control subjects.
Conclusion
Our results suggested that MDA‐5, but not RIG‐I, may play a critical role in airway inflammation in COPD.
Keywords: airway inflammation, nic obstructive pulmonary disease, RIG‐I‐like receptors, RIG‐I, MDA‐5
Introduction
Chronic obstructive pulmonary disease (COPD) is a disease characterized by airflow limitation and is a progressive disease that is not fully reversible 1. The World Health Organization has predicted that COPD will be the third leading cause of death worldwide in 2030 2, 3. Tissues from patients with COPD are characterized by chronic inflammation, mucus metaplasia, alveolar destruction and structural cell apoptosis 4. Inflammation is an important factor in the development of COPD 5. Inflammation in patients may be causally related to emphysema or other pathological alterations in the lungs and worsens with disease progression 6, 7. Pulmonary inflammation might have an adverse impact on various extrapulmonary organs, such as blood vessels and the heart 8, 9, 10. During the progression of pulmonary inflammation in COPD, both the innate immune and adaptive immune systems are involved. However, their specific mechanisms are still elusive. This is particularly true for the innate immune system, whose role is unknown during the inflammation response in COPD.
Innate immunity is the first line of defense against foreign pathogens. Its role in the airways involves the detection of pathogen‐ or damage‐associated molecular patterns (PAMPs or DAMPs) by pattern recognition receptors (PRRs) including Toll‐like receptors (TLRs) and retinoic acid inducible gene I (RIG‐I)‐like receptors (RLRs) 11. Latent infection of adenoviruses in childhood can promote chronic inflammation and thus gradually lead to COPD 12. Recent studies have demonstrated that a variety of viruses can cause exacerbation of the disease 13, 14. Compared with exacerbation due to other causes, viral‐induced COPD exacerbation is more severe, lasts longer and is associated with airway and systemic inflammatory responses 15, 16. RLRs are newly discovered pattern recognition receptors, which play a critical role in the elimination of viral infections 17, 18, 19, 20, 21, 22. RLRs include RIG‐I, melanoma differentiation associated gene 5 (MDA‐5) and laboratory of genetics and physiology 2 (LGP2) 23, 24, 25. These RLRs localize within the cytoplasm. When viral RNA binds to the C‐terminal regulatory domain of RIG‐I or MDA‐5, it can initiate a signaling cascade leading to activation and nuclear translocation of the transcription factors NF‐κB and IRF‐3, which are needed to turn on transcription of interferons (IFNs) 26, 27, 28. The LGP2 lacks a caspase activation and recruitment domain and incapable of interferon production 29.
Once triggered, RIG‐I and MDA‐5 activate intracellular signaling pathways that culminate in the induction of antiviral cytokines and pro‐inflammatory mediators. This raises the pivotal question of whether RIG‐I and MDA‐5 are involved in COPD pathogenesis. In this study, we assessed the expression of RIG‐I and MDA‐5 in peripheral blood mononuclear cells (PBMCs) and bronchial tissue of COPD patients. We demonstrate that COPD patients have significantly different expression levels of RLRs and inflammatory cytokines, thus may define a critical role for RLRs in COPD airway inflammation.
Materials and methods
Study subjects
Peripheral blood of 29 COPD patients and 24 age‐matched health control subjects were respectively obtained from outpatients and a physical examination center of Ruijin Hospital, affiliated with Shanghai Jiaotong University, China. Endobronchial biopsies (8 COPD patients and 12 non‐COPD control subjects) were recruited from the ward of respiratory medicine, Ruijin Hospital. Bronchoalveolar lavage (BAL) in 25 COPD patients and 15 non‐COPD control subjects was performed by infusing 50 mL of sterile sodium chloride in either the middle or lingula lobe. The diagnosis of COPD was made according to the guidelines of the Global Initiative for Chronic Obstructive Lung Disease. All subjects were in a stable condition at the time of the study, and all control subjects have normal lung function. Exclusion criteria included a current or recent (past month) respiratory tract infection, exacerbation of respiratory disease or a course of oral steroids or antibiotics in the previous month. Participant details are shown in Table 1. This study was reviewed and approved by the Ethics Committees of Ruijin Hospital, and written informed consent was obtained from all subjects.
Table 1.
