Abstract
Morpholino oligonucleotides are stable, uncharged, water‐soluble molecules that bind to complementary sequences of RNA, thereby inhibiting mRNA processing, read‐through, and protein binding at those sites. Morpholinos are typically used to inhibit translation of mRNA, splicing of pre‐mRNA, and maturation of miRNA, although they can also inhibit other interactions between biological macromolecules and RNA. Morpholinos are effective, specific, and lack non‐antisense effects. They work in any cell that transcribes and translates RNA. However, unmodified Morpholinos do not pass well through plasma membranes and must therefore be delivered into the nuclear or cytosolic compartment to be effective. Morpholinos form stable base pairs with complementary nucleic acid sequences but apparently do not bind to proteins to a significant extent. They are not recognized by proteins and do not undergo protein‐mediated catalysis; nor do they mediate RNA cleavage by RNase H or the RISC complex. This work focuses on techniques and background for using Morpholinos. Curr. Protoc. Mol. Biol. 83:26.8.1‐26.8.29. © 2008 by John Wiley & Sons, Inc.
Keywords: Morpholino, antisense, oligo, knockdown, splicing, zebrafish, Xenopus
Introduction
Morpholino oligos (Morpholinos) are synthetic uncharged P‐chiral analogs of nucleic acids. They are typically constructed by linking together 25 subunits, each bearing one of the four nucleic acid bases. Figure 1 illustrates the structure of three Morpholino subunits joined by inter‐subunit linkages. The morpholino phosphorodiamidate backbone of Morpholinos consists of morpholine rings that bear methylene groups and are bound through modified phosphates in which the anionic oxygen is replaced by a nonionic dimethylamino group. The substituted phosphate is bound through an oxygen atom to the morpholine's exocyclic methylene group, and through a phosphorus‐nitrogen bond to the nitrogen atom of another morpholine ring. One standard DNA nucleobase (adenine, guanine, cytosine, or thymine) is bound to each morpholine ring. The ends of Morpholinos are conventionally named 3′ and 5′ by analogy with the nomenclature for nucleic acids (although if one were to number the atoms of a morpholino oligonucleotide backbone by IUPAC rules, the numbers assigned to the ends would be different). The secondary amine of the morpholine ring at the end of an unmodified morpholino oligonucleotide is called the 3′ end of the oligo, whereas the 5′ end terminates with a chiral carboxamidated phosphorodiamidate group (Fig. 1).
Figure 1.
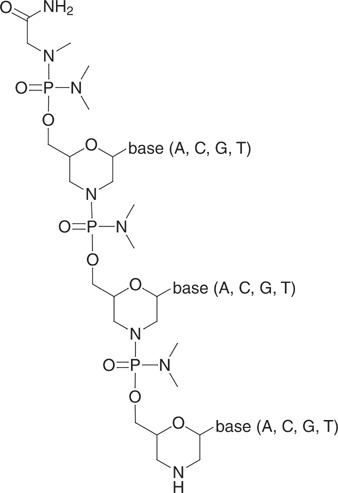
Structure of a Morpholino 3‐mer.
Antisense Morpholinos inhibit the interactions of macromolecules with mRNA by base pairing with the targeted mRNA in a complementary fashion, thus preventing initiation complex read‐through or modifying splicing in cells ranging from bacterial (Geller et al., 2005) to human (Suwanmanee et al., 2002). In particular, antisense Morpholinos have become a standard tool for developmental biologists to manipulate gene expression in embryos such as zebrafish and Xenopus sp. (Ekker and Larson, 2001). These modified oligonucleotides combine efficacy, specificity, stability, lack of non‐antisense effects, and good water‐solubility properties.
This unit presents four protocols for the design of a knockdown experiment: planning a Morpholino experiment (Basic Protocol 1), preparation of Morpholino solutions (Basic Protocol 2), introduction of Morpholinos into cells by endocytosis in the presence of an amphiphilic protonatable peptide (Basic Protocol 3), and injection of Morpholinos into embryos of fish or amphibians (Basic Protocol 4). The Commentary provides a thorough discussion of conditions and considerations for the application of Morpholinos.
Basic Protocol 1. Design of a Morpholino Knockdown Experiment
This protocol outlines the steps and choices commonly made while designing a Morpholino knockdown experiment (based on translation inhibition or splice modification). Considerations for the steps are addressed in the Commentary.
-
1
Choose the target gene.
-
2
Choose the cells or organism into which the oligo will be delivered.
-
3
Choose between splice inhibiting or translation inhibiting, which determines the molecular assays available for measuring antisense activity.
-
4
Obtain the sequence of the target RNA. Use the mRNA 5′‐UTR and the first 25 coding bases for translation inhibitors, or pre‐mRNA with introns and exons defined for splice inhibitors.
-
5
Choose a delivery method.
-
6
Select control oligos.
-
7
Decide whether end‐modifications on any oligos are necessary (see Commentary).
-
8
To inhibit splicing, select which pre‐mRNA splice boundary (intron‐exon or exon‐intron) to block.
-
9
Select the oligo target (following the targeting rules described in the Commentary) and determine the Morpholino sequence (the inverse complement of the target).
-
10
Use a transcript database and a homology search tool such as BLAST to test the selected target for homologies with other RNAs.
If the selected target is very homologous with a region of an off‐target mRNA, a partially complementary Morpholino might affect the expression of that mRNA. Another target on the desired mRNA should be selected to prevent off‐site Morpholino interaction.
-
11
Order the synthesis of the selected Morpholinos.
Basic Protocol 2. Preparation and Verification of Morpholino Stock Solutions
This protocol describes the preparation of a 1 mM aqueous Morpholino stock solution (or a 0.5 mM solution if solubility dictates). If higher concentrations are required for an experiment, take care to ensure that the oligo is completely dissolved.
Materials
Lyophilized Morpholino oligonucleotide (Gene Tools)
Distilled autoclaved water without DEPC, sterile
0.1 M HCl (aqueous)
65°C water bath
Glass or polypropylene/polyethylene tubes with labels
Quartz spectrophotometer cell (1‐cm path length)
Parafilm
Lint‐free lab tissues
UV spectrophotometer (or UV colorimeter) capable of measurements at 265 nm
Morpholino product information sheet
Prepare Morpholino solution
-
1
Read the amount of Morpholino given on the vial label and, using sterile technique, add the appropriate volume of distilled sterile water to make a 1 mM stock solution (e.g., 0.1 ml water for a vial containing 100 nmol Morpholino). Cap the vial, shake it, and wait 5 min.
The aqueous solubility of Morpholinos is sequence‐dependent, but most Morpholino sequences with G content below 36% will dissolve in water at the recommended stock concentration of 1 mM. Do not keep Morpholino solutions of <1 µM because submicromolar concentrations can lose significant activity by binding to glass and plastic surfaces.
It is strongly recommended that Morpholino stock solutions be made with distilled water, but isotonic buffers (e.g., Ringer's solution, Danieau buffer) can also be used. The use of distilled water facilitates the processes of lyophilizing Morpholinos and analyzing them by MALDI‐TOF mass spectrometry, should either of these be required.
If water must be treated with diethylpyrocarbonate (DEPC), it is very important to autoclave the treated water to destroy residual DEPC before using it to dissolve Morpholinos. Otherwise, DEPC reacts with adenines and compromises the ability of Morpholinos to bind to their targets (Henderson et al., 1973).
-
2
Swirl and inspect the solution to see whether the oligo has dissolved. If it has not dissolved, warm the vial for 5 min in a water bath at 65°C.
-
3
If the oligo has not dissolved, repeat steps 1 and 2, adding the same volume of water, to make a 0.5 mM stock solution.
-
4
If desired, dispense into several tubes. Label the tube(s) with the concentration and oligo name, and store any tubes that will not be used immediately.
Scrupulously avoid microbial contamination of the stock solutions. Store fluorescent‐tagged Morpholinos in a closed opaque box so that light will not bleach fluorescent moieties.
Morpholinos are stable in stock solutions stored at 25°C or 4°C. Room temperature is best for storing Morpholinos in solution. Morpholinos can also be stored frozen; however, ice crystal formation during slow freezing can cause the concentration of the oligos in the bulk solution phase to increase until the Morpholinos precipitate.
After thawing and prior to use, Morpholino solutions should be heated for 10 min in a water bath at 65°C to aid in dissolution. Morpholinos precipitated onto the inside surfaces of containers can be difficult to redissolve; if necessary, autoclave a solution of unsubstituted, fluorescent, or Vivo‐Morpholino to redissolve the oligo (see Commentary).
Check Morpholino concentration by UV absorbance
-
5
Turn on the UV spectrophotometer and let it warm up for a few minutes. Set the spectrophotometer to report absorbance at 265 nm.
-
6
Clean the quartz spectrophotometer cell, if needed, and rinse the inside twice with 0.1 M HCl. Carefully shake excess liquid from cell.
Do not touch the outside of the quartz spectrophotometer cell on the surfaces where light will pass through, as skin oils can contain materials that skew the measurements.
-
7
Pipet 995 µl of 0.1 M HCl into the quartz cell and place the cell in the spectrophotometer. Blank the spectrophotometer at 265 nm.
-
8
Remove the cell from the spectrophotometer and add 5 µl aqueous Morpholino solution into the quartz cell containing the 0.1 M HCl.
Like natural nucleic acids, the nucleobases of a Morpholino are stacked, and stacking produces a hypochromic effect. Without unstacking the bases, the use of the molar absorptivity of an individual nucleobase to calculate the concentration of the oligo would lead to an erroneously low value. Oligos with A, C, and G bases can be unstacked by dissolving the oligos in 0.1 M HCl. Under these conditions, A, C, and G bases are protonated and are out of the stacked state due to electrostatic repulsion. When the nucleobases of the oligo are unstacked, the molar absorptivity of each nucleobase can be applied to determine the concentration of the oligo.
-
9
Place a piece of Parafilm over the open end of the cell, placing a thumb over the Parafilm to seal the cell, and invert several times to mix.
Avoid touching surfaces that will be in the spectrophotometer's light path.
-
10
Remove the Parafilm. Wipe the outside of the cell with a lint‐free tissue, if needed.
-
11
Place the cell in the spectrophotometer and read the absorbance at 265 nm (A 265).
-
12
Calculate the molar concentration (C) of the original Morpholino solution as:
C = (A 265 × 200)/(ε b)
where 200 is the factor for dilution in HCl, ε is the molar absorptivity, and b is the path length of the cell (1 cm).
The molar absorptivity (ε) of the Morpholino is provided on the product information sheet. Alternatively, ε can be calculated by multiplying the molar absorptivity of each nucleobase (A, C, G, and T) by the number of instances that the nucleobase is present in the oligo, and adding these products.
This Beer's law calculation works when absorbance ≤2, where the relationship of absorbance to concentration is linear. If the measured absorbance is >2, the sample should be diluted and remeasured.
Basic Protocol 3. Delivery of Morpholinos into Cells Using Endo‐Porter
Endo‐Porter is an amphiphilic peptide. After co‐endocytosis with Morpholinos, Endo‐Porter is protonated as the endosome becomes acidic, and in this form Endo‐Porter permeabilizes endosomal membranes, releasing the Morpholino from the endosomes to the cytosol (Summerton, 2005). Endo‐Porter was optimized using a HeLa cell line. Because tolerance of other cell types toward Endo‐Porter often varies, a range of Endo‐Porter concentrations should be tested before beginning knockdown experiments.
Materials
Cell cultures in plates or flasks at 80% to 100% confluence
1 mM fluoresceinated dextran (10 kDa; e.g., Invitrogen) or 1 mM fluoresceinated Morpholino stock solution (Basic Protocol 2)
Cell culture medium with 10% (v/v) serum
1 mM Endo‐Porter solution (aqueous or DMSO formulation, Gene Tools)
Fluorescence microscope with fluorescein filter cube (e.g., with filters for 501.5‐nm excitation and 524.5‐nm emissions)
Select amount of Endo‐Porter for cell type
-
1
Add 10 µM fluorescently labeled Morpholino (10 µl of 1 mM stock per 990 µl cell culture) or fluoresceinated dextran (10 µl of 1 mM stock per 990 µl cell culture) to cell culture medium with 10% serum.
Fluoresceinated dextran has been a useful proxy for predicting the behavior of an unlabeled Morpholino.
-
2
Change the medium on the cultured cells, replacing spent medium with the fresh medium containing the fluorescent Morpholino or dextran.
-
3
Introduce concentrations of 2, 4, 6, and 8 µM Endo‐Porter by pipetting 2, 4, 6, and 8 µl of a 1 mM Endo‐Porter solution into 1‐ml aliquots of cell culture and immediately swirl to mix.
-
4
Allow endocytotic uptake to proceed over a period of 24 hr.
-
5
Observe intracellular fluorescence using a fluorescence microscope.
See discussion on assessing delivery in the Commentary section.
-
6
Observe cells 72 hr after delivery to determine any cellular toxicity.
For subsequent Morpholino delivery to the selected cell type, use the concentration of Endo‐Porter that gave the best delivery without toxicity.
Deliver Morpholinos to cells
-
7
Using a cell culture not previously exposed to Endo‐Porter, replace spent culture medium with fresh medium containing up to 10% serum.