Characteristics of COPD patients and control subjects
| Control | COPD | |
|---|---|---|
| Endobronchial biopsies | ||
| Age (year) | 57.00 ± 3.22 | 59.75 ± 7.42 |
| Sex (M : F) | 8:4 | 6:2 |
| FEV1% | 82.47 ± 5.73 | 62.31 ± 6.60 |
| FEV1/FVC (%) | 82.30 ± 2.61 | 67.18 ± 5.44 |
| GOLD stage (I/II/III/IV) | 0/0/0/0 | 1/4/3/0 |
| BALF | ||
| Age (year) | 56.32 ± 6.02 | 60.00 ± 7.38 |
| Sex (M : F) | 9:6 | 15:10 |
| FEV1% | 81.30 ± 5.17 | 65.37 ± 4.75 |
| FEV1/FVC (%) | 84.24 ± 2.70 | 62.30 ± 4.67 |
| GOLD stage (I/II/III/IV) | 0/0/0/0 | 8/6/9/2 |
| Blood | ||
| Age (year) | 56.00 ± 6.22 | 54.00 ± 6.01 |
| Sex (M : F) | 13:11 | 17:12 |
| FEV1% | 84.27 ± 5.63 | 68.36 ± 2.11 |
| FEV1/ FVC (%) | 88.30 ± 2.74 | 69.30 ± 2.89 |
| GOLD stage (I/II/III/IV) | 0/0/0/0 | 6/12/9/2 |
Data are represented as mean ± standard error.
BALF, bronchoalveolar lavage fluid; COPD, chronic obstructive pulmonary disease; FEV1: Forced expiratory volume in 1 s, FVC: Forced vital capacity; GOLD, The Global Initiative for Chronic Obstructive Lung Disease.
Immunohistochemical analysis
Immunohistochemistry was used to analyze the protein levels of RIG‐I and MDA‐5 in bronchial tissues of COPD and control patients. Forty biopsies were obtained from 20 subjects, and 40 specimens were used for immunohistochemistry analysis. Briefly, lung sections (endobronchial biopsies) were embedded in paraffin and sectioned to 5 μm, followed by dewaxing in xylene and rehydration. Endogenous peroxidases were inhibited with 0.5% hydrogen peroxide in methanol for 10 min, then slides were incubated overnight at 4°C with a rabbit polyclonal immunoglobulin G (IgG) antibody against RIG‐I or MDA‐5. Immunodetection was performed with biotinylated goat anti‐rabbit IgG reagent, horseradish peroxidase (HRP) reagent and diaminobenzidine.
Isolation of PBMCs
Blood samples were collected in heparinized, 10‐mL evacuated test tube (BD Vacutainer system, Plymouth, UK) from all the subjects. PBMCs were separated from the peripheral blood using Ficoll‐Paque Plus (Sigma, Shanghai, China) for 30 min at 1800 rpm. The band containing PBMCs was collected and transferred to new tubes. Phosphate buffered saline (PBS) was added to the PBMCs and the PBMCs were washed three times by centrifugation for 10 min at 1000 rpm. In COPD patients, the cell count is higher but the percentage of monocytes, lymphocytes, neutrophils of PBMCs is the same as that of control group.
RNA extraction and real‐time PCR
Total RNA from PBMCs and bronchial tissue was extracted using an RNeasy kit (Qiagen, Hilden, Germany), and the quality and quantity of the obtained RNA was determined by spectrophotometry based on the absorbance A260/A280. One microgram of total RNA for each sample was used with oligo(dT) as primers for the synthesis of cDNA (RevertAidTM First Strand cDNA Synthesis Kit, Fermentas, Burlington, ON, Canada). Real‐time PCR was performed with a 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) with FastStart Universal SYBR Green (Roche, Basel, Switzerland) after cDNA synthesis. PCR conditions were as follows: initial denaturation at 95°C (10 min), followed by 40 cycles of amplification 95°C (15 s), 60°C (60 s). The standard curve for quantification was made using a modified procedure as previously described 30. The sequences of all primers are shown in Table 2. Fold change of the gene expression were calculated 2‐ΔΔCt relative to the internal reference gene [glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH)] and the mean of all sample.
Table 2.
Primer sequences for real‐time PCR
| Gene of interest | Primer sequence (5′ to 3′) |
|---|---|
| RIG‐I |
Forward primer: GGCAAGTCCCGCTGTAAAC Reverse Primer: TTGGTATCTCCTAATCGCAAAAG |
| MDA‐5 |
Forward primer: TGGACAAGCTTCTAGTTAGAGACG Reverse Primer: GATTCATTTCCATTGTTTTCTGC |
| IFN‐α |
Forward primer: CCATAGCCTGGATAACCGTCG Reverse Primer: TCTTGGGGAAAGCCAAAGTCA |
| IL‐1β |
Forward primer: TACAGTGGCAATGAGGAT Reverse Primer: ATGAAGGGAAAGAAGGTG |
| IL‐8 |
Forward primer: ATGACTTCCAAGCTGGCCGTGGCT Reverse Primer: TCTCA GCCCTCTTCAAAAACTTCTC |
| GAPDH |
Forward primer: GAAGGTGAAGGTCGGAGTC Reverse Primer: GAAGATGGTGATGGGATTTC |
IFN‐α, interferon alpha; IL, interleukin; GADPH, glyceraldehyde‐3‐phosphate dehydrogenase; MDA‐5, melanoma differentiation associated gene 5; RIG‐I, retinoic acid inducible gene I.
Western blots
PBMCs isolated from peripheral blood were lysed in 100‐μL lysis buffer (150 mM NaCl, 50 mM Tris‐Cl pH 7.4, 1 mM ethylene diamine tetraacetic acid, 0.1% sodium dodecyl sulfate, 1.0% deoxycholatic, 1.0% Tritonx‐100, 1 mM phenylmethanesulfonyl fluoride). Protein concentrations were quantified using the BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) according to the manufacturer's instructions. Equal amounts of proteins (30 μg) were resolved by sodium dodecyl sulfate‐polyacrylamide gelelectrophoresis and transferred onto a poly(vinylidene fluoride) membrane. The membrane was blocked with PBS containing 5% nonfat milk for 2 h at room temperature, then incubated overnight at 4°C with an anti‐RIG‐I antibody (1:500, Cell Signaling, Beverly, MA, USA), an anti‐MDA5 antibody (1.5 μg/mL, Abcam, Cambridgeshire, UK), or a β‐actin antibody (1:1000, Cell Signaling). The membrane was then washed three times for 5 min with 15‐mL Tris‐buffered saline Tween‐20, and incubated with HRP‐conjugated goat anti‐rabbit IgG antibody (Sigma, 1:10000 dilution) for 40 min at room temperature. After washing, 1‐mL chemiluminescent substrate (Thermo Scientific) was added to the membrane. The signal was detected and quantified with an enhanced chemiluminescence system (ImageQuant LAS‐4000 MINI, GE, Fairfield, CT, USA). All samples were normalized to β‐actin. Data were represented as mean ± standard error (SEM).
Cytokine enzyme‐linked immunosorbent assays (ELISAs)
IL‐8 and tumor necrosis factor (TNF)‐α concentrations in the bronchoalveolar lavage fluid (BALF) were determined using DuoSet ELISA kits (R&D Systems, Minneapolis, MN, USA), according to the manufacturer's instructions.
Statistical analysis
Results are presented as mean ± SEM. A two‐tailed t‐test was performed with each group. Associations between variables were assessed using Spearman's rank correlation test. Statistical analyses were performed using SPSS 18.0 and P values ≤0.05 were considered statistically significant.
Results
Immunohistochemistry analysis of MDA‐5 and RIG‐I in bronchial mucosa of COPD patients
MDA‐5 and RIG‐I were mainly present in the airway epithelium cells (Fig. 1). COPD patients had a higher expression level of MDA‐5 in bronchial mucosa compared with control subjects (Fig. 1A and B). No significant differences in RIG‐I was observed between the two groups (Fig. 1C and D).
Figure 1.

Immunohistochemistry analysis of the expression of melanoma differentiation associated gene 5 (MDA‐5) and retinoic acid inducible gene I (RIG‐I) in bronchial mucosa. Representative images of immunohistochemistry in bronchial biopsy tissue for MDA‐5 in a chronic obstructive pulmonary disease (COPD) subject (A) compared with a control subject (B), and for RIG‐I in a COPD subject (C) and a control subject (D). Arrows indicate positive staining in the epithelium cells. Magnification: 400×.
MDA‐5 and RIG‐I mRNA expression in the bronchial mucosa of COPD patients
Total RNA was isolated from bronchial biopsies samples of 12 non‐COPD and 8 COPD subjects, and then were analyzed for MDA‐5 and RIG‐I mRNA expression by real‐time PCR (Fig. 2). Fold change in RIG‐I mRNA levels between COPD patients and control subjects was 0.767 (COPD/control), and showed no significant difference (P = 0.7871) (Fig. 2B). The ratio of MDA‐5 expression to the housekeeping gene GAPDH was significantly higher in COPD subjects than in control subjects (COPD/control fold change = 2.427, P = 0.0023) (Fig. 2A). These results were consistent with immunohistochemistry results illustrated in Fig. 1.