-
8
Add the Morpholino stock solution to produce the desired concentration and swirl well to mix.
For functional experiments (e.g., gene knockdown, splice blocking), Morpholinos are typically effective at concentrations as low as 1 µM. However, it is recommended that a range of concentrations be tested (such as 1, 4, and 10 µM Morpholino) to define optimal conditions.
-
9
Add Endo‐Porter to produce the optimized concentration for the cell type and immediately swirl to mix.
-
10
Place the plates or flasks in the incubator. Wait at least 16 hr before assessing uptake by fluorescence, and at least 24 hr before measuring knockdowns by molecular assays.
The delay needed when using an antibody to assay a knockdown depends on the stability of any pre‐existing protein encoded by the targeted mRNA; a protein with a long half‐life will take longer to disappear from the cells. In contrast, assaying splice modification of RNA by RT‐PCR does not require a long delay and can be done 16 hr after commencing delivery.
Basic Protocol 4. Injection of Morpholinos into Embryos of Fish or Amphibians
When Morpholino oligos are used in embryos, they are typically delivered by microinjection into zygotes. Daughters of the injected cell will also contain the oligos and, depending on the species‐specific permeability characteristics of the embryonic cells, the oligo might diffuse into noninjected lines. This diffusion is commonly observed in the early zebrafish embryo, while the cells of Xenopus embryos do not release Morpholinos to their neighbors.
In placental organisms, an embryo increases its overall volume when the placenta forms and provides nutrients to the embryo, so Morpholino activity is diluted at an earlier developmental stage. In contrast, embryos that grow enclosed within eggs do not increase their overall volume (yolk + embryo) until they begin to feed, so Morpholino activity persists for a relatively longer duration through development. Because of this, fish and frogs are good model organisms for studying development using unmodified microinjected Morpholino oligos.
This protocol provides a procedure for injecting Morpholinos into fish or frog embryos. Additional details about microinjection techniques can be found in The Zebrafish Book (Westerfield, 2007). This protocol also describes making an agarose block to gently hold the embryo during injections, but plastic blocks for this purpose are commercially available.
Materials
Embryo medium (with antibiotic, if desired; see recipe)
Agarose (e.g., Amresco)
Morpholino stock solution (Basic Protocol 2)
1% (w/v) aqueous phenol red, sodium salt (e.g., Sigma)
Embryos: e.g., single‐cell Zebrafish embryos, one to eight cell frog embryos
1.2‐mm o.d., 0.94‐mm i.d. glass capillary tubes (e.g., FHC, Harvard Apparatus, or Sutter Instrument)
Needle puller (e.g., Sutter Instrument)
100 × 20−mm transparent plastic dish, sterile
1‐mm wide plastic mold to form linear wedge‐shaped troughs atop the agarose block (Adaptive Science Tools)
1.5‐ml microcentrifuge tubes
0.5‐ to 10‐µl pipet tip for filling microinjection capillaries (e.g., microloaders, Eppendorf)
Storage jar with electrode gasket to hold injection needle (World Precision Instruments), optional
-
Microinjection system including:
Nitrogen gas and regulator (if required)
Pressure injector with foot switch (e.g., Applied Scientific Instrumentation or Eppendorf)
Micromanipulator (e.g., Märzhäuser)
Microelectrode holder for 1.2‐mm‐o.d. glass, with port (e.g., World Precision Instrument)
Dissecting microscope (e.g., Leica S4E)
Controlled‐drop borosilicate Pasteur pipet, capable of delivering 22 to 26 drops per ml (e.g., Fisher)
Pipet pump with thumbwheel (e.g., Scienceware, Fisher Scientific)
Forceps
Prepare needle for microinjection
-
1
Fit a glass capillary tube (o.d. 1.2 mm, i.d. 0.94 mm) into the needle puller.
-
2
Pull the capillary using the desired program.
Preferred needle characteristics vary, but the pulled needle should have a short shank for sufficient stiffness. A good length to try from shoulder to tip is ∼9 mm. Later the tip will be snapped off to open a tip i.d. of 0.02 to 0.03 mm.
Prepare embryo medium and agarose block
-
3
Prepare fresh embryo medium, with antibiotics, if desired.
-
4
Mix agarose with embryo medium at 2 g agarose per 100 ml medium.
-
5
Microwave the mixture until it boils and swirl while hot to ensure that the agarose is dissolved.
-
6
Pour the hot 2% agarose into a sterile transparent dish.
-
7
Suspend the mold over the agarose so that the mold is immersed in the hot agarose and will form 1‐mm‐wide wedge‐shaped linear slots when withdrawn from the cooled agarose.
-
8
Allow the agarose to cool and set.
-
9
Remove the molds gently after the agarose is set.
Plates of molded agar can be made in advance and stored tightly covered in the refrigerator.
Prepare Morpholino solution and microinjector
-
10
Make a dilution from the Morpholino stock solution in a microcentrifuge tube so that a 1‐ng to 10‐ng dose of Morpholino can be delivered in a 2‐nl to 5‐nl injection.
-
11
Add 1 volume of 1% aqueous phenol red solution to 9 volumes of Morpholino injection solution (final 0.1%).
-
12
Centrifuge the mixture briefly in a microcentrifuge before the injection to prevent needle clogging.
Note that if precipitated Morpholino spins out of solution, the actual concentration will be lower than the calculated concentration.
This centrifugation step is commonly performed to prevent clogging. For critical quantitative work, the solution concentration should be checked by measuring UV absorbance, but this would become an expensive drain on the Morpholino solution if done routinely.
-
13
Load 0.5 to 1.0 µl of the supernatant solution into the glass microinjection needle using a pipettor fitted with a very fine microloader pipet tip.
There are two commonly used techniques for loading the microinjection needle: (1) pipetting into the tip of the needle or (2) placing the needle vertically into a micropipet storage jar with an electrode gasket and loading into the back of the needle, letting gravity and capillary action draw the solution into the tip.
-
14
Switch on the nitrogen gas connected to the pressure injector.
-
15
Switch on the pressure injector connected to the micromanipulator through a tube.
-
16
Fit the needle into the microelectrode holder and connect it to the micromanipulator.
-
17
Break the tip of the glass injection needle by grasping the tip at a right angle with fine tweezers, or by touching the tip gently to a hard surface. Snap off the end at a point where the needle has an inside diameter of ∼0.02 to 0.03 mm.
Calibrate injection volume
-
18
Determine the drop size delivered by the microinjection system.
Using the Morpholino solution, a rough estimate can be made by comparing the drop size diameter, normally ∼100 to 150 µm, to the diameter of a zebrafish embryo, normally ∼600 µm. Observe the drop size just after the drop has been injected into an embryo (following the steps below in the Microinject section).
Injection volumes can be accurately calibrated by injecting aqueous dye solution into mineral oil, measuring the diameter of the aqueous droplet with an ocular micrometer, and calculating its volume. Performing this measurement and calculation on multiple droplets allows one to assay the reproducibility of the injection volume from a particular microinjection apparatus (Riedel‐Kruse et al., 2007; online supplementary information). The absolute volume delivered by the microinjector can be adjusted.
Set up embryos for microinjections
-
19
Place the transparent dish containing the molded block of 2% agarose onto the stage of the dissecting microscope.
-
20
Turn on the light below the microscope stage.
-
21
Place embryos onto the slots in the agarose block using a controlled‐drop glass Pasteur pipet fitted with a pipet pump. Gently press the embryos into the slot in the agarose block using forceps.
-
22
Add room temperature or 28°C embryo medium (optionally with antibiotics) to the dish so that the embryos are covered.
-
23
Fit the needle loaded with Morpholino solution into the port of the microelectrode holder, and connect the microelectrode holder to the micromanipulator attached to the pressure injector.
-
24
Orient the slots in the agarose at right angles to the needle.
Microinject Morpholino solution into embryo
-
25
Use the micromanipulator to force the needle through the chorion of a single‐celled embryo, placing the tip of the needle in the cytosol.
In zebrafish embryos, injections can be done into the yolk close to the cell or into the cytosol of a one‐cell or two‐cell embryo. Morpholinos distribute fairly evenly through the embryo after injections as late as the eight‐cell stage.
For frog embryos, inject only at the single cell stage if distribution throughout the cells of the growing embryo is desired.
-
26
Press the foot switch to deliver the Morpholino solution into the embryo.
Typically, microinjections are done at 1 to 10 ng of Morpholino delivered in 2 to 5 nl per embryo.
-
27
After the injection, withdraw the needle from the embryo. If the embryo adheres to the needle, use forceps to gently press the embryo with force parallel to the needle so that the needle and embryo are separated.
Move embryos to tank for incubation
-
28
Using a dissecting needle, forceps, or a pipet tip, gently dislodge the embryos from the agarose block and pipet with a controlled‐drop pipet into a dish filled with embryo medium (optionally with antibiotics) for incubation.
-
29
Incubate embryos at 28.5°C for later observations.
Reagents and Solutions
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see appendix mba02; for suppliers, see appendix mba04.
Embryo medium
Prepare stock solutions:
Stock 1: 8.0 g NaCl and 0.4 g KCl in 100 ml H2O
Stock 2: 0.358 g anhydrous Na2HPO4 and 0.60 g KH2PO4 in 100 ml H2O
Stock 3: 0.72 g CaCl2 in 50 ml H2O
Stock 4: 1.23 g MgSO4 ·7H2O in 50 ml H2O
Stock 5: 0.35 g NaHCO3 in 10.0 ml H2O (prepare fresh)
Sterilize the stock solutions by passing through a 0.2‐µm filter and store solutions 1 to 4 up to several years at 4°C (in the absence of microbial contamination).
Prepare medium:
95.9 ml H2O
1.0 ml stock 1
0.1 ml stock 2
1.0 ml stock 3
1.0 ml stock 4
1.0 ml fresh stock 5
Adjust the pH to 7.2 with ∼10 drops of 1 M NaOH. If desired, add 20 ml of 50× penicillin‐streptomycin (e.g., Sigma) to 1 liter of embryo medium to make a final concentration of 1× antibiotic. Store up to 1 month at room temperature.
A 0.5× antibiotic solution is also usually sufficient to inhibit infection.
This recipe is from Westerfield (2007).
Commentary
Background Information
Morpholino structure
The morpholino phosphorodiamidate backbone of a Morpholino oligonucleotide has no significant ionic charge at neutral pH, in contrast to the polyanionic phosphodiester backbone of a natural nucleic acid. This favors the interaction of Morpholinos with nucleic acids, since there is no repulsion between anionic backbones as there is in duplexes of natural nucleic acids. When dissolved in pure water, nucleic acids lose their ability to form stable Watson‐Crick bonds due to anionic repulsion between strands, whereas Morpholinos will still bind to complementary nucleic acid sequences (Summerton, 2004). Because Morpholinos are uncharged, they have no strong electrostatic interactions with proteins. Unmodified Morpholinos have little or no affinity for bovine or human serum albumin (H.M. Moulton, unpub. observ.). In contrast, interactions of anionic phosphorothioate oligos with proteins cause multiple physiological, non‐antisense effects (Lebedeva and Stein, 2001). Proteins that bind nucleic acids generally interact electrostatically with the anionic phosphates of nucleic acids, stabilizing binding. Morpholinos appear to have little or no interaction with nucleic acid–binding proteins (Hudziak et al., 1996).
Morpholinos are very stable to nucleolytic enzymes. There are no known enzymes that degrade Morpholinos. Specifically, Morpholinos have been exposed to a range of nucleases (e.g., DNase I, DNase II, Benzonase, S1 nuclease, mung bean nuclease, Bal 31 nuclease, RNase A, RNase T1, phosphodiesterase I, and phosphodiesterase II) and proteases (e.g., pronase E, proteinase K, and pig liver esterase) under conditions where lytic enzymes would degrade their substrates. In no case was degradation of the Morpholinos detected (Hudziak et al., 1996). Morpholinos were incubated in serum and in liver homogenate without degradation (Summerton and Weller, 1997). When peptide‐Morpholino conjugates were extracted from cells and analyzed by MALDI‐TOF mass spectrometry, the Morpholino oligo entity was not degraded in the cells (Youngblood et al., 2007).
No crystal structure or high‐resolution NMR structural analysis of phosphorodiamidate Morpholinos has been published. However, study of a morpholino phosphorodiamidate ApA dimer using circular dichroic spectroscopy showed stacking of bases in aqueous phosphate buffer (Kang et al., 1992). On the basis of molecular modeling, the bases of Morpholinos should stack in a fashion analogous to those of natural nucleic acids, allowing strong interactions with complementary nucleic acid sequences by Watson‐Crick base pairing. A 400 MHz 1H NMR analysis of a carbamate‐linked Morpholino found the morpholine ring in the chair conformation (Stirchak et al., 1989). Molecular modeling of a Morpholino with the morpholine rings in the chair conformation suggests that a Morpholino and an RNA form an A‐form heteroduplex with a helical pitch similar to that of an A‐form RNA‐RNA duplex (J.E. Summerton, unpub. observ.).