Figure 2.

mRNA transcript levels of melanoma differentiation associated gene 5 (MDA‐5) and retinoic acid inducible gene I (RIG‐I) in bronchial mucosa. Total RNA was isolated from bronchial biopsies samples and followed by reverse transcription and real‐time polymerase chain reaction. Results are expressed as fold change over housekeeping genes. Chronic obstructive pulmonary disease (COPD) patients showed significant high level of MDA‐5 mRNA compared with control sample (A). In contrast, RIG‐I mRNA level did not show significant difference between two groups (B). Data represent mean ± standard error (n = 8–12; **P values < 0.01 vs the control group).
Expression of MDA‐5 and RIG‐I in PBMCs
Detected by real‐time PCR, MDA‐5 mRNA levels were significantly higher in COPD subjects than in control subjects (COPD/control fold change = 3.690, P = 0.0051) (Fig. 3A). Similar results were obtained with MDA‐5 protein levels, assessed by Western blotting (Fig. 3C). MDA‐5 protein levels in PBMCs were threefold higher in COPD subjects than control subjects (Fig. 3E). Whereas the RIG‐I mRNA level was almost identical in both groups (COPD/control fold change = 1.650, P = 0.0759) (Fig. 3B). Similar results were obtained with the RIG‐I protein level in PBMCs (COPD/control fold change = 0.977, P = 0.4726) (Fig. 3D and F).
Figure 3.
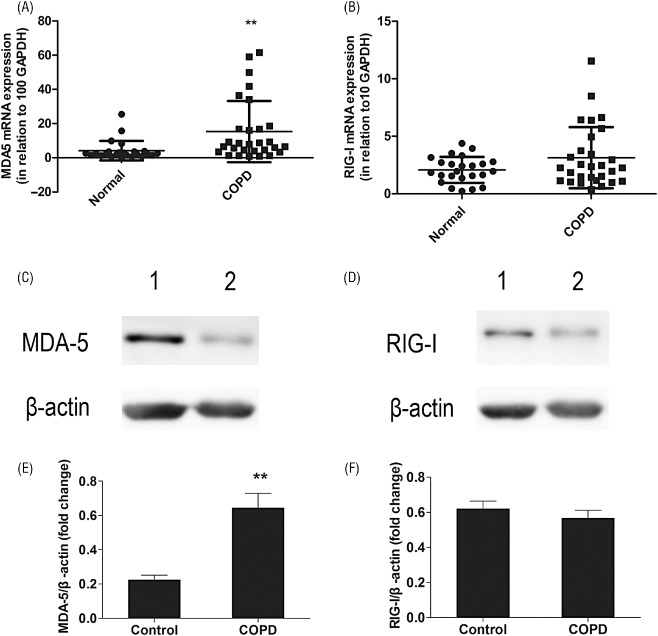
Expression of mRNA and protein of melanoma differentiation associated gene 5 (MDA‐5) and retinoic acid inducible gene I (RIG‐I) in peripheral blood mononuclear cells (PBMCs). Using real‐time polymerase chain reaction, the expressions of MDA‐5 and RIG‐I were evaluated in PBMCs of chronic obstructive pulmonary disease (COPD) patients and control subjects. Results are expressed as fold change over housekeeping genes. COPD patients showed significant high level of MDA‐5 mRNA compared with control sample (A). In contrast, RIG‐I mRNA level did not show significant difference between two groups (B). Data represent mean ± standard error (SEM) (n = 24–29; **P values < 0.01 vs the control group). Representative western blot results of MDA‐5 and RIG‐I protein levels in PBMCs of COPD patients and control subjects (C, D). Western blots analysis revealed an increased level of MDA‐5 protein expression in COPD patients (C, E). RIG‐I protein level did not show significant difference between two groups (D, F). Quantification of the Western blots was expressed as a relative ratio of individual samples normalized to β‐actin. Data represent mean ± SEM (n = 6–7; **P values < 0.01 vs the control group).
Cytokine expression in PBMCs and BALF
Compared with control subjects, COPD patients showed significantly higher mRNA expression levels of IL‐1β, IL‐8 and IFN‐α in peripheral blood (Fig. 4 A–C).
Figure 4.

Inflammatory cytokine expression levels in peripheral blood mononuclear cells (PBMCs) and bronchoalveolar lavage fluid (BALF). RNA isolated from PBMCs of chronic obstructive pulmonary disease (COPD) patients and control subjects was subjected to real‐time polymerase chain reaction, and the results were expressed as fold change over housekeeping genes. Compared with control group, COPD patients showed significant high level of interleukin (IL)‐1β (A), IL‐8 (B) and interferon (IFN)‐α (C). Data are represented as mean ± standard error (SEM) (n = 24–29; **P values < 0.01 vs the control group). BALF were assayed by enzyme‐linked immunosorbent assay, and COPD patients showed significant high level of IL‐8 (D) and tumor necrosis factor‐α (E) compared with control group. Data are represented as mean ± SEM (n = 15–25; **P values < 0.01 vs the control group, *P values < 0.05 vs the control group).
The levels of IL‐8 and TNF‐α, assessed by ELISA, were also higher in BALF of COPD patients than control subjects (Fig. 4D and E). These results demonstrate a persistent inflammation in the system and airway of COPD patients, consistent with previous reports 31.
Relationships between RLRs mRNA expression level, cytokine expression level and clinical outcomes
In this study we found a negative correlation between MDA‐5 mRNA level of PBMCs and forced expiratory volume in 1 s (FEV1) %pred in COPD patients (r = −0.6339, P = 0.0036; Fig. 5A). However, there was no significant correlation between RIG‐I mRNA level of PBMCs and FEV1 %pred (r = −0.0702, P = 0.7751; Fig. 5B). There was no significant correlation between the mRNA levels of MDA‐5 in bronchial mucosa and FEV1 %pred (r = −0.2619, P = 0.5364; data not shown), nor between mRNA levels of RIG‐I in bronchial mucosa and FEV1 %pred (r = −0.3264, P = 0.4302; data not shown). Furthermore, PBMCs IL‐1β and IL‐8 mRNA levels positively correlated with MDA‐5 mRNA levels in PBMCs of COPD patients (r = 0.7691, P = 0.0001; r = 0.4815, P = 0.0316, respectively, Fig. 5C and D).
Figure 5.
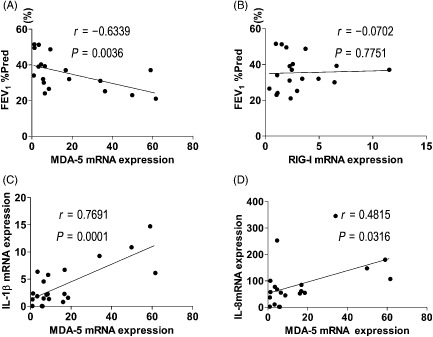
Correlations between retinoic acid inducible gene I‐like receptors mRNA levels, cytokine expression level and forced expiratory volume in 1 s (FEV 1) %pred. There were significant correlations between melanoma differentiation associated gene 5 (MDA‐5) mRNA level of peripheral blood mononuclear cells (PBMCs) and FEV 1 %pred in chronic obstructive pulmonary disease (COPD) patients (A). However, there was no significant correlation between RIG‐I mRNA level of PBMCs and FEV 1 %pred (B). Correlation between PBMCs IL‐1β mRNA levels and MDA‐5 mRNA levels in PBMCs of COPD patients was significant (C), and correlation between PBMCs IL‐8 mRNA levels and MDA‐5 mRNA levels in PBMCs of COPD patients was also significant (D).
Discussion
Viral infection is a significant cause of chronic obstructive pulmonary disease (COPD) and acute exacerbation of COPD 13, 32, 33. Compared with exacerbation due to other causes, viral‐induced COPD exacerbation is more severe, lasts longer and is associated with airway and systemic inflammatory responses 15, 16. Structural cells, especially epithelial cells, endothelium and fibroblasts, take part in the airway inflammation. Patients with COPD are more susceptible to viral infections, and up to one half of cases of COPD exacerbations are associated with viral infections 16, 34. An epidemiology survey illustrates that viral infection is the main cause of COPD exacerbation, and the top four causes are rhinovirus (RV), coronavirus, influenza virus and respiratory syncytial virus 13.
RLRs are essential for virus recognition in the cytoplasm. Both RIG‐I and MDA‐5 detect dsRNA in the cytoplasm, where they are linked to downstream‐signaling molecules via MAVS 35. Previous studies have shown that human RIG‐I and MDA‐5 are implicated in the recognition of different groups of RNA viruses, and subsequently activate the NF‐κB‐ and IRF3/7‐signaling pathways, resulting in production of pro‐inflammatory cytokines and type‐I interferons 36.