Various types of antisense oligos are ranked by their affinity for binding to single strands of sense RNA based on their dissociation temperatures in physiological salt buffers (Table 1; Stein et al., 1997). The affinity of RNA for RNA is greater than the affinity of Morpholinos for RNA. However, single strands of mRNA folded into secondary structures contain single‐stranded regions, such as the loops of stem‐loops, with which Morpholinos can readily hybridize. Because double‐stranded regions of most RNA secondary structures are shorter than 25 base pairs, the overall binding affinity of Morpholinos for RNA is usually sufficient to invade and displace those short double‐stranded regions (Summerton, 1999).
Table 1.
RNA Binding Affinity of Various Oligo Types Ranked by Dissociation Temperature in Physiological Isotonic Buffers
|
Affinity |
Type of oligoa |
|---|---|
|
Strongest |
RNA:RNA, PNA:RNA, 2′‐O‐methyl‐RNA:RNA (all very similar) |
|
Strong |
Morpholino:RNA |
|
Medium |
DNA:RNA |
|
Weakest |
Phosphorothioate:RNA |
Abbreviation: PNA, protein nucleic acid.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Modes of action
Antisense oligos such as DNA, RNA, and phosphorothioate (S‐DNA) oligos recruit RNase H to degrade their mRNA targets (Summerton, 1999). RNAi and siRNA (see unit mb2601) also employ an antisense mechanism to recognize a sense mRNA through interaction with a RISC complex, which leads to enzymatic degradation of complementary mRNA and translation inhibition of partially complementary mRNA (Scacheri et al., 2004). In contrast, instead of degrading mRNA, antisense Morpholinos were designed to block the translation of mRNA into protein (Summerton and Weller, 1997). Figure 2 compares steric blocking, RNase H–dependant, and RISC‐dependant oligos.
Figure 2.
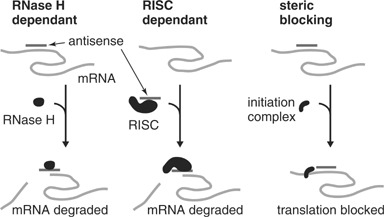
Comparison of RNase H–dependant, RISC‐dependant, and steric blocking oligos.
When comparing an RNase H–dependant oligo (a methylphosphonate diester/phosphodiester chimera) with a Morpholino, a CpGNNN motif was shown to induce apoptosis and cell cycle arrest when present in the RNase H–dependent oligo but not when present in the Morpholino (Tidd et al., 2001). There have been no reports of Morpholinos inducing either interferon production or induction of NF‐κB mediated inflammation, and Morpholinos containing CpG motifs do not stimulate immune responses (J.E. Summerton and A. Krieg, unpub. observ.), suggesting that Morpholino‐RNA heteroduplexes do not stimulate Toll‐like receptors.
Morpholinos complementary to sequences in the 5′‐UTR and the first 25 coding bases of an mRNA can halt the progression of the initiation complex toward the start codon, preventing assembly of the entire ribosome. This inhibits the translation of the mRNA sequence into a polypeptide. Morpholinos targeted downstream of the start codon are usually ineffective for inhibiting translation (Summerton, 1999).
In addition to their application to knock‐down gene expression, Morpholinos are also widely used to inhibit splicing of pre‐mRNA because steric‐blocking oligos do not trigger degradation of RNA. Splicing in eukaryotes is directed by small nuclear ribonucleoproteins (snRNPs) that bind to introns and mark the intron‐exon boundaries. Morpholinos targeted to these snRNP‐binding sites can modify splicing (Sazani et al., 2001), either preventing splicing and causing an intron inclusion (Giles et al., 1999) or redirecting splicing and causing an exon excision (Draper et al., 2001). Blocking a splice site can cause activation of a cryptic splice site, complicating interpretation of the splice modification by producing partial deletions of exons (Draper et al., 2001) or partial inclusions of introns.
Morpholinos can inhibit miRNA activity by binding to the miRNA and preventing it from binding its mRNA target (Kloosterman et al., 2004) or by binding to the site on the mRNA where the miRNA would otherwise bind (Choi et al., 2007). Morpholinos can inhibit maturation of pre‐miRNA by binding at the RNA processing enzymes Drosha or Dicer processing sites (Kloosterman et al., 2007).
Although Morpholinos are most often used to inhibit activity of the translation initiation complex or the snRNPs that direct splicing, there are other mRNA sequences that are attractive targets for steric blocking. Morpholinos targeted across the cleavage site of a hammerhead ribozyme inhibited auto‐cleavage, leading to over two orders of magnitude increase in the expression of a downstream reporter gene (Yen et al., 2004). Morpholinos stimulate site‐specific ribosome frame‐shifting when bound just downstream of a shift site on an mRNA, and they do so with far higher efficiency than RNA, phosphorothioate oligos, or 2′‐O‐methyl RNA oligos (Howard et al., 2004). While Morpholinos have also been shown to inhibit intronic splice silencers (Bruno et al., 2004) and exonic splice enhancers (McClorey et al., 2006), no publications have yet explored other potential regulatory targets such as zipcode binding sites, riboswitches, or binding sites for elements of the nonsense‐mediated mRNA decay pathway.
In vivo uses of Morpholinos
Morpholinos are commonly microinjected into embryos at the single‐cell or few‐cell stages to inhibit genes involved in development (Heasman et al., 2000; Nasevicius and Ekker, 2000; Nutt et al., 2001). Morpholinos are also commonly used in cell cultures (Tyson‐Capper and Europe‐Finner, 2006). Combinations of several oligonucleotide sequences can bind to several different RNA targets simultaneously if introduced together into embryos (Ekker, 2000) or cell cultures (Summerton, 2005), allowing multiple simultaneous knockdowns or synergistic targeting of a single messenger. Applications in intact adult organisms have until recently been limited by poor in vivo delivery into the cytosol of cells (Summerton, 1999; Sazani et al., 2002). However, conjugation of Morpholinos to cell‐penetrating peptides (Nelson et al., 2005; Moulton and Moulton, 2008) allows effective systemic delivery into adult organisms (Alonso et al., 2005; Kinney et al., 2005; Neuman et al., 2005; Enterlein et al., 2006), as does conjugation of a Morpholino with an octaguanidinium dendrimer, forming a Vivo‐Morpholino (Li and Morcos, 2008).
Targeting of viral RNA with Morpholinos has been reported for hepatitis C (Jubin et al., 2000; McCaffrey et al., 2003), dengue virus (Kinney et al., 2005), ebola virus (Enterlein et al., 2006; Warfield et al., 2006), SARS virus (Neuman et al., 2005), West Nile virus (Deas et al., 2005), equine arterivirus (van den Born et al., 2005), mouse hepatitis virus (Neuman et al., 2004), novirhabdovirus (Alonso et al., 2005), and vesivirus (Stein et al., 2001). In addition to translation start sites, successful targets for inhibition of viral replication include cyclization sequences (Deas et al., 2005), terminal stem loops (Deas et al., 2005), and internal ribosomal entry sites (IRES; Jubin et al., 2000).
Radioisotope delivery into organisms can be pretargeted using Morpholinos (Mang'era et al., 2001). Practitioners of nuclear medicine strive to minimize radiation exposure of a patient while delivering radionuclides to target tissues for imaging or for therapeutic applications. By attaching radioisotopes to antibodies that are specific for target tissues, the antibodies can anchor isotopes on these tissues. Because the large antibody molecules diffuse slowly, the isotopes must be maintained in the plasma at high concentrations or for long durations to achieve good delivery of radioisotope‐linked antibodies to their targets. Pretargeting with Morpholinos involves introducing an antibody‐Morpholino conjugate into the bloodstream; this can be done using high concentrations or re‐dosing to saturate the target without exposing the patient to radiation during this pretargeting stage. Next, a conjugate of a radioisotope (possibly chelated) with a complementary Morpholino is added to the blood. Because the Morpholino has a much smaller molecular mass than an antibody, the radionuclide‐Morpholino conjugate diffuses relatively quickly and is captured at the target tissue more rapidly through Morpholino‐Morpholino pairing. Unbound radionuclide‐Morpholino conjugate is rapidly eliminated through the kidneys. This technique allows delivery of radioisotopes to the targeted tissue while exposing the organism to lower doses of radiation away from the targeted region. In the process of developing these techniques, pharmacokinetics of Morpholino‐radionuclide conjugates have been studied in vivo (Liu et al., 2002a,b; He et al., 2003). Signals can be amplified by binding a polymer bearing many complementary Morpholinos to each Morpholino‐conjugated antibody fragment, followed by delivering radioisotope‐labeled Morpholino complementary to the polymer‐linked Morpholinos (He et al., 2003, 2004).
Critical Parameters
Choosing Morpholino sequences
The parameters considered when selecting oligonucleotide target sequences include percent CG, percent G, self‐complementarity, tetra‐G moieties, length of the oligo, and the intended temperature at which the oligo will be used. The targeting recommendations are summarized below and in Table 2.
Table 2.
Summary of Targeting Recommendations for 37°C Systems
|
Parameter |
Recommendation |
Comments |
|---|---|---|
|
CG range |
40%‐60% |
At lower GC, affinity may be too low to inhibit processes; higher GC favors nonspecific binding of subsequences. |
|
G content |
Up to 36% G |
Higher G causes loss of water solubility; avoid upper end of acceptable range, if possible. |
|
Self‐complementarity |
16 contiguous H‐bonds maximum |
For intermolecular (complementary palindrome) and intramolecular (stem loop) binding. Example: AGCGCT has 16 H‐bonds (2+3+3+3+3+2 = 16). Check for non‐Watson‐Crick G‐T pairing, which can participate in self‐complementarities. |
|
Consecutive G |
3 consecutive Gs maximum |
Runs of ≥4 G can associate through Hoogsteen bonding to form oligo tetramers. |
|
Oligo length |
25 bases or shorter by only a few bases |
Using shorter oligos can decrease the chance of off‐target interaction for high CG oligos. |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
CG range. A range of 40% to 60% CG is considered ideal for 25‐base Morpholinos in 37°C systems. Oligos with <40% CG may lack the affinity needed for effective steric blocking, while oligos with >60% CG are more likely to interact with off‐target messengers through high‐affinity subsequences.
G content. G content affects aqueous solubility of an oligo, with higher G contents being less soluble, particularly when the oligo is dissolved in isotonic salt solutions. Oligos with G contents up to 36% should be soluble in the millimolar range in pure water or aqueous buffer. However, freeze‐thaw cycles are likely to cause high‐G oligos to precipitate and the oligos must be heated to redissolve (see Basic Protocol 2).
Self‐complementarity. Self‐complementary sequences can cause either intramolecular interactions, forming stem‐loops, or intermolecular interactions, forming dimeric Morpholinos. When a short sequence of one part of an oligo is complementary to another short sequence separated by an intervening sequence, stem‐loops can form. If small self‐complementary sequences are separated by zero to a few bases, formation of a stable stem‐loop is unlikely because a hairpin with a small loop is not energetically favored. To prevent loss of oligo activity through competition between self‐pairing and target binding, it is prudent to limit self‐complementary sequences in oligo designs to 16 contiguous hydrogen bonds or less, where CG pairs contribute 3 hydrogen bonds and AT pairs contribute 2 hydrogen bonds. For instance, the short sequences ATGGC and GCCAT can form 13 contiguous hydrogen bonds (2+2+3+3+3 = 13). When analyzing sequences for self‐complementarity, check for both Watson‐Crick base‐pairing and for GT base‐pairing. Like an AT pair, a GT pair also forms two hydrogen bonds. However, because the overall stability of the GT pair is far lower than an AT pair, a GT pair can be scored as a single hydrogen bond when calculating its contribution to the stability of a self‐complementary moiety (Aboul‐ela et al., 1985).
An oligo containing a self‐complimentary sequence can form dimers. To prevent loss of oligo activity through competition between dimer formation and target binding, it is prudent to limit complimentary palindromes to 16 contiguous hydrogen bonds or less. For instance, if two oligos bearing the self‐complimentary sequence ATGCATGCGT encounter each other, they can form 22 contiguous hydrogen bonds (2+1+3+3+2+2+3+3+1+2 = 22, taking into account the GT pairs) and would likely have poor antisense activity.
G tetrads. Nucleic acids containing GGGG moieties can interact through Hoogsteen bonding to form oligo tetramers (Cheong and Moore, 1992). Morpholinos containing G tetrads have reduced activity, likely through the same mechanism. Because of this, contiguous stretches of four or more G bases should be avoided when designing Morpholinos.
MIL and oligo length. The minimum inhibitory length (MIL) of an antisense oligo is the length needed to achieve 50% reduction in translation of a targeted gene at a concentration typically achieved in cells. The MIL of Morpholinos varies somewhat between targets, but averages about 14 bases for 37°C cell cultures (Summerton, 1999). To ensure good affinity between Morpholinos and their RNA targets, the oligos are usually synthesized as 25‐mers.
CG content can influence the MIL of an oligo, with a higher CG oligo having a shorter MIL. Oligos with high CG content have an increased chance of interacting with off‐target RNA; these oligos can be shortened by a few bases to lessen the likelihood of off‐target interactions. The marginal loss of affinity resulting from shortening a high‐CG oligo will not ruin activity but will slightly improve specificity. A more effective way to improve specificity is to choose a target with a lower CG content.
Temperature and oligo selection
The targeting guidelines were developed for oligos to be used at 37°C. Many embryos are grown at lower temperatures. When temperatures are decreased appreciably, stability of base‐pairing increases. The ideal CG content for oligos designed for use at lower temperatures is lower than the 40% to 60% CG recommended for 37°C systems. Oligos with CG contents down to 32% are usually effective in zebrafish or Xenopus embryos. The ideal CG content for colder systems must be determined experimentally. Similarly, the allowable number of base pairs in self‐complementary sequences should be reduced for colder systems. Solubility is also decreased at lower temperatures, so it is prudent to select oligos with lower G contents for use in colder systems.
Targetable region for translation inhibitors
To inhibit cap‐dependent translation, a 25‐mer Morpholino can target anywhere between the 5′ cap to 25 nucleotides into the coding sequence. The target can extend downstream into the coding sequence as long as the start codon is covered. In the first steps of translation, the initiation complex forms at the 5′ cap and then scans through the UTR to the start codon (Fig. 3A). At the start codon, the large ribosomal subunit binds, the initiation factors dissociate, and translation proceeds through the coding region. If a Morpholino gets in the way of the initiation complex before the initiation complex reaches the start codon, it prevents assembly of the ribosome and translation of the mRNA. Nonetheless, it is preferable to target the start codon instead of upstream sequences for two reasons. First, the quality of sequence deposited in public databases is often poor in the UTR, especially for older sequence records. Second, although rare in vertebrate genomes, internal ribosome entry sites (IRES) do exist and can allow a ribosome to enter and assemble downstream of a Morpholino bound in the 5′‐UTR.
Figure 3.
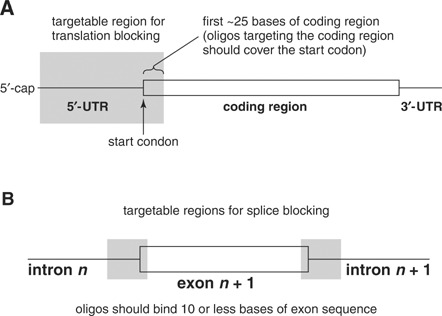
Targetable regions for translation (A) and splice (B) inhibition.
Targetable region for splice inhibitors
To inhibit splicing, Morpholinos are targeted to pre‐mRNA across or near the boundaries between exons and introns. A pre‐mRNA that undergoes splicing has two flanking exons (the first and last exon) and an arbitrary number of internal exons. The first exon has a single splice site, i.e., a splice donor where it contacts intron 1. The internal exons have two junctions each, a splice acceptor at the upstream end and a splice donor at the downstream end. The last exon has only a splice acceptor at its upstream end. Targeting the splice sites of the internal exons usually causes exon excision, resulting in an mRNA missing the exon with the inhibited splice site (Fig. 3B). Targeting splice sites of the flanking (first or last) exons usually causes intron inclusion, resulting in an mRNA containing the first or last intron. Sometimes inhibiting a splice site activates a cryptic splice site, resulting in an mRNA with an unexpected mass.
The snRNPs that direct splicing bind at the intronic sides of the splice junctions, so Morpholinos are chosen that are complementary to more intronic sequence than exonic sequence. Morpholinos can have good activity if targeted entirely to intronic sequence near the splice junction, but activity decreases as the target is moved farther into the intron (Morcos, 2007).
Splicing can also be modified by preventing excision of an arbitrary intron by blocking the nucleophilic adenosine that closes the splicing lariat (Morcos, 2007) or by targeting splice‐regulatory sequences (Bruno et al., 2004, Melton et al., 2007).
It is often the goal of a splice‐inhibiting experiment to eliminate activity of a protein. If the active site of the protein is known, a straightforward strategy is to target a Morpholino to the exon encoding the active site, causing the loss of that exon and of the active site. When the active site is not known, other useful strategies are available. One is to eliminate an upstream exon that has a number of nucleotides not evenly divisible by three, causing downstream translation to be frameshifted. Another is to trigger inclusion of the first intron in the coding region, especially useful if that intron contains an in‐frame stop codon or if its number of nucleotides is not evenly divisible by three. Sometimes causing a random exon exclusion or intron inclusion is sufficient to eliminate activity of a protein, perhaps due to a resulting change in the protein's tertiary structure.
Quality of sequence
Since a few mismatches can seriously decrease the activity of a Morpholino, the quality of the target sequence is an important consideration when designing Morpholinos. There are sometimes errors in sequence database files. Variations in sequence between strains of an organism can also present a problem. The most definitive way to ensure the correct target sequence is to sequence the target region of the gene in the strain that will be used in the experiments.
Mismatched unintentional targets and Morpholinos
When a 25‐base Morpholino is used near its lowest effective concentration, its effects are very specific. Under such conditions the oligo might also interact with sequences containing one or two mismatches when compared to the oligo's perfectly complementary target, although even a single mismatch can decrease activity (Khokha et al., 2002). However, few to no such sequences are expected to occur randomly in a base pool the size of the Morpholino‐targetable sites in the human transcriptome (Summerton, 1999).
Effect of concentration on specificity
When the concentration of any antisense oligo is increased well above its minimum effective concentration, it can interact with targets containing more mismatches; at some concentration a Morpholino will begin knocking down expression of off‐target mRNAs. Therefore it is important that the oligo concentration be kept as low as practicable while still eliciting the desired targeted knockdown. The concentration at which off‐target effects occur, the concentration at which targeted knockdown occurs, and the ratio of these concentrations are all sequence‐specific. In most cases, an effective and specific concentration window exists such that, for complementary mRNA and off‐target mispaired mRNA at similar concentrations, the onset of the targeted knockdown will occur at a lower concentration than the onset of the off‐target knockdown. However, knocking down high‐copy‐number mRNAs requires higher oligo concentrations, increasing the probability of knocking down low‐copy‐number off‐target mRNAs; such a situation can narrow or even close the effective and specific concentration window.
Acceptable off‐target homology
A single mismatch in a Morpholino 25‐mer may cause a significant decrease in antisense activity (Khokha et al., 2002), although many single‐mismatched oligos have retained good activity. When used near the concentration at which a perfectly complementary oligo elicits a knockdown, five mismatches distributed throughout a 25‐mer usually decreases activity of the mismatched oligo to near undetectable levels (Kamachi et al., 2008).
It is prudent to check the target sequence of a proposed oligo against a nucleotide sequence database in order to identify regions where the Morpholino might bind to off‐target mRNA. When searching for homologous targets, keep in mind that 25‐base Morpholinos will only inhibit translation when targeted to the 5′‐UTR and first 25 bases of coding sequence. Morpholinos can modify splicing if targeted mostly in introns at or near intron‐exon boundaries. If the Morpholino has homology to an off‐target mRNA outside of these limited regions, binding of the oligo to the mRNA is not likely to affect expression of the off‐target mRNA (although inhibiting miRNA targets or regulatory sequences such as exonic splice enhancers may affect expression).
When comparing a 25‐base Morpholino against an off‐target sequence in a region where a Morpholino might have a biological effect, the fraction of homologous bases should always be below 80%. However, that percentage ignores important considerations about the distribution of the mismatches throughout the oligo. About 14 contiguous bases of homology is the minimum inactivating length for a Morpholino (Summerton, 1999). However, if 10 bases of perfect homology are flanked with a mismatch at either side and some runs of homologous bases are just beyond the flanking single mismatches, the oligo may still bind sufficiently to inhibit translation or splicing. High CG content can make shorter homologous sequences active, since CG pairs are more stable than AT pairs. Distributing five mismatches throughout a 25‐mer almost always results in loss of knockdown at low concentrations, so 5‐mispair oligos are commonly used as specificity controls. If all five mismatches are at one end of the oligo, there are still 20 contiguous complementary bases in a 25‐mer, and those 20 bases would retain considerable antisense activity. When checking a Morpholino target against a sequence database and finding a partially homologous region, following a rule of thumb like “<80% homology won't cause off‐target knockdown” can lead to trouble; it is important to consider the distribution of the mismatches.
Additional factors to consider when analyzing partially homologous targets are that losing a CG pair due to a mismatch impacts the oligo activity more than losing an AT pair (three H‐bonds compared to two), and that mismatches sometimes form GT pairs, which contribute about half the stability of an AT pair (Aboul‐ela et al., 1985).
Delivery of Morpholinos to the cytosol/nuclear compartment of cells
Unmodified Morpholinos. Since unmodified Morpholinos diffuse between the cytosol and the nucleus, delivery of Morpholinos to the cytosol is sufficient to ensure entry into the nucleus (Morcos, 2001). However, unmodified Morpholinos do not readily diffuse across the plasma membrane of most cell types. If unmodified Morpholinos are added to cell cultures without delivery reagents, high concentrations and long exposure times must be used to achieve minimal delivery (Sazani et al., 2001). Further demonstrating plasma membrane impermeability, when Morpholinos are microinjected into one blastomere of a Xenopus laevis embryo at the two‐cell stage, daughter cells of the injected cell will contain Morpholino activity while daughter cells of the uninjected cell contain no detectable Morpholino activity (Nutt et al., 2001). There have been some reports of particular cell types in tissue explants that take up experimentally useful concentrations of unmodified Morpholinos. These cell types include epithelial cells in mouse embryo pancreatic explant cultures during E11 through E13 (Prasadan et al., 2002) and liver cells in mouse embryo E10 liver explants (Monga et al., 2003). Using an engineered mouse with a stably integrated green fluorescent protein (GFP) up‐regulation splice‐correction reporter system (see Up‐regulation system, below), Sazani showed that there is scant uptake of unmodified Morpholinos into most tissues from the blood of adult mice (Sazani et al., 2002).
Scrape loading. Scrape loading of Morpholinos into adherent cell cultures was an early method for introducing Morpholinos into cultured cells (Partridge et al., 1996). When adherent cells are gently lifted from the bottom of a well using a soft rubber scraper, the cells become transiently permeable, allowing Morpholinos to diffuse into the cytosol from the medium. This technique will not deliver oligos to all cells in a culture, and reproducibility depends on the technique of the experimenter. This method has fallen out of favor as more reproducible techniques producing more homogeneous delivery have been developed.
Microinjection. Microinjection of Morpholinos into early embryos is a widely used technique for knocking down gene expression. Microinjection introduces Morpholinos directly into the cytosol. As the cytoplasm is apportioned into daughter cells at cell division, both daughter cells will contain Morpholinos. Some embryos, such as Xenopus sp., have strong permeability barriers that prevent appreciable leakage to the daughters of uninjected cells (Nutt et al., 2001). Other embryos such as the zebrafish, Danio rerio , allow diffusion of Morpholinos between cells through the first few cell divisions (for a good model of Morpholino diffusion in zebrafish embryos, see Kimmel and Law, 1985a,b).
Electroporation. Electroporation has become a standard method for delivery of Morpholinos into chick embryos (Kos et al., 2003), especially for studies of neural tube development (Tucker, 2004). Electroporation has also been used to deliver Morpholinos into other embryos including mice (Mellitzer et al., 2002), into brains of developing rats (Takahashi et al., 2002), into zebrafish (Cerda et al., 2006), into clipped fins of zebrafish (Thummel et al., 2005), and into cell cultures (Jubin, 2005). Uncharged Morpholinos can be electroporated; the electroporation procedure makes cells transiently permeable so that Morpholinos can diffuse across the plasma membrane.
Endo‐Porter. Endo‐Porter is a reagent developed to deliver Morpholino oligos conveniently and reproducibly to the cytosol of cultured cells through an endocytotic pathway. Endo‐Porter is an amphiphilic peptide that becomes cationic at low pH. In culture medium at neutral pH, Endo‐Porter is uncharged but sticks to the surface of cells. Upon endocytosis, Endo‐Porter is protonated in the acidic endosome and permeabilizes the endosomal membrane, releasing the endosomal contents into the cytoplasm. Morpholinos co‐endocytosed with membrane‐associated Endo‐Porter are released into the cytoplasm when the endosome is permeabilized (Summerton, 2005). Endo‐Porter allows simultaneous delivery of multiple Morpholinos. The concentration of Morpholinos can be varied independently of the Endo‐Porter concentration, allowing dose‐response antisense studies while holding the delivery reagent concentration constant. Cells treated with a 5 µM carboxyfluoresceinated Morpholino and 8 µM Endo‐Porter gave transfection efficiencies of 82% for human amnion‐derived WISH cells and 78% for human myometrial cells when assayed by confocal microscopy (Tyson‐Capper and Europe‐Finner, 2006), although concentrations too low to be detected by fluorescence might still be sufficient to have measurable antisense activity. Endo‐Porter has been used successfully with traditionally hard‐to‐transfect cells such as cardiomyocytes (Masaki et al., 2005). It works well with unmodified Morpholinos or carboxyfluoresceinated Morpholinos, but best delivery is achieved with lissaminated Morpholinos (S.T. Knuth, unpub. observ; see Critical Parameters, End modifications). Endo‐Porter is commercially available in neat DMSO or in a less‐effective aqueous formulation for cells sensitive to DMSO.
The recommended concentration of Endo‐Porter is 6 µM, achieved by using 6 µl of a 1 mM Endo‐Porter solution per milliliter of cell culture; this concentration gives good delivery without toxicity to many cell types. However, cell types vary in their tolerance to Endo‐Porter, with some cells tolerating higher exposures while other cells are harmed by a 6 µM Endo‐Porter solution. When trying Endo‐Porter with a new cell type, it is prudent initially to test a range of Endo‐Porter concentrations (e.g., 2, 4, 6, and 8 µM) to assess delivery and to check the tolerance of the cells for the reagent.