It has been shown that MDA5 and RIG‐I recognize widely distinct groups of viruses. RIG‐I preferentially detects variety of positive‐ and negative‐strand viruses through recognition of a 5′ triphosphate group of genomic RNAs such as influenza, paramyxovirus and flavivirus 37. In contrast, MDA5 recognizes long dsRNAs through detection of dsRNA replication intermediates and genomic dsRNAs, and likely detects positive‐strand and dsRNA viruses including RV and reoviruses 38, 39, 40. An epidemiological survey has illustrated that RV, a positive and single‐stranded RNA virus, is responsible for the majority of virus‐induced COPD exacerbation 13, 34. In COPD patients, RV infection causes a rapid decline in lung function. Recent studies by Wang and colleagues provide evidence that MDA‐5, but not RIG‐I, is responsible for recognizing RV, and subsequently activates the signaling pathways, causing an exaggerated inflammatory response in COPD patients, with increased levels of IL‐8, IL‐6, CXCL1, TNF‐α, IL‐1β and monocyte chemotactic protein 1 41. Baines and colleagues found that compared with healthy primary bronchial epithelial (pBECs), pBECs from COPD patients express large amounts of antiviral and pro‐inflammatory agents and were highly susceptible to apoptosis when responded to RV infection 22.
In this study, we found that compared with control subjects, the basal levels of TNF‐α, IL‐1β and IL‐8 in patients with COPD were elevated in the BALF or PBMCs. These results are consistent with previously reported increases in systemic and airway cytokine in COPD patients 42. However, we show for the first time that the basal level of MDA‐5 is elevated in the bronchial mucosa and PBMCs from patients with COPD. Our data implies that the exaggerated inflammatory response to RV infection in COPD patients may be associated with the high level of MDA‐5 in airway epithelial cells. Excess MDA‐5 in COPD patients activated by RV could lead to induction of a ‘cytokine flood’. This ‘cytokine flood’ is not only effective in facilitating viral clearance, but also may further contribute to airway inflammation.
Furthermore, we found that in COPD patients, there were strong correlations among MDA‐5 mRNA levels, pro‐inflammatory cytokine mRNA levels and airflow obstruction severity in PBMCs. These correlations indicate that MDA‐5 may be an important receptor that influences disease outcome or are the result of permanent stimulation of high levels of pro‐inflammatory cytokines. There was no significant correlation between the mRNA levels of MDA‐5 in bronchial biopsy and FEV1 %pred, which may have been due to the small number of bronchial biopsies samples in this study or the particular cell types present. As we all know, the major cell type in PBMCs are lymphocytes and monocytes, while the major cell type in bronchial biopsies samples is epithelial cells. As the lymphocytes and monocytes are a critical component in the immune system to fight infection and adapt to intruders, the MDA‐5 produced in these cells can induce higher inflammatory response than in epithelial cells, which may lead to different correlation between bronchial biopsy and PBMCs.
On the other hand, there were no changes in both mRNA and protein levels of RIG‐I in COPD patients. The different outcome of the basal level of MDA‐5 and RIG‐I may due to the divergent reorganization mechanism and feedback regulation of these two receptors.
Recently, Schneider and colleagues reported that the mRNA levels of MDA‐5 and RIG‐I are enhanced in the airway epithelial cells of COPD patients, and it is RIG‐I rather than MDA‐5 that has statistical significant 43. These findings were inconsistent with our results. The heterogeneity may be due to the differences in genetic background, different exposure to the viruses, cigarette smoke and other environmental agents.
In summary, our findings indicate that MDA‐5 is upregulated in bronchial tissues and PBMCs of COPD patients, and that this upregulation may participate in the high expression levels of inflammatory cytokines. Importantly, MDA‐5 expression levels negatively correlate with FEV1 %pred, which indicated that MDA‐5 may play a critical role in the disease progression. Thus, it also identifies a new target, against which therapies can be developed to modulate the severity of viral‐induced responses in clinical settings. Understanding the molecular mechanisms underlying these processes will open novel avenues for the treatment of COPD.
Acknowledgements
We would like to acknowledge the invaluable contribution of Dr. Yi Tao and Dr. AijiaoYe in Health Examination Center, Ruijin Hospital. This work was supported in part by the National Natural Science Foundation of China (Grant Numbers 30970125 and 81200027), the Natural Science Foundation of Shanghai Program (Grant Number 13ZR1423000) and Scientific Research Foundation for Returned Scholars, Ministry of Education of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship and contributorship
Qiurui Zhang performed research, participated in the collection of data and wrote the paper. Huanying Wan and Shaoguang Huang participated in the design of the study and reviewed the manuscript. Yan Zhang participated in the collection of data. Yanchun Wang performed the statistical analyses. Xiaokui Guo conceived of the study, participated in the design of the study and revised the manuscript. Ping He and Min Zhou participated in the design of the study, helped write the paper and revised the manuscript. All authors read and approved the final manuscript.