Special Delivery. Morpholinos are sometimes delivered using cationic delivery reagents, such as ethoxylated polyethylenimine (EPEI) or Lipofectamine. However, since Morpholinos are not charged they will not form electrostatic complexes with cationic delivery reagents. Without such complexation, the Morpholinos are poorly delivered to the cytosol of treated cell cultures. To overcome this limitation, Morpholinos can be annealed to complementary or partially complementary strands of anionic nucleic acids. Special Delivery oligos are heteroduplexes of Morpholinos and partially complementary DNA, and are delivered after complexation with a cationic delivery reagent, usually EPEI (Morcos, 2001). Special Delivery oligos were designed as a replacement for scrape loading of adherent cells, but can also be used with cells in suspension. Special Delivery oligos provide a more homogeneous delivery than scrape loading, and many studies have been published using them. However, several problems are inherent in the system: (1) EPEI is somewhat toxic to cells; (2) the concentration ratio of heteroduplex to EPEI is fixed; (3) only a single oligo sequence can be delivered at an effective concentration at any one time; and (4) the complexation procedure, which must be done prior to each delivery, adds complexity and variability to the experiment. Special Delivery oligos can be made by following a fairly simple design and hybridization protocol. This approach has mostly been supplanted by Endo‐Porter, which is simpler to use, more versatile, more effective, and less toxic in most cell types.
Peptide conjugates. Cell‐penetrating peptides covalently conjugated to Morpholinos are in development to enhance cytosolic delivery of Morpholinos in cell culture (Neuman et al., 2005)and in vivo (Kinney et al., 2005; Neuman et al., 2005). Most published research describing Morpholino‐peptide conjugates has used arginine‐rich peptides (Moulton et al., 2004; Neuman et al., 2004; Deas et al., 2005; Kinney et al., 2005; Nelson et al., 2005; McClorey et al., 2006). Conjugation with arginine‐rich peptides alters the specificity, target affinity, and toxicity of Morpholinos (Nelson et al., 2005).
Due to the high density of cationic charges on the peptide moiety, Morpholinos conjugated with arginine‐rich peptides associate with subcellular structures and with outer cell surfaces. This property might lead to false‐positive artifacts when assessing delivery of arginine‐conjugated peptides by fluorescence‐based methods, such as fluorescence microscopy, fluorometry, or flow cytometry. To determine the concentration of an internalized conjugate using fluorescence‐based methods, the membrane‐associated conjugate should be removed in order to avoid overestimation. Trypsin treatment has been effective for eliminating binding of Morpholino‐peptide conjugates to the outside of cells (Moulton and Moulton, 2003).
Vivo‐Morpholinos. An octaguanidinium group attached to the 3′‐end of a Morpholino oligo has delivered splice‐inhibiting antisense activity to the nuclei of cells in mice after intravenous administration (Li and Marcos, 2008).
Minimum effective Morpholino concentration
To avoid off‐target knockdowns, the lowest concentration of Morpholino producing the desired knockdown should be determined. When delivering Morpholinos to cell cultures using Endo‐Porter, starting with a 10 µM Morpholino concentration for both fluorescent delivery assays and functional experiments increases the chances that the fluorescence will be visible in the cytosol and that the first functional experiment will produce measurable results. Because a Morpholino concentration of 10 µM might cause nonspecific effects due to interaction with nontarget genes, functional assays should be performed using a range of Morpholino concentrations. Determining the minimum Morpholino concentration that produces measurable results allows one, subsequently, to avoid off‐target knockdowns and to conserve oligo. Effective Morpholino concentrations in culture medium for knockdown experiments are typically in the 1 to 10 µM range.
Simultaneous oligo strategy
Oligos can sometimes be delivered together to enhance their effects. Pairs of nonoverlapping translation‐inhibiting Morpholinos targeting the same mRNA can be used simultaneously in order to decrease the concentration required for a knockdown (Ekker and Larson, 2001; Kamachi et al., 2008). If the paired oligos are simultaneously introduced into the same cells, they are sometimes effective at much lower concentrations than for either oligo alone. If oligos are individually toxic in zebrafish, their use in combination at concentrations below their toxicity thresholds might elicit the desired phenotype without toxicity. Efficiency of splice inhibition can be increased by inhibiting both donor and acceptor splice sites flanking a single exon (Morcos, 2007). Targeting several exons simultaneously is an effective way to deplete a wild‐spliced mRNA (Draper et al. 2001). When designing oligos intended for co‐delivery, check for complementarity between oligos that may cause them to form Morpholino heterodimers and lose activity (see Troubleshooting, Oligo activity decreases with pairs of oligos).
Assessing oligo delivery
It is best to begin a set of Morpholino experiments in a cell line by confirming and optimizing delivery. Most experimental problems involving Morpholinos in cell culture are due to insufficient delivery of oligo and can be solved by optimizing delivery to the particular type of cells used. Checking whether good cytosolic delivery can be achieved before starting to use custom‐made Morpholinos is usually the most efficient use of time and resources.
By fluorescence. Fluorescence can be measured by fluorescence microscopy, flow cytometry, or fluorometry. Only fluorescence microscopy can distinguish cytosolic and nuclear fluorescence (indicating successful delivery of a fluorescent Morpholino) from endosomal or surface‐bound fluorescence (which does not contribute to antisense activity). A fluorescence microscope and a fluorescently labeled marker such as a Morpholino or a 10‐kDa dextran are required for a reliable delivery assay. Using a 10‐kDa fluoresceinated dextran or a carboxyfluoresceinated standard Morpholino control before using a more expensive, custom‐made Morpholino produces reliable uptake assays at reduced cost. After delivery, live cells may be conveniently observed using an inverted epifluorescence microscope. Fixing cells can lead to false positives for delivery due to permeabilization of the plasma membrane and release of the oligo from endosomes during fixation. Using an objective with a higher numerical aperture increases the amount of light gathered from a cell and helps reveal dim fluorescence. If diffuse fluorescence is seen throughout the cytosol of the cells, the Morpholino has been delivered successfully. Bright punctate spots are likely labeled oligos trapped in endosomes. Punctate fluorescence does not indicate delivery, but it does not preclude it either.
For delivery with Endo‐Porter, start by assaying a range of Endo‐Porter concentrations for delivery efficacy and cell tolerance (see Basic Protocol 3) or by trying a concentration of 6 µM Endo‐Porter in the selected cell culture. After Endo‐Porter delivery, antisense activity can be detected using as little as 1 µM Morpholino. However, although antisense activity can be achieved at Morpholino concentrations that do not produce detectable fluorescence, visual proof‐of‐delivery assays do require detectable fluorescence. To accumulate enough fluorescence for microscopy, a concentration of about 10 µM Morpholino is needed. The Endo‐Porter and labeled Morpholino should be left on the cells overnight to allow time for endocytotic uptake and accumulation.
By measuring antisense activity. If delivery is successful and a Morpholino targeting translation or splicing works as designed, a decrease in protein concentration or a shift in RT‐PCR product mass (respectively) can be measured (Draper et al., 2001; Stancheva et al., 2003). Successful delivery might also be indicated by phenotypic effects, such as a decrease in targeted enzyme activity (Hayashi et al., 2005) or a change in morphology (Ekker, 2000). However, assaying only for a phenotypic effect becomes problematic if the expected change in phenotype does not occur; if antisense activity is not separately assessed at the level of protein concentration or mRNA mass, the experimenter will not be able to discern whether (1) the oligo failed to reach and interact with its target mRNA to produce the knockdown or splice inhibition or (2) the knockdown or splice inhibition was successful but did not cause the expected phenotypic change.
Assaying translation‐inhibition activity
Activity of translation‐inhibiting Morpholinos can be assayed using immunoblots. However, while Morpholinos can halt new translation, they do not cause degradation of existing protein; it therefore takes some time after Morpholino treatment before immunoblots will show evidence of a knockdown. The time required will vary with the half‐life of the protein.
If no antibody is available for the protein product when targeting an mRNA for translation inhibition, then indirect assays such as the change in phenotype of an embryo must sometimes be used to assess the effectiveness of translation inhibition. Morpholinos can phenocopy (mimic the phenotype of) many known mutations that affect morphology during development; embryos with phenotypes modified by Morpholino treatment are known as morphants (Ekker, 2000).
In some cases, the enzymatic activity of a target protein can be assayed (Hayashi et al., 2005). An enzyme activity assay may serve as an assay for Morpholino activity, although there must be a delay between application of the Morpholino and the enzyme activity assay to allow for degradation of pre‐existing protein (see also the discussion of complementation in Troubleshooting).
The effect of a Morpholino on target RNA stability varies with the sequence. Target mRNA concentrations in cells treated with translation‐inhibiting Morpholinos may be decreased, unchanged, or increased relative to untreated cells. Changes in mRNA concentrations may be due to changes in the secondary structure of the mRNA on binding a Morpholino, thereby altering the availability of the mRNA for nucleolytic degradation. Consequently, mRNA assays such as Northern blots or RT‐PCR are not suitable for assaying the activity of a translation‐inhibiting Morpholino.
Assaying splice‐inhibiting activity
Because inhibiting splicing changes the mass of the mRNA produced, RT‐PCR with appropriate choice of primers is a good molecular assay for detecting the activity of splice‐inhibiting Morpholinos. However, it is important to keep in mind that it cannot be predicted with certainty whether a splice‐inhibiting Morpholino will cause an exon deletion (most common), an intron insertion, or activation of a cryptic splice site (which can cause a partial insertion or deletion). Because cryptic sites often redirect splicing of only a fraction of the targeted pre‐mRNA population, splice inhibition might produce a mixture of RT‐PCR product masses (Draper et al., 2001). To detect any of these changes, it is best to use primers targeted to the two exons flanking and closest to, but not including, the Morpholino's splice junction. Targeting a splice junction on an internal exon is likely to cause exon deletion. Primers should be chosen so that, if an exon deletion occurs, the RT‐PCR product will be large enough to detect easily on a gel (one hundred to several hundred bases). That means for the system exon1–intron1–(splice inhibitor target)–exon2–intron2–exon3, the RT‐PCR primers should be targeted to exon 1 and exon 3 in order to detect either intron 1 insertions (unusual) or exon 2 deletions (common).
If the first (most 5′) or last (most 3′) splice junction in an mRNA is targeted, the usual result is an intron insertion instead of an exon deletion. However, targeting the first splice junction might activate a cryptic splice site in the first exon or intron, resulting in deletion of the 3′ end of the first exon or inclusion of a 5′ fragment of the first intron. When targeting the last exon, an intron insertion is a more likely outcome. This is because consensus sequences of splice acceptors are more complex than those of splice donors, so it is less likely that the last exon will contain a near‐consensus cryptic splice acceptor.
When assaying the activity of a splice‐inhibiting Morpholino at the molecular level using RT‐PCR, it is important to compare the expected size of the RT‐PCR product after the splice modification with the size of the RT‐PCR product produced by an untreated cell or organism. For easiest detection, splice‐inhibited RT‐PCR products would be about half or twice the size of the native‐spliced product (for exon deletion or intron insertion, respectively). A real system usually will not allow such a tidy result, but it is necessary for the change in mass to be clearly visible on the gel (e.g., a 5% change in mass can be difficult to detect).
When possible, primers should amplify RT‐PCR products with lengths of hundreds of bases to ensure full‐length replication and visible bands. Since fragments should be large enough that they are clearly visible, it is prudent to select primer targets set back from the splice junctions into the exons flanking the targeted exon.
Splice modifications can cause downstream frame shifts or inclusion of intronic sequence in the mature messenger. Either can cause a range of complicating effects, including truncation of the protein product by the appearance of in‐frame stop codons, translation suppression by the appearance of a miRNA target site, degradation or suppression of the messenger through siRNA or miRNA activity, and nonsense‐mediated mRNA decay. Rapid decay of a splice‐modified messenger may suppress the appearance of an electrophoretic band corresponding to the RT‐PCR product from the splice‐modified transcript; in this case, the band corresponding to the wild‐type spliced RT‐PCR product may be dimmed or disappear when splice modified without the appearance of the mass‐shifted band expected from splice modification.
Splice modification may or may not cause a change detectable by an immunochemical assay such as an ELISA or immunoblot, since the conformation of the modified protein around the antigenic site may or may not be changed by a splice modification. A large insertion or deletion might result in loss of antibody binding or at least a significant shift in the band position on a western blot, but a small insertion or deletion could be difficult to detect.
It is possible that inserting an intron or deleting an exon will cause the protein product to lose function, but it is far from certain. If the active site of the protein is known and the exon encoding the active site is targeted, a loss of function is likely. However, if the active site is not known, then splice‐inhibition might not change the protein's activity. A protein might be made that retains the conformation of its active site even though it has an inserted or deleted polypeptide moiety at a different part of the protein. This means that looking for a phenotypic change in an embryo or assaying enzyme activity is often inadequate for assessing the splice‐inhibiting activity of a Morpholino. This also means that while RT‐PCR is a useful tool to confirm splice‐inhibiting activity, one should independently assay for protein function before concluding that a targeted gene is not required for a biological process, because successful splice inhibition may not alter the activity of the protein in the process.
Up‐regulation system
Assaying antisense activity by knocking down a protein can lead to false positives, because toxicity can cause a decrease in gene expression and this can be misinterpreted as targeted gene knockdown unless careful controls are used. To address this problem and to provide an increased signal‐to‐noise ratio for antisense activity assays, Ryzard Kole's group developed a set of signal up‐regulation reporter systems based on splice modification. These systems use a mutation in human β‐globin that creates a new splice site and causes thalessemia. The splice mutant has a stop codon in‐frame in the mRNA as well as a frameshift in the downstream coding region; inhibiting the mutant site splices out the stop and restores the correct reading frame. Constructs coupling this mutation to luciferase or GFP have been engineered. Of particular interest are the pLuc705 HeLa cell line (Schmajuk et al., 1999), which expresses luciferase when the mutant splice site is splice inhibited with control oligo, and the Sazani mouse (Sazani et al., 2002), which expresses GFP when splice inhibited with the appropriate oligo.
Controls
When an oligo is used to target an mRNA, a parallel experiment should be done using a negative control oligo. Negative control oligos include the standard control oligo, the oligo‐N control, and an invert oligo (see below). A 5‐mispair specificity control oligo is also sometimes used as a negative control. The negative control shows that the effects observed during the antisense experiment are due to the sequence of the targeting oligo and not to the backbone chemistry of the Morpholino or the cytosolic delivery method used.
Standard control oligo. A standard control Morpholino with the sequence CCTCCTACCTCAGTTACAATTTATA has been used in many organisms as a negative control sequence without triggering off‐target or non‐antisense effects. This negative control produced no toxic or teratogenic effects even when administered at considerably higher concentrations than typically used for specific knockdown experiments. Any custom‐sequence control oligo has some risk of interacting with off‐target RNA; in contrast, the standard control has an established history of inactivity and is a reliable choice for a negative control oligo. The standard control Morpholino is designed to inhibit the mutant splice site used in the pLuc705 up‐regulation reporter system.
Oligo‐N control. A mixture of oligo sequences formed by synthesizing a 25‐base Morpholino oligo using a mixture of all four subunits delivered at each synthetic cycle is called an oligo‐N. These mixed oligo preparations, of sequence NNNNNNNNNNNNNNNNNNNNNNNNN, can be used as negative controls because the concentration of any given sequence in the mixture is too low to have biological activity.
Invert control. If a negative control oligo needs to have the same base composition as the custom‐made targeting oligo, the invert oligo is a good choice of sequence. The invert has the same base sequence as the targeting oligo, but the sequence is reversed in the 5′‐to‐3′ orientation (i.e., 5′‐ACGGTGC would become 5′‐CGTGGCA). The advantage of an invert over a scrambled sequence is that the invert sequence can be conveniently generated by a simple algorithm and will have the same CG content, G content, and self‐complementarities as the targeting oligo. However, there is always a risk with any custom‐made oligo that the oligo may interact with unintended RNAs.
Sense control. Sense Morpholinos have sometimes caused an increase in concentration of the mRNA targeted by an antisense oligo (P.A. Morcos, unpub. observ.). Thus, a sense sequence is not a good choice for a negative control Morpholino.
5‐Mispair oligo. Off‐target knockdown by any antisense molecule increases with increasing concentration. A 5‐mispair oligo is used to define the effective and specific concentration window for a targeting oligo. Using the targeting oligo within its effective and specific concentration range decreases the chance of causing experimental artifacts by interaction with off‐target RNA. A 5‐mispair control oligo has nearly the same sequence as a targeting oligo, but has five mismatched bases distributed through the sequence. The mismatches should be distributed fairly evenly through a 25‐mer oligo, starting a few bases in from each end. Ideally, the mismatches should be formed by exchanging C for G and G for C, since these mismatches disrupt the formation of three hydrogen bonds per base pair. When a targeting oligo is used near the lowest concentration that produces a discernable effect, most 5‐mispair oligos used at the same concentration will not produce the effect. However, as with any custom‐sequence control oligo, there is a possibility that the 5‐mispair oligo will interact with an untargeted RNA, triggering an off‐target effect. If an oligo targets a high‐copy‐number transcript, requiring relatively high Morpholino concentration for knockdown, and the mispair oligo interacts with a low‐copy‐number transcript, the mispair oligo might cause effects even at concentrations below the concentration at which the targeting oligo becomes effective.
The 5‐mispair oligo can be used in an experiment that determines the effective and specific window of concentrations for a targeting oligo, which is the concentration range between the onset of measurable activity for the targeting oligo and the onset of measurable activity for its 5‐mispair oligo. The definition of the effective and specific concentration range based on a 5‐mispair oligo evolved through trial and error. Originally a 4‐mispair specificity control was recommended, but a 4‐mispair oligo sometimes measurably decreased the target protein concentration at concentrations low enough that the corresponding targeting oligo was just becoming effective, so there was not a wide enough effective and specific concentration range to be consistently useful. Many investigators use the 5‐mispair oligo as a negative control, but that was not its intended purpose. Assuming that adding the mismatches does not create too much complementarity to an important off‐target mRNA and trigger an off‐target knockdown, the 5‐mispair oligo usually behaves as a negative control when used at concentrations low enough that the targeting oligo is just becoming effective. However, the 5‐mispair oligo is intended as a specificity control to be used in an experiment to determine the effective and specific concentration window. The 5‐mispair oligo is intended as a specificity control that shows the targeting oligo is being used in its effective and specific range. To demonstrate specificity of the targeting oligo, the targeting oligo, a 5‐mispair control oligo, and a true negative control oligo (such as the standard control) are used at the same concentration in parallel treatments. If the 5‐mispair and negative control oligos produce the same results, and the targeting oligo produces a different result, the experiment indicates that the targeting oligo has been used within its effective and specific concentration range. Appearance of an effect due to interaction of a target RNA with the 5‐mispair control oligo suggests that, at that same concentration, the targeting oligo might also interact with off‐target RNAs.
Two nonoverlapping translation inhibitors. Another strategy for showing that the effect of a translation‐inhibiting Morpholino is due to the knockdown of its targeted mRNA is to use a second oligo targeted to a different and nonoverlapping site in the 5′‐UTR of the targeted mRNA. If the second oligo has the same effect on the cells (or organism) as the first, this supports the hypothesis that the effect observed is due to the knockdown of the targeted gene.
Two splice inhibitors targeting one internal exon. If a Morpholino targeting a splice donor site produces the same result as a Morpholino targeting the splice acceptor of the same exon, this supports the hypothesis that the effect observed is due to the excision of the targeted exon. However, failure of the two oligos to produce the same result may be due to activation of a cryptic splice site(s) by one or both of the oligos.
mRNA rescue. A very strong proof of specificity involves the use of a rescue mRNA. A rescue mRNA codes for the same protein targeted by the Morpholino knockdown, but has a modified 5′‐UTR that is not targeted by the Morpholino. For this experiment, the rescue mRNA and Morpholino are delivered to the cytosol together. If the co‐delivered rescue mRNA and Morpholino produce the same wild‐type phenotype as untreated cells or organisms, this supports the hypothesis that the morphant phenotype elicited by the Morpholino alone is due to interaction with the targeted RNA. Unfortunately, the mRNA rescue experiment cannot work for some genes when used in embryos. The timing of the onset of translation is crucial for some developmental genes, and the early onset of translation resulting from co‐injection of Morpholino and rescue mRNA in the early zygote may alter the developmental process so that these embryos never recapitulate the wild‐type phenotype. Furthermore, the location of gene expression is often crucial for development, but oocyte microinjection causes rescue mRNAs to be present in all cells of the embryo until degraded or diluted by growth.
End modifications
Several optional modifications attached to the ends of Morpholinos are commercially available. Carboxyfluorescein, lissamine, and primary amines (Fig. 4) are the most commonly used. Optional groups are usually added to the secondary amine on the 3′‐end of the oligo and are assumed to be 3′ modifications unless explicitly declared to be 5′ modifications. Fluorophores and biotin are attached to Morpholinos through flexible polyethylene‐glycol spacers. The length of the spacers was chosen based on antisense activity studies to ensure that the fluorophores would not interfere with binding of the Morpholinos to their target RNA sequences. The primary amine modification includes a short spacer of two methylenes.
Figure 4.
Commercially available 3′‐end modifications of Morpholino oligos.
Carboxyfluorescein. Carboxyfluorescein is a green‐emitting fluorophore that was chosen from among the fluoresceins for its good chemical stability. While its photostability is better than that of many of the fluoresceins, all of the fluoresceins are subject to photobleaching, so carboxyfluorescein should not be exposed to intense light unnecessarily. The excitation wavelength of a carboxyfluoresceinated Morpholino in water is 502 nm, and its emission wavelength is 525 nm. Carboxyfluorescein has two negative charges at neutral pH.
Lissamine. Lissamine is a red‐emitting sulforhodamine B. The excitation wavelength of a lissaminated Morpholino in water is 575 nm, and the emission wavelength is 593 nm. Lissamine is a zwitterion at neutral pH, with one positive and one negative charge. Adding a lissamine to a Morpholino increases its delivery efficiency with Endo‐Porter, but adding lissamine to a Morpholino can decrease its aqueous solubility. It is therefore recommended to use a carboxyfluorescein tag when a fluorochrome is needed, especially for Morpholino sequences with relatively high G contents (>30% G).
Primary amine. Morpholinos may be modified with a primary amine to provide a reactive site for attachment of other moieties to the oligo. An unmodified Morpholino has a secondary amine on the 3′ end, the pK a of which is 6.5. The primary amine, with its pK a of 10.2, provides a more reactive site. When a primary amine is attached to the 3′ end of the oligo, this converts the 3′‐secondary amine of the morpholine ring to a tertiary amine as a consequence of the attachment of a short spacer tethering the new primary amine. When a primary amine is attached to the 5′ end of the oligo, the 3′‐secondary amine is acetylated so that a reagent added to react with the primary amine will not react with the 3′ end of the oligo. When reacting a primary amine with a derivatizing reagent, it is prudent to include an additional short spacer to prevent steric hindrance between the moiety being added and the Morpholino.
Morpholino stock solutions and reconcentrating Morpholinos
Morpholino stock solutions in distilled water should be kept sterile; they can be autoclaved. Do not use water containing diethylpyrocarbonate (DEPC), which can modify Morpholino bases. Morpholino stock solutions can be dissolved in buffers such as Ringer's solution or Danieau buffer, but this can cause problems later if the stock solution must be reconcentrated because lyophilization can be more difficult from a buffer. The presence of additional solutes makes analysis of Morpholino mass by MALDI‐TOF mass spectrometry more difficult. Finally, Morpholinos are substantially more soluble in distilled water than in isotonic salt solutions.
A solution of Morpholino in water can be concentrated by using a Speedvac or by lyophilization (freeze‐drying). Lyophilizing Morpholinos from water produces a fluffy solid that dissolves fairly readily if the sequence has good solubility properties. However, dissolution of Morpholinos concentrated with a Speedvac may be more difficult and will likely at least require patience and heating to 65°C.
Temperature during handling
Morpholinos are not degraded by nucleolytic enzymes. Solutions of DNA and RNA are normally kept on ice during experiments to prevent nucleolytic degradation, but this is not a concern with Morpholinos. However, some Morpholino solutions have low enough solubility that icing a solution may cause a loss of activity, due to the oligo coming out of solution. Therefore, icing Morpholino solutions is not only unnecessary, but it can cause problems; Morpholinos should be kept at room temperature during experiments.
Material affinity
Morpholinos have some affinity for plastics, so passaging very dilute (submicromolar) solutions through plastic containers may cause appreciable decreases in activity. Similarly, filter sterilization may cause Morpholino solutions to lose some activity as some oligo binds to the filter. When put through the same procedures with the same exposure to plastic surfaces, high‐concentration Morpholino solutions have a smaller fractional decrease in concentration than low‐concentration Morpholino solutions. Therefore, if exposure to plastic surfaces is required, it is best to do the procedures with Morpholinos in a relatively concentrated state (>1 µM). Similarly, if Morpholinos are to be stored in solution for more than a few days, it is best to store them at high concentration. Since solutions of Morpholinos at very low concentrations (<1 µM) may lose activity over a time scale of minutes to hours, dilutions should be made just before use. If Morpholino solutions of less than ∼100 µM are filtered, the concentration may be affected appreciably given the large surface area of filters. As the oligo bound is proportional to the surface to which it is exposed, a small‐diameter filter should be used to minimize oligo losses. Pall Acrodisc HT Tuffryn 0.2‐µm membrane filters were found to bind less Morpholino per area than other filters tested (J.E. Summerton, unpub. observ.). The concentration of oligo in a solution can be measured spectrophotometrically just before and after performing a procedure to determine the loss of oligo caused by the procedure.
Troubleshooting
Loss of antisense activity over time
Morpholinos can be safely stored at temperatures ranging from room temperature to −80°C. Some Morpholino solutions lose activity when stored frozen, not due to degradation of the oligos but simply to aggregation. The activity can sometimes be recovered by heating the solution to 65°C for 10 min prior to use. If necessary, stock solutions of unmodified, fluorescent‐tagged, or Vivo‐Morpholino can be autoclaved to help redissolve the oligos (turn off the autoclave vacuum cycle to prevent liquid loss).