Ethics
This study has been performed in accordance with the ethical standards laid down in the Declaration of Helsinki. All patients provided written informed consent prior to their inclusion in the study.
Conflict of interest
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
References
- 1. Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS, GOLD Scientific Committee . Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163: 1256–1276. [DOI] [PubMed] [Google Scholar]
- 2. Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176: 532–555. [DOI] [PubMed] [Google Scholar]
- 3. Diaz‐Guzman E, Mannino DM. Epidemiology and prevalence of chronic obstructive pulmonary disease. Clin Chest Med. 2014;35: 7–16. [DOI] [PubMed] [Google Scholar]
- 4. Yokohori N, Aoshiba K, Nagai A, Respiratory Failure Research Group in Japan . Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest. 2004;125: 626–632. [DOI] [PubMed] [Google Scholar]
- 5. Sethi S, Mahler DA, Marcus P, Owen CA, Yawn B, Rennard S. Inflammation in COPD: implications for management. Am J Med. 2012;125: 1162–1170. [DOI] [PubMed] [Google Scholar]
- 6. Sohal SS, Ward C, Danial W, Wood‐Baker R, Walters EH. Recent advances in understanding inflammation and remodeling in the airways in chronic obstructive pulmonary disease. Expert Rev Respir Med. 2013;7: 275–288. [DOI] [PubMed] [Google Scholar]
- 7. Hogg JC, Chu F, Utokaparch S, et al. The nature of small‐airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350: 2645–2653. [DOI] [PubMed] [Google Scholar]
- 8. Pinto‐Plata VM, Livnat G, Girish M, et al. Systemic cytokines, clinical and physiological changes in patients hospitalized for exacerbation of COPD. Chest. 2007;131: 37–43. [DOI] [PubMed] [Google Scholar]
- 9. Agusti AG, Noguera A, Sauleda J, Sala E, Pons J, Busquets X. Systemic effects of chronic obstructive pulmonary disease. Eur Respir J. 2003;21: 347–360. [DOI] [PubMed] [Google Scholar]
- 10. Eid AA, Ionescu AA, Nixon LS, et al. Inflammatory response and body composition in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;164: 1414–1418. [DOI] [PubMed] [Google Scholar]
- 11. Gerlier D, Lyles DS. Interplay between innate immunity and negative‐strand RNA viruses: towards a rational model. Microbiol Mol Biol Rev. 2011;75: 468–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morimoto K, Gosselink J, Kartono A, Hogg JC, Hayashi S, Ogawa E. Adenovirus E1A regulates lung epithelial ICAM‐1 expression by interacting with transcriptional regulators at its promoter. Am J Physiol Lung Cell Mol Physiol. 2009;296: L361–371. [DOI] [PubMed] [Google Scholar]
- 13. Wark P, Tooze M, Powell H, Parsons K. Viral and bacterial infection in acute asthma & chronic obstructive pulmonary disease increases the risk of readmission. Respirology. 2013;18: 996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perotin JM, Dury S, Renois F, et al. Detection of multiple viral and bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a pilot prospective study. J Med Virol. 2013;85: 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sapey E, Stockley RA. COPD exacerbations · 2: aetiology. Thorax. 2006;61: 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seemungal T, Harper‐Owen R, Bhowmik A, et al. Respiratory viruses, symptoms, and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;164: 1618–1623. [DOI] [PubMed] [Google Scholar]
- 17. Fullam A, Schroder M. DExD/H‐box RNA helicases as mediators of anti‐viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta. 2013;1829: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu W, Patel KB, Booth JL, Zhang W, Metcalf JP. Cigarette smoke extract suppresses the RIG‐I‐initiated innate immune response to influenza virus in the human lung. Am J Physiol Lung Cell Mol Physiol. 2011;300: L821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Triantafilou K, Vakakis E, Richer EAJ, Evans GL, Villiers JP, Triantafilou M. Human rhinovirus recognition in non‐immune cells is mediated by Toll‐like receptors and MDA‐5, which trigger a synergetic pro‐inflammatory immune response. Virulence. 2011;2: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma B, Dela Cruz CS, Hartl D, et al. RIG‐like helicase innate immunity inhibits vascular endothelial growth factor tissue responses via a type I IFN‐dependent mechanism. Am J Respir Crit Care Med. 2011;183: 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wornle M, Sauter M, Kastenmuller K, et al. Role of toll‐like receptor 3, RIG‐I, and MDA5 in the expression of mesothelial IL‐8 induced by viral RNA. Appl Biochem Biotechnol. 2010;160: 1179–1187. [DOI] [PubMed] [Google Scholar]