Loss of fluorescence over time
Fluorescent tags can be photobleached by exposure to bright light or prolonged exposure to dim light. Always store fluorescent materials in the dark. Wrapping aluminum foil around tubes containing fluorescent materials is an easy and prudent method for protecting fluorophores. Fluorescent materials can autoquench at high concentrations and decrease their light emission, so do not check for fluorescence at very high concentration. Labeled Morpholinos at 10 µM are well below the concentration at which their fluorophores autoquench.
No apparent activity
If a Morpholino does not produce the anticipated result, there are several possibilities to consider. Has delivery been confirmed? If the oligo is not reaching the cytosol of the cells, no antisense activity will be observed. Is the activity checked by a molecular assay? If the selected activity assay depends upon observation of a phenotype, such as a change in embryo morphology or in enzyme activity, the oligo may be successfully knocking down translation or modifying splicing, but a second protein may be complementing the lost activity of the target protein, thereby confounding the assay. Assaying translation inhibition by immunoblot and splice inhibition by RT‐PCR can help determine whether the oligo is not interacting with its target or has not been delivered, or whether there is a more subtle reason for the failure to produce the expected phenotype, such as complementation by another protein. Feedback up‐regulation can also cause a knockdown to fail; greatly increased transcription of the targeted mRNA in response to an attempted knockdown can overwhelm the ability of the oligo to inhibit all of the targeted messengers.
Oligo activity decreases with pairs of oligos
When two or more oligos are together in a cell, they may hybridize with each other if they share complementary sequences. If a pair of oligos has less activity than each individual oligo, check the sequences for complementarities. Sixteen contiguous hydrogen bonds of complementarity is the maximum recommended for oligos used together in cells or organisms at 37°C.
Clogging microinjectors
If a Morpholino solution causes a microinjector to clog, one can: (1) heat the solution to disrupt tiny clumps (65°C for 10 min), (2) filter‐sterilize the solution (although some oligo may be lost on the filter), (3) try injecting a higher volume of a less concentrated solution, or (4) give the oligo solution a quick spin in a centrifuge to remove the precipitate.
Anticipated Results
Translation inhibitors
If a translation‐inhibiting Morpholino knocks down expression of a protein, this activity can be revealed by a delayed decrease in the protein signal on an immunoblot using an antibody to the protein. The successful knockdown should also decrease the activity of the targeted protein, although an assay for the activity of that protein can be confounded by complementation by another protein.
Splice inhibitors
If a splice‐inhibiting Morpholino changes the mass of an mRNA, this activity can be revealed soon after delivery by a change in the mass of an RT‐PCR product produced using appropriately chosen primers. A successful exon excision should also result in a delayed decrease of the activity encoded by the deleted exon as pre‐existing protein degrades. Splice inhibition may also decrease activities encoded on untargeted exons of a target RNA due to frameshifts, inserted stop codons, or changes in tertiary structure.
Time Considerations
After delivery, wait for antisense effects to be measurable
When a Morpholino is delivered into the cytosol of a cell, pre‐existing protein is not altered by the Morpholino. For a translation‐inhibiting oligo, this means that even if translation of a protein is immediately and completely halted on Morpholino delivery, an assay for protein concentration will not immediately reveal a successful knockdown. A considerable fraction of the population of the existing protein molecules must be degraded before the knockdown will be evident on an immunoblot. Similarly, although a splice‐inhibiting oligo may cause a rapid change in the mass of an RT‐PCR product, the protein produced prior to splice inhibition will persist in the cell until degraded.
Delivery systems using endocytotic uptake, such as Endo‐Porter or Special Delivery, increase the lag between the start of delivery and the appearance of a knockdown or splice‐inhibition signal. An overnight wait is generally sufficient to allow for endocytotic uptake.
For embryonic studies, the presence of maternal transcripts in a zygote may delay the loss of protein activity when splice‐inhibiting Morpholinos are used. Although a splice inhibitor can modify splicing of pre‐mRNA transcribed in the zygote, maternal transcripts are already spliced before the onset of zygotic transcription and will be expressed in their unmodified form. Translation inhibitors can inhibit both maternal and zygotic transcripts, and can thus provide more rapid knockdown of protein activity than splice inhibitors. Often inhibition of translation of maternal and zygotic transcripts produces a more severe phenotype than a splice‐inhibiting Morpholino targeting the pre‐mRNA of the same transcript.
Redelivery
When translation of a protein is inhibited by a Morpholino, existing protein in the cell persists until broken down. After delivery, as cells grow and divide, the concentration of Morpholino oligos in their cytoplasm decreases due to dilution. Because of these two processes, when Morpholinos are used to knock down genes that code for unusually abundant or stable proteins, redelivery of the oligos may be required before a significant decrease in protein levels can be detected by immunoblotting. After an initial treatment with a Morpholino at the start of day 1, redelivery on day 4 usually suffices to produce a clear knockdown of stable and abundant proteins by day 6. However, attempts to inhibit translation of actin have so far failed to produce a decrease in actin levels on immunoblots, suggesting that Morpholinos cannot knockdown some very abundant proteins (P.A. Morcos, unpub. observ.).
Acknowledgements
Thanks to Dr. Shannon T. Knuth for editing and for information from protocols she has written, and to Conrad Shultz and Drs. John Postlethwait, James E. Summerton, Hong M. Moulton, Yongfu Li, and Paul A. Morcos for their editing and suggestions.
Literature Cited
- Aboul‐ela, F. , Koh, D. , Tinoco, I. Jr. , and Martin, F.H. 1985. Base‐base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 13:4811‐4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, M. , Stein, D.A. , Thomann, E. , Moulton, H.M. , Leong, J.C. , Iversen, P. , and Mourich, D.V. 2005. Inhibition of infectious haematopoietic necrosis virus in cell cultures with peptide‐conjugated morpholino oligomers. J. Fish Dis. 28:399‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, I.G. , Jin, W. , and Cote, G.J. 2004. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum. Mol. Genet. 13:2409‐2420. [DOI] [PubMed] [Google Scholar]
- Cerda, G.A. , Thomas, J.E. , Allende, M.L. , Karlstrom, R.O. , and Palma, V. 2006. Electroporation of DNA, RNA, and morpholinos into zebrafish embryos. Methods 39:207‐211. [DOI] [PubMed] [Google Scholar]
- Cheong, C. and Moore, P.B. 1992. Solution structure of an unusually stable RNA tetraplex containing G‐ and U‐quartet structures. Biochemistry 31:8406‐8414. [DOI] [PubMed] [Google Scholar]
- Choi, W.Y. , Giraldez, A.J. , and Schier, A.F. 2007. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR‐430. Science 318:271‐274. [DOI] [PubMed] [Google Scholar]
- Deas, T.S. , Binduga‐Gajewska, I. , Tilgner, M. , Ren, P. , Stein, D.A. , Moulton, H.M. , Iversen, P.L. , Kauffman, E.B. , Kramer, L.D. , and Shi, P.Y. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599‐4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, B.W. , Morcos, P.A. , and Kimmel, C.B. 2001. Inhibition of zebrafish fgf8 pre‐mRNA splicing with Morpholino oligos: A quantifiable method for gene knockdown. Genesis 30:154‐156. [DOI] [PubMed] [Google Scholar]
- Ekker, S.C. 2000. Morphants: A new systematic vertebrate functional genomics approach. Yeast 17:302‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker, S.C. and Larson, J.D. 2001. Morphant technology in model developmental systems. Genesis 30:89‐93. [DOI] [PubMed] [Google Scholar]
- Enterlein, S. , Warfield, K.L. , Swenson, D.L. , Stein, D.A. , Smith, J.L. , Gamble, C.S. , Kroeker, A.D. , Iversen, P.L. , Bavari, S. , and Muhlberger, E. 2006. VP35 knockdown inhibits ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50:984‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller, B.L. , Deere, J. , Tilley, L. , and Iversen, P.L. 2005. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J. Antimicrob. Chemother. 55:983‐988. [DOI] [PubMed] [Google Scholar]
- Giles, R.V. , Spiller, D.G. , Clark, R.E. , and Tidd, D.M. 1999. Antisense Morpholino oligonucleotide analog induces missplicing of c‐myc mRNA. Antisense Nucleic Acid Drug Dev. 9:213‐220. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y. , Horibata, Y. , Sakaguchi, K. , Okino, N. , and Ito, M. 2005. A sensitive and reproducible assay to measure the activity of glucosylceramide synthase and lactosylceramide synthase using HPLC and fluorescent substrates. Anal. Biochem. 345:181‐186. [DOI] [PubMed] [Google Scholar]
- He, J. , Liu, G. , Zhang, S. , Rusckowski, M. , and Hnatowich, D.J. 2003. Pharmacokinetics in mice of four oligomer‐conjugated polymers for amplification targeting. Cancer Biother. Radiopharm. 18:941‐947. [DOI] [PubMed] [Google Scholar]
- He, J. , Liu, G. , Gupta, S. , Zhang, Y. , Rusckowski, M. , and Hnatowich, D.J. 2004. Amplification targeting: A modified pretargeting approach with potential for signal amplification‐proof of a concept. J. Nucl. Med. 45:1087‐1095. [PubMed] [Google Scholar]
- Heasman, J. , Kofron, M. , and Wylie, C. 2000. Beta‐catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev. Biol. 222:124‐134. [DOI] [PubMed] [Google Scholar]
- Henderson, R.E. , Kirkegaard, L.H. , and Leonard, N.J. 1973. Reaction of diethylpyrocarbonate with nucleic acid components. Adenosine‐containing nucleotides and dinucleoside phosphates. Biochim. Biophys. Acta 294:356‐364. [DOI] [PubMed] [Google Scholar]
- Howard, M.T. , Gesteland, R.F. , and Atkins, J.F. 2004. Efficient stimulation of site‐specific ribosome frameshifting by antisense oligonucleotides. RNA 10:1653‐1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudziak, R.M. , Barofsky, E. , Barofsky, D.F. , Weller, D.L. , Huang, S.B. , and Weller, D.D. 1996. Resistance of Morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug. Dev. 6:267‐272. [DOI] [PubMed] [Google Scholar]
- Jubin, R. 2005. Optimizing electroporation conditions for intracellular delivery of Morpholino antisense oligonucleotides directed against the hepatitis C virus internal ribosome entry site. Methods Mol. Med. 106:309‐322. [DOI] [PubMed] [Google Scholar]
- Jubin, R. , Vantuno, N.E. , Kieft, J.S. , Murray, M.G. , Doudna, J.A. , Lau, J.Y. , and Baroudy, B.M. 2000. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 74:10430‐10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi, Y. , Okuda, Y. , and Kondoh, H. 2008. Quantitative assessment of the knockdown efficiency of Morpholino antisense oligonucleotides in zebrafish embryos using a luciferase assay. Genesis 46:1‐7. [DOI] [PubMed] [Google Scholar]
- Kang, H. , Chou, P.J. , Johnson, W.C. Jr. , Weller, D. , Huang, S.B. , and Summerton, J.E. 1992. Stacking interactions of ApA analogues with modified backbones. Biopolymers 32:1351‐1363. [DOI] [PubMed] [Google Scholar]
- Khokha, M.K. , Chung, C. , Bustamante, E.L. , Gaw, L.W. , Trott, K.A. , Yeh, J. , Lim, N. , Lin, J.C. , Taverner, N. , Amaya, E. , Papalopulu, N. , Smith, J.C. , Zorn, A.M. , Harland, R.M. , and Grammer, T.C. 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 225:499‐510. [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B. and Law, R.D. 1985a. Cell lineage of zebrafish blastomeres. II. Formation of the yolk syncytial layer. Dev. Biol. 108:86‐93. [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B. and Law, R.D. 1985b. Cell lineage of zebrafish blastomeres. III. Clonal analyses of the blastula and gastrula stages. Dev. Biol. 108:94‐101. [DOI] [PubMed] [Google Scholar]
- Kinney, R.M. , Huang, C.Y. , Rose, B.C. , Kroeker, A.D. , Dreher, T.W. , Iversen, P.L. , and Stein, D.A. 2005. Inhibition of dengue virus serotypes 1 to 4 in vero cell cultures with Morpholino oligomers. J. Virol. 79:5116‐5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman, W.P. , Wienholds, E. , Ketting, R.F. , and Plasterk, R.H. 2004. Substrate requirements for let‐7 function in the developing zebrafish embryo. Nucleic Acids Res. 32:6284‐6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman, W.P. , Lagendijk, A.K. , Ketting, R.F. , Moulton, J.D. , and Plasterk, R.H. 2007. Targeted inhibition of miRNA maturation with Morpholinos reveals a role for miR‐375 in pancreatic islet development. PLoS Biol. 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos, R. , Tucker, R.P. , Hall, R. , Duong, T.D. , and Erickson, C.A. 2003. Methods for introducing Morpholinos into the chicken embryo. Dev. Dyn. 226:470‐477. [DOI] [PubMed] [Google Scholar]
- Lebedeva, I. and Stein, C.A. 2001. Antisense oligonucleotides: Promise and reality. Annu. Rev. Pharmacol. Toxicol. 41:403‐419. [DOI] [PubMed] [Google Scholar]
- Li, Y.F. and Morcos, P.A. 2008. Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo. Bioconjug. Chem. In press. [DOI] [PubMed] [Google Scholar]
- Liu, G. , He, J. , Zhang, S. , Liu, C. , Rusckowski, M. , and Hnatowich, D.J. 2002a. Cytosine residues influence kidney accumulations of 99mTc‐labeled Morpholino oligomers. Antisense Nucleic Acid Drug. Dev. 12:393‐398. [DOI] [PubMed] [Google Scholar]
- Liu, G. , Zhang, S. , He, J. , Liu, N. , Gupta, S. , Rusckowski, M. , and Hnatowich, D.J. 2002b. The influence of chain length and base sequence on the pharmacokinetic behavior of 99mTc‐Morpholinos in mice. Q. J. Nucl. Med. 46:233‐243. [PubMed] [Google Scholar]
- Mang'era, K.O. , Liu, G. , Yi, W. , Zhang, Y. , Liu, N. , Gupta, S. , Rusckowski, M. , and Hnatowich, D.J. 2001. Initial investigations of 99mTc‐labeled Morpholinos for radiopharmaceutical applications. Eur. J. Nucl. Med. 28:1682‐1689. [DOI] [PubMed] [Google Scholar]
- Masaki, M. , Izumi, M. , Oshima, Y. , Nakaoka, Y. , Kuroda, T. , Kimura, R. , Sugiyama, S. , Terai, K. , Kitakaze, M. , Yamauchi‐Takihara, K. , Kawase, I. , and Hirota, H. 2005. Smad1 protects cardiomyocytes from ischemia‐reperfusion injury. Circulation 111:2752‐2759. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P. , Meuse, L. , Karimi, M. , Contag, C.H. , and Kay, M.A. 2003. A potent and specific Morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology 38:503‐508. [DOI] [PubMed] [Google Scholar]
- McClorey, G. , Moulton, H.M. , Iversen, P.L. , Fletcher, S. , and Wilton, S.D. 2006. Antisense oligonucleotide‐induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 13:1373‐1381. [DOI] [PubMed] [Google Scholar]
- Mellitzer, G. , Hallonet, M. , Chen, L. , and Ang, S.L. 2002. Spatial and temporal ‘knock down’ of gene expression by electroporation of double‐stranded RNA and Morpholinos into early postimplantation mouse embryos. Mech. Dev. 118:57‐63. [DOI] [PubMed] [Google Scholar]
- Melton, A.A. , Jackson, J. , Wang, J. , and Lynch, K.W. 2007. Combinatorial control of signal‐induced exon repression by hnRNP L and PSF. Mol. Cell Biol. 27:6972‐6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monga, S.P. , Monga, H.K. , Tan, X. , Mule, K. , Pediaditakis, P. , and Michalopoulos, G.K. 2003. Beta‐catenin antisense studies in embryonic liver cultures: Role in proliferation, apoptosis, and lineage specification. Gastroenterology 124:202‐216. [DOI] [PubMed] [Google Scholar]
- Morcos, P.A. 2001. Achieving efficient delivery of Morpholino oligos in cultured cells. Genesis 30:94‐102. [DOI] [PubMed] [Google Scholar]
- Morcos, P.A. 2007. Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem. Biophys. Res. Commun. 358:521‐527. [DOI] [PubMed] [Google Scholar]
- Moulton, H.M. and Moulton, J.D. 2003. Peptide‐assisted delivery of steric‐blocking antisense oligomers. Curr. Opin. Mol. Ther. 5:123‐132. [PubMed] [Google Scholar]
- Moulton, H.M. and Moulton, J.D. 2008. Antisense morpholino oligomers and their peptide conjugates In Therapeutic Oligonucleotides (Kurreck J., ed.) pp. 43‐79. Royal Society of Chemistry, London. In press. [Google Scholar]
- Moulton, H.M. , Nelson, M.H. , Hatlevig, S.A. , Reddy, M.T. , and Iversen, P.L. 2004. Cellular uptake of antisense Morpholino oligomers conjugated to arginine‐rich peptides. Bioconjug. Chem. 15:290‐299. [DOI] [PubMed] [Google Scholar]
- Nasevicius, A. and Ekker, S.C. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216‐220. [DOI] [PubMed] [Google Scholar]
- Nelson, M.H. , Stein, D.A. , Kroeker, A.D. , Hatlevig, S.A. , Iversen, P.L. , and Moulton, H.M. 2005. Arginine‐rich peptide conjugation to Morpholino oligomers: Effects on antisense activity and specificity. Bioconjug. Chem. 16:959‐966. [DOI] [PubMed] [Google Scholar]
- Neuman, B.W. , Stein, D.A. , Kroeker, A.D. , Paulino, A.D. , Moulton, H.M. , Iversen, P.L. , and Buchmeier, M.J. 2004. Antisense Morpholino‐oligomers directed against the 5′ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 78:5891‐5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B.W. , Stein, D.A. , Kroeker, A.D. , Churchill, M.J. , Kim, A.M. , Kuhn, P. , Dawson, P. , Moulton, H.M. , Bestwick, R.K. , Iversen, P.L. , and Buchmeier, M.J. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense Morpholino oligomers. J. Virol. 79:9665‐9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt, S.L. , Bronchain, O.J. , Hartley, K.O. , and Amaya, E. 2001. Comparison of morpholino based translational inhibition during the development of Xenopus laevis and Xenopus tropicalis . Genesis 30:110‐113. [DOI] [PubMed] [Google Scholar]
- Partridge, M. , Vincent, A. , Matthews, P. , Puma, J. , Stein, D. , and Summerton, J. 1996. A simple method for delivering Morpholino antisense oligos into the cytoplasm of cells. Antisense Nucleic Acid Drug Dev. 6:169‐175. [DOI] [PubMed] [Google Scholar]
- Prasadan, K. , Daume, E. , Preuett, B. , Spilde, T. , Bhatia, A. , Kobayashi, H. , Hembree, M. , Manna, P. , and Gittes, G.K. 2002. Glucagon is required for early insulin‐positive differentiation in the developing mouse pancreas. Diabetes 51:3229‐3236. [DOI] [PubMed] [Google Scholar]
- Riedel‐Kruse, I.H. , Muller, C. , and Oates, A.C. 2007. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science 317:1911‐1915. [DOI] [PubMed] [Google Scholar]
- Sazani, P. , Kang, S.H. , Maier, M.A. , Wei, C. , Dillman, J. , Summerton, J. , Manoharan, M. , and Kole, R. 2001. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 29:3965‐3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazani, P. , Gemignani, F. , Kang, S.H. , Maier, M.A. , Manoharan, M. , Persmark, M. , Bortner, D. , and Kole, R. 2002. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 20:1228‐1233. [DOI] [PubMed] [Google Scholar]
- Scacheri, P.C. , Rozenblatt‐Rosen, O. , Caplen, N.J. , Wolfsberg, T.G. , Umayam, L. , Lee, J.C. , Hughes, C.M. , Shanmugam, K.S. , Bhattacharjee, A. , Meyerson, M. , and Collins, F.S. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101:1892‐1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmajuk, G. , Sierakowska, H. , and Kole, R. 1999. Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J. Biol. Chem. 274:21783‐21789. [DOI] [PubMed] [Google Scholar]
- Stancheva, I. , Collins, A.L. , Van Den Veyver, I.B. , Zoghbi, H. , and Meehan, R.R. 2003. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol. Cell. 12:425‐435. [DOI] [PubMed] [Google Scholar]
- Stein, D. , Foster, E. , Huang, S.B. , Weller, D. , and Summerton, J. 1997. A specificity comparison of four antisense types: Morpholino, 2′‐O‐methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151‐157. [DOI] [PubMed] [Google Scholar]
- Stein, D.A. , Skilling, D.E. , Iversen, P.L. , and Smith, A.W. 2001. Inhibition of Vesivirus infections in mammalian tissue culture with antisense Morpholino oligomers. Antisense Nucleic Acid Drug Dev. 11:317‐325. [DOI] [PubMed] [Google Scholar]
- Stirchak, E.P. , Summerton, J.E. , and Weller, D.D. 1989. Uncharged stereoregular nucleic acid analogs: 2. Morpholino nucleoside oligomers with carbamate internucleoside linkages. Nucleic Acids Res. 17:6129‐6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton, J. 1999. Morpholino antisense oligomers: The case for an RNase H‐independent structural type. Biochim. Biophys. Acta 1489:141‐158. [DOI] [PubMed] [Google Scholar]
- Summerton, J. 2004. Morpholinos and PNAs compared. Lett. Pept. Sci. 10:215‐236. [Google Scholar]
- Summerton, J. 2005. Endo‐Porter: A novel reagent for safe, effective delivery of substances into cells. Ann. N.Y. Acad. Sci. 1058:1‐14. [DOI] [PubMed] [Google Scholar]
- Summerton, J. and Weller, D. 1997. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187‐195. [DOI] [PubMed] [Google Scholar]
- Suwanmanee, T. , Sierakowska, H. , Fucharoen, S. , and Kole, R. 2002. Repair of a splicing defect in erythroid cells from patients with beta‐thalassemia/HbE disorder. Mol. Ther. 6:718‐726. [DOI] [PubMed] [Google Scholar]
- Takahashi, M. , Sato, K. , Nomura, T. , and Osumi, N. 2002. Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation 70:155‐162. [DOI] [PubMed] [Google Scholar]
- Thummel, R. , Bai, S. , Sarras, M.P. Jr. , Song, P. , McDermott, J. , Brewer, J. , Perry, M. , Zhang, X. , Hyde, D.R. , and Godwin, A.R. 2005. Inhibition of zebrafish fin regeneration using in vivo electroporation of Morpholinos against fgfr1 and msxb. Dev. Dyn. 235:336‐346. [DOI] [PubMed] [Google Scholar]
- Tidd, D.M. , Giles, R.V. , Broughton, C.M. , and Clark, R.E. 2001. Expression of c‐myc is not critical for cell proliferation in established human leukemia lines. BMC Mol. Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, R.P. 2004. Antisense knockdown of the beta1 integrin subunit in the chicken embryo results in abnormal neural crest cell development. Int. J. Biochem. Cell Biol. 36:1135‐1139. [DOI] [PubMed] [Google Scholar]
- Tyson‐Capper, A.J. and Europe‐Finner, G.N. 2006. Novel targeting of cyclooxygenase‐2 (COX‐2) pre‐mRNA using antisense morpholino oligonucleotides directed to the 3′ acceptor and 5′ donor splice sites of exon 4: Suppression of COX‐2 activity in human amnion‐derived WISH and myometrial cells. Mol. Pharmacol. 69:796‐804. [DOI] [PubMed] [Google Scholar]
- van den Born, E. , Stein, D.A. , Iversen, P.L. , and Snijder, E.J. 2005. Antiviral activity of Morpholino oligomers designed to block various aspects of equine arteritis virus amplification in cell culture. J. Gen. Virol. 86:3081‐3090. [DOI] [PubMed] [Google Scholar]
- Warfield, K.L. , Swenson, D.L. , Olinger, G.G. , Nichols, D.K. , Pratt, W.D. , Blouch, R. , Stein, D.A. , Aman, M.J. , Iversen, P.L. , and Bavari, S. 2006. Gene‐specific countermeasures against ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield, M. 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish ( Danio rerio ), 5th ed. University of Oregon Press, Eugene, Oregon. [Google Scholar]
- Yen, L. , Svendsen, J. , Lee, J.S. , Gray, J.T. , Magnier, M. , Baba, T. , D'Amato, R.J. , and Mulligan, R.C. 2004. Exogenous control of mammalian gene expression through modulation of RNA self‐cleavage. Nature 431:471‐476. [DOI] [PubMed] [Google Scholar]
- Youngblood, D.S. , Hatlevig, S.A. , Hassinger, J.N. , Iversen, P.L. , and Moulton, H.M. 2007. Stability of cell‐penetrating peptide‐Morpholino oligomer conjugates in human serum and in cells. Bioconjug. Chem. 18:50‐60. [DOI] [PubMed] [Google Scholar]
Key References
First description of splice inhibition in a zebrafish, including an analysis of a cryptic splice site.
Description of peptide‐Morpholino conjugates now in use for in vivo experiments.
Review article presenting data determining the effective region for targeting translation inhibition oligos and presenting a detailed discussion of Morpholino specificity and minimum inhibitory length.
Structure and early synthetic scheme for Morpholino oligos.
- Draper et al., 2001. See above.
- Nelson et al., 2005. See above.
- Summerton, 1999. See above.
- Summerton and Weller, 1997. See above.
Internet Resources
Commercial source for Morpholinos.
Morpholino publication database. As of printing, >2000 publications have reported experiments with Morpholino oligos in a broad range of systems. Citations and many abstracts are searchable here.
Discussion board for Morpholino users.
Zebrafish Information Network. References related to Morpholino use in zebrafish are searchable in an annotated database.
Annotated database of zebrafish Morpholino sequences by gene name.
AVI BioPharma, Inc., Morpholino therapeutics company.