- 22. Baines KJ, Hsu AC, Tooze M, Gunawardhana LP, Gibson PG, Wark PA. Novel immune genes associated with excessive inflammatory and antiviral responses to rhinovirus in COPD. Respir Res. 2013;14: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saito T, Hirai R, Loo YM, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG‐I and LGP2. PNAS. 2007;104: 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nat Immunol. 2004;5: 730–737. [DOI] [PubMed] [Google Scholar]
- 25. Andrejeva J, Childs KS, Young DF, et al. The V proteins of paramyxoviruses bind the IFN‐inducible RNA helicase, mda‐5, and inhibit its activation of the IFN‐beta promoter. PNAS. 2004;101: 17264–17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kawai T, Takahashi K, Sato S, et al. IPS‐1, an adaptor triggering RIG‐I‐ and Mda5‐mediated type I interferon induction. Nat Immunol. 2005;6: 981–988. [DOI] [PubMed] [Google Scholar]
- 27. Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus‐triggered IFN‐beta signaling. Mol Cell. 2005;19: 727–740. [DOI] [PubMed] [Google Scholar]
- 28. Fitzgerald KA, McWhirter SM, Faia KL, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4: 491–496. [DOI] [PubMed] [Google Scholar]
- 29. Satoh T, Kato H, Kumagai Y, et al. LGP2 is a positive regulator of RIG‐I‐ and MDA5‐mediated antiviral responses. PNAS. 2010;107: 1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y, Lou XL, Yang HL, et al. Establishment of a leptospirosis model in guinea pigs using an epicutaneous inoculations route. BMC Infect Dis. 2012;12: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chung KF. Inflammatory mediators in chronic obstructive pulmonary disease. Curr Drug Targets Inflamm Allergy. 2005;4: 619–625. [DOI] [PubMed] [Google Scholar]
- 32. Hogg JC. Childhood viral infection and the pathogenesis of asthma and chronic obstructive lung disease. Am J Respir Crit Care Med. 1999;160: S26–28. [DOI] [PubMed] [Google Scholar]
- 33. Svanes C, Sunyer J, Plana E, et al. Early life origins of chronic obstructive pulmonary disease. Thorax. 2010;65: 14–20. [DOI] [PubMed] [Google Scholar]
- 34. Hutchinson AF, Ghimire AK, Thompson MA, et al. A community‐based, time‐matched, case‐control study of respiratory viruses and exacerbations of COPD. Respir Med. 2007;101: 2472–2481. [DOI] [PubMed] [Google Scholar]
- 35. Yoneyama M, Fujita T. Function of RIG‐I‐like receptors in antiviral innate immunity. J Biol Chem. 2007;282: 15315–15318. [DOI] [PubMed] [Google Scholar]
- 36. Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14: 86–92. [DOI] [PubMed] [Google Scholar]
- 37. Cui S, Eisenacher K, Kirchhofer A, et al. The C‐terminal regulatory domain is the RNA 5′‐triphosphate sensor of RIG‐I. Mol Cell. 2008;29: 169–179. [DOI] [PubMed] [Google Scholar]
- 38. Peisley A, Lin C, Wu B, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. PNAS. 2011;108: 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loo YM, Fornek J, Crochet N, et al. Distinct RIG‐I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature. 2006;441: 101–105. [DOI] [PubMed] [Google Scholar]
- 41. Wang Q, Nagarkar DR, Bowman ER, et al. Role of double‐stranded RNA pattern recognition receptors in rhinovirus‐induced airway epithelial cell responses. J Immunol. 2009;183: 6989–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stankiewicz W, Dabrowski MP, Chcialowski A, Plusa T. Cellular and cytokine immunoregulation in patients with chronic obstructive pulmonary disease and bronchial asthma. Mediators Inflamm. 2002;11: 307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider D, Ganesan S, Comstock AT, et al. Increased cytokine response of rhinovirus‐infected airway epithelial cells in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182: 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]


